Abstract
Historically, poor data management has hampered the establishment and operation of wildlife health surveillance (WHS) systems and limited the integration of environmental data into One Health frameworks. Effective WHS purpose-built databases are key to solve this problem, yet the few options available remain inaccessible or narrow in scope.
To address this gap, an international partnership is developing the Health and Wildlife Knowledge (HAWK) database. HAWK supports the management of diverse data generated by multiple actors and methodologies, all within a harmonized structure and vocabulary facilitating data access, analysis, communication, and reuse. Data are secured through compartmentalization across organizations and users, while supporting compliance of FAIR and CARE data principles.
Slated for release in late 2025, HAWK is envisioned as a global public good to encourage data compatibility and best practices in the wildlife conservation and One Health communities, independent of languages and location, with minimal to no cost for users.
Keywords: Wildlife health, Surveillance, Wildlife health surveillance, Database, Data, Data management, Wildlife, Disease, Pathogen, Toxin
1. Wildlife health surveillance and data
Wildlife health surveillance (WHS) is critical for addressing health hazards that threaten wildlife and undermine One Health [1]. It involves a comprehensive set of activities aimed at continuously and rapidly generating and analyzing information on the health of wildlife and associated hazards [2,3] to prevent, mitigate, control, or eliminate exposure to these hazards and support population resilience [2,3].
WHS systems rely on diverse data, including information on environmental features, animal observations, specimens collected, necropsy findings, and diagnostic test results, generated by many actors through various methodologies targeting different hazards, vectors, and hosts [4,5]. These diverse data need to be managed properly so they remain safe, secured, centralized, available, standardized, and comprehensible in the short- and long-term. In this manner, WHS data can be rapidly assessed, communicated, and responded to utilizing an evidence-based decision making process [[6], [7], [8], [9], [10]].
2. Current issues with wildlife health surveillance data management
Despite its relevance, WHS data management is a historical weakness [5,[10], [11], [12]] that has impacted the response to health crises and the establishment of One Health surveillance systems [8,9,13]. Furthermore, as ∼50 % of countries report no WHS [12,14,15], collaborative networks among key in-country partners can be developed to expand its coverage [7]. Efficient data management is key to ensure their operation as a functionally cohesive unit [7].
3. Proposing solutions
Well-designed databases promote and facilitate data safety, security, centralization, harmonization, quality, availability, communication, and sharing. Moreover, purpose-built databases are key for improving data governance and the implementation and operation of effective WHS systems.
While a few databases exist, they are private, costly, restricted to specific agencies, or limited in scope. WHS is generally underfunded, limiting the development and maintenance of system-specific databases. In line with this reality, the World Organization for Animal Health (WOAH) recently reported that only 61 % of surveyed countries maintain wildlife health data in a “sustainable and reliable information system” [12] and most WOAH national focal points for wildlife recently expressed the need for a centralized wildlife health national database [15].
4. The HAWK database
To address this gap, Wildlife Conservation Society (WCS) is coordinating the Wildlife Health Intelligence Network (WHIN), a broad community of practice supporting WHS implementation from local to global scales (https://impacthealth.azurewebsites.net/About-WHIN), to develop the WHS purpose-built and accessible web-based Health and Wildlife Knowledge (HAWK) database. Rather than serving as another data repository, HAWK functions as a data service that allows users to properly manage WHS data they generate or collate, applying self-defined methodologies to achieve objectives such as pathogen detection or establishing disease freedom.
HAWK structures WHS data following a main hierarchy, capturing relevant epidemiological information at each step (Fig. 1 and https://dmontecino.github.io/WH_Database/). It includes Projects that identify a specific leader under which Surveillance Activities, sets of surveillance goals and methodology documented following extensive metadata, are pursued. Surveillance Activities are conducted through Field Visits that encompass a time period during which Locations (e.g., study areas) are surveyed. These Locations link to Events, point epidemiological units with spatiotemporal coordinates. Events can be further grouped into areas or tracks placed within Locations (e.g., a sector within a Protected Area or a ranger patrol track; Collection Areas or Collection Tracks). Events can contain four types of Sources: groups of animals of the same species (Group) and individual animals (Animal) that can be observed, examined and sampled (Animal can be linked to Necropsy); arthropod capture sites (Arthropod); and environmental sampling sites (e.g., ponds or feces in the field; Environmental). Material (Specimens) or observations from Sources are obtained cross-sectionally or longitudinally (Source Records) through an effort (e.g., trap hours; Collection Points) or opportunistically. These main units can be grouped within further temporal and spatial nested and non-nested clusters when needed, based on the requirements of a study design. Groups, Animals, and Specimens can be tested using Diagnostics for biological, physical, chemical, and physiological targets. Diagnostics can be conducted in a Laboratory and they contain one or more Results. Tested Specimens and Source Records can receive an Interpretation following documented case definitions. The data model also supports the administration of Specimens or Animal and Group Sources storage and shipments.
Fig. 1.
Basic structure of the Wildlife Health Intelligence Network data model.
The engagement of a community of practice (WHIN) throughout the design resulted in a data model that incorporates real-world WHS data generation and management experiences. For example, HAWK contains elements of the Wildlife Health Intelligence Platform [16], a 30+ year-old database used by the Canadian Wildlife Health Cooperative to manage wildlife health information from across Canada, and VectorSurv [17], a data system used by more than 380 agencies in 36 states and territories of the United States of America to manage vector-borne disease surveillance information (https://vectorsurv.org/).
HAWK employs a modular approach to the data model, enabling components to be added based on the specific WHS needs. When data comes from opportunistic Source observations (e.g. observed-only dead animals) then Collections, Necropsy, Specimens, Diagnostics, and Results are not enabled (Fig. 1).
The terminology employed (e.g., hazards, anatomical parts) follows controlled vocabularies (e.g., Agrovoc, National Agricultural Library Thesaurus) [[18], [19], [20]] advancing data harmonization, comparability over time and space, and comprehension.
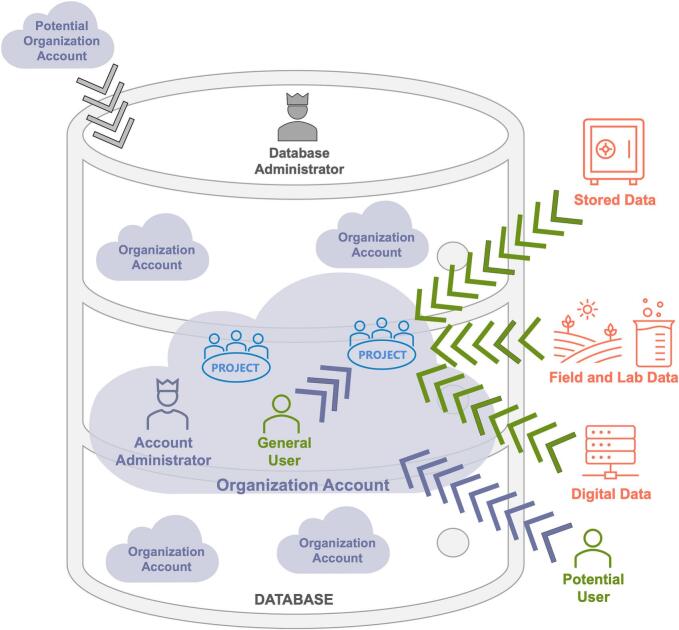
Data safety, security, and ownership are safeguarded through strictly private organization accounts. Organizations have account-specific users with distinct permission levels as determined by organization account administrators, and two-factor authentication access (Fig. 2).
Fig. 2.
Conceptual representation of the accounts, users, projects, and data within and outside the database and their potential integration into the database for those elements not yet created or included. The database, the database administrator, and the authorizations granted by the database administrator, are shown in grey. The potential and accepted organization accounts, account administrators, and the authorizations granted by the account administrator (e.g., new users in the organization account and their assignment to specific Projects) are shown in purple. To-be-accepted general users and accepted general users and the addition of data to a Project by them is shown in green. Projects belonging to an organization account are shown in blue and potential data sources for a Project are shown in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Requested data and metadata in HAWK match or exceed those required by DataCite Metadata Schema, standards for reporting arthropod studies and wildlife health data, and recommendations to record mortality events [[21], [22], [23], [24]]. In this manner, HAWK simplifies the comprehension of data generation methods and supports more detailed assessment of diseases impacts, associations, and trends.
HAWK, currently in North American English, can be easily adapted to local languages and cultural conventions (e.g., commas vs. periods for decimals) through its localization functionality with DeepL integration and optional manual translation entry. Moreover, a forthcoming API will allow interoperability with other systems for data collection (e.g., mobile applications), storage (e.g., online repositories), visualization (e.g., dashboard applications), analysis (e.g., AI and machine learning providers), and external reporting, fostering its global use.
5. Why use HAWK?
HAWK is optimized to meet the needs of public agencies, private organizations, academia, and other interested actors conducting WHS or related monitoring of different hosts, hazards, and diseases, using variable methods, effectively managing diverse data. For example, an organization can structure and centralize data from mortality investigations, opportunistic findings, tested samples from handled or harvested animals, community-based or systematic monitoring (e.g., mortality recorded via iNaturalist, ranger patrols), collated news articles, and research projects using advanced designs (Fig. 2).
HAWK's data structure is compatible with repositories such as the Global Biodiversity Information Facility (GBIF) and WOAH's World Animal Health Information System (WAHIS). Therefore, data in these repositories considered relevant for WHS can be brought into a HAWK account (for a Surveillance Activity), whilst data in a HAWK account relevant for global-level wildlife health reporting (e.g., WAHIS) or biodiversity monitoring (e.g., GBIF) could be shared with these platforms. WOAH's early engagement with HAWK can enhance collaboration and synergies to support interoperability with future WOAH data systems and could help improve current limitations in global wildlife health reporting [13,25]. The upcoming API will streamline interoperability and expand it to include WHS-relevant information collected or stored in widely distributed tools (e.g., Spatial Monitoring and Reporting Tool, District Health Information Software 2), allowing stakeholders to continue using their primary platforms. By structuring, harmonizing, centralizing, and securing a broader range of WHS information, including data that typically remain scattered, HAWK supports better decision-making within and across organizations, administrative units, or study areas (Fig. 2).
By facilitating WHS data safety, centralization, accessibility, and interoperability, plus its detailed metadata, HAWK supports FAIR and CARE compliance [26,27] (Supplementary Material Table 1 and 2). Balance between data protection and availability is achieved by establishing Surveillance Activity data ownership and by organizations agreeing to comply with self-selected Project data embargoes, ranging from immediate data availability to obligatory long-term release under open license, except in the case of Indigenous-sourced data which may remain confidential.
HAWK is powered by WCS, a 130-year old independent nonprofit conservation organization with global presence and a longstanding mission that relies on worldwide distributed funding from public and private sources (https://www.wcs.org/about-us/literature/annual-reports). Through WCS institutional support, HAWK is being developed for long-term sustainability, with data safety and security independent of government regulations, political contexts, or national priorities. If needed, HAWK and its contents can be deployed in any country with Microsoft web services, while remaining under WCS hosting and oversight.
HAWK is a ready-to-adopt global public good, available for any organization in any country. It is meant to encourage best data practices whose costs can be shared across many parties with funds allocated for data management and storage, alleviating the burden on isolated and resource-limited stakeholders. Critically, when local funding is unavailable, Global South users and Indigenous people will have access to HAWK at no charge, within reasonable usage limits. This approach, together with multilanguage user interfaces, interoperability, and the worldwide reach of WCS and WHIN will foster a global WHS network.
Many funding agencies mandate data management plans but long-term safety and accessibility are not universally requested yet. It is foreseeable that private funders, as well as taxpayers who indirectly support WHS activities conducted by public agencies and through publicly funded research, could soon demand to safeguard the data output of these investments. We argue that HAWK will be essential to comply with these potential requests. In the meantime, HAWK's adoption offers the facilitation of WHS implementation and operation.
6. Conclusion
While numerous barriers to WHS remain (e.g., expertise, siloed actors, funding, and unclear policy frameworks), HAWK, slated for release in late 2025, aims to fill a historical gap in the availability of a sustainable purpose-built database to support the adequate management of WHS data. We invite the WHS and One Health communities to lay the foundation of their data practices on this database and contribute to its iterative improvement.
CRediT authorship contribution statement
Diego Montecino-Latorre: Writing – review & editing, Writing – original draft, Visualization, Supervision, Software, Resources, Project administration, Methodology, Conceptualization. Mathieu Pruvot: Writing – review & editing, Writing – original draft, Supervision, Methodology, Funding acquisition, Conceptualization. Paloma H.F. Shimabukuro: Writing – review & editing, Validation, Resources, Conceptualization. Christopher M. Barker: Writing – review & editing, Validation, Resources, Methodology, Conceptualization. Steve Gallo: Writing – review & editing, Software, Conceptualization. Jonathan Palmer: Writing – review & editing, Supervision, Methodology, Conceptualization. Susan J. Kutz: Writing – review & editing, Validation, Conceptualization. Claire Cayol: Writing – review & editing, Validation, Conceptualization. Fernanda Dórea: Writing – review & editing, Validation. Liz P. Noguera: Writing – review & editing, Validation. Damien O. Joly: Writing – review & editing, Validation. Chris Walzer: Writing – review & editing, Supervision. Lucy Keatts: Writing – review & editing, Validation. Amanda E. Fine: Writing – review & editing, Validation. Sarah H. Olson: Writing – original draft, Validation, Supervision, Funding acquisition, Conceptualization.
Funding
The work by the Wildlife Health Intelligence Network has been supported by Science for Nature and People Partnership [SNAPP055]. The development of the WildHealth Database is currently supported by the IUCN World Commission on Protected Areas' Task Force on Protected Areas and One Health through a grant from the Gordon and Betty Moore Foundation [SMA-G00-00990] and by the Science for Nature and People Partnership [SNAPP055]. This work was supported by the National Science Foundation grant 2412446 and some funding for this project was provided by the United States Department of Agriculture (USDA) - Animal and Plant Health Inspection Service. The findings and conclusions in this publication are those of the author(s) and should not be construed to represent any official USDA or United States Government determination or policy.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Our thanks to the Gordon and Betty Moore Foundation, the IUCN World Commission on Protected Areas, and the Science for Nature and People Partnership, for supporting funding to develop WHIN and/or the HAWK database web-application.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2025.101227.
Appendix A. Supplementary data
Supplementary material
Data availability
No data was used for the research described in the article.
References
- 1.World Organisation for Animal Health, International Union Conservation of Nature . 2024. General Guidelines for Surveillance of Diseases, Pathogens and Toxic Agents in Free-ranging Wildlife: An Overview for Wildlife Authorities and Others Working with Wildlife. [Google Scholar]
- 2.Artois M., Bengis R., Delahay R.J., Duchêne M.-J., Paul Duff J., Ferroglio E., Gortazar C., Hutchings M.R., Kock R.A., Leighton F.A., Mörner T., Smith G.C. Management of Disease in Wild Mammals. Springer; Tokyo: 2009. Wildlife disease surveillance and monitoring; pp. 187–213. [Google Scholar]
- 3.Stephen C., Berezowski J. In: Wildlife Population Health. Stephen C., editor. Springer International Publishing; Cham: 2022. Wildlife health surveillance and intelligence. Challenges and opportunities; pp. 99–111. [Google Scholar]
- 4.Stallknecht D.E. In: Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission. Childs J.E., Mackenzie J.S., Richt J.A., editors. Springer; Berlin, Heidelberg: 2007. Impediments to wildlife disease surveillance, research, and diagnostics; pp. 445–461. [Google Scholar]
- 5.Sleeman J.M., Brand C.J., Wright S.D. In: New Directions in Conservation Medicine: Applied Cases in Ecological Health. Aguirre A.A., Ostfield R.S., Daszak P., editors. Oxford University Press; 2012. Strategies for wildlife disease surveillance; pp. 539–551. [Google Scholar]
- 6.Stephen C., Sleeman J., Nguyen N., Zimmer P., Duff J.P., Gavier-Widén D., Grillo T., Lee H., Rijks J.M., Ryser-Degiorgis M.P., Tana T., Uhart M. Proposed attributes of national wildlife health programmes. Rev. Sci. Tech. 2018;37 doi: 10.20506/37.3.2896. [DOI] [PubMed] [Google Scholar]
- 7.Pruvot M., Denstedt E., Latinne A., Porco A., Montecino-Latorre D., Khammavong K., Milavong P., Phouangsouvanh S., Sisavanh M., Nga N.T.T., Ngoc P.T.B., Thanh V.D., Chea S., Sours S., Phommachanh P., Theppangna W., Phiphakhavong S., Vanna C., Masphal K., Sothyra T., San S., Chamnan H., Long P.T., Diep N.T., Duoc V.T., Zimmer P., Brown K., Olson S.H., Fine A.E. WildHealthNet: supporting the development of sustainable wildlife health surveillance networks in Southeast Asia. Sci. Total Environ. 2023;863 doi: 10.1016/j.scitotenv.2022.160748. [DOI] [PubMed] [Google Scholar]
- 8.Hayman D.T.S., Adisasmito W.B., Almuhairi S., Behravesh C.B., Bilivogui P., Bukachi S.A., Casas N., Becerra N.C., Charron D.F., Chaudhary A., Ciacci Zanella J.R., Cunningham A.A., Dar O., Debnath N., Dungu B., Farag E., Gao G.F., Khaitsa M., Machalaba C., Mackenzie J.S., Markotter W., Mettenleiter T.C., Morand S., Smolenskiy V., Zhou L., Koopmans M. Developing one health surveillance systems. One Health. 2023;17:100617. doi: 10.1016/j.onehlt.2023.100617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmichael C. Coordinating an effective response to wildlife diseases. Wildl. Soc. Bull. 2012;36:204–206. [Google Scholar]
- 10.Heiderich E., Keller S., Pewsner M., Origgi F.C., Zürcher-Giovannini S., Borel S., Marti I., Scherrer P., Pisano S.R.R., Friker B., Adrian-Kalchhauser I., Ryser-Degiorgis M.-P. Analysis of a European general wildlife health surveillance program: chances, challenges and recommendations. PLoS One. 2025;19(5) doi: 10.1371/journal.pone.0301438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suwanpakdee S., Sangkachai N., Wiratsudakul A., Wiriyarat W., Sakcamduang W., Wongluechai P., Pabutta C., Sariya L., Korkijthamkul W., Blehert D.S., White C.L., Walsh D.P., Stephen C., Ratanakorn P., Sleeman J.M. Wildlife health capacity enhancement in Thailand through the world organisation for animal health twinning program. Front. Vet. Sci. 2024;11 doi: 10.3389/fvets.2024.1462280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Organization for Animal Health In-country Wildlife Disease Surveillance Survey Report. 2023. https://www.woah.org/app/uploads/2023/03/survey-report-wildlife-2023.pdf
- 13.Nerpel A., Käsbohrer A., Walzer C., Desvars-Larrive A. Data on SARS-CoV-2 events in animals: mind the gap! One Health. 2023;17 doi: 10.1016/j.onehlt.2023.100653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machalaba C., Uhart M., Ryser-Degiorgis M.-P., Karesh W.B. Gaps in health security related to wildlife and environment affecting pandemic prevention and preparedness, 2007-2020. Bull. World Health Organ. 2021;99:342–350B. doi: 10.2471/BLT.20.272690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Organisation of Animal Health Wildlife Health Data Management and Information Systems WOAH Survey 2023. 2023. https://app.powerbi.com/view?r=eyJrIjoiMWRjMTBkNmUtNzA5OC00ZTY4LWI3ZmItOGQ2OGI4MDliZDUzIiwidCI6ImYxZmFmNTYzLWIwNmQtNGMzNS04NzM5LTM0Y2NjMjgwZGNhZiIsImMiOjh9&pageName=ReportSectionc23ecdac85d480b9de31 (accessed February 16, 2025)
- 16.Stephen C. The Canadian wildlife health cooperative: addressing wildlife health challenges in the 21st century. Can. Vet. J. 2015;56:925–927. [PMC free article] [PubMed] [Google Scholar]
- 17.Barker C.M. Models and surveillance systems to detect and predict West Nile virus outbreaks. J. Med. Entomol. 2019;56:1508–1515. doi: 10.1093/jme/tjz150. [DOI] [PubMed] [Google Scholar]
- 18.Lipscomb C.E. Medical subject headings (MeSH) Bull. Med. Libr. Assoc. 2000;88:265–266. [PMC free article] [PubMed] [Google Scholar]
- 19.Subirats-Coll I., Kolshus K., Turbati A., Stellato A., Mietzsch E., Martini D., Zeng M. AGROVOC: the linked data concept hub for food and agriculture. Comput. Electron. Agric. 2022;196 [Google Scholar]
- 20.U.S. Department of Agriculture National Agricultural Library, National Agricultural Library Thesaurus and Glossary (NALT) 2024. https://agclass.nal.usda.gov (accessed August 28, 2025)
- 21.Schwantes C.J., Sánchez C.A., Stevens T., Zimmerman R., Albery G., Becker D.J., Brookson C.B., Kading R.C., Keiser C.N., Khandelwal S., Kramer-Schadt S., Krut-Landau R., McKee C.D., Montecino-Latorre D., O’Donoghue Z., Olson S.H., O’Shea M., Poisot T., Robertson H., Ryan S., Seifert S., Simons D., Vicente-Santos A., Wood C.L., Graeden E., Carlson C.J. A minimum data standard for wildlife disease research and surveillance. Sci Data. 2025;12 doi: 10.1038/s41597-025-05332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rund S.S.C., Braak K., Cator L., Copas K., Emrich S.J., Giraldo-Calderón G.I., Johansson M.A., Heydari N., Hobern D., Kelly S.A., Lawson D., Lord C., MacCallum R.M., Roche D.G., Ryan S.J., Schigel D., Vandegrift K., Watts M., Zaspel J.M., Pawar S. MIReAD, a minimum information standard for reporting arthropod abundance data. Sci Data. 2019;6:40. doi: 10.1038/s41597-019-0042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DataCite Metadata Working Group . DataCite e.V; 2023. DataCite Metadata Schema Documentation for the Publication and Citation of Research Data and other Research Outputs. [DOI] [Google Scholar]
- 24.Harvey J., Ramey A., Avery S., Robertson G., Romano M., Mullinax J., Boldenow M., Atkinson P., Prosser D. A practical decision tool for marine bird mortality assessments. EcoEvoRxiv. 2025 doi: 10.32942/x23s6r. [DOI] [Google Scholar]
- 25.Thompson L., Cayol C., Awada L., Muset S., Shetty D., Wang J., Tizzani P. Role of the world organisation for animal health in global wildlife disease surveillance. Front. Veterin. Sci. 2024;11 doi: 10.3389/fvets.2024.1269530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkinson M.D., Dumontier M., Aalbersberg I.J.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E., Bouwman J., Brookes A.J., Clark T., Crosas M., Dillo I., Dumon O., Edmunds S., Evelo C.T., Finkers R., Gonzalez-Beltran A., Gray A.J.G., Groth P., Goble C., Grethe J.S., Heringa J., Hooft R., Kuhn T., Kok R., Kok J., Lusher S.J., Martone M.E., Mons A., Packer A.L., Persson B., Rocca-Serra P., Roos M., van Schaik R., Sansone S.-A., Schultes E., Sengstag T., Slater T., Strawn G., Swertz M.A., Thompson M., van der Lei J., van Mulligen E., Velterop J., Waagmeester A., Wittenburg P., Wolstencroft K., Zhao J., Mons B. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3 doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carroll S.R., Garba I., Figueroa-Rodríguez O.L., Holbrook J., Lovett R., Materechera S., Parsons M., Raseroka K., Rodriguez-Lonebear D., Rowe R., et al. Open Scholarship Press Curated Volumes: Policy. 2023. The CARE principles for indigenous data governance. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
No data was used for the research described in the article.