Abstract
In 2000 the United States launched the National Nanotechnology Initiative and, along with it, a well-defined set of goals for nanomedicine. This Perspective looks back at the progress made toward those goals, within the context of the changing landscape in biomedicine that has occurred over the past 15 years, and considers advances that are likely to occur during the next decade. In particular, nanotechnologies for health-related genomics and single-cell biology, inorganic and organic nanoparticles for biomedicine, and wearable nanotechnologies for wellness monitoring are briefly covered.
Keywords: nanotechnology, biotechnology, nanomedicine
When rowing a boat across a big lake, you can paddle for a long time, and the far shore will continue to appear almost as far away as when you started. However, when you look back to your starting point, the amount of distance traveled can be startling. In 2000, Neal Lane, then President Clinton’s Science Advisor, presented the first Implementation Plan for the US National Nanotechnology Initiative (NNI) (1). That document laid out some clear goals for nanomedicine:
-
i)
Rapid, more efficient genome sequencing enabling a revolution in diagnostics and therapeutics
-
ii)
Effective and less expensive health care using remote and in vivo devices
-
iii)
New formulations and routes for drug delivery that enormously broaden their therapeutic potential by targeting the delivery of new types of medicine to previously inaccessible sites in the body
-
iv)
More durable rejection-resistant artificial tissues and organs
-
v)
Devices that enable vision and hearing aids
-
vi)
Sensor systems that detect emerging disease in the body, which ultimately will shift the focus of patient care from disease treatment to early detection and prevention.
The goals of the 2000 NNI have largely stood the test of time and even capture some of today’s grand challenges in the health sciences. I’ll look back at selected items from this list and provide brief descriptions of how far we have come. For most of these goals, nanotechnologies have played supporting, albeit gradually increasing, roles. For example, any discussion of diagnostics, early disease detection, and disease prevention would likely lead with the amazing progress in the “-omics” fields and the emerging efforts to define “wellness” quantitatively that ultimately will provide the baseline against which disease is measured (2). However, a little digging reveals that nanotech is playing increasingly important roles in the emerging generation of omics tools. For other goals, such as those associated with new formulations and routes for drug delivery, nanotech has played a major role, even if widespread clinical applications are still on the horizon. Just like the rower’s view from the lake, the various end goals of the NNI are still a ways off, but the distance covered is impressive.
Although the goals of the 2000 NNI still appear prescient, there have been major breakthroughs in medicine and biotechnology that weren’t anticipated 15 y ago. For example, single-cell biology was not really a field in 2000, but now it is one of the most rapidly evolving assay biotechnologies, with potentially disruptive implications in terms of how we think about biological systems and how we understand human disease and health states. As a second example, until a couple of years ago the idea of harnessing a patient’s immune system as a powerful anticancer drug was more or less a backwater of cancer research. Immune checkpoint inhibitors, which now are dominating new cancer clinical trials, with several recent Food and Drug Administration approvals, were only being identified in 2000 (3). However, cancer immunotherapy (4) in the forms of both checkpoint inhibitors and cell-based therapies is increasingly altering the dialogue of “cancer treatments” to “cancer cures.” These two breakthrough areas relate somewhat to the Year 2000 NNI goals 1 and 3, respectively, although the supporting roles that nanotech is playing in the development of these new fields are quite distinct from those anticipated 15 y ago.
This paper is a perspective rather than a proper review, so many topics are necessarily not covered. Of the six original NNI goals, I don’t discuss numbers 4 and 5, and progress toward a couple of the others is mentioned only briefly. Even for those topics I do discuss, I am forced to neglect a great deal of great science, and for that I apologize in advance! In the following pages I stress both the fundamental scientific advances that are enabling the current state of the art of nanomedicine and the conceptual advances and biomedical needs that are driving the field. In the “bottom-up” spirit of nanotechnology, I begin with discussions of nanotech contributions to genomics and other -omics measurements, because those measurements are increasingly providing a foundation for modern medicine. Finally I briefly peer into the bright future of nanomedicine.
Genomics and Nanotechnology
Goal no. 1 of the 2000 NNI forecasts an important role for nanotech in enabling inexpensive sequencing. The rapid advancement in next-generation genome sequencing (NGS) technologies has been a triumph of modern biotechnology, so that the $1000 genome is now a reality (5). In fact, the NGS tools have advanced to the point that the technology is effectively a given, and the intellectual effort within a genomics study now is centered on the scientific design and interpretation of population-based studies (6). However, NGS has major deficiencies. First, most NGS methods are limited to short reads, although alternative technologies are emerging that permit longer reads (7). Analyzing an NGS dataset is, by analogy, similar to reading a book in which short sentence fragments are placed in random order relative to how the text actually was written. To make it understandable, the book must be reassembled correctly from those short fragments. Second, NGS requires amplification. This requirement, combined with short reads, means that the identification of many natural or disease-associated molecular lesions, such as repeat-rich regions, gene amplifications and deletions, or chromosomal translocations, can be challenging. Finally, NGS requires expensive reagents. Thus, new sequencing or mapping technologies are emerging based on long reads of single DNA molecules, without the need for amplification or even reagents (8). These approaches are enabled by nanotechnologies (Fig. 1) with specific design parameters that draw from many years of polymer-based physics, biophysics, and physical chemistry.
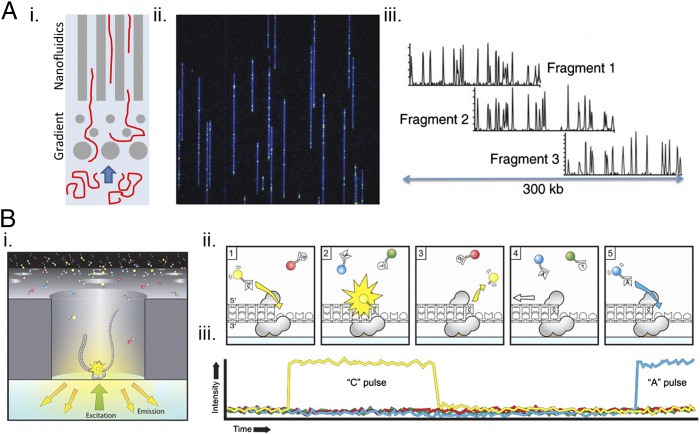
Fig. 1.
Nanotechnologies for genome mapping and genome sequencing. (A) Nanochannel-based genome mapping. (i) The microchip is designed to untangle the DNA and guide long fragments into nanochannels with diameters <100 nm. (ii) The DNA fragments are fluorescently labeled at specific sites to provide a spatial map of those sites. (iii) Different fragments are lined up according to the spatially resolved fluorescent signatures to provide the genome map, shown here for a 300-kb segment. Adapted from ref. 9, with permission from Macmillan Publishers Ltd: Nature Biotechnology, copyright 2012. (B) The Pacific Biosciences nanopore-based genome-sequencing platform, in which single-molecule, real-time sequencing data are obtained from a DNA polymerase that is isolated within a 100-nm-diameter pore in an aluminum film. (i) The pore serves as a zero mode waveguide for optical analysis of uninterrupted template-directed DNA synthesis using four distinguishable fluorescently labeled dNTPs. (ii) The temporal order of enzymatic incorporation of the dNTPs into a growing DNA strand is illustrated. At each step a fluorescent dNTP is incorporated, generating a fluorescent signal that is collected efficiently within the nanowaveguide. The fluorophore then is cleaved and diffuses out of the waveguide. (iii) An optical readout for a single channel is illustrated. Reproduced from ref. 19, with permission from AAAS.
Nanochannel-based single-molecule genome mapping (Fig. 1A) (9) is used to help assemble genomes and fill in information gaps that are not captured by NGS. The origins of the technique were methods in which single DNA molecules, 102–104 kb long, were stretched out on a microscope slide and cleaved using site-specific enzymes. In this way a 400-kb DNA molecule might be cut into 20–40 components (10). The physical length of the resulting segments, averaged over many DNA molecules similarly analyzed, provided a crude genome map (11). For nanochannels, the concept illustrated in Fig. 1A relies on fluorophores that are introduced by using nicking endonucleases that cut the dsDNA at specific restriction sites. These locations provide a map that includes deletions, amplifications, and translocations and provides guides for NGS genome assembly. Of course, many physical chemistry issues, such as the interplay of the persistence length of the DNA molecules, the ionic strength of the solvent, the conditions for uncoiling the DNA, and the dimensions of the nanochannel, have all provided a scientific foundation for this nanotechnology (12, 13).
Long, accurate sequencing reads of unamplified DNA would negate the need for mapping. Nanopore-based sequencing (14) is such a method, although it has been technically challenging to develop and only very recently has permitted some sequence determination of an actual genome (15, 16) The basic idea dates back some 20 y and draws from fundamental work in surface science, molecular biology, nanofabrication, and electrochemistry (17). In one manifestation, two aqueous electrolyte solutions are connected through a single protein nanopore, such as such as Mycobacterium smegmatis porin A (MspA). A bias is applied, and current is measured as DNA is drawn through a bilayer-entrained pore protein at a rate that is controlled through the use of molecular motors such as Φ 29 DNA polymerase (DNAP) (18). A variant developed by Pacific Biosciences (19) is shown in Fig. 1B. Four distinct fluorophores are used to read out the action of the polymerase, which is isolated within a nanofabricated waveguide. The polymerase, which also is used for whole-genome amplification, permits long, relatively unbiased reads. Each polymerase permits a read rate of about four to five bases per minute, with thousands of single polymerases in nanowaveguides used in parallel. A third variant, which was released by Oxford Nanopore Technologies as the MinION nanopore sequencing product, is about the size of a USB memory stick. Early literature reports on the MinION imply that it may not yet be ready for wholesale sequencing (20), but it does have some compelling applications (21).
Although genomics has led the -omics revolution, nanotechnologies (and microfluidics) are playing increasingly important roles in reducing -omics technologies to the level of a single cell, and I turn to this area next.
Single-Cell -Omics in Biology and Biomedicine
Single-cell biology holds the promise of unraveling the heterogeneity that often confounds the interpretation of biological or biomedical measurements. For example, single-cell analysis is increasingly applied toward understanding immune responses triggered during various cancer immunotherapies (22–25) or for unraveling the functional behaviors that emerge from heterogeneous healthy or diseased tissues (26, 27). Aside from flow- and mass-cytometry methods (28, 29), virtually every single-cell -omics platform involves some level of microfluidics and/or nanotech. Specifically, a single cell has only a certain number of copies of any given analyte. By isolating a single cell within a nanoliter or subnanoliter volume, those copy numbers can correspond to detectable concentrations, and the concentrations of contaminants are minimized. Microchip methods also can uniquely permit analysis of very small tissue samples.
Single-cell methods have been reviewed recently (30–32), so only a general technology overview plus a brief discussion of what is learned uniquely through single-cell assays is given here. Single cells are, from a physico-chemical point of view, finite systems. That is, quantitative measurements of transcripts, proteins, metabolites, and so forth in different single cells will yield different copy numbers of those analytes. Thus, the most useful single-cell methods are designed to assay a panel of analytes from statistically significant numbers of single cells. Added value that is uniquely provided by such single-cell analyses includes
-
•
Histograms of the abundance distributions of a given analyte across a population of cells help define outliers, such as cells that overexpress a particular protein. This information can be particularly important for immunology applications (23, 33). For certain analytes, the abundance distributions also may be interpreted as the fluctuations of the cell (similar to positional fluctuations in a Brownian particle), thus providing a bridge to statistical thermodynamics models (34).
-
•
Correlations (and anticorrelations) between the measured analytes are statistical relationships that provide important clues regarding the structure of gene or protein signaling networks and how that structure is altered by some perturbation (28, 35–37). Correlations derived from single-cell assays are not the same as those derived from bulk assays. In a bulk assay, the average levels of two analytes may both be repressed by a drug and thus exhibit correlated behavior. For single-cell analyses, statistical correlations between any two analytes are determined through an x,y scatter plot of the assayed values for each single cell. The average analyte abundance is not a factor in such plots (Fig. 2A).
-
•
Detailed lineage tracing is enabled, permitting diseased tissues or cells to be traced back to the originating healthy tissue or cell types (26, 38) or circulating tumor cells to be traced back to the originating lesion (39).
-
•
Some recent studies capture multiple levels of biological information (i.e., proteins and metabolites) from the same single cells (Fig. 2A). Such studies have many applications, ranging from associating a particular T-cell receptor gene with a T-cell antigen (22, 25) to associating the functional behaviors of cells [e.g., cellular motility (40) and glycolytic rate (41), among others] with genetics, drug response, and so forth.
-
•
Several platforms permit an -omics analysis of defined cell populations (i.e., zero, one, two, three, or more cells). Such platforms provide unique insights into cellular behaviors, such as how pairwise cell interactions influence bulk cellular architectures or quantitative analysis of immune cell–disease cell interactions (36, 42–44).
-
•
A technologically distinct class of microchip designs permits individual cells to be isolated within droplets with a volume 0.05 pL to 1 nL (45, 46), and those droplets are entrained within a fluidic flow that can be coupled with an optical analysis platform (Fig. 2B) (47, 48). The chemical composition of individual droplets can be manipulated to permit rapid and high-throughput screening assays. Applications related to screening antimicrobial or antiviral agents or for various binding assays have been reported with very significant throughput advantages over traditional assays. For a technique known as Drop-seq, individual cells are similarly entrained within nanodrops on microfluidic chips. Each droplet also contains a microparticle that is encoded with a unique DNA barcode address. Cells are lysed within the droplet, the mRNAs are captured on the barcoded microbead, and then all microbeads are analyzed in parallel. The barcode address allows the transcriptome analysis to be reassociated with individual cells (49). A related molecular barcoding microwell transcriptomics approach was recently reported by Fan et al. (50)
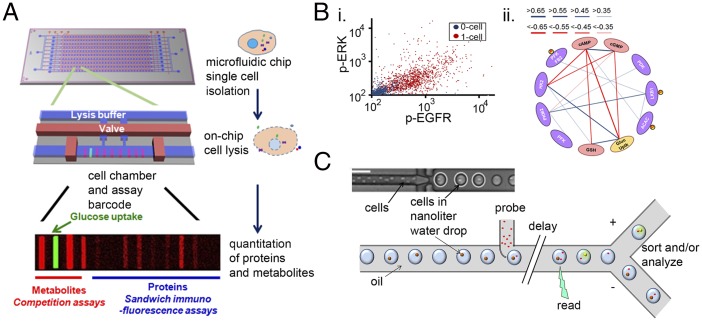
Fig. 2.
Microfluidics and nanotech tools for single-cell analysis. (A) Illustration of a single-cell barcode chip (SCBC) in which individual cells are isolated within nanoliter or smaller volume microchambers within a microfluidics chip mounted on a microscope slide. The glass slide is patterned with a high-density barcode for protein and/or metabolite assays from isolated single cells. Cells are lysed using the valved microchamber structure shown in the middle drawing, and the contents are captured on specific locations within the barcode array. The fluorescence intensities of the developed barcode stripes are related to calibration curves to yield the level of the specific analytes. (B) Sample of single-cell data taken on an SCBC. (i) The scatter plot shows the correlated levels of two phosphoproteins as measured from single cells (red dots) or zero-cell chambers (blue dots). (ii) A protein–protein correlation matrix from a multiplex SCBC protein and metabolite assay. Adapted from ref. 41. (C) An illustration of a microfluidics platform for building a regular array of single cells within nanoliter droplets of water, entrained in oil. The optical micrograph is reproduced from ref. 45, with permission from The Royal Society of Chemistry. The drawing illustrates some of the flexible design parameters that are used in this type of high-throughput assay. Cells can be probed with antibodies, viruses, mRNA-encoded beads, NPs, and so forth for a controllable amount of time, using a delay line or related method. Cells may be interrogated optically and sorted or otherwise analyzed at the protein, transcript, or functional level.
Most single-cell analysis tools, aside from cytometry methods, are young and so are only now being commercialized.
Inorganic Nanoparticles and Related Nanomaterials in Biomedicine
Metal nanoparticles (NPs) (51) and semiconductor quantum dots (QDs) (52, 53) and related materials, such as single-wall carbon nanotubes (SWNTs) (54, 55), are increasingly used for preclinical in vitro and in vivo (imaging) diagnostics (56–58). Therapeutic applications have been slower to develop (59). The basic concept is that the size, shape, and composition of the inorganic core provide a useful physical property that enables a colorimetric, electrochemical, Raman, or other class of assays. The NP surface chemistry is tailored for the specific assay, including biomolecular recognition, solubility, and other characteristics that translate into a very large matrix of materials properties. For in vitro applications, there is substantial flexibility in exploiting this matrix. For in vivo applications, the task is much more challenging. Each of the elements of this properties matrix [which includes the Stokes radius, the surface charge (60, 61), the density of surface chemical groups (62, 63), and the elastic modulus (64), among others] can strongly influence in vivo pharmacokinetics (PK) (i.e., circulation times, uptake within specific cells or within specific organs or disease sites, clearance route, and toxicology) (65, 66). In particular, understanding which basic properties of nanomaterials can lead to nonspecific and potentially harmful biological interactions (67–69) in vivo is a very active field of study. In any case, this matrix of materials properties for optimization represents a sort of “opportunity hurricane” for the scientist or engineer. Efforts to elucidate how best to optimize NPs for specific tasks comprise much of the basic science of these NPs. Organizations such as the National Cancer Institute’s Nanotechnology Characterization Laboratory have played important roles in guiding such efforts (70). Although approved clinical applications are beginning to appear, the bulk of the science is still maturing through mouse model studies. In following paragraphs, I provide a very brief overview of this underlying science and highlight illustrative examples in Figs. 3 and 4.
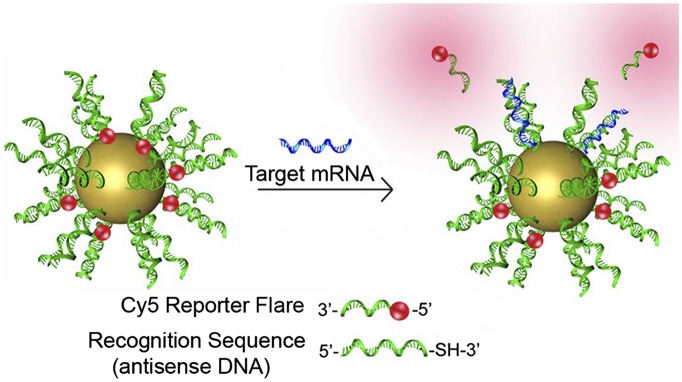
Fig. 3.
The gold NP-based nanoflare construct is used for detecting specific mRNAs in live cells. The gold NPs are coated with a dense layer of DNA, which promotes cell penetration. The DNA shown comprises a fluorescent reporter (the Cy5 flare), which is nonfluorescent when bound to the Au NP. This nanoflare is hybridized with an antisense DNA. When the nanoflare encounters the target mRNA, the flare is released, thus activating fluorescence within the cell and permitting live-cell sorting based on the expression of a specific gene. Adapted from ref. 75.
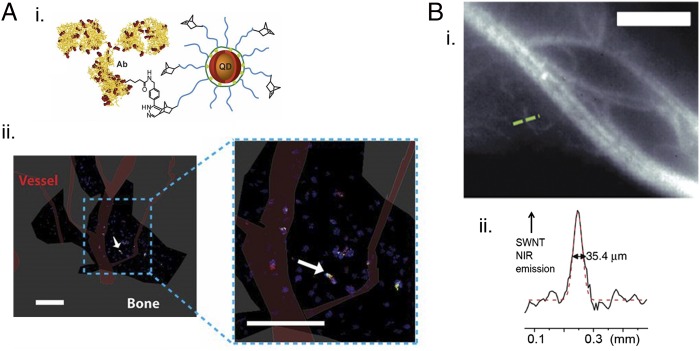
Fig. 4.
Semiconductor QD and SWNT in vivo imaging probes. (A) Semiconductor QD imaging of stem cells in bone marrow. (i) Polyimidazole incorporating norbornene provides a bio-compatible surface functionalization for highly luminescent semiconductor core-shell QDs as well as a chemical handle for preparing QD–antibody conjugates, as shown (not to scale). Lyscine (amine-presenting) residues on the antibody are highlighted in red. The QD–antibody particles exhibit a moderate negative surface charge, which is generally favorable for improved in vivo circulation. (ii) Use of the QD–antibody particles as in vivo imaging probes for single-cell imaging in the bone marrow of a live murine model, viewed through a calvarial window. The arrow points to single Sca-1+c-Kit+ cell, which is a late-stage hematopoietic stem cell. The red and green cells represent Sca-1+ and c-Kit+ cells, respectively. Adapted from ref. 89. (B) Polymer-stabilized SWNTs used as in vivo near-infrared (NIR) fluorescent probes of vasculature. (i) Near-IR fluorescence of mouse vasculature. (ii) Fluorescence intensity taken along the dashed green line drawn on the image in i is plotted and reveals an imaging resolution of a few tens of micrometers. Adapted from ref. 90, with permission from Macmillan Publishers Ltd: Nature Medicine, copyright 2012.
Noble-metal NPs have been used for several years for the point-of-care detection of blood-based biomarkers from droplets of blood; gold NPs provide the colorimetric agents for analyte detection. The basic exploited physical property is the surface plasmon resonance (SPR), which is in the visible or near-visible part of the spectrum for noble-metal NPs. The SPR is a collective resonance that carries a very high oscillator strength, with a peak wavelength, line shape, and intensity that strongly depend upon NP size, shape, and local dielectric environment. The basic optical properties of colloidal gold were described more than 100 y ago by Mie using Maxwell’s equations (71). Current models can capture the remarkable linear and nonlinear (72) optical properties achievable through modern synthetic methods in which surface chemistry, local chemical environment (73), and NP size and shape (74) are controlled. For in vitro biological applications, synthetic methods for surface-loading high-density coverages of DNA have enabled efficient cell delivery for various in-cell, live-cell assays (Fig. 3) (75). Other surface chemistries have enabled a myriad of electrochemical, chemical, and optical platforms for sensing and in vitro diagnostics (76–78). For in vivo applications, that synthetic control has been harnessed for minimizing toxicity [exhibited by some very small gold NPs (79)] and for targeting delivery of gold nanorods to specific tissues (80). Gold nanorods have shown promise in animal models for photothermal therapy applications. In such a therapy (81), the particles are delivered to the disease site, and laser light is used to penetrate into the tissue, excite the NPs, and transfer thermal energy to—and thus kill—the diseased tissue. This concept has had only limited success in human trials (82) and so may need more time to mature.
Remarkable advances in nanomaterials synthesis have occurred for a broad variety of NPs, including magnetic NPs (83, 84). These advances have enabled applications of ferromagnetic and superparamagnetic NPs for both in vitro and in vivo (imaging) diagnostic assays (85, 86). For in vitro assays, biofunctionalized magnetic NPs have a history of use for magnet-assisted cell sorting. A recent variant of that application is the use of antibody-coated magnetic NPs for detecting (using a custom-built microchip-NMR tool) and sorting microvesicles released by certain solid tumors (87). Microvesicles are 10- to 100-fold smaller than the tumor cells from which they are released but can carry many of the biomarkers that identify the originating cell as a diseased cell (88). For certain tumors, such as glioblastomas, microvesicles are released into the blood, although circulating tumor cells are not.
Fig. 4 illustrates two in vivo live-animal imaging applications of highly fluorescent semiconductor QDs (Fig. 4A) (89) and near-IR fluorescing SWNTs (Fig. 4B) (90). In both cases, the nanomaterial exhibits a compelling optical property that enables the imaging application and also is highly engineered, through surface chemistry control, for useful PK and pharmacodynamics (PD) properties. These types of probes, although not yet clinical tools, are providing powerful approaches for preclinical investigations.
Organic Nanoparticles for Drug Delivery
Goal no. 3 of the 2000 NNI, which describes the promise of NP drug-delivery systems, was largely inspired by the promising performance of liposomal delivery vehicles (91), at least four of which had received approval (one from outside the United States) before 2000 (92, 93). Liposomes are spherical vesicles with at least one lipid bilayer and can be used to carry hydrophobic drug molecules that associate within the membrane or hydrophilic molecules that are trapped in the core. The approval of the protein-drug conjugate Abraxane (94) in 2005 gave further impetus to the field.
Nanotherapeutics such as liposomes are based on a concept in which many copies of a drug molecule are loaded into a delivery vehicle (the NP) that is optimized for improved PK and PD relative to the drug by itself. Nanotherapeutics have broad applications for many disease conditions; their use in oncology illustrates their general value (59). For almost all metastatic cancers, the frontline molecular treatments are small-molecule chemotherapies. High systemic exposure of patients to those drugs frequently leads to dose-limiting toxicity. The nature of small-molecule drugs is that they exhibit short (<1 h) circulation times and indiscriminate tissue penetration, thus affecting both healthy and diseased tissue with a PK and PD profile that is defined largely by the nature of the drug rather than the needs of the patient. Early work on polymer–drug conjugates showed that large-molecular-weight particles, or macromolecules, could avoid such rapid clearance and, in fact, accumulate in tumor sites over a period of many hours (95, 96). This effect became known as the “enhanced permeability and retention” (EPR) effect. EPR is not a general characteristic of all nanotherapeutics; early liposomal formulations were found to clear rapidly from the blood and often to accumulate in the liver. The variations in PK and PD across different formulations suggested the compelling bioengineering challenge that drives the current science and translational progress in the field.
Organic nanotherapeutics under development today include liposomes with single or multiple bilayered membrane structures built from natural or synthetic lipids (97), dendrimer constructs, albumin-bound formulations (94), and polymeric nanoparticles engineered from biocompatible and biodegradable polymers. A common theme across these systems is that the NP delivery vehicle itself contains the engineering handles for controlling the drug PK and PD. These handles include ligands for tumor targeting (98) and mechanisms for triggered drug release based on pH (99) or enzymatic signals (100) as well as molecular or nanofabrication engineering to control surface charge (zeta potential), NP size, NP shape (101), and NP elastic modulus (102). Of course, as with inorganic NPs, the available design space is vast. A measure of progress in this field is the rapidly increasing number of NP formulations that have advanced into Phase I–III clinical trials (92).
In Figs. 5 and 6 I highlight two very different examples of organic NP preparations that reflect unique aspects of the state of the art and which are being translated through human trials. Fig. 5 illustrates the use of the particle replication in nonwetting templates (PRINT) technology, developed by DeSimone’s group (102). This example highlights the role that the particle modulus can play in organ delivery. The highly versatile PRINT technology has been used to prepare NP vaccines that have been translated into an initial Phase I/IIa clinical trial for influenza (103).
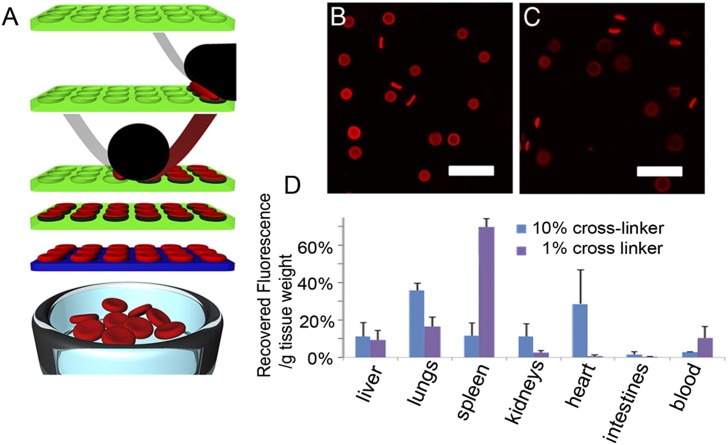
Fig. 5.
Illustration of the PRINT method for making nano- and microparticles. (A) PRINT uses the micro- and nanolithography fabrication tools of the semiconductor industry to build molds (shown in green). These are coupled with roll-to-roll processing to prepare size- and shape-controlled particles that then are released from the molds. Different polymer and hydrogel chemistries are used to control the chemical and physical properties of those particles. (B and C) Two batches of fluorescent hydrogel microparticles are prepared with different elastic moduli, based on the extent of cross-linking (10% in B; 1% in C). The particles with the lowest cross-linking have elastic moduli designed to emulate that of a red blood cell. (D) Illustration of the organ distribution of the PRINT particles with high and low numbers of elastic moduli 2 h after tail injection into a mouse. Adapted from ref. 102. The particles illustrated here are being investigated, in a preclinical setting, as a component of synthetic blood.
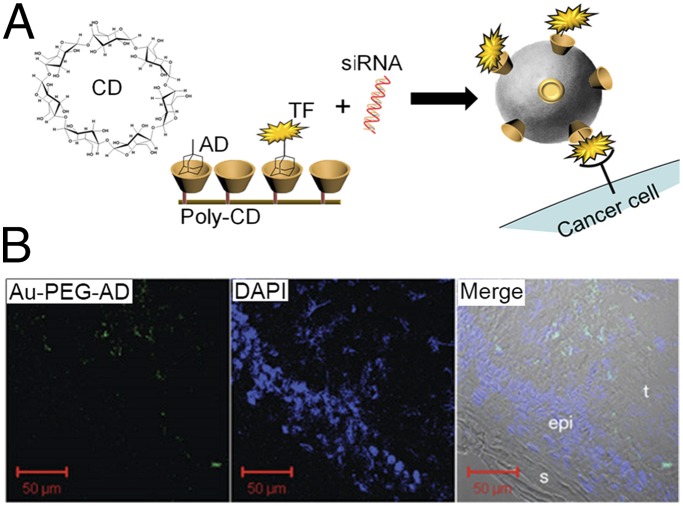
Fig. 6.
A polymer-based 70-nm nanotherapeutic for siRNA delivery in humans. (A) Cyclodextran (CD) forms a conical binding pocket for the supramolecular assembly of adamantane (AD). A poly-CD oligomer has several such binding pockets, which can be used for the assembly of adamantane-labeled drugs or, in the example shown, TF ligands that can target cancer cells. When poly-CD is combined with adamantane-labeled TF and siRNA, the TF is presented on the surface of the NP, and the siRNA is localized within the hydrophilic interior, thus providing directed delivery of the siRNA to cancer cells. The nanotherapeutic is administered to patients intravenously. (B) Data from a clinical trial on melanoma cancer patients. Five-nanometer adamantane-labeled gold NPs (Au-PEG-AD) are used for tissue labeling of the poly-CD NPs. The three images show that the NPs (green color in left image) are not in the skin (s) or the epidermis (epi) but instead are localized within the tumor (t). Adapted from ref. 104, with permission from Macmillan Publishers Ltd, Nature, copyright 2012. Related poly-CD nanotherapeutics are being tested in several clinical trials for various cancer indications.
Fig. 6 illustrates the use of polycyclodextrin (poly-CD) NPs as tumor-targeting delivery agents for siRNA therapies. In the example, a human transferrin (TF) protein-targeting ligand provides the NP with tumor-targeting characteristics. This NP was translated into a Phase I clinical trial for melanoma cancer patients. Tumor biopsies, collected after therapy, revealed that the NPs localized to the tumor (Fig. 6B) and successfully repressed the levels of the target protein in the tumor cells (104). Related NP formulations serving as carriers of the chemotherapeutic camptothecin are being explored currently in a number of human trials (92).
Nanomechanical and Nanoelectronic Devices and Wellness Monitoring
Goals nos. 2 and 6 of the 2000 NNI, which emphasized remote sensor systems and in vivo devices for early disease detection and diagnosis, represented interrelated grand challenges with somewhat similar roadmaps to success. Recent influences on these goals are the proliferation of smart phones and other smart devices as well as the emerging emphasis on wellness (105). A relevant emerging wellness trend is that an individual is provided with regular measurements of his/her health status. The individual can make lifestyle, exercise, and diet adjustments informed by those measurements so as to maximize health benefit. The technology challenge is to provide accurate, informative, and readily interpretable diagnostic measurements at low cost. The emerging science and technology of wearable health monitoring devices illustrates a dominant approach toward meeting this challenge. Nanotech does not play a role in the early-stage examples of such wearable devices, although it is clear that nanotech will play important roles in the near future. Current smart devices monitor a panel of basic body functions, such as heart rate, body temperature, and exercise but, with few exceptions (106), do not monitor specific biomarker analytes. This current functionality is enabled by a menu of microelectromechanical machines (MEMs) such as microphones, accelerometers, and gyroscopes. The uptake of MEMs in consumer products was young in 2000, but of the three waves of proliferation initially envisioned for MEMs—automotive, consumer electronics, and home integration—the first two are now considered complete (107). A fourth wave, relevant to this discussion, is likely personal integration.
Recent advances (108–115) in integrating electronics onto low-elastic modulus polymer substrates and other moldable materials that are conformal, biocompatible, and, in some instances, even self-powered (116) are opening up a large menu of wearable devices based on mechanical, electronic, or electrochemical signal transduction (117, 118). A large fraction of these devices contain some nanotechnology, with a scientific basis that can be traced back to early demonstrations of nanotube (119) or nanowire (120, 121) sensors and the integration of nanowires and nanotubes onto plastic substrates (122, 123). The net result is that biomarker-rich liquids, such as sweat or saliva (124), can be used for continuous multiplex monitoring of health status-relevant analytes. An example of a nanotech-enabled tooth “decal” sensor is shown in Fig. 7. This device straddles the line between an implant and a wearable device. Although the presented data are from a benchtop experiment, they clearly illustrate the potential for real-time monitoring of both bodily functions (breathing) and analytes (bacteria in saliva).
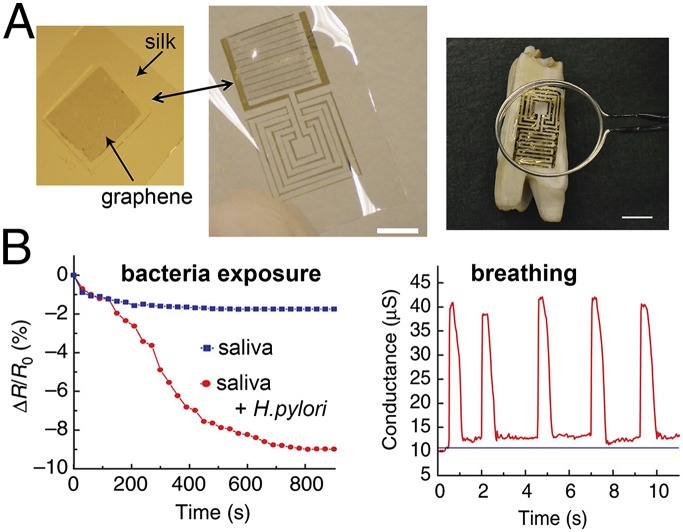
Fig. 7.
Graphene nanotechnology integrated into flexible electronics yields a potentially wearable sensor platform. (A) Graphene is printed onto bioresorbable silk and then is electrically contacted with a small electrical circuit before transfer onto the surface of a tooth. The flexible circuit consists of interdigitated capacitive electrodes (to sense the graphene electrical conductivity) and a planar meander line inductor. The graphene is chemically modified with a bifunctional peptide to bind to the graphene and to exhibit affinity for specific bacteria. Exposure to bacteria modulates the electrical conductivity of the graphene, which is measured by the interdigitating electrodes and is transmitted wirelessly to an inductively coupled receiver. (B) Benchtop experiments show the ability of this sensor system to detect a specific biological agent (Helicobacter pylori) (Left) and a periodic physiological process (breathing) (Right). Adapted from ref. 124, with permission from Macmillan Publishers Ltd: Nature Communications, copyright 2012.
Concluding Remarks: Nanomedicine and Emerging Challenges
In this Perspective, I have provided a partial view of the state of the art of nanomedicine. Unlike applications in fields such as energy or non–health-related biotech, biomedicine has a unique and justifiably conservative set of rules for adopting new technologies for human use. Witness, for example, the remarkably rapid rise of CRISPR genome-editing technologies (and associated biotech companies) over the past 2 y. Witness also, however, the healthy scientific debate (125) and public (126) controversy that emerged when gene editing was carried out on nonviable human embryos (127). This response was warranted, but it means that gene-editing technologies have a long road to travel from the benchtop to the bedside. Nanomedicine faced similar challenges 15–20 y ago. It was a brand-new field, and the ability to tune physical or PK properties by tuning NP size and shape was a sparkling new addition to the synthetic toolkit. However, that addition was not well understood, and current good manufacturing practice (cGMP)-type manufacturing, which is a critical first step toward human health applications, was virtually unknown in nanotech. In addition, there were challenges unique to the field. The pool of young scientific talent that could work across the disciplines of chemistry, physics, materials science, engineering, and biomedicine didn’t exist, and most research institutions didn’t have the infrastructure to support that talent. All these challenges have been fully or partially addressed over the past 15 y. The number of nanotechnologies that currently are undergoing some level of human testing and the several areas in medicine in which nanotech is providing unique solutions are important measures of that success. Although the impact of this progress on the human health condition is still marginal, it will be felt increasingly over the next decade. Thus, as we look back at the distance traveled, it is indeed startling to find how far we have come.
Acknowledgments
Some of the work described in this review was supported by National Cancer Institute Grants 5R01CA170689 and 5U54 CA119347, the Ben and Catherine Ivy Foundation, and the Jean Perkins Foundation.
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
This article is part of the special series of PNAS 100th Anniversary articles to commemorate exceptional research published in PNAS over the last century.
References
- 1. National Science and Technology Council Committee on Technology. National Nanotechnology Initiative: leading to the next industrial revolution. A report by the Interagency Working Group on Nanoscale Science, Engineering, and Technology. Washington, DC: US Government; 2000. Available at: https://www.whitehouse.gov/files/documents/ostp/NSTC%20Reports/NNI2000.pdf.
- 2.Gibbs WW. Medicine gets up close and personal. Nature. 2014;506(7487):144–145. doi: 10.1038/506144a. [DOI] [PubMed] [Google Scholar]
- 3.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271(5256):1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 4.Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342(6165):1432–1433. doi: 10.1126/science.342.6165.1432. [DOI] [PubMed] [Google Scholar]
- 5.Hayden EC. Technology: The $1,000 genome. Nature. 2014;507(7492):294–295. doi: 10.1038/507294a. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein DB, et al. Sequencing studies in human genetics: Design and interpretation. Nat Rev Genet. 2013;14(7):460–470. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 8.Persson F, Tegenfeldt JO. DNA in nanochannels--directly visualizing genomic information. Chem Soc Rev. 2010;39(3):985–999. doi: 10.1039/b912918a. [DOI] [PubMed] [Google Scholar]
- 9.Lam ET, et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 2012;30(8):771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou S, et al. Validation of rice genome sequence by optical mapping. BMC Genomics. 2007;8(1):278. doi: 10.1186/1471-2164-8-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin J, et al. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science. 1999;285(5433):1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 12.Reinhart WF, et al. Distribution of distances between DNA barcode labels in nanochannels close to the persistence length. J Chem Phys. 2015;142(6):064902. doi: 10.1063/1.4907552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reisner W, et al. Statics and dynamics of single DNA molecules confined in nanochannels. Phys Rev Lett. 2005;94(19):196101. doi: 10.1103/PhysRevLett.94.196101. [DOI] [PubMed] [Google Scholar]
- 14.Branton D, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4(4):265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 16.Laszlo AH, et al. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32(8):829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30(4):349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 20.Mikheyev AS, Tin MMY. A first look at the Oxford Nanopore MinION sequencer. Mol Ecol Resour. 2014;14(6):1097–1102. doi: 10.1111/1755-0998.12324. [DOI] [PubMed] [Google Scholar]
- 21.Ashton PM, et al. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol. 2015;33(3):296–300. doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 22.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32(7):684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma C, et al. Multifunctional T-cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer Discovery. 2013;3(4):418–429. doi: 10.1158/2159-8290.CD-12-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 25.van Buuren MM, Calis JJ, Schumacher TN. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. OncoImmunology. 2014;3:e28836. doi: 10.4161/onci.28836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci USA. 2014;111(26):E2770–E2777. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, et al. Microfluidics-based single-cell functional proteomics for fundamental and applied biomedical applications. Annu Rev Anal Chem (Palo Alto, Calif) 2014;7:275–295. doi: 10.1146/annurev-anchem-071213-020323. [DOI] [PubMed] [Google Scholar]
- 31.de Bourcy CF, et al. A quantitative comparison of single-cell whole genome amplification methods. PLoS One. 2014;9(8):e105585. doi: 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11(1):41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat Biotechnol. 2006;24(6):703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 34. Anonymous (2013) Nonequilibrium Statistical Physics of Small Systems: Fluctuation Relations and Beyond (Wiley-VCH, Berlin), 1st Ed.
- 35.Irish JM, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118(2):217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 36.Kravchenko-Balasha N, Wang J, Remacle F, Levine RD, Heath JR. Glioblastoma cellular architectures are predicted through the characterization of two-cell interactions. Proc Natl Acad Sci USA. 2014;111(17):6521–6526. doi: 10.1073/pnas.1404462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Q, et al. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc Natl Acad Sci USA. 2012;109(2):419–424. doi: 10.1073/pnas.1110865109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29(12):1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohr JG, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32(5):479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y, et al. High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal Chem. 2013;85(4):2548–2556. doi: 10.1021/ac400082e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue M, et al. Chemical methods for the simultaneous quantitation of metabolites and proteins from single cells. J Am Chem Soc. 2015;137(12):4066–4069. doi: 10.1021/jacs.5b00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamanaka YJ, et al. Single-cell analysis of the dynamics and functional outcomes of interactions between human natural killer cells and target cells. Integr Biol (Camb) 2012;4(10):1175–1184. doi: 10.1039/c2ib20167d. [DOI] [PubMed] [Google Scholar]
- 43.Elitas M, Brower K, Lu Y, Chen JJ, Fan R. A microchip platform for interrogating tumor-macrophage paracrine signaling at the single-cell level. Lab Chip. 2014;14(18):3582–3588. doi: 10.1039/c4lc00676c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liadi I, et al. Individual motile CD4(+) T cells can participate in efficient multikilling through conjugation to multiple tumor cells. Cancer Immunol Res. 2015;3(5):473–482. doi: 10.1158/2326-6066.CIR-14-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edd JF, et al. Controlled encapsulation of single-cells into monodisperse picolitre drops. Lab Chip. 2008;8(8):1262–1264. doi: 10.1039/b805456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joensson HN, Andersson Svahn H. Droplet microfluidics--a tool for single-cell analysis. Angew Chem Int Ed Engl. 2012;51(49):12176–12192. doi: 10.1002/anie.201200460. [DOI] [PubMed] [Google Scholar]
- 47.Guo MT, Rotem A, Heyman JA, Weitz DA. Droplet microfluidics for high-throughput biological assays. Lab Chip. 2012;12(12):2146–2155. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 48.Mazutis L, et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc. 2013;8(5):870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan HC, Fu GK, Fodor SPA. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347(6222):1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 51.Dykman L, Khlebtsov N. Gold nanoparticles in biomedical applications: Recent advances and perspectives. Chem Soc Rev. 2012;41(6):2256–2282. doi: 10.1039/c1cs15166e. [DOI] [PubMed] [Google Scholar]
- 52.Chan WCW, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281(5385):2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 53.Bruchez M, Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281(5385):2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 54.Mundra RV, Wu X, Sauer J, Dordick JS, Kane RS. Nanotubes in biological applications. Curr Opin Biotechnol. 2014;28:25–32. doi: 10.1016/j.copbio.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 55.Hong G, et al. Through-skull fluorescence imaging of the brain in a new near-infrared window. Nat Photonics. 2014;8(9):723–730. doi: 10.1038/nphoton.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22(8):969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 57.Massey M, Wu M, Conroy EM, Algar WR. Mind your P’s and Q’s: The coming of age of semiconducting polymer dots and semiconductor quantum dots in biological applications. Curr Opin Biotechnol. 2014;34C:30–40. doi: 10.1016/j.copbio.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 58.Jin Z, Hildebrandt N. Semiconductor quantum dots for in vitro diagnostics and cellular imaging. Trends Biotechnol. 2012;30(7):394–403. doi: 10.1016/j.tibtech.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 59.Wang AZ, Langer R, Farokhzad OC. Nanoparticle delivery of cancer drugs. Annu Rev Med. 2012;63(1):185–198. doi: 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- 60.Weissleder R, Nahrendorf M, Pittet MJ. Imaging macrophages with nanoparticles. Nat Mater. 2014;13(2):125–138. doi: 10.1038/nmat3780. [DOI] [PubMed] [Google Scholar]
- 61.Heath JR, Davis ME. Nanotechnology and cancer. Annu Rev Med. 2008;59(1):251–265. doi: 10.1146/annurev.med.59.061506.185523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-flares: Probes for transfection and mRNA detection in living cells. J Am Chem Soc. 2007;129(50):15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Townson JL, et al. Re-examining the size/charge paradigm: Differing in vivo characteristics of size- and charge-matched mesoporous silica nanoparticles. J Am Chem Soc. 2013;135(43):16030–16033. doi: 10.1021/ja4082414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anselmo AC, et al. Elasticity of nanoparticles influences their blood circulation, phagocytosis, endocytosis, and targeting. ACS Nano. 2015;9(3):3169–3177. doi: 10.1021/acsnano.5b00147. [DOI] [PubMed] [Google Scholar]
- 65.Albanese A, Tang PS, Chan WCW. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng. 2012;14(1):1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 66.Nel A, Xia T, Mädler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311(5761):622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 67.Treuel L, et al. Impact of protein modification on the protein corona on nanoparticles and nanoparticle-cell interactions. ACS Nano. 2014;8(1):503–513. doi: 10.1021/nn405019v. [DOI] [PubMed] [Google Scholar]
- 68.Rauch J, Kolch W, Laurent S, Mahmoudi M. Big signals from small particles: Regulation of cell signaling pathways by nanoparticles. Chem Rev. 2013;113(5):3391–3406. doi: 10.1021/cr3002627. [DOI] [PubMed] [Google Scholar]
- 69.Chen KL, Bothun GD. Nanoparticles meet cell membranes: Probing nonspecific interactions using model membranes. Environ Sci Technol. 2014;48(2):873–880. doi: 10.1021/es403864v. [DOI] [PubMed] [Google Scholar]
- 70.Crist RM, et al. Common pitfalls in nanotechnology: Lessons learned from NCI’s Nanotechnology Characterization Laboratory. Integr Biol (Camb) 2013;5(1):66–73. doi: 10.1039/c2ib20117h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mie G. Contributions for the optics of turbid media, especially colloidal metal solutions. Ann Phys. 1908;25:377–345. [Google Scholar]
- 72.Le Ru EC, Etchegoin PG. Single-molecule surface-enhanced Raman spectroscopy. Annu Rev Phys Chem. 2012;63(1):65–87. doi: 10.1146/annurev-physchem-032511-143757. [DOI] [PubMed] [Google Scholar]
- 73.Eustis S, el-Sayed MA. Why gold nanoparticles are more precious than pretty gold: Noble metal surface plasmon resonance and its enhancement of the radiative and nonradiative properties of nanocrystals of different shapes. Chem Soc Rev. 2006;35(3):209–217. doi: 10.1039/b514191e. [DOI] [PubMed] [Google Scholar]
- 74.Grzelczak M, Pérez-Juste J, Mulvaney P, Liz-Marzán LM. Shape control in gold nanoparticle synthesis. Chem Soc Rev. 2008;37(9):1783–1791. doi: 10.1039/b711490g. [DOI] [PubMed] [Google Scholar]
- 75.Halo TL, et al. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc Natl Acad Sci USA. 2014;111(48):17104–17109. doi: 10.1073/pnas.1418637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Llevot A, Astruc D. Applications of vectorized gold nanoparticles to the diagnosis and therapy of cancer. Chem Soc Rev. 2012;41(1):242–257. doi: 10.1039/c1cs15080d. [DOI] [PubMed] [Google Scholar]
- 77.Zhou W, Gao X, Liu D, Chen X. Gold nanoparticles for in vitro diagnostics. Chem Rev. 2015 doi: 10.1021/acs.chemrev.5b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dreaden EC, Alkilany AM, Huang X, Murphy CJ, El-Sayed MA. The golden age: Gold nanoparticles for biomedicine. Chem Soc Rev. 2012;41(7):2740–2779. doi: 10.1039/c1cs15237h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan Y, et al. Size-dependent cytotoxicity of gold nanoparticles. Small. 2007;3(11):1941–1949. doi: 10.1002/smll.200700378. [DOI] [PubMed] [Google Scholar]
- 80.Xiao Z, et al. DNA self-assembly of targeted near-infrared-responsive gold nanoparticles for cancer thermo-chemotherapy. Angew Chem Int Ed Engl. 2012;51(47):11853–11857. doi: 10.1002/anie.201204018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kennedy LC, et al. A new era for cancer treatment: Gold-nanoparticle-mediated thermal therapies. Small. 2011;7(2):169–183. doi: 10.1002/smll.201000134. [DOI] [PubMed] [Google Scholar]
- 82.Maier-Hauff K, et al. Efficacy and safety of intratumoral thermotherapy using magnetic iron-oxide nanoparticles combined with external beam radiotherapy on patients with recurrent glioblastoma multiforme. J Neurooncol. 2011;103(2):317–324. doi: 10.1007/s11060-010-0389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Puntes VF, Krishnan KM, Alivisatos AP. Colloidal nanocrystal shape and size control: The case of cobalt. Science. 2001;291(5511):2115–2117. doi: 10.1126/science.1057553. [DOI] [PubMed] [Google Scholar]
- 84.Jana NR, Chen Y, Peng X. Size- and shape-controlled magnetic (Cr, Mn, Fe, Co, Ni) oxide nanocrystals via a simple and general approach. Chem Mater. 2004;16(20):3931–3935. [Google Scholar]
- 85.Colombo M, et al. Biological applications of magnetic nanoparticles. Chem Soc Rev. 2012;41(11):4306–4334. doi: 10.1039/c2cs15337h. [DOI] [PubMed] [Google Scholar]
- 86.Gallo J, Long NJ, Aboagye EO. Magnetic nanoparticles as contrast agents in the diagnosis and treatment of cancer. Chem Soc Rev. 2013;42(19):7816–7833. doi: 10.1039/c3cs60149h. [DOI] [PubMed] [Google Scholar]
- 87.Shao H, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18(12):1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han H-S, et al. Quantum dot/antibody conjugates for in vivo cytometric imaging in mice. Proc Natl Acad Sci USA. 2015;112(5):1350–1355. doi: 10.1073/pnas.1421632111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hong G, et al. Multifunctional in vivo vascular imaging using near-infrared II fluorescence. Nat Med. 2012;18(12):1841–1846. doi: 10.1038/nm.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bangham AD, Standish MM, Weissmann G. The action of steroids and streptolysin S on the permeability of phospholipid structures to cations. J Mol Biol. 1965;13(1):253–259. doi: 10.1016/s0022-2836(65)80094-8. [DOI] [PubMed] [Google Scholar]
- 92.Wicki A, Witzigmann D, Balasubramanian V, Huwyler J. Nanomedicine in cancer therapy: Challenges, opportunities, and clinical applications. J Control Release. 2015;200:138–157. doi: 10.1016/j.jconrel.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 93.Rivera E. Liposomal anthracyclines in metastatic breast cancer: Clinical update. Oncologist. 2003;8(Suppl 2):3–9. doi: 10.1634/theoncologist.8-suppl_2-3. [DOI] [PubMed] [Google Scholar]
- 94.Hawkins MJ, Soon-Shiong P, Desai N. Protein nanoparticles as drug carriers in clinical medicine. Adv Drug Deliv Rev. 2008;60(8):876–885. doi: 10.1016/j.addr.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 95.Maeda H, Takeshita J, Kanamaru R. A lipophilic derivative of neocarzinostatin. A polymer conjugation of an antitumor protein antibiotic. Int J Pept Protein Res. 1979;14(2):81–87. doi: 10.1111/j.1399-3011.1979.tb01730.x. [DOI] [PubMed] [Google Scholar]
- 96.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46(12 Pt 1):6387–6392. [PubMed] [Google Scholar]
- 97.Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nat Rev Drug Discov. 2005;4(2):145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 98.Brannon-Peppas L, Blanchette JO. Nanoparticle and targeted systems for cancer therapy. Adv Drug Deliv Rev. 2012;64(Supplement):206–212. [Google Scholar]
- 99.Cheng J, Khin KT, Davis ME. Antitumor activity of β-cyclodextrin polymer-camptothecin conjugates. Mol Pharm. 2004;1(3):183–193. doi: 10.1021/mp049966y. [DOI] [PubMed] [Google Scholar]
- 100.Andresen TL, Davidsen J, Begtrup M, Mouritsen OG, Jørgensen K. Enzymatic release of antitumor ether lipids by specific phospholipase A2 activation of liposome-forming prodrugs. J Med Chem. 2004;47(7):1694–1703. doi: 10.1021/jm031029r. [DOI] [PubMed] [Google Scholar]
- 101.Perry JL, Herlihy KP, Napier ME, Desimone JM. PRINT: A novel platform toward shape and size specific nanoparticle theranostics. Acc Chem Res. 2011;44(10):990–998. doi: 10.1021/ar2000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Merkel TJ, et al. Using mechanobiological mimicry of red blood cells to extend circulation times of hydrogel microparticles. Proc Natl Acad Sci USA. 2011;108(2):586–591. doi: 10.1073/pnas.1010013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Galloway AL, et al. Development of a nanoparticle-based influenza vaccine using the PRINT technology. Nanomedicine (Lond) 2013;9(4):523–531. doi: 10.1016/j.nano.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 104.Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Japsen B. Employers boost wellness spending 17% from yoga to risk assessments. Forbes March 26, 2015 [Google Scholar]
- 106.Wang J. Electrochemical glucose biosensors. Chem Rev. 2008;108(2):814–825. doi: 10.1021/cr068123a. [DOI] [PubMed] [Google Scholar]
- 107.Lammel G. 2015. The future of MEMS sensors in our connected world. 28th IEEE International Conference on Micro Electro Mechanical Systems (MEMS). (IEEE, New York) pp 61–64.
- 108.Kim D-H, et al. Epidermal electronics. Science. 2011;333(6044):838–843. doi: 10.1126/science.1206157. [DOI] [PubMed] [Google Scholar]
- 109.Xu S, et al. Soft microfluidic assemblies of sensors, circuits, and radios for the skin. Science. 2014;344(6179):70–74. doi: 10.1126/science.1250169. [DOI] [PubMed] [Google Scholar]
- 110.McAlpine M, Amad H, Wang D, Heath JR. Highly ordered nanowire arrays on plastic substrates for ultra-sensitive flexible chemical sensors. Nat Mater. 2007;6(5):379–383. doi: 10.1038/nmat1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takei K, et al. Nanowire active-matrix circuitry for low-voltage macroscale artificial skin. Nat Mater. 2010;9(10):821–826. doi: 10.1038/nmat2835. [DOI] [PubMed] [Google Scholar]
- 112.Wu W, et al. Piezoelectricity of single-atomic-layer MoS2 for energy conversion and piezotronics. Nature. 2014;514(7523):470–474. doi: 10.1038/nature13792. [DOI] [PubMed] [Google Scholar]
- 113.Lipomi DJ, et al. Skin-like pressure and strain sensors based on transparent elastic films of carbon nanotubes. Nat Nanotechnol. 2011;6(12):788–792. doi: 10.1038/nnano.2011.184. [DOI] [PubMed] [Google Scholar]
- 114.Kaltenbrunner M, et al. An ultra-lightweight design for imperceptible plastic electronics. Nature. 2013;499(7459):458–463. doi: 10.1038/nature12314. [DOI] [PubMed] [Google Scholar]
- 115.Wang C, et al. User-interactive electronic skin for instantaneous pressure visualization. Nat Mater. 2013;12(10):899–904. doi: 10.1038/nmat3711. [DOI] [PubMed] [Google Scholar]
- 116.Xu S, et al. Self-powered nanowire devices. Nat Nanotechnol. 2010;5(5):366–373. doi: 10.1038/nnano.2010.46. [DOI] [PubMed] [Google Scholar]
- 117.Hammock ML, Chortos A, Tee BCK, Tok JBH, Bao Z. 25th anniversary article: The evolution of electronic skin (e-skin): A brief history, design considerations, and recent progress. Adv Mater. 2013;25(42):5997–6038. doi: 10.1002/adma.201302240. [DOI] [PubMed] [Google Scholar]
- 118.Bandodkar AJ, Wang J. Non-invasive wearable electrochemical sensors: A review. Trends Biotechnol. 2014;32(7):363–371. doi: 10.1016/j.tibtech.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 119.Kong J, et al. Nanotube molecular wires as chemical sensors. Science. 2000;287(5453):622–625. doi: 10.1126/science.287.5453.622. [DOI] [PubMed] [Google Scholar]
- 120.Cui Y, Wei Q, Park H, Lieber CM. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293(5533):1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 121.Bunimovich YL, et al. Quantitative real-time measurements of DNA hybridization with alkylated nonoxidized silicon nanowires in electrolyte solution. J Am Chem Soc. 2006;128(50):16323–16331. doi: 10.1021/ja065923u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McAlpine MC, et al. High-performance nanowire electronics and photonics on glass and plastic substrates. Nano Lett. 2003;3(11):1531–1535. [Google Scholar]
- 123.Cao Q, Rogers JA. Ultrathin films of single-walled carbon nanotubes for electronics and sensors: A review of fundamental and applied aspects. Adv Mater. 2009;21(1):29–53. [Google Scholar]
- 124.Mannoor MS, et al. Graphene-based wireless bacteria detection on tooth enamel. Nat Commun. 2012;3:763. doi: 10.1038/ncomms1767. [DOI] [PubMed] [Google Scholar]
- 125.Baltimore D, et al. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348(6230):36–38. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Corbyn Z. May 10, 2015. Crispr: Is it a good idea to 'upgrade' our DNA. The Guardian, US Edition (The Guardian, New York)
- 127.Liang P, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]