Abstract
This work aims to understand how normouricemic levels of uric acid can induce endothelial dysfunction seeking global proteomic alterations in Human Umbilical Vein cells (HUVEC). It reveals significant alterations in redox-sensitive and antioxidant proteins, chaperones, and proteins associated with cell migration and adhesion in response to uric acid exposure. Monitoring cellular oxidation with the roGFP2-Grx1 probe proved increased oxidation levels induced by uric acid, which can be attenuated by peroxidasin (PXDN) inhibition, suggesting a regulatory role for PXDN in mitigating oxidative stress induced by uric acid. As a consequence of uric acid oxidation and the formation of reactive intermediate, we identified adducts in proteins (+140 kDa) in a novel post-translation modification named uratylation. Increased misfolded protein levels and p62 aggregation were also found, indicating disturbances in cellular proteostasis. Furthermore, uric acid promoted monocyte adhesion and upregulated ICAM and VCAM protein levels, implicating a pro-inflammatory response in endothelial cells. These findings provide critical insights into the molecular mechanisms underlying vascular damage associated with uric acid.
Keywords: Uric acid, Proteomics, HUVEC, Endothelial cell damage, PXDN, Inflammation
Graphical abstract
Highlights
-
•
First proteomics study revealing the modulation of protein abundance in HUVEC upon physiological concentrations of uric acid.
-
•
Chaperones, redox signaling proteins, proteins related to protein degradation and to the inflammatory processes were changed.
-
•
Uric acid induced an increase in oxidation levels in cells.
-
•
PXDN inhibition or silencing decreased urate-dependent oxidation.
-
•
Uric acid increased protein misfolding and monocyte adhesion.
1. Introduction
Uric acid is the final product of purine metabolism in humans, due to the absence of the enzyme uricase, which typically converts uric acid into allantoin [[1], [2], [3]]. It can also be referred to as urate, since in plasma, uric acid predominantly exists as the mono-anion urate (pKa 5.4). Its plasma concentrations range from 50 to 400 μM under normouricemic conditions but can reach millimolar concentrations in hyperuricemic patients [2,4]. With its notable abundance in plasma and low one-electron reduction potential (E° = 0.56 V, pH 7.0), urate has been considered the primary antioxidant in this fluid [[3], [4], [5]]. Uric acid can chelate transition metals and effectively scavenges oxidants like hydroxyl radical, singlet oxygen, peroxynitrite (ONOO−), and hypochlorous acid (HOCl) [5,6]. Despite its antioxidant capacity, elevated levels of uric acid in the bloodstream have been associated with various physiological and pathological conditions. The assumption that uric acid is one of the main antioxidants in plasma remains a paradox since the reaction of uric acid with these oxidants can produce intermediates that cause oxidative damage [5,6]. For instance, the reaction of uric acid with products of ONOO− decomposition produces carbon-centered radicals that are potentially toxic [7]. It can also oxidize low-density lipoproteins (LDLs) in the presence of copper ions and lipid hydroperoxides [8,9]. Moreover, uric acid may impair nitric oxide (NO) bioavailability, a key regulator of vascular tone and function, contributing to hypertension [10,11].
Uric acid crystals are formed when uric acid levels exceed the saturation point in the blood, and soluble uric acid can trigger the activation of the NLRP3 inflammasome [[12], [13], [14]]. As a result, uric acid is considered a DAMP (damage-associated molecular pattern) [15]. Beyond its well-known role in gout, hyperuricemia is also a marker for metabolic syndrome and vascular damage, and it has been associated with cardiovascular diseases, endothelial dysfunction, and atherosclerosis [[15], [16], [17], [18], [19], [20], [21], [22], [23]]. Of note, plasma uric acid, within normouricemic range, was positively associated with carotid intima media thickness (c-IMT) in subclinical atherosclerosis. The significant and positive correlation between uric acid levels and c-IMT was independent of traditional risk factors in a cohort of 45–60 years old men [24]. Additionally, in a broader cohort study, patients were stratified in quintiles according to the levels of uric acid and a significant correlation between c-IMT and serum uric acid was found in those quintiles where uric acid varied from 275 to 300 μM (Q3) and 301–350 μM (Q4) [25]. These clinical evidences pointed that uric acid, at concentrations nowadays considered within a normal range, could be involved in vascular damage and atherosclerosis progression. Not surprisingly, some researchers have proposed revising the current established normal range for uric acid [26].
The mechanisms underlying the association of uric acid with cardiovascular disease in absence of gout or hyperuricemia are still unclear. Uric acid is the main organic substrate for peroxidases in plasma [27], and our hypothesis is that the reactive intermediates of this reaction are involved in the pathogenesis of the endothelial dysfunction. We and other groups have shown that, even under normouricemic conditions, uric acid is oxidized by myeloperoxidase (MPO) [28,29], lactoperoxidase (LPO) [30], and peroxidasin (PXDN) [[31], [32], [33]]. The peroxidase-catalyzed oxidation of uric acid produces urate free radicals that can add to superoxide forming urate hydroperoxide (HOOU) (Fig. 1). This peroxide oxidizes cysteine in biomolecules, including peroxiredoxins and protein disulfide isomerase, generating hydroxyisourate (HOU) [[34], [35], [36]], leading to an imbalance in the cellular redox state [29]. In addition, urate-derived electrophiles can post-translationally modify lysine residues, a reaction called uratylation [37], but the consequence of this modification to different proteins is still unknown. Uratylated albumin appears to be increased in patients with heart failure [38] and increases the secretion of pro-inflammatory cytokines and monocyte adhesion to endothelial cells (under submission).
Fig. 1.
– Oxidation of urate by peroxidases can produce urate-radical, 5-hydroxyisourate, urate hydroperoxide, adducts in lysine residues (R) and allantoin [28,29,31,34].
We have recently shown that normouricemic concentrations of urate decrease HUVEC migration and adhesion, effects that were correlated with the alternative oxidation of urate by PXDN [31]. Additionally, endothelial cells are sensitive to a variety of inflammatory stimuli and can alter their secreted content according to this activation. In this sense, uric acid and/or its oxidation products may modify the content secreted by endothelial cells as well as the overall cellular proteome, contributing to the alteration of vascular homeostasis. In this study, we performed a global proteomic analysis of the secretome and whole-cell lysate from HUVECs, specifically investigating the effects of uric acid at concentrations traditionally considered within the normal range. Importantly, our findings demonstrate that, even at normouricemic levels, uric acid can significantly impact endothelial function. While our data confirmed the involvement of uric acid in regulating cell migration and adhesion, we also identified additional pathways affected by urate, including protein folding and redox homeostasis. Based on the results obtained by proteomics, we conducted a series of validation experiments, which not only reinforced our initial findings but also provided novel insights into the complex mechanisms by which uric acid, even at physiologically normal levels, disrupts endothelial function.
2. Material and methods
2.1. Reagents and antibodies
RPMI 1640 medium (with and without phenol red), Trypsin/EDTA 0.1 %, 100x Antibiotic Solution, complete™ protease inhibitor cocktail (Roche), and Hank's Balanced Salt Solution (HBSS) were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Fetal bovine serum (FBS) was purchased from Vitrocell (Campinas, Brazil). Lipofectamine™ RNAiMAX Transfection Reagent and Lipofectamine™ LTX Plus Transfection Reagent were obtained from Invitrogen (Life Technologies, Waltham, MA, USA). Solvents were purchased from JTBaker (Thermo Fisher Scientific, Life Technologies, Waltham, MA, USA). Mass Spectrometry Grade Trypsin was purchased from Promega (Madison, WI, USA). Mass spectrometry grade iodoacetamide and dithiothreitol (DTT) were obtained from Bio-Rad Laboratories (Hercules, CA, USA). Amicon® Ultra-15 mL centrifugal filters with 10 kDa cutoff were acquired from Merck Millipore (Merck KGaA, Darmstadt, Germany). Plasmid pCDNA5/FRT/TO + roGFP-GRX1 and E. coli dh5α were kindly provided by Professor Luis Eduardo Soares Netto from the University of Sao Paulo, Brazil. Rabbit Monoclonal CD54/ICAM-1c(67836) antibody was purchased from Cell Signaling Technology (Massachusetts, USA). VCAM Mouse Monoclonal SQSTM1/p62 antibody (ab56416) was purchased from Abcam (Cambridge, UK). Mouse Monoclonal Anti-α-Tubulin antibody (T5168), anti-rabbit, and anti-mouse IgG HRP-linked antibodies were acquired from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). PXDN primary antibody was kindly provided by Dr. Miklos Geiszt at Semmelweis University, Hungary. Small interfering (si) RNAs targeting PXDN (siPXDN), and control siRNA (siRNA scramble) were obtained from Invitrogen (Life Technologies, Waltham, MA, USA). All other reagents were purchased from Sigma Aldrich (Merck KGaA, Darmstadt, Germany).
2.2. Cell culture and transfection
A selection of immortalized human umbilical vein endothelial cell line (HUVEC) was kindly provided by Prof. Francisco Laurindo INCOR (São Paulo – Brazil). Cells were maintained in RPMI 1640 medium containing 10 % fetal bovine serum (FBS), streptomycin (100 μg/mL), and penicillin (30 μg/mL) at 37 °C in a 5 % CO2 atmosphere. For experiments, confluent adherent cells were harvested with Trypsin 0.1 %. After centrifugation and counting, cells were seeded on culture plates in the appropriate concentration. HUVECs were used between passages 4 and 10.
2.3. Isolation of secretome
Cells were grown at 80 % confluence in 100 mm culture plates. To isolate the secretome, cells were washed twice with PBS (10 mM, pH 7.4) and incubated with urate for 1h 30 min or 24h with RPMI 1640 (without FBS and phenol red). The culture media was collected after the incubation period, centrifuged at 1400 rpm for 10 min, and filtered through disposable filters of 0.22 μm to remove cellular debris and floating cells. Next, the secretome was concentrated 200-fold on Amicon-Ultra 15 mL - 10 kDa filters.
2.4. Cell lysate
After urate treatment, cells were washed twice with PBS and scrapped in lysis buffer (8 M urea, 100 mM ammonium bicarbonate, 1 mM PMSF, 1x protease inhibitor cocktail). Samples were sonicated for 30 s, incubated for 30 min on ice, and spun down (4 °C, 20 min, 12,000 rpm).
2.5. Protein quantification from cell extracts
Protein concentration was calculated using Pierce Bicinchoninic Acid (BCA) Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA) with bovine albumin as standard carried out following the manufacturer's instructions.
2.6. Protein identification by proteomics
An amount of 10 μg protein was solubilized in 100 mM ammonium bicarbonate with 0.4 % sodium deoxycholate, reduced with DTT (5 mM), alkylated with iodoacetamide (15 mM), and digested with Sequencing Grade Modified Trypsin (Promega), protein/trypsin ratio 1:40 w/w for 4 h at 37°C. A second aliquot of trypsin (1:50 w/w) was added and samples were incubated overnight at 37°C. After acidic hydrolysis with 2% trifluoroacetic acid, samples were desalted using the StageTip protocol [39].
2.7. Data dependent acquisition (DDA) proteomics
Digested and desalted samples were suspended in 0.1% formic acid (25 ng/μL final protein concentration) and submitted to MS analyses. Angiotensin (0.2 pmol/μL) was used as a global internal standard to monitor MS variability, and iRT peptides (Pierce Biotechnology, Rockford, 0.1 pmol/μL) were used to normalize the retention time of all peptides. An Easy-nLC 1200 UHPLC (Thermo Scientific, Bremen, Germany) was used for peptide separation with a linear gradient of solvent A (0.1% formic acid) and solvent B (0.1% formic acid in 80% acetonitrile). Each sample was loaded onto a trap column (nanoViper C18, 3 μm, 75 μm × 2 cm, Thermo Scientific) with 12 μL solvent A at 980 bar. After this period, the trapped peptides were eluted onto a C18 column (nanoViper C18, 2 μm, 75 μm × 15 cm, Thermo Scientific) at a flow rate of 300 nL/min. Peptides were eluted from the column using a linear gradient of 5–28% B for 25 min followed by a linear gradient of 28–40% B for 5 min. Finally, the percentage of solvent B was increased to 95% in 2 min and the column was washed for 10 min with this solvent proportion. Re-equilibration of the system with 100% A was performed before each injection. Acquisition of the data was performed using an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific, Bremen, Germany) with a nanospray Flex NG ion source (Thermo Scientific, Bremen, Germany). A full MS scan was followed by data-dependent MS2 scans in a 3 s cycle time. Precursor ions selected for MS2 were excluded for subsequent MS2 scans for 20 s. The resolution for the full scan mode was set as 120,000 (at m/z 200) and the automatic gain control (AGC) target at 4 × 105. The m/z range 400–1600 was monitored. Each full scan was followed by a data-dependent MS2 acquisition with a resolution of 30,000 (at m/z 200), maximum fill time of 54 ms, isolation window of 1.2 m/z, and normalized collision energy of 30.
2.8. Protein identification
Raw files of all proteomic experiments were processed using MaxQuant software. Tandem mass spectrometry (MS/MS) spectra were searched against the reviewed UniProt human database, using the MaxQuant search engine with fixed Cys carbamidomethylation, variable Met oxidation, and variable N-terminal acetylation. MaxQuant default mass tolerance was used for precursors and product ions. Trypsin/P was selected as the enzyme, and two missed cleavages were allowed. Error mass tolerance for precursors and fragments was set to 4.5 ppm and 20 ppm. The results were processed by label-free quantification. The protein was considered present if at least two peptides (one of them being unique) were detected. The match between runs option was enabled and the other parameters were kept as default.
2.9. PTM identification
Uratylation in lysine residues (composition: C(4)N(4)O(2)H(4), +140.0334 Da [37]) was set as variable modification. Ubiquitination was considered as a sum of ubiquitination residue (GlyGly (K) + 114.0429 Da), and small ubiquitin-like modifier (SUMO) (QQTGG (K) 471.2077 Da). Since the modifications occur on lysine residues, a maximum of 5 missed cleavages was allowed.
2.10. Differently abundance protein analysis
Cell lysate and supernatant from HUVEC from two independent experiments containing there replicates per group were subject to DDA proteomic analysis. The relative abundance of the proteins was obtained by a label-free method using the MaxQuant software, in which the peak area of the protein precursor ions is integrated [40]. The result is a relative quantification of the most abundant proteins in the system under study, being more accurate for major differences in abundance between proteins [41]. The values of intensities from the LFQ analysis were normalized between all experimental groups and their technical replicates. After the MaxQuant search, Label-free quantification (LFQ) intensities were loaded in Perseus software [42]. Reverse and only identified by site peptides and potential contaminants were removed via filtering, and the data was transformed to log2. To filter missing values, a percentage of 90% of valid values per group was considered. Hierarchical clustering and principal component analysis (PCA) were also performed using Perseus default settings. ClustVis (biit.cs.ut.ee/clustvis) was used to plot heatmap and PCA graphs using data imported from Perseus. After clustering rows on Perseus, data was imported, and Gene Ontology analysis was performed by PANTHER Classification System (pantherdb.org). The top 6 (for secretome) and top 10 (for lysate) terms with higher significance (−log10 p-value) and the respective number of identified proteins from each term are shown for each process. P-values were considered significant when they were lower than 0.05 after FDR (False Discovery Rate) correction.
2.11. Redox sensor (roGFP2-Grx1) monitoring
Competent E. coli dh5α cells were transfected with the pCDNA5/FRT/TO + roGFP-GRX1 plasmid. Bacteria were grown overnight in LB medium, and plasmid was isolated by PureLink™ HiPure Plasmid Maxiprep Kit (Invitrogen, Thermo).
HUVEC were transfected with 5 ng isolated DNA using Lipofectamine LTX + Plus reagent according to the manufacturer's instructions. Media was changed 24 h post-transfection. 48 h after transfection, cells were harvested, washed with PBS, and resuspended in HBSS buffer (750 cells/μL). Cells were transferred to black 96 well plates with clear bottoms in a final volume of 200 μL per well. Fluorescence was read in a microplate reader (Tecan Infinite M1000) using an excitation wavelength of 405 (oxidized sensor) and 485 (reduced sensor) nm for a 535 nm emission wavelength every 2 min with 10 s agitation in between for 6 min at 37°C. Reading was paused, and 5 μL stock solutions were added to yield a final concentration of 100 μM H2O2, 2 mM DTT, and 50, 100, and 200 μM urate. Fluorescence was read until 2 h of final incubation, and the data were plotted using the 405/485 fluorescence intensity ratio normalized by fluorescence obtained by DTT-treated cells (total reduced sensor). For the end-point imaging of the redox sensor, cells were plated in 8 well Nunc™ Lab-Tek™ II Chambered Coverglass (Thermo). A confocal microscope (Leica SP8 laser scanning confocal microscope on a DMI8 microscope base) using LAS X software, and a 63 × oil objective were used to acquire images of excitation wavelength of 405 (oxidized sensor) and 485 (reduced sensor) nm for a 535 nm emission wavelengths. Images were acquired after 2 h of total incubation. Ratiometric images were analyzed by Image J Fiji.
2.12. Proteostat aggregation assay
The PROTEOSTAT aggresome detection kit (ENZ-51035 - Enzo Life Sciences, Farmingdale, New York, USA) was used to detect aggregating proteins. Cells were plated in 24 well plates (104 cells/well) containing 12 mm glass coverslips and incubated for an additional 24 h. After incubation, cells were washed with PBS and treated with urate (50, 100, and 200 μM) for 24 h, and MG132 1 μM for 6h in RPMI 1640 medium. After incubation, cells were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.3% Triton X-100 in PBS, and blocked with 2% BSA in PBS for 1h, with three PBS washes between the steps. Coverslips were incubated with α-tubulin primary antibody overnight at room temperature. After washing with PBS, coverslips were incubated with a secondary antibody conjugated with Alexa Fluor 488 for 2 h at room temperature. Cells were again washed twice with PBS and the Proteostat dye was added at 1:2000 dilution with 2.5 μg/ml DAPI in PBS for 30 min at room temperature. Cells were mounted in ProLongGold Antifade Reagent (Cell Signaling, Danvers, Massachusetts, USA) after washing twice in PBS. Images were acquired with a Leica SP8 laser scanning confocal microscope (Leica GmbH, Mannheim Germany) on a DMI8 microscope base using LAS X software, a 63 × oil objective, and a 405 nm diode laser, in addition to a multi-line white light laser, with excitation wavelengths of 488 nm (for α-tubulin) and 594 nm (for Proteostat). Spectral detection using a PMT from 410 to 450 nm was utilized for DAPI, a HyD detector from 490 to 560 nm for AlexaFluor 488, and a HyD detector from 604 to 640 nm for Proteostat probe. Single-channel images were obtained with LASX Office software (v1.4.7 28921, Leica Microsystems). Fluorescence intensity was quantified using the RGB Measure plugin in Image J software.
2.13. Immunofluorescence staining of p62
HUVEC were harvested and seeded (104 cells/well) into 12 mm glass coverslips placed in 24 well plates. After 48 h, cells were washed with PBS, and incubated with urate (50, 100, and 200 μM) in RPMI 1640 medium. After 24 h, cells were fixed with 4% paraformaldehyde in PBS for 20 min, permeabilized with 0.3% Triton X-100 in PBS, and blocked with 2% BSA in PBS for 1 h, with three PBS washes between the steps. Coverslips were incubated with primary antibodies diluted (1:1000) in BSA 2% overnight at room temperature. After incubation, cells were 4-fold washed with PBS and incubated with secondary antibody Alexa Fluor 488 at room temperature for 2 h. Cells were 3-fold washed with PBS and incubated with 2.5 μg/ml DAPI for 15 min. After 2 more PBS washes, coverslips were mounted on microscope objective slides using ProLongGold Antifade Reagent (9071, Cell Signaling, Danvers, Massachusetts, USA). Images were acquired with a Leica SP8 laser scanning confocal microscope (Leica GmbH, Mannheim Germany) on a DMI8 microscope base using LAS X software, a 63 × oil objective, and a 405 nm diode laser, in addition to a multi-line white light laser set to 488 excitation wavelengths. Spectral detection using a PMT from 410 to 450 nm was utilized for DAPI and a HyD detector from 490 to 560 nm for AlexaFluor 488. Single channel images were obtained with LASX software and p62 puncta was analyzed by Image J Particle Analysis.
2.14. Monocyte (THP-1) adhesion in HUVECs
Confluent adherent cells were harvested using a solution containing trypsin (0.1%) and EDTA (0.5 mM) prepared in phosphate-buffered saline solution (PBS; 10 mM, pH 7.4). After centrifugation and counting, cells were seeded on 24-well plates at an initial confluence of 5 × 104 cells/well. After 3 days, cells were washed with PBS and incubated with urate (50–200 μM) in RPMI medium without phenol red and fetal bovine serum. LPS (1 μg/mL) and TNF-α (5 ng/mL) were used as a positive control. After 24 h, cells were washed with PBS twice and incubated with THP-1 cells (4 × 105 cells per well) for 1 h. THP-1 cells were previously stained with 10 nM Calcein-AM (Invitrogen) for 30 min. After incubation, cells were washed with PBS until the complete removal of non-adherent cells. Images were taken by EVOS FLoid Imaging System (Thermo Fisher Scientific). THP-1 cells/optical field were counted by Image J.
2.15. Western blot
HUVEC were seeded on 24-well plates at an initial confluence of 5 × 104 cells/well. After 3 days, cells were washed with PBS and incubated with urate (50–200 μM) in RPMI medium without phenol red and fetal bovine serum. LPS (1 μg/mL) and TNF-α (5 ng/mL) were used as a positive control. After 24 h, cells were washed with PBS twice and lysed with RIPA buffer (10 mM Tris-HCl, pH 8.0, 140 mM NaCl 1 mM EDTA, 0.5 mM EGTA, 1 % Triton X-100, 0.1 % Sodium Deoxycholate, 0.1 % SDS, 1 mM PMSF, 1x Complete Protease Inhibitor Cocktail (Roche)). After protein quantification (as stated in 2.5), proteins from HUVEC secretome and lysate were mixed with reducing Laemmli buffer (31.5 mM Tris-HCl, pH 6.8, 10 % glycerol, 1 % SDS, 0.005 % Bromophenol Blue, 50 mM DTT) and heated at 95 °C for 10 min. A total amount of 20 μg total protein content was loaded on 10 % SDS-PAGE gels. Proteins were transferred to PVDF membranes with the Trans-Blot Turbo Transfer System (Bio-Rad). Blot membranes were blocked for 1 h with 5% milk in PBS and incubated with primary antibodies overnight at 4°C. HRP-conjugated secondary antibodies were incubated at 1:10,000 dilution for 1 h at room temperature. Membranes were incubated with Clarity Western ECL Substrate (Bio-Rad Laboratories) and proteins were detected using chemiluminescence mode on the ChemiDoc Imaging System (Bio-Rad Laboratories).
2.16. Cell viability
Cell viability was evaluated with differential staining by the DNA-binding dyes propidium iodide (PI) and Hoechst 33342 (HO) in the absence and presence of 10% FBS with or without urate. HUVECs were grown in 6-well plates with an initial density of 0.5 x 106 cells/well for 24 h. Cells were washed with PBS and incubated in RPMI culture medium with and without 10% FBS in the presence of 50 and 100 μM urate for 48 h or 24 h. After treatment, cells were incubated with at final concentration of 1 μg/ml for each dye. After 10 min of incubation, viable or dead cells were verified using an inverted fluorescence microscope (Nikon Eclipse Ti) with 10 × magnification. The cultures were evaluated according to the total number of cells (determined by counting the nuclei stained with HO) and number of dead cells (determined by the number of nuclei stained with PI or brightly HO (condensed chromatin).
Additionally, cell viability was evaluated by flow cytometry with the Annexin V-FITC Apoptosis Detection Kit (APOAF-20TST, Sigma Aldrich) in urate-treated HUVECs cells with or without 10% FBS. HUVECs were grown in 6-well plates with an initial density of 0.5 x 106 cells/well for 24 h. The wells were washed with PBS and incubated in RPMI culture medium with and without 10 % FBS in the presence of urate at concentrations of 50 and 100 μM and at 24 or 48 h. After incubations, cell suspension was collected, and adhered cells were detached. Fractions were pooled and the resulting cell pellet was subjected to flow cytometry analysis after the addition of Annexin V-FTIC and PI according to manufacturing structures. Treatment with 1 μM staurosporine for 2 h in RPMI complete medium was considered positive control for cell death. Cells were subjected to flow cytometry analysis in a BD- FACS Verse (BD Biosciences). FITC Annexin V and PI negative cells were considered viable and not undergoing apoptosis, and cells undergoing apoptosis (FITC Annexin V positive and PI negative) or end-stage apoptosis or already dead (FITC Annexin V and PI positive) were considered not viable. Data was analyzed by FlowJo v10 (BD Biosciences), were total cell number and percentage were calculated.
2.17. Statistical analysis
All data are expressed as mean ± standard error of the mean (SEM) of at least 3 independent experiments. All analyses were performed using GraphPad Prism v.10. Data were analyzed by one-way ANOVA or two-way ANOVA and post-test, as indicated in each legend.
3. Results
3.1. Proteomic analysis of HUVEC upon uric acid treatment
The effects of uric acid in HUVECs were analyzed by DDA proteomics in lysate and secretome. To collect and concentrate the secretome it was necessary to incubate cells in a serum-free medium. Therefore, we first performed viability assays to ensure FBS depletion would not be cytotoxic within 24 h and 48 h. There was no difference in cell viability among groups looking at propidium iodide/Hoechst 33342 nuclei labeling (Suppl. information Fig. S1) nor at Annexin V/propidium iodide (Suppl. information Fig. S2), confirming that the absence of FBS did not alter cell viability at these time and conditions. Uric acid also did not induce cell death within the concentrations and time scale tested. Likewise, there was no visible change in cell morphology.
To ensure the secretome fraction was enriched with extracellular proteins we performed a Gene Ontology (GO) enrichment analysis with all identified proteins from the control groups (Supplementary Table 1). This analysis revealed the predominance of extracellular proteins and components of vesicles and extracellular matrix, indicating that during the secretome collection process, there was no relevant cell lysis. Differently expressed proteins from secretome and lysates are shown in Fig. 2A–D, with hierarchical grouping clustering and principal component analysis (PCA) [42]. One and a half and 24 h after urate (50–200 μM) incubation led to 159 and 147 differently expressed proteins in the secretome (Fig. 2A and B). Analysis of whole-cell lysate revealed 356 and 814 proteins differently expressed 1h30 (Figs. 2C) and 24 h (Fig. 2D) after urate (50–200 μM) incubation. Proteins were clustered concerning their abundance profile, as indicated in each figure. Functional annotation was made for the enrichment in terms of GO (Gene Ontology), to better understand the function of the proteins that were regulated by the presence of urate, as shown in Table 1, Table 2, Table 3, Table 4. To highlight the major changes in protein abundance between control and urate 200 μM, the data were also plotted as a scatterplot of significance (p-value) vs. fold change (Suppl. information Fig. S3).
Fig. 2.
– Label-free proteomics data from HUVEC untreated (green) or treated with three different concentrations of urate 50 μM (orange), 100 μM (purple), and 200 μM (pink) in secretome 1:30 (A) or 24 h (B) after urate incubation or whole-cell lysate 1:30 (C) or 24 h (D) after urate incubation. Hierarchical clustering (left panel) of significantly altered proteins was obtained by ANOVA correcting for multiple comparisons using FDR<0.05, and proteins were grouped into 2 clusters (1 – green and 2 – light purple). Relative expression values are displayed as a gradient from red (higher amount) to blue (lower amount). Rows (proteins) are centered; unit variance scaling is applied to rows (treatment). Both rows and columns are clustered using Euclidean distance and average linkage. Principal component analysis (PCA) (right panel) from the studied groups. Unit variance scaling is applied to rows; SVD with imputation is used to calculate principal components. X and Y axis show principal component 1 and principal component 2 that explain the shown percentages of the total variance, respectively. Prediction ellipses are such that with probability 0.95. Representative data from two independent experiments in technical replicates. Images were obtained by ClustVis (https://biit.cs.ut.ee/clustvis/).
Table 1.
Gene ontology (GO) enrichment analysis of the biological processes, molecular functions, and cellular components from the analyzed clusters containing secretome proteins that were regulated by incubation with urate for 1:30 h.
| Cluster 1 (n = 37) |
Cluster 2 (n = 122) |
||||
|---|---|---|---|---|---|
| Biological process | # of proteins | -log(p-value) | Biological process | # of proteins | -log(p-value) |
| response to external stimulus | 15 | 4.88 | gene expression | 55 | 16.57 |
| cellular component biogenesis | 16 | 4.69 | ribosome biogenesis | 17 | 8.38 |
| chromosome organization | 8 | 1.96 | protein folding | 9 | 2.54 |
| translational initiation | 4 | 1.74 | response to cytokine | 15 | 1.54 |
| DNA replication initiation | 3 | 1.55 | 'de novo' protein folding | 4 | 1.45 |
| double-strand break repair | 5 | 1.30 | response to chemical | 39 | 1.43 |
| Molecular function | # of proteins | -log(p-value) | Molecular Function | # of proteins | -log(p-value) |
| ATP hydrolysis activity | 11 | 5.75 | RNA binding | 58 | 27.42 |
| nucleic acid binding | 23 | 4.25 | cell adhesion molecule binding | 20 | 7.22 |
| ATP binding | 13 | 3.21 | protein binding | 110 | 6.65 |
| translation regulator activity | 5 | 2.75 | laminin binding | 5 | 3.23 |
| cadherin binding | 6 | 2.24 | cadherin binding | 11 | 2.94 |
| unfolded protein binding | 4 | 1.87 | oxidoreductase activity | 5 | 2.32 |
| Cellular component | # of proteins | -log(p-value) | Cellular component | # of proteins | -log(p-value) |
| cytosol | 30 | 6.18 | ribosomal subunit | 21 | 26.41 |
| extracellular exosome | 18 | 4.91 | extracellular space | 69 | 22.48 |
| vesicle | 23 | 4.45 | focal adhesion | 27 | 17.47 |
| extracellular space | 19 | 3.27 | cytosol | 73 | 12.82 |
| nucleus | 29 | 3.00 | cell junction | 44 | 11.37 |
| microtubule | 6 | 1.66 | endoplasmic reticulum | 33 | 5.72 |
Table 2.
Gene ontology (GO) enrichment analysis of the biological processes, molecular functions, and cellular components from the analyzed clusters containing secretome proteins that were regulated by incubation with urate for 24 h.
| Cluster 1 (n = 123) |
Cluster 2 (n = 21) |
||||
|---|---|---|---|---|---|
| Biological process | # of proteins | -log(p-value) | Biological process | # of proteins | -log(p-value) |
| inflammatory response regulation | 26 | 6.39 | extracellular matrix protein | 6 | 5.94 |
| cellular component organization | 34 | 4.05 | cell adhesion | 9 | 2.52 |
| cytokine signaling | 14 | 2.43 | response to wounding | 6 | 2.05 |
| RNA processing | 17 | 2.39 | extracellular matrix | 5 | 1.92 |
| regulation of proteolysis | 12 | 1.48 | cell projection | 8 | 1.83 |
| protein folding | 5 | 1.45 | cell migration | 7 | 1.72 |
| Molecular function | # of proteins | -log(p-value) | Molecular Function | # of proteins | -log(p-value) |
| cadherin binding | 21 | 11.66 | extracellular matrix | 6 | 4.12 |
| cell adhesion molecule binding | 22 | 8.60 | calcium ion binding | 7 | 2.06 |
| protein binding | 114 | 7.92 | proteoglycan binding | 3 | 1.95 |
| nucleic acid binding | 51 | 5.15 | fibronectin binding | 3 | 1.91 |
| actin binding | 15 | 4.45 | peptidase activator activity | 3 | 1.62 |
| unfolded protein binding | 6 | 1.81 | laminin interactions | 2 | 1.34 |
| Cellular component | # of proteins | -log(p-value) | Cellular component | # of proteins | -log(p-value) |
| extracellular space | 78 | 29.44 | extracellular region | 18 | 6.43 |
| cytoplasm | 108 | 10.72 | basement membrane | 5 | 4.77 |
| focal adhesion | 20 | 9.68 | lysosomal lumen | 4 | 3.28 |
| cell junction | 36 | 6.17 | vesicle | 13 | 2.52 |
| secretory vesicle | 24 | 6.03 | cell adhesion | 3 | 2.46 |
| nuclear lumen | 50 | 4.26 | laminin complex | 2 | 2.25 |
Table 3.
Gene ontology (GO) enrichment analysis of the biological processes, molecular functions, and cellular components from the analyzed clusters containing lysate proteins that were regulated by incubation with urate for 1:30 h.
| Cluster 1 (n = 61) |
Cluster 2 (n = 297) |
||||
|---|---|---|---|---|---|
| Biological process | # of proteins | -log(p-value) | Biological process | # of proteins | -log(p-value) |
| DNA repair | 11 | 5.40 | translation | 40 | 17.83 |
| response to stress | 22 | 4.84 | ribosome biogenesis | 15 | 5.50 |
| DNA replication initiation | 4 | 3.41 | protein folding | 17 | 4.96 |
| RNA transport | 5 | 2.01 | cell adhesion | 34 | 3.71 |
| response to chemical | 18 | 1.80 | cell migration | 31 | 2.75 |
| recombinational repair | 4 | 1.44 | extracellular matrix organization | 14 | 2.22 |
| regulation of DNA replication | 4 | 1.40 | response to unfolded protein | 8 | 1.53 |
| NADP metabolic process | 3 | 1.53 | |||
| cell redox homeostasis | 5 | 1.51 | |||
| proteolysis | 19 | 1.47 | |||
| Molecular Function | # of proteins | -log(p-value) | Molecular function | # of proteins | -log(p-value) |
| RNA nuclease activity | 7 | 5.79 | structural constituent of ribosome | 32 | 21.20 |
| helicase activity | 5 | 4.36 | cadherin binding | 37 | 18.16 |
| cadherin binding | 7 | 3.31 | oxidoreductase activity | 30 | 4.28 |
| nucleic acid binding | 18 | 1.92 | DNA binding | 3 | 4.09 |
| peroxiredoxin activity | 2 | 1.68 | laminin binding | 7 | 3.90 |
| protein folding chaperone | 7 | 3.19 | |||
| protein disulfide isomerase activity | 4 | 2.76 | |||
| extracellular matrix binding | 7 | 2.54 | |||
| ubiquitin-like protein binding | 14 | 1.68 | |||
| peroxiredoxin activity | 3 | 1.58 | |||
| Cellular component | # of proteins | -log(p-value) | Cellular component | # of proteins | -log(p-value) |
| extracellular organelle | 13 | 1.95 | cytosol | 175 | 31.66 |
| extracellular vesicle | 13 | 1.91 | focal adhesion | 52 | 29.13 |
| extracelular exosome | 13 | 1.83 | ribosome | 34 | 19.72 |
| secretory granule | 8 | 1.71 | endoplasmic reticulum | 61 | 6.32 |
| vesicle lumen | 5 | 1.60 | cytoplasmic vesicle | 66 | 5.03 |
| basement membrane | 10 | 4.33 | |||
| cell-cell junction | 21 | 3.32 | |||
| proteasome complex | 7 | 2.87 | |||
| nucleus | 134 | 2.05 | |||
| cell surface | 25 | 1.64 | |||
Table 4.
Gene ontology (GO) enrichment analysis of the biological processes, molecular functions, and cellular components from the analyzed clusters containing lysate proteins that were regulated by incubation with urate for 24 h.
| Cluster 1 (n = 525) |
Cluster 2 (n = 289) |
||||
|---|---|---|---|---|---|
| Biological process | # of proteins | -log(p-value) | Biological process | # of proteins | -log(p-value) |
| translation | 73 | 34.03 | intracellular transport | 55 | 8.81 |
| protein folding | 41 | 18.38 | proton transmembrane transport | 16 | 6.59 |
| chaperone-mediated protein folding | 24 | 14.58 | vesicle-mediated transport | 44 | 4.90 |
| protein stabilization | 34 | 12.71 | glucose metabolic process | 7 | 4.51 |
| ribosome assembly | 20 | 11.42 | oxidative phosphorylation | 11 | 3.55 |
| actin filament-based process | 41 | 5.48 | mRNA stabilization | 8 | 3.14 |
| proteasome-mediated ubiquitin-dependent protein catabolic process | 27 | 4.08 | gluconeogenesis | 7 | 2.84 |
| protein refolding | 8 | 4.01 | ATP synthesis | 8 | 2.18 |
| cellular response to stress | 27 | 3.34 | tricarboxylic acid cycle | 4 | 2.11 |
| nucleosome assembly | 11 | 2.76 | mRNA processing | 10 | 1.99 |
| Molecular Function | # of proteins | -log(p-value) | Molecular function | # of proteins | -log(p-value) |
| structural constituent of ribosome | 48 | 28.77 | protein binding | 262 | 13.53 |
| cadherin binding | 59 | 26.73 | oxidoreductase activity | 38 | 8.32 |
| unfolded protein binding | 35 | 20.49 | cadherin binding | 21 | 5.45 |
| cell adhesion molecule binding | 63 | 19.05 | cell adhesion molecule binding | 27 | 5.09 |
| protein folding chaperone | 22 | 13.27 | GTPase activity | 20 | 4.68 |
| ATP-dependent protein folding chaperone | 16 | 10.25 | electron transfer activity | 12 | 4.49 |
| ubiquitin protein ligase binding | 34 | 9.01 | double-stranded RNA binding | 9 | 3.67 |
| heat shock protein binding | 19 | 6.20 | aldehyde dehydrogenase [NAD(P)+] activity | 5 | 3.00 |
| protein-folding chaperone binding | 18 | 5.39 | pre-mRNA binding | 6 | 2.54 |
| ubiquitin-protein transferase regulator activity | 9 | 4.60 | AMP binding | 4 | 2.07 |
| Cellular component | # of proteins | -log(p-value) | Cellular component | # of proteins | -log(p-value) |
| cytosol | 344 | 75.27 | cytoplasm | 259 | 27.90 |
| ribonucleoprotein complex | 110 | 46.16 | extracellular space | 121 | 22.00 |
| intracellular organelle | 470 | 45.02 | secretory granule | 52 | 14.81 |
| nucleus | 350 | 43.14 | membrane-enclosed lumen | 138 | 10.69 |
| extracellular region | 239 | 35.60 | mitochondrion | 63 | 10.20 |
| ribosome | 61 | 35.23 | membrane | 198 | 9.74 |
| focal adhesion | 65 | 27.16 | focal adhesion | 28 | 8.78 |
| proteasome complex | 23 | 15.52 | cell-substrate junction | 28 | 8.59 |
| cell junction | 124 | 14.97 | membrane protein complex | 53 | 8.33 |
| protein folding chaperone complex | 16 | 10.92 | ribonucleoprotein complex | 36 | 7.97 |
In the secretome, 1:30 h uric acid treatment led to a decrease (Fig. 2A - cluster 2) in the abundance of endoplasmic reticulum (ER) folding proteins (GO:0034975), such as Protein Disulfide-isomerase (P4HB and PDIA3) and the endoplasmic reticulum chaperone BiP (HSPA5), and other proteins related to protein folding (GO:0006457): 60 kDa heat shock protein (HSPD1), 10 kDa heat shock protein (HSPE1) and peptidyl-prolyl cis-trans isomerases (FKBP1A, PPIL1, PPIA). We also found a decrease in cell adhesion-related proteins (GO:0050839) such as Reelin (RELN), Desmoglein-2 (DSG2), Coronin-1B (CORO1B), Profilin-1 (PFN1) Tubulointerstitial nephritis antigen-like (TINAGL1), Galectin-1 (LGALS1), and Agrin (AGRN). On the other hand, urate increased the abundance (Fig. 2 A - cluster 1) of translational-related proteins (GO:0006413), including Eukaryotic Translation Elongation Factor 1 alpha 1 (EEF1A1), Eukaryotic Elongation Factor 2 (EIF2A), Eukaryotic translation initiation factor 3 (EIF3), in addition to the Initiation Factor 4A1 (EIF4A1). Another interesting finding was the increase in proteins related to cellular response to stress (GO:0033554), as Ras GTPase-activating-like protein (IQGAP1), Annexin A1 (ANXA1), Ras GTPase-activating protein-binding protein 1 (G3BP1), Heat shock protein HSP 90 (HSP90), T-complex protein 1 (CCT), Stress-70 protein (HSPA9) and S100 calcium-binding protein A1 (S100A11).
Twenty-four hours after urate treatment there was a decrease in proteins related to cell adhesion (GO:0007155), migration (GO:0016477), and extracellular matrix (GO: GO:0031012) in the secretome (Fig. 2B, cluster 2), including Laminin (LAMB1 and LAMC1), Vascular endothelial growth factor A (VEGFA), Tenascin (TNC), Fibulin-1 (FBLN1), Protein jagged-1 (JAG1), Midkine (MDK), Syndecan-4 (SDC4), Desmocollin (DSC2 and DSC3) and Calsyntenin-1 (CLSTN1). While there was an increase (Fig. 2B - cluster 2) in folding-related proteins (GO:0006457) given by 60 kDa heat shock protein (HSPD1), T-complex protein 1 (CCT6A), Stress-70 protein (HSPA9), Heat shock protein beta-1 (HSPB1), Heat shock protein HSP 90 (HSP90A1), Heat shock 70 kDa (HSPA4), Endoplasmin (HSP90B1). Likewise, proteins that take part in the regulation of proteolysis (GO:0030162) such as 26S proteasome regulatory subunit (PSMC3 and PSMC5), Ubiquilin-1 (UBQLN1), Ephrin-A1 (EFNA1) and Proteasome activator complex subunit 1 (PSME1). Surprisingly, even at these low physiological concentrations, urate increased proteins of inflammatory response (GO:0050727) and cytokine signaling GO:0019221) (Fig. 2B, cluster 1), such as Progranulin (PGRN), Midkine (MDK), Spectrin beta chain 1 (SPTBN1), Beta-2-microglobulin (B2M), and A-kinase anchor protein 12 (AKAP12).
Analyzing cell lysate after 1:30 h urate treatment proteins related to cell adhesion (GO:0007155) like Filamin-A (FLNA), Calsyntenin-1 (CLSTN1), Reelin (RELN), Vinculin (VCL), Desmocollin-3 (DSC3), Laminin (LAMA5 and LAMC1), Talin-1 (TLN1), Integrin alpha (ITGA3), Integrin beta-1 (TGB1), Tenascin (TNC), Nidogen-1 (NID1), and Calsyntenin-3 (CLSTN3) were down-regulated in the presence of urate (Fig. 2C, cluster 2). Additionally, proteins that take part in cell redox homeostasis (GO:0045454), such as Glutaredoxin-3 (GLRX3), Peroxiredoxin 3 (PRDX3), Peroxiredoxin-5 (PRDX5), Peroxiredoxin-6 (PRDX6), and Thioredoxin reductase 1 (TXNRD1) were decreased (Fig. 2C, cluster 2). On the other hand, we detected an increase in the abundance of proteins related to response to stress (GO:0006950), such as Vascular endothelial growth factor A (VEGFA, 60 kDa heat shock protein (HSP60), Annexin A1 (ANXA1), Peroxiredoxin-1 (PRDX1) and Protein disulfide-isomerase (PDIA3) (Fig. 2C, cluster 1).
Finally, analysis of the cell lysate 24 h after urate incubation showed a decrease in proteins that take part in ATP synthesis (GO:0006754) such as NADH dehydrogenase (NDUFB4, NDUFA, and NDUFS2) and ATP synthase (ATP5F1, ATP5I, and ATP5L) (Fig. 2D, cluster 2). Most relevantly, we detected an increase in proteins involved in protein folding (GO:0006457), such as Prefoldin (PFDN1, VBP1, PFDN4, PFDN5 and PFDN6), T-complex protein 1 (CCT3, CCT6A, CCT4, CCT5, CCT2, TCP1, CCT7), 60 kDa heat shock protein (HSPD1), Stress-70 protein (HSPA9), peptidyl-prolyl cis-trans isomerase (FKBP4, PPID, FKBP1A and PPIA), Heat shock protein beta-1 (HSPB1), Endoplasmic reticulum chaperone BiP (HSPA5), Calreticulin (CALR), Heat shock protein HSP 90 (HSP90AA1 and HSP90AB1), Heat shock protein 105 kDa (HSPH1), Heat shock cognate 71 kDa protein (HSPA8), Peroxiredoxin-4 (PRDX4), DnaJ homolog subfamily (DNAJB11 and DNAJA1). Oppositely from what was found in 1:30 h lysate, there was an increase in proteins related to cell redox homeostasis (GO:0045454), such as Peroxiredoxin-1 (PRDX1), Glutaredoxin-3 (GLRX3), Peroxiredoxin-4 (PRDX4), Peroxiredoxin-6 (PRDX6) and Thioredoxin reductase 1 (TXNRD1) (Fig. 2D, cluster 1), looking like cells were adapting to initial redox stress by overexpressing these antioxidant enzymes.
Taking the above results, we carried out experiments to examine specific pathways aiming to validate the proteomic findings and also to elucidate the relationship between uric acid and endothelial cell dysfunction. The proteomic assays revealed that many of the differentially regulated proteins were involved in redox metabolism and the proteostasis network, leading us to hypothesize that uric acid may contribute to oxidative stress in the vascular environment, potentially resulting in increased protein misfolding.
3.2. Uric acid increased oxidation and modified proteins in HUVEC
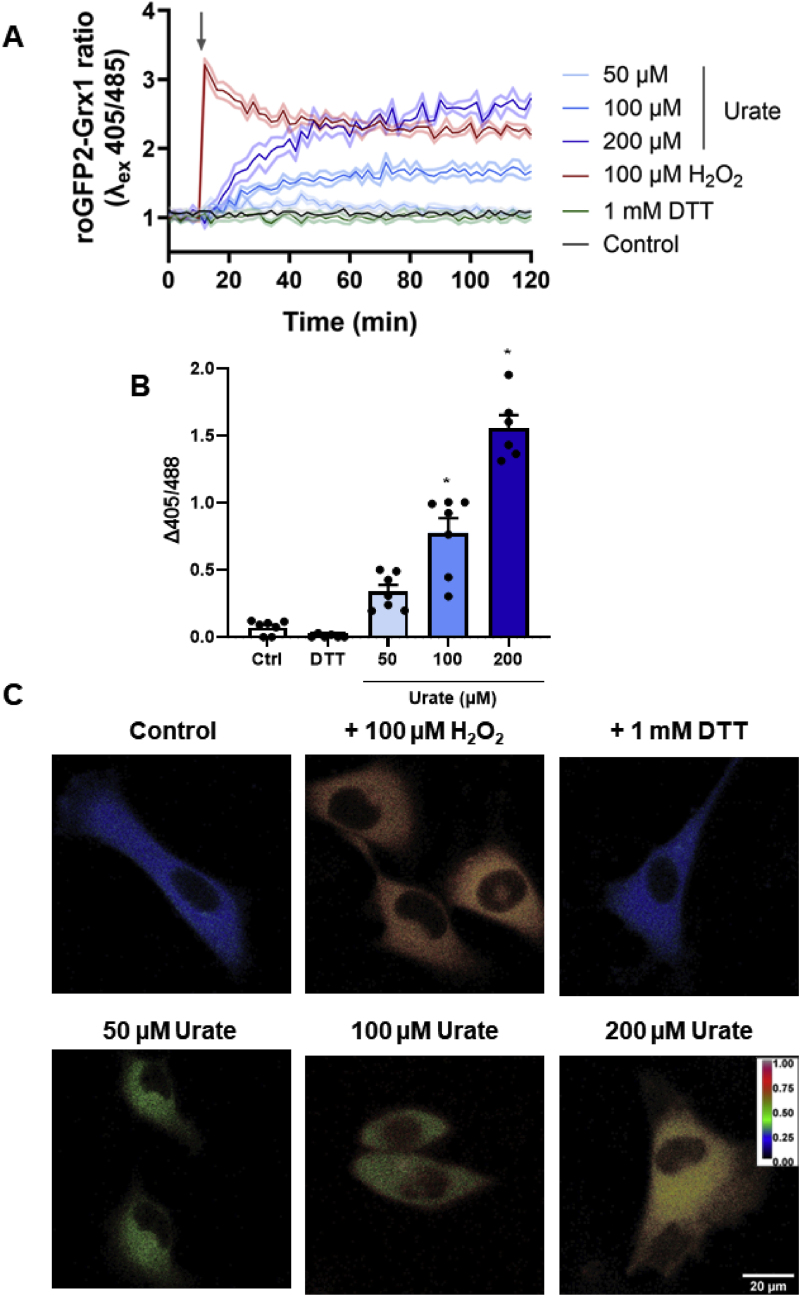
Since urate induced an initial (1:30 h) decrease with a latter (24 h) increase in proteins related to redox homeostasis, we assessed, by mass spectrometry, the reduced (GSH) and oxidized (GSSG) glutathione ratio after 24 h from urate incubation. However, we found no significant difference in this parameter (Suppl information Fig. S4). Because this approach provides endpoint measurements rather than a dynamic process, we expressed the redox sensor roGFP2-Grx1 into HUVECs to estimate the GSSG/GSH ratio indirectly. The glutaredoxin 1 (Grx1) in the roGFP2-Grx1 probe reacts with GSSG, transferring the oxidizing equivalents to the fluorescent sensor roGFP2, and fluorescence emission (λ510nm) is expected to increase when excited at 405 nm (oxidized sensor) and decrease when excited at 488 nm (reduced sensor) [43]. Incubation with uric acid at 100 and 200 μM increased probe oxidation over time (Fig. 3A), either indicating that it is being processed by the cells to generate oxidizing metabolites [29] or is indirectly activating cells to produce oxidants [44]. Both mechanisms corroborate the pro-oxidant effect of uric acid. By adding H2O2, cells reached a peak of oxidation that was partially recovered over time, whereas DTT displayed no effect, indicating that the probe is majority reduced in baseline conditions. Fig. 3B shows the delta (final – initial) of the roGFP2-Grx1 ratio in 2 h. Uric acid significantly increased the oxidation of the cells. Fig. 3C shows the ratiometric images of HUVEC expressing roGFP2-Grx1 upon DTT, H2O2, and 50–200 μM urate after 2 h treatment. Taken together, it was possible to show that uric acid had a pro-oxidant effect in HUVEC.
Fig. 3.
– Uric acid increases roGFP2-Grx1 oxidation in HUVEC. A) Fluorescence kinetics of HUVEC transfected with roGFP2-Grx1 in the absence (control) or presence of 100 μM H2O2, urate (50–200 μM), or 1 mM DTT. Fluorescence was monitored using the excitation wavelengths of 405 and 485 nm for a 510 nm emission for 2h in a plate reader. Representative image from a single experiment with 5 replicates per group. Data was plotted as roGFP-Grx1 ratio (405/485) ± SEM (light shadow). B) Difference between the initial and final fluorescence 405/485 ratio from 2 h incubation time. Bar graphs show the mean ± SEM from seven independent experiments. Statistical analysis was performed by ordinary one-way ANOVA and Tukey's multiple comparisons test, with p < 0.05 (∗) when compared to control. C) Confocal microscopy imaging of HUVEC roGFP2-Grx1 cells. A confocal microscope was used to acquire images using an excitation wavelength of 405 (oxidized) and 485 (reduced) nm for a 510–540 nm emission wavelength. Fluorescence ratio images from the two channels were constructed from ImageJ. Images have been treated to show a color scheme that reflects the oxidation level of the probe from blue (reduced) to red (oxidized).
Uric acid can be oxidized by heme-peroxidases, such as MPO, LPO, or PXDN producing oxidizing intermediates [[28], [29], [30], [31], [32]]. We previously showed that PXDN is the main peroxidase expressed in HUVEC cells, and its reaction with uric acid produces oxidizing intermediates that further decompose to hydroxyisourate [31]. To evaluate whether the overall imbalance toward oxidation was dependent on urate oxidation by heme-peroxidases, we compared the redox status of HUVECs treated with uric acid (200 μM) in the presence of phloroglucinol (PHG, 50 μM), a known peroxidase inhibitor [45]. No significant probe oxidation was detected in the absence of urate. Pre-treatment with PHG significantly decreases urate-induced oxidation (Fig. 4), indicating that peroxidases play a role in the pro-oxidation effects of uric acid. Other studies have also suggested that uric acid could activate NADPH oxidases (NOX) and increase superoxide production [29,46,47]. Therefore, we tested whether urate-induced roGFP2-Grx1 oxidation was related to NOX activation/superoxide production. Because there are no specific inhibitors for NOX, we used two common compounds to this end, the flavoprotein inhibitor, diphenyleneiodonium chloride (DPI - 1 μM) and apocynin (10 μM). Apocynin slightly decreased roGFP2-Grx1 oxidation while, DPI, could not alter the probe oxidation after the addition of urate (Fig. 4).
Fig. 4.
– roGFP2-Grx1 oxidation in HUVEC after urate treatment. Cells were pre-treated with Apocynin (10 μM), PHG (50 μM), or DPI (1 μM) for 20 min before 200 μM urate treatment. (A) Confocal microscopy imaging of HUVEC-Grx1-roGFP2 cells after 2 h incubation. A confocal microscope was used to acquire images using an excitation wavelength of 405 (oxidized) and 485 (reduced) nm for a 510–540 nm emission wavelength. Fluorescence ratio images from the two channels were constructed from ImageJ, generating a color scale from blue to red, that corresponds to the relative oxidation level of the probe. B) Difference between the initial and final fluorescence 405/485 ratio with 2 h incubation time. Bar graphs show mean ± SEM from seven independent experiments. Statistical analysis was performed by two-way ANOVA and Tukey's multiple comparisons test. Statistical significance is shown as p < 0.001 (∗∗). Cells without uric acid did not have significant differences between oxidation levels.
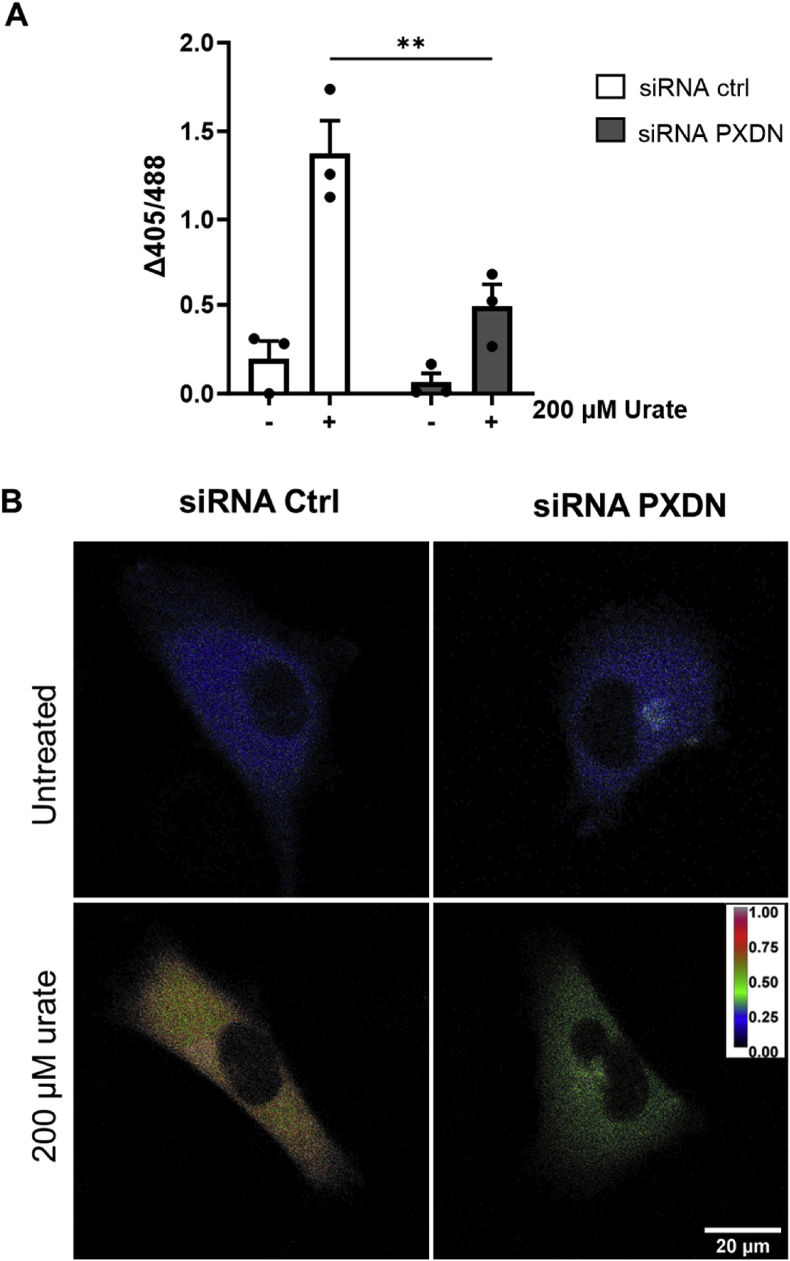
To confirm PXDN dependence on the pro-oxidant effect of urate, PXDN knockdown HUVECs transfected with roGFP2-Grx1 (Suppl information Fig. S5) were challenged with 200 μM urate. There was no significant difference in probe oxidation between wild-type and PXDN knockdown HUVECs in the absence of urate. However, in the presence of urate, the overall oxidation was decreased by 3-fold in PXDN-lacking cells (Fig. 5). As mentioned before, oxidation of urate can produce stable adducts on lysine residues [37], this newly described modification, named uratylation, can alter protein function, even leading to protein misfolding and aggregation. In our proteomic data, it was possible to identify uratylation sites in secretome and lysate samples (Table 5). Extracellular matrix and associated proteins such as Rho-associated protein kinase 2 (ROCK2), Zinc finger CCCH-type containing 15 (ZC3H15), Matrix-remodeling-associated protein 5 (MXRA5) were found to be exclusively modified by the urate treatments.
Fig. 5.
– PXDN silencing prevents roGFP2-Grx1 oxidation in HUVEC. HUVEC cells were harvested and resuspended in HBSS buffer (750 cells/μL) in a final volume of 200 μL. Cells were incubated in a black 96-well plate in the plate reader. The experiment was performed 48 h-post siRNA transfection. A) Difference between the initial and final fluorescence 405/485 ratio with 2 h incubation time. Bar graphs are the mean ± SEM of three independent experiments. Statistical analysis was performed by ordinary two-way ANOVA and Tukey's multiple comparisons test. Statistical significance is shown as p < 0.001 (∗∗) B) Confocal microscopy imaging of HUVEC-Grx1-roGFP2 cells after 2 h incubation. 24 h post-transfection cells were plated in 8 well-coverslips in a density of 10 A confocal microscope was used to acquire images using an excitation wavelength of 405 (oxidized) and 485 (reduced) nm for a 510–540 nm emission wavelength. Fluorescence ratio images from the two channels were constructed from ImageJ, generating a color scale from blue to red, that corresponds to the relative oxidation level of the probe.
Table 5.
– Uratylation sites (∗ = +140 Da adduct in lysine (K) residues) were identified in HUVEC secretome and lysate after 1:30 and 24h exclusively in urate treated samples (50–200 μM). PTM analysis was performed with MaxQuant using raw files from initial proteomic experiments.
| Protein name | Gene name | Sequence | Type of sample/incubation time |
|---|---|---|---|
| Serine/arginine repetitive matrix protein 2 | SRRM2 | TKQPSSPYEDKDKDKK∗EK∗SATRPSPSPERSS | Secretome – 24 h, Lysate – 24 h |
| Matrix-remodeling-associated protein 5 | MXRA5 | MRWFLEWDAKSRGILK∗CK∗K∗DK∗AYEGGQLCAM | Lysate – 24 h |
| Rho-associated protein kinase 2 | ROCK2 | EKRQLQERFTDLEKEK∗SNMEIDMTYQLKVIQ | Secretome – 1:30 and 24 h |
| Zinc finger CCCH domain-containing protein 15 | ZC3H15 | SKKAEQKKKEKIIEDK∗TFGLK∗NK∗K∗GAKQQKF | Lysate – 1:30 and 24 h |
| Tubulin polyglutamylase | TTLL6 | ICRKDLLARNMSRMLK∗MFPKDFRFFPRTWCL | Secretome 1:30, Lysate – 24 h |
| Sorting nexin-5 | SNX5 | AAFRKNLIEMSELEIK∗HARNNVSLLQSCIDL | Lysate – 24 h |
| Myosin-15 | MYH15 | TNQVREGTKNLTEMEK∗VK∗K∗LIEEEKTEVQVT | Lysate – 24 h |
3.3. Uric acid increases misfolding protein levels and disturbs cellular proteostasis
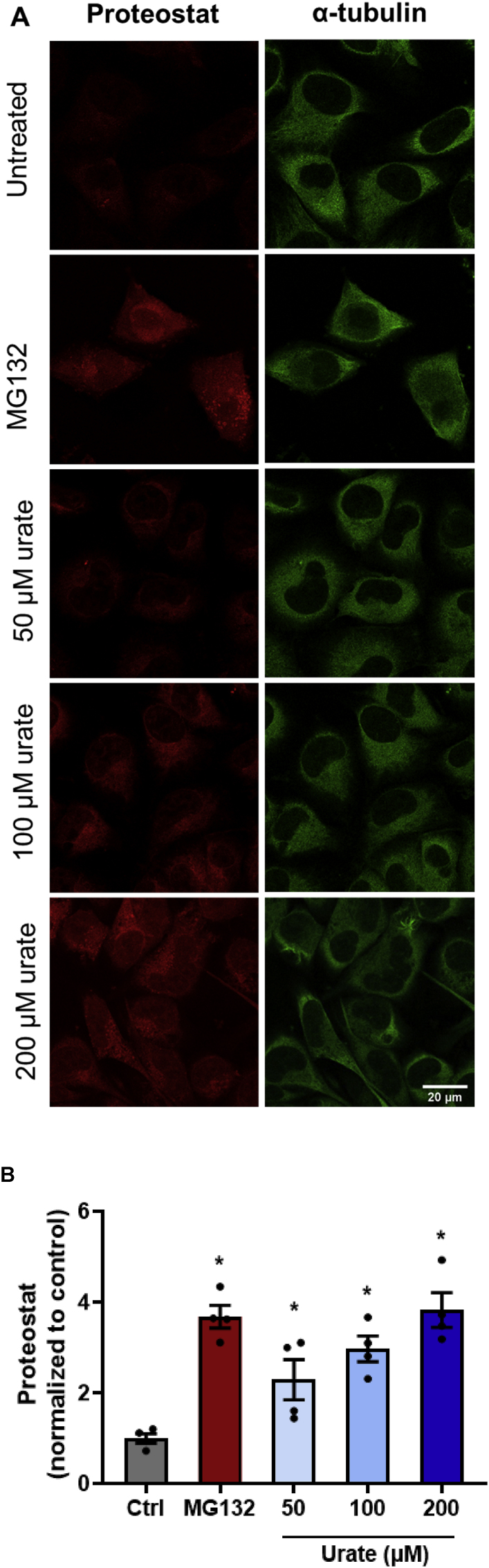
Since the protein-folding biological process, proteins from the endoplasmic reticulum and protein folding chaperone complex were down-regulated in secretome and lysate within 1:30 h urate treatment and this was followed by a possible adaptive response, with an increase in protein levels 24 h after urate treatment (Table 1, Table 2, Table 3, Table 4), we evaluated the degree of protein misfolding and aggregation by the Proteostat kit (Enzo Biosciences). This novel fluorescence dye allows the detection and relative quantification of aggregated protein cargo within aggresomes and other inclusion bodies [48]. MG-132 (1 μM), a proteasome inhibitor was used as a positive control for protein aggregation [49]. After treating cells with MG-132 for 6 h, it was possible to identify aggregated protein cargo accumulating within cytoplasmic structures. As observed by fluorescence microscopy (Fig. 6A), 24 h uric acid treatment also induced the accumulation of aggresome bodies and misfolding proteins. We then quantified the levels of misfolding proteins stained by the Proteostat probe, using α-tubulin as a marker for cell surface (Fig. 6B). Proteostat signal was strongly increased in cells treated with uric acid, indicating that uric acid induces misfolding and aggregation of proteins.
Fig. 6.
– Urate treatment increased misfolding proteins and aggregates in HUVEC. A) Confocal microscopy analysis of Proteostat dye (red) and α-tubulin (green) of HUVEC cells upon incubation with 1 μM MG132 for 6 h (positive control) and urate (50–200 μM) for 24 h. Images were obtained by confocal microscopy using the default Rhodamine set filter and a FITC filter for α-tubulin (Leica Microsystems – LAX Office software). B) Quantification of Proteostat staining normalized by α-tubulin intensity was obtained with ImageJ for 4 independent experiments, using 10 pictures per experiment that contained 4–7 cells per image field. Statistical analysis was performed by the Kruskal-Walli's test followed by Dunn's multiple comparisons test. Statistical significance is shown as p < 0.05 (∗) when compared to control.
We also observed that the aggresome bodies in the urate-treated HUVEC were near p62 foci (Fig. 7A). p62 is an autophagy marker that binds to damaged proteins and can reflect autophagic status and disturb proteostasis [50]. Immunofluorescence staining (Fig. 7B) showed an increase in p62-positive punctuated structures in uric acid-treated cells. We also quantified the number of foci per cell and p62 foci intensity (Fig. 7C), which were both increased in urate-treated cells. These results corroborate the data obtained by the proteomic analysis, in which there was an increase in chaperones, and the GO enrichment analysis showed that pathways related to protein folding and ubiquitination are increased in the treated groups. Considering that, we also performed an analysis of ubiquitinated proteins in the proteomic sample. Ubiquitinated peptides (GlyGly(K) + QQTGG (K)) were exclusively found in urate-treated samples and included the Heat shock protein HSP90-beta and Heat shock protein 105 kDa, Rho-associated protein kinase 2 and Annexin A4 (Table 6).
Fig. 7.
– Urate treatment increased p62 foci expression. A) Confocal microscopy of the co-colocalization of protein aggregates stained with Proteostat probe (red) and p62 (green) MG132 1 μM was used as a positive control for protein aggregation. B) Confocal microscopy imaging of p62 expression and accumulation forming foci upon uric acid treatment for 24h. Nuclei were stained with DAPI (blue) and p62 with Alexa-Fluor-488 conjugated antibody (green). C) Foci signal total area (upper panel) and foci signal count/nucleus (lower panel) of control, MG132, and urate treatment. Quantification was performed by Particle Analysis by ImageJ plugin. Bar graphs show mean ± SEM from images containing 10–20 cells per field from four independent experiments. Statistical analysis was performed by one-way ANOVA followed by Dunnett's multiple comparisons test. Statistical significance is shown as p < 0.05 (∗) when compared to control.
Table 6.
Ubiquitination sites were identified in HUVEC secretome and lysate after 1:30 and 24 h exclusively in urate treated samples (50–200 μM). PTM analysis was performed with MaxQuant using war data obtained by initial proteomic experiments.
| GlyGly (K)∗ | ||
|---|---|---|
| Protein name | Gene name | Sequence |
| Heat shock protein HSP 90-beta | HSP90AB1 | CLELFSELAEDKENYK∗K∗FYEAFSKNLKLGIH |
| Calnexin | CANX | FDRGTLSGWILSKAKK∗DDTDDEIAKYDGKWE |
| Heat shock protein 105 kDa | HSPH1 | TSEENKIPDADK∗ANEK∗KVDQPPEAKKPKIKV |
| Rho-associated protein kinase 2 | ROCK2 | RKEPVKRGNDTDVRRK∗EK∗ENRK∗LHMELK∗SER |
| Eukaryotic translation initiation factor 5B | EIF5B | DASFKIKTVAQKKAEK∗K∗ERERKK∗RDEEKAKL |
| Annexin A4 | ANXA4 | MATK∗GGTVK∗AASGFNAMEDAQTLR |
| Elongation factor 1-alpha 1 | EEF1A1 | FLKSGDAAIVDMVPGK∗PMCVESFSDYPPLGR |
| Titin | TTN | GSKITNYIIEK∗K∗EVGK∗DVWMPVTSASAKTTC |
| QQTGG (K)∗ | ||
| Protein name | Gene name | Sequence |
| Rho-associated protein kinase 2 | ROCK2 | ELEKRQLQERFTDLEK∗EKSNMEIDMTYQLKV |
| Microtubule-associated protein 4 | MAP4 | KPMSLASGLVPAAPPK∗RPAVASARPSILPSK |
| Cytoskeleton-associated protein 2 | CKAP2 | PKETSEERKARLSEWK∗AGKGRVLKRPPNSVV |
| Caprin-1 | CAPRIN1 | KELQRSFMALSQDIQK∗TIKKTARREQLMREE |
3.4. Uric acid increases monocyte adhesion in HUVEC cells
The proteomic experiments also revealed an increase in proteins from the inflammatory response and cytokines signaling in the secretome 24 h after urate treatment. Additionally, the Intercellular Adhesion Molecule-1 (ICAM-1), a glycoprotein responsible for the adhesion of circulating monocytes onto endothelial cells [51,52], was found exclusively in the secretome and lysate of 24 h urate-treated groups (Suppl information Fig. S6). Corroborating this data, the western blot revealed higher protein levels of ICAM-1 and VCAM-1 (Vascular Cell Adhesion Molecule-1) in the presence of urate or the positive controls (Fig. 8B and C). Next, we evaluated the adhesion of THP-1 cells (monocytic leukemic lineage) to HUVEC cells previously treated with uric acid for 24 h. The results (Fig. 8D and E) show a higher adhesion of THP-1 cells (labeled in green, with calcein-AM) upon HUVECs that were treated with uric acid, an effect comparable to LPS and TNF-α positive controls.
Fig. 8.
– Urate increases the expression of ICAM and VCAM and increases monocyte adhesion. A) HUVEC were seeded on 24-well plates at an initial confluence of 5 × 104 cells/well. After 3 days, cells were washed with PBS and incubated with urate (50–200 μM) in RPMI medium without phenol red and fetal bovine serum for 24 h. LPS (1 μg/mL) and TNF-α (5 ng/mL) were used as a positive control. Western blot analysis of ICAM1 and VCAM expression in HUVECs lysate after 24 h urate treatment. B) Bands densitometry was quantified by Image J. Bars are mean ± S.E.M of relative protein level quantification of 4 independent experiments (protein signal/loading control). D) Assessment of THP-1 cell adhesion to HUVEC cells after urate treatment. HUVECs were seeded on 24-well plates at an initial confluence of 5 × 104 cells/well. After 3 days, cells were washed with PBS and incubated with urate (50–200 μM), LPS (1 μg/mL) or TNF-α (5 ng/mL) for 24 h. THP-1 cells previously stained with calcein-AM were added (4 × 105 cells per well) over the monolayer of HUVEC. After 1 h incubation, non-adhered cells were removed, followed by washing with PBS. The images were captured using a fluorescence microscope and the quantification of the number of attached cells (E) was performed using the Image-J software. Bar graphs represent the mean ± S.E.M. of four independent experiments, using five pictures for each experiment. Statistical analysis was performed by ordinary one-way ANOVA and Tukey's multiple comparisons test for control and urate treatments. Statistical significance is shown as p < 0.05 (∗) when compared to control. Statistical analysis was performed by ordinary one-way ANOVA for control and urate treatments. Statistical significance is shown as p < 0.05 (∗) when compared to control.
4. Discussion
Clinical studies have progressively highlighted the association between elevated uric acid levels and an increased risk of cardiovascular diseases (CVD), including hypertension, coronary artery disease, heart failure, and stroke. Hyperuricemia has been indicated as an independent risk factor for CVD, even when traditional risk factors such as hypertension, obesity, and dyslipidemia are accounted for [23,53]. Hyperuricemia can also exacerbate insulin resistance, promote renal dysfunction, and enhance platelet aggregation, further favoring cardiovascular risk [[54], [55], [56]]. Patients with higher UA levels often have worse cardiovascular outcomes, including a higher incidence of heart attacks and strokes [57,58]. Furthermore, uric acid-lowering therapies, such as allopurinol, febuxostat, and probenecid, have been associated with improvement in endothelial function and reduction in cardiovascular events [[59], [60], [61], [62]], suggesting that managing uric acid levels may be beneficial in decreasing cardiovascular risk. However, the precise role of hyperuricemia in CVD pathogenesis remains complex, and ongoing research is needed to fully understand whether lowering uric acid levels can directly reduce cardiovascular risk.
While there is no consensus on the optimal range for serum uric acid levels, targets of <6.0 mg/dL (357 μM) for women and <7.0 mg/dL (416 μM) for men are commonly accepted for the treatment of hyperuricemia [63]. However, some studies have shown that serum uric acid, even within the traditionally accepted normal range, is correlated with an increased risk for hypertension, dyslipidemia, and chronic kidney disease [[64], [65], [66], [67]]. Literature has shown that it is important to consider uric acid levels as part of a broader assessment of cardiovascular risk, rather than relying solely on whether they fall within the traditionally defined "normal" range [26,63]. The present study reinforces this idea by showing that uric acid, even at physiological concentrations, led to alterations in endothelial cells.
A clinical study in collaboration with the ELSA-Brasil project (Longitudinal Study of Adult Health) [24], demonstrated a significant positive association between uric acid and allantoin levels with the thickness of the carotid intima-media layer (c-IMT). The correlation found was independent of other cardiovascular risk factors, such as the LDL/HDL ratio, pulse pressure, and neck circumference, indicating that uric acid oxidation may occur at the beginning of the atherosclerotic process and could play a role in disease progression, with allantoin as an early marker of atherosclerosis development. The authors raised the hypothesis that the oxidation of uric acid by vascular peroxidases, such as PXDN, and the generation of the reactive intermediates urate free radical and urate hydroperoxide may propagate the endothelial injury described for uric acid. Interestingly, uric acid levels were associated with the progression of atherosclerosis but were still under the normal range.
Besides these clinical studies, several investigations have linked disturbances in endothelial cell homeostasis to uric acid. However, the uric acid concentrations typically studied are much higher than normouricemic levels, at which urate crystals can precipitate. Uric acid appears to be taken up by HUVEC cells via the URAT1 [68] and GLUT9 transporter [69], and the expression of URAT1 is not regulated by the presence of uric acid [68]. Under hyperuricemia, uric acid has been reported to cause endothelial dysfunction in HUVECs by generating pro-inflammatory cytokines and producing adhesion molecules by activating the NF-κB transcription factor [[68], [69], [70]], by inducing oxidants production, decreasing the levels of the vasodilator nitric oxide, promoting apoptosis and cellular senescence [10,11,71]. A proteomic study in HAEC endothelial cell line also showed that uric acid, in concentrations greater than 300 μM, alters the abundance of proteins from the ubiquitin-proteasome pathway, hypoxia signaling pathways, NO synthesis, proteins related to oxidative stress and alters the levels of translation initiation pathways, mainly eIF4 [72].
Uric acid, even without forming crystals, can activate inflammatory pathways such as the NLRP3 inflammasome [14], and can alter cell proliferation and induction of cell senescence and apoptosis, via activation of the renin-angiotensin system [73,74], by triggering reactive oxygen and nitrogen species and endoplasmic reticulum stress [71]. We and others have demonstrated that uric acid can have a pro-oxidant effect when reacting with heme-peroxidases, such as MPO, LPO, and PXDN [[28], [29], [30], [31], [32]]. As an alternative substrate for PXDN, physiological concentrations of uric acid decreased the production of hypobromous acid, impaired collagen IV crosslinks, and the basement membrane establishment, altering migration and adhesion of HUVECs [31]. Additionally, the product of uric acid oxidation, urate hydroperoxide, directly oxidized extracellular cell surface thiol proteins in HUVECs, impairing cell adhesion [35]. Taking together, all these effects could induce an adaptive response in these endothelial cells and, therefore, we performed a global proteomic analysis to find out which pathways were being affected by uric acid treatment.
Overall, proteomic analysis of HUVECs lysate and secretome revealed alterations in the abundance of proteins related to redox processes, cell adhesion and migration, protein folding and proteostasis, translation, intracellular transport, and inflammatory response by uric acid. The proteomic data set corroborates the previously reported functional disruptions induced by uric acid in HUVECs [31]. The enrichment of the proteomic data showed a decrease in proteins related to general metabolism, mainly to the glycolytic pathway (lysate 24 h - cluster 2), a known consequence of oxidative stress, resulting in a decrease in ATP levels [75,76]. This substantial decrease in energy levels can be attributed to the oxidative inactivation of redox-regulated metabolic enzymes involved in ATP-generating pathways, mainly GAPDH. Of note, products of uric acid oxidation have been described by inhibiting GAPDH [37]. The inactivation of glycolytic enzymes is also associated with a shift from glycolysis to the pentoses phosphate pathway, increasing NAPDH levels and decreasing ATP synthesis [75]. A drop in ATP levels can disrupt ATP-dependent chaperone pathways, leading to proteolysis, but NAPDH can help restore redox homeostasis through the thioredoxin and glutaredoxin systems [75].
Interestingly, urate induced a decrease in antioxidant proteins within 1:30 h treatment, followed by an increase in thioredoxin family proteins (Peroxiredoxin-PRDX I, IV, VI and thioredoxin reductase 1 - TXNRD1) and Glutaredoxin-3 (GLRX3) after 24 h treatment, consistent with an adaptive response to protect against oxidative stress. In neutrophils, the oxidation of uric acid by MPO led to a simultaneous decrease in the GSH/GSSG ratio, demonstrating that the products of uric acid oxidation favor a pro-oxidant environment [29]. As assessed by the ro-GFP-Grx1 probe, uric acid also displayed a pro-oxidant effect in HUVECs a few minutes after incubation, which was greatly dependent on PXDN activity. The peroxidase-dependent oxidation of uric acid in HUVECs, with the production of hydroyisourate, was previously demonstrated by our group [31]. Therefore, a primary pro-oxidant effect, due to the oxidation of uric acid, could indeed promote a later adaptive antioxidant response. In agreement, the transcription factor Nuclear Factor Erythroid 2–Related Factor 2 (Nrf2), a master regulator of the antioxidant response, is activated by soluble uric acid [77]. In this present study, data dispersion did not allow us to find a significant increase in total glutathione, which would corroborate an antioxidant adaptive response dependent on Nrf2 activation. However, a 1.5-fold increase of GSH was found 24 h after urate treatment (Fig. S4). Additionally, proteomics revealed an up-regulation of the Ras GTPase-activating-like protein (IQGAP1) by uric acid, a protein that mediates Nrf2 protein activation [78]. It is noteworthy to mention that other transcription factors like HIF-1, FOXO, and p53 could be indirectly activated by the oxidative status induced by uric acid and, consequently, contribute to the antioxidant adaptive response [[79], [80], [81], [82], [83], [84]].
Products from uric acid oxidation can react with amine groups to form adducts in proteins, a post-translational modification called uratylation [37]. In agreement with the hypothesis that uric acid is being oxidized in HUVECs and that it could alter the cellular response, we found proteins that were exclusively uratylated in the presence of uric acid. The Matrix-remodeling-associated protein 5 (MXRA5), an important enzyme involved in cell adhesion and matrix remodeling [85,86], also known by its role in tumorigenesis [87]; the Rho-associated protein kinase 2 (ROCK2), a protein involved in actin cytoskeleton organization, cell adhesion, and motility, remodeling of the extracellular matrix and smooth muscle cell contraction, highly involved in CVD [88]; the Serine/arginine repetitive matrix 2 (SRRM2), a nuclear speckle protein involved in RNA splicing that was recently identified on the surface of malignant cells [89], and the Zinc finger CCCH-type containing 15 (ZC3H15), a protein involved in cell proliferation, epithelial-mesenchymal transition, invasion, and metastasis [90] were uratylated. These results reinforce that some of the effects caused by uric acid are indeed dependent on its oxidation and strongly corroborate our previous findings on alterations in adhesion/migration of HUVECs by peroxidase-dependent oxidation of uric acid [31,35]. The identified post-translational modification in these proteins also opens the question of whether it could play a role in tumorigenesis.
The pro-oxidant effects of soluble uric acid have also been attributed to the activation of NADPH oxidase (NOX), with an increase in superoxide and hydrogen peroxide [29,46,47]. Here, we have tested the possible contribution of NOXs by using the unspecific inhibitors, apocynin and DPI [91]. We found conflicting results since apocynin, one of the most used NOX inhibitors [92], significantly protected from uric acid-induced roGFP2-Grx1 oxidation, while DPI, a broader inhibitor of electron transference in flavin-containing proteins, did not. Perhaps, apocynin protection was rather due to a global antioxidant effect as previously observed in endothelial and vascular smooth muscle cells [93]. Therefore, these results do not allow us to ensure whether NOX is being activated by uric acid in the conditions of our study and if this activation is contributing to roGFP2-Grx1 oxidation. Previous studies have also shown that hyperuricemia can induce oxidative stress by inducing mitochondria dysfunction [94,95]. However, we did not test mitochondrial quality in our assays.
Endothelial dysfunction by uric acid has already been associated with an induction of a pro-oxidant environment and endoplasmic reticulum stress in HUVECs upon uric acid concentration as in hyperuricemia [71]. High levels of uric acid also induced phosphorylation of ERK/Akt and heat shock factor 1 (HSF1), activates several chaperones in the cells [96]. Additionally, by activating inflammasome [[12], [13], [14]], uric acid can also indirectly affect the clearance of misfolded proteins [97]. High levels of uric acid induced endoplasmic reticulum stress and activation of the unfolded protein response (UPR) in different types of cells [21,71,98]. During UPR activation, three main signaling pathways are initiated: the inositol-requiring enzyme 1 (IRE1), protein kinase RNA-like ER kinase (PERK), and activating transcription factor 6 (ATF6) pathways [99]. These pathways work together to restore endoplasmic reticulum homeostasis by reducing protein synthesis, enhancing protein folding capacity, and promoting the degradation of misfolded proteins [74]. Interestingly, these studies focus on the effects of uric acid on hyperuricemic concentrations. Even using lower concentrations of urate, our proteomic data strongly corroborates these previous findings as a protein-folding biological process, proteins from the endoplasmic reticulum and protein folding chaperone complex were down-regulated in both secretome and lysate within 1:30 h urate treatment, followed by a possible adaptive response, with an increase of levels of this group of proteins 24 h after urate treatment (Table 1, Table 2, Table 3, Table 4). In agreement, uric acid induced the accumulation of aggresome bodies and misfolded proteins to the same extent as the proteasome inhibitor MG132. In addition, the ubiquitination of important heat shock proteins was found exclusively in urate-treated cells. Uric acid increases p62 puncta, which is usually due to an increase in protein aggregation. The p62 protein, also known as sequestosome 1 (SQSTM1), targets ubiquitinated protein aggregates, damaged organelles, and other cellular debris for degradation via autophagy [50]. p62 regulates the degradation of Keap1, a negative regulator of Nrf2, promoting Nrf2 activation and enhancing cellular antioxidant responses [100].
Uric acid, even at the normal range concentrations used in this study, increased proteins of inflammatory response and cytokine signaling in the secretome. It also increased the abundance of proteins related to endothelial dysfunction, including the S100 protein family, which has been described by activating the pro-inflammatory transcription factor kappa B (NF-κB) and by inducing a pro-inflammatory phenotype in a variety of cell types [80]. Urate also induced an up-regulation of Annexin A1. This protein promotes monocyte recruitment and is involved in the modulation and resolution of inflammation through multiple mechanisms [101]. Annexin-A1 is also an important regulator of VEGF-mediated endothelial cell migration and angiogenesis [102]. Likewise, ICAM-1 was exclusively present in urate-treated cells, at the three tested concentrations. Complementing these proteomic findings, western blot analysis confirmed HUVECs activation with an enhanced abundance of ICAM-1 and VCAM by urate. Consequently, there was a higher adhesion of monocytes to these endothelial cells. Activation of endothelial cells with an increase in monocyte adhesion is a crucial step in the development of atherosclerosis [103,104]. Our results corroborate with some effects of uric acid in hyperuricemic conditions already known in the literature but they contribute to bring light to the pro-inflammatory effects of uric acid in normouricemic conditions, situation that has been neglected in clinic.
Several studies have performed proteomic analysis in different cell types, particularly in the context of hyperuricemia. Sutthimethakorn et al. (2020) demonstrated that a high-dose uric acid alters the renal tubular cell proteome, altering the expression of proteins related to cell adhesion, response to unfolding proteins, and ATP metabolism. Similarly, Oberbach et al. (2014) analyzed the proteomic response of human aortic endothelial cells (HAEC) to different uric acid concentrations and identified that hyperuricemia alters multiple cellular pathways, including the ubiquitin-proteasome system, superoxide signaling, nitric oxide signaling, and hypoxia-inducible factor 1-alpha (HIF-1α) pathways. Proteomic and metabolomic studies have also identified molecular markers and pathways associated with hyperuricemia and gout. Wu & You (2023) [105] reviewed multiple studies indicating that hyperuricemia is linked to systemic inflammation, complement activation, and metabolic disturbances. Their work suggests that prolonged exposure to high uric acid concentrations contributes to chronic kidney disease and cardiovascular dysfunction, supporting the concept that uric acid plays a more complex role in disease pathophysiology than previously recognized.
While these studies provide valuable insights into the impact of uric acid on cellular function, they primarily focus on uric acid levels that are already known to be harmful. In contrast, our study demonstrates that uric acid, even at concentrations within the normouricemic range, can significantly alter endothelial cell function. Proteomics analysis from secretome and lysate of urate-treated HUVECs showed changes in protein abundance related to adhesion, migration, redox processes, proteostasis and inflammation. The results indicate that the presence of urate in HUVECs alters migration and adhesion without inducing cell death. Additionally, we found a real-time increase in the oxidation status of HUVECs induced by uric acid. We also propose the involvement of PXDN and urate-derived oxidants, as the oxidation was decreased by PXDN inhibition or silencing. Uratylation, a post-translational modification caused by the addition of urate-oxidized products upon lysine residues, also confirmed the association of uric acid oxidation with its harmful effects upon the endothelium. We also observed disturbances in proteostasis by detecting increases in misfolding proteins and p62 puncta, and HUVEC activation, by monitoring monocyte adhesion. Overall, this research provides valuable insights into the molecular pathways through which normouricemic concentrations of uric acid exert damaging effects on endothelial cells and may raise the discussion whether the considered normal range of uric acid should be reviewed.
CRediT authorship contribution statement
Bianca Dempsey: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Beatriz Pereira da Silva: Methodology. Litiele Cezar Cruz: Methodology. Danielle Vileigas: Methodology. Amanda R.M. Silva: Methodology. Railmara Pereira da Silva: Methodology, Formal analysis. Flavia Carla Meotti: Writing – review & editing, Supervision, Resources, Project administration, Investigation, Funding acquisition, Conceptualization.
Funding
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP): CEPID Redoxoma 2013/07937-8; Young Investigator—2, grant number 2018/14898-2. B.D., B.P.S., D.V., A.P.S., and R.P.S. received a scholarship from FAPESP. Proteomic analysis was performed at the Redox Proteomics Core of the Mass Spectrometry Resource at Chemistry Institute, University of Sao Paulo (FAPESP grant numbers 2012/12663-1, CEPID Redoxoma 2013/07937-8; 2023/00995-4) led by Prof. Graziella Eliza Ronsein and Prof. Paolo Di Mascio.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We thank MSc. Izaura N. Toma (in memoriam), Dr. Mariana P. Massafera and Prof. Dr. Graziella Eliza Ronsein for their technical assistance in the proteomics analysis. We thank Professor Luis Netto (University of Sao Paulo) for kindly providing plasmids and transformed bacteria, Professor Francisco Laurindo (Incor) for providing the HUVEC cell line and Prof. Mikloz Geiszt (Semmelweis University) by kindly providing the PXDN antibody. We acknowledge BioRender.com for the creation of the Graphical Abstract Image.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2025.103625.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Hediger M.A., Johnson R.J., Miyazaki H., Endou H. Molecular physiology of urate transport. Physiology. 2005;20:125–133. doi: 10.1152/physiol.00039.2004. [DOI] [PubMed] [Google Scholar]
- 2.Mandal A.K., Mount D.B. The molecular physiology of uric acid homeostasis. Annu. Rev. Physiol. 2015;77:323–345. doi: 10.1146/annurev-physiol-021113-170343. [DOI] [PubMed] [Google Scholar]
- 3.Barsoum R., El-Khatib M. Uric acid and life on earth. J. Adv. Res. 2017;8:471–474. doi: 10.1016/j.jare.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker B.F. Towards the physiological function of uric acid. Free Radic. Biol. Med. 1993;14:615–631. doi: 10.1016/0891-5849(93)90143-i. [DOI] [PubMed] [Google Scholar]
- 5.Kaur H., Halliwell B. Action of biologically-relevant oxidizing species upon uric acid. Identification of uric acid oxidation products. Chem. Biol. Interact. 1990;73:235–247. doi: 10.1016/0009-2797(90)90006-9. [DOI] [PubMed] [Google Scholar]
- 6.Simic M.G., Jovanovic S.V. Antioxidation mechanisms of uric acid. J. Am. Chem. Soc. 1989;111:5778–5782. doi: 10.1021/ja00197a042. [DOI] [Google Scholar]
- 7.Sautin Y.Y., Johnson R.J. Uric acid: the oxidant-antioxidant paradox. Nucleosides Nucleotides Nucleic Acids. 2008;27:608–619. doi: 10.1080/15257770802138558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filipe P., Haigle J., Freitas J., Fernandes A., Mazière J.C., Mazière C., et al. Anti- and pro-oxidant effects of urate in copper-induced low-density lipoprotein oxidation. Eur. J. Biochem. 2002;269:5474–5483. doi: 10.1046/j.1432-1033.2002.03245.x. [DOI] [PubMed] [Google Scholar]
- 9.Bagnati M., Perugini C., Cau C., Bordone R., Albano E., Bellomo G. When and why a water-soluble antioxidant becomes pro-oxidant during copper-induced low-density lipoprotein oxidation: a study using uric acid. Biochem. J. 1999;340:143–152. doi: 10.1042/0264-6021:3400143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J.H., Jin Y.M., Hwang S., Cho D.H., Kang D.H., Jo I. Uric acid attenuates nitric oxide production by decreasing the interaction between endothelial nitric oxide synthase and calmodulin in human umbilical vein endothelial cells: a mechanism for uric acid-induced cardiovascular disease development. Nitric Oxide. 2013;32:36–42. doi: 10.1016/j.niox.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Papežíková I., Pekarová M., Kolářová H., Klinke A., Lau D., Baldus S., et al. Uric acid modulates vascular endothelial function through the down regulation of nitric oxide production. Free Radic. Res. 2013;47:82–88. doi: 10.3109/10715762.2012.747677. [DOI] [PubMed] [Google Scholar]
- 12.Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 13.Martinon F. Mechanisms of uric acid crystal-mediated autoinflammation. Immunol. Rev. 2010;233:218–232. doi: 10.1111/J.0105-2896.2009.00860.X. [DOI] [PubMed] [Google Scholar]
- 14.Braga T.T., Forni M.F., Correa-Costa M., Ramos R.N., Barbuto J.A., Branco P., et al. Soluble uric acid activates the NLRP3 inflammasome. Sci. Rep. 2017;7 doi: 10.1038/srep39884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghaemi-Oskouie F., Shi Y. The role of uric acid as an endogenous danger signal in immunity and inflammation. Curr. Rheumatol. Rep. 2011;13:160–166. doi: 10.1007/s11926-011-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kono H., Chen C.-J., Ontiveros F., Rock K.L. Uric acid promotes an acute inflammatory response to sterile cell death in mice. J. Clin. Investig. 2010;120:1939–1949. doi: 10.1172/JCI40124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timofte D., Mandita A., Balcangiu-Stroescu A.E., Balan D., Raducu L., Tanasescu M.D., et al. Hyperuricemia and cardiovascular diseases. Rev. Chem. 2019;70:1045–1046. doi: 10.2174/1381612825666190408122557. [DOI] [Google Scholar]
- 18.Ndrepepa G. Uric acid and cardiovascular disease. Clin. Chim. Acta. 2018;484:150–163. doi: 10.1016/j.cca.2018.05.046. [DOI] [PubMed] [Google Scholar]
- 19.Iliesiu A., Campeanu A., Dusceac D. Serum uric acid and cardiovascular disease. Maedica (Bucur) 2010;5:186–192. [PMC free article] [PubMed] [Google Scholar]
- 20.Hare J.M., Johnson R.J. Uric acid predicts clinical outcomes in heart failure: insights regarding the role of xanthine oxidase and uric acid in disease pathophysiology. Circulation. 2003;107:1951–1953. doi: 10.1161/01.CIR.0000066420.36123.35. [DOI] [PubMed] [Google Scholar]
- 21.Choi Y.-J., Shin H.-S., Choi H.S., Park J.-W., Jo I., Oh E.-S., et al. vol. 94. Laboratory Investigation; 2014. pp. 1114–1125. (Uric Acid Induces Fat Accumulation via Generation of Endoplasmic Reticulum Stress and SREBP-1c Activation in Hepatocytes). [DOI] [PubMed] [Google Scholar]
- 22.Gaubert M., Marlinge M., Alessandrini M., Laine M., Bonello L., Fromonot J., et al. Uric acid levels are associated with endothelial dysfunction and severity of coronary atherosclerosis during a first episode of acute coronary syndrome. Purinergic Signal. 2018;14:191–199. doi: 10.1007/s11302-018-9604-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feig D.I., Kang D.-H., Johnson R.J. Uric acid and cardiovascular risk. N. Engl. J. Med. 2008;359:1811–1821. doi: 10.1056/NEJMra0800885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santana M.S., Nascimento K.P., Lotufo P.A., Benseñor I.M., Meotti F.C. Allantoin as an independent marker associated with carotid intima-media thickness in subclinical atherosclerosis. Braz. J. Med. Biol. Res. 2018;51 doi: 10.1590/1414-431x20187543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mello F.M., Bensenor I.M., Santos I.S., Bittencourt M.S., Lotufo P.A., Fuller R. Serum uric acid levels and subclinical atherosclerosis: results from the Brazilian longitudinal study of Adult health (ELSA-Brasil) Curr. Probl. Cardiol. 2023;48 doi: 10.1016/j.cpcardiol.2022.101525. [DOI] [PubMed] [Google Scholar]
- 26.Desideri G., Castaldo G., Lombardi A., Mussap M., Testa A., Pontremoli R., et al. Is it time to revise the normal range of serum uric acid levels? Eur. Rev. Med. Pharmacol. Sci. 2014;18:1295–1306. [PubMed] [Google Scholar]
- 27.Padiglia A., Medda R., Longu S., Pedersen J.Z., Floris G. Uric acid is a main electron donor to peroxidases in human blood plasma. Med. Sci. Monit. 2002;8:BR454–B459. [PubMed] [Google Scholar]
- 28.Meotti F.C., Jameson G.N.L., Turner R., Harwood D.T., Stockwell S., Rees M.D., et al. Urate as a physiological substrate for myeloperoxidase. J. Biol. Chem. 2011;286:12901–12911. doi: 10.1074/jbc.M110.172460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva R.P., Carvalho L.A.C., Patricio E.S., Bonifacio J.P.P., Chaves-Filho A.B., Miyamoto S., et al. Identification of urate hydroperoxide in neutrophils: a novel pro-oxidant generated in inflammatory conditions. Free Radic. Biol. Med. 2018;126:177–186. doi: 10.1016/j.freeradbiomed.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Seidel A., Parker H., Turner R., Dickerhof N., Khalilova I.S., Wilbanks S.M., et al. Uric acid and thiocyanate as competing substrates of lactoperoxidase. J. Biol. Chem. 2014;289:21937–21949. doi: 10.1074/jbc.M113.544957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dempsey B., Cruz L.C., Mineiro M.F., da Silva R.P., Meotti F.C. Uric acid reacts with peroxidasin, decreases collagen IV crosslink, impairs human endothelial cell migration and adhesion. Antioxidants (Basel) 2022;11 doi: 10.3390/ANTIOX11061117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sevcnikar B., Paumann-Page M., Hofbauer S., Pfanzagl V., Furtmüller P.G., Obinger C. Reaction of human peroxidasin 1 compound I and compound II with one-electron donors. Arch. Biochem. Biophys. 2020;681 doi: 10.1016/j.abb.2020.108267. [DOI] [PubMed] [Google Scholar]
- 33.Bathish B., Turner R., Paumann-Page M., Kettle A.J., Winterbourn C.C. Characterisation of peroxidasin activity in isolated extracellular matrix and direct detection of hypobromous acid formation. Arch. Biochem. Biophys. 2018;646:120–127. doi: 10.1016/j.abb.2018.03.038. [DOI] [PubMed] [Google Scholar]
- 34.Patrício E.S., Prado F.M., da Silva R.P., Carvalho L.A.C., Prates M.V.C., Dadamos T., et al. Chemical characterization of urate hydroperoxide, A pro-oxidant intermediate generated by urate oxidation in inflammatory and photoinduced processes. Chem. Res. Toxicol. 2015;28:1556–1566. doi: 10.1021/acs.chemrestox.5b00132. [DOI] [PubMed] [Google Scholar]
- 35.Mineiro M.F., Patricio E. de S., Peixoto Á.S., Araujo T.L.S., da Silva R.P., Moretti A.I.S., et al. Urate hydroperoxide oxidizes endothelial cell surface protein disulfide isomerase-A1 and impairs adherence. Biochim. Biophys. Acta Gen. Subj. 2020;1864 doi: 10.1016/j.bbagen.2019.129481. [DOI] [PubMed] [Google Scholar]
- 36.Carvalho L.A.C.C., Truzzi D.R., Fallani T.S., Alves S.V., Toledo J.C., Augusto O., et al. Urate hydroperoxide oxidizes human peroxiredoxin 1 and peroxiredoxin 2. J. Biol. Chem. 2017;292:8705–8715. doi: 10.1074/jbc.M116.767657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner R., Brennan S.O., Ashby L.V., Dickerhof N., Hamzah M.R., Pearson J.F., et al. Conjugation of urate-derived electrophiles to proteins during normal metabolism and inflammation. J. Biol. Chem. 2018;293:19886–19898. doi: 10.1074/jbc.RA118.005237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva RP da. Universidade de São Paulo; 2020. Investigation of Oxidative Metabolism of Uric Acid and its Role in Redox Processes in Inflammation. Doctoral Thesis. [DOI] [Google Scholar]
- 39.Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/NPROT.2007.261. [DOI] [PubMed] [Google Scholar]
- 40.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 41.O'Connell J.D., Paulo J.A., O'Brien J.J., Gygi S.P. Proteome-Wide evaluation of two common protein quantification methods. J. Proteome Res. 2018;17:1934–1942. doi: 10.1021/acs.jproteome.8b00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 43.Gutscher M., Pauleau A.L., Marty L., Brach T., Wabnitz G.H., Samstag Y., et al. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods. 2008;5(6):553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 44.Baldwin W., McRae S., Marek G., Wymer D., Pannu V., Baylis C., et al. Hyperuricemia as a mediator of the proinflammatory endocrine imbalance in the adipose tissue in a murine model of the metabolic syndrome. Diabetes. 2011;60:1258–1269. doi: 10.2337/db10-0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paumann-Page M., Obinger C., Winterbourn C.C., Furtmüller P.G. Peroxidasin inhibition by phloroglucinol and other peroxidase inhibitors. Antioxidants. 2024;13 doi: 10.3390/antiox13010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sautin Y.Y., Nakagawa T., Zharikov S., Johnson R.J. Adverse effects of the classic antioxidant uric acid in adipocytes: NADPH oxidase-mediated oxidative/nitrosative stress. Am. J. Physiol.: Cell Physiol. 2007;293:584–596. doi: 10.1152/AJPCELL.00600.2006/SUPPL_FILE/FIGURES. [DOI] [PubMed] [Google Scholar]
- 47.Verzola D., Ratto E., Villaggio B., Parodi E.L., Pontremoli R., Garibotto G., et al. Uric acid promotes apoptosis in human proximal tubule cells by oxidative stress and the activation of NADPH oxidase NOX 4. PLoS One. 2014;9 doi: 10.1371/JOURNAL.PONE.0115210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen D., Coleman J., Chan E., Nicholson T.P., Dai L., Sheppard P.W., et al. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem. Biophys. 2011;60:173–185. doi: 10.1007/S12013-010-9138-4/FIGURES/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakawa Y., Sonoda E., Barber L.J., Zeng W., Yokomori K., Kimura H., et al. Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res. 2007;67:8536–8543. doi: 10.1158/0008-5472.CAN-07-1166. [DOI] [PubMed] [Google Scholar]
- 50.Liu W.J., Ye L., Huang W.F., Guo L.J., Xu Z.G., Wu H.L., et al. p62 links the autophagy pathway and the ubiqutin–proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 2016;21:29. doi: 10.1186/s11658-016-0031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchini T., Mitre L.S., Wolf D. Inflammatory cell recruitment in cardiovascular disease. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.635527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huo Y., Ley K. Adhesion molecules and atherogenesis. Acta Physiol. Scand. 2001;173:35–43. doi: 10.1046/j.1365-201X.2001.00882.x. [DOI] [PubMed] [Google Scholar]
- 53.Gagliardi A.C.M., Miname M.H., Santos R.D. Uric acid: a marker of increased cardiovascular risk. Atherosclerosis. 2009;202:11–17. doi: 10.1016/j.atherosclerosis.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 54.Abbott R.D., Brand F.N., Kannel W.B., Castelli W.P. Gout and coronary heart disease: the framingham study. J. Clin. Epidemiol. 1988;41:237–242. doi: 10.1016/0895-4356(88)90127-8. [DOI] [PubMed] [Google Scholar]
- 55.Johnson R.J., Bakris G.L., Borghi C., Chonchol M.B., Feldman D., Lanaspa M.A., et al. Hyperuricemia, acute and chronic kidney disease, hypertension, and cardiovascular disease: report of a scientific workshop organized by the national kidney foundation. Am. J. Kidney Dis. 2018;71:851–865. doi: 10.1053/J.AJKD.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiner D.E., Tighiouart H., Elsayed E.F., Griffith J.L., Salem D.N., Levey A.S. Uric acid and incident kidney disease in the community. J. Am. Soc. Nephrol. 2008;19:1204–1211. doi: 10.1681/ASN.2007101075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dyrbuś M., Desperak P., Pawełek M., Możdżeń M., Gąsior M., Hawranek M. Serum uric acid is an independent risk factor of worse mid- and long-term outcomes in patients with non-ST-segment elevation acute coronary syndromes. Cardiol. J. 2021 doi: 10.5603/CJ.a2021.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borghi C., Agabiti-Rosei E., Johnson R.J., Kielstein J.T., Lurbe E., Mancia G., et al. Hyperuricaemia and gout in cardiovascular, metabolic and kidney disease. Eur. J. Intern. Med. 2020;80:1–11. doi: 10.1016/j.ejim.2020.07.006. [DOI] [PubMed] [Google Scholar]
- 59.Kim S.C., Neogi T., Kang E.H., Liu J., Desai R.J., Zhang M., et al. Cardiovascular risks of probenecid versus allopurinol in older patients with gout. J. Am. Coll. Cardiol. 2018;71:994–1004. doi: 10.1016/j.jacc.2017.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taheraghdam A.A., Sharifipour E., Pashapour A., Namdar S., Hatami A., Houshmandzad S., et al. Allopurinol as a preventive contrivance after acute ischemic stroke in patients with a high level of serum uric acid: a randomized, controlled trial. Med. Princ. Pract. 2014;23:134–139. doi: 10.1159/000355621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larsen K.S., Pottegård A., Lindegaard H.M., Hallas J. Effect of allopurinol on cardiovascular outcomes in hyperuricemic patients: a cohort study. Am. J. Med. 2016;129:299–306.e2. doi: 10.1016/j.amjmed.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 62.Singh J.A., Yu S. Allopurinol and the risk of stroke in older adults receiving medicare. BMC Neurol. 2016;16:164. doi: 10.1186/s12883-016-0692-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Copur S., Demiray A., Kanbay M. Uric acid in metabolic syndrome: does uric acid have a definitive role? Eur. J. Intern. Med. 2022;103:4–12. doi: 10.1016/j.ejim.2022.04.022. [DOI] [PubMed] [Google Scholar]
- 64.Borghi C., Rosei E.A., Bardin T., Dawson J., Dominiczak A., Kielstein J.T., et al. Serum uric acid and the risk of cardiovascular and renal disease. J. Hypertens. 2015;33:1729–1741. doi: 10.1097/HJH.0000000000000701. [DOI] [PubMed] [Google Scholar]
- 65.Kanbay M., Jensen T., Solak Y., Le M., Roncal-Jimenez C., Rivard C., et al. Uric acid in metabolic syndrome: from an innocent bystander to a central player. Eur. J. Intern. Med. 2016;29:3–8. doi: 10.1016/j.ejim.2015.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mazzali M., Hughes J., Kim Y.-G., Jefferson J.A., Kang D.-H., Gordon K.L., et al. Elevated uric acid increases blood pressure in the rat by a novel crystal-independent mechanism. Hypertension. 2001;38:1101–1106. doi: 10.1161/hy1101.092839. [DOI] [PubMed] [Google Scholar]
- 67.Johnson R.J., Lanaspa M.A., Gaucher E.A. Uric acid: a danger signal from the RNA world that may have a role in the epidemic of obesity, metabolic syndrome, and cardiorenal disease: evolutionary considerations. Semin. Nephrol. 2011;31:394–399. doi: 10.1016/j.semnephrol.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang W.Y., Zhu X.Y., Zhang J.W., Feng X.R., Wang Y.C., Liu M.L. Uric acid promotes chemokine and adhesion molecule production in vascular endothelium via nuclear factor-kappa B signaling. Nutr. Metabol. Cardiovasc. Dis. 2015;25:187–194. doi: 10.1016/j.numecd.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 69.Liu S., Yuan Y., Zhou Y., Zhao M., Chen Y., Cheng J., et al. Phloretin attenuates hyperuricemia-induced endothelial dysfunction through co-inhibiting inflammation and GLUT9-mediated uric acid uptake. J. Cell Mol. Med. 2017;21:2553–2562. doi: 10.1111/jcmm.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai W., Duan X.M., Liu Y., Yu J., Tang Y.L., Liu Z.L., et al. Uric acid induces endothelial dysfunction by activating the HMGB1/RAGE signaling pathway. BioMed Res. Int. 2017;2017:1–11. doi: 10.1155/2017/4391920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li P., Zhang L., Zhang M., Zhou C., Lin N. Uric acid enhances PKC-dependent eNOS phosphorylationand mediates cellular ER stress: a mechanism for uric acid-induced endothelial dysfunction. Int. J. Mol. Med. 2016;37:989–997. doi: 10.3892/ijmm.2016.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oberbach A., Neuhaus J., Jehmlich N., Schlichting N., Heinrich M., Kullnick Y., et al. A global proteome approach in uric acid stimulated human aortic endothelial cells revealed regulation of multiple major cellular pathways. Int. J. Cardiol. 2014;176:746–752. doi: 10.1016/j.ijcard.2014.07.102. [DOI] [PubMed] [Google Scholar]
- 73.Corry D.B., Eslami P., Yamamoto K., Nyby M.D., Makino H., Tuck M.L. Uric acid stimulates vascular smooth muscle cell proliferation and oxidative stress via the vascular renin-angiotensin system. J. Hypertens. 2008;26:269–275. doi: 10.1097/HJH.0b013e3282f240bf. [DOI] [PubMed] [Google Scholar]
- 74.Yu M.-A., Sánchez-Lozada L.G., Johnson R.J., Kang D.-H. Oxidative stress with an activation of the renin-angiotensin system in human vascular endothelial cells as a novel mechanism of uric acid-induced endothelial dysfunction. J. Hypertens. 2010;28:1234–1242. [PubMed] [Google Scholar]
- 75.Jakob U., Reichmann D. vol. 9789400757. Springer; Netherlands: 2013. (Oxidative Stress and Redox Regulation). [DOI] [Google Scholar]
- 76.Reichmann D., Voth W., Jakob U. Maintaining a healthy proteome during oxidative stress. Mol. Cell. 2018;69:203–213. doi: 10.1016/j.molcel.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin Y., Xie Y., Hao Z., Bi H., Liu Y., Yang X., et al. Protective effect of uric acid on ox-LDL-induced HUVECs injury via Keap1-Nrf2-ARE pathway. J. Immunol. Res. 2021;2021:1–19. doi: 10.1155/2021/5151168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheung K.L., Lee J.H., Shu L., Kim J.-H., Sacks D.B., Kong A.-N.T. The Ras GTPase-activating-like protein IQGAP1 mediates Nrf2 protein activation via the mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)-ERK pathway. J. Biol. Chem. 2013;288:22378–22386. doi: 10.1074/jbc.M112.444182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sablina A.A., Budanov A.V., Ilyinskaya G.V., Agapova L.S., Kravchenko J.E., Chumakov P.M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Budanov A.V. 2014. The Role of Tumor Suppressor P53 in the Antioxidant Defense and Metabolism; pp. 337–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bernardo V.S., Torres F.F., da Silva D.G.H. FoxO3 and oxidative stress: a multifaceted role in cellular adaptation. J. Mol. Med. 2023;101:83–99. doi: 10.1007/s00109-022-02281-5. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y., Zhou Y., Graves D.T. FOXO transcription factors: their clinical significance and regulation. BioMed Res. Int. 2014;2014:1–13. doi: 10.1155/2014/925350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sastre J., Pérez S., Sabater L., Rius-Pérez S. Redox signaling in the pancreas in health and disease. Physiol. Rev. 2025;105:593–650. doi: 10.1152/physrev.00044.2023. [DOI] [PubMed] [Google Scholar]
- 84.Xi H., Gao Y.-H., Han D.-Y., Li Q.-Y., Feng L.-J., Zhang W., et al. Hypoxia inducible factor‐1α suppresses Peroxiredoxin 3 expression to promote proliferation of CCRCC cells. FEBS Lett. 2014;588:3390–3394. doi: 10.1016/j.febslet.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 85.Ding L., Li S., Zhang Y., Gai J., Kou J. MXRA5 is decreased in preeclampsia and affects trophoblast cell invasion through the MAPK pathway. Mol. Cell. Endocrinol. 2018;461:248–255. doi: 10.1016/j.mce.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 86.Xiao H., Jiang Y., He W., Xu D., Chen P., Liu D., et al. Identification and functional activity of matrix-remodeling associated 5 (MXRA5) in benign hyperplastic prostate. Aging. 2020;12:8605–8621. doi: 10.18632/aging.103175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiong D., Li G., Li K., Xu Q., Pan Z., Ding F., et al. Exome sequencing identifies MXRA5 as a novel cancer gene frequently mutated in non–small cell lung carcinoma from Chinese patients. Carcinogenesis. 2012;33:1797–1805. doi: 10.1093/carcin/bgs210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hartmann S., Ridley A.J., Lutz S. The function of Rho-associated kinases ROCK1 and ROCK2 in the pathogenesis of cardiovascular disease. Front. Pharmacol. 2015;6 doi: 10.3389/fphar.2015.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kellner M., Hörmann J., Fackler S., Hu Y., Zhou T., Lu L., et al. The nuclear speckles protein SRRM2 is exposed on the surface of cancer cells. Cells. 2024;13:1563. doi: 10.3390/cells13181563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hou J., Huang P., Lan C., Geng S., Xu M., Liu Y., et al. ZC3H15 promotes gastric cancer progression by targeting the FBXW7/c-Myc pathway. Cell Death Dis. 2022;8:32. doi: 10.1038/s41420-022-00815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reis J., Massari M., Marchese S., Ceccon M., Aalbers F.S., Corana F., et al. A closer look into NADPH oxidase inhibitors: validation and insight into their mechanism of action. Redox Biol. 2020;32 doi: 10.1016/j.redox.2020.101466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Petrônio M.S., Zeraik M.L., Da Fonseca L.M., Ximenes V.F. Apocynin: chemical and biophysical properties of a NADPH oxidase inhibitor. Molecules. 2013;18:2821. doi: 10.3390/MOLECULES18032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heumüller S., Wind S., Barbosa-Sicard E., Schmidt H.H.H.W., Busse R., Schröder K., et al. Apocynin is not an inhibitor of vascular NADPH oxidases but an antioxidant. Hypertension. 2008;51:211–217. doi: 10.1161/HYPERTENSIONAHA.107.100214. [DOI] [PubMed] [Google Scholar]
- 94.Lanaspa M.A., Sanchez-Lozada L.G., Choi Y.-J., Cicerchi C., Kanbay M., Roncal-Jimenez C.A., et al. Uric acid induces hepatic steatosis by generation of mitochondrial oxidative stress. J. Biol. Chem. 2012;287:40732–40744. doi: 10.1074/jbc.m112.399899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang Y., Zhou Y., Cheng S., Sun J.L., Yao H., Ma L. Effect of uric acid on mitochondrial function and oxidative stress in hepatocytes. Genet. Mol. Res. 2016;15 doi: 10.4238/gmr.15028644. [DOI] [PubMed] [Google Scholar]
- 96.Taufiq F., Li P., Miake J., Hisatome I. Hyperuricemia as a risk factor for atrial fibrillation due to soluble and crystalized uric acid. Circ Rep. 2019;1:469–473. doi: 10.1253/circrep.CR-19-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shi F., Kouadir M., Yang Y. NALP3 inflammasome activation in protein misfolding diseases. Life Sci. 2015;135:9–14. doi: 10.1016/j.lfs.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 98.Ebrahimi R., Pasalar P., Shokri H., Shabani M., Emamgholipour S. Evidence for the effect of soluble uric acid in augmenting endoplasmic reticulum stress markers in human peripheral blood mononuclear cells. J. Physiol. Biochem. 2022;78:343–353. doi: 10.1007/s13105-021-00869-y. [DOI] [PubMed] [Google Scholar]
- 99.Chen X., Shi C., He M., Xiong S., Xia X. Endoplasmic reticulum stress: molecular mechanism and therapeutic targets. Signal Transduct. Targeted Ther. 2023;8:352. doi: 10.1038/s41392-023-01570-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jiang T., Harder B., Rojo de la Vega M., Wong P.K., Chapman E., Zhang D.D. p62 links autophagy and Nrf2 signaling. Free Radic. Biol. Med. 2015;88:199–204. doi: 10.1016/j.freeradbiomed.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sugimoto M.A., Vago J.P., Teixeira M.M., Sousa L.P. Annexin A1 and the resolution of inflammation: modulation of neutrophil recruitment, apoptosis, and clearance. J. Immunol. Res. 2016;2016:1–13. doi: 10.1155/2016/8239258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pin A.-L., Houle F., Fournier P., Guillonneau M., Paquet É.R., Simard M.J., et al. Annexin-1-mediated endothelial cell migration and angiogenesis are regulated by vascular endothelial growth factor (VEGF)-induced inhibition of miR-196a expression. J. Biol. Chem. 2012;287:30541–30551. doi: 10.1074/jbc.M112.393561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cybulsky M.I., Gimbrone M.A. Endothelial expression of a mononuclear leukocyte adhesion molecule during atherogenesis. Science (1979) 1991;251:788–791. doi: 10.1126/science.1990440. [DOI] [PubMed] [Google Scholar]
- 104.Gimbrone M.A., García-Cardeña G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ. Res. 2016;118:620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu X., You C. The biomarkers discovery of hyperuricemia and gout: proteomics and metabolomics. PeerJ. 2022;11 doi: 10.7717/peerj.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.