Abstract
Introduction
Bacteremia, a common disease but difficult to diagnose early, may result in significant morbidity and mortality without prompt treatment. We aimed to develop machine-learning (ML) algorithms to predict patients with bacteremia from febrile patients presenting to the emergency department (ED) using data that is readily available at the triage.
Methods
We included all adult patients (≥18 years of age) who presented to the emergency department (ED) of National Taiwan University Hospital (NTUH), a tertiary teaching hospital in Taiwan, with the chief complaint of fever or measured body temperature more than 38°C, and who received at least one blood culture during the ED encounter. We extracted data from the Integrated Medical Database of NTUH from 2009–2018.The dataset included patient demographics, triage details, symptoms, and medical history. The positive blood culture result of at least one potential pathogen was defined as bacteremia and used as the binary classification label. We split the dataset into training/validation and testing sets (60-to-40 ratio) and trained five supervised ML models using K-fold cross-validation. The model performance was evaluated using the area under the receiver operating characteristic curve (AUC) in the testing set.
Results
We included 80,201 cases in this study. Of them, 48120 cases were assigned to the training/validation set and 32,081 to the testing set. Bacteremia was identified in 5,831 (12.1%) and 3,824 (11.9%) cases of the training/validation set and test set, respectively. All ML models performed well, with CatBoost achieving the highest AUC (.844, 95% confidence interval [CI] .837–.850), followed by extreme gradient boosting (.843, 95% CI .836–.849), gradient boosting (.842, 95% CI .836–.849), light gradient boosting machine (.841, 95% CI .834–.847), and random forest (.828, 95% CI .821–.834).
Conclusion
Our machine-learning model has shown excellent discriminatory performance to predict bacteremia based only on clinical features at ED triage. It has the potential to improve care quality and save more lives if successfully implemented in the ED.
INTRODUCTION
Bacteremia, defined as the existence of bacteria in the bloodstream, is a serious and potentially life-threatening condition associated with mortality rates ranging from 13–35%.1–3 The timely and accurate diagnosis of bacteremia is crucial for appropriate treatment and management of patients who present with fever to the emergency department (ED). Emergency physicians often order blood culture tests for patients suspected of bacterial infections based on the presence of fever. However, excessive and unnecessary blood culture tests can lead to extended hospital length of stay, increased antibiotic use, additional laboratory testing, and even higher rates of side effects related to blood collection and antibiotic administration.4
Despite the advancements in modern medical technology, blood culture remains the gold standard for diagnosing bacteremia and determining antimicrobial susceptibility. However, it often takes more than 24 hours to achieve a detectable colony size, preventing emergency physicians from obtaining real-time blood culture results. This delay can lead to the administration of ineffective antibiotics or the premature discharge of patients with bacteremia, which may become evident later on. While detailed evaluations by emergency physicians may not always lead to irreparable consequences, bacteremia caused by specific bacteria can indeed result in unfavorable outcomes.5–7
Previous studies have identified various variables, including vital signs, clinical symptoms, comorbid conditions, and laboratory values, which are independently associated with bacteremia.8–10 Several prediction models have also been developed, with area under the receiver operating characteristic curve (AUC) values ranging from .71–.854.11–15 In recent years, machine-learning (ML) techniques have been applied to predict bacteremia, but most studies have primarily focused on hospitalized patients,16–19 with limited research involving ED patients.20,21 Furthermore, many of these studies rely on laboratory results that are not immediately available in the ED. In the ED setting, physicians typically rely solely on demographics, ED triage data, symptoms, and past medical history to make early predictions of bacteremia, emphasizing the critical role of ordering necessary blood cultures.21,22
In light of these challenges, we sought to create a predictive model that can assist emergency physicians in making timely and informed decisions regarding the need for blood culture tests and appropriate treatment strategies. By harnessing the power of ML and incorporating a wide range of clinical, demographic, and other accessible variables during the ED encounter collected from electronic medical records (EMR), our study aimed to develop the ML algorithms that can predict bacteremia in febrile patients presenting to the ED using data readily available during the triage and history-taking stages. Additionally, we discuss the potential implications of our findings, acknowledge the study’s limitations, and outline future directions for research in this critical field of emergency medicine.
Population Health Research Capsule.
What do we already know about this issue?
Early diagnosis of bacteremia is challenging, yet delayed treatment increases morbidity and mortality in febrile emergency department (ED) patients.
What was the research question?
Can machine-learning methods predict bacteremia in febrile ED patients using triage data?
What was the major finding of the study?
CatBoost achieved the highest AUC (.844, 95% CI .837–.850) for bacteremia prediction at ED triage.
How does this improve population health?
Early bacteremia prediction by machine-learning at triage has the potential to enable timely treatment, improving outcomes and reducing mortality in ED patients.
METHODS
Study Design and Setting
We conducted this retrospective cohort study using EMR data extracted from the integrated Medical Database (iMD) of National Taiwan University Hospital (NTUH), a 2,500-bed, university-affiliated teaching hospital providing primary, secondary, and tertiary care in northern Taiwan. The hospital has various departments covering all major specialties, including transplantation and oncology. The ED of this hospital sees an annual average of 110,000 patient visits, with approximately 91,600 visits when excluding pediatric (<18 years of age) cases. This study was approved by the Institutional Review Board of NTUH (202104109RINC) with a waiver of informed consent. This study protocol followed the guidelines of “Minimum information about clinical artificial intelligence modeling: the MI-CLAIM checklist”.23
Study Population
We included all adult patients (≥18 years of age) who presented to the ED of NTUH over a 10-year period (January 2009–December 2018) with the chief complaint of fever or measured body temperature of more than 38° Celsius at triage, and received at least one blood culture during the ED encounter. Repeat ED visits by the same patient within a 30-day period were considered as the same index visit, and duplicate visits were eliminated from the analysis.
Collection of Variables (Features)
From the iMD, we obtained patient demographics, including age, sex, height, and body mass index (BMI), that are readily available during triage. We also collected ED triage data, which encompassed the five-level Taiwan Triage and Acuity Scale (TTAS), details of emergency medical services (EMS) transport, and transfer status.24 The initial ED presenting vital signs, including the Glasgow Coma Scale (GCS) score, body temperature, pulse rate, respiratory rate, oxygen saturation (SpO2), pain index, and acute changes in consciousness, were also retrieved. The GCS scores were further categorized into clear consciousness (GCS 15), minor coma (GCS 13–14), moderate coma (GCS 9–12), severe coma (GCS 3–8), and other (intubated, tracheostomy, or aphasia). Furthermore, we extracted clinical symptoms from the TTAS documentation, which encompassed a total of 179 structured chief complaints (CC) recorded during patient encounters by the triage nurse. Incorporating past medical histories (PMH) as input features for this study involved two different sources of information. First, PMHs were obtained through direct patient reporting during their current ED encounters as part of the interview conducted by the triage nurse while enquiring their CCs. The second source of PMH data was derived from patients’ prior chart records, spanning outpatient, inpatient, or previous ED admissions, and subsequently coded using the International Classification of Diseases, 10th Rev (ICD-10) codes.25
Outcome Measures
The primary outcome of this study —the presence of true bacteremia—was also employed as the classification label for our binary classification task. Patients were labeled as having true bacteremia when either a single blood culture yielded pathogenic bacteria or when two or more sets of blood cultures collected from distinct sites revealed the same bacterial species. Contaminants, including coagulase-negative staphylococci, Corynebacterium species, Bacillus species (except for B anthracis), Propionibacterium species, Micrococcus species, and Clostridium perfringens, were identified and excluded based on prior studies, except in cases where a significant indwelling catheter was present.26
Data Analysis and Feature Selection
We used Microsoft Excel 2016 (Microsoft Corporation, Redmond, WA) for data entry and processing. The data was further analyzed with SPSS Statistics for Windows v24.0 (IBM Corp, Armonk, NY). The variables with missing values were imputed by replacing them with the mean, median, or mode of their respective class if the missing rate was less than 20% for that variable. We presented the results as percentages for categorical variables, standard deviations for continuous variables, and medians with interquartile ranges (IQR) for time variables. Our feature selection approach involved univariate analyses, where we assessed outcome differences between groups using statistical tests such as Student t-test, chi-squared test, Fisher exact test, or Mann-Whitney U test, depending on the distribution of the data. We selected variables with a significance level of P<.05 in the training cohort as the input features for constructing the ML models.
Machine-Learning Model Construction
We employed supervised ML algorithms using random forest (RF), gradient boosting (GB), CatBoost (CB), light gradient boosting machine (LGBM) and extreme gradient boosting (XGB) to construct the prediction models. The dataset was randomly split into the training/validation and testing cohorts at a ratio of 60:40. To train our model, we employed K-fold cross-validation, with the value of K ranging from 7–10. The model’s performances were evaluated through the area under the receiver operating characteristic curve (AUC) on the test set. We selected the K value that resulted in the highest AUC performance as our final choice. In addition to AUC, we also reported the classification performances on the testing cohort using F1-score, precision (or positive predictive value [PPV]), recall (sensitivity), specificity, negative predictive value (NPV), and area under the precision-recall curve (AUPRC) for each model. Furthermore, we incorporated SHAP (Shapley additive explanations) value analysis alongside feature importance to enhance the interpretability and transparency of the ML models we developed.27 All ML analyses were performed using Python 3.8 programming language (Python Software Foundation, Wilmington, DE) with package scikit-learn 0.23.1 installed.
RESULTS
During the study period, a total of 124,158 adult ED visits with at least one blood culture performed were retrieved from the iMD. After excluding records of repeat ED visits within 30 days and patients without fever, we included 80,201 records for analysis. The flow chart of case inclusion and exclusion process is shown in Figure 1. We assigned 48,120 cases to the training/validation cohort and 32,081 to the testing cohort. True bacteremia was identified in 5,831 (12.1%) and 3,824 (11.9%) cases of the training/validation cohort and testing cohort, respectively.
Figure 1.

The case inclusion and exclusion flow chart.
ED, emergency department.
The characteristics of the study population, including patient demographics and ED triage data for both the training/validation and testing cohorts, are presented in Table 1. A comprehensive breakdown of population characteristics, including past medical histories (PMH) and ED presenting symptoms across groups, can be found in Supplementary Table S1. Univariate analyses of features distinguishing patients with and without bacteremia are partially summarized in Table 2 for the training/validation and testing sets. Full details are provided in Supplementary Table S2.
Table 1.
Patient demographics, ED triage data of the training/validation cohort and testing cohort.
| Variables (features) | Total (N=80,201) | Training cohort (n=48,120) | Testing cohort (n=32,081) | P-value | |||
|---|---|---|---|---|---|---|---|
| Sex | 0.87 | ||||||
| Female | 40,220 | (50.1) | 24,143 | (50.2) | 16,077 | (50.1) | |
| Male | 39,981 | (49.9) | 23,977 | (49.8) | 16,004 | (49.9) | |
| Age, Mean (SD) | 59.3 | (20.0) | 59.3 | (20) | 59.4 | (20.1) | 0.36 |
| Temperature, Mean (SD) | 38.4 | (1.4) | 38.4 | (1.3) | 38.4 | (1.5) | 0.25 |
| Acute change | 0.47 | ||||||
| No | 71,876 | (89.6) | 43,142 | (89.7) | 28,734 | (89.6) | |
| Yes | 2,570 | (3.2) | 1,561 | (3.2) | 1,009 | (3.1) | |
| NA | 5,755 | (7.2) | 3,417 | (7.1) | 2,338 | (7.3) | |
| EMS transfer | 0.12 | ||||||
| No | 64,486 | (80.4) | 38,686 | (80.4) | 25,800 | (80.4) | |
| Yes | 68 | (0.1) | 47 | (0.1) | 21 | (0.1) | |
| NA | 15,647 | (19.5) | 9,387 | (19.5) | 6,260 | (19.5) | |
| Triage | 0.83 | ||||||
| 1 | 3,477 | (4.3) | 2,118 | (4.4) | 1,359 | (4.2) | |
| 2 | 20,770 | (25.9) | 12,448 | (25.9) | 8,322 | (25.9) | |
| 3 | 54,769 | (68.3) | 32,851 | (68.3) | 21,918 | (68.3) | |
| 4 | 1,127 | (1.4) | 669 | (1.4) | 458 | (1.4) | |
| 5 | 58 | (0.1) | 34 | (0.1) | 24 | (0.1) | |
| Systolic BP, Mean (SD) | 132.5 | (27.1) | 132.5 | (27) | 132.6 | (27.2) | 0.88 |
| DiastolicC BP, Mean (SD) | 75.7 | (15.1) | 75.7 | (15.1) | 75.7 | (15.2) | 0.97 |
| Pulse, Mean (SD) | 106.2 | (19.7) | 106.4 | (19.7) | 105.9 | (19.7) | 0.003 |
| Oxygen, Miean (SD) | 95.9 | (3.6) | 95.9 | (3.5) | 95.9 | (3.7) | 0.97 |
| Respiration rate, Mean (SD) | 19.6 | (5.5) | 19.6 | (4.8) | 19.6 | (6.4) | 0.77 |
| Pain Index | 0.19 | ||||||
| 0 | 58,424 | (72.8) | 35,085 | (72.9) | 23,339 | (72.8) | |
| 1 | 49 | (0.1) | 22 | (0.0) | 27 | (0.1) | |
| 2 | 314 | (0.4) | 187 | (0.4) | 127 | (0.4) | |
| 3 | 847 | (1.1) | 510 | (1.1) | 337 | (1.1) | |
| 4 | 2,430 | (3.0) | 1,444 | (3) | 986 | (3.1) | |
| 5 | 5,315 | (6.6) | 3,182 | (6.6) | 2,133 | (6.6) | |
| 6 | 2,749 | (3.4) | 1,659 | (3.4) | 1,090 | (3.4) | |
| 7 | 3,867 | (4.8) | 2,387 | (5) | 1,480 | (4.6) | |
| 8 | 2,549 | (3.2) | 1,488 | (3.1) | 1,061 | (3.3) | |
| 9 | 494 | (0.6) | 289 | (0.6) | 205 | (0.6) | |
| 10 | 596 | (0.7) | 351 | (0.7) | 245 | (0.8) | |
| NA | 2,567 | (3.2) | 1516 | (3.2) | 1051 | (3.3) | |
| Height, Mean (SD) | 161.8 | (10.4) | 161.8 | (10.5) | 161.8 | (10.2) | 0.88 |
| Weight, Mean (SD) | 61.1 | (19.4) | 61.1 | (22.5) | 61.1 | (13.5) | 0.73 |
| BMI, Mean (SD) | 23.2 | (4.4) | 23.2 | (4.4) | 23.2 | (4.4) | 0.79 |
| GCS TYPE | 0.32 | ||||||
| Clear consciousness | 65,917 | (82.2) | 39,514 | (82.1) | 26,403 | (82.3) | |
| Minor coma | 1,240 | (1.5) | 766 | (1.6) | 474 | (1.5) | |
| Moderate coma | 3,553 | (4.4) | 2,155 | (4.5) | 1,398 | (4.4) | |
| Severe coma | 1,747 | (2.2) | 1,047 | (2.2) | 700 | (2.2) | |
| others | 1,686 | (2.1) | 1,043 | (2.2) | 643 | (2.0) | |
| NA | 6,058 | (7.6) | 3,595 | (7.5) | 2,463 | (7.7) | |
ED, emergency department; EMS, emergency medical service; BP, blood pressure; BMI, body mass index; GCS, Glasgow Coma Scale; NA, not available
Table 2.
Univariate analyses of features between patients with or without bacteremia on the on the training/validation and testing cohorts.
| Cohort | Training/validation | Testing cohort | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| Variables (features) | Bacteremia (−) (n=42,298) | Bacteremia (+) (n=5,822) | P-value | Bacteremia (−) (n=28,248) | Bacteremia (+) (n=3,833) | P-value | ||||
| Sex | 0.04 | 0.04 | ||||||||
| Female | 21,147 | (50.0) | 2,996 | (51.5) | 14,097 | (49.9) | 1,980 | (51.7) | ||
| Male | 21,151 | (50.0) | 2,826 | (48.5) | 14,151 | (50.1) | 1,853 | (48.3) | ||
| Age, Mean (SD) | 58.3 | (20.3) | 66.4 | (16.3) | <.001 | 58.4 | (20.3) | 66.6 | (16.4) | <0.001 |
| Body temperature, Mean (SD) | 38.4 | (1.3) | 38.8 | (1.4) | <.001 | 38.4 | (1.5) | 38.8 | (1.3) | <0.001 |
| Acute change | <.001 | <0.001 | ||||||||
| No | 38,090 | (90.1) | 5,052 | (86.8) | 25,425 | (90.0) | 3,309 | (86.3) | ||
| Yes | 1,229 | (2.9) | 332 | (5.7) | 780 | (2.8) | 229 | (6.0) | ||
| NA | 2,979 | (7.0) | 438 | (7.5) | 2,043 | (7.2) | 295 | (7.7) | ||
| EMS | 0.005 | 0.72 | ||||||||
| No | 33,974 | (80.3) | 4,712 | (80.9) | 22,687 | (80.3) | 3,113 | (81.2) | ||
| Yes | 35 | (0.1) | 12 | (0.2) | 19 | (0.1) | 2 | (0.1) | ||
| NA | 8289 | (19.6) | 1098 | (18.9) | 5542 | (19.6) | 718 | (18.7) | ||
| Triage | <0.001 | <0.001 | ||||||||
| 1 | 1,616 | (3.8) | 502 | (8.6) | 1,038 | (3.7) | 321 | (8.4) | ||
| 2 | 10,618 | (25.1) | 1,830 | (31.4) | 7,133 | (25.3) | 1,189 | (31.0) | ||
| 3 | 29,392 | (69.5) | 3,459 | (59.4) | 19,619 | (69.5) | 2,299 | (60.0) | ||
| 4 | 639 | (1.5) | 30 | (0.5) | 434 | (1.5) | 24 | (0.6) | ||
| 5 | 33 | (0.1) | 1 | (0.0) | 24 | (0.1) | 0 | (0.0) | ||
| Systolic BP, Mean (SD) | 132.7 | (26.4) | 131.2 | (30.8) | <0.001 | 132.8 | (26.7) | 130.7 | (30.6) | <0.001 |
| Diastolic BP, Mean (SD) | 76.1 | (14.9) | 72.8 | (16.2) | <0.001 | 76.1 | (14.9) | 72.4 | (16.4) | <0.001 |
| Pulse, Mean (SD) | 105.7 | (19.3) | 111.1 | (21.5) | <0.001 | 105.3 | (19.4) | 110.5 | (21.6) | <0.001 |
| Oxygen, Mean (SD) | 96 | (3.5) | 95.6 | (4.1) | <.001 | 96 | (3.6) | 95.7 | (4.2) | <0.001 |
| Respiration, Mean (SD) | 19.5 | (4.7) | 20.1 | (5.2) | <.001 | 19.5 | (6.7) | 20 | (3.8) | <0.001 |
| Pain index | <.001 | <0.001 | ||||||||
| 0 | 30,657 | (72.5) | 4,428 | (76.1) | 20,397 | (72.2) | 2942 | (76.8) | ||
| 1 | 21 | (0.0) | 1 | (0.0) | 25 | (0.1) | 2 | (0.1) | ||
| 2 | 179 | (0.4) | 8 | (0.1) | 114 | (0.4) | 13 | (0.3) | ||
| 3 | 460 | (1.1) | 50 | (0.9) | 315 | (1.1) | 22 | (0.6) | ||
| 4 | 1,303 | (3.1) | 141 | (2.4) | 897 | (3.2) | 89 | (2.3) | ||
| 5 | 2,879 | (6.8) | 303 | (5.2) | 1,939 | (6.9) | 194 | (5.1) | ||
| 6 | 1,494 | (3.5) | 165 | (2.8) | 974 | (3.4) | 116 | (3.0) | ||
| 7 | 2,112 | (5.0) | 275 | (4.7) | 1,333 | (4.7) | 147 | (3.8) | ||
| 8 | 1,323 | (3.1) | 165 | (2.8) | 940 | (3.3) | 121 | (3.2) | ||
| 9 | 246 | (0.6) | 43 | (0.7) | 184 | (0.7) | 21 | (0.5) | ||
| 10 | 311 | (0.7) | 40 | (0.7) | 203 | (0.7) | 42 | (1.1) | ||
| NA | 1,313 | (3.1) | 203 | (3.5) | 927 | (3.3) | 124 | (3.2) | ||
| Height, Mean (SD) | 162 | (10.4) | 160.4 | (11.5) | <.001 | 162.1 | (10.2) | 160.3 | (10.6) | <0.001 |
| Weight, Mean (SD) | 61.2 | (23.5) | 60.3 | (13.3) | 0.01 | 61.1 | (13.6) | 60.4 | (12.9) | 0.001 |
| BMI, Mean (SD) | 23.2 | (4.4) | 23.4 | (4.6) | 0.003 | 23.2 | (4.4) | 23.3 | (4.4) | 0.04 |
| GCS type | <.001 | <0.001 | ||||||||
| NA | 3,134 | (7.4) | 461 | (7.9) | 2,157 | (7.6) | 306 | (8.0) | ||
| Clear consciousness | 34,958 | (82.6) | 4,556 | (78.3) | 23,390 | (82.8) | 3013 | (78.6) | ||
| Minor coma | 634 | (1.5) | 132 | (2.3) | 384 | (1.4) | 90 | (2.3) | ||
| Moderate coma | 1,768 | (4.2) | 387 | (6.6) | 1,147 | (4.1) | 251 | (6.5) | ||
| Other | 929 | (2.2) | 114 | (2.0) | 575 | (2.0) | 68 | (1.8) | ||
| Severe coma | 875 | (2.1) | 172 | (3.0) | 595 | (2.1) | 105 | (2.7) | ||
EMS, emergency medical service; BP, blood pressure; BMI, body mass index; GCS, Glasgow Coma Scale; NA, not available
A total of 395 clinical features were selected by setting the P-value <0.05 from the training/validation cohort. These features comprised six demographic factors, 10 triage-related variables, and 25 symptoms. Additionally, there were 354 PMHs sourced from two distinct origins: 98 features were gathered through direct patient interviews conducted by triage nurses, while 256 features were extracted from EMR-based ICD-10 codes. Five ML models, including RF, LGBM, XGB, CB, and GB, were constructed for predicting bacteremia. By selecting 9-fold cross validation, the constructed models showed excellent discrimination ability on the training/validation cohort and maintained their discriminatory performances on the testing cohort in terms of AUC (Figure 2).
Figure 2.
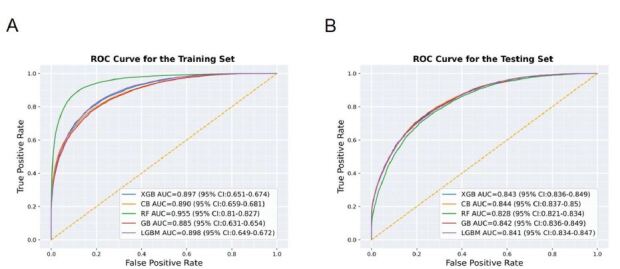
Results and the comparisons of the five machjine-learning models on the training/validation (A), and Testing (B) cohorts, based on the performances of area under the receiver operating characteristic (ROC) curves (AUC).
RF, random forest; GB, gradient boosting; CB, CatBoost; LGBM, light gradient boosting machine; XGB, extreme gradient boosting; ROC, receiver operating characteristic; AUC, area under the curve.
As illustrated in Table 3, the ML model constructed using 9-fold cross-validation in the testing cohort achieved the better area under the curve (AUC) with CB at 0.844 (95% CI 0.837–0.850), closely followed by XGB at 0.843 (95% CI 0.836–0.839), GB at 0.842 (95% CI 0.836–0.849), LGBM at 0.841 (95% CI 0.834–0.847), and RF at 0.828 (95% CI 0.821–0.834). When considering the balance between precision and recall by calculating the AUPRC, CB exhibited performance at 0.540 (95% CI 0.525–0.555), followed by XGB at 0.537 (95% CI 0.522–0.552), GB at 0.536 (95% CI 0.521–0.550), LGBM at 0.533 (95% CI 0.518–0.548), and RF at 0.473 (95% CI 0.458–0.489). Except for RF, the differences in AUC and AUPRC among the ML models were not statistically significant. Additional performance metrics, including sensitivity (or recall), specificity, negative predictive value (NPV), and positive predictive value (PPV, or precision), are shown in Table 3.
Table 3.
Comparison between model performances on the training/validation and testing cohorts.
| Model | AUC (95% CI) | AUPRC (95% CI) | F1 | Kappa | Sensitivity (Recall) | Specificity | PPV (Precision) | NPV |
|---|---|---|---|---|---|---|---|---|
| Training/validation cohort | ||||||||
| CatBoost | 0.890 (0.886–0.895) | 0.670 (0.659–0.681) | 0.462 | 0.428 | 0.307 | 0.997 | 0.932 | 0.913 |
| XGBoost | 0.897 (0.893–0.901) | 0.663 (0.651–0.674) | 0.439 | 0.404 | 0.289 | 0.996 | 0.909 | 0.911 |
| Gradient boosting | 0.885 (0.880–0.889) | 0.643 (0.631–0.654) | 0.440 | 0.404 | 0.292 | 0.995 | 0.891 | 0.911 |
| Light GBM | 0.898 (0.894–0.903) | 0.661 (0.649–0.672) | 0.412 | 0.379 | 0.266 | 0.997 | 0.914 | 0.908 |
| Random forest | 0.955 (0.952–0.957) | 0.818 (0.810–0.827) | 0.699 | 0.649 | 0.839 | 0.923 | 0.599 | 0.977 |
| Testing cohort | ||||||||
| CatBoost | 0.844 (0.837–0.850) | 0.540 (0.525–0.555) | 0.378 | 0.339 | 0.249 | 0.990 | 0.779 | 0.906 |
| XGBoost | 0.843 (0.836–0.849) | 0.537 (0.522–0.552) | 0.372 | 0.334 | 0.244 | 0.991 | 0.785 | 0.906 |
| Gradient boosting | 0.842 (0.836–0.849) | 0.536 (0.521–0.550) | 0.380 | 0.341 | 0.252 | 0.990 | 0.773 | 0.907 |
| LightGBM | 0.841 (0.834–0.847) | 0.533 (0.518–0.548) | 0.361 | 0.325 | 0.233 | 0.992 | 0.803 | 0.905 |
| Random forest | 0.828 (0.821–0.834) | 0.473 (0.458–0.489) | 0.455 | 0.370 | 0.518 | 0.897 | 0.406 | 0.932 |
Note: The best parameters for CatBoost are {‘depth’: 4, ‘learning_rate’: 0.12, ‘random_seed’: 0, ‘loss_function’: ‘Logloss’, ‘iterations’: 700}; for XGBoost are {‘learning_rate’: 0.14, ‘max_depth’: 4, ‘n_estimators’: 300}; for Gradient boosting are {‘max_depth’: 3, ‘learning_rate’: 0.1, ‘n_estimators’: 600, ‘random_state’: 0}: for Light GBM are {‘learning_rate’: 0.1, ‘n_estimators’: 100, ‘max_depth’: 30, ‘random_state’: 0}: for Random forest are {‘max_depth’: 35, ‘n_estimators’: 1000, ‘random_state’: 0, ‘max_leaf_nodes’: 1000, ‘class_weight’: ‘balanced’}.
AUC, area under the receiver operating characteristic curve; AUPRC, area under the precision recall curve; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; GBM, gradient boosting machine; XGBoost, eXtreme Gradient Booting.
A heat map of the computed top 30 features ordered by median normalized importance across all models of the constructed five different ML models are shown in Figure 3A. The top 100 important features visualized as a heat map are available in Supplementary Figure S1. Of them, the eight most important features selected for constructing the ML models are respiratory rate, body temperature, height, BMI, age, cough, weight, and pulse rate. Using the SHAP values approach, we present the outcomes of our analysis conducted with a 9-fold cross-validation in Figure 3B. This figure visually represents the top 30 importance scores assigned to each feature in predicting outcomes within the CB model we constructed. For the results pertaining to the other four models, please refer to Supplementary Figure S2.
Figure 3.

A heat map of the computed top 30 features ordered by median normalized importance across all models of the constructed five different machine-learning (ML) models (A). The Shapley additive explanations of the top 30 important features as a way to explain the output of the constructed ML models by selecting 9-fold cross validation using CB classifiers (B).
PHx, past history; ICD-10, International Classification of Diseases 10th Rev; DM, diabetes mellitus; COPD, chronic obstructive pulmonary disease; BMI, body mass index; CB, CatBoost; GIB, gastrointestinal bleeding; GCS, Glasgow Coma Scale.
DISCUSSION
In this study, we used ML methods to predict the presence of bacteremia in febrile patients presenting to the ED. Using 395 clinical features that are readily available at the ED triage, all five ML models we constructed showed excellent discriminatory performance in predicting bacteremia, with AUC ranging from 0.828 to 0.844.
Bacteremia is a serious and potentially life-threatening condition that can lead to severe complications and increased mortality. The diagnosis of bacteremia relies on a high index of suspicion by the emergency physician to order for blood culture tests based on the patient’s demographics, triage data, CCs, PMH, and physical examination. Predicting bacteremia has been studied for decades. However, previous studies have found that the accuracy of clinician impressions was poor,28 leading to subsequent studies to include laboratory testing results as predicting variables. However, laboratory results are not readily available at the initial triage and history-taking stage. Even in studies that adopted laboratory values as predicting features, these prediction models still exhibit suboptimal performance in predicting bacteremia with small sample sizes and often lack a validation group for confirmation.11–14
In the era of big data and ML, Choi et al developed and validated ML models to predict bacteremia in the ED during triage stages.20 The best performing model, the “Triage XGB model,” demonstrated only acceptable discrimination performance with an AUC of 0.718 by using demographics, triage data, and CCs as predicting variables. However, comorbidity is also an important reference for emergency physicians to assess patients’ prognosis and whether they have bacteremia.29 Our study included 80,201 ED visits during a 10-year period, including demographics, triage data, CCs, and PMH obtained through patient interviews by the triage nurse and retrieved from the patients’ EMRs. The AUC of our five ML models all demonstrated excellent results, with the CB model achieving the best performance (0.844).
Furthermore, by using SHAP analysis, our study offers additional evidence highlighting the importance of triage vital signs such as respiratory rate, body temperature, and pulse rate in predicting bacteremia, as depicted in both Figure 3B and Supplementary Figure S2. Notably, our findings reveal that advanced patient age and the presence of chills are also associated with positive predictors of bacteremia. These observations emphasize the importance of vigilance among emergency physicians when encountering such patients, warranting prompt action, including the ordering of blood culture tests.13 In contrast, the symptom of cough is a negative predictor of bacteremia possibly from patients experiencing upper respiratory infection due to viral infection.30
In addition to the AUC, we also evaluated the performance of our models using a range of available performance metrics, including sensitivity, specificity, PPV, and NPV. Prediction models from previous studies always showed high sensitivity and NPV but low specificity and PPV; thus, they can only be used clinically to rule out the possibility of bacteremia. The low specificity and PPV may lead to many false positive results, resulting in antibiotic overuse, prolonged length of stay in the ED, and even higher hospitalization rates. While the models we developed exhibit low F1 and recall scores, indicating challenges in identifying true positive cases, they can still serve valuable roles when carefully contextualized. For instance, the models can act as screening tools for bacteremia or provide supporting diagnostic insights. Additionally, our models demonstrate excellent specificity and NPV while also achieving good PPV and AUPRC, which suggests robust overall diagnostic performance, particularly in their ability to balance ruling out negative cases and identifying positive cases effectively.
LIMITATIONS
There are several limitations in this study. Firstly, this was a retrospective database analysis relying on data collected exclusively from patients who underwent blood culture examinations. We could have missed those patients who might have had bacteremia but did not receive blood culture examination, potentially introducing selection bias. Secondly, the acquisition of patients’ PMHs was reliant on interviews conducted by triage nurses and the EMR. In cases where patients had no prior hospital visits, the PMH was solely obtained through interviews with triage nurses, which might have introduced recall bias.
Thirdly, our models achieved low F1 score and recall, indicating the potential of missed diagnoses. Further strategies—such as addressing class imbalance with techniques like oversampling or undersampling, or optimizing decision thresholds—could be implemented to enhance these metrics and make the models even more reliable for diagnostic tasks. Fourthly, it’s important to note that the dataset used in this study was derived from a single, tertiary teaching hospital’s ED, and external validation from other healthcare settings is lacking. Our future works will expand our study to a regional, multicenter cohort to validate our model’s performance and improve its generalizability.
CONCLUSION
We have developed ML models to predict bacteremia, achieving excellent discrimination performance by using clinical features readily accessible during ED triage. These models demonstrate potential for identifying low-risk patients, which could help reduce unnecessary healthcare costs. Furthermore, they may assist emergency physicians in making more informed decisions regarding blood cultures orders and antibiotics administration for high-risk patients, potentially improving patient safety. However, as this study lacks external validation, the generalizability of our findings to other settings remains uncertain. Future work should focus on validating these models with external datasets to confirm their robustness and applicability across diverse clinical environments.
Supplementary Information
Footnotes
Section Editor: Ioannis Koutroulis, MD, MBA, PhD
Full text available through open access at http://escholarship.org/uc/uciem_westjem
Conflicts of Interest: By the WestJEM article submission agreement, all authors are required to disclose all affiliations, funding sources and financial or management relationships that could be perceived as potential sources of bias. This work was supported by the National Taiwan University Hospital (113-EKN0007; NTUH.111-UN0066); the National Health Research Institutes Taiwan (NHRI-EX113-11137PI). The funders had no role in the study design, in the collection, analysis and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication. No other author has professional or financial relationships with any companies that are relevant to this study. There are no other conflicts of interest or sources of funding to declare.
REFERENCES
- 1.Laupland KB, Gregson DB, Flemons WW, et al. Burden of community-onset bloodstream infection: a population-based assessment. Epidemiol Infect. 2007;135:1037–42. doi: 10.1017/S0950268806007631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Søgaard M, Nørgaard M, Dethlefsen C, et al. Temporal changes in the incidence and 30-day mortality associated with bacteremia in hospitalized patients from 1992 through 2006: a population-based cohort study. Clin Infect Dis. 2011 Jan;52:61–9. doi: 10.1093/cid/ciq069. [DOI] [PubMed] [Google Scholar]
- 3.Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect. 2013;19:501–9. doi: 10.1111/1469-0691.12195. [DOI] [PubMed] [Google Scholar]
- 4.Fabre V, Sharara SL, Salinas AB, et al. Does this patient need blood cultures? A scoping review of indications for blood cultures in adult nonneutropenic inpatients. Clin Infect Dis. 2020;71:1339–47. doi: 10.1093/cid/ciaa039. [DOI] [PubMed] [Google Scholar]
- 5.Epstein D, Raveh D, Schlesinger Y, et al. Adult patients with occult bacteremia discharged from the emergency department: epidemiological and clinical characteristics. Clin Infect Dis. 2001;32:559–65. doi: 10.1086/318699. [DOI] [PubMed] [Google Scholar]
- 6.Fu CM, Tseng WP, Chiang WC, et al. Occult Staphylococcus aureus bacteremia in adult emergency department patients: rare but important. Clin Infect Dis. 2012;54:1536–44. doi: 10.1093/cid/cis214. [DOI] [PubMed] [Google Scholar]
- 7.Chang EK, Kao KL, Tsai MS, et al. Occult Klebsiella pneumoniae bacteremia at emergency department: a single center experience. J Microbiol Immunol Infect. 2015;48:684–91. doi: 10.1016/j.jmii.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 8.de Jager CP, van Wijk PT, Mathoera RB, et al. Lymphocytopenia and neutrophil-lymphocyte count ratio predict bacteremia better than conventional infection markers in an emergency care unit. Crit Care. 2010;14:R192. doi: 10.1186/cc9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wildi K, Tschudin-Sutter S, Dell-Kuster S, et al. Factors associated with positive blood cultures in outpatients with suspected bacteremia. Eur J Clin Microbiol Infect Dis. 2011;30:1615–9. doi: 10.1007/s10096-011-1268-0. [DOI] [PubMed] [Google Scholar]
- 10.Müller F, Christ-Crain M, Bregenzer T, et al. Procalcitonin levels predict bacteremia in patients with community-acquired pneumonia: a prospective cohort trial. Chest. 2010;138:121–9. doi: 10.1378/chest.09-2920. [DOI] [PubMed] [Google Scholar]
- 11.Shapiro NI, Wolfe RE, Wright SB, et al. Who needs a blood culture? A prospectively derived and validated prediction rule. J Emerg Med. 2008;5:255–64. doi: 10.1016/j.jemermed.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Su CP, Chen TH, Chen SY, et al. Predictive model for bacteremia in adult patients with blood cultures performed at the emergency department: a preliminary report. J Microbiol Immunol Infect. 2011;44:449–55. doi: 10.1016/j.jmii.2011.04.006. ht. [DOI] [PubMed] [Google Scholar]
- 13.Lee CC, Wu CJ, Chi CH, et al. Prediction of community-onset bacteremia among febrile adults visiting an emergency department: rigor matters. Diagn Microbiol Infect Dis. 2012;73:168–73. doi: 10.1016/j.diagmicrobio.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Chase M, Klasco RS, Joyce NR, et al. Predictors of bacteremia in emergency department patients with suspected infection. Am J Emerg Med. 2012;30:1691–7. doi: 10.1016/j.ajem.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Laukemann S, Kasper N, Kulkarni P, et al. Can we reduce negative blood cultures with clinical scores and blood markers? Results from an observational cohort study. Medicine (Baltimore) 2015;94:e2264. doi: 10.1097/MD.0000000000002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhavani SV, Lonjers Z, Carey KA, et al. The development and validation of a machine learning model to predict bacteremia and fungemia in hospitalized patients using electronic health record data. Crit Care Med. 2020;48:e1020–8. doi: 10.1097/CCM.0000000000004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roimi M, Neuberger A, Shrot A, et al. Early diagnosis of bloodstream infections in the intensive care unit using machine-learning algorithms. Intensive Care Med. 2020;46:454–62. doi: 10.1007/s00134-019-05876-8. [DOI] [PubMed] [Google Scholar]
- 18.Mahmoud E, Al Dhoayan M, Bosaeed M, et al. Developing machine-learning prediction algorithm for bacteremia in admitted patients. Infect Drug Resist. 2021;14:757–65. doi: 10.2147/IDR.S293496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee KH, Dong JJ, Kim S, et al. Prediction of bacteremia based on 12-year medical data using a machine learning approach: effect of medical data by extraction time. Diagnostics (Basel) 2022;12:102. doi: 10.3390/diagnostics12010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi DH, Hong KJ, Park JH, et al. Prediction of bacteremia at the emergency department during triage and disposition stages using machine learning models. Am J Emerg Med. 2022;53:86–93. doi: 10.1016/j.ajem.2021.12.065. [DOI] [PubMed] [Google Scholar]
- 21.Goh V, Chou YJ, Lee CC, et al. Predicting Bacteremia among septic patients based on ED Information by machine learning methods: a comparative study. Diagnostics (Basel) 2022;12:2498. doi: 10.3390/diagnostics12102498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coburn B, Morris AM, Tomlinson G, et al. Does this adult patient with suspected bacteremia require blood cultures? JAMA. 2012;308:502–11. doi: 10.1001/jama.2012.8262. [DOI] [PubMed] [Google Scholar]
- 23.Norgeot B, Quer G, Beaulieu-Jones BK, et al. Minimum information about clinical artificial intelligence modeling: the MI-CLAIM checklist. Nat Med. 2020;26:1320–4. doi: 10.1038/s41591-020-1041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng CJ, Yen ZS, Tsai JC, et al. Validation of the Taiwan triage and acuity scale: a new computerised five-level triage system. Emerg Med J. 2011;28:1026–31. doi: 10.1136/emj.2010.094185. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. International Statistical Classification of Diseases and Related Health Problems 10th Revision. 2019. [Accessed September 9, 2023]. Available at: https://icd.who.int/browse10/2019/en.
- 26.Hall KK, Lyman JA. Updated review of blood culture contamination. Clin Microbiol Rev. 2006;19:788–802. doi: 10.1128/CMR.00062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst. 2017;30:4765–74. [Google Scholar]
- 28.Pfitzenmeyer P, Decrey H, Auckenthaler R, et al. Predicting bacteremia in older patients. J Am Geriatr Soc. 1995;43:230–5. doi: 10.1111/j.1532-5415.1995.tb07327.x. [DOI] [PubMed] [Google Scholar]
- 29.Schuttevaer R, Boogers W, Brink A, et al. Predictive performance of comorbidity for 30-day and 1-year mortality in patients with bloodstream infection visiting the emergency department: a retrospective cohort study. BMJ Open. 2022;12:e057196. doi: 10.1136/bmjopen-2021-057196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CH, Lien CJ, Huang YS, et al. A simplified scoring model for predicting bacteremia in the unscheduled emergency department revisits: the SADFUL score. J Microbiol Immunol Infect. 2023;56:793–801. doi: 10.1016/j.jmii.2023.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


