Abstract
Background
Hepatitis B virus associated hepatocellular carcinoma (HBV-HCC) have been a serious global health problem. This study aimed to uncover the key genes in HBV-HCC, and clarity their function, interaction, diagnostic and prognostic value, impacts on immune infltration and potential drugs targeting these genes.
Methods
Four gene expression datasets totally containing 117 paired tumor tissues and adjacent control tissues were selected from the GEO database and used to screen the differentially expressed genes (DEGs). Function analysis were performed by using GO and KEGG enrichment. STRING and cytoscape were used to analyze protein-protein interaction (PPI) and screen hub gene. Survival analysis and receiver operator characteristic (ROC) curve were used to explore the prognostic and diagnostic value of key genes. Immune infiltration analysis were performed by CIBERSORT algorithm. Drug–Gene Interaction Database (DGIdb) was used to screen the potential drug that affect hub genes.
Results
Overall, 234 shared DEGs were screened from four GSE datasets, which were mainly enrichment in cell growth regulation, epoxygenase P450 pathway, cellular response to multiple ion, xenobiotic metabolic process and complement activation. Six hub genes (HMMR, NDC80, CDK1, EZH2, ESR1, FOXM1) were screen by PPI analysis. ESR1 was down-regulated and associated with favorable prognosis in HBV-HCC, while HMMR, NDC80, CDK1 and EZH2 were up-regulated and correlation with shorter overall survival. Furthermore, ROC analysis and nomogram demonstrated the high diagnostic performance of NDC80, CDK1 and EZH2. Immune infiltration analysis showed that there were significant difference of several immune cell types between tumor and control tissues, including T cells, monocyte/macrophage and dendritic cells. There were significant correlation between hub genes with immune infiltration. Finally, DGIdb analysis showed there were several approved or new drugs that interaction with HMMR, CDK1, ESR1 and EZH2.
Conclusion
Six hug-genes are closely related to the HBV-HCC development, which involved in multiple biological progress and immune infiltration. Among them, NDC80, CDK1, EZH2 could severed as markers with good diagnostic and prognostic value. Notably, several approved drugs interaction with hub genes might be potential drug used for HBV-HCC therapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-025-03320-6.
Keywords: HBV-HCC, Differentially expressed genes, Diagnose, Prognosis, Immune infiltration
Introduction
Hepatocellular carcinoma (HCC) is the sixth most commonly diagnosed cancer and third leading cause of cancer-related deaths globally [1]. Hepatitis virus infection, excessive drinking and Aflatoxicosis are the important risk factors in HCC pathogenesis. Chronic infection with hepatitis B virus (HBV) is considered a major risk factor for the HCC occurrence and progression, accounting for more than half of global HCC cases [2]. About 240 million people worldwide were estimated to be chronically infected with HBV, presenting a serious global health problem [3]. The influnces of HBV infection on host cells were complex, including enhanced genomic instability, chromosomal and epigenetic reshaping, abnormal expression of key genes involved in HCC development, change of immune microenviroment and etc [4, 5]. Therefore, there is an urgent need to screen the important molecules in HBV-HCC, which might be key pathogenic factors, prognostic markers or novel therapeutic targets.
In this study, we aimed to identify HBV-HCC related genes with significant expression change compared with paired pericarcinomatous tissue. Based on Comprehensive analysis of RNA sequencing data from HBV-HCC patients was performed, including differentially expressed genes (DEGs) screening, function enrichment, molecule network construction, hub gene screening, prognostic analysis, diagnostic performance analysis, immune infltration analysis and etc. As a results, we screened six hub genes assciated with HBV-HCC. These hub genes might be play inmportant roles in HBV-HCC development, and used for prognosis evaluation. Notably, there have been FDA approved drugs which targets to several hub genes decoved in this study, which offer possible features for HBV-HCC treament.
Methods
Data sources and differential expression gene screening
Gene expression datasets of HBV related HCC were searched in the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database. The datasets which contained more than 10 HBV positive paired specimen (i.e. tumor tissues and adjacent normal tissue) were included in this study. The information of GSE47197, GSE55092, GSE84402 and GSE121248 datasets were listed in Table 1. Then the The expression difference of genes between tumor tissues and adjacent control tissue were analyzed by using R software. Differentially expressed genes (DEGs) were screened with |Log2FC|>1 and P < 0.05. P < 0.05 represents significant statistical significance. |Log2FC|>1 represents the difference of expression levels reached more than twice.
Table 1.
GSE datasets information
Function analysis for DEGs
GO and KEGG pathway analysis of the DEGs were performing by using DAVID bioinformatics Resources (https://david.ncifcrf.gov/tools.jsp) [6]. R Bubble diagrams are used for visualization the enriched GO term and KEGG pathway.
Protein-protein interaction (PPI) network analysis
The DEGs were used to construct PPI networks. The interactions among proteins encode by the DEGs were predict base on The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, and the confidence score threshold is set to 0.4, which is the medium confidence of interaction score. The Cytoscape software were used to reconstruct, analyze and visualize the PPI network and screen hub genes, based on the distinct algorithm, including Betweenness, Closeness and Degree.
Prognostic value of hub genes
Kaplan-Meier (KM) Plotter was used to perform survival analysis [7]. To divide the HCC patients into two groups with high-expression and low-expression, the “auto select best cutoff” model was used to choose the best performing threshold. Survival curves of each hub genes were plotted, and The hazard ratio (HR) and log-rank P-value were used to evaluate the prognostic value of genes.
Diagnostic value of hub genes
The diagnostic performance of hub genes were investigated by receiver operator characteristic curve (ROC) analysis. Multiple index diagnosis model was constructed by using logistic regression analysis. Nomogram was generated by using R “rms” package, assigning scores to genes based on their expression levels.
Drug - gene interaction
The Drug–Gene Interaction Database (DGIdb; https://dgidb.org/) is for precision medicine and drug discovery. DGIdb 5.0 was used to examine the potential therapeutic targets of the selected hub genes.
Gene-gene, TFs-gene and microRNA-gene interaction
GeneMANIA database (https://genemania.org/) was used to analyze the gene-gene interaction network of hub genes [8]. The transcription factors (TFs)-gene regulatory network is constructed using the Networkanalyst platform based on JASPAR database. The microRNA-gene interaction is anlysis by Networkanalyst based on miRTarBase database.
Immune cell infiltration analysis
Immune cell infiltration analysis was performed by using CIBERSORT algorithm. The original expression matrix from four GEO datasets (GSE47197, GSE55092, GSE84402 and GSE121248) were combined. To compare differences in immune cell infiltration between HBV-HCC and control tissues, we generated box plots by using “ggplot2” R package to visualize these differences. Additionally, the correlation between hub genes and immune cell subsets in HBV-HCC were calculated using the R “corr.plot” package, and the results were visualized by bubble diagram.
Statistical analysis
In this present study, R version 4.4.3 was employed to perform statistical analyses. The comparison of continuous variables between the two groups was performed using the Student’s t-test. The pearson correlation analysis was used to analyzed the correlation between continuous variables. P < 0.05 was considered statistically significant.
Results
DEGs screening and function analysis
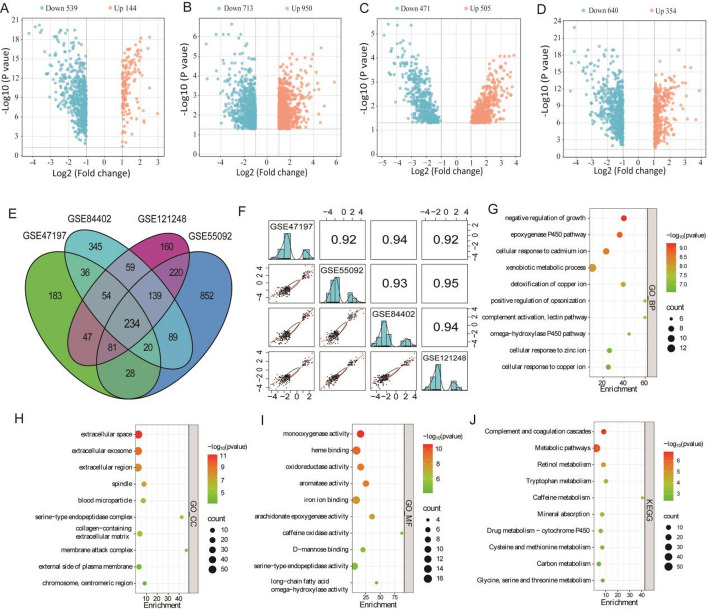
The workflow of this study was shown in supplementary Fig. 1. Firstly, gene expression profile of HBV-HCC and paired adjacent control tissue were obtained from GEO datasets. The results showed that 683 (144 up; 539 down), 1663 (950 up; 713 down), 976 (505 up; 471 down) and 994 (354 up; 640 down) DEGs were obtained from GSE47197, GSE55092, GSE84402 and GSE121248 respectively (Fig. 1A–D). Subsequently, 234 common DEGs (53 up; 181 down) were screened (Fig. 1E). Notably, the correlation of expression change of the 234 common DEGs among four datasets were high (r > 0.9) (Fig. 1F).
Fig. 1.
DEGs screening and function analysis based on GSE datasets. A The DEGs of GSE47197 dataset. B The DEGs of GSE55092 dataset. C The DEGs of GSE84402 dataset. D The DEGs of GSE121248 dataset. E Venn diagram showed the number of shared DEGs among four datasets. F The correlation matrix of 234 shared DEGs among four datasets. G–I GO analysis for 234 shared DEGs. J KEGG analysis for 234 shared DEGs
Then, function analysis of the 234 DEGs were performed by using GO and KEGG enrichment. For biological process (GO_BP), these DEGs were mainly enrichment in growth regulation, epoxygenase P450 pathway, cellular response to ion (including Ca, zinc and copper), xenobiotic metabolic process, opsonization regulation and complement activation (Fig. 1G). GO_CC (cellular component) analysis showed that these DEGs were wildly expressed in the extracellular, spindle, blood microparticle, collagen-containing extracellular matrix and external side of plasma membrane (Fig. 1H). GO_MF (molecular function) analysis showed that these DEGs related to monooxygenase activity, heme binding, oxidoreductase activity, aromatase activity, iron ion binding and arachidonate epoxygenase activity (Fig. 1I). KEGG pathway analysis showed that these DEGs were involved in complement and coagulation cascades, metabolic pathways (including metabolism of Retinol, Tryptophan, Caffeine, Drug, Cysteine and methionine, etc.), and mineral absorption (Fig. 1J).
PPI conduction and hub gene selection
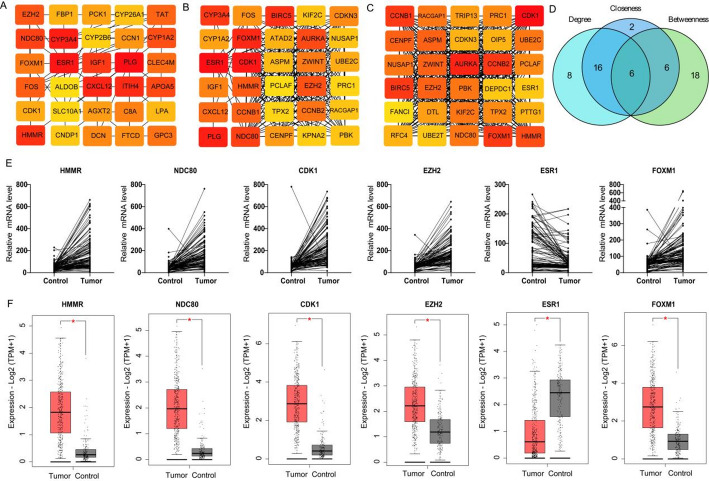
To explore the hub genes in HBV-HCC, the 234 DEGs were used to construct PPI network. By using k-means clustering, the comprehensive PPI network comprising 192 DEGs, the others were excluded due to a lack of interaction. Then, Cytoscape was used to visualize the PPI network. Firstly, three distinct algorithms were used to screen top 30 interactive DEGs respectively (Fig. 2A–C). Then, 6 shared DEGs among these algorithms were screened (Fig. 2D). Among 6 hub DEGs, five genes were up-regulated (i.e. HMMR, NDC80, FOXM1, CDK1 and EZH2) and ESR1 was down-regulated (Fig. 2E). These expression change of the 6 hub DEGs were validated base on TCGA data, which is in line with the results from GSE datasets (Fig. 2F).
Fig. 2.
PPI network construction and hub genes screening. A–C PPI network construction based on Betweenness, Closeness and Degree algorithm using Cytoscape. D Venn diagram showed the number of shared hub genes. E The expression changes of hub genes in paired tumor and adjacent control tissues based on GSE datasets. F The expression changes of hub genes in TCGA database
Prognostic analysis of hub DEGs
KM plotter was used to explore the correlation between HBV-HCC patients’ overall survival (OS) and expression of hub DEGs. Survival curve showed the correlation between poor prognosis and higher expression of HMMR, NDC80, CDK1 and EZH2 (HR > 2, P < 0.05; Fig. 3A–D). However, high ESR1 expression were associated with the longer OS (HR = 0.4, P < 0.05; Fig. 3E). In addition, there was no significant prognostic value of FOXM1 in HBV-HCC (HR = 1.87, P = 0.062; Fig. 3F). For progression free survival (PFS), As shown in Fig. 3G–L, high HMMR and EZH2 expression was correlated to poor PFS. However, these were no significant correlation between PFS and expression of NDC80, CDK1, ESR1 and FOXM1.
Fig. 3.
Prognostic value of hub genes in HBV-HCC patients. A–F Overall survival of patients with high or low expression of HMMR, NDC80, CDK1, EZH2, ESR1 and FOXM1. G–L Progression Free Survival of patients with high or low expression of HMMR, NDC80, CDK1, EZH2, ESR1 and FOXM1
The diagnostic value of hub genes for HBV-HCC
We further investigated the diagnostic values of 6 hub DEGs in HBV-HCC. Based on ROC analysis (Fig. 4A–F), the results indicated good diagnostic performance of CDK1 (AUC 0.929, 95% CI: 0.894–0.964), EZH2 (AUC: 0.900, 95% CI: 0.857–0.943) and NDC80 (AUC: 0.898, 95% CI: 0.857–0.939). ROC analysis showed the ordinary diagnostic performance of HMMR (AUC: 0.859, 95% CI: 0.812–0.907) and FOXM1 (AUC: 0.832, 95% CI: 0.778 to 0.887), and the poor diagnostic value of ESR1 (AUC: 0.631, 95% CI: 0.560 to 0.703). Furthermore, we established a combine diagnostic models based on logistic regression analysis. Subsequently, the nomogram was generated (Fig. 4G). ROC analysis showed that the diagnostic performance of combine model higher than the single gene (AUC: 0.934, 95% CI: 0.900-0.968; Fig. 4H).
Fig. 4.
Diagnostic value of hub genes. A–F ROC analysis showed the diagnostic performance of HMMR, NDC80, CDK1, EZH2, ESR1 and FOXM1. G, H Nomogram showed the diagnostic value of multi-gene combined diagnostic model
Drug-targets analysis
As the significant expression change and prognostic value of the five hub genes, i.e. HMMR, NDC80, CDK1, EZH2 and ESR1. We further explore the potential clinical drug interaction with above gene. Base on DGI database, there were 4 approved drugs interaction with HMMR, 6 approved drugs interaction with CDK1, 2 approved drugs interaction with EZH2 and 105 approved drugs interaction with with ESR1 (Supplementary Table 1).
Gene-gene and TF-genes interaction network analysis
By using GeneMANIA tool, the Genes-Genes interaction network of hub genes was construction. The top 20 related genes that changed most frequently with hub genes was shown in Fig. 5A. HMMR, NDC80, CDK1, EZH2, ESR1 and FOXM1 interact with the surrounding shared genes through co-expression, physical interaction, co-localization, prediction, genetic interaction and pathways. These genes were mainly correlated with mitotic nuclear division, microtubule cytoskeleton organization, spindle, cell cycle G2/M phase transition, chromosomal region and cell cycle checkpoint (Fig. 5B). By using Networkanalyst, the TFs-genes interaction network was constructed to find potential regulatory molecules for hub genes (Fig. 5C–H). These results indicated that FOXC1, NRF1, YY1 and POU2F2 are the key transcription factors, which are related to the regulation of at least three of the hub genes. Furthermore, the potential microRNA related to hub genes was found by Networkanalyst based on miRTarBase database (Supplementary Table 2). hsa-miR-193b-3p was correlated with NDC80, CDK1, EZH2 and ESR1.
Fig. 5.
The interaction network of hub genes. A Gene-gene network of hub genes was analyzed via GeneMANIA. B GO enrichment of genes interaction with hub genes. C–H Networkanalyst showed the TFs-gene network of HMMR, NDC80, CDK1, EZH2, ESR1 and FOXM1
Correlation between hub genes and immune infiltration
To further explore the differences of various immune cells infiltration in the HBV-HCC and control groups, the proportions of each sample in 22 kinds of immune cells were obtained based on the “CIBERSORT” algorithm. The results showed that compared with control, the proportion of native CD4 T cells, regulatory T cells (Treg), M0 macrophage and activated dendritic cells (DCs) were significant higher in HBV-HCC tissues, and the proportion of CD8 T cells, γδT cells, monocytes and activated mast cells were significantly down-regulated (Fig. 6A). Furthermore, we investigated the associated hub genes and immune cells in HBV-HCC. The results showed that HMMR, NDC80 and FOXM1 were significantly positively correlated with eosinophils, Treg cells and activated DCs, while negatively correlated with CD8+ T cells, monocytes and M1 macrophage. EZH2 was significantly positively correlated with DCs and Tregs, while negatively correlated with CD8+ T and M1 macrophage. ESR1 was significantly positively correlated with eosinophils, activated DCs and resting NK cells, while negatively correlated with macrophage, plasma cells and neutrophils. CDK1 was positively correlated with activated DCs and negatively correlated with resting mast cells (Fig. 6B–G). As the immune checkpoint play important roles in immune regulation. We further examined the correlation between hub genes and immune checkpoints. The results showed that there were no significant correlation between HMMR, NDC80, CDK1 and most of the immune checkpoint. However, ESR1 and FOXM1 was positively correlated with multiple immune checkpoint (Fig. 6H).
Fig. 6.
Immune infiltration analysis of HBV-HCC. A The difference of 22 types of immune cells infiltration between tumor tissues and adjacent control tissues based on four GSE datasets. B–G The correlation between hub genes expression (HMMR, NDC80, CDK1, EZH2, ESR1 and FOXM1) and immune cell infiltration in tumor tissues. (H) The correlation between the expression of hub genes and immune checkpoint
Discussion
HBV infection is one of the main contributor in HCC development. The number of patients infected with HBV is huge worldwide, which is a significant threat to global public health. Study of the key genes involved in HBV-HCC development was helpful for discovering the tumorigenic mechanism and therapeutic targets. In this study, we screened the key DEGs based on GSE datasets of HBV-HCC, and further explore the prognostic value, diagnostic performance, molecule network and the immune correlation of these key genes.
Due to the high heterogeneity of tumors, only the paired samples (tumor and pericarcinomatous tissue) were selected to screened the DEGs in this study. Based on gene expression data of 117 paired samples in four GSE datasets, 234 shared DEGs were screened, which were involved in with cell growth, xenobiotic metabolism, P450 pathway, complement activation and cell response to ion (cadmium, copper, zinc). By PPI analysis, 6 hug genes were found, including 5 genes with up-regulated expression in tumor tissues (HMMR, NDC80, CDK1, EZH2, FOXM1) and one down regulated gene -ESR1.
Hyaluronic acid is the markers of liver fibrosis and cirrhosis [9]. HMMR, a receptor for hyaluronic acid, is widely expressed in various cell types and plays important role in cell migration, growth and differentiation [10]. Study have showed that HMMR expression was increased and related to poor prognosis in multiple cancer [11, 12]. In this study, we showed the HMMR expression was significant increased and correlation with poor OS in HBV-HCC patients. He L et al.’s study have demonstrated that HMMR could provide suitable conditions for tumor formation during HBV-HCC via alleviating endoplasmic reticulum stress [13]. Wu H and colleagues found that HMMR triggers immune evasion of HCC by inactivation of phagocyte killing, and targeting HMMR could enhance anti-PD-1 treatment efficiency by recruiting CD8+ T cells [14]. Consistently, immune infiltration in this study showed HMMR was positively correlation with Tregs and negatively correlation with CD8+ T cells. Notably, there have been FDA-approved drug which have interaction with HMMR.
NDC80, CDK1 and FOXM1 are involved in cell cycle and proliferation. NDC80 organize and stabilize microtubule-kinetochore interactions and is required for proper chromosome segregation. Ju LL and colleagues have found that NDC80 knockdown reduced HCC cell proliferation and colony formation [15]. CDK1 and FOXM1 are key regulator for G2/M phase in cell cycle. Study has reported that FOXM1 could activate HMMR transcription and induced bladder cancer cells growth and invasion [16]. Uncontrollable cell proliferation is an important cause and characteristic of tumorigenesis. In this study, we showed that NDC80 and CDK1 were significant up-regulated in HBV-HCC and correlated with poor prognosis. A number of small molecules targeting CDK1 or multiple CDKs have been developed and evaluated in preclinical studies, such as BEY1107, Flavopiridol, Roniciclib and ect [17]. Moreover, PHA-793,887 is a novel CDK inhibitor, which has not been used in HCC therapy. However, Caputo WL et al.’s study also presenting the potential application in HCC [18]. EZH2 encodes a member of the Polycomb-group (PcG) family, regulate the methylation of H3 histone 27-Lys (H3K27), leading to transcriptional repression of the affected target gene. EZH2 has been regarded as a promising therapeutic targets for lymphoma [19, 20]. However, EZH2 target drug has not been used for solid tumor treatment. Overexpression of EZH2 in solid tumors is associated with aggressive tumor behavior and poor prognosis, due to epigenetic silencing of tumor suppressor genes which drives uncontrolled cell growth and metastasis [21–23]. This study showed the increase expression of EZH2 in HBV-HCC and associated with the shorter survival, which indicated the potential therapeutical effect of EZH2 blockage. Recently, Bai J et al.’s study have showed the silence of EZH2 by using antisense oligonucleotides could suppresses tumor growth in a xenograft mouse model of HCC [24]. The selective EZH2 inhibitor is currently under investigation in various solid tumors and cases of advanced cancer with hepatic impairment [25]. Lee J et al.’s study showed that tazemetostat (a EZH2 inhibitor) treatment could reducing levels of ferroptosis markers FSP1, NFS1, and SLC7A11 in HCC cells, reduced cell viability and increased lipid peroxidation in dose-dependently, which indicated the potential application of EHZ2 targeted therapy in HCC [26].
Notably, NDC80, CDK1 and EZH2 showed the good diagnostic performance for HBV-HCC. Nomogram analysis showed the better diagnostic value of three-gene combine model. Notably, besides molecular genetic factors, multiple other factors were correlation with survival of HCC patients. Li YK and colleagues’ study have showed the portal venous and hepatic arterial coefficients is valuable for independently predicting postoperative survival of HCC patients [27]. Li J et al.’s study have showed that a higher level of lymphatic vessel density was significantly associated with poor prognosis of HCC [28].
In this study, ESR1 was down-regulated in HBV-HCC and correlated with favorable prognosis, which indicated that ESR1 might be a protective factor. Wang DH and colleagues found that ESR1 could suppresses HCC by restricting M2 macrophage infiltration through the YAP-CCL2 axis [29]. Supportively, This study showed the negative correlation between ESR1 expression and M2 macrophage infiltration in HBV-HCC.
There are some limitation of this study. Firstly, the expression levels of hub genes were not verified in clinical tissues. We will investigate these genes expression on clinical samples from HBV-HCC in subsequent study. Secondly, there were still unclear about that whether the expression change of these hub genes are only concomitant markers of tumor occurrence, or these hub genes contribute to HBV-HCC progression. Previous evidence have indicated the potential function of EZH2, ESR1 and HMMR on HCC progress. We will follow up with cellular and animal experiments to clarify the potential function and mechanism of other hub genes. Thirdly, we observed a significant correlation between hub genes expression and patients’ survival. However, other factors were not included in survival analysis, such as caner stage, drink condition and etc. Further study will perform a more detailed stratified analysis for prognostic value of these hub genes in HBV-HCC patients. Fourthly, ESR1 expression was decreased and severed as a favourable prognostic marker in HBV-HCC, which indicated the protective effects ESR1 during HCC progression. However, ESR1 expression showed a negative correlation with cells with immune killing activity (including M1 macrophage and CD8 T cells), and a positive correlation with immunosuppressive Treg cells. This is not consistent with the fundamentals of tumor immune microenvironment in general. The answer to this issue still needs to be based on the accumulation of more study on ESR1. Finally, we have screened several drugs target hub genes that may contribute to therapeutic sensitization of HBV-HCC patients. However, these drugs have yet to be validated for their effects and potential toxicity by animal experiments and clinical trail. We will improve this work subsequently at the cellular and animal experimental levels.
In conclusion, we identified six hub genes that play important roles in HBV-HCC, with significant expression change. These hug-genes involved in cell growth and immune infiltration regulation. NDC80, CDK1, and EZH2 might served as the diagnostic and prognostic marker in HBV-HCC. ESR1 might be a protective molecule in HBV-HCC. Several new or approved drugs have potentially therapeutic value against HBV-HCC, such as tazemetostat, clofibrate, flavopiridol and ect.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Material 1—The drug-gene interaction via DGIdb analysis
Supplementary Material 2—The microRNA related to hub genes found by Networkanalyst based on miRTarBase database
Supplementary Material 3—The flowchart of this work
Acknowledgements
Not applicable.
Author contributions
LR: Conceptualization, Methodology, Writing-original draft, Writing-review and editing. LY: Conceptualization, Data curation, Investigation, Supervision, Writing-original draft. LS: Formal Analysis and Supervision, Writing-review and editing. JH and QH: Visualization, Writing-original draft. All authors reviewed the manuscript.
Funding
This work was supported by the Youth Project Natural Science Foundation of Shaanxi Province (2024JC-YBQN-0878).
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information.
Declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical approval
Not applicable to this type of manuscript.
Consent to publication
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. 10.3322/caac.21834. [DOI] [PubMed] [Google Scholar]
- 2.Jia L, Gao Y, He Y, Hooper JD, Yang P. HBV induced hepatocellular carcinoma and related potential immunotherapy. Pharmacol Res. 2020;159:104992. 10.1016/j.phrs.2020.104992. [DOI] [PubMed] [Google Scholar]
- 3.Fu MX, Lambert G, Cook A, Ndow G, Haddadin Y, Shimakawa Y, et al. Quality of life in patients with HBV infection: A systematic review and meta-analysis. JHEP Rep. 2025;7:101312. 10.1016/j.jhepr.2024.101312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y, Han Q, Zhao H, Zhang J. The mechanisms of HBV-Induced hepatocellular carcinoma. J Hepatocell Carcinoma. 2021;8:435–50. 10.2147/JHC.S307962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’souza S, Lau KC, Coffin CS, Patel TR. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J Gastroenterol. 2020;26:5759–83. 10.3748/wjg.v26.i38.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 7.Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warde-Farley D, Donaldson SL, Comes O, Ndow G, Haddadin Y, Shimakawa Y, et al. The genemania prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20. 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzudzor B, Hammond H, Tachi K, Alisi A, Vento S, Gyasi RK, et al. Serum 25-hydroxyvitamin D and hyaluronic acid levels as markers of fibrosis in patients with chronic liver disease at the main tertiary referral hospital in ghana: A case-control study design. Health Sci Rep. 2023;6:e1101. 10.1002/hsr2.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Z, Mei L, Connell M, Maxwell CA. Hyaluronan mediated motility receptor (HMMR) encodes an evolutionarily conserved homeostasis, mitosis, and meiosis regulator rather than a hyaluronan receptor. Cells. 2020;9:819. 10.3390/cells9040819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mateo F, He Z, Mei L, de Garibay GR, Herranz C, García N, et al. Modification of BRCA1-associated breast cancer risk by HMMR overexpression. Nat Commun. 2022;13:1895. 10.1038/s41467-022-29335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HG, Kim WJ, Kang HG, Chun KH, Kim SJ. Galectin-3 interacts with C/EBPβ and upregulates Hyaluronan-Mediated motility receptor expression in gastric cancer. Mol Cancer Res. 2020;18:403–13. 10.1158/1541-7786.MCR-19-0811. [DOI] [PubMed] [Google Scholar]
- 13.He L, Li H, Li C, Liu ZK, Lu M, Zhang RY, et al. HMMR alleviates Endoplasmic reticulum stress by promoting autophagolysosomal activity during Endoplasmic reticulum stress-driven hepatocellular carcinoma progression. Cancer Commun (Lond). 2023;43:981–1002. 10.1002/cac2.12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, Liu Y, Liu Q, Li Z, Wan Y, Cao C, et al. HMMR triggers immune evasion of hepatocellular carcinoma by inactivation of phagocyte killing. Sci Adv. 2024;10(eadl6083). 10.1126/sciadv.adl6083. [DOI] [PMC free article] [PubMed]
- 15.Ju LL, Chen L, Li JH, Wang YF, Lu RJ, Bian ZL, et al. Effect of NDC80 in human hepatocellular carcinoma. World J Gastroenterol. 2017;23:3675–83. 10.3748/wjg.v23.i20.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang D, Ma Y, Zhao P, Ma J, He C. HMMR is a downstream target of FOXM1 in enhancing proliferation and partial epithelial-to-mesenchymal transition of bladder cancer cells. Exp Cell Res. 2021;408:112860. 10.1016/j.yexcr.2021.112860. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Bode AM, Zhang T. Targeting CDK1 in cancer: mechanisms and implications. NPJ Precis Oncol. 2023;7:58. 10.1038/s41698-023-00407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caputo WL, de Souza MC, Basso CR, Pedrosa VA, Seiva FRF. Comprehensive profiling and therapeutic insights into differentially expressed genes in hepatocellular carcinoma. Cancers (Basel). (2023)15:5653. 10.3390/cancers15235653 [DOI] [PMC free article] [PubMed]
- 19.Sarkozy C, Molina TJ, Dubois S, Portugues C, Bohers E, Ysebaert L, et al. Efficacy of Tazemetostat in combination with R-CHOP in elderly patients newly diagnosed with diffuse large B cell lymphoma: results of the epirchop phase II study of the LYSA. EClinicalMedicine. 2025;82:103157. 10.1016/j.eclinm.2025.103157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen VTM, Namba H, Porter H, Shlyueva D, Lopez E, Melcher A, et al. Synergistic antitumor effect of combined EZH2 and DOT1L Inhibition in B-cell lymphoma. Blood. 2025;2024026534. 10.1182/blood.2024026534. [DOI] [PubMed]
- 21.Verma A, Khan MA, Satrusal SR, Datta D. Emerging role of EZH2 in solid tumor metastasis. Biochim Biophys Acta Rev Cancer. 2025;1880:189253. 10.1016/j.bbcan.2024.189253. [DOI] [PubMed] [Google Scholar]
- 22.Porazzi P, Nason S, Yang Z, Carturan A, Ghilardi G, Guruprasad P, et al. EZH1/EZH2 Inhibition enhances adoptive T cell immunotherapy against multiple cancer models. Cancer Cell. 2025;43:537–51. 10.1016/j.ccell.2025.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Jones BA, Varambally S, Arend RC. Histone methyltransferase EZH2: A therapeutic target for ovarian cancer. Mol Cancer Ther. 2018;17:591–602. 10.1158/1535-7163.MCT-17-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai J, Zhao B, Ma Y, Wang L, Feng P, Hua Y. Antisense-mediated exon skipping targeting EZH2 suppresses tumor growth in a xenograft mouse model of hepatocellular carcinoma. Mol Ther. 2025;33:1485–501. 10.1016/j.ymthe.2025.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol. 2020;13(1):104. 10.1186/s13045-020-00937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J, You C, Kwon G, Noh J, Lee K, Kim K, et al. Integration of epigenomic and transcriptomic profiling uncovers EZH2 target genes linked to cysteine metabolism in hepatocellular carcinoma. Cell Death Dis. 2024;15(11):801. 10.1038/s41419-024-07198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li YK, Wu S, Wu YS, Zhang WH, Wang Y, Li YH, et al. Portal venous and hepatic arterial coefficients predict Post-Hepatectomy overall and Recurrence-Free survival in patients with hepatocellular carcinoma: A retrospective study. J Hepatocell Carcinoma. 2024;11:1389–402. 10.2147/JHC.S462168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Liang YB, Wang QB, Li YK, Chen XM, Luo WL, Lakang Y, Yang ZS, Wang Y, Li ZW, Ke Y. Tumor-associated lymphatic vessel density is a postoperative prognostic biomarker of hepatobiliary cancers: a systematic review and meta-analysis. Front Immunol. 2025;15:7. 10.3389/fimmu.2024.1519999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang DH, He DW, Lv TT, Zhang XK, Li ZJ, Wang ZY. Estrogen receptor α suppresses hepatocellular carcinoma by restricting M2 macrophage infiltration through the YAP-CCL2 axis. BMC Cancer. 2025;25:550. 10.1186/s12885-025-13676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1—The drug-gene interaction via DGIdb analysis
Supplementary Material 2—The microRNA related to hub genes found by Networkanalyst based on miRTarBase database
Supplementary Material 3—The flowchart of this work
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information.