Abstract
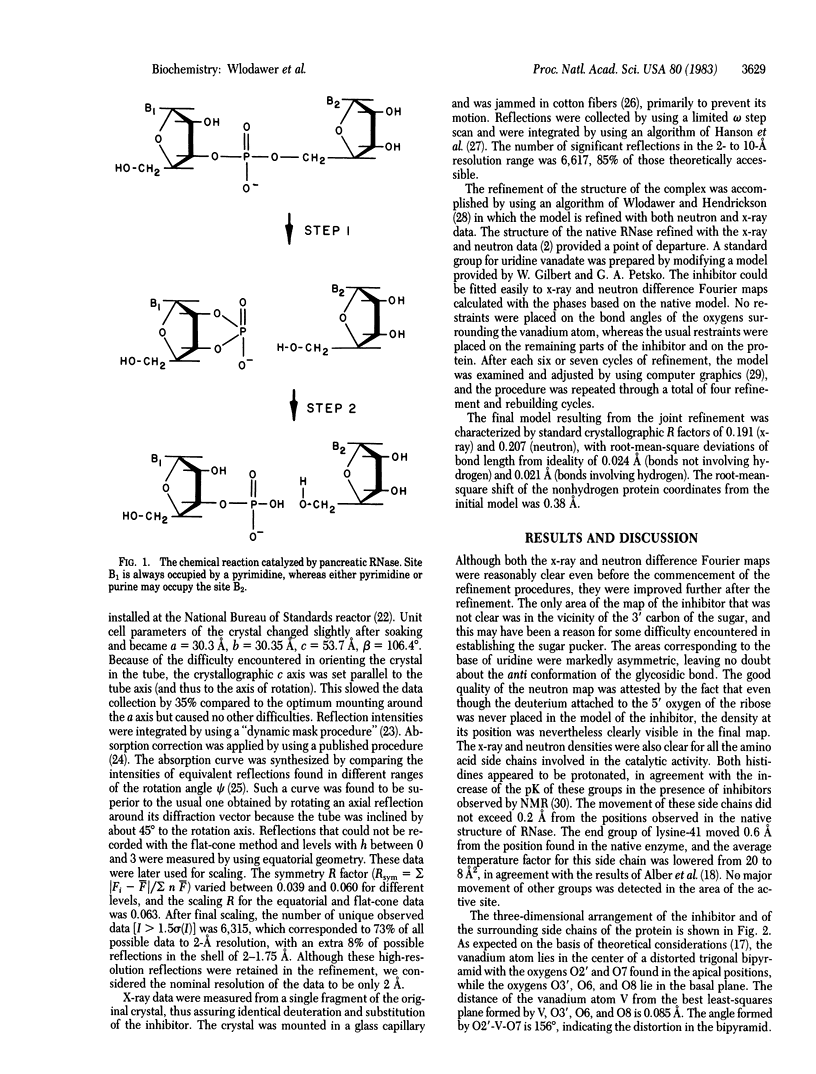
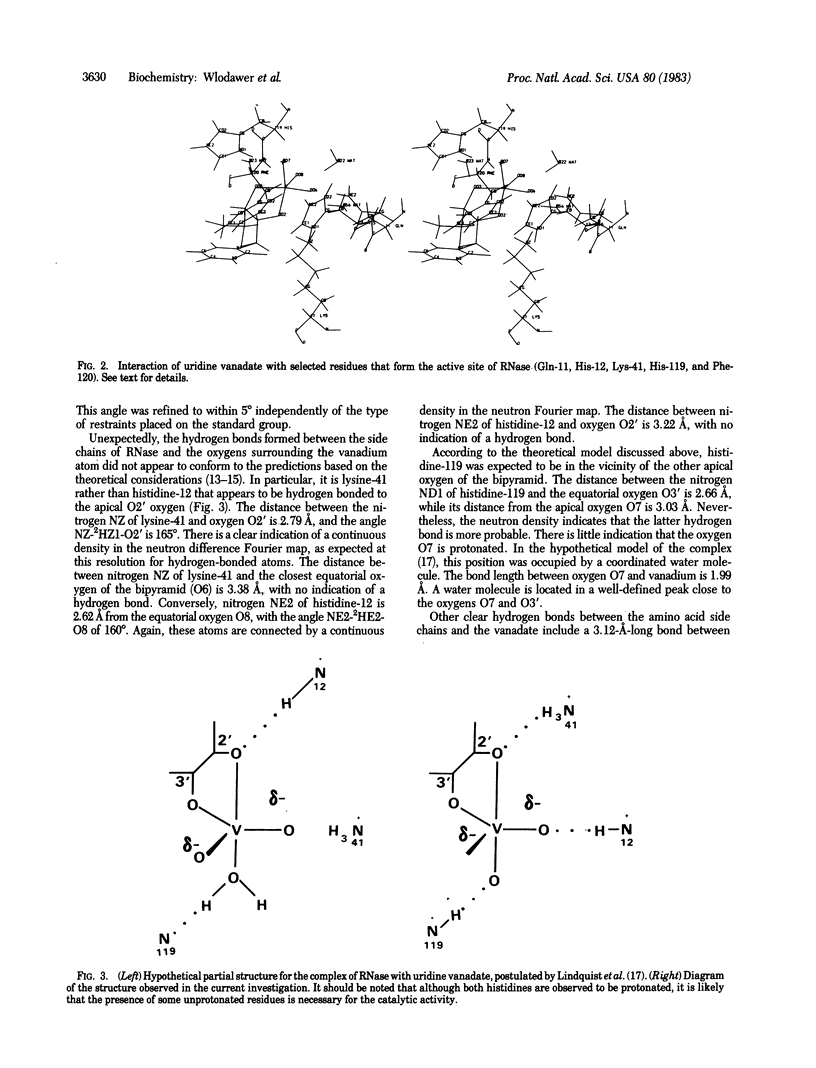
A complex of RNase A with a transition-state analog, uridine vanadate, has been studied by a combination of neutron and x-ray diffraction. The vanadium atom occupies the center of a distorted trigonal bipyramid, with the ribose oxygen O2' at the apical position. Contrary to expectations based on the straightforward interpretation of the known in-line mechanism of action of RNase, nitrogen NE2 of histidine-12 was found to form a hydrogen bond to the equatorial oxygen O8, while nitrogen NZ of lysine-41 makes a clear hydrogen bond to the apical oxygen O2'. Nitrogen ND1 of histidine-119 appears to be within a hydrogen-bond distance of the other apical oxygen, O7. Two other hydrogen bonds between the vanadate and the protein are made by nitrogen NE2 of glutamine-11 and by the amide nitrogen of phenylalanine-120. The observed geometry of the complex may necessitate reinterpretation of the mechanism of action of RNase.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Gilbert W. A., Ponzi D. R., Petsko G. A. The role of mobility in the substrate binding and catalytic machinery of enzymes. Ciba Found Symp. 1983;93:4–24. doi: 10.1002/9780470720752.ch2. [DOI] [PubMed] [Google Scholar]
- Breslow R. Artificial enzymes. Science. 1982 Nov 5;218(4572):532–537. doi: 10.1126/science.7123255. [DOI] [PubMed] [Google Scholar]
- Jentoft J. E., Gerken T. A., Jentoft N., Dearborn D. G. [13C]Methylated ribonuclease A. 13C NMR studies of the interaction of lysine 41 with active site ligands. J Biol Chem. 1981 Jan 10;256(1):231–236. [PubMed] [Google Scholar]
- Lindquist R. N., Lynn J. L., Jr, Lienhard G. E. Possible transition-state analogs for ribonuclease. The complexes of uridine with oxovanadium(IV) ion and vanadium(V) ion. J Am Chem Soc. 1973 Dec 26;95(26):8762–8768. doi: 10.1021/ja00807a043. [DOI] [PubMed] [Google Scholar]
- Pavlovsky A. G., Borisova S. N., Borisov V. V., Antonov I. V., Karpeisky M. Y. The structure of the complex of ribonuclease S with fluoride analogue of UpA at 2.5 A resolution. FEBS Lett. 1978 Aug 15;92(2):258–262. doi: 10.1016/0014-5793(78)80766-2. [DOI] [PubMed] [Google Scholar]
- Umeyama H., Nakagawa S., Fujii T. Simulation of the charge relay structure in ribonuclease A. Chem Pharm Bull (Tokyo) 1979 Apr;27(3):974–980. doi: 10.1248/cpb.27.974. [DOI] [PubMed] [Google Scholar]
- Usher D. A., Erenrich E. S., Eckstein F. Geometry of the first step in the action of ribonuclease-A (in-line geometry-uridine2',3'-cyclic thiophosphate- 31 P NMR). Proc Natl Acad Sci U S A. 1972 Jan;69(1):115–118. doi: 10.1073/pnas.69.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher D. A., Richardson D. I., Jr, Eckstein F. Absolute stereochemistry of the second step of ribonuclease action. Nature. 1970 Nov 14;228(5272):663–665. doi: 10.1038/228663a0. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Bott R., Sjölin L. The refined crystal structure of ribonuclease A at 2.0 A resolution. J Biol Chem. 1982 Feb 10;257(3):1325–1332. [PubMed] [Google Scholar]
- Wlodawer A. Neutron diffraction of crystalline proteins. Prog Biophys Mol Biol. 1982;40(1-2):115–159. doi: 10.1016/0079-6107(82)90012-8. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Sjölin L. Orientation of histidine residues in RNase A: neutron diffraction study. Proc Natl Acad Sci U S A. 1981 May;78(5):2853–2855. doi: 10.1073/pnas.78.5.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodak S. Y. The structure of cytidilyl(2',5')adenosine when bound to pancreatic ribonuclease S. J Mol Biol. 1977 Nov;116(4):855–875. doi: 10.1016/0022-2836(77)90275-3. [DOI] [PubMed] [Google Scholar]
- Wyckoff H. W., Tsernoglou D., Hanson A. W., Knox J. R., Lee B., Richards F. M. The three-dimensional structure of ribonuclease-S. Interpretation of an electron density map at a nominal resolution of 2 A. J Biol Chem. 1970 Jan 25;245(2):305–328. [PubMed] [Google Scholar]


