Abstract
Human cytomegalovirus extensively alters nuclear organization and the cellular transcriptome, yet understanding of these genome-wide events remains relatively limited. Here, chromatin conformation capture (Hi-C) revealed how cytomegalovirus alters chromosome organization at both large- and small-scales. Nascent transcriptomics further revealed how transcriptional changes correlate with genomic reorganization, while also uncovering infection-induced transcriptional dysregulation that contributes to the induction of neuronal gene signatures in infected fibroblasts. Combining Hi-C and Cleavage Under Targets & Release Using Nuclease (CUT&RUN) we find that viral genomes preferentially localize to highly euchromatic compartments, further dysregulating transcription of host genes. Finally, RNAi-mediated depletion of two key effectors of microtubule-based forces that are exerted on the nucleus provides insights into their diverging roles in regulating compartment-scale contacts and viral genomic interactions with host chromatin. Combined, we reveal the extent to which HCMV interacts with and alters host chromatin and transcription, and the influence of microtubule mechanotransduction on these processes.
Subject terms: Herpes virus, Nuclear organization, Genomics
Using chromatin conformation capture and nascent transcriptomics, the authors reveal the extent to which HCMV interacts with and alters host chromatin and transcriptional programs, along with the role played by microtubule-derived mechanical forces in shaping these events.
Introduction
The human genome is organized into a highly complex three-dimensional structure that involves extensive intra- and inter-chromosomal contacts1. This structural complexity involves the formation of active and repressive compartments of various scales, which both influence and are influenced by the transcriptional activity of genes within these regions. Larger-scale active “A” or repressive “B” compartments form contacts within themselves and with other active or repressive compartments at both local and long-distance ranges. Within these compartments, topologically associating domains (TADs) form smaller regulatory units that contain active and repressed genes or gene clusters. This complex, compartmentalized genome organization and the transcriptional activity of genes within each compartment is carefully regulated by an array of proteins ranging from the central lamina scaffold on the inner nuclear membrane to histones, insulators and transcription factors, as well as by external cues such as kinase signaling pathways and cytoskeletal mechanotransduction1–3. In the latter process, assembly of cytoplasmic filaments can influence the levels of actin monomers in the nucleus that control transcription, while microtubules can exert forces on the nuclear surface to control its spatial organization and transcriptional activity4–6. Yet, in many cases, we are only beginning to understand the underlying complexity of these events.
One such instance is infection by human cytomegalovirus (HCMV), a member of the β-herpesvirus family of DNA viruses. In fact, HCMV represents an intriguing case of virus-host interactions and genome regulation for several reasons. Most viruses have a relatively short replication cycle and kill cells rapidly, and many induce a phenomenon termed “host shut-off” by globally destabilizing host mRNAs to favor viral protein synthesis. In stark contrast, HCMV does not cause shut-off and instead, it slowly replicates over a 5–10 day period7,8. During this time, HCMV extensively remodels cellular architecture and the host transcriptome. A wide range of genomic approaches have been used to study HCMV infection, many of which have focused on understanding the organization and transcriptional regulation of viral genomes9,10. From these studies, we now know, for example, that unlike latent (non-productive) infection of myeloid cells, lytic (productive) replication in permissive cell types such as fibroblasts is accompanied by dynamic changes in the chromatinization of viral genomes9. None-the-less, viral DNA remains less chromatinized and more open or accessible than the cellular genome, and undergoes a remarkably high level of pervasive transcription across both strands11–20. However, the effects of HCMV on the host genome are less well understood21. During initial infection of myeloid cells leading to latency establishment, there are transient host responses but few, if any, large-scale changes in host chromatin accessibility20. By contrast, early studies showed that productive replication of HCMV in fibroblasts causes up- and down-regulation of thousands of host mRNAs22–25. Recently, techniques such as chromatin immunoprecipitation with sequencing (ChIPseq) and Precision Run-On Sequencing (PROseq) have also been used to study promoter occupancy of modified histones and formation of RNA Polymerase II (Pol II) pre-initiation complexes (PICs) on host versus viral promoters11,16. These studies highlight the differences in promoter usage between human and viral genes in the first 24–48 h of infection and further revealed that Pol II has limited nucleosome interactions on viral genomes. Follow-up studies extending through 72 h of infection further showed that, beyond the expected transcriptional downregulation of many host genes, upregulated host genes were associated with increased release of Pol II into productive elongation26. Moreover, a current eLife pre-print employs an assay for transposase-accessible chromatin with sequencing (ATACseq) to define changes in host genome accessibility in the first 48 hours of infection27. In doing so, it was found that ∼139,000 regions remained unchanged, while ∼ 38,651 became less accessible and ∼49,000 became newly accessible across the human genome. Using Hi-ChIP, changes in chromatin looping interactions were also defined, revealing ∼75,000 that open and ∼121,000 that close during infection. Of regions that closed, there was enrichment for TEA domain (TEAD) motifs. In addition, infection was found to reduce TEAD1 protein levels as well as the abundance and phosphorylation of regulatory factors such as YAP1, suggesting that infection affects TEAD-associated developmental programs in multiple ways. Notably, Immediate Early (IE) proteins IE1 or IE2, which directly control viral transcription, do not interact extensively with the host genome but instead appear to affect host transcription indirectly28–30. This includes repression of innate response pathways, while a recent study also implicates IE1 interactions with the histone variant MacroH2.A in the gradual induction of neuronal genes whose protein products support the maturation of new virions31.
In addition to these genetic changes, HCMV also causes visual changes in the shape and organization of the nucleus. This occurs through multiple effects of infection, ranging from disruption of intranuclear Lamin A/C organization7 to an increase in external microtubule-based forces applied to the nucleus32,33. Central to the latter is the formation of a Golgi-derived structure in the cytoplasm, termed the Assembly Compartment (AC)7,8. While the AC serves as a maturation site for progeny virions after they exit the nucleus, it also doubles as a microtubule-organizing center (MTOC)33. The AC forms next to the nucleus early in infection, and as it grows and matures, its MTOC function generates mechanically strong acetylated microtubules that connect to and pull on the nucleus through Linker of Nucleoskeleton and Cytoskeleton (LINC) complexes that contain the inner nuclear protein, SUN132,33. The forces applied by these microtubules are strong enough to cause the nucleus to rotate or rock while they pull on SUN1-containing LINC complexes, thereby drawing a large portion of the cell’s repressive histones towards the region of the nucleus that sits adjacent to the AC32. This helps to segregate heterochromatic regions from viral DNA, which is contained within Replication Compartments (RCs) that form partially phase-separated lobular structures at the opposing side of the nucleus. Nuclear rotation and polarization is evident over the first 24–96 h of infection, which then ceases concurrent with the appearance of mature virus particles in the AC and the onset of cell migration32,33. Despite these advances, however, we continue to have a relatively limited understanding of the extent to which HCMV affects host genome organization and nascent transcriptional activity on a global scale, along with the impact that microtubule forces may have on these processes.
Here, we use High-throughput Chromatin Conformation Capture (HiC) and Cleavage Under Targets & Release Using Nuclease (CUT&RUN)34,35 to determine the organization of both host and viral genomes, and their interactions. We further employ nascent and total transcriptomic approaches, which, when combined, reveal the extent to which infection alters chromatin organization and its correlations with transcriptional reprogramming and dysregulation genome-wide. This includes spurious transcription of neuronal genes in infected fibroblasts, while we further find that HCMV genomes accumulate near the most euchromatic regions of their host genome. Finally, applying these approaches to cells depleted of acetylated tubulin or SUN1 reveals the distinct roles of each factor in mediating microtubule-based separation of host and viral genomes and in controlling chromatin contacts, providing detailed genetic insights into how mechanotransduction refines these virus-host interactions.
Results
HCMV globally alters host chromatin contacts and compartment organization
To determine the effects of HCMV on host genome organization, we mock-infected or infected primary Normal Human Dermal Fibroblasts (NHDFs) with the clinical strain, TB40/E, at a multiplicity of infection (MOI) 5. We then cross-linked and processed samples for subsequent sequencing and Hi-C analysis36,37 at 72, 96 and 120 h post-infection (h.p.i.). These timepoints span the peak of nuclear rotation and polarization (72-96 h.p.i.) into the subsequent switch to cell motility that coincides with later stages of progeny virion production and spread32,33,38. Two independent biological replicates were performed for each infected-cell timepoint, taking two independent uninfected samples at both the start and end points of the time course (72 and 120 h.p.i.) (Supplementary Fig. 1A). Starting from a whole-genome perspective, Hi-C interaction mapping revealed a relative reduction in long-range inter-chromosomal contacts in infected compared to uninfected samples (Supplementary Fig. 1B). Whole genome maps were further visualized in HiGlass39, following alignment to the standard human genome sequence, GRCh38 and the viral TB40/E reference sequence40 (Supplementary Fig. 1C). Differences in chromatin contact strengths were visualized using division maps, which again highlighted the relative reduction in inter-chromosomal contacts as well as reduced interactions between p- and q-arms of individual chromosomes that occurred by 72 h.p.i. (Fig. 1A). Viewing these changes at different genomic resolutions highlighted how the most robust increases in contacts occurred at the intra-chromosomal level within chromosome arms, as seen along the central diagonal of the division map (Fig. 1A and Supplementary Fig. 1D). Using Chr 2, 5 and 17 as examples of chromosomes of varying sizes and gene densities further highlighted these differences between uninfected and infected samples at the chromosome level at each timepoint (Supplementary Fig. 2A). Uninfected and infected samples showed a high degree of similarity across timepoints, in line with uninfected primary cells being relatively stable in their genome organization and infection causing widespread changes by 72 h.p.i. that persist through 120 h.p.i. (Supplementary Figs. 1A, B and 2A). Given this similarity in samples and our focus on the nuclear rotation period of infection, to increase sequence depth and analytical power, we combined replicates for 72 h and 96 h infected datasets for subsequent comparisons with all four 72 h and 120 h uninfected control samples. Using these datasets to generate division maps further highlighted the extent to which relative contacts within p- or q-arms increased, while inter-arm contacts decreased (Fig. 1B and Supplementary Fig. 2B). GENOVA analysis of the Relative Contact Probability (RCP)41 further showed that long-range contacts across individual chromosomes decreased in infected samples, while shorter-range cis-contacts increased (Supplementary Fig. 2C).
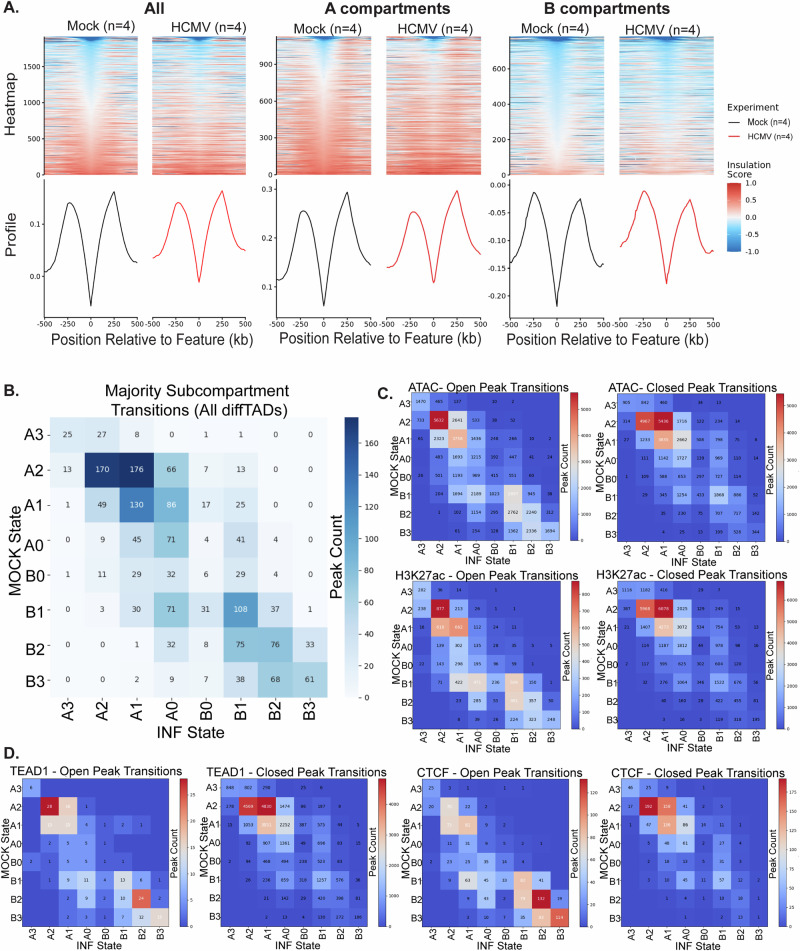
Fig. 1. HCMV remodels host chromatin contacts and compartment organization.
NHDFs were mock-infected or infected with HCMV (MOI 5) for the indicated times. A Genome-wide changes in chromatin interactions displayed by dividing Hi-C matrices for mock versus infected cells at 72.h.p.i. TB40/E genome added as an extra chromosome. HiGlass resolution = 5.12 Mb. Division = ratio of normalized contact frequencies. n = 2 per sample. B Intrachromosomal contact alterations shown by division comparison of combined Hi-C datasets from 72/96 h.p.i. for infected samples and 72/120 h for mock conditions. Juicebox resolution = 500 kb. Normalization = Balanced. Division = log[Observed/Control*(AvgC/OvgO)]. Scale = − 9 to 9 log enrichment. n = 4 per sample. C Pearson correlation matrices and corresponding eigenvectors (PC1) of Chr 2 between mock and infected combined datasets reveal large-scale cis-chromosomal reorganization. Juicebox resolution = 500 kb. Scale = − 1 to + 1. n = 4 per condition. D Saddle plots illustrating compartment interaction frequencies. Infection affects global compartmentalization, as contacts between A compartments or between B compartments decrease while interactions between A and B compartments increase. Difference saddle plot (lower right); green colors in A-A or B-B corners indicate loss of interactions and gain of A-B interactions shown as pink. GENOVA resolution = 100 kb. Bins = 20. n = 4 per condition. E, F. Chromosomal compartment level changes in (E). were identified by dcHiC for Chr 17, displayed with IGV, performed at 100 kb resolution. Top rows, dcHiC subcompartment segmentations set to default calls = 6. Red = A compartments. Blue = B compartments. Resolution = 100 kb. Middle rows, dcHiC PCA tracks. Bottom row (green), Log2Pvalue of differential compartments called by dcHiC (DC stat). Zooms in F. show examples of compartment changes and spreading. G Alluvial diagram showing starting subcompartment state in mock and end state in infection, using 100 kb binned subcompartments (not merged, unlike Bedtools approaches in S3C-D). Note that in each case, compartment numbers refer to the total number before or after infection, while the thickness of lines traces starting compartments to their final compartment state after infection. Generated with Flourish, and an interactive version is available at https://public.flourish.studio/visualisation/21041814/.
We next examined the effects of infection at the compartment level. Whole chromosome Pearson’s correlation Hi-C maps and corresponding Eigenvectors revealed extensive changes in active “A” or repressive “B” compartments and their interactions across chromosomes, again with notable differences in effects within versus between p- and q-arms (Fig. 1C and Supplementary Fig. 3A). GENOVA-generated saddle plots further showed that infection caused an increase in interactions between A and B compartments, and a reduction in A-A or B-B compartment interactions (Fig. 1D). This suggested that localized changes in compartment strengths and contacts were occurring in infected cells. Using dcHiC42 to identify compartments and visualizing examples using Integrative Genomics Viewer (IGV) (Fig. 1E, F and Supplementary Fig. 3B), instances of enlarged A or B compartments could be observed at 100 kb. Compartments can be further sub-classified into strengths, with A0 and B0 being the weakest and A3 or B3 being the strongest active or repressive region, respectively. Subcompartment calls were made using dcHiC and showed that infection reduced the number of stronger A compartments and increased that of weaker A0 compartments (Supplementary Fig. 3C). By contrast, all B compartments were reduced in number. Compartment size analysis further showed that while total numbers decreased, this arose due to increases in the sizes of all compartments except A2’s (Supplementary Fig. 3D). To understand this in more detail, we mapped the fate of compartments before and after infection using interactive Sankey plots (Fig. 1G and https://public.flourish.studio/visualisation/21041814/). Interestingly, while weak A0 and B0 compartments changed considerably and were prone to flipping from active to repressive states, and vice versa, stronger A or B compartments often changed in strength but rarely flipped.
On smaller organizational scales, tornado plots at TAD boundaries made using GENOVA showed that TAD insulation was reduced in infected cells (Fig. 2A). Interestingly, when divided by compartment types, TADs within A compartments of uninfected cells were more insulated that those in B compartments, which likely reflects the generally heterochromatinized and repressed state of the latter. Infection resulted in a loss of TAD insulation in both compartments, but this was particularly evident in A compartments. Overall, using diffDomain analysis43, we identified a total of 7268 reorganized TADs, of which 1799 were significantly affected by infection. Subcompartment transition heat maps showed that changes in TAD structure were most prevalent in A compartments, particularly in A1 and A2 compartments, although changes also occurred in B compartments (Fig. 2B). To relate our findings to those of others, we compared our compartment and TAD change analyses with published ATACseq as well as CCCTC-binding factor (CTCF), TEAD1 and H3K27ac ChIPseq datasets from cells also infected with TB40/E at MOI 527. Despite being performed at a slightly earlier point in the nuclear polarization window (48 h), transition matrices of changes in genome accessibility and in CTCF, TEAD1 or H3K27ac peaks showed striking similarities to our called TAD changes. In particular, opening (gains upon infection) and closing (loss upon infection) of ATACseq and H3K27ac peaks again occurred predominantly in A1 and A2 compartments, with a degree of opening also occurring in B1 and B2 (Fig. 2C). Interestingly, while TEAD1 and CTCF peaks showed similar patterns, opening CTCF peaks (binding gained during infection) were notably enriched in B compartments (Fig. 2D). Moreover, closing TEAD1 and CTCF peaks were enriched in A compartments, suggesting that there may be widespread transcriptional repression of TEAD-associated genes in A compartments. Combined, these analyses suggested that most transcriptional changes caused by infection were likely to be controlled through changes in compartment strengths rather than compartment-scale flips, and in particular through changes in TAD organization and the activity of insulators and specific transcription factors within these compartments.
Fig. 2. Local contact alterations and features occur within changing and stable compartments during infection.
Topologically associating domain (TAD) insulation and reorganization was assessed globally and locally, while TAD changes and other chromatin features associated with different subcompartments. A Global TAD insulation (left) is reduced during infection, both in A and B compartments (right). B Subcompartment transition state localization of reorganized TADs identified with diffDomain at 10 kb, comparing dcHiC compartment calls in original mock and final infected states. C, D Subcompartment transition state localizations were assigned for open (gained on infection) or closed (lost on infection) peaks sourced from Sayeed et al. for various chromatin features; accessibility (ATACseq), transcriptional activity (H3K27ac), transcription factor binding (TEAD1), and boundary elements (CTCF), comparing dcHiC compartment calls in original mock and final infected states.
HCMV both reprograms and dysregulates nascent transcription
While HCMV-induced host transcriptomic changes are well characterized22–25, this reflects the combined effects of both transcription and post-transcriptional events. As such, we next employed transient transcriptome sequencing (TTseq)44 to better understand transcriptional and transcriptomic changes at 72 and 96 h.p.i. In this approach, nascent transcripts are labeled by pulsing with 4-thiouridine (4sU) followed by their selective isolation by 4sU-biotinylation for subsequent sequencing alongside corresponding total RNA samples (Supplementary Fig. 4A). To do this rigorously and to allow for later comparisons with the effects of ATAT1 or SUN1 depletion, these experiments were performed in control non-targeting siRNA-treated NHDFs alongside corresponding control siRNA-treated uninfected samples. Two independent control or targeting siRNAs were used in each case, performing replicate experiments that generated two independent samples per condition for 72 h and four independent samples per condition for 96 h analyses (Supplementary Fig. 4B). As expected from this approach, nascent transcriptomic reads captured a larger fraction of intronic than exonic regions while total RNA samples were more enriched in exonic reads (Supplementary Fig. 4C). Interestingly, infection appeared to result in an increase in detection of intergenic reads particularly in our TTseq datasets, which offered an initial indication of infection-induced dysregulation of transcription.
To begin understanding the effects of infection on the nascent transcriptome, we analyzed changes in gene expression using DEseq245. Comparing 72 and 96 h.p.i. nascent transcriptomic datasets provided kinetic information and identified a core set of HCMV-regulated genes. Comparisons showed that only a small number of either upregulated or downregulated genes were unique to the 72 h timepoint, while 2710 upregulated and 2431 downregulated genes were common to both timepoints (Fig. 3A). In line with ongoing reprogramming of the host transcriptome between 72 and 96 h.p.i., a larger number of genes were unique to 96 h.p.i. samples. We also sequenced total RNA isolated prior to biotin-based enrichment of nascent RNA from our TTseq samples and found that a core set of 3233 upregulated and 2664 downregulated transcripts were shared across timepoints. Moreover, 1723 upregulated and 1566 downregulated genes were common to both timepoints and to both TTseq and RNAseq approaches. This revealed that approximately two-thirds of core transcriptional changes result in corresponding transcript-level changes, while non-overlapping genes reflect the added complexity of post-transcriptional control during infection. This, in turn, underscores the rationale for our primary focus on nascent transcriptomics to determine transcriptional correlations with chromatin remodeling.
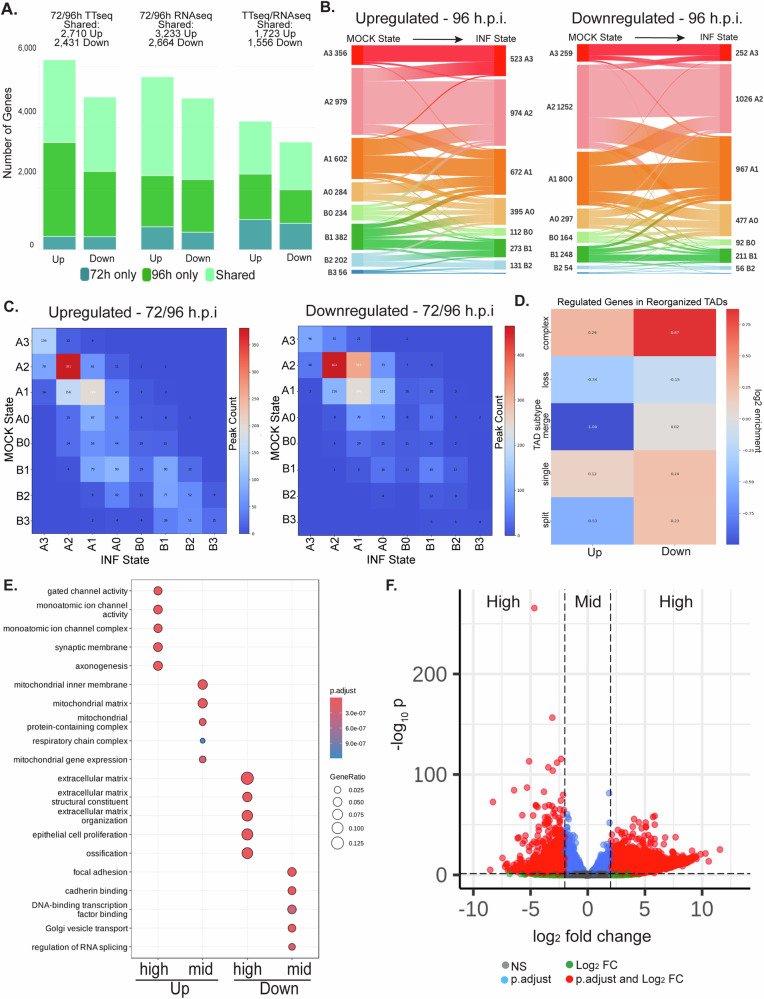
Fig. 3. HCMV extensively reprograms host nascent and steady state transcriptomes.
Mock or infected samples taken at 72 h.p.i. (n = 2) or 96 h.p.i. (n = 4) were pulsed with 4sU and nascent RNA was isolated for TTseq, or total RNA was sequenced (RNAseq). A Comparisons of DE genes identified at the indicated timepoints showing shared and unique genes in TTseq, RNAseq and across both TTseq and RNAseq datasets. The number of shared genes at both times is indicated for each condition. B, C Compartment distributions of DE genes. Genes were assigned to 100 kb binned subcompartment calls, requiring an overlap of 75% of the gene. Alluvial diagram of DE genes showing start (mock) and end (inf) compartment state for up-regulated or down-regulated genes at 96 h.p.i. is shown in B. Interactive version is available at https://public.flourish.studio/visualisation/20359025/. Compartment transition states of core 72/96 h.p.i. DE genes are shown in (C). D diffDomain was used to identify several types of TAD reorganization upon infection (split, single, merge, loss and complex subtypes) and enrichment in each subtype of up- or down-regulated genes identified by DEseq2. See also Supplementary Fig. 5. E, F DEseq2 was used to identify differentially expressed genes between mock and infected samples at 96 h.p.i. n = 4 per condition. Differentially expressed genes were subdivided by Log2FC below (mid) or above 2 (high) (Log2FC > 2 & < − 2). E Gene Ontology analysis of statistically significant upregulated or downregulated genes at 96 h.p.i., plotted using clusterProfiler. F EnhancedVolcano plot displaying mid versus high (Log2FC > 2 & < − 2) expression changes.
Combining nascent transcriptomics and Hi-C, we next examined the chromatin subcompartment locations of the top 1000 log2FC up- or down-regulated genes at either 72 or 96 h.p.i. (Supplementary Fig. 4D). At both timepoints, only ∼ 19% of downregulated genes were found in B compartments in infected cells, with most located in A compartments. Many upregulated genes were also found in A compartments, while ∼ 35% were found in B compartments by 96 h.p.i. Examining the more extensive reprogramming that occurred by 96 h.p.i., we further tracked the compartment location of all DE genes before and after infection using Alluvial Diagrams (Fig. 3B and https://public.flourish.studio/visualisation/20359025/). Similar trends were observed for core genes that were commonly regulated at both 72 and 96 h.p.i., adding high confidence to our compartment calls and transcriptional activity comparisons (Supplementary Fig. 4E and https://public.flourish.studio/visualisation/20400069/). Although most DE genes remained within their original A or B compartment categories, 308 significantly downregulated genes were associated with compartment-scale A-to-B flips. Meanwhile, 330 significantly upregulated genes were associated with B compartments strengthening to A compartments, and these were notably enriched in neuronal gene signatures discussed below. However, most regulations appeared to occur at more localized levels. Indeed, heatmaps plotting up- or down-regulated genes relative to their original uninfected- versus infected-cell compartment locations (Fig. 3C) showed a very similar distribution to TAD, ATACseq, TEAD1 and CTCF peak changes (Fig. 2B–D). Most up- and down-regulated genes were found in A1 or A2 compartments, while some upregulated genes were also found in B compartments. TAD changes can be classified into various states ranging from straightforward losses to more complex changes (Supplementary Fig. 5A, B). Interestingly, we found that downregulated genes were enriched in complex TAD changes (Fig. 3D). Although not so abundant, complex TAD changes were also predominant in A2 compartments where downregulated genes were found (Supplementary Fig. 5C). Indeed, CTCF1 and TEAD1 peak closures were highly enriched in the same A compartments (Fig. 2D), suggesting these may be key drivers of gene suppression. By contrast, upregulated genes were moderately depleted in merged TAD states, but overall, they were not enriched in any specific TAD subtype. This suggested that upregulated genes are associated with multiple types of TAD changes or gross dysregulation of TAD organization during infection (Fig. 3D). Overall, these data suggested that many repressed genes were likely specifically regulated by factors such as TEAD1, while upregulated genes appeared to be associated with broader changes in TAD structures or infection-induced dysregulation.
To determine the nature of these changes in nascent transcription, we next performed Gene Ontology (GO) analysis. In line with prior mRNA transcriptomics studies22–25, downregulated nascent transcripts were highly enriched in GO terms associated with actin and adhesion processes while upregulated transcripts included expected associations with biosynthetic processes such as DNA replication and mitochondrial activity, but also genes associated with neuronal processes (Fig. 3E). Activation of neuronal genes in fibroblasts was surprising but agrees with a recent report detailing extensive upregulation of neuronal mRNAs as infection progresses31. Indeed, a clear distinction in GO terms associated with more commonly reported biosynthetic processes versus neuronal processes becomes evident when using log2FC thresholds to define high versus moderate changes in expression, highlighting the large relative fold-change in neuronal genes (Fig. 3E, F).
To better understand these transcriptional changes, we plotted reads from our TTseq and paired RNAseq datasets across core commonly changed genes and flanking regions, size-normalizing the variation in gene body sizes for averaged comparisons using Deeptools46 (Fig. 4A, B). This revealed that genes that were normally highly expressed in uninfected cells were those that were strongly downregulated upon infection. By contrast, genes that were most strongly upregulated by infection were largely those that were originally expressed at relatively low levels in uninfected cells, below the average expression of all genes. Interestingly, infection did not induce their expression beyond average gene expression levels in uninfected cells. This shows that although these genes were induced by a relatively large magnitude, this was, in part, because their basal expression in uninfected cells was low, which makes sense for biosynthetic and, in particular, neuronal genes in primary fibroblasts.
Fig. 4. HCMV causes transcriptional dysregulation.
A, B Averaged read coverage within and nearby gene bodies for upregulated or downregulated genes between mock and infected conditions, plotted for nascent RNA (TTseq) (A) or total RNA levels (RNAseq) (B). Read counts for “all genes” is also plotted, highlighting the effects of infection on gene expression overall versus up- or down-regulated genes. Data were normalized and replicates combined using Deeptools before display with plotProfile. Region body length = 100 kb. Bin size = 10. C–F ARTDeco was used to characterize read-in (RI) and read-through (RT) in mock versus infected TTseq and RNAseq samples described in A-B above. Rates of RI or RT for the top 1000 expressed genes (C) in individual 72 (n = 2) and 96 (n = 4) h datasets, as well as averaged datasets for effects of infection over both timepoints (dark green, n = 6). Transcriptional profile (D) of upregulated gene categories, high (logFC > 2), mid (0;< logFC > 2), and read-in (log2FC > 2, read-in rate > − 2). E Subcompartment transition state localization of read-in genes shows affected gene flow from the original mock to the final infected states. F Examples of read-in genes visualized in IGV; PLAC8L1 is found in an A compartment that is altered by infection, and C3orf20 is in a weak B (white box) that becomes A upon infection. Both are accompanied by TAD changes during infection. Shown in each are compartments in uninfected and infected cells determined by dcHiC together with DC stats and viral reads (n = 4), H3K4me3 (promoter, n = 2) and H3K9me3 (repressive mark, n = 2) CUT&RUN tracks, average read counts from TTseq at 72 (n = 2) or 96 h (n = 4), up- or down-regulated genes called by DEseq2 (green and orange, respectively), genes upregulated by read-in as identified by ARTDeco (light green) and 20 kb TADs in mock (blue) or infected (purple) cells identified using HiCExplorer.
The unexpected induction of neuronal signatures prompted us to determine if infection caused transcriptional read-in to genes due to aberrant termination and read-through from active upstream genes, which can be analyzed using ARTDeco47. In first examining the top 1000 upregulated genes, we found that, as expected, read-in and read-through occurred at very low frequencies in uninfected cells, but their rates increased during infection between 72−96 h.p.i. (Fig. 4C). This was evident in both TTseq and paired RNAseq samples, although effects were more robustly detected in direct measurements of transcription using TTseq. This difference is likely due to post-transcriptional control of some of these read-in transcripts, such as degradation, but in general, read-in and read-through remained detectable in our total RNAseq datasets, as we did not use polyA selection for mature transcripts. We then used ARTDeco to identify read-in or read-through genes within our datasets that were significantly upregulated. Overall, infection resulted in elevated expression (Log2FC > 2) of 1416 genes with an increase in read-in and 972 genes with an increase in read-through rates (Log2 RI or RT/Gene > − 2) from a total of 6142 upregulated genes identified using ARTDeco analysis of TTseq datasets at 96 h.p.i. However, we found no robust correlation between read-through from upstream genes and read-in of downstream genes during HCMV infection (Supplementary Fig. 6A). Moreover, read-in and read-through occurred on the same gene in 760 cases. This was also evident in read plots wherein the read counts upstream and downstream of read-in genes were notably higher than in gene bodies compared to other highly or moderately upregulated genes (Fig. 4D). This suggested that these transcripts arise due to spurious transcriptional events rather than previously reported forms of read-through into downstream genes (for simplicity, given their higher frequency, we refer to these spurious events as read-in going forward). These genes were found across both A and B subcompartments called by dcHiC (Supplementary Fig. 6B). Alluvial diagrams at the individual gene level revealed the underlying complexity in their subcompartment-level associations, wherein some read-in genes switched compartments but many did not (Supplementary Fig. 6C). Heat maps of their locations relative to uninfected versus infected-cell compartment calls further showed that many read-in genes clustered in A1 and A2 compartments (Fig. 4E), which in turn clustered with the most extensive TAD and ATAC-Seq changes (Fig. 2B, C). Viewing examples of read-in genes illustrated how they were often found in either A or B compartments where local TADs were altered (Fig. 4F and Supplementary Fig. 6D). Overall, these findings suggest that read-in and read-through events reflect spurious transcriptional events on neuronal and other repressed genes that are likely driven by infection-induced dysregulation of TAD organization.
GO term analysis showed that read-in genes were enriched for plasma membrane and neuronal processes but that these only accounted for a proportion of the high Log2FC genes that drive neuronal signatures in infected-cell transcriptomics (Supplementary Fig. 7A). To explore this further, we compared our nascent and total transcriptomic datasets with published proteomic datasets also generated from fibroblasts infected with the same HCMV strain, similar MOI of 3 and sampled at the same timepoints48. In doing so, we found strong correlations across TTseq, RNAseq and proteomic changes both in general GO terms and gene-specific comparisons across our moderately induced gene sets (Supplementary Fig. 7B–D). By contrast, there was a much weaker correlation with proteomics datasets across our high Log2FC genes and a near total lack of proteins detected from read-in genes. While this was in part because there was a higher proportion of long non-coding RNAs (lncRNAs) amongst read-in genes (Supplementary Fig. 7C), this also suggested that many of these read-in events were indeed spurious in nature and not productive drivers of changes in the host proteome. However, 97 proteins were detected as overlapping with high Log2FC changes in both TTseq and RNAseq datasets. Moreover, in our TTseq analyses, we detected 9 out of the 12 neuronal mRNAs recently reported to be upregulated by the lab-adapted HCMV strain, Towne31, of which at least 3 were also detected as upregulated in proteomics. This includes Kif1A, the primary neuronal gene studied for its functional contribution to virus maturation31, which we found to be induced across our TTseq and RNAseq as well as proteomic analyses, but is not identified as a read-in gene. As such, our analyses suggest that infection drives a robust neuronal gene signature through a complex mixture of spurious transcriptional events and induction of lncRNAs, while distinct subsets of neuronal protein-encoding genes are induced specifically to support virus replication.
HCMV genomes localize to subsets of euchromatic A compartments
Beyond changes in host chromatin organization, we also noted viral contacts with specific regions of the host genome in our Hi-C analysis (Fig. 5A and Supplementary Fig. 8A). Approximately 23% of all reads were viral, and of these, we found that 3.4% (namely, 15% of total viral reads) contacted the host genome. This algins with the idea that viral DNA at the periphery of RCs is proximal and accessible to host DNA. More closely inspecting total viral read distributions, we noted that they were highly enriched in A but far less so in B compartments, particularly when normalized for differences in compartment sizes (Fig. 5B). We further tested our dcHiC compartment calls by performing H3K9me3 CUT&RUN to independently define repressive B compartments. Deeptools was then used to plot viral and H3K9me3 CUT&RUN reads against compartment calls, size-normalizing compartments for genome-wide comparisons. This highlighted the enrichment of viral reads in A compartments and de-enrichment in B compartments, which contrasted with the expected enrichment of host reads in H3K9me3 CUT&RUN in B compartments (Fig. 5C, D). Further highlighting the specificity of these virus-host genomic interactions, we did not detect significant levels of association or enrichment in specific compartments for mitochondrial DNA in our analyses. These genetic approaches align with prior observations that repressive histone marks, such as H3K9me3, are spatially separated from viral RCs32, while revealing that viral DNA remains intermingled with euchromatin.
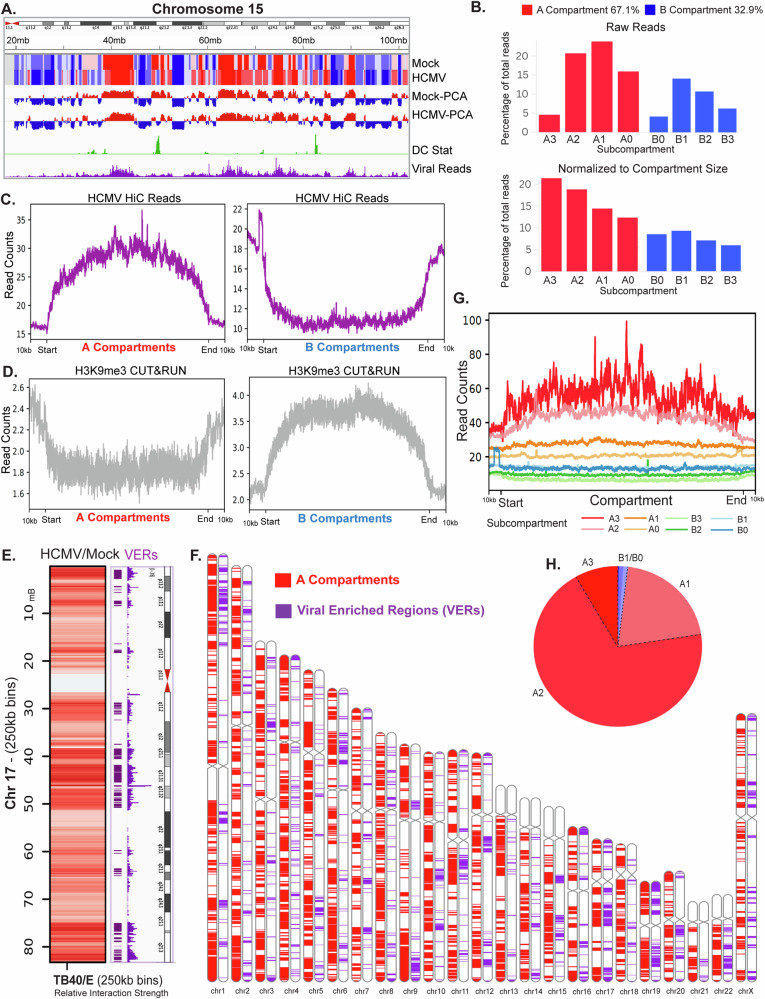
Fig. 5. HCMV genomes accumulate at euchromatic regions of the host genome.
Viral-host interactions were extracted using HiCPro’s makeviewpoints from combined 72 and 96 h (n = 4) infected HiC datasets to identify regions of enrichment. A Viral-host contacts visualized with dcHiC compartment analysis shows overlap with A compartments using Chromosome 15 as an example. B Viral genome interactions across subcompartments. Top; percent of viral-host reads mapped to subcompartments. Bottom; subcompartments were normalized by size and viral genome coverage. C, D. Enrichment of viral reads in (A) compartments and de-enrichment in (B) compartments (C) is evident in the aggregated signal profile controlling for compartment size. H3K9me3 (repressive mark) CUT&RUN performed on mock-infected or infected NHDFs at 96 h.p.i. (MOI 5, n = 2 per sample) shows the opposing depletion of H3K9me3 reads in A compartments and enrichment in (B) compartments (D). E–H Identification of viral interaction hotspots (VERs) and their interactions with the host genome. E GENOVA-derived heatmaps of trans contacts highlight viral-host reads across Chromosome 17. Parameters = znorm. Mode = difference. The top 10% of 100 kb bins of the host genome most enriched for viral reads, termed VERs, are highlighted in purple on the accompanying chromosome map with viral read plots on the right. F Distribution of A compartments and VERs across all chromosomes displayed with an Ideogram. GDeeptools plotProfile of VERs across subcompartments. Region body length = 100 kb. Bin size = 10. H Percent of Viral Enriched Regions (VERs) within each subcompartment.
To explore this further, we determined the primary virus-host interaction “hotspots” by taking the top 10% of 100 kb bins of the host genome containing the most viral reads, which we term “virus enriched regions” (VERs) (Fig. 5E and Supplementary Fig. 8B). Ideograms demonstrated that VERs were found in subsets of A compartments across all chromosomes (Fig. 5F and Supplementary Fig. 8C). Notably, VERs did not simply accumulate at gene rich regions (Supplementary Fig. 8C). Moreover, viral reads did not accumulate in all A compartments to the same extent, suggesting further levels of specificity in their localization. Plotting viral reads across size-normalized subcompartments using Deeptools, viral genomes were found to be most enriched in strong A3 compartments, followed by A2 and A1 (Fig. 5G). This revealed that viral DNA has a preference towards more open or active chromatin states. Moreover, the overall distribution of VERs showed a strong enrichment in A2 compartments (Fig. 5H). This aligns with the fact that there are many more A2 than A3 compartments in uninfected and infected cells (Supplementary Fig. 3C). However, considering the higher abundance of A0 and A1 compartments, this combined analysis suggests that VERs predominantly accumulate in A2 compartments due to the combination of their abundance and euchromatic state.
As a brief aside, we also used our datasets to generate a Hi-C map of the viral genome, which showed a high level of localized contacts along the diagonal and far fewer longer-range self-contacts (Supplementary Fig. 9). This agrees with studies using ChIP or ATACseq approaches, which suggest that the viral genome is sparsely chromatinized and largely accessible. Indeed, the intensely red nature of the overall map likely reflects the proximity of many copies of viral genomes to one another within RCs, as well as distinct forms of transcribing versus replicating genomes, rather than a high level of specific longer-range self-interactions. Interestingly, we did observe what appear to be more specific interactions between the Origin of Lytic Replication (OriLyt) and two distal regions of the viral genome, and enrichment of active H3K4me3 at the OriLyt. We could not detect any common sequence elements at this site of H3K4me3 enrichment on the virus compared to the host. This also aligns with reports that the OriLyt contains a strong enhancer element16,18 and that these are sequence-independent interactions28. Moreover, while the OriLyt was slightly more enriched in host reads, this was not unique to the OriLyt and in general, host reads were evenly distributed across the HCMV genome (Supplementary Fig. 9B). As such, we could not detect any strong indications of specific regions on the viral genome that might mediate contacts with the host genome. Overall, our data support the idea that viral genomes are generally open and accessible but may also form limited higher-order contacts within itself.
We next examined the genomic location of DE genes identified in TTseq relative to VERs to determine whether proximity to virus replication sites correlates with specific changes in transcription. Ideograms showed that, like VERs, DE genes did not simply cluster at gene-rich regions and were found on all chromosomes (Supplementary Fig. 10A). Upregulated and downregulated genes were also relatively evenly distributed in terms of their numbers within versus outside of VERs (Supplementary Fig. 10B, C). Amongst downregulated genes, those within and outside of VERs showed similar enrichment in developmental transcription factor pathways and cell signaling and adhesion GO terms (Supplementary Fig. 10B–D). This included TWIST, TEAD, and Hippo pathways previously reported to be downregulated during infection27,49,50, and in line with this being specific regulation, we also find that downregulated genes cluster in A2 subcompartments enriched in TAD changes and the closing of TEAD and CTCF peaks in infected cells (Figs. 2B–D, 3C). By contrast, upregulated genes exhibited more diversity depending on their location relative to VERs. Upregulated genes outside of VERs were highly enriched in neuronal GO terms and transcription factor pathways, while genes within VERs were more associated with mitochondrial regulation and lacked significant transcription factor pathway enrichment. This suggested that while infection is likely to directly control specific developmental programs, it may also indirectly dysregulate the transcriptional activity of some genes. This includes mitochondrial genes within A compartments that are proximal to sites of virus replication, as well as normally repressed neuronal genes outside of VERs, which simply give rise to predicted roles for neuronal transcription factor pathways.
Microtubule-derived forces regulate viral and host compartment interactions
Our prior work showed that microtubule forces on the nucleus help to maximize the segregation of repressive histones and viral DNA, at least on a visual level32. However, the true nature of this phenomenon and whether it contributes to HCMV’s effects on host gene expression remain unknown. We therefore used RNAi to deplete ATAT1, thereby blocking tubulin acetylation and mechanical strengthening of microtubules, or SUN1, an inner membrane protein that mediates the effects of microtubule-associated LINC complexes5,6,32 (Supplementary Fig. 11A). We first examined the broader effects of target depletion on virus-host genome interactions and host chromatin organization using Hi-C. In infected cells or infected cells treated with control non-targeting siRNAs, hotspots of viral contacts representing VERs were found across each chromosome (Fig. 6A and Supplementary Fig. 11B). We then compared the effects of ATAT1 or SUN1 depletion using subtraction maps, which showed that under both conditions, some of the primary VER hotspots remained stronger in control siRNA-treated cells than in target-depleted cells. However, patterns of VER contacts were not only visibly altered, but differences in the effects of ATAT1 or SUN1 knockdown were also observed. Notably, some primary VERs became difficult to discern while regions where viral contacts were normally weaker in control conditions now became stronger in ATAT1-depleted cells, as illustrated by blue colors in subtraction maps (Fig. 6A and Supplementary Fig. 11B). This supports the idea that without microtubule forces to maximize viral-host genomic separation, more intermingling occurs. Interestingly, depletion of SUN1 resulted in a different phenotype wherein viral contacts remained generally stronger in control conditions (red colors in subtraction maps). However, primary VER distributions were changed, and their intensity was more similar to other normally weaker viral interactions, suggesting less specific virus-host interactions were occurring. On the host side, we did not observe notable effects of ATAT1 or SUN1 depletion on either genome-wide or long-range chromatin interactions (Supplementary Fig. 11C, D), but changes in global compartment contacts were evident in Saddle plots41. As seen earlier in non-siRNA-treated cells (Fig. 1D), under control siRNA conditions, HCMV infection reduced short-range A-A or B-B interactions, while increasing A-B interactions (Fig. 6B). However, depletion of either SUN1 or ATAT1 resulted in a further decrease in B-B interactions during infection, and a specific increase in A-A interactions. This demonstrated that microtubule forces on the nucleus enhance those B-B interactions that continue to occur during infection, likely through pulling and concentrating heterochromatin towards the AC, while helping to reduce A-A interactions in the euchromatic space.
Fig. 6. Microtubule forces alter virus-host genomic interactions and effects on host chromatin compartments.
A, B Mock or HCMV-infected (MOI 5) NHDFs were treated with control non-targeting siRNAs or siRNAs targeting SUN1 or ATAT1. Samples were harvested at 96 h.p.i. and then processed for HiC. n = 3 independent experiments. A GENOVA-derived heatmaps of trans contacts highlight viral-host reads across Chromosomes 17 and 19, as examples. Shown are mock versus infected (left) alongside mock versus control siRNA (siC)-treated infected samples or infected samples treated with control siRNA versus ATAT1 or SUN1 siRNAs. Parameters = znorm. Mode = difference. VERs are highlighted in purple on chromosome maps with viral read plots on the right. Note that VERs distributions are altered, and many normally weak viral contacts become stronger, resulting in blue colors in subtraction plots of infected control versus ATAT1 knockdown samples. A more broadly mixed phenotype is seen for SUN1 depletion. B Differences in local A-A, B-B or A-B contacts visualized using saddle plots, with A-A and B-B contacts decreasing during infection. Depletion of SUN1 or ATAT1 results in stronger A-A contacts and weaker B-B contacts in infected cells compared to control conditions. GENOVA resolution = 100 kb. Bins = 25. C HCMV-infected NHDFs were treated with either of two independent control non-targeting siRNAs or siRNAs targeting SUN1 or ATAT1. Samples were pulsed with 4sU and then harvested at 72 h.p.i. (n = 2) or 96 h.p.i. (n = 4) for TTSeq. Volcano plots illustrate the effects of target depletion on host gene expression compared to control siRNA-treated infected cells identified using DESeq. Adjusted p-value cutoff = 0.05, Log2FC cutoff = 0. D Ideograms displaying the chromosome locations of Viral Enriched Regions (VERs) and significantly affected ATAT1 and SUN1 knockdown-sensitive genes identified in TTseq datasets at 96 h.p.i. n = 4 per condition.
To determine whether these forces solely control the spatial organization and interactions of viral and host genomes, or also impact host transcriptional reprogramming during infection, we performed ATAT1 or SUN1 depletions alongside the TTseq time courses detailed earlier. Specifically, two independent siRNAs were used for control non-targeting, SUN1 or ATAT1 knockdown, performed at 72 h.p.i. and 96 h.p.i., with a second set of replicates performed at 96 h.p.i. While uninfected samples clustered from infected, those from infected samples treated with control or targeting siRNAs were intermingled (Supplementary Fig. 12A), suggesting a high degree of similarity and limited effects of target depletion. DESeq was then used to determine genes that were significantly affected by target depletion. In line with our earlier kinetic comparisons, progressively more DE genes were significantly affected at 96 h.p.i. than at 72 h.p.i. in each case (Fig. 6C). However, while ATAT1 or SUN1 depletion similarly affected ∼50% of all DE genes arising during infection, these changes were not statistically significant, suggesting that mechanotransduction only indirectly influences how HCMV alters host gene expression, and its loss primarily results in noise. Indeed, this noise may reflect the effects of target depletion on VERs interactions rather than direct effects on transcription. Notably, only a few hundred non-overlapping genes were significantly up- or down-regulated under each condition. Combining these genes, GO term analysis showed that knockdown-sensitive genes consisted largely of mitotic and ribosomal genes (Supplementary Fig. 12B) while ideograms further showed that they were broadly distributed across chromosomes and did not specifically cluster within or outside of VERs (Fig. 6D). Combined, these findings suggest that microtubule forces do not directly or robustly affect transcriptional reprogramming during infection but instead, primarily serve to control the segregation of host and viral DNA (Fig. 7).
Fig. 7. Model of the role of microtubule mechanotransduction in host and viral genome organization during HCMV infection.
A Under normal infection conditions, acetylated microtubules not only serve to pull heterochromatic B compartments, but also euchromatic A compartments towards the viral Assembly Compartment (AC) to maximize their separation from partially phase-separated viral replication compartments (RCs). This results in interactions between viral genomes and specific, nearby euchromatic regions of the host genome. These pulling forces, effects of infection and the presence of expanding RCs result in increased A-B compartment interactions. B Loss of tubulin acetylation and microtubule-based mechanotransduction results in reduced heterochromatic B-B interactions and increased euchromatic A-A interactions, along with greater intermingling of RCs with host euchromatin. In the case of SUN1, its loss results in even greater intermingling due to its dual roles in mechanotransduction and direct control of genome organization. Created in BioRender. Rosencrance, C. (2025) https://BioRender.com/ dgwdiy8.
Discussion
How nuclear-replicating herpesviruses, in particular HCMV with its unusually protracted replication cycle and lack of host shutoff, interact with and affect the human genome is complex and only beginning to be understood.
In terms of effects on the host genome, infection with the γ-herpesvirus Epstein-Barr Virus (EBV) results in immortalization of resting B lymphocytes, and this is associated with global changes in 3D genome organization, including the merging and subdividing of compartments51,52, similar to our observations for HCMV. However, these events are associated with latent infection and specific EBV nuclear antigen (EBNA) proteins driving immortalization. By contrast, entry of HCMV into latency is not associated with gross changes in chromatin accessibility as assessed using ATACseq20. Instead, as discussed earlier, recent studies of HCMV lytic replication also using ATACseq revealed extensive changes in chromatin accessibility by 48 h.p.i27. Complementing and extending on these findings, our Hi-C approaches provide insights into the global changes in chromatin organization that occur as infection progresses. This includes a relative reduction in long-range contacts at both the inter- and intra-chromosomal level, while shorter-range contacts within chromosome arms increase. It is important to note that these are relative differences that likely reflect a broad loss of long-range interactions or compaction of chromosomes that result in increased local contacts, or a combination of both processes. Indeed, HCMV disrupts Lamin A/C7, which plays a crucial role in anchoring and organizing genomes in 3D space2, while the expansion of RCs undoubtedly creates compressive forces on host chromatin. Moreover, by comparing our compartment and TAD calls with ATACseq and ChIPseq datasets from others, we present a unified view of how infection alters host genome organization at large and small scales. We further combine TTseq to understand how these changes correlate with changes in nascent transcription. In doing so, we find that transcriptional up- or down-regulation does not show a robust correlation with absolute compartment flips and only modest correlations with weakening or strengthening of compartments. Instead, many changes appear to be controlled at local levels through complex changes in TADs, insulation and specific transcription factor programs, while others reflect infection-induced dysregulation. Our global analyses reveal robust transcriptional downregulation of genes associated with developmental processes and transcription factor pathways. Our combined analyses also suggest that TEAD1 and CTCF peaks close in conjunction with complex changes in TADs to suppress active genes within specific A subcompartments, which aligns with prior reports that HCMV inhibits TEAD1, Hippo and WNT pathways, for example27,49,50. Associated genes were found both within and outside of VERs, further suggesting these are actively manipulated pathways during infection. Interestingly, within VERs, there was little to no enrichment of transcription factor pathways for upregulated genes, which were largely associated with metabolic processes. This raises the possibility that viral replication proximal to these loci recruits host or viral factors that activate these genes, rather than the activation of a broader host transcriptional program.
Outside of VERs, we find that infection increases the transcription of many genes that are associated with neuronal processes. Approximately 300 of these were associated with changes from B to A compartments, while most were regulated at more local levels, like other upregulated genes. Almost 1500 of these genes had signatures of upstream read-in events that were particularly evident through our use of TTseq. Interestingly, several stresses and viral infections, including by the α-herpesvirus herpes simplex type 1 (HSV−1), cause defects in termination that result in read-through and downstream of gene (DoG) transcription of neighboring genes53. However, we find that this does not arise in the case of HCMV and instead, aberrant read-in and read-through events often occur on the same gene. These events were only detected for high log2FC genes found outside of VERs and, in most cases, were either associated with lncRNAs or spurious transcriptional events that did not lead to protein production. As such, many of these events are likely caused by the broader disruption of host chromatin organization during infection. However, a recent study found that at least some of the neuronal genes that are induced at later stages do play a role in promoting virus maturation31. Moreover, the viral transcription factor IE1 was found to function with the histone variant, MacroH2A, to induce expression of neuronal genes. Notably, we and many others have shown that although IE1 concentrates in viral RC, it is also more broadly distributed throughout the nucleus32, meaning that it can likely induce host genes both within and outside of the sphere of influence arising from proximity to viral RCs. Indeed, our data aligns with these findings and provides complementary insights into the complexity of how neuronal genes are induced. We find that read-in genes only account for a proportion of the neuronal signature that is induced upon infection, supporting the idea that distinct IE1-driven mechanisms are also at play and serve a purpose in promoting virion maturation. Clearly, much remains to be understood about the many neuron-associated genes that become expressed during infection, the different factors driving their expression, and which of them are functionally important to infection.
In terms of the viral genome, an emerging concept is that some viruses replicate in phase-separated RCs. However, while several herpesviruses form RC’s that exhibit key features of phase separation54, studies suggest that these membrane-less compartments non-the-less remain highly accessible to the nuclear environment and undergo non-specific interactions55–58. Our Hi-C data aligns with this concept, whereby ∼ 15% of HCMV reads map to the host, which likely represents viral DNA at the periphery of RCs. Indeed, we do not find evidence for any specific region of the viral genome in mediating contacts with the host. Our Hi-C maps of the viral genome itself also support the idea that it is largely open and accessible, but data suggest that specific longer-range self-interactions may occur. From the perspective of its interactions with the host, our combined Hi-C and CUT&RUN analyses show that viral contacts occur primarily within euchromatic A compartments, in line with prior visual observations that viral DNA is spatially separated from repressive histone marks32. Genomes of the γ-herpesviruses, EBV and Kaposi’s Sarcoma-associated Herpesvirus (KSHV), localize to B compartments during latency but switch to A compartments during virus reactivation59, which may be linked to global changes in chromatin architecture during B-cell activation in the case of EBV51,52. α-herpesviruses, such as HSV-1, and γ-herpesviruses, such as EBV, also replicate in RCs whose expansion pushes heterochromatin to the nuclear periphery56,60–65. This suggests that many herpesviruses productively replicate in semi-accessible phase-separated compartments that form or at least optimally replicate and expand within euchromatic regions. Here, we extend our understanding of this concept and show that HCMV genomes not only localize to euchromatic regions but show a strong preference towards the most euchromatic A2 and A3 subcompartments.
Finally, HCMV also uses microtubule-derived mechanical forces to further optimize the partitioning of host and viral genomes. While this has been observed from a large-scale visual perspective32, our genomic approaches provide deeper insights into both the nature of this separation and the specific contributions of acetylated microtubules and SUN1. In terms of the host genome, we find that large-scale chromosome organization is severely disrupted by infection, and microtubule forces have little impact at this scale. This is perhaps unsurprising given that, as mentioned earlier, HCMV alters many aspects of nuclear organization and function, including gross disruption of Lamin A/C7. Loss of microtubule-based forces also had no significant impact on host transcriptional reprogramming by HCMV. This too makes sense considering that genome organization and transcriptional activity is further regulated by a complex array of histones, insulators and other factors that are bound to DNA. Instead, our findings reveal that mechanical forces play a role in spatially organizing DNA in what is undoubtedly a more genetically fluid environment and serve to refine compartment-scale genomic interactions. Of the chromatin interactions that continue to occur in infected cells, loss of microtubule-derived forces on the nucleus decreased B-B compartment interactions and increased A-A interactions. This supports the idea that microtubules pull and concentrate heterochromatic regions towards the AC, resulting in more B-B interactions while reducing euchromatic A-A organization in the infected cell. Our data further suggests that the primary purpose for doing this is to limit non-specific or suboptimal interactions between viral RCs and host DNA. Interestingly, depletion of ATAT1 resulted in stronger viral interactions with regions of chromosomes that are normally weaker sites of interaction, in line with the loss of this optimal partitioning. By contrast, SUN1 depletion resulted in more complex effects that included broad changes in the distribution of virus-host contact sites. This too makes sense considering that SUN1 is located at the inner nuclear membrane, where it not only mediates the forces applied by microtubules on the nucleus as a component of LINC complexes but also independently contributes to genomic organization, including that of mitotic chromosomes5,6,66–69. As such, these findings demonstrate that although depleting either factor results in a visually similar phenotype of reduced polarization of viral DNA and host heterochromatin, at a deeper genetic level, the loss of these factors results in distinct forms of intermingling of viral and host genomes. In terms of why the virus uses mechanical forces in this way, most likely, HCMV attempts to create spaces as free of host DNA as possible, with proximity to and spurious interactions with the least condensed euchromatic regions of the genome being more optimal than condensed genetic spaces. It is important to note that ultimately, disrupting these microtubule-based forces has no effect on viral gene expression32. As we show here, there is also no significant effect on host transcriptional reprogramming. Moreover, loss of ATAT1 or SUN1 only results in a 2-fold reduction in DNA replication rates and infectious virion production32, yet as we show here, each factor has a distinct effect on virus-host genomic intermingling. Combined, this suggests that microtubule mechanotransduction simply optimizes the nuclear environment for RCs to from and replicate viral DNA, but in its absence, HCMV still replicates efficiently in a manner that is more intermingled with euchromatin and more akin to other herpesviruses.
Methods
Cell culture and viruses
Primary normal human dermal fibroblasts (NHDFs) were isolated from human male neonatal foreskin (Lonza: CC-2509). Cells were authenticated by the supplier and were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Fisher Scientific) supplemented with 2 mM L-glutamine, penicillin/streptomycin and 5% Fetal Bovine Serum (FBS), incubated at 37 °C with 5% CO2. Cultures were routinely screened and verified to be free of mycoplasma using commercial mycoplasma test kits and by screening for mycoplasma sequences in genomic datasets. HCMV strain TB40/E expressing mCherry-tagged UL99 was grown and titrated on NHDFs, as described previously32,33. Briefly, virus stocks were titrated by serial dilution and infection of NHDFs, measuring numbers of cells infected by immunofluorescence staining for the early IE1/2 protein at 24–48 h.p.i. and counting plaques formed at 12 d d.p.i. NHDFs were infected at an MOI of 5 for all experiments, visually inspecting cultures for UL99-mCherry expression to confirm all cells were infected prior to harvesting.
RNA interference (RNAi) and Western Blotting
The siRNAs used in this study were obtained from ThermoFisher Scientific; Control non-targeting siRNA-A (Cat#: AM4635), Control non-targeting siRNA-B (Cat#: AM4637); siRNA αTAT1-A (Cat#: AM16708A, ID: 130789), siRNA αTAT1-B (Cat#: 4392420, ID: s36739), siRNA SUN1-A (Cat#: AM16708A, ID: 222696), siRNA SUN1-B (Cat#: AM16708A, ID: 241855). Confluent NHDFs were transfected with 150 pmol/ml siRNAs using Lipofectamine siRNA max (Invitrogen #13778075), following the manufacturers protocols and scaled appropriately for the surface area of the dish used. For experiments involving infections, NHDFs were infected at MOI 5 for 12 h to establish infection prior to siRNA treatment, as described previously32,33. For 96 and 120 timepoints, knockdowns were performed a second time at 72 h to maintain depletion efficiency throughout the course of infection, as described previously32,33. Western blots were performed as previously described32,33. Briefly, cells were lysed in laemmli buffer (62.5 mM Tris-HCl, pH 6.8, 10% (v/v) glycerol, 2% (w/v) SDS, 0.7 M β-mercaptoethanol) and set in a heat block at 100 °C for 3 min. Samples were then resolved through a SDS-PAGE gel (10% polyacrylamide), transferred to a nitrocellulose membrane (GE Healthcare Life Sciences), washed three times using tris-buffered saline (TBS) with 0.1% (v/v) Tween 20 (TBS-T) and blocked in TBS-T containing 5% (w/v) non-fat milk for 1 h at room temperature. Membranes were then incubated overnight with primary antibodies diluted in TBS-T containing 3% (w/v) bovine serum albumin (BSA) at 4 °C. The next day, membranes were rinsed and then washed three times in TBS-T for 5 min, followed by incubation with the appropriate horseradish peroxidase (HRP) conjugated secondary antibody (GE Healthcare Life Sciences) diluted in TBS-T with 5% (w/v) non-fat milk for 1 h at room temperature. Membranes were then rinsed and washed three times in TBS-T for 5 min, then visualized using ECL detection substrate and x-ray film (Thermo Fisher Scientific). Primary antibodies used for immunoblotting were: Ac-K40 tubulin (Sigma-Aldrich: T6793); SUN1 (Novus Biologicals: NBP1–87396); β-actin (3700; Cell Signaling Technology).
Arima Hi-C Library preparation
Hi-C data was generated using the Arima HiC + Kit (#A410231), according to the manufacturer’s protocols. Briefly, cells were detached from 1-2 10 cm plates using 0.05% Trypsin (Corning #25-052-CI), washed and resuspended in PBS, then crosslinked with 2% final concentration of formaldehyde (Fisher #F79-500). After adding Stop Solution, samples were washed, pelleted, and flash frozen before processing for Hi-C. Approximately 1-2 million cells were used per experiment. Libraries were made using a modified NEB Ultra II DNA Library Prep Kit (#E7645S) and sent to Admera (LLC) for sequencing. Samples were sequenced 2 × 150 bp on an Illumina NovaSeq X Plus.
Hi-C Data processing
Samples were processed using the Nextflow nfcore-hic (v2.1.0) pipeline with a few modifications. Briefly, HiCPro was used to align reads to the concatenated hg38 (GCA_000001405.15_GRCh38) and TB40E (MW439039) assemblies of the human and HCMV genomes, respectively. Raw and normalized matrices were generated at several resolutions, duplicates were removed, mapping statistics were generated, and, separately, replicate fastqs were concatenated and processed to generate combined datasets. Genome-wide interaction maps were visualized using normalized mcool files in HiGlass (higlass-docker:v0.6.1) at a genome-wide scale39. For visualization per chromosome, HiCPro matrices were converted to.hic files with hicpro2juicer.sh and loaded into Juicebox (v2.17.00) with balanced normalization70. Using the Python implementation of HiCRep (v0.2.6)71, stratum-adjusted correlation coefficient (SCC) scores were calculated for all pairs of 100 kb HiC datasets and visualized with heatmaps.
Viral-host Interactions and VERs
Using HiCPro’s makeviewpoints.py function, interactions between viral DNA and host DNA were extracted. Briefly, fragment delimited maps of both host and viral genomes were generated based on the Arima enzyme combination, and TB40E regions were used as trans-baits for makeviewpoints.py. Interactions from the combined viral Hi-C data were then merged using bedtools merge -d 100 and converted to bigwig. Viral-enriched regions (VERs) were determined by mapping viral reads to a 100-kb binned genome and taking the top 10% of interacting bins.
dcHiC Differential compartments
The 100 kb replicate matrices and files generated by nfcore-hic’s HiCPro were used as input to the dcHiC (v2.1) pipeline42. dcHiC was used to call differential compartments and identify subcompartment changes between Mock and Infected samples at 100 kb resolution. Because of the effect infection had on global chromatin conformation (including loss of interactions between chromosome arms), p and q arms were processed separately, assigned PCA scores accordingly, and subsequently combined. Subcompartment number was set to 6, the default.
TAD Identification and analysis
TAD boundary calls from HiCExplorer were generated from the Nextflow pipeline at 40 kb and 20 kb and visualized with examples of genes in IGV. To better identify TAD reorganization during infection, TADs were first called by a compatible approach, Arrowhead (juicer v2.17.00)37, using combined matrices at 10 kb and subsequently used as input for diffDomain (py3)43. Reorganized TADs were identified, filtered (BH < 0.05), and classified into strength-change, loss, split, merge, zoom, and complex subtypes using default parameters.
GENOVA
Combined balanced cool were used for downstream GENOVA analysis in R with parameters, scale_bp = 1e9 and scale_cis = TRUE unless otherwise indicated. Relative contact probability maps (RCP) were calculated genome-wide, excluding chrM and chrY, at 100 kb resolution. Trans-contact maps between host chromosomes and the viral TB40E genome were generated at 250 kb using z-score normalized matrices. Insulation scores generated using window sizes of 25 and 10 kb matrices were used in identifying TAD boundaries and calling TADs, which were then visualized via tornado insulation plots.
External data peak analysis
Open (gained during infection) and closed (lost during infection) peaks for ATAC, H3K27ac, TEAD1, and CTCF identified from Sayeed et al., 202427. ChIPseq data were downloaded from their supplement datasets and saved as bed files, then converted from hg19 to hg38 genome coordinates using CrossMap72.
Deeptools enrichment
To determine enrichment on a more global scale, Deeptools (v3.5.1) computematrix scaleregions function was used to scale regions of interest to the same size for both compartment and gene-level analysis46. Average signal over regions was visualized using Deeptools plotProfile. Average signal from two replicate CUT&RUN experiments for H3K9me3 and viral enrichment extracted from combined, infected Hi-C data (n = 4) were plotted over compartments called by dcHiC. Transcriptional coverage tracks were generated from TTseq or RNAseq data BAM files, normalized using RPGC, and plotted over different identified gene sets.
CUT&RUN Library preparation
Samples were processed using Epicypher’s CUT&RUN Kit (#14-1048) following the manufacturers protocol. Briefly, cells were harvested by trypsinization (0.05% Corning #25-052-CI), washed once with PBS and resuspended in Wash Buffer to bind to ConA Beads. Once bound to beads and immobilized, cells were permeabilized with 0.01% digitonin and incubated with a target-specific antibody. Targets for CUT&RUN were H3K4me3 (CST #9751) and H3K9me3 (CST #4658), as well as kit-provided IgG controls. pAG-MNase was added to cleave chromatin fragments, and the resulting DNA purified for library prep with the NEB Ultra II DNA Library Prep Kit. Spike-in of E.Coli DNA was included for downstream normalization. Samples were sequenced 2 × 150 bp on an Illumina NovaSeq 6000.
CUT&RUN Data processing
Samples were processed using the Nextflow nfcore-cutrun pipeline (v3.2.2) with default parameters: --bowtie2 hg38 + TB40E --normalisation_mode Spikein, --use_control true, --igg_scale_factor 0.5, --spikein_genome K12-MG1655. Replicates were combined using Deeptools bigwigAverage function.
TTseq Library preparation
TTseq was performed as described previously35. Briefly, one hour before collection at each infection timepoint, media was removed from cells and replaced with media containing 500 μM 4-thiouridine (Sigma-Aldrich #T4509) prewarmed to 37 °C. Cells were incubated for 60 minutes at 37 °C with 5% CO2. The media was then removed, and TRIzol reagent (Thermo Fisher #15596018) was immediately added to lyse the cells and extract total RNA. Sample concentration was determined by Qubit 3.0 fluorometer and Qubit RNA BR Assay Kit (Invitrogen #Q10210), and RNA integrity was confirmed by Qubit RNA Integrity Assay (Invitrogen #Q33221).
RNA was fragmented using NEBNext RNA Fragmentation Buffer (New England Biolabs #E6150) and cleaned up by RNAClean XP beads (Beckman Colter #A63987). RNA was then denatured and biotinylated by incubating with MTSEA-biotin-XX (Biotium #90066). Dynabeads MyOne Streptavidin C1 beads (Life Technologies #65001) were added and used to enrich for biotinylated products. After several bead washes, 4sU-enriched RNA was eluted, and rRNA was depleted from the sample using a NEBNext rRNA Depletion Kit v2 (New England Biolabs #E7400), and concentration was determined with a Qubit RNA HS Assay. DNA libraries (from reverse transcribed RNA) were prepared for high-throughput sequencing following chapter 5 in the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs #E7760) with minor modifications to account for small insert sizes. Final sample concentration was determined with a Qubit dsDNA HS Assay and sent to Admera for QC and sequencing. Samples were sequenced 2 × 150 bp on an Illumina NovaSeq 6000.
RNAseq Library preparation
RNA was isolated from TTseq samples prior to biotin enrichment of nascent transcripts. RNAseq libraries were prepared using an NEBNext rRNA Depletion Kit v2 to deplete rRNA and the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs #E7760). Samples were sequenced at 2 × 150 bp on an Illumina NovaSeq 6000.
TTseq/RNAseq Alignment and differential gene expression analysis
Reads were trimmed for adapter sequences using trimmomatic (v0.36) and then aligned to concatenated hg38 (GCA_000001405.15_GRCh38) and TB40E (MW439039) assemblies of the human and HCMV genomes, respectively, using STAR(v2.7.5c). Genes were counted using htseq-count (v2.0.3)73. Expression levels and differentially expressed genes were determined using DESeq2 (v1.46.0)45, excluding genes with less than 4 counts between all samples. For coverage tracks, Deeptools bamCoverage was used on BAM files with the parameter --normalizeUsing RPGC and resulting bigwigs for replicates were combined using Deeptools bigwigAverage function.
Gene enrichment analysis and visualization
Gene ontology (GO) enrichment was performed with clusterProfiler (v4.14.0) or ShinyGO (v0.81)74 using default parameters and matched background gene lists. In clusterProfiler, settings were pool=TRUE, ont = “ALL”, and the simplify function was used to generate dotplots. Volcano plots were generated with EnhancedVolcano (v1.24.0) with parameters pCutoff = 0.05, FCcutoff = 2.0. UpSet plots were generated with ComplexHeatmap (v2.22.0).
ARTDeco Analysis
Transcriptional read-through was measured using ARTDeco (v0.4) with default parameters for stranded libraries47. Briefly, a GTF file for gene read-in/through identification and aligned BAM files were used as input. Read-in genes were classified as previously described using the parameter of having a log2 fold change > 2, adjusted p-value < 0.05, and read-in rate > − 2, readthrough genes met the same parameters with readthrough rate > − 2. Relationships between upstream and downstream genes were determined using custom python code, briefly, genes with adjusted p-value < 0.05 were matched to annotations and paired with their nearest non-overlapping downstream neighbor within 10 kb. Pearson correlations were computed between upstream RT and downstream RI or log2FC, as well as within-gene RI, RT, and expression.
Illustrations
Illustrations were generated under license using Biorender at Biorender.com.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Elizabeth Bartom for generously providing guidance on bioinformatics and Eain Murphy for reagents and advice. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under grant number R01AI141470 to D.W. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This manuscript is the result of funding in whole or in part by the National Institutes of Health (NIH). It is subject to the NIH Public Access Policy. Through acceptance of this federal funding, NIH has been given the right to make this manuscript publicly available in PubMed Central upon the Official Date of Publication, as defined by NIH. Additional funding was provided by the National Science Foundation to C.R. (DGE-2234667) and the Northwestern Sequencing (NU-seq) Core (2022 Illumina Pilot Project Program) to D.W.
Author contributions
Experimental procedures, sample preparation and data analysis were performed by C.R. C.R. and D.W. analyzed results, prepared figures and wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Lars Dolken and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its supplementary files. Sequencing data is deposited to the Gene Expression Omnibus (GEO) repository under the following accession codes; TTseq Datasets - GSE284780 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE284780. RNAseq Datasets - GSE284781. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE284781. CUT&RUN Datasets - GSE284782 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE284782. HiC Datasets - GSE284783 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE284783 Source data are provided in this paper.
Code availability
Public nfcore pipelines were used for HiC and CUT&RUN analysis as detailed in the methods. No custom code was used in this manuscript. Standard code for TTseq, RNAseq, GENOVA and related analysis is deposited on Github at https://github.com/crosencr12/walsh2024 or 10.5281/zenodo.16237191.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-025-62921-5.
References
- 1.Rowley, M. J. & Corces, V. G. Organizational principles of 3D genome architecture. Nat. Rev. Genet.19, 789–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Steensel, B. & Belmont, A. S. Lamina-associated domains: Links with chromosome architecture, heterochromatin, and gene repression. Cell169, 780–791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke, B. & Stewart, C. L. The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol.14, 13–24 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Dupont, S. & Wickstrom, S. A. Mechanical regulation of chromatin and transcription. Nat. Rev. Genet.23, 624–643 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Lele, T. P., Dickinson, R. B. & Gundersen, G. G. Mechanical principles of nuclear shaping and positioning. J. Cell Biol.217, 3330–3342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartwright, S. & Karakesisoglou, I. Nesprins in health and disease. Semin Cell Dev. Biol.29, 169–179 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Sanchez, V. & Britt, W. Human cytomegalovirus egress: Overcoming barriers and Co-opting cellular functions. Viruses 14, 10.3390/v14010015 (2021). [DOI] [PMC free article] [PubMed]
- 8.Alwine, J. C. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog.8, e1002878 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews, S. M., Groves, I. J. & O’Connor, C. M. Chromatin control of human cytomegalovirus infection. mBio14, e0032623 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marti-Carreras, J. & Maes, P. Human cytomegalovirus genomics and transcriptomics through the lens of next-generation sequencing: revision and future challenges. Virus Genes55, 138–164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spector, B. M. et al. Differences in RNA polymerase II complexes and their interactions with surrounding chromatin on human and cytomegalovirus genomes. Nat. Commun.13, 2006 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nitzsche, A., Paulus, C. & Nevels, M. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J. Virol.82, 11167–11180 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuevas-Bennett, C. & Shenk, T. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J. Virol.82, 9525–9536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groves, I. J., Reeves, M. B. & Sinclair, J. H. Lytic infection of permissive cells with human cytomegalovirus is regulated by an intrinsic ‘pre-immediate-early’ repression of viral gene expression mediated by histone post-translational modification. J. Gen. Virol.90, 2364–2374 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Zalckvar, E. et al. Nucleosome maps of the human cytomegalovirus genome reveal a temporal switch in chromatin organization linked to a major IE protein. Proc. Natl. Acad. Sci. USA110, 13126–13131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parida, M. et al. Nucleotide resolution comparison of transcription of human cytomegalovirus and host genomes reveals universal use of RNA polymerase II elongation control driven by dissimilar core promoter elements. mBio 10, 10.1128/mBio.02047-18 (2019). [DOI] [PMC free article] [PubMed]
- 17.Kent, J. R. et al. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol.78, 10178–10186 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forte, E. et al. Critical role for the human cytomegalovirus major immediate early proteins in recruitment of RNA polymerase II and H3K27Ac to an enhancer-like element in OriLyt. Microbiol. Spectr.11, e0314422 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter, R. M., Majumder, K. & Kalejta, R. F. ATRX restricts Human Cytomegalovirus (HCMV) viral DNA replication through heterochromatinization and minimizes unpackaged viral genomes. PLoS Pathog.20, e1012516 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forte, E. et al. Epigenetic reprogramming of host and viral genes by Human Cytomegalovirus infection in Kasumi-3 myeloid progenitor cells at early times post-infection. J. Virol. 95, 10.1128/JVI.00183-21 (2021). [DOI] [PMC free article] [PubMed]
- 21.Friedman, M. J., Lee, H., Kwon, Y. C. & Oh, S. Dynamics of viral and host 3D genome structure upon infection. J. Microbiol. Biotechnol.32, 1515–1526 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hertel, L. & Mocarski, E. S. Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of Pseudomitosis independent of US28 function. J. Virol.78, 11988–12011 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tirosh, O. et al. The transcription and translation landscapes during human cytomegalovirus infection reveal novel host-pathogen interactions. PLoS Pathog.11, e1005288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu, H., Cong, J. P., Mamtora, G., Gingeras, T. & Shenk, T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA95, 14470–14475 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Browne, E. P., Wing, B., Coleman, D. & Shenk, T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol.75, 12319–12330 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ball, C. B. et al. Human cytomegalovirus infection elicits global changes in host transcription by RNA polymerases I, II, and III. Viruses 14, 10.3390/v14040779 (2022). [DOI] [PMC free article] [PubMed]
- 27.Sayeed, K. et al. Human cytomegalovirus infection coopts chromatin organization to diminish TEAD1 transcription factor activity. ELife13, RP101578 (2024). [Google Scholar]
- 28.Ball, C. B. et al. Human cytomegalovirus IE2 both activates and represses initiation and modulates elongation in a context-dependent manner. mBio13, e0033722 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, X. et al. Human virus transcriptional regulators. Cell182, 24–37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulus, C. & Nevels, M. The human cytomegalovirus major immediate-early proteins as antagonists of intrinsic and innate antiviral host responses. Viruses1, 760–779 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelnhofer-Millevolte, L. E. et al. Human cytomegalovirus induces neuronal gene expression through IE1 for viral maturation. Nat. Commun. 16, 7316 (2025). [DOI] [PMC free article] [PubMed]
- 32.Procter, D. J., Furey, C., Garza-Gongora, A. G., Kosak, S. T. & Walsh, D. Cytoplasmic control of intranuclear polarity by human cytomegalovirus. Nature587, 109–114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Procter, D. J. et al. The HCMV assembly compartment is a dynamic Golgi-derived MTOC that controls nuclear rotation and virus spread. Dev. Cell45, 83–100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge, T., Rosencrance, C. D. & Eagen, K. P. Contact mapping to unravel chromosome folding. Trends Biochem. Sci.44, 1089–1090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosencrance, C. D. et al. Chromatin hyperacetylation impacts chromosome folding by forming a nuclear subcompartment. Mol. Cell78, 112–126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh, D., Perez, C., Notary, J. & Mohr, I. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol.79, 8057–8064 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerpedjiev, P. et al. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol.19, 125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karamitros, T., van Wilgenburg, B., Wills, M., Klenerman, P. & Magiorkinis, G. Nanopore sequencing and full genome de novo assembly of human cytomegalovirus TB40/E reveals clonal diversity and structural variations. BMC Genom.19, 577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Weide, R. H. et al. Hi-C analyses with GENOVA: a case study with cohesin variants. NAR Genom. Bioinform.3, lqab040 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty, A., Wang, J. G. & Ay, F. dcHiC detects differential compartments across multiple Hi-C datasets. Nat. Commun.13, 6827 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hua, D. et al. DiffDomain enables identification of structurally reorganized topologically associating domains. Nat. Commun.15, 502 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwalb, B. et al. TT-seq maps the human transient transcriptome. Science352, 1225–1228 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res.42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth, S. J., Heinz, S. & Benner, C. ARTDeco: automatic readthrough transcription detection. BMC Bioinform.21, 214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hofstadter, W. A., Park, J. W., Lum, K. K., Chen, S. & Cristea, I. M. HCMV strain- and cell type-specific alterations in membrane contact sites point to the convergent regulation of organelle remodeling. J. Virol.98, e0109924 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong, Q. et al. Human cytomegalovirus inhibits the proliferation and invasion of extravillous cytotrophoblasts via Hippo-YAP pathway. Virol. J.18, 214 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angelova, M. et al. Human cytomegalovirus infection dysregulates the canonical Wnt/beta-catenin signaling pathway. PLoS Pathog.8, e1002959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, C. et al. A DNA tumor virus globally reprograms host 3D genome architecture to achieve immortal growth. Nat. Commun.14, 1598 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.SoRelle, E. D. et al. Epstein-Barr virus evades restrictive host chromatin closure by subverting B cell activation and germinal center regulatory loci. Cell Rep.42, 112958 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hennig, T. et al. HSV-1-induced disruption of transcription termination resembles a cellular stress response but selectively increases chromatin accessibility downstream of genes. PLoS Pathog.14, e1006954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caragliano, E. et al. Human cytomegalovirus forms phase-separated compartments at viral genomes to facilitate viral replication. Cell Rep.38, 110469 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McSwiggen, D. T. et al. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife 8, 10.7554/eLife.47098 (2019). [DOI] [PMC free article] [PubMed]
- 56.Kobiler, O. & Weitzman, M. D. Herpes simplex virus replication compartments: From naked release to recombining together. PLoS Pathog.15, e1007714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mahmutefendic Lucin, H. et al. Membraneless compartmentalization of nuclear assembly sites during murine cytomegalovirus infection. Viruses 15, 766 (2023). [DOI] [PMC free article] [PubMed]
- 58.Caragliano, E., Brune, W. & Bosse, J. B. Herpesvirus replication compartments: Dynamic biomolecular condensates? Viruses 14, 960 (2022). [DOI] [PMC free article] [PubMed]
- 59.Moquin, S. A. et al. The Epstein-Barr virus episome maneuvers between nuclear chromatin compartments during reactivation. J. Virol. 92, 10.1128/JVI.01413-17 (2018). [DOI] [PMC free article] [PubMed]
- 60.Myllys, M. et al. Herpes simplex virus 1 induces egress channels through marginalized host chromatin. Sci. Rep.6, 28844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aho, V. et al. Chromatin organization regulates viral egress dynamics. Sci. Rep.7, 3692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang, L. et al. Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proc. Natl. Acad. Sci. USA108, E136–E144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor, T. J., McNamee, E. E., Day, C. & Knipe, D. M. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology309, 232–247 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Monier, K., Armas, J. C., Etteldorf, S., Ghazal, P. & Sullivan, K. F. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol.2, 661–665 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Rosemarie, Q., Kirschstein, E. & Sugden, B. How Epstein-Barr virus induces the reorganization of cellular chromatin. mBio14, e0268622 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crabbe, L., Cesare, A. J., Kasuboski, J. M., Fitzpatrick, J. A. & Karlseder, J. Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell Rep.2, 1521–1529 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding, X. et al. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell12, 863–872 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Bhargava, A. et al. Inhibition of HIV infection by structural proteins of the inner nuclear membrane is associated with reduced chromatin dynamics. Cell Rep.36, 109763 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Chi, Y. H., Haller, K., Peloponese, J. M. Jr & Jeang, K. T. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J. Biol. Chem.282, 27447–27458 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst.3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, T. et al. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res.27, 1939–1949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao, H. et al. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics30, 1006–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anders, S., Pyl, P. T. & Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ge, S. X., Jung, D. & Yao, R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics36, 2628–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article and its supplementary files. Sequencing data is deposited to the Gene Expression Omnibus (GEO) repository under the following accession codes; TTseq Datasets - GSE284780 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE284780. RNAseq Datasets - GSE284781. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE284781. CUT&RUN Datasets - GSE284782 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE284782. HiC Datasets - GSE284783 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE284783 Source data are provided in this paper.
Public nfcore pipelines were used for HiC and CUT&RUN analysis as detailed in the methods. No custom code was used in this manuscript. Standard code for TTseq, RNAseq, GENOVA and related analysis is deposited on Github at https://github.com/crosencr12/walsh2024 or 10.5281/zenodo.16237191.