Abstract
Innate lymphoid cells (ILC) distribution and compartmentalization in human lymphoid tissues are incompletely described. Through combined multiplex immunofluorescence, multispectral imaging, and advanced computer vision methods, we provide a map of ILCs at the whole-slide single-cell resolution level, and study their proximity to T helper (Th) cells. The results show that ILC2 predominates in thymic medulla; by contrast, immature Th cells prevail in the cortex. Unexpectedly, we find that Th2-like and Th17-like phenotypes appear before complete T cell receptor gene rearrangements in these immature thymocytes. In the periphery, ILC2 are more abundant in lymph nodes and tonsils, penetrating lymphoid follicles. NK cells are uncommon in lymphoid tissues but abundant in the spleen, whereas ILC1 and ILC3 predominate in the ileum and appendix. Under pathogenic conditions, a deep perturbation of both ILC and Th populations is seen in follicular lymphoma compared with non-neoplastic conditions. Lastly, all ILCs are preferentially in close proximity to their Th counterparts. In summary, our histopathology tool help present a spatial mapping of human ILCs and Th cells, in normal and neoplastic lymphoid tissues.
Subject terms: Innate lymphoid cells, Systems analysis, Lymphopoiesis, Gene regulation in immune cells
Innate lymphoid cells (ILC) are an important modulator of immunity in many tissues, including lymphoid tissues, but their spatial information is still scarce. Here, the authors use multiplexed and multispectral imaging methods to provide a spatial map of ILCs in human thymus, spleen, lymph nodes, intestinal lymphoid tissue, and lymphoma, and observe ILC/T helper cells colocalization.
Introduction
Innate lymphoid cells (ILCs) are tissue resident cells that play an important role in host protection, tissue remodeling and homeostasis1–6. ILCs are highly plastic modulators of immunity3 and represent a first-line defense system against invading pathogens at barrier surfaces, including mucosal surfaces and skin7,8. They adapt to the organ microenvironment9 and establish close interactions with hematopoietic cells but also with stromal cells, epithelial cells, neurons, amongst other cell types2. ILCs represent transcriptional and functional equivalents of T lymphocytes but lack clonally distributed antigen-specific receptors2,10,11 and are instead activated by cytokines7. Based on the cytokines they produce and transcription factors they express, ILCs have distinct functional outputs9,10,12,13. The discovery of the different ILC subsets, their functional similarity to the T lymphocytes of the adaptive immune system and their potential as therapeutic target in different types of disease including tumors14–17, has generated great interest in the mechanisms of ILC development3,9,18 and in their tissue distribution13,19–23. Although the function of circulating innate immune cells has been studied extensively24–26, our understanding of tissue-based innate immunity is limited, particularly in humans27. Besides limitations in terms of access to human peripheral tissues, the discrimination of ILCs is particularly complicated on a biological level. Depending on the state of cell maturity and activation, and the microenvironment in which they reside, ILC phenotype and function change, thus complicating their classification9,10,27,28. It is generally accepted that ILCs can be categorized into three groups. Group 1 comprises ILCs that depend on EOMES and/or TBX21 (T-Bet) and produce IFNγ in response to the driver cytokine interleukin IL-12. Group 2 ILCs proliferate in response to IL-25 and IL-33, and produce preferentially the type 2 cytokines IL-5 and IL-13, and require GATA-binding protein 3 (GATA-3) and retinoic acid receptor-related orphan receptor-α (RORα) for correct function and development. Group 3 ILCs produce IL-17, IL-22, or both upon stimulation with IL-23 and IL-1b and depend on RORγT (RORC) and AHR (Aryl Hydrocarbon Receptor) for function and development2,28–31. ILC nomenclature has since been reviewed and standardized28,32,33, which led to the recognition of five groups of ILCs now approved by the International Union of Immunological Societies: NK cells, ILC1, ILC2, ILC3, and LTi (Lymphoid Tissue inducer)9. ILCs share phenotypic and functional characteristics with CD4+ T helper cells, where ILC1 mirror Th1, ILC2 mirror Th2, and ILC3 mirror Th17 cells. NK cells, originally assigned to the innate immune system, are cells with adaptive properties34 and mirror the CD8+ cytotoxic T cells of the adaptive immune system9,35 through specific gene expression profiles5,24,26. In addition to their organ-specific signatures and heterogeneity24,36, NK cells are particularly difficult to discriminate from ILC1 cells due to their high degree of similarity and plasticity5.
While ILCs are functionally important in health and disease6, their detection in human tissues remains problematic, and to date, little is known about the topographic characteristics of ILCs throughout human tissues. Our current knowledge is based on global morphological studies13,19–23, using different positive and negative selection criteria, and on flow cytometry and single-cell RNAseq experiments, which unfortunately lack spatial information. Among numerous clinical applications, a comprehensive topography of human ILCs in their biological context is vital to study inflammatory disease onset and treatment follow-up16,17,37, immunosurveillance in cancer patients16,29,38,39, risk management in obesity40, pathological pregnancies41, and innate immunity early in life to determine ILC targeting preventive strategies for inflammatory disease42. To our knowledge, no study has described the ILCs’ spatial distribution in human lymphoid tissues with accurate cellular mapping by using standard histopathology techniques. As already reported28,43, the heterogeneity and tissue-specific transcriptional imprinting of human ILCs confirm that this approach is very challenging. The high plasticity of human ILCs was recently confirmed and further elucidated by Mazzurana et al.44, highlighting the complexity of defining discriminative marker combinations. Phenotypic analysis of human ILCs by flow cytometry is well described45 and can serve as a reference. ILC subtypes are all positive for CD45 and all lack protein surface expression for rearranged antigen-specific receptors and lineage markers of other leukocyte cell populations2,9,10,20,27,28,35,43,45,46. Lineage markers, which are typically present on T, B and dendritic cells (DC), serve as exclusion markers for ILCs (CD3− CD14− CD19− FcεRIα− CD123− CD11c− BDCA3−). Mature ILCs can be further classified as IL-7Rα (CD127) negative (most NK, intra-epithelial ILC1, small subsets of ILC2) and IL-7Rα (CD127) positive cells (some NK, ILC1, most ILC2, ILC3, LTi)2,9,27,44,47,48. Several groups proposed to further differentiate ILC cell populations with transcription factors (TF) or receptors on which the different ILC subtypes depend to reach their adequate effector state19–21,49. As clearly stated by Klose and Artis2, the expression of lineage-specific TF is not fully selective and previously reported attempts for human ILC in situ characterization have proposed different methodologies and inclusion/exclusion markers for ILC subtype characterization19–21,49–51.
Here, we show a detailed and accurate spatial mapping of ILC compartmentalization in human lymphoid organs, in relation to their T helper counterparts by proximity analysis and mature B and T cells and immature thymic T cells. The results show that ILCs are in close proximity to their T helper counterparts and that a large IL-7Rαpool is present in the studied lymphoid tissues, especially in the thymus, lymph node and tonsil, where ILC2 are predominant. NK cells are abundant in spleen, whereas ILC1 and ILC3 are the principal ILC-phenotypes of the appendix and ileum. We propose a new tool, using conventional automated staining strategies and standard fluorescent whole-slide imaging and analysis methods, to study ILCs and Ths in reactive and neoplastic conditions, which can be instrumental to elucidate both the protective and adverse functions of these highly responsive lymphoid cells.
Results
Whole-slide conventional histopathology ILC mapping
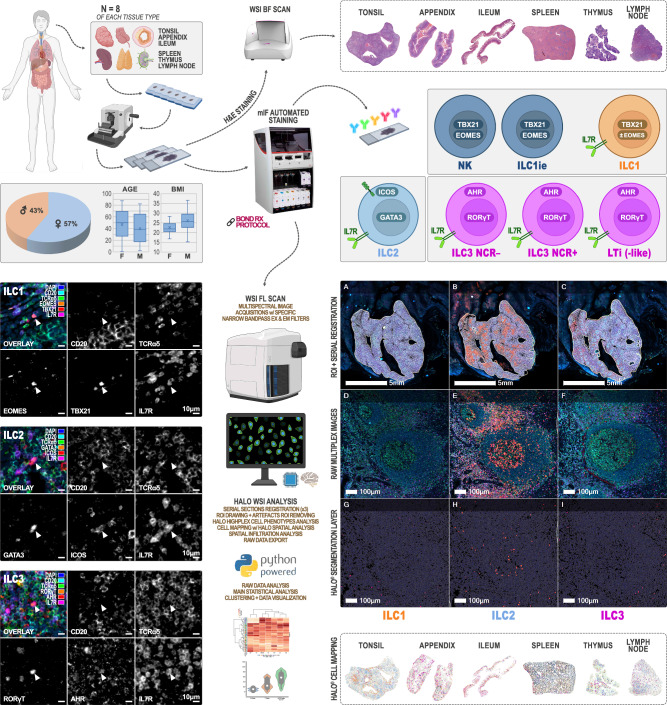
To study the detailed topography of ILCs, we developed multiplex immunofluorescent (mIF) panels using conventional, automated, and validated staining technologies, in combination with standardized whole-slide fluorescent imaging and analysis workflows. We studied a primary lymphoid organ, the thymus, and different secondary lymphoid organs such as lymph node, spleen and tonsil. Additionaly, we studied the intestinal mucosa, and Peyer’s patches in the ileum and mucosa-associated lymphoid tissue (MALT) in appendix. For each lymphoid tissue type, we studied 8 independent biological specimens. On the basis of an extensive literature review and complementary experiments (summarized in Supplementary Methods 4 of the Supplementary Information file), we have defined three discriminative mIF panels (Methods and Fig. 1) for the ILC phenotypes based on TF expression combined with proteins involved in environmental responses and activation (AF)22,28,43,52–54, while being aware however of the phenotypic and functional plasticity of these cells2,5,24.
Fig. 1. Wet and dry lab work up for high-resolution computational mapping of Innate Lymphoid Cells (ILCs) in human primary and secondary lymphoid organs.
After tissue selection (n = 8 independent biological samples per lymphoid tissue type), 4 to 6 serial FFPE slides were cut at 4 µm. The first slide was stained with H&E for morphological quality control. mIF was performed for the 3 panels (NK/ILC1ie + ILC1, ILC2, and ILC3) in parallel for each tissue sample (7 batches, 1 staining cycle of 3 mIF per lymphoid tissue type) on the BOND RX automated staining platform (Leica Biosystems). 6-channels multispectral fluorescence imaging was performed using a Zeiss AxioScan Z1 whole-slide scanner. Object-oriented whole-slide image analysis was carried out using the Halo framework (Indica Labs). Each image was manually annotated by a certified pathologist with three regions of interest (ROI: “interface”, “extra-interface”, and “interface + extra-interface”). In tonsil, appendix, ileum, lymph node, and follicular lymphoma images, the “interface” and “extra-interface” ROIs matched respectively the follicular and extra-follicular zones, the white and red pulp in the spleen, and the medulla and cortex in thymus. Morphometric features and OPAL dye-intensities were quantified, and each object was tagged and classified for a specific phenotype based on its marker content. Spatial analysis and mapping was based on their X-Y location coordinates. Raw data were exported from the Halo database as.csv text files to be analyzed and visualized using Panda, Matplotlib, and Seaborn (Python packages). Partially created in BioRender. Bezombes, C. (2025) https://BioRender.com/shbs5qf.
The experiments were performed in 7 staining batches (1 staining batch equals 3 mIF ILC-panels for 1 lymphoid tissue type) including 8 independent biological samples (different patients) per tissue type. We submitted the three 4 µm serial whole tissue sections of each lymphoid organ (n = 56, Supplementary Table 1, Fig. 1) to mIF assays (Supplementary Methods 1, 2, and 3), after selection based on morphological quality of the corresponding Hematoxylin & Eosin (H&E) slide (Methods). Lineage exclusion was performed using TCRαδ (conventional αβ and γδ T cells, innate like T cells29) and CD20 (B cells) combined with the ILC inclusion marker IL-7Rα (CD127) to ensure ILC enrichment as documented in the Supplementary Methods 4 report22,43,45,52–54. We identified IL-7Rα+ (CD127+) cells as being a pool of ILC1, ILC2, ILC3 NCR± (natural cytotoxicity receptor), and LTi in human FFPE (formalin fixed paraffin embedded) tissue sections. Group 1 ILCs are discriminated from the other ILC subtypes by TBX21 (T-Bet) and EOMES markers. Aware of the findings5,24 and the comprehensive review33 of the group of Vivier E., we accepted a differentiation between NK/ILC1ie and ILC1 based on these more conventional marker combinations13,43,55–58 (TBX21+ EOMES+ for NK/ILC1ie; TBX21+ EOMES± for ILC1) and absence or presence of IL-7Rα+ (CD127+), respectively. For ILC2 differentiation, two additional markers were selected (ICOS and GATA-3), as for ILC3 (RORγT and AHR). Individual ILC phenotypes were thus the following (Methods, Fig. 1 and Supplementary Fig. 1): CD20− TCRαδ− IL-7Rα− TBX21+ EOMES+ for NK/ILC1ie, CD20− TCRαδ− IL-7Rα+ TBX21+ EOMES± for ILC1, CD20− TCRαδ− IL-7Rα+ GATA-3+ ICOS+ for ILC2 and CD20− TCRαδ− IL-7Rα+ RORγT+ AHR+ for ILC3. They were analyzed together with their respective T-helper counterpart: Th1 (CD20− TCRαδ+ TBX21+ EOMES+), Th2 (CD20− TCRαδ+ GATA-3+ ICOS+) and Th17 (CD20− TCRαδ+ RORγT+ AHR+). It should be noted that Th1 cell density as defined here might be biased by some effector memory CD8+ T cells (CD8+ Tmem), known to express the same TF59,60. We considered this bias negligible after quantifying Tmem as CD8+ TCRαδ+ TBX21+ EOMES+ (Supplementary Methods 4) and showing extremely low level densities of these cells. The strategy allowed us to study differentiated ILCs and T helper cells in relation to the pool of IL-7Rα+ cells (TCRαδ− CD20−) further referred to as IL-7Rαpool, undifferentiated IL-7Rα+ cells (TCRαδ− CD20− TF−/AF−) further referred to as IL-7Rαundiff, B cells (CD20+ TCRαδ−) and T cells (TCRαδ+ CD20−).
To study the thymus, the primary function of which is the differentiation and maturation of T lymphocytes in coordination with ILC development61–63, we needed to adjust our ILC identification strategy. Using the initially defined mIF panels (Supplementary Fig. 1A, B: classic mIF), we confirmed a very low B cell density in this organ (0.62 ± 0.01/mm2) and concluded that exclusion of anti-CD20 antibody from the mIF panels would not interfere with correct identification of the ILCs, nor Th phenotypes. To avoid TCRαδ− thymocytes from contaminating ILC counts, we included anti-terminal deoxynucleotidyl transferase (TdT) instead (Supplementary Fig. 1B: alternative “TdT” mIF panel), to identify thymocytes64,65. With this strategy we were able to identify: TCRαδ+ TdT− T cells (primarily post DN4 stage according to Sun et al.64 and Matsutani et al.66) and further referred to as mature T cells, and TCRαδ− TdT+ T cells (pre DN4 stage according to Sun et al.64 and Matsutani et al.66), further referred to as immature T cells. ILCs were identified as TCRαδ− TdT− IL-7Rα+, except for the NK/ILC1ie phenotype not always exhibiting IL-7Rα expression24 (Supplementary Methods 4).
Inspired by Hue et al.67 and Ettersperger and colleagues68, we explored the T cell-like innate intra-epithelial lymphocytes (IE-ILC) expressing intracytoplasmic CD3ε (icCD3ε+) in intestinal mucosa-associated lymphoid tissue. To do so, we replaced CD20 by CD3ε in the initial (classic) mIF panel (Supplementary Fig. 1B: alternative “CD3” mIF panel) and determined the following ILC phenotypes: icCD3ε+ ILC1-like cells (CD3ε+ TCRαδ− IL-7Rα+ TBX21+ EOMES±), icCD3ε+ NK/ieILC1-like cells (CD3ε+ TCRαδ− IL-7Rα− TBX21+ EOMES+) and icCD3ε+ ILC3-like cells (CD3ε+ TCRαδ− IL-7Rα+ RORγT+ AHR+). This alternative mIF panel allowed us to study the T lymphocyte pool (CD3ε+ TCRαδ+) and Th1 (CD3ε+ TCRαδ+ TBX21+ EOMES±) and Th17 phenotypes (CD3ε+ TCRαδ+ RORγT+ AHR+) as well (Supplementary Fig. 1B).
For cell mapping and spatial analyses, we studied 208 whole-tissue 6-channels fluorescent slides in total, resulting in the analysis of 91,257,351 cells in a total area of approximately 9000 mm2 (8,866,305,074 µm2), representing nearly 28 billion of pixels (27,280,938,689). For each of the 8 patients and the 7 selected tissue types, 3 scanned serial sections (classic mIF panels or alternative “TdT” mIF panels depending on the tissue) were analyzed, plus 2 additional serial sections for the ileum (alternative “CD3” mIF panel) (Methods, Fig. 1 and Supplementary Fig. 1). Morphometric features (DAPI-based nuclear detection) and multiple OPAL dye grey-level intensities were quantified allowing detection of several cell phenotypes in lymphoid follicular and extra-follicular regions of tonsil, appendix, ileum, reactive lymph node and follicular lymphoma, and in functionally/morphologically different structures such as white/red pulp of spleen or cortex/medulla in thymus. Each detected object was tagged and classified for a specific phenotype based on its marker content (Supplementary Fig. 1), and spatial analysis was based on their X-Y location coordinates allowing merged spatial plot mapping for each case on registered images (Supplementary Fig. 2) and quantitative distribution analysis (around the ROI interface) of individual cell phenotypes. Cell densities for several phenotypes or percentages relative to specific cell populations were computed and plotted using Panda, Matplotlib, and Seaborn (Python packages, refer to Methods). An Unpaired T-test was performed to compare ILC subtypes detected by Flow Cytometry and mIF. Non-parametric statistical analyses (Mann-Whitney tests) were performed to compare ILC and T helper phenotype densities in follicular lymphoma, tonsil, and lymph node.
Cluster analysis reveals distinct ILC and T helper subset partnership
To better understand the immune architecture and cellular distribution in the different peripheral lymphoid tissues included in this study, we aimed to cluster cell densities of the ILC subsets and of their T helper counterparts with patient annotated data and tissue features (Fig. 2A). Neither gender (42% female and 58% male patients, self-reported) nor age of the patients (Supplementary Table 1) altered the density of different lymphoid cell populations per organ. Our study-population showed an heterogenous age-distribution related to tissue selection (<33 years for thymus, > 64 for ileum), covering a rather wide age distribution of 44 ± 26 years. It is known, however, that innate immunity decreases with age69, thus we would expect an inverse correlation between ILC-density and age22, which needs to be confirmed in a larger cohort. Conversely, we observed tissue-specific clustering, in particular for spleen, which exhibited few ILC2 and Th2 cells, yet a remarkably high frequency of NK/ILC1ie cells, contrasting with other organs analyzed. Other clusters were unrelated to tissue types, but unveiled ILC/T helper cell co-distribution, confirmed by proximity analysis. A close interaction between the two cell populations has been extensively described, where cytokines and activating or inhibiting receptors of ILCs are considered pivotal for regulating the Th immune responses70. ILC1 and ILC3 clustered altogether, which was expected since an ILC1/ILC3 balance is crucial to prevent inflammation and cancer development71. The latter clustered with Th17 cells described to be inhibited by ILC3 through AHR signaling and IL-22 release70,72,73. NK/ILC1ie were associated with Th1 cells, which in turn clustered with the previously mentioned group (Th17, ILC1, ILC3). ILC2, that were clustering with Th2, appeared independent in their distribution from the other Th and ILC subtypes. This finding supports the fact that ILC2, which drives the initiation of type 2 innate immune responses74, colocalize with Th275.
Fig. 2. Cluster analysis reveals distinct ILC and T helper subset partnership.
An important IL-7Rαpool resides within lymphoid organs and ILC2 are the main ILC-subtype penetrating lymphoid follicles (FZ). A A Standardized cell densities clustermap of ILCs and T helper cells in relation to patient characteristics (gender and age range) and tissue types (tonsil, appendix, ileum, lymph node, spleen, and thymus) (n = 8 independent biological samples per lymphoid tissue type). Four clusters as determined using the elbow method and the K-means algorithm: Blue, Green, Beige, Deep red. B B, T, IL-7Rαpool, and ILCs quantification in lymphoid organs. Split-violin plots representing B, T and IL-7Rαpool cell densities (number/mm2) inside (left) and outside (right) of the follicular zone of lymph node, tonsil, appendix and ileum, in the white (left) and red (right) pulp of spleen, and in the medulla (left) and cortex (right) of thymus (n = 8 independent biological samples per lymphoid tissue type). C Split-violin plots representing the percentage of ILC phenotypes (per IL-7Rαpool) inside (left) and outside (right) of the follicular zone of lymph node, tonsil, appendix and ileum, in the white (left) and red (right) pulp of spleen, and in the medulla (left) and cortex (right) of thymus. D–G Classic mIF of tonsil for CD20 (aqua), TCRαδ (green), and IL-7Rα (red). Interface between follicular and extra-follicular zones: dashed line. Scale bar: 100 µm. Exemple of ILC1 (CD20− TCRαδ− IL-7Rα+ TBX21+ EOMES±) (H), ILC2 (CD20− TCRαδ− IL-7Rα+ GATA-3+ ICOS+) (I), and ILC3 (CD20− TCRαδ− IL-7Rα+ RORγT+ AHR+) (J) identified cells. Scale bar: 10 µm. Box-plots within Violin Plots, created using Seaborn in Python, present the median value (white dot, 50th percentile) in between the first quartile (the middle value between “minimum non-outlier” and median (marked as Q1, portrays the 25th percentile) and third quartile (the middle value between “maximum non-outlier” and median (marked as Q3, portrays the 75th percentile)). Source data is provided as a Source Data file.
An important IL-7Rαpool resides within lymphoid organs
In line with the function of the thymus63,66,76, immature T cells were the most represented in this organ (overall density: 2528.81 ± 369.71/mm2), followed by IL-7Rαpool (overall density: 2171.55 ± 284.08/mm2) and mature T cells (overall density: 1800.71 ± 337.03/mm2) (Fig. 2B, Supplementary Table 2). The majority of these mature T cells showed IL-7Rα+ expression (1630.03 ± 325.32/mm2), most likely inducing pro-survival signaling and T cell fate cues63. About 60% of the immature T cells (2528.81 ± 369.71/mm2) had IL-7Rα (1454.88 ± 195.12/mm2), presumably coinciding with the start of IL-7 signaling at DN3-stage together with TCRβ selection63. The immature T cell population described here corresponds to thymocytes entering DN2 stage not going beyond DN4, as deduced from what has been reported previously63. As expected, immature T cells resided mainly in cortical thymus (3143.65 ± 479.87/mm2), whereas mature T cells predominantly occupied the medulla (3829.16 ± 536.54/mm2) (Fig. 2B). IL-7Rαpool reached equal densities in medulla as mature T cells (3317.40 ± 291.81/mm2 and 3829.16 ± 536.54/mm2 respectively) and exceeded mature T cells (1061.98 ± 250.87/mm2), but not immature T cells (3143.65 ± 479.87/mm2) in cortical areas (IL-7Rαpool: 1741.69 ± 282.16/mm2) (Supplementary Table 2).
The spleen, the largest secondary lymphoid organ in the human body, showed the lowest density of IL-7Rαpool of all studied lymphoid tissues (344.10 ± 23.98/mm2) (Fig. 2B, Supplementary Table 3). They were however evenly present in white pulp (WP) and red pulp (RP) (302.60 ± 27.50/mm2 and 347.45 ± 25.24/mm2 respectively), largely dominated by T and B cells, occupying mainly WP regions (T: 3261.82 ± 258.61/mm2 and B: 2002.35 ± 253.65/mm2).
The appendix (Supplementary Table 3), known as an adaptive immunity induction site77, showed an overall variable but dominant T cell population (1175.52 ± 232.21/mm2), slightly overruling the IL-7Rαpool (783.16 ± 86.75/mm2). Although present in extra-follicular areas (B cell density: 497.85 ± 48.48/mm2), B cells were concentrated within the germinal centers of the mucosa-associated lymphoid follicles (5917.33 ± 273.24/mm2) as expected (Fig. 2B). When studying the mucosa and its Peyer’s patches composed of lymphoid follicles in the ileum, the IL-7Rαpool (787.18 ± 119.66/mm2) slightly prevailed over T and B cells (500.51 ± 115.10/mm2 and 309.81 ± 70.68/mm2 respectively) especially in extra-follicular areas. As expected, B cells largely occupied germinal centers (4594.19 ± 399.86/mm2), whereas IL-7Rαpool density (802.24 ± 123.32/mm2) exceeded that of T cells in extra-follicular areas (506.00 ± 120.01/mm2) (Fig. 2B).
In LN, IL-7Rαpool (3011.16 ± 263.58/mm2) was present at comparable densities as T cell (2692.13 ± 299.11/mm2) but largely dominated the B cells (1360.19 ± 103.44/mm2) (Fig. 2B, Supplementary Table 3). Their number doubled as compared to the IL-7Rαpool in tonsil (1501.46 ± 182.31/mm2), whereas T and B cells were equally present in LN compared to tonsil (overall density in tonsil: 2735.40 ± 262.14/mm2 and 1411.54 ± 123.40/mm2, respectively). Spatial distribution study of the IL-7Rαpool, showed that the majority was present in extra-follicular regions of the LN (3058.41 ± 267.69/mm2) while less represented in lymphoid follicles (682.32 ± 152.35/mm2). Outside the follicles, they slightly dominated T (2723.46 ± 306.83/mm2) but especially B cells (1299.09 ± 98.97/mm2). In primary follicles, lacking germinal centers (GC), B cells predominated as expected (4562.38 ± 282.48/mm2) (Supplementary Table 3). In contrast to reactive LN, palatine tonsils were mainly composed of secondary B cell follicles with GC. In palatine tonsil (Fig. 2B, Supplementary Table 3), IL-7Rαpool cells were present at a comparable density (1501.46 ± 182.31/mm2) as B cells (1411.54 ± 123.40/mm2), distributed mainly through extra-follicular regions (1682.53 ± 177.66/mm2) but also present in lymphoid follicles (591.00 ± 97.93/mm2) (Fig. 2B). This compartmentalization has been previously described in mice78 and humans21,79.
Only a small percentage of IL-7Rαpool differentiated into specific ILC-subsets in the human lymphoid tissues examined (Fig. 2C, Supplementary Tables 2 and 3). ILC-specialization (% of IL-7Rαpool) was found to be highest in ileum (26.15 ± 2.03%), appendix (25.75 ± 1.69%) and tonsil (24.43 ± 2.69%) (Fig. 2D–J). Less ILC differentiation was detected in spleen (15.51 ± 1.60%), lymph node (12.41 ± 1.10%) and thymus (6.65 ± 0.65%) (Supplementary Tables 2 and 3, sum ILCs). These results are expected since tonsil and gut-associated lymphoid tissue serve as a protective barrier against inhaled and/or ingested pathogens77,80. ILC2 dominated in lymphoid follicles of tonsil (29.15 ± 5.72%) and lymph node (25.56 ± 6.22%), and in thymic medulla (6.90 ± 1.61%). ILC1 and ILC3 were the most important intra-epithelial (extra-follicular) ILC phenotypes of ileum (9.50 ± 2.26 and 14.76 ± 2.58% respectively) and appendix (9.98 ± 1.64 and 12.63 ± 2.38% respectively). ILC2 was enriched in the lymphoid follicles of Peyer’s patches (9.83 ± 4.02%) and MALT of appendix (8.92 ± 2.07%), accompanied by ILC3 in lymphoid follicles of ileal Peyer’s patches (14.86 ± 3.23%). Finally, ILC1 was the dominant population in red pulp of spleen (10.86 ± 2.68%), at percentages similar to extra-follicular regions of ileum and appendix (9.50 ± 2.26% and 9.98 ± 1.64%, respectively).
Flow cytometry validates histological ILC mapping and confirms an in situ stock of IL-7Rαundiff in tonsil
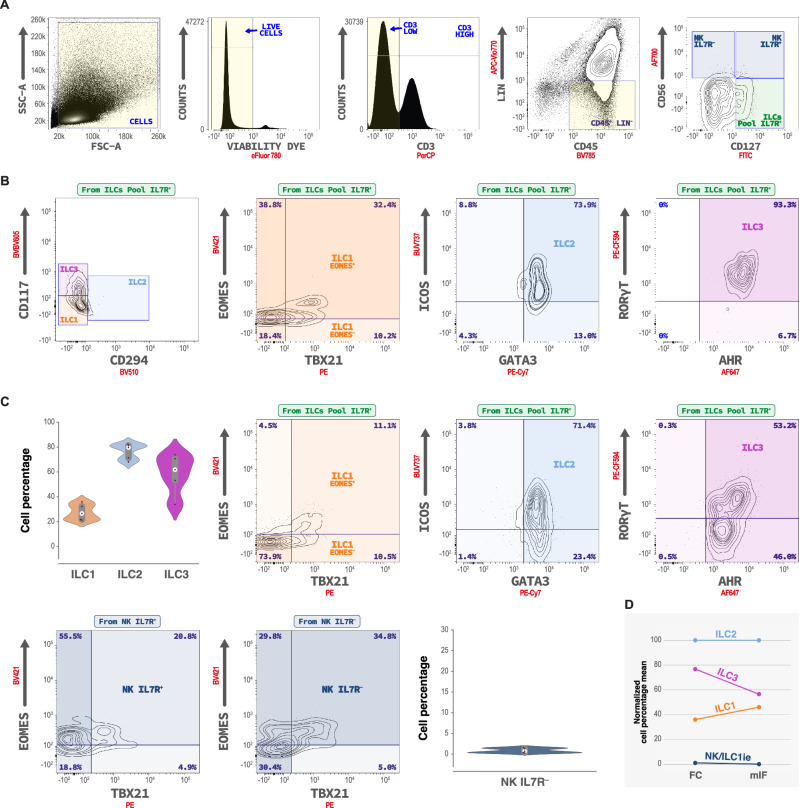
Before moving forward to study the tissue-specific topography of ILCs, we validated our histological gating strategy using flow cytometry (FC) applied to frozen human tonsil tissue (Methods), since this technology was mainly used to study ILC populations until now. We used a gating strategy for ILCs as described by Yudanin et al.22, Björklund et al.43, and for NK cells as described by E. Vivier and colleagues5,24. Following viability assessment, unwanted lineage markers (CD141− CD14− CD11c− CD20− FcεRIα− CD3− CD34− CD123− CD1a−) were excluded and the ILC pool population was defined through the expression of CD45, IL-7Rα and absence of expression of CD56 NK cell marker (Fig. 3A). The different ILC phenotypes, gated following the work of Yudanin et al., confirmed that our histological subset identification strategy (ILC1 (CD20− TCRαδ− IL-7Rα+ TBX21+ EOMES±), ILC2 (CD20− TCRαδ− IL-7Rα+ GATA-3+ ICOS+) and ILC3 (CD20− TCRαδ− IL-7Rα+ RORγT+ AHR+)) captures the same ILC populations (Fig. 3B, C, Supplementary Table 4A). Data confirmed that the identification of ILC1 is the most challenging. Overall ILC percentages were not significantly different in human tonsil, whether determined by mIF or FC. The same proportions of ILC-phenotypes were found in human tonsil, with a predominance of ILC2, followed by ILC3 and ILC1. Both methods confirmed a nearly absence of NK cells (Fig. 3D) in the studied tonsils. NK cells, gated alternatively according to Yudanin et al.22 and Vivier and colleagues5,24, were confirmed to be at very low densities in human tonsil. Altogether, FC confirmed and validated our mIF panels applied on FFPE tissue sections (Supplementary Tables 4A and 5).
Fig. 3. Flow cytometric ILCs identification and quantification in human tonsils (n = 5 independent biological samples performed in a single batch).
A Example of the gating strategy used for identification of ILCs in tonsil, with ILCs defined as lineage negative (Lin−) (CD141− CD14− CD11c− CD20− FcεRIα− CD3− CD34− CD123− CD1a−) and CD45+ CD127(IL-7Rα)+ CD56−. Following viability assessment, unwanted lineage markers were excluded, and ILCs pool population was defined through the expression of IL-7Rα and the absence of expression of CD56 NK cell marker. B ILCs identification according to Crinier A. et al.24, Yudanin et al.22, Bjorklünd et al.43 based on the relative co-expression of CD117 and CD294 (CRTH2) markers. ILC1 were defined as CD117− CRTH2−, ILC2 were defined as CD117± CRTH2+, ILC3 were defined as CD117+ CRTH2−. Co-expression of T-Bet/EOMES, GATA3/ICOS, and AHR/RORγT assessment in each determined ILCs subset. C ILCs identification according to mIF gating strategy. ILCs pool was defined as LIN− CD45+ CD3− CD56− CD127+ and the populations were defined as T-Bet+ EOMES± for ILC1, GATA3+ ICOS+ for ILC2 and RORγT+ AHR+ for ILC3. Violin plot represents the quantification of ILC1, ILC2, and ILC3 subsets according to the mIF gating strategy in tonsil. T-Bet and EOMES expression (“ILC1 profile”) were assessed in NK cells populations either positive or negative for IL-7Rα expression. D FC and mIF quantification obtained values (% relative to IL-7Rα+ population) were compared through normalized means of cell percentages observed. Source data are provided as a Source Data file.
As mentioned above, our data indicate that a large proportion of IL-7Rαpool remains undifferentiated (IL-7Rαundiff), suggesting that a large pool, ready to differentiate upon correct microenvironmental cues, can serve as a local reservoir to produce specific ILC subsets as previously reported81. We therefore used FC data to investigate the phenotype of the IL-7Rαundiff cells (Supplementary Table 4B) using the gating strategy conform to the work of Yudanin et al.22 and following widely accepted guidelines54. We explored the multipotency of tissue-resident ILCs and the presence of ILC precursors (ILCP) according to Lim et al.82, and reviewed by Ruf et al.29. When analyzing the Lin− CD3− CD45+ IL-7Rα+ cells using the FC gating strategy used in tissue (Fig. 3), we identified the presence of GATA-3+ ICOS− cells (mean 15.77 ± 2.17%). Using mIF, we found 32.47 ± 2.13% of CD20− TCRαδ− IL-7Rα+ GATA-3+ cells (Supplementary Methods 4), higher than the ILC2 content (12.06 ± 3.18%). We hypothesized that a common ILC precursor1,32,60 (CHILP) might be present in situ. CHILPs express GATA-3, a TF crucial for ILC development and function83–85. Our results, together with previously reported data, prompted us to investigate GATA-3 expression by the entire Lin− CD3− CD45+ IL-7Rα+ population. 92.29 ± 1.75% of this population showed GATA-3 expression, confirming its importance for ILCs as described by Zhu et al.83. Practically all of these GATA-3+ cells also expressed AHR (98.64 ± 1.03%), a ligand-dependent TF that helps ILCs to sense the environmental cues86. Interestingly, a relatively high but variable proportion of these cells were RORγT positive (56.10 ± 7.12%,). Thus, a relatively large pool of so-called CHILP resides in human tonsil, with the potential to differentiate into any ILC phenotype as proposed by Montaldo87, Lim82 and Scoville88. IL-7Rαundiff includes this precursor population, which is most likely as important as described by other groups9,82. To further explore the phenotype of IL-7Rαundiff, we subsequently investigated CD117+ (c-kit) in Lin− CD3− CD45+ IL-7Rα+82,89. First of all, CD117 was expressed by 27.28 ± 3.50% of the CHILP (Lin− CD3− CD45+ IL-7Rα+ GATA-3+ RORγT+) described above. We additionally confirmed the presence of CD117+ ILCs in human tonsil, representing 19.15 ± 2.13% of the overall Lin− CD3− CD45+ IL-7Rα+ cells. Interestingly, 66.64 ± 10.06% of this CD117+ population represented an ILC3 profile (RORγT+ AHR+). These cells express NKp44 (confirmed at 55.14 ± 9.62%) and might thus represent the more mature90, cytotoxic ILC3 variant. Additionally, practically all of them were GATA3+ (98.93 ± 0.47%), as published91. The other cytotoxicity receptor, NKp46, was barely expressed (14.04 ± 2.63%) as it was the case for TBX21 (3.60 ± 0.63%). Our data indicate that ILCPs are present in situ in human tonsil and that these cells are characterized by the absence of NKp44-expression as suggested by Lim82 (Supplementary Table 4B).
ILC2 predominate in thymus and are mainly located in the medulla
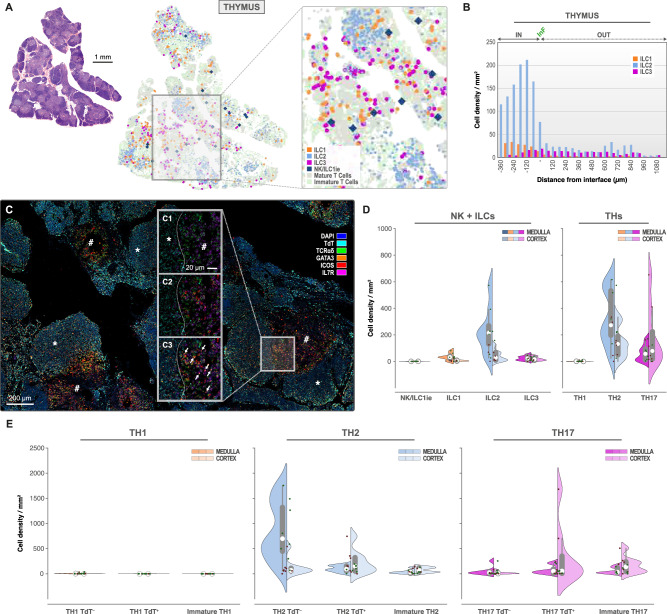
ILC phenotyping confirmed that the ILC2 subset was the most abundant in thymus62,92 (Fig. 4A–E). This is consistent with findings reported earlier in mice93. 4.98 ± 1.09% of the ILC pool was represented by the ILC2 population, whereas only 1.12 ± 0.27% and 0.55 ± 0.15% of ILC3 and ILC1 were detected, respectively (Supplementary Table 2). ILC2 was preferentially located in medulla (232.69 ± 62.95/mm2) versus cortex (42.35 ± 15.14/mm2) and was exceeded in density by Th2 cells, which were TdT+ mainly in cortex (226.39 ± 86.42/mm2) and TdT− mainly in medulla (846.52 ± 211.63/mm2) (Fig. 4D–E). Engagement of Th cells towards Th2 seems the most recurrent, representing 21.29 ± 4.25% of the T cell pool, present evenly in medulla (24.52 ± 4.64%) and cortex (18.58 ± 4.46%). Although present at very low numbers, IL-7Rα+ ILC1 (overall density 14.89 ± 4.09/mm2) resided primarily in medulla (34.01 ± 9.49/mm2 versus 7.08 ± 2.48/mm2 in cortex) and were dominant to any stage of Th1 cells (TdT−: 2.70 ± 1.94/mm2, TdT+: 0.12 ± 0.05/mm2) (Fig. 4D), representing only 0.07 ± 0.03% of the T cell pool. Only very low numbers of NK/ILC1ie cells were observed (0.46 ± 0.14/mm2). ILC3 seemed equally distributed throughout cortex (19.38 ± 5.52/mm2) and medulla (24.27 ± 6.42/mm2) and were less represented than Th17 cells, all stages predominantly present in thymic cortex (342.73 ± 208.86/mm2 for TCRαδ+ TdT+, 170.43 ± 59.07/mm2 for TCRαδ− TdT+) (Fig. 4B, D, E), except for the mature TCRαδ+ TdT− Th17 cells confirmed to reside in medulla (50.90 ± 30.10/mm2). The overall Th17 population represented 11.69 ± 3.26% of the T cell pool. In thymic cortex, immature Th cells showed a Th2 (4.05 ± 0.93%) and more importantly, Th17 (6.40 ± 2.33%) profile prior to expression of the rearranged TCR. No clear Th1 phenotype profile was detected prior to TCR-rearrangement (0.01 ± 0.00%). Additionally, we confirmed that the majority of mature (TCRαδ+) Th2 cells do not have TdT when residing in medulla (846.52 ± 211.63/mm2) and that mature (TCRαδ+) Th17-cells still express TdT in thymic cortex (342.73 ± 208.86/mm2) (Supplementary Fig. 8A).
Fig. 4. ILC2 and Th2 predominate thymic medulla, whereas Th17 is predominant in thymic cortex.
Topographic distribution of NK/ILC1ie (dark blue), ILC1 (orange), ILC2 (light blue) and ILC3 (pink) in thymus (n = 8 independent biological samples, 1 mIF staining batch). A H&E overview of thymus with corresponding ILC distribution map (detailed in caption). Scale bar: 1 mm. B Infiltration analysis plot with interface (InF) between medulla (IN) and cortex (OUT) for IL-7Rα+ ILC1 (orange), ILC2 (light blue) and ILC3 (pink) in thymus. C mIF staining of TdT (aqua), TCRαδ (green), GATA-3 (orange), ICOS (red) and IL-7Rα (pink). DAPI nuclear counterstain (DAPI, blue). Scale bar: 200 µm. C1–3. Detailed image of mIF in “C”, at medulla/cortex interface (dashed line) presenting TdT (aqua), TCRαδ (green) and IL-7Rα (pink) in “C1”, with additional ICOS (red) in “C2” and GATA-3 (orange) in “C3”. ILC2 indicated by white arrows in “C3”. Scale bar: 20 µm. D Split-violin plots representing the ILC and T helper phenotype densities (number/mm2) in medulla (left) and cortex (right) of thymus (alternative “TdT” mIF panel). E Split-violin plots representing the T-helper phenotype densities (number/mm2) in medulla (left) and cortex (right) of thymus. Box-plots within Violin Plots, created using Seaborn in Python, present the median value (white dot, 50th percentile) in between the first quartile (the middle value between “minimum non-outlier” and median (marked as Q1, portrays the 25th percentile) and third quartile (the middle value between “maximum non-outlier” and median (marked as Q3, portrays the 75th percentile)). Source data are provided as a Source Data file.
NK and ILC1 predominate in spleen and occupy primarily red pulp
In spleen (Fig. 5A, B, Supplementary Table 3), NK/ILC1ie (IL-7Rα− ILC1 subtype) cell densities (89.71 ± 24.82/mm2) exceeded that of other IL-7Rα+ ILCs (48.74 ± 14.12/mm2 for ILC1, 0.18 ± 0.05/mm2 for ILC2, and 16.08 ± 4.40/mm2 for ILC3). Although identified differently, Simoni and colleagues94 confirm NK/ILC1ie predominance in spleen. Also, in this organ, the ILC1 population (10.34 ± 2.57%) prevailed over ILC3 (5.11 ± 1.06%) and ILC2 (0.06 ± 0.02%), the latter being nearly absent (Fig. 5C). Spatial distribution study of the different ILC subsets indicated that ILC1 and ILC3 were found in white and red pulp of the spleen, with ILC3 evenly distributed over the spleen (15.50 ± 4.10/mm2 in WP and 15.92 ± 4.57/mm2 in RP). ILC1 preferentially resided in RP (52.55 ± 15.62/mm2 in RP and 17.00 ± 5.65/mm2 in WP). Regarding ILCs’ Th equivalents, Th2 presence was low (2.97 ± 0.80/mm2), whereas Th1 and Th17 were equally represented (overall density of 51.95 ± 20.58/mm2 for Th1 and 46.66 ± 20.69/mm2 for Th17) (Fig. 5C). Th1 cells were predominantly detected in RP areas (55.74 ± 22.18/mm2 in RP versus 13.43 ± 4.36/mm2 in WP), while Th17 mainly occupied WP regions (40.69 ± 21.51/mm2 in RP and 96.23 ± 25.16/mm2 in WP). WP infiltration studies (Fig. 5D) showed a minor accumulation of ILC1 and ILC3 subtypes at the white and red pulp interface, with an even distribution throughout RP compartments.
Fig. 5. NK/ILC1ie and ILC1 dominate red pulp of spleen, accompagnied by Th1. Th17 is the dominant population in white pulp.
Topographic distribution of NK/ILC1ie (dark blue), ILC1 (orange), ILC2 (light blue), and ILC3 (pink) in spleen (n = 8 independent biological samples, 1 mIF staining batch). A H&E overview of the spleen with corresponding ILCs distribution map (detailed in caption). Scale bar: 2 mm. B Classic mIF staining of CD20 (aqua), TCRαδ (green), EOMES (orange), TBX21 (red) and IL-7Rα (pink). DAPI nuclear counterstain (DAPI, blue). Scale bar: 250 µm. B1–3. Detailed image of mIF in “B”, at white pulp (hashtag)/red pulp interface (dashed line) presenting CD20 (aqua), TCRαδ (green) and IL-7Rα (pink) in “B1”, with additional TBX21 (red) in “B2” and EOMES (orange) in “B3”. ILC1 (TBX21+/EOMES−) indicated by white arrows and ILC1 (TBX21+/EOMES+) indicated by white triangles. Scale bar: 20 µm. C Split-violin plots representing the ILC and T helper phenotype densities (number/mm2) in white (left) and red pulp (right) of the spleen (classic mIF panel). D Infiltration analysis plot with interface (InF) between white (IN) and red pulp (OUT) for IL-7Rα+ ILC1 (orange), ILC2 (light blue), and ILC3 (pink) in spleen. E Spatial Transcriptomics (GEO Accession N° GSE228056) with mapping of ILCs and NK signatures in reactive spleen. Box-plots within Violin Plots, created using Seaborn in Python, present the median value (white dot, 50th percentile) in between the first quartile (the middle value between “minimum non-outlier” and median (marked as Q1, portrays the 25th percentile) and third quartile (the middle value between “maximum non-outlier” and median (marked as Q3, portrays the 75th percentile)). Source data are provided as a Source Data file.
Spatial Transcriptomics (ST) (Fig. 5E) was used as an alternative approach to evaluate ILC infiltration in human reactive spleen. A total of 807 spots was obtained and normalized for this ST analysis. ILC-specific published signatures were defined from Mazzurana44 and were scored across this dataset. By defining a threshold for each score and using Virtual Cytometer95, we established a splenic infiltration “gradient” depending on the ILC subtype. Indeed, NK cells predominated in the spleen (through the positivity of 316 spots/807), followed by ILC1 (121 spots/807), ILC2 (67 spots/807) and ILC3 cells (27 spots/807) (Fig. 5E).
Differential location of ILC subtypes within sub-areas of intestinal lymphoid tissues
In appendix (Fig. 6A, D, E), we found that 12.51% of the ILCs (±2.34%) were of the ILC3 phenotype, followed by 9.74% (±1.55 %) of ILC1 and 3.50% (±0.81 %) of ILC2. ILC3 and ILC1 subsets preferentially occupied extra-follicular mucosal tissue (12.63 ± 2.38% and 9.98 ± 1.64% respectively) confirming the work of Hepworth et al.96, whereas ILC2 infiltrated more efficiently the germinal center (8.92 ± 2.07% compared to 3.38 ± 0.78% in extra-follicular regions), representing 31.32% of all ILC2 detected (Supplementary Table 3). As shown by germinal center infiltration analysis (Fig. 6E), ILCs were enriched close to the germinal center of Peyer’s patches and extra-follicular interface, and decreased progressively when moving away from this interface. Th1 cells were largely outnumbered by ILC1, especially outside follicles (14.84 ± 12.34/mm2 and 95.80 ± 20.7/mm2, respectively). The same was found for ILC3 (88.13 ± 20.39/mm2) which dominated their Th17 counterpart (36.33 ± 7.07/mm2) in these extra-follicular regions (Fig. 6D). ILC2 was the only ILC subpopulation not reaching densities of Th2 (23.40 ± 9.75/mm2 and 240.78 ± 113.40/mm2 respectively). It is worth noting that Th2 were found in lymphoid follicles and that they were preferentially located at the extra-follicular interface together with Th17 (Supplementary Fig. 8C). Again, NK/ILC1ie were nearly absent (1.18 ± 0.44/mm2) but when detected, mainly located in extra-follicular areas (Supplementary Table 3). To the best of our knowledge, ILCs are thus far not studied in the appendix and we therefore provide new elements on innate immunity in this organ.
Fig. 6. ILC1 and ILC3 are the dominant ILC-populations in intestinal mucosal tissue, where they are colocalizing with Th2.
Topographic distribution of NK/ILC1ie (dark blue), ILC1 (orange), ILC2 (light blue), and ILC3 (pink) in appendix (A) and ileum (B, C) (n = 8 independent biological samples for each tissue type, 1 mIF staining batch per tissue type). A H&E overview of appendix with corresponding ILC distribution map (detailed in caption). B cells (light grey) are used to highlight the follicular areas. Scale bar: 2 mm. B H&E overview of ileum with corresponding ILC distribution map (detailed in caption). B cells (light grey) are used to highlight the follicular areas. Scale bar: 2 mm. C Classic mIF staining for CD20 (aqua), TCRαδ (green), EOMES (orange), TBX21 (red), and IL-7Rα (pink) in ileum. DAPI nuclear counterstain (blue). Dashed line and hashtag indicating the follicular zone. ILC1s indicated by white arrows. Scale bar: 200 µm. D Split-violin plots representing the ILC phenotype densities (number/mm2) in the follicular (left) and extra-follicular (right) zones of the appendix and ileum (classic mIF using CD20). E Infiltration analysis plot with interface (InF) between extra-follicular zone (OUT) and follicular zone (IN) for IL-7Rα+ ILC1 (orange), ILC2 (light blue) and ILC3 (pink) in appendix (upper) and ileum (lower). Box-plots within Violin Plots, created using Seaborn in Python, present the median value (white dot, 50th percentile) in between the first quartile (the middle value between “minimum non-outlier” and median (marked as Q1, portrays the 25th percentile) and third quartile (the middle value between “maximum non-outlier” and median (marked as Q3, portrays the 75th percentile)). Source data are provided as a Source Data file.
In the ileum (Fig. 6B–E), 14.76% (± 2.58%) of the IL-7Rαpool were of the ILC3 phenotype, 9.41% (± 2.23%) had differentiated into ILC1 and a minor population presented the ILC2 profile (1.97 ± 0.85%). These data are conform to the work of Yudanin and coworkers22, showing a predominance for ILC3, followed by ILC1 and ILC2 in ileum using flow cytometry methods. The same tendency was reported by Song et al.97 who used single cell RNA-sequencing and pseudo-bulk analysis across different tissues. All ILCs seemed to favor perifollicular regions, as confirmed by a follicle infiltration study (Fig. 6E). ILC1 (114.94 ± 36.38/mm2) and ILC3 (80.76 ± 29.24/mm2) seemed to prevail over ILC2 (21.85 ± 15.54/mm2). ILC2 were less represented than their Th2 equivalents (148.57 ± 102.33/mm2), whereas conversely the ILC1 and ILC3 densities exceeded the ones of their Th counterparts (4.74 ± 3.19/mm2 for Th1 and 42.43 ± 12.11/mm2 for Th17). As seen in appendix, Th2 were found in lymphoid follicles and that they were preferentially located at the extra-follicular interface together with Th17 (Supplementary Fig. 8C). Again, NK/ILC1ie cells were nearly undetected (0.64 ± 0.24/mm2) (Supplementary Table 3).
Since we have found a predominance for ILC1 and ILC3 in ileum, we examined whether these populations were composed of icCD3ε+ intra-epithelial ILC (IE-ILC)67,68 (Supplementary Table 6, Supplementary Fig. 4). IE-ILC1, named here icCD3ε+ ILC1-like, were indeed variably present in the ileum at a density of 17.86 ± 14.62/mm2. Although predominantly present in extra-follicular regions (18.74 ± 15.49/mm2), they were found in germinal centers of the ileal Peyer’s patches (12.00 ± 4.96/mm2). We found a small subset of cells displaying the ILC3 phenotype while expressing CD3ε in their cytoplasm (icCD3ε+ ILC3-like). Despite their very low density (3.37 ± 1.36/mm2), they correspond to an ILC subpopulation preferentially residing in germinal centers of Peyer’s patches (6.78 ± 3.56/mm2) and even at mucosal sites (3.38 ± 1.38/mm2) (Supplementary Table 6). Whether these icCD3ε+ ILCs correspond to the Id2-independent lymphoid precursors programmed to engage ILC cell fate68 is unknown, we confirmed however their existence and characterized their specific location.
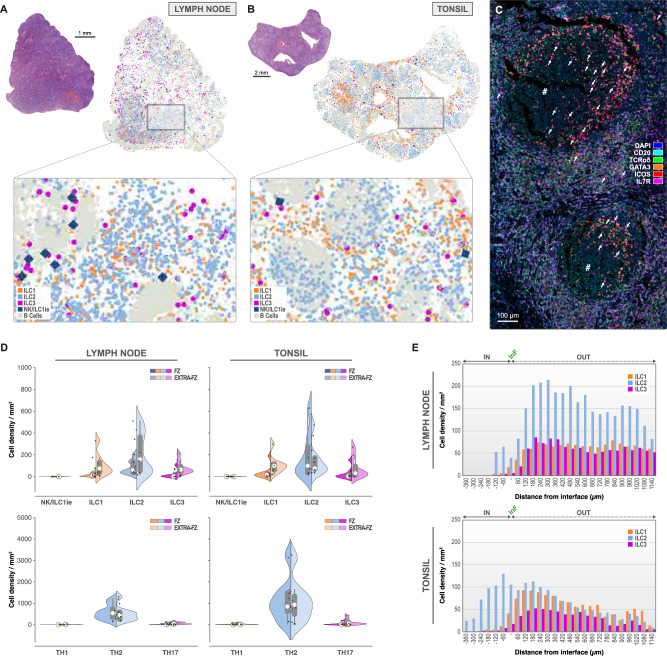
Hyperplastic lymph nodes contain high IL-7Rαpool in extra-follicular regions, where ILC2 are most represented
We examined 8 human lymph nodes (Fig. 7A, D, E, Supplementary Table 3), 7 of them were axillary lymph nodes, 1 was isolated from the abdominal region. Morphologically, all samples presented unstimulated primary follicles as a consequence of the lymphadenopathy, only few germinal centers were observed. ILC2 was the most represented (214.09 ± 68.06/mm2) in the extra-follicular areas and seemed outnumbered by the Th2 cells (505.83 ± 128.73/mm2), a phenomenon that was confirmed in follicles (87.58 ± 23.06/mm2 for ILC2 versus 588.37 ± 126.16/mm2 for Th2) (Fig. 7D, E, Supplementary Fig. 8D). In lymph nodes, ILC1 and ILC3 seemed unfavorable for follicular infiltration (15.58 ± 5.84/mm2 for ILC1 and 17.00 ± 12.89/mm2 for ILC3), whereas ILC2 penetrated into lymphoid follicles (87.58 ± 23.06/mm2), as was observed for Th2 (Supplementary Fig. 8D). ILC1 was primarily identified in extra-follicular regions (107.32 ± 36.40/mm2) and reached higher densities than its Th1 counterparts (8.88 ± 3.87/mm2). The same was true for ILC3, predominantly occupying the same regions (84.37 ± 22.35/mm2), equally accompanied by their Th17 counterparts (60.21 ± 17.23/mm2). As for all the previous organs described except for the spleen, lymph nodes did not show an important presence of the NK/ILC1ie phenotype (0.92 ± 0.18/mm2).
Fig. 7. ILC2 and Th2 dominate the lymph node and tonsil.
Topographic distribution of NK/ILC1ie (dark blue), ILC1 (orange), ILC2 (light blue) and ILC3 (pink) in lymph node (A) and tonsil (B, C) (n = 8 independent biological samples for each tissue type, 1 mIF staining batch per tissue type). A H&E overview of lymph node with corresponding ILC distribution map (detailed in the caption). B cells (light grey) are used to highlight the follicular areas. Scale bar: 1 mm. B H&E overview of tonsil with corresponding ILC distribution map (detailed in caption). B cells (light grey) are used to highlight the follicular areas. Scale bar: 2 mm. C Classic mIF staining for CD20 (aqua), TCRαδ (green), GATA-3 (orange), ICOS (red), and IL-7Rα (pink) in tonsil. DAPI nuclear counterstain (blue). Hashtag indicating the germinal center. ILC2 indicated by white arrows. Scale bar: 100 µm. D Split-violin plots representing the ILC phenotype densities (number/mm2) in the follicular (left) and extra-follicular (right) zones of the lymph node and tonsil (classic mIF using CD20). E Infiltration analysis plot with interface (InF) between extra-follicular zone (OUT) and follicular zone (IN) for IL-7Rα+ ILC1 (orange), ILC2 (light blue), and ILC3 (pink) in lymph node (upper) and tonsil (lower). Box-plots within Violin Plots, created using Seaborn in Python, present the median value (white dot, 50th percentile) in between the first quartile (the middle value between “minimum non-outlier” and median (marked as Q1, portrays the 25th percentile) and third quartile (the middle value between “maximum non-outlier” and median (marked as Q3, portrays the 75th percentile)). Source data is provided as a Source Data file.
ILC2 predominate tonsil and infiltrate germinal centers of lymphoid follicles
Among ILCs in tonsil (Supplementary Table 3, Fig. 7B–E), 12.06% (± 3.18%) were identified as ILC2, whereas ILC1 and ILC3 were present in lower percentages (5.55 ± 1.45% and 6.82 ± 3.07%, respectively). The density of the ILC2 was slightly higher in germinal centers compared to extra-follicular regions (192.19 ± 76.00/mm2 and 141.95 ± 50.74/mm2, respectively) and was largely dominated by Th2 in both studied areas (overall density of 1083.61 ± 352.56/mm2). This ILC2 predominance in tonsil is in contrast to what is described21,98,99. CRTH2 in combination with GATA-3 is often used to identify ILC2. CRTH2 however, detects conventional ILC2 only48 and seems more specific for circulating ILC2 than for tissue-resident ones44, therefore underestimating their true density in situ. In accordance to Liu and colleagues48, and to Simoni et al.94 we used ICOS along with GATA-3 to identify ILC2 (Supplementary Fig. 1A, B). IL-7Rα+ ILC1 seemed to be mostly localized in the extra-follicular regions (109.28 ± 29.56/mm2) where they outnumber their Th1 counterpart (23.32 ± 12.10/mm2). The ILC3 density (66.67 ± 30.41/mm2) however roughly matched the one of the Th17 population (93.49 ± 43.53/mm2) in both germinal centers and extra-follicular regions, but preferentially occupied extra-follicular areas of the tonsil (119.79 ± 51.99/mm2). T cells were abundant (overall density of 2735.40 ± 262.14/mm2), whereas NK/ILC1ie remained low (1.49 ± 0.61/mm2). Germinal center infiltration study (Fig. 7E) confirmed that ILC2 penetrated the germinal centers of follicles, as was the case for Th2 cells (Suplpementary Fig. 8D) and were preferentially enriched in perifollicular regions together with ILC1 and ILC3 (Fig. 7E) confirming published data21.
Fig. 8. Loss of IL-7Rαpool and ILC subtypes, and altered T helper cell subsets in follicular lymphoma.
Topographic distribution of NK/ILC1ie (dark blue), ILC1 (orange), ILC2 (light blue), and ILC3 (pink) in follicular lymphoma (FL) (n = 8 independent biological samples, 1 mIF staining batch). A H&E overview of FL with corresponding ILC distribution map (detailed in caption). B cells (light grey) are used to highlight the follicular areas. Scale bar: 2 mm. B Infiltration analysis plot with interface (InF) between extra-follicular regions (OUT) and follicular regions (IN) for IL-7Rα+ ILC1 (orange), ILC2 (light blue) and ILC3 (pink) in FL. C Classic mIF staining for CD20 (aqua), TCRαδ (green), EOMES (orange), TBX21 (red), and IL-7Rα (pink) showing ILC1s (white arrows) in follicular regions (dashed line and hashtag) and outside the follicles. DAPI nuclear counterstain (blue). Hashtag indicating the follicles. Scale bar: 100 µm. D Same as “C” but for classic mIF staining using GATA-3 (orange) and ICOS (red) (instead of EOMES and TBX21) and showing the spatial distribution of ILC2 around the follicle interface (dashed line). Scale bar: 100 µm. E Split-violin plots (left panel) representing the ILC and T helper densities (number/mm2) in the follicular (left) and extra-follicular (right) zones of the FL. Bubble plots (right panel) comparing the overall relative cell densities (bubble size) and the spatial distribution of each ILCs with their T helper counterparts around the follicular interfaces in the lymph node, tonsil, and FL. X-axis: mean cell density % inside the interface (log10 scale). Y-axis: mean cell density % outside the interface. LN: Lymph Node (yellow). TS Tonsil (green). FL Follicular Lymphoma (orange). Box-plots within Violin Plots, created using Seaborn in Python, present the median value (white dot, 50th percentile) in between the first quartile (the middle value between “minimum non-outlier” and median (marked as Q1, portrays the 25th percentile) and third quartile (the middle value between “maximum non-outlier” and median (marked as Q3, portrays the 75th percentile)). Source data are provided as a Source Data file.
Loss of IL-7Rαpool and ILC subtypes, and altered T helper cell subsets in follicular lymphoma
To test our mIF panels for ILC detection in a pathologic condition, we studied the ILC population in 8 follicular lymphoma (FL) patients, with a disease morphologically classified as grade 1-2 (Fig. 8A–E, Supplementary Tables 3 and 7). These samples were characterized by a lymphomatous proliferation with follicular architecture presenting small to moderate-sized cells with hyperchromatic nuclei, primarily of centrocyte-type with variable enrichment of centroblasts and germinal center-like formation. Inherent to the differences in function and cell composition between reactive LN and palatine tonsil, secondary follicles characterized by germinal centers were more abundant in the tonsil than LN. We studied therefore the differences of ILC and Th distribution in FL compared to both LN and tonsil by studying germinal center infiltraton and extra-follicular occupation.
In FL, a significant loss of IL-7Rαpool (608.54 ± 158.28/mm2) was observed as compared to LN (3011.16 ± 263.58/mm2, p < 0.0001) and tonsil (1501.46 ± 182.31/mm2, p < 0.001). The difference of IL-7Rαpool in LN and tonsil was of the same statistical strength (p < 0.001). The overall ILC differentiation rate (% sum ILCs of IL-7Rαpool, Supplementary Table 3) was lowest in LN (12.41 ± 1.10%) and comparable in FL and tonsil (17.34 ± 1.86% and 24.43 ± 2.69%, respectively). Remarkably, ILC differentiation occurred preferentially in follicular regions (characterized by germinal center-like presence) where IL-7Rαpool density was the lowest. In LN and tonsil, ILC2 was the best-represented phenotype both in intra and extra-follicular regions (overall density of 211.85 ± 67.27/mm2 and 154.41 ± 57.54/mm2 respectively). FL on the contrary, presented a significant drop of ILC2 (43.37 ± 22.43/mm2) when compared to LN (p = 0.028) and tonsil (p = 0.021), and this inside and outside the follicular regions of tonsil (p = 0.028 and p = 0.038 respectively), and especially extra-follicular when compared to LN (p = 0.021) (Supplementary Table 7). ILC3 dropped significantly, especially in extra-follicular regions of FL as compared to LN (p = 0.005). This difference was however not significant when compared to tonsil (p = 0.065). NK/ILC1ie, a cell type present at very low density in the studied LN and tonsils (overall density of 0.92 ± 0.18/mm2 and 1.49 ± 0.61/mm2 respectively) but residing preferentially in extra-follicular regions, appeared more frequently (overall density of 3.72 ± 1.24/mm2) in neoplastic follicles of FL (p = 0.0002 as compared to LN and p = 0.1009 as compared to tonsil). In contrast, no significant changes were observed for ILC1 densities (p > 0.05 for both LN and tonsil, Supplementary Table 7). However, these cells appeared to have relocated in and around the neoplastic follicles as shown by the follicular infiltration analysis (Fig. 8B), a phenomenon that was observed for ILC2 in LN.
In the FL T cell microenvironment (Supplementary Table 3), the T cell pool was quantitatively unchanged but a T helper shift (Th1/Th17) occurred when compared to LN and tonsil. While Th2-like cells seemed relatively unaffected (506.79 ± 127.30/mm2 for LN, 1083.61 ± 352.56/mm2 in tonsil and 634.35 ± 209.24/mm2 for FL), Th17 decreased (p < 0.05) (59.69 ± 17.19/mm2 for LN, 93.49 ± 43.53/mm2 in tonsil and 20.02 ± 8.61/mm2 for FL). Conversely, the Th1 lymphocyte density increased (p < 0.01) in tumoral follicles as compared to LN and tonsil (0.58 ± 0.58/mm2 in LN, 21.16 ± 11.13/mm2 in tonsil and 73.08 ± 46.58/mm2 in FL) (Supplementary Fig. 8E).
Bubble plots in Fig. 8E (right panel) draw a visual comparison of the overall relative cell densities (bubble size) with the spatial distribution of each ILC and T helper counterparts around the follicular interfaces in the lymph node, tonsil, and FL. This method of data visualization showed an equivalent distribution gradient of these cells with primarily extra-follicular localization in LN and tonsil, whereas these cells were evenly distributed in follicular and extra-follicular regions in FL (except for ILC3). A clear intra-follicular relocation of ILC2 and Th2 cells in FL (more than 50% of the overall cell populations are located inside the follicular zone) was confirmed as compared to LN and tonsil (Supplementary Fig. 8E). Th2 cells were by far the dominant phenotype in the three tissue types, followed by ILC2 then ILC1 in lymph node and tonsil. ILC1 density was relatively equivalent across the three tissue types, but a drop was seen for ILC2 and ILC3 in FL as compared to LN and tonsil (4- and 5-fold, respectively). ILC3 and Th17 phenotypes represented only 5% of the biggest cell population (Th2) and showed the same distribution pattern with relatively equal cell densities in LN and tonsil and much lower densities in FL. Th1 cells were barely present in all cases, but showed higher cell densities in FL compared to tonsil and LN. This clearly illustrated the Th1/Th17 switch in FL described above.
ILCs are close to their Th counterparts and participate in thymus homeostasis
To further explore the ILCs tissue-specific distribution in relation to their Th counterparts, we performed cellular proximity analysis. Close interactions between ILCs and Th cells are widely described and studied100, but a detailed cartography of both innate and adaptive immune cells lacked until now. A fundamental question is how ILC/T cell interactions sustain and promote tissue homeostasis70. Indeed, we found that in thymus, ILCs and Th cells occupy the same regions and seem to be present in close proximity (Fig. 9A, C, and Supplementary Fig. 9A). Even if the different ILCs seem to be surrounded more or less by all Th subtypes, ILC1, ILC2 and ILC3 cells are preferentially close to their specific Th counterpart. 34% of the detected Th1 seem to be present within a 50 µm radius of ILC1, supporting the evidence that type 1 immune cells have an impact on thymic-resident innate lymphoid cells (ILC1)101. The same proximity was seen for ILC3, surrounded by 33% of Th17 cells, and for ILC2 which are accompanied by 65% of identified Th2 cells (Fig. 9C). As mentioned before, ILC2 was the most abundant ILC phenotype detected in thymus, suggesting that they have an important role for T cell development. With this proximity analysis, we show that they are in close contact with Th1 (56%) and Th17 (27%) as well.
Fig. 9. Proximity analysis of Th cells within 50 µm of each ILC subtype in thymus (A, C) and spleen (B, D) (n = 8 independent biological samples for each tissue type, 1 mIF staining batch per tissue type).
Heatmaps show the percentage of Th1, Th2 or Th17 (red gradient) according to the ILC1, ILC2 or ILC3 distance (blue gradient, 5 µm steps) in thymus (A) and spleen (B, left panel). Bars show the percentage of Th cells within 50 µm of a single ILC1, ILC2 or ILC3 in the thymus (A) and spleen (B, left panel). In the spleen, where NK/ILC1ie cells were abundant, proximity analysis was also computed for NK/ILC1ie cells versus the three ILC and Th subtypes (B, right panel). C, D Schematic overview of the proximity analysis data. Each circular diagram represents the proximity of each “small” cell (Th cells for thymus and spleen, and ILCs for spleen only) within a 50 µm radius (2 × 25 µm concentric) of the center of the target cell nucleus drawn in the middle (ILC1, ILC2, and ILC3 for thymus and ILC1, ILC3 and NK/ILC1ie for spleen). The mean number and location of each “small” cell are shown schematically within the 2 × 25 µm radius proximity of the target cells (ILC1, ILC2, and ILC3 for thymus and ILC1, ILC3 and NK/ILC1ie for spleen). The small mirrored pie charts represent the percentages of each “small” cell population (Th cells for thymus and spleen, and ILCs for spleen only) within (when located inside the circular diagram, dark grey pie part) or over (when located outside the circular diagram, dark grey pie part) the 50 µm radius of the target cells (ILC1, ILC2, and ILC3 for thymus and ILC1, ILC3 and NK/ILC1ie for spleen). Caption in “C”. Detail of IndicaLabs Halo distribution map with proximity analysis visualization output in thymus. Source data is provided as a Source Data file. Partially created in BioRender. Bezombes, C. (2025) https://BioRender.com/shbs5qf.
As reviewed by Shin and Mc Nagny62, T cell development depends on a strict maturation process of thymocytes in the thymic microenvironment before departure to peripheral tissues. In contrast to T cells, ILCs can develop in the absence of the thymic niche62, and several non-immune functions were attributed to ILCs residing in the thymus92. The thymus appears, however, to play a role in providing ILC-pools to the peripheral tissues102. To better understand cellular interactions between ILCs and other lymphoid and non-lymphoid cells, we explored the publicly available dataset produced by Li et al.103 (Supplementary Fig. 10). We need to stress the fact that within the clusters identified, ILC3 and NK cells could be extracted. No specific data is present, however, on the other ILC phenotypes, nor on the Th subsets. Interestingly, ILC3 and NK cells seem to receive incoming signals from especially fibroblasts, epithelial cells, blood vessels, and lymphatic vessels. Recently, Yayon et al.104 showed that T cell development is directly dependent on and supported by the resident stromal and immune cells, with key roles for thymic epithelial cells (TEC). ILC3 and NK cells receive incoming signals (Supplementary Fig. 10C) from these epithelial cells through FN1 (Fibronectin 1), MK (Midkine), CXCL (chemokine (C-X-C motif) ligand), and ICAM (intra-cellular adhesion molecule) pathways. All shared ILC incoming signaling pathways coming from lymphatic vessels, blood vessels, and fibroblasts as well. FN1, one of the main extra-cellular matrix components, defines the spatial distribution of the basal laminae of the postnatal human thymus105, participates in cell adhesion, migration, and movement of various cells106, and plays an important role in the morphogenesis of the thymus107. MK promotes the cell migration of inflammatory cells108 and CXCL signaling, which comprises chemokines are indispensable for the immune system since they regulate directional migration and activation of leukocytes109. Finally, ICAM boosts the homing of progenitor cells and lymphocyte output from the thymus110. It is thus likely that all of these pathways are involved in the homing and departure of ILCs and NK cells towards or from the thymus, as we identified them primarily in the medulla and thus in close contact with the medullar epithelial, vascularized compartment. Interestingly, all cell clusters identified from the Li et al.103 dataset show a bidirectional signaling through MIF (macrophage migration inhibitory factor). The importance of the MIF-pathway was already reported by Yang et al.111 and reviewed by Calandra and Roger112 as a regulator of innate immunity expressed by a wide variety of cells, helping to override immunosuppressive mechanisms, negatively regulating cell cycle arrest, and thought to be mainly produced by Th2 cells112.
When looking at lymphoid cell interactions, a strong interaction has been identified coming from NK cells towards ILC3 (Supplementary Fig. 10C) through the CLEC (C-type lectin receptors) and ITGB2 (integrin subunit beta 2)-pathway. CLEC seem to be involved in signaling coming from most of the lymphoid cell clusters identified in the Li et al.103 paper, towards ILC3 and NK cells and is known to have diverse functions in immunity113. These receptors ensure critical discrimination between self and non-self and assure correct balance between immune activation and inhibition114. The same is true for ITGB2 signaling, apparently involved in lymphoid cell contact in the thymus including for ILC and NK cells, reported to be important for cell adhesion115. Other strong interactions are revealed between B cells (CD22 signaling), dendritic cells (DC, Galectin signaling) and DN T lymphocytes (Galectin signaling) on one hand and ILC3 and NK cells on the other. Galectins act on both positive and negative thymocyte selection116 and seem to influence thymus homeostasis117. How galectin-pathways influence ILC/NK cells in the thymus remains unclear but galectins are described to be involved in leukocyte and innate immune cell recruitment by affecting chemoattraction and/or cell adhesion118. What the impact of B cell interactions might be in the thymus is not clear to us since we barely detected them in the studied fragments, but it is well known that CD22 signaling affects innate immune signals and might assure self-recognition in the thymus119.
When focusing on the outgoing signals from ILC3 in the thymus (Supplementary Fig. 10B), almost all identified cell clusters receive lymphotoxin (LT)-signaling, which seems to ensure the generation of a diverse and self-tolerant T cell repertoire and plays a role in thymic homeostasis120. A strong outgoing signaling using MHC-II and IL16 pathways is sent to DP, Treg lymphocytes, to dendritic cells, and myeloid cells. Hepworth et al.96 demonstrated that ILC3 intrinsic expression of MHC-II is regulated similarly to thymic epithelial cells and that they may induce cell death in activated commensal bacteria-specific T cells. To et al.121 reviewed the topic and indicated that MHC-II+ ILC3 mediate a negative selection process through antigen presentation and withdrawal of IL-2 from the local milieu, resulting in deletion of activated commensal bacteria-specific T cells121. IL-16, known to be a chemoattractant cytokine for CD4+ T cells, macrophages, and eosinophils, might be used by ILC3 to recruit these cells and assure a correct balance between immune activation and inhibition122. They might attract transendothelial DCs by IL-16 production inducing a mechanism by which the thymus can actively acquire blood-borne antigens to induce and maintain central tolerance123. ILC3 seem to send pro-inflammatory signals to all cell clusters except DP lymphocytes through the APP (amyloid precursor protein) pathway124,125. ILC3-derived BAFF has been found to promote IL-15 generation by B cells126,127. Finally, ILC3 interact with structural cells in the thymus as the epithelial cells, fibroblasts and lymphatic vessels. Fibroblasts receive EGF (epidermal growth factor) signaling, stimulating their proliferation128, and epithelial cells and lymphatic vessels receive PECAM1 signaling, most likely assuring that they form a tight barrier through cell-contact129.
Alltogether these data suggest that, even in low numbers, ILCs play an important role in thymic homeostasis, and we bring here important information about their distribution in this primary lymphoid organ being in close proximity to their Th counterparts.
ILCs closely interact with Th1 and Th17 cells after traumatic injury of the spleen
In spleen, as a consequence of the traumatic nature of the fragments studied here, a predominance was found for NK/ILC1ie and ILC1. They were especially located in red pulp areas associated to an apparent Th1 response (Fig. 5C). By proximity analysis, we clearly show that in red pulp, NK/ILC1ie and ILC1 are in close contact to Th1 cells (Fig. 9B, Supplementary Fig. 9A). Indeed, 56% of Th1 cells detected seem to reside within a 50 µm radius from ILC1 and 70% of them have been found in close contact with NK/ILC1ie (Fig. 9D). Remarkably, 64% of ILC1 were found next to (within this 50 µm radius) NK/ILC1ie (Fig. 9B, D). Both innate lymphoid cells were closely surrounded by 30 to 39% of Th17 cells respectively (Fig. 9D). The traumatic spleen fragments showed an important Th17 response, infiltrating in white pulp (Fig. 5D, Supplementary Fig. 8B), for which 39% was seen in close proximity of their ILC counterparts (ILC3) and 32% of them seemed to approach closely NK/ILC1ie (Fig. 9D). It is well known that after tissue injury, the innate system is the first to activate130,131. Injured cells release danger-associated molecular signals (DAMPs) and activate ILC1 and NK cells, responding by IFNγ−production, activating both macrophages and CD8+ T lymphocytes, to clear pathogens and contain tissue damage. They act before Th1 adaptive responses are up and work together closely afterwards. To protect the tissue and activate its repair, type 3 immunity is then activated with ILC3 being engaged first to recruit neutrophils a the injured site by IL-17 and IL-22 release. Th17 cells join ILC3 with similar mechanisms to respond to tissue damage and prevent subsequent infections35,130–132. The low presence of ILC2 and Th2 cells in the studied spleen fragments is consistent since it is typically associated with later phases of immunological reactions after injury, such as tissue repair, and thus less prominent in this context133.
Predominant Th2 immune cells in MALT are in close proximity to the 3 ILC phenotypes to assure intestinal homeostasis
Intestinal mucosa-associated lymphoid tissue (MALT) is dominated by a Type 2 immune surveillance to assure intestinal barrier integrity mainly by promoting mucus production by goblet cells and epithelial cell turnover134,135 in homeostatic conditions. Indeed, we found a predominance of Th2 cells in extra and intrafollicular regions of MALT (Fig. 6, Supplementary Fig. 8C) where they promote B cell clonal expansion and antibody production136. Type 2 immunity needs good coordination between innate and adaptive immune responses to protect the mucosal sites from pathogens134,137. By proximity analysis we show that Th2 cells are in close proximity (within 50 µm radius) with ILC2 in appendix and ileum (35% and 37% of the Th2 detected respectively) (Fig. 10A, C, Supplementary Fig. 9B) and seem to occupy inter- and extrafollicular regions (Supplementary Fig. 8C). Th2 cells seem to exert a surveillance function in these mucosal associated lymphoid tissues133, and are found in close proximity with ILC1 and ILC3 in both appendix (51% and 47% of Th2 respectively) and ileum (66% and 52% of Th2 respectively) (Fig. 10A, C). Due to the non-pathological nature of the studied fragments, appendix and ileum show a low Th1 and Th17 occupation. They were even dominated by their ILC counterparts, especially in mucosal (extra-follicular) regions, but were located in close proximity (Fig. 10B, Supplementary Fig. 9B). 67% and 66% of the detected Th17 cells were present within a 50 µm radius of ILC3 in the appendix and ileum, respectively. Similarly, 68% (appendix) and 65% (ileum) of Th1 cells seemed to approach ILC1 within this radius. All T helper subtypes seemed to favor close proximity with ILC2 (Fig. 10C). Both ILC1 and ILC3, outnumbering their Th couterparts (Fig. 6D), are known to be vital regulators of gastrointestinal homeostasis through close interactions with structural, stromal and other lymphoid cells at the epithelial barriers138. They arrive in the intestine following a homing receptor switch regulated by a gut-specific retinoic acid tissue factor139. Locally, ILC3 stimulates epithelial cells to produce anti-microbial peptides upon IL-22 release, closely sustained by ILC1 and NK cells to fight intra-cellular infection through IFNγ production140. Finally, ILC2, equipped with a gut-homing receptor during development in the bone marrow139, appear to integrate an epithelial remodeling circuit together with tuft cells, supporting the intestinal defense and homeostasis141–143.
Fig. 10. Proximity analysis of Th cells within 50 µm of each ILC subtype in appendix, ileum, lymph node, tonsil, and follicular lymphoma (n = 8 independent biological samples for each tissue type, 1 mIF staining batch per tissue type).
Schematic overview of the proximity analysis data for appendix, ileum, lymph node, tonsil, and follicular lymphoma. Each circular diagram represents the proximity of each “small” cell (Th2 cells in A, Th1 and Th17 cells in B) within a 50 µm radius (2 × 25 µm concentric) of the center of the target cell nucleus, drawn in the middle (ILC1, ILC2, and ILC3). The mean number and location of each “small” cell are shown schematically within the 2 × 25 µm radius proximity of the target cells (ILC1, ILC2, and ILC3). The small pie charts represent the percentages of each “small” cell population (Th cells) within (dark grey pie part) and over (light grey pie part) the 50 µm radius of the target cells (ILC1, ILC2, and ILC3). Only one circular diagram is shown in (B) because the number and location pattern of Th1 and Th17 cells is the same for all the tissue types analyzed, whereas each diagram for each ILC subtype is divided by 5 (for appendix, ileum, lymph node, tonsil and follicular lymphoma) in (A) to display per quadrant the proximity analysis data overview that are different for each tissue type. C Heatmaps show the percentage of Th1, Th2 or Th17 (red gradient) according to the ILC1, ILC2, or ILC3 distance (blue gradient; 5 µm steps) in tonsil, lymph node, and follicular lymphoma, appendix and ileum. Bars show the percentage of Th cells within 50 µm of an ILC1, ILC2, or ILC3 in tonsil, lymph node, follicular lymphoma, appendix, and ileum. Source data are provided as a Source Data file. Partially created in BioRender. Bezombes, C. (2025) https://BioRender.com/shbs5qf.
Predominant Th2 immune cells in the lymph node and tonsil are in close proximity to ILC2
In both lymph nodes (LN) and tonsil, we found a Th2 dominance, appearing in close proximity to ILC2. Comparative diagram chords for proximity analysis of ILCs with their T helper counterparts in LN and tonsil are shown in Supplementary Fig. 9C. 71% of the detected Th2 cells are located within a 50 µm radius of ILC2 (Fig. 10A), in both LN and tonsil. Both Th2 and ILC2 seemed favorable for germinal center infiltration (Fig. 7E, Supplementary Fig. 8D). Although ILC2 and Th2 exhibit similar effector functions, their relative contributions and interdependency is still to be elucidated144. ILC2 appears to be closely surrounded by Th1 and Th17 as well (Tonsil: 60%, LN: 56%) (Fig. 10B, C). Interestingly, despite their lower presence, Th17 cells seem to be localized near all three ILC phenotypes in tonsil, and especially ILC1 and ILC3 in LN (Fig. 10C). For Th1, close proximity is seen with ILC1 in LN, and both ILC1 and ILC2 in the tonsil (Fig. 10B, C).
To elucidate the function of the ILC populations, and to clarify possible cellular interactions with non-lymphoid and other lymphoid cells residing in tonsil, we explored the publically available dataset of Massoni-Badosa et al.79 (Supplementary Fig. 11). They conducted several analyses, resulting in 34 cell clusters going from general cell phenotypes (ILC pool, NK amongst others), to more detailed clustering of CD4+ T cells and Th cells, CD8+ T cells and ILC (NK, ILC1, ILC3), B cells, plasma cells, and myeloid cells. It is worth noting that within the clusters identified, ILC2 and Th subsets were absent. The data reveal however, a close interaction between dendritic cells (DCs) and clustered ILC phenotypes. ILC1 and ILC3 receive incoming signaling (Supplementary Fig. 11C) from DCs through APP, Galectin, ICAM1, ITGB2 pathways amongst others. Galectins are known to exhibit different roles in innate and adaptive immune responses145, and play a role in innate immune cell recruitment by affecting chemo attraction and/or cell adhesion118. DCs might attenuate or calm down the innate immune reponses of ILC1 and ILC3 cells146 through galectin signaling. ITGB2 signaling between DCs and ILCs is likely to be important for cell adhesion and migration147, whereas ICAM1 plays an important role in the cell/cell interaction and migration of leukocytes and assures that DCs enter lymph nodes148. As extracted from the data of Massoni-Badosa et al.79, DCs receive a strong incoming signal from both ILC1 and ILC3 through MIF signaling (Supplementary Fig. 11B). As discussed elsewhere, this pathway is an important regulator of innate immunity, helping to override immunosuppressive mechanisms112. Another attenuating pathway existing between ILCs and DCs is the annexin signaling. Annexin is described to inhibit activation of ILC2 by an intrinsic metalloregulatory mechanism, which might exist for ILC1 and ILC3 as well149. A close interaction between ILC2 and DCs is also described150, where IL-13 produced by activated ILC2 is promoting the migration of activated DCs to draining LNs, with subsequent differentiation of naive CD4+ T cells into Th2 cells150. Finally, some of the pathways of cellular interactions between ILCs and non-lymphoid or other lymphoid cells (BAFF (B-cell activating factor), IL-16, and MHC-II) appear to be in common with those found in thymus103.
Alltogether, these data emphasize that exploring the spatial proximity of Th cells and their ILC counterparts together with the signaling pathways existing between them, underline the importance of ILCs in tonsil homeostasis and their potential impact in orchestrating innate and adaptive immune responses.
Discussion
ILCs have been described using flow cytometry methods in embryonic tissues, bone marrow, secondary lymphoid organs, and many non-lymphoid tissues, including at mucosal sites50. Nevertheless, how these populations are maintained in these organs, and whether they recirculate from lymphoid to non-lymphoid organs, remains unclear51. One of the most important studies on tissue distribution of ILCs in non-neoplastic lymphoid organs was carried out by Yudanin et al.22, who reported spatial and temporal mapping of ILC subtypes in human fat tissue, lymph node, spleen, lung, and intestinal tissues. They used so-called global techniques based on a combination of flow cytometry and transcriptional analyses (RNA sequencing)22. This seminal work provided a comprehensive map of the spatial and temporal dynamics regulating the anatomical distribution, subset heterogeneity, and functional potential of ILCs in non-diseased human tissues. Only a few studies have been published in which tissue distribution of one or more ILC subtypes is described in human tissue by applying immunohistochemical approaches, with or without accompanying techniques as flow cytometry and single cell mass spectrometry, RNA sequencing, and single cell RNA sequencing19–21,49,78,151–153. Despite the high quality of the different studies published on this topic, none of them expanded the analysis to the global whole-slide level, leaving us with unanswered questions about the spatial distribution of human ILC populations in reactive and pathological conditions. Determining the accurate densities and spatial distribution of ILCs in human lymphoid organs and their co-distribution with other lymphocytes, especially with their Th counterparts, using low-cost and routine-compatible technologies became therefore our main objective.
Recent advances in multiple protein in situ imaging techniques has enabled many researchers to bring new insights into the complex interactions between different cell types and their spatial distribution154–156. These valuable methodologies, recently reviewed by de Souza et al.157, shed new light on spatially resolved expression of up to 100 proteins and more on a single slide. These techniques are however, expensive, limited in terms of area examined, and therefore not suitable for a daily pathology lab practice nor large batch analyses157,158. Until recently, another important drawback of such methods was the reduced resolution compared to conventional microscopy techniques, which hampers an accurate cell segmentation159, leading scientists to find alternative and complementary workflows to solve this issue159,160. Although this resolutive issue is now fully addressed with the latest microscopy-based spatial transcriptomics technologies offering a subcellular resolution using probe-based readouts, the cost of such methodologies still remains incompatible with routine diagnosis161. In this study, we therefore provide a precise map of ILC and Th compartmentalization in lymphoid organs in relation to B and T cell populations, through conventional spatial quantitative multiplex immunofluorescence techniques and whole-slide fluorescent multispectral imaging, combined with deep automated image and data analysis at the entire tissue and single-cell resolution. Several non-neoplastic reactive lymphoid organs are studied in a fast, reproducible and cost-effective way. Our histological gating strategy has been validated by correlating data with flow cytometry and NK and ILC signatures obtained from in-house162 and publicly available spatial transcriptomics experiments and datasets24,44.
Our approach allowed intriguing observations on the different Th cells and ILC subpopulations in the thymus. The initial mIF panel (without anti-TdT antibody) did not allow us to separate immature T helper (Th) cells from the different ILC sub-populations, both negative for TCRαδ. Indeed, we clearly show here that TCRαδ− TdT+ “ILC like” cells were actually immature T cells, predominating in the thymic cortex. Interestingly, the latter display a Th2 or Th17 differentiation profile prior to T cell receptor maturation and expression. No such profile is observed for immature T cells showing a Th1 phenotype. T cell differentiation in the thymus is well characterized64,66,76, progressing from the CD4− CD8− double-negative through the rearrangement of the TCR genes towards peripheral CD4+ or CD8+ T cells. Our findings raise the question whether Th cell subsets could be selected so early in the thymic cortex. This intriguing finding requires further experimental investigation to determine the clear nature of these immature Th2 and Th17-like cells. Evidence was found, however, for the essential functions of GATA-3 before and after Th2 differentiation163. Although ICOS expression was shown on DN thymocytes in mice, Dong et al. stated that it remains unclear which developing thymocyte population expresses this costimulatory protein164. Interestingly, Wang and colleagues165 demonstrated that pre-DN4 stage thymocytes express RORγT. Whether this coincides with the expression of AHR, shown to be involved in the development of early thymocyte progenitors, remains unclear166. While T cell maturation pathways are well depicted167, the development of ILC subpopulations, in particular where this occurs and whether early T cell progenitors are involved, is still a matter of debate60,61,168. However, our study provides a precise cartography of the different ILC subsets in the thymus, which completes the work of Park et al.167, and Yayon et al.104,169. A predominance of ILC2 subtypes was observed in the thymus, preferentially confined to the medulla. This makes sense and is in line with recent studies on mouse models suggesting that functional ILC2 can arise in the thymus from an early T cell progenitor in parallel with pro-T cells and that ILC2 are predominant after birth93,170. ILC2 also appears more abundant in lymph nodes and tonsils, as compared to ILC1 and ILC3. It is worth noting that in most of the peripheral lymphoid structures, ILC2 is the principal phenotype to penetrate into follicles towards germinal centers and to be especially present at the mantle zone interface under normal conditions. ILCs are primarily found in extra-follicular regions, confirming what has been described previously in lymph nodes21,96,171. As stated before by Sonnenberg and Hepworth172, this particular spatial distribution suggests that ILCs are in contact with recently migrated lymphocytes from tissues, and might influence T/B cell interactions or the initiation of humoral immune response172. The interplay between B cells and ILC2 has been documented in experimental models173 where inflammation induced IL-33 release by stromal cells, resulting in ILC2 and B cell activation, and antigen-specific IgM production. Interestingly, after exploration of the public available molecular dataset provided by Björklund and coworkers43, we found that CXCR5 is expressed by a subset of ILC2 in human tonsil. It might thus be that CXCR5 provides a mechanism for ILC2 to penetrate B cell follicles as it is the case for T cells174–177. The precise role of ILC2 within the follicular microenvironment remains unclear but they might represent the follicular regulatory ILC type recently put forth by O’Connor et al.178. These ILCs express the same TF/AF as regular ones and inhibit Th cells in their supportive role of B cells178. Nevertheless, we provide important complementary information to the work of Massoni-Badosa and coworkers79 and to the work of Pascual-Reguant et al.21 on ILC spatial distribution in tonsil. As expected, NK cells are of low abundance in most of the examined lymphoid tissues34, except in the spleen162 that is known as a major homing site for NK cells179. Their entry and distribution in the red pulp is controlled180, but in our cases, traumatic spleens, NK cell density might be overestimated in some areas following hemorrhagic changes.
We further investigated the presence of ILC subsets in the gastrointestinal mucosa-associated lymphoid tissue, more specifically in appendix (related to the colon) and ileum, and found a dominant ILC3 population at mucosal sites (extra-follicular), followed by ILC1 and a minority of ILC2, concordant to the findings of Yudanin et al.22. They analyzed ILC distribution at intra-epithelial (IE) sites and in lamina propria of human ileum and colon, and confirmed that ILCs reside primarily in mucosa (IE). Some of these ILCs, more precisely ILC2, might interact with T cells locally through an ICAM2-ITGAL/ITGB2 pathway as identified from the publicly available dataset from Björklund et al43. Some IE-ILCs express cytoplasmic CD3ε 67 and are known to present primarily an ILC1 phenotype68 and mimic in rare cases the ILC3 phenotype67. Because of phenotypic similarities between ILC1 and icCD3ε+ IE-ILC1-like cells, ILC3 and icCD3ε+ IE-ILC3-like lymphocytes, we adapted our mIF panel to allow a distinction between these subsets of innate lymphoid cells. Very low levels of icCD3ε+ IE-ILC1-like cells were detected, most likely of the Id2 independent type as previously described by Ettersperger68, but interestingly, we identified a small icCD3ε+ IE-ILC3-like population, favoring however germinal centers of MALT.
We challenged our assay in follicular lymphoma (FL) tissues where we highlighted a reduction of ILC2 and ILC3, as compared to reactive lymph nodes and tonsil, with a sustained ILC1 population being dominant in FL, and a NK/ILC1ie population enriched in the neoplastic germinal centers, as do ILC2. Rare studies have investigated the role of ILCs in lymphoma development. In a recent one181, the authors designed a twelve-color multiparametric flow cytometry panel to investigate the distribution of the ILC family members in non-Hodgkin lymphoma (NHL) and healthy donors’ peripheral blood and LN. No differences were highlighted in LN of both groups, in terms of ILC frequency or subset distribution, but ILCs appeared activated with higher cytotoxicity in the peripheral blood of NHL181. Interestingly, they showed that the immunosuppressive microenvironment set by the tumor might negatively regulate ILCs. Considering this, FL tumoral cells might disturb ILC differentiation and function, explaining the drop of ILC-subtypes while IL-7Rαpool was sustained and even higher than in LN. Remarkably, the T cell pool was maintained, but a T helper shift occurred in FL compared to reactive LN. Th2 cells seem unaffected or slightly increased in lymphoma samples, but an important overall decrease of the Th17 phenotype is observed in FL conditions. In parallel, Th1 lymphocyte density increases significantly in tumor follicles when compared to LN.
The observations presented here raise important questions on the functional cooperation between Th cells and ILCs in the lymphoma microenvironment and beyond. The bidirectional ILC/T cell cross-talk is actively studied and reviewed70,172,182,183, for which molecular processes remain to be elucidated184. Close potential interactions between ILC1, ILC3, and Th17 cells, between ILC2 and Th2 cells, and finally NK/ILC1ie and Th1 cells, were evidenced by our cluster analysis and confirmed by proximity analysis and could have relevant clinical implications. Additionnaly, ILC1 dominate the corresponding Th1 phenotypes in all examined lymphoid tissues except in spleen, whereas conversely, ILC2 are exceeded in density by Th2 in all cases. In intestinal MALT and LN, ILC3 are more represented than Th17 cells, whereas in thymus, tonsil, and spleen, Th17 take over from ILC3. Only minor follicular infiltration was seen for ILCs (Supplementary Fig. 5) in appendix, ileum, LN and tonsil, except for ILC2. NK/ILC1ie, ILC1, and ILC3 do not display a strong affinity for the follicular regions in these lymphoid tissues. Under FL conditions, however, NK/ILC1ie and ILC1 join ILC2 in the lymphomatous follicles. In the spleen, ILCs preferentially occupy the white pulp and the medulla in the thymus. Our data clearly show that ILC2 behaves differently from the other ILC phenotypes, and appears, as identified by cluster analysis, an independent ILC cell type, characterized by follicular infiltration and predominant splenic white pulp and thymic medulla occupation. A concordant spatial gradient is seen for the different Th subtypes following their ILC counterparts as confirmed by spatial proximity analysis.
Altogether, the results presented herein demonstrate the feasibility and the potential of determining an Innate Lymphoid Index (ILIx), precisely depicting the spatial distribution of human ILC populations in relation to lymphocytes of the adaptive immune system, especially to their Th counterparts. The daily-practice compatible high-resolution computational mapping of ILCs is now available to be applied in human reactive and pathological conditions. It provides a simple and versatile method that puts forward the ILIx as a promising biomarker in clinical settings.
Methods
Patient and tissue characteristics
Patients’ samples were obtained after written informed consent in accordance with the Declaration of Helsinki and stored at the “CRB Cancer des Hôpitaux de Toulouse (BB-0033-00014)” collection. According to the French law, CRB Cancer collection has been declared to the Ministry of Higher Education and Research (DC-2008-463) and obtained a transfer agreement (AC-2013-1955) after approval by the ethics committee (Comité de Protection des Personnes Sudouest et outre mer II, CPP). Clinical and biological annotations of the samples have been declared to the CNIL (Comité National Informatique et Libertés). We studied the following formalin-fixed-paraffin embedded (FFPE) lymphoid tissues (n = 8, eight independent biological samples for each tissue type): palatine tonsil, lymph nodes (7 axillary plus 1 abdominal), thymus, spleen, appendix and ileum. All tissue fragments were treated using standard histopathology procedures. For each tissue fragment, 4 serial slides were cut at 4 µm on a standard microtome (LEICA Biosystems, Nussloch, Germany). The first slide was used to validate morphological quality using Hematoxylin-Eosin (H&E) stains and to determine the regions of interest (ROI). The remaining three slides were used for multiplex immunofluorescence assays. Careful selection of samples was based on the overall morphological quality of the FFPE tissue samples. Tissue fragments lacked pre-fixation, fixation, and tissue-processing artefacts. Detailed morphological quality assessment was performed on H&E stained slides, and samples were accepted for inclusion when morphological tissue-specific characteristics were classified as normal, except for spleen since these samples are only obtained after traumatic injury. Nuclei didn’t show any artefacts such as crushing, nuclear bubbling, indistinct chromatin, distortion, or aberrant or heterogenous hematoxylin-affinity within the morphologically equivalent groups of cell phenotypes. Cells lacked artefacts such as shrinkage, cytoplasmic clumping, detached appearance from neighboring cells, and heterogenous affinity for eosin within the morphologically equivalent groups of cell phenotypes. Tissue samples in which cell layers seemed detached from underlying layers were excluded. Consistency was strived for in a good representation of the principal, functionally important tissue compartments characterizing a certain tissue type. For tonsil, secondary follicles with germinal centers, crypts, epithelium, connective tissue and inter-follicular T-zones must be represented. Lymph nodes contained capsule, sinus, trabeculae and lymphoid follicles with or without germinal centers, less represented in reactive lymph nodes selected. In spleen, a clear distinction between white and red pulp was set as a criterium. For appendix and ileum, all functional layers of the intestinal tract (mucosa, muscularis mucosa, submucosa, muscularis) and lymphoid follicles needed to be represented on the cutting plane. In thymus fragments, distinction between cortex and medulla was set as a selection-criterium. Follicular lymphoma samples were characterized by a lymphomatous proliferation with follicular architecture presenting small to moderate-sized cells with hyperchromatic nuclei, primarily of centrocyte-type, with variable enrichment of centroblasts and germinal center-like formation. We included the specimens, independently of patient-heterogeneity, independently of their gender (self-reported), medical history, age (ranging from 4 months up to 88 years); and thus sample-heterogeneity independently on size, orientation of the tissue, or exact in situ position of the fragment in the studied organ before surgery. The multiplex immunofluorescence assays (mIF) were challenged on follicular lymphoma samples graded 1–2 (n = 8) according to the WHO classification185. Patient and tissue information is summarized in Supplementary Table 1. Ten out of 48 patients received chemotherapy and/or radiotherapy prior to surgery. Tissue fragments studied came from 43% female and 57% male patients. Five patients died prior to study closure. The entire experimental set-up and workflow is displayed in Fig. 1.
Multiplex Immunofluorescence
The BOND RX staining platform (LEICA Biosystems, Nussloch, Germany) was used to automate the mIF staining procedure on 3 serial 4 µm FFPE tissue sections (software version 7.0.9.267 RX). One batch represented the staining of the 3 mIF panels (3 times 5 markers and nuclear counterstain) for ILC discrimination in parallel on 8 independent biological samples of each lymphoid tissue type (total of 7 batches). After baking and dewaxing, tissue slides were heat-pretreated for 20 minutes at 100 °C using ER2 pretreatment solution (pH9, LEICA Biosystems, Nussloch, Germany). The slides were blocked for endogenous peroxidase activity using the Discovery inhibitor (15 minutes at room temperature) (07017944001, Roche Diagnostics, Basel, Switzerland). The slides were then stained for 5-plex immunofluorescence using the OPAL Technology (AKOYA Biosciences, Marlborough, USA) with sequential denaturation using ER1 pretreatment buffer (pH6, LEICA Biosystems, Nussloch, Germany), for 20 minutes at 97 °C between each antibody-staining cycle. The sequential application of all reagents (including catalog numbers and suppliers) on the 3 serial slides is detailed in the Supplementary Information (Supplementary Methods 1: Protocol NK-ILC1 mIF BRX; 2: ILC2 mIF BRX; 3: ILC3 mIF BRX). Primary antibody details are provided in Supplementary Table 8. Three mIF panels (Fig. 1 and Supplementary Methods 1 to 3) were used on serial sections for each selected tissue in which three markers were in common: CD20 (clone L26, ¼ in Ventana Antibody Diluent, Roche Diagnostics, Basel, Switzerland), TCRαδ cocktail (TCRδ, clone BSB-127, ready to use; TCRα, clone BSB-126, 1/100 in TCRδ, BioSB, Diagomics, Toulouse, France) and IL-7Rα (CD127, clone EPR23747-333, at 5.98 µg/ml in Ventana Antibody Diluent (Roche Diagnostics, Basel, Switzerland), ABCAM Netherlands, Amsterdam, Netherlands). Experimental conditions such as pretreatment, denaturation times and temperatures, concentrations of primary/secondary antibodies and OPAL Dyes, incubation times and temperatures were unchanged for all tissue types stained. Internal control of repeatability was performed by comparing the two common markers used in the 3 mIF panels, i.e., TCRαδ (T cells) and IL-7Rα (non-B/T), for all samples analyzed and is documented in Supplementary Fig. 7 validating intra-batch concordance. Inter-batch variation is related to tissue type heterogeneity and thus accepted as such. For thymic tissue examination, CD20 was replaced by Terminal Deoxynucleotidyl Transferase (TdT, clone SEN28, LEICA Biosystems, Nussloch, Germany) (Supplementary Fig. 1B, alternative “TdT” mIF panel) (additional staining batch). Ileum was examined using a mIF panel (additional staining batch) in which CD20 was replaced by CD3 (clone 2GV6, Roche Diagnostics, Basel, Switzerland) to investigate intra-epithelial CD3ε+ lymphoid cells (IE-ILC) (Supplementary Fig. 1B, alternative “CD3” mIF panel).
Two additional markers were included in each mIF panel to allow for ILC subtype differentiation. EOMES (TBR2, clone EPR21950-241, at 1.34 µg/ml in Ventana Antibody Diluent (Roche Diagnostics, Basel, Switzerland), ABCAM Netherlands, Amsterdam, Netherlands) and TBX21 (T-Bet, clone EPR9301, at 2.8 µg/ml in Ventana Antibody Diluent (Roche Diagnostics, Basel, Switzerland), ABCAM Netherlands, Amsterdam, Netherlands) were used for ILC1/ILC1ie/cNK differentiation. ICOS (CD278, clone RM417, ready to use, BioSB by Diagomics, Toulouse, France) and GATA-3 (clone L50-823, at 1 µg/ml in Ventana Antibody Diluent (Roche Diagnostics, Basel, Switzerland), BioSB, Diagomics, Toulouse, France) were used for ILC2 differentiation. For ILC3/LTi differentiation, AHR (clone ARC53212, at 1/500 in Ventana Antibody Diluent (Roche Diagnostics, Basel, Switzerland), ABclonal by Diagomics, Toulouse, France) and RORγT (clone 6F3.1, at 1 µg/ml in Ventana Antibody Diluent (Roche Diagnostics, Basel, Switzerland), BIOCARE Medical, Danaher, USA) were applied. Five OPAL dyes with excitation and emission wavelengths compatible with our whole slide imaging system were used: OPAL Polaris 480, OPAL 520, OPAL 570, OPAL 620 (1/300 in 1x Plus Automation Amplification Diluent, AKOYA Biosciences, Marlborough, USA) and OPAL 690 (1/150 in 1x Plus Automation Amplification Diluent, AKOYA Biosciences, Marlborough, USA). The tissue slides were counterstained using Spectral DAPI (AKOYA Biosciences, Marlborough, USA) and mounted with Invitrogen ProLong Gold Antifade Mounting medium (Life Technologies, ThermoFisher Scientific, California, USA) (Supplementary Methods 1–3).
Whole-slide imaging
Whole Hematoxylin-Eosin stained slides were digitized with a Pannoramic 250 Flash II digital microscope (3DHISTECH, Budapest, Hungary) equipped with a Zeiss Plan-Apochromat 20X NA 0.8 objective and a CIS VCC-FC60FR19CL 4-megapixel CMOS sensor (unit cell size 5.5 × 5.5 µm) mounted on a 1.6X optical adaptor, to achieve a scan resolution of 0.24 μm/pixel in the final image (corresponds to 41.1X magnification at the highest optical resolution in traditional microscopy). For mIF slides, 6-channels multispectral fluorescence imaging was performed using an AxioScan Z1 (Carl Zeiss Microscopy, Oberkochen, Germany) whole-slide scanner with appropriate narrow band-pass excitation and emission filters and specific dichroic mirrors (Semrock Inc., Rochester, NY, USA), and with a multi-channel solid-state light engine (Colibri 7, Carl Zeiss Microscopy, Oberkochen, Germany) equipped with 7 LEDs covering the entire visible spectrum from UV to far-red (370–648 nm) (Supplementary Fig. 3). Analog to digital image sampling was performed at 16-bit (65 536 grey levels, 37 000:1 dynamic range) with a high-resolution scientific complementary metal oxide semiconductor (sCMOS sensor with 2048 × 2048 cells of size 6.5 × 6.5 µm each) Peltier-cooled monochrome camera (Orca Flash 4.0 V3, Hamamatsu Photonics K.K., Japan), to achieve a final scan resolution of 0.32 µm/pixel. Fluorescence acquisition settings were established for each multiplex panel during the methods validation process and not modified afterwards.
Whole-slide image analysis and data visualization
Object-oriented whole-slide image analysis was carried out using the Halo framework (Indica Labs, Albuquerque, New Mexico, USA) with different image analysis modules and algorithms. For each selected tissue and each patient, multispectral images from the three mIF panels (serial sections) were registered using the Halo Image Registration module, which uses a “grid deformation” process translating position data from the target image to a grid, and from the grid to the source image. This module allowed for parallel, synchronized navigation and simultaneous annotation of slides within a registration project. This ensures to match sister slides at the structural level for a whole tissue slide (Supplementary Fig. 2). Each registered set of images was then manually annotated by a certified pathologist with three regions of interest (“interface”, “extra-interface”, and “interface + extra-interface” ROIs) in which stained cells were studied using the Halo HighPlex algorithm. In tonsil, appendix, ileum, lymph node, and follicular lymphoma images, the “interface” and “extra-interface” ROIs matched respectively the follicular and extra-follicular zone, the white pulp and red pulp in the spleen, and the medulla and cortex in thymus. With the Halo HighPlex algorithm, nuclear contour detection and segmentation were optimized based on the DAPI counterstain as well as various parameters and morphological features in real-time fine-tuning mode using graphical overlays (signal intensity and contrast, nuclear size and roundness, segmentation aggressiveness). Individual cell objects were then simulated by growing nuclei objects boundaries, and OPAL dye signal intensities for each marker were detected in their corresponding cell compartments (membrane + cytoplasm and/or nucleus) by thresholding grey level signal intensities. Several cell phenotypes were finally defined by selecting various marker combinations (Supplementary Fig. 1). As for the image acquisition process, all image analysis parameter settings were established for each multiplex panel during the methods validation process and not modified afterwards for the batch analysis process. For each ROI analyzed, morphometric features and fluorescence intensities were quantified in each cell compartment, and each detected object was tagged and classified for a specific phenotype based on their marker content, allowing calculation of positive cell densities and percentages. Individual X-Y location coordinates were also recorded for each detected object allowing merged spatial plot mapping for each case on registered images. The 3 serial slides of 4 µm do not necessarily contain the same cells, and all the biomarkers used to phenotype our cells of interest (ILCs and Ths) are always located on the same section (1 serial section for 1 mIF panel). The whole slide image registration process helped us to plot the different cellular phenotypes spatially on the same map in order to study the relative population distribution at a tissue structure level (Supplementary Fig. 2). Quantitative distribution analysis (around the interface ROIs) of individual cell phenotypes was performed using the Halo spatial analysis module, which was also used to analyze the putative cell/cell interactions in each tissue by quantifying the spatial proximity of Th cells within a 50 µm radius around each ILC nuclei. Raw data were finally exported from the Halo database as.csv text files, filtered using Power Query in Microsoft Excel (version 16.86), to be analyzed and visualized using GraphPad Prism (10.4.1), Panda (version 2.2.1), Matplotlib (version 3.8.3), and Seaborn (version 0.13.2) (Python packages). For proximity analysis, heatmaps and diagram cords were produced using the R packages “ComplexHeatmap” and “Circlize” respectively186,187. Public dataset scRNAseq from thymus (Li et al.103) and tonsil (Badosa et al.79) were downloaded and analysed with “CellChat” R package (version 2.1.2) to infer cell-cell communication.
Clustering analysis
ILC and Th cell density measurements were scaled using the Box-Cox power transformation implemented in the Scikit-learn Python library (version 1.4.1.post1)188. Four clusters were determined using the elbow method and the K-means algorithm. Cluster map representation of scaled data was done using the Seaborn Python library (version 0.13.2), assisted by ward hierarchical clustering calculated from distances computed with the SciPy (version 1.12.0) “pdist” Euclidean metric189.
Software and hardware used
Image visualization was performed using SlideViewer (version 2.5, 3DHISTECH) in brightfield mode and Zen Blue Edition (version 3.4.91, Carl Zeiss Microscopy) in fluorescence mode. Multispectral fluorescent images were analysed using the IndicaLabs Halo framework (version 3.6.4134) and the HighPlex algorithm (version 4.2.5). Data was visualized using R (version 4.3.2 “Eye Holes”), Panda (version 2.2.1), Matplotlib (version 3.8.3), and Seaborn (version 0.13.2) (Python packages). Non-parametric statistical analyses were performed using GraphPad Prism (6.0 h and 10.4.1). Image analysis algorithms ran on a Dell PowerEdge R740XD computing cluster (bi-CPU Intel Xeon Platinum 8280L 2.7 G 28C/56T, 64 GB RAM, GPU NVIDIA Tesla V100S 32 GB) and data were stored on a Dell PowerEdge R640 server (Intel Xeon Gold 5222 3.8G 4C/8T, 32 GB) running MySQL Server 8.0 + IndicaLabs gRPC and GraphQL APIs.
Flow cytometry and gating strategy
Fresh control lymphoid tissues (n = 5 independent biological samples from tonsil) were cut in 2 mm2 pieces and frozen in Cryostor CS10 Cell Cryopreservation Medium (Sigma-Aldrich, Saint Louis, MO, USA) until analysis. At thawing, tissue pieces were rinsed with complete medium, and cell suspensions were obtained following enzymatic digestion using the Tumor Dissociation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany, 130-095-929). Cell suspensions were filtered (70 µm) and rinsed once with PBS. Viability was assessed with the LIVE/DEAD Fixable Blue Viability Dye (Thermo Fisher Scientific, Waltham, MA, USA), following manufacturer’s instructions. All antibodies used for staining are listed in Supplementary Tables 8 and 9 (including clones and suppliers). Cells isolated from healthy donor tonsils were incubated with the following mixes of antibodies (Supplementary Table 9): (i) isotype controls; (ii-a to ii-e) standard FMO controls, one for each separate intracellular marker (including anti cell surface markers antibodies and individual isotypes controls for GATA-3, EOMES, TBX21, RORγT and AHR); (iii) anti cell surface markers antibodies and isotype controls mix for GATA-3, EOMES, TBX21, RORγT and AHR (i.e. specific control, SC); (iv) anti- cell surface marker antibodies and anti- GATA-3, EOMES, TBX21, RORγT and AHR antibodies. Peaks and Mean Fluorescence Intensities (MFI) corresponding to each individual FMO were strictly superposable to peaks and MFI of SC in all the populations considered for the analysis, confirming that either FMO or SC can be performed for FC experiments.
Cell surface markers (Supplementary Table 9) were stained with antibodies directed against CD141, CD11c, FcεRIα, CD14, CD20 and CD123, CD3, CD45, IL-7Rα (CD127), CRTH2 (CD294), CD56 (NCAM-1), CD117 (ckit), ICOS (CD278), NKp46 (CD335) and NKp44 (CD336). Cells were fixed and permeabilized with Transcription factor staining buffer set (eBiosciences, Thermo Fisher Scientific, Waltham, MA, USA) following manufacturer’s instructions. Intracellular saturation was performed by incubating cells for 10 minutes in permeabilisation buffer/4% FCS (Gibco, Thermo Fisher Scientific, Waltham MA, USA). Intracellular staining (Supplementary Table 9) was performed with the following antibodies: GATA-3, EOMES, TBX21 (T-Bet), RORγT, and AHR.
A minimum of 200,000 CD3− cells (defined through gating strategy) were registered for the acquisition. Flow cytometry data were analyzed following the presented gating strategy (Fig. 3A, Supplementary Fig. 6). Briefly, debris were excluded, and LIVE/DEAD negative cells were considered as viable. CD3− cells were then selected. CD45+/LIN− cells were subsequently gated, based on concomitant expression of CD45 and absence of staining with the following monoclonal antibodies: CD141, CD11c, FcεRIα, CD14, CD20, and CD123 (Supplementary Table 9). ILC pool IL-7Rα+ cells were then selected based on the absence of expression of CD56 and positivity for CD127 (IL-7Rα) (Fig. 3A, Supplementary Fig. 6). Positivity for each marker, boundaries, and thresholds were defined compared to isotypes, FMO, and SC considering different subpopulations of cells, for single and multiplex labeling (Supplementary Fig. 6).
Spatial transcriptomics
Frozen section of a reactive spleen sample was mounted onto the Visium spatial slide, fixed using methanol, stained using H&E stain, and digitized using LSM 710 videomicroscope. Then, the tissue was permeabilized on the slide and reverse transcription was performed in situ. Finally, second-strand synthesis, denaturation, DNA amplification, library construction and sequencing were performed following the manufacturer’s instructions (Visium Spatial Gene Expression Reagent Kits User Guide - Rev C) and as previously described162.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank the clinical repository tissue bank “CRB Cancer des Hôpitaux de Toulouse (BB-0033-00014)”, and especially Sophie Peries and Anne Gomez-Mascard. We thank Christine Bezombes, member of our team (Cancer Research Center of Toulouse (INSERM), Team 9 NoLymIT) for contributing to figure registration.
Author contributions
P.B. conceived the project and provided the necessary resources for project execution. P.B. and C.L. selected patient samples for inclusion as certified pathologists. N.V.A. and F.X.F. designed experiments and developed mIF panels. F.X.F. and N.V.A. designed and performed data acquisition, analysis, and visualization. F.X.F. and N.V.A. composed figures and tables. N.V.A. and F.X.F. equally contributed to this article. P.G. designed Flow Cytometry panels and performed gating. P.G. performed spatial transcriptomics analysis. P.G. and M.T. performed the public datasets analysis. N.V.A., F.X.F., and P.B. wrote the paper with input from C.L., P.G. and C.S. N.V.A., F.X.F., P.G., C.S., C.L. and P.B. discussed results and reviewed the article.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw data produced for this study, used for data analysis and visualization, are available as a public GitHub repository here: https://github.com/ouatataz/innate-lymphoid-cells. The spatial transcriptomics raw data produced in this study are available at Gene Expression Omnibus (GEO) with accession number GSE228056, spatial transcriptomics raw data produced by Björklund et al.43 are available at Gene Expression Omnibus (GEO) with accession number GSE70580. Publicly available datasets were downloaded from the following sources: https://zenodo.org/records/13207776 and https://bioconductor.org/packages/release/data/experiment/html/HCATonsilData.html. All data are included in the Supplementary Information file or are available from the authors, as are the unique reagents used in this article. This file contains: general specifications of marker combinations and resulting cell phenotypes of the used mIF panels (Supplementary Fig. 1), image registration (Supplementary Fig.2), spectral band settings for image acquisition (Supplementary Fig. 3), split violin plots of alternative “CD3” mIF panel cell phenotypes (Supplementary Fig. 4), gauges representing the NK/ILCs/Th cell densities within interface for all tissue types analyzed (Supplementary Fig. 5), control plots justifying gating strategy thresholds used for ILCs analysis in human tonsils (Supplementary Fig. 6), correlation matrix to validate intra-batch (7 batches, 1 batch per tissue type studied) concordance for two common markers used in the mIF panels, i.e. TCRαδ (T cells) and IL-7Rα (non-B/T) (Supplementary Fig. 7), comparative infiltration analysis plots for Th1, Th2 and Th17 cells location for all tissue types (Supplementary Fig. 8), and comparative diagram chords for proximity analysis for all tissue types (Supplementary Fig. 9). Public dataset analysis are presented on Supplementary Figs. 10 and 11 for thymus and tonsil respectively. Supplementary Table 1 provides information about the patients and tissues, Supplementary Tables 2 to 7 provide descriptive statistics, and Supplementary Tables 8 and 9 show the antibody panels used for mIF and flow cytometry analysis. The raw numbers for charts and graphs are available in the corresponding Source Data file whenever possible and as indicated in the corresponding legend. FACS data are only available after official request, to avoid uncontrolled reproduction and publication of patient data, by e-mailing the corresponding author, who will respond within two weeks. The request must be accompanied by a short description of the project, the reason, and aim for using our FACS data. It is the corresponding author who will decide whether the external use of the FACS data is allowed, depending on the compatibility of the initial question and internal scientific scopes. Source data are provided with this paper.
Code availability
The python scripts used for data analysis and visualization are available as a public GitHub repository here: https://github.com/ouatataz/innate-lymphoid-cells.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nathalie Van Acker, François-Xavier Frenois.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-025-59811-1.
References
- 1.Artis, D. & Spits, H. The biology of innate lymphoid cells. Nature517, 293–301 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Klose, C. S. N. & Artis, D. Innate lymphoid cells control signaling circuits to regulate tissue-specific immunity. Cell Res.30, 475–491 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bal, S. M., Golebski, K. & Spits, H. Plasticity of innate lymphoid cell subsets. Nat. Rev. Immunol.20, 552–565 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Spits, H. & Di Santo, J. P. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol.12, 21–27 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Lopes, N. et al. Tissue-specific transcriptional profiles and heterogeneity of natural killer cells and group 1 innate lymphoid cells. Cell Rep. Med.3, 100812 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, I. S. & Van Dyken, S. J. Both Horatio and Polonius: Innate lymphoid cells in tissue homeostasis and repair. ImmunoHorizons7, 729–736 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan, X. & Rudensky, A. Y. Hallmarks of tissue-resident lymphocytes. Cell164, 1198–1211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan, Z. et al. Innate lymphoid cells are double-edged swords under the mucosal barrier. J. Cell. Mol. Med. 25, 8579–8587 (2021). [DOI] [PMC free article] [PubMed]
- 9.Lim, A. I. & Santo, J. P. D. ILC-poiesis: Ensuring tissue ILC differentiation at the right place and time. Eur. J. Immunol.49, 11–18 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Mjösberg, J. & Spits, H. Human innate lymphoid cells. J. Allergy Clin. Immunol.138, 1265–1276 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Mazzurana, L., Rao, A., Van Acker, A. & Mjösberg, J. The roles for innate lymphoid cells in the human immune system. Semin. Immunopathol.40, 407–419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenzie, A. N. J., Spits, H. & Eberl, G. Innate lymphoid cells in inflammation and immunity. Immunity41, 366–374 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Moreno-Nieves, U. Y. et al. Landscape of innate lymphoid cells in human head and neck cancer reveals divergent NK cell states in the tumor microenvironment. Proc. Natl Acad. Sci.118, e2101169118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, S., Wu, P., Chen, Y. & Chai, Y. Ambiguous roles and potential therapeutic strategies of innate lymphoid cells in different types of tumor. Oncol. Lett.20, 1513–1525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinrich, B. et al. The tumour microenvironment shapes innate lymphoid cells in patients with hepatocellular carcinoma. Gut gutjnl-2021-325288. 10.1136/gutjnl-2021-325288. (2021). [DOI] [PMC free article] [PubMed]
- 16.Bennstein, S. B. & Uhrberg, M. Biology and therapeutic potential of human innate lymphoid cells. FEBS J.289, 3967–3981 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Kortekaas Krohn, I. et al. Emerging roles of innate lymphoid cells in inflammatory diseases: Clinical implications. Allergy73, 837–850 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Spits, H. TOX sets the stage for innate lymphoid cells. Nat. Immunol.16, 594–595 (2015). [DOI] [PubMed] [Google Scholar]
- 19.de Boer, O. J. et al. Comparison of two different immunohistochemical quadruple staining approaches to identify innate lymphoid cells in formalin-fixed paraffin-embedded human tissue. J. Histochem. Cytochem.68, 127–138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brüggen, M.-C. et al. In situ mapping of innate lymphoid cells in human skin: evidence for remarkable differences between normal and inflamed skin. J. Invest. Dermatol.136, 2396–2405 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Pascual-Reguant, A. et al. Multiplexed histology analyses for the phenotypic and spatial characterization of human innate lymphoid cells. Nat. Commun.12, 1737 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yudanin, N. A. et al. Spatial and temporal mapping of human innate lymphoid cells reveals elements of tissue specificity. Immunity50, 505–519.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, C. et al. Delineating spatiotemporal and hierarchical development of human fetal innate lymphoid cells. Cell Res. 10.1038/s41422-021-00529-2. (2021). [DOI] [PMC free article] [PubMed]
- 24.Crinier, A. et al. High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity49, 971–986.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.High-dimensional single-cell analysis of natural killer cell heterogeneity in human blood. https://www.researchsquare.com10.21203/rs.3.rs-3870228/v1. (2024).
- 26.Ercolano, G. et al. Distinct and shared gene expression for human innate versus adaptive helper lymphoid cells. J. Leukoc. Biol.108, 723–737 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meininger, I. et al. Tissue-specific features of innate lymphoid cells. Trends Immunol.41, 902–917 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Spits, H. et al. Innate lymphoid cells — a proposal for uniform nomenclature. Nat. Rev. Immunol.13, 145–149 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Ruf, B., Greten, T. F. & Korangy, F. Innate lymphoid cells and innate-like T cells in cancer — at the crossroads of innate and adaptive immunity. Nat. Rev. Cancer23, 351–371 (2023). [DOI] [PubMed] [Google Scholar]
- 30.Surace, L. & Di Santo, J. P. Local and systemic features of ILC immunometabolism. Curr. Opin. Hematol.29, 209–217 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Nabekura, T. & Shibuya, A. Type 1 innate lymphoid cells: Soldiers at the front line of immunity. Biomed. J. 10.1016/j.bj.2020.10.001. (2020). [DOI] [PMC free article] [PubMed]
- 32.Vivier, E. et al. Innate lymphoid cells: 10 years on. Cell174, 1054–1066 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Vivier, E. et al. Natural killer cell therapies. Nature626, 727–736 (2024). [DOI] [PubMed] [Google Scholar]
- 34.Tomasello, E. et al. Mapping of NKp46(+) cells in healthy human lymphoid and non-lymphoid tissues. Front. Immunol.3, 344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Annunziato, F., Romagnani, C. & Romagnani, S. The 3 major types of innate and adaptive cell-mediated effector immunity. J. Allergy Clin. Immunol.135, 626–635 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Crinier, A. et al. Multidimensional molecular controls defining NK/ILC1 identity in cancers. Semin. Immunol. 101424 10.1016/j.smim.2020.101424. (2020). [DOI] [PubMed]
- 37.Ryu, S., Lim, M., Kim, J. & Kim, H. Y. Versatile roles of innate lymphoid cells at the mucosal barrier: from homeostasis to pathological inflammation. Exp. Mol. Med.55, 1845–1857 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, Y. et al. Innate immunity in cancer biology and therapy. Int. J. Mol. Sci.24, 11233 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi, M. et al. Exploiting innate immunity for cancer immunotherapy. Mol. Cancer22, 187 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, H., Sun, L., Feng, L., Yin, Y. & Zhang, W. Role of innate lymphoid cells in obesity and insulin resistance. Front. Endocrinol.13, 855197 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katirci, E., Kendirci-Katirci, R. & Korgun, E. T. Are innate lymphoid cells friend or foe in human pregnancy? Am. J. Reprod. Immunol.91, e13834 (2024). N. Y. N 1989. [DOI] [PubMed] [Google Scholar]
- 42.Orimo, K. et al. Characteristics of tissue–resident ILCs and their potential as therapeutic targets in mucosal and skin inflammatory diseases. Allergy76, 3332–3348 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Björklund, Å. K. et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol.17, 451–460 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Mazzurana, L. et al. Tissue-specific transcriptional imprinting and heterogeneity in human innate lymphoid cells revealed by full-length single-cell RNA-sequencing. Cell Res31, 554–568 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cossarizza, A. et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur. J. Immunol.49, 1457–1973 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serafini, N., Vosshenrich, C. A. J. & Di Santo, J. P. Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol.15, 415–428 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Spits, H. & Cupedo, T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol.30, 647–675 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Liu, S. et al. Optimal identification of human conventional and nonconventional (CRTH2-IL7Rα-) ILC2s using additional surface markers. J. Allergy Clin. Immunol.146, 390–405 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lundgren, S. et al. Topographical distribution and spatial interactions of innate and semi-innate immune cells in pancreatic and other Periampullary Adenocarcinoma. Front. Immunol.11, 558169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gasteiger, G., Fan, X., Dikiy, S., Lee, S. Y. & Rudensky, A. Y. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science350, 981–985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, N. et al. Mass cytometry reveals innate lymphoid cell differentiation pathways in the human fetal intestine. J. Exp. Med.215, 1383–1396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barros-Martins, J., Bruni, E., Fichtner, A. S., Cornberg, M. & Prinz, I. OMIP-084: 28-color full spectrum flow cytometry panel for the comprehensive analysis of human γδ T cells. Cytom. Part J. Int. Soc. Anal. Cytol. 10.1002/cyto.a.24564. (2022). [DOI] [PubMed]
- 53.Doyle, C. M. et al. OMIP 082: A 25-color phenotyping to define human innate lymphoid cells, natural killer cells, mucosal-associated invariant T cells, and γδ T cells from freshly isolated human intestinal tissue. Cytom. Part J. Int. Soc. Anal. Cytol.101, 196–202 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Cossarizza, A. et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (third edition). Eur. J. Immunol.51, 2708–3145 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, J. et al. T-bet and Eomes govern differentiation and function of mouse and human NK cells and ILC1. Eur. J. Immunol.48, 738–750 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Huang, C. & Bi, J. Expression regulation and function of T-Bet in NK cells. Front. Immunol.12, 761920 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conlon, T. M., Knolle, P. A. & Yildirim, A. Ö. Local tissue development of type 1 innate lymphoid cells: guided by interferon-gamma. Signal Transduct. Target. Ther.6, 287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuchs, A. et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity38, 769–781 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li, G. et al. T-Bet and Eomes regulate the balance between the effector/central memory T cells versus memory stem like T cells. PLoS ONE8, e67401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cherrier, D. E., Serafini, N. & Di Santo, J. P. Innate lymphoid cell development: A T cell perspective. Immunity48, 1091–1103 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Kogame, T., Egawa, G., Nomura, T. & Kabashima, K. Waves of layered immunity over innate lymphoid cells. Front. Immunol.13, 957711 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shin, S. B. & McNagny, K. M. ILC-You in the Thymus: A fresh look at innate lymphoid cell development. Front. Immunol.12, 681110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong, C., Luckey, M. & Park, J.-H. Intrathymic IL-7: The where, when, and why of IL-7 signaling during T cell development. Semin. Immunol.24, 151–158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun, L., Su, Y., Jiao, A., Wang, X. & Zhang, B. T cells in health and disease. Signal Transduct. Target. Ther.8, 235 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Müller, V., de Boer, R. J., Bonhoeffer, S. & Szathmáry, E. An evolutionary perspective on the systems of adaptive immunity. Biol. Rev.93, 505–528 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Matsutani, T. et al. Alteration of T-cell receptor repertoires during thymic T-cell development. Scand. J. Immunol.64, 53–60 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Hue, S. S.-S., Ng, S.-B., Wang, S. & Tan, S.-Y. Cellular origins and pathogenesis of gastrointestinal NK- and T-Cell Lymphoproliferative disorders. Cancers14, 2483 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ettersperger, J. et al. Interleukin-15-dependent T-cell-like innate intraepithelial lymphocytes develop in the intestine and transform into lymphomas in celiac disease. Immunity45, 610–625 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Liu, Z. et al. Immunosenescence: molecular mechanisms and diseases. Signal Transduct. Target. Ther.8, 200 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.von Burg, N., Turchinovich, G. & Finke, D. Maintenance of immune homeostasis through ILC/T cell interactions. Front. Immunol.6, 416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schroeder, J.-H., Howard, J. K. & Lord, G. M. Transcription factor-driven regulation of ILC1 and ILC3. Trends Immunol.43, 564–579 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiu, J. et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity36, 92–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qiu, J. et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity39, (2013). 10.1016/j.immuni.2013.08.002 [DOI] [PMC free article] [PubMed]
- 74.Qiu, J. et al. Tissue signals imprint Aiolos expression in ILC2s to modulate type 2 immunity. Mucosal. Immunol.14, 1306–1322 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Naito, M. & Kumanogoh, A. Group 2 innate lymphoid cells and their surrounding environment. Inflamm. Regen.43, 21 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chopp, L., Redmond, C., O’Shea, J. J. & Schwartz, D. M. From thymus to tissues and tumors: A review of T-cell biology. J. Allergy Clin. Immunol.151, 81–97 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mörbe, U. M. et al. Human gut-associated lymphoid tissues (GALT); diversity, structure, and function. Mucosal. Immunol.14, 793–802 (2021). [DOI] [PubMed] [Google Scholar]
- 78.Mackley, E. C. et al. CCR7-dependent trafficking of RORγ+ ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat. Commun.6, 5862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Massoni-Badosa, R. et al. An atlas of cells in the human tonsil. Immunity57, 379–399.e18 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andreas, N., Geißler, K., Priese, J., Guntinas-Lichius, O. & Kamradt, T. Age-related changes of the innate immune system of the palatine tonsil in a healthy cohort. Front. Immunol.14, 1183212 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qin, M. et al. Tissue microenvironment induces tissue specificity of ILC2. Cell Death Discov.10, 1–10 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lim, A. I. et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell168, 1086–1100.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Zhu, J. GATA3 regulates the development and functions of innate lymphoid cell subsets at multiple stages. Front. Immunol.8, 1571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhong, C. et al. Differential expression of the transcription factor GATA3 specifies lineage and functions of innate lymphoid cells. Immunity52, 83–95.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yagi, R. et al. The Transcription Factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity40, 378–388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Helm, E. Y. & Zhou, L. Transcriptional regulation of innate lymphoid cells and T cells by aryl hydrocarbon receptor. Front. Immunol.14, 1056267 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Montaldo, E. et al. Human RORγt(+)CD34(+) cells are lineage-specified progenitors of group 3 RORγt(+) innate lymphoid cells. Immunity41, 988–1000 (2014). [DOI] [PubMed] [Google Scholar]
- 88.Scoville, S. D. et al. A progenitor cell expressing transcription factor RORγt generates all human innate lymphoid cell subsets. Immunity44, 1140–1150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hazenberg, M. D. & Spits, H. Human innate lymphoid cells. Blood124, 700–709 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Bernink, J. H. et al. Interleukin-12 and −23 control plasticity of CD127(+) Group 1 and Group 3 innate lymphoid cells in the intestinal Lamina Propria. Immunity43, 146–160 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Boyd, A., Ribeiro, J. M. C. & Nutman, T. B. Human CD117 (cKit)+ innate lymphoid cells have a discrete transcriptional profile at homeostasis and are expanded during filarial infection. PLoS ONE9, e108649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cupedo, T. ILC2: at home in the thymus. Eur. J. Immunol.48, 1441–1444 (2018). [DOI] [PubMed] [Google Scholar]
- 93.Jones, R. et al. Dynamic changes in intrathymic ILC populations during murine neonatal development. Eur. J. Immunol.48, 1481–1491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Simoni, Y. et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity46, 148–161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pont, F. et al. Single-Cell Virtual Cytometer allows user-friendly and versatile analysis and visualization of multimodal single cell RNAseq datasets. NAR Genom. Bioinforma.2, lqaa025 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hepworth, M. R. et al. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4+ T cells. Science348, 1031–1035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Song, P. et al. Tissue specific imprinting on innate lymphoid cells during homeostasis and disease process revealed by integrative inference of single-cell transcriptomics. Front. Immunol.14, 1127413 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Çakıcı, K., Gökdoğan, O., Genç, D., Günaydın, B. & Üçüncü, H. Comparison of innate lymphoid cells from tissue and blood in chronic tonsillitis and tonsillar hypertrophy. Int. J. Pediatr. Otorhinolaryngol.174, 111740 (2023). [DOI] [PubMed] [Google Scholar]
- 99.Poposki, J. A. et al. Group 2 innate lymphoid cells are elevated and activated in chronic rhinosinusitis with nasal polyps. Immun. Inflamm. Dis.5, 233–243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Symowski, C. & Voehringer, D. Interactions between innate lymphoid cells and cells of the innate and adaptive immune system. Front. Immunol.8, 1422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tougaard, P. et al. Type 1 immunity enables neonatal thymic ILC1 production. Sci. Adv.10, eadh5520 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bajana, S. et al. Correlation between circulating innate lymphoid cell precursors and thymic function. iScience25, 103732 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li, Y. et al. Unraveling the spatial organization and development of human thymocytes through integration of spatial transcriptomics and single-cell multi-omics profiling. Nat. Commun.15, 7784 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yayon, N. et al. A spatial human thymus cell atlas mapped to a continuous tissue axis. Nature635, 708–718 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ragazzini, R. et al. Defining the identity and the niches of epithelial stem cells with highly pleiotropic multilineage potency in the human thymus. Dev. Cell58, 2428–2446.e9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou, Y., Cao, G., Cai, H., Huang, H. & Zhu, X. The effect and clinical significance of FN1 expression on biological functions of gastric cancer cells. Cell. Mol. Biol. Noisy–le-Grand.66, 191–198 (2020). [PubMed] [Google Scholar]
- 107.Wang, X. et al. Fn1 regulates the third pharyngeal pouch patterning and morphogenesis. J. Dent. Res.101, 1082–1091 (2022). [DOI] [PubMed] [Google Scholar]
- 108.Jono, H. & Ando, Y. Midkine: A novel prognostic biomarker for cancer. Cancers2, 624–641 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cambier, S., Gouwy, M. & Proost, P. The chemokines CXCL8 and CXCL12: molecular and functional properties, role in disease and efforts towards pharmacological intervention. Cell. Mol. Immunol.20, 217–251 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Emre, Y. et al. Thymic epithelial cell expansion through matricellular protein CYR61 boosts progenitor homing and T-cell output. Nat. Commun.4, 2842 (2013). [DOI] [PubMed] [Google Scholar]
- 111.Yang, X. et al. Transcriptional profile of human thymus reveals IGFBP5 is correlated with age-related thymic involution. Front. Immunol.15, 1322214 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Calandra, T. & Roger, T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat. Rev. Immunol.3, 791–800 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scur, M., Parsons, B. D., Dey, S. & Makrigiannis, A. P. The diverse roles of C-type lectin-like receptors in immunity. Front. Immunol.14, 1126043 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Redelinghuys, P. & Brown, G. D. Inhibitory C-type lectin receptors in myeloid cells. Immunol. Lett.136, 1–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rebuffet, L. et al. High-dimensional single-cell analysis of human natural killer cell heterogeneity. Nat. Immunol.25, 1474–1488 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Earl, L. A., Bi, S. & Baum, L. G. N- and O-glycans modulate Galectin-1 Binding, CD45 signaling, and T cell death*. J. Biol. Chem.285, 2232–2244 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oliveira-de-Abreu, E. et al. Lack of Galectin-3 disrupts thymus homeostasis in association to increase of local and systemic glucocorticoid levels and steroidogenic machinery. Front. Endocrinol.9, 365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu, F.-T. & Stowell, S. R. The role of galectins in immunity and infection. Nat. Rev. Immunol. 23, 479–494 (2023). [DOI] [PMC free article] [PubMed]
- 119.Clark, E. A. & Giltiay, N. V. CD22: A regulator of innate and adaptive B cell responses and autoimmunity. Front. Immunol.9, 2235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Borelli, A. & Irla, M. Lymphotoxin: from the physiology to the regeneration of the thymic function. Cell Death Differ.28, 2305–2314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.To, T. T., Oparaugo, N. C., Kheshvadjian, A. R., Nelson, A. M. & Agak, G. W. Understanding Type 3 innate lymphoid cells and crosstalk with the microbiota: a skin connection. Int. J. Mol. Sci.25, 2021 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chupp, G. L. et al. Tissue and T cell distribution of precursor and mature IL-16. J. Immunol. Baltim. Md 1950161, 3114–3119 (1998). [PubMed] [Google Scholar]
- 123.Vollmann, E. H. et al. Specialized transendothelial dendritic cells mediate thymic T-cell selection against blood-borne macromolecules. Nat. Commun.12, 6230 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Puig, K. L. et al. Amyloid precursor protein modulates macrophage phenotype and diet-dependent weight gain. Sci. Rep.7, 43725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sondag, C. M. & Combs, C. K. Amyloid precursor protein mediates proinflammatory activation of monocytic lineage cells. J. Biol. Chem.279, 14456–14463 (2004). [DOI] [PubMed] [Google Scholar]
- 126.Komlósi, Z. I. et al. Human CD40 ligand–expressing type 3 innate lymphoid cells induce IL-10–producing immature transitional regulatory B cells. J. Allergy Clin. Immunol.142, 178–194.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 127.Hu, Y., Liu, X., Zhang, Y. & Guo, X. The interaction network between group 3 innate lymphoid cells and other cells. Fundam. Res. 10.1016/j.fmre.2023.10.021. (2023).
- 128.Wee, P. & Wang, Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers9, 52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hu, M., Zhang, H., Liu, Q. & Hao, Q. Structural basis for human PECAM-1-mediated trans-homophilic cell adhesion. Sci. Rep.6, 38655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chang, T.-D. et al. The different paradigms of NK cell death in patients with severe trauma. Cell Death Dis.15, 1–14 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Osuka, A., Ogura, H., Ueyama, M., Shimazu, T. & Lederer, J. A. Immune response to traumatic injury: harmony and discordance of immune system homeostasis. Acute Med. Surg.1, 63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yu, H. et al. The “Dialogue” between central and peripheral immunity after ischemic stroke: focus on spleen. Front. Immunol.12, 792522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gieseck, R. L., Wilson, M. S. & Wynn, T. A. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol.18, 62–76 (2018). [DOI] [PubMed] [Google Scholar]
- 134.Gamez-Belmonte, R., Erkert, L., Wirtz, S. & Becker, C. The regulation of intestinal inflammation and cancer development by Type 2 immune responses. Int. J. Mol. Sci.21, 9772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Luo, X. & Villablanca, E. J. Type 2 immunity in intestinal homeostasis and inflammatory bowel disease. Biochem. Soc. Trans.49, 2371–2380 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Smith, K. M. et al. Th1 and Th2 CD4+ T cells provide help for B cell clonal expansion and antibody synthesis in a similar manner in vivo. J. Immunol.165, 3136–3144 (2000). [DOI] [PubMed] [Google Scholar]
- 137.Shinkai, K., Mohrs, M. & Locksley, R. M. Helper T cells regulate type-2 innate immunity in vivo. Nature420, 825–829 (2002). [DOI] [PubMed] [Google Scholar]
- 138.Fan, H. et al. Innate lymphoid cells: regulators of gut barrier function and immune homeostasis. J. Immunol. Res.2019, 2525984 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim, M. H., Taparowsky, E. J. & Kim, C. H. Retinoic acid differentially regulates the migration of innate lymphoid cell subsets to the gut. Immunity43, 107–119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Panda, S. K. & Colonna, M. Innate lymphoid cells in mucosal immunity. Front. Immunol.10, 861 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hayakawa, Y. & Wang, T. C. The Tuft cell-ILC2 circuit integrates intestinal defense and homeostasis. Cell174, 251–253 (2018). [DOI] [PubMed] [Google Scholar]
- 142.von Moltke, J., Ji, M., Liang, H.-E. & Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2–epithelial response circuit. Nature529, 221–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schneider, C., O’Leary, C. E. & Locksley, R. M. Regulation of immune responses by tuft cells. Nat. Rev. Immunol.19, 584–593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gurram, R. K. et al. Crosstalk between ILC2s and Th2 cells varies among mouse models. Cell Rep.42, 112073 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rabinovich, G. A. & Toscano, M. A. Turning ‘sweet’ on immunity: galectin–glycan interactions in immune tolerance and inflammation. Nat. Rev. Immunol.9, 338–352 (2009). [DOI] [PubMed] [Google Scholar]
- 146.Ilarregui, J. M. et al. Tolerogenic signals delivered by dendritic cells to T cells through a galectin-1-driven immunoregulatory circuit involving interleukin 27 and interleukin 10. Nat. Immunol.10, 981–991 (2009). [DOI] [PubMed] [Google Scholar]
- 147.Jacquelot, N. et al. Immune checkpoints and innate lymphoid cells—new avenues for cancer immunotherapy. Cancers13, 5967 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xu, H. et al. The role of ICAM-1 molecule in the migration of Langerhans cells in the skin and regional lymph node. Eur. J. Immunol.31, 3085–3093 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Irie, M. et al. Annexin A1 is a cell-intrinsic metalloregulator of zinc in human ILC2s. Cell Rep.42, 112610 (2023). [DOI] [PubMed] [Google Scholar]
- 150.Halim, T. Y. F. et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T Helper 2 cell-mediated allergic lung inflammation. Immunity40, 425–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Blomme, E. E. et al. Quantification and role of innate lymphoid cell subsets in Chronic Obstructive Pulmonary Disease. Clin. Transl. Immunol.10, e1287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.He, Y. et al. Single-cell profiling of human CD127+ innate lymphoid cells reveals diverse immune phenotypes in hepatocellular carcinoma. Hepatology. 76, 1013–1029 (2022). [DOI] [PMC free article] [PubMed]
- 153.Brouwer, T. P. et al. Local and systemic immune profiles of human pancreatic ductal adenocarcinoma revealed by single-cell mass cytometry. J. Immunother. Cancer10, e004638 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Navikas, V. et al. Semi-automated approaches for interrogating spatial heterogeneity of tissue samples. Sci. Rep. 14, 5025 (2024). [DOI] [PMC free article] [PubMed]
- 155.Millian, D. E. et al. Cutting-edge platforms for analysis of immune cells in the hepatic microenvironment—focus on tumor-associated macrophages in hepatocellular carcinoma. Cancers14, 1861 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Scheuermann, S. et al. Unveiling spatial complexity in solid tumor immune microenvironments through multiplexed imaging. Front. Immunol.15, 1383932 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.de Souza, N., Zhao, S. & Bodenmiller, B. Multiplex protein imaging in tumour biology. Nat. Rev. Cancer24, 171–191 (2024). [DOI] [PubMed] [Google Scholar]
- 158.Mungenast, F. et al. Next-generation digital histopathology of the tumor microenvironment. Genes12, 538 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim, E. N. et al. Dual-modality imaging of immunofluorescence and imaging mass cytometry for whole-slide imaging and accurate segmentation. Cell Rep. Methods3, 100595 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lin, J.-R. et al. High-plex immunofluorescence imaging and traditional histology of the same tissue section for discovering image-based biomarkers. Nat. Cancer4, 1036–1052 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tian, L., Chen, F. & Macosko, E. Z. Moving genomics into tissues. Nat. Biotechnol.41, 773–782 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pont, F. et al. Single-cell spatial explorer: easy exploration of spatial and multimodal transcriptomics. BMC Bioinforma.24, 30 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ho, I.-C., Tai, T.-S. & Pai, S.-Y. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol.9, 125–135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dong, M. et al. The ICOS–ICOSL pathway tunes thymic selection. Immunol. Cell Biol.100, 205–217 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang, R. et al. TCF-1 regulates thymocyte survival via a RORγt-dependent pathway. J. Immunol. Baltim. Md 1950187, 5964–5973 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Laiosa, M. D. et al. Identification of stage-specific gene modulation during early thymocyte development by whole-genome profiling analysis after aryl hydrocarbon receptor activation. Mol. Pharmacol.77, 773–783 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Park, J.-E. et al. A cell atlas of human thymic development defines T cell repertoire formation. Science367, eaay3224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hernández-Torres, D. C. & Stehle, C. Embryonic ILC-poiesis across tissues. Front. Immunol.13, 1040624 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Symmank, D., Richter, F. C. & Rendeiro, A. F. Navigating the thymic landscape through development: from cellular atlas to tissue cartography. Genes Immun. 1–3 10.1038/s41435-024-00257-8. (2004). [DOI] [PMC free article] [PubMed]
- 170.Ferreira, A. C. F. et al. RORα is a critical checkpoint for T cell and ILC2 commitment in the embryonic thymus. Nat. Immunol.22, 166–178 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kim, S. et al. CD117+ CD3− CD56− OX40Lhigh cells express IL-22 and display an LTi phenotype in human secondary lymphoid tissues. Eur. J. Immunol.41, 1563–1572 (2011). [DOI] [PubMed] [Google Scholar]
- 172.Sonnenberg, G. F. & Hepworth, M. R. Functional interactions between innate lymphoid cells and adaptive immunity. Nat. Rev. Immunol.19, 599–613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jackson-Jones, L. H. et al. Fat-associated lymphoid clusters control local IgM secretion during pleural infection and lung inflammation. Nat. Commun.7, 12651 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hardtke, S., Ohl, L. & Förster, R. Balanced expression of CXCR5 and CCR7 on follicular T helper cells determines their transient positioning to lymph node follicles and is essential for efficient B-cell help. Blood106, 1924–1931 (2005). [DOI] [PubMed] [Google Scholar]
- 175.Junt, T. et al. CXCR5-dependent seeding of follicular niches by B and Th cells augments antiviral B cell responses. J. Immunol.175, 7109–7116 (2005). [DOI] [PubMed] [Google Scholar]
- 176.Kim, C. H. et al. Subspecialization of Cxcr5+ T Cells. J. Exp. Med.193, 1373–1382 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bunse, M. et al. CXCR5 CAR-T cells simultaneously target B cell non-Hodgkin’s lymphoma and tumor-supportive follicular T helper cells. Nat. Commun.12, 240 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.O’Connor, M. H. et al. A follicular regulatory Innate Lymphoid Cell population impairs interactions between germinal center Tfh and B cells. Commun. Biol.4, 563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dogra, P. et al. Tissue determinants of human NK cell development, function, and residence. Cell180, 749–763.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Grégoire, C. et al. The trafficking of natural killer cells. Immunol. Rev.220, 169–182 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Roma, S. et al. Dampening of cytotoxic innate lymphoid cells: A new tumour immune escape mechanism in B cell non-Hodgkin’s lymphoma. Cell. Immunol.382, 104615 (2022). [DOI] [PubMed] [Google Scholar]
- 182.Cherrier, M., Ramachandran, G. & Golub, R. The interplay between innate lymphoid cells and T cells. Mucosal Immunol.13, 732–742 (2020). [DOI] [PubMed] [Google Scholar]
- 183.Kiniwa, T. & Moro, K. Localization and site-specific cell-cell interactions of group 2 innate lymphoid cells. Int. Immunol.33, 251–259 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Frech, M. et al. Btn2a2 regulates ILC2–T cell cross talk in Type 2 immune responses. Front. Immunol.13, 757436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kurz, K. S. et al. Follicular lymphoma in the 5th Edition of the WHO-classification of haematolymphoid neoplasms—updated classification and new biological data. Cancers15, 785 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize Implements and enhances circular visualization in R. Bioinforma. Oxf. Engl.30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 187.Gu, Z. Complex heatmap visualization. iMeta1, e43 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pedregosa, F. et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res.12, 2825–2830 (2011). [Google Scholar]
- 189.Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data produced for this study, used for data analysis and visualization, are available as a public GitHub repository here: https://github.com/ouatataz/innate-lymphoid-cells. The spatial transcriptomics raw data produced in this study are available at Gene Expression Omnibus (GEO) with accession number GSE228056, spatial transcriptomics raw data produced by Björklund et al.43 are available at Gene Expression Omnibus (GEO) with accession number GSE70580. Publicly available datasets were downloaded from the following sources: https://zenodo.org/records/13207776 and https://bioconductor.org/packages/release/data/experiment/html/HCATonsilData.html. All data are included in the Supplementary Information file or are available from the authors, as are the unique reagents used in this article. This file contains: general specifications of marker combinations and resulting cell phenotypes of the used mIF panels (Supplementary Fig. 1), image registration (Supplementary Fig.2), spectral band settings for image acquisition (Supplementary Fig. 3), split violin plots of alternative “CD3” mIF panel cell phenotypes (Supplementary Fig. 4), gauges representing the NK/ILCs/Th cell densities within interface for all tissue types analyzed (Supplementary Fig. 5), control plots justifying gating strategy thresholds used for ILCs analysis in human tonsils (Supplementary Fig. 6), correlation matrix to validate intra-batch (7 batches, 1 batch per tissue type studied) concordance for two common markers used in the mIF panels, i.e. TCRαδ (T cells) and IL-7Rα (non-B/T) (Supplementary Fig. 7), comparative infiltration analysis plots for Th1, Th2 and Th17 cells location for all tissue types (Supplementary Fig. 8), and comparative diagram chords for proximity analysis for all tissue types (Supplementary Fig. 9). Public dataset analysis are presented on Supplementary Figs. 10 and 11 for thymus and tonsil respectively. Supplementary Table 1 provides information about the patients and tissues, Supplementary Tables 2 to 7 provide descriptive statistics, and Supplementary Tables 8 and 9 show the antibody panels used for mIF and flow cytometry analysis. The raw numbers for charts and graphs are available in the corresponding Source Data file whenever possible and as indicated in the corresponding legend. FACS data are only available after official request, to avoid uncontrolled reproduction and publication of patient data, by e-mailing the corresponding author, who will respond within two weeks. The request must be accompanied by a short description of the project, the reason, and aim for using our FACS data. It is the corresponding author who will decide whether the external use of the FACS data is allowed, depending on the compatibility of the initial question and internal scientific scopes. Source data are provided with this paper.
The python scripts used for data analysis and visualization are available as a public GitHub repository here: https://github.com/ouatataz/innate-lymphoid-cells.