Summary
The molecular pathogenesis of many Staphylococcus aureus infections involves growth of bacteria as biofilm. In addition to polysaccharide intercellular adhesin (PIA) and extracellular DNA, surface proteins appear to mediate the transition of bacteria from planktonic growth to sessile lifestyle as well as biofilm growth, and can enable these processes even in the absence of PIA expression. However, the molecular mechanisms by which surface proteins contribute to biofilm formation are incompletely understood. Here we demonstrate that self-association of the serine-aspartate repeat protein SdrC promotes both bacterial adherence to surfaces and biofilm formation. However, this homophilic interaction is not required for the attachment of bacteria to abiotic surfaces. We identified the subdomain that mediates SdrC dimerization and subsequent cell-cell interactions. In addition, we determined that two adjacently located amino acid sequences within this subdomain are required for the SdrC homophilic interaction. Comparative amino acid sequence analysis indicated that these binding sites are conserved. In summary, our study identifies SdrC as a novel molecular determinant in staphylococcal biofilm formation and describes the mechanism responsible for intercellular interactions. Furthermore, these findings contribute to a growing body of evidence suggesting that homophilic interactions between surface proteins present on neighboring bacteria induce biofilm growth.
Introduction
Bacterial biofilms are communities of microorganisms growing attached to biotic or abiotic surfaces. Within the biofilm, bacteria encase themselves in a self-secreted matrix with precise micro-architectural properties that allow free circulation of nutrients, water and metabolites (Costerton et al., 1978). Besides acting as a scaffold, the matrix enables microbes to survive and persist as reservoirs, avoid desiccation or antimicrobials, and subvert host defenses. In clinical settings, biofilms allow pathogens to establish chronic and/or infections that are refractory to treatment (Costerton et al., 1999). Thus, it has long been recognized that their effective therapy management is challenging.
Widely known as an opportunistic pathogen, Staphylococcus aureus is a frequent cause of biofilm-related infections (Otto, 2008). The transition of staphylococci from planktonic organisms to sessile growth is triggered by environmental cues or receptor availability (Donlan, 2002). Overall bacterial surface hydrophobicity determines the degree of attachment to inert materials (Pasqual et al., 1986). In the case of S. aureus, colonization of abiotic components is attributed to teichoic acids and autolysin Atl (Gross et al., 2001; Houston et al., 2011). Attachment to host tissues and synthetic surfaces coated with plasma proteins is facilitated by members of the MSCRAMM sub-family of cell-wall-anchored (CWA) proteins (Foster et al., 2014; Ponnuraj et al., 2003). For example, fibrinogen-binding proteins, such as clumping factor A ClfA, ClfB, fibronectin (Fn)-binding protein FnBPA, FnBPB (McDevitt et al., 1994; Ni Eidhin et al., 1998, Schwartz-Linek et al., 2003; Keane et al., 2007), and collagen-binding protein (Cna) (Patti et al., 1993) initiate adherence, and as a result, favor biofilm formation. Upon attachment, bacteria proliferate and synthesize a scaffolding matrix composed of intercellular polysaccharide adhesin (PIA) (Cramton et al., 1999), CWA proteins (Cucarella et al., 2001; Abraham and Jefferson, 2012; O’Neill et al., 2008; Schroeder et al., 2009; Corrigan et al., 2007; Merino et al., 2009), and extracellular DNA (eDNA) (Izano et al., 2008). Accumulating evidence suggest that CWA proteins important for biofilm growth include biofilm associated protein (Bap) (Cucarella et al., 2001), ClfB (Abraham and Jefferson, 2012), FnBPA, FnBPB (O’Neill et al., 2008), Staphylococcus aureus surface protein SasC (Schroeder et al., 2009), SasG (Corrigan et al., 2007), and surface protein A (SpA) (Merino et al., 2009). However, Bap has been found only in bovine mastitis isolates (Cucarella et al., 2001). While ClfB, FnBPA, FnBPB, SpA and SasC are found in the majority of strains, SasG is present only in a subset of isolates (Schroeder et al., 2009; Corrigan et al., 2007). The molecular mechanism by which these proteins contribute to intercellular interaction is largely unknown, but recent studies suggest the involvement of metal-ion dependent interactions between CWA proteins present on adjacent cells (Conrady et al., 2008; Geoghegan et al., 2010; Abraham and Jefferson, 2012; Geoghegan et al., 2013; Conrady et al., 2013).
The initiation and development of biofilm appears to be chiefly regulated by the quorum sensing accessory gene regulator (Agr) system in response to cell density. Typically, at low density staphylococci express surface proteins involved in cell attachment and biofilm accumulation (Periasamy et al., 2012). When the population reaches high density, Agr activation triggers the upregulation of toxin, proteases and surfactants involved in biofilm dispersal and bacterial dissemination. Conversely, inactivation of Agr leads to an augmented biofilm phenotype, in part due to the deregulation of surface proteins involved in adherence and biofilm growth. In agreement, recent studies have indicated that these mutants form a thicker and more resistant biofilms presumably due to perpetual expression of CWA proteins that engage newborn cells within the structure. Notably, agr spontaneous mutants are frequently isolated from biofilms (Shopsin et al., 2010).
Despite their lack of sequence similarity, MSCRAMMs share common structural organization. An amino terminal signal sequence is followed by the ligand-binding A region subdivided into three subdomains (N1, N2, and N3), where N2 and N3 each adopt an IgG-like fold. In a subset of MSCRAMMs, the A-region is followed by a B-region containing repeated β-sandwich modules of unknown function (Fig 1A). In the case of the Sdr subfamily (serine-aspartate repeats) of MSCRAMMs, the B-region is accompanied by a repeat (R) domain composed of multiple Ser-Asp dipeptide repeats. The carboxyl terminal section of the proteins contains the LPXTG motif required for cell wall anchoring (reviewed in Foster et al., 2014) (Fig. 1A). The ligand-binding activity of MSCRAMMs to host molecules has been shown to proceed via a “dock, lock and latch” mechanism in which the ligand peptide “docks” into the groove formed between the N2 and N3 subdomains. The ligand is “locked” in place by interacting with amino acids residues in the C-terminal extension of the N3 domain. The complex is stabilized by the insertion of the subsequent “latch” region into the N2 domain through a β-strand complementation (Ponnuraj et al., 2003).
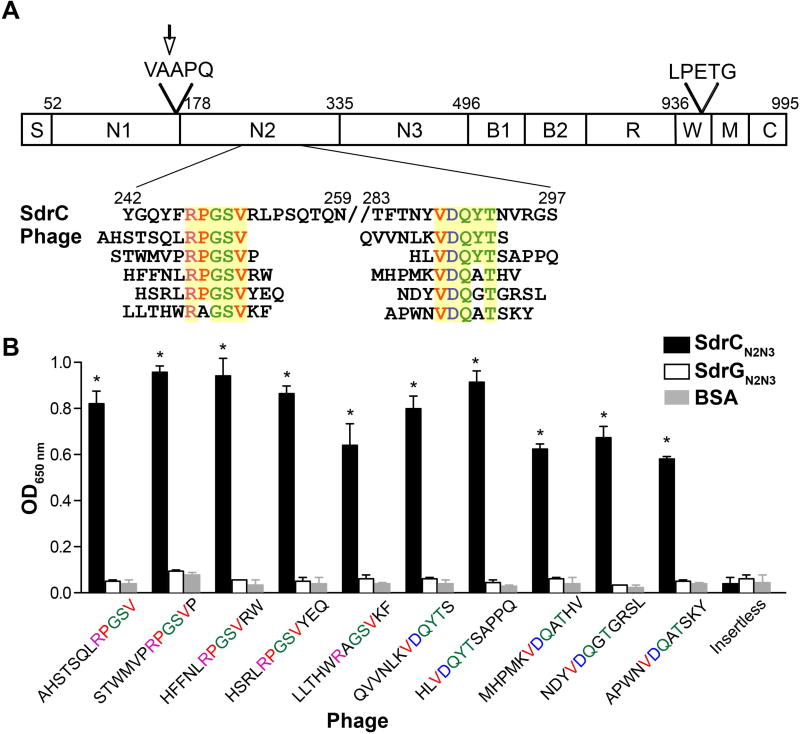
Fig. 1. Specificity of enriched peptide-displaying phage.
A. Schematic overall representation of SdrC domains. S, signal sequence; A region, composed of N1, N2 and N3; B repeats, B1 and B2; R, serine-aspartic acid repeat region; W, wall-spanning fragment; M, transmembrane domain; C, cytoplasmic tail; LPETG, cell wall anchoring motif; VAAPQ, cleavage site; the arrow indicates the enzymatic cleavage position. Also shown, are the enriched phage-displayed peptides aligned with the relevant SdrC sequence. Consensus sequences are highlighted in yellow.
B. Purified phage clones binding to immobilized SdrCN2N3, SdrGN2N3 and BSA. Binding was detected with an anti-M13-HRP antibody. The data shown is representative of three individual experiments performed in triplicate. Bars represent mean ± standard error of the mean (SEM); *p<0.001.
Using phage display, we previously identified peptides targeting the putative ligand-binding region of SdrC. Further analysis of one peptide motif identified β-neurexin as a host ligand for SdrC (Barbu et al., 2010). In this study, we report that two other consensus sequences identified by our phage display screen are responsible for SdrC self-association and that this interaction is inhibited by Mn2+. As a result, SdrC promotes bacterial intercellular interactions and contributes to biofilm formation. Exploiting heterologous expression in the non-pathogenic bacterium Lactococcus lactis, we show that SdrC also triggers bacterial adhesion to abiotic surfaces. However, bacterial attachment to uncoated inert surfaces is independent of self-association. Moreover, our results indicate that the SdrC contribution to biofilm formation is dependent on the strain background.
Results
The N2 subdomain mediates SdrC self-association
We demonstrated previously the feasibility of screening phage display libraries to identify binding partners for MSCRAMMs. We identified peptides binding to the orphan MSCRAMM SdrC after three consecutive rounds of selection with a 12 amino acids random phage library. Although several consensus sequences were identified, only one motif matched a protein found in the human protein sequence database (β-neurexin) (Barbu et al., 2010). In an effort to identify ligands corresponding to the other consensus amino acids (RPGSV and VDQXT) (Fig. 1A), we repeated the similarity search using a staphylococcal protein sequence database. We found that these sequences are located adjacent to each other (positions 247–251 and 288–292) in the C-terminal region of the N2 subdomain of SdrC itself (Fig. 1A). Selected purified phage clones specifically bound recombinant SdrCN2N3 (r SdrCN2N3), whereas they did not bind to rSdrGN2N3, a similar MSCRAMM from S. epidermidis (Fig. 1B).
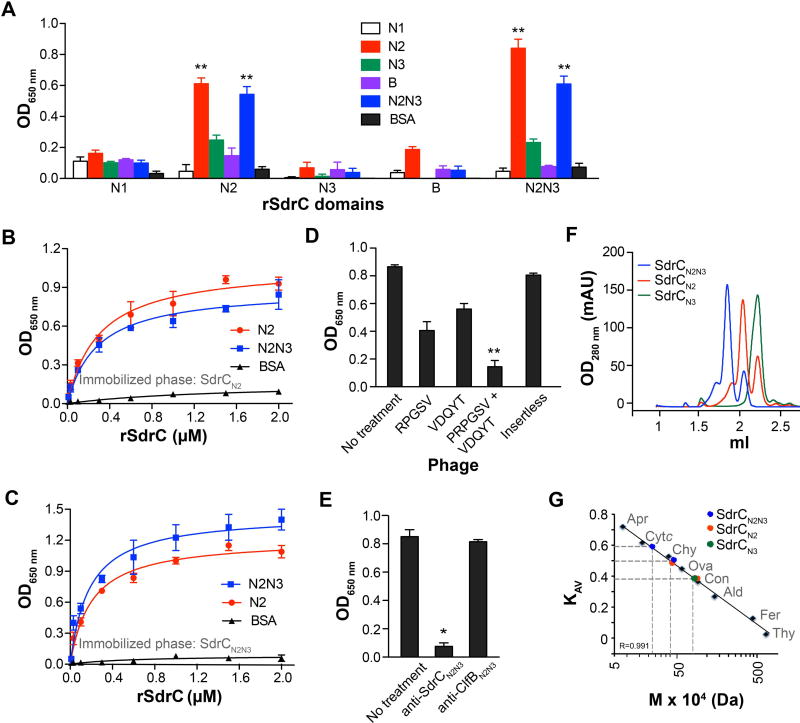
Considering that the peptide sequences identified by phage display are found in the bait protein, we reasoned that SdrC interacts with itself. We compared the binding of SdrC subdomains to each other using a solid-phase binding assay. Recombinant polypeptides containing the N2 subdomain showed significant levels of binding to N2 subdomain-containing proteins relative to N1 and B segments (Fig. 2A) (p<0.01). To establish binding specificity, either N2 or N2N3 recombinant proteins were immobilized on microtiter plates and probed with increasing concentrations of biotin-labeled subdomains. We found that N2-containing proteins bound in a dose-dependent and saturable manner to both proteins. The apparent dissociation constant (corresponding to the concentration needed for half maximum binding) ranged between 0.2 and 0.3 µM (Fig. 2 B and C). To further assess the binding specificity, we tested the ability of consensus peptides to inhibit N2-mediated SdrC self-association. Purified phage clones displaying each of the self-binding motifs inhibited the binding of biotin-labeled rSdrCN2 to immobilized rSdrCN2N3 by nearly 50% (Fig. 2D). Of importance, the binding was completely inhibited only when both phage clones displaying consensus sequences were combined, suggesting that the identified binding sites act cooperatively (Fig. 2D). An insertless phage had no effect on the N2 subdomain interaction with its partner. Similarly, antibodies against the N2N3 domain of SdrC inhibited the N2-containing domains binding to each other (Fig. 2E). In contrast, antibodies against ClfBN2N3, a related staphylococcal MSCRAMM, had no effect on rSdrCN2 binding to rSdrCNN2N3 (Fig. 2E).
Fig. 2. The N2 subdomain mediates SdrC dimerization.
A. Biotin-labeled recombinant SdrC subdomains (1µM) binding to unlabeled segments. The interaction was detected with avidin-HRP.
B and C. Dose-dependent and saturable binding of N2 (red) and N2N3 (blue) subdomains to one another. Increasing concentrations of biotin-labeled subdomains were incubated with either immobilized (B) unlabeled N2 or (C) N2N3. The apparent dissociation constant (KD) calculated using the equation ΔP = ΔPmax [protein]/ (KD + [protein]) ranged between 0.2 – 0.3 µM.
D and E. Inhibition of N2N3 subdomain dimerization by phage displaying consensus peptide sequences (D) or anti-SdrCN2N3 antibodies (E). Immobilized recombinant SdrCN2N3 protein was first allowed to interact with phage or antibodies and then incubated with biotin-labeled recombinant SdrCN2 protein. Insertless phage and anti-ClfB antibodies were used as negative controls. Binding was detected with an anti-M13-HRP antibody or avidin-HRP. The data shown is representative of three individual experiments performed in triplicate. Bars represent mean ± SEM; *p<0.001, **p<0.01.
F. N2 subdomain-mediated dimer formation demonstrated by gel permeation chromatography. Pure recombinant SdrC subdomains were separated on a Sephadex 200 5/150 GL column at a flow rate of 0.3 ml min−1.
G. Interpolated relative molecular masses of SdrC species from the linear regression based on column calibration with thyroglobulin (Thy, 659 KDa), ferritin (Fer, 440 KDa), aldolase (Ald, 158 KDa), conalbumin (Con, 75 KDa), ovalbumin (Ova, 44 KDa), chymotrypsin (Chy, 29 KDa), cytochrome c (Cytc, 12.4 KDa), aprotinin (Fer, 6.5 KDa). Gel phase distribution coefficients (KAV) were calculated from the respective elution volumes (Ve) and represented as a function of molecular mass
To determine the effects of N2 subdomain association, we compared the gel-permeation profiles of rSdrCN2 (calculated theoretical mass 19 KDa), rSdrCN3 (19,1 KDa), and rSdrCN2N3 (36.8 KDa) subdomains. Both rSdrCN2 and rSdrCN2N3 eluted as two major distinct peaks (Fig. 2F) where the peaks corresponding to the larger isoforms contained most of the protein. The relative molecular masses calculated from the linear regression based on column calibration with globular proteins revealed a 40 KDa dimeric species and an 18 KDa form indicative of a monomeric state for rSdrCN2 (Fig. 2 F and G). Similarly, rSdrCN2N3 eluted as a 76 KDa dimer and a 36 KDa monomeric form (Fig. 2 F and G). In contrast, rSdrCN3 eluted from the column as a single peak with a relative molecular mass of 19 KDa, which is similar to the calculated theoretical mass of the monomer (Fig. 2 F and G). These results indicate that the N2 subdomain mediates SdrC self-association.
SdrC contributes to staphylococcal biofilm formation
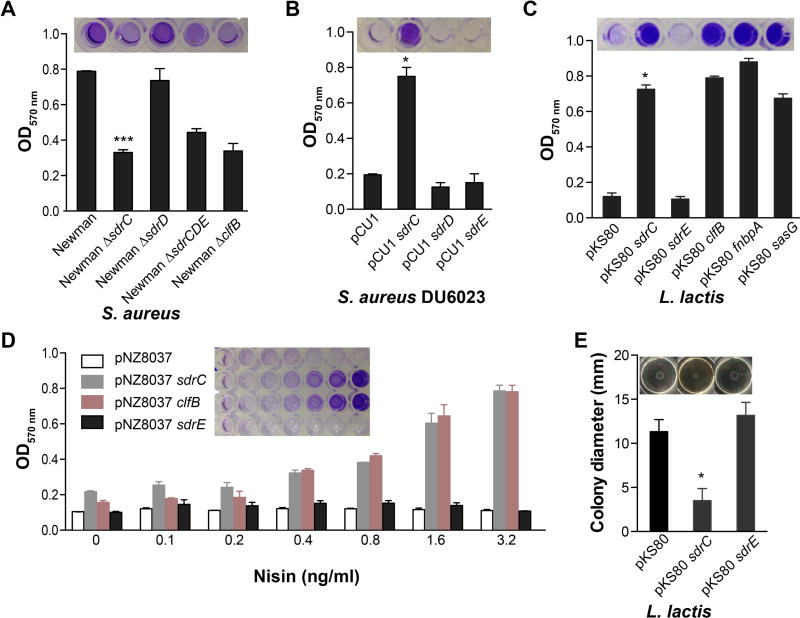
An earlier study investigating biofilm formation by S. aureus SA113, an Agr-defective isolate, revealed that sdrC is highly expressed in mature biofilm (Resch et al., 2005). Since surface molecule self-association may lead to intercellular interactions, we hypothesized that SdrC may be involved in biofilm formation. We compared the biofilm growth of the parent strain S. aureus Newman (a strain which does not express SasG, and does not anchor FnBPA and FnBPB on the cell-wall), and clfB, sdrC, sdrD, sdrCDE mutants. A mutation in sdrC significantly inhibited biofilm formation, whereas an sdrD single mutant had no detectable effect (Fig. 3A). Biofilm formation by Newman clfB (Abraham and Jefferson, 2012) or ΔsdrCDE was reduced compared to the wild-type strain (Fig. 3A). Complete elimination of biofilm was not detected because these strains likely express additional factors involved in biofilm accumulation. To clarify this, we assessed the biofilm formed by S. aureus Newman mutant (DU6023) (Corrigan et al., 2009), a strain defective in the CWA proteins ClfA, ClfB, IsdA, IsdB, SdrC, SdrD, and SdrE, complemented with SdrC on a multi-copy plasmid. We found that SdrC expression restored biofilm formation (Fig. 3B). To determine whether additional staphylococcal factors are necessary for SdrC activity, we exploited the apathogenic bacterium L. lactis. Biofilm accumulation by lactococci constitutively expressing CWA proteins known to promote this type of growth was significant relative to the parent strain carrying the empty vector pKS80. SdrC conferred biofilm-forming activity similar to that of ClfB, FnBPA and SasG, whereas SdrE expression did not contribute to biofilm growth (Fig. 3C). Furthermore, a strong correlation between inducible surface protein expression and biofilm formation was observed for both SdrC and ClfB (Fig. 3D).
Fig. 3. SdrC contributes to staphylococcal biofilm formation.
A and B. Biofilm formation on plastic plates in TSB 1% glucose medium (TBSG) by (A) S. aureus Newman, Newman ΔsdrC, Newman ΔsdrD, Newman ΔsdrCDE, and Newman ΔclfB or (B) S. aureus Newman DU6023 pCU1, DU6023 pCU1 sdrC, DU6023 pCU1 sdrD, and DU6023 pCU1 sdrE. Bacteria were added to microtiter wells at OD600 nm = 0.01.
C and D. Biofilm formation in M17 medium containing 0.5% glucose (GM17) by (C) L. lactis pKS80 or L. lactis pKS80 constitutively expressing SdrC, SdrE, ClfB, FnBPA or SasG or (D) L. lactis pNZ8037, L. lactis pNZ8037 sdrC, and L. lactis pNZ8037 clfB. Bacteria were added to microtiter wells at OD600 nm = 0.01. Heterologous protein expression was induced with increasing concentrations of nisin. Static biofilm formation was measured by staining with 0.5% crystal violet (CV). The data shown is representative of three individual experiments performed in triplicate. Bars represent mean ± SEM; *p<0.001, ***p<0.05. Representative images of wells containing the CV extracted from previously stained biofilm are shown above the graphs.
E. L. lactis pKS80, pKS80 sdrC, pKS80 sdrE colony spreading motility overnight incubation at 30°C. Bacteria from overnight cultures (2µl) were spotted on soft agar GM17 plates. The data shown is representative of six individual experiments. Bars represent mean ± SEM; *p<0.001. Representative images of colony swarming motility are shown above the graphs.
Previous work showed that MSCRAMMs, such as FnBPA and ClfB, involved in cell-cell aggregation and biofilm formation antagonize colony spreading motility on soft agar consistent with the hypothesis that these proteins promote intercellular adherence (Tsompanidou et al., 2012). Similarly, SdrC also reduced the efficiency of spreading of lactococci on wet surfaces, whereas SdrE had no effect (Fig. 3E). These data demonstrate that SdrC contributes to staphylococcal intercellular interactions and subsequent biofilm formation in vitro.
Inhibition of SdrC self-association inhibits biofilm accumulation
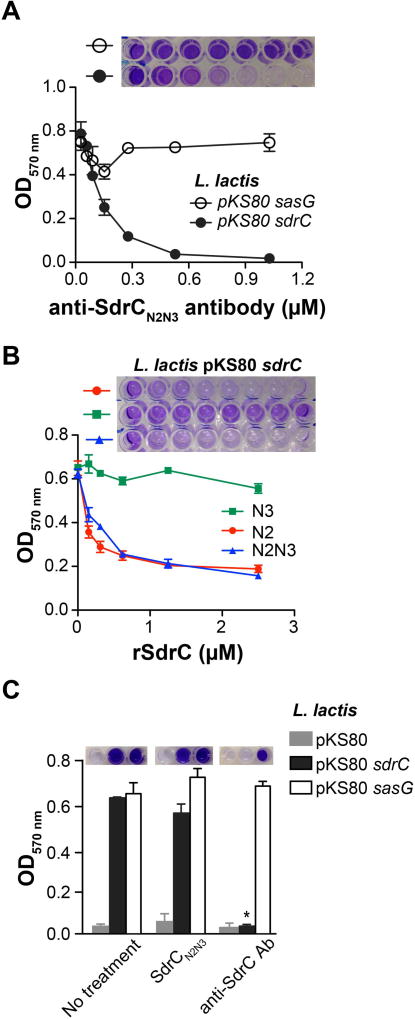
To further investigate the mechanism of SdrC self-association, we determined the effect of anti-SdrCN2N3 antibodies on biofilm formation. SasG-driven lactococcal biofilm was not affected by the antiserum, whereas biofilm formation by L. lactis pKS80 sdrC was completely eliminated (Fig. 4A). Next, we compared the abilities of different recombinant protein segments to inhibit biofilm formation. The rSdrCN3 protein had no effect on L. lactis pKS80 sdrC biofilm (Fig. 4B). Both rSdrCN2 and rSdrCN2N3 inhibited biofilm accumulation in a dose-dependent manner (Fig. 4B). Despite repeated attempts, we were unable to determine a concentration at which biofilm was completely eradicated by the addition of recombinant SdrC fragments. We hypothesized that the residual biofilm is due to an SdrC-dependent bacterial adherence to plastic. Thus, we assessed the ability of L. lactis pKS80 and L. lactis pKS80 sdrC resuspended in PBS to adhere to plastic for one hour in the presence of either rSdrCN2N3 or anti-SdrCN2N3 antibodies. We detected complete inhibition of bacterial attachment to plastic in the presence of antibodies, whereas rSdrCN2N3 had no significant effect (Fig. 4C). These results suggest that SdrC promotes both bacterial adherence to plastic and biofilm growth. However, SdrC self-association is important only for biofilm accumulation.
Fig. 4. Inhibition of SdrC dimerization disrupts biofilm formation.
A. L. lactis pKS80 sdrC biofilm inhibition by anti-SdrCN2N3 serum (range, 0 – 1 µM). L. lactis pKS80 sasG was used as negative control.
B. L. lactis pKS80 sdrC biofilm inhibition by either recombinant SdrCN2, SdrCN3 SdrCN2N3 proteins (range, 0 – 2.4 µM). Increasing concentrations of proteins were added to the plates at the same time as bacteria and incubated for 24 hours a 30°C.
C. Inhibition of L. lactis pKS80 sdrC initial adherence to 96-well microtiter plates by anti-SdrC antibodies. Bacteria were grown overnight, washed in PBS, resuspended at OD600 nm = 1 and added to plastic plates in the presence of either recombinant SdrCN2N3 (3 µM) or antibodies (1 µM). After one hour, unbound bacteria were removed by washing with PBS. L. lactis pKS80 and L. lactis pKS80 sasG was used as negative control. Bound bacteria were stained with 0.5% CV. The data shown is representative of three individual experiments performed in triplicate. Bars represent mean ± SEM; *p<0.001. Representative images of wells containing the CV extracted from previously stained biofilm are shown above the graphs.
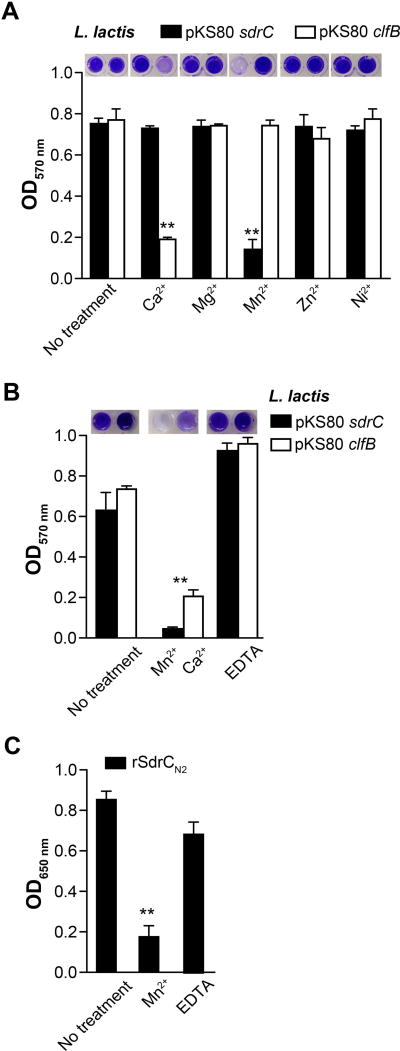
Mn2+ inhibits SdrC-mediated biofilm formation
Metal ions often influence CWA-mediated staphylococcal biofilm formation. While Zn2+ is required for FnBPA and SasG promoted biofilm (Geoghegan et al., 2010; Geoghegan et al., 2013; Conrady et al., 2008; Conrady et al., 2013) ClfB-dependent biofilm formation is inhibited by Ca2+ (Abraham and Jefferson, 2012). To determine whether SdrC self-association is metal ion dependent, we conducted an initial screen where biofilm accumulation was assessed in the presence of selected metal ions (1 mM). In this assay, Mn2+ abolished biofilm formation by L. lactis pKS80 sdrC, whereas other metal ions tested had no effect (Fig. 5A). As expected, Ca2+ eliminated lactococcal ClfB-promoted biofilm (Abraham and Jefferson, 2012). Chelation with EDTA (1 mM) restored Mn2+ inhibited SdrC-driven biofilm formation (Fig. 5B). Furthermore, Mn2+ (1 µM) inhibited the binding of biotin-labeled rSdrCN2 to immobilized rSdrCN2N3 (Fig. 5C). These results indicate that the self-association of SdrC and its contribution to biofilm is inhibited by Mn2+.
Fig. 5. Manganese inhibits SdrC-mediated biofilm formation.
A. L. lactis pKS80 sdrC biofilm formation in the presence of metal ions. Bacteria (OD600 nm = 0.01) and metal ions (1 mM) were added to microtiter wells at the same time and incubated for 24 hours a 30°C. L. lactis pKS80 clfB was used a positive control.
B. EDTA metal chelation restores the ability of both SdrC and ClfB to promote biofilm growth of heterologous host L. lactis. The data shown is representative of two individual experiments performed in triplicate.
C. Biotin-labeled rSdrCN2 binding to immobilized rSdrCN2N3 in the presence of Mn2+ (1µM) or Mn2+ (1µM) chelated with EDTA (10 µM). Bars represent mean ± SEM; **p<0.01. Representative images of wells containing the CV extracted from previously stained biofilm are shown above the graphs.
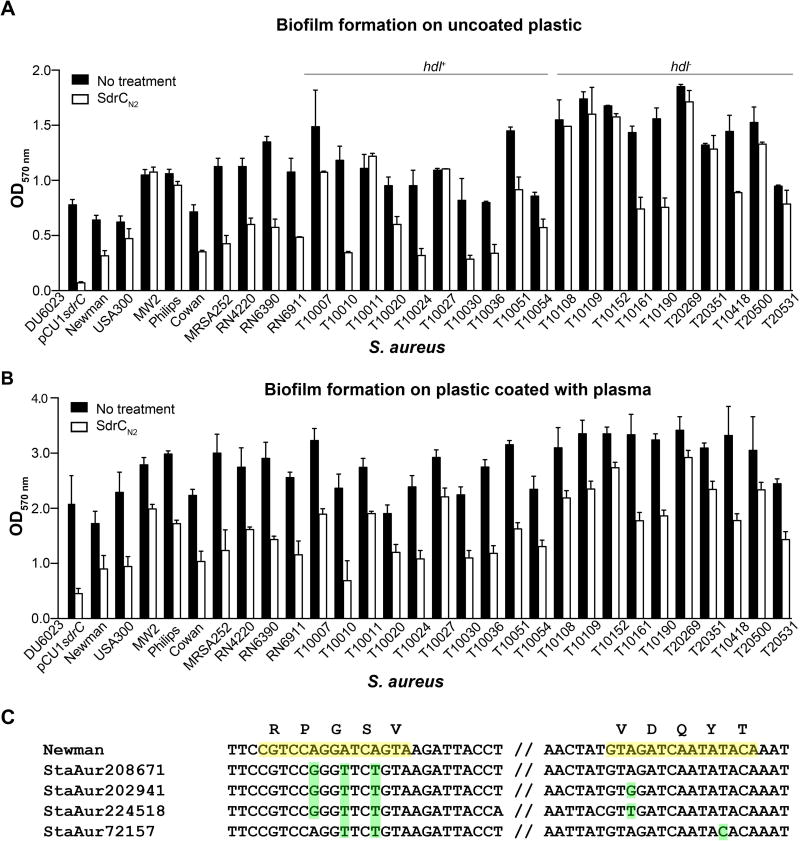
The contribution of SdrC to biofilm formation is strain-dependent
We compared the biofilm formation on plasma-coated and uncoated plastic by common laboratory strains and clinical isolates in the presence of rSdrCN2 subdomain (3 µM). When the biofilm was grown on bare plastic, we detected a reduction of nearly 50% in biofilm accumulation by strains Newman, Cowan, MRSA252, RN4220, RN6390 and RN691. Biofilm formation in vitro by strains USA300, MW2 and Phillips was not affected by the incubation with rSdrCN2 (Fig. 6A). In addition, we investigated SdrC-mediated biofilm formation by 20 clinical isolates. Similar to the strains commonly used in the laboratory, SdrC contributed to biofilm formation in approximately 50% of the clinical isolates, suggesting that the contribution of SdrC to biofilm formation is dependent on the strain background (Fig. 6A). To mimic the conditions that arise in vivo when implanted devices are rapidly covered with plasma proteins, a low amount of plasma (20 ng total protein ml−1) was added to microtiter plates, and biofilm was allowed to accumulate on the surfaces of the coated wells. Overall, we found that biofilm development was more robust on coated plastic (Fig. 6B). Interestingly, rSdrCN2 was able to reduce biofilm formation on plasma-coated plastic even by strains such as USA300 and MW2. A similar trend was observed for biofilm accumulation by the clinical isolates tested (Fig. 6B). Notably, half of the clinical isolates used in our study were δ hemolysin negative consistent with an Agr-deficient phenotype. Indeed, biofilm formation by these strains was enhanced and only three isolates were affected by the exogenous addition of SdrCN2. In contrast, inhibition of SdrC self-association reduced biofilm formation in seven of the δ hemolysin positive strains.
Fig. 6. SdrC contribution to biofilm formation is strain-dependent.
A and B. Biofilm formation by staphylococcal laboratory and clinical strains in the presence of recombinant SdrCN2 protein (3 µM), (A) plastic and (B) immobilized plasma (20 ng protein ml−1). The data shown is representative of four individual experiments performed in triplicate. Bars represent mean ± SEM.
C. SdrC nucleic acid sequences from 134 staphylococcal genomes deposited in the PATRIC database were analyzed for mutations in the dimerization site identified by phage display. The consensus sequence present in S. aureus Newman is highlighted in yellow; examples of polymorphisms are highlighted in green.
To determine whether polymorphisms in the primary amino acid sequence are related to the differential involvement of SdrC in biofilm formation, we analyzed the corresponding nucleotide sequences of all strains described above. We detected no differences in the sequence of the identified SdrC dimerization sites (Fig. 6C). We also compared the nucleic acids and the amino acids sequences of the SdrC dimerization sites from 134 sequenced S. aureus strains deposited in the PathoSystems Resource Integration Center (PATRIC) database (http://patric.vbi.vt.edu). DNA alignments revealed several single nucleotide polymorphisms (SNPs) corresponding to amino acids residues P248, G249, S250, V288 and Y291 (Fig. 6C). Multiple polymorphisms occurred very often in the same S. aureus isolate (89%), while single SNPs were very rare (3%). Importantly, all SNPs resulted in synonymous codons suggesting that the site of SdrC dimerization is conserved.
Discussion
Biofilms are important for human health because they protect microorganisms from both antimicrobials and host immune defenses (Costerton et al., 1999). S. aureus is a common cause of biofilm formation on indwelling medical devices or implants (Otto, 2008). Although significant progress has been made in understanding how staphylococci adhere to inert surfaces and develop sessile communities, the identification of molecular factors involved in this process is still underway. In addition to exopolysaccharides (Cramton et al., 1999) and eDNA (Izano et al., 2008), several CWA proteins have been shown to participate in biofilm accumulation. Interestingly, biofilm formation by staphylococcal clinical isolates could be completely eradicated after treatment with either carbohydrate-degrading enzymes or proteases, indicating that surface proteins and polysaccharides are equally important (Rohde et al., 2007).
In this study, we demonstrate that the surface protein SdrC contributes to staphylococcal biofilm formation in vitro. Using a combinatorial peptide screening approach, we identified two amino acid sequences located adjacent to each other within the N2 subdomain, which appear to act cooperatively to promote SdrC dimerization and, as a result, biofilm expansion. Interestingly, both peptides were necessary to completely inhibit SdrC self-association. We speculate that rather than binding to each other, these sequences either have an as yet unidentified binding site located within the N2 subdomain or bind to the same residues from another SdrC molecule. Although the mechanistic details are poorly understood, the N2N3 subdomains of three other CWA proteins (FnBPA, FnBPB and SasC) have been implicated in biofilm formation (Schroeder et al., 2009; Geoghegan et al., 2013). In addition, SasG mediates biofilm accumulation via Zn2+-dependent B repeat dimerization (Geoghegan et al., 2010; Conrady et al., 2008; Conrady et al., 2013). Thus, homophilic interactions between either N2N3 or B repeat subdomains appear to be a common mechanism by which CWA proteins promote biofilm growth. Intriguingly, the N2N3 domain is also the ligand-binding domain of FnBPs for Fg and of SdrC for β-neurexin (Schwartz-Linek et al., 2003; Patti et al., 1993; Barbu et al., 2010). In the case of FnBPA, the “dock, lock and latch” mechanism of Fg-binding is not essential for biofilm formation (Geoghegan et al., 2013). Thus, it is possible that SdrC follows the same trend. Nonetheless, it is still unknown if the presence of the host ligand in solution interferes with biofilm growth.
Precisely why staphylococci require several CWA proteins with presumably similar mechanisms of action to maintain intercellular interactions is still unclear. It is possible that these molecules are expressed at different stages during infection. In vitro, FnBPs, ClfB and SasG are displayed during early exponential phase regardless of the growth medium (Novick and Jiang, 2003; Ythier et al., 2012), while SdrC expression is more robust during late exponential to stationary phase in RPMI or glucose-containing TSB (Ythier et al., 2012; and our data). Thus, temporal regulation of CWA protein expression may be beneficial for bacteria depending on the environmental conditions (e.g. the type of infection or stage of growth). Conflicting with previously reported data, recent studies have shown that FnBPs are expressed at high levels throughout the growth cycle in methicillin-resistant clinical isolates and this correlates with their contribution to biofilm formation (Geoghegan et al., 2013). Consistent with these observations, our attempts to eliminate biofilm accumulation by commonly used laboratory strains and clinical isolates by interfering with SdrC dimerization was also strain-dependent and not caused by polymorphisms in the self-association site. We speculate that the extent of SdrC contribution to biofilm formation depends on strain-specific genetic and molecular factors.
It is also possible that that co-ordinate expression of MSCRAMMs involves a spatial component. A recent study investigating the role of surface adhesins in Vibrio cholerae biofilm formation has demonstrated that two molecules, Bap1 and RbmA, have distinct roles in agreement with their localization (Absalon et al., 2011). While RbmA reinforces interactions within the biofilm, Bap1 is expressed at the surface interphase where it stabilizes adhesion and recruits cells that have not yet made the transition from planktonic living to sessile lifestyle. We conclude that further investigation is necessary to understand the genetic and spatially segregated factors involved in SdrC regulation in vivo and in vitro.
Heterologous expression in L. lactis indicated that SdrC is involved in both initial bacterial adherence to plastic as well as biofilm expansion. However, initiation of biofilm is independent of dimerization and is likely related to the overall hydrophobicity of the CWA proteins. The ability of S. epidermidis AtlE and S. aureus SasC to promote surface attachment has been attributed to their overall hydrophobicity (Heilmann et al., 1996; Gross et al., 2001; Houston et al., 2011). Our analysis revealed that the percent of hydrophobic amino acids in SdrC (21.2%) is similar to that of that of AtlE (26%) (Heilmann et al., 1996) or SasC (25%) (Schroeder et al., 2009). Furthermore, the ability of SdrC to promote lactococcal biofilms demonstrates that the contribution of SdrC to this process is independent of both autolysin and PIA activity.
Previous studies have shown that chelation of Zn2+ inhibits biofilm formation by FnBPs and SasG expressing strains (Geoghegan et al., 2010; Geoghegan et al., 2013). Thus, the use of metal ion-depleting agents, such as trisodium citrate, as catheter lock solutions has become more widespread (Shanks et al., 2006; O’Grady et al., 2011). More recent work has demonstrated that only a subset of strains is susceptible to chelators, whereas biofilm formation by a significant number of clinical isolates is enhanced by metal ion-depletion (Abraham et al., 2012). Our data reveled that, similar to ClfB (Abraham and Jefferson, 2013), SdrC-mediated biofilm formation is inhibited by Mn2+ and restored by chelating agents. The selective advantage, if any, of differential susceptibility to metal ions remains to be determined. It is possible that the presence of these cofactors may vary depending on the environmental conditions at the site of infection. On one hand, Ca2+ efflux and persistence in the skin is essential for rapid and effective wound healing (Xu and Chisholm, 2011). On the other hand, Mn2+ and Zn2+ chelation by the host innate immune protein calprotectin limits S. aureus growth in the abscess (Corbin et al., 2008). Moreover, within the biofilm eDNA acts as a metal chelator (Mulcahy et al., 2008). Thus, it is tempting to speculate that staphylococci have evolved their surface proteins to exploit and modulate environmental conditions, such as the availability of divalent metal ions.
In summary, we have demonstrated that SdrC promotes biofilm formation through homophilic interactions between the N2 subdomains likely occurring on neighboring bacteria. We have also shown that additional genetic or molecular factors, and environmental conditions affect the biofilm-forming activity of SdrC. How our in vitro studies will correlate with biofilm formation in vivo remains to be determined, but a recent study investigating differential gene expression in biofilm-forming clinical isolates from skin indicated that ica operon and sdrC are highly expressed suggesting that SdrC may be important for in vivo biofilm (Shin et al., 2013). Nevertheless, our study identified and characterized a novel factor involved in staphylococcal biofilm formation. In addition, the mechanistic aspects described in this work contribute to the accumulating evidence that the self-association MSCRAMMs mediates biofilm formation.
Experimental procedures
Media and growth conditions
S. aureus (Table 1) was cultured in tryptic soy broth (TSB) at 37°C with shaking at 250 rpm. L. lactis pKS80 (Table 1) was cultured in M17 containing 0.5% glucose (GM17, Oxoid) and erythromycin (10 µg ml−1) at 30°C without shaking. L. lactis pNZ8037 (Table 1) was cultured in GM17 supplemented with chloramphenicol (10 µg ml−1). Overnight cultures were diluted 1:100, grown for another three hours and surface protein expression was induced with nisin (0 – 3.2 ng ml−1), unless otherwise mentioned (Corrigan et al., 2009).
Table 1.
Bacterial strains, plasmids and oligonucleotides used in this study
| Strain | Description | Source or RE+ |
|---|---|---|
| E.coli | ||
| XL1Blue | recA1 endA1, gyrA96, thi-1, hsdR17 (rk mk+) supE44 relA1 lac [F´ proAB+lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| TOPP3 | Rifr, [F’ proAB+ lacIqZΔM15 Tn10 (Tetr) (Kanr)] | Stratagene |
| S. aureus | ||
| Newman | ST8, human clinical MSSA++ isolate | Duthie et al., 1952 |
| Newman DU5943 | clfB::Tetr | McDevitt et al., 1994 |
| Newman DU5988 | sdrC::pG+Host | O’Brien et al., 2002 |
| Newman DU5989 | sdrD::pG+Host | O’Brien et al., 2002 |
| Newman DU5973 | ΔsdrCDE::Tetr | O’Brien et al., 2002 |
| Newman DU6023 | clfA5 isdA clfB::Ermr ΔsdrCDE::Tetr | Corrigan et al., 2009 |
| LAC | ST8, human clinical MRSA isolate, USA300 | Voyich et al., 2005 |
| MW2 | ST1, human clinical MRSA isolate | Baba et al., 2002 |
| Philips | Human clinical isolate from patient with osteomyelitis; cna+ | Patti et al., 1993 |
| Cowan | ST30, MSSA, ATCC12598 | Cowan et al., 1954 |
| MRSA252 | ST36, MRSA | Holden et al., 2004 |
| RN4220 | ST8, chemically mutagenized derivative of 8325-4, transformable with E. coli DNA | Kreiswirth et al., 1983 |
| RN6390 | 8325-4, rsbU | Peng et al., 1988 |
| RN6911 | RN6390 Δagr::tetM | Novick et al., 1993 |
| T10007, T10010, T10011, T10020, T10024, T10027, T10030, T10036, T10051, T10054 | Clinical isolate; hld positive | |
| T10108, T10109, T10152, T10161, T10190, T20269, T20351, T10418, T20500, T20531 | Clinical isolate; hld negative | |
| L. lactis | ||
| MG1363 | Plasmid-free derivative of NCD 0712; host for constitutive expression vector pKS80 and its derivatives | Gasson et al., 1983 |
| NZ9000 | MG1363 nisA; host strain for nisin-inducible expression vector pNZ8037 and its derivatives | Kuipers et al., 1993 |
| Plasmids | ||
| pQE30 | E. coli plasmid for protein expression with an N-terminal His-tag; Ampr | Qiagen |
| pQE30 SdrCN1 | SdrCN1 with an N-terminal His-tag; Ampr | This study |
| pQE30 SdrCN2 | SdrCN2 with an N-terminal His-tag; Ampr | This study |
| pQE30 SdrCN3 | SdrCN3 with an N-terminal His-tag; Ampr | This study |
| pQE30 SdrCN23 | SdrCN23 with an N-terminal His-tag; Ampr | Barbu et al., 2011 |
| pQE30 SdrCB | SdrCB with an N-terminal His-tag; Ampr | This study |
| pCU1 | E. coli – S. aureus shuttle vector; Ampr, Camr | Augustin et al., 1992 |
| pCU1 sdrC | Derivative of pCU1 encoding sdrC; Ampr, Camr | Barbu et al., 2011 |
| pCU1 sdrD | Derivative of pCU1 encoding sdrD; Ampr, Camr | Corrigan et al., 2009 |
| pCU1 sdrE | Derivative of pCU1 encoding sdrE; Ampr, Camr | Corrigan et al., 2009 |
| pKS80 | L. lactis constitutive expression vector expression; Ermr | Wells et al., 1993 |
| pKS80 clfB | Derivative of pKS80 encoding clfB; Ermr | O’Brien et al., 2002 |
| pKS80 sdrC | Derivative of pKS80 encoding sdrC; Ermr | O’Brien et al., 2002 |
| pKS80 sdrE | Derivative of pKS80 encoding SdrE; Ermr | O’Brien et al., 2002 |
| pKS80 fnbpA | Derivative of pKS80 encoding fnbpA; Ermr | Massey et al., 2001 |
| pKS80 sasG | Derivative of pKS80 encoding sasG; Ermr | Roche et al., 2003 |
| pNZ8037 | L. lactis nisin-inducible expression vector; Camr | de Ruyter et al., 1996 |
| pNZ8037 clfB | Derivative of pNZ8037encoding clfB; Camr | Miajlovic et al., 2007 |
| pNZ8037 sdrC | Derivative of pNZ8037encoding sdrC; Camr | Barbu et al., 2011 |
| pNZ8037 sdrE | Derivative of pNZ8037 encoding sdrE; Camr | This study |
| Oligonucleotides | Restriction site | |
| SdrCN1 forward | GCACGGATCCGGTGACTATGTATGGGAAGATACAA | BamHI |
| SdrCN1 reverse | GCACAAGCTTTTATACATCAACTTCGCCACCCAT | HindIII |
| SdrCN2 forward | GCACGGATCCCATACGAATGGAGAATTAAATCAA | BamHI |
| SdrCN2 reverse | GCACAAGCTTTTAAGCTGCAACAGTATTCACTGC | HindIII |
| SdrCN3 forward | GCACGGATCCCACAACAAGAACAAATGTTAATG | BamHI |
| SdrCN3 reverse | GCACAAGCTTTTAATAATCGACAATGATTTCTTCGCTAT | HindIII |
| SdrCN2N3 forward | CCCGGATCCGGAACAAATGTTAATGATAAAGTACAT | BamHI |
| SdrCN2N3 reverse | CCCAAGCTTTTATTTCTTTTGGTCGCCATTAG | HindIII |
| SdrCB forward | GCACGGATCCAAAGCACAACCGCTTATTTCA | BamHI |
| SdrCB reverse | GCACAAGCTTTTATTTCTTTTGGTCGCCATTAGC | HindIII |
RE, restriction site
MSSA, methicillin sensitive S. aureus
MRSA, methicillin resistant S. aureus
For recombinant protein expression, plasmids pQE30-SdrC52–178 (SdrCN1), pQE30-SdrC178–335 (SdrCN2), SdrC335–496 (SdrCN3), SdrC178–496 (SdrCN2N3) were transformed into E. coli TOPP3 (Stratagene). Overnight starter cultures were diluted 1:50 in Luria-Bertani medium (LB) containing ampicillin (100 µg ml−1) and incubated with shaking until mid-exponential phase was reached (OD600 0.6 – 0.8). Protein expression was induced by adding 1 mM isopropyl-1-thio-β-D-galactopyranoside (final concentration) and growth was continuing for 4 hours, after which bacteria were harvested by centrifugation, resuspended in PBS and frozen at −80°C.
Plasmid construction
Fragments encoding different domains of SdrC were amplified by PCR from S. aureus Newman genomic DNA, and oligonucleotide primers listed in Table 1. The PCR products were analyzed by agarose gel electrophoresis and purified using a QIAquick gel extraction kit (Qiagen). To construct SdrC expression plasmids, a BamHI-HindIII fragment containing the appropriate gene segment was cloned into pQE30 (Qiagen). All plasmid constructs were sequenced to ensure the integrity of the amplified fragments (Baylor College of Medicine DNA Sequencing Core Facility).
Protein expression and purification
The His-tagged recombinant SdrC subdomains were purified by affinity chromatography with a 5 ml nickel-charged HiTrap column (GE Healthcare) and 5 ml anion or cation exchange Sepharose column (GE Healthcare) as described by Barbu et al. (Barbu et al., 2010). Fractions containing >95% pure recombinant were dialyzed against Tris-buffered saline (TBS), 10 mM EDTA, pH 7.4.
Phage display
The phage display screen was described earlier (Barbu et al., 2010).
Phage binding assays
Binding of phage displaying enriched peptides to immobilized rSdrCN2N3 was determined as described (Barbu et al., 2010). Briefly, microtiter wells were coated with recombinant proteins overnight (1µg per well in carbonate buffer pH 9.3). After blocking with PBS containing 3% BSA for one hour, wells were incubated with 109 phage. Unbound phage were removed by washing with PBS six times and subsequently probed with an anti-M13-HRP antibody. Color development was performed using TMB (Calbiochem) and the binding was measured using a microtiter plate reader (Molecular Devices) at 650 nm.
Solid phase binding assays
To detect self-association, SdrC recombinant subdomains were labeled with biotin according to the manufacturer’s instruction (EZ-Link-NHS-Biotin, Pierce). Microtiter wells were coated with unlabeled SdrC subdomains overnight at 4°C (1 µg per well in carbonate buffer pH 9.3). Coated wells were blocked for one hour at room temperature with 2% BSA in TBS buffer. Increasing concentrations of biotin-labeled SdrC subdomains were added to the wells and incubated for one hour at room temperature. Wells were then probed with avidin-HRP. Color development was performed using TMB (Calbiochem) and the binding was measured using a microtiter plate reader (Molecular Devices) at 650 nm. For inhibition assays, either phage (109 TU) or 1 µM anti-SdrCN2N3 antibodies were added to the wells before incubating with 1 µM biotin labeled SdrC N2N3. Data presented represent the mean ± SD of three independent experiments performed in triplicate. The apparent dissociation constant (KD) calculated using the equation ΔP = ΔPmax [protein]/ (KD + [protein]).
Gel permeation chromatography
To assess SdrC oligomerization, pure SdrC subdomains (1 mg ml−1) were analyzed on a gel filtration column (Superdex 200 5/150 GL, GE Healthcare) attached to an AKTA FPLC at a flow rate of 0.3 ml min−1 in TBS pH 7.4. The column was calibrated with the following proteins: thyroglobulin (660 KDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (4 kDa), chymotrypsin (29 kDa), cytochrome c (12.4 kDa) and aprotinin (6.5 kDa) (Gel Filtration Calibration Kits HMW & LMW, GE Healthcare). Gel phase distribution coefficients (KAV) were calculated from the respective elution volumes (Ve), represented as a function of molecular mass and analyzed by linear regression (R=0.9905). The relative molecular mass of SdrC species eluted from the column was calculated by interpolation.
Biofilm formation
S. aureus was grown overnight in TSB and diluted 1:100 in TSB supplemented with 1% glucose and dispensed into sterile non-treated microtiter plates (100 µl per well). Overnight cultures of L. lactis were diluted in GM17 and added to sterile non-treated microtiter plates (275 µl per well). After 24 h incubation, plates were washed three times with PBS, dried and stained with 0.5% crystal violet (CV). Subsequently, the plates were rinsed with water and dried. The dye was dissolved in 200 µl of 95% ethanol and the absorbance was measured at OD570 nm (Corrigan et al., 2007). For specific accumulation assays, microtiter wells were coated with plasma (20 ng protein ml−1 in PBS) at 4°C for 18 h and washed twice with sterile PBS. For competition assays, rSdrC subdomains, anti-SdrC antibodies or metal ions (1mM for biofilm and 1µM for recombinant proteins) were added to the plates at the same time as bacteria. For inhibition of biofilm by clinical isolates, 3 µM SdrCN2 was added to the well at the same time as bacterial cells.
Adherence of bacteria to plastic
L. lactis was grown overnight in GM17 supplemented with the appropriate antibiotic, washed and resuspended in PBS at OD600 nm =1. Bacteria were added to the plates (100 µl) and incubated at room temperature for one hour. Unattached cells were removed by washing three times with PBS. Bacterial adherence was measured as described above. For inhibition assays, either 1 µM anti-SdrCN2N3 serum or 3 µM SdrC N2N3 were added to the wells before bacteria were allowed to adhere to plates. Data presented represent the mean ± SD of three independent experiments performed in triplicate.
Colony spreading
The colony spreading assay was performed as described (Kaito and Sekimizu, 2007). Briefly, a 2 µl aliquot of bacteria from overnight cultures was spotted on soft agar GM17 (0.24% agar) after plates were dried for approximately 10 minutes in a laminar flow cabinet. Dishes were then dried for an additional 5 minutes and incubated overnight at 37°C. Colony spreading assays were repeated six times.
Statistical analysis
For all assays, three independent experiments were carried out in triplicate. Comparisons of multiple treatment groups were performed by using two-way analysis of variance with post-hoc paired comparisons by Dunnett’s test.
Calculations were made with InStat (GraphPad Software). Two-tailed P values of less than 0.05 were considered statistically significant.
Acknowledgments
S. aureus clinical isolates were a gift from Vance Fowler. This work was supported by grant AI020624 from NIAID to M.H.
References
- Abraham NM, Jefferson KK. Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology. 2012;158:1504–1512. doi: 10.1099/mic.0.057018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham NM, Lamlertthon S, Fowler VG, Jefferson KK. Chelating agents exert distinct effects on biofilm formation in Staphylococcus aureus depending on strain background: role for clumping factor B. J Med Microbiol. 2012;61:1062–1070. doi: 10.1099/jmm.0.040758-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absalon C, Van Dellen K, Watnick PI. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 2011;7:e1002210. doi: 10.1371/journal.ppat.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin J, Rosenstein R, Wieland B, Schneider U, Schnell N, Engelke G, et al. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. FEBS. 1992;204:1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- Barbu EM, Ganesh VK, Gurusiddappa S, Mackenzie RC, Foster TJ, Sudhof TC, Höök M. beta-Neurexin is a ligand for the Staphylococcus aureus MSCRAMM SdrC. PLoS Pathog. 2010;6:e1000726. doi: 10.1371/journal.ppat.1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA. 2008;105:19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrady DG, Wilson JJ, Herr AB. Structural basis for Zn2+-dependent intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA. 2013;110:E202–211. doi: 10.1073/pnas.1208134110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- Corrigan RM, Miajlovic H, Foster TJ. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol. 2009;9:22. doi: 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Rigby D, Handley P, Foster TJ. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153:2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Geesey GG, Cheng KJ. How bacteria stick. Scientific American. 1978;238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Cowan ST, Shaw C, Williams RE. Type strain for Staphylococcus aureus Rosenbach. J Gen Microbiol. 1954;10:174–176. doi: 10.1099/00221287-10-1-174. [DOI] [PubMed] [Google Scholar]
- Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penadés JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ruyter P, Kupiers OP, De Vos WM. Controlled gene expression systems for Lactococcus lactis with the food grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duthie ES, Lorenz LL. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12(1):49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson MJ. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M, Cramton SE, Gotz F, Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JA, Corrigan RM, Gruszka DT, Speziale P, O’Gara JP, Potts JR, Foster TJ. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J Bacteriol. 2010;192:5663–5673. doi: 10.1128/JB.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JA, Monk IR, O’Gara JP, Foster TJ. Subdomains N2N3 of fibronectin binding protein A mediate Staphylococcus aureus biofilm formation and adherence to fibrinogen using distinct mechanisms. J Bacteriol. 2013;195:2675–2683. doi: 10.1128/JB.02128-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Gerke C, Perdreau-Remington F, Gotz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun. 1996;64:277–282. doi: 10.1128/iai.64.1.277-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston P, Rowe SE, Pozzi C, Waters EM, O’Gara JP. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun. 2011;79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaito C, Sekimizu K. Colony spreading in Staphylococcus aureus. J Bacteriol. 2007;189:2553–2557. doi: 10.1128/JB.01635-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane FM, Loughman A, Valtulina V, Brennan M, Speziale P, Foster TJ. Fibrinogen and elastin bind to the same region within the A domain of fibronectin binding protein A, an MSCRAMM of Staphylococcus aureus. Mol Microbiol. 2007;63:711–723. doi: 10.1111/j.1365-2958.2006.05552.x. [DOI] [PubMed] [Google Scholar]
- Kreiswirth BN, Lofdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Kuipers OP, de Ruyter PG, Kleerebezem M, de Vos WM. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotech. 1997;15:135–140. doi: 10.1016/S0167-7799(97)01029-9. [DOI] [PubMed] [Google Scholar]
- Massey RC, Kantzanou MN, Fowler T, Day NP, Schofield K, Wann ER, et al. Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell Microbiol. 2001;3:839–851. doi: 10.1046/j.1462-5822.2001.00157.x. [DOI] [PubMed] [Google Scholar]
- McDevitt D, Francois P, Vaudaux P, Foster TJ. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, et al. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol. 2009;191:832–843. doi: 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miajlovic H, Loughman A, Brennan M, Cox D, Foster TJ. Both complement- and fibrinogen-dependent mechanisms contribute to platelet aggregation mediated by Staphylococcus aureus clumping factor B. Infect Immun. 2007;75:3335–43. doi: 10.1128/IAI.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Eidhin D, Perkins S, Francois P, Vaudaux P, Höök M, Foster TJ. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol Microbiol. 1998;30:245–257. doi: 10.1046/j.1365-2958.1998.01050.x. [DOI] [PubMed] [Google Scholar]
- Novick RP, Jiang D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology. 2003;149:2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien L, Kerrigan SW, Kaw G, Hogan M, Penades J, Litt D, et al. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol Microbiol. 2002;44:1033–1044. doi: 10.1046/j.1365-2958.2002.02935.x. [DOI] [PubMed] [Google Scholar]
- O’Grady NP, Alexander M, Burns LA, Dellinger EP, Garland J, Heard SO, et al. Guidelines for the prevention of intravascular catheter-related infections. Clin Infect Dis. 2011;52:e162–193. doi: 10.1093/cid/cir257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O’Gara JP. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual A, Fleer A, Westerdaal NA, Verhoef J. Modulation of adherence of coagulase-negative staphylococci to Teflon catheters in vitro. Eur J Clin Microbiol. 1986;5:518–522. doi: 10.1007/BF02017694. [DOI] [PubMed] [Google Scholar]
- Patti JM, Boles JO, Höök M. Identification and biochemical characterization of the ligand binding domain of the collagen adhesin from Staphylococcus aureus. Biochemistry. 1993;32:11428–11435. doi: 10.1021/bi00093a021. [DOI] [PubMed] [Google Scholar]
- Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, Otto M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci USA. 2012;109:1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, et al. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115:217–228. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- Resch A, Rosenstein R, Nerz C, Gotz F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol. 2005;71:2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Roche FM, Massey R, Peacock SJ, Day NPJ, Visai L, Speziale P, et al. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology. 2003;149:649–654. doi: 10.1099/mic.0.25996-0. [DOI] [PubMed] [Google Scholar]
- Schroeder K, Jularic M, Horsburgh SM, Hirschhausen N, Neumann C, Bertling A, et al. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS One. 2009;4:e7567. doi: 10.1371/journal.pone.0007567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Linek U, Werner JM, Pickford AR, Gurusiddappa S, Kim JH, Pilka ES, et al. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature. 2003;423:177–181. doi: 10.1038/nature01589. [DOI] [PubMed] [Google Scholar]
- Shanks RM, Sargent JL, Martinez RM, Graber ML, O’Toole GA. Catheter lock solutions influence staphylococcal biofilm formation on abiotic surfaces. Nephrol, dialys, transpl. 2006;21:2247–2255. doi: 10.1093/ndt/gfl170. [DOI] [PubMed] [Google Scholar]
- Shin K, Yun Y, Yi S, Lee HG, Cho JC, Suh KD, et al. Biofilm-forming ability of Staphylococcus aureus strains isolated from human skin. J Dermatol Sci. 2013;71:130–137. doi: 10.1016/j.jdermsci.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Shopsin B, Eaton C, Wasserman GA, Mathema B, Adhikari RP, Agolory S, et al. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2010;202:1593–1599. doi: 10.1086/656915. [DOI] [PubMed] [Google Scholar]
- Tsompanidou E, Denham EL, Sibbald MJ, Yang XM, Seinen J, Friedrich AW, et al. The sortase A substrates FnbpA, FnbpB, ClfA and ClfB antagonize colony spreading of Staphylococcus aureus. PLoS One. 2012;7:e44646. doi: 10.1371/journal.pone.0044646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- Ythier M, Resch G, Waridel P, Panchaud A, Gfeller A, Majcherczyk P, Quadroni M, Moreillon P. Proteomic and transcriptomic profiling of Staphylococcus aureus surface LPXTG-proteins: correlation with agr genotypes and adherence phenotypes. Mol Cell Prot: MCP. 2012;11:1123–1139. doi: 10.1074/mcp.M111.014191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JM, Robinson K, Chamberlain LM, Schofield KM, Le Page RW. Lactic acid bacteria as vaccine delivery vehicles. Antonie van Leeuwenhoek. 1996;70:317–330. doi: 10.1007/BF00395939. [DOI] [PubMed] [Google Scholar]
- Xu S, Chisholm AD. A Galphaq-Ca2+ signaling pathway promotes actin-mediated epidermal wound closure in C. elegans. Curr Biol: CB. 2011;21:1960–1967. doi: 10.1016/j.cub.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]