Abstract
Background
Pseudomonas aeruginosa is a major pathogen in cystic fibrosis (CF), where chronic and intermittent infections significantly affect patient outcomes. This study aimed to investigate the molecular epidemiology of P. aeruginosa in CF patients from the Brazilian Amazon, focusing on genotypic diversity, resistance profiles, and virulence factors.
Methods
A cross-sectional study included 72 P. aeruginosa isolates from 44 CF patients treated at a regional reference center between 2018 and 2019. Antimicrobial susceptibility patterns were determined using VITEK-2 system and Kirby-Bauer disk diffusion. Virulotypes were defined by molecular detection of exoS, exoU, exoT, exoY, algU, and algD genes. Genetic diversity was assessed using multilocus sequence typing (MLST). Demographic data, clinical severity, and spirometry results were also collected.
Results
Among the patients, 54.55% experienced intermittent infections, while 45.45% had chronic infections. Chronic infections were associated with older age, lower FEV1, and reduced Shwachman-Kulczycki scores. Multidrug resistance was observed in 15.3% of isolates, particularly against ciprofloxacin and piperacillin/tazobactam. The exoU gene was present in 55.56% of isolates, an uncommon finding in CF populations. High genetic diversity was evident, with 37 sequence types (STs), including 14 novel STs. High-risk clones (HRCs) constituted 25% of isolates, with ST274 being the most prevalent (12.5%). Longitudinal analysis revealed transient colonization in intermittent infections, while chronic infections were dominated by stable clones.
Conclusion
This study highlights the molecular and clinical dynamics of P. aeruginosa in CF patients from the Brazilian Amazon. Chronic infections were linked to severe lung impairment , while intermittent infections were dominated by HRCs. These findings underscore the need for robust genotypic surveillance to mitigate the burden of P. aeruginosa in CF populations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-025-03920-w.
Keywords: Genotyping, Antimicrobial Resistance, Virulence, Pseudomonas aeruginosa, Brazil
Background
Cystic fibrosis (CF) is a severe autosomal recessive genetic disorder characterized by mutations in the CFTR gene, which leads to the production of thick, viscous mucus, particularly in the lungs [1]. This pathological condition predisposes individuals to chronic respiratory infections and progressive lung deterioration. Among the most critical pathogens associated with persistent infections in CF patients is Pseudomonas aeruginosa, an opportunistic Gram-negative bacterium notorious for its remarkable ability to develop multidrug resistance (MDR) and adapt to the highly selective environment of the CF lung, making clinical management exceedingly complex [2].
One of the main challenges in treating P. aeruginosa infections in CF is its high genetic and phenotypic diversity, which is crucial for its adaptability and persistence in chronic infections. This pathogen possesses a wide array of virulence factors which aid in adhesion and biofilm formation, such as the secretion of alginate, which provides protection against both the host immune system and antibiotic therapies [3, 4]. Additionally, the type III secretion system (T3SS), a needle-like apparatus used to inject effector proteins into host cells, plays a major role in disrupting host cell functions and evading immune responses, thus contributing to the pathogen's ability to establish infections [5, 6]. Indeed, several studies have highlighted the role of virulence factors such as exoU in bloodstream infections, lung impairment and acute damage, and antimicrobial non-susceptibility [7–13].
Antimicrobial resistance (AMR) presents significant challenges in the management of P. aeruginosa infections in CF, as repeated and prolonged antibiotic treatments in these patients often promote the selection of MDR strains. Notably, P. aeruginosa employs various resistance mechanisms, including the production of β-lactamases, efflux pumps, and biofilm formation, which collectively impair antibiotic efficacy and complicate treatment strategies [2, 14, 15].
The application of multilocus sequence typing (MLST) in the molecular epidemiology of P. aeruginosa has enabled more precise characterization of strains isolated from CF patients [16, 17]. MLST has revealed significant genetic diversity among isolates, with certain sequence types (STs) being particularly prevalent in this context. Among these, high-risk clones (HRCs) have been associated with worse clinical outcomes, increased rates of AMR, and treatment failures [18–21]. Epidemic strains are of particular concern in the CF population, as their persistence and adaptability often correlate with exacerbated disease and additional difficulty in clinical management [22–25]. However, it is important to note the scarcity of MLST-based studies on P. aeruginosa in the CF context, particularly in regions such as Brazil. Indeed, most of studies genotyping P. aeruginosa isolates from CF patients in the country are outdated and utilized alternative genotyping methods rather than MLST, such as pulsed field gel electrophoresis (PFGE) and ribotyping [26–30]. This highlights a significant gap in the literature, emphasizing the need for more comprehensive molecular epidemiological studies using standardized MLST protocols to better understand the genetic landscape of P. aeruginosa in CF populations.
In the present study, we conducted a comprehensive analysis of P. aeruginosa isolates from CF patients at a referral center in state of Pará, Brazilian Amazon region. This region presents unique challenges due to its geographic isolation, the potential influence of environmental factors on pathogen evolution, and low prevalence of CF. This study addresses existing gaps by profiling the AMR patterns, virulence, and genetic diversity of P. aeruginosa isolates from CF patients in the Amazon region, providing essential insights into the local epidemiology of this critical pathogen in the Brazilian context, contributing to a broader understanding of the pathogen's role in disease progression and management strategies in such patients.
Methods
Study population and adopted criteria
This cross-sectional study evaluated the clinical and epidemiological data of CF patients, along with the phenotypic and genotypic characteristics of P. aeruginosa isolates responsible for infections in this population. Between January 2018 and March 2019, a total of 136 CF patients were treated at the Cystic Fibrosis Clinic of the João de Barros Barreto University Hospital, Federal University of Pará (HUJBB/UFPA), a reference center in the state of Pará, Brazilian Amazon. During this period, 49 (36.05%) patients had positive cultures for P. aeruginosa. However, due to incomplete demographic and microbiological data, only 44 patients (32.35%) were included in the study.
Demographic data, clinical severity, and spirometry results were collected from the included patients when available. The type of infection, categorized as either chronic or intermittent, was determined based on medical follow-up on patients over the years (clinical records) and microbial culture results. Intermittent infection was defined as a patient with a positive culture for P. aeruginosa, followed by treatment with ciprofloxacin (CIP) and tobramycin (TOB) and three subsequent consecutive negative cultures, indicating successful eradication of the infection after treatment. Chronic infection was defined as having more than 50% positive cultures for P. aeruginosa over the past 12 months, or three or more positive cultures within a six-month period, in accordance with the Brazilian and reference center’s protocol [31, 32]. Patients with a positive culture for P. aeruginosa who had other lung diseases/infections (asthma, sequelae of tuberculosis, mycobacteriosis, Burkholderia cepacia, resistant Staphylococcus aureus and other pathogens) were excluded.
The clinical severity of CF patients was assessed using the Shwachman-Kulczycki score, which includes four components: general activity evaluation, radiological findings, physical examination, and nutritional status, each scored from 0 to 25. The total score was interpreted as follows: 86–100 points indicated an "excellent" condition, 71–85 points "good," 56–70 points "mild," 41–55 points "moderate," and below 40 points "severe." Spirometry data were extracted from medical records to evaluate lung function in the study population, including Forced Expiratory Volume in one second (FEV1), Forced Vital Capacity (FVC), the FEV1/FVC ratio (Tiffeneau Index), and Forced Expiratory Flow at 25–75% of lung volume (FEF25-75%) [32, 33].
Ethical statement
The samples included in this study were obtained during routine laboratory procedures at the CF reference center. Consent for participation and publication was obtained from all included patients upon written signature of informed consent form. All data collection and experiments related to this study were performed in accordance with Brazilian guidelines and regulations (Resolution CNS nº 196/96), as well as the Declaration of Helsinki. This study was approved by the Ethics Committee of the HUJBB/UFPA (Approval no. 1.910.716) and was registered with the National System for the Management of Genetic Heritage and Associated Traditional Knowledge (SisGen; registration no. AF44CCB).
Bacterial isolates
Bacterial isolates were obtained from oropharyngeal swabs and/or sputum samples from CF patients, collected during routine clinical visits, which occur every 2 to 4 months at the reference center. All P. aeruginosa samples were initially processed by the Bacteriology Service at the Reference Center (HUJBB/UFPA) and subsequently forwarded to the Molecular Biology Laboratory at the Bacteriology and Mycology Section, Evandro Chagas Institute (SEBAC/IEC) for further testing and storage.
For preliminary identification, all isolates were cultured on MacConkey and Luria–Bertani agar plates and incubated at 37°C for 24 to 48 h to observe colony morphology, the presence or absence of a mucoid phenotype, and Gram staining. Bacterial suspensions for each sample were prepared to correspond to the 0.5 McFarland standards and processed using the automated VITEK-2 system (bioMérieux, Marcy l'Etoile, France).
Antimicrobial Susceptibility Testing (AST)
The antimicrobial susceptibility testing (AST) was performed by determining the minimum inhibitory concentrations (MICs) using the automated VITEK-2 system (bioMérieux, Marcy l'Etoile, France) according to the manufacture’s standards for six antimicrobials, including piperacillin + tazobactam (TZP), ceftazidime (CAZ), cefepime (FEP), imipenem (IMP), meropenem (MEM) and amikacin (AMK). Also, two antimicrobials applied in patients’ treatment CIP and TOB were tested by employing the Kirby–Bauer disk diffusion method on Mueller–Hinton Agar. Isolates were classified as sensitive (S), intermediate (I), or resistant (R) based on the Clinical and Laboratory Standards Institute (CLSI) breakpoints [34]. Non-susceptible isolates include both I and R isolates. Isolates were also phenotypically categorized as MultiS if susceptible to all tested antimicrobial classes, moderately resistant (ModR) if non-susceptible to ≥ 1 drug in < 3 antimicrobial classes, multidrug-resistant (MDR) if non-susceptible to ≥ 1 drug in ≥ 3 antimicrobial classes, and extensively drug-resistant (XDR) if not susceptible to one agent in all tested antimicrobial classes except ≤ 2, according to previously described criteria [35, 36].
DNA extraction and detection of virulence-related markers
Bacterial DNA was extracted from overnight cultures for each P. aeruginosa isolate using the Easy Pure® Buccal Swab Genomic DNA Kit, following the manufacturer’s instructions. The DNA was then quantified using the Picodrop PICO100 spectrophotometer (Picodrop Limited, Hinxton, UK). DNA concentrations were adjusted to 25–50 ng/μl for all posterior molecular experiments.
The detection of six virulence-related markers was performed via PCR, targeting adhesion genes (algU and algD) and type III secretion system (T3SS) genes (exoS, exoU, exoT, and exoY). The primers and reaction parameters used were based on previously described methods [37–39]. PCR products were analyzed under ultraviolet light on a 1% agarose gel stained with Syber®Safe dye (Invitrogen™, Carlsbad, USA). P. aeruginosa strains ATCC 27853 (exoS +) and PA14 (exoU +) were used as positive controls. Based on the detection of exoS and exoU virulotypes, the strains were classified as invasive (exoS + /exoU-), or cytotoxic (exoS-/exoU +), as previously described [40].
Genetic diversity assessment via MLST
Molecular typing based on MLST was performed according to the protocol previously described by Curran et al. [41], with slight modifications utilizing primers described by Santos et al. [42]. The seven housekeeping genes included in the scheme (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) were amplified by PCR, followed by sequencing of the reaction products using the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA, USA) on an ABI Prism 3500xL genetic analyzer (Applied Biosystems, Foster City, CA, USA). Allele profiles and STs were determined by comparing the obtained sequences with the data documented in the Public databases for molecular typing and microbial genome diversity (PubMLST) database (https://pubmlst.org/organisms/pseudomonas-aeruginosa) (Accessed on August 20, 2024) [43]. Undefined profiles were considered as new alleles and STs and were submitted to the PubMLST database for curation and validation. Data management and the analysis of clonal complexes (CCs) were conducted using the PHYLOViZ 2.0 and Bionumerics v6.6 platforms. Clonal complexes were identified as clusters of closely related sequence types (STs) that differ at a single genetic locus (single locus variant—SLV) and double locus variants (DLVs). The minimum spanning tree (MST) was constructed following the goeBURST Full algorithm [44].
Statistical analysis
Descriptive statistics, including means, standard deviations, and percentages, were calculated for demographic variables (age, sex, and geographic origin) as well as clinical variables (type of infection, clinical severity, spirometric parameters, and annual pulmonary exacerbations). To compare groups, various statistical tests were employed. A T-test was used to evaluate differences in mean ages between patients with intermittent and chronic infections. Chi-square and G tests were conducted to assess associations between infection type and categorical variables such as sex, clinical severity, and HRC status. The Mann–Whitney U test was applied for comparisons of clinical severity scores and spirometric parameters between groups. Additionally, one-way ANOVA was utilized to analyze differences in mean ages across clinical severity categories. A normality test was performed to determine whether parametric or non-parametric tests were appropriate for each dataset. To further investigate associations, a binary logistic regression model was constructed, with high-risk clone status as the dependent variable and independent covariates selected based on clinical relevance. All statistical analyses were conducted using the Statistical Package for the Social Sciences (SPSS) software, with a significance level set at p ≤ 0.05.
Results
Infection profile of P. aeruginosa in CF patients
A total of 72 P. aeruginosa isolates were recovered from 44 CF patients monitored at a reference center in the state of Pará, Brazilian Amazon. Most of the patients were male (61.36%; 27/44), with ages ranging from 1 to 63 years and a mean age of 15.41 years. The highest frequency of patients was observed in the age group of 6 to 12 years (38.64%; 17/44). The mean age at diagnosis was 8.96 years, with a minimum of 0.1 years and a maximum of 62 years (Table 1). Regarding the geographic origin, 50.0% (22/44) of patients were from the metropolitan region of Belém, including 29.55% (13/44) from Belém/PA, 11.36% (5/44) from Ananindeua/PA and 9.09% (4/44) from Barcarena/PA (Supplementary Material 1).
Table 1.
Distribution of sex and clinical severity among CF patients with chronic and intermittent P. aeruginosa infections
| Chronic Infection n (%) | Intermittent Infection n (%) | P-value | |
|---|---|---|---|
| Sex | |||
| Male | 10 | 17 | 0.2703* |
| Female | 10 | 7 | |
| Clinical Severity | |||
| Excellent | 0 | 11 (45.83%) | 0.004# |
| Good | 5 (25%) | 8 (33.33%) | |
| Mild | 11 (55%) | 3 (12.5%) | |
| Moderate | 2 (10%) | 2 (8.33%) | |
| Severe | 2 (10%) | 0 | |
Source: Authors study, 2025. Legend: * Chi-square test, # G-Test
In terms of infection type, 54.55% (24/44) of the patients had intermittent infection and 45.45% had chronic infections (20/44). Among patients with intermittent infections, 29.1% (7/24) were female and 70.9% (17/24) were male, while patients with chronic infections included 50% (10/20) male and 50% (10/20) female. The distribution of infection types by sex did not show a significant difference, as indicated by the Chi-square test (χ2 = 1.215, p = 0.27) (Table 1). Patients with intermittent infection were mostly between 1 and 12 years old, while patients with chronic infections were predominantly older, within the age range of 19 to 29 years. A T-test confirmed a significant difference in the mean ages between patients with chronic and intermittent infections, indicating that chronic infections are associated with older age (t = 2.31, p = 0.029).
The assessment of clinical severity revealed significant differences between patients with intermittent and chronic infections. Among those with intermittent infections, 45.83% (11/24) were classified as "Excellent," and 33.33% (8/24) as "Good." Instead, most patients with chronic infections were classified as "Mild" (55%, 11/20), and as "Severe" (10%, 2/20). The G test confirmed a significant association between infection type and clinical severity (χ2 = 18.27, p = 0.004) (Table 1).
We evaluated the differences between infection types and variables such as age, spirometry parameters, and Shwachman-Kulczycki scores. Intermittent infections were associated with a significantly lower median age (p = 0.004) and a mean difference of 9 years. Furthermore, spirometric parameters consistently demonstrated better outcomes in the intermittent group. Specifically, FEV1, FVC% and FEF25-75% (p = 0.004) and the Shwachman-Kulczycki scores (p < 0.001) were markedly higher in the intermittent group (Table 2).
Table 2.
Comparison of age, spirometry parameters, and Shwachman-Kulczycki scores by infection type in cystic fibrosis patients
| Group | Mean | Median | Standard deviation | P-value* | Mean difference | |
|---|---|---|---|---|---|---|
| Age | Chronic | 24.4 | 22.5 | 15.4 | 0.004 | 9 |
| Intermittent | 13.4 | 10 | 7.4 | |||
| FEV1 | Chronic | 56.6 | 49 | 30.2 | 0.004 | −28.38 |
| Intermittent | 85 | 86 | 24.42 | |||
| FVC | Chronic | 67.1 | 63 | 28.4 | 0.004 | −28.25 |
| Intermittent | 95.3 | 102 | 25.9 | |||
| FEV1/FVC | Chronic | 81.2 | 84.5 | 14.5 | 0.064 | −7.5 |
| Intermittent | 88.7 | 89 | 8.27 | |||
| FEF25-75% | Chronic | 47.9 | 33 | 35.7 | 0.004 | −37.18 |
| Intermittent | 85.1 | 86 | 35.09 | |||
| Shwachman-Kulczycki scores | Chronic | 61.8 | 65 | 14.2 | < 0 .001 | −24.75 |
| Intermittent | 83.1 | 85 | 13.22 |
Source: Authors study, 2025. Legend: *Mann–Whitney U test
Significant correlations were observed between age and certain spirometric parameters, indicating that age is an important factor affecting lung function in CF patients. The Spearman correlation analysis showed that FEF25-75% had a significant negative correlation with age (ρ = − 0.41, p = 0.014), indicating a decline in this parameter with increasing age. These results highlight the importance of considering age-related decline in lung function, particularly for parameters like FEF25-75%, in CF patients. Other parameters did not show statistically significant correlations (FEV1: ρ = − 0.28, p = 0.105; FVC: ρ = − 0.29, p = 0.086; FEV1/FVC: ρ = − 0.24, p = 0.158).
AST and resistance features
The AST revealed that the isolates were predominantly non-susceptible to CIP (16/72–22.2%), PIP/TAZ (13/72–18.1%), CPM (11/72–15.3%), IMP (11/72–15.3%), and AMK (9/72–12.5%). The antibiotics with the lowest rates of non-susceptibility were meropenem MER (5/72–6.9%) and CAZ (4/72–5.6%). All tested isolates presented susceptibility to TOB (72/72 – 100.0%) According to the phenotypic classification, the most of isolates were categorized as MultiS (54.1%; 39/72), followed by ModR (30.6%; 22/72) and MDR (15.3%; 11/72).
The association between AMR phenotypes and clinical parameters in CF patients were analyzed, revealing no statistically significant differences between the resistance phenotypes (MDR, ModR, and MultiS) for any of the evaluated parameters. Spirometric measures, clinical assessments, likewise, the Shwachman-Kulczycki total showed similar mean values across the phenotypes indicating comparable clinical severity among CF patients with different resistance phenotypes. These findings highlight the lack of a clear relationship between resistance phenotypes and the clinical parameters evaluated in this cohort (Supplementary Material 2).
Virulence related-data
The T3SS exoS and exoY genes were detected in all isolates (72/72; 100.0%), while exoU and exoT gene were detected in 40 (55.56%) and 68 (94.44%) isolates, respectively. Alginate production-related genes (algD and algU) were both detected in all isolates (72/72 – 100.0%). Based on the presence of exoS-exoU genes, an unusual 40 isolates (55.56%) were classified as cytotoxic-invasive (exoS + /exoU +), while the remaining 32 isolates (44.44%) were classified as invasive (exoS +). Also, three distinct virulotypes where identified: V1: exoS ( +); exoU ( +); exoT ( +); exoY ( +); algD ( +); algU ( +); V2: exoS ( +); exoU (-); exoT ( +); exoY ( +); algD ( +); algU ( +); V3: exoS ( +); exoU (-); exoT (-); exoY ( +); algD ( +); algU ( +). The relationship between T3SS virulotypes, infection status, and clinical data was extensively investigated in a previous study by our research group [13].
Fisher's exact test revealed significant and negative associations between exoU presence and non-susceptibility to CPM (p = 0.0017), IMP (p = 0.0089), and MER (p = 0.0144). No significant associations were observed for PIP/TAZ (p = 0.7617), CAZ (p = 1.0000), AMK (p = 0.2821), or CIP (p = 0.7764). Also, no significant association between exoU and the susceptibility phenotypes (Table 3).
Table 3.
Association between AMR features and cytotoxic virulotype (exoU +) in P. aeruginosa isolates from CF patients
| Antibiotic/Resistance Phenotype | exoU + | exoU- | P-value |
|---|---|---|---|
| CPM | 1 | 10 | 0.0017* |
| IMP | 2 | 9 | 0.0089* |
| MER | 0 | 5 | 0.0144* |
| PIP/TAZ | 8 | 5 | 0.7617* |
| CAZ | 2 | 2 | 1.0000* |
| AMK | 3 | 6 | 0.2821* |
| CIP | 8 | 8 | 0.7764* |
| MDR Phenotype | 3 | 8 | 0.0992# |
| MultiS Phenotype | 25 | 14 | |
| ModR Phenotype | 12 | 10 |
Source: Authors study, 2025. Legend: * Chi-square test, # G-Test
Among the 72 isolates analyzed, the non-mucoid phenotype was the most common, seen in 63.89% (46/72) of cases. Of these non-mucoid isolates, 43.48% (20/46) were linked to chronic infections, while 56.52% (26/46) were associated with intermittent infections. In contrast, the mucoid phenotype appeared in 36.11% (26/72) of isolates, with the majority, 92.30% (24/26), coming from chronic infections. Statistical analysis confirmed a significant association between colony phenotype and infection type (χ2 = 16.80, p < 0.001). The mucoid phenotype was much more common in chronic infections, while the non-mucoid phenotype was more frequently found in intermittent infections. Also, G-test revealed no statistical evidence for a significant association between the resistance phenotype and the mucoid/non-mucoid phenotype (p = 0.9992).
Molecular epidemiology by MLST
A remarkable genetic diversity was observed among the investigated P. aeruginosa population with a total of 37 identified STs, including 14 newly identified STs and 23 previously reported and curated at the PubMLST dabase for P. aeruginosa. Furthermore, the novel allele (mutL – 395) was associated with the novel genotype ST4993. The isolates demonstrated low relatedness overall, with only four showing closer genetic relationships, differing by a SLV between them (Fig. 1). Among the isolates, 18 (25.0%) were identified as HRCs (ST235, ST274, ST357 and ST446) while the remaining 54 (75.0%) were classified as non-HRCs.
Fig. 1.
goeBURST full MST of P. aeruginosa isolated from CF patients demonstrating unique STs and clonal complexes. Each node represents a unique ST and node size is proportional to the isolates related with that ST. Solid lines between STs shaded in grey represents SLVs and belong to CCs, and dashed lines between STs represents double locus variants DLVs
ST274 was the most prevalent and associated with nine isolates (12.50%), followed by the novel ST4978, linked to eight isolates (11.11%), and ST357, with seven isolates (9.72%). Other notable STs included ST2140, identified in six isolates (8.33%), while ST2476, ST4256, and ST3118 were each found in three isolates (4.17%). Additionally, the novel ST4979, as well as ST1284 and ST2021, were each linked to two isolates (2.78%). Several STs were identified in only one isolate (1.39%), 15 of which had been previously reported: ST9, ST109, ST235, ST252, ST256, ST273, ST446, ST569, ST900, ST1490, ST1685, ST2069, ST2603, ST2747, and ST3090. The remaining STs were newly discovered in this study: ST4981, ST4983, ST4985, ST4986, ST4988, ST4990, ST4991, ST4992, ST4993, ST4995, ST4996, and ST4997 (Fig. 1).
The association between the type of infection and the presence of HRCs was statistically significant (Fisher’s exact test, p = 0.0016), suggesting that HRCs are more frequently associated with intermittent infections, while non-HRCs are predominantly linked to chronic infections. We also conducted a analysis to investigate the differences between HRCs groups and infection types, comparing numerical variables such as age, spirometry parameters, and Shwachman-Kulczycki total scores. For HRCs, only Shwachman-Kulczycki total scores were significant based on the Student’s T-test (p = 0.004) (Table 4).
Table 4.
Comparison of age, spirometry parameters, and Shwachman-Kulczycki total scores between HRC and non-HRC groups in CF patients
| Group | Mean | Median | Standard deviation | P-value* | Mean difference | |
|---|---|---|---|---|---|---|
| Age | Non-HRC | 21.5 | 18 | 14.8 | 0.074 | 5 |
| HRC | 13.1 | 10 | 6.06 | |||
| FEV1 | Non-HRC | 70 | 80 | 33.8 | 0.623 | −5.34 |
| HRC | 75.4 | 81 | 24.62 | |||
| FVC | Non-HRC | 79.7 | 83.5 | 32.1 | 0.497 | −7.32 |
| HRC | 87 | 97 | 27.4 | |||
| VEF1/CVF | Non-HRC | 84.4 | 89 | 13.5 | 0.720 | 1 |
| HRC | 86.7 | 86 | 9.2 | |||
| FEF25-75% | Non-HRC | 65 | 70.5 | 42.3 | 0.566 | −8.11 |
| HRC | 73.2 | 80 | 35.56 | |||
| Shwachman-Kulczycki Total Scores | Non-HRC | 67.2 | 67.5 | 17.3 | 0.004* | −16.51 |
| HRC | 83.7 | 85 | 11.54 |
Source: Authors study, 2024. * Student's t-test
Chi-square test was conducted to assess the association between HRCs and AST data. For AMR, the only significant association observed was for CIP, where HRC isolates were significantly less likely to be non-susceptible (p = 0.0015). In terms of resistance phenotypes, no significant associations were found. Overall, these findings suggest that HRC status is not significantly associated with resistance to most antibiotics or resistance phenotypes, accordingly to the evaluated data (Supplementary Material 3).
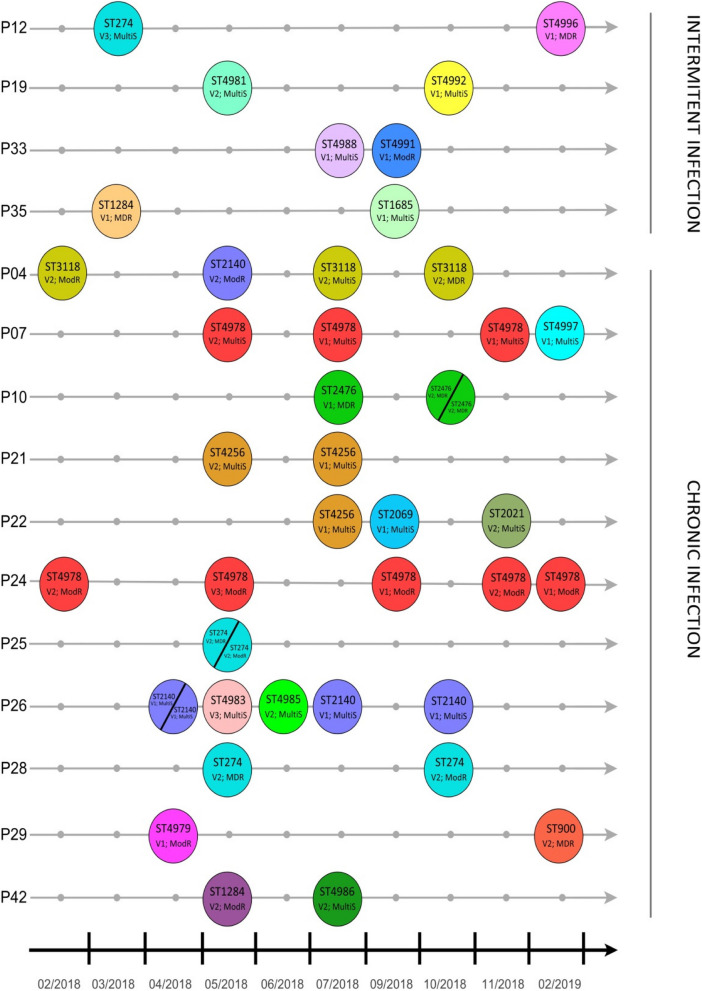
Our data also revealed significant variability in STs among isolates recovered from the same patient, highlighting the genetic diversity of P. aeruginosa in CF infections. A total of 15 patients had more than one isolated collected in the period of study (Fig. 2). Among patients with intermittent infections (P12, P19, P33 and P35), a higher diversity of STs was observed, with distinct STs identified across isolates. For instance, ST274, ST1284, and ST4996 were found in different patients. There were few recurrent STs, reflecting the transient nature of colonization in these cases. For patients with chronic infections, several STs persisted over time, indicating clonal stability. Notably, ST4978 was frequently isolated in multiple samples. Other recurring STs, such as ST3118 and ST2476, were also prominent. The figure stresses the differences in ST dynamics between infection types, with intermittent infections characterized by high ST variability and chronic infections dominated by stable, recurrent clones and provides insights into the longitudinal behavior of P. aeruginosa in CF patients.
Fig. 2.
Longitudinal distribution of P. aeruginosa Sequence Types (STs) in CF patients with intermittent and chronic infections. Legend: ST: sequence type; Virulotype. V1: exoS ( +); exoU ( +); exoT ( +); exoY ( +); algD ( +); algU ( +); V2: exoS ( +); exoU (-); exoT ( +); exoY ( +); algD ( +); algU ( +); V3: exoS ( +); exoU (-); exoT (-); exoY ( +); algD ( +); algU ( +). Resistance pattern: MultiS: Multi susceptive; ModR: Moderate resistant; MDR: Multidrug resistant
We conducted a final multivariable logistic regression model with independent variables to identify factors associated with HRCs among all 72 isolates. The analysis revealed that intermittent infection (p = 0.001, OR = 10.40, 95% CI = 2.584–41.855) was associated with a tenfold higher likelihood of being linked to HRCs. (Supplementary Material 4).
Discussion
This study investigated the molecular epidemiology and clinical impact of P. aeruginosa infections in CF patients, with particular focus on genetic diversity, AMR patterns, and associated virulence factors. The findings reveal significant differences between chronic and intermittent infections, which are closely linked to patient demographics, clinical severity, spirometric parameters, and microbial-related factors. Importantly, the study highlights the infection dynamics and high prevalence of novel STs and the widespread presence of HRCs within the CF population in the Brazilian Amazon, highlighting the evolving nature of P. aeruginosa in this patient group.
The proportion of patients registered in the studied service with positive cultures for P. aeruginosa (36.0%—49/136) is lower than the prevalence reported in the U.S. Cystic Fibrosis Patient Registry (46.4%) but slightly higher than the European Cystic Fibrosis Society Patient Registry (32.8%) [45, 46]. In both the U.S. and Europe, reductions in P. aeruginosa prevalence over the past decade have been attributed to improved CF care strategies, such as enhanced infection control measures and the adoption of highly effective CFTR modulators. Although trends were not assessed in this study, the observed prevalence aligns with global patterns, reflecting comparable standards of CF management within the studied cohort, however, variations in healthcare infrastructure, access to CF-specific therapies, and microbiological surveillance protocols, as well as differences in diagnostic criteria and definitions of chronic infections across regions, might influence such rates [46].
There is strong evidence suggesting that the clinical status of CF patients deteriorates soon after the first isolation of P. aeruginosa from the respiratory tract. This decline is particularly alarming as infections become chronic, leading to rapid lung function impairment, frequent pulmonary exacerbations, and significantly increased mortality rates [47–49]. It is important to emphasize that FEV1 is the gold-standard measure for assessing the severity and prognosis of pulmonary disease in CF. It is also a key criterion for lung transplant eligibility and access to high-cost medications, making it a critical focus in studies evaluating clinical outcomes across reference centers. Factors such as poor nutritional status, chronic P. aeruginosa infection, and CF-related diabetes are strongly associated with a more rapid decline in FEV1 [50]. Our study reinforces these findings as the clinical severity analysis further demonstrated stark differences between intermittent and chronic infections. The presented data emphasize the critical role of P. aeruginosa in the clinical deterioration of CF patients, especially those with chronic infections and older age, which were associated with a higher degree of lung function impairment and the profound impact of chronic P. aeruginosa infections on the overall health and quality of life of CF patients.
The rising prevalence of P. aeruginosa infection in adolescence and early adulthood can be attributed to repeated environmental exposure and the frequent use of antibiotics treatments aimed at eradicating other early airway pathogens. Analyses of CF patient registry data have shown a notable increase in P. aeruginosa prevalence between the ages of 6 to 10 years and 18 to 24 years [51]. The early colonization by P. aeruginosa observed in our cohort adds further urgency to these findings. Among the patients studied, 6.82% (3/44) of children aged 1 to 5 years had already developed chronic infections, a notable finding given that early P. aeruginosa infections are typically intermittent and often eradicated with antibiotic therapy in more than 69% of the cases [52, 53]. This early colonization has also been linked to elevated pulmonary inflammation markers, early lung function decline, and irreversible airway changes [49]. Furthermore, early colonization predisposes individuals to chronic colonization, as previously reported [31, 54]. Despite these observations, our data reveal that most chronic infections occurred in older patients, demonstrating the progressive nature of P. aeruginosa infections in CF patients, with a transition from intermittent infections during childhood to chronic infections in adulthood, accompanied by worsening clinical outcomes.
While antibiotic therapies have significantly extended survival and improved the quality of life for CF patients, their extensive and repeated use has also accelerated the development of AMR [55]. The progressive nature of CF-related lung infections often necessitates cycles of "suppressive" and "curative" antibiotic strategies, which, while essential for managing exacerbations, inadvertently select for resistant strains [56]. Of particular concern are chronically infected patients, where prolonged treatment facilitates the emergence of chromosomal mutations, leading to stable resistance mechanisms. Over time, this dynamic has driven the increasing prevalence of MDR/XDR P. aeruginosa clones within CF populations worldwide, a trend consistently observed over recent decades [57, 58]. While most isolates were categorized as fully susceptible, a considerable proportion demonstrated ModR and MDR phenotypes, and non-susceptibility rates were observed for ciprofloxacin, piperacillin/tazobactam, cefepime and imipenem, and amikacin. These findings align with the known resistance mechanisms of P. aeruginosa, including efflux pump activity and β-lactamase production [3, 4, 59]. Also, the prevalence of MDR P. aeruginosa strains observed in this study (15.3%) is consistent with global reports, which recorded a slightly higher rate of 18.1% [45, 46]. In particular, the occurrence of MDR in patients from rural (far from capital areas) of Pará raises significant concerns. Factors such as delayed identification of P. aeruginosa infections, limited access to specialized CF care centers, challenges in obtaining appropriate antibiotic therapy, and repeated courses of intravenous antibiotics may contribute to this trend.
The exoS, exoU, exoT, and exoY genes encode toxins that are directly injected into eukaryotic host cells, mediating distinct pathogenic effects, including antifagocytic activity, necrotic damage, disruption of wound healing, and edema formation [3]. These genes have been strongly associated with invasive acute infections and high mortality rates [5, 9, 40]. Previous studies have also reported that CF isolates are less likely to carry exoU, as the chronic nature of pulmonary infections in CF does not favor strains producing this highly cytotoxic enzyme. Such strains can cause rapid tissue damage, often leading to either early death or infection clearance—outcomes that are inconsistent with the persistence typically observed in CF infections. Also, harboring multiple virulence and resistance mechanisms simultaneously imposes significant metabolic and selective pressures on the bacterium, often leading to downregulation of certain genes. This trade-off may explain the reduced frequency of highly virulent and resistant strains [60].
Our findings showed a higher prevalence of the exoS + /exoU + virulotype than previously reported in CF contexts, raising concerns that more virulent strains may be emerging in this population. Our research group previously evaluated the relationship between P. aeruginosa virulence factors and infection outcomes in CF patients. In Sarges et al. [13], the atypical exoS + /exoU + virulotype was found at a higher frequency than previously reported, predominantly associated with intermittent infections, pulmonary exacerbations, and clinical changes, though not linked to worse outcomes in chronic infections. Conversely, the exoS + /exoU − virulotype was strongly associated with chronic infections and worse clinical outcomes, including lower spirometric measures. Expanding on this, our study tested 72 isolates from 44 patients, confirming the presence of the exoS + /exoU + virulotype at an uncommon prevalence. These results further support the importance of detecting P. aeruginosa virulotypes, given their decisive impact on pulmonary impairment, clinical progression, and disease severity in CF patients. Our study also revealed that exoU + isolates were negatively associated with CPM and carbapenems and mainly related to non-sucesptibility to CIP AND PIP/TAZ. Such findings agree with previous negative association of virulence of T3SS and AMR, as maintaining both traits impose a fitness cost the bacteria [12, 61]. Also, such finding highlights the potential for these strains to evolve into HRCs clones with both increased virulence and moderate resistance, posing a significant threat to CF patient outcomes.
The algD and algU genes were detected in all isolates (72/72–100%), which is in line with previous data of high prevalence of those genes [62, 63]. These genes encode proteins crucial for alginate production, a key exopolysaccharide involved in biofilm formation, which is one of the main adhesins found in the respiratory tracts of patients with acute and CF infections [3]. In our study, the mucoid phenotype, was less frequent, found in 25 (34.72%) of the 72 isolates analyzed. However, mucoid isolates were predominantly from chronic infections (96%; n = 24/25). The non-mucoid phenotype was more common (63.89%; n = 46/72), yet the number of non-mucoid isolates in chronic infections (43.47%; 20 out of 46 isolates) raised concerns. This support that the mucoid form may play a role in long-term, persistent infections and suggests a potential conversion to the mucoid phenotype, which remains a feared complication in CF treatment due to the increased difficulty in eradicating infections and the greater damage to CF airways [64].
Typically, P. aeruginosa isolates recovered from diverse environments, including CF patients, reveal notable clonal diversity, with most isolates represented by unique genotypes [19, 20]. This clonal diversity was further highlighted by MLST genotyping in our study, revealing impressive genetic diversity and low genetic relatedness among P. aeruginosa populations in CF patients (Fig. 1). The clonal complex ST274 (CC/ST274) has been reported as a major clone among CF patients and associated with high mutation rates in AMR markers [22, 65]. In our study, ST274 strains isolated from seven patients, in which six were under nine years displaying heterogeneous genotypes and phenotypes. All ST274 related with chronic infections were exoU-, but were MDR or ModR.
The HRCs ST357 and ST235 are more prevalent in hospital environments and are among the top 10 global high-risk P. aeruginosa clones due to their prevalence, global dissemination, and association with MDR/XDR profiles, particularly in relation to horizontally acquired β-lactamases such as ESBLs (Extended-Spectrum Beta-Lactamases) and carbapenemases [21, 66]. ST357 is frequently associated with XDR strains and outbreaks of fluoroquinolone-resistant, potentially virulent strains carrying the exoU + genotype in various countries [12, 67, 68]. In our study, ST357 was related to seven unique isolates from distinct patients under 16 years, all non-mucoid, from intermittent infections, positive for all virulence genes tested, but presenting non-MDR phenotypes (three ModR isolates and the remaining MultiS). This homogeneity among the isolates may suggest possible spread of this clone in the region and possible cross-transmission between these patients, underscoring the importance of continuous and personalized monitoring of P. aeruginosa infections. In our study, ST235 showed no resistance to the antimicrobials tested but exhibited high virulence potential, carrying all tested virulence markers (exoU +), and isolated from a 19-year patient with an intermittent infection. Similarly, ST446 was identified as ModR and invasive-cytotoxic and has previously been linked to outbreaks of carbapenem-resistant P. aeruginosa strains carrying the IMP-1 gene among kidney transplant recipients [69]. Finally, these findings highlight the evolving dynamics of hospital-disseminated HCR like ST357, ST235 and ST446 within CF infections.
The non-HRC group predominated among the isolates analyzed (54/72; 75.0%). In this group, a new lineage, ST4978, was the most prevalent, found in eight isolates from two patients, both with chronic infections and classified as ModR, carrying virulence genes variably. Another new lineage, ST4979, was recovered from two patients, both with chronic infections and classified as MultiS and ModR, with one being invasive and the other invasive-cytotoxic. The other new lineages were found in only one isolate each, notable among them were ST4993, related to the new mutL 395 allele, and ST4996. ST4993 was recovered from a chronic infection, displaying an MDR phenotype and an invasive virulotype. ST4996, also MDR, came from an intermittent infection and exhibited an invasive-cytotoxic virulotype. In this group contrasting finding was the higher association with increased non-susceptibility rates compared to the HRC group, which challenges the common association of HRCs with MDR/XDR phenotypes [70]. These findings underscore the high heterogeneity of the P. aeruginosa population and potentially suggest local transmission events due to occurrence of new lineages in more than one patient.
Several non-HRC STs, previously associated with potentially resistant and virulent profiles, were identified in this study, including ST1284, ST2603, ST9, ST109, ST252, ST273, and ST900, as detailed in Supplementary Material 5. The remaining STs, to our knowledge, were only recorded in the PubMLST database [43] in different countries (ST2140, ST4256, ST1685, ST2021, ST3090, ST569, ST256, ST1490, ST2069, ST2747, ST2476, and ST3118) [71-76]. Of note are STs 2476 and 3118, which, in our study, were associated with MDR isolates (Supplementary Material 5).
Another particularly interesting finding in our study was the significant variability in STs among samples collected from the same patient over time. Additionally, virulotypes, resistance profiles, and colony morphologies (mucoid and non-mucoid) also showed temporal variability. On this matter, Parkins, Somayaji e Waters [17] explain that, although patients with chronic infections are typically infected by a single strain of P. aeruginosa, in some cases, multiple genetically unrelated strains may coinfect CF patients either temporarily or permanently. This can lead to complete strain replacements or the coexistence of distinct strains, resulting in persistent, genetically distinct lineages with varying functional characteristics [77]. Furthermore, the genotypic and phenotypic changes within genetically related bacterial populations can be strongly attributed to the selective pressures imposed by the hostile CF airway environment. The continuous presence of antibiotics used in treatment and the intrinsic components of the host immune system drive the adaptive evolution of bacterial populations in CF patients. This process results in genomic alterations and phenotypic changes within the bacteria over time [58].
While this study provides valuable insights, certain limitations should be acknowledged. First, the cross-sectional and retrospective design limits the ability to assess long-term trends to a better picture of the evolution of infection dynamics over time. Second, the relatively small sample size and regional focus may not fully capture the genetic and phenotypic diversity of P. aeruginosa across other regions in Brazil or globally. Third, while genotypic methods like MLST were employed to identify STs and clonal diversity, whole-genome sequencing (WGS) could have provided a more comprehensive understanding of resistance mechanisms and virulence factors. Finally, the study's findings are constrained to a single reference center, and broader multicenter studies are needed to validate these results and generalize them to other CF populations in diverse geographic and clinical contexts. Despite these limitations, the study demonstrates critical trends in Brazil and highlights the need for continued surveillance and tailored treatment strategies for P. aeruginosa infections in CF patients.
Conclusion
This study offers a comprehensive exploration of the molecular epidemiology of P. aeruginosa in CF patients from the Brazilian Amazon, emphasizing its genetic diversity, AMR patterns, and virulence-associated traits. The identification of novel STs and HRCs, along with unique regional infection dynamics, highlight the pathogen's adaptability and the challenges it poses in clinical management. Importantly, the findings highlight significant associations between chronic and intermittent infections, resistance phenotypes, and patient clinical data. The detection of uncommon virulotypes, such as exoS + /exoU + , further underscores the evolving pathogenic landscape in this population. These results emphasize the critical need for continuous surveillance, tailored therapeutic approaches, and multidisciplinary strategies to mitigate the progression of chronic infections, improve patient outcomes, and address AMR in CF care. Future research should focus on longitudinal studies and whole-genome analyses to unravel the mechanisms driving resistance and clinical severity in this context.
Supplementary Information
Acknowledgements
The present study will be presented by Maria Isabel Montoril Gouveia to obtain her master’s within the Program in Epidemiology and Health Surveillance (PPGEVS), Evandro Chagas Institute (IEC), Ministry of Health, Brazil.
Authors’ contributions
Conceptualization, M.I.M.G., Y.C.R., E.S.N.F.S and K.V.B.L; Formal Analysis and Acquisition of Data, M.I.M.G., H.S.d.R., P.A.S.d.S., D.M.S., L.R.R., M.J.A.S., M.V.H.M, A.J.P.G.Q.; Writing—Original Draft Preparation, M.I.M.G., Y.C.R., E.S.N.F.S., H.S.d.R., D.M.S.; Writing—Review & Editing, M.I.M.G., Y.C.R., H.S.d.R., P.A.S.d.S., D.M.S., L.R.R., M.J.A.S., A.J.P.G.Q., L.N.G.C.L., D.M.B. and K.V.B.L; Resources and Funding Acquisition, A.J.P.G.Q., L.N.G.C.L., D.M.B., K.V.B.L. and Y.C.R; Supervision and Projet Administration, Y.C.R. and K.V.B.L. All authors have read and agreed to the published version of the manuscript.
Funding
The study and article processing charges were funded by Evandro Chagas Institute (IEC/PA). Yan Corrêa Rodrigues scholarship was funded by PDPG—Pós-Doutorado Estratégico (PDPG-POSDOC), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES/Edital 16/2022—File Nº 88887.759036/2022–00). Maria Isabel Montoril Gouveia scholarship is funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). Pabllo Antonny Silva dos Santos scholarship is funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). Marcos Jessé Abrahão Silva scholarship is funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. The MLST genotyping data generated and analyzed during the current study are available in the repository, PubMLST—Pseudomonas aeruginosa isolates database [https://pubmlst.org/bigsdb?db = pubmlst_paeruginosa_isolates]”. Further inquiries must be addressed to the corresponding authors.
Declarations
Ethics approval and consent to participate
The samples included in this study were obtained during routine laboratory procedures at the CF reference center. Consent for participation was obtained from all included patients upon written signature of informed consent form. All data collection and experiments related to this study were performed in accordance with Brazilian guidelines and regulations (Resolution CNS nº 196/96), as well as the Declaration of Helsinki. This study was approved by the Ethics Committee of the HUJBB/UFPA (Approval no. 1.910.716) and was registered with the National System for the Management of Genetic Heritage and Associated Traditional Knowledge (SisGen; registration no. AF44CCB).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Karla Valéria Batista Lima, Email: karlalima@iec.gov.br.
Yan Corrêa Rodrigues, Email: yanrodrigues@iec.gov.br.
References
- 1.Trouvé P, Saint Pierre A, Férec C. Cystic Fibrosis: A Journey through Time and Hope. Int J Mol Sci. 2024;25(17):9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nickerson R, Thornton CS, Johnston B, Lee AHY, Cheng Z. Pseudomonas aeruginosa in chronic lung disease: untangling the dysregulated host immune response. Front Immunol. 2024;15:1405376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin S, Xiao W, Zhou C, Pu Q, Deng X, Lan L, et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct Target Ther. 2022;7(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cepas V, Soto SM. Relationship between virulence and resistance among gram-negative bacteria. Antibiot Basel Switz. 2020;9(10):719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu T, Zhang Z, Li T, Dong X, Wu D, Zhu L, et al. The type III secretion system facilitates systemic infections of Pseudomonas aeruginosa in the clinic. Microbiol Spectr. 2024;12(1):e0222423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horna G, Ruiz J. Type 3 secretion system as an anti-Pseudomonal target. Microb Pathog. 2021;155:104907. [DOI] [PubMed] [Google Scholar]
- 7.Cho HH, Kwon KC, Kim S, Koo SH. Correlation between virulence genotype and fluoroquinolone resistance in carbapenem-resistant Pseudomonas aeruginosa. Ann Lab Med. 2014;34(4):286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan E, Bensman J, Lou M, Agnello M, Shriner K, Wong-Beringer A. Risk of developing pneumonia is enhanced by the combined traits of fluoroquinolone resistance and type III secretion virulence in respiratory isolates of Pseudomonas aeruginosa. Crit Care Med. 2014;42(1):48–56. [DOI] [PubMed] [Google Scholar]
- 9.Peña C, Cabot G, Gómez-Zorrilla S, Zamorano L, Ocampo-Sosa A, Murillas J, et al. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis Off Publ Infect Dis Soc Am. 2015;60(4):539–48. [DOI] [PubMed] [Google Scholar]
- 10.Sánchez-Diener I, Zamorano L, López-Causapé C, Cabot G, Mulet X, Peña C, et al. Interplay among resistance profiles, high-risk clones, and virulence in the caenorhabditis elegans pseudomonas aeruginosa infection model. Antimicrob Agents Chemother. 2017;61(12):e01586-e1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Recio R, Mancheño M, Viedma E, Villa J, Orellana MÁ, Lora-Tamayo J, et al. Predictors of mortality in bloodstream infections caused by pseudomonas aeruginosa and impact of antimicrobial resistance and bacterial virulence. Antimicrob Agents Chemother. 2020;64(2):e01759-e1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horna G, Amaro C, Palacios A, Guerra H, Ruiz J. High frequency of the exoU+/exoS+ genotype associated with multidrug-resistant “high-risk clones” of Pseudomonas aeruginosa clinical isolates from Peruvian hospitals. Sci Rep. 2019;9(1):10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarges Edo SNF, Rodrigues YC, Furlaneto IP, de Melo MVH, da C Brabo GL, Lopes KCM, et al. Pseudomonas aeruginosa type III secretion system virulotypes and their association with clinical features of cystic fibrosis patients. Infect Drug Resist. 2020;13:3771–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossitto M, Fox V, Vrenna G, Tuccio Guarna Assanti V, Essa N, Lepanto MS, et al. The challenging life of mutators: how pseudomonas aeruginosa survives between persistence and evolution in cystic fibrosis lung. Microorganisms. 2024;12(10):2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidd TJ, Canton R, Ekkelenkamp M, Johansen HK, Gilligan P, LiPuma JJ, et al. Defining antimicrobial resistance in cystic fibrosis. J Cyst Fibros Off J Eur Cyst Fibros Soc. 2018;17(6):696–704. [DOI] [PubMed] [Google Scholar]
- 16.Waine DJ, Honeybourne D, Smith EG, Whitehouse JL, Dowson CG. Cross-sectional and longitudinal multilocus sequence typing of pseudomonas aeruginosa in cystic fibrosis sputum samples. J Clin Microbiol. 2009;47(11):3444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkins MD, Somayaji R, Waters VJ. Epidemiology, biology, and impact of clonal pseudomonas aeruginosa infections in cystic fibrosis. Clin Microbiol Rev. 2018;31(4):e00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigues YC, Silva MJA, Dos Reis HS, Dos Santos PAS, Sardinha DM, Gouveia MIM, et al. Molecular epidemiology of pseudomonas aeruginosa in brazil: a systematic review and meta-analysis. Antibiot Basel Switz. 2024;13(10):983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliver A, Rojo-Molinero E, Arca-Suarez J, Beşli Y, Bogaerts P, Cantón R, et al. Pseudomonasaeruginosa antimicrobial susceptibility profiles, resistance mechanisms and international clonal lineages: update from ESGARS-ESCMID/ISARPAE Group. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2024;30(4):469–80. [DOI] [PubMed] [Google Scholar]
- 20.Sawa T, Momiyama K, Mihara T, Kainuma A, Kinoshita M, Moriyama K. Molecular epidemiology of clinically high-risk Pseudomonas aeruginosa strains: Practical overview. Microbiol Immunol. 2020;64(5):331–44. [DOI] [PubMed] [Google Scholar]
- 21.Kocsis B, Gulyás D, Szabó D. Diversity and Distribution of Resistance Markers in Pseudomonas aeruginosa International High-Risk Clones. Microorganisms. 2021;9(2):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupont C, Aujoulat F, Benaoudia M, Jumas-Bilak E, Chiron R, Marchandin H. Highly diverse dynamics of Pseudomonas aeruginosa colonization from initial detection in cystic fibrosis patients: A 7-year longitudinal genetic diversity study. Infect Genet Evol. 2023;115:105513. [DOI] [PubMed] [Google Scholar]
- 23.van Mansfeld R, de Vrankrijker A, Brimicombe R, Heijerman H, Teding van Berkhout F, Spitoni C, et al. The effect of strict segregation on pseudomonas aeruginosa in cystic fibrosis patients. PloS One. 2016;11(6):e0157189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Mansfeld R, Willems R, Brimicombe R, Heijerman H, van Berkhout FT, Wolfs T, et al. Pseudomonas aeruginosa genotype prevalence in Dutch cystic fibrosis patients and age dependency of colonization by various P. aeruginosa sequence types. J Clin Microbiol. 2009;47(12):4096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.López-Causapé C, de Dios-Caballero J, Cobo M, Escribano A, Asensio Ó, Oliver A, et al. Antibiotic resistance and population structure of cystic fibrosis Pseudomonas aeruginosa isolates from a Spanish multi-centre study. Int J Antimicrob Agents. 2017;50(3):334–41. [DOI] [PubMed] [Google Scholar]
- 26.Caçador NC, Capizzani CP da C, Torres LAGMM, Galetti R, Ciofu O, Darini AL da C, et al. Adaptation of Pseudomonas aeruginosa to the chronic phenotype by mutations in the algTmucABD operon in isolates from Brazilian cystic fibrosis patients. PLOS ONE. 2018;13(11):e0208013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira AG, Leão RS, Carvalho-Assef APD, da Silva ÉADSR, de Cássia Firmida M, Folescu TW, et al. Low-level resistance and clonal diversity of Pseudomonas aeruginosa among chronically colonized cystic fibrosis patients. APMIS. 2015;123(12):1061–8. [DOI] [PubMed] [Google Scholar]
- 28.Silbert S, Barth AL, Sader HS. Heterogeneity of Pseudomonas aeruginosa in Brazilian cystic fibrosis patients. J Clin Microbiol. 2001;39(11):3976–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stehling EG, Leite DS, Silveira WD. Molecular typing and biological characteristics of Pseudomonas aeruginosa isolated from cystic fibrosis patients in Brazil. Braz J Infect Dis Off Publ Braz Soc Infect Dis. 2010;14(5):462–7. [PubMed] [Google Scholar]
- 30.Almeida MM, Freitas MT, Folescu TW, Firmida MC, Carvalho-Assef APD, Marques EA, et al. Carbapenem-Resistant Pseudomonas aeruginosa in Chronic Lung Infection: Current Resistance Profile and Hypermutability in Patients with Cystic Fibrosis. Curr Microbiol. 2021;78(2):696–704. [DOI] [PubMed] [Google Scholar]
- 31.da Silva LVRF, de A Ferreira F, Reis FJC, de Britto MCA, Levy CE, Clark O, et al. Pseudomonas aeruginosa infection in patients with cystic fibrosis: scientific evidence regarding clinical impact, diagnosis, and treatment. J Bras Pneumol Publicaça̋o Of Soc Bras Pneumol E Tisilogia. 2013;39(4):495–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brazil. Conass Informa n. 75/2024 – Publicada a Portaria Conjunta SAES/SECTICS n. 5 que aprova o Protocolo Clínico e Diretrizes Terapêuticas da Fibrose Cística. 2024. Available from: https://www.conass.org.br/conass-informa-n-75-2024-publicada-a-portaria-conjunta-saes-sectics-n-5-que-aprova-o-protocolo-clinico-e-diretrizes-terapeuticas-da-fibrose-cistica/. Cited 2024 Nov 30.
- 33.Pereira C, Neder J. Diretrizes para Testes de Função Pulmonar. Vol. Vol 28 (Suppl 3). Sociedade Brasileira de Pneumologia e Tisiologia; 2002.
- 34.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 34th ed. Malvern, PA: Clinical and Laboratory Standards Institute; 2024. (CLSI supplement M100).
- 35.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2012;18(3):268–81. [DOI] [PubMed] [Google Scholar]
- 36.Mulet X, Cabot G, Ocampo-Sosa AA, Domínguez MA, Zamorano L, Juan C, et al. Biological markers of Pseudomonas aeruginosa epidemic high-risk clones. Antimicrob Agents Chemother. 2013;57(11):5527–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finnan S, Morrissey JP, O’Gara F, Boyd EF. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J Clin Microbiol. 2004;42(12):5783–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu H, Conibear TCR, Bandara R, Aliwarga Y, Stapleton F, Willcox MDP. Type III secretion system-associated toxins, proteases, serotypes, and antibiotic resistance of Pseudomonas aeruginosa isolates associated with keratitis. Curr Eye Res. 2006;31(4):297–306. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigues YC, Furlaneto IP, Maciel AHP, Quaresma AJPG, de Matos ECO, Conceição ML, et al. High prevalence of atypical virulotype and genetically diverse background among Pseudomonas aeruginosa isolates from a referral hospital in the Brazilian Amazon. PLoS ONE. 2020;15(9):e0238741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nolasco-Romero CG, Prado-Galbarro FJ, Jimenez-Juarez RN, Gomez-Ramirez U, Cancino-Díaz JC, López-Marceliano B, et al. The exoS, exoT, exoU and exoY Virulotypes of the type 3 secretion system in multidrug resistant pseudomonas aeruginosa as a death risk factor in pediatric patients. Pathogens. 2024;13(12):1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol. 2004;42(12):5644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dos Santos PAS, Rodrigues YC, Marcon DJ, Lobato ARF, Cazuza TB, Gouveia MIM, et al. Endemic high-risk clone ST277 is related to the spread of SPM-1-producing pseudomonas aeruginosa during the COVID-19 pandemic period in Northern Brazil. Microorganisms. 2023;11(8):2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carriço JA. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics. 2012;13:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cystic Fibrosis Foundation. Patient Registry 2021 Annual Data Report. Bethesda, Maryland: Cystic Fibrosis Foundation; 2022. [Google Scholar]
- 46.Zolin A, Adamoli A, Bakkeheim E, van Rens J et al. Annual Report 2022. European Cystic Fibrosis Society; 2024.
- 47.Langton Hewer SC, Smith S, Rowbotham NJ, Yule A, Smyth AR. Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis. Cochrane Database Syst Rev. 2023;6(6):CD004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarro J, Rainisio M, Harms HK, Hodson ME, Koch C, Mastella G, et al. Factors associated with poor pulmonary function: cross-sectional analysis of data from the ERCF. European Epidemiologic Registry of Cystic Fibrosis. Eur Respir J. 2001;18(2):298–305. [DOI] [PubMed] [Google Scholar]
- 49.Malhotra S, Hayes D, Wozniak DJ. Cystic Fibrosis and Pseudomonas aeruginosa: the Host-Microbe Interface. Clin Microbiol Rev. 2019;32(3):e00138-e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerem E, Konstan MW, De Boeck K, Accurso FJ, Sermet-Gaudelus I, Wilschanski M, et al. Ataluren for the treatment of nonsense-mutation cystic fibrosis: a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Respir Med. 2014;2(7):539–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thornton CS, Parkins MD. Microbial Epidemiology of the Cystic Fibrosis Airways: Past, Present, and Future. Semin Respir Crit Care Med. 2023;44(2):269–86. [DOI] [PubMed] [Google Scholar]
- 52.Hu H, Harmer C, Anuj S, Wainwright CE, Manos J, Cheney J, et al. Type 3 secretion system effector genotype and secretion phenotype of longitudinally collected Pseudomonas aeruginosa isolates from young children diagnosed with cystic fibrosis following newborn screening. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2013;19(3):266–72. [DOI] [PubMed] [Google Scholar]
- 53.Reynolds D, Kollef M. The Epidemiology and Pathogenesis and Treatment of Pseudomonas aeruginosa Infections: An Update. Drugs. 2021;81(18):2117–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh J, Hunt S, Simonds S, Boyton C, Middleton A, Elias M, et al. The changing epidemiology of pulmonary infection in children and adolescents with cystic fibrosis: an 18-year experience. Sci Rep. 2024;14(1):9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vitiello A, Blasi F, Sabbatucci M, Zovi A, Miele F, Ponzo A, et al. The Impact of Antimicrobial Resistance in Cystic Fibrosis. J Clin Med. 2024;13(6):1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perikleous EP, Gkentzi D, Bertzouanis A, Paraskakis E, Sovtic A, Fouzas S. Antibiotic Resistance in Patients with Cystic Fibrosis: Past, Present, and Future. Antibiotics. 2023;12(2):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonyadi P, Saleh NT, Dehghani M, Yamini M, Amini K. Prevalence of antibiotic resistance of Pseudomonas aeruginosa in cystic fibrosis infection: A systematic review and meta-analysis. Microb Pathog. 2022;165:105461. [DOI] [PubMed] [Google Scholar]
- 58.Rossi E, La Rosa R, Bartell JA, Marvig RL, Haagensen JAJ, Sommer LM, et al. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol. 2021;19(5):331–42. [DOI] [PubMed] [Google Scholar]
- 59.Dos Santos PAS, Silva MJA, Gouveia MIM, Lima LNGC, Quaresma AJPG, De Lima PDL, et al. The prevalence of metallo-beta-lactamese-(MβL)-Producing Pseudomonas aeruginosa Isolates in Brazil: a systematic review and meta-analysis. Microorganisms. 2023;11(9):2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bradbury RS, Roddam LF, Merritt A, Reid DW, Champion AC. Virulence gene distribution in clinical, nosocomial and environmental isolates of Pseudomonas aeruginosa. J Med Microbiol. 2010;59(Pt 8):881–90. [DOI] [PubMed] [Google Scholar]
- 61.Sawa T, Shimizu M, Moriyama K, Wiener-Kronish JP. Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: a review. Crit Care. 2014;18(6):668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elmouaden C, Laglaoui A, Ennanei L, Bakkali M, Abid M. Virulence genes and antibiotic resistance of Pseudomonas aeruginosa isolated from patients in the Northwestern of Morocco. J Infect Dev Ctries. 2019;13(10):892–8. [DOI] [PubMed] [Google Scholar]
- 63.Mitov I, Strateva T, Markova B. Prevalence of virulence genes among bulgarian nosocomial and cystic fibrosis isolates of pseudomonas aeruginosa. Braz J Microbiol. 2010;41(3):588–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo M, Li S, Luo W. Comparative analysis of antibiotic susceptibility patterns and clinical features of mucoid and non-mucoid Pseudomonas aeruginosa infections: a retrospective study. Front Public Health. 2024;12. Available from: https://www.frontiersin.org/journals/public-health/articles/10.3389/fpubh.2024.1333477/full. Cited 2024 Nov 30. [DOI] [PMC free article] [PubMed]
- 65.Chichón G, López M, de Toro M, Ruiz-Roldán L, Rojo-Bezares B, Sáenz Y. Spread of pseudomonas aeruginosa ST274 clone in different niches: resistome, virulome, and phylogenetic relationship. Antibiot Basel Switz. 2023;12(11):1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Del Barrio-Tofiño E, López-Causapé C, Oliver A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents. 2020;56(6):106196. [DOI] [PubMed] [Google Scholar]
- 67.Kainuma A, Momiyama K, Kimura T, Akiyama K, Inoue K, Naito Y, et al. An outbreak of fluoroquinolone-resistant Pseudomonas aeruginosa ST357 harboring the exoU gene. J Infect Chemother Off J Jpn Soc Chemother. 2018;24(8):615–22. [DOI] [PubMed] [Google Scholar]
- 68.Strateva T, Stratev A, Peykov S. Genomic insights into vietnamese extended-spectrum β-lactamase-9-producing extensively drug-resistant pseudomonas aeruginosa isolates belonging to the high-risk clone ST357 obtained from bulgarian intensive care unit patients. Pathog Basel Switz. 2024;13(9):719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Freire MP, Camargo CH, Yamada AY, Nagamori FO, Reusing Junior JO, Spadão F, et al. Critical points and potential pitfalls of outbreak of IMP-1-producing carbapenem-resistant Pseudomonas aeruginosa among kidney transplant recipients: a case-control study. J Hosp Infect. 2021;115:83–92. [DOI] [PubMed] [Google Scholar]
- 70.Cabot G, Ocampo-Sosa AA, Domínguez MA, Gago JF, Juan C, Tubau F, et al. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother. 2012;56(12):6349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Balero de Paula S, Cayô R, Streling AP, Silva Nodari C, Pereira Matos A, Eches Perugini MR, et al. Detection of blaVIM-7 in an extensively drug-resistant Pseudomonas aeruginosa isolate belonging to ST1284 in Brazil. Diagn Microbiol Infect Dis. 2017;89(1):80–2. [DOI] [PubMed] [Google Scholar]
- 72.Baleivanualala SC, Matanitobua S, Soqo V, Smita S, Limaono J, Sharma SC, et al. Molecular and clinical epidemiology of carbapenem resistant Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacterales in Fiji: a multicentre prospective observational study. Lancet Reg Health – West Pac [Internet]. 2024 Jun 1;47. Available from: https://www.thelancet.com/journals/lanwpc/article/PIIS2666-6065(24)00089-0/fulltext. Cited 2024 Nov 30. [DOI] [PMC free article] [PubMed]
- 73.Cabrera R, Fernández-Barat L, Vázquez N, Alcaraz-Serrano V, Bueno-Freire L, Amaro R, et al. Resistance mechanisms and molecular epidemiology of Pseudomonas aeruginosa strains from patients with bronchiectasis. J Antimicrob Chemother. 2022;77(6):1600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deroche L, Aranzana-Climent V, Rozenholc A, Prouvensier L, Darnaud L, Grégoire N, et al. Characterization of Pseudomonas aeruginosa resistance to ceftolozane-tazobactam due to ampC and/or ampD mutations observed during treatment using semi-mechanistic PKPD modeling. Antimicrob Agents Chemother. 2023;67(10):e00480-e523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Urbanowicz P, Izdebski R, Baraniak A, Żabicka D, Hryniewicz W, Gniadkowski M. Molecular and genomic epidemiology of VIM/IMP-like metallo-β-lactamase-producing Pseudomonas aeruginosa genotypes in Poland. J Antimicrob Chemother. 2021;76(9):2273–84. [DOI] [PubMed] [Google Scholar]
- 76.Ramos MS, Furlan JPR, Dos Santos LDR, Rosa R da S, Savazzi EA, Stehling EG. Patterns of antimicrobial resistance and metal tolerance in environmental Pseudomonas aeruginosa isolates and the genomic characterization of the rare O6/ST900 clone. Environ Monit Assess. 2023;195(6):713. [DOI] [PubMed] [Google Scholar]
- 77.Burkett A, Vandemheen KL, Giesbrecht-Lewis T, Ramotar K, Ferris W, Chan F, et al. Persistency of Pseudomonas aeruginosa in sputum cultures and clinical outcomes in adult patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2012;31(7):1603–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The MLST genotyping data generated and analyzed during the current study are available in the repository, PubMLST—Pseudomonas aeruginosa isolates database [https://pubmlst.org/bigsdb?db = pubmlst_paeruginosa_isolates]”. Further inquiries must be addressed to the corresponding authors.