Abstract
Since their serendipitous discovery in nematodes, microRNAs (miRNAs) have emerged as key regulators of biological processes in animals. These small RNAs form complex regulatory networks in cell development, differentiation and homeostasis. Deregulation of miRNA function is associated with an increasing number of human diseases, particularly cancer. Recent discoveries have expanded our understanding of how miRNAs are regulated. Here we review the mechanisms that modulate miRNA activity, their stability and their localization through alternative processing, sequence editing, post-translational modifications of Argonaute proteins, viral factors, transport from the cytoplasm and regulation of miRNA–target interactions. We conclude by discussing intriguing open questions to be answered by future research.
Introduction
microRNAs (miRNAs) are short non-coding RNAs of ~22 nucleotides that mediate gene silencing by guiding Argonaute (AGO) proteins to target sites in the 3’ untranslated region (UTR) of mRNAs. AGOs constitute a large family of proteins that use single-stranded small nucleic acids as guides to complementary sequences in RNA or DNA targeted for silencing1. The miRNA-loaded AGO forms the targeting module of the miRNA-induced silencing complex (miRISC), which promotes translation repression and degradation of targeted mRNAs2. The first miRNA was discovered in Caenorhabditis elegans as a short RNA produced of the lin-4 gene, which post-transcriptionally represses the lin-14 mRNA3,4. Such small RNAs were widely thought to be unique to nematodes, until they were shown to be abundant in diverse animal phyla5. This new class of regulators was subsequently named ‘microRNAs’6–8.
miRNAs are involved in virtually every cellular process and are essential for animal development, cell differentiation and homeostasis; deletions of the fundamental miRNA biogenesis factors Dicer9 and Drosha10 are lethal in mouse embryos. Although the importance of miRNAs for embryonic development is well established11, Drosha and Dicer are involved in other nuclear processes, such as pre-mRNA splicing, which may also contribute to their deletion phenotypes12. The miRNA repository miRbase (www.mirbase.org) currently lists 1,917 precursor miRNAs (pre-miRNAs) and 2,654 mature miRNAs in Homo sapiens13, and more than 60% of human protein-coding genes harbor predicted miRNA target sites14. Deregulation of miRNA function is associated with numerous diseases15, particularly cancer16,17: miRNAs can be both oncogenes (called oncomirs) 18 or tumor suppressors19, although overall down regulation of miRNA expression is a hallmark of cancer20. Some miRNAs are prognostic markers21,22 or potential targets for novel cancer therapies23. Plant miRNAs24,25, which are not discussed here, differ considerably from animal miRNAs in their evolution26, biogenesis27 and function28(reviewed in 29,30).
In this Review, we first provide an overview of miRNA function and regulation. We than discuss in detail the regulation of miRNA function through the formation of isomirs[G, glossary term] Where you agree please provide succinct, one-sentence definitions for these specialist terms.], the addition of non-templated nucleotides, post-translational modification (PTM) of miRNA-binding proteins, miRNA sequestration, modulation by viral factors, transport from the cytoplasm, and the regulation of interactions between miRNAs and their target mRNAs (miRNA–target interactions).
Overview of miRNA function
miRNA genes are transcribed into primary miRNA (pri-miRNA) transcripts and undergo multi-step biogenesis, in which they are processed first into pre-miRNAs and finally into mature miRNAs (BOX 1). miRNAs exhibit tissue-specific expression patterns31, which are primarily regulated transcriptionally32. They are transcribed mainly by RNA polymerase II33–35 and can be derived from introns or from long non-coding RNAs. Pri-miRNAs can consist of a single mature miRNA or of clusters of often related miRNAs6,36. miRNAs are grouped into families37 based on the similarity of their seed sequences; the seed comprises nucleotides 2–8 (counting from the 5’ end) and is primarily responsible for miRNA targeting of mRNAs38.
Box 1: miRNA biogenesis.
The biogenesis of microRNAs (miRNAs) is a multi-step process. They are transcribed mainly by RNA polymerase II 33–35 as structured primary miRNAs (pri-miRNAs) and processed into precursor miRNAs (pre-miRNAs) and finally into mature miRNA duplexes (see the figure). The mature miRNA comprises a 5p strand (arising from the 5’ arm of the pre-miRNA hairpin) and a 3p strand. The sequence of pri-miRNAs can be altered by A-to-I editing by double-stranded RNA-specific adenosine deaminase (ADAR) proteins, which may affect further biogenesis and the sequence of the mature miRNA, or promote degradation of the pri-miRNA99.
The pri-miRNA hairpin is excised in the nucleus by the Microprocessor complex comprising the RNase III enzyme Drosha252 and the protein DiGeorge syndrome critical region gene 8 (DGCR8) 253,254. Drosha recognizes the double-strand RNA–single strand RNA junction at the hairpin base, whereas two DGCR8 proteins bind the stem and ensure correct cleavage255,256. Alternative cleavage by Drosha leads to the production of isomirs97,98 (FIGURE 2; Supplementary Table S1).
Pre-miRNAs are hairpins of ~70 nucleotides6–8. The hairpin end features a 2-nucleotide overhang at the 3’, a 5’ phosphate and a 3’ hydroxyl, which are typical of RNase III products257. Exportin-5 recognizes the overhang258 and transports the pre-miRNA to the cytoplasm259–261. XPO5 knockout in a human cell line reduced but did not eliminate nuclear export of pre-miRNAs, which suggested that alternative modes of pre-miRNA nuclear export exist262.
In the cytoplasm, the RNase III enzyme Dicer259–261 binds the pre-miRNA by recognizing the 5’ phosphate, 3’ overhang and loop structure263–266. Dicer is a ‘molecular ruler’263,267 that cleaves pre-miRNAs at a species-specific length268 and yields a mature-miRNA duplex with another typical 2-nucleotide 3’ overhang257,269. Alternative cleavage by Dicer can also lead to the production of isomirs98 (FIGURE 2; Supplementary Table S1). In vertebrates, cleavage by Dicer is modulated by TAR RNA-binding protein (TARBP) and protein activator of the interferon-induced protein kinase (PACT); in flies, by Loquacious proteins103–106,270.
One strand of the mature miRNA (the ‘guide’ strand) is loaded into Ago, whereas the other strand (‘passenger strand’) is discarded271–273. Loading preference is given to the strand possessing the less stably paired 5’ end274–276; Ago2 was also reported to prefer an A or U as the 5’-terminal nucleotide277.
The mature miRNA forms a functional unit with an AGO protein (FIGURE 1A), which is a single polypeptide chain composed of four characteristic domains: the amino-terminal (N) domain, the Piwi-Argonaute-Zwille (PAZ) domain, the middle (MID) domain and the P-element induced wimpy testes (PIWI) domain. Two linker domains (L1 and L2) connect the N and PAZ domains (L1) and the PAZ and MID domains (L2). The N and PAZ domains form one lobe of AGO and the MID and PIWI domains form the second lobe. The MID and PIWI domains hold the 5’ end of the miRNA, whereas the PAZ domain binds its 3’ nucleotide39–42. The mammalian genome encodes four AGO proteins (Ago1–4). Ago2 is the most highly expressed and the only AGO protein able to cleave a target that is fully complementary to the guide strand[G] of the miRNA43,44, which is a feature important for the biogenesis of miR-45145,46 and for regulation of a subset of miRNAs that are extensively paired with their targets47,48. Although some human miRNAs are preferentially loaded into specific AGO proteins, many associate with all AGO proteins49–52.
Figure 1: Overview of miRNA function and its regulation.
(A) Mature miRNAs operate as functional units that include an Argonaute protein. Argonaute proteins have four domains, the amino-terminal domain (N), the Piwi-Argonaute-Zwille (PAZ) domain, the middle (MID) domain and the P-element induced wimpy testes (PIWI) domain, and two linker regions (L1 and L2). The MID and PIWI domain hold the 5’ end of the miRNA and arrange it in a helical conformation; the MID domain has a binding pocket for the 5’-terminal nucleotide. Another binding pocket, in the PAZ domain, holds the 3’-terminal nucleotide 39–41. Nucleotides 2–8 from the 5’ end of the miRNA form the seed, which is crucial for target-mRNA recognition38. Seed interactions involve nucleotides 2–8, 2–7 and 2–6 54, and can be supplemented by the binding to the MID domain of an adenine in the target mRNA opposite miRNA nucleotide 1 (t1) 54,56,57, or through additional, base-pairing to nucleotides ~13–16 of the miRNA, which are termed ‘supplemental region’65.
(B) miRNAs silence gene expression by inhibiting translation at the initiation step, likely through release of eukaryotic initiation factor 4A-1 (eIF4A1) and eIF4A279–81, and by mediating mRNA decay2 through interactions with glycine-tryptophan protein of 182 kDa (GW182) proteins68–71. GW182 binds polyadenylate-binding protein (PABPC) and the deadenylation complexes poly(A) nuclease 2 (PAN2)–PAN3 and carbon catabolite repressor protein 4 (CCR4)–NOT72–76. Deadenylation is followed by decapping by the complex mRNA-decapping enzyme subunit 1 (DCP1)–DCP273 and 5’−3’ mRNA degradation (not shown) 77.
(C) miRNAs form complex networks of interactions, as one miRNA can target many different mRNAs, 86 and one mRNA can be regulated by many different miRNAs87, with cooperative repression achieved by binding closely-spaced target sites65,89,90.
miRNA target sites are generally located in the 3’ UTR of mRNAs; they possess strong complementarity to the seed region38, which is the main criterion for target-site prediction53–55. The strongest canonical (seed-matching) target sites are those that complement miRNA nucleotides 2–8 and have an adenine opposite miRNA nucleotide 1 (known as ‘t1A’), followed by those complementing nucleotides 2–8 without a t1A and nucleotides 2–7 with a t1A54. t1A is not recognized by the miRNA guide strand, but by a binding pocket within AGO56,57 (FIGURE 1A). Target sites with complementarity to nucleotides 2–7 or 3–8 of the miRNA are much weaker, but still considered canonical54. Structural and single-molecule studies suggest that target recognition is achieved by a two-step mechanism where first nucleotides 2–6 of the seed are pre-organized by the MID and PIWI domains in a stacked, helical conformation, with nucleotides 2–4 exposed to the solvent41. This conformation allows for rapid initial binding, which is weak and metastable and only becomes long-lived if the site presents complementarity to nucleotides 7–8 of the guide strand. Otherwise, AGO disengages the site and either laterally diffuses along the mRNA or dissociates from it completely58. Thus, the mRNA-binding behavior of a miRNA is similar to that of an RNA-binding protein (RBP) 58–60.
AGO–target mRNA crosslinking has enabled the identification of non-canonical (not seed-matching) binding sites61–63, and a recent large scale microarray-based survey in HeLa cells reported sites with minimal seed-pairing64. However, the biological function of non-canonical sites, when considered as a whole, has been disputed as these sites imparted no detectable repression in meta-analyses of miRNA and small RNA transfection datasets 54. In addition to the seed, the 3’ half of the miRNA can also contribute to target recognition (particularly nucleotides 13–16, which are termed ‘supplemental region’) in a subset of sites54,65, and can direct miRNA family members with the same seed to different target sites, as recently shown in C. elegans66 and in mouse brains63.
AGO–miRNA binding to the 3’ UTR leads to gene silencing through translation repression and mRNA decay[G] 2,67 (FIGURE 1B). The latter involves recruitment by AGO of a member of the glycine-tryptophan protein of 182 kDa (GW182) protein family (in humans, trinucleotide repeat-containing gene 6A protein (TNRC6A), TNRC6B and TNRC6C) 68–71. GW182 interacts with polyadenylate-binding protein (PABPC), thereby promoting mRNA deadenylation by recruiting the poly(A) nuclease 2 (PAN2)–PAN3 and carbon catabolite repressor protein 4 (CCR4)–NOT complexes72–76. Deadenylation promotes decapping by the mRNA-decapping enzyme subunit 1 (DCP1)–DCP2 complex73, thereby making the mRNA susceptible to rapid degradation by 5’−3’ exoribonuclease 1 (XRN1) 77. GW182-mediated recruitment of CCR4–NOT also leads to translation repression through recruitment of the RNA helicase DDX62,78.
Inhibition of translation initiation is caused also by interfering with the function of eukaryotic initiation factor 4A-1 (eIF4A1) and eIF4A22,79–81. In Drosophila melanogaster S2 cell lysates this activity was independent of GW18280, suggesting the existence of multiple mechanisms of translation inhibition. There is disagreement on the mechanism of interference with eIF4A1 and eIF4A2 function79–81, but most data indicate that miRISC induces their dissociation from target mRNAs and thereby inhibits ribosome scanning and assembly of the eIF4F translation initiation complex. A recent investigation in human and D. melanogaster cells reported the AGO Trp-binding pockets that mediate GW182 binding41,82 are required for translation inhibition83. Thus, miRISC-mediated translation inhibition is still incompletely understood.
Both modes of miRISC-mediated gene silencing are thought to be interconnected2, and ribosome profiling assays revealed that mRNA decay is generally responsible for 66–90% of silencing67,84. The observation that translation inhibition can be rescued85 but that mRNA degradation is irreversible, raises the possibility that the regulated pauses, or blocks, in the molecular cascade leading to mRNA degradation could allow translation repression without mRNA decay.
One miRNA can silence hundreds of genes, although the effect on each gene is generally mild86, and multiple miRNAs can regulate the same gene87 (FIGURE 1C). Furthermore, entire cellular pathways can be regulated by individual miRNAs87 or miRNA clusters[G] 88. miRNA binding of neighboring target sites on a target mRNA can result in cooperative repression65,89,90, which might explain why the function of non-canonical sites can depend on the occupancy of neighboring canonical sites91. Cooperativity is explained, in part, by the formation of multivalent interactions of GW182 with AGO proteins82. miRNAs can either switch off, or function as fine tuners of protein expression92, and thereby buffer against noise in gene expression93.
Modification of miRNA sequence
The interaction of miRNAs with their targets is largely based on their seed sequence 38,54, and miRNA biogenesis if affected by RNA secondary structures that mediate interactions with RBPs94. Cellular processes that alter the sequence of a miRNA or of its precursor therefore can affect miRNA biogenesis, activity and turnover.
Isomir formation is a regulated process that determines miRNA activity
Isomirs are mature-miRNA variants that differ in length, sequence or both31,95–97. The maturation, stability and turnover, activity or targetome of isomirs can vary (FIGURE 2). Isomirs are classified into 5’, 3’ or polymorphic (internal), depending on the site of variation, and can be derived from alternative Drosha or Dicer processing, RNA editing or non-templated nucleotide addition (NTA) 98,99. A recent study in a human breast cancer cell line reported that many miRNAs had no isoforms whereas miR-21–5p had 43 isoforms100. Isomir expression patterns have been found useful in distinguishing different types of breast cancer and represent potential biomarkers101.
Figure 2: Isomirs differ in length and sequence and expand the functional repertoire of miRNAs.
Isomirs are classified as 5’, 3’ or polymorphic, with combinations possible. Depending on the arm of the miRNA precursors (5p or 3p; see inset) used to produce the mature miRNA, cleavage by either Drosha or Dicer can result in the formation of the isomir 98. 5’ isomirs have shifted seeds and can thereby target a different set of genes109. The functions of 3’ isomirs are less clear, but there is increased evidence for their differential activity113,114. Polymorphic isomirs are generated by RNA editing, mainly by ADAR. The editing can affect miRNA biogenesis, either by preventing it, or by leading to the formation of 5’ isomirs or 3’ isomirs; if editing alters the seed, it could retarget a miRNA to other mRNAs98,99. nt, nucleotides.
The primary determinant of miRNA length and sequence is cleavage by Drosha and Dicer32 (Supplementary Table S1). Alternative processing by Drosha was first reported for miR-142 and miR-342 in mouse CD8 T cells102 and shown to be widespread in human cells97. Cleavage by Dicer has been more extensively studied; it is modulated by TAR RNA-binding protein (TARBP; also known as TRBP) and protein activator of the interferon-induced protein kinase (PACT) in vertebrates (Box 1) and by loquacious in flies103–106. Variation in the Dicer cleavage site directly modulates the seed sequence of 3p miRNA strands, and can alter guide-strand selection106,107 (FIGURE 2; Supplementary Table S1). Dicer binds TARBP and PACT by the same domain, as revealed by a recent crystal structure of the binding interface, suggesting Dicer is regulated by a mechanism based on binding–competition between TARBP and PACT, which produce specific isomirs for some miRNAs106.
Alternative cleavage by Drosha or Dicer can shift the position of the seed and is the primary mechanism of 5’ isomir production (BOX 1; FIGURE 2), as nucleotide addition or removal at the 5’ end of mature miRNAs is rare98. Cleavage-directed 5’ end variation can retarget a miRNA, as observed in flies104 and humans108–111, and 5’ isomirs can target the same biological pathways together109,112 (Supplementary Table S1). The abundance of 5’ isomirs can vary between cell types, with certain isomirs being predominant in specific cell types96. By contrast, 3’ isomirs mostly differ in their stability and turnover (see below), although recent findings point also to length-related effects on targeting and activity. For example, binding to the RNA of Hepatitis C virus (see below) of a 21-mer 3’ isomir of miR-122 is weaker compared to binding of longer miR-122 isomirs113, and longer 3’ isomirs of miR-222 have increased apoptotic activity114.
Editing the sequence of miRNA precursors
RNA editing can change miRNA sequence, generating isomirs, and can also affect biogenesis, leading to 5’ or 3’ variation98,99 (BOX 1; FIGURE 2). Deamination is the most commonly observed type of miRNA precursor editing. Adenosine deaminase acting on RNA (ADAR) converts adenosine to inosine99, which is read as guanosine during splicing and translation. Similarly, cytidine deaminase acting on RNA (CDAR) converts cytidine to uracil115. In vertebrates ADAR1 and ADAR2 act on double-stranded RNA and edit miRNA precursors99. A-to-I editing sites appear widespread116, and in the human brain ~16% of pri-miRNAs were predicted to be edited (based on the analysis of 209 pri-miRNAs). However, editing levels at different miRNAs vary considerably99,117.
A-to-I editing of miRNA precursors can interfere with cleavage by Drosha 117–121 or Dicer 117,122, or, less frequently, with AGO loading, and editing of the seed can redirect a miRNA to a new set of targets120,123–126. A-to-I editing of miRNAs is involved in miRNA deregulation in cancer121,127,128. For example, in melanoma, overexpression of cAMP-responsive element-binding protein (CREB) reduces ADAR1 expression, thereby reducing the editing of miR-455–5p; unedited miR-455–5p represses cytoplasmic polyadenylation element-binding protein 1 (CPEB1) and other tumor suppressors, thereby promoting tumor growth and metastasis121. Changes in seed sequences can also be mediated by the more elusive C-to-U editing129. miRNA A-to-I editing was first observed in pre-miR-22 in human and mouse tissues at species-specific positions, and was predicted to alter miRNA targeting and activity130. In summary, RNA editing is a versatile regulatory mechanism that can control the abundance and target specificity of miRNAs, but in light of the relatively low frequency of editing99, further research is required to characterize its biological importance.
Non-templated nucleotide addition
Non-templated addition of nucleotides (NTA) was first suggested to be biologically regulated during D. melanogaster embryogenesis131. NTAs predominantly comprise of adenylation or uridylation at 3’ ends, and are miRNA-specific across tissue types, developmental stages, disease states and different species 132.
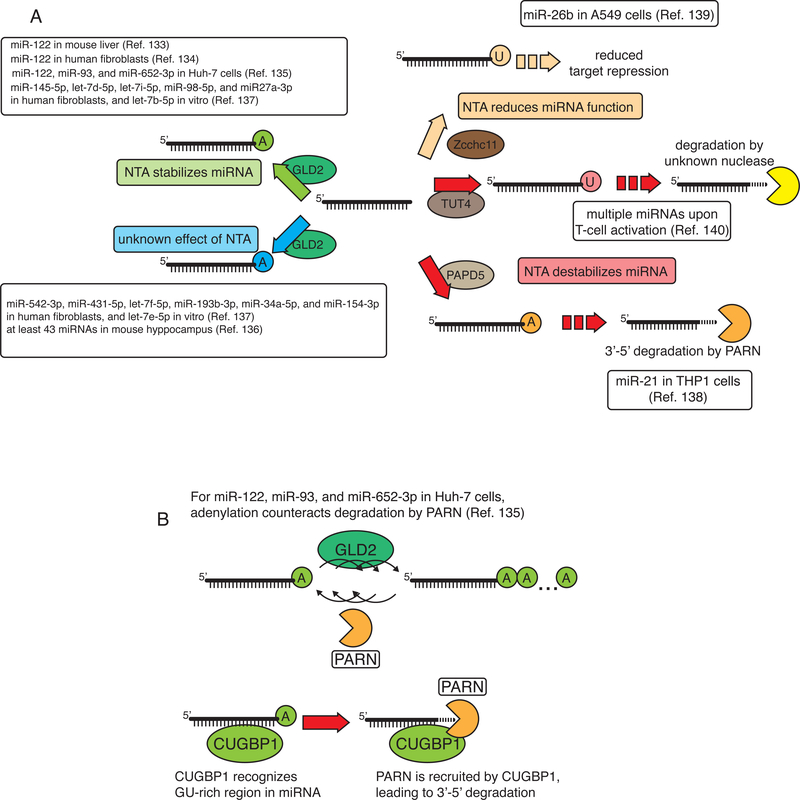
NTAs are carried out by several enzymes and can modulate miRNA stability (FIGURE 3A). Poly(A) RNA polymerase Germ Line Development 2 (GLD2)-mediated 3’-end mono-adenylation stabilizes miR-122 in the liver133 and in mouse embryonic fibroblasts (MEF) 134. Poly(A)-specific ribonuclease (PARN) counteracts this as its depletion in a human liver cancer cell line leads to the appearance of miR-122 with 3’-oligo adenylation and to increased miR-122 stability. CUG triplet repeat RNA-binding protein 1 (also known as CELF1) binds miR-122 and mediates its degradation by recruiting PARN (FIGURE 3B). A similar mechanism regulates miR-93 and miR-652–3p135. By contrast, a recent analysis of miRNAs in the hyppocampus of GLD2 knockout mice revealed reduced levels of 3’ adenylation, but no effect on miRNA stability136; similarly, only about half of the investigated miRNAs were destabilized in human fibroblasts when GLD2-mediated adenylation was suppressed 137. Moreover, 3’ adenylation by the poly(A) RNA polymerase PAP-associated domain-containing protein 5 (PAPD5) initiates degradation of miR-21138.
Figure 3: Non-templated nucleotide addition and miRNA turnover.
(A) Non-templated nucleotide addition (NTA) by GLD2 can stabilize some miRNAs, but has no effect on others. Terminal 3’ adenylation of miR-21 by PAP-associated domain-containing protein 5 (PAPD5) was even found to promote exonucleolytic cleavage by poly(A)-specific ribonuclease (PARN) 138. Terminal 3’ uridylation by terminal uridylyltransferase 4 (TUT4) can reduce the activity of miR-26b139, or prime miRNAs for degradation following T-cell activation140.
(B) Terminal 3’ adenylation stabilizes some miRNAs by counteracting 3’−5’ exonucleolytic activity by PARN. CUG triplet repeat RNA-binding protein 1 (CUGBP1) interacts with miR-122 and recruits PARN135.
(C) Interactions of an Argonaute (AGO)-bound miRNA with a target mRNA through the miRNA seed sequence result in translation repression and mRNA degradation. Targets with extensive pairing to the 3’ end promote tailing144,151 and trimming148, and target-directed miRNA degradation142,149,150. A target mRNA that is fully complementary to a miRNA is cleaved when bound by a catalytic Argonaute, such as mammalian Ago244,49, but such pairing can also result in unloading of the miRNA from AGO154. Cell-lines: A549 (adenocarcinomic human alveolar basal epithelial cell line), Huh-7 (human hepatocellular carcinoma cell line), human cellular carcinoma, THP1 (human leukemic monocyte cell line).
Compared to 3’ adenylation, 3’ uridylation more consistently negatively regulates miRNA activity. Uridylation by terminal uridylyltransferase 4 (TUT4; also known as ZCCHC11) of miR-26b in a human adenocarcinoma cell line reduces target repression139. Moreover, TUT4-mediated uridylation primes numerous miRNAs for degradation during T-cell activation140 (FIGURE 3A).
To date, the enzymes reported to mediate NTA include GLD2133,141, PAPD5141, poly(A) polymerase γ and poly(A) RNA polymerase, mitochondrial132 (adenylation) TUT7, TUT4139, TUT1 (uridylation). Species-dependent differences in NTAs have been observed, with uridylation being more common in C. elegans and adenylation more common in mouse and human132. In summary NTA appears to influence, both positively and negatively, the stability of specific miRNAs in specific cell types. Recent findings demonstrate that interactions between the miRNA 3’ region and target RNAs can substantially affect miRNA modification and turnover (see below), thereby providing a potential mechanism for the cellular-context-dependence of NTA.
miRNA turnover
miRNAs are generally thought to be stable in vivo, but turnover rates are regulated by multiple factors and across different miRNAs can range from minutes to days142. Turnover rates are miRNA-specific and isomir-specific and thus linked to miRNA sequence143,144; indeed, a 5’ guanine or cytosine is associated with faster turnover rates compared to a uracil in this position143. Stability can also be miRNA-specific. For example, miRNAs in mouse fibroblast 3T3 cells are generally stable, except for miR-16 family members, which are intrinsically unstable145. This instability allows miR-16 levels to vary with, and thereby help regulate, the progression of the cell cycle. Some miRNAs are stable unless specifically degraded in response to developmental cues; for example, miR-150 is rapidly lost when murine naive T cells differentiate into Th1 and Th2 lymphocytes146. Turnover rates can depend also on tissue context: fast turnover rates are generally observed in neuronal miRNAs compared to miRNAs in other tissues147.
One mechanism of miRNA destabilization is termed target RNA–directed miRNA degradation (TDMD) 148. In TDMD, target RNAs with extensive complementarity to both 5’ and 3’ regions of a miRNA promote its turnover (FIGURE 3C). TDMD is often associated with 3’ NTA, or “tailing”, and with 3’−5’ trimming of the degraded miRNA142,149,150. It has been proposed that 3’ tailing extends the AGO-loaded miRNA144,151 until a 3’−5’ nuclease can bind and degrade it148. This hypothesis is consistent with the observation that 3’-remodeled isomirs generally coincide with TDMD. However, the hypothesis has recently been challenged by the observation that tailing of miR-7 by GLD2, which coincides with TDMD of miR-7 in mouse neurons, is not required for efficient miR-7 degradation152. Similarly, the 3’−5’ exoribonuclease DIS3-like exonuclease 2 (DIS3L2) was implicated in 3’ trimming associated with TDMD of miR-27a, but DIS3L2 depletion did not impact TDMD efficiency151. Thus, it is possible that TDMD and miRNA 3’ remodeling are distinct processes, and the molecular mechanism of TDMD therefore remains poorly understood.
Although viral inducers of TDMD have been known for several years (see below), endogenous inducers of TDMD have only recently been reported. In zebrafish, miR-29b is selectively degraded in the cerebellar granule cell layer by a long non-coding RNA (lncRNA)and in mice by the 3’ UTR of a protein-coding gene153. Both transcripts show extensive sequence similarity around a highly complementary target site for miR-29b. Similarly, the lncRNA Cyrano can induce TDMD of miR-7, thereby affecting the stability of a miR-7-interacting circular RNA (circRNA) (see below) 152. Furthermore, miR-503, which is inherently unstable in 3T3 cells, can be stabilized by mutating either its seed or 3’ regions145, raising the possibility that its inherent instability is mediated by TDMD. It is possible that many inherently unstable miRNAs are subjected to TDMD through yet-to-be-identified target RNAs.
A biochemical screen using HEK239 cell (a human embryonic kidney cell line) lysate revealed that miRNA degradation is promoted particularly by seedless targets with high complementarity to miRNA 3’ ends, with complementarity of the 3’-terminal nucleotide being crucial for degradation150. Combined with the observation that the sequences of both the 5’-end and the 3’-end influence turnover rates143,144, this might partially explain the tissue-specific and miRNA-specific effects of NTA on stability. Alternatively, highly complementary targets can destabilize the AGO–miRNA interaction and promote release of the guide strand154, although miRNAs appear to be stabilized by high abundance of seed-matching targets 155.
In addition to exonucleolytic miRNA turnover, Tudor staphylococcal nuclease, which is responsible for degradation of A-to-I edited double-strand RNA99, was found to target miR-31–5p, miR-29b-3p and miR-125a-5p in HEK293T cells. Other tested miRNAs did not appear to be affected, suggesting this is a miRNA-specific mechanism, though how selectivity is established remains to be determined156. Finally, phosphorylation-dependent regulation of miRNAs was recently discovered for the tumor suppressor miR-34. The nuclei of four human cancer cell lines were found to contain pools of inactive, mature single-stranded miR-34 lacking a 5’ phosphate and not bound by AGO157. Irradiation of the cells led to 5’ phosphorylation, export to the cytoplasm and loading into AGO of miR-34. The serine-protein kinase ATM and the RNA kinase Clp1 were required for miR-34 phosphorylation. It is unclear how widespread this phenomenon is, but these findings suggest that miRNAs can be maintained in inactive form and rapidly activated in response to stimuli.
Post-translational modification of AGO
AGO proteins undergo PTM on multiple residues (FIGURE 4). Phosphorylation is the best characterized AGO PTM and occurs at three main sites: at the L2 linker, on Ser387 and Tyr393 of human Ago2; at the miRNA 5’-end binding region in the MID domain, on Tyr529; and at a surface-exposed loop in the PIWI domain known as the eukaryotic insertion or the S824–S834 cluster, which includes Ser824, Ser828, Thr830, Ser831 and Ser834.
Figure 4: The activity and the stability of miRISC is modulated by post-translational modifications (PTMs) of Argonaute proteins.
(A) Phosphorylation of Ser387 (S387) in the L2 region of Argonaute (AGO) was found to be mediated by MAP kinase-activated protein kinase 2 (MAPKAPK2) 158 and AKT3159 in vitro. Ser387 phosphorylation increases miRNA activity by stimulating the assembly of miRNA-induced silencing complexes, and reduces translocation of Ago2 to multivesicular endosomes and secretion of exosomes162. Phosphorylation of the nearby Tyr393 (Y393), also in the L2 region, decreases the miRNA–Ago2 association, thereby reducing miRNA activity164,165. Tyr 529 (Y529) is located in the middle (MID) domain, near the miRNA 5’-nucleotide binding pocket, and its phosphorylation prevents miRNA loading160. No function has yet been assigned to phosphorylation sites in the Piwi-Argonaute-Zwille (PAZ) domain (S253, T303, T307) and the P-element induced wimpy testes (PIWI) domain (S798) 160. Additional AGO PTMs include Pro700 (P700) 4-hydroxylation, which increases Ago2 stability171. Lys402 (K402) SUMOylation, which was reported to either destabilize Ago2177 or be required for full siRNA activity178; and poly(ADP-ribosylation) (PARylation), which inhibits miRNA activity173,174, presumably by decreasing target accessibility.
(B) The S824–S834 cluster in the eukaryotic insertion region of human Ago2 undergoes a phosphorylation cycle, which regulates AGO–target interactions. Phosphorylation of the Ser residues in the cluster by casein kinase I isoform α (CSNK1A1) favors target release. Subsequent dephosphorylation by the serine/threonine-protein phosphatase 6 complex ANKRD52–PPP6C primes Ago2 for the next round of target binding169,170.
Several studies report phosphorylation of Ser387 in the L2 region of human Ago2 (FIGURE 4A) 158–160. Ser387 phosphorylation is stimulated by the p38 MAPK pathway-mediated stress response, for example by MAP kinase-activated protein kinase 2 in vitro158. The proto-oncogene serine/threonine kinase AKT3 also phosphorylates Ser387 in vitro and in HeLa cells and its depletion or the expression of a S387A Ago2 mutant (which cannot be phosphorylated) led to de-repression of a luciferase reporter and weakened the Ago2 interactions with TNRC6A159. Similarly, Ser387 phosphorylation is necessary for interactions between Ago2 and LIM domain-containing protein 1, which is a protein suggested to bridge Ago2 and GW182161. Blocking Ser387 phosphorylation also reduced accumulation of Ago2 in cytoplasmic foci with GW182158,159 and was suggested to reduce trafficking of Ago2–miRNA complexes to multivesicular endosomes[G] and reduce the secretion of exosomes[G] 162. Notably however, a recent investigation found that S387A Ago2 did not reduce co-localization GW182 in HeLa cells, indicating that additional factors may be involved in enabling the Ago2–GW182 interaction163. The combined data indicate that Ser387 phosphorylation promotes miRNA function by stimulating the assembly of miRISC.
Tyr393 is adjacent to Ser387 in the L2 region and its phosphorylation is also well-documented160,164,165 (FIGURE 4A). It appears to be mediated by epidermal growth factor receptor (EGFR) and stimulated by hypoxic stress164. Overexpression of oncogenic RAS reversibly inhibits protein tyrosine phosphatase 1B (also known as tyrosine-protein phosphatase non-receptor type 1), resulting in Ago2 hyperphosphorylation at Tyr393165. Both studies reported diminished interactions between Ago2 and Dicer and reduced levels of Ago2-associated miRNAs following Tyr393 phosphorylation. Thus, in contrast to Ser387, Tyr393 phosphorylation appears to negatively regulate miRNA activity by inhibiting the loading of miRNAs into Ago2, thereby promoting tumorigenesis.
Tyr529 phosphorylation also potentially blocks miRNA loading160 (FIGURE 4A). Tyr529 binds the 5’ phosphate and first nucleotide of miRNAs loaded into AGO41,166,167 Tyr529 phosphorylation is therefore expected to preclude miRNA binding, as indeed was demonstrated in Tyr529 Ago2 mutants160. Tyr529 phosphorylation was also associated with decreased miRNA binding to Ago2 during macrophage activation168.
A phosphorylation cycle of residues S824–S834 in Ago2 regulates the release of target mRNAs169,170 (FIGURE 4B). Target-binding leads to phosphorylation of these residues by casein kinase I isoform α, which reduces the affinity of Ago2 for mRNA and enables target release. The serine/threonine-protein phosphatase 6 complex ANKRD52–PPP6C dephosphorylates the residues, which primes Ago2 for binding a new target. Interrupting this cycle strongly inhibited miRNA activity, and the cycle was suggested to prevent overly long association with mRNA targets170.Alternatively, the phosphorylation cycle may represent an AGO-regulation mechanism that is mediated by mRNA-binding proteins. The cluster is conserved in human AGO proteins, mouse Ago2, rat Ago2, zebrafish Ago2, the C. elegans argonaute ALG1169, and in the fruit fly Ago1 (but not in the fruit fly Ago2, which is used for siRNA-mediated target cleavage) 170.
Additional Ago2 phosphorylation sites have been reported in the PAZ domain (Ser253, Thr303, Thr307) and the PIWI domain (Ser798), but their function is currently unknown160 (FIGURE 4A).
In addition to phosphorylation, other AGO PTMs are known (FIGURE 4A). Hydroxylation of Ago2 Pro700 appears to increase the stability of Ago2 in mouse and human cells171. Interestingly, Pro700 is adjacent to a Trp-binding pocket in Ago2 that is used to interact with GW18241,82, and its 4-hydroxylation thus could potentially alter the assembly of miRISC, though this has not yet been demonstrated.
Ago1–4 undergo poly(ADP-ribosylation) (PARylation) in human cell lines172. Poly(ADP-ribose) (PAR) is a polymer of ADP-ribose units that can be enzymatically added to Asp, Glu and Lys residues. PARylation regulates the cellular stress response, particularly the formation of cytoplasmic stress granules, which regulate mRNA translation and stability172. PARylation of Ago1–4 reduced translation repression and endonucleolytic cleavage and was hypothesized to impair target accessibility173. PARylation-induced down regulation of miRNA activity was observed in antiviral response in HEK293 cells: cytotoxic interferon-stimulated genes are generally heavily regulated by miRNAs, and miRNA inhibition through AGO PARylation might enable a cytotoxic response to viral infection174.
The ubiquitin–proteasome system reduces the abundance of the fruit fly Ago1 and mouse Ago2175, and drives global miRNA down-regulation during T cell activation176. Finally, two groups have reported Ago2 SUMOylation at Lys402, with contradicting consequences: SUMOylation might destabilize Ago2177, or be required for full Ago2 siRNA-mediated activity178.
Compared to AGO proteins, less is known about the regulation of other proteins involved in miRNA function. Tripartite motif-containing protein 65 ubiquitylates TNRC6 in HEK293 cells, leading to proteasomal degradation and de-repression of miRNA targets179. TNRC6A has long been known to be phosphorylated in mammalian cells180, and phosphorylation of the PAM2 motif [G] of TNRC6C was suggested to reduce its interactions with PABPC1181. A recent mass-spectrometry study of the TNRC6A interactome in HeLa cells revealed a large number of binding partners, but it is currently unknown whether any of these regulate TNRC6A182.
Regulation of miRNAs by sequestration
The competing endogenous RNA (ceRNA) hypothesis 183 postulates that an increase in the cellular concentration of a miRNA target RNA would reduce the cytoplasmic availability of the specific miRNA by binding it, thereby de-repressing other mRNAs that are targets of the same miRNA; this is similar to the proposed function of miRNA sponges[G] 184. Thus, according to the ceRNA model, gene expression would be shaped by the global competition of target RNAs for miRNAs185 (FIGURE 5A). The potential efficacy of ceRNAs is controversial, as the increased expression of any single ceRNA is expected to increase only slightly the total number of target sites of a miRNA, and thus be unlikely to meaningfully affect miRNA activity186–189. Two studies measuring the cellular abundance of miRNAs and miRNA target sites proposed that the ability of a putative ceRNA to de-repress the expression of other target mRNAs depends on miRNA abundance190 and/or target site abundance186. Both studies agreed that, in most cases, unphysiologically high levels of a ceRNA would generally be necessary to yield a biologically meaningful effect186,190. Mathematical modeling of miRNA distribution in the targetome indicates that a ceRNA must increase the cellular abundance of target sites at least two fold to be effective188. On the other hand, it was also noted that ceRNA efficacy could be enhanced if local concentrations of ceRNAs and/or miRNAs deviated strongly from the cytoplasmic average188. The discovery of the phosphorylation cycle regulating AGO activity also raises interesting new possibilities, as AGO–target binding might be modulated by mRNA-associated RBPs170. With respect to the ceRNA hypothesis, such a mechanism could alter the affinities of different target sites that otherwise seem equivalent based on miRNA-complementarity alone. Additionally, although this has yet to be demonstrated, ceRNAs that promote TDMD could function at lower concentrations by reducing miRNA abundance189. Thus, although the original ceRNA hypothesis may be implausible under general cellular conditions, ceRNA mechanisms may still have a biological role 188.
Figure 5: miRNA sequestration by endogenous and viral RNAs.
(A) The competing endogenous RNA (ceRNA) hypothesis states that a newly expressed RNA can compete with the already present microRNA (miRNA) targets for cytoplasmic miRNA-induced silencing complexes (miRISCs), potentially leading to de-repression of certain genes185. However, a ceRNA is unlikely to lead to gene de-repression when it is expressed at typical physiological levels186–190.
(B) Long non-coding RNAs (lncRNAs), pseudogenes and mRNAs can have ceRNA activity. In mice, long intergenic non-protein coding RNA of muscle differentiation 1 (Linc-md1) contains one binding site for miR-133 (blue) and two for miR-135 (green). By sequestering these miRNAs, the muscle-specific transcription factors mastermind-like protein 1(Maml1) and myocyte-specific enhancer factor 2C (Mef2c) are de-repressed, thereby promoting myoblast differentiation191. The pseudogene phosphatase and tensin homolog pseudogene 1 (PTENP1) shares many miRNA target sites with the tumor suppressor PTEN, and can de-repress PTEN in human cells 183. Similarly, the mouse zinc finger E-box binding homeobox 2 (Zeb2) mRNA can de-repress PTEN193.
(C) Different viral mechanisms affect miRNA function. Hepatitis C virus (HCV) harbors two binding sites for miR-122 at the very end of the 5’ untranslated region (UTR) of its RNA genome203. These recruit argonaute2 (AGO2)–miR-122 complexes204 to protect the viral RNA from the cellular antiviral response and the activity of exonucleases205, and functionally sequester miR-122 and de-repress hepatic miR-122 target mRNAs207.
Bovine viral diarrhea virus (BVDV) is an RNA virus that contains a binding site for miR-17 in its 3’ UTR; miR-17 binding enhances the stability of the viral RNA. The site functionally sequesters the miRNA and de-represses its cellular targets208.
Herpesvirus saimiri (HVS) produces short non-coding RNAs termed herpesvirus saimiri uracyl-rich RNAs (HSURs), two of which are known to modulate miRNA function. HSUR1 binds miR-27a and promotes its TDMD, which leads to de-repression of miR-27a cellular targets and promotes T cell activation212. HSUR2 binds miR-142–3p and miR-16 and tethers them to cellular target mRNAs, which prevents apoptosis214.
The controversy notwithstanding, the list of potential ceRNAs is growing and includes lncRNAs, pseudogenes, mRNAs (FIGURE 5B) and specific circRNAs187. In mice, long intergenic non-protein coding RNA of muscle differentiation 1 (Linc-md1) is proposed to drives myoblast differentiation by sequestering miR-133 and miR-135, which target muscle-specific transcription factors191. Similarly, in human cells the untranslated pseudogene phosphatase and tensin homolog pseudogene 1 (PTENP1) de-represses PTEN183. This was suggested to be a common function of pseudogenes187, as they regularly share miRNA target sites with their parent genes. Recently the overexpression of the Braf pseudogene was found to promote B cell lymphoma in mice through a possible ceRNA mechanism192. mRNAs have also been suggested to function as ceRNAs; for example, PTEN expression is regulated not just by pseudogenes, but also by numerous protein-coding ceRNAs193,194 (FIGURE 5B). Different types of ceRNAs were found to crosstalk in a network of various oncogenic pathways in glioblastoma195.
circRNAs are enigmatic ncRNAs with dynamic and complex expression patterns, whose function is still poorly understood196. The circRNA CDR1 antisense RNA (CDR1as; also known as ciRS-7), is expressed in human and mouse brains197,198. Because CDR1as contains many binding sites (63–74) for miR-7, it acts as a miR-7 sponge when overexpressed197. In support of the sponge mechanism, depletion of CDR1as in HEK293 cells led to downregulation of miR-7 target mRNAs198. However, knockout of CDR1as in mice led to reduction in miR-7 levels and increase in the levels of miR-7 target genes in the brain, suggesting the sponge effect stabilizes miR-7, as none of its binding sites in CDR1as possesses the extensive 3’ complementarity required for TDMD199. The miR-7 molecules can be released from CDR1as through its slicing by miR-671, for which CDR1as contains a highly-complementary target site. CRD1as may thus bind to miR-7 in order help to localize it to specific subcellular compartments197–199. The lncRNA Cyrano was proposed to be responsible for the increased turnover of miR-7 upon CDR1as depletion199, and has now been reported to induce TDMD of miR-7152. Interestingly, Cyrano depletion increased miR-7 levels and led to a decrease in CDR1as levels, in part due to an enhanced miR-671-mediated degradation of CDR1as152.
Although thousands of circRNAs have been identified, only a handful stand out as harboring large numbers of miRNA target sites200. In addition to CDR1as, ten different circRNAs derived from zinc finger genes contain 7–24 target sites for miR-23, miR-181 or miR-199 families200. Another proposed sponge, the testis-specific circRNA sex-determining region Y, possesses 16 sites for miR-138 in mice197, but only one in humans200. Thus, whereas some circRNAs may interact with miRNAs, this does not appear to be a major function of most of them.
Viral modulation of miRNA activity
Viruses have evolved to repurpose or modulate host miRNAs for their replication, which often affects miRNA function (FIGURE 5C). The best studied interaction is between the Hepatitis C virus (HCV) and miR-122. HCV is a positive-sense single-stranded RNA virus of the Flaviviridae family, which causes acute and chronic liver infection201. Its highly structured 5’ UTR contains an internal ribosomal entry site202, and upstream of it, at the very 5’ end of the genomic RNA, two binding sites for the liver-abundant miR-122203 (FIGURE 5C). The binding sites recruit Ago2–miR-122 to the uncapped 5’ end of the viral RNA204 to protect it from the cellular antiviral response and exonuclease activity205,206. This functionally sequesters cytoplasmic miR-122, which is a tumor suppressor miRNA responsible for hepatic maintenance, and potentially explains the link between chronic HCV infection and an increased risk of developing hepatocellular carcinoma207.
Another virus from the Flaviviridae family that binds host miRNAs is bovine viral diarrhea virus, which is an economically important cattle virus. Binding of let-7 and miR-17 at a structured region of the viral RNA 3’ UTR promotes viral replication. miR-17 also stabilizes the viral RNA upon binding and enhances its translation through an unknown mechanism. The binding of miR-17 to the viral RNA de-represses cellular targets of the miRNA, an effect also observed with classical swine fever virus208. The detection of similar interactions between miRNAs and viral genomic RNA of different members of the Flaviviridae family raises the question of whether this mechanism might be even more widespread in this family, which includes other health-relevant members such as dengue virus and zika virus.
Hepatitis B virus (HBV), which is a partially double-stranded DNA virus unrelated to HCV, directly down-regulates miR-122 in liver cells, as this miRNA appears to inhibit viral infection209. A hybrid viral–human transcript210 contains multiple miR-122 binding sites, one of which resembles a TDMD element that depletes cellular miR-122 levels and promotes loss of hepatic function and liver damage in mice211.
Several members of the Herpesviridae family also modulate host miRNA function. Herpesvirus saimiri (HVS) expresses H. saimiri uracyl-rich RNAs (HSURs), which are short ncRNAs with structural similarity to small nuclear RNAs[G]. Two HSURs harbor binding sites for host miRNAs: miR-16 (HSUR2), miR-27a (HSUR1) and miR-142–3p (both HSUR1 and HSUR2). HSUR1 reduces miR-27a levels in infected marmoset T cells through TDMD, thereby de-repressing miR-27a cellular target mRNAs and promoting T cell activation212. TDMD of miR-27a is required for efficient HVS replication, as viral strains with HUSR1 bearing a mutated miR-27a binding site have reduced titers213. HSUR2 does not deplete the miRNAs it binds, but instead acts as a tether that recruits AGO–miR-142–3p and AGO–miR-16 complexes to cellular mRNAs that encode pro-apoptosis factors, thereby inducing silencing of the tethered mRNAs and preventing apoptosis214.
Analogously to HSUR1 function, TDMD and reduction in miR-27a levels was observed in mouse cell lines and primary macrophages upon murine cytomegalovirus infection215, mediated by a transcript harboring a miR-27a target site with substantial 3’-end complementarity216. Similarly, human cytomegalovirus (HCMV) targets miR-17 and miR-20a, two members of the miR-17–92 cluster. Degradation of these miRNAs, which is mediated by complementarity with sites in viral non-coding RNA, accelerates virus production during HCMV infection217.
miRNA transport from the cytoplasm
As miRNAs function in the cytoplasm, their activity can be modulated by transferring them to the nucleus or to extracellular vesicles, where they potentially have location-specific functions218.
Nuclear localization of miRNAs
miR-21 was the first miRNA to be found in the nucleus, as 20% of total miR-21 is present in nuclear extracts49; miR-29b was found to be predominantly localized to the nucleus owing to a 3’ hexanucleotide nuclear localization signal219. Ago2, Dicer, TARBP and GW182 are present in the nucleus and form complexes. In C. elegans, the AGO homolog ALG-1 uses mature let-7 to bind pri-let-7 transcripts in the nucleus and promote their biogenesis, thereby creating a positive feedback loop220. Recent single molecule studies in mammalian cells indicated that nuclear miRNAs do not repress complementary targets. The importance of the miR-29b hexanucleotide localization signal was also questioned, as miRNA nuclear localization was found to be primarily dependent on the presence of nuclear targets155. These findings suggest that our understanding of nuclear miRNA activity is still very partial.
Circulating miRNAs
Circulating miRNAs[G] are potential cancer biomarkers221; their discovery in exosomes led to the hypothesis that they might contribute to inter-cellular signaling222 (FIGURE 6A). It remains unclear if only a small fraction of circulating miRNAs travel within exosomes (~10% or less in plasma) 223,224, or if exosomes contain the majority (83–99% in serum) 225 of circulating miRNAs. These apparently contrasting observations could be due to technical differences in exosome isolation, or differences between serum and plasma225.
Figure 6: Mechanisms of sorting miRNAs into exosomes.
(A) miRNAs can be packaged into exosomes and thus may contribute to inter-cellular signaling. Uptake of exosomes can be receptor-mediated or receptor-indepedent278. Upon entering a target cell, exosome-delivered miRNAs are speculated to regulate target mRNAs222.
(B) Although the biological function of exosomal miRNAs is still incompletely understood, multiple mechanisms direct miRNAs into exosomes. Argonaute 2 (Ago2)-dependent sorting of specific miRNAs has been reported in isogenic colon cancer cells, and phosphorylation of Ago2 Ser387 inhibited loading of some miRNAs into exosomes162. Sorting based on miRNA-sequence complementary of exosomal long non-coding RNAs (lncRNA) was shown for miR-149–3p in prostate cancer cells232. Exosomal RNA-binding proteins (RBP) can direct miRNAs into exosomes by binding ‘exomotifs’ at the miRNA 3’ ends. The exomotif GGAG promotes exosomal sorting of miR-198 by heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNPA2B1) in human primary T cells233, and the exomotif GGCU promotes hnRNPQ -mediated exosomal sorting of miRNAs in murine hepatocytes234. No exomotifs are known for Y-box-binding protein 1 (YBX1)-mediated sorting of miR-223 in HEK293T cells235 and for major vault protein (MVP)-mediated sorting of miR-193a236. Finally, in human B cells 3’-adenylated miRNAs are depleted in exosomes whereas 3’-uridylated miRNAs enriched in exosomes237. Exosomes have been reported to carry proteins, different RNAs and miRNA biogenesis components 226, but also Ago2–miRNA complexes162 and AGO-free miRNAs235.
Circulating miRNAs may have regulatory functions. Breast-cancer cell derived exosomes contain pre-miRNAs and the proteins required for cell-independent miRNA maturation, can induce cell proliferation in culture226, and might have prometastatic properties in vivo227,228. A major concern with these studies is the technical challenge of clearly distinguishing between miRNA-mediated effects and changes caused by other exosomal components229. Moreover, exosomes from five different sources were found to have less than one miRNA molecule per exosome on average230. Nevertheless, a recent study showed that brown adipose tissue is a major source of exosomal miRNAs in humans and mice, in support of a functional role of exosomal miRNAs231. Specifically, exosomes derived from a donor mouse expressing an exogenous miRNA in brown adipose tissue were found to repress a reporter gene in the liver of an acceptor mouse231.
A biological function of exosomal miRNAs would require miRNA-specific mechanisms of sorting into exosomes (FIGURE 6B). One proposed sorting mechanism is through exosomal lncRNAs, based on the observed enrichment in exosomes of prostate cancer cell lines of miR-149–3p with lncRNAs harboring its target sites232. Exosomal RBPs could also promote sorting by directly interacting with miRNA motifs termed ‘exomotifs’. The exomotif GGAG located at the miRNA 3’ end was suggested to mediate binding of miR-198 and other exosomal miRNAs to the exosomal RBP heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNPA2B1) in human primary T cells, thereby directing the miRNAs to exosomes233. Similarly, the exomotif GGCU, which is also preferentially located at miRNA 3’ ends, was suggested to recruit hnRNPQ (also known as SYNCRIP) in murine hepatocytes, yielding exosomal secretion of the bound miRNAs234. Other RBPs proposed to mediate miRNA sorting into exosomes, such as Y-box-binding protein 1 (also known as nuclease-sensitive element-binding protein 1) for miR-223 in HEK293T cells235 and major vault protein for miR-193a in colon tumor cells in mice236, lack specific associated recognition motifs. The tumor suppressor miR-193a is an interesting case, because the main reason for its sorting into exosomes appears to be its removal from the cytoplasm236. NTA at miRNA 3’ ends may also influence sorting, as reported for human B cells and B-cell-derived exosomes, where adenylation was associated with miRNA depletion from and uridylation with miRNA enrichment in exosomes237. Finally, Ago2 Ser387 phosphorylation by the KRAS–MAPK pathway antagonizes exosomal sorting of Ago2 in colon cancer cells162. Although free, single-stranded miRNAs are rapidly degraded in the cell and thus are hardly functional155, some reported sorting mechanisms233,234 focus on miRNAs, with no mentioning of AGO proteins, or do not detect AGO proteins in exosomes232. Clearly, more studies are required to fully understand the mechanistic and functional aspects of exosomal miRNAs.
Regulation of miRNA target sites
The activity of miRNAs can also be modulated through changes in their target sites. RNA editing can alter target site complementarity and deregulation of target site editing is associated with cancer238,239. Similarly, the t1A binding site on AGO does not recognize N6-methyladenosine modification in RNA, raising the possibility that adenosine methylation of target mRNAs could subtly modulate their targeting by miRNAs, though this idea has yet to be tested in a cellular context57. Less subtly, formation of mRNA 3’ UTR isoforms can add or remove miRNA target sites, thereby altering mRNA susceptibility to miRNAs in a cell-type-specific or tissue-specific manner240.
RBPs also can modulate miRNA–target interactions. Pumilio promotes cell-cycle re-entry of quiescent cells by binding the 3’ UTR of the mRNA encoding the tumor suppressor p27 (also known as CDKN1B) and remodeling it, thereby exposing target sites for miR-221 and miR-222241. AU-binding factor 1 (AUF1) facilitates interactions of Ago2 with mRNAs in HeLa cells242. The AU-rich element binding protein human antigen R (HuR; also known as ELAV-like protein 1) de-represses the miR-122-targeted mRNA encoding cationic amino acid transporter 1 (CAT1) by binding an AU and U rich region in the mRNA, thereby preventing miRNA function but apparently not miRNA binding85. Regulation by HuR appears to be widespread, as over 75% of human mRNAs that harbor miRNA binding sites also possess HuR binding sites, often in overlap or in close proximity243. HuR prevents repression of p53 by miR-125b by binding to the mRNA and causing miRISC dissociation244, and of programmed cell death protein 4 (PDCD4) by miR-21 by binding to targets in the mRNA and displacing miRISC and by directly binding to miR-21 in MCF cells245. RBPs have also been suggested to regulate the Ago2 phosphorylation cycle, but no specific RBP has been discovered yet to do so170.
Interestingly, in extracts of most adult mouse tissues, miRNAs are found in low molecular weight miRISC of ~100 kDa, which is approximately the molecular mass of AGO with its miRNA guide and not binding mRNA. By contrast, in cell lines most miRNAs are found in high molecular weight miRISC (up to 2 MDa) containing GW182, other proteins and target mRNAs. Consequently, in transformed cells miRNA abundance may correlate more strongly with miRNA activity than in primary tissues, where additional regulation of AGO activity by the cell appears present246. Findings obtained in cell lines should therefore be considered with caution.
Future perspective
A number of interesting questions remain unanswered regarding the regulation of miRNA function. Non-templated nucleotide addition at the 3’ end of miRNAs has been recognized as a widespread process131,132. However, neither the tissue-specific significance of the added nucleotides that promote miRNA stability or degradation, nor the mechanism by which the cell selects which miRNAs to modify are known. Moreover, an extended miRNA 3’ end has been shown to improve interactions between miR-122 and HCV genomic RNA113, and to alter the function of miR-222 to promote apoptosis in a breast cancer cell line114, suggesting that 3’-isomir variation may affect miRNA targeting in ways that remain to be understood. Thus, whereas altered seed-targeting has been explored in 5’ isomirs, very little is known about the functions and properties of 3’ isomirs.
The role of mRNA secondary and tertiary structure in miRNA–target recognition is also largely unexplored. Structures that render target sites inaccessible to miRISC clearly inhibit silencing247,248. However, miRNA binding sites have been identified within heavily structured segments of RNA viruses203,208, raising the possibility that some structures may not be detrimental for targeting or might even enable recognition by miRNAs. Indeed, effective miR-159 target sites in Arabidopsis thaliana require an adjoining structural element composed of two stem loops249. The interactions between RNA structures and AGO and the degree to which they shape miRNA targeting remain unknown.
Although the activity of exosomal miRNAs and the presence of miRNA-selective exosomal sorting mechanisms suggest exosomal miRNAs participate in intercellular communication, evidence in a physiological context remains elusive. The reported scarcity of miRNAs in exosomes230 and the difficulty of disentangling miRNA-mediated effects from effects of other exosomal cargoes add to this challenge229. Similarly, although exosomal Ago2-loaded miRNAs have been detected162,226, so were single-stranded miRNAs, which would require loading into AGO in the target cells235. Our understanding of exosomal miRNAs is thus far from complete.
Finally, the recent report of molecular condensation properties of Argonaute and TNRC6250 connects miRNA regulation to the growing field of biological phase separation251. The data demonstrate that miRISC can form large molecular condensates in vitro and in living cells and it was hypothesized that the ability to form higher order complexes through molecular condensation may allow miRISC to organize miRNA–target interactions within the cytoplasm and thereby modulate rates of mRNA translation and decay. This raises the possibility that miRNA activity is regulated through the assembly of miRISC itself, by modulation of the biophysical properties of miRISC components.
In conclusion, although the first evidence of miRNAs was discovered over 25 years ago and major advances have been made since, many aspects surrounding the complex mechanisms that govern the activity of these tiny regulators remain to be discovered and explored.
Supplementary Material
Acknowledgments
LFRG is supported by an Advanced Postdoc Mobility fellowship from the Swiss National Science Foundation, project number P300PA_177860. IJM is supported by NIH grants R01-GM104475 and R01-GM115649.
Glossary
- Isomirs
Variant forms of a canonical miRNA, generated by alternative cleavage during biogenesis, RNA editing, or non-templated nucleotide addition
- guide strand
The strand in the mature miRNA duplex generated by Dicer cleavage, which is loaded into AGO and used to identify complementary sites in target mRNAs
- Metastable
A stable state in a system that is not the state of least energy
- mRNA decay
Controlled nucleolytic degradation of mRNA, usually starting with de-adenylation, and then progressing either with 3’−5’ exonucleolytic processing, or de-capping and 5’−3’ exonucleolytic processing
- multivalent interactions
Interactions between binding partners mediated by multiple individual, often times relatively weak, binding events or points of contact
- multivesicular body
A type of late endosome that contains intraluminal vesicles formed by budding into the lumen of the endosome. Its content can be degraded by fusion with lysosomes, or released into the extra-cellular space through fusion with the cell membrane
- exosomes
A type of extracellular vesicles, ~50–150 nm in diameter, which contain proteins, lipids and RNA and can carry cargo to target cells
- PAM2 motif
Poly A binding protein interacting motif 2, which mediates the interaction between GW182 and PABP
- small nuclear RNAs
Small non-coding RNAs localized to the nucleus that form complexes with proteins and are part of the splicing machinery
- circulating miRNAs
miRNAs present in circulation and found either as AGO:miRNA complexes or as vesicle (exosome) cargo
- miRNA cluster
Multiple miRNAs located in close proximity on the genome and transcribed as a single pri-miRNA
- seedless target
A miRNA target with severely reduced complementarity to the miRNA seed
- Trp-binding pocket
Ago possesses three pockets located in the PIWI domain, that bind Trp and mediate the interaction with GW182
- Stress granules
Granules formed by untranslating mRNAs, initiation factors, RBPs and other proteins, when translation stops as part of the cellular stress response.
- miRNA sponge
A transcript expressed from a strong promoter in a cell, which contains multiple target sites for a specific miRNA, thereby derepressing the miRNAs targets
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Swarts DC et al. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol 21, 743–753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jonas S & Izaurralde E Towards a molecular understandingof microRNA-mediated gene silencing. Nat. Rev. Genet 16, 421–433 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL & Ambros V The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Wightman B, Ha I & Ruvkun G Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Pasquinelli AE et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Lau NC, Lim LP, Weinstein EG & Bartel DP An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, Rauhut R, Lendeckel W & Tuschl T Identification of novel genes coding for small expressed RNAs. Science 294, 853–858 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Lee RC & Ambros V An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E et al. Dicer is essential for mouse development. Nat. Genet 35, 215–217 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Chong MMW et al. Canonical and alternate functions of the microRNA biogenesis machinery. Genes & Development 24, 1951–1960 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pauli A, Rinn JL & Schier AF Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet 12, 136–149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burger K & Gullerova M Swiss army knives: non-canonical functions of nuclear Drosha and Dicer. Nat Rev Mol Cell Biol 16, 417–430 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Kozomara A & Griffiths-Jones S miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman RC, Farh KK-H, Burge CB & Bartel DP Most mammalian mRNAs are conserved targets of microRNAs. Genome Research 19, 92–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteller M Non-coding RNAs in human disease. Nat. Rev. Genet 12, 861–874 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Lin S & Gregory RI MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15, 321–333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bracken CP, Scott HS & Goodall GJ A network-biology perspective ofmicroRNA function and dysfunctionin cancer. Nat. Rev. Genet 17, 719–732 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Ventura A et al. Targeted Deletion Reveals Essential and Overlapping Functions of the miR-17∼92 Family of miRNA Clusters. Cell 132, 875–886 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takamizawa J et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Research 64, 3753–3756 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Lu J et al. MicroRNA expression profiles classify human cancers. Nature Cell Biology 435, 834–838 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Schwarzenbach H, Nishida N, Calin GA & Pantel K Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 11, 145–156 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Goodison S, Li X & Hu H Prognostic cancer gene signatures share common regulatory motifs. Sci Rep 7, 1183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rupaimoole R & Slack FJ MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16, 203–221 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Llave C, Kasschau KD, Rector MA & Carrington JC Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B & Bartel DP MicroRNAs in plants. Genes & Development 16, 1616–1626 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee K, Campos H & Kolaczkowski B Evolution of Animal and Plant Dicers: Early Parallel Duplications and Recurrent Adaptation of Antiviral RNA Binding in Plants. Molecular Biology and Evolution 30, 627–641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurihara Y & Watanabe Y Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U.S.A 101, 12753–12758 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llave C, Xie Z, Kasschau KD & Carrington JC Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Rogers K & Chen X Biogenesis, Turnover, and Mode of Action of Plant MicroRNAs. Plant Cell 25, 2383–2399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borges F & Martienssen RA The expanding world of small RNAsin plants. Nat Rev Mol Cell Biol 16, 727–741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landgraf P et al. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 129, 1401–1414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha M & Kim VN Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15, 509–524 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Lee Y et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai X, Hagedorn CH & Cullen BR Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957–1966 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bracht J, Hunter S, Eachus R, Weeks P & Pasquinelli AE Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA 10, 1586–1594 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berezikov E Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet 12, 846–860 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Lim LP The microRNAs of Caenorhabditis elegans. Genes & Development 17, 991–1008 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartel DP MicroRNAs: Target Recognition and Regulatory Functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Sheng G, Juranek S, Tuschl T & Patel DJ Structure of the guide-strand-containing argonaute silencing complex. Nature 456, 209–213 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakanishi K, Weinberg DE, Bartel DP & Patel DJ Structure of yeast Argonaute with guide RNA. Nature 486, 368–374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schirle NT & MacRae IJ The Crystal Structure of Human Argonaute2. Science 336, 1037–1040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheu-Gruttadauria J & MacRae IJ Structural Foundations of RNA Silencing by Argonaute. Journal of Molecular Biology 429, 2619–2639 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diederichs S & Haber DA Dual Role for Argonautes in MicroRNA Processing and Posttranscriptional Regulation of MicroRNA Expression. Cell 131, 1097–1108 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Liu J et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Cheloufi S, Santos, dos CO, Chong MMW & Hannon GJ A Dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cifuentes D et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328, 1694–1698 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yekta S, Shih I-H & Bartel DP MicroRNA-directed cleavage of HOXB8 mRNA. Science 304, 594–596 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Karginov FV et al. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Molecular Cell 38, 781–788 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meister G et al. Human Argonaute2 Mediates RNA Cleavage Targeted by miRNAs and siRNAs. Molecular Cell 15, 185–197 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Burroughs AM et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol 8, 158–177 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dueck A, Ziegler C, Eichner A, Berezikov E & Meister G microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 40, 9850–9862 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D et al. Quantitative functions of Argonaute proteins in mammalian development. Genes & Development 26, 693–704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Betel D, Koppal A, Agius P, Sander C & Leslie C Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11, R90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agarwal V, Bell GW, Nam J-W & Bartel DP Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong N & Wang X miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43, D146–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schirle NT, Sheu-Gruttadauria J & MacRae IJ Structural basis for microRNA targeting. Science 346, 608–613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schirle NT, Sheu-Gruttadauria J, Chandradoss SD, Joo C & MacRae IJ Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ & Joo C A Dynamic Search Process Underlies MicroRNA Targeting. Cell 162, 96–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salomon WE, Jolly SM, Moore MJ, Zamore PD & Serebrov V Single-Molecule Imaging Reveals that Argonaute Reshapes the Binding Properties of Its Nucleic Acid Guides. Cell 162, 84–95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klum SM, Chandradoss SD, Schirle NT, Joo C & MacRae IJ Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. EMBO J. 37, 75–88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helwak A, Kudla G, Dudnakova T & Tollervey D Mapping the Human miRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell 153, 654–665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grosswendt S et al. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Molecular Cell 54, 1042–1054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moore MJ et al. miRNA-target chimeras reveal miRNA 3’-end pairing as a major determinant of Argonaute target specificity. Nature Communications 6, 8864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim D et al. General rules for functional microRNA targeting. Nat. Genet 48, 1517–1526 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Grimson A et al. MicroRNA Targeting Specificity in Mammals: Determinants beyond Seed Pairing. Molecular Cell 27, 91–105 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Broughton JP, Lovci MT, Huang JL, Yeo GW & Pasquinelli AE Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Molecular Cell 64, 320–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo H, Ingolia NT, Weissman JS & Bartel DP Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding L, Spencer A, Morita K & Han M The Developmental Timing Regulator AIN-1 Interacts with miRISCs and May Target the Argonaute Protein ALG-1 to Cytoplasmic P Bodies in C. elegans. Molecular Cell 19, 437–447 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Liu J et al. A role for the P-body component GW182 in microRNA function. Nature Cell Biology 7, 1261–1266 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meister G et al. Identification of Novel Argonaute-Associated Proteins. Current Biology 15, 2149–2155 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Rehwinkel J, Behm-Ansmant I, Gatfield D & Izaurralde E A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11, 1640–1647 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Braun JE, Huntzinger E, Fauser M & Izaurralde E GW182 Proteins Directly Recruit Cytoplasmic Deadenylase Complexes to miRNA Targets. Molecular Cell 44, 120–133 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Chen C-YA, Zheng D, Xia Z & Shyu A-B Ago–TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol 16, 1160–1166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Behm-Ansmant I mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Development 20, 1885–1898 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chekulaeva M et al. miRNA repression involves GW182-mediated recruitment of CCR4–NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol 18, 1218–1226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fabian MR et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4–NOT. Nat. Struct. Mol. Biol 18, 1211–1217 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Braun JE et al. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5’ exonucleolytic degradation. Nat. Struct. Mol. Biol 19, 1324–1331 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Mathys H et al. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Molecular Cell 54, 751–765 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Meijer HA et al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 340, 82–85 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Fukaya T, Iwakawa H-O & Tomari Y MicroRNAs Block Assembly of eIF4F Translation Initiation Complex in Drosophila. Molecular Cell 56, 67–78 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Fukao A et al. MicroRNAs Trigger Dissociation of eIF4AI and eIF4AII from Target mRNAs in Humans. Molecular Cell 56, 79–89 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Elkayam E et al. Multivalent Recruitment of Human Argonaute by GW182. Molecular Cell 67, 646–658.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuzuoğlu-Öztürk D et al. miRISC and the CCR4-NOT complex silence mRNA targets independently of 43S ribosomal scanning. EMBO J. 35, 1186–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eichhorn SW et al. mRNA Destabilization Is the Dominant Effect of Mammalian MicroRNAsby the Time Substantial Repression Ensues. Molecular Cell 56, 104–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhattacharyya SN, Habermacher R, Martine U, Closs EI & Filipowicz W Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124 (2006). [DOI] [PubMed] [Google Scholar]
- 86.Selbach M et al. Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Uhlmann S et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Molecular Systems Biology 8, 1–15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mestdagh P et al. The miR-17–92 MicroRNA Cluster Regulates Multiple Componentsof the TGF-b Pathway in Neuroblastoma. Molecular Cell 40, 762–773 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saetrom P et al. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 35, 2333–2342 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Broderick JA, Salomon WE, Ryder SP, Aronin N & Zamore PD Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA 17, 1858–1869 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Flamand MN, Gan HH, Mayya VK, Gunsalus KC & Duchaine TF A non-canonical site reveals the cooperative mechanisms of microRNA-mediated silencing. Nucleic Acids Res. 45, 7212–7225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsang J, Zhu J & van Oudenaarden A MicroRNA-Mediated Feedback and Feedforward Loops Are Recurrent Network Motifs in Mammals. Molecular Cell 26, 753–767 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ebert MS & Sharp PA Roles for MicroRNAs in Conferring Robustness to Biological Processes. Cell 149, 515–524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim VN, Han J & Siomi MC Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10, 126–139 (2009). [DOI] [PubMed] [Google Scholar]
- 95.Morin RD et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Research 18, 610–621 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tan GC et al. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 42, 9424–9435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim B, Jeong K & Kim VN Genome-wide Mapping of DROSHA Cleavage Sites on Primary MicroRNAs and Noncanonical Substrates. Molecular Cell 66, 258–269.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 98.Neilsen CT, Goodall GJ & Bracken CP IsomiRs – the overlooked repertoire in the dynamic microRNAome. Trends in Genetics 28, 544–549 (2012). [DOI] [PubMed] [Google Scholar]
- 99.Nishikura K A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol 17, 83–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Telonis AG, Loher P, Jing Y, Londin E & Rigoutsos I Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 43, 9158–9175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Telonis AG et al. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 45, 2973–2985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu H, Ye C, Ramirez D & Manjunath N Alternative Processing of Primary microRNA Transcripts by Drosha Generates 5′ End Variation of Mature microRNA. PLoS ONE 4, e7566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee HY & Doudna JA TRBP alters human precursor microRNA processing in vitro. RNA 18, 2012–2019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fukunaga R et al. Dicer Partner Proteins Tune the Length of Mature miRNAs in Flies and Mammals. Cell 151, 533–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim Y et al. Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. CellReports 9, 1061–1074 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Wilson RC et al. Dicer-TRBP Complex Formation Ensures Accurate Mammalian MicroRNA Biogenesis. Molecular Cell 57, 397–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee CH, Kim JH, Kim HW, Myung H & Lee S-W Hepatitis C virus replication-specific inhibition of microRNA activity with self-cleavable allosteric ribozyme. Nucleic Acid Ther 22, 17–29 (2012). [DOI] [PubMed] [Google Scholar]
- 108.Llorens F et al. A highly expressed miR-101 isomiR is a functional silencing small RNA. BMC Genomics 14, 104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Manzano M, Forte E, Raja AN, Schipma MJ & Gottwein E Divergent target recognition by coexpressed 5′-isomiRs of miR-142–3p and selective viral mimicry. RNA 21, 1606–1620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karali M et al. High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 44, 1525–1540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mercey O et al. Characterizing isomiR variants within the microRNA-34/449 family. FEBS Letters 591, 693–705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cloonan N et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 12, R126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yamane D et al. Differential hepatitis C virus RNA target site selection and host factor activities of naturally occurring miR-122 3΄ variants. Nucleic Acids Res. 45, 4743–4755 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu F et al. Naturally existing isoforms of miR-222 have distinct functions. Nucleic Acids Res. 45, 11371–11385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Salter JD, Bennett RP & Smith HC The APOBEC Protein Family:United by Structure, Divergentin Function. Trends in Biochemical Sciences 41, 578–594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Blow MJ et al. RNA editing of human microRNAs. Genome Biol. 7, R27 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kawahara Y et al. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 36, 5270–5280 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang W et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol 13, 13–21 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chawla G & Sokol NS ADAR mediates differential expression of polycistronic microRNAs. Nucleic Acids Res. 42, 5245–5255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vesely C et al. ADAR2 induces reproducible changes in sequence and abundance of mature microRNAs in the mouse brain. Nucleic Acids Res. 42, 12155–12168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shoshan E et al. Reduced adenosine-to-inosine miR-455–5p editing promotes melanoma growth and metastasis. Nature Cell Biology 17, 311–321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R & Nishikura K RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8, 763–769 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kawahara Y et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315, 1137–1140 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nigita G et al. microRNA editing in seed region aligns with cellular changes in hypoxic conditions. Nucleic Acids Res. 44, 6298–6308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vitsios DM, Davis MP, van Dongen S & Enright AJ Large-scale analysis of microRNA expression, epi-transcriptomic features and biogenesis. Nucleic Acids Res. 45, 1079–1090 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Paul D et al. A-to-I editing in human miRNAs is enriched in seed sequence, influenced by sequence contexts and significantly hypoedited in glioblastoma multiforme. Sci Rep 7, D195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tomaselli S et al. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 16, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang Y et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Research 27, 1112–1125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Negi V et al. Altered expression and editing of miRNA-100 regulates iTreg differentiation. Nucleic Acids Res. 43, 8057–8065 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Luciano DJ, Mirsky H, Vendetti NJ & Maas S RNA editing of a miRNA precursor. RNA 10, 1174–1177 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fernandez-Valverde SL, Taft RJ & Mattick JS Dynamic isomiR regulation in Drosophila development. RNA 16, 1881–1888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wyman SK et al. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Research 21, 1450–1461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Katoh T et al. Selective stabilization of mammalian microRNAs by 3’ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & Development 23, 433–438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Burns DM, D’Ambrogio A, Nottrott S & Richter JD CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 473, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Katoh T, Hojo H & Suzuki T Destabilization of microRNAs in human cells by 3′ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res. 43, 7521–7534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mansur F et al. Gld2-catalyzed 3′ monoadenylation of miRNAs in the hippocampus has no detectable effect on their stability or on animal behavior. RNA 22, 1492–1499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.D’Ambrogio A, Gu W, Udagawa T, Mello CC & Richter JD Specific miRNA stabilization by Gld2-catalyzed monoadenylation. CellReports 2, 1537–1545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boele J et al. PAPD5-mediated 3’ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc. Natl. Acad. Sci. U.S.A 111, 11467–11472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jones MR et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nature Cell Biology 11, 1157–1163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gutiérrez-Vázquez C et al. 3′ Uridylation controls mature microRNA turnover during CD4 T-cell activation. RNA 23, 882–891 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Burroughs AM et al. A comprehensive survey of 3’ animal miRNA modification events and a possible role for 3’ adenylation in modulating miRNA targeting effectiveness. Genome Research 20, 1398–1410 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rüegger S & Großhans H MicroRNA turnover: when, how, and why. Trends in Biochemical Sciences 37, 436–446 (2012). [DOI] [PubMed] [Google Scholar]
- 143.Guo Y et al. Characterization of the mammalian miRNA turnover landscape. Nucleic Acids Res. 43, 2326–2341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Marzi MJ et al. Degradation dynamics of microRNAs revealed by a novel pulse-chase approach. Genome Research 26, 554–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rissland OS, Hong S-J & Bartel DP MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Molecular Cell 43, 993–1004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Monticelli S et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 6, R71 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Krol J et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631 (2010). [DOI] [PubMed] [Google Scholar]
- 148.Ameres SL et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328, 1534–1539 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.la Mata, de M et al. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep 16, 500–511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Park JH, Shin S-Y & Shin C Non-canonical targets destabilize microRNAs in human Argonautes. Nucleic Acids Res. gkx029 (2017). doi: 10.1093/nar/gkx029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Haas G et al. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res. 44, 2873–2887 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kleaveland B, Shi CY, Stefano J & Bartel DP A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 1–31 (2018). doi: 10.1016/j.cell.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bitetti A et al. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol 25, 244–251 (2018). [DOI] [PubMed] [Google Scholar]
- 154.De N et al. Highly Complementary Target RNAs Promote Release of Guide RNAs from Human Argonaute2. Molecular Cell 50, 344–355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pitchiaya S, Heinicke LA, Park JI, Cameron EL & Walter NG Resolving Subcellular miRNA Trafficking and Turnover at Single-Molecule Resolution. CellReports 19, 630–642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Elbarbary RA et al. Tudor-SN-mediated endonucleolytic decay of human cell microRNAs promotes G1/S phase transition. Science 356, 859–862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Salzman DW et al. miR-34 activity is modulated through 5’-end phosphorylation in response to DNA damage. Nature Communications 7, 10954 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zeng Y, Sankala H, Zhang X & Graves PR Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem. J 413, 429–436 (2008). [DOI] [PubMed] [Google Scholar]
- 159.Horman SR et al. Akt-Mediated Phosphorylation of Argonaute 2 Downregulates Cleavage and Upregulates Translational Repression of MicroRNA Targets. Molecular Cell 50, 356–367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rüdel S et al. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 39, 2330–2343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bridge KS et al. Argonaute Utilization for miRNA Silencing Is Determined by Phosphorylation-Dependent Recruitment of LIM-Domain-Containing Proteins. CellReports 20, 173–187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.McKenzie AJ et al. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. CellReports 15, 978–987 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lopez-Orozco J et al. Functional analyses of phosphorylation events in human Argonaute 2. RNA 21, 2030–2038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shen J et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497, 383–387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yang M et al. Dephosphorylation of tyrosine 393 in argonaute 2 by protein tyrosine phosphatase 1B regulates gene silencing in oncogenic RAS-induced senescence. Molecular Cell 55, 782–790 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ma J-B et al. Structural basis for 5’-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666–670 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Parker JS, Roe SM & Barford D Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434, 663–666 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mazumder A, Bose M, Chakraborty A, Chakrabarti S & Bhattacharyya SN scientific report. EMBO Rep 14, 1008–1016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Quévillon Huberdeau M et al. Phosphorylation of Argonaute proteins affects mRNA binding and is essential for microRNA‐guided gene silencing in vivo. EMBO J. 36, 2088–2106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Golden RJ et al. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 542, 197–202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Qi HH et al. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 455, 421–424 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Leung A, Todorova T, Ando Y & Chang P Poly(ADP-ribose) regulates post-transcriptional gene regulation in the cytoplasm. RNA Biol 9, 542–548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Leung AKL et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular Cell 42, 489–499 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Seo GJ et al. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host & Microbe 14, 435–445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Smibert P, Yang J-S, Azzam G, Liu J-L & Lai EC Homeostatic control of Argonaute stability by microRNA availability. Nat. Struct. Mol. Biol 20, 789–795 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bronevetsky Y et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med 210, 417–432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sahin U, Lapaquette P, Andrieux A, Faure G & Dejean A Sumoylation of Human Argonaute 2 at Lysine-402 Regulates Its Stability. PLoS ONE 9, e102957 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Josa-Prado F, Henley JM & Wilkinson KA Biochemical and Biophysical Research Communications. Biochemical and Biophysical Research Communications 464, 1066–1071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li S et al. TRIM65 regulates microRNA activity by ubiquitination of TNRC6. Proc. Natl. Acad. Sci. U.S.A 111, 6970–6975 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Eystathioy T et al. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Molecular Biology of the Cell 13, 1338–1351 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Huang KL, Chadee AB, Chen CYA, Zhang Y & Shyu AB Phosphorylation at intrinsically disordered regions of PAM2 motif-containing proteins modulates their interactions with PABPC1 and influences mRNA fate. RNA 19, 295–305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Suzawa M et al. Comprehensive Identification of Nuclear and Cytoplasmic TNRC6A-Associating Proteins. Journal of Molecular Biology 429, 3319–3333 (2017). [DOI] [PubMed] [Google Scholar]
- 183.Poliseno L et al. A coding-independent function of geneand pseudogene mRNAs regulatestumour biology. Nature 465, 1033–1038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ebert MS, Neilson JR & Sharp PA MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Salmena L, Poliseno L, Tay Y, Kats L & Pandolfi PP A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 146, 353–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Denzler R, Agarwal V, Stefano J, Bartel DP & Stoffel M Assessing the ceRNA Hypothesis with Quantitative Measurements of miRNA and Target Abundance. Molecular Cell 54, 766–776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Thomson DW & Dinger ME Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet 17, 272–283 (2016). [DOI] [PubMed] [Google Scholar]
- 188.Jens M & Rajewsky N Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat. Rev. Genet 16, 113–126 (2015). [DOI] [PubMed] [Google Scholar]
- 189.Denzler R et al. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Molecular Cell 64, 565–579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bosson AD, Zamudio JR & Sharp PA Endogenous miRNA and Target Concentrations Determine Susceptibility to Potential ceRNA Competition. Molecular Cell 56, 347–359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cesana M et al. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Karreth FA et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 161, 319–332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Karreth FA et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147, 382–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tay Y et al. Coding-Independent Regulationof the Tumor Suppressor PTENby Competing Endogenous mRNAs. Cell 147, 344–357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sumazin P et al. An Extensive MicroRNA-Mediated Network of RNA-RNA Interactions Regulates Established Oncogenic Pathways in Glioblastoma. Cell 147, 370–381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Barrett SP & Salzman J Circular RNAs: analysis, expression and potential functions. Development 143, 1838–1847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hansen TB et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013). [DOI] [PubMed] [Google Scholar]
- 198.Memczak S et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013). [DOI] [PubMed] [Google Scholar]
- 199.Piwecka M et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017). [DOI] [PubMed] [Google Scholar]
- 200.Guo JU, Agarwal V, Guo H & Bartel DP Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15, 101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Webster DP, Klenerman P & Dusheiko GM Hepatitis C. Lancet 385, 1124–1135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Otto GA & Puglisi JD The pathway of HCV IRES-mediated translation initiation. Cell 119, 369–380 (2004). [DOI] [PubMed] [Google Scholar]
- 203.Jopling CL, Schütz S & Sarnow P Position-Dependent Function for a Tandem MicroRNA miR-122-Binding Site Located in the Hepatitis C Virus RNA Genome. Cell Host & Microbe 4, 77–85 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Shimakami T et al. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. U.S.A 109, 941–946 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sedano CD & Sarnow P Short Article. Cell Host & Microbe 16, 257–264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Amador-Cañizares Y, Bernier A, Wilson JA & Sagan SM miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5’ end from the cellular pyrophosphatases, DOM3Z and DUSP11. Nucleic Acids Res. 46, 5139–5158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Luna JM et al. Hepatitis C Virus RNA Functionally Sequesters miR-122. Cell 160, 1099–1110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Scheel TKH et al. A Broad RNA Virus Survey Reveals Both miRNA Dependence and Functional Sequestration. Cell Host & Microbe 19, 409–423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bandiera S, Pfeffer S, Baumert TF & Zeisel MB miR-122--a key factor and therapeutic target in liver disease. Journal of Hepatology 62, 448–457 (2015). [DOI] [PubMed] [Google Scholar]
- 210.Lau C-C et al. Viral-Human Chimeric Transcript Predisposes Risk to Liver Cancer Development and Progression. Cancer Cell 25, 335–349 (2014). [DOI] [PubMed] [Google Scholar]
- 211.Liang H-W et al. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. Journal of Hepatology 64, 278–291 (2016). [DOI] [PubMed] [Google Scholar]
- 212.Cazalla D, Yario T, Steitz JA & Steitz J Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328, 1563–1566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Marcinowski L et al. Degradation of Cellular miR-27 by a Novel, Highly Abundant Viral Transcript Is Important for Efficient Virus Replication In Vivo. PLoS Pathog. 8, e1002510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gorbea C, Mosbruger T & Cazalla D A viral Sm-class RNA base-pairs with mRNAs and recruits microRNAs to inhibit apoptosis. Nature 550, 275–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Buck AH et al. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA 16, 307–315 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Libri V et al. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl. Acad. Sci. U.S.A 109, 279–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lee S et al. Selective Degradation of Host MicroRNAs by an Intergenic HCMV NoncodingRNA Accelerates Virus Production. Cell Host & Microbe 13, 678–690 (2013). [DOI] [PubMed] [Google Scholar]
- 218.Leung AKL The Whereabouts of microRNA Actions: Cytoplasm and Beyond. Trends Cell Biol. 25, 601–610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hwang H-W, Wentzel EA & Mendell JT A Hexanucleotide Element Directs MicroRNA Nuclear Import. Science 315, 97–100 (2007). [DOI] [PubMed] [Google Scholar]
- 220.Zisoulis DG, Kai ZS, Chang RK & Pasquinelli AE Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature 486, 541–544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mitchell PS et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A 105, 10513–10518 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Valadi H et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology 9, 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- 223.Arroyo JD et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A 108, 5003–5008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Turchinovich A, Weiz L, Langheinz A & Burwinkel B Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gallo A, Tandon M, Alevizos I & Illei GG The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 7, e30679 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Melo SA et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell 26, 707–721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Fong MY et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nature Cell Biology 17, 183–194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhang L et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527, 100–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Tkach M & Théry C Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 164, 1226–1232 (2016). [DOI] [PubMed] [Google Scholar]
- 230.Chevillet JR et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. U.S.A 111, 14888–14893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Thomou T et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ahadi A, Brennan S, Kennedy PJ, Hutvágner G & Tran N Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci Rep 6, 24922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Villarroya-Beltri C et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature Communications 4, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Santangelo L et al. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. CellReports 17, 799–808 (2016). [DOI] [PubMed] [Google Scholar]
- 235.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S & Schekman R Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Teng Y et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nature Communications 8, 14448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Koppers-Lalic D et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. CellReports 8, 1649–1658 (2014). [DOI] [PubMed] [Google Scholar]
- 238.Zhang L, Yang C-S, Varelas X & Monti S Altered RNA editing in 3’ UTR perturbs microRNA-mediated regulation of oncogenes and tumor-suppressors. Sci Rep 6, 23226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Nakano M, Fukami T, Gotoh S & Nakajima M A-to-I RNA Editing Up-regulates Human Dihydrofolate Reductase in Breast Cancer. Journal of Biological Chemistry 292, 4873–4884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Nam J-W et al. Global Analyses of the Effect of Different Cellular Contexts on MicroRNA Targeting. Molecular Cell 53, 1031–1043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kedde M et al. A Pumilio-induced RNA structure switch in p27–3’ UTR controls miR-221 and miR-222 accessibility. Nature Cell Biology 12, 1014–1020 (2010). [DOI] [PubMed] [Google Scholar]
- 242.Min K-W et al. AUF1 facilitates microRNA-mediated gene silencing. Nucleic Acids Res. 45, 6064–6073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mukherjee N et al. Integrative Regulatory Mapping Indicates that the RNA-Binding Protein HuR Couples Pre-mRNA Processing and mRNA Stability. Molecular Cell 43, 327–339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ahuja D, Goyal A & Ray PS Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biol 13, 1152–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Poria DK, Guha A, Nandi I & Ray PS RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene 35, 1703–1715 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.La Rocca G et al. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc. Natl. Acad. Sci. U.S.A 112, 767–772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ameres SL, Martinez J & Schroeder R Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130, 101–112 (2007). [DOI] [PubMed] [Google Scholar]
- 248.Kertesz M, Iovino N, Unnerstall U, Gaul U & Segal E The role of site accessibility in microRNA target recognition. Nat. Genet 39, 1278–1284 (2007). [DOI] [PubMed] [Google Scholar]
- 249.Zheng Z et al. Target RNA Secondary Structure Is a Major Determinant of miR159 Efficacy. Plant Physiol. 174, 1764–1778 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sheu-Gruttadauria J & MacRae IJ Phase Transitions in the Assembly and Function of Human miRISC. Cell 173, 946–957.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Lee Y et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003). [DOI] [PubMed] [Google Scholar]
- 253.Denli AM, Tops BBJ, Plasterk RHA, Ketting RF & Hannon GJ Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235 (2004). [DOI] [PubMed] [Google Scholar]
- 254.Gregory RI et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004). [DOI] [PubMed] [Google Scholar]
- 255.Nguyen TA et al. Functional Anatomy of the Human Microprocessor. Cell 161, 1374–1387 (2015). [DOI] [PubMed] [Google Scholar]
- 256.Kwon SC et al. Structure of Human DROSHA. Cell 164, 81–90 (2016). [DOI] [PubMed] [Google Scholar]
- 257.Nicholson AW Ribonuclease III mechanisms of double-stranded RNA cleavage. WIREs RNA 5, 31–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Okada C et al. A High-Resolution Structure of the Pre-microRNA Nuclear Export Machinery. Science 326, 1275–1279 (2009). [DOI] [PubMed] [Google Scholar]
- 259.Bernstein E, Caudy AA, Hammond SM & Hannon GJ Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001). [DOI] [PubMed] [Google Scholar]
- 260.Grishok A et al. Genes and Mechanisms Related to RNA Interference Regulate Expression of the Small Temporal RNAs that Control C. elegans Developmental Timing. Cell 106, 23–34 (2001). [DOI] [PubMed] [Google Scholar]
- 261.Hutvagner G et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 (2001). [DOI] [PubMed] [Google Scholar]
- 262.Kim Y-K, Kim B & Kim VN Re-evaluation of the roles of DROSHA, Exportin 5, and DICERin microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A 113, E1881–E1889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.MacRae IJ, Zhou K & Doudna JA Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol 14, 934–940 (2007). [DOI] [PubMed] [Google Scholar]
- 264.Park J-E et al. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475, 201–205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Tsutsumi A, Kawamata T, Izumi N, Seitz H & Tomari Y Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat. Struct. Mol. Biol 18, 1153–1158 (2011). [DOI] [PubMed] [Google Scholar]
- 266.Tian Y et al. A Phosphate-Binding Pocket within the Platform-PAZ-Connector Helix Cassette of Human Dicer. Molecular Cell 53, 606–616 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.MacRae IJ et al. Structural basis for double-stranded RNA processing by Dicer. Science 311, 195–198 (2006). [DOI] [PubMed] [Google Scholar]
- 268.Lau P-W et al. The molecular architecture of human Dicer. Nat. Struct. Mol. Biol 19, 436–440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zhang H, Kolb FA, Jaskiewicz L, Westhof E & Filipowicz W Single Processing Center Models for Human Dicer and Bacterial RNase III. Cell 118, 57–68 (2004). [DOI] [PubMed] [Google Scholar]
- 270.Lee HY, Zhou K, Smith AM, Noland CL & Doudna JA Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 41, 6568–6576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Rand TA, Petersen S, Du F & Wang X Argonaute2 Cleaves the Anti-Guide Strand of siRNA during RISC Activation. Cell 123, 621–629 (2005). [DOI] [PubMed] [Google Scholar]
- 272.Matranga C, Tomari Y, Shin C, Bartel DP & Zamore PD Passenger-Strand Cleavage Facilitates Assembly of siRNA into Ago2-Containing RNAi Enzyme Complexes. Cell 123, 607–620 (2005). [DOI] [PubMed] [Google Scholar]
- 273.Leuschner PJF, Ameres SL, Kueng S & Martinez J RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. EMBO Rep 101, 314–320 (2006). [DOI] [PubMed] [Google Scholar]
- 274.Schwarz DS et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003). [DOI] [PubMed] [Google Scholar]
- 275.Khvorova A, Reynolds A & Jayasena SD Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 (2003). [DOI] [PubMed] [Google Scholar]
- 276.Suzuki HI et al. 2015 Suzuki NSMB - Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat. Struct. Mol. Biol 22, 512–521 (2015). [DOI] [PubMed] [Google Scholar]
- 277.Frank F, Sonenberg N & Nagar B Structural basis for 5. Nature 465, 818–822 (2010). [DOI] [PubMed] [Google Scholar]
- 278.van Niel G, D’Angelo G & Raposo G Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.