Abstract
The human gut microbiota has been the interest of extensive research in recent years and our knowledge on using the potential capacity of these microbes are growing rapidly. Microorganisms colonized throughout the gastrointestinal tract of human are coevolved through symbiotic relationship and can influence physiology, metabolism, nutrition and immune functions of an individual. The gut microbes are directly involved in conferring protection against pathogen colonization by inducing direct killing, competing with nutrients and enhancing the response of the gut-associated immune repertoire. Damage in the microbiome (dysbiosis) is linked with several life-threatening outcomes viz. inflammatory bowel disease, cancer, obesity, allergy, and auto-immune disorders. Therefore, the manipulation of human gut microbiota came out as a potential choice for therapeutic intervention of the several human diseases. Herein, we review significant studies emphasizing the influence of the gut microbiota on the regulation of host responses in combating infectious and inflammatory diseases alongside describing the promises of gut microbes as future therapeutics.
Keywords: Gut microbiota, Inflammatory disorders, Infectious diseases, Homeostasis, Probiotics
1. Introduction
The human gut microbiota encompasses the entire population of microorganisms viz. bacteria, fungi, archaea, viruses and protozoa that are colonized in the gastrointestinal tract [1], [2]. The gut microbes are tenfold higher than human cells and the coding capacity of the gut microbiota commonly referred to as “microbiome”, is almost hundred times higher in numbers [3]. The gut microbiota in the human colon is primarily comprised of bacteria of four phyla namely Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria in the relative abundance order of Firmicutes≥Bacteroidetes>Proteobacteria≥Actinobacteria [4]. Alongside these, Verrucomicrobia, Fusobacteria, Bacteroides, Ruminococcus and Prevotella are considered as minor contributors [4]. The gut microbiota also contains several bacterial communities that include Clostridium spp, Roseburia spp, Butyricicoccus and lactic acid bacteria (Lactobacillus and Bifidobacterium) [4]. Apart from the bacterial communities, the human gut microbiota is embodied with the virome principally comprised of different phages such as double-stranded and single-stranded DNA bacteriophages belonging to Myoviridae, Podoviridae, Siphoviridae and Microviridae family [5], [6], [7], [8], [9]. These phages infect bacteria of Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria families and regulate their activity by altering the microbial population ratio [9]. Interestingly, each individual has its own phage community and is important for maintaining normal physiology, immunity, and prevention of disease progression in the host body [10]. The notion about the presence of a beneficial gut virome in humans was introduced lately though the presence of the pathogenic ones (Rotavirus, Astrovirus, Hepatitis E virus, Coronavirus, Adenovirus, Torovirus) has been widely accepted for decades [10].
These microbes stay in the human body from birth to death and regulate several important physiological processes like protective, metabolic, trophic and immune functions [11], [12], [13]. The major metabolic functions of the colonic microbes (Escherichia, Bacteroides, Eubacterium, Propionibacterium, Fusobacterium, Bifidobacterium) include fermentation of non-digestible carbohydrates (polysaccharides, oligosaccharides and sugar alcohols) [14], [15], synthesis of certain vitamins (e.g., vitamin B12) [16] and also synthesis of short chain fatty acids (SCFAs, butyrate, propionate andacetate to stream energy to the colon and host) [17], [18]. SCFAs released from the gut microflora regulate colonic physiology by modulating epithelial cell proliferation and differentiation in the intestine and also influence intestinal barrier function [18], [19], [20]. The metabolites released by gut microbes also modulate the metabolic activity of different tissues and organs in the human host such as upregulation of lipid metabolism in adipocytes by inhibiting lipases [21], [22], [23] and lipid digestion in the pancreas by upregulating pancreatic lipase activity [1]. The microbial protein metabolizing machinery constituting proteinases and peptidases plays an important role in regulating the amino acid pool and can supplement human proteinases [1], [17], [24]. The microbial amino acid metabolism pathways yield various bioactive small signaling molecules and antimicrobial peptides [17], [25]. Various signaling molecules from gut microbes can efficiently regulate the function of important human organs viz. cognitive functions of the brain, metabolic functions of the liver and the pancreas [26]. The capability of metabolizing xenobiotics and drugs is another important functional attribute of the gut microbiota which exerts a profound impact on various human diseases including diarrhea, inflammation, and anorexia as well as cancer [25], [27], [28]. The gut microbiota potentially functions in conferring protection to exogenous pathogenic microorganisms by inhibiting their colonization employing multiple mechanisms out of which competition for limited nutrients and modulation of host immunity are crucial [29], [30], [31], [32].The resident bacteria resist colonization of gut pathogens to prevent their invasion through intestinal mucosa/epithelial cells [29], [33], [34], [35] while commensal bacteria compete with nutrients in ecological niches to hamper the microenvironment of the invading pathogens [36]. In addition, the gut microbiota inhibits the passage of the harmful foreign antigens, microbes, and their toxins by promoting the formation of the intestinal epithelial barrier [37]. Another striking feature of the gut microbiota is the immune functioning that is, in part, executed through the regulation of mucosal immunity and the impact of these immune responses are extended to almost all parts of the human body [38].
The composition and functions of human gut microbes are greatly influenced by genetic, physiological, nutritional, environmental and geographical parameters, particularly age, diet, stress, and drugs [39]. The protective functions of the gut microbiota often damaged due to the perturbation by any of the aforementioned factors or by other external factors lead to one or several pathogenic transformations in the host body that result in several life-threatening diseases [40], [41], [42]. Dysbiosis, i.e., breakdown of microbial communities, has been implicated as a cause for several acute and chronic problems such as inflammatory bowel disease, irritable bowel syndrome, metabolic diseases such as obesity, diabetes, allergic diseases, neuro-developmental illness and life-threatening infectious diseases ranging from microbial infections to multicellular parasitic infections [43], [44], [45], [46], [47]. The reestablishment of a healthy microbiota in the gut (eubiosis) by supplementing with the pre- and probiotics is now considered as an effective choice to treat several disorders resulting from the damage of the gut microbiota [48], [49]. Herein, the breakthrough findings achieved by presenting the gut microbes as future therapeutics for treating pathogenic infections and current trends in gut microbial research have been reviewed by incorporating the existing and upcoming approaches.
2. Gut microbiota and human diseases
The extraordinary progress in gut microbiota research has clarified the essential roles of the gut microbial community in governing the fundamental physiological roles in the human physiology and disease which have provided a new dimension for basic and translational research to understand the complexity of host-microbe interactions and to adopt effective means to manipulate it for treating the human health problems. Several human health issues have been found closely associated with the perturbations of the composition of the gut microbiota especially the intra- and interpersonal variations in the microbial consortium amongst the individuals. Alike genetic polymorphism, alterations in the genetic makeup of microbial communities of the human microflora can cause a disease or facilitate its pathogenesis (as reviewed in [50]). Therefore, a clear understanding of microbial communities is of major importance. The changes in the composition of the gut microbiota i.e. the increased or decreased abundance of some of the specialized microbes, can lead to the immune dysfunction and changes in the regulatory responses causing several inflammatory and infectious disorders. The composition of the normal gut microbiota of humans, their functions, and associated health issues have been discussed in Fig. 1 and Table 1 .
Fig. 1.
Human gut microbes and their physiological functions. Gut microbes and their metabolites regulate physiology, development and immune functioning of the human gut epithelium. [1] Gut microbiota confers protection against pathogens through four different mechanisms viz. [i.] direct inhibition of growth and colonization of the pathogens by competing with the nutrients and available space; [ii.] modulation of the immune responses that act against the pathogens but not against the beneficial gut microbes of the human host; [iii.] formation and maintenance of the mucosal barrier that restricts the entry of the invading pathogens; [iv.] utilization of nutrients available in the gut to control the growth of pathogenic organisms and repair of the damaged tissue [2] Gut microbiota regulates the brain function through the gut-brain axis governing modification of nervous system and [3] cognitive function; [4] Gut microbiota influences the metabolic and the biochemical functions of the liver through establishment of the liver-microbiome axis; [5] Gut microbiota can regulate the cardiac function and physiology; [6] Gut microbes potentially regulate the immune surveillance of the lungs through the gut-lung axis; [7] Gut microbes play essential roles in the maintenance of renal physiology and immunity; [8] Gut microbiota maintains healthy prostate and prevents development of cancer; [9] Gut microbiota regulates the tone of lipid metabolism and insulin homeostasis.
Table 1.
Organ specific localization of gut microbes in human gastrointestinal tract, their functions and associated diseases.
| Organ | Name | Normal function within GI tract | Associated Disease | References |
|---|---|---|---|---|
| Esophagus |
Bacteroides sp Pseudomonas sp Rothia sp Streptococcus sp Veillonella sp Streptococcus sp Gemella sp |
Flavanols, Lignins degradation | Type II diabetes, ulcerative colitis pouchitis, Crohn’s disease, Coeliac disease | [51] |
| Stomach | Lactobacillus sp | Folic acid, lactase production, stabilization of intestinal microflora and exclusion of pathogenic bacteria. De-conjugation of bile acids and its hydrolysis, reduction of cholesterol level, decrease risk of colon cancer, tumor suppressor function, modulation of HDL and LDL ratio. | Infection of urogenital and intestinal tracts, prevents Candida albicans and other microbes. | [52] |
| Prevotella sp | Glucose metabolism | Chronic inflammation in individual infected with HIV, Rheumatoid arthritis | [53], [54], [55] | |
| Enterococcus sp | Flavanols, Flavones degradation | Urinary tract infection, bacteremia, endocarditis and meningitis | [56] | |
| Helicobacter pylori | Prevents gastro esophageal reflux disease, esophageal adenocarcinoma. | Chronic inflammation in stomach and duodenum. | [57] | |
| Bacteroides sp | Flavanols, Lignins degradation | Type II diabetes, ulcerative colitis pouchitis, Crohn’s disease, Coeliac disease | [58] | |
| Bifidobacterium sp | Carbohydrate digestion by glycosyl hydrolases, metabolize milk oligosaccharides in infants Colon regularity, prevents gastrointestinal infections. | Colorectal cancer, diarrhea, necrotizing colitis, Inflammatory bowel disease | [59], [60], [61] | |
| Small intestine | Clostridium sp | Flavanones, Flavan-3-ols, Lignins degradation Synthesis of vitamin K and B12, biotransformation of bile acids, degradation of dietary oxalates, carbohydrate and amino acid metabolism, release butyrate, anti-inflammatory environment of the intestine, normal function of colonocytes generation of catecholamines nor epinephrine and dopamine generation of αβ T cell receptor intraepithelial lymphocytes, IgA. |
Colitis, colorectal cancer, Inflammatory bowel disease, microbial dysbiosis | [62] |
| Streptococcus sp | - | Sepsis, diabetes mellitus, cardiovascular diseases, congestive heart failure, obesity | [63] | |
| Lactobacillus sp | Production of folic acid, lactase, stabilization of intestinal microflora, exclusion of pathogenic bacteria, Deconjugation of bile acids and its hydrolysis, reduction of cholesterol level, decrease risk of colon cancer, tumor suppressor function, modulation of HDL and LDL ratio. |
Infection of urogenital and intestinal tracts, prevents Candida albicans and other microbes | [64] | |
| Proteobacteria | - | Diarrhoea-predominant IBS, dysbiosis, Crohn’s disease | [65], [66] | |
| Lachnospira sp | pectin degraders, colonic fermentation of dietary fibers. | Colon cancer, asthma | [67] | |
| Escherichia coli | Prevent the colonization of pathogenic strains of microorganisms, Vitamin K production. |
Opportunistic diseases, gastroenteritis, urinary tract infections, neonatal meningitis, hemorrhagic colitis, and Crohn's disease | [68] | |
| Cecum | Roseburia sp | saccharolytic, butyrate-producing bacterium, prevents ulcerative colitis. | Weight loss and reduced glucose intolerance, irritable bowel syndrome, obesity, Type 2 diabetes, nervous system conditions and allergies | [69], [70] |
| Butyrivibrio sp | Tannins degradation, butyrate production, Fibre degradation, protein breakdown, bio hydrogenation of lipids and the production of microbial inhibitors. |
Enterocolitis | [71], [72], [73] | |
| Ruminococcus sp | digest resistant starches, reduce risk of diabetes and colon cancer. | Irritable bowel syndrome | [74], [75], [76] | |
| Fecallibacterium sp | butyrate and other short-chain fatty acids generation through the fermentation of dietary fiber. | Crohn's Disease, obesity, asthma and Major Depressive Disorder | [77], [78], [79] | |
| Fusobacteria sp | - | Colon cancer, ulcerative colitis. | [80] | |
| Colon | Porphyromonas sp | - | Atherosclerosis | [81] |
| Eubacterium sp | Flavanols, Lignins degradation, cleavage of polyphenolic glycoside, deglycosylating certain flavone C-glucosides and of hydrolyzing O-glucosides of flavones and isoflavones, glucose utilization and fermentation of intermediates acetate and lactate, to form butyrate and hydrogen. Oxalate catabolizing species. | Bacterial vaginosis in women, UC pouchitis | [82], [83], [84] | |
| Enterobacterium sp | - | Maternally transmitted colitis, Acute cholecystitis. | [85], [86] | |
| Peptostreptococcus sp | Lignins degradation | Brain, liver, breast, and lung abscesses, as well as generalized necrotizing soft tissue infections. They participate in mixed anaerobic infections. | [87] |
2.1. Gastrointestinal diseases
Dysregulation of the immunological homeostasis is the cause of more than 100 life-threatening human diseases to date [67], [88]. Since gut microbiota controls the immunological homeostasis, ‘dysbiosis’ is therefore proven to induce and promote several gut-associated diseases primarily alongside diseases in other body parts of the human [67], [88]. Inflammatory bowel disease (IBD) is one of the very common and debilitating problems of modern age humans characterized by the induction of inflammation and subsequent development of mucosal lesions [89], [90]. The pathogenesis of IBD is mainly caused by the perturbation of normal gut microbial communities [1], [91]. The loss of bacteria belonging to the Firmicutes phylum and abundance of some Gammaproteobacteria, collectively impair the mucosal immune response to normal bacterial flora and disrupt regulatory functions of T-lymphocytes playing a key role in inducing chronic intestinal inflammation to develop IBD [1], [91], [92], [93]. The loss of diversity in fecal communities has been consistently shown in IBD cases and on an average, 25% fewer microbial genes could be detected in the fecal community of IBD patients [94], [95], [96]. The most prominent change in the microbial diversity associated with IBD is the decreased abundance of Firmicutes phylum that includes loss of Faecalibacterium prausnitzii, a bacterium that maintains inflammatory homeostasis of the gut with its potential anti-inflammatory effect [91]. Bacteroides fragilis, B. vulgatus, Ruminococcus albus, R. callidus and R. bromii are also considered as the influential members of the gut microbiota in healthy subjects, however, IBD patients display more than fivefold depletion in these microbial loads [97]. Conversely, the specific taxa that are increased in IBD patients include Enterobacteriaceae, such as Escherichia/Shigella [91], [98].
Irritable bowel syndrome (IBS) is characterized by abdominal pain or discomfort and altered bowel habits. Although the etiology is multifactorial, recent understanding of the pathophysiology of IBS revealed that variations in the normal gut microbiota may play a critical role in the low-grade intestinal inflammation associated with the syndrome [66], [99], [100]. Gut dysbiosis is thought to be involved in IBS pathogenesis through facilitating the adhesion of pathogens to the bowel wall [101]. IBS patients are characterized by an increase in Firmicutes (Ruminococcus, Clostridium, and Dorea), depletion in Bifidobacterium and Faecalibacterium spp. [102] along with an abundance of Firmicutes to Bacteroidetes ratio [103], [104], [105]. Pediatric IBS patients demonstrate alterations amongst the members of Firmicutes and Proteobacteria like adults. In addition, low levels of the genus Bacteroides and increases in Alistipes were also linked with a greater frequency of pain in the pediatric patients [66], [106]. Celiac disease (CD) is another inflammatory systemic disorder of gut characterized by high proinflammatory responses following tissue damage in response to gluten proteins present in the human diet [107]. The gut microbiota plays a profound role in regulating the pathogenesis of CD. Intriguingly, a change in the microbial consortium in infants is considered as a key reason to develop CD in adulthood [107].
2.2. Cancer
The ability of the gut microbiota to modulate host physiology, metabolism as well as differentiation and functioning of immune cells, is linked with the establishment of a pro-inflammatory or anti-tumor milieu [108]. Certain strains of the gut microbes and their by-products (hydrogen sulfide) have been reported to be associated with the development and progression of cancers in intestinal and extra intestinal tissues [109], [110]. For example, pathogenic cyclomoduling-positive E. coli strain was reported for promoting the pathogenesis of colorectal cancer [111]. Similarly, the enterotoxigenic Bacteroides fragilis strain possesses direct oncogenic effects on intestinal epithelial cells [112]. The factors corresponding to dysbiosis viz. pathogenic foreign organisms in the gut, commensals, environmental factors (antibiotics, xenobiotics, smoking, hormones, and dietary influences etc.) are accompanied with the development and progression of intestinal or extraintestinal neoplasms [113]. Moreover, intra-abdominal infections, robust use of antibiotics leading to the alteration of gut microbial composition may work in concert for the incidence and progression of colorectal carcinoma, and extraintestinal cancers (breast and hepatocellular carcinoma) [109], [111], [113], [114], [115], [116], [117] predominantly through the dysregulation of inflammatory and metabolic circuits. Several current studies that assessed beta diversity through 16S rRNA sequencing have illustrated structural differences amongst the gut microbiota, where samples belonging to different disease status (cancer, adenoma, or control/normal adjacent tissue) cluster in different two-dimensional spaces indicating the presence of dysbiosis [118], [119], [120]. Analysis of community diversity/richness indices has also shown a significant reduction in the microbial diversity in the feces of colorectal patients in comparison to the healthy controls [119]. The mechanism of cancer progression relies on the manifestation of the genomic and epigenomic instability [121]. The genetic abnormalities in the intestinal immune system also play a significant role in the neoplastic transformations [29]. Deficiency or mutation of activation-induced cytidine deaminase results in deficiency or dysfunction of IgA promoting dysbiosis [29]. On the otherside, the epigenetic modification (hypermethylation) of CXXC-chemokine ligand 16 gene results in the accumulation of intestinal NKT cells that promote the development of colitis [122]. Microbial metabolites (H2S, N-nitroso compounds, polyamines, and equol) have direct effects on DNA such as the development of microsatellite unstable or chromosomal unstable tumors and modulation of DNA repair system through modifications of histone proteins by acetylation or CpG island methylation [123], [124]. The microbial products also possess the potential to suppress cancer progression. 3-methylbutyrolactone, kynurenic acid, and 3-methyladenine from Lactobacillus johnsonii have been reported to reduce the gene damage and inflammation by altering the central carbon metabolism that ultimately plays a beneficial role in cancer prevention [125].
Gut microbes are essential regulators of the functions of the anticancer drugs, especially the platinum-based chemotherapeutics that primarily acts through the formation of DNA adducts and breakage of the double stranded DNA. In GF mice or mice depleted of gut commensals, the platinum-based anti-cancer drug can form platinum-DNA adducts but the DNA damaging effect of the drug is dramatically reduced [108]. Moreover, the activity of several anticancer drugs significantly decreases in absence of the commensal microbes (reviewed in [108]).
2.3. Metabolic diseases
Obesity and associated inflammatory disorders, as well as metabolic abnormalities collectively known as metabolic syndrome comprised of hyperglycemia, hyperlipidemia, insulin resistance/diabetes, obesity and hepatic steatosis, are currently considered as the major threats to human civilization [126], [127]. An imbalance in the energy uptake and energy expenditure is considered as the major cause behind these complex metabolic abnormalities [128], [129]. Although the lifestyle and genetic factors are also considered as the influential determinants of obesity, recent research exploring physiological significance on the gut microbiota suggested it to be a key environmental factor that influences the metabolic syndrome [128], [129]. Another striking feature of the gut microbiota is the heritability that links abundance of the specific gut microbial taxa to the host genetics [130] and amongst the heritable taxa, Firmicutes phylum is associated with leanness [47]. Composition of the gut microbiota is influenced by host genetics and can impact host metabolism [131]. Diet-induced perturbation of gut microbial composition [132] results in an increase in the ratio of Firmicutes to Bacteroidetes as well as a marked reduction in the bacterial diversity. These features of the gut microbiota promote obesity in both genetic- and diet-induced mouse models [133], [134] and in clinical obesity [134], [135], [136]. In particular, antibiotic-induced disruption of the gut microbiota in early life increases the risk of childhood obesity [137], [138], [139]. Hitherto, studies conducted on the relative influence of the gut microbes in obesity indicated that the relationship between the obesity and human gut microbiota is highly complex and difficult to interpret [46]. However, it is clear that a change in the composition of the gut microbiota is associated with obesity [134], [140].
2.4. Allergies
Allergic diseases are global health concerns affecting over half a billion people worldwide. Several genetic, molecular and environmental risk factors are associated with the development of allergies including the gut microbiota. The gut microbiota is considered a crucial environmental factor playing a key role in regulating the severity of allergic diseases [141]. Reduced gut microbial diversity has been reported to increase the risk of allergy in school children and in most of the cases, it is food allergy [142]. A recent study has shown that depletion of bacteria like Bifidobacterium, Akkermansia, and Faecalibacterium, along with an abundance of fungi such as Candida and Rhodotorula in neonates, may predispose to allergic susceptibility by influencing T-cell differentiation [47]. Furthermore, a study by the Canadian Healthy Infant Longitudinal Development (CHILD) identified a transient alteration in the gut microbiota associated with the reduced abundance of bacteria belonging to Lachnospira, Veillonella, Faecalibacterium and Rothia genera during the first 100 days of life in the infants found to increase the risk of asthma [46]. Moreover, induction of allergic responses (mostly in children) after consumption of antibiotics like penicillin has also been documented [143]. The depletion of gut microbiota in mice through the oral administration of antibiotic vancomycin has been reported to promote susceptibility to allergic airway inflammation (AAI) and serum IgE response [47], [144], [145]. Colonization of GF mice with the aforementioned microbes ameliorated AAI in offspring which demonstrates a causal role of the bacterial communities in reducing susceptibility to allergy [47]. The gut microbiota of infants having cow’s milk allergy is different in composition and diversity than that of the non-allergic infants [146]. Studies have suggested that dysbiosis also plays a key role in the onset and pathogenesis of several inflammatory autoimmune disorders including rheumatoid arthritis, multiple sclerosis, systemic lupus erythematosus (SLE), type-I diabetes, and Crohn’s disease [147], [148], [149]. The functional role of the gut microbiota in regulating pathogenesis of inflammatory diseases has been depicted in Figs. 2 and 3 .
Fig. 2.
Gut microbes induced modulation/perturbation of the inflammatory homeostasis in human. Gut microbes can directly sensitize the gut epithelium associated antigen presenting cells for inducing proinflammatory response directly by enhancing the proinflammatory cytokine production and/or polarizing the naive T cell towards Th1 and Th17 phenotypes. The metabolites (largely the short chain fatty acids (SCFAs)) released from the gut microbes bind to the intestinal free fatty acid receptors (like GPR43) and induces the expression of FoxP3 to signal polarization of regulatory T cell (Treg) response. This Treg response plays a crucial role in inhibiting the inflammatory responses resulted from the pathogenic infections or the inflammatory disorders/autoimmunity and also maintain the immune homeostasis in the gut. SCFA like butyrate acts as a ligand for the immunoregulatory receptors (like GPR109a) on the macrophages and drives the polarization of M2 macrophages (anti-inflammatory phenotype of macrophage) that secrete anti-inflammatory cytokines (IL-10, TGF-β). These anti-inflammatory cytokines suppress the effects of proinflammatory cytokines (IL-12, IL-17, IFN-γ) released from the antigen presenting cells (macrophages and dendritic cells) and sensitized T cells (Th1 and Th17) induced from dysbiosis and/or microbiota-pathogen interactions. SCFA also induces IL-18 expression through inflammasome activation in the intestinal epithelial cells (IECs). IL-18 released from the IECs further triggers activation of the innate lymphoid cells to secrete IL-22 that guides synthesis of antimicrobial peptides and mucin to maintain intestinal homeostasis.
Fig. 3.
Mechanism of the function of gut microbes against inflammatory, metabolic and allergic diseases. Gut microbes confer protection against the physiological abnormalities principally via two different approaches. The gut microbes itself and the metabolites released from the microbes can function separately or together to restore the homeostasis. Short-chain fatty acids (SCFAs) released by the gut bacteria potentially interact with the receptors on the dendritic cells (DCs) resulting in the secretion of anti-inflammatory cytokines that ameliorate or suppress proinflammatory milieu associated with the pathological outcomes of dysbiosis, inflammatory bowel disease, and cancer. SCFA alone and the cytokines released from SCFA-educated DCs inhibit inflammasome activation. Moreover, SCFA sensitized DCs promote the induction and expansion of the anti-inflammatory immune cell types the regulatory T cells (Treg) that regulate the detrimental effects of the proinflammatory cytokines. SCFAs can directly inhibit DC activation and concomitant upregulation of the Th2 responses that subdue the pulmonary inflammation. The Tregs alongside other anti-inflammatory cytokines are also indispensable for preventing the hypersensitivity. Gut microbes potentially prevent the trans-epithelial entry and the translocation of pathogenic microorganisms to prevent a human from systemic inflammation or sepsis. The gut microbes also maintain the normal metabolic activity of the adipocyte by upregulating the browning of white adipose tissue and inhibiting the lipogenesis. In addition, gut microbes block the entry of LPS from pathobionts and inhibit TLR4 activation. This inhibition of TLR4 ceases systemic inflammatory responses and plays a pivotal role to maintain the normal insulin activity required for the metabolic homeostasis in the human host.
3. Mechanism of the function of gut microbiota
The gut microbiota can efficiently prevent pathogen infections and overgrowth of harmful pathobionts to reduce the chance of developing inflammatory and infectious diseases [150]. In homeostatic conditions, a healthy gut microbiota puts the gut mucosal immune system in a challenging situation that is tolerant to the beneficial commensals but prevents the overgrowth of the resident pathogens.
3.1. Physicochemical mechanisms: Human-Gut microbiota interactions
The gut microbiota limits the infectivity of the pathobionts in the gut epithelium by competitive exclusion [29]. Colonization resistance (CR) is one of the key mechanisms by which the human gut microbiota confers protection to infectious microbes [30], [33]. CR is known to be executed via four inter-related mechanisms, viz. [i] direct inhibition of the surrounding bacteria by producing toxic metabolites; [ii] maintaining the mucus barrier as well as the underlying intestinal epithelium; [iii] manipulation of the host’s immune responses; [iv] competent utilization of available host nutrients [151]. Out of these, the foremost is the competition for space and nutrients along the mucus layer and within the lumen [151]. Commensals uptake mucopolysaccharides and limit the expansion of less well-adapted invading microbes [151]. Metabolites released by the gut microbes also exert potential antimicrobial action [152]. SCFAs, synthesized by commensal bacteria, possess bactericidal action against some enteric pathogens [30]. For instance, Lactobacilli sp. generates reactive oxygen species to kill the pathogenic microbes [153]. Numerous commensals produce bacteriocin and bacteriocin-like molecules with different types of antimicrobial properties viz. nucleases (DNases), inhibitors of biosynthesis of cell wall component (eg, peptidoglycan) and pore formers [154], [155]. Furthermore, gut microbes exert a different degree of influences over the pathogens and beneficial microbes. Hosts with high Escherichia coli densities are more susceptible to Salmonella enterica infection, while hosts having high Lactobacilli show the reverse [156]. The molecular relationship between the disease susceptibility and the features of microbial communities leading to the protection against the infectious diseases remains an open question to the scientific communities to date.
Another simple mechanism of antimicrobial protection is the presence of the two-tiered mucus layer, which keeps luminal and mucosal microbes away from the epithelial contact, predominantly in the large intestine [1], [157], [158]. The inner epithelial layer is denser and does not permit any organism while the outer layer is more dynamic and provides glycans as nutrients for the gut organisms [1]. The mucus layer in the small intestine is discontinuous and inadequate and therefore antimicrobial proteins from the gut microbes play the major role in maintaining gut health [1]. Especially, the structural components and metabolites are known to induce synthesis of the antimicrobial proteins (AMP) such as cathelicidins, C-type lectins, and (pro) defensins by the host Paneth cells via pattern recognition receptor (PRR)- mediated mechanism [1], [159], [160], [161]. The membrane-associated TLRs, C-type lectin receptors (CLRs) viz. dectin-1, and the cytosolic nucleotide-binding and oligomerization domains (NOD) like receptors (NLRs) constitute PRR family in the gut [162]. Moreover, metabolites released from gut microbes can enhance the host’s antimicrobial humoral immunity. Lactate from Lactobacillus sp. augments the antimicrobial activity of the host lysozyme by disrupting the outer membrane of the bacterial cell wall [1]. Enteric viruses present in the gut microbiota also exert immunomodulatory effects on the host [163]. The first evidence on the role of enteric viruses in chronic GIT inflammation came from the studies in mice [164]. Alike the commensals, gut viruses also exploit TLR3 and TLR7 mediated pathways to confer protection against the inflammatory pathology of colitis and cancer [165], [166].
3.2. Manipulation of human immune response
Signaling molecules derived from the gut microbiota are critical for the development of the immune system [167]. Comparative studies on the germ-free animal models [168], [169], mice with disrupted microbiota and mice with healthy microbiota clearly revealed the indispensable roles of the gut microbiota in shaping innate and adaptive immunity and maintaining the immune homeostasis in the human [168], [169]. The localized immune crosstalk observed between the gut microbiota and local immune cells constitute the first line of interactions behind eliciting the mucosal immune responses [170], [171] and these interactions not only regulate the physiological functions of the host but also play a key role in building the protective response against pathogens [172], [173].
During a pathogenic infection, gut microbiota promotes the production of IgA that binds to the invading microbes at mucosal surfaces, neutralizes toxins and contributes to the microbial tolerance [174], [175]. Besides these, the microbial molecular patterns are recognized by the pattern recognition receptors (PRR) likewise toll-like receptors (TLRs) or nod-like receptors (NLRs) located on the APCs co-evolved with the microbiota and play an important role epithelial cell homeostasis [176]. The microbiota further contributes in priming the signal of the inflammasome pathway by inducing the transcription of the proinflammatory cytokines viz. tumor necrosis factor (TNF)-α, interleukin (IL)-6, as well as pro-IL-1β and pro-IL-18 that are further converted to their activated forms by proteolytic cleavage [177]. The gut microbiota modulates functions of the APCs (macrophages and dendritic cells) to protect the host body from pathogenic infections and maintain immune tolerance against gut microbes itself [167]. For example, dendritic cells (DCs) of Peyer’s patches exhibit a high level of IL-10 compared to the splenic DCs under similar conditions [178]. Neutrophil activity is also influenced by the microbiota as GF rats are mainly neutropenic and characterized by an impairment in the generation of superoxide anion and nitric oxide alongside decreased phagocytic function [179].
3.3. Microbiota and adaptive immune response
The gut microbiota regulates the polarization of the primary immune signal towards different arms of T cell responses from naïve intestinal CD4+T cells [180], [181]. Th1 and Th17 polarized cells are critical for the host defense against the invading pathogenic microbes while Th2 cells act against the parasitic infections [182]. Segmented filamentous bacteria (SFB) adhere to the IEC cells to activate IL-23 pathway leading to IL-22 production by the innate lymphoid cells (ILC3) for inducing a Th17 response to confer protection against bacterial and fungal infections [183]. On the other side, polysaccharide A from Bacteroides fragilis has been found to stimulate the induction of CD25 FoxP3+ regulatory T cells (Tregs) via TLR2 activation [184], [185]. This Treg response is essential to ameliorate pathological outcomes of the proinflammatory milieu resulted from Th1 and Th17 responses. Moreover, released SCFAs (propionate and butyrate) can potentially suppress the Th1 response by inhibiting the activation of nuclear factor kappa B (NF-κB) to subdue the inflammatory cytokine production [186]. Amongst the SCFAs, butyratepotentially induces the FoxP3+ Treg differentiation [187]. Furthermore, the SCFAs have been demonstrated to promote mucus production by intestinal goblet cells, induction of secretory IgA and inflammasome activation to result in IL-18 secretion [188], [189]. Gut microbe-induced immunomodulation strategies for therapeutic intervention of inflammatory diseases have been presented in Figs. 2 and 3.
4. Gut microbes and infectious diseases
The gut microbiota plays a pivotal role in regulating the pathogenesis of infectious micro- and macro-organisms. The relative influence of the gut microbiota against pathogenic infections is not only limited within the gut but also extended to the other parts of the human body. However, the mode of interaction varies with the nature of pathogenic infections. For example, the interaction between the gut microbiota and a bacterial pathogen is different than that of the gut microbiota- parasite interaction.
Gut microbes prevent the growth of the invading gut pathogens largely by competing with space and nutrients available in the gut [47]. On the other hand, manipulation of the host immunity is another key strategy behind the gut microbiota-induced protection against infectious pathogens within and outside the gut [47]. Despite the tremendous progress made in gut microbiota research throughout the last decade, the molecular insights of the gut microbiota-mediated regulation of pathogenesis of infectious pathogens are still considered as a shaded area in our understanding. In this section, we have discussed the potential regulatory role of gut microbes in altering the pathogenesis of different infectious diseases.
The gut microbiota explores multiple mechanisms to regulate the infectivity and pathogenesis of several gut pathogens, including bacteria, protozoa, viruses, and parasites. A healthy gut microbiota protects entry (translocation) of the pathogens inside the host body, whereas a leaky gut due to dysbiosis results in the reverse leading to pathogenic outcomes. Gut microbes protect the host from infectious pathogens through the selective inhibition of growth, killing and modulation of the infectivity of the pathogens [190]. The mechanism of direct inhibition is executed through competition between the gut microbes and invading pathogenic microbes. For example, the virulence and pathogenesis of several protozoan parasites such as Entamoeba histolytica [191], Giardia sp. [192], pathogenic bacteria like Helicobacter pylori [12], Salmonella sp. and Enterococcus sp. [193] are regulated by the gut microbes principally through direct inhibition. A similar mechanism of protection is also conferred against other enteric infections like rotavirus, Shiga toxin-producing E. coli (STEC), enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), Salmonella enterica Typhi and Vibrio cholerae [194]. The gut E. coli communities inhibit STEC largely by competing with the nutrient source (carbon). Moreover, metabolites released from the gut commensals such as deoxycholic acid (Clostridium scindens) and post-translationally modified peptides namely bacteriocins and microcins (E. coli, E. faecalis) directly inhibit the growth of the enteropathogens [194]. Gut microbial metabolites can also selectively alter the pathogenicity of infectious microorganisms. For example, Ruminococcus obeum releases mutagenic autoinducer (AI)-2 that attenuates expression of the genes encoding the virulence factor of V. cholerae [194]. The bioactive SCFAs (acetate, butyrate, and propionate) from gut microbial (Lachnospiraceae) metabolome possess direct influence over the pathogenesis of the enteropathogens such as Salmonella through repressing the expression of the virulence genes [195]. Similarly, the SCFAs secreted from human gut microbes like Roseburia spp. and Bacteroides ovatus also reported to inhibit the growth and virulence of the opportunistic yeast, Candida albicans through triggering the TOR signaling pathway [196].
Intriguingly, the gut microbiota and their metabolites can regulate several other pathogens infecting distantly from the gut. Pathogenesis of Trichomonas vaginalis, a major vaginal pathogen, and its endosymbiont Mycoplasma hominis has been reported to be regulated by the gut microbial composition [197], [198]. Interestingly, recent studies conducted on mouse models suggested that the composition of the gut microbiota modulates the host’s susceptibility to several enteropathogens [134], [190].
Gut microbes, largely the commensal bacteria, exert multiple facets of regulation to modulate and shape the anti-pathogen host immunity as well as for facilitating their own survival and fitness. SCFAs, specifically butyrate drives polarization of the host macrophages typically towards M2 via activation of H3K9 acetylation to promote STAT-6 mediated transcription [199]. M2 macrophages reduce the inflammatory pathology resulting from the host-pathogen interaction and facilitate the repair of damaged tissues [199], [200]. The commensal community controls pathogens beyond the gut through modulating the adaptive immunity specifically augmenting the differentiation of T cell subsets not only through self-/non-self-recognition processes but also directly educating them to induce anti-infectious immunity [201], [202]. The commensal bacteria also induce a low degree of IgA response to establish a noninflammatory gut environment essential for maintaining the normal physiological processes regulated by the host-gut microbiota relationship [203]. In addition, gut bacteria also enhance specific antimicrobial response. Bacteroides thetaiotaomicron induces synthesis of the antibacterial peptides namely angiogenins (Human angiogenin (Ang), Ang1 and Ang4 in mouse) that selectively act on the pathogenic microbes [204]. Moreover, metabolites secreted from gut commensals simultaneously induce a selective lethal response to invading pathogens and also promote regulatory responses to protect beneficial microbes in the gut [205]. For example, the polysaccharide A (PSA) produced by Bacteroides fragilis provides protection from inflammation resulted from Helicobacter hepaticus infection [205]. The PSA sensitizes DCs to prime T cell responses by presenting PSA on the MHC class II to CD4+ T helper (Th) cells for inducing an anti-inflammatory response (IL-10) that inhibits pro-inflammatory cytokines (IL-17, IL-23, and TNF-α) to suppress H. hepaticus induced immunopathology [205]. Intriguingly, such IL-10 producing T cells are the Foxp3+ Tregs commonly referred to as “inducible Tregs” [205].
Recent experimental investigations revealed exciting results in favor of adopting the microbiota-based strategy as a future therapeutic option for treating the life-threatening infectious diseases viz. tuberculosis, malaria, HIV through microbiota-based solutions [194]. Administration of the engineered Lactobacillus isolates (L. jensenii, L. casei, L. reuteri) expressing anti-HIV proteins found efficacious in inhibiting the HIV infection in in vitro cell culture models [206], [207]. Although several medications are available for tuberculosis and malaria, due to the several failures viz. the emergence of resistant strains and the lack of personalized treatment, the gut microbiota-based treatment is now considered as an effective approach. Considering the regulatory role of the gut microbiota on host immunity for maintaining the pulmonary physiology (lung-gut axis), prompted the use of the gut microbes and/or probiotics-based therapy for treating tuberculosis (TB) [208]. Gut microbes confer protection to TB by enhancing the synthesis of the antimicrobial components by Prevotella and Veillonella in the lungs and by inducing the pro-inflammatory response in the pulmonary lymphocytes [209]. However, dysfunction of the gut microbiota could cause an abundance of anaerobes (Prevotella, Veillonella, and Haemophilus) in the lung which in turn results in the risk of M. tuberculosis infection in the humans by lowering TH1 and TH17 responses [194]. On the other hand, an understanding of gut microbiota-induced protection against malaria is relatively poor. It has been postulated that gut bacteria (Bifidobacteria and Lactobacillus) can efficiently regulate the pathogenesis of the malarial parasite by modulating the anti-malarial immunity of the host [210]. Induction of anti-α-gal antibodies by a gut pathobiont E. coli O86:B7 has been reported to cross-react with the sporozoites of human and rodent Plasmodium spp. and result in the impairment in the transmission of the parasites between the vector and the vertebrate host [211]. However, it remains unclear how the gut microbiota regulates the pathogenesis of Plasmodium during cerebral malaria. But the widely accepted fact is that a change in the gut microbiota increases the risk of the malarial infection [194], [210], [212], [213]. The current scenarios of malarial research have demonstrated the complication in the disease pathology due to the parasitic co-infections existing with malaria [214]. Intriguingly, mild treatment with antibiotics followed by a probiotic supplementation through the diet displayed a reduction in the parasite burden and in the disease severity [210]. Toxoplasma gondii, a gut pathogenic protozoan, alters the gut microbial composition by increasing the Gram-negative commensals such as Enterobacteria, Bacteroides and Prevotella sp [215]. T. gondii was also found to exist as a co-infection in the malarial subjects and regulate the pathological manifestation of malaria [216]. Therefore, the T. gondii co-infection could be responsible for the alteration of the gut microbial composition in malaria and this may have a connection with the malarial pathogenesis. However, this postulation still remains an open question in the scientific community. Gut microflora can protect the host from inflammatory damages due to T. gondii driven hyperactivation of the DCs, over secretion of the pro-inflammatory cytokines (IL-12, IFN-γ) and TLR activation [217]. Different gut bacteria viz. Clostridia, Bacteroides, Lactobacillus, Campylobacter, Eubacterium, Bifidobacterium and segmented filamentous bacteria also provide a similar kind of protection to the other parasitic protozoa such as E. histolytica, Cryptosporidium parvum and Giardia spp that infect human host through the gut [218]. Perturbation of the gut microbiota displaying an abundance of the pathobiont Prevotella copri and induction of inflammation are considered as the cardinal markers of E. histolytica infection [219]. Dietary supplementation of the probiotic bacteria like Lactobacillus acidophilus, Lactobacillus johnsonii, and Lactobacillus casei showed potential in restoring gut function by stimulating the mucosal and humoral immune responses to eliminate the enteropathogens [218].
On the other hand, the protective effects of the gut virome have been recently reported in the case of patients with Clostridium difficile infection [211], [220]. The marked abundance of the certain microbial genes in the study cohorts suggests that the viruses may have a regulatory role in different diseases such as diarrhea and pneumonitis [10], [221], [222]. The metagenomic evidence on the abundance of Adenoviridae and Picornaviridaein the stool samples of children with diarrhea indicated a clear involvement of the gut virome [221]. An interesting study by Kernbauer et al, [223] demonstrated that murine norovirus (MNV), an enteric RNA virus is capable of replacing the beneficial functions of the commensal bacteria in the intestine, can support the intestinal homeostasis and can regulate the mucosal immune response likewise the commensals. Apart from the viruses present in the gut, the intestinal bacteriophages have also been taken under consideration [224]. Lim et al. [225] have studied that Siphoviridae and Anelloviridae are prevalent in the early life but gradually decrease with age while Microviridae gradually increases with the age. In addition to metabolic disorders and cardiovascular disease, changes in the virome community are often associated with several infectious and autoimmune diseases [226], [227]. Human stool derived phage-based therapies are emerging as new hopes for preventing the pathogenesis of multi-drug resistant Enterobacteria [228], and, burn wound-causing Escherichia coli and Pseudomonas aeruginosa [229]. This target specificity of phages provides the opportunity for treating specific bacterial pathogen/community.
Apart from pathogenic microorganisms, the gut microbiota also controls the macro-parasite loads in the human gut and also invading pathogenic parasites. Helminths and bacteria are the co-habitants in the human intestine and interact with each other. These interactions not only maintain the gut homeostasis but also inhibit the pathogenic functions of each other. The gut microbiota potentially modulates the pathogenesis of soil-transmitted helminthiasis (STH), a debilitating problem in the developing countries [230]. Comparative studies in GF model, gut microbes especially Lactobacillus sp. has been demonstrated for inhibiting the infection and/or reducing the burden of Trichuris muris, Heligmosomoides polygyrus/ bakeri/Nematospiroides dubius, Nippostrongylus brasiliensis, Trichinella spiralis and Ascaridia spp. whilst Bifidobacterium animalis found conferring protection against Strongyloides venezuelensis (reviewed in [230]). Interestingly, immunopathological outcomes associated with some ofthe intestinal parasitic infections, likewise in the case of Schistosoma mansoni (a trematode), are promoted by the gut microbiota [231]. However, many of these parasites play essential roles to inhibit the hypersensitivity in the human. Therefore, the intriguing question is whether the reduction of the eukaryotic parasite load could lead to the increase of autoimmune responses or not. Recent progress has also indicated the possible significance of gut microbial action over the devastating parasitic diseases viz. malaria, filariasis as well as other parasitic infections which need to be addressed in the near future. The role of gut microbes in regulating pathogenic infections has been demonstrated in Figs. 4 and 5 .
Fig. 4.
Mechanism of gut microbiota induced protection against the microbial infections. The gut microbes exert a protective action on the microbial infections by direct inhibition, competition with nutrient and inducing antimicrobial immunity. The antimicrobial immune response comprises excess mucus secretion from goblet cells, synthesis of antimicrobial peptides and induction of pro-inflammatory response. The gut microbes trigger the Paneath cells to secrete antimicrobial peptides that control the growth of pathogenic microorganisms. Moreover, the gut microbes also result in the copious production of mucosal IgA to act against the pathobionts especially by blocking their adherence to the intestinal epithelium. Polysaccharide A (PSA) from gut bacteria sensitizes the DCs via TLR2 activation and subsequently stimulates the induction of Tregs. Anti-inflammatory cytokines secreted from the PSA-educated DCs also inhibit the immunopathological effects of the proinflammatory cytokines secreted in response to the various infectious agents. On other side, gut microbes efficiently induce and shape the host immunity against the invading pathogens by directly activating the proinflammatory T cell responses (TH1 and TH17) through the activation of antigen presenting cells (macrophages and DCs). The gut microbes, especially the commensals, restrict the entry of gut pathogens through the formation of mucosal barrier and prevent the bacterial translocation in the circulation.
Fig. 5.
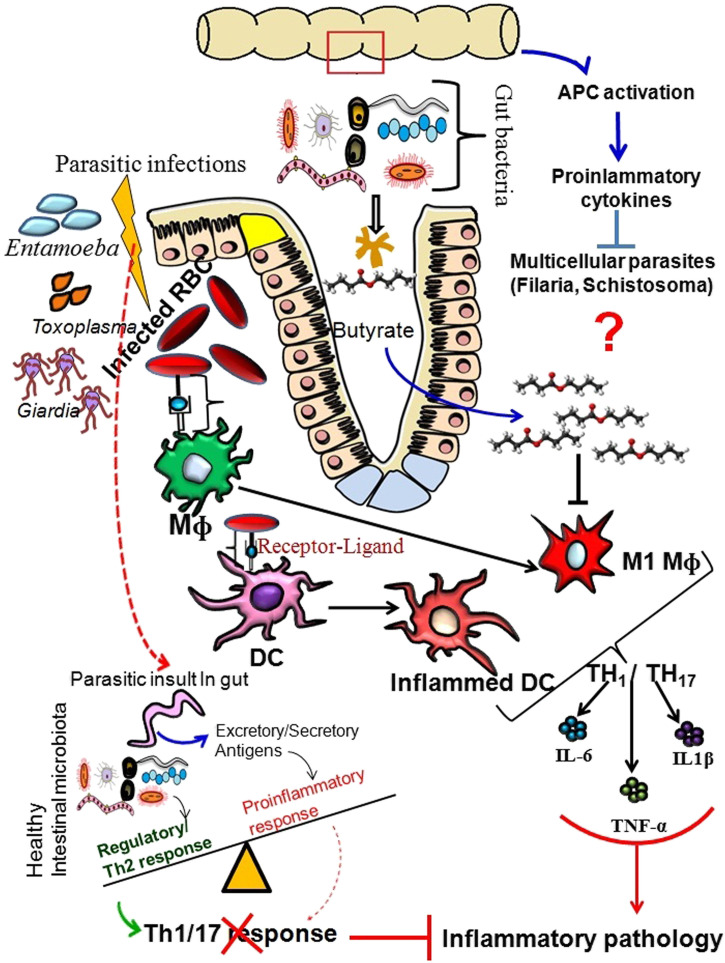
Mechanism of the gut microbe induced protection against the parasitic infections. The gut microbes release short-chain fatty acids (such as butyrate) that block polarization of the M1 macrophages and activation of the DCs to suppress the overt immunopathology associated with the different parasitic infections (Malaria, Giardia, Entamoeba, and Toxoplasma). Multicellular parasites in the gut, especially the gut helminths, also drive Th2 response to strengthen the anti-inflammatory responses and reduction of tissue damage. A healthy gut microbiota directs the major antigen presenting cells (APCs) to secrete anti-inflammatory cytokines that suppress immunopathogenesis associated with the different parasitic infections (Malaria, Toxoplasma, Giardia, Entamoeba, and helminths). The exact role of the gut microbiome in the control of multicellular parasitic infection is yet not clear. However, APC activation leading to the secretion of proinflammatory cytokines is considered one of the key mechanisms to inhibit the pathogenesis of multicellular parasites.
5. Gut microbes as therapeutics: Current trends of using gut microbes, prospects and challenges
The recent advancements in exploring the therapeutic possibility of gut microbes in treating inflammatory and infectious diseases have suggested that most of the human diseases discussed in earlier sections are due to the breakdown of the normal microflora and therefore, reconstruction of the microbial community through the administration of appropriate microbes is now considering as an effective mean [232]. In this context, development of the advanced tools and techniques played a major part.
5.1. Molecular approaches in gut microbiota research
The molecular identification and phylogenetic analyses through the sequencing of the bacterial 16S rDNA and internal transcribed spacer region 2 (ITS2) of the fungal 26S rDNA are commonly used for studying the human gut microbiome [47], [233], [234]. Shotgun sequencing of the microbial metagenomes and metatranscriptomes from fecal material and/or biopsy are particularly used for determining personalized and community-based variation amongst gut microbiomes [47], [233], [234]. The metabolomic profiling and metaproteomic studies through mass spectrometric analysis and nuclear magnetic resonance spectroscopic studies have provided additional advantages in investigating changes in the gut microbiota especially in pathogenic/diseased conditions [47], [233], [234]. The characterization of the gut virome showed a dearth of 16S or 18S rRNA genes, thus the empirical metataxonomic approaches to characterize virus-like particles (VLPs) is dependent on the isolation and sequencing of DNA from VLPs along with the classical techniques like microscopy and cultivation [235]. The gut-associated phages are isolated directly from the human microbiota rather being isolated from the sewage or water sources [226] for proper characterization, which will surely play a promising role in improving the knowledge on the gut virome and will serve as a key for the metagenomic studies that will further furnish the idea about the influence of the phages on the human gut. Lately, the Virome Capture Sequencing platform for Vertebrate viruses (VirCapSeq-VERT) system has been introduced to study the viruses in the clinical settings [236]. Furthermore, the metagenomic analysis that collects different gene sequences from the genetic materials from the human gut is potent enough to classify the viral families, taxa and generato serve as a better alternative to the professional perspectives of microbiologists and translational researchers in the study of gut virome [10]. The tool protects against contamination or other confounding factors that occur during culturing of viruses [237]. The integration of the modern applications to assess the microbiota in the preclinical and clinical studies will help to clarify this complex interplay and yield promise to identify the novel treatments for beneficial modulation of the gut microbiota for improving metabolic health.
5.2. Gut microbes as therapeutics
Most of the studies conducted so far have demonstrated that gut microbes exert their effects on the host through regulating their physiology, metabolic and immunological processes [66], [137], [190]. In particular, the indispensable contributions of gut microbes in ameliorating inflammation found in the aforementioned studies using experimental animal models [47], [238] and/or human based data [7], [239] clearly revealed the utility of gut microbes as a future medicine for treating the diseases resulting from inflammation [47]. Anti-inflammatory roles of the gut microbiota have shown promising results in treating colorectal cancer [240], [241], [242], ulcers [243], [244] and chemical-induced damages in the small intestine [245], [246]. Modulation of the gut microbiota can be efficiently used to manage several gastrointestinal disorders [247]. Different potential strategies viz. administration of fecal microbiota and treatment with probiotics are now being developed for modulating the gut microbiota for the therapeutic purposes. Manipulation of the microbiota through fecal microbiota transplantation (FMT) is now a widespread treatment option for treating the aforesaid ailments along with infectious and inflammatory disorders [248]. Several trials on the administration of fecal materials containing the distal gut microbiota from a healthy person to a subject having an altered gut microbiota have shown promises, especially in re-establishing the microbial consortium after dysbiosis [82], [249]. FMT has been successful in combating the Clostridium difficile infection (CDI) and IBD [247] by supplementing the microbiota instability through enhancing the abundance of Firmicutes and reducing Actinobacteria and Enterobacteriaceae as well as reducing inflammatory responses [250]. However, a mechanistic exploration is necessary to optimize and improve the applicability of the FMT to treat microbiota associated human diseases beyond IBD and CDI [250]. A recent investigation revealed that gut microbiota can efficiently prevent non-steroidal anti-inflammatory drugs (NSAIDs)-induced ulcers and erosions in the small intestine [251]. Similarly, understandings on the crosstalk amongst the gut microbes, barrier function, and the inflammatory responses have provided novel dimensions in targeting the pathology of colorectal cancer and colitis-associated cancer [118], [240].
Microbial amelioration of the allergic and autoimmune responses is considered as an important finding in the field of gut microbiota research [167]. The investigation on the experimental colonization of the GF mice with the gut microbiota was found sufficient in protecting the mice from the allergic inflammation in the lungs in early life, however, allergic airway inflammation (AAI) persisted in mice during the adult stage [252], [253]. The efficacy of the gut microbiota during early life colonization in the host is exerted through the inhibition of the serum IgE responses [47], [254] that eventually confer protection against the sensitization to the food allergens [255]. Oral supplementation with Lactobacillus probiotic has been shown as a promising mean to accelerate the tolerance to cow’s milk in the infants having cow’s milk allergy [256]. In fact, the Lactobacillus treatment is also effective in restoring the normal levels of the fecal butyrate-producing bacteria and butyrate in the allergic subjects [256]. Previously, a study by Gaudreau et al. [257] indicated that the gut immune system may play a key role in the initiation and progression of the systemic lupus erythematosus (SLE) and the related gender bias responses. However, the underlying mechanistic insight of the differences in the composition and function of the gut microbiota of the males and the females influencing the functions of hormones and shaping the immune responses are yet unknown.
The gut microbiota exerts a substantial effect on the metabolic abnormalities by influencing the efficiency of energy harvest and storage, low-grade inflammation and browning of adipose tissue [47], [258]. Notably, beyond gut microbes, human lifestyle factors such as energy intake and expenditure as well as the genetic determinants are also considered as key risk factors for these metabolic abnormalities [47]. Studies on this complex interplay of all these factors including the gut microbiota might be the strategy to understand the disease pathology. In recent times, the intestinal SCFA-binding receptors, particularly the G-protein-coupled receptors (GPRs) like GPR41, GPR43, GPR109a and GPR119 [259], [260], [261], have been emphasized for their potential as future therapeutic targets for the various inflammatory disorders, obesity, and cancer [262]. These receptors are activated by the SCFAs (propionate, acetate, butyrate, valerate, formate) prepared by the gut microbes [262] and induces M2 polarization of the macrophages that indeed regulate the immune surveillance during dysbiosis [263]. Therefore, selective targeting of the GPRs with the microbial metabolites through the administration of the probiotics could be an efficacious choice of treating human diseases by modulating mucosal immunity [200], [232]. Recently a study by Cohen et al. [264] demonstrated the presence of a family of genes coding for the enzymes driving the biosynthesis of N-acyl amides which act as a ligand for the human intestinal GPR119 to modulate the metabolic tone by influencing the levels of blood glucose and metabolic hormones. In light of high rates of childhood obesity, pre- and probiotics have been documented as the potential therapeutic options to prevent obesity in the children from overweight and obese mother by restoring the energy homeostasis [265], [266]. Such pre- and probiotic treatments have been found to be promoted by the low dose antibiotics that shift the taxonomic composition towards an increased Firmicutes to Bacteroides ratio without affecting the healthy microbial consortium [267], [268]. Probiotics are microbes that confer beneficial health outcomes for the host [269]. Twenty-two probiotics are currently in use as living drugs and exert effects on the epigenetic alterations. On the other hand, antibiotics can significantly change the intestinal microbiome of an individual and consequently affect the corresponding metagenome [270]. Antibiotics are efficacious as they prevent the growth of noxious microbes or kill them and these actions are necessary to treat the bacterial/viral/protozoan infections that cannot be cured by other means [271]. However, the common consumption of the antibiotics during medication especially those with broad-spectrum activities caused perturbation of the human microbiota to undertake considerable changes in their therapeutic potential as well as biochemical adaptation [135], [136]. In this regard, resistance to the antimicrobials is the most vulnerable outcome associated with antibiotic consumption in humans [261], [262]. Antibiotic resistance is responsible for the three fundamental outcomes: [i] inactivation of enzyme production, [ii] a change of the target site, and [iii] antibiotic exclusion from the natural target site (e.g. vancomycin against Gram-negative organisms) [272]. Modern age antibiotics for promoting growth and protection of the crops can cause resistance elements to appear in saprophytic bacteria and these resistance mechanisms may transform human bacteria to yield pathogenic effects when the crops are eaten [273]. Resistant microorganisms transferred from one person to another remain resistant and can further promote emergence of the multidrug-resistant microbes [274], [275]. Therefore, microbial resistance to available antibiotics has come out as a major public-health threat to date. Considering the alarming stipulation, the combinatorial treatment comprising antibiotics along with the pre- and probiotics have come out as an effective option in restoring the healthy gut microbiota in the disease condition, especially against the increasing number of emerging infections around the globe. Probiotics efficiently aid the naturally occurring gut microbiota. Recent studies have demonstrated the therapeutic efficacy of the conventional and new age probiotics. The probiotics are also implemented to stop the Antibiotic Associated Disease (AAD). A meta-analysis of the probiotics in controlling AAD through nine controlled trials showed that both Lactobacilli sp. and Saccharomyces boulardii can reduce the risk of AAD from 28.5 to 11.9% especially in children [276], [277]. The administration of the probiotics can efficiently prevent diarrhea commonly occur after the antibiotic treatment [278]. A sustained probiotic treatment on various gastrointestinal and extraintestinal disorders such as irritable bowel syndrome [279]; vaginal infections [280], [281]; inflammatory bowel disease [282] and immune enhancement [283] are in practice worldwide. Interestingly, the different probiotic treatment studies conducted on human and other animal models showed satisfactory results in reducing the burden of protozoan and helminths without producing any health hazard [284]. Probiotics have also been evidenced to reduce the risk of colon cancer by destroying the activity of the particular bacterial enzymes involved in raising the levels of pro-carcinogens [285]. Although rare, the administration of probiotics may pose adverse effects sometimes out of which majority of complications occur only in the severely immune-compromised subjects or in the patients with other life-threatening illness managed in the intensive-care areas [286]. Thus, the therapeutic applicability of the probiotics in the immune-competent subjects seems to be safe but their use in the prevention of AAD in the immune-compromised patients is yet to be tested [287], [288]. The promising probiotics used in controlling parasitic infections of human have been shown in Table 2 .
Table 2.
Gut microbes and probiotics in controlling parasitic infections
| Parasite | Gut microbe/Probiotic | References |
|---|---|---|
| Plasmodium sp | Eschereria coli O86:B7 | [213] |
| Giardia lambia |
Lactobacillus johnsonii La1 Lactobacillus casei MTCC 1423 Enterococcus faecium SF68 |
[289], [290], [291] |
| Ascaris suum | Bifidobacterium lactis | [284], [292] |
| Babesia microti | Lactobacillus casei ATCC7469 | [284], [293] |
| Cryptosporidium parvum |
Lactobacillus reuteri 4000, 4020 Lactobacillus acidophilus NCFM Lactobacillus rhamnosus GG Lactobacillus casei shirota |
[284], [294], [295], [296] |
| Schistosoma mansoni | Zymomonas mobilis | [284], [297] |
| Toxocara canis | Enterococcus faecalis CECT7121 | [284], [298] |
| Trichinella spiralis | Lactobacillus casei ATCC7469 | [284], [299] |
| Trypanosoma cruzi | Lactobacillus casei ATCC7469 | [284], [300] |
Besides the prebiotics and probiotics, synbiotics can also be an efficient approach in the gut microbiota targeted therapy. Synbiotic interventions involve administration of new microbe(s) in a consortium along with providing an ample amount of specific substrate to facilitate the growth of a new community for the beneficial outcomes [268]. The application of small molecule inhibitors designed against the specific microbial metabolic activities, especially the rate-limiting enzymes of different metabolic pathways is considered advantageous in protecting the invading organisms in the gut microbiota [301]. In particular, small molecules have been found to inhibit the bacterial β-glucuronidase enzyme that plays a crucial role in metabolizing the glucuronide drug-conjugates induced by host metabolism. This intervention strategy found effective in preventing the toxicity of irinotecan (an anticancer drug) in mice [302]. Recently, perturbation of gutmicrobial communities through the synbiotic approach by providing Lactobacillus plantarum (probiotic) in combination with the fructo-oligosaccharide (prebiotic) reduced the number of newborn deaths caused by sepsis [303]. The mechanism of protection involves restoration of gut epithelial barrier function through upregulating mucus production and inducing host immunity leading to the secretion of antimicrobial peptide and inhibiting pathogen translocation [304].
Genetically engineered microbes can confer selective protection to a number of pathogenic infections. In recent years, genetically engineered Escherichia coli Nissle with N-acylhomoserinelactone has been found effective in preventing Pseudomonas aeruginosa infection by inducing lysis through secreting the anti-P. aeruginosa toxin [305]. This approach seems to be effective for other diseases too. For example, the pathogenesis of E. histolytica is linked with Escherichia coli or Shigella dysenteriae [191] and therefore engineered microbes lacking the virulence promoting gene could effectively treat the amoebiasis. Interestingly, new age gut microbiota research has shown the promise of personalized utilization of the human gut microbes as pharmabiotics (“bugs to drugs”) [4], [305], [306]. Although studied less, but considering the increased interest exploring the gut phages, it is expected that trials on using the gut phages for controlling the pathogenesis of pathogenic bacteria to treat gut infections as well as cancer will definitely provide a new hope.
6. Conclusion and future directions
Research on gut microbes provide evidence that the gut microbiota is essentially another organ that plays a vital role in human physiology and disease. Co-existence of the microbiota within humans throughout the lifespan and crosstalk between the two have prompted basic researchers to understand the biology of host-microbe interactions and to investigate the underlying mechanisms of the functions of the human microbiota in order to manipulate and use these microbes to improve human health. Recent research findings on the human gut microbiota revealed the crucial contribution of gut microbes in human physiology and therapeutic potential in treating the health problems. In particular, progress in understanding the complex interplay between the gut microbial composition and the host immune responses has demonstrated that microbes are indispensable for maintaining the immuno-homeostasis in the host and disease outcomes. In this review article, we have discussed the correlation between alterations in human gut microbial composition and the outcome of inflammatory and infectious diseases. Therefore, the targeted modulation of the gut microbiota is now becoming a powerful strategy in anti-inflammatory and anti-infectious disease therapies. Therapeutic intervention strategies involving the gut microbiota have now been implicated using several approaches, including antibiotics, probiotics, postbiotics and FMT. Especially the dietary consumption or administration of the probiotics can modulate the intestinal microbial composition by increasing the abundance of the beneficial microbial load that functions as an excellent means of remediation and deterrence against a variety of intestinal disorders and infections. Moreover, the co-administration of the nutraceuticals (chemical entity having nutritional and therapeutic properties) could act in synergy for re-establishing the functional gut microbiota. The advances in current experimental models and methods allow us to obtain the scientific understanding of the interplay amongst the gut microbiota, barrier function, and host responses. These insights will lead to uncovering new therapeutic targets in complex human diseases including cancer. Despite these gains, many challenges lie ahead that make it difficult to close the gap between the basic sciences and clinical applications. Identification of the components of the microbiota and elucidation of the mechanisms of their action in inducing the pathological changes or exerting the beneficial effects along with disease-protective activities could aid in our ability to influence the composition of the microbiota. Particularly the recent understanding of the immense importance of the gut microbiota in controlling the pathogenic infections in insect model paved the way for investigating the molecular insights of the gut microbiota induced protection against human infectious diseases. Thus, the suitable exploration of the beneficial gut microbes and their mode of function could guide us in combating the more complex diseases. However, the ‘bench to bed’ translation of the research findings through the clinical trials should be aimed to check the potency of a specific microbe or microbial consortium in therapy against a particular disease. The success of these approaches will direct future scientists or physicians to adopt gut microbiome-based strategies for the diagnosis, therapy, and prevention of the infectious and inflammatory diseases of humans in the near future.
Acknowledgment
The authors acknowledge The Department of Science & Technology (DST), Govt. of India for funding (SR/SO/AS-006/2014) and providing research fellowship to NJ. Due to the limitation of the space, we have cited only selected works without the intention of ignoring the contributions made by others in this field. Our grateful thanks are due to Prof. Goutam Ghosal, Department of English & Other Modern European Languages, Visva-Bharati University, India for critically reviewing the language of the manuscript. We are grateful to Dr. Kristina Martinez-Guryn, Editor of the special issue, The Journal of Nutritional Biochemistry for her generous help in critical evaluation of the manuscript.
Footnotes
Potential conflict of interest: None.
References
- 1.Jandhyala S.M., Talukdar R., Subramanyam C., Vuyyuru H., Sasikala M., Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grice E.A., Segre J.A. The human microbiome:Our second genome. Annu Rev Genomics Hum Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley R.E., Peterson D.A., Gordon J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Sun J., Chang B. Exploring gut microbes in human health and disease: pushing the envelope. Genes Dis. 2014;1:132–139. doi: 10.1016/j.gendis.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogilvie L.A., Jones B.V. The human gut virome: A multifaceted majority. Front Microbiol. 2015;6:918. doi: 10.3389/fmicb.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitbart M., Hewson I., Felts B., Mahaffy J.M., Nulton J., Salamon P. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyes A., Haynes M., Hanson N., Angly F.E., Heath A.C., Rohwer F. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minot S., Sinha R., Chen J., Li H., Keilbaugh S.A., Wu G.D. The human gut virome, inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirzaei M.K., Maurice C.F. Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol. 2017;15:397–408. doi: 10.1038/nrmicro.2017.30. [DOI] [PubMed] [Google Scholar]
- 10.Scarpellini E., Ianiro G., Attili F., Bassanelli C., De Santis A., Gasbarrini A. The human gut microbiota and virome: potential therapeutic implications. Dig Liver Dis. 2015;47:1007–1012. doi: 10.1016/j.dld.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekirov I., Finlay B.B. The role of the intestinal microbiota in enteric infection. J Physiol. 2009;587:4159–4167. doi: 10.1113/jphysiol.2009.172742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekirov I., Russell S.L., Antunes L.C., Finlay B.B. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 13.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 14.Flint H.J., Scott K.P., Duncan S.H., Louis P., Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabernero M., Cedron M.G. Microbial metabolites derived from colonic fermentation of non-digestible compounds. Curr Opin Food Sci. 2017;13:91–96. [Google Scholar]
- 16.Rowland I., Gibson G., Heinken A., Scott K., Swann J., Thiele I. Gut microbiota functions: Metabolism of nutrients and other food components. Eur J Nutr. 2017 doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neis E.P.J.G., Dejong C.H.C., Rensen S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients. 2015;7:2930–2946. doi: 10.3390/nu7042930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison D.J., Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puddu A., Sanguineti R., Montecucco F., Viviani G.L. Evidence for the gut microbiota short chain fatty acids as key pathophysiological molecules improving diabetes. Mediat Inflamm. 2014;2014:162021. doi: 10.1155/2014/162021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rios-Covian D., Ruas-Madiedo P., Margolles A., Gueimonde M., de los Reyes-Gavilan C.G., Salazar N. Intestinalshort chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulange C.L., Neves A.L., Chilloux J., Nicholson J.K., Dumas M.E. Impact of the gut microbiota on inflammation,obesity, and metabolic disease. Genome Med. 2016;8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho B.M., Saad M.J.A. Influence of gut microbiota on subclinical inflammation and insulin resistance. Mediat Inflamm. 2013 doi: 10.1155/2013/986734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musso G., Gambino R., Cassader M. Obesity, diabetes, and gut microbiota the hygiene hypothesis expanded? Diabetes Care. 2010;33:2277–2284. doi: 10.2337/dc10-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao M.B., Tanksale A.M., Ghatge A.S., Deshpande V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev. 1998;62:597–635. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koppel N., Rekdal V.M., Balskus E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science. 2017;356:2770. doi: 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korecka A., Arulampalam V. The gut microbiome: Scourge, sentinel or spectator? J Oral Microbiol. 2012;4:10. doi: 10.3402/jom.v4i0.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanson H. Drug metabolism by the host and gut microbiota: A partnership or rivalry? Drug Metab Dispos. 2015;43:1499–1504. doi: 10.1124/dmd.115.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jourova L., Azenbacher P., Azenbacherova E. Human gut microbiota plays a role in the metabolism of drugs. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016;160:317–326. doi: 10.5507/bp.2016.039. [DOI] [PubMed] [Google Scholar]
- 29.Kamada N., Seo S.U., Chen G.Y., Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 30.Ubeda C., Djukovic A., Isaac S. Roles of the intestinal microbiota in pathogen protection. Clin Transl Immunol. 2017;6 doi: 10.1038/cti.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaiko G.E., Stappenbeck T.S. Host-microbe interactions shaping the gastrointestinal environment. Trends Immunol. 2014;35:538–548. doi: 10.1016/j.it.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegel S.J., Weiser J.N. Mechanisms of bacterial colonization of the respiratory tract. Annu Rev Microbiol. 2015;69:425–444. doi: 10.1146/annurev-micro-091014-104209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buffie C.G., Pamer E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson M.K., Pesesky M.W., Dantas G. The Yin and Yang of bacterial resilience in the human gut microbiota. J Mol Biol. 2014;426:3866–3876. doi: 10.1016/j.jmb.2014.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim M., Ashida H., Oqawa M., Yoshikawa Y., Mimuro H., Sasakawa C. Bacterial interactions with the host epithelium. Cell Host Microbe. 2010;8:20–35. doi: 10.1016/j.chom.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Pereira F.C., Berry D. Microbial nutrient niches in the gut. Environ Microbiol. 2017;19:1366–1378. doi: 10.1111/1462-2920.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pastorelli L., De Salvo C., Mercado J.R., Vechhi M., Pizarro T.T. Central role of gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: Lessons learned from animal models and human genetics. Front Immunol. 2013;4:280. doi: 10.3389/fimmu.2013.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montalto M., D’Onofrio F., Gallo A., Cazzato A., Gasbarrini G. Intestinal microbiota and its functions. Dig Liver Dis Suppl. 2009;3:30–34. [Google Scholar]
- 39.Rawls J.F., Mahowald M.A., Ley R.E., Gordon J.I. Reciprocal gut microbiota transplants from zebra fish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kau A.L., Ahern P.P., Griffin N.W., Goodman A.L., Gordon J.I. Human nutrition, the gut microbiome and immune system: envisioning the future. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Littman D.R., Pamer E.G. Role of commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krezalek M.A., Skowron K.B., Guyton K.L., Shakhsheer B., Hyoju S., Alverdy J.C. The intestinal microbiome and surgical disease. Curr Probl Surg. 2016;53:257–293. doi: 10.1067/j.cpsurg.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warner B.B., Deych E., Zhou Y., Hall-Moore C., Weinstock G.M., Sodergren E. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet. 2016;387(10031):1928–1936. doi: 10.1016/S0140-6736(16)00081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joossens M., Huys G., Cnockaert M., De Preter V., Verbeke K., Rutgeerts P. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60(5):631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 45.Cani P.D., Bibiloni R., Knauf C., Waget A., Neyrinck A.M., Delzenne N.M. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 46.Arrieta M.C., Stiemsma L.T., Dimitriu P.A., Thorson L., Russell S., Yurist-Doutsch S. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 47.van den Elsen L.W.J., Poyntz H.C., Weyrich L.S., Young W., Forbes-Blom E.E. Embracing the gut microbiota: the new frontier for inflammatory and infectious diseases. Clin Transl Immunol. 2017;6 doi: 10.1038/cti.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemarajata P., Versalovic J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Therap Adv Gastroenterol. 2013;6:39–51. doi: 10.1177/1756283X12459294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erejuwa O.O., Sulaiman S.A., Wahab M.S.A. Modulation of gut microbiota in the management of metabolic disorders: The prospects and challenges. Int J Mol Sci. 2014;15:4158–4188. doi: 10.3390/ijms15034158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonnenberg J.L., Fischbach M.A. Community health care: Therapeutic opportunities in the human microbiome. Sci Transl Med. 2011;3(78):78. doi: 10.1126/scitranslmed.3001626. (ps12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pilato V.D., Chiarelli A., Boinett C.J., Riccobono E., Harris S.R., D’Andrea M.M. Complete genome sequence of the first KPC-type carbapenemase-positive Proteus mirabilis strain from a bloodstream infection. Genome Announc. 2016;4:e00607–e00616. doi: 10.1128/genomeA.00607-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reid G., Burton J. Use of Lactobacillus to prevent infection by pathogenic bacteria. Microbes Infect. 2002;4:319–324. doi: 10.1016/s1286-4579(02)01544-7. [DOI] [PubMed] [Google Scholar]
- 53.Ley R.E. Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol. 2016;13:69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 54.Scher J.U., Sczesnak A., Longman R.S., Segata N., Ubeda C., Bielski C. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. elife. 2013;5 doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kovatcheva-Datchary P., Nilsson A., Akrami R., Lee Y.S., De Vadder F., Arora T. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Agudelo Higuita N.I., Huycke M.M. Enterococcal disease, epidemiology, and implications for treatment. In: Gilmore M.S., Clewell D.B., Ike Y., Shankar N., editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary; Boston, MA: 2014. [Google Scholar]
- 57.Abadi A.T.B. Helicobacter pylori: emergence of a superbug. Front Med. 2014;1:34. doi: 10.3389/fmed.2014.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cardona F., Andrés-Lacueva C., Tulipani S., Tinahones F.J., Queipo-Ortuño M.I. Benefits of polyphenols on gut microbiota and implications in human health. J Nutr Biochem. 2013;24:1415–1422. doi: 10.1016/j.jnutbio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 59.O'Callaghan A., van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El Kaoutari A., Armougom F., Gordon J.I., Raoult D., Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 61.Sela D.A., Chapman J., Adeuya A., Kim J.H., Chen F., Whitehead T.R. The genome sequence of Bifidobacterium longumsubsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bien J., Palagani V., Bozko P. The intestinal microbiota dysbiosis and Clostridium difficileinfection: is there a relationship with inflammatory bowel disease? Therap Adv Gastroenterol. 2013;6:53–68. doi: 10.1177/1756283X12454590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skoff T.H., Farley M.M., Petit S., Craig A.S., Schaffner W., Gershman K. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990-2007. Clin Infect Dis. 2009;49:85–92. doi: 10.1086/599369. [DOI] [PubMed] [Google Scholar]
- 64.Mokoena M.P. Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules. 2017;22:1255. doi: 10.3390/molecules22081255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown K., DeCoffe D., Molcan E., Gibson D.L. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 2012;4:1095–1119. doi: 10.3390/nu4081095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guinane C.M., Cotter P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap Adv Gastroenterol. 2013;6:295–308. doi: 10.1177/1756283X13482996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y.J., Li S., Gan R.Y., Zhou T., Xu D.P., Li H.B. Impacts of gut bacteria on human health and diseases. Int J Mol Sci. 2015;16:7493–7519. doi: 10.3390/ijms16047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.E. coli - Symptoms and causes - Mayo Clinic. http://www.mayoclinic.org/diseases-conditions/e-coli/basics/definition/con-20032105
- 69.Duncan S.H., Aminov R.I., Scott K.P., Louis P., Stanton T.B., Flint H.J. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol. 2006;56:2437–2441. doi: 10.1099/ijs.0.64098-0. [DOI] [PubMed] [Google Scholar]
- 70.Tamanai-Shacoori Z., Smida I., Bousarghin L., Loreal O., Meuric V., Fong S.B. Roseburia spp.: a marker of health? Future Microbiol. 2017;12:157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 71.Ohkawara S., Furuya H., Nagashima K., Asanuma N., Hino T. Effect of oral administration of Butyrivibrio fibrisolvens MDT-1 on experimental enterocolitis in mice. Clin Vaccine Immunol. 2006;13:1231–1236. doi: 10.1128/CVI.00267-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miller T.L., Jenesel S.E. Enzymology of butyrate formation by Butyrivibrio fibrisolvens. J Bacteriol. 1979;138:99–104. doi: 10.1128/jb.138.1.99-104.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalmokoff M.L., Teather R.M. Isolation and characterization of a bacteriocin (butyrivibriocin AR10) from the ruminal anaerobe Butyrivibrio fibrisolvens AR10: evidence in support of the widespread occurrence of bacteriocin-like activity among ruminal isolates of B.fibrisolvens. Appl Environ Microbiol. 1997;63:394–402. doi: 10.1128/aem.63.2.394-402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malinen E., Krogius-Kurikka L., Lyra A., Nikkilä J., Jääskeläinen A., Rinttilä T. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol. 2010;16:4532–4540. doi: 10.3748/wjg.v16.i36.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ze X., Duncan S.H., Louis P., Flint H.J. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lagier J.C., Million M., Hugon P., Armougom F., Raoult D. Human gut microbiota: repertoire and variations. Front Cell Infect Microbiol. 2012;2:136. doi: 10.3389/fcimb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Newton R.J., McLellan S.L., Dila D.K., Vineis J.H., Morrison H.G., Eren A.M. Sewage reflects the microbiomes of human populations. MBio. 2015;6 doi: 10.1128/mBio.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang H., Ling Z., Zhang Y., Mao H., Ma Z., Yin Y. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 79.Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L.G., Gratadoux J.J. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCoy A.N., Araújo-Pérez F., Azcárate-Peril A., Yeh J.J., Sandler R.S., Keku T.O. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kramer C.D., Genco C.A. Microbiota, immune subversion, and chronic inflammation. Front Immunol. 2017;8:255. doi: 10.3389/fimmu.2017.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gerritsen J., Smidt H., Rijkers G.T., de Vos W.M. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011;6:209–240. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Braune A., Blaut M. Intestinal bacterium Eubacterium cellulosolvens deglycosylates flavonoid C- and O-glucosides. Appl Environ Microbiol. 2012;78:8151–8153. doi: 10.1128/AEM.02115-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Engels C., Ruscheweyh H.J., Beerenwinke N., Lacroix C., Schwab C. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front Microbiol. 2016;7:713. doi: 10.3389/fmicb.2016.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garrett W.S., Gallini C.A., Yatsunenko T., Michaud M., DuBois A., Delaney M.L. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 87.Mader J.T., Calhoun J. Bone, Joint, and Necrotizing Soft Tissue Infections. In: Baron S., editor. Medical Microbiology. 4th edition. University of Texas Medical Branch at Galveston; Galveston (TX): 1996. [PubMed] [Google Scholar]
- 88.Forbes J.D., Domselaar G.V., Bernstein C.N. The gut microbiota in immune-mediated inflammatory diseases. Front Microbiol. 2016;7:1081. doi: 10.3389/fmicb.2016.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Y.Z., Li Y.Y. Inflammatory bowel disease: Pathogenesis. World J Gastroenterol. 2014;20:91–99. doi: 10.3748/wjg.v20.i1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim D.H., Cheon J.H. Pathogenesis of inflammatory bowel disease and recent advances in biological therapies. Immune Netw. 2017;17(1):25–40. doi: 10.4110/in.2017.17.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hold G.L., Smith M., Grange C., Watt E.R., El-Omar E.M., Mukhopadhyay I. Role of gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World J Gastroenterol. 2014;20:1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu X.R., Liu C.Q., Feng B.S., Liu Z.J. Dysregulation of mucosal immune response in pathogenesis of inflammatory bowel disease. World J Gastroenterol. 2014;20:3255–3264. doi: 10.3748/wjg.v20.i12.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matricon J., Barnich N., Ardid D. Immunopathogenesis of inflammatory bowel disease. Self Nonself. 2010;1(4):299–309. doi: 10.4161/self.1.4.13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ahmed I., Roy B.C., Khan S.A., Septer S., Umar S. Microbiome, Metabolome and inflammatory bowel disease. Microorganisms. 2016;4:20. doi: 10.3390/microorganisms4020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matsuoka K., Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015;37:47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lane E.R., Zisman T.L., Suskind D.L. The microbiota in inflammatory bowel disease: Current and therapeutic insights. J Inflamm Res. 2017;10:63–73. doi: 10.2147/JIR.S116088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nagao-Kitamoto H., Kamada N. Host-microbial crosstalk in inflammatory bowel disease. Immune Netw. 2017;17:1–12. doi: 10.4110/in.2017.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martinez-Medina M., Garcia-Gil L.J. Escherichiacoli in chronic inflammatory bowel diseases: An update on adherent invasive Escherichiacolipathogenicity. World J Gastrointest Pathophysiol. 2014;5:213–227. doi: 10.4291/wjgp.v5.i3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brint E.K., MacSharry J., Fanning A., Shanahan F., Quigley E.M. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol. 2011;106:329–336. doi: 10.1038/ajg.2010.438. [DOI] [PubMed] [Google Scholar]
- 100.Ponnusamy K., Choi J., Kim J., Lee S., Lee C. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol. 2011;60:817–827. doi: 10.1099/jmm.0.028126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ghosal U., Shukla R., Gwee K., Ng S., Quigley E. The gut microbiota and irritable bowel syndrome: friend or foe? Int. J Inflamm. 2012;2012:151085. doi: 10.1155/2012/151085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rajilic-Stojanovic M., Biagi E., Heiliq H.G., Kajander K., Kekkonen R.A., Tims S. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 103.Jeffery I., Claesson M., O’Toole P., Shanahan F. Categorization of the gut microbiota: Enterotypes or gradients? Nat Rev Microbiol. 2012;10:591–592. doi: 10.1038/nrmicro2859. [DOI] [PubMed] [Google Scholar]
- 104.Jeffery I., O’ Toole P., Ohman L., Claesson M., Deane J., Quigley E. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 105.Jeffery I., Quigley E., Ohman L., Simren M., O’ Toole P. The microbiota link to irritable bowel syndrome: an emerging story. Gut Microbes. 2012;3:572–576. doi: 10.4161/gmic.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saulnier D.M., Riehle K., Mistretta T.A., Diaz M.A., Mandal D., Raza S. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Olivares M., Walker A.W., Capilla A., Benítez-Páez A., Palau F., Parkhill J. Gut microbiota trajectory in early life may predict development of celiac disease. Microbiome. 2018;6(1):36. doi: 10.1186/s40168-018-0415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dzutsev A., Badger J.H., Perez-Chanona E., Roy S., Salcedo R., Smith C.K. Microbes and Cancer. Annu Rev Immunol. 2017;35:199–228. doi: 10.1146/annurev-immunol-051116-052133. [DOI] [PubMed] [Google Scholar]
- 109.Roy S., Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;5:271–285. doi: 10.1038/nrc.2017.13. [DOI] [PubMed] [Google Scholar]
- 110.Zitovgel L., Galluzzi L., Viaud S., Vetizou M., Daillere R., Merad M. Cancer and the gut microbiota: An unexpected link. Sci Transl Med. 2015;7:271. doi: 10.1126/scitranslmed.3010473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bonnet M., Buc E., Sauvanet P., Darcha C., Dubois D., Pereira B. Colonization of human gut by E.coli and colorectal cancer risk. Clin Cancer Res. 2014;20:859–867. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 112.Goodwin A.C., Destefano Shields C.E., Wu S., Huso D.L., Wu X., Murray-Stewart T.R. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108(37):15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang J.L., Chang C.H., Lin J.W., Wu L.C., Chuang L.M., Lai M.S. Infection, antibiotic therapy and risk of colorectal cancer: A nationwide nested case-control study in patients with Type-2 diabetes mellitus. Int J Cancer. 2014;20:859–867. doi: 10.1002/ijc.28738. [DOI] [PubMed] [Google Scholar]
- 114.Arthur J.C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J.M., Fan T.J. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhan Y., Chen P.J., Sadler W.D., Wang F., Poe S., Núñez G. Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer Res. 2013;73:7199–7210. doi: 10.1158/0008-5472.CAN-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yoshimoto S., Loo T.M., Atarashi K., Kanda H., Sato S., Oyadomari S. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 117.Dapito D.H., Mencin A., Gwak G.Y., Pradere J.P., Jang M.K., Mederacke I. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sun J., Kato I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016;3:130–143. doi: 10.1016/j.gendis.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peters B.A., Dominianni C., Shapiro J.A., Church T.R., Wu J., Miller G. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4:69. doi: 10.1186/s40168-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lu Y., Chen J., Zheng J., Hu G., Wang J., Huang C. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci Rep. 2016;6:26337. doi: 10.1038/srep26337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ferguson L.R., Chen H., Collins A.R., Connell M., Damia G., Dasgupta S. Genomic instability in human cancer: molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition. Semin Cancer Biol. 2015;35:S5–S24. doi: 10.1016/j.semcancer.2015.03.005. [Suppl.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Olszak T., An D., Zeissig S., Vera M.P., Richter J., Franke A. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336(6080):489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Maneval M.L., Eckert K.A. Effects of oxidative and alkylating damage on microsatellite instability in nontumorigenic human cells. Mutat Res. 2004;546:29–38. doi: 10.1016/j.mrfmmm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 124.Soreide K. Proteinase-activated receptor 2 (PAR-2) in gastrointestinal and pancreatic pathophysiology, inflammation and neoplasia. Scand J Gastroenterol. 2008;43:902–909. doi: 10.1080/00365520801942141. [DOI] [PubMed] [Google Scholar]
- 125.Cheema A.K., Maier I., Dowdy T., Wang Y., Singh R., Ruegger P.M. Chemopreventive metabolites are correlated with a change in intestinal microbiota measured in A-T mice and decreased carcinogenesis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0151190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roberts C.K., Hevener A.L., Barnard R.J. Metabolic syndrome and insulin resistance: underlying causes and modification by exercise training. Compr Physiol. 2013;3:1–58. doi: 10.1002/cphy.c110062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lonardo A., Ballestri S., Marchesini G., Angulo P., Loria P. Nonalcoholic fatty liver disease: a precursor of the metabolic syndrome. Dig Liver Dis. 2015;47:181–190. doi: 10.1016/j.dld.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 128.Hill J.O., Wyatt H.R., Peters J.C. Energy balance and obesity. Circulation. 2012;126:126–132. doi: 10.1161/CIRCULATIONAHA.111.087213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jung U.J., Choi M.S. Obesity and its metabolic complications: the role of adipokines and the relationship between obesity, inflammation, insulin resistance, dyslipidemia and nonalcoholic fatty liver disease. Int J Mol Sci. 2014;15:6184–6223. doi: 10.3390/ijms15046184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lim M.Y., You H.J., Yoon H.S., Kwon B., Lee J.Y., Lee S. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut. 2017;66:1031–1038. doi: 10.1136/gutjnl-2015-311326. [DOI] [PubMed] [Google Scholar]
- 131.Wong A.C.N., Vanhove A.S., Watnick P.I. The interplay between intestinal bacteria and host metabolism in health and disease: lessons from Drosophila melanogaster. Dis Model Mech. 2016;9:271–281. doi: 10.1242/dmm.023408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Murphy E.A., Velazquez K.T., Herbert K.M. Influence of high-fat-diet on gut microbiota: a driving force for chronic disease risk. Curr Opin Clin Nutr Metab Care. 2015;18:515–520. doi: 10.1097/MCO.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang X., Zhao Y., Zhang M., Pang X., Xu J., Kang C. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Conlon M.A., Bird A.R. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2015;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ravussin Y., Koren O., Spor A., LeDuc C., Gutman R., Stombaugh J. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring) 2012;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moreno-Indias I., Cardona F., Tinahones F.J., Queipo-Ortuño M.I. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front Microbiol. 2014;5:190. doi: 10.3389/fmicb.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Francino M.P. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol. 2015;6:1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Langdon A., Crook N., Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016;8:39. doi: 10.1186/s13073-016-0294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Turta O., Rautava S. Antibiotics, obesity and the link to microbes - what are we doing to our children? BMC Med. 2016;14:57. doi: 10.1186/s12916-016-0605-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Baothman O.A., Zamzami M.A., Taher I., Abubaker J., Abu-Farha M. The role of Gut microbiota in the development of obesity and diabetes. Lipids Health Dis. 2016;15:108. doi: 10.1186/s12944-016-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim B.J., Lee S.Y., Kim H.B., Lee E., Hong S.J. Environmental changes, microbiota, and allergic diseases. Allergy, Asthma Immunol Res. 2014;6:389–400. doi: 10.4168/aair.2014.6.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Prince B.T., Mandel M.J., Nadeau K., Singh A.M. Gut microbiome and the development of food allergy and allergic disease. Pediatr Clin N Am. 2015;62(6):1479–1492. doi: 10.1016/j.pcl.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Langley J.M., Halperin S. Allergy to antibiotics in children: perception versus reality. Can J Infect Dis. 2002;13:160–163. doi: 10.1155/2002/767068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Russell S.L., Gold M.J., Hartmann M., Willing B.P., Thorson L., Wlodarska M. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Russell S.L., Gold M.J., Willing B.P., Thorson L., McNagny K.M., Finlay B.B. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4:158–164. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wopereis H., Sim K., Shaw A., Warner J.O., Knol J., Kroll S. Intestinal microbiota in infants at high risk for allergy: effects of prebiotics and role in eczema development. J Allergy Clin Immunol. 2018;141:1334–1342. doi: 10.1016/j.jaci.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 147.Campbell A.W. Autoimmunity and the gut. Autoimmune Dis. 2014;2014:152428. doi: 10.1155/2014/152428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ignacio A., Morales C.I., Câmara N.O.S., Almeida R.R. Innate sensing of the gut microbiota: modulation of inflammatory and autoimmune diseases. Front Immunol. 2016;7:54. doi: 10.3389/fimmu.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mu Q., Kirby J., Reilly C.M., Luo X.M. Leaky gut as a danger signal for autoimmune diseases. Front Immunol. 2017;8:598. doi: 10.3389/fimmu.2017.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shen W., Gaskins H.R., McIntosh M.K. Influence of dietary fat on intestinal microbes, inflammation, barrier function and metabolic outcomes. J Nutr Biochem. 2014;25:270–280. doi: 10.1016/j.jnutbio.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 151.McKenney P.T., Pamer E.G. From hype to hope: The gut microbiota in enteric infectious disease. Cell. 2015;163:1326–1332. doi: 10.1016/j.cell.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sharon G., Garg N., Debelius J., Knight R., Dorrestein P.C., Mazmanian S.K. Specialized metabolites from the microbiome in health and disease. Cell Metab. 2014;20:719–730. doi: 10.1016/j.cmet.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jones R.M., Mercante J.W., Neish A.S. Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr Med Chem. 2012;19:1519–1529. doi: 10.2174/092986712799828283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Perez R.H., Zendo T., Sonomoto K. Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb Cell Factories. 2014;13(Suppl. 1):S3. doi: 10.1186/1475-2859-13-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wardwell L.H., Huttenhower C., Garrett W.S. Current concepts of the intestinal microbiota and the pathogenesis of infection. Curr Infect Dis Rep. 2011;13:28–34. doi: 10.1007/s11908-010-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Burkholder K.M., Bhunia A.K. Salmonella enterica serovar Typhimurium adhesion and cytotoxicity during epithelial cell stress is reduced by Lactobacillus rhamnosus GG. Gut Pathog. 2009;1:14. doi: 10.1186/1757-4749-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yang Y., Jobin C. Microbial imbalance and intestinal pathologies: connections and contributions. Dis Model Mech. 2014;7:1131–1142. doi: 10.1242/dmm.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rodríguez J.M., Murphy K., Stanton C., Ross R.P., Kober O.I., Juge N. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis. 2015;26 doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kerrigan A.M., Brown G.D. C-type lectins and phagocytosis. Immunobiology. 2009;214:562–575. doi: 10.1016/j.imbio.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Willing B.P., Russell S.L., Finlay B.B. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol. 2011;9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 161.Ostaff M.J., Stange E.F., Wehkamp J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol Med. 2013;5:1465–1483. doi: 10.1002/emmm.201201773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Plato A., Willment J.A., Brown G.D. C-type lectin-like receptors of the dectin-1 cluster: ligands and signaling pathways. Int Rev Immunol. 2013;32:134–156. doi: 10.3109/08830185.2013.777065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gorski A., Wazna E., Dabrowska B.W., Dabrowska K., Switała-Jeleń K., Miedzybrodzki R. Bacteriophage translocation. FEMS Immunol Med Microbiol. 2006;46:313–319. doi: 10.1111/j.1574-695X.2006.00044.x. [DOI] [PubMed] [Google Scholar]
- 164.Cadwell K., Patel K.K., Maloney N.S., Liu T.C., Ng A.C., Storer C.E. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yang J.Y., Kim M.S., Kim E., Cheon J.H., Lee Y.S., Kim Y. Enteric viruses ameliorate gut inflammation via Toll-like receptor 3 and Toll-like receptor 7- mediated interferon-b production. Immunity. 2016;44:889–900. doi: 10.1016/j.immuni.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 166.Eriksson F., Tsagozis P., Lundberg K., Parsa R., Mangsbo S.M., Persson M.A. Tumor-specific bacteriophages induce tumor destruction through activation of tumor-associated macrophages. J Immunol. 2009;182:3105–3111. doi: 10.4049/jimmunol.0800224. [DOI] [PubMed] [Google Scholar]
- 167.Wu H.J., Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kostic A.D., Howitt M.R., Garrett W.S. Exploring host-microbiota interactions in animal models and humans. Genes Dev. 2013;27:701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Weng M., Walker W.A. The role of gut microbiota in programming the immune phenotype. J Dev Orig Health Dis. 2013;4:203–214. doi: 10.1017/S2040174412000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Round J.L., Mazmanian S.K. The gut microbiome shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shi N., Li N., Duan X., Niu H. Interaction between the gut microbiome and mucosal immune system. Mil Med Res. 2017;4:14. doi: 10.1186/s40779-017-0122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Belkaid Y., Hand T. Role of the Microbiota in Immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mantis N.J., Rol N., Corthésy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kaetzel C.S. Cooperativity among secretory IgA, the polymeric immunoglobulin receptor, and the gut microbiota promotes host-microbial mutualism. Immunol Lett. 2014;162:10–21. doi: 10.1016/j.imlet.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mukherjee S., Karmakar S., Babu S.P.S. TLR2 and TLR4 mediated host immune responses in major infectious diseases: a review. Braz J Infect Dis. 2016;20:193–204. doi: 10.1016/j.bjid.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kiseleva E.P., Novik G.I. In: Probiotics as immunomodulators: substances, mechanisms and therapeutic benefits microbial pathogens and strategies for combating them: science, technology and education. Mendez-Vilas A., editor. 2013. [Google Scholar]
- 178.Davies J.M., MacSharry J., Shanahan F. Differential regulation of Toll-like receptor signalling in spleen and Peyer's patch dendritic cells. Immunology. 2010;131:438–448. doi: 10.1111/j.1365-2567.2010.03317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kanther M., Tomkovich S., Sun X., Grosser M.R., Koo J., Flynn E.J. Commensal microbiota stimulate systemic neutrophil migration through induction of Serum amyloid A. Cell Microbiol. 2014;16:1053–1067. doi: 10.1111/cmi.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Molloy M., Bouladoux N., Belkaid Y. Intestinal Microbiota: Shaping local and systemic immune responses. Semin Immunol. 2012;24:58–66. doi: 10.1016/j.smim.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Alexander K.L., Targan S.R., Elson C.O. Microbiota activation and regulation of innate and adaptive immunity. Immunol Rev. 2014;260:206–220. doi: 10.1111/imr.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kaiko G.E., Horvat J.C., Beagley K.W., Hansbro P.M. Immunological decision-making: how does the immune system decide to mount a helper T-cell response? Immunology. 2008;123:326–338. doi: 10.1111/j.1365-2567.2007.02719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kempski J., Brockmann L., Gagliani N., Huber S. TH17 cell and epithelial cell crosstalk during inflammatory bowel disease and carcinogenesis. Front Immunol. 2017 doi: 10.3389/fimmu.2017.01373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mazmanian S.K., Round J.L., Kasper D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;7195:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 185.Jun J.C., Jones M.B., Oswald D.M., Sim E.S., Jonnalagadda A.R., Kreisman L.S.C. T cell-intrinsic TLR2 stimulation promotes IL-10 expression and suppressive activity by CD45RbHi T cells. PLoS One. 2017;12 doi: 10.1371/journal.pone.0180688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Vinolo M.A., Rodrigues H.G., Hatanaka E., Sato F.T., Sampaio S.C., Curi R. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J Nutr Biochem. 2011;22(9):849–855. doi: 10.1016/j.jnutbio.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 187.Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 188.Paassen N., Vincent A., Puiman P.J., van der Sluis M., Bouma J., Boehm G. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 2009;420(2):211–219. doi: 10.1042/BJ20082222. [DOI] [PubMed] [Google Scholar]
- 189.Kalina U., Koyama N., Hosoda T., Nuernberger H., Sato K., Hoelzer D. Enhanced production of IL-18 in butyrate-treated intestinal epithelium by stimulation of the proximal promoter region. Eur J Immunol. 2002;32(9):2635–2643. doi: 10.1002/1521-4141(200209)32:9<2635::AID-IMMU2635>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 190.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Galván-Moroyoqui J.M., Domínguez-Robles M.C., Franco E., Meza I. The interplay between Entamoeba and enteropathogenic bacteria modulates epithelial cell damage. PLoS Negl Trop Dis. 2008;2 doi: 10.1371/journal.pntd.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nemanic P.C., Owen R.L., Stevens D.P., Mueller J.C. Ultrastructural observations on giardiasis in a mouse model. II. Endosymbiosis and organelle distribution in Giardia muris and Giardia lamblia. J Infect Dis. 1979;140:222–228. doi: 10.1093/infdis/140.2.222. [DOI] [PubMed] [Google Scholar]
- 193.Deatherage Kaiser B.L., Li J., Sanford J.A., Kim Y.-M., Kronewitter S.R., Jones M.B. A multi-omic view of host-pathogen-commensal interplay in Salmonella-mediated intestinal infection. PLoS One. 2013;8 doi: 10.1371/journal.pone.0067155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Waldman A.J., Balskus E.P. The human microbiota, infectious disease, and global health: challenges and opportunities. ACS Infect Dis. 2017 doi: 10.1021/acsinfecdis.7b00232. [DOI] [PubMed] [Google Scholar]
- 195.Antunes L.C., McDonald J.A., Schroeter K., Carlucci C., Ferreira R.B., Wang M. Antivirulence activity of the human gut metabolome. mBio. 2014;5 doi: 10.1128/mBio.01183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.García C., Tebbji F., Daigneault M., Liu N.-N., Köhler J.R., Allen-Vercoe E. The human gut microbial metabolome modulates fungal growth via the TOR signaling pathway. mSphere. 2017;2 doi: 10.1128/mSphere.00555-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Soper D. Trichomoniasis: under control or undercontrolled? Am J Obstet Gynecol. 2004;190:281–290. doi: 10.1016/j.ajog.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 198.Rappelli P., Carta F., Delogu G., Addis M.F., Dessì D., Cappuccinelli P. Mycoplasma hominis and Trichomonas vaginalis symbiosis: multiplicity of infection and transmissibility of M. hominis to human cells. Arch Microbiol. 2001;175:70–74. doi: 10.1007/s002030000240. [DOI] [PubMed] [Google Scholar]
- 199.Ji J., Shu D., Zheng M., Wang J., Luo C., Wang Y. Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Sci Rep. 2016;6:24838. doi: 10.1038/srep24838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Spiljar M., Merkler D., Trajkovski M. The Immune System Bridges the Gut Microbiota with Systemic Energy Homeostasis: Focus on TLRs, Mucosal Barrier, and SCFAs. Front Immunol. 2017;8:1353. doi: 10.3389/fimmu.2017.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lathrop S.K., Bloom S.M., Rao S.M., Nutsch K., Lio C.W., Santacruz N. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lee Y.K., Mazmanian S.K. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Peterson D.A., McNulty N.P., Guruge J.L., Gordon J.I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 204.Hooper L.V., Stappenbeck T.S., Hong C.V., Gordon J.I. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 205.Round J.L., Mazmanian S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Vangelista L., Secchi M., Liu X., Bachi A., Jia L., Xu Q. Engineering of Lactobacillus jensenii to secrete RANTES and a CCR5 antagonist analogue as live HIV-1 blockers. Antimicrob Agents Chemother. 2010;54(7):2994–3001. doi: 10.1128/AAC.01492-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chang T.L., Chang C.H., Simpson D.A., Xu Q., Martin P.K., Lagenaur L.A. Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proc Natl Acad Sci U S A. 2003;100(20):11672–11677. doi: 10.1073/pnas.1934747100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wood M.R., Yu E.A., Mehta S. The human microbiome in the fight against tuberculosis. Am J Trop Med Hyg. 2017;96:1274–1284. doi: 10.4269/ajtmh.16-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Adami A.J., Cervantes J.L. The microbiome at the pulmonary alveolar niche and its role in Mycobacterium tuberculosis infection. Tuberculosis. 2015;95:651–658. doi: 10.1016/j.tube.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Villarino N.F., LeCleir G.R., Denny J.E., Dearth S.P., Harding C.L., Sloan S.S. Composition of the gut microbiota modulates the severity of malaria. Proc Natl Acad Sci U S A. 2016;113:2235–2240. doi: 10.1073/pnas.1504887113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ott S.J., Waetzig G.H., Rehman A., Moltzau-Anderson J., Bharti R., Grasis J.A. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology. 2017;152:799–811. doi: 10.1053/j.gastro.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 212.Yooseph S., Kirkness E.F., Tran T.M., Harkins D.M., Jones M.B., Torralba M.G. Stool microbiota composition is associated with the prospective risk of Plasmodium falciparum infection. BMC Genomics. 2015;16:631. doi: 10.1186/s12864-015-1819-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yilmaz B., Portugal S., Tran T.M., Gozzelino R., Ramos S., Gomes J. Gut microbiota elicits a protective immune response against malaria transmission. Cell. 2014;159:1277–1289. doi: 10.1016/j.cell.2014.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Njunda A.L., Fon S.G., Assob J.C., Nsagha D.S., Kwenti T.D., Kwenti T.E. Coinfection with malaria and intestinal parasites, and its association with anaemia in children in Cameroon. Infect Dis Poverty. 2015;4:43. doi: 10.1186/s40249-015-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.von Klitzing E., Ekmekciu I., Kühl A.A., Bereswill S., Heimesaat M.M. Intestinal, extra-intestinal and systemic sequelae of Toxoplasma gondii induced acute ileitis in mice harboring a human gut microbiota. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0176144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Cumber S.N., Yvonne-Estelle K.N., Jaila S. The effects of toxoplasmosis and malaria coinfection on malaria parasite density and hematological parameters in children (0-6 Years) in the Nkolbisson health district, Cameroon. J Family Med Health Care. 2016;2:81–88. [Google Scholar]
- 217.Benson A., Pifer R., Behrendt C.L., Hooper L.V., Yarovinsky F. Gut commensal bacteria direct a protective immune response against the human pathogen Toxoplasma gondii. Cell Host Microbe. 2009;6:187–196. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Berrilli F., Di Cave D., Cavallero S., D’Amelio S. Interactions between parasites and microbial communities in the human gut. Front Cell Infect Microbiol. 2012;2:141. doi: 10.3389/fcimb.2012.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Burgess S.L., Petri W.A., Jr. The intestinal bacterial microbiome and E. histolytica Infection. Curr Trop Med Rep. 2016;3:71–74. doi: 10.1007/s40475-016-0083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zuo T., Wong S.H., Lam K., Lui R., Cheung K., Tang W. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2017 doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Holtz L.R., Cao S., Zhao G., Bauer I.K., Denno D.M., Klein E.J. Geographic variation in the eukaryotic virome of human diarrhea. Virology. 2014;468-470:556–564. doi: 10.1016/j.virol.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Colsona P., Fancelloa L., Gimeneza G., Armougom F., Desnues C., Fournous G. Evidence of the megavirome in humans. J Clin Virol. 2013;57:191–200. doi: 10.1016/j.jcv.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 223.Kernbauer E., Ding Yi, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516(7529):94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Reyes A., Semenkovich N.P., Whiteson K., Rohwer F., Gordon J.I. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lim E.S., Zhou Y., Zhao G., Bauer I.K., Droit L., Ndao I.M. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Carding S.R., Davis N., Hoyles L. Review article: the human intestinal virome in health and disease. Aliment Pharmacol Ther. 2017;46:800–815. doi: 10.1111/apt.14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Norman J.M., Handley S.A., Baldridge M.T., Droit L., Liu C.Y., Brian C.K. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.ClincalTrials.gov . National Library of Medicine (US); Bethesday (MD): 2017. Identifier NCT03231267, Existence in the human digestive flora of phages able to prevent the acquisition of multiresistant Enterobacteria (PHAGO-BMR)https://clinicaltrials.gov/ct2/show/study/NCT03231267?recrs=ab&cond=phage&rank=4#contacts [Google Scholar]
- 229.ClincalTrials.gov . National Library of Medicine (US); Bethesday (MD): 2014. Identifier NCT02116010, Evaluation of phage therapy for the treatment of Escherichia coli and Pseudomonas aeruginosa wound infections in burned patients (PHAGOBURN)https://clinicaltrials.gov/ct2/show/NCT02116010 [Google Scholar]
- 230.Zaiss M.M., Harris N.L. Interactions between the intestinal microbiome and helminth parasites. Parasite Immunol. 2016;38:5–11. doi: 10.1111/pim.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Holzscheiter M., Layland L.E., Loffredo-Verde E., Mair K., Vogelmann R., Langer R. Lack of host gut microbiota alters immune responses and intestinal granuloma formation during schistosomiasis. Clin Exp Immunol. 2013;175:246–257. doi: 10.1111/cei.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Iacono A., Raso G.M., Canani R.B., Calignano A., Meli R. Probiotics as an emerging therapeutic strategy to treat NAFLD: focus on molecular and biochemical mechanisms. J Nutr Biochem. 2011;22:699–711. doi: 10.1016/j.jnutbio.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 233.Fujimura K.E., Lynch S.V. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. 2015;17:592–602. doi: 10.1016/j.chom.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Laukens D., Brinkman B.M., Raes J., De Vos M., Vandenabeele P. Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol Rev. 2016;40:117–132. doi: 10.1093/femsre/fuv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Howe A., Ringus D.L., Williams R.J., Choo Z.N., Greenwald S.M., Owens S.M. Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. ISME J. 2016;5:1217–1227. doi: 10.1038/ismej.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Briese T., Kapoor A., Mishra N., Jain K., Kumar A., Jabado O.J. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio. 2015;6 doi: 10.1128/mBio.01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Li J., Jia H., Cai X., Zhong H., Feng Q., Sunagawa S. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 238.Kiesler P., Fuss I.J., Strober W. Experimental Models of Inflammatory Bowel Diseases. Cell Mol Gastroenterol Hepatol. 2015;1:154–170. doi: 10.1016/j.jcmgh.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lynch S.V., Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 240.Gagnière J., Raisch J., Veziant J., Barnich N., Bonnet R., Buc E. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22:501–518. doi: 10.3748/wjg.v22.i2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Brennan C.A., Garrett W.S. Gut microbiota, inflammation, and colorectal cancer. Annu Rev Microbiol. 2016;70:395–411. doi: 10.1146/annurev-micro-102215-095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gao R., Gao Z., Huang L., Qin H. Gut microbiota and colorectal cancer. Eur J Clin Microbiol Infect Dis. 2017;36:757–769. doi: 10.1007/s10096-016-2881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Foxx-Orenstein A.E., Chey W.D. Manipulation of the gut microbiota as a novel treatment strategy for gastrointestinal disorders. Am J Gastroenterol Suppl. 2012;1:41–46. [Google Scholar]
- 244.Khoder G., Al-Menhali A.A., Al-Yassir F., Karam S.M. Potential role of probiotics in the management of gastric ulcer. Exp Ther Med. 2016;12:3–17. doi: 10.3892/etm.2016.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Dukowicz A.C., Lacy B.E., Levine G.M. Small intestinal bacterial overgrowth a comprehensive review. Gastroenterol Hepatol (NY) 2007;3:112–122. [PMC free article] [PubMed] [Google Scholar]
- 246.Xiao X., Nakatsu G., Jin Y., Wong S., Yu J., Lau J.Y.W. Gut microbiota mediates protection against enteropathy induced by indomethacin. Sci Rep. 2017;7:40317. doi: 10.1038/srep40317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Vieira A.T., Fukumori C., Ferreira C.M. New insights into therapeutic strategies for gut microbiota modulation in inflammatory diseases. Clin Transl Immunol. 2016;5 doi: 10.1038/cti.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lopez J., Grinspan A. Fecal Microbiota Transplantation for Inflammatory Bowel Disease. Gastroenterol Hepatol (NY) 2016;12:374–379. [PMC free article] [PubMed] [Google Scholar]
- 249.Hirsch B.E., Saraiya N., Poeth K., Schwartz R.M., Epstein M.E., Honig G. Effectiveness of fecal-derived microbiota transfer using orally administered capsules for recurrent Clostridium difficile infection. BMC Infect Dis. 2015;15:191. doi: 10.1186/s12879-015-0930-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Choi H.H., Cho Y.S. Fecal microbiota transplantation: current applications, effectiveness, and future perspectives. Clin Endosc. 2016;49(3):257–265. doi: 10.5946/ce.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Otani K., Tanigawa T., Watanabe T., Shimada S., Nadatani Y., Nagami Y. Microbiota plays a key role in Non-Steroidal Anti-Inflammatory Drug-Induced small intestinal damage. Digestion. 2017;95:22–28. doi: 10.1159/000452356. [DOI] [PubMed] [Google Scholar]
- 252.Huang Y.J. Asthma microbiome studies and the potential for new therapeutic strategies. Curr Allergy Asthma Rep. 2013;13:453–461. doi: 10.1007/s11882-013-0355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Herbst T., Sichelstiel A., Schär C., Yadava K., Bürki K., Cahenzli J. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 254.Cahenzli J., Koller Y., Wyss M., Geuking M.B., McCoy K.D. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Stefka A.T., Feehley T., Tripathi P., Qiu J., McCoy K., Mazmanian S.K. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Berni Canani R., Sangwan N., Stefka A.T., Nocerino R., Paparo L., Aitoro R. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016;10:742–750. doi: 10.1038/ismej.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Gaudreau M.C., Johnson B.M., Gudi R., Al-Gadban M.M., Vasu C. Gender bias in lupus: does immune response initiated in the gut mucosa have a role? Clin Exp Immunol. 2015;180:393–407. doi: 10.1111/cei.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Suárez-Zamorano N., Fabbiano S., Chevalier C., Ozren S., Colin D.J., Ana S. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat Med. 2015;21:1497–1501. doi: 10.1038/nm.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Maslowski K.M., Vieira A.T., Ng A., Kranich J., Sierro F., Yu D. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Thorburn A.N., McKenzie C.I., Shen S., Stanley D., Macia L., Mason L.J. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 261.Sun M., Wu W., Liu Z., Cong Y. Microbiota metabolite short chain fatty acids, GCPR, and inflammatory bowel diseases. J Gastroenterol. 2017;52(1):1–8. doi: 10.1007/s00535-016-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Bindels L.B., Dewulf E.M., Delzenne N.M. GPR43/FFA2: physiopathological relevance and therapeutic prospects. Trends Pharmacol Sci. 2013;34:226–232. doi: 10.1016/j.tips.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 263.Macia L., Tan J., Vieira A.T., Leach K., Stanley D., Luong S. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 264.Cohen L.J., Esterhazy D., Kim S.H., Lemetre C., Aguilar R.R., Gordon E.A. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature. 2017;549:48–53. doi: 10.1038/nature23874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Sanchez M., Panahi S., Tremblay A. Childhood obesity: a role for gut microbiota? Int J Environ Res Public Health. 2015;12:162–175. doi: 10.3390/ijerph120100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Slattery J., MacFabe D.F., Frye R.E. The significance of the enteric microbiome on the development of childhood disease: a review of prebiotic and probiotic therapies in disorders of childhood. Clin Med Insights Pediatr. 2016;10:91–107. doi: 10.4137/CMPed.S38338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Verna E.C. Use of probiotics in gastrointestinal disorders: what to recommend? Therap Adv Gastroenterol. 2010;3:307–319. doi: 10.1177/1756283X10373814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Pandey K.R., Naik S.R., Vakil B.V. Probiotics, prebiotics and synbiotics- a review. J Food Sci Technol. 2015;52:7577–7587. doi: 10.1007/s13197-015-1921-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Howarth G.S., Wang H. Role of endogenous microbiota, probiotics and their biological products in human health. Nutrients. 2013;5:58–81. doi: 10.3390/nu5010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Nami Y., Haghshenas B., Haghshenas M., Khosroushahi A.Y. Antimicrobial activity and the presence of virulence factors and bacteriocin structural genes in Enterococcus faecium CM33 isolated from ewe colostrums. Front Microbiol. 2015;6:782. doi: 10.3389/fmicb.2015.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Costelloe C., Metcalfe C., Lovering A., Mant D., Hay A.D. Effect of antibiotic prescribing in primary care on antimicrobial resistance in individual patients: systematic review and meta-analysis. BMJ. 2010;340 doi: 10.1136/bmj.c2096. [DOI] [PubMed] [Google Scholar]
- 272.Scherer A., Vogt H.R., Vilei E.M., Frey J., Perreten V. Enhanced antibiotic multi-resistance in nasal and faecal bacteria after agricultural use of streptomycin. Environ Microbiol. 2013;15:297–304. doi: 10.1111/1462-2920.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Founou L.L., Founou R.C., Essack S.Y. Antibiotic resistance in the food chain: a developing country-perspective. Front Microbiol. 2016;7:1881. doi: 10.3389/fmicb.2016.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lee C.R., Cho I.H., Jeong B.C., Lee S.H. Strategies to Minimize Antibiotic Resistance. Int J Environ Res Public Health. 2013;10:4274–4305. doi: 10.3390/ijerph10094274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Munita J.M., Arias C.A. Mechanisms of Antibiotic Resistance. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.McFarland L.V. Meta-analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol. 2006;101:812–822. doi: 10.1111/j.1572-0241.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 277.Szajewska H., Ruszczyński M., Radzikowski A. Probiotics in the prevention of antibiotic-associated diarrhea in children: a meta-analysis of randomized controlled trials. J Pediatr. 2006;149:367–372. doi: 10.1016/j.jpeds.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 278.Hickson M. Probiotics in the prevention of antibiotic-associated diarrhoea and Clostridium difficile infection. Therap Adv Gastroenterol. 2011;4:185–197. doi: 10.1177/1756283X11399115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Hoveyda N., Heneghan C., Mahtani K.R., Perera R., Roberts N., Glasziou P. A systematic review and meta-analysis: probiotics in the treatment of irritable bowel syndrome. BMC Gastroenterol. 2009;9:15. doi: 10.1186/1471-230X-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Reid G., Younes J.A., Van der Mei H.C., Gloor G.B., Knight R., Busscher H.J. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat Rev Microbiol. 2011;9:27–38. doi: 10.1038/nrmicro2473. [DOI] [PubMed] [Google Scholar]
- 281.Reid G. The development of probiotics for women’s health. Can J Microbiol. 2017;63:269–277. doi: 10.1139/cjm-2016-0733. [DOI] [PubMed] [Google Scholar]
- 282.Damaskos D., Kolios G. Probiotics and prebiotics in inflammatory bowel disease: microflora 'on the scope'. Br J Clin Pharmacol. 2008;65:453–467. doi: 10.1111/j.1365-2125.2008.03096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Gill H., Prasad J. Probiotics, immunomodulation, and health benefits. Adv Exp Med Biol. 2008;606:423–454. doi: 10.1007/978-0-387-74087-4_17. [DOI] [PubMed] [Google Scholar]
- 284.Travers M.A., Florent I., Kohl L., Grellier P. Probiotics for the control of parasites: an overview. J Parasitol Res. 2011;2011:610769. doi: 10.1155/2011/610769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Markowiak P., Śliżewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients. 2017;9:1021. doi: 10.3390/nu9091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Llor C., Bjerrum L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf. 2014;5:229–241. doi: 10.1177/2042098614554919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Botina S.G., Poluéktova E.U., Glazkova A.A., Zakharevich N.V., Koroban N.V., Zinchenko V.V. Antibiotic resistance of potential probiotic bacteria of the genus Lactobacillus from human gastrointestinal microbiome. Mikrobiologiia. 2011;80:175–183. [PubMed] [Google Scholar]
- 288.Lönnermark E., Friman V., Lappas G., Sandberg T., Berggren A., Adlerberth I. Intake of Lactobacillus plantarum reduces certain gastrointestinal symptoms during treatment with antibiotics. J Clin Gastroenterol. 2010;44:106–112. doi: 10.1097/MCG.0b013e3181b2683f. [DOI] [PubMed] [Google Scholar]
- 289.Humen M.A., De Antoni G.L., Benyacoub J., Costas M.E., Cardozo M.I., Kozubsky L. Lactobacillus johnsonii La1 antagonizes Giardia intestinalisin vivo. Infect Immun. 2005;73:1265–1269. doi: 10.1128/IAI.73.2.1265-1269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Shukla G., Devi P., Sehgal R. Effect of Lactobacillus casei as a probiotic on modulation of giardiasis. Dig Dis Sci. 2008;53:2671–2679. doi: 10.1007/s10620-007-0197-3. [DOI] [PubMed] [Google Scholar]
- 291.Benyacoub J., Pérez P.F., Rochat F., Saudan K.Y., Reuteler G., Antille N. Enterococcus faecium SF68 enhances the immune response to Giardia intestinalis in mice. J Nutr. 2005;135:1171–1176. doi: 10.1093/jn/135.5.1171. [DOI] [PubMed] [Google Scholar]
- 292.Solano-Aguilar G., Shea-Donohue T., Madden K., Dawson H., Ledbetter T., Urban J.J., Gasbarre L.C., editors. Symposium (Veterinary Parasitology): New Approaches in the Study of Animal Parasites. Vol. 125. 2004. The effect of human-derived probiotic bacteria on the intestinal function of pigs; pp. 147–161. [DOI] [PubMed] [Google Scholar]
- 293.Bautista C.R., Sandoval A., Aguilar B.R. Effect of highand low-molecular-weight components of Lactobacillus casei on resistance against Babesia microti in NIH mice. Ann N Y Acad Sci. 2008;1149:152–154. doi: 10.1196/annals.1428.037. [DOI] [PubMed] [Google Scholar]
- 294.Alak J.I.B., Wolf B.W., Mdurvwa E.G., Pimentel-Smith G.E., Adeyemo O. Effect of Lactobacillus reuteri on intestinal resistance to Cryptosporidium parvum infection in a murine model of acquired immunodeficiency syndrome. J Infect Dis. 1997;175:218–221. doi: 10.1093/infdis/175.1.218. [DOI] [PubMed] [Google Scholar]
- 295.Alak J.I.B., Wolf B.W., Mdurvwa E.G., Pimentel-Smith G.E., Kolavala S., Abdelrahman H. Supplementation with Lactobacillus reuteri or L. acidophilus reduced intestinal shedding of Cryptosporidium parvum oocysts in immunodeficient C57BL/6 mice. Cell Mol Biol. 1999;45:855–863. [PubMed] [Google Scholar]
- 296.Pickerd N., Tuthill D. Resolution of cryptosporidiosis with probiotic treatment. Postgrad Med J. 2004;80:112–113. doi: 10.1136/pmj.2003.014175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.de Fátima Macedo Santos J., Vasconcelos J., de Souza J.R., de Medeiros Coutinho E., Montenegro S.M., Azevedo-Ximenes E. The effect of Zymomonas mobilis culture on experimental Schistosoma mansoni infection. Rev Soc Bras Med Trop. 2004;37:502–504. doi: 10.1590/s0037-86822004000600015. [DOI] [PubMed] [Google Scholar]
- 298.Kato I., Tanaka K., Yokokura T. Lactic acid bacterium potently induces the production of interleukin-12 and interferon-γ by mouse splenocytes. Int J Immunopharmcol. 1999;21:121–131. doi: 10.1016/s0192-0561(98)00072-1. [DOI] [PubMed] [Google Scholar]
- 299.Basualdo J., Sparo M., Chiodo P., Ciarmela M., Minvielle M. Oral treatment with a potential probiotic (Enterococcus faecalis CECT 7121) appears to reduce the parasite burden of mice infected with Toxocara canis. Ann Trop Med Parasitol. 2007;101:559–562. doi: 10.1179/136485907X193824. [DOI] [PubMed] [Google Scholar]
- 300.Garfias C.R.B., Alvarez M.C.T., Gómez F.M. The inoculation of Lactobacillus casei in NIH mice induces a protective response against Trypanosoma cruzi (Ninoa strain) infection. Vet Mex. 2008;39:139–144. [Google Scholar]
- 301.Wallace B.D., Redinbo M.R. The human microbiome is a source of therapeutic drug targets. Curr Opin Chem Biol. 2013;17:379–384. doi: 10.1016/j.cbpa.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Wallace B.D., Wang H., Lane K.T., Scott J.E., Orans J., Koo J.S. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Panigrahi P., Parida S., Nanda N.C., Satpathy R., Pradhan L., Chandel D.S. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548:407–412. doi: 10.1038/nature23480. [DOI] [PubMed] [Google Scholar]
- 304.Hwang I.Y., Koh E., Wong A., March J.C., Bentley W.E., Lee Y.S. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat Commun. 2017;8:15028. doi: 10.1038/ncomms15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Vangay P., Ward T., Gerber J.S., Knights D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe. 2015;17:553–564. doi: 10.1016/j.chom.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Zmora N., Zeevi D., Korem T., Segal E., Elinav E. Taking it personally: personalized utilization of the human microbiome in health and disease. Cell Host Microbe. 2016;19:12–20. doi: 10.1016/j.chom.2015.12.016. [DOI] [PubMed] [Google Scholar]