Abstract
Glioblastoma (GBM) remains the most aggressive and lethal brain tumor in adults and poses significant challenges to patient survival. This review provides a comprehensive exploration of the molecular and genetic landscape of GBM, focusing on key oncogenic drivers, such as epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor (PDGFR), and the PI3K/AKT/mTOR pathway, which are critical for tumorigenesis and progression. We delve into the role of epigenetic alterations, including DNA methylation and histone modifications, in driving therapy resistance and tumor evolution. The tumor microenvironment is known for its pivotal role in immune evasion, with tumor-associated macrophages, myeloid-derived suppressor cells, and regulatory T cells creating an immunosuppressive niche that sustains GBM growth. Emerging therapies, such as immunotherapies, oncolytic viral therapies, extracellular vesicle-based approaches, and non-coding RNA interventions, are highlighted as promising avenues to disrupt GBM pathogenesis. Advances in precision medicine and innovative technologies, including electric field therapy and locoregional treatments, are discussed for their potential to overcome the blood‒brain barrier and treatment resistance. Additionally, this review underscores the importance of metabolic reprogramming, particularly hypoxia-driven adaptations and altered lipid metabolism, in fueling GBM progression and influencing the therapeutic response. The role of glioma stem cells in tumor recurrence and resistance is also emphasized, highlighting the need for targeted therapeutic approaches. By integrating molecular targeting, immune energetics, and technological advancements, this review outlines a multidisciplinary framework for improving GBM treatment outcomes. Ultimately, the convergence of genetic, metabolic, and immune-based strategies offers transformative potential in GBM management, paving the way for increased patient survival and quality of life.
Subject terms: Drug development, Prognostic markers
Introduction
Glioblastoma (GBM) is the most prevalent and aggressive malignant brain tumor in adults and presents a formidable challenge in oncology due to its rapid progression, therapeutic resistance, and poor prognosis. Despite extensive research, the median survival remains dismal at 12–15 months.1 The latest classification of central nervous system (CNS) tumors categorizes gliomas into a diverse group of glial-derived brain tumors, with GBM being the most aggressive grade IV subtype, characterized by an isocitrate dehydrogenase (IDH) wild-type status. GBM is further distinguished by key molecular alterations, including epidermal growth factor receptor (EGFR) amplification, telomerase reverse transcriptase (TERT) promoter mutations, and distinct chromosomal abnormalities.2,3 These features contribute to the highly invasive nature and resistance of tumors to conventional therapies. In contrast, IDH mutant gliomas, which are commonly found in lower-grade gliomas and secondary GBMs, exhibit distinct epigenetic landscapes and are associated with better clinical outcomes. These tumors exhibit the glioma-CpG island methylator phenotype (G-CIMP),4,5 influencing tumor behavior and therapeutic response, highlighting the importance of epigenetic regulation in gliomagenesis. Additional genetic alterations, such as mutations in the alpha-thalassemia mental retardation X-linked (ATRX) gene and DNA methylation profiles, further refine tumor classification and influence treatment strategies.6,7
A major obstacle in GBM treatment is its cellular and molecular heterogeneity, comprising differentiated tumor cells, glioma stem-like cells (GSCs), and a dynamic tumor microenvironment (TME). Advanced sequencing technologies have identified diverse GBM subtypes and cellular states, emphasizing the need for therapeutic strategies targeting both molecular drivers and the TME. GSCs, in particular, play pivotal roles in tumor progression, therapeutic resistance, and recurrence due to their self-renewal capabilities and adaptability.8,9 However, their resilience poses a major barrier to effective treatment. Additionally, genomic instability and oncogenic signaling pathways, such as the EGFRvIII-driven dysregulation of the receptor tyrosine kinase/mitogen-activated protein kinase (RTK/RAS/MAPK) pathway, fuel aggressive tumor behavior. The frequently altered phosphoinositide-3 kinase/protein kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR) axis, which regulates tumor growth and survival, is a promising therapeutic target, although clinical trials of mTOR inhibitors have shown limited success.10 A comprehensive understanding of the interplay between molecular alterations, GSC biology, and the TME is essential for developing innovative, more effective treatment strategies.
The TME significantly contributes to tumor progression by fostering tumor growth, immune evasion, and resistance to therapy.11 Interactions among tumor-associated macrophages (TAMs), neutrophils, myeloid-derived suppressor cells (MDSCs), and T cells within the TME create an immunosuppressive niche that enables tumor survival and proliferation. In recurrent GBMs, these dynamics intensify, with increased immune cell infiltration and the upregulation of checkpoint proteins such as programmed death-ligand 1 (PD-L1) and PD-1, underscoring the importance of precision immunotherapy to improve outcomes.12 In addition to cellular components, extracellular vesicles (EVs), microRNAs (miRNAs), and long non-coding RNAs (lncRNAs) have emerged as both molecular biomarkers and therapeutic targets in GBM.13 For example, miRNA-21, which is frequently upregulated in GBM, is correlated with poor survival and higher tumor grades, whereas other miRNAs, such as miR-128 and miR-342-3p, exhibit therapy-induced expression changes and are linked to glioma grade.14 Similarly, circulating lncRNAs and circular RNAs (circRNAs) have shown potential for predicting patient outcomes,15 further emphasizing their value in GBM treatment strategies. Targeting the TME and integrating molecular markers into therapeutic approaches represent crucial steps toward enhancing treatment efficacy. By addressing these intricate interactions and leveraging molecular insights, GBM management can progress toward more personalized and effective strategies.
This review delves into the cellular heterogeneity of GBM, emphasizing the genetic, epigenetic and oncogenic signaling pathways that drive tumor progression, therapy resistance and recurrence. This highlights the crucial role of GSCs in tumor persistence, as well as the impact of the TME in fostering immune evasion and therapeutic resistance. Additionally, key molecular alterations, including EGFR amplification, IDH mutations, O6-methylguanine-DNA methyltransferase (MGMT) modifications, histone epigenetic changes and signaling pathway dysregulation, are being examined for their contributions to the aggressive behavior and treatment challenges of GBM. This review critically evaluates current and emerging therapeutic strategies, including locoregional treatments, systemic chemotherapy, and combination therapies, alongside innovative approaches such as oncolytic viral therapy, EV-based therapies, non-coding RNA (ncRNA) interventions, electric field therapy, and precision medicine advancements (Fig. 1). These approaches are discussed for their potential to overcome existing limitations, such as therapeutic resistance, tumor recurrence, immune adaptation, metabolic reprogramming and blood‒brain barrier (BBB) delivery challenges. By addressing these persistent hurdles and highlighting promising research directions, this review aims to inspire innovative strategies that could transform GBM treatment, improve patient outcomes, and advance the therapeutic landscape for this devastating disease.
Fig. 1.
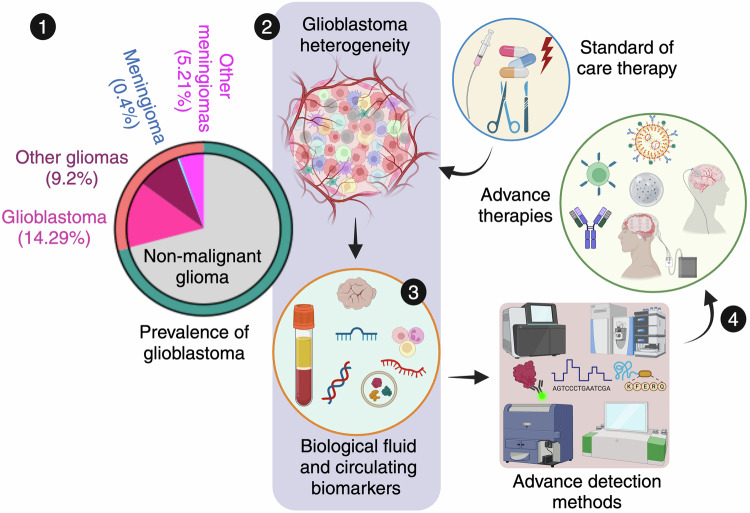
Glioblastoma landscape and path towards targeted therapies. 1. The pie chart illustrates glioma trends, with a focus on glioblastoma (GBM) prevalence in the United States. Data source: Cancer Stat Facts: Brain and other nervous system cancers identified by the National Cancer Institute’s Surveillance, Epidemiology, and End Results Program, 2014–2020. 2. GBM, marked by its pronounced molecular, genetic, and cellular heterogeneity, presents substantial obstacles for accurate diagnosis and effective treatment. 3. Advanced diagnostic methods, leveraging biofluid biomarkers such as liquid biopsies and circulating biomolecules, alongside high-definition detection technologies, are crucial for precise detection. 4. These innovations are driving the development of targeted and more effective therapies for GBM treatment
Molecular characterization of GBM and diagnostic biomarkers
Clinical grading of GBM
GBM is classified as a World Health Organization (WHO) grade IV glioma, distinguished by aggressive behavior, high recurrence rates, and resistance to conventional therapies. Its histopathological hallmarks include nuclear atypia, cellular pleomorphism, mitotic activity, microvascular proliferation, and necrosis. In addition to these defining features, several histologic variants, such as gliosarcomas, giant-cell GBM, small-cell GBM, and epithelioid GBM, present distinct molecular and clinical implications. Notably, epithelioid GBM is characterized by v-raf murine sarcoma viral oncogene homolog B1 (BRAF) V600E mutations,16 highlighting the genetic heterogeneity within GBM. GBM falls within the diffuse glioma category, which presents significant treatment challenges because of its highly infiltrative nature. Unlike circumscribed gliomas, which have well-defined margins and generally better prognosis, diffuse gliomas are characterized by extensive invasion into normal brain tissue, limiting the effectiveness of surgical resection.17 As the most aggressive form of diffuse glioma, GBM accounts for nearly 50% of all primary malignant brain tumors and represents the most lethal intrinsic brain tumor.18
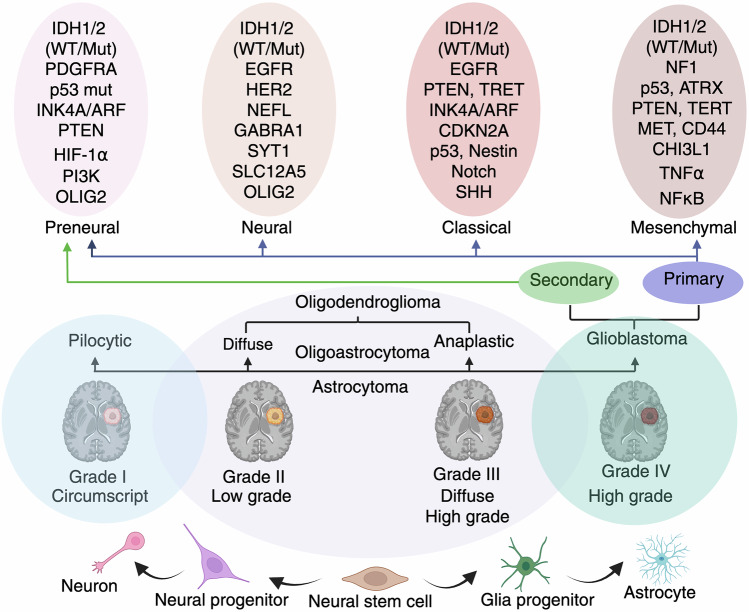
The evolution of molecular classification has refined GBM subtyping, moving beyond histological grading to a deeper understanding of its genetic and epigenetic landscape (Table S1). The classification system proposed by Phillips et al. divides GBM into three subtypes with distinct prognostic and therapeutic implications. 1) Proneural GBM, which is predominantly observed in younger patients, is associated with lower pathological severity and relatively better survival outcomes. It is characterized by neural-like gene expression patterns, including those of the neural cell adhesion molecules GABR1 and SNAP91, which resemble those of normal brain tissue. 2) Proliferative GBM is associated with high levels of cellular proliferation, with significant upregulation of the expression of markers such as TOP2A and PCNA, indicating a more aggressive tumor biology. 3) Mesenchymal GBM is the most invasive subtype and is characterized by the overexpression of angiogenesis markers (e.g., vascular endothelial growth factor {VEGF}, PECAM1), the loss of phosphatase and tensin homolog (PTEN) and neurofibromin 1 (NF1), and the activation of PI3K/AKT signaling, which are correlated with a poor prognosis.19
Verhaak et al. further expanded the classification into four subtypes: proneural, neural, classical, and mesenchymal. While proneural GBM is enriched in platelet-derived growth factor receptor alpha (PDGFR-α) expression and IDH1 mutations, which confer a potential survival advantage, it remains resistant to conventional therapy. Neural subtypes, which share gene expression similarities (SYT1, GABRA1 and NEFL) with normal neurons, exhibit enhanced sensitivity to radiation and chemotherapy.20 In contrast, the classical subtype is characterized by EGFR amplification, RB pathway alterations, chromosome 7 amplification, chromosome 10 loss, and high activation of the sonic hedgehog (SMO, GAS1, GLI2) and Notch signaling (NOTCH3, JAG1, LFNG) pathways, making it more responsive to aggressive treatment. Mesenchymal GBM, characterized by extensive necrosis, inflammatory markers, the upregulation of interstitial and angiogenesis genes, frequent deletions of the tumor suppressor genes tumor protein 53 (p53), PTEN and NF1 and highly expressed genes such as VEGF-A, VEGF-B, ANG1, and ANG2, represents the most aggressive subtype with limited treatment success21 (Fig. 2).
Fig. 2.
Clinical and molecular grading of gliomas. Schematic representation of the molecular classification and histopathological grading of gliomas, along with their cellular origins and progression. The bottom panel shows a developmental lineage from neural stem cells to neurons, astrocytes, and glial progenitors. Pilocytic astrocytomas (Grade I) are typically circumscribed and low grade, whereas diffuse astrocytomas (Grade II), anaplastic astrocytomas (Grade III), and glioblastomas (Grade IV) represent progressive stages of malignancy and infiltrative behavior. The top panel highlights the molecular subtypes of glioblastoma: proneural, neural, classical, and mesenchymal, each defined by distinct genetic alterations such as IDH1/2, EGFR, p53, PTEN, NF1, and others. These subtypes correlate with the primary (de novo) or secondary (progression from lower-grade gliomas) origins of glioblastoma. This classification underscores the integration of molecular and clinical parameters for diagnosis, prognosis, and therapeutic decision-making in gliomas
In addition to transcriptomic profiling, DNA methylation-based classification provides further granularity in GBM subtyping. Large-scale sequencing studies have identified six methylation clusters (M1–M6), each with distinct prognostic and biological implications. Among them, the G-CIMP subtype (cluster M5) is characterized by hypermethylation and frequent IDH1 mutations, which are correlated with improved survival outcomes and a less aggressive clinical course. In contrast, Cluster M6, characterized by relative hypomethylation and a predominance of IDH1 wild-type tumors, represents a more aggressive phenotype with a poorer prognosis. Further molecular refinement revealed the enrichment of missense mutations and deletions in histone-lysine N-methyltransferase 2A (KMT2A) or MLL and histone deacetylase (HDAC) family genes within Cluster M2, underscoring the role of chromatin remodeling in GBM pathogenesis.22 Additionally, Cluster 3 has a greater frequency of p53 mutations, along with IDH1 wild-type and 1p/19q deletions, further distinguishing high-risk subgroups with aggressive tumor behavior.23
The integration of DNA methylation patterns with genetic alterations offers a comprehensive framework for patient stratification, refining prognostic predictions and informing therapeutic decision making. These molecular subtypes not only highlight the heterogeneity of GBM but also provide potential targets for precision medicine. Future research should focus on unraveling the regulatory mechanisms driving these epigenetic changes, which is essential for overcoming the inherent therapeutic resistance of GBM and improving patient outcomes.
Diagnostic biomarkers
IDH mutation
IDH mutations are pivotal in glioma classification and influence tumor metabolism, epigenetic regulation, redox balance, DNA repair and cellular differentiation. These mutations, which primarily affect IDH1 (R132), which is localized in the cytosol and peroxisomes, and IDH2 (R172), which is located in the mitochondria, lead to the accumulation of D-2-hydroxyglutarate metabolites, driving oncogenesis through widespread epigenetic dysregulation.24 These mutations serve as key molecular discriminators between glioma subtypes and are highly prevalent in lower-grade diffuse gliomas (WHO grades II–III) and secondary GBMs25 but are largely absent in primary GBMs, which are predominantly IDH wild-type.26 This distinction has led to the integration of the IDH status into the WHO glioma classification.
IDH mutations are correlated with improved survival and treatment response, distinguishing IDH-mutant gliomas from their more aggressive IDH wild-type counterparts. Large-scale analysis confirmed a high prevalence of IDH mutations in oligodendrogliomas (71%) and diffuse astrocytomas (58.6%), with a decreasing frequency in anaplastic astrocytomas (27.6%) and GBMs (10.4%).27 These patterns reinforce IDH mutation status as a key factor in glioma stratification and prognosis. Patients with Grade III gliomas lacking 1p/19q codeletion and harboring IDH mutations have significantly prolonged progression-free survival (PFS) and overall survival (OS), with similar trends observed in secondary high-grade gliomas (HGGs).28 A comprehensive meta-analysis further validated the strong correlation between IDH1/2 mutations and improved survival in patients with GBM.29
In addition to survival outcomes, IDH mutations define a distinct epigenetic subclass, G-CIMP, that is linked to a better prognosis. G-CIMP+ tumors, which frequently harbor IDH1 mutations, align with a proneural gene expression profile and are diagnosed at a younger age, whereas G-CIMP− tumors, including most primary GBMs, exhibit a more aggressive phenotype.30 IDH status also influences surgical decisions, with supramaximal resection showing significant benefits in IDH mutant gliomas but a limited impact in IDH wild-type GBMs.31 At the molecular level, IDH mutations frequently cooccur with p53 mutations and 1p/19q codeletions32 but remain mutually exclusive with EGFR amplification and chromosome 10 loss,33 further reinforcing their role in glioma subtyping. The formal integration of the IDH status into the WHO glioma classification solidifies IDH mutations as essential diagnostic and prognostic molecular markers,17,29,34 emphasizing the need for molecularly driven therapeutic approaches to improve GBM patient outcomes.
MGMT promoter methylation
The MGMT gene is crucial for DNA repair and cellular defense, counteracting alkylating chemotherapy-induced damage by removing alkyl groups from the O6 position of guanine. Methylation status critically influences the GBM treatment response by regulating gene expression. Hypermethylation leads to transcriptional silencing, impairing the ability of tumors to repair alkylating agent-induced DNA damage and thereby increasing their sensitivity to temozolomide (TMZ). This epigenetic alteration is more prevalent in secondary GBMs than in primary GBMs or their precursor low-grade gliomas (LGGs) and serves as a robust predictive biomarker for chemotherapy efficacy, particularly in GBM.35 Clinical trials, including NOA-08, the Nordic trial, and RTOG 0525, have demonstrated that patients with MGMT-methylated tumors benefit significantly from TMZ treatment, resulting in prolonged PFS and OS.36–38 This predictive value is especially evident in elderly patients, where the MGMT status guides therapeutic decisions between chemotherapy and radiotherapy.39 Conversely, unmethylated MGMT tumors maintain their DNA repair capacity, diminishing the effectiveness of alkylating agents and correlating with poor outcomes.40 With approximately 50% of GBMs exhibiting MGMT promoter methylation, routine testing is increasingly recognized as essential for tailoring personalized treatment strategies. Notably, the MGMT methylation status outperforms conventional prognostic indicators such as tumor grade, performance status, and patient age in predicting therapeutic response, underscoring its clinical relevance. Future research should focus on strategies to overcome resistance in MGMT-unmethylated tumors, exploring novel therapeutic approaches to increase treatment efficacy. The continued evolution of molecular classification in GBM highlights the MGMT status as a crucial determinant of personalized treatment approaches, paving the way for improved patient outcomes.
Imaging biomarkers
Although no clinically approved imaging biomarkers currently exist for GBM, advanced functional imaging techniques hold significant potential in tumor characterization and treatment planning. Methods such as diffusion-weighted magnetic resonance imaging (DW-MRI), dynamic susceptibility contrast-enhanced perfusion imaging, MR spectroscopy, and positron emission tomography (PET) offer valuable insights into tumor biology, genetic alterations, and therapeutic response.41 However, variations in sensitivity and specificity across studies highlight the need for standardized acquisition protocols and validation in clinical settings. Among promising imaging biomarkers, proton MR spectroscopy can detect 2-HG levels, which are correlated with IDH1/2 mutations, making it a noninvasive diagnostic and prognostic marker.42 Additionally, MRI-derived parameters such as apparent diffusion coefficient values,43 the T2-to-contrast-enhancing volume ratio, and relative cerebral blood volume have demonstrated predictive value for genetic alterations such as EGFR amplification and clinical outcomes such as PFS.44 Notably, increased tumor blood volume is strongly associated with an unmethylated MGMT status, further reinforcing the role of imaging biomarkers in guiding treatment response and prognosis.45
PET imaging plays a crucial role in GBM assessment, but the commonly used 18F-fluorodeoxyglucose (18F-FDG) PET has limited sensitivity because of high baseline glucose uptake in the brain, reducing its accuracy in detecting early recurrences and low-grade tumors. Consequently, alternative PET tracers, such as radiolabeled amino acids (11C-methionine [11C-MET], 18F-fluoroethyltyrosine [18F-FET], and 18F-fluoro-L-DOPA [18F-FDOPA]), along with hypoxia agents such as 18F-fluoromisonidazole (18F-FMISO), have gained prominence for their ability to visualize gliomas independent of BBB integrity. Studies indicate that higher 11C-MET uptake correlates with poorer survival, whereas 18F-FET and 18F-FDOPA effectively differentiate glioma grades and predict tumor proliferation,46,47 Hypoxia imaging using 18F-FMISO has emerged as a potential predictive biomarker that is correlated with tumor progression and decreased survival in GBM patients. Its ability to identify radiation-resistant tumor regions suggests applications in radiotherapy planning and treatment adaptation, providing critical insights for optimizing therapeutic strategies.48 These findings emphasize the growing relevance of molecular imaging in refining GBM prognosis and guiding personalized treatment approaches.
Despite promising results, further prospective validation is necessary before the integration of imaging biomarkers into routine clinical practice. The potential of these techniques to predict treatment response, detect early recurrence, and guide therapeutic strategies highlights their growing importance in GBM management. Future research should focus on optimizing imaging protocols, validating biomarkers across large patient cohorts, and integrating imaging data with molecular classification systems to enhance precision oncology approaches in GBM treatment.
Circulating biomarkers
Biochemical biomarkers
Circulating biochemical biomarkers have emerged as potential noninvasive tools for GBM diagnosis and prognosis, reflecting the molecular and immunological landscape of the disease. These biomarkers include proteins, cytokines, and traditional cancer markers, many of which have altered expression levels in GBM patient body fluids. Notable proteins, such as glial fibrillary acidic protein, brain-derived neurotrophic factor, protein S100B, and neural cell adhesion molecules, have been identified as neuronal markers linked to GBM pathology. Additionally, metabolic and inflammatory biomarkers such as 2-HG, chitinase-3-like protein 1, interleukin-2 (IL-2), transforming growth factor-β (TGF-β), tumor necrosis factor-α (TNF-α), and matrix metalloproteinases (MMPs) have been implicated in GBM progression and immune modulation.49 Despite extensive research, many circulating biochemical biomarkers lack tumor specificity, limiting their diagnostic utility. However, advancements in proteomic profiling have led to the identification of more promising biomarker candidates. Recent studies have highlighted a panel of biomarkers with high diagnostic accuracy, with six markers demonstrating over 80% efficiency in distinguishing GBM from nontumor conditions. Among the most promising biomarkers, the overexpression of complement component C9 (C9), C-reactive protein, and leucine-rich α-2-glycoprotein (LRG1) is strongly correlated with GBM tumor burden and progression. Conversely, low expression of gelsolin, apolipoprotein A-IV and the Ig α-1 chain C region has also shown diagnostic significance. Importantly, the concentrations of C9, CRP, and LRG1 are significantly associated with tumor size, reinforcing their potential role in GBM prognosis and clinical stratification.50
The identification of circulating biochemical biomarkers represents a promising avenue for noninvasive GBM detection and monitoring. However, further validation in large-scale clinical studies is essential to establish their diagnostic reliability and prognostic value. Future research should focus on standardizing biomarker panels, integrating multiomics approaches, and improving specificity to increase the clinical utility of biochemical biomarkers in GBM management.
Circulating tumor cells
Circulating tumor cells (CTCs) play a pivotal role in GBM progression, offering valuable insights into tumor behavior, treatment response, and prognosis. The presence of these genes in the bloodstream correlates with tumor progression, recurrence, and the GBM subtype, establishing them as promising biomarkers for disease monitoring. As a noninvasive alternative to conventional biopsies, CTC-based liquid biopsy allows real-time tracking of tumor dynamics, enabling repeated assessments over time without the need for invasive procedures.51 The prevalence of CTCs in GBM exceeds 75%, with their levels directly reflecting the tumor burden and therapeutic response.52 A decrease in CTC counts post-therapy indicates treatment efficacy, whereas persistent or rising levels may suggest resistance to therapy. Additionally, CTC genetic profiling can be used to determine drug sensitivity, paving the way for personalized treatment strategies in GBM.53
CTCs represent a critical diagnostic and prognostic tool with potential applications in therapy selection and disease monitoring. Their quantification and molecular analysis provide insights into tumor evolution, facilitating precision oncology approaches.53 The integration of CTC assessment into routine clinical practice could enhance treatment personalization, improve early detection of therapeutic resistance, and optimize GBM management strategies. However, further standardization and validation in large-scale clinical studies are essential to establish their full clinical utility.
Circulating RNA
Circulating RNA biomarkers, including circRNAs, miRNAs, and lncRNAs, serve as powerful, noninvasive diagnostic tools in GBM, enabling early detection, precise prognosis assessment, and real-time monitoring of treatment response. Their presence in the bloodstream and cerebrospinal fluid (CSF) offers a unique opportunity to track tumor dynamics, paving the way for personalized therapeutic strategies and improved clinical outcomes in GBM management.
circRNAs
Dysregulated circRNA expression is a defining feature of GBM progression, influencing cell proliferation, metastasis, angiogenesis, and oncogenesis. High-throughput RNA sequencing and microarray analysis have identified numerous differentially expressed circRNAs in tumor tissues, highlighting their potential as diagnostic and prognostic biomarkers.54 Table S2 lists the circRNAs that serve as biomarkers for GBM and are involved in pathogenesis (Table S3). Studies have revealed that the expression of most circRNAs is greater in normal brain tissues than in GBM tissues, with only a few displaying elevated levels in tumor samples.55 Notably, circ-SMARCA5 is significantly downregulated in GBM, whereas circ-CFH and circ_0012129 are upregulated,56 indicating their distinct roles in tumor progression. Additionally, circRNA_0037655 and circ-MAPK4 promote tumor survival and invasion,57 whereas circ-E-cadherin and circ-XRCC5 are linked to GBM aggressiveness and poor prognosis.58 In contrast, circ-DCL1 suppresses tumor proliferation through METTL3-mediated m6A modification,59 highlighting the dual role of circRNAs as oncogenes and tumor suppressors.
In addition to promoting tumor proliferation, circRNAs interact with the TME to increase GBM progression. circ-NEIL3 stabilizes insulin-like growth factor (IGF)-2 mRNA binding protein 3, facilitating exosomal transfer to TAMs and thereby reinforcing their immunosuppressive functions.60 Moreover, circ-LGMN, which is significantly upregulated in HGGs, drives GBM malignancy by regulating legumain.61 The identification of circRNAs as potential biomarkers presents promising opportunities for noninvasive GBM diagnosis and personalized treatment strategies. Their expression profiles provide critical insights into tumor behavior, prognosis, and therapeutic response. However, further large-scale validation and functional studies are necessary to standardize circRNA-based biomarker panels, paving the way for their integration into clinical GBM management.
lncRNAs
lncRNAs have emerged as key prognostic biomarkers in GBM, offering insights into tumor progression, survival prediction, and therapy resistance. Table S4 presents the lncRNAs that serve as biomarkers for GBM and its pathogenesis (Table S5). Studies have revealed that several lncRNAs are strongly correlated with tumor grade, survival rates and treatment response, highlighting their clinical relevance. Among the most significant lncRNAs, the lncRNA MAGI2-AS3 is upregulated in GBM, and its expression is positively correlated with tumor grade and the Karnofsky performance score (KPS). Lower levels of the lncRNA MAGI2-AS3 are associated with poorer survival outcomes, making it an independent predictor of OS.62 Similarly, the lncRNA ELF3-AS1 is significantly elevated in tumor tissues, reinforcing its potential as a GBM-specific biomarker.63 Additionally, the lncRNA PXN antisense RNA-1 is overexpressed in GBMs and serves as an indicator of poor prognosis.64 The diagnostic value of N6-methylandenosine (m6A)-related lncRNAs has also been demonstrated in prognostic models incorporating m6A-LPS, age, and WHO grade, effectively predicting OS in LGG patients.65 Furthermore, elevated levels of the lncRNA HOTAIR in GBM patient serum further support the diagnostic utility of lncRNAs.66
lncRNAs also contribute to therapy resistance and immune regulation in GBM. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) expression is linked to TMZ resistance, positioning it as a prognostic marker for chemoresistant GBMs.67 Immune-related lncRNAs, such as DiGeorge syndrome critical region gene 5, are associated with immune and stromal cell infiltration, highlighting their role in regulating the tumor immune response.68 Additionally, upregulation of the lncRNA CRNDE in GBMs is linked to tumor size, recurrence risk, and chemosensitivity to TMZ,69 reinforcing its role in predicting therapeutic response. Some lncRNAs, such as the lncRNA GAS5, are expressed at higher levels in LGGs than in GBM,70 suggesting their role in monitoring tumor progression. Conversely, the zinc finger E-box-binding homeobox 1 (ZEB1)-lncRNA AS1 and the lncRNA ANRIL are highly expressed in GBM and are correlated with tumor size and malignancy grade.71 However, further large-scale validation and functional studies are necessary to establish their clinical applicability. Integrating lncRNA-based biomarker panels into GBM diagnosis and personalized treatment strategies could enhance patient stratification, therapeutic decision-making, and overall clinical outcomes.
miRNAs
miRNAs have demonstrated significant potential as biomarkers for diagnosis, tumor grading, and monitoring treatment response in GBM.72 Among these, miR-21 is consistently upregulated in GBMs, with elevated levels detected in the CSF and serum of HGG patients, making it a reliable biomarker for early detection and disease progression.73 Additionally, the levels of miR-21, miR-222, and miR-124-3p are significantly elevated in HGGs compared with those in LGGs and healthy patients, with levels decreasing post-surgery,74 reinforcing their prognostic and diagnostic importance. Several miRNAs, such as miR-128 and miR-342-3p, are downregulated in GBM, increasing after surgery and chemoradiation, suggesting their potential as indicators of therapeutic efficacy. Similarly, miR-20a-5p, miR-106a-5p, and miR-181b-5p are associated with tumor progression, whereas miR-19a-3p, miR-106a-5p, and miR-181b-5p are linked to poor prognosis.75 Furthermore, miR-1238 is elevated in recurrent GBM, highlighting its role in disease monitoring and predicting recurrence risk.76
miR-301a expression is correlated with tumor progression and a reduced KPS, with exosomal levels dynamically changing following tumor resection and recurrence, making it a valuable biomarker for disease monitoring.77 Additionally, exosomal miR-210, miR-5194, and miR-449 target key genes in the EGFR and cellular mesenchymal epithelial transition (c-MET) signaling pathways and are correlated with histopathological grade and GBM aggressiveness.78 Table S6 presents the miRNAs that serve as biomarkers associated with GBM pathogenesis. Some miRNAs, such as miR-524-3p and miR-524-5p, are downregulated in GBM and associated with EGFR overexpression and EGFRvIII mutation, while their overexpression inhibits tumor proliferation and migration, improving OS through the TGF-β, Notch, and Hippo pathways.79 Similarly, low miR-133 levels correlate with poor prognosis, as its overexpression inhibits EGFR mRNA translation, suppresses GBM growth and induces apoptosis.80 Conversely, miR-148a functions as an oncogene, negatively impacting survival through its regulation of BIM, MIG6, and EGFR,81 making it a potential therapeutic target.
miRNA expression profiling offers a noninvasive and dynamic approach for GBM diagnosis, prognosis, and treatment monitoring. miR-34a deletion and EGFR amplification are linked to poor survival, whereas high miR-340 and miR-615 expression are correlated with longer overall and recurrence-free survival,82,83 reinforcing their potential as independent prognostic factors. The identification of circulating miRNAs in serum and plasma provides a powerful tool for personalized GBM management, allowing early detection, prediction of therapeutic response, and disease monitoring. However, further validation in large-scale clinical studies is essential to fully integrate miRNAs into routine GBM diagnostics and treatment planning.
Circulating DNA
The analysis of circulating tumor DNA (ctDNA) provides a noninvasive approach for disease monitoring and treatment response assessment in GBM. Studies have demonstrated that circulating cell-free DNA levels fluctuate throughout treatment, with elevations before surgery and at disease progression, reinforcing its potential as a dynamic biomarker.84 Importantly, next-generation sequencing and methylation assays have identified key genetic alterations in ctDNA, including mutations in genes such as p53, EGFR, MET, PIK3CA, and NOTCH1, highlighting the feasibility of liquid biopsies in molecular profiling and personalized therapy selection.85 The detection rates of ctDNA in GBM remain variable, with 51% of advanced primary GBM patients exhibiting detectable ctDNA, some of whom have genomically targetable mutations.86 Notably, somatic alterations in genes such as p53, JAK2, NF1, EGFR, BRAF, IDH1, NRAS, GNAS and ataxia telangiectasia mutated (ATM) further illustrate the genetic heterogeneity of GBM,87 underscoring the importance of ctDNA in tumor characterization. Additionally, CSF-based ctDNA analysis has shown higher sensitivity than plasma ctDNA analysis,87 suggesting that CSF-based ctDNA analysis is a more reliable method for tumor-specific genetic assessment.
ctDNA has demonstrated potential in detecting drug resistance mutations in patients receiving kinase inhibitor therapy, aiding in treatment adaptation and precision oncology approaches. Furthermore, integrated platforms analyzing key genes such as IDH1, IDH2, p53, ATRX, TERT, and H3 histone family 3 A (H3F3A) enable more efficient subclassification of diffuse gliomas.88 However, ctDNA detection remains challenging in localized tumors such as GBMs, emphasizing the need for further optimization of ctDNA extraction and analysis methods. As liquid biopsy technology advances, refining ctDNA-based assays will be crucial in enhancing early detection, disease monitoring, and therapeutic decision making in GBM.
Extracellular vesicles
EVs have emerged as promising noninvasive biomarkers for GBM and play critical roles in tumor progression, intercellular communication, and treatment response monitoring. GBM and stromal cells release tumor-associated EVs into bodily fluids such as plasma, serum, CSF, and urine, providing an accessible liquid biopsy tool for disease monitoring and molecular profiling.89 Elevated EV concentrations in the peripheral blood of GBM patients, independent of specific molecular alterations (EGFR amplification, PTEN deletion, MGMT expression, and IDH mutations),90 suggest their broad applicability in GBM detection, prognosis, and relapse prediction. Additionally, fluctuations in EV concentrations are correlated with surgical resection and recurrence,91 reinforcing their potential as dynamic biomarkers. Table S7 lists the EVs that serve as biomarkers for GBM diagnosis.
In addition to their presence in the circulation, EVs carry molecular cargo, including DNA, RNA, and proteins, reflecting the genetic and epigenetic landscape of tumors. Plasma EV-based markers such as EGFR, EGFRvIII, and IDH1-R132H mutations have demonstrated high specificity for GBM classification and subtyping. The tumor progression index, which incorporates EV counts and molecular cargo,92,93 effectively differentiates treatment responders from nonresponders, offering a refined tool for therapy monitoring. The detection of IDH1 mutations in EV-derived DNA from plasma and CSF provides a minimally invasive alternative to conventional tissue biopsies, enabling a comprehensive molecular assessment of GBM. The presence of EGFRvIII in CSF-derived EVs, even when it is absent in tissue biopsies,94 underscores the superiority of EVs in capturing tumor heterogeneity,95 offering insights into oncogenic signaling and tumor progression.
EV-based biomarkers show potential for assessing treatment response and predicting patient outcomes. Studies indicate that PTEN and MGMT mRNA levels in GBM-derived EVs (GDEVs) correlate with tumor grade and therapy response,96 whereas miR-21 in CSF-derived EVs is linked to poor prognosis.73 Moreover, EV-associated epigenetic modifications, including DNA methylation, reflect the molecular profile of tumors,97 supporting their role in real-time GBM monitoring. With increasing evidence supporting the use of EV-based biomarkers, their integration into clinical GBM management could revolutionize diagnosis, treatment response assessment, and personalized therapy strategies. However, further validation through large-scale studies is essential to standardize EV-based assays for routine clinical application in GBM.
Regulatory mechanisms in GBM pathogenesis
Epigenetic characteristics of GBM
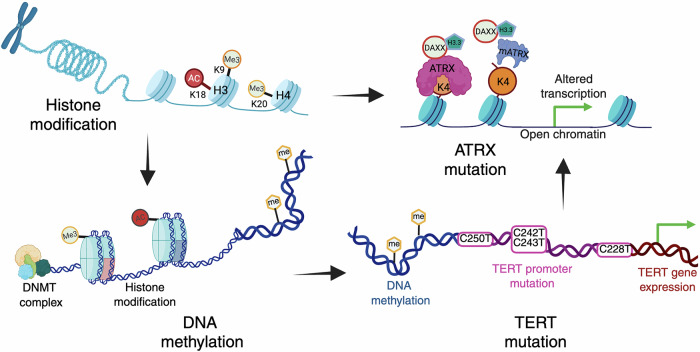
GBM pathogenesis is driven by a combination of extensive genetic and epigenetic alterations that regulate gene expression and tumor progression. Among these, epigenetic changes, such as histone modifications, DNA methylation, and chromatin remodeling, play a central role in tumor biology. Aberrant histone methylation and acetylation, ATRX mutations impacting chromatin stability, and widespread promoter hypermethylation, including MGMT, disrupt the balance between tumor suppressor genes and oncogenic pathways. Furthermore, TERT promoter mutations activate telomerase, enabling replicative immortality, whereas copy number alterations exacerbate the dysregulation of key cellular pathways. The intricate crosstalk between these epigenetic mechanisms drives genomic instability, tumor proliferation, and therapy resistance, highlighting their importance in GBM pathogenesis and their potential as promising therapeutic targets (Fig. 3).
Fig. 3.
Epigenetic characteristics of glioblastoma and their role in pathogenesis. The figure depicts the key epigenetic mechanisms contributing to glioblastoma (GBM) development, including histone modifications, DNA methylation, ATRX mutations, and TERT promoter mutations. Histone modifications such as methylation (Me3) and acetylation (Ac) at specific lysine residues regulate chromatin accessibility and gene expression. DNA methylation, which is catalyzed by DNA methyltransferases (DNMTs), further influences gene silencing or activation. ATRX mutations impair chromatin remodeling by disrupting the ATRX-DAXX complex, which is responsible for H3.3 deposition, leading to altered transcription and increased chromatin accessibility. TERT promoter mutations result in aberrant telomerase expression, contributing to tumor cell immortality
Histone modification
Histone modifications are crucial regulators of gene expression, influencing GBM progression, tumor proliferation, and therapy resistance. Histones undergo various modifications, including acetylation and methylation. Acetylation typically promotes gene activation, whereas methylation can either enhance or repress transcription, depending on the specific histone site.98 Disruptions in these processes contribute to GBM aggressiveness and treatment resistance.99 Distinct histone modification patterns are correlated with prognosis; for example, lower H3K18 acetylation (H3K18Ac) is linked to improved survival in primary GBM, whereas higher H4K20 trimethylation (H4K20me3) is associated with better outcomes in secondary GBM. Additionally, H3K9 trimethylation (H3K9me3), a transcriptional repressor, is linked to IDH mutant gliomas, distinguishing them from wild-type GBM.100
Mutations in H3F3A, including H3.3 and H3.1, frequently occur in pediatric GBM and drive distinct epigenetic alterations. The K27M mutation disrupts histone methylation and acetylation, whereas the G34R/G34V mutations alter transcription regulation. These mutations alone do not initiate tumor formation but act alongside additional genetic changes.101 The H3K27M mutation inhibits the polycomb repressive complex 2 chromatin-modifying complex, influencing the transcriptional programs associated with pediatric GBM.102 Enhancer of zeste homolog 2 (EZH2) overexpression further drives oncogenic pathways, including c-Myc activation, which is correlated with poor prognosis. Targeting EZH2 suppresses tumor growth, enhances radiation sensitivity, and disrupts GSC maintenance, making it a promising therapeutic approach. Similarly, dysregulation of protein arginine methyltransferase 1 (PRMT1) and PRMT2 alters histone methylation, further driving GBM progression and therapy resistance.103
Lysine demethylases (KDMs) and HDACs regulate tumor proliferation, cell death, and therapy resistance in GBM. KDM5A overexpression contributes to TMZ resistance, and its inhibition enhances the treatment response.104 HDAC9, a regulator of Hippo signaling via TAZ activation, promotes GBM progression, highlighting HDAC9 inhibition as a therapeutic strategy.105 Ras-related protein on chromosome 22 (RRP22) functions as a tumor suppressor, with low expression linked to increased tumor grade and reduced survival. Its downregulation is associated with 5’-CpG island hypermethylation and altered histone acetylation (H3/H4 acetylation loss). In primary GBM, elevated H3K9me3 levels and reduced pan-Ac-H3-bound RRP22 expression further implicate epigenetic dysregulation in tumor progression.106 Targeting EZH2, KDMs, and HDACs offers promising avenues for overcoming treatment resistance and improving therapeutic outcomes. Understanding the interplay between histone modifications and transcriptional regulation is essential for advancing precision medicine strategies in GBM treatment.
ATRX mutation
ATRX mutations play a critical role in GBM pathogenesis by disrupting chromatin remodeling, telomere maintenance, and DNA repair. ATRX loss is associated with alternative lengthening of telomeres, a hallmark of genomic instability, and is predominantly observed in LGGs with IDH mutations and without 1p/19q codeletion.107 Although rare in adult primary GBM, ATRX mutations are more prevalent in younger patients and secondary GBMs, where they correlate with improved prognosis. Their presence offers potential as prognostic biomarkers and therapeutic targets in GBM. ATRX deficiency has been shown to accelerate GBM growth and reduce survival, linking ATRX loss to DNA repair deficiencies, particularly impaired nonhomologous end joining.108 These findings suggest that ATRX-deficient GBMs are vulnerable to therapies targeting DNA damage repair pathways.
In pediatric GBM, ATRX mutations contribute to genetic instability, influencing mutation rates and molecular subtypes. Studies have revealed that ATRX mutations in grade II–III astrocytomas, oligoastrocytomas, and secondary GBMs often cooccur with IDH1 mutations and ALT activation.109 Additionally, the H3.3–ATRX–DAXX chromatin remodeling complex is frequently altered in pediatric GBMs, underscoring the role of ATRX in tumor development.101 These findings emphasize that ATRX mutations are key molecular markers for glioma classification and potential therapeutic intervention. Further exploration of ATRX-related pathways may provide novel treatment strategies for ATRX-mutated GBMs, particularly through targeted approaches that disrupt ALT and DNA repair mechanisms.
DNA methylation
DNA methylation, which is mediated by DNA methyltransferases, is a critical epigenetic modification that influences gene expression, tumor progression, and therapeutic response in GBM. Advances in genome-wide methylation profiling have significantly improved tumor classification, prognosis, and treatment stratification.110 DNA methylation patterns provide insights into molecular subtypes, with studies demonstrating their accuracy in predicting key glioma features such as IDH mutations and 1p/19q codeletions.111 These findings highlight methylation profiling as a powerful diagnostic and prognostic tool that complements histopathological classification. The identification of methylation signatures, such as those distinguishing IDH mutant and IDH wild-type GBMs, provides a framework for personalized treatment strategies. The integration of methylation biomarkers, including three-gene signatures (EMP3, GSX2, and EMILIN3), has demonstrated prognostic potential in GBM patients,112 allowing for more precise risk assessment and therapeutic decision-making.
In addition to classification, the DNA methylation status is instrumental in predicting therapy response. Recent studies have linked low DNMT1 expression with TMZ resistance, suggesting that alterations in DNA methylation may serve as indicators of treatment efficacy.113 Emerging research has also identified methylation patterns in DNA damage response (DDR) genes, including MGMT, MLH3, RAD21, and SMC4, as potential biomarkers for therapy response prediction.114 Findings from the EORTC 22033 phase III trial further underscore the clinical relevance of molecular stratification in GBM treatment. While no overall difference in progression-free survival was observed between dose-dense TMZ and radiotherapy, IDH mutant, 1p/19q codeleted tumors responded more favorably to chemotherapy.115 This finding reinforces the role of DNA methylation profiling in optimizing treatment regimens. The continued exploration of DNA methylation in GBM pathogenesis highlights its potential for refining diagnostic models, improving prognostic assessments, and guiding personalized therapies. Future research should focus on integrating methylation-based classifiers into clinical practice, further validating their predictive utility, and exploring novel epigenetic targets for therapeutic intervention.
Copy number alterations
Copy number alterations (CNAs) significantly impact genomic integrity, leading to the emergence of driver amplifications and deletions that disrupt crucial genes. Widespread chromosomal abnormalities, including losses on chromosomes 9 and 10 and polysomy of chromosomes 7, 19, and 20, are recurrently observed in GBM. Key focal alterations include CDKN2A/B deletions and high-level EGFR amplifications, which contribute to tumor progression and therapy resistance.116 Recent studies highlight the importance of CNA profiling in stratifying GBM patients and guiding clinical decision-making. Molecular characterization of CNAs improves the selection of treatment strategies, emphasizing the need for integrating CNA data into clinical trial designs to ensure more representative patient cohorts.117 Additionally, emerging findings suggest that both frequent and patient-specific CNAs influence survival outcomes, underscoring their potential for refining prognostic models.118 Computational analyses, such as those utilizing Oncoscape, have further demonstrated the prognostic significance of CNAs in GBM and diffuse gliomas. Multidimensional molecular grouping has enabled visualization of glioma classifications on the basis of CNAs, correlating specific chromosomal alterations with distinct survival outcomes. The identification of CNA-driven molecular subtypes reinforces their predictive value, highlighting critical genomic variations that could inform targeted therapeutic strategies.119 The incorporation of CNA profiling into routine clinical practice holds promise for improving patient stratification, treatment selection, and outcome prediction. Future research should focus on leveraging CNA data to refine GBM classification systems and develop personalized therapeutic approaches.
TERT promoter mutation
Mutations in the promoter region of the TERT gene have emerged as key molecular alterations in gliomas, influencing tumor progression, prognosis, and treatment response. These mutations, which primarily occur at C228T and C250T, create novel Ets/TCF binding sites, leading to aberrant TERT expression and sustained telomerase activity.120 The high prevalence of these genes in GBMs and other diffuse GBMs highlights their role in tumor maintenance and resistance to apoptosis. Clinical studies emphasize the prognostic importance of TERT promoter (TERTp) mutations, particularly in the context of other molecular alterations. In diffuse gliomas, TERTp mutations are associated with worse OS, with distinct prognostic implications depending on tumor grade and cooccurring mutations. For example, in Grade II and III gliomas, survival outcomes vary significantly on the basis of the interplay between TERTp mutations, MGMT methylation, IDH mutation, and 1p/19q codeletion.121 Notably, patients with IDH mutant gliomas and concurrent TERTp mutations have poorer prognoses than those with IDH mutations alone, underscoring the complex molecular interactions governing glioma progression.
The frequency and prognostic impact of TERTp mutations differ across glioma subtypes. Oligodendrogliomas, characterized by IDH mutation and 1p/19q codeletion, present the highest prevalence of TERTp mutations. In contrast, anaplastic astrocytomas and IDH wild-type GBMs also harbor these mutations but have varying prognostic outcomes. IDH wild-type GBMs with TERTp mutations exhibit particularly poor survival, reinforcing their value as prognostic biomarkers in this aggressive glioma subtype.122 These findings highlight the necessity of integrating the TERTp mutation status into glioma classification and clinical decision-making. Beyond prognostication, ongoing research into the mechanistic role of TERTp mutations may provide insights into novel therapeutic targets, potentially leading to the development of telomerase-directed therapies aimed at improving outcomes for GBM patients.
Loss of heterozygosity
Loss of heterozygosity (LOH) is a common genomic alteration in GBM that drives tumor progression by disrupting tumor suppressor genes. LOH occurs across several chromosomal regions, including 9p, 10q, 17p, 19q, and 22, with LOH at chromosome 10q being one of the most frequent and significant events in primary GBM, affecting approximately 70% of cases.123 Notably, LOH at 10q is more prevalent in older patients, suggesting a potential age-related influence on GBM tumorigenesis. The prognostic significance of LOH 10q is well established, particularly in differentiating primary from secondary GBM.124 LOH at 10q25-qter is highly specific for secondary GBM, whereas broader loss of 10q is associated with both primary and secondary subtypes. In contrast, LOH at 1p and 19q, although key molecular markers for oligodendrogliomas, lacks prognostic or predictive relevance in GBM.125 The tumor suppressor genes affected by LOH 10q, particularly PTEN, p53, and NF1, play crucial roles in regulating cell survival and proliferation. Among these, PTEN loss is particularly consequential, as it leads to dysregulation of the PI3K/AKT pathway, promoting unchecked cell growth and therapy resistance.126 Given the role of LOH 10q in GBM pathogenesis, integrating LOH analysis into molecular profiling could enhance prognostic assessment and guide targeted therapeutic strategies aimed at restoring tumor suppressor function or counteracting downstream oncogenic pathways.
1p/19q codeletion
The 1p/19q codeletion is a well-established prognostic biomarker in gliomas, particularly in oligodendrogliomas, where it is correlated with prolonged PFS and OS. This genetic alteration defines a distinct molecular glioma subtype, aiding in tumor classification and therapeutic decision-making. In LGGs, the iso-deletion of chromosome 1p alone is associated with a prognosis comparable to that of the full 1p/19q codeletion, whereas the iso-deletion of 19q alone also confers prolonged PFS.127 The frequency of 1p/19q codeletion varies among glioma subtypes, with the highest prevalence in oligodendrogliomas (WHO grade III) and a lower occurrence in astrocytomas. This alteration is strongly associated with IDH mutations and is almost mutually exclusive with ATRX mutations, reinforcing its role as a key molecular marker in glioma classification. Clinically, 1p/19q codeletion is linked to increased chemosensitivity, particularly in LGGs that respond favorably to TMZ-based therapy.128 Studies have demonstrated that patients with 1p/19q codeletion derive significant survival benefits from combined treatment with procarbazine, lomustine, and vincristine (PCV) chemotherapy alongside radiotherapy compared with radiotherapy alone.129 These findings emphasize its predictive role in optimizing treatment strategies. Given its strong association with favorable treatment response and prolonged survival, integrating the 1p/19q codeletion status into routine GBM management enhances personalized treatment planning and improves patient outcomes.
Fusion genes
Advances in sequencing technologies have led to the identification of oncogenic fusion genes in GBM, including those encoding FGFR, ALK, and EGFR, and neurotrophic tyrosine receptor kinase fusions. FGFR fusions are the most common, present in 8.33% of cases, followed by EGFR (4%) and ALK (1.9%), with the latter being more prevalent in pediatric GBM.130 NTRK1 fusions, although rare (1.2%), may contribute to GBM oncogenesis.131 Clinically, inhibitors such as lorlatinib and larotrectinib show promise in targeting fusion-positive GBMs.132,133 FGFR3-TACC3, a recurrent fusion protein, drives tumorigenesis by promoting kinase transphosphorylation and disrupting chromosomal stability. This fusion is mutually exclusive with IDH1/2 mutations and EGFR amplification, suggesting its role as an independent driver of GBM progression.130 Additionally, the PTPRZ1-MET fusion represents another oncogenic event, warranting further investigation as a potential therapeutic target.134 The identification of these fusions highlights the importance of personalized treatment strategies in GBM, emphasizing the need for continued research into targeted therapies that exploit these unique molecular alterations.
Genetic alterations in GBM
Genetic alterations are fundamental to GBM pathogenesis, driving its aggressive growth and therapeutic resistance. Amplifications and mutations in RTKs, such as EGFR, PDGFR, and fibroblast growth factor receptors, lead to dysregulated signaling, promoting tumor proliferation and survival. Oncogenes such as MYB (myeloblastosis transcription factor), meningioma 1 (MN1), progranulin (PGRN) and amphiregulin (AREG) contribute to abnormal transcriptional activity and tumor progression. Concurrently, the loss or mutation of critical tumor suppressor genes, including p53 and PTEN, disrupts cell cycle regulation and DNA repair, fostering genomic instability. Deletions in CDKN2A (cyclin-dependent kinase inhibitor 2A) impair the cell cycle checkpoint, whereas aberrant activation of stem cell markers such as SRY-Box transcription factor 2 (SOX2) supports tumor cell self-renewal and invasion. These genetic changes collectively form the backbone of the highly malignant nature of GBM, underscoring the complexity of its molecular landscape and the challenges in developing effective treatments (Fig. 4).
Fig. 4.
Genetic alterations driving glioblastoma pathogenesis. The schematic illustrates key oncogenic genetic alterations contributing to glioblastoma (GBM) development and progression. Receptor tyrosine kinases (RTKs), such as EGFR, PDGFR, FGFR, and VGFR, initiate downstream signaling cascades, including the PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways. Loss of tumor suppressors (e.g., PTEN, CDKN2A, RB1, p53) and overactivation of oncogenes (e.g., EGFR, MDM2, CDK4/6, TERT, MYB, SOX2, AREG) promote cell cycle progression, proliferation, stemness, survival, angiogenesis, and resistance to apoptosis. DNA damage response elements (ATM/ATR-Chk1/Chk2) are activated by radiation and chemotherapy (TMZ) but are frequently bypassed in GBM. Downstream transcriptional regulators such as MYB and SOX2 further enhance tumor cell plasticity and malignancy. Collectively, these alterations reprogram the tumor cell phenotype, driving GBM progression and therapy resistance
EGFR
The amplification and mutation of EGFR, particularly the EGFRvIII variant, are defining characteristics of GBM, especially in the classical subtype and primary GBM cases. EGFR, a key tyrosine kinase receptor, regulates critical growth factors involved in tumor proliferation and survival. The EGFRvIII mutation results from a deletion of exons 2 and 7, eliminating the extracellular ligand-binding domain and leading to constitutive receptor activation. This alteration, driven by histone modifications at the EGFR enhancer region on chromosome 7p12, contributes to uncontrolled tumor growth and resistance to apoptosis.135 EGFR-mediated activation of the RTK/RAS/PI3K signaling axis disrupts the G1/S checkpoint, facilitating unchecked cell cycle progression.136 Patients with EGFRvIII mutations exhibit worse survival outcomes than those with wild-type EGFR, with coexpression of both forms further exacerbating tumor aggressiveness. This is attributed to cross-phosphorylation between EGFRvIII and wild-type EGFR, amplifying oncogenic signaling cascades.137 Notably, EGFRvIII expression is correlated with increased tumor heterogeneity, complicating treatment responses and limiting the efficacy of targeted therapies, including tyrosine kinase inhibitors and immunotherapy.138
In addition to the classical RTK/RAS/PI3K pathway, EGFRvIII activation has been linked to alternative tumorigenic mechanisms involving forkhead box G1 protein (FOXG1) and SOX9,139 which contribute to GBM stemness and invasive potential. These findings underscore the complexity of EGFR-driven oncogenesis and the necessity for precision-based therapeutic strategies. Given the resistance of EGFR-altered tumors to conventional EGFR inhibitors, ongoing research into combination therapies and novel targeted approaches remains critical for improving treatment efficacy in EGFR-mutant GBM. Understanding the molecular interplay between EGFR mutations and tumor behavior is essential for advancing therapeutic interventions and optimizing patient outcomes.
PDGFR
A distinct subset of GBMs, classified as the PDGFR subclass, accounts for approximately 25–30% of cases and is characterized by aberrant PDGFR signaling. The dysregulation of PDGFR in these tumors arises through various genetic mechanisms, including PDGFRA gene amplification, chromosomal rearrangements, and the overexpression of PDGF ligands.140 These alterations contribute to enhanced tumor cell proliferation, survival, and invasion. Age-related differences in PDGF signaling have been observed in GBM. Tumors in patients over 65 years of age exhibit significantly higher PDGFA expression levels than those in younger individuals do, with an increased PDGFA/PDGFRA expression ratio.141 In contrast, pediatric GBM patients show a greater prevalence of PDGFRA amplification than adult GBM patients do.142 This amplification is notably associated with tumors affecting the corpus callosum and is frequently linked to the aggressive H3K27M mutation found in diffuse midline gliomas.143 Despite its frequent occurrence, the prognostic significance of PDGFRA amplification remains uncertain. While some studies have associated PDGFRA amplification with poor survival (PS) outcomes, particularly in diffuse midline gliomas, its predictive value as an independent biomarker in GBM remains debated. Further research is needed to clarify its role in disease progression and response to targeted therapies. Given the therapeutic challenges associated with PDGFR-driven GBM, ongoing investigations into PDGFR inhibitors and combination treatment strategies could provide new avenues for improving patient outcomes.
FGFR
Lesions with FGFR1-TKDD mutations are primarily diffuse gliomas located in the cerebral cortex. Duplications of the FGFR1 TKD have also been found in low-grade astrocytomas, including pilocytic astrocytomas and dysembryoplastic neuroepithelial tumors (DNETs), which are typically located outside the cerebellum.144 These mutations are notable features of low-grade neuroepithelial tumors (LGNTs), occurring in 7.4% to 24% of cases, but they are rare in HGGs. In a study screening 33 HGG cases for FGFR1 region duplication in the tyrosine kinase domain, only one tumor was found to be positive for FGFR1-TKDD. This tumor, which was diagnosed as an anaplastic oligoastrocytoma (WHO grade III) that had progressed from a grade II tumor, exhibited FGFR1-TKDD positivity. Notably, FGFR1-TKDD has not been identified in adult-type oligodendrogliomas with IDH mutations and 1p/19q codeletion.145,146 Additionally, there was a case report of a glioneuronal tumor with features of both pilocytic astrocytoma and pleomorphic xanthoastrocytoma, which also carried FGFR1-TKDD and showed focal increases in mitotic activity.147 These findings highlight the range of gliomas associated with FGFR1-TKDD mutations and the need for further study to understand their clinical significance and potential treatment approaches.
MYB
MYB transcription factors, including MYBL1, function as proto-oncogenes that regulate progenitor cell proliferation and differentiation. In GBMs, MYB gene alterations are more common in young children and primarily affect tumors in the cerebral hemispheres. cIMPACT-Now Update 4 highlights the importance of integrated diagnostics in assessing WHO grade II IDH wild-type/H3-wild-type diffuse gliomas, particularly those with MYB or MYBL1 rearrangements.148 These mutations are associated with a favorable prognosis, with gliomas harboring MYB or MYBL1 alterations demonstrating prolonged disease stability and high OS rates. Reports indicate a 10-year OS rate of 90% and a 10-year PFS rate of 95%. The WHO CNS5 classification introduced diffuse astrocytoma, MYB- or MYBL1-altered, as a distinct entity within pediatric-type diffuse LGGs, designating it as a CNS WHO grade I tumor. MYB alterations are also highly prevalent in angiocentric gliomas, reinforcing their role in glioma subtyping.149 Future research will likely focus on MYB- and MYBL1-driven oncogenic mechanisms, particularly in pediatric LGGs, to refine diagnostic classification and identify targeted therapeutic strategies. Understanding MYB-driven pathways may lead to more personalized treatment approaches, potentially minimizing the need for aggressive therapies while maintaining favorable survival outcomes.
MN1
The MN1 gene, located on chromosome 22q, functions as a transcriptional coregulator and is frequently altered in astroblastomas, a rare glioma subtype predominantly affecting pediatric and young adult populations. The WHO CNS5 classification designates astroblastomas with MN1 alterations as a distinct molecular entity, yet further research is needed to differentiate them from other neuroepithelial tumors with overlapping genetic features. MN1 alterations have emerged as potential prognostic markers, with studies indicating improved PSF and OS in gliomas exhibiting MN1 rearrangements.150,151 Compared with BRAF V600E-mutated pleomorphic xanthoastrocytomas, MN1-rearranged astroblastomas have a more favorable prognosis.152 However, the mechanistic role of MN1 in tumorigenesis remains unclear, necessitating further studies to elucidate its functional impact on glioma biology and its potential utility in guiding clinical decision-making. Expanding the molecular characterization of MN1-altered gliomas could increase diagnostic accuracy and inform targeted therapeutic approaches.
PGRN and AREG
PGRN and AREG have emerged as critical players in GBM pathogenesis, each contributing uniquely to tumorigenesis, progression, and therapeutic resistance. PGRN, a member of the adipokine family, has gained attention for its elevated expression in GBM tissues compared with that in normal brain tissue, where it is correlated with increased tumor cell proliferation, pathological grading, and disease severity.153 Notably, PGRN levels in patient serum mirror those in tumor tissues, with higher expression linked to poorer overall and disease-free survival, particularly in LGGs.154 Multivariate analysis has identified PGRN as an independent prognostic factor, emphasizing its potential as a diagnostic and prognostic biomarker.155 Similarly, AREG, an EGFR ligand, plays crucial roles in GBM progression, drug resistance, and oncogenesis. AREG knockdown enhances doxorubicin (DOX)-induced endoplasmic reticulum stress, triggering autophagy and apoptosis and leading to GBM cell death. Bioinformatics analysis revealed that AREG is highly expressed in GBM and is correlated with PS.156 Additionally, AREG expression and methylation levels vary with astrocytoma grade, with GBM exhibiting higher mRNA expression but lower protein levels and increased methylation. Survival analysis revealed that AREG expression and methylation significantly impact patient prognosis, independent of astrocytoma grade.157 Furthermore, AREG is upregulated in microglia via colony-stimulating factor 1 receptor (CSF-1R) signaling, promoting GBM cell invasion. Blocking AREG through RNA interference or antibodies significantly reduces invasion, and the CSF-1R-MAPK/ERK pathway regulates its expression. Inhibiting ERK prevents microglia-stimulated invasion, and microglia require cell‒cell contact to increase invasion.158 Both PGRN and AREG are being explored as therapeutic targets, with preclinical studies investigating monoclonal antibodies, small-molecule inhibitors, and combination therapies to overcome resistance and improve outcomes. The dual roles of these genes as prognostic biomarkers and drivers of tumorigenesis make them promising candidates for advancing GBM research and treatment strategies.
SOX2
SOX2 is a critical regulator in GBM that influences key developmental pathways and contributes to tumor progression. Its overexpression is associated with increased proliferation, invasion, and self-renewal, particularly in GSCs.159 SOX2 is widely overexpressed across GBM but absent in normal central nervous system tissues,160 reinforcing its potential as a diagnostic and prognostic biomarker. High SOX2 levels are correlated with tumor aggressiveness and poor prognosis, making it a target of interest for therapeutic intervention. Studies have revealed a strong correlation between SOX2 expression and GBM malignancy, with the highest levels detected in aggressive GBM and oligodendrogliomas. SOX2 is particularly overexpressed in GBM, distinguishing malignant tissues from normal brain and nonmalignant tissues. SOX2-expressing cells are resistant to TMZ, but targeting SOX2 with inhibitors such as rapamycin has been shown to sensitize GBM cells to treatment,159 suggesting a potential strategy to increase therapeutic efficacy. Molecular profiling of GBM samples revealed frequent SOX2 amplification and overexpression, supporting its role in gliomagenesis. High SOX2 expression alone is sufficient to drive GBM cell invasion and migration. Additionally, silencing SOX2 in tumor-initiating cells (TICs) reduces tumor proliferation and tumorigenicity, emphasizing its functional importance in GBM progression.161,162 These findings underscore the importance of SOX2 as a biomarker for glioma classification and prognosis while highlighting its potential as a therapeutic target to improve treatment outcomes in aggressive brain tumors.
p53
p53 plays a crucial tumor-suppressive role in regulating cell cycle arrest, apoptosis, and DNA repair. Its function is tightly controlled by murine double minute (MDM) 2 and MDM4, which regulate p53 stability and activity through negative feedback mechanisms. While p53 alterations are less emphasized than other GBM markers, they are still significant in tumor pathogenesis. p53 mutations frequently occur early in gliomagenesis and accumulate as tumors progress. These alterations are particularly prevalent in the proneural GBM subtype, in contrast with the lower frequency in the classical subtype.19,20,163 The ARF-MDM2-p53 pathway is a major regulatory axis in GBM. The deletion of the CDKN2A/ADP-ribosylation factor (ARF) locus, which is observed in approximately 60% of GBM cases, contributes to p53 inactivation by impairing ARF-mediated MDM2 degradation. This disruption promotes tumor proliferation, invasion, and resistance to apoptosis.164 Additionally, MDM2 and MDM4 overexpression further suppresses p53 activity, leading to impaired DNA repair and enhanced tumor progression.165 Notably, MDM4-mediated p53 suppression is more common in classical GBM. Collectively, genetic alterations within the p53/MDM2/p14ARF pathway, including p53 mutations, MDM2 amplification, and p14ARF deletions, constitute major drivers of GBM pathogenesis.166
Targeting the p53/MDM2/p14ARF pathway represents a promising therapeutic avenue. Strategies aimed at restoring p53 function, including MDM2/MDM4 inhibitors and gene-editing approaches, could reactivate its tumor-suppressive role. Understanding how p53 mutations vary across GBM subtypes may enable more tailored therapeutic interventions. Given the high frequency of p53-related alterations, therapies targeting this pathway could improve GBM treatment outcomes by reinstating p53-driven tumor suppression.
CDKN2A
CDKN2A is a critical tumor suppressor gene that is frequently deleted or inactivated in GBM and LGGs. Its loss is associated with tumor progression, poor prognosis, and resistance to therapy. CDKN2A inactivation, primarily through homozygous deletion or promoter methylation, disrupts cell cycle regulation by impairing p16INK4a and p14ARF functions, leading to unchecked proliferation and reduced apoptosis. Genome-wide association studies have identified CDKN2A as a susceptibility locus for GBM, further highlighting its role in tumorigenesis.167,168 CDKN2A deletion is strongly linked to worse OS in astrocytoma patients, suggesting its utility as a prognostic biomarker.169 Lower CDKN2A expression is correlated with higher tumor grade and aggressive disease, reinforcing its relevance in glioma classification. Additionally, CDKN2A mRNA levels have been proposed as independent predictors of PFS and OS, supporting their potential clinical application in GBM management.170
Although targeting CDKN2A loss remains a challenge, its role in gliomagenesis underscores the need for therapeutic strategies aimed at restoring cell cycle control. Approaches such as CDK4/6 inhibitors, which compensate for p16INK4a loss, are being explored in GBM with CDKN2A deletion. Further research into CDKN2A-related pathways may provide new avenues for personalized GBM treatment, improving patient outcomes by integrating molecular diagnostics with targeted therapies.
PTEN
PTEN loss in GBM drives tumor progression and therapeutic resistance by dysregulating the PI3K/AKT/mTOR pathway, leading to uncontrolled cell growth, immune evasion, and an immunosuppressive TME. This is marked by increased PD-L1 expression, impaired T cell activation, and resistance to immune-mediated cell death, underscoring PTEN deficiency as a key factor in GBM immune escape.171,172 Additionally, PTEN loss alters the extracellular matrix (ECM) through the yes-associated protein 1 (YAP1) and lysyl oxidase (LOX) axes, facilitating angiogenesis and macrophage infiltration, which further supports tumor growth.173 Key mediators in this process include LOX and olfactomedin-like 3, which regulate macrophage and microglia recruitment. Inhibiting LOX in PTEN-deficient GBM enhances OLFML3 expression, promoting microglial infiltration via the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)-POZ/BTB and AT hook containing zinc finger 1 pathway. Targeting both macrophages and microglia through LOX inhibition and modulation of the CLOCK-OLFML3 axis, in combination with anti-PD-1 therapy, has demonstrated significant antitumor effects, highlighting a promising therapeutic strategy for GBM.174 This mechanism underscores the role of PTEN in modulating both cellular and microenvironmental factors in GBM progression.
The PTEN status of GBM has prognostic and therapeutic implications. Its loss is correlated with poor survival outcomes and resistance to standard treatments, including radiotherapy and chemotherapy. Given its central role in gliomagenesis, strategies aimed at restoring PTEN function or targeting downstream effectors, such as PI3K inhibitors or immune checkpoint blockade, are being explored as potential therapeutic approaches. Further research into PTEN-related pathways may enhance precision medicine strategies, offering new avenues for the effectiveness of GBM therapies.
Tumor microenvironment
Tumor heterogenicity
GBM exhibits significant intratumor heterogeneity, driven by clonal evolution and cancer stem cell models. The clonal evolution model suggests that cumulative genetic and epigenetic alterations drive tumor progression, whereas the cancer stem cell model posits that a subset of tumor-initiating cells sustains growth and therapeutic resistance.175 These mechanisms contribute to glioma diversity, with tumor clones adapting to distinct brain regions, metabolic environments, and microarchitectures.176 TICs, a subset of GSCs, play a central role in GBM progression and resistance to therapy. They interact with TAMs and tumor-infiltrating lymphocytes (TILs), modulating immune evasion and tumor survival. TAMs constitute a significant proportion of the TME, promoting vascularization and resistance to immune clearance.177,178 The concept of interclonal cooperativity highlights how tumor subpopulations and stromal components create a supportive microenvironment that enhances tumor adaptability and malignancy.179,180 GBM rarely metastasizes outside the brain but frequently recurs locally. Whole-exome sequencing of recurrent GBM suggests that these tumors arise from residual primary tumor cells, supporting a model of evolutionary adaptation to treatment.181 Tumor heterogeneity influences differential treatment responses, particularly the expression of key biomarkers such as MGMT and RTKs.182,183 Studies have revealed that mixed tumor cell populations with distinct RTK amplifications, including EGFR, MET, and PDGFRA, contribute to therapeutic resistance.184
Cellular communication within the tumor niche occurs through EVs and tunneling nanotubes (TNTs), which facilitate the intercellular transfer of oncogenic signals, metabolic factors, and resistance-conferring molecules. TNTs, which are composed of F-actin extensions, allow tumor cells to exchange mitochondrial DNA and other critical components, driving tumor repopulation following therapy.185 Additionally, the role of Bruton’s tyrosine kinase (BTK) in GBM core cells suggests that BTK is a potential biomarker for distinguishing intratumor spatial heterogeneity, with implications for targeted therapies.186 The complexity of GBM heterogeneity presents challenges for treatment, necessitating strategies that target multiple tumor subpopulations and their interactions with the microenvironment. Overcoming therapy resistance requires a deeper understanding of GBM cell plasticity, metabolic adaptations, and immune modulation. Future therapeutic approaches must integrate precision medicine strategies that account for the dynamic evolution of GBM for better clinical outcomes.
GBM stem cells
GSCs exhibit key features, such as treatment resistance, low proliferative activity, and tumor recurrence potential. These stem-like cells are categorized into mesenchymal and proneural subtypes, with evidence suggesting that proneural GSCs can transition into mesenchymal GSCs upon recurrence, contributing to GBM heterogeneity and therapeutic resistance.187,188 GSCs play crucial roles in tumor invasion and recurrence by migrating along the vasculature and white matter tracts, where they utilize cadherins, integrins, and MMPs.189,190 The invasive potential of these cells is driven by upregulated signaling pathways, including L1CAM, ephrin-B2,191 and epithelial‒mesenchymal transition (EMT)-associated factors such as twist-related protein 1 (TWIST1), SOX2, and signal transducer and activator of transcription 3 (STAT3).192 Additionally, GSCs exhibit heightened DNA repair capabilities, relying on Rad3-related kinase (ATR), ATM, poly(ADP‒ribose) polymerase 1 (PARP1), and other repair proteins, which contribute to their resistance to radiation and chemotherapy.193 Replication stress in GSCs, associated with prolonged transcription of long neural genes, results in increased reliance on DNA damage response pathways, including ATR and checkpoint kinase 1 (Chk1) activation. These adaptations increase GSC survival under genotoxic stress, suggesting potential therapeutic targets.194
GSCs modulate the TME by promoting immunosuppressive mechanisms. They induce M2 differentiation in glioma-associated macrophages (GAMs) through periostin secretion and IL-10 signaling, contributing to immune evasion.195 Additionally, GSC-derived pericytes support angiogenesis, promoting vascular abnormalities and BBB disruption.190 The Wnt and Sonic hedgehog signaling pathways maintain GSC self-renewal and therapy resistance. Aberrant Wnt activation, influenced by FAT atypical cadherin 1 (FAT1) mutations, enhances tumor progression, whereas sonic hedgehog signaling promotes Nanog expression and drug efflux transporter activity, further increasing chemoresistance.196 GSCs contribute to GBM relapse by resisting conventional therapies. The ability of these cells to persist in a quiescent state, evade apoptosis, and promote tumor angiogenesis underscores the need for targeted strategies.197 Future research should focus on disrupting GSC-specific pathways, enhancing tumor immunogenicity, and integrating novel therapies to support improved disease management in GBM.
Angiogenesis
Angiogenesis is a key process in GBM progression and is driven by multiple growth factors and signaling pathways. VEGF is a primary regulator, and its expression increases with tumor grade, promoting vascular proliferation and tumor progression.198 VEGFR-1 and VEGFR-2 activation play distinct roles in GBM initiation and malignancy.199 The overexpression of VEGF and VEGFR-1 in low-grade astrocytomas is correlated with poor prognosis, indicating their potential as prognostic biomarkers.200 Angiogenic factors in GBM are regulated by oncogene activation, tumor suppressor loss, and hypoxia. FGFR signaling, which is mediated by FGF ligands, supports endothelial migration, proliferation, and angiogenesis through PI3K/AKT/mTOR activation.201,202 FGF-2 enhances ECM remodeling and cooperates with VEGF and PDGF to promote neovascularization, indicating their combined role in tumor vascularization.203 The HGF/c-MET axis further drives tumor growth, invasion, and angiogenesis, with increased expression linked to increased tumor grade and poor prognosis.204 The inhibition of MET and VEGF has synergistic effects on suppressing tumor growth, suggesting a viable therapeutic strategy.205
Angiopoietins (Ang-1, Ang-2, Ang-4) contribute to GBM vascularization. Ang-2 disrupts vessel stability, promoting neovascularization, whereas Ang-4 enhances tumor angiogenesis.206 The Tie-2 receptor, which is expressed in GBM, regulates VEGF expression, and dual inhibition of VEGF and Ang-2 improves survival outcomes.207 TGF-β modulates angiogenesis through context-dependent effects, promoting VEGF, FGF, and PDGF expression while also inducing EMT in GBM-derived endothelial cells.208 MMPs degrade the endothelial basement membrane, facilitating angiogenic switching. MMP-9-mediated VEGF release contributes to tumor vascularization.209,210 Targeting these angiogenic pathways offers potential therapeutic avenues, with combination therapies addressing VEGF resistance through simultaneous inhibition of complementary pathways. Further research is needed to refine antiangiogenic strategies and improve patient outcomes in GBM treatment.
Autophagy
Autophagy plays a complex role in GBM, influencing tumor progression, treatment response, and patient prognosis.211 While it contributes to tumor survival by providing metabolic substrates under hypoxic conditions, excessive autophagy can also lead to cell death and suppress invasion. In GBM, the expression of autophagy-related genes such as autophagy-related (ATG) 7, ATG13, and UNC-51, such as autophagy-activating kinase 1 (ULK1), is often downregulated, impairing the autophagic capacity of tumors as they progress.212 However, high levels of autophagic markers such as microtubule-associated protein 1 light chain 3 (LC3) and Beclin-1 (BECN1) correlate with better patient outcomes, suggesting a potential tumor-suppressive function in certain contexts.213 The interplay between autophagy and key oncogenic pathways further complicates its role. The mTOR pathway inhibits autophagy and supports GSC proliferation, whereas autophagy suppression enhances EGFR overexpression, promoting tumor progression.213 Additionally, miR-224-3p downregulation under hypoxic GBM conditions reduces autophagy by targeting ATG5 and FAK family kinase-interacting protein of 200 kDa (FIP200), linking miRNA regulation to tumor metabolism.214 Conversely, the upregulation of Bcl-2 interacting protein 3 (BNIP3) under hypoxic conditions promotes autophagy, supporting GBM cell survival.215
Autophagy also regulates EMT and treatment resistance in GBM. It suppresses tumor invasion by increasing N-cadherin membrane localization and degrading EMT transcription factors such as Snail.216 However, stress-induced autophagy can also contribute to therapy resistance, enhancing GBM cell survival following radiation or chemotherapy. For example, autophagy promotes resistance to TMZ by maintaining GSCs, while targeting ATG4C has been shown to increase TMZ sensitivity.217 Autophagy-related proteins such as p62 and transcription factor EB (TFEB) are linked to GBM prognosis, with high p62 expression correlating with PS and tumor recurrence.218 In contrast, BECN-1 expression is associated with IDH1 mutation and 1p/19q codeletion, suggesting a context-dependent impact on GBM biology.219 Overall, autophagy represents a double-edged sword in GBM, with both tumor-promoting and tumor-suppressing effects. Targeting autophagic pathways may offer novel therapeutic strategies, but a deeper understanding of their dual role is necessary to optimize treatment approaches.
Hypoxia
Hypoxia, regulated primarily by hypoxia-inducible factor 1 (HIF-1), plays a critical role in GBM progression, influencing angiogenesis, metabolic reprogramming, immunosuppression, and therapy resistance.220 HIF-1 expression is strongly associated with increased tumor grade and poor prognosis, as it drives the adaptation of GBM cells to hypoxic stress.221 A meta-analysis confirmed that elevated HIF-1 levels correlate with reduced OS in GBM patients.222 However, additional hypoxia-related markers, such as carbonic anhydrase IX (CA9) and osteopontin, have emerged as potentially superior indicators of tumor aggressiveness.223 HIF-1 plays a pivotal role in the metabolic shift to aerobic glycolysis, known as the Warburg effect, facilitating glucose conversion to lactate despite sufficient oxygen availability. This metabolic reprogramming supports tumor proliferation and enhances malignancy by promoting lactate production and extracellular acidification, which in turn stabilizes HIF-1α and sustains tumor hypoxia.224 Additionally, HIF-1 regulates glutamine metabolism, shifting it toward α-ketoglutarate (α-KG) production, which fuels fatty acid biosynthesis and prevents lipotoxicity.225
In addition to its role in metabolism, HIF-1 contributes to GBM invasiveness by promoting EMT through the activation of the Snail and ZEB1 transcription factors, downregulating E-cadherin, and enhancing ECM remodeling.226 HIF-1 also upregulates matrix MMPs (MMP-2, MMP-9, and MMP-14), cathepsins, and fibronectin, facilitating basement membrane degradation and tumor cell migration.227 Furthermore, it fosters an immunosuppressive microenvironment by increasing lactate production and adenosine accumulation, which suppress T cell function and enhance regulatory T cell (Treg) activity. HIF-1 plays a key role in treatment resistance, particularly in radiotherapy, by activating antioxidant systems that mitigate ROS-induced DNA damage.228 It stabilizes DNA strand breaks, promoting survival under oxidative stress. Additionally, HIF-1 supports the maintenance of GSCs by upregulating stemness-associated genes such as KLF4, MYC, OCT4, SOX2, and NANOG, sustaining their self-renewal and resistance to conventional therapies.229,230
Therapeutically, targeting HIF-1α has shown promise in sensitizing GBM cells to TMZ, particularly in patients with MGMT promoter methylation. By decreasing MGMT expression, HIF-1 inhibition enhances the cytotoxic effects of alkylating agents, offering a potential strategy to improve patient outcomes. Given the extensive role of HIF-1 in GBM progression, metabolic adaptation, and therapy resistance, it remains a critical target for novel therapeutic interventions aimed at disrupting tumor survival mechanisms in hypoxic microenvironments.
Metabolic reprogramming in GBM
The metabolic characterization of GBM, particularly in relation to IDH1/2 mutations, provides critical insights into tumor adaptation and progression. IDH mutant GBMs exhibit distinct metabolic alterations, including the accumulation of the oncometabolite 2-HG, which inhibits α-KG-dependent dioxygenases and disrupts DNA repair and cellular differentiation. This metabolic reprogramming contributes to tumor maintenance and therapeutic vulnerability, particularly in the context of targeting NAD+ metabolism and PARP inhibitors. GBMs demonstrate remarkable metabolic plasticity, relying on glucose metabolism while adapting to alternative carbon sources under stress. Increased expression of glucose transporters (GLUT1/3) and hexokinase 2 (HK2), which are regulated by HIF-1α and HIF-2α, supports glycolysis, even under hypoxic conditions. Loss of PTEN function further enhances glycolysis via AKT1 activation, stabilizing phosphofructokinase (PFKP).231,232 Additionally, MYC-driven metabolic reprogramming promotes a shift toward aerobic glycolysis and lactate production, limiting mitochondrial oxidative phosphorylation.
Lipid metabolism plays a crucial role in GBM heterogeneity, with GSCs displaying distinct metabolic dependencies. GSCs utilize fatty acids and ketone bodies for energy, allowing survival in nutrient-limited environments.233 The activation of EGFR-PI3K-AKT signaling regulates sterol regulatory element-binding protein-1 (SREBP-1), driving lipid biosynthesis and promoting tumor progression. Pseudopalisading regions in GBM accumulate fatty acids via FABP3/7, supporting tumor invasion and angiogenesis.234 Emerging evidence suggests that targeting fatty acid synthase (FASN)235 and polyunsaturated fatty acid (PUFA) synthesis may provide therapeutic benefits.236 Nitrogen metabolism is also altered in GBM, with dysregulated glutamine and cysteine metabolism contributing to tumor growth and resistance. Increased glutaminase activity and amino acid transport (SLC7A11) promote glutathione synthesis, enhancing redox homeostasis and therapy resistance.237 Targeting glutaminase with inhibitors such as telaglenastat (CB-839) in combination with radiotherapy and TMZ is a promising strategy that is currently under clinical investigation.238,239
These metabolic adaptations highlight potential therapeutic targets in GBM. Inhibiting PTEN loss-driven glucose metabolism, disrupting PUFA biosynthesis, and blocking 2-HG production in IDH mutant tumors represent viable strategies. Understanding the metabolic vulnerabilities of GBM patients offers new opportunities for precision medicine, emphasizing the need for continued research into metabolic-targeted therapies to increase the survival and quality of life of GBM patients.
Impact of immune cells
TAMs play crucial roles in GBM progression by promoting tumor growth, immune evasion, and therapy resistance. TAMs secrete factors such as EGF, TGF-β, and MMP-2, which enhance GBM proliferation and invasiveness.240 They also drive GSC renewal via cytokines such as IL-6 and IL-12.241 Additionally, TAMs support an immunosuppressive TME by recruiting Tregs and suppressing effector T cells and natural killer (NK) cells through chemokines such as chemokine (CC-motif) ligand 2 (CCL2) and C-C motif chemokine ligand 22 (CCL22).242 The overexpression of indoleamine 2,3-dioxygenase 1 (IDO1) and TDO2 in GBM leads to the production of L-kynurenine, which interacts with the aryl hydrocarbon receptor (AHR) on TAMs, further inhibiting immune responses.243 Recent studies have highlighted how TAM infiltration reshapes GBM transcriptional profiles, promoting mesenchymal transformation and therapy resistance.244 Neutrophils also contribute to GBM progression, with increased peripheral neutrophil-to-lymphocyte ratios correlated with poor prognosis.245 GBM cells recruit neutrophils through IL-8 and granulocyte–macrophage colony-stimulating factor (GM-CSF) signaling, extending their survival and promoting tumor invasiveness. The receptor for advanced glycation end products (RAGE) on GBM enhances neutrophil infiltration and NF-κB activation, leading to increased tumor-supportive inflammation.246 While early-stage neutrophils can exert antitumor effects via ROS production, their tumor-promoting functions dominate in advanced disease. Notably, a subset of tumor-associated neutrophils (TANs) can differentiate into antigen-presenting cells (APCs), activating T cells and counteracting tumor progression.247
MDSCs contribute to GBM immune evasion by inhibiting T cell activation, NK-cell function, and antigen-presenting cells. Tumor-derived cytokines such as IL-6, IL-8, IL-10, CSF-1, CCL2, CXCL2, PGE2 and TGF-β promote MDSC expansion and recruitment,248,249 whereas hypoxia shifts MDSC metabolism toward fatty acid oxidation,250 reinforcing their immunosuppressive properties. MDSCs release nitric oxide and arginase 1 (Arg1), depleting essential metabolites and suppressing T cell proliferation.251 The ability of MDSCs to induce Tregs and impair cytotoxic immune responses makes them key targets for immune-modulatory therapies. T cells play diverse roles in GBM and are influenced by tumor genetics and immune interactions. CD8+ cytotoxic T lymphocytes (CTLs) can induce tumor cell apoptosis, but their infiltration and activation vary among glioma subtypes. LGGs with NF1 mutations show greater T cell infiltration, whereas mesenchymal GBMs demonstrate substantial but often ineffective T cell presence. GBM exploits T cell regulatory mechanisms, such as PD-L1 upregulation and TGF-β signaling, to evade immune surveillance. While T cell infiltration has been linked to improved survival in some GBM patients, tumor-driven immunosuppression often limits its effectiveness.252,253
T cell exhaustion is a key immunosuppressive mechanism in GBM that is characterized by diminished effector function and elevated inhibitory receptor expression. Unlike memory T cell differentiation in acute immune responses, exhausted T cells in GBM fail to sustain long-term antitumor immunity due to persistent antigen exposure, metabolic stress, and an immunosuppressive TME.254 The TME of GBM suppresses T cell activation through inhibitory cytokines such as TGF-β, IL-10, and IL-35,255,256 along with an abundance of regulatory immune cells, including Tregs. MDSCs, and TAMs. These factors contribute to an immune-desert phenotype, limiting T cell infiltration and function.257 Chronic T cell receptor stimulation and nutrient deprivation further promote exhaustion, particularly in CD8+ T effector memory (Tem) cells, which are critical for long-term immune surveillance. The high expression of PD-L1 on MDSCs, immature dendritic cells (DCs), and plasmacytoid DCs reinforces immune suppression by impairing T cell activation.258,259 The accumulation of GAMs, comprising up to 30–50% of the GBM tumor mass, further skews the immune landscape.260 While M1 macrophages exhibit antitumor properties, M2-polarized macrophages secrete IL-10 and PD-L1, enhancing T cell dysfunction and promoting tumor progression.261
Targeting myeloid cells has emerged as a strategy to restore T cell function. CSF-1R inhibitors, aimed at blocking M2-macrophage polarization, have shown promise in preclinical models but have failed to improve survival in clinical trials.262 In contrast, CD47-blocking antibodies have demonstrated potential in reprogramming macrophages toward the M1 phenotype, enhancing CD8+ Tem cell-mediated immunity.263 Advanced epigenetic and single-cell transcriptomic analyses revealed that GAMs exhibit plasticity, adapting to environmental stimuli to either suppress or enhance immune responses. Understanding these dynamic interactions between T cells and myeloid populations may provide novel therapeutic avenues for reversing T cell exhaustion and overcoming immune evasion in GBM. Future research should prioritize strategies that modulate GAM polarization, suppress MDSCs, and reinvigorate exhausted T cells to strengthen antitumor immunity and improve therapeutic outcomes.
Dysregulated signaling pathways in GBM progression
Dysregulated signaling pathways are central to GBM pathogenesis and promote tumor growth, invasion, and resistance to therapy. Aberrant activation of key molecular and immune signaling pathways leads to uncontrolled cell proliferation, enhanced tumor cell survival, and maintenance of cancer stem-like cells. These alterations not only promote aggressive tumor growth but also contribute to resistance to standard therapies by enhancing DNA repair mechanisms and evasion of apoptosis. Targeting these dysregulated pathways represents a fundamental approach for therapeutic interventions and disruption of the TME, offering the potential to improve GBM treatment outcomes (Fig. 5).
Fig. 5.
Deregulated molecular signaling pathways and crosstalk in glioblastoma. This illustration highlights key oncogenic signaling pathways and their interconnected roles in glioblastoma (GBM) pathogenesis. Dysregulated pathways such as the PI3K/AKT/mTOR, MAPK/ERK, p53, NF-κB, JAK/STAT, β-catenin, and Notch pathways collectively drive tumor progression, survival, and resistance to therapy. NF-κB activation, triggered by TNFα/TNFR1 and the IKK complex, integrates inflammatory signaling, whereas cytokine-mediated activation of the JAK/STAT pathway promotes the transcription of survival genes. The Wnt/β-catenin and Notch pathways further support stemness, angiogenesis, and immune modulation. The convergence and crosstalk among these pathways contribute to the complexity and aggressiveness of GBM
Molecular signaling pathways
PI3K/AKT/mTOR
The PI3K/AKT/mTOR pathway is a central regulator of GBM progression and influences cell survival, proliferation, and metabolic adaptation. Its activation, triggered by tyrosine kinase receptors, Ras, and integrins, promotes tumor growth and treatment resistance.10 Dysregulation of this pathway is observed in approximately 70% of GBM patients and is correlated with poor prognosis, highlighting its relevance as a therapeutic target. PTEN, a critical tumor suppressor, negatively regulates the PI3K/AKT/mTOR pathway, and its loss further exacerbates GBM aggressiveness.264 The inhibition of mTOR with rapamycin has demonstrated promising effects in vitro, but clinical trials have shown limited efficacy,265 suggesting the need for combination therapies. Studies using orthotopic GBM models highlight a strong correlation between AKT activation and increased tumor growth, invasion, and resistance to therapy,266 reinforcing its role as a therapeutic target. However, AKT also plays a role in astrocytic differentiation, adding complexity to its function in GBM.
mTOR, a key effector of PI3K/AKT signaling, is implicated in cell survival, metabolic reprogramming, and GBM cell proliferation.267 Both mTORC1 and mTORC2 contribute to GBM progression, with mTORC1 promoting glycolysis via HIF-1 activation and mTORC2 enhancing tumor cell motility through RICTOR overexpression.10,268 Additionally, alternative activation pathways, such as PKCα-mediated EGFR-mTOR signaling, indicate that multiple regulatory inputs sustain mTOR activity in GBM.269 The complexity of PI3K/AKT/mTOR signaling in GBM necessitates a multifaceted therapeutic approach. Combination strategies targeting mTOR, PI3K, and associated compensatory mechanisms may enhance treatment efficacy. Future research should focus on identifying resistance pathways and refining targeted therapies to improve patient outcomes.
NF-κB
NF-κB activation is a hallmark of GBM and is driven by oncogenic pathways such as the EGFR and PDGFR signaling pathways, as well as genetic alterations in PTEN, NF1, and ARF. The inflammatory TME further amplifies NF-κB activity, reinforcing its role in tumor progression. Additionally, NF-κB signaling is sustained by epithelial V-like antigen 1 (Eva1), which maintains GSC characteristics through the regulation of stemness-associated genes.270,271 In addition to its role in tumor maintenance, NF-κB promotes the mesenchymal phenotype of GBM by activating key transcription factors, including STAT3, CCAAT/enhancer-binding protein β (C/EBPβ), and TAZ. This process is reinforced by a feedback loop involving fibroblast growth factor-inducible 14, further enhancing GBM invasion.272 NF-κB also plays a critical role in angiogenesis by upregulating VEGF and IL-8, contributing to tumor vascularization.273 Metabolic reprogramming in GBM is influenced by NF-κB, particularly through its regulation of pyruvate kinase M2, a key glycolytic enzyme upregulated in response to EGFR signaling.274 Moreover, NF-κB is implicated in therapy resistance, enhancing DNA damage repair to promote radioresistance and regulate MGMT expression, contributing to chemoresistance.275 Given its multifaceted role in tumor invasion, angiogenesis, metabolism, and therapy resistance, NF-κB represents a promising therapeutic target in GBM. Future strategies should focus on disrupting NF-κB signaling to increase treatment sensitivity and inhibit tumor progression.
STAT3
STAT3 activation in GBM is driven by multiple receptor tyrosine kinase pathways, including the EGFR, PDGFR, and c-MET pathways, along with the loss of negative regulators such as protein tyrosine phosphatases, suppressors of cytokine signaling, and protein inhibitors of activated STAT3.276 This sustained activation promotes tumor growth by upregulating the expression of oncogenic transcription factors such as c-Myc, cyclin D1, and Bcl-xl. STAT3 also plays a crucial role in maintaining GSC properties through its regulation of SOX2, OLIG2, OCT4, and NANOG,277 reinforcing the self-renewal and invasive capabilities of tumors. Additionally, STAT3 facilitates hypoxia-driven angiogenesis and tumor migration by modulating HIF-1, VEGF, MMP2, and TWIST.278 In addition to promoting tumor proliferation, STAT3 contributes to GBM aggressiveness by promoting EGFRvIII-mediated invasion through JAK2/STAT3 signaling and stabilizing focal adhesion complexes.279 It also establishes an immunosuppressive microenvironment, enabling tumor immune evasion.280 Importantly, STAT3 is a major player in therapy resistance and regulates MGMT expression, conferring TMZ resistance, interacting with FOXM1 to promote radioresistance, and mediating resistance to anti-VEGF and MET inhibitors.281,282 Interestingly, the role of STAT3 in GBM is context dependent. In PTEN-deficient tumors, STAT3 may act as a tumor suppressor, inhibiting proliferation and invasion.283 This complexity underscores the need for a nuanced therapeutic approach targeting STAT3. Given its involvement in multiple oncogenic processes and therapy resistance, STAT3 represents a key target for improving GBM treatment outcomes.
Wnt/β-catenin
The Wnt/β-catenin pathway plays a key role in glioma progression by maintaining tumor stem cell populations, inhibiting differentiation, and promoting invasion. While essential for normal brain development, its dysregulation in GBM is linked to increased malignancy and poor prognosis.284 Aberrant activation of this pathway contributes to treatment resistance and tumor aggressiveness, making it a critical therapeutic target. Epigenetic alterations further regulate Wnt signaling in GBMs. Hypermethylation-mediated silencing of Wnt inhibitors is a common event, particularly in astrocytic gliomas, that influences tumor progression. Distinct patterns of Wnt pathway gene hypermethylation in primary and secondary GBMs suggest subtype-specific regulatory differences.285 Studies have also reported that the overexpression of Wnt ligands (Wnt2, Wnt3a, and Wnt5a), Frizzled receptors, and β-catenin in GBM correlates with tumor grade and poor patient outcomes. Knockdown of Wnt2 and β-catenin has been shown to suppress tumor growth, reduce invasion, and induce apoptosis in tumor cells.286 Targeting the Wnt pathway offers a promising therapeutic strategy for GBMs, with potential applications in overcoming radioresistance and chemoresistance. Further research into subtype-specific alterations and regulatory mechanisms is essential for the development of effective Wnt-targeted therapies tailored to GBM heterogeneity.
IGFR
Dysregulated IGF signaling contributes to GBM progression and therapy resistance. Elevated IGF ligands and IGF1R overexpression are linked to increased tumor growth, poor prognosis, and a reduced response to TMZ therapy.287 IGF1R activates key oncogenic pathways, including the PI3K/AKT and RAS/RAF/MAPK pathways, with ligand-driven activation playing a primary role in tumor cell proliferation.288 Targeting IGF signaling has shown therapeutic potential. IGF1R inhibitors such as IMC-A12 and picropodophyllin effectively suppress GBM growth in preclinical models, reducing tumor proliferation and angiogenesis.288,289 These findings highlight IGF1R as a promising therapeutic target, warranting further investigation into its role in chemoresistance and the potential benefits of combination therapies integrating IGF1R inhibitors with standard GBM treatments.
NOTCH
Notch signaling plays a complex role in GBM, exhibiting both tumor-promoting and tumor-suppressive effects depending on the molecular and cellular context.290 While Notch1 overexpression is correlated with PS in some cases, it is also linked to better prognosis in specific GBM subtypes. Notch pathway activity varies across tumor regions, with higher expression in peritumoral GSCs than in the tumor core, suggesting a role in maintaining stemness and therapeutic resistance. Notch2 and Notch4 also influence GBM differentiation and aggressiveness, reinforcing the impact of these pathways on tumor heterogeneity.291,292 Hypoxia-driven Notch activation further promotes tumor progression by increasing the expression of TRPC6, which stimulates NFAT activity and GBM cell proliferation.293 The interplay between Notch and STAT3 signaling in mesenchymal GBMs suggests a cooperative mechanism in driving tumor aggressiveness.20,294 Additionally, Notch activation in GSCs contributes to perivascular niche remodeling and angiogenesis, supporting tumor vascularization and therapy resistance.295 Targeting Notch signaling represents a potential therapeutic avenue, particularly in combination with hypoxia or angiogenesis inhibitors. Inhibiting Notch1 has shown promise in reducing tumor hypoxia and sensitizing GBM to radiotherapy.296 The convergence of the Notch pathway with the PDGF and nitric oxide signaling pathways highlights additional regulatory mechanisms that sustain GSCs and GBM progression.295 Understanding the context-dependent role of Notch in GBM could facilitate the development of more precise therapeutic interventions and prognostic markers for patient stratification.
Hedgehog pathway
The Hedgehog (Hh) signaling pathway plays a critical role in GBM, influencing tumor growth, stemness, angiogenesis, and treatment resistance. Its key effectors, particularly GLI family zinc finger 1 (GLI1), regulate cell proliferation through interactions with p53 and are essential for GSC maintenance. A truncated GLI1 variant (tGLI1), detected in most GBM cases but absent in normal brain cells, promotes tumor progression by activating genes not regulated by canonical GLI1, including VEGFR1, VEGF-C, TEM7, HPSE, CD24, and CD44.297 tGLI1 drives GBM invasion by upregulating CD24 and contributes to angiogenesis through VEGF signaling. It also induces the mesenchymal GBM subtype by increasing the expression of CD44, a key marker of mesenchymal GSCs.297,298 Additionally, tGLI1 enhances EMT by modulating miRNAs such as miR-21, miR-128, and miR-200. Recent findings revealed that metabotropic glutamate receptor 4 (mGluR4) negatively regulates GLI1, suppressing proliferation and inducing apoptosis, suggesting a potential therapeutic target.299
In addition to its role in tumor growth, GLI1 contributes to treatment resistance. It enhances the replicative potential of GBM cells by activating TERT and promotes resistance to TMZ and radiotherapy by upregulating MGMT expression.300 Given its multifaceted role in GBM progression, targeting aberrant Hh signaling—particularly tGLI1—may offer promising therapeutic strategies to counteract metastasis and treatment resistance. Further research into Hh pathway dysregulation could pave the way for more effective, targeted therapies for GBM management.
Ceramide signaling
Acid ceramidase (ASAH1) plays a critical role in GBM metabolism by converting ceramides into sphingosine and free fatty acids. This shift promotes the production of sphingosine-1-phosphate (S1P), a key driver of GBM survival, proliferation, and resistance to apoptosis.301 Elevated ASAH1 expression in GBM has been linked to increased tumor cell viability, migration, and recurrence, highlighting its potential as a prognostic biomarker. Additionally, ASAH1 secretion into interstitial tissues facilitates tumor progression by modifying the surrounding microenvironment. Targeting ASAH1 represents a promising therapeutic strategy. Inhibitors of ASAH1 have demonstrated efficacy in preclinical studies, reducing tumor cell growth and potentially overcoming resistance to standard treatments such as TMZ.302 While no clinical trials currently focus on ceramide signaling in GBM, further research into ASAH1 inhibition could provide novel approaches to restoring ceramide-induced apoptosis and improving patient outcomes. Expanding our understanding of the role of ASAH1 in GBM progression may lead to the development of targeted therapies that disrupt its protumorigenic effects.
TEAD transcription factors
TEA domain (TEAD) transcription factors, in coordination with YAP1 and TAZ, play crucial roles in GBM pathogenesis. The TAZ-TEAD2 complex drives mesenchymal differentiation by binding to mesenchymal gene promoters, whereas TEAD1 and TEAD4 contribute to various tumorigenic processes.303 TEAD1 enhances EGFR-mediated c-Myc expression and regulates migration through aquaporin 4 (AQP4).79 TEAD4, in partnership with TAZ, regulates cell proliferation, apoptosis, invasion, and EMT by modulating key genes such as cyclin D1, KI67, c-Myc, Bcl2, MMP-9, vimentin, and N-cadherin.304 These findings highlight the TEAD family as critical mediators of GBM progression, with implications for tumor aggressiveness and treatment resistance. Targeting TEAD signaling, particularly its interaction with YAP1 and TAZ, may offer new therapeutic strategies to disrupt mesenchymal transition and GBM proliferation, paving the way for improved patient outcomes.
C/EBPβ
C/EBPβ is a key transcription factor implicated in GBM pathogenesis, particularly in the mesenchymal subtype. Its activation is linked to KLHL9 deletions and EGFR signaling, positioning it as a central player in tumor progression.305 In conjunction with STAT3, C/EBPβ drives mesenchymal differentiation, enhancing GBM cell invasion, proliferation, and survival.192 C/EBPβ contributes to GBM pathobiology by regulating DNA damage responses and inducing genes associated with invasion and metastasis. It also promotes angiogenesis via IL-6 and IL-8 and fosters an immunosuppressive TME by upregulating tryptophan-2,3-dioxygenase (TDO2), which enhances kynurenine production. Additionally, C/EBPβ modulates antioxidative defense mechanisms by regulating NAD(P)H quinone oxidoreductase 1 (NQO1) and glutathione S-transferase pi 1 (GSTP1), protecting GBM cells from oxidative stress.306,307 Given its multifaceted role in GBM progression, targeting C/EBPβ presents a promising therapeutic strategy. Inhibiting its activity could mitigate tumor aggressiveness, disrupt immunosuppression, and enhance treatment responses, making it a viable candidate for future GBM therapies.
c-Myc
c-Myc is a key transcription factor in GBM that influences tumor growth, stemness, invasion, and resistance to therapy. Its dysregulation, driven by gene amplification and epigenetic modifications, promotes GBM cell proliferation and mitotic activity. Additionally, c-Myc enhances tumor vascularization by upregulating miR-9 and facilitates GBM cell invasion through RhoA activation.308,309 In addition to its role in tumor progression, c-Myc is a central regulator of GBM metabolism, driving a shift toward glycolysis to sustain energy production under hypoxic conditions. Importantly, it contributes to resistance against radiation and TMZ by upregulating DNA repair proteins such as Nibrin (NBS1) and Reversionless 3-like (REV3L), enabling tumor cells to withstand genotoxic stress.310 Given its broad oncogenic influence, targeting c-Myc represents a promising therapeutic strategy in GBM. Inhibiting its activity could disrupt tumor metabolism, angiogenesis, and therapy resistance, providing a potential approach to improve treatment efficacy in this aggressive malignancy.
PKC
Dysregulated protein kinase C (PKC) signaling contributes to GBM growth, proliferation, and invasion. Elevated PKC activity is correlated with aggressive tumor behavior, with specific isoforms playing distinct roles in GBM progression.311 PKCα drives mitogenic and prosurvival signaling, enhances GBM migration via the ERK/NF-κB pathways, and regulates FGF expression for tumor cell proliferation. Other isoforms, such as PKCε and PKCη, facilitate cell adhesion, motility, and survival. Given the multifaceted role of PKC in glioma biology, targeting its isoforms offers a potential therapeutic approach to disrupt tumor growth and invasion.312,313 Inhibiting PKC-mediated pathways could improve treatment efficacy, providing a rationale for further exploration of the use of PKC inhibitors in GBM therapy.
Immune signaling pathways
CSF-1R
The CSF-1R pathway critically influences macrophage polarization and contributes significantly to immunosuppression and tumor progression in GBM. CSF-1R, which is predominantly expressed on macrophages and microglia, binds to CSF-1, driving the activation, proliferation, and survival of these immune cells. Within the GBM microenvironment, elevated CSF-1 signaling promotes the recruitment and polarization of macrophages toward an immunosuppressive, protumorigenic M2 phenotype, increasing tumor growth, invasion, and immune evasion314 (Fig. 6). Targeting the CSF-1R pathway has demonstrated potential in shifting macrophage polarization from an M2-like immunosuppressive phenotype toward an M1-like proinflammatory phenotype, thereby facilitating antitumor responses. Preclinical models of GBM have shown that CSF-1R inhibition can significantly reduce TAMs, resulting in reduced tumor growth and improved survival outcomes. Additionally, combining CSF-1R blockade with ICIs has shown enhanced therapeutic efficacy by overcoming macrophage-mediated immunosuppression.315 Thus, targeting CSF-1R signaling represents a promising strategy for GBM immunotherapy.
Fig. 6.
Key immune signaling pathways regulating tumor-associated immunosuppression. In macrophages, CSF-1 or IL-34 binds to the CSF-1 receptor, inducing rapid dimerization and autophosphorylation of tyrosine residues. This activation triggers downstream signaling through the PI3K/AKT and JAK/STAT pathways, regulating macrophage polarization. CTLA-4, which is expressed on activated T cells, binds to CD80/CD86 on APCs. Upon engagement, CTLA-4 signaling dephosphorylates TCR signaling components, inhibiting CD3 and ZAP70 activation and suppressing the RAS signaling pathway. CTLA-4 signaling disrupts AKT phosphorylation, negatively regulating the cell cycle and suppressing key transcription factors such as NF-κB, AP-1, and NF-AT. PD-1 interacts with its ligands, leading to the phosphorylation of two tyrosine residues on its cytoplasmic tail. This phosphorylation recruits SHP-1 and SHP-2 to the ITSM motif, inhibiting the PI3K/AKT/mTOR pathway, reducing metabolic activity, and promoting T cell exhaustion. In the case of TGF-βR2 ligand binding, the receptor activates and facilitates PI3K and AKT signaling through physical interaction with the PI3K subunit. This cascade leads to mTOR kinase activation, which drives translational responses. Collectively, these signaling pathways induce IDO1 activation, which converts tryptophan to kynurenine, thereby enhancing tumor immune evasion through immune suppression. The CD39/CD73 pathway hydrolyzes extracellular ATP into adenosine, an immunosuppressive metabolite. Adenosine prevents tyrosine phosphorylation of ZAP70, AKT, and ERK1/2 in naive αCD3/CD28-stimulated CD8+ T cells, impairing their activation
CTLA-4
Cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) represents another pivotal checkpoint molecule that negatively regulates T cell activation. Tregs in GBM express high levels of CTLA-4, which competitively bind to B7 ligands (CD80/CD86) on antigen-presenting cells. Upon binding to its ligands, CD80 or CD86, CTLA-4 recruits phosphatases such as SHP-2 and protein phosphatase 2A (PP2A) to its cytoplasmic domain. These phosphatases dephosphorylate key signaling molecules downstream of the T cell receptor (TCR), leading to attenuation of the PI3K/AKT signaling pathway.315 This interaction significantly reduces the availability of essential costimulatory signals for effector T cells, leading to diminished activation, proliferation, and antitumor function of these cells (Fig. 6). Thus, CTLA-4 engagement reinforces the immunosuppressive TME, promoting tumor progression and resistance to checkpoint blockade therapies.316
PD-1/PD-L1
The PD-1/PD-L1 axis is a critical immune checkpoint pathway exploited by GBM cells to evade immune surveillance. In addition to MDSCs, tumor cells upregulate PD-L1 expression, which interacts with PD-1 on activated T cells, leading to T cell exhaustion and reduced antitumor activity. Upon engagement of PD-1 by its ligand PD-L1, the immunoreceptor tyrosine-based inhibitory motif (ITIM) and immunoreceptor tyrosine-based switch motif (ITSM) within the PD-1 cytoplasmic domain become phosphorylated. This phosphorylation recruits SH2 domain-containing phosphatases, specifically SHP-1 and SHP-2, which dephosphorylate key signaling molecules downstream of the TCR, leading to attenuation of T cell activation. Consequently, this inhibits pathways such as the PI3K/AKT pathway, reducing T cell proliferation and cytokine production, thereby contributing to an immunosuppressive environment317 (Fig. 6). This immunosuppressive signaling results in impaired cytokine secretion, decreased proliferation, and diminished cytotoxic functions of T cells, significantly undermining the effectiveness of ICIs in GBM.318
TGF-β signaling
Transforming growth factor-beta (TGF-β) signaling plays a pivotal role in immune modulation and tumor progression in GBM.319 This signaling pathway begins when TGF-β binds to type I and type II serine/threonine kinase receptors, triggering the phosphorylation and activation of receptor-regulated SMAD proteins (SMAD2 and SMAD3). Activated SMAD2/3 complexes with SMAD4 are translocated into the nucleus, where they modulate gene transcription linked to proliferation, differentiation, apoptosis, and immune regulation.320 In GBM, TGF-β critically contributes to immune evasion by suppressing the cytotoxic activity of CTLs and NK cells, thereby hindering the host immune response against tumor cells.321 Concurrently, TGF-β signaling promotes the proliferation and immunosuppressive functions of Tregs, further dampening immune surveillance.319 Additionally, TGF-β drives macrophages toward the M2 phenotype, which is characterized by the secretion of immunosuppressive cytokines such as IL-10 and additional TGF-β, reinforcing the suppressive TME.322 TGF-β induces EMT, which increases tumor invasiveness and metastatic potential.323 Furthermore, TGF-β signaling positively affects the NF-κB and MAPK pathways, amplifying immunosuppressive and protumorigenic signals in GBM. This interaction enhances tumor progression by promoting immune evasion, T cell suppression, and increased tumor cell survival and invasion321,323 (Fig. 6).
CD39/CD73-adenosine
The CD39/CD73-adenosine pathway is a critical immunoregulatory mechanism in GBM. CD39 and CD73 are ectonucleotidases that sequentially hydrolyze extracellular ATP to adenosine, an immunosuppressive metabolite. Under physiological conditions, this pathway helps maintain tissue homeostasis by modulating inflammation and preventing excessive immune responses.324 In GBM, however, the upregulation of CD39 and CD73 contributes significantly to increased levels of adenosine. This increased adenosine prevents rapid phosphorylation of the ZAP70 kinase as well as AKT and ERK1/2 in T cells.325 This leads to the inhibition of effector T cell and NK-cell functions, enhances Treg function and proliferation, reducing the ability of Tregs to mount effective antitumor responses.326 Consequently, targeting the CD39/CD73-adenosine pathway has emerged as a promising therapeutic strategy to reverse immunosuppression, enhance antitumor immunity, and potentially improve clinical outcomes in GBM (Fig. 6).
IDO1 and kynurenine
IDO1 is a heme-containing enzyme that catalyzes the initial step of tryptophan catabolism through the kynurenine pathway, generating the immunosuppressive metabolite kynurenine. Under physiological conditions, IDO1 modulates immune tolerance by regulating T cell function.327 In GBM, IDO1 expression is significantly elevated.328 Elevated IDO1 activity in GBM leads to tryptophan depletion and increased kynurenine production, which leads to GCN activation, PD-1/PD-L1 upregulation, AHR activation, and increased kynurenine metabolite production.329 This leads to the inhibition of effector T cell and NK cell functions while promoting Treg differentiation, thus impairing antitumor immunity330 (Fig. 6). Consequently, targeting the IDO1-kynurenine pathway with specific inhibitors represents a promising therapeutic approach to restore immune function and improve GBM treatment outcomes.331
GBM therapeutics for clinical management
Current standard care for GBM patients offers modest survival benefits, but the prognosis remains poor due to tumor recurrence and therapy resistance. Advances in GBM therapeutics have introduced novel approaches, including targeted therapies (e.g., EGFR inhibitors), immunotherapies such as ICIs and CAR-T cell therapies, and noninvasive modalities such as the tumor-treating field (TTF) (Fig. 7). Despite these developments, clinical management remains challenging due to the genetic heterogeneity, invasive nature, and ability of GBM to evade treatment, emphasizing the urgent need for innovative and effective therapeutic strategies.
Fig. 7.
Current and emerging glioblastoma therapeutics for clinical management. The figure provides an overview of current therapeutic strategies used in the clinical management of glioblastoma (GBM). Locoregional treatments include surgical resection, laser interstitial thermal therapy (LITT), and radiation, which aim to eliminate tumor tissue through direct cytotoxic effects. Tumor-treating fields (TTFs) and directional nonrotating electric field therapy (dnEFTs) promote mitotic disruption and apoptosis. Chemical interventions include chemotherapy (TMZ), which induces G2/M arrest and apoptosis, and immunotherapy, which enhances T cell-mediated tumor killing. Oncolytic viruses induce immunogenic cell death, whereas therapeutic vaccines such as peptides, mRNAs, viral vectors, and dendritic cell-based platforms stimulate immune surveillance. Combination therapies leverage multiple modalities to simultaneously target diverse oncogenic pathways, offering a promising route toward overcoming resistance and advancing personalized GBM treatment
Locoregional treatments
Surgery
Surgical resection remains a primary treatment for gliomas, contributing to both diagnostic accuracy and therapeutic efficacy. Extensive resection is linked to improved survival in both LGGs and HGGs,332 although the precise correlation between the extent of resection and patient outcomes requires further study. In patients with metastatic brain lesions, resection offers survival benefits and enhances quality of life. The evidence suggests that for single brain metastases, surgery is more effective than radiation therapy alone. Fluorescence-guided surgery, particularly with 5-aminolevulinic acid (5-ALA), has significantly improved the extent of resection (EOR) in HGGs.333 Clinical trials indicate that 5-ALA enhances gross total resection rates, outperforming conventional surgical methods.334 Studies have reported that 5-ALA-guided resection increases EOR, extends OS, and improves PFS. Additionally, compared with intraoperative MRI alone, combining 5-ALA with intraoperative imaging in eloquent brain regions enhances resection success.335 These advancements underscore the critical role of fluorescence-guided techniques in neurosurgical oncology, improving tumor visualization, maximizing resection, and ultimately enhancing clinical outcomes.
Radiosurgery
The treatment of brain metastases has evolved with a shift toward targeted radiation approaches that enhance tumor control while minimizing neurocognitive decline. Whole-brain radiation therapy, once a standard for patients ineligible for surgical resection or stereotactic radiosurgery (SRS), is now being reevaluated owing to its impact on cognitive function.336 SRS techniques, particularly hypofractionated stereotactic radiosurgery, have demonstrated efficacy in treating larger tumors and lesions in eloquent brain regions while potentially engaging immune mechanisms.337 Gamma knife (GK) radiosurgery remains a highly precise modality for treating localized tumors, with emerging combinations of GK and bevacizumab showing promise in improving therapeutic outcomes. However, further clinical validation is necessary to refine protocols and mitigate potential treatment biases.338 Leading-edge radiosurgery represents an evolving strategy to increase the safety and efficacy of GBM treatment. Gamma tiles or brachytherapy, particularly with cesium-131 isotopes, have shown potential advantages over traditional iodine-125 implants by offering improved tumor control with reduced radiation necrosis,339 making it a viable option for recurrent GBM treatment. These advancements reflect a growing emphasis on personalized, precision-driven radiation strategies that aim to optimize both survival and quality of life in patients with brain metastases.
Laser-interstitial thermotherapy
Laser-interstitial thermotherapy (LITT) provides a minimally invasive alternative for GBM patients ineligible for surgical resection. By inducing localized hyperthermia, LITT effectively eradicates tumor cells while preserving surrounding tissue, offering advantages over other thermal ablation techniques.340 MRI-guided LITT has demonstrated safety and efficacy, with potential benefits such as enhanced BBB permeability for improved drug delivery. While rapid recovery is a key advantage, patient selection and monitoring remain critical to mitigate risks.341 LITT shows promise in managing recurrent GBM and hard-to-access HGGs, potentially extending PFS.342 Ongoing clinical trials (NCT02880410, NCT03022578, NCT03341806, and NCT03277638) are evaluating its efficacy, safety, and potential synergy with chemotherapy. Similarly, 18F-DOPA PET-guided, dose-escalated, hypofractionated proton beam therapy has shown potential survival benefits with manageable adverse effects,343 warranting further investigation in phase 2 trials (NCT05781321). These studies will be crucial in defining LITT’s long-term impact and integration into GBM treatment protocols.
Focused ultrasound
Focused ultrasound (FU), including high-intensity (HIFU) and low-intensity (LIFU) modalities, has emerged as a potential GBM treatment strategy.344 HIFU induces thermal ablation, effectively destroying tumor cells, whereas LIFU, combined with microbubbles, transiently disrupts the BBB, enhancing targeted drug delivery.345 This approach significantly increases therapeutic concentrations within tumor tissue while lowering systemic drug exposure.346,347 LIFU with MB-mediated delivery improves GBM treatment by increasing drug permeability, downregulating efflux transporters such as P-glycoprotein, and increasing apoptosis.347,348 Studies have demonstrated that this method enhances the efficacy of chemotherapeutic agents such as etoposide, paclitaxel (PTX), and DOX, reducing tumor growth and extending survival. In preclinical models, PTX liposomes with anti-PD-1 increased survival by 40%, whereas cabazitaxel treatment reduced tumor size by two-thirds.349,350 Additionally, nanoparticle-based drug carriers, including mesoporous silica nanoparticles (MSNs) and shRNA-loaded liposomes, further optimize therapeutic delivery and tumor suppression.351 The optimization of the acoustic pressure and treatment parameters plays a critical role in maximizing the treatment efficacy. Compared with a lower pressure, a higher acoustic pressure (0.80 MPa) improves tumor inhibition by 64%.352 Despite the promising results, challenges remain in standardizing treatment protocols and translating preclinical findings into clinical applications. Future research should focus on refining acoustic parameters, evaluating long-term safety, and integrating focused ultrasound with combination therapies to improve GBM treatment outcomes.
Tumor-treating field
The approval of TTF as an adjuvant therapy for newly diagnosed GBM in 2015 introduced a novel approach to treatment. TTF, delivered via the Optune® device, applies low-intensity, intermediate-frequency alternating electric fields to disrupt GBM cell proliferation by interfering with mitotic spindle formation. This noninvasive therapy has shown clinical efficacy in extending OS and PFS, particularly when combined with TMZ.353 For recurrent GBM, TTFs can be used as monotherapies, whereas in newly diagnosed patients, TTFs can be combined with the standard chemotherapy TMZ. Ongoing clinical trials (NCT01925573) continue to assess TTFs in combination with bevacizumab and hypofractionated stereotactic irradiation for recurrent GBM. Completed clinical trials have demonstrated that TTFs significantly improve patient outcomes without severe adverse effects, in addition to mild skin irritation. A phase III randomized clinical trial revealed that the combination of TTF with TMZ extended the median survival to 19.2 months in newly diagnosed patients,353,354 supporting its integration into standard GBM treatment. However, the high cost of TTFs remains a potential barrier to their widespread adoption.
Directionally nonrotating electric field therapy
Directionally nonrotating electric field therapy (dnEFT) employs implanted electrodes to deliver continuous, targeted electric fields directly to GBM-affected brain regions. Preclinical studies have demonstrated that dnEFT, which is applied via a clinical-grade spinal cord stimulator or a custom two-electrode system, induces apoptosis in GBM cells and significantly reduces the tumor burden in vivo. dnEFTs exhibited prolonged survival and an immune shift toward an antitumor response in a preclinical model, marked by increased M1 macrophages and reduced M2-associated gene expression.355 dnEFTs offer potential advantages over TTFs by maintaining a consistent directional field, potentially enhancing tumor disruption while minimizing resistance development.355 Further refinement of electrode placement and field modulation could optimize precision, particularly with real-time adaptation to tumor evolution. Future studies should explore the integration of dnEFTs with immunotherapies, CAR-T cell therapy, and gene editing to increase therapeutic efficacy. Additionally, the development of wearable dnEFT devices may improve patient compliance and accessibility, paving the way for their clinical translation in GBM management.
Chemical interventions
Chemotherapy
TMZ is the standard chemotherapy used for GBM. When radiotherapy is added, the median survival time increases from 12.1 months to 14.6 months. This combination also improved the two-year survival rate from 10.4% to 26.5%, leading to its adoption as the “Stupp protocol”. TMZ, an alkylating agent, exerts cytotoxic and mutagenic effects predominantly by alkylating the O6 position of guanine in DNA. The cytotoxic effects of O6-methylguanine induce replication arrest, and the accumulation of single-stranded DNA breaks ultimately leads to G2‒M cell cycle arrest and apoptosis. However, its efficacy is significantly limited by DNA repair mechanisms, particularly the activity of MGMT, which reverses guanine methylation and reduces TMZ-induced cytotoxicity.356 Patients with MGMT promoter methylation receive the greatest benefit, as their tumors have a diminished capacity to repair TMZ-induced DNA damage. Despite its clinical utility, TMZ resistance remains a major challenge, affecting more than 50% of GBM patients through both intrinsic and acquired mechanisms. In addition to MGMT, multiple molecular pathways contribute to resistance, complicating treatment strategies.357 Addressing these mechanisms is crucial for improving GBM therapy, highlighting the need for novel approaches such as combination therapies, MGMT inhibitors, and alternative drug delivery systems to increase TMZ efficacy.
Small molecules
Small molecules such as LP-184, LMP400, and Azeliragon are emerging as promising therapeutic candidates in GBM, particularly for overcoming resistance mechanisms. LP-184 has potent anti-GBM activity, including in TMZ-resistant and MGMT-expressing tumors, with effective brain and tumor penetration. Its efficacy is linked to PTGR1 expression, EGFR signaling, and low NER/ERCC3 levels, while spironolactone enhances its cytotoxic effects, suggesting a potential combination therapy for GBM.358 Similarly, LMP400 shows high efficacy in PTEN-null GBM, inducing G2/M cell cycle arrest, DNA damage, and apoptosis. Combining the small molecule LMP400 with niraparib enhances cytotoxicity, evades ABC transporters, and extends survival in GBM models, supporting its therapeutic potential.359 Azeliragon, currently under evaluation in dose-escalation trials, aims to determine its recommended phase 2 dose while maintaining dose-limiting toxicities below 33% within 28 days and assessing its impact on PSF and OS.360 Furthermore, early clinical evaluation of Azeliragon (20 mg/day) with radiotherapy indicates a favorable safety profile, with no dose-limiting toxicities or treatment-related discontinuations.361
Additional targeted therapies show promise in GBM management. Dacomitinib effectively penetrates GBM tumors, with 14.3% of patients experiencing clinical benefit and 8.9% achieving PFS for at least one year.362 Napabucasin disrupts STAT3 and NF-κB signaling, inhibiting GBM cell proliferation, colony formation, and invasion, while in vivo studies have confirmed its efficacy in impairing GBM growth in xenograft models.363 Infigratinib demonstrates superior efficacy over larotrectinib in GBM patients with tyrosine kinase alterations, despite a greater adverse effect profile. Initial bevacizumab therapy has been associated with improved OS, reinforcing its potential role in GBM treatment.364 Ogremorphin, a GPR68 inhibitor, induces ferroptosis and cytotoxicity in GBM cells with minimal toxicity, highlighting its potential as a therapeutic strategy. Suppression of ATF4 via GPR68 inhibition further disrupts GBM survival, reinforcing its viability as a target for treatment.365,366 Additionally, epigenetic drug treatment of GSCs results in transposable element-derived transcripts that are selectively expressed in cancer cells, generating antigens with potential for targeted immunotherapy. However, the risk of unintended genomic activation raises safety concerns. CRISPR-mediated strategies may help mitigate these risks while optimizing antigen targeting for therapy.367 Vorasidenib, a BBB-penetrant IDH1/2 inhibitor, significantly reduces 2-HG levels, reversing gene expression and epigenetic changes in IDH mutant gliomas, highlighting its therapeutic potential.368
CSF-1R inhibitors, such as PLX3397, have demonstrated potential in reprogramming TAMs in GBM. TAMs predominantly exhibit an immunosuppressive M2-like phenotype, promoting tumor growth and immune evasion. Inhibiting CSF-1R shifts TAMs toward a proinflammatory M1-like state, enhancing phagocytosis and reducing immune suppression.314 Additionally, IDO1 inhibitors, such as epacadostat, target metabolic immunosuppression by blocking the conversion of tryptophan to kynurenine, a pathway that suppresses effector T cell activity while promoting regulatory T cell expansion. By inhibiting IDO1, these therapies restore T cell proliferation and function, enhancing antitumor immunity328 (Fig. 6). In combination with TMZ, the PARP inhibitor niraparib enhances immune recognition by upregulating NKG2DL, leading to increased ULBP1/Mult-1 mRNA expression and improved gamma-delta T cell-mediated cytotoxicity in GBM patient-derived xenografts.369
These small-molecule inhibitors, either alone or in combination with immune checkpoint blockade, hold promise for overcoming GBM immune resistance. Ongoing clinical trials are evaluating their therapeutic potential, emphasizing the need for synergistic treatment strategies to improve patient outcomes. Continued exploration of combination therapies and immune-targeting agents will be essential for advancing GBM immunotherapy and overcoming resistance mechanisms.
Immunotherapy
The highly immunosuppressive TME of GBM significantly limits the efficacy of immune ICIs. Despite PD-L1 expression in tumor cells ranging from 61% to 88%, clinical trials such as CheckMate-143 have failed to show significant survival benefits.370 This limited efficacy is linked to poor BBB penetration, low tumor-infiltrating lymphocyte levels, and PTEN mutations, which are prevalent in nonresponders. However, neoadjuvant PD-1 blockade has shown potential in stimulating tumor-specific T cell activation and modulating tumor cell cycle-associated gene expression,371 indicating that optimizing treatment timing and combination strategies may improve outcomes. In addition to PD-1/PD-L1 blockade, CTLA-4 inhibition is being explored, with the anti-CTLA-4 monoclonal antibody ipilimumab, which shows promise in the treatment of melanoma brain metastases. Early trials combining ipilimumab, GM-CSF, and bevacizumab in recurrent GBM reported partial responses in 31% of patients.372 Ongoing studies are evaluating ipilimumab in combination with TMZ, bevacizumab, and other ICIs.373 Additionally, IDO inhibition, which targets metabolic immunosuppression, has shown synergy with anti-PD-L1 and anti-CTLA-4 therapies in preclinical GBM models, resulting in 100% long-term survival.374 Furthermore, the current immunotherapeutic drugs in clinical trials are presented in Table 1.
Table 1.
Clinical trials of immune therapies for GBM treatment
| Drug | Target | Clinical trial identifier | Condition | Combination | Phase | Status |
|---|---|---|---|---|---|---|
| Durvalumab | Anti-PD-L1 | NCT02336165 | GBM | Bevacizumab and radiotherapy | II | Completed |
| NCT02794883 | Recurrent GBM | Tremelimumb | II | Completed | ||
| NCT02866747 | Recurrent GBM | Hypofractionated stereotactic radiotherapy | I/II | Active, not recruiting | ||
| Avelumab | NCT03341806 | Recurrent GBM | MRI-guided LITT therapy | I | Completed | |
| NCT03291314 | Recurrent GBM | Axitinib | II | Completed | ||
| NCT02968940 | IDH-mutant GBM | Hypofractionated stereotactic radiotherapy | II | Completed | ||
| NCT03047473 | Newly diagnosed GBM | TMZ | II | Completed | ||
| NCT03750071 | Progressive GBM | VXM01 | I/II | Active, not recruiting | ||
| Atezolizumab | NCT04160494 | Recurrent GBM | D2C7-IT | I | Active, not recruiting | |
| NCT03158389 | MGMT unmethylated GBM | Radiotherapy | I/II | Completed | ||
| NCT03174197 | Newly diagnosed GBM | Radiotherapy and TMZ | I/II | Active, not recruiting | ||
| NCT03673787 | GBM | Ipatasertib | I/II | Recruiting | ||
| INCMGA00012 | Anti-PD-1 | NCT04225039 | Recurrent GBM | INCAGN01876 and stereotactic radiosurgery | II | Active, not recruiting |
| NCT03532295 | Recurrent GBM | Bevacizumab and radiotherapy | II | Active, not recruiting | ||
| Nivolumab | NCT02529072 | GBM | Dendritic cell vaccine | I | Completed | |
| NCT02658981 | Recurrent GBM | BMS-986016 | I | Completed | ||
| NCT02311920 | Newly diagnosed GBM | Ipilimumab and TMZ | I | Completed | ||
| NCT03233152 | Recurrent GBM | Ipilimumab | I | Active, not recruiting | ||
| NCT04047706 | Newly diagnosed GBM | BMS-986205, TMZ and radiotherapy | I | Active, not recruiting | ||
| NCT04003649 | Recurrent GBM | IL-13Ralpha2-CAR T-cells and Ipilimumab | I | Recruiting | ||
| NCT04323046 | Recurrent GBM | Ipilimumab | I | Recruiting | ||
| NCT03636477 | Recurrent or progressive GBM | Ad-RTS-hIL-12 and Veledimex | II | Completed | ||
| NCT03367715 | Newly diagnosed, MGMT unmethylated GBM | Ipilimumab and radiotherapy | II | Completed | ||
| NCT02617589 | Newly diagnosed, MGMT unmethylated GBM | TMZ | III | Completed | ||
| NCT03452579 | Recurrent GBM | Bevacizumab | II | Active, not recruiting | ||
| NCT03743662 | Recurrent MGMT methylated GBM | Bevacizumab and radiotherapy | II | Active, not recruiting | ||
| NCT04195139 | Newly diagnosed elderly patients with GBM | TMZ | II | Active, not recruiting | ||
| NCT02667587 | Newly diagnosed, MGMT methylated GBM | TMZ and radiotherapy | II | Active, not recruiting | ||
| NCT03890952 | Recurrent GBM | Bevacizumab | II | Active, not recruiting | ||
| NCT03718767 | IDH-mutant GBM with and without hypermutator phenotype | - | II | Recruiting | ||
| NCT04396860 | Newly diagnosed, MGMT unmethylated GBM | Ipilimumab | II/III | Active, not recruiting | ||
| NCT04116658 | Progressive GBM | Therapeutic vaccine EO2401 | I/II | Active, not recruiting | ||
| Spartalizumab | NCT03961971 | Recurrent GBM | MBG453 | I | Active, not recruiting | |
| Pembrolizumab | NCT03726515 | Newly diagnosed, MGMT unmethylated GBM | EGFRvIII-CAR T-cells | I | Completed | |
| NCT03426891 | Newly diagnosed GBM | Vorinostat, TMZ and radiotherapy | I | Completed | ||
| NCT02852655 | Recurrent or progressive GBM | - | I | Completed | ||
| NCT02313272 | Recurrent GBM | Bevacizumab and radiotherapy | I | Completed | ||
| NCT03722342 | Recurrent GBM | TTAC-0001 | I | Active, not recruiting | ||
| NCT04201873 | Recurrent or progressive GBM | Dendritic cell tumor cell lysate vaccine | I | Recruiting | ||
| NCT03277638 | Recurrent GBM | Laser interstitial thermotherapy | I | Recruiting | ||
| NCT02287428 | MGMT unmethylated, newly diagnosed GBM | Radiotherapy, TMZ and neoantigen vaccine | I | Recruiting | ||
| NCT02798406 | Recurrent GBM | Adenovirus (DNX-2401) | II | Completed | ||
| NCT03018288 | Newly diagnosed GBM | HSPPC-96 and TMZ | II | Completed | ||
| NCT04013672 | Recurrent GBM | SurVaxM, sargramostim, montanide, and ISA51 | II | Active, not recruiting | ||
| NCT03661723 | Recurrent GBM | Bevacizumab and radiotherapy | II | Active, not recruiting | ||
| NCT03899857 | Newly diagnosed GBM | - | II | Active, not recruiting | ||
| NCT03405792 | Newly diagnosed GBM | Tumor treating field (TTF) | II | Active, not recruiting | ||
| NCT02337686 | Recurrent GBM | - | II | Active, not recruiting | ||
| NCT03797326 | GBM | Lenvatinib | II | Active, not recruiting | ||
| NCT03347617 | Newly diagnosed GBM | Ferumoxytol | II | Active, not recruiting | ||
| NCT04479241 | Recurrent GBM | Oncolytic polio/rhinovirus recombinant (PVSRIPO) | II | Active, not recruiting | ||
| NCT01174121 | Progressive GBM | Cyclophosphamide, fludarabine, aldesleukin and TIL | II | Recruiting | ||
| NCT03412877 | GBM | Cyclophosphamide, fludarabine, aldesleukin and TCR | II | Recruiting | ||
| NCT02311582 | Recurrent GBM | MRI-guided laser ablation | I/II | Completed | ||
| NCT03665545 | Recurrent GBM | IMA950/poly-ICLC | I/II | Active, not recruiting | ||
| NCT02658279 | Recurrent GBM with a hypermutator phenotype | - | - | Active, not recruiting | ||
| Cemiplimab | NCT04006119 | Recurrent or progressive GBM | Ad-RTS-hIL-12 and Veledimex | II | Completed | |
| NCT03491683 | Newly diagnosed GBM | INO-5401 and INO-9012 | I/II | Active, not recruiting | ||
| BMS-986205 (Linrodostat) | Anti-IDO1 | NCT04047706 | Newly diagnosed GBM | Nivolumab, TMZ and radiotherapy | I | Active, not recruiting |
| Indoximod | NCT02502708 | Newly diagnosed GBM | TMZ, radiotherapy, cyclophosphamide and etoposide | I | Completed | |
| NCT04049669 | Progressive GBM | TMZ, radiotherapy, cyclophosphamide, etoposide and Lomustine | II | Recruiting | ||
| NCT02052648 | GBM | Bevacizumab, TMZ and stereotactic radiation | I/II | Completed | ||
| Tremelimumab | Anti-CLTA4 | NCT02794883 | Recurrent GBM | Durvalumab | II | Completed |
| Ipilimumab | NCT02311920 | newly diagnosed GBM | Nivolumab and TMZ | I | Completed | |
| NCT03233152 | Recurrent GBM | Nivolumab | I | Active, not recruiting | ||
| NCT04003649 | Recurrent GBM | IL13Ralpha2-CAR T-cells and Nivolumab | I | Recruiting | ||
| NCT04323046 | Recurrent GBM | Nivolumab | I | Recruiting | ||
| NCT03367715 | newly diagnosed, MGMT unmethylated GBM | Nivolumab and radiotherapy | II | Completed | ||
| NCT02052648 | GBM | Bevacizumab, TMZ and stereotactic radiation | I/II | Completed | ||
| NCT04396860 | Newly diagnosed, MGMT unmethylated GBM | Nivolumab | II/III | Active, not recruiting | ||
| APX005M | Phage based therapy | NCT03389802 | GBM | - | I | Active, not recruiting |
| NK-92/5.28.z | NK cell therapy | NCT03383978 | Recurrent HER2-positive GBM | - | I | Active, not recruiting |
| Adoptive cell therapy | ||||||
| EGFRvIII CAR-T | NCT03726515 | Newly diagnosed, MGMT unmethylated GBM | Pembrolizumab | I | Completed | |
| IL-13Ralpha2- autologous T-lymphocytes | NCT02208362 | Recurrent GBM | - | I | Active, not recruiting | |
| B7-H3 CAR-T | NCT04385173 | Recurrent GBM | TMZ | I | Recruiting | |
| Chlorotoxin (EQ)-CD28- CD3zeta-CD19T | NCT04214392 | Recurrent GBM | - | I | Recruiting | |
| IL-13Ralpha2, CAR-T | NCT04003649 | Recurrent GBM | Nivolumab and Ipilimumab | I | Recruiting | |
| TIL | NCT01174121 | Progressive GBM | Cyclophosphamide, fludarabine, aldesleukin and Pembrolizumab | II | Recruiting | |
| Autologous T-cells, neoantigens | NCT03412877 | GBM | Cyclophosphamide, fludarabine, aldesleukin and Pembrolizumab | II | Recruiting |
Strategies targeting TAMs focus on blocking recruitment via CCL2-CCR2 inhibition, promoting M1 polarization via CD47-SIRPα blockade, or depleting M1 polarization via CSF-1R inhibitors.375,376 These approaches have demonstrated preclinical efficacy but require further validation in GBM patients. NK cell-based therapies, including CYNK-001, are in early clinical trials, with CAR-NK cells engineered to target GBM-specific antigens such as EGFRvIII, HER2, IL-13Rα2, and CD133 showing preclinical efficacy.377,378 CAR-T cell therapy also presents potential, with intrathecal bivalent CAR-T cells targeting EGFR and IL-13Rα2 demonstrating early tumor reduction.379 Additionally, PTP4A2 regulates GBM recurrence via roundabout guidance receptor 1 (ROBO1), and CAR-T cell targeting of ROBO1 improves survival in recurrent GBM models, highlighting a potential therapeutic strategy for GBM.380
Overall, the failure of single-agent ICIs underscores the necessity of combination strategies addressing immune evasion and TME constraints. Continued research and clinical trials are essential for refining immunotherapy approaches and overcoming resistance in GBM treatment.
Targeted therapy
Targeted therapy, which is designed to selectively inhibit molecular pathways critical for tumor progression while minimizing systemic toxicity, has become a cornerstone of GBM treatment. Unlike conventional therapies that broadly affect both malignant and normal cells, targeted approaches aim to improve efficacy while mitigating adverse effects. Bevacizumab, a VEGF inhibitor, is approved for recurrent GBM and effectively delays disease progression; however, its impact on OS remains limited.381 Similarly, RTK inhibitors targeting PDGFR (e.g., olaratumab and crenolanib) and c-KIT (avapritinib) show promising BBB penetration but are hindered by resistance mechanisms.382 EGFR inhibitors such as gefitinib, cetuximab, and ABT-414 have demonstrated variable efficacy, primarily due to tumor heterogeneity and adaptive resistance. In addition to growth factor receptors, the c-MET/HGF pathway has emerged as a key driver of GBM invasion and therapy resistance. Dacomitinib, an EGFR inhibitor, reduces tumor viability and self-renewal in EGFR-amplified GBM, although its effectiveness is influenced by the PTEN status,383,384 Despite their initial promise, c-MET inhibitors such as onartuzumab and rilotumumab have failed to significantly improve survival outcomes. Onartuzumab tended to reduce tumor growth but lacked clinical efficacy when combined with bevacizumab. Similarly, rilotumumab did not show notable antitumor activity as a monotherapy or in combination with bevacizumab. These findings highlight the need for refined therapeutic targeting strategies in c-MET-driven GBMs.
The PI3K/AKT/mTOR pathway, which is frequently dysregulated in GBM, remains a key therapeutic target. However, inhibitors such as buparlisib have not demonstrated significant clinical benefits, reinforcing the need for dual mTORC1/2 inhibitors such as vistusertib.385,386 Epigenetic modulators, including DNMT and BET inhibitors, are being explored as potential alternatives. PARP inhibitors have gained traction in GBM therapy, particularly in combination with agents that disrupt DNA repair mechanisms. Stellettin B sensitizes GBM cells to PARP inhibitors (e.g., rucaparib and olaparib) by downregulating BRCA1/2 and RAD51, leading to synthetic lethality and tumor apoptosis.387 BET inhibitors, such as Birabresib, further enhance this effect by impairing DNA repair and disrupting cell cycle progression. Notably, compared with monotherapies, sequential PARP-BET inhibitor treatment maintains sustained antitumor activity while minimizing toxicity.388 These findings underscore the potential of targeting chromatin regulators alongside DNA damage response pathways.
Resistance to targeted therapies remains a major barrier in GBM treatment. Although promising for disrupting tumor proliferation and immune evasion, STAT3 and JAK inhibitors face significant limitations due to poor BBB penetration.279 Overcoming these obstacles requires innovative drug delivery approaches, such as nanoparticle-based carriers and convection-enhanced delivery, to enhance the therapeutic reach. The complex and adaptive nature of GBM necessitates combination strategies that disrupt compensatory pathways while improving drug retention in tumor cells. The integration of targeted agents with immunotherapy or radiation has potential for overcoming resistance. As research progresses, precision medicine approaches and biomarker-driven strategies will be critical in refining targeted therapy regimens.
While targeted therapies have made significant strides in GBM management, their clinical efficacy remains inconsistent due to tumor heterogeneity and acquired resistance. Refining therapeutic combinations, improving drug delivery mechanisms, and leveraging biomarker-based treatment selection are critical for advancing GBM treatment.
Targeted combination therapies
GBM treatment resistance often arises from extensive intra- and intertumoral heterogeneity, necessitating combination therapies targeting multiple pathways. Table 2 outlines clinical trials evaluating combination strategies alongside radiotherapy and/or chemotherapy. The combination of radiotherapy with PCV has shown promising long-term outcomes. A 140-month follow-up study demonstrated significantly prolonged OS and PFS compared with adjuvant radiotherapy alone.389 Bevacizumab has also been evaluated in combination with TTFs and as an early intervention at the first recurrence. However, studies comparing bevacizumab-radiation combinations with bevacizumab monotherapy have yielded mixed results. Some retrospective analyses have suggested improved OS in patients with recurrent GBM,338,390 whereas others have reported that the addition of resurgery significantly enhances survival compared with bevacizumab alone. However, conflicting data from other studies indicate no significant survival advantage.391 The variability in the GBM response to combination therapies underscores the importance of personalized treatment strategies. Ongoing clinical trials continue to explore the integration of targeted agents with immunotherapies and standard treatments to improve patient survival. Advancements in biomarker-driven therapy selection, adaptive resistance monitoring, and novel drug delivery technologies are essential for more effective and personalized treatment regimens for GBM patients.
Table 2.
Clinical trials of targeted and combination therapies for GBM treatment
| Drug | Target | Condition | Clinical trial identifier | Combination | Phase | Status |
|---|---|---|---|---|---|---|
| Erlotinib | EGFR | Relapsed/refractory GBM | NCT00301418 | - | I/II | Completed |
| Newly diagnosed GBM | NCT00720356 | Bevacizumab and TMZ | II | Completed | ||
| Progressive or recurrent GBM | NCT00445588 | Sorafenib | II | Completed | ||
| Cetuximab | Relapsed/refractory GBM | NCT02800486 | Mannitol and radiotherapy | II | Recruiting | |
| Newly diagnosed GBM | NCT02861898 | Mannitol | I/II | Recruiting | ||
| Osimertinib | Recurrent GBM | NCT03732352 | Fludeoxyglucose F-18 PET | II | Active, not recruiting | |
| Nimotuzumab | Newly diagnosed GBM | NCT03388372 | Radiotherapy and TMZ | II | Completed | |
| Newly diagnosed GBM | NCT00753246 | - | III | Completed | ||
| Rindopepimut | Newly diagnosed, surgically resected, EGFRvIII-positive GBM | NCT01480479 | TMZ | III | Completed | |
| Newly diagnosed GBM | NCT00458601 | Radiotherapy and TMZ | II | Completed | ||
| Depatuxizumab | Recurrent GBM | NCT02343406 | TMZ | II | Completed | |
| GBM | NCT01800695 | Radiotherapy and TMZ | I | Completed | ||
| Newly diagnosed GBM With EGFR amplification | NCT02573324 | Radiotherapy and TMZ | III | Completed | ||
| Newly diagnosed or recurrent GBM | NCT02590263 | Radiotherapy and TMZ | I/II | Completed | ||
| AZD4547 | FGFR | GBM with FGFR-TACC gene fusion | NCT02824133 | - | I/II | Completed |
| Cediranib | VEGFR | Recurrent GBM | NCT02974621 | Bevacizumab and Olaparib | II | Active, not recruiting |
| Pazopanib | Recurrent GBM | NCT01931098 | Topotecan | II | Completed | |
| Newly diagnosed GBM | NCT02331498 | - | I/II | Recruiting | ||
| Vandetanib | Recurrent GBM | NCT00821080 | Sirolimus | I | Completed | |
| Sorafenib | Recurrent GBM | NCT01434602 | Everolimus | I/II | Completed | |
| Lenvatinib | GBM | NCT03797326 | Pembrolizumab | II | Active, not recruiting | |
| Temsirolimus | PI3K/AKT/mTOR | Recurrent GBM | NCT00329719 | Sorafenib and Tosylate | I/II | Completed |
| Recurrent GBM | NCT00335764 | Sorafenib and Tosylate | I/II | Completed | ||
| Recurrent GBM | NCT0223849 | Perifosine | I | Active, not recruiting | ||
| Recurrent GBM | NCT02343406 | TMZ | II | Completed | ||
| Everolimus | Recurrent GBM | NCT03834740 | Ribociclib | Early I | Completed | |
| Newly diagnosed GBM | NCT00553150 | Radiotherapy and TMZ | I/II | Completed | ||
| Newly diagnosed GBM | NCT01062399 | Radiotherapy and TMZ | I/II | Completed | ||
| Recurrent GBM | NCT01434602 | Sorafenib | I/II | Completed | ||
| Buparlisib | Relapsed/refractory GBM | NCT01349660 | Bevacizumab | I/II | Completed | |
| Newly diagnosed GBM | NCT01473901 | Radiotherapy and TMZ | I | Completed | ||
| Recurrent GBM | NCT01934361 | Lomustine or Carboplatin | I | Completed | ||
| Recurrent GBM | NCT01339052 | - | II | Completed | ||
| BBI608 | STAT-3 | Recurrent or progressed GBM | NCT02315534 | TMZ | I/II | Completed |
| Palbociclib | CDK | Newly diagnosed GBM without MGMT promoter methylation | NCT03158389 | Radiotherapy | I/II | Completed |
| Ribociclib | Recurrent GBM | NCT03834740 | Everolimus | Early I | Completed | |
| Preoperative GBM | NCT02933736 | - | Early I | Active, not recruiting | ||
| Abemaciclib | Recurrent GBM | NCT04074785 | Bevacizumab | Early I | Active, not recruiting | |
| Recurrent GBM | NCT02981940 | - | II | Active, not recruiting | ||
| Recurrent GBM | NCT04391595 | LY3214996 | Early I | Recruiting | ||
| GBM | NCT02977780 | TMZ | II | Recruiting | ||
| Olaparib | PARP | Recurrent GBM | NCT02974621 | Bevacizumab and Cediranib | II | Active, not recruiting |
| Newly diagnosed and recurrent GBM | NCT04614909 | Radiotherapy, Pamiparib and TMZ | I | Recruiting | ||
| GBM | NCT03212274 | - | II | Active, not recruiting | ||
| Pamiparib | Newly diagnosed and recurrent GBM | NCT03150862 | Radiotherapy and TMZ | I/II | Completed | |
| Recurrent GBM | NCT03914742 | TMZ | I/II | Completed | ||
| Recurrent GBM | NCT03749187 | TMZ | I | Recruiting | ||
| Niraparib | Newly diagnosed and recurrent GBM | NCT05076513 | Fractionated radiotherapy | 0/II | Active, not recruiting | |
| Rilotumumab | MET | Recurrent GBM | NCT01113398 | Bevacizumab | II | Completed |
| Onartuzumab | Recurrent GBM | NCT01632228 | Bevacizumab | II | Completed | |
| Capmatinib | GBM | NCT02386826 | Bevacizumab | I | Completed | |
| Vorinostat | HDAC | Newly diagnosed GBM | NCT03426891 | Radiotherapy, Pembrolizumab and TMZ | I | Completed |
| Recurrent GBM | NCT00555399 | Isotretinoin and TMZ | I/II | Active, not recruiting | ||
| Newly diagnosed GBM | NCT00731731 | Radiotherapy and TMZ | I/II | Completed | ||
| Recurrent GBM | NCT01738646 | Bevacizumab | II | Completed | ||
| Fimepinostat | Recurrent GBM | NCT03893487 | Surgery | Early I | Active, not recruiting | |
| AMG232 | MDM2 | Newly diagnosed and recurrent GBM | NCT03107780 | Radiotherapy | I | Recruiting |
| RG7388 | Newly diagnosed GBM without MGMT promoter methylation | NCT03158389 | Radiotherapy | I/II | Completed | |
| BCA101 | TGF-β | GBM | NCT04429542 | Pembrolizumab | II | Recruiting |
| Galunisertib | Recurrent GBM | NCT01582269 | Lomustine | I/II | Active, not recruiting | |
| AZD1390 | ATM | Newly diagnosed and recurrent GBM | NCT03423628 | Radiotherapy | I | Recruiting |
| Veliparib | GBM | NCT01514201 | Radiotherapy and TMZ | I/II | Completed | |
| GBM | NCT03581292 | Radiotherapy and TMZ | II | Active, not recruiting | ||
| Newly diagnosed GBM with MGMT promoter hypermethylation | NCT02152982 | TMZ | II/III | Active, not recruiting | ||
| Bortezomib | Proteasome | Recurrent GBM with unmethylated MGMT promoter | NCT03643549 | TMZ | I/II | Recruiting |
| Recurrent GBM | NCT01435395 | Bevacizumab and TMZ | I | Completed | ||
| Ixazomib | GBM | NCT02630030 | - | Early I | Completed | |
| Marizomib | Newly diagnosed GBM | NCT03345095 | Radiotherapy and TMZ | III | Completed | |
| GBM | NCT02330562 | Bevacizumab | I/II | Completed | ||
| Newly diagnosed GBM | NCT02903069 | Radiotherapy, TMZ and TTF | I | Completed | ||
| Newly diagnosed GBM | NCT03463265 | Nab-rapamycin | II | Completed | ||
| Azeliragon | RAGE | Newly diagnosed GBM | NCT05635734 | Radiotherapy and TMZ | Ib/II | Active, not recruiting |
| NCT05986851 | Radiotherapy | II | Active, not recruiting | |||
| Imipramine | Serotonin, Norepinephrine | Recurrent GBM | NCT04863950 | - | II | Recruiting |
| Anlotinib | TKI | Recurrent GBM | NCT04004975 | - | I/II | Unknown |
| Ponatinib | c-KIT | Bevacizumab-refractory GBM | NCT02478164 | - | II | Completed |
| Erdafinitib | FGFR fusion | IDH-wild type GBM | NCT05859334 | - | II | Recruiting |
| BGJ398 | Recurrent GBM | NCT01975701 | - | II | Completed | |
| Entrectinib | NTRK fusion | Primary brain tumors | NCT02568267 | - | II | Active, not recruiting |
| Advanced or metastatic solid or primary brain tumors | NCT02650401 | - | I/II | Active, not recruiting | ||
| Larotrectinib | NTRK-fusion positive solid tumors | NCT02576431 | - | II | Active, not recruiting | |
| PLB1001 | PTPRZ1-MET fusion | Recurrent high-grade gliomas | NCT02978261 | - | I | Completed |
| Vorasidenib | IDH | Residual or recurrent grade II glioma | NCT04164901 | - | III | Active, not recruiting |
Combining ICIs with complementary therapies is a promising approach for improving GBM treatment efficacy. When integrated with ICIs, radiotherapy enhances immunogenic cell death, leading to the release of tumor-associated antigens that activate DCs and prime T cells. This process improves T cell infiltration and activity, potentially overcoming immune resistance mechanisms inherent to GBM.392,393 Another emerging strategy involves personalized cancer vaccines that target neoantigens unique to GBM and are designed to elicit robust tumor-specific immune responses. These vaccines, when combined with ICIs, significantly increase T cell activation and amplify antitumor immunity, leading to increased tumor rejection rates and potentially improved clinical outcomes.394
The integration of personalized vaccines with ICIs represents a highly promising approach that is currently undergoing extensive research and clinical validation. These targeted combination strategies hold significant potential in overcoming GBM immune barriers, emphasizing the need for continued investigation and clinical development to refine their effectiveness in GBM immunotherapy.
Vaccines
Vaccine-based immunotherapy has emerged as a promising strategy for GBM treatment, with the aim of enhancing tumor-specific immune responses. By leveraging tumor antigens, these vaccines activate adaptive immunity and promote sustained immune surveillance against GBM cells. Currently, four primary vaccine-based strategies are under investigation for GBM: peptide vaccines, DNA vaccines, cell-based vaccines, and mRNA vaccines.395 Peptide and DNA vaccines introduce tumor-specific antigens or DNA sequences encoding tumor-associated proteins to elicit an adaptive immune response.396 Peptide vaccines target well-defined tumor antigens, whereas DNA vaccines utilize plasmid DNA to drive antigen expression in host cells. Cell vaccines, particularly DC vaccines, involve priming DCs derived from peripheral blood mononuclear cells with tumor antigens.397 rWTC-MBTA is an autologous vaccine that induces complete tumor regression in GBM models through T cell activation, long-term immune memory, and minimal toxicity. Its ability to enhance DC activation and T cell cytotoxicity suggests its potential for combination with other immunotherapies to improve GBM treatment.398 On the other hand, mRNA vaccines utilize viral vectors loaded with mRNAs encoding tumor antigens to induce robust immune responses.399 This strategy has gained attention because of its ability to induce strong immune responses and its adaptability in targeting multiple GBM-associated antigens.
Despite encouraging preclinical and early-phase clinical trial results, vaccine efficacy in GBM remains inconsistent. Key challenges include antigenic variability among tumors, limited infiltration of immune cells into the CNS, and the presence of immunosuppressive factors such as Tregs and MDSCs.395 Combination strategies integrating vaccines with ICIs, cytokine adjuvants, or personalized neoantigen approaches are being explored to enhance vaccine-induced immune responses. Another exciting avenue involves the development of personalized cancer vaccines targeting neoantigens unique to GBM tumor cells. These tailored vaccines aim to induce strong, tumor-specific immune responses, particularly those that enhance T cell activation. When utilized alongside ICIs, personalized cancer vaccines can significantly amplify immune responses, increasing tumor rejection rates and potentially leading to superior clinical outcomes. The combined strategy of personalized vaccines and ICIs represents a highly promising approach that is currently undergoing extensive research and clinical validation.394,400 Ongoing clinical trials (Table S8) continue to assess the therapeutic potential of GBM vaccines, with an emphasis on optimizing antigen selection, delivery methods, and immune modulation strategies. Further research is essential to refine vaccine-based immunotherapy and integrate it into multimodal GBM treatment paradigms.
Precision and personalized therapy
Advancements in drug screening and precision medicine are shaping the future of GBM treatment. A novel 3D brain cancer chip constructed from a photopolymerizable poly(ethylene) glycol diacrylate (PEGDA) hydrogel represents a significant breakthrough in drug testing. This platform mimics the TME by enabling controlled chemical release and replicating cell-to-cell and cell-to-matrix interactions. Its application in evaluating the combined effects of pitavastatin and irinotecan underscores its potential for high-throughput drug screening and personalized therapy, requiring minimal tumor biopsy samples.401 Gene expression profiling and mutation analysis further enhance the ability to develop targeted and personalized therapies. Despite the challenges posed by the spatial and temporal heterogeneity of tumors, this approach allows for the identification of effective therapeutic responses on the basis of genetic similarity. Multifocal tumors with PIK3CA mutations exhibit variable drug responses,402 emphasizing the necessity for comprehensive genomic analysis across multiple tumor regions to refine treatment strategies.
Additionally, induced neural stem cells (iNSCs) derived from patient skin cells present a promising avenue for personalized cell therapy. Engineered iNSCs can selectively induce apoptosis in GBM cells while retaining their differentiation potential. In preclinical models, iNSCs successfully target distant tumor sites and deliver therapeutic molecules such as TRAIL, improving survival rates by overcoming the BBB and minimizing systemic toxicity.403 While this approach holds potential, further validation is needed to establish its safety and efficacy for clinical application. Together, these advancements in drug screening technology, genomic profiling, and cell-based therapy highlight the shift toward more precise and effective GBM treatment strategies. Integrating these approaches could lead to improved therapeutic outcomes and personalized treatment regimens tailored to individual tumor characteristics.
Exploration of new horizons in GBM therapy
Stem cell therapy
The emergence of stem cell-based therapy represents a transformative approach in GBM treatment, offering a promising solution to major therapeutic challenges such as BBB penetration, tumor heterogeneity, and immune evasion.404 Neural stem cells (NSCs) and mesenchymal stem cells (MSCs) have garnered attention for their intrinsic tumor-homing ability, allowing them to serve as efficient vehicles for targeted drug delivery and immunomodulation in GBM. Their ability to migrate toward tumor sites is mediated by chemokine receptors such as CXCR1, CXCR2, CXCR4, and CCR2, which respond to glioma-secreted signals such as IL-8, stromal cell-derived factor 1, and monocyte chemoattractant protein-1 (MCP-1).405,406 This glioma-tropic migration enables direct therapeutic intervention within the TME, significantly improving drug bioavailability and reducing systemic toxicity. In addition to their innate migratory properties, genetically engineered NSCs and MSCs provide a versatile platform for delivering cytotoxic agents, cytokines, and OVs to GBMs. Patient-derived human-induced NSCs (hi-NSCs) offer a personalized therapeutic strategy, further enhancing compatibility and reducing the risk of immune rejection. In preclinical studies, TRAIL-expressing hi-NSCs have been shown to selectively induce apoptosis in GBM cells, leading to improved survival outcomes.407 Additionally, NSCs have been modified to secrete immunostimulatory cytokines such as IL-7, IL-12, and IL-23, promoting immune cell recruitment and antitumor activity.408
Stem cells also serve as delivery vehicles in enzyme/prodrug-based therapy and OV therapy. The FDA-approved HB1.F3. The CD NSC line, which converts 5-fluorocytosine into the active chemotherapeutic agent 5-fluorouracil (5-FU), has shown promising tumor localization and safety profiles in early-phase clinical trials.409 Similarly, carboxylesterase-releasing NSCs are being tested in combination with irinotecan to enhance its active metabolite, SN-38, for improved efficacy against HGGs. In virotherapy, NSC-mediated delivery of glioma-restricted adenoviruses (CRAd-S-pk7) enhances viral distribution while reducing immune clearance, demonstrating significant survival benefits in clinical trials.410,411 Overall, stem cell therapy represents a paradigm shift in GBM treatment, leveraging the ability of NSCs and MSCs to overcome therapeutic barriers, enhance precision drug delivery, and modulate the tumor immune microenvironment. While ongoing clinical trials continue to assess their safety and efficacy, stem cell-based strategies have the potential to redefine GBM management, paving the way for personalized and more effective treatment modalities for this aggressive form of brain cancer.
Oncolytic viruses
Oncolytic viruses (OVs) represent a promising therapeutic strategy for GBM, leveraging their ability to selectively infect and lyse rapidly proliferating tumor cells while transforming the immunosuppressive TME into an immune-responsive state. Unlike other malignancies, GBM lacks distant metastases, making it an ideal candidate for OV therapy, as the virus remains localized, maximizing its tumor-specific effects. In addition to direct oncolysis, OVs trigger immunogenic cell death, releasing tumor-associated antigens, damage-associated molecular patterns, and viral pathogen-associated molecular patterns, which enhance antigen presentation and stimulate immune activation.412 This process reverses the “cold” tumor phenotype of GBM by promoting APC recruitment, activating CD8+ CTLs, and counteracting the immunosuppressive influence of TAMs and Tregs. The highly immunosuppressive TME of GBM, characterized by M2-polarized TAMs and T cell exhaustion, limits the effectiveness of conventional immunotherapies.413 OVs counteract these suppressive mechanisms by inducing an inflammatory response and increasing immune infiltration, facilitating sustained antitumor immune attack. OVs fall into two major categories: replication-competent viruses, which selectively replicate within tumor cells, and replication-deficient viral vectors, which deliver therapeutic genes. Engineered viruses, such as adenoviruses (Ads), herpes simplex viruses (HSVs), vaccinia viruses (VVs), vesicular stomatitis viruses (VSVs), polioviruses, and measles viruses (MVs), have been optimized for tumor selectivity, enhanced oncolysis, and immune modulation. The oncolytic HSV-G47∆ agent demonstrated promising therapeutic potential in GBM, achieving a 1-year survival rate of 84.2% and a median OS of 20.2 months posttreatment, with a favorable safety profile. Its ability to induce TIL recruitment and repeated lesion responses on imaging contributed to its approval as Japan’s first OV therapy for GBM.414 More than 20 different OVs, including HSV-1, Ad, reovirus, NDV, MV, and poliovirus, have progressed to clinical trials for GBM,415–419 underscoring their therapeutic potential.
Effective OV delivery remains a critical challenge, with intratumoral administration preferred to avoid immune clearance.420 Convection-enhanced delivery, which uses a pressure gradient to bypass the BBB, has shown success in delivering recombinant nonpathogenic polio-rhinovirus chimeras into the CNS. Furthermore, innovative biological vectors such as NSCs and lymphocytes are being explored for OV delivery, improving viral biodistribution and persistence. Phase I clinical trials using NSC-mediated OV delivery (NSC-CRAd-S-pk7) in GBM patients have demonstrated enhanced safety and efficacy with minimal toxicity.410 Similarly, the use of lymphocytes modified with the herpesvirus saimiri represents a novel approach for OV transport.421 Table S9 presents the OVs used in clinical trials for GBM treatment. Advancing OV therapy for GBM requires continued optimization of viral engineering, immune modulation, and delivery strategies. The integration of OVs with ICIs, CAR-T cell therapy, and radiation is under investigation to further enhance therapeutic efficacy. With ongoing clinical trials and novel bioengineering approaches, OV-based therapies hold great potential for transforming GBM treatment, offering a multifaceted approach that combines direct tumor lysis with potent immune activation.
Combination of OVs with chemo-, radio- and immunotherapy
The combination of OVs with ICIs has shown promising results in the treatment of GBM. Studies have shown that MV infection upregulates PD-L1 expression in GBM models, increasing the susceptibility of tumors to anti-PD-1 therapy and significantly improving survival compared with monotherapy.422,423 Similarly, engineered reovirus expressing GM-CSF demonstrated enhanced survival with anti-PD-1 therapy.424 Another OV, DNX-2401, combined with anti-PD-1 therapy has led to a substantial shift in the TME and prolonged survival in preclinical GBM models.425 Strong synergy is also observed with IL-12-expressing oHSVs combined with anti-PD-1 and anti-CTLA-4 therapies, which effectively eliminate GSCs and boost immune activity.426,427 Similarly, the efficacy of the combination of VSV engineered to express tumor-specific antigens such as HIF-2α, Sox-10, and c-Myc with dual checkpoint blockade was improved.428 Clinical trials using DNX-2401 with pembrolizumab (anti-PD-1) reported a 100% nine-month survival rate in GBM patients.429
Genetically modified OVs expressing cytokines or fusion proteins have also demonstrated improved outcomes. When combined with agents such as rapamycin and GBM-specific neoantigens, vaccinia virus or Myxoma virus expressing the IL-15Rα-IL-15 fusion enhances survival.430 VSV encoding IFN-β has been explored alongside CAR-T cell therapy targeting EGFRvIII, highlighting the need for further optimization to fully understand immunological interactions.431 Combining OVΔ-24-RGD OVs with TMZ increased CD8+ T cell infiltration and prolonged survival.432 Other OVs, such as Toca 511 and TG6002, serve as prodrug-converting agents, transforming 5-FC into cytotoxic 5-FU433 and offering alternative therapeutic options. In addition to direct oncolysis and immune activation, engineered OVs are being leveraged to enhance adoptive cell therapies. HER2-CAR virus-specific T cells (HER2-CAR-VSTs) have shown safety and clinical efficacy in GBM patients.434 Bispecific T cell engagers (BiTEs) represent another innovative strategy, linking T cells to tumor antigens, preventing antigen escape, and amplifying antitumor responses.435 The continuous development of OVs as combinatorial immunotherapies, particularly with ICIs, CAR-T cells, and BiTEs, holds great promise for overcoming GBM’s immunosuppressive barriers and improving patient survival.
Extracellular vesicles for GBM treatment
EVs as therapeutic targets
EVs play a crucial role in GBM, facilitating tumor progression by increasing proliferation, invasiveness, chemoresistance, and immune evasion. Disrupting EV release, uptake, and circulation represents a promising therapeutic strategy for mitigating GBM progression. Several approaches have been identified, including targeting EVs in transit through hemodialysis, inhibiting their release via agents such as berberine and ketoconazole, or repurposing existing drugs such as heparin and reserpine.436 Berberine not only enhances photodynamic therapy sensitization but also inhibits GBM proliferation by suppressing fatty acid synthesis and reducing EV secretion.437 Additionally, heparan sulfate proteoglycans (HSPGs) modulate EV uptake in GBM cells, and strategies targeting HSPGs have been demonstrated to reduce EV internalization.438 However, the lack of cancer cell specificity in heparin-mediated EV inhibition poses a challenge for clinical application. Notably, GDEVs can activate glycolysis in human bone marrow mesenchymal stem cells (hBMSCs), leading to tumor-supportive transformation. This interaction between exosomes and hBMSCs highlights the potential of targeting EV-mediated signaling in GBM therapy.439
Gene and RNA therapies have gained traction in GBM treatment, with emerging research identifying multiple lncRNAs, miRNAs, and circRNAs within GDEVs that contribute to tumor progression. Key lncRNAs such as POUF3F3 and TALC significantly reshape the GBM microenvironment. POUF3F3 drives angiogenesis and tumor expansion, whereas TALC induces M2-macrophage polarization and upregulates the complement components C5/C5a, fostering chemoresistance.440 Other oncogenic lncRNAs, including MALAT1, MEG3, NEAT1, and HOTAIR, promote EMT and contribute to the aggressive phenotype of GBM.441 Moreover, targeting the mTOR pathway to suppress GDEV production offers a potential strategy to disrupt the supportive TME and curb tumor progression.442 Understanding the specific cargo within GDEVs is vital for designing targeted therapies. Proteomic analysis of GDEVs revealed that EGFRvIII, PDGFR, and HER2 are linked to enhanced tumor cell proliferation. In addition, proteins such as L1CAM, ANXA1, ITB1, and ACTR3 have been associated with increased tumor invasiveness. Furthermore, MRP1 has been shown to contribute to chemoresistance. Additionally, GDEVs are enriched with proangiogenic factors such as VEGF, TGF-β1, and CXCR4, which facilitate endothelial proliferation and vascular remodeling, as well as immunosuppressive mediators such as PD-L1 and MDSCs, which contribute to immune evasion.443 By selectively targeting these GDEV-associated proteins and pathways, novel therapeutic strategies can be developed to inhibit tumor growth, modulate the TME, and improve GBM treatment efficacy. Research into the molecular composition and functional impact of GDEVs is essential for refining these therapeutic strategies, with the potential to develop more precise and personalized treatments for GBM patients.
EVs as therapeutic candidates
EVs have emerged as promising therapeutic candidates for GBM because of their ability to influence key biological processes, including cell proliferation, apoptosis, differentiation, and immune modulation. Unlike viral vectors, EVs exhibit minimal adverse gene expression effects, enhancing their therapeutic potential for GBM treatment.444 Exosomes derived from MSCs engineered to carry tumor-suppressive miRNAs offer a targeted strategy to modulate GBM progression.445 Studies have demonstrated that MSC-derived exosomes loaded with miR-146b effectively reduce GBM cell proliferation and invasion in vitro while significantly decreasing tumor volume and improving survival in vivo.446 Similarly, the delivery of miR-124 and miR-145 via exosomes has been shown to suppress tumor growth by inhibiting GBM cell migration and altering the TME. Specifically, miR-124a-loaded exosomes (Exo-miR124a) suppress the clonogenicity of patient-derived GBM stem cells and reduce the tumor burden in intracranial xenograft models. Mechanistic studies have identified FOXA2 as a key target of miR-124a, linking its downregulation to apoptotic pathways and tumor suppression.447 Additionally, engineering GBM cells to express miR-302 and miR-367 profoundly affects the surrounding tumor environment, leading to decreased proliferation, reduced tumorigenicity, and the modulation of stemness markers in neighboring GBM cells.448 When implanted alongside GBM stem cells, these engineered cells significantly inhibited tumor growth in vivo.
In addition to miRNA-based approaches, exosome-mediated gene silencing has demonstrated efficacy in targeting oncogenic pathways in GBM. Studies have shown that exosomes engineered to carry a miR-21 sponge can effectively downregulate miR-21 while upregulating the expression of the tumor suppressors programmed cell death protein 4 and reversion-inducing cysteine-rich protein with Kazal motifs, key regulators of apoptotic and metastatic pathways. These effects have been validated in preclinical models, where modified exosomes suppressed tumor growth and enhanced the therapeutic response.449 Additionally, exosomes containing anti-miR-9 derived from hBMSCs, when combined with TMZ, significantly increased caspase activation and reduced GBM cell viability compared with those derived from TMZ alone, suggesting their potential to overcome chemoresistance.450 A landmark study using exosomes derived from rat bone marrow MSCs demonstrated their direct cytotoxic effects against GBM, indicating a shift from their traditional role as drug carriers to standalone therapeutic agents. These exosomes induced apoptosis in GBM cells and exhibited dose-dependent antitumor activity. Functional assays further revealed their ability to impair GBM cell migration and invasion, underscoring their potential in mitigating tumor progression and metastasis.444 Collectively, these findings highlight the growing importance of EV-based therapies in GBM treatment, providing a novel approach for targeted and personalized therapeutic interventions.
EVs as a drug delivery tool
EVs have emerged as a transformative drug delivery system for GBM therapy, offering a targeted and efficient approach to overcoming the challenges posed by the BBB and tumor resistance mechanisms. These vesicles efficiently transport chemotherapeutic agents such as DOX and PTX across the BBB, increasing drug accumulation within tumor cells while reducing systemic toxicity.13 Similarly, selumetinib-loaded EVs have demonstrated precise targeting capabilities, selectively delivering the drug to GBM cells while sparing healthy tissues, underscoring their potential for precision medicine.451 Methotrexate-loaded EVs modified with LDL and KLA peptides exhibited superior uptake in GBM spheroids,452 whereas yeast cytosine deaminase uracil phosphoribosyl transferase-engineered MSC EVs in combination with 5-FC effectively inhibited GBM growth.453 Neutrophil-derived EVs loaded with DOX demonstrated chemotactic migration toward tumor-infiltrating inflammatory cells, efficiently crossed the BBB and suppressed GBM progression.454 The adaptability of EVs for various administration routes, including intranasal and intraperitoneal delivery, further highlights their therapeutic flexibility.
In addition to conventional chemotherapy, EVs have been engineered to deliver novel therapeutic agents, including gene-editing tools and immunomodulatory molecules. EV-based systems integrating nanoparticle imaging agents with curcumin therapy have demonstrated dual functionality in GBM diagnosis and treatment, enhancing both detection and therapeutic outcomes.455 Their role in targeting GBM angiogenesis has also been explored, with miRNA-29a-3p-enriched MSC-derived EVs suppressing vasculogenic mimicry and angiogenesis independently of VEGF, suggesting a promising antiangiogenic strategy.456 Furthermore, DC-derived EVs loaded with dexamethasone exhibited immunomodulatory properties, promoting T cell activation and enhancing antitumor responses.457 Innovative surface modifications have further improved EV-based therapies. Arginylglycyl aspartic acid polypeptide-engineered EVs exhibit enhanced internalization into GBM cells, significantly improving drug delivery efficiency.458 Similarly, EVs loaded with small interfering RNAs targeting the FGFR3-TACC3 fusion gene effectively inhibited tumor cell viability while sparing adjacent normal tissues, demonstrating precision in gene-targeted therapy.459 Additionally, Cas9/sgRNA complexes encapsulated within Angiopep-2 (Ang)- and TAT-modified EVs achieved high-efficiency gene editing within GBM cells with minimal off-target effects,460 highlighting their potential in precision gene therapy.
Collectively, these advancements underscore EV-based therapies as novel and promising strategies for GBM treatment. Their ability to traverse the BBB, selectively target tumor cells and modulate the TME positions them as transformative tools for improving GBM outcomes. However, challenges such as optimizing targeting specificity, dosing, and long-term safety remain critical hurdles. Bridging the gap between experimental success and clinical application requires further research to establish standardized EV-based treatments, ultimately advancing personalized and effective GBM therapies.
Nanoparticles
Nanoparticle-based therapies offer a promising strategy for GBM treatment by improving drug delivery, enhancing BBB penetration, and overcoming tumor resistance. Their ability to precisely target tumors while minimizing systemic toxicity has led to significant advancements. Curcumin in the nanomicellar form, combined with TMZ, reduces GBM cell invasion and modulates apoptotic and autophagy pathways.461 Aptamer-conjugated polyamidoamine dendrimer nanoparticles loaded with PTX and TMZ effectively suppressed tumors by decreasing autophagy and drug resistance.462 A nose-to-brain delivery system using nanoparticles conjugated with alpha-cyano-4-hydroxycinnamic acid and cetuximab has been shown to reduce tumor size by inhibiting EGFR activation.463 Transferrin-modified liposomes with cell-penetrating peptides significantly increase DOX and erlotinib transport across the BBB, leading to tumor cell apoptosis.464
Liposomal delivery systems integrating transferrin and penetrating peptides have further optimized receptor-mediated transcytosis, improving drug translocation and extending survival in GBM models.465 Codelivery of PTX and methotrexate via PLGA nanoparticles has outperformed free drug formulations. Chlorotoxin-conjugated PLGA nanoparticles effectively target and irradiate tumor cells, reducing ECM MMP-2 activity.466 When combined with radiation therapy, this approach results in increased nanovector accumulation and tumor suppression.467,468
The combination of gold nanoparticles with SI306 and radiotherapy improved tumor inhibition, while pH-sensitive polymersomes loaded with DOX exhibited excellent ability to cross the BBB.469 Magnetic nanoparticles loaded with camptothecin, TMZ, and indocyanine green have shown strong anti-GBM effects, as validated through imaging techniques.470 Composite microbowls that integrate curcumin, DOX, and amino acids have successfully delivered dual chemophotodynamic therapy, showing potential in 3D glioma spheroids.471 Gold–silver nanotriangles stabilized with polyethylene glycol have been demonstrated to be effective photothermal therapies, significantly reducing GBM cell viability with brief laser irradiation.472
Advanced nanotechnologies such as anti-EphA3-modified gold nanoparticles loaded with TMZ have been effective in overcoming TMZ resistance while enhancing photothermal therapy.473 A multifunctional phototheranostic agent incorporating dicysteamine-modified hypocrellin and cyclic peptides has enabled efficient tumor targeting via near-infrared absorption for chemo/photodynamic/photothermal therapy.474 Similarly, the indocyanine green-conjugated peptide AE105, which targets the urokinase plasminogen activator receptor, has improved the ability of fluorescence-guided surgery475 and photothermal therapy to prolong survival.476 Bradykinin aggregation-induced emission nanoparticles have shown high photothermal conversion efficiency, enabling deep-tissue tumor suppression and immune activation involving CD8+ T cells and NK cells.477 Gold nanorods conjugated with MCP-1 and iron-based frameworks significantly reduce the tumor volume after laser therapy.478 Immune-responsive nanoscale drug carriers, such as DOX-MSN-SS-iRGD&1MT nanoparticles, have been developed to codeliver chemotherapy and ICIs across the BBB.479 Compared with conventional DOX formulations, damage-associated molecular pattern-emitting nano-DOX formulations have exhibited superior immunogenicity, enhancing DC activation and CD8+ T cell responses in GBM. The administration of docetaxel-sHDL-CpG nanodiscs with radiotherapy has resulted in long-term tumor remission in GBM patients.480 Further innovations include Angiopep LipoPCB nanoparticles and poly(L-malic acid)-based nanoscale immunoconjugates, which have demonstrated enhanced BBB penetration and immune modulation.
Nanotechnology is advancing GBM treatment by integrating chemotherapy, photothermal therapy, immune modulation, and gene targeting. The ability of nanoparticles to cross the BBB, selectively target tumors, and stimulate immune responses highlights their clinical potential. Further research is needed to optimize formulations, reduce toxicity, and evaluate long-term efficacy to improve GBM therapy.
Challenges and prospects in GBM therapy
The treatment of GBM remains profoundly challenging in oncology because of the resilience and plasticity of GSCs, extensive tumor heterogeneity, the highly immunosuppressive TME, metabolic adaptability, and the BBB. Traditional therapies focused solely on cancer cell destruction often fail, as GSCs exploit β-catenin-mediated signaling pathways to evade apoptosis while simultaneously reinforcing immunosuppressive mechanisms that support tumor survival.481 Addressing these barriers requires a paradigm shift toward integrated molecular, immune, and metabolic interventions. The future of GBM therapy involves multifaceted strategies that incorporate immunotherapy, precision medicine, metabolic targeting, and advanced drug delivery systems to overcome resistance mechanisms and enhance treatment efficacy (Fig. 8).
Fig. 8.
Major challenges and future therapeutic prospects in glioblastoma treatment. Key aspects, such as glioma stem cells (GSCs), therapy resistance, the blood‒brain barrier (BBB), metabolic reprogramming, and immune adaptation, are highlighted. 1. The glioblastoma (GBM) tumor microenvironment (TME) contributes to therapy resistance and disease progression. GSCs exhibit self-renewal capacity and plasticity, driving tumor recurrence and treatment failure. The proneural-to-mesenchymal transition underscores the heterogeneity of GBM, further complicating treatment strategies. 2. Therapy resistance mechanisms, including genetic mutations, epigenetic modifications, and adaptive survival pathways, are key obstacles to effective treatment. These mechanisms enable GBM cells to evade chemotherapy, radiotherapy, and targeted therapies. 3. This study highlights the challenges of overcoming the BBB, which restricts drug penetration and limits the efficacy of systemic therapies. Prospects involve strategies such as engineered EV-mediated drug delivery, efflux pump inhibitors, and modified pericytes and astrocytes to increase therapeutic access to the tumor site. 4. Metabolic reprogramming involves altered ATP production, lipid metabolism, and glycolysis, which provide energy for rapid tumor growth. Targeting metabolic vulnerabilities through the use of mitochondrial inhibitors, glycolysis modulators, and lipid metabolism disruptors is an emerging therapeutic approach. 5. In GBM, tumor-associated macrophages (TAMs), regulatory T cells (Tregs), and exhausted CD8+ T cells (Tex) contribute to an immunosuppressive environment. Immunotherapy strategies, including checkpoint inhibitors, dendritic cell (DC)-based vaccines, and the reprogramming of macrophage phenotypes (M2 to M1), aim to restore antitumor immunity and improve therapeutic responses. This schematic underscores the multifaceted nature of GBM pathophysiology and emphasizes the need for multimodal approaches integrating targeted therapy, immunotherapy, metabolic intervention, and BBB-modulating strategies to increase treatment efficacy and improve patient outcomes
Targeting GSCs
GSC-driven resistance significantly contributes to tumor recurrence and therapeutic failure by regulating DNA repair mechanisms, promoting proneural-to-mesenchymal transition (PMT), and enhancing invasive pathways.12 Therapeutic targeting of these resistance mechanisms with PARP, ATR, and ATM inhibitors has been shown to increase radiosensitivity,482 whereas STAT3 and TGF-β inhibitors prevent PMT-driven resistance.363 Additionally, tumor invasion can be mitigated by disrupting adhesion molecules such as L1CAM and inhibiting matrix remodeling enzymes such as MMPs, thereby improving therapeutic outcomes. Pharmacological inhibition of β-catenin and Wnt signaling further disrupts GSC self-renewal, ultimately reducing tumor progression.
Metabolic targeting
Metabolic reprogramming in GBM represents another key therapeutic target. Inhibiting glycolysis through GLUT1/3, HK2, and HIF-1α blockade, modulating lipid metabolism via FASN and SREBP-1 inhibitors, and disrupting amino acid metabolism via glutaminase (GLS) and SLC7A11 inhibitors have shown promise in limiting tumor growth.483 Exploiting IDH1/2 mutations with 2-HG inhibitors reverses metabolic and epigenetic dysregulation,484 whereas combination therapies incorporating metabolic inhibitors with standard treatments block metabolic plasticity and enhance therapeutic responses. The identification of compensatory metabolic pathways and the use of AI-driven analysis to predict resistance patterns further refine personalized treatment approaches.
Overcoming the immunosuppressive TME
A critical limitation of immunotherapy in GBM is the highly immunosuppressive nature of the TME, which actively restricts T cell infiltration and function.485 A dysfunctional BBB exacerbates this issue by permitting the secretion of immunosuppressive cytokines such as TGF-β and IL-10 while promoting the accumulation of MDSCs and TAMs, both of which inhibit immune activation.316 To overcome these barriers, ICIs combined with TME-modulating agents, such as CSF-1R inhibitors and anti-TGF-β therapies, are being explored. However, clinical trials, including CheckMate-143, CheckMate-498,486 and CheckMate-548,487 have demonstrated limited efficacy, largely due to the low tumor mutational burden and adaptive immune resistance of GBM. These findings underscore the need for novel combination approaches that integrate epigenetic modulation and metabolic reprogramming to reinvigorate immune responses and improve therapeutic outcomes.
Reinvigorating T cell exhaustion
The major limitation in GBM immunotherapy is T cell exhaustion, which results from chronic antigen exposure and leads to a progressive decline in CTL function. This exhaustion is driven by transcription factors such as FOXO1,488 FOXO3, and TOX and is further reinforced by epigenetic modifications involving DNMT3A, EZH2, and HBO1489,490 (Fig. 9). Addressing this issue through the use of FOXO1 modulators, PI3K/AKT inhibitors, and epigenetic therapies presents a promising strategy to restore T cell function and improve responsiveness to ICIs. Additionally, metabolic constraints within the TME, including glucose deprivation,491 amino acid competition,492 and lipid accumulation, further impair T cell activity. Strategies targeting these metabolic disruptions, such as GLUT1 inhibition, IDO blockade, and FASN inhibitors, have demonstrated potential in restoring T cell function.493 Emerging evidence also suggests that sodium chloride modulates T cell exhaustion by enhancing TCR signaling, metabolic fitness, and cytotoxicity,494,495 suggesting that sodium chloride is an innovative adjunct to existing immunotherapies.
Fig. 9.
Transcriptional and epigenetic regulation and functional dynamics in CD8+ T cell exhaustion. The illustration depicts the intricate interplay between signaling pathways and transcriptional regulators that drive CD8+ T cell exhaustion in the TME. Key pathways include the PI3K/AKT signaling pathway and the modulation of the activity of FOXO1/3 transcription factors. In the nucleus, TCF-1 promotes stemness by upregulating genes such as ID-3, EOMES, Bcl-2/6, and c-Myb. Together with TCF-1, FOXO1/3 represses effector T cell (Teff) functions by regulating exhaustion-associated genes (ID-2, Tbet, Blimp-1, RUNX3, and TCF-7). Exhausted T cells progress through a continuum, transitioning from progenitor-like (pro/stem-like Tex) states (PD-1low, TCF-1+, and CXCR3+) to terminally exhausted (terminal Tex) states (PD-1hi, TOXhi, and TCF-1−). FOXO1/3 also govern antioxidant and proapoptotic genes and regulate cell cycle arrest genes, maintaining cellular integrity and upregulating PD-1 and TOX. PD-1 and TOX function as central mediators of epigenetic regulation, influencing chromatin accessibility and transcriptional programming to stabilize exhaustion phenotypes. This PD-1 epigenetic regulation shapes T cell function and metabolic fitness within the tumor microenvironment
CAR-T cell therapy
CAR-T cell therapy has emerged as a promising approach for GBM treatment, yet its efficacy is hindered by antigen heterogeneity and the immunosuppressive microenvironment. Optimizing CAR-T cell therapy requires multiple antigen-targeting strategies, such as dual- and trivalent CAR constructs directed against EGFRvIII, IL13Rα2, and EphA2, reducing the likelihood of immune escape.496 Advances in switch-controlled CAR-T cell systems, including synthetic Notch circuits, enable selective activation in high-antigen-density environments while minimizing off-target effects.497 Additionally, hypoxia-sensitive CAR-T cells, engineered to adapt to the oxygen-deprived microenvironment of GBM, offer a novel way to increase specificity while reducing systemic toxicity. Improving CAR-T cell persistence through metabolic engineering, including the modulation of SIRT1 and PRMT5, has also demonstrated promise in sustaining antitumor activity. Furthermore, engineering CAR-T cells to express chemokine receptors such as CCR6 enhances their ability to infiltrate the dense stromal architecture of GBM,498 improving overall treatment efficacy (Fig. 10).
Fig. 10.
Increased therapeutic potential of CAR-T cells. The figure illustrates key strategies to optimize CAR-T cell therapy for GBM treatment. a. Multiantigen targeting improves CAR-T cell precision and efficacy against heterogeneous tumors. b. Advanced receptor designs, including costimulatory domain modifications and switch-controlled circuits such as synNotch CAR-T cells, enhance activation, persistence, and adaptability while sparing normal cells. c. Genome engineering introduces transcriptional and epigenetic changes to reduce exhaustion, improve memory, and increase cytokine production for sustained therapeutic effects. d. Inhibiting ubiquitin ligase-mediated degradation enhances CAR-T cell therapeutic potential
Overcoming the BBB
The BBB remains a formidable obstacle in GBM therapy, preventing efficient drug delivery and limiting the efficacy of systemic treatments. To overcome this barrier, advanced drug delivery systems such as nanoparticle-based carriers, focused ultrasound, and convection-enhanced delivery are being explored. Gene therapies utilizing CRISPR-based genome editing and OVs offer promising approaches for modifying the BBB or directly delivering therapeutic agents to tumor cells. Additionally, efflux pump inhibitors targeting P-glycoprotein and ABC transporters prevent premature drug elimination, whereas tumor vasculature normalization strategies enhance drug distribution.
Drug delivery technology
EVs and ncRNAs are emerging as novel therapeutic tools for crossing the BBB in GBM therapy. Engineered EVs carrying therapeutic ncRNAs such as miRNAs and lncRNAs offer precise targeting of GSCs and the TME,278 although their clinical translation requires further validation and standardization. Electric field therapy, particularly TTF, has gained attention as a noninvasive strategy to disrupt mitotic processes in GBM cells and prolong patient survival.353,354 However, challenges such as tumor resistance and electrode placement issues necessitate further refinement. The next generation of dynamic dnEFTs aims to enhance immune modulation, reduce tumor resistance, and improve penetration for deep-seated tumors.355 Combining dnEFTs with immunotherapies and ferroptosis-inducing agents may amplify their therapeutic impact and increase their long-term efficacy.
Precision medicine
Advances in precision medicine and adaptive therapy are reshaping GBM treatment by leveraging single-cell sequencing, AI-driven resistance prediction, and liquid biopsy technologies for real-time monitoring and personalized interventions. CRISPR-based genome editing and RNA interference technologies offer novel avenues for correcting oncogenic mutations and silencing tumor-promoting genes. AI-driven computational models optimize therapy selection and predict resistance mechanisms, facilitating more effective and tailored treatment regimens.
The future of GBM therapy lies in the seamless integration of diverse strategies targeting both tumor-intrinsic and microenvironmental resistance mechanisms. A comprehensive approach encompassing GSC eradication, immune reprogramming, metabolic modulation, CAR-T cell advancements, and innovative drug delivery technologies holds promise for improving GBM treatment efficacy. With continued research into synergistic treatment combinations, the translation of novel scientific advancements into effective clinical interventions offers new hope for prolonged survival and improved quality of life for GBM patients.
Conclusion
Despite significant advancements in understanding GBM pathogenesis, effective treatments remain elusive because of tumor heterogeneity, adaptability, and complex interactions with the TME. Future research must prioritize novel drug combination therapies that simultaneously target multiple oncogenic pathways, disrupting the adaptive mechanisms of GBM and overcoming therapeutic resistance. Personalized and precision medicine offers promising strategies by integrating genomic, transcriptomic, metabolomic, and epigenomic insights with biomarker-driven treatment selection and AI-powered predictive models. These approaches optimize treatment regimens by identifying patient-specific vulnerabilities. However, challenges such as biomarker validation, refining treatment paradigms, and ensuring accessibility to advanced diagnostics must be addressed to realize their full clinical potential. Immunotherapy holds great promise but faces barriers such as T cell exhaustion, checkpoint inhibitor resistance, and antigenic heterogeneity. Future directions should focus on reprogramming the immunosuppressive TME, enhancing T cell infiltration and function, and developing next-generation immunotherapies. Innovations such as improved CAR-T cell designs, OV-based therapies, and mRNA-based cancer vaccines combined with metabolic and epigenetic modifications may significantly increase immune responses. Additionally, overcoming key obstacles such as the BBB and drug efflux is crucial for improving drug delivery and minimizing tumor recurrence. Emerging technologies, including focused ultrasound, nanomaterial-based drug carriers, and electric field therapy, offer novel solutions to enhance therapeutic penetration and efficacy. The integration of precision therapeutics, molecular targeting, immunomodulation, and metabolic interventions provides a comprehensive framework for tackling GBM. Interdisciplinary collaboration and innovative clinical trial designs will be vital in translating these scientific advances into transformative clinical interventions. By leveraging these innovative strategies, the field has moved closer to achieving significant improvements in survival and quality of life for GBM patients.
Supplementary information
Acknowledgements
We thank Dr. Manish Sharma, Mayo Clinic, Mankato, MN, for careful review of this manuscript. We also acknowledge the Department of Neurosurgery and Department of Surgery at the University of Minnesota–Twin Cities for providing start-up funding to G.S. and A.S. All figures were created via BioRender.com.
Author contributions
Conceptualization: G.S, A.S.; original draft preparation: G.S., S.S., D.D., A.S.; review and editing: G.S., S.S., A.S., D.D., D.B., I.M., S.K., M.S., S.P., P.W.; visualization: G.S., A.S.; funding acquisition: G.S., A.S. All the authors have read and approved the article.
Competing interests
The authors declare no competing interests. The funders had no role in the design or writing of the manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Amar Singh, Email: singh423@umn.edu.
Gatikrushna Singh, Email: gsingh@umn.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41392-025-02299-4.
References
- 1.Ostrom, Q. T. et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013-2017. Neuro Oncol.22, iv1–iv96 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torp, S. H., Solheim, O. & Skjulsvik, A. J. The WHO 2021 classification of central nervous system tumours: a practical update on what neurosurgeons need to know-a minireview. Acta Neurochir.164, 2453–2464 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brás, J. P. et al. TERTmonitor-qPCR detection of TERTp mutations in glioma. Genes14, 1693 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller, J. J. Targeting IDH-mutant glioma. Neurotherapeutics19, 1724–1732 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han, S. et al. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br. J. Cancer122, 1580–1589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClellan, B. L. et al. Impact of epigenetic reprogramming on antitumor immune responses in glioma. J. Clin. Investig.133, e163450 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu, Q., Berglund, A. E. & Etame, A. B. The impact of epigenetic modifications on adaptive resistance evolution in glioblastoma. Int. J. Mol. Sci.22, 8324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan, H. et al. The heterogeneous cellular states of glioblastoma stem cells revealed by single cell analysis. Stem Cells41, 111–125 (2023). [DOI] [PubMed] [Google Scholar]
- 9.Guo, X. et al. Neuronal activity promotes glioma progression by inducing proneural-to-mesenchymal transition in glioma stem cells. Cancer Res84, 372–387 (2024). [DOI] [PubMed] [Google Scholar]
- 10.Singh, S. et al. Unveiling novel avenues in mTOR-targeted therapeutics: advancements in glioblastoma treatment. Int. J. Mol. Sci.24, 14960 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma, P., Aaroe, A., Liang, J. & Puduvalli, V. K. Tumor microenvironment in glioblastoma: current and emerging concepts. Neurooncol. Adv.5, vdad009 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, L., Jiang, Y., Zhang, G. & Wei, S. The diversity and dynamics of tumor-associated macrophages in recurrent glioblastoma. Front. Immunol.14, 1238233 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh, S., Chen, C. C., Kim, S., Singh, A. & Singh, G. Role of Extracellular vesicle microRNAs and RNA binding proteins on glioblastoma dynamics and therapeutics development. Extracell. Vesicle4, 100049 (2024). [Google Scholar]
- 14.Ratti, M. et al. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Target. Oncol.15, 261–278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uppaluri, K. R. et al. Unlocking the potential of non-coding RNAs in cancer research and therapy. Transl. Oncol.35, 101730 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakajima, N. et al. BRAF V600E, TERT promoter mutations and CDKN2A/B homozygous deletions are frequent in epithelioid glioblastomas: a histological and molecular analysis focusing on intratumoral heterogeneity. Brain Pathol.28, 663–673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louis, D. N. et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol.131, 803–820 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Louis, D. N. et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol.23, 1231–1251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips, H. S. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell9, 157–173 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Verhaak, R. G. W. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell17, 98–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma, A. et al. Angiogenic gene signature derived from subtype specific cell models segregate proneural and mesenchymal glioblastoma. Front. Oncol.7, 146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, H. et al. Specific glioblastoma multiforme prognostic-subtype distinctions based on DNA methylation patterns. Cancer Gene Ther.27, 702–714 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Molenaar, R. J., Maciejewski, J. P., Wilmink, J. W. & van Noorden, C. J. F. Wild-type and mutated IDH1/2 enzymes and therapy responses. Oncogene37, 1949–1960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, H., Ye, D., Guan, K.-L. & Xiong, Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin. Cancer Res.18, 5562–5571 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen, A. L., Holmen, S. L. & Colman, H. IDH1 and IDH2 mutations in gliomas. Curr. Neurol. Neurosci. Rep.13, 345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukasa, A. et al. Significance of IDH mutations varies with tumor histology, grade, and genetics in Japanese glioma patients. Cancer Sci.103, 587–592 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juratli, T. A. et al. The prognostic value of IDH mutations and MGMT promoter status in secondary high-grade gliomas. J. Neurooncol.110, 325–333 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Ramos-Fresnedo, A. et al. The survival outcomes of molecular glioblastoma IDH-wildtype: a multicenter study. J. Neurooncol.157, 177–185 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Duncan, C. G. et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res22, 2339–2355 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beiko, J. et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol.16, 81–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichimura, K. et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol.11, 341–347 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med.360, 765–773 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weller, M. et al. Molecular neuro-oncology in clinical practice: a new horizon. Lancet Oncol.14, e370–e379 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Hegi, M. E. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med.352, 997–1003 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Wick, W. et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomized, phase 3 trial. Lancet Oncol.13, 707–715 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Malmström, A. et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomized, phase 3 trial. Lancet Oncol.13, 916–926 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Armstrong, T. S. et al. Net clinical benefit analysis of radiation therapy oncology group 0525: a phase III trial comparing conventional adjuvant temozolomide with dose-intensive temozolomide in patients with newly diagnosed glioblastoma. J. Clin. Oncol.31, 4076–4084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinslow, C. J. et al. MGMT promoter methylation predicts overall survival after chemotherapy for 1p/19q-codeleted gliomas. Clin. Cancer Res.29, 4399–4407 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang, Q. The versatile attributes of MGMT: its repair mechanism, crosstalk with other DNA repair pathways, and itS ROLE IN CANCer. Cancers16, 331 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barajas, R. F. et al. Differentiation of recurrent glioblastoma multiforme from radiation necrosis after external beam radiation therapy with dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology253, 486–496 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi, C. et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat. Med.18, 624–629 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsen, V. A., Simonsen, H. J., Law, I., Larsson, H. B. W. & Hansen, A. E. Evaluation of dynamic contrast-enhanced T1-weighted perfusion MRI in the differentiation of tumor recurrence from radiation necrosis. Neuroradiology55, 361–369 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Mabray, M. C., Barajas, R. F. & Cha, S. Modern brain tumor imaging. Brain Tumor Res. Treat.3, 8–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korfiatis, P. et al. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med. Phys.43, 2835–2844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singhal, T., Narayanan, T. K., Jacobs, M. P., Bal, C. & Mantil, J. C. 11C-methionine PET for grading and prognostication in gliomas: a comparison study with 18F-FDG PET and contrast enhancement on MRI. J. Nucl. Med.53, 1709–1715 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Kim, S. et al. 11C-methionine PET as a prognostic marker in patients with glioma: comparison with 18F-FDG PET. Eur. J. Nucl. Med. Mol. Imaging32, 52–59 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Fueger, B. J. et al. Correlation of 6-18F-fluoro-L-DOPA PET uptake with proliferation and tumor grade in newly diagnosed and recurrent gliomas. J. Nucl. Med.51, 1532–1538 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Best, M. G. et al. Liquid biopsies in patients with diffuse glioma. Acta Neuropathol.129, 849–865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller Bark, J., Kulasinghe, A., Chua, B., Day, B. W. & Punyadeera, C. Circulating biomarkers in patients with glioblastoma. Br. J. Cancer122, 295–305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui, H. et al. Frosted slides decorated with silica nanowires for detecting circulating tumor cells from prostate cancer patients. ACS Appl. Mater. Interfaces10, 19545–19553 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Zhang, H., Yuan, F., Qi, Y., Liu, B. & Chen, Q. Circulating tumor cells for glioma. Front. Oncol.11, 607150 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pantel, K. & Speicher, M. R. The biology of circulating tumor cells. Oncogene35, 1216–1224 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Gao, X. et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR–STAT3 signaling. Nat. Cell Biol.23, 278–291 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Zhang, M. et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene37, 1805–1814 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Ahmed, S. P., Castresana, J. S. & Shahi, M. H. Role of circular RNA in brain tumor development. Cells11, 2130 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiao, J., Liu, M., Tian, Q. & Liu, X. Microarray analysis of circRNAs expression profile in gliomas reveals that circ_0037655 could promote glioma progression by regulating miR-214/PI3K signaling. Life Sci.245, 117363 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Chen, P. et al. CircXRCC5, as a potential novel biomarker, promotes glioma progression via the miR-490-3p/XRCC5/CLC3 competing endogenous RNA network. Neuroscience494, 104–118 (2022). [DOI] [PubMed] [Google Scholar]
- 59.Wu, Q., Yin, X., Zhao, W., Xu, W. & Chen, L. Molecular mechanism of m6A methylation of circDLC1 mediated by RNA methyltransferase METTL3 in the malignant proliferation of glioma cells. Cell Death Discov.8, 1–11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan, Z. et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer21, 16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen, B. et al. Circular RNA circLGMN facilitates glioblastoma progression by targeting miR-127-3p/LGMN axis. Cancer Lett.522, 225–237 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Yang, G. et al. lncRNA MAGI2-AS3 suppresses castration-resistant prostate cancer proliferation and migration via the miR-106a-5p/RAB31 axis. Genomics115, 110599 (2023). [DOI] [PubMed] [Google Scholar]
- 63.Mei, J.-C., Yan, G. & Mei, S.-Q. Diagnostic and prognostic potentials of long noncoding RNA ELF3-AS1 in glioma patients. Dis. Markers2020, 8871746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen, H. et al. SOX9-activated PXN-AS1 promotes the tumorigenesis of glioblastoma by EZH2-mediated methylation of DKK1. J. Cell. Mol. Med.24, 6070–6082 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tu, Z. et al. N6-Methylandenosine-related lncRNAs are potential biomarkers for predicting the overall survival of lower-grade glioma patients. Front. Cell Dev. Biol.8, 642 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan, S. K. et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol. Cancer17, 74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baspinar, Y., Elmaci, I., Ozpinar, A. & Altinoz, M. A. Long non-coding RNA MALAT1 as a key target in pathogenesis of glioblastoma. Janus faces or Achilles’ heal?. Gene739, 144518 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Xue, C., Chen, C., Gu, X. & Li, L. Progress and assessment of lncRNA DGCR5 in malignant phenotype and immune infiltration of human cancers. Am. J. Cancer Res.11, 1–13 (2021). [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao, Z. et al. Knockdown lncRNA CRNDE enhances temozolomide chemosensitivity by regulating autophagy in glioblastoma. Cancer Cell Int.21, 456 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, S. et al. LncRNA GAS5 represses stemness and malignancy of gliomas via elevating the SPACA6-miR-125a/let-7e axis. Front. Oncol.12, 803652 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lv, Q.-L. et al. A long noncoding RNA ZEB1-AS1 promotes tumorigenesis and predicts poor prognosis in glioma. Int. J. Mol. Sci.17, 1431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, L. et al. MicroRNA-21 expression is associated with overall survival in patients with glioma. Diagn. Pathol.8, 200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aloizou, A.-M. et al. The role of MiRNA-21 in gliomas: hope for a novel therapeutic intervention? Toxicol. Rep.7, 1514–1530 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santangelo, A. et al. A microRNA signature from serum exosomes of patients with glioma as complementary diagnostic biomarker. J. Neurooncol.136, 51–62 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Zhi, F. et al. Identification of 9 serum microRNAs as potential noninvasive biomarkers of human astrocytoma. Neuro Oncol.17, 383–391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yin, J. et al. Exosomal transfer of miR-1238 contributes to temozolomide-resistance in glioblastoma. EBioMedicine42, 238–251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lan, F. et al. Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell. Oncol.41, 25–33 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Tabibkhooei, A. et al. Profiling of novel circulating microRNAs as a non-invasive biomarker in diagnosis and follow-up of high and low-grade gliomas. Clin. Neurol. Neurosurg.190, 105652 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Zhao, K. et al. EGFR/c-myc axis regulates TGFβ/Hippo/Notch pathway via epigenetic silencing miR-524 in gliomas. Cancer Lett.406, 12–21 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Mahinfar, P. et al. The role of microRNAs in multidrug resistance of glioblastoma. Cancers14, 3217 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim, J. et al. microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer Res.74, 1541–1553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yin, D. et al. miR-34a functions as a tumor suppressor modulating EGFR in glioblastoma multiforme. Oncogene32, 1155–1163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim, S. et al. miR-340-5p suppresses aggressiveness in glioblastoma multiforme by targeting Bcl-w and Sox2. Mol. Ther. Nucleic Acids17, 245–255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jarmuzek, P., Wawrzyniak-Gramacka, E., Morawin, B., Tylutka, A. & Zembron-Lacny, A. Diagnostic and prognostic value of circulating DNA fragments in glioblastoma multiforme patients. Int. J. Mol. Sci.25, 4221 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seyhan, A. A. CirculatinG Liquid Biopsy Biomarkers In Glioblastoma: Advances And Challenges. Int. J. Mol. Sci.25, 7974 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zill, O. A. et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin. Cancer Res.24, 3528–3538 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Piccioni, D. E. et al. Analysis of cell-free circulating tumor DNA in 419 patients with glioblastoma and other primary brain tumors. CNS Oncol.8, CNS34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller, A. M. et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature565, 654–658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cilibrasi, C. et al. Definition of an inflammatory biomarker signature in plasma-derived extracellular vesicles of glioblastoma patients. Biomedicines10, 125 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Osti, D. et al. Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin. Cancer Res.25, 266–276 (2019). [DOI] [PubMed] [Google Scholar]
- 91.Cumba Garcia, L. M., Peterson, T. E., Cepeda, M. A., Johnson, A. J. & Parney, I. F. Isolation and analysis of plasma-derived exosomes in patients with glioma. Front. Oncol.9, 651 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shao, H. et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med.18, 1835–1840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ricklefs, F. L. et al. Circulating extracellular vesicles as biomarker for diagnosis, prognosis, and monitoring in glioblastoma patients. Neuro Oncol.26, 1280–1291 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen, W. W. et al. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol. Ther. Nucleic Acids2, e109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Figueroa, J. M. et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol.19, 1494–1502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Manda, S. V. et al. Exosomes as a biomarker platform for detecting epidermal growth factor receptor-positive high-grade gliomas. J. Neurosurg.128, 1091–1101 (2018). [DOI] [PubMed] [Google Scholar]
- 97.Vaidya, M. & Sugaya, K. DNA associated with circulating exosomes as a biomarker for glioma. Genes11, 1276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim, Y. Z. Altered histone modifications in gliomas. Brain Tumor Res. Treat.2, 7–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kreth, S., Thon, N. & Kreth, F. W. Epigenetics in human gliomas. Cancer Lett.342, 185–192 (2014). [DOI] [PubMed] [Google Scholar]
- 100.Venneti, S. et al. Histone 3 lysine 9 trimethylation is differentially associated with isocitrate dehydrogenase mutations in oligodendrogliomas and high-grade astrocytomas. J. Neuropathol. Exp. Neurol.72, 298–306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature482, 226–231 (2012). [DOI] [PubMed] [Google Scholar]
- 102.Lewis, P. W. et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bryant, J.-P., Heiss, J. & Banasavadi-Siddegowda, Y. K. Arginine methylation in brain tumors: tumor biology and therapeutic strategies. Cells10, 124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Romani, M., Daga, A., Forlani, A., Pistillo, M. P. & Banelli, B. Targeting of histone demethylases KDM5A and KDM6B inhibits the proliferation of temozolomide-resistant glioblastoma cells. Cancers11, 878 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen, R. et al. The application of histone deacetylases inhibitors in glioblastoma. J. Exp. Clin. Cancer Res.39, 138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schmidt, N., Windmann, S., Reifenberger, G. & Riemenschneider, M. J. DNA hypermethylation and histone modifications downregulate the candidate tumor suppressor gene RRP22 on 22q12 in human gliomas. Brain Pathol.22, 17–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Haase, S. et al. Mutant ATRX: uncovering a new therapeutic target for glioma. Expert Opin. Ther. Targets22, 599–613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nandakumar, P., Mansouri, A. & Das, S. The role of ATRX in glioma biology. Front. Oncol.7, 236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Koschmann, C., Lowenstein, P. R. & Castro, M. G. ATRX mutations and glioblastoma: impaired DNA damage repair, alternative lengthening of telomeres, and genetic instability. Mol. Cell. Oncol.3, e1167158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jones, P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet.13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- 111.Ferreyra, V. S. et al. DNA methylation profiling for molecular classification of adult diffuse lower-grade gliomas. Clin. Epigenetics13, 102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dejaegher, J. et al. DNA methylation based glioblastoma subclassification is related to tumoral T-cell infiltration and patient survival. Neuro Oncol.23, 240–250 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou, D. et al. DNMT1 mediates chemosensitivity by reducing methylation of miRNA-20a promoter in glioma cells. Exp. Mol. Med.47, e182–e182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bady, P. et al. The DNA methylome of DDR genes and benefit from RT or TMZ in IDH mutant low-grade glioma treated in EORTC 22033. Acta Neuropathol.135, 601–615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baumert, B. G. et al. Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033-26033): a randomized, open-label, phase 3 intergroup study. Lancet Oncol.17, 1521–1532 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nawaz, Z. et al. Impact of somatic copy number alterations on the glioblastoma miRNome: miR-4484 is a genomically deleted tumour suppressor. Mol. Oncol.11, 927–944 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cimino, P. J. et al. Copy number profiling across glioblastoma populations has implications for clinical trial design. Neuro Oncol.20, 1368–1373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Seifert, M., Friedrich, B. & Beyer, A. Importance of rare gene copy number alterations for personalized tumor characterization and survival analysis. Genome Biol.17, 204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cimino, P. J. et al. Multidimensional scaling of diffuse gliomas: application to the 2016 World Health Organization classification system with prognostically relevant molecular subtype discovery. Acta Neuropathol. Commun.5, 39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Spiegl-Kreinecker, S. et al. Prognostic quality of activating TERT promoter mutations in glioblastoma: interaction with the rs2853669 polymorphism and patient age at diagnosis. Neuro Oncol.17, 1231–1240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim, H. S. et al. Clinical implications of TERT promoter mutation on IDH mutation and MGMT promoter methylation in diffuse gliomas. Pathol. Res. Pract.214, 881–888 (2018). [DOI] [PubMed] [Google Scholar]
- 122.Lee, Y. et al. The frequency and prognostic effect of TERT promoter mutation in diffuse gliomas. Acta Neuropathol. Commun.5, 62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ryland, G. L. et al. Loss of heterozygosity: what is it good for?. BMC Med. Genom.8, 45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kakkar, A. et al. Loss of heterozygosity on chromosome 10q in glioblastomas, and its association with other genetic alterations and survival in Indian patients. Neurol. India59, 254–261 (2011). [DOI] [PubMed] [Google Scholar]
- 125.Hata, N. et al. Allelic losses of chromosome 10 in glioma tissues detected by quantitative single-strand conformation polymorphism analysis. Clin. Chem.52, 370–378 (2006). [DOI] [PubMed] [Google Scholar]
- 126.Ohgaki, H. & Kleihues, P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci.100, 2235–2241 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reuss, D. E. et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an ‘integrated’ diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol.129, 133–146 (2015). [DOI] [PubMed] [Google Scholar]
- 128.Vogazianou, A. P. et al. Distinct patterns of 1p and 19q alterations identify subtypes of human gliomas that have different prognoses. Neuro Oncol.12, 664–678 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Erdem-Eraslan, L. et al. Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. J. Clin. Oncol.31, 328–336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Di Stefano, A. L. et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin. Cancer Res.21, 3307–3317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang, Y., Long, P., Wang, Y. & Ma, W. NTRK fusions and TRK inhibitors: potential targeted therapies for adult glioblastoma. Front. Oncol.10, 593578 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bagchi, A. et al. Lorlatinib in a child with ALK-fusion-positive high-grade glioma. N. Engl. J. Med.385, 761–763 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Drilon, A. et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov.7, 400–409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bao, Z.-S. et al. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res.24, 1765–1773 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vogel, T. W. et al. Proteins and protein pattern differences between glioma cell lines and glioblastoma multiforme. Clin. Cancer Res.11, 3624–3632 (2005). [DOI] [PubMed] [Google Scholar]
- 136.Liu, F. et al. EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol. Cell60, 307–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Taylor, T. E., Furnari, F. B. & Cavenee, W. K. Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr. Cancer Drug Targets12, 197–209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Padfield, E., Ellis, H. P. & Kurian, K. M. Current therapeutic advances targeting EGFR and EGFRvIII in glioblastoma. Front. Oncol.5, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bulstrode, H. et al. Elevated FOXG1 and SOX2 in glioblastoma enforces neural stem cell identity through transcriptional control of cell cycle and epigenetic regulators. Genes Dev.31, 757–773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Assanah, M. et al. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J. Neurosci.26, 6781–6790 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Higa, N. et al. Prognostic impact of PDGFRA gain/amplification and MGMT promoter methylation status in patients with IDH wild-type glioblastoma. Neurooncol. Adv.4, vdac097 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Phillips, J. J. et al. PDGFRA amplification is common in pediatric and adult high-grade astrocytomas and identifies a poor prognostic group in IDH1 mutant glioblastoma. Brain Pathol.23, 565–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dufour, C. et al. Identification of prognostic markers in diffuse midline gliomas H3K27M-mutant. Brain Pathol.30, 179–190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jones, D. T. W. et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat. Genet.45, 927–932 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang, J. et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet.45, 602–612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Johnson, A. et al. Comprehensive genomic profiling of 282 pediatric low- and high-grade gliomas reveals genomic drivers, tumor mutational burden, and hypermutation signatures. Oncologist22, 1478–1490 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ballester, L. Y., Penas-Prado, M., Leeds, N. E., Huse, J. T. & Fuller, G. N. FGFR1 tyrosine kinase domain duplication in pilocytic astrocytoma with anaplasia. Cold Spring Harb. Mol. Case Stud.4, a002378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ellison, D. W. et al. cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAFV600E mutation. Acta Neuropathol.137, 683–687 (2019). [DOI] [PubMed] [Google Scholar]
- 149.Chiang, J. et al. A single-center study of the clinicopathologic correlates of gliomas with a MYB or MYBL1 alteration. Acta Neuropathol.138, 1091–1092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Saini, M., Jha, A. N., Tangri, R., Qudratullah, M. & Ali, S. MN1 overexpression with varying tumor grade is a promising predictor of survival of glioma patients. Hum. Mol. Genet.29, 3532–3545 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mhatre, R. et al. MN1 rearrangement in astroblastoma: study of eight cases and review of literature. Brain Tumor Pathol.36, 112–120 (2019). [DOI] [PubMed] [Google Scholar]
- 152.Lehman, N. L. et al. Genomic analysis demonstrates that histologically defined astroblastomas are molecularly heterogeneous and that tumors with MN1 rearrangement exhibit the most favorable prognosis. Acta Neuropathol. Commun.7, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liau, L. M. et al. Identification of a human glioma-associated growth factor gene, granulin, using differential immuno-absorption. Cancer Res.60, 1353–1360 (2000). [PubMed] [Google Scholar]
- 154.Wang, M., Li, G., Yin, J., Lin, T. & Zhang, J. Progranulin overexpression predicts overall survival in patients with glioblastoma. Med. Oncol.29, 2423–2431 (2012). [DOI] [PubMed] [Google Scholar]
- 155.Bandey, I., Chiou, S.-H., Huang, A.-P., Tsai, J.-C. & Tu, P. h. Progranulin promotes temozolomide resistance of glioblastoma by orchestrating DNA repair and tumor stemness. Oncogene34, 1853–1864 (2015). [DOI] [PubMed] [Google Scholar]
- 156.Lee, I.-N. et al. Knockdown of amphiregulin triggers doxorubicin-induced autophagic and apoptotic death by regulating endoplasmic reticulum stress in glioblastoma cells. J. Mol. Neurosci.70, 1461–1470 (2020). [DOI] [PubMed] [Google Scholar]
- 157.Steponaitis, G. et al. Significance of Amphiregulin (AREG) for the outcome of low and high grade astrocytoma patients. J. Cancer10, 1479–1488 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Desai, B., Rath, U., Segall, J. E. & Coniglio, S. J. Abstract 2391: microglia-stimulated glioblastoma cell invasion is dependent on the EGFR ligand amphiregulin. Cancer Res75, 2391 (2015). [Google Scholar]
- 159.Garros-Regulez, L. et al. Targeting SOX2 as a therapeutic strategy in glioblastoma. Front. Oncol.6, 222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Annovazzi, L., Mellai, M., Caldera, V., Valente, G. & Schiffer, D. SOX2 expression and amplification in gliomas and glioma cell lines. Cancer Genom. Proteom.8, 139–147 (2011). [PubMed] [Google Scholar]
- 161.Gangemi, R. M. R. et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells27, 40–48 (2009). [DOI] [PubMed] [Google Scholar]
- 162.Lopez-Bertoni, H. et al. Sox2 induces glioblastoma cell stemness and tumor propagation by repressing TET2 and deregulating 5hmC and 5mC DNA modifications. Signal Transduct. Target Ther.7, 1–12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Furnari, F. B. et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev.21, 2683–2710 (2007). [DOI] [PubMed] [Google Scholar]
- 164.Wang, J., Su, H., Zhao, H., Chen, Z. & To, S. T. Progress in the application of molecular biomarkers in gliomas. Biochem. Biophys. Res. Commun.465, 1–4 (2015). [DOI] [PubMed] [Google Scholar]
- 165.Network, T. C. G. A. R. Corrigendum: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature494, 506 (2013). [DOI] [PubMed] [Google Scholar]
- 166.Zhang, Y. et al. The p53 pathway in glioblastoma. Cancers10, 297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Romagosa, C. et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene30, 2087–2097 (2011). [DOI] [PubMed] [Google Scholar]
- 168.Serrano, M. The tumor suppressor protein p16INK4a. Exp. Cell Res.237, 7–13 (1997). [DOI] [PubMed] [Google Scholar]
- 169.Reis, G. F. et al. CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II-III) astrocytomas. J. Neuropathol. Exp. Neurol.74, 442–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lu, V. M. et al. The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: a systematic review of the contemporary literature. J. Neurooncol.148, 221–229 (2020). [DOI] [PubMed] [Google Scholar]
- 171.Parsa, A. T. et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med.13, 84–88 (2007). [DOI] [PubMed] [Google Scholar]
- 172.Waldron, J. S. et al. Implications for immunotherapy of tumor-mediated T-cell apoptosis associated with loss of the tumor suppressor PTEN in glioblastoma. J. Clin. Neurosci.17, 1543–1547 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen, P. et al. Symbiotic macrophage-glioma cell interactions reveal synthetic lethality in PTEN-Null glioma. Cancer Cell35, 868–884.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu, Y. et al. Dual targeting macrophages and microglia is a therapeutic vulnerability in models of PTEN-deficient glioblastoma. J. Clin. Investig.134, e178628 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Prasetyanti, P. R. & Medema, J. P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer16, 41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Larjavaara, S. et al. Incidence of gliomas by anatomic location. Neuro Oncol.9, 319–325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bradshaw, A. et al. Cancer stem cells in glioblastoma multiforme. Front. Surg.3, 48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Becker, A. P., Sells, B. E., Haque, S. J. & Chakravarti, A. Tumor heterogeneity in glioblastomas: from light microscopy to molecular pathology. Cancers13, 761 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bonavia, R., Inda, M.-M., Cavenee, W. K. & Furnari, F. B. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res.71, 4055–4060 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Marusyk, A., Almendro, V. & Polyak, K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer12, 323–334 (2012). [DOI] [PubMed] [Google Scholar]
- 181.Johnson, B. E. et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science343, 189–193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Szerlip, N. J. et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl. Acad. Sci. USA109, 3041–3046 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Little, S. E. et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res.72, 1614–1620 (2012). [DOI] [PubMed] [Google Scholar]
- 184.Snuderl, M. et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell20, 810–817 (2011). [DOI] [PubMed] [Google Scholar]
- 185.Dominiak, A., Chełstowska, B., Olejarz, W. & Nowicka, G. Communication in the cancer microenvironment as a target for therapeutic interventions. Cancers12, 1232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ibrahim, A. N. et al. Intratumoral spatial heterogeneity of BTK kinomic activity dictates distinct therapeutic response within a single glioblastoma tumor. J. Neurosurg.133, 1683–1694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Āboliņš, A. et al. Molecular subtype shift in breast cancer upon trastuzumab treatment: a case report. Pol. J. Pathol.62, 65–68 (2011). [PubMed] [Google Scholar]
- 188.Guardia, G. D. A. et al. Proneural and mesenchymal glioma stem cells display major differences in splicing and lncRNA profiles. npj Genom. Med.5, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ortensi, B., Setti, M., Osti, D. & Pelicci, G. Cancer stem cell contribution to glioblastoma invasiveness. Stem Cell Res. Ther.4, 18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cheng, L. et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell153, 139–152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Krusche, B. et al. EphrinB2 drives perivascular invasion and proliferation of glioblastoma stem-like cells. Elife5, e14845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Carro, M. S. et al. The transcriptional network for mesenchymal transformation of brain tumors. Nature463, 318–325 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ahmed, S. U. et al. Selective inhibition of parallel DNA damage response pathways optimizes radiosensitization of glioblastoma stem-like cells. Cancer Res.75, 4416–4428 (2015). [DOI] [PubMed] [Google Scholar]
- 194.Carruthers, R. D. et al. Replication stress drives constitutive activation of the DNA damage response and radioresistance in glioblastoma stem-like cells. Cancer Res78, 5060–5071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lin, C., Wang, N. & Xu, C. Glioma-associated microglia/macrophages (GAMs) in glioblastoma: Immune function in the tumor microenvironment and implications for immunotherapy. Front. Immunol.14, 1123853 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Biserova, K., Jakovlevs, A., Uljanovs, R. & Strumfa, I. Cancer stem cells: significance in origin, pathogenesis and treatment of glioblastoma. Cells10, 621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Prager, B. C., Bhargava, S., Mahadev, V., Hubert, C. G. & Rich, J. N. Glioblastoma stem cells: driving resilience through chaos. Trends Cancer6, 223–235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hatva, E. et al. Expression of endothelial cell-specific receptor tyrosine kinases and growth factors in human brain tumors. Am. J. Pathol.146, 368–378 (1995). [PMC free article] [PubMed] [Google Scholar]
- 199.Plate, K. H. & Risau, W. Angiogenesis in malignant gliomas. Glia15, 339–347 (1995). [DOI] [PubMed] [Google Scholar]
- 200.Yao, Y. et al. Prognostic value of vascular endothelial growth factor and its receptors Flt-1 and Flk-1 in astrocytic tumours. Acta Neurochir.143, 159–166 (2001). [DOI] [PubMed] [Google Scholar]
- 201.Sooman, L. et al. FGF2 as a potential prognostic biomarker for proneural glioma patients. Acta Oncol.54, 385–394 (2015). [DOI] [PubMed] [Google Scholar]
- 202.Batchelor, T. T., Reardon, D. A., de Groot, J. F., Wick, W. & Weller, M. Antiangiogenic therapy for glioblastoma: current status and future prospects. Clin. Cancer Res.20, 5612–5619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kanda, S., Landgren, E., Ljungström, M. & Claesson-Welsh, L. Fibroblast growth factor receptor 1-induced differentiation of endothelial cell line established from tsA58 large T transgenic mice. Cell Growth Differ.7, 383–395 (1996). [PubMed] [Google Scholar]
- 204.Abounader, R. & Laterra, J. Scatter factor/hepatocyte growth factor in brain tumor growth and angiogenesis. Neuro Oncol.7, 436–451 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Okuda, T. et al. Efficacy of combination therapy with MET and VEGF inhibitors for MET-overexpressing glioblastoma. Anticancer Res.37, 3871–3876 (2017). [DOI] [PubMed] [Google Scholar]
- 206.Brunckhorst, M. K., Wang, H., Lu, R. & Yu, Q. Angiopoietin-4 promotes glioblastoma progression by enhancing tumor cell viability and angiogenesis. Cancer Res.70, 7283–7293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Scholz, A. et al. Endothelial cell-derived angiopoietin-2 is a therapeutic target in treatment-naive and bevacizumab-resistant glioblastoma. EMBO Mol. Med.8, 39–57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bruna, A. et al. High TGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell11, 147–160 (2007). [DOI] [PubMed] [Google Scholar]
- 209.Bergers, G. et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol.2, 737–744 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ahir, B. K., Engelhard, H. H. & Lakka, S. S. Tumor development and angiogenesis in adult brain tumor: glioblastoma. Mol. Neurobiol.57, 2461–2478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Cj, P. et al. High LC3/beclin expression correlates with poor survival in glioma: a definitive role for autophagy as evidenced by in vitro autophagic flux. Pathol. Oncol. Res.25, 137–148 (2019). [DOI] [PubMed] [Google Scholar]
- 212.Gammoh, N. et al. Suppression of autophagy impedes glioblastoma development and induces senescence. Autophagy12, 1431–1439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tini, P. et al. Combined epidermal growth factor receptor and beclin1 autophagic protein expression analysis identifies different clinical presentations, responses to chemo- and radiotherapy, and prognosis in glioblastoma. Biomed. Res. Int.2015, 208076 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Pourhanifeh, M. H. et al. Autophagy in cancers including brain tumors: role of MicroRNAs. Cell Commun. Signal.18, 88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Azad, M. B. et al. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy4, 195–204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Catalano, M. et al. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Mol. Oncol.9, 1612–1625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wen, Z. et al. Knockdown ATG4C inhibits gliomas progression and promotes temozolomide chemosensitivity by suppressing autophagic flux. J. Exp. Clin. Cancer Res.38, 298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Galavotti, S. et al. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene32, 699–712 (2013). [DOI] [PubMed] [Google Scholar]
- 219.Zhang, L., Fritah, S., Nazarov, P. V., Kaoma, T. & Dyck, E. V. Impact of IDH mutations, the 1p/19q co-deletion and the G-CIMP status on alternative splicing in diffuse gliomas. Int. J. Mol. Sci.24, 9825 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Grimes, D. R., Jansen, M., Macauley, R. J., Scott, J. G. & Basanta, D. Evidence for hypoxia increasing the tempo of evolution in glioblastoma. Br. J. Cancer123, 1562–1569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Persano, L. et al. BMP2 sensitizes glioblastoma stem-like cells to Temozolomide by affecting HIF-1α stability and MGMT expression. Cell Death Dis.3, e412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Liu, Q. & Cao, P. Clinical and prognostic significance of HIF-1α in glioma patients: a meta-analysis. Int. J. Clin. Exp. Med.8, 22073–22083 (2015). [PMC free article] [PubMed] [Google Scholar]
- 223.Domènech, M., Hernández, A., Plaja, A., Martínez-Balibrea, E. & Balañà, C. Hypoxia: the cornerstone of glioblastoma. Int. J. Mol. Sci.22, 12608 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Reszec, J., Rutkowski, R. & Chyczewski, L. The expression of hypoxia-inducible factor-1 in primary brain tumors. Int. J. Neurosci.123, 657–662 (2013). [DOI] [PubMed] [Google Scholar]
- 225.Bouthelier, A. & Aragonés, J. Role of the HIF oxygen sensing pathway in cell defense and proliferation through the control of amino acid metabolism. BBA- Mol. Cell. Res.1867, 118733 (2020). [DOI] [PubMed] [Google Scholar]
- 226.Li, X., He, S. & Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer19, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Schito, L. & Semenza, G. L. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer2, 758–770 (2016). [DOI] [PubMed] [Google Scholar]
- 228.Zhang, J. et al. The roles of HIF-1α in radiosensitivity and radiation-induced bystander effects under hypoxia. Front. Cell Dev. Biol.9, 637454 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Colwell, N. et al. Hypoxia in the glioblastoma microenvironment: shaping the phenotype of cancer stem-like cells. Neuro Oncol.19, 887–896 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Tang, J.-H. et al. Downregulation of HIF-1a sensitizes U251 glioma cells to the temozolomide (TMZ) treatment. Exp. Cell Res.343, 148–158 (2016). [DOI] [PubMed] [Google Scholar]
- 231.Wolf, A. et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med.208, 313–326 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lee, J.-H. et al. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat. Commun.8, 949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sperry, J. et al. Glioblastoma utilizes fatty acids and ketone bodies for growth allowing progression during ketogenic diet therapy. iScience23, 101453 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Guo, D. et al. EGFR signaling through an Akt-SREBP-1 dependent, rapamycin-resistant pathway sensitizes glioblastomas to anti-lipogenic therapy. Sci. Signal2, ra82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ricklefs, F. L. et al. FASN is a biomarker enriched in malignant glioma-derived extracellular vesicles. Int. J. Mol. Sci.21, 1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Gimple, R. C. et al. Glioma stem cell-specific superenhancer promotes polyunsaturated fatty-acid synthesis to support EGFR signaling. Cancer Discov.9, 1248–1267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Robert, S. M. et al. SLC7A11 expression is associated with seizures and predicts poor survival in patients with malignant glioma. Sci. Transl. Med.7, 289ra86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yang, C. et al. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res.69, 7986–7993 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cano-Galiano, A. et al. Cystathionine-γ-lyase drives antioxidant defense in cysteine-restricted IDH1-mutant astrocytomas. Neurooncol. Adv.3, vdab057 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Carvalho da Fonseca, A. C. et al. Increased expression of stress inducible protein 1 in glioma-associated microglia/macrophages. J. Neuroimmunol.274, 71–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zhu, X. et al. Interaction of tumor-associated microglia/macrophages and cancer stem cells in glioma. Life Sci.320, 121558 (2023). [DOI] [PubMed] [Google Scholar]
- 242.Korbecki, J. et al. CC chemokines in a tumor: a review of pro-cancer and anti-cancer properties of the ligands of receptors CCR1, CCR2, CCR3, and CCR4. Int. J. Mol. Sci.21, 8412 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Takenaka, M. C. et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat. Neurosci.22, 729–740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ravi, V. M. et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell40, 639–655.e13 (2022). [DOI] [PubMed] [Google Scholar]
- 245.Fossati, G. et al. Neutrophil infiltration into human gliomas. Acta Neuropathol.98, 349–354 (1999). [DOI] [PubMed] [Google Scholar]
- 246.Zha, C. et al. Neutrophil extracellular traps mediate the crosstalk between glioma progression and the tumor microenvironment via the HMGB1/RAGE/IL-8 axis. Cancer Biol. Med.17, 154–168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Singhal, S. et al. Origin and role of a subset of tumor-associated neutrophils with antigen presenting cell features (Hybrid TANs) in early-stage human lung cancer. Cancer Cell30, 120–135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Gielen, P. R. et al. Elevated levels of polymorphonuclear myeloid-derived suppressor cells in patients with glioblastoma highly express S100A8/9 and arginase and suppress T cell function. Neuro Oncol.18, 1253–1264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Mi, Y. et al. The emerging role of myeloid-derived suppressor cells in the glioma immune suppressive microenvironment. Front. Immunol.11, 737 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Corzo, C. A. et al. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J. Exp. Med.207, 2439–2453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Rodriguez, P. C., Quiceno, D. G. & Ochoa, A. C. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood109, 1568–1573 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Mathewson, N. D. et al. Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell184, 1281–1298.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lohr, J. et al. Effector T-cell infiltration positively impacts survival of glioblastoma patients and is impaired by tumor-derived TGF-β. Clin. Cancer Res.17, 4296–4308 (2011). [DOI] [PubMed] [Google Scholar]
- 254.Kim, P. S. & Ahmed, R. Features of responding T cells in cancer and chronic infection. Curr. Opin. Immunol.22, 223–230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Thommen, D. S. & Schumacher, T. N. T cell dysfunction in cancer. Cancer Cell33, 547–562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Guedan, S., Ruella, M. & June, C. H. Emerging cellular therapies for cancer. Annu. Rev. Immunol.37, 145–171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Daubon, T., Hemadou, A., Romero Garmendia, I. & Saleh, M. Glioblastoma immune landscape and the potential of new immunotherapies. Front. Immunol.11, 585616 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Yi, J. S., Cox, M. A. & Zajac, A. J. T-cell exhaustion: characteristics, causes and conversion. Immunology129, 474–481 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Kim, Y.-J., Park, S.-J. & Broxmeyer, H. E. Phagocytosis, a potential mechanism for myeloid-derived suppressor cell regulation of CD8+ T cell function mediated through programmed cell death-1 and programmed cell death-1 ligand interaction. J. Immunol.187, 2291–2301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Markwell, S. M., Ross, J. L., Olson, C. L. & Brat, D. J. Necrotic reshaping of the glioma microenvironment drives disease progression. Acta Neuropathol.143, 291–310 (2022). [DOI] [PubMed] [Google Scholar]
- 261.Mills, C. D., Kincaid, K., Alt, J. M., Heilman, M. J. & Hill, A. M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol.164, 6166–6173 (2000). [DOI] [PubMed] [Google Scholar]
- 262.Butowski, N. et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro Oncol.18, 557–564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.von Roemeling, C. A. et al. Therapeutic modulation of phagocytosis in glioblastoma can activate both innate and adaptive antitumour immunity. Nat. Commun.11, 1508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sheppard, K., Kinross, K. M., Solomon, B., Pearson, R. B. & Phillips, W. A. Targeting PI3 kinase/AKT/mTOR signaling in cancer. Crit. Rev. Oncog.17, 69–95 (2012). [DOI] [PubMed] [Google Scholar]
- 265.Mendiburu-Eliçabe, M., Gil-Ranedo, J. & Izquierdo, M. Efficacy of rapamycin against glioblastoma cancer stem cells. Clin. Transl. Oncol.16, 495–502 (2014). [DOI] [PubMed] [Google Scholar]
- 266.Neshat, M. S. et al. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl Acad. Sci. USA98, 10314–10319 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Castedo, M., Ferri, K. F. & Kroemer, G. Mammalian target of rapamycin (mTOR): pro- and anti-apoptotic. Cell Death Differ.9, 99–100 (2002). [DOI] [PubMed] [Google Scholar]
- 268.Masri, J. et al. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res.67, 11712–11720 (2007). [DOI] [PubMed] [Google Scholar]
- 269.Fan, Q.-W. et al. EGFR signals to mTOR through PKC and independently of Akt in glioma. Sci. Signal2, ra4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Cahill, K. E., Morshed, R. A. & Yamini, B. Nuclear factor-κB in glioblastoma: insights into regulators and targeted therapy. Neuro Oncol.18, 329–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ohtsu, N. et al. Eva1 maintains the stem-like character of glioblastoma-initiating cells by activating the noncanonical NF-κB signaling pathway. Cancer Res.76, 171–181 (2016). [DOI] [PubMed] [Google Scholar]
- 272.Tran, N. L. et al. Increased fibroblast growth factor-inducible 14 expression levels promote glioma cell invasion via Rac1 and nuclear factor-kappaB and correlate with poor patient outcome. Cancer Res.66, 9535–9542 (2006). [DOI] [PubMed] [Google Scholar]
- 273.Kiriakidis, S. et al. VEGF expression in human macrophages is NF-kappaB dependent: studies using adenoviruses expressing the endogenous NF-kappaB inhibitor IkappaBalpha and a kinase-defective form of the IkappaB kinase 2. J. Cell Sci.116, 665–674 (2003). [DOI] [PubMed] [Google Scholar]
- 274.Yang, W. et al. EGFR-induced and PKCε monoubiquitylation-dependent NF-κB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell48, 771–784 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lavon, I. et al. Novel mechanism whereby nuclear factor kappaB mediates DNA damage repair through regulation of O(6)-methylguanine-DNA-methyltransferase. Cancer Res.67, 8952–8959 (2007). [DOI] [PubMed] [Google Scholar]
- 276.Ou, A., Ott, M., Fang, D. & Heimberger, A. B. The role and therapeutic targeting of JAK/STAT signaling in glioblastoma. Cancers13, 437 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Guryanova, O. A. et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell19, 498–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kang, S.-H. et al. Activated STAT3 regulates hypoxia-induced angiogenesis and cell migration in human glioblastoma. Neurosurgery67, 1386–1395 (2010). discussion 1395. [DOI] [PubMed] [Google Scholar]
- 279.Zheng, Q. et al. JAK2/STAT3 targeted therapy suppresses tumor invasion via disruption of the EGFRvIII/JAK2/STAT3 axis and associated focal adhesion in EGFRvIII-expressing glioblastoma. Neuro Oncol.16, 1229–1243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Piperi, C., Papavassiliou, K. A. & Papavassiliou, A. G. Pivotal role of STAT3 in shaping glioblastoma immune microenvironment. Cells8, 1398 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Kohsaka, S. et al. STAT3 inhibition overcomes temozolomide resistance in glioblastoma by downregulating MGMT expression. Mol. Cancer Ther.11, 1289–1299 (2012). [DOI] [PubMed] [Google Scholar]
- 282.Gong, A.-H. et al. FoxM1 drives a feed-forward STAT3-activation signaling loop that promotes the self-renewal and tumorigenicity of glioblastoma stem-like cells. Cancer Res75, 2337–2348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.de la Iglesia, N. et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev.22, 449–462 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.McCord, M., Mukouyama, Y., Gilbert, M. R. & Jackson, S. Targeting WNT signaling for multifaceted glioblastoma therapy. Front. Cell. Neurosci. 11, (2017). [DOI] [PMC free article] [PubMed]
- 285.Götze, S., Wolter, M., Reifenberger, G., Müller, O. & Sievers, S. Frequent promoter hypermethylation of Wnt pathway inhibitor genes in malignant astrocytic gliomas. Int. J. Cancer126, 2584–2593 (2010). [DOI] [PubMed] [Google Scholar]
- 286.Pu, P. et al. Downregulation of Wnt2 and beta-catenin by siRNA suppresses malignant glioma cell growth. Cancer Gene Ther.16, 351–361 (2009). [DOI] [PubMed] [Google Scholar]
- 287.Maris, C. et al. IGF-IR: a new prognostic biomarker for human glioblastoma. Br. J. Cancer113, 729–737 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Yin, S. et al. Targeting the insulin-like growth factor-1 receptor by picropodophyllin as a treatment option for glioblastoma. Neuro Oncol.12, 19–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Zamykal, M. et al. Inhibition of intracerebral glioblastoma growth by targeting the insulin-like growth factor 1 receptor involves different context-dependent mechanisms. Neuro Oncol.17, 1076–1085 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Xing, Z., Sun, L. & Guo, W. Elevated expression of Notch-1 and EGFR induced apoptosis in glioblastoma multiforme patients. Clin. Neurol. Neurosurg.131, 54–58 (2015). [DOI] [PubMed] [Google Scholar]
- 291.Tchorz, J. S. et al. Constitutive Notch2 signaling in neural stem cells promotes tumorigenic features and astroglial lineage entry. Cell Death Dis.3, e325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Dell’albani, P. et al. Differential patterns of NOTCH1-4 receptor expression are markers of glioma cell differentiation. Neuro Oncol.16, 204–216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Qiang, L. et al. HIF-1α is critical for hypoxia-mediated maintenance of glioblastoma stem cells by activating Notch signaling pathway. Cell Death Differ.19, 284–294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Cheng, W. et al. Bioinformatic analyses reveal a distinct Notch activation induced by STAT3 phosphorylation in the mesenchymal subtype of glioblastoma. J. Neurosurg.126, 249–259 (2017). [DOI] [PubMed] [Google Scholar]
- 295.Jeon, H.-M. et al. Crosstalk between glioma-initiating cells and endothelial cells drives tumor progression. Cancer Res.74, 4482–4492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Hai, L. et al. Notch1 is a prognostic factor that is distinctly activated in the classical and proneural subtype of glioblastoma and that promotes glioma cell survival via the NF-κB(p65) pathway. Cell Death Dis.9, 158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Doheny, D., Manore, S. G., Wong, G. L. & Lo, H.-W. Hedgehog signaling and truncated GLI1 in cancer. Cells9, 2114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Zhu, H., Carpenter, R. L., Han, W. & Lo, H.-W. The GLI1 splice variant TGLI1 promotes glioblastoma angiogenesis and growth. Cancer Lett.343, 51–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Rimkus, T. K. et al. Truncated glioma-associated oncogene homolog 1 (tGLI1) mediates mesenchymal glioblastoma via transcriptional activation of CD44. Cancer Res78, 2589–2600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Wang, K. et al. Hedgehog/Gli1 signaling pathway regulates MGMT expression and chemoresistance to temozolomide in human glioblastoma. Cancer Cell Int.17, 117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Morad, S. A. F. & Cabot, M. C. Ceramide-orchestrated signaling in cancer cells. Nat. Rev. Cancer13, 51–65 (2013). [DOI] [PubMed] [Google Scholar]
- 302.Nguyen, H. S., Awad, A. J., Shabani, S. & Doan, N. Molecular targeting of acid ceramidase in glioblastoma: a review of its role, potential treatment, and challenges. Pharmaceutics10, 45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Bhat, K. P. L. et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev.25, 2594–2609 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Vigneswaran, K. et al. YAP/TAZ transcriptional coactivators create therapeutic vulnerability to verteporfin in EGFR-mutant glioblastoma. Clin. Cancer Res.27, 1553–1569 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Selagea, L. et al. EGFR and C/EBP-β oncogenic signaling is bidirectional in human glioma and varies with the C/EBP-β isoform. FASEB J.30, 4098–4108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Kudo, T. et al. Constitutive expression of the immunosuppressive tryptophan dioxygenase TDO2 in glioblastoma is driven by the transcription factor C/EBPβ. Front. Immunol.11, 657 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Lei, K. et al. C/EBPβ mediates NQO1 and GSTP1 anti-oxidative reductases expression in glioblastoma, promoting brain tumor proliferation. Redox Biol.34, 101578 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Kozono, D. et al. Dynamic epigenetic regulation of glioblastoma tumorigenicity through LSD1 modulation of MYC expression. Proc. Natl. Acad. Sci. USA112, E4055–E4064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Chen, X. et al. MiR-9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. J. Exp. Clin. Cancer Res.38, 99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Tateishi, K. et al. Myc-driven glycolysis is a therapeutic target in glioblastoma. Clin. Cancer Res.22, 4452–4465 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Geribaldi-Doldán, N. et al. Targeting protein kinase c in glioblastoma treatment. Biomedicines9, 381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Dey, A., Islam, S. M. A., Patel, R. & Acevedo-Duncan, M. The interruption of atypical PKC signaling and Temozolomide combination therapy against glioblastoma. Cell. Signal.77, 109819 (2021). [DOI] [PubMed] [Google Scholar]
- 313.Kenchappa, R. S. et al. Protein kinase Cι and SRC signaling define reciprocally related subgroups of glioblastoma with distinct therapeutic vulnerabilities. Cell Rep.37, 110054 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Pyonteck, S. M. et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med.19, 1264–1272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Cannarile, M. A. et al. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother. Cancer5, 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Quail, D. F. & Joyce, J. A. The microenvironmental landscape of brain tumors. Cancer Cell31, 326–341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Chemnitz, J. M., Parry, R. V., Nichols, K. E., June, C. H. & Riley, J. L. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J. Immunol.173, 945–954 (2004). [DOI] [PubMed] [Google Scholar]
- 318.Majc, B., Novak, M., Kopitar-Jerala, N., Jewett, A. & Breznik, B. Immunotherapy of glioblastoma: current strategies and challenges in tumor model development. Cells10, 265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.David, C. J. & Massagué, J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol.19, 419–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Hata, A. & Chen, Y.-G. TGF-β signaling from receptors to smads. Cold Spring Harb. Perspect. Biol.8, a022061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Mariathasan, S. et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature554, 544–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Caja, L. et al. TGF-β and the tissue microenvironment: relevance in fibrosis and cancer. Int. J. Mol. Sci.19, 1294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Derynck, R., Turley, S. J. & Akhurst, R. J. TGFβ biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol.18, 9–34 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Antonioli, L., Pacher, P., Vizi, E. S. & Haskó, G. CD39 and CD73 in immunity and inflammation. Trends Mol. Med.19, 355–367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Linnemann, C. et al. Adenosine regulates CD8 T-cell priming by inhibition of membrane-proximal T-cell receptor signaling. Immunology128, e728–e737 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Young, A. et al. Co-inhibition of CD73 and A2AR adenosine signaling improves anti-tumor immune responses. Cancer Cell30, 391–403 (2016). [DOI] [PubMed] [Google Scholar]
- 327.Munn, D. H. & Mellor, A. L. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol.34, 137–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Wainwright, D. A. et al. IDO expression in brain tumors increases the recruitment of regulatory T cells and negatively impacts survival. Clin. Cancer Res.18, 6110–6121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Platten, M., Nollen, E. A. A., Röhrig, U. F., Fallarino, F. & Opitz, C. A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov.18, 379–401 (2019). [DOI] [PubMed] [Google Scholar]
- 330.Opitz, C. A. et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature478, 197–203 (2011). [DOI] [PubMed] [Google Scholar]
- 331.Ladomersky, E. et al. IDO1 Inhibition synergizes with radiation and PD-1 blockade to durably increase survival against advanced glioblastoma. Clin. Cancer Res.24, 2559–2573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Sanai, N. & Berger, M. S. Glioma extent of resection and its impact on patient outcome. Neurosurgery62, 753–764 (2008). discussion 264-266. [DOI] [PubMed] [Google Scholar]
- 333.Stepp, H. & Stummer, W. 5-ALA in the management of malignant glioma. Lasers Surg. Med.50, 399–419 (2018). [DOI] [PubMed] [Google Scholar]
- 334.Gandhi, S. et al. Survival outcomes among patients with high-grade glioma treated with 5-aminolevulinic acid-guided surgery: a systematic review and meta-analysis. Front. Oncol.9, 620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Haider, S. A., Lim, S., Kalkanis, S. N. & Lee, I. Y. The impact of 5-aminolevulinic acid on extent of resection in newly diagnosed high grade gliomas: a systematic review and single institutional experience. J. Neurooncol.141, 507–515 (2019). [DOI] [PubMed] [Google Scholar]
- 336.Brown, P. D. et al. Postoperative stereotactic radiosurgery compared with whole brain radiotherapy for resected metastatic brain disease (NCCTG N107C/CEC·3): a multicenter, randomized, controlled, phase 3 trial. Lancet Oncol.18, 1049–1060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Marchan, E. M. et al. Postoperative cavity stereotactic radiosurgery for brain metastases. Front. Oncol.8, 342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Morris, S.-A. L. et al. Gamma knife stereotactic radiosurgery in combination with bevacizumab for recurrent glioblastoma. World Neurosurg.127, e523–e533 (2019). [DOI] [PubMed] [Google Scholar]
- 339.Gessler, D. J. et al. GammaTile® brachytherapy in the treatment of recurrent glioblastomas. Neurooncol. Adv.4, vdab185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Morello, A. et al. Laser Interstitial Thermotherapy (LITT) in recurrent glioblastoma: what window of opportunity for this treatment?. Technol. Cancer Res. Treat.23, 15330338241249026 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Kamath, A. A. et al. Glioblastoma treated with magnetic resonance imaging-guided laser interstitial thermal therapy: safety, efficacy, and outcomes. Neurosurgery84, 836–843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Mohammadi, A. M. et al. The role of laser interstitial thermal therapy in enhancing progression-free survival of difficult-to-access high-grade gliomas: a multicenter study. Cancer Med.3, 971–979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Vora, S. et al. Short-course hypofractionated proton beam therapy, incorporating 18F-DOPA PET and contrast-enhanced MRI targeting, for patients aged 65 years and older with newly diagnosed glioblastoma: a single-arm phase 2 trial. Lancet Oncol.25, 1625–1634 (2024). [DOI] [PubMed] [Google Scholar]
- 344.Meng, Y., Hynynen, K. & Lipsman, N. Applications of focused ultrasound in the brain: from thermoablation to drug delivery. Nat. Rev. Neurol.17, 7–22 (2021). [DOI] [PubMed] [Google Scholar]
- 345.Mungur, R. et al. Low-intensity focused ultrasound technique in glioblastoma multiforme treatment. Front. Oncol.12, 903059 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Coluccia, D. et al. Enhancing glioblastoma treatment using cisplatin-gold-nanoparticle conjugates and targeted delivery with magnetic resonance-guided focused ultrasound. Nanomedicine14, 1137–1148 (2018). [DOI] [PubMed] [Google Scholar]
- 347.McDannold, N. et al. Acoustic feedback enables safe and reliable carboplatin delivery across the blood‒brain barrier with a clinical focused ultrasound system and improves survival in a rat glioma model. Theranostics9, 6284–6299 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Arvanitis, C. D. et al. Mechanisms of enhanced drug delivery in brain metastases with focused ultrasound-induced blood–tumor barrier disruption. Proc. Natl. Acad. Sci. USA115, E8717–E8726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Wei, H.-J. et al. Focused ultrasound-mediated blood‒brain barrier opening increases delivery and efficacy of etoposide for glioblastoma treatment. Int. J. Radiat. Oncol. Biol. Phys.110, 539–550 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Li, Y. et al. Neutrophil delivered hollow titania covered persistent luminescent nanosensitizer for ultrosound augmented chemo/immuno glioblastoma therapy. Adv. Sci.8, 2004381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Wu, M. et al. Focused ultrasound-augmented delivery of biodegradable multifunctional nanoplatforms for imaging-guided brain tumor treatment. Adv. Sci.5, 1700474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Li, Y. et al. Mechanisms of enhanced antiglioma efficacy of polysorbate 80-modified paclitaxel-loaded PLGA nanoparticles by focused ultrasound. J. Cell Mol. Med.22, 4171–4182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Regev, O., Merkin, V., Blumenthal, D. T., Melamed, I. & Kaisman-Elbaz, T. Tumor-treating fields for the treatment of glioblastoma: a systematic review and meta-analysis. Neurooncol. Pract.8, 426–440 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Stupp, R. et al. NovoTTF-100A versus physician’s choice chemotherapy in recurrent glioblastoma: a randomized phase III trial of a novel treatment modality. Eur. J. Cancer48, 2192–2202 (2012). [DOI] [PubMed] [Google Scholar]
- 355.Ma, J. et al. Directionally non-rotating electric field therapy delivered through implanted electrodes as a glioblastoma treatment platform: a proof-of-principle study. Neurooncol. Adv.6, vdae121 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Jezierzański, M. et al. Temozolomide (TMZ) in the treatment of glioblastoma multiforme—a literature review and clinical outcomes. Curr. Oncol.31, 3994–4002 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Singh, N., Miner, A., Hennis, L. & Mittal, S. Mechanisms of temozolomide resistance in glioblastoma—a comprehensive review. Cancer Drug Resist4, 17–43 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Lal, B. et al. Preclinical efficacy of LP-184, a tumor site activated synthetic lethal therapeutic, in glioblastoma. Clin. Cancer Res.29, 4209–4218 (2023). [DOI] [PubMed] [Google Scholar]
- 359.Kim, O. et al. Combined inhibition of topoisomerase I and poly(ADP-ribose) polymerase: a synergistic therapeutic strategy for glioblastoma with phosphatase and tensin homolog deficiency. Neurooncol. Adv.5, vdad102 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Joseph-Thomas, J. et al. A prospective study comparing AI-based clinical trial eligibility screening with traditional EMR-based screening. J. Clin. Oncol.42, e13636–e13636 (2024). [Google Scholar]
- 361.Sepulveda, J. M. et al. Azeliragon, a RAGE inhibitor, in combination with temozolomide and radiotherapy in patients with newly diagnosed glioblastoma: phase Ib/II CAN-201 NDG trial design. J. Clin. Oncol.42, TPS2096–TPS2096 (2024). [Google Scholar]
- 362.Chi, A. S. et al. Exploring predictors of response to dacomitinib in EGFR-amplified recurrent glioblastoma. JCO Precis. Oncol.4, PO.19.00295 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Han, D. et al. Napabucasin, a novel STAT3 inhibitor suppresses proliferation, invasion and stemness of glioblastoma cells. J. Exp. Clin. Cancer Res.38, 289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Chen, Y. et al. Larotrectinib versus infigratinib for adult patients with both glioma and tyrosine kinase alterations after failure of initial therapies: Efficacy and safety analysis. Clinics79, 100329 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Neitzel, L. R., Fuller, D. T., Williams, C. H. & Hong, C. C. Inhibition of GPR68 kills glioblastoma in zebrafish xenograft models. BMC Res. Notes17, 235 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Williams, C. H. et al. GPR68-ATF4 signaling is a novel prosurvival pathway in glioblastoma activated by acidic extracellular microenvironment. Exp. Hematol. Oncol.13, 13 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Jang, H. J. et al. Epigenetic therapy potentiates transposable element transcription to create tumor-enriched antigens in glioblastoma cells. Nat. Genet56, 1903–1913 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Mellinghoff, I. K. et al. Vorasidenib in IDH1- or IDH2-mutant low-grade glioma. N. Engl. J. Med.389, 589–601 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Jones, A. B. et al. Temozolomide and the PARP inhibitor Niraparib enhance expression of natural killer group 2D ligand ULBP1 and gamma-delta T cell cytotoxicity in glioblastoma. Cancers16, 2852 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Caccese, M., Indraccolo, S., Zagonel, V. & Lombardi, G. PD-1/PD-L1 immune-checkpoint inhibitors in glioblastoma: a concise review. Crit. Rev. Oncol. Hematol.135, 128–134 (2019). [DOI] [PubMed] [Google Scholar]
- 371.Cloughesy, T. F. et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat. Med.25, 477–486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Carter, T., Shaw, H., Cohn-Brown, D., Chester, K. & Mulholland, P. Ipilimumab and Bevacizumab in Glioblastoma. Clin. Oncol.28, 622–626 (2016). [DOI] [PubMed] [Google Scholar]
- 373.Xu, S., Tang, L., Li, X., Fan, F. & Liu, Z. Immunotherapy for glioma: current management and future application. Cancer Lett.476, 1–12 (2020). [DOI] [PubMed] [Google Scholar]
- 374.Li, M. et al. The indoleamine 2,3-dioxygenase pathway controls complement-dependent enhancement of chemo-radiation therapy against murine glioblastoma. J. Immunother. Cancer2, 21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Grégoire, H. et al. Targeting tumor associated macrophages to overcome conventional treatment resistance in glioblastoma. Front. Pharmacol.11, 368 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Singh, S. et al. Small molecule targeting immune cells: a novel approach for cancer treatment. Biomedicines11, 2621 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Burger, M. C. et al. CAR-engineered NK cells for the treatment of glioblastoma: turning innate effectors into precision tools for cancer immunotherapy. Front. Immunol.10, 2683 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Morimoto, T. et al. Natural killer cell-based immunotherapy against glioblastoma. Int. J. Mol. Sci.24, 2111 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Bagley, S. J. et al. Intrathecal bivalent CAR T cells targeting EGFR and IL13Rα2 in recurrent glioblastoma: phase 1 trial interim results. Nat. Med.30, 1320–1329 (2024). [DOI] [PubMed] [Google Scholar]
- 380.Chokshi, C. R. et al. Targeting axonal guidance dependencies in glioblastoma with ROBO1 CAR T cells. Nat. Med.30, 2936–2946 (2024). [DOI] [PubMed] [Google Scholar]
- 381.Dewdney, B. et al. From signaling pathways to targeted therapies: unraveling glioblastoma’s secrets and harnessing two decades of progress. Signal Transduct. Target Ther.8, 400 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Lane, R. et al. PDGF-R inhibition induces glioblastoma cell differentiation via DUSP1/p38MAPK signaling. Oncogene41, 2749–2763 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Sepúlveda-Sánchez, J. M. et al. Phase II trial of dacomitinib, a pan-human EGFR tyrosine kinase inhibitor, in recurrent glioblastoma patients with EGFR amplification. Neuro Oncol.19, 1522–1531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Zahonero, C. et al. Preclinical test of dacomitinib, an irreversible EGFR inhibitor, confirms its effectiveness for glioblastoma. Mol. Cancer Ther.14, 1548–1558 (2015). [DOI] [PubMed] [Google Scholar]
- 385.Wen, P. Y. et al. Buparlisib in patients with recurrent glioblastoma harboring phosphatidylinositol 3-kinase pathway activation: an open-label, multicenter, multi-arm, phase II trial. J. Clin. Oncol.37, 741–750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Kahn, J. et al. The mTORC1/mTORC2 inhibitor AZD2014 enhances the radiosensitivity of glioblastoma stem-like cells. Neuro Oncol.16, 29–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Peng, X. et al. Stellettin B renders glioblastoma vulnerable to poly (ADP-ribose) polymerase inhibitors via suppressing homology-directed repair. Signal Transduct. Target. Ther.8, 1–4 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Peng, X. et al. Sequential inhibition of PARP and BET as a rational therapeutic strategy for glioblastoma. Adv. Sci.11, e2307747 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Parasramka, S., Talari, G., Rosenfeld, M., Guo, J. & Villano, J. L. Procarbazine, lomustine and vincristine for recurrent high-grade glioma. Cochrane Database Syst. Rev.7, CD011773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Palmer, J. D. et al. Bevacizumab and re-irradiation for recurrent high grade gliomas: does sequence matter? J. Neurooncol.140, 623–628 (2018). [DOI] [PubMed] [Google Scholar]
- 391.Azoulay, M. et al. Benefit of re-operation and salvage therapies for recurrent glioblastoma multiforme: results from a single institution. J. Neurooncol.132, 419–426 (2017). [DOI] [PubMed] [Google Scholar]
- 392.Sharabi, A. B., Lim, M., DeWeese, T. L. & Drake, C. G. Radiation and checkpoint blockade immunotherapy: radiosensitization and potential mechanisms of synergy. Lancet Oncol.16, e498–e509 (2015). [DOI] [PubMed] [Google Scholar]
- 393.Demaria, S., Coleman, C. N. & Formenti, S. C. Radiotherapy: changing the Game in Immunotherapy. Trends Cancer2, 286–294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Keskin, D. B. et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature565, 234–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Xiong, Z. et al. Glioblastoma vaccines: past, present, and opportunities. eBioMedicine100, (2024). [DOI] [PMC free article] [PubMed]
- 396.Yang, T. et al. Peptide vaccine against glioblastoma: from bench to bedside. Holist. Integ. Oncol.1, 21 (2022). [Google Scholar]
- 397.Santos, P. M. & Butterfield, L. H. Dendritic cell-based cancer vaccines. J. Immunol.200, 443–449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Wang, H. et al. rWTC-MBTA vaccine induces potent adaptive immune responses against glioblastomas via dynamic activation of dendritic cells. Adv. Sci.11, e2308280 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Liu, C. et al. mRNA-based cancer therapeutics. Nat. Rev. Cancer23, 526–543 (2023). [DOI] [PubMed] [Google Scholar]
- 400.Ott, P. A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Xie, Z., Chen, M., Lian, J., Wang, H. & Ma, J. Glioblastoma-on-a-chip construction and therapeutic applications. Front. Oncol. 13, (2023). [DOI] [PMC free article] [PubMed]
- 402.Lee, J.-K. et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat. Genet.49, 594–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Chakraborty, S., Schneider, J. & Boockvar, J. A. Transdifferentiation induced neural stem cells for the treatment of malignant gliomas. Neurosurgery79, N17–N18 (2016). [DOI] [PubMed] [Google Scholar]
- 404.Bagó, J. R. et al. Therapeutically engineered induced neural stem cells are tumour-homing and inhibit progression of glioblastoma. Nat. Commun.7, 10593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Magge, S. N. et al. Role of monocyte chemoattractant protein-1 (MCP-1/CCL2) in migration of neural progenitor cells toward glial tumors. J. Neurosci. Res.87, 1547–1555 (2009). [DOI] [PubMed] [Google Scholar]
- 406.Benmelouka, A. Y. et al. Neural stem cell-based therapies and glioblastoma management: current evidence and clinical challenges. Int. J. Mol. Sci.22, 2258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Satterlee, A. B., Dunn, D. E., Lo, D. C., Khagi, S. & Hingtgen, S. Tumoricidal stem cell therapy enables killing in novel hybrid models of heterogeneous glioblastoma. Neuro Oncol.21, 1552–1564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Gunnarsson, S. et al. Intratumoral IL-7 delivery by mesenchymal stromal cells potentiates IFN gamma-transduced tumor cell immunotherapy of experimental glioma. J. Neuroimmunol.218, 140–144 (2010). [DOI] [PubMed] [Google Scholar]
- 409.Portnow, J. et al. Feasibility of intracerebrally administering multiple doses of genetically modified neural stem cells to locally produce chemotherapy in glioma patients. Cancer Gene Ther.28, 294–306 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Fares, J. et al. Neural stem cell delivery of an oncolytic adenovirus in newly diagnosedmalignant glioma: a first-in-human, phase 1 clinical trial. Lancet Oncol.22, 1103–1114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Metz, M. Z. et al. Neural stem cell-mediated delivery of irinotecan-activating carboxylesterases to glioma: implications for clinical use. Stem Cells Transl. Med.2, 983–992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Gujar, S., Bell, J. & Diallo, J.-S. SnapShot: cancer immunotherapy with oncolytic viruses. Cell176, 1240–1240.e1 (2019). [DOI] [PubMed] [Google Scholar]
- 413.Frederico, S. C. et al. Making a cold tumor hot: the role of vaccines in the treatment of glioblastoma. Front. Oncol.11, 672508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Todo, T. et al. Intratumoral oncolytic herpes virus G47∆ for residual or recurrent glioblastoma: a phase 2 trial. Nat. Med.28, 1630–1639 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Markert, J. M. et al. A phase 1 trial of oncolytic HSV-1, G207, given in combination with radiation for recurrent GBM demonstrates safety and radiographic responses. Mol. Ther.22, 1048–1055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Chiocca, E. A. et al. A phase I open-label, dose-escalation, multi-institutional trial of injection with an E1B-Attenuated adenovirus, ONYX-015, into the peritumoral region of recurrent malignant gliomas, in the adjuvant setting. Mol. Ther.10, 958–966 (2004). [DOI] [PubMed] [Google Scholar]
- 417.Kicielinski, K. P. et al. Phase 1 clinical trial of intratumoral reovirus infusion for the treatment of recurrent malignant gliomas in adults. Mol. Ther.22, 1056–1062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Allen, C. et al. Oncolytic measles virus strains have significant antitumor activity against glioma stem cells. Gene Ther.20, 444–449 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Freeman, A. I. et al. Phase I/II trial of intravenous NDV-HUJ oncolytic virus in recurrent glioblastoma multiforme. Mol. Ther.13, 221–228 (2006). [DOI] [PubMed] [Google Scholar]
- 420.Desjardins, A. et al. Recurrent glioblastoma treated with recombinant poliovirus. N. Engl. J. Med.379, 150–161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Hamad, A. & Chumakov, S. P. Engineering a recombinant Herpesvirus saimiri strain by co-culturing transfected and permissive cells. Bull. RSMU 37–44 10.24075/brsmu.2019.079 (2019).
- 422.Azambuja, J. H. et al. CD73 Downregulation decreases in vitro and in vivo glioblastoma growth. Mol. Neurobiol.56, 3260–3279 (2019). [DOI] [PubMed] [Google Scholar]
- 423.Goswami, S. et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat. Med.26, 39–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Sharma, P. & Allison, J. P. The future of immune checkpoint therapy. Science348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 425.Pérez-Ruiz, E., Etxeberria, I., Rodriguez-Ruiz, M. E. & Melero, I. Anti-CD137 and PD-1/PD-L1 antibodies en route toward clinical synergy. Clin. Cancer Res.23, 5326–5328 (2017). [DOI] [PubMed] [Google Scholar]
- 426.Segal, N. H. et al. Results from an integrated safety analysis of urelumab, an agonist anti-CD137 monoclonal antibody. Clin. Cancer Res.23, 1929–1936 (2017). [DOI] [PubMed] [Google Scholar]
- 427.Ribas, A. et al. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell170, 1109–1119.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Shi, T., Song, X., Wang, Y., Liu, F. & Wei, J. Combining oncolytic viruses with cancer immunotherapy: establishing a new generation of cancer treatment. Front. Immunol.11, 683 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Hardcastle, J. et al. Immunovirotherapy with measles virus strains in combination with anti-PD-1 antibody blockade enhances antitumor activity in glioblastoma treatment. Neuro Oncol.19, 493–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Samson, A. et al. Intravenous delivery of oncolytic reovirus to brain tumor patients immunologically primes for subsequent checkpoint blockade. Sci. Transl. Med.10, eaam7577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Saha, D., Martuza, R. L. & Rabkin, S. D. Oncolytic herpes simplex virus immunovirotherapy in combination with immune checkpoint blockade to treat glioblastoma. Immunotherapy10, 779–786 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Migliorini, D. et al. CAR T-cell therapies in glioblastoma: a first look. Clin. Cancer Res.24, 535–540 (2018). [DOI] [PubMed] [Google Scholar]
- 433.Guedan, S. & Alemany, R. CAR-T cells and oncolytic viruses: joining forces to overcome the solid tumor challenge. Front. Immunol.9, 2460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Mitchell, L. A. et al. Toca 511 gene transfer and treatment with the prodrug, 5-fluorocytosine, promotes durable antitumor immunity in a mouse glioma model. Neuro Oncol.19, 930–939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Maggs, L., Cattaneo, G., Dal, A. E., Moghaddam, A. S. & Ferrone, S. C. A. R. T. Cell-based immunotherapy for the treatment of glioblastoma. Front. Neurosci.15, 662064 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Wang, S. E. Extracellular vesicles in cancer therapy. Semin. Cancer Biol.86, 296–309 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Gu, S. et al. Berberine inhibits cancer cells growth by suppressing fatty acid synthesis and biogenesis of extracellular vesicles. Life Sci.257, 118122 (2020). [DOI] [PubMed] [Google Scholar]
- 438.Atai, N. A. et al. Heparin blocks transfer of extracellular vesicles between donor and recipient cells. J. Neurooncol.115, 343–351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Joo, H. S., Suh, J. H., Lee, H. J., Bang, E. S. & Lee, J. M. Current knowledge and future perspectives on mesenchymal stem cell-derived exosomes as a new therapeutic agent. Int. J. Mol. Sci.21, 727 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Guo, H., Wu, L., Yang, Q., Ye, M. & Zhu, X. Functional linc-POU3F3 is overexpressed and contributes to tumorigenesis in glioma. Gene554, 114–119 (2015). [DOI] [PubMed] [Google Scholar]
- 441.Mukherjee, S. & Pillai, P. P. Current insights on extracellular vesicle-mediated glioblastoma progression: Implications in drug resistance and epithelial–mesenchymal transition. Biochim. Biophys. Acta Gen. Subj.1866, 130065 (2022). [DOI] [PubMed] [Google Scholar]
- 442.Liguori, G. L. & Kralj-Iglič, V. Pathological and therapeutic significance of tumor-derived extracellular vesicles in cancer cell migration and metastasis. Cancers15, 4425 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Macedo-Pereira, A., Martins, C., Lima, J. & Sarmento, B. Digging the intercellular crosstalk via extracellular vesicles: may exosomes be the drug delivery solution for target glioblastoma? J. Control Release358, 98–115 (2023). [DOI] [PubMed] [Google Scholar]
- 444.Galardi, A. et al. Recent advancements on the use of exosomes as drug carriers for the treatment of glioblastoma. Life13, 964 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Dalmizrak, A. & Dalmizrak, O. Mesenchymal stem cell-derived exosomes as new tools for delivery of miRNAs in the treatment of cancer. Front. Bioeng. Biotechnol.10, 956563 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Katakowski, M. et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett.335, 201–204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Lang, F. M. et al. Mesenchymal stem cells as natural biofactories for exosomes carrying miR-124a in the treatment of gliomas. Neuro Oncol.20, 380–390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Fareh, M. et al. Cell-based therapy using miR-302-367 expressing cells represses glioblastoma growth. Cell Death Dis.8, e2713–e2713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Monfared, H., Jahangard, Y., Nikkhah, M., Mirnajafi-Zadeh, J. & Mowla, S. J. Potential therapeutic effects of exosomes packed with a mir-21-sponge construct in a rat model of glioblastoma. Front. Oncol.9, 782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Munoz, J. L. et al. Delivery of functional anti-miR-9 by mesenchymal stem cell-derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol. Ther. Nucleic Acids2, e126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Lee, H. et al. Glioblastoma-derived exosomes as nanopharmaceutics for improved glioma treatment. Pharmaceutics14, 1002 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Ye, Z. et al. Methotrexate-loaded extracellular vesicles functionalized with therapeutic and targeted peptides for the treatment of glioblastoma multiforme. ACS Appl. Mater. Interfaces10, 12341–12350 (2018). [DOI] [PubMed] [Google Scholar]
- 453.Tibensky, M. et al. Gene-directed enzyme/prodrug therapy of rat brain tumor mediated by human mesenchymal stem cell suicide gene extracellular vesicles in vitro and in vivo. Cancers14, 735 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Wang, J. et al. Inflammatory tumor microenvironment responsive neutrophil exosomes-based drug delivery system for targeted glioma therapy. Biomater273, 120784 (2021). [DOI] [PubMed] [Google Scholar]
- 455.Jia, G. et al. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomater178, 302–316 (2018). [DOI] [PubMed] [Google Scholar]
- 456.Z, Z. et al. MicroRNA-29a-3p delivery via exosomes derived from engineered human mesenchymal stem cells exerts tumour suppressive effects by inhibiting migration and vasculogenic mimicry in glioma. Aging13, 5055–5068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Bu, N. et al. Exosomes from dendritic cells loaded with chaperone-rich cell lysates elicit a potent T cell immune response against intracranial glioma in mice. J. Mol. Neurosci.56, 631–643 (2015). [DOI] [PubMed] [Google Scholar]
- 458.Gečys, D. et al. Internalization of RGD-engineered extracellular vesicles by glioblastoma cells. Biology11, 1483 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Parker Kerrigan, B. C. et al. RNAi technology targeting the FGFR3-TACC3 fusion breakpoint: an opportunity for precision medicine. Neurooncol. Adv.2, vdaa132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Liu, X. et al. Engineered extracellular vesicle-delivered CRISPR/Cas9 for radiotherapy sensitization of glioblastoma. ACS Nano17, 16432–16447 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Bagherian, A. et al. Combination therapy with nanomicellar-curcumin and temozolomide for in vitro therapy of glioblastoma multiforme via Wnt signaling pathways. J. Mol. Neurosci.70, 1471–1483 (2020). [DOI] [PubMed] [Google Scholar]
- 462.Behrooz, A. B. et al. Tailoring drug co-delivery nanosystem for mitigating U-87 stem cells drug resistance. Drug Deliv. Transl. Res.12, 1253–1269 (2022). [DOI] [PubMed] [Google Scholar]
- 463.Ferreira, N. N. et al. Nose-to-brain co-delivery of drugs for glioblastoma treatment using nanostructured system. Int. J. Pharm.603, 120714 (2021). [DOI] [PubMed] [Google Scholar]
- 464.Lakkadwala, S. & Singh, J. Co-delivery of doxorubicin and erlotinib through liposomal nanoparticles for glioblastoma tumor regression using an in vitro brain tumor model. Colloids Surf. B Biointerfaces173, 27–35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Lakkadwala, S., dos Santos Rodrigues, B., Sun, C. & Singh, J. Dual functionalized liposomes for efficient co-delivery of anti-cancer chemotherapeutics for the treatment of glioblastoma. J. Control Release307, 247–260 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Madani, F. et al. Paclitaxel/methotrexate coloaded PLGA nanoparticles in glioblastoma treatment: Formulation development and in vitro antitumor activity evaluation. Life Sci.256, 117943 (2020). [DOI] [PubMed] [Google Scholar]
- 467.Tamborini, M. et al. A combined approach employing chlorotoxin-nanovectors and low dose radiation to reach infiltrating tumor niches in glioblastoma. ACS Nano10, 2509–2520 (2016). [DOI] [PubMed] [Google Scholar]
- 468.Yang, J., Shi, Z., Liu, R., Wu, Y. & Zhang, X. Combined-therapeutic strategies synergistically potentiate glioblastoma multiforme treatment via nanotechnology. Theranostics10, 3223–3239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.He, C. et al. LRP1-mediated pH-sensitive polymersomes facilitate combination therapy of glioblastoma in vitro and in vivo. J. Nanobiotechnol.19, 29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Qiao, R. et al. Magnetic iron oxide nanoparticles for brain imaging and drug delivery. Adv. Drug Deliv. Rev.197, 114822 (2023). [DOI] [PubMed] [Google Scholar]
- 471.Chibh, S., Katoch, V., Singh, M., Prakash, B. & Panda, J. J. Miniatured fluidics-mediated modular self-assembly of anticancer drug-amino acid composite microbowls for combined chemo-photodynamic therapy in glioma. ACS Biomater. Sci. Eng.7, 5654–5665 (2021). [DOI] [PubMed] [Google Scholar]
- 472.Maturi, M. et al. Synthesis of ultrasmall single-crystal gold–silver alloy nanotriangles and their application in photothermal therapy. Nanomater11, 912 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Yu, Y. et al. Efficacy of temozolomide-conjugated gold nanoparticle photothermal therapy of drug-resistant glioblastoma and its mechanism study. Mol. Pharm.19, 1219–1229 (2022). [DOI] [PubMed] [Google Scholar]
- 474.Zhang, C. et al. Hypocrellin-based multifunctional phototheranostic agent for nir-triggered targeted chemo/photodynamic/photothermal synergistic therapy against glioblastoma. ACS Appl. Bio Mater.3, 3817–3826 (2020). [DOI] [PubMed] [Google Scholar]
- 475.Schupper, A. J. et al. Fluorescence-guided surgery: a review on timing and use in brain tumor surgery. Front. Neurol.12, 682151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Wang, J. et al. Brain-targeted aggregation-induced-emission nanoparticles with near-infrared imaging at 1550 nm boosts orthotopic glioblastoma theranostics. Adv. Mater.34, e2106082 (2022). [DOI] [PubMed] [Google Scholar]
- 477.Zhang, M. et al. Upregulating aggregation-induced-emission nanoparticles with blood-tumor-barrier permeability for precise photothermal eradication of brain tumors and induction of local immune responses. Adv. Mater.33, e2008802 (2021). [DOI] [PubMed] [Google Scholar]
- 478.Chien, W. C. et al. MCP-1-Functionalized, core-shell gold nanorod@iron-based metal-organic framework (MCP-1/GNR@MIL-100(Fe)) for photothermal therapy. ACS Appl. Mater. Interfaces13, 52092–52105 (2021). [DOI] [PubMed] [Google Scholar]
- 479.Kuang, J. et al. iRGD Modified Chemo-immunotherapeutic Nanoparticles For Enhanced Immunotherapy Against Glioblastoma. Adv. Funct. Mater.28, 1800025 (2018). [Google Scholar]
- 480.Kadiyala, P. et al. High Density Lipoprotein-mimicking Nanodiscs For Chemo-immunotherapy Against Glioblastoma Multiforme. ACS Nano13, 1365–1384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Chen, Z. et al. Disruption of β-catenin-mediated negative feedback reinforces cAMP-induced neuronal differentiation in glioma stem cells. Cell Death Dis.13, 1–13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Huang, R.-X. & Zhou, P.-K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target Ther.5, 60 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.He, Y. et al. The roles and mechanisms of SREBP1 in cancer development and drug response. Genes Dis.11, 100987 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Hvinden, I. C., Cadoux-Hudson, T., Schofield, C. J. & McCullagh, J. S. O. Metabolic adaptations in cancers expressing isocitrate dehydrogenase mutations. Cell Rep. Med.2, 100469 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Lim, M., Xia, Y., Bettegowda, C. & Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol.15, 422–442 (2018). [DOI] [PubMed] [Google Scholar]
- 486.Omuro, A. et al. Radiotherapy combined with nivolumab or temozolomide for newly diagnosed glioblastoma with unmethylated MGMT promoter: An international randomized phase III trial. Neuro Oncol.25, 123–134 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Lim, M. et al. Phase III trial of chemoradiotherapy with temozolomide plus nivolumab or placebo for newly diagnosed glioblastoma with methylated MGMT promoter. Neuro Oncol.24, 1935–1949 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Staron, M. M. et al. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity41, 802–814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Khan, O. et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature571, 211–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Zeng, Z., Wei, F. & Ren, X. Exhausted T cells and epigenetic status. Cancer Biol. Med.17, 923–936 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Chang, C.-H. et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell162, 1229–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Qiu, J. et al. Acetate Promotes T cell effector function during glucose restriction. Cell Rep.27, 2063–2074.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Franco, F., Jaccard, A., Romero, P., Yu, Y.-R. & Ho, P.-C. Metabolic and epigenetic regulation of T-cell exhaustion. Nat. Metab.2, 1001–1012 (2020). [DOI] [PubMed] [Google Scholar]
- 494.Soll, D. et al. Sodium chloride in the tumor microenvironment enhances T cell metabolic fitness and cytotoxicity. Nat. Immunol.25, 1830–1844 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Scirgolea, C. et al. NaCl enhances CD8+ T cell effector functions in cancer immunotherapy. Nat. Immunol.25, 1845–1857 (2024). [DOI] [PubMed] [Google Scholar]
- 496.Mishra, A., Maiti, R., Mohan, P. & Gupta, P. Antigen loss following CAR-T cell therapy: mechanisms, implications, and potential solutions. Eur. J. Hematol.112, 211–222 (2024). [DOI] [PubMed] [Google Scholar]
- 497.Hernandez-Lopez, R. A. et al. T cell circuits that sense antigen density with an ultrasensitive threshold. Science371, 1166–1171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Liu, G. et al. CXCR2-modified CAR-T cells have enhanced trafficking ability that improves treatment of hepatocellular carcinoma. Eur. J. Immunol.50, 712–724 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.