Abstract
Accurate forecasting of diabetes burden is essential for healthcare planning, resource allocation, and policy-making. While deep learning models have demonstrated superior predictive capabilities, their real-world applicability is constrained by computational complexity and data quality challenges. This study evaluates the trade-offs between predictive accuracy, robustness, and computational efficiency in diabetes forecasting. Four forecasting models were selected based on their ability to capture temporal dependencies and handle missing healthcare data: Transformer with Variational Autoencoder (VAE), Long Short-Term Memory (LSTM), Gated Recurrent Unit (GRU), and AutoRegressive Integrated Moving Average (ARIMA). Annual data on Disability-Adjusted Life Years (DALYs), Deaths, and Prevalence from 1990 to 2021 were used to train (1990–2014) and evaluate (2015–2021) the models. Performance was measured using Mean Absolute Error (MAE) and Root Mean Squared Error (RMSE). Robustness tests introduced noise and missing data, while computational efficiency was assessed in terms of training time, inference speed, and memory usage. Statistical significance was analyzed using ANOVA and Tukey’s post-hoc tests. The Transformer-VAE model achieved the highest predictive accuracy (MAE: 0.425, RMSE: 0.501) and demonstrated superior resilience to noisy and incomplete data (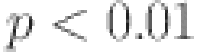 ). LSTM effectively captured short-term patterns but struggled with long-term dependencies, while GRU, though computationally efficient, exhibited higher error rates. ARIMA, despite being resource-efficient, showed limited capability in modeling long-term trends, indicating potential benefits in hybrid approaches. While Transformer-VAE provides the most accurate diabetes burden forecasting, its high computational cost and interpretability challenges limit its scalability in resource-constrained settings. These findings highlight the potential of deep learning models for healthcare forecasting, while underscoring the need for further validation before integration into real-world public health decision-making.
). LSTM effectively captured short-term patterns but struggled with long-term dependencies, while GRU, though computationally efficient, exhibited higher error rates. ARIMA, despite being resource-efficient, showed limited capability in modeling long-term trends, indicating potential benefits in hybrid approaches. While Transformer-VAE provides the most accurate diabetes burden forecasting, its high computational cost and interpretability challenges limit its scalability in resource-constrained settings. These findings highlight the potential of deep learning models for healthcare forecasting, while underscoring the need for further validation before integration into real-world public health decision-making.
Keywords: Forecasting, Deep learning, Artificial intelligence, Diabetes mellitus, Global burden of disease, Robustness to noisy and incomplete data, Disability-adjusted life years, Public health forecasting applications
Subject terms: Diabetes, Statistics, Health care economics
Introduction
Diabetes remains a major global health challenge, affecting over 9% of the world’s population and placing a substantial burden on healthcare systems and economies1. The World Health Organization (WHO) projects that by 2030, diabetes will be the seventh leading cause of death, disproportionately impacting low- and middle-income countries due to healthcare disparities and resource limitations2–5. From an economic perspective, the cost of diabetes management is expected to surpass $2.5 trillion annually, highlighting the urgent need for accurate forecasting models to inform policy decisions and optimize healthcare resources6. The global outbreak of COVID-19 posed unprecedented public health challenges and highlighted the need for accurate epidemic forecasting and preparedness strategies7.
Effective forecasting of diabetes trends is critical for public health planning, early intervention, and resource allocation2,8. Traditional statistical models such as AutoRegressive Integrated Moving Average (ARIMA) and rule-based approaches, while computationally efficient, often struggle to capture the complex, multifactorial nature of diabetes progression. Traditional statistical models, such as ARIMA and linear regression, are often constrained in capturing nonlinear relationships, long-range temporal dependencies, and the complex interactions of socioeconomic and environmental factors1,9–11. These limitations can hinder their effectiveness in modeling the multifaceted dynamics of chronic disease trends such as diabetes. As a result, more robust and adaptive predictive frameworks are required to handle large-scale, real-world healthcare data with missing values and noisy inputs.
Deep learning models, particularly Long Short-Term Memory (LSTM) and Gated Recurrent Units (GRU), have emerged as powerful tools for disease forecasting, demonstrating improved performance over traditional methods by capturing intricate temporal dependencies and nonlinear patterns in health data12,13. However, these models still face challenges related to data sparsity, sensitivity to missing data, and high computational costs, which may hinder their deployment in real-world settings.
A key innovation in this study is the Transformer with Variational Autoencoder (VAE) for diabetes burden forecasting. The Transformer architecture utilizes a self-attention mechanism to effectively model long-range dependencies in time-series data, while the VAE component enhances robustness by learning meaningful latent representations, making it particularly useful for handling missing or incomplete healthcare data. Unlike alternative hybrid models such as CNN-LSTM or Attention-based RNNs, Transformer-VAE offers a strong balance between accuracy and adaptability to data irregularities. Despite these advantages, its computational complexity and scalability require further investigation, particularly for deployment in resource-constrained healthcare environments.
This study conducts a systematic and comparative evaluation of Transformer-VAE, LSTM, and GRU, benchmarking their performance against ARIMA as a statistical baseline. The models are assessed based on three key criteria: predictive accuracy, robustness to missing and noisy data, and computational efficiency. Unlike previous studies that focus primarily on model accuracy, this research also emphasizes interpretability and real-world applicability, addressing challenges in deploying deep learning models for public health forecasting.
The remainder of this paper is structured as follows: “Related work” outlines the methodology, detailing data sources, preprocessing techniques, and model architectures. “Results” presents the experimental results and performance comparisons. “Discussion” discusses the implications of the findings, particularly in relation to real-world deployment and previous research. “Conclusion” concludes with recommendations for future research, including potential optimizations and the integration of deep learning models into healthcare forecasting frameworks.
Related work
Forecasting the burden of diabetes using Artificial Intelligence (AI) has been an evolving field, with significant advances attributed to deep learning (DL) techniques. Compared to traditional statistical models, deep learning approaches have demonstrated superior performance in several key areas related to diabetes management and prediction.
Deep learning systems have been shown to accurately estimate the prevalence and systemic risk factors of diabetic complications, such as diabetic retinopathy, with a performance comparable to expert human graders, but at a fraction of the time14. Similarly, deep learning models developed to predict diabetic retinopathy progression from fundus images have achieved promising results, highlighting the potential of AI in early intervention and monitoring strategies15.
Several recent studies have examined diabetes forecasting using statistical and machine learning approaches. For example, Almutairi et al.16 conducted a national-level analysis in Saudi Arabia using models such as Artificial Neural Networks (ANN), Support Vector Regression (SVR), and Adaptive Neuro-Fuzzy Inference Systems (ANFIS). Their results showed excellent performance (e.g., RMSE as low as 0.02), but their dataset was limited to behavioral risk factors and covered data only up to 2013. No deep learning or sequence-based models were evaluated.
Khan et al.17, on the other hand, analyzed the global burden of type 2 diabetes using GBD 2017 data. Their forecasting relied on SPSS’s classical time series models, focusing primarily on incidence and prevalence. However, they did not utilize deep learning techniques, nor did they explore stratification by income groups or multidimensional burden metrics such as YLLs, YLDs, or DALYs.
In contrast, our study applies deep learning architectures-including Transformer-based VAE, LSTM, and GRU models-on global data spanning up to 2021. We forecast five key indicators of diabetes burden and further stratify the predictions across four World Bank income groups. To the best of our knowledge, this is the first global study combining modern deep learning with income-based stratification and multidimensional disease burden forecasting.
In a systematic review, deep learning approaches outperformed traditional machine learning models across tasks such as diabetes diagnosis, glucose management, and complication detection, although challenges related to data availability and model interpretability persist18. Studies using wide and deep learning architectures based on electronic health records have further demonstrated that hybrid models can improve the prediction of type 2 diabetes onset compared to purely statistical methods19. Moreover, deep learning models have shown better predictive performance than Cox proportional hazards models in forecasting all-cause mortality among patients with type 2 diabetes, particularly using architectures like DeepHit20. Predictive tools based on longitudinal health records and deep recurrent neural networks (LSTM, GRU) have also demonstrated enhanced ability in forecasting blood glucose fluctuations and severe hypoglycemic events21,22.
Recent research has applied deep learning models to disease forecasting with promising results. Ullah et al.23 compared LSTM, GRU, and RNN models for predicting dengue outbreaks in Bangladesh using a two-decade multivariate time series. LSTM achieved the highest accuracy (87.98%), followed by GRU (79.81%). Similarly, Sah et al.24 employed a stacked LSTM-GRU model for COVID-19 forecasting in India, which outperformed traditional models like ARIMA and Prophet in terms of RMSE and  . In the context of diabetes, Rochman et al.25 found that GRU yielded lower RMSE than LSTM on clinical data from a local health center. While ARIMA-based models have been widely used in health forecasting, their seasonal variants like SARIMA have shown promising results in domains such as crime prediction26.
. In the context of diabetes, Rochman et al.25 found that GRU yielded lower RMSE than LSTM on clinical data from a local health center. While ARIMA-based models have been widely used in health forecasting, their seasonal variants like SARIMA have shown promising results in domains such as crime prediction26.
While these studies highlight the strengths of recurrent architectures, they are typically confined to single diseases, localized datasets, or short-term forecasting horizons. Furthermore, robustness to missing data and cross-population generalization are rarely addressed. Our study extends this line of work by introducing a Transformer-VAE model that combines attention-based temporal modeling with latent variable inference. Distinctly, we conduct stratified forecasting across income groups, incorporate external validation using WHO data, and evaluate performance across multiple global health indicators (DALYs, deaths, prevalence). This provides a more comprehensive and equitable framework for forecasting the global diabetes burden.
In addition, as noted in previous literature18,27, enhancing model interpretability and ensuring clinical relevance remain critical challenges. Our methodology, by integrating deep learning with transparent comparative baselines and structured evaluation, addresses both predictive performance and practical applicability.
Methods
Accurate forecasting of the future burden of diabetes requires addressing the complex temporal dependencies in time-series data. These dependencies emerge from interactions among factors such as prevalence, mortality rates, and Disability-Adjusted Life Years (DALYs). Figure 1 illustrates the proposed approach. The process involves several steps: first, the diabetes burden dataset is collected. Second, the dataset undergoes preprocessing, where missing data are imputed, followed by normalization and formatting adjustments. Finally, model parameters are initialized. In the third step, a deep learning model is used for forecasting. Details of the proposed model are presented in the next section. In the fourth step, the model’s performance is evaluated using appropriate metrics.
Fig. 1.
Proposed framework for forecasting diabetes burden using deep learning, illustrating preprocessing, modeling, and evaluation stages.
To tackle the challenges posed by complex temporal dependencies, incomplete data, and the need for robust forecasting, we conduct a comparative evaluation of three advanced machine learning models: Transformer with Variational Autoencoder (VAE), Long Short-Term Memory (LSTM), and Gated Recurrent Unit (GRU). These models were selected based on their ability to capture long-term temporal dependencies, handle missing data effectively, and enhance predictive robustness.
Dataset description
This study utilized data from two publicly available sources: the Global Burden of Disease (GBD) Results Tool developed by the Institute for Health Metrics and Evaluation (IHME), and the World Health Organization (WHO) Global Health Observatory (GHO) data repository. Both datasets include longitudinal health indicators relevant to diabetes and are categorized by World Bank income groups.
The GBD dataset provides annual time series data from 1990 to 2021 for three key health metrics: Disability-Adjusted Life Years (DALYs), deaths attributed to diabetes, and crude prevalence of diabetes. Each record corresponds to one health metric for a specific income group and year. The four income groups considered are: High, Upper-Middle, Lower-Middle, and Low income. For each metric and income group combination, the time series includes one scalar value per year, with no additional features.
For model development, the data were divided into training and testing sets. Data from 1990 to 2014 were used for training, while data from 2015 to 2021 were reserved for testing. Table 1 summarizes the sample counts for each health metric and income group across the two subsets.
Table 1.
Training and testing sample distribution across DALYs, deaths, and prevalence, categorized by World Bank income groups. Each row represents one univariate time series.
| Health metric | Income group | Train (1990–2014) | Test (2015–2021) |
|---|---|---|---|
| DALYs | High income | 25 | 7 |
| DALYs | Upper-middle income | 25 | 7 |
| DALYs | Lower-middle income | 25 | 7 |
| DALYs | Low income | 25 | 7 |
| Deaths | High income | 25 | 7 |
| Deaths | Upper-middle income | 25 | 7 |
| Deaths | Lower-middle income | 25 | 7 |
| Deaths | Low income | 25 | 7 |
| Prevalence | High income | 25 | 7 |
| Prevalence | Upper-middle income | 25 | 7 |
| Prevalence | Lower-middle income | 25 | 7 |
| Prevalence | Low income | 25 | 7 |
To evaluate the model’s generalization capability, we used an external dataset from the WHO, which provides crude prevalence estimates of diabetes in adults aged 18+ for the same set of income groups, spanning the years 1990 to 2022. Income group labels (e.g., “World Bank High Income” in GBD and “High-income” in WHO) were harmonized to allow matching across datasets. For each group, we extracted a univariate time series of diabetes prevalence, aligned by year with the corresponding GBD data.
Data preprocessing
Before model training, the following preprocessing steps were applied:
Handling missing data: To simulate real-world missing data, random values were removed from the dataset. The missing values were imputed using the Simple Imputer from the scikit-learn library, where the mean of the respective feature was used to fill the gaps28.
Normalization: All input features and target variables (e.g., DALYs, mortality rates) were normalized to the range [0, 1] using the MinMaxScaler to ensure that the models could efficiently process the data without being influenced by differences in scale29. Importantly, the scaler was fitted exclusively on the training portion of the GBD dataset to avoid data leakage. The same scaling parameters (i.e., the minimum and maximum values derived from the training set) were then applied to the validation and test subsets. For external validation using the WHO dataset, we reused the same scaler fitted on the GBD training data to maintain consistency in feature representation across datasets.
Data split: The data was divided into training (1990–2014) and testing (2015–2021) sets. This allows for training the models on historical data and evaluating them on future unseen data, which mimics real-world prediction tasks30.
Model selection
We selected three models based on their ability to capture complex temporal dependencies and handle missing data:
Transformer with variational autoencoder (VAE)
The Transformer with VAE model is designed to capture long-term dependencies in sequential data and handle data sparsity by generating synthetic data, but it does not generate new synthetic samples for training31. This capability allows the model to handle incomplete sequences more effectively during inference, thereby improving its resilience to data sparsity. The VAE consists of two main components: an encoder and a decoder. The encoder compresses the input data into a latent representation, while the decoder reconstructs the original data from this representation32,33. The encoder learns two distributions34:
where  and
and  are the mean and logarithmic variance of the latent space representation. The latent variable z is sampled from a normal distribution using the reparameterization trick:
are the mean and logarithmic variance of the latent space representation. The latent variable z is sampled from a normal distribution using the reparameterization trick:
The decoder then reconstructs the input  from z:
from z:
 |
where  is the decoding function. The loss function of the VAE combines a reconstruction loss and a Kullback-Leibler (KL) divergence:
is the decoding function. The loss function of the VAE combines a reconstruction loss and a Kullback-Leibler (KL) divergence:
where  is the dimensionality of the latent space.
is the dimensionality of the latent space.
The Transformer module captures the temporal dependencies within the reconstructed data35. The architecture consists of:
- Multi-head attention: Computes the attention across all input features. The output is:
where Q, K, and V represent the query, key, and value matrices.
- Feed-forward layers: Applies a point-wise feed-forward network:
where
 ,
,  ,
,  , and
, and  are learnable parameters.
are learnable parameters. - Layer normalization: This technique stabilizes and accelerates training by normalizing the inputs to each layer across features rather than across batch instances, which is particularly effective for sequence models and non-batch-dependent architectures such as Transformers. The output of layer normalization is computed as follows36:
Here,
 is the activation of the
is the activation of the  feature in a given layer,
feature in a given layer,  is the total number of hidden units (features),
is the total number of hidden units (features),  and
and  are the mean and variance computed over all features of that particular layer instance, and
are the mean and variance computed over all features of that particular layer instance, and  is a small constant added for numerical stability. Layer normalization differs from batch normalization in that it does not depend on batch statistics, making it well-suited for recurrent and attention-based models where sequence length varies or batch sizes are small.
is a small constant added for numerical stability. Layer normalization differs from batch normalization in that it does not depend on batch statistics, making it well-suited for recurrent and attention-based models where sequence length varies or batch sizes are small.
The reconstructed data  from the VAE is passed to the Transformer for further processing. The final output y is computed as:
from the VAE is passed to the Transformer for further processing. The final output y is computed as:
The combined model optimizes the loss function  , which includes the VAE loss and the task-specific loss (e.g., mean squared error for regression):
, which includes the VAE loss and the task-specific loss (e.g., mean squared error for regression):
This hybrid approach ensures that the VAE extracts meaningful latent features while the Transformer effectively models temporal relationships. Below is a simplified algorithm for training the Transformer with VAE model (Algorithm 1):
Algorithm 1.
Training architecture of the transformer-VAE model, showing the encoder, latent space, and forecasting components.
The Transformer with VAE model excels particularly when dealing with missing or sparse data. The integration of the Transformer’s ability to capture long-term dependencies and the VAE’s capability to reconstruct missing values makes this model particularly robust37. By simultaneously learning temporal dependencies and generating synthetic data points, the model improves both its prediction accuracy and its robustness against noisy or incomplete data. This combination enhances the model’s ability to forecast future trends, even in situations where the data is incomplete or noisy, making it highly suitable for real-world applications involving sparse or missing data.
Long short-term memory (LSTM)
LSTM is a type of recurrent neural network (RNN) designed to model long-term dependencies in sequential data38. LSTM networks are equipped with gating mechanisms-specifically the input, forget, and output gates-that regulate the flow of information through the network39. This allows LSTM to selectively retain important information over time and discard irrelevant data, addressing the vanishing gradient problem encountered by traditional RNNs in long sequences40.
The state of the memory cell  and the hidden state
and the hidden state  at each time step are updated as follows:
at each time step are updated as follows:
Where  is the memory cell at time step
is the memory cell at time step  , representing the long-term memory of the model.
, representing the long-term memory of the model.  is the hidden state at time step
is the hidden state at time step  , which contains the model’s output for the current time step.
, which contains the model’s output for the current time step.  is the forget gate, which decides how much of the previous memory cell
is the forget gate, which decides how much of the previous memory cell  should be carried forward.
should be carried forward.  is the input gate, which controls how much of the candidate memory
is the input gate, which controls how much of the candidate memory  should be stored in the memory cell.
should be stored in the memory cell.  is the output gate, determining how much of the memory cell
is the output gate, determining how much of the memory cell  should be output to the hidden state
should be output to the hidden state  .
.  is the candidate memory, which represents new information that could potentially be added to the memory cell. These gating mechanisms allow the model to effectively learn long-term dependencies and avoid issues like gradient vanishing, making LSTM well-suited for time-series forecasting and sequence modeling tasks.
is the candidate memory, which represents new information that could potentially be added to the memory cell. These gating mechanisms allow the model to effectively learn long-term dependencies and avoid issues like gradient vanishing, making LSTM well-suited for time-series forecasting and sequence modeling tasks.
The training of the LSTM network involves updating the model parameters through backpropagation over time. Algorithm 2 outlines the general steps for training an LSTM model:
Algorithm 2.
Training flow of the LSTM model for time-series forecasting of diabetes burden.
LSTM is particularly effective in tasks where long-term dependencies are crucial. A prime example is forecasting diabetes-related trends, where the progression of the disease is influenced by a variety of long-term factors such as medical history, lifestyle, and treatment regimens. LSTM’s ability to capture these complex temporal dependencies makes it an ideal model for predicting trends in chronic diseases, financial data, and other domains where historical context plays a significant role in forecasting future events41.
The LSTM model excels in capturing long-range temporal dependencies, making it highly effective for sequence modeling and time-series forecasting tasks. Its gating mechanisms ensure that relevant information is preserved over time while irrelevant data is forgotten, allowing for more accurate predictions over long sequences.
Gated recurrent unit (GRU)
The GRU is a variant of the LSTM network, designed to simplify the architecture while retaining the ability to model long-term dependencies in sequential data42. GRU combines the forget and input gates of LSTM into a single update gate  , which controls the flow of information43. Additionally, the reset gate
, which controls the flow of information43. Additionally, the reset gate  is used to determine how much of the previous hidden state should be remembered when computing the candidate hidden state
is used to determine how much of the previous hidden state should be remembered when computing the candidate hidden state  44.
44.
The final hidden state  at each time step is computed as a weighted sum of the previous hidden state
at each time step is computed as a weighted sum of the previous hidden state  and the candidate hidden state
and the candidate hidden state  , as shown in the following equation:
, as shown in the following equation:
Where  is the update gate, determining how much of the previous hidden state
is the update gate, determining how much of the previous hidden state  should be retained and how much of the candidate hidden state
should be retained and how much of the candidate hidden state  should be used to update the current hidden state.
should be used to update the current hidden state.  is the reset gate, which controls the influence of the previous hidden state on the candidate hidden state
is the reset gate, which controls the influence of the previous hidden state on the candidate hidden state  .
.  is the candidate hidden state, calculated using both the input data and the reset gate, representing the potential new information for the current time step.
is the candidate hidden state, calculated using both the input data and the reset gate, representing the potential new information for the current time step.
The GRU’s architecture allows it to model temporal dependencies with fewer parameters compared to LSTM, making it a computationally efficient model for many sequence-based tasks. Training a GRU network involves iteratively updating the model parameters using backpropagation through time (BPTT). Algorithm 3 outlines the general training procedure for a GRU model:
Algorithm 3.
Training process of the GRU model, including sequence handling and prediction.
GRU models are particularly beneficial for sequence modeling tasks where computational efficiency is important, without sacrificing performance. For example, in applications such as time-series forecasting, speech recognition, and natural language processing, GRUs offer a simpler and faster alternative to LSTM, while still capturing essential temporal dependencies.
The GRU network is an effective alternative to LSTM that simplifies the gating mechanism while maintaining the ability to model long-range dependencies in sequential data. Its computational efficiency and simpler structure make it well-suited for real-time applications and resource-constrained environments.
Model training and hyperparameter optimization
Beyond dropout and batch normalization, additional regularization techniques were applied to further prevent overfitting. To mitigate overfitting due to limited sequence length per subgroup, we employed regularization techniques including dropout, KL annealing, and early stopping based on validation loss. L2 regularization (weight decay) was incorporated into the loss function to penalize excessively large model weights, enhancing generalization. Furthermore, hyperparameter tuning was conducted using a grid search approach to optimize key parameters, including learning rates, batch sizes, and layer configurations. These strategies collectively ensured the models maintained strong predictive performance while avoiding excessive reliance on training data, thereby improving their applicability in real-world forecasting scenarios.
To ensure a fair comparison, deep learning models underwent hyperparameter tuning via grid search, while ARIMA parameters were selected based on time-series diagnostics. The grid search procedure explored various combinations of hidden layers, attention heads, dropout rates, and learning rates to identify the most optimal settings. Mean Absolute Error (MAE) was used as the primary evaluation metric during hyperparameter tuning, ensuring that model configurations were optimized for predictive accuracy. The Adam optimizer was employed for deep learning models due to its adaptive learning capabilities. The final selected hyperparameters for each model are summarized in Table 2.
Table 2.
Final tuned hyperparameters used for each model, including learning rate, layers, and latent dimensions.
| Model | Layers | Hidden units | Attention heads | Learning rate | Dropout | Batch size | Optimizer |
|---|---|---|---|---|---|---|---|
| Transformer-VAE | 4 | – | 8 | 0.001 | 0.1 | 32 | Adam |
| LSTM | 2 | 128 | – | 0.0005 | 0.2 | 32 | Adam |
| GRU | 2 | 128 | – | 0.0005 | 0.2 | 32 | Adam |
| ARIMA | – | – | – | – | – | – | AIC-based selection |
For ARIMA, preprocessing steps were applied to improve forecasting performance. The Augmented Dickey-Fuller (ADF) test was conducted to assess stationarity, and differencing was applied where necessary to transform non-stationary data into a stationary series. The optimal autoregressive order (p), differencing order (d), and moving average order (q) parameters were selected using Akaike Information Criterion (AIC) minimization, ensuring an optimal balance between model complexity and goodness of fit.
As ARIMA is inherently a univariate time series model, it cannot directly model multivariate inputs. Therefore, in this study, we applied separate ARIMA models to each health indicator-DALYs, deaths, and prevalence-independently. For each metric, we constructed and tuned an individual ARIMA configuration based solely on its historical time series within each income group. This allowed us to benchmark ARIMA’s predictive performance alongside multivariate deep learning models, while maintaining consistency in evaluation across indicators.
The Transformer-VAE model was configured with four encoder layers, each containing eight attention heads, and employed a dropout rate of 0.1 to prevent overfitting. The learning rate was set to 0.001 based on validation performance. The LSTM and GRU models were structured with two hidden layers of 128 units each, using a learning rate of 0.0005 and a dropout rate of 0.2 to enhance generalization. The ARIMA model’s parameters were determined dynamically through AIC minimization after ensuring stationarity with the ADF test.
Evaluation metrics
The performance of each model is evaluated using a comprehensive set of metrics, focusing on both prediction accuracy and computational efficiency, as well as the models’ ability to handle incomplete and variable data. The following metrics were used to assess model performance:
- Prediction accuracy: The primary metric for evaluating prediction accuracy is the MAE, which measures the average absolute difference between the predicted and actual values. It is calculated as:
where
 represents the actual values,
represents the actual values,  represents the predicted values, and
represents the predicted values, and  is the number of data points. A lower MAE indicates better accuracy in the model’s predictions. Additionally, we use Root Mean Squared Error (RMSE) to evaluate model performance by penalizing large errors more heavily. RMSE is calculated as:
is the number of data points. A lower MAE indicates better accuracy in the model’s predictions. Additionally, we use Root Mean Squared Error (RMSE) to evaluate model performance by penalizing large errors more heavily. RMSE is calculated as:
RMSE provides a measure of how well the model captures deviations from the actual values, with larger values indicating poorer model performance.
Stability with variable data: The stability of the models is assessed by evaluating their ability to maintain prediction accuracy when exposed to changing trends in the data. A smaller accuracy drop indicates that the model is better at handling shifts in the data distribution, demonstrating its stability in the presence of evolving trends.
- Stability analysis: This metric evaluates the model’s ability to maintain prediction accuracy across different conditions, including varying data characteristics. Stability is assessed by analyzing the residuals (the differences between predicted and actual values), with the goal of ensuring that these residuals remain small and consistent under different conditions. A model that demonstrates low variability in residuals is considered stable. We use the following residual analysis to measure stability:
where
 is the actual value and
is the actual value and  is the predicted value. The lower the spread of residuals, the more stable the model is.
is the predicted value. The lower the spread of residuals, the more stable the model is. - Robustness to incomplete data: The ability of each model to handle missing or incomplete data is assessed using a combination of residual analysis and the model’s performance on datasets with missing values. The residuals from models trained on incomplete data are compared to those from models trained on complete data. Models that perform better with incomplete data are considered more robust. Specifically, we measure how well the model can adapt when missing values are introduced by calculating the increase in residuals:
A lower residual increase indicates better robustness to missing data.
- Computational efficiency: The efficiency of the models is evaluated in terms of their computational cost. This includes three key aspects:
- Training time: The time taken to train the model on the training dataset, typically measured in minutes or seconds.
- Inference time: The time taken by the model to make predictions on unseen test data, measured in seconds.
- Memory usage: The amount of memory consumed by the model during both training and inference phases, typically measured in megabytes.
By evaluating these metrics, we gain a comprehensive understanding of how well each model performs not only in terms of prediction accuracy but also in terms of computational efficiency, robustness to missing data, and stability under varying conditions.
Results
This section presents the evaluation results for four models: VAE, LSTM, GRU, and ARIMA.
Prediction accuracy
The predictive performance of the models is illustrated in Fig. 2, which presents the relative error of predictions against the actual values for three key diabetes-related indicators: DALYs, Deaths, and Prevalence. Each subplot corresponds to one health metric and displays the relative errors for different models. This visualization highlights not only the dispersion of prediction errors but also how model performance varies across metrics.
Fig. 2.
Relative prediction error versus actual values on IHME data for DALYs (a), deaths (b), and prevalence (c), across different models.
Figures 3 and 4 provide a comparative analysis of forecasting model performance across three major health indicators-(a) DALYs, (b) deaths, and (c) prevalence-based on Mean Absolute Error (MAE) and Root Mean Squared Error (RMSE), respectively. Each boxplot illustrates the distribution of errors for four models: ARIMA, GRU, LSTM, and Transformer with VAE.
Fig. 3.
Comparison of mean absolute error (MAE) across forecasting models (ARIMA, GRU, LSTM, and transformer-VAE) on IHME Data and three health indicators: (a) DALYs, (b) deaths, and (c) prevalence.
Fig. 4.
Comparison of Root Mean Squared Error (RMSE) across forecasting models (ARIMA, GRU, LSTM, and Transformer-VAE) on IHME Data and three health indicators: (a) DALYs, (b) deaths, and (c) prevalence.
The ANOVA test results indicated statistically significant differences between the models ( ), confirming that Transformer with VAE consistently outperforms other models. Tukey’s post-hoc analysis further revealed that differences between Transformer with VAE and ARIMA were highly significant (
), confirming that Transformer with VAE consistently outperforms other models. Tukey’s post-hoc analysis further revealed that differences between Transformer with VAE and ARIMA were highly significant ( ), while LSTM and GRU differences were not statistically significant.
), while LSTM and GRU differences were not statistically significant.
External validation
To further evaluate the generalizability of the Transformer-VAE model, we conducted an external validation using diabetes prevalence data from the World Health Organization (WHO) across four World Bank income groups: High, Upper-Middle, Lower-Middle, and Low Income (see Data Availability section for details). The model, trained exclusively on GBD data from 1990 to 2014, was tested-without any retraining-on WHO data covering the period 2015 to 2022. The resulting MAE and RMSE values for each income group are presented in Table 3.
Table 3.
External validation of the transformer-VAE model on WHO data (2015–2022) by income group.
| Income group | MAE (%) | RMSE (%) |
|---|---|---|
| High income | 0.581 | 0.639 |
| Upper-middle income | 0.604 | 0.673 |
| Lower-middle income | 0.598 | 0.658 |
| Low income | 0.632 | 0.703 |
Stability with variable data
Figure 5 presents the predicted values compared to actual data for DALYs and Deaths in high- and low-income countries. These figures highlight the ability of the models to track variations across different economic settings.
Fig. 5.
Predicted vs. actual trends for DALYs and deaths in high- and low-income countries, showing model performance across regions on IHME Data.
Stability analysis
The residual analysis of predictions under noisy and non-noisy conditions is illustrated in Fig. 6. This analysis evaluates the consistency of the models when subjected to variations in input data.
Fig. 6.
Residual error comparison of Transformer-VAE, LSTM, GRU, and ARIMA models under clean and noisy input conditions on IHME Data.
Robustness to incomplete data
Figure 7 demonstrates the robustness of the forecasting models against missing data by plotting the relative error for each model across three key health indicators: (a) DALYs, (b) Deaths, and (c) Prevalence. In each subplot, the models were evaluated under two conditions: using the complete dataset and using a dataset with 20% randomly introduced missing values, imputed using a simple mean strategy.
Fig. 7.
Relative error of model predictions under complete data and 20% missing data conditions for three health indicators: (a) DALYs, (b) deaths, and (c) prevalence on IHME data.
Distinct markers were used to differentiate between models and conditions: filled markers represent model performance on the complete dataset, while hollow markers indicate performance under the missing data condition.
Training and testing loss dynamics
Figure 8 illustrates the evolution of training and testing loss (measured as Mean Squared Error) across 100 epochs for the GRU, LSTM, and Transformer with VAE models. As shown, the Transformer model achieves faster convergence and lower final loss values for both training and testing, indicating its superior generalization capability. In contrast, GRU and LSTM converge more slowly and stabilize at higher error levels. The early-stage oscillations, particularly in training loss, reflect the natural adjustment dynamics of the models during initial learning.
Fig. 8.
Training and testing loss curves (MSE) over 100 epochs for GRU, LSTM, and Transformer with VAE.
Computational efficiency
The computational efficiency of the models, including training time, inference time, and memory usage, is presented in Fig. 9. ARIMA demonstrates significantly lower computational resource requirements compared to deep learning models.
Fig. 9.
Comparison of computational efficiency (training time and inference speed) across all evaluated models.
Discussion
Global diabetes trends indicate a significant rise in prevalence, mortality, and economic burden. Sun et al.45 and Steinmetz et al.46 emphasized the increasing global burden of diabetes, particularly type 2, driven by behavioral factors such as poor diet and physical inactivity. High BMI alone accounted for 52% of diabetes cases in 2021, highlighting the need for improved forecasting methods to support evidence-based interventions47.
This study evaluated four predictive models-Transformer with VAE, LSTM, GRU, and ARIMA-for forecasting the global burden of diabetes. The selection of these models was based on their effectiveness in time-series forecasting, their proven success in health informatics, and their ability to capture nonlinear relationships in data. While other architectures such as CNN-LSTM or hybrid deep learning models exist, the focus was placed on sequential models due to their suitability for long-term health trend prediction. The results demonstrated that Transformer with VAE achieved the highest predictive accuracy, particularly in handling complex, multifactorial relationships. It effectively modeled long-term dependencies in DALYs, Deaths, and Prevalence across different income groups. These findings align with previous research on the importance of advanced deep learning techniques in health forecasting2. However, its superior performance comes at the cost of increased computational requirements, which may limit its applicability in resource-constrained settings.
Although ARIMA required significantly fewer computational resources, its predictive performance was limited, especially for long-term forecasting. Traditional statistical models such as ARIMA assume stationarity and struggle with capturing complex nonlinear trends in diabetes progression. However, ARIMA remains valuable for short-term forecasting and baseline comparisons. The results showed that its accuracy was considerably lower compared to deep learning models, which suggests that it may not be suitable for long-term trend predictions. To improve ARIMA’s effectiveness, preprocessing steps such as the Augmented Dickey-Fuller (ADF) test for stationarity and differencing were applied, but the model still underperformed compared to deep learning approaches. Future research could explore hybrid models that integrate ARIMA’s lightweight computational efficiency with the deep learning models’ ability to capture complex dependencies.
An important aspect of model evaluation is its ability to generalize beyond the dataset on which it was trained. In this study, the Transformer-VAE model, initially trained solely on GBD data, was externally validated using WHO-reported diabetes prevalence data for the period 2015–2022. The model demonstrated strong alignment with the external dataset, yielding consistently low MAE and RMSE values when calculated on unscaled data. For instance, in the DALY prediction task, the Transformer-VAE achieved an average MAE of 47 and RMSE of 72 across all World Bank income groups. This suggests that the model captures underlying temporal trends in diabetes prevalence robustly, despite potential differences in data collection methodologies or reporting standards between GBD and WHO sources. Notably, the model performed slightly better in high and lower-middle income groups, which may reflect more stable epidemiological patterns or higher data consistency in those regions. These findings strengthen the argument for the model’s applicability in real-world public health forecasting scenarios.
Compared to prior deep learning approaches in disease forecasting, our Transformer-VAE framework demonstrates meaningful advancements in both methodological depth and practical utility. Studies such as Ullah et al.23 and Rochman et al.25 highlighted the effectiveness of LSTM and GRU models for forecasting dengue and diabetes at local levels. However, these models did not explore generalization across diverse populations, nor did they address robustness to data incompleteness or inconsistencies. Similarly, while Sah et al.24 proposed a stacked LSTM-GRU model with promising results for COVID-19 forecasting in India, their approach was reliant on large training datasets and lacked latent representation learning or cross-context validation.
Our study contributes to and extends this literature in several key ways. First, unlike Almutairi et al.16, who focused on diabetes prevalence prediction using traditional machine learning models and data limited to a single country (Saudi Arabia) up to 2013, we utilize a comprehensive global dataset extending to 2021. Moreover, we model not only prevalence but also incidence and DALYs, capturing multiple dimensions of diabetes burden. Khan et al.17 offered valuable global projections using classical statistical models, but their focus was restricted to incidence and prevalence and did not leverage deep learning architectures capable of capturing complex temporal patterns.
Our model further introduces a novel integration of Transformer-based attention with the variational latent space of a VAE, allowing for both fine-grained temporal modeling and resilience to data sparsity. Importantly, we disaggregate forecasts across four income groups, revealing socioeconomic gradients in disease burden-an analytical perspective largely absent in previous studies.
Finally, in addition to benchmarking our framework against traditional baselines (ARIMA, LSTM, GRU), we validated its generalizability using an independent dataset from the WHO. The proposed model consistently performed well across metrics and income strata, positioning it as a scalable and policy-relevant tool for long-term diabetes forecasting.
One of the key challenges in health forecasting is the presence of noise and inconsistencies in data reporting. Residual analysis indicated that Transformer with VAE maintained stable predictions even under noisy conditions, whereas LSTM and GRU exhibited greater deviations. This highlights the importance of robust models in public health forecasting, particularly when dealing with incomplete or inconsistent datasets. Additionally, missing data remains a critical challenge in health informatics. The VAE component of Transformer enabled plausible data generation, preserving model accuracy even with incomplete datasets. In contrast, LSTM and GRU showed performance declines when faced with missing values. ARIMA, which relies on structured time-series assumptions, demonstrated further limitations in handling data gaps. To quantitatively assess the significance of these differences, statistical tests such as ANOVA and Tukey’s post-hoc analysis confirmed that Transformer with VAE significantly outperformed ARIMA ( ), while differences between LSTM and GRU were not statistically significant. Future studies should investigate advanced data imputation techniques and their impact on forecasting accuracy across different models.
), while differences between LSTM and GRU were not statistically significant. Future studies should investigate advanced data imputation techniques and their impact on forecasting accuracy across different models.
Although the Transformer with VAE demonstrated the highest prediction performance (e.g., lowest RMSE and highest  on unscaled data), its computational demands raise concerns about scalability, particularly in low-resource healthcare settings. The model required significantly more training time and memory compared to LSTM, GRU, and ARIMA. While GRU showed relatively lower predictive performance, it offered a computationally efficient alternative, making it a viable option for real-time applications. In practice, balancing predictive power with computational feasibility is essential. For instance, the Transformer-VAE may be suited for large-scale policy analysis, whereas GRU or LSTM could be more practical for use in hospital-level decision-making with limited hardware capacity. Future research should explore optimization techniques-such as pruning, quantization, and model distillation-to reduce computational overhead without sacrificing predictive accuracy. Cloud-based deployment may also help enable the use of Transformer-based models in settings with limited on-site resources.
on unscaled data), its computational demands raise concerns about scalability, particularly in low-resource healthcare settings. The model required significantly more training time and memory compared to LSTM, GRU, and ARIMA. While GRU showed relatively lower predictive performance, it offered a computationally efficient alternative, making it a viable option for real-time applications. In practice, balancing predictive power with computational feasibility is essential. For instance, the Transformer-VAE may be suited for large-scale policy analysis, whereas GRU or LSTM could be more practical for use in hospital-level decision-making with limited hardware capacity. Future research should explore optimization techniques-such as pruning, quantization, and model distillation-to reduce computational overhead without sacrificing predictive accuracy. Cloud-based deployment may also help enable the use of Transformer-based models in settings with limited on-site resources.
Despite demonstrating strong predictive capabilities, the models in this study did not incorporate external socioeconomic or environmental factors such as income distribution, healthcare accessibility, or pollution levels, which have been shown to influence diabetes prevalence. Future studies should consider integrating these variables to improve the contextual relevance of forecasts. Furthermore, assessing the generalizability of these models across different geographic regions is necessary, as regional variations in diabetes trends could impact forecasting accuracy. Expanding the dataset to include country-specific policy interventions or healthcare expenditure data may enhance the ability to forecast the effectiveness of public health measures.
From a policy perspective, accurate forecasting is essential for addressing the rising prevalence of diabetes, particularly among aging populations1,48. Predictive models can support national health agencies in planning preventive interventions, optimizing healthcare budgets, and allocating medical resources efficiently. Governments can adjust funding for diabetes prevention programs based on predicted prevalence rates. Hospitals can optimize the distribution of medical supplies by forecasting demand in different regions. Public health officials can target high-risk populations through early intervention programs informed by predictive trends. The growing economic burden of diabetes, projected to exceed $2.5 trillion annually by 203049, further underscores the need for precise forecasting tools. Transformer with VAE’s ability to analyze demographic, socioeconomic, and behavioral factors enhances its role in shaping targeted public health policies.
Although the Transformer-VAE demonstrates strong predictive performance, it poses interpretability challenges that can hinder its adoption in healthcare settings. The use of a latent variable layer in the VAE component obscures the direct relationship between inputs and outputs, making it difficult to trace how specific features influence predictions. Similarly, while attention mechanisms offer some transparency, interpreting multi-head attention across layers remains complex. In this study, we did not employ interpretability tools such as SHAP or attention heatmaps, but future work should explore these methods to better understand the model’s decision-making process and increase trustworthiness in clinical applications.
While the Transformer-VAE model exhibits strong predictive performance in retrospective evaluations, its real-world applicability is challenged by high computational costs (Fig. 9), limited data accessibility, and interpretability issues. Future research should prioritize the development of efficient, scalable, and hybrid models that balance deep learning capabilities with the simplicity of statistical approaches. Additionally, validation in clinical environments is essential to assess usability and real-world impact, paving the way for broader adoption in public health systems
Conclusion
This study compared four predictive models-Transformer with VAE, LSTM, GRU, and ARIMA-for forecasting the global diabetes burden. Transformer with VAE significantly outperformed LSTM and GRU, particularly in handling long-term dependencies and missing data, making it more suitable for real-world health forecasting. While ARIMA was computationally efficient, its limited ability to capture long-term trends reduces its applicability in dynamic health forecasting.
The findings highlight the importance of advanced predictive models in public health planning, resource allocation, and intervention strategies. Despite its strong predictive performance, the Transformer-VAE’s high computational cost and limited interpretability hinder its practical use in healthcare. Future work should apply techniques like SHAP and attention visualization to improve transparency and trust. Future research should focus on improving its efficiency through model compression, hybrid approaches such as ARIMA-LSTM or Bayesian deep learning models, and real-time implementation. Additionally, integrating socioeconomic and environmental factors could enhance forecasting accuracy and support more targeted public health interventions.
Acknowledgements
This project was financially supported by Shiraz University of Medical Sciences with grant number 31032. The funder had no role in the study design, data collection, analysis, and interpretation, and writing of the manuscript.
Author contributions
M.B. was responsible for data acquisition, study supervision, and validation of the results. He also reviewed and revised the manuscript. R.E. conducted data analysis, developed predictive models, wrote the main manuscript text, and prepared figures and tables. Both authors contributed to conceptualization and methodology design. All authors reviewed and approved the final version of the manuscript.
Data availability
The primary dataset used in this study was obtained from the Global Burden of Disease (GBD) Results Tool provided by the Institute for Health Metrics and Evaluation (IHME). The specific subset used for analysis can be accessed at the following permanent link: https://vizhub.healthdata.org/gbd-results?params=gbd-api-2021-permalink/e623d782766fef97b561e237794e9c90 For external validation, we utilized publicly available data on the prevalence of diabetes from the World Health Organization (WHO), accessible at: https://www.who.int/data/gho/data/indicators/indicator-details/GHO/prevalence-of-diabetes-age-standardized The source code is available at https://github.com/res-sudo/diabetes-burden-forecasting/ under a CC BY-NC 4.0 license. Commercial use requires author permission.
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was found to be in accordance to the ethical principles and the national norms and standards for conducting medical research. The research protocol was approved by the Ethics Committee of Shiraz University of Medical Sciences with code IR.SUMS.REC.1403.261.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lin, X. et al. Global, regional, and national burden and trend of diabetes in 195 countries and territories: An analysis from 1990 to 2025. Sci. Rep.10, 13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye, J. et al. The global, regional and national burden of type 2 diabetes mellitus in the past, present and future: A systematic analysis of the global burden of disease study 2019. Front. Endocrinol.14, 1192629 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahrouseh, N., Kovács, N. & Varga, O. Diabetes mellitus EU comorbidity index: Using data from EHIS 2019 and GBD disability weights. Eur. J. Public Health34, 144–2114 (2024). [Google Scholar]
- 4.Xie, J. et al. Global burden of type 2 diabetes in adolescents and young adults, 1990–2019: Systematic analysis of the global burden of disease study 2019. BMJ379, e072385 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ampofo, A., & Boateng, E. B. Beyond 2020: Modelling obesity and diabetes prevalence. Diabetes Res. Clin. Pract. 108362 (2020). [DOI] [PubMed]
- 6.Moreira, P. et al. Predicting the prevalence of type 2 diabetes in Brazil: A modeling study. Front. Public Health12, 1275167 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai, C.-C., Shih, T.-P., Ko, W.-C., Tang, H.-J. & Hsueh, P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents55(3), 105924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felizardo, V., Machado, D., Garcia, N., Pombo, N. & Brandão, P. Hypoglycaemia prediction models with auto explanation. IEEE Access1, 1–1 (2021). [Google Scholar]
- 9.Fayaz, L., Joseph, R., Ankayarkanni, B., Princemary, S., Asha, P. & Student, U. Multi-scale and context aware optimized glucose prediction using neural networks. In 2023 7th International Conference on Trends in Electronics and Informatics (ICOEI). 820–825 (2023).
- 10.Talirongan, F. J. B., Talirongan, H. & Orong, M. Y. Modeling national trends on health in the Philippines using ARIMA. arXiv. (Computers and Society, 2021).
- 11.Zhang, Y., Ning, Y., Li, B., Liu, Y. & Dang, Y. Short-term prediction for dynamic blood glucose trends based on ARIMA-LSSVM-GRU model. J. Phys. Conf. Ser.2030, 133 (2021). [Google Scholar]
- 12.Bian, Q., As’arry, A., Cong, X., Rezali, K. A. & Ahmad, R. M. K. B. R. A hybrid transformer-LSTM model apply to glucose prediction. PLOS ONE 19 (2024). [DOI] [PMC free article] [PubMed]
- 13.Prendin, F., Favero, S. D., Vettoretti, M., Sparacino, G. & Facchinetti, A. Forecasting of glucose levels and hypoglycemic events: Head-to-head comparison of linear and nonlinear data-driven algorithms based on continuous glucose monitoring data only. Sensors (Basel, Switzerland)21 (2021). [DOI] [PMC free article] [PubMed]
- 14.Ting, D. et al. Deep learning in estimating prevalence and systemic risk factors for diabetic retinopathy: A multi-ethnic study. NPJ Digit. Med.2, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arcadu, F. et al. Deep learning algorithm predicts diabetic retinopathy progression in individual patients. NPJ Digit. Med.2, 92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almutairi, M. et al. A comparative study of forecasting models for predicting the prevalence of diabetes in Saudi Arabia. Algorithms18(3), 145 (2025). [Google Scholar]
- 17.Khan, M. A. B. et al. Epidemiology of type 2 diabetes - Global burden of disease and forecasted trends. J. Epidemiol. Glob. Health10(1), 107–111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu, T., Li, K., Herrero, P. & Georgiou, P. Deep learning for diabetes: A systematic review. IEEE J. Biomed. Health Inform.25(8), 2744–2757 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Nguyen, B. P. et al. Predicting the onset of type 2 diabetes using wide and deep learning with electronic health records. Comput. Methods Prog. Biomed.182, 105055 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Zhang, T. et al. Machine learning and statistical models to predict all-cause mortality in type 2 diabetes: Results from the UK biobank study. Diabetes Metab. Syndr.18(9), 103135 (2024). [DOI] [PubMed] [Google Scholar]
- 21.Lee, G., Ko, S., Ahn, Y. et al. Assessment of a deep learning model based on three-year longitudinal electronic health record data for predicting severe hypoglycemia in patients with type 2 diabetes. Diabetes (2021).
- 22.Faruqui, S. H. A., Du, Y., Meka, R. et al. Development of a deep learning model for dynamic forecasting of blood glucose level for type 2 diabetes mellitus. JMIR mHealth uHealth7 (2019). [DOI] [PMC free article] [PubMed]
- 23.Ullah, M. A., Mim, A. S., Hasan, M. N. & Sadik, M. R. Deep learning based forecasting models of dengue outbreak in Bangladesh: Comparative analysis of LSTM, RNN, and GRU models using multivariate variables with a two-decade dataset. In 2024 International Conference on Smart Systems for applications in Electrical Sciences (ICSSES), pp. 1–6 (2024).
- 24.Sah, S., Surendiran, B., Dhanalakshmi, R., Mohanty, S., Alenezi, F. S. & Polat, K. Forecasting COVID-19 pandemic using prophet, ARIMA, and hybrid stacked LSTM-GRU models in India. In Computational and Mathematical Methods in Medicine. Vol. 2022 (2022). [DOI] [PMC free article] [PubMed]
- 25.Rochman, E., Miswanto, Suprajitno, H., Rachmad, A., Nindyasari, R. & Rachman, F. H. Comparison of LSTM and GRU in predicting the number of diabetic patients. In 2022 IEEE 8th Information Technology International Seminar (ITIS), pp. 145–149 (2022).
- 26.Noor, T. H., Almars, A. M., Alwateer, M., Almaliki, M., Gad, I. & Atlam, E.-S. Sarima: A seasonal autoregressive integrated moving average model for crime analysis in Saudi Arabia. Electronics11(23) (2022).
- 27.García-Jaramillo, M., Luque, C. & León-Vargas, F. Machine learning and deep learning techniques applied to diabetes research: A bibliometric analysis. J. Diabetes Sci. Technol. (2023). [DOI] [PMC free article] [PubMed]
- 28.Jia, L., Wang, Z., Lv, S. & Xu, Z. PE-DIM: An efficient probabilistic ensemble classification algorithm for diabetes handling class imbalance missing values. IEEE Access10, 107459–107476 (2022). [Google Scholar]
- 29.Smith, G. J. et al. Impact of missing data on the accuracy of glucose metrics from continuous glucose monitoring assessed over a two-week period. Diabetes Technol. Ther. (2023). [DOI] [PubMed]
- 30.Yang, H., Chen, Z., Huang, J. & Li, S. Awd-stacking: An enhanced ensemble learning model for predicting glucose levels. PLOS One19 (2024). [DOI] [PMC free article] [PubMed]
- 31.Akuzawa, K., Iwasawa, Y. & Matsuo, Y. Information-theoretic regularization for learning global features by sequential VAE. Mach. Learn.110, 2239–2266 (2021). [Google Scholar]
- 32.Akkem, Y., Biswas, S. K. & Varanasi, A. A comprehensive review of synthetic data generation in smart farming by using variational autoencoder and generative adversarial network. Eng. Appl. Artif. Intell.131, 107881 (2024). [Google Scholar]
- 33.Barrej’on, D., Olmos, P. & Artés-Rodríguez, A. Medical data wrangling with sequential variational autoencoders. IEEE J. Biomed. Health Inform.26, 2737–2745 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Qiu, Y., Zheng, H. & Gevaert, O. Genomic data imputation with variational auto-encoders. GigaScience9 (2020). [DOI] [PMC free article] [PubMed]
- 35.Deihim, A., Alonso, E. & Apostolopoulou, D. STTRE: A spatio-temporal transformer with relative embeddings for multivariate time series forecasting. Neural Netw.168, 549–559 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Ba, J., Kiros, J. & Hinton, G. E. Layer normalization. arXiv:1607.06450 (2016).
- 37.Jing, T., Zheng, P., Xia, L. & Liu, T. Transformer-based hierarchical latent space VAE for interpretable remaining useful life prediction. Adv. Eng. Inform.54, 101781 (2022). [Google Scholar]
- 38.Gao, R. et al. Time-distanced gates in long short-term memory networks. Med. Image Anal.65, 101785 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He, T., Mao, H. & Yi, Z. Subtraction gates: Another way to learn long-term dependencies in recurrent neural networks. IEEE Trans. Neural Netw. Learn. Syst.33, 1740–1751 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Tolic, A., Boshkoska, B. M. & Skansi, S. Chrono initialized LSTM networks with layer normalization. IEEE Access12, 115219–115236 (2024). [Google Scholar]
- 41.Shastri, S., Singh, K., Kumar, S., Kour, P. & Mansotra, V. Deep-LSTM ensemble framework to forecast covid-19: An insight to the global pandemic. Int. J. Inf. Technol.13(4), 1291–1301 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmadzadeh, E., Kim, H., Jeong, O., Kim, N. & Moon, I. A deep bidirectional LSTM-GRU network model for automated ciphertext classification. IEEE Access1, 1–1 (2022). [Google Scholar]
- 43.Fanta, H., Shao, Z. & Ma, L. Sitgru: Single-tunnelled gated recurrent unit for abnormality detection. arXiv:2003.13528 (2020).
- 44.Chen, Z., Hu, J., Min, G., Zomaya, A. Y. & El-Ghazawi, T. Towards accurate prediction for high-dimensional and highly-variable cloud workloads with deep learning. IEEE Trans. Parallel Distrib. Syst.31, 923–934 (2020). [Google Scholar]
- 45.Sun, J., Hu, W., Ye, S., Deng, D. & Chen, M. The description and prediction of incidence, prevalence, mortality, disability-adjusted life years cases, and corresponding age-standardized rates for global diabetes. J. Epidemiol. Glob. Health13(3), 566–576 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinmetz, J. D. et al. Global, regional, and national burden of osteoarthritis, 1990–2020 and projections to 2050: A systematic analysis for the global burden of disease study 2021. Lancet Rheumatol.5(9), e508–e522 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, X., Wang, X., Wang, M. ran, Hu, B. yue, Tang, W.-C., Wu, Y., Gu, J., Ni, T. & Li, Q. The global burden of type 2 diabetes attributable to high body mass index in 204 countries and territories, 1990-2019: An analysis of the global burden of disease study. Front. Public Health10 (2022). [DOI] [PMC free article] [PubMed]
- 48.Jiang, S., Yu, T., Di, D., Wang, Y. & Li, W. Worldwide burden and trends of diabetes among people aged 70 years and older, 1990-2019: A systematic analysis for the global burden of disease study 2019. Diabetes/Metab. Res. Rev.40 (2023). [DOI] [PubMed]
- 49.Bommer, C. et al. Global economic burden of diabetes in adults: Projections from 2015 to 2030. Diabetes Care41, 963–970 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The primary dataset used in this study was obtained from the Global Burden of Disease (GBD) Results Tool provided by the Institute for Health Metrics and Evaluation (IHME). The specific subset used for analysis can be accessed at the following permanent link: https://vizhub.healthdata.org/gbd-results?params=gbd-api-2021-permalink/e623d782766fef97b561e237794e9c90 For external validation, we utilized publicly available data on the prevalence of diabetes from the World Health Organization (WHO), accessible at: https://www.who.int/data/gho/data/indicators/indicator-details/GHO/prevalence-of-diabetes-age-standardized The source code is available at https://github.com/res-sudo/diabetes-burden-forecasting/ under a CC BY-NC 4.0 license. Commercial use requires author permission.














