Abstract
Suicidal ideation (SI) and behavior (SB) are major public health concerns, but risk factors for their development and progression are poorly understood. We used ICD codes and a natural language processing algorithm to identify individuals in a hospital biobank with SI-only, SB, and controls without either. We compared the profiles of SB and SI-only patients to controls, and each other, using phenome-wide association studies (PheWAS) and polygenic risk scores (PRS). PheWAS identified many risk factors for SB and SI-only, plus specific psychiatric disorders which may be involved in progression from SI-only to SB. PRS for suicide attempt were only associated with SB, and even after accounting for psychiatric disorder PRS. SI PRS were only associated with SI-only, although not after accounting for psychiatric disorder PRS. These findings advance understanding of distinct genetic and clinical risk factors for SB and SI-only, which will aid in early detection and intervention efforts.
Subject terms: Genomics, Psychiatric disorders
Introduction
Suicide is the 11th leading cause of death in the United States [1]. Furthermore, for every suicide, it is estimated there are 38 individuals who make a nonfatal suicide attempt, and 265 who seriously consider suicide [1]. Suicidality is a growing public health concern, and the suicide rate has risen almost every year since 2000, but importantly, it is preventable with proper detection and intervention. Detection of those at risk for suicide is challenging, as risk factors for suicide outcomes remain limited in predictive ability [2]. Amongst the strongest predictors of suicide death is prior suicidal behavior (SB) [2–4], which encompasses suicide attempt (SA), preparatory behavior, and aborted or interrupted attempts; although, still, only 6.7% of individuals with previous SB die by suicide in the next 5–14 years [3, 4]. Nonfatal SB is somewhat better predicted by risk factors, with 29% of individuals experiencing SI attempting suicide in the future [5]. Even more elusive are risk factors which differentially contribute to SI and SB and/or mediate the progression from SI to SB. Several “ideation-to-action” theories [6–10], regarding factors that contribute to progression from SI to SB, suggest phenotypes more associated with SB than SI-only are likely those marked by impulsivity or habituation to pain, fear, and death. However, substantiating these theories with real-world data has been a challenge due to the limitations of epidemiological studies.
While epidemiological studies are often limited to demographic variables or a small selection of readily available risk factors for which prior hypotheses exist, electronic health record (EHR)-linked biobank data provides an opportunity to examine an abundance of clinical risk factors for association with suicide outcomes. The phenome-wide association study (PheWAS) [11, 12] method maps International Classification of Diseases (ICD) codes to “phecodes” encapsulating broad groups of clinical phenotypes, which can then be tested for association with a diagnosis or independent variable of interest. Such EHR-based studies have been able to identify more comprehensive risk factors for diseases than ever before. For example, a PheWAS of SB in veterans with schizophrenia and bipolar disorder identified associations with many psychiatric disorders, physical health conditions and laboratory test results [13]. Also, a PheWAS of SA polygenic risk scores (PRS) in the UK Biobank found associations with over 400 behavior-related and physiological phenotypes, many of which may have causal effects on SA as suggested by Mendelian Randomization analyses [14]. Furthermore, PheWAS can elucidate how related, yet distinct phenotypes may differ in their associations across the phenome. This has been studied for anxiety and depression [15] but not yet for SB vs. SI.
Genetic liability is also a crucial factor contributing to risk for SI and SB. Family and twin studies have estimated the heritability of suicide outcomes to be between 30–55% [16]. PRS studies have found PRS generated from GWAS of SA and SI predict up to 1.1% [17] and 0.6% [18] of the variance in their respective phenotypes. Various suicidality PRS have also shown associations with suicide outcomes in clinically ascertained cohorts [19], veteran research cohorts [13], and even a population-based study of children [20]. Furthermore, SA PRS, after conditioning on the genetic contributions of depression, has also been found to remain associated with SA [19, 21]. It is unknown whether associations with SA PRS remain after conditioning on PRS for psychiatric disorders other than depression, or whether SA or SI PRS exhibit associations with other suicide outcomes, given the strong yet incomplete genetic correlations between them (rg = 0.77–0.82) [18, 21].
Using the diverse and large EHR-linked BioMe Biobank, we identified individuals with SI-only and SB in their EHRs using a combination of ICD codes and a natural language processing (NLP) algorithm applied to clinical psychiatric notes. We detected differences in the demographic and behavioral characteristics of each group versus controls, and between individuals with SB versus SI-only. We performed PheWAS to test whether these groups had different patterns of associations with mental and physical health conditions. Lastly, we examined whether polygenic liability to SA and SI were associated with SB and SI-only in BioMe individuals, and whether these associations were independent from genetic liabilities to severe mental illnesses.
Methods
Data
Study population
The BioMe Biobank is an EHR-linked biobank of >50,000 patients from the Mount Sinai Health System (MSHS) [22]. Available data include demographic information, ICD codes, clinical notes, questionnaires, and genetic data. The current study restricted analyses to unrelated individuals with at least one recorded encounter with MSHS and a genetic ancestry similar to one of four genetic ancestry populations (African (AFR), East Asian (EAS), European (EUR), Latino (LAT)), resulting in 45,956 individuals. Genetic data for BioMe individuals had already undergone quality control, imputation, and genetically determined ancestry assignment, as previously described [22, 23]. Relevant study activities for the current report were approved by the Icahn School of Medicine at Mount Sinai Institutional Review Board (Institutional Review Board 07 0529) and all study participants provided written informed consent.
Suicide phenotypes
SI and SB were determined using a combination of two methods. First, we identified instances of SI and SB using lists of ICD 9th and 10th edition codes which have been validated to specifically capture SI or SB by the PGC Suicide Working Group [24] (Supplementary Tables 1, 2). Next, we used an NLP algorithm (https://github.com/wcmc-research-informatics/SI_Ideation; downloaded on April 18th, 2022) to detect SI or SB from unstructured clinical notes. These comprised 426,300 inpatient, outpatient, and emergency room psychiatric notes for 3565 individuals in our analytic sample. The NLP algorithm works by searching the notes for mentions of terms in a lexicon developed based on the Columbia-Suicide Severity Rating Scale [25], then determines whether the note contains an affirmative mention, which is marked as indicating SI or SB, accordingly. An important feature of this algorithm is that non-suicidal self-injury (NSSI), and thoughts of NSSI, are excluded from the definitions of SB and SI. Versions of this NLP algorithm [26] have been validated in several hospital systems including the King’s College London medical center, Weill Cornell Medicine [26], University of Utah Health Sciences Center [27] and MSHS [28]. The NLP algorithm used in the current study has been validated in MSHS (SI: accuracy = 0.88, precision = 0.85; SB: accuracy = 0.86, precision = 0.86) by comparing the classifications from the algorithm to gold standard classifications from notes read and annotated by trained assessors [28]. Prior to applying the algorithm, we cleaned the notes to remove standard clinical templates from screening assessments that could result in false positives.
SB cases comprised individuals with at least one SB ICD code or instance of SB detected in their notes by the NLP algorithm. SI-only cases were defined as individuals with at least one instance of SI detected by ICD codes or the NLP algorithm, but who did not have evidence of SB from either method, thus making the SB and SI-only case groups distinct. A single control group was constructed for comparison with both SI-only and SB, comprising individuals screened for the absence of both suicide phenotypes.
Sociodemographic and behavioral characteristics
BioMe participants completed several self-report questionnaires assessing demographic, lifestyle, and behavioral characteristics. Using these data, we derived seven characteristics of interest relating to age, sex, marital status, education level (“College”, whether an individual completed college), religiosity (whether or not an individual reported a religious affiliation), tobacco use (“Ever used tobacco”) and illicit drug use (“Ever used illicit drugs”). More specific descriptions and methods for deriving specific variables are described in the Supplementary Note. Age and sex variables were self-reported and unaltered. Some patients completed multiple surveys, and, in that case, the most recent responses were used, except for lifetime measures in which case all responses were assessed.
GWAS discovery datasets
We used summary statistics from published ancestry-specific GWAS of SA, SI, bipolar disorder (BD), depression (DEP), and schizophrenia (SCZ) to calculate PRS for BioMe individuals. Phenotypes used for PRS construction were selected based on the criteria that 1) ancestry-specific GWAS summary statistics were available for at least two of the genetic ancestry groups in BioMe 2) there was no sample overlap between the GWAS data and BioMe and 3) the phenotype was either a suicidal thought or behavior (SA and SI) or was significantly genetic correlated with suicidal thoughts or behaviors (BD, DEP, SCZ). Discovery datasets are described briefly below and in further detail in Supplementary Table 3:
SA: GWAS summary statistics were generated by the Psychiatric Genomics Consortium (PGC) Suicide Working Group comprising 43,871 SA cases of AFR, EAS, EUR, and LAT genetic ancestries [17].
SI: GWAS summary statistics were derived from ancestry-specific GWAS of 99,814 SI cases without SA performed in the Million Veteran Program in AFR, EAS, EUR, and LAT samples [18].
BD: GWAS summary statistics were generated by the PGC Bipolar Disorder Working Group’s meta-analysis of 41,917 BD cases from 57 cohorts of EUR genetic ancestry [29] and a separate meta-analysis of 2964 BD cases from 2 Japanese cohorts [30].
DEP: GWAS summary statistics were derived from a bi-ancestral (AFR and EUR) GWAS of 366,434 depression cases in the Million Veteran Program [31].
SCZ: GWAS summary statistics were generated by the PGC Schizophrenia Working Group’s meta-analysis [32] of 74,776 SCZ cases from 90 EUR and/or EAS cohorts and 9 LAT and/or AFR cohorts from the Genomic Psychiatry Cohort Consortium [33].
Analyses
Sociodemographic and behavioral differences
We performed Chi-squared tests seeking differences in proportions of six variables (sex, marital status, college, religiosity, ever used tobacco, and ever used illicit drugs) between SB cases and controls, SI-only cases and controls, and SB cases and SI-only cases. We used a t-test to test for a significant difference in average age between each of these comparison groups. We used a Bonferroni-corrected significance threshold of p < 0.05/21 tests = 2.38 × 10−3.
PheWAS
We performed three phenotype-based PheWAS, testing the associations between phecodes and SB cases vs. controls, SI-only cases vs. controls, and SB cases vs. SI-only cases. We used PheCodeX [34] to map ICD codes in BioMe to sets of “phecodes”. We followed the commonly used “Rule of Two” [35], such that cases were defined as having a phecode on two or more unique encounter dates and controls did not have the phecode present in their EHR. Individuals with only one instance of a phecode were not included in the analysis of that phecode. We recognize that this “Rule of Two” may be a limitation in that it may be too conservative for rare conditions; however, given our conservative case count threshold (described below), rare phecodes were unlikely to be included in the analysis. PheCodeX has several improvements from past PheCode versions. In addition to revisions to the labeling system and phecode categories, it allows for the incorporation of ICD-10 codes by implementing multi-mapping, such that each ICD code can map to multiple phecodes (e.g., the ICD code for “Cannabis dependence” can map to both the “Cannabis misuse or dependence” and “Psychoactive substance dependence” phecodes). To ensure sufficient power, we only tested associations with phecodes with 200 cases or more in each model [36], resulting in 894 phecodes tested in the PheWAS of SB versus controls, 888 phecodes tested in SI-only versus controls, and 161 phecodes tested in SB versus SI-only. We also removed phecodes which had substantial overlap with the ICD codes used to define SB and SI in these analyses, specifically MB_284.1 “Suicidal ideations” and MB_284.2 “Suicide and self-inflicted harm”. We performed the association tests using the PheWAS R package [37], specifying a logistic regression model in which one of the binary suicide phenotypes (e.g., SB vs. controls) predicted each of the binary phecodes. We controlled for age, age2, sex, genetically determined ancestry group, number of encounters and lifetime record coverage (calculated as length of their medical record divided by age). As an exception, sex was not used as a covariate when testing associations with sex-specific phecodes. Significant associations in each PheWAS were identified using a Bonferroni-corrected p-value threshold (0.05/number of phecodes tested). We favored the use of the association tests implemented in the PheWAS package over a traditional comparison of risk factor prevalences in cases and controls, as it allows for the inclusion of important covariates in the models and the published “phecode map” enables the assessment of many more clinical risk factors than when manual phenotype construction is necessary. Lastly, to investigate whether SB cases with low genetic liability had a higher burden of phecodes than SB cases with high genetic liability, we tested if the number of phecodes individuals had significantly differed between SB cases with SA PRS in the bottom 10% and SB cases with SA PRS in the top 10%. We did this using a logistic regression and covaried for age, age2, sex, genetic ancestry, and record coverage.
Associations with primary care utilization
Given that many BioMe patients with suicide outcomes in their EHR were identified through psychiatric care at MSHS, we investigated whether cases also received primary care through MSHS, and whether they did so as regularly as controls. To approximate whether and how regularly an individual received routine primary care at MSHS, we used a variable representing the number of annual wellness checks recorded in their EHR. Annual wellness checks are appointments with a primary care provider intended for a general examination rather than the treatment of specific conditions. However, individuals may still have specific medical issues/conditions that are assessed or treated during these visits. We created the variable by counting the number of years an individual had an ICD code for “Encounter for general adult medical examination” (ICD-10-CM code Z00.0) or “Encounter for newborn, infant and child health examinations” (ICD-10-CM code Z00.1). The distribution of number of annual wellness checks was zero-inflated, thus we tested the associations with SI-only and SB vs. controls, using a zero-inflated Poisson regression and the “pscl” R package [38]. To account for multiple tests, we used a Bonferroni-corrected p-value threshold (0.05/4 = 0.0125) to determine significance.
Polygenic risk scores
We used PRS-CSx [39], to calculate multi-ancestry PRS for SA, SI, BD, DEP, and SCZ in BioMe individuals. PRS-CSx uses GWAS summary statistics, i.e. discovery datasets, from multiple populations to estimate meta-analyzed SNP weights to calculate multi-ancestry PRS for individuals in a target cohort. PRS-CSx default settings were used except for increasing the number of MCM iterations to 10,000 and the number of burn-in iterations to 5000, as this has been shown to increase reproducibility of the posterior effect sizes generated [40].
We then tested if each of the suicide outcome PRS (SI, SA) were significantly associated with 1) SB versus controls, 2) SI-only versus controls, or 3) SB versus SI-only in BioMe using logistic regression models which covaried for the first 10 genetic principal components (PCs). Associations were first tested within ancestry, then meta-analyzed in a fixed-effects model using the metafor R package [41]. To ensure sufficient power, association tests were only performed when the effective sample size was > 100 and as such, the EAS ancestry group (Neff = 25.4–59.5) was not included in PRS analyses. To determine the proportion of phenotypic variance explained by the PRS in each model, we calculated R2 on the liability scale using the population prevalence for each phenotype (KSI = 9%, KSB = 2%, KSB in SI = 29%). To calculate the multi-ancestry meta-R2 we transformed the within-ancestry Rs to “Zr”s using the Fisher Z method [42], calculated the effective sample size weighted mean Zr, then converted it back to R2 on the liability scale. To account for multiple tests, we used a Bonferroni-corrected p-value threshold (0.05/6 = 0.0083) to determine significance.
Furthermore, as suicide outcomes are strongly genetically correlated with severe mental illnesses, for each significant PRS-suicide outcome association in BioMe, we also tested if the PRS explained unique variance in suicide outcomes not explained by polygenic liability to severe mental illnesses. To do so, we used multi-PRS models that tested the association with either the SA or SI PRS, while including BD, DEP, and SCZ PRS as additional predictors.
Results
Detection of SI and SB in BioMe
Table 1 displays the number of individuals of each phenotype identified according to each phenotyping method. Supplementary Figure 1 details the phenotyping procedure and provides information on overlap between phenotypes and by method. In total, combining information from both NLP and ICD code methods, we identified 1066 SB cases, 894 SI-only cases (individuals with SI but no evidence of SB) and 43,996 controls who had neither SB nor SI. Of the 1066 SB cases, 54 were identified by both ICD codes and the NLP algorithm, 62 were identified by only ICD codes, and 950 were identified by only the NLP algorithm. For the 894 SI-only cases, 33 were identified by both ICD codes and the NLP algorithm, 60 were identified by only ICD codes, and 801 were identified by only the NLP algorithm (Table 1, Supplementary Fig. 1). Overall, the NLP identified about 10 times more cases of SB and SI-only compared with ICD codes. Number of cases and controls in each genetic ancestry are available in Supplementary Table 4.
Table 1.
Number of individuals of each phenotype identified according to each phenotyping method.
| Phenotyping Method | N Suicidal Behavior Cases (prevalence) | N Suicidal Ideation-only Cases (prevalence) | N Controls (prevalence) |
|---|---|---|---|
| ICD Code Only | 62 (0.13%) | 60 (0.13%) | 43,996 (95.73%) |
| Note Only | 950 (2.07%) | 801 (1.74%) | |
| Both ICD Code and Note | 54 (0.12%) | 33 (0.07%) | |
| Unique Total | 1066 (2.32%) | 894 (1.95%) |
Suicidal ideation (SI)-only case counts represent those after excluding suicidal behavior (SB). Controls are screened for the absence of both SB and SI. The prevalence in BioMe (N = 45,956) for each group is in parentheses. More details are provided in Supplementary Figure 1.
ICD International Classification of Diseases.
Sociodemographic and behavioral differences
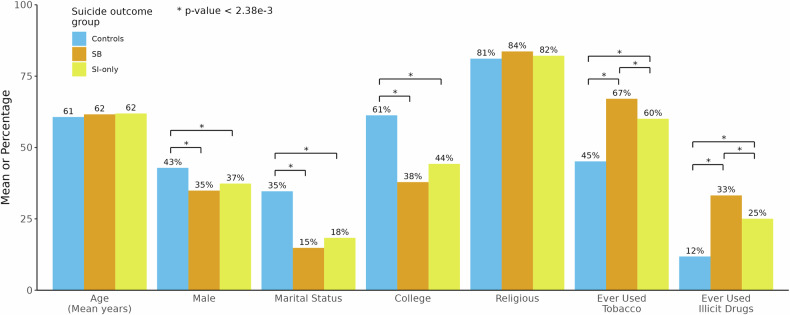
SB and SI-only cases compared to controls, were on average significantly (all p-values < 2.38 × 10−3) less likely to be male (SB = 35%, SI-only = 37%, controls = 43%), married (SB = 15%, SI-only = 18%, controls = 35%) or college educated (SB = 38%, SI-only = 44%, controls = 61%) (Fig. 1, Supplementary Table 5). Both SB and SI-only cases were more likely to have ever used tobacco (SB = 67%, SI = 60%, controls=45%) or illicit drugs (SB = 33%, SI-only = 25%, controls = 12%) compared with controls. SB cases and SI-only cases did not differ from controls in age or religiosity (p-values = 0.031–0.502). SI-only and SB cases did not differ from each other on most sociodemographic and behavioral characteristics, with the exceptions of SB cases being more likely than SI-only cases to have ever used tobacco (p = 1.53 × 10−3) or illicit drugs (p = 1.57 × 10−4) (Fig. 1, Supplementary Table 5).
Fig. 1. Comparison of sociodemographic and behavioral characteristics between individuals with and without different suicide outcomes.
Suicidal behavior (SB) cases and suicidal ideation (SI)-only cases were compared to determine their differences from controls and from each other. Values represent either means (for age) or percentages (for all others). Bonferroni-corrected significance threshold: p < 2.38 × 10−3. * indicates a statistically significant difference between two groups.
Phenome-wide association studies
PheWAS of SB vs. controls
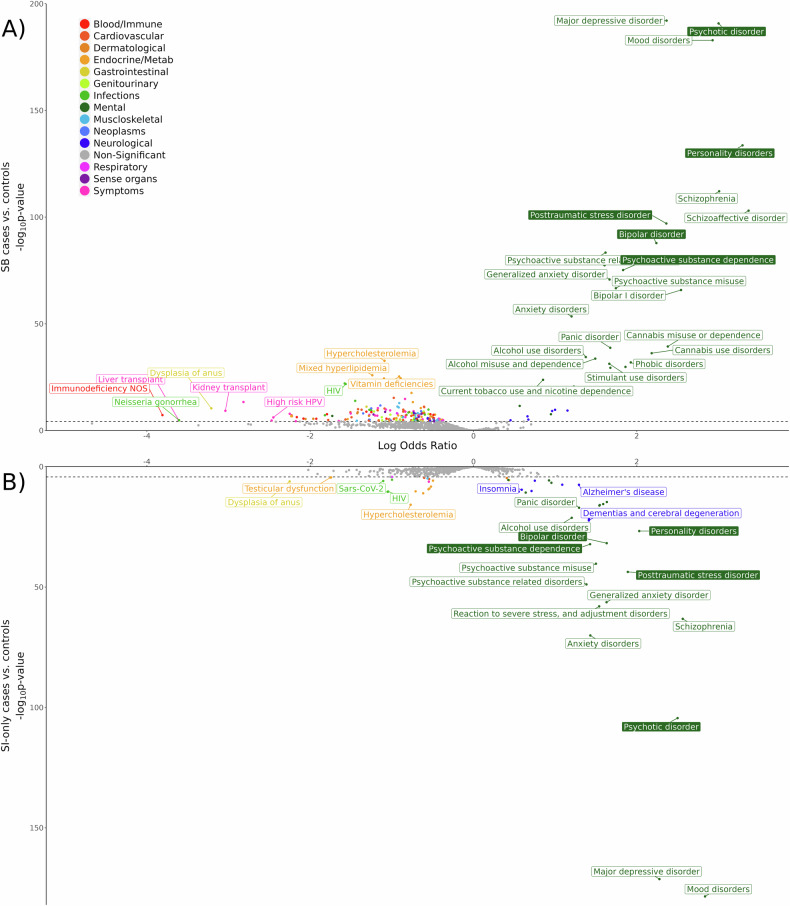
There were 216 phecodes with significantly different frequencies between SB cases and controls controlling for covariates (Fig. 2A, Supplementary Table 6). Mental health phecodes showed the strongest positive associations with SB (ORs = 1.76–29.06, all p-values < 5.59 × 10−5). For example, individuals with SB when compared with controls had a 29-times higher likelihood of schizoaffective disorder diagnoses (OR = 29.06, 95% CI:[21.42–39.44], p = 9.36 × 10−104), 27-times higher likelihood of personality disorder diagnoses (OR = 26.99, 95% CI:[20.77–35.06], p = 2.30 × 10−134), and 9.40–20.28 increased odds of diagnoses of other severe mental illnesses like major depressive disorder, bipolar disorder, and schizophrenia (Supplementary Table 6). Individuals with SB also had significantly increased odds of various substance use disorder diagnoses (e.g., cannabis misuse or dependence: OR = 10.81, 95% CI:[7.60–15.37], p = 4.25 × 10−40). Adverse physical health conditions were typically negatively associated with SB and those most depleted in SB versus controls were primarily phecodes related to symptoms, blood/immune diseases, endocrine/metabolic diseases, and infections (ORs = 0.02–0.12, all p-values < 5.59 × 10−5).
Fig. 2. PheWAS of suicide outcomes in BioMe.
PheWAS results comparing (A) SB cases and controls in the upper plot and (B) SI-only cases and controls in the lower plot. Color filled boxes indicate phecodes that were also significantly associated in the comparison between SB cases and SI-only cases. Some significant associations are not labeled for legibility, but full results can be found in Supplementary Tables 6–8. The dashed lines on the y-axes indicate the Bonferroni-corrected significance thresholds: SB vs. controls p = 5.59 × 10−5 and SI-only vs. controls p = 5.63 × 10−5.
PheWAS of SI-only vs. controls
Comparing SI-only versus controls, there were 64 phecodes with significant frequency differences (Fig. 2B, Supplementary Table 7) and the patterns of association were similar to those identified in the PheWAS of SB versus controls. Positive associations were found primarily with mental health and neurological phecodes; the diagnoses most associated with SI-only were mood disorders (OR = 17.06, 95% CI:[14.04–20.73], p = 3.32 × 10−179), schizophrenia (OR = 12.98, 95% CI:[9.64–17.48], p = 6.39 × 10−64), and psychotic disorders (OR = 12.19, 95% CI:[9.73–15.26], p = 3.80 × 10−105). Notably, diagnoses relating to insomnia and sleep disorders were not associated with SB but did show significant associations with SI-only (ORs = 1.74–1.81, p < 2.73 × 10−10). Like BioMe individuals with SB, we observed that individuals with SI-only had decreased likelihood of 20 adverse physical health conditions, specifically phecodes related to infections or endocrine/metabolic diseases (ORs = 0.11–0.61, all p-values < 5.63 × 10−5) such as dysplasia of anus, testicular dysfunction and hypofunction, coronavirus, and retrovirus and HIV (Supplementary Table 7).
PheWAS of SB vs. SI-only
To identify conditions associated with SB versus SI-only, and potentially involved in the progression from SI to SB, we performed a PheWAS comparing these two groups (Supplementary Table 8). There were five phecodes significantly elevated in SB versus SI-only cases (Fig. 2): personality disorders (OR = 3.09, 95% CI:[2.22–4.30], p = 1.88 × 10−11), bipolar disorder (OR = 1.92, 95% CI:[1.43–2.57], p = 1.46 × 10−5), posttraumatic stress disorder (OR = 1.67, 95% CI:[1.27–2.20], p = 2.52 × 10−4), psychoactive substance dependence (OR = 1.66, 95% CI:[1.28–2.15], p = 1.30 × 10−4), and psychotic disorder (OR = 1.60, 95% CI:[1.25–2.04], p = 1.78 × 10−4).
Phecode burden in SB cases by genetic liability
SB cases with SA PRS in the bottom 10% of the distribution did not have a significantly different number of phecodes compared with SB cases with SA PRS in the top 10% (beta = −0.003, se = 0.004, p = 0.452).
Associations with primary care utilization
When comparing the association with ever having received an annual wellness check at Mount Sinai, we found no differences between controls and SB cases (beta = −0.099, se = 0.102, p = 0.332) or SI-only cases (beta = −0.225, se = 0.103, p = 0.028). However, among those with at least one annual wellness check recorded in their EHR, SB and SI-only cases had significantly fewer wellness checks compared to controls (SB: beta = −0.137, se = 0.019, p = 1.52 × 10−12; SI-only: beta = –0.120, se = 0.021, p = 8.31 × 10−9).
Multi-ancestry polygenic risk score association models
The SA PRS was significantly associated with SB versus controls and explained 0.33% of phenotypic variance (liability scale) (beta = 0.178, se = 0.040, p = 9.93 × 10−6, Table 2). There was no significant difference in the SA PRS between SI-only cases and controls (beta = 0.092, se = 0.044, p = 0.035). Conversely, the SI PRS was not significantly associated with SB versus controls (beta = 0.122, se = 0.053, p = 0.021) but was significantly associated with and explained 0.26% of the variance in SI-only versus controls (beta = 0.155, se = 0.057, p = 6.82 × 10−3). Neither the SA nor SI PRS showed significantly different associations with SB cases vs. SI-only cases, however effects were still in the expected direction, with higher SA PRS in SB cases than SI-only cases (beta = 0.089, se = 0.06, p = 0.137, R2 = 0.16%, Table 2) and lower SI PRS in SB cases compared with SI-only cases (beta = −0.036, se = 0.075, p = 0.633, R2 = 0.18%).
Table 2.
PRS Associations.
| Outcome | PRS | Beta | SE | P | Liability R2% | N Cases | N Controls | Effective N |
|---|---|---|---|---|---|---|---|---|
| SB vs. controls | SA | 0.178 (0.153) | 0.04 (0.053) | 9.93 × 10−6* (1.81 × 10−4)** | 0.325 (0.240) | 1051 | 42,330 | 4,079.991 |
| SB vs. controls | SI | 0.122 | 0.053 | 0.021 | 0.066 | 1051 | 42,330 | 4,079.991 |
| SI-only vs. controls | SA | 0.092 | 0.044 | 0.035 | 0.146 | 883 | 42,330 | 3,446.367 |
| SI-only vs. controls | SI | 0.155 (0.141) | 0.057 (0.058) | 6.82 × 10−3* (0.015) | 0.257 (0.207) | 883 | 42,330 | 3,446.367 |
| SB vs. SI-only | SA | 0.089 | 0.060 | 0.137 | 0.160 | 1051 | 883 | 1,918.224 |
| SB vs. SI-only | SI | −0.036 | 0.075 | 0.633 | 0.178 | 1051 | 883 | 1,918.224 |
Test statistics from logistic regression testing the associations between suicide outcomes and PRS for suicide outcomes. Values in parentheses represent test statistics after controlling for BD, DEP, and SCZ PRS (multi PRS model). Bold indicates significance, * indicates significant associations in the single PRS model, and ** indicates significant associations in the multi PRS model. We used a Bonferroni-corrected p-value threshold of 8.33 × 10−3.
PRS polygenic risk score, SB suicidal behavior, SI suicidal ideation, SA suicide attempt.
The two significant associations (SA PRS with SB cases vs. controls and SI PRS with SI-only cases vs. controls) were tested again in a model which included PRS for bipolar disorder (BD), depression (DEP), and schizophrenia (SCZ) as additional predictors. The SA PRS remained significantly associated with SB after controlling for the psychiatric disorder PRS in the model, although the phenotypic variance explained was attenuated (beta = 0.153, se = 0.053, p = 1.81 × 10−4, R2 = 0.24%, Table 2). The SI PRS was no longer significantly associated with SI after accounting for the psychiatric disorder PRS (beta = 0.141, se = 0.058, p = 0.015, R2 = 0.21%). Results of all PRS models in the African (AFR), European (EUR), and Latino (LAT) ancestry groups separately were consistent in directions of effect but not significant (Supplementary Table 9).
Discussion
We applied novel methods for detecting suicidality in the EHRs of an ancestrally diverse group of BioMe Biobank patients. We defined and compared groups with SB, SI-only, and controls with no suicidality, and identified clinical and genetic risks for SB and SI-only which are distinguishable both from each other and from controls. Additionally, our findings highlight plausible risk factors involved in the progression from SI-only to SB and add to the mounting evidence that genetic liabilities to specific suicide severities have components which are distinct from each other and psychiatric disorders.
ICD codes for suicidality were relatively rare in the BioMe Biobank, with only 116 (0.25%) and 93 (0.20%) individuals having ICD codes for SB and SI-only, respectively. A majority of SB and SI-only ICD code cases with psychiatric notes available also had the respective phenotype identified by the NLP algorithm (76% for SB and 90% for SI). Notably, the NLP algorithm identified an additional 950 SB and 801 SI-only cases who did not have these ICD codes in their EHR. Overlap in case detection between the two methods was imperfect, with many more cases being detected by the NLP algorithm alone, but still some being detected only by the ICD data. One explanation may be the rarity and underutilization of SI/SB ICD codes due to coding practices, billing purposes, and administrative/legal issues related to the use of such codes [43]. Thus, as has been seen in other studies, ICD codes may poorly detect suicidality cases [13, 44]. Additionally, SI/SB occurrences may be documented in one data source but not the other, depending on differences in the timing of the records relative to the SI/SB. However, the imperfect convergence of the two methods underscores the importance of a combined approach, as cases missed by one method can potentially be detected by the other.
Consistent with prior studies [45–47], individuals with SB or SI-only were less likely to be males, married, and college educated, and more likely to have ever used tobacco or illicit drugs than controls. Individuals with SB did not significantly differ from individuals with SI-only for most sociodemographic characteristics examined, in line with reports of similar sociodemographic profiles between these groups [48]. As an exception, individuals with SB were more likely to have ever used tobacco or illicit drugs compared to SI-only cases. Previous reports have found mixed answers as to whether substance use significantly differs between individuals with SB and SI-only [48–52].
PheWAS indicated that specific diagnoses were associated with SB or SI-only compared to controls, most notably diagnoses relating to mental and neurological health. For both suicide outcomes, the strongest associations were with diagnoses such as personality, psychotic, and mood disorders, which are known to increase risk for SI and SB [52–54]. Both suicide outcomes were also associated with decreased odds of many adverse physical health diagnoses such as infections and blood/immune diseases, contradicting established notions that suicide outcomes are associated with adverse physical health conditions [55, 56]. We found that individuals with SB/SI-only had significantly fewer annual wellness checks in their EHR compared with controls but had no difference in ever having received an annual wellness check at the Mount Sinai Hospital System (MSHS). These results suggest that individuals with SB/ SI-only are less likely to seek or receive proper routine care for physical health conditions, which would in turn mean somatic diagnoses would be underrepresented in their EHR. This has previously been documented for conditions commonly co-occurring with suicide outcomes, such as psychiatric disorders or lower socioeconomic status [57–59].
The PheWAS comparing SB and SI-only revealed six phecodes associated with increased likelihood of SB compared to SI-only. The strongest association was with personality disorders (OR = 3.09, 95% CI = 2.22–4.30). Individuals with personality disorders, specifically borderline personality disorder, have higher rates of non-suicidal self-injury (14.4%) compared to other psychiatric disorders (6.8–10.9%) [60]. Non-suicidal self-injury may desensitize individuals to pain from self-injury, increasing their capacity to act on suicidal thoughts and engage in lethal self-injury [8, 10]. The next strongest association with SB versus SI-only was bipolar disorder (OR = 1.92, 95% CI = 1.43–2.57). Amongst common psychiatric disorders, impulsivity is most prevalent in personality and bipolar disorders [61]. Impulsivity may increase risk for acting on suicidal thoughts and risk of exposure to more painful events (e.g., drug use, physical fights) which contribute to an acquired capability for escalation of SI to SB [10]. Exposure to trauma (e.g., abuse, combat, physical and sexual violence) can also contribute to this capability by habituating an individual to painful and life threatening events [62, 63], potentially explaining the stronger association of posttraumatic stress disorder with SB compared to SI-only. Furthermore, psychotic disorder diagnoses were also elevated in individuals with SB compared to SI-only and psychotic experiences may elevate risk for SB amongst individuals with SI [64, 65]. Lastly, individuals with SB were more likely to have a diagnosis of psychoactive substance dependence than those with SI-only. Psychoactive substance use is linked with many of the mentioned risk factors (psychotic experiences [66], impulsivity, aggression [67], trauma [68]) and may also have different genetic relationships with distinct suicide outcomes [69].
Lastly, in PRS analyses, we observed phenotype-specific associations between polygenic liability and suicide severity. While SA PRS associated significantly with SB, it did not associate with SI-only, and SI PRS associated with SI-only but not SB. These results provide further evidence that polygenic liabilities to different suicide severities are somewhat distinct, as has been inferred by their incomplete genetic correlations [18, 21]. When testing these associations while controlling for PRS for BD, DEP, and SCZ, the SA PRS remained significantly associated with SB, although to a lesser extent; however, the SI PRS was no longer significantly associated with SI. This mirrors results from family studies indicating that relatives of suicidal individuals are more likely to exhibit SI or attempt suicide, yet only their risk for SA, and not SI, remains elevated after accounting for familial psychiatric disorders [70, 71]. Comorbid mental illnesses likely partially mediate genetic contributions to both suicide outcomes; however, they may play a more substantial role in the development of SI, whereas SB/SA likely have larger components of their genetic etiology which are independent from their shared genetics with BD, DEP, and SCZ [21].
Several limitations of the current study are worth noting. First, we may have missed cases due to incomplete coverage of patients’ medical histories in their EHR and because the NLP algorithm was applied only to psychiatric notes, as they were the only types of notes the algorithm was validated on in MSHS. However, our observed prevalences of SB (2.32%) and SI (3.85%) were similar to, although on the lower end of, documented lifetime prevalences of 0.5–5% for SB and 3.0–15.9% for SI [5]. Another explanation for the slightly lower prevalence in BioMe may be that all individuals in our study had to voluntarily enroll in BioMe, so patients with chronic mental health conditions like SI/SB may be underrepresented in our study due to participation bias in population-based and biobank studies [72–74]. Second, we only constructed PRS for a limited number of mental illnesses to assess independent genetic effects of suicide outcome PRS, given the current availability of multi-ancestry GWAS. As multi-ancestry GWAS of other relevant disorders become available, future studies should consider other psychiatric disorders’ involvement in these relationships. Third, instances of SI/SB were not linked to specific time points, and as a result we were not able to investigate when associated diagnoses occurred in relation to suicide outcomes. Finally, analyses were likely somewhat underpowered by small case sample sizes, which additionally prevented inclusion of individuals of other genetic ancestries that did not reach sufficient sample sizes.
Conclusions
In summary, this study characterizes the diagnoses associated with suicide outcomes in a real-world diverse hospital population, highlighting strong associations with psychiatric diagnoses, specifically personality, psychotic, mood, and substance use disorders. Underrepresentation of physical health phecodes in the EHRs of individuals with SI/SB suggests potential biases in care-seeking behaviors or healthcare provision for those with SI/SB. Furthermore, our findings that diagnoses for psychiatric disorders marked by impulsivity or exposure to painful experiences are more prevalent in individuals with SB than SI-only, suggest they may be involved in the progression from SI-only to SB. Finally, we show that PRS for SI and SA specifically predict SI-only and SB, respectively, and polygenic liability to SA is still independently associated with SB when accounting for BD, DEP, and SCZ PRS. These insights underscore the need for integrated approaches to suicide research that leverage the breadth of information available in diverse, large-scale biobanks to identify factors that can be used to detect high-risk individuals and subsequently reduce the incidence of suicide outcomes.
Supplementary information
Acknowledgements
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. 1842169 (PI Colbert), the National Institute of Mental Health R01MH123489 (PI Coon), and the Brain and Behavior Research Foundation NARSAD Young Investigator Award No: 29551 (PI Mullins). This work was supported in part through the computational and data resources and staff expertise provided by Scientific Computing and Data at the Icahn School of Medicine at Mount Sinai and supported by the Clinical and Translational Science Awards (CTSA) grant UL1TR004419 from the National Center for Advancing Translational Sciences. Research reported in this publication was also supported by the Office of Research Infrastructure of the National Institutes of Health under award number S10OD026880 and S10OD030463. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Work for the Japanese cohort was supported by Japan Agency for Medical Research and Development (AMED) grants JP24dk0307123, 22wm0425008, 21ek0109555, 21tm0424220, 21ck0106642, 23ek0410114 and 23tm0424225, Japan Society for the Promotion of Science (JSPS) KAKENHI grant JP22H03003, JP21H02854 and JP20H00462.
Author contributions
N.M., H.C., and S.M.C.C. designed the project. S.M.C.C. performed the analyses. N.M. supervised the analyses. M.P. and B.F. performed pre-processing of the BioMe data. L.L., J.P., and J.J.M. provided advice on the NLP algorithm. N.I., M.I., T.S., C.T. provided access to data. N.M., H.C., and S.M.C.C. contributed to interpretation of results. S.M.C.C. wrote the manuscript with contributions and approval from all authors.
Data availability
Researchers can apply for BioMe data access through dbGaP (dbGaP study accession: phs001644.v1.p1). GWAS summary statistics from the Psychiatric Genomics Consortium are publicly available (https://pgc.unc.edu/for-researchers/download-results/), or can be obtained through a data access application (https://pgc.unc.edu/for-researchers/data-access-committee/data-access-portal/). Summary statistics from the depression GWAS in MVP are also publicly available (https://medicine.yale.edu/lab/gelernter/stats/#project-1).
Code availability
The scripts used to run the NLP algorithm were downloaded from GitHub (https://github.com/wcmc-research-informatics/SI_Ideation). The PheWAS was run using the phewas R package (https://github.com/PheWAS/PheWAS).
Competing interests
Dr. Mann receives royalties for commercial use of the C-SSRS from the Research Foundation for Mental Hygiene and from Columbia University for the Columbia Pathways App. The other authors have no conflicts of interest to declare.
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. Relevant study activities for the current report were approved by the Icahn School of Medicine at Mount Sinai Institutional Review Board (Institutional Review Board 07 0529) and all study participants provided written informed consent.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-025-03287-6.
References
- 1.Centers for Disease Control and Prevention. Facts About Suicide. Available from: https://www.cdc.gov/suicide/facts. Accessed March 2024.
- 2.Franklin JC, Ribeiro JD, Fox KR, Bentley KH, Kleiman EM, Huang X, et al. Risk factors for suicidal thoughts and behaviors: a meta-analysis of 50 years of research. Psychol Bull. 2017;143:187–232. [DOI] [PubMed] [Google Scholar]
- 3.Beautrais AL. Further suicidal behavior among medically serious suicide attempters. Suicide Life Threat Behav. 2004;34:1–11. [DOI] [PubMed] [Google Scholar]
- 4.Suokas J, Suominen K, Isometsä E, Ostamo A, Lönnqvist J. Long-term risk factors for suicide mortality after attempted suicide–findings of a 14-year follow-up study. Acta Psychiatr Scand. 2001;104:117–21. [DOI] [PubMed] [Google Scholar]
- 5.Nock MK, Borges G, Bromet EJ, Alonso J, Angermeyer M, Beautrais A, et al. Cross-national prevalence and risk factors for suicidal ideation, plans and attempts. Br J Psychiatry. 2008;192:98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joiner T. Why people die by suicide. Harvard University Press; 2005. [Google Scholar]
- 7.Klonsky ED, May AM. Differentiating suicide attempters from suicide ideators: a critical frontier for suicidology research. Suicide Life Threat Behav. 2014;44:1–5. [DOI] [PubMed] [Google Scholar]
- 8.Klonsky ED, May AM. The three-step theory (3ST): a new theory of suicide rooted in the “ideation-to-action” framework. Int J Cognit Ther. 2015;8:114–29. [Google Scholar]
- 9.O’Connor RC. Towards an integrated motivational–volitional model of suicidal behaviour. Int Handb Suicide Prevention: Research, Policy Pract. 2011;1:181–98. [Google Scholar]
- 10.Van Orden KA, Witte TK, Cukrowicz KC, Braithwaite SR, Selby EA, Joiner TE Jr. The interpersonal theory of suicide. Psychol Rev. 2010;117:575–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastarache L, Denny JC, Roden DM. Phenome-wide association studies. JAMA. 2022;327:75–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bigdeli TB, Barr PB, Rajeevan N, Graham DP, Li Y, Meyers JL, et al. Correlates of suicidal behaviors and genetic risk among United States veterans with schizophrenia or bipolar I disorder. Mol Psychiatry. 2024;29:2399–407. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B, You J, Rolls ET, Wang X, Kang J, Li Y, et al. Identifying behaviour-related and physiological risk factors for suicide attempts in the UK Biobank. Nat Hum Behav. 2024;8:1784–97. [DOI] [PubMed] [Google Scholar]
- 15.Gao YN, Coombes B, Ryu E, Pazdernik V, Jenkins G, Pendegraft R, et al. Phenotypic distinctions in depression and anxiety: a comparative analysis of comorbid and isolated cases. Psychol Med. 2023;53:7766–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voracek M, Loibl LM. Genetics of suicide: a systematic review of twin studies. Wien Klin Wochenschr. 2007;119:463–75. [DOI] [PubMed] [Google Scholar]
- 17.Docherty AR, Mullins N, Ashley-Koch AE, Qin X, Coleman JRI, Shabalin A, et al. GWAS meta-analysis of suicide attempt: identification of 12 genome-wide significant loci and implication of genetic risks for specific health factors. Am J Psychiatry. 2023;180:723–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashley-Koch AE, Kimbrel NA, Qin XJ, Lindquist JH, Garrett ME, Dennis MF, et al. Genome-wide association study identifies four pan-ancestry loci for suicidal ideation in the Million Veteran Program. PLoS Genet. 2023;19:e1010623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colbert SMC, Mullins N, Chan G, Meyers JL, Schulman J, Kuperman S, et al. Polygenic contributions to suicidal thoughts and behaviors in a sample ascertained for alcohol use disorders. Complex Psychiatry. 2023;9:11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee PH, Doyle AE, Silberstein M, Jung JY, Liu RT, Perlis RH, et al. Associations between genetic risk for adult suicide attempt and suicidal behaviors in young children in the US. JAMA Psychiatry. 2022;79:971–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullins N, Kang J, Campos AI, Coleman JRI, Edwards AC, Galfalvy H, et al. Dissecting the shared genetic architecture of suicide attempt, psychiatric disorders, and known risk factors. Biol Psychiatry. 2022;91:313–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abul-Husn NS, Soper ER, Odgis JA, Cullina S, Bobo D, Moscati A, et al. Exome sequencing reveals a high prevalence of BRCA1 and BRCA2 founder variants in a diverse population-based biobank. Genome Med. 2019;12:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belbin GM, Cullina S, Wenric S, Soper ER, Glicksberg BS, Torre D, et al. Toward a fine-scale population health monitoring system. Cell. 2021;184:2068–2083.e2011. [DOI] [PubMed] [Google Scholar]
- 24.Monson ET, Colbert SMC, Andreassen OA, Ayinde OO, Bejan CA, Ceja Z et al. Defining suicidal thought and behavior phenotypes for genetic studies. medRxiv 2024.2007.2027.24311110 [Preprint]. 2024 https://www.medrxiv.org/content/10.1101/2024.07.27.24311110v1.
- 25.Posner K, Brown GK, Stanley B, Brent DA, Yershova KV, Oquendo MA, et al. The Columbia-Suicide Severity Rating Scale: initial validity and internal consistency findings from three multisite studies with adolescents and adults. Am J Psychiatry. 2011;168:1266–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cusick M, Velupillai S, Downs J, Campion TR Jr., Sholle ET, Dutta R, et al. Portability of natural language processing methods to detect suicidality from clinical text in US and UK electronic health records. J Affect Disord Rep. 2022;10:100430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coon H, Shabalin A, DiBlasi E, Monson ET, Han S, Kaufman EA et al. Absence of nonfatal suicidal behavior preceding suicide death reveals differences in clinical risks. medRxiv 2024.2006.2005.24308493 [Preprint]. 2024 https://www.medrxiv.org/content/10.1101/2024.06.05.24308493v1.
- 28.Lepow LA, Adekkanattu P, Cusick M, Coon H, Fennessy B, OConnell S et al. A natural language processing pipeline based on the columbia- suicide severity rating scale. medRxiv 2024.2012.2019.24319352 [Preprint]. 2024 https://www.medrxiv.org/content/10.1101/2024.12.19.24319352v1.
- 29.Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry. 2018;23:639–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levey DF, Stein MB, Wendt FR, Pathak GA, Zhou H, Aslan M, et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat Neurosci. 2021;24:954–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bigdeli TB, Genovese G, Georgakopoulos P, Meyers JL, Peterson RE, Iyegbe CO, et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol Psychiatry. 2020;25:2455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shuey MM, Stead WW, Aka I, Barnado AL, Bastarache JA, Brokamp E, et al. Next-generation phenotyping: introducing phecodeX for enhanced discovery research in medical phenomics. Bioinformatics. 2023;39:btad655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bastarache L. Using phecodes for research with the electronic health record: from PheWAS to PheRS. Annu Rev Biomed Data Sci. 2021;4:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verma A, Bradford Y, Dudek S, Lucas AM, Verma SS, Pendergrass SA, et al. A simulation study investigating power estimates in phenome-wide association studies. BMC Bioinforma. 2018;19:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30:2375–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeileis A, Kleiber C, Jackman S. Regression models for count data in R. J Stat Softw. 2008;27:1–25. [Google Scholar]
- 39.Ruan Y, Lin YF, Feng YA, Chen CY, Lam M, Guo Z, et al. Improving polygenic prediction in ancestrally diverse populations. Nat Genet. 2022;54:573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz LM, Merikangas AK, Ruparel K, Jacquemont S, Glahn DC, Gur RE, et al. Stability of polygenic scores across discovery genome-wide association studies. HGG Adv. 2022;3:100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1–48. [Google Scholar]
- 42.Fisher RA. Frequency Distribution of the Values of the Correlation Coefficient in Samples from an Indefinitely Large Population. Biometrika. 1915;10:507–21. [Google Scholar]
- 43.Anderson HD, Pace WD, Brandt E, Nielsen RD, Allen RR, Libby AM, et al. Monitoring suicidal patients in primary care using electronic health records. J Am Board Fam Med. 2015;28:65–71. [DOI] [PubMed] [Google Scholar]
- 44.Bejan CA, Ripperger M, Wilimitis D, Ahmed R, Kang J, Robinson K, et al. Improving ascertainment of suicidal ideation and suicide attempt with natural language processing. Sci Rep. 2022;12:15146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olfson M, Blanco C, Wall M, Liu SM, Saha TD, Pickering RP, et al. National trends in suicide attempts among adults in the United States. JAMA Psychiatry. 2017;74:1095–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schrijvers DL, Bollen J, Sabbe BG. The gender paradox in suicidal behavior and its impact on the suicidal process. J Affect Disord. 2012;138:19–26. [DOI] [PubMed] [Google Scholar]
- 47.Poorolajal J, Haghtalab T, Farhadi M, Darvishi N. Substance use disorder and risk of suicidal ideation, suicide attempt and suicide death: a meta-analysis. J Public Health. 2016;38:e282–e291. [DOI] [PubMed] [Google Scholar]
- 48.May AM, Klonsky ED. What distinguishes suicide attempters from suicide ideators? a meta-analysis of potential factors. Clin Psychology: Sci Pract. 2016;23:5–20. [Google Scholar]
- 49.Han B, Compton WM, Gfroerer J, McKeon R. Prevalence and correlates of past 12-month suicide attempt among adults with past-year suicidal ideation in the United States. J Clin Psychiatry. 2015;76:295–302. [DOI] [PubMed] [Google Scholar]
- 50.Kessler RC, Borges G, Sampson N, Miller M, Nock MK. The association between smoking and subsequent suicide-related outcomes in the National Comorbidity Survey panel sample. Mol Psychiatry. 2009;14:1132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mars B, Heron J, Klonsky ED, Moran P, O’Connor RC, Tilling K, et al. What distinguishes adolescents with suicidal thoughts from those who have attempted suicide? A population-based birth cohort study. J Child Psychol Psychiatry. 2019;60:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nock MK, Hwang I, Sampson N, Kessler RC, Angermeyer M, Beautrais A, et al. Cross-national analysis of the associations among mental disorders and suicidal behavior: findings from the WHO World Mental Health Surveys. PLoS Med. 2009;6:e1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baldessarini RJ, Tondo L. Suicidal risks in 12 DSM-5 psychiatric disorders. J Affect Disord. 2020;271:66–73. [DOI] [PubMed] [Google Scholar]
- 54.Xu YE, Barron DA, Sudol K, Zisook S, Oquendo MA. Suicidal behavior across a broad range of psychiatric disorders. Mol Psychiatry. 2023;28:2764–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calati R, Laglaoui Bakhiyi C, Artero S, Ilgen M, Courtet P. The impact of physical pain on suicidal thoughts and behaviors: meta-analyses. J Psychiatr Res. 2015;71:16–32. [DOI] [PubMed] [Google Scholar]
- 56.Scott KM, Hwang I, Chiu WT, Kessler RC, Sampson NA, Angermeyer M, et al. Chronic physical conditions and their association with first onset of suicidal behavior in the world mental health surveys. Psychosom Med. 2010;72:712–9. [DOI] [PubMed] [Google Scholar]
- 57.De Hert M, Cohen D, Bobes J, Cetkovich-Bakmas M, Leucht S, Ndetei DM, et al. Physical illness in patients with severe mental disorders. II. Barriers to care, monitoring and treatment guidelines, plus recommendations at the system and individual level. World Psychiatry. 2011;10:138–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Druss BG, Rosenheck RA, Desai MM, Perlin JB. Quality of preventive medical care for patients with mental disorders. Med Care. 2002;40:129–36. [DOI] [PubMed] [Google Scholar]
- 59.Happell B, Scott D, Platania-Phung C. Provision of preventive services for cancer and infectious diseases among individuals with serious mental illness. Arch Psychiatr Nurs. 2012;26:192–201. [DOI] [PubMed] [Google Scholar]
- 60.Ose SO, Tveit T, Mehlum L. Non-suicidal self-injury (NSSI) in adult psychiatric outpatients - A nationwide study. J Psychiatr Res. 2021;133:1–9. [DOI] [PubMed] [Google Scholar]
- 61.Moeller FG, Barratt ES, Dougherty DM, Schmitz JM, Swann AC. Psychiatric aspects of impulsivity. Am J Psychiatry. 2001;158:1783–93. [DOI] [PubMed] [Google Scholar]
- 62.Christensen H, Batterham PJ, Mackinnon AJ, Donker T, Soubelet A. Predictors of the risk factors for suicide identified by the interpersonal-psychological theory of suicidal behaviour. Psychiatry Res. 2014;219:290–7. [DOI] [PubMed] [Google Scholar]
- 63.Ryan AT, Daruwala SE, Perera KU, Lee-Tauler SY, Tucker J, Grammer G, et al. The relationship between trauma exposure and psychiatric hospitalization for suicide ideation or suicide attempt among patients admitted to a military treatment setting. Int J Env Res Public Health. 2020;17:2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeVylder JE, Lukens EP, Link BG, Lieberman JA. Suicidal ideation and suicide attempts among adults with psychotic experiences: data from the Collaborative Psychiatric Epidemiology Surveys. JAMA Psychiatry. 2015;72:219–25. [DOI] [PubMed] [Google Scholar]
- 65.Kelleher I, Devlin N, Wigman JT, Kehoe A, Murtagh A, Fitzpatrick C, et al. Psychotic experiences in a mental health clinic sample: implications for suicidality, multimorbidity and functioning. Psychol Med. 2014;44:1615–24. [DOI] [PubMed] [Google Scholar]
- 66.Degenhardt L, Saha S, Lim CCW, Aguilar-Gaxiola S, Al-Hamzawi A, Alonso J, et al. The associations between psychotic experiences and substance use and substance use disorders: findings from the World Health Organization World Mental Health surveys. Addiction. 2018;113:924–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brady KT, Myrick H, McElroy S. The relationship between substance use disorders, impulse control disorders, and pathological aggression. Am J Addict. 1998;7:221–30. [PubMed] [Google Scholar]
- 68.Konkolÿ Thege B, Horwood L, Slater L, Tan MC, Hodgins DC, Wild TC. Relationship between interpersonal trauma exposure and addictive behaviors: a systematic review. BMC Psychiatry. 2017;17:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Colbert SMC, Hatoum AS, Shabalin A, Li QS, Coon H, Nelson EC, et al. Exploring the genetic overlap of suicide-related behaviors and substance use disorders. Am J Med Genet B Neuropsychiatr Genet. 2021;186:445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brent DA, Bridge J, Johnson BA, Connolly J. Suicidal behavior runs in families. A controlled family study of adolescent suicide victims. Arch Gen Psychiatry. 1996;53:1145–52. [DOI] [PubMed] [Google Scholar]
- 71.Kim CD, Seguin M, Therrien N, Riopel G, Chawky N, Lesage AD, et al. Familial aggregation of suicidal behavior: a family study of male suicide completers from the general population. Am J Psychiatry. 2005;162:1017–9. [DOI] [PubMed] [Google Scholar]
- 72.Drivsholm T, Eplov LF, Davidsen M, Jørgensen T, Ibsen H, Hollnagel H, et al. Representativeness in population-based studies: a detailed description of non-response in a Danish cohort study. Scand J Public Health. 2006;34:623–31. [DOI] [PubMed] [Google Scholar]
- 73.Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, et al. Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taylor AE, Jones HJ, Sallis H, Euesden J, Stergiakouli E, Davies NM, et al. Exploring the association of genetic factors with participation in the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2018;47:1207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Researchers can apply for BioMe data access through dbGaP (dbGaP study accession: phs001644.v1.p1). GWAS summary statistics from the Psychiatric Genomics Consortium are publicly available (https://pgc.unc.edu/for-researchers/download-results/), or can be obtained through a data access application (https://pgc.unc.edu/for-researchers/data-access-committee/data-access-portal/). Summary statistics from the depression GWAS in MVP are also publicly available (https://medicine.yale.edu/lab/gelernter/stats/#project-1).
The scripts used to run the NLP algorithm were downloaded from GitHub (https://github.com/wcmc-research-informatics/SI_Ideation). The PheWAS was run using the phewas R package (https://github.com/PheWAS/PheWAS).