Abstract
MiRNAs are single-stranded short noncoding sequences which display crucial roles on gene transcription regulation in many biological processes especially such as embryonic development, cell proliferation or apoptosis. Also, they are recognized for triggering the host’s internal defence mechanisms and immune cell responses thereby playing crucial role in host-parasite interactions. In the present study, a snap-shot of miRNAs, referred to as the miRNome, is described from the hemolymph, the main immune-related compartment of Biomphalaria glabrata snails, one of the intermediate hosts of the trematode parasite Schistosoma mansoni, the causative agent of schistosomiasis. A high throughput sequencing approach of small RNAs has revealed the presence of 63 miRNAs in the hemolymphatic compartment. Mollusc-specific miRNAs including bgl-miR-1985-5p and bgl-miR-1984-5p were identified, along with 25 novel miRNAs. Bioinformatic predictions, thanks to multiple software tools, helped us to identify more than 6000 potential miRNA target gene candidates. Among them is BgTEP1, a complement-like factor involved in parasite clearance. Interestingly, this factor appeared to be targeted by a newly identified miRNA, named bgl-miR-22707-5p. Our study underscores the inherent diversity of miRNAs in the hemolymph of B. glabrata and discusses their potential role in the regulation of the snail’s innate immune response.
Keywords: Biomphalaria glabrata, Hemocytes, miRNA, Small RNAs, Host-parasite interaction
Graphical abstract
Highlights
-
•
Constitutive hemolymphatic microRNAs from Biomphalaria glabrata.
-
•
Characterization of 25 new miRNAs over 63 yet identified.
-
•
More than 6000 potential miRNA target gene candidates identified.
-
•
MicroRNAs potential regulation of immune genes.
1. Introduction
MiRNAs are small regulatory RNAs, ranging from 18 to 24 nucleotides in length, known to regulate gene expression by binding to specific target mRNAs, thereby inhibiting their translation or promoting their degradation (Berezikov, 2011). Since their discovery in 1993 in Caenorhabditis elegans (Lee et al., 1993), miRNAs have been extensively studied in various invertebrate organisms. For instance, the initial lin-4 and let-7 miRNAs were identified as key regulators as of the timing and specification of neuronal and hypodermal cell fate during C. elegans larval development (Ying et al., 2008). Additionally, certain miRNAs such as mir-184 and mir-277 in Drosophila melanogaster are known to play roles in the development and maintenance of the nervous system (Faggins, 2013). Concerning their implications in immunity, numerous reviews have synthetized their functions in the regulation of the immune blood cell response of vertebrates (O’Connell et al., 2010; Chen et al., 2013; Hadj-Moussa et al., 2022) and in innate immune response of invertebrates (Yang et al., 2012; Burgos-Aceves et al., 2018). As an example, miR-100, miR-1984, miR-184 and the miR-9/200/8 families have been highlighted as miRNAs involved in immune-response processes in marine invertebrates (Yang et al., 2012). These miRNAs regulate immune defence and capacities through their own transcriptional regulation, leading to changes in the immune system’s effectiveness (Zhou et al., 2014). In the immune cells of molluscs and arthropods named hemocytes, some miRNAs are conserved and known to be involved in immune response activation through the regulation of pro-inflammatory cytokines (Weitzel et al., 2009). These miRNAs appear to positively regulate phagocytic activity, as seen with miR-184 or miR-190 (Yang et al., 2012; Martín-Gómez et al., 2014), and are involved in processes such as apoptosis, migration and other immune cellular processes, exemplified by miR-92a (Martín-Gómez et al., 2012). In coelomocyte of Apostichopus japonicas, miRNAs are involved in regulating immune pathways, miR-133 modulates Toll-like receptor (TLR) signaling by regulating the IRAK-1 protein, resulting in increased phagocytic activity (Lu et al., 2015). Additionally, in the European flat oyster (Ostrea edulis) the overexpression of miR-18 (Lee and Hyun, 2014) or miR-335 has been associated with the increase of reactive oxygen species (ROS) levels (Morga et al., 2011, 2012).
Biomphalaria glabrata is a Neotropical planorbid snail that transmits in South America and Caribbean islands the trematode parasite Schistosoma mansoni, the etiological agent of schistosomiasis (Kokaliaris et al., 2022). Schistosomiasis is considered as a neglected tropical disease, however responsible for almost 200,000 human deaths each year (Lo et al., 2022). Consequently, this parasitic disease remains a major global public health problem despite considerable efforts to prevent and/or treat it. To find new ways to fight and control this disease, it appeared necessary to enhance our current knowledge and understanding of the molecular interactions between Schistosoma spp. and their hosts, including especially its intermediate mollusc snail vectors, Biomphalaria glabrata (Basch, 1976). Biomphalaria glabrata develops some complex innate immune responses to eliminate this trematode parasite. Its internal defence system includes an innate immune cellular and humoral response resulting in parasite encapsulation and killing following the recognition of the parasite by pathogen recognition receptors, the activation of immune signalling pathway and the activation of the cellular immune response in which hemocytes are involved. Indeed, local co-evolution between the two organisms has led to a compatibility polymorphism. It had been observed that the intensity and rate of infection depend on the co-evolution between the two organisms. Théron et al. (2014) showed that with its sympatric strain, BgBRE snails can reach 100% infection rate with 20 miracidia compared to an allopatric strain such as smGUA (Guadeloupe strain) where only 5% of snails were infected. This difference is also evident in the intensity of mother sporocyst (MSp) development, with an average of 3.58 MSp developed with SmBRE and 1 MSp with SmGUA (Théron et al., 2014; Galinier et al., 2017; Mitta et al., 2017; Portet et al., 2019). Comparative approaches, along with genomic, transcriptomic, proteomic and epigenetics studies, have mainly demonstrated strong gene cluster regulation during parasitic infection (Hanington et al., 2010; Coustau et al., 2015; Tennessen et al., 2015; Pinaud et al., 2016; Galinier et al., 2017; Pila et al., 2017; Portet et al., 2017, 2019; Wu et al., 2017; Allan et al., 2019; Mendes et al., 2019; Lu et al., 2020, 2022; Li et al., 2022; Simphor et al., 2024). In this context, while the role of miRNAs in the response of the vertebrate host to exposure to the parasite has been studied (El-Taweel et al., 2022; Hamway et al., 2022), the involvement of miRNAs in the immune response of the intermediate host has been largely neglected (Queiroz et al., 2017, 2020; Alves et al., 2023).
To date, most studies have focused on whole snails rather than hemocytes, probably due to the low abundance of these immune cells and the difficulties associated with their recovery, isolation and enrichment. However, recent advances in massive single-cell sequencing approaches have revealed a remarkable and intricate landscape within hemocytes, key producers of immune effectors and known to play crucial roles in immune defence through encapsulation, phagocytosis and antimicrobial activities (Mitta et al., 2017; Li et al., 2022; Pichon et al., 2022). Moreover, when the miracidium enters the snail host, the parasite comes into direct contact with the hemolymph that bathes all mollusc tissues, as the mollusc has an open circulatory system. The immune response against the parasite is then extremely rapid and in just 3 to 6 hours, the first hemocytes are already activated and arrive in contact with the parasite to eliminate it. This suggests the constitutive presence and very quick regulation of key immune response molecules directly present in the snail hemolymph. Thus, in the present study, we propose to characterize for the first time the miRNome present in the hemolymph compartment of the intermediate snail host B. glabrata.
2. Materials and methods
2.1. Biological material
Snails of the albino strain of Biomphalaria glabrata (BgBRE2) were used. This strain was recovered in the field in 1975 from the locality of Recife, Brazil, and maintained since then under laboratory conditions. All individuals lived in tanks filled with pond water at 25 °C with a 12:12 h light/dark cycle and were supplied with fresh lettuce ad libitum. To determine miRNA sets by deep sequencing, hemolymph (plasma + hemocytes) of adult snails (sexually mature) with a size ranging from 7 to 12 mm was collected using the retraction defence mechanism (Sminia and Barendsen, 1980) and maintained at −80 °C. Two biological replicates corresponding to different batches were performed, each consisting of hemolymph collected from a pool of 20 individuals. The objective of the present study was to describe the hemolymphatic miRNome from uninfected snails. Using hemolymph, we have both the miRNAs freely or transported into exosomes and/or micro-vesicles.
2.2. RNA-seq sequencing
Total RNA from the hemolymph pools was extracted using TRI® reagent (Sigma Life Science, Saint Louis, USA), according to the manufacturer’s instructions. Size selection was performed using the Truseq cDNA kit (Illumina, USA) to obtain only reads below 60 bp, and HiSeq SR 50-bp sequencing was performed by Genome Quebec, Canada.
2.3. Small RNA analysis
The sequenced reads were filtered according to the quality score using FASTQC software, and the adapter sequences were trimmed using Trim Galore software on the Galaxy server. Only read sizes of 15–50 nucleotides were selected, and those considered to be of low quality (Phred score < 30) were discarded from the analysis. A very stringent quality score was imposed in this study to remove low-quality sequences due to sequencing errors and to ensure a reliable set of sequences given their small size, particularly in the context of isomiR analysis. All reads sequenced have been analysed by the Kraken tool on the Galaxy server to clean all reads belonging to bacterial and protozoan communities present in the snail microbiota (Supplementary file 1: Table S1).
2.4. miRNA analysis
Combined tools for miRNAs identification and their target’s prediction are necessary to prevent any false positive candidates. The pipeline of analysis was used with the B. glabrata genome BB02 assembly (BglaB1.7 (Adema et al., 2017) from the VectorBase database) (Supplementary file 1: Fig. S1).
2.4.1. Identification of miRNAs
All 15–50 nucleotide reads were mapped on the B. glabrata genome using Bowtie2 (Langmead and Salzberg, 2012) with 1 mismatch permitted and MiRDeep2 Mapper software (v.2.0.0.8.1) (for which no mismatch has been authorized). For the prediction of mature miRNAs, MiRDeep2 (v.2.0.1.2) (Friedländer et al., 2012) and ShortStack (3.8.5) (Axtell, 2013) software were used. MiRDeep2 predicted miRNA read clusters (i.e. precursor elements) with a score ≥ 10 or 100% sequence identity to known miRNAs in the available database (MirBase). While MirDeep2 offers high accuracy (98.6–99.9%) and sensitivity (66–76%), ShortStack enhances the sensitivity and specificity of miRNA identification (Axtell, 2013). ShortStack can annotate miRNA and hairpin-association, with strandedness, small RNA size distribution, phasing, repetitiveness, and quantification. For ShortStack analysis, one mismatch and a size range of 20–24 nucleotides were allowed. miRNA read clusters with near and exact hairpin structure (“N15” (missing part of miRNA∗ (star strand)) and “Y” (considering as a true miRNA) were retained for further analysis. The predictions common to both software packages were retained in addition to the new miRNAs predicted by ShortStack.
2.4.2. miRNAs abundance
Reads per million (RPM) were derived and normalized as counts per million (CPM) using ShortStack, while MiRDeep2 provided the number of unique and total reads for both precursor and mature sequences. The RPM for each miRNA predicted by both MiRDeep2 and ShortStack was calculated using the formula: No. of reads mapped to a mature sequence × 106/Total no. of mapped reads from library. Linear regression analysis is available in Supplementary file 1: Fig. S2. Known miRNAs annotated in the B. glabrata genome, available in RFAM (v.14.9) and those identified by Queiroz et al. (2020), were used as input data read sets.
2.4.3. miRNA gene target prediction
miRNA target prediction was conducted using 4 software packages: MiRanda (v.3.3a) (Enright et al., 2003), PITA (v.6) (Kertesz et al., 2007), RNAhybrid (v.2.1.2) (Kruger and Rehmsmeier, 2006) and RNA22 (v.2) (Loher and Rigoutsos, 2012). For MiRanda, only predictions with an alignment score threshold of 140 or higher and a minimum energy threshold of −10 kcal/mol were considered. For PITA and RNA22, predictions were performed using default settings, while for RNAhybrid, a P-value cut-off < 0.05 was requested. All target predictions were performed on 3′UTR, Protein Coding Gene (PCG) and 5′UTR. To ensure consistency, clusters of common predictions among all tools were created. MiRanda, PITA, RNA22 and RNAhybrid were used to predict potential targets based on the four commonly used features in miRNA target prediction: seed match, conservation, free energy, and site accessibility. Miranda (script last update: 2020) employs a three-step analysis focusing on seed match as the primary feature, followed by free energy and conservation. PITA (script last update: 2008) prioritizes target-site accessibility but initially applies with the seed match criterion, then evaluates site accessibility through free energy scoring and considers target-site abundance. RNA22 (script last update: 2016) can predict targets in the 3′UTRs, 5′UTRs and PCGs, whereas PITA, MiRanda and RNAhybrid typically focus on 3′UTR target sites. For the 3′UTR targets, only predictions common to all four tools were retained. For the 5′UTRs and PCGs, predictions from MiRanda, PITA and RNAhybrid on the RNA22 were overlapped with RNA22 results to identify shared targets. Using multiple software tools is more particularly beneficial for predicting unconventional targets (Wang et al., 2016; Hassan et al., 2022) or for identifying non-conserved interactions and rare sites (Riffo-Campos et al., 2016).
2.5. miRNAs nomenclature
To prevent incorrect annotations, the nomenclature described by Budak et al. (2016) has been adopted. Lettered suffixes indicate closely related mature miRNA sequences derived from different precursors or genomic loci, signifying members of the same miRNA family (e.g. bgl-miR-216a and bgl-miR-216b). Identical mature miRNA sequences that are encoded by different gene in the genome are differentiated by a number following a dash (e.g. bgl-miR-87b-1 and bgl-miR-87b-2). Additionally, the designations 3p or 5p specify miRNAs originating from the 3′- and 5′-arms of the precursor (Griffiths-Jones, 2004). Several variants by length and/or sequences from the same miRNA are considered as isomiRs and are identified with a number suffix after a dot (e.g. canonical miRNA: bgl-miR-92b-3p.1; isomiR1: bgl-miR-92b-3p.1.1; isomiR2: bgl-miR-92b-3p.1.2) to associate them with their respective canonical miRNA (Neilsen et al., 2012).
3. Results and discussion
3.1. Small RNAs from the hemolymph of Biomphalaria glabrata BgBRE2 strain
To deepen the sequencing, reads from two replicates were combined and analysed. Over 67 million of small RNA reads were sequenced from the snail hemolymph. Of these, 38.8% of reads mapped to B. glabrata genome (769,104 unique reads; 26,175,564 count reads). Kraken analysis using bacterial and viral genome databases on the Galaxy server assigned 11.19% (7,567,679 count reads) to bacterial and 2.78% (1,882,966 count reads) to viral sequences. The small RNA sequencing from hemolymph samples revealed a high level of diversity of different types of small RNAs with high-quality scores (PHRED score > 30). Sequences ranging from 15 to 50 nucleotides were retained for small RNA analysis to distinguish between mature miRNAs and their precursors.
The percentage of total mapped reads may seem low and could be attributed to the incomplete genomic assembly and/or SNP between the B. glabrata genome and the BgBRE2 strain used in this study. Nevertheless, this result aligns with those obtained by Queiroz et al. (2020) from the same biological model, which reported with 344,624 unique reads out of 1,123,762 total unique reads, representing 30.66% of reads mapped to the reference genome.
Here, 28.32% of the aligned reads were identified as sequences annotated as small RNAs in the B. glabrata genome corresponding to 7,414,351 count reads, with 491,924 of these identified as unique reads. Mature miRNAs represent approximately 10% of the aligned reads, with 2,601,481 total reads and 2658 unique reads, making them the least abundant. rRNA and tRNA account for 27% of the identified small-RNAs with 22,625 and 16,949 unique reads, respectively. Nearly 5 million unique reads remain unidentified in detail. Although these reads have been annotated as non-coding RNA in the B. glabrata genome, their precise characterization is still lacking. Detailed information on small-RNAs identifications is summarized in Supplementary file 1: Table S1.
3.2. miRNAs present in the hemolymph of B. glabrata BgBRE2 strain
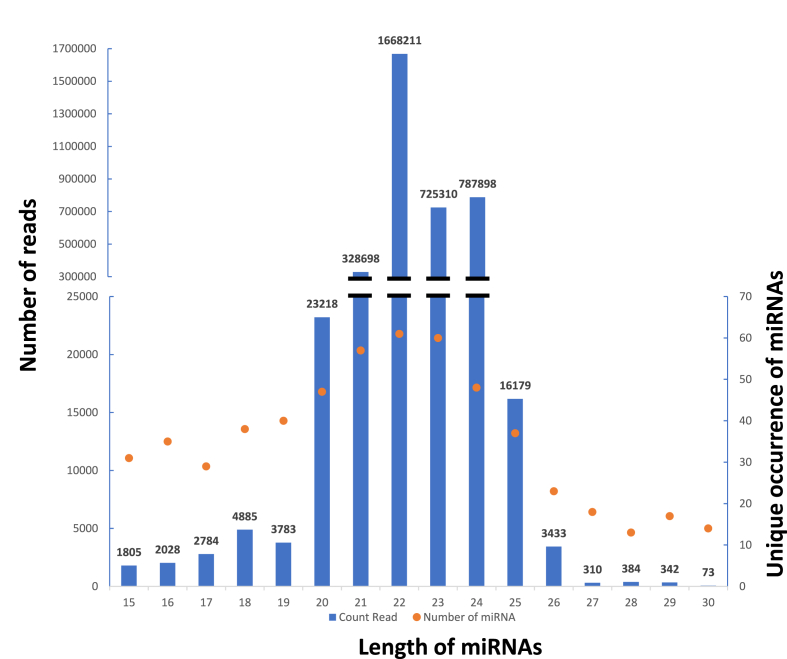
In this study, a total of 3,569,341 count reads (2,601,481 unique reads) aligned to mature miRNAs sequences or pre-miRNAs with a length of 15–30 nucleotides. The length distribution of these reads matches the typical size range for mature miRNAs (Fig. 1). All mature miRNA forms are between 15 and 30 nucleotides, with about 97% of them falling between 21 and 24 nucleotides (3,569,341 count reads).
Fig. 1.
Distribution of count reads (blue histograms - left axis) and unique occurrence (orange points - right axis) for mature miRNAs by size.
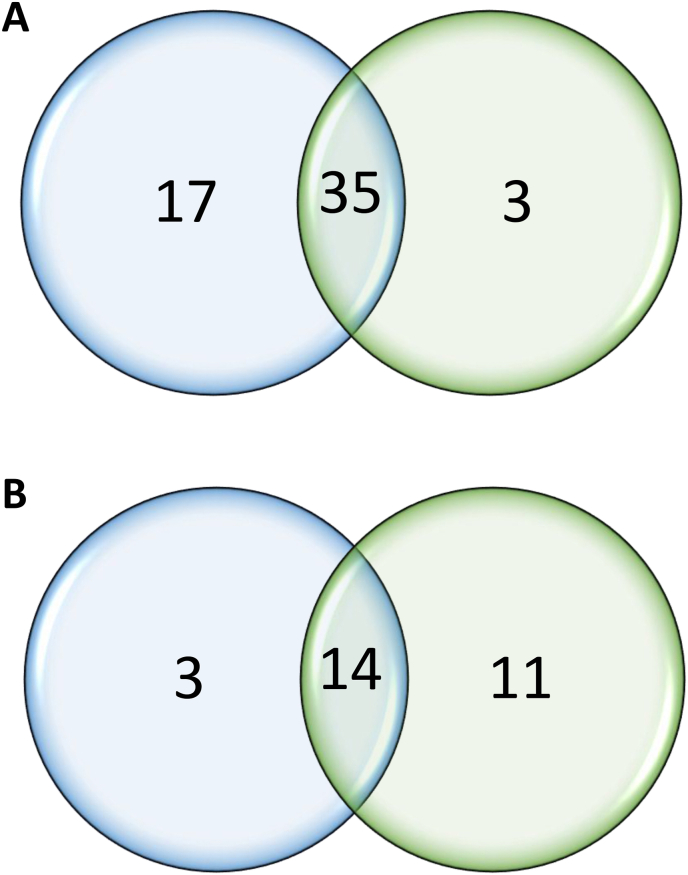
MiRNAs were identified using a comparative tool approach (Berezikov, 2011). Analysis with MiRDeep2 (shown in blue in Fig. 2) identified 52 known miRNAs and 17 new miRNAs. In comparison, ShortStack (shown in green in Fig. 2) detected 38 miRNAs already annotated in the B. glabrata genome and 25 new miRNAs (Fig. 2). Both tools commonly identified 35 known miRNAs and 14 new miRNAs. As described in Section 2, predictions commonly from ShortStack and MirDeep2 software (35 known miRNAs and 14 new miRNAs) and the predictions of ShortStack software (3 known miRNAs and 11 new miRNAs) only were retained; the ones strictly predicted only by MirDeep2 were not considered in this study. In total, 63 miRNAs were identified. Of the 25 new miRNAs, 12 did not have corresponding entries in the MiRBase or RFAM databases for any other species with significant P-values. Following the consensus nomenclature, these new miRNAs were assigned temporary numbers pending their inclusion in the MiRBase registry. Details of all identified miRNAs are provided in Table 1, Table 2.
Fig. 2.
Venn diagram of miRNAs predicted by ShortStack (green) and MirDeep2 (blue). A Known miRNAs. B. New miRNAs.
Table 1.
Known miRNAs identified by ShortStack and MiRDeep2.
| Mature sequence | Length (bp) | RPM ShortStack | Scaffold ID | Start-End position in scaffold | Gene location | Name |
|---|---|---|---|---|---|---|
| aacccguagaaccgaacuugug | 22 | 17620.732 | KE714233 | 93682–93749 | Intergenic | bgl-miR-100-5p |
| uggacggagaacugauaagggc | 22 | 7504.299 | APKA01103429 | 1551–1691 | Intergenic | bgl-miR-184-3p |
| uucguugucgucgaaaccugccu | 23 | 2777.740 | KE712523 | 35130–35194 | Intergenic | bgl-miR-981-3p |
| ugcccuauccgucaggaacugu | 22 | 1867.472 | KE719960 | 139821–139951 | Intergenic | bgl-miR-1984-5p |
| uaauacugucagguaaagauguc | 23 | 1432.879 | KE719932 | 179103–179262 | PCG - Intronic | bgl-miR-8-3p |
| uaaaugcauuaucugguaucuga | 23 | 1291.194 | KE720423 | 46611–46862 | Intergenic | bgl-miR-277a-3p |
| ugccauuuuuaucagucacugug | 23 | 1261.939 | KE719960 | 136001–136167 | Intergenic | bgl-miR-1985-5p |
| ucccugagaccauaauuugugc | 22 | 932.109 | KE714233 | 104289–104401 | Intergenic | bgl-miR-125-5p |
| ugagguaguagguuguauuguu | 22 | 647.388 | KE714233 | 100237–100421 | Intergenic | bgl-miR-let-7-5p |
| cuuggcacuggcggaauagucac | 23 | 557.631 | KE714275 | 468616–468693 | PCG - Intronic | bgl-miR-96a-5p |
| ccagaucuaacucuuccagcuc | 22 | 466.315 | KE714253 | 178310–178415 | PCG - Intronic | bgl-miR-750-3p |
| ugacuagauccacacucaucc | 21 | 331.940 | KE713120 | 16198–16305 | Intergenic | bgl-miR-279-3p |
| ugagacagugcguccucccuca | 22 | 317.751 | KE711897 | 46379–46444 | Intergenic | bgl-miR-1994b-3p |
| uggcagugugguuagcugguugu | 23 | 266.387 | KE720423 | 58888–59175 | PCG - Intronic | bgl-miR-449-5p |
| aagggagcauccgucgacagu | 21 | 187.180 | KE720378 | 29219–29292 | Intergenic | bgl-miR-281-5p |
| ugaaagacauggguagugagaug | 23 | 169.350 | KE720056 | 15367–15605 | PCG - Intronic | bgl-miR-71-5p |
| ucuuugguuaucuagcuguauga | 23 | 142.338 | KE713467 | 127633–127761 | Intergenic | bgl-miR-9-5p |
| aggcaagauguuggcauagcuga | 23 | 136.975 | KE720379 | 50449–50534 | Intergenic | bgl-miR-31-5p |
| ugagauucaacuccuccaacug | 22 | 104.481 | KE714253 | 170944–171010 | PCG - Intronic | bgl-miR-1175-3p |
| ugagaucauugugaaaacugauu | 23 | 86.785 | KE717415 | 2175–2236 | Intergenic | bgl-miR-bantam-1-3p |
| ugagaucauugugaaaacugauu | 23 | 84.304 | APKA01398116 | 2064–2124 | Intergenic | bgl-miR-bantam-2-3p |
| uuuguucguucggcucgcguua | 22 | 82.595 | KE716631 | 129311–129370 | Intergenic | bgl-miR-375-3p |
| cuaaguacuggugccgcggga | 21 | 70.367 | APKA01096956 | 3003–3106 | Intergenic | bgl-miR-252a-5p |
| agauauguuugauauauuuggug | 23 | 59.521 | KE719849 | 62697–62811 | PCG - Intronic | bgl-miR-190-5p |
| ugagacaguguguccucccuug | 22 | 59.268 | KE711897 | 47019–47082 | Intergenic | bgl-miR-1994a-3p |
| uaauaucagcugguaauccuga | 22 | 23.371 | KE711606 | 757139–757244 | PCG - Intronic | bgl-miR-216b-5p |
| uauuaugcugcuauucacgaga | 22 | 21.202 | KE719985 | 231681–231740 | Intergenic | bgl-miR-1993-1-3p |
| uauuaugcugcuauucacgaga | 22 | 20.296 | APKA01101172 | 817–877 | Intergenic | bgl-miR-1993-2-3p |
| uggcgccguggaaacaucuacc | 22 | 20.132 | KE711817 | 53444–53502 | Intergenic | bgl-miR-2722-3p |
| gagcugccaaaugaagggcugu | 22 | 14.992 | KE707076 | 13220–13338 | PCG - Intronic | bgl-miR-745b-3p |
| uauugcacuuuuccaggccuuu | 22 | 14.813 | KE713921 | 19826–19893 | PCG - Intronic | bgl-miR-92a-3p |
| uugguccccuucaaucaguugu | 22 | 12.689 | KE712896 | 111967–112030 | PCG - Intronic | bgl-miR-133-3p |
| uugugaccguuauaaugggcauu | 23 | 9.880 | KE716868 | 11312–11379 | Intergenic | bgl-miR-2001-5p |
| uagcaccauuugaaaucaguuu | 22 | 9.613 | KE715109 | 101169–101333 | Intergenic | bgl-miR-29-3p |
| gugagcaaaguuucagguguau | 22 | 3.046 | KE715168 | 21952–22013 | PCG - Intronic | bgl-miR-87b-2-3p |
| gugagcaaaguuucagguguau | 22 | 2.704 | KE715168 | 17506–17589 | PCG - Intronic | bgl-miR-87b-1-3p |
| uggaauguaaagaaguauguau | 22 | 1.887 | KE712896 | 141942–142050 | PCG - Intronic | bgl-miR-1a-3p |
| caaugucucugcagugcaauca | 22 | 0.787 | KE711726 | 29962–30064 | PCG - Intronic | bgl-miR-33-3p |
Abbreviation: PCG, Protein Coding Gene.
Table 2.
Novel miRNAs.
| Mature sequence | Length (bp) | RPM ShortStack | Scaffold ID | Start-End position in scaffold | Gene location | Best Hit on miRbase/Queiroz et al. (2020) (P < 0.05) | Name |
|---|---|---|---|---|---|---|---|
| uacccuguagauauccgaauuugu | 24 | 11621.477 | KE719220 | 17337–17405 | Intergenic | mle-miR-10a | bgl-miR-10a-5p |
| aauugcacuucgaccggccugc | 22 | 1262.296 | KE713921 | 19641–19700 | PCG - Intronic | bgl-miR-92-3p | bgl-miR-92-3p.1.1 |
| aauugcacuuaccccggccugu | 22 | 654.816 | KE713921 | 19999–20139 | PCG - Intronic | bgl-miR-92a-1-3p | bgl-miR-92a-3p.1.2 |
| cauugcacuuuucccggccugu | 22 | 596.380 | KE713921 | 20227–20286 | PCG - Intronic | bgl-miR-92a-1-3p | bgl-miR-92a-3p.1.3 |
| uaaugcccccucaaacccuaaa | 22 | 98.077 | KE711626 | 184519–184591 | Intergenic | mle-miR-12097-3p | bgl-miR-12097-1-3p |
| uaaugcccccucaaacccuaaa | 22 | 96.665 | APKA01388332 | 813–876 | Intergenic | mle-miR-12097-3p | bgl-miR-12097-2-3p |
| uaucacagccaguauaccccug | 22 | 52.775 | KE720464 | 153508–153566 | PCG - Intronic | lgi-miR-2c | bgl-miR-2c-3p |
| ugcuccaggauaaagcugcauc | 22 | 15.051 | KE707776 | 2504–2611 | PCG - Intronic | NC | bgl-miR-12604-5p |
| uaucacagccaauaauccccac | 22 | 10.401 | KE720464 | 152823–153010 | PCG - Intronic | lgi-miR-2c | bgl-miR-2c-3p.1 |
| uuuuguucggguaguuugaaga | 22 | 10.222 | KE714890 | 12969–13203 | Intergenic | bmo-miR-3216 | bgl-miR-3216-5p |
| aaucacaaucauuggauggguu | 22 | 7.964 | KE720464 | 153672–153843 | PCG - Intronic | mle-miR-2f | bgl-miR-2f-3p |
| caucuaccuauccuucuucuuc | 22 | 5.765 | KE714719 | 130732–130795 | Intergenic | mle-miR-122104-3p | bgl-miR-122104-3p |
| uuuaccuacaguuaauacgagug | 23 | 3.937 | KE712501 | 31367–31426 | Intergenic | NC | bgl-miR-16599-5p |
| uuuaccugcagauaauaugaac | 22 | 1.590 | KE712501 | 37204–37258 | Intergenic | NC | bgl-miR-21775-3-5p |
| uuuaccugcagauaauaugaac | 22 | 1.367 | APKA01302483 | 52–107 | Intergenic | NC | bgl-miR-21775-2-5p |
| uuuaccugcagauaauaugaac | 22 | 1.278 | APKA01110979 | 89–144 | Intergenic | NC | bgl-miR-21775-1-5p |
| uugaucaguagcuucaaagaga | 22 | 1.189 | KE714890 | 14783–14841 | Intergenic | NC | bgl-miR-22707-5p |
| cgcgggcguugggggccccacg | 22 | 1.085 | KE718106 | 164257–164314 | Intergenic | mle-miR-1986-5p | bgl-miR-1986-5p |
| uuuggcaccaaagaauucacuga | 23 | 0.862 | KE720231 | 207209–207265 | Intergenic | mle-miR-263b-5p | bgl-miR-263b-5p |
| uggccacgcgcuaucggucucc | 22 | 0.431 | APKA01099832 | 1954–2015 | Intergenic | NC | bgl-miR-12705-3p |
| cgcgacgucacaaccugugggc | 22 | 0.163 | KE713001 | 5787–5883 | Intergenic | NC | bgl-miR-22107-5p |
| agcggaaaggaacuaucucacu | 22 | 0.149 | KE713634 | 6346–6432 | Intergenic | NC | bgl-miR-91194-5p |
| cucaggcugugacguugcaggu | 22 | 0.119 | KE710729 | 100165–100254 | Intergenic | NC | bgl-miR-11705-5p |
| accgguuuacucguc | 15 | 0.104 | KE720447 | 88449–88536 | Intergenic | NC | bgl-miR-21902-5p |
| accgguuuacucguc | 15 | 0.104 | KE720447 | 88449–88536 | Intergenic | NC | bgl-miR-21902-3p |
Note: MiRNAs published by Queiroz et al. (2020) have been recovered and added to our workflow as known miRNAs for our studies. The list of miRNAs is available in the article.
Abbreviations: PCG, Protein Coding Gene; NC, Not characterized.
The genomic locations of the 63 mature miRNAs were investigated. Of these, 65% (41 miRNAs) are located in intergenic region, while 22 miRNAs are found within introns of Protein Coding Gene (PCG) sequences. Among the 63 miRNAs, 4 are IsomiRNAs. IsomiRs can be classified into 3′ and 5′ types, with the 3′ isomiR being the most common. When the dominant form is the 5′ isomiR, it can lead to changes in the seed region, also called seed shifting (Berezikov, 2011). Here, the bgl-miR-92-3p.1.1 has an additional nucleotide at the 3′-end and bgl-miR-1175-3p.1.1 exhibits trimming at the 3′-end. Additionally, bgl-miR-92a-3p.1.2 and bgl-miR-92a-3p.1.3 are isomiRs with nucleotide substitutions in the core and 5′-end, respectively, compared to bgl-miR-92a-1-3p as reported by Queiroz et al. (2020), which may result in seed shifting. To adhere to consensus nomenclature, it is proposed to rename these isomiRs to reflect their distinction: bgl-miR-92a and bgl-miR-92a-1 are on the same loci with different mature sequences and should be categorized as isomiRs of bgl-miR-92a, designated as bgl-miR-92a-3p.1.1.
The three most abundant miRNAs were bgl-miR-100-5p, bgl-miR-10a-5p, and bgl-miR-184-3p, with 744,443, 689,715, and 360,156 count reads, respectively (count per million (CPM) are indicated in Table 3). Together, the 10 mature miRNAs presented in Table 3 accounted for 90.9% of the total read counts among the 63 identified mature miRNAs. Hereafter, we provide descriptions of these 10 more abundant miRNAs of Biomphalaria glabrata.
-
(i)
Bgl-miR-100-5p: miR-100 is a conserved miRNA found across all bilaterians (Berezikov, 2011). It has been implicated in human cancers, where it influences tumour growth and apoptosis by targeting the mTOR pathway (Zheng et al., 2012; Li et al., 2013, 2015; Sun et al., 2013). Similar functions have been observed in both vertebrates and invertebrates (Yang et al., 2014). Additionally, miR-100 is present in the hemolymph of various invertebrates, including, insects (Dhahbi et al., 2016), oysters (Martín-Gómez et al., 2014) and shrimps (Wang and Zhu, 2017) in which miR-100 has been associated with broader effects on superoxide dismutase, phenol oxidase, apoptosis, and phagocytosis processes (Wang and Zhu, 2017).
-
(ii)
Bgl-miR-10a-5p: MiR-10a is a member of the highly conserved miR-10 family, which plays significant role involved in cancer and immune-related processes across vertebrates and invertebrates (Huang et al., 2017). In vertebrates, miR-10a regulates autoimmune diseases like rheumatoid arthritis. Its downregulation leads to the degradation of IκB and activation of NF-κB by targeting IRAK4, TAK1 and BTRC (Mu et al., 2016). In invertebrates, miR-10a has been detected in the hemocytes of Ostrea edulis following infection with the protozoan parasite Bonamia ostreae (Martín-Gómez et al., 2014) and in C. gigas in response to bacterial infections or heat stress (Zhou et al., 2014).
-
(iii)
Bgl-miR-184-3p: In vertebrates, miR-184 regulates immune response by reducing the expression of pro-inflammatory cytokines (Weitzel et al., 2009). In Drosophila melanogaster, miR-184 is crucial for the development and maintenance of post-embryonic nervous systems (Li et al., 2011). In the shrimp Marsupenaeus japonicus, miR-184 is involved in both cellular immunity (such as phagocytosis and apoptosis) and humoral immunity (e.g. phenoloxidase activity) (Yang et al., 2012). In the pea aphid, miR-184 regulates the JNK pathway following bacterial challenges, a pathway known for mediating and controlling phagocytosis, prophenoloxidase (PPO) activation and ROS metabolism (Ma et al., 2020). In B. glabrata, bgl-miR-184-3p is one of the most abundant and may act as a regulator of apoptosis and proteolysis processes (Chen and Stallings, 2007; Queiroz et al., 2020).
-
(iv)
Bgl-miR-981-3p: In Drosophila, miR-981 acts as a negative regulator of antibacterial defences by inhibiting the expression of the antimicrobial peptide diptericin expression within the Immune Deficiency (IMD) pathway (Li et al., 2017; Lu and Chtarbanova, 2022). In the shrimp Penaeus vannamei, miR-981 modulates the expression of C-type lectins expression in hemocytes, playing a role in the antiviral response against WSSV (White Spot Syndrome Virus) (Shekhar et al., 2019). In Aedes aegypti, miR-981 regulates importin β-4, which controls the translocation of AGO1 and its carried miRNAs between the nucleus and the cytoplasm during infection with the endosymbiotic gram-negative bacteria Wolbachia (Hussain et al., 2013).
-
(v)
Bgl-miR-1984-5p: miR-1984 is recognized as a mollusc-specific miRNA (Im and Kim, 2019) and has also been found occasionally in insects (Zha et al., 2016). It was first described in the gastropods Lottia gigantea and Haliotis rufescens (Wheeler et al., 2009). In C. gigas, miR-1984 is known to play a role in immune response, as it is present in hemocytes and involved in redox regulation and energy metabolism (Zhou et al., 2014; Zhao et al., 2016). In B. glabrata, bgl-miR-1984-5p is regulated during developmental processes (Queiroz et al., 2020) and upregulated following Schistosoma infection in B. tenagophila (Alves et al., 2023).
-
(vi)
Bgl-miR-8-3p: Commonly observed in both vertebrates and invertebrates, miR-8 is involved in neural development, cell cycle regulation and cell differentiation (Trümbach and Prakash, 2015). In Drosophila, Bolin et al. (2016) have shown that loss of miR-8 can protect cells from apoptosis following UV irradiation, and it may also regulate apoptosis in shrimp (Yang et al., 2012). Finally, miR-8 is frequently found in hemocytes of various invertebrates (Martín-Gómez et al., 2014).
-
(vii)
Bgl-miR-277a-3p: In D. melanogaster, mir-277 is involved in the development and maintenance of the post-embryonic nervous systems (Faggins, 2013). In mosquitoes, CRISPR-Cas9 knockout of mir-277 results in defects in lipid storage and ovarian development (Ling et al., 2017). It was also identified in shrimp hemocytes, where it targets FAD-dependent oxidoreductase (Shekhar et al., 2019). In B. glabrata snails, a high level of bgl-miR-277a-3p has been observed (Queiroz et al., 2020), suggesting potential roles in endogenous biological processes, immune functions or adaptation to environmental stress.
-
(viii)
Bgl-miR-92-3p.1.1: The miR-17-92 family is conserved across all metazoans. Dysregulation of its members can lead to lymphoproliferative diseases and systemic autoimmunity in vertebrates by enhancing the activation, proliferation and survival of T and B cells (Chen et al., 2013). The cluster miR-17-92 includes multiple isomiRs with cancer-regulatory functions in vertebrates (Tong et al., 2012). In oysters, miR-92 regulates the proliferation and development of immune cells by targeting G-protein expression following infection with Bonamia sp. (Martín-Gómez et al., 2014). In shrimp, miR-92 levels are modulated in response to apoptotic signals, either being down- or upregulated depending on the context (Yang et al., 2012). In our study, Bgl-miR-92-3p.1 and one of its isomiRs bgl-miR-92-3p.1.1 were highly abundant in the hemolymph (Supplementary file 2) assuming for a potential involvement in immune response.
-
(ix)
Bgl-miR-1985-5p: Like miR-1984, miR-1985 is also known as a mollusc-specific miRNA (Wheeler et al., 2009). This miRNA has been found to be highly expressed in the hemocytes of Mytilus galloprovincialis and has been shown to target cyp-like proteins involved in immune responses against pathogens (Moreira et al., 2020). In B. glabrata, bgl-miR-1985-5p has been detected in whole snails (Adema et al., 2017; Queiroz et al., 2020) and is also differentially regulated following S. mansoni infection (Alves et al., 2023).
-
(x)
Bgl-miR-125-5p: miR-125 is highly conserved across the animal kingdom and regulates key processes such as apoptosis, innate immunity, inflammation and hematopoietic differentiation in vertebrates (Martín-Gómez et al., 2014). In the mud crab, miR-125 is expressed in hemocytes and its levels increase following pathogen infection. Knockdown studies of miR-125 reveal that it positively regulates phagocytosis and apoptosis (Qian et al., 2024). In comparison to the study by Queiroz et al. (2020), which explored the miRNome of B. glabrata in whole snails, our study reveals that bgl-miR-981-3p is more abundant in the hemolymph, with 174,273 unique reads compared to 153,740 count reads in whole snails without shells.
Table 3.
Abundance of miRNAs by counts per million (CPM): A list of the 10 most abundant miRNAs in the snail immune compartment. The quantification of miRNA abundance using CPM provides insight into their relative representation in the mollusc hemolymph.
| miRNA | Counts per million (CPM) |
|---|---|
| bgl-miR-100-5p | 17620.73 |
| bgl-miR-10a-5p | 11621.47 |
| bgl-miR-184-3p | 7504.29 |
| bgl-miR-981-3p | 2777.74 |
| bgl-miR-1984-5p | 1867.47 |
| bgl-miR-8-3p | 1432.87 |
| bgl-miR-277a-3p | 1291.19 |
| bgl-miR-92-3p.1.1 | 1262.29 |
| bgl-miR-1985-5p | 1261.93 |
| bgl-miR-125-5p | 932.10 |
3.3. miRNAs gene target prediction
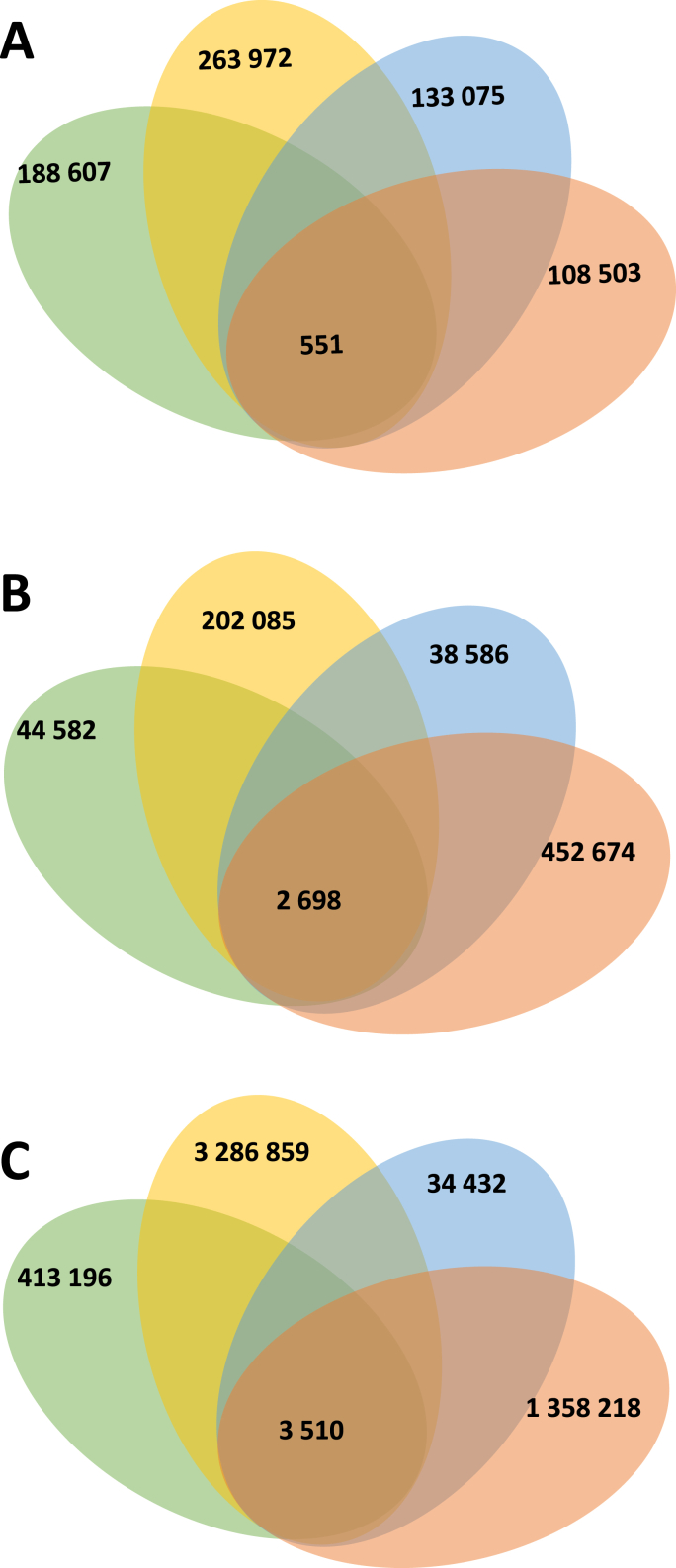
For identifying miRNA gene targets, 3′UTR and 5′UTR regions and Protein Coding Gene (PCG) were collected from GFF3 annotation files of B. glabrata genome. To improve prediction accuracy and minimize false positives, it is advisable to combine results from multiple prediction tools (Akhtar et al., 2019). Therefore, we employed a multi-tool approach using four prediction software tools: MiRanda, PITA, RNA22 and RNAhybrid. More than 6759 potential miRNA target genes were predicted (Fig. 3; Supplementary file 1). Among these, 551 hits were identified by all prediction tools and correspond to 441 genes for which predicted miRNA-target duplexes are located on the 3′UTR localisation (Fig. 3A). Interestingly, about 2698 hits are presents on the 5′UTR and correspond to 1979 snail genes (Fig. 3 (b)). An even larger number of targets were predicted for the PCG with 3510 hits corresponding to 914 genes (Fig. 3C). In total, 3078 unique miRNAs downregulate genes were identified. Interestingly, some mRNAs are targeted along their entire length by various miRNAs. Specifically, 15 target genes were identified with multiple hits within their 3′UTR, 5′UTR and PCG regions. These hits were contributed by the same miRNAs at different locations or by different miRNAs (Table 4 and Fig. 4).
Fig. 3.
Venn diagram of miRNAs predicted hits on mRNA target. One hit corresponds to the mapping of one miRNA for one mRNA predicted target. The software tools MiRanda (green), PITA (yellow), RNA22 (blue) and RNAhybrid (orange) were used. A Target’s predictions on 3′UTR. B Target’s predictions on 5′UTR. C Target’s predictions on Protein Coding Gene (PCG).
Table 4.
Fifteen genes (BB02 and laboratory internal annotation) predicted to be targeted on their total length (UTRs and Protein Coding Gene part) by miRNAs.
| BB02 Genome Gene ID | Gene annotation from IHPE Laboratory |
|---|---|
| BGLB000051 | Baculoviral IAP repeat-containing protein 2-like isoform |
| BGLB000786 | ATP-binding cassette sub-family D member 2-like |
| BGLB000846 | Not annotated |
| BGLB000876 | Ras guanine nucleotide exchange factor L-like |
| BGLB001150 | Receptor-type tyrosine-protein phosphatase kappa-like isoform |
| BGLB001225 | Transcriptional activator Myb-like isoform |
| BGLB016762 | Synaptotagmin 7 |
| BGLB017474 | Bridge-like lipid transfer protein family member 1 isoform |
| BGLB020718 | Integrator complex subunit 13-like |
| BGLB022562 | Verprolin-like |
| BGLB027526 | Adhesion G-protein coupled receptor D1-like |
| BGLB028227 | D(2) dopamine receptor A-like isoform |
| BGLB030287 | Ankyrin repeat and KH domain-containing protein 1-like |
| BGLB036645 | RWD domain-containing protein 2A |
| BGLB038139 | Gem-associated protein 5-like |
Fig. 4.
Venn diagram of occurrence of gene targeted by miRNAs on three localisations: 3′UTR (green); 5′UTR (orange); and Protein Coding Gene (PCG) (blue).
An enrichment analysis showed an expected result such as no up- or under-biological, molecular or cellular function are more regulated by miRNAs. We decided to investigate in the literature the biological implications of the 15 targeted genes. All miRNAs targeting this cluster of genes are detailed in Supplementary file 1: Table S4. One of these genes (BGLB000846) is not annotated in the genome. The 14 remaining genes can be categorized into 7 groups.
The first group of gene pertains to protein interaction, specifically the RWD Domain Containing 2A (RWDD2A-BGLB036645). This gene encodes proteins with Ring finger-domains, WD repeat-, and Dead-like helicases (Doerks et al., 2002). While their functions are not well established, it is suggested to be involved in protein interactions (Puthiyedth et al., 2016) and may serve as a substrate for E2 dependent ubiquitination (Lorick et al., 1999).
Two genes (BGLB001225 and BGLB038139), targeted by the bgl-miR-92a-3p family, bgl-miR-9-5p, or by bgl-miR-bantam-3p, constitute the second class associated with transcriptional regulation functions. For example, Gem-associated protein 5 (Gemin5-BGLB038139) identified as a large tryptophan-aspartic acid (WD) repeat protein serves as an RNA-binding protein to deliver small nuclear RNAs to the survival of the motor neurons complex (Gubitz et al., 2002). While its role in the snRNPs (small nuclear ribonucleoproteins) biogenesis is well characterized, recent studies have also highlighted a function as a modulator of translation activity (Pacheco et al., 2009). The second gene, BGLB001225, encodes a Myb transcriptional protein homologous to b-myb in vertebrates. This protein is recognized for its role in DNA replication in mitotically dividing larval brain cells and endocycling larval fat body cells in D. melanogaster (Davidson et al., 2005). Its role in the JAK/STAT pathway, which mediates hemocytes hyperproliferation and cellular differentiation in hematopoietic organs, has been demonstrated in D. melanogaster and mammals (Graf, 1992; Davidson et al., 2005).
Signal transduction can be likened to a game of “telephone” between cells where receptors binding to chemical messengers as circulating hormones and neurotransmitters, pass on the information to a series of intracellular middlemen that ultimately pass on the orders to the final executors (Linder and Gilman, 1992). In our study, miRNAs such Bgl-miR-9-5p, bgl-miR-2c-3p, bgl-miR-449-5p or bgl-miR-12604-5p may regulate the mRNA of several genes implicated in this signalling. One notable gene involved in signal transduction is the receptor-type tyrosine-protein phosphatase к (PTPRK-BGLB001150). The tyrosine phosphatases family, including PTPRK, is present across all vertebrate taxa and in the invertebrate chordate Ciona intestinalis (Chen et al., 2017). This gene, targeted by several miRNAs including bgl-miR-2c-3p and bgl-miR-12604-5p, play a critical role in cell adhesion. Loss-of-function in PTPRK has been linked to decreased junctional integrity in mammary epithelial cells (Young et al., 2021) and is particularly significant in the nervous system (Craig and Brady-Kalnay, 2015). Additionally, PTPRK directly regulates the signal transducer and activator of transcription 3 (STAT3) in human’s nasal KT/T-cell lymphoma leading to an increase in patient mortality (Chen et al., 2015). A second gene, BGLB027256, potentially regulated by bgl-miR-2001-5p, bgl-miR-71-5p, bgl-miR-let-7-5p, and bgl-miR-9-5p, is adhesion G-coupled protein receptor D1-like (GPCR). This receptor is known for its role in signalling against pathogens and has been implicated in vertebrate immune responses (Wang, 2018). GPCRs are upregulated following exposure to S. mansoni in susceptible (11 GPCRs) and resistant (52 GPCRs) snails, highlighting their importance in parasite-snail interactions (Lu et al., 2022). A third gene, BGLB028227, is a (D2) dopamine receptor A-like, a member of the rhodopsin-like GPCR family. This receptor regulates adenylyl cyclase and modulates calcium and potassium channels in cells (Mustard et al., 2005). Phylogenetic analysis shows that in B. glabrata D2 receptor is closely related to the those of Aplysia california and Sinonovacula constricta. In these species, the dopamine 2 receptor has been involved in immune responses to bacterial challenges by modulating SOD (superoxide dismutase) and CAT (catalase) activities (Niu et al., 2019). The last gene involved in signal transduction is an ankyrin repeat and KH domain-containing protein 1 (ANKHD1-BGLB030287), targeted by bgl-miR-449-5p but also bgl-miR-2c-3p. ANKHD1 contains an ankyrin repeat domain that mediates protein-protein interactions in signalling pathways such as JAK/STAT (Müller et al., 2005), Hippo (Sansores-Garcia et al., 2013), and PINK/PARKIN (M. Zhu et al., 2015b) as well as a KH (K-Homology) domain that binds RNAs, miRNAs or single-stranded DNA (Mullenger et al., 2023). The interaction between miRNAs and circ-ANKHD1 has been investigated revealing that circ-ANKHD1 inhibits miR-27a-3p, which positively regulates SFRP1 (Secreted Frizzled Related Protein 1) expression in granulosa cells in ovarian sows. This modulation promotes granulosa cell proliferation (Li et al., 2021).
Another gene revealed to be targeted by miRNAs (bgl-miR-11705-5p, bgl-miR-449-5p, and bgl-miR-71-5p) corresponds to an integrator complex subunit 13 (BGLB020718). Integrator subunits (INTSs) are a metazoan-specific protein family comprising 15 subunits (Kirstein et al., 2021), that play crucial roles in biological processes such as the cleavage of the extended 3′-end of Uridine-rich small nuclear RNAs, essential for the biogenesis of spliceosomal snRNPs and the 3′-end formation of enhancer RNA (Barbieri et al., 2018). While its role in transcriptional regulation is well documented, its impact on cellular homeostasis, cell proliferation and apoptosis in hepatocellular carcinoma in humans (HCC) has also been highlighted (Wang et al., 2024). Additionally, its association with EGR1/2 TFs, which regulate enhancer regions during the differentiation of progenitor cells into monocytes and macrophages, underscores its significance (Barbieri et al., 2018). This class of cellular functions also includes another gene, BGLB000051, which corresponds to a BIRC2, a member of the anti-apoptotic gene family. This encoded protein is composed of three domains: Baculovirus Inhibitor of Apoptosis Repeat (BIR), Really Interesting New Gene (RING) and Caspase Recruitment Domain (CARD). Studies have highlighted BIRC2’s role in inhibiting apoptosis by interfering with the activation of caspases (Yang and Li, 2000). Several miRNAs are known to target this gene as miR-29c in the cerebral ischemia/reperfusion in rats (Lingjie et al., 2021), or miR-5195-3p in glioma cells in humans (Yang et al., 2020). In our study, BIRC2 is targeted by multiple miRNAs, such as bgl-miR-bantam-1-3p, bgl-miR-bantam-2-3p, bgl-miR-let-7-5p, bgl-miR-33-3p, and bgl-miR-71-5p. Interestingly, during schistosomiasis, the vertebrate host exhibits a high level of BIRC2 expression during infection in lung endothelial cell (Oliveira et al., 2021). The last gene in this functional category is a ras guanine nucleotide exchange factor L-like (Ras GEF-BGLB000876). Ras is a small G-protein primarily involved in assembling intracellular downstream cell signalling pathways, including those related to proliferation, differentiation, apoptosis, senescence and metabolism (Schlessinger, 2000). GEFs are key regulatory elements in the RAS-GDP/GTP exchange process (Hennig et al., 2015). Activation of GTP triggers a signal transduction cascade by the extracellular signal-regulated kinase (ERK) and mitogen activate protein kinase (MAPK). The Ras/MAPK pathway influences several biological processes, such as the Insulin/IGF-1 signalling (ISS) pathway, the mTOR pathway involved in longevity in both invertebrates and vertebrates, and the AMPK (AMP-activated protein kinase) pathway, a major regulator of energy metabolism, stress resistance and proteostasis (Slack, 2017).
The verprolin protein (BGLB022562) is targeted by a single miRNA, bgl-miR-9-5p, across its entire length (5′UTR, PCG and 3′UTR). This protein is involved in cellular processes and more precisely in cellular migration. Verprolin is essential for actin polymerization during polarized growth and endocytosis. In invertebrates, only one verprolin gene has been identified, compared to three in vertebrates, i.e. WASP-interacting protein (WIP), glucocorticoid-regulated gene product (CR16) and WIP-related (WIRE) (Aspenström, 2005).
Another functional class identified through our predictions relates to intracellular lipid transport. Lipids are essential for the homeoviscous adaptation (HVA) enabling cells to adjust to developmental, physiological and environmental changes. Two genes, BGLB017474 and BGLB00786, correspond to a bridge-like lipid transfer protein member 1 (BLTP1) and ATP-binding cassette sub-family D respectively. The BLTP protein family has been characterized for its ability to connect two organelle membranes facilitating non-vesicular lipid transfer in vitro (Neuman et al., 2022). BLTP1 is recognized by the HUGO Gene Nomenclature Committee and is found in both vertebrates (known as KIAA1109 in humans) and in invertebrates (as ldp-3 in C. elegans and tweek in the genus Drosophila). In C. elegans, ldp-3 gene has been highlighted through CRISPR-GFP studies as a crucial link between plasma membrane and the endoplasmic reticulum for lipid trafficking (Pandey et al., 2023). The second gene in this group, ABCD2-BGLB000786, is an ATP-binding cassette transporter and targeted by bgl-miR-8-3p. ABCD family transporters are known to be located in peroxisomal compartment and play a role in fatty acid metabolism by importing proteins (Ford and Beis, 2019; Parreira de Aquino et al., 2021). Studies have showed that depletion of ABCD2 lead to oxidative stress (Fourcade et al., 2009). Moreover, ABCD2 appears to share a similar function with ABCD1 in the import/catabolism of Very Long Chain of Fatty Acid (VLCFA) such as C26:0 (Hexacosanoic acid), where an excess can result in the production of ROS (Fourcade et al., 2010).
The last class of gene highlighted in our study pertains to immunity-related gene functions. One such gene is Synaptotagmin 7 (BGLB016762), which is predicted to be targeted by multiple miRNAs, including bgl-miR-981-3p, bgl-miR-2c-3p, and bgl-miR-9-5p. Notably, bgl-miR-9-5p is predicted to target Synaptotagmin 7 on both the 3′UTR and PCG regions. The Synaptotagmin family has been studied for their role in inhibition of cytokine secretions and phagocytosis in macrophages (Du et al., 2017). Additionally, in human thyroid cancer cells, it plays a role in regulation (cell migration and autophagy, processes regulated by miRNA-363-3p) (Zhang et al., 2024).
In general, most of the genes targeted by miRNA characterized in hemolymphatic compartment are involved in immune pathways. Bgl-miR-100-5p, bgl-miR-10a-5p, and bgl-miR-1985-5p target several glycosidase molecules found in the snail’s hemolymph, which can bind to oligosaccharides (such as mannose and galactose as terminal surface carbohydrates) present on the tegument of schistosome sporocysts (Zelck, 1999). Interestingly, the predicted miRNAs could regulate immune genes including toxins like biomphalysins known for their lytic activity against the membrane of the parasite S. mansoni (Galinier et al., 2013; Pinaud et al., 2021). Our predictions suggest that several genes encoding immune factors such as interleukin receptors (BGLB40335, BGLB31074, and BGLB007388) who are humoral factors and are targeted by bgl-miR-1985-5p, bgl-miR-981-3p, bgl-miR-449-5p or bgl-miR-184-3p in diverse locations (3′UTR, PCG, and 5′UTR). The interleukin factor has been directly associated with the ability of snail hemocytes to kill S. mansoni sporocysts (Granath Jr. et al., 2001). Moreover, others immune molecules such as toxins could be targeted by miRNAs. For instance, Biomphalysin 11 (BGLB000108) is targeted by bgl-miR-184-3p and bgl-miR-10a-5p; Biomphalysin 19 (BGLB000001) is targeted by bgl-miR-279-3p; Biomphalysin 18 (BGLB000010) is targeted by bgl-miR-216-5p; and Biomphalysin 17 (BGLB000119) is targeted bgl-miR-11705-5p. These miRNAs specifically target biomphalysin genes at their 5′UTR. These toxins are known to have cytotoxic activity on the sporocyst, and to be expressed in immune-competent cells (Galinier et al., 2013; Pinaud et al., 2021). Given that B. glabrata genome is not fully assembled, particularly for highly polymorphic and diversified multigenic molecules, RNA-seq studies which have been instrumental in reconstructing these molecules’ sequences specific to the BgBRE strain from Recife, Brazil (Dheilly et al., 2015), and in identifying scaffolds in B. glabrata genome BB02 (Duval et al., 2020), were used to identify if these immune molecules could be targeted by miRNAs.
3.4. Did FREP and TEP are targets for miRNAs?
FREPs (fibrinogen-related proteins) are a highly diversified family of immune pathogen recognition receptors (PRRs). The combination of one or two N-terminal immunoglobulin superfamily (IgSF) domain with a C-terminal fibrinogen-related (FBG) domain defines FREP molecule. Their roles in binding the parasite and leading the immune response for the host have been described and characterized (Leonard et al., 2001; Hertel et al., 2005; Hanington and Zhang, 2011; Galinier et al., 2017; Portet et al., 2017). Co-immunoprecipitation experiments have raised immune complex interactions that associate three partners: FREPs of B. glabrata, SmPoMucs of the parasite (polymorphic mucins) and TEPs (thioester-containing protein) molecules (Roger et al., 2008a, 2008b, 2008c; Portet et al., 2017). This antiprotease has been characterised to be involved in innate immune response of B. glabrata against the S. mansoni (Portet et al., 2018; Pinaud et al., 2019; Duval et al., 2020). Genes from the multigenic families FREPs and TEPs are not well annotated and fully assembled in B. glabrata genomes. Therefore, the domains of FREPs and TEPs molecules are predicted to be regulated by miRNAs. Notably, the immunoglobulin domain (IgSF), MAM domain, and LDL-receptor class A related to alpha 2-macroglobulin appear to be targeted by miRNAs such as bgl-miR-2001-5p, bgl-miR-29-3p, bgl-miR-12707-5p, and bel-miR-31-5p.
Thus, to assess whether miRNAs might target some members of the BgTEP and FREPs families, we selected their full-length assembled sequences, including the 5′UTR and 3′UTR if this information was available (Duval et al., 2020) (Table 5; Supplementary file 1: Table S2) for inclusion in the present analysis.
Table 5.
Thioester-containing proteins (TEPs) targeted by miRNAs in hemolymph of Biomphalaria glabrata (PCG: Protein Coding Gene).
| TEP name | Nos of localisation | miRNA | Localisation |
|---|---|---|---|
| bgA2M | 5 | bgl-miR-22707-5p | PCG |
| bgC3-2 | 1 | bgl-miR-252a-5p | PCG |
| bgC3-2 | 3 | bgl-miR-31-5p | PCG |
| bgTEP1 | 2 | bgl-miR-22707-5p | PCG |
| bgTEP2 | 3 | bgl-miR-252a-5p | PCG |
| bgTEP3 | 4 | bgl-miR-22707-5p | PCG |
| bgTEP3 | 3 | bgl-miR-8-3p | PCG |
| bgTEP3 | 10 | bgl-miR-96a-5p | PCG |
| bgTEP4 | 3 | bgl-miR-216b-5p | PCG |
| bgTEP4 | 8 | bgl-miR-96a-5p | PCG |
Members of the BgTEP family are targeted by several miRNAs, including the newly described bgl-miR-22707-5p. Some miRNAs exhibited multiple hits towards the same TEP gene; for example, bgl-miR-96a-5p had 10 hits targeting the PCG of BgTEP3 (Table 5). Among the FREPs sequences referring to BgBRE (strain from Recife, Brazil) (Supplementary file 1: Table S3), curiously, no common miRNAs were predicted to target FREPs by the four tools. This result may be explained by three hypotheses: (i) FREP expression may not be regulated by miRNAs; (ii) FREP may be regulated by another gene expression regulator; or (iii) miRNAs implicated in FREP regulation have not been detected in this study.
To go further, a comparative small RNA sequencing of hemocytes from different B. glabrata snail strains, infected or not, could provide valuable insights into the roles of miRNAs in host-pathogen interaction, similar to the approaches conducted for B. tenagophila highlighting the differential expression of miRNAs between infected/not infected but also between resistant/susceptible strains (Alves et al., 2023). Another prospect is that in response to the miRNAs produced by the hosts, the parasites themselves produce small RNAs to interfere potentially with host immunity (Meningher et al., 2016, 2020). Indeed, in the literature, it has been demonstrated that parasites, especially the intracellular or endo-parasites, can also utilize miRNAs to manipulate gene expression of their hosts, either by secreting their own miRNAs into host cells or by hijacking the host’s miRNAs for their benefit (Weiberg et al., 2013; Cui et al., 2019; Villares et al., 2020). For example, the intracellular parasite Nosema ceranae releases exogenous miRNAs that could target host mRNAs in honeybee cells (Fan et al., 2022). Surprisingly, similar strategies have been reported for endoparasites, i.e. Trichuris suis (Entwistle and Wilson, 2017), miRNAs and miRNAs from exosomes vesicles secreted by the nematode Heligmosomoides polygyrus have been detected in Sus scrofa domesticus (pig) (Hansen et al., 2015) and mouse cells, respectively (Buck et al., 2014). Interestingly, the miRNA sma-miR-10-5p, from S. mansoni is internalized in vertebrate host’s T-cells, where it targets MAP3K7 involved in NF-κB activity regulation, thereby disrupting Th2 lineage and impairing the host’s immune responses (Hamway et al., 2022). Also, the presence of two miRNAs from S. mansoni, sma-miR-bantam and sma-miR-10 has been observed in situ within the gastrointestinal tract and the mesenteric lymph nodes of its vertebrate murine host during chronic infection (Meningher et al., 2020). Additionally, these miRNAs have also been detected in T lymphocytes (both Th1 and Th2) isolated from exosomes released by adult worms in the serum of infected patients. The success of the parasite or its elimination by the host could therefore also involve parasite miRNAs altering host immune response, or impacting directly the host miRNAs involved in an effective defence against the pathogen (Cai et al., 2015; Zheng et al., 2013; Zhu et al., 2015a; Kifle et al., 2020; Mu et al., 2021). This molecular dialogue between Biomphalaria spp. and Schistosoma spp. remains to be elucidated even though some indications suggest that following parasite exposure, some parasite miRNAs are present in snails and may interfere with the snail immune response or with snail miRNAs abundance (Phan et al., 2024). For example, 24 h after S. mansoni infection in B. glabrata snails, 11 mature miRNAs from the parasite were detected, including sma-miR-2d-3p and sma-miR-190-3p (Portet et al., 2019). Notably, the sma-miR-190-3p was found in both sympatric and allopatric conditions and is predicted to target biomphalysin transcript (Portet et al., 2019).
4. Conclusions
Herein, we describe the miRNome expressed in the hemolymphatic compartment of the snail B. glabrata. This first description should lay the foundation for a deeper understanding of the snail’s immune response to pathogens. With this computational approach, we identified 63 miRNAs of which 25 new miRNAs. Several isomiRs have been founded such as bgl-miR-92-3p.1.1; bgl-miR-92a-3p.1.2 or bgl-miR-92a-3p.1.3. The miR-92 family is highly conserved in animals and known to regulate multiple immune pathways in organisms; it could be a good candidate to investigate the implications in the immunity in B. glabrata against its parasite S. mansoni. MiRNAs would play a key role in this interaction, drive immune responses and be involved in compatibility polymorphism. The newly identified miRNAs in our study suggest that specific targets related to immune pathways could be regulated by these small RNAs. Indeed, in our study, we predicted in silico the potential regulation of immune molecules by miRNAs identified. Humoral factors such as interleukin receptors are predicted to be target by several miRNAs (bgl-miR-1985-5p; bgl-miR-184-3p or bgl-miR-449-5p) who are highly expressed in hemolymph. Immune molecules such biomphalysins, BgTEP and others have been identified as potential targets for miRNAs with multiples sites of binding. These discoveries could lead to immune regulation mechanisms studies by miRNAs in B. glabrata. To further investigate their roles in the snail-parasite interaction, we propose to elucidate their specific functions and effects on snail extreme-selected phenotypes using antagomiR or mimicmiR in snails in response to S. mansoni. Knock-down experiments would reveal whether specific miRNAs could be responsible in an efficient immune response or if the parasite’s miRNAs could thwart the snail defence. Taken together, further investigations involving a well-characterized miRNome of the host are essential to determine whether the findings in vertebrate systems also apply to invertebrate systems. Indeed, a key question has been raised as to whether parasite miRNAs might function as xeno-miRNAs to circumvent host immunity. We consider that the Biomphalaria-parasite molecular dialogue could play a significant role in their interaction, establishing a specific molecular cross-talk through the modulation of key gene players.
CRediT authorship contribution statement
Sarah Dametto: Conceptualization, Methodology, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing. Benjamin Gourbal: Conceptualization, Writing – original draft, Writing – review & editing. Cristian Chaparro: Methodology, Data curation, Writing – review & editing. Silvain Pinaud: Conceptualization, Investigation, Writing – review & editing. David Duval: Conceptualization, Investigation, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition.
Ethical approval
Not applicable.
Funding
This work was funded by BQR (Name: mimicSNAIL) from UPVD. This study is set within the framework of the “Laboratoires d’Excellences (LABEX)” TULIP (ANR-10-LABX-41) and CeMEB (ANR-10-LABX-04-01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors want to thank Damien Pouzol and Olivier Portela for the snail breeding facilities. The authors are grateful to Pierre-Vincent Bonhoure for his help in correcting the English. We acknowledge Damien Lassalle and Gurvan Meignen for their work performed during their MSc internships.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crpvbd.2025.100262.
Contributor Information
Sarah Dametto, Email: sarah.dametto@univ-perp.fr.
David Duval, Email: david.duval@univ-perp.fr.
Appendix. ASupplementary data
The following are the Supplementary data to this article.
Data availability
The data supporting the conclusions of this article are included within the article and its supplementary files.
References
- Adema C.M., Hillier L.W., Jones C.S., Loker E.S., Knight M., Minx P., et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 2017;8 doi: 10.1038/ncomms15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M.M., Micolucci L., Islam M.S., Olivieri F., Procopio A.D. In: MicroRNA Target Identification, Methods in Molecular Biology. Laganà A., editor. Springer; New York, New York, NY: 2019. A practical guide to miRNA target prediction; pp. 1–13. [DOI] [PubMed] [Google Scholar]
- Allan E.R.O., Yang L., Tennessen J.A., Blouin M.S. Allelic variation in a single genomic region alters the hemolymph proteome in the snail Biomphalaria glabrata. Fish Shellfish Immunol. 2019;88:301–307. doi: 10.1016/j.fsi.2019.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves T.C., Queiroz F.R., De Melo Neto A.B., Da Rocha Fernandes G., Pais F.S.-M., De Jesus Jeremias W., et al. Identification and characterization of microRNAs in Biomphalaria tenagophila and comparative analysis of their expression in Schistosoma mansoni-resistant and -susceptible snail populations. Gene. 2023;884 doi: 10.1016/j.gene.2023.147742. [DOI] [PubMed] [Google Scholar]
- Aspenström P. The verprolin family of proteins: regulators of cell morphogenesis and endocytosis. FEBS Lett. 2005;579:5253–5259. doi: 10.1016/j.febslet.2005.08.053. [DOI] [PubMed] [Google Scholar]
- Axtell M.J. ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri E., Trizzino M., Welsh S.A., Owens T.A., Calabretta B., Carroll M., et al. Targeted enhancer activation by a subunit of the integrator complex. Mol. Cell. 2018;71:103–116.e7. doi: 10.1016/j.molcel.2018.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch P.F. Intermediate host specificity in Schistosoma mansoni. Exp. Parasitol. 1976;39:150–169. doi: 10.1016/0014-4894(76)90022-9. [DOI] [PubMed] [Google Scholar]
- Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- Bolin K., Rachmaninoff N., Moncada K., Pula K., Kennell J., Buttitta L. miR-8 modulates cytoskeletal regulators to influence cell survival and epithelial organization in Drosophila wings. Dev. Biol. 2016;412:83–98. doi: 10.1016/j.ydbio.2016.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck A.H., Coakley G., Simbari F., McSorley H.J., Quintana J.F., Le Bihan T., et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 2014;5:5488. doi: 10.1038/ncomms6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budak H., Bulut R., Kantar M., Alptekin B. MicroRNA nomenclature and the need for a revised naming prescription. Brief Funct. Genomics. 2016;15:65–71. doi: 10.1093/bfgp/elv026. [DOI] [PubMed] [Google Scholar]
- Burgos-Aceves M.A., Cohen A., Smith Y., Faggio C. A potential microRNA regulation of immune-related genes in invertebrate haemocytes. Sci. Total Environ. 2018;621:302–307. doi: 10.1016/j.scitotenv.2017.11.285. [DOI] [PubMed] [Google Scholar]
- Cai P., Gobert G.N., You H., Duke M., McManus D.P. Circulating miRNAs: Potential novel biomarkers for hepatopathology progression and diagnosis of schistosomiasis japonica in two murine models. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-Z., Schaffert S., Fragoso R., Loh C. Regulation of immune responses and tolerance: The microRNA perspective. Immunol. Rev. 2013;253:112–128. doi: 10.1111/imr.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.J., Dixon J.E., Manning G. Genomics and evolution of protein phosphatases. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aag1796. [DOI] [PubMed] [Google Scholar]
- Chen Y., Stallings R.L. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–983. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- Chen Y.-W., Guo T., Shen L., Wong K.-Y., Tao Q., Choi W.W.L., et al. Receptor-type tyrosine-protein phosphatase κ directly targets STAT3 activation for tumor suppression in nasal NK/T-cell lymphoma. Blood. 2015;125:1589–1600. doi: 10.1182/blood-2014-07-588970. [DOI] [PubMed] [Google Scholar]
- Coustau C., Gourbal B., Duval D., Yoshino T.P., Adema C.M., Mitta G. Advances in gastropod immunity from the study of the interaction between the snail Biomphalaria glabrata and its parasites: A review of research progress over the last decade. Fish Shellfish Immunol. 2015;46:5–16. doi: 10.1016/j.fsi.2015.01.036. [DOI] [PubMed] [Google Scholar]
- Craig S.E.L., Brady-Kalnay S.M. Regulation of development and cancer by the R2B subfamily of RPTPs and the implications of proteolysis. Semin. Cell Dev. Biol. 2015:108–118. doi: 10.1016/j.semcdb.2014.09.004. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C., Wang Y., Liu J., Zhao J., Sun P., Wang S. A fungal pathogen deploys a small silencing RNA that attenuates mosquito immunity and facilitates infection. Nat. Commun. 2019;10:4298. doi: 10.1038/s41467-019-12323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson C.J., Tirouvanziam R., Herzenberg L.A., Lipsick J.S. Functional evolution of the vertebrate Myb gene family. Genetics. 2005;169:215–229. doi: 10.1534/genetics.104.034132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhahbi J.M., Atamna H., Li R., Yamakawa A., Guerrero N., Lam H.T., Mote P., Spindler S.R. MicroRNAs circulate in the hemolymph of Drosophila and accumulate relative to tissue microRNAs in an age-dependent manner. Genomics Insights. 2016;9 doi: 10.4137/GEI.S38147. GEI.S38147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheilly N.M., Duval D., Mouahid G., Emans R., Allienne J.-F., Galinier R., et al. A family of variable immunoglobulin and lectin domain containing molecules in the snail Biomphalaria glabrata. Dev. Comp. Immunol. 2015;48:234–243. doi: 10.1016/j.dci.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T., Copley R.R., Schultz J., Ponting C.P., Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Wang Y., Zhang F., Yan S., Guan Y., Gong X., et al. Synaptotagmin-11 inhibits cytokine secretion and phagocytosis in microglia. Glia. 2017;65:1656–1667. doi: 10.1002/glia.23186. [DOI] [PubMed] [Google Scholar]
- Duval D., Pichon R., Lassalle D., Laffitte M., Gourbal B., Galinier R. A new assessment of thioester-containing proteins diversity of the freshwater snail Biomphalaria glabrata. Genes. 2020;11:69. doi: 10.3390/genes11010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Taweel H.A., Issa Y.A., Mady R.F., Shehata G.A., Youssef E.A., Tolba M.M. Expression of microRNA-223 and microRNA-146b in serum and liver tissue of mice infected with Schistosoma mansoni. Parasitol. Res. 2022;121:1963–1972. doi: 10.1007/s00436-022-07542-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entwistle L.J., Wilson M.S. Micro RNA‐mediated regulation of immune responses to intestinal helminth infections. Parasite Immunol. 2017;39 doi: 10.1111/pim.12406. [DOI] [PubMed] [Google Scholar]
- Faggins A. Elucidation of the role of miR-184 in the development and maintenance of the Drosophila melanogaster post-embryonic nervous system. 2013. https://www.proquest.com/openview/7a486b192b056a0cff7133ddf04effbb/1?pq-origsite=gscholar&cbl=18750
- Fan X., Zhang W., Zhang Kaiyao, Zhang J., Long Q., et al. In-depth investigation of microRNA-mediated cross-kingdom regulation between Asian honeybee and microsporidian. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford R.C., Beis K. Learning the ABCs one at a time: Structure and mechanism of ABC transporters. Biochem. Soc. Trans. 2019;47:23–36. doi: 10.1042/BST20180147. [DOI] [PubMed] [Google Scholar]
- Fourcade S., Ruiz M., Camps C., Schlüter A., Houten S.M., Mooyer P.A.W., et al. A key role for the peroxisomal ABCD2 transporter in fatty acid homeostasis. Am. J. Physiol. Endocrinol. Metab. 2009;296:E211–E221. doi: 10.1152/ajpendo.90736.2008. [DOI] [PubMed] [Google Scholar]
- Fourcade S., Ruiz M., Guilera C., Hahnen E., Brichta L., Naudi A., et al. Valproic acid induces antioxidant effects in X-linked adrenoleukodystrophy. Hum. Mol. Genet. 2010;19:2005–2014. doi: 10.1093/hmg/ddq082. [DOI] [PubMed] [Google Scholar]
- Friedländer M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinier R., Portela J., Moné Y., Allienne J.F., Henri H., Delbecq S., et al. Biomphalysin, a new β pore-forming toxin Involved in Biomphalaria glabrata immune defense against Schistosoma mansoni. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinier R., Roger E., Moné Y., Duval D., Portet A., Pinaud S., et al. A multistrain approach to studying the mechanisms underlying compatibility in the interaction between Biomphalaria glabrata and Schistosoma mansoni. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T. Myb: A transcriptional activator linking proliferation and differentiation in hematopoietic cells. Curr. Opin. Genet. Dev. 1992;2:249–255. doi: 10.1016/S0959-437X(05)80281-3. [DOI] [PubMed] [Google Scholar]
- Granath W.O., Jr., Connors V.a., Raines A.E. Effects of exogenous interleukin-1β on primary sporocysts of Schistosoma mansoni (Trematoda) incubated with plasma and hemocytes from schistosome-susceptible and resistant Biomphalaria glabrata (Gastropoda) Invertebr. Biol. 2001;120:365–371. doi: 10.1111/j.1744-7410.2001.tb00044.x. [DOI] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32(Database issue) doi: 10.1093/nar/gkh023. 109D-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubitz A.K., Mourelatos Z., Abel L., Rappsilber J., Mann M., Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- Hadj-Moussa H., Hawkins L.J., Storey K.B. In: miRNomics: MicroRNA Biology and Computational Analysis, Methods in Molecular Biology. Allmer J., Yousef M., editors. Springer US; New York, NY: 2022. Role of microRNAs in extreme animal survival strategies; pp. 311–347. [DOI] [PubMed] [Google Scholar]
- Hamway Y., Zimmermann K., Blommers M.J.J., Sousa M.V., Häberli C., Kulkarni S., et al. Modulation of host-parasite interactions with small molecules targeting Schistosoma mansoni microRNAs. ACS Infect. Dis. 2022;8:2028–2034. doi: 10.1021/acsinfecdis.2c00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanington P.C., Lun C.-M., Adema C.M., Loker E.S. Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei. Int. J. Parasitol. 2010;40:819–831. doi: 10.1016/j.ijpara.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanington P.C., Zhang S.-M. The primary role of fibrinogen-related proteins in invertebrates is defense, not coagulation. J. Innate Immun. 2011;3:17–27. doi: 10.1159/000321882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen E.P., Kringel H., Williams A.R., Nejsum P. Secretion of RNA-containing extracellular vesicles by the porcine whipworm, Trichuris suis. J. Parasitol. 2015;101:336–340. doi: 10.1645/14-714.1. [DOI] [PubMed] [Google Scholar]
- Hassan M., Iqbal M.S., Naqvi S., Alashwal H., Moustafa A.A., Kloczkowski A. Prediction of site directed miRNAs as key players of transcriptional regulators against influenza C virus infection through computational approaches. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.866072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig A., Markwart R., Esparza-Franco M., Ladds G., Rubio I. Ras activation revisited: Role of GEF and GAP systems. Biol. Chem. 2015 doi: 10.1515/hsz-2014-0257. 0. [DOI] [PubMed] [Google Scholar]
- Hertel L.A., Adema C.M., Loker E.S. Differential expression of FREP genes in two strains of Biomphalaria glabrata following exposure to the digenetic trematodes Schistosoma mansoni and Echinostoma paraensei. Dev. Comp. Immunol. 2005;29:295–303. doi: 10.1016/j.dci.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Huang J.-Y., Kang S.-T., Chen I.-T., Chang L.-K., Lin S.-S., Kou G.-H., et al. Shrimp miR-10a is co-opted by white spot syndrome virus to increase viral gene expression and viral replication. Front. Immunol. 2017;8:1084. doi: 10.3389/fimmu.2017.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., O'Neill S.L., Asgari S. Wolbachia interferes with the intracellular distribution of Argonaute 1 in the dengue vector Aedes aegypti by manipulating the host microRNAs. RNA Biol. 2013;10:1868–1875. doi: 10.4161/rna.27392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im J., Kim H.-S. Genetic features of Haliotis discus hannai by infection of vibrio and virus. Genes Genomics. 2019;42 doi: 10.1007/s13258-019-00892-w. [DOI] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kifle D.W., Chaiyadet S., Waardenberg A.J., Wise I., Cooper M., Becker L., et al. Uptake of Schistosoma mansoni extracellular vesicles by human endothelial and monocytic cell lines and impact on vascular endothelial cell gene expression. Int. J. Parasitol. 2020;50:685–696. doi: 10.1016/j.ijpara.2020.05.005. [DOI] [PubMed] [Google Scholar]
- Kirstein N., Gomes Dos Santos H., Blumenthal E., Shiekhattar R. The Integrator complex at the crossroad of coding and noncoding RNA. Curr. Opin. Cell Biol. 2021;70:37–43. doi: 10.1016/j.ceb.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokaliaris C., Garba A., Matuska M., Bronzan R.N., Colley D.G., Dorkenoo A.M., et al. Effect of preventive chemotherapy with praziquantel on schistosomiasis among school-aged children in sub-Saharan Africa: A spatiotemporal modelling study. Lancet Infect. Dis. 2022;22:136–149. doi: 10.1016/S1473-3099(21)00090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.J., Hyun S. Multiple targets of the microRNA miR-8 contribute to immune homeostasis in Drosophila. Dev. Comp. Immunol. 2014;45:245–251. doi: 10.1016/j.dci.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Leonard P.M., Adema C.M., Zhang S.-M., Loker E.S. Structure of two FREP genes that combine IgSF and fibrinogen domains, with comments on diversity of the FREP gene family in the snail Biomphalaria glabrata. Gene. 2001;269:155–165. doi: 10.1016/s0378-1119(01)00444-9. [DOI] [PubMed] [Google Scholar]
- Li C., Gao Y., Zhang K., Chen J., Han S., Feng B., et al. Multiple roles of MicroRNA-100 in human cancer and its therapeutic potential. Cell. Physiol. Biochem. 2015;37:2143–2159. doi: 10.1159/000438572. [DOI] [PubMed] [Google Scholar]
- Li H., Gharamah A.A., Hambrook J.R., Wu X., Hanington P.C. Single-cell RNA-seq profiling of individual Biomphalaria glabrata immune cells with a focus on immunologically relevant transcripts. Immunogenetics. 2022;74:77–98. doi: 10.1007/s00251-021-01236-3. [DOI] [PubMed] [Google Scholar]
- Li P., Peng J., Hu J., Xu Z., Xie W., Yuan L. Localized expression pattern of miR-184 in Drosophila. Mol. Biol. Rep. 2011;38:355–358. doi: 10.1007/s11033-010-0115-1. [DOI] [PubMed] [Google Scholar]
- Li S., Shen L., Sun L., Xu J., Jin P., Chen L., Ma F. Small RNA-Seq analysis reveals microRNA-regulation of the Imd pathway during Escherichia coli infection in Drosophila. Dev. Comp. Immunol. 2017;70:80–87. doi: 10.1016/j.dci.2017.01.008. [DOI] [PubMed] [Google Scholar]
- Li X., Gao F., Fan Y., Xie S., Li C., Meng L., et al. A novel identified circ-ANKHD1 targets the miR-27a-3p/SFRP1 signaling pathway and modulates the apoptosis of granulosa cells. Environ. Sci. Pollut. Res. 2021;28:57459–57469. doi: 10.1007/s11356-021-14699-4. [DOI] [PubMed] [Google Scholar]
- Li X.-J., Luo X.-Q., Han B.-W., Duan F.-T., Wei P.-P., Chen Y.-Q. MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br. J. Cancer. 2013;109:2189–2198. doi: 10.1038/bjc.2013.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M.E., Gilman A.G. G proteins. Sci. Am. 1992;267:56–65. doi: 10.1038/scientificamerican0792-56. [DOI] [PubMed] [Google Scholar]
- Ling L., Kokoza V.A., Zhang C., Aksoy E., Raikhel A.S. MicroRNA-277 targets insulin-like peptides 7 and 8 to control lipid metabolism and reproduction in Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. USA. 2017;114:E8017–E8024. doi: 10.1073/pnas.1710970114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingjie M., Yuanpu Q., Ke D., Xiaoyu Z. Lnc01296 regulates apoptosis genes birc2 and bak1 by targeting mirna-29c and participates in neuroprotection during cerebral ischemia-reperfusion injury in rats. Turk. Neurosurg. 2021 doi: 10.5137/1019-5149.JTN.36564-21.1. [DOI] [PubMed] [Google Scholar]
- Lo N.C., Bezerra F.S.M., Colley D.G., Fleming F.M., Homeida M., Kabatereine N., et al. Review of 2022 WHO guidelines on the control and elimination of schistosomiasis. Lancet Infect. Dis. 2022;22:e327–e335. doi: 10.1016/S1473-3099(22)00221-3. [DOI] [PubMed] [Google Scholar]
- Loher P., Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics. 2012;28:3322–3323. doi: 10.1093/bioinformatics/bts615. [DOI] [PubMed] [Google Scholar]
- Lorick K.L., Jensen J.P., Fang S., Ong A.M., Hatakeyama S., Weissman A.M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Bu L., Zhang S.-M., Buddenborg S.K., Loker E.S. An overview of transcriptional responses of Schistosome-susceptible (M line) or -resistant (BS-90) Biomphalaria glabrata exposed or not to Schistosoma mansoni infection. Front. Immunol. 2022;12 doi: 10.3389/fimmu.2021.805882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Loker E.S., Adema C.M., Zhang S.-M., Bu L. Genomic and transcriptional analysis of genes containing fibrinogen and IgSF domains in the schistosome vector Biomphalaria glabrata, with emphasis on the differential responses of snails susceptible or resistant to Schistosoma mansoni. PLoS Negl. Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Zhang P.-J., Li C.-H., Lv Z.-M., Zhang W.-W., Jin C.-H. miRNA-133 augments coelomocyte phagocytosis in bacteria-challenged Apostichopus japonicus via targeting the TLR component of IRAK-1 in vitro and in vivo. Sci. Rep. 2015;5 doi: 10.1038/srep12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M.Y., Chtarbanova S. The role of micro RNAs (miRNAs) in the regulation of Drosophila melanogaster's innate immunity. Fly (Austin) 2022;16:382–396. doi: 10.1080/19336934.2022.2149204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Liu L., Zhao Y., Yang L., Chen C., Li Z., Lu Z. JNK pathway plays a key role in the immune system of the pea aphid and is regulated by microRNA-184. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Gómez L., Villalba A., Abollo E. Identification and expression of immune genes in the flat oyster Ostrea edulis in response to bonamiosis. Gene. 2012;492:81–93. doi: 10.1016/j.gene.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Martín-Gómez L., Villalba A., Kerkhoven R.H., Abollo E. Role of microRNAs in the immunity process of the flat oyster Ostrea edulis against bonamiosis. Infect. Genet. Evol. 2014;27:40–50. doi: 10.1016/j.meegid.2014.06.026. [DOI] [PubMed] [Google Scholar]
- Mendes T.M.F., Carrilho E., Afonso A.J.P.F.S., Galinaro C.A., Cabral F.J., Allegretti S.M. Proteomic, metabolic and immunological changes in Biomphalaria glabrata infected with Schistosoma mansoni. Int. J. Parasitol. 2019;49:1049–1060. doi: 10.1016/j.ijpara.2019.08.001. [DOI] [PubMed] [Google Scholar]
- Meningher T., Barsheshet Y., Ofir‐Birin Y., Gold D., Brant B., Dekel E., et al. Schistosomal extracellular vesicle‐enclosed miRNAs modulate host T helper cell differentiation. EMBO Rep. 2020;21 doi: 10.15252/embr.201947882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meningher T., Lerman G., Regev-Rudzki N., Gold D., Ben-Dov I., Sidi Y., et al. Schistosomal miRNAs isolated from extracellular vesicles in sera of infected patients; a new tool for diagnosis and follow-up of human schistosomiasis. J. Infect. Dis. 2016 doi: 10.1093/infdis/jiw539. [DOI] [PubMed] [Google Scholar]
- Mitta G., Gourbal B., Grunau C., Knight M., Bridger J.M., Théron A. The compatibility between Biomphalaria glabrata snails and Schistosoma mansoni. Adv. Parasitol. 2017;97:111–145. doi: 10.1016/bs.apar.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Moreira R., Romero A., Rey-Campos M., Pereiro P., Rosani U., Novoa B., Figueras A. Stimulation of Mytilus galloprovincialis hemocytes with different immune challenges induces differential transcriptomic, miRNomic, and functional responses. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.606102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morga B., Arzul I., Faury N., Segarra A., Chollet B., Renault T. Molecular responses of Ostrea edulis haemocytes to an in vitro infection with Bonamia ostreae. Dev. Comp. Immunol. 2011;35:323–333. doi: 10.1016/j.dci.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Morga B., Renault T., Faury N., Arzul I. New insights in flat oyster Ostrea edulis resistance against the parasite Bonamia ostreae. Fish Shellfish Immunol. 2012;32:958–968. doi: 10.1016/j.fsi.2012.01.026. [DOI] [PubMed] [Google Scholar]
- Mu N., Gu J., Huang T., Zhang C., Shu Z., Li M., et al. A novel NF-κB/YY1/microRNA-10a regulatory circuit in fibroblast-like synoviocytes regulates inflammation in rheumatoid arthritis. Sci. Rep. 2016;6 doi: 10.1038/srep20059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y., McManus D.P., Gordon C.A., Cai P. Parasitic helminth-derived microRNAs and extracellular vesicle cargos as biomarkers for helminthic infections. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.708952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenger J.L., Zeidler M.P., Fragiadaki M. Evaluating the molecular properties and function of ANKHD1, and its role in cancer. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms241612834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P., Kuttenkeuler D., Gesellchen V., Zeidler M.P., Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- Mustard J.A., Beggs K.T., Mercer A.R. Molecular biology of the invertebrate dopamine receptors. Arch. Insect Biochem. Physiol. 2005;59:103–117. doi: 10.1002/arch.20065. [DOI] [PubMed] [Google Scholar]
- Neilsen C.T., Goodall G.J., Bracken C.P. IsomiRs – the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012;28:544–549. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Neuman S.D., Levine T.P., Bashirullah A. A novel superfamily of bridge-like lipid transfer proteins. Trends Cell Biol. 2022;32:962–974. doi: 10.1016/j.tcb.2022.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D., Li Z., Du Y., He S., Jiale L.J. Identification of a dopamine receptor in Sinonovacula constricta and its antioxidant responses. Dev. Comp. Immunol. 2019;103 doi: 10.1016/j.dci.2019.103512. [DOI] [PubMed] [Google Scholar]
- O'Connell R.M., Rao D.S., Chaudhuri A.A., Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- Oliveira S.D., Schwartz A., Williams D., Bonini M., Silva C.L.M., Minshall R.D. Abstract P138: Schistosoma mansoni-induced shedding of extracellular vesicles from lung microvascular endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2021;41(Suppl. 1) doi: 10.1161/atvb.41.suppl_1.P138. [DOI] [Google Scholar]
- Pacheco A., De Quinto S.L., Ramajo J., Fernández N., Martínez-Salas E. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res. 2009;37:582–590. doi: 10.1093/nar/gkn979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey T., Zhang J., Wang B., Ma D.K. Bridge-like lipid transfer proteins (BLTPs) in C. elegans: From genetics to structures and functions. Contact (Thousand Oaks) 2023;6 doi: 10.1177/25152564231186489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parreira de Aquino G., Mendes Gomes M.A., Köpke Salinas R., Laranjeira-Silva M.F. Lipid and fatty acid metabolism in trypanosomatids. Microb. Cell. 2021;8:262–275. doi: 10.15698/mic2021.11.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan P., Fogarty C.E., Eamens A.L., Duke M.G., McManus D.P., Wang T., Cummins S.F. ARGONAUTE2 localizes to sites of sporocysts in the Schistosome-infected snail, Biomphalaria glabrata. Genes. 2024;15:1023. doi: 10.3390/genes15081023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon R., Pinaud S., Vignal E., Chaparro C., Pratlong M., Portet A., et al. Single cell RNA sequencing reveals hemocyte heterogeneity in Biomphalaria glabrata: Plasticity over diversity. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.956871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pila E.A., Peck S.J., Hanington P.C. The protein pheromone temptin is an attractant of the gastropod Biomphalaria glabrata. J. Comp. Physiol. 2017;203:855–866. doi: 10.1007/s00359-017-1198-0. [DOI] [PubMed] [Google Scholar]
- Pinaud S., Portela J., Duval D., Nowacki F.C., Olive M.-A., Allienne J.-F., et al. A shift from cellular to humoral responses contributes to innate immune memory in the vector snail Biomphalaria glabrata. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud S., Portet A., Allienne J.-F., Belmudes L., Saint-Beat C., Arancibia N., et al. Molecular characterisation of immunological memory following homologous or heterologous challenges in the schistosomiasis vector snail, Biomphalaria glabrata. Dev. Comp. Immunol. 2019;92:238–252. doi: 10.1016/j.dci.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Pinaud S., Tetreau G., Poteaux P., Galinier R., Chaparro C., Lassalle D., et al. New insights into biomphalysin gene family diversification in the vector snail Biomphalaria glabrata. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.635131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portet A., Galinier R., Pinaud S., Portela J., Nowacki F., Gourbal B., Duval D. BgTEP: An antiprotease involved in innate immune sensing in Biomphalaria glabrata. Front. Immunol. 2018;9:1206. doi: 10.3389/fimmu.2018.01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portet A., Pinaud S., Chaparro C., Galinier R., Dheilly N.M., Portela J., et al. Sympatric versus allopatric evolutionary contexts shape differential immune response in Biomphalaria/Schistosoma interaction. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portet A., Pinaud S., Tetreau G., Galinier R., Cosseau C., Duval D., et al. Integrated multi-omic analyses in Biomphalaria-Schistosoma dialogue reveal the immunobiological significance of FREP-SmPoMuc interaction. Dev. Comp. Immunol. 2017;75:16–27. doi: 10.1016/j.dci.2017.02.025. [DOI] [PubMed] [Google Scholar]
- Puthiyedth N., Riveros C., Berretta R., Moscato P. Identification of differentially expressed genes through integrated study of Alzheimer's disease affected brain regions. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Zhang Y., Zhou X., Zhu F. MicroRNA-125 modulates hemocyte proliferation through regulating Astakine expression and participate the disease resistant in Scylla paramamosain. Aquaculture. 2024;582 doi: 10.1016/j.aquaculture.2023.740491. [DOI] [Google Scholar]
- Queiroz F.R., Portilho L.G., Jeremias W.D.J., Babá É.H., Do Amaral L.R., Silva L.M., et al. Deep sequencing of small RNAs reveals the repertoire of miRNAs and piRNAs in Biomphalaria glabrata. Mem. Inst. Oswaldo Cruz. 2020;115 doi: 10.1590/0074-02760190498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz F.R., Silva L.M., Jeremias W.D.J., Babá É.H., Caldeira R.L., Coelho P.M.Z., Gomes M.D.S. Differential expression of small RNA pathway genes associated with the Biomphalaria glabrata/Schistosoma mansoni interaction. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffo-Campos Á., Riquelme I., Brebi-Mieville P. Tools for sequence-based miRNA target prediction: What to choose? Int. J. Mol. Sci. 2016;17:1987. doi: 10.3390/ijms17121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger E., Gourbal B., Grunau C., Pierce R.J., Galinier R., Mitta G. Expression analysis of highly polymorphic mucin proteins (Sm PoMuc) from the parasite Schistosoma mansoni. Mol. Biochem. Parasitol. 2008;157:217–227. doi: 10.1016/j.molbiopara.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Roger E., Grunau C., Pierce R.J., Hirai H., Gourbal B., Galinier R., et al. Controlled chaos of polymorphic mucins in a metazoan parasite (Schistosoma mansoni) interacting with its invertebrate host (Biomphalaria glabrata) PLoS Negl. Trop. Dis. 2008;2 doi: 10.1371/journal.pntd.0000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger E., Mitta G., Moné Y., Bouchut A., Rognon A., Grunau C., et al. Molecular determinants of compatibility polymorphism in the Biomphalaria glabrata/Schistosoma mansoni model: new candidates identified by a global comparative proteomics approach. Mol. Biochem. Parasitol. 2008;157:205–216. doi: 10.1016/j.molbiopara.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Sansores-Garcia L., Atkins M., Moya I.M., Shahmoradgoli M., Tao C., Mills G.B., Halder G. Mask is required for the activity of the Hippo pathway effector yki/YAP. Curr. Biol. 2013;23:229–235. doi: 10.1016/j.cub.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Shekhar M.S., Karthic K., Kumar K.V., Kumar J.A., Swathi A., Hauton C., et al. Comparative analysis of shrimp (Penaeus vannamei) miRNAs expression profiles during WSSV infection under experimental conditions and in pond culture. Fish Shellfish Immunol. 2019;93:288–295. doi: 10.1016/j.fsi.2019.07.057. [DOI] [PubMed] [Google Scholar]
- Simphor E., Rognon A., Vignal E., Henry S., Allienne J.-F., Turtoi A., et al. Combining a transcriptomic approach and a targeted metabolomics approach for deciphering the molecular bases of compatibility phenotype in the snail Biomphalaria glabrata toward Schistosoma mansoni. Acta Trop. 2024;255 doi: 10.1016/j.actatropica.2024.107212. [DOI] [PubMed] [Google Scholar]
- Slack C. Ras signaling in aging and metabolic regulation. Nutr. Healthy Aging. 2017;4:195–205. doi: 10.3233/NHA-160021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sminia T., Barendsen L. A comparative morphological and enzyme histochemical study on blood cells of the freshwater snails Lymnaea stagnalis, Biomphalaria glabrata, and Bulinus truncatus. J. Morphol. 1980;165:31–39. doi: 10.1002/jmor.1051650104. [DOI] [PubMed] [Google Scholar]
- Sun J., Chen Z., Tan X., Zhou F., Tan F., Gao Y., et al. MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinoma. Med. Oncol. 2013;30:411. doi: 10.1007/s12032-012-0411-9. [DOI] [PubMed] [Google Scholar]
- Tennessen J.A., Théron A., Marine M., Yeh J.-Y., Rognon A., Blouin M.S. Hyperdiverse gene cluster in snail host conveys resistance to human schistosome parasites. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théron A., Rognon A., Gourbal B., Mitta G. Multi-parasite host susceptibility and multi-host parasite infectivity: A new approach of the Biomphalaria glabrata/Schistosoma mansoni compatibility polymorphism. Infect. Genet. Evol. 2014;26:80–88. doi: 10.1016/j.meegid.2014.04.025. [DOI] [PubMed] [Google Scholar]
- Tong M.-H., Mitchell D.A., McGowan S.D., Evanoff R., Griswold M.D. Two miRNA clusters, Mir-17-92 (Mirc1) and Mir-106b-25 (Mirc3), are involved in the regulation of spermatogonial differentiation in mice. Biol. Reprod. 2012;86:1–10. doi: 10.1095/biolreprod.111.096313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trümbach D., Prakash N. The conserved miR-8/miR-200 microRNA family and their role in invertebrate and vertebrate neurogenesis. Cell Tissue Res. 2015;359:161–177. doi: 10.1007/s00441-014-1911-z. [DOI] [PubMed] [Google Scholar]
- Villares M., Berthelet J., Weitzman J.B. The clever strategies used by intracellular parasites to hijack host gene expression. Semin. Immunopathol. 2020;42:215–226. doi: 10.1007/s00281-020-00779-z. [DOI] [PubMed] [Google Scholar]
- Wang B., Du Z., Lin C., Liu D., Guo J., Shi J., Wang X. Comprehensive analysis of INTS family related to expression, prognosis, diagnosis and immune features in hepatocellular carcinoma. Heliyon. 2024;10 doi: 10.1016/j.heliyon.2024.e30244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. The essential role of G protein-coupled receptor (GPCR) signaling in regulating T cell immunity. Immunopharmacol. Immunotoxicol. 2018;40:187–192. doi: 10.1080/08923973.2018.1434792. [DOI] [PubMed] [Google Scholar]
- Wang S., Pan Y., Zhang R., Xu T., Wu W., Zhang R., et al. Hsa-miR-24-3p increases nasopharyngeal carcinoma radiosensitivity by targeting both the 3′UTR and 5′UTR of Jab1/CSN5. Oncogene. 2016;35:6096–6108. doi: 10.1038/onc.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhu F. MicroRNA-100 is involved in shrimp immune response to white spot syndrome virus (WSSV) and Vibrio alginolyticus infection. Sci. Rep. 2017;7 doi: 10.1038/srep42334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F.-M., Zhao H., Zhang Z., Kaloshian I., et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel R.P., Lesniewski M.L., Haviernik P., Kadereit S., Leahy P., Greco N.J., Laughlin M.J. microRNA 184 regulates expression of NFAT1 in umbilical cord blood CD4+ T cells. Blood. 2009;113:6648–6657. doi: 10.1182/blood-2008-09-181156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler B.M., Heimberg A.M., Moy V.N., Sperling E.A., Holstein T.W., Heber S., Peterson K.J. The deep evolution of metazoan microRNAs. Evol. Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- Wu X.-J., Dinguirard N., Sabat G., Lui H., Gonzalez L., Gehring M., et al. Proteomic analysis of Biomphalaria glabrata plasma proteins with binding affinity to those expressed by early developing larval Schistosoma mansoni. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Yang L., Zhao Z., Wang J., Zhang X. Signature miRNAs involved in the innate immunity of invertebrates. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan D.-M., Xhu L.-X., Si D.-M., Liang Q.-H. MiR-5195-3p inhibits the proliferation of glioma cells by targeting BIRC2. Eur. Rev. Med. Pharmacol. Sci. 2020;24:267–273. doi: 10.26355/eurrev_202001_19921. [DOI] [PubMed] [Google Scholar]
- Yang L., Yang G., Zhang X. The miR-100-mediated pathway regulates apoptosis against virus infection in shrimp. Fish Shellfish Immunol. 2014;40:146–153. doi: 10.1016/j.fsi.2014.06.019. [DOI] [PubMed] [Google Scholar]
- Yang Y.L., Li X.M. The IAP family: Endogenous caspase inhibitors with multiple biological activities. Cell Res. 2000;10:169–177. doi: 10.1038/sj.cr.7290046. [DOI] [PubMed] [Google Scholar]
- Ying S.-Y., Chang D.C., Lin S.-L. The microRNA (miRNA): Overview of the RNA genes that modulate gene function. Mol. Biotechnol. 2008;38:257–268. doi: 10.1007/s12033-007-9013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.A., Biggins L., Sharpe H.J. Protein tyrosine phosphatases in cell adhesion. Biochem. J. 2021;478:1061–1083. doi: 10.1042/BCJ20200511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelck U.E. Glycosidase activities in plasma of naive and schistosome-infected Biomphalaria glabrata (Gastropoda) Parasitology. 1999;119:563–568. doi: 10.1017/S0031182099005028. [DOI] [PubMed] [Google Scholar]
- Zha W., Zhou L., Li S., Liu K., Yang G., Chen Z., et al. Characterization and comparative profiling of the small RNA transcriptomes in the hemipteran insect Nilaparvata lugens. Gene. 2016;595:83–91. doi: 10.1016/j.gene.2016.09.042. [DOI] [PubMed] [Google Scholar]
- Zhang J., Ren G., Huang T., Sang Y., Zhong Y., Yi Y. miRNA-363-3p hinders proliferation, migration, invasion and autophagy of thyroid cancer cells by controlling SYT1 transcription to affect NF-κB. Endocr. Metab. Immune Disord. Drug Targets. 2024;24:153–162. doi: 10.2174/1871530323666230504112553. [DOI] [PubMed] [Google Scholar]
- Zhao X., Yu H., Kong L., Liu S., Li Q. High throughput sequencing of small RNAs transcriptomes in two Crassostrea oysters identifies microRNAs involved in osmotic stress response. Sci. Rep. 2016;6 doi: 10.1038/srep22687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Cai X., Bradley J.E. microRNAs in parasites and parasite infection. RNA Biol. 2013;10:371–379. doi: 10.4161/rna.23716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.-S., Zhang H., Zhang X.-J., Feng D.-D., Luo X.-Q., Zeng C.-W., et al. MiR-100 regulates cell differentiation and survival by targeting RBSP3, a phosphatase-like tumor suppressor in acute myeloid leukemia. Oncogene. 2012;31:80–92. doi: 10.1038/onc.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Wang L., Song L., Liu R., Zhang H., Huang M., Chen H. The identification and characteristics of immune-related microRNAs in haemocytes of oyster Crassostrea gigas. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Dao J., Du X., Li H., Lu K., Liu J., Cheng G. Altered levels of circulating miRNAs are associated Schistosoma japonicum infection in mice. Parasites Vectors. 2015;8:196. doi: 10.1186/s13071-015-0806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Li X., Tian X., Wu C. Mask loss-of-function rescues mitochondrial impairment and muscle degeneration of Drosophila pink1 and parkin mutants. Hum. Mol. Genet. 2015;24:3272–3285. doi: 10.1093/hmg/ddv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the conclusions of this article are included within the article and its supplementary files.