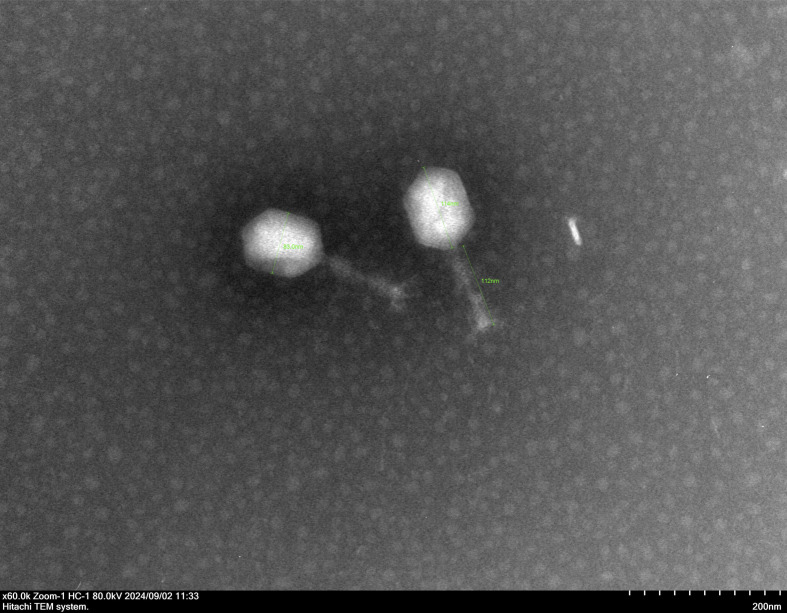ABSTRACT
We describe the genome of a lytic phage isolated from sewage, which is capable of lysing ST2 KL104-type carbapenem-resistant Acinetobacter baumannii strains. The genome is 167,208 bp in length, has a guanine-cytosine (GC) content of 37%, and includes 266 protein-coding sequences and five tRNAs.
KEYWORDS: Acinetobacter baumannii, carbapenem resistance, capsular polysaccharide, phage, phage therapy
ANNOUNCEMENT
Carbapenem-resistant Acinetobacter baumannii (CRAb) has been designated by the World Health Organization as a critical pathogen of concern (1). Treatment options for CRAb are severely limited (2, 3), making phage therapy a promising alternative (4). While phages targeting sequence type 2 (ST2) and capsular type KL2 CRAb have been reported (5–7), no phages specific to ST2 KL104 CRAb have been described. Expanding the library of lytic phages for precise targeting of specific strain types is of significant clinical and practical importance. Here, we present the genome of phi1_092060, a phage capable of effectively lysing ST2 KL104 CRAb clinical isolates.
In July 2024, phi1_092060 was isolated from untreated sewage at the wastewater treatment plant of West China Hospital using the CRAb clinical strain 091006 (accession no. JBLLSI000000000), following the methodology described in previous studies (8). Strain 091006 belongs to sequence type 2 and capsular type KL104. Individual phage plaques were purified three times to ensure phage purity. Genomic DNA of phi1_092060 was extracted from purified phage particles using the Phage DNA Isolation Kit (Norgen Biotek, Thorold, Canada) according to the manufacturer’s instructions. Sequencing libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA), and sequencing was performed on the HiSeq X10 platform. Raw sequencing reads underwent quality control with Trimmomatic v0.39 (9) to remove adapter sequences and discard reads shorter than 130 bases. Genome assembly was performed using Unicycler v0.5.0 (10). Contamination was assessed using CheckV, and non-phage contigs were discarded (11). Genome annotation was conducted using Pharokka v1.7.2 (12), and BLAST was used to identify the phage with the highest overall DNA similarity (identity × coverage) (13). Unless specified otherwise, the software was run with default parameters.
The genome sequencing of phi1_092060 produced 4,217,862 pairs of 150 bp reads, totaling 1.27 Gb (accession no. SRR32014619). The genome of phi1_092060 is 167,208 bp in length, with a GC content of 37%, 266 predicted coding sequences (CDSs), and five tRNAs. BLAST analysis showed that phi1_092060 shares the highest DNA similarity (96.38%, identity × coverage) with Acinetobacter phage AB-Navy97 (accession no. OL770261.1), which belongs to the genus Lazarusvirus of the family Straboviridae within the class Caudoviricetes.
Negative staining of the phi1_092060 phage particles was performed using 2.0% (wt/vol) uranyl acetate. The morphology of phi1_092060 was observed under a Hitachi transmission electron microscope (Hitachi, Japan) at an accelerating voltage of 80 kV. phi1_092060 features a polyhedral head structure, approximately 83 nm in diameter, and a long, rod-shaped, contractile tail approximately 112 nm in length (Fig. 1). The host range of phi1_092060 was determined as described previously (14). phi1_092060 was able to lyse ST2-KL52, ST2-KL104, and ST195-KL45 CRAb isolates. No known antimicrobial resistance or virulence genes were identified in the phi1_092060 genome using the CARD v4.0.0 (15) and VFDB 2025 (16) databases. The high BACPHLIP (17) score (83.7%) suggested a virulent lifestyle for phi1_092060. As such, phi1_092060 meets the standards for clinical phage therapy applications, thereby expanding the phage arsenal against difficult-to-treat CRAb pathogens.
Fig 1.
The morphology of phage phi1_092060 under transmission electron microscopy. The scale was 200 nm.
ACKNOWLEDGMENTS
The work was supported by the National Key Research and Development Program of China (grant numbers: 2023YFC2308800 and 2024YFE0106200) and grants from the West China Hospital of Sichuan University (1.3.5 project for disciplines of excellence, grant number: ZYGD22001).
Contributor Information
Zhiyong Zong, Email: zongzhiy@scu.edu.cn.
John J. Dennehy, Department of Biology, Queens College, Queens, New York, USA
DATA AVAILABILITY
The complete genome sequence of Acinetobacter baumannii phage phi1_092060 has been deposited in GenBank under accession number PQ885451.1 and Sequence Read Archive (SRA) accession number SRR32014619. The version described in this paper is the first version.
REFERENCES
- 1. World Health Organization . 2024. WHO bacterial priority pathogens list, 2024: bacterial pathogens of public health importance, to guide research, development, and strategies to prevent and control antimicrobial resistance. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed. [Google Scholar]
- 2. Shields RK, Paterson DL, Tamma PD. 2023. Navigating available treatment options for carbapenem-resistant Acinetobacter baumannii-calcoaceticus complex infections. Clin Infect Dis 76:S179–S193. doi: 10.1093/cid/ciad094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iovleva A, Fowler VG, Doi Y. 2025. Treatment approaches for carbapenem-resistant Acinetobacter baumannii infections. Drugs (Abingdon Engl) 85:21–40. doi: 10.1007/s40265-024-02104-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koncz M, Stirling T, Hadj Mehdi H, Méhi O, Eszenyi B, Asbóth A, Apjok G, Tóth Á, Orosz L, Vásárhelyi BM, et al. 2024. Genomic surveillance as a scalable framework for precision phage therapy against antibiotic-resistant pathogens. Cell 187:5901–5918. doi: 10.1016/j.cell.2024.09.009 [DOI] [PubMed] [Google Scholar]
- 5. Evseev PV, Sukhova AS, Tkachenko NA, Skryabin YP, Popova AV. 2024. Lytic capsule-specific Acinetobacter bacteriophages encoding polysaccharide-degrading enzymes. Viruses 16:771. doi: 10.3390/v16050771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Soontarach R, Srimanote P, Arechanajan B, Nakkaew A, Voravuthikunchai SP, Chusri S. 2024. Characterization of a novel bacteriophage endolysin (LysAB1245) with extended lytic activity against distinct capsular types associated with Acinetobacter baumannii resistance. PLoS One 19:e0296453. doi: 10.1371/journal.pone.0296453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soontarach R, Srimanote P, Enright MC, Blundell-Hunter G, Dorman MJ, Thomson NR, Taylor PW, Voravuthikunchai SP. 2022. Isolation and characterisation of bacteriophage selective for key Acinetobacter baumannii capsule chemotypes. Pharmaceuticals (Basel) 15:443. doi: 10.3390/ph15040443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li J, Fang Q, Luo H, Feng Y, Feng Y, Zong Z. 2025. “Sichuanvirus”, a novel bacteriophage viral genus, able to lyse carbapenem-resistant Klebsiella pneumoniae. BMC Microbiol 25:17. doi: 10.1186/s12866-024-03736-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC. 2021. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol 39:578–585. doi: 10.1038/s41587-020-00774-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bouras G, Nepal R, Houtak G, Psaltis AJ, Wormald PJ, Vreugde S. 2023. Pharokka: a fast scalable bacteriophage annotation tool. Bioinformatics 39:btac776. doi: 10.1093/bioinformatics/btac776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 14. Fang Q, Feng Y, McNally A, Zong Z. 2022. Characterization of phage resistance and phages capable of intestinal decolonization of carbapenem-resistant Klebsiella pneumoniae in mice. Commun Biol 5:48. doi: 10.1038/s42003-022-03001-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, Edalatmand A, Petkau A, Syed SA, Tsang KK, et al. 2023. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res 51:D690–D699. doi: 10.1093/nar/gkac920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou S, Liu B, Zheng D, Chen L, Yang J. 2025. VFDB 2025: an integrated resource for exploring anti-virulence compounds. Nucleic Acids Res 53:D871–D877. doi: 10.1093/nar/gkae968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hockenberry AJ, Wilke CO. 2021. BACPHLIP: predicting bacteriophage lifestyle from conserved protein domains. PeerJ 9:e11396. doi: 10.7717/peerj.11396 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of Acinetobacter baumannii phage phi1_092060 has been deposited in GenBank under accession number PQ885451.1 and Sequence Read Archive (SRA) accession number SRR32014619. The version described in this paper is the first version.