Abstract
DNA and RNA can adopt various secondary structures. Four-stranded G-quadruplex (G4) structures form through self-recognition of guanines into stacked tetrads, and considerable biophysical and structural evidence exists for G4 formation in vitro. Computational studies and sequencing methods have revealed the prevalence of G4 sequence motifs at gene regulatory regions in various genomes, including in humans. Experiments using chemical, molecular and cell biology methods have demonstrated that G4s exist in chromatin DNA and in RNA, and have linked G4 formation with key biological processes ranging from transcription and translation to genome instability and cancer. In this Review, we first discuss the identification of G4s and evidence for their formation in cells using chemical biology, imaging and genomic technologies. We then discuss possible functions of DNA G4s and their interacting proteins, particularly in transcription, telomere biology and genome instability. Roles of RNA G4s in RNA biology, especially in translation, are also discussed. Furthermore, we consider the emerging relationships of G4s with chromatin and with RNA modifications. Finally, we discuss the connection between G4 formation and synthetic lethality in cancer cells, and recent progress towards considering G4s as therapeutic targets in human diseases.
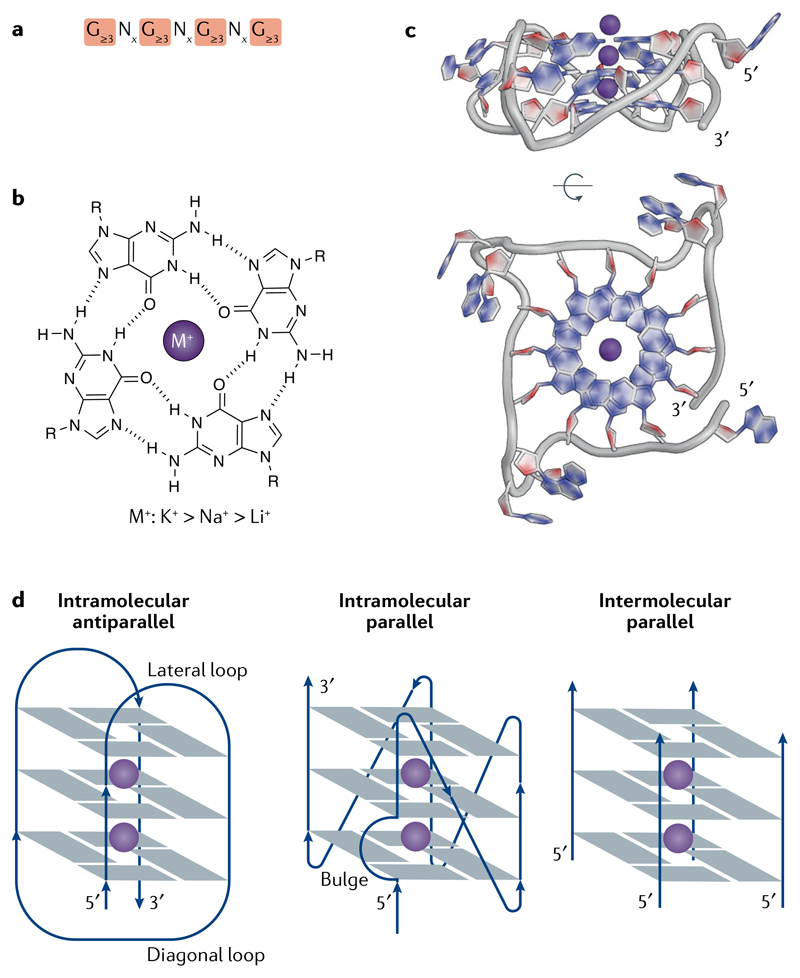
Nucleic acids have considerable potential to fold into three-dimensional, ‘secondary’ structures. This can happen through the formation of non-Watson–Crick hydrogen bonds between nucleobases. Early observations on the self-assembly of guanylic acid1 led to the elucidation of the guanine tetrad-forming sequence motif2 (FIG. 1a), in which guanines are mutually bonded by Hoogsteen hydrogen base-pairing to form a planar array that is further stabilized by interactions between positively charged ions and the O-6 lone-pair electrons of each guanine (FIG. 1b,c). Initial evidence for the assembly of four-stranded G-quadruplex (G4) structures from natural sequences was provided by the formation in vitro of higher-order secondary structures from oligonucleotides resembling G-rich sequences from immunoglobulin switch regions3. Biophysical and structural biology methods subsequently provided substantial physical evidence for the formation of intermolecular and intramolecular G4s from DNA and RNA in vitro, including a framework for recognizing sequences likely to fold into G4s.
Fig. 1. The structure and topologies of G-quadruplexes.
a | The G-quadruplex (G4) consensus sequence. x denotes the number of nucleotides in the loops (see part d). b | A guanine tetrad is stabilized by Hoogsteen base-pairing and by a central cation (M+), with a preference for monovalent cations in the order of potassium (K+) > sodium (Na+) > lithium (Li+). c | X-ray crystal structure of an intramolecular, parallel G4 from a human telomere sequence (PDB: 1KF1)214. d | Schematic representation of some G4 topologies.
Although G4s are related to each other in primary sequence, they in fact comprise a diverse family of structures that can fold into various topologies, which are dictated by the pattern of strand polarities and also the orientation of interconnecting loops4 (FIG. 1d). The extent to which distinct topologies can influence G4 formation and function in cells is unknown. There has been a recent surge in research activity directed towards understanding G4 formation in living cells and organisms. Considerable attention has focused on the detection and occurrence of G4 structures in genomes and in RNA with a view to elucidating how these elements might regulate key biological processes, such as transcription, telomere homeostasis and translation. Identifying specific proteins that directly interact with G4 structures and elucidating their influence on such processes is an important step towards increasing our understanding of G4 biology. Detailed structural investigations into the mechanisms of G4 unfolding by helicases are providing insights into the control of G4 folding at the biochemical level, although further work is needed to fully understand the regulation of G4 formation in cells. The fact that G4s are linked with DNA damage and genome instability in addition to key cancer-associated genes has prompted investigations into possible roles of G4s in cancer biology and an evaluation of small-molecule G4 ligands as potential therapeutic agents.
In this Review, we discuss the evidence for G4 formation in DNA and RNA in biological systems, factors that regulate G4 formation and biological processes that are influenced by G4s. We also discuss important links between G4s and cancer, and the progress made towards using G4s as therapeutic targets. Other reviews provide extensive details on G4 prediction5, biophysics and structure4,6, and roles in DNA replication7, human disease8 and therapeutic possibilities9.
Identification of G4s
Biophysical studies using oligonucleotides were the first to establish that many DNA and RNA sequences featuring G-tracts separated by other bases (loops) can fold into G4 structures. Rules for predicting G4 structure formation emerged on the basis of data from circular dichroism, ultraviolet melting and NMR spectroscopy studies on different G4-forming oligonucleotides4,10. G4s have been identified as cellular features through a combination of computational sequence analyses and experiments that detect G4s in cellular genomes and in purified nucleic acids using chemical and molecular biology and imaging methods.
Circular dichroism
A spectroscopic technique to investigate structure based on the interaction of plane-polarized light with a structurally asymmetric molecule.
Bayesian predictions
Statistical methods to infer probabilities for a hypothesis, which can be updated when new information becomes available.
G-fraction
The proportion of G bases in a sequence, that is, G-richness.
G-skew
The under-representation or over-representation of G bases in a sequence.
Polytene chromosomes
Giant chromosomes found in particular tissues of various eukaryotes, which are formed following several rounds of DNA replication without cell division.
Fragile telomeres
Aberrant or discontinuous appearance of telomere chromatin in metaphase chromosomes, identified by fluorescence in situ hybridization and indicative of telomere replication defects.
Common fragile sites
Specific chromosomal regions that are intrinsically hard to replicate and preferentially form chromatin gaps or breaks during metaphase following replication stress.
Stress granules
Cytoplasmic membraneless bodies of proteins and RNAs that appear in response to conditions of cellular stress.
CpG island
A genomic region with CG:GC content higher than 60%.
Computational identification of G4s
The use of early algorithms to search for the relatively simple G3–5N1–7G3–5N1–7G3–5N1–7G3–5 consensus sequence suggested that the human genome may have over 300,000 sequences with the potential to form G4s11,12. Such computational tools have helped to identify potential G4s associated with key genes and reveal enrichment of G4-forming sequences in genomic regions associated with gene regulation, specific cellular functions and disease states13,14. The early search algorithms were unable to account for structural variants, such as G4s with longer loops, bulges or mismatches, for two-tetrad G4s or for the importance of flanking sequences. More recent computational tools accommodate some of these factors15, use Bayesian predictions 16, account for flanking sequence effects based on G-fraction and G-skew 17 or consider possibilities of higher-order assemblies18. Although machine learning approaches have helped to identify G4s that are likely to form in genomic contexts19 and those likely to fold in RNA20, computational methods that account for chromatin contexts and protein binding are yet to be developed.
DNA and RNA chain-extension stalling
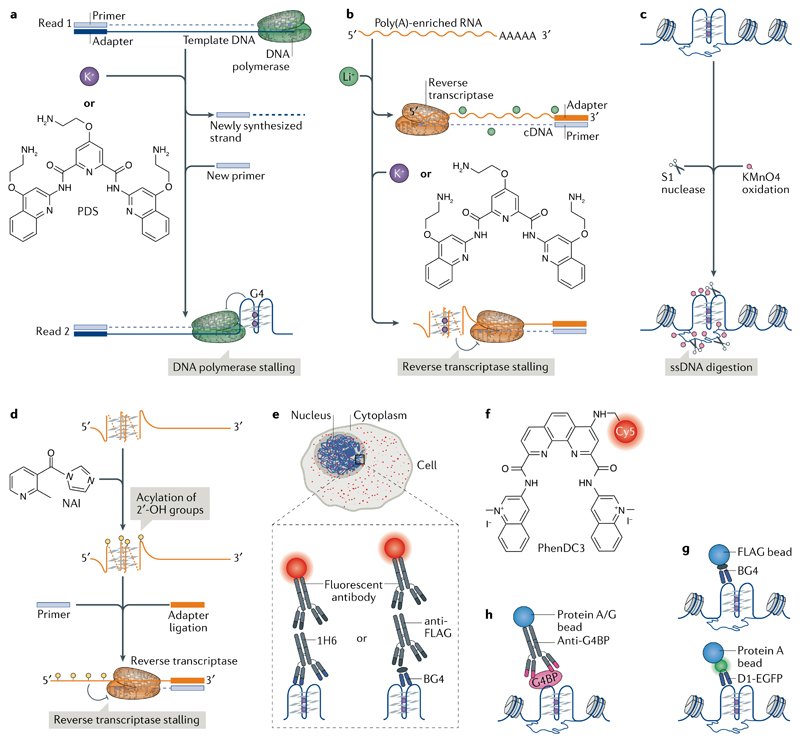
Experimental approaches have been developed to detect G4 structures and complement computational prediction. G4s in DNA or RNA can stall a DNA polymerase21,22 or a reverse transcriptase23, respectively. Comparison of polymerase pause sites in G4-stabilizing conditions (for example, in the presence of potassium ions (K+) and/or a stabilizing ligand) and in conditions that do not stabilize G4s (for example, in the presence of lithium ions (Li+)) enables the detection of the 5′ end of G4s in vitro. This method was initially applied using sequence analysis on a polyacrylamide gel and, more recently, adapted into a genome-wide DNA polymerase-stop assay followed by high-throughput sequencing (G4-seq; FIG. 2a). G4-seq identified more than 700,000 DNA G4 sites in the human genome24, which included various non-canonical G4 structures that are difficult to predict; conversely, G4s predicted by some search algorithms were not observed, highlighting the advantages of experimental G4 mapping. Subsequently, G4-seq reference maps have been made available for model organisms25. An analogous approach using reverse transcriptase stalling (rG4-seq) on poly(A)-enriched RNAs mapped RNA G4 structures in more than 3,000 human mRNAs26,27 (FIG. 2b). These techniques provide important in vitro reference maps of G4-forming sites, although these data represent cell-population averages and do not account for the effect that proteins may have on G4 formation.
Fig. 2. Approaches to detect and map DNA and RNA G-quadruplexes.
a,b | Mapping G-quadruplexes (G4s) by chain-extension stalling followed by high-throughput sequencing (G4-seq). G4s formed in the genome are mapped using G4-seq and G4s formed in the transcriptome are mapped using an analogous approach with reverse transcriptase stalling (rG4-seq). a | In G4-seq, a library of fragmented genomic DNA is sequenced twice, first in non-G4-forming conditions (Read 1) to provide a reference and then in G4-stabilizing conditions (for example, in the presence of K+ or the G4-stabilizing ligand pyridostatin (PDS; Read 2)) to determine the positions of G4-dependent DNA polymerase stalling. b | In rG4-seq, poly(A)-enriched RNA is reverse transcribed in the presence of Li+ (a non-G4-forming condition) as a reference and with K+ or PDS to map RNA G4-dependent reverse transcriptase stalling. c,d | Chemical methods for mapping G4s. DNA G4s are mapped by potassium permanganate (KMnO4)–single-strand nuclease (S1 nuclease) footprinting and RNA G4s are mapped by selective 2′-OH acylation analysed by primer extension (SHAPE). c | In KMnO4–S1 nuclease footprinting, KMnO4 selectively oxidizes and traps single-stranded DNA (ssDNA), thereby allowing its digestion by S1 nuclease. Subsequent computational analyses infer the formation of DNA G4s based on the nuclease footprints. d | SHAPE utilizes differences in acylation kinetics of RNA 2′-hydroxyl groups treated with 2-methylnicotinic acid imidazolide (NAI) and the ability of these groups to stall reverse transcription, to determine the formation of RNA G4s. e | Visualization of G4s (red foci) by immunofluorescence in the nucleus and cytoplasm using G4 antibodies (for example, the IgG antibody 1H6 or the single-chain variable fragment antibody (ScFv) BG4) together with fluorescently conjugated secondary or tertiary antibodies. f | PhenDC3 is an example of a fluorescence-labelled G4-targeting ligand. g | Mapping of DNA G4s using chromatin immunoprecipitation and next-generation sequencing (ChIP-seq) with G4-specific ScFv antibodies. BG4 precipitates DNA G4 structures from chromatin isolated using G4 ChIP-seq, whereas D1 is used in a ChIP-seq-like approach by expressing the antibody in cells. h | DNA G4 formation can be inferred indirectly by mapping the location of G4-binding proteins (G4BPs) using ChIP-seq.
Chemical mapping of G4s
G4 chemical mapping exploits the different reactivity of nucleobases following the formation of G4 structures. Use of potassium permanganate-dependent single-strand nuclease (S1 nuclease) footprinting provided a snapshot of single-stranded DNA (ssDNA) regions of the genome in mouse B cell chromatin (FIG. 2c). The combination of ssDNA mapping with computational prediction of non-B-DNA sequence motifs identified putative G4 (and other non-B-DNA) structures in gene regulatory regions28. Alternatively, the location of G4 structures can be deduced from their relative protection (compared with guanines in double-stranded DNA) from methylation by dimethyl sulfate (DMS) and subsequent cleavage by piperidine, which is provided by the Hoogsteen hydrogen interactions between the guanines in G-tetrads29. Another method of G4 chemical mapping is selective 2′-OH acylation analysed by primer extension (SHAPE; FIG. 2d), which exploits structure-dependent differences in acylation kinetics of RNA 2′-hydroxyl groups30. SHAPE performed in lithium versus potassium conditions was used to reveal RNA G4s in vitro31. Global analysis by DMS and SHAPE did not detect RNA G4s in eukaryotic cells26, prompting the hypothesis that RNA G4 structures are unfolded. However, caution is required when interpreting such data because of the limitations of chemical mapping, such as a map based on the averaging of dynamic structural states over time and across cell populations, along with the possibility of shifting the dynamic equilibrium of structural states during an experiment. These considerations have been discussed in more detail elsewhere32.
Imaging G4s
G4s have been visualized by immunofluorescence in cells with G4 structure-specific antibodies (FIG. 2e). Use of the single-chain variable fragment antibody (scFv) Sty49 revealed G4 formation at the telomeres of a ciliate33. The scFv antibody BG4 revealed G4s in telomeric and non-telomeric DNA in fixed human cells34; this finding was corroborated by related observations using the G4-specific antibodies IgG 1H6 (REF.35) and scFv D1 (REF.36). BG4 was also employed to visualize RNA G4s in the cytoplasm of human cells37. An increased G4-antibody signal has been observed following the depletion of known G4-interacting proteins38–40, during S phase of the cell cycle and after treatment of live cells with G4-stabilizing ligands34,41. Although the specificity of each antibody may not be absolute42, and their sensitivity of detecting a single G4 structure as opposed to G4 clusters, is unproven in cells, there is an increasing level of cross-validation of G4 observations using these antibodies with observations of natural G4-interacting proteins.
Synthetic small molecules that recognize and stabilize G4s have also been used to probe cellular G4 structures. Derivatives of pyridostatin (PDS)43 and PhenDC3 (REF.44) (FIG. 2f) incubated with live cells have subsequently been conjugated to a fluorescence probe using ‘click chemistry’ after cell fixation to visualize G4s. G4 ligands with intrinsic fluorescence, such as DAOTA-M2, support the existence of G4s in cells45. Real-time, live cell imaging provided by such ligands has advantages over antibodies, which require cell fixation and permeabilization, although in live cells the sensitivity of fluorescence to G4-independent changes in pH, redox status, intracellular polarity and viscosity require careful consideration to validate G4-specific binding.
Genomic and transcriptomic techniques
DNA G4s have been detected and mapped in the chromatin of human cells using chromatin immunoprecipitation followed by high-throughput sequencing (G4 ChIP-seq) using BG4 (REFS46,47) (FIG. 2g). Strong overlap between these data and the human G4-seq reference map (of all genomic sequences that can form a G4) can provide cross-validation24, as does a G4 map generated from the expression of the G4-specific D1 antibody in cells36 (FIG. 2g). Alternatively, G4s can be inferred using antibodies against known G4-binding proteins. For example, the helicases α-thalassemia mental retardation X-linked protein (ATRX)48, xeroderma pigmentosum group B (XPB) and XPD49 bind folded G4 oligonucleotides in vitro, and have been mapped to G4 motifs in human chromatin using ChIP-seq (FIG. 2h). In yeast, the G4-associated ATP-dependent DNA helicase Pif1 (REF.50), ribosome biogenesis protein SLX9 (REF.51) and Rap1-interacting factor 1 (Rif1)52 have been mapped to G4 motifs. Limitations of ChIP-seq include the averaging of results obtained from cell populations, the inability to resolve temporal dynamics and the dependence on accessibility of the target to the antibody. There are also potential biases introduced by antibody specificity, sample fixation and fragmentation that must be eliminated by carefully designed control experiments53. Single-cell genomics approaches, super-resolution microscopy and live cell imaging by single-molecule light sheet microscopy may, in due course, provide additional insights into the nature and dynamics of DNA G4s in cells.
Recent studies have used RNA immunoprecipitation techniques, such as individual-nucleotide resolution ultraviolet crosslinking and immunoprecipitation (iCLIP) and photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (parCLIP), to identify transcriptome-wide binding sites of known RNA G4 protein interactors54–56. These techniques provide a valid strategy for identifying RNA G4s that exist in a protein-bound state and the biological processes they influence. However, they do not provide explicit proof of the existence of an RNA G4-folded structure at these sites.
G4 occurrence in living cells has now been demonstrated using chemical mapping, antibody-based or small molecule-based imaging and sequencing techniques. The endogenous landscape of G4s, revealed by these methods, is only a small fraction of the total number of possible G4s in the genome. Although this may be a fair reflection of biological reality, it remains possible that existing technologies do not detect the full repertoire of G4s in cells. In particular, transient and/or dynamic G4s states may not be accurately detected with current approaches (TABLE 1 summarizes the advantages and limitations of current technologies). The development of new tools and methodologies, alongside technical advances in live cell imaging and genome editing, will provide further insights in the future.
Table 1. Advantages and limitations of techniques to map and detect G-quadruplexes (G4s).
| Methodology | Technique | Uses and advantages | Limitations |
|---|---|---|---|
| Mapping by chain-extension stalling | G4-seq RNA G4-seq (rG4-seq) | Identifies in vitro nucleic acid sequences with the potential to form G4s in the genome (G4-seq) or transcriptome (rG4-seq) | Performed on extracted DNA or RNA. Thus, the influence of the cellular environment, for example proteins or chromatin structure, on the G4 landscape is not considered |
| Chemical mapping | Potassium permanganate–S1 nuclease footprinting | Maps genome-wide multiple types of non-B DNA structures in the chromatin context | Relies on motif-annotation algorithms to map the non-B-DNA structures (including G4 formation) Cannot accurately discern individual non-B DNA structures at sites containing large clusters of non-B DNA Readout is an averaging of structural states Can shift the dynamic equilibrium of structural states and hence may not reflect true intracellular structures. Readout is not specific to G4 structures |
| Selective 2’-OH acylation analysed by primer extension (SHAPE) | Provides quantitative, single-nucleotide-resolution RNA structural information | Structural information is lost at both the 5´ and 3´ ends of an RNA because the technology depends on primer extension Readout is not specific to G4 structures |
|
| In vivo dimethyl-sulphate (DMS) footprinting | Determines nucleic acid (DNA and RNA) secondary and tertiary structures at single-nucleotide resolution DMS easily and rapidly penetrates cells and all cellular compartments |
Readout is an averaging of structural states High cellular toxicity Can shift the dynamic equilibrium of structural states and, hence, does not reflect true cellular structures DMS reactivity depends on solvent accessibility and local electrostatic environment |
|
| Antibody-based mapping | G4 chromatin immunopre-cipitation sequencing (G4 ChIP-seq) | Genome-wide mapping of DNA G4s in the chromatin context | Possible biases introduced by sample fixation and fragmentation Relies on antibody specificity, target accessibility and cell-population averaging Cannot determine on which DNA strand G4s are located Antibodies against G4-binding proteins provide indirect evidence of DNA G4s, which relies on the specificity of the protein for G4s |
| ChIP-seq of G4-binding proteins | Genome-wide mapping of G4 DNA binding proteins in the chromatin context | ||
| Individual-nucleotide resolution ultraviolet crosslinking and immuno-precipitation (iCLIP) | Identifies all RNA sequences bound to the RNA binding protein (RBP) of interest | Relies on the specificity of the RBP to bind RNA G4 Cannot account for protein binding to unfolded G4 sequence motifs Relies on cell-population averaging |
|
| Imaging | Immunofluorescence | Single-cell resolution of G4 abundance Possible to detect DNA and RNA G4s simultaneously |
Requires cellular fixation and permeabilization Relies on the specificity of the antibody Does not provide sequence context Undetermined resolution: do detected G4 foci represent one or several G4s? |
| Fluorescent G4-stabilizing ligands | Allow the study of dynamic formation of G4s in fixed and live cells | Fluorescence is sensitive to cellular changes in pH, polarity (hydrophilic versus hydrophobic compartments) and viscosity, making discrimination of G4-specific from non-specific binding events difficult The dynamic equilibrium of G4 formation may be shifted by the experiment and thus will not reflect the true cellular state Lack of sequence context Relies on ligand specificity and half-life |
G4-seq, genome-wide DNA polymerase-stop assay followed by high-throughput sequencing; rG4-seq, transcriptome-wide reverse transcriptase stalling assay followed by high-throughput sequencing; S1 nuclease, a single-strand nuclease.
Control of G4 formation and unwinding
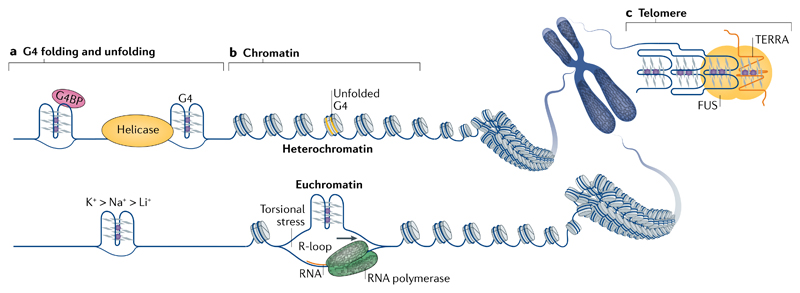
Sequences that form G4s are prevalent in genomes, and it is becoming clear that not all sequences with G4 potential form structures in cells and that different cell types or cell states have distinct patterns of G4 formation. Understanding how G4 formation is regulated in a cellular context is therefore a question of fundamental importance. Biophysical approaches have been used to evaluate properties that influence DNA and RNA G4 formation in oligonucleotides in near-physiological conditions57. G4 stability is affected by numerous factors, including the number of G-tetrads, loop length and topology58,59 and the sequence composition, both within the G4 motif and flanking regions19. RNA G4s are generally more thermally stable than their DNA counterparts57. G4s are stabilized by centrally located, monovalent cations in the order K+ > Na+ > Li+ (REF.60) (FIG. 1), which may be physiologically relevant because K+ is the most abundant metal ion in mammalian cells61. Furthermore, G4 formation can be favoured by the induction of negative torsional stress behind RNA and DNA polymerases62 and by molecular crowding63, which are both relevant in the cellular context of genomic DNA.
Helicases and other G4-binding proteins
G4s can impede nucleic acid functions (for example, DNA replication, transcription or translation) and proteins exist that can resolve them (FIG. 3a). Many such proteins belong to canonical helicase families, such as the RecQ-like and DEAD box or DEAH box helicase families. The RecQ-like helicases Bloom syndrome protein (BLM) and Werner syndrome ATP-dependent helicase (WRN) were the first recognized G4-resolving mammalian helicases64,65. DEAH-box helicase DHX36 is able to resolve both RNA and DNA G4s66. In vitro, these three helicases unwind G4s formed within oligonucleotides by binding the 3′ tail-containing DNA substrate and performing a repetitive 3′–5′, ATP-independent unfolding of the structure67–69. In the case of BLM and WRN, this activity is a result of cooperative binding of their helicase-RNaseD domain to the 3′ ssDNA and their RecQ domain to the G467. For DHX36, G4 binding induces rearrangements in the helicase core, which pulls the single-stranded region, thereby tugging the G4 one nucleotide out of alignment68. This is sufficient to destabilize a DNA G4 in the presence of its complementary strand69. DHX36 is also able to fully unwind an RNA G4, but then refolds it in the absence of a complementary strand, giving rise to dynamic G4 unfolding and refolding70. Pif1 (in budding yeast) and Fanconi anaemia group J (FANCJ) unwind G4s 5′–3′ in an ATP-dependent manner, requiring (in vitro) a 5′ tail-containing DNA71,72.
Fig. 3. Regulation of G-quadruplex structure formation.
a | Physical factors, such as the presence of stabilizing cations (bottom), length and sequence composition (not shown) of the loops and flanking sequences, determine the thermodynamic stability of G-quadruplexes (G4s). In cells, specialized proteins that unwind G4s (for example, helicases) or bind and stabilize G4s (for example, some G4-binding proteins (G4BPs)) can shape the G4 landscape. b | The genomic structural context, such as the negative superhelicity behind the RNA polymerase complex, as well as the crosstalk with other structural phenomena, like RNA–DNA hybrids known as R-loops that arise from hybridization of the nascent RNA with the template DNA, can contribute to G4 formation (bottom). Chromatin structure seems to have a strong influence on G4 formation, as the majority of endogenous G4s have been mapped to nucleosome-depleted, open chromatin regions. c | Telomere heterochromatin homeostasis can be influenced by the simultaneous binding of proteins, such as fused in sarcoma (FUS), to G4s formed in telomere DNA and in the long non-coding RNA TERRA (telomeric repeat RNA).
Other non-helicase proteins, such as cellular nucleic acid-binding protein (CNBP)73 and G-rich sequence factor 1 (REF.74), have been reported to sequester unfolded form of G4s. The ssDNA-binding proteins protection of telomeres 1 (POT1) and replication protein A (RPA) use a Brownian ratchet-like mechanism and unfold G4s in a multistep process75. However, additional mechanisms such as simple trapping of the unfolded G4 have also been proposed for POT1 (REF.76), suggesting that its mechanism of function remains somewhat elusive. Conversely, proteins such as nucleolin77 and LARK (also known as RNA-binding protein 4 (RBM4))78 were shown to stabilize G4 structures. Several studies have applied affinity enrichment, using G4 baits, to identify G4 interacting proteins, most of which require further studies to elucidate the nature and biological relevance of the interaction54,79. For many proteins identified to bind G4 oligonucleotides, evidence for specific recognition of the G4 structure in vivo is lacking.
Local features favouring DNA G4s
G4 formation in genomic DNA competes with Watson–Crick base-pairing; however, this base-pairing is necessarily disrupted during replication, transcription and DNA damage repair, thereby favouring G4 formation. During transcription, negative torsional stress induced behind the RNA polymerase complex can be relayed upstream (in the opposite direction of transcription) and promote melting of duplex DNA, which contributes to G4 formation, for example as proposed for the far upstream element of the human MYC promoter80 (FIG. 3b). The formation of RNA–DNA hybrids known as R-loops from the hybridization of the nascent RNA with the template DNA may contribute to G4 formation on the displaced DNA strand, as observed by electron microscopy81. G4s and R-loops are favoured by similar DNA features, such as GC-richness and negative torsional tension, and genome-wide profiling of R-loops in human embryonic kidney cells revealed considerable overlap with G4-forming sequences identified by G4-seq82. C-rich DNA can form an intercalated motif (i-motif) secondary structure by stacking intercalated and hemi-protonated cytosine base pairs (C+:C)83. Biophysical studies have shown that i-motifs are favoured by acidic pH, although recent antibody-based experiments have shown i-motif formation in nuclei of fixed human cells84. Although i-motifs can in principle occur on the C-rich strand opposite G4, cell cycle analysis shows that i-motif formation is maximal in late G1, whereas G4 formation peaks in S phase, indicating that the situation may be more complex85.
Studies to detect and map DNA G4s in a chromatin context have shown that G4s occur primarily at regulatory, nucleosome-depleted regions and promoters of actively transcribed genes in human cancer cells using G4 ChIP-seq46,47 (FIG. 3b) and in mouse B cells using permanganate footprinting28. Furthermore, in human cells, G4s co-localized with RNA polymerase II (Pol II) and with trimethylated histone H3 Lys4 (H3K4me3), which is a histone modification associated with active genes but not with the heterochromatin modification H3K9me3 (REF.46). Overall, G4 structures were detected in relatively accessible chromatin46,47. By contrast, in Drosophila melanogaster polytene chromosomes, G4s were found in the heterochromatin86, suggesting the existence of species-specific differences in G4 formation.
Oxidative base damage and G4 formation
G4s are sensitive to redox chemistry and early work showed that G4s in complex with porphyrins, such as haem, have peroxidase and peroxygenase enzyme-like activity87. Free haem is potentially toxic as it can catalyse the formation of reactive oxygen species, leading to oxidative stress. It has thus been hypothesized that G4s act as a sink for free haem to prevent DNA damage88. Reactive oxygen species can induce DNA damage in the form of 8-oxoguanine (8-oxoG), particularly in G-rich regions. For example, at telomeres, 8-oxoG disrupts G4 formation, stimulates telomerase function and promotes telomere instability89. Considerable transcriptional and DNA damage responses are also observed following oxidative damage at promoter G4s, such as in the vascular endothelial growth factor (VEGF; also known as VEGFA) gene90.
Cellular functions of G4s
A major and largely unanswered question is what G4s do. G4s are found in so many different cellular contexts that they can be considered either physical obstacles that must be overcome to enable some nucleic-acid related process or useful for normal cellular functions.
G4 structures at telomeres
DNA and RNA G4s have roles in telomere biology. Telomeres are nucleoprotein structures located at chromosome ends, which maintain genome integrity by suppressing aberrant DNA repair of the DNA ends through binding of telomere-specific protein complexes and formation of higher-order DNA secondary structures91. In the somatic nucleus of the ciliate Stylonychia lemnae, protection is provided by intermolecular telomere DNA G4s, which are stabilized by telomere-binding protein-α (TEBPα) and TEBPβ92. In yeast and vertebrates, protection is provided by the ‘lasso-like’ telomere loop, in which the telomere ssDNA overhang invades the upstream double-stranded telomere DNA93. G4s may also cap chromosome ends, as G4s have been detected at telomeres34–36 and several telomeric proteins (for example, RIF1 (REF.52) and telomeric repeat-binding factor 2 (TRF2)94) bind G4s in vitro. Indeed, G4s can act as rudimentary protective structures when the normal telomere capping structure is compromised95. Chromatin homeostasis at telomeres and at sub-telomeric regions is also dependent on the long non-coding RNA TERRA (telomeric repeat RNA) forming a G4 structure, which was proposed to be a protein docking scaffold96 (FIG. 3c). The human proteins TRF2, Ewing sarcoma breakpoint region 1 protein (EWS) and fused in sarcoma (FUS) can co-bind the TERRA G4 and telomere DNA G4 in vitro94,96,97. By co-binding TERRA G4 and telomere G4, FUS can recruit histone methyltransferases that are important for telomere and sub-telomere heterochromatin maintenance96.
Persistent formation of G4s at telomeres during DNA replication is problematic. Depletion of many proteins known to interact with telomeric G4 oligonucleotides (for example, the CTC1–STN1–TEN1 (CST) complex39 and helicases like regulator of telomere elongation helicase 1 (RTEL1)98) results in telomere shortening, altered telomere replication rate and/or formation of fragile telomeres, which arise from stalling of replication forks at telomeres during lagging strand synthesis99. Furthermore, these phenotypes are exacerbated by the presence of G4-stabilizing ligands; for example, sudden telomere loss occurs only in cells subjected to the combination of CST depletion and PDS treatment39.
G4 formation can control access to telomeres of telomerase, the non-coding RNA–reverse transcriptase complex that extends 3′ ends of chromosomes in cancer cells, stem cells and cells of the germline to prevent telomere shortening and genome instability. Formation of anti-parallel intramolecular telomere DNA G4s (FIG. 1d), prevents telomere extension by limiting access to the 3′ end of the telomere to telomerase100, whereas parallel intermolecular telomere DNA G4s can be extended due to partial G4 resolution by telomerase in vitro101. Supporting these findings, in Saccharomyces cerevisiae, parallel telomeric G4 stabilization by the telomere elongation protein Est1 is essential for telomerase recruitment102. Furthermore, in S phase, when G4s can co-localize with human telomerase, intermolecular G4s between sister chromatids have been hypothesized to form101. However, the complex POT1–tripeptidyl-peptidase 1 (TPP1), which is responsible for the processivity and recruitment to telomeres of human telomerase, is known to destabilize G4 structures76,103. The recent observation in vitro that G4 folding within the active pocket of human telomerase supports POT1–TPP1-dependent telomerase processivity104 suggests that G4 formation is important during human telomere DNA synthesis. Telomerase is also influenced by the unfolding of a G4 at the 5′ end of the RNA component of telomerase by DHX36 (REF.105).
Although the natural function of telomere G4s with regards to telomerase is still unclear in vivo, ligand-stabilized telomere G4s inhibit telomerase-mediated telomere extension106. The additional ability of G4 ligands to displace components of the telomere protection complex shelterin (for example, TRF2 and POT1) results in telomere DNA damage and cell death, and has led to the development of a plethora of G4 ligands as potential chemotherapeutic agents9,107. Furthermore, although G4-induced replication stress in certain cancer cells has been proposed to promote alternative lengthening of telomeres, which is a mechanism activated in 15% of cancers108, treatment with G4 ligands is still effective in killing these cells109, thereby supporting the consideration of G4 ligands as pan-anticancer compounds.
Transcription
A K+-dependent G4 structure was initially detected in the promoter of the chicken β-globin-encoding gene21, and subsequently other G4-forming sequences were noted in the promoters of human genes, most notably in MYC and several other oncogenes110. Computational predictions indicated that G4 motifs are prevalent and enriched in human gene promoters compared with the rest of the genome13, suggesting that G4s are involved in transcription regulation. Notably, sequencing-derived genomic G4 maps of 12 organisms confirmed increased G4-forming potential at gene promoters and 5′ untranslated regions (5′ UTRs) in human, mouse and Trypanosoma brucei, which indicates the existence of functional similarities in their G4 biology24,25. By contrast, no enrichment or even depletion was found in lower eukaryotes and in bacteria. Recent studies have mapped G4 structures in chromatin to regulatory regions upstream of the transcription start sites of actively transcribing genes in human cells28,46, further supporting the link between G4s and transcription.
Many publications report that transcription can be regulated by small molecules that target G4s. Treatment of cells with the G4-stabilizing ligand TMPyP4 resulted in reduced MYC expression111, with comparable observations subsequently reported for other gene promoters, such as KRAS 112 and KIT 113. Furthermore, transcriptome-wide changes have been reported at genes with promoter G4s114. Although such correlations are consistent with the existence of a G4–transcription link, more explicit evidence of small-molecule binding to folded promoter G4s in cellular DNA, along with an improved consideration of the potential consequences of indirect, network effects on transcription, would help to better characterize the link.
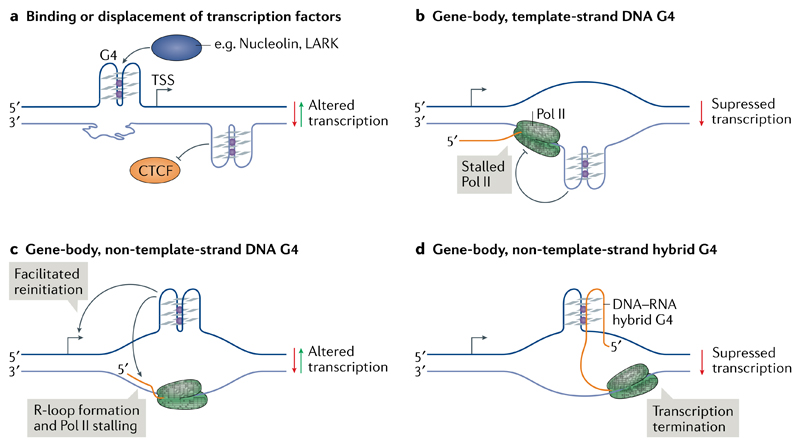
Computational analysis of the binding motifs of several transcription factors showed that they are strongly enriched in certain promoter G4 motifs115 and in endogenous G4s as observed by G4 ChIP-seq116. This enrichment suggests that positive or negative interactions exist between transcription factors and G4 structures at gene promoters. An early study reported the binding in vitro of the G4 motif from the MYC promoter by recombinant nucleolin77 (FIG. 4a). By contrast, Förster resonance energy transfer experiments indicated the unfolding of a MYC G4 oligonucleotide upon binding of NM23-H2 (also known as NDK-B)117, prompting the hypothesis that nucleolin and NM23-H2 are involved in G4 stabilization and unwinding, respectively, to regulate MYC transcription. Since then, in vitro interactions with G4 oligonucleotides have been shown for several other transcription factors, including CNBP118, SP1 (REF.119) and LARK78. Additional experiments confirming G4 structure formation as part of protein–DNA complexes, ideally using structural biology, and explicit evidence for binding at endogenous G4s would help strengthen the link between protein binding and G4 status. Conversely, G4 formation in the first exon of the human telomerase reverse transcriptase (hTERT) gene has been suggested to disrupt binding of the gene repressor CCCTC-binding factor, resulting in elevated plasmid-encoded hTERT transcription120 (FIG. 4a).
Fig. 4. Models of G-quadruplex involvement in transcription.
a | DNA G-quadruplexes (G4s) upstream of the transcription start site (TSS) or in the gene body could bind or displace transcription factors, resulting in altered transcription. b | During transcription elongation, the separation of DNA strands in the transcription bubble may result in the formation of G4s in gene bodies. G4 formation on the template strand can block the progression of RNA polymerase II (Pol II). c | Gene-body G4s on the non-template strand may facilitate transcription re-initiation. Conversely, such G4s may favour the stable association of nascent RNA (orange) with the template DNA, resulting in the formation of RNA–DNA hybrids known as R-loops and in Pol II stalling. d | Formation of DNA–RNA hybrid G4s between the non-template DNA and the nascent RNA can lead to premature transcription termination.
Several studies have investigated the effects of G4 motifs in gene bodies on transcription elongation. A substantial inhibitory effect was observed when a G4 motif was present on the template strand, both in human embryonic kidney cells121 and in Escherichia coli 122, which is consistent with impairment of Pol II progression by G4 structures in the template strand (FIG. 4b). By contrast, G-rich sequences inserted in the non-template strand (but not in the template strand) impaired in vitro transcription by T7 polymerase123. In this case, transcription blockage was ascribed to R-loop formation independent of G4 formation, as no major transcriptional differences were observed between G4-stabilizing and G4-destabilizing buffer conditions. Computational analysis of human genes has observed a correlation between increased promoter-proximal Pol II pausing and the presence of downstream G4 motifs on the template and non-template strands124. In addition, human genes with a greater number of G4s on the non-template strand up to 500 base pairs downstream of the transcription start site are associated with higher than average steady-state transcription levels and Pol II occupancy, suggesting that G4s on the non-template strand could maintain the DNA in an open state and, thus, aid transcription reinitiation125 (FIGS 3b,4c).
Transcription of the mitochondrial gene CSB II was investigated in vitro using 7-deaza-dGTP or 7-deaza-GTP nucleotides, which cannot form Hoogsteen base-pairing and thus cannot stabilize G4s. The nascent RNA and non-template DNA strand were shown to co-transcriptionally form a stable DNA–RNA hybrid G4, which was suggested to promote transcription termination126 (FIG. 4d). Similarly, transcription suppression was observed when a hybrid-G4 forming sequence was inserted into reporter plasmids127.
Models linking G4s and transcription have been largely based on computationally predicted G4 sequences, supporting correlations and the manipulation of isolated G4 structures in plasmid constructs. It is also evident that the specific G4 positions (for example, in regulatory regions or gene bodies and in template strand or non-template strand) may contribute to different regulatory mechanisms. Furthermore, the local chromatin context appears to have a substantial effect on G4 formation and function46. Additional work is needed to elucidate the molecular mechanistic details of how G4s influence transcription in a chromatin context.
The effects of G4s on genome stability
G4s can cause replication stress by obstructing the progression of DNA replication forks and causing replication-fork collapse128,129, which generates DNA double-strand breaks that can lead to genome instability. Computational analyses of large cancer datasets associated G4s with breakpoints that accompany somatic copy-number alterations130. Another large cancer association study found that G4 motifs, particularly thermodynamically more stable G4s, were enriched at sites of somatic mutations, implying that G4 structures increase the probability of recurrent mutations and may be important determinants of mutagenesis131. The link between G4s and genome instability has been strengthened by sequencing of G4s in the human genome, which revealed a notable association of G4s with gene amplifications commonly observed in cancers24,46.
Studies in model organisms provided substantial support in vivo for G4s being a direct cause of genome instability. Caenorhabditis elegans lacking dog-1 (also known as helicase ATP-binding domain-containing protein), which is the ortholog of the helicase FANCJ, accumulate deletions at G-rich regions, including in predicted G4 motifs132. Experiments using plasmids as replication templates in Xenopus laevis egg extracts showed that the absence of FANCJ or the presence of G4-stabilizing ligands cause replication stalling at G4 structures133. In C. elegans, genetic analyses have also demonstrated that site-specific genome deletions can originate from a single predicted G4 sequence motif134. Genetic analyses in S. cerevisiae have also shown that Pif1, a potent G4-unwinding helicase, suppresses DNA damage and gross chromosomal rearrangements mediated by G4s135. Human minisatellite tandem repeats comprising G4 motifs also show increased instability when introduced into S. cerevisiae lacking Pif1 or in the presence of G4-stabilizing ligands136,137; thermodynamically stable G4s with short loops preferentially caused rearrangements138.
Helicases prevent G4-induced genome instability in humans
G4 helicases protect the genome by unfolding G4s that can cause DNA breakage and subsequent aberrant recombination; failure to resolve G4s owing to loss of helicase activity may induce genome instability. Sister chromatid exchanges are common in cells of individuals with Bloom syndrome and are enriched at predicted G4 sites, particularly in transcribed genes139. In glioma cells, ATRX loss promotes G4 formation, somatic copy-number alterations and increased occupancy of BLM at DNA damage sites38. Chromosomal regions known as common fragile sites are predisposed to breakage and undergoing rearrangements during replication stress. ATRX localizes to common fragile sites during replication stress and ATRX loss is associated with increased numbers of chromosomal breaks at common fragile sites140. FANCJ together with the ssDNA-binding protein RPA enable S-phase progression by facilitating G4 unwinding72. BLM and WRN also form complexes with RPA, which are mediated by the BRCA1-interacting E3 ubiquitin-protein ligase HERC2 (REF.40). Similar to BLM and WRN loss, HERC2 depletion or inhibition of its ubiquitylation activity increases G4 formation. In a functional genomic screen for G4-interacting factors, HERC2 loss was found to promote cell death in cells treated with G4-stabilizing ligands141. BLM and WRN may operate independently or on different G4s in the genome, whereas HERC2 seems to be epistatic to BLM and WRN and a master regulator of G4 suppression40.
G4 stabilization induces DNA damage
Small molecules, such as PDS, can stabilize G4s142 and cause replication-dependent and transcription-dependent DNA double-strand breaks (detected by γ-H2AX ChIP-seq), which map to G4-rich regions at loci that include several oncogenes43. Cells compromised in their ability to process G4s are particularly sensitive to G4-stabilizing ligands. For example, loss of FANCJ, HERC2 or ATRX sensitizes cells to different G4 ligands, including to telomestatin, PDS and CX-3543 (REFS38,40,72). In mice, ATRX-deficient glioma xenografts are growth impaired by CX-3543 and the host mice show increased survival38, highlighting the therapeutic potential of G4 ligands.
G4s and R-loops
R-loops and G4s can form on opposite DNA strands (FIGS 3b,4c), and then threaten genome stability by blocking DNA and RNA polymerases and causing transcription–replication conflicts. Immunoprecipitation of such DNA–RNA hybrids with an antibody or using an inactive version of the R-loop processing enzyme RNase H1 identified R-loops in GC-rich promoter regions143,144, including regions enriched in G4s motifs82. It is striking that an immediate response to G4 stabilization by PDS is an increase in R-loops, particularly opposite G4 sites, which results in DNA damage followed by the formation of micronuclei145. Although the underlying mechanism of this process is unclear, overexpression of RNase H1 counteracts these effects of G4 ligands, suggesting that R-loops are required for the induction of DNA damage by G4 stabilization145. G4 and R-loop induction by PDS were shown in HeLa and U20S cancer cell lines, but are absent in immortalized fibroblasts, suggesting that aberrant G4-related pathways are present in cancer cells. Notably, ATRX loss also results in increased R-loop formation at telomeres146, consistent with coupling of G4 regulation with R-loop stability.
Translation
G4-mediated translation inhibition was first reported for an RNA G4 from the coding region of the mRNA of fragile X mental retardation protein (FMRP; also known as synaptic functional regulator FMR1); when inserted into a luciferase reporter, this RNA G4 caused a 1.5-fold reduction in translation in vitro in reticulocyte lysates147. A similar in vitro reporter assay showed that a G4 from the 5′ UTR of the NRAS mRNA caused a fourfold reduction in translation148. Subsequently, G4s from numerous 5′ UTRs, including the mRNAs of BCL2 (REF.149) and ADAM10 (REF.150), were shown to inhibit translation in cell-free or cell-based reporter assays. The G4-forming triplet repeat expansion (CGG)60–90 from the 5′ UTR of the FMR1 gene also inhibits translation of a reporter construct and of exogenously expressed FMR1 mRNA in cells, but, despite folding into G4 structures, the shorter (CGG)30 repeats elevate translation151. RNA G4 density, thermodynamic stability and position relative to the 5′ cap have all been shown to differentially influence translation151,152.
G4 motifs are often located near the beginning of 5′ UTRs, suggesting they have a role in translation initiation153. The helicase eukaryotic initiation factor 4A (eIF4A) unwinds structured 5′ UTRs to facilitate the recruitment of the 43S pre-initiation complex and the subsequent scanning for the start codon154. Depletion of eIF4A or its inhibition by silvestrol reduced the translation efficiency of mRNAs with longer 5′ UTRs enriched in two-tetrad G4s, indicating that RNA G4s directly influence recruitment of, or scanning by, the ribosome155,156. Unresolved G4s in 5′ UTRs can promote the formation of 80S ribosomes on alternative, upstream start codons, thus inhibiting the translation of the main open reading frame55 (FIG. 5a). RNA G4s in the mRNAs of FGF2 (REF.157), α-synuclein158 and VEGF159 stimulate internal ribosome entry site (IRES)-mediated translation, potentially by recruiting the 40S ribosome160. However, RNA G4s in the IRES of VEGF and in α-synuclein were subsequently found to be functionally dispensable, and the role of RNA G4s in IRES-dependent translation is far from being clear158,161,162.
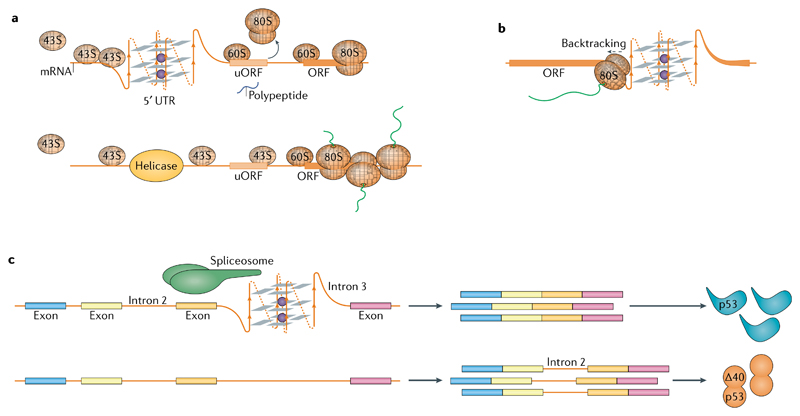
Fig. 5. G-quadruplexes in RNA biology.
a | Formation of RNA G-quadruplexes (G4s) impedes scanning of the 5′ untranslated region (UTR) by 43S ribosomes and leads to translation initiation at an upstream open reading frame (uORF) at the expense of translation of the main ORF (top). Helicases, such as DHX36 or DHX9, resolve the G4s and facilitate translation of the main ORF (bottom). b | The 80S ribosomes engaged in translation elongation stall 6–7 nucleotides prior to a G4 within the ORF. Stalling can cause ribosome backtracking and synthesis of an alternative peptide. c | Recognition of RNA G4s by spliceosome-associated RNA-binding proteins directs splicing of nearby introns, for example the second intron of p53.
G4s occur at a lower abundance in mRNA coding sequences when compared with 5′ UTRs153. In the coding sequence, ribosomes stall 6–7 nucleotides before a G4, which is the distance of the tRNA acceptor site from the RNA entry site163. Thus, despite the helicase activity associated with the 80S ribosome, RNA G4s are problematic for translation elongation and, therefore, evolutionarily selected against through the use of synonymous codons164 (FIG. 5b).
As indicated above, interactions of RNA G4s with RNA-binding proteins (RBPs) can influence translation. A prime example is FMRP, which regulates translation of hundreds of mRNAs and is reviewed in detail elsewhere165. Other examples include CNBP, which binds RNA G4s in 4,178 different mRNAs and elevates translation by preventing the formation of RNA G4s73, and DHX36, depletion of which reduces mRNA translation56.
Our current knowledge of the effects of RNA G4s on translation relies substantially on reporter assays, which are a helpful tool, but care must be taken when interpreting results obtained from such non-native systems. For example, an isolated G4 derived from the 5′ UTR of the TGFβ2 mRNA reduces translation when embedded in a reporter mRNA, but conversely enhances translation in the context of the entire UTR166. In another example, a G4 from BCL2 mRNA was shown to suppress translation in vitro, but its genomic disruption by CRISPR–Cas9 failed to increase translation of the native mRNA167. Interestingly, translation output is unaltered when G4s from the mRNAs of VEGF and TGFβ2 associated with translation stimulation are replaced with G4s associated with translation repression from the mRNAs of NRAS and MT3-MMP, and vice versa168. Therefore, the context in which RNA G4s form, their dynamic relationship with surrounding alternative structures169,170 and their interactions with RBPs should all be considered when evaluating translational output.
Other roles of G4s in RNA biology
RNA G4s can influence the subcellular localization of mRNAs in neurons171. Interaction of RNA G4s with RBPs, such as FMRP172, regulates their localization and local translation in dendrites. Recent studies have suggested a potential function of RNA G4s in the formation of stress granules; for example, RNA G4-binding proteins, such as DHX36 and DDX3X, associate with stress granules173,174. Moreover, the C9ORF72 mRNA, which contains G4-forming repeats, the extension of which causes amyotrophic lateral sclerosis and frontotemporal dementia, promotes phase separation of stress granule proteins and granule assembly175. The propensity of mRNAs to localize to stress granules correlates with longer UTRs and coding regions and with poor translation, which are features of G4-containing mRNAs176. Finally, translation interfering tRNAs (tiRNAs) are tRNA fragments formed in stress conditions that may have roles in cancer progression177. A G4 structure in tiRNAala or tetramolecular G4s (formed from four individual tiRNAs) appear to mediate stress granule formation178,179 and translation inhibition by interacting with Y-box binding protein 1 and, subsequently, displacing eIF4F from mRNAs180. DNA analogues of tiRNAala may trigger a neuroprotective response in motor neurons, suggesting new possibilities for interventions in neurodegenerative diseases180.
RNA G4-mediated recruitment of splicing-associated RBPs, such as heterogeneous ribonucleoprotein H (hnRNPH)181 and hnRNPF182, regulates alternative splicing. In the context of its native sequence, the FMRP mRNA G4 is a potent splicing enhancer147,183. A G4 in the third intron of the p53 mRNA promotes the splicing of intron 2, and mutating the guanines of this G4 increases intron retention and synthesis of the truncated protein Δ40p53 (REF.184) (FIG. 5c). Likewise, a G4 promotes the correct splicing of intron 1 of the PAX9 mRNA185 and another promotes the correct splicing of exon 3 of BACE-1 mRNA186. Recently, an RNA G4-binding ligand was found to cause thousands of alternative splicing events in cells187. Additional transcriptomics studies are required to elucidate the rule set by which G4s regulate splicing. RNA G4s are also involved in mRNA polyadenylation, piRNA biogenesis and form in ribosomal RNA (reviewed in REF.188).
Nucleic acid and histone modifications
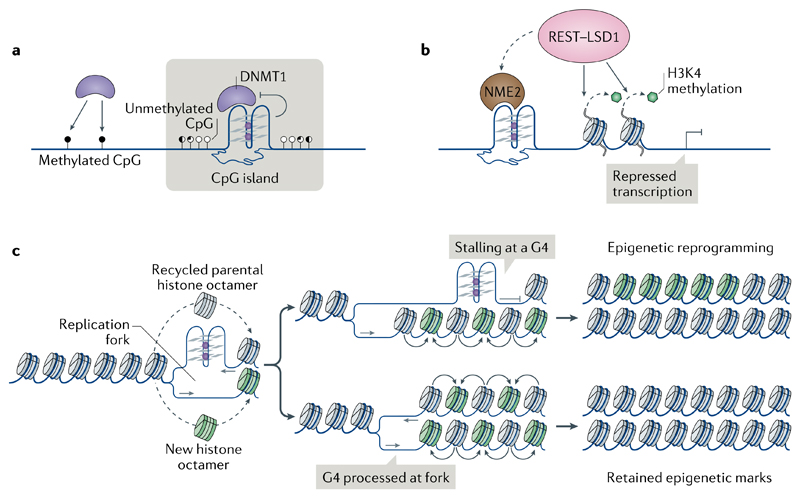
Emerging observations link G4s to covalent chemical modifications of DNA and of histones. DNA methyltransferases, which catalyse the formation of 5-methylcytosine, principally at CpG dinucleotides in mammalian cells, have a biophysical preference to bind G4 DNA over double-stranded DNA189,190 (FIG. 6a). G4 binding inhibits the activity of DNA (cytosine-5)-methyltransferase 1 (DNMT1), and DNMT1 binding sites in chromatin that are marked by G4 structures are strongly hypomethylated in human leukaemia cells, prompting the hypothesis that DNMT1 is sequestered at G4 sites to inhibit methylation of proximal CpG island promoters190 (FIG. 6a). In addition to DNA methylation, a recent study reported G4-dependent transcription repression of hTERT through non-metastatic 2 (NME2)-dependent recruitment to the promoter G4s of the RE1-silencing transcription factor (REST)–lysine-specific histone demethylase 1A (LSD1; also known as KDM1A) repressor complex, which removes the gene-activating monomethylation and dimethylation of histone H3 Lys4 (H3K4me1 and H3K4me2, respectively)191 (FIG. 6b). A similar mechanism was postulated for the promoter of cyclin-dependent kinase inhibitor 1 (which encodes p21Cip1), where a TRF2–G4 interaction is required to mediate REST–LSD1 activity192.
Fig. 6. The involvement of G-quadruplexes in epigenetic control.
a | Binding to G-quadruplexes (G4s) inactivates DNA (cytosine-5)-methyltransferase 1 (DNMT1), thereby contributing to hypomethylation at CpG islands. b | Promoter G4s and their associated proteins, such as non-metastatic 2 (NME2), recruit the RE1-silencing transcription factor (REST)–lysine-specific histone demethylase 1A (LSD1) repressor complex to remove the gene-activating methylation of histone H3 Lys4 (H3K4) and repress gene expression. c | Stalling of DNA replication forks at G4s (for example, owing to impaired activity of helicases or the presence of G4-stabilizing ligands) may impair histone recycling through the formation of a post-replicative gap (top). Parental histones with their established modifications (grey) are lost and replaced with new histones with no or with different modifications (green), resulting in local epigenetic reprogramming.
During replication, DNA synthesis and histone recycling (into the newly formed sister chromatids) are closely coordinated to ensure the maintenance of parental histone modifications in the daughter cells. In DT40 chicken cells, impaired activity of proteins required for replication of G4-forming sequences193,194, depletion of nucleotide pools (causing replication stress)195 or G4 stabilization by small molecules196 resulted in local alteration of epigenetic marks, including histone modifications and cytosine methylation (FIG. 6c). These observations suggest that replication fork pausing at G4 sites in conditions of replication stress can uncouple the replication machinery from histone recycling. Notably, loss of H3K4me3 at a defined G4 site led to proximal DNA cytosine methylation and heritable inactivation of BU-1 gene expression196.
RNA G4s can influence gene expression in multiple ways. For example, the formation of a G4 instead of the canonical stem-loop in certain precursor microRNAs can inhibit their maturation by Dicer31,197. Production of mature microRNAs can therefore be influenced by RNA G4-stabilizing factors, such as high K+ or Mg2+ levels, or N-7 methylation of guanosines198,199. Additionally, G4 formation in a mature microRNA or in its target mRNA sequence can alter the regulation of the target mRNA200,201. Cross-talk between RNA G4s and chromatin modification is exemplified by the binding of RNA G4s by Polycomb repressive complex 2 (PRC2), which catalyses the gene-repressive histone modification H3K27me3 (REF.202). An association of N 6-methyladenosine (m6A) in viral RNA with predicted RNA G4s has suggested that RNA G4s may also function to guide RNA base modifications203. Multiple retrotransposons in the human genome harbour G4s, which may promote their transposition, as seen in the case of long interspersed element 1, although the underlying mechanism remains unclear204.
Higher-order chromatin architecture
As DNA G4 structures form in chromatin46, they might have a role in regulating higher-order genome architecture, for example in mediating promoter–enhancer interactions205. G4s may be involved in orchestrating long-range interactions, for example G4 encoding sequences that are split over long distances may come together to form a G4. In fission yeast, binding by the Rif1 protein to such half G4s is proposed to create local chromosomal compartments by enabling chromatin looping at the nuclear lamina, which may be involved in suppression of DNA replication origin firing in late S phase52. A recent computational study using a ChIP-seq dataset from human cells indicates that DNA G4s help define higher-hierarchy chromatin domains called topological associated domains116. These studies are suggestive, and more experimentation and direct evidence is required to determine whether DNA G4s are involved in active chromatin looping.
The therapeutic relevance of G4s
The multiple functions of G4s in DNA and in RNA collectively present opportunities for interference by small molecule-mediated manipulation of folded G4s. In this section we highlight some examples, primarily from cancer, where considerable progress towards demonstrating therapeutic potential has been reported.
Targeting G4s
Established modes of targeting DNA with small molecules, for example through intercalation or covalent modification, have led to the development of therapeutic agents against pathogens and cancers206. Although many of these drugs are used effectively, including as first-line therapies, their use is limited owing to their toxicity and side effects. G4s could offer a new modality for targeting DNA. The distinct molecular features of G4s, in particular the G-tetrads and loops, enable structure-selective recognition by small molecules207. The functional links between G4s and gene regulation (particularly regulation of cancer genes), DNA replication and genome instability and telomere biology have prompted exploration of G4-targeting therapeutics. Initial efforts focused on targeting G4 structures at telomeres with a view to inhibiting telomere extension by telomerase in cancer cells106,208. Subsequently, it emerged that G4-binding molecules can cause DNA damage at telomeres and also at G4s throughout the genome43,209. Targeting of G4s in genes by PDS can inhibit gene expression, including numerous important oncogenes43. The naphthalene diimide G4-targeting ligand CM03 has shown promising activity against cancer cell lines and in a mouse xenograft model of pancreatic ductal adenocarcinoma, including a notable reduction in the expression of many G4-rich genes implicated in vital pathways of cancer-cell survival, metastasis and drug resistance114. Whereas early studies focused on modulating individual cancer genes by targeting their G4s111–113, the prevalence of G4s in many cancer-promoting genes suggests that collectively targeting multiple G4s, and thus inhibiting the expression of many such genes, as exemplified by CM03, would be a feasible strategy114. The observed increase in G4s in the chromatin of cancer tissues210 and cell line models34,46, in comparison with normal cells and tissues, also favours targeting G4s as a general anticancer strategy.
Synthetic lethality
The capacity of G4 ligands to specifically create synthetic lethality in tumour cells provides another potential G4-based therapeutic avenue. Synthetic lethality refers to the cell-lethal combination of two or more non-lethal genetic perturbations. This can also be mimicked chemically by pharmacological inhibition of key genes that phenocopy genetic sensitivities211. G4 ligands enhance killing of BRCA1-deficient or BRCA2-deficient cancer cells by exploiting their deficiency in homologous-directed DNA repair41,212,213. A recent genomic RNAi screen has expanded the cancer genotypes and pathways sensitive to G4-ligand treatment141. Thus, there is scope for exploring genotype-specific G4-targeting strategies, as exemplified by the G4-targeting clinical compound CX-5461, which has recently entered clinical trials for BRCA-deficient tumours41 (see NCT02719977 at ClinicalTrials.gov). G4 ligands can be also used in combination with other agents. For example, the cytotoxic activities of PDS synergize with the compound NU7441, which inhibits the essential non-homologous end-joining factor DNA-dependent protein kinase213; with MK1775, which inhibits the cell cycle kinase WEE1; and with pimozide, which inhibits the deubiquitylating enzyme USP1 (also known as ubiquitin carboxyl-terminal hydrolase 1)141. These findings demonstrate the potential of G4 ligands as therapeutic agents in multiple cancer types.
Conclusions
The study of G4s originated from curiosity-driven structural investigations, and has progressed to the point where G4s should now be considered a fundamental feature of the genome. G4s are implicated in numerous important cellular processes, in particular transcription, but also in translation and maintenance of genome stability. The key future challenge is to elucidate the details of how G4 formation is regulated, especially at gene promoters and UTRs, and the specific mechanisms underlying their biological roles. Specifically, the many G4–protein interactions that have been revealed need to be characterized in greater detail to generate a robust molecular understanding of how G4s influence protein function. Such insight will naturally lead to clearer understanding of the role G4s in disease and could ultimately be exploited in a clinical context. The study of G4s has progressed considerably over the past decade and, consequently, a framework of computational reference data and experimental tools and methodologies now exists to help drive the elucidation of the functions of G4s in finer detail over the coming years.
Acknowledgements
The Balasubramanian laboratory is supported by Cancer Research UK core and programme award funding (C14303/A17197; C9681/A18618), S.B. is a Senior Investigator of the Wellcome Trust (099232/Z/12/Z) and D.V. is a Herchel Smith postdoctoral fellow. J.S. gratefully acknowledges EU H2020 Framework Programme funding (H2020-MSCA-IF-2016, ID: 747297-QAPs).
Footnotes
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
S.B. is a founder and shareholder of Cambridge Epigenetix Ltd.
Peer review information
Nature Reviews Molecular Cell Biology thanks Cyril Dominguez, Sua Myong and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bang I. Untersuchungen über die Guanylsäure. Biochemische. 1910;26:293–311. [Google Scholar]
- 2.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [ This paper is an early demonstration of a G4 comprising stacked tetrads with interconnecting loop sequences performed using chemical mapping and providing biological insight. ] [DOI] [PubMed] [Google Scholar]
- 4.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwok CK, Merrick CJ. G-Quadruplexes: prediction, characterization, and biological application. Trends Biotechnol. 2017;35:997–1013. doi: 10.1016/j.tibtech.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerner LK, Sale JE. Replication of G Quadruplex DNA. Genes. 2019;10:95. doi: 10.3390/genes10020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maizels N. G4-associated human diseases. EMBO Rep. 2015;16:910–922. doi: 10.15252/embr.201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J Med Chem. 2016;59:5987–6011. doi: 10.1021/acs.jmedchem.5b01835. [DOI] [PubMed] [Google Scholar]
- 10.Mergny JL, Lacroix L. UV melting of G-quadruplexes. Curr Protoc Nucleic Acid Chem. 2009;37:17.1.1–17.1.15. doi: 10.1002/0471142700.nc1701s37. [DOI] [PubMed] [Google Scholar]
- 11.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [ This work presents the earliest computational predictions showing that sequences encoding G4s are widespread in the human genome. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varizhuk A, et al. The expanding repertoire of G4 DNA structures. Biochimie. 2017;135:54–62. doi: 10.1016/j.biochi.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Stegle O, Payet L, Mergny JL, MacKay DJ, Leon JH. Predicting and understanding the stability of G-quadruplexes. Bioinformatics. 2009;25:i374–i382. doi: 10.1093/bioinformatics/btp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedrat A, Lacroix L, Mergny JL. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016;44:1746–1759. doi: 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belmonte-Reche E, Morales JC. G4-iM Grinder: when size and frequency matter. G-Quadruplex, i-Motif and higher order structure search and analysis tool. NAR Genom Bioinform. 2020;2:1–12. doi: 10.1093/nargab/lqz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahakyan AB, et al. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci Rep. 2017;7 doi: 10.1038/s41598-017-14017-4. 14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garant JM, Perreault JP, Scott MS. Motif independent identification of potential RNA G-quadruplexes by G4RNA screener. Bioinformatics. 2017;33:3532–3537. doi: 10.1093/bioinformatics/btx498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodford KJ, Howell RM, Usdin K. A novel K+-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J Biol Chem. 1994;269:27029–27035. [PubMed] [Google Scholar]
- 22.Han H, Hurley LH, Salazar M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999;27:537–542. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwok CK, Balasubramanian S. Targeted detection of G-quadruplexes in cellular RNAs. Angew Chem Int Ed Engl. 2015;54:6751–6754. doi: 10.1002/anie.201500891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chambers VS, et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 25.Marsico G, et al. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019;47:3862–3874. doi: 10.1093/nar/gkz179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo JU, Bartel DP. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016;353 doi: 10.1126/science.aaf5371. aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwok CK, Marsico G, Sahakyan AB, Chambers VS, Balasubramanian S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat Methods. 2016;13:841–844. doi: 10.1038/nmeth.3965. [DOI] [PubMed] [Google Scholar]
- 28.Kouzine F, et al. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 2017;4:344–356. doi: 10.1016/j.cels.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 31.Kwok CK, Sahakyan AB, Balasubramanian S. Structural analysis using SHALiPE to reveal RNA G-quadruplex formation in human precursor microRNA. Angew Chem Int Ed Engl. 2016;55:8958–8961. doi: 10.1002/anie.201603562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwok CK, Marsico G, Balasubramanian S. Detecting RNA G-quadruplexes (rG4s) in the transcriptome. Cold Spring Harb Perspect Biol. 2018;10 doi: 10.1101/cshperspect.a032284. a032284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaffitzel C, et al. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [ This work is the first demonstration of G4s in human cells by imaging using a structure-specific antibody. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson A, et al. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu HY, et al. Conformation selective antibody enables genome profiling and leads to discovery of parallel G-quadruplex in human telomeres. Cell Chem Biol. 2016;23:1261–1270. doi: 10.1016/j.chembiol.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Biffi G, Di Antonio M, Tannahill D, Balasubramanian S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat Chem. 2014;6:75–80. doi: 10.1038/nchem.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, et al. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat Commun. 2019;10 doi: 10.1038/s41467-019-08905-8. 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M, et al. Mammalian CST averts replication failure by preventing G-quadruplex accumulation. Nucleic Acids Res. 2019;47:5243–5259. doi: 10.1093/nar/gkz264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu W, et al. HERC2 facilitates BLM and WRN helicase complex interaction with RPA to suppress G-quadruplex DNA. Cancer Res. 2018;78:6371–6385. doi: 10.1158/0008-5472.CAN-18-1877. [DOI] [PubMed] [Google Scholar]
- 41.Xu H, et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun. 2017;8 doi: 10.1038/ncomms14432. 14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kazemier HG, Paeschke K, Lansdorp PM. Guanine quadruplex monoclonal antibody 1H6 cross-reacts with restrained thymidine-rich single stranded DNA. Nucleic Acids Res. 2017;45:5913–5919. doi: 10.1093/nar/gkx245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lefebvre J, Guetta C, Poyer F, Mahuteau-Betzer F, Teulade-Fichou MP. Copper−alkyne complexation responsible for the nucleolar localization of quadruplex nucleic acid drugs labeled by click reactions. Angew Chem Int Ed Engl. 2017;56:11365–11369. doi: 10.1002/anie.201703783. [DOI] [PubMed] [Google Scholar]
- 45.Shivalingam A, et al. The interactions between a small molecule and G-quadruplexes are visualized by fluorescence lifetime imaging microscopy. Nat Commun. 2015;6 doi: 10.1038/ncomms9178. 8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansel-Hertsch R, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48:1267–1272. doi: 10.1038/ng.3662. [ This paper presents the first maps of G4s generated in an endogenous chromatin context using G4 ChIP-seq and demonstrating G4 enrichment in active promoters linked with elevated transcription. ] [DOI] [PubMed] [Google Scholar]
- 47.Hansel-Hertsch R, Spiegel J, Marsico G, Tannahill D, Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 2018;13:551–564. doi: 10.1038/nprot.2017.150. [DOI] [PubMed] [Google Scholar]
- 48.Law MJ, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 49.Gray LT, Vallur AC, Eddy J, Maizels N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat Chem Biol. 2014;10:313–318. doi: 10.1038/nchembio.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [ This paper presents genetic experiments in yeast showing that the helicase Pif1 resolves G4s in vivo to prevent replication-fork stalling and DNA breaks. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gotz S, Pandey S, Bartsch S, Juranek S, Paeschke K. A novel G-quadruplex binding protein in yeast-Slx9. Molecules. 2019;24:1774. doi: 10.3390/molecules24091774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanoh Y, et al. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat Struct Mol Biol. 2015;22:889–897. doi: 10.1038/nsmb.3102. [DOI] [PubMed] [Google Scholar]
- 53.Meyer CA, Liu XS. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat Rev Genet. 2014;15:709–721. doi: 10.1038/nrg3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herdy B, et al. Analysis of NRAS RNA G-quadruplex binding proteins reveals DDX3X as a novel interactor of cellular G-quadruplex containing transcripts. Nucleic Acids Res. 2018;46:11592–11604. doi: 10.1093/nar/gky861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murat P, et al. RNA G-quadruplexes at upstream open reading frames cause DHX36- and DHX9-dependent translation of human mRNAs. Genome Biol. 2018;19:229. doi: 10.1186/s13059-018-1602-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauer M, et al. DHX36 prevents the accumulation of translationally inactive mRNAs with G4-structures in untranslated regions. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10432-5. 2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joachimi A, Benz A, Hartig JS. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg Med Chem. 2009;17:6811–6815. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 58.Bugaut A, Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J Am Chem Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 60.Sen D, Gilbert W. A sodium–potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 61.Zoroddu MA, et al. The essential metals for humans: a brief overview. J Inorg Biochem. 2019;195:120–129. doi: 10.1016/j.jinorgbio.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 62.Selvam S, Koirala D, Yu Z, Mao H. Quantification of topological coupling between DNA superhelicity and G-quadruplex formation. J Am Chem Soc. 2014;136:13967–13970. doi: 10.1021/ja5064394. [DOI] [PubMed] [Google Scholar]
- 63.Shrestha P, et al. Confined space facilitates G-quadruplex formation. Nat Nanotechnol. 2017;12:582–588. doi: 10.1038/nnano.2017.29. [DOI] [PubMed] [Google Scholar]
- 64.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 65.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Creacy SD, et al. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J Biol Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chatterjee S, et al. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat Commun. 2014;5 doi: 10.1038/ncomms6556. 5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen MC, et al. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature. 2018;558:465–469. doi: 10.1038/s41586-018-0209-9. [ This work presents the first X-ray crystallography structure of a G4-resolving helicase bound to a G4, proposing a structural mechanism for G4 unfolding. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tippana R, Hwang H, Opresko PL, Bohr VA, Myong S. Single-molecule imaging reveals a common mechanism shared by G-quadruplex-resolving helicases. Proc Natl Acad Sci USA. 2016;113:8448–8453. doi: 10.1073/pnas.1603724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tippana R, Chen MC, Demeshkina NA, Ferre-D’Amare AR, Myong S. RNA G-quadruplex is resolved by repetitive and ATP-dependent mechanism of DHX36. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09802-w. 1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Byrd AK, Raney KD. Structure and function of Pif1 helicase. Biochem Soc Trans. 2017;45:1159–1171. doi: 10.1042/BST20170096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benhalevy D, et al. The human CCHC-type zinc finger nucleic acid-binding protein binds G-rich elements in target mRNA coding sequences and promotes translation. Cell Rep. 2017;18:2979–2990. doi: 10.1016/j.celrep.2017.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pietras Z, et al. Dedicated surveillance mechanism controls G-quadruplex forming non-coding RNAs in human mitochondria. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05007-9. 2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ray S, Bandaria JN, Qureshi MH, Yildiz A, Balci H. G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc Natl Acad Sci USA. 2014;111:2990–2995. doi: 10.1073/pnas.1321436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gonzalez V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J Biol Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niu K, et al. Identification of LARK as a novel and conserved G-quadruplex binding protein in invertebrates and vertebrates. Nucleic Acids Res. 2019;47:7306–7320. doi: 10.1093/nar/gkz484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Serikawa T, et al. Comprehensive identification of proteins binding to RNA G-quadruplex motifs in the 5′ UTR of tumor-associated mRNAs. Biochimie. 2018;144:169–184. doi: 10.1016/j.biochi.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 80.Kouzine F, Liu J, Sanford S, Chung HJ, Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat Struct Mol Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 81.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen L, et al. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol Cell. 2017;68:745–757. doi: 10.1016/j.molcel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gehring K, Leroy JL, Gueron M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 84.Zeraati M, et al. I-motif DNA structures are formed in the nuclei of human cells. Nat Chem. 2018;10:631–637. doi: 10.1038/s41557-018-0046-3. [DOI] [PubMed] [Google Scholar]
- 85.Cui Y, Kong D, Ghimire C, Xu C, Mao H. Mutually exclusive formation of G-quadruplex and i-motif is a general phenomenon governed by steric hindrance in duplex DNA. Biochemistry. 2016;55:2291–2299. doi: 10.1021/acs.biochem.6b00016. [DOI] [PubMed] [Google Scholar]
- 86.Hoffmann RF, et al. Guanine quadruplex structures localize to heterochromatin. Nucleic Acids Res. 2016;44:152–163. doi: 10.1093/nar/gkv900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sen D, Poon LC. RNA and DNA complexes with hemin [Fe(III) heme] are efficient peroxidases and peroxygenases: how do they do it and what does it mean? Crit Rev Biochem Mol Biol. 2011;46:478–492. doi: 10.3109/10409238.2011.618220. [DOI] [PubMed] [Google Scholar]
- 88.Gray LT, et al. G-quadruplexes sequester free heme in living cells. Cell Chem Biol. 2019;26:1681–1691. doi: 10.1016/j.chembiol.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 89.Fouquerel E, et al. Targeted and persistent 8-oxoguanine base damage at telomeres promotes telomere loss and crisis. Mol Cell. 2019;75:117–130. doi: 10.1016/j.molcel.2019.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fleming AM, Zhu J, Ding Y, Burrows CJ. 8-Oxo-7,8-dihydroguanine in the context of a gene promoter G-quadruplex is an on−off switch for transcription. ACS Chem Biol. 2017;12:2417–2426. doi: 10.1021/acschembio.7b00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shay JW, Wright WE. Telomeres and telomerase: three decades of progress. Nat Rev Genet. 2019;20:299–309. doi: 10.1038/s41576-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 92.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 93.de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 94.Biffi G, Tannahill D, Balasubramanian S. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J Am Chem Soc. 2012;134:11974–11976. doi: 10.1021/ja305734x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith JS, et al. Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae . Nat Struct Mol Biol. 2011;18:478–485. doi: 10.1038/nsmb.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Takahama K, et al. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem Biol. 2013;20:341–350. doi: 10.1016/j.chembiol.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 97.Takahama K, Kino K, Arai S, Kurokawa R, Oyoshi T. Identification of Ewing’s sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. FEBS J. 2011;278:988–998. doi: 10.1111/j.1742-4658.2011.08020.x. [DOI] [PubMed] [Google Scholar]
- 98.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 99.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [ This paper is the earliest demonstration that the activity of telomerase can be affected by a G4 structure in its telomere DNA substrate, suggesting that G4s might regulate telomere elongation. ] [DOI] [PubMed] [Google Scholar]
- 101.Moye AL, et al. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat Commun. 2015;6 doi: 10.1038/ncomms8643. 7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang ML, et al. Yeast telomerase subunit Est1p has guanine quadruplex-promoting activity that is required for telomere elongation. Nat Struct Mol Biol. 2010;17:202–209. doi: 10.1038/nsmb.1760. [DOI] [PubMed] [Google Scholar]
- 103.Hwang H, Buncher N, Opresko PL, Myong S. POT1–TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20:1872–1880. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jansson LI, et al. Telomere DNA G-quadruplex folding within actively extending human telomerase. Proc Natl Acad Sci USA. 2019;116:9350–9359. doi: 10.1073/pnas.1814777116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Booy EP, et al. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res. 2012;40:4110–4124. doi: 10.1093/nar/gkr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun D, et al. Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 107.Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 108.Clynes D, et al. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat Commun. 2015;6 doi: 10.1038/ncomms8538. 7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gowan SM, Heald R, Stevens MF, Kelland LR. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol Pharmacol. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 110.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [ This study of the G4 structure in the MYC promoter shows that a small-molecule G4 ligand can inhibit transcription. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bejugam M, et al. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. J Am Chem Soc. 2007;129:12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marchetti C, et al. Targeting multiple effector pathways in pancreatic ductal adenocarcinoma with a G-quadruplex-binding small molecule. J Med Chem. 2018;61:2500–2517. doi: 10.1021/acs.jmedchem.7b01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kumar P, et al. Zinc-finger transcription factors are associated with guanine quadruplex motifs in human, chimpanzee, mouse and rat promoters genome-wide. Nucleic Acids Res. 2011;39:8005–8016. doi: 10.1093/nar/gkr536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hou Y, et al. Integrative characterization of G-Quadruplexes in the three-dimensional chromatin structure. Epigenetics. 2019;14:894–911. doi: 10.1080/15592294.2019.1621140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thakur RK, et al. Metastases suppressor NM23–H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Borgognone M, Armas P, Calcaterra NB. Cellular nucleic-acid-binding protein, a transcriptional enhancer of c-Myc, promotes the formation of parallel G-quadruplexes. Biochem J. 2010;428:491–498. doi: 10.1042/BJ20100038. [DOI] [PubMed] [Google Scholar]
- 119.Raiber EA, Kranaster R, Lam E, Nikan M, Balasubramanian S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012;40:1499–1508. doi: 10.1093/nar/gkr882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li PT, et al. Expression of the human telomerase reverse transcriptase gene is modulated by quadruplex formation in its first exon due to DNA methylation. J Biol Chem. 2017;292:20859–20870. doi: 10.1074/jbc.M117.808022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Agarwal T, Roy S, Kumar S, Chakraborty TK, Maiti S. In the sense of transcription regulation by G-quadruplexes: asymmetric effects in sense and antisense strands. Biochemistry. 2014;53:3711–3718. doi: 10.1021/bi401451q. [DOI] [PubMed] [Google Scholar]
- 122.Holder IT, Hartig JS. A matter of location: influence of G-quadruplexes on Escherichia coli gene expression. Chem Biol. 2014;21:1511–1521. doi: 10.1016/j.chembiol.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 123.Belotserkovskii BP, Soo Shin JH, Hanawalt PC. Strong transcription blockage mediated by R-loop formation within a G-rich homopurine-homopyrimidine sequence localized in the vicinity of the promoter. Nucleic Acids Res. 2017;45:6589–6599. doi: 10.1093/nar/gkx403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Eddy J, et al. G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res. 2011;39:4975–4983. doi: 10.1093/nar/gkr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wanrooij PH, et al. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012;40:10334–10344. doi: 10.1093/nar/gks802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zheng KW, et al. Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic Acids Res. 2013;41:5533–5541. doi: 10.1093/nar/gkt264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Puget N, Miller KM, Legube G. Non-canonical DNA/RNA structures during transcription-coupled double-strand break repair: roadblocks or bona fide repair intermediates? DNA Repair. 2019;81 doi: 10.1016/j.dnarep.2019.102661. 102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Techer H, Koundrioukoff S, Nicolas A, Debatisse M. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat Rev Genet. 2017;18:535–550. doi: 10.1038/nrg.2017.46. [DOI] [PubMed] [Google Scholar]
- 130.De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Georgakopoulos-Soares I, Morganella S, Jain N, Hemberg M, Nik-Zainal S. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res. 2018;28:1264–1271. doi: 10.1101/gr.231688.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kruisselbrink E, et al. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans . Curr Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 133.Castillo Bosch P, et al. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014;33:2521–2533. doi: 10.15252/embj.201488663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lemmens B, van Schendel R, Tijsterman M. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat Commun. 2015;6 doi: 10.1038/ncomms9909. 8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Paeschke K, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Piazza A, et al. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae . Nucleic Acids Res. 2010;38:4337–4348. doi: 10.1093/nar/gkq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Piazza A, et al. Non-canonical G-quadruplexes cause the hCEB1 minisatellite instability in Saccharomyces cerevisiae . eLife. 2017;6:e26884. doi: 10.7554/eLife.26884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Piazza A, et al. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015;34:1718–1734. doi: 10.15252/embj.201490702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.van Wietmarschen N, et al. BLM helicase suppresses recombination at G-quadruplex motifs in transcribed genes. Nat Commun. 2018;9 doi: 10.1038/s41467-017-02760-1. 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pladevall-Morera D, et al. Proteomic characterization of chromosomal common fragile site (CFS)-associated proteins uncovers ATRX as a regulator of CFS stability. Nucleic Acids Res. 2019;47:8004–8018. doi: 10.1093/nar/gkz510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zyner KG, et al. Genetic interactions of G-quadruplexes in humans. eLife. 2019;8:e46793. doi: 10.7554/eLife.46793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Muller S, et al. Pyridostatin analogues promote telomere dysfunction and long-term growth inhibition in human cancer cells. Org Biomol Chem. 2012;10:6537–6546. doi: 10.1039/c2ob25830g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sanz LA, et al. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol Cell. 2016;63:167–178. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ginno PA, Lim YW, Lott PL, Korf I, Chedin F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23:1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.De Magis A, et al. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc Natl Acad Sci USA. 2019;116:816–825. doi: 10.1073/pnas.1810409116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nguyen DT, et al. The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 2017;18:914–928. doi: 10.15252/embr.201643078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schaeffer C, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shahid R, Bugaut A, Balasubramanian S. The BCL-2 5′ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry. 2010;49:8300–8306. doi: 10.1021/bi100957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lammich S, et al. Translational repression of the disintegrin and metalloprotease ADAM10 by a stable G-quadruplex secondary structure in its 5′-untranslated region. J Biol Chem. 2011;286:45063–45072. doi: 10.1074/jbc.M111.296921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Khateb S, Weisman-Shomer P, Hershco-Shani I, Ludwig AL, Fry M. The tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance the in vivo translation of fragile X premutation mRNA. Nucleic Acids Res. 2007;35:5775–5788. doi: 10.1093/nar/gkm636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kumari S, Bugaut A, Balasubramanian S. Position and stability are determining factors for translation repression by an RNA G-quadruplex-forming sequence within the 5′ UTR of the NRAS proto-oncogene. Biochemistry. 2008;47:12664–12669. doi: 10.1021/bi8010797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 155.Wolfe AL, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [ This study presents a comprehensive investigation into the control of translation by the helicase eIF4A–RNA G4 interactions in the 5′ UTR of mRNAs. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Modelska A, et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015;6:e1603. doi: 10.1038/cddis.2014.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bonnal S, et al. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J Biol Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Koukouraki P, Doxakis E. Constitutive translation of human alpha-synuclein is mediated by the 5′-untranslated region. Open Biol. 2016;6 doi: 10.1098/rsob.160022. 160022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 160.Bhattacharyya D, Diamond P, Basu S. An independently folding RNA G-quadruplex domain directly recruits the 40S ribosomal subunit. Biochemistry. 2015;54:1879–1885. doi: 10.1021/acs.biochem.5b00091. [DOI] [PubMed] [Google Scholar]
- 161.Cammas A, et al. Stabilization of the G-quadruplex at the VEGF IRES represses cap-independent translation. RNA Biol. 2015;12:320–329. doi: 10.1080/15476286.2015.1017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wu Y, et al. Stabilization of VEGF G-quadruplex and inhibition of angiogenesis by quindoline derivatives. Biochim Biophys Acta. 2014;1840:2970–2977. doi: 10.1016/j.bbagen.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 163.Endoh T, Kawasaki Y, Sugimoto N. Suppression of gene expression by G-quadruplexes in open reading frames depends on G-quadruplex stability. Angew Chem Int Ed Engl. 2013;52:5522–5526. doi: 10.1002/anie.201300058. [DOI] [PubMed] [Google Scholar]
- 164.Mirihana Arachchilage G, Hetti Arachchilage M, Venkataraman A, Piontkivska H, Basu S. Stable G-quadruplex enabling sequences are selected against by the context-dependent codon bias. Gene. 2019;696:149–161. doi: 10.1016/j.gene.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 165.Hagerman RJ, et al. Fragile X syndrome. Nat Rev Dis Prim. 2017;3 doi: 10.1038/nrdp.2017.65. 17065. [DOI] [PubMed] [Google Scholar]
- 166.Agarwala P, Pandey S, Mapa K, Maiti S. The G-quadruplex augments translation in the 5′ untranslated region of transforming growth factor β2. Biochemistry. 2013;52:1528–1538. doi: 10.1021/bi301365g. [DOI] [PubMed] [Google Scholar]
- 167.Serikawa T, Eberle J, Kurreck J. Effects of genomic disruption of a guanine quadruplex in the 5′ UTR of the Bcl-2 mRNA in melanoma cells. FEBS Lett. 2017;591:3649–3659. doi: 10.1002/1873-3468.12855. [DOI] [PubMed] [Google Scholar]
- 168.Bhattacharyya D, et al. Engineered domain swapping indicates context dependent functional role of RNA G-quadruplexes. Biochimie. 2017;137:147–150. doi: 10.1016/j.biochi.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 169.Endoh T, Sugimoto N. Conformational dynamics of the RNA G-quadruplex and its effect on translation efficiency. Molecules. 2019;24:1613. doi: 10.3390/molecules24081613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bugaut A, Murat P, Balasubramanian S. An RNA hairpin to G-quadruplex conformational transition. J Am Chem Soc. 2012;134:19953–19956. doi: 10.1021/ja308665g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Subramanian M, et al. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 173.Valentin-Vega YA, et al. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci Rep. 2016;6 doi: 10.1038/srep25996. 25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chalupnikova K, et al. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J Biol Chem. 2008;283:35186–35198. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fay MM, Anderson PJ, Ivanov P. ALS/FTD-associated C9ORF72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 2017;21:3573–3584. doi: 10.1016/j.celrep.2017.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Khong A, et al. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell. 2017;68:808–820. doi: 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tao EW, Cheng WY, Li WL, Yu J, Gao QY. tiRNAs: a novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J Cell Physiol. 2020;235:683–690. doi: 10.1002/jcp.29057. [DOI] [PubMed] [Google Scholar]
- 178.Lyons SM, Achorn C, Kedersha NL, Anderson PJ, Ivanov P. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 2016;44:6949–6960. doi: 10.1093/nar/gkw418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lyons SM, Gudanis D, Coyne SM, Gdaniec Z, Ivanov P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01278-w. 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ivanov P, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci USA. 2014;111:18201–18206. doi: 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Conlon EG, et al. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife. 2016;5:e17820. doi: 10.7554/eLife.17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huang H, Zhang J, Harvey SE, Hu X, Cheng C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017;31:2296–2309. doi: 10.1101/gad.305862.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Didiot MC, et al. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008;36:4902–4912. doi: 10.1093/nar/gkn472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Marcel V, et al. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis. 2011;32:271–278. doi: 10.1093/carcin/bgq253. [DOI] [PubMed] [Google Scholar]
- 185.Ribeiro MM, et al. G-quadruplex formation enhances splicing efficiency of PAX9 intron 1. Hum Genet. 2015;134:37–44. doi: 10.1007/s00439-014-1485-6. [DOI] [PubMed] [Google Scholar]
- 186.Fisette JF, Montagna DR, Mihailescu MR, Wolfe MS. A G-rich element forms a G-quadruplex and regulates BACE1 mRNA alternative splicing. J Neurochem. 2012;121:763–773. doi: 10.1111/j.1471-4159.2012.07680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang J, Harvey SE, Cheng C. A high-throughput screen identifies small molecule modulators of alternative splicing by targeting RNA G-quadruplexes. Nucleic Acids Res. 2019;47:3667–3679. doi: 10.1093/nar/gkz036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kharel P, Balaratnam S, Beals N, Basu S. The role of RNA G-quadruplexes in human diseases and therapeutic strategies. Wiley Interdiscip Rev RNA. 2020;11:e1568. doi: 10.1002/wrna.1568. [DOI] [PubMed] [Google Scholar]
- 189.Cree SL, et al. DNA G-quadruplexes show strong interaction with DNA methyltransferases in vitro. FEBS Lett. 2016;590:2870–2883. doi: 10.1002/1873-3468.12331. [DOI] [PubMed] [Google Scholar]
- 190.Mao SQ, et al. DNA G-quadruplex structures mold the DNA methylome. Nat Struct Mol Biol. 2018;25:951–957. doi: 10.1038/s41594-018-0131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Saha D, et al. Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion. J Biol Chem. 2017;292:15205–15215. doi: 10.1074/jbc.M117.792077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hussain T, et al. Transcription regulation of CDKN1A (p21/CIP1/WAF1) by TRF2 is epigenetically controlled through the REST repressor complex. Sci Rep. 2017;7 doi: 10.1038/s41598-017-11177-1. 11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sarkies P, et al. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sarkies P, Reams C, Simpson LJ, Sale JE. Epigenetic instability due to defective replication of structured DNA. Mol Cell. 2010;40:703–713. doi: 10.1016/j.molcel.2010.11.009. [ This paper presents genetic experiments that provide the first demonstration of a link between G4s and cellular epigenetic inheritance. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Papadopoulou C, Guilbaud G, Schiavone D, Sale JE. Nucleotide pool depletion induces G-quadruplex-dependent perturbation of gene expression. Cell Rep. 2015;13:2491–2503. doi: 10.1016/j.celrep.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Guilbaud G, et al. Local epigenetic reprogramming induced by G-quadruplex ligands. Nat Chem. 2017;9:1110–1117. doi: 10.1038/nchem.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mirihana Arachchilage G, Dassanayake AC, Basu S. A potassium ion-dependent RNA structural switch regulates human pre-miRNA 92b maturation. Chem Biol. 2015;22:262–272. doi: 10.1016/j.chembiol.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 198.Pandey S, Agarwala P, Jayaraj GG, Gargallo R, Maiti S. The RNA stem-loop to G-quadruplex equilibrium controls mature microRNA production inside the cell. Biochemistry. 2015;54:7067–7078. doi: 10.1021/acs.biochem.5b00574. [DOI] [PubMed] [Google Scholar]
- 199.Pandolfini L, et al. METTL1 promotes let-7 microRNA processing via m7G methylation. Mol Cell. 2019;74:1278–1290.e9. doi: 10.1016/j.molcel.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chan KL, et al. Structural analysis reveals the formation and role of RNA G-quadruplex structures in human mature microRNAs. Chem Commun. 2018;54:10878–10881. doi: 10.1039/c8cc04635b. [DOI] [PubMed] [Google Scholar]
- 201.Rouleau S, Glouzon JS, Brumwell A, Bisaillon M, Perreault JP. 3′ UTR G-quadruplexes regulate miRNA binding. RNA. 2017;23:1172–1179. doi: 10.1261/rna.060962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wang X, et al. Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol Cell. 2017;65:1056–1067.e5. doi: 10.1016/j.molcel.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 203.Fleming AM, Nguyen NLB, Burrows CJ. Colocalization of m6A and G-quadruplex-forming sequences in viral RNA (HIV, Zika, hepatitis B, and SV40) suggests topological control of adenosine N 6-methylation. ACS Cent Sci. 2019;5:218–228. doi: 10.1021/acscentsci.8b00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sahakyan AB, Murat P, Mayer C, Balasubramanian S. G-quadruplex structures within the 3′ UTR of LINE-1 elements stimulate retrotransposition. Nat Struct Mol Biol. 2017;24:243–247. doi: 10.1038/nsmb.3367. [DOI] [PubMed] [Google Scholar]
- 205.Hegyi H. Enhancer−promoter interaction facilitated by transiently forming G-quadruplexes. Sci Rep. 2015;5 doi: 10.1038/srep09165. 9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Silverman RB, Holladay MW. The Organic Chemistry of Drug Design and Drug Action. 3rd edn. Academic; 2015. [Google Scholar]
- 207.Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat Rev Chem. 2017;1 0041. [Google Scholar]
- 208.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat Rev Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 209.Salvati E, et al. PARP1 is activated at telomeres upon G4 stabilization: possible target for telomere-based therapy. Oncogene. 2010;29:6280–6293. doi: 10.1038/onc.2010.344. [DOI] [PubMed] [Google Scholar]
- 210.Biffi G, Tannahill D, Miller J, Howat WJ, Balasubramanian S. Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLOS ONE. 2014;9:e102711. doi: 10.1371/journal.pone.0102711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.O’Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat Rev Genet. 2017;18:613–623. doi: 10.1038/nrg.2017.47. [DOI] [PubMed] [Google Scholar]
- 212.Zimmer J, et al. Targeting BRCA1 and BRCA2 deficiencies with G-quadruplex-interacting compounds. Mol Cell. 2016;61:449–460. doi: 10.1016/j.molcel.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.McLuckie KI, et al. G-quadruplex DNA as a molecular target for induced synthetic lethality in cancer cells. J Am Chem Soc. 2013;135:9640–9643. doi: 10.1021/ja404868t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]