Abstract
Background
We recently described the utility of the “TB Concentration & Transport” kit for biosafe, ambient temperature transport of dried sputum on Trans-Filter, and the “TB DNA Extraction” kit for DNA extraction from Trans-Filter for the early diagnosis of drug-resistant tuberculosis (TB). This study aimed to assess the feasibility and compatibility of these kits with line probe assay (LPA) under National Tuberculosis Elimination Programme (NTEP) settings.
Methods
Patients with presumptive pulmonary TB, multidrug-resistant (MDR) TB, or extensively drug-resistant TB (N = 8491) who attended Designated Microscopy Centers (DMCs, n = 13) under National Reference Laboratories (NRLs) at Bhopal, New Delhi, Chennai, and Bhubaneswar were screened by smear microscopy. The performance of Trans-Filter extracted DNA–based LPA (Kit-LPA) was assessed against Direct-LPA on smear-positive sputum (n = 681), and feedback was obtained from scientists (n = 10) and laboratory technicians (n = 42) regarding logistics, kit usage, training, and troubleshooting. A scoring questionnaire was used to assess (i) the TB Concentration & Transport kit versus conventional sputum transport and (ii) the TB DNA Extraction kit versus Hain GenoLyse kit (statistical significance of the scores was calculated using paired t test).
Results
Kit-LPA showed a sensitivity and specificity in the range of 89%–96% and 99%–100%, respectively, for rifampicin and isoniazid resistance detection and was comparable to Direct-LPA (concordance = 99%–100%; κ = 0.94–0.97). Overall scores indicated that (i) sputum transport on Trans-Filters was more convenient as compared to conventional sputum transport (P < .0001) and (ii) Trans-Filter extracted DNA was easily amalgamated with LPA testing for MDR-TB detection.
Conclusions
These findings provide evidence for incorporation of these kits into the NTEP. Their use can facilitate sputum transport from DMCs to NRLs and provide universal drug susceptibility testing to people with TB residing in remote areas.
Keywords: biosafe sputum transport, diagnosis, LPA, MDR-TB, tuberculosis
This study aimed to assess the feasibility and compatibility of the “TB Concentration & Transport” and “TB DNA Extraction” kits with line probe assay under the National Tuberculosis Elimination Programme. Their use can provide universal drug susceptibility testing to people with tuberculosis residing in remote or distant areas.
Out of 8.2 million new tuberculosis (TB) cases notified worldwide in 2023, around 0.4 million cases were multidrug resistant (MDR)/rifampicin resistant [1]. This poses a daunting challenge to the effective treatment of TB and underscores the importance of early diagnosis of drug-resistant TB. The diagnosis of drug-resistant TB is mainly dependent on World Health Organization (WHO)–endorsed nucleic acid amplification–based tests (NAATs) such as line probe assay (LPA), Xpert MTB/RIF, Xpert MTB/RIF Ultra, Xpert MTB/XDR, Truenat MTB plus, and Truenat MTB-RIF Dx [2].
Under the National Tuberculosis Elimination Programme (NTEP) of India, sputum is transported from designated microscopy centers (DMCs) and NAAT sites (Xpert/Truenat sites) to National Reference Laboratories and Intermediate Reference Laboratories (NRL/IRLs) for drug susceptibility testing (DST) using culture-based DST and LPA because these facilities are restricted to the latter, except for rifampicin (RIF) resistance detection, which is done using Xpert/Truenat assay [3]. Sputum transport utilizes triple-layer containers under cold conditions as per NTEP guidelines, which poses logistic challenges, incurs a high cost, and is associated with bio-safety concerns for health workers at DMCs. In 2023, LPA was the single most frequently used assay for DST in India; 450 992 and 68 046 samples were tested for resistance to first-line drugs and second-line drugs, respectively, at NRLs/IRLs [3].
We recently described the development and evaluation of the TB Concentration & Transport kit (hereafter “Transport kit”) for bio-safe, ambient temperature transportation of dried sputum on Trans-Filter device, and the TB DNA Extraction kit for DNA extraction from Trans-Filter for use in drug resistance assays (Supplementary Figure 1) [4]. The Transport kit exhibited a disinfection efficacy of up to 8 logs of Mycobacterium tuberculosis and mycobacterial DNA was stable on Trans-Filter at 50°C for up to 4 weeks (last time point tested). DNA extracted from Trans-Filter using the TB DNA Extraction kit was shown to be compatible with polymerase chain reaction (PCR) and DNA sequencing for direct drug resistance testing on sputum [4]. A subsequent study established the compatibility of Trans-Filter extracted DNA with LPA (GenoType MTBDRplus and GenoType MTBDRsl) for first-line and second-line drugs on sputum samples. The performance of Kit-LPA (kit-extracted DNA amalgamated with LPA) was comparable to the endorsed test Direct-LPA (LPA from direct sputum) for the detection of MDR-TB and extensively drug-resistant (XDR) TB. The disinfection of sputum samples using the Transport kit was also confirmed [5].
As these 2 studies [4, 5] were performed at central laboratories and transport of sputum on Trans-Filters from distant DMCs was not performed, assessing the kits’ robustness and performance under the NTEP was imperative. Therefore, in the present study, the operational feasibility of both these kits was assessed under the NTEP. Sputum transport on Trans-Filters from DMCs to NRLs/IRLs in field settings and the integration of Trans-Filter extracted DNA into LPA testing was assessed by (i) evaluating the performance of Kit-LPA versus Direct-LPA and (ii) obtaining user feedback.
MATERIALS AND METHODS
Study Population and Patient Consent
A total of 8491 consecutive patients including presumptive TB cases (treatment-naive) and presumptive MDR-TB/XDR-TB cases (as per the NTEP guidelines [6, 7]) were enrolled in the study after ethical approval. Ethical approval was obtained from the institutional ethics committees of all NRL sites (Bhopal Memorial Hospital Research Centre, Bhopal [BMHRC; BMHRC/IEC/43/Micro/20]; National Institute of Tuberculosis and Respiratory Diseases, New Delhi [NITRD; NITRD/EC/2020/A85]; Indian Council of Medical Research [ICMR]–National Institute for Research in Tuberculosis, Chennai [NIRT; 154/NIRT-IEC/2020]; and ICMR–Regional Medical Research Centre, Bhubaneswar [RMRC; ICMR/-RMRCB/IHEC-2020/10]) for the collection of sputum samples at their associated DMCs (n = 13). All sputum samples were collected at DMCs (n = 13) between May 2022 and December 2022 after obtaining an informed written consent (consent was taken from the guardian in the case of a minor; Supplementary Appendix 1), and clinical data were collected from each patient (Supplementary Appendix 2). This study was conducted according to Standards for Reporting Diagnostic Accuracy (STARD) guidelines (Supplementary Appendix 3).
Study Design
This study was supervised by the Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh. At the start of the study, on-site, hands-on training on the usage of the Transport kit was provided to laboratory technicians at all DMCs and the TB DNA Extraction kit at NRLs/IRLs by PGIMER. In addition, audio-visual clips were used to demonstrate the kits’ protocols. All laboratory technicians and scientists confirmed that they obtained adequate training for the use of both the kits before the start of the study.
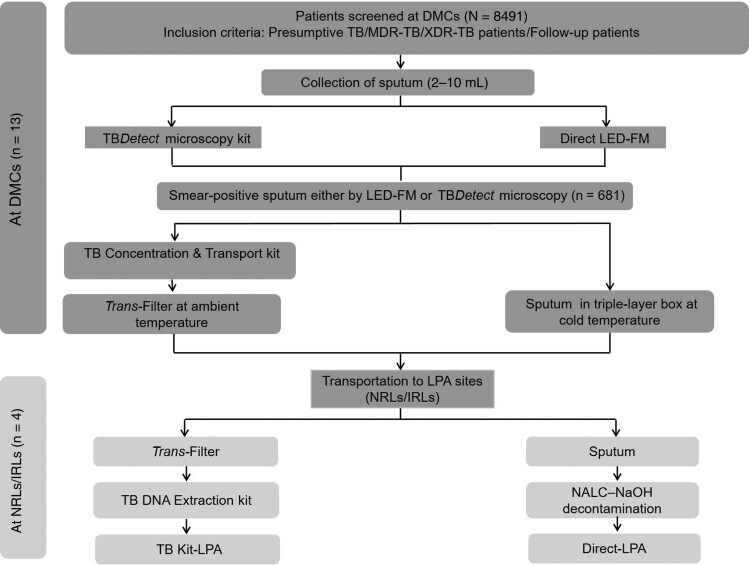
This study was conducted at 13 sites along with the operational feasibility study of the TBDetect smear microscopy kit [8]. All sputum samples were screened by smear examination (light-emitting diode fluorescent microscopy [LED-FM] or TBDetect microscopy) and samples that were positive by either method were included in the present study (n = 681, Figure 1), as NTEP guidelines recommend Direct-LPA testing only on smear-positive sputum. It is to be noted that for smear-negative sputum samples, indirect LPA testing (ie, LPA on M tuberculosis clinical isolates from smear-negative sputum samples) has been recommended, for which transport of sputum samples containing live bacteria is required for culturing, and therefore indirect LPA testing cannot be performed on bio-safe or inactivated sputum (on Trans-Filter). Smear-positive sputa were processed by Transport kit at the DMCs and then paired Trans-Filters and sputum samples were transported to the associated NRL/IRLs (Figure 1 and Supplementary Table 1). At the NRLs/IRLs, a unique 3-digit code was assigned to each sample for Kit-LPA and the results were decoded and analyzed at the completion of the study.
Figure 1.
Workflow of the present study. Abbreviations: DMC, Designated Microscopy Center; LED-FM, Light-emitting diode fluorescent microscopy; LPA, Line probe assay; NALC-NaOH, N-acetyl L-cysteine- sodium hydroxide; NRL/IRL, National Reference Laboratory/Intermediate Reference Laboratory; TB, Tuberculosis; MDR-TB, Multi drug resistant-TB; XDR-TB, Extensively drug resistant-TB.
LED-FM and TBDetect Microscopy
All sputum samples were screened by smear examination (LED-FM and/or TBDetect microscopy) at DMCs using a previously described protocol [4, 8, 9].
Sputum Processing by TB Concentration & Transport Kit
Smear-positive sputum samples were processed using Transport kit as described previously [4, 5]. In brief, ‘Dissolving solution’ (400 µL) was added to a sputum dissolution tube. Then, 100 µL of smear-positive sputum was added, mixed, and incubated for 30 minutes. Liquefied sputum (300 µL) was filtered through the Trans-Filter device followed by the addition of ‘Sterilizing solution’ and ‘Wash solution’. The Trans-Filter membrane was then removed, air-dried at room temperature, and inserted into a zip-lock bag for transportation.
Transportation of Trans-Filter and Sputum
Dried Trans-Filter membranes were transported at ambient temperature and sputum samples were transported in triple-layer packaging at cold temperature from the DMCs to their associated NRLs/IRLs.
DNA Extraction From Transported Sputum and Trans-Filter Membrane
At NRLs/IRLs, the sputum samples were decontaminated by the N-acetyl L-cysteine- sodium hydroxide (NALC-NaOH) method [10] and DNA extraction from decontaminated pellets was performed using GenoLyse DNA Extraction Kit version 1.0 (Hain Lifesciences, Nehren, Germany). Simultaneously, DNA was extracted from dried Trans-Filter membranes using TB DNA Extraction kit using the protocol described previously [4, 5].
Line Probe Assay
Extracted DNA from sputum samples and Trans-Filter membranes were subjected to PCR amplification in parallel followed by reverse hybridization in GT-Blot 48 system (Hain Lifesciences) using GenoType MTBDRplus version 2.0 (Hain Lifesciences) as per the manufacturer's instructions at the NRLs/IRLs [11].
Data Compilation and Analysis
All patients’ data (clinical information and test results) were collected on the study clinical proforma (Supplementary Appendix 2) at each DMC and were compiled in a data collection sheet at the associated NRL. The data collection sheets were communicated to PGIMER, Chandigarh, on a weekly basis through email (Supplementary Appendix 4). All results were analyzed at the end of the study.
Operational Feasibility Under Field Conditions
The operational feasibility of the kits was evaluated in 2 ways: (i) by assessing the performance of Kit-LPA versus Direct-LPA and (ii) by obtaining feedback from users. For (i), the diagnostic accuracy of Kit-LPA was assessed using Direct-LPA as the reference standard for MDR-TB detection on each sputum sample. For (ii), feedback was obtained from scientists/study-in-charge (n = 10) and laboratory technicians (n = 42) using a semi-structured questionnaire that assessed the following parameters: feasibility of adopting the Transport kit in DMCs and TB DNA Extraction kit in NRLs/IRLs, on-site training for kit protocol implementation, time to result, logistics, and troubleshooting. The questionnaire also included the assessment of the kit user manual and its components and user feedback on the feasibility of amalgamating both of these kits in the NTEP. In addition, a scoring questionnaire (on a scale of 1–5) was also used to obtain feedback from users at each DMC on the transport of sputum by the conventional method used under NTEP versus dried Trans-Filter membrane in terms of ease of (i) performing the protocol, (ii) use and packaging, (iii) handling and storage, (iv) time saving, and (v) cost-effectiveness (Supplementary Appendix 5). Feedback scores were also obtained at each NRL/IRL site on DNA extraction methods (ie, Hain GenoLyse kit versus TB DNA Extraction kit), in terms of ease of (i) performing the protocol, (ii) handling and storage, and (iii) operator fatigue (Supplementary Appendix 5).
Statistical Analysis
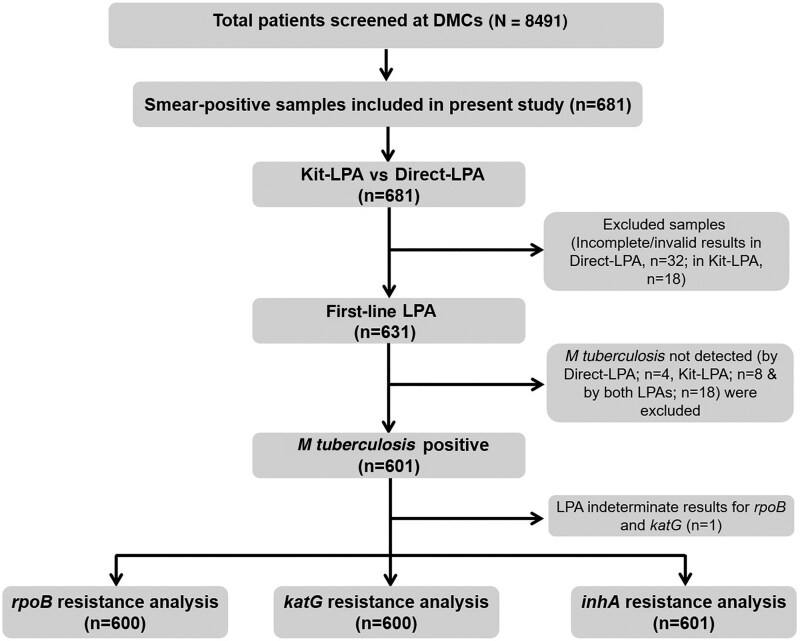
Patients with incomplete data and invalid or indeterminate results were excluded from the study (Figure 2). In case of any discrepancy, Kit-LPA and Direct-LPA were repeated and DNA sequencing of kit-extracted DNA was also performed to resolve the discrepancy. Kit-LPA results were assessed versus Direct-LPA (gold standard) for the detection of RIF and isoniazid (INH) drug resistance. The sensitivity of Kit-LPA was calculated as true-positive [(TP) / (TP + false-negative (FN)], where TP was mutant (MUT) strains identified by both Kit-LPA and Direct-LPA, and FN was wild-type (WT) strains by Kit-LPA but MUT strains by Direct-LPA. The specificity of Kit-LPA was defined as [true-negative (TN) / (TN + false-positive (FP)], where TN was samples identified as WT strains by both Kit-LPA and Direct-LPA and FP was samples showing MUT strains by Kit-LPA but WT strains by Direct-LPA. Concordance between Kit-LPA and Direct-LPA results was calculated as (TP + TN) / (total number of samples), and the degree of agreement was calculated by Cohen kappa (κ) (https://www.graphpad.com/quickcalcs/kappa1/). The statistical significance of scoring feedback was calculated for each parameter using paired t test (GraphPad Prism software version 5.01 for Windows).
Figure 2.
Workflow of final analysis (Kit-LPA vs Direct-LPA) for drug resistance detection. Abbreviations: Direct-LPA, line probe assay from direct sputum; DMC, designated microscopy center; Kit-LPA, kit-extracted DNA amalgamated with line probe assay; LPA, line probe assay.
RESULTS
Study Participants
Sputum samples from patients with presumptive TB/MDR-TB/XDR-TB (n = 8491) who visited DMCs from May 2022 to December 2022 were screened, of which 681 patients were included in the study based on smear-positive results (either by LED-FM or TBDetect microscopy; Figure 2). The included patients were in the age range of 5–85 years (including 22 children aged 1–18 years). Around 62% (422/681) of patients were male. The most common clinical symptoms were cough (∼79% [384/485]), fever (∼67% [325/485]), weakness (∼65% [317/485]), weight loss (∼49% [237/485]), and loss of appetite (34% [165/485]). The human immunodeficiency virus (HIV) status of 343 of 681 subjects included in this study was available and none of them were HIV positive. Clinical characteristic data of enrolled patients (n = 196) at the ICMR-NIRT, Chennai site were not available for this analysis (Supplementary Table 2).
On analysis, Direct-LPA results were not available for 32 samples and Kit-LPA results were not available for 18 samples; therefore, these samples (incomplete/invalid results) were excluded (Figure 2). In drug resistance analysis, M tuberculosis–negative samples either by Direct-LPA (n = 4) or Kit-LPA (n = 8) or by both LPAs (n = 18) were excluded from the analysis, including 1 sample that showed indeterminate results for rpoB and katG in Direct-LPA. Most of the M tuberculosis–negative samples in Direct-LPA and/or Kit-LPA were found to had lower smear grade status (scanty/1+). Finally, 600 samples were included in rpoB and katG gene analysis for RIF and INH drug resistance detection, respectively, and 601 samples were included for inhA promoter region (referred as inhA later) analysis, which is also associated with INH drug resistance (Figure 2).
Operational Feasibility
Performance of Kit-LPA vs Direct-LPA
Kit-LPA showed an overall sensitivity (at all sites) of 94.1% (95% confidence interval [CI], 80.3%–99.2%), 95.7% (95% CI, 84.6%–98.8%), and 88.8% (95% CI, 51.7%–99.7%) and specificity of 100% (95% CI, 99.4%–100%), 99.4% (95% CI, 98.4%–99.9%), and 100% (95% CI, 99.4%–100%) for RIF, katG-INH, and inhA-INH resistance detection, respectively (Table 1). A concordance of 99%–99.8% (κ = 0.94–0.97) was noted between the results of Kit-LPA and Direct-LPA.
Table 1.
Performance of Kit Line Probe Assay vs Direct Line Probe Assay for Drug Resistance Detection at All 4 Sites
| Site | Drug | Target Gene | Kit-LPA vs Direct-LPA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TP | FP | FN | TN | Sensitivity, % (95% CI)* | Specificity, % (95% CI)* | Concordance* %, (κ) | |||
| Combined | RIF | rpoB (n = 600) | 32 | 0 | 2 | 566 | 94 (80–99) | 100 (99–100) | 100 (0.97) |
| INH | katG (n = 600) | 45 | 3 | 2 | 550 | 96 (85–99) | 99 (98–100) | 99 (0.94) | |
| inhA (n = 601) | 8 | 0 | 1 | 592 | 89 (52–100) | 100 (99–100) | 100 (0.94) | ||
| BMHRC, Bhopal | RIF | rpoB (n = 135) | 7 | 0 | 0 | 128 | 100 (59–100) | 100 (97–100) | 100 (1.0) |
| INH | katG (n = 135) | 7 | 1 | 0 | 127 | 100 (59–100) | 99 (96–100) | 99 (0.93) | |
| inhA (n = 136) | 4 | 0 | 0 | 132 | 100 (40–100) | 100 (97–100) | 100 (1.0) | ||
| NITRD, New Delhi | RIF | rpoB (n = 231) | 16 | 0 | 2 | 213 | 89 (65–99) | 100 (98–100) | 99.1 (0.93) |
| INH | katG (n = 231) | 23 | 2 | 2 | 204 | 96 (80–100) | 99 (96–100) | 99 (0.96) | |
| inhA (n = 231) | 3 | 0 | 1 | 227 | 75 (19–99) | 100 (98–100) | 99 (0.85) | ||
| ICMR-NIRT, Chennai | RIF | rpoB (n = 188) | 9 | 0 | 0 | 179 | 100 (66–100) | 100 (98–100) | 100 (1.0) |
| INH | katG (n = 188) | 15 | 0 | 0 | 173 | 100 (78–100) | 100 (98–100) | 100 (1.0) | |
| inhA (n = 188) | 1 | 0 | 0 | 187 | 100 (2–100) | 100 (98–100) | 100 (1.0) | ||
| ICMR-RMRC, Bhubaneswar | RIF | rpoB (n = 46) | 0 | 0 | 0 | 46 | NaN | 100 (92–100) | 100 (NaN) |
| INH | katG (n = 46) | 0 | 0 | 0 | 46 | NaN | 100 (92–100) | 100 (NaN) | |
| inhA (n = 46) | 0 | 0 | 0 | 46 | NaN | 100 (92–100) | 100 (NaN) | ||
Abbreviations: BMHRC, Bhopal Memorial Hospital Research Centre; NITRD, National Institute of Tuberculosis and Respiratory Diseases; ICMR-NIRT, Indian Council of Medical Research-National Institute for Research in Tuberculosis; ICMR-RMRC, Indian Council of Medical Research-Regional Medical Research Centre; CI, confidence interval; Direct-LPA, line probe assay from direct sputum; Kit-LPA, kit-extracted DNA amalgamated with line probe assay; FN, false negative; FP, false positive; RIF, Rifampicin; INH, Isoniazid; TN, true negative; TP, true positive; NaN, non-estimable (no true-positive sample was detected at Regional Medical Research Centre, Bhubaneswar site).
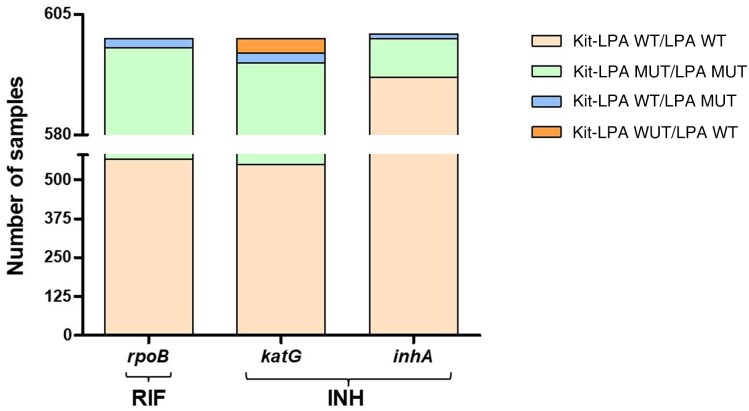
On a head-to-head comparison of Kit-LPA versus Direct-LPA, 566 samples were WT, 32 were MUT, and 2 were FN for the rpoB gene target. For the katG gene target, 550 samples were WT, 45 were classified as MUT, 3 were FP, and 2 samples were FN. For the inhA gene target, 592 samples were WT, 8 were MUT, and 1 sample was FN (Figure 3 and Supplementary Figure 2).
Figure 3.
Concordance/discordance between Kit-LPA and Direct-LPA for detection of rifampicin and isoniazid resistance in a combined analysis of all 4 sites. Abbreviations: INH, isoniazid; Kit-LPA, kit-extracted DNA amalgamated with line probe assay; LPA, line probe assay; MUT, mutant; RIF, rifampicin; WT, wild-type.
Discrepant results (n = 8) between Kit-LPA and Direct-LPA remained unchanged after repeating both the LPA tests. For further resolution, DNA sequencing of kit-extracted DNA was performed and 7 of 8 discrepant results were concordant with the result from Kit-LPA, and 1 sample (L-66888) was concordant with Direct-LPA (Supplementary Table 3).
User Feedback for the TB Concentration & Transport Kit
Feedback for the Transport kit was obtained from scientists (n = 10) and laboratory technicians (n= 42) at DMCs and NRLs/IRLs. Scientists and laboratory technicians had a median work experience of 21 years (interquartile range [IQR], 10.5–23.7 years) and 12 years (IQR, 4.0–18.0 years), respectively. It took a median of 5 samples (IQR, 4.0–5.0) for the laboratory technicians to become comfortable using the Transport kit. All laboratory technicians and scientists confirmed that they obtained adequate training for the use of Transport kit before the start of the study.
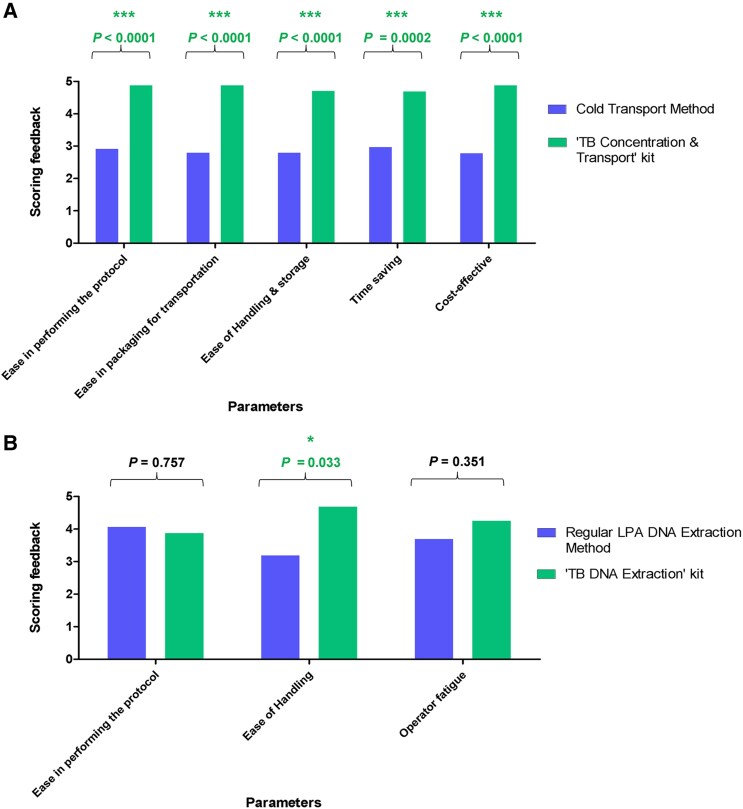
Based on the feedback obtained from scientists and laboratory technicians, we concluded that (i) the Transport kit is self-sufficient (all the reagents and materials are provided in the kit) and convenient to use; (ii) sputum transport on Trans-Filter was more convenient compared to regular transport; (iii) it was feasible to adopt Transport kit in DMCs; (iv) minimal training was required for kit protocol; and (v) the kit user manual is self-explanatory and easy to follow. An examination of the compiled scores indicated that the Transport kit was considered to be superior to regular triple-layer cold chain transport of sputum (Figure 4A and Supplementary Figure 3), and all users concluded that it can easily replace the conventional sputum transport method. Analysis of the feedback indicated that there was a significant difference in the scores for all 5 parameters; namely, ease in performing the protocol (P < .0001), ease of use and packaging (P < .0001), ease of handling and storage (P < .0001), time saving (P = .0002), and cost-effectiveness (P < .0001) between the Transport kit and conventional triple-layer cold chain packaging for sputum transport.
Figure 4.
Scoring feedback on sputum transport using the TB Concentration & Transport kit vs conventional sputum cold transport method (A) and the TB DNA Extraction kit vs regular line probe assay (LPA) DNA extraction method (GenoLyse kit) (B). *Statistically significant.
User Feedback for the TB DNA Extraction Kit
Feedback for the TB DNA Extraction kit was obtained only at NRL/IRL sites from 7 scientists and 14 laboratory technicians. It took the laboratory technicians a median of 5 samples (IQR, 3.0–9.0) to become comfortable using the TB DNA Extraction kit. All scientists and laboratory technicians confirmed that they obtained adequate training for the use of TB DNA Extraction kits before the start of the study and after familiarization, healthcare workers could easily perform the test.
Summary of feedback from scientists and laboratory technicians indicated that the TB DNA Extraction kit is also self-contained and requires minimal training, the kit user manual is easy to follow, and Trans-Filter extracted DNA was easily amalgamated with LPA testing. However, the users felt that the time taken for the TB DNA Extraction kit was slightly longer than for the GenoLyse kit. On analyzing the scoring feedback, only 1 parameter (ie, ease in handling the Trans-Filter in comparison to sputum) was scored as significantly different (P = 0.033). For other parameters, namely, ease of performing the protocol (P = .757) and operator fatigue (P = .351), the difference in scores between the TB DNA Extraction kit and the Hain LPA GenoLyse kit were not significant (Figure 4B and Supplementary Figure 4).
DISCUSSION
Significant challenges are associated with the transportation of infectious sputum samples. We developed a Transport kit for use at point-of-care settings (DMCs/Peripheral Healthcare Centres) to enable ambient temperature transport of sputum on dried Trans-Filter to a higher-level laboratory for molecular DST [4]. This kit fulfils all of the specifications stated in WHO's target product profile for the transport of sputum with the exception of “compatibility with culture methods” [12]. Trans-Filter enables biosafe and cost-effective transportation of inactivated bacteria in comparison to sputum transport kits available in the market (a detailed comparison was reported in our previous study) [5]. The TB DNA Extraction kit extracts good-quality M tuberculosis DNA from dried sputum on Trans-Filter and is compatible with molecular DST approaches, such as DNA sequencing [4] and LPA for the diagnosis of drug resistance [5]. In the present study, we evaluated the operational feasibility of (i) transporting dried sputum on Trans-Filter in field settings and (ii) the compatibility of Trans-Filter extracted DNA using TB DNA Extraction kit with first-line LPA (GenoType MTBDRplus version 2.0).
Upon site-wise analysis, Kit-LPA showed sensitivity in the range of 89% to 100%, for RIF resistance detection; 96% to 100%, for katG-INH resistance detection; and 75% to 100%, for inhA-INH resistance detection (Table 1). Kit-LPA showed specificity in the range of 99%–100% with a concordance of 99%–100% (κ = 0.85–1.0; Table 1) with Direct-LPA at all sites. The sensitivity and specificity of Kit-LPA were highly similar at all sites (99%–100%), except at NITRD. At NITRD, the sensitivity of RIF (89%) and INH (katG [96%] and inhA [75%]) were comparatively lower than those at other sites (100%; Table 1), due to some discrepancies between Kit-LPA and LPA results (Supplementary Figure 2 and Supplementary Table 3). On DNA sequencing, 7 of 8 discrepant samples showed concordant results with Kit-LPA findings. A possible reason for this discordance might be a small variation in band intensity observed with Kit-LPA versus Direct-LPA (LPA result interpretation is based on the band intensity of WT or MUT probes corresponding to the amplification control band, as per manufacturer's instructions). The combined sensitivity of Kit-LPA against Direct-LPA was lower than in our previous study (96%–100% [5]), and this might be due to this study being a multicenter study, wherein the sensitivity of Kit-LPA versus Direct-LPA was 100% at 3 sites and comparable to the previous study (96%–100%), but lower (75%–96%) at 1 site (New Delhi). Importantly, the diagnostic accuracy of Kit-LPA at all 4 NRL sites (sensitivity and specificity for RIF [94% and 100%] and for INH [katG and inhA combined, ∼96% and ∼99%] drugs, respectively) was comparable to the pooled sensitivity and specificity of Direct-LPA for RIF (∼96% and ∼98%, respectively) and for INH (∼94% and ∼99%), Xpert MTB/RIF (96% and 98%), and Xpert MTB/XDR for INH (94% and 98%) that has been reported previously [2].
The key findings of this feasibility study were that the Transport kit was employed successfully for the bio-safe transportation of dried sputum at ambient temperature from DMCs to associated NRLs/IRLs under field conditions, and was given excellent user feedback compared to the conventional triple-layer packaging method of sputum transport (Figure 4A and Supplementary Figure 3). The feedback for the TB DNA Extraction kit indicated that the Trans-Filter was easy to handle in comparison to the sputum sample for DNA extraction, and Trans-Filter extracted DNA was easily amalgamated with LPA testing. However, this kit received comparatively lower score in “time saving” and “operator fatigue” parameters (Figure 4B and Supplementary Figure 4) due to the protocol time of approximately 55 minutes for DNA extraction in comparison to the GenoLyse kit protocol (∼25 minutes). A noteworthy point, though, is that before DNA extraction by GenoLyse kit, an essential sputum processing step using the NALC-NaOH method of approximately 40 minutes makes the overall process longer than the TB DNA Extraction kit protocol. Both the kits are cost-effective (ie, the cost/sample is 120 Indian rupees or 1.37 US dollars for each kit). Our study had certain limitations. First, clinical data including HIV testing results were not available for all the subjects included in the study, and second, the performance of Kit-LPA was not evaluated directly on smear-negative sputum samples as the study was performed as per the NTEP guidelines where only smear-positive sputum samples are subjected to Direct-LPA testing and smear-negative sputum samples need to be cultured prior to LPA testing.
CONCLUSIONS
On the completion of this feasibility study, the Indian TB Research Consortium, government of India, recommended these kits for incorporation into the NTEP. The Transport kit is expected to improve sputum transport in the nationwide network of DMCs (n = 24 573) to NRLs/IRLs. This approach will not only eliminate the need for a cold chain transport of infectious sputum samples but also enhance the safety of healthcare workers. Importantly, by integrating these kits with LPA testing, drug resistance testing services can be efficiently provided and extended to patients in remote and peripheral areas.
Supplementary Material
Contributor Information
Keerti Chauhan, Department of Experimental Medicine and Biotechnology, Postgraduate Institute of Medical Education and Research, Chandigarh, India.
Rakesh Kumar Gupta, Department of Experimental Medicine and Biotechnology, Postgraduate Institute of Medical Education and Research, Chandigarh, India.
Divya Anthwal, Department of Experimental Medicine and Biotechnology, Postgraduate Institute of Medical Education and Research, Chandigarh, India.
Nikita Panwalkar, Department of Microbiology, Bhopal Memorial Hospital and Research Centre, Bhopal, India.
Prabha Desikan, Department of Microbiology, Bhopal Memorial Hospital and Research Centre, Bhopal, India.
Ritu Singhal, Department of Microbiology, National Institute of Tuberculosis and Respiratory Diseases, New Delhi, India.
Manpreet Bhalla, Department of Microbiology, National Institute of Tuberculosis and Respiratory Diseases, New Delhi, India.
Vithal Prasad Myneedu, Department of Microbiology, National Institute of Tuberculosis and Respiratory Diseases, New Delhi, India.
Khalid Umar Khayyam, Department of Microbiology, National Institute of Tuberculosis and Respiratory Diseases, New Delhi, India.
Siva Kumar Shanmugam, Department of Bacteriology, Indian Council of Medical Research (ICMR)-National Institute for Research in Tuberculosis, Chennai, India.
K Silambu Chelvi, Department of Bacteriology, Indian Council of Medical Research (ICMR)-National Institute for Research in Tuberculosis, Chennai, India.
Padmapriyadarsini Chandrasekaran, Department of Bacteriology, Indian Council of Medical Research (ICMR)-National Institute for Research in Tuberculosis, Chennai, India.
Sidhartha Giri, National Reference Laboratory (NRL) for Tuberculosis, ICMR-Regional Medical Research Centre, Bhubaneshwar, India.
Jyotirmayee Turuk, National Reference Laboratory (NRL) for Tuberculosis, ICMR-Regional Medical Research Centre, Bhubaneshwar, India.
Dasarathi Das, National Reference Laboratory (NRL) for Tuberculosis, ICMR-Regional Medical Research Centre, Bhubaneshwar, India.
Sanghamitra Pati, National Reference Laboratory (NRL) for Tuberculosis, ICMR-Regional Medical Research Centre, Bhubaneshwar, India.
Abhinav Goyal, Advanced Microdevices Pvt Ltd, Ambala Cantt, India.
Ashawant Gupta, Advanced Microdevices Pvt Ltd, Ambala Cantt, India.
Nalini Kant Gupta, Advanced Microdevices Pvt Ltd, Ambala Cantt, India.
Manjula Singh, India Tuberculosis Research Consortium, Indian Council of Medical Research, V. Ramalingaswami Bhawan, Ansari Nagar, New Delhi, India.
Jaya Sivaswami Tyagi, Department of Biotechnology, All India Institute of Medical Sciences, Ansari Nagar, New Delhi, India.
Sagarika Haldar, Department of Experimental Medicine and Biotechnology, Postgraduate Institute of Medical Education and Research, Chandigarh, India.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We acknowledge the guidance of Sivaramakrishnan N. Gomathi and assistance of A. Radhakrishnan, E. Jayaseelan, J. Hemalatha, J. Priy, P. Saravana, N. Ravi Rajan, S. Durga Devi, S. Sarala, S. Ulagu, C. Geetha Priya, and Gayathri from the NIRT site; Shubham Sahu, Umesh Singh Lodhi, Abhay Kumar Mishra, Dr Puneet Maheshwari, Dr Sanjay Purohit, Dr Sanjay Joshi, Dr Manoj Verma, Amrit Lal, Prakash Morvi, Shubham Singh, Chandra Prakash, Gomati Sahu, Jeevanlal Katare, Mahesh Jarman, Mamta Soni, Manoj Kumar Dhahana, Mohan Mondre, Prakash Kumar, Savitri Yadav, Sunita Kanashe, Vikram Patel, Arun Kumar Sharma, Rajeev Ranjan, Ragini Kushwaha, and Swapnil Dhobale from the BMHRC site; Prakasha Kumar Nayak, Tapaswini Hota, Soumya Nayak, Satya Narayan Singh, Sadruddin Khan, Devi Prasad Das, Chinmayee Mohanty, Baikuntha Bihari Sahoo, Triyanbakesh Mohanty, and Sunil Swick Rout from the RMRC site; and Aditi Goyal, Kunjbihari Rajput, Ankita, Mauj Kumar, Mukesh Kumar, Pooja Madhan, and Ravi from NITRD site. We also acknowledge all of the enrolled patients for providing consent to participate in the study.
Author contributions. K. C. performed the methodology, investigation, data curation, and formal analysis and wrote the original draft of the manuscript. R. K. G. performed the methodology, investigation, data curation, and formal analysis. D.A. conceptualized the study, performed formal analysis, and wrote the original draft of the manuscript. N. P., R. S., K. U. K., P.C., K. S. C., J. S. T., and D. D. performed data curation and project administration, supervised the study, and provided resources. P. D., M. B., V. P. M., S. K. S., S. G., and S. P. designed the study, acquired funding, performed project administration, provided resources, and supervised the study. A. Go. was involved in the project implementation and training. A. Gu., N. K. G., and M. S. conceptualized the study, provided resources, and supervised the study. J. S. T. and S. H. conceptualized the study, acquired funding, performed formal analysis and project administration, provided resources, supervised the study, and wrote the original draft of the manuscript. All authors reviewed and approved the final manuscript.
Data availability. Data can be made available upon request to the corresponding author.
Financial support. This work was supported by the Indian Council of Medical Research (ICMR), government of India (grant number 5/8/5/7/Adhoc/2020/ECD-I) received jointly by S. H., P. D., M. B., S. K. S., and S. G.; in addition, J. S. T. is thankful to the National Academy of Sciences (India) for a Senior Scientist Fellowship. The Senior Research Fellowship of K. C. and R. K. G. was from the ICMR grant awarded to S. H.
References
- 1. World Health Organization . Global tuberculosis report. 2024. Available at: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2024. Accessed 12 April 2024.
- 2. World Health Organization . WHO consolidated guidelines on tuberculosis. Module 3: diagnosis—rapid diagnostics for tuberculosis detection 2021 update. 2021. Available at: https://iris.who.int/bitstream/handle/10665/342331/9789240029415-eng.pdf. Accessed 10 May 2024.
- 3. National Tuberculosis Elimination Programme . India TB report. 2024. Available at: https://tbcindia.mohfw.gov.in/wp-content/uploads/2024/10/TB-Report_for-Web_08_10-2024-1.pdf. Accessed 8 October 2024.
- 4. Anthwal D, Lavania S, Gupta RK, et al. Development and evaluation of novel bio-safe filter paper–based kits for sputum microscopy and transport to directly detect Mycobacterium tuberculosis and associated drug resistance. PLoS One 2019; 14:e0220967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anthwal D, Gupta RK, Singhal R, et al. Compatibility of a novel filter paper–based bio-safe sputum transport kit with line probe assay for diagnosing drug-resistant tuberculosis: a single-site evaluation study. ERJ Open Res 2021; 7:00137-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Revised National Tuberculosis Control Programme . Technical and operational guidelines of tuberculosis control in India. 2016. Available at: https://tbcindia.mohfw.gov.in/technical-and-operational-guidelines-for-tb-control-in-india-2016/. Accessed 9 November 2024.
- 7. Revised National Tuberculosis Control Programme . Guidelines on programmatic management of drug-resistant TB (PMDT) in India. 2017. Available at: https://tbcindia-wp.azurewebsites.net/guideline-for-pmdt-in-india-2017-3/. Accessed 17 November 2024.
- 8. Chauhan K, Gupta RK, Anthwal D, et al. Operational feasibility and multi-centric evaluation of ‘TBDetect sputum microscopy kit’ for the direct detection of Mycobacterium tuberculosis in field settings. Infect Dis (Lond) 2024; 56:1040–8. [DOI] [PubMed] [Google Scholar]
- 9. Anthwal D, Gupta RK, Gomathi NS, et al. Evaluation of ‘TBDetect’ sputum microscopy kit for improved detection of Mycobacterium tuberculosis: a multi-centric validation study. Clin Microbiol Infect 2021; 27:911.e1–7. [DOI] [PubMed] [Google Scholar]
- 10. Global Laboratory Initiative . Mycobacteriology laboratory manual. 2014. Available at: https://docslib.org/doc/6773056/gli-mycobacteriology-laboratory-manual. Accessed 14 September 2024.
- 11. Hain Lifesciences . GenoType MTBDRplus VER 2.0: molecular genetic assay for identification of the M. tuberculosis complex and its resistance to rifampicin and isoniazid from clinical specimens and cultivated samples. 2012. Available at: https://www.hain-lifescience.de/include_datei/kundenmodule/packungsbeilage/download.php?id=936. Accessed 28 September 2024.
- 12. World Health Organization . Technical expert group meeting report: commercial products for preserving clinical specimens for the diagnosis of tuberculosis. 2017. Available at: https://www.who.int/publications/i/item/WHO-HTM-TB-2017.19. Accessed 7 October 2024.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.