Abstract
N-glycosylation can increase the rate of protein folding, enhance thermodynamic stability, and slow protein unfolding; however, the molecular basis for these effects is incompletely understood. Without clear engineering guidelines, attempts to use N-glycosylation as an approach for stabilizing proteins have resulted in unpredictable energetic consequences. Here we review the recent development of three “enhanced aromatic sequons”, which appear to facilitate stabilizing native-state interactions between Phe, Asn-GlcNAc and Thr when placed in an appropriate reverse turn context. It has proven to be straightforward to engineer a stabilizing enhanced aromatic sequon into glycosylation-naïve proteins that have not evolved to optimize specific protein-carbohydrate interactions. Incorporating these enhanced aromatic sequons into appropriate reverse turn types within proteins should enhance the well-known pharmacokinetic benefits of N-glycosylation-based stabilization by lowering the population of unfolded protease-susceptible and misfolded aggregation-prone states, thereby making such proteins more useful in research and pharmaceutical applications.
Introduction
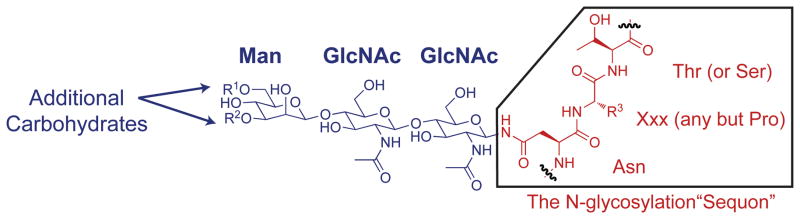
Cellular glycosylation of Asn residues (N-glycosylation) generally occurs co-translationally as the ribosome inserts nascent polypeptides into the endoplasmic reticulum (ER). The oligosaccharyl transferase enzyme complex transfers the Glc3Man9GlcNAc2 triantennary oligosaccharide (Figure 1; where Glc is D-Glucose, Man is D-mannose, and GlcNAc is N-acetyl-D-glucosamine) from a membrane-anchored dolichol phosphate to the side-chain amide nitrogen of an Asn residue within the Asn-Xxx-Thr/Ser recognition sequence (or sequon, where Xxx is any amino acid except Pro, Figure 2).1–3
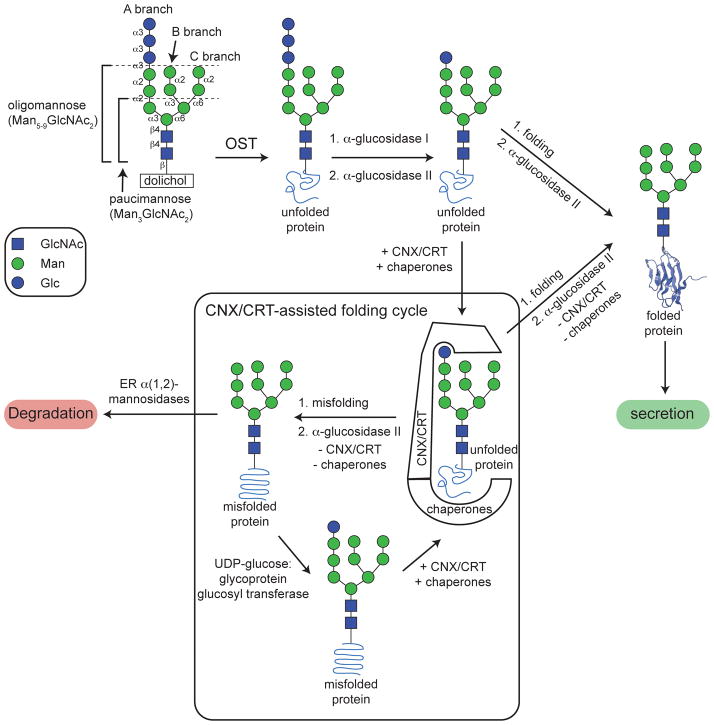
Figure 1.
Cellular N-glycosylation allows a glycoprotein to enter the calnexin/calreticulin-assisted folding vs. degradation cycle in the ER, allowing the N-glycoprotein to fold and be secreted or to be targeted for degradation if attempts to fold the N-glycoprotein are unsuccessful.
Figure 2.
The oligosaccharyl transferase enzyme transfers the Glc3Man9GlcNAc2 glycan en bloc to the side-chain amide nitrogen of an Asn residue within the consensus sequence Asn-Xxx-Thr/Ser, called the N-glycosylation “sequon”; Xxx is any amino acid but proline.
The Asn-linked oligosaccharide (N-glycan) mediates key interactions between the nascent polypeptide and components of the ER proteostasis network, which promote proper folding of N-glycoproteins (Figure 1). Removal of the outer two Glc residues from the A-branch of the N-glycan by glucosidase I and glucosidase II unmasks the inner Glc residue, which binds to the lectins calnexin and calreticulin (CNX/CRT), allowing N-glycosylated polypeptides to participate in the CNX/CRT-assisted folding vs. degradation cycle.4,5 CNX and CRT promote intramolecular folding interactions over aggregation, and also recruit the oxidoreductase ERp57 (which catalyzes disulfide bond isomerization) to help N-glycoproteins acquire the proper fold.5 Upon release from CNX/CRT, glucosidase II removes the inner Glc residue from the A-branch of the N-glycan, thereby preventing N-glycoproteins from re-associating with CNX/CRT.
Properly folded N-glycoproteins then engage trafficking receptors and are exported from the ER via vesicular trafficking.6–8 In contrast, non-native N-glycoproteins are bound by ER-retained chaperones including Hsp40’s and 70’s, which prevent them from being secreted.5 These non-native proteins are recognized by UDP-glucose:glycoprotein glucosyltransferase, which adds a single Glc residue back to the A-branch of the N-glycan, thereby restoring the ability of the N-glycoprotein to bind CNX and/or CRT (Figure 1).9–12 Non-native N-glycoproteins can then re-enter the CNX/CRT cycle, where they have additional time to acquire the proper fold. Persistently misfolded N-glycoproteins eventually encounter ER-resident α(1–2)-mannosidases,13 which remove mannose residues from the N-glycan. This modification reduces the binding affinity of the N-glycan for CNX/CRT (by blocking reglucosylation of the A branch),14,15 ultimately leading to dislocation of the misfolded protein from the ER to the cytosol for ER-associated degradation (mediated by ubiquitination, and proteasomal endoproteolysis).13,16,17 Thus, the N-glycan promotes protein folding, trafficking and function vs. ER-associated degradation18,19 via an extrinsic mechanism, i.e., one that depends on intermolecular interactions between the N-glycan and components of the ER proteostasis network.
In addition to these extrinsic roles, the N-glycan can also strongly influence the folding free energy landscape of individual glycoproteins via intrinsic mechanisms that depend on the N-glycan and the protein sequence. N-glycosylation is established to (1) promote the formation of secondary structure in small peptide model systems,20–33 (2) accelerate protein folding,34 (3) facilitate disulfide formation,31 (4) reduce aggregation,34–36 and (5) increase protein thermodynamic stability.37–41 However, the molecular basis for these effects is incompletely understood. Understanding how an N-glycan remodels a protein’s folding energy landscape at the molecular level is important from a basic science perspective, but could also provide engineering guidelines that yield more stable glycoproteins for research and pharmaceutical applications. N-glycosylation has been used often in protein drug development42–45 to increase the shelf-life and serum half-life46–48 of proteins, and to decrease their aggregation propensity37,49 and proteolytic susceptibility,50 via a mechanism that appears to depend simply on shielding of hydrophobic protein surface residues by the large N-glycan. If the N-glycan could also increase protein thermodynamic stability predictably, the pharmacokinetic properties of the protein could be enhanced even further, due to the resulting lower population of the protease-susceptible unfolded states and aggregation-prone partially denatured conformational ensembles. Installing N-glycans at naïve sites in proteins to achieve these pharmacokinetic benefits is typically done by trial and error,48,51 a time-consuming process that can have unpredictable energetic consequences.52–54 There is a need for clear engineering principles to guide placement of N-glycans within proteins at positions which are predisposed to be stabilized by glycosylation or which, at the very least, will not be strongly destabilized by N-glycosylation.
Learning from the adhesion domain of human CD2
We began our search for these engineering guidelines by studying the adhesion domain of the human protein CD2 (HsCD2ad).55 HsCD2ad is a N-glycosylated member of the immunoglobulin super family,56 featuring a single N-glycan flanked by strands D and E of the classic beta-sandwich fold (Figure 3A).57–59 Previous studies reveal that HsCD2ad cannot fold or function properly when deglycosylated with PNGase F,60 or when the N-glycosylation sequon is removed by mutation,60,61 suggesting that the N-glycan plays an essential role in HsCD2ad folding.
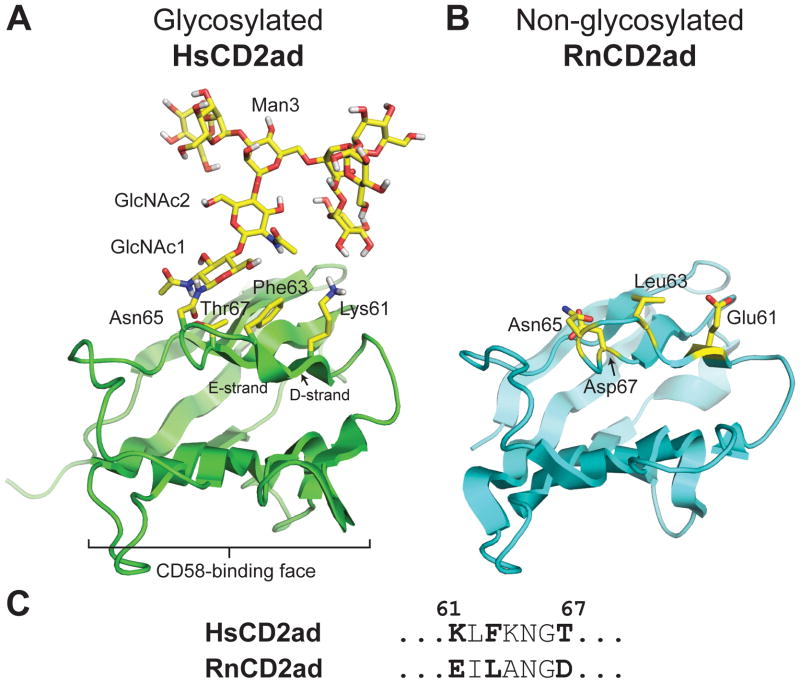
Figure 3.
(A) Ribbon diagram of the adhesion domain of the human protein CD2, HsCD2ad (PDB code: 1GYA), which requires glycosylation at Asn65 to fold properly. The N-glycan and HsCD2ad side chains at positions 61, 63, and 67 are highlighted in yellow. Wyss et al. (ref 59) observed NOEs between the N-glycan and these side chains. (B) Ribbon diagram of the rat ortholog of HsCD2ad, RnCD2ad (PDB code: 1HNG), which is not glycosylated at Asn65, but is still able to fold properly. RnCD2ad side chains at positions 61, 63, and 67 are highlighted in yellow. (C) Comparison of the HsCD2ad amino acid sequence near the glycosylation site with the homologous sequence from RnCD2ad. All structures rendered in Pymol.
Elegant 2- and 3-dimensional NMR experiments on 15N-labelled HsCD2ad (expressed in CHO cells) from the Wagner Laboratory facilitated a structural determination of both the polypeptide and the high mannose N-glycan substructures.57,59,62 Several NOEs between the N-glycan and the amino acid side chains on the surface of HsCD2ad opposite the binding site for CD58, the ligand of CD2, suggest that the N-glycan indirectly promotes binding between CD58 and HsCD2ad via structural stabilization, possibly through stabilizing interactions between these side chains and the N-glycan.59 Wagner and coworkers probed the impact of the N-glycan on the stability of HsCD2ad via binding assays with CD58. An HsCD2ad variant with a single GlcNAc retains substantial binding affinity for CD58, whereas fully deglycosylated HsCD2ad does not. The CD spectrum of the fully deglycosylated HsCD2ad variant is dramatically different from the spectrum of HsCD2ad variants bearing a high mannose N-glycan or GlcNAc at Asn65, suggesting substantial unfolding and structural instability in HsCD2ad in the absence of the N-glycan.59
The rat ortholog of HsCD2ad (RnCD2ad)63,64 lacks the glycosylation site at Asn65 (because the residue at position 67 is Asp instead of Thr), but is able to fold into a stable CD2ad structure nonetheless (Figure 3B). Comparison of the amino acid sequence of HsCD2ad with that of RnCD2ad at positions where the HsCD2ad NMR data indicate interactions with the N-glycan revealed the following differences: RnCD2ad has Glu at position 61 and Leu at position 63, whereas HsCD2ad has Lys at position 61 and Phe at position 63 (Figure 3C). Wagner and coworkers hypothesized that Lys61 and Phe63 in HsCD2ad might be responsible for HsCD2ad’s dependence on the N-glycan for proper folding and function. To test this hypothesis, they created a non-glycosylated variant of HsCD2ad (via a T67D mutation, which abolishes the glycosylation sequon), and then mutated Lys61 and Phe63 to Glu61 and Leu63, respectively.59 This non-glycosylated HsCD2ad variant binds CD58, unlike the non-glycosylated wild-type HsCD2ad sequence, supporting the hypothesis that Lys61 and Phe63 are responsible for the dependence of HsCD2ad on N-glycosylation for folding. However, the specific role played by each residue was unclear. By itself, the Phe63Leu mutation fails to restore substantial CD58-binding affinity to non-glycosylated HsCD2ad, and the authors did not test the impact of the Lys61Glu mutation by itself. Moreover, replacing Lys61 or Phe63 with Ala in the glycosylated HsCD2ad does not substantially decrease CD58-binding affinity, in contrast to what one would expect if Phe and Lys were engaging in essential stabilizing interactions with the N-glycan. The authors conclude that no single interaction between the N-glycan and nearby protein side chains is crucial for C58-binding affinity, and by extension, HsCD2ad structural stability.59 Instead, they hypothesize that the large N-glycan-dependent stabilization of HsCD2ad comes from an ill-defined combination of hydrogen bonds, van der Waals contacts, and possibly entropic contributions.59
It is likely that some of the ambiguity regarding the extent to which individual amino acid residues contribute to the glycan-dependent stabilization of HsCD2ad stems from the use of a binding assay as a read-out for HsCD2ad conformational stability. Conformational stability is generally required for proper function (i.e., CD58-binding), and functional assays are good at differentiating between folded proteins, which function properly, and unfolded proteins, which do not. However, functional assays may not be sensitive enough to detect more subtle differences in conformational stability. Suppose, for example, that one protein undergoes a thermal denaturation transition at 37 °C, whereas a sequence variant of that protein undergoes a thermal denaturation transition at 58 °C. A functional assay conducted at 25 °C may not be able to detect the significant difference in thermodynamic stability between these two proteins, because both proteins are mostly folded at 25 °C.
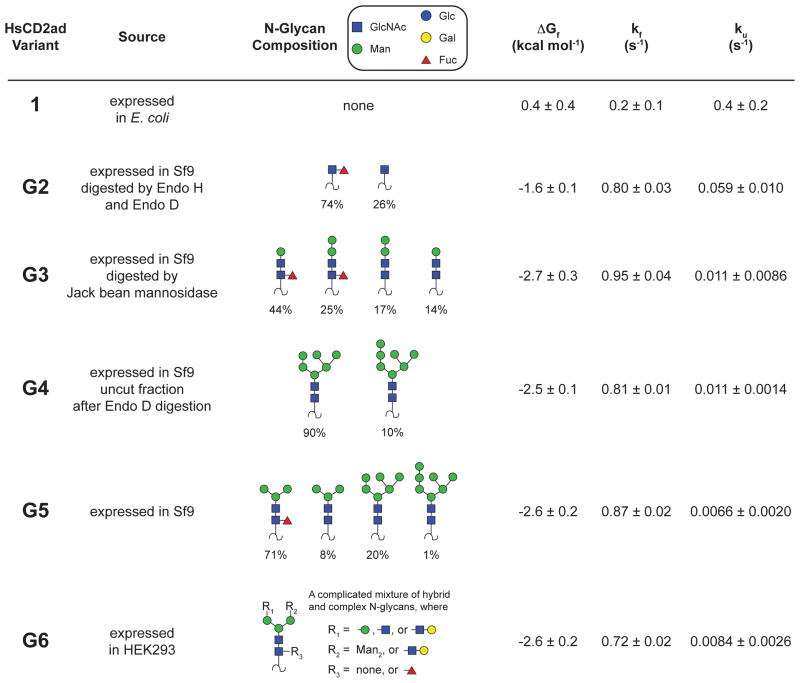
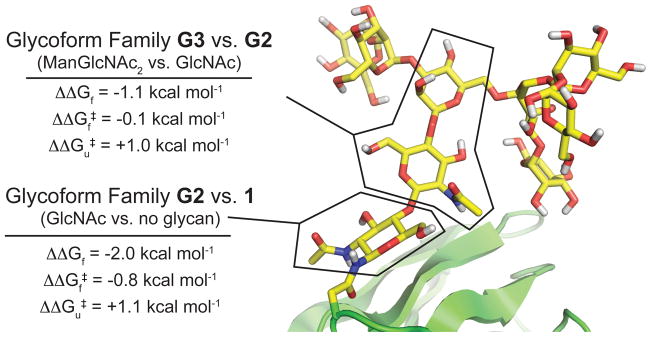
We sought to gain a clearer understanding of why N-glycosylation stabilizes HsCD2ad. The substantial CD58-binding activity of a HsCD2ad variant with a truncated N-glycan59 led us to hypothesize that specific substructures of the conserved oligomannose glycan (Figures 1, 3) are responsible for the glycan-dependent stabilization of HsCD2ad. To test this hypothesis, we generated several glycoforms of HsCD2ad via expression in mammalian and insect cell lines (Figure 4).65 These N-glycoproteins share an invariant HsCD2ad amino acid sequence, but differ in the composition of the N-glycan attached to Asn65. Non-glycosylated variant 1 was expressed in E. coli. Glycoform family G6 was expressed in HEK293 cells as a complicated mixture of hybrid, complex, and oligomannose glycoforms, whereas G5 was expressed in Sf9 cells as a mixture of fucosylated and non-fucosylated paucimannose and oligomannose glycoforms. Treatment of G5 with endoglycosidase H and endoglycosidase D generated glycoform family G2, in which the glycan is mostly fucosylated GlcNAc. In contrast, treatment of G5 with jack bean mannosidase generated glycoform family G3, in which the glycan is a mixture of fucosylated and non-fucosylated ManGlcNAc2 and Man2GlcNAc2 glycoforms. Finally, glycoform family G4 is a mixture of the Man6GlcNAc2 and Man7GlcNAc2 glycoforms that remain uncut after digestion of G5 with endoglycosidase D. We anticipated that measuring the folding energetics of these variants, which are enriched in certain N-glycan substructures, would provide insight into which portions of the oligosaccharide are important for the glycan-dependent stabilization of HsCD2ad.65
Figure 4.
Folding free energies and folding and unfolding rates for HsCD2ad glycoforms generated via enzymatic remodeling (ref 65). Variants G2–G6 are predominantly mixtures of the indicated glycoforms as determined by mass spectrometry. Only species with abundance >5% are shown, except for variant G2, where only species with relative abundance > 0.5 are shown (owing to the complexity of the heterogeneous mixture of glycans obtained upon expression of HsCD2ad in HEK293 cells).
We analyzed the folding energy landscapes of these HsCD2ad glycoform families by monitoring folding and unfolding as a function of time and denaturant concentration using tryptophan fluorescence.65 Conformational changes are induced either by rapidly mixing a solution of protein in buffer with a concentrated urea solution (a kinetic unfolding experiment), or by mixing a solution of protein in concentrated urea with buffer (a kinetic folding experiment). Analysis of the resulting chevron plots provides folding and unfolding rate constants (kf and ku, respectively) and the folding free energy ΔGf for each variant, because ΔGf = −RTln(kf/ku). Non-glycosylated 1 is too unstable to obtain a complete set of folding data, so we used a combination of equilibrium and kinetic methods to characterize its folding energetics.65 After correcting for photobleaching, the kinetic data for 1 and G2–G6 were monoexponential, indicating that glycan heterogeneity within each glycoform family does not significantly impact the folding or unfolding rate (if this were not the case, the kinetics would necessarily involve additional exponential terms). For example, the N-glycan in glycoform family G2 is 26% GlcNAc and 74% fucosylated GlcNAc. The monoexponential nature of the kinetics observed during the folding and unfolding experiments on G2 indicates that fucosylation of GlcNAc does not significantly affect the folding or unfolding rate relative to GlcNAc alone.65 We use a similar approach for G3–G6, attributing the observed energetic data to the simplest N-glycan present in each glycoform family.
The results of this analysis are shown in Figure 4.65 Non-glycosylated 1 has ΔGf = 0.4 ± 0.4 kcal mol−1, meaning that its unfolded ensemble is roughly as stable as the folded state. G2, in which the glycan is a single GlcNAc residue (ΔGf = −1.6 ± 0.1 kcal mol−1), is substantially more stable than non-glycosylated 1 (ΔΔGf = −2.0 ± 0.4 kcal mol−1), in agreement with the observation by Wagner and coworkers that a single GlcNAc at Asn65 restores CD58-binding affinity to HsCD2ad.59 This GlcNAc-associated increase in stability comes from a four-fold increase in folding rate (corresponding to a −0.8 kcal mol−1 change in folding activation energy ΔΔGf‡), and a 6.8-fold decrease in the unfolding rate (corresponding to a +1.1 kcal mol−1 change in unfolding activation energy ΔΔGu‡). Elongation of the N-glycan from GlcNAc (as in G2) to the ManGlcNAc2 triose (as in glycoform family G3) also stabilizes HsCD2ad (ΔΔGf = −1.1 ± 0.3 kcal mol−1). This increase in stability appears to come mostly from a decrease in unfolding rate (G3 unfolds 5.4 times more slowly than G2; ΔΔGu‡ = +1.0 kcal mol−1) and from a small increase in folding rate (G3 folds 1.2 times faster than G2; ΔΔGf‡ = −0.1 kcal mol−1). The increased folding rate associated with the first GlcNAc residue of the glycan either results from destabilization of the HsCD2ad denatured ensemble, or from stabilization of the folding transition state, or from some combination of these two factors. The decreased unfolding rate associated with both the first GlcNAc and elongation to the ManGlcNAc2 triose is most easily explained by stabilizing native-state interactions between the N-glycan and the HsCD2ad protein.
Comparison of the folding energetics of glycoform families G3–G6 reveals that further elongation of the ManGlcNAc2 triose core to more elaborate paucimannose and oligomannose glycans in G4 and G5 or to the complex and hybrid glycans in G6 does not substantially change the folding energetics of HsCD2ad.65 G3–G6 have similar folding rates, unfolding rates, and free energies of folding, which can be attributed to the simplest N-glycan substructure that the G3–G6 glycoform families have in common with each other, but not with GlcNAcylated G2: the ManGlcNAc2 triose (Figure 4). Interestingly, this ManGlcNAc2 triose is the conserved core of all eukaryotic N-glycans. The picture that emerges is that all of the energetic impact of glycosylation on HsCD2ad (ΔΔGf = −3.1 kcal mol−1) comes from the ManGlcNAc2 triose, with −2.0 kcal mol−1 of stabilization coming from the first GlcNAc, and the remaining −1.1 kcal mol−1 of stabilization coming from addition of the ManGlcNAc disaccharide (Figure 5). Furthermore, the first GlcNAc is responsible for most of the increased folding rate associated with the ManGlcNAc2 triose relative to the non-glycosylated HsCD2ad, and addition of the ManGlcNAc disaccharide affects mostly the unfolding rate (Figure 5).65 That the energetic impact of N-glycosylation on HsCD2ad can be entirely attributed to the highly conserved ManGlcNAc2 triose core of the glycan led us to speculate that the energetic benefits conferred by glycosylation of HsCD2ad might apply to other proteins as well. Moreover, we hypothesized that the intrinsic energetic contributions of the evolutionarily-conserved triose core might explain why N-glycosylation is so common among eukaryotic proteins (approximately one-third of the proteome traverses the cellular secretory pathway,66 and most of these secreted proteins are N-glycosylated).
Figure 5.
The energetic contributions of N-glycan substructures to the change in HsCD2ad folding free energy (ΔΔGf), folding activation energy (ΔΔGf‡), and unfolding activation energy (ΔΔGu‡) upon N-glycosylation. Structure of glycosylated HsCD2ad (PDB code: 1GYA) rendered in Pymol.
Does N-glycosylation Stabilize Proteins by Destabilizing the Denatured Ensemble?
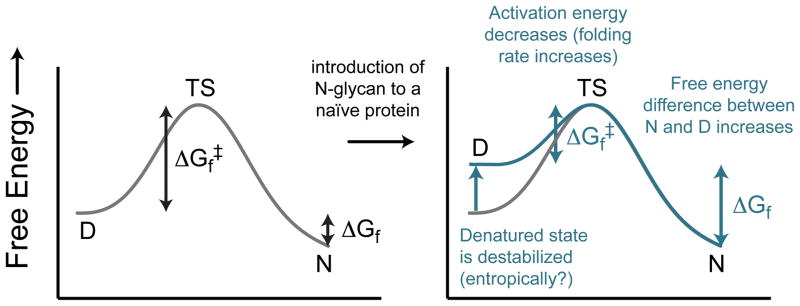
We explored whether the introduction of an N-glycan into a glycosylation-naïve protein (via a mutation that installs the canonical glycosylation sequon) might generally confer an energetic advantage to the protein. We anticipated that glycosylation would accelerate protein folding and increase thermodynamic stability by destabilizing the unfolded ensemble via an excluded volume mechanism that would not depend strongly on the location of the glycosylation site or on interactions between the N-glycan and nearby protein side chains (Figure 6).65 Others have rationalized the conformational impacts of N-glycosylation using similar explanations.20,24,26,28,29,31,67 Levy and coworkers40 tested this hypothesis computationally using a coarse-grained native-topology model of glycosylation in which the glycan is represented as a chain of excluded volume beads. Consistent with this hypothesis, they found that glycosylation of the SH3 domain in silico was generally stabilizing, provided that the proposed glycosylation site was not buried, sterically crowded, or highly structured in the wild-type protein, and that the increase in stability derived from destabilization of the denatured ensemble.40
Figure 6.
Energy diagram view of the hypothesis that introduction of an N-glycosylation site into a glycosylation-naïve protein would restrict the conformational freedom of the denatured ensemble, thereby destabilizing it relative to the native state, resulting in a more negative free energy of folding (i.e., overall stabilization).
We wondered whether destabilization of the denatured ensemble might be a general consequence of N-glycosylation. We tested this hypothesis in collaboration with Levy and coworkers employing experimental and computational methodology to analyze the energetic impact of introducing an N-glycan at various positions within a glycosylation-naïve protein.52 We chose the WW domain of the human Pin1 protein (hereafter called WW), a triple-stranded anti-parallel β-sheet protein (Figure 7) whose structure and energetics have been extensively characterized.68–87 We used Asn-linked GlcNAc (Asn-GlcNAc) as a surrogate for the large triantennary glycan present in secreted glycoproteins for synthetic simplicity (we prepared these glycosylated WW variants chemically via solid-phase peptide synthesis), and because of previous observations that even a single GlcNAc residue can have a substantial impact on glycopeptide structure20–25,27–31 and/or energetics.59,65,88 Unlike biological N-glycosylation, which occurs only at Asn residues within the canonical Asn-Xxx-Thr/Ser sequon, the chemical N-glycosylation approach allows us to replace any residue in WW with Asn-GlcNAc, whether or not there is a Thr or Ser at the +2 position. This approach neglects the possibility that interactions between the GlcNAc and Thr might be energetically significant, but such interactions were not observed previously in several conformational studies of glycopeptides,27–31 and we expected that the hypothesized energetically favorable excluded volume effects of glycosylation would not depend strongly on the identities of the side-chains near the glycosylation site. Consequently, we chose to replace wild-type residues in WW with Asn-GlcNAc without any additional changes to nearby residues.
Figure 7.
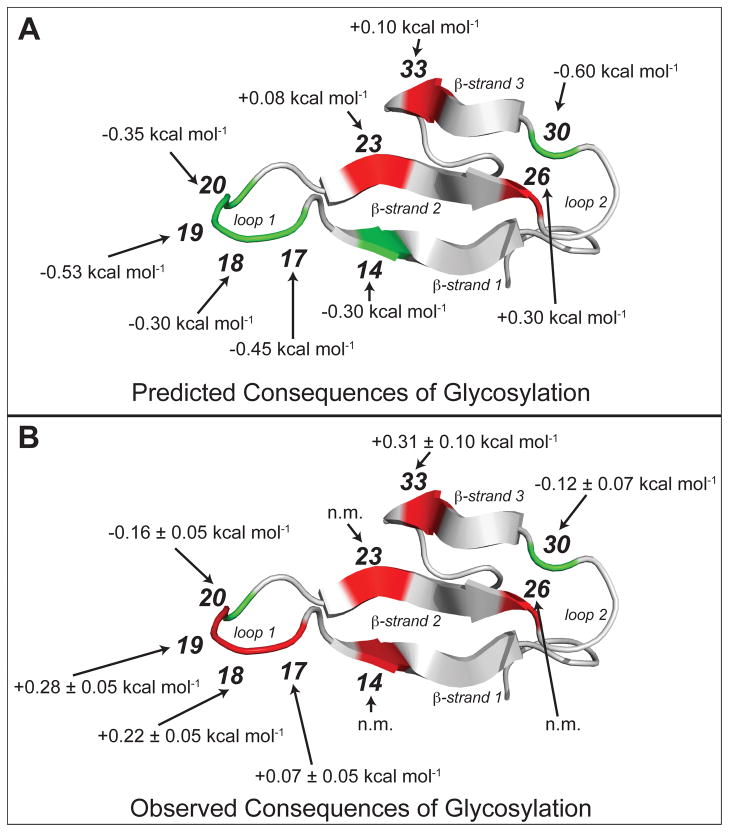
(A) Predicted and (B) observed energetic consequences of glycosylation (i.e., Asn to Asn-GlcNAc mutation) at each of the indicated positions within the WW domain of the human protein Pin 1 (WW). Positions where glycosylation is predicted or observed to stabilize WW are highlighted in green; positions where glycosylation is predicted or observed to destabilize WW are highlighted in red (ref. 52). WW domain structure rendered in Pymol (PDB code: 1PIN).
The coarse-grained native topology model predicts that conjugation of an excluded volume glycan to Asn will be stabilizing to Pin WW, provided that the conjugation site is not highly-structured, and thus prone to steric clashes between the glycan and protein side-chain or backbone groups (Figure 7A).52 For example, the model predicted that glycosylation would stabilize WW at position 14 in β-strand 1, at positions 17, 18, 19, and 20 in the flexible reverse turn 1, and at position 30 in reverse turn 2. In contrast, the model predicted that glycosylation would destabilize WW at buried and/or structured positions like positions 23 and 26 in β-strand 2 and position 33 in β-strand 3.
We replaced wild-type residues at each of these positions in WW with either Asn or Asn-GlcNAc, and evaluated the energetic consequences of the Asn to Asn-GlcNAc mutation at each position (Figure 7B).52 In agreement with the model, GlcNAcylation at positions 23, 26, and 33 is destabilizing to WW, possibly due to disruption of important stabilizing interactions in the native-state structural ensemble via generic glycan-based excluded volume effects. In contrast, the model was less accurate at predicting positions within the WW domain at which glycosylation would be stabilizing. GlcNAcylation at positions 20 and 30 was stabilizing to WW (as predicted by the model),52 though notably by a much smaller amount than the stability gained by HsCD2ad upon GlcNAcylation.65 However, GlcNAcylation at positions 17, 18, and 19 destabilized WW, even though the model predicted the opposite. Positions 17, 18, and 19 are in a flexible reverse turn, exactly the sort of place where one would predict that excluded volume effects would be most stabilizing. One can debate the possible explanations for this observation, but the results are clear: increased thermodynamic stability and increased folding rate (whether from denatured state destabilization or from simultaneous stabilization of the native and transition states) are not general consequences of glycosylation, even when glycosylation occurs within a flexible loop, where generic glycan-based excluded volume effects would be predicted to be most influential.52 Clearly, factors other than glycan-based excluded volume effects strongly influence the energetic consequences of glycosylation. We hypothesized that these factors include the presence or absence of specific stabilizing interactions between the glycan and protein backbone or side-chain groups.89
Testing the hypothesis that the sequence and structural context of N-glycosylation is important for stabilization
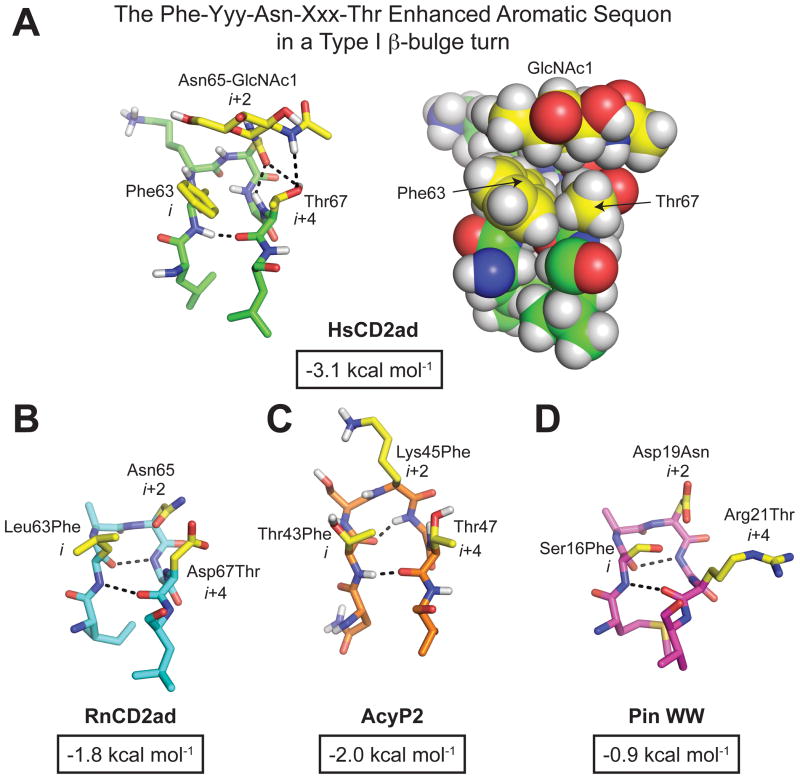
Because generic excluded volume effects failed to accurately predict the energetic consequences of N-glycosylation, we returned to the hypothesis that specific stabilizing interactions between the protein and the glycan contribute substantially to the stabilizing impact of glycosylation on HsCD2ad. A closer look at the published NMR-based structure of HsCD2ad59 reveals that the glycosylated Asn65 occupies the i+2 position of a five-residue type I β-turn with a G1 bulge, hereafter called a type I β-bulge turn (Figure 8A). Phe63 occupies the i position, Gly66 resides at the i+3 position (and adopts the G1 bulge conformation), and Thr67, a component of the sequon, occupies the i+4 position (Figure 8A). The type I β-bulge turn places the Phe63, Asn65, and Thr67 side chains in proximity on the same face of the reverse turn, where, according to the NMR data, the Phe63 side chain interacts with the hydrophobic α face of the first GlcNAc of the glycan (GlcNAc1), and with the Thr67 side chain (Figure 8A; right). The NMR data also show that Lys61, which occupies the i−2 position (two residues before the start of the five-residue type I β-bulge turn), is close to the second GlcNAc (i.e., GlcNAc2) of the glycan (Figure 3A). As discussed above, the energetic contributions of these interactions had not previously been assessed directly, although mutational studies and functional assays suggested that they might play a key role in mediating the large increase in stability experienced by HsCD2ad upon glycosylation.59,65
Figure 8.
(A) Stick and space-filling representations of the Phe-Yyy-Asn-Xxx-Thr enhanced aromatic sequon in the type I β-turn of HsCD2ad (PDB code: 1GYA). We also installed the Phe-Yyy-Asn-Xxx-Thr enhanced aromatic sequon in similar reverse turns in (B) RnCD2ad (PDB code: 1HNG), (C) AcyP2 (PDB code: 1APS), and (D) WW (PDB code: 2F21) via the indicated mutations at the i, i+2, and i+4 positions (side chains at these positions in each structure are highlighted in yellow). The change in folding free energy (ΔΔGf) upon glycosylation of the Phe-Yyy-Asn-Xxx-Thr enhanced aromatic sequon in each structural context is shown below each structure in an outlined box (refs. 90, 93). All structures rendered in Pymol.
We tested the hypothesis that these interactions contribute to the large stabilizing impact of N-glycosylation on HsCD2ad by making analogs of RnCD2ad, which does not require glycosylation to fold (so that the energetics of non-glycosylated variants could be directly assessed). Structural data from non-glycosylated RnCD2ad63,64 show that residues 63–67 adopt a type I β-bulge turn strictly analogous to the one observed in HsCD2ad (Figure 8B), making RnCD2ad an ideal system for testing the hypothesis that interactions between the glycan and nearby side-chains stabilize proteins. We installed the glycosylation sequon from HsCD2ad (Asn65-Gly66-Thr67) into RnCD2ad via an Asp67 to Thr mutation (Asn65 and Gly66 are already present in RnCD2ad).90 To facilitate expression of RnCD2ad variants that were glycosylated at a single position (thus simplifying our biophysical analysis), we also removed three other glycosylation sequons present at other locations in RnCD2ad via Asn to Gln and/or Asn to Asp mutations. We call this modified RnCD2ad variant RnCD2*.90 We expressed RnCD2* in E. coli to obtain the non-glycosylated form, and in Sf9 cells to obtain the glycosylated form, g-RnCD2* (“g” for N-glycosylated). We analyzed the folding energetics of these variants by fluorescence-monitored stopped-flow folding and unfolding experiments, as we had done previously for HsCD2ad.65 The resulting chevron plots reveal that RnCD2*, which has Glu at position 61 instead of Lys and Leu at position 63 instead of Phe, is not significantly stabilized by glycosylation (ΔΔGf = −0.6 ± 0.6 kcal mol−1). However, grafting in residues from HsCD2ad, at positions 61 and 63 respectively, changes the energetic impact of glycosylation on RnCD2ad. In RnCD2*-K, Glu61 has been replaced by Lys. In RnCD2*-F, Leu63 has been replaced by Phe. In RnCD2*-K,F, both Lys61 and Phe63 have been incorporated. Glycosylated g-RnCD2*-K is −1.5 ± 0.6 kcal mol−1 more stable than non-glycosylated RnCD2*-K. This effect is substantially larger than the effect of glycosylation on RnCD2*, and is consistent with the presence of a stabilizing native-state interaction between the N-glycan and Lys61. Similarly, glycosylated g-RnCD2*-F is −1.8 ± 0.4 kcal mol−1 more stable than nonglycosylated RnCD2*-F, suggesting a favorable native-state interaction between the N-glycan and Phe63. Notably, when both modifications are made, g-RnCD2*-K,F is −3.7 ± 0.6 kcal mol−1 more stable than non-glycosylated RnCD2*-K,F, suggesting that the interaction between the N-glycan and Phe and the interaction between the N-glycan and Lys contribute synergistically to stabilize RnCD2*.90 The unfolding rates of g-RnCD2*-K, g-RnCD2*-F and g-RnCD2*-K,F are 9-, 20- and 200-fold slower, respectively, than the corresponding non-glycosylated sequences, suggesting that interactions between Lys61, Phe63, and the N-glycan stabilize the native state of RnCD2ad. Notably, RnCD2* variants appear to be more efficiently glycosylated when Phe is present at position 63. These observations led us to hypothesize that the sequence Phe-Yyy-Asn(glycan)-Xxx-Thr in the context of a type I β-bulge turn forms the basis for a compact stabilizing structural motif, which we call an enhanced aromatic sequon, because of its increased propensity to be stabilized by glycosylation and to be glycosylated in cells more efficiently than conventional Asn-Xxx-Thr sequons.90
The Enhanced Aromatic Sequon: A Portable Stabilizing Structural Motif
We tested the hypothesis that the Phe-Yyy-Asn(glycan)-Xxx-Thr sequence (in the context of a type I β-bulge turn) is a portable stabilizing structural module by installing the enhanced aromatic sequon into a glycosylation-naïve five-residue reverse turn in the cytoplasmic protein human muscle acylphosphatase (AcyP2).90 This turn (comprising residues 43 to 47) is not well enough defined in the NMR structure of horse AcyP2 to discern its precise conformation;91 however, corresponding residues in the X-ray crystal structure of the homologous human common-type acylphosphatase (AcyP1) adopt a type I β-bulge turn conformation.92 After removing additional glycosylation sequons present in wild-type AcyP2 protein (via Ser to Ala mutations), we installed the enhanced aromatic sequon into human AcyP2 (Figure 8C) by placing Phe at the i position (residue 43) and Asn at the i+2 position (residue 45) (Thr is already present at the i+4 position (residue 47) in wild-type AcyP2) to generate non-glycosylated variant AcyP2*-F (expressed in E. coli) and glycosylated g-AcyP2*-F (expressed in Sf9 cells).90 We characterized the folding energetics of these AcyP2 variants via equilibrium chaotrope denaturation experiments. Glycosylated g-AcyP2*-F is −2.0 ± 0.7 kcal mol−1 more stable than non-glycosylated AcyP2*-F. To determine whether an interaction between the N-glycan and Phe contributes to this large stabilizing effect, we replaced Phe43 with Thr (the residue that occupies position 43 in wild-type AcyP2), thus generating non-glycosylated AcyP2* (expressed in E. coli) and glycosylated g-AcyP2* (expressed in Sf9 cells). Without Phe43, glycosylation has little effect on the stability of AcyP2: g-AcyP2* is +0.5 ± 0.8 kcal mol−1 less stable than AcyP2*, suggesting that the interaction between the N-glycan and Phe43 contributes −2.5 ± 1.1 kcal mol−1 to the stability of g-AcyP2*-F, even greater than its contribution to the stability of g-RnCD2*-F (−1.2 ± 0.7 kcal mol−1). These observations support the hypothesis that the Phe-Yyy-Asn-Xxx-Thr enhanced aromatic sequon is a portable stabilizing structural module that, when incorporated into an appropriate reverse turn context, allows proteins to be stabilized by glycosylation because of stabilizing native-state interactions between the N-glycan, Phe, and likely Thr. Also, as was observed for RnCD2*, the enhanced aromatic sequon in g-AcyP2*-F appears to be glycosylated more efficiently by cells than the conventional Asn-Xxx-Thr sequon in g-AcyP2*.90
The structure of the Phe-Yyy-Asn-Xxx-Thr enhanced aromatic sequon in the context of the type I β-bulge turn of HsCD2ad (Figure 8A) implies that the i+4 Thr engages in stabilizing interactions with Phe and the N-glycan. However, we were unable to test this hypothesis directly by site-directed mutagenesis in g-RnCD2*-F and g-AcyP2*-F because the i+4 Thr residue is required for efficient cellular glycosylation of these expressed glycoproteins by the oligosaccharyl transferase enzyme complex. However, structure-activity relationships involving the i+4 Thr of the Phe-Yyy-Asn-Xxx-Thr enhanced aromatic sequon (Figure 8A) can be explored by chemical synthesis, using variants of a previously characterized engineered WW domain that contains a type I β-bulge turn.79
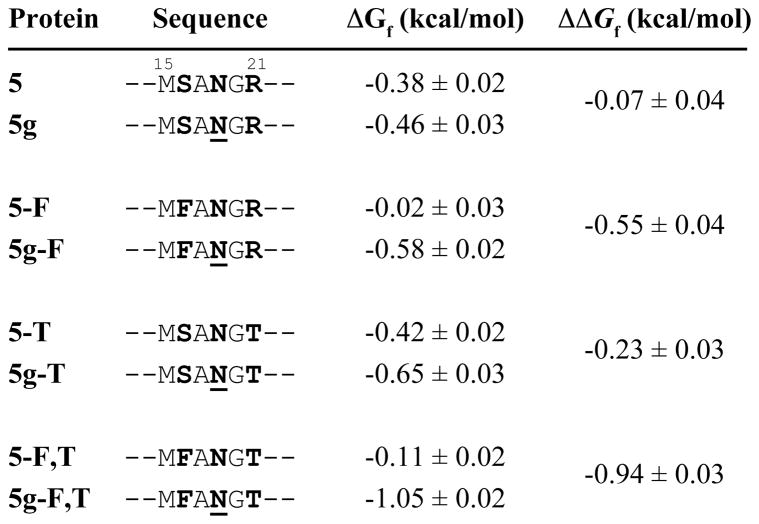
We prepared eight WW variants in which we replaced the original i, i+2, and i+4 residues of the type I β-bulge turn (Ser, Asn, and Arg, respectively) with Phe, Asn-GlcNAc, and Thr, respectively, in every possible combination (Figure 9).93 As we had done previously,52 we used a single GlcNAc residue as a surrogate for the larger oligosaccharides present in g-RnCD2*-F and g-AcyP2*-F because of experiments described above,59,65 which showed that a single GlcNAc can generate stabilizing native-state interactions. We named these variants according to the following conventions: the number five indicates that reverse turn 1 in this WW domain has five residues. Proteins in which the number 5 is followed by “g” have Asn-GlcNAc at position 19; proteins without “g” have Asn at position 19. Proteins 5, 5g, 5-T, and 5g-T have Ser at position 16. Proteins 5-F, 5g-F, 5-F,T, and 5g-F,T have Phe at position 16. Proteins 5, 5g, 5-F, and 5g-F have Arg at position 21. Proteins 5-T, 5g-T, 5-F,T, and 5g-F,T have Thr at position 21. We analyzed the folding energetics of these proteins by variable temperature far-UV circular dichroism spectropolarimetry.93
Figure 9.
Folding free energies of WW-derived glycoproteins and their non-glycosylated counterparts along with the amino acid sequence of the five-residue reverse turn for each variant (ref. 93). The i, i+2, and i+4 positions where we installed the enhanced aromatic sequon are highlighted in bold font. Tabulated data are given as mean ± standard error at 65 °C for WW variants at 10 μM in 20 mM aqueous sodium phosphate, pH 7. N = Asn-GlcNAc.
Glycosylated 5g-F,T is −0.94 ± 0.03 kcal mol−1 more stable than non-glycosylated 5-F,T at 65 °C (Figure 9). Replacing Phe16 or Thr21 with Ser16 or Arg21, respectively, dramatically reduces the stabilizing impact of glycosylation: 5g-F is only −0.55 ± 0.04 kcal mol−1 more stable than non-glycosylated 5-F, and 5g-T is only −0.23 ± 0.03 kcal mol−1 more stable than non-glycosylated 5-T. When both Phe16 and Thr21 are simultaneously replaced by Ser16 and Arg21, respectively, glycosylation is no longer stabilizing (5g and 5 are energetically indistinguishable: ΔΔGf = −0.07 ± 0.04 kcal mol−1).93
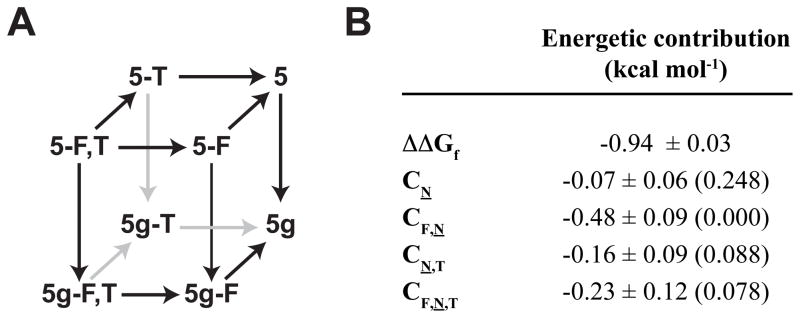
The data from 5, 5g, 5-F, 5g-F, 5-T, 5g-T, 5-F,T, and 5g-F,T comprise a triple mutant cycle (Figure 10A),94 which contains information about the stabilizing interactions between the i-position (Phe16), the i+2 position (Asn19-GlcNAc), and the i+4 position (Thr21). We extracted this information by fitting the folding free energy data for these eight WW variants (at 65 °C) to the following equation:94
Figure 10.
(A) Triple mutant cycle formed by 5g-F,T and its derivatives (ref. 93). (B) Triple mutant cycle analysis of folding free energy data at 65 °C for 5g-F,T and its derivatives. Parameters are given as mean ± standard error. P values in parentheses indicate the probability that random sampling error accounts for the difference between zero and the observed value of the parameter.
| (1) |
Equation 1 shows how the ΔGf of a given variant of 5 is related to the average ΔGf° of 5, plus a series of correction terms that account for the interactions among the amino acids at positions 16, 19, and 21. Each correction term is a product of one or more indicator variables W (which reflect whether a mutation is present in the given variant) and a free energy contribution C. WF is 0 when position 16 is Ser or 1 when it is Phe; WN is 0 when position 19 is Asn or 1 when it is Asn-GlcNAc; WT is 0 when position 21 is Arg or 1 when it is Thr. CF, CN, and CT describe the energetic consequences of the Ser16 to Phe16, Asn19 to Asn19-GlcNAc, and Arg21 to Thr21 mutations, respectively. These energies probably primarily reflect the difference in conformational preferences between Ser and Phe at position 16, Asn and Asn-GlcNAc at position 19, and Arg and Thr at position 21. CF,N, CF,T, and CN,T describe the free energies of the two-way interactions between Phe16 and Asn19-GlcNAc, between Phe16 and Thr21, and between Asn19-GlcNAc and Thr21, respectively. CF,N,T describes the energetic impact of the three-way interaction between Phe16, Asn19-GlcNAc, and Thr21. This analysis assumes that the Ser16 side chain does not interact with the side chains at positions 19 or 21, and that the Arg21 side chain does not interact with the side chains at positions 16 or 19, in any variant. This assumption is, to a first approximation, consistent with the available structural data.68,79
According to equation 1, the stabilizing effect of glycosylating the Phe-Yyy-Asn-Xxx-Thr enhanced aromatic sequon in the context of the type I β-bulge turn of 5-F,T [ΔΔGf = ΔGf(5g-F,T) − ΔGf(5-F,T)] is equal to the sum of CN, CF,N, CN,T, and CF,N,T (Figure 10B). Changing Asn19 to Asn19-GlcNAc has no substantial energetic effect (CN = −0.07 ± 0.06 kcal mol−1) on its own. In contrast, most of the stability from glycosylating this enhanced aromatic sequon comes from energetically favorable two-way interactions between Phe and Asn-GlcNAc (CF,N = −0.48 ± 0.09 kcal mol−1) and between Asn-GlcNAc and Thr (CN,T = −0.16 ± 0.09 kcal mo−1), and an energetically favorable three way interaction among Phe, Asn-GlcNAc and Thr (CF,N,T = −0.23 ± 0.11 kcal mol−1). These results reveal that the i+4-position Thr plays an important role in stabilizing the enhanced aromatic sequon by engaging in favorable native-state interactions with the i-position Phe and the i+2-position Asn-GlcNAc.93
Extending the Enhanced Aromatic Sequon Concept to Additional Reverse Turn Types
Nearly 9% of the reverse turns in the Protein Data Bank (PDB) are type I β-bulge turns.95,96 Installing the Phe-Yyy-Asn-Xxx-Thr enhanced aromatic sequon could be an attractive strategy for increasing the stability of the many proteins that already harbor type I β-bulge turns. We wondered whether other types of reverse turns might also serve as scaffolds for allowing stabilizing interactions between an N-glycan and nearby Phe and Thr side chains. Identifying such reverse turn types would expand the number of proteins that could benefit from the increased stability and possibly the increased glycosylation efficiency afforded by the enhanced aromatic sequon without having to resort to reverse turn type engineering.
The WW domain is a convenient system for testing this hypothesis, because WW variants harboring different reverse turn types in loop 1 have been structurally68,79,85 and energetically71,72,79,85 characterized. In addition to the structural data for the WW variant containing the five-residue type I β-bulge turn discussed above (Figures 8D, 11A),79 crystal structures also exist for WW domains harboring a type II β-turn within a six-residue hydrogen-bonded loop (hereafter called a six-residue turn-within-a-turn; Figure 11B),68 or a four-residue type I′ β-turn (Figure 11C).79
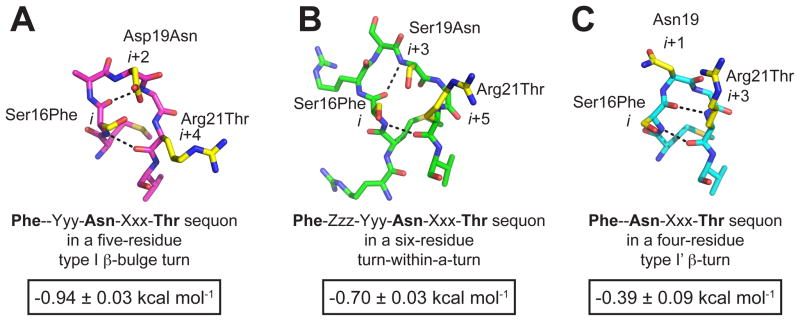
Figure 11.
We installed enhanced aromatic sequons (A) in a type I β-bulge turn (PDB code: 2F21) in WW variant 5 using the sequence Phe-Yyy-Asn-Xxx-Thr; (B) in a six-residue turn-within-a-turn (PDB code: 1PIN) in WW variant 6 using the sequence Phe-Zzz-Yyy-Asn-Xxx-Thr; and (C) in a type I′ β-turn (PDB code: 1ZCN) in WW variant 4 using the sequence Phe-Asn-Xxx-Thr, using the mutations indicated at each position. The stick representation of each reverse turn was rendered in Pymol, with hydrogen bonds shown as black dashed lines. Positions where we introduced mutations to install the enhanced aromatic sequons are highlighted in yellow. The change in folding free energy (ΔΔGf) upon glycosylation of each enhanced aromatic sequon is shown below each structure in a black outlined box.
The six-residue turn-within-a-turn is an unusual reverse turn; only 0.1% of all reverse turns in the PDB have this conformation.95 In contrast, 11% of the reverse turns in the PDB are four-residue type I′ β-turns.95 Notably, both of these turn types appear to be suitable scaffolds for the enhanced aromatic sequon. The β carbon’s (Cβ’s) of the side chains at the i, i+3, and i+5 positions of the six-residue turn-within-a-turn are within 5 Å of each other (Figure 11B), as are the Cβ’s of the side chains at the i, i+1, and i+3 positions of the four-residue type I′ β-turn (Figure 11C). We hypothesized that installing Phe, the N-glycan, and Thr, respectively, at these positions would enable stabilizing tripartite interactions in each reverse turn, reminiscent of those observed in g-RnCD2*-F, g-AcyP2*-F, and 5g-F,T. The enhanced aromatic sequon applied to the six-residue turn-within-a-turn would have the sequence Phe-Zzz-Yyy-Asn-Xxx-Thr93 (note that Phe is three positions before Asn, instead of two as in the five-residue type I β-bulge turn90). The enhanced aromatic sequon applied to the four-residue type I′ β-turn would have the sequence Phe-Asn-Xxx-Thr.93 If successful, applying the enhanced aromatic sequon concept to these reverse turn types would essentially double the number of proteins that could be stabilized by glycosylated enhanced aromatic sequons without resorting to reverse turn type reengineering.
We installed the Phe-Zzz-Yyy-Asn-Xxx-Thr enhanced aromatic sequon in the six-residue turn-within-a-turn by preparing variant 6-F,T, which has Phe at the i position, Asn at the i+3 position, and Thr at the i+5 position (Figure 11B). Chemical GlcNAcylation of 6-F,T at the i+3-position Asn gives variant 6g-F,T, which is −0.70 ± 0.03 kcal mol−1 more stable than non-glycosylated 6-F,T. As was the case for the Phe-Yyy-Asn-Xxx-Thr enhanced aromatic sequon in the type I β-bulge turn, replacement of the i-position Phe with Ser and/or replacement of the i+5-position Thr with Arg significantly decreases the stabilizing impact of glycosylation. In fact without Phe and Thr, glycosylation of Asn in the six-residue turn is actually destabilizing.90
We installed the Phe-Asn-Xxx-Thr enhanced aromatic sequon in the four-residue type I′ β-turn by preparing variant 4-F,T, which has Phe at the i-position, Asn at the i+1-position, and Thr at the i+3-position (Figure 11C). Chemical GlcNAcylation of 4-F,T affords variant 4g-F,T, which is −0.39 ± 0.09 kcal mol−1 more stable than 4-F,T.93 This effect decreases substantially when the i-position Phe is replaced by Ser, though the stabilization remains significant when the i+3-position Thr is replaced by Arg, suggesting that a two-way interaction between Thr and Asn-GlcNAc does not play as strong a role in the stabilizing effect of glycosylating the Phe-Asn-Xxx-Thr enhanced aromatic sequon in the type I′ β-turn as it does for the other enhanced aromatic sequons in their correlated reverse turn contexts.
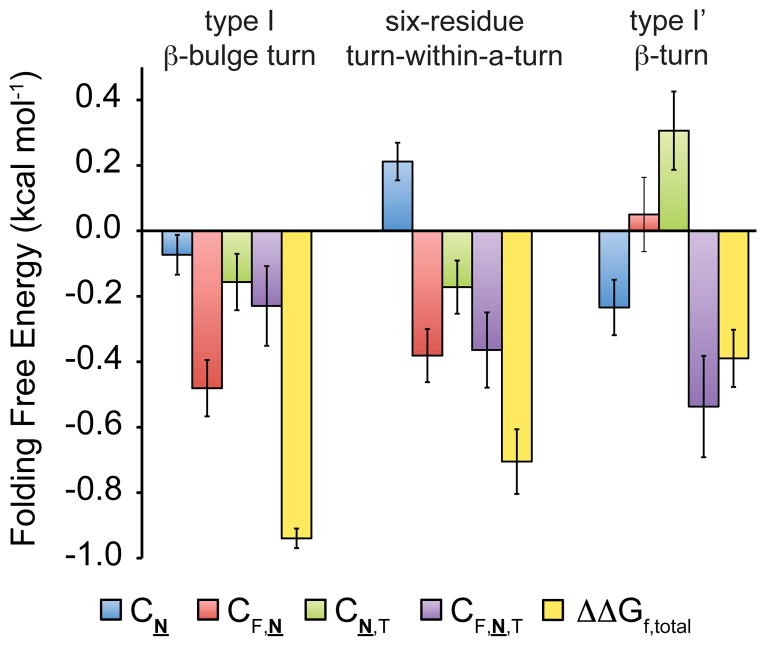
We performed a triple mutant cycle analysis on 6g-F,T and 4g-F,T and their derivatives, as we described above for 5g-F,T and its derivatives (Figure 12).90,93 Comparison of the results reveals differences in the two- and three-way interactions that stabilize each enhanced aromatic sequon in its correlated reverse turn type (Figure 12). Changing Asn to Asn-GlcNAc affects each turn type differently: it stabilizes the four-residue type I′ β-turn (CN = −0.23 ± 0.08 kcal mol−1), does not affect the five-residue type I β-bulge turn substantially (CN = −0.07 ± 0.06 kcal mol−1), and destabilizes the type II β-turn within a six-residue loop (CN = 0.21 ± 0.06 kcal mol−1). Asn-GlcNAc may have backbone dihedral angle preferences that are more compatible with the i+1 position of a type I′ β-turn than with the i+2 position of a type I β-bulge turn or with the i+3 position of a type II β-turn. Such preferences would differ substantially from those of Asn itself,97 which is favored at i+1 in a type I′ β-turn, and at i+2 in a type I β-turn, but not at i+3 in a type II β-turn 98.
Figure 12.
Triple mutant cycle analysis of data from 4g-F,T, 5g-F,T, 6g-F,T, and their derivatives (ref. 93). The change in folding free energy (ΔΔGf,total) upon glycosylation of each enhanced aromatic sequon in its correlated reverse turn type can be attributed to energetic contributions from the Asn19 to Asn-GlcNAc mutation (ΔΔGN19N), the two-way interaction between Phe and Asn-GlcNAc, the two-way interaction between Asn-GlcNAc and Thr, and the three way interaction between Phe, Asn-GlcNAc, and Thr.
The two-way interaction between Phe and Asn-GlcNAc stabilizes the five-residue type I β-bulge turn (CF,N = −0.48 ± 0.09 kcal mol−1) and the six-residue turn-within-a-turn (CF,N = −0.38 ± 0.08 kcal mol−1), but does not substantially change the stability of the four-residue type I′ β-turn (CF,N = 0.05 ± 0.11 kcal mol−1) (Figure 12). It is possible that the backbone flexibility and/or direction of the Cα-Cβ bond vectors in the five- and six-residue turns allow for better two-way interactions between Phe and Asn-GlcNAc than are possible in the four-residue turn.
The two-way interaction between Asn-GlcNAc and Thr stabilizes the five- and six-residue turns (CN,T = −0.16 ± 0.09 kcal mol−1 and −0.17 ± 0.08 kcal mol−1, respectively), but substantially destabilizes the four-residue reverse turn (CN,T = 0.31 ± 0.12 kcal mol−1) (Figure 12). The glycosylated enhanced aromatic sequon in an analogous type I β-bulge turn in HsCD2ad involves three hydrogen bonds between Thr and Asn-GlcNAc: one between the Thr side-chain oxygen and the amide proton of the 2-acetamido group of GlcNAc, and two between the Asn side-chain amide carbonyl oxygen and the backbone amide and side-chain hydroxyl protons of Thr.59 The differences we observe between the CN,T values in the four-, five-, and six-residue turn contexts could reflect the presence of analogous hydrogen bonds in the type I β-bulge turn of 5g-F,T and in six-residue loop of 6g-F,T, but not in the type I′ β-turn of 4g-F,T (Figure 12).
Despite these differences, the CF,N,T values for the four-residue type I′ β-turn (CF,N,T = −0.54 ± 0.15 kcal mol−1), the five-residue type I β-bulge turn (CF,N,T = −0.23 ± 0.12 kcal mol−1), and the six-residue turn-within-a-turn (CF,N,T = −0.36 ± 0.11 kcal mol−1) suggest that the three-way interaction between Phe, Asn-GlcNAc, and Thr stabilizes each reverse turn by similar amounts (Figure 12). By successfully applying the new Phe-Asn-Xxx-Thr enhanced aromatic sequon to the type I′ β-turn (which comprises nearly 11% of all reverse turns in the PDB), we have doubled the number of candidate proteins in which enhanced aromatic sequons can be employed without altering the conformation or the number of residues comprising the native reverse turn.99,100 It remains to characterize the structure of the enhanced aromatic sequons in the context of 4-, 5- and 6-residue turns to gain further insight into the underlying structure-free energy relationships.
Conclusions
Adding N-glycans to naïve sites within proteins can be an attractive strategy for increasing their thermodynamic stability. This approach has been used in the development of protein-based drugs,42,43,45,101 where new N-glycans can extend serum half-life 46–48 and shelf-life, owing in part to increased protease resistance,50 decreased aggregation propensity, and increased compensation for the destabilizing effect of methionine oxidation.102 Historically, efforts to increase protein stability via N-glycosylation have depended on a trial-and-error approach,48,51 which has resulted in unpredictable energetic consequences.52–54 By pairing each enhanced aromatic sequon with an appropriate reverse turn context (Figure 13), we have provided engineering guidelines by which N-glycosylation can reliably stabilize the native states of proteins. These stabilizing enhanced-aromatic-sequon/reverse-turn pairings include the Phe-Asn-Xxx-Thr sequon for type I′ β-turns, the Phe-Yyy-Asn-Xxx-Thr sequon for type I β-bulge turns, and the Phe-Yyy-Zzz-Asn-Xxx-Thr sequon for the six-residue turn-within-a-turn, Figure 13. Each enhanced-aromatic-sequon/reverse-turn pairing in Figure 13 appears to facilitate stabilizing native-state interactions between Phe, Asn-GlcNAc and Thr, even in glycosylation-naïve proteins that have not evolved to optimize specific protein-carbohydrate interactions.90 However, it remains to provide direct structural evidence for these native state stabilizing tripartite interactions in each enhanced aromatic sequence/reverse turn context. Incorporating these stabilizing interactions into proteins should lower the concentration of misfolded or unfolded protein, thereby enhancing the previously described pharmacokinetic benefits of N-glycosylation, and making the modified proteins more useful in research and pharmaceutical applications.
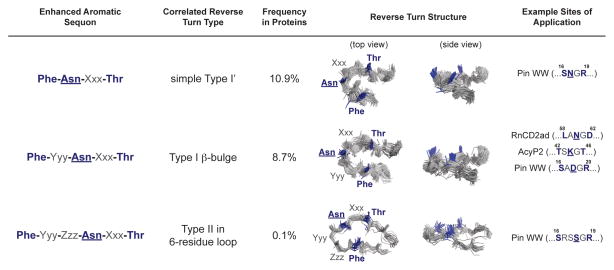
Figure 13.
The enhanced aromatic sequons in their correlated reverse turn types. The first three columns list the sequence of each enhanced aromatic sequon, its correlated reverse turn type, and the frequency of that reverse turn type in the non-redundant protein structural database (<40% sequence homology, see ref. 95). In the fourth and fifth columns, the sequences are mapped onto an overlay of the main chains of members of each reverse turn family. The side chains where Phe, Asn(glycan), and Thr should be incorporated are highlighted in blue. The last column lists examples of proteins that we have studied that contain the reverse turn types that are compatible with the indicated enhanced aromatic sequons.
Acknowledgments
This work was supported in part the Skaggs Institute for Chemical Biology, the Lita Annenberg Hazen Foundation, and by National Institutes of Health (NIH) grant GM051105 to J.W.K. and E.T.P., and AI072155 to C-H. W. J.L.P. was supported in part by an NIH post-doctoral fellowship (F32 GM086039) while this work was carried out at The Scripps Research Institute.
Contributor Information
Joshua L. Price, Email: jlprice@chem.byu.edu.
Evan T. Powers, Email: epowers@scripps.edu.
Jeffery W. Kelly, Email: jkelly@scripps.edu.
References
- 1.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Yan A, Lennarz WJ. J Biol Chem. 2005;280:3121–3124. doi: 10.1074/jbc.R400036200. [DOI] [PubMed] [Google Scholar]
- 3.Kelleher DJ, Gilmore R. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- 4.Helenius A, Aebi M. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 5.Molinari M. Nat Chem Biol. 2007;3:313–320. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- 6.Gürkan C, Stagg SM, LaPointe P, Balch WE. Nat Rev Mol Cell Biol. 2006;7:727–738. doi: 10.1038/nrm2025. [DOI] [PubMed] [Google Scholar]
- 7.Sekijima Y, Wiseman RL, Matteson J, Hammarström P, Miller SR, Sawkar AR, Balch WE, Kelly JW. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 8.Wiseman RL, Powers ET, Buxbaum JN, Kelly JW, Balch WE. Cell. 2007;131:809–821. doi: 10.1016/j.cell.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 9.Caramelo JJ, Castro OA, de Prat-Gay G, Parodi AJ. J Biol Chem. 2004;279:46280–46285. doi: 10.1074/jbc.M408404200. [DOI] [PubMed] [Google Scholar]
- 10.Taylor SC, Ferguson AD, Bergeron JJM, Thomas DY. Nat Struct Mol Biol. 2004;11:128–134. doi: 10.1038/nsmb715. [DOI] [PubMed] [Google Scholar]
- 11.Molinari M, Galli C, Vanoni O, Arnold SM, Kaufman RJ. Mol Cell. 2005;20:503–512. doi: 10.1016/j.molcel.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 12.Ritter C, Quirin K, Kowarik M, Helenius A. EMBO J. 2005;24:1730–1738. doi: 10.1038/sj.emboj.7600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lederkremer GZ, Glickman MH. Trends Biochem Sci. 2005;30:297–303. doi: 10.1016/j.tibs.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Sousa MC, Ferrero-Garcia MA, Parodi AJ. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- 15.Spiro RG, Zhu Q, Bhoyroo V, Söling H-D. J Biol Chem. 1996;271:11588–11594. doi: 10.1074/jbc.271.19.11588. [DOI] [PubMed] [Google Scholar]
- 16.Cabral CM, Liu Y, Sifers RN. Trends Biochem Sci. 2001;26:619–624. doi: 10.1016/s0968-0004(01)01942-9. [DOI] [PubMed] [Google Scholar]
- 17.Moremen KW, Molinari M. Curr Opin Chem Biol. 2006;16:592–599. doi: 10.1016/j.sbi.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong DST, Mu T-W, Palmer AE, Kelly JW. Nat Chem Biol. 2010;6:424–432. doi: 10.1038/nchembio.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parthasarathy R, Subramanian S, Boder ET, Discher DE. Biotechnol Bioeng. 2006;93:159–168. doi: 10.1002/bit.20684. [DOI] [PubMed] [Google Scholar]
- 20.Wormald MR, Wooten EW, Bazzo R, Edge CJ, Feinstein A, Rademacher TW, Dwek RA. Eur J Biochem. 1991;198:131–139. doi: 10.1111/j.1432-1033.1991.tb15995.x. [DOI] [PubMed] [Google Scholar]
- 21.Laszlo O, Jr, Jan T, Emma K, Laszlo U, Henry HM, Miklos H. Int J Pept Protein Res. 1991;38:476–482. [Google Scholar]
- 22.Laczko I, Hollosi M, Urge L, Ugen KE, Weiner DB, Mantsch HH, Thurin J, Otvos L. Biochemistry. 1992;31:4282–4288. doi: 10.1021/bi00132a019. [DOI] [PubMed] [Google Scholar]
- 23.Urge L, Gorbics L, Otvos L., Jr Biochem Biophys Res Commun. 1992;184:1125–1132. doi: 10.1016/0006-291x(92)90709-t. [DOI] [PubMed] [Google Scholar]
- 24.Rickert KW, Imperiali B. Chem Biol. 1995;2:751–759. doi: 10.1016/1074-5521(95)90103-5. [DOI] [PubMed] [Google Scholar]
- 25.Imperiali B, Rickert KW. Proc Natl Acad Sci USA. 1995;92:97–101. doi: 10.1073/pnas.92.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Live DH, Kumar RA, Beebe X, Danishefsky SJ. Proc Natl Acad Sci USA. 1996;93:12759–12761. doi: 10.1073/pnas.93.23.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Connor SE, Imperiali B. Chem Biol. 1996;3:803–812. doi: 10.1016/s1074-5521(96)90064-2. [DOI] [PubMed] [Google Scholar]
- 28.O’Connor SE, Imperiali B. J Am Chem Soc. 1997;119:2295–2296. [Google Scholar]
- 29.O’Connor SE, Imperiali B. Chem Biol. 1998;5:427–437. doi: 10.1016/s1074-5521(98)90159-4. [DOI] [PubMed] [Google Scholar]
- 30.Imperiali B, O’Connor SE. Curr Opin Chem Biol. 1999;3:643–649. doi: 10.1016/s1367-5931(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 31.O’Connor SE, Pohlmann J, Imperiali B, Saskiawan I, Yamamoto K. J Am Chem Soc. 2001;123:6187–6188. doi: 10.1021/ja010094s. [DOI] [PubMed] [Google Scholar]
- 32.Wormald MR, Petrescu A-J, Pao Y-L, Glithero A, Elliott T, Dwek RA. Chem Rev. 2002;102:371–386. doi: 10.1021/cr990368i. [DOI] [PubMed] [Google Scholar]
- 33.Bosques CJ, Tschampel SM, Woods RJ, Imperiali B. J Am Chem Soc. 2004;126:8421–8425. doi: 10.1021/ja049266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi H. Trends Glycosci Glycotech. 2002;14:139–151. [Google Scholar]
- 35.Mitra N, Sinha S, Ramya TNC, Surolia A. Trends Biochem Sci. 2006;31:156–163. doi: 10.1016/j.tibs.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Petrescu A-J, Milac A-L, Petrescu S, Dwek RA, Wormald MR. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 37.Narhi LO, Arakawa T, Aoki KH, Elmore R, Rohde MF, Boone T, Strickland TW. J Biol Chem. 1991;266:23022–23026. [PubMed] [Google Scholar]
- 38.Wang C, Eufemi M, Turano C, Giartosio A. Biochemistry. 1996;35:7299–7307. doi: 10.1021/bi9517704. [DOI] [PubMed] [Google Scholar]
- 39.Wormald MR, Dwek RA. Struct Fold Des. 1999;7:R155–R160. doi: 10.1016/s0969-2126(99)80095-1. [DOI] [PubMed] [Google Scholar]
- 40.Shental-Bechor D, Levy Y. Proc Natl Acad Sci USA. 2008;105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shental-Bechor D, Levy Y. Curr Opin Struct Biol. 2009;19:524–533. doi: 10.1016/j.sbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 42.Walsh G, Jefferis R. Nat Biotechnol. 2006;24:1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 43.Sinclair AM, Elliott S. J Pharm Sci. 2005;94:1626–1635. doi: 10.1002/jps.20319. [DOI] [PubMed] [Google Scholar]
- 44.Li HJ, d’Anjou M. Curr Opin Biotech. 2009;20:678–684. doi: 10.1016/j.copbio.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 45.Sola RJ, Griebenow K. Biodrugs. 2010;24:9–21. doi: 10.2165/11530550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Egrie JC, Dwyer E, Browne JK, Hitz A, Lykos MA. Exp Hematol. 2003;31:290–299. doi: 10.1016/s0301-472x(03)00006-7. [DOI] [PubMed] [Google Scholar]
- 47.Su DM, Zhao HL, Xia HZ. Int J Hematol. 2010;91:238–244. doi: 10.1007/s12185-010-0496-x. [DOI] [PubMed] [Google Scholar]
- 48.Ceaglio N, Etcheverrigaray M, Kratje R, Oggero A. Biochimie. 2008;90:437–449. doi: 10.1016/j.biochi.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 49.Endo Y, Nagai H, Watanabe Y, Ochi K, Takagi T. J Biochem. 1992;112:700–706. doi: 10.1093/oxfordjournals.jbchem.a123961. [DOI] [PubMed] [Google Scholar]
- 50.Raju TS, Scallon BJ. Biochem Biophys Res Commun. 2006;341:797–803. doi: 10.1016/j.bbrc.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 51.Elliott S, Chang D, Delorme E, Eris T, Lorenzini T. J Biol Chem. 2004;279:16854–16862. doi: 10.1074/jbc.M311095200. [DOI] [PubMed] [Google Scholar]
- 52.Price JL, Shental-Bechor D, Dhar A, Turner MJ, Powers ET, Gruebele M, Levy Y, Kelly JW. J Am Chem Soc. 2010;132:15359–15367. doi: 10.1021/ja106896t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hackenberger CPR, Friel CT, Radford SE, Imperiali B. J Am Chem Soc. 2005;127:12882–12889. doi: 10.1021/ja051855k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen MM, Bartlett AI, Nerenberg PS, Friel CT, Hackenberger CPR, Stultz CM, Radford SE, Imperiali B. Proc Natl Acad Sci USA. 2010;107:22528–22533. doi: 10.1073/pnas.1015356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tangye SG, Phillips JH, Lanier LL. Semin Immunol. 2000;12:149–157. doi: 10.1006/smim.2000.0217. [DOI] [PubMed] [Google Scholar]
- 56.Williams AF, Barclay AN. Annual Rev of Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 57.Wyss DF, Withka JM, Knoppers MH, Sterne KA, Recny MA, Wagner G. Biochemistry. 1993;32:10995–11006. doi: 10.1021/bi00092a008. [DOI] [PubMed] [Google Scholar]
- 58.Bodian DL, Jones EY, Harlos K, Stuart DI, Davis SJ. Structure. 1994;2:755–766. doi: 10.1016/s0969-2126(94)00076-x. [DOI] [PubMed] [Google Scholar]
- 59.Wyss DF, Choi JS, Li J, Knoppers MH, Willis KJ, Arulanandam ARN, Smolyar A, Reinherz EL, Wagner G. Science. 1995;269:1273–1278. doi: 10.1126/science.7544493. [DOI] [PubMed] [Google Scholar]
- 60.Recny MA, Luther MA, Knoppers MH, Neidhardt EA, Khandekar SS, Concino MF, Schimke PA, Francis MA, Moebius U, Reinhold BB. J Biol Chem. 1992;267:22428–22434. [PubMed] [Google Scholar]
- 61.Arulanandam AR, Withka JM, Wyss DF, Wagner G, Kister A, Pallai P, Recny MA, Reinherz EL. Proc Natl Acad Sci USA. 1993;90:11613–11617. doi: 10.1073/pnas.90.24.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wyss DF, Choi JS, Wagner G. Biochemistry. 1995;34:1622–1634. doi: 10.1021/bi00005a019. [DOI] [PubMed] [Google Scholar]
- 63.Driscoll PC, Cyster JG, Campbell ID, Williams AF. Nature. 1991;353:762–765. doi: 10.1038/353762a0. [DOI] [PubMed] [Google Scholar]
- 64.Jones EY, Davis SJ, Williams AF, Harlos K, Stuart DI. Nature. 1992;360:232–239. doi: 10.1038/360232a0. [DOI] [PubMed] [Google Scholar]
- 65.Hanson SR, Culyba EK, Hsu T-L, Wong C-H, Kelly JW, Powers ET. Proc Natl Acad Sci USA. 2009;106:3131–3136. doi: 10.1073/pnas.0810318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 67.Reinherz EL, Li J, Smoylar A, Wyss DF, Knoppers MH, Willis KJ, Arulanandam ARN, Choi JS, Wagner G. Science. 1996;273:1242–1242. doi: 10.1126/science.273.5279.1242. [DOI] [PubMed] [Google Scholar]
- 68.Ranganathan R, Lu KP, Hunter T, Noel JP. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 69.Koepf EK, Petrassi HM, Sudol M, Kelly JW. Protein Sci. 1999;8:841–853. doi: 10.1110/ps.8.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koepf EK, Petrassi HM, Ratnaswamy G, Huff ME, Sudol M, Kelly JW. Biochemistry. 1999;38:14338–14351. doi: 10.1021/bi991105l. [DOI] [PubMed] [Google Scholar]
- 71.Jäger M, Nguyen H, Crane JC, Kelly JW, Gruebele M. J Mol Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 72.Kaul R, Angeles AR, Jäger M, Powers ET, Kelly JW. J Am Chem Soc. 2001;123:5206–5212. doi: 10.1021/ja0102890. [DOI] [PubMed] [Google Scholar]
- 73.Deechongkit S, Kelly JW. J Am Chem Soc. 2002;124:4980–4986. doi: 10.1021/ja0123608. [DOI] [PubMed] [Google Scholar]
- 74.Kaul R, Deechongkit S, Kelly JW. J Am Chem Soc. 2002;124:11900–11907. doi: 10.1021/ja020675x. [DOI] [PubMed] [Google Scholar]
- 75.Kowalski JA, Kiu K, Kelly JW. Biopolymers. 2002;63:111–121. doi: 10.1002/bip.10020. [DOI] [PubMed] [Google Scholar]
- 76.Nguyen H, Jäger M, Moretto A, Gruebele M, Kelly JW. Proc Natl Acad Sci USA. 2003;100:3948–3953. doi: 10.1073/pnas.0538054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deechongkit S, Nguyen H, Powers ET, Dawson PE, Gruebele M, Kelly JW. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 78.Nguyen H, Jäger M, Kelly JW, Gruebele M. J Phys Chem B. 2005;109:15182–15186. doi: 10.1021/jp052373y. [DOI] [PubMed] [Google Scholar]
- 79.Jäger M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, Noel JP, Gruebele M, Kelly JW. Proc Natl Acad Sci USA. 2006;103:10648–10653. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jäger M, Dendle M, Fuller AA, Kelly JW. Protein Sci. 2007;16:2306–2313. doi: 10.1110/ps.072904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharpe T, Jonsson AL, Rutherford TJ, Daggett V, Fersht AR. Protein Sci. 2007;16:2233–2239. doi: 10.1110/ps.073004907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jäger M, Nguyen H, Dendle M, Gruebele M, Kelly JW. Protein Sci. 2007;16:1495–1501. doi: 10.1110/ps.072775507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jäger M, Deechongkit S, Koepf EK, Nguyen H, Gao J, Powers ET, Gruebele M, Kelly JW. Biopolymers. 2008;90:751–758. doi: 10.1002/bip.21101. [DOI] [PubMed] [Google Scholar]
- 84.Liu F, Du D, Fuller AA, Davoren JE, Wipf P, Kelly JW, Gruebele M. Proc Natl Acad Sci USA. 2008;105:2369–2374. doi: 10.1073/pnas.0711908105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fuller AA, Du D, Liu F, Davoren JE, Bhabha G, Kroon G, Case DA, Dyson HJ, Powers ET, Wipf P, Gruebele M, Kelly JW. Proc Natl Acad Sci USA. 2009;106:11067–11072. doi: 10.1073/pnas.0813012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao J, Bosco DA, Powers ET, Kelly JW. Nat Struct Mol Biol. 2009;16:684–690. doi: 10.1038/nsmb.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jäger M, Dendle M, Kelly JW. Protein Sci. 2009;18:1806–1813. doi: 10.1002/pro.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang VT, Crispin M, Aricescu AR, Harvey DJ, Nettleship JE, Fennelly JA, Yu C, Boles KS, Evans EJ, Stuart DI, Dwek RA, Jones EY, Owens RJ, Davis SJ. Structure. 2007;15:267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barb AW, Borgert AJ, Liu MA, Barany G, Live D. Methods in Enzymology, Vol 478: Glycomics. 2010;478:365–388. doi: 10.1016/S0076-6879(10)78018-6. [DOI] [PubMed] [Google Scholar]
- 90.Culyba EK, Price JL, Hanson SR, Dhar A, Wong CH, Gruebele M, Powers ET, Kelly JW. Science. 2011;331:571–575. doi: 10.1126/science.1198461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pastore A, Saudek V, Ramponi G, Williams RJP. Journal of Molecular Biology. 1992;224:427–440. doi: 10.1016/0022-2836(92)91005-a. [DOI] [PubMed] [Google Scholar]
- 92.Yeung RCY, Lam SY, Wong KB. Acta Crystallographica Section F-Structural Biology and Crystallization Communications. 2006;62:80–82. doi: 10.1107/S174430910504145X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Price JL, Powers DL, Powers ET, Kelly JW. Proc Nat Acad Sci USA. 2011;108:14127–14132. doi: 10.1073/pnas.1105880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Horovitz A, Fersht AR. J Mol Biol. 1992;224:733–740. doi: 10.1016/0022-2836(92)90557-z. [DOI] [PubMed] [Google Scholar]
- 95.Oliva B, Bates PA, Querol E, Aviles FX, Sternberg MJE. Journal of Molecular Biology. 1997;266:814–830. doi: 10.1006/jmbi.1996.0819. [DOI] [PubMed] [Google Scholar]
- 96.Sibanda BL, Blundell TL, Thornton JM. Journal of Molecular Biology. 1989;206:759–777. doi: 10.1016/0022-2836(89)90583-4. [DOI] [PubMed] [Google Scholar]
- 97.Hovmoller S, Zhou T, Ohlson T. Acta Crystallographica Section D-Biological Crystallography. 2002;58:768–776. doi: 10.1107/s0907444902003359. [DOI] [PubMed] [Google Scholar]
- 98.Hutchinson EG, Thornton JM. Protein Science. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. Annual Review of Biochemistry. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 100.Gellman SH. Curr Opin Chem Biol. 1998;2:717–725. doi: 10.1016/s1367-5931(98)80109-9. [DOI] [PubMed] [Google Scholar]
- 101.Li HJ, d’Anjou M. Current Opinion in Biotechnology. 2009;20:678–684. doi: 10.1016/j.copbio.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 102.Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, Li L, Brems DN, Remmele RL. Biochemistry. 2008;47:5088–5100. doi: 10.1021/bi702238b. [DOI] [PubMed] [Google Scholar]