Abstract
Background
Despite the critical role of endocytosis-related genes in oncogenic processes, research exploring their potential for prognosticating hepatocellular carcinoma (HCC) remains limited. Establishing a connection between endocytosis and HCC is imperative. This study aimed to create a gene signature related to endocytosis to identify HCC subtypes and predict outcomes.
Methods
RNA sequencing and clinical data of 371 HCC patients were obtained from The Cancer Genome Atlas (TCGA)-HCC dataset. Subtypes of HCC were identified through endocytosis-associated genes through consistent clustering analysis, and prognosis was assessed using an endocytosis-associated HCC model. Construction and validation of a prognostic endocytosis-related risk scoring system were created for HCC.
Results
A univariate Cox regression analysis was performed using the TCGA-HCC dataset, resulting in the identification of 4,354 genes significantly associated with patient prognosis. Subsequent Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of these genes identified several biologically relevant pathways, particularly those related to endocytosis, autophagy, and cell cycle regulation. Through the application of consensus clustering methods, patients with TCGA-HCC were stratified into two distinct subtypes based on a selection of 82 genes associated with endocytosis. Importantly, the overall survival rate for the high-risk subtype (C1) was significantly higher than that of the low-risk subtype (C2). KEGG analysis indicated that the upregulated genes in the high-risk C1 subtype were predominantly related to various pathways, including the p53 signaling pathway, proteoglycans in cancer, cell cycle regulation, interactions between the extracellular matrix and receptors, and cellular senescence. In contrast, in the comparison between the C1 and C2 HCC samples, the genes exhibiting downregulation were predominantly linked to metabolic pathways, including tyrosine metabolism and steroid hormone biosynthesis. Boxplots showed significant differences in immune cell populations, including CD4+ T lymphocytes, endothelial cells, natural killer cells, and macrophages. From a pool of 82 endocytosis-related genes, 14 genes were identified through least absolute shrinkage and selection operator and Cox regression, including CLTA, STAM, RAB10, DAB2, VPS45, AGAP3, ARPC4, VPS29, HSPA8, DNAJC6, PARD6B, ACTR3B, PSD4, and ARRB2. Based on these genetic markers, patients were stratified into low-risk and high-risk categories. The prognostic performance of the model was validated using receiver operating characteristic curve analysis, which produced area under the curve values of 0.807, 0.757, and 0.716 for 1-, 3-, and 5-year survival predictions, respectively. The model of endocytosis-related genes was validated by external International Cancer Genome Consortium (ICGC)-HCC datasets.
Conclusions
Genes linked to endocytosis strongly correlate with tumor classification in patients with HCC. The related expression profiles may be valuable for predicting HCC prognosis and informing diagnosis and treatment.
Keywords: Hepatocellular carcinoma (HCC), endocytosis, prognostic model, biomarkers
Highlight box.
Key findings
• Endocytosis is critical to the development and management of hepatocellular carcinoma (HCC), influencing processes such as nutrient absorption, receptor internalization, and signal transmission, making it a reliable prognostic marker.
What is known, and what is new?
• Assessment of genes involved in endocytosis in HCC is feasible.
• This study identified specific HCC subtypes and biomarkers associated with endocytosis.
What is the implication, and what should change now?
• Our predictive model for HCC, composed of endocytosis-related gene expression, could facilitate the creation of innovative targeted treatments.
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent primary hepatic malignancies, presenting substantial challenges to global health (1). The 5-year survival rate for patients with liver cancer serves as a crucial indicator for assessing treatment efficacy and prognostic outcomes among patients diagnosed with this condition (1-3). Research indicates that survival rates exhibit considerable variation due to influences from a diversity of factors, including demographic traits, clinical parameters, and treatment modalities (1-3). The therapeutic landscape for liver cancer is undergoing continuous transformation, driven by ongoing investigations into novel systemic therapies and combinations of locoregional treatments (2-4). The incorporation of molecular targeted therapies and immunotherapies, such as nivolumab and pembrolizumab, into treatment protocols is a focal point of current research, and has the potential to improve the outcomes of patients with advanced HCC (4). As the comprehension of the molecular mechanisms underlying liver cancer deepens, precision medicine approaches may facilitate the development of more personalized and efficacious treatment strategies.
Endocytosis is critically involved in HCC progression and treatment, affecting nutrient uptake, receptor internalization, and signaling (5-7). Its dysregulation is a cancer hallmark, impacting tumor growth, metastasis, and therapy response. The clathrin-mediated endocytosis pathway is vital for receptor and nutrient internalization, influencing cancer cell survival and proliferation; for example, hepatitis B virus (HBV) entry into HepG2-NTCP cells relies on this pathway (5). Similarly, clathrin-mediated endocytosis of epidermal growth factor receptor (EGFR) is key to modulating EGFR signaling and is often altered in HCC (6). Other endocytic processes, such as macropinocytosis, also contribute to HCC progression (6). In addition to clathrin-mediated pathways, macropinocytosis also promotes HCC progression by allowing cancer cells to absorb extracellular nutrients, supporting growth in nutrient-poor conditions. Phospholipid flippase ATP9A is crucial for regulating macropinocytosis in HCC, enhancing nutrient starvation tolerance and cancer cell proliferation (7). Endocytosis also influences immune responses in the tumor microenvironment, with the zinc transporter ZNT1 and Zn2+ levels in macrophages affecting Toll like receptor 4 (TLR4) and programmed cell death ligand 1 (PD-L1) endocytosis, thus promoting inflammation and immune suppression (8). This regulation can alter the efficacy of chemotherapy, indicating that targeting endocytic pathways can improve therapeutic outcomes. Additionally, modulating endocytosis can improve anticancer drug delivery and efficacy. Valproic acid, a histone deacetylase inhibitor, promotes the endocytosis of doxorubicin, enhancing its cytotoxicity in HCC cells. This synergy highlights the potential of pairing endocytosis-modulating agents with standard chemotherapy to enhance treatment efficacy (9).
HCC is a complex and heterogeneous disease, which presents significant challenges in clinical management, particularly in prognosis prediction and treatment stratification. The development of new molecular subtypes and prognostic models is crucial to address these challenges and improve patient outcomes (10-12). Recent studies have focused on identifying molecular markers and constructing predictive models that can better stratify patients based on their risk and guide personalized treatment strategies (10-12).
In this study, we systematically evaluated the molecular changes and clinical significance of endocytosis-associated genes through the construction of two unique molecular signatures. Our work is the first of its kind to design an endocytosis-based prognostic model for forecasting the survival of patients with HCC. This novel scoring framework offers a valuable clinical perspective that may enhance therapeutic decision-making and personalized treatment approaches. We present this article in accordance with the TRIPOD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-359/rc).
Methods
Acquisition of HCC data from The Cancer Genome Atlas (TCGA)
The RNA-sequencing datasets and the relevant clinical details from 371 patients with HCC were obtained from TCGA database (https://portal.gdc.cancer.gov/). From the TCGA database, we acquired STAR-counts data and clinical information for HCC, extracted transcripts per million (TPM) data, and used log2(TPM +1) normalization. For further analysis, we ultimately chose HCC samples after retaining those with RNAseq data and clinical information. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Identification of prognostic HCC-related genes
Through the implementation of a univariate Cox regression analysis, we successfully identified prognostic genes with a P value below 0.05 in individuals diagnosed with HCC. To further clarify the biological significance of these prognostic genes, we employed the “clusterProfiler” package in R (The R Foundation for Statistical Computing) to conduct enrichment analyses of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and sought to delineate the signaling pathways associated with the identified HCC prognostic genes.
Subtypes of HCC identified through endocytosis-associated genes
The relationship between genes associated with endocytosis and subtypes of HCC was examined. The “ConsensusClusterPlus” package (version 1.54.0) was applied to TCGA-HCC cohort for clustering analysis and to calculate the cumulative distribution function (CDF) and consensus matrix. The clustering variable (k) was tested across a range from 2 to 6, and a heatmap was created using the “Pheatmap” package (version 1.0.12). Additionally, the “GGalluvial” package in R was used to determine the associations between different subtypes, overall survival (OS) status, and risk scores.
Differentially expressed gene (DEG) and enrichment analyses of the HCC subtypes
The evaluation of gene expression variability across the different subtypes was performed via the “limma” package in R. Genes that exhibited a fold change greater than 2 and a P value less than 0.05 were classified as DEGs. Additionally, enrichment analyses were conducted for the KEGG and Gene Ontology (GO) through the “clusterProfiler” package to identify the functional pathways associated with the different subtypes.
Immune activity of two endocytosis-related clusters in HCC
Immunogenomic analysis was employed to evaluate the immune functionality of groups associated with endocytosis. To analyze the infiltration of immune cells and the activation of immune pathways across the two clusters, the Wilcoxon test was implemented. A P value <0.05 indicated statistical significance.
Construction and validation of a prognostic endocytosis-related risk scoring system for HCC
The genes associated with endocytosis that were correlated with survival outcomes were subsequently refined via the least absolute shrinkage and selection operator (LASSO) penalty method. The optimal parameter λ was determined through a 10-fold cross-validation process. The candidate genes identified were categorized into high- and low-expression groups based on the optimal cutoff values derived from the “survminer” package, and their prognostic significance was validated through survival analysis. Each cohort of patients was stratified into high-risk and low-risk groups according to the median values pertinent to the given dataset, and differences in survival rates were assessed with Kaplan-Meier survival curves. In addition, a time-dependent receiver operating characteristic (ROC) curve was developed to assess the sensitivity and specificity of the multigene risk score (MRS) across the different datasets. The model of endocytosis-related genes was validated by external International Cancer Genome Consortium (ICGC)-HCC datasets. The genes associated with endocytosis that were correlated with survival outcomes were further refined via the LASSO penalty method.
Statistical analysis
Statistical analyses were performed via R software version 3.62. The primary metric of this study was OS, which was considered to be the period from diagnosis to death. ROC curves were generated via the “survivalROC” package, and the nomogram was developed with the “rms” package. All tests were two-tailed, with the significance threshold being P<0.05 unless otherwise indicated.
Results
Prognostic gene signature analysis and pathway enrichment in HCC
In order to identify the prognostic biomarkers associated with HCC, univariate Cox regression modeling was first applied. This method effectively identified 4,352 genes that demonstrated significant correlations with survival in patients with HCC. The top 20 most significant genes are displayed in Figure 1A. Further analysis through KEGG pathway enrichment of these genes identified several biologically significant pathways that may contribute to the pathogenesis of HCC, including those involved in endocytosis, lysosomal function, autophagy, and cell cycle regulation (Figure 1B).
Figure 1.
Prognostic gene signature analysis and pathway enrichment in hepatocellular carcinoma. (A) Survival-associated genes in hepatocellular carcinoma. Number outside parentheses represents the point estimate of the hazard ratio and number in parentheses represents the 95% CI of the HR. (B) Functional pathway enrichment of prognostic genes in hepatocellular carcinoma. Data are presented as the number of survival genes enriched in the signaling pathway (statistical P value). CI, confidence interval; HR, hazard ratio.
HCC subtypes delineated through endocytosis-associated genes
A strong correlation between endocytosis and tumor progression has been established (7-9). However, the specific mechanisms by which endocytosis affects HCC remain unclear. To conduct a more refined classification of patients with HCC from TCGA, we performed consensus clustering analysis with the above-mentioned endocytosis-related genes.
The consistency cluster analysis conducted on the TCGA-HCC patient cohort delineated two distinct clusters (Figure 2A). Heatmap analyses revealed that the expression profiles of 82 endocytosis-associated genes exhibited significant differentiation between the two patient groups within TCGA lung adenocarcinoma (LUAD) cohort (Figure 2B). Notably, a statistically significant discrepancy in OS rates was observed between cluster 1 (C1) and cluster 2 (C2) [hazard ratio (HR) =2.0, 95% confidence interval (CI): 1.404–2.848; P<0.001; Figure 2C].
Figure 2.
Subtypes of hepatocellular carcinoma identified through endocytosis-associated genes. (A) Survival-associated genes in hepatocellular carcinoma. Purple indicates high clustering stability, white indicates poor clustering stability, and the higher the consensus score, the more stable it is. (B) A heatmap of endocytosis-related genes in hepatocellular carcinoma. (C) Kaplan-Meier survival curves of the two endocytosis-related clusters. C1, cluster 1; C2, cluster 2; CI, confidence interval; HR, hazard ratio.
DEG and functional enrichment analyses of the two HCC clusters
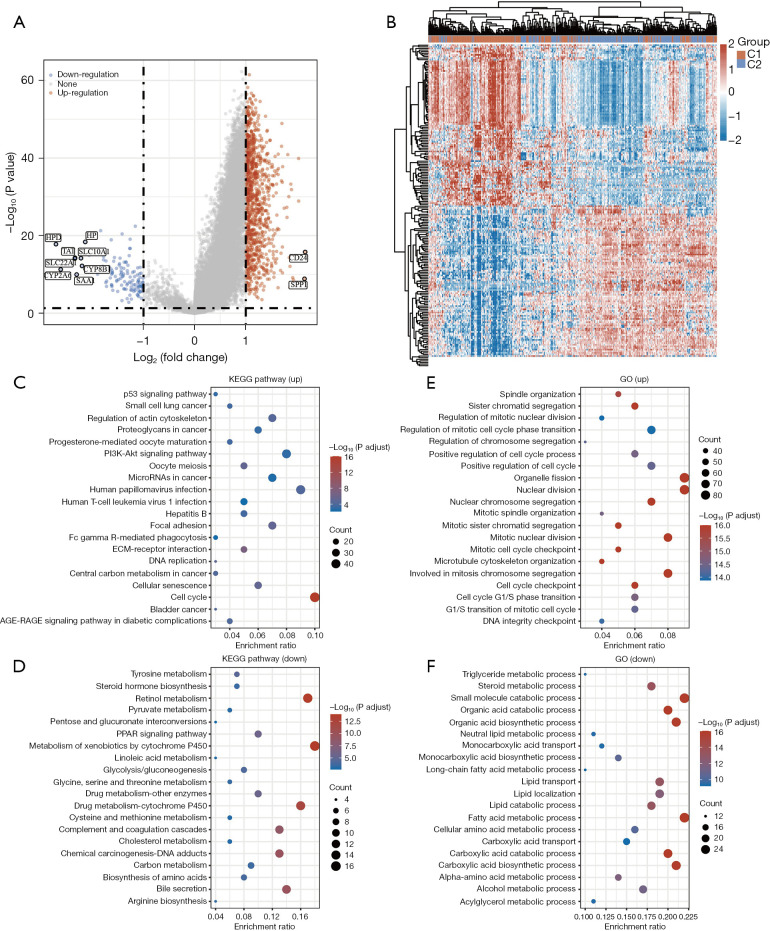
To clarify the underlying differences between the two HCC groups, we identified DEGs with P<0.05 and |log2 fold change| >1, as illustrated in the volcano plot (C1 vs. C2) (Figure 3A). The expression profiles of these DEGs revealed notable differences between the two clusters, as represented in the heatmap (Figure 3B).
Figure 3.
Differentially expressed gene and enrichment analyses of the HCC subtypes. (A) The volcano plot of the DEGs between C1 and C2 (the red color indicates upregulated genes, and the blue color indicates downregulated genes). (B) Heatmap of the DEGs between C1 and C2. (C) KEGG pathway enrichment analysis of the upregulated DEGs. (D) KEGG pathway enrichment analysis of the downregulated DEGs. (E) GO biological process enrichment for upregulated DEGs. (F) GO biological process enrichment for downregulated DEGs. C1, cluster 1; C2, cluster 2; DEG, differentially expressed gene; GO, Gene Ontology; HCC, hepatocellular carcinoma; KEGG, Kyoto Encyclopedia of Genes and Genomes.
To determine the biological implications of these DEGs, we conducted KEGG and GO enrichment analyses. The KEGG analysis revealed that the upregulated genes were related to a range of pathways, most notably the p53 signaling pathway, regulation of the actin cytoskeleton, proteoglycans in cancer, PI3K-Akt signaling pathway, cell cycle regulation, extracellular matrix (ECM)-receptor interaction, and cellular senescence (Figure 3C). In contrast, the downregulated genes were primarily associated with processes such as tyrosine metabolism, steroid hormone biosynthesis, retinol metabolism, PPAR signaling pathway, and the metabolism of xenobiotics via cytochrome P450 (Figure 3D).
The GO analysis of biological processes (BPs) indicated that the upregulated genes were significantly linked to critical cancer-related functions, such as spindle organization, sister chromatid segregation, positive regulation of cell cycle processes, cell cycle checkpoints, and chromosome segregation (Figure 3E). On the other hand, the downregulated genes were primarily associated with steroid metabolic processes, small molecule catabolic processes, organic acid catabolic processes, lipid catabolic processes, and alcohol metabolic processes (Figure 3F). Several studies have demonstrated that chromosome segregation, proteoglycans in cancer, and cell cycle checkpoints serve as tumor markers (13,14). Collectively, these findings suggest that tumor cells within the C1 HCC subtype may possess an enhanced capacity for tumor migration and proliferation.
Immune activity between two endocytosis-related groups in HCC
There is a robust correlation between endocytosis and immune function across different cancer types (7-9). In our study, we evaluated the immune responses within two distinct clusters related to endocytosis in patients diagnosed with HCC. Boxplots comparing the C1 and C2 HCC specimens revealed notable differences in the populations of immune cells, including CD4+ T cells, endothelial cells, natural killer (NK) cells, and macrophages (Figure 4). These results indicate that the mechanisms of endocytosis and immune activity are highly related.
Figure 4.

Comparison of the immune activity between the two endocytosis-related hepatocellular carcinoma groups. The enrichment scores of six distinct immune cell types in HCC were analyzed and compared between the two groups associated with endocytosis. *, P<0.05; **, P<0.01; ***, P<0.001; ns, not significant. C1, cluster 1; C2, cluster 2; EPIC, estimating the proportion of immune and cancer cells; HCC, hepatocellular carcinoma; NK, natural killer.
Construction of a prognostic model based on endocytosis-related genes
LASSO and Cox regression analyses identified 82 genes associated with endocytosis in HCC. A gene signature comprising 14 genes was established based on the optimal λ value (Figure 5A,5B). The risk score formula was as follows: risk score = 0.0433 × CLTA + 0.1331 × STAM + 0.0708 × RAB10 + 0.0271 × DAB2 + 0.0215 × VPS45 + 0.1411 × AGAP3 + 0.1564 × ARPC4 + 0.1143 × VPS29 + 0.14 × HSPA8 + 0.0389 × DNAJC6 + 0.0851 × PARD6B + 0.1783 × ACTR3B − 0.3408 × PSD4 + (5e−04) × ARRB2.
Figure 5.
Construction of a prognostic model based on endocytosis-related genes. (A) LASSO regression of endocytosis-related genes. (B) Cross-validation of the LASSO regression. (C) Patients with HCC from TCGA were stratified into low- (blue) and high-risk (red) groups based on the expression levels of endocytosis-related genes. (D) A Kaplan-Meier survival analysis was conducted to compare the survival outcomes between the high- and low-risk groups. (E) ROC curve analysis was performed to determine the AUC values for the stratification of risk groups. AUC, area under the curve; CI, confidence interval; HCC, hepatocellular carcinoma; HR, hazard ratio; ROC, receiver operating characteristic; TCGA, The Cancer Genome Atlas.
According to this gene signature, patients from the TCGA-HCC cohort were stratified into low- and high-risk categories (Figure 5C). Subsequent OS analysis indicated that individuals in the low-risk group exhibited superior survival outcomes as compared to those in the high-risk group (HR =2.554, 95% CI: 1.773–3.68; P<0.001; Figure 5D). To evaluate the predictive accuracy of the prognostic model, an ROC curve was generated, revealing AUC values of 0.807, 0.757, and 0.716 for 1-, 3-, and 5-year survival (Figure 5E), respectively, thus demonstrating the prognostic utility of the model.
Validation of a prognostic model based on endocytosis-related genes
LASSO and Cox regression analyses were further validated 14 genes associated with endocytosis in HCC. A gene signature comprising 6 genes was established. The risk score formula was as follows: risk score = 0.0965 × CLTA + 0.502 × RAB10 + 0.1751 × ARPC4 + 0.2106 × VPS29 + 0.1766 × ACTR3B − 0.1368 × ARRB2.
According to this gene signature, patients from the ICGC-HCC cohort were stratified into low- and high-risk categories (Figure 6A). Subsequent OS analysis indicated that individuals in the low-risk group exhibited superior survival outcomes as compared to those in the high-risk group (HR =3.233, 95% CI: 1.626–6.428; P<0.001; Figure 6B). To evaluate the predictive accuracy of the prognostic model, an ROC curve was generated, revealing AUC values of 0.729, 0.727, and 0.631 for 1-, 3-, and 5-year survival (Figure 6C).
Figure 6.
Validation of a prognostic model based on endocytosis-related genes. (A) Patients with HCC from ICGC were stratified into low- (blue) and high-risk (red) groups based on the expression levels of endocytosis-related genes. (B) A Kaplan-Meier survival analysis was conducted to compare the survival outcomes between the high- and low-risk groups. (C) ROC curve analysis was performed to determine the AUC values for the stratification of risk groups. AUC, area under the curve; CI, confidence interval; HCC, hepatocellular carcinoma; HR, hazard ratio; ROC, receiver operating characteristic; ICGC, International Cancer Genome Consortium.
Discussion
In this study, univariate Cox regression analysis was performed on TCGA-HCC dataset, resulting in the identification of 4,354 genes correlated with patient prognosis. The KEGG pathway enrichment analysis of these genes highlighted several biologically relevant pathways, notably those related to endocytosis, autophagy, and the regulation of the cell cycle. Using consensus clustering techniques, we categorized TCGA-HCC patients into two distinct subtypes based on a set of 82 genes associated with endocytosis. The OS of the high-risk subtype (C1) was found to be significantly superior to that of the low-risk subtype (C2). KEGG analysis indicated that the upregulated genes in the high-risk C1 subtype were associated with various pathways, including the p53 signaling pathway, proteoglycans in cancer, regulation of the cell cycle, interactions between the ECM and receptors, and cellular senescence. Conversely, the downregulated genes were primarily linked to metabolic processes such as tyrosine metabolism and steroid hormone biosynthesis. Boxplot analyses comparing the C1 and C2 HCC specimens revealed significant differences in immune cell populations, including CD4+ T lymphocytes, endothelial cells, NK cells, and macrophages. Through LASSO and Cox regression methodologies, 14 genes were selected from the 82 endocytosis-related genes, including CLTA, STAM, RAB10, DAB2, VPS45, AGAP3, ARPC4, VPS29, HSPA8, DNAJC6, PARD6B, ACTR3B, PSD4, and ARRB2. Based on these gene markers, patients were stratified into low-risk and high-risk categories. The prognostic performance of the model was validated via ROC curve analysis, yielding AUC values of 0.807, 0.757, and 0.716 for 1-, 3-, and 5-year survival, respectively.
Endocytosis is critically involved in the progression and treatment of HCC, influencing nutrient uptake, receptor internalization, and signaling. Its dysregulation is a hallmark of cancer, affecting tumor growth, metastasis, and therapy response. Moreover, clathrin-mediated endocytosis is critical for receptor and nutrient internalization, promoting cancer cell survival and proliferation. The expression of endocytosis-related genes can be significantly influenced by various factors, including the status of hepatitis virus infection, tumor stage, and treatment history. Hepatitis viruses, such as hepatitis C virus (HCV) and hepatitis B virus (HBV), have been shown to interact with host cell receptors and influence cellular processes, including endocytosis (15). For instance, HBV entry into HepG2-NTCP cells and EGFR signaling in HCC rely on this pathway. Additionally, macropinocytosis supports HCC progression by enabling cancer cells to absorb extracellular nutrients in nutrient-poor conditions, with the phospholipid flippase ATP9A playing a key role in regulating this process. Endocytosis impacts immune responses in the tumor microenvironment by affecting TLR4 and PD-L1 endocytosis in macrophages, which are influenced by ZNT1 and Zn2+ levels, thus reducing the efficacy of chemotherapy. Therefore, targeting endocytic pathways may improve therapeutic outcomes and drug delivery. For example, valproic acid enhances doxorubicin’s cytotoxicity in HCC cells by promoting its endocytosis, suggesting that combining endocytosis-modulating agents with chemotherapy could enhance treatment efficacy. In our study, we identified CLTA, STAM, RAB10, DAB2, VPS45, AGAP3, ARPC4, VPS29, HSPA8, DNAJC6, PARD6B, ACTR3B, PSD4, and ARRB2 as key genes involved in various cellular processes relevant to cancer biology.
DAB2, a multifunctional signaling molecule, has been shown to be deregulated in various cancers, including HCC. Its role in tumor progression is significant, as it is involved in multiple receptor-mediated signaling pathways. The frequent downregulation of DAB2 due to promoter hypermethylation in cancers such as nasopharyngeal carcinoma suggests its potential tumor-suppressor role, which may also be relevant in HCC (16). Additionally, DAB2’s involvement in skin squamous cell carcinoma highlights its broader implications in cancer biology (17). VPS45, a member of the Sec1/Munc18 family, is crucial for maintaining intracellular organization and trafficking through the endosomal system. Its role in regulating endosomal trafficking and receptor recycling is vital for cellular homeostasis and may influence the progression of various cancers, including HCC (18). The requirement of VPS45 in proper cell growth and development in model organisms such as Arabidopsis thaliana further establishes its importance to cellular processes (19). ARRB2 (β-arrestin2) has been demonstrated to promote cancer growth and progression. In colorectal cancer, ARRB2 enhances cell growth and motility by regulating the pathways mediated by proteins such WTAP, which could have parallels in HCC (20). Its role in endothelial progenitor cell-mediated neovascularization through the activation of ERK and Akt signaling pathways also suggests potential therapeutic applications in cancer treatment (21). The expression of genes such as ARRB2 in HCC has been associated with poor prognosis and could serve as a biomarker for disease progression. Comprehensive genomic analyses have identified ARRB2 as a hub gene in various cancers, including prostate cancer, where it contributes to poor prognosis and could be a target for immunotherapy (22). This suggests that ARRB2 may also be a therapeutic target in HCC.
Overall, the substantial involvement of endocytosis in HCC indicates its potential as a therapeutic target. Through manipulating endocytic pathways, it may be possible to develop novel strategies to combat HCC, improve drug delivery, and overcome resistance to existing therapies. Further research into the specific mechanisms and regulatory factors involved in endocytosis in HCC is essential for advancing these therapeutic approaches.
Conclusions
In patients with HCC, the genes involved in endocytosis were significantly correlated with tumor classification. The related expression profiles may be valuable for predicting HCC prognosis and informing diagnosis and treatment.
Supplementary
The article’s supplementary files as
Acknowledgments
None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Footnotes
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-359/rc
Funding: This work was supported by Gansu Provincial Joint Research Fund Project (Nos. 23JRRA1489, 24JRRA911).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-359/coif). The authors have no conflicts of interest to declare.
References
- 1.Chen JG, Zhu J, Zhang YH, et al. Liver Cancer Survival: A Real World Observation of 45 Years with 32,556 Cases. J Hepatocell Carcinoma 2021;8:1023-34. 10.2147/JHC.S321346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Zhang L, Han F, et al. Using Period Analysis to Timely Assess and Predict 5-Year Relative Survival for Liver Cancer Patients From Taizhou, Eastern China. Front Oncol 2022;12:920094. 10.3389/fonc.2022.920094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarveazad A, Agah S, Babahajian A, et al. Predictors of 5 year survival rate in hepatocellular carcinoma patients. J Res Med Sci 2019;24:86. 10.4103/jrms.JRMS_1017_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X, Yang C, Zhang S, et al. Precision treatment in advanced hepatocellular carcinoma. Cancer Cell 2024;42:180-97. 10.1016/j.ccell.2024.01.007 [DOI] [PubMed] [Google Scholar]
- 5.Herrscher C, Pastor F, Burlaud-Gaillard J, et al. Hepatitis B virus entry into HepG2-NTCP cells requires clathrin-mediated endocytosis. Cell Microbiol 2020;22:e13205. 10.1111/cmi.13205 [DOI] [PubMed] [Google Scholar]
- 6.Gong C, Zhang J, Zhang L, et al. Dynamin2 downregulation delays EGFR endocytic trafficking and promotes EGFR signaling and invasion in hepatocellular carcinoma. Am J Cancer Res 2015;5:702-13. [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Li Y, Xiao Y, et al. The phospholipid flippase ATP9A enhances macropinocytosis to promote nutrient starvation tolerance in hepatocellular carcinoma. J Pathol 2023;260:17-31. 10.1002/path.6059 [DOI] [PubMed] [Google Scholar]
- 8.Yang D, Tian T, Li X, et al. ZNT1 and Zn 2+ control TLR4 and PD-L1 endocytosis in macrophages to improve chemotherapy efficacy against liver tumor. Hepatology 2024;80:312-29. 10.1097/HEP.0000000000000629 [DOI] [PubMed] [Google Scholar]
- 9.Saha SK, Yin Y, Kim K, et al. Valproic Acid Induces Endocytosis-Mediated Doxorubicin Internalization and Shows Synergistic Cytotoxic Effects in Hepatocellular Carcinoma Cells. Int J Mol Sci 2017;18:1048. 10.3390/ijms18051048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xian J, Jin A, Zhou M, et al. Identification of the MEX3 family as potential biomarkers of hepatocellular carcinoma based on bioinformatics and experiments. Transl Cancer Res 2025;14(5):2626-2647. 10.21037/tcr-24-2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, Li B, Liu H, et al. In silico development and validation of a novel six-gene-derived signature in hepatocellular carcinoma. Transl Cancer Res 2025;14:2940-55. 10.21037/tcr-2024-2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Xu H, Chen H, et al. The high expression of Long noncoding RNA TEX41 promotes the proliferation, migration, and invasion of hepatocellular carcinoma. Transl Cancer Res 2025;14:3175-85. 10.21037/tcr-2025-812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santaguida S, Richardson A, Iyer DR, et al. Chromosome Mis-segregation Generates Cell-Cycle-Arrested Cells with Complex Karyotypes that Are Eliminated by the Immune System. Dev Cell 2017;41:638-651.e5. 10.1016/j.devcel.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colombo R, Moll J. Destabilizing aneuploidy by targeting cell cycle and mitotic checkpoint proteins in cancer cells. Curr Drug Targets 2010;11:1325-35. 10.2174/1389450111007011325 [DOI] [PubMed] [Google Scholar]
- 15.Kapur N, Thakral D, Durgapal H, et al. Hepatitis E virus enters liver cells through receptor-dependent clathrin-mediated endocytosis. J Viral Hepat 2012;19:436-48. 10.1111/j.1365-2893.2011.01559.x [DOI] [PubMed] [Google Scholar]
- 16.Tong JH, Ng DC, Chau SL, et al. Putative tumour-suppressor gene DAB2 is frequently down regulated by promoter hypermethylation in nasopharyngeal carcinoma. BMC Cancer 2010;10:253. 10.1186/1471-2407-10-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy S, Mehta D, Sawant S, et al. Disabled 2 (Dab2) Regulates Tumour Progression in Skin Squamous Cell Carcinoma. Exp Dermatol 2024;33:e70009. 10.1111/exd.70009 [DOI] [PubMed] [Google Scholar]
- 18.Frey L, Ziętara N, Łyszkiewicz M, et al. Mammalian VPS45 orchestrates trafficking through the endosomal system. Blood 2021;137:1932-44. 10.1182/blood.2020006871 [DOI] [PubMed] [Google Scholar]
- 19.Mugume Y, Roy R, Agbemafle W, et al. VPS45 is required for both diffuse and tip growth of Arabidopsis thaliana cells. Front Plant Sci 2023;14:1120307. 10.3389/fpls.2023.1120307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang H, Lin Z, Ye Y, et al. ARRB2 promotes colorectal cancer growth through triggering WTAP. Acta Biochim Biophys Sin (Shanghai) 2021;53:85-93. 10.1093/abbs/gmaa151 [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Huang G, Mu J, et al. Arrb2 promotes endothelial progenitor cell-mediated postischemic neovascularization. Theranostics 2020;10:9899-912. 10.7150/thno.45133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou B, Song H, Xu W, et al. The Comprehensive Analysis of Hub Gene ARRB2 in Prostate Cancer. Dis Markers 2022;2022:8518378. 10.1155/2022/8518378 [DOI] [PMC free article] [PubMed] [Google Scholar]