Abstract
Chronically increased echogenicity on renal ultrasound is a sensitive early finding of chronic kidney disease that can be detected before manifestation of other symptoms. Increased echogenicity, however, is not specific for a certain etiology of chronic kidney disease. Here, we performed whole exome sequencing in 79 consanguineous or familial cases of suspected nephronophthisis in order to determine the underlying molecular disease cause. In 50 cases, there was a causative mutation in a known monogenic disease gene. In 32 of these cases whole exome sequencing confirmed the diagnosis of a nephronophthisis-related ciliopathy. In 8 cases it revealed the diagnosis of a renal tubulopathy. The remaining 10 cases were identified as Alport syndrome (4), autosomal-recessive polycystic kidney disease (2), congenital anomalies of the kidney and urinary tract (3), and APECED syndrome (1). In 5 families, in whom mutations in known monogenic genes were excluded, we applied homozygosity mapping for variant filtering, and identified 5 novel candidate genes (RBM48, FAM186B, PIAS1, INCENP, and RCOR1) for renal ciliopathies. Thus, whole exome sequencing allows the detection of the causative mutation in 2/3 of affected individuals, thereby presenting the etiologic diagnosis and allows identification of novel candidate genes.
Keywords: chronic kidney disease, pediatric nephrology, genetic kidney disease, whole exome sequencing, mutation analysis, monogenic diseases, increased renal echogenicity, nephronophthisis
INTRODUCTION
Renal ultrasound imaging (RUS) represents a simple and broadly accessible tool for the non-invasive early diagnosis of chronic kidney disease (CKD) in children and young adults. Often abnormal findings on RUS are detectable years before the kidney function deteriorates and before other symptoms develop. A typical abnormal finding in early stages of CKD is chronically increased echogenicity on RUS. It is frequently accompanied by loss of cortico-medullary differentiation and renal cysts. Increased echogenicity is easily detected as a degree of echogenicity that is equal to or more pronounced than the echogenicity of the liver. Unfortunately, chronically increased echogenicity is not specific to certain types of kidney disease1–3. Particularly in early stages, in which other symptoms are not yet present, a correct diagnosis can be challenging. In these cases, whole exome sequencing (WES) provides a novel means of establishing an etiologic diagnosis. By revealing the causative monogenic mutation, it provides affected individuals and their families with an unequivocal, early diagnosis1. As a result, a targeted therapeutic regimen can be initiated early if available.
Chronically increased echogenicity on RUS is often found in the early stages of nephronophthisis-related ciliopathies (NPHP-RC). NPHP-RC represent a group of cystic and fibrotic kidney diseases with an autosomal recessive mode of inheritance that typically progress to end-stage renal failure within the first three decades of life4, 5. Nephronophthisis can present as isolated renal disease (MIM #613550), or together with extrarenal symptoms such as retinal degeneration (Senior Loken syndrome MIM #266900), cerebellar vermis hypoplasia (Joubert syndrome MIM #213300), and hepatic fibrosis. The renal manifestation ranges from severe, early onset cystic kidney disease6 to slowly progressive, fibrotic remodeling of the kidney with CKD starting in adolescence7. Interestingly, the genotype-phenotype correlation in NPHP-RC is dependent on the gene and the specific mutation involved, which can both give rise to a broad phenotypic disease spectrum. NPHP-RC are a very heterogeneous disease group as by now, mutations in more than 90 genes have been identified as causative for renal ciliopathies in humans4. Mutations in some of these genes are very rare, accounting for only two8, or three9 families worldwide. WES with direct inspection of the coding regions of these genes therefore represents the most rational and currently, most cost-effective approach for mutation analysis in these patients1, 10–12. So far, no more than 13 ciliopathy genes have been systematically studied in a larger patient cohort13, 14.
Here, we performed WES combined with homozygosity mapping in an international cohort of 79 families with pediatric onset of CKD and suspected nephronophthisis based on renal ultrasound presentation with chronically increased echogenicity, loss of cortico-medullary differentiation, and/ or ≥2 cysts. All individuals were born of consanguineous union, or represented familial cases of CKD, and were therefore at high risk for recessive, monogenic diseases. In summary, we were able to identify a mutation in a known monogenic disease gene in 50 families (63.3%). In 32 of these families (64%) WES identified mutations in NPHP-RC genes as the molecular disease cause, and confirmed the suspected clinical diagnosis. However, in 18 families (36%) we discovered a molecular diagnosis of a monogenic kidney disease that was not NPHP-RC, specifically renal tubulopathies (n=8, 16%), Alport syndrome (n=4, 8%), congenital anomalies of the kidney and urinary tract (CAKUT) (n=3, 6%), autosomal recessive polycystic kidney disease (ARPKD) (n=2, 4%), and autoimmune nephropathy (APECED syndrome) (n=1, 2%). In 5 consanguineous families, in whom we excluded mutations in known monogenic disease genes, we identified 5 novel candidate genes for NPHP-RC (RBM48, FAM168B, PIAS1, INCENP, and RCOR1).
RESULTS
WES identifies the molecular disease cause in 63% of cases
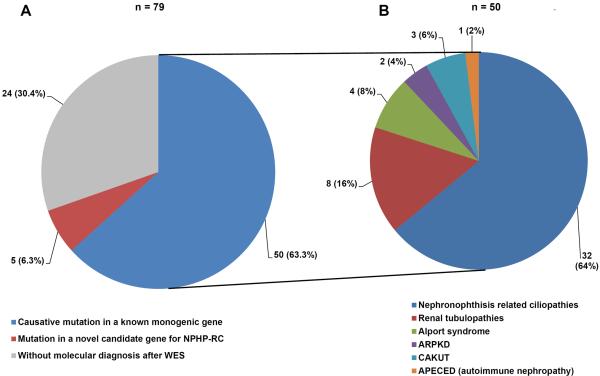
We performed WES in 79 families with suspected NPHP-RC based on renal ultrasound criteria (chronically increased echogenicity, loss of cortico-medullary differentiation, and/or ≥2 renal cysts). In 50/79 families (63.3%) we identified a mutation in a gene that is known to cause monogenic renal disease when mutated (Fig. 1A, Suppl. Table 1 / 2).
Figure 1. Relative number of cases molecularly diagnosed by WES in 79 families with child-hood onset chronically increased echogenicity or ≥2 cysts on renal ultrasound.
A) We performed whole exome sequencing in 79 consanguineous or sibling cases with increased echogenicity and/or ≥2 cysts on renal ultrasound. In 50 families (63.3%) we identified a mutation in a known monogenic disease gene as causative. In 5 families (6.3%) we identified a mutation in a novel candidate gene for NPHP-RC, and 24 families (30.4%) remained without a molecular diagnosis after WES.
B) Fractional contribution of different disease entities to the molecular diagnosis of 50 families in whom a causative mutation in a known monogenic disease gene was detected. (See Table 1 for the underlying monogenic causes in each disease group.)
NPHP-RC account for 64% of molecularly diagnosed individuals
32 of the 50 families with established molecular diagnosis after WES (64.0%) harbored a mutation in a known NPHP-RC gene (Fig. 1B). Of the 90 genes that are known to cause renal ciliopathies when mutated which were systematically analyzed in this study, mutations in 18 genes contributed to this result. Mutations in the genes NPHP3, NPHP4, and NPHP5 accounted for the majority of NPHP-RC cases (Table 1, Suppl. Table 1/2). In addition to mutations in NPHP-RC genes, we detected causative mutations in monogenic genes of renal tubulopathies (8/50), Alport syndrome (4/50), CAKUT (3/50), and ARPKD (2/50). Furthermore, we established the molecular diagnosis of APECED (autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy) in one previously undiagnosed individual (Fig. 1B, Table 1, Suppl. Table 1/2). In 10 individuals, the molecular diagnosis after WES was different from the previous clinical diagnosis. In particular, in one case of previously undiagnosed nephropathic cystinosis the molecular finding changed the therapeutic regimen (Suppl. Table 1/2).
Table 1.
Diagnostic groups and distribution among specific genes with causative recessive mutations in 50 families with increased echogenicity and/or ≥ 2 cysts on renal ultrasound in whom the disease-causing mutation was detected by WES.
| Disease group | Patients within the disease group | Gene with causative mutation (disease, OMIM#a) |
|---|---|---|
|
| ||
| NPHP-RC | 32/50 (64.0%) | |
| 2/50 (4.0%) | NPHP1 (Nephronophthisis type, juvenile, #256100) | |
| 3/50 (6.0%) | NPHP3 (Nephronophthisis type 3, #604387) | |
| 5/50 (10 %) | NPHP4 (Nephronophthisis type 4, #606966) | |
| 3/50 (6.0%) | IQCB1 (Senior-Loken syndrome type 5, #609254) | |
| 1/50 (2.0%) | CEP290 (Senior-Loken syndrome 6, #610189) | |
| 2/50 (4.0%) | SDCCAG8 (Senior-Loken syndrome type 7, #613615) | |
| 1/50 (2.0%) | TMEM67 (Nephronophthisis 11, #613550) | |
| 2/50 (4.0%) | TTC21B (Nephronophthisis 12, #613820) | |
| 2/50 (4.0%) | WDR19 (Nephronophthisis 13, #614377) | |
| 3/50 (6.0%) | ANKS6 (Nephronophthisis 16, #615382) | |
| 1/50 (2.0%) | TMEM138 (Joubert syndrome 16, #614465) | |
| 1/50 (2.0%) | TMEM231 (Joubert syndrome 20, #614970) | |
| 1/50 (2.0%) | ARL6 (Bardet-Biedl syndrome 3, #600151) | |
| 1/50 (2.0%) | BBS4 (Bardet-Biedl syndrome 4, #615982) | |
| 1/50 (2.0%) | MKKS (Bardet-Biedl syndrome 6, #605231) | |
| 1/50 (2.0%) | BBS12 (Bardet-Biedl syndrome 12, #615989) | |
| 1/50 (2.0%) | DYNC2H1 (Short-rib thoracic dysplasia 3, #613091) | |
| 1/50 (2.0%) | IFT43 (Cranioectodermal dysplasia 3, #614099) | |
|
| ||
| Renal tubulopathies | 8/50 (16.0%) | |
| 3/50 (6.0%) | CLDN16 (Hypomagnesemia 3, renal, #248250) | |
| 2/50 (4.0%) | CLCNKB (Bartter syndrome, type 3, #607364) | |
| 1/50 (2.0%) | ATP6V0A4 (Renal tubular acidosis, distal, #602722) | |
| 1/50 (2.0%) | BCS1L (Mitochondrial complex III deficiency, #124000) | |
| 1/50 (2.0%) | CTNS (Cystinosis, nephropathic, #219800) | |
|
| ||
| Alport syndrome | 4/50 (8.0%) | |
| 2/50 (4.0%) | COL4A3 (Alport syndrome, #203780) | |
| 1/50 (2.0%) | COL4A4 (Alport syndrome, #203780) | |
| 1/50 (2.0%) | COL4A5 (Alport syndrome, #301050) | |
|
| ||
| CAKUT | 3/50 (6.0%) | |
| 2/50 (4.0%) | HNF1b (Renal cysts and diabetes syndrome, #137920) | |
| 1/50 (2.0%) | FREM2 (Fraser syndrome, #219000) | |
|
| ||
| Autosomal recessive polycystic kidney disease | 2/50 (4.0%) | PKHD1 (Polycystic kidney and hepatic disease, #263200) |
|
| ||
| APECED | 1/50 (2.0%) | AIRE (Autoimmune polyendocrinopathy syndrome type I, #240300) |
APECED, autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy; CAKUT, congenital anomalies of the kidney and urinary tract; NPHP-RC, nephronophthisis related ciliopathies
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org;
Mutations in more than one recessive disease gene can be present in consanguineous families
In one example of a highly inbred family (412 Mb of cumulative homozygosity) we identified homozygous mutations in 7 known monogenic disease genes. 4 of them are known to cause diseases with renal involvement (Suppl. Table 3). The affected child showed a complex phenotype suggestive of a renal ciliopathy including Caroli's disease with massive cystic dilation of intrahepatic bile ducts, CKD that progressed to end-stage renal failure within the 3rd year of life, post axial polydactyly, nystagmus, and red cone dystrophy. In addition to chronically increased echogenicity as the classical symptom of NPHP-RC, the child showed congenital hydronephrosis due to uretero-pelvic junction obstruction (UPJO) that required corrective surgery, as well as renal tubular acidosis type 4. (Suppl. Table 3). An older brother, from whom no DNA sample was available for mutation analysis, deceased at 15 months of age due to end-stage renal and liver disease. Additionally, he showed post axial polydactyly and blindness at 5 months of age due to progressive retinal dystrophy. A younger brother presented with Caroli's disease and retinitis pigmentosa. Interestingly, no polydactyly was present, and the renal function was preserved at 3 years of age. The right kidney showed mild pelvicalyceal dilation.
The percentage of molecularly solved cases in consanguineous families and familial cases of suspected NPHP-RC is comparable (~63%)
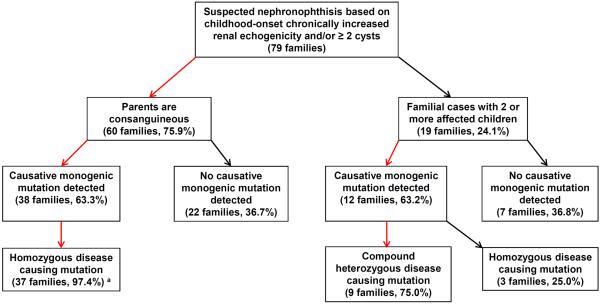
When performing WES in 79 families with suspected nephronophthisis based on renal ultrasound imaging with childhood-onset chronically increased renal echogenicity and/or ≥2 renal cysts, we detected a causative mutation in a known monogenic disease gene in 38 of 60 consanguineous families (63.3%) (Fig. 2). As postulated15, 16 in the vast majority of consanguineous families the causative mutation was present in the homozygous state (37 of 38 families (97.4%)) (Fig. 2). The only exception was a family in whom we identified a single heterozygous mutation in the dominant gene HNF1ß as causative. In 12 of 19 non-consanguineous families with 2 or more affected children (63.2%) we identified a causative monogenic mutation, showing a comparable percentage of molecularly solved cases in both cohorts. However, as expected in non-consanguineous families16 only in 3 of 12 families (25.0%) the disease-causing mutation was present in the homozygous state. The remaining 9 of 12 families (75.0%) harbored two compound heterozygous disease-causing mutations (Fig. 2).
Figure 2. Algorithm for molecular diagnostics in consanguineous or familial cases of suspected nephronophthisis based on renal ultrasound presentation.
Of 79 families with childhood-onset increased renal echogenicity and/or ≥ 2 cysts on RUS, 60 individuals were born of consanguineous unions, and 19 families were non-consanguineous with two or more affected children. In 63.3% of consanguineous families, we identified a mutation in a known monogenic disease gene as causative. The majority of these mutations were, as postulated, present in the homozygous state. In 63.2% of familial cases, we identified a causative mutation in a recessive, monogenic disease gene. 3/4 of these mutations were compound heterozygous.
a In one consanguineous family a single heterozygous mutation in the dominant gene HNF1ß was identified as the molecular disease cause.
Identification of 5 novel candidate genes for NPHP-RC
By applying homozygous peak regions as a filter for WES data, we identified 5 novel candidate genes for monogenic, recessive NPHP-RC (Table 2). In these 5 consanguineous families we had excluded mutations in known monogenic disease genes with renal phenotypes through evaluation and direct inspection of the WES data. The 5 novel candidate genes (RBM48, FAM168B, PIAS1, INCENP, and RCOR) represented the most deleterious alleles within the homozygous regions (Suppl. Fig 2) after variant filtering as outlined in Suppl. Fig 1. In the example of family A2621 with 181 Mb of cumulative homozygosity (Suppl. Table 4), we started with 482,406 variants from normal reference sequence. Excluding common variants (minor allele frequency >1% in healthy control cohorts), synonymous variants, and heterozygous variants reduced the number to 1,699. Considering only variants that were positioned within homozygous peak regions reduced the number of remaining variants by 11 fold to 156. Exclusion of artifacts by direct inspection of the sequence alignment, left us with 3 potentially disease causing variants in this family (Suppl. Table 4). Subsequently, these 3 variants were ranked based on their predicted likelihood to be deleterious for the function of the encoded protein following the criteria as outlined in Suppl. Fig. 1. At the end of the filtering process the gene INCENP (Inner centromere protein) represented the strongest remaining variant in this family. All mutations in novel candidate genes were confirmed by Sanger sequencing in original patient DNA, and segregated with the affected status.
Table 2.
Novel candidate genes for NPHP-RC identified in 5 families with increased echogenicity and/or ≥ 2 cysts on renal ultrasound in whom a causative mutation in a known monogenic disease gene was excluded. Each candidate gene represents the most deleterious mutation within the homozygous peak regions of the respective family.
| Family | Gene | Zygosity | Accession # | c.Position | p.Position | Continuosly conserved to | Polyphen-2/MutTast/SIFT | ExAc | EVSa | Clinical diagnosis |
|---|---|---|---|---|---|---|---|---|---|---|
| A2621 | INCENP | Hom | NM_020238.2 | c.2403G>C | p.Gln801His | Danio rerio | 0.96/ DC/ del | 0/337/120826 | 0/32/4267 | NPHP |
| A1833 | RBM48 | Hom | NM_032120.2 | c.835A>G | p.Thr279Ala | Ciona intestinalis | 0.71/ DC/ del | 0/5/120562 | not present | NPHP + JATD |
| A2275 | FAM186B | Hom | NM_032130.2 | c.506-2A>G | Obligatory splice site mutation | 0/1/111410 | not present | NPHP | ||
| A2287 | PIAS 1 | Hom | NM_016166.1 | C.317C>T | p.Ser106Leu | Danio rerio | 0.77/ DC/ del | 0/3/120766 | not present | NPHP |
| A1239 | RC0R1 | Hom | NM_015156.2 | C.437-3C>T | Splice site mutation | 0/3/121306 | not present | JBTS | ||
DC, disease causing (MutationTaster); del, deleterious (SIFT); EVS, Exome variant server; ExAc, Exome Aggregation Consortium; JATD, Jeune asphyxiating thoracic dystrophy; JBTS: Joubert syndrome; MutTast, MutationTaster; NPHP, nephronophthisis; SIFT, Sorting Intolerant From Tolerant (SIFT)
EVS: genotypes in European Americans.
24 families (30.4%) remained without a molecular diagnosis after WES. In these families, no convincing biallelic variants in known monogenic genes, or candidate genes were identified that segregated with the affected status.
DISCUSSION
We performed WES combined with homozygosity mapping in 79 consanguineous or familial cases of childhood onset chronically increased renal echogenicity or the presence of ≥2 cysts on renal ultrasound. Based on renal ultrasound presentation, clinical presentation with childhood-onset of disease, and if available renal histology, a nephronophthisis-related ciliopathy (NPHP-RC) was suspected as the primary disease cause. We show that in this patient cohort, WES allows detecting the specific causative mutation in about 2/3 of affected individuals, thereby presenting the etiologic diagnosis. In addition, we identified 5 potential novel renal ciliopathy genes in consanguineous families in whom mutations in 90 known monogenic NPHP-RC genes had been excluded.
Chronically increased echogenicity on renal ultrasound represents a sensitive diagnostic criterion for early stages of CKD in children and young adults. Typically, this results in the suspected diagnosis of nephronophthisis or related ciliopathies. However, we and others1–3 have shown that other kidney diseases can phenocopy the presentation of NPHP-RC on renal ultrasound. Here, we study 79 consanguineous or familial cases with suspected NPHP-RC based on renal ultrasound presentation with childhood-onset increased renal echogenicity and/or more than one cyst, and perform WES in order to determine the percentage in which NPHP-RC and various other monogenic, recessive kidney diseases contribute to this phenotypic spectrum (Figure 2). As a second aspect, we aimed to systematically examine all 90 genes that are known monogenic, recessive causes of NPHP-RC or other CKD that can phenocopy the presentation of NPHP-RC in a large cohort of affected individuals. So far, this has not been done for more than 13 renal ciliopathy genes13, 14 at a time. As we identified the disease-causing mutation in ~ 2/3 (50/79) of individuals with increased echogenicity on RUS, our data shows that WES is an efficient tool for molecular diagnostics in this patient cohort. WES provides affected individuals and their families with an unequivocal diagnosis that avoids unnecessary diagnostic and therapeutic interventions. WES is a non-invasive diagnostic tool with little risk, and recently, the cost has dropped to a level that makes it accessible to a broader group of affected individuals in a clinical rather than a research setting. Considering the genetic heterogeneity of NPHP-RC, WES is the only available technique that can reliably establish the correct molecular diagnosis. The use of targeted sequencing panels as utilized for other disease entities in clinical genetics is technically not feasible and/or not economical for such a large number of potentially causative genes. However, WES yields a large number of variants from normal reference sequence, most of which are functionally irrelevant. Therefore an efficient strategy for variant filtering is indispensable for the work with WES data. In addition, variant calling should follow an indication-driven strategy: After thorough clinical evaluation, a subset of potentially causative genes should be defined prior to WES, and the evaluation should primarily focus on variants in these genes. For example in a patient with increased echogenicity and/or ≥2 cysts on renal ultrasound, primarily variants in the ~90 monogenic causes of renal ciliopathies, and genes that can phenocopy NPHP-RC, should be taken into consideration.
In our cohort, WES molecularly confirmed the clinically suspected diagnosis of NPHP-RC in 64% of cases. However, 18/50 individuals (36%) harbored a mutation in a gene that causes a monogenic kidney disease different from NPHP-RC. As already described in a smaller cohort1, the majority of these cases (8/18) were diagnosed as renal tubulopathies. In addition, we identified 4 individuals with Alport syndrome, 3 individuals with mutations in CAKUT genes, 2 individuals with ARPKD, and 1 individual with autoimmune nephropathy in the context of APECED syndrome. In 10 of these 18 cases the molecular diagnosis differed from the initial clinical diagnosis. We therefore suggest that WES helps to distinguish between NPHP-RC and other kidney diseases that phenocopy the presentation of NPHP-RC on renal ultrasound. This can be particularly advantageous in very early stages of disease progression in which additional characteristic symptoms might not yet be present, as well as for very rare genetic syndromes. This observation underlines an additional benefit of WES: The genetic diagnostic is not restricted to a limited subset of monogenic diseases that were already suspected based on the clinical presentation, but a broader spectrum of monogenic causes can be taken into consideration. In addition, the very high rate of detecting a causative mutation in genes that are known to be mutated in renal disorders with increased echogenicity, confirms that increased renal echogenicity, even if performed in numerous centers worldwide, is a reproducible and reliable criterion for non-invasive diagnostic in pediatric kidney disease. Interestingly, the percentage of molecularly diagnosed individuals was not considerably different in consanguineous families as compared to outbred sibling cases. We therefore suggest that the relevance of our findings reaches beyond consanguineous cases.
As we and others have shown10, 12, 17 combining WES and homozygosity mapping in consanguineous families represents a powerful tool for the identification of novel human disease genes. We applied this technique in 5 families in whom mutations in known human disease genes had been excluded, and we identified RBM48, FAM168B, PIAS1, INCENP, and RCOR1 as novel candidate genes for renal ciliopathies. So far, mutation analysis has not yielded additional families with mutations in these genes. However, this rarity is not unexpected as for the majority of recently identified NPHP-RC genes less than 10 families with mutations have been described worldwide9–12, 18–20 At this point, additional functional evidence to proof the pathogenicity of these mutations is lacking. Further genetic and experimental evidence in the future will help to determine whether mutations in these newly identified candidate genes are indeed disease-causing in humans with renal ciliopathies.
24 of 79 families in our cohort (30.4%) remained without a molecular diagnosis after WES. This is not unexpected as in recessive monogenic diseases only about 85% of all causative mutations are located within the coding sequence or the adjacent splice sites21, 22. The remaining 15% however, are complex deletion-insertion variants, copy-number variants, or reside within a promotor and other intronic region. As none of these variants can be detected by WES, this technical limitation explains why some cases cannot be solved by WES. Furthermore, WES might miss a subset of causative variants due to low coverage in the respective target region. Considering the fact that about 20% of all individuals with NPHP-RC harbor a homozygous deletion in the NPHP1 gene, the percentage of solved cases in this study had been even higher than the reported 63% if these cases were included. As it has been postulated that only 85% of recessive mutations can be technically detected with currently available techniques22, we show that in the targeted patient cohort virtually all cases with detectable recessive mutations are molecularly solved after WES.
We here present WES as a rapid and reliable tool for molecular diagnostics in consanguineous individuals or familial cases with clinically suspected NPHP-RC based on renal ultrasound that allows establishing an etiologic diagnosis in ~2/3 of affected individuals. This represents a major advance in a diagnostic finding that is frequently interpreted as “medical renal disease” rather than leading to an etiologic diagnosis.
MATERIALS AND METHODS
Human subjects
The study was approved by the institutional review board (IRB) of the University of Michigan and Boston Children's Hospital. After obtaining informed consent, clinical data, and pedigree information, DNA samples were collected from individuals with suspected nephronophthisis based on clinical findings, renal ultrasound criteria, and if present, renal histology. Subjects were either of consanguineous families, or sibling cases with multiple affected children in one family. 107 individuals from 82 families were included in the study. 4 individuals (from 3 families) were excluded for technical reasons, leading to a final cohort of 103 individuals from 79 families. 60 families were consanguineous singlets, and 19 families were non-consanguineous familial cases with more than one affected child (16 families with 2 affected children, 2 families with 3 affected children, and 1 family with 5 affected children). In all families a homozygous deletion in the NPHP1 gene had been excluded prior to study inclusion. This was necessary as homozygous deletions in the NPHP1 gene represent the most frequently observed mutation in individuals with nephronophthisis, but cannot be detected by whole-exome sequencing.
Linkage analysis
For genome-wide homozygosity mapping the GeneChip® Human Mapping 250k Sty Array from Affymetrix was used. Non-parametric LOD scores were calculated using a modified version of the program GENEHUNTER2.123, 24 through stepwise use of a sliding window with sets of 110 SNPs and the program ALLEGRO25 in order to identify regions of homozygosity as described15, 26 using a disease allele frequency of 0.0001 and Caucasian marker allele frequencies. The genetic mapping was based on multipoint analysis. A non-parametric LOD score of 2 was applied as the threshold for relevant homozygous peak regions, and a nonparametric LOD cut-off of 0.5 as threshold for linkage in sibling cases.
Whole Human Exome Capture, Next Gen Sequencing, Sequence Alignment, and Variant Calling
Genomic DNA was isolated from blood lymphocytes and subjected to whole exome capture using a customized Agilent SureSelect All Exome Kit v2.0 (Agilent Technologies, Santa Clara, CA, USA) according to manufacturer's protocol. The library was sequenced on an Illumina HighSeq™ sequencing platform. Image analysis and base calling were generated by the Illumina pipeline using default parameters. Sequence reads were mapped to the human reference genome assembly (NCBI build 3/hg19) using CLC Genomics Workbench™ (version 6.5.2) software (CLC bio, Aarhus, Denmark). In consanguineous cases, only variants with a common allele frequency of more than 80% were called for further evaluation in order to identify homozygous variants. Trimmed sequence reads were mapped to the human reference genome (hg19) using the Map Reads to Reference program with the following settings: mismatch cost = 2, insertion cost = 3, deletion cost =3, length fraction = 0.5, similarity fraction = 0.9 and map to nonspecific reads = “randomly”. The non-specific reads were then ignored for count and coverage. All variants with a minimum coverage of 2 were used. The variations in the samples were called using probabilistic variant detection using CLC bio. All the called variants were then annotated and evaluated using mutation calling criteria as outlined in Suppl. Fig. 1.
Variant Annotation and Evaluation
After alignment to the human reference genome, variants were filtered as previously described1, 27, and as summarized in Suppl. Fig. 1. The ranking and calling of alleles as disease-causing mutations follows accepted standards in molecular diagnostics28–31, and with regards to the composition of our cohort, we applied criteria that are even stricter than the ones typically used in the field. In the first step variants with minor allele frequencies >1% in the dbSNP (version 135) or the 1,000 Genomes (1,094 subjects of various ethnicities; May, 2011 data release) databases were excluded. In the second step only homozygous and biallelic variants (in non-consanguineous cases) were kept, whereas single heterozygous variants were excluded from the further evaluation. Subsequently, in step 3 synonymous variants and intronic variants that were not located within splice site regions were excluded. In step 4 genetic mapping data (homozygosity mapping in consanguineous families and linkage analysis in sibling cases) was applied for variant filtering15. In the final step, remaining variants were ranked based on their probable impact on the function of the encoded protein. We considered only mutations as disease-causing that full-filled the following criteria: a) protein-truncating, or b) previously reported as disease-causing in individuals with a similar phenotype (HGMD biobase), or c) missense mutations were only included if they affected highly conserved amino acid residues (conserved in orthologues below vertebrate evolution) and were not reported with a minor variant frequency >0.5% in healthy control individuals, and were predicted to be deleterious for the protein function (PolyPhen-2 score32 >0.7). Variants that were present in the homozygous state in any publicly available control cohort were excluded. Mutation calling was performed by geneticists/cell biologists, who had knowledge of the clinical phenotypes and pedigree structure, as well as experience with homozygosity mapping and exome evaluation. Remaining variants were confirmed in original patient DNA by Sanger sequencing as previously described33. Whenever parental DNA was available, segregation analysis was performed.
In a second evaluation process, variants in all 90 genes that are known monogenic, recessive causes of NPHP-RC or other CKD that can phenocopy the presentation of NPHP-RC were systematically evaluated.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the physicians and the participating families for their contribution. F.H. is an Investigator of the Howard Hughes Medical Institute, a Doris Duke Distinguished Clinical Scientist, and the Warren E. Grupe Professor of Pediatrics. This research was supported by grants from the National Institutes of Health (DK1069274, DK1068306, and DK064614 to FH; DK099434 to RA, 5U54HG006504 to RPL), and by the March of Dimes Foundation (6-FY11-241 to FH). H.Y.G. is supported by the ASN-NephCure Foundation grant. B.B.B. acknowledges funding from university endowments of the Faculty of Medicine (Stiftungsgelder Imhoff-Stifung).
Footnotes
Web Resources UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway.
1000 Genomes Browser, http://browser.1000genomes.org;
Ensembl Genome Browser, http://www.ensembl.org;
Exome Variant Server, http://evs.gs.washington.edu/EVS;
Exome Aggregation Consortium, exac.broadinstitute.org;
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org;
Polyphen2, http://genetics.bwh.harvard.edu/pph2;
Sorting Intolerant From Tolerant (SIFT), http://sift.jcvi.org/
MutationTaster http://www.mutationtaster.org/
STATEMENT OF DISCLOSURE None of the authors has competing financial interests to disclose.
REFFERENCES
- 1.Gee HY, Otto EA, Hurd TW, et al. Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney international. 2014;85:880–887. doi: 10.1038/ki.2013.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraus RA, Gaisie G, Young LW. Increased renal parenchymal echogenicity: causes in pediatric patients. Radiographics : a review publication of the Radiological Society of North America, Inc. 1990;10:1009–1018. doi: 10.1148/radiographics.10.6.2259758. [DOI] [PubMed] [Google Scholar]
- 3.Moghazi S, Jones E, Schroepple J, et al. Correlation of renal histopathology with sonographic findings. Kidney international. 2005;67:1515–1520. doi: 10.1111/j.1523-1755.2005.00230.x. [DOI] [PubMed] [Google Scholar]
- 4.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. The New England journal of medicine. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hildebrandt F, Attanasio M, Otto E. Nephronophthisis: disease mechanisms of a ciliopathy. Journal of the American Society of Nephrology : JASN. 2009;20:23–35. doi: 10.1681/ASN.2008050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otto EA, Schermer B, Obara T, et al. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nature genetics. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olbrich H, Fliegauf M, Hoefele J, et al. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nature genetics. 2003;34:455–459. doi: 10.1038/ng1216. [DOI] [PubMed] [Google Scholar]
- 8.Attanasio M, Uhlenhaut NH, Sousa VH, et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nature genetics. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- 9.Otto EA, Trapp ML, Schultheiss UT, et al. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. Journal of the American Society of Nephrology : JASN. 2008;19:587–592. doi: 10.1681/ASN.2007040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaki M, Airik R, Ghosh Amiya K, et al. Exome Capture Reveals ZNF423 and CEP164 Mutations, Linking Renal Ciliopathies to DNA Damage Response Signaling. Cell. 2012;150:533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto EA, Hurd TW, Airik R, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nature genetics. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schueler M, Braun DA, Chandrasekar G, et al. DCDC2 Mutations Cause a Renal-Hepatic Ciliopathy by Disrupting Wnt Signaling. American journal of human genetics. 2015;96:81–92. doi: 10.1016/j.ajhg.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halbritter J, Diaz K, Chaki M, et al. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. Journal of medical genetics. 2012;49:756–767. doi: 10.1136/jmedgenet-2012-100973. [DOI] [PubMed] [Google Scholar]
- 14.Halbritter J, Porath JD, Diaz KA, et al. Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Human genetics. 2013;132:865–884. doi: 10.1007/s00439-013-1297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hildebrandt F, Heeringa SF, Ruschendorf F, et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS genetics. 2009;5:e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ten Kate LP, Scheffer H, Cornel MC, et al. Consanguinity sans reproche. Human genetics. 1991;86:295–296. doi: 10.1007/BF00202413. [DOI] [PubMed] [Google Scholar]
- 17.Zariwala MA, Gee HY, Kurkowiak M, et al. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. American journal of human genetics. 2013;93:336–345. doi: 10.1016/j.ajhg.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaki M, Hoefele J, Allen SJ, et al. Genotype-phenotype correlation in 440 patients with NPHP-related ciliopathies. Kidney international. 2011 doi: 10.1038/ki.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoff S, Halbritter J, Epting D, et al. ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nature genetics. 2013;45:951–956. doi: 10.1038/ng.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Failler M, Gee HY, Krug P, et al. Mutations of CEP83 cause infantile nephronophthisis and intellectual disability. American journal of human genetics. 2014;94:905–914. doi: 10.1016/j.ajhg.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. The New England journal of medicine. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lifton RP. Individual genomes on the horizon. The New England journal of medicine. 2010;362:1235–1236. doi: 10.1056/NEJMe1001090. [DOI] [PubMed] [Google Scholar]
- 23.Kruglyak L, Daly MJ, Reeve-Daly MP, et al. Parametric and nonparametric linkage analysis: a unified multipoint approach. American journal of human genetics. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 24.Strauch K, Fimmers R, Kurz T, et al. Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. American journal of human genetics. 2000;66:1945–1957. doi: 10.1086/302911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudbjartsson DF, Jonasson K, Frigge ML, et al. Allegro, a new computer program for multipoint linkage analysis. Nature genetics. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 26.Sayer JA, Otto EA, O'Toole JF, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nature genetics. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 27.Sadowski CE, Lovric S, Ashraf S, et al. A Single-Gene Cause in 29.5% of Cases of Steroid-Resistant Nephrotic Syndrome. Journal of the American Society of Nephrology : JASN. 2014 doi: 10.1681/ASN.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. Jama. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. Jama. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. The New England journal of medicine. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otto EA, Ramaswami G, Janssen S, et al. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. Journal of medical genetics. 2010 doi: 10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.