Abstract
Lung macrophages play a pivotal role in pulmonary fibrosis, with monocyte-derived alveolar macrophages driving disease progression. However, the mechanisms regulating their pro-fibrotic behavior and survival remain unclear, and effective therapeutic strategies are lacking. Here we show that triggering receptors expressed on myeloid cells 2 are predominantly expressed on monocyte-derived alveolar macrophages in fibrotic mouse lungs and are significantly elevated in lung macrophages from patients with idiopathic pulmonary fibrosis. Deletion or knockdown of this receptor disrupts intracellular survival signaling, promotes macrophage apoptosis, and attenuates their pro-fibrotic phenotype. We further demonstrate that a lipid mediator and a high-avidity ligand of this receptor, encountered by macrophages in the alveolar milieu, enhance macrophage survival and activity. Ablation of TREM2 or blocking this receptor with soluble receptors or specific antibodies effectively alleviates lung fibrosis in male mice. These findings identify this receptor as a critical regulator of macrophage-mediated fibrosis and a promising therapeutic target for intervention.
Subject terms: Respiratory tract diseases, Alveolar macrophages
Monocyte-derived alveolar macrophages (Mo-AMs) are involved in lung injury and fibrosis through incompletely understood mechanisms. Here, the authors show that TREM2 is a critical regulator of lung fibrosis progression mediated by Mo-AMs and a potential target for therapeutic intervention.
Introduction
Recent genetic lineage tracing, immunohistochemistry, and immunocytometry studies have established that tissue-resident alveolar macrophages (TR-AMs) arise from fetal origins, while postnatal circulating monocytes are recruited to the alveolar space and differentiate into alveolar macrophages (AMs) during the course of lung injury and repair1–7. The latter is commonly known as monocyte-derived alveolar macrophages (Mo-AMs). Mo-AMs and TR-AMs can be readily separated by flow cytometry based on a set of typical cell surface markers8–12. A separation in the transcriptome between these two subsets of macrophages was also evident in RNA sequencing (RNA-seq) analyses1,5,6,11,13–16. Furthermore, there is an appreciable level of similarity between the differentially expressed transcripts in Mo-AMs compared to TR-AMs from mouse fibrotic lungs and the altered transcripts in IPF AMs compared to those from healthy donors, especially certain pro-fibrotic mediators1,11,13,17,18, suggesting a common pathogenic mechanism shared by human and mouse AMs in pulmonary fibrosis.
AMs have long been recognized as important players in the pathogenesis of lung fibrosis2,3,19–27. However, it was only recently that a series of studies established that Mo-AMs, but not TR-AMs, are responsible for the pro-fibrotic activity of this group of cells1,3,10,13. These studies showed that depletion of Mo-AMs via several strategies conferred protection from lung fibrosis in multiple mouse models1,3,10,11,13,20. The pathogenic role of Mo-AMs in lung fibrosis has been largely attributed to their activity in producing a number of pro-fibrotic mediators, such as TGF-β, PDGF-α, osteopontin (OPN), and MMPs1,3,11,13,15,18,28.
Despite these inspiring progresses, knowledge gaps remain in understanding how the Mo-AM pro-fibrotic phenotype and its fate are regulated during fibrogenesis, how intervention in a potential regulatory mechanism in the Mo-AMs may affect the progression of this pathology, and how to target the regulatory mechanism in the Mo-AMs to treat lung fibrosis. A better understanding of these unanswered questions will likely lead to novel Mo-AM-based therapeutics for this pathology.
TREM2 belongs to a family of receptors referred to as the triggering receptors expressed on myeloid cells (TREM)29–31. This receptor consists of an extracellular V-type Ig domain followed by a short stalk and a single transmembrane helix that interacts with the immunoreceptor tyrosine-based activation motif (ITAM) containing DNAX-activation protein 12 (DAP12)29,30. In the presence of a disintegrin and metalloproteinase (ADAM) activity, the extracellular domain undergoes shedding and becomes soluble TREM2 (sTREM2). TREM2 engagement with ligands triggers the phosphorylation of the DAP12 ITAM, subsequently initiating a cascade of activation of downstream kinases, such as AKT, MAPK, and mTOR29,30. These kinases are centrally involved in controlling cell metabolism, survival, proliferation, and phagocytosis29,30. TREM2 ligands include a variety of molecules, such as bacterial wall products, Aβ oligomers, lipoproteins, and anionic lipids, such as phospholipids and sphingolipids29,32–35, reflective of a diversified function of this receptor in different pathological settings.
Recent studies on TREM2 have been heavily concentrated in the realm of Alzheimer’s disease since several TREM2 variants were found to be closely associated with the onset of this pathology29,30,33,36,37. However, high TREM2 expression has also been found in subpopulations of peripheral macrophages in other pathological conditions, such as those in the cancer stroma29,38. New evidence implicating TREM2 in cancer immunity, atherosclerosis, and steatosis is rapidly emerging29,38,39. TREM2 expression is frequently associated with scar-associated macrophages (SAMs)40–42. However, except for the mere recognition as a marker of fibrosis-associated macrophages39,43–45, the role of TREM2 in regulating the pro-fibrotic AM phenotype in lung fibrosis remains unclear.
In this study, we found that TREM2 was highly expressed on Mo-AMs in the fibrotic mouse lungs. TREM2 promoted macrophage survival and the pro-fibrotic phenotype. We found that TREM2 mediated the anti-apoptotic effect of lipid ligands, to which the Mo-AMs would be exposed when entering the new milieu in the fibrotic lungs. We also showed that TREM2 deficiency protected mice from lung fibrosis. More importantly, we found that interference of TREM2 engagement with its lipid ligands by sTREM2 neutralization or by a specific TREM2 blocking antibody attenuated bleomycin-induced lung fibrosis. Our data suggest that Mo-AM TREM2 is critical to its pathogenic activity in lung fibrosis. Targeting TREM2 may be an effective strategy to treat lung fibrosis.
Results
Mo-AMs show higher expression of TREM2 than TR-AMs in mice with bleomycin-induced pulmonary fibrosis
Recent evidence has compellingly shown that Mo-AMs, but not TR-AMs, play a key role in the pathogenesis of lung fibrosis1,3,10,13. Given our long-standing interest in delineating how lung macrophages contribute to this pathology, our initial findings, as well as those from studies on microglia and cancer-associated macrophages32,38, led us to recognize that TREM2, a receptor primarily expressed on myeloid cells, may be a key regulator of the pro-fibrotic activity of Mo-AMs in lung fibrosis. To identify the cell type in the lung that expresses TREM2, we interrogated several lung scRNA-seq databases46,47 and found that TREM2 was not significantly expressed in any other type of cells, except for some moderate levels on AMs and classical monocytes, in normal mouse lungs (Fig. 1A, B).
Fig. 1. Mo-AMs demonstrate markedly higher expression of TREM2 than TR-AMs in the lungs of mice with bleomycin (BLM)-induced pulmonary fibrosis.
A UMAP with TREM2 expression shown was re-produced from scRNA-seq dataset (lungendothelialcellatlas) on normal mouse lung. B Boxplots representing the average normalized TREM2 expression for each subject, grouped by cell type, was re-produced from dataset lungendothelialcellatlas. 25th and 75th percentiles (horizontal box edges), median (center line), minimum and maximum values (outer whiskers); each dot represents one subject; whiskers are 1.5x IQR. n = 29 mouse lungs. C 8 week male C57BL/6 mice were intratracheally (i.t.) instilled with saline or BLM (1.5 U/kg in 50 µl saline). Three weeks after treatment, lung sections were prepared and immunofluorescence staining was performed to determine the expression of TREM2 and F4/80. Scale bar: 100 µm. The experiment was repeated three times independently with similar results. D Mice were treated as in “C”. Bronchoalveolar lavages (BALs) were harvested and BAL cells stained with a cocktail of antibodies as detailed in Methods. TREM2 levels on F4/80 + / Siglec Fhigh/ CD11blow TR-AMs and F4/80 + / Siglec Flow/CD11bhigh Mo-AMs were determined by flow cytometric analysis. n = 3, 4, and 4 mice, respectively; mean ± SD; one-way ANOVA with Bonferroni’s multiple comparisons test. MFI, mean fluorescence intensity. E Mice were i.t. instilled with BLM as in “C”. 6 days after treatment, BAL cells were harvested and stained with a cocktail of antibodies as detailed in Methods. Cell surface TREM2 levels on MERTK + /CD11blow TR-AMs, MERTK + /CD11high Mo-AMs, MERTK-/CD11b + /Ly-6G+ neutrophils and other BAL cells were determined by flow cytometric analysis. The experiments for “D, E” were repeated twice with similar results. Source data are provided as a Source Data file.
To characterize TREM2 expression in the lungs of mice with or without bleomycin-induced pulmonary fibrosis, we first performed immunofluorescence microscopy and found that a few cells expressing the pan-macrophage marker F4/80 had some weak TREM2 expression in normal mouse lungs (Fig. 1C). In contrast, there were substantially more macrophages in the lungs of bleomycin-treated mice, which also demonstrated elevated TREM2 levels (Fig. 1C). To better quantify TREM2 expression in AMs, we collected bronchoalveolar lavage (BAL) cells and gated F4/80 + /CD64 + /Siglec Fhigh TR-AMs and F4/80 + /CD64 + /CD11bhigh Mo-AMs, finding that there was a markedly higher level of TREM2 on Mo-AMs compared to TR-AMs (Fig. 1D), with neutrophils and other BAL cells showing undetectable TREM2 expression (Fig. 1E). These findings indicate that TREM2 may play a role in shaping the phenotype of Mo-AMs in the context of lung fibrosis.
IPF AMs demonstrate considerably greater TREM2 expression than those from normal control lungs
Previous studies have shown that there is an increased number of AMs in IPF lungs compared to normal controls1. Furthermore, the altered transcripts in IPF AMs, compared to those from normal human donors, and the differentially expressed transcripts in Mo-AMs compared to TR-AMs from mouse fibrotic lungs, share a number of common genes, especially some pro-fibrotic mediators1,11,13,17,18. To characterize TREM2 expression in human lungs, we interrogated human lung scRNA-seq databases and found that, similar to mouse lungs, TREM2 expression was predominantly restricted to AMs and classical monocytes in normal human lungs (Fig. 2A, B). We also performed immunofluorescence microscopy and found that, similar to what was observed in mouse lungs, IPF lungs had more macrophages that also expressed greater TREM2 than normal subjects (Fig. 2B). To further assess TREM2 levels in IPF AMs, we interrogated the IPF scRNA-seq database and found that there was markedly increased TREM2 expression in IPF AMs compared to normal lungs (Fig. 2C), with all other cell types in both normal and diseased lungs expressing minimal TREM2 (Fig. 2C). Together, these data suggest that TREM2 may contribute to the pro-fibrotic phenotype of Mo-AMs and IPF AMs.
Fig. 2. IPF AMs demonstrate considerably greater TREM2 expression than those from normal control lungs.
A UMAP with TREM2 expression shown was re-produced from scRNA-seq dataset (GSE164829) on normal human lung. B Immunofluorescence staining was performed on human normal control and idiopathic pulmonary fibrosis (IPF) lung sections to determine the expression and localization of TREM2 and CD68. Scale bar: 100 µm. The experiment was repeated three times independently with similar results. C Boxplots representing the average normalized TREM2 expression for each subject, grouped by cell type, was re-produced from dataset GSE128033. 25th and 75th percentiles (horizontal box edges), median (center line), minimum and maximum values (outer whiskers); each dot represents one subject; whiskers are 1.5x IQR. n = 10 control (normal) lungs, n = 8 IPF lungs. CTRL, control (normal).
Global TREM2 knockout, as well as conditional TREM2 knockout in Mo-AMs, protects mice from bleomycin-induced lung fibrosis
To determine if TREM2 participates in the development of lung fibrosis, we first tested the hypothesis in a strain with global TREM2 knockout. As shown in Fig. 3A, bleomycin-induced lung fibrosis was significantly decreased in TREM2 knockout (KO) mice compared to TREM2 wild-type controls, as indicated by the comparison of lung hydroxyproline levels among the groups. Consistently, the expression of pro-fibrotic mediators was also diminished in the lungs of bleomycin-treated TREM2 KO mice (Fig. 3B). Despite being a global TREM2 KO strain, these data indicate an important role of Mo-AM TREM2 in the pathogenesis of lung fibrosis because TREM2 was primarily expressed in the Mo-AMs and minimally in other types of lung cells (Fig. 1). Notably, we also confirmed the status of TREM2 KO in Mo-AMs of TREM2 (-/-) mice (Fig. 3C).
Fig. 3. Global TREM2 knockout, as well as conditional TREM2 knockout in Mo-AMs, protects mice from BLM-induced lung fibrosis.
A Eight to ten week TREM2 wild-type (+/+) and TREM2 knockout (-/-) mice were i.t. instilled with saline or BLM (1.5 U/kg in 50 µl saline). Three weeks later, lung hydroxyproline levels were determined. n = 3, 3, 8, and 6 mice, respectively; mean ± SEM; *p < 0.05, one-way ANOVA with Turkey’s multiple comparisons test. B Total lung RNAs from the BLM treated mice in “A” were isolated and the expression of the indicated genes was determined by real-time PCR. mean ± SEM; unpaired two-tailed Student’s t-test. C Mo-AMs from the BLM treated mice in “A” were isolated by flow sorting and levels of TREM2 in the cells determined by real-time PCR. mean ± SEM; unpaired two-tailed Student’s t-test. D, E Eight to ten week Mo-AM TREM2 (+/+) and Mo-AM TREM2 (-/-) mice were intraperitoneally (i.p.) injected with tamoxifen (75 mg/kg) for 5 consecutive days. 3 days later, the mice were i.t. instilled with saline or BLM (1.5 U/kg in 50 µl saline). The mice continued to receive tamoxifen injection, twice/week. 3 weeks after BLM treatment, Mo-AMs were isolated by flow sorting from the BLM treated mice and levels of TREM2 in the cells determined by real-time PCR (D). n = 6 and 5 mice, respectively; mean ± SEM; unpaired two-tailed Student’s t-test. The lungs in the above experiment were harvested and lung hydroxyproline levels were determined (E). n = 3, 3, 5, and 5 mice, respectively; mean ± SEM; one-way ANOVA with Bonferroni’s multiple comparisons test. F Mo-AM TREM2 (+/+) and Mo-AM TREM2 (-/-) mice were treated as in “D, E”. Lung sections were prepared and histological analysis performed by Masson’s trichrome staining. Scale bar: 500 µm. The experiment was repeated three times independently with similar results obtained. G Mo-AM TREM2 (+/+) and Mo-AM TREM2 (-/-) mice were treated as in “D-F”. The severity of pulmonary fibrosis was assessed by noninvasive lung micro-CT scan. Representative axial micro-CT images of mouse lungs (gray scale or quad-prism color scale (blue to red represents low to high density)) and quantification of lung density (Hounsfield unit at the 85th percentile of the lung density) are shown. n = 3 mice per group; mean ± SEM; one-way ANOVA with Bonferroni’s multiple comparisons test. Source data are provided as a Source Data file.
To provide direct evidence for TREM2 mediation of the pro-fibrotic activity of Mo-AMs in lung fibrosis, we generated a strain (Mo-AM TREM2 (-/-)) by crossing CX3CR1-Cre-ERT2 mice with TREM2 (fl/fl) mice. The inducible CX3CR1-driven Cre activity is robust only in monocytes and Mo-AMs, but not in TR-AMs13. We confirmed that Mo-AM TREM2 was markedly downregulated in the Mo-AM TREM2 (-/-) mice compared to the control Mo-AM TREM2 (+/+) animals (Fig. 3D). More importantly, the ablation of TREM2 in Mo-AMs protected mice from bleomycin-induced lung fibrosis. This protection was evidenced by reduced hydroxyproline levels, decreased trichrome-stained collagen deposition, and lower micro-computed tomography (CT)-measured lung densities in bleomycin-treated Mo-AM TREM2 (-/-) mice compared to their Mo-AM TREM2 (+/+) counterparts (Fig. 3E–G), consistent with findings in the global TREM2 KO mice. These findings underscore the important role of Mo-AM TREM2 in the pathogenesis of lung fibrosis.
Bleomycin-induced lung fibrosis is initiated by alveolar injury and is accompanied by an acute inflammatory phase within the first week post-treatment48. The intensity of the early inflammatory response generally correlates with the severity of lung fibrosis in this model48. To determine whether the reduced lung fibrosis observed in Mo-AM TREM2 (-/-) mice could simply result from dampened inflammation, we assessed several parameters reflecting the inflammatory response in the lungs three days after bleomycin injection. We found no significant differences in lung inflammation between bleomycin-treated Mo-AM TREM2 (+/+) and Mo-AM TREM2 (-/-) mice, as evidenced by comparable levels of BAL proteins and inflammatory cytokine expression (Supplementary Fig. 1A–D). These findings suggest that the attenuation of fibrosis in Mo-AM TREM2 (-/-) mice is unlikely due to reduced bleomycin-induced inflammation.
To further support the pro-fibrotic role of TREM2 in bleomycin-treated mice, we employed a second lung fibrosis model49. In this experiment, Mo-AM TREM2 (+/+) and Mo-AM TREM2 (-/-) mice were intratracheally instilled with adenovirus expressing constitutively active TGF-β1. As shown in Supplementary Fig. 2A–C, Mo-AM TREM2 (-/-) mice were protected from TGF-β1-induced lung fibrosis. This protection was evidenced by reduced hydroxyproline levels, decreased expression of pro-fibrotic mediators, and improved lung histology compared to Mo-AM TREM2 (+/+) controls.
TREM2 deficiency promotes apoptosis and diminishes pro-fibrotic gene expression in macrophages
During an active fibrotic phase, not only do Mo-AMs significantly outnumber TR-AMs, they also appear more pro-fibrotic due to the elevated expression of certain pro-fibrotic genes19. These might be the two main characteristics responsible for the dominating role of Mo-AMs in the pathogenesis of lung fibrosis. Mo-AM numbers decline steadily through the late fibrotic to the resolution stages, which might be due to apoptosis, with their transcriptome becoming increasingly indistinguishable from TR-AMs1,50.
To determine the underlying mechanism by which TREM2 mediates the pro-fibrotic actions of Mo-AMs, we assessed the apoptosis and number of these cells in Mo-AM TREM2 (+/+) and Mo-AM TREM2 (-/-) mice. As shown in Fig. 4A, TREM2 (-/-) Mo-AMs demonstrated increased apoptosis compared to TREM2 (+/+) Mo-AMs, which likely contributed to the decreased number of TREM2 (-/-) Mo-AMs in the mouse fibrotic lungs (Fig. 4B). These data suggest that TREM2 promotes Mo-AM survival in fibrotic lungs. To provide additional evidence supporting this notion, we also determined the effect of TREM2 on the survival of bone marrow-derived macrophages (BMDMs). As shown in Fig. 4C and concordant with a recent study51, TREM2 knockdown rendered BMDMs more susceptible to apoptosis upon macrophage colony-stimulating factor (M-CSF) withdrawal. Consistently, there was diminished general survival signaling, such as the activation of AKT, ERK, and mTOR, and expression of the pro-survival mediator, in TREM2 knockdown BMDMs (Fig. 4D). Furthermore, TREM2 (-/-) Mo-AMs also showed less expression of SPP1, PDGFA, and MMP12 than TREM2 (+/+) Mo-AMs (Fig. 4E), suggesting that TREM2 is also critical to the pro-fibrotic phenotype of these cells. Given that AMs are also a major source of TGF-β in fibrotic lungs52, a decrease in the number of TREM2 (-/-) Mo-AMs might also contribute to the reduced TGF-β1 observed in the BALs from bleomycin-treated Mo-AM TREM2 (-/-) mice compared to those from the Mo-AM TREM2 (+/+) controls (Fig. 4F-G). To further delineate the effect of TREM2 on the pro-fibrotic and pro-survival phenotype of macrophages in fibrotic lungs, we re-analyzed published scRNA-seq data. We separated macrophages in bleomycin-induced fibrotic lungs into two subsets, TREM2-low and TREM2-high, and found greater expression of a previously defined set of scar-associated macrophage markers and anti-apoptotic mediators in TREM2-high mouse lung macrophages (Fig. 4H). Consistent with the above findings, TREM2-high macrophages in IPF lungs also demonstrated considerably increased expression of scar-associated macrophage markers and anti-apoptotic mediators compared to TREM2-low macrophages in the same lungs (Fig. 4I). Taken together, these data suggest that TREM2 confers a survival advantage to macrophages and promotes their production of pro-fibrotic mediators, thereby promoting lung fibrosis upon infiltration into the lungs.
Fig. 4. TREM2 deficiency promotes apoptosis and diminishes pro-fibrotic gene expression in macrophages.
A TREM2 (+/+) and TREM2 (-/-) Mo-AMs were isolated by flow sorting from mice that were treated BLM for 3 weeks, and apoptosis evaluated. n = 4 mice per group; mean ± SEM; unpaired two-tailed Student’s t-test. B Numbers of Mo-AMs in the BALs. n = 6 and 5 mice, respectively; mean ± SEM; unpaired two-tailed Student’s t-test. C BMDMs were transfected with control (con) and TREM2 siRNAs and then cultured in the presence of 10 ng/ml M-CSF for 2 days. Cell apoptosis was determined 6 h after M-CSF withdrawal. n = 4 independent culture of cells per group; mean ± SD; one-way ANOVA with Bonferroni’s multiple comparisons test. The experiment was repeated three with similar results. D BMDMs were transfected with control and TREM2 siRNAs and then cultured in the presence of 10 ng/ml M-CSF for 2 days. Cells were harvested 6 h after M-CSF withdrawal and the expression of the indicated proteins were determined by Western blotting. The experiment was repeated three times with similar results. E The expression of pro-fibrotic mediators in Mo-AMs from mice treated in “A” was determined by real-time PCR. n = 4 mice per group; mean ± SEM; unpaired two-tailed Student’s t-test. F TGF-β1 levels in BALs from mice treated in “A” were determined by ELISA. n = 6 and 5 mice, respectively; mean ± SEM; unpaired two-tailed Student’s t-test. G BAL TGF-β1 levels in TREM2 (+/+) and TREM2 (-/-) mice instilled with saline or BLM for 3 weeks. n = 3, 3, 7, and 6 mice, respectively; mean ± SEM; one-way ANOVA with Bonferroni’s multiple comparisons test. H, I Bubble plots for the indicated pro-fibrotic and anti-apoptotic genes in the macrophages from BLM treated mouse lungs (H) and human IPF lungs (I) were re-produced from scRNA-seq datasets GSE132771 (n = 2 mouse lungs) and GSE128033 (n = 5 IPF lungs), respectively. Source data are provided as a Source Data file.
Mo-AM TREM2 inhibits alveolar type II (ATII) cells regeneration and differentiation into alveolar type I (ATI) cells
We have shown that TREM2 promotes AM survival and pro-fibrotic gene expression. We were then intrigued to investigate the effect of Mo-AM TREM2 on ATII cell phenotypic presentation given the critical role of ATII dysfunction in the pathogenesis of lung fibrosis and interactions of ATII and macrophages in fibrotic niches. To address this question, flow-sorted ATIIs from Sftpc-Cre/tdTomato mice were mixed with isolated TREM2 (+/+) or TREM2 (-/-) Mo-AMs (1:5 ratio) in Matrigel and cultured in ATII maintenance medium (AMM) for 10–14 days to allow organoid formation. As shown in Fig. 5A-B, we found significantly more and larger organoids when ATIIs were co-cultured with TREM2 (-/-) Mo-AMs than with the TREM2 (fl/fl) counterparts, suggesting improved ATII regeneration with the TREM2 (-/-) Mo-AMs. To further characterize the phenotype of ATIIs in the co-culture system, we harvested the organoids and performed immunofluorescent staining. As shown in Fig. 5C, ATII cells demonstrated greater proliferation, as evidenced by Ki-67 expression, when co-cultured with TREM2 (-/-) Mo-AMs compared to TREM2 (fl/fl) counterparts, suggesting that TREM2 (+/+) Mo-AMs inhibit ATII regeneration. Furthermore, we found that ATIIs co-cultured with TREM2 (-/-) Mo-AMs in the organoids demonstrated greater expression of AGER (advanced glycation end-products receptor), an ATI marker, and diminished KRT17, a marker of the recently defined alveolar epithelial transitional state, compared to those co-cultured with TREM2 (+/+) Mo-AMs (Fig. 5D-E). Interestingly, the expression of the ATII marker SPC seemed comparable in the two groups (Fig. 5E). These data suggest that TREM2 (+/+) Mo-AMs inhibit ATII regeneration and differentiation into ATI. Together with our findings that TREM2 (-/-) Mo-AMs produce fewer pro-fibrotic mediators than their wild-type counterparts, these data indicate that Mo-AM TREM2 plays a critical role in regulating the activity of this subset of pathological macrophages, leading to ATII alveolar regeneration failure.
Fig. 5. Mo-AM TREM2 inhibits ATII regeneration and differentiation into ATI.
A, B TREM2 (+/+) and TREM2 (-/-) Mo-AMs were isolated by flow sorting from mice treated with i.t. BLM for 1 week, respectively. The AMs were mixed with sorted ATIIs (5:1 ratio) in Matrigel and cultured for 12 days. ATII organoid number and size in each group were determined by ImageJ. n = 12 independent culture of cells per group; mean ± SD; unpaired two-tailed Student’s t-test. Scale bar: 2 mm. C–E ATII organoids from “A” were harvested and frozen sections prepared. Immunofluorescence microscopy was performed for the indicated proteins. Scale bar: 20 µm. All experiments were repeated twice with similar results obtained. Source data are provided as a Source Data file.
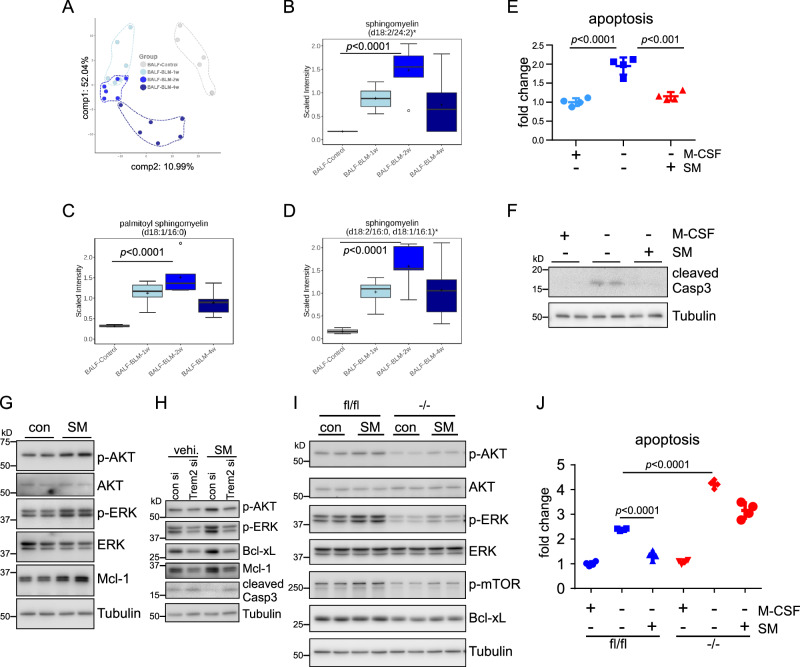
Sphingomyelin protects macrophages from apoptosis
Circulating monocytes enter the alveolar space via the interstitium and differentiate into alveolar macrophages in response to lung injuries. The Mo-AMs contribute to lung fibrogenesis in a new milieu to which these cells were not previously exposed. We have shown that there is considerably higher expression of TREM2 on Mo-AMs. We reasoned that Mo-AM TREM2, as a receptor, may promote the pathological activity of these cells in lung fibrosis by engaging ligands in their new environment. Of the known ligands of TREM2, we are particularly interested in lipid mediators since this class of molecules is enriched as components of pulmonary surfactant in the alveolar space. We performed lipidomic analyses on BALs and found that although abundantly present in normal mouse BAL, many species of sphingolipids and phospholipids, such as sphingomyelin (SM), phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI), demonstrated a marked elevation in the BALs from mice treated with bleomycin (Fig. 6A–D). Given that these sphingolipids and phospholipids are well-characterized high-avidity ligands of TREM232, the data suggest that TREM2 activation by these sphingolipids and phospholipids may sustain the survival and pro-fibrotic secretion of Mo-AMs when the cells encounter these molecules in the new milieu, thereby promoting the progression of lung fibrosis.
Fig. 6. Sphingomyelin (SM) protects macrophages from apoptosis.
A–D C57BL/6 mice were i.t. instilled with BLM. At 0, 1, 2, and 4 weeks post-treatment, BALs were collected and lipidomics was performed. A PCA analysis shows segregation of BALF samples based on groups. B–D The levels of various forms of sphingomyelins in the BALs. The box-and-whisker plots depict the 25th and 75th percentiles (horizontal box edges), median (center line), mean (+), minimum and maximum values (outer whiskers), and outliers (dots outside the whiskers). n = 6 mice per group; one-way ANOVA with Bonferroni’s multiple comparisons test. E BMDMs were cultured in the absence or presence of 10 ng/ml M-CSF, and treated with or without 10 µg/ml SM for 6 h. Cell apoptosis was determined. The average luminescent intensity in the control group ( + M-CSF, -SM) was regarded as “1”. The intensity of other groups to the control group was shown as fold change. n = 4 independent culture of cells per group; mean ± SD; one-way ANOVA with Bonferroni’s multiple comparisons test. F Experiments were done as in “E”. The expression of the indicated protein was determined by Western blotting. G BMDMs were treated with or without 10 µg/ml SM in the absence of M-CSF for 6 h. The expression of the indicated protein was determined by Western blotting. H BMDMs were transfected with control and TREM2 siRNAs and then cultured in the presence of 10 ng/ml M-CSF for 2 days. The cells were then treated with or without 10 µg/ml SM in the absence of M-CSF for 6 h. The expression of the indicated protein was determined by Western blotting. I TREM2 (+/+) and TREM2 (-/-) BMDMs were treated with or without 10 µg/ml SM in the absence of M-CSF for 6 h. The expression of the indicated protein was determined by Western blotting. J TREM2 (+/+) and TREM2 (-/-) BMDMs were cultured in the absence or presence of 10 ng/ml M-CSF, and treated with or without 10 µg/ml SM for 6 h. Cell apoptosis was determined. The average luminescent intensity in the control group (fl/fl +M-CSF, -SM) was regarded as “1”. The intensity of other groups to the control group was shown as fold change. n = 4 independent culture of cells; mean ± SD; one-way ANOVA with Bonferroni’s multiple comparisons test. The experiments for “E–J” were repeated three times with similar results. Source data are provided as a Source Data file.
To test if these BAL lipid mediators enhance the survival of macrophages, we initially treated BMDMs with or without SM. As shown in Fig. 6E, F, SM protected macrophages from apoptosis upon M-CSF withdrawal, as evidenced by the diminished Annexin V staining and reduced Caspase 3 cleavage. SM’s promotion of macrophage survival might be attributable to its elevation of survival signaling (Fig. 6G). SM activation of survival signaling and promotion of macrophage survival were dependent on TREM2 because SM-induced AKT and ERK phosphorylation, Bcl-xL, and Mcl-1, as well as SM-inhibited apoptosis, were attenuated in macrophages with TREM2 knockdown or knockout (Fig. 6H–J). Taken together, these data suggest that the sphingolipids, such as SM, in the alveolar space, which are further increased in lung fibrosis, activate Mo-AM TREM2 to sustain their survival once they are exposed to the new surroundings, thereby promoting the development, and/or impeding the resolution, of lung fibrosis.
sTREM2 blunts the protective activity of sphingomyelin on macrophages and alleviates bleomycin-induced lung fibrosis
The extracellular domain of TREM2 can be shed to form soluble TREM2 (sTREM2), which has been shown to function as a decoy receptor. As we found that SM, a high-avidity TREM2 ligand, protected macrophages from apoptosis in a TREM2-dependent manner, we were curious if sTREM2 might neutralize the protective effect of SM. As anticipated, pre-incubation of SM with sTREM2 did abrogate its activation of the survival signaling events and abolished its protection of Mo-AMs from apoptosis (Fig. 7A, B). Consistent with its cancellation of SM’s pro-survival activity in macrophages, we found that intratracheal delivery of sTREM2 protected mice from bleomycin-induced lung fibrosis (Fig. 7C). Notably, sTREM2 in BAL of bleomycin-treated mice was considerably increased compared to saline-treated controls (Fig. 7D), indicating that macrophage TREM2 shedding is a negative feedback mechanism the host develops to control fibrotic progression. Despite this protective mechanism, additional sTREM2 is still beneficial in curbing the development of lung fibrosis.
Fig. 7. sTREM2 blunts the protective activity of sphingomyelin on macrophage survival and alleviates BLM-induced lung fibrosis.
A BMDMs were treated with vehicle, 10 μg/ml SM, or 10 μg/ml SM that was pre-incubated with 2.5 µg/ml mouse sTREM2, for 6 h in the absence of M-CSF. The levels of the indicated proteins were determined by Western blotting. The experiment was repeated three times with similar results. B Plated Mo-AMs were treated with SM (5 µg/ml), pre-incubated for 1 h with vehicle or 4 μg/ml mouse sTREM2, for 2 days and apoptosis determined. n = 4 independent culture of cells per group; mean ± SD; one-way ANOVA with Bonferroni’s multiple comparisons test. The experiment was repeated twice times with similar results. C, D Eight week C57BL/6 mice were i.t. instilled with saline or BLM (1.5 U/kg). One week later, mice were administered vehicle or sTREM2 (65 μg/kg), once a day for 8 days. 3 weeks after BLM treatment, lungs and BALs were collected, and whole lung hydroxyproline (C) and BAL TREM2 protein levels (D) were determined. n = 3, 5, and 5 mice, respectively for “C”; n = 3, 5, and 4 mice, respectively for “D”; mean ± SEM; one-way ANOVA with Bonferroni’s multiple comparisons test. Source data are provided as a Source Data file.
To extend the significance of our findings from mouse cells to human models, we investigated the effects of SM and sTREM2 on human macrophages. In peripheral blood mononuclear cell (PBMC)-derived macrophages, withdrawal of M-CSF led to reduced pro-survival signaling and increased apoptosis. These effects were reversed by SM treatment (Supplementary Fig. 3A). However, the protective effects of SM were largely abolished when it was pre-incubated with sTREM2 (Supplementary Fig. 3A). Similarly, in AMs from IPF patients, SM reduced apoptosis following M-CSF withdrawal. As observed in PBMC-derived macrophages, the protective effect of SM was blocked by sTREM2 in IPF AMs (Supplementary Fig. 3B).
TREM2 blocking antibody abolishes the protective effect of SM and attenuates lung fibrosis in bleomycin-treated mice
sTREM2 abrogation of the pro-survival effect of SM on macrophages indicates that interfering with the receptor-ligand engagement promotes Mo-AM apoptosis. This prompted us to ask if an antibody that blocks TREM2 engagements with its lipid ligands would produce a similar effect to sTREM2 on macrophages and lung fibrosis. In our previous study, a mouse TREM2 blocking antibody, with mutations in the Fc domain to prevent its recognition by the Fc receptor and complement, was developed and shown to block the binding of TREM2 and its high-avidity ligands38. This antibody is particularly useful because it can specifically demonstrate the effect of receptor TREM2 blockade without the confounding activity of antibody-dependent cellular cytotoxicity or antibody-dependent phagocytosis in vivo. We first tested its effect on receptor TREM2 in vitro. As shown in Fig. 8A, the TREM2 blocking antibody markedly attenuated the pro-survival activity of SM on TREM2 (+/+) Mo-AMs. Notably, SM had minimal impact on the apoptosis of TREM2 (-/-) Mo-AMs, which was not affected by the TREM2 blocking antibody either, reinforcing the notion that SM’s pro-survival effect is dependent on TREM2 in Mo-AMs (Fig. 8B).
Fig. 8. TREM2 blocking antibody abolishes the protective effect of SM and attenuates lung fibrosis in BLM-treated mice.
A, B TREM2 (+/+) or (-/-) Mo-AMs were pre-treated for 1 h with control (con Ab) or TREM2 blocking antibody (TREM2 Ab) (4 μg/ml), followed by incubation with SM (5 μg/ml) for 2 days and apoptosis determined. n = 3, 4, 4 (for “A”) and n = 4 (for “B”) independent culture of cells for each group; mean ± SD; one-way ANOVA with Bonferroni’s multiple comparisons test. n.s., non-significant. Experiments were repeated twice times with similar results. C–E Eight week C57BL/6 mice were i.t. instilled with saline or BLM (1.5 U/kg). One day later, mice were administered i.p. with control antibody (200 µg/mouse) or the TREM2 blocking antibody (200 µg/mouse), once every five days for a total of 4 times. Three weeks after BLM treatment, lungs were collected and whole lung hydroxyproline levels were determined (C). n = 3, 6, and 6 mice, respectively; mean ± SEM; one-way ANOVA with Bonferroni’s multiple comparisons test. Mo-AMs from the BLM treated mice were flow sorted (D). n = 6 mice per group; mean ± SEM; TGF-β1 in the BALs from the BLM treated mice was determined (E). n = 6 mice per group; mean ± SEM; unpaired two-tailed Student’s t-test. Source data are provided as a Source Data file.
Logically, we asked if this antibody would be beneficial in lung fibrosis in vivo. To test this, mice were treated with a control antibody or the TREM2 blocking antibody, one day after bleomycin administration, once every five days for a total of four times. As shown in Fig. 8C, the TREM2 blocking antibody significantly diminished bleomycin-induced lung fibrosis. Consistently, there were also decreased Mo-AM numbers and TGF-β1 levels in the fibrotic lungs of mice treated with the TREM2 blocking antibody (Fig. 8D, E). To minimize the potential confounding effects of the TREM2 blocking antibody on bleomycin-induced lung inflammation, we initiated the antibody treatment one week after bleomycin administration. As shown in Supplementary Fig. 4, this delayed treatment was equally effective in mitigating bleomycin-induced lung fibrosis. Together, these data suggest that the TREM2 blocking antibody inhibits lung fibrosis by promoting Mo-AM apoptosis. This finding also indicates that TREM2 blocking is a novel and effective therapeutic strategy for lung fibrosis.
Discussion
Although it has long been recognized that macrophages play a crucial role in the pathogenesis of lung fibrosis, it remains largely unclear what regulates their pro-fibrotic potency in the lung2,3,19–27. As a result, there is still a lack of effective strategies to target the pathogenic activity of these cells in lung fibrosis. Recent studies have made inroads into this challenge by demonstrating that it is only a subgroup, the Mo-AMs, and not the entire type of cells, that are responsible for the detrimental role of lung macrophages in pulmonary fibrosis1,3,10,13. In this study, we specifically focused on the Mo-AM subgroup and found an important mechanism by which these cells promote lung fibrogenesis. Our data suggest that TREM2, which is highly expressed in Mo-AMs and markedly induced in IPF macrophages, is a key mediator in this pathology and a valuable target for developing strategies to neutralize the pro-fibrotic effect of this pathologically significant group of cells in lung fibrosis.
Macrophage phenotype is not only decided by ontogeny but also reshaped by the residing environment53. As monocytes enter the alveolar space in response to various injuries and differentiate into macrophages, these cells encounter various stimuli to which they are not normally exposed in the new milieu. As a receptor prominently expressed on Mo-AMs, but not TR-AMs, TREM2 is likely a key sensor employed by these cells to receive environmental input, adjust their phenotypic manifestations, and thereby exert their impact on surrounding cells. We found that SM promoted Mo-AM survival in a TREM2-dependent manner. Given that phospholipid and sphingolipid species are abundant in the alveolar space, and their levels are further elevated in fibrotic lungs, this finding suggests that Mo-AM TREM2 likely serves as a key receptor to sense new cues and sustain their presence in the new alveolar settings. TREM2 may be a key factor necessary for the pathogenic activity of this subset of cells in lung fibrosis. On the other hand, this property of Mo-AMs also presents an opportunity for TREM2 interference as a new therapeutic strategy to treat lung fibrosis.
As we showed TREM2-mediated SM pro-survival activity in macrophages, we also found that TREM2 knockdown or knockout promoted macrophage apoptosis in the absence of obvious ligands. These findings suggest that TREM2 may have an intrinsic regulatory activity independent of its receptor function. Although the underlying mechanism remains unclear, this is not totally surprising because previous studies also found that TREM2-deficient macrophages demonstrated aberrant cellular metabolism and mitochondrial activities. This might also hold true for Mo-AMs. However, distinct from other types of tissue macrophages, Mo-AMs are exposed to a totally different and unique new environment in the alveolar space, where there is an abundance of TREM2 ligands. Therefore, the most likely scenario is that the pro-fibrotic phenotype of Mo-AMs is shaped by both intrinsic and extrinsic TREM2-mediated molecular events.
We showed that sTREM2 abrogated the anti-apoptotic effect of SM on macrophages and protected mice from lung fibrosis. However, we also found a considerable amount of sTREM2 in the BAL of bleomycin-treated mice. This result appears counterintuitive as it raises the question of why mice still develop lung fibrosis, as well as it argues against the demonstrated anti-fibrotic role of sTREM2. However, a plausible explanation is that sTREM2 shedding is a protective feedback mechanism developed by hosts to limit fibrotic development. The anti-fibrotic effect of sTREM2 in vivo nevertheless suggests an interesting strategy, which is to enhance TREM2 shedding, which may be effective in treating this pathology. Indeed, several proteases have been identified as TREM2 sheddases, such as ADAM1054,55.
We showed that a TREM2 blocking antibody abolished the protective effect of SM on macrophages and alleviated lung fibrosis. This suggests that blocking the engagement of TREM2 with its ligands is an efficacious therapeutic strategy to treat lung fibrosis. Although antibodies as remedies are not uncommon in disease treatment, the better strategy to block this receptor-ligand engagement may lie in a small molecular compound. However, special attention should be paid to identifying an effective compound that has a broad activity in blocking the binding of TREM2 with lipid ligands and perhaps other reported ligands that may also play a role in regulating the Mo-AM phenotype in a TREM2-dependent manner.
While conducting this study, a recently published report revealed that mice deficient in Mo-AM TREM2 were protected from bleomycin-induced lung fibrosis56. Although the underlying mechanism identified in that study differs from what is demonstrated here, both studies suggest that Mo-AM TREM2 represents a promising therapeutic target for lung fibrosis. However, it is important to note that monocyte-derived macrophage TREM2 has been shown to play a beneficial role in other contexts of organ fibrosis, particularly in metabolic liver diseases57–59. The apparently divergent roles of monocyte-derived macrophage TREM2, and even monocyte-derived macrophages themselves60, in influencing the progression and resolution of tissue fibrosis across different organs underscore the evolving understanding that macrophage phenotype, function, and adaptation are shaped by both their ontogeny and the local environment.
In summary, we found that Mo-AM TREM2 is a key mediator of the pro-fibrotic activity of these cells in lung fibrosis. Our findings improve the mechanistic understanding of the crucial role of Mo-AMs in the pathogenesis of this disease.
Methods
Study approval
Protocols for all experiments involving mice in this study were approved by the University of Alabama at Birmingham (UAB) Institutional Animal Care and Use Committee (IACUC). We have complied with all relevant ethical regulations for animal testing. The human protocol was approved by the UAB Institutional Review Board for Human Use (IRB), and written informed consent was obtained from all subjects involved in the human studies.
Reagents
Bleomycin was from Hospira. RNA isolation kit RNeasy Mini was from Qiagen. Type I collagenase, Dispase II and DNase I were from Worthington. Growth Factor Reduced Matrigel was from Corning. Recombinant mouse sTREM2 was from R&D systems. Recombinant human sTREM2 was from Sino Biological. N-palmitoyl-D-erythro-sphingosylphosphorylcholine (16:0 SM (d18:1/16:0)) was from Avanti Polar Lipids.
scRNA-seq data processing
Previously reported unique molecular identifier (UMI) count data of scRNA-seq datasets were downloaded from GEO database and the respective accession numbers were indicated in the figure legends. Analysis was performed in R version 3.6.1 using the Seurat package 3.0.4, base R functions and the plotting package ggplot2 2.3.4. UMI counts were scaled to 10,000 UMIs per cell, then log transformed with a pseudocount of 1. Macrophages with expression level of TREM2 greater than or equal to 0.3 are considered TREM2-high cells.
Mice
C57BL/6 mice, myeloid lineage Cre mice CX3CR1 Cre and Lyz2 Cre, TREM2 (-/-), TREM2 (fl/fl), Sftpc Cre and tdTomato reporter mice were purchased from The Jackson Laboratory. Mice with ablation of monocytic TREM2 (Mo-AM TREM2 (-/-)) were established by cross-breeding TREM2 (fl/fl) with the CX3CR1 Cre line. Mice with ablation of myeloid TREM2 were established by cross-breeding TREM2 (fl/fl) with the Lyz2 Cre line. ATII-specific tdTomato reporter mice (ATII tdTomato + ) were established by cross-breeding Sftpc Cre line with the tdTomato reporter line. The animals were housed under constant temperature and humidity ( ~ 21 °C, 40–60% humidity), 12 h light/12 h dark cycle and with free access to a standard diet and water. 8–10 weeks male mice were used for all experiments.
Cell apoptosis assay
Cell apoptosis was evaluated by Annexin V based assays using RealTime-Glo™ Annexin V Apoptosis and Necrosis Assay kit (Promega) or Pacific Blue™ Annexin V Apoptosis Detection Kit with PI (BioLegend), according to the manufacturers’ instructions.
Mouse BMDMs and human PBMC-derived macrophages
Mouse BMDMs were established as previously described61. Briefly, bone marrow cells were cultured in DMEM supplemented with 10% FBS and 50 ng/ml murine M-CSF (R&D Systems) for 5 days. Human PBMC-derived macrophages were established by culturing human PBMCs (Zen-Bio) in DMEM containing 10% FBS and 50 ng/ml human M-CSF (Sino Biological) for 6 days. The differentiated cells were then split and plated for following experiments.
Human lung and cell samples
Human lung tissues and BAL samples were obtained according to protocols approved by the UAB IRB. IPF and failed donor normal lung tissues were obtained from the University of Alabama at Birmingham Tissue Procurement and Cell Culture Core. BAL samples were obtained from IPF patients at the UAB clinic. To prepare AMs, BAL cells were collected by centrifugation and red blood cells were lysed. Cells were then plated for 1 h at 37 °C with 5% CO2 in DMEM containing 10% FBS, followed by extensive wash to remove non-adherent cells. The adherent AMs were used for following experiments.
Flow cytometry
Primary mouse ATII cells with tdTomato+ expression were isolated from ATII tdTomato+ reporter mice by fluorescence-activated cell sorting (FACS) using FACSAria II instrument (BD Biosciences). Briefly, lungs were minced and digested in Hank’s Balanced Salt Solution containing 0.1% type I collagenase, 0.1% dispase II, and 0.01% DNase I. Lung digests were passed through a 40-μm mesh cell strainer and red blood cells were lysed. Suspended single cells were used for FACS.
To isolate Mo-AMs and TR-AMs, BAL cells were blocked with TruStain FcX™ PLUS (anti-mouse CD16/32) antibody (BioLegend, clone S17011E) and stained with a cocktail of antibodies including PerCP/Cy5.5 anti-CD64 (BioLegend, clone X54-5/7.1), eFluor® 450 anti-F4/80 (clone BM8), Alexa Fluor® 488 anti-CD11b (clone M1/70) and eFluor® 660 anti-CD170 (Siglec F) (clone 1RNM44N), all from eBioscience. TR-AMs (CD64+ F4/80+ Siglec Fhigh CD11blow) and Mo-AMs (CD64+ F4/80+ Siglec Flow CD11bhigh) were isolated by FACS.
Alternatively, BAL cells were co-stained with BV421 anti-MERTK (BioLegend, clone 2B10C42) or eFluor® 450 anti-F4/80 (eBioscience, clone BM8), FITC anti-CD11b (BioLegend, clone M1/70), APC anti-Siglec F (BioLegend, clone S17007L) or APC anti-Ly-6G (BioLegend, clone 1A8), and rabbit anti-TREM2 (Abcam, cat # ab245227), followed by PE anti-rabbit 2nd antibody (BioLegend, cat # 406421) staining. Cell surface TREM2 levels in BAL cells were determined by flow cytometry using FACSymphony A3 instrument (BD Biosciences) and FlowJo software (version 10, Tree Star).
Alveolosphere culture of mouse ATII cells
FACS sorted tdTomato+ ATII cells and Mo-AMs were cultured together in a Matrigel three-dimensional (3D) culture system. Briefly, 60 µl Matrigel/medium mixture (1:1 ratio) containing 5,000 ATII cells and 25,000 Mo-AMs were plated in 96-well plates. After 1 h of solidification at 37 °C, 150 µl warm modified ATII maintenance medium (AMM)62 (without SB431542, BIRB796, DMH-1, with 5% FBS) was added. The organoids were cultured in humidified incubators at 37 °C with 5% CO2 and medium was changed every other day. After 10–14 days of culture, the organoid formation was visualized with fluorescent microscope. Alternatively, organoids were fixed with 4% paraformaldehyde (PFA) at room temperature for 1 h, embedded in O.C.T compound and cryosectioned (10 μm thick).
Hydroxyproline determinations
The right three (superior, middle and inferior) lobes and/or the left lobe of mouse lungs were homogenized in 2- or 3 ml H2O. 100 µl homogenates were incubated with 100 µl 12 N HCl at 100 °C for 24 h. Hydroxyproline levels were determined with the Abcam’s Hydroxyproline Assay Kit.
Histological staining
Masson’s trichrome staining for collagen deposition was performed using Trichrome Stain (Masson) Kit (Sigma-Aldrich) according to the manufacturer’s instructions.
Micro-CT scanning
The in vivo mouse lung micro-CT was performed using the µSPECT7/µCT instrument (MILabs) at the University of Alabama at Birmingham Small Animal Imaging Facility. During acquisition, the mice were under isoflurane anesthesia and images were acquired with the following parameters: magnification (ultra focus), scan angle (360 degrees), tube current (0.17 mA), tube voltage (55 kV), and with 75 ms exposure. After acquisition, the images were reconstructed in MILabs software at a voxel size of 40 µm. Quantitative image analysis was performed using MIM software (MIM Software Inc.) by a blinded observer to preserve objectivity and minimize bias. The entire lung area was delineated as regions of interest (ROIs) using a semi-automated threshold contour tool, applying specific Hounsfield unit (HU) thresholds to differentiate lung tissue from surrounding structures. This approach provided consistent and objective ROI definition across all images. The mean HU value within the ROI was calculated and used to statistically evaluate the lung density changes associated with fibrosis.
Immunofluorescence staining
Paraffin sections of human or mouse lungs were deparaffinized, rehydrated and antigen retrieved by boiling in citric acid-based antigen unmasking solution (Vector Laboratories, H-3300) for 15 min. Tissue sections or organoid cryosections were blocked in TBS containing 3% Bovine Serum Albumin (BSA) and 0.1% Triton X-100, flowed by blocking with Fc Receptor Blocker reagent (Innovex). Sections were then stained with primary antibodies (1:100-1:500 dilution) and fluorochrome conjugated secondary antibodies (1:500 dilution, ThermoFisher). After wash, tissue slides were selectively treated with Vector® TrueVIEW™ Autofluorescence Quenching kit (Vector Laboratories). The slides were mounted with VECTASHIELD antifade mounting medium (Vector Laboratories) and imaged using the Olympus IX73 Microscope System. Antibodies information is listed in Supplementary Table 1.
Untargeted metabolomics
Untargeted metabolomics was performed on BAL fluid (BALF) samples from normal mouse lungs and bleomycin-injured mouse lungs. Following centrifugation to remove cells, the supernatants were analyzed using Metabolon Inc.’s global untargeted metabolomics platform (HD4). Data extraction, compound identification, and quantification were conducted utilizing Metabolon’s proprietary hardware and software.
siRNA transfection
siRNA transfection was performed using HiPerFect reagents (Qiagen) according to the manufacturer’s instructions. ON-TARGETplus negative control siRNA (catalog number D-001810-01-05) and specific mouse TREM2 siRNA pool (catalog number L-040918-01-0005) were from Dharmacon.
RNA isolation and real-time PCR
Total RNA was extracted using Qiagen RNeasy Mini Kit. After reverse transcription, real-time PCR was performed using SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad) on Bio-Rad CFX384 Real-Time PCR System. Primer sequences are listed in Supplementary Table 2. To calculate fold change in the expression of these genes, ΔCt = Ct of Tubulin - Ct of individual genes was first obtained. ΔΔCt = ΔCt of treated groups - ΔCt of untreated control groups was then obtained. Fold change was calculated as 2ΔΔCt, with control groups as 1.
Western blotting
Western blotting was performed as previously described63. Briefly, proteins were separated by 6-15% SDS-PAGE and electro-transferred to PVDF membranes. The membranes were blocked in TBST with 5% BSA for 30 min at room temperature, incubated with primary antibodies overnight at 4°C, followed by incubation with HRP-conjugated secondary antibodies (Bio-Rad) for 1 h at room temperature. After washing, blots were developed using the SuperSignal™ West Dura Extended Duration Substrate (Thermo Scientific) and imaged on a Bio-Rad ChemiDoc Imaging system. Antibodies information is listed in Supplementary Table 1. All antibodies were used at a dilution of 1:2000. The uncropped/unedited western blot images are included in the Source Data file.
Enzyme-linked immunosorbent assay (ELISA)
Levels of TGF-β1, IL-1β and KC in the BALs were determined using the respective DuoSet ELISA kit (R&D Systems) according to the manufacturer’s instructions.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism 8. One-way ANOVA followed by the Bonferroni or Turkey test was used for multiple group comparisons. The unpaired two-tailed Student’s t-test was used for comparison between two groups. All comparisons were two-sided, and p < 0.05 was considered statistically significant. n = 3-4 biological replicates for all experiments unless indicated otherwise in the Figure Legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by NIH grants R35HL135830 and R01AI170913 to G.L. and United States Department of Defense grant W81XWH-20-1-0226 to G.L.
Author contributions
G.L. designed the study; H.C., S.B., N.X., M.H., A.J., H.L. performed the cell and animal experiments and analyzed the data; V.B.A. provided human lung sections; T.K. provided human alveolar macrophages; R.M.L. provided TGF-β1 adenovirus; M.C. provided the control and TREM2 blocking antibodies, and edited the manuscript; H.C. and G.L. wrote the manuscript. H.C. and S.B. contributed equally.
Peer review
Peer review information
Nature Communications thanks Lars Knudsen, and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data supporting the results of this study are presented within the Article, Supplementary Information, or the Source Data file. The metabolomics data generated in this study have been deposited to the Metabolomics Workbench under accession number ST003580. Additional information is available from the corresponding author upon reasonable request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Huachun Cui, Sami Banerjee.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-025-57024-0.
References
- 1.Misharin, A. V. et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J. Exp. Med.214, 2387–2404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morales-Nebreda, L., Misharin, A. V., Perlman, H. & Budinger, G. R. The heterogeneity of lung macrophages in the susceptibility to disease. Eur. Respir. Rev.24, 505–509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe, S., Alexander, M., Misharin, A. V. & Budinger, G. R. S. The role of macrophages in the resolution of inflammation. J. Clin. Invest.129, 2619–2628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen, W. J. et al. Fas determines differential fates of resident and recruited macrophages during resolution of acute lung injury. Am. J. Respir. Crit. Care Med.184, 547–560 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibbings, S. L. et al. Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood126, 1357–1366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mould, K. J. et al. Cell origin dictates programming of resident versus recruited macrophages during acute lung injury. Am. J. Respir. Cell Mol. Biol.57, 294–306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan, S. Y. & Krasnow, M. A. Developmental origin of lung macrophage diversity. Development143, 1318–1327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misharin, A. V., Morales-Nebreda, L., Mutlu, G. M., Budinger, G. R. & Perlman, H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol.49, 503–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bharat, A. et al. Flow cytometry reveals similarities between lung macrophages in humans and mice. Am. J. Respir. Cell Mol. Biol.54, 147–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCubbrey, A. L. et al. Selective and inducible targeting of CD11b+ mononuclear phagocytes in the murine lung with hCD68-rtTA transgenic systems. Am. J. Physiol. Lung Cell Mol. Physiol.311, L87–L100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCubbrey, A. L. et al. Deletion of c-FLIP from CD11b(hi) macrophages prevents development of bleomycin-induced lung fibrosis. Am. J. Respir. Cell Mol. Biol.58, 66–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King, E. M., Hume, P. S., Janssen, W. J. & McCubbrey, A. L. Isolation and analysis of macrophage subsets from the mouse and human lung. Methods Mol. Biol.2506, 257–267 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi, N. et al. A spatially restricted fibrotic niche in pulmonary fibrosis is sustained by M-CSF/M-CSFR signalling in monocyte-derived alveolar macrophages. Eur. Respir. J.55, 1900646 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe, S. et al. Resetting proteostasis with ISRIB promotes epithelial differentiation to attenuate pulmonary fibrosis. Proc. Natl Acad. Sci. USA118, e2101100118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McQuattie-Pimentel, A. C. et al. The lung microenvironment shapes a dysfunctional response of alveolar macrophages in aging. J. Clin. Invest.131, e140299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander, M. J., Budinger, G. R. S. & Reyfman, P. A. Breathing fresh air into respiratory research with single-cell RNA sequencing. Eur. Respir. Rev.29, 200060 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reyfman, P. A. et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med.199, 1517–1536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol.20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi, N., Walter, J. M. & Misharin, A. V. Alveolar macrophages. Cell Immunol.330, 86–90 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Gibbons, M. A. et al. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am. J. Respir. Crit. Care Med.184, 569–581 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Redente, E. F. et al. Tumor necrosis factor-alpha accelerates the resolution of established pulmonary fibrosis in mice by targeting profibrotic lung macrophages. Am. J. Respir. Cell Mol. Biol.50, 825–837 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan, S. H. & Kunkel, S. L. Inhibition of bleomycin-induced pulmonary fibrosis by nordihydroguaiaretic acid. The role of alveolar macrophage activation and mediator production. Am. J. Pathol.124, 343–352 (1986). [PMC free article] [PubMed] [Google Scholar]
- 23.Murray, L. A. et al. TGF-beta driven lung fibrosis is macrophage dependent and blocked by serum amyloid P. Int. J. Biochem. Cell Biol.43, 154–162 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Carre, P. C. et al. Increased expression of the interleukin-8 gene by alveolar macrophages in idiopathic pulmonary fibrosis. A potential mechanism for the recruitment and activation of neutrophils in lung fibrosis. J. Clin. Invest.88, 1802–1810 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasse, A. et al. A vicious circle of alveolar macrophages and fibroblasts perpetuates pulmonary fibrosis via CCL18. Am. J. Respir. Crit. Care Med.173, 781–792 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Moore, B. B. et al. GM-CSF regulates bleomycin-induced pulmonary fibrosis via a prostaglandin-dependent mechanism. J. Immunol.165, 4032–4039 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Baran, C. P. et al. Important roles for macrophage colony-stimulating factor, CC chemokine ligand 2, and mononuclear phagocytes in the pathogenesis of pulmonary fibrosis. Am. J. Respir. Crit. Care Med.176, 78–89 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young, L. R. et al. Epithelial-macrophage interactions determine pulmonary fibrosis susceptibility in Hermansky-Pudlak syndrome. JCI Insight1, e88947 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deczkowska, A., Weiner, A. & Amit, I. The physiology, pathology, and potential therapeutic applications of the TREM2 signaling pathway. Cell181, 1207–1217 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Ulland, T. K. & Colonna, M. TREM2 - a key player in microglial biology and Alzheimer disease. Nat. Rev. Neurol.14, 667–675 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Colonna, M. The biology of TREM receptors. Nat. Rev. Immunol.23, 580–594 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Y. et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell160, 1061–1071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaitin, D. A. et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell178, 686–698.e614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iizasa, E. et al. TREM2 is a receptor for non-glycosylated mycolic acids of mycobacteria that limits anti-mycobacterial macrophage activation. Nat. Commun.12, 2299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poliani, P. L. et al. TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Invest125, 2161–2170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulland, T. K. et al. TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell170, 649–663.e613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell169, 1276–1290.e1217 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Molgora, M. et al. TREM2 modulation remodels the tumor myeloid landscape enhancing anti-PD-1 immunotherapy. Cell182, 886–900.e817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramachandran, P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature575, 512–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fabre, T. et al. Identification of a broadly fibrogenic macrophage subset induced by type 3 inflammation. Sci. Immunol.8, eadd8945 (2023). [DOI] [PubMed] [Google Scholar]
- 41.Henlon, Y. et al. Single-cell analysis identifies distinct macrophage phenotypes associated with prodisease and proresolving functions in the endometriotic niche. Proc. Natl Acad. Sci. USA121, e2405474121 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, J. et al. Multiplexed digital spatial protein profiling reveals distinct phenotypes of mononuclear phagocytes in livers with advanced fibrosis. Cells11, 3387 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lv, J. et al. Dynamic atlas of immune cells reveals multiple functional features of macrophages associated with progression of pulmonary fibrosis. Front Immunol.14, 1230266 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Muynck, K. et al. Osteopontin characterizes bile duct-associated macrophages and correlates with liver fibrosis severity in primary sclerosing cholangitis. Hepatology79, 269–288 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daemen, S. et al. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep.34, 108626 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schupp, J. C. et al. Integrated single-cell atlas of endothelial cells of the human lung. Circulation144, 286–302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaddis, N. et al. LungMAP portal ecosystem: systems-level exploration of the lung. Am J Respir Cell Mol Biol70, 129–139 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore, B. B. & Hogaboam, C. M. Murine models of pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol.294, L152–L160 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Xie, N. et al. Glycolytic reprogramming in myofibroblast differentiation and lung fibrosis. Am. J. Respir. Crit. Care Med.192, 1462–1474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allawzi, A. et al. Redistribution of EC-SOD resolves bleomycin-induced inflammation via increased apoptosis of recruited alveolar macrophages. FASEB J.33, 13465–13475 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui, Y. et al. TREM2 deficiency aggravates renal injury by promoting macrophage apoptosis and polarization via the JAK-STAT pathway in mice. Cell Death Dis.15, 401 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soroosh, P. et al. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J. Exp. Med.210, 775–788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mass, E., Nimmerjahn, F., Kierdorf, K. & Schlitzer, A. Tissue-specific macrophages: how they develop and choreograph tissue biology. Nat. Rev. Immunol.23, 563–579 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schlepckow, K. et al. An Alzheimer-associated TREM2 variant occurs at the ADAM cleavage site and affects shedding and phagocytic function. EMBO Mol Med.9, 1356–1365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wunderlich, P. et al. Sequential proteolytic processing of the triggering receptor expressed on myeloid cells-2 (TREM2) protein by ectodomain shedding and gamma-secretase-dependent intramembranous cleavage. J. Biol. Chem.288, 33027–33036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu, X., Kang, H., Cao, S., Tong, Z. & Song, N. Blockade of TREM2 ameliorates pulmonary inflammation and fibrosis by modulating sphingolipid metabolism. Transl. Res.275, 1–17 (2025). [DOI] [PubMed] [Google Scholar]
- 57.Ganguly, S. et al. Lipid-associated macrophages’ promotion of fibrosis resolution during MASH regression requires TREM2. Proc. Natl Acad. Sci. USA121, e2405746121 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shan, S. et al. TREM2 protects against inflammation by regulating the release of mito-DAMPs from hepatocytes during liver fibrosis. Free Radic. Biol. Med.220, 154–165 (2024). [DOI] [PubMed] [Google Scholar]
- 59.Hendrikx, T. et al. Soluble TREM2 levels reflect the recruitment and expansion of TREM2(+) macrophages that localize to fibrotic areas and limit NASH. J. Hepatol.77, 1373–1385 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Cruz Tleugabulova, M. et al. Induction of a distinct macrophage population and protection from lung injury and fibrosis by Notch2 blockade. Nat. Commun.15, 9575 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banerjee, S. et al. MicroRNA let-7c regulates macrophage polarization. J. Immunol.190, 6542–6549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katsura, H. et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell27, 890–904.e898 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui, H. et al. CD38 mediates lung fibrosis by promoting alveolar epithelial cell aging. Am. J. Respir. Crit. Care Med.206, 459–475 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the results of this study are presented within the Article, Supplementary Information, or the Source Data file. The metabolomics data generated in this study have been deposited to the Metabolomics Workbench under accession number ST003580. Additional information is available from the corresponding author upon reasonable request. Source data are provided with this paper.