Abstract
We isolated and characterized traD mutants with an altered specificity of interaction with relaxosomes of various conjugative (F and R388) and mobilizable (RSF1010 and ColE1) plasmids. The change in specificity was due to a loss of some amino acids in the carboxyl terminus of TraD that resulted in a broadening of the range of mobilizable relaxosomes at the expense of a decrease in the efficiency of F-plasmid transfer.
Bacterial conjugation is a process of DNA transfer that is widespread among bacteria. Essential functions for conjugation include mating-pair formation (Mpf) and conjugative DNA processing. In every conjugative system there is a protein that is essential for conjugation but is not required for any of these processes; this protein presumably connects the relaxosome (a nucleoprotein complex for conjugative DNA processing that forms at oriT) and the membrane-spanning protein complex for DNA translocation (encoded by Mpf genes), and so it has been called the “coupling protein” (5). Mobilizable plasmids (such as RSF1010 and ColE1) bear functions required for relaxosome formation but encode neither Mpf proteins nor the coupling protein, which have to be provided by a conjugative plasmid (3–5, 12). Conjugative plasmids bear all functions required to promote their own transfer, including a gene for a coupling protein, which has been found in different conjugative plasmids from gram-negative bacteria (traG in RP4, trwB in R388, and traD in F) and also in the Agrobacterium tumefaciens T-DNA transfer system (virD4). Proteins encoded by these genes, which are termed the TraG protein family, show significant similarities among their amino acid sequences and share transmembrane domains and sequence signatures for nucleoside triphosphate binding (1, 13). Interactions of the TraD coupling protein with different components of the F-plasmid relaxosome have already been suggested, both in vivo (TraD with TraI) (6) and in vitro (TraD with TraM) (8).
E. coli strains and plasmids.
Escherichia coli strains and plasmids used in this study are listed in Tables 1 and 2, respectively.
TABLE 1.
E. coli strains
| Strain | Genotype | Reference or source |
|---|---|---|
| DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 relA1 Δ(argF-lacZYA)U169 φ80d lacZΔM15 gyrA96 | 10 |
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA11 Δ(lac-proAB) (F′ traD36 proAB lacIqZΔM15) | 17 |
| N100 | F− galK lac+ recA pro | 14 |
| UB1637 | F− his lys trp rspL recA56 λ− | 7 |
| XL1-Red | endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac mutD5 mutS mutT Tn10(Tetr) | Stratagene |
TABLE 2.
Plasmids
| Plasmid | Relevant featuresa | Replicon | Reference or source |
|---|---|---|---|
| pSU18 | Cmr, cloning vector | p15A | 2 |
| pSU1456 | Tpr, Sur, R388(trwB) | R388 | 13 |
| pSU4305 | Cmr, pSU18::(traD+oriTW+) | p15A | This work |
| pSU4309 | Cmr, pSU18::(traD680fs oriTW+) | p15A | This work |
| pSU4314 | Cmr, pSU18::(traD664fs) | p15A | This work |
| pSU4316 | Cmr, pSU18::(traD518) | p15A | This work |
| pSU4327 | Cmr, pSU18::(traD576) | p15A | This work |
| pSU4601 | Kmr, ColE1::kan | pMB1 | 4 |
| pSU4619 | Cmr, pSU18::traD | p15A | 4 |
| pSU4622 | Cmr, pSU18::(trwA+trwB+) | p15A | 4 |
| RSF1010K | Kmr, RSF1010::kan | RSF1010 | 16 |
Cmr, chloramphenicol resistant; Sur, sulfamide resistant; Tpr, trimethoprim resistant.
Effects on conjugation of a deletion in the carboxyl terminus of protein TraD.
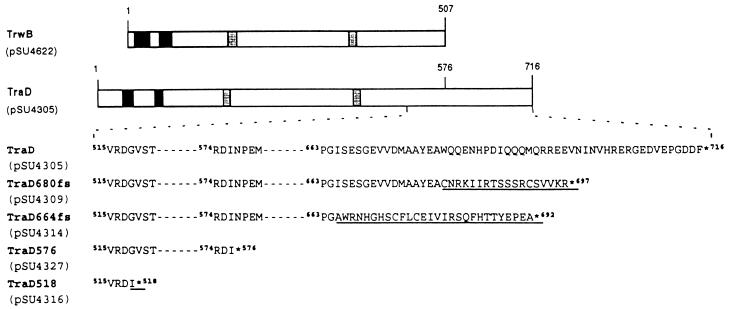
Because of the homology among coupling proteins, some attempts have been made to interchange them for conjugative transfer and mobilization. Cabezón et al. (4), working with R388, demonstrated that efficient transfer of mobilizable plasmid RSF1010 could be achieved when R388 trwB was replaced by RP4 traG. However, F traD only very inefficiently complemented R388 trwB for RSF1010 mobilization (5). Amino acid sequence alignment of TrwB, TraG, and TraD indicated that TraD displays a carboxyl terminus longer than those of TrwB or TraG (Fig. 1). We suspected that differences in RSF1010 mobilization could be due to this feature, so the TraD carboxyl terminus was deleted by introducing a stop codon in traD by site-directed mutagenesis. The stop codon was introduced just after amino acid 576, which corresponds to the point in the TraD sequence where homology with TrwB ends. The resulting protein (TraD576 [Fig. 1]) lacks the last 140 amino acids of TraD (nearly 20% of the whole protein).
FIG. 1.
Comparison of TrwB with TraD and a series of TraD mutants in the carboxyl-terminal region. The upper part of the figure shows a schematic alignment of TrwB and TraD. Solid boxes represent predicted transmembrane segments, and shaded segments represent theoretical nucleoside triphosphate-binding motifs. When the amino acid sequences of TrwB and TraD were aligned, C-terminal residue 507 of TrwB aligned with residue 576 of TraD. The lower part of the figure shows the amino acid sequences of the carboxyl-terminal regions of TraD and its derivatives. The plasmids that encode each of the proteins are shown in parentheses. Superscript numbers correspond to the positions of the corresponding amino acids in the sequence of TraD. Amino acids that differ from those of TraD are underlined. Asterisks represent the ends of the proteins.
Plasmid pSU4327, containing traD576, was used to complement pSU1456, a trwB mutant of R388, for self-transfer and for mobilization of either ColE1 or RSF1010. The results are shown in Table 3. While self-transfer of pSU1456 was complemented by TraD to very low levels (10−8 transconjugants/donor), this frequency increased 1,000-fold with TraD576. The frequency of mobilization of ColE1, however, remained unaffected. Interestingly, the traD576 mutant also increased pSU1456-mediated mobilization of RSF1010 by 1,000-fold, resulting in a frequency equivalent to that obtained when RSF1010 mobilization was complemented by trwB (10−4 transconjugants/donor). This result suggested that the presence of the TraD carboxyl terminus was hindering mobilization of RSF1010 by the MpfW system. It seems likely that the TraD C terminus led to deficient interaction either with the RSF1010 relaxosome or with the R388 MpfW apparatus. To decide between both possibilities, we analyzed RSF1010 mobilization while employing the MpfF apparatus (provided by strain JM109). With TraD as the complementing protein, RSF1010 mobilization was poor (5 × 10−6 transconjugants/donor [Table 3]). However, conjugation of F itself occurred at high frequencies (1 transconjugant/donor [Table 3]), so it can be assumed that TraD is able to interact properly with MpfF but fails to interact with the RSF1010 relaxosome. If, as we supposed, the TraD carboxyl terminus was hindering the interaction with the RSF1010 relaxosome, TraD576 should lead to a higher RSF1010 mobilization frequency than TraD when complementing JM109. In fact that was the case, and the results showed that mobilization of RSF1010 by MpfF also increased 1,000-fold with TraD576 (Table 3). Thus, the RSF1010 mobilization frequency was 103-fold higher with TraD576 than with TraD, regardless of the Mpf system employed, and the lack of the TraD carboxyl terminus in TraD576 allowed better interaction of TraD576 with components of the RSF1010 and R388 relaxosomes.
TABLE 3.
Transfer frequencies of mobilizable plasmids, or of conjugative plasmids deficient in the coupling protein, when complemented by TraD variantsa
| Plasmid (coupling protein) | Transfer frequency
|
|||||
|---|---|---|---|---|---|---|
| DH5α(pSU1456) (R388 trwB) with mobilizable plasmid:
|
JM109 (F traD) with mobilizable plasmid:
|
|||||
| None | pSU4601 (ColE1 Kmr) | RSF1010K | Noneb | pSU4601 (ColE1 Kmr) | RSF1010K | |
| pSU4305 (TraD)c | 3 × 10−8 | 2 × 10−4 | 3 × 10−7 | 1 | 5 × 10−2 | 5 × 10−6 |
| pSU4327 (TraD576) | 4 × 10−5 | 2 × 10−4 | 2 × 10−4 | 1 × 10−4 | 1 × 10−2 | 2 × 10−3 |
| pSU4309 (TraD680fs) | 2 × 10−5 | 3 × 10−3 | 5 × 10−4 | 1 × 10−4 | 1 × 10−1 | 3 × 10−3 |
| pSU4314 (TraD664fs) | 4 × 10−5 | 2 × 10−4 | 1 × 10−4 | 1 × 10−4 | 5 × 10−2 | 6 × 10−4 |
| pSU4316 (TraD518) | <1 × 10−8 | <1 × 10−8 | <1 × 10−8 | 1 × 10−5 | 2 × 10−6 | <1 × 10−8 |
| pSU4622 (TrwB) | 4 × 10−2 | 1 × 10−4 | 3 × 10−4 | 2 × 10−6 | 8 × 10−6 | 2 × 10−7 |
| None | <1 × 10−8 | <1 × 10−8 | <1 × 10−8 | <1 × 10−8 | <1 × 10−8 | <1 × 10−8 |
Donor strains were derivatives either of strain DH5α(pSU1456) or of strain JM109 carrying one of the plasmids providing a coupling protein with or without a mobilizable plasmid. Transfer frequencies are expressed as the number of transconjugants per donor cell obtained when donor strains were mated with UB1637 (except as indicated in footnote b) for 1 h at 37°C on solid media. Reported figures are means of at least three experiments.
Recipient strain was N100.
Frequencies obtained with pSU4619 (equivalent to pSU4305 but not containing oriTW) were the same as for pSU4305.
To this point, our results showed only a hindering function for the TraD carboxyl terminus, but its physiological significance in F conjugation was suggested by an experiment in which we used TraD576 to complement JM109 (F traD) for F transfer. In this case, the frequency of conjugative transfer of F traD upon complementation by TraD576 was reduced 104-fold compared with the frequency obtained when F traD was complemented by wild-type TraD (Table 3). The conclusion was that the TraD carboxyl terminus is responsible for efficient coupling with the F relaxosome, at the cost of hindering the interaction with unrelated relaxosomes.
It is worth noting here that the frequencies of ColE1 mobilization with MpfW (pSU1456) or MpfF (JM109) did not vary when TraD576 instead of TraD was employed (Table 3). These results suggest that interaction of TraD with the ColE1 relaxosome probably does not involve the carboxyl region. These data reinforce the concept that the Mpf functions of both F and R388 can interact properly with TraD576, and so the results reported above should be a consequence of altered interactions with the relevant relaxosomes.
The C-terminal 37 amino acids of TraD are responsible for the change in specificity.
Using a different experimental approach, we randomly mutagenized traD to improve its ability to complement conjugative transfer of R388 trwB. A method that combined in vivo mutagenesis and selection steps was used. Random mutagenesis was carried out by introducing pSU4305, a recombinant plasmid containing F traD together with R388 oriT, into E. coli XL1-Red (catalog no. 200129; Stratagene). This strain is damaged in three different DNA repair systems (mutD mutS mutT), so its mutation rate is about 5,000-fold higher than that of common E. coli laboratory strains. After 200 generations of growth of XL1-Red(pSU4305), plasmid DNA was isolated and introduced into strain DH5α(pSU1456). About 104 transformant colonies were pooled, diluted in Luria broth to 2 × 109 cells/ml, and used as donors for mating with the recipient strain, UB1637. The presence of R388 oriT in pSU4305 allowed conjugative DNA processing of this plasmid by pSU1456-encoded proteins and thus its transfer to the recipient strain. Donor cells (107) were mated with 109 recipient cells at 37°C on a solid surface (0.22-μm-pore-size Millipore nitrocellulose filter on prewarmed nutritive agar plates) for 1 h. Transconjugants harboring pSU4305 derivatives were isolated. In that way, a selection step that eliminated nonconjugative mutants and enriched the population of those traD mutants that increased complementation of R388 oriTW transfer was carried out. Resulting transconjugants were pooled, and plasmid DNA was obtained from them and reintroduced into XL1-Red cells to carry out another mutagenesis-selection cycle. After five of these cycles, a mutant plasmid named pSU4309, which produced a frequency of transfer of R388 trwB that was 103-fold higher than that obtained with the original plasmid, pSU4305, was isolated (Table 3).
The mutation present in pSU4309 that was responsible for the effect on the frequency of transfer of R388 trwB was localized by DNA heteroduplex analysis in mutation detection enhancement (MDE) polyacrylamide gels (Hydrolink). Briefly, wild-type and mutant plasmid DNAs were endonuclease digested, mixed, heat denatured, and then renatured and run on an MDE gel. Heteroduplex DNA molecules carrying mismatches were detected by means of their reduced electrophoretic mobility, allowing detection of single-base substitutions. In this way, we limited mutations to a segment of 36 bp located 128 bp upstream from the end of traD. This region and adjacent DNA were sequenced, revealing that the only mutation detected was a deletion of a G at position 2358 in the published DNA sequence of traD (11). This point deletion caused a frameshift that changed the TraD amino acid sequence from position 680 onward and resulted in the appearance of a premature stop signal at amino acid 698. The resulting protein, named TraD680fs, had lost the last 37 amino acids of the carboxyl terminus of TraD (Fig. 1). That small deletion produced an effect on plasmid transfer frequencies equivalent to that observed with TraD576: a 103-fold increase in the transfer frequencies of both R388 and the mobilizable plasmid RSF1010 and a 104-fold reduction in F plasmid transfer frequency. A 10-fold increase in the transfer frequency of plasmid ColE1 may also be significant (Table 3). Therefore, the determinant for efficient interaction with the F relaxosome that hindered the interaction with R388 and RSF1010 relaxosomes was located at the very end of TraD, its last 37 amino acids. The remaining amino acid sequence deleted in TraD576 apparently had no additional effect, and its role could be simply to position properly the specificity determinant in the whole protein.
We next wondered if further deletion of the TraD carboxyl terminus beyond amino acid residue 576 would maintain the same properties. Therefore, we constructed the TraD derivative TraD518, which ends with the substitution G518I and lacks the C-terminal 199 amino acids of TraD (Fig. 1). When plasmid pSU4316, containing traD518, was used to complement the transfer of either R388 trwB or F traD, the transfer frequencies obtained were very low (Table 3), suggesting that the region between amino acids 518 and 576 contains sequences of TraD that are essential for function.
Altogether, our results showed that the loss of even a small fragment of the TraD carboxyl terminus causes a change in the specificity of its interaction with different relaxosomes, resulting in a moderate frequency of conjugative transfer for a series of them (Table 3). Thus, the effect can be described as a widening of the range of mobilizable relaxosomes at the expense of a reduction in the efficiency of F relaxosome mobilization. This result could be interpreted as the presence in F TraD of a carboxyl-terminal arm, not present in other coupling proteins (such as TrwBR388 and TraGRP4), that constitutes a high-affinity site for interaction with its own relaxosome. The existence of this C-terminal arm hinders the interaction of a second, less efficient, site with affinity for a broader range of relaxosomes. Evolutionarily, this kind of specialization typically occurs in stable environments, where it is advantageous to sacrifice a capacity for adaptation to different conditions for an increase in functional efficiency (called a K-strategy); in contrast, under unstable conditions, specialization is a burden instead of an advantage, and an r-strategy based on a high reproduction rate is more convenient for ecological success (15). In nature, F exists usually as a single-copy plasmid exclusively in bacteria belonging to the Enterobacteriaceae, which proliferate in animal digestive tracts. This ecosystem probably constitutes a stable environment, so F may have followed a K-strategy consisting of the development of a specific and efficient transfer apparatus to promote its own transfer. This is consistent with the higher genetic complexity of the F transfer apparatus compared with other conjugative systems (9). Meanwhile, plasmids dwelling in more unstable environments, as do promiscuous plasmids within IncN, IncP, IncW, or IncQ, may have developed a simpler, less specialized, and less efficient but more versatile conjugative apparatus.
Acknowledgments
This work received financial support from the Spanish Ministry of Education (DGICYT) through project PB95-1269. J.I.S. was a recipient of grants from the Department of Education of the Basque Government and from the Marqués de Valdecilla Foundation (in different periods), and E.C. was a recipient of a grant from the Department of Education of the Basque Government.
REFERENCES
- 1.Balzer D, Pansegrau W, Lanka E. Essential motifs of relaxase TraI and TraG proteins involved in conjugative transfer of plasmid RP4. J Bacteriol. 1994;176:4285–4295. doi: 10.1128/jb.176.14.4285-4295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartolomé B, Jubete Y, Martínez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors, compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 3.Beijersbergen A, Dulk-Ras A D, Schilperoort R A, Hooykaas P J J. Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science. 1992;256:1324–1327. doi: 10.1126/science.256.5061.1324. [DOI] [PubMed] [Google Scholar]
- 4.Cabezón E, de la Cruz F, Lanka E. Requirements for mobilization of plasmid RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J Bacteriol. 1994;176:4455–4458. doi: 10.1128/jb.176.14.4455-4458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabezón E, Sastre J I, de la Cruz F. Genetic evidence of a coupling role for the TraG protein-family in bacterial conjugation. Mol Gen Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 6.Dash P K, Traxler B A, Panicker M M, Hackney D D, Minkley E G. Biochemical characterization of Escherichia coli DNA helicase I. Mol Microbiol. 1992;6:1163–1172. doi: 10.1111/j.1365-2958.1992.tb01555.x. [DOI] [PubMed] [Google Scholar]
- 7.De La Cruz F, Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982;151:222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Disqué-Kochem C, Dreiseikelmann B. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J Bacteriol. 1997;179:6133–6137. doi: 10.1128/jb.179.19.6133-6137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firth N, Ippen-Ihler K, Skurray R A. Structure and function of the F factor and mechanism of conjugation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2377–2401. [Google Scholar]
- 10.Grant S G N, Jesee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jalajakumari M B, Manning P A. Nucleotide sequence of the traD region in Escherichia coli F sex factor. Gene. 1989;81:195–202. doi: 10.1016/0378-1119(89)90179-0. [DOI] [PubMed] [Google Scholar]
- 12.Lessl M, Balzer D, Lanka E. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J Bacteriol. 1993;175:6415–6425. doi: 10.1128/jb.175.20.6415-6425.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llosa M, Bolland S, de la Cruz F. Genetic organization of the conjugal DNA processing region of the IncW plasmid R388. J Mol Biol. 1994;235:448–464. doi: 10.1006/jmbi.1994.1005. [DOI] [PubMed] [Google Scholar]
- 14.McKenney K, Shimatake H, Court D, Schmeissner U, Rosenberg M. A system to study promoter and terminator signals recognised by Escherichia coli DNA polymerase. In: Chirikjian J G, Papas T, editors. Gene amplification and analysis. Vol. 2. Amsterdam, The Netherlands: Elsevier/North-Holland; 1981. pp. 383–415. [PubMed] [Google Scholar]
- 15.Pianka E R. On r- and K-selection. Am Nat. 1970;104:592–597. [Google Scholar]
- 16.Scherzinger E, Lurz R, Otto S, Dobrinski B. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 1992;20:41–48. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC18 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]