Abstract
Abiotic stress is a primary threat to fulfill the demand of agricultural production to feed the world in coming decades. Plants reduce growth and development process during stress conditions, which ultimately affect the yield. In stress conditions, plants develop various stress mechanism to face the magnitude of stress challenges, although that is not enough to protect them. Therefore, many strategies have been used to produce abiotic stress tolerance crop plants, among them, abscisic acid (ABA) phytohormone engineering could be one of the methods of choice. ABA is an isoprenoid phytohormone, which regulates various physiological processes ranging from stomatal opening to protein storage and provides adaptation to many stresses like drought, salt, and cold stresses. ABA is also called an important messenger that acts as the signaling mediator for regulating the adaptive response of plants to different environmental stress conditions. In this review, we will discuss the role of ABA in response to abiotic stress at the molecular level and ABA signaling. The review also deals with the effect of ABA in respect to gene expression.
Keywords: abiotic stress, abscisic acid, ABA signaling, gene expression, phytohormone
Introduction
The world population is increasing and is projected to rise by more than one billion by 2030 and over 2.4 billion by 2050 (United Nations Department of Economic and Social Affairs, and Population Division, 2015). Therefore, to feed the increasing population, agricultural food production must be increased by 70 percent by 2050 (Tilman et al., 2011; Wani and Sah, 2014). The current and projected changes in abiotic stresses such as drought, salinity, cold, and heat will adversely affect the plant growth that ultimately limits the productivity and is the leading cause of crop losses worldwide (Boyer, 1982; Tuteja, 2007; Qin et al., 2011; Bailey-Serres et al., 2012), and thus our ability to feed the population.
According to various studies, abiotic stresses trigger many physiological, biochemical, and molecular responses that influence various cellular processes in plants (Wang et al., 2001, 2003; Hasanuzzaman et al., 2013). To combat various environmental stresses novel and dynamic approaches should be devised, and phytohormone engineering could be a method of choice to improve the productivity. Phytohormones are the key regulators of plant growth and development as well as mediators of environmental stress responses (Sreenivasulu et al., 2012). Among various phytohormones, abscisic acid (ABA), which is the central regulator of abiotic stress resistance in plants and coordinates an array of functions (Finkelstein, 2013; Wani and Kumar, 2015), enabling plants to cope with different stresses. In the plant, when environmental conditions are harsh, the level of ABA increases via ABA biosynthesis. The increased ABA binds to its receptor to initiate signal transduction leading to cellular responses to stresses (Ng et al., 2014); therefore, ABA is also called a stress hormone (Mehrotra et al., 2014). ABA is a weak acid that was first isolated from young cotton fruits as an abscission-accelerating substance by Frederick Addicott and his associates (Ohkuma et al., 1963). Initially, it was thought that ABA had a significant role in the process of fruit abscission (Ohkuma et al., 1963; Cracker and Abeles, 1969). However, later intensive studies show that ABA was not directly involved in abscission process. The precise role of ABA in abscising organs is to promote senescence and stress responses, which are the processes preceding abscission (Finkelstein, 2013). The environmental stresses like drought, salt, and low temperature that lead to decreased water availability (Cutler et al., 2010; Kim et al., 2010), also called osmotic stress, promote the synthesis of ABA. ABA is significantly increased under drought or salinity stress conditions, stimulating stomatal closure, change in gene expression, and adaptive physiological responses (Seki et al., 2002; Yamaguchi-Shinozaki and Shinozaki, 2006; Shinozaki and Yamaguchi-Shinozaki, 2007; Cutler et al., 2010; Kim et al., 2010). ABA also plays an important role in many cellular processes including seed development, dormancy, germination, vegetative growth (Xiong and Zhu, 2003; Finkelstein et al., 2008) and modulation of root architecture (Harris, 2015). Since the discovery of ABA, several efforts have been devoted to understanding how ABA is synthesized under stress conditions.
For ABA perception and signaling, two breakthroughs were achieved in 2009 by the discoveries of the soluble ABA receptor proteins and the core signaling complexes that perceive ABA and transmit cues to subsequent molecular events (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009), which adds the essences of more research with new ideas in ABA signaling. Recently, Park et al. (2015) engineered ABA receptor using agrochemicals, which provide new possibilities for the better development of the crop. Thus, a better understanding of ABA regulatory mechanisms will contribute to engineered stress tolerant crop plants, which is one of the primary goals of plant molecular biologists. This review focuses on the recent development of the role of ABA in understanding cellular networks of biotechnological relevance in abiotic stress responses of crop plants.
Role of ABA in Plants
Abscisic acid is the most important phytohormone that confers abiotic stress tolerance in crop plants (Shinozaki and Yamaguchi-Shinozaki, 2000; Schroeder et al., 2001). In stress conditions like drought, extreme temperature, and high salinity, content in plants increases considerably, inspiring stress-tolerance effects that help plants, adapt, and survive under these stressful situations (Ng et al., 2014). ABA is also required for plant growth and development under non-stress conditions. ABA has multiple functions in plants. Among them, primary features are as follows.
Seed Dormancy and Germination
Seed is a crucial organ in higher plants and the transition to seed dormancy and germination signifies a key stage in the plant life cycle, which is an important ecological and economical traits. Two hormones play a role in controlling the mechanism of seed dormancy and germination, i.e., ABA and gibberellins (GAs). Both hormones, ABA, and GAs monitor the equilibrium between seed dormancy and germination. The ABA plays a central role in the induction and maintenance of seed dormancy. It also inhibits the transition from embryonic to germination growth. During the phase of desiccation tolerance, ABA metabolism must be regulated. Sometimes, ABA translocates from the roots through xylem or phloem (Rodríguez-Gacio et al., 2009; Miransari and Smith, 2014) to the shoot system to exert its action there.
Modulation of Root Architecture
In roots, three main factors control the architecture of roots, i.e., lateral root (positions of branch roots), the angle form with parent root, and root length. Root structure is determined through interactions between the roots and its environment during their lifetime (Harris, 2015). One of the primary functions of ABA is altering root architecture, and thus changing the pattern of growth and quiescence in plant roots. For the root system, the major abiotic stress occurs when there is a scarcity of water, or the availability of water is inconsistent. Under this scenario, the levels of ABA is altered in response to the intensity of water stress. Subsequently, changes in the root environment will have both local and systemic effects on ABA-mediated responses (Dodd et al., 2008; Dodd et al., 2010; Puertolas et al., 2015). Similarly, ABA is an important mediator of drought, salt, and osmotic stress. All these three stresses reduce soil water potential whereas, salt stress has the added the component of ionic stress. Both salt and osmotic stresses lead to increased osmotic strength.
ABA stimulates water flow and ion flux in root tissues suggesting that ABA regulates turgor by decreasing transpiration as well as by increasing water influx into roots (Glinka and Reinhold, 1971). In the presence of ABA, at low water potentials, the ratio of root growth to shoot growth is higher than in the absence of ABA (Saab et al., 1990). Similarly, Spollen et al. (2000) also reported that, at low water potential, ABA accumulation maintains maize primary root elongation by restricting the production of ethylene and demonstrated that ethylene plays a main role in inhibition of root elongation. At low water potential, the maize primary root became thinner and adaptive toward resource utilization by searching new soil for water (Sharp et al., 1988; Liang et al., 1997), whereas ethylene increases lateral expansion or roots. It is also speculated that later root-specific activation of ABA signaling will shift the balance of root growth toward soil exploration, away from resource utilization (Duan et al., 2013). The accumulation of ABA in maize roots helps to elongate the roots, allowing for the exploitation of surrounding soil environments and contributing to the plants ability to cope of with water stress.
Also, ABA may act in root meristem maintenance (Liang et al., 2007). The evidence was supported by Zhang et al. (2010a), reported that ABA can promote the maintenance of stem cell by promoting of QC (quiescent center) quiescence and the suppression of stem cell differentiation (Ortega-Martínez et al., 2007; Sarkar et al., 2007; Zhang et al., 2010a). All the effects of ABA on roots appear to modulate root architecture to adopt the environmental stress conditions.
Senescence
Leaf senescence is an essential part of the final stage of plant development, which is regulated by a complex range of endogenous and environmental factors (van der Graaff et al., 2006; Lim et al., 2007). Among these factors, ABA has been put forward to affect leaf senescence significantly (Xue-Xuan et al., 2010). Foliar spraying with ABA has been revealed to promote leaf senescence in rice (Ray et al., 1983) and similar results were shown in maize by He and Jin (1999). In another study, He et al. (2005) reported that the translocation of ABA from roots to shoots may be blocked in the stay green cultivar (Maize P3845) in which lower ABA level might be significant for delaying leaf senescence. Further, it has been reported that with ABA can induce leaf yellowing, which is an indicator of leaf senescence (Yang et al., 2003; Fang et al., 2008).
An increase in endogenous ABA appears to coincide with senescence of leaves (Yang et al., 2002), which supports the notion that ABA functions in the leaf senescence. Likewise, ABA can promote senescence and induce expression of several specific senescence-associated genes (Xue-Xuan et al., 2010; Finkelstein, 2013). Many studies also reported that, in Arabidopsis mutants with deficiencies in ABA biosynthesis or signaling, exhibit altered or delayed senescence (Gan, 2003; Lim and Nam, 2005; Passioura, 2007). Lee et al. (2011) identified a receptor kinase (RPK1) that mediates age-and ABA-induced senescence in old leaves. The four different independent studies reported that NAP (NAC-like, activated by apetala3/pistillata) is a significant positive regulator that controls leaf senescence in rice and Arabidopsis (Guo and Gan, 2006; Zhou et al., 2013; Liang et al., 2014; Yang et al., 2014). Moreover, AtNAP, an ABA-inducible NAC family transcription factor, has been shown to play a fundamental role in leaf senescence (Guo and Gan, 2006). Zhang and Gan (2012) reported that senescence-associated gene 113 (SAG113) positively regulates ABA-induced leaf senescence by inhibition of stomata closure, leads to accelerates water loss in senescing leaves. In rice, overexpression of an NAC-like gene (OsNAP) can promote leaf senescence while knockdown of OsNAP delay leaf senescence (Liang et al., 2014). Further, the expression of OsNAP can be induced by ABA and is reduced in ABA-deficient mutants. They concluded that ABA-mediated leaf senescence is dependent primarily on the modulation of OsNAP expression and that OsNAP controls the ABA synthesis via a feedback mechanism (Liang et al., 2014). Recently, Takasaki et al. (2015) reported that SNAC-A, a subfamily of stress responsive NAC transcription factors, plays critical roles in ABA-induced leaf senescence signaling in Arabidopsis (Takasaki et al., 2015). Similarly, a cotton NAP-like transcription factor (GhNAP) has been shown to regulate leaf senescence through ABA-mediated pathways (Fan et al., 2015). All these studies point to the significance of NAC-type transcription factors in ABA-mediated leaf senescence.
Stomata Regulation
Stomata are small pores on the leaf surfaces formed by guard cells, which control plant gas exchange processes. Light usually stimulates stomatal opening whereas ABA and elevated CO2 levels promote partial or complete closure of stomata (Kim et al., 2010). During stomatal closure, decreased gas exchange results in the reduction of photosynthate production while decreased transpiration can reduce water loss from leaves (Suzuki et al., 2013; Mittler and Blumwald, 2015). In drought conditions, ABA alteration of guard cell ion transport, which promotes stomatal closure and prevents stomatal opening, reducing water loss (Kim et al., 2010). The ABA-activated protein kinase (AAPK) from Vicia faba is a guard cell-specific protein kinase whose catalytic activity is activated by ABA (Li and Assmann, 1996). Further, Li et al. (2000) showed that AAPK is a positive regulator of ABA-induced stomatal closure through activating plasma membrane anion channels. Many studies have shown that two types of anion channels that mediate anion release from guard cells, activated by elevated cytosolic Ca2+ levels, i.e., slow-acting sustained (S-type) and rapid transient (R-type) (Schroeder and Keller, 1992; Schroeder et al., 2001). Only S-type channel is responsible for the ABA-mediated stomatal closure (Joshi-Saha et al., 2011). Two different groups reported that Slow Anion Channel-Associated 1 (SLAC1), is the S-type channel that triggers membrane depolarization required for stomatal closure (Negi et al., 2008; Vahisalu et al., 2008). The OST1 (open stomata 1) kinase is an AAPK-related protein kinase that mediates ABA-induced stomatal closure in Arabidopsis (Mustilli et al., 2002). Recently, Grondin et al. (2015) identified OST1 as a novel regulator of plasma membrane intrinsic proteins (PIPs, a type of plant aquaporins) through phosphorylating a distinct phosphorylation site. They performed assays in epidermal peels which showed that pip2;1 knockout mutants in Arabidopsis have a defect in stomatal closure in response to ABA. This study has determined the novel function of aquaporins, which contribute to ABA-triggered stomata closure that requires an increase in guard cell permeability to water via OST1 dependent phosphorylation of PIP2;1 at Ser-121 (Grondin et al., 2015). Several studies revealed that activated OST1 kinase binds directly to and phosphorylates the anion channel SLAC1, intervening anion release from the guard cells and promoting stomatal closure (Geiger et al., 2009, 2010; Lee et al., 2009; Brandt et al., 2012). Similarly, Sato et al. (2009) reported that K+ channel KAT1 is also a target of SnRK2.6/OST1 and phosphorylation occurs in the C-terminal region of KAT1. These results suggest that phosphorylation is important for regulating ion channels. Zou et al. (2015) reported that the functions of CALCIUM-DEPENDENT PROTEIN KINASE8 (CPK8) in ABA-mediated stomatal regulation in response to drought stress via regulating CATALASE3 (CAT3) activity. They also reported that CPK8 can phosphorylate CAT3 at Ser-261 and regulate its activity (Zou et al., 2015). Constant efflux of both anions and K+ from guard cells drives water efflux and contributes toward to the loss of guard cell turgor, leading to stomatal closure (Fan et al., 2004; Munemasa et al., 2015). For depolarization of the guard cell membrane, ABA also inhibits the activity of guard cell plasma membrane H+-ATPase (Hayashi et al., 2011). ABA can induce the accumulation of reactive oxygen species (ROS) in guard cells, causing stomata to close. Two NADPH oxidases, AtrbohD, and AtrbohF are responsible for ABA-promoted ROS production in Arabidopsis guard cells (Kwak et al., 2003). OST1 can phosphorylate AtrbohF, presumably affecting its enzymatic activity important for ROS production (Sirichandra et al., 2009).
Reactive oxygen species could enhance ABA biosynthesis (Zhao et al., 2001). ABA treatment enhances the ROS production in guard cells (Pei et al., 2000). Some studies also suggested that ROS accumulation is involved in the induction of stomatal closure (An et al., 2008; Khokon et al., 2011). Thus, enhanced ROS production in the guard cells creates a positive feedback loop to mediate stomatal closure (Mittler and Blumwald, 2015). The level of ABA in guard cells can increase by de novo synthesis, transporter-mediated import, and recycling from inactive conjugates (Merilo et al., 2015). Rapid stomatal responses to environmental stimuli might mainly rely on guard cell-synthesized ABA whereas ABA synthesized in the vasculature might contribute more to stomatal regulation during long-term soil water deficit (Merilo et al., 2015). In this way, ABA has a very useful role in regulating stomatal aperture, which helps plants to adapt and survive stress conditions.
Abiotic Stress Response
Abscisic acid is believed to be the key hormone that mediates plant responses to adverse environmental stimuli since the level of ABA in plants usually increases during abiotic stress conditions, and elevated ABA can enhance plant adaptation to various abiotic stresses (Swamy and Smith, 1999; Tuteja, 2007). Since the first observation of ABA accumulation in drought-stressed wheat (Wright, 1969), increased levels of endogenous ABA under drought stress conditions have been reported in many plant species which include maize (Beardsell and Cohen, 1975; Wang et al., 2008), sorghum (Kannangara et al., 1983), rice (Henson, 1984), barley (Stewart and Voetberg, 1985; Thameur et al., 2011), soybean (Bensen et al., 1988), and wheat (Guoth et al., 2009). Some earlier studies have shown endogenous ABA accumulation in response to cold stress (Daie and Campbell, 1981; Eze et al., 1983; Lalk and Dörffling, 1985). Increased level of endogenous ABA has also been observed in salt-stressed tobacco cells and alfalfa seedlings (Singh et al., 1987; Luo et al., 1992). Exogenous application of ABA to plants can increase their adaptive responses to abiotic stresses. External application of ABA has been revealed to increase drought tolerance in some plant species (Waterland et al., 2010; Du et al., 2013; Yadegari et al., 2014; Wei et al., 2015). ABA treatments could increase cold resistance in cucumber (Flores et al., 1988) and alfalfa (Mohapatra et al., 1988). It is also shown that ABA can alleviate salt stress in common bean and potato (Khadri et al., 2006; Etehadnia et al., 2008).
In addition to its role in regulating stomatal aperture, which is required to limit water loss from leaves under drought conditions, ABA induces the expression of many genes whose products are important for stress responses and tolerance such as enzymes for osmoprotectant synthesis (Fujita et al., 2011). Transcriptome studies have shown that over 50% of the genes regulated by ABA are also governed by drought or salinity, whereas cold-regulated transcriptome shows less overlap with those induced by other stresses. Seki and his associates identified 245 ABA-inducible genes in Arabidopsis. Among the ABA-inducible genes, 63% (155 genes) were induced by drought, 54% (133 genes) by high salinity, 10% (25 genes) by cold treatment (Seki et al., 2002). Rabbani et al. (2003) identified 73 stress-inducible genes in rice, among them, 43 genes were induced by ABA. These results indicate significant crosstalk between ABA response and abiotic stress signaling pathways, especially for drought and high-salinity.
The main goal of studying plant stress responses is to develop crops with improved tolerance to abiotic stresses. The knowledge of ABA being a key regulator of abiotic stress responses has been utilized for developing crops with enhances tolerance under stress conditions. Molecular manipulation of ABA synthesis or signaling has been done in different crop plants for improved stress tolerance (Table 1). Transgenic crops overexpressing the key ABA synthesis gene NCED (9-cis-epoxycarotenoid dioxygenase) have been shown to exhibit improved drought tolerance (Table 1). Similarly, crops overexpressing the ABA receptor gene PYR (pyrabactin resistance) have been reported to confer drought tolerance (Table 1). However, in many studies, the effects of ABA manipulated transgenics were not tested for its performance in yield though they reported tolerance toward abiotic stress. Some studies reported improved biomass and nutritional quality at yield level (Sivamani et al., 2000; Aswath et al., 2005; Bao et al., 2016). Wang et al. (2009) reported that RNAi-mediated suppression of the alpha subunit of protein farnesyltransferase (protein farnesyltransferase had been shown to be a key negative regulator controlling ABA sensitivity in Arabidopsis guard cells; Pei et al., 1998) leads to drought tolerance in canola under drought conditions. In well water conditions, they were not able to find yield difference but in drought conditions, they found better yield than non-transgenics. Recently, Wang et al. (2016) reported that the major latex protein-like protein 43 (MLP43) is a positive regulator of ABA response and overexpression of MLP43 conferred drought tolerance in Arabidopsis. It remains to be seen whether genetic manipulation of MLP43 expression could improve the drought tolerance of crops.
Table 1.
List of transgenic plants that used genes for improved abiotic stress tolerance through manipulating ABA signaling or synthesis in crops.
| Gene transferred/targeted | Gene product | Transgenic plant | Abiotic stress | Yield∗ | Reference |
|---|---|---|---|---|---|
| PYR/PYL/RCAR | ABA receptors | Tomato | Drought | NA | Gonzalez-Guzman et al., 2014 |
| OsPYL3 and OsPYL9 | ABA receptors | Rice | Drought and cold | NA | Tian et al., 2015 |
| SIAREB1 | AREB/ABF subfamily of bZIP transcription factors | Tomato | Drought and salt | NA | Orellana et al., 2010 |
| OsbZIP46CA1 | bZIP transcription factor | Rice | Drought and osmotic stress | NA | Tang et al., 2012 |
| OsbZIP72 | bZIP transcription factor | Rice | Drought | NA | Lu et al., 2009 |
| OsSLI1 | Homeodomain-leucine zipper type I | Rice | Multiple abiotic stresses and panicle development | NA | Huang et al., 2014 |
| HVA1 | ABA-responsive late embryogenesis abundant protein | Wheat | Drought | Improved biomass | Sivamani et al., 2000 |
| BnFTA | Farnesyl transferase | Canola | Drought | Improved during drought conditions | Wang et al., 2009 |
| LeNCED1 | 9-cis-epoxycarotenoid dioxygenase | Tomato | Drought | NA | Thompson et al., 2000 |
| PvNCED1 | 9-cis-Epoxycarotenoid Dioxygenase | Tobacco | Drought | NA | Qin and Zeevaart, 2002 |
| VuNCED1 | 9-cis-epoxycarotenoid dioxygenase | Bent grass | Drought and salt | Biomass increased with 3–4 fold as compared to control | Aswath et al., 2005 |
| SgNCED1 and ALO | co-expressing stylo 9-cis-epoxycarotenoid dioxygenase and yeast D-arabinono-1,4-lactone oxidase genes | Tobacco | Drought and chilling | Improved nutritional quality, i.e., vitamin C | Bao et al., 2016 |
| SgNCED1 | 9-cis-epoxycarotenoid dioxygenase | Tobacco | Drought and salinity | NA | Zhang et al., 2008a, 2009 |
| LeNCED1 | 9-cis-epoxycarotenoid dioxygenase (NCED) | Petunia | Drought | NA | Estrada-Melo et al., 2015 |
∗NA, not applicable.
ABA Biosynthesis, Catabolism, and Transport
ABA Biosynthesis
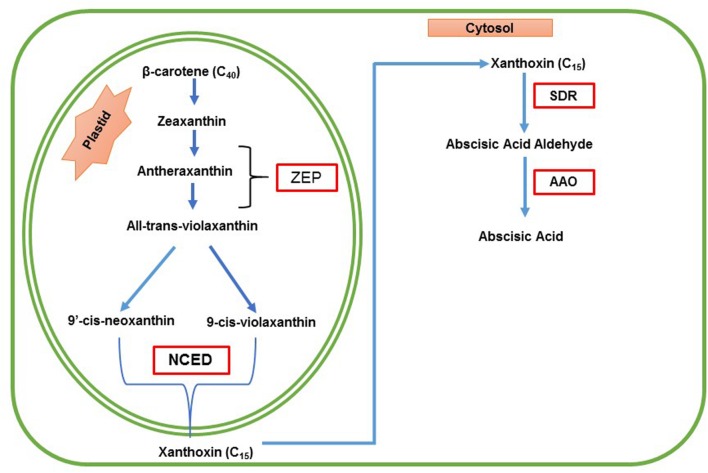
Abscisic acid biosynthesis occurs in two places; it starts from plastids and ends in the cytosol. ABA in higher plants synthesized via the mevalonic acid-independent pathway also called indirect pathway. In this pathway, ABA is synthesized through cleavage of a C40 carotenoid precursor, followed by a two-step conversion of the intermediate xanthoxin to ABA via ABA aldehyde, which will be oxidized into ABA. Mutants defective in ABA biosynthesis have been isolated in many plant species from maize, tomato, tobacco, potato, barley and Arabidopsis (Xiong and Zhu, 2003). The primary mechanism of ABA biosynthesis pathway is shown in Figure 1.
FIGURE 1.
Schematic representation of biosynthesis of ABA in plants. ABA is derived from β-carotene (C40) through an oxidative cleavage reaction in plastids. The first step of ABA biosynthesis pathway is the conversion of zeaxanthin and antheraxanthin to all trans-violaxanthin, which will be catalyzed by zeaxanthin epoxidase (ZEP). Antheraxanthin is the intermediate product. All–trans-violaxanthin is converted to 9-cis-violaxanthin or 9′-cis-neoxanthin by the 9-cis-epoxy carotenoid dioxygenase (NCED), which yields a C15 intermediate product called xanthoxin. Then the product xanthoxin is exported to the cytosol (Nambara and Marion-Poll, 2005) where xanthoxin is converted to ABA. Xanthoxin is then converted into ABA by two enzymatic reactions. Finally, xanthoxin is converted to an ABA aldehyde by the enzyme, short-chain alcohol dehydrogenase/reductase (SDR), and then oxidation of the abscisic aldehyde to ABA is catalyzed by the abscisic aldehyde oxidase (AAO). (Modified from, Taylor et al., 2000; Finkelstein and Rock, 2002; Tuteja, 2007; Mehrotra et al., 2014).
The first step of ABA biosynthesis pathway is the conversion of zeaxanthin and antheraxanthin to all trans-violaxanthin, which will be catalyzed by zeaxanthin epoxidase (ZEP) in the plastid. The ZEP was identified by Marin et al. (1996). In this reaction, antheraxanthin is the intermediate formed. After that, all trans-violaxanthin converted to 9-cis-violaxanthin or 9′-cis-neoxanthin. The enzyme involved in this reaction is unknown (Seiler et al., 2011). After that, oxidative cleavage of 9-cis-violaxanthin and 9-cis-neoxanthin, catalyzed by the enzyme called 9-cis-epoxy carotenoid dioxygenase (NCED), which yields a C15 intermediate product called xanthoxin and C25 metabolite (Schwartz et al., 1997). ZmNCED gene was first isolated using the maize viviparous14 mutant, and NCED is the key enzyme in ABA biosynthesis (Tan et al., 1997). Then the product xanthoxin is exported to the cytosol (Nambara and Marion-Poll, 2005) where xanthoxin is converted to ABA. In this step, xanthoxin is converted into ABA by two enzymatic reactions. First of all, xanthoxin is converted to an ABA aldehyde by an enzyme called short-chain alcohol dehydrogenase/reductase (SDR) encoded by the AtABA2 gene in Arabidopsis thaliana (Rook et al., 2001; Cheng et al., 2002; Gonzalez-Guzman et al., 2002). The next and final step of ABA biosynthesis is oxidation of the abscisic aldehyde to ABA, catalyzed by the abscisic aldehyde oxidase (AAO).
ABA Catabolism
When stress signals are diminished ABA is metabolized into inactive products (Ng et al., 2014). That is accomplished by two pathways called hydroxylation and conjugation (Nambara and Marion-Poll, 2005). In hydroxylation, ABA is hydroxylated via oxidation of three methyl groups (C-7′, C-8′, and C-9′) of the ring structure. Among these C-8′ is known as the dominant catalytic pathway (Zeevaart and Creelman, 1988; Cutler and Krochko, 1999; Okamoto et al., 2009). In the 8′-hydroxylation pathway, phaseic acid (PA) and dihydro phaseic acid (DPA) are the amplest ABA catabolites (Gillard and Walton, 1976; Kushiro et al., 2004; Saito et al., 2004; Nambara and Marion-Poll, 2005). Cytochrome P450 type enzyme (CYP707A) is a crucial enzyme for ABA metabolism (Kushiro et al., 2004; Saito et al., 2004). The metabolism is controlled by the expression of CYP707A gene in the stomata and vascular tissues of leaves under rehydrated conditions (Nambara and Marion-Poll, 2005; Okamoto et al., 2009, 2011). According to Lee et al. (2006), the activation of glucosidase via stress-induced polymerization also rapidly increases the active pools of ABA. Similarly, Okamoto et al. (2009) reported that CYP707A3 functions in vascular tissues to reduce systematic ABA levels. On the other hand, CYP707A1 catabolizes local ABA pools in guard cells in response to high humidity. The second pathway for inactivating ABA is conjugation. ABA conjugation plays a significant role in the regulation of the amounts under both normal and dehydration conditions (Lee et al., 2006; Xu et al., 2012). ABA and its hydroxylated catabolites can be conjugated to glucose. ABA glucosyl ester (ABA-GE) is synthesize by glycosyltransferase. ABA-GE is synthesized in the cytosol and stored in vacuoles (Lim et al., 2005; Boursiac et al., 2013). Under abiotic stress conditions, ABA glucosyl ester could be converted to ABA by enzyme-catalyzed hydrolysis. The glycosidase enzyme catalyzing the hydrolysis of ABA-GE to free ABA was first demonstrated in barley (Dietz et al., 2000). Subsequently, two ABA-GE hydrolyzing enzymes, BG1 (beta-glycosidase homolog 1) and BG2, have been isolated in Arabidopsis (Lee et al., 2006; Xu et al., 2012). BG1 is found in endoplasmic reticulum whereas BG2 is present in the vacuole (Lee et al., 2006; Xu et al., 2012). The ABA released from ABA-GE may not be required for fast stomatal responses, as the loss-of-function mutant of BG1 and BG2, bg1bg2, show wild-type like stomatal responses to reduced air humidity, elevated CO2, and ABA (Merilo et al., 2015).
ABA Transport
The translocation of ABA between cells, tissues and organs also play important roles in whole plant physiological response to stress conditions. ABA, being a weak acid, can diffuse passively across biological membranes when it is protonated (Wilkinson and Davies, 2010; Ng et al., 2014). ABA can also be transported across membranes by transporters. ABA transporters were first identified in Arabidopsis, and they are ATP-binding cassette (ABC)-containing transporter proteins (Kang et al., 2010; Kuromori et al., 2010). AtABCG25 encodes a half-size ABC transporter protein and which is responsible for exporting ABA from vascular tissues, the main sites of ABA synthesis in plants (Kuromori et al., 2010); AtBCG40, a full-size ABC transporter, acts as an ABA importer in plant cells (Kang et al., 2010). Subsequently, AtABCG22 was shown to be required for stomatal regulation and proposed to function as ABA transporter (Kuromori et al., 2011). Later, Ji et al. (2014) reported that ABCG16 is involved in ABA tolerance. Currently, Kang et al. (2015) have demonstrated four AtABCG protein functions together to supply ABA in mature imbibed seeds. They showed that AtABCG25 and AtABCG31 export ABA from the endosperm to the embryo, whereas AtABCG30 and AtABCG40, transport ABA into the embryo. These studies demonstrate that certain ATP-binding cassette (ABC)-containing transporter proteins are important for ABA transport and responses. In addition to ABA transporters belonging to the ABC family, other types of ABA transporters are also found. Kanno et al. (2012) identified an ABA-importing transporter (AIT1) by transport assay in yeast and insect cells. AIT1 belongs to the NRT1 (nitrate transporter 1) transporter family. ait1 mutants were less sensitive to ABA during germination and post-germination growth and stomata remained open whereas overexpression of AIT1 resulted in ABA hypersensitivity (Kanno et al., 2012). Recently, AtDTX50 (Detoxification Efflux Carrier 50) has been found to participate in ABA transport in Arabidopsis (Zhang et al., 2014). AtDTX50 is a membrane protein in the MATE (Multidrug and Toxic Compound Extrusion) transporter family (Zhang et al., 2014). The atdtx50 mutant showed faster ABA-induced stomatal closure, indicating increased ABA accumulation in the guard cells that confer drought tolerance. AtDTX50 is primarily expressed in vascular tissues and guard cells. These results indicate that AtDTX50 mediates ABA efflux from the cytosol of vascular and guard cells located in the plasma membrane (Zhang et al., 2014). It appears that multiple types of transporters are involved in ABA transport in plants.
ABA Signaling
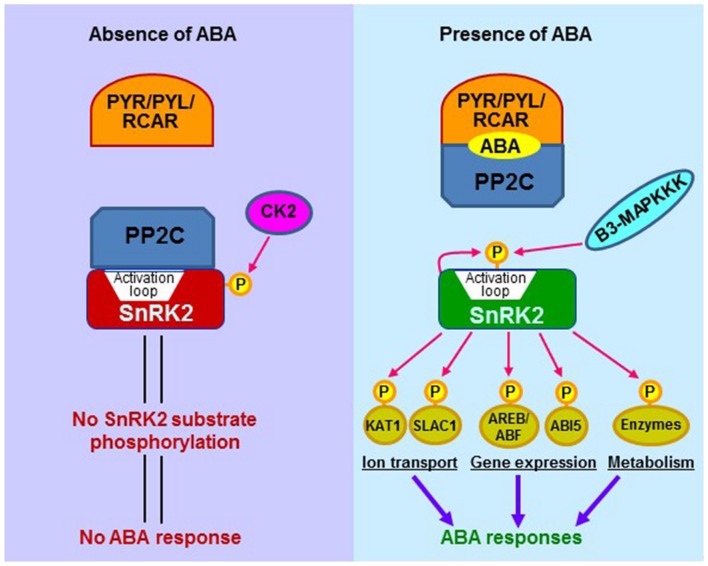
Abscisic acid is a key endogenous messenger in plants, and it has a crucial role in various plant stresses. Therefore, understanding the signaling mechanism of ABA is critical for improving plant performance under stress environments and most needed in projected warmer and drier future climate. There are three core components of ABA signaling; pyrabactin resistance (PYR)/pyrabactin resistance-like (PYL)/regulatory component of ABA receptors (RCAR), protein phosphatase 2C (PP2C: acts as negative regulators) and (Sucrose non-fermenting) SNF1-related protein kinase 2 (SnRK2: acts as positive regulators). In the presence of ABA, PYR/PYL/RCAR-PP2C complex formation leads to inhibition of PP2C activity, which allows the activation of SnRK2. Activated SnRK2 then phosphorylates downstream substrate proteins such as transcription factors, and thus facilitating transcription of ABA-responsive genes (Figure 2).
FIGURE 2.
The schematic representation of major ABA signaling pathway in plants with and without ABA presence. The core components in ABA signaling include ABA receptors (PYR/PYL/RCAR), PP2C phosphatases (negative regulators), and SnRK2 kinases (positive regulators). In the absence of ABA, PP2Cs associate with SnRK2s and prevent the activation of SnRK2s. The inactive SnRK2s are unable to phosphorylate downstream substrates, and thus signal transduction is not occurring. In the presence of ABA, PYR/PYL/RCAR receptors bind to ABA and interact with PP2Cs, which release SnRK2s. The SnRK2s are then activated by autophosphorylation of the activation loop. The active SnRK2s can phosphorylate downstream substrate proteins, including transcription factors, ion channels, and enzymes such as NADPH oxidases, thereby inducing ABA responses. SnRK2s are subjected to regulation by other protein kinases. A Raf-like kinase (B3-MAPKKK) has been shown to activate SnRK2 through phosphorylating the activation loop, whereas casein kinase 2 (CK2) can phosphorylate SnRK2’s carboxyl-terminal serine residues, thereby enhancing SnRK2-PP2C interaction and inactivating SnRK2. Catalytically active SnRK2 is shown in green and inactive SnRK2 is in red. ABF, ABA-responsive element binding factor; ABI5, ABA insensitive 5; AREB, ABA-responsive element binding protein; B3-MAPKKK, B3-group Raf-like MAP kinase kinase kinase; KAT1, potassium channel in Arabidopsis thaliana 1; PP2C, Protein phosphatase 2C; PYR, pyrabactin resistance; PYL, PYR-related; RCAR, regulatory component of ABA receptor; SLAC1, slow anion channel 1; SnRK2, sucrose nonfermenting-1-related protein kinase 2.
The PYR/PYL/RCAR ABA Receptors
Two independent research groups (Ma et al., 2009; Park et al., 2009) made remarkable discovery of an ABA receptor. They identified Arabidopsis PYR1 (pyrabactin resistance 1), PYL (PYR-related) or RCAR (Regulatory Component of ABA Receptor) proteins as ABA receptors. The Arabidopsis genome contains 14 PYR/PYL/RCAR genes, which encode highly conserved small proteins with 159-211 amino acid residues (Ma et al., 2009; Park et al., 2009; Nishimura et al., 2010). Many of PYR/PYL/RCARs including PYR1, PYL1 and PYL2 have been shown to directly bind ABA (Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009; Yin et al., 2009). Once bound to ABA this receptor can bind to group A protein phosphatases (PP2C), including ABI1 (ABA-insensitive 1) and ABI2, the negative regulator of ABA signaling (Ma et al., 2009; Park et al., 2009; Nishimura et al., 2010). These ABA receptors played a critical role in ABA perception, as quadruple mutants pyr1, pyl1, pyl2 and pyl4 (recently six i.e., pyr1/pyl1/pyl2/pyl4/pyl5/pyl8) are insensitive toward ABA (Park et al., 2009; Gonzalez-Guzman et al., 2012). They also showed hyposensitivity in germination and root growth response to ABA (Park et al., 2009; Gonzalez-Guzman et al., 2012). Overexpression of PYL5, PYL8 or PYL9 enhanced ABA responses or conferred drought resistance to Arabidopsis thaliana (Ma et al., 2009; Santiago et al., 2009; Saavedra et al., 2010). The PYR/PYL/RCAR family in Arabidopsis (except PYL13) is capable of activating ABA signaling response, suggesting that nearly all members can function as ABA receptors (Fujii et al., 2009). Antoni et al. (2013) reported that ABA-dependent inhibition of PP2Cs by PYR/PYLs is essential for the perception of moisture gradient. The structural and molecular studies conducted by many scientists have convincingly shown that PYR/PYL/RCAR receptors play a central role in ABA perception (Ma et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Park et al., 2009; Santiago et al., 2009; Melcher et al., 2010).
PP2C: The Negative Regulators
Reversible protein phosphorylation catalyzed by protein kinases and phosphatases play a significant role in cellular signal transduction, which helps in the transmission of signals from external to the internal environment of the cell. There are 112 phosphatases encoded in the Arabidopsis genome, among them 76 genes encode for PP2Cs (Schweighofer et al., 2004). At least 6 of the nine members of the group A PP2Cs have been shown to involved in ABA signaling, among them, ABI1, ABI2, and HAB1 are the well characterized. The members of the group A have defined roles in different tissues with tissue-specific expression patterns (Ng et al., 2014). The PP2C group A genes first characterized are ABI1 and ABI2, which was isolated from a genetic screen in Arabidopsis. Both abi1 and abi2 mutants show insensitivity in various tissues, developmental stages, suggesting that they act as a negative regulator of ABA signaling. Both of these mutants display reduced seedling growth, seed dormancy, drought tolerance and stomatal regulations (Leung et al., 1994, 1997; Meyer et al., 1994; Rodriguez et al., 1998; Saez et al., 2004; Schweighofer et al., 2004; Yoshida T. et al., 2006). Few years later, HAB1 (homolog to ABI1) and HAB2 were isolated based on their sequence similarity to ABI1 (Saez et al., 2004). Similarly, other members of A group PP2Cs such as AHG1 and AHG3/AtPP2CA were identified in genetic screens of Arabidopsis and yeast complementation test. It has been shown that all of the loss-of-function mutants of group A PP2Cs exhibit significant ABA hypersensitivity, which establishes them as the major negative regulators of ABA signaling (Kuromori and Yamamoto, 1994; Kuhn et al., 2006; Yoshida R. et al., 2006; Nishimura et al., 2007; Umezawa et al., 2010; Antoni et al., 2012). ABI1, ABI2, and HAB1 showed a dominant role in ABA signaling pathways in both seeds and vegetative tissues; the evidence is supported by gene expression data and genetic analysis. Recessive hab1-1 mutants showed enhanced ABA-responsive gene expression, increased ABA-mediated stomatal closure and ABA-hypersensitivity in seed germination. These results indicate that HAB1 negatively regulates ABA signaling (Gosti et al., 1999; Saez et al., 2004; Hirayama and Shinozaki, 2007). These results are consistent with the fact that PP2Cs function as negative regulators of ABA signaling (Mehrotra et al., 2014). Recently, Singh et al. (2015) reported a group A PP2C from rice gene (OsPP108), whose expression is highly inducible under salt and drought stresses. The overexpression of OsPP108 confers ABA insensitivity and tolerance to stresses like high salt and mannitol (Singh et al., 2015).
SnRK2 Protein Kinases: The Positive Regulators
The identification of PP2Cs showed that reversible protein phosphorylation process is primary in ABA signaling pathway. Protein kinases are known to be involved in ABA signaling pathway. The sucrose non-fermenting 1 (SNF1)-related protein kinase2 (SnRK2) family of protein kinases are plant specific serine/threonine kinases participating in plant response and has a central role in cellular responses to drought and dehydration (Hrabak et al., 2003; Saruhashi et al., 2015). The first protein kinase having ABA-stimulated catalytic activity was identified as an AAPK in Vicia faba (Li and Assmann, 1996; Li et al., 2000). AAPK is a positive regulator of ABA-induced stomatal closure (Li et al., 2000). Subsequently, the AAPK ortholog in Arabidopsis, OST1 (Open Stomata 1)/SnRK2.6, was shown to function in ABA-mediated stomatal regulation (Mustilli et al., 2002; Yoshida et al., 2002). Similar to AAPK, ABA-activated protein kinases have been identified in Arabidopsis (Boudsocq et al., 2004) and rice (Kobayashi et al., 2004); they belong to the SnRK2 family. These ABA-activated protein kinases mediate ABA signaling by phosphorylating their substrate proteins such as transcription factors, ion channels, and metabolic enzymes (Umezawa et al., 2010; Fujita et al., 2013). According to Gómez-Cadenas et al. (1999), transcription of PKABA1 is induced by ABA and occurs in embryos and seedlings. The function of PKABA1 is to mediate ABA-suppressed gene expression. PKABA1 phosphorylates and activates the ABA-responsive basic-domain leucine-zipper (bZIP) transcription factor TaABF, which help in regulation of ABA signaling in seed development and germination (Shen et al., 2001; Johnson et al., 2002). Similarly, AAPK modulates an RNA-binding protein AKIP1 by phosphorylating it and inducing its translocation into subnuclear speckles in guard cells (Li et al., 2000, 2002).
SnRK2 family is the key regulator of plant response to abiotic stress and is divided into three sub-groups depending upon the affinity toward ABA. Subgroup-I kinases do not respond to ABA whereas subgroup-III actively respond to ABA and believed to be the key regulator of ABA-dependent pathway gene expression. Subgroup-II weakly responds or did not provide a response to ABA (Hrabak et al., 2003; Boudsocq et al., 2004; Kulik et al., 2011). There are altogether 10 SnRK2 members found in Arabidopsis thaliana, i.e., SnRK2.1-SnRK2.10 or SnRK2A-SnRK2J. Among them, 5 SnRK2 members (SnRK2.2., SnRK2.3, SnRK2.6, SnRK2.7, and SnRK2.8) can be activated by ABA. The all of the SnRK2s except SnRK2.9 can be activated by osmotic stress, which indicated that most SnRK2 members are involved in ABA and stress signaling (Boudsocq et al., 2004; Furihata et al., 2006; Yoshida T. et al., 2006). Among them, SnRK2.2, SnRK3.3, and SnRK2.6 (Subgroup-III) are known as primary regulators of ABA since they exhibit the strongest activation of ABA (Boudsocq et al., 2004). The first member of the Arabidopsis SnRK2 family is OST1/SnRK2.6 which identified by forward genetic approach and act as a regulator of ABA signaling. The mutant snrk2.6 (ost1) cannot exhibit ABA-induced stomatal closure, indicating that it positively regulates the stomatal response to ABA. The SnRK2.6 gene mainly expressed in guard cells, and the vascular system whereas seed germination and post-germination growth were not affected in snrk2.6 loss-of-function mutant, which indicated that the SnRK2.6 protein kinase may not function in seed germination (Mustilli et al., 2002; Yoshida et al., 2002). Moreover, SnRK2.2 and SnRK2.3 function redundantly in ABA-induced inhibition of seed germination and post-germination growth (Fujii et al., 2007). According to Fujii et al. (2007), snrk2.2 and 2.3 single mutants show weak, while snrk2.2 and snrk2.3 double mutant exhibits high ABA-insensitive phenotypes regarding seed germination and seedling growth. The triple mutation (snrk2.2/2.3/2.6), nearly block all the main ABA responses (Fujii and Zhu, 2009; Fujita et al., 2009; Nakashima et al., 2009a; Waadt et al., 2015). In Arabidopsis proteins extract, these 3 SnRK2s, along with 9 of the 14 members of PYR/PYL/RCARs, could be co-immunoprecipitated with ABI. Though the composition of co-purified proteins was ABA-independent, these data suggest that at least ABI1, the 3 SnRK2s and at least 9 of the 14 receptor proteins may constitute a core signaling complex (Fujii and Zhu, 2009; Nishimura et al., 2010; Joshi-Saha et al., 2011). The expression profile of these three SnRK2s genes supports their redundant, supportive roles in the ABA responses (Fujii et al., 2007, 2011). Double mutant snrk2.7/2.8 affect the expression of ABA and drought responsive genes, indicating that SnRK2.7/2.8 has potential roles in ABA signaling although snrk2.7 and snrk2.8 single mutation show wild-type phenotypes due to functional redundancy (Mizoguchi et al., 2010).
Many studies reported that phosphorylation at the activation loop is critical for the activation of SnRK2s. Several phosphorylation sites have been identified in SnRK2.6; ABA stimulates phosphorylation of Ser 175 in the activation loop, whereas PP2C can dephosphorylate the Ser175 of SnRK2.6 in the absence of ABA, resulting in the deactivation of SnRK2.6 (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009; Vlad et al., 2010). Quantitative analysis confirmed that ABA treatment increases the phosphorylation of the site in vivo (Kline et al., 2010) and phosphorylation of the site is also increased by osmotic stress (Vlad et al., 2010). SnRK2 activation loop phosphorylation may also implicate unidentified upstream kinases in plants (Burza et al., 2006; Boudsocq et al., 2007). ABA- or stress-activated SnRK2 family members have been found in Vicia faba, soybean, Arabidopsis, tobacco, rice, Chlamydomonas reinhardtii, maize, sorghum, and wheat (Li et al., 2000; Monks et al., 2001; Boudsocq et al., 2004; Kelner et al., 2004; Kobayashi et al., 2004; Gonzalez-Ballester et al., 2008; Huai et al., 2008; Li et al., 2010; Mao et al., 2010; Zhang et al., 2010b).
Understanding how SnRK2 kinases exert their effects in ABA signaling network requires knowledge of SnRK2 targets. Several phosphorylation substrates of SnRK2 kinases have been identified using biochemical and molecular approaches. The ABFs (ABA-responsive-element Binding Factor)/AREBs (ABA-Responsive Element Binding factors) transcription factors involving in regulating ABA-responsive genes are SnRK2 substrates as ABA-activated SnRK2 kinases have been shown to phosphorylate ABF/AREB proteins in vitro and in vivo (Kobayashi et al., 2005; Furihata et al., 2006; Fujii et al., 2007; Sirichandra et al., 2010). Another transcription factor involved in ABA signaling, ABI5, can be phosphorylated in vitro by ABA-activated SnRK2 kinases (Fujii et al., 2007; Nakashima et al., 2009a; Wang et al., 2013b). In addition to nuclear transcription factors, SnRK2 can also phosphorylate plasma membrane proteins. The guard cell OST1/SnRK2.6 has been shown to phosphorylate the anion channel SLAC1, the potassium channel KAT1, and the NADPH oxidase AtrbohF, all of which are plasma-membrane proteins important in controlling stomatal aperture (Geiger et al., 2009; Lee et al., 2009; Sato et al., 2009; Sirichandra et al., 2009). To identify more potential SnRK2 targets, phosphoproteomic approaches have been used to survey ABA-responsive phosphoproteins. Six ABA-regulated phosphoproteins (ascorbate peroxidase, Ca2+/H+ antiporter regulator protein, G protein beta subunit-like protein, glyoxysomal malate dehydrogenase, manganese superoxide dismutase, and triosephosphate isomerase) have been identified from rice leaves by using 2-D gel electrophoresis, phospho-antibody immunoblotting, and mass spectrometry (He and Li, 2008). Using snrk2.2/2.3/2.6 triple knockout mutant and comparative phosphoproteomics, two research groups have identified respectively 35 and 58 putative SnRK2 substrates (Umezawa et al., 2013; Wang et al., 2013a). The putative SnRK2 substrates identified by the two groups show little overlap, suggesting that the three ABA-activated protein kinases (SnKR2.2, SnKR2.3, and SnKR2.6) can target and regulate a range of proteins via ABA-mediated phosphorylation.
Possible Regulator of SnRK2
Recently, a novel regulatory component ARK is identified by Saruhashi et al. (2015). They reported that ARK was an essential signaling component for the regulation of SnRK2 in basal land plants. A single gene encodes ARK (ABA, and abiotic stress-responsive Raf-like kinase) in moss (Physcomitrella patens) which belongs to the family of group B3 Raf-like MAP kinase kinases (B3-MAPKKKs). Phosphorylation and activation events in these kinases are modulated through their non-kinase domain by dimerization, binding of other regulatory proteins or phosphorylation by upstream MAPKKK kinases (Kolch, 2000). However, Saruhashi et al. (2015) reported that ARK might be activated itself by autophosphorylation in the activation loop by using transient assay of phosphopeptide mapping and mutational analysis. Their study showed that ARK provides positive regulation of ABA signaling in addition to the negative relation by group A PP2Cs in bryophytes, which provides new insights into a signaling pathway for a possible connection between these kinases through unknown mechanisms operating in plant ABA response. They also revealed that hyperosmosis induced activation of plant SnRK2s mediated by an upstream of ARK (Saruhashi et al., 2015). Recently the other study also reported that hyperosmosis mediated response by both ABA-dependent and independent mechanism using ABA-deficient mutant of P. patens (Takezawa et al., 2015). To understand the molecular mechanism, they identified one mutant AR7 (ARK) and microarray profiles showed that the expression of a majority of the ABA-responsive genes was affected in AR7. The expression of 518 out of 579 ABA upregulated genes in the wild-type line was reduced significantly in AR7. These genes are included 27 LEA-like genes which are responsible for dehydration stress tolerance. RNA gel blot analysis also confirmed impaired ABA-induced gene expression in AR7 (Saruhashi et al., 2015). Furthermore, levels of 150 of 165 ABA down-regulated transcripts were higher in AR7 than in wild type. They showed that ARK could phosphorylate serine residues in the activation loop of SnRK2B thus activating SnRK2B and ABA signaling (Saruhashi et al., 2015). These results indicated that gene impaired in AR7 encodes an important positive regulator of ABA-responsive gene expression in P. patens. All these findings suggested that B3-MAPKKKs in angiosperms are involved in positive regulation of SnRK2 in ABA response and SnRK2 activation.
In contrast to ARK, another new negative regulator of SnRK2 has been reported recently Vilela et al. (2015) identified Casein Kinase 2 (CK2) as a negative regulator of SnRK2 in maize. CK2 is an evolutionary multi-subunit serine/threonine kinase and found in all eukaryotes (Meggio and Pinna, 2003; Vilela et al., 2015). Among all kinase, CK2 is unique because it can use either ATP or GTP as phosphoryl donors (Niefind et al., 1999). CK2 is involved in plant growth and development, light-regulated gene expression, hormone responses, cell-cycle regulation, flowering time, DNA repair or responses to abiotic stress and biotic stresses in plants (Lee et al., 1999; Riera et al., 2004; Portolés and Más, 2007; Moreno-Romero et al., 2008; Riera et al., 2013; Mulekar and Huq, 2014; Vilela et al., 2015). In the case of abiotic stresses, CK2 acts as an ABA regulator because ck2α mutants are hypersensitive to ABA concerning seed germination, cotyledon greening, and stomatal opening (Mulekar et al., 2012; Wang et al., 2014). These effects were attributed to the down-regulation of ABA-related genes, including OST1, but the biochemical relationship between CK2 and OST1 was not clear. In Arabidopsis, OST1 is the best genetically and biochemically characterized SnRK2, functioning as a key regulator in the core of ABA signaling module (Cutler et al., 2010). CK2 can phosphorylate the ABA box of ZmOST1 to accelerate the latter’s turnover and to promote binding to PP2Cs, suggesting that CK2 is a regulator of OST1 protein stability (Vilela et al., 2015). They also showed that overexpression of ZmOST1 in transgenic Arabidopsis plants exhibit highly resistant to drought and hypersensitive to ABA at the level of stomata. These two newly discovered SnRK2 regulators (B3-MAPKKK and CK2) will add more focused research toward ABA signaling pathway (Figure 2).
ABA Signaling Response at Gene Expression Levels
Transcription Factors Involved in ABA-Regulated Gene Networks
Transcription factors are master regulators that integrate, balance, and coordinate hormonal, developmental and environmental signals in plant systems (Jaradat et al., 2013). A single transcription factor can regulate the expression of many target genes through specific binding of a transcription factor to cis-acting elements in promoters of respective target genes. Plant genomes assign approximately 7% of their coding sequences to transcription factors, which provides the complexity of transcriptional regulation (Udvardi et al., 2007). Transcription factors significant for the regulation of ABA-related gene network include AREBs (ABA-responsive element binding proteins)/ABFs (ABRE binding factors), ABI5 (ABA insensitive 5), MYB (myeloblastosis), MYC (myelocytomatosis), NAC (NAM: no apical meristem; ATAF: Arabidopsis transcription activation factor; CUC: cup-shaped cotyledon), and ERF (ethylene response factor).
AREBs/ABFs belong to the bZIP (basic leucine zipper) transcription factor subfamily. They were isolated based on their interaction with ABRE (ABA-responsive element) in vitro and regulatory function in ABA and or stress responses (Choi et al., 2000; Uno et al., 2000). For example, AREB1/ABF2, AREB2/ABF4, and ABF3 can be induced by high salt, dehydration or ABA treatment in vegetative tissues (Riechmann et al., 2000; Yoshida et al., 2010). Kim et al. (2004) reported that overexpression of ABF3 confers tolerance to chilling, freezing, high temperature, and drought stress. Similarly, ABP9 (ABA-responsive-element binding protein 9) is also a member of the bZIP family found to be associated with drought and heat stress. Constitutively expression of ABP9 in Arabidopsis improve photosynthetic capacity under water deficit and heat shocks, suggesting that ABP9 has a significant role in the regulation of plant photosynthesis under stress conditions (Zhang et al., 2008b).
ABI5 is also a member of Arabidopsis bZIP transcription factor subfamily and has four conserved domains (Jakoby et al., 2002). The ABI5 expression is higher in mature seeds and young seedling exposed to ABA or dehydration stress (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001). ABI5 can bind to ABRE in the promoters of its target genes and is a positive regulator of ABA-regulated transcription networks (Shen et al., 1996; Finkelstein and Lynch, 2000). Similar to Arabidopsis ABI5, overexpression and upregulation of OsABI5 in rice under ABA and salinity conditions confers salt tolerance (Zou et al., 2008).
MYC and MYB, belonging to the bHLH (basic helix loop helix) subfamily, also play a regulatory role in ABA signaling by activating some stress-inducible genes (Abe et al., 2003). MYB in Arabidopsis namely AtMYB60, AtMYB44, and AtMYB15 have been shown to be involved in the regulation of stomatal closure and ABA-mediated response to drought and salt stresses (Jaradat et al., 2013). The Arabidopsis MYC transcription factor, AtMYC2, is an ABA-responsive gene induced by drought and salt (Abe et al., 1997).
NAC transcription factors appear to be involved in ABA transcriptional network. The Arabidopsis NAC-type transcription factor, AtNAC, has been shown to play a fundamental role in the regulation of ABA- and senescence-induced genes (Zhang and Gan, 2012). Furthermore, ABA can induce the expression of seven stress-induced NAC transcription factors in Arabidopsis (Takasaki et al., 2015). Loss-of-function mutation of these NAC transcription factor genes confers retardation of leaf senescence induced by ABA, suggesting that NAC transcription factors play a key role ABA-induced leaf senescence. Recently, Jiang et al. (2014) reported that the RhNAC3 transcription factor is involved in regulating ABA signaling pathway in Arabidopsis and rose. Overexpression of RhNAC3 in Arabidopsis resulted in ABA hypersensitivity during seed germination and enhanced stomatal closure in response to ABA or drought treatments. Furthermore, the expression of downstream genes of ABA signaling pathways was repressed in RhNAC3-silenced petals in a rose, suggesting that the RhNAC3 transcription factor plays a role in regulating ABA-related genes.
Finally, ERF-type transcription factors have been implicated in regulating ABA response. Overexpression of AtERF7 (Arabidopsis ERF 7) caused reduced sensitivity to ABA in guard cells and increased transpiration water loss, whereas repression of AtERF7 via RNA interference resulted in increased sensitivity to ABA (Song et al., 2005). These findings show that the ERF7 transcription factor can play a significant role in regulating ABA responses. Similarly, a tomato ERF-type transcription factor, JERF3, was shown to be transcriptionally induced by salt and ABA and overexpression of JERF3 leads to tobacco plants with enhanced tolerance to salinity (Li et al., 2008). Therefore, the transcription factors mentioned in the above sections illustrate the regulation of the complex ABA-dependent transcription networks in plants under stress conditions.
ABA-Induced Gene Expression by Activation of SnRK2
Drought and salt stress-induced the accumulation of ABA, which then act as a stress signal (Ng et al., 2014). According to Cutler et al. (2010), ABA signaling leads to substantial changes in gene expression, which implicate changes in transcription, transcript processing, and stability. Many studies reported that almost 10% of the Arabidopsis genes are regulated by ABA (Hoth et al., 2002; Nemhauser et al., 2006; Yang et al., 2008). In the other study, Böhmer and Schroeder (2011) identified 3494 ABA-responsive transcripts and 50 ABA-responsive proteins using microarrays and quantitative proteomics. Among the identified ABA-responsive genes, 1512 genes were upregulated, and 1982 genes were downregulated by ABA treatment.
Abscisic acid-dependent gene expression requires the binding of transcription factors to cis-regulatory elements in target genes promoters. The major cis-regulatory element associated with expression of ABA-responsive-genes is the ABA-response element (ABRE: PyACGTGG/TC), which may require an ABRE-coupling element (CE) for full function (Hobo et al., 1999; Zhang et al., 2005; Nakashima et al., 2014). ABRE-binding proteins (AREBs) or ABRE-binding factors (ABFs) are the transcription factors that can bind to the ABREs of ABA-responsive genes and activate their gene expression (Choi et al., 2000; Uno et al., 2000). Further, ABA-dependent phosphorylation of AREBs/ABFs in multiple RXXS/T sites is necessary for their activation (Uno et al., 2000; Kagaya et al., 2002). ABA-activated SnRK2 kinases have been shown to be responsible for ABA-dependent phosphorylation of AREBs/ABFs (Johnson et al., 2002; Kobayashi et al., 2005; Furihata et al., 2006; Fujii et al., 2007). In Arabidopsis, ABA-dependent upregulation of gene expression could be stamped out in the mutant lacking all three ABA-activated SnRK2 kinases (Fujii and Zhu, 2009; Nakashima et al., 2009a), suggesting that ABA-activated SnRK2s play an essential role in regulating the expression of ABA-responsive genes through phosphorylating AREBs/ABFs. Moreover, comparative transcriptome analyzes reveal that the majority but not all of ABA-responsive genes down-regulated in the triple mutant of ABA-activated SnRK2s are also down-regulated in the quadruple mutant of ABREs/ABFs (Yoshida et al., 2015). These results indicate that AREB1, AREB2, and ABF3 and ABF1 are the major transcription factors downstream of the ABA-activated SnRK2s in ABA signal transduction during vegetative growth. Nevertheless, these results also suggest that other transcription factors targeted by ABA-activated SnRK2s may contribute to transcriptional regulation of ABA-responsive genes.
ABA insensitive 5 (ABI5) is an important regulator of ABA responses during seed germination and seedling growth (Finkelstein and Lynch, 2000; Brocard et al., 2002). Like AREBs/ABFs, the basic leucine zipper (bZIP)-type transcription factor ABI can also bind to ABREs and regulate the expression of some ABA-responsive genes (Lopez-Molina et al., 2001; Kim et al., 2002; Yang et al., 2011). ABA-activated SnRKs can phosphorylate ABI5 (Nakashima et al., 2009a; Wang et al., 2013b) and phosphorylation of ABI5 is required for activating the transcription factor (Wang et al., 2013b). Dephosphorylation of ABI5 by serine/threonine protein phosphatase 6 can negatively regulate ABA signaling (Dai et al., 2013). These results indicate that ABA could regulate some ABRE-dependent gene expression via SnRK2-mediated phosphorylation and activation of ABI5 family transcription factors during cellular dehydration in seeds (Fujita et al., 2011).
The RAV1 (Related to ABI3/VP1) transcription factor, which belongs to the Ethylene Responsive Factor (ERF) family (Feng et al., 2005), is another transcriptional regulator targeted by the ABA-activated SnRK2s in Arabidopsis (Feng et al., 2014). Feng et al. (2014) demonstrated that RAV1 is a negative regulator of ABA signaling, and it suppresses the expression of ABA-responsive genes such as ABI3, ABI4, ABI5, Em1, and Em6. Biochemical analysis showed that the ABA-activated SnRK2s (SnRK2.2, SnRK2.3, and SnRK2.6) interact with and phosphorylates the RAV1 protein (Feng et al., 2014). Genetic analysis revealed that the ABA-activated SnRK2s could repress the activity of RAV1. Taken together, this work shows that ABA-activated SnRK2s can inactivate a transcription factor via phosphorylating it. Another study has shown that the ABA-activated SnRK2.6 is involved in phosphorylating and inactivating basic helix-loop-helix (bHLH)-type transcription factors, which have critical roles in stomatal regulation (Takahashi et al., 2013). It is interesting to see whether SnRK2s mediated phosphorylation of transcriptional regulators would contribute to regulating ABA-repressive genes. Earlier studies demonstrate that bZIP transcription factors (ABF1, ABF2/AREB1, and ABI5) play a positive role in ABA signaling and the transcriptional activity of these factors are activated via SnRK2s-mediated phosphorylation (Fujii et al., 2007; Fujii and Zhu, 2009; Nakashima et al., 2009b; Wang et al., 2013b). The work by Feng et al. (2014) showed that RAV1 is a negative regulator of ABA signaling and SnRK2s-mediated phosphorylation repress the activity of RAV1. Thus, SnRK2s, as core components in ABA signaling, can phosphorylate both positive and negative regulators in ABA signaling network. Not all ABA-responsive genes contain ABRE(s) in their promoter regions (Wang et al., 2011a; Kim, 2014). Therefore, transcription factors recognizing non-ABRE regulatory elements such as NAC and MYC/MYB may be involved in regulating these ABA-responsive genes. It remains to see if the ABA-activated SnRK2s have a role in the regulation of the activity of these transcription factors.
Post-transcriptional Control in Regulating ABA Response
RNA-binding proteins, which interact directly with RNA molecules, have crucial roles in many aspects of post-transcriptional control of gene expression, including RNA splicing and processing, mRNA stabilization, mRNA localization and translation (Glisovic et al., 2008).
RNA-binding proteins typically contain one or more RNA-binding domains which include RNA-recognition motif, K homology domain, zinc finger, cold-shock domain, aspartate-glutamate-alanine-aspartate (DEAD) box, and double-stranded RNA-binding domain (Ambrosone et al., 2012). The current understanding suggests that RNA-binding protein-mediated post-transcriptional gene regulation plays a significant role in ABA response. The mutation in the hyponastic leaves 1 (HYL1) gene encoding a double-stranded RNA-binding protein causes hypersensitivity to ABA during seed germination in Arabidopsis (Lu and Fedoroff, 2000). Similarly, the mutation in the supersensitive to ABA and drought 1 (SAD1) gene encoding a polypeptide similar to Sm-like small nuclear ribonucleoproteins (snRNP) involving nuclear mRNA processing increases plant sensitivity to ABA and drought stress (Xiong et al., 2001). Arabidopsis mutant defective in the mRNA cap-binding protein, ABH1 (ABA hypersensitive 1), shows ABA-hypersensitive regulation of seed germination and stomatal closing (Hugouvieux et al., 2001, 2002). Mutation in the DEAD box RNA helicase gene LOS4 (low expression of osmotically responsive genes 4) in Arabidopsis confers ABA hypersensitive and cold tolerance phenotype (Gong et al., 2005). The accumulation of poly (A)+ RNA in the nuclei in this mutant suggests that this RNA helicase plays a crucial role in mRNA export from the nucleus (Gong et al., 2005). Two other Arabidopsis DEAD-box RNA helicase genes have been shown to be down-regulated by ABA and multiple abiotic stresses (Kant et al., 2007). Insertional mutants of the two genes exhibit improved tolerance to salt, osmotic, and heat stresses, suggesting that these two DEAD-box RNA helicases are involved in regulating abiotic stress responses (Kant et al., 2007). In maize, a DEAD box RNA helicase (ZmDRH1) has been shown to interact with a glycine-rich RNA-binding protein (MA16), which can be induced by ABA and dehydration although their functions in ABA and stress responses are not understood (Gomez et al., 1988; Gendra et al., 2004). Another example of the involvement of RNA-binding proteins in ABA response in crop plants is ABA-activated protein kinase (AAPK)-interacting protein 1 (AKIP1), a heterogeneous nuclear ribonucleoprotein (hnRNP) in Vicia faba (Li et al., 2000, 2002). ABA treatment induces phosphorylation of AKIP1 and its relocation into the subnuclear structures resembling splicing speckles (Li et al., 2002). Similarly to AKIP1, its homolog in Arabidopsis called UBA2a [poly(U)-Binding Associated 2a] has also been shown to be translocated into nuclear speckles in response to ABA treatment (Riera et al., 2006). However, it is not certain whether the AKIP1 and UBA2a relocate into the genuine splicing speckles upon ABA treatment, the functional significance of nuclear re-localization of AKIP1 and UBA2a remain unclear (Lorković, 2009).
Besides hnRNPs, another class of RNA-binding proteins playing a key role in regulating alternative splicing is serine/arginine-rich (SR) proteins (Long and Caceres, 2009). Several studies have provided a link between plant SR proteins and ABA response. Two Arabidopsis SR genes (SR1 and SR33) have been shown to change their alternative splicing patterns in response to ABA treatment (Palusa et al., 2007). Also, gene expression analysis has identified six SR genes and the two SR-like genes from Arabidopsis to be involved in ABA-related responses (Cruz et al., 2014). Furthermore, a genetic study has pinpointed that the Arabidopsis SR45 protein as a negative regulator of glucose and ABA signaling in early seedling development (Carvalho et al., 2010). These studies indicate that post-transcriptional control of gene expression at the stages of pre-mRNA splicing and processing, mRNA stabilization, and mRNA export from the nucleus to the cytoplasm by RNA-binding proteins is critical for ABA signaling. Further identification of the direct targets of these RNA-binding proteins will be necessary to fully understand the molecular mechanisms underlying ABA action.
Besides post-transcriptional regulation, post-translational control of targeted proteins by ubiquitination, the covalent ligation of proteins to the highly conserved small protein called ubiquitin, has been shown to be involved in ABA signaling (Yu et al., 2016). Ubiquitination is carried out by three types of enzymes: ubiquitin-activating enzymes (E1s), ubiquitin conjugating enzymes (E2s), and ubiquitin ligases (E3s). Ubiquitination can affect proteins in different ways: it can tag proteins for degradation via the proteasome, change their cellular location, and promote or prevent interactions of proteins (Komander and Rape, 2012). Several key proteins in ABA signaling have been shown to be targeted by ubiquitination for proteasomal degradation. Low ABA levels enable recognition and ubiquitination of ABA receptors (e.g., PYL8) by the ubiquitin ligase substrate adaptor DDA1 and associated ubiquitin ligases (Irigoyen et al., 2014). Proteasomal degradation of ubiquitinated ABA receptors allows the release of PP2C phosphatases, negatively regulating SnRK2-mediated ABA signaling. On the other hand, increased ABA levels can stabilize the ABA receptors by restricting their ubiquitination (Irigoyen et al., 2014). Furthermore, the ubiquitin ligase RSL1 can target PYL4 and PYR1 ABA receptors in the plasma membrane and promote ubiquitination-mediated degradation of the ABA receptors (Bueso et al., 2014). The PP2C phosphatase ABI1 can also be ubiquitinated when it is associated the ABA receptor (e.g., PYR1). Proteasomal degradation of ABI1 may allow ABA signal to be transmitted more efficiently under stress conditions by eliminating the negative regulator of ABA signaling (Kong et al., 2015). Also, ABI5, a key regulator of ABA signaling, can be targeted by ubiquitination for proteasomal degradation, which could be important for inhibiting the expression of ABA-responsive genes regulated by ABI5 (Liu and Stone, 2013; Seo et al., 2014). Similarly, the abundance of the ABF1 and ABF3 transcription factor proteins is regulated by ubiquitination-mediated degradation, and ABA can stabilize ABF1 and ABF3 proteins (Chen et al., 2013). Two dozen ubiquitin ligases are known to participate in ABA signaling (Yu et al., 2016). For example, the endoplasmic reticulum-localized ubiquitin ligase SDIR1 (Salt-and Drought-Induced Really interesting new gene finger1) regulates ABA-mediated seed germination and salt stress response through ubiquitinating its substrate, SDIRIP1 (SDIR1-Interacting Protein 1) and subsequent proteasomal degradation (Zhang et al., 2015). The substrate proteins of some ubiquitin ligases implicated in ABA signaling are still not known. For instance, the ubiquitin ligase CHYR1 (CHY zinc-finger and Ring protein 1) promotes ABA-induced stomatal closure and improves plant drought tolerance (Ding et al., 2015). However, the substrate of CHYR1 is unknown. Nevertheless, substantial studies have demonstrated the important role of ubiquitination in many steps of ABA signal transduction (Yu et al., 2016).
Epigenetic Regulation of ABA Response
Epigenetics are the changes in gene function that are not due to changes in DNA nucleotide sequence, but rather due to chemical modification of DNA and its associated proteins (Bonasio et al., 2010). Epigenetic modifications comprise DNA methylation, histone modifications, and the production of microRNA (Yaish et al., 2011). As environmental stress factors can cause epigenetic modification of the genome, which provides an important mechanism for mediating gene-environment interplay (King, 2015). Recent studies suggest that epigenetic modification plays a significant role in plant response to abiotic stresses such as drought, salinity, heat, and cold (Kim et al., 2015). Here, we provide insights on epigenetic regulation of ABA-related stress signaling networks.
DNA methylation is the addition of a methyl group to the cytosine residues in DNA by DNA methyltransferases. In plants, cytosines in the contexts of CG, CHG, and CHH (H = A, C, or T) can be methylated (Law and Jacobsen, 2010). While CG methylation can occur in active genes, CHG and CHH methylation is almost exclusively present in heterochromatin (Stroud et al., 2013). DNA methylation in promoter regions is accompanying with repression of gene expression. The effect of DNA methylation in the gene body varies according to the level of methylation: extreme low or high level of DNA methylation correlates with lower gene expression while modest DNA methylation is associated with higher gene expression (Zemach et al., 2010). Two recent studies suggested that ABA can regulate gene expression through DNA methylation. Khraiwesh et al. (2010) showed that ABA represses gene expression in P. patens via DNA methylation of gene promoters. In another study, ABA was shown to increase DNA methylation of promoters in three ABA-repressive genes in Arabidopsis crown galls, indicating the importance of DNA methylation in gene regulation by ABA (Gohlke et al., 2013). Further studies are needed to elucidate more ABA-responsive genes for DNA methylation changes in response to ABA to understand the role of ABA in DNA methylation-dependent gene regulation.
Among histone modifications, histone acetylation has been shown to be important in regulating ABA-responsive genes and stress responses. Overexpression of AtHD2C (an Arabidopsis histone deacetylase gene) in Arabidopsis resulting in enhanced expression of ABA-responsive genes and improved drought tolerance (Sridha and Wu, 2006) suggest that histone deacetylation could play a fundamental role in activating ABA-responsive genes in acclimating plants to drought. Conversely, mutation or repression of a histone deacetylase gene HDA6 causing decreased expression of ABA and abiotic stress-responsive genes confers ABA and salt stress-hypersensitive phenotypes (Chen et al., 2010). Similarly, HDA19 histone deacetylase mutants also showed hypersensitive to ABA and salt stress (Chen and Wu, 2010). Furthermore, mutation of the HOS15 gene, which encodes a protein similar a component in the human nucleosome deacetylation complex, confers ABA and salt stress-hypersensitive phenotypes (Zhu et al., 2008; Chinnusamy et al., 2010). These studies indicate that histone acetylation and deacetylation play a critical role in the regulation of ABA-responsive genes and plant responses to abiotic stresses. In addition, chromatins remodeling factors and nucleosome assembly proteins have also been implicated in ABA responses. The chromatin remodeling protein BRM in Arabidopsis has been shown to repress ABA responses by remodeling the nucleosomes at the ABI5 (ABA insensitive 5) locus and thus inactivating expression of the ABA-related transcription factor ABI5 (Han et al., 2012). Three nucleosome assembly proteins in Arabidopsis (AtNAP1;1, AtNAP1;2, and AtNAP1; 3) have been reported to function as positive regulators of ABA signaling responses (Liu et al., 2009a).
MicroRNAs (miRNAs) are short non-coding RNA that regulates gene expression post-transcriptionally by complementing the target mRNAs causing target mRNA degradation or translational repression (Bartel, 2009). miRNAs can exert epigenetic regulation of gene expression by downregulation of key epigenetic regulators such as DNA methyltransferases and histone deacetylases (Sato et al., 2011). miRNA genes with changed expression in response to ABA have been identified in plants. In Arabidopsis, ABA upregulates the expression of miR159, miR393, and miR402, whereas miR169a can be downregulated by ABA (Sunkar and Zhu, 2004; Reyes and Chua, 2007; Li et al., 2008). The predicted target of miR402 is the transcript of Demeter-like protein 3 (DML3), a DNA glycosylase involved in DNA demethylation. It has been hypothesized that miR402 under ABA induction reduces the transcript level of DML3 resulting in repression of DML3-targeted genes (Sunkar and Zhu, 2004; Chinnusamy et al., 2008). ABA-induced miR159 accumulation can cause MYB33 and MYB101 transcript degradation, which is believed to desensitize ABA signaling during stress responses (Reyes and Chua, 2007). miR169a targets the transcript of NFYA5, a transcription factor vital in regulating plant development and stress responses. Downregulation of miR169 by ABA can contribute to lead to increased expression of NFYA5 and promote drought resistance (Li et al., 2008). In rice, ABA downregulates miR167 expression (Liu et al., 2009b). Interestingly, drought treatment can decrease miR167 expression and increase the expression of PLD (phospholipase D), which is the potential target of miR167 (Wei et al., 2009). As PLD is an important regulator of ABA response and stress signaling in plants (Guo and Wang, 2012), miR167 is likely to be involved in regulating ABA-dependent stress signaling networks. Global expression profiling in rice has identified 34 miRNAs whose expression is induced or suppressed by ABA treatment (Shen et al., 2010). Most of the ABA-responsive miRNAs (32) were responsive to at least one of the stress (drought, salt, or cold) treatments, suggesting that they are crucial for ABA-related stress pathways (Shen et al., 2010).
The current knowledge indicates that epigenetic modulation through DNA methylation, histone acetylation, and microRNA action, is critical to ABA-mediated stress signaling. A better understanding of epigenetics-mediated genome-environment interaction might provide new avenues for improving plant tolerance to abiotic stress.
Crosstalk
Crosslinks between ABA and MAPK Signaling Pathways
The mitogen-activated protein kinase (MAPK) cascade is an important signaling mechanism for regulating cellular responses to various stimuli including osmotic stress (Kelkar et al., 2000; Hamel et al., 2012). It is composed of an MAPK, MAPK kinase (MAPKK), and MAPKK kinase (MAPKKK). These three kinases can carry out phosphorylation relay from MAPKKK to MAPKK to MAPK. Twenty of MAPKs, 10 MAPKKs, and 80 MAPKKKs have been identified in Arabidopsis genome by structural and functional genome analysis (Colcombet and Hirt, 2008). A similar number of genes encoding MAPKs, MAPKKs, and MAPKKKs have been found in other plant’s genomes (Ichimura et al., 2002; Hamel et al., 2006; Colcombet and Hirt, 2008). For example, rice genome contains 17 MAPK genes, 8 MAPKK genes, and 74 MAPKKK genes (Reyna and Yang, 2006; Rao et al., 2010). Earlier study (Mizoguchi et al., 1996) reported that the MEKK1 (MAPKKK) transcript is up-regulated in response to various environmental stresses such as high salinity, low temperature, and mechanical stress. Teige et al. (2004), proposed the complete MAPK cascade functioning model MEKK1-MEKK2-MPK4/MPK6 for abiotic stress signaling. Few years later, some study showed that this model has a significant role in abiotic stress signaling (Ichimura et al., 2000; Teige et al., 2004; Nakagami et al., 2006; Ortiz-Masia et al., 2007; Xing et al., 2008). Many studies reported that MAPK cascades are involved in several ABA responses such as seed germination, guard cell signaling, and antioxidant defense (Xing et al., 2008; Jammes et al., 2009; Zhang et al., 2012).
It has been shown that ABA can affect the transcriptional regulation, protein accumulation, and stability of different component in MAPK cascade. For example, transcriptional expression of MAPKs (MPK3, MPK5, MPK7, MPK18, MPK20, MKK9), MAPKKKs (Raf6, Raf12, and Raf35), MAPKKK1(ANP1) and MAPKKK14-19 in Arabidopsis is induced by ABA treatment, suggesting that they play a significant role in ABA signaling. However, the functions of many MAPK pathway genes are still unknown (Menges et al., 2008; Wang et al., 2010; Danquah et al., 2014). Similarly in case of rice, ABA induces transcriptional activation of several MAPKs like OsMAP1(OsMAPK5), OsMAPK2, OsMSRMK2, OsMSRMK3, OsMAPK44, OsBIMK1, DMS1, OsEDR1, OsSIPK, OsWJUMK1 and OmMKK1 (Huang et al., 2002; Song and Goodman, 2002; Wen et al., 2002; Agrawal et al., 2003; Xiong and Yang, 2003; Jeong et al., 2006; You et al., 2007; Lee et al., 2008; De Vleesschauwer et al., 2010; Ning et al., 2010). Similar to Arabidopsis and rice, ABA also induces MAPKs in maize like ZmMPK7, ZmPK17, ZmSIMK1, and ZmMPK3 (Zong et al., 2009; Gu et al., 2010; Wang et al., 2010; Pan et al., 2012). Recently Zhang et al. (2012) reported that ZmMKK3 is involved in osmotic stress and ABA responses. In pea, p45MAPK is ABA-activated MAPK involved in guard cell signaling (Burnett et al., 2000; Schroeder et al., 2001). Brock et al. (2010) reported that a double knockout mutant (pp2c5/ap2c1) of the MAP-interacting phosphatases showed increased ABA-dependent activation of MPK3 and MPK6, which gives ABA sensitivity and suggests that MAPK cascade involving MPK3/MPK6 adversely regulates ABA signaling in plants.
Recently Danquah et al. (2015) demonstrated that a complete MAPK cascade (comprised of MAP3K17/18, MKK3, and the MAPKs MPK1/2/7/14) can be regulated by ABA core signaling module. They performed yeast two-hybrid analysis against all MAP2Ks and identified that only MAP2K is interacting with MAP3K17 and 18. All plants genome contain one gene coding for such an MKK3-like MAP2K, suggesting that this fusion occurred early and were conserved during evolution. Danquah et al. (2015) showed that a loss of function of MKK3 totally abolished ABA-dependent MPK17 activation, suggesting that this MAP2k is a significant factor of the ABA-triggered MAPK pathway. They also reported that ABA-triggered MAPK pathway depends on the transcription and protein synthesis of the MAP3Ks, MAP3K17, and MAP3K18. They also showed that ABA core machinery is required for the activation of the MAP3k17/18-MKK3-MPK1/2/7/14. The knowledge of crosslinks between MAPK cascade and ABA signaling should provide valuable information on the complex mechanism of plant stress adaptation, which helps to enhance crop stress tolerance that will be helpful for future sustainable agriculture.
Crosstalk between ABA and Other Hormones
Abscisic acid and auxin control many aspects of plant growth and development together, mostly in opposing directions (Sun and Li, 2014). For examples, the expression of AUXIN RESPONSE FACTOR 2 (ARF2) induced by ABA and arf2 mutants exhibited enhanced ABA sensitivity in seed germination and primary root growth (Wang et al., 2011b). ABA treatment reduces the cell division and alters the auxin distribution more in arf2 mutants as compared to wildtypes. Mutant arf2 showed shorter roots under ABA application. They also suggested that there is a significant mechanism in ABA inhibiting the primary root growth through mediating cell division in root tips (Wang et al., 2011b). ABA inhibits seedling growth through enhancing auxin signaling (Belin et al., 2009). ABA and auxin also control seed dormancy through gene expression network involving ABSCISIC ACID INSENSITIVE 3 (ABI3) transcription factor (Liu et al., 2013). Auxin acts upstream of the major regulator of seed dormancy, ABI3, by recruiting AUXIN RESPONSE FACTOR 10 and AUXIN RESPONSE FACTOR 16 to control the expression of ABI3 during seed germination. They also showed that auxin is essential for seed dormancy and ABA inhibition of seed germination, signifying that the roles of auxin and ABA in seed dormancy are mutually dependent. In the ABA-mediated process, auxin functions in two ways: auxin activates the ABA response or stimulates the ABA biosynthesis response (Liu et al., 2013). In radical protrusion assayed, both exogenous and endogenous indole acetic acid (IAA) inhibited seed germination, indicating that ABA and auxin act synergistically to inhibit seed germination (Liu et al., 2013). They also observed that equal amount of IAA did not inhibit the seed germination in the absence of ABA, indicating that the auxin-mediated inhibition of seed germination is dependent on ABA. Further, osmotically induced ABA increases basipetal auxin transport through elevated AUX1 (an auxin influx transporter) and PIN2 (an auxin efflux transporter) levels (Xu et al., 2013). Therefore, ABA and auxin interact with crosstalk network via auxin response pathways in regulating plant growth.
Abscisic acid and cytokinin also appear to crosstalk each other (El-Showk et al., 2013). Cytokinin degradation is mediated by cytokinin oxidase enzymes which are encoded by CYTOKININ OXIDASE (CKX) multigene families with a varying number of members. For example, the Arabidopsis genome contains 7 CKX genes (CKX1-CKX7). Microarray and RT-PCR analysis showed that ABA downregulates CKX genes (CKX1, CKX3, CKX4, and CKX6) (Werner et al., 2006). During stress conditions, cytokinin receptor kinases (AHK2 and AHK3) negatively regulate ABA as well as osmotic stress responsive gene expression, whereas, cytokinin-deficient mutants have increased survival rates under stress (Tran et al., 2007). In addition, AHK2 and AHK3 are involved in mediating cold stress response by inhibiting ABA signaling (Jeon et al., 2010). Therefore, ABA and cytokinin have crosstalk to regulate stress responses.
Abscisic acid and ethylene interact to control the regulation of stomatal closure (Harrison, 2012). ABA and ethylene also control the increased ethylene concentration in leaves. The ethylene delays stomata closure by inhibiting the ABA signaling pathway (Tanaka et al., 2005). ABA inhibits ethylene biosynthesis by positively regulating ethylene response factor 11 (ERF11) via LONG HYPOCOTYL5 (HY5) transcription factor to repress expression of ethylene biosynthesis gene (ACS5), which encodes the rate-limiting enzyme in ethylene biosynthesis. The HY5 transcription factor serves as an important molecular link between ABA and ethylene biosynthesis (Li et al., 2011). They proposed a model suggesting that the HYF-AtERF11 regulon is a key factor modulating ABA-regulated ethylene biosynthesis (Li et al., 2011).
There is a complex interplay between ABA-signaling pathway and jasmonate (JA) regulating plant defense and gene expression (Anderson et al., 2004). For example, exogenous ABA treatment could suppress both basal and JA activated transcription from defense gene. In contrast, mutation of the ABA synthesis genes aba1 and aba2 resulted in upregulation of JA-responsive defense gene. These results indicate that the antagonistic interactions between multiple components of ABA and the JA-ethylene signaling pathways to modulate defense and stress-responsive gene expression in response to abiotic and biotic stresses (Anderson et al., 2004). Transcription factor MYC2 is a master regulator of most aspects of the JA-signaling pathway in Arabidopsis (Chen et al., 2011; Fernandez-Calvo et al., 2011). Transgenic plants overexpressing MYC2 gene had higher sensitivity to ABA and several ABA-inducible genes upregulated in the transgenic plants whereas Ds insertion mutant myc2 was less sensitive to ABA and showed reduced ABA-induced gene expression (Abe et al., 2003). Thus, MYC2 is proposed as an integrative hub for the regulation of both JA and ABA signaling in Arabidopsis (Sun and Li, 2014).
Thus, ABA is an important phytohormone which regulates plant growth and development in response to environmental stresses interacting through other plant hormones. The molecular mechanisms for the complex signaling networks between ABA and other hormones still need to study for further details in future.
Conclusion and Future Perspectives
Due to growing population and global climate change food demand is increasing day by day. Thus, to fulfill the required demand, food production must be increased. Therefore, understanding abiotic stress tolerance in the plant has important, and broad spread implications that can not be understated. ABA is a vital hormone and acts as a central regulator of different plant stresses like drought, low temperature, and salinity. Due to the advancement of molecular genetic and genome-wide technologies, a deeper understanding of the underlying mechanisms of the involvement of ABA in stress tolerance is achieved, although more is yet to be revealed. The different physiological reactions that ABA regulates have been described at molecular level like stomatal closure, change in gene expression and accumulation of osmoprotectants. Understanding of ABA signal transduction pathways and epigenetic modifications can be further exploited to produce better transgenic plants with improved stress tolerance without a yield penalty that can withstand adverse climatic conditions. Genetic regulation through cis-engineering can be accomplished by designing promoter targeted at tissue-specific expression through which plant dynamics can be further explored. Many efforts are still required to uncover in details of each product of a gene induced by ABA and their interacting partners to understand the complexity of stress signal transduction pathways. The source of ABA in guard cells is still not fully understood. The mechanisms by which the abiotic stress upregulate ABA biosynthesis genes are still needed to know. Furthermore, the molecular mechanisms of crosstalk between ABA and other phytohormone signaling pathways remain to be elucidated. Although many questions are still open, the current advances in ABA signaling in Arabidopsis pave the way to address the molecular events underlying stress responses in other plant species, with the prospect to improve the abiotic stress performance of crop plants. Recently, Park et al. (2015) engineered ABA receptors by agrochemical engineering that provides the lots of opportunity in crop improvement program.
Author Contributions
SS, JL, and KR drafted the manuscript. SS and JL designed the figures. SS, JL, and KR revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SS is supported by an MAFES/FWRC Directors’ Doctoral Fellowship awarded to JL and KR. This material is based upon work that is supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch/Multi-State Project (Multistate No. NC1200; Project No. MIS-153180) under Accession No. 232044. We acknowledge Mississippi Soybean Promotion Board for supporting our research.
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78. 10.1105/tpc.006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinizaki K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9 1859–1868. 10.1105/tpc.9.10.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal G. K., Tamogami S., Iwahashi H., Agrawal V. P., Rakwal R. (2003). Transient regulation of jasmonic acid-inducible rice MAP kinase gene (OsBWMK1) by diverse biotic and abiotic stresses. Plant Physiol. Biochem. 41 355–361. 10.1016/S0981-9428(03)00030-5 [DOI] [Google Scholar]
- Ambrosone A., Costa A., Leone A., Grillo S. (2012). Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci. 182 12–18. 10.1016/j.plantsci.2011.02.004 [DOI] [PubMed] [Google Scholar]
- An Z., Jing W., Liu Y., Zhang W. (2008). Hydrogen peroxide generated by copper amine oxidase is involved in abscisic acid-induced stomatal closure in Vicia faba. J. Exp. Bot. 59 815–825. 10.1093/jxb/erm370 [DOI] [PubMed] [Google Scholar]
- Anderson J. P., Badruzsaufari E., Schenk P. M., Manners J. M., Desomnd O. J., Ehlert C., et al. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistant in Arabidopsis. Plant Cell 16 3460–3479. 10.1105/tpc.104.025833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R., Gonzalez-Guzman M., Rodriguez L., Peirats-Llobet M., Pizzio G. A., Fernandez M. A., et al. (2013). PYRABACTIN RESISTANCE1-LIKE8 plays an important role in the regulation of abscisic acid signaling in root. Plant Physiol. 161 931–941. 10.1104/pp.112.208678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R., Gonzalez-Guzman M., Rodriguez L., Rodrigues A., Pizzio G. A., Rodriguez P. L. (2012). selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 158 970–980. 10.1104/pp.111.188623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswath C. R., Kim S. H., Mo S. Y., Kim D. H. (2005). Transgenic plants of creeping bent grass harboring the stress-inducible gene, 9-cis-epoxycarotenoid dioxygenase, are highly tolerant to drought and NaCl Stress. Plant Growth Regul. 47 129–139. 10.1007/s10725-005-3380-6 [DOI] [Google Scholar]
- Bailey-Serres J., Lee S. C., Brinton E. (2012). Waterproofing crops: effective flooding survival strategies. Plant Physiol. 160 1698–1709. 10.1104/pp.112.208173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao G., Zhuo C., Qian C., Xiao T., Guo Z., Lu S. (2016). Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol. J. 14 206–214. 10.1111/pbi.12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardsell M. F., Cohen D. (1975). Relationships between leaf water status, abscisic acid levels, and stomatal resistance in maize and sorghum. Plant Physiol. 56 207–212. 10.1104/pp.56.2.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin C., Megies C., Hauserová E., Lopez-Molina L. (2009). Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21 2253–2268. 10.1105/tpc.109.067702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen R. J., Boyer J. S., Mullet J. E. (1988). Water deficit-induced changes in abscisic acid, growth, polysomes, and translatable RNA in soybean hypocotyls. Plant Physiol. 88 289–294. 10.1104/pp.88.2.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhmer M., Schroeder J. I. (2011). Quantitative transcriptomic analysis of abscisic acid-induced and reactive oxygen species-dependent expression changes and proteomic profiling in Arabidopsis suspension cells. Plant J. 67 105–118. 10.1111/j.1365-313X.2011.04579.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R., Tu S., Reinberg D. (2010). Molecular signals of epigenetic states. Science 330 612–616. 10.1126/science.1191078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Barbier-Brygoo H., Lauriere C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279 41758–41766. 10.1074/jbc.M405259200 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Droillard M. J., Barbier-Brygoo H., Lauriere C. (2007). Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol. 63 491–503. 10.1007/s11103-006-9103-1 [DOI] [PubMed] [Google Scholar]
- Boursiac Y., Leran S., Corratge-Faillie C., Gojon A., Krouk G., Lacombe B. (2013). ABA transport and transporters. Trends Plant Sci. 18 325–333. 10.1016/j.tplants.2013.01.007 [DOI] [PubMed] [Google Scholar]
- Boyer J. S. (1982). Plant productivity and environment. Science 218 443–448. 10.1126/science.218.4571.443 [DOI] [PubMed] [Google Scholar]
- Brandt B., Brodsky D. E., Xue S., Negi J., Iba K., Kangasjarvi J., et al. (2012). Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. U.S.A. 109 10593–10598. 10.1073/pnas.1116590109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard I. M., Lynch T. J., Finkelstein R. R. (2002). Regulation and role of the Arabidopsis abscisic acid-insensitive 5 genes in abscisic acid, sugar, and stress response. Plant Physiol. 129 1533–1543. 10.1104/pp.005793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock A. K., Willmann R., Kolb D., Grefen L., Lajunen H. M., Bethke G., et al. (2010). The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 153 1098–1111. 10.1104/pp.110.156109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E., Rodriguez L., Lorenzo-Orts L., Gonzalez-Guzman M., Sayas E., Munoz-Bertomeu J., et al. (2014). The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 80 1057–1071. 10.1111/tpj.12708 [DOI] [PubMed] [Google Scholar]
- Burnett E. C., Desikan R., Moser R. C., Neill S. J. (2000). ABA activation of an MBP kinase in Pisum sativum epidermal peels correlates with stomatal responses to ABA. J. Exp. Bot. 51 197–205. 10.1093/jexbot/51.343.197 [DOI] [PubMed] [Google Scholar]
- Burza A. M., Pekala I., Sikora J., Siedlecki P., Malagocki P., Bucholc M., et al. (2006). Nicotiana tabacum osmotic stress-activated kinase is regulated by phosphorylation of Ser-154 and Ser-158 in the kinase activation loop. J. Biol. Chem. 281 34299–34311. 10.1074/jbc.M601977200 [DOI] [PubMed] [Google Scholar]
- Carvalho R. F., Carvalho S. D., Duque P. (2010). The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol. 154 772–783. 10.1104/pp.110.155523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. T., Luo M., Wang Y. Y., Wu K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61 3345–3353. 10.1093/jxb/erq154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. T., Wu K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5 1318–1320. 10.4161/psb.5.10.13168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Sun J., Zhai Q., Zhou W., Qi L., Xu L., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23 3335–3352. 10.1105/tpc.111.089870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. T., Liu H. X., Stone S., Callis J. (2013). ABA and the ubiquitin E3 ligase keep on going affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J 75 965–976. 10.1111/tpj.12259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W. H., Endo A., Zhou L., Penney J., Chen H. C., Arroyo A., et al. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14 2723–2743. 10.1105/tpc.006494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Gong Z., Zhu J.-K. (2008). Abscisic acid-mediated epigenetic processes in plant development and stress responses. J. Integr. Plant. Biol. 50 1187–1195. 10.1111/j.1744-7909.2008.00727.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J. K., Sunkar R. (2010). Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 639 39–55. 10.1007/978-1-60761-702-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. I., Hong J. H., Ha J. O., Kang J. Y., Kim S. Y. (2000). ABFs, a Family of ABA-responsive element binding factors. J. Biol. Chem. 275 1723–1730. 10.1074/jbc.275.3.1723 [DOI] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: a complex signaling network involved in multiple biological processes. Biochem. J. 413 217–226. 10.1042/bj20080625 [DOI] [PubMed] [Google Scholar]
- Cracker L. E. A., Abeles F. B. (1969). Abscission: role of abscisic acid. Plant Physiol. 44 1144–1149. 10.1104/pp.44.8.1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz T. M. D., Carvalho R. F., Richardson D. N., Duque P. (2014). Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression. Int. J. Mol. Sci. 15 17541–17564. 10.3390/ijms151017541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler A. J., Krochko J. E. (1999). Formation and breakdown of ABA. Trends Plant. Sci. 4 472–478. 10.1016/S1360-1385(99)01497-1 [DOI] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61 651–679. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- Dai M., Xue Q., Mccray T., Margavage K., Chen F., Lee J. H., et al. (2013). The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25 517–534. 10.1105/tpc.112.105767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daie J., Campbell W. F. (1981). Response of tomato plants to stressful temperatures. Increase in abscisic acid concentrations. Plant Physiol. 67 26–29. 10.1104/pp.67.1.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danquah A., de Zelicourt A., Colcombet J., Hirt H. (2014). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32 40–52. 10.1016/j.biotechadv.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Danquah A., Zélicourt A., Boudsocq M., Neubauer J., Frei dit Frey N., Leonhardt N., et al. (2015). Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 82 232–244. 10.1111/tpj.12808 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Yang Y., Vera Cruz C., Höfte M. (2010). Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol. 152 2036–2052. 10.1104/pp.109.152702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K. J., Sauter A., Wichert K., Messdaghi D., Hartung W. (2000). Extracellular beta-glucosidase activity in barley involved in the hydrolysis of ABA-glucose conjugate in leaves. J. Exp. Bot. 51 937–944. 10.1093/jexbot/51.346.937 [DOI] [PubMed] [Google Scholar]
- Ding S., Zhang B., Qin F. (2015). Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27 3228–3244. 10.1105/tpc.15.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. C., Egea G., Davies W. J. (2008). Abscisic acid signaling when soil moisture is heterogeneous: decreased photoperiod sap flow from drying roots limits abscisic acid export to the shoots. Plant Cell Environ. 31 1263–1274. 10.1111/j.1365-3040.2008.01831.x [DOI] [PubMed] [Google Scholar]
- Dodd I. C., Egea G., Watts C. W., Whalley W. R. (2010). Root water potential integrates discrete soil physical properties to influence ABA signaling during partial rootzone drying. J. Exp. Bot. 61 3543–3551. 10.1093/jxb/erq195 [DOI] [PubMed] [Google Scholar]
- Du Y. L., Wang Z. Y., Fan J. W., Turner N. C., He J., Wang T., et al. (2013). Exogenous abscisic acid reduces water loss and improves antioxidant defence, desiccation tolerance and transpiration efficiency in two spring wheat cultivars subjected to a soil water deficit. Funct. Plant Biol. 40 494–506. 10.1071/FP12250 [DOI] [PubMed] [Google Scholar]
- Duan L., Dietrich D., Ng C. H., Chan P. M. Y., Bhalerao R., Bennett M. J., et al. (2013). Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25 324–341. 10.1105/tpc.112.107227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Showk S., Ruonala R., Helariutta Y. (2013). Crossing paths: cytokinin signalling and crosstalk. Development 140 1373–1383. 10.1242/dev.086371 [DOI] [PubMed] [Google Scholar]
- Estrada-Melo A. C., Chao, Reid M. S., Jiang C.-Z. (2015). Overexpression of an ABA biosynthesis gene using a stress-inducible promoter enhances drought resistance in petunia. Hortic. Res. 2:15013 10.1038/hortres.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etehadnia M., Waterer D. R., Tanino K. K. (2008). The method of ABA application affects salt stress responses in resistant and sensitive potato lines. J. Plant Growth Regul. 27 331–341. 10.1007/s00344-008-9060-9 [DOI] [Google Scholar]
- Eze J. M., Dumbroff E. B., Thompson J. E. (1983). Effects of temperature and moisture stress on the accumulation of abscisic acid in bean. Physiol. Plant. 58 179–183. 10.1111/j.1399-3054.1983.tb04165.x [DOI] [Google Scholar]
- Fan K., Bibi N., Gan S., Li F., Yuan S., Ni M., et al. (2015). A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. J. Exp. Bot. 66 4669–4682. 10.1093/jxb/erv240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L. M., Zhao Z., Assmann S. M. (2004). Guard cells: a dynamic signaling model. Curr. Opin. Plant Biol. 7 537–546. 10.1016/j.pbi.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Fang J., Chai C., Qian Q., Li C., Tang J., Sun L., et al. (2008). Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 54 177–189. 10.1111/j.1365-313X.2008.03411.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C. Z., Chen Y., Wang C., Kong Y. H., Wu W. H., Chen Y. F. (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI 3, ABI 4, and ABI5 during seed germination and early seedling development. Plant J. 80 654–668. 10.1111/tpj.12670 [DOI] [PubMed] [Google Scholar]
- Feng J. X., Liu D., Pan Y., Gong W., Ma L. G., Luo J. C., et al. (2005). An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 59 853–868. 10.1007/s11103-005-1511-0 [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvo P., Chini A., Fernandez-Barbero G., Chico J. M., Gimenez-Ibanez S., Geerinck J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23 701–715. 10.1105/tpc.110.080788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. (2013). Abscisic acid synthesis and response. Arabidopsis Book 11:e0166 10.1199/tab.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu Rev. Plant Biol. 59 387–415. 10.1146/annurev.arplant.59.032607.092740 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. R., Lynch T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. 10.1105/tpc.12.4.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R., Rock C. D. (2002). Abscisic acid biosynthesis and response. Arabidopsis Book 1:e0058 10.1199/tab.0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A., Grau A., Laurich F., Dorffling K. (1988). Effect of new terpenoid analogues of abscisic acid on chilling and freezing resistance. J. Plant Physiol. 132 362–369. 10.1016/S0176-1617(88)80121-4 [DOI] [Google Scholar]
- Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.-Y., et al. (2009). In vitro reconstitution of an abscisic acid signaling pathway. Nature 462 660–664. 10.1038/nature08599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P. E., Zhu J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19 485–494. 10.1105/tpc.106.048538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P. E., Zhu J. K. (2011). Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. U.S.A. 108 1717–1722. 10.1073/pnas.1018367108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Zhu J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106 8380–8385. 10.1073/pnas.0903144106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124 509–525. 10.1007/s10265-011-0412-3 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Nakashima K., Yoshida T., Katagiri T., Kidokoro S., Kanamori N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50 2123–2132. 10.1093/pcp/pcp147 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Yoshida T., Yamaguchi-Shinozaki K. (2013). Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 147 15–27. 10.1111/j.1399-3054.2012.01635.x [DOI] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103 1988–1993. 10.1073/pnas.0505667103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S. (2003). Mitotic and postmitotic senescence in plants. Sci. Aging Knowledge. Environ 2003:RE7 10.1126/sageke.2003.38.re7 [DOI] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Marten I., Ache P., Matschi S., et al. (2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. U.S.A. 107 8023–8028. 10.1073/pnas.0912030107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., et al. (2009). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U.S.A. 106 21425–21430. 10.1073/pnas.0912021106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendra E., Moreno A., Alba M. M., Pages M. (2004). Interaction of the plant glycine-rich RNA-binding protein MA16 with a novel nucleolar DEAD box RNA helicase protein from Zea mays. Plant J. 38 875–886. 10.1111/j.1365-313X.2004.02095.x [DOI] [PubMed] [Google Scholar]
- Gillard D. F., Walton D. C. (1976). Abscisic acid metabolism by a cell-free preparation from Echinocystis lobata liquid endoserum. Plant Physiol. 58 790–795. 10.1104/pp.58.6.790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka Z., Reinhold L. (1971). Abscisic acid raises the permeability of plant cells to water. Plant Physiol. 48 103 10.1104/pp.48.1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T., Bachorik J. L., Yong J., Dreyfuss G. (2008). RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582 1977–1986. 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke J., Scholz C.-J., Kneitz S., Weber D., Fuchs J., Hedrich R., et al. (2013). DNA methylation mediated control of gene expression is critical for development of crown gall tumors. PLoS Genet. 9:e1003267 10.1371/journal.pgen.1003267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J., Sanchez-Martinez D., Stiefel V., Rigau J., Puigdomenech P., Pages M. (1988). A gene induced by the plant hormone abscisic acid in response to water stress encodes a glycine-rich protein. Nature 334 262–264. 10.1038/334262a0 [DOI] [PubMed] [Google Scholar]
- Gómez-Cadenas A., Verhey S. D., Holappa L. D., Shen Q., Ho T.-H. D., Walker-Simmons M. K. (1999). An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc. Natl. Acad. Sci. U.S.A. 96 1767–1772. 10.1073/pnas.96.4.1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Dong C. H., Lee H., Zhu J., Xiong L., Gong D., et al. (2005). A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17 256–267. 10.1105/tpc.104.027557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ballester D., Pollock S. V., Pootakham W., Grossman A. R. (2008). The central role of an SNRK2 kinase in sulfur deprivation responses. Plant Physiol. 147 216–227. 10.1104/pp.108.116137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Apostolova N., Belles J. M., Barrero J. M., Piqueras P., Ponce M. R., et al. (2002). The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14 1833–1846. 10.1105/tpc.002477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Pizzio G. A., Antoni R., Vera-Sirera F., Merilo E., Bassel G. W., et al. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24 2483–2496. 10.1105/tpc.112.098574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Rodriguez L., Lorenzo-Orts L., Pons C., Sarrion-Perdigones A., Fernandez M. A., et al. (2014). Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 65 4451–4464. 10.1093/jxb/eru219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F., Beaudoin N., Serizet C., Webb A. A., Vartanian N., Giraudat J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11 1897–1909. 10.2307/3871085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin A., Rodrigues O., Verdoucq L., Merlot S., Leonhardt N., Maurel C. (2015). Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 27 1945–1954. 10.1105/tpc.15.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Liu Y., Zong X., Liu L., Li D.-P., Li D.-Q. (2010). Overexpression of maize mitogen-activated protein kinase gene, ZmSIMK1 in Arabidopsis increases tolerance to salt stress. Mol. Biol. Rep. 37 4067–4073. 10.1007/s11033-010-0066-6 [DOI] [PubMed] [Google Scholar]
- Guo L., Wang X. (2012). Crosstalk between phospholipase D and sphingosine kinase in plant stress signaling. Front. Plant Sci. 3:51 10.3389/fpls.2012.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2006). AtNAP, NAC family transcription factor, has an important role in leaf senescence. Plant J. 46 601–612. 10.1111/j.1365-313X.2006.02723.x [DOI] [PubMed] [Google Scholar]
- Guoth A., Tari I., Galle A., Csiszar J., Pecsvaradi A., Cseuz L., et al. (2009). Comparison of the drought stress responses of tolerant and sensitive wheat cultivars during grain filling: changes in flag leaf photosynthetic activity, ABA levels, and grain yield. J. Plant Growth Regul. 28 167–176. 10.1007/s00344-009-9085-8 [DOI] [Google Scholar]
- Hamel L.-P., Nicole M.-C., Duplessis S., Ellis B. E. (2012). Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers. Plant Cell 24 1327–1351. 10.1105/tpc.112.096156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel L. P., Nicole M. C., Sritubtim S., Morency M. J., Ellis M., Ehlting J., et al. (2006). Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11 192–198. 10.1016/j.tplants.2006.02.007 [DOI] [PubMed] [Google Scholar]
- Han S. K., Sang Y., Rodrigues A., Wu M. F., Rodriguez P. L., Wagner D. (2012). The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24 4892–4906. 10.1105/tpc.112.105114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. (2015). Abscisic acid: hidden architect of root system structure. Plants 4 548–572. 10.3390/plants4030548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. A. (2012). “Cross-talk between phytohormone signaling pathways under both optimal and stressful environmental conditions,” in Phytohormones and Abiotic Stress Tolerance in Plants, Chap. 2 eds Khan N. A., Nazar R., Iqbal N., Anjum N. A. (Berlin: Springer link; ). [Google Scholar]
- Hasanuzzaman M., Nahar K., Alam M. M., Roychowdhury R., Fujita M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14 9643–9684. 10.3390/ijms14059643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Inoue S., Takahashi K., Kinoshita T. (2011). Immunohistochemical detection of blue light-induced phosphorylation of the plasma membrane H+-ATPase in stomatal guard cells. Plant Cell Physiol. 52 1238–1248. 10.1093/pcp/pcr072 [DOI] [PubMed] [Google Scholar]
- He H., Li J. (2008). Proteomic analysis of phosphoproteins regulated by abscisic acid in rice leaves. Biochem. Biophy. Res. Commun. 371 883–888. 10.1016/j.bbrc.2008.05.001 [DOI] [PubMed] [Google Scholar]
- He P., Jin J. Y. (1999). Relationships among hormone changes, transmembrane flux of Ca2+ and lipid peroxidation during leaf senescing in spring maize. Acta Bot. Sin. 41 1221–1225. [Google Scholar]
- He P., Osaki M., Takebe M., Shinano T., Wasaki J. (2005). Endogenous hormones and expression of senescence-related genes in different senescent types of maize. J. Exp. Bot. 56 1117–1128. 10.1093/jxb/eri103 [DOI] [PubMed] [Google Scholar]
- Henson I. E. (1984). Effects of atmospheric humidity on abscisic acid accumulation and water status in leaves of rice. Ann. Bot. 54 569–582. [Google Scholar]
- Hirayama T., Shinozaki K. (2007). Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12 343–351. 10.1016/j.tplants.2007.06.013 [DOI] [PubMed] [Google Scholar]
- Hobo T., Asada M., Kowyama Y., Hattori T. (1999). ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19 679–689. 10.1046/j.1365-313x.1999.00565.x [DOI] [PubMed] [Google Scholar]
- Hoth S., Morgante M., Sanchez J.-P., Hanafey M. K., Tingey S. V., Chua N.-H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115 4891–4900. 10.1242/jcs.00175 [DOI] [PubMed] [Google Scholar]
- Hrabak E. M., Chan C. W. M., Gribskov M., Harper J. F., Choi J. H., Halford N., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132 666–680. 10.1104/pp.102.011999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai J., Wang M., He J., Zheng J., Dong Z., Lv H., et al. (2008). Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep. 27 1861–1868. 10.1007/s00299-008-0608-8 [DOI] [PubMed] [Google Scholar]
- Huang H.-J., Fu S.-F., Tai Y.-H., Chou W.-C., Huang D.-D. (2002). Expression of Oryza sativa MAP kinase gene is developmentally regulated and stress-responsive. Physiol. Plant. 114 572–580. 10.1034/j.1399-3054.2002.1140410.x [DOI] [PubMed] [Google Scholar]
- Huang X., Duan M., Liao J., Yuan X., Chen H., Feng J., et al. (2014). OsSLI1, a Homeodomain-containing transcription activator, involves abscisic acid-related stress response in rice (Oryza sativa L.). Sci. World J. 2014:809353 10.1155/2014/809353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V., Kwak J. M., Schroeder J. I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106 477–487. 10.1016/S0092-8674(01)00460-3 [DOI] [PubMed] [Google Scholar]
- Hugouvieux V., Murata Y., Young J. J., Kwak J. M., Mackesy D. Z., Schroeder J. I. (2002). Localization, ion channel regulation, and genetic interactions during abscisic acid signaling of the nuclear mRNA cap-binding protein, ABH1. Plant Physiol. 130 1276–1287. 10.1104/pp.009480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K., Mizoguchi T., Yoshida R., Yuasa T., Shinozaki K. (2000). Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 24 655–665. 10.1046/j.1365-313x.2000.00913.x [DOI] [PubMed] [Google Scholar]
- Ichimura K., Shinozaki K., Tena G., Sheen J., Henry Y., Champion A., et al. (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7 301–308. [DOI] [PubMed] [Google Scholar]
- Irigoyen M. L., Iniesto E., Rodriguez L., Puga M. I., Yanagawa Y., Pick E., et al. (2014). Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 26 712–728. 10.1105/tpc.113.122234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111. 10.1016/S1360-1385(01)02223-3 [DOI] [PubMed] [Google Scholar]
- Jammes F., Song C., Shin D., Munemasa S., Takeda K., Gu D., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. U.S.A. 106 20520–20525. 10.1073/pnas.0907205106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaradat M. R., Feurtado J. A., Huang D., Lu Y., Cutler A. J. (2013). Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 13:1 10.1186/1471-2229-13-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., Kim N. Y., Kim S., Kang N. Y., Novák O., Ku S. J., et al. (2010). A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 285 23371–23386. 10.1074/jbc.M109.096644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong M.-J., Lee S.-K., Kim B.-G., Kwon T.-R., Cho W.-S., Park Y.-T., et al. (2006). A rice (Oryza sativa L.) MAP kinase gene, OsMAPK44, is involved in response to abiotic stresses. Plant Cell Tiss. Org. Cult. 85 151–160. 10.1007/s11240-005-9064-0 [DOI] [Google Scholar]
- Ji H., Peng Y., Meckes N., Allen S., Stewart C. N., Traw M. B. (2014). ATP-dependent binding cassette transporter G family member 16 increases plant tolerance to abscisic acid and assists in basal resistance against Pseudomonas syringae DC3000. Plant Physiol. 166 879–888. 10.1104/pp.114.248153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Zhang C., Lü P., Jiang G., Liu X., Dai F., et al. (2014). RhNAC3, a stress-associated NAC transcription factor, has a role in dehydration tolerance through regulating osmotic stress-related genes in rose petals. Plant Biotechnol. J. 12 38–48. 10.1111/pbi.12114 [DOI] [PubMed] [Google Scholar]
- Johnson R. R., Wagner R. L., Verhey S. D., Walker-Simmons M. K. (2002). The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 130 837–846. 10.1104/pp.001354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Saha A., Valon C., Leung J. (2011). A brand new START: abscisic acid perception and transduction in the guard cell. Sci. Signal. 4:re4 10.1126/scisignal.2002164 [DOI] [PubMed] [Google Scholar]
- Kagaya Y., Hobo T., Murata M., Ban A., Hattori T. (2002). Abscisic acid–induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14 3177–3189. 10.1105/tpc.005272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Hwang J.-U., Lee M., Kim Y.-Y., Assmann S. M., Martinoia E., et al. (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107 2355–2360. 10.1073/pnas.0909222107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Yim S., Choi H., Kim A., Lee K. P., Lopez-Molina L., et al. (2015). Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 6:8113 10.1038/ncomms9113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannangara T., Seetharama N., Durley R. C., Simpson G. M. (1983). Drought resistance of sorghum bicolor: 6. changes in endogenous growth regulators of plants grown across an irrigation gradient. Can. J. Plant Sci. 63 147–155. 10.4141/cjps83-014 [DOI] [Google Scholar]
- Kanno Y., Hanada A., Chiba Y., Ichikawa T., Nakazawa M., Matsui M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U.S.A. 109 9653–9658. 10.1073/pnas.1203567109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant P., Kant S., Gordon M., Shaked R., Barak S. (2007). STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol. 145 814–830. 10.1104/pp.107.099895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar N., Gupta S., Dickens M., Davis R. J. (2000). Interaction of a mitogen-activated protein kinase signaling module with the neuronal protein JIP3. Mol. Cell. Biol. 20 1030–1043. 10.1128/MCB.20.3.1030-1043.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelner A., Pekala I., Kaczanowski S., Muszynska G., Hardie D. G., Dobrowolska G. (2004). Biochemical characterization of the tobacco 42-kD protein kinase activated by osmotic stress. Plant Physiol. 136 3255–3265. 10.1104/pp.104.046151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadri M., Tejera N. A., Lluch C. (2006). Alleviation of salt stress in common bean (Phaseolus vulgaris) by exogenous abscisic acid supply. J. Plant Growth Regul. 25 110–119. 10.1007/s00344-005-0004-3 [DOI] [Google Scholar]
- Khokon M., Okuma E. I. J. I., Hossain M. A., Munemasa S., Uraji M., Nakamura Y., et al. (2011). Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 34 434–443. 10.1111/j.1365-3040.2010.02253.x [DOI] [PubMed] [Google Scholar]
- Khraiwesh B., Arif M. A., Seumel G. I., Ossowski S., Weigel D., Reski R., et al. (2010). Transcriptional control of gene expression by microRNAs. Cell 140 111–122. 10.1016/j.cell.2009.12.023 [DOI] [PubMed] [Google Scholar]
- Kim J.-M., Sasaki T., Ueda M., Sako K., Seki M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114 10.3389/fpls.2015.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kang J.-Y., Cho D.-I., Park J. H., Kim S. Y. (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling, and its overexpression affects multiple stress tolerance. The Plant J. 40 75–87. 10.1111/j.1365-313X.2004.02192.x [DOI] [PubMed] [Google Scholar]
- Kim S., Ma J., Perret P., Li Z., Thomas T. (2002). Arabidopsis ABI5 subfamily members have distinct DNA binding and transcriptional activities. Plant Physiol. 130 688–697. 10.1104/pp.003566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y. (2014). “Transcription factors involved in ABA signaling,” in Abscisic Acid: Metabolism, Transport, and Signaling, ed. Zhang D-P. (Berlin: Springer:), 225–242. [Google Scholar]
- Kim T.-H., Böhmer M., Hu H., Nishimura N., Schroeder J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ Signaling. Annu. Rev. Plant Biol. 61 561–591. 10.1146/annurev-arplant-042809-112226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. J. (2015). Crop epigenetics and the molecular hardware of genotype × environment interactions. Front Plant Sci. 6:968 10.3389/fpls.2015.00968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline K. G., Barrett-Wilt G. A., Sussman M. R. (2010). In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107 15986–15991. 10.1073/pnas.1007879107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., et al. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44 939–949. 10.1111/j.1365-313X.2005.02583.x [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Yamamoto S., Minami H., Kagaya Y., Hattori T. (2004). Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16 1163–1177. 10.1105/tpc.019943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W. (2000). Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 351 289–305. 10.1042/bj3510289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M. (2012). The ubiquitin code. Annu. Rev. Biochem. 81 203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Kong L., Cheng J., Zhu Y., Ding Y., Meng J., Chen Z., et al. (2015). Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 6:8630 10.1038/ncomms9630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J. M., Boisson-Dernier A., Dizon M. B., Maktabi M. H., Schroeder J. I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140 127–139. 10.1104/pp.105.070318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik A., Wawer I., Krzywinska E., Bucholc M., Dobrowolska G. (2011). SnRK2 protein kinases-key regulators of plant response to abiotic stresses. OMICS 15 859–872. 10.1089/omi.2011.0091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T., Miyaji T., Yabuuchi H., Shimizu H., Sugimoto E., Kamiya A., et al. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. U.S.A. 107 2361–2366. 10.1073/pnas.0912516107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T., Sugimoto E., Shinozaki K. (2011). Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 67 885–894. 10.1111/j.1365-313X.2011.04641.x [DOI] [PubMed] [Google Scholar]
- Kuromori T., Yamamoto M. (1994). Cloning of cDNAs from Arabidopsis thaliana that encode putative protein phosphatase 2C and a human Dr1-like protein by transformation of a fission yeast mutant. Nucleic Acids Res. 22 5296–5301. 10.1093/nar/22.24.5296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., et al. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8’-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23 1647–1656. 10.1038/sj.emboj.7600121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J. M., Mori I., Pei Z. M., Leonhardt N., Torres M. A., Dangl J. L., et al. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22 2623–2633. 10.1093/emboj/cdg277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalk I., Dörffling K. (1985). Hardening, abscisic acid, proline and freezing resistance in two winter wheat varieties. Physiol. Plant. 63 287–292. 10.1111/j.1399-3054.1985.tb04267.x [DOI] [Google Scholar]
- Law J. A., Jacobsen S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11 204–220. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I. C., Hong S. W., Whang S. S., Lim P. O., Nam H. G., Koo J. C. (2011). Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 52 651–662. 10.1093/pcp/pcr026 [DOI] [PubMed] [Google Scholar]
- Lee K. H., Piao H. L., Kim H. Y., Choi S. M., Jiang F., Hartung W., et al. (2006). Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126 1109–1120. 10.1016/j.cell.2006.07.034 [DOI] [PubMed] [Google Scholar]
- Lee M.-O., Cho K., Kim S.-H., Jeong S.-H., Kim J.-A., Jung Y.-H., et al. (2008). Novel rice OsSIPK is a multiple stress responsive MAPK family member showing rhythmic expression at mRNA level. Planta 227 981–990. 10.1007/s00425-007-0672-2 [DOI] [PubMed] [Google Scholar]
- Lee S. C., Lan W., Buchanan B. B., Luan S. (2009). A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. U.S.A. 106 21419–21424. 10.1073/pnas.0910601106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Lloyd A. M., Roux S. J. (1999). Antisense expression of the CK2 α-subunit gene in Arabidopsis. Effects on light-regulated gene expression and plant growth. Plant Physiol. 119 989–1000. 10.1104/pp.119.3.989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J., Bouvier-Durand M., Morris P. C., Guerrier D., Chefdor F., Giraudat J. (1994). Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264 1448–1452. 10.1126/science.7910981 [DOI] [PubMed] [Google Scholar]
- Leung J., Merlot S., Giraudat J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771. 10.1105/tpc.9.5.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Assmann S. M. (1996). An abscisic acid-activated and calcium-independent protein kinase from guard cells of fava bean. Plant Cell 8 2359–2368. 10.1105/tpc.8.12.2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Kinoshita T., Pandey S., Ng C. K., Gygi S. P., Shimazaki K., et al. (2002). Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418 793–797. 10.1038/nature00936 [DOI] [PubMed] [Google Scholar]
- Li J., Wang X.-Q., Watson M. B., Assmann S. M. (2000). Regulation of Abscisic acid-induced stomatal closure and anion channels by guard cell AAPK Kinase. Science 287 300–303. 10.1126/science.287.5451.300 [DOI] [PubMed] [Google Scholar]
- Li L.-B., Zhang Y.-R., Liu K.-C., Ni Z.-F., Fang Z.-J., Sun Q.-X., et al. (2010). Identification and bioinformatics analysis of SnRK2 and CIPK family genes in sorghum. Agric. Sci. China 9 19–30. 10.1016/S1671-2927(09)60063-8 [DOI] [Google Scholar]
- Li W. X., Oono Y., Zhu J., He X. J., Wu J. M., Iida K., et al. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20 2238–2251. 10.1105/tpc.108.059444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang L., Yu Y., Quan R., Zhang Z., Zhang H., et al. (2011). The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J. 68 88–89. 10.1111/j.1365-313X.2011.04670.x [DOI] [PubMed] [Google Scholar]
- Liang B. M., Sharp R. E., Baskin T. I. (1997). Regulation of growth anisotropy in well-watered and water-stressed maize roots (I. Spatial distribution of longitudinal, radial, and tangential expansion rates). Plant Physiol. 115 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Wang Y., Zhu Y., Tang J., Hu B., Liu L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U.S.A. 111 10013–10018. 10.1073/pnas.1321568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Mitchell D. M., Harris J. M. (2007). Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Dev. Biol. 304 297–307. 10.1016/j.ydbio.2006.12.037 [DOI] [PubMed] [Google Scholar]
- Lim E.-K., Jackson R. G., Bowles D. J. (2005). Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Lett. 579 2802–2806. 10.1016/j.febslet.2005.04.016 [DOI] [PubMed] [Google Scholar]
- Lim P. O., Kim H. J., Nam H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58 115–136. 10.1146/annurev.arplant.57.032905.105316 [DOI] [PubMed] [Google Scholar]
- Lim P. O., Nam H. G. (2005). The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr. Top. Dev. Biol. 67 49–83. 10.1016/S0070-2153(05)67002-0 [DOI] [PubMed] [Google Scholar]
- Liu H. X., Stone S. L. (2013). Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON Going. J. Biol. Chem. 288 20267–20279. 10.1074/jbc.M113.465369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhang Y. C., Wang C. Y., Luo Y. C., Huang Q. J., Chen S. Y., et al. (2009b). Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett. 583 723–728. 10.1016/j.febslet.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Liu X., Zhang H., Zhao Y., Feng Z., Li Q., Yang H. Q., et al. (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110 15485–15490. 10.1073/pnas.1304651110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. Q., Gao J., Dong A. W., Shen W. H. (2009a). A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3-2 mutant. Mol. Plant 2 688–699. 10.1093/mp/ssp026 [DOI] [PubMed] [Google Scholar]
- Long J. C., Caceres J. F. (2009). The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417 15–27. 10.1042/BJ20081501 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98 4782–4787. 10.1073/pnas.081594298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković Z. J. (2009). Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 14 229–236. 10.1016/j.tplants.2009.01.007 [DOI] [PubMed] [Google Scholar]
- Lu C., Fedoroff N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12 2351–2365. 10.1105/tpc.12.12.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Gao C., Zheng X., Han B. (2009). Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229 605–615. 10.1007/s00425-008-0857-3 [DOI] [PubMed] [Google Scholar]
- Luo M., Liu J. H., Mahapatra S., Hiu R. D., Hill R. D., Mahapatra S. S. (1992). Characterization of a gene family encoding abscisic acid and environmental stress-inducible proteins of alfalfa. J. Biol. Chem. 267 15367–15374. [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324 1064–1068. 10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- Mao X., Zhang H., Tian S., Chang X., Jing R. (2010). TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multi stress tolerance in Arabidopsis. J. Exp. Bot. 61 683–696. 10.1093/jxb/erp331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E., Nussaume L., Quesada A., Gonneau M., Sotta B., Hugueney P., et al. (1996). Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15 2331–2342. [PMC free article] [PubMed] [Google Scholar]
- Meggio F., Pinna L. A. (2003). One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17 349–368. [DOI] [PubMed] [Google Scholar]
- Mehrotra R., Bhalothia P., Bansal P., Basantani M. K., Bharti V., Mehrotra S. (2014). Abscisic acid and abiotic stress tolerance - different tiers of regulation. J. Plant Physiol. 171 486–496. 10.1016/j.jplph.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Melcher K., Zhou X. E., Xu H. E. (2010). Thirsty plants and beyond: structural mechanisms of abscisic acid perception and signaling. Curr. Opin. Struct. Biol. 20 722–729. 10.1016/j.sbi.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M., Dóczi R., Ökrész L., Morandini P., Mizzi L., Soloviev M., et al. (2008). Comprehensive gene expression atlas for the Arabidopsis MAP kinase signaling pathways. New Phytol. 179 643–662. 10.1111/j.1469-8137.2008.02552.x [DOI] [PubMed] [Google Scholar]
- Merilo E., Jalakas P., Kollist H., Brosche M. (2015). The role of ABA recycling and transporter proteins in rapid stomatal responses to reduced air humidity elevated CO2, and exogenous ABA. Mol. Plant 8 657–659. 10.1016/j.molp.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Meyer K., Leube M. P., Grill E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264 1452–1455. 10.1126/science.8197457 [DOI] [PubMed] [Google Scholar]
- Miransari M., Smith D. L. (2014). Plant hormones and seed germination. Environ. Exp. Bot. 99 110–121. 10.1016/j.envexpbot.2013.11.005 [DOI] [Google Scholar]
- Mittler R., Blumwald E. (2015). The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27 64–70. 10.1105/tpc.114.133090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K.-I., Miyakawa T., Sawano Y., Kubota K., Kang H.-J., Asano A., et al. (2009). Structural basis of abscisic acid signaling. Nature 462 609–614. 10.1038/nature08583 [DOI] [PubMed] [Google Scholar]
- Mizoguchi M., Umezawa T., Nakashima K., Kidokoro S., Takasaki H., Fujita Y., et al. (2010). Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 51 842–847. 10.1093/pcp/pcq041 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Irie K., Hirayama T., Hayashida N., Yamaguchi-Shinozaki K., Matsumoto K., et al. (1996). A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 93 765–769. 10.1073/pnas.93.2.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra S. S., Poole R. J., Dhindsa R. S. (1988). Abscisic acid-regulated gene expression in relation to freezing tolerance in alfalfa. Plant Physiol. 87 468–473. 10.1104/pp.87.2.468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks D. E., Aghoram K., Courtney P. D., DeWald D. B., Dewey R. E. (2001). Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell 13 1205–1219. 10.2307/3871374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Romero J., Carme Espunya M., Platara M., Ariño J., Carmen Martínez M. (2008). A role for protein kinase CK2 in plant development: evidence obtained using a dominant-negative mutant. Plant J. 55 118–130. 10.1111/j.1365-313X.2008.03494.x [DOI] [PubMed] [Google Scholar]
- Mulekar J. J., Bu Q., Chen F., Huq E. (2012). Casein kinase II α subunits affect multiple developmental and stress-responsive pathways in Arabidopsis. Plant J. 69 343–354. 10.1111/j.1365-313X.2011.04794.x [DOI] [PubMed] [Google Scholar]
- Mulekar J. J., Huq E. (2014). Expanding roles of protein kinase CK2 in regulating plant growth and development. J. Exp. Bot. 65 2883–2893. 10.1093/jxb/ert401 [DOI] [PubMed] [Google Scholar]
- Munemasa S., Hauser F., Park J., Waadt R., Brandt B., Schroeder J. I. (2015). Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 28 154–162. 10.1016/j.pbi.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A.-C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099. 10.1105/tpc.007906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Soukupová H., Schikora A., Zárskı V., Hirt H. (2006). A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281 38697–38704. 10.1074/jbc.M605293200 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., et al. (2009a). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50 1345–1363. 10.1093/pcp/pcp083 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Ito Y., Yamaguchi-Shinozaki K. (2009b). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149 88–95. 10.1104/pp.108.129791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K., Shinozaki K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170 10.3389/fpls.2014.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56 165–185. 10.1146/annurev.arplant.56.032604.144046 [DOI] [PubMed] [Google Scholar]
- Negi J., Matsuda O., Nagasawa T., Oba Y., Takahashi H., Kawai-Yamada M., et al. (2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452 483–486. 10.1038/nature06720 [DOI] [PubMed] [Google Scholar]
- Nemhauser J. L., Hong F., Chory J. (2006). different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475. 10.1016/j.cell.2006.05.050 [DOI] [PubMed] [Google Scholar]
- Ng L. M., Melcher K., Teh B. T., Xu H. E. (2014). Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacol. Sin. 35 567–584. 10.1038/aps.2014.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niefind K., Putter M., Guerra B., Issinger O. G., Schomburg D. (1999). GTP plus water mimic ATP in the active site of protein kinase CK2. Nat. Struct. Biol. 6 1100–1103. 10.1038/70033 [DOI] [PubMed] [Google Scholar]
- Ning J., Li X., Hicks L. M., Xiong L. (2010). A Raf-Like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 152 876–890. 10.1104/pp.109.149856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Hitomi K., Arvai A. S., Rambo R. P., Hitomi C., Cutler S. R., et al. (2009). Structural mechanism of abscisic acid binding and signaling by Dimeric PYR1. Science 326 1373–1379. 10.1126/science.1181829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Sarkeshik A., Nito K., Park S. Y., Wang A., Carvalho P. C., et al. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61 290–299. 10.1111/j.1365-313X.2009.04054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Yoshida T., Kitahata N., Asami T., Shinozaki K., Hirayama T. (2007). ABA-hypersensitive germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50 935–949. 10.1111/j.1365-313X.2007.03107.x [DOI] [PubMed] [Google Scholar]
- Ohkuma K., Lyon J. L., Addicott F. T., Smith O. E. (1963). Abscisin II, an abscission-accelerating substance from young cotton fruit. Science 142 1592–1593. 10.1126/science.142.3599.1592 [DOI] [PubMed] [Google Scholar]
- Okamoto M., Kushiro T., Jikumaru Y., Abrams S. R., Kamiya Y., Seki M., et al. (2011). ABA 9’-hydroxylation is catalyzed by CYP707A in Arabidopsis. Phytochemistry 72 717–722. 10.1016/j.phytochem.2011.02.004 [DOI] [PubMed] [Google Scholar]
- Okamoto M., Tanaka Y., Abrams S. R., Kamiya Y., Seki M., Nambara E. (2009). High humidity induces abscisic acid 8’-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiol. 149 825–834. 10.1104/pp.108.130823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana S., Yañez M., Espinoza A., Verdugo I., González E., Ruiz-Lara S., et al. (2010). The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 33 2191–2208. 10.1111/j.1365-3040.2010.02220.x [DOI] [PubMed] [Google Scholar]
- Ortega-Martínez O., Pernas M., Carol R. J., Dolan L. (2007). Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317 507–510. 10.1126/science.1143409 [DOI] [PubMed] [Google Scholar]
- Ortiz-Masia D., Perez-Amador M. A., Carbonell J., Marcote M. J. (2007). Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett. 581 1834–1840. 10.1016/j.febslet.2007.03.075 [DOI] [PubMed] [Google Scholar]
- Palusa S. G., Ali G. S., Reddy A. S. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49 1091–1107. 10.1111/j.1365-313X.2006.03020.x [DOI] [PubMed] [Google Scholar]
- Pan J., Zhang M., Kong X., Xing X., Liu Y., Zhou Y., et al. (2012). ZmMPK17, a novel maize group D MAP kinase gene, is involved in multiple stress responses. Planta 235 661–676. 10.1007/s00425-011-1510-0 [DOI] [PubMed] [Google Scholar]
- Park S.-Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., et al. (2009). Abscisic acid inhibits PP2Cs via the PYR/PYL family of ABA-binding START proteins. Science 324 1068–1071. 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-Y., Peterson F. C., Mosquna A., Yao J., Volkman B. F., Cutler S. R. (2015). Agrochemical control of plant water use using engineered abscisic acid receptors. Nature 520 545–548. 10.1038/nature14123 [DOI] [PubMed] [Google Scholar]
- Passioura J. (2007). The drought environment: physical, biological and agricultural perspectives. J. Exp. Bot. 58 113–117. 10.1093/jxb/erl212 [DOI] [PubMed] [Google Scholar]
- Pei Z. M., Ghassemian M., Kwak C. M., McCourt P., Schroeder J. I. (1998). Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282 287–290. 10.1126/science.282.5387.287 [DOI] [PubMed] [Google Scholar]
- Pei Z. M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signaling in guard cells. Nature 406 731–734. 10.1038/35021067 [DOI] [PubMed] [Google Scholar]
- Portolés S., Más P. (2007). Altered oscillator function affects clock resonance and is responsible for the reduced day-length sensitivity of CKB4 overexpressing plants. Plant J. 51 966–977. 10.1111/j.1365-313X.2007.03186.x [DOI] [PubMed] [Google Scholar]
- Puertolas J., Conesa M. R., Ballester C., Dodd I. C. (2015). Local root abscisic acid (ABA) accumulation depends on the spatial distribution of soil moisture in potato: implications for ABA signalling under heterogeneous soil drying. J. Exp. Bot. 66 2325–2334. 10.1093/jxb/eru501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., Shinozaki K., Yamaguchi-Shinozaki K. (2011). Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 52 1569–1582. 10.1093/pcp/pcr106 [DOI] [PubMed] [Google Scholar]
- Qin X., Zeevaart J. A. D. (2002). Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 128 544–551. 10.1104/pp.010663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani M. A., Maruyama K., Abe H., Khan M. A., Katsura K., Ito Y., et al. (2003). Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 133 1755–1767. 10.1104/pp.103.025742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao K. P., Richa T., Kumar K., Raghuram B., Sinha A. K. (2010). In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 17 139–153. 10.1093/dnares/dsq011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S., Mondal W. A., Choudhuri M. A. (1983). Regulation of leaf senescence, grain-filling and yield of rice by kinetin and abscisic acid. Physiol. Plant. 59 343–346. 10.1111/j.1399-3054.1983.tb04212.x [DOI] [Google Scholar]
- Reyna N. S., Yang Y. (2006). Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Plant Microbe. Interact. 19 530–540. 10.1094/MPMI-19-0530 [DOI] [PubMed] [Google Scholar]
- Reyes J. L., Chua N. H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49 592–606. 10.1111/j.1365-313X.2006.02980.x [DOI] [PubMed] [Google Scholar]
- Riechmann J. L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110. 10.1126/science.290.5499.2105 [DOI] [PubMed] [Google Scholar]
- Riera M., Figueras M., López C., Goday A., Pagès M. (2004). Protein kinase CK2 modulates developmental functions of the abscisic acid responsive protein Rab17 from maize. Proc. Natl. Acad. Sci. U.S.A. 101 9879–9884. 10.1073/pnas.0306154101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera M., Redko Y., Leung J. (2006). Arabidopsis RNA-binding protein UBA2a relocalizes into nuclear speckles in response to abscisic acid. FEBS Lett. 580 4160–4165. 10.1016/j.febslet.2006.06.064 [DOI] [PubMed] [Google Scholar]
- Riera M., Vélez-Bermúdez I. C., Legnaioli T., Pagès M. (2013). Specific Features of Plant CK2 Protein Kinase CK2. New York, NY: John Wiley and Sons, Inc, 267–289. [Google Scholar]
- Rodriguez P. L., Benning G., Grill E. (1998). ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 421 185–190. 10.1016/S0014-5793(97)01558-5 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Gacio M. C., Matilla-Vázquez M. A., Matilla A. J. (2009). Seed dormancy and ABA signaling: the breakthrough goes on. Plant Signal. Behav. 4 1035–1048. 10.4161/psb.4.11.9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F., Corke F., Card R., Munz G., Smith C., Bevan M. W. (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 26 421–433. 10.1046/j.1365-313X.2001.2641043.x [DOI] [PubMed] [Google Scholar]
- Saab I. N., Sharp R. E., Pritchard J., Voetberg G. S. (1990). Increased endogenous abscisic acid maintains primary root growth and inhibits shoot growth of maize seedlings at low water potentials. Plant Physiol. 93 1329–1336. 10.1104/pp.93.4.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra X., Modrego A., Rodríguez D., González-García M. P., Sanz L., Nicolás G., et al. (2010). The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol. 152 133–150. 10.1104/pp.109.146381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A., Apostolova N., Gonzalez-Guzman M., Gonzalez-Garcia M. P., Nicolas C., Lorenzo O., et al. (2004). Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 37 354–369. 10.1046/j.1365-313X.2003.01966.x [DOI] [PubMed] [Google Scholar]
- Saito S., Hirai N., Matsumoto C., Ohigashi H., Ohta D., Sakata K., et al. (2004). Arabidopsis CYP707As encode (+)-abscisic acid 8’-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134 1439–1449. 10.1104/pp.103.037614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., et al. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60 575–588. 10.1111/j.1365-313X.2009.03981.x [DOI] [PubMed] [Google Scholar]
- Sarkar A. K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., et al. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446 811–814. 10.1038/nature05703 [DOI] [PubMed] [Google Scholar]
- Saruhashi M., Ghosh T. K., Arai K., Ishizaki Y., Hagiwara K., Komatsu K., et al. (2015). Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. U.S.A. 112 E6388–E6396. 10.1073/pnas.1511238112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Sato Y., Fukao Y., Fujiwara M., Umezawa T., Shinozaki K., et al. (2009). Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 424 439–448. 10.1042/bj20091221 [DOI] [PubMed] [Google Scholar]
- Sato F., Tsuchiya S., Meltzer S. J., Shimizu K. (2011). MicroRNAs and epigenetics. FEBS J. 278 1598–1609. 10.1111/j.1742-4658.2011.08089.x [DOI] [PubMed] [Google Scholar]
- Schroeder J. I., Keller B. U. (1992). Two types of anion channel currents in guard cells with distinct voltage regulation. Proc. Natl. Acad. Sci. U.S.A. 89 5025–5029. 10.1073/pnas.89.15.7287-b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J. I., Kwak J. M., Allen G. J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410 327–330. 10.1038/35066500 [DOI] [PubMed] [Google Scholar]
- Schwartz S. H., Tan B. C., Gage D. A., Zeevaart J. A., McCarty D. R. (1997). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276 1872–1874. 10.1126/science.276.5320.1872 [DOI] [PubMed] [Google Scholar]
- Schweighofer A., Hirt H., Meskiene I. (2004). Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9 236–243. 10.1016/j.tplants.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Seiler C., Harshavardhan V. T., Rajesh K., Reddy P. S., Strickert M., Rolletschek H., et al. (2011). ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 62 2615–2632. 10.1093/jxb/erq446 [DOI] [PubMed] [Google Scholar]
- Seki M., Ishida J., Narusaka M., Fujita M., Nanjo T., Umezawa T., et al. (2002). Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genomics 2 282–291. 10.1007/s10142-002-0070-6 [DOI] [PubMed] [Google Scholar]
- Seo K. I., Lee J. H., Nezames C. D., Zhong S. W., Song E., Byun M. O., et al. (2014). ABD1 is an Arabidopsis DCAF substrate receptor for Cul4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 26 695–711. 10.1105/tpc.113.119974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp R. E., Silk W. K., Hsiao T. C. (1988). Growth of the maize primary root at low water potentials I. Spatial distribution of expansive growth. Plant Physiol. 87 50–57. 10.1104/pp.87.1.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Xie K., Xiong L. (2010). Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol. Genet. Genomics 284 477–488. 10.1007/s00438-010-0581-0 [DOI] [PubMed] [Google Scholar]
- Shen Q., Gomez-Cadenas A., Zhang P., Walker-Simmons M. K., Sheen J., Ho T. H. (2001). Dissection of abscisic acid signal transduction pathways in barley aleurone layers. Plant Mol. Biol. 47 437–448. [DOI] [PubMed] [Google Scholar]
- Shen Q., Zhang P., Ho T. H. (1996). Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient or ABA induction of gene expression in barley. Plant Cell 8 1107–1119. 10.1105/tpc.8.7.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3 217–223. 10.1016/S1369-5266(00)00067-4 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58 221–227. 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- Singh A., Jha S. K., Bagri J., Pandey G. K. (2015). ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS ONE 10:e0125168 10.1371/journal.pone.0125168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. K., LaRosa P. C., Handa A. K., Hasegawa P. M., Bressan R. A. (1987). Hormonal regulation of protein synthesis associated with salt tolerance in plant cells. Proc. Natl. Acad. Sci. U.S.A. 84 739–743. 10.1073/pnas.84.3.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C., Davanture M., Turk B. E., Zivy M., Valot B., Leung J., et al. (2010). The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS ONE 5:e13935 10.1371/journal.pone.0013935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C., Gu D., Hu H., Davanture M., Lee S., Djaoui M., et al. (2009). Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583 2982–2986. 10.1016/j.febslet.2009.08.033 [DOI] [PubMed] [Google Scholar]
- Sivamani E., Bahieldin A., Wraith J. M., Al-Niemi T., Dyer W. E., Ho T. D., et al. (2000). Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Plant Sci. 155 1–9. 10.1016/S0168-9452(99)00247-2 [DOI] [PubMed] [Google Scholar]
- Song C.-P., Agarwal M., Ohta M., Guo Y., Halfter U., Wang P., et al. (2005). Role of an Arabidopsis AP2/EREBP-Type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396. 10.1105/tpc.105.033043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F., Goodman R. (2002). OsBIMK1, a rice MAP kinase gene involved in disease resistance responses. Planta 215 997–1005. 10.1007/s00425-002-0794-5 [DOI] [PubMed] [Google Scholar]
- Spollen W. G., LeNoble M. E., Samuels T. D., Bernstein N., Sharp R. E. (2000). Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 122 967–976. 10.1104/pp.122.3.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N., Harshavardhan V. T., Govind G., Seiler C., Kohli A. (2012). Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506 265–273. 10.1016/j.gene.2012.06.076 [DOI] [PubMed] [Google Scholar]
- Sridha S., Wu K. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46 124–133. 10.1111/j.1365-313X.2006.02678.x [DOI] [PubMed] [Google Scholar]
- Stewart C. R., Voetberg G. (1985). Relationship between stress-induced ABA and proline accumulations and ABA-induced proline accumulation in excised barley leaves. Plant Physiol. 79 24–27. 10.1104/pp.79.1.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M. V., Feng S., Bernatavichute Y. V., Jacobsen S. E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152 352–364. 10.1016/j.cell.2012.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Li C. (2014). “Crosstalk of signaling pathways between ABA and other phytohormones,” in Abscisic Acid: Metabolism, Transport, and Signaling, ed. Zhang D-P. (Berlin: Springer; ), 243–253. [Google Scholar]
- Sunkar R., Zhu J.-K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16 2001–2019. 10.1105/tpc.104.022830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Miller G., Salazar C., Mondal H. A., Shulaev E., Cortes D. F., et al. (2013). Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell 25 3553–3569. 10.1105/tpc.113.114595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy P. M., Smith B. (1999). Role of abscisic acid in plant stress tolerance. Curr. Sci. 76 1220–1227. [Google Scholar]
- Takasaki H., Maruyama K., Takahashi F., Fujita M., Yoshida T., Nakashima K., et al. (2015). SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 84 1114–1123. 10.1111/tpj.13067 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Ebisu Y., Kinoshita T., Doi M., Okuma E., Murata Y.et al. (2013). bHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal. 6:ra48 10.1126/scisignal.2003760 [DOI] [PubMed] [Google Scholar]
- Takezawa D., Watanabe N., Ghosh T. K., Saruhashi M., Suzuki A., Ishiyama K., et al. (2015). Epoxycarotenoid-mediated synthesis of abscisic acid in Physcomitrella patens implicating conserved mechanisms for acclimation to hyperosmosis in embryophytes. New Phytol. 206 209–219. 10.1111/nph.13231 [DOI] [PubMed] [Google Scholar]
- Tan B. C., Schwartz S. H., Zeevaart J. A. D., McCarty D. R. (1997). Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. U.S.A. 94 12235–12240. 10.1073/pnas.94.22.12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Sano T., Tamaoki M., Nakajima N., Kondo N., Hasezawa S. (2005). Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 138 2337–2343. 10.1104/pp.105.063503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., Zhang H., Li X., Xiao J., Xiong L. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158 1755–1768. 10.1104/pp.111.190389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I. B., Burbidge A., Thompson A. J. (2000). Control of abscisic acid synthesis. J. Exp. Bot. 51 1563–1574. 10.1093/jexbot/51.350.1563 [DOI] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Doczi R., Ichimura K., Shinozaki K., et al. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15 141–152. 10.1016/j.molcel.2004.06.023 [DOI] [PubMed] [Google Scholar]
- Thameur A., Ferchichi A., López-Carbonell M. (2011). Quantification of free and conjugated abscisic acid in five genotypes of barley (Hordeum vulgare L.) under water stress conditions. S. Afr. J. Bot. 77 222–228. 10.1016/j.sajb.2010.08.004 [DOI] [Google Scholar]
- Thompson A. J., Jackson A. C., Parker R. A., Morpeth D. R., Burbidge A., Taylor I. B. (2000). Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol. Biol. 42 833–845. 10.1023/A:1006448428401 [DOI] [PubMed] [Google Scholar]
- Tian X., Wang Z., Li X., Lv T., Liu H., Wang L., et al. (2015). Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice 8:28 10.1186/s12284-015-0061-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D., Balzer C., Hill J., Befort B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U.S.A. 108 20260–20264. 10.1073/pnas.1116437108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.-S. P., Urao T., Qin F., Maruyama K., Kakimoto T., Shinozaki K., et al. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl Acad. Sci. U.S.A. 104 20623–20628. 10.1073/pnas.0706547105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N. (2007). Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2 135–138. 10.4161/psb.2.3.4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M. K., Kakar K., Wandrey M., Montanari O., Murray J., Andriankaja A., et al. (2007). Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol. 144 538–549. 10.1104/pp.107.098061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Nakashima K., Miyakawa T., Kuromori T., Tanokura M., Shinozaki K., et al. (2010). Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51 1821–1839. 10.1093/pcp/pcq156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 17588–17593. 10.1073/pnas.0907095106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Takahashi F., Anderson J. C., Ishihama Y., Peck S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6 rs8. 10.1126/scisignal.2003509 [DOI] [PubMed] [Google Scholar]
- United Nations Department of Economic and Social Affairs and Population Division (2015). World Population Prospects: The 2015 Revision, Key Findings and Advance Tables. Working Paper, No. ESA/P/WP.241. New York, NY: United Nations Department of Economic and Social Affairs. [Google Scholar]
- Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 97 11632–11637. 10.1073/pnas.190309197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T., Kollist H., Wang Y.-F., Nishimura N., Chan W.-Y., Valerio G., et al. (2008). SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452 487–491. 10.1038/nature06608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E., Schwacke R., Schneider A., Desimone M., Flugge U. I., Kunze R. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141 776–792. 10.1104/pp.106.079293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela B., Nájar E., Lumbreras V., Leung J., Pagès M. (2015). Casein kinase 2 negatively regulates abscisic acid-activated SnRK2s in the core abscisic acid-signaling module. Mol. Plant 8 709–721. 10.1016/j.molp.2014.12.012 [DOI] [PubMed] [Google Scholar]
- Vlad F., Droillard M. J., Valot B., Khafif M., Rodrigues A., Brault M., et al. (2010). Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 63 778–790. 10.1111/j.1365-313X.2010.04281.x [DOI] [PubMed] [Google Scholar]
- Waadt R., Manalansan B., Rauniyar N., Munemasa S., Booker M. A., Brandt B., et al. (2015). Identification of open stomata1-interacting proteins reveals interactions with sucrose non-fermenting1-related protein Kinases2 and with Type 2A protein phosphatases that function in abscisic acid responses. Plant Physiol. 169 760–779. 10.1104/pp.15.00575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yang A., Yin H., Zhang J. (2008). Influence of water stress on endogenous hormone contents and cell damage of maize seedlings. J. Integr. Plant Biol. 50 427–434. 10.1111/j.1774-7909.2008.00638.x [DOI] [PubMed] [Google Scholar]
- Wang J., Ding H., Zhang A., Ma F., Cao J., Jiang M. (2010). A novel mitogen-activated protein kinase gene in maize (Zea mays), ZmMPK3, is involved in response to diverse environmental cues. J. Integr. Plant Biol. 52 442–452. 10.1111/j.1744-7909.2010.00906.x [DOI] [PubMed] [Google Scholar]
- Wang L., Hua D., He J., Duan Y., Chen Z., Hong X., et al. (2011b). Auxin response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 7:e1002172 10.1371/journal.pgen.1002172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Xue L., Batelli G., Lee S., Hou Y., Oosten M. J. V., et al. (2013a). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U.S.A. 110 11205–11210. 10.1073/pnas.1308974110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.-S., Pandey S., Li S., Gookin T. E., Zhao Z., Albert R., et al. (2011a). Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12:216 10.1186/1471-2164-12-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Altman A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218 1–14. 10.1007/s00425-003-1105-5 [DOI] [PubMed] [Google Scholar]
- Wang W. X., Vinocur B., Shoseyov O., Altman A. (2001). Biotechnology of plant osmotic stress toelrance and physiological and moleucalr considerations. Acta Hort. 560 285–292. 10.17660/ActaHortic.2001.560.54 [DOI] [Google Scholar]
- Wang Y., Beaith M., Chalifoux M., Ying J., Uchacz T., Sarvas C., et al. (2009). Shoot-specific down-regulation of protein farnesyltransferase (α-subunit) for yield protection against drought in canola. Mol. Plant 2 191–200. 10.1093/mp/ssn088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chang H., Hu S., Lu X., Yuan C., Zhang C., et al. (2014). Plastid casein kinase 2 knockout reduces abscisic acid (ABA) sensitivity, thermotolerance, and expression of ABA- and heat-stress-responsive nuclear genes. J. Exp. Bot. 65 4159–4175. 10.1093/jxb/eru190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li L., Ye T., Lu Y., Chen X., Wu Y. (2013b). The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 64 675–684. 10.1093/jxb/ers361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yang L., Chen X., Ye T., Zhong B., Liu R., et al. (2016). Major latex protein-like protein 43 (MLP43) functions as a positive regulator during abscisic acid responses and confers drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 67 421–434. 10.1093/jxb/erv477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani S. H., Kumar V. (2015). Plant stress tolerance: engineering ABA: a Potent phytohormone. Transcriptomics: An Open Access 3:1000113 10.4172/2329-8936.1000113 [DOI] [Google Scholar]
- Wani S. H., Sah S. K. (2014). Biotechnology and abiotic stress tolerance in rice. J. Rice Res. 2:1000e1105 10.4172/jrr.1000e105 [DOI] [Google Scholar]
- Waterland N. L., Campbell C. A., Finer J. J., Jones M. L. (2010). Abscisic acid application enhances drought stress tolerance in bedding plants. Hort Sci. 45 409–413. [Google Scholar]
- Wei L., Wang L., Yang Y., Wang P., Guo T., Kang G. (2015). Abscisic acid enhances tolerance of wheat seedlings to drought and regulates transcript levels of genes encoding ascorbate-glutathione biosynthesis. Front. Plant Sci. 6:458 10.3389/fpls.2015.00458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Zhang D., Xiang F., Zhang Z. (2009). Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int. J. Plant Sci. 170 979–989. 10.1086/605122 [DOI] [Google Scholar]
- Wen J.-Q., Oono K., Imai R. (2002). Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol. 129 1880–1891. 10.1104/pp.006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Köllmer I., Bartrina I., Holst K., Schmülling T. (2006). New insights into the biology of cytokinin degradation. Plant Biol. 8 371–381. 10.1055/s-2006-923928 [DOI] [PubMed] [Google Scholar]
- Wilkinson S., Davies W. J. (2010). Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ. 33 510–525. 10.1111/j.1365-3040.2009.02052.x [DOI] [PubMed] [Google Scholar]
- Wright S. T. C. (1969). An increase in the “inhibitor-β” content of detached wheat leaves following a period of wilting. Planta 86 10–20. 10.1007/BF00385299 [DOI] [PubMed] [Google Scholar]
- Xing Y., Jia W., Zhang J. (2008). AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54 440–451. 10.1111/j.1365-313X.2008.03433.x [DOI] [PubMed] [Google Scholar]
- Xiong L., Zhu J. K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133 29–36. 10.1104/pp.103.025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Gong Z., Rock C. D., Subramanian S., Guo Y., Xu W., et al. (2001). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1 771–781. 10.1016/S1534-5807(01)00087-9 [DOI] [PubMed] [Google Scholar]
- Xiong L., Yang Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid–inducible mitogen-activated protein kinase. Plant Cell 15 745–759. 10.1105/tpc.008714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Jia L., Shi W., Liang J., Zhou F., Li Q., et al. (2013). Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytol. 197 139–150. 10.1111/nph.12004 [DOI] [PubMed] [Google Scholar]
- Xu Z. Y., Lee K. H., Dong T., Jeong J. C., Jin J. B., Kanno Y., et al. (2012). A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24 2184–2199. 10.1105/tpc.112.095935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue-Xuan X., Hong-Bo S., Yuan-Yuan M., Gang X., Jun-Na S., Dong-Gang G., et al. (2010). Biotechnological implications from abscisic acid (ABA) roles in cold stress and leaf senescence as an important signal for improving plant sustainable survival under abiotic-stressed conditions. Crit. Rev. Biotechnol. 30 222–230. 10.3109/07388551.2010.487186 [DOI] [PubMed] [Google Scholar]
- Yadegari L. Z., Heidari R., Rahmani F., Khara J. (2014). Drought tolerance induced by foliar application of abscisic acid and sulfonamide compounds in tomato. J. Stress Physiol. Biochem. 10 327–334. [Google Scholar]
- Yaish M. W., Colasanti J., Rothstein S. J. (2011). The role of epigenetic processes in controlling flowering time in plants exposed to stress. J. Exp. Bot. 62 3727–3735. 10.1093/jxb/err177 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803. 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- Yang J., Worley E., Udvardi M. (2014). A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26 4862–4874. 10.1105/tpc.114.133769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhang J., Wang Z., Zhu Q., Liu L. (2002). Abscisic acid and cytokinins in the root exudates and leaves and their relationship to senescence and remobilization of carbon reserves in rice subjected to water stress during grain filling. Planta 215 645–652. 10.1007/s00425-002-0789-2 [DOI] [PubMed] [Google Scholar]
- Yang J. C., Zhang J. H., Wang Z. Q., Zhu Q. S., Liu L. J. (2003). Involvement of abscisic acid and cytokinins in the senescence and remobilization of carbon reserves in wheat subjected to water stress during grain filling. Plant Cell Environ. 26 1621–1631. 10.1046/j.1365-3040.2003.01081.x [DOI] [PubMed] [Google Scholar]
- Yang X., Yang Y. N., Xue L. J., Zou M. J., Liu J. Y., Chen F., et al. (2011). Rice ABI5-like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol. 156 1397–1409. 10.1104/pp.111.173427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Costa A., Leonhardt N., Siegel R. S., Schroeder J. I. (2008). Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4 1 10.1186/1746-4811-4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., Fan H., Hao Q., Yuan X., Wu D., Pang Y., et al. (2009). Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 16 1230–1236. 10.1038/nsmb.1730 [DOI] [PubMed] [Google Scholar]
- Yoshida R., Hobo T., Ichimura K., Mizoguchi T., Takahashi F., Aronso J., et al. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43 1473–1483. 10.1093/pcp/pcf188 [DOI] [PubMed] [Google Scholar]
- Yoshida R., Umezawa T., Mizoguchi T., Takahashi S., Takahashi F., Shinozaki K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 Interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281 5310–5318. 10.1074/jbc.M509820200 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Maruyama K., Mogami J., Todaka D., Shinozaki K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38 35–49. 10.1111/pce.12351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61 672–685. 10.1111/j.1365-313X.2009.04092.x [DOI] [PubMed] [Google Scholar]
- Yoshida T., Nishimura N., Kitahata N., Kuromori T., Ito T., Asami T., et al. (2006). ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 140 115–126. 10.1104/pp.105.070128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M. K., Oh S. I., Ok S. H., Cho S. K., Shin H. Y., Jeung J. U., et al. (2007). Identification of putative MAPK kinases in Oryza minuta and O. sativa responsive to biotic stresses. Mol. Cells 23 108–114. [PubMed] [Google Scholar]
- Yu F., Wu Y., Xie Q. (2016). Ubiquitin-proteasome system in ABA signaling: from perception to action. Mol. Plant 9 21–33. 10.1016/j.molp.2015.09.015 [DOI] [PubMed] [Google Scholar]
- Zeevaart J. A. D., Creelman R. A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39 439–473. 10.1146/annurev.pp.39.060188.002255 [DOI] [Google Scholar]
- Zemach A., McDaniel I. E., Silva P., Zilberman D. (2010). Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328 916–919. 10.1126/science.1186366 [DOI] [PubMed] [Google Scholar]
- Zhang H., Han W., De Smet I., Talboys P., Loya R., Hassan A., et al. (2010a). ABA promotes quiescence of the quiescent center and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 64 764–774. 10.1111/j.1365-313X.2010.04367.x [DOI] [PubMed] [Google Scholar]
- Zhang H., Mao X., Wang C., Jing R. (2010b). Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS ONE 5:e16041 10.1371/journal.pone.0016041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhu H., Pan Y., Yu Y., Luan S., Li L. (2014). A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 7 1522–1532. 10.1093/mp/ssu063 [DOI] [PubMed] [Google Scholar]
- Zhang H. W., Cui F., Wu Y. R., Lou L. J., Liu L. J., Tian M. M., et al. (2015). The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-Interacting Protein1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27 214–227. 10.1105/tpc.114.134163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Gan S.-S. (2012). An Abscisic Acid-AtNAP Transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol. 158 961–969. 10.1104/pp.111.190876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Pan J., Kong X., Zhou Y., Liu Y., Sun L., et al. (2012). ZmMKK3, a novel maize group B mitogen-activated protein kinase kinase gene, mediates osmotic stress and ABA signal responses. J. Plant Physiol. 169 1501–1510. 10.1016/j.jplph.2012.06.008 [DOI] [PubMed] [Google Scholar]
- Zhang W., Ruan J., Ho T.-H. D., You Y., Yu T., Quatrano R. S. (2005). Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics 21 3074–3081. 10.1093/bioinformatics/bti490 [DOI] [PubMed] [Google Scholar]
- Zhang X., Wollenweber B., Jiang D., Liu F., Zhao J. (2008b). Water deficits and heat shock effects on photosynthesis of a transgenic Arabidopsis thaliana constitutively expressing ABP9, a bZIP transcription factor. J. Exp. Bot. 59 839–848. 10.1093/jxb/erm364 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Tan J., Guo Z., Lu S., He S., Shu W., et al. (2009). Increased abscisic acid levels in transgenic tobacco over-expressing 9 cis-epoxycarotenoid dioxygenase influence H2O2 and NO production and antioxidant defences. Plant Cell Environ. 32 509–519. 10.1111/j.1365-3040.2009.01945.x [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yang J., Lu S., Cai J., Guo Z. (2008a). Overexpressing SgNCED1 in Tobacco increases ABA level, antioxidant enzyme activities, and stress tolerance. J. Plant Growth Regul. 27 151–158. 10.1007/s00344-008-9041-z [DOI] [Google Scholar]
- Zhao Z., Chen G., Zhang C. (2001). Interaction between reactive oxygen species and nitric oxide in drought-induced abscisic acid synthesis in root tips of wheat seedlings. Aust. J. Plant Physiol. 28 1055–1061. [Google Scholar]
- Zhou Y., Huang W., Liu L., Chen T., Zhou F., Lin Y. (2013). Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 13:132 10.1186/1471-2229-13-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Jeong J. C., Zhu Y., Sokolchik I., Miyazaki S., Zhu J. K., et al. (2008). Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. U.S.A. 105 4945–4950. 10.1073/pnas.0801029105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong X. J., Li D. P., Gu L. K., Li D. Q., Liu L. X., Hu X. L. (2009). Abscisic acid and hydrogen peroxide induce a novel maize group C MAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta 229 485–495. 10.1007/s00425-008-0848-4 [DOI] [PubMed] [Google Scholar]
- Zou J. J., Li X. D., Ratnasekera D., Wang C., Liu W. X., Song L. F., et al. (2015). Arabidopsis calcium-dependent protein kinase8 and catalase3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27 1445–1460. 10.1105/tpc.15.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., Guan Y., Ren H., Zhang F., Chen F. (2008). A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 66 675–683. 10.1007/s11103-008-9298-4 [DOI] [PubMed] [Google Scholar]