ABSTRACT
Autophagy is a process of cellular self-eating, which allows organisms to eliminate and recycle unwanted components and damaged organelles to maintain cellular homeostasis. It is an important process in the development of eukaryotic organisms. Autophagy plays a critical role in many physiological processes in plants such as nutrient remobilization, cell death, immunity, and abiotic stress responses. Autophagy thus represents an obvious target for generating resilient crops. During plant development, autophagy is also implicated in the differentiation and maturation of various cell types and plant organs, including root cap cells, tracheary elements, gametes, fruits and seeds. Here, we review our current understanding and recent advances of plant autophagy including insight into autophagy regulation and signaling as well as autophagosome membrane biogenesis. In addition, we describe how autophagy contributes to development, metabolism, biotic and abiotic stress tolerance and where the autophagic field is heading in terms of applied research for crop improvement.
KEYWORDS: Plant autophagy, regulation and signalling, endomembrane trafficking, quality control, cargo receptors, development, stress tolerance, immunity, metabolism, crop improvement
Introduction
Autophagy is a highly conserved process in which cells deliver cytoplasmic components to their lysosomes or vacuoles for recycling or storage, particularly under stressful conditions. This process is crucial for maintaining cellular homeostasis and plays a key role in responses to biotic and abiotic stress, nutrient deprivation, and development. In plants, two main autophagy modalities have been identified: macroautophagy and microautophagy [1,2]. Macroautophagy is the best studied of the two pathways and is commonly referred to simply as autophagy. Activation of macroautophagy results in the formation of a cup-shaped double-membrane structure, called the phagophore, that typically develops from the endoplasmic reticulum (ER) in plants. As the phagophore extends and closes, it sequesters cytoplasmic material in an autophagosome. Autophagosomes loaded with cytoplasmic cargo such as organelles, ribosomes, and protein aggregates fuse with the vacuolar membrane, releasing the inner core or macroautophagic body for breakdown by hydrolytic vacuolar enzymes. In microautophagy, the cytoplasmic proteins or organelles closely associate with the vacuolar membrane, which directly engulf the cargo by invagination, forming a microautophagic body that is then degraded or stored in the vacuolar lumen. A third and less known form of autophagy reported in plants is mega-autophagy, in which the vacuole bursts and releases the hydrolytic enzyme to degrade the entire cytoplasm, as the final steps of developmental programmed cell death in cell types such as tracheary elements in the xylem [3].
In recent years, substantial progress has been achievd in the understanding of the mechanisms and functions of autophagy in plants, and it is now evident that autophagy is part of most if not all aspects of the plant’s life cycle. Primary discoveries are therefore of outstanding interest for the development of many agricultural traits (reviewed in [4-6]) and have also strong implications for the wider field of autophagy [7-9]. In this review, we will give insight into the recent advances of autophagy research in plants by focussing on the regulation of the autophagy machinery, autophagosome biogenesis and selective autophagy processes. We will further highlight the emerging roles of autophagy in metabolism, development, immunity, and abiotic stress responses, and outline the growing efforts to dissect and utilize autophagy mechanisms for the improvement of productivity and resilience in crop species.
Autophagy core machinery – regulation and signaling
The core genes encoding the autophagy machinery, termed ATG (autophagy-related) genes, were initially identified in yeast [10-12] and are highly conserved throughout eukaryotes, including plants [7] (Figure 1). Autophagy is initiated upon activation of the ATG1 kinase complex [13], consisting of the ATG1 catalytic subunit together with additional ATG13, ATG11 and ATG101 subunits, which phosphorylates and activates downstream autophagy components. A phosphatidylinositol 3-kinase complex (PI3K) produces phosphatidylinositol 3-phosphate (PI3P) at the phagophore [14], recruiting PI3P binding proteins including ATG18 [15]. Expansion and maturation of the phagophore requires delivery of lipids by the lipid scramblase ATG9 [16] and lipid transfer protein ATG2 [17], and two ubiquitin-like conjugation cascades culminating in the conjugation of the ubiquitin-related protein ATG8 to phosphatidylethanolamine (PE) on the autophagosome membrane [18]. The critical role of the autophagy pathway in controlling development, stress responses and cell death necessitates tight control of autophagy activity in response to environmental and hormonal cues. This regulation is most prominently by post-translational modification of the core autophagy machinery, although transcriptional regulation of ATG genes is also important under a number of conditions (Figure 1).
Figure 1.

Overview of the autophagy pathway and the major mechanisms regulating the autophagy core machinery in plants. Plant genomes contain more than 40 ATG genes that encode the functional units of the autophagy machinery to drive autophagosome biogenesis. Autophagosomes are initiated at ER-localised preautophagosomal structures (phagophore assembly sites, PAS) and mature after recruitment of cellular content to the expanding phagophore. Upon autophagosomal fusion with the vacuole, the sequestered cargo is released for lytic degradation and subsequent recycling. Transcription factors (purple) activate or repress ATG genes in response to environmental or endogenous cues. Protein kinases and phosphatases (orange) modify autophagy core components to control their activity. Post-translational modifications (grey) of autophagy proteins by persulfidation inhibit or by acetylation activate autophagy. Stability of the autophagy machinery is modulated by ubiquitination and subsequent degradation (blue). See text for details.
Transcriptional regulation of autophagy genes
Some knowledge is now being accumulated on transcriptional regulation of ATG genes, although it is certain that much more is yet to be discovered. Earlier work in tomato showed that heat and drought stress induce autophagy, and transcription factors that upregulate ATG genes are key components of the stress response, allowing tolerance of these conditions. These include HSFA1a (heat shock factor A1a), a transcription factor that activates genes encoding proteins of the autophagy core machinery during drought [19], including ATG10 and ATG18, and WRKY-family transcription factors, which upregulate several ATG genes during heat stress [20].
More recently, several transcription factors that regulate ATG gene expression, and therefore autophagy, have been identified in Arabidopsis. For example, the MADS box-family transcription factor SOC1 (suppressor of overexpression of constans 1) has been shown to repress the expression of several ATG genes. A soc1 mutant has increased autophagy and tolerance of fixed-carbon starvation, and SOC1 may repress autophagy to prevent its inappropriate activation [21]. Similarly, HY5 (elongated hypocotyl 5) represses the expression of a subset of ATG genes, and a hy5 mutant has increased autophagy. HY5 acts by recruiting HDA9 (histone deacetylase 9) to ATG genes to suppress their expression, revealing that autophagy can also be regulated epigenetically [22]. HY5 is degraded by the proteasome in the dark or upon nitrogen deficiency, leading to release of its repressive activity and activation of autophagy in response to these stresses [22].
In plants, ATG8 is encoded by a multiple gene family, with the ATG8 isoforms having distinct spatial and temporal expression patterns. Numerous transcription factors that bind to ATG8 promoters were identified by yeast 1-hybrid screening, and the bZIP family factor TGA9 (TGACG motif-binding protein 9) was confirmed as a positive regulator of autophagy via the upregulation of a range of ATG genes [23].
Autophagy has recently been shown to be controlled by the circadian clock, and two circadian-related transcription factors have been identified as negative regulators of ATG gene expression. TOC1 (timing of cab expression 1) is a core oscillator component, and directly binds to the promoters of several autophagy genes to repress their expression [24]. In addition, LUX (lux arrhythmo) represses the expression of a different set of ATG genes and prevents rhythmic autophagy activity [25].
Each of these transcription factors appears to regulate a distinct subset of ATG genes, and the rationale behind this complex regulation is unclear. Whether each ATG promoter binds a different set of transcription factors to provide gene-specific regulation of its transcript levels, or whether this reflects a limitation of the experimental approaches used, is not yet known.
Post-translational regulation of core autophagy components
ATG1 complex
Autophagosome formation is initiated by the ATG1 protein kinase complex. In Arabidopsis, ATG1, in complex with ATG13, activates autophagy, and in turn these proteins are packaged into autophagosomes and degraded by autophagy during starvation [26]. This feedback presumably controls the extent of autophagy activation and prevents excessive autophagic degradation of cell contents. Activity of the ATG1 complex is under the negative control of the protein kinase TORC (target of rapamycin complex) [13], which regulates growth and metabolism in response to nutrient conditions [27,28]. A phosphoproteomic analysis of Arabidopsis cell culture in the presence or absence of a TORC inhibitor identified putative TORC phosphorylation targets [29]. ATG13 and ATG1 were found as likely major TOR substrates, and their sites of phosphorylation identified; phosphorylation at these sites may regulate autophagy activation. A second nutrient and energy sensing kinase, SnRK1 (snf1-related kinase 1), also phosphorylates ATG1 [30] . SnRK1 is a positive regulator of autophagy [31], and therefore phosphorylation by TORC and SnRK1 may act antagonistically to determine the extent of autophagy activation. Dephosphorylation of ATG13 by TOPP (type one protein phosphatase) is also required for autophagy induction, probably by counteracting phosphorylation by the negative regulator TORC. TOPP is encoded by a large gene family, and mutation of multiple members of this family leads to decreased autophagy. TOPP interacts with and can dephosphorylate ATG13, in turn leading to increased ATG1 complex formation and activation of autophagy [32].
While ATG1 and ATG13 can be degraded by autophagy upon induction [26], they are also degraded by the 26S proteasome during prolonged starvation and recovery [33]. This degradation is mediated by the SINAT (seven in absentia of Arabidopsis thaliana) 1 and 2 E3 RING-type ubiquitin ligases, together with the TRAF (tumor necrosis factor receptor-associated factor) adaptors, which ubiquitinate ATG13 and regulate ATG1 complex activity. The authors propose that appropriate autophagy levels are maintained upon prolonged starvation by degradation of ATG13 via SINAT1 and 2. In contrast, SINAT6, which has a truncated RING domain and is catalytically inactive, interacts with ATG13 and suppresses its ubiquitination and degradation, thus promoting autophagy [33]. Autophagy therefore is controlled by competition between SINAT1 and 2 as negative regulators and SINAT6 as a positive regulator. Further complexity is added by a recent report of a role for 14-3-3 proteins in ATG13 stability [34]. 14-3-3 proteins bind both to SINAT1 and ATG13, and are required for the degradation of ATG13 upon ubiquitination by SINAT1. 14-3-3 mutants have increased ATG1 and ATG13 levels and increased starvation tolerance, placing them as negative regulators of autophagy. Consistent with the general function of 14-3-3 proteins in binding to phosphorylated peptide sequences [35], phosphorylation of ATG13 is required for its interaction with 14-3-3, and for ATG13 ubiquitination and ATG1 complex dissociation.
PI3K complex
The PI3K complex produces PI3-phosphate (PI3P) at expanding autophagosomes, which enables recruitment of other autophagy components to the membrane. The stability and phosphorylation state of the non-catalytic subunit ATG6 regulates the complex. SINAT1 and 2, along with TRAF adaptors, control ATG6 stability similarly to that of ATG13 described above. SINAT1 and 2 ubiquitinate and degrade ATG6 upon starvation, in a reaction that also requires TRAFs, whereas SINAT6 has the opposite effect [36]. The SINATs therefore coordinately regulate the activities of the ATG1 and PI3K complexes to maintain the required autophagy levels. ATG6 is phosphorylated by SnRK1 during prolonged fixed-carbon starvation [37,38], and this appears to directly activate autophagy, with no requirement for ATG1 complex activation under these conditions [38]. Multiple initiation pathways for autophagy may therefore exist, dependent on the precise environmental conditions to which the plants are exposed.
PI3P at the nascent phagophore recruits the PI3P-binding protein ATG18, which functions in membrane elongation [39]. Arabidopsis ATG18a can be phosphorylated by the BAK1 (BRI1 [brassinosteroid insensitive 1]-associated kinase 1), suppressing autophagosome formation and decreasing resistance to necrotrophic pathogens [40,41], although whether it acts under other conditions is not yet known. ATG18a is also modified by persulfidation, again suppressing autophagy. Persulfidation increases binding of ATG18a to PI3P, leading to production of larger but fewer autophagosomes upon ER stress [42]. By contrast, acetylation of ATG18a by the acetyltransferase HOOKLESS1 increases autophagy, and blocking this acetylation inhibits binding to PI3P [43]. Post-translational modification of ATG18a therefore may control the release of ATG18a from the membrane, determining the size versus number of autophagosomes produced and thus, regulating the extent of autophagic degradation.
ATG8-PE conjugation
A substantial fraction of the core conserved ATG factors function in ubiquitin-like conjugation reactions that ultimately result in conjugation of ATG8 to PE in the phagophore membrane, which is required for autophagosome expansion and maturation [7]. The protease ATG4 is involved in two steps of this pathway, maturation of ATG8 by removing its C-terminus after synthesis and removal of ATG8 from the outer membrane of the autophagosome prior to fusing with the vacuole [44]. ATG4 was isolated in a screen for proteins persulfidated in response to abscicic acid (ABA), with persulfidation occurring at the proteolytic active site Cys. This modification reversibly inactivated the protease, thereby inhibiting autophagy [45]. The authors hypothesized that ATG4 is persulfidated in normal growth conditions, keeping autophagy activity low. Under stress conditions, a decrease in ATG4 persulfidation increases its activity, allowing autophagy activation via lipidation of ATG8. The redox-sensitive active site of ATG4 also makes it a candidate for redox regulation. While the role of redox regulation in vivo in response to stress in plants is not yet clear, ATG4 protease activity has been shown to be reversibly inhibited by H2O2 [46], and in the green alga Chlamydomonas reinhardtii, ATG4 is activated by disulfide bond reduction, as disulfide bond formation inhibits its activity [47].
Other mechanisms of regulation of the ubiquitin-like conjugation pathways include interactions with other proteins, regulation of protein stability, and regulation of ATG8 lipidation. For example, CML24 (calmodulin-like 24), a calmodulin-related protein, interacts with ATG4b and controls its activity [48]; GAPC (cytosolic glyceraldehyde-3-phosphate dehydrogenase) interacts with ATG3 and regulates autophagy activity [49,50], in a phosphatidic acid-dependent manner [51]; phospholipase Dɛ hydrolyzes ATG8-PE, although paradoxically this appears to activate autophagy [52]; ACBP3 (acyl-Co-A binding protein 3) may compete with ATG8 for binding to phosphatidylethanolamine (PE) [53]; COST1 (constitutively stressed 1) interacts with ATG8 and regulates its stability [54]. The coordination between these modes of regulation is unclear, and much remains to be determined regarding how they interact to control the overall activity of the autophagy pathway.
In summary, autophagy in plants is tightly regulated in response to environmental and developmental cues, and this regulation often involves the post-translational modification of core components of the autophagy machinery to control their activity. Transcriptional regulation and degradation of autophagy proteins are also critical in activation of autophagy and in modulating its activity upon prolonged stress. How these regulatory mechanisms work together to enable growth, development and stress tolerance remains an exciting area of research.
Autophagosome biogenesis and endomembrane trafficking
During autophagosome biogenesis, internal and external signals such as nutrient deprivation, stresses, abnormal protein aggregates or damaged organelles trigger the initiation of autophagosomes by hierarchical ATG complex recruitment [55] (Figure 1 and 2). An “autophagosome precursor” (preautophagosomal structure/phagophore assembly site [PAS]), which later transforms into the phagophore after membrane nucleation, is generated upon autophagy induction from the ER with the recruitment of the ATG1/ULK1 complex [56], ATG9 vesicles [57], and COPII (coat protein complex II) vesicles [58]. Subsequently, the phagophore matures into the autophagosome after PI3K-complex-mediated membrane expansion [59] and later membrane closure depending on ATG8 in yeast or LC3 (light chain 3)/GABARAP (GABA type A receptor-associated protein) in mammals (herafter referred to as ATG8 proteins) [60]. The last step of autophagic flux is the autophagosome-lysosome/vacuole fusion mediated by specific soluble SNARE (N-ethylmaleimide-sensitive factor attachment protein receptor) proteins leading to degradation and recycling of cargos [61].
Figure 2.

Updated interactions between the endomembrane system and autophagosome biogenesis in plants. The plant endomembrane system contains multiple membrane-bound organelles including the endoplasmic reticulum (ER), the Golgi apparatus (GA), trans-Golgi network/early endosome (TGN/EE), multivesicular body/prevacuolar compartment/late endosome (MVB/PVC/LE), and vacuole. The endomembrane system contributes to autophagosome biogenesis via multiple new mechanisms or pathways (A-F) for subsequent autophagosome-vacuole fusion and degradation in plants, with mechanistic details highlighted in the corresponding enlarged boxes shown below. (A) Autophagosome biogenesis is regulated by AtEH/Pan1, F-actin and endocytic machinery at the ER-PM contact site (EPCS) for the degradation of endocytic components in Arabidopsis. (B) The Arabidopsis ORP2A coordinates with both the ER residential VAP27-1 and ATG8 on the autophagosome to mediate ER-autophagosomal MCS for autophagosome biogenesis. (C) The Arabidopsis SNARE family proteins AtVAMP724/AtVAMP726 regulate the trafficking of ATG9 vesicles. (D) Distinct AtSAR1d-positive COPII population is formed to regulate autophagic flux via the AtSAR1D-AtRABD2a nexus. FYVE2 interacts with AtSar1b and ATG18-ATG2 complex to regulate autophagosome biogenesis. (F) Phosphorylated FREE1 coordinates with both the ESCRT and ATG machinery to mediate autophagosome closure in Arabidopsis. (G) CFS1 interacts with ATG8 and VPS23A in mediating autophagosome-MVB fusion to facilitate the formation of amphisome.
In recent years, plant-unique components and mechanisms involved in autophagosome biogenesis have also been revealed, including membrane contact sites (MCSs) and functionally diverse COPII components participating in autophagosome biogenesis. Crosstalk between the endocytic pathway and autophagosome closure linked by the plant-unique ESCRT (endosomal sorting complex required for transport) component FREE1 (FYVE domain protein required for endosomal sorting 1), and autophagy-related plant SNARE proteins as well as their interactions with ATG9 in autophagosome maturation has also been established.
Membrane contact sites in plant autophagosome biogenesis
MCSs are formed between heterologous or homologous membranes of two closely apposed organelles (10nm-30nm) [62]. ER forms MCSs with other organelles to mediate dynamic lipid or membrane exchange and trafficking activities [62,63]. In mammalian cells, ER-organelle contacts are crucial for autophagosome biogenesis. For example, ER-mitochondria contact sites participate in autophagosome formation [64]. The ER-localized protein STX17 (syntaxin 17) recruits VPS34 (vacuolar protein sorting 34) PI3K complex to ER-mitochondria MCS, where the PI3P is generated for autophagosome formation [64]. In contrast, the ER-plasma membrane (PM) contact sites (EPCSs) are involved in autophagosome assembly, where extended synaptotagmins (E-syts) form complexes with VPS34 and VMP1 (vacuolar membrane protein 1) for autophagosome formation [65].
The Arabidopsis EH proteins (EH1/Pan1) are components of the TPLATE complex (TPC) that regulate endocytosis and actin-mediated autophagy (Figure 2A). EH1/Pan1 localizes on both PM and autophagosomes, interacting with the ER-resident protein VAP27-1 (VAMP [vesicle-associated membrane protein]-associated protein 27-1] as a complex to bridge the EPCSs and regulate autophagosome biogenesis. Upon nutrient starvation conditions, F-actin and the endocytic machinery, including the TPC, AP-2 (adaptor protein-2) protein and clathrin, are recruited to autophagosomes by EH1/Pan1 at the EPCSs, and contribute to autophagosome formation [66]. In mammalian cells, ER-isolation membrane (IM, precursor of the autophagosome) is modulated by VAPs interacting with multiple ATG proteins, contributing to autophagosome formation. Furthermore, ATG2 has been reported to mediate the ER-autophagosome MCSs (EACSs) with ATG18, serving as a lipid-transfer protein in forming autophagosomes [65]. The Arabidopsis ORP2A (oxysterol-biding protein related protein 2A) forms a complex with VAP27-1 and ATG8e to mediate EACSs upon autophagic induction (Figure 2B). In ORP2A knock down (KD) plants, autophagic proteins and PI3P accumulated on the ER membrane, while autophagosome formation is impaired, indicating that OPR2A may serve as a PI3P regulator in autophagosome initiation [67]. These findings highlight the roles of MCSs in regulating plant autophagosome formation and organelle communication in plants.
Crosstalk between ATG9 vesicles and distinct plant SNAREs in autophagosome formation
In yeast and mammals, specific SNAREs are required for the homotypic fusion of ATG9 vesicles and phagophore precursors for autophagosome biogenesis [68,69]. In plants, only one Arabidopsis SNARE, VTI12 (vesicle transport v-SNARE 12), was shown to play a potential role in mediating autophagosome-vacuole fusion [70]. In Arabidopsis, depletion of ATG9 affects autophagic flux where abnormal tubular autophagosome-ER structure was observed in the atg9 mutant [71]. Cryo-EM structures of the ATG9 revealed that ATG9 oligomerization is crucial and may promote ATG9 vesicle budding to the PAS [72].
Two plant SNARE proteins, VAMP724 and VAMP726, were recently shown to regulate autophagosome formation as they colocalize with the autophagosome marker ATG8e and may function together with ATG9 vesicles [73] (Figure 2C). Interestingly, similar tubular autophagosome-ER structures found in the atg9 mutants [71] were also found in the vamp724 vamp726 double mutants, indicating these two SNARE proteins and ATG9 vesicles likely function in the same pathway [73]. Indeed, the distribution of ATG9 puncta was affected in the vamp mutants while the subcellular localization of VAMP724 and VAMP726 remained normal in the atg9 mutant, supporting the notion that VAMP724 and VAMP726 regulate ATG9 trafficking to contribute to autophagosome biogenesis.
Distinct COPII populations contribute to phagophore initiation and expansion
Conventionally, COPII vesicles transport proteins and lipids from the ER to the Golgi apparatus (Figure 2, top panel), while they also play roles in autophagosome biogenesis. Studies in yeast showed that the phosphorylation of COPII cargo adaptor SEC24 reprograms COPII vesicles to fuse with the phagophore by interacting with ATG9 [74]. In mammalian cells, COPII vesicles that are specifically derived from ER–Golgi intermediate compartment (ERGIC) contain SEC23B protein and contribute as a membrane source to the phagophore precursor [75,76]. Plants harbor functionally diverse COPII components, including multiple homologs of SAR1 (secretion associated RAS 1), SEC23, SEC24, SEC13, and SEC31 [77]. Those plant-unique homologs play multiple functions in plants, including the SAR1a-mediated giant COPII vesicle formation enriched with membrane transporters in response to drought stresses [77], pollen development [78], and autophagy [79,80] in Arabidopsis.
SAR1D, a plant-specific SAR1 homolog, was found to interact with RABD2a (a plant-unique RAB1/YPT1 homolog) and the ATG machinery to regulate a specific COPII population in autophagy [81] (Figure 2D). The RABD2a-AtSAR1D nexus serves as a molecular switch to redirect COPII vesicles from the canonical pathway to the autophagosome biogenesis pathway, contributing to autophagosome initiation as a membrane source. Interestingly, SAR1D, but not other SAR1 paralogs, specifically regulates this autophagosome biogenesis process. In addition to SAR1D, a large-scale proteomic analysis has identified several other COPII components such as Sec24-like CEF and SEC23f involved in the process [81]. SEC23 homologs function diversely in the formation of ER exit sites [82], yet which SEC23 or SEC24 homolog functions in autophagy-related COPII population remains elusive. The COPII machinery is also involved in autophagosome maturation steps. The interaction between FYVE2 and the COPII component SAR1B in autophagosome elongation was revealed recently [80] (Figure 2E). Taken together, functionally diverse COPII components mediate distinct pathways and developmental stages in specific environments in plants.
Crosstalk between autophagosome maturation and endocytic pathway
The ESCRT complex is involved in the endocytic pathway, mediating endosome maturation and cargo degradation (Figure 2, top panel). In both mammalian cells and yeast, ESCRT components have been shown to contribute to autophagosome closure [83,84]. In plants, several ESCRTIII components such as CHMP1 (charged multivesicular body protein 1) [85] and AMSH3 (associated molecule with the SH3 domain of STAM3) [86] were previously shown to play a role in autophagosome closure.
The identification of the plant unique ESCRT component FREE1 revealed direct crosstalk between autophagosome biogenesis and endocytic pathway. FREE1 is a unique plant ESCRT component that regulates multivesicular body (MVB) biogenesis and vacuolar protein transport as well as autophagosome degradation [87]. The Arabidopsis free1 mutant showed defects in both intralumenal vesicle (ILV) formation in MVB and central vacuole formation, as well as accumulations of autophagosomes and lipid droplets [88,89]. Further investigations showed that FREE1 directly participated in the autophagy pathway via interacting with autophagosome membrane-located autophagy regulator SH3P2 (SH3 domain-containing protein 2) [90].
Recently, Zeng et al. (2023) [79] investigated how FREE1 shuttles between the traditional endocytic pathways and autophagosomes (Figure 2F). Under non-stress condition, FREE1 works together with other ESCRT components to regulate endosomal sorting and MVB biogenesis. During starvation, SnRK1 (KIN10) suppresses the TOR signaling pathway and phosphorylates FREE1, which interacts with ATG8 and the ATG12-ATG5-ATG16 conjugation system, to recruit ESCRTIII components to regulate autophagosome closure. KIN10 serves as a switch turning FREE1 into a new role via its phosphorylation status. Indeed, both FREE1 protein per se and its phosphorylation process are necessary for autophagosome closure, as the free1 mutant and FREE1S530AS533A/free1-ct phosphorylation-deficient mutant showed accumulation of “unclosed” autophagosomes [79].
Role of autophagosome-MVB fusion in amphisome formation
The process of autophagosome biogenesis and maturation varies among yeast, metazoans, and plants. In yeast, autophagosomes form and mature near the vacuole, benefiting from their close spatial relationship [91]. In contrast, metazoans generate autophagosomes at diverse cellular locations, prior to their fusion with endosomes/lysosomes to produce amphisomes [92]. In plants, autophagosomes distribute throughout the cell and are destined to the central vacuole [93]. However, it remains elusive whether plant autophagosomes undergo amphisome formation before reaching the central vacuole. Recently, the role of CFS1 (cell death related endosomal FYVE/SYLF protein 1) as an autophagy adaptor for amphisome formation in plants has been investigated [94] (Figure 2G). CFS1 interacted with both ATG8 via AIM and VPS23A (vacuolar protein sorting 23A), a MVB-localized ESCRT-I complex component, playing a bridging role in recruiting autophagosome and MVB together for the formation of an amphisome [94] (Figure 2G).
In summary, recent studies have shed new lights on the crosstalk and participation of the endomembrane trafficking in autophagosome biogenesis with distinct mechanisms in plants. However, several outstanding questions are waiting for answers in future studies: (i) In addition to the known ER- and PM-autophagosome connections, is there other membrane contact site playing role in regulating autophagosome biogenesis? (ii) The plant unique ESCRT component FREE1 plays multiple roles in regulating membrane trafficking and organelle biogenesis in both endomembrane system and autophagic pathway with distinct mechanisms in plants, are there other plant unique components playing multiple roles? (iii) What are the spatio-temporal resolutions in regulating the precise steps of autophagosome biogenesis from phagophore growth to autophagosome closure?
Autophagy meets receptor biology
Eukaryotic cells have evolved receptor proteins to sense and respond to environmental cues and intrinsic demands. In plant biology, receptor proteins are heavily associated with developmental or immune responses, where, for example, cell surface localized receptors sense damage associated or developmentally regulated small peptides to induce an immune or developmental signaling cascade [95]. The common feature of these receptors is that they are specific to certain stimuli and trigger an adaptive signaling response [96]. Studies in the last decade have shown that autophagy is selective, and this selectivity emerges from dedicated receptor proteins known as selective autophagy receptors or cargo receptors [97-100]. Cargo receptors are modular proteins. They (i) have cargo binding or recognition domains, which confer selectivity, and (ii) interact with ATG8 and other core autophagy proteins to bridge the cargo with the growing autophagosomes [101,102]. Similar to the cell surface receptors, depending on the cargo and the nature of the selective autophagy response induced under stress or developmental transitions, each cargo receptor is connected to a distinct homeostatic response that works harmoniously with the rest of the cellular signaling and metabolism [103,104]. Therefore, identification and characterization of cargo receptors are key to understand the role of autophagy in plant homeostasis [105].
Mechanistic details of how cargo receptors recruit cargo to the autophagosomes mostly come from studies in human cells. Even though cargo receptor studies are still in their infancy in plants, several studies in the last few years have discovered new cargo receptors that mediate protein or organelle degradation. Some of these mechanisms turned out to be also conserved in mammalian cells or yeast, illustrating the power of plants as organismal model systems to study autophagy mediated cellular quality control.
Plant cargo receptors that degrade proteins and protein aggregates
The best studied cargo receptor in plants is NBR1 (neighbor of brca1), which is a hybrid form of mammalian p62 and NBR1 proteins [106,107]. The Arabidopsis NBR1 can form oligomeric structures via the PB1 domain and interact with ubiquitinated aggregates and ATG8 via to the ubiquitin-associated (UBA) domains and the AIM, respectively [106,107]. Whether it also interacts with FIP200 homolog ATG11 or cooperates with other hitherto unknown receptors is unknown. Functionally, NBR1 is crucial for clearing protein aggregates that arise during abiotic or biotic stress conditions [108-113]. Although the specific cargo degraded by NBR1 during stress conditions haven’t been catalogued in depth, NBR1 was shown to degrade small heat shock proteins to regulate heat stress memory [114]. Further studies are necessary to understand NBR1-mediated stress tolerance mechanisms.
In addition to NBR1, another UBA domain containing protein, DSK2 was also shown to function as a cargo receptor. DSK2 degrades a key regulator of brassinosteroid signaling, BES1 (BRI1-EMS suppressor 1), upon drought stress or carbon starvation [115]. BIN2 (brassinosteroid insensitive 2), another key regulator of brassinosteroid hormone signaling, phosphorylates DSK2 residues around the AIM, strengthening ATG8 interaction and leading to the activation of DSK2 cargo receptor activity [115]. Activated DSK2 could in turn recruit BES1 into autophagosomes for degradation [115] (Figure 3A). Since BES1 is also degraded via the proteasome, it could serve as a model substrate to dissect how plant cells decide to employ autophagy over the proteasome [116].
Figure 3.

Examples of selective autophagy targeting proteins, organelles, and pathogens in plants. (A) During drought or carbon starvation BIN2 phosphorylates DSK2 which targets BES1 to ATG8 for subsequent autophagic degradation. (B) Upon immune activation MPK3 phosphorylates Exo70B2 which binds to ATG8. (C) Ubiquitinated proteasome binds RPN10 under proteotoxic stress or nitrogen starvation leading to proteophagy. (D) Photodamaged chloroplast gets ubiquitinated leading to chlorophagy. (E) NBR1 targets ABI transcription factors for autophagic degradation when overexpressed. (F) NBR1 targets the capsid protein P4 and entire viral particle leading to xenophagy of cauliflower mosaic virus (CaMV). See text for details.
Another cargo receptor that is regulated by phosphorylation is the exocyst subunit EXO70B2 [117]. Upon elicitor or salicylic acid analog BTH treatment, EXO70B2 gets phosphorylated by MPK3 (mitogen-activated protein [MAP] kinase 3) around the AIM residues [117]. This increases ATG8 affinity and activates autophagic degradation of EXO70B2 [117] (Figure 3B). Since EXO70B2 normally localizes at sites of active secretion such as the tip region of root hairs, autophagic degradation is proposed to dampen exocyst mediated secretion [117]. Although some other exocyst subunits such as EXO70A1 or SEC6 also undergo vacuolar degradation, whether their degradation is mediated by EXO70B2 remains unknown. Also, if EXO70B2 mediates the degradation of other proteins is not studied so far. However, another EXO70 isoform, EXO70D1-3 has been shown to function as a cargo receptor that selectively degrades type A response regulators in Arabidopsis [118]. Type-A ARRs (Arabidopsis thaliana response regulators) are negative regulators of cytokinin signaling [119]. They are also well-known proteasome substrates [119]. Upon phosphorylation, they interact with EXO70D and become an autophagy substrate to regulate cytokinin outputs [118].
In addition to phosphorylation, ATG8-cargo receptor interaction is also regulated by S-nitrosylation in Arabidopsis [120]. During hypoxia, GSNOR (S-nitroglutathione reductase), a key regulator of nitric oxide signaling gets S-nitrosylated. This leads to a conformational change, where a normally buried AIM becomes accessible for ATG8 interaction and autophagic degradation [120]. Whether GSNOR also mediates the degradation of other proteins remains unknown.
These three examples highlight how plants employ selective autophagy to regulate various signaling and trafficking mechanisms. In contrast to the conceptual model that we described above, how the cargo forms bulky aggregates to become autophagy substrates and how other members of the core autophagy machinery except ATG8 are brought to the aggregation site need further investigation.
Nevertheless, these examples highlight the close crosstalk between the proteasome and the autophagy pathway. Accumulating evidence suggests that aggregates are initially channelled to the proteasomes, since it is rapid and does not involve the biogenesis of a vesicle. Once these aggregates become too bulky to be handled by the proteasome, autophagy is employed to clear them out [121]. Autophagy-proteasome crosstalk is not limited to the substrates. Upon proteotoxic stress or nitrogen starvation, the 26S proteasome has been shown to be degraded via autophagy in Arabidopsis, known as proteaphagy [122]. The ubiquitin receptor RPN10 (regulatory particle non-ATPase 10) can bind ATG8 to recruit proteasomes to the autophagosome [122] (Figure 3C). Similarly, another ubiquitin binding protein family, PUX (plant UBX domain-containing protein) was shown to recruit CDC48 (cell devision cycle 48) complexes to the autophagosomes [123]. Disease associated mutations increased the autophagic flux of CDC48, suggesting autophagy clears out non-functional CDC48 complexes to prevent cytotoxicity [123].
Interestingly, both RPN10 and PUX proteins contain ubiquitin interacting motifs (UIMs) and were suggested to interact with ATG8 via a non-canonical site, defined as ubiquitin docking site (UDS) [123]. UDS is in close proximity to the ATG8 C-terminus that gets lipidated to attach to the phagophore [123]. How receptors could fit into this region without disrupting membrane interaction remained puzzling. Furthermore, in solution NMR studies of yeast ATG8 have shown that the region defined as UDS interacts with the phagophore membrane to cause membrane bending, which is crucial for autophagosome biogenesis [124]. Therefore, further studies are necessary to elucidate how proteasome and CDC48 complexes are degraded via selective autophagy and how UIM-type cargo receptors could function without disrupting ATG8 function.
Another UIM-containing protein was discovered from virus infected Arabidopsis plants. A 71 residues long peptide, VISP1 (virus induced small peptide 1), was identified as a viral induced small open reading frame. Overexpression of VISP1 induced autophagy and triggered the degradation of phase separated RNA-protein granules known as SGS3 (suppressor of gene silencing 3)-RDR6 (RNA-dependent RNA polymerase 6) bodies [125]. Whether VISP1 could interact with UDS due its small size or there are additional adaptors involved in VISP1 induced autophagy remains unknown. In addition to viral infection, SGS3 bodies are also induced and degraded via autophagy upon hypoxia, and this degradation requires CML38 (calmodulin-like protein 38) [126]. However, it is not clear if CML38 functions as a cargo receptor or triggers SGS3 autophagic degradation in another way.
Organelle recycling via selective cargo receptors
In contrast to soluble proteins, we know less about selective autophagy of organelles. There is accumulating evidence suggesting that, similar to yeast and metazoans, organelles are degraded via autophagy in plants, but the cargo receptors that mediate organelle recycling and particularly their mechanism of action is far less understood.
For endoplasmic reticulum, similar to mammalian cells, reticulon proteins were shown to mediate ER-recycling in maize. Particularly, in maize endosperm Rtn1 (reticulon 1) and Rtn2 proteins remodel the endoplasmic reticulum to maintain homeostasis [127]. Another conserved ER-phagy receptor is SEC62. It was initially discovered as an cargo receptor in human cells that removes excess ER formed during stress [128]. Following studies in Arabidopsis have shown that the recoverophagy function of SEC62 is also conserved in plants [129]. Finally, C53 was initially discovered in plants to function as an ER-phagy cargo receptor that is activated upon stalling of ER-bound ribosomes [130]. Further studies have shown that C53 also functions as an cargo receptor in human cells and its activity is regulated by UFM1 (ubiquitin fold modifier 1) [130,131]. Another family of cargo receptors is the ATI (ATG8 interacting protein) family proteins [132]. They have transmembrane domains and localize to ER and plastid associated bodies [132-134]. Recent studies have shown that ATI family could mediate the degradation of ER-associated AGO1 (argonaute 1) protein and ER-inserted MSBP1 (membrane steroid binding protein 1), during viral infection and starvation, respectively [133,134]. How these receptors remodel the ER, fragment it and recruit the cargo into autophagosomes remains for further investigation.
Besides ER-recycling, there is a growing interest in chloroplast and mitochondria autophagic recycling in plants. There are several reports demonstrating chloroplast degradation via macro- or microautophagy pathways [135]. Recent studies have shown that NBR1 plays a role in degradation of chloroplasts, plausibly upon loss of chloroplast integrity [112,136] (Figure 3D). However, whether plants have evolved dedicated chlorophagy receptors remain unknown. For mitochondria, it was shown that FMT (friendly), a REC-family protein involved in the regulation of mitochondrial abundance and division, is essential for mitophagy [137-139]. In fmt mutants, mitochondria aggregate and uncoupler induced mitophagy is prevented [137-139]. Excitingly, FMT-mediated mitophagy was also crucial for de-etiolation, a major cellular reprogramming response that involves the maturation of chloroplasts from etioplasts [137]. Whether mitophagy is crucial for deetiolation due to the energy demand required for reprogramming, or mitochondria and chloroplasts form interconnected networks that will require both organelles to be reprogrammed during deetiolation are exciting hypotheses that need to be tested. In addition, other members of the REC family regulate chloroplast distribution and abundance [140]. Whether they are involved in chlorophagy remains unknown. In addition to FMT, TRAB proteins were recently shown to regulate mitophagy [141]. TRAB proteins localize to the ER-mitochondria contact sites, where they colocalize with the known ER-plasma membrane contact site protein VAP27-1 [141]. Similar to FMT, TRAB proteins regulate mitochondrial abundance and morphology. Furthermore, they were shown to interact with ATG8 via an AIM and regulate uncoupler induced mitophagy [141]. Previous studies have shown that VAP27 proteins also regulate endocytosis associated autophagy, cytoskeletal dynamics, and lipid trafficking [66]. As such they form a dynamic signaling hub where biogenesis, energy production, and degradation mechanisms could crosstalk to maintain cellular homeostasis, despite changing environmental conditions and intrinsic demands.
In summary, despite the growing list of selective cargo receptors in plants, their molecular mechanism of action, cargo, and crosstalk with other homeostatic pathways remain largely unknown. Since the cargo is playing an active role in activating selective autophagy, cargo receptor discovery approaches should focus on comparative analysis of physiological stress conditions that would trigger different selective autophagy responses. Also, the cargo receptor catalogue will differ in each cell type. Developmentally regulated mitophagy that happens during sperm maturation in mitochondria may not be similar to the mitophagy pathways triggered during de-etiolation [142]. Nevertheless, comparative analyses of these responses will reveal the extent to which plants employ cargo receptors to shape and remodel their cytoplasm.
Autophagy and development
Since the discovery of the first autophagy-deficient plant mutants in Arabidopsis, autophagy has, in plants as well as in animals, been considered as a pro-longevity process. While autophagy deficient plants thus exhibit early onset of chlorosis, they develop remarkedly normal at the macroscopic level under controlled growth conditions [44,143]. Nevertheless, autophagy affects most, if not all aspect of plant development including seed, vasculature tissue, root, and reproductive development [144,145]. In line with this, plant hormones are also implicated in the regulation of autophagy and strongly affected by autophagy [146,147]. A well-studied example includes the plant growth hormone brassinosteroid (BR) that may both regulate autophagy [148] and is regulated by autophagy [115,149].
Thus, as testified by these and numerous other reviews and reports, we are digging in on autophagy in development. A recent example includes the study of root cap development in Arabidopsis. The root cap is kept at a constant size by cell death and shedding and autophagic activity increase in pre-programmed cell death (PCD) of lateral root cap cells [150,151]. To study more directly the role of autophagy in developmental PCD in the root cap, Feng and colleagues generated transgenic lines with root cap specific KOs of ATG2 or ATG5. Sloughed root cap cells in both complete atg5 and cell type specific atg2 and atg5 KOs remained viable significantly longer than in wild type [150], thus directly demonstrating an autonomous function of autophagy in the developmental PCD required to form normal root caps.
In addition to studying autophagy deficient plants, Wang and colleagues recently reported increased autophagic activity in Arabidopsis feronia mutants [152]. FERONIA (FER) encodes a receptor-like kinase which functions together with co-receptors to regulate many aspects of plant development including pollen tube growth, optimal vegetative growth, and root hair development [153]. Autophagic activity is markedly increased in fer mutants while TOR kinase activity is severely compromised, and this is also true when overexpressing TOR in fer mutants. Oppositely, overexpression of FER is sufficient to inhibit autophagy induction by sucrose starvation but only in wild type plants, not in raptor1b mutants placing FER directly upstream of TOR kinase function [152].
The GSK3 (glycogen synthase kinase 3)-like kinase BIN2 functions as a negative regulator of BR responses. Thus, in the presence of BR, BIN2 is kept inactive to favor growth. However, in contrast to FER, BIN2 was recently shown to positively regulate autophagy. More precisely, BIN2 can both be found in proximity and to directly phosphorylate RAPTOR1B [148]. RAPTOR1B functions to recruit substrates to TOR and Liao and colleagues [148] found that when BIN2 is inhibited by BR, RAPTOR1B phosphorylation is decreased. This promotes phosphorylation of ATG13a by TOR and inhibition of autophagy. Thus, when BR is absent, BIN2 can phosphorylate and suppress the TOR complex through RAPTOR1B to induce autophagy. With these two examples the authors speculate that FER and BIN2 are involved in regulating TOR function to balance nutrient and energy levels via autophagy. Nevertheless, it is still unknown how directly or indirectly FER is linked to autophagy.
Recent evidence also points to the involvement of autophagy in seed development in Arabidopsis since autophagy deficient plants exhibit a variation of abnormality of embryo development [154]. In support of this, ATG8 transcripts accumulate in mature green embryo-bearing siliques compared to earlier developmental stages and autophagic activity also increases in the developing siliques [154]. ATG gene expression has also recently been reported to increase during fruit ripening in strawberry, and tomato ATG4 RNAi lines showed reduced fruit growth, supporting the notion that autophagy contributes to fruit development [155,156].
To study how autophagy in the maternal tissue may affect seed development Erlichman and co-workers [157] elegantly performed reciprocal crosses between atg mutants and wild type plants. These crosses revealed that F1 seedlings derived from an autophagy-deficient mother exhibited shorter hypocotyls than seedlings originating from a wild type mother. Since the protein content was also significantly reduced in F1 seeds originating from autophagy deficient mothers, the authors concluded that autophagy is involved in the regulation of carbon and nitrogen allocation from the mother plant to the seed [157]. Importantly, the authors could demonstrate that the shorter hypocotyl was independent of the senescence associated phenotype seen in atg mutants.
Recent findings also indicate that autophagy modulates lateral root (LR) formation. Ebstrup and co-workers found that atg mutants display reduced numbers of LRs and that NBR1 mediates the selective autophagic degradation of AUXIN RESPONSIVE FACTOR 7 (ARF7), a key regulator of LR formation [158]. The authors further showed that auxin promotes the co-localsation of ARF7 with NBR1 and ATG8a leading to its autophagic turnover. Concordantly, when autophagy was impaired either chemically or genetically, ARF7 accumulated in nuclei and cytoplasmic condensates, causing decreased auxin responsiveness and consequent defects in LR pre-positioning and formation.
However, a major obstacle in studying autophagy deficient mutants can be the pleiotropic and secondary effects of knocking out such a central degradation system at the organismal level. Rodrigues and colleagues [9], for example, recently demonstrated that practically all plant hormones, microbial and danger associated molecular patterns induced a rapid activation of autophagy. Thus, autophagic deficient plants accumulate vast amounts of proteins representing signatures of earlier endogenous and exogenous signalling events which they are unable to degrade. Consequently, autophagic deficient mutants first struggle to make cellular changes exemplified by reduced and delayed callus formation. However, at later stages the responses may become exaggerated as illustrated by bulky callus development when explants are placed on shoot inducing media [9]. Oppositely, plants overexpressing autophagy are simply better at adapting and make cellular changes, probably because they rapidly degrade signatures from previous cellular programs which smoothen transitions [159,160].
In summary, and as also mentioned above, much of what we have learned about autophagy in development is based upon observations done in autophagy-deficient plants. However, it cannot be surprising that loss of autophagic activity from fertilization can mask the role of this process in plant development. Autophagy itself may thus not directly regulate plant development. The “blurred proteome” in autophagic deficient plants can have a negative impact on plant development. Its role in maintaining proteostasis is essential for ensuring that cells function properly during multiple developmental transitions. Thus, tissue-specific knockout mutants as for example the ones used by Feng and co-workers [150] may help further clarify the developmental consequences of autophagic deficiencies, maybe in combination with conditional KOs.
Autophagy and metabolism
While autophagy delivers macromolecules and even entire organelles to a lytic organelle [161], the interaction of autophagy and metabolism displays considerable differences with that observed in yeast and mammals. These are due both to inherent differences in the metabolism of the species as well as the different requirements each organism requires to maintain optimal fitness [162]. Mega-autophagy, in which the disintegration of the tonoplast results in the release of a suite of enzymes including proteases and hydrolases into the cytosol, results in the degradation of cytosolic material and eventually cell death [163,164]. It has also been characterized to be important during the formation of secondary xylem in poplar [165]. That said both the interaction of megaautophagy and microautophagy with metabolism are relatively poorly studied so we will concentrate this section solely on the interaction of macroautophagy (hereafter simply referred to as autophagy) and metabolism. Moreover, given that their roles have been extensively reviewed elsewhere [166–168] we will not cover the roles of TOR or SnRK1 in this interaction.
An obvious place to start discussing the interaction of autophagy and metabolism is at the level of transcription. While transcriptional regulation of the core machinery is described above its role in the interface of autophagy and metabolism is considerable. Indeed, the majority of ATG8 genes in both Arabidopsis and soybean (Glycine max) exhibits rapid increases in gene expression following nitrogen deprivation [169,170], yet by contrast only a small number of the ATG8 genes are enhanced in expression following several days of carbon starvation [169,171,172]. Beyond ATG8 several other of the ATG genes are induced on nutrient stress with ATG18 being responsive to both carbon and nitrogen starvation [173] and several ATG genes being induced by prolonged darkness in N. benthamiana [174]. Although the transcription factors responsible for regulating ATG gene expression are well characterized in yeast and mammalian cells [175], in plants knowledge is currently restricted to that concerning HSFA1A, ERF5 (ethylene responsive element binding factor 5), BZR1 (brassinazole resistant 1) and WRKY 33 and 45 [20,169,176-178] as well as the transcriptional regulator HY5 [179]. However, despite the identification of these transcription factors alongside the detailed characterization of the expression of the autophagy genes themselves neither the gene-regulatory networks nor more importantly their metabolic triggers are yet fully understood in plants and their study remains an important priority.
The importance of autophagy both under nutrient-rich and -deficient conditions has been well documented for multiple plant species. Under nutrient-rich conditions Arabidopsis and maize autophagy mutants display considerable metabolic changes with a compromised nitrogen remobilization with increased protein levels and altered lipid metabolism being a characteristic of both species, while amino acids and ammonia were elevated only in Arabidopsis [180-183]. Similarly, detailed characterization of tomato plants that were deficient in the autophagy-regulating protease ATG4 revealed that these displayed an early senescence phenotype yet relatively mild changes in the foliar metabolome when grown under nutrient-rich conditions [156]. Indeed, the levels of many fruit primary metabolites exhibited decreases in the ATG4-RNAi lines, such as proline, tryptophan and phenylalanine, whilst representative secondary metabolites were present at substantially higher levels in ATG4-RNAi green fruits than in wild type. Furthermore, integration analysis of the metabolome, transcriptome and proteome data indicated that ATG4 significantly affected lipid metabolism, chlorophyll binding proteins and chloroplast function [156].
A common plant stress is the lack of carbon availability caused by insufficient light irradiance. Autophagy is one of three mechanisms which promotes the degradation of carbon sinks, the other two processes being proteasome-mediated degradation and chloroplast vesiculation. Autophagy has for example been suggested to contribute to transitory starch degradation in Arabidopsis [174] and been found to be induced by C starvation in several other species. For instance, ATG transcripts and ATG-PE conjugates being elevated in maize on C stress [184], whilst overexpression of ATG8 in apple lead to an increased tolerance to C stress [185]. Similarly, metabolic profiling of etiolated Arabidopsis atg mutants displayed reduced levels of amino acids and elevated protein contents [186], with a similar phenotype observed when mature plants were exposed to continuous darkness [187] and in atg mutants grown under short-day conditions [188]. In maize atg mutants the changes described above in nutrient rich conditions were exacerbated with major increases in sugar, organic and amino acid levels as well as in starch degradation [183]. Such effects of autophagy are not, however, confined to the core central metabolism with a clear role for the process emerging in lipid homeostasis [161]. Indeed, this was observed both in the etiolated Arabidopsis seedling experiment described above [186] and has been the subject of a recent comprehensive review which we refer the reader to [189]. It is worth mentioned that the changes observed in both central metabolites and lipids are not all conserved across species with considerable differences being observed between for example Arabidopsis [190] and maize [183] suggesting that it will be highly important to expand the range of such studies in the future to accommodate a greater number of species and tissue types in order to better understand the different metabolic contexts in which autophagy operates.
The picture is further muddied by recent findings of the significance of S and P stress in the interaction between autophagy and metabolism. Given that considerably less research has focused on these nutrients and they have been extensively reviewed recently [161], we will only discuss them briefly together here. Considerable further research is needed to understand the interaction of both S and P and autophagy. Sulfide induced persulfidation of specific cysteine residues on ATG4 and ATG18a [45,191,192], however, this effect is independent of the role of S as a nutrient since it also occurs under S sufficiency [193]. The central metabolite which coordinates S, C and N flux in plants – cysteine – has also been demonstrated to play an important role in Glucose-TOR signaling [194] which has considerable overlap with autophagy. Similarly, P starvation induces autophagy in several species causing ER stress in for example tobacco [195,196] while Arabidopsis mutants are hypersensitive to P limitation [197]. Many studies have centered on the role of autophagy in nitrogen (N) mobilization in several crop species and will be discussed in detail in the section “Autophagy in crops” below.
Although we have learned a lot about the extensive interaction between metabolism and autophagy many open questions remain. While the study of constitutive mutants and transgenics have largely supplied the above knowledge, future experiments that shift towards inducible manipulations of autophagy and/or metabolism will likely issue in a more detailed understanding of this fascinating yet highly complex interplay. The very recent findings that three consecutive enzymes of glycolysis and FCS-like zinc finger (FLZ) proteins regulate autophagic flux [198,199] is an intriguing novel aspect at the interface of autophagy and metabolism, and how metabolism influences autophagy represents a key future research front.
Autophagy and abiotic stress
One of the earliest phenotypes described for Arabidopsis atg mutants is hypersensitivity to abiotic stresses, such as heat, drought (osmotic), salt, ER, and oxidative stress [200]. In addition, overexpression of ATG genes in Arabidopsis resulted in increased resistance to oxidative stress [159]. These data indicate that an increased understanding of the role of autophagy in abiotic stress resistance might aid in developing novel agricultural solutions, specifically in light of the current climate change. Several excellent reviews have been published in recent years describing the roles of autophagy in abiotic stress and its agricultural importance [201]. That said, common themes have emerged from manuscripts published in the last few years. This section will describe them.
The study of autophagy in plants has expanded to many plant species in recent years, including wild plants, specifically regarding abiotic stress response. Autophagosomes were shown to accumulate in shoots of the resurrection plant Tripogon loliiformis during desiccation. Autophagy induction was mediated by trehalose accumulation [202], suggesting a possible role for regulatory sugars in autophagy induction. Interestingly, trehalose accumulation was also observed in Paspalum vaginatum, a wild relative of maize and sorghum, which is resistant to many abiotic stresses. Inhibiting trehalose breakdown in maize resulted in increased biomass in an autophagy-dependent manner [203]. Other works have highlighted autophagy as a survival strategy for plants acclimated to harsh environments. Samples of the shrub Caragana korshinskii collected from locations with various drought conditions demonstrated the existence of autophagosomes under drought conditions as well as increased expression of ATG genes. The authors correlated this expression with sugar concentrations in the plants. However, a direct connection has not been established [204]. In addition, increased expression of ATG3, ATG4, ATG7 and ATG8 was observed in leaves of the halophyte Eutrema salsugineum under salt stress [205].
Studies from Arabidopsis have shown that atg mutants are hypersensitive to abiotic stress. This phenotype has been expanded to additional plant species, specifically crops (Figure 4). Down-regulation of ATG genes in tomato (Solanum lycopersicum) resulted in increased sensitivity to heat stress [20]. In addition, inhibition of autophagy in wheat (Triticum aestivum) seedlings increased their sensitivity to drought and salt stress [206,207]. Moreover, as opposed to testing the phenotype of whole plants under stress conditions, the effect of autophagy at the tissue level was also investigated. Arabidopsis atg mutants exposed to heat stress displayed aberrant pollen development and increased male sterility [208]. Alternatively, examples from various plant species demonstrate improved abiotic stress tolerance due to the overexpression of ATG genes. As mentioned above, overexpression of AtATG5 and AtATG7 in Arabidopsis resulted in increased resistance to oxidative stress [159]. Similar results were observed in additional plant species, such as apple (Malus domestica) and sweet orange (Citrus sinensis), in response to a variety of abiotic stresses, such as drought, heat, salt, and cold stress. Improved phenotypes were observed when ATG genes were overexpressed in the same species or in Arabidopsis [209-213] (Figure 4).
Figure 4.

The contribution of autophagy to abiotic stress tolerance in plants. The figure summarizes the observed effects of altered autophagy levels on abiotic stress tolerance in different plant species. Autophagy inhibition was achieved by loss-of-function mutations in ATG genes and the NBR1 cargo receptor or by chemical means. Autophagy activation was mediated by overexpression (OE) of ATG, NBR1 and other genes, as well as loss-of-function mutations in negative autophagy regulators or chemical treatment. See text for details. The figure was created with BioRender.com.
More insight was gained regarding the mechanisms behind autophagy induction during abiotic stress. Accumulating evidence from past and recent studies point to the involvement of reactive oxygen species (ROS), such as H2O2. Research from the model alga Chlamydomonas reinhardtii revealed that autophagy induction during ER stress results from oxidative stress. Addition of the antioxidant glutathione partially suppressed ER stress-induced autophagy [214]. In addition, metabolic analysis of developing maize seeds revealed that atg mutants hyper-accumulated metabolites associated with oxidative stress, suggesting this stress is alleviated by autophagy [215].
The source of ROS is still under debate. ROS were shown to mediate autophagy induction by spermidine in cucumber (Cucumis sativus) exposed to salt stress. Treatment with spermidine induced the expression of ATG genes and increased plant stress resistance [216]. In Arabidopsis, an alkaline ceramidase (AtACER) possibly functions in ROS-dependent autophagy induction [217]. Research in tomato revealed that alternative oxidase (AOX) induces autophagy during ethylene-dependent drought response. Plants overexpressing AOX1a displayed enhanced resistance to drought stress and increased autophagosome numbers upon treatment with the ethylene precursor ACC (1-aminocyclopropane-1-carboxylate). The authors attributed this increase to ROS accumulation, followed by ethylene-dependent expression of SlATG8d and SlATG18h [218]. Interestingly, quantitative trait loci (QTL) analysis revealed that ATG18 also functions in drought stress in maize (Zea mays, ZmATG18b) [219] and rice (Oryza sativa, OsATG18a) [220]. Autophagy is also implicated in ROS scavenging. Overexpression of MdATG8a in apple led to reduced ROS accumulation upon heat stress [211]. This dual function was also demonstrated in wheat plants exposed to waterlogging. On the one hand, ATG gene expression was induced by waterlogging and ROS treatment. On the other hand, chemical induction or inhibition of autophagy led to increased or decreased ROS clearance, respectively. This was more pronounced in a waterlogging-resistant cultivar [221].
Further contributing to our understanding on the regulation of autophagy, plant autophagy inhibitors have also been described in recent years. Hydrogen sulfide (H2S) negatively regulates autophagy induction [222]. Recent studies revealed that this inhibition is exerted by persulfidation of Cys residues of ATG proteins. This was shown for ATG4 in plants exposed to osmotic stress and ATG18a in plants exposed to ER stress [42]. During ER stress, the protein IRE1B functions in autophagy induction. The protein has both kinase and ribonuclease activities. An examination of both functions revealed that the endonuclease activity of IRE1B is necessary for autophagy induction, and this is presumably regulated by the transcript degradation of proteins that inhibit autophagy upon ER stress. Overexpressing several of the degradation targets of IRE1B resulted in autophagy inhibition upon ER stress [223]. COST1 was recently identified as a protein that inhibits autophagy under favorable conditions in Arabidopsis. cost1 mutants display constitutive autophagy under normal conditions and increased drought tolerance. Interestingly, drought tolerance of cost1 mutants is partially reliant on H2O2 signaling, further strengthening the connection between autophagy and ROS. The amounts of COST1 are negatively regulated by autophagy [54].
Selective autophagy has been implicated as the executor of abiotic stress response. Of note is the autophagy receptor NBR1. nbr1 mutant plants are hypersensitive to drought and salt stress, demonstrating the accumulation of damaged proteins. Moreover, NBR1 overexpression in Populus resulted in increased salt tolerance [224]. Recent studies implicated NBR1 in the degradation of photodamaged chloroplasts either by microautophagy [112] or by macroautophagy of the translocon at the outer envelope membrane of chloroplasts (TOC) components, impacting chloroplast import [225]. In addition, novel selective autophagy receptors have been identified as abiotic stress modulators. One example is MtCAS31 (cold acclimation specific 31) from Medicago truncatula. The protein induces autophagy during drought stress via ATG8 binding to promote the autophagic degradation of the aquaporin MtPIP2;7 [226]. Other examples include the ER-phagy receptors, which alleviate ER stress caused by misfolded protein accumulation. These are C53, found in plant and animal systems, and the reticulon proteins Rtn1 and Rtn2 from maize [127,130]. In addition, the dicot-specific ATI3 was shown to bind UBAC2A and UBAC2B (ubiquitin-associated protein 2A/B) to facilitate heat and ER stress response [227].
Selective autophagy also modulates the transition from growth to survival through the regulation of plant hormones. NBR1 is presumed to modulate the ABA response to abiotic stress, as overexpression of NBR1 resulted in transcriptional changes of genes related to ABA signaling. The authors demonstrated binding of NBR1 to the ABI (ABA insensitive) 3, ABI4, and ABI5 transcription factors involved in ABA signaling [228] (Figure 3E). In Arabidopsis, the BR-related transcription factor BES1 is degraded by selective autophagy under starvation and drought stress. Surprisingly, autophagy was shown to be induced by BR during cold stress in tomato plants. The authors claim this is due to the transcriptional induction of NBR1 and ATG genes by BZR1, another type of transcription factor functioning in BR signaling [229]. These findings raise the question whether the interplay between BR and autophagy is stress-specific. A recent publication revealed shared signaling elements between BR and TOR, highlighting the complexity of the interplay between hormonal and autophagy regulation [230].
An exciting emerging facet of studying autophagy and abiotic stress is the investigation of recovery from stress. NBR1 was shown to degrade proteins related to heat stress response during recovery from heat stress. Thus, the resulting phenotype of nbr1 and atg mutants is increased thermal memory, stemming from the “lingering” of heat response proteins in the cell [114,231]. The ATI1 cargo receptor was also implicated in the degradation of proteins during recovery from heat stress and, thus, thermal memory regulation [232]. In addition, autophagy was shown to contribute to the production of dipeptides (pairs of amino acids stemming from the proteolytic activity and serving in regulatory roles) during recovery from heat stress [233]. Surprisingly, a recent publication demonstrated that ATG8 translocates to swollen Golgi membranes in an autophagy-independent manner to facilitate their recovery from heat stress, suggesting that autophagy components might serve independent functions during stress [234].
Another advance linking autophagy to field conditions is investigating its role in stress combinations rather than individual stresses. A recent publication revealed that combining high light and heat stress results in different physiological and metabolic responses than individual stresses. These differences were attributed to an accumulation of GABA (γ-aminobutyric acid), possibly inducing autophagy. Indeed, atg mutants and gad3 mutants displayed hypersensitivity to the combined stress compared to control plants [156]. Another research investigated the interplay between drought stress and tomato yellow leaf curl virus (TYLCV) infection. Infection with TYLCV improved the drought tolerance of tomato plants. However, drought stress induced the expression of the transcription factor HSFA1, which was previously shown to promote autophagy induction by the transcriptional regulation of ATG10 and ATG18f [19]. Expression of both genes was indeed enhanced following drought stress, and the authors postulate this increased autophagy promoted the degradation of TYLCV [235]. Further work is needed to uncover this fascinating topic.
In summary, autophagy plays a major role in plant adaptation to abiotic stress, regulated under stress at the transcriptional and post-transcriptional levels. Yet, further research regarding the factors inducing stress-related autophagy is required, as well as studies identifying autophagy targets under stress. Moreover, additional investigations regarding the role of autophagy in stress combinations and recovery from stress will help us get closer to an understanding of the role of autophagy in the field. Recent studies suggest that increased autophagic activity can improve plant performance under abiotic stress. These encouraging results, coupled with a greater understanding of autophagy induction, could have far-reaching implications in improving crop resistance to changing environmental conditions.
Autophagy and immunity
Research over more than a decade has provided compelling evidence that autophagy plays multifaceted roles in plant immunity and that pathogens have evolved sophisticated measures to manipulate autophagy processes for their own benefit. Pioneering work focused primarily on the functions of autophagy in immunity- and disease-related cell death, revealing both pro-survival and pro-death activities of autophagy in response to pathogens with different lifestyles. Recent efforts showcased the importance of selective autophagy components and pathways as part of the host immune weaponry or, if hijacked by pathogens, as powerful tools for counterdefence and alteration of cell functions. In this section, we will highlight the latest advances in understanding the complex roles of autophagy in plant immunity and disease (Figure 5), and discuss major knowledge gaps and outstanding questions for future research.
Figure 5.
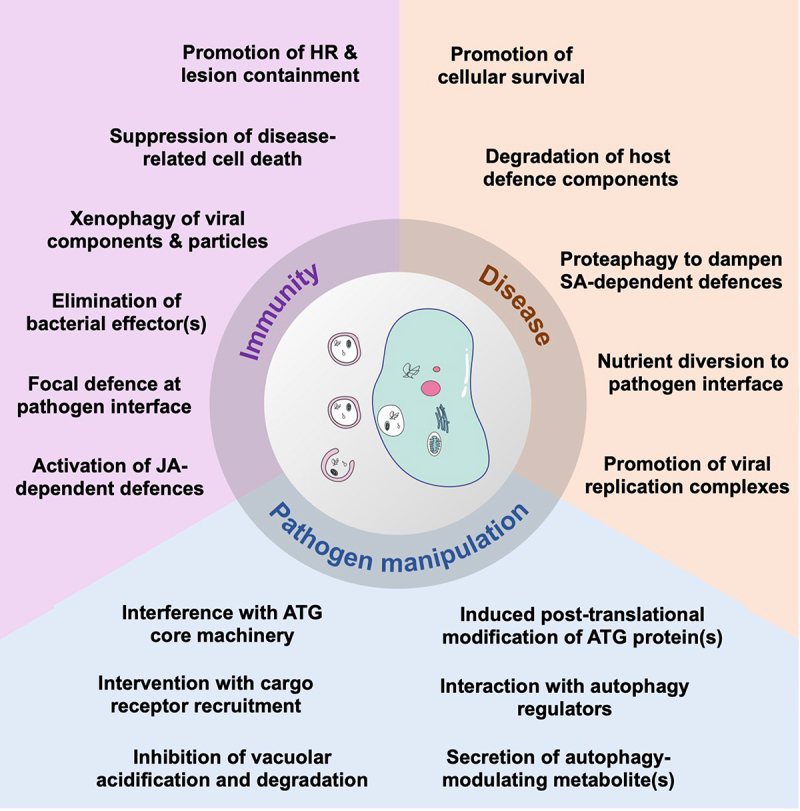
Overview of known functions of autophagy in immunity and disease, and strategies of pathogens to manipulate autophagy processes for their own benefit. Autophagy plays a dual role in the regulation of the immunity-related hypersensitive response (HR) upon infection with various pathogens and suppresses disease-related cell death of necroptrophic fungi. Selective autophagy pathways target viral proteins and particles as well as bacterial effector proteins for degradation. Furthermore, autophagosomes are diverted towards haustoria to mediate focal defence responses against an oomycete pathogen, and autophagy mechanisms are involved in the activation of jasmonic acid (JA)-dependent defences against nematodes. In contrast, cytoprotective functions of autophagy benefit infection by increasing host cell survival, and selective autophagy pathways are hijacked by pathogens to eliminate defence components and to target the proteasome involved in salicylic-dependent immune responses. Autophagic structures are also likely involved in nutrient diversion to the haustorial feeding sites of oomycetes and promote formation of viral replication complexes. Pathogens are able to manipulate autophagy processes by various strategies. These include effector-mediated interactions with and/or modifications of ATG proteins and autophagy regulators as well as interruption of vacuolar functions required for autophagic cargo degradation. Autophagy levels are also modulated by fungal secretion of secondary metabolites. See text for further details.
Autophagy and immunity-associated cell death
The plant innate immune system builds on the two interconnected branches of pattern- and effector-triggered immunity (PTI/ETI) to recognize and respond to pathogen challenge (for an detailed overview, see [236]). ETI is activated by intracellular nucleotide-binding, leucine-rich repeat containing (NLR) immune receptors and often associated with a localized PCD reaction, known as the hypersensitive response (HR). Autophagy has previously shown to promote the HR conditioned by certain NLRs during bacterial, oomycete, and viral infection [49,237,238], but is also required to confine the HR and prevent spreading of cell death into healthy tissues [239-241]. Although such dual role of autophagy in HR regulation has long been established in the Arabidopsis and N. benthamiana models, the underlying mechanisms and the spatial-temporal control of the opposing activities remain largely unknown. The pro-survival function of autophagy outside of HR lesions has been mainly assigned to its homeostatic role in removing harmful components and attenuating cellular stress associated with salicylic acid (SA)- and NPR1 (non-expressor of pathogenesis-related protein 1)-dependent systemic immune responses [242,243]. In this context, the recently discovered formation of SA-induced NPR1 condensates (SINCs) was shown to directly contribute to cell survival by sequestering ETI- and stress-related components for subsequent degradation [244]. Since the autophagy protein ATG8a and cargo receptor NBR1 co-localise with SINCs, it is tempting to speculate that autophagy processes are involved in either condensate formation and/or turnover. Likewise, it remains to be investigated whether autophagy is engaged in the selective removal of NPR1 and/or other negative PCD regulators at the initial infection site to facilitate HR induction. A pro-death function of autophagy could also be connected to the activation and recycling of immune receptors involved in HR. Based on the recently emerged PTI-ETI crosstalk, this may not only apply to specific NLRs, as discussed before [245], but also to PTI-related pattern recognition receptors like FLS2 (flagellin sensitive 2), which is required for full HR activation upon bacterial infection [246]. Strikingly, FLS2 homeostasis is controlled by autophagy through the selective cargo receptors ORM1 (orosomucoid 1) and ORM2 [247], yet the relevance of these processes for the HR remains to be addressed.
In addition to its catabolic function, two recent reports suggest a novel role of the membrane trafficking activities of autophagy in the dual regulation of cell death. On the one hand, spatial HR restriction in Arabidopsis upon Pseudomonas syringae pv. tomato (Pst) recognition was found to rely on the autophagy-dependent secretory transport of monolignols, which serve as lignin precursors in cell wall lignification resulting in the physical isolation of infection sites [248]. On the other hand, promotion of carbon starvation-induced PCD in potato was shown to involve the autophagy-mediated translocation of the VPE (vacuolar processing enzyme) to the vacuole [249]. VPE exhibits caspase-like activities and is a well-known executioner of developmental and stress-related PCD including the HR in response to tobacco mosaic virus (TMV) and other elicitors [250,251]. However, whether the autophagic trafficking of VPE is indeed decisive for the autophagy dependency of some NLR-triggered HR pathways requires further investigation.
Autophagy and disease resistance
Consistent with the frequently observed uncoupling of NLR-conditioned HR from growth restriction of (hemi)biotrophic pathogens [252-254], the autophagy-dependent control of immune cell death was found to influence only occasionally ETI-associated disease resistance [49,237]. In contrast, autophagy plays an important role in the defence against necrotrophs, which appears to be tightly linked to the suppression of disease-associated ‘necrotic’ cell death induced by the pathogens to retrieve nutrients from host tissue [41,159,255]. However, the exact mechanisms of defensive autophagy during plant-necrotroph interactions are far from being understood. Previous reports suggested the interplay and/or cooperation of autophagy with hormone and metabolic defence networks [40], for instance through the interaction of ATG18a with WRKY33, a key transcriptional regulator of disease resistance against necrotrophic fungi [40]. Remarkably, the activation of autophagy-dependent ‘immune’ cell death was found to counteract disease progression by Botrytis cinerea and Sclerotinia sclerotiorum [255,256], supporting the view that autophagy primarily acts during a short (“cryptic”) biotrophic phase required for the pathogenic fungi to initiate growth and establish infection [257]. Likewise, cytoprotective, pro-survival activities of autophagy could negatively impact the transition to necrosis by mediating the selective removal of host- or pathogen-derived compounds associated with cell death activation.
The importance of selective autophagy mechanisms in plant immunity became particularly evident by the analysis of autophagy functions during compatible plant-virus interactions. Similar to the situation in animals, a diverse set of DNA and RNA viruses were shown to be targeted by autophagy as part of the antiviral host response. Thus far, the elimination of certain virulence factors, including various viral suppressors of RNA silencing (VSR) [258-261], a replicase [262] and a movement protein [263], has emerged as the predominant autophagic activity to limit virus infection in plants. In addition, the NBR1-mediated degradation of viral particles and non-assembled capsid protein (P4) of cauliflower mosaic virus (CaMV) highlighted the plant’s capacity to remove an entire intracellular pathogen and/or its structural component [264] (Figure 3F), closely resembling ‘xenophagy’ and ‘virophagy’ pathways in animal cells [265]. Besides NBR1, a few other host proteins (e.g. rgs-CaM [266], P3IP [260], Beclin1/ATG6 [262], and VISP1 [267]) have been implicated as cargo receptors in antiviral autophagy, while some viral proteins were shown to be degraded via direct ATG8-binding [261,263,268].
Notably, NBR1-based selective autophagy has also been linked to the resistance response against virulent bacterial and oomycete infections. NBR1 was found to dampen Pst virulence by counteracting the establishment of a pathogen-induced aqueous microenvironment in the apoplast [269]. Yet, it remained unknown whether this anti-bacterial pathway is mediated by the NBR1-dependent degradation of a bacterial effector, such as HopM1, or a specific host protein involved in disease-promoting `water-soaking´. Intriguingly, a recent report demonstrated that NBR1/Joka2 is able to directly target the XopL effector protein from Xanthomonas campestris pv. vesicatoria (Xcv) [270], indicating that NBR1 has evolved as a cargo receptor for the xenophagic degradation of virulence factors across different pathogen classes. In contrast, NBR1-stimulated disease resistance against the oomycete Phytophthora infestans has been linked to a focal immune response at the plant-pathogen haustorium interface, which was proposed to rely on the targeted delivery of as yet unknown anti-microbial cargo rather than the removal of secreted effectors [271].
Recently, selective autophay has been also reported to promote jasmonic acid (JA)-dependent resistance against root-knot nematode (RKN) infection [272]. Autophagy stimulation in RKN-challenged tomato plants mediates the selective degradation of the negative JA signalling regulator JAM1 (jasmonate-associated myc2-like 1) via direct interaction with ATG8 proteins. Autophagic removal of JAM1 leads to the activation of the ERF1 transcription factor, which induces JA-dependent defence genes and positively feedbacks on autophagy regulation by enhancing ATG gene expression. It remains to be investigated whether autophagy is similarly integrated in the ERF1-branch of the JA signalling pathway during the resistance response against necrotrophic pathogens [273].
Autophagy manipulation by pathogens
Mounting evidence suggests that pathogens translocate effector proteins or other virulence factors to target the autophagy machinery, its regulators or associated cellular pathways, leading to the activation or suppression of autophagy to the favour of infection. A recent interaction screen with a large collection of effectors from bacterial, fungal, oomycete and nematode pathogens revealed that a significant proportion of the pathogen’s effector repertoire has the potential to interfere with autophagy via direct binding with core ATG proteins [274]. However, the mechanistic details and relevance of these interactions during infection remain to be largely explored. Nonetheless, the initial analysis of three selected Pst effectors (HrpZ1, HopF1, AvrPtoB) suggested that both the inhibition and enhancement of autophagy can promote bacterial virulence. In support of this notion, the Pst effector HopM1 was previously shown to induce autophagy and proteasome degradation, indicating that Pst hijacks the proteaphagy pathway to suppress SA-dependent defence responses for enhanced pathogenicity [269]. On the contrary, the Xcv effector XopL inhibits host autophagy by targeting SH3P2, a central regulator of autophagosome biogenesis, to counteract the antibacterial functions of NBR1 [270]. Together, these findings suggest that certain autophagy activities and/or pathways have different functions during the bacterial infection cycle and that effectors are likely secreted in a temporally coordinated fashion to precisely alter autophagy levels for growth promotion.
Due to their intracellular and obligate biotrophic lifestyle, viruses are particularly challenged to activate or maintain beneficial activities of autophagy while evading or suppressing antiviral xenophagy. Several reports have indicated that virus propagation relies on a functional cytoprotective autophagy pathway to dampen disease-related cellular stress and promote host survival [258,259,264]. Besides being activated as a “by-product” of infection and triggered e.g. by SA-mediated defence signalling [259], autophagy can be directly induced and hijacked by viral proteins. For instance, the geminiviral virulence factor βC1 activates autophagy by interfering with the interaction between the negative autophagy regulator GAPC and the core autophagy protein ATG3 [275]. Furthermore, the poleroviral P0 protein triggers an ER-derived autophagy pathway involving the cargo receptors ATI1 and ATI2 to mediate the degradation of AGO1, the central element of the RNA-induced silencing complex (RISC) [133]. Viral proteins were also proposed to induce the selective removal of other host defence components like the cell-to-cell transport regulator REM1 (remorin 1) [276,277] and the silencing pathway component SGS3 [125,278]. In the latter case, cucumber mosaic virus (CMV) co-opts the peptide cargo receptor VISP1 to target the SGS3/RDR6-formed small-interfering RNA bodies for autophagic degradation [125]. Notably, at later stages of CMV infection, VISP1 mediates the turnover of the increasing amounts of the VSR proteins 2b instead of SGS3 [267], which limits viral accumulation and adds to the self-attenuation strategy of the virus to dampen disease severity and enhance plant tolerance [259,279].
In order to cope with autophagy-mediated antiviral defences, viruses have evolved various strategies to subvert autophagic processes. Barley stripe mosaic virus (BSMV), for instance, exploits the γb protein to disrupt the ATG7-ATG8 interaction required for autophagosome formation [280]. In addition, the BSMV γa replicase inhibits vacuolar acidification and autophagosomal degradation through interaction with a subunit of the vacuolar ATPase [281]. A recent report further demonstrated that the C4 protein of the geminivirus cotton leaf curl multan virus (CLCuMuV) interacts with the negative autophagy regulator eIF4A to stabilize its association with ATG5, thus preventing the function of ATG5 in the core autophagy machinery [282]. Turnip mosaic virus (TuMV) utilizes the VPg and 6K2 proteins to block the NBR1-mediated degradation of the silencing suppressor HCpro; however, the exact mechanisms and host targets underlying the inhibitory effect remain to be resolved [258]. Strikingly, the interference of TuMV with NBR1 flux seems to be accompanied by the viral replicase (NIb)-mediated recruitment of NBR1 and its interacting ATG8f isoform for the formation of viral replication complexes on the tonoplast [283].
The effector-induced inhibition of NBR1-dependent autophagy defences and diversion of autophagic processes for microbial pathogenesis are also illustrated during infection with Phytophthora infestans. The pathogen deploys the RXLR effector PexRD54 to specifically bind the potato isoform ATG8CL, thereby outcompeting Joka2/NBR1 and, presumably, its antimicrobial cargo from ATG8CL-containing complexes [271,284]. Intriguingly, PexRD54 stimulates autophagosome formation at haustoria by recruiting the vesicle trafficking regulator Rab8a to ATG8CL structures. PexRD54-mediated autophagy activation resembles a carbon starvation-induced autophagy response and is assumed to redirect plant nutrients and/or other resources to the pathogen interface for growth and proliferation [285].
Due to the importance of autophagy in the resistance response against necrotrophic fungi, the capacity of these pathogens to subvert autophagy during early stages of infection likely influences disease progression. However, surprisingly little is known about fungal strategies to interfere with autophagy processes. For instance, the secretion of oxalic acid by S. sclerotiorum was shown to induce necrotic cell death by the suppression of autophagy and autophagy-dependent hypersensitive cell death [256], yet the molecular details of this interplay remain to be investigated. Similarly, autophagy is suppressed during B. cinerea infection upon phosphorylation of the autophagy protein ATG18a by the receptor kinase BAK1 [41], a key regulator of PRR-mediated immune signaling. Whether this effect is triggered by fungal virulence factors is unclear, but it is tempting to speculate that several necrosis-inducing effectors (e.g. Botrytis BcXYG1 [xyloglucanase 1]) hijack the recognition by BAK1 and/or associated receptor complexes to overcome autophagy-mediated defences for cell death and disease promotion [286,287].
Finally, a recent study showed that an effector from parasitic cyst nematodes (Heterodera and Globodera ssp.), termed NMAS1 (nematode manipulator of autophagy system 1), interacts with ATG8 proteins to suppress immunity-related ROS production [288]. Together with the enhanced resistance of atg mutants to Heterodera schachtii infection [288], these findings suggest that cyst nematodes require autophagy to promote disease, which is strikingly different from the positive role of autophagy in the resistance against root-knot nematodes [272].
In summary, numerous studies have now demonstrated the enormous complexity of the autophagy-pathogen interplay which is in particular reflected in the often overlapping operation of autophagy defences, their effector-mediated subversion and co-option of autophagic components for microbial pathogenesis (Figure 5). Hence, future research faces the main challenge to untangle the specific mechanisms underlying the pro- and antimicrobial autophagic activities and to distinguish them from more general functions of autophagy in cellular homeostasis and stress tolerance. To achieve this, advanced tools and methods need to be further developed to allow e.g. (i) the assessment of autophagy processes at spatio-temporal resolution, (ii) the systematic search for autophagy-related effector targets, (iii) the comprehensive inventory of the selective receptor repertoire and autophagic cargoes, and (iv) the analysis of autophagy processes beyond their involvement in canonical catabolic functions. While most of these approaches will likely continue to explore well-established pathosystems in the Arabidopsis and N. benthamiana models, an increasing attention has recently been directed to the analysis of autophagy interactions with economically relevant pathogens in crop species [289,290]. Such research will add to the growing interest in translational approaches to assess the potential of autophagy modulation for agricultural trait development including disease resistance.
Autophagy in crops
Genome-wide identification of ATG genes in crop species
In contrast to yeast, which typically have only one gene encoding each ATG protein, plants generally have a few to several isoforms of core ATG components (Table 1). While the precise number of ATG isoforms may vary among plant genomes depending on the quality of their gene annotations, the largest ATG families in crop species are ATG8 and ATG18. The ATG8 protein family is represented by four isoforms in foxtail millet (Setaria italica), five in maize (Zea mays), seven in rice (Oryza sativa), 13 in wheat (Triticum aestivum), eight in alfalfa (Medicago truncatula), 14 in cabbage (Brassica oleraceae), 22 in rapeseed (Brassica napus), and nine in Arabidopsis thaliana (AtATG8a-i) [291,292], whereas the ATG18 family consists of four isoforms in wheat, six in rice, eight in alfalfa, cabbage, and Arabidopsis (AtATG18a-h), and ten in maize [293-295], with one study reporting 31 ATG18 isoforms in maize [296].
Table 1.
Number of ATG genes identified in different plant species, including several major crops.
 |
The expansion and diversification of ATG8 isoforms have occurred repeatedly in plant lineages. [291]. A phylogenetic analysis on 59 plant genomes showed that plant ATG8 isoforms group into two major clades: clade I (including AtATG8a-g) with similarities to fungal ATG8 proteins, and clade II (comprising AtATG8h and i) with similarity metazoan ATG8 homologs [102,291]. Both clades have well-supported subclades specific to plant groups, including Brassicales, Solanaceae, and Poaceae (including all cereals) [291]. The ATG8 family is believed to have expanded through functional specialization, although the specific roles of individual plant ATG8 proteins remain largely unknown. In wheat, TaATG8a, g (clade I), and h (clade II) are upregulated in response to a wide range of abiotic stressors, including high salt, drought, low temperature/darkness, and nutrient deficiency. However, TaATG8a is particularly responsive to high salinity, as shown by its most pronounced upregulation under this stress condition [297]. Consistently, overexpression of TaATG8a alone confers enhanced tolerance to high salt in wheat [292], suggesting a specific role for this isoform in salt stress response. Further supporting the notion that plant ATG8 isoforms have evolved specific function, the silencing of one of the 13 ATG8 isoforms in wheat, TaATG8j (clade I, clustering close AtATG8a), is enough to enhance susceptibility to stripe rust [298].
The ATG18 family in Arabidopsis consists of eight isoforms (ATG18a-h), which are grouped into three subfamilies based on the presence of different structural motifs, and are believed to be present in all vascular plants [296]. In Arabidopsis, ATG18a plays a role in autophagosome formation in response to starvation and senescence [299] as well as in the turnover of the ER through a process called ERphagy [42]. ATG18a is expressed in response to nitrogen (N) starvation whereas both ATG18a and f are upregulated in seedlings transferred to sucrose-lacking medium [299]. In tomato (Solanum lycopersicum), ATG18a, b, and f are upregulated in response to drought and ATG18f is required for autophagosome formation under drought conditions [19]. In common bean (Phaseolus vulgaris), the expression of the eight ATG18 isoforms varies among tissues and between nodulated and mycorrhized roots, suggesting functional specialization [296].
As mentioned above, the overexpression of ATG18a in apple (Malus domestica) seems enough to confer enhanced protection against both biotic and abiotic stresses [209,300]. The fact that a single isoform can confer enhanced resistance to various stresses is remarkable, considering the coordination required among numerous ATG components to execute autophagy. These findings highlight the crucial roles of ATG8 and ATG18 subunits and make them prime candidates for breeding and genetic engineering programs aimed at generating stress-tolerant plants.
The ATG8 and ATG18 protein families are well-known for their large and diversified nature within vascular plants. However, some cereal crops have also shown an expansion in other ATG components. For instance, while Arabidopsis, rice, and alfalfa have two or three ATG13 isoforms, maize and wheat contain six and nine ATG13 proteins, respectively [301]. The complexity of the wheat genome can explain the high number of ATG genes due to two hybridization events and massive local rearrangement. However, it is unknown whether the multiple ATG13 isoforms have specific functions or are entirely redundant.
Although the molecular and cellular roles of many ATG components have been initially characterized in Arabidopsis thaliana, studies in crop species have revealed the crucial function of autophagy in grain yield [302,303], biotic stress tolerance [304], nutrient recycling [302,305-307], fruit ripening [155], programmed cell death in cereal grain pericarps [308], and overall crop fitness. Below we discuss some relevant studies related to the role of autophagy in N recycling, crop yield, and development.
Autophagy during nitrogen remobilization in crops
Nutrient recycling through cellular catabolic pathways is important for crop yield [309], particularly under nutrient-limiting conditions. As autophagy is a key cellular recycling pathway, understanding how it is modulated is fundamental to enable sustainable production of cost-competitive crops by better understanding nutrient recycling and increasing yields in low-nutrient soils while minimizing fertilizer requirements. Several studies have focused on how autophagy controls N recycling. Approximately 75% of the N present in a plant is in the leaf chloroplasts [310], with Rubisco constituting between 20 and 50% of the total leaf protein content [311,312]; thus turnover of chloroplasts and other organelles to mobilize assimilated N from old vegetative tissues into new growing organs and seeds during leaf senescence is important for crop yield [313]. Given that, as indicated by 15N flux experiments [180], autophagy plays the major role in remobilizing nitrogen captured in Rubisco it is perhaps unsurprising that autophagy deficient mutants are hypersensitive to N deficiency [180,182,314-316].
In maize, autophagy is activated under N-limiting conditions and during senescence, as evidenced by the accumulation of lipidated forms of ATG8 in leaf tissues [317]. Maize atg12 mutants with reduced ATG8 lipidation develop normally but produce smaller seeds even when grown under nutrient-rich conditions. This is consistent with reduced N mobilization from vegetative tissues into seeds in the atg12 mutant, as demonstrated by 15NO3− labeling experiments [307]. Under limited N availability, growth of maize atg12 mutant plants is severely reduced with enhanced leaf senescence [307].
Multi-omic analyses performed in maize atg12 mutant plants under either N-rich or N-deficient conditions revealed some intriguing and unanticipated insights. For example, although atg12 plants grow normally when N is available, their proteomic, transcriptomic, and metabolic profiles are drastically altered compared to wild type, suggesting a deep reprograming needed to overcome the lack of macroautophagy. The leaves of atg12 mutant plants either under N-rich or N-low conditions, showed drastic changes in metabolic profiles related to fatty acid catabolism, including free fatty acids, lysolipids, oxylipins, glycerolipids, phospholipids, sphingolipids, and galactolipids, suggesting that other catabolic pathways increased lipolysis and membrane turnover when autophagy is impaired [305].
In rice, plants lacking OsATG7 function showed reduced biomass production even when grown under N-rich conditions and reduced N remobilization compared to wild type plants [318]. Among the eight OsATG8 isoforms in rice, OsATG8b is strongly upregulated in vegetative tissues during seed development and under N deficiency, consistent with a role in N remobilization. Rice plants with reduced or null expression of OsATG8b display reduced vegetative growth and seed yield, while overexpression of OsATG8b results in increased seed production [302,319]. Moreover, a 15NO3− partition analysis confirms the role of OsATG8b in N mobilization from vegetative tissues into seeds [302,319]. Although disruption of OsATG8b function is enough to reduce plant fitness, suggesting reduced or no redundancy with other OsATG8 isoforms, the overexpression of OsATG8c in rice is also able to enhance N mobilization with higher grain yields and plant biomass [320], with consistent phenotypes observed by the overexpression of SiATG8a in foxtail millet [321] and CsATG8a in tea [322].
In barley, the expression of several ATG genes (e.g. HvATG3, HvATG5, HvATG7, HvATG8a, HvATG8c, HvATG9, HvATG18f) increases during senescence and nutrient mobilization from the flag leaf, which is critical for grain filling [323,324]. This further supports the role of autophagy in N recycling and grain filling in cereals.
Autophagy in crop development
Autophagy plays diverse roles in the development of different plant species, as revealed by various mutant studies. Mutations in ZmATG12 lead to reduced seed size without impairing vegetative growth or reproductive development under nutrient-rich conditions [307]. Similarly, mutations in AtATG7 and AtATG5, which also participate in ATG8 lipidation in Arabidopsis, do not block reproductive development [325]. However, in rice, mutations in OsATG7 cause abnormal pollen development, male sterility, reduced pollen germination, and impaired anther dehiscence due to reduced autophagic activity in the tapetum, abnormal lipid metabolism, and reduced phytohormone content (gibberellins and cytokinins) in anthers [326,327]. In tobacco (Nicotiana tabacum), silencing of NtATG2, NtATG5, and NtATG7 drastically reduce pollen germination. Pollen grains with silenced ATG genes show normal lipid and mitochondrial metabolism and fail to degrade a cytoplasmic layer located under the pollen wall germination aperture, which might need to be removed via autophagy for successful extrusion of the vegetative cell and formation of the pollen tube [328].
As mentioned above, maize atg12 mutant plants develop smaller seeds even when plants are grown under nutrient rich conditions, suggesting that autophagy may play additional roles in seed development besides nutrient recycling. For instance, maize plants lacking Rtn2 proteins, which are highly expressed in the endosperm and act as selective autophagy receptor for ERphagy, show exacerbated ER stress in aleurone cells of the endosperm [127], suggesting that autophagy may be required during endosperm development to mitigate ER stress during the intense synthesis of storage proteins. However, based on the analysis of atg12 mutants, autophagy does not seem to be required for programmed cell death in the maize endosperm [307]. The delivery of storage proteins into vacuoles of aleurone cells is mediated by microautophagy [329] but this autophagic process is not dependent on the ATG8 conjugation pathway [307]. Rice plants lacking Osatg7 produce seeds with extremely low germination rate, chalky appearance, smaller starch granules, and abnormal sugar metabolism [302], while in wheat, silencing of ATG8 results in delayed PCD, increased pericarp thickness, premature seed maturation, and smaller grains [308].
The differences in developmental phenotypes (pollen development and viability; seed and fruit development) seen in different plant species deficient in autophagy are likely due to different functional specializations and requirements for autophagy. Thus, the genetic and molecular analysis of more specific ATG components across plant species will be critical to understand the multiple roles of autophagy in crop yield and fitness.
Perspectives
The field of plant autophagy is expanding rapidly. However, while the conserved autophagy machinery is well-documented, its interaction with the endomembrane system, selective catabolic pathways, hormonal signaling, and developmental roles together with autophagy’s crucial function in carbon and nitrogen starvation, requires further exploration which should include inducible manipulations of autophagy. Moreover, interactions between autophagy and hormonal signaling pathways, potentially using advanced techniques like tissue-specific knockout mutants, will be essential to clarify specific roles of autophagy in diverse developmental programs.
In the context of abiotic stress, Arabidopsis atg mutants’ heightened sensitivity versus enhanced resistance upon ATG overexpression underscore the significance of autophagy in agricultural solutions amid climate change. Recent findings also indicate that pathogens, including bacteria, fungi, oomycetes, and nematodes, manipulate autophagy through direct binding with core ATG proteins. However, mechanistic details and the relevance of these interactions during infection are yet to be fully explored. Nevertheless, unraveling autophagy induction mechanisms are crucial frontiers and has the potential to contribute significantly to advancements in agriculture with new ideas for agronomic interventions.
Acknowledgement
This work was supported by a grant from Novo Nordisk Fonden (NNF190C0055222) to M.P., by grant #1942/19 from the Israeli Science Foundation and grant #0005816 from the Israeli Ministry of Science and Technology to T.A-W., by grant #MCB-2040582 from the US National Science Foundation to D.C.B., by grants from the U.S. National Science Foundation IOS-1840687 to M.S.O., by a grant from the Danish Research Council (DFF1- 1032-00249B) to E.R. and by grants from the research councils FORMAS (2017-01596; 2022-01583) and Vetenskapsrådet (2020-05327; 2023-05429) as well as from the Novo Nordisk (NNF22OC0079970) and Carl-Tryggers (CTS23:2928) foundations to D.H.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was supported by the Carl Tryggers Stiftelse för Vetenskaplig Forskning [CTS23:2928 to D.H.]; Novo Nordisk Fonden [NNF22OC0079970 to D.H., NNF190C0055222 to M.P.]; Danmarks Frie Forskningsfond [DFF1-1032-00249B to E.R.]; U.S. National Science Foundation [#MCB-2040582 to D.C.B.]; U.S. National Science Foundation [IOS-1840687 to M.S.O.]; Israeli Ministry of Science and Technology [0005816 to T.A.-W.]; Israeli Science Foundation [1942/19 to T.A.-W.]; Svenska Forskningsrådet Formas [2017-01596, 2022-01583 to D.H.]; Vetenskapsrådet [2020-05327, 2023-05429 to D.H.].
List of abbreviations
- ARR:
Arabidopsis thaliana response regulator
- ABA:
abscicic acid
- ABI:
ABA insensitive
- ACBP3:
acyl-Co-A binding protein 3
- ACC:
1-aminocyclopropane-1-carboxylate
- AGO1:
argonaute 1
- AIM:
ATG8-interacting motif
- AMSH3:
associated molecule with the SH3 domain of STAM 3
- AOX:
alternative oxidase
- ARF:
Auxin responsive factor
- ATG:
Autophagy-related
- ATI:
ATG8 interacting protein
- BAK1:
BRI1-associated kinase 1
- BSMV:
barley stripe mosaic virus
- BES1:
BRI1-EMS suppressor 1
- BIN2:
brassinosteroid insensitive 2
- BR:
brassinosteroid
- BRI1:
brassinosteroid insensitive 1
- BZR1:
brassinazole resistant 1
- CAS31:
cold-acclimation-specific 31
- CaMV:
cauliflower mosaic virus
- CDC48:
cell devision cycle 48
- CFS1:
cell death related endosomal FYVE/SYLF protein 1
- CHMP1:
charged multivesicular body protein 1
- CML24/38:
calmodulin-like 24/38
- CMV:
cucumber mosaic virus
- COST1:
constitutively stressed 1
- CLCuMuV:
cotton leaf curl multan virus
- COPII:
coat protein complex II
- EACS:
ER-autophagosome membrane contact site
- EE:
early endosome
- EPCS:
ER-PM contact site
- ER:
endoplasmic reticulum
- ERGIC:
ER–Golgi intermediate compartment
- ERF:
ethylene responsive element binding factor
- ESCRT:
endosomal sorting complex required for transport
- E-syts:
extended synaptotagmins
- ETI:
effector-triggered immunity
- FER:
feronia
- FLS2:
flagellin sensitive 2
- FLZ:
FCS-like zinc finger
- FMT:
friendly
- FREE1:
FYVE domain protein required for endosomal sorting 1
- GA:
golgi apparatus
- GABA:
γ-aminobutyric acid
- GABARAP:
GABA type A receptor-associated protein
- GAPC:
cytosolic glyceraldehyde-3-phosphate dehydrogenase
- GSNOR:
S-nitroglutathione reductase
- GSK3:
glycogen synthase kinase 3
- HDA9:
histone deacetylase 9
- HR:
hypersensitive response
- H2S:
hydrogen sulfide
- HY5:
elongated hypocotyl 5
- IM:
ER isolation membrane
- ILV:
intralumenal vesicle
- IRE1:
inositol-requiring enzyme 1
- JA:
jasmonic acid
- JAM1:
jasmonate-associated myc2-like 1
- LE:
late endosome
- LR:
lateral root
- LUX:
lux arrhythmo
- MCS:
membrane contact site
- MPK3:
mitogen-activated protein (MAP) kinase 3
- MSBP1:
membrane steroid binding protein 1
- MVB:
multivesicular body
- NBR1:
neighbor of brca1
- NLR:
nucleotide-binding, leucine-rich repeat containing
- NMAS1:
nematode manipulator of autophagy system 1
- NPR1:
non-expressor of pathogenesis-related protein 1
- NUE:
nutrient use efficiency
- ORM:
orosomucoid
- ORP2A:
oxysterol-binding protein related protein 2A
- PAS:
preautophagosomal structure/phagophore assembly site
- PCD:
programmed cell death
- PE:
phosphatidylethanolamine
- PI3P:
phosphatidylinositol 3-phophate
- PI3K:
phosphatidylinositol 3-kinase
- PM:
plasma membrane
- Pst:
Pseudomonas syringae pv. tomato
- PTI:
pattern-triggered immunity
- PUX:
plant UBX domain-containing protein
- PVC:
prevacuolar compartment
- RDR6:
RNA-dependent RNA polymerase 6
- REM1:
remorin 1
- RISC:
RNA-induced silencing complex
- RKN:
root-knot nematode
- ROS:
reactive oxygen species
- RPN10:
regulatory particle non-ATPase 10
- Rtn1:
reticulon 1
- SA:
salicylic acid
- SAR1:
secretion associated RAS 1
- SGS3:
suppressor of gene silencing 3
- SH3P2:
SH3 domain-containing protein 2
- SINAT:
seven in absentia of Arabidopsis thaliana
- SINCs:
SA-induced NPR1 condensates
- SNARE:
N-ethylmaleimide-sensitive factor attachment protein receptor
- SnRK1:
snf1-related kinase 1
- SOC1:
suppressor of overexpression of constans 1
- STX17:
syntaxin 17
- TGA9:
TGACG motif-binding protein 9
- TGN:
trans-Golgi network
- TOC:
translocon at the outer envelope membrane of chloroplasts
- TRAF:
tumor necrosis factor receptor-associated factor
- TOC1:
timing of cab expression 1
- TOPP:
type one protein phosphatase
- TORC:
target of rapamycin complex
- TPC:
TPLATE complex
- TuMV:
Turnip mosaic virus
- TYLCV:
tomato yellow leaf curl virus
- UBAC2A/B:
ubiquitin-associated protein 2A/B
- UDS:
ubiquitin docking site
- UFM1:
ubiquitin fold modifier 1
- UIM:
Ubiquitin interacting motif
- VAMP:
vesicle-associated membrane protein
- VAP27:
VAMP-associated protein 27
- VISP1:
virus induced small peptide 1
- VMP1:
vacuolar membrane protein 1
- VPE:
vacuolar processing enzyme
- VPS23A:
vacuolar protein sorting 23A
- VPS34:
vacuolar protein sorting 34
- VSR:
viral suppressors of RNA silencing
- VTI12:
vesicle transport v-SNARE 12
- Xcv:
Xanthomonas campestris pv. vesicatopria
- XYG:
xyloglucanase 1
References
- [1].Su T, Li X, Yang M, et al. Autophagy: an intracellular degradation pathway regulating plant survival and stress response. Frontiers in plant science. 2020;11:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ding X, Zhang X, Otegui MS.. Plant autophagy: new flavors on the menu. Current opinion in plant biology. 2018. Dec;46:113–61. [DOI] [PubMed] [Google Scholar]
- [3].van Doorn WG, Papini A.. Ultrastructure of autophagy in plant cells: a review. Autophagy. 2013. Dec;9(12):1922–1936. [DOI] [PubMed] [Google Scholar]
- [4].Thanthrige N, Bhowmik SD, Ferguson BJ, et al. Potential Biotechnological Applications of Autophagy for Agriculture. Frontiers in plant science. 2021;12:760407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Avin-Wittenberg T, Baluska F, Bozhkov PV, et al. Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J Exp Bot. 2018 Mar 14;69(6):1335–1353. [DOI] [PubMed] [Google Scholar]
- [6].Cao JJ, Liu CX, Shao SJ, et al. Molecular Mechanisms of Autophagy Regulation in Plants and Their Applications in Agriculture. Frontiers in plant science. 2020;11:618944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Marshall RS, Vierstra RD.. Autophagy: The Master of Bulk and Selective Recycling. Annu Rev Plant Biol. 2018 Apr 29;69:173–208. [DOI] [PubMed] [Google Scholar]
- [8].Stephani M, Picchianti L, Dagdas Y.. C53 is a cross-kingdom conserved reticulophagy receptor that bridges the gap betweenselective autophagy and ribosome stalling at the endoplasmic reticulum. Autophagy. 2021. Feb;17(2):586–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rodriguez E, Chevalier J, Olsen J, et al. Autophagy mediates temporary reprogramming and dedifferentiation in plant somatic cells. EMBO J. 2020 Feb 17;39(4):e103315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tsukada M, Ohsumi Y.. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993 Oct 25;333(1–2):169-174. [DOI] [PubMed] [Google Scholar]
- [11].Thumm M, Egner R, Koch B, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994 Aug 1;349(2):275–280. [DOI] [PubMed] [Google Scholar]
- [12].Harding TM, Morano KA, Scott SV, et al. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995. Nov;131(3):591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000 Sep 18;150(6):1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kihara A, Noda T, Ishihara N, et al. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001 Feb 5;152(3):519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, et al. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 2007 Jul 24;581(18):3396–3404. [DOI] [PubMed] [Google Scholar]
- [16].Matoba K, Kotani T, Tsutsumi A, et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nature structural & molecular biology. 2020. Dec;27(12):1185–1193. [DOI] [PubMed] [Google Scholar]
- [17].Valverde DP, Yu S, Boggavarapu V, et al. ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol. 2019 Jun 3;218(6):1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000 Nov 23;408(6811):488-492. [DOI] [PubMed] [Google Scholar]
- [19].Wang Y, Cai S, Yin L, et al. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy. 2015 Nov 2;11(11):2033–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhou J, Wang J, Yu JQ, et al. Role and regulation of autophagy in heat stress responses of tomato plants. Frontiers in plant science. 2014;5:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li X, Liao J, Bai H, et al. Arabidopsis flowering integrator SOC1 transcriptionally regulates autophagy in response to long-term carbon starvation. J Exp Bot. 2022 Nov 2;73(19):6589–6599. [DOI] [PubMed] [Google Scholar]
- [22].Yang C, Shen W, Yang L, et al. HY5-HDA9 Module Transcriptionally Regulates Plant Autophagy in Response to Light-to-Dark Conversion and Nitrogen Starvation. Molecular plant. 2020 Mar 2;13(3):515–531. [DOI] [PubMed] [Google Scholar]
- [23].Wang P, Nolan TM, Yin Y, et al. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy. 2020. Jan;16(1):123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen W, Hu Z, Yu M, et al. A molecular link between autophagy and circadian rhythm in plants. Journal of integrative plant biology. 2022. May;64(5):1044–1058. [DOI] [PubMed] [Google Scholar]
- [25].Yang MK, Zhu XJ, Chen CM, et al. The plant circadian clock regulates autophagy rhythm through transcription factor LUX ARRHYTHMO. Journal of integrative plant biology. 2022. Nov;64(11):2135–2149. [DOI] [PubMed] [Google Scholar]
- [26].Suttangkakul A, Li F, Chung T, et al. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011. Oct;23(10):3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Menand B, Desnos T, Nussaume L, et al. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci U S A. 2002 Apr 30;99(9):6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Quilichini TD, Gao P, Pandey PK, et al. A role for TOR signaling at every stage of plant life. J Exp Bot. 2019 Apr 15;70(8):2285–2296. [DOI] [PubMed] [Google Scholar]
- [29].Van Leene J, Han C, Gadeyne A, et al. Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat Plants. 2019. Mar;5(3):316–327. [DOI] [PubMed] [Google Scholar]
- [30].Chen L, Su ZZ, Huang L, et al. The AMP-Activated Protein Kinase KIN10 Is Involved in the Regulation of Autophagy in Arabidopsis. Frontiers in plant science. 2017;8:1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Soto-Burgos J, Bassham DC.. SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLOS ONE. 2017;12(8):e0182591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang Q, Qin Q, Su M, et al. Type one protein phosphatase regulates fixed-carbon starvation-induced autophagy in Arabidopsis. Plant Cell. 2022 Oct 27;34(11):4531–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Qi H, Li J, Xia FN, et al. Arabidopsis SINAT Proteins Control Autophagy by Mediating Ubiquitylation and Degradation of ATG13. Plant Cell. 2020. Jan;32(1):263–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Qi H, Lei X, Wang Y, et al. 14-3-3 proteins contribute to autophagy by modulating SINAT-mediated degradation of ATG13. Plant Cell. 2022 Nov 29;34(12):4857–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Muslin AJ, Tanner JW, Allen PM, et al. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996 Mar 22;84(6):889–897. [DOI] [PubMed] [Google Scholar]
- [36].Qi H, Xia FN, Xie LJ, et al. TRAF Family Proteins Regulate Autophagy Dynamics by Modulating AUTOPHAGY PROTEIN6 Stability in Arabidopsis. Plant Cell. 2017. Apr;29(4):890–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Van Leene J, Eeckhout D, Gadeyne A, et al. Mapping of the plant SnRK1 kinase signalling network reveals a key regulatory role for the class II T6P synthase-like proteins. Nat Plants. 2022. Nov;8(11):1245–1261. [DOI] [PubMed] [Google Scholar]
- [38].Huang X, Zheng C, Liu F, et al. Genetic Analyses of the Arabidopsis ATG1 Kinase Complex Reveal Both Kinase-Dependent and Independent Autophagic Routes during Fixed-Carbon Starvation. Plant Cell. 2019. Dec;31(12):2973–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Obara K, Ohsumi Y.. Dynamics and function of PtdIns(3)P in autophagy. Autophagy. 2008. Oct;4(7):952–954. [DOI] [PubMed] [Google Scholar]
- [40].Lai Z, Wang F, Zheng Z, et al. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011. Jun;66(6):953–968. [DOI] [PubMed] [Google Scholar]
- [41].Zhang B, Shao L, Wang J, et al. Phosphorylation of ATG18a by BAK1 suppresses autophagy and attenuates plant resistance against necrotrophic pathogens. Autophagy. 2021. Sep;17(9):2093–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Aroca A, Yruela I, Gotor C, et al. Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proc Natl Acad Sci U S A. 2021 May 18;118(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Huang L, Wen X, Jin L, et al. HOOKLESS1 acetylates AUTOPHAGY-RELATED PROTEIN18a to promote autophagy during nutrient starvation in Arabidopsis. Plant Cell. 2023 Dec 21;36(1):136–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yoshimoto K, Hanaoka H, Sato S, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004. Nov;16(11):2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Laureano-Marin AM, Aroca A, Perez-Perez ME, et al. Abscisic Acid-Triggered Persulfidation of the Cys Protease ATG4 Mediates Regulation of Autophagy by Sulfide. Plant Cell. 2020. Dec;32(12):3902–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Woo J, Park E, Dinesh-Kumar SP.. Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc Natl Acad Sci U S A. 2014 Jan 14;111(2):863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Perez-Perez ME, Lemaire SD, Crespo JL.. Control of Autophagy in Chlamydomonas Is Mediated through Redox-Dependent Inactivation of the ATG4 Protease. Plant Physiol. 2016. Dec;172(4):2219–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tsai YC, Koo Y, Delk NA, et al. Calmodulin-related CML24 interacts with ATG4b and affects autophagy progression in Arabidopsis. Plant J. 2013. Jan;73(2):325–35. [DOI] [PubMed] [Google Scholar]
- [49].Han S, Wang Y, Zheng X, et al. Cytoplastic Glyceraldehyde-3-Phosphate Dehydrogenases Interact with ATG3 to Negatively Regulate Autophagy and Immunity in Nicotiana benthamiana. Plant Cell. 2015. Apr;27(4):1316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Henry E, Fung N, Liu J, et al. Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLOS genetics. 2015. Apr;11(4):e1005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Guan B, Jiang YT, Lin DL, et al. Phosphatidic acid suppresses autophagy through competitive inhibition by binding GAPC (glyceraldehyde-3-phosphate dehydrogenase) and PGK (phosphoglycerate kinase) proteins. Autophagy. 2022. Nov;18(11):2656–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yao S, Peng S, Wang X.. Phospholipase Depsilon interacts with autophagy-related protein 8 and promotes autophagy in Arabidopsis response to nitrogen deficiency. Plant J. 2022. Mar;109(6):1519–1534. [DOI] [PubMed] [Google Scholar]
- [53].Xiao S, Chye ML.. The Arabidopsis thaliana ACBP3 regulates leaf senescence by modulating phospholipid metabolism and ATG8 stability. Autophagy. 2010. Aug;6(6):802–804. [DOI] [PubMed] [Google Scholar]
- [54].Bao Y, Song WM, Wang P, et al. COST1 regulates autophagy to control plant drought tolerance. Proc Natl Acad Sci U S A. 2020 Mar 31;117(13):7482–7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nakatogawa H. Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol. 2020. Aug;21(8):439–458. [DOI] [PubMed] [Google Scholar]
- [56].Yamamoto H, Fujioka Y, Suzuki SW, et al. The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes. Dev Cell. 2016 Jul 11;38(1):86–99. [DOI] [PubMed] [Google Scholar]
- [57].Judith D, Jefferies HBJ, Boeing S, et al. ATG9A shapes the forming autophagosome through Arfaptin 2 and phosphatidylinositol 4-kinase IIIbeta. J Cell Biol. 2019 May 6;218(5):1634–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tan D, Cai Y, Wang J, et al. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc Natl Acad Sci U S A. 2013 Nov 26;110(48):19432–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Simonsen A, Tooze SA.. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009 Sep 21;186(6):773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nguyen TN, Padman BS, Usher J, et al. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol. 2016 Dec 19;215(6):857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Itakura E, Kishi-Itakura C, Mizushima N.. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012 Dec 7;151(6):1256–1269. [DOI] [PubMed] [Google Scholar]
- [62].Scorrano L, De Matteis MA, Emr S, et al. Coming together to define membrane contact sites. Nature communications. 2019 Mar 20;10(1):1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhang X, Man Y, Zhuang X, et al. Plant multiscale networks: charting plant connectivity by multi-level analysis and imaging techniques. Sci China Life Sci. 2021. Sep;64(9):1392–1422. [DOI] [PubMed] [Google Scholar]
- [64].Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013 Mar 21;495(7441):389–393. [DOI] [PubMed] [Google Scholar]
- [65].Ye H, Ji C, Guo R, et al. Membrane Contact Sites and Organelles Interaction in Plant Autophagy. Frontiers in plant science. 2020;11:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang P, Pleskot R, Zang J, et al. Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nature communications. 2019 Nov 13;10(1):5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ye H, Gao J, Liang Z, et al. Arabidopsis ORP2A mediates ER-autophagosomal membrane contact sites and regulates PI3P in plant autophagy. Proc Natl Acad Sci U S A. 2022 Oct 25;119(43):e2205314119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Nair U, Jotwani A, Geng J, et al. SNARE proteins are required for macroautophagy. Cell. 2011 Jul 22;146(2):290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Moreau K, Ravikumar B, Renna M, et al. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011 Jul 22;146(2):303–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Surpin M, Zheng H, Morita MT, et al. The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell. 2003. Dec;15(12):2885–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhuang X, Chung KP, Cui Y, et al. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci U S A. 2017 Jan 17;114(3):E426-E435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lai LTF, Yu C, Wong JSK, et al. Subnanometer resolution cryo-EM structure of Arabidopsis thaliana ATG9. Autophagy. 2020. Mar;16(3):575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].He Y, Gao J, Luo M, et al. VAMP724 and VAMP726 are involved in autophagosome formation in Arabidopsis thaliana. Autophagy. 2023. May;19(5):1406–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Davis S, Wang J, Zhu M, et al. Sec24 phosphorylation regulates autophagosome abundance during nutrient deprivation. eLife. 2016. Nov 18; 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ge L, Zhang M, Schekman R.. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife. 2014 Nov 28;3:e04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ge L, Zhang M, Kenny SJ, et al. Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 2017. Sep;18(9):1586–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Li B, Zeng Y, Jiang L.. COPII vesicles in plant autophagy and endomembrane trafficking. FEBS Lett. 2022. Sep;596(17):2314–2323. [DOI] [PubMed] [Google Scholar]
- [78].Liang X, Li SW, Gong LM, et al. COPII Components Sar1b and Sar1c Play Distinct Yet Interchangeable Roles in Pollen Development. Plant Physiol. 2020. Jul;183(3):974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zeng Y, Li B, Huang S, et al. The plant unique ESCRT component FREE1 regulates autophagosome closure. Nature communications. 2023 Mar 30;14(1):1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kim JH, Lee HN, Huang X, et al. FYVE2, a phosphatidylinositol 3-phosphate effector, interacts with the COPII machinery to control autophagosome formation in Arabidopsis. Plant Cell. 2022 Jan 20;34(1):351–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zeng Y, Li B, Ji C, et al. A unique AtSar1D-AtRabD2a nexus modulates autophagosome biogenesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2021 Apr 27;118(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chang M, Wu SZ, Ryken SE, et al. COPII Sec23 proteins form isoform-specific endoplasmic reticulum exit sites with differential effects on polarized growth. Plant Cell. 2022 Jan 20;34(1):333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Takahashi Y, He H, Tang Z, et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nature communications. 2018 Jul 20;9(1):2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Zhou F, Wu Z, Zhao M, et al. Rab5-dependent autophagosome closure by ESCRT. J Cell Biol. 2019 Jun 3;218(6):1908–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Spitzer C, Li F, Buono R, et al. The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell. 2015. Feb;27(2):391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Katsiarimpa A, Kalinowska K, Anzenberger F, et al. The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell. 2013. Jun;25(6):2236–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Gao C, Luo M, Zhao Q, et al. A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol. 2014 Nov 3;24(21):2556–2563. [DOI] [PubMed] [Google Scholar]
- [88].Gao C, Zhuang X, Cui Y, et al. Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc Natl Acad Sci U S A. 2015 Feb 10;112(6):1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Huang S, Liu Z, Cao W, et al. The plant ESCRT component FREE1 regulates peroxisome-mediated turnover of lipid droplets in germinating Arabidopsis seedlings. Plant Cell. 2022 Oct 27;34(11):4255–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhuang X, Wang H, Lam SK, et al. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell. 2013. Nov;25(11):4596–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Zhao YG, Zhang H.. Autophagosome maturation: An epic journey from the ER to lysosomes. Journal of Cell Biology. 2019;218(3):757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zhao YG, Codogno P, Zhang H.. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nature Reviews Molecular Cell Biology. 2021. 2021/11/01;22(11):733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Marshall RS, Vierstra RD. Autophagy: The Master of Bulk and Selective Recycling. Annual Review of Plant Biology. 2018;69(1):173–208. [DOI] [PubMed] [Google Scholar]
- [94].Zhao J, Bui MT, Ma J, et al. Plant autophagosomes mature into amphisomes prior to their delivery to the central vacuole. Journal of Cell Biology. 2022;221(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Bender KW, Zipfel C.. Paradigms of receptor kinase signaling in plants. Biochem J. 2023 Jun 28;480(12):835–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Chen L, Torii KU.. Signaling in plant development and immunity through the lens of the stomata. Curr Biol. 2023 Jul 10;33(13):R733-R742. [DOI] [PubMed] [Google Scholar]
- [97].Vargas JNS, Hamasaki M, Kawabata T, et al. The mechanisms and roles of selective autophagy in mammals. Nat Rev Mol Cell Biol. 2023. Mar;24(3):167–185. [DOI] [PubMed] [Google Scholar]
- [98].Luong AM, Koestel J, Bhati KK, et al. Cargo receptors and adaptors for selective autophagy in plant cells. FEBS Lett. 2022. Sep;596(17):2104–2132. [DOI] [PubMed] [Google Scholar]
- [99].Adriaenssens E, Ferrari L, Martens S.. Orchestration of selective autophagy by cargo receptors. Curr Biol. 2022 Dec 19;32(24):R1357-R1371. [DOI] [PubMed] [Google Scholar]
- [100].Lamark T, Johansen T.. Mechanisms of Selective Autophagy. Annu Rev Cell Dev Biol. 2021 Oct 6;37:143–169. [DOI] [PubMed] [Google Scholar]
- [101].Eickhorst C, Licheva M, Kraft C.. Scaffold proteins in bulk and selective autophagy. Prog Mol Biol Transl Sci. 2020;172:15–35. [DOI] [PubMed] [Google Scholar]
- [102].Rogov VV, Nezis, IP, Tsapras P, et al. Atg8 family proteins, LIR/AIM motifs and other interaction modes. Autophagy Reports. 2023. 2023/12/31;2(1):2188523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Chang C, Jensen LE, Hurley JH.. Autophagosome biogenesis comes out of the black box. Nat Cell Biol. 2021. May;23(5):450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Klionsky DJ, Petroni G, Amaravadi RK, et al. Autophagy in major human diseases. The EMBO Journal. 2021;40(19):e108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Stephani M, Dagdas Y.. Plant Selective Autophagy-Still an Uncharted Territory With a Lot of Hidden Gems. Journal of molecular biology. 2020 Jan 3;432(1):63–79. [DOI] [PubMed] [Google Scholar]
- [106].Svenning S, Lamark T, Krause K, et al. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1 [Research Support, Non-U.S. Gov’t]. Autophagy. 2011. Sep;7(9):993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Zientara-Rytter K, Lukomska J, Moniuszko G, et al. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors [Research Support, Non-U.S. Gov’t]. Autophagy. 2011. Oct;7(10):1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Zhou J, Wang J, Cheng Y, et al. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses [Research Support, Non-U.S. Gov’t]. PLOS genetics. 2013;9(1):e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Zhou J, Zhang Y, Qi J, et al. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLOS Genet. 2014. Jan;10(1):e1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Luo L, Zhang P, Zhu R, et al. Autophagy Is Rapidly Induced by Salt Stress and Is Required for Salt Tolerance in Arabidopsis. Frontiers in plant science. 2017;8:1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Jung H, Lee HN, Marshall RS, et al. Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J Exp Bot. 2020 Jan 1;71(1):73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Lee HN, Chacko JV, Gonzalez Solis A, et al. The autophagy receptor NBR1 directs the clearance of photodamaged chloroplasts. eLife. 2023. Apr 18; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Saeed B, Deligne F, Brillada C, et al. K63-linked ubiquitin chains are a global signal for endocytosis and contribute to selective autophagy in plants. Curr Biol. 2023. Apr 10;33(7):1337–1345 e5. [DOI] [PubMed] [Google Scholar]
- [114].Thirumalaikumar VP, Gorka M, Schulz K, et al. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90.1 and ROF1. Autophagy. 2021. Sep;17(9):2184–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Nolan TM, Brennan B, Yang M, et al. Selective Autophagy of BES1 Mediated by DSK2 Balances Plant Growth and Survival. Dev Cell. 2017 Apr 10;41(1):33–46 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wang P, Nolan TM, Clark NM, et al. The F-box E3 ubiquitin ligase BAF1 mediates the degradation of the brassinosteroid-activated transcription factor BES1 through selective autophagy in Arabidopsis. Plant Cell. 2021 Nov 4;33(11):3532–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Brillada C, Teh OK, Ditengou FA, et al. Exocyst subunit Exo70B2 is linked to immune signaling and autophagy. Plant Cell. 2021 Apr 17;33(2):404–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Acheampong AK, Shanks C, Cheng CY, et al. EXO70D isoforms mediate selective autophagic degradation of type-A ARR proteins to regulate cytokinin sensitivity. Proc Natl Acad Sci U S A. 2020 Oct 27;117(43):27034–27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Argueso CT, Raines T, Kieber JJ.. Cytokinin signaling and transcriptional networks. Curr Opin Plant Biol. 2010. Oct;13(5):533–539. [DOI] [PubMed] [Google Scholar]
- [120].Zhan N, Wang C, Chen L, et al. S-Nitrosylation Targets GSNO Reductase for Selective Autophagy during Hypoxia Responses in Plants. Mol Cell. 2018 Jul 5;71(1):142–154 e6. [DOI] [PubMed] [Google Scholar]
- [121].Pohl C, Dikic I.. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019 Nov 15;366(6467):818-822. [DOI] [PubMed] [Google Scholar]
- [122].Marshall RS, Li F, Gemperline DC, et al. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol Cell. 2015 Jun 18;58(6):1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Marshall RS, Hua Z, Mali S, et al. ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell. 2019 Apr 18;177(3):766–781 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Maruyama T, Alam JM, Fukuda T, et al. Membrane perturbation by lipidated Atg8 underlies autophagosome biogenesis. Nat Struct Mol Biol. 2021. Jul;28(7):583–593. [DOI] [PubMed] [Google Scholar]
- [125].Tong X, Liu SY, Zou JZ, et al. A small peptide inhibits siRNA amplification in plants by mediating autophagic degradation of SGS3/RDR6 bodies. EMBO J. 2021 Aug 2;40(15):e108050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Field S, Conner WC, Roberts DM.. Arabidopsis CALMODULIN-LIKE 38 Regulates Hypoxia-Induced Autophagy of SUPPRESSOR OF GENE SILENCING 3 Bodies. Front Plant Sci. 2021;12:722940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Zhang X, Ding X, Marshall RS, et al. Reticulon proteins modulate autophagy of the endoplasmic reticulum in maize endosperm. Elife. 2020 Feb 3;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Fumagalli F, Noack J, Bergmann TJ, et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016. Nov;18(11):1173–1184. [DOI] [PubMed] [Google Scholar]
- [129].Hu S, Ye H, Cui Y, et al. AtSec62 is critical for plant development and is involved in ER-phagy in Arabidopsis thaliana. J Integr Plant Biol. 2020. Feb;62(2):181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Stephani M, Picchianti L, Gajic A, et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. eLife. 2020 Aug 27;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Picchianti L, Sanchez de Medina Hernandez V, Zhan N, et al. Shuffled ATG8 interacting motifs form an ancestral bridge between UFMylation and autophagy. EMBO J. 2023 May 15;42(10):e112053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Michaeli S, Honig A, Levanony H, et al. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell. 2014. Oct;26(10):4084–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Michaeli S, Clavel M, Lechner E, et al. The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1. Proc Natl Acad Sci U S A. 2019 Nov 5;116(45):22872–22883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Wu J, Michaeli S, Picchianti L, et al. ATI1 (ATG8-interacting protein 1) and ATI2 define a plant starvation-induced reticulophagy pathway and serve as MSBP1/MAPR5 cargo receptors. Autophagy. 2021. Nov;17(11):3375–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Izumi M, Nakamura S.. Chloroplast Protein Turnover: The Influence of Extraplastidic Processes, Including Autophagy. Int J Mol Sci. 2018 Mar 12;19(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Wan C, Zhang H, Cheng H, et al. Selective autophagy regulates chloroplast protein import and promotes plant stress tolerance. EMBO J. 2023 May 30:e112534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Ma J, Liang Z, Zhao J, et al. Friendly mediates membrane depolarization-induced mitophagy in Arabidopsis. Curr Biol. 2021 May 10;31(9):1931–1944 e4. [DOI] [PubMed] [Google Scholar]
- [138].Nakamura S, Hagihara S, Otomo K, et al. Autophagy Contributes to the Quality Control of Leaf Mitochondria. Plant Cell Physiol. 2021. May 11;62(2):229–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Kacprzak SM, Van Aken O.. FRIENDLY is required for efficient dark-induced mitophagy and controlled senescence in Arabidopsis. Free Radic Biol Med. 2023 Aug 1;204:1–7. [DOI] [PubMed] [Google Scholar]
- [140].Larkin RM, Stefano G, Ruckle ME, et al. REDUCED CHLOROPLAST COVERAGE genes from Arabidopsis thaliana help to establish the size of the chloroplast compartment. Proc Natl Acad Sci USA. 2016 Feb 23;113(8):E1116-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Li C, Duckney P, Zhang T, et al. TraB family proteins are components of ER-mitochondrial contact sites and regulate ER-mitochondrial interactions and mitophagy. Nat Commun. 2022. Sep 26;13(1):5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Norizuki T, Minamino N, Sato M, et al. Dynamic rearrangement and autophagic degradation of mitochondria during spermiogenesis in the liverwort Marchantia polymorpha. Cell Rep. 2022. Jun 14;39(11):110975. [DOI] [PubMed] [Google Scholar]
- [143].Doelling JH, Walker JM, Friedman EM, et al. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002 Sep 6;277(36):33105–33114. [DOI] [PubMed] [Google Scholar]
- [144].Tang J, Bassham DC.. Autophagy in crop plants: what’s new beyond Arabidopsis? Open Biol. 2018 Dec 5;8(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Pettinari G, Finello J, Plaza Rojas M, et al. Autophagy modulates growth and development in the moss Physcomitrium patens. Front Plant Sci. 2022;13:1052358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Gou W, Li X, Guo S, et al. Autophagy in Plant: A New Orchestrator in the Regulation of the Phytohormones Homeostasis. Int J Mol Sci. 2019 Jun 14;20(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Liao CY, Wang P, Yin Y, et al. Interactions between autophagy and phytohormone signaling pathways in plants. FEBS Lett. 2022. Sep;596(17):2198–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Liao CY, Pu Y, Nolan TM, et al. Brassinosteroids modulate autophagy through phosphorylation of RAPTOR1B by the GSK3-like kinase BIN2 in Arabidopsis. Autophagy. 2023. Apr;19(4):1293–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Zhang Z, Zhu JY, Roh J, et al. TOR Signaling Promotes Accumulation of BZR1 to Balance Growth with Carbon Availability in Arabidopsis. Curr Biol. 2016 Jul 25;26(14):1854–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Feng Q, De Rycke R, Dagdas Y, et al. Autophagy promotes programmed cell death and corpse clearance in specific cell types of the Arabidopsis root cap. Curr Biol. 2022 Oct 24;32(20):4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Goh T, Sakamoto K, Wang P, et al. Autophagy promotes organelle clearance and organized cell separation of living root cap cells in Arabidopsis thaliana. Development. 2022 Jun 15;149(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Wang P, Clark NM, Nolan TM, et al. FERONIA functions through Target of Rapamycin (TOR) to negatively regulate autophagy. Front Plant Sci. 2022;13:961096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Chen J, Zhu S, Ming Z, et al. FERONIA cytoplasmic domain: node of varied signal outputs. aBIOTECH. 2020. Apr;1(2):135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Yu P, Hua Z.. The ubiquitin-26S proteasome system and autophagy relay proteome homeostasis regulation during silique development. Plant J. 2022. Sep;111(5):1324–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Sanchez-Sevilla JF, Botella MA, Valpuesta V, et al. Autophagy Is Required for Strawberry Fruit Ripening. Front Plant Sci. 2021;12:688481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Alseekh S, Zhu F, Vallarino JG, et al. Autophagy modulates the metabolism and growth of tomato fruit during development. Hortic Res. 2022;9:uhac129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Erlichman OA, Weiss S, Abu Arkia M, et al. Autophagy in maternal tissues contributes to Arabidopsis seed development. Plant Physiol. 2023 Aug 31;193(1):611–626. [DOI] [PubMed] [Google Scholar]
- [158].Ebstrup E, Ansbol J, Paez-Garcia A, et al. NBR1-mediated selective autophagy of ARF7 modulates root branching. EMBO Rep. 2024. Jun;25(6):2571–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Minina EA, Moschou PN, Vetukuri RR, et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J Exp Bot. 2018 Mar 14;69(6):1415–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Kanne JV, Ishikawa M, Bressendorff S, et al. Overexpression of ATG8/LC3 enhances wound-induced somatic reprogramming in Physcomitrium patens. Autophagy. 2022. Jun;18(6):1463–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Magen S, Seybold H, Laloum D, et al. Metabolism and autophagy in plants-a perfect match. FEBS Lett. 2022 Apr 25. [DOI] [PubMed] [Google Scholar]
- [162].Feng Y, Chen Y, Wu X, et al. Interplay of energy metabolism and autophagy. Autophagy. 2023. Aug 18:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].van Doorn WG, Papini A.. Ultrastructure of autophagy in plant cells: A review. Autophagy. 2013;9(12):1922–1936. [DOI] [PubMed] [Google Scholar]
- [164].Marshall RS, Vierstra RD. Autophagy: The Master of Bulk and Selective Recycling. In: Merchant SS, editor. Annual Review of Plant Biology, Vol 69. Annual Review of Plant Biology. Vol. 692018. p. 173–208. [DOI] [PubMed] [Google Scholar]
- [165].Wojciechowska N, Smugarzewska I, Marzec-Schmidt K, et al. Occurrence of autophagy during pioneer root and stem development in Populus trichocarpa. Planta. 2019. Dec;250(6):1789–1801. [DOI] [PubMed] [Google Scholar]
- [166].Wang PP, Wang TT, Han JY, et al. Plant Autophagy: An Intricate Process Controlled by Various Signaling Pathways. Frontiers in Plant Science. 2021 Sep; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Mugume Y, Kazibwe Z, Bassham DC.. Target of Rapamycin in Control of Autophagy: Puppet Master and Signal Integrator. International Journal of Molecular Sciences. 2020. Jan;21(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Signorelli S, Tarkowski LP, Van den Ende W, et al. Linking Autophagy to Abiotic and Biotic Stress Responses. Trends in Plant Science. 2019. May;24(5):413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Wang Y, Cao JJ, Wang KX, et al. BZR1 Mediates Brassinosteroid-Induced Autophagy and Nitrogen Starvation in Tomato. Plant Physiology. 2019. Feb;179(2):671–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Bedu M, Marmagne A, Masclaux-Daubresse C, et al. Transcriptional Plasticity of Autophagy-Related Genes Correlates with the Genetic Response to Nitrate Starvation in Arabidopsis Thaliana. Cells. 2020. Apr;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Slavikova S, Shy G, Yao YL, et al. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. Journal of Experimental Botany. 2005. Nov;56(421):2839–2849. [DOI] [PubMed] [Google Scholar]
- [172].Xia T, Xiao D, Liu D, et al. Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLOS ONE. 2012;7(5):e37217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Xiong Y, Contento AL, Bassham DC.. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant Journal. 2005. May;42(4):535–546. [DOI] [PubMed] [Google Scholar]
- [174].Wang Y, Yu BJ, Zhao JP, et al. Autophagy Contributes to Leaf Starch Degradation. Plant Cell. 2013. Apr;25(4):1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Pietrocola F, Izzo V, Niso-Santano M, et al. Regulation of autophagy by stress-responsive transcription factors. Seminars in Cancer Biology. 2013. Oct;23(5):310–322. [DOI] [PubMed] [Google Scholar]
- [176].Lai ZB, Wang F, Zheng ZY, et al. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant Journal. 2011. Jun;66(6):953–968. [DOI] [PubMed] [Google Scholar]
- [177].Zhu T, Zou LJ, Li Y, et al. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnology Journal. 2018. Dec;16(12):2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Barros JAS, Cavalcanti JHF, Pimentel KG, et al. The significance of WRKY45 transcription factor in metabolic adjustments during dark-induced leaf senescence. Plant Cell Environ. 2022. Sep;45(9):2682–2695. [DOI] [PubMed] [Google Scholar]
- [179].Yang C, Shen WJ, Yang LM, et al. HY5-HDA9 Module Transcriptionally Regulates Plant Autophagy in Response to Light-to-Dark Conversion and Nitrogen Starvation. Molecular Plant. 2020. Mar;13(3):515–531. [DOI] [PubMed] [Google Scholar]
- [180].Guiboileau A, Yoshimoto K, Soulay F, et al. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytologist. 2012. May;194(3):732–740. [DOI] [PubMed] [Google Scholar]
- [181].Huang X, Zheng CY, Liu F, et al. Genetic Analyses of the Arabidopsis ATG1 Kinase Complex Reveal Both Kinase-Dependent and Independent Autophagic Routes during Fixed-Carbon Starvation. Plant Cell. 2019. Dec;31(12):2973–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].McLoughlin F, Augustine RC, Marshall RS, et al. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nature Plants. 2018. Dec;4(12):1056–1070. [DOI] [PubMed] [Google Scholar]
- [183].McLoughlin F, Marshall RS, Ding XX, et al. Autophagy Plays Prominent Roles in Amino Acid, Nucleotide, and Carbohydrate Metabolism during Fixed-Carbon Starvation in Maize(OPEN). Plant Cell. 2020. Sep;32(9):2699–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Chung T, Suttangkakul A, Vierstra RD.. The ATG Autophagic Conjugation System in Maize: ATG Transcripts and Abundance of the ATG8-Lipid Adduct Are Regulated by Development and Nutrient Availability. Plant Physiology. 2009. Jan;149(1):220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Wang P, Sun X, Jia X, et al. Characterization of an Autophagy-Related Gene MdATG8i from Apple. Frontiers in Plant Science. 2016. May;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Avin-Wittenberg T, Bajdzienko K, Wittenberg G, et al. Global Analysis of the Role of Autophagy in Cellular Metabolism and Energy Homeostasis in Arabidopsis Seedlings under Carbon Starvation. Plant Cell. 2015. Feb;27(2):306–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Barros JAS, Cavalcanti JHF, Medeiros DB, et al. Autophagy Deficiency Compromises Alternative Pathways of Respiration following Energy Deprivation in Arabidopsis thaliana. Plant Physiology. 2017. Sep;175(1):62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Izumi M, Hidema J, Makino A, et al. Autophagy Contributes to Nighttime Energy Availability for Growth in Arabidopsis. Plant Physiology. 2013. Apr;161(4):1682–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Barros JAS, Magen S, Lapidot-Cohen T, et al. Autophagy is required for lipid homeostasis during dark-induced senescence. Plant Physiology. 2021. Apr;185(4):1542–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Barros JAS, Siqueira JAB, Cavalcanti JHF, et al. Multifaceted Roles of Plant Autophagy in Lipid and Energy Metabolism. Trends in Plant Science. 2020. Nov;25(11):1141–1153. [DOI] [PubMed] [Google Scholar]
- [191].Aroca A, Yruela I, Gotor C, et al. Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2021. May;118(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Gotor C, Aroca A, Romero LC.. Persulfidation is the mechanism underlying sulfide-signaling of autophagy. Autophagy. 2022. Mar;18(3):695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Alvarez C, Garcia I, Moreno I, et al. Cysteine-Generated Sulfide in the Cytosol Negatively Regulates Autophagy and Modulates the Transcriptional Profile in Arabidopsis. Plant Cell. 2012. Nov;24(11):4621–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Dong YH, Silbermann M, Speiser A, et al. Sulfur availability regulates plant growth via glucose-TOR signaling. Nature Communications. 2017 Oct; 8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [195].Liu YM, Burgos JS, Deng Y, et al. Degradation of the Endoplasmic Reticulum by Autophagy during Endoplasmic Reticulum Stress in Arabidopsis. Plant Cell. 2012. Nov;24(11):4635–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Naumann C, Mueller J, Sakhonwasee S, et al. The Local Phosphate Deficiency Response Activates Endoplasmic Reticulum Stress-Dependent Autophagy. Plant Physiology. 2019. Feb;179(2):460–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Yoshitake Y, Nakamura S, Shinozaki D, et al. RCB-mediated chlorophagy caused by oversupply of nitrogen suppresses phosphate-starvation stress in plants. Plant Physiology. 2021. Feb;185(2):318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Lee DH, Choi I, Park SJ, et al. Three consecutive cytosolic glycolysis enzymes modulate autophagic flux. Plant Physiol. 2023 Oct 26;193(3):1797–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Yang C, Li X, Yang L, et al. A positive feedback regulation of SnRK1 signaling by autophagy in plants. Molecular plant. 2023 Jul 3;16(7):1192–1211. [DOI] [PubMed] [Google Scholar]
- [200].Avin-Wittenberg T. Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 2019. Mar;42(3):1045–1053. [DOI] [PubMed] [Google Scholar]
- [201].Tang J, Bassham DC.. Autophagy during drought: function, regulation, and potential application. Plant J. 2022. Jan;109(2):390–401. [DOI] [PubMed] [Google Scholar]
- [202].Williams B, Njaci I, Moghaddam L, et al. Trehalose Accumulation Triggers Autophagy during Plant Desiccation. PLOS Genet. 2015. Dec;11(12):e1005705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Sun G, Wase N, Shu S, et al. Genome of Paspalum vaginatum and the role of trehalose mediated autophagy in increasing maize biomass. Nature communications. 2022. Dec 13;13(1):7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Luo X, Zhang Y, Wu H, et al. Drought stress-induced autophagy gene expression is correlated with carbohydrate concentrations in Caragana korshinskii. Protoplasma. 2020. Jul;257(4):1211–1220. [DOI] [PubMed] [Google Scholar]
- [205].Li C, Qi Y, Zhao C, et al. Transcriptome Profiling of the Salt Stress Response in the Leaves and Roots of Halophytic Eutrema salsugineum. Front Genet. 2021;12:770742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Li YB, Cui DZ, Sui XX, et al. Autophagic Survival Precedes Programmed Cell Death in Wheat Seedlings Exposed to Drought Stress. Int J Mol Sci. 2019. Nov 16;20(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Yue JY, Wang YJ, Jiao JL, et al. Silencing of ATG2 and ATG7 promotes programmed cell death in wheat via inhibition of autophagy under salt stress. Ecotoxicol Environ Saf. 2021. Dec 1;225:112761. [DOI] [PubMed] [Google Scholar]
- [208].Dundar G, Shao Z, Higashitani N, et al. Autophagy mitigates high-temperature injury in pollen development of Arabidopsis thaliana. Dev Biol. 2019 Dec 15;456(2):190–200. [DOI] [PubMed] [Google Scholar]
- [209].Sun X, Wang P, Jia X, et al. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol J. 2018. Feb;16(2):545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Jia X, Mao K, Wang P, et al. Overexpression of MdATG8i improves water use efficiency in transgenic apple by modulating photosynthesis, osmotic balance, and autophagic activity under moderate water deficit. Hortic Res. 2021 Apr 1;8(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Huo L, Sun X, Guo Z, et al. MdATG18a overexpression improves basal thermotolerance in transgenic apple by decreasing damage to chloroplasts. Hortic Res. 2020;7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Fu XZ, Zhou X, Xu YY, et al. Comprehensive Analysis of Autophagy-Related Genes in Sweet Orange (Citrus sinensis) Highlights Their Roles in Response to Abiotic Stresses. Int J Mol Sci. 2020 Apr 13;21(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Wang P, Sun X, Jia X, et al. Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 2017. Mar;256:53–64. [DOI] [PubMed] [Google Scholar]
- [214].Perez-Martin M, Perez-Perez ME, Lemaire SD, et al. Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in Chlamydomonas reinhardtii. Plant Physiol. 2014. Oct;166(2):997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Barros JAS, Chatt EC, Augustine RC, et al. Autophagy during maize endosperm development dampens oxidative stress and promotes mitochondrial clearance. Plant Physiol. 2023 Sep 22;193(2):1395–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Zhang Y, Wang Y, Wen W, et al. Hydrogen peroxide mediates spermidine-induced autophagy to alleviate salt stress in cucumber. Autophagy. 2021. Oct;17(10):2876–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Zheng P, Wu JX, Sahu SK, et al. Loss of alkaline ceramidase inhibits autophagy in Arabidopsis and plays an important role during environmental stress response. Plant Cell Environ. 2018. Apr;41(4):837–849. [DOI] [PubMed] [Google Scholar]
- [218].Zhu T, Zou L, Li Y, et al. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol J. 2018. Dec;16(12):2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Feng X, Liu L, Li Z, et al. Potential interaction between autophagy and auxin during maize leaf senescence. J Exp Bot. 2021 May 4;72(10):3554–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Selamat N, Nadarajah KK.. Meta-Analysis of Quantitative Traits Loci (QTL) Identified in Drought Response in Rice (Oryza sativa L.). Plants (Basel). 2021 Apr 7;10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Lin Z, Wang YL, Cheng LS, et al. Mutual regulation of ROS accumulation and cell autophagy in wheat roots under hypoxia stress. Plant Physiol Biochem. 2021. Jan;158:91–102. [DOI] [PubMed] [Google Scholar]
- [222].Laureano-Marin AM, Moreno I, Romero LC, et al. Negative Regulation of Autophagy by Sulfide Is Independent of Reactive Oxygen Species. Plant Physiol. 2016. Jun;171(2):1378–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Bao Y, Pu Y, Yu X, et al. IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana. Autophagy. 2018;14(9):1562–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Su W, Bao Y, Lu Y, et al. Poplar Autophagy Receptor NBR1 Enhances Salt Stress Tolerance by Regulating Selective Autophagy and Antioxidant System. Front Plant Sci. 2020;11:568411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Wan C, Zhang H, Cheng H, et al. Selective autophagy regulates chloroplast protein import and promotes plant stress tolerance. EMBO J. 2023 Jul 17;42(14):e112534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Li X, Liu Q, Feng H, et al. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy. 2020. May;16(5):862–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Zhou J, Wang Z, Wang X, et al. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy. 2018;14(3):487–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Tarnowski L, Rodriguez MC, Brzywczy J, et al. A selective autophagy cargo receptor NBR1 modulates abscisic acid signalling in Arabidopsis thaliana. Sci Rep. 2020 May 8;10(1):7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Chi C, Li X, Fang P, et al. Brassinosteroids act as a positive regulator of NBR1-dependent selective autophagy in response to chilling stress in tomato. J Exp Bot. 2020 Jan 23;71(3):1092–1106. [DOI] [PubMed] [Google Scholar]
- [230].Montes C, Wang P, Liao CY, et al. Integration of multi-omics data reveals interplay between brassinosteroid and Target of Rapamycin Complex signaling in Arabidopsis. New Phytol. 2022. Nov;236(3):893–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Sedaghatmehr M, Thirumalaikumar VP, Kamranfar I, et al. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019. Mar;42(3):1054–1064. [DOI] [PubMed] [Google Scholar]
- [232].Sedaghatmehr M, Thirumalaikumar VP, Kamranfar I, et al. Autophagy complements metalloprotease FtsH6 in degrading plastid heat shock protein HSP21 during heat stress recovery. J Exp Bot. 2021 Jun 29. [DOI] [PubMed] [Google Scholar]
- [233].Thirumalaikumar VP, Wagner M, Balazadeh S, et al. Autophagy is responsible for the accumulation of proteogenic dipeptides in response to heat stress in Arabidopsis thaliana. FEBS J. 2021. Jan;288(1):281–292. [DOI] [PubMed] [Google Scholar]
- [234].Zhou J, Ma J, Yang C, et al. A non-canonical role of ATG8 in Golgi recovery from heat stress in plants. Nat Plants. 2023. May;9(5):749–765. [DOI] [PubMed] [Google Scholar]
- [235].Mishra R, Shteinberg M, Shkolnik D, et al. Interplay between abiotic (drought) and biotic (virus) stresses in tomato plants. Mol Plant Pathol. 2022. Apr;23(4):475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Ngou BPM, Ding P, Jones JDG.. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell. 2022 Apr 26;34(5):1447–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Hofius D, Schultz-Larsen T, Joensen J, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009 May 15;137(4):773–783. [DOI] [PubMed] [Google Scholar]
- [238].Hackenberg T, Juul T, Auzina A, et al. Catalase and NO CATALASE ACTIVITY1 Promote Autophagy-Dependent Cell Death in Arabidopsis. Plant Cell. 2013. Nov;25(11):4616–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Yoshimoto K, Jikumaru Y, Kamiya Y, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis [Research Support, Non-U.S. Gov’t]. Plant Cell. 2009. Sep;21(9):2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Liu Y, Schiff M, Czymmek K, et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005 May 20;121(4):567–577. [DOI] [PubMed] [Google Scholar]
- [241].Patel S, Dinesh-Kumar SP.. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008 Jan 1;4(1):20–27. [DOI] [PubMed] [Google Scholar]
- [242].Munch D, Rodriguez E, Bressendorff S, et al. Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy. 2014. Sep;10(9):1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Coll NS, Smidler A, Puigvert M, et al. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ. 2014 May 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Zavaliev R, Mohan R, Chen T, et al. Formation of NPR1 Condensates Promotes Cell Survival during the Plant Immune Response. Cell. 2020 Sep 3;182(5):1093–1108 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Leary AY, Savage Z, Tumtas Y, et al. Contrasting and emerging roles of autophagy in plant immunity. Curr Opin Plant Biol. 2019. Dec;52:46–53. [DOI] [PubMed] [Google Scholar]
- [246].Yuan M, Jiang Z, Bi G, et al. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 2021. Apr;592(7852):105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Yang F, Kimberlin AN, Elowsky CG, et al. A Plant Immune Receptor Degraded by Selective Autophagy. Molecular plant. 2019 Jan 7;12(1):113–123. [DOI] [PubMed] [Google Scholar]
- [248].Jeon HS, Jang E, Kim J, et al. Pathogen-induced autophagy regulates monolignol transport and lignin formation in plant immunity. Autophagy. 2023. Feb;19(2):597–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Teper-Bamnolker P, Danieli R, Peled-Zehavi H, et al. Vacuolar processing enzyme translocates to the vacuole through the autophagy pathway to induce programmed cell death. Autophagy. 2021. Oct;17(10):3109–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Hatsugai N, Kuroyanagi M, Yamada K, et al. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science. 2004 Aug 6;305(5685):855-858. [DOI] [PubMed] [Google Scholar]
- [251].Hatsugai N, Yamada K, Goto-Yamada S, et al. Vacuolar processing enzyme in plant programmed cell death. Frontiers in plant science. 2015;6:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Coll NS, Vercammen D, Smidler A, et al. Arabidopsis type I metacaspases control cell death [Research Support, N.I.H. Extramural; [Research Support, Non-U.S. Gov’t]. Science. 2010 Dec 3;330(6009):1393-1397. [DOI] [PubMed] [Google Scholar]
- [253].Heidrich K, Wirthmueller L, Tasset C, et al. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses [Research Support, Non-U.S. Gov’t]. Science. 2011 Dec 9;334(6061):1401-1404. [DOI] [PubMed] [Google Scholar]
- [254].Bai S, Liu J, Chang C, et al. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLOS Pathog. 2012;8(6):e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Li Y, Kabbage M, Liu W, et al. Aspartyl Protease-Mediated Cleavage of BAG6 Is Necessary for Autophagy and Fungal Resistance in Plants. Plant Cell. 2016. Jan;28(1):233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Kabbage M, Williams B, Dickman MB.. Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLOS Pathog. 2013;9(4):e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Rajarammohan S. Redefining Plant-Necrotroph Interactions: The Thin Line Between Hemibiotrophs and Necrotrophs. Front Microbiol. 2021;12:673518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Hafren A, Ustun S, Hochmuth A, et al. Turnip Mosaic Virus Counteracts Selective Autophagy of the Viral Silencing Suppressor HCpro. Plant Physiol. 2018. Jan;176(1):649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Shukla A, Hoffmann G, Kushwaha NK, et al. Salicylic acid and the viral virulence factor 2b regulate the divergent roles of autophagy during cucumber mosaic virus infection. Autophagy. 2022. Jun;18(6):1450–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Jiang L, Lu Y, Zheng X, et al. The plant protein NbP3IP directs degradation of Rice stripe virus p3 silencing suppressor protein to limit virus infection through interaction with the autophagy-related protein NbATG8. New Phytol. 2021. Jan;229(2):1036–1051. [DOI] [PubMed] [Google Scholar]
- [261].Haxim Y, Ismayil A, Jia Q, et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife. 2017 Feb 28; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Li F, Zhang C, Li Y, et al. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nature communications. 2018 Mar 28;9(1):1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Niu E, Liu H, Zhou H, et al. Autophagy Inhibits Intercellular Transport of Citrus Leaf Blotch Virus by Targeting Viral Movement Protein. Viruses. 2021 Oct 30;13(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Hafren A, Macia JL, Love AJ, et al. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci USA. 2017 Mar 07;114(10):E2026-E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Levine B, Mizushima N, Virgin HW.. Autophagy in immunity and inflammation. Nature. 2011 Jan 20;469(7330):323-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Nakahara KS, Masuta C, Yamada S, et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc Natl Acad Sci USA. 2012 Jun 19;109(25):10113–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Tong X, Zhao JJ, Feng YL, et al. A selective autophagy receptor VISP1 induces symptom recovery by targeting viral silencing suppressors. Nature communications. 2023 Jun 29;14(1):3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Li F, Zhang M, Zhang C, et al. Nuclear autophagy degrades a geminivirus nuclear protein to restrict viral infection in solanaceous plants. New Phytol. 2020. Feb;225(4):1746–1761. [DOI] [PubMed] [Google Scholar]
- [269].Ustun S, Hafren A, Liu Q, et al. Bacteria Exploit Autophagy for Proteasome Degradation and Enhanced Virulence in Plants. Plant Cell. 2018. Mar;30(3):668–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Leong JX, Raffeiner M, Spinti D, et al. A bacterial effector counteracts host autophagy by promoting degradation of an autophagy component. EMBO J. 2022 Jul 4;41(13):e110352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Dagdas YF, Pandey P, Tumtas Y, et al. Host autophagy machinery is diverted to the pathogen interface to mediate focal defense responses against the Irish potato famine pathogen. eLife. 2018 Jun 22;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Zou J, Chen X, Liu C, et al. Autophagy promotes jasmonate-mediated defense against nematodes. Nature communications. 2023 Aug 8;14(1):4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Hickman R, Van Verk MC, Van Dijken AJH, et al. Architecture and Dynamics of the Jasmonic Acid Gene Regulatory Network. Plant Cell. 2017. Sep;29(9):2086–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Lal NK, Thanasuwat B, Huang PJ, et al. Phytopathogen Effectors Use Multiple Mechanisms to Manipulate Plant Autophagy. Cell Host Microbe. 2020 Oct 7;28(4):558–571 e6. [DOI] [PubMed] [Google Scholar]
- [275].Ismayil A, Yang M, Haxim Y, et al. Cotton leaf curl Multan virus betaC1 Protein Induces Autophagy by Disrupting the Interaction of Autophagy-Related Protein 3 with Glyceraldehyde-3-Phosphate Dehydrogenases. Plant Cell. 2020. Apr;32(4):1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Cheng G, Yang Z, Zhang H, et al. Remorin interacting with PCaP1 impairs Turnip mosaic virus intercellular movement but is antagonised by VPg. New Phytol. 2020. Mar;225(5):2122–2139. [DOI] [PubMed] [Google Scholar]
- [277].Fu S, Xu Y, Li C, et al. Rice Stripe Virus Interferes with S-acylation of Remorin and Induces Its Autophagic Degradation to Facilitate Virus Infection. Molecular plant. 2018 Feb 5;11(2):269–287. [DOI] [PubMed] [Google Scholar]
- [278].Cheng X, Wang A.. The Potyvirus Silencing Suppressor Protein VPg Mediates Degradation of SGS3 via Ubiquitination and Autophagy Pathways. J Virol. 2017 Jan 1;91(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Zhang XP, Liu DS, Yan T, et al. Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta. PLOS Pathog. 2017. Jul;13(7):e1006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Yang M, Zhang Y, Xie X, et al. Barley stripe mosaic virus gammab Protein Subverts Autophagy to Promote Viral Infection by Disrupting the ATG7-ATG8 Interaction. Plant Cell. 2018. Jul;30(7):1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Yang M, Ismayil A, Jiang Z, et al. A viral protein disrupts vacuolar acidification to facilitate virus infection in plants. EMBO J. 2022 Dec 17;41(2):e108713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Yang M, Ismayil A, Gao T, et al. Cotton Leaf Curl Multan virus C4 protein suppresses autophagy to facilitate viral infection. Plant Physiol. 2023. Apr 19. [DOI] [PubMed] [Google Scholar]
- [283].Li F, Zhang C, Tang Z, et al. A plant RNA virus activates selective autophagy in a UPR-dependent manner to promote virus infection. New Phytol. 2020. Oct;228(2):622–639. [DOI] [PubMed] [Google Scholar]
- [284].Dagdas YF, Belhaj K, Maqbool A, et al. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife. 2016 Jan 14; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Pandey P, Leary AY, Tumtas Y, et al. An oomycete effector subverts host vesicle trafficking to channel starvation-induced autophagy to the pathogen interface. eLife. 2021 Aug 23;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Zhu W, Ronen M, Gur Y, et al. BcXYG1, a Secreted Xyloglucanase from Botrytis cinerea, Triggers Both Cell Death and Plant Immune Responses. Plant Physiol. 2017. Sep;175(1):438–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Seifbarghi S, Borhan MH, Wei Y, et al. Receptor-Like Kinases BAK1 and SOBIR1 Are Required for Necrotizing Activity of a Novel Group of Sclerotinia sclerotiorum Necrosis-Inducing Effectors. Frontiers in plant science. 2020;11:1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Chen J, Chen S, Xu C, et al. A key virulence effector from cyst nematodes targets host autophagy to promote nematode parasitism. New Phytol. 2023. Feb;237(4):1374–1390. [DOI] [PubMed] [Google Scholar]
- [289].Shi J, Gong Y, Shi H, et al. ‘Candidatus Liberibacter asiaticus’ secretory protein SDE3 inhibits host autophagy to promote Huanglongbing disease in citrus. Autophagy. 2023. Sep;19(9):2558–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Che R, Liu C, Wang Q, et al. The Valsa Mali effector Vm1G-1794 protects the aggregated MdEF-Tu from autophagic degradation to promote infection in apple. Autophagy. 2023. Jun;19(6):1745–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Kellner R, De la Concepcion JC, Maqbool A, et al. ATG8 Expansion: A Driver of Selective Autophagy Diversification? Trends in plant science. 2017. Mar;22(3):204–214. [DOI] [PubMed] [Google Scholar]
- [292].Yue W, Nie X, Cui L, et al. Genome-wide sequence and expressional analysis of autophagy Gene family in bread wheat (Triticum aestivum L.). Journal of Plant Physiology. 2018. 2018/10/01/;229:7–21. [DOI] [PubMed] [Google Scholar]
- [293].Bu F, Yang M, Guo X, et al. Multiple functions of ATG8 family proteins in plant autophagy. Frontiers in cell and developmental biology. 2020;8:466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Chung T, Suttangkakul A, Vierstra RD.. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant physiology. 2009. Jan;149(1):220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Xia K, Liu T, Ouyang J, et al. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA research: an international journal for rapid publication of reports on genes and genomes. 2011. Oct;18(5):363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Quezada-Rodríguez EH, Gómez-Velasco H, Arthikala MK, et al. Exploration of autophagy families in legumes and dissection of the ATG18 family with a special focus on Phaseolus vulgaris. Plants (Basel, Switzerland). 2021 Nov 29;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Pei D, Zhang W, Sun H, et al. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Reports. 2014. 2014/10/01;33(10):1697–1710. [DOI] [PubMed] [Google Scholar]
- [298].Mamun MA, Tang C, Sun Y, et al. Wheat gene TaATG8j contributes to stripe rust resistance. International journal of molecular sciences. 2018 Jun 5;19(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Xiong Y, Contento AL, Bassham DC.. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005. May;42(4):535–546. [DOI] [PubMed] [Google Scholar]
- [300].Sun X, Jia X, Huo L, et al. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 2018. Feb;41(2):469–480. [DOI] [PubMed] [Google Scholar]
- [301].Yue W, Zhang H, Sun X, et al. The landscape of autophagy-related (ATG) genes and functional characterization of TaVAMP727 to autophagy in wheat. International journal of molecular sciences. 2022 Jan 14;23(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Zhen X, Zheng N, Yu J, et al. Autophagy mediates grain yield and nitrogen stress resistance by modulating nitrogen remobilization in rice. PlOS one. 2021;16(1):e0244996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Sera Y, Hanamata S, Sakamoto S, et al. Essential roles of autophagy in metabolic regulation in endosperm development during rice seed maturation. Scientific reports. 2019. 2019/12/06;9(1):18544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Sun X, Pan B, Xu W, et al. Genome-wide identification and expression analysis of the pear autophagy-related gene PbrATG8 and functional verification of PbrATG8c in Pyrus bretschneideri Rehd. Planta. 2021 Jan 13;253(2):32. [DOI] [PubMed] [Google Scholar]
- [305].McLoughlin F, Augustine RC, Marshall RS, et al. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat Plants. 2018. Dec;4(12):1056–1070. [DOI] [PubMed] [Google Scholar]
- [306].McLoughlin F, Marshall RS, Ding X, et al. Autophagy Plays Prominent Roles in Amino Acid, Nucleotide, and Carbohydrate Metabolism during Fixed-Carbon Starvation in Maize. Plant Cell. 2020. Sep;32(9):2699–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Li F, Chung T, Pennington JG, et al. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell. 2015. May;27(5):1389–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Li YB, Yan M, Cui DZ, et al. Programmed degradation of pericarp cells in wheat grains depends on autophagy. Frontiers in genetics. 2021;12:784545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Avila-Ospina L, Moison M, Yoshimoto K, et al. Autophagy, plant senescence, and nutrient recycling. Journal of Experimental Botany. 2014;65(14):3799–3811. [DOI] [PubMed] [Google Scholar]
- [310].Mae T, Makino A, Ohira K.. Changes in the amounts of Ribulose Bisphosphate Carboxylase synthesized and degraded during the life span of rice leaf (Oryza sativa L.). Plant and Cell Physiology. 1983;24(6):1079–1086. [Google Scholar]
- [311].Sage RF, Pearcy RW, Seemann JR.. THE NITROGEN USE EFFICIENCY OF C-3 AND C-4 PLANTS.3. LEAF NITROGEN EFFECTS ON THE ACTIVITY OF CARBOXYLATING ENZYMES IN CHENOPODIUM-ALBUM (L) AND AMARANTHUS-RETROFLEXUS (L). Plant Physiology. 1987. Oct;85(2):355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Masclaux C, Valadier MH, Brugiere N, et al. Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta. 2000. Sep;211(4):510–518. [DOI] [PubMed] [Google Scholar]
- [313].Gregersen PL, Culetic A, Boschian L, et al. Plant senescence and crop productivity. Plant molecular biology. 2013. Aug;82(6):603–622. [DOI] [PubMed] [Google Scholar]
- [314].Suttangkakul A, Li FQ, Chung T, et al. The ATG1/ATG13 Protein Kinase Complex Is Both a Regulator and a Target of Autophagic Recycling in Arabidopsis. Plant Cell. 2011. Oct;23(10):3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Li FQ, Chung T, Pennington JG, et al. Autophagic Recycling Plays a Central Role in Maize Nitrogen Remobilization. Plant Cell. 2015. May;27(5):1389–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Phillips AR, Suttangkakul A, Vierstra RD.. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008. Mar;178(3):1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Chung T, Phillips AR, Vierstra RD.. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 2010;62(3):483–493. [DOI] [PubMed] [Google Scholar]
- [318].Wada S, Hayashida Y, Izumi M, et al. Autophagy Supports Biomass Production and Nitrogen Use Efficiency at the Vegetative Stage in Rice Plant physiology. 2015;168(1):60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Fan T, Yang W, Zeng X, et al. A rice autophagy gene OsATG8b is Involved in nitrogen remobilization and control of grain quality. Frontiers in plant science. 2020;11:588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Zhen X, Li X, Yu J, et al. OsATG8c-Mediated Increased Autophagy Regulates the Yield and Nitrogen Use Efficiency in Rice. International journal of molecular sciences. 2019;20(19):4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Li WW, Chen M, Zhong L, et al. Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochemical and Biophysical Research Communications. 2015. Dec;468(4):800–806. [DOI] [PubMed] [Google Scholar]
- [322].Huang W, Ma DN, Liu HL, et al. Genome-Wide Identification of CsATGs in Tea Plant and the Involvement of CsATG8e in Nitrogen Utilization. International Journal of Molecular Sciences. 2020. Oct;21(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Hollmann J, Gregersen PL, Krupinska K.. Identification of predominant genes involved in regulation and execution of senescence-associated nitrogen remobilization in flag leaves of field grown barley. Journal of Experimental Botany. 2014;65(14):3963–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Avila-Ospina L, Marmagne A, Soulay F, et al. Identification of barley (Hordeum vulgare L.) autophagy genes and their expression levels during leaf senescence, chronic nitrogen limitation and in response to dark exposure. Agronomy. 2016;6(1):15. [Google Scholar]
- [325].Thompson AR, Doelling JH, Suttangkakul A, et al. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005. Aug;138(4):2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Kurusu T, Koyano T, Hanamata S, et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy. 2014. May;10(5):878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Kurusu T, Koyano T, Kitahata N, et al. Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant signaling & behavior. 2017 Sep 2;12(9):e1365211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Zhao P, Zhou XM, Zhao LL, et al. Autophagy-mediated compartmental cytoplasmic deletion is essential for tobacco pollen germination and male fertility. Autophagy. 2020. Dec;16(12):2180–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Ding X, Zhang X, Paez-Valencia J, et al. Microautophagy mediates vacuolar delivery of storage proteins in maize aleurone cells. Frontiers in plant science. 2022;13:833612. [DOI] [PMC free article] [PubMed] [Google Scholar]


