Abstract
Background
Cefiderocol, a newly introduced siderophore cephalosporin, exhibits activity against various multidrug-resistant (MDR) Gram-negative bacilli (GNB), including producers of Ambler class A, B and D carbapenemases. The TROJAN-MDR study aimed to (i) compare the in vitro activity of cefiderocol with other last-resort antibiotics against a well-characterized collection of Enterobacterales and Pseudomonas aeruginosa strains from Southern France, and (ii) assess the performance of available cefiderocol antimicrobial susceptibility testing (AST) methods.
Methods
The collection comprised 127 Enterobacterales from various clones, including 119 carbapenemase producers (93.7%), and 53 MDR P. aeruginosa. The minimum inhibitory concentrations (MICs) of cefiderocol were determined using the UMIC® broth microdilution method (BMD) as the reference. Comparators MICs were measured using Sensititre™ EUMDRXXF plates and Liofilchem strips for aztreonam-avibactam. Results were interpreted according to EUCAST breakpoints, with CLSI breakpoints also used for cefiderocol. The performance of the ComASP® BMD and disk diffusion on two different Mueller–Hinton media (Bio-Rad and BD) were evaluated according to ISO 20776-2:2007 and 2021.
Results
Cefiderocol demonstrated potent activity on Enterobacterales (81.9% susceptible) and P. aeruginosa (84.9%) using EUCAST breakpoints. Among Enterobacterales, the most effective comparators were colistin, aztreonam-avibactam, meropenem-vaborbactam, and amikacin, with susceptibility rates of 99.2%, 98.4%, 85%, and 76.4%, respectively. For P. aeruginosa, only colistin exhibited better activity (100%). The disk diffusion method showed superior performance on BD medium compared to Bio-Rad. The ComASP® method did not provide sufficient performance to be considered reliable.
Conclusions
Cefiderocol was highly active against a large collection of MDR GNB, including high-risk clones. It is crucial to assess susceptibility to this last-resort antibiotic using a validated method when considering clinical use.
Keywords: Cefiderocol, Multi-drug resistance, Enterobacterales, Pseudomonas aeruginosa, Difficult-to-treat, Carbapenemase, Aztreonam-avibactam, Antimicrobial susceptibility testing, UMIC®, ComASP®
Background
Carbapenemase- and/or extended spectrum β-lactamase (ESBL)-producing Enterobacterales, carbapenem-resistant Pseudomonas aeruginosa, and Acinetobacter baumannii are on the WHO's list of priority bacteria for which new active antibiotics are urgently needed [1]. The incidence of infections caused by these bacteria is increasing, and with treatment options rapidly depleting, they are associated with increased morbidity, mortality, and associated costs [2].
Cefiderocol, a recently developed siderophore β-lactam, is used as a last-resort antibiotic in the treatment of infections caused by aerobic Gram-negative bacteria in adult patients with limited therapeutic options [3]. Its innovative structure, a hybrid of cefepime and ceftazidime combined with a siderophore, enables it to penetrate bacterial cells like a Trojan horse, via both porins- and siderophores-mediated pathways [4]. Additionally, this unique structure increases its stability against β-lactamases and efflux pumps, enhancing its antibacterial activity [4].
Consequently, cefiderocol has a broad spectrum of activity against Gram-negative bacilli (GNB) such as Enterobacterales, P. aeruginosa, and A. baumannii, including multidrug-resistant (MDR) strains. It remains active in cases of porin deficiency, upregulation of efflux pumps, and production of various β-lactamases, including active serine carbapenemases and metallo-enzymes [5]. This molecule joins the list of last-resort antibiotics that can be used in the treatment of MDR-GNB infections, including ceftolozane-tazobactam, ceftazidime-avibactam, imipenem-relebactam, meropenem-vaborbactam, aztreonam-avibactam, colistin, tigecycline, and eravacycline [3].
To ensure treatment efficacy and increase the chances of clinical success, it is crucial to assess the in vitro susceptibility of cefiderocol using a reliable method [5]. Broth microdilution (BMD) for MIC determination is the commonly accepted reference method for antimicrobial susceptibility testing (AST) [6, 7]. Given the importance of iron in the mechanism of action of cefiderocol, iron-depleted Mueller–Hinton broth is required to mimic in vivo conditions. Commercially available BMD and classical agar diffusion methods using disks and MIC strips have also been proposed, but some studies have reported that performance failed to meet the recommendations of the International Organization for Standardization (ISO) [8–10]. In France, the UMIC® BMD (Bruker) is the only commercial method currently validated for both Enterobacterales and non-fermenting GNB by the National Reference Centers (NRC) for Antibiotic Resistance [11, 12]. In this study, we aimed to (i) compare the activity of cefiderocol with that of last-resort antibiotics on a panel of various clones of MDR-GNB strains isolated in the South of France and (ii) evaluate the performance of different AST methods to assess the in vitro activity of cefiderocol.
Material and methods
Bacterial isolates
We selected 180 MDR-GNB isolates from the Regional Reference Laboratory of Occitania in the South of France (LBMR BHRe, Nîmes University Hospital) whose genome has been well characterized by whole genome MultiLocus Sequence Typing. Strains were selected on the basis of their relevance (mechanism of resistance, high-risk clone, context of infection, non-duplicate isolates). The panel comprised (i) 127 Enterobacterales, including 119 carbapenemase-producing Enterobacterales (CPE) of classes A (n = 13), B (n = 32), D (n = 63), B + D (n = 10) or A + D (n = 1), as well as 8 non-CPE isolates (ESBL-producing Enterobacterales (ESBL-E), derepressed AmpC ± decreased permeability), and (ii) 53 MDR P.aeruginosa strains, including 6 metallo-β-lactamase (MBL) producers (Tables 1 and 2).
Table 1.
Characteristics of the Enterobacterales isolates
| Resistance mechanism (n. of strains) | Species (n. of strains) | Associated β-lactamases (n. of strains) | STs (n. of strains) | |
|---|---|---|---|---|
| Class A (13) | KPC-2 (9) | K. pneumoniae (1) | KLUY-1 (1) | 258 (1) |
| E. coli (1) | None | 744 (1) | ||
| C. freundii (3) | CMY-48 (3), CTX-M-15 (1) | 22 (3) | ||
| E. cloacae complex (4) | ACT-15 (1), ACT-23 (1), ACT-59 (1), ACT-67 (1) | 106 (1), 286 (1), 873 (1), new ST (1) | ||
| KPC-3 (4) | K. pneumoniae (4) | CTX-M-15 (1) | 11 (1), 147 (3) | |
| Class A + D (1) | KPC-2 + OXA-48 (1) | C. freundii (1) | CMY-48 (1) | 22 (1) |
| Class B (32) | NDM-1 (20) | K. pneumoniae (16) | CTX-M-15 (10) | 11 (1), 17 (1), 147 (6), 247 (5), 273 (1), 307 (1), 2084 (1) |
| C. freundii (1) | CMY-65 (1) | 91 (1) | ||
| E. cloacae complex (3) | ACT-23 (2), CMY-4 (1), CTX-M-15 (1) | 102 (2), 749 (1) | ||
| NDM-4 (1) | K. pneumoniae (1) | CTX-M-15 (1) | 16 (1) | |
| NDM-5 (5) | K. pneumoniae (2) | CTX-M-15 (1) | 219 (1), 785 (1) | |
| C. farmeri (1) | CTX-M-15 (1) | NA | ||
| E. coli (2) | CTX-M-15 (2) | 167 (1), 361 (1) | ||
| NDM-14 (2) | K. pneumoniae (2) | CTX-M-15 (2) | 147 (2) | |
| NDM-19 (1) | E. coli (1) | None | 38 (1) | |
| VIM-1 (1) | E. cloacae complex (1) | ACT-24 (1), CTX-M-9 (1) | 118 (1) | |
| VIM-4 (2) | E. cloacae complex (2) | ACT-23 (2) | 78 (1), 168 (1) | |
| Class B + D (10) | NDM-1 + OXA-48 (4) | K. pneumoniae (4) | CTX-M-15 (3) | 147 (2), 2084 (2) |
| NDM-5 + OXA-48 (2) | K. pneumoniae (2) | CTX-M-15 (1) | 383 (2) | |
| NDM-5 + OXA-181 (3) | K. pneumoniae (2) | CTX-M-15 (2) | 147 (1), new ST (1) | |
| E. cloacae complex (1) | ACT-73 (1), CTX-M-15 (1) | 116 (1) | ||
| NDM-7 + OXA-48 (1) | E. cloacae complex (1) | ACT-25 (1), CTX-M-15 (1) | 121 (1) | |
| Class D (63) | OXA-48 (59) | K. pneumoniae (18) | CTX-M-15 (6), DHA-1 (1) | 12 (1), 13 (3), 15 (1), 29 (1), 37 (1), 147 (1), 307 (4), 405 (1), 2074 (1), 2084 (2), 2674 (1), 3167 (1) |
| E. coli (2) | None | 38 (1), 648 (1) | ||
| C. freundii (11) | ACT-25 (1), CMY-48 (8), CMY-79 (1), CTX-M-15 (8), DHA-7 (1), SHV-12 (1) | 8 (1), 22 (4), 107 (1), 216 (4), 261 (1) | ||
| C. farmeri (1) | None (1) | NA | ||
| E. cloacae complex (27) | ACT-15 (6), ACT-16 (1), ACT-23 (1), ACT-24 (6), ACT-25 (2), ACT-45 (4), ACT-46 (1), ACT-59 (3), ACT-72 (2), ACT-73 (1), DHA-7 (1), MIR-6 (1), CTX-M-15 (12), KLUB-1 (4) | 66 (1), 78 (6), 90 (2), 106 (3), 109 (1), 113 (1), 114 (1), 116 (1), 121 (1), 168 (1), 171 (3), 279 (1), 418 (1), 595 (1), 873 (3) | ||
| OXA-181 (4) | K. pneumoniae (3) | CTX-M-15 (2) | 16 (1), 4988 (2) | |
| E. cloacae complex (1) | ACT-25 (1) | 418 (1) | ||
| Non-CPE (8) | K. pneumoniae (4) | CTX-M-15 (3), outer membrane decreased permeability (1) | 11 (1), 13 (1), 247 (1), new ST (1) | |
| C. freundii (1) | CMY-109 (1), CTX-M-15 (1), outer membrane decreased permeability (1) | 98 (1) | ||
| E. cloacae complex (2) | ACT-3 (1), ACT-59 (1), CTX-M-15 (1), KLUB-1 (1), outer membrane decreased permeability (2) | 252 (1), new ST (1) | ||
| K. aerogenes (1) | AmpC (1), outer membrane decreased permeability (1) | 137 (1) | ||
CPE Carbapenemase-Producing Enterobacterales, ST Sequence Type
Table 2.
Characteristics of the Pseudomonas aeruginosa isolates
| Resistance mechanism (n. of strains) | Associated β-lactamases (n. of strains) | STs (n. of strains) | |
|---|---|---|---|
| Class B CP (6) | IMP-1 (2) | OXA-35 (1), OXA-488 (1), OXA-904 (1), PDC-35 (1), PDC-119 (1) | 235 (1) |
| IMP-26 (1) | OXA-488 (1), PDC-35 (1) | 235 (1) | |
| VIM-2 (1) | OXA-488 (1), PDC-35 (1) | 35 (1) | |
| VIM-4 (1) | OXA-10 (1), OXA-488 (1), PDC-19a (1) | 308 (1) | |
| NDM-1 (1) | OXA-488 (1), PDC-19a (1) | 308 (1) | |
| Non-CP (47) | OXA-35 (1), OXA-50 (6), OXA-395 (6), OXA-396 (9), OXA-488 (9), OXA-494 (1), OXA-846 (1), OXA-847 (9), OXA-902 (1), OXA-904 (3), OXA-913 (2), PDC-3 (2), PDC-15 (1), PDC-16 (2), PDC-19a (3), PDC-19b (1), PDC-24 (1), PDC-25 (1), PDC-30 (1), PDC-34 (1), PDC-35 (5), PDC-37 (2), PDC-43 (3), PDC-58 (9), PDC-60 (2), PDC-80 (1), PDC-109 (1), PDC-114 (1), PDC-120 (1), PDC-172 (1), PDC-189 (1), PDC-198 (1), PDC-303 (2), PDC-321 (3), PDC-394 (1) | 27 (1), 175 (3), 207 (1), 235 (5), 244 (2), 253 (1), 267 (1), 308 (2), 309 (1), 313 (2), 446 (2), 609 (1), 611 (2), 654 (9), 679 (1), 1182 (1), 1184 (1), 1330 (1), 1613 (1), 1616 (1), 1755 (1), 2128 (2), 2475 (1), 2844 (1), 2996 (1), new ST (2) | |
CP Carbapenemase-Producers, ST Sequence Type
The vast majority of strains (n = 128, 71.1%) were isolated from diagnostic samples: respiratory samples (n = 42, 23.3%), urines (n = 33, 18.3%), blood (n = 26, 14.4%), abscesses (n = 21, 11.7%), bone (n = 4, 2.2%), catheter (n = 1, 0.6%), and cerebrospinal fluid (n = 1, 0.6%) (Table 3). Among the Enterobacterales, the most represented species were Klebsiella pneumoniae (n = 59, 46.5%), followed by the Enterobacter cloacae complex (n = 42, 33.1%), Citrobacter freundii (n = 17, 13.4%), Escherichia coli (n = 6, 4.7%), Citrobacter farmeri (n = 2, 1.6%), and Klebsiella aerogenes (n = 1, 0.8%).
Table 3.
Origin of the strains included in the study
| Specimen | Enterobacterales | Pseudomonas aeruginosa | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KPC | KPC + OXA-48-like | NDM | VIM | NDM + OXA-48-like | OXA-48-like | Non-CP | NDM | VIM | IMP | Non-CP | ||
| Rectal swab | 6 | 1 | 12 | 2 | 4 | 24 | 2 | 1 | 52 | |||
| Respiratory samples | 4 | 6 | 3 | 29 | 42 | |||||||
| Urine | 3 | 7 | 1 | 2 | 20 | 33 | ||||||
| Blood | 2 | 4 | 2 | 6 | 3 | 1 | 1 | 7 | 26 | |||
| Abcess | 2 | 2 | 2 | 5 | 1 | 1 | 1 | 7 | 21 | |||
| Bone | 1 | 3 | 4 | |||||||||
| Cerebrospinal fluid | 1 | 1 | ||||||||||
| Catheter | 1 | 1 | ||||||||||
| Total | 13 | 1 | 29 | 3 | 10 | 63 | 8 | 1 | 2 | 3 | 47 | 180 |
CP Carbapenemase-Producers
Control strains were associated with each set of AST: E. coli from American Type Culture Collection (ATCC) 25922 for Enterobacterales and P. aeruginosa ATCC 27853 for P. aeruginosa.
Comparators susceptibility testing
The susceptibility of the strains to various antibiotics was determined by the classic agar diffusion method on Mueller–Hinton (MH) medium (Bio-Rad, Marne-la-Coquette, France), in accordance with EUCAST recommendations. The MICs of comparators (ceftolozane-tazobactam, ceftazidime-avibactam, imipenem-relebactam, meropenem-vaborbactam, tigecycline, eravacycline, amikacin, fosfomycin and colistin) were measured by BMD using the Sensititre™ EUMDRXXF plate (ThermoFisher, Les Ulys, France), following the manufacturer's recommendations. The MICs of aztreonam-avibactam were determined with the recently marketed strips (Liofilchem, Roseto degli Abruzzi, Italia).
Results were interpreted according to EUCAST breakpoints [13]. In the absence of specific aztreonam-avibactam breakpoint for bacilli other than Enterobacterales, the aztreonam breakpoint (16 mg/L) was applied for P. aeruginosa isolates.
Cefiderocol susceptibility testing
The MICs of cefiderocol were determined by BMD using two different commercial tests, with the same 0.5 McFarland bacterial inoculum prepared for agar diffusion. For the UMIC® test (Bruker), 25 µL of the prepared suspension were added to the iron-depleted, cation-adjusted Mueller–Hinton broth medium provided (ID-CAMHB, 5 mL), and then 100 µL was distributed to each well of the strip. For the ComASP® BMD (Liofilchem), the 0.5 McFarland suspension was diluted to 1:20 in sterile 0.9% NaCl, then 400 µL of this dilution was added to the ID-CAMHB medium provided (3.6 mL), and 100 µL was dispensed into each well of the plate. In both cases, the MIC value was read by two different readers after 18–20 h of incubation at 35 °C. Results were interpreted according to both EUCAST and CLSI breakpoints [7, 13].
Furthermore, susceptibility to cefiderocol was assessed by the diffusion method using 30 µg cefiderocol disks (Mast group) on two different MH media: Bio-Rad MH Agar and BD BBL™ MH II Agar.
The UMIC® testing, approved by the French NRCs for Antibiotic Resistance, was considered as the gold standard [10–12].
Clinical performance of cefiderocol AST
The performances of the ComASP® BMD method and disk diffusion using Bio-Rad and BD BBL™ MH agar media were evaluated according to EUCAST 2024 breakpoints [13]. Performance parameters were calculated as follow: Categorical Agreement (CA) rate of isolates tested that yielded the same categorical interpretation as UMIC® BMD result, Essential Agreement (EA) rate of MIC values within 1 log2 dilution of the reference method), Major Errors (ME) number of isolates that yielded false-resistant results from number of isolates susceptible by the reference method), Very Major Errors (VME) number of isolates that tested false-susceptible from number of isolates resistant by the reference method, bias (percentage of MICs higher than the reference MIC subtracted by the percentage of MICs lower).
Acceptable performance was defined as recommended by the ISO 20776-2:2007 and 2021 standards: CA and EA ≥ 90%, VME ≤ 1.5%, ME ≤ 3%, and the difference for bias within ± 30% [14, 15].
Results
Overall in vitro activity of cefiderocol against MDR-GNB
Cefiderocol showed potent in vitro activity when tested using the BMD approach, which is approved by the French NRC (UMIC®) as the reference AST method. When interpreted using the EUCAST breakpoints, susceptibility was observed in 81.9% (104/127) of Enterobacterales and 84.9% (45/53) of P. aeruginosa MDR strains. Using the CLSI breakpoints, the susceptibility increased to 87.4% (111/127) for Enterobacterales and 94.3% (51/53) for P. aeruginosa (Table 4).
Table 4.
Overall in vitro activity of cefiderocol against MDR Enterobacterales and P. aeruginosa
| Strains (n. of isolates) | MICs range (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | Clinical categorization | ||||
|---|---|---|---|---|---|---|---|---|
| Breakpoints | S | I | R | |||||
| Enterobacterales (127) | ≤ 0.03 to > 32 | 1 | 8 | N. of isolates (%) | EUCAST | 104 (81.9) | NA | 23 (18.1) |
| CLSI | 111 (87.4) | 8 (6.3) | 8 (6.3) | |||||
| P. aeruginosa (53) | 0.125 to 32 | 1 | 4 | N. of isolates (%) | EUCAST | 45 (84.9) | NA | 8 (15.1) |
| CLSI | 50 (94.3) | 1 (1.9) | 2 (3.8) | |||||
S Susceptible, I susceptible, Increased exposure, R Resistant, NA Non-Applicable
In vitro activity of cefiderocol and comparators against Enterobacterales
Table 5 presents the in vitro activity of cefiderocol and its comparators against the 127 strains of Enterobacterales included in this study. Table 6 details the characteristics of cefiderocol-resistant strains.
Table 5.
Activity of cefiderocol and its comparators against MDR Enterobacterales and P. aeruginosa from South of France
| Resistance mechanism (Number of strains) | Antibiotic | MIC range (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | Clinical interpretation (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| S | I | R | |||||||
| Enterobacterales (n = 127) | |||||||||
| Total (127) | Cefiderocol | ≤ 0.03 > 32 | 1 | 8 | EUCAST | 81.9 | NA | 18.1 | |
| CLSI | 87.4 | 6.3 | 6.3 | ||||||
| Piperacillin-tazobactam | > 32 | > 32 | > 32 | 0 | NA | 100 | |||
| Cefepime | ≤ 1 to > 16 | 16 | > 16 | 19.7 | 12.6 | 67.7 | |||
| Ceftolozane-tazobactam | 0.5 to > 8 | > 8 | > 8 | 15.7 | NA | 84.3 | |||
| Ceftazidime-avibactam | ≤ 0.25 to > 16 | 1 | > 16 | 65.4 | NA | 34.6 | |||
| Aztreonam | ≤ 1 to > 32 | > 32 | > 32 | 25.2 | 1.6 | 73.2 | |||
| Aztreonam-avibactam | 0.023 to 24 | 0.125 | 0.75 | 98.4 | NA | 1.6 | |||
| Imipenem | ≤ 1 to > 8 | 2 | > 8 | 59.8 | 11.8 | 28.4 | |||
| Imipenem-relebactam | 0.12 to > 8 | 1 | > 8 | 68.5 | NA | 31.5 | |||
| Meropenem | ≤ 0.12 to > 16 | 2 | > 16 | 54.3 | 29.9 | 15.8 | |||
| Meropenem-vaborbactam | ≤ 0.06 to > 16 | 1 | 16 | 85 | NA | 15 | |||
| Amikacin | ≤ 2 to > 32 | 2 | > 32 | 76.4 | NA | 23.6 | |||
| Tobramycin | ≤ 0.5 to > 4 | > 4 | > 4 | 34.6 | NA | 65.4 | |||
| Fosfomycina | ≤ 16 to > 64 | 32 | > 64 | 69.3 | NA | 30.7 | |||
| Tigecyclineb | ≤ 0.5 to > 1 | ≤ 0.5 | > 1 | 63.8 | NA | 36.4 | |||
| Eravacyclinec | 0.06 to > 0.5 | 0.5 | > 0.5 | 66.1 | NA | 33.9 | |||
| Colistin | 0.25 to > 16 | ≤ 0.5 | ≤ 0.5 | 99.2 | NA | 0.8 | |||
| Class A | KPC (13) | Cefiderocol | 0.125 to 2 | 1 | 2 | EUCAST | 100 | NA | 0 |
| CLSI | 100 | 0 | 0 | ||||||
| Piperacillin-tazobactam | > 32 | > 32 | > 32 | 0 | NA | 100 | |||
| Cefepime | 2 to > 16 | 8 | > 16 | 0 | 23.1 | 76.9 | |||
| Ceftolozane-tazobactam | 4 to > 8 | > 8 | > 8 | 0 | NA | 100 | |||
| Ceftazidime-avibactam | ≤ 0.25 to 2 | 1 | 1 | 100 | NA | 0 | |||
| Aztreonam | 16 to > 32 | > 32 | > 32 | 0 | 0 | 100 | |||
| Aztreonam-avibactam | 0.032 to 0.75 | 0.19 | 0.5 | 100 | 0 | 0 | |||
| Imipenem | ≤ 1 to > 8 | 4 | 8 | 38.4 | 30.8 | 30.8 | |||
| Imipenem-relebactam | 0.12 to 0.5 | 0.25 | 0.5 | 100 | NA | 0 | |||
| Meropenem | 1 to > 16 | 4 | 16 | 38.4 | 38.4 | 23.2 | |||
| Meropenem-vaborbactam | ≤ 0.06 to 0.5 | ≤ 0.06 | 0.12 | 100 | NA | 0 | |||
| Amikacin | ≤ 2 to > 32 | 8 | > 32 | 69.2 | NA | 30.8 | |||
| Tobramycin | ≤ 0.5 to > 4 | > 4 | > 4 | 23.1 | NA | 76.9 | |||
| Fosfomycina | ≤ 16 to > 64 | ≤ 16 | 64 | 69.2 | NA | 30.8 | |||
| Tigecyclineb | ≤ 0.5 to > 1 | ≤ 0.5 | 1 | 76.9 | NA | 23.1 | |||
| Eravacyclinec | 0.25 to > 0.5 | 0.5 | > 0.5 | 76.9 | NA | 23.1 | |||
| Colistin | ≤ 0.5 to > 16 | ≤ 0.5 | ≤ 0.5 | 92.3 | NA | 7.7 | |||
| Class A + D | KPC + OXA-48-like (1) | Cefiderocol | NC | NC | NC | EUCAST | 100 | NA | 0 |
| CLSI | 100 | 0 | 0 | ||||||
| Piperacillin-tazobactam | NC | NC | NC | 0 | NA | 100 | |||
| Cefepime | NC | NC | NC | 0 | 0 | 100 | |||
| Ceftolozane-tazobactam | NC | NC | NC | 0 | NA | 100 | |||
| Ceftazidime-avibactam | NC | NC | NC | 100 | NA | 0 | |||
| Aztreonam | NC | NC | NC | 0 | 0 | 100 | |||
| Aztreonam-avibactam | NC | NC | NC | 100 | 0 | 0 | |||
| Imipenem | NC | NC | NC | 100 | 0 | 0 | |||
| Imipenem-relebactam | NC | NC | NC | 100 | NA | 0 | |||
| Meropenem | NC | NC | NC | 100 | 0 | 0 | |||
| Meropenem-vaborbactam | NC | NC | NC | 100 | NA | 0 | |||
| Amikacin | NC | NC | NC | 100 | NA | 0 | |||
| Tobramycin | NC | NC | NC | 0 | NA | 100 | |||
| Fosfomycina | NC | NC | NC | 100 | NA | 0 | |||
| Tigecyclineb | NC | NC | NC | 0 | NA | 100 | |||
| Eravacyclinec | NC | NC | NC | 0 | NA | 100 | |||
| Colistin | NC | NC | NC | 100 | NA | 0 | |||
| Class B | NDM (29) | Cefiderocol | 0.5 to > 32 | 2 | 8 | EUCAST | 79.3 | NA | 20.7 |
| CLSI | 86.2 | 6.9 | 6.9 | ||||||
| Piperacillin-tazobactam | > 32 | > 32 | > 32 | 0 | NA | 100 | |||
| Cefepime | 2 to > 16 | > 16 | > 16 | 0 | 6.9 | 93.1 | |||
| Ceftolozane-tazobactam | > 8 | > 8 | > 8 | 0 | NA | 100 | |||
| Ceftazidime-avibactam | > 16 | > 16 | > 16 | 0 | NA | 100 | |||
| Aztreonam | ≤ 1 to > 32 | > 32 | > 32 | 34.5 | 0 | 65.5 | |||
| Aztreonam-avibactam | 0.032 to 1.5 | 0.25 | 0.75 | 100 | NA | 0 | |||
| Imipenem | ≤ 1 to > 8 | 8 | > 8 | 17.2 | 20.7 | 62.1 | |||
| Imipenem-relebactam | 0.5 to > 8 | 8 | > 8 | 17.2 | NA | 82.8 | |||
| Meropenem | 0.25 to > 32 | 8 | > 16 | 17.3 | 37.9 | 44.8 | |||
| Meropenem-vaborbactam | 0.25 to > 16 | 8 | > 16 | 58.6 | NA | 41.4 | |||
| Amikacin | ≤ 2 to > 32 | 16 | > 32 | 44.8 | NA | 55.2 | |||
| Tobramycin | ≤ 0.5 to > 4 | > 4 | > 4 | 13.8 | NA | 86.2 | |||
| Fosfomycina | ≤ 16 to > 64 | 32 | 64 | 65.5 | NA | 34.5 | |||
| Tigecyclineb | ≤ 0.5 to > 1 | ≤ 0.5 | > 1 | 72.4 | NA | 27.6 | |||
| Eravacyclinec | 0.06 to > 0.5 | 0.5 | > 0.5 | 82.8 | NA | 17.2 | |||
| Colistin | ≤ 0.5 to 2 | ≤ 0.5 | 1 | 100 | NA | 0 | |||
| VIM (3) | Cefiderocol | 1 to 2 | NC | NC | EUCAST | 100 | NA | 0 | |
| CLSI | 100 | 0 | 0 | ||||||
| Piperacillin-tazobactam | > 32 | NC | NC | 0 | NA | 100 | |||
| Cefepime | 4 to > 16 | NC | NC | 0 | 33.3 | 66.7 | |||
| Ceftolozane-tazobactam | > 8 | NC | NC | 0 | NA | 100 | |||
| Ceftazidime-avibactam | 16 to > 16 | NC | NC | 0 | NA | 100 | |||
| Aztreonam | ≤ 1 to 16 | NC | NC | 33.3 | 33.3 | 33.3 | |||
| Aztreonam-avibactam | 0.125 to ≤ 1 | NC | NC | 100 | 0 | 0 | |||
| Imipenem | 2 to 4 | NC | NC | 66.7 | 33.3 | 0 | |||
| Imipenem-relebactam | 2 to 4 | NC | NC | 33.3 | NA | 66.7 | |||
| Meropenem | 1 to 2 | NC | NC | 100 | 0 | 0 | |||
| Meropenem-vaborbactam | 1 to 2 | NC | NC | 100 | NA | 0 | |||
| Amikacin | ≤ 2 to 16 | NC | NC | 66.7 | NA | 33.3 | |||
| Tobramycin | > 4 | NC | NC | 0 | NA | 100 | |||
| Fosfomycina | ≤ 16 to 32 | NC | NC | 100 | NA | 0 | |||
| Tigecyclineb | ≤ 0.5 | NC | NC | 100 | NA | 0 | |||
| Eravacyclinec | 0.25 to > 0.5 | NC | NC | 66.7 | NA | 33.3 | |||
| Colistin | ≤ 0.5 | NC | NC | 100 | NA | 0 | |||
| Overall class B (32) | Cefiderocol | 0.5 to > 32 | 1 | 8 | EUCAST | 81.3 | NA | 18.7 | |
| CLSI | 87.6 | 6.2 | 6.2 | ||||||
| Piperacillin-tazobactam | > 32 | > 32 | > 32 | 0 | NA | 100 | |||
| Cefepime | 2 to > 16 | > 16 | > 16 | 0 | 9.4 | 90.6 | |||
| Ceftolozane-tazobactam | 8 to > 8 | > 8 | > 8 | 0 | NA | 100 | |||
| Ceftazidime-avibactam | 16 to > 16 | > 16 | > 16 | 0 | NA | 100 | |||
| Aztreonam | > 32 | > 32 | > 32 | 34.4 | 3.1 | 62.5 | |||
| Aztreonam-avibactam | 0.032 to 1.5 | 0.125 | 0.75 | 100 | NA | 0 | |||
| Imipenem | ≤ 1 to > 8 | 8 | > 8 | 21.9 | 21.9 | 56.2 | |||
| Imipenem-relebactam | 0.5 to > 8 | 8 | > 8 | 18.7 | NA | 81.3 | |||
| Meropenem | 0.25 to 32 | 8 | > 16 | 28.1 | 40.6 | 31.2 | |||
| Meropenem-vaborbactam | 0.25 to > 16 | 4 | > 16 | 62.5 | NA | 37.5 | |||
| Amikacin | ≤ 2 to > 32 | 16 | > 32 | 46.9 | NA | 53.1 | |||
| Tobramycin | ≤ 0.5 to > 4 | > 4 | > 4 | 12.5 | NA | 87.5 | |||
| Fosfomycina | ≤ 16 to > 64 | 32 | 64 | 68.8 | NA | 31.2 | |||
| Tigecyclineb | ≤ 0.5 to > 1 | ≤ 0.5 | 1 | 75 | NA | 25 | |||
| Eravacyclinec | 0.06 to > 0.5 | 0.5 | > 0.5 | 81.2 | NA | 18.8 | |||
| Colistin | ≤ 0.5 to 2 | ≤ 0.5 | 0.5 | 100 | NA | 0 | |||
| Class B + D | NDM + OXA-48-like (10) | Cefiderocol | 0.125 to 4 | 2 | 2 | EUCAST | 90 | NA | 10 |
| CLSI | 100 | 0 | 0 | ||||||
| Piperacillin-tazobactam | > 32 | > 32 | > 32 | 0 | NA | 100 | |||
| Cefepime | 8 to > 16 | > 16 | > 16 | 0 | 0 | 100 | |||
| Ceftolozane-tazobactam | > 8 | > 8 | > 8 | 0 | NA | 100 | |||
| Ceftazidime-avibactam | > 16 | > 16 | > 16 | 0 | NA | 100 | |||
| Aztreonam | ≤ 1 to > 32 | > 32 | > 32 | 20 | 0 | 80 | |||
| Aztreonam-avibactam | 0.064 to 2 | 0.125 | 0.5 | 100 | NA | 0 | |||
| Imipenem | 2 to > 8 | 8 | > 8 | 10 | 10 | 80 | |||
| Imipenem-relebactam | 2 to > 8 | 8 | > 8 | 10 | NA | 90 | |||
| Meropenem | 4 to > 32 | 16 | > 32 | 0 | 40 | 60 | |||
| Meropenem-vaborbactam | 2 to > 16 | 16 | > 16 | 40 | NA | 60 | |||
| Amikacin | ≤ 2 to > 32 | 4 | > 32 | 60 | NA | 40 | |||
| Tobramycin | ≤ 0.5 to > 4 | > 4 | > 4 | 10 | NA | 90 | |||
| Fosfomycina | ≤ 16 to > 64 | 32 | > 64 | 60 | NA | 40 | |||
| Tigecyclineb | ≤ 0.5 to > 1 | 1 | 1 | 20 | NA | 80 | |||
| Eravacyclinec | 0.12 to > 0.5 | > 0.5 | > 0.5 | 30 | NA | 70 | |||
| Colistin | ≤ 0.5 | ≤ 0.5 | ≤ 0.5 | 100 | NA | 0 | |||
| Class D | OXA-48-like (63) | Cefiderocol | ≤ 0.03 to 16 | 1 | 8 | EUCAST | 84.1 | NA | 15.9 |
| CLSI | 87.2 | 6.4 | 6.4 | ||||||
| Piperacillin-tazobactam | > 32 | > 32 | > 32 | 0 | NA | 100 | |||
| Cefepime | ≤ 1 to > 16 | 4 | > 16 | 34.9 | 19.1 | 46 | |||
| Ceftolozane-tazobactam | 0.5 to > 8 | > 8 | > 8 | 28.6 | NA | 71.4 | |||
| Ceftazidime-avibactam | ≤ 0.25 to 16 | 0.5 | 2 | 98.4 | NA | 1.6 | |||
| Aztreonam | ≤ 1 to > 32 | 32 | > 32 | 30.2 | 1.6 | 68.2 | |||
| Aztreonam-avibactam | 0.023 to 24 | 0.125 | 0.75 | 98.4 | NA | 1.6 | |||
| Imipenem | ≤ 1 to > 8 | ≤ 1 | 4 | 87.3 | 4.8 | 7.9 | |||
| Imipenem-relebactam | 0.12 to > 8 | 1 | 2 | 92.1 | NA | 7.9 | |||
| Meropenem | 0.5 to > 16 | 1 | 4 | 76.2 | 22.2 | 1.6 | |||
| Meropenem-vaborbactam | 0.25 to > 16 | 1 | 4 | 98.4 | NA | 1.6 | |||
| Amikacin | ≤ 2 to > 32 | ≤ 2 | 8 | 92.1 | NA | 7.9 | |||
| Tobramycin | ≤ 0.5 to > 4 | 2 | > 4 | 52.4 | NA | 47.6 | |||
| Fosfomycina | ≤ 16 to > 64 | ≤ 16 | > 64 | 76.2 | NA | 23.8 | |||
| Tigecyclineb | ≤ 0.5 to > 1 | ≤ 0.5 | > 1 | 63.5 | NA | 36.5 | |||
| Eravacyclinec | 0.12 to > 0.5 | 0.5 | > 0.5 | 63.5 | NA | 36.5 | |||
| Colistin | ≤ 0.25 to 1 | ≤ 0.5 | ≤ 0.5 | 100 | NA | 0 | |||
| Non CPE (8) | Cefiderocol | 0.5 to > 32 | NC | NC | EUCAST | 25 | NA | 75 | |
| CLSI | 50 | 25 | 25 | ||||||
| Piperacillin-tazobactam | > 32 | NC | NC | 0 | NA | 100 | |||
| Cefepime | 4 to > 16 | NC | NC | 0 | 12.5 | 87.5 | |||
| Ceftolozane-tazobactam | 0.5 to > 8 | NC | NC | 25 | NA | 75 | |||
| Ceftazidime-avibactam | ≤ 0.25 to > 16 | NC | NC | 87.5 | NA | 12.5 | |||
| Aztreonam | 8 to > 32 | NC | NC | 0 | 0 | 100 | |||
| Aztreonam-avibactam | 0.032 to 16 | NC | NC | 87.5 | NA | 12.5 | |||
| Imipenem | ≤ 1 to 8 | NC | NC | 87.5 | 0 | 12.5 | |||
| Imipenem-relebactam | 0.25 to 2 | NC | NC | 100 | NA | 0 | |||
| Meropenem | ≤ 0.12 to 8 | NC | NC | 75 | 25 | 0 | |||
| Meropenem-vaborbactam | ≤ 0.06 to 4 | NC | NC | 100 | NA | 0 | |||
| Amikacin | ≤ 2 to 8 | NC | NC | 100 | NA | 0 | |||
| Tobramycin | ≤ 0.5 to > 4 | NC | NC | 37.5 | NA | 62.5 | |||
| Fosfomycina | ≤ 16 to > 64 | NC | NC | 37.5 | NA | 62.5 | |||
| Tigecyclineb | ≤ 0.5 to > 1 | NC | NC | 62.5 | NA | 37.5 | |||
| Eravacyclinec | 0.25 to > 0.5 | NC | NC | 62.5 | NA | 37.5 | |||
| Colistin | ≤ 0.5 | NC | NC | 100 | NA | 0 | |||
| Pseudomonas aeruginosa (n = 53) | |||||||||
| Overall (53) | Cefiderocol | 0.125 to 32 | 1 | 4 | EUCAST | 84.9 | NA | 15.1 | |
| CLSI | 94.3 | 1.9 | 3.8 | ||||||
| Piperacillin-tazobactam | 16 to > 32 | > 32 | > 32 | 0 | 7.5 | 92.5 | |||
| Cefepime | 4 to > 16 | > 16 | > 16 | 0 | 15.1 | 84.9 | |||
| Ceftolozane-tazobactam | 0.5 to > 8 | 4 | > 8 | 56.6 | NA | 43.4 | |||
| Ceftazidime-avibactam | 1 to > 16 | 8 | > 16 | 52.8 | NA | 47.2 | |||
| Aztreonam | 2 to > 32 | 32 | > 32 | 0 | 18.9 | 81.1 | |||
| Aztreonam-avibactamd | 2 to > 256 | 16 | > 256 | 0 | 52.8 | 47.2 | |||
| Imipenem | ≤ 1 to > 8 | > 8 | > 8 | 0 | 30.2 | 69.8 | |||
| Imipenem-relebactam | 0.25 to > 8 | 2 | > 8 | 56.6 | NA | 43.4 | |||
| Meropenem | 0.5 to > 32 | 16 | > 16 | 13.2 | 35.8 | 51 | |||
| Meropenem-vaborbactam | 0.5 to > 16 | 16 | > 16 | 47.2 | NA | 52.8 | |||
| Amikacin | ≤ 2 to > 32 | 16 | > 32 | 67.9 | NA | 32.1 | |||
| Tobramycin | ≤ 0.5 to > 4 | 2 | > 4 | 52.8 | NA | 47.2 | |||
| Fosfomycine | ≤ 16 to > 64 | > 64 | > 64 | NA | NA | NA | |||
| Colistin | ≤ 0.5 to 2 | 2 | 2 | 100 | NA | 0 | |||
| Class B carbapenemase producers (6) | Cefiderocol | 0.5 to 16 | NC | NC | EUCAST | 66.7 | NA | 33.3 | |
| CLSI | 83.3 | 0 | 16.7 | ||||||
| Piperacillin-tazobactam | 32 to > 32 | NC | NC | 0 | 0 | 100 | |||
| Cefepime | > 16 | NC | NC | 0 | 0 | 100 | |||
| Ceftolozane-tazobactam | > 8 | NC | NC | 0 | NA | 100 | |||
| Ceftazidime-avibactam | > 16 | NC | NC | 0 | NA | 100 | |||
| Aztreonam | 4 to > 32 | NC | NC | 0 | 50 | 50 | |||
| Aztreonam-avibactamd | 3 to 256 | NC | NC | 0 | 66.7 | 33.3 | |||
| Imipenem | > 8 | NC | NC | 0 | 0 | 100 | |||
| Imipenem-relebactam | > 8 | NC | NC | 0 | NA | 100 | |||
| Meropenem | > 16 | NC | NC | 0 | 0 | 100 | |||
| Meropenem-vaborbactam | > 16 | NC | NC | 0 | NA | 100 | |||
| Amikacin | 16 to > 32 | NC | NC | 16.7 | NA | 83.3 | |||
| Tobramycin | > 4 | NC | NC | 0 | NA | 100 | |||
| Fosfomycine | 32 to > 64 | NC | NC | NA | NA | NA | |||
| Colistin | 1 to 2 | NC | NC | 100 | NA | 0 | |||
S Susceptible, I susceptible Increased exposure, R Resistant, NA Non-Applicable, NC Not Calculated because the number of strains was less than 10
aIntravenous breakpoints
bE. coli and Citrobacter koseri breakpoints
cE. coli breakpoints
dAztreonam breakpoint (16 mg/L) applied for P. aeruginosa strains
eNo clinical breakpoints
Table 6.
Characteristics of the resistant strains according to EUCAST breakpoints
| Species (number of strains) | Resistance mechanism | Strain | β-lactamases content and other β-lactams resistance proteins | ST | Cefiderocol MIC (mg/L) |
|---|---|---|---|---|---|
| K. pneumoniae (9) | Class B | 1866 | NDM-5, SHV-77, LAP-2, TEM-1, OmpK36 mutations | 25 | 16 |
| 1976 | NDM-5, CTX-M-15, SHV-223, TEM-1, OmpK36 mutations | 219 | 8 | ||
| Class B + D | 1724 | NDM-1, OXA-48, CTX-M-15, OXA-1, SHV-11, TEM-1, OmpK36 mutations | 2084 | 4 | |
| Class D | 1966 | OXA-181, CTX-M-15, SHV-28, OmpK36 mutations | 4988 | 16 | |
| 2069 | OXA-181, CTX-M-15, SHV-28, OmpK36 mutations | 4988 | 16 | ||
| 2205 | OXA-48, SHV-40, OmpK36 mutations | 2074 | 4 | ||
| Non-CP | 1802 | CTX-M-15, OXA-1, SHV-101, OmpK36 mutations | 13 | 4 | |
| 1957 | SHV-1, OmpK36 mutations | New ST | 4 | ||
| 2081 | CTX-M-15, OXA-1, SHV-11, TEM-1 | 11 | 8 | ||
| E. cloacae complex (7) | Class B | 1248 | NDM-1, ACT-23, CTX-M-15, OXA-1, OXA-10 | 102 | 4 |
| 1742 | NDM-1, ACT-23, CMY-4, OXA-1, SHV-12, TEM-1 | 102 | 8 | ||
| Class D | 1618 | OXA-48, ACT-15, CTX-M-15 | 106 | 16 | |
| 1787 | OXA-48, ACT-43 | 90 | 8 | ||
| 1967 | OXA-48, ACT-59, DHA-7 | 873 | 4 | ||
| Non-CP | 1585 | ACT-3, KLUB-1, TEM-1, OmpE35 and OmpE36 mutations | 252 | 16 | |
| 2082 | ACT-59, CTX-M-15, OXA-1, OmpE35 and OmpE36 mutations | New ST | 8 | ||
| C. freundii (5) | Class B | 1926 | NDM-1, CMY-65, TEM-1 | 91 | 4 |
| Class D | 1545 | OXA-48, CMY-48, OXA-4, TEM-1 | 216 | 8 | |
| 1583 | OXA-48, CMY-48, CTX-M-15, OXA-1, TEM-1 | 216 | 8 | ||
| 1668 | OXA-48, CMY-48, DHA-7, CTX-M-15, SHV-12, OXA-1, TEM-1 | 216 | 16 | ||
| 1928 | OXA-48, CMY-48, OXA-13 | 216 | 8 | ||
| K. aerogenes (1) | Non-CP | 2265 | AmpC, Omp36 mutations | 137 | > 32 |
| E. coli (1) | Class B | 2280 | NDM-5, EC-15, CTX-M-15, TEM-1 | 167 | > 32 |
| P. aeruginosa (8) | Class B | 1113 | IMP-26, PDC-35, OXA-488, OprD mutations | 235 | 16 |
| 2309 | NDM-1, PDC-19a, OXA-488, OprD mutations | 308 | 4 | ||
| Non-CP | 1764 | PDC-120, OXA-50, OprD mutations | 2996 | 8 | |
| 1820 | PDC-394, OXA-847, OprD mutations | 1613 | 4 | ||
| 1869 | PDC-35, OXA-488, OXA-35, OprD mutations | 235 | 4 | ||
| 2010 | PDC-321, OXA-50, OprD mutations | 175 | 4 | ||
| 2111 | PDC-321, OXA-50, OprD mutations | 175 | 4 | ||
| 2124 | PDC-43, OXA-488 | 267 | 32 |
CP Carbapenemase-Producers, ST Sequence Type
Overall, cefiderocol demonstrated high activity against Enterobacterales, with 81.9% (104 isolates) and 87.4% (111 isolates) of the strains deemed susceptible according to the EUCAST and CLSI breakpoints, respectively. The MIC50/MIC90 was 1/8 mg/L. Among the comparators, colistin, aztreonam-avibactam, and meropenem-vaborbactam were the most effective agents, with susceptibility rates of 99.2%, 98.4%, and 85%, respectively. The MIC50/MIC90 values were ≤ 0.5/ ≤ 0.5 mg/L, 0.125/0.75 mg/L, and 1/16 mg/L, respectively. Amikacin's activity was comparable to that of cefiderocol, with 76.4% of the strains being susceptible and a MIC50/MIC90 of 2/ > 32 mg/L. Fosfomycin, imipenem-relebactam, eravacycline, ceftazidime-avibactam, tigecycline, imipenem, meropenem, exhibited lower activity, with susceptibility rates of 69.3%, 68.5%, 66.1%, 65.4%, 63.8%, 59.8%, and 54.3%, respectively. Their MIC50/MIC90 values were 32/ > 64 mg/L, 1/ > 8 mg/L, 0.5/ > 0.5 mg/L, 1/ > 16 mg/L, ≤ 0.5/ > 1 mg/L 2/ > 8 mg/L, and 2/ > 16 mg/L, respectively.
The distribution of cefiderocol MICs varied with the resistance mechanism of Enterobacterales strains (Fig. 1A). The nine KPC-2 producers and four KPC-3 producers were all susceptible to cefiderocol, with MIC50/MIC90 of 1/2 mg/L. These strains were also all susceptible to ceftazidime-avibactam, aztreonam-avibactam, imipenem-relebactam, and meropenem-vaborbactam, with MIC50/MIC90 of 1/1 mg/L, 0/19/0.5 mg/L, 0.25/0.5 mg/L, and ≤ 0.06/0.12 mg/L, respectively. Colistin, tigecycline, eravacycline, amikacin, and fosfomycin were active against 92.3%, 76.9%, 76.9%, 69.2%, and 69.2% of the isolates, respectively.
Fig. 1.
Distribution of cefiderocol MICs among Enterobacterales (A) and Pseudomonas aeruginosa (B) isolates and clinical categorization according to EUCAST and CLSI breakpoints. S, Susceptible; I, Intermediate (susceptible increased exposure); R Resistant, CPE Carbapenemase-Producing Enterobacterales
Cefiderocol was active against 79.3% of the 29 NDM-producing isolates, with MIC50/MIC90 values of 2/8 mg/L. In more detail, 15 of the NDM-1 producers (n = 20) were susceptible, two of the NDM-5 producers (n = 5) were susceptible, and the NDM-4 (n = 1), NDM-14 (n = 2), and NDM-19 producers (n = 1) were susceptible. When cefiderocol MICs were interpreted using CLSI breakpoints, 86.2% of the isolates were susceptible, 6.9% were intermediate, and 6.9% were resistant (two NDM-5 producers). The only potent comparator agents were colistin (100% of susceptibility; MIC50/MIC90 ≤ 0.5/1 mg/L), aztreonam-avibactam (100%; 0.25/0.75 mg/L), eravacycline (82.8%; 0.5/ > 0.5 mg/L), tigecycline (72.4%; ≤ 0,5/ > 1 mg/L), and fosfomycin (65.5%; 32/64 mg/L). The VIM-1 and the two VIM-4 producing isolates were susceptible to cefiderocol regardless of the breakpoints used. Among the 10 NDM + OXA-48-like producers, only cefiderocol (90%), aztreonam-avibactam (100%), and colistin (100%) demonstrated efficacy greater than 75% with MIC50/MIC90 of 2/2 mg/L, 0.125/0.5 mg/L, and ≤ 0.5/ ≤ 0.0, 5 mg/L, respectively. The only isolate of C. freundii co-producing KPC-2 + OXA-48 had a low cefiderocol MIC of 0.5 mg/L.
Concerning the main resistance mechanism of the strains included, i.e. OXA-48-like production, 84.1% and 87.2% of the 63 isolates were susceptible to cefiderocol according to EUCAST and CLSI breakpoints, respectively. The MICs distribution was wide, ranging from ≤ 0.03 to 16 mg/L, with MIC50/MIC90 of 1/8 mg/L. Other comparators with high activity rates included colistin (100% susceptible), ceftazidime-avibactam (98.4%), aztreonam-avibactam (98.4%), imipenem ± relebactam (87.3% and 92.1%, respectively), meropenem ± vaborbactam (76.2% and 92.4%, respectively), amikacin (92.1%), and fosfomycin (76.2%).
Within the eight non-CPE isolates, 25% and 50% were susceptible to cefiderocol according to EUCAST and CLSI breakpoints, respectively, with MIC values ranging between 0.5 and > 32 mg/L. The six resistant isolates included three K. pneumoniae showing OmpK36 mutation (associated with CTX-M-15 production for two of them), two E. cloacae complex with decreased permeability of the outer membrane (associated with KLUB-1 or CTX-M-15 ESBL production in addition to the constitutive ACT-type AmpC), and one K. aerogenes with overproduction of AmpC associated with decreased permeability of the outer membrane. Many of the comparators had superior activity, with susceptibility rates of 100% to colistin and amikacin, 75% to meropenem alone and 100% when combined with vaborbactam, 87.5% to imipenem alone and 100% when combined to relebactam, 87.5% to ceftazidime-avibactam and aztreonam-avibactam, and 62.5% to tigecycline and eravacycline.
In vitro activity of cefiderocol and comparators against P. aeruginosa
Table 5 shows the in vitro activity of cefiderocol and its comparators against the 53 strains of MDR P. aeruginosa included in this study. Table 6 details the characteristics of cefiderocol-resistant strains.
Cefiderocol was active against 84.9% (n = 45) and 96.2% (n = 51) of the 53 P. aeruginosa isolates, according to EUCAST and CLSI breakpoints, respectively. The MIC50/MIC90 ratio was 1/4 mg/L. The distribution of cefiderocol MICs is illustrated in Fig. 1B. Among the comparators, only colistin had better rates of susceptibility (100%; MIC50/MIC90 2/2 mg/L). The susceptibility rates and MIC50/MIC90 for other antibiotics tested were as follows: 67.9% and 16/ > 32 mg/L for amikacin, 56.6% and 4/ > 8 mg/L for ceftolozane-tazobactam, 56.6% and 2/ > 8 for imipenem-relebactam, 52.8% and 8/ > 16 mg/L for ceftazidime-avibactam, 52.8% and 16/ > 256 mg/L for aztreonam-avibactam, 52.8% and 2/ > 4 mg/L for tobramycin. The remaining antibiotics tested (piperacillin-tazobactam, cefepime, aztreonam, imipenem, and meropenem) had susceptibility rates below 50%.
Among the six MBL producers, 66.7% and 83.3% were susceptible to cefiderocol according to EUCAST and CLSI breakpoints, respectively, with MIC values between 0.5 and 16 mg/L. Only aztreonam (high exposure), aztreonam-avibactam (high exposure), and colistin had susceptibility rates above 50% (50%, 66.7%, and 100%, respectively).
Comparison of ComASP® and UMIC® BMD to evaluate cefiderocol MIC
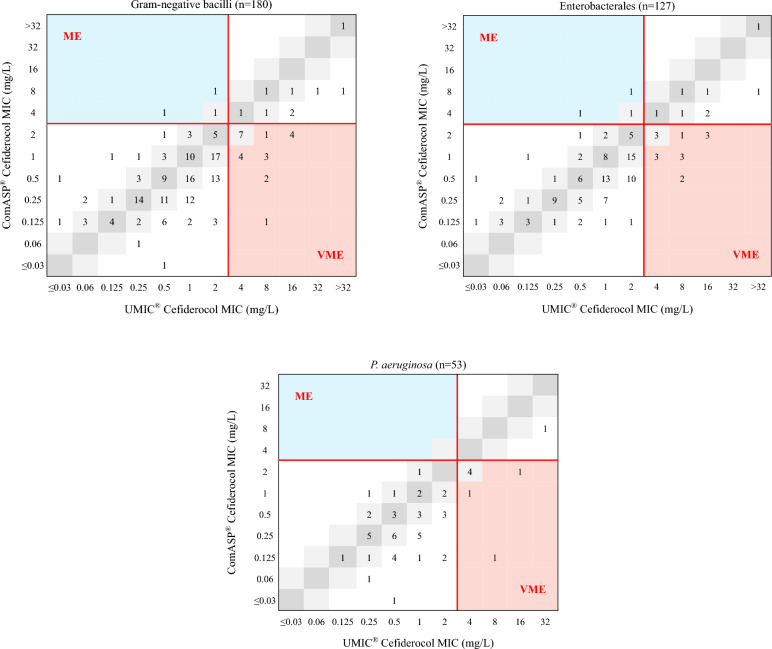
The MICs of cefiderocol for the 127 Enterobacterales and 53 P. aeruginosa strains ranged between ≤ 0.03 and > 32 mg/L. Overall, the ComASP® method had an EA of 63.3% (95% CI: 56.2%–70.3%), a CA of 86.1% (95% CI: 81%–91.2%), and a bias of −50%, with 22 VME (71%) and three ME (2%) compared to the UMIC® method. Figure 2 shows the distribution of cefiderocol MICs using the UMIC® and ComASP® methods. Table 7 shows the performance of ComASP® BMD method in comparison with UMIC® reference method.
Fig. 2.
MICs of cefiderocol using ComASP® BMD method compared to UMIC®. ME Major Error, VME Very Major Error. MICs corresponding to Categorical Agreement (CA) are in white, to Essential Agreement (EA) in grey (light + dark), to ME in blue and to VME in salmon-pink. Red lines correspond to EUCAST breakpoints
Table 7.
Performance of cefiderocol AST methods in comparison with UMIC® reference method
| AST method | Enterobacterales (n = 127) | P. aeruginosa (n = 53) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CA (%) | EA (%) | VME (%) | ME (%) | Difference of bias (%) | CA (%) | EA (%) | VME (%) | ME (%) | Difference of bias (%) | |
| ComASP® BMD | 85.8 | 65.4 | 65.2 | 2.9 | − 49.6 | 86.8 | 58.5 | 87.5 | 0 | − 60.2 |
| BD BBL™ MH II agar DD | 89 (73.2) | NA | 21.1 (17.4) | 8.6 (28.8) | NA | 96 (94.3) | NA | 33.3 (25) | 0 (2.2) | NA |
| Bio-Rad MH agar DD | 87 (74) | NA | 37.5 (26.1) | 8.3 (26) | NA | 88.2 (88.7) | NA | 100 (75) | 0 (0) | NA |
EUCAST 2024 clinical breakpoints were used for the interpretation of the results. Values in brackets are the performance when the ATU is ignored
CA Categorical Agreement, EA Essential Agreement, VME Very MajorEerrors, ME Major Errors, BMD Broth MicroDilution, DD Disk Diffusion, NA Non-Applicable
Focusing on Enterobacterales, the EA and CA rates of the ComASP® method were 65.4% (95% CI 57.1–73.6%) and 85.8% (95% CI: 79.7–91.9%), respectively, with a bias of − 49.6%, with 15 VME (65.2%) and three ME (2.9%). Among the 15 VME, six isolates had a MIC of 4 mg/L with the reference method, and the obtained MICs using ComASP® were 1 mg/L for three of them and 2 mg/L for three of them.
Among the P. aeruginosa isolates, the ComASP® method had an EA of 58.5% (95% CI: 45.2–71.8%), a CA of 86.8% (95% CI 77.8–96%), and a bias of −60.2%. Seven VME (87.5%) were noticed with five isolates displaying a reference MIC of 4 mg/L, and no ME were identified.
Comparison of disk diffusion to UMIC® BMD to assess cefiderocol susceptibility
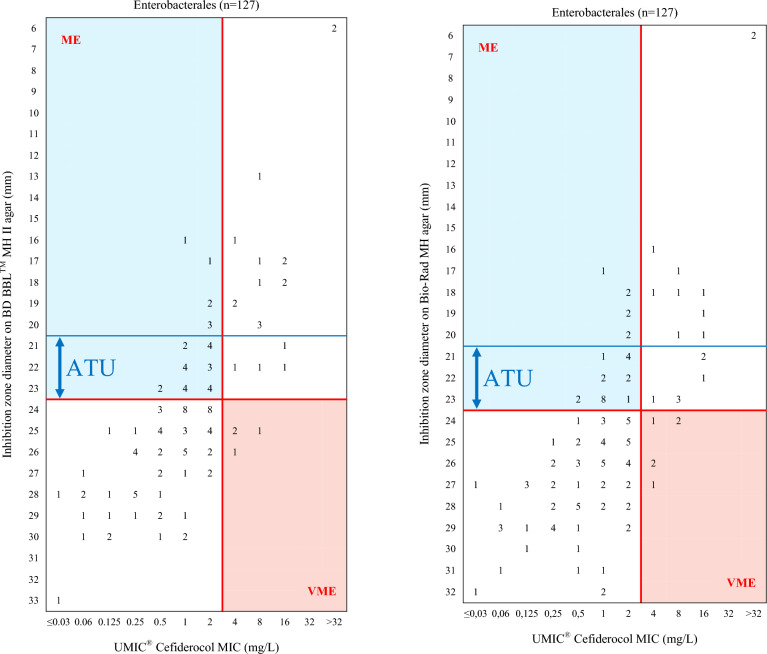
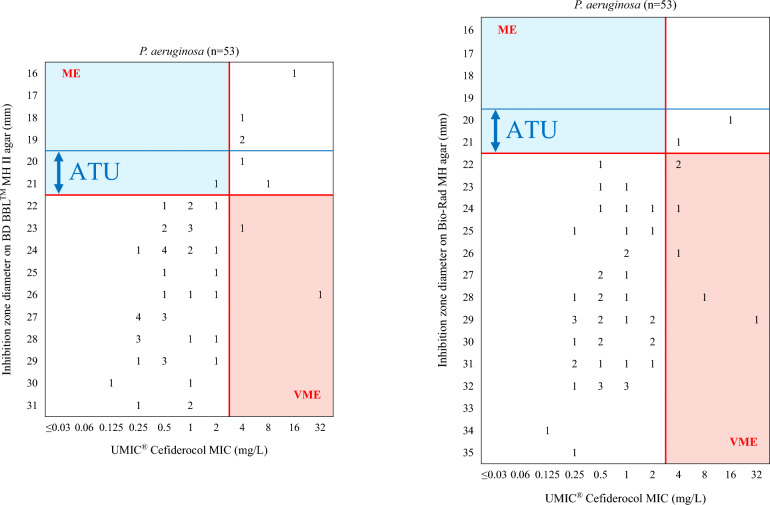
Figures 3 and 4 illustrate the distribution of inhibition zone diameter of cefiderocol on BD BBL™ MH II and Bio-Rad MH agar compared to the MICs determined using the UMIC® method. The performance of disk diffusion using BD BBL™ MH II and Bio-Rad MH agar, in comparison with the MICs obtained using UMIC® method, is presented in Table 7.
Fig. 3.
Cefiderocol inhibition zone diameter on BD BBL™ MH II agar (left) and Bio-Rad MH agar (right) compared to MICs using UMIC® for Enterobacterales. ME Major Error, VME Very Major Error, ATU Area of Technical Uncertainty (21–23 mm). MICs corresponding to Categorical Agreement (CA) are in white, to ME in blue and to VME in salmon-pink. Red lines correspond to EUCAST breakpoints, blue lines correspond to EUCAST ATU
Fig. 4.
Cefiderocol inhibition zone diameter on BD BBL™ MH II agar (left) and Bio-Rad MH agar (right) compared to MICs using UMIC® for P. aeruginosa. ME Major Error, VME Very Major Error, ATU Area of Technical Uncertainty (20–21 mm). MICs corresponding to Categorical Agreement (CA) are in white, to ME in blue and to VME in salmon-pink. Red lines correspond to EUCAST breakpoints, blue lines correspond to EUCAST ATU
For Enterobacterales, when using the BD BBL™ MH medium, the CA rate was 89% (95% CI 82.9%–95.1%), with four VME (21.1%) and seven ME (8.6%), excluding isolates in the Area of Technical Uncertainty (ATU, 21.3% of the isolates, n = 27). When the EUCAST ATU (21–23 mm) was not considered, the CA decreased to 73.2% (95% CI 65.5–90.9%), with four VME (17.4%) and 30 ME (28.8%). For P. aeruginosa, CA rates were 96% (95% CI 90.5–100%) with two VME (33.3%) and no ME, excluding isolates in the ATU (5.7% of the isolates, n = 3), and 94.3% (95% CI: 88–100%) with two VME (25%) and one ME (2.2%) when the ATU (20–21 mm) was not considered.
When applying the Bio-Rad MH, the CA for Enterobacterales was 87% (95% CI: 80.4–93.6%) with six VME (37.5%) and seven ME (8.3%), excluding the 27 strains with an inhibition zone diameter in the ATU. Ignoring the ATU, the CA was 74% (95% CI 66.4–81.6%) with six VME (26.1%) and 27 ME (26%) among the 127 Enterobacterales isolates. For P. aeruginosa isolates, the CA rate was 88.2% (95% CI 79.3–97.1%) when the isolates with an inhibition zone diameter in the ATU were excluded, with six VME (100%) and no ME. The CA rate remained at 88.7% (95% CI 80.2–97.1%) when the ATU was ignored, also with six VME (75%) and no ME.
Discussion
The escalating challenge of antibiotic resistance has expedited the development of new drugs and novel β-lactam/β-lactamase inhibitor combinations. In the TROJAN-MDR study, we assessed the in vitro susceptibility of cefiderocol and other last-resort antibiotics against a variety of well-characterized MDR Enterobacterales and P. aeruginosa isolates from the South of France. These isolates were primarily collected from clinical samples.
Our findings revealed that cefiderocol exhibited significant activity against MDR Enterobacterales and P. aeruginosa, with 82.8% and 89.4% of the isolates respectively being categorized as susceptible according to EUCAST and CLSI breakpoints. Compared to other tested compounds, cefiderocol emerged as one of the most potent antibiotics. These findings align with those from similar studies [16–22].
When focusing on the KPC producers, the 100% susceptibility rate obtained is higher than those reported by other authors [23]. This could be attributed to the low number of KPC-producing isolates (n = 13) in our study, and the absence of variants responsible for resistance to ceftazidime-avibactam in our panel [4]. Indeed, recent reports have indicated higher cefiderocol MICs among ceftazidime-avibactam resistant strains harboring KPC-3 variants [4].
The cefiderocol MIC50 for NDM-producing Enterobacterales was found to be 2 mg/L, which is the value of the EUCAST breakpoint. This aligns with the MICs50 measured in other studies [18, 24, 25]. As described by Mushtaq et al. the addition of the MBL inhibitor dipicolinic acid reduced cefiderocol MICs in NDM-producing Enterobacterales indicating that these carbapenemases have the capacity to hydrolyze cefiderocol [23]. This finding may explain why MICs50 values for NDM-producers are higher than to those of other resistance mechanisms. Although NDM production alone does not seem to be sufficient to cause cefiderocol resistance, its overproduction or the association with mutations of siderophore receptors are some of the described mechanisms that can lead to resistance [26–28]. The susceptibility rates among NDM-producing Enterobacterales vary across studies. When considering only studies that have applied the EUCAST breakpoints, susceptibility rates of 41% (n = 61 NDM-producing Enterobacterales), 48% (n = 21), 48.1% (n = 27), 51.4% (n = 37), 53.1% (n = 96), 70% (n = 118), 82.5% (n = 97) and 90.6% (n = 53) have been reported by various authors [8, 18, 20, 21, 23–25, 29]. The susceptibility rates obtained in our study (79.3% for NDM-producing isolates and 90% of NDM + OXA-48 producers susceptible) are closer to those of Bonnin et al., Malisova et al. and Delgado-Valverde et al. [8, 20, 29] Seven strains were resistant to cefiderocol according to EUCAST breakpoints. Four isolates were NDM-1 producers (one ST2084 K. pneumoniae coproducing OXA-48, two ST102 E. cloacae complex, one ST91 C. freundii) and three isolates were NDM-5 producers (one ST25 and one ST219 K. pneumoniae, one E. coli belonging to ST167 international high-risk clone for which cefiderocol resistance has already been reported, due to modified PBP3 and overexpression of NDM-5 and mutation of CirA iron transporter [28]). Finally, concerning NDM-producing Enterobacterales, colistin, aztreonam-avibactam, and eravacycline were the only comparators with susceptibility rates superior to cefiderocol (100%, 100%, and 82.8%, respectively).
A majority of the OXA-48-like enzymes do not hydrolyze extended-spectrum cephalosporins [30], but in cases of ESBL production or AmpC overexpression, the isolates become resistant to these drugs. Ceftazidime-avibactam, which is the treatment of choice for infections involving this type of isolates, had a very high susceptibility rate in our study (98.4%), as expected [3, 31]. The selection pressure following the use of ceftazidime-avibactam has led to the emergence of OXA-48 variants resistant to this combination [32]. Given that OXA-48-like are the most common carbapenemases in France [12], we cannot rule out the future emergence and diffusion of ceftazidime-avibactam resistant isolates. Cefiderocol, with a susceptibility rate of 84.1% on OXA-48-like CPE, which is globally comparable to other studies, may be an interesting alternative to ceftazidime-avibactam [18, 25]. The resistant isolates in this study were eight OXA-48 producers (one ST2074 K. pneumoniae, one ST90, one ST106 and one ST873 E. cloacae complex, four ST216 C. freundii) and two OXA-181 producers (ST4988 K. pneumoniae), also producers of diverse β-lactamases and in some cases with outer membrane porin mutations (Table 6). The blaSHV-12 gene has been detected in one of the C. freundii resistant isolates. The production of SHV-12 could explain the elevated MIC (16 mg/L) as Poirel et al. showed that production of this “old” EBSL can lead to cefiderocol resistance [33]. Since OXA-48 does not significantly hydrolyze cefiderocol [34], further investigations such as sequence analysis of PBP3 or iron transport encoding genes would be necessary to explain the mechanism of resistance to cefiderocol in the other isolates. Despite good susceptibility rates related to the weaker hydrolysis of carbapenems by OXA-48-like enzymes compared with class A or B carbapenemases, imipenem ± relebactam and meropenem ± vaborbactam are not recommended for the treatment of OXA-48 CPE infections [31].
For the non-CPE isolates included in this study, six (75%) were resistant to cefiderocol (MICs range 4 to > 32 mg/L). The β-lactamase genes harbored by these isolates (blaSHV-1, blaSHV-11, blaSHV-101, blaOXA-1, blaTEM-1 and blaCTX-M-15 for the three K. pneumoniae strains; blaACT-3, blaACT-59, blaOXA-1, blaTEM-1, blaCTX-M-15, blaKLUB-1 for the two E. cloacae complex strains and only blaAmpC for the K. aerogenes isolate) have not been described as related to cefiderocol resistance [4]. The exhibited phenotype may result from decreased membrane permeability, as observed for five isolates, or from alterations in PBP3 or iron transport systems such as CirA. Mushtaq et al. described fewer susceptibility rates among Enterobacterales isolates expressing ESBL + porin loss phenotype [23].
The high susceptibility rate of aztreonam-avibactam (98.4% susceptible) among Enterobacterales of this study highlights the potential contribution of this new combination in the treatment arsenal, regardless of the resistance mechanism of MDR strains. Other studies reported similar susceptibility rates (94–100%) [35, 36].
Even though the susceptibility rate of meropenem-vaborbactam on Enterobacterales was high (85%), this combination is almost exclusively reserved for the treatment of infections involving KPC-producing isolates [3]. The high susceptibility rate can be explained by its breakpoint being 8 mg/L, which is higher than that of meropenem (2 mg/L). If the breakpoints were identical, meropenem-vaborbactam would have the same susceptibility rate as meropenem alone (54.3%). This underlines the importance of interpreting antimicrobial susceptibility results by the clinical microbiologist according to the genotypic profile to avoid inappropriate use, which can compromise clinical success [37].
Concerning MDR P. aeruginosa isolates, cefiderocol was the second most potent agent in vitro (84.9% and 96.2% of susceptibility rate by applying EUCAST and CLSI breakpoints, respectively), following colistin (100%) and preceding other β-lactams of last resort. These results are consistent with those observed in other studies [18, 20, 23], and highlight the key role of cefiderocol in overcoming severe MDR P. aeruginosa infections [38]. Notably, two of the eight cefiderocol resistant isolates produced a MBL (one ST235 IMP-26 producer and one ST308 NDM-1 producer). Aztreonam-avibactam inhibited only 52.8% of P. aeruginosa isolates in this study, which is lower compared with Enterobacterales. This may be in link with the diversity of resistance mechanisms in P. aeruginosa (efflux, production of diverse β-lactamases) [39].
An interesting finding of our study was that cefiderocol was active against 93% of the 57 isolates belonging to high-risk clones spreading worldwide (25 K. pneumoniae from ST11, ST15, ST147, ST258 and ST307; 12 E. cloacae complex from ST66, ST78, ST114 and ST171; eight C. freundii from ST22 and ST98; and 12 P. aeruginosa from ST175, ST235, ST244 and ST253) [19, 30, 39–41].
Different AST methods have been developed to assess cefiderocol in vitro activity [8–11, 42]. The BMD method using Iron Depleted Cation Adjusted MH Broth (ID-CAMHB) is considered as the gold standard [43]. UMIC® (Bruker) is a commercial unitary BMD test in which cefiderocol is dried into the wells (concentration range: 0.03–32 mg/L). A matching ID-CAMHB is available separately from the same manufacturer. UMIC®, the only commercial method currently validated by French NRC for both Enterobacterales and non-fermenting GNB, was considered as the reference method in our study [10, 11]. The ComASP® (Liofilchem) cefiderocol BMD test is another commercial assay on which two isolates are tested on every plate (concentration range: 0.008–128 mg/L), an ID-CAMHB is provided with the kit. Our results showed that ComASP® BMD method did not fulfill ISO 20776-2:2007 and 2021 criteria (CA and EA ≥ 90%, VME ≤ 1.5%, ME ≤ 3% and—30% ≤ bias ≤ + 30%) on Enterobacterales and P. aeruginosa isolates, as EA rate was below 90% (90% was outside of 95% CI), VME rate was above 1.5% and biais below − 30%. This low rate of EA and the bias lower than −30% illustrate the tendency of this method to underestimate MICs. Forty percent of the 15 VME identified among Enterobacterales and 71.4% of the seven VME on P. aeruginosa concerned isolates with a MIC of 4 mg/L. Bianco et al. compared ComASP® method with a reference BMD method on 50 MDR-GNB (Enterobacterales, P. aeruginosa and A. baumannii) that had an inhibition zone diameter in the former EUCAST ATU [44]. They obtained CA and EA rates of 94% and 84% respectively, with one VME and two ME. Emeraud et al. compared ComASP® and UMIC® tests with a reference BMD method on 60 carbapenem-resistant Enterobacterales [10]. ComASP® had 76.7% of EA and 83.3% of CA with 34.5% of VME and no ME, while UMIC® method had an EA of 91.7% and a CA of 83.3% with 24.1% of VME and 9.7% of ME. The findings in our study are in line with those of Emeraud et al. who conclude that ComASP® BMD assay is not a reliable method to assess cefiderocol susceptibility [10].
Easy-to-perform diffusion techniques with MIC strips and disks have also been developed on conventional MH agar. However, the cefiderocol-impregnated MIC strips initially drawn up for P. aeruginosa are no longer recommended, regardless of the species, due to the major underestimation of MICs that lead to a high number of VME [8, 9]. Matuschek et al. concluded that disk diffusion on conventional MH agar is a robust method to assess cefiderocol susceptibility in Enterobacterales and P. aeruginosa [43]. In addition to the breakpoint of 23 mm (Enterobacterales) and 22 mm (P. aeruginosa), an ATU has been proposed by EUCAST for Enterobacterales (21–23 mm) and P. aeruginosa (21–22 mm) to increase the performance of disk diffusion method [13]. Devoos et al. compared the performance of disk diffusion on 150 MDR P. aeruginosa isolates using disks from three manufacturers on MH agar plates from six manufacturers [9]. When isolates with an inhibition zone diameter in the ATU of EUCAST 2023 (14–22 mm) were excluded, CA rates were ≥ 90% for MH agar from BD, bioMérieux, and Mast group regardless of the disk manufacturer, while Bio-Rad did not reach the target. Nevertheless, in the same study, the authors demonstrated that the iron concentration was lower for Bio-Rad MH agar. Moreover, Bonnin et al. evaluated the performance of disk diffusion on Bio-Rad MH-agar compared to BMD and concluded that this method did not meet the CA of 90% [8]. It would therefore suggest that the different iron concentrations alone do not explain the differences in performance between the different MH media, since the agar medium mimics an iron-depleted medium (the iron being bound to the agar) [9]. Currently, in France, the CA-SFM and the Antimicrobial Resistance NRC do not recommend disk diffusion to assess cefiderocol susceptibility in Enterobacterales, while it can still be used on P. aeruginosa and A. baumannii to screen susceptible isolates when inhibition diameter is ≥ 27 mm and ≥ 22 mm, respectively [12].
In our study, we evaluated the cefiderocol inhibition zone diameter on two MH agar from two manufacturers (BD and Bio-Rad) using Mast disks in comparison with UMIC® BMD. The inhibition diameters were on average 1.39 mm (95% CI 1.11–1.67) smaller on BD BBL™ MH II agar than on Bio-Rad. On Enterobacterales, the BD BBL™ MH II agar (CA 89%, VME 21.1%) was more reliable than Bio-Rad MH medium (CA 87%, VME 37.5%). Similar results were found for P. aeruginosa isolates with CA of 96% and VME rate of 25% for BD BBL™ MH II agar while Bio-Rad’s MH agar had CA of 88.2% and VME rate of 100%. It is noteworthy that none of the VME strains had an inhibition diameter ≥ 27 mm on BD BBL™ MH II agar, but these results must be interpreted with caution given the lower number of P. aeruginosa strains.
We acknowledge that this study has some limitations. Firstly, the reference method used was not the gold standard in-house BMD method but the UMIC® BMD which displayed approximatively 90% CA and EA [11]. Additionally, discrepant results between the evaluated methods were not retested. Lastly, fosfomycin AST was not assessed with the reference agar dilution method, which may have resulted in an underestimation of its susceptibility.
Conclusions
The TROJAN-MDR study demonstrated the potent activity of cefiderocol against a large collection of Enterobacterales and P. aeruginosa, both harboring various resistance mechanisms. This reaffirms the importance of cefiderocol in the treatment of difficult-to-treat infections. Constant surveillance of antimicrobial resistance and further investigation into resistant strains are keys to understanding the various mechanisms that may contribute to cefiderocol resistance.
AST according to validated methods is crucial to ensure the efficacy of cefiderocol. The two disk diffusion methods and ComASP® BMD did not meet the ISO 20776-2:2007 and 2021 criteria. Furthermore, antimicrobial stewardship is essential to ensure the appropriate use of this siderophore cephalosporin and to preserve its activity.
Acknowledgements
The authors wish to thank Dr. Florian Salipante for his assistance with the statistical analysis and Dr Matthieu Curtil Dit Galin and Romain Durello for technical assistance.
Abbreviations
- AST
Antimicrobial Susceptibility Testing
- ATU
Area of Technical Uncertainty
- BMD
Broth MicroDilution
- CA
Categorical Agreement
- CA-SFM
Comité de l’Antibiogramme de la Société Française de Microbiologie
- CLSI
Clinical & Laboratory Standards Institute
- CPE
Carbapenemase-Producing Enterobacterales
- EA
Essential Agreement
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- ID-CAMHB
Iron Depleted Cation Adjusted MH Broth
- MDR
Multi-Drug Resistance
- ME
Major Error
- MIC
Minimal Inhibitory Concentration
- MH
Mueller Hinton
- NRC
National Reference Center
- ST
Sequence Type
- VME
Very Major Error
Author contributions
Concept and design: AP, MB Acquisition, analysis, or interpretation of data: MB, AP, SL, ABD, MG, CM, MM Drafting of the manuscript: MB, AP Revision of the manuscript: JPL, LR, HM, VJP, ABD.
Funding
This research was partially supported by SHIONOGI & Co. which provided supplies for antimicrobial susceptibility testing (disk, medium and commercial BMD assays) and for open access fees. We thank the Nîmes University hospital for its structural, human and financial support through the award obtained by our team during the internal call for tenders « Thématiques phares ». However, the funders did not influence the study design, data collection and analysis, decision to publish, or the preparation of the manuscript in any way.
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
This research was partially supported by SHIONOGI & Co. which provided supplies for antimicrobial susceptibility testing (disk, medium and commercial BMD assays) and for open access fees. AP received fees as Advisory Board member from SHIONOGI & Co., Ltd., Osaka, Japan. However, the funder did not influence the study design, data collection and analysis, decision to publish, or the preparation of the manuscript in any way.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO Bacterial Priority Pathogens List, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Geneva: World Health Organization; 2024.
- 2.JI Barrasa-Villar C Aibar-Remón P Prieto-Andrés R Mareca-Doñate J Moliner-Lahoz 2017 Impact on morbidity, mortality, and length of stay of hospital-acquired infections by resistant microorganisms Clin Infect Dis 65 644 652 [DOI] [PubMed] [Google Scholar]
- 3.R Cantón P Ruiz-Garbajosa 2023 Treatment guidelines for multidrug-resistant Gram-negative microorganisms Rev Espanola Quimioter 36 Suppl 1 46 51 [DOI] [PMC free article] [PubMed]
- 4.S Karakonstantis M Rousaki EI Kritsotakis 2022 Cefiderocol: systematic review of mechanisms of resistance, heteroresistance and in vivo emergence of resistance Antibiotics 11 723 [DOI] [PMC free article] [PubMed]
- 5.PJ Simner R Patel 2020 Cefiderocol antimicrobial susceptibility testing considerations: the Achilles’ heel of the Trojan horse? J Clin Microbiol 59 e00951 e1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Available online: https://www.eucast.org/. Accessed 21 Oct 2024.
- 7.Clinical & Laboratory Standards Institute: CLSI Guidelines. Available online: https://clsi.org/. Accessed 21 Oct 2024.
- 8.RA Bonnin C Emeraud AB Jousset T Naas L Dortet 2022 Comparison of disk diffusion, MIC test strip and broth microdilution methods for cefiderocol susceptibility testing on carbapenem-resistant Enterobacterales Clin Microbiol Infect 28 1156.e1 1156.e5 [DOI] [PubMed] [Google Scholar]
- 9.L Devoos A Biguenet J Rousselot M Bour P Plésiat D Fournier 2023 Performance of discs, Sensititre EUMDROXF microplates and MTS gradient strips for the determination of the susceptibility of multidrug-resistant Pseudomonas aeruginosa to cefiderocol Clin Microbiol Infect 29 652.e1 652.e8 [DOI] [PubMed] [Google Scholar]
- 10.C Emeraud C Gonzalez L Dortet 2023 Comparison of ComASP® and UMIC® methods with the reference method for cefiderocol susceptibility testing on carbapenem-resistant Enterobacterales J Antimicrob Chemother 78 1800 1801 [DOI] [PubMed] [Google Scholar]
- 11.L Dortet C Niccolai N Pfennigwerth S Frisch C Gonzalez A Antonelli 2023 Performance evaluation of the UMIC® Cefiderocol to determine MIC in Gram-negative bacteria J Antimicrob Chemother 78 1672 1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centre National de Référence de la Résistance aux Antibiotiques. Available online: https://www.cnr-resistance-antibiotiques.fr/. Accessed 21 Oct 2024.
- 13.EUCAST. Clinical breakpoints and dosing of antibiotics. Available online: https://www.eucast.org/clinical_breakpoints. Accessed 21 Oct 2024.
- 14.ISO. EN ISO 20776-2:2007—Clinical laboratory testing and in vitro diagnostic test systems—Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices—Part 2: Evaluation of performance of antimicrobial susceptibility test devices (ISO 20776-2:2007). ITeh Standards. Available online: https://standards.iteh.ai/catalog/standards/cen/e58114a5-4af0-488d-addf-f410a8445ee0/en-iso-20776-2-2007. Accessed 21 Oct 2024.
- 15.ISO. EN ISO 20776-2:2021—Clinical laboratory testing and invitro diagnostic test systems - Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices—Part 2: Evaluation of performance of antimicrobial susceptibility test devices against reference broth micro-dilution (ISO 20776-2:2021). ITeh Standards. Available online: https://standards.iteh.ai/catalog/standards/cen/92fc193a-5135-4d74-b3b5-a6962a16e505/en-iso-20776-2-2022. Accessed 21 Oct 2024.
- 16.D Shortridge JM Streit R Mendes M Castanheira 2020 In vitro activity of cefiderocol against U.S. and European Gram-negative clinical isolates collected in 2020 as part of the SENTRY antimicrobial surveillance program Microbiol Spectr 2022 10 e0271221 [DOI] [PMC free article] [PubMed]
- 17.JA Karlowsky MA Hackel M Takemura Y Yamano R Echols DF Sahm 2022 In vitro susceptibility of Gram-negative pathogens to cefiderocol in five consecutive annual multinational SIDERO-WT surveillance studies, 2014 to 2019 Antimicrob Agents Chemother 66 e0199021 [DOI] [PMC free article] [PubMed]
- 18.S Oueslati P Bogaerts L Dortet S Bernabeu H Ben Lakhal C Longshaw 2022 In vitro activity of cefiderocol and comparators against carbapenem-resistant Gram-negative pathogens from France and Belgium Antibiotics 11 1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M Delgado-Valverde MDC Conejo L Serrano F Fernández-Cuenca Á Pascual 2020 Activity of cefiderocol against high-risk clones of multidrug-resistant Enterobacterales, Acinetobacter baumannii, Pseudomonas aeruginosa and Stenotrophomonas maltophilia J Antimicrob Chemother 75 1840 1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.L Malisova I Vrbova K Pomorska V Jakubu H Zemlickova 2023 In vitro activity of cefiderocol against carbapenem-resistant Enterobacterales and Pseudomonas aeruginosa Microb Drug Resist 29 10 485 491 [DOI] [PMC free article] [PubMed]
- 21.A Santerre Henriksen F Arena M Attwood R Canton S Gatermann T Naas 2024 In vitro activity of cefiderocol against european Enterobacterales, including isolates resistant to meropenem and recent β-lactam/β-lactamase inhibitor combinations Microbiol Spectr. 2024 e0418123 [DOI] [PMC free article] [PubMed]
- 22.A Santerre Henriksen K Jeannot A Oliver JD Perry MW Pletz S Stefani 2024 In vitro activity of cefiderocol against european Pseudomonas aeruginosa and Acinetobacter spp., including isolates resistant to meropenem and recent β-lactam/β-lactamase inhibitor combinations Microbiol Spectr. 12 e0383623 [DOI] [PMC free article] [PubMed]
- 23.S Mushtaq Z Sadouki A Vickers DM Livermore N Woodford 2020 In vitro activity of cefiderocol, a siderophore cephalosporin, against multidrug-resistant Gram-negative bacteria Antimicrob Agents Chemother 64 e01582 e1620 [DOI] [PMC free article] [PubMed]
- 24.C Longshaw D Manissero M Tsuji R Echols Y Yamano 2020 In vitro activity of the siderophore cephalosporin, cefiderocol, against molecularly characterized, carbapenem-non-susceptible Gram-negative bacteria from Europe JAC Antimicrob Resist. 2 dlaa060 [DOI] [PMC free article] [PubMed]
- 25.MG Wise JA Karlowsky MA Hackel M Takemura Y Yamano R Echols 2023 In vitro activity of cefiderocol against meropenem-nonsusceptible Gram-negative bacilli with defined β-Lactamase carriage: SIDERO-WT surveillance studies, 2014–2019 Microb Drug Resist 29 360 370 [DOI] [PMC free article] [PubMed]
- 26.A Ito T Sato M Ota M Takemura T Nishikawa S Toba 2018 In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against Gram-negative bacteria Antimicrob Agents Chemother 62 e01454 e1517 [DOI] [PMC free article] [PubMed]
- 27.N Kohira MA Hackel Y Ishioka M Kuroiwa DF Sahm T Sato 2020 Reduced susceptibility mechanism to cefiderocol, a siderophore cephalosporin, among clinical isolates from a global surveillance programme (SIDERO-WT-2014) J Glob Antimicrob Resist 22 738 741 [DOI] [PubMed] [Google Scholar]
- 28.PJ Simner Y Bergman R Conzemius E Jacobs T Tekle S Beisken 2023 An NDM-producing Escherichia coli clinical isolate exhibiting resistance to cefiderocol and the combination of ceftazidime-avibactam and aztreonam: another step toward pan-β-lactam resistance Open Forum Infect Dis 10 ifad276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M Delgado-Valverde I Portillo-Calderón E Recacha P Pérez-Palacios A Pascual 2023 In vitro activity of cefiderocol compared to other antimicrobials against a collection of metallo-beta-lactamase-producing Gram-negative bacilli from southern Spain Microbiol Spectr 11 e0493622 [DOI] [PMC free article] [PubMed]
- 30.JDD Pitout G Peirano MM Kock K-A Strydom Y Matsumura 2019 The global ascendency of OXA-48-type carbapenemases Clin Microbiol Rev 33 e00102 e119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.SE Boyd A Holmes R Peck DM Livermore W Hope 2022 OXA-48-like β-lactamases: global epidemiology, treatment options, and development pipeline Antimicrob Agents Chemother 66 e0021622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.C Fröhlich V Sørum AM Thomassen PJ Johnsen H-KS Leiros Ø Samuelsen 2019 OXA-48-mediated ceftazidime-avibactam resistance is associated with evolutionary trade-offs mSphere. 4 e0002419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.L Poirel J-M Ortiz de la Rosa M Sadek P Nordmann 2022 Impact of acquired broad-spectrum β-lactamases on susceptibility to cefiderocol and newly developed β-lactam/β-lactamase inhibitor combinations in Escherichia coli and Pseudomonas aeruginosa Antimicrob Agents Chemother 66 e0003922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.L Poirel N Kieffer P Nordmann 2018 Stability of cefiderocol against clinically significant broad-spectrum oxacillinases Int J Antimicrob Agents 52 866 867 [DOI] [PubMed] [Google Scholar]
- 35.S Vasoo SA Cunningham NC Cole PC Kohner SR Menon KM Krause 2015 In vitro activities of ceftazidime-avibactam, aztreonam-avibactam, and a panel of older and contemporary antimicrobial agents against carbapenemase-producing Gram-negative bacilli Antimicrob Agents Chemother 59 7842 7846 [DOI] [PMC free article] [PubMed]
- 36.Y-S Huang P-Y Chen P-C Chou J-T Wang 2023 In vitro activities and inoculum effects of cefiderocol and aztreonam-avibactam against metallo-β-lactamase-producing Enterobacteriaceae Microbiol Spectr 11 e0056923 [DOI] [PMC free article] [PubMed]
- 37.CM Gill TE Asempa DP Nicolau 2021 Efficacy of human-simulated exposures of meropenem/vaborbactam and meropenem against OXA-48 β-lactamase-producing Enterobacterales in the neutropenic murine thigh infection model J Antimicrob Chemother 76 184 188 [DOI] [PubMed] [Google Scholar]
- 38.R Larcher P Laffont-Lozes C Roger R Doncesco C Groul-Viaud A Martin 2022 Last resort beta-lactam antibiotics for treatment of New-Delhi Metallo-beta-lactamase producing Enterobacterales and other difficult-to-treat resistance in Gram-negative bacteria: a real-life study Front Cell Infect Microbiol 12 1048633 [DOI] [PMC free article] [PubMed]
- 39.Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32. [DOI] [PMC free article] [PubMed]
- 40.P Lumbreras-Iglesias M Toro de X Vázquez E García-Carús MR Rodicio J Fernández 2023 High-risk international clones ST66, ST171 and ST78 of Enterobacter cloacae complex causing blood stream infections in Spain and carrying blaOXA-48 with or without mcr-9 J Infect Public Health 16 272 279 [DOI] [PubMed] [Google Scholar]
- 41.D Nobrega G Peirano Y Matsumura JDD Pitout 2023 Molecular epidemiology of global carbapenemase-producing Citrobacter spp. (2015–2017) Microbiol Spectr. 11 e04144 e4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.V Jean-Pierre P Sorlin K Jeannot R Chiron J-P Lavigne A Pantel 2024 Commercially available tests for determining cefiderocol susceptibility display variable performance in the Achromobacter genus Ann Clin Microbiol Antimicrob 23 78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.E Matuschek C Longshaw M Takemura Y Yamano G Kahlmeter 2022 Cefiderocol: EUCAST criteria for disc diffusion and broth microdilution for antimicrobial susceptibility testing J Antimicrob Chemother 77 1662 1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.G Bianco M Boattini S Comini G Banche R Cavallo C Costa 2023 Disc diffusion and ComASP® cefiderocol microdilution panel to overcome the challenge of cefiderocol susceptibility testing in clinical laboratory routine Antibiotics 12 604 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.