Abstract
Bipolar disorder is a heritable mental illness with complex etiology. While the largest published genome-wide association study identified 64 bipolar disorder risk loci, the causal SNPs and genes within these loci remain unknown. We applied a suite of statistical and functional fine-mapping methods to these loci and prioritized 17 likely causal SNPs for bipolar disorder. We mapped these SNPs to genes and investigated their likely functional consequences by integrating variant annotations, brain cell-type epigenomic annotations, brain quantitative trait loci and results from rare variant exome sequencing in bipolar disorder. Convergent lines of evidence supported the roles of genes involved in neurotransmission and neurodevelopment, including SCN2A, TRANK1, DCLK3, INSYN2B, SYNE1, THSD7A, CACNA1B, TUBBP5, FKBP2, RASGRP1, FURIN, FES, MED24 and THRA among others in bipolar disorder. These represent promising candidates for functional experiments to understand biological mechanisms and therapeutic potential. Additionally, we demonstrated that fine-mapping effect sizes can improve performance of bipolar disorder polygenic risk scores across diverse populations and present a high-throughput fine-mapping pipeline.
Subject terms: Bipolar disorder, Genomics, Genome-wide association studies
This study used fine-mapping to analyze genetic regions associated with bipolar disorder, identifying specific risk genes and providing new insights into the biology of the condition that may guide future research and treatment approaches.
Main
Bipolar disorder (BD) is a heritable mental illness with complex etiology1. Heritability estimates from twin studies range between 60% and 90%2–4, while SNP-based heritability (h2SNP) calculations suggest that common genetic variants can explain up to 20% of the phenotypic variance of BD5. Genome-wide association studies (GWAS) of common variants have been successful in identifying associated genetic risk loci for BD5–15. For example, the largest published BD GWAS to date, conducted by the Psychiatric Genomics Consortium (PGC), comprised more than 40,000 BD cases and 370,000 controls from 57 cohorts of European ancestry, and identified 64 genome-wide significant (GWS) risk loci16. However, identifying the causal SNPs within these loci (that is, SNPs responsible for the association signal at a locus and with a biological effect on the phenotype, as opposed to those associated owing to linkage disequilibrium (LD) with a causal variant) is a major challenge.
Computational fine-mapping methods aim to identify independent causal variants within a genomic locus by modeling LD structure, SNP association statistics, number of causal variants and/or prior probabilities of causality based on functional annotations. There are a variety of fine-mapping models ranging from regression to Bayesian methods, with different strengths and limitations17–19. For example, the sum of single effects (SuSiE) model uses iterative Bayesian selection with posterior probabilities20, FINEMAP uses a stochastic search algorithm for SNP combinations21, and POLYgenic FUNctionally-informed fine-mapping (PolyFun) computes functional priors to improve fine-mapping accuracy18,22. Bayesian fine-mapping methods typically generate a posterior inclusion probability (PIP) of causality per SNP and ‘credible sets’ of SNPs, which represent the minimum set of SNPs with a specified probability of including the causal variant(s). Many methods can assume one or multiple causal variants per locus and can now be applied to GWAS summary statistics from large and well-powered studies. This is highly advantageous for fine-mapping GWAS meta-analyses; however, the specification of appropriate LD structure is crucial for accurate fine-mapping. When LD cannot be obtained from the original cohort(s) (for example, owing to data access restrictions), it should instead be obtained from a sufficiently large sample that is ancestrally similar to the GWAS population23.
Fine-mapping methods have recently been applied to GWAS of psychiatric disorders. For example, a recent study using FINEMAP and integrating functional genomic data identified more than 100 genes likely to underpin associations in risk loci for schizophrenia24. Several fine-mapped candidates had particularly strong support for their pathogenic role in schizophrenia owing to convergence with rare variant associations24. Here we use a suite of tools to conduct statistical and functional fine-mapping of 64 GWS risk loci for BD16 and assess the impact of the LD reference panel and fine-mapping window specifications. We link the likely causal SNPs to their relevant genes and investigate their potential functional consequences by integrating functional genomic data, including brain cell-type-specific epigenomic annotations and quantitative trait loci data. We also fine-mapped the major histocompatibility complex (MHC) separately by imputing human leukocyte antigen (HLA) variants, and assessed the effect of fine-mapping on polygenic risk score (PRS) predictions. Finally, we present a comprehensive fine-mapping pipeline implemented via Snakemake25 as a rapid, scalable and cost-effective approach to prioritize likely variants from GWS risk loci. This strategy yielded promising candidate genes for future experiments to understand the mechanisms by which they increase the risk of BD.
Results
Fine-mapping identifies likely causal BD variants
Stepwise conditional analyses using the COJO tool from the Genome-Wide Complex Trait Analysis software (GCTA-COJO) were performed in each of the 64 PGC3 BD GWS loci (Supplementary Table 2), conditioning associations on their top lead SNP and any subsequent conditionally independent associations, to identify loci that contained independent signals (conditional P < 5 × 10−6). This analysis supported the existence of one association signal at 62 loci (Supplementary Table 3) and two independent association signals within the MSRA locus on chromosome 8 and the RP1-84O15.2 locus on chromosome 8 (Supplementary Table 3).
Excluding the MHC, GWS loci were fine-mapped via a suite of Bayesian fine-mapping tools (SuSiE, FINEMAP, PolyFun + SuSiE and PolyFun + FINEMAP) to prioritize SNPs likely to be causal for BD and examine the impact of different LD reference options (Methods and Fig. 1). Figure 2 shows the number of SNPs with a PIP > 0.95 and PIP > 0.50 in each fine-mapping analysis, alongside the Jaccard index of concordance in results between each pair of the 16 fine-mapping analyses, calculated based on SNPs with PIP > 0.5 and part of a 95% credible set. Jaccard indices ranged from 0.25 to 1 (mean = 0.54, s.d. = 0.20), with higher values indicating more similar fine-mapping results (Fig. 2). A breakdown of the Jaccard indices for analyses grouped by LD option, statistical or functional fine-mapping and fine-mapping method are provided in the Supplementary Fig. 2.
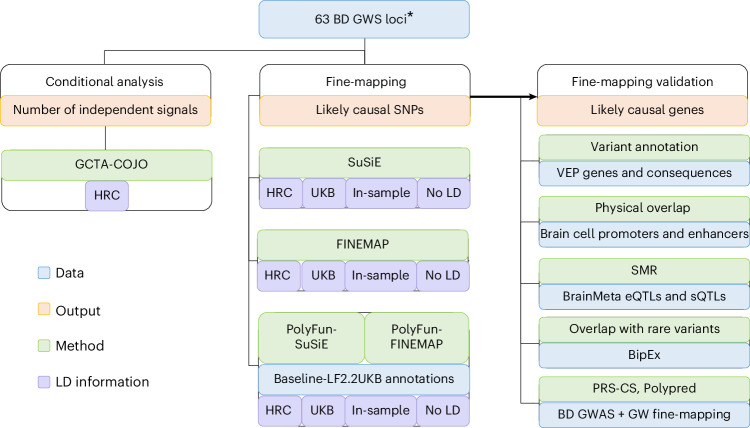
Fig. 1. Schematic workflow of the fine-mapping pipeline developed for PGC3 BD GWAS risk loci.
Conditional analyses were performed within GWS loci using GCTA-COJO, based on the LD structure of the Haplotype Reference Consortium (HRC) reference panel. Fine-mapping was conducted using statistical (SuSiE and FINEMAP) and functionally-informed (PolyFun) methods, according to the LD structure of the HRC, UK Biobank (UKB) and a subset of the GWAS data (‘in-sample LD’), as well as implementing single-variant (no LD) fine-mapping. PolyFun functional priors were based on the published baseline-LF2.2 UKB model22. Fine-mapping results were validated computationally via VEP annotations and functional consequences, overlap with epigenomic peaks from brain cell types, SMR with brain expression, splicing and methylation QTL data, convergence with rare variant associations from the BipEx sequencing collaboration and testing whether fine-mapping effect sizes improve PRSs (PRS-CS and PolyPred). Asterisk indicates that the MHC was fine-mapped using separate procedures (see ‘Fine-mapping the MHC locus’ section). VEP, variant effect predictor.
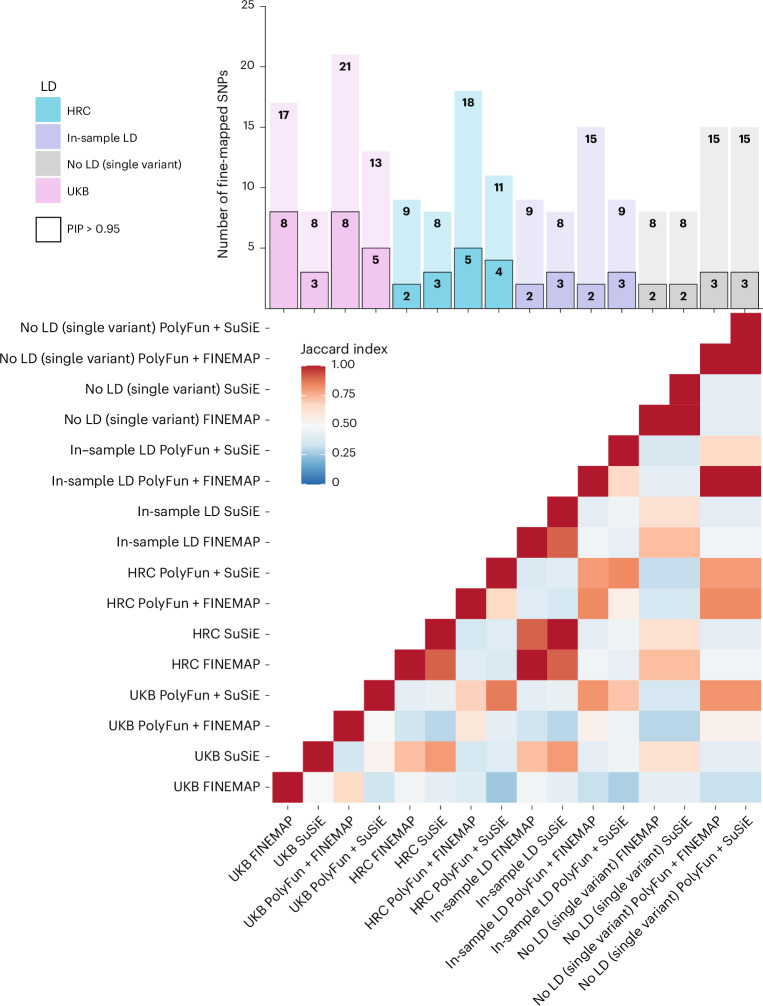
Fig. 2. Results and comparison of 16 fine-mapping analyses conducted.
The barplot displays the number of SNPs fine-mapped with PIP > 0.5 and part of a 95% credible set on the y axis and each fine-mapping analysis on the x axis. The black bordered bars indicate the number of SNPs fine-mapped with PIP > 0.95 and part of a 95% credible set. Each analysis is named according to (LD option)_(fine-mapping method). The heatmap displays the Jaccard index of concordance in results between each pair of fine-mapping analyses, calculated based on SNPs with PIP > 0.5 and part of a 95% credible set. Jaccard indices ranged from 0.25 to 1 (mean = 0.54, s.d. = 0.20), with higher values indicating more similar fine-mapping results.
Functional fine-mapping analyses yielded significantly more fine-mapped SNPs compared to the corresponding statistical fine-mapping analyses at PIP > 0.95 and PIP > 0.5 (P = 6.47 × 10−4 and P = 0.03, respectively; Fig. 2). There were no significant differences in the numbers of SNPs fine-mapped between the four LD options, between the two statistical fine-mapping methods or between the two functional fine-mapping methods. Approximately one-quarter of GWS loci (n = 16) had high PIP SNPs (>0.50). Using different fine-mapping methods and LD reference panels revealed a substantial number of consensus SNPs with PIP > 0.50 (17 SNPs), but fewer met the stricter threshold of PIP > 0.95 (6 SNPs; Fig. 3). The number of 95% credible sets per locus varied based on the fine-mapping method (Supplementary Fig. 3).
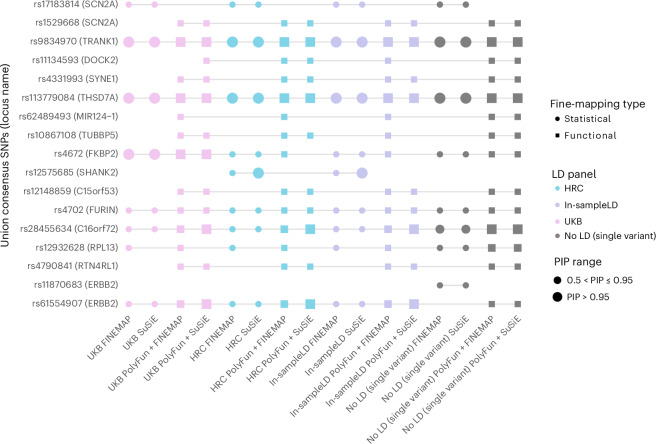
Fig. 3. Plot of union consensus SNPs across all 16 fine-mapping analyses, including different LD options and fine-mapping methods.
The color of the points corresponds to the LD option used—UKB (pink), HRC (blue), in-sample LD (purple) and no LD (single-variant fine-mapping; gray). Circles indicate statistical fine-mapping methods and squares indicate functional fine-mapping methods. Small shapes denote SNPs with PIP between 0.50 and 0.90, while large shapes denote SNPs with PIP above 0.95. On the x axis, analyses are named according to (LD option)_(fine-mapping method). On the y axis, the PGC3 locus name is displayed in parentheses after each fine-mapped SNP and indicates the name assigned to identify the locus in the original PGC3 BD GWAS publication, which is not necessarily the causal gene.
The smallest 95% CS per locus for every fine-mapping method and LD reference panel (Supplementary Fig. 3) was also calculated. Approximately one of five (n = 10–19) or half (n = 32–41) of the 63 fine-mapped loci had 95% CSs with a small number of SNPs (nSNPs < 10). The percentage of fine-mapped loci harboring 95% CSs with nSNPs < 10 was dependent on the fine-mapping method, with FINEMAP and PolyFun + FINEMAP harboring smaller 95% CSs and SuSiE and PolyFun + SuSiE larger 95% CSs.
The union consensus set (PIP > 0.5) comprised 17 SNPs (from 16 GWS loci), indicating that many of the same SNPs were prioritized regardless of which LD reference panel was used (Fig. 3). There were 15 SNPs consistently prioritized as the likely causal variant across all LD options (Fig. 3 and Supplementary Fig. 4). Notably, while rs11870683 met consensus SNP criteria, it was only prioritized using single-variant (no LD) fine-mapping, and the multivariant fine-mapping methods were unable to resolve the signal in this locus (Fig. 3). The distribution of SNPs with PIP > 0.50 for each GWS locus across different methods and LD options is provided in the Supplementary Fig. 4.
Variant annotation of the union consensus SNPs via Variant Effect Predictor26 indicated that 5 of the 17 fall in intronic regions (Supplementary Table 4). Two of the union consensus SNPs are missense variants—rs17183814 in SCN2A (Combined Annotation Dependent Depletion (CADD): 20, ClinVar benign for seizures and developmental and epileptic encephalopathy) and rs4672 in FKBP2 (CADD: 22.5, not in ClinVar). More details about the variant annotations of the union consensus SNPs through different online databases are provided in Supplementary Table 4.
QTL integrative analyses and overlap with epigenomic peaks
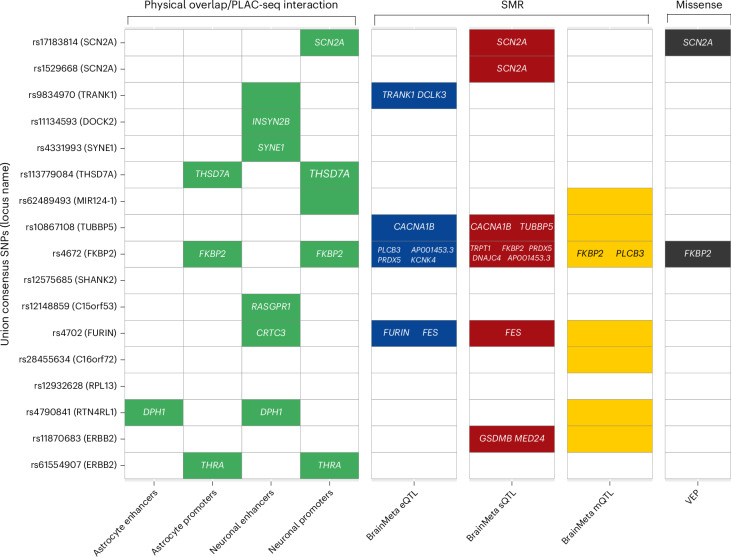
Summary data-based Mendelian randomization (SMR)27,28 was used to identify putative causal relationships between union consensus SNPs and BD via gene expression, splicing or methylation, by integrating the BD GWAS association statistics with brain expression quantitative trait locus (eQTL), splicing QTL (sQTL) and methylation QTL (mQTL) summary statistics. eQTL and sQTL data were based on the BrainMeta study (2,865 brain cortex samples from 2,443 unrelated individuals of EUR ancestry)29 and mQTL data were from the Brain-mMeta study (adult cortex or fetal brain samples in 1,160 individuals)30. Union consensus SNPs with GWS cis-QTL P values (P < 5 × 10−8) and their corresponding gene expression, slicing or methylation probes were selected as target SNP–probe pairs for SMR, yielding 13, 57 and 40 SNP–probe pairs for eQTL, sQTL and mQTL analyses, respectively. In the eQTL analyses, five union consensus SNPs with significant PSMR passed the HEIDI (heterogeneity in dependent instruments) test for nine different genes, suggesting that their effect on BD is mediated via gene expression in the brain (Fig. 4 and Supplementary Table 5). Three of the union consensus SNPs showed evidence of causal effects on BD via expression of more than one gene in their cis region. In the sQTL analyses, there were six union consensus SNPs with significant PSMR results, and passing the HEIDI test, implicating 11 genes (Fig. 4 and Supplementary Table 5). In the mQTL analyses, there were 20 SNP–probe pairs passing the PSMR and PHEIDI thresholds, of which two methylation probes were annotated to specific genes (FKBP2 and PLCB3; Fig. 4 and Supplementary Table 5).
Fig. 4. Summary of analyses performed to link each fine-mapped SNP to the relevant gene(s).
The y axis shows the 17 union consensus SNPs with the PGC3 locus name displayed in parentheses after each one, which indicates the name assigned to identify the locus in the original PGC3 BD GWAS publication and not necessarily the causal gene. On the x axis, the columns depict the results of eight analyses performed to link the fine-mapped SNPs to the relevant gene(s). The analysis method and the dataset used are labeled above and below the figure, respectively. Colored cells denote significant results and the relevant gene names are printed within each cell. For fine-mapped SNPs located in active enhancers, the relevant genes were obtained using data on PLAC-seq interactions with gene promoters. A colored cell includes no gene name when there was no known interaction between the enhancer and a promoter, or when the methylation probe was not annotated to any gene. Empty cells are those with nonsignificant results or where the SNP was not present in the dataset used.
There were 11 union consensus SNPs that physically overlapped with active enhancers or promoters of gene expression in brain cell types31, particularly neurons (Fig. 4). Four union consensus SNPs were located in active promoters of the SCN2A, THSD7A, FKBP2 and THRA genes. We explored enhancer–promoter interactions using PLAC-seq data, specifically for enhancers in which there is a physical overlap with the union consensus SNPs, and prioritized their genes (Fig. 4). Among the implicated target genes through enhancer–promoter interactions are INSYN2B, SYNE1, RASGRP1, CRTC3, DPH1 and THRA.
Candidate risk genes based on convergence of evidence
By aggregating multiple lines of fine-mapping validation evidence, we present results for high-confidence genes for BD. Specifically, a gene was characterized as high-confidence if it was linked to a fine-mapped SNP via active promoters or enhancers, brain gene expression, splicing or methylation, or if the fine-mapped SNP was a missense variant (Fig. 4 and Supplementary Fig. 5). Assuming that a single variant may act through multiple risk genes, we took the union of the prioritized genes across the different lines of evidence described above. Together, the data support the roles of the following 23 genes in BD: SCN2A, TRANK1, DCLK3, INSYN2B, SYNE1, THSD7A, CACNA1B, TUBBP5, PLCB3, AP001453.3, PRDX5, KCNK4, CRTC3, TRPT1, FKBP2, DNAJC4, RASGRP1, FURIN, FES, DPH1, GSDMB, MED24 and THRA (Supplementary Table 6). Supplementary Fig. 5 provides multitrack locus plots depicting GWAS association statistics, fine-mapping results, overlap with epigenomic peaks from neurons or astrocytes and gene tracks for the majority of GWS loci. We assessed the high-confidence genes for evidence of rare variant associations with BD, using data from the Bipolar Exome (BipEx) collaboration study32. Among the 23 genes examined, THSD7A, CACNA1B, SCN2A and TRANK1 had a significant burden (P < 0.05) of damaging missense or loss-of-function (LoF) variants in BD versus controls. Many high-confidence genes were classified as druggable based on the Open Targets platform (SCN2A, CACNA1B, PRDX5, THRA, MED24, SYNE1, KCNK4, FKBP2, RASGRP1, PLCB3, DCLK3, FURIN and FES). Detailed literature information about the biological relevance of the high-confidence genes can be found in the Supplementary Table 6.
Dissecting the MHC locus
In the original GWAS, the most significant SNP in the extended MHC was rs13195402 (26.4 Mb, P = 5.8 × 10−15), which is a missense variant in BTN2A1. Conditional analysis on this SNP suggested a single association signal across the extended MHC, and there were no associations between structural haplotypes of the complement component four genes (C4A/C4B; ~31.9 Mb) and BD16. Here, we performed association analyses of variants in the MHC region (chromosome 6, 29–34 Mb) including HLA alleles, amino acids, SNPs and insertion/ deletion variants, in a sample of 33,781 BD cases and 53,869 controls. The most significant variant in the classical MHC was rs1541269 (30.1 Mb, P = 6.71 × 10−12, LD r2 = 0.55 with the original index SNP rs13195402 in European populations)16. While initially some variants in HLA genes reached GWS (Supplementary Table 7), none remained after conditioning on rs1541269, suggesting the associations were driven by LD with more strongly associated variants located upstream (Supplementary Fig. 6 and Supplementary Table 8).
Leveraging fine-mapping to improve BD PRSs
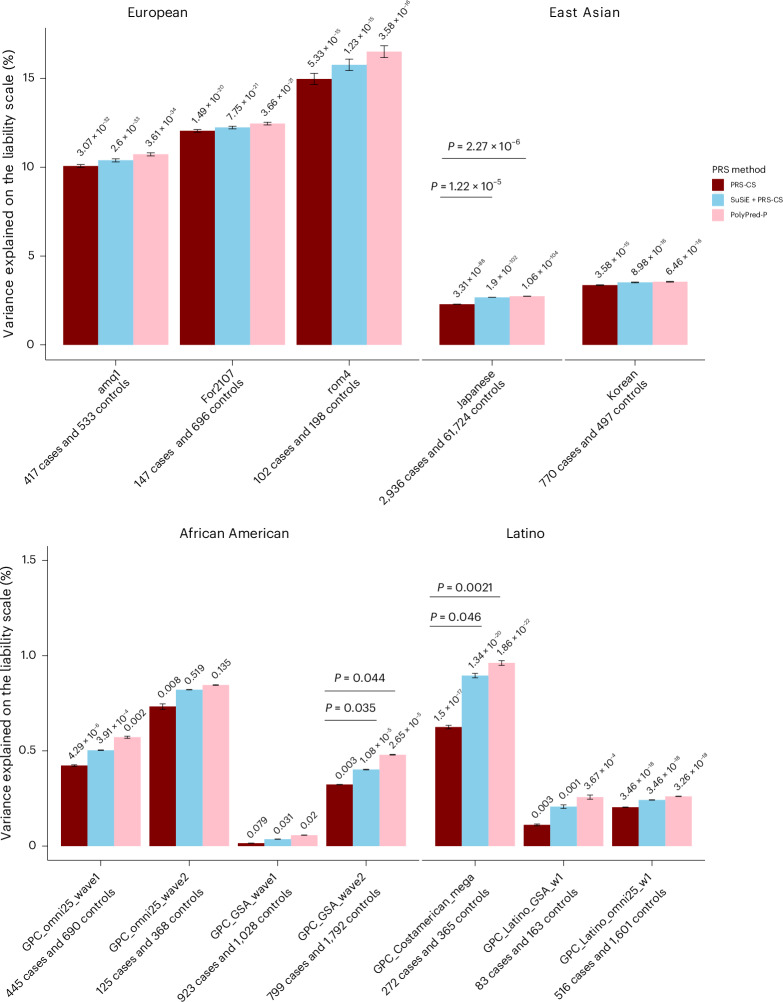
We assessed whether fine-mapping results could be used to improve the performance of BD PRS in twelve testing cohorts: three EUR cohorts that were independent of the PGC3 BD GWAS, two East Asian cohorts, four admixed African American cohorts, and three Latino cohorts33–35. Standard PRS were calculated using the PRS-CS method, and fine-mapping informed PRS were calculated via PolyPred, to integrate statistical fine-mapping results (SuSiE + PRS-CS) or functional fine-mapping results (PolyPred-P). Across PRS methods, PRS were substantially higher in BD cases versus controls in all EUR target cohorts and most non-EUR cohorts (Fig. 5 and Supplementary Tables 9). Using PRS-CS, the effective sample size-weighted phenotypic variance explained on the liability scale was 12.26% in EUR ancestries, 2.41% in East Asian ancestries, 0.20% in African American ancestries and 0.28% in Latino ancestries (Fig. 5 and Supplementary Table 10). Examining fine-mapping-informed PRS, SuSiE + PRS-CS or PolyPred-P explained more phenotypic variance than PRS-CS in all cohorts, with PolyPred-P showing the best performance (Fig. 5). However, increased variance explained by SuSiE + PRS-CS or PolyPred-P compared with PRS-CS was only statistically significant in the Japanese BD cohort (P = 1.22 × 10−5 and P = 2.29 × 10−6, respectively), one African American (P = 0.035 and P = 0.044, respectively) and one Latino cohort (P = 0.046 and P = 0.002, respectively; Supplementary Table 9 and Fig. 5).
Fig. 5. Phenotypic variance in BD explained by standard PRS (PRS-CS) and fine-mapping-informed PRS (SuSiE + PRS-CS and PolyPred-P) in target cohorts of diverse genetic ancestries.
The x axis displays the target cohorts, grouped by genetic ancestry, and the PRS method used. The name of each cohort and the number of BD cases and controls are shown below each barplot. The y axis shows the percentage variance explained on the liability scale (assuming a 2% population prevalence of BD) with error bars indicating the 95% confidence interval around each R2 value. P values for the association of PRS with case versus control status are printed on top of each bar. Significant P values (P < 0.05) for the test of difference in variance explained by the fine-mapping informed PRS versus PRS-CS are provided above the horizontal lines, using the F test for nested models.
Discussion
In the most comprehensive fine-mapping study of BD GWAS risk loci to date, we applied a suite of statistical and functional fine-mapping methods to prioritize 17 likely causal SNPs for BD in 16 genomic loci. We linked these SNPs to genes and investigated their likely functional consequences by integrating variant annotations, brain cell-type epigenomic annotations and brain QTLs. Convergence of evidence across these analyses prioritized 23 high-confidence genes, which are strong candidates for functional validation experiments to understand the mechanisms by which they increase the risk of BD.
We defined a union consensus set of SNPs representing those likely causal for BD based on the convergence between fine-mapping methods and LD reference panels. This comprised 17 SNPs (from 16 GWS loci), indicating that many of the same SNPs were prioritized across fine-mapping analyses (Fig. 3). Linking these SNPs to genes and investigating their likely functional consequences using computational approaches and relevant datasets, prioritized 23 high-confidence genes (Fig. 4). Overall, we hypothesized that a single putative causal SNP may influence multiple genes due to various factors, such as the impact of enhancer elements on multiple genes’ expression, overlap of eQTLs and sQTLs with epigenomic annotations and missense variants, and overlapping genomic coordinates of genes29,36,37.
This study uncovered new insights into BD. Six of the genes prioritized have synaptic functions, including two with presynaptic and four with postsynaptic annotations. The functions of these genes encompass both cellular excitability (regulation of neurotransmitter levels and membrane potential) and cellular organization (arrangement of the actin cytoskeleton, endocytosis, and the postsynaptic specialization). Prioritized genes implicate a variety of neurotransmitters, both excitatory and inhibitory. These findings highlight the impact of BD risk variants on diverse aspects of synaptic signaling. Although all prioritized genes are expressed in the brain and most display enrichment of expression in several brain cell types, three of the genes prioritized have enhanced expression in cells of the gut, including gastric mucous secreting cells and proximal and distal enterocytes. These cells have roles in intestinal permeability, inflammation and the enteric nervous system, and our findings lend genetic support to the involvement of the microbiota–gut–brain axis in BD38. The PLCB3, KCNK4 and DPH1 genes prioritized have previously been linked to neurodevelopmental delay39–41, but not BD. Our study also provides new insights into the potential molecular mechanisms underlying known BD risk genes. For example, results suggest that fine-mapped variants impact BD through alternate splicing of SCN2A and CACNA1B in the brain, findings which may inform functional laboratory experiments.
In the MHC, there were several polymorphic alleles and amino acid variants in the HLA-C and HLA-B genes associated with BD at GWS (chromosome 6, 31.2–31.3 Mb). The HLA-C*07:01 and HLA-B*08:01 alleles were negatively associated with BD, in line with previous studies reporting their protective effects on schizophrenia42,43. However, these associations were removed after conditioning on the top lead variant in the MHC (rs1541269, 30 Mb), suggesting the effects were driven by LD with more strongly associated variants located upstream. This is consistent with published findings in the PGC BD data, showing no association between the structural variants in the complement component four genes (C4A/C4B, ~31.9 Mb) and BD, either before or after conditioning on the most associated MHC SNP (rs13195402, 26.4 Mb)16. Overall, this analysis of HLA variation in BD again suggests a single association signal across the MHC, and that the causal variants and genes are outside the classical MHC locus, in contrast to findings in schizophrenia44.
Fine-mapping-informed PRS, developed by combining GWAS effect sizes and genome-wide fine-mapping effect sizes using PolyPred, explained a greater proportion of phenotypic variance compared with PRS based on GWAS effect sizes alone. This adds support to our fine-mapping results, as leveraging information on causal effect sizes rather than relying solely on association statistics should improve genetic risk prediction. Under the assumption that the causal variants are shared across ancestries, we anticipated that fine-mapping-informed PRS would improve the transferability of BD PRS into diverse genetic ancestries. Indeed, there was a modest increase in the phenotypic variance explained relative to standard PRS in all genetic ancestry groups. However, the performance of all PRS in non-European cohorts still lagged greatly behind that in Europeans (Fig. 5 and Supplementary Tables 9 and 10), emphasizing the need for larger studies in diverse genetic ancestries and further development of methods to improve PRS transferability between ancestries.
Our strategy of applying a suite of fine-mapping methods and examining the convergence of the results was driven by the variety of the underlying fine-mapping algorithms and their corresponding strengths and limitations. Consistent with previous literature, we detected more SNPs with high PIPs when incorporating functional priors using PolyFun18. FINEMAP, using a shotgun stochastic algorithm, refines promising SNP sets efficiently by focusing on a subset with higher PIPs, making it well-suited for dense genomic data. By contrast, SuSiE’s Bayesian algorithm accommodates LD structure and identifies multiple causal signals within loci, offering credible sets that increase confidence in the discovered variants. As expected, the specification of LD structure, fine-mapping window and number of causal variants impacted fine-mapping results. Considering ‘in-sample’ LD from the PGC BD data (albeit a subset of available cohorts) as the gold standard, using the HRC reference panel yielded the most similar fine-mapping results. This observation may be explained by the HRC being used as an imputation reference panel for almost all cohorts in the GWAS (53/57 cohorts). Results suggest that a large and well-matched LD reference panel to the GWAS sample can be used to achieve high-quality fine-mapping results. This has advantageous implications in scenarios when calculating in-sample LD is not possible owing to data sharing restrictions, or when obtaining LD information from many cohorts becomes increasingly challenging as GWAS meta-analyses grow.
Although there were some differences in the number of SNPs fine-mapped (threshold of PIP > 0.5 and in a 95% credible set) by the same method using different LD options (Fig. 2), our strategy of requiring SNPs to be fine-mapped using two methods was used to safeguard against false positives. Moreover, although conditional analysis indicated one causal variant per GWS locus, our results are highly consistent when using LD reference panels and allowing up to five causal variants per GWS locus. The latter analyses also yielded a greater number of likely causal SNPs. As an exception, we note that one consensus SNP (rs11870683) was prioritized using single-variant (no LD) fine-mapping only, and we caution that there may be an additional or different causal SNP at this locus, since multivariant fine-mapping methods were unable to resolve the signal. To facilitate rapid and scalable fine-mapping of GWAS loci, we developed a fine-mapping pipeline (GitHub, https://github.com/mkoromina/SAFFARI) with options to specify multiple fine-mapping methods, GWAS summary statistics, fine-mapping windows and LD reference panels.
Several limitations of this study and future directions must be noted. First, our fine-mapping focused exclusively on EUR ancestry data, owing to the composition of the PGC3 BD GWAS. However, this enabled us to investigate the impact of LD reference panels on fine-mapping, which would be challenging for diverse ancestry data, given the limited availability of such panels at present. Increasing ancestral diversity in BD GWAS is an active area of research33 and in future, the differences in LD structure between populations could be leveraged to aid fine-mapping45 and PRS predictions46. Second, we approximated ‘in-sample LD’ of the GWAS as we only had access to a subset of the individual-level data (73% of the total effective sample size), we used best guess genotypes to represent imputed dosages and we merged genotypes across cohorts and calculated LD, in contrast to the GWAS, which was a meta-analysis between cohorts. Third, we applied a conservative approach focusing on SNPs with high PIPs (>0.50) that were part of credible sets and were supported by different fine-mapping methods. Thus, we prioritized likely causal variants or genes at 16 of the 64 GWS loci. The improvements in PRS performance after integrating genome-wide fine-mapping results suggest that our analyses capture meaningful information on causality in other genomic regions that did not meet the stringent criteria we applied to fine-map GWS loci. Fourth, these statistical analyses prioritize variants and genes with high probabilities of being causal risk factors for BD; however, computational approaches fall short of proving causality and have limited capacity to uncover mechanisms. Finally, the enhancer, promoter and QTL data used may be incomplete owing to cell-type or context-specific effects, or incomplete mapping of active enhancers to their target genes, and therefore some union consensus SNP effects may not have been detected in our analysis.
In summary, we conducted a comprehensive statistical and functional fine-mapping analysis of BD genomic loci, yielding a resource of likely causal genes and variants for the disorder. These genes and variants now require investigation in functional laboratory experiments to validate their roles, understand mechanisms of risk, and examine opportunities for therapeutic intervention in BD.
Methods
Ethics statement
Ethics approval was obtained from the ethics committees of the Medical Schools of the Universities of Marburg (approval identifier studie 07/2014) and Münster, in accordance with the Declaration of Helsinki, with all participants providing written informed consent.
GWAS summary statistics and BD risk loci
Summary statistics from the latest published BD GWAS by the Psychiatric Genomics Consortium (‘PGC3’ study) were used as input to the fine-mapping pipeline16. This GWAS comprised 41,917 BD cases and 371,549 controls of European (EUR) ancestry from 57 cohorts (Supplementary Table 1). Of these cohorts, 53 were imputed using the HRC EUR ancestry reference panel v1.0 (ref. 47). GWAS summary statistics were cleaned using DENTIST software48, yielding a total of 7,598,903 SNPs. The GWAS meta-analysis identified 64 independent loci associated with BD at GWS, which were selected for fine-mapping. Each GWS locus window was established around the GWS significant ‘top lead’ SNP (P < 5 × 10−8), with boundaries defined by the positions of the 3′-most and 5′-most SNPs, requiring an LD r2 > 0.1 with the top lead SNP within a 3 Mb range, according to the LD structure of the HRC EUR reference panel16. Owing to the complexity and long-range LD of the MHC/HLA region, this locus was analyzed separately (see ‘Fine-mapping the MHC locus’ section). Supplementary Table 2 shows the top lead SNP from each GWS locus, association statistics, locus boundaries, locus size, and locus names (as defined in the original GWAS)16. Excluding the MHC, GWS locus windows ranged between 14,960–3,730,000 bp in size.
Conditional analysis
Figure 1 shows an overview of the fine-mapping pipeline. First, conditional analyses were conducted using a stepwise selection procedure (--cojo-slct) via GCTA49,50 to explore potential independent association signals within each locus, according to the LD structure of the HRC EUR reference panel. In brief, this procedure iteratively adds SNPs to a conditional model until no conditional tests are significant (conditional P > 5 × 10−6)50 to estimate the number of independent association signals per locus.
LD reference panels
Statistical and functional fine-mapping methods require information on LD between variants and selection of a genomic region (‘window’) to fine-map. To examine the impact of LD on fine-mapping, analyses were performed using LD information from the HRC EUR reference panel, published LD matrices based on EUR ancestry individuals in the UK Biobank18, and ‘in-sample’ LD calculated from a subset of 48 BD cohorts in the PGC BD GWAS for which individual-level genetic data were available within the PGC (33,781 cases, 53,869 controls, all of EUR ancestry), representing 73% of the total effective sample size of the GWAS. In brief, HRC-imputed dosage data were converted to hard calls with a genotype call probability cut-off of 0.8 and PLINK binary files were merged across cohorts, restricting to the set of unrelated individuals included in the GWAS, using PLINK v1.90 (ref. 51). Missingness rates per SNP were calculated in each cohort, and SNPs absent in all individuals from any one cohort were excluded from the merged dataset, yielding 7,594,494 SNPs overlapping with the GWAS summary statistics. Individual-level genetic data per chromosome were used as an ‘in-sample’ LD reference panel for fine-mapping. We also performed single-variant fine-mapping without any LD.
Statistical and functional fine-mapping
GWS loci were fine-mapped using a suite of Bayesian fine-mapping methods that can be applied to GWAS summary statistics—SuSiE, FINEMAP, PolyFun + SuSiE and PolyFun + FINEMAP (Fig. 1). SuSiE and FINEMAP are statistical fine-mapping methods, while PolyFun incorporates functional annotations as prior probabilities to improve subsequent fine-mapping accuracy18,20,21. Since these methods have different underlying assumptions, strengths and limitations, results were compared to examine convergence of evidence across methods. Briefly, each Bayesian method generates SNP-wise posterior inclusion probabilities of causality (PIP), and a 95% credible set (95% CS), defined as the minimum subset of SNPs that cumulatively have at least 95% probability of containing the causal SNP(s). PIP refers to the marginal probability that a SNP is included in any causal model, conditional on the observed data, hence providing weight of evidence that a SNP should be considered potentially causal.
First, single-variant fine-mapping, which makes the simple assumption of one causal variant per locus (K = 1) and does not require LD information18,20,21, was performed within each GWS locus fine-mapping window. FINEMAP and SuSiE can assume multiple causal variants per locus, modeling the LD structure between them. Fine-mapping was additionally performed assuming the default maximum of five causal variants per locus (K = 5) and separately using the HRC, UKB and ‘in-sample’ LD structures. Finally, PolyFun was used to incorporate 187 published functional annotations from the baseline-LF2.2.UKB model22 to compute prior causal probabilities (priors) via an L2-regularized extension of stratified LD-score regression52, and subsequently perform fine-mapping using FINEMAP and SuSiE18. Briefly, functional annotations included epigenomic and genomic annotations, minor allele frequency (MAF) bins, binary or continuous functional annotations, LD-related annotations such as LD level, predicted allele age, recombination rate, and CpG content22. Functionally-informed fine-mapping was also performed using the three LD reference panels.
In total, 16 fine-mapping analyses were conducted (12 multivariant analyses using four fine-mapping methods and three LD reference panels and four LD-independent single-variant fine-mapping analyses), varying parameters to examine their impact and the convergence of results. We used the Jaccard index (or Jaccard similarity coefficient) to summarize the concordance in the results between pairs of fine-mapping analyses. The Jaccard index was calculated as the number of fine-mapped SNPs (PIP > 0.5 and in a 95% CS) in both fine-mapping methods (intersection), divided by the total number of fine-mapped SNPs across either method (union) and ranges from 0 (no concordance between the methods) to 1 (complete concordance between the methods). ‘Consensus SNPs’ were defined as those in the 95% CS from at least two methods (either statistical and/or functional fine-mapping) that used the same LD option and with a PIP > 0.95 or >0.50 (24 opportunities for a SNP to be a consensus SNP). The ‘union consensus’ set of SNPs was defined as all consensus SNPs across LD options PIP > 0.50, excluding SNPs identified only with the UKB LD reference panel. The number of SNPs fine-mapped at PIP > 0.95 and PIP > 0.50 between different methods and different LD options was compared using two-sided paired t tests.
All steps of the statistical and functional fine-mapping analyses have been compiled into a high-throughput pipeline named Statistical and Functional Fine-mapping Applied to GWAS Risk Loci (SAFFARI). SAFFARI is implemented through Snakemake in a Linux environment25, with options to provide sets of GWAS summary statistics, lists of fine-mapping windows, and to specify LD reference panels, in the form of LD matrices or individual-level genetic data (GitHub, https://github.com/mkoromina/SAFFARI).
Effect of LD options and locus windows on fine-mapping
We aimed to investigate the impact of using an LD reference panel for fine-mapping or performing single-variant fine-mapping with no LD compared with using LD information calculated from the original GWAS data. The latter is typically considered the gold-standard approach; however, it is difficult in practice due to data availability and sharing restrictions. We performed several comparative analyses, including calculating Jaccard indices and correlation of PIP values for fine-mapped SNPs, and found that the HRC reference panel, a panel that closely resembles the genetic ancestry of the GWAS, achieves comparable fine-mapping resolution with in-sample LD estimates (Supplementary Note). We also compared results from fine-mapping the GWS locus windows versus fixed 3 Mb windows, which indicated substantial differences between them, and that the GWS locus windows best represent the GWS association signals from the original GWAS (Supplementary Note).
Annotation of union consensus SNPs
Union consensus SNPs were characterized using the Variant Effect Predictor (GRCh37) Ensembl release 109 (ref. 26). When SNPs were mapped to multiple transcripts, the most severe variant consequence was retained for annotation, and when SNPs fell within intergenic or regulatory regions, no genes were annotated26. If annotated genes overlapped and the SNP had the same severity consequence, then both genes were annotated. Additional annotations included the CADD scores (https://cadd.gs.washington.edu/), which denote the likelihood of the variant being deleterious or disease-causing (CADD ≥ 20) and ClinVar annotations (https://www.ncbi.nlm.nih.gov/clinvar/) describing the association of variants with diseases (that is, benign, pathogenic, etc). Union consensus SNPs were further annotated with RegulomeDB (version 2.2) to determine whether they have functional consequences and lie in noncoding regions and to annotate them to the relevant regulatory elements53. RegulomeDB probability and ranking scores are positively correlated and predict functional variants in regulatory elements. Probability scores closer to 1 and ranking scores below 2 provide increased evidence of a variant to be in a functional region53. Probability of being LoF intolerant (pLI) and LoF observed/expected upper bound fraction (LOEUF) scores were retrieved from the Genome Aggregation Database (gnomAD) v4.0.0. Genes were classified as intolerant to LoF variants if LOEUF < 0.6 or pLI ≥ 0.9. We also used the Open Targets platform54 to detect druggable genes among our set of high-confidence genes for BD risk.
QTL integrative analyses
Union consensus SNPs were investigated for putative causal relationships with BD via brain gene expression, splicing or methylation, using SMR (version 1.03)27,28. Data on eQTLs and sQTLs were obtained from the BrainMeta study (version 2), which comprised RNA-sequencing data of 2,865 brain cortex samples from 2,443 unrelated individuals of EUR ancestry with genome-wide SNP data29. Data on mQTLs were obtained from the Brain-mMeta study30, a meta-analysis of adult cortex or fetal brain samples, comprising 1,160 individuals with methylation levels measured using the Illumina HumanMethylation450K array. We analyzed cis-QTLs, which were defined as those within 2 Mb of each gene29. Of the union consensus SNPs, ten were present in the BrainMeta QTL data and ten were present in the Brain-mMeta data. Using the BD GWAS16 and QTL summary statistics29, each union consensus SNP was analyzed as the target SNP for probes within a 2 Mb window on either side using the --extract-target-snp-probe option in SMR. Only probes for which the union consensus SNP was a GWS QTL (P < 5 × 10−8) were analyzed, to ensure robustly associated instruments for the SMR analysis27,28. A Bonferroni correction was applied for 13 tests in the eQTL (PSMR < 3.84 × 10−3), 57 tests in the sQTL (PSMR < 8.77 × 10−4) and 40 tests in the mQTL analyses (PSMR < 1.25 × 10−3). The significance threshold for the HEIDI test (heterogeneity in dependent instruments) was PHEIDI ≥ 0.01 (ref. 28). The HEIDI test is used to identify potential violations of the Mendelian randomization assumptions, specifically the assumption of no horizontal pleiotropy. An SNP passing the Bonferroni-corrected PSMR and the PHEIDI thresholds indicates either a direct causal role or a pleiotropic effect of the BD-associated SNPs on gene expression, splicing or methylation level.
Overlap with epigenomic peaks and rare variant association signal
Union consensus SNPs were examined for physical overlap with promoters or enhancers of gene expression in human brain cell types. Data on epigenomic peaks were obtained from purified bulk, H3K27ac and H3K4me3 ChIP–seq of neurons and astrocytes previously published and used to detect active promoters and enhancers31. Physical overlap was visually examined via locus plots using R (version 4.1.2). For SNPs located in promoters, we assigned the corresponding gene name. For active enhancers, the target gene was assigned based on PLAC-seq data31 on enhancer–promoter interactions. Genes linked to union consensus SNPs via overlap with epigenomic peaks, SMR, or missense annotations, were further assessed for convergence with findings from an exome sequencing study of BD published by the BipEx collaboration32. Using the BipEx browser32, genes annotated to union consensus SNPs were compared for an overlap against BipEx genes characterized by a significant (P < 0.05) burden of either damaging missense or LoF variants.
Fine-mapping the MHC locus
The MHC locus was fine-mapped separately due to its complex genetic variation and long-range LD structure55. The HLA alleles and amino acid variants were imputed in the PGC BD data, using the 1000 Genomes phase 3 reference panel comprising 503 EUR individuals56 with HLA alleles determined via sequencing. This reference was obtained from the CookHLA GitHub repository57 (CookHLA version 1.0.1) and included 151 HLA alleles (65 two-digit and 86 four-digit) with a MAF > 0.01 and <0.99, 1,213 amino acid variants, and 1,268 SNPs within the MHC region (chromosome 6, 29–34 Mb).
Variation in the MHC was imputed for 48 BD cohorts where individual-level genotyped SNP data were available within the PGC (33,827 BD and 53,953 controls), using IMPUTE2, implemented via the Rapid Imputation and Computational Pipeline for GWAS (RICOPILI)58. RICOPILI was used to perform association analysis, under an additive logistic regression model in PLINK v1.90 (ref. 51), covarying for the first five principal components of genetic ancestry and any others associated with case-control status within each cohort, as per the BD GWAS16. To control test statistic inflation at variants with low MAF in small cohorts, variants were retained only if cohort MAF was greater than 1% and minor allele count was greater than ten in either cases or controls (whichever had smaller n). Meta-analysis of the filtered association statistics was conducted using an inverse-variance-weighted fixed-effects model in METAL (version 25 March 2011) via RICOPILI59.
Conditional analysis of the MHC-association results was performed to identify whether there are any additional independent associations, by conditioning on the top lead variant within the locus. In brief, the dosage data for the top lead variant in the meta-analysis were extracted for each cohort, converted into a single value representing the dosage of the A1 allele (range = 0–2) and added as a covariate in the analysis. Association testing, filtering of results per cohort, and the meta-analysis were carried out as described above.
Polygenic risk scoring
Fine-mapping results were further evaluated by testing whether fine-mapping effect sizes could improve the performance of PRS in independent cohorts using PolyPred46, a method that combines effect sizes from fine-mapping with those from a standard PRS approach, such as PRS-CS60. PRS were calculated for individuals in 12 testing cohorts of BD cases and controls that were independent of the PGC3 BD GWAS: three new PGC cohorts of EUR ancestries, two cohorts of East Asian ancestries, four cohorts of admixed African American ancestries, and three cohorts of Latino ancestries, some of which have been described previously16 (Supplementary Note).
An analytical workflow outlining the steps of the PolyPred pipeline that we followed is shown in Supplementary Fig. 1. First, the standard approach used was PRS-CS, which uses a Bayesian regression framework to place continuous shrinkage priors on effect sizes of SNPs in the PRS, adaptive to the strength of their association signal in the BD GWAS16, and the LD structure from an external reference panel60. The UKB EUR ancestry reference panel was used to estimate LD between SNPs, matching the ancestry of the discovery GWAS16. PRS-CS yielded weights for approximately 1 million SNPs to be included in the PRS. Second, genome-wide fine-mapping was performed on the BD GWAS summary statistics16, using both SuSiE and PolyFun-SuSiE as previously described, with LD information obtained from the HRC reference panel, to derive causal effect sizes for all SNPs across the genome. Third, PolyPred was used to combine the SNP weights from PRS-CS with SuSiE effect sizes (SuSiE + PRS-CS) and SNP weights from PRS-CS with PolyFun-SuSiE effect sizes (PolyPred-P). In brief, Polypred ‘mixes’ the effect sizes from the two predictors via the non-negative least squares method, assigning a weight to each predictor that yields the optimally performing PRS in a specific testing cohort. Each testing cohort was used to tune the optimal PolyPred weights. Fourth, three PRS were calculated for each individual in the testing cohorts, using PLINK v1.90 (ref. 51) to weight SNPs by their effect sizes from PRS-CS, SuSiE + PRS-CS and PolyPred-P, respectively, and sum across all SNPs in each PRS. Finally, PRS were tested for association with case versus control status in each testing cohort using a logistic regression model including principal components as necessary to control for genetic ancestry33. In each testing cohort, the amount of phenotypic variance explained by the PRS (R2) and the 95% confidence intervals were calculated on the liability scale61, using the r2redux R package62, assuming a lifetime prevalence of BD in the general population of 2%. The R2 of each fine-mapping-informed PRS was statistically compared against the R2 of PRS-CS using the r2redux package (r2_diff function)62. In addition, we computed the effective sample size-weighted combined R2 values from PRS across different ancestries. Specifically, we transformed each R2 to a correlation coefficient, applied the Fisher z transformation, computed the effective sample size (neff)-weighted mean of the Fisher z values, and then back-transformed to obtain a combined R2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41593-025-01998-z.
Supplementary information
Supplementary Figs. 1–6 and Supplementary Note.
Supplementary Tables 1–10.
Acknowledgements
For the purposes of open access, the author has applied a Creative Commons Attribution (CC BY) license to any accepted author manuscript version arising from this submission. We thank the participants who contributed their time, life experiences and DNA to this research, and the clinical and scientific teams that worked with them. Statistical analyses were carried out on the NL Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and the Mount Sinai high-performance computing cluster (http://hpc.mssm.edu), which is supported by the Office of Research Infrastructure of the National Institutes of Health (awards S10OD018522 and S10OD026880). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Full acknowledgements are included in the Supplementary Note. The list of grants supporting authors of the current work is as follows: Baszucki Brain Research Fund via the Milken Institute Center for Strategic Philanthropy (to N.M.); US National Institute of Mental Health (PGC4: R01MH124839 to N.M. and O.A.A.); Medical Research Foundation (grant MRF-001-0012-RG-COLE-C0930 to J.R.I.C.); NIHR Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London (to J.R.I.C.); Consorcio Centro de Investigación Biomédica en Red (CIBER; CB07/09/0004 to E.V.); Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation (grants PI18/00805 and PI21/00787 (integrated into the Plan Nacional de I + D + I and cofinanced by ISCIII—Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER)) to E.V.); Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement (2021 SGR 01358 to E.V.); CERCA Programme (to E.V.); Department de Salut de la Generalitat de Catalunya for the PERIS (grant SLT006/17/00357 to E.V.); European Union Horizon 2020 research and innovation program—EU.3.1.1 (understanding health, wellbeing and disease—grant 754907 to E.V.) and EU.3.1.3 (treating and managing disease—grant 945151 to E.V.); CIBER (CB15/00154), Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation and grants PI18/01788 and PI22/00464: Integrated into the Plan Nacional de I + D + I and cofinanced by the ISCIII—Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER; to J.A.R.Q.); Instituto de Salud Carlos III (to J.A.R.Q.); Instituto de Salud Carlos III and NEURON BIOPHARMA, S.A (grant JTC2021 to J.A.R.Q.); Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement (2021 SGR 00840 to J.A.R.Q.); European Union Horizon 2020 research and innovation program (Eat2beNICE, grant 728018 and Timespan, grant 965381 to J.A.R.Q.); Australian National Health and Medical Research Council Investigator Grant (1177991 and 1176716 to P.B.M. and P.R.S., respectively); Japan Agency for Medical Research and Development (AMED; grants 22wm0425008, 21ek0109555, 21tm0424220, 21ck0106642, 23ek0410114 and 23tm0424225 to X.L., C.T., M.I. and N.I.); Japan Society for the Promotion of Science (JSPS) KAKENHI (grants 21H02854 and JP20H00462 to X.L., C.T., M.I. and N.I.) and German Research Foundation (DFG; grants FOR2107 KI588/14-1, KI588/14-2, KI588/20-1 and KI588/22-1 to T.L.; DA1151/5-1, DA1151/5-2, DA1151/9-1, DA1151/10-1 and DA1151/11-1 to U.D.; SFB-TRR393 to T.L. and U.D.). We also gratefully acknowledge the investigators who comprise the PGC and note that biosamples and corresponding data were sampled, processed and stored in the Marburg Biobank CBBMR.
Author contributions
M. Koromina conducted all fine-mapping and QTL analyses. A.R. replicated the results and assisted with pipeline parallelization. J.H., B.S. and T.R. provided useful feedback on the fine-mapping analyses. T.B., C.C., X.L. and J.K. computed PRSs for the non-European cohorts. G.P., A.B. and S.R. assisted with the analytical fine-mapping design of the MHC locus. N.M. conceived and supervised the study. The remaining authors have contributed data to the current project. M. Koromina and N.M. were responsible for the primary drafting and editing of the paper. All authors reviewed the manuscript critically for important intellectual content and approved the final version of the manuscript for publication.
Peer review
Peer review information
Nature Neuroscience thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Data availability
GWAS data were retrieved from ref. 16 from the following figshare link: https://figshare.com/articles/dataset/PGC3_bipolar_disorder_GWAS_summary_statistics/14102594. The PGC’s policy is to make genome-wide summary results public. All results are made available through the Figshare open access repository at the following DOI links: 10.6084/m9.figshare.27871677.v2, 10.6084/m9.figshare.27880524.v1, 10.6084/m9.figshare.27886110.v1. Data provided include MHC fine-mapping analyses of the PGC3 BIP study, as well as aggregated fine-mapping results using various methods (PolyFun + SuSiE, PolyFun + FINEMAP, SuSiE, FINEMAP) across four LD reference panels (UKB, HRC, LD, no LD) and GWS locus windows, provided in both .txt.gz and .merged.csv formats. Additional files include genome-wide fine-mapping results from SuSiE and PRS-CS protocols, and a detailed Excel file on credible sets for 12 fine-mapping analyses, specifying the SNPs and loci involved (10.6084/m9.figshare.28027706.v1).
Individual-level genetic data are accessible via secondary analysis proposals to the Bipolar Disorder Working Group of the PGC (https://www.med.unc.edu/pgc/shared-methods/how-to/). This study included some publicly available datasets accessed through dbGaP—PGC bundle phs001254.v1.p1.
Additional annotations were retrieved from the following databases: gnomAD database v4.0.0 (https://gnomad.broadinstitute.org), CADD (https://cadd.gs.washington.edu/) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
Code availability
Analysis scripts are available online at GitHub (https://github.com/mkoromina/SAFFARI). Additional scripts to recreate the visuals/graphs are available online at GitHub (https://github.com/Mullins-Lab/Post-finemap_processing/). Other software used include DENTIST (GitHub: https://github.com/Yves-CHEN/DENTIST), PolyPred (GitHub: https://github.com/omerwe/polyfun/wiki/6.-Trans-ethnic-polygenic-risk-prediction-with-PolyPred), PRS-CS (GitHub: https://github.com/getian107/PRScs), r2redux (GitHub: https://github.com/mommy003/r2redux) and RICOPILI (GitHub: https://github.com/Ripkelab/ricopili). All software used is publicly available at the URLs or references cited.
Competing interests
O.A.A. has served as a speaker for Janssen, Lundbeck and Sunovion, and as a consultant for Cortechs.ai. S.K.S. has served as speaker for Janssen, Takeda and Medice Arzneimittel Puetter GmbH & CoKG. E.V. has received grants and served as consultant, advisor or CME speaker for the following entities (unrelated to the present work): AB-Biotics, Abbott, AbbVie, Adamed, Angelini, Biogen, Biohaven, Boehringer Ingelheim, Casen-Recordati, Celon, Compass, Dainippon Sumitomo Pharma, Ethypharm, Ferrer, Gedeon Richter, GH Research, Glaxo Smith-Kline, Idorsia, Janssen, Johnson & Johnson, Lundbeck, Newron, Novartis, Organon, Otsuka, Rovi, Sage, Sanofi-Aventis, Sunovion, Takeda and Viatris. P.B.M. has received remuneration from Janssen (Australia) and Sanofi (Hangzhou) for lectures, and Janssen (Australia) for advisory board membership. M.O.D. and M.J.O. have received grants from Akrivia Health and Takeda Pharmaceuticals for work unrelated to this project. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/27/2025
A Correction to this paper has been published: 10.1038/s41593-025-02133-8
Contributor Information
Maria Koromina, Email: maria.koromina@mssm.edu.
Niamh Mullins, Email: niamh.mullins@mssm.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41593-025-01998-z.
References
- 1.O’Connell, K. S. & Coombes, B. J. Genetic contributions to bipolar disorder: current status and future directions. Psychol. Med.51, 2156–2167 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craddock, N. & Sklar, P. Genetics of bipolar disorder. Lancet381, 1654–1662 (2013). [DOI] [PubMed] [Google Scholar]
- 3.McGuffin, P. et al. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry60, 497–502 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Smoller, J. W. & Finn, C. T. Family, twin, and adoption studies of bipolar disorder. Am. J. Med. Genet. C Semin. Med. Genet.123C, 48–58 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Stahl, E. A. et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet.51, 793–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, D. T. et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol. Psychiatry18, 195–205 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Charney, A. W. et al. Evidence for genetic heterogeneity between clinical subtypes of bipolar disorder. Transl. Psychiatry7, e993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cichon, S. et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am. J. Hum. Genet.88, 372–381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreira, M. A. R. et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet.40, 1056–1058 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, E. K. et al. Association at SYNE1 in both bipolar disorder and recurrent major depression. Mol. Psychiatry18, 614–617 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Hou, L. et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum. Mol. Genet.25, 3383–3394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mühleisen, T. W. et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat. Commun.5, 3339 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Scott, L. J. et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc. Natl Acad. Sci. USA106, 7501–7506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulze, T. G. et al. Two variants in Ankyrin3 (ANK3) are independent genetic risk factors for bipolar disorder. Mol. Psychiatry14, 487–491 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith, E. N. et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol. Psychiatry14, 755–763 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet.53, 817–829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaid, D. J., Chen, W. & Larson, N. B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet.19, 491–504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weissbrod, O. et al. Functionally informed fine-mapping and polygenic localization of complex trait heritability. Nat. Genet.52, 1355–1363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schilder, B. M., Humphrey, J. & Raj, T. echolocatoR: an automated end-to-end statistical and functional genomic fine-mapping pipeline. Bioinformatics38, 536–539 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, G., Sarkar, A., Carbonetto, P. & Stephens, M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. Ser. B Stat. Methodol.82, 1273–1300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benner, C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics32, 1493–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazal, S. et al. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet.50, 1600–1607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanai, M. et al. Insights from complex trait fine-mapping across diverse populations. Preprint at medRxiv10.1101/2021.09.03.21262975 (2021).
- 24.Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature604, 502–508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mölder, F. et al. Sustainable data analysis with Snakemake. F1000Res.10, 33 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol.17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet.48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Wu, Y. et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat. Commun.9, 918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi, T. et al. Genetic control of RNA splicing and its distinct role in complex trait variation. Nat. Genet.54, 1355–1363 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi, T. et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun.9, 2282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nott, A. et al. Brain cell type-specific enhancer–promoter interactome maps and disease risk association. Science366, 1134–1139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer, D. S. et al. Exome sequencing in bipolar disorder identifies AKAP11 as a risk gene shared with schizophrenia. Nat. Genet.54, 541–547 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Connell, K. S. et al. Genomics yields biological and phenotypic insights into bipolar disorder. Nature639, 968–975 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda, M. et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol. Psychiatry23, 639–647 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moon, S. et al. The Korea Biobank Array: design and identification of coding variants associated with blood biochemical traits. Sci. Rep.9, 1382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet.20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Schoenfelder, S. & Fraser, P. Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet.20, 437–455 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Ortega, M. A. et al. Microbiota–gut–brain axis mechanisms in the complex network of bipolar disorders: potential clinical implications and translational opportunities. Mol. Psychiatry28, 2645–2673 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bayoumi, R. et al. Localisation of a gene for an autosomal recessive syndrome of macrocephaly, multiple epiphyseal dysplasia, and distinctive facies to chromosome 15q26. J. Med. Genet.38, 369–373 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gripp, K. W. et al. Syndromic disorders caused by gain-of-function variants in KCNH1, KCNK4, and KCNN3—a subgroup of K+ channelopathies. Eur. J. Hum. Genet.29, 1384–1395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urreizti, R. et al. DPH1 syndrome: two novel variants and structural and functional analyses of seven missense variants identified in syndromic patients. Eur. J. Hum. Genet.28, 64–75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andreassen, O. A. et al. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol. Psychiatry20, 207–214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature460, 748–752 (2009). [DOI] [PMC free article] [PubMed]
- 44.Sekar, A. et al. Schizophrenia risk from complex variation of complement component 4. Nature530, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan, K. et al. Fine-mapping across diverse ancestries drives the discovery of putative causal variants underlying human complex traits and diseases. Nat. Genet.56, 1841–1850 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weissbrod, O. et al. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet.54, 450–458 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet.48, 1279–1283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen, W. et al. Improved analyses of GWAS summary statistics by reducing data heterogeneity and errors. Nat. Commun.12, 7117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet.88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet44, 369–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet.81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet.47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong, S. et al. Annotating and prioritizing human non-coding variants with RegulomeDB v.2. Nat. Genet.55, 724–726 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ochoa, D. et al. The next-generation Open Targets Platform: reimagined, redesigned, rebuilt. Nucleic Acids Res.51, D1353–D1359 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet.50, 668–681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature526, 68–74 (2015). [DOI] [PMC free article] [PubMed]
- 57.Cook, S. et al. Accurate imputation of human leukocyte antigens with CookHLA. Nat. Commun.12, 1264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lam, M. et al. RICOPILI: Rapid Imputation for Consortias Pipeline. Bioinformatics36, 930–933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ge, T., Chen, C.-Y., Ni, Y., Feng, Y.-C. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun.10, 1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee, S. H., Goddard, M. E., Wray, N. R. & Visscher, P. M. A better coefficient of determination for genetic profile analysis. Genet. Epidemiol.36, 214–224 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Momin, M. M., Lee, S., Wray, N. R. & Lee, S. H. Significance tests for R of out-of-sample prediction using polygenic scores. Am. J. Hum. Genet.110, 349–358 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–6 and Supplementary Note.
Supplementary Tables 1–10.
Data Availability Statement
GWAS data were retrieved from ref. 16 from the following figshare link: https://figshare.com/articles/dataset/PGC3_bipolar_disorder_GWAS_summary_statistics/14102594. The PGC’s policy is to make genome-wide summary results public. All results are made available through the Figshare open access repository at the following DOI links: 10.6084/m9.figshare.27871677.v2, 10.6084/m9.figshare.27880524.v1, 10.6084/m9.figshare.27886110.v1. Data provided include MHC fine-mapping analyses of the PGC3 BIP study, as well as aggregated fine-mapping results using various methods (PolyFun + SuSiE, PolyFun + FINEMAP, SuSiE, FINEMAP) across four LD reference panels (UKB, HRC, LD, no LD) and GWS locus windows, provided in both .txt.gz and .merged.csv formats. Additional files include genome-wide fine-mapping results from SuSiE and PRS-CS protocols, and a detailed Excel file on credible sets for 12 fine-mapping analyses, specifying the SNPs and loci involved (10.6084/m9.figshare.28027706.v1).
Individual-level genetic data are accessible via secondary analysis proposals to the Bipolar Disorder Working Group of the PGC (https://www.med.unc.edu/pgc/shared-methods/how-to/). This study included some publicly available datasets accessed through dbGaP—PGC bundle phs001254.v1.p1.
Additional annotations were retrieved from the following databases: gnomAD database v4.0.0 (https://gnomad.broadinstitute.org), CADD (https://cadd.gs.washington.edu/) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
Analysis scripts are available online at GitHub (https://github.com/mkoromina/SAFFARI). Additional scripts to recreate the visuals/graphs are available online at GitHub (https://github.com/Mullins-Lab/Post-finemap_processing/). Other software used include DENTIST (GitHub: https://github.com/Yves-CHEN/DENTIST), PolyPred (GitHub: https://github.com/omerwe/polyfun/wiki/6.-Trans-ethnic-polygenic-risk-prediction-with-PolyPred), PRS-CS (GitHub: https://github.com/getian107/PRScs), r2redux (GitHub: https://github.com/mommy003/r2redux) and RICOPILI (GitHub: https://github.com/Ripkelab/ricopili). All software used is publicly available at the URLs or references cited.