Abstract
Posttranscriptional modifications are involved in cancer progression. However, the function and regulatory mechanism of mRNA acetylation modification remains largely unknown. Here, we discover an unexpected role of N4-acetylcytidine (ac4C) RNA acetyltransferase NAT10 in reshaping the tumor immune microenvironment. By analyzing patients’ data, we find that NAT10 is upregulated in tumor tissues, and negatively correlated with immune cell infiltration and overall survival. Loss of tumoral NAT10 enhances tumor-specific cellular immune response and suppresses tumor growth. Mechanistically, MYC is identified as a key downstream target of NAT10 via enhancing mRNA ac4C modification. Inhibition of NAT10 blocks the MYC/CDK2/DNMT1 pathway, enhances double-stranded RNA (dsRNA) formation, which triggers type I interferon response and improves tumor specific CD8+ T cell response in vivo. More importantly, the inhibition of NAT10, using either small molecule inhibitor (Remodelin) or PEI/PC7A/siNAT10 nanoparticles, synergize PD-1 blockade in elevating anti-tumor immune response and repressing tumor progression. Our findings thus uncover the crucial role of tumor-intrinsic NAT10 in tumor immune microenvironment, which represents a promising target for enhancing cancer immunotherapy.
Subject terms: Tumour immunology, Cancer microenvironment, Cancer epigenetics, Cancer therapy
The function and regulatory mechanism of mRNA acetylation modification in cancer progression remains largely unknown. Here the authors identify that targeting tumor N4-acetylcytidine (ac4C) RNA acetyltransferase NAT10 improves anti-tumor response via MYC/CDK2/DNMT1/dsRNA/type I IFN pathway, and show the synergic anti-tumor effect of NAT10 inhibition with aPD-1.
Introduction
The significant advancements and successes in cancer immunotherapy over the past decade have markedly improved the clinical management of numerous malignancies that previously exhibited poor prognoses. Immune checkpoint inhibitors (ICI) represent the forefront of tumor immunotherapy strategies1,2. However, the limited infiltration and activation of immune cells in the tumor microenvironment undermine their efficacy3,4.
Interferons (IFN) play a significant role in promoting the anti-tumor response by activating immune cells5,6. IFNs are primarily released by immune and stromal cells, with tumor cells also contributing to the release of IFNs. Various genotoxic anticancer treatments, such as radiation and chemotherapy, small-molecule kinase inhibitors, induce DNA damage or produce double-stranded RNA (dsRNA), thereby activating IFN pathways to stimulate an immune response against cancer7. Recent transcriptomic studies have revealed that metazoan cells express different types of endogenous dsRNA, including endogenous retroviral elements (ERV), repetitive RNA elements, mitochondrial dsRNAs, mRNAs with inverted Alu-containing 3′UTRs, and structural dsRNA with long dsRNA stems8. Malignant cells have increased levels of dsRNAs due to the loss of suppressive epigenetic modifications in repetitive elements, genomic instability or oxidative stress-induced mitochondrial damage, thereby increasing the dsRNAs load in tumor cells9. In certain cancers, loss of ERV DNA methylation leads to aberrant overexpression of ERVs, and bidirectional transcription of ERVs has been demonstrated to upregulate dsRNA formation10,11. Accumulating evidence indicates that intracellular deposition of dsRNA in tumor cells stimulates the production of type I IFN, thereby enhancing antitumor immunity12. Abnormal accumulation of endogenous dsRNAs triggers an innate antiviral and antitumor immune response by activating dsRNA-sensing pathways, including retinoic acid-inducible gene I (RIG-I)13–15. Further studies on these gene signatures and their associated changes in the anti-tumor immune response could provide valuable insights into predicting patients’ outcomes and their responses or resistance to chemotherapy and immunotherapy.
Recent studies have revealed that tumor cells achieve immune evasion through epigenetic reprogramming catalyzed by related enzymes16. Inhibition of activity of epigenetic-modifying enzyme promotes the infiltration and activation of CD8+ T cells17–19. However, whether and how RNA epitranscriptome modifications are involved in immune evasion remains to be fully understood. In a recent study, Arango and colleagues identified N4-acetylcytidine (ac4C) as a new mRNA acetylation modification catalyzed by N-acetyltransferase 10 (NAT10) to increase the mRNA ac4C modification20. NAT10 has recently been shown to regulate tumor progression and drug resistance21,22; however, its role in tumor immunity remains unknown.
In this study, we demonstrate that inhibition of tumor-intrinsic NAT10 can regulate mRNA acetylation modifications, effectively enhance the type I IFN response to stimulate adaptive immune responses and thereby inhibit tumor progression. We also succeed in establishing PEI/PC7A/siNAT10 nanoparticles that specifically target and degrade NAT10 in tumor cells. The nanoparticles not only exhibit potent antitumor effects alone but also display enhanced efficacy when combined with programmed cell death 1 (PD-1) blockade. Together, our findings indicate that NAT10 is a tumor-intrinsic regulator that orchestrates immune evasion.
Results
Elevated NAT10 correlates to low immune cell infiltration and poor survival in cancer
Immunosuppression is the main obstacle for clinical tumor therapy. NAT10, the only writer for ac4C modification on mRNAs, has been reported to have many critical functions; however, its role in the immunosuppressive tumor microenvironment has not been reported20. We first utilized the GEPIA database23 to analyze the possible role of NAT10 in lung adenocarcinoma (LUAD). The result showed that NAT10 expression was increased in the TCGA-LUAD cohort and positively associated with tumor stage (Fig. 1A). We further analyzed the raw data to generate a Kaplan-Meier survival curve, which showed that higher NAT10 expression is significantly correlated to shorter overall survival (Fig. 1B). Furthermore, we collected lung cancer samples and performed NAT10 immunohistochemical staining (Fig. S1A, Supplementary Data 1). Consistent with the TCGA database, higher NAT10 expression was also associated with shorter survival in our cohort (Fig. 1C). Moreover, the ROC curve was utilized to validate the prognostic efficiency of NAT10, and the results indicated that NAT10 has strong predictive value in lung cancer (Fig. 1D). These results suggest that NAT10 is a cancer-promoting gene and associated with the progression of lung cancer.
Fig. 1. Elevated NAT10 expression is associated with decreased survival and reduced infiltration of immune cells in cancer.
A The expression of NAT10 was assessed in different stages of LUAD using the GEPIA website (http://gepia.cancer-pku.cn/). B Kaplan-Meier survival curve comparing the high- and low-NAT10 expression groups (optimal cut-off) in the TCGA-LUAD cohort. C Kaplan-Meier survival curve comparing the high- and low-NAT10 expression groups in 37 patients with lung cancer. NAT10 expression was quantified using immunohistochemistry and Image Pro Plus. Statistical significance was determined using the log-rank test. D ROC curves for survival prediction with corresponding AUC values. The area under the curve (AUC) was calculated, and the statistical significance of AUC comparison between groups was determined using the DeLong test. E, F Tumor weight and growth curves for C57BL/6 N mice inoculated with TC1 (E) or MCA205 tumor cells (F). 2 × 106 WT cells were subcutaneously inoculated into the back of C57BL/6 N mice; n = 5 mice per group. Mice received Remodelin or saline via oral gavage for the first 5 days at a dose of 100 mg/kg. Tumor size was measured daily using calipers to generate growth curves. Tumor growth curves were analyzed by two-way ANOVA with the tumor size at the final day used for significance testing. From left to right, **P = 0.004; ***P < 0.001; **P = 0.0015; ***P < 0.001, respectively. G, H Tumor weight and growth curves for nude/nude mice inoculated with TC1 (G) or MCA205 tumor cells (H). 2 × 106 WT cells were subcutaneously inoculated into the back of C57BL/6 N mice; n = 5 mice per group. Mice received Remodelin or saline via oral gavage for the first 5 days at a dose of 100 mg/kg. I Analysis of immune cell infiltration using the CIBERSORT algorithm between high- and low-NAT10 expression groups in the TCGA-LUAD cohort. J Immunohistochemical analysis of NAT10 expression and CD8+ T cell infiltration in patient-derived lung cancer samples (n = 37). CD8+ T cell counts in the high- and low-NAT10 expression groups are presented on the right. The arrow indicates CD8+ T cells; Scale bar: 50 µm; *P = 0.0484. Unless specified otherwise, the data are presented as means ± SEM (error bar) and compared using the two-sided Student’s t test; ns, no significance; Source data are provided as a Source Data file.
Next, we investigated the effects of NAT10 inhibition on tumor cells. We first treated tumor cells with Remodelin, a well-established inhibitor of NAT1024, and measured cell proliferation by a colony formation assay. The results showed that Remodelin significantly suppressed the proliferation of tumor cells (Fig. S1B, S1C). Then we established syngeneic tumor models by injecting murine lung tumor cells (TC1) or murine fibrosarcoma cells (MCA205) into immunocompetent (C57BL/6 N) mice subcutaneously. The mice were administrated with Remodelin, with saline used as the control. Our results showed that both the weight and size of tumor masses were significantly reduced in mice exposed to low-dose Remodelin compared to those exposed to saline control (Fig. 1E, F). However, in the immunodeficient (nude/nude) murine tumor model, no significant difference in tumor size or weight was observed between the low-dose Remodelin group and the saline control group (Fig. 1G, H). These results suggest that NAT10 has a pro-tumor effect, which is partly mediated by suppressing immunity.
We then analyzed the immune cell infiltration in cancers using the CIBERSORT algorithm on the related TCGA-LUAD cohort25. As expected, a negative correlation was found between NAT10 mRNA and the infiltration of antitumor immune cell subsets, such as CD8+ T cells and DCs (Fig. 1I). Moreover, scRNAseq data (GSE148071) were applied for the expression analysis of NAT10 in individual tumor patients26. Consistently, the results also indicated a negative correlation between NAT10 expression and the proportion of T cells or DCs, while a positive relationship between NAT10 expression and the malignant cell percentage (Fig. S1D). Furthermore, our immunohistochemical staining results showed the similar negative association between NAT10 and CD8+ T cells in the lung cancer samples (n = 37) (Fig. 1J). Given the important roles of EGFR, ALK, and KRAS in LUAD, we downloaded the mutation data of the TCGA-LUAD cohort and performed a multivariate Cox regression analysis incorporating EGFR and ALK gene mutations, as well as NAT10 expression levels. The results indicated that NAT10’s impact on lung cancer prognosis is independent of these gene mutations (Fig. S1E, S1F). Collectively, these data suggest a potential role for tumor-intrinsic NAT10 in tumor progression, particularly in regulating immune cell infiltration.
NAT10 deficiency suppresses tumor growth via eliciting immunological protection
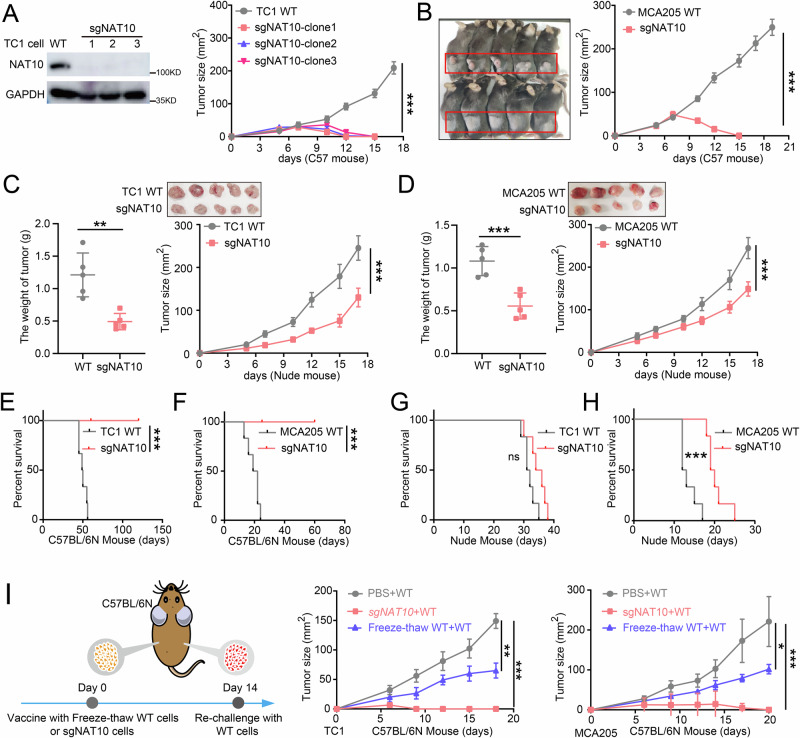
To elucidate the effect of tumor-intrinsic NAT10 on tumor immunology, we first genetically deleted the Nat10 gene in TC1 and MCA205 cells using CRISPR/Cas9 technology with Nat10-specific sgRNA (sgNAT10) pairs, while Wild-type (WT) cells transfected with an empty vector were used as controls (Fig. 2A, S2A). Syngeneic tumor models were then established in C57BL/6 N mice and immunodeficient nude mice by transplanting either sgNAT10 or WT tumor cells. All immunocompetent C57BL/6 N mice bearing sgNAT10 or WT tumor cells developed apparent tumors after transplantation, whereas the tumor masses rapidly disappeared in the mice bearing sgNAT10 tumor cells (Fig. 2A, B). However, in immunodeficient nude mice, both WT and sgNAT10 groups developed large tumors while no tumor disappeared during the study period, with sgNAT10 group bearing smaller tumor weight and size (Fig. 2C, D). These results suggest that NAT10 might inhibit tumor growth in vivo in an immune-dependent manner. As sgNAT10 group exhibited slightly smaller tumors compared to WT group in nude mice, we speculated that NAT10 deficiency might have additional tumor-suppressive effects except for strengthening immune responses. Thus, we performed a colony formation assay and CCK8 assay, and the results showed that NAT10 deficiency significantly hindered the colony formation and proliferation of tumor cells (Fig. S2B–E). Taken together, tumor-intrinsic NAT10 promotes tumor growth, at least in part, by facilitating tumor immune evasion.
Fig. 2. NAT10 deficiency suppresses tumor growth via immune-dependent mechanisms.
A, B Tumor growth curves for C57BL/6 N mice bearing TC1 (A) or MCA205 (B) tumor cells transplants. 2 × 106 WT or sgNAT10 tumor cells were subcutaneously transplanted into the back and flanks of C57BL/6 N mice (n = 5 mice per group), and tumor growth was monitored with calipers at the indicated time points. Tumor growth curves were analyzed by two-way ANOVA with the tumor size at the final day used for significance testing; ***P < 0.001; ***P < 0.001. C, D Tumor weight and growth curves for nude mice (n = 5 mice per group) inoculated with TC1 (C) or MCA205 tumor cells (D). 2 × 106 WT or sgNAT10 tumor cells were subcutaneously transplanted into nude mice that lacked mature T lymphocytes. Tumor growth was monitored at the indicated time points. Tumor growth curves were analyzed by two-way ANOVA with the tumor size at the final day used for significance testing; **P = 0.0021; ***P < 0.001; ***P = 0.0009; ***P < 0.001. E, F Kaplan-Meier survival curves for C57BL/6 N mice injected with TC1 (E) and MCA205 (F) tumor cells (n = 6 mice per group). 2 × 106 WT or sgNAT10 tumor cells were injected intravenously into C57BL/6 N mice, and the number of dead mice was recorded every day. Statistical significance was determined using Log-rank test; ***P = 0.0005 (E); ***P = 0.0006 (F). G, H Kaplan-Meier survival curves for nude/nude mice injected with TC1 (G) and MCA205 (H) tumor cells (n = 6 mice per group). 2 × 106 WT or sgNAT10 tumor cells were injected intravenously into C57BL/6 N mice, and the number of dead mice was recorded every day. Statistical significance was determined using Log-rank test. ns, no significance, P = 0.0524 (G); ***P = 0.0004 (H). I C57BL/6 N mice were subcutaneously immunized at the left flank with equal numbers of sgNAT10 tumor cells, freeze-thawed WT tumor cells, or PBS (control). Freezing and thawing were performed three times. Fourteen days after immunization, an equivalent number of live WT tumor cells were subcutaneously transplanted into the right flank of immunized mice. A schematic representation of the vaccination experiment with sgNAT10 tumor cells is provided in the left panel. Tumor growth was monitored at the specified time point points; n = 5 mice per group. Tumor growth curves were analyzed by two-way ANOVA with the tumor size at the final day used for significance testing; **P = 0.006; ***P = 0.0003; *P = 0.026; ***P < 0.0008. Unless specified otherwise, the data are presented as means ± SEM (error bar) and compared using the two-sided Student’s t test. Source data are provided as a Source Data file.
Moreover, we investigated whether NAT10 could affect survival via the host immune system. C57BL/6 N mice bearing sgNAT10 tumor cells exhibited significantly prolonged survival compared to mice with WT tumor cells (Fig. 2E, F). However, in nude mice, all mice died within 40 days (Fig. 2G, H). These findings further support that NAT10 inhibition may prolong mouse survival by activating the immune response.
To exclude the possible non-specific effects of the CRISPR/Cas9 system, we tried to restore NAT10 expression in sgNAT10 tumor cells. We first constructed a plasmid VP64-NAT10-GFP, in which the base sequence corresponding to the NAT10 sgRNA position was modified, while the encoded amino acids remained unchanged to avoid cleavage by the CRISPR-Cas9 enzyme (Fig. S2F, upper panel). Western blot results showed that NAT10 expression was well restored when transfected with VP64-NAT10 vector compared to the control vector group (Fig. S2F, middle panel). We further injected these tumor cells into C57BL/6 N mice. Compared to the disappeared tumors in sgNAT10 group on day 16, NAT10-restored group displayed much larger tumors, which were even larger than those of WT group (Fig. S2F, lower panel). These results validate the reliability of our CRISPR-Cas9 system and confirm the oncogenic effect of NAT10.
Immune memory is important for the anti-tumor immune responses. To assess the possible involvement of immune memory, we subcutaneously immunized C57BL/6 N mice with either live sgNAT10 tumor cells or freeze-thawed WT tumor cells on the left side, followed by re-challenging with comparable numbers of live WT tumor cells on the right side after 2 weeks (Fig. 2I, left panel). Intriguingly, immunization with live sgNAT10 tumor cells completely inhibited WT tumor growth on the right side, resulting in tumor-free mice, while immunization with freeze-thawed WT tumor cells showed a significantly weaker effect (Fig. 2I). We also tested whether T cell depletion in C57BL/6 N mice could abolish the preventive effects of NAT10-deficient tumor vaccines. In immunized mice, live WT tumor cells were inoculated while T cell function was blocked using CD8 antibody. The results showed that CD8+ T cell depletion in immunized C57BL/6 N mice completely abrogated the preventive effects of the NAT10-deficient tumor vaccine, as tumors developed on the left flank (Fig. S2G, H). These results further suggest that NAT10 deficiency in tumor cells suppresses tumor growth via eliciting immunological protection.
NAT10 deficiency in tumor cells triggers immune responses of CD8+ T cells in vivo
To explore the molecular mechanism of NAT10 deficiency in activating the immune response, we conducted transcriptomic RNA-sequencing (RNA-seq) analysis by using murine tumor tissues inoculated with sgNAT10 or WT tumor cells (Supplementary Data 2). Gene Set Enrichment Analysis (GSEA) revealed significant upregulation of “hallmark” signatures including “interferon-gamma (IFN-γ) response”, “interferon-alpha (IFN-α) response”, and “inflammatory response” in sgNAT10 tumor tissues. Heatmap analysis showed increased expression of T cells activation related chemokines in sgNAT10 tumor tissues, such as C-X-C motif chemokine ligand 9, 10, and 11 (CXCL9, 10, 11) (Fig. 3A). Moreover, our results showed that antigen presentation machinery (APM) and CD8+ Teff associated genes were significantly upregulated (Fig. 3B, C). Therefore, these results indicate that NAT10 deficiency might be involved in regulating CD8+ T cells associated immune response.
Fig. 3. NAT10 deficiency triggers immune-response signaling and induces cellular immune responses in vivo.
A Gene Set Enrichment Analysis (GSEA) was conducted on the differentially expressed genes between WT and sgNAT10 TC1 tumor tissues (n = 3 biologically independent samples). Three positively regulated ‘hallmark’ signatures were identified: interferon-alpha response, interferon-gamma response, and inflammatory response (left panel). The gene list was ranked based on the signed likelihood ratio (from log2 fold change [log2FC]) comparing sgNAT10 tumors versus WT TC1 tumors (right panel). B Heatmaps illustrating core biological pathways, such as antigen presentation machinery (APM) and CD8+ T effector cells (Teff), and depicting gene expression (color-coded by log2FC) in columns. C Heatmaps depicting biological pathways related to cell cycle and illustrating gene expression (color-coded by log2FC) in columns. D Multichannel imaging and image analysis were employed to investigate immune cell infiltration in the tumor microenvironment. C57BL/6 N mice were subcutaneously transplanted with either WT or sgNAT10 TC1 tumor cells. On day 8, tumor tissues were subjected to a six-color immunofluorescence analysis. The arrow indicates CD8+ T cells, DCs and Treg cells; n = 3 biologically independent samples; ***P < 0.0001. E C57BL/6 N mice (n = 5 mice per group) were subcutaneously inoculated with 2×10^6 WT or sgNAT10 TC1 tumor cells. They were intravenously administered with 200 µg of anti-CD8 antibodies per mouse on days -1, 3, and 5. Red arrows indicate the time points of anti-CD8 antibody injections. Tumor growth was monitored at specified time points, starting on day 0. Tumor growth curves were analyzed by two-way ANOVA with the tumor size at the final day used for significance testing; ***P = 0.0002. F, G Flow cytometry was used to analyze the proportions of major immune cell populations in TC1 (F) and MCA205 (G) tumor tissues (n = 5 mice per group). Tumor tissues from C57BL/6 N mice, transplanted as described in (E), underwent flow cytometry to identify IFN-γ+CD8+ T and GZMB+CD8+ T cells; ***P = 0.0005; *P = 0.0103; *P = 0.0124; **P = 0.0015. H mRNA expression levels of CD8a, IFN-γ, GZMA, GZMB, Cxcl19, and Cxcl10 genes were analyzed using RT-qPCR in TC1 (left panel) and MCA205 (right panel) tumor tissue. Tumor tissues from C57BL/6 N mice transplanted as described in (E) underwent RT-qPCR analysis. Data are presented as fold changes relative to WT tumor (n = 5 mice per group). From left to right, *P = 0.03; ***P < 0.001; ***P < 0.001; ***P < 0.001; *P = 0.038; **P = 0.0077; **P = 0.0036; **P = 0.0069; ***P < 0.001; *P = 0.0284; *P = 0.049; *P = 0.0283; respectively. I ELISpot assay was conducted to measure IFN-γ secretion in TC1 (left panel) and MCA205 (right panel) tumors. Tumor tissues from C57BL/6 N mice (n = 5 mice per group) transplanted as described in (E) underwent ELISpot analysis. The number of spots was quantified using an ELISpot reader. The results are expressed as spot-forming units (SFU); **P = 0.0018 (left); ***P = 0.0003 (right). J, K FACS analysis was performed to assess the proliferation of CD8+ (J) and CD4+ (K) T cells co-cultured with TC1 and MCA205 tumor cells, with or without NAT10 deficiency. The percentage of proliferating (CFSE-low) cells among all labeled CD4+ or CD8+ T cells is shown on the right of (J) and (K) (n = 3 biologically independent samples). From left to right, ***P < 0.001; ***P < 0.001; **P = 0.0057; **P = 0.0018; respectively. Unless specified otherwise, the data are presented as means ± SEM (error bar) and compared using the two-sided Student’s t test. Source data are provided as a Source Data file.
Next, we further applied multi-color immunofluorescence experiments to detect the immune cells infiltration in tumor tissues. Our results showed that the frequency of CD8+ T cells and DCs were increased in sgNAT10 tumor tissues compared to WT tumor tissues, while no noticeable difference was observed for the frequency of Tregs (Fig. 3D). Considering the importance of CD8+ T cells on antitumor immunity, we further measured CD8+ T cell infiltration by immunofluorescence and flow cytometry assays, and the results also revealed an increased tumor-infiltrating CD8+ T cells in sgNAT10 tumor tissues (Fig. S3A, B). We next performed CD8+ T-depleted murine experiments to confirm the above phenomenon. We first depleted CD8+ T cells in receipt mice using an anti-CD8 antibody or an IgG control, and then injected sgNAT10 or WT tumor cells. The results showed that depletion of CD8+ T cells slightly promoted tumor progression in mice bearing WT tumor cells compared to the IgG-treated controls. More importantly, compared to the lower sgNAT10 tumor burden in mice treated with IgG antibody, sgNAT10 tumor size was significantly increased in CD8+ T-depleted mice (Fig. 3E). The observed significant difference in tumor size between WT and sgNAT10 tumors in CD8+ T cell-depleted mice could be attributed to the intrinsic proliferative capabilities of NAT10.
Furthermore, we determined CD8+ T cell function in tumor tissues, and the results showed that IFN-γ and granzyme B (GZMB) levels in tumor-infiltrating CD8+ T cells were elevated in the sgNAT10 tumor group compared to WT tumor group (Fig. 3F, G). We also measured inguinal lymph nodes, critical for anti-tumor immunity, to investigate the activation of CD8+ T cells. Our results demonstrated the significant upregulation of IFN-γ+CD8+ T cells in the NAT10-deficient group (Fig. S3C, D). Additionally, gene expression analysis using the real-time PCR confirmed the apparent upregulation of CD8a, IFN-γ, granzyme A (GZMA), GZMB, CXCL9, and CXCL10 in the sgNAT10 group (Fig. 3H). IFN-γ ELISpot assay indicated that IFN-γ secretion was increased in the sgNAT10 tumor tissues (Fig. 3I). Moreover, T-cell proliferation assay revealed the enhanced proliferation of both CD4+ and CD8+ T cells in NAT10-deficient tumor tissues (Fig. 3J, K and S3E). We performed additional analyses using flow cytometry to characterize various immune cell populations, including DCs, NK cells, neutrophils, monocytes, Th1 cells, Th17 cells, and Treg cells (Fig. S3F–L). Although some immune cell populations in the sgNAT10 group demonstrated slightly increased infiltration compared to the WT group, these differences, with the exception of DCs, did not reach statistical significance. Collectively, these findings suggest that NAT10 deficiency in tumor cells activated the adaptive immune response in vivo, mainly through CD8+ T cell-mediated antitumor immunity.
NAT10 deficiency induces the type I interferon (IFN-I) response in tumor cells
The above results showed an enhanced IFN response and increased tumor-infiltrating lymphocytes (TIL) in the NAT10-deficient tumor microenvironment, suggesting a possible link between IFN-mediated chemokines secreted by tumor cells and TIL infiltration. To validate this hypothesis, RNA-seq was performed with total mRNA extracted from sgNAT10 and WT tumor cells (Supplementary Data 3 and 4). Utilizing the GSEA algorithm, we identified gene sets and pathways that were associated with “IFN-I” signaling in sgNAT10 tumor cells (Fig. S4A, B). Furthermore, NAT10 deletion induced the expression of genes involved in IFN-I response (Fig. 4A, B). We further applied RT-qPCR to detect the expression of IFN-stimulated genes (ISG), and the results confirmed a significant upregulation of ISGs in sgNAT10 tumor cells, including type I IFN gene Ifnb1 itself, transcription factor Stat1, antiviral gene Mx2, pattern recognition receptor genes Tlr3 and Ddx58, antigen presentation-related gene Tap1, and chemokine-encoding genes Ccl5 and Ccl7 (Fig. 4C, D). These results suggest the involvement of IFN-I response in NAT10 deficiency-induced tumor suppression.
Fig. 4. NAT10 deficiency induces IFN-I responses in tumor cells.
A, B GSEA was conducted on DEGs between the WT group and the sgNAT10 group of TC1 (A) and MCA205 (B) tumor cells (n = 3 biologically independent samples). Two positively regulated ‘hallmark’ signatures were identified: interferon-alpha response and interferon-gamma response. The heatmaps of the gene list of ‘hallmark’ signatures are shown on the right. C, D The mRNA expression levels of Ifnb, Stat1, Tlr3, Ddx58, Ccl5, Ccl7, Tap1, and Mx2 in WT and sgNAT10 of TC1 (C) and MCA205 (D) tumor cells were detected by RT-qPCR, with normalization to GAPDH (n = 3 biologically independent samples). From left to right, *P = 0.028; ***P < 0.001; **P = 0.0034; ***P < 0.001; ***P < 0.001; ***P < 0.001; **P = 0.002; ***P < 0.001; ***P < 0.001; *P = 0.0213; ***P < 0.001; ***P < 0.001; ***P < 0.001; ***P < 0.001; respectively. E, F WT or sgNAT10 TC1 (E) and MCA205 (F) tumor cells were subcutaneously transplanted into ifnar-/- C57BL/6 N mice (n = 5 mice per group). From left to right, ***P < 0.001; ***P = 0.0003; ***P < 0.001; **P = 0.0058. The tumor weight and growth were monitored at the indicated time points. Unless specified otherwise, the data are presented as means ± SEM (error bar) and compared using the two-sided Student’s t test. Source data are provided as a Source Data file.
Therefore, we next investigate the effect of NAT10 deficiency-induced IFN-I response in tumor inhibition. We transplanted sgNAT10 tumor cells into C57BL/6 N mice with IFN-I receptor knockout (Ifnar1 KO) while using WT tumor cells as controls. Interestingly, sgNAT10 tumor cells developed apparent tumors in Ifnar1 KO mice, increased gradually and did not disappear during the experiment period. Moreover, WT tumor cells development much larger tumor compared to sgNAT10 tumor cells in Ifnar1 KO mice, which was consistent with the phenomenon of IFNAR1 antibodies blockage (Fig. 4E, F and S4C). These findings suggest that the antitumor immune effects of NAT10 deficiency are dependent on the IFN-I response.
NAT10 enhances mRNA stability of Myc via ac4C modification in tumor cells
Next, we explored the mechanism by which NAT10 deletion induced interferon production. Given that NAT10 is the only gene currently known to undergo mRNA ac4C acetylation modification, we conducted acetylation RIP-seq (acRIP-seq) analysis in WT and sgNAT10 cell lines using an ac4C-specific acetylation antibody (Supplementary Data 5). This approach aimed to preliminarily identify genes potentially involved in anti-tumor immune responses mediated by NAT10 deficiency. Acetylated mRNAs enriched in WT cells were significantly much more abundant compared to those in sgNAT10 cells. Sequential analysis of the ac4C peaks revealed a strong enrichment of the characteristic GAGGAGA motif at ac4C sites within mRNA (Fig. 5A). This finding suggested that NAT10 may mediate mRNA acetylation through binding to this specific nucleic acid motif. Further analysis showed that ac4C peaks predominantly occurred in coding sequences (CDS) and 3’untranslated regions (3’UTR) (Fig. 5B, C). As reported, the acetyltransferase NAT10 can enhance mRNA stability, and ac4C peaks in wobble sites can increase mRNA ac4C modification27. Therefore, to investigate the potential modified target genes, we further performed label-free quantitative proteomics and combined analysis with acRIP-seq data (Supplementary Data 6). Seven candidate genes (Phf2, Myc, Wwc2, Kmt2a, Gigyf1, Timeless, and Nufip2) were identified with decreased mRNA acetylation and reduced protein levels in sgNAT10 tumor cells (Fig. 5D, S5A).
Fig. 5. NAT10 regulates MYC protein expression.
A The highly enriched motif within ac4C peaks was analyzed using acRIPseq. B The proportion of ac4C peak distribution in the TSS, 5’UTR, start codon, stop codon, and 3’UTR regions across the entire set of mRNA transcripts. C Density distribution of ac4C peaks across mRNA transcripts. D Seven candidate genes, including Phf2, Myc, Wwc2, Kmt2a, Gigyf1, Timeless, and Nufip2, were identified using acRIP-seq and label-free quantitative proteomics. E The expression levels of MYC in sgNAT10 TC1 and MCA205 tumor cells were analyzed by Western blotting. F The peaks of myc in WT and sgNAT10 TC1 cells from acRIP-seq data were visualized using the IGV software. G Schematic representation of the positions of ac4C motifs in Myc mRNA (upper panel). The ac4C sites in the 3’UTR of Myc mRNA were mutated to eliminate ac4C sites as much as possible. The lower panel shows the schematic representation of the mutated 3’UTR of the pEZX-MT06 vector for studying the roles of ac4C in Myc mRNA stability. H Effect of NAT10 on pEZX-MT06-Myc reporter. TC1 tumor cells were cultured in 24-well plates and transfected with Lipofectamine 3000 reagent according to the manufacturer’s instructions. Specifically, 100 ng/well of pEZX-MT06-Myc and either 0, 150, or 300 ng/well of VP64-NAT10 or empty vector were co-transfected. Additionally, Renilla luciferase plasmids (30 ng/well) were co-transfected as a normalization control for transcription efficiency. Luciferase activity was measured 24 h after transfection. The results are presented as relative luciferase activity (luciferase activity normalized to Renilla activity); ***P < 0.001; ***P < 0.001. I Anti-NAT10 antibody-based RIP-PCR analysis of Myc mRNA in TC1 cells. J The mRNA levels of MYC were detected in sgNAT10 TC1 cells after treatment with Act-D. The statistical method used was two-way ANOVA; **P = 0.0023. Unless specified otherwise, the data are presented as means ± SEM (error bar). Source data are provided as a Source Data file.
The aforementioned experiments demonstrated that NAT10 not only regulates cell proliferation but also influences the tumor immune response. Among the 7 candidate genes, MYC, as an oncogenic transcription factor, has been linked to both cell proliferation and antitumor immunity28,29. We then performed Western blotting, and our results showed that NAT10 deficiency decreased the protein expression of MYC in tumor cells (Fig. 5E). To identify the key ac4C sites that regulate mRNA stability, we analyzed the acetylation peaks of Myc mRNA. AcRIP-seq data showed that the ac4C peaks were distributed in the CDS and 3’/5’UTR regions of Myc mRNA (Fig. 5F). Importantly, we found that the 3’UTR region of Myc mRNA contains a nucleic acid sequence consistent with typical GAGGAGA motifs (Fig. 5A), suggesting that this ac4C site might be more dynamic in regulating Myc mRNA stability. Subsequently, we constructed 3’UTR reporters containing WT or mutant Myc 3’UTR using the firefly luciferase reporter system (Fig. 5G). The dual-luciferase assay showed significantly attenuated fluorescence activity in the mut-3’UTR groups compared to WT-3’UTR groups, suggesting NAT10 mainly bound to Myc 3’UTR positions (Fig. 5H). Moreover, we used a plasmid containing full-length Myc cDNA with mutations in the 3’UTR to disrupt the NAT10 binding site, along with a plasmid carrying full-length WT Myc. Using anti-NAT10 antibody-based RIP-PCR with primers targeting the 3’UTR of MYC, we found that NAT10 could bind to WT MYC but not to the mutant with the 3’UTR mutation, indicating that NAT10 primarily binds to the 3’UTR (Fig. 5I). Furthermore, we applied mRNA stability assay with 5 µg/ml actinomycin D and found that the half-life of Myc mRNA was nearly 16 h in WT tumor cells, whereas it was significantly reduced in sgNAT10 tumor cells. These findings suggested that reduced ac4C enrichment was accompanied by increased destruction of Myc mRNA (Fig. 5J). We also performed Ribo-seq analysis to investigate whether NAT10 regulates the translation of MYC mRNA (Supplementary Data 7). Our findings showed a reduction in footprint levels on MYC mRNA (Fig. S5B), indicating an overall decrease in MYC protein expression. Taken together, these results suggest that NAT10 upregulates MYC protein expression by enhancing mRNA ac4C modification.
Considering the critical expression-regulating role of NAT10 for MYC, we verified whether NAT10 modulates anti-tumor immunity via MYC. Firstly, CRISPR/Cas9 technology utilizing MYC-specific sgRNA (sgMYC) pairs was employed to successfully knock out Myc gene in tumor cells (Fig. S5C). We then established syngeneic tumor models in C57BL/6 N mice transplanted with either WT or sgMYC tumor cells to evaluate whether tumor-intrinsic MYC deficiency inhibits tumor growth by inducing the immune response. Our results showed that sgMYC tumor cells developed much smaller tumor masses compared with WT tumor cells in C57BL/6 N mice (Fig. S5C). Since we mainly focus on the host immune response, we further determined the CD8+ T cell immune infiltration by flow cytometry and found a significantly higher percentage of CD8+ T cells in sgMYC tumor tissues compared to WT tumors (Fig. S5D), suggesting that adaptive immunity is partly involved in MYC deficiency-induced tumor regression. Furthermore, we rescued Myc (VP64-Myc) in sgNAT10 tumor cells and subsequently transplanted these cells into C57BL/6 N mice. The results showed that restoration of MYC protein by Myc overexpression in sgNAT10 tumor cells significantly attenuated NAT10-deficiency-induced tumor suppression, similar to the effect of WT tumor cells (Fig. S5E). Moreover, Myc overexpression in sgNAT10 cells significantly attenuated NAT10 deficiency-induced IFN-γ secretion, the key cytokine for immune response, causing its level near to WT cells as determined by ELIspot assay (Fig. S5F). These findings suggest that inhibition of tumor-intrinsic NAT10 promotes anti-tumor immune response by down-regulating MYC expression.
NAT10 depletion induces dsRNA-mediated RIG-I-dependent IFN-I signaling via the MYC/CDK2/DNMT1 pathway
We further investigated the molecular mechanism by which NAT10 deficiency promoted anti-tumor immunity via downregulating of MYC protein levels. As a well-established oncogene, MYC regulates numerous genes involved in various cellular processes. To explore this broader regulatory network, RNA-seq analysis on sgMyc TC1 cells were performed and revealed significant transcriptional changes, with 767 genes upregulated and 851 genes downregulated (Supplementary Data 8). To screen downstream key target genes, we reanalyzed RNA-seq data from NAT10-deficient cells to identify downregulated genes and performed a cross-analysis with RNA-seq data from MYC-deficient cells. CDK2 emerged as a common target, highlighting its critical role as a downstream effector of the NAT10 and Myc (Fig. S6A). CDK2, a member of the cyclin-dependent kinase family30, was the most significantly down-regulated gene in NAT10-deficient cells (Fig. 6A, B). As CDK2 expression has been reported to be directly regulated by MYC31, we further determined the CDK2 expression in sgMYC tumor cells. Western blotting showed that MYC deletion significantly inhibited the expression of CDK2 in tumor cells (Fig. S6C). Moreover, we used CRISPR/Cas9 to knockout CDK2 (sgCDK2) in tumor cells and the sgCDK2 cells were then transplanted into C57BL/6 N mice. The results showed that sgCDK2 tumor cells developed significantly smaller tumors in mice compared to WT tumor cells, indicating that CDK2 deficiency suppressed tumor growth, which was consistent with the tumor-inhibitory effect of NAT10 deficiency (Fig. S6D). Additionally, CDK2 deficiency in tumor cells significantly enhanced CD8+ T cell infiltration and IFN-γ secretion in tumor tissues (Fig. S6E, F). These findings suggest that NAT10 deficiency enhances the antitumor immune response via inhibiting MYC-regulated CDK2 expression.
Fig. 6. Depletion of NAT10 induces dsRNA-mediated RIG-I-dependent signaling through the Myc/CDK2/DNMT1 pathway.
A The FPKM of individual genes of the CDK family from RNAseq data originating from WT and sgNAT10 TC1 tumor cells. Heatmap depicting the CDKs and illustrating gene expression (color-coded by log2FC) (n = 3 biologically independent samples); **P = 0.0065. B The FPKM of individual genes of the DNMT family from RNAseq data originating from WT and sgNAT10 TC1 tumor cells. Heatmap depicting DNMTs and illustrating gene expression (color-coded by log2FC) (n = 3 biologically independent samples); ***P = 0.0004. C Western blotting of DNMT1, CDK2, and NAT10 in matched WT, sgNAT10 TC1 (upper panel), and MCA205 (lower panel) tumor cells. β-actin was used as a loading control. D Representative immunofluorescence staining of dsRNA in WT, sgNAT10, sgCDK2, sgNAT10 Myc-rescued, and sgNAT10 Cdk2-rescued TC1 tumor cells was detected using confocal microscopy. Antibody J2 targeted dsRNA (labeled in red). The corresponding statistical diagrams are shown on the right. Statistical analysis was conducted using one-way ANOVA (n = 15 biologically independent samples); ***P < 0.001; **P = 0.0055. E Representative immunofluorescence staining of dsRNA in WT, sgNAT10, sgCDK2, sgNAT10 Myc-rescued, and sgNAT10 Cdk2-rescued MCA205 tumor cells was detected using confocal microscopy. The corresponding statistical diagrams are shown on the right. Statistical analysis was conducted using one-way ANOVA (n = 15 biologically independent samples); ***P < 0.001; **P = 0.0057. F Protein expression levels of NAT10 and RIG-I in vector, sgNAT10, and sgNAT10/RIG-I TC1 tumor cells determined by Western blotting (upper panel). mRNA expression levels of Ifnb1, Stat1, Tlr3, Ddx58, Ccl5, Ccl7, Tap1, and Mx2 by RT-PCR in vector, sgNAT10, and sgNAT10/RIG-I TC1 tumor cells (n = 3 biologically independent samples). From left to right, all *** indicate P < 0.001; *P = 0.0451. Unless specified otherwise, the data are presented as means ± SEM (error bar) and compared using the two-sided Student’s t test. Source data are provided as a Source Data file.
Next, we explored the signaling pathway of CDK2 on NAT10 deficiency-induced IFN-I responses. As CDK2-deficient tumor cells have shown DNA methyltransferases (DNMT) family-inhibitory activity and loss of DNMT activity can induce IFN-I responses32, we then reanalyzed our RNA-seq data of sgNAT10 tumor cells. The results showed that, among the DNMT family, DNMT1 had the highest expression in tumor cells, and its expression significantly decreased following NAT10 deletion, which was consistent with the downregulation of CDK2 (Fig. 6B, S6G). Western blotting results also verified that the protein levels of CDK2 and DNMT1 were significantly reduced in NAT10-deletion murine tumor cells and human lung cancer A549 tumor cells compared to WT tumor cells (Fig. 6C, S6H). Furthermore, when we successfully rescued CDK2 in sgNAT10 tumor cells, DNMT1 expression was also restored to high level (Fig. S6I). More importantly, CDK2 overexpression in sgNAT10 tumor cells resulted in the development of detectable tumor masses, which did not regress during the experiment period, though they were smaller in weight than tumors formed by WT cells (Fig. S6I). In addition, correlation analysis between NAT10 and several downstream genes was performed in the GEPIA website23. The results revealed a significant and positive correlation between NAT10 and MYC, NAT10 and CDK2, NAT10 and DNMT1, or CDK2 and DNMT1 (Fig. S6J). Collectively, all these data demonstrate that NAT10 deficiency suppresses tumor by inhibiting MYC/CDK2/DNMT1 pathway.
We further explored the mechanism of NAT10-regulated MYC/CDK2/DNMT1 in tumor-intrinsic IFN-I response. As DNMT1 inhibition has been proven to trigger IFN-I response by inducing dsRNA formation13,33,34, we quantified cytoplasmic dsRNA using dsRNA-specific J2 antibody. The result showed a significantly higher abundance of dsRNA in sgNAT10 or sgCDK2 cells compared to that in WT cells. Restoration of MYC or CDK2 significantly reduced NAT10 deficiency-induced cytoplasmic dsRNA production (Fig. 6D, E). Therefore, cytoplasmic dsRNA formation could be regulated by NAT10 and its downstream genes.
How did dsRNA induce the production of type I IFN? It has been reported that dsRNA can be recognized by the cytoplasmic RNA sensors RIG-I and MDA-5, thereby triggering the IFN-I response35. GO enrichment analysis revealed that “RIG-I-like receptor signaling pathways” were significantly upregulated in sgNAT10 tumor cells compared to WT cells (Fig. S6K–L). Therefore, we further silenced RIG-I in sgNAT10 tumor cells and assessed the activation of the IFN-I signaling pathway. Our results showed that deletion of RIG-I partly down-regulated the high expression of ISGs induced by NAT10 deficiency (Fig. 6F). Taken together, our results demonstrate that NAT10 deficiency activates IFN-I signaling pathway via RIG-I-mediated signaling in response to cytoplasmic dsRNA.
Inhibition of NAT10 enhances the therapeutic effect of ICIs
Our findings demonstrate that NAT10 inhibition enhances the tumor-intrinsic IFN-I response through RIG-I-mediated cytoplasmic dsRNA sensing. This leads to the upregulation of numerous ISGs, which facilitates CD8+ T-cell infiltration and activation. Previous studies have reported that activation of the IFN-I pathway and enhancing T cell infiltration could promote the therapeutic effect of ICIs36,37. Therefore, we next investigated whether inhibition of NAT10 could also promote the efficacy of ICIs by using PD-1 inhibitor, a typical ICI, in mice models. Generally, mice were orally given Remodelin once a day for 7 consecutive days. On day 8, mice were treated with anti-PD-1 mAb (10 mg/kg) or isotype control (vehicle) twice a week for 2 weeks. A humane endpoint was reached on day 29 (Fig. 7A). The results showed that either PD-1 mAb or Remodelin significantly inhibited tumor growth compared to the control group. More importantly, the group that received combination therapy bore the smallest tumor size compared to the single PD-1 mAb or Remodelin group (Fig. 7B, C), suggesting that combined inhibition of NAT10 and PD-1 can synergistically suppress tumor growth in vivo.
Fig. 7. Inhibition of NAT10 enhances the response to ICIs.
A Schematic diagram showing combination therapy of C57BL/6 N mice with Remodelin and ICIs. B, C Tumor weight for C57BL/6 N mice (n = 5 mice per group) inoculated with TC1 (B) or MCA205 (C) tumor cells treated with Remodelin and/or anti-PD-1 antibodies. TC1 or MCA205 tumor cells were inoculated subcutaneously into C57BL/6 N mice. Mice received Remodelin or saline via oral gavage for the first 7 days at a dose of 100 mg/kg. On day 8, mice were treated with IgG control or anti-PD-1 antibodies. After sacrificing the mice, tumor tissues were excised and representative images are shown in the left panel. Statistical significance was determined using One-way ANOVA; from left to right, all *** indicate P < 0.001. D, E Representative immunofluorescence staining of CD8+ T cells in TC1 (D) and MCA205 (E) tumor tissues. Tumor tissues from C57BL/6 N mice (B) were subjected to immunostaining for CD8+ T cells (red) and nuclei (blue). CD8+ T cells were quantified by counting positive signals in 3 randomly selected fields (20×) per tumor section using Image J. Statistical analysis was conducted using One-way ANOVA, n = 15 biologically independent samples. Scale bar, 100 µm; from left to right, all *** indicate P < 0.001. F, G FACS analysis of the proportions of IFN-γ+CD8+ immune cell populations in TC1 (F) and MCA205 (G) tumor tissues. Tumor tissues from C57BL/6 N mice (B) were subjected to FACS analysis for IFN-γ+CD8+ immune cells (n = 5 mice per group). Statistical significance was determined using One-way ANOVA; from left to right, ** P = 0.0027; ** P = 0.0063; ***P < 0.001; *** P < 0.001. H, I ELISpot assay was conducted to measure IFN-γ secretion in TC1 (H) and MCA205 (I) tumor tissues after different treatments. The number of spots and the results were quantified (n = 5 mice per group). Statistical significance was determined using One-way ANOVA; from left to right, ** P = 0.0039; ***P < 0.001; ** P = 0.0011; *** P = 0.0002. Unless specified otherwise, the data are presented as means ± SEM (error bar). Source data are provided as a Source Data file.
Furthermore, tumor tissue immunofluorescence assay showed that the frequency of tumor-infiltrating CD8+ T cells was significantly increased in PD-1mAb or Remodelin group compared to controls, with the highest frequency observed in the combination group (Fig. 7D, E). Moreover, a remarkable increase was observed in the frequency of IFN-γ-positive CD8+ T cells in both the PD-1 mAb and Remodelin single treatment group, with the effects being significantly enhanced in the combination treatment group (Fig. 7F, G). We also tested the secretion of IFN-γ in tumor tissues and found that the combination treatment resulted in significantly more IFN-γ secretion compared to either single treatment (Fig. 7H, I). Considering the clinical setting, we further analyzed the relationship between NAT10 and PD-L1 in the tumor tissues from lung cancer patients. Our results suggested that samples with high NAT10 expression were accompanied by elevated PD-L1 expression levels (Fig. S7A). Overall, these data suggest that inhibition of NAT10 significantly strengthens the therapeutic effect of ICIs on tumors.
Intratumoral delivery of siNAT10-lipid nanoparticles (LNP) induces immune response and improves tumor immunotherapy
Our above findings demonstrate that inhibition of NAT10 could enhance the therapeutic effect on tumor, indicating that NAT10 may serve as a potentially effective target for cancer treatment. While Remodelin is convenient and well-studied, its efficacy is limited in vivo due to poor absorption. In this view, we developed two efficiently delivery systems, SM102 and PEI/PC7A nanoparticles, to inhibit NAT1038. SM102 nanoparticle, a cationic amino lipid approved for mRNA delivery in the Moderna COVID-19 vaccine, functions as LNPs for RNAi-based therapeutics, and offers a safer alternative with minimal off-target effects39. PEI/PC7A nanoparticle, composed of polyethyleneimine (PEI) and a pH-responsive PC7A polymer, was developed for highly-efficient siRNA transfection40. We first determined the particle sizes of the two siNAT10 LNPs by using dynamic light scattering (DLS, Malvern), and we confirmed that their sizes were approximately 160 nm (Fig. 8A, S8A). Then, confocal laser scanning microscopy (CLSM) was applied to observe the entry of nanoparticles into tumor cells. We found that siRNAs nanoparticles (shown as green fluorescence) overlapped with lysosomes (shown as red fluorescence) within transfection of 4 h, indicating that nanoparticles successfully entered into tumor cells. Moreover, a significantly abundant amount of green fluorescence (siRNAs nanoparticles) was observed outside the lysosomes, indicating siRNA successfully escaped from lysosomes and released into cytosol (Fig. 8B, S8B). Therefore, successful release of siRNA from lysosomes indicated the formation of RNA-induced silencing complex in the cytosol.
Fig. 8. Intratumoral delivery of PEI/PC7A/siNAT10 nanoparticles for cancer immunotherapy.
A The diagram of PEI/PC7A and its size distribution. B Confocal image showing the uptake of PEI/PC7A. TC1 tumor cells were cultured in chamber slides overnight, and then added with 20 nM FAM-labeled siRNA for 4 h. Cells were stained with 50 nM Lyso-Tracker Red (Beyotime, catalog C1046) and 10 µg/mL Hoechst (Beyotime, catalog C1022) for 30 min. Immunofluorescence images were acquired using a Nikon A1 fluorescence microscope. C The mRNA expression levels of NAT10 were measured using RT-PCR in TC1 tumor cells with or without siRNA. 2×105 TC1 tumor cells were seeded in 12-well plates overnight. The medium was then replaced with Opti-MEM, and PEI/PC7A was added with a final siRNA concentration of 20 nM (n = 3 biologically independent samples). The data are compared using the two-sided Student’s t test; **P = 0.006. D Protein expression levels of NAT10 were determined by Western blotting in TC1 (left panel) and MCA205 (right panel) tumor cells treated with or without PEI/PC7A/siNAT10 nanoparticles. E Tumor weight for C57BL/6 N mice (n = 5 mice per group) inoculated with TC1 tumor cells treated with Remodelin or PEI/PC7A/siNAT10 nanoparticles. TC1 tumor cells were inoculated subcutaneously into C57BL/6 N mice. Mice received Remodelin via oral gavage for the first 7 days at a dose of 100 mg/kg. PEI/PC7A containing siRNA (5 nmol/kg) was dissolved in PBS and injected into the tumor on days 4, 7, and 9. Tumor tissues were harvested after sacrificing the mice. Representative images are shown in the left panel. Statistical significance was determined using One-way ANOVA; from left to right, ** P = 0.0006; *P = 0.0183. F Tumor weight for C57BL/6 N mice (n = 5 mice per group) inoculated with TC1 tumor cells treated with PEI/PC7A/siRNA nanoparticles and/or anti-PD-1 antibodies. TC1 tumor cells were inoculated subcutaneously into C57BL/6 N mice. PEI/PC7A/siNAT10 nanoparticles or saline were injected into the tumor on days 4, 7, and 9. On day 8, mice were treated with IgG control or anti-PD-1 antibodies. Statistical significance was determined using One-way ANOVA; from left to right, *** P < 0.001; **P = 0.0092. G Representative immunofluorescence staining of CD8+ T cells in TC1 tumor tissues. Tumor tissues from C57BL/6 N mice (F) were subjected to immunostaining analysis for CD8+ T cells (red) and nucleus (blue). CD8+ T cells were quantified by counting positive signals in 3 randomly selected fields (20×) per tumor section using Image J. Statistical analysis was conducted using One-way ANOVA, Scale bar, 100 µm; n = 15 biologically independent samples. Statistical significance was determined using One-way ANOVA; from left to right, *** P < 0.001; ***P = 0.0007. H FACS analysis of the proportions of IFN-γ+CD8+ immune cells in TC1 tumor tissues. Tumor tissues from C57BL/6 N mice (F) were subjected to FACS analysis for IFN-γ+CD8+ immune cell populations (n = 5 mice per group). Statistical significance was determined using One-way ANOVA; from left to right, *** P = 0.0007; **P = 0.0046. I Diagram illustrating how tumor-intrinsic NAT10 orchestrates immune evasion and regulates antitumor immunity. Tumor-intrinsic NAT10 directly acetylated Myc mRNA, enhancing Myc transcription and subsequently promoting CDK2 expression, which in turn upregulates DNMT1 and drives cell proliferation. However, inhibition of NAT10 downregulates the MYC/CDK2/DNMT1 pathway, leading to increased formation of dsRNA and triggering RIG-I-mediated IFN-I response. This activation of the innate immune response enhances CD8+ T cell mediated antitumor immunity, offering a potential therapeutic avenue for boosting immune surveillance in cancer. Unless specified otherwise, the data are presented as means ± SEM (error bar). Source data are provided as a Source Data file.
Next, RT-qPCR analysis was applied to assess the inhibitory effect of nanoparticles on NAT10 expression. The results demonstrated a significant reduction in NAT10 mRNA expression levels with both SM102 and PEI/PC7A/siNAT10 nanoparticles, compared to siNAT10 transfection alone (Fig. 8C, S8C). Furthermore, our in vivo experiments revealed that PEI/PC7A/siNAT10 nanoparticles inhibited tumor growth more effectively than SM102 (Fig. S8D, E). Consequently, we employed PEI/PC7A/siNAT10 nanoparticles to evaluate its tumor inhibitory effect for the following experiments.
Western blot analysis confirmed that PEI/PC7A/siNAT10 nanoparticles significantly suppressed the protein expression of NAT10 in vitro (Fig. 8D). In subsequent in vivo experiments, intratumoral delivery of PEI/PC7A/siNAT10 nanoparticles significantly reduced tumor growth in C57/BL6N mice, showing much better inhibitory effects compared to Remodelin (Fig. 8E). Next, we combined PEI/PC7A/siNAT10 nanoparticles with PD-1mAb, and observed both PD-1mAb and PEI/PC7A/siNAT10 nanoparticles effectively suppressed tumor growth. However, combination therapy resulted in substantially smaller tumors compared to either PD-1 mAb or PEI/PC7A/siNAT10 monotherapy (Fig. 8F). Furthermore, a significant increase in tumor-infiltrating CD8+ T cells was observed following administering PEI/PC7A/siNAT10 nanoparticles or combination therapy. Notably, the combination group exhibited a markedly higher abundance of CD8+ T cells within tumor microenvironment (Fig. 8G). Additionally, a notable increase in the abundance of IFN-γ-positive, active CD8+ T cells was observed in the PEI/PC7A/siNAT10 nanoparticle group, with combination therapy further enhancing this effect (Fig. 8H). Overall, these findings suggest that NAT10 inhibition by nanoparticles enhances the antitumor efficacy of ICIs by promoting tumor immune responses.
Discussion
Over the past few decades, researchers have increasingly focused on the epigenetic regulation of DNA-templated processes. Both DNA and RNA epigenetic modification play pivotal roles in modulating a wide array of biological processes that are crucial for cancer development, progression and acquisition of drug resistance41,42. Dysregulation of the epigenome drives aberrant transcriptional programs in human cancers and represents a promising target for tumor treatment. Epigenetic modifications have also been shown to influence tumor immunogenicity and the function of immune cells involved in antitumor responses43–45. Thus, the development and application of epigenetic therapies, particularly in combination with cancer immunotherapy, holds significant potential for improving cancer treatment outcomes46. Here, we identified a pivotal role for NAT10-mediated mRNA ac4C modification in tumor immune evasion. Our finding showed that tumor-intrinsic NAT10 not only promotes tumor cell proliferation but also suppresses the antitumor effector function of CD8+ T cells within the tumor microenvironment. Specifically, our approach leverages NAT10 inhibition to reduce Myc expression, thereby downregulating the MYC/CDK2/DNMT1 pathway and inhibiting tumor growth. This disruption leads to increased formation of dsRNA, which cross-primes T cells through a RIG-I-mediated IFN-I response (Fig. 8I). Moreover, when combined with PD-1 blockage, NAT10 inhibition via PEI/PC7A/siRNA nanoparticles significantly enhances antitumor immunity, providing a compelling strategy for improving cancer treatment outcomes. These results underscore the potential of combining epigenetic therapies with ICIs to achieve more effective therapeutic responses.
In this study, we demonstrated that NAT10 has a pro-tumor effect, which is partly mediated by suppressing immunity. Specifically, there was no significant difference in tumor size or weight between NAT-10 specific inhibitor Remodelin-treated group and the control group in immunodeficient mice. This outcome contrasts with the significant reduction in tumor growth observed in immunocompetent mice. Similarly, when we established tumor model by transplanting sgNAT10 tumor cells, the tumor masses rapidly disappeared in the mice bearing sgNAT10 tumor cells in immunocompetent mice. In contrast, no tumor regression was observed in immunodeficient nude mice, where the sgNAT10 group exhibited smaller tumor weight and size throughout the study period. Notably, there were inconsistencies between the in vitro and in vivo effects of NAT10 inhibition. For instance, the NAT10 inhibitor Remodelin suppressed clonogenicity of murine tumor cell lines in vitro, yet it only inhibited tumor growth in immunocompetent mice, not in immunocompromised mice. This disparity can be attributed to the different concentrations of Remodelin used in the two settings. In vitro, higher concentrations of remodelin were employed, which significantly suppressed clonogenicity in murine tumor cell lines. In contrast, in vivo experiments were conducted with a low dose of Remodelin, primarily aimed at investigating its immunomodulatory effects, which resulted in limited cytotoxicity. This explains why Remodelin effectively inhibited tumor growth in immunocompetent mice through immune activation, but showed no significant effect in immunocompromised mice. This observation aligns with previous study, such as those reporting that low-dose HDAC inhibitors activate immune responses in immunocompetent mice but show little efficacy in immunodeficient models47.
Our RNA-seq analysis of NAT10-deficient tumor tissue revealed significant upregulation of type I IFN signaling pathways, as well as increased expression chemokines related to T cell activation, such as CXCL9, CXCL10, and CXCL11. Type I IFNs are known to enhance DC antigen presentation and activate adaptive immunity48. Consistent with these findings, we observed increased DC and CD8+ T cell populations in NAT10-deficient tumors, while no significant changes in NK cells, macrophages, Treg cells, neutrophils, monocytes, Th1 cells or Th17 cells. Furthermore, we observed that tumor size was significantly increased in CD8+ T cell-depleted mice bearing sgNAT10 tumors. Additionally, levels of IFN-γ and GZMB in tumor-infiltrating CD8+ T cells were elevated in the sgNAT10 tumor group compared to the WT tumor group. This suggests that NAT10 deficiency may enhance antigen presentation and cytotoxic T cell activation.
NAT10, the only known ac4C “writer” protein and a predominantly nuclear protein, contains a unique RNA cytosine acetyltransferase domain49. As expected, acRIP-seq revealed a significant decrease in enriched acetylated mRNAs following NAT10 deletion. Together with proteomics analyses in NAT10-deficient cells, 125 genes with reduced mRNA acetylation and 323 with decreased protein expression were identified. Among these, seven candidate genes- Phf2, Myc, Wwc2, Kmt2a, Gigyf1, Timeless, and Nufip2- showed reductions in both mRNA acetylation and protein levels. Given MYC plays a dual role in promoting cell proliferation and modulating antitumor immunity50, a phenomenon similar to the effect observed with NAT10 inhibition, we further focus on MYC as the primary target regulated by NAT10. Further study showed NAT10 binds to the 3’UTR of MYC as dual luciferase assay confirmed a significant reduction in fluorescence activity in the mut-3’UTR groups compared to WT-3’UTR groups at the MYC gene locus, which is further verified by anti-NAT10 antibody-based RIP-PCR. Since MYC as a well-known oncogene impacts a much broader range of genes and cellular processes, we then performed RNA-seq analysis on sgMyc TC1 cells and screen downstream key target genes. Cross-analysis between RNA-seq data from NAT10-deficient cells and RNA-seq data from MYC-deficient cells revealed 120 common target genes, including CDK2, Psd4, Nsg1, Smpd3, Psph, Edn1, and Batf3, and so on. As CDK2 has been reported to be directly regulated by MYC31 and similar to MYC, CDK2 plays a critical role in regulating cell cycle progression and modulating tumor immune responses51, we thereafter focus on CDK2 to explore the antitumor immunity mediated by NAT10 deletion-MYC axis. Western blot analysis further confirmed reduced CDK2 expression in both sgNAT10 and sgMYC cell lines. Future studies should also investigate the broader impacts of NAT10-MYC on other cellular processes.
As CDK2-deficient tumor cells have shown DNMTs family-inhibitory activity and loss of DNMTs activity can induce IFN-I responses32, we are curious if one member of DNMTs is the potential target of NAT-10/Myc/CDK2 axis. RNA-seq data and revealed DNMT1 had the highest expression among the DNMTs family in TC1 tumor cells and its expression significantly decreased following NAT10 deletion, which was consistent with the downregulation of CDK2. Research have revealed DNMT1 knockout can induce the expression of ERV genes, which can form dsRNA and contribute to the interferon response13,34. Abnormal accumulation of the endogenous dsRNA triggers an antitumor immune response through activation of dsRNA-sensing pathways52,53. MDA5 and RIG-I recognize specific features of dsRNA. Binding of either RIG-I or MDA5 to dsRNA drives induction of IFN signaling, which is critical for T cell priming7,54. In this study, NAT10 deficiency led to the accumulation of cytosolic dsRNA in tumor cells, which provided a substantial substrate for RIG-I signaling. This activation of RIG-I-mediated pathways contributed to the induction of IFN-I responses, enhancing antitumor immunity. It is reported that dsRNA is also responsible for epigenetic changes involving DNA methylation and histone modification55. Interestingly, RNA-seq showed significant downregulation of DNMT1 after NAT10 deficiency, confirming an association between NAT10 inhibition and DNA demethylation. DNMT1 is required to maintain CpG methylation, and its inhibitors induce ERV demethylation and expression helping trigger the dsRNA response13. Therefore, such changes in dsRNA are partly due to DNA demethylation induced by the down-regulation of DNMT1 after NAT10 inhibition.
Tumor-intrinsic IFN-I production triggered by radiotherapy or chemotherapy is mainly dependent on cytosolic dsDNA-mediated RIG-I signaling, which increases cancer immunogenicity and enhances the antitumor immune response, thereby improving therapeutic efficacy56. Immunotherapy has been hailed as a significant advance in cancer treatment. In particular, combination of PD-1/PD-L1 blockage and chemotherapy and/or radiotherapy has extremely prolonged the survival of tumor patients. However, its effectiveness heavily relies on immune cells infiltration, with patients who have low or no infiltration often showing poorer responses to treatment57. Therefore, enhancing the effectiveness of immunotherapy could greatly benefit these patients. Given NAT10 inhibition enhanced CD8+ T cells infiltration and the critical role of CD8+ T cells and checkpoint blockade in immunotherapy, we tested the combination of NAT10 inhibition (either molecule inhibitor or genetically knockout) and anti-PD1 therapy. This combination significantly suppressed tumor growth and enhanced the anti-tumor immune response compared to either treatment administered individually. Additionally, effective inhibition of NAT10 in vivo can enhance the anti-tumor immune response. Remodelin, an orally active and selective NAT10 inhibitor58, inhibits tumor growth by activating host immunity upon intragastric administration. However, its low oral bioavailability limited its efficacy. Lipid nanoparticles (LNPs), approved for siRNA and mRNA delivery, have been explored to improve RNA delivery59. Among the components of LNPs, ionizable lipids are pivotal for efficient RNA delivery. In this study, we developed two delivery systems: SM-102 and PEI/PC7A/siNAT10 nanoparticles. Our findings indicated that PEI/PC7A/siNAT10 effectively penetrates cell membranes, inhibits NAT10 expression, and suppresses tumor growth. Notably, PEI/PC7A/siNAT10 outperformed SM-102 and Remodelin. When combined with ICIs, PEI/PC7A/siNAT10 nanoparticles stimulated potent anti-tumor immunity, effectively suppressing tumor growth.
It is important to note that in our rescue experiments, it is quite surprising that overexpression of either Myc or CDK2 substantially restores the phenotypes (eg. cytoplasmic dsRNA production) associated with NAT10 deficiency back to those of the parental cells. The restoration of specific phenotypes observed in sgNAT10 cancer cells through Myc and CDK2 overexpression can be attributed to two key factors. First, NAT10 regulates the expression of these genes, and their reintroduction partly reverses the phenotypic alterations caused by NAT10 deficiency. Second, Myc and CDK2 inherently possess pro-proliferative and immunosuppressive properties, and their overexpression in sgNAT10 cancer cells further facilitates the restoration of these phenotypes. Although the MYC-CDK2 axis plays an important role, we acknowledge that it is not the only downstream mechanism through which NAT10 regulates the phenotype. There are likely other potentially significant downstream factors that warrant further exploration in future studies.
There are some limitations in this study. The initial purpose of our study is to investigate the effects and mechanisms of NAT10 inhibition in antitumor immunity. Therefore, we chose the cancers with the following characteristics: a “cold” tumor type with low immune cell infiltration, high tumorigenic potential, and poor therapeutic response. We initially selected lung cancer for our study, as it meets the relevant characteristics and is the most prevalent cancer type, and subsequently expanded our research to evaluate its relevance in other cancer types. Fibrosarcoma was selected as a model, given its classification as a “cold” tumor type. Given the limited availability of human sarcoma samples, our findings were primarily validated in lung cancer patients. Further research should investigate the function of NAT10 inhibitors in other cancer types, beyond lung cancer and fibrosarcoma.
Collectively, our work highlights the central role of the mRNA ac4C writer NAT10 in tumor immune evasion, specifically through modulation of CD8+ T cell activity. We demonstrate that PEI/PC7A/siNAT10 nanoparticles effectively block NAT10 activity both in vitro and in vivo. Our findings suggest that the impact of anti-cancer therapies on immune cells should be considered, and provide a foundation for evaluating NAT10 inhibition in combination with PD-1/PD-L1 blockade as a therapeutic strategy for “cold” tumors.
Methods
Analysis of tumor-infiltrating immune and prognostic model
We analyzed the patterns of immune cell infiltration according to the immune cell biomarker previously reported. The algorithm was operated with the R-package and the data were visualized with R package ggplot2. TCGA-LUAD (n = 590) were applied to illustrate the potential prognostic significance of NAT10. Patients were divided into high- or low-expression group based on the expression levels of NAT10. The CIBERSORT algorithms was used to calculate the proportion of immune cell infiltration of different groups. The diagram was drawn by using the ggplot2 package. Additionally, survival analysis was performed using R ‘survival’ package. The ggplot2 and survminer R packages were used to create survival curves between different groups. In addition, the ROC curve was generated using the R package survival ROC to detect the prognostic value for NAT10 expression.
Patients’ tumor tissues and clinical data
37 lung cancer samples collected from surgical patients were fixed with formalin and embedded with paraffin and then examined. These tumors represented a range of stages from I to IV, ensuring a comprehensive disease spectrum. Detailed patient information, including age, sex, smoking history, pathological classification, and tumor stage, has been provided in the supplementary data. Immunohistochemical (IHC) staining was performed to assess NAT10 expression in these samples. The staining was predominantly localized in the nucleus, with intensity levels correlating with tumor stage and degree of differentiation. The study was conducted according to the principles of the Declaration of Helsinki and approved by the Human Research Ethics Committee of Qilu Hospital of Shandong University with the ethic number of KYLL-202311-045. All ethical regulations relevant to human research participants were followed.
Cell culture
MCA205 fbrosarcoma cell line (SCC173) was purchased from Millipore Sigma. Human embryonic kidney 293 T cell line (CRL-3216), A549 human lung adenocarcinoma (CCL-185) and TC-1 lung epithelial tumor cell line (CRL-2785) were purchased from American Type culture collection (ATCC). They were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Gibco). Cultures were maintained in a humidified atmosphere containing 5% CO2 in 37°C. All cell lines were tested to confirm that they were free of mycoplasma and authenticated by short-tandem repeat analysis.
Construction of stable cell lines with CRISPR/Cas9 system
Deletion of NAT10, MYC, RIG-I or CDK2 was achieved using LentiCRISPR v2 (Addgene, Cambridge, MA, USA), which carried expression cassettes for Streptococcus pyogenes CRISPR-Cas9 and a chimeric guide RNA selected from the Guide Design Resources (http://crispr.mit.edu). HEK293 cells were co-transfected with three plasmids: pMD2.G (Addgene, catalog12259), psPAX2 (Addgene, catalog12260), and either LentiCRISPR v2 or a control vector, using Lipofectamine 3000 (Thermo Fisher Scientific) for 48 h. Viral stocks generated were used to infect target cells. Post-infection, cells were cultured in puromycin (4 µg/ml, InvivoGen, catalog ant-pr-1) for at least seven days. Monoclonal cells were collected using FACSAria III cell sorter (Becton Dickinson, San José, CA, USA) and plated in a 96-well plate. Sequences synthesized in this study are provided in Supplementary Table S1. NAT10 heterozygous knockout, MYC, CDK2 and RIG-I knockout, and control cell lines were further validated through Western blot analysis of NAT10 expression.
Overexpression vectors and transfection
To achieve overexpression of NAT10, MYC-HA, and CDK2-GFP, the dCAS9-VP64-GFP plasmid (Addgene, catalog 61422) was digested with BamH I (NEB, catalog R0136S) and Nhe I (NEB, catalog no. R0131), and the VP64 sequence was replaced with the cDNA sequences corresponding to the genes of interest. Subsequently, 293 T cells were transfected with dCAS9-VP64-GFP plasmid along with packaging plasmids psPAX2 (Addgene, plasmid catalog 12260) and envelope pMD2G (Addgene, catalog 12259) using Lipofectamine 3000 (Invitrogen, catalog no. L3000-015) according to the manufacturer’s instructions. After 48 h, lentivirus was harvested from the cell culture medium, followed by collection of lentiviral particles via centrifugation (3000 g/10 min) and filtration through a 0.45 µm sterile filter (Merck Millipore Ltd. PR05543). The collected lentivirus was stored at −80°C. Transfected cells were subsequently isolated using fluorescence-activated cell sorting (FACS).
Tumor models
Female C57BL/6 N background mice (strain code 213) and BALB/c athymic nude mice (nude/nude, strain code 403), aged 6–8 weeks, were purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China; license number SCXK [Jing] 2021-0011). Mice were housed in a specific pathogen-free (SPF) facility under controlled conditions with a 12-hour light/dark cycle (lights on at 7:00 a.m. and off at 7:00 p.m.), ambient temperature maintained at 22 ± 2°C, and relative humidity of 50% ± 10%. Food and water were provided ad libitum. All animal experiments were approved by the Institutional Animal Care and Use Committee of Qilu Hospital of Shandong University (DWLL-2024-176). All mice used in this study were age-matched littermates. The number of mice in each experiment was indicated in the corresponding figure legend. All animals were handled strictly according to the Principles for the Utilization and Care of Vertebrate Animals and the Guide for the Care and Use of Laboratory Animals.
Following grouping, cells (2 × 10^6 cells per mouse) were subcutaneously implanted. Tumor dimensions were measured daily using vernier calipers, and tumor size was calculated by multiplying the length by the width. After 9–13 days, tumors were harvested for RNA sequencing, flow cytometric analysis, tissue immunofluorescence staining, and ELISpot analysis. Survival analysis involved intravenous injection of cancer cells, was conducted with daily recording of mouse mortality. In the CD8+ T cell and PD-1 blocking assay, anti-CD8 antibodies (10 mg/kg, BioXCell, catalog BE0004-1) and PD-1 antibodies (10 mg/kg, BioXCell, catalog CP162) were intravenously injected at the specified time point. In the IFNAR1 blocking assay, mice were treated with blocking IFNAR1 antibody (2.5 mg/kg, InVivoMAb, Clone: MAR1-5A3, BioXcell) by tail vein injection on days 0, 3, 6, 9, 12, 14, and 18 after subcutaneous injection with cancer cells. Tumor growth curves were presented with error bars representing mean ± SEM at each time point. Kaplan-Meier survival curves were generated. Sex was not considered in the study design and analysis because previous studies and our preliminary data did not indicate significant sex-based differences in the tumor models used. Early termination criteria were as follows: (1) the maximum cumulative tumor burden of 2.0 cm in diameter; (2) the tumor impedes eating, urination, defecation, or ambulation; and (3) very poor body condition.
Multichannel imaging and Image analysis
Multichannel imaging was conducted using a Vectra Polaris Imaging System (Akoya Biosciences). Slides were captured at ×200 magnification. Image analysis was performed using QuPath version 0.4.3 (Queen’s University)60. Tissue sections were divided into tumor and stroma regions based on pan-CK staining. Cell segmentation employed an algorithm based on nuclear DAPI staining. Fluorescence intensity of cells was quantified for each marker. Cells were classified into distinct phenotypic classes using positivity thresholds for individual markers, determined by cytoplasmic or nuclear staining intensity, and evaluated across all samples. Cell count, density, and percentage in different regions were calculated for each phenotype.
4D Label-free Proteomics and data analysis
The LC-MS data were analyzed for data interpretation and protein identification against the Mus musculus database from Uniprot (downloaded on 01/02/2021, and including 88069 protein sequences), which is sourced from the protein database at https://www.uniprot.org/uniprot/?query=taxonomy:10090.
The MS spectra were searched using MSFragger version 2.4 and FragPipe version 13.1 with mass calibration and parameter optimization enabled. Tryptic cleavage specificity was applied, along with variable methionine oxidation, variable protein N-terminal acetylation, and fixed carbamidomethyl cysteine modifications. The allowed peptide length and mass ranges were 6–50 residues and 500–5000 Da, respectively. PeptideProphet and ProteinProphet in Philosopher (version 2.2.0; https://philosopher.nesvilab.org/) were used to filter all peptide-spectrum matches (PSMs), peptides, and proteins with <1% false discovery rate. Entries from decoy proteins was removed. Label-free quantification analysis was performed with IonQuant (version 1.1.0). The differentially expressed proteins were selected with fold change >2 or fold change <0.5 and with Student’s t-test (p value < 0.05).
RNA sequencing (RNA-Seq) and data analysis (GSE275283)
Cells and tumor tissues were lysed directly after grinding, and total RNA extraction was carried out using the RNeasy Mini Kit (QIAGEN, catalog 74104). Six hundred nanograms of total RNA were reversely transcribed into cDNA using ProtoScript II Reverse Transcriptase (New England BioLabs, catalog E7420L). The resulted double-stranded cDNA was purified with Agencourt AMPure XP Beads (Beckman, catalog A63881) and then ligated with paired-end adaptors using Multiplex Oligos for RNA sequencing. Sequencing was conducted on an Illumina HiSeq 10X platform, and data analysis was performed using the Linux system. Fastp software (https://github.com/OpenGene/fastp) were used to remove the reads that contained adaptor contamination, low quality bases and undetermined bases with default parameter. Then sequence quality was also verified using fastp. We used HISAT2 to map reads to the reference genome of Homo sapiens GRCh38. The mapped reads of each sample were assembled using StringTie (https://ccb.jhu.edu/software/stringtie) with default parameters. Then, all transcriptomes from all samples were merged to reconstruct a comprehensive transcriptome using gffcompare (https://github.com/gpertea/gffcompare/). After the final transcriptome was generated, StringTie and was used to estimate the expression levels of all transcripts. StringTie was used to perform expression level for mRNAs by calculating FPKM (FPKM = [total_exon_fragments/mapped_reads(millions) × exon_length(kB)]). The deferentially expressed mRNAs were selected with fold change > 2 or fold change <0.5 and with parametric F-test comparing nested linear models (p value < 0.05) by R package edgeR.
Gene set enrichment analysis (GSEA)
GSEA analysis was conducted using GSEA 4.1.0 software following the guidelines provided on the official website. The complete normalized RNA expression count matrix, including all genes rather than just differentially expressed ones, was utilized as input. The matrix was partitioned into two groups: (1) KO-High group; (2) WT-Low group. Hallmarks were chosen from the gene sets database, and 1,000 permutations were performed based on default weighted enrichment statistics.
RNA extraction and RT-qPCR
Following the manufacturer’s protocol, cell pellets were collected and subjected to total RNA extraction using NucleoZol (MNG, catalog 740404.200). The extracted RNA was then reversely transcribed into cDNA using the One Step PrimeScript RT-PCR kit (TaKaRa, catalog 36110 A). SYRB Green qPCR mix (Bimake. cn, catalog B21202) were employed for PCR amplification and detected on the Light Cycler Real-Time PCR System (Roche). RT-qPCR data were normalized to GAPDH and presented as fold changes in gene expression relative to the control sample. Gene-specific primers were listed in Table S2.
Protein extraction and Western blot analysis
Cells, tumor tissues or paired adjacent tissues were collected and lysed using the lysis buffer from Bestbio Company (China). Protein concentration was determined with the BCA kit (Beyotime, catalog P0011). An equal amount of protein from each sample was loaded onto SDS-PAGE gels and subsequently transferred to PVDF membranes. The PVDF membranes were then blocked in 5% non-fat milk for 1 h at room temperature. After washed with PBST, the membranes were incubated with the primary antibodies overnight as following: anti-NAT10 (1:1000, Abcam, catalog ab194297), anti-Myc (1:1000, CST, catalog 18583), anti-CDK2 (1:1000, CST, catalog 2546), anti-DNMT1 (1:1000, CST, catalog 5032), anti-RIG-I (1:1000, CST, catalog 3743), anti-HA-tag (1:1000, CST, catalog 3724), anti-Tubulin (1:1000, CST, catalog 2146), anti-β-Actin (1:1000, CST, catalog 4970). On the next day, the members were washed with TBST three times and incubated with HRP-linked secondary antibodies (1:2000, CST, catalog 7074) at room temperature for 50 min. Membranes were washed with TBST three times before being imaged using the ChemiDoc XRS+ system (Bio-Rad, USA).
CCK8 and colony formation assay
After chemical inhibition of NAT10 by Remodelin (10 µM or 20 µM, MCE, catalog HY-16706A) or genomic depletion, the proliferation assays of cancer cells were detected by CCK8 and colony formation assays. For CCK8 assay, 10000 cancer cells were seeded in 96-well plates one day in advance to achieve 50% confluency. The absorbance was detected by a microplate reader (Biotek, HIMFD, USA) according to the manufacturers’ instructions (Bestbio Company, China) 4 h later after CCK8 detection reagent was added. For the colony formation assays, 200 cells were plated in the six-well plates and 7 days later, colonies were fixed with 4% PFA, stained with a 0.5% crystal violet staining solution (Beyotime Company, China) for 30 min and counted with microscopy.
Flow cytometry
Mice were sacrificed at the appropriate time, and the tumors were collected and separated into single cells. Briefly, tumors were excised and minced. Then Liberase TL Research Grade 10 (2 µg/mL, Roche, catalog 05401020001) and DNase I (Roche, catalog 10104159001) was used for the digestion. After being filtered with strainer, the cell suspensions were stimulated with brefeldin A (BFA, PeproTech, 10 mg/mL), phorbol myristate acetate (PMA, 100 µg/mL, PeproTech), and ionomycin (1 mg/mL, PeproTech) at 37 °C for 4 h. For the cell surface staining, cells were stained for cell markers including cell death dye (1:300, Invitrogen, eBioscience Fixable Viability Dye eFluor 780, catalog 2633409), CD45.2 (1:100, BioLegend, catalog 109814), CD8a (1:100, BioLegend, catalog B373965), CD4 (1:100, Invitrogen, catalog 2286404), CD25 (1:100, Invitrogen, catalog 2151388), LY6G (1:100, Tonbo, catalog SKU 65-1276-U100), CD11B (1:100, Tonbo, catalog SKU 65-1276-U100), LY6C (1:100, Biolegend, catalog 128008), CD11c (1:100, Invitrogen, catalog 2400633), and IA/IE (1:100, BioLegend, catalog 107608) for 30 min. As for intracellular staining, cells were stained with IFN-γ (1:100, Invitrogen, catalog 2481435), IL-17A (1:100, Invitrogen, catalog 2384219), Foxp3 (1:100, Invitrogen, catalog 2513557) and Granzyme B (1:100, BioLegend, catalog 515406) after being treated with fixation/permeabilization kit at 4°C for 30 min. Cells were analyzed using a Gallios flow cytometer (Beckman Coulter, USA) and the results were analyzed by Flowjo.
Immunohistochemical (IHC) analysis
The pathology sections of patients was obtained from Qilu Hospital of Shandong University. After dewaxing, dehydration, and antigen retrieval, paraffin-embedded slides (4 µm) were blacked and labeled with anti-NAT10 (1:250, Abcam, catalog ab182744), anti-CD8a (1:250, Abcam, catalog ab237709), and anti-PD-L1 (1:250, Abcam, catalog ab213524) antibody. The next day, the slides were incubated with the secondary antibody labeled with HRP (1:2000, Shanghai Gene Company, catalog GK500705) for 1 h after washing, stained with DAB, and counterstained with hematoxylin. The images were detected by a microscope.
Immunofluorescence staining and imaging
Tumors were collected at the appropriate time and fixed in 4% paraformaldehyde for 24 h. Subsequently, the tumors were embedded in OCT after dehydration in a 30% (wt/vol) sucrose solution. Following sectioned into 4.5 mm thick slices, the sections were blocked in 10% goat serum in PBS. Primary antibodies against CD8a (1:100, Abcam, catalog ab217344) were then used to incubate the tumor sections. The next day, secondary antibodies (1:500, Invitrogen, catalog A32732) were applied to the sections for 1 h. Nuclei were stained with DAPI, and the results were visualized using a confocal microscope and analyzed with ImageJ software. dsRNA was detected by the J2 antibody [1:250 dilution, 4 mg/ml, English and Scientific Consulting Kft (SCICONS), catalog 10010200].
T-cell proliferation assay
Freshly purified splenocytes were isolated from C57BL/6 N mice. Splenocytes were labeled with cell proliferation Dye eFluor 670 (eBioscience, catalog 65-0840-85) at 5 µM for 10 min at 37°C, and then resuspended in the RPMI media containing 10% FBS, 1% penicillin/streptomycin, 0.5 µg/ml purified anti-mouse CD3 Antibody (Biolegend, catalog 100238) and 0.5 µg/ml anti-mouse CD28 Antibody (Biolegend, catalog 102112). 3^105 purified splenocytes were then co-cultured with 1^104 WT or NAT10 deficient TC1/MCA205 cancer cells in 96-well round-bottom plates. The unstimulated splenocytes were used as a negative control, and those stimulated with CD3 and CD28 antibodies were used as a positive control. After 72 h, cells were collected and stained with cell proliferation Dye eFluor 670 in CD4+ T (1:100, Biolegend, catalog 100428) or CD8+ T (1:100, Biolegend, catalog 100708) cells was determined by flow-cytometric analysis.
Enzyme-linked immune spot (ELISpot) assay
IFN-γ secretion was assessed using BD ELISpot assay kits (BD Biosciences, catalog 551881) according to the manufacturer’s instructions. Tumors were aseptically harvested and processed into a single-cell suspension. Cells were then plated at a density of 2 × 10^6 per well in ELISpot plates precoated with capture antibodies and incubated in a humidified 5% CO2 incubator at 37 °C for 20 h. After incubation, cells were removed, and the plate was washed for three times. IFN-γ production was detected by incubating with a detection antibody for 2 h, followed by three times wash and incubation with an HRP-linked secondary antibody (1:1000, BD Biosciences, catalog 557630) for 2 h. Color development was achieved by adding 100 µL of Final Substrate Solution (AEC). Red dot signals were visualized using the CTL ImmunoSpot S6 Analyzers (LLC, OH, USA).
Dual-luciferase reporter assay
A dual-luciferase assay was employed to investigate the mRNA acetylation modification sites mediated by NAT10. In brief, pEZX-MT06-MYC-WT-Luc or pEZX-MT06-MYC--Mut-Luc were cloned into pEZX-MT06 Reporter Vector pGL4.0 (GeneCopoeia, catalog NM_001177354.1). Nat10 coding DNA sequence was cloned into dCAS9-VP64-GFP (Addgene, catalog 61422). TC1 WT cells were seeded at a density of 2^105 cells per well in 24-well plates and incubated overnight prior to transfection. Subsequently, cells were co-transfected with pEZX-MT06-MYC, Renilla luciferase plasmids, and either VP64-NAT10 plasmids or empty plasmids using Lipofectamine 3000 (Invitrogen, catalog L3000-015). After 24 h, firefly luciferase and Renilla luciferase activities were assessed using the Dual-Luciferase reporter system (Promega, catalog E1960), and efficacy was determined by calculating the ratio of firefly luciferase to Renilla luciferase activity.
Acetylated RNA immunoprecipitation sequencing (acRIP-seq) (GSE285518) and anti-NAT10 antibody-based RIP-PCR
acRIP-seq and data analysis was conducted by Guangzhou Epibiotek Co., Ltd. The WT and sgNAT10 TC1 cells were subjected to acRIP-seq. Total RNA was extracted and purified from WT and sgNAT10 TC1 cells using TRIzol reagent (Invitrogen). One hundred micrograms of total RNA was fragmented into 100–200 nt RNA fragments using 10X RNA Fragmentation Buffer (100 mm Tris-HCl, 100 mm ZnCl2 in nuclease-free H2O), followed by termination of the reaction with 10XEDTA. Immunoprecipitated RNA fragments were obtained by incubating fragmented RNA with anti-ac4C monoclonal antibody (Abcam, ab252215) for 3 h at 4 °C, followed by incubation with protein A/G magnetic beads (Invitrogen, catalog 8880210002D/10004D) for 2 h at 4 °C, as per the EpiTM ac4C immunoprecipitation kit protocol (Epibiotek, R1815). The library was prepared using the smart-seq method. Both the input samples without IP and the ac4C IP samples were subjected to 150-bp, paired-end sequencing on an Illumina NovaSeq 6000 sequencer. Identified ac4C peaks which p < 0.05 were chosen for the de novo motif analysis using homer (v4.10.4) under parameters:“-len 12 -rna”.
The Anti-NAT10 antibody-based RIP-PCR assay was conducted to confirm the interaction between NAT10 and Myc mRNA using the RIP Kit (BersinBio, catalog Bes5101). In addition to the full-length MYC cDNA plasmids, a mutant full-length MYC gene with targeted mutations in the 3’ UTR was constructed. WT MYC-V64 or Mut MYC-V64, along with the NAT10-V64 plasmid, were transfected into 293 T cells. TC1 cells were lysed using a polysome lysis buffer containing protease and RNase inhibitors. DNase was added to degrade the DNA at 37 °C for 10 min. NAT10 or IgG antibodies were added to the samples and incubated at 4 °C for 16 h in a vertical mixer. Subsequently, the samples were incubated with protein A/G beads for 1 h. Following the manufacturer’s instructions, the beads containing the immunoprecipitated RNA-protein complex were treated with proteinase K to remove proteins. The target RNAs were then extracted using the phenol-chloroform method, amplified by PCR (primers sequences listed in Table S3), and detected using DNA gel electrophoresis with normalization to their input group.
Ribosome Profiling Sequencing (Ribo-seq) (GSE285518)
library preparation: A total of 107 cells were prepared for cell lysis. Then, the cell medium was discarded, and the cells were washed twice with ice-cold PBS containing 100 µg/mL cycloheximide (Adooq, 66-81-9). 100 mg tissue were pulverized manually under liquid nitrogen and treated by ice-cold PBS containing 100 µg/mL cycloheximide. Ribosome profiling was performed using Epi Ribosome Profiling Kit (Epibiotek, R1814). Subsquently, RPFs (ribosome-protected RNA fragments) was extracted using RNA clean&ConcentratorTM-5 kit (ZYMO, R1016). EpiTM RiboRNA Depletion Kit (Human/Mouse/Rat) (Epibiotek, R1805) was used for rRNA depletion. Sequencing libraries were constructed using QIAseq miRNA Library kit (QIAGEN, 1103679).
Ribo-seq data processing: Adapter sequences were removed from raw sequencing data using cutadapt software. Meanwhile, reads with length between 25 and 35 bp were kept for downstream analysis. Then reads were aligned to rRNA and tRNA sequences so as to remove rRNA and tRNA reads using bowtie software, remaining reads were used to align to reference genome and transcriptome (Ensembl Version 91) using hisat2 and bowtie software separately. The trinucleotide periodicity of ribosomes and codon usage frequency were estimated using revised riboWaltz package. ORF identification was performed using Price software. Read counts were calculated using featureCounts software. Raw counts were further normalized as RPKM values using fpkm function in DESeq2 package. Translational efficiencies were determined as the ratio of (normalized abundance determined by ribosome profiling)/(normalized abundance determined by RNA-seq) as previously repored. GO and KEGG analysis were performed using clusterProfiler package in R software.
mRNA stability assay
TC1 cells were cultured in complete DMEM medium supplemented with 5 µg/ml actinomycin D (Sigma, catalog A9415) for 0, 4, 8, 12, 16, and 24 h. At the specified time points, cells were harvested, and total RNA was extracted following the protocol outlined in the “RNA extraction” section for subsequent real-time PCR analysis (primers sequences listed in Table S2).
Synthesis of SM-102
NAT10 siRNA and FAM-labeled siRNA-NC were synthesized by Atantares. Ionizable lipids SM-102 (catalog O02010), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC, catalog S01005) and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2000, catalog O02005) were purchased from AVT (Shanghai) Pharmaceutical Tech Co., Ltd. Cholesterol (catalog A90286) was purchased from Innochem. Polyetherimide (catalog 61128-46-9) was purchased from MACKLIN. 2-(azepan-1-yl) ethanol (catalog 35984E), Methacryloyl chloride (catalog 90100B) and 2-Bromoisobutyryl Bromide were purchased from Adamas.
Synthesis of PC7A
For synthesis of 2-(Hexamethyleneimino) ethyl Methacrylate Monomer (C7A-MA), 2-(azepan-1-yl) ethanol (5.0 g) and triethylamine (7.0 g) were dissolved in 100 mL of dried tetrahydrofuran and cooled to 0 °C in an ice bath. Methacryloyl chloride (4.0 g) was dissolved in 15 mL THF and subsequently dropped into previous solution. This reaction was conducted at room temperature under stirring for 8 h. For synthesis of PC7A polymer, 0.5 g C7A-MA, 8.5 g CuBr, and 11.6 mg initiator were dissolved in 0.5 mL of dried THF. After undergoing three rounds of freeze−pump−thaw, 10.3 mg N,N,N′,N″,N″-pentamethyldiethylenetriamine was introduced. Subsequently, the polymerization process was conducted at a temperature of 70 °C for a duration of 10 h. The resulting reaction mixture was then dissolved in acidic water with a pH of 4 and dialyzed in distilled water, utilizing a cut-off molecular weight of 3500 Da, to eliminate any unreacted monomers and copper. Finally, the product was obtained through the process of lyophilization.
Preparation of lipid nanoparticles
siRNA was encapsulated lipid nanoparticles (LNPs)38. Briefly, siRNAs were dissolved in sodium acetate (pH=4) and combined with a lipid solution at an amine-to-phosphate (N/P) ratio of 8. The lipid stock solutions were prepared with a total lipid concentration of 12.5 mM by dissolving SM102, DSPC, cholesterol, and DMG-PEG-2000 in ethanol at a molar ratio of 50:10:38.5:1.5. Ultrafiltration centrifugation (3500 G, 40 min) was used to remove unentrapped siRNA from the LNPs.
Preparation of PEI/PC7A
PEI and PC7A were dissolved in sterile water. Subsequently, mix the PEI, PC7A and siRNA in a weight ratio of 1.3:1:1 to form nanoparticles through electrostatic interaction with the negatively charged siRNA. Leave the nanoparticle for 5 min before using.
Size distribution of nanoparticles
Nanoparticles are dispersed in PBS to ensure a stable and uniform suspension. Dynamic Light Scattering (DLS) is employed to measure the intensity-weighted size distribution. The size distribution is calculated using algorithms such as CONTIN or cumulant analysis. The results are presented as a histogram, illustrating the proportion of particles within different size ranges. The polydispersity index (PDI) serves as a measure of the size distribution’s breadth; a lower PDI (approaching 0) indicates a narrow and uniform distribution, while a higher PDI reflects greater variability in particle size. Size distribution and PDI were measured by Malvern Nano Sizer (Malvern Instruments Ltd) in double-distilled water.
LNPs and PEI/PC7A transfection
Cells were counted using trypan blue dye. For Real-time PCR and Western blotting, 2 × 105 cells were placed in 12-well plates overnight. Replace the medium with Opti-MEM and add LNPs or PEI/PC7A with a final concentration of siRNA of 20 nM. For immunofluorescence, cells were cultured in chamber slides overnight, and then added with 20 nM FAM-labeled siRNA for 4 h. Cells were stained with 50 nM Lyso-Tracker Red (Beyotime, catalog C1046) and 10 µg/mL Hoechst (Beyotime, catalog C1022) for 30 min. Immunofluorescence images were acquired on a Nikon A1 fluorescence microscope.
Ethics approval and consent to participate
Animal studies were approved by the Animal Ethics Committee of Qilu Hospital of Shandong University. Fresh tumor tissues and paired adjacent tissues were collected from patients in Qilu Hospital. All patients provided informed consent, and our study was approved by the Medical Ethics Committee of Qilu Hospital (KYLL-202311-043).
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 9.1.2. Data are presented as means ± SEM unless otherwise indicated. Statistical comparisons of continuous variables between two groups were performed using two-sided Student’s t test, as appropriate. Tumor growth curves were analyzed by two-way ANOVA with the tumor size. Kaplan-Meier survival curves were plotted, and the significance of differences between curves was estimated using the log-rank test. A p value below 0.05 was considered statistically significant.
Study approval
This project is financially supported by grants from the National Natural Science Foundation of China (No. 32241005, 82370173, 82300219), China Postdoctoral Science Foundation (No. 2023M742117), and Natural Science Foundation of Shandong Province (Youth Program, ZR2023QH249).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Author contributions
D.-x.M. conceptualized this study. W.-c.L. and Yi-h. W. acquired public samples and analyzed data. W.-c.L., J.-f.C., X.-y.Y., X.-m.L., C.C., X.-d.G., and H.-x.J. performed the experiments. Y.-j.H., X.-l.X., N.H., H.-l.Z., and K.X. collected clinical specimens. W.-c.L., Y.-h.W., A.-m.Z., and J.-f.C. analyzed and interpreted the data. W.-c.L., X.-l.X., Yi-h.W., and D.-x.M. wrote and revised the manuscript. All authors studied and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks Seyed Mehdi, Muh-Hwa Yang and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The RNA-seq, acRIP-seq and Ribo-seq datasets have been deposited in the GEO database under the GSE285518 and GSE275283. The Proteomics data have been deposited in PRIDE database under the PXD059463. All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Wan-cheng Liu, Yi-hong Wei, Jin-feng Chen.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-025-60293-4.
References
- 1.Salik, B., Smyth, M. J. & Nakamura, K. Targeting immune checkpoints in hematological malignancies. J. Hematol. Oncol.13, 111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu, S. et al. Combination strategies to maximize the benefits of cancer immunotherapy. J. Hematol. Oncol.14, 156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Miguel, M. & Calvo, E. Clinical challenges of immune checkpoint inhibitors. Cancer Cell38, 326–333 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Li, B., Chan, H. L. & Chen, P. Immune checkpoint inhibitors: Basics and challenges. Curr. Med Chem.26, 3009–3025 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Benci, J. L. et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell167, 1540–1554.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alicea-Torres, K. et al. Immune suppressive activity of myeloid-derived suppressor cells in cancer requires inactivation of the type I interferon pathway. Nat. Commun.12, 1717 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y. G. & Hur, S. Cellular origins of dsRNA, their recognition and consequences. Nat. Rev. Mol. Cell Biol.23, 286–301 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, R. X. & Zhou, P. K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target Ther.5, 60 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, B. et al. Recent advances in DDR (DNA damage response) inhibitors for cancer therapy. Eur. J. Med Chem.230, 114109 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Cañadas, I. et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med24, 1143–1150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, E. et al. WEE1 inhibition induces anti-tumor immunity by activating ERV and the dsRNA pathway. J. Exp. Med219, e20210789 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorrity, T. J. et al. Long 3’UTRs predispose neurons to inflammation by promoting immunostimulatory double-stranded RNA formation. Sci. Immunol.8, eadg2979 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsrna including endogenous retroviruses. Cell162, 974–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im, J. H. et al. Mechanisms of length-dependent recognition of viral double-stranded RNA by RIG-I. Sci. Rep.13, 6318 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu, B. et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell152, 276–289 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Dai, E. et al. Epigenetic modulation of antitumor immunity for improved cancer immunotherapy. Mol. Cancer20, 171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karin, M. & Shalapour, S. Regulation of antitumor immunity by inflammation-induced epigenetic alterations. Cell Mol. Immunol.19, 59–66 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin, J. et al. Epigenetic modulation of antitumor immunity and immunotherapy response in breast cancer: biological mechanisms and clinical implications. Front Immunol.14, 1325615 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, W. et al. Dual inhibitors of DNMT and HDAC induce viral mimicry to induce antitumour immunity in breast cancer. Cell Death Discov.10, 143 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo, J. et al. Emerging role of RNA acetylation modification ac4C in diseases: Current advances and future challenges. Biochem Pharm.213, 115628 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Li, K. J. et al. NAT10 promotes prostate cancer growth and metastasis by acetylating mRNAs of HMGA1 and KRT8. Adv. Sci. (Weinh.)11, e2310131 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie, R. et al. NAT10 drives cisplatin chemoresistance by enhancing ac4C-associated DNA repair in bladder cancer. Cancer Res83, 1666–1683 (2023). [DOI] [PubMed] [Google Scholar]
- 23.Tang, Z. et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res47, W556–w560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larrieu, D. et al. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science344, 527–532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, B. et al. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol.1711, 243–259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu, F. et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun.12, 2540 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, X., Peng, J. & Yi, C. Acetylation enhances MRNA stability and translation. Biochemistry58, 1553–1554 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Li, J. et al. The effects of MYC on tumor immunity and immunotherapy. Cell Death Discov.9, 103 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmerli, D. et al. MYC promotes immune-suppression in triple-negative breast cancer via inhibition of interferon signaling. Nat. Commun.13, 6579 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tadesse, S. et al. Cyclin-dependent kinase 2 inhibitors in cancer therapy: An update. J. Med Chem.62, 4233–4251 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Jian, Y. et al. Actin-like protein 6A/MYC/CDK2 axis confers high proliferative activity in triple-negative breast cancer. J. Exp. Clin. Cancer Res40, 56 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, J. et al. Effect of CDK4/6 inhibitors on tumor immune microenvironment. Immunol. Invest53, 437–449 (2024). [DOI] [PubMed] [Google Scholar]
- 33.Huang, K. C. et al. DNMT1 constrains IFNβ-mediated anti-tumor immunity and PD-L1 expression to reduce the efficacy of radiotherapy and immunotherapy. Oncoimmunology10, 1989790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao, H. et al. PRMT1 inhibition activates the interferon pathway to potentiate antitumor immunity and enhance checkpoint blockade efficacy in melanoma. Cancer Res84, 419–433 (2024). [DOI] [PubMed] [Google Scholar]
- 35.Barral, P. M. et al. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharm. Ther.124, 219–234 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, H. et al. Metabolic reprograming mediated by tumor cell-intrinsic type I IFN signaling is required for CD47-SIRPα blockade efficacy. Nat. Commun.15, 5759 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, L. et al. RIG-I is a key antiviral interferon-stimulated gene against hepatitis E virus regardless of interferon production. Hepatology65, 1823–1839 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Ball, R. L. et al. Lipid nanoparticle formulations for enhanced co-delivery of siRNA and mRNA. Nano Lett.18, 3814–3822 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Huang, C. et al. Liver-specific ionizable lipid nanoparticles mediated efficient RNA interference to clear “bad cholesterol. Int J. Nanomed.18, 7785–7801 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye, M. et al. Double-network nanogel as a nonviral vector for DNA delivery. ACS Appl Mater. Interfaces11, 42865–42872 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Yang, J. et al. Epigenetic regulation in the tumor microenvironment: molecular mechanisms and therapeutic targets. Signal Transduct. Target Ther.8, 210 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orsolic, I., Carrier, A. & Esteller, M. Genetic and epigenetic defects of the RNA modification machinery in cancer. Trends Genet39, 74–88 (2023). [DOI] [PubMed] [Google Scholar]
- 43.Kong, Y. et al. Novel insight into RNA modifications in tumor immunity: Promising targets to prevent tumor immune escape. Innov. (Camb.)4, 100452 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen, L. et al. The roles and mechanism of m(6)A RNA methylation regulators in cancer immunity. Biomed. Pharmacother.163, 114839 (2023). [DOI] [PubMed] [Google Scholar]
- 45.Hogg, S. J. et al. Targeting the epigenetic regulation of antitumour immunity. Nat. Rev. Drug Discov.19, 776–800 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Topper, M. J. et al. The emerging role of epigenetic therapeutics in immuno-oncology. Nat. Rev. Clin. Oncol.17, 75–90 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, L. et al. HDAC3 inhibition promotes antitumor immunity by enhancing CXCL10-mediated chemotaxis and recruiting of immune cells. Cancer Immunol. Res11, 657–673 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duong, E. et al. Type I interferon activates MHC class I-dressed CD11b(+) conventional dendritic cells to promote protective anti-tumor CD8(+) T cell immunity. Immunity55, 308–323.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie, L. et al. Mechanisms of NAT10 as ac4C writer in diseases. Mol. Ther. Nucleic Acids32, 359–368 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dhanasekaran, R. et al. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol.19, 23–36 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tadesse, S. et al. Targeting CDK2 in cancer: challenges and opportunities for therapy. Drug Discov. Today25, 406–413 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Choi, H. et al. Targeting DDX3X triggers antitumor immunity via a dsRNA-mediated tumor-intrinsic type i interferon response. Cancer Res81, 3607–3620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen, Z. et al. ISG20 stimulates anti-tumor immunity via a double-stranded RNA-induced interferon response in ovarian cancer. Front Immunol.14, 1176103 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cottrell, K. A., Andrews, R. J. & Bass, B. L. The competitive landscape of the dsRNA world. Mol. Cell84, 107–119 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ku, Y. et al. Noncanonical immune response to the inhibition of DNA methylation by Staufen1 via stabilization of endogenous retrovirus RNAs. Proc. Natl Acad. Sci. USA118, e2016289118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang, K. C. et al. Colorectal cancer-specific IFNβ delivery overcomes dysfunctional dsRNA-mediated type I interferon signaling to increase the abscopal effect of radiotherapy. J. Immunother. Cancer12, e2016289118 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pauken, K. E. et al. Emerging concepts in PD-1 checkpoint biology. Semin Immunol.52, 101480 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dalhat, M. H. et al. Remodelin, a N-acetyltransferase 10 (NAT10) inhibitor, alters mitochondrial lipid metabolism in cancer cells. J. Cell Biochem122, 1936–1945 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Lu, Q. et al. Nanoparticles in tumor microenvironment remodeling and cancer immunotherapy. J. Hematol. Oncol.17, 16 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bankhead, P. et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep.7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The RNA-seq, acRIP-seq and Ribo-seq datasets have been deposited in the GEO database under the GSE285518 and GSE275283. The Proteomics data have been deposited in PRIDE database under the PXD059463. All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.