Abstract
Background
This study aimed to describe the transmission of drug-resistant HIV-1 variants and phylogenetic characterisation of viral strains in mother–child pairs from the cities of Recife and São Luís, located in the Northeast region of Brazil, in 2007–2022.
Methods
This study included 15 mother–child pairs with confirmed vertical transmission of HIV-1. The genotyping sequences were provided by the Brazilian Ministry of Health. The analyses included descriptions of antiretroviral resistance mutation profiles of the mothers and children, subtype determination, and phylogenetic analyses.
Results
Seven mother–child pairs exhibited similar mutation profiles, with three showing no mutations and four displaying similar resistance mutations to the nucleoside reverse transcriptase inhibitor (NRTI) and non-nucleoside reverse transcriptase inhibitor (NNRTI) drug classes. Among four pairs, mutation similarities were observed for only one antiretroviral drug class. In the remaining four pairs, distinct mutation profiles were noted, with two children having mutations in two or three drug classes and their mothers exhibiting no or one mutation. Among the eight pairs with tests obtained within the first four years after birth, six of them had very similar mutation profiles. Among the seven pairs with exams obtained five years or more after birth, four pairs presented very different DRAM profiles. Mutations conferring resistance to efavirenz, nevirapine, lamivudine, abacavir, and didanosine were frequently observed in children and mothers. Thirteen pairs (86.6%) were identified as having HIV-1 subtype B, while two (13.3%) were identified as having HIV-1 subtype F1.
Conclusions
Differences in mutation profiles and antiretroviral resistance for NRTI and NNRTI drug classes were observed in half of the mother–child pairs, emphasising the importance of individualised therapeutic strategies.
Keywords: Antiretroviral resistance, Genotyping, Antiretroviral therapy (ART), HIV variants, Genetic sequencing
Background
Multiple lines of evidence have demonstrated a significant reduction in the vertical transmission of HIV, with many developed countries achieving a 0% annual transmission rate [1]. Despite this success, eliminating vertical HIV transmission globally remains challenging, particularly in low- and middle-income countries. The World Health Organization estimates that, in 2023, approximately 120,000 new HIV infections occurred in children worldwide; of them, 86% were in Sub-Saharan Africa [2].
In a meta-analysis of Brazilian studies published in 2019, the vertical HIV transmission rates in different regions of the country over the previous 10 years were 1.8–27.8%, reducing significantly over time. Regional variations reflect socioeconomic differences and the healthcare system of a country with vast territorial dimensions [3]. To date, only two Brazilian states (São Paulo and Paraná) have achieved the goal of eliminating vertical HIV transmission [4].
The impact of vertical HIV transmission extends beyond the simple transfer of the virus from the mother to the child; rather, it also involves the transmission of HIV variants that may be resistant to antiretroviral drugs. The presence of these drug-resistant variants in children is a complicating factor because it can limit antiretroviral therapy (ART) options and increase the risk of therapeutic failure [5].
Regarding drug resistance–associated mutations (DRAMs), Vaz et al. [6] reported a prevalence of 43.9% in the pol region among mother–child pairs with vertical HIV transmission in a study conducted in Northeast Brazil. The study also demonstrated the predominance of HIV-1 subtype B (82.1%) and recombinant BF (17.9%). Despite the high prevalence of DRAMs in mother–child pairs, Schultze et al. [7] and Santos-Pereira et al. [8] demonstrated that these mutations in children were associated with lower CD4 + T lymphocyte counts, higher HIV-1 viral loads, and a greater number of therapeutic regimens.
Although the transmission of drug-resistant HIV variants is a concern, evidence suggests that not all transmitted variants have been established in children. This phenomenon may be related to the effect of ART administered after HIV-1 transmission, which suppresses the replication of resistant variants and thereby preventing resistance progression [9]. The hypothesis that ART administered after HIV-1 transmission may have a suppressive effect on resistant variants is an area of growing interest with potential implications for optimising therapeutic regimens in HIV-exposed newborns. However, this hypothesis requires further validation through additional studies employing more sensitive sequencing techniques and analyses of proviral DNA and circulating viral RNA [10, 11].
This study aimed to characterise the antiretroviral resistance profiles of HIV-infected mother–child pairs. Individuals were monitored at HIV/AIDS referral services in Recife, Pernambuco, and São Luís, Maranhão, Brazil, in 2007–2022. Using genetic analyses of HIV variants, this study aimed to understand the transmission of resistant variants and how such dynamics influence the evolution of resistance in children.
Methods
Study design, location, and period
This study was conducted from October 2021 to September 2023 in São Luís, Maranhão, and Recife, Pernambuco, at the High-Risk Maternity Hospital in Imperatriz, Pediatric Infectious Disease Outpatient Clinic at the University Hospital of the Federal University of Maranhão, and Pediatric Infectious Disease Outpatient Clinic at the University Hospital Oswaldo Cruz of the University of Pernambuco.
Population
The study included 15 mother–child pairs diagnosed with HIV-1 infections who were under therapeutic follow-up at the selected reference units. The mothers included both treatment-naïve women diagnosed during pregnancy who underwent pretreatment genotyping and those living with HIV-1 during prior treatment who required genotyping due to virological failure.
Among the 15 mother–child pairs, eight (53%) were from Maranhão and seven (47%) from Pernambuco. Of the children, eight (53%) were male, and 10 (66.7%) started ART within the first 2 years of life. In one case (6.7%), no record of ART initiation was found. Nine children (60%) had initiated ART, including zidovudine (ZDV) + lamivudine (3TC) + nevirapine (NVP) in three (20%), ZDV + 3TC + lopinavir/ritonavir in two (13.3%), and ZDV + 3TC + efavirenz (EFV) in two (6.7%), while no treatment was described for three (20%). Mother–child pairs 1, 2, 5, and 7 received the same ART regimen (ZDV + 3TC + NVP). Mothers 3, 6, 8, 9, and 15 received EFV-based regimens (Supplementary Material).
Data collection
Data were obtained from secondary sources, including medical records from hospital and outpatient services where patients were followed. Genotypic sequences of the HIV pol region (protease/reverse transcriptase-PR/RT) were obtained using the ViroSeq Genotyping System, carried out in the laboratories of the National Genotyping Network (RENAGENO) coordinated by the Department of HIV/AIDS of the Brazilian Ministry of Health (GenBank accession number PV526713-PV526742).
Ethical aspects
This study was approved by the Research Ethics Committees of the Clinical Hospital of the Federal University of Pernambuco (HC/UFPE; CAAE 51758621.7.1001.8807) and the Federal University of Maranhão (UFMA; CAAE 57505422.6.0000.5086). Informed consent to participate was obtained from all of the participants or their guardians in the study.
Determination of HIV-1 subtypes and antiretroviral resistance
Preliminary determination of HIV-1 subtypes was performed using online reference tools such as HIV-1 COMET [12], HIVdb from Stanford University [13, 14], and the REGA HIV-1 Subtyping Tool [15]. HIV-1 resistance mutations were analysed using the HIV Drug Resistance Database platform from Stanford University (http://hivdb.stanford.edu) [16].
Phylogenetic analysis
The highest similarity was calculated considering the top 20 hits for each sequence used in the phylogenetic analysis. Since redundant sequences were removed, the final number of sequences per pair may vary. However, the similarity was determined by comparing the non-redundant sequences within this selected set. An initial alignment was performed using MAFFT [17] that incorporated the query sequences and those obtained from HIV BLAST [18] that exhibited the highest similarity. Maximum likelihood phylogenetic trees were constructed for each subtype, with clade support obtained through 1,200 bootstrap replications using the IQtree Web Server [19].
Recombination analysis
Recombination between subtypes was assessed using SIMPLOT software version 3.5.1 [11]. The following parameters were applied: nucleotide substitution model F84, 1,000 bootstrap replications, a sliding window of 200 bp with 20-bp steps, and a 90% confidence level. The transition/transversion ratio was empirically determined for each alignment.
Results
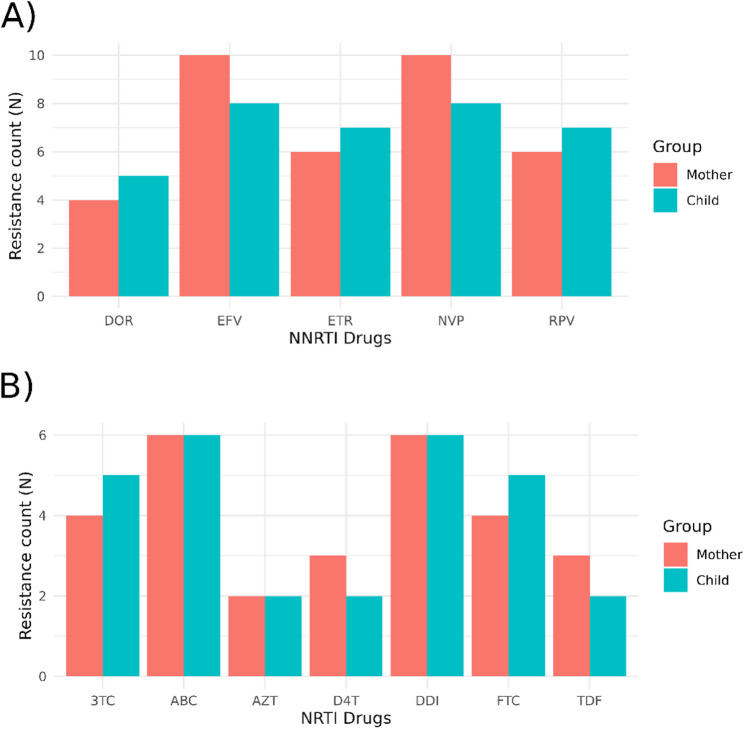
A comparison of resistance mutations to nucleoside reverse transcriptase inhibitors (NRTIs) and non-nucleoside reverse transcriptase inhibitors (NNRTIs) between the mothers and children did not reveal significant differences in their occurrence (Fig. 1).
Fig. 1.
Number of individuals (mothers and children) with resistance to NNRTIs (A) and NRTIs (B) by study group. 3TC, lamuvidine; ABC, abacavir; D4T, stavudine; DDI, didanosine; DOR, doravirine; EFV, efavirenz; ETR, etravirine; FTC, emtricitabine; NNRTIs, non-nucleoside reverse transcriptase inhibitors; NRTIs, nucleoside reverse transcriptase inhibitors; NVP, nevirapine; RPV, rilpovirine; TDF, tenofovir disoproxil fumarate; ZDV, zidovudine
Among the NRTI resistance–associated mutations, M184V was the most frequent (26.7%) (Fig. 2). The K219KR and K70E mutations were observed in only one mother (6.7%) (Fig. 2). The M184I, M41L, and T215FY mutations were identified in only one child each (6.7%). Regarding NNRTI resistance–associated mutations, K103N was identified in four patients (26.7%) in both groups. The G190A mutation was more frequent in the child group (n = 3 patients [20%]) than in the mother group (n = 2 [13.3%]) (Fig. 2). Mutations at amino acid positions 106 and 188 on reverse transcriptase were found only in mothers, whereas mutations at positions 101 and 230 were observed only in children.
Fig. 2.
Proportion of individuals (mother and child) with NNRTI (a) and NRTI (b) DRAMs. The absence of a bar indicates that the respective mutation was not found in that group. DRAM, drug resistance–associated mutation; NNRTI, non-nucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor
An analysis for antiretroviral therapy resistance revealed varied patterns between the mothers and children. Except for three mother–child pairs that did not exhibit antiretroviral resistance mutations, all pairs showed the presence of mutations that conferred high-level resistance in at least one member of the pair (Fig. 3). Among the analysed pairs, five (33%) showed the presence of mutations conferring high-level resistance in only one individual (mothers: pairs 6, 9, and 14; children: pairs 1 and 15). In four pairs (27%), higher antiretroviral resistance was detected in the children versus mothers.
Fig. 3.
Levels of antiretroviral resistance in mothers and children classified by Stanford Drug Resistance Database resistance score. The dotted line separates NRTIs on the left and NNRTIs on the right. 3TC, lamuvidine; ABC, abacavir; BIC, bictegravir; D4T, stavudine; DDI, didanosine; DOR, doravirine; EFV, efavirenz; ETR, etravirine; FTC, emtricitabine; NVP, nevirapine; RPV, rilpovirine; TDF, tenofovir disoproxil fumarate; ZDV, zidovudine
The 184 V mutation was present in four mothers (26.6%) and three (75%) of their children (pairs 3, 5 and 7). Mutations at position 103 were present in five mothers (33%) and four children (27%), showing a high correlation between them. In addition to the three mother–child pairs without DRAMs, some showed the presence of mutations that conferred high-level resistance in only one individual (pairs 6, 9, and 14 for mothers; pairs 1 and 15 for children). Children in pairs 1, 5, 7, and 15 exhibited broader resistance to various antiviral drugs (Table 1).
Table 1.
Main drug resistance–associated mutations and genetic difference detected between pair in HIV-1 sequences isolated from mother–child pairs in Maranhão and Pernambuco (Northeast region of Brazil), 2007–2022. PI, protease inhibitors; NNRTIs, non-nucleoside reverse transcriptase inhibitors; NRTIs, nucleoside reverse transcriptase inhibitors
| Pair | Time between child birth and genotyping (years) | Drug resistance–associated mutations | Distance between pair | ||
|---|---|---|---|---|---|
| NRTIs | NNRTIs | PI | |||
| M1 | 15 | None | V108I | None | 0.0314 |
| C1 | 13 | M184V | K103N, G190GE | M46ML | |
| M2 | 01 | None | None | None | 0.0070 |
| C2 | 01 | None | None | None | |
| M3 | 06 | K70E, M184V | V106I, Y188L | None | 0.0152 |
| C3 | 04 | M184V | None | None | |
| M4 | 02 | None | None | None | 0.0071 |
| C4 | 02 | None | None | None | |
| M5 | 06 | M184V | K103N | None | 0.0207 |
| C5 | 08 | M184I | K103T, M230L | Q58E | |
| M6 | 13 | None | K103N, E138A | None | 0.0313 |
| C6 | 13 | None | E138A | None | |
| M7 | 04 | S68G, M184V | K103N | None | 0.0207 |
| C7 | 03 | M41L, S68G, M184V, T215FY | A98AG, K101KE, V108VI, G190A | None | |
| M8 | 02 | None | None | None | 0.0071 |
| C8 | 01 | None | None | None | |
| M9 | 14 | None | G190GRS | None | 0.0143 |
| C9 | 12 | None | None | None | |
| M10 | 09 | K219KR | L100LIV, F227L | None | 0.0388 |
| C10 | 09 | None | L100I, K103N, E138EK | Q58E | |
| M11 | 03 | None | K103N | None | 0.0060 |
| C11 | 01 | None | K103N | None | |
| M12 | 04 | None | G190A | None | 0.0033 |
| C12 | 01 | None | G190A | None | |
| M13 | 01 | S68G, L210W, T215D | G190A | None | 0.0081 |
| C13 | 03 | S68G, L210W, T215D | V179IT, G190A | None | |
| M14 | 03 | M184V | None | None | 0.0232 |
| C14 | 01 | None | None | None | |
| M15 | 02 | None | None | None | 0.0046 |
| C15 | 05 | M184V | K103N, V108I | None | |
The time between birth and genotyping varied significantly between pairs (ranging from 1 to 15 years). Among the eight pairs with tests obtained within the first four years after birth (pairs 2, 4, 7, 8, 11, 12, 13 and 14), six of them had very similar mutation profiles. The remaining two pairs, one had a similar profile for S68G and 184 V mutations and, in the other pair, the mother had only 184 V and the child had no mutations. Among the seven pairs with exams obtained five years or more after birth (pairs 1, 3, 5, 6, 9, 10 and 15), four pairs presented very different DRAM profiles and the other three showed low similarities (Table 1).
The genetic divergence between maternal and child viral sequences is quantified as a pairwise genetic distance, with values ranging from 0.0033 to 0.0388. Higher genetic distances tend to be associated with longer time intervals between childbirth and genotyping, which may reflect within-host viral evolution over time. Cases with shorter time intervals generally exhibit lower divergence, suggesting limited viral evolution post-transmission.
In the phylogenetic inference, it was possible to find other sequences originating from Brazil that formed several monophyletic groups (Figs. 4 and 5). The mother–child pairs identified in the phylogenetic trees were not closely correlated from an evolutionary standpoint. Thirteen pairs with HIV-1 subtype B (86.6%) and two pairs with HIV-1 subtype F1 (13.3%) were identified; however, no recombinant sequences were confirmed on similarity-based analyses (Simplot). The clustering patterns indicate potential vertical transmission events, with mother-child pairs forming closely related clades. Additionally, the presence of reference sequences interspersed among the clinical isolates provides insights into the genetic diversity and phylogeographic distribution of the studied viral strains. These findings underscore the importance of phylogenetic surveillance in understanding mother-to-child transmission dynamics and viral evolution.
Fig. 4.
Maximum likelihood tree of HIV-1 subtype B showing the phylogenetic relationship between the sequences from mothers (blue) and their respective children (green). Sequences with the highest similarity on a BLAST search are highlighted in red
Fig. 5.
Maximum likelihood tree of HIV-1 subtype F1 showing the phylogenetic relationship between the sequences from mothers (blue) and their respective children. Sequences with the highest similarity on a BLAST search are highlighted in red
Discussion
The results of the present study evaluating antiretroviral resistance in HIV-1 mother–child pairs suggest that three situations may occur: (a) transmission of the wild-type virus without mutations; (b) transmission of resistance mutations from mothers to children, either partially or fully; and (c) late acquisition of resistance by children after their antiretroviral therapy. Only half the children in this study had resistance mutation profiles like those of their mothers. The great majority of this group was detected in those pairs that had their genotyping in the interval of one to four years of the childbirth. Understanding the intergroup differences in mutation profiles is important and requires a review of the dynamics of vertical HIV transmission. Evidence supports the hypothesis of restricted viral heterogeneity in the blood of infants at the time of infection, with only one or very few viral variants being transmitted from mother to child, although the transmission of multiple variants to infants has been described. Transmission during childbirth is associated with a single HIV-1 variant, whereas intrauterine transmission more likely involves sone or more predominant maternal viral variants [20]. Another explanation for the difference in mutation profiles between mothers and children could involve horizontal HIV-1 transmission, which involves the reversion of resistance mutations due to immune pressure in the new host [21]. Finally, it has been shown that the use of antiretrovirals in children may limit the transmission of DRAMs [22].
However, we must consider that the methodology used herein may not have identified minority variants among the resistance mutations derived from the mothers. In the Swiss Mother-Child Cohort with HIV, which evaluated 22 pregnant women who transmitted HIV to their children, 10 had DRAMs; however, only one child presented with this type of resistance mutation, specifically M184V, from their mother. In the same group of children, an in-depth analysis of antiretroviral resistance mutations using next-generation sequencing was conducted of three children who were born with HIV but lacked mutations. In two cases, DRAMs at frequencies < 20%, which were not detected by Sanger sequencing, were detected by next-generation sequencing. In the third case, mutations emerged as the predominant variant after 2 years [23]. When evaluating 190 acute infections acquired by horizontal transmission, Faraci et al. [24] found that all resistance mutations in the plasma also appeared in proviral DNA; however, nine high-frequency mutations were detected only in the latter. Additionally, considering a DRAM rate < 20%, 11 NNRTI, seven NRTI, and six protease inhibitor mutations were exclusively located in proviral DNA.
Similarly, a study of 72 South African children with therapeutic failure to respond to regimens containing NRTIs and protease inhibitors showed that those whose viruses contained mutations for the NNRTI class had not been previously used, suggesting that these mutations were acquired from their mothers with therapeutic failure [25]. These findings demonstrate that the transmission of resistance mutations in acute infections is more common than that detected by conventional genotyping methods and suggest their potential for their emergence after selective pressure from treatment or irregular use of antiretrovirals.
In another study, DRAM transmission from mother to child occurred in only one of 22 cases (4.3% of children), whereas acquired resistance after treatment occurred in 16 of 22 cases (73%) [23]. Acquired resistance to antiretrovirals likely occurred in children from pairs 1, 3,10 and 15, (sample collected after four or more years after childbirth), in whom the resistance mutation profiles differed between the children and mothers, indicating that in addition to transmission, other factors such as poor treatment adherence and selective pressure from antiretrovirals strongly influence the evolution of resistance in children. Considering the NRTI-related mutation profiles of the mothers, we found that M184V was the most common and transmitted to half of their children, while the thymidine analogue mutation pathway 1 mutation, present in only one mother, was also identified in her child.
Regarding NNRTIs, the K103N mutation was found in four mothers who transmitted it to two (50%) of their children. The G190A mutation associated with prolonged NVP resistance was identified in two mothers and transmitted to their two children. Our results are similar to those found in India in 72 children with perinatal infections and virological failure of antiretrovirals after 12 months of therapy. Pretreatment genotyping showed that 14% of the children had pretreatment viral resistance mutations, with NNRTI-related mutations identified in 9.7% of cases and NRTI-related K103N and M184V mutations identified in 2.7% of cases [26].
The mutational profile of the cases studied here, which were collected through 2023, showed resistance to NRTIs, particularly 3TC, conferred by the 184 V mutation present in approximately 26% of mothers, and to NNRTIs, such as EFV and NVP, conferred by the K103N and G190A mutations, which were present in 26.6% and 13.3% of the mothers, respectively. This finding suggests that the vertical HIV-1 transmission prophylaxis regimens used for pregnant women in Brazil until 2022, consisting of ZDV + 3TC + EFV or NVP, are exhausted and no longer aligned with the need for drugs with genetic barriers against the most prevalent resistance mutations. Since then, recognising the superiority of dolutegravir (DTG)–containing regimens over those containing EFV, as evidenced by the VESTED randomised, multicentre clinical trial conducted in nine countries that evaluated the efficacy of antiretroviral regimens for the prevention of vertical HIV transmission [27], the Brazilian Ministry of Health discontinued the preferential use of NNRTIs in the treatment of pregnant women and established the tenofovir (TFV) + 3TC + DTG regimen for pregnant women and the ZDV + 3TC + DTG regimen for prophylaxis in exposed newborns [28]. Notably, the 184 V mutation, induced by 3TC, reduces viral fitness and hypersensitises the virus to TFV and ZDV, components of the current therapeutic protocols for pregnant women and infants.
A low genetic distance (< 0.01 substitutions per site) between maternal and child sequences, especially when genotyping occurs within the first one to two years, suggests that the virus in the child closely resembles the transmitted maternal strain. This supports the hypothesis that the child’s virus was acquired at birth with minimal subsequent evolution. However, if the genetic distance is higher (> 0.02 substitutions per site), it suggests that the virus has undergone significant divergence, which is more consistent with intra-host evolution driven by viral replication, immune pressure, and ART exposure [29].
Another factor influencing genetic distance is the transmission of a minor maternal variant. If the transmitted virus was already divergent at birth, the genetic distance may appear large even at early time points, as observed in pair C14-M14. In contrast, if the transmitted virus was initially similar to the maternal consensus strain but diverged significantly over several years, intra-host evolution is a more plausible explanation, as seen in pairs C1-M1, C6-M6, and C10-M10.
A limitation of our study is the lack of longitudinal sampling of the child’s viral population over time, which could provide greater resolution in distinguishing these scenarios. Early viral sequences closely related to the maternal strain, followed by increasing divergence over time, would suggest ongoing intra-host evolution. Conversely, if the child’s virus is already highly divergent at the earliest time points, this would support the hypothesis of selective transmission of a distinct maternal subpopulation rather than extensive postnatal evolution.
Conclusions
In conclusion, the analysis of resistance mutations performed herein revealed differences in the mutation profiles and resistance to antiretrovirals in NRTI and NNRTI drug classes between mothers and children, reinforcing the importance of individualised therapeutic strategies and an integrated approach to preventing HIV-1 transmission.
Acknowledgements
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)– Brazil.
Abbreviations
- 3TC
Lamuvidine
- ABC
Abacavir
- ART
Antiretroviral therapy
- BIC
Bictegravir
- D4T
Stavudine
- DDI
Didanosine
- DOR
Doravirine
- DRAMs
Drug resistance–associated mutations
- DTG
Dolutegravir
- EFV
Efavirenz
- ETR
Etravirine
- FTC
Emtricitabine
- LPV/r
Lopinavir/ritonavir
- NNRTI
Non-nucleoside reverse transcriptase inhibitor
- NRTI
Nucleoside reverse transcriptase inhibitor
- NVP
Nevirapine
- RPV
Rilpovirine
- TDF
Tenofovir disoproxil fumarate
- TFV
Tenofovir
- ZDV
Zzidovudine
Authors’ contributions
Aldicléya Lima Luz, Kledoaldo Lima, Élcio Leal and Heloisa Ramos Lacerda contributed to the conception and design of the study. Aldicléya Lima Luz, Kledoaldo Lima, Geovani de Oliveira Ribeiro, and Heloisa Ramos Lacerda wrote the first draft and/or revised the manuscript. Aldicléya Lima Luz, Fabrício Silva Pessôa, Cláudia Regina de Andrade Arrais Rosa, Marcos Davi Gomes de Sousa and Pablo Cantalice Santos Farias organized the database. Kledoaldo Lima, Geovani de Oliveira Ribeiro and Pablo Cantalice Santos Farias performed the analysis. Aldicléya Lima Luz, Kledoaldo Lima, Élcio Leal and Heloisa Ramos Lacerda contributed equally to the current work. All authors contributed to the manuscript revision and read and approved the submitted version.
Funding
No funding was received for the development of this study.
Data availability
Genotypic sequences of the HIV pol region (protease/reverse transcriptase-PR/RT) were obtained using the ViroSeq Genotyping System, carried out in the laboratories of the National Genotyping Network (RENAGENO) coordinated by the Department of HIV/AIDS of the Brazilian Ministry of Health (GenBank accession number PV526713-PV526742).
Declarations
Ethics approval and consent to participate
This study complied with the Declaration of Helsinki for research involving human subjects, data and/or materials. Informed consent to participate was obtained from all of the participants or their guardians in the study. This study was submitted to and approved in accordance with Resolution 466/2012 by the Research Ethics Committee of the Hospital das Clínicas, Federal University of Pernambuco (HC/UFPE), under CAAE 51758621.7.1001.8807, and the Research Ethics Committee of UFMA, under CAAE 57505422.6.0000.5086.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cardenas MC, Farnan S, Hamel BL, Mejia Plazas MC, Sintim-Aboagye E, Littlefield DR, et al. Prevention of the vertical transmission of HIV; A recap of the journey so far. Viruses. 2023;15:849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joint United Nations Programme on HIV/AIDS (UNAIDS). Transforming vision into reality: the 2024 Global Alliance progress report on ending AIDS in children by 2030. 2024. https://www.unaids.org/en/resources/documents/2024/transforming-vision-into-reality. Acessed 25 Dec 2024.
- 3.Guimarães MF, Lovero KL, de Avelar JG, Pires LL, de Oliveira GRT, Cosme EM, et al. Review of the missed opportunities for the prevention of vertical transmission of HIV in brazil. Clin (Sao Paulo). 2019;74:e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ministério da Saúde (Brasil). Aumenta em 70% o número de municípios reconhecidos pela eliminação da transmissão vertical de HIV e sífilis. 2023. https://www.gov.br/saude/pt-br/assuntos/noticias/2023/dezembro/aumenta-em-70-o-numero-de-municipios-reconhecidos-pela-eliminacao-da-transmissao-vertical-de-hiv-e-sifilis. Accessed 25 Dec 2024.
- 5.World Health Organization (WHO). HIV drug resistance report 2021. 2021. https://www.who.int/publications/i/item/9789240038608. Accessed 25 Dec 2024.
- 6.Vaz SN, Giovanetti M, Rego FF, Oliveira Td, Danaviah S, Gonçalves ML, et al. Molecular characterization of the human immunodeficiency virus type 1 in women and their vertically infected children. AIDS Res Hum Retroviruses. 2015;31:1046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultze A, Torti C, Cozzi-Lepri A, Vandamme AM, Zazzi M, Sambatakou H, et al. The effect of primary drug resistance on CD4 + cell decline and the viral load set-point in HIV-positive individuals before the start of antiretroviral therapy. AIDS. 2019;33:315–26. [DOI] [PubMed] [Google Scholar]
- 8.Santos-Pereira A, Triunfante V, Araújo PMM, Martins J, Soares H, Poveda E, et al. Nationwide study of drug resistance mutations in HIV-1 infected individuals under antiretroviral therapy in brazil. Int J Mol Sci. 2021;22:5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nazziwa J, Andrews SM, Hou MM, Bruhn CAW, Garcia-Knight MA, Slyker J, et al. Higher HIV-1 evolutionary rate is associated with cytotoxic T lymphocyte escape mutations in infants. J Virol. 2024;98:e0007224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koay WLA, Kose-Otieno J, Rakhmanina N. HIV drug resistance in children and adolescents: always a challenge?? Curr Epidemiol Rep. 2021;8:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silver N, Paynter M, McAllister G, Atchley M, Sayir C, Short J, et al. Characterization of minority HIV-1 drug resistant variants in the united kingdom following the verification of a deep sequencing-based HIV-1 genotyping and tropism assay. AIDS Res Ther. 2018;15:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Struck D, Lawyer G, Ternes A-M, Schmit J-C, Bercoff DP. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res. 2014;42:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.HIV Drug Resistance Database. Stanford University. 2024. http://hivdb.stanford.edu. Accessed 25 Dec 2024.
- 14.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, et al. Full-Length human immunodeficiency virus type 1 genomes from subtype C-Infected seroconverters in india, with evidence of intersubtype recombination. J Virol. 1999;73:152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanford University HIV Drug Resistance Database. REGA HIV-1 Subtyping Tool– Version 3.0. 2024. http://dbpartners.stanford.edu:8080/RegaSubtyping/stanford-hiv/typingtool. Accessed 25 Dec 2024.
- 16.Major HIV-1 Drug Resistance Mutations. 2024.https://cms.hivdb.org/prod/downloads/resistance-mutation-handout/resistance-mutation-handout.pdf. Accessed 24 Dec 2024.
- 17.Nakamura T, Yamada KD, Tomii K, Katoh K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 2018;34:2490–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.HIV Sequence Database, Basic Local Alignment Search Tool (BLAST). Los Alamos National Laboratory. https://www.hiv.lanl.gov/contentsequence/BASIC_BLAST/basic_blast.html. Accessed 25 Dec 2024.
- 19.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucl Acids Res. 2016;44:W232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kourtis AP, Bulterys M. Mother-to-child transmission of HIV: pathogenesis, mechanisms and pathways. Clin Perinatol. 2010;37:721–37. [DOI] [PubMed] [Google Scholar]
- 21.Ryland EG, Tang Y, Christie CD, Feeney ME. Sequence evolution of HIV-1 following mother-to-child transmission. J Virol. 2010;84:12437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koay WLA, Kose-Otieno J, Rakhmanina N. HIV Drug Resistance in Children and Adolescents: Always a Challenge? Curr Epidemiol Rep. 2021;8(3):97–107. 10.1007/s40471-021-00268-3 [DOI] [PMC free article] [PubMed]
- 23.Compagno F, Naegele K, Kahlert CR, Hösli I, Aebi-Popp K, de Tejada BM, et al. The rate of mother-to-child transmission of antiretroviral drug-resistant HIV strains is low in the swiss mother and child HIV cohort study. Swiss Med Wkly. 2019;149:w20059. [DOI] [PubMed] [Google Scholar]
- 24.Faraci G, Park SY, Dubé MP, Lee HY. Full-spectrum HIV drug resistance mutation detection by high-resolution complete pol gene sequencing. J Clin Virol. 2023;164:105491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hackett S, Teasdale CA, Pals S, Muttiti A, Mogashoa M, Chang J, et al. Drug resistance mutations among south african children living with HIV on WHO-recommended ART regimens. Clin Infect Dis. 2021;73:e2217–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karunaianantham R, Nesa Kumar M, Gopalan B, Haribabu H, Hanna LE, Sanjeeva GN, et al. Molecular characterization of the pol gene of vertically transmitted HIV-1 strains in children with virological failure. AIDS Res Hum Retroviruses. 2022;38:491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lockman S, Brummel SS, Ziemba L, Stranix-Chibanda L, McCarthy K, Coletti A, et al. Efficacy and safety of dolutegravir with emtricitabine and tenofovir alafenamide fumarate or tenofovir disoproxil fumarate, and efavirenz, emtricitabine, and tenofovir disoproxil fumarate HIV antiretroviral therapy regimens started in pregnancy (IMPAACT 2010/VESTED): a multicentre, open-label, randomised, controlled, phase 3 trial. Lancet. 2021;397:1276–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ministério da Saúde (Brasil), Protocolo clínico e diretrizes terapêuticas para prevenção da transmissão vertical de HIV, sífilis e hepatites virais. 2022. https://bvsms.saude.gov.br/bvs/publicacoes/protocolo_clinico_hiv_sifilis_hepatites.pdf. Accessed 25 Dec 2024.
- 29.Ryland EG, Tang Y, Christie CD, Feeney ME. Sequence evolution of HIV-1 following mother-to-child transmission. J Virol. 2010;2010(84):12437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genotypic sequences of the HIV pol region (protease/reverse transcriptase-PR/RT) were obtained using the ViroSeq Genotyping System, carried out in the laboratories of the National Genotyping Network (RENAGENO) coordinated by the Department of HIV/AIDS of the Brazilian Ministry of Health (GenBank accession number PV526713-PV526742).