Abstract
Aims
This study evaluated hematological parameters and cell population data (CPDs) to assess their ability to discriminate between moderate and severe COVID-19 in hospitalized female patients, and to identify potential markers associated with worse outcomes.
Patients & methods
A retrospective study was conducted on 84 adult female COVID-19 patients hospitalized at CHC-UFPR (Brazil) between March 2020 and July 2021. Patients were stratified into moderate (n = 46) and severe (n = 38) disease groups. A control group included 100 healthy female outpatients. Parameters analyzed included D-dimer, WBC count, neutrophil-to-lymphocyte ratio (NLR), and CPDs (LY-X, LY-Y). RT-qPCR was used for SARS-CoV-2 confirmation and variant identification.
Results
Significant differences (p < 0.05) in LY-X, LY-Y, NLR, and WBC were found between moderate and severe groups. D-dimer was elevated in severe cases. Among deceased patients (n = 17), WBC and NLR were markedly increased. ROC curve analysis confirmed the discriminatory power of these markers. No significant association was found between viral genotype and severity (p = 0.9602).
Conclusions
Hematological parameters, particularly CPDs and NLR, are valuable for early stratification of COVID-19 severity. These automated, rapid, and cost-effective measures can support clinical decision-making. However, CPD usage depends on analyzer availability and lacks standardization across platforms.
Keywords: SARS-CoV-2, COVID-19, CPD (cell population data), D-dimer, sysmex XN-3100, severe disease, moderate disease, women
HIGHLIGHTS
Introduction
COVID-19 overview: COVID-19 is a highly transmissible and potentially severe infectious disease caused by SARS-CoV-2, associated with a complex systemic inflammatory response.
Relevance of hematological parameters: Traditional biomarkers have shown limitations in disease stratification. Recent evidence supports the use of hematological indicators such as cell population data (CPD) and D-dimer in evaluating disease severity.
Study objective: To assess the diagnostic and prognostic potential of CPD parameters, neutrophil-to-lymphocyte ratio (NLR), white blood cell count (WBC), and D-dimer levels in female COVID-19 patients.
Methods
Design and population: A retrospective analysis was conducted at CHC-UFPR, involving 84 female COVID-19-positive patients (38 with severe and 46 with moderate disease) and 100 female controls without COVID-19.
Laboratory procedures: Complete blood count (CBC) and CPDs were analyzed using the Sysmex XN-3100. D-dimer measurements were performed on the Sysmex CS Series. RT-qPCR was used for SARS-CoV-2 variant genotyping.
Statistical analysis: Data were evaluated using R software with GCMR package and ROC curves generated with MedCalc. A 5% significance level was adopted.
Results
Key biomarkers: LY-X and LY-Y (CPDs), NLR, WBC, and D-dimer levels were significantly associated with disease severity (p < 0.05).
Prognostic indicators: Elevated NLR and WBC levels were strongly correlated with fatal outcomes.
Severity vs. outcome: A statistically significant correlation (p < 0.001) was observed between clinical severity and mortality, reinforcing the value of hematological parameters in predicting outcomes.
Variant analysis: Although the Gamma variant predominated among severe cases and deaths, genotyping results did not statistically influence severity (p = 0.9602).
ROC performance: LY-X, LY-Y, NLR, and WBC showed statistically significant diagnostic accuracy in distinguishing between moderate and severe disease.
Discussion
Inflammatory dysregulation: Severe COVID-19 patients exhibited leukocytosis, lymphopenia, and elevated NLR, indicative of systemic immune dysregulation.
Lymphocyte markers: LY-X and LY-Y effectively represented lymphocyte depletion and inflammatory activity, correlating with disease progression.
NLR as a predictor: NLR values were markedly elevated in severe cases, reinforcing its utility as an accessible inflammatory marker in COVID-19.
Role of D-dimer: While elevated in severe cases and useful in early risk assessment, D-dimer showed limited utility in monitoring treatment response.
Variant considerations: Despite the predominance of the Gamma variant among severe cases, the sample size limited definitive conclusions on variant-specific outcomes.
Final considerations
Clinical applicability: Hematological parameters, particularly CPDs and NLR, are promising tools for early severity stratification in COVID-19, providing fast, cost-effective, and minimally invasive support for clinical decision-making.
Advantages of CPDs: These parameters offer real-time insights during routine blood analysis, without requiring additional samples or testing time.
Limitations and future directions: The need for specialized equipment and the lack of standardization may restrict widespread CPD implementation. Further studies with larger, more diverse populations are warranted.
1. Introduction
COVID-19 is an acute respiratory infection caused by SARS-CoV-2, characterized by its potential severity, high transmissibility, and global distribution [1]. SARS-CoV-2, a betacoronavirus (subgenus Sarbecovirus, family Coronaviridae), was initially identified in bronchoalveolar lavage samples from patients with pneumonia of unknown etiology in Wuhan, Hubei Province, China, in December 2019 [2].
Clinical manifestations of coronavirus disease include symptoms such as fever, dyspnea, cough, and anosmia. The incubation period is typically 14 days after exposure and symptomatic infections range from mild to critical, with the majority being non-sever [3–12].
Severe disease can occur in otherwise healthy individuals of any age but is predominantly observed in older adults with specific underlying medical comorbidities. COVID-19 vaccination significantly reduces the risk of severe disease even in susceptible populations [13].
Specific demographic characteristics and laboratory abnormalities have also been associated with severe disease among hospitalized COVID-19 patients such as lymphopenia, elevated transaminases, lactate dehydrogenase, and elevated inflammatory markers (ferritin, C-reactive protein), along with coagulation test abnormalities [8,14,15].
The gold standard for biomolecular diagnosis of COVID-19 is the RT-qPCR, complemented by readily available rapid tests, ensuring widespread access to the general population at an affordable cost. Given the limitations of many laboratory biomarkers, recent years have seen the proposal of numerous new markers related to hematological parameters for infectious diseases [16]. However, none of these markers can be recommended as a sole criterion for COVID-19 or any other infection, independent of the etiological agent. It is advocated the use of a combination of biological parameters alongside clinical and imaging assessments to compose a comprehensive severity score for the disease.
A complete blood count (CBC) is widely employed for diagnosing acute infections and the modern hematology analyzers provide precise and cost-effective leukocyte differential counts swiftly. These enhanced technologies facilitate the inclusion of cell population data parameters (CPD) along with the basic CBC. CPDs offer quantitative and qualitative insights into the morphological and functional characteristics of leukocytes, enabling a more detailed study of cells in response to stimuli such as infections, providing valuable information on cellular activation status [17].
Thus, the central premise of our investigation contends that the measurement of CPD, and hematological parameters and D-dimer assays, could represent a robust approach, enhancing our understanding of the multifaceted implications of the complex pathophysiology induced by COVID-19 on the hematological dynamics. Sex-based differences in physiology, hormones, and behavior can significantly influence study outcomes. Focusing on a female-only cohort enhances analytical specificity and minimizes biological variability. Additionally, women have historically been underrepresented in biomedical research. By examining a homogeneous female sample, this study aims to generate findings that are both biologically meaningful and clinically applicable to women.
The aim of this study is to analyze the correlation between hematological markers and the severity of SARS-CoV-2 infection, proposing their application in the early stratification of the disease.
2. Methods
2.1. Study site
This retrospective study was conducted at the Hematology Laboratory of the Clinical Hospital Complex of University Federal do Paraná (CHC-UFPR), following thorough review and approval by the Research Ethics Committee, under protocol number CAAE 36697020.7.0000.0096. The research was carried out in close collaboration with the Molecular Virology Laboratory at UFPR, Curitiba, Paraná, Brazil. As this was a retrospective study, the ethics committee released the need for a free and informed consent form. This study adheres to the ethical principles contained in the Declaration of Helsinki, ensuring that all research procedures and methods are carried out in accordance with the rights of the participants and with respect for their dignity.
2.2. Specimens and patients
The subjects enrolled in the study were attended to in the screening, emergency, and/or hospitalized departments from March 2020 to July 2021 at the CHC-UFPR. The selection of study participants adhered to the following inclusion criteria: female patients aged >18 years with a positive RT-qPCR test for SARS-CoV-2, attended to and hospitalized in the wards or Intensive Care Unit (ICU) of CHC. Exclusion criteria encompassed female patients with a positive test for SARS-CoV-2 who concurrently presented hospital-acquired infection with COVID-19.
2.3. Study design
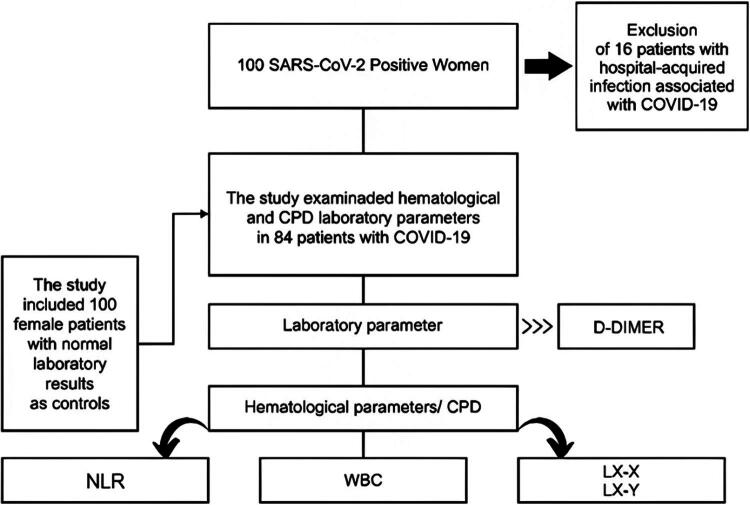
The study included two distinct groups (Figure 1), the first consisting of 84 female patients diagnosed with COVID-19, of whom 38 had severe cases requiring hospitalization in intensive care units (ICUs) and 46 had moderate cases requiring hospitalization in general wards. The second group consisted of 100 female patients without a diagnosis of COVID-19, a control group whose objective was to validate the hematological laboratory parameters proposed in the study, establishing a standard normal range for the population under investigation and analyzing the Receiver Operating Characteristic (ROC) curves to obtain the diagnostic accuracy of the relevant parameters in the study. It is important to note that the patients in the control group were outpatients with CBC and D-dimer results within the normal and reference criteria established for these tests. Both groups included individuals between the ages of 20 and 90 years, with a median age of 54 years for the control group and 56 years for the COVID-19 group. This study evaluated five continuous response variables, namely D-dimer, white blood cell (WBC) count, neutrophil-to-lymphocyte ratio (NLR), and cell population data (CPDs) related to WBC count and lymphocyte subpopulations (LY-X, LY-Y). Categorical covariates were hospital admission and outcome. Samples were collected in tubes containing K2-EDTA and processed on the Sysmex XN-3100 hematology analyzer. For D-dimer dosing, samples were collected in 3.2% sodium citrate and processed on Sysmex CS Series automated coagulometric analyzers, models 2100 and 2500.
Figure 1.
Flowchart. CPD: cell population data; D-dimer: mg/FEU: milligrams/fibrinogen equivalent units; WBC: white blood cell count expressed as × 1000/mm3; NLR: neutrophil to lymphocyte ratio; LY-X: lymphocyte complexity; LY-Y: lymphocyte fluorescence intensity.
2.4. Automated hematology analyzer
We used the Sysmex XN-3100 hematology analyzer (Sysmex Corporation, Kobe, Japan), which is an automated analyzer that uses different principles based on fluorescence flow cytometry to provide information on the shape, size and internal granularity of cells, thus generating total and differential leukocyte counts, as well as CPDs. It has a leukocyte differentiation channel that discriminates leukocytes by virtue of signals that are plotted on a scatter graph. The optical signals along the X-axis (lateral dispersion) are proportional to internal complexity; fluorescence along the Y-axis correlates with nucleic acid content (cell activation), while forward dispersion (Z-axis) is related to cell size. These parameters represent the distribution ranges of cell population clouds within the analysis channels (WDF: WBC differential and WNR: White cell nucleated). Among the various parameters of CPDs reported along the X, Y axes for all normal leukocyte populations, the parameters related to lymphocytes stand out in this study: X-axis the internal complexity of lymphocytes (LY-X), Y-axis the fluorescence intensity of lymphocytes (LY-Y) [17,18].
2.5. Automated coagulometric analyzer
This study used Sysmex CS series coagulometric analyzers, models 2100 and 2500 (Sysmex Corporation, Japan), which have more than one analytical module available, including coagulometry, immunoturbidimetry and colorimetry. D-dimer dosing was carried out using immunoturbidimetry, which is an analytical test that uses immunoprecipitation and light scattering to quantify the D-dimers present in plasma (polystyrene particles covered with monoclonal antibody undergo aggregation when in contact with samples containing D-dimer and the turbidity generated is proportional to the concentration).
2.6. RT-qPCR genotyping
Two probe-based genotyping protocols were used in sequence to identify variant of concern (VoC) [19]. The first used two deletion regions as targets – 69–70del in the spike and 3675–3677 del and in ORF1-a/b to identify Alpha, Beta/Gamma, without discriminating between the latter two [20]. The other protocol used primers 73 from the Thermo Fischer Scientific panel (Thermo Fisher Scientific, Applied Biosystems, US), which targets mutations K417T, E484K, N501Y, P681R, to identify Delta and Zeta variants, as well as differentiating Beta from Gamma.
2.7. Statistical analysis
Statistical analysis of the data was conducted using R Core Team software version (2021) and the GCMR (Gaussian Copula Marginal Regression) package. The significance level of 5% was adopted. To assess the normality of the data, the Shapiro-Wilk test was used. A normal (Gaussian) distribution was assumed for the response variable when p > 0.05. Otherwise, when p < 0.05, a Gamma distribution was assumed for the response variable. The null hypothesis H0 assumed normality for the proposed data. To evaluate the response performance of hematological laboratory parameters, statistical analyses were conducted using MedCalc software version 20.218 (MedCalc Software, Mariakerke, Belgium). The results were presented as ROC curves, indicating the sensitivity, specificity, and accuracy percentages of the parameters.
3. Results
The statistical analysis focused on three main parameters: D-dimer, WBC and NLR. Furthermore, the analysis of disease severity highlighted the relevance of CPDs of LY-X and LY-Y lymphocyte subsets in differentiating between moderate and severe disease. Table 1 summarizes the analysis of hematologic parameters in relation to COVID-19 severity.
Table 1.
Statistical analysis of hematological parameters in COVID-19 severity, comparing moderate and severe disease.
| *Hospital admission | Patient COVID-19 (n = 84) |
|||||||
|---|---|---|---|---|---|---|---|---|
| COVID-19 moderate (n = 46) IIQ | COVID-19 severe (n = 38) IIQ | |||||||
| Parameters | Med | P25 | P75 | Med | P25 | P75 | p-Value | Reference values |
| D-dimer | 1.31 | 0.63 | 3.32 | 2.19 | 1.0 | 4.24 | <0.001 | <0.55 mg/FEU |
| WBC | 7.1 | 5.5 | 10.6 | 10.7 | 7.2 | 12.4 | <0.001 | 3.8–11.0 × 10³/mm³ |
| NLR | 6.05 | 3.23 | 11.1 | 10.15 | 6.13 | 17.9 | <0.001 | <2.1 (1.5–2.0) |
| LY-X | 78.1 | 76.4 | 81.1 | 77.3 | 75.3 | 78.8 | 0.023 | 80.1 (79.6–80.4) |
| LY-Y | 71.9 | 70.7 | 75.3 | 70.2 | 67.7 | 72.7 | 0.002 | 72.8 (72.3–73.5) |
| **Hospital outcomes | ||||||||
| D-dimer | 1.44 | 0.62 | 5.91 | 1.09 | 0.63 | 4.62 | 0.128 | <0.55 mg/FEU |
| WBC | 7.9 | 5.9 | 11.2 | 10.9 | 7.7 | 15.6 | <0.001 | 3.8–11.0 × 10³/mm³ |
| NLR | 3.95 | 2.3 | 7.4 | 4.70 | 2.4 | 14.5 | <0.001 | <2.1 (1.5–2.0) |
| LY-X | 79.6 | 77.5 | 81.2 | 78.0 | 76.5 | 80.4 | 0.026 | 80.1 (79.6–80.4) |
| LY-Y | 74.9 | 72.1 | 77.0 | 72.9 | 67.5 | 75.7 | 0.024 | 72.8 (72.3–73.5) |
Legend Tables 1 and 2: Med.: median; (IIQ): interquartile range; P25: 25th percentile; P75: 75th percentile; D-dimer: mg/FEU: milligrams/fibrinogen equivalent units; WBC: blood cell count expressed as × 1000/mm3; NLR: neutrophil to lymphocyte ratio; LY-X: lymphocyte complexity; LY-Y: lymphocyte fluorescence intensity; *hospital admission: tests performed in the first 24 hours of hospitalization; **hospital outcome: last test performed before hospital discharge or death.
Table 2 presents the analysis of hematologic parameters at the time of outcome for the group of 84 will patients, with 20% of cases resulting in death and 80% of cases in recovery. The parameters that stood out were WBC and NLR in cases that resulted in death.
Table 2.
Statistical analysis of hematological parameters in the clinical outcome of COVID-19.
| *Hospital admission | Patient COVID-19 (n = 84) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Clinical outcome at discharge (n = 67) IIQ | Clinical outcome: mortality (n = 17) IIQ | |||||||
| Parameters | Med | P25 | P75 | Med | P25 | P75 | p-Value | Reference values |
| D-dimer | 1.20 | 0.74 | 3.24 | 3.70 | 2.31 | 11.0 | <0.01 | <0.55 mg/FEU |
| WBC | 8.6 | 6.0 | 10.9 | 11.5 | 7.8 | 13.0 | <0.001 | 3.8–11.0 × 10³/mm³ |
| NLR | 7.0 | 4.0 | 12.6 | 15.2 | 4.5 | 22.0 | 0.022 | <2.1 (1.5–2.0) |
| **Hospital outcomes | ||||||||
| D-dimer | 1.09 | 0.58 | 3.34 | 2.33 | 1.01 | 5.9 | <0.001 | <0.55 mg/FEU |
| WBC | 7.9 | 6.3 | 10.9 | 16.1 | 12.5 | 20.8 | <0.001 | 3.8–11.0 × 10³/mm³ |
| NLR | 3.3 | 2.3 | 5.5 | 16.6 | 6.8 | 42.5 | 0.002 | <2.1 (1.5–2.0) |
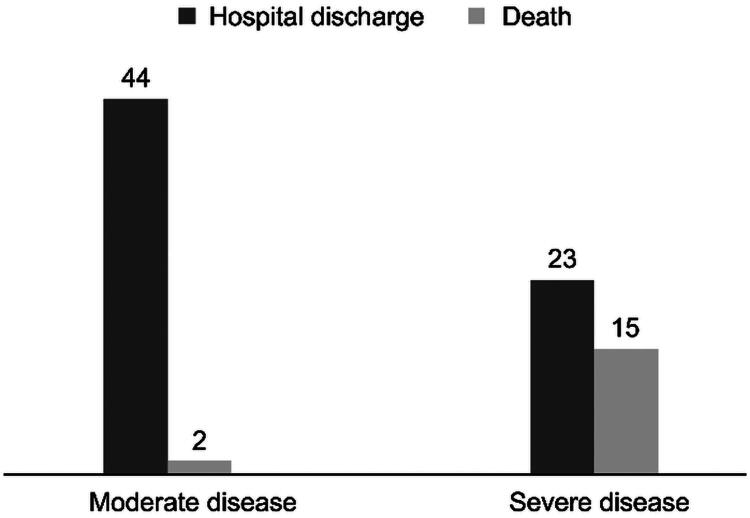
Figure 2 displays the correlation between severity and outcome of the 84 patients with COVID-19 was established using Fisher’s exact test, which indicated a statistically significant difference (p < 0.001) in cases that progressed to the worst outcome, with (n = 15) severe disease and (n = 2) moderate disease in cases that died. In addition, the analysis of the outcome shows that 67 patients recovered from the disease, including 23 patients with severe COVID-19.
Figure 2.
COVI-19 severity vs. outcome clinical.
Regarding genotyping, no significant results were observed when comparing the severity of the cases studied with the respective genotypes tested (p = 0.9602, Fisher’s test). The genotypes detected in the samples were Wild (13%), Alpha (1%), Gamma (34%) and Zeta (1%), totaling approximately 50% of the 84 patients. Of the 84 samples, 60 were tested and 11% were inconclusive, where the genotype could not be defined. Of the 17 patients who died, 82% were affected by the Gamma variant. This includes 12 severe and 2 moderate cases of COVID-19.
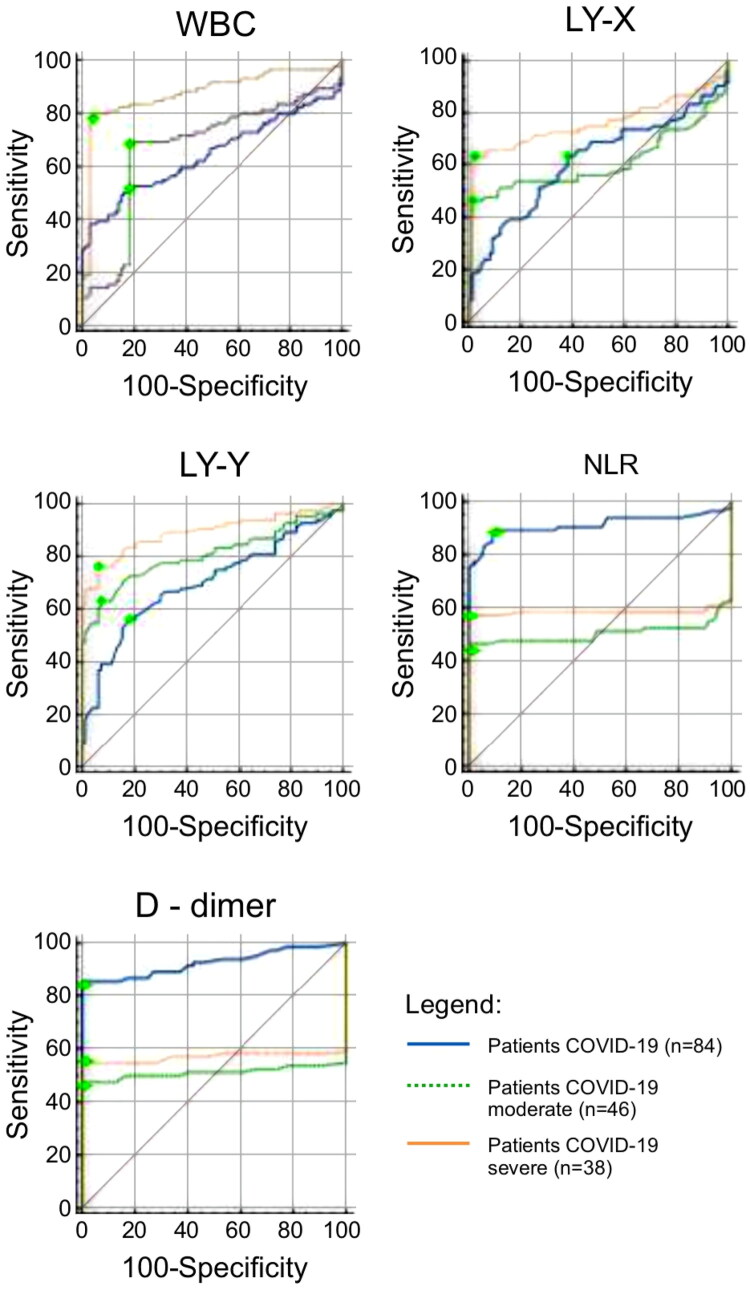
Tables 3 and 4 and Figure 3 show the results of the ROC curves, which confirm some of the statistical findings described above. In the comparison between moderate and severe COVID-19 in the ROC curves, the following hematological parameters: LY-X, LY-Y, NLR and WBC presented p-values <0.05, which corroborates the statistical results reported in Table 1 and demonstrates a discriminatory character for the severity of the disease.
Table 3.
ROC curves comparing COVID-19 groups.
| Parameters | Patient (n = 84) versus moderate (n = 46) | Patient (n = 84) versus severe (n = 38) | Moderate (n = 46) versus severe (n = 38) |
|---|---|---|---|
| p-Value | |||
| LY–X | 0.0243 | <0.0001 | 0.0074 |
| LY–Y | 0.9223 | 0.0176 | 0.0006 |
| NLR | <0.0001 | <0.0001 | 0.0284 |
| WBC | 0.6415 | <0.0001 | <0.0001 |
Note: Bolded values indicate a statistically significant difference between severe and moderate COVID-19 cases p < 0.05 for LY-X, LY-Y, NRL and p < 0,001 for WBC.
Legend table 3,4. D-dimer: mg/FEU: milligrams/fibrinogen equivalent units; WBC: white blood cell count expressed as × 1000/mm3; NLR: neutrophil to lymphocyte ratio; LY-X: lymphocyte complexity; LY-Y: lymphocyte fluorescence intensity.
Table 4.
Comparative statistical analysis of ROC curves in relation to parameters: WBC, NRL, D-dimer and LY-X, LY-Y.
| Parameters | COVID-19 | Accuracy | Sensitivity (%) | Specificity (%) | Youden index | Cut off |
|---|---|---|---|---|---|---|
| WBC | Patients (84) | 0.643 | 38.1 | 97 | 0.35 | >10420 |
| Moderate (46) | 0.669 | 57.5 | 72 | 0.29 | >8480 | |
| NLR | Severe (38) | 0.876 | 77.3 | 91.7 | 0.69 | >10420 |
| Patients (84) | 0.913 | 89.29 | 90 | 0.79 | >2.7 | |
| Moderate (46) | 0.504 | 27.6 | 68.5 | 0.04 | >2.7 | |
| D-DIMER | Severe (38) | 0.584 | 35.8 | 77 | 0.13 | >3.5 |
| Patients (84) | 0.928 | 92 | 93.5 | 0.85 | >0.55 | |
| Moderate (46) | 0.512 | 34.9 | 64.9 | 0 | >0.6 | |
| LY-X | Severe (38) | 0.570 | 41.5 | 63.35 | 0.04 | >1.0 |
| Patients (84) | 0.702 | 68.9 | 71.4 | 0.40 | ≤77.9 | |
| Moderate (46) | 0.808 | 57.5 | 99 | 0.56 | ≤77.9 | |
| LY-Y | Severe (38) | 0.894 | 76 | 97.6 | 0.74 | ≤78.2 |
| Patients (84) | 0.617 | 63.1 | 62 | 0.25 | >70.6 | |
| Moderate (46) | 0.610 | 54.8 | 66.4 | 0.21 | >70.6 | |
| Severe (38) | 0.765 | 48.7 | 98 | 0.47 | >65.8 |
Note: This table presents the diagnostic accuracy of the analyzed parameters. Not all parameters demonstrated good accuracy, as reflected in the tabulated results. These findings are further explored in the Discussion and Conclusion, where the most reliable indicators for screening COVID-19 severity are highlighted.
Figure 3.
Shows the ROC curves for WBC (leukocytes), lymphocyte subpopulation (LY-X and LY-Y), NLR (neutrophil/lymphocyte ratio), and D-dimer.
Hematological parameters, including D-dimer, suggest differentiation between moderate and severe disease, particularly during the assessment of patients at hospital admission. This indicates changes in the body’s innate cellular response to SARS-CoV-2 and disease progression in severe cases.
4. Discussion
COVID-19 presents clinical and laboratory characteristics that result from local and systemic inflammatory processes. These processes are potentiated by preexisting comorbidities, such as diabetes, obesity, systemic arterial hypertension, cardiovascular, renal, and pulmonary diseases, as well as biological factors such as gender and age [21]. However, investigations into the critical factors that profoundly influence the severity of the disease and death were prompted by the discrepancies in the clinical signs of COVID-19 observed among these patients [22].
The studies conducted in China reported that changes in peripheral blood inflammatory cells can indicate the severity and spread of COVID-19 [23,24]. They found that the majority of patients with severe COVID-19 symptoms had increased serum levels of cytokines and chemokines, as well as high neutrophil production and low lymphocyte production. This indicates that the hyperinflammatory response was responsible for the severity of the disease [25].
A meta-analysis revealed that 35–75% of patients developed lymphopenia [26]. The study found that 45% of severe cases had marked lymphopenia at the onset of SARS-COV-2 infection, which supports previous research. The LY-X and LY-Y parameters, which represent lymphopenia in this study, were found to be discriminatory between severe and moderate COVID-19, as shown by both logistic statistical analysis (p < 0.05).
The neutrophil-to-lymphocyte ratio (NLR) is a widely used marker for predicting bacterial infections, including respiratory syndromes and pneumonia. The study found that patients with a more severe condition had an increase in leukocytes and NLR compared to the mild-moderate group [27]. The NLR median value on admission was approximately 5 times higher than the reference value. Furthermore, a statistically significant difference between moderate and severe disease was observed (p = 0.0284).
It was observed that several COVID-19 patients experienced an increase in neutrophil count and a decrease in lymphocyte count during the severe phase, indicating significant dysregulation of the immune system and a critical condition in the most severe cases of the infection [8].
A retrospective multicenter study conducted in China during the first two months of the pandemic found that 46.4% of patients with SARS-COV-2 infection had elevated D-dimer values (>0.5 mg/L), with the highest results observed in the most severe cases [14]. The study results indicate an average 2.4-fold increase in the D-dimer values of patients with severe COVID-19. In the multivariate analysis, D-dimer was found to be an effective parameter for screening disease severity. However, although it has been identified that patients with higher initial levels of inflammatory markers such as ferritin, CRP, procalcitonin, and D-dimer are more likely to progress to critical illness and have a worse outcome, monitoring the treatment of patients using these markers did not prove to be a reliable tool for clinical decision-making [6,28].
Additionally, genotypes did not have any influence on the outcome of the most severe cases. However, it is important to note that the sample size for this analysis was small. Out of the 84 samples included, only 60 were tested and 11 were inconclusive. The study revealed that 34 (57%) patients had the gamma variant, with 70% of them being between the ages of 20 and 60. This indicates that a younger population was infected at a time when vaccination was beginning in the country for the population most at risk, which until then had been represented by the elderly (>65 years) and health professionals.
The study suggests that combined use of CPD, D-dimers, and NLR offers complementary, accessible, and cost-effective tools for early risk stratification in COVID-19. These markers can support clinical decision-making, optimize triage, and improve resource allocation, particularly in settings with limited capacity.
Although initial reports suggested higher mortality and severity of COVID-19 among men, subsequent epidemiological data have shown that women constitute a significant proportion of global COVID-19 cases. Nevertheless, studies focusing specifically on the female population particularly with regard to long-term outcomes, symptomatology, and comorbidities remain limited in the current literature. The present study aims to address this gap by providing sex-specific data that may support more equitable and targeted public health strategies.
The importance of sex-balanced research is recognized, and comparative studies involving both sexes are essential for a comprehensive understanding of COVID-19. However, research specifically focused on women is also considered necessary to identify potential differences in disease presentation, treatment response, and recovery patterns.
5. Conclusions
This study suggests that hematological laboratory parameters are effective screening tools for COVID-19 cases. Data collected at patient admission can aid in the classification of disease severity. Genotyping did not show any discernible influence on disease severity or outcomes. The significance of CPDs in this study cannot be overstated. They are generated during automated differential counting without the need for additional samples and provide a quantitative, objective, and highly accurate alternative to manual differential counts. Furthermore, CPDs have fast turnaround times and are cost-effective. However, their widespread use may be limited by the dependence on specific instruments. Despite their advantages, the lack of standardization is a limitation that requires cautious consideration of CPDs in clinical decisions. It is important to note that early diagnosis is crucial, as rapid intervention has the potential to reduce mortality.
Acknowledgment
To the Postgraduate Program in Tocogynecology and Women’s Health and to the Clinical Hospital Complex of the Federal University of Paraná, including the Virology and Hematology laboratories. To the Municipal Laboratory of Curitiba of the Municipal Government of Curitiba. To the clinical research team, including multidisciplinary resident Ludmilla Amadeu and doctors Lucas Bochnia Bueno and Bruna Amaral Lapinscki. To Siemens Diagnostics and Sysmex Roche Diagnostics. And to all the collaborators and professors who guided me during the research. This paper was not funded.
Appendix A.
Table A1.
Table of additional data: controls samples.
| No Covid-19 | Control samples (100) iIQ |
Reference values | |
|---|---|---|---|
| Parameters | Median | P25–P75 | |
| Age | 54 | 44–67 | |
| D-dimer | 0.36 | 0.27–0.45 | **<0.55 mg/FEU |
| WBC | 6.885 | 5.962–8.040 | 4.0–12.0 × 10³/mm³ |
| NLR | 1.69 | 1.20–2.30 | <2.0% |
| WNR-X | 149.5 | 148.2–152 | 141.40 − 159.20 ch |
| WNR-Y | 102.6 | 99.9–109.2 | 95.0 − 113.80 ch |
| WNR-WX | 300 | 276–407 | 227–491 ch |
| WNR-WY | 670 | 635–699 | 585–782 ch |
| NE-SSC | 151.4 | 146.8–153.6 | 150.6 − 52.0 ch |
| NE-SFL | 47.85 | 46.3–49.6 | 49.4–50.3 ch |
| WDF-WX | 456 | 441–533 | 297–772 ch |
| WDF-WY | 1.580 | 1410–1753 | 1.070–3.109 ch |
| LY-X | 79.95 | 78.4–81.7 | 79.6–80.4 ch |
| LY-Y | 69.80 | 68.2–72 | 72.3–73.5 ch |
| RBC | 4.67 | 4.36–4.87 | 4.05–5.25 × 106 |
| HB | 13.80 | 13–14.5 | 12.5–15.7 g/dl |
| HCT | 40.90 | 38.9–43 | 36.7–46.3% |
| RDW | 12.7 | 12.3–13.2 | <14.8% |
| PLQ | 265 | 222–291 | 140–400/mm³ |
Reference value established by the test manufacturer.
Note: The other reference values represent the average female outpatient population attended at CHC-UFPR (Curitiba, PR, Brazil), established according to the recommendations of CLSI document EP28-A3c.10Control group consisted of 100 asymptomatic women undergoing elective exams (age range 18–98).
These are results of the laboratory markers listed in patients who do not have COVID-19. These values are consistent with normal reference ranges for a healthy population matched by age to the studied COVID-19 cohort.
Med.: Median (IIQ): interquartile range; P25: 25th percentile; P75: 75th percentile; D-dimer: in milligrams/fibrinogen equivalent units; WBC: white blood cell count expressed as × 103/mm3; NLR: neutrophil to lymphocyte ratio; WNR-X: leukocyte complexity reported in arbitrary units of light scatter(ch); WNR-Y: leukocyte fluorescence intensity (ch); WNR-WX: width of dispersion of leukocyte complexity(ch); WNR-WY: width of dispersion of leukocyte fluorescence intensity(ch); NE-SSC: neutrophils complexity (ch); NE-SFL: neutrophils fluorescence intensity(ch); WDF-WX: differential of leukocyte channel complexity and width of dispersion(ch); WDF-WY: differential of leukocyte channel fluorescence intensity and width of dispersion(ch); LY-X: lymphocyte complexity(ch); LY-Y: lymphocyte fluorescence intensity (ch); RBC: red blood cells count expressed as × 106/mm3; HB: hemoglobin g/dl; HCT: hematocrit in percentile; RDW: red cell distribution width or anisocytosis coefficient; PLQ: Platelets count expressed as × 1000/mm3.
Author contributions
Conceptualization, S.R.C; S.C.C; M.B.N; methodology, S.C.C; S.M.L.; L.L.M.A.; L.BBC; and B.A.L software, S.C.C; S.R.C.; R.R.P; validation; S.C.C; S.R.C; M.B.N; formal analysis, S.C.C; S.R.C M.B.N.; investigation, S.C.C; S.M.L.; L.L.M.A.; L.BBC; and B.A.L; writing—original draft preparation, S.C.C; writing—review and editing, S.C.C; M.B.N.; S.R.C.; S.M.R; visualization, S.C.C; S.R.C; M.B.N; S.M.R; R.R.P; S.M.L.; L.L.M.A.; L.BBC; and B.A.L; supervision, S.R.C and M.B.N.; project administration, M.B.N.; resources and funding acquisition, S.R.C. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and additional data can be accessed at the URL:<https://hdl.handle.net/1884/87527> or the data will be shared by the corresponding author translation.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Hu B, Guo H, Zhou P, et al. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya A, Baker S, Baric RS, et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishak A, Mehendale M, AlRawashdeh MM, et al. The association of COVID-19 severity and susceptibility and genetic risk factors: A systematic review of the literature. Gene. 2022;836:146674. doi: 10.1016/j.gene.2022.146674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kousathanas A, Pairo-Castineira E, Rawlik K, et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature. 2022;607(7917):97–103. doi: 10.1038/s41586-022-04576-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juzėnas S. Genomewide association study of severe covid-19 with respiratory failure/severe Covid-19 GWAS Group: David Ellinghaus, Frauke Degenhardt, Luis Bujanda, Maria Buti, Agustín Albillos, Pietro Invernizzi,[…] Simonas Juzenas. N Engl J Med. 2020;383(16):1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z, McGoogan JM.. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- 11.Stokes EK, Zambrano LD, Anderson KN, et al. Coronavirus disease 2019 case surveillance – United States, January 22–May 30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(24):759–765. doi: 10.15585/mmwr.mm6924e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oran DP, Topol EJ.. The proportion of SARS-CoV-2 infections that are asymptomatic: a systematic review. Ann Intern Med. 2021;174(5):655–662. doi: 10.7326/M20-6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Paulson KR, Pease SA, et al. Estimating excess mortality due to the COVID-19 pandemic: a systematic analysis of COVID-19-related mortality, 2020–21. Lancet. 2022;399(10334):1513–1536. doi: 10.1016/S0140-6736(21)02796-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan WJ, Ni ZY, Hu Y, et al. China medical treatment expert group for covid-19. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goyal P, Choi JJ, Pinheiro LC, et al. Clinical characteristics of COVID-19 in New York City. N Engl J Med. 2020;382(24):2372–2374. doi: 10.1056/NEJMc2010419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan SL, Miller NS, Lee J, et al. Diagnosing sepsis – the role of laboratory medicine. Clin Chim Acta. 2016;460:203–210. doi: 10.1016/j.cca.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaves F, Tierno B, Xu D.. Neutrophil volume distribution width: a new automated hematologic parameter for acute infection. Arch Pathol Lab Med. 2006;130(3):378–380. doi: 10.5858/2006-130-378-NVDWAN [DOI] [PubMed] [Google Scholar]
- 18.Urrechaga E, Bóveda O, Aguirre U.. Role of leucocytes cell population data in the early detection of sepsis. J Clin Pathol. 2018;71(3):259–266. doi: 10.1136/jclinpath-2017-204524 [DOI] [PubMed] [Google Scholar]
- 19.Adamoski D, de Oliveira JC, Bonatto AC, et al. Large-scale screening of asymptomatic persons for SARS-CoV-2 variants of concern and gamma takeover, Brazil. Emerg Infect Dis. 2021;27(12):3124–3127. doi: 10.3201/eid2712.211326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogels CBF, Brito AF, Wyllie AL, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pradhevi L, Soegiarto G, Wulandari L, et al. More severe comorbidities, advanced age, and incomplete vaccination increase the risk of COVID-19 mortality. Narra J. 2024;4(2):e949. doi: 10.52225/narra.v4i2.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilger-Casagrande M, de Barros CT, Antunes VAN, et al. Perspectives and challenges in the fight against COVID-19: the role of genetic variability. Front Cell Infect Microbiol. 2021;11:598875. doi: 10.3389/fcimb.2021.598875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun S, Cai X, Wang H, et al. Abnormalities of peripheral blood system in patients with COVID-19 in Wenzhou, China. Clin Chim Acta. 2020;507:174–180. doi: 10.1016/j.cca.2020.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oktiviyari A, Hayati Z, Zanaria T.. Cytokine storm in SARS-CoV-2 infection and the potential treatments. Trends Infect Global Health. 2022;2(1):1–13. doi: 10.24815/tigh.v2i1.25901 [DOI] [Google Scholar]
- 26.Lippi G, Plebani M.. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020;58(7):1131–1134. doi: 10.1515/cclm-2020-0198 [DOI] [PubMed] [Google Scholar]
- 27.Fleury MK. COVID-19 and the clinical hematology laboratory: a review of recent literature. Rev Bras Anal Clin. 2020;52(2):131–137. DOI: 10.21877/2448-3877.20200003 [DOI] [Google Scholar]
- 28.Pfister F, Vonbrunn E, Ries T, et al. Complement activation in kidneys of patients with COVID-19. Front Immunol. 2021;11:594849. doi: 10.3389/fimmu.2020.594849 [DOI] [PMC free article] [PubMed] [Google Scholar]
Reference annotations
- *1.Hu B, Guo H, Zhou P, et al. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]; The study provides a comprehensive overview of the salient characteristics of SARS-CoV-2 and the resulting condition, known as "COVID-19". It undertakes a thorough examination of the virology, zoonosis, pathology, epidemiology, antiviral treatments, and animal models associated with the virus.
- **6.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]; The article provides information about SARS-CoV-2 infection in Wuhan, which caused severe cases of pneumonia with high rates of ICU admission and mortality, presenting respiratory symptoms, pulmonary abnormalities in imaging tests, and elevated inflammatory markers, especially in critically ill patients.
- *18.Urrechaga E, Bóveda O, Aguirre U.. Role of leucocytes cell population data in the early detection of sepsis. J Clin Pathol. 2018;71(3):259–266. doi: 10.1136/jclinpath-2017-204524 [DOI] [PubMed] [Google Scholar]; The article addresses cellular population data (CPD) parameters, especially MO-X and NE-SFL, in the early diagnosis of sepsis. The creation of the NEMO score allows for effective risk stratification and facilitates rapid and reliable clinical decisions.
- **23.Sun S, Cai X, Wang H, et al. Abnormalities of peripheral blood system in patients with COVID-19 in Wenzhou, China. Clin Chim Acta. 2020;507:174–180. doi: 10.1016/j.cca.2020.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]; The present study demonstrates that dynamic analysis of blood counts, especially eosinophils, NLR, and MLR, exhibited significant predictive value for the severity of cases of severe acute respiratory syndrome (SARS) caused by the novel coronavirus (SARS-CoV-2). This dynamic analysis is useful in the early identification and monitoring of severe cases.
- **24.Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]; The study indicates that the novel coronavirus primarily affects T lymphocytes, and that changes in the neutrophil-to-lymphocyte ratio (NLR) and lymphocyte subsets may aid in the early identification and management of severe cases.
- **26.Lippi G, Plebani M.. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020;58(7):1131–1134. doi: 10.1515/cclm-2020-0198 [DOI] [PubMed] [Google Scholar]; The article underscores the pivotal role of laboratory medicine in combating the pandemic, and it presents a comparative analysis of the severity of the known coronaviruses: SARS, MERS, and SARS-CoV-2. The analysis also highlights frequent laboratory findings, such as elevated levels of CRP, ESR, LDH, and D-dimer, which play a crucial role in the diagnosis, monitoring, and stratification of disease severity.
- **27.Fleury MK. COVID-19 and the Clinical Hematology Laboratory: a Review of Recent Literature. Bras Anal Clin. 2020;52(2). [Google Scholar]; The study underscores the multifaceted nature of the disease, asserting that while it is predominantly characterized as a respiratory illness, it also has the potential to impact multiple physiological systems, manifesting in hematological and coagulation changes that hold significant prognostic value. These alterations serve as crucial indicators, facilitating the timely identification of severe cases and the development of targeted therapeutic interventions.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and additional data can be accessed at the URL:<https://hdl.handle.net/1884/87527> or the data will be shared by the corresponding author translation.