Abstract
Introduction
Nephronophthisis (NPH) comprises a group of rare disorders accounting for up to 10% of end-stage kidney disease (ESKD) in children. Prediction of kidney prognosis poses a major challenge. We assessed differences in kidney survival, impact of variant type, and the association of clinical characteristics with declining kidney function.
Methods
Data was obtained from 3 independent sources, namely the network for early onset cystic kidney diseases clinical registry (n = 105), an online survey sent out to the European Reference Network for Rare Kidney Diseases (n = 60), and a literature search (n = 218).
Results
A total of 383 individuals were available for analysis: 116 NPHP1, 101 NPHP3, 81 NPHP4 and 85 NPHP11/TMEM67 patients. Kidney survival differed between the 4 cohorts with a highly variable median age at onset of ESKD as follows: NPHP3, 4.0 years (interquartile range 0.3–12.0); NPHP1, 13.5 years (interquartile range 10.5–16.5); NPHP4, 16.0 years (interquartile range 11.0–25.0); and NPHP11/TMEM67, 19.0 years (interquartile range 8.7–28.0). Kidney survival was significantly associated with the underlying variant type for NPHP1, NPHP3, and NPHP4. Multivariate analysis for the NPHP1 cohort revealed growth retardation (hazard ratio 3.5) and angiotensin-converting enzyme inhibitor (ACEI) treatment (hazard ratio 2.8) as 2 independent factors associated with an earlier onset of ESKD, whereas arterial hypertension was linked to an accelerated glomerular filtration rate (GFR) decline.
Conclusion
The presented data will enable clinicians to better estimate kidney prognosis of distinct patients with NPH and thereby allow personalized counseling.
Keywords: end-stage kidney disease, genetic variant severity, genotype-phenotype correlations, kidney survival, nephronophthisis, prognostic factors
Graphical abstract
NPH comprises a clinically and genetically heterogeneous group of autosomal recessive tubulointerstitial cystic kidney disorders representing the most frequent monogenic cause of ESKD in children and adolescents.1,2 Incidence ranges from 1 per 1,000,000 to 1 per 50,000 depending on ethnicity and underlying disease-causing variant.3,4 Variants in 25 genes have been identified to be associated with NPH, still leaving about 40% of patients genetically unsolved.4, 5, 6, 7 A large homozygous deletion on chromosome 2q12-q13 including the entire NPHP1 gene is the most frequent disease-causing variant accounting for 20% to 40% of all cases. Variants in the other known NPHP genes makeup for 3% or less each.5,8, 9, 10 Three clinical subtypes have been established based on age at onset of ESKD as follows: Infantile, juvenile, and adolescent NPH.11,12 The so called “classical juvenile NPH” is the most common entity associated with ESKD at a median age of 13 years.13 Conversely, in infantile NPH, ESKD is reached before the age of 5 years (mean age 8 months) whereas in adolescent forms kidney function is usually preserved until adulthood (mean age 19 years).11,14,15 Variants in NPHP2 and NPHP3 rather predispose for infantile NPH, whereas other genes are linked to late onset forms (e.g., NPHP4).5,16 Nevertheless, the more widespread use of modern sequencing techniques in combination with clinical information gained from large-scale databases opened up new perspectives to define more precisely the age spectrum for the onset of ESKD associated with distinct gene variants. This particularly applies to the homozygous NPHP1 deletion.17,18 Similarly, distinctive NPHP3 variants have been reported to cause late onset NPH in contrast to the majority of patients displaying an infantile disease course.19, 20, 21
Despite the large number of identified NPHP genes, only a few of them are of relevance in routine clinical practice, namely NPHP1, NPHP3, NPHP4, and NPHP11/TMEM67. These 4 genes account for 75% of all identified disease-causing variants.9 In addition, NPHP5/IQCB1 and NPHP6/CEP290 represent the 2 main causes for the clinical spectra of Senior-Løken and Joubert syndrome.16,22,23
NPH-related diagnostic approaches have shifted from a predominantly clinical assessment to the early use of multigene panels and exome sequencing as a result of technical progress.24, 25, 26 This allows for a detailed molecular diagnosis early in life, sometimes even before the onset of kidney symptoms. Nevertheless, because of the lack of clear-cut genotype-phenotype correlations, phenotypic variability and the consequential unpredictability of individual disease progression, clinical management and personalized counseling remain major challenges. The prognostic uncertainty poses an enormous psychological burden on pediatric patients and their families facing a potentially life-threatening disease of unclear onset and extent.
Herein, we analyzed differences in gene-related kidney survival based on large patient cohorts of >80 each. Furthermore, we assessed potential prognostic factors for the NPHP1 cohort, representing the clinically most relevant group. This is one of the largest studies addressing kidney survival for NPH in genetically characterized individuals.
Methods
Patient Recruitment
Phenotypic data of 383 genetically characterized individuals was obtained from 3 independent sources, namely the Network of Early Onset Cystic Kidney Diseases (NEOCYST) clinical registry (n = 105),27 an online survey sent out to the members of the European Reference Network for Rare Kidney Diseases (ERKNet; n = 60) and a complementary literature search (n = 218) (Figure 1). In addition, clinical information of 44 genetically unsolved individuals was available from the NEOCYST registry. Patients were enrolled in the registry from February 1, 2010, through December 31, 2020. The ERKNet-based online survey asked for a limited data set containing information on sex, age, genetic background, GFR decline, hypertension, proteinuria, and a medical history of ACEI treatment. Patient baseline characteristics at study entry are outlined in Supplementary Table S1. All patients and/or their parents gave written informed consent. The study was approved by the Ethics Committee of the University of Münster (2016-284-f-S) in accordance with the Declaration of Helsinki.
Figure 1.
Patient recruitment. Phenotypic data of 383 genetically characterized individuals was obtained from 3 independent sources: the Network of Early Onset Cystic Kidney Diseases clinical registry (n = 105), an online survey sent out to the members of the ERKNet (n = 60) and a complementary literature search (n = 218). Homogeneous data from 116 NPHP1 patients obtained from the Network of Early Onset Cystic Kidney Diseases registry (n = 80) and the online survey (n = 36) was fused for analyzes of impact of clinical factors. Gene-specific and variant-related kidney survival was analyzed including all 383 genetically characterized individuals (NPHP1: n = 116; NPHP3: n = 101; NPHP4: n = 81; NPHP11/TMEM67: n = 85) originating from the Network of Early Onset Cystic Kidney Diseases registry (n = 105), the online survey (n = 60) and a comprehensive literature search (n = 218). ERKNet, European Reference Network for Rare Kidney Diseases.
For the literature search, the MEDLINE library was screened for original articles including case reports up to March 2020, providing data on kidney survival or onset of ESKD for the following genotypes: NPHP3, NPHP4, and NPHP11/TMEM67. In total, data from 218 individuals (86 NPHP3, 64 NPHP4, and 68 NPHP11/TMEM67) were identified and extracted from the literature containing information on the age at ESKD and the underlying disease-causing variants (Supplementary Table S2). No literature-based data was generated for the NPHP1 cohort. For the NPHP1 cohort, solely information from the NEOCYST registry (n = 80) or reported via questionnaire (n = 36) were used for analysis (Figure 1). For the NPHP1 cohort, duplicate inclusions were thoroughly excluded by crosschecking identifying information (initials, month and year of birth, and sex). In the case of all other genotypes, combined information on individual genetic variants, sex and age were used to avoid duplicate inclusions from different data sources. A total of 3 duplicates were identified and withdrawn before analysis, resulting in phenotypic data of a total of 383 genetically characterized individuals (Figure 1).
Genetic Testing
Because of the study design, various methods for genetic testing were used, including PCR-based gel electrophoresis or MLPA for detection of homozygous NPHP1 deletions (n = 97/165), targeted Sanger sequencing on the basis of the patient’s phenotype (n = 11/165), as well as massively parallel sequencing based approaches such as multigene panel sequencing and exome sequencing (n = 57/165).28,29
Variables
Estimated GFR (eGFR) was assessed by the FAS equation for the full age spectrum.30 Annual eGFR decline was determined in patients with at least 2 subsequent eGFR values in intervals of at least 3 months and displayed as absolute values (ml/min per 1.73 m2 body surface area (BSA) per year).
ESKD was defined as either the start of renal replacement therapy (chronic dialysis treatment or kidney transplantation) or an eGFR value <10 ml/min per 1.73 m2 BSA. Proteinuria was defined as >300 mg/24 h (>0.2 g/g). Growth retardation was defined by a baseline height below the third percentile or height development crossing the third percentile.
Variant Classification
Interpretation of the clinical significance of variants was completed following the American College of Medical Genetics and Genomics and the Association for Molecular Pathology guidelines31 using the VarSome32 online tool (VarSome The Human Genomics Community). In addition to the automatically generated criteria, manual adjustment for the following 2 criteria was made: PM3 (recessive disorders, detected in trans with a pathogenic variant) and PP4 (patient’s phenotype or family history is highly specific for a disease with a single genetic etiology). Further interpretation of functional effects of human nsSNPs was conducted using the PolyPhen-233 (http://genetics.bwh.harvard.edu/pph2/), SIFT34 (https://sift.bii.a-star.edu.sg/), PROVEAN35 (http://provean.jcvi.org/about.php), varSEAK Online (https://varseak.bio, developed by JSI medical systems GmbH, Ettenheim, Germany) and ClinVar (release 13./31.07.2021)36 (https://www.ncbi.nlm.nih.gov/clinvar/) online tools.
Statistical Analysis
Based on the detailed phenotypic data provided by the NEOCYST registry and complemented by information gained from the online survey, cross-sectional and longitudinal statistical analyses were performed on the NPHP1 cohort (n = 116). Kidney survival was defined as the time from birth to the age at onset of ESKD. Patients who did not reach ESKD were censored at their age of last follow-up. Kidney survival was depicted using Kaplan-Meier estimator and the association of potential prognostic factors was estimated by Cox regression analysis in a univariate and multivariate fashion. The P ≤ 0.05 were considered noticeable (“significant”). Results are considered exploratory, not confirmatory. No adjustment for multiple testing was performed. An overall significance level was not determined and cannot be calculated. Statistical analyses were performed using R software and SAS (SAS Institute Inc., Cary, NC, USA).
Results
Kidney Survival
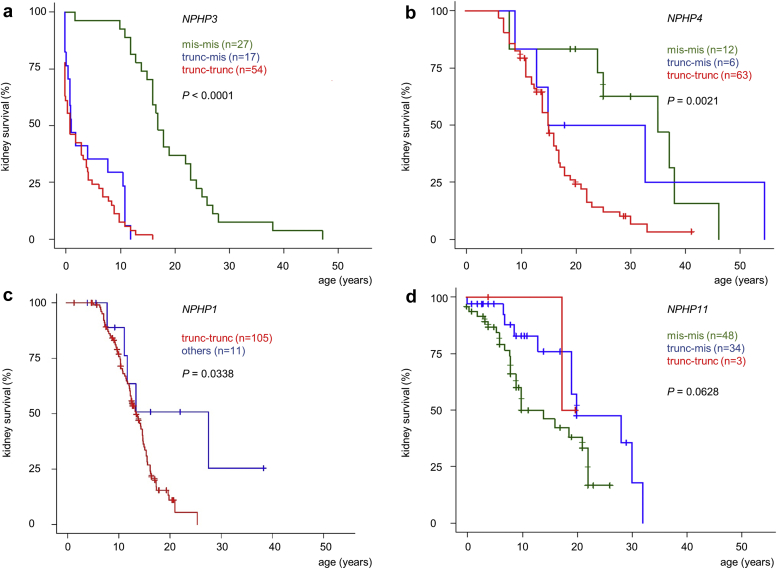
Using the data of all 383 genetically determined and 44 genetically unsolved patients, Kaplan–Meier analysis revealed remarkable differences between the genetically defined subgroups (Figure 2). A rapid decline of kidney survival between the age of 8 and 16 years was observed in the NPHP1 subgroup (Figure 2a). By 16.5 years of age, 75% of affected teenagers received kidney replacement therapy (Figure 2b). Among the patients, 19% retained residual kidney function beyond the age of 20 years (5 individuals with a homozygous NPHP1 deletion and 3 carrying other biallelic NPHP1 variants) (Supplementary Table S2). Kidney survival for patients carrying NPHP4 variants was characterized by a later onset and a slower decline compared with NPHP1 (Figure 2a). The main differences between both groups were illustrated by the age when 50% (13.5 years vs. 16 years) and 75% (16.5 years vs. 25 years) of patients reached ESKD. Almost no age difference was observed for the onset of 25% receiving renal replacement therapy (Figure 2b). Notably, kidney survival of the genetically unsolved individuals was comparable to survival curves observed for NPHP1 and NPHP4 (Figure 2a).
Figure 2.
Gene-related kidney survival. Differences in gene-related kidney survival displayed as Kaplan–Meier survival curve (a) and median age (black line)/interquartile range for the onset of ESKD in 50%, 25% and 75% of participants (b). Significant statements: NPHP1 vs. NPHP3: P < 0.0001; NPHP1 versus NPHP4: P < 0.003; NPHP1 versus NPHP11: P = 0.057; NPHP3 versus NPHP4: P < 0.0001; NPHP3 versus NPHP11: P < 0.0001; NPHP4 versus NPHP11: P = 0.539; NPHP1 versus NPHP3 vs. NPHP4 versus NPHP11: P < 0.0001. ESKD, end-stage kidney disease.
NPHP11/TMEM67 related kidney survival was characterized by an overall lower percentage of ESKD as well as a flatter decline in survival, particularly in the second decade of life. Two distinct groups could be identified, namley those with severe kidney involvement represented by 25% having reached ESKD as early as 8.7 years, and those with mild or no obvious kidney involvement with another 25% of patients not requiring kidney replacement therapy at the age of 28 (Figure 2a). An even more pronounced picture emerged for the NPHP3 group. Though approximately half of the patients experienced infantile NPH with an early onset of ESKD before the age of 4 years, a minority of 15% retained residual kidney function beyond the age of 20 (Figure 2a). Gene-related differences in mean age, median age, and interquartile age ranges for the onset of ESKD are summarized in Table 1.
Table 1.
Kidney survival of genetically defined NPH cohorts
| Patient characteristics | NPHP1 | NPHP3 | NPHP4 | NPHP11 | Unsolved |
|---|---|---|---|---|---|
| Total number of participants | 116 | 101 | 81 | 85 | 44 |
| Participants with ESKD (%) | 70% | 81% | 100% | 45% | 57% |
| Mean age at onset of ESKD (years/ range) | 12.5 (5.3–27.6) | 7.7 (0–47) | 17.1 (6–54) | 11.9 (0–32) | 11.1 (2.1–18.6) |
| Median age and interquartile range (IQR) for the onset of ESKD (years) | 13.5 (10.5–16.5) | 4.0 (0.25–12.0) | 16.0 (11.0–25.0) | 19.0 (8.7–28.0) | 15.4 (9.7–n.a.) |
ESKD, end-stage kidney disease; IQR, interquartile range; n.a., not assessed; NPH, nephronophthisis.
Impact of Variant Type
We subclassified the genetic variants into 3 different categories as follows: truncating-truncating (trunc/trunc), truncating-missense (trunc/mis) and missense-missense (mis/mis). Truncating variants were defined as predicted loss of function (Supplementary Tables S1 and S2). Because of the nature of the presentation of variants in the NPHP1 group, a different classification was applied to this cohort as follows: biallelic truncating variants including a homozygous deletion (n = 105), and other genetic NPHP1 variants (n = 11).
For NPHP1, NPHP3 and NPHP4, the type of underlying disease-causing variant was significantly associated with kidney survival. In the NPHP3 cohort, the presence of at least 1 truncating variant (trunc/trunc or trunc/mis) led to an early onset of ESKD before the age of 2 years in >50% (Figure 3a). In contrast, the presence of 2 predicted missense variants was associated with a significantly better kidney survival (P < 0.0001). For NPHP4, only the presence of 2 truncating variants was associated with a significantly (P = 0.0021) worse kidney outcome compared with patients carrying at least 1 missense variant (Figure 3b). In individuals carrying biallelic truncating NPHP1 variants including those with a homozygous deletion, a significantly worse kidney survival was observed compared with individuals with at least 1 missense variant (P = 0.03) (Figure 3c). Nevertheless, analyzing only individuals with a homozygous deletion (n = 97) and comparing them with the rest of the cohort (n = 19), no significant difference in kidney survival was observed (P = 0.20; Supplementary Figure S1). In addition, no association between variant type and kidney survival was apparent for NPHP11/TMEM67 (Figure 3d).
Figure 3.
Variant-related kidney survival. Genetic variants for NPHP3, NPHP4, and NPHP11/TMEM67 were subclassified into truncating/truncating, truncating/missense or missense/missense, defined by the presence of either 2 loss of function, 1 loss of function and 1 missense mutation or 2 missense mutations. The NPHP1 group was divided into biallelic truncating including a homozygous deletion and others. NPHP3 (n = 98) (a); NPHP4 (n = 81) (b); NPHP1 (n = 116) (c); NPHP11 (n = 85) (d).
Impact of Nongenetic Factors
A cross-sectional multivariate analysis was applied to the NPHP1 cohort, representing the only group providing sufficient information on the clinical variables used for analysis (Figure 4). In total, 116 patients with NPHP1 (54 females) with a median age of 10.5 years (0.6–31.5 years) at study onset were included, comprising a phenotypic spectrum of isolated kidney disease in 101 individuals, whereas the remaining 15 children displayed characteristic features for Senior-Løken syndrome (n = 7), Joubert syndrome (n = 3), congenital oculomotor apraxia type Cogan II (n = 4), or Bardet-Biedl syndrome (n = 1). Details of the extrarenal phenotype are outlined in Supplementary Table S4. Arterial hypertension was reported in 62 individuals (52%), accompanied by significant proteinuria in 26%. Either of both conditions led to ACEI treatment in 27 individuals. Patients were characterized ‘hypertensive’ based on the start of antihypertensive treatment (Table 2).
Figure 4.
Cross-sectional analysis of NPHP1 patients identifying clinical factors. Multivariate cross-sectional analysis identifying clinical factors for an early onset of ESKD in patients with NPHP1 gene variations (a). Univariate cross-sectional analysis of NPHP1 patients displaying the differences in annual deltaGFR (e.g., eGFR slope = deltaGFR male - deltaGFR female) for multiple clinical characteristics: ACEI treatment and most strikingly arterial hypertension were associated with an accelerated GFR decline; however, in a multivariate approach adjusted for each characteristic, only the influence of arterial hypertension remained significant (b). ACEI, angiotensin-converting enzyme inhibitor; eGFR, estimated glomerular filtration rate; ESKD, end-stage kidney disease; HR, hazard ratio.
Table 2.
Clinical characteristics of NPHP1 patients at the time point when classified “hypertensive”
| Characteristics | Patients |
|---|---|
| Sex | |
| Female | 27 |
| Male | 35 |
| Age at diagnosis (yrs) | 11.2 (2.4–33.4) |
| CKD stage | |
| CKD I | 2 |
| CKD II | 1 |
| CKD III | 16 |
| CKD IV | 17 |
| CKD V | 26 |
| Average time from diagnosis hypertension to ESKD (yrs) | 1.52 (0–7.1) |
| Pre-RRT antihypertensive treatment | |
| No | 14 |
| Mono | 32 |
| Dual | 8 |
| Triple | 8 |
| Calcium antagonist | 23 |
| Beta blocker | 18 |
| ACEIs | 27 |
| Other | 4 |
ACEI, angiotensin-converting enzyme inhibitor; CKD, chronic kidney disease; EKSD, end-stage kidney disease; RRT, renal replacement therapy.
Arterial hypertension was either defined by the start of antihypertensive treatment before the onset of ESKD (n = 48) or simultaneously with the start of RRT.
Cross-sectional multivariate analysis revealed growth retardation (hazard ratio 3.52; CI 1.67–7.41) and ACEI treatment (hazard ratio 2.80; CI 1.13–6.91) as 2 significant (P < 0.05) and independent factors correlated with an earlier onset of ESKD (Figure 4a).
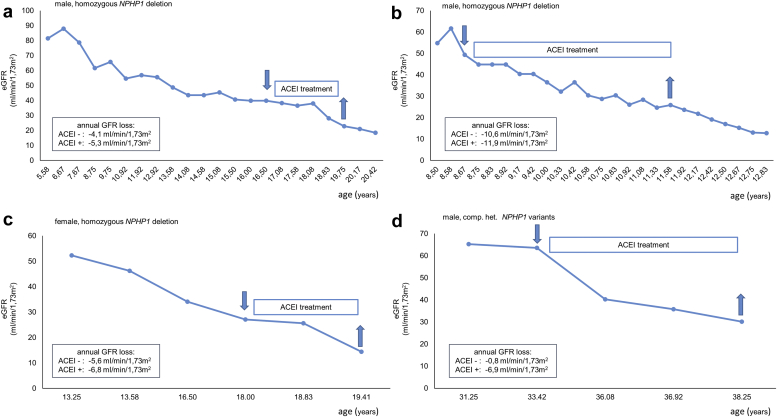
Furthermore, univariate analysis on the GFR trajectory revealed arterial hypertension and ACEI treatment to be significantly (P < 0.01) associated with an accelerated annual GFR decline (Figure 4b and Supplementary Table S5). Based on 47 deltaGFR values from 23 individuals treated with ACEIs against 125 values from 62 individuals without treatment, we observed a difference in annual GFR loss of 7.2 ± 0.4 versus 5.2±0.6 ml/min per 1.73 m2 BSA. Patients with and without ACEI treatment were comparable regarding sex, age, and CKD stage (Table 3). In single cases with only temporary ACEI treatment, intraindividual GFR decline was higher under the influence of ACEIs compared with no treatment (Figure 5). However, in a subsequent multivariate approach adjusted for each individual factor, only the impact of arterial hypertension remained significant with an annual GFR loss of 7.0 ± 0.5 versus 4.6 ± 0.4 ml/min per 1.73 m2 BSA (P < 0.01).
Table 3.
Clinical characteristics of 85 NPHP1 patients with subsequent eGFR values on and off ACEI treatment.
| Patient characteristics | ACEI− | ACEI+ |
|---|---|---|
| Number of patients (n) | 62 | 23 |
| DeltaGFR values (n) | 125 | 47 |
| Male/female | 33/29 | 12/11 |
| Age at study entry (yrs) | 10.2 ± 3.7 (2.4–20.8) | 13.9 ± 7.9 (4.4–33.4) |
| CKD stage, n (%) | ||
| CKD1 | 3 (4) | 2 (9) |
| CKD2 | 7 (10) | 1 (4) |
| CKD3 | 26 (38) | 11 (48) |
| CKD4 | 23 (34) | 10 (43) |
| CKD5 | 9 (13) | 3 (13) |
| Absolute GFR loss/yr (ml/min per 1.73 m2cBSA) | 5.2 ± 0.6 | 7.2 ± 0.4 |
ACEI, angiotensin-converting enzyme inhibitor; BSA, body surface area; CKD, chronic kidney disease; GFR, glomerular filtration rate.
DeltaGFR values were determined for individuals with at least 2 subsequent eGFR values in intervals of minimum 3 months. This way we were able to generate 125 deltaGFR values from 62 patients without and 47 deltaGFR values from 23 patients with the influence of ACEI treatment.
Figure 5.
Representative eGFR trajectories from 4 patients with biallelic NPHP1 variants and temporary ACEI treatment. In all 4 cases, eGFR decline was more pronounced under the influence of ACEIs compared with no treatment-irrespective of CKD stage and age. However, no statistical significance was reached. ACEI, angiotensin-converting enzyme inhibitor; CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate.
Discussion
Since the identification of a large homozygous 290 kb deletion covering the entire NPHP1 gene in the 1990s,7 tremendous progress has been made in the molecular understanding of NPH and related ciliopathies.1,37, 38, 39, 40, 41 However, the focus of most scientific efforts has been on the discovery of the genetic background and molecular pathways.5,7,9,42 Consequently, a lack of clear-cut genotype-phenotype correlations and prognostic factors is preventing reliable prognosis of kidney survival, and individual counseling. Herein, we present one of the largest studies addressing kidney survival in genetically characterized NPH cohorts.
In 1997, Hildebrandt et al.13 established creatinine based, age–dependent quartiles for the NPHP1-related progression of chronic kidney failure. The survey was based on a large number of 308 serial serum creatinine values originating from 19 individuals only. The present study covered a significantly higher number of patients including 97 individuals with a homozygous NPHP1 deletion. Still, results were consistent with those of the historical study. Median age at ESKD was 13.5 years versus 13.1 years in the historical cohort and 12.9 years in a recent survey of 20 Chinese children.13,43 Quartiles of 25% and 75% reaching ESKD were similar to 10.5 years and 16.5 years compared with 11.3 years and 17.3 years.13 In a series of 20 Egyptian children with NPHP1 variants including 5 with a homozygous deletion, Soliman et al.44 reported a significantly earlier onset of ESKD (7.9 years). Surprisingly, in that survey, children without NPHP1 deletion showed a worse kidney outcome compared with those with a homozygous deletion whereas in our study it was the other way around (P = 0.03). Eight NPHP1 patients (5 with a homozygous deletion) retained residual kidney function beyond the age of 20 years. This is in line with previous case reports and the recent awareness of NPHP1 variants being responsible for a remarkable number of adult-onset ESKD.12,13,16, 17, 18
Biallelic variants in NPHP3 were originally described in a Venezuelan pedigree presenting with late onset ESKD.15,19 Recent reports identified NPHP3 variants as one of the most relevant genetic causes for infantile NPH.9,20,45,46 In a worldwide cohort of 1056 patients displaying a NPH phenotype, 17 were identified with either a homozygous or compound heterozygous variant in NPHP3, of whom 9 presented an onset of ESKD before the age of 2 and only 3 individuals reached ESKD beyond 5 years of age.9 Similarly, ESKD in our NPHP3 cohort (n = 15) occurred at a median age of 3.3 years. No association between the NPHP3 genotype and the age at ESKD has been revealed so far.15,42 In this study, we found a significant association between the time of ESKD and variant type. Individuals carrying 2 missense variants presented a significantly better kidney survival compared with those with at least 1 truncating variant. However, it needs to be mentioned that 24 of 27 patients with 2 missense variants belonged to the same Venezuelan pedigree.19
For NPHP4, there was significant phenotypic overlap with the NPHP1 cohort, particularly when presenting as isolated kidney disease and reaching ESKD in the early teens. However, the overall kidney survival was much better as compared with other NPHP groups. Even the ‘trunc/trunc’ scenario was characterized by juvenile NPH mimicking the situation of most NPHP1 patients. Comparable survival rates were observed among large international NPH cohorts comprising 45 patients with NPHP4 variants.42,47 In contrast, Halbritter et al.9 described 22 NPHP4 patients with 19 experiencing rather early ESKD (mean 12.5 years). None of the studies revealed a correlation between variant type and the onset of ESKD. We observed an earlier onset of ESKD in individuals with 2 truncating NPHP4 alleles. Supporting this observation, a case series on a consanguineous Filipino family carrying 2 NPHP4 missense variants reported a consistently late onset kidney failure (median age 36.2 years).48
NPHP11/TMEM67 related kidney survival was characterized by an overall lower percentage of ESKD accounting for 35% of NEOCYST patients and 45% when including published cases. This is in line with a large genotype-phenotype study including 22 individuals with Joubert syndrome caused by biallelic NPHP11/TMEM67 variants, of whom only 50% showed kidney involvement.49 Though the overall onset of ESKD in our cohort was 11.9 years, 2 groups could be distinguished, namely those with severe and early kidney involvement, and those with mild or no obvious kidney phenotype. This reflects the wide phenotypic spectrum covered by NPHP11/TMEM67 variants comprising isolated liver disease, Joubert syndrome with and without kidney involvement, COACH syndrome, RHYNS syndrome, and lethal Meckel-Gruber syndrome.50, 51, 52, 53 Previous genotype-phenotype correlation studies identified the association of variant and phenotypic severity, observing mostly truncating variants or missense variants clustering within exons 8 to 15 to be causative for lethal Meckel-Gruber syndrome phenotypes.54 Nevertheless, this did not apply to kidney survival in the current study population. The presence of disease-causing variants in exons 8 to 15 was not associated with age at ESKD. In patients with such variants in 1 allele, onset of ESKD was 11.8 years (n = 19) compared with 11.3 years in patients without a disease-causing variant in exons 8 to 15 (n = 18). Biallelic exon 8 to 15 variants (n = 4) were associated with ESKD at a median age of 9.2 years (n = 3) (Supplementary Table S6). This observation underpins the idea of the organ-specific impact of distinct genotypes, which is an important point to keep in mind for the clinical management of affected patients.
So far, neither molecular markers nor clinical factors associated with kidney prognosis have been unraveled for NPH. In this study, applying an in-depth phenotyping registry-based approach, we were able to identify 2 independent factors associated with an earlier onset of ESKD in NPHP1 patients, namely growth retardation and ACEI treatment. Whereas growth retardation was observed in 34% consistent with previous data (11%–40%),47,55 information on the effects of ACEI treatment in the setting of NPH is extremely sparse. The association of ACEIs with a negative effect on kidney survival observed in our study appeared peculiar at first glance considering the nephroprotective benefits of ACEI treatment in the setting of chronic kidney disease.56,57 However, a detailed look at the original pediatric data reveals that these benefits were mainly limited to glomerulopathies and congenital anomalies of the kidney and urinary tract disorders, but did not apply to cystic kidney diseases.58,59 Regarding individuals with autosomal dominant polycystic kidney disease, clear benefits of ACEI treatment have been described in the literature. Yet, many of these benefits rather applied to blood pressure control and cardiac outcome whereas evidence for a direct benefit to the decline of kidney function remains sparse.60 In fact, there is only 1 prospective observational study directly showing a slower GFR decline in autosomal dominant polycystic kidney disease patients treated with ACEIs compared with a matched group treated with diuretics.61 Further evidence is either based on indirect conclusions drawn from an epidemiologic study or on the presence of increased plasma renin activity levels.62,63 In contrast, there have also been studies on autosomal dominant polycystic kidney disease cohorts that failed to show beneficial effects of ACEI treatment on the decline of kidney function.64 The limited amount of data included in our study (n = 27) does not allow us to draw final conclusions at this stage. Yet, the univariate analysis of individual GFR trajectories revealed that the use of ACEIs was also significantly associated with a faster GFR decline (Supplementary Table S5 and Figure 5). Further investigation beyond the scope of this study is necessary, including more data and prospective studies to confirm the current impression because it would have a major impact on clinical practice recommendations.
In addition to ACEI treatment, both in a univariate and multivariate analysis revealed a significant negative impact of arterial hypertension on the annual GFR decline. This appears contradictory at first because both arterial hypertension and antihypertensive ACEI treatment had the same negative effect on the GFR trajectory. A potential explanation for this phenomenon might be the content overlap expressed by 27 of 62 hypertensive individuals receiving ACEI treatment. It is noteworthy that after adjusting for each individual factor in the multivariate analysis, only the impact of hypertension remained significant. The high number of hypertensive patients in our cohort appeared striking considering the absence of elevated blood pressure levels being referred to as a typical feature of NPH.5,11,12,65 In fact, different cohort studies reported predialysis arterial hypertension in up to 30%, exceeded by the ratio in our cohort of 62:116.47,55 Against this background it must be mentioned that, in most cases arterial hypertension was closely related to ESKD reflected by an average interval between the diagnosis of hypertension and the onset of ESKD of 1.5 years (Table 2). This again raises the question of whether the arterial hypertension is a consequence of advanced chronic kidney disease rather than an independent risk factor for the early onset of ESKD. Supporting this assumption, predialysis antihypertensive treatment was started in 48 cases only. The remaining 14 individuals were diagnosed hypertensive concomitant with ESKD at their first clinical presentation (Table 2). Neither concomitant extrarenal organ involvement nor the sonographic detection of kidney cysts did have an impact on kidney prognosis.
The current study was limited by the fact that different genetic methodologies were included. Unfortunately, the registry-based approach did not allow a universal use of state-of-the-art genetic analysis because many of the patients were no longer in pediatric care and there was no access to current blood samples. In addition, the classification of genetic missense variants as ‘predicted mild’ without proof of functional data may be vulnerable. However, we tried to address these concerns as best as possible by the application of the American College of Medical Genetics and Genomics criteria on all variants to (re)assess the validity of all included and partly formerly published variants (Supplementary Table S3).
In this study, we provide evidence that NPH-related kidney survival is not only determined by the underlying affected gene but may also varies depending on variant type. Growth retardation and the use of ACEIs were identified as independent factors associated with an earlier onset of ESKD in the setting of an underlying NPHP1 genotype. Furthermore, arterial hypertension was linked to an accelerated GFR decline regardless of the kind of antihypertensive treatment. The presented data enables clinicians to better estimate individual kidney prognosis and thereby facilitate personalized counseling.
Disclosure
All the authors declared no competing interests.
Acknowledgments
The authors contributing their patients: Ulrike Walden, Katharina Hohenfellner, Ludwig Patzer, Henry Fehrenbach, Hagen Staude, Martin Koemhoff and Heiko Billing. NEOCYST (Network of Early Onset Cystic Kidney Diseases; www.neocyst.de)27 is funded by the German Federal Ministry of Education and Research—grant code 01GM1515A. This study was also supported by the European Reference Network for Rare Kidney Diseases, which is partly co-funded by the European Union within the framework of the Third Health Program “ERN-2016-Framework Partnership Agreement 2017-2021.”
Author Contributions
JCK and MK designed the study; JCK, RK, KPS, MD-H, SK and AKT collected data; CB performed most of the genetic analysis; JCK, JG, RK, KPS, MDH, AKT and MK analyzed data, JG carried out statistical analysis, JCK and MDH made the figures and drafted the manuscript; all authors contributed their patients, gave valuable intellectual input and critically revised and approved the final version of the manuscript.
Footnotes
Members of NEOCYST consortium are listed in the Appendix.
Figure S1. Kidney survival of individuals with a homozygous NPHP1 deletion compared with other NPHP1 genotypes.
Table S1. Patient baseline characteristics at study entry (NEOCYST registry cohort and ERKNet survey cohort). Supplementary File (Excel)
Table S2. Genotype, variant type and data source of all individuals carrying biallelic variants in NPHP1, NPHP3, NPHP4 or NPHP11/TMEM67 included in the study.
Table S3. Characterization of disease-causing variants of all included 383 patients following ACMG criteria.
Table S4. Clinical characterization of 15 individuals with a homozygous NPHP1 deletion presenting an extrarenal phenotype.
Table S5. Univariate analysis of patient characteristics on the annual eGFR slope.
Table S6. Impact of the genetic involvement of exon 8-15 in NPHP11/TMEM67 on the onset of ESKD.
Contributor Information
Jens Christian König, Email: jens.koenig@ukmuenster.de.
NEOCYST consortium:
P. Antczak, J. Birtel, C. Bergmann, M. Cetiner, M. Dahmer-Heath, J. Drube, J. Gerß, D. Haffner, T. Illig, I. Kamp-Becker, N. Klopp, S. Kollmann, J. König, M. Konrad, M.C. Liebau, C. Nittel, C. Okorn, H. Omran, L. Pape, P. Pennekamp, F. Schäfer, B. Schermer, H. Storf, J. Vasseur, S. Weber, K. Wohlgemuth, W. Ziegler, C. Gimpel, J. Göbel, and B. Schlevogt
Appendix
List of NEOCYST Consortium
NEOCYST members: P. Antczak, J. Birtel, C. Bergmann, M. Cetiner, M. Dahmer-Heath, J. Drube, J. Gerß,D. Haffner, T. Illig, I. Kamp-Becker, N. Klopp, S. Kollmann, J. König, M. Konrad, MC. Liebau, C. Nittel, C. Okorn, H. Omran, L. Pape, P. Pennekamp, F. Schäfer, B. Schermer, H. Storf, J. Vasseur, S. Weber, K. Wohlgemuth, W. Ziegler, C. Gimpel, J. Göbel, B. Schlevogt
Supplementary Material
Figure S1. Kidney survival of individuals with a homozygous NPHP1 deletion compared with other NPHP1 genotypes.
Table S1. Patient baseline characteristics at study entry (NEOCYST registry cohort and ERKNet survey cohort).
Table S2. Genotype, variant type and data source of all individuals carrying biallelic variants in NPHP1, NPHP3, NPHP4 or NPHP11/TMEM67 included in the study.
Table S3. Characterization of disease-causing variants of all included 383 patients following ACMG criteria.
Table S4. Clinical characterization of 15 individuals with a homozygous NPHP1 deletion presenting an extrarenal phenotype.
Table S5. Univariate analysis of patient characteristics on the annual eGFR slope.
Table S6. Impact of the genetic involvement of exon 8-15 in NPHP11/TMEM67 on the onset of ESKD. Supplementary File (Excel)
References
- 1.Stokman M.F., Saunier S., Benmerah A. Renal ciliopathies: sorting out therapeutic approaches for nephronophthisis. Front Cell Dev Biol. 2021;9:653138. doi: 10.3389/fcell.2021.653138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harambat J., van Stralen K.J., Kim J.J., Tizard E.J. Epidemiology of chronic kidney disease in children. Pediatr Nephrol. 2012;27:363–373. doi: 10.1007/s00467-011-1939-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.König J, Ermisch-Omran B, Omran H. Nephronophthisis and autosomal dominant interstitial kidney disease (ADIKD). Pediatric Kidney Disease. In: Geary, D., Schaefer, F. (eds) Pediatric Kidney Disease. Springer; 2021:369-388.
- 4.Titieni A., König J. Nephronophthise und assoziierte Ziliopathien. med genet. 2018;30:461–468. doi: 10.1007/s11825-018-0213-3. [DOI] [Google Scholar]
- 5.Luo F., Tao Y.H. Nephronophthisis: a review of genotype-phenotype correlation. Nephrology (Carlton) 2018;23:904–911. doi: 10.1111/nep.13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stokman M., Lilien M., Knoers N. In: GeneReviews. Adam M.P., Mirzaa G.M., Pagon R.A., et al., editors. University of Washington; Seattle: 2016. Nephronophthisis. [PubMed] [Google Scholar]
- 7.Adamiok-Ostrowska A., Piekiełko-Witkowska A. Ciliary genes in renal cystic diseases. Cells. 2020;9:907. doi: 10.3390/cells9040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konrad M., Saunier S., Heidet L., et al. Large homozygous deletions of the 2q13 region are a major cause of juvenile nephronophthisis. Hum Mol Genet. 1996;5:367–371. doi: 10.1093/hmg/5.3.367. [DOI] [PubMed] [Google Scholar]
- 9.Halbritter J., Porath J.D., Diaz K.A., et al. Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum Genet. 2013;132:865–884. doi: 10.1007/s00439-013-1297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konig J., Kranz B., Konig S., et al. Phenotypic spectrum of children with nephronophthisis and related ciliopathies. Clin J Am Soc Nephrol. 2017;12:1974–1983. doi: 10.2215/CJN.01280217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf M.T. Nephronophthisis and related syndromes. Curr Opin Pediatr. 2015;27:201–211. doi: 10.1097/MOP.0000000000000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salomon R., Saunier S., Niaudet P. Nephronophthisis. Pediatr Nephrol. 2009;24:2333–2344. doi: 10.1007/s00467-008-0840-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hildebrandt F., Strahm B., Nothwang H.G., et al. Molecular genetic identification of families with juvenile nephronophthisis type 1: rate of progression to renal failure. APN Study Group. Arbeitsgemeinschaft für Pädiatrische Nephrologie. Kidney Int. 1997;51:261–269. doi: 10.1038/ki.1997.31. [DOI] [PubMed] [Google Scholar]
- 14.Gagnadoux M.F., Bacri J.L., Broyer M., Habib R. Infantile chronic tubulo-interstitial nephritis with cortical microcysts: variant of nephronophthisis or new disease entity? Pediatr Nephrol. 1989;3:50–55. doi: 10.1007/BF00859626. [DOI] [PubMed] [Google Scholar]
- 15.Omran H., Fernandez C., Jung M., et al. Identification of a new gene locus for adolescent nephronophthisis, on chromosome 3q22 in a large Venezuelan pedigree. Am J Hum Genet. 2000;66:118–127. doi: 10.1086/302705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergmann C. Educational paper: ciliopathies. Eur J Pediatr. 2012;171:1285–1300. doi: 10.1007/s00431-011-1553-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snoek R., van Setten J., Keating B.J., et al. NPHP1 (Nephrocystin-1) gene deletions cause adult-onset ESRD. J Am Soc Nephrol. 2018;29:1772–1779. doi: 10.1681/ASN.2017111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimaru T., Kawanishi K., Mori T., et al. Genetic background and clinicopathologic features of adult-onset nephronophthisis. Kidney Int Rep. 2021;6:1346–1354. doi: 10.1016/j.ekir.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olbrich H., Fliegauf M., Hoefele J., et al. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat Genet. 2003;34:455–459. doi: 10.1038/ng1216. [DOI] [PubMed] [Google Scholar]
- 20.Sun L., Tong H., Wang H., et al. High mutation rate of NPHP3 in 18 Chinese infantile nephronophthisis patients. Nephrology (Carlton) 2016;21:209–216. doi: 10.1111/nep.12563. [DOI] [PubMed] [Google Scholar]
- 21.Molinari E., Decker E., Mabillard H., et al. Human urine-derived renal epithelial cells provide insights into kidney-specific alternate splicing variants. Eur J Hum Genet. 2018;26:1791–1796. doi: 10.1038/s41431-018-0212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otto E.A., Loeys B., Khanna H., et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 23.Parisi M.A. The molecular genetics of Joubert syndrome and related ciliopathies: the challenges of genetic and phenotypic heterogeneity. Transl Sci Rare Dis. 2019;4:25–49. doi: 10.3233/TRD-190041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braun D.A., Schueler M., Halbritter J., et al. Whole exome sequencing identifies causative mutations in the majority of consanguineous or familial cases with childhood-onset increased renal echogenicity. Kidney Int. 2016;89:468–475. doi: 10.1038/ki.2015.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raina R., Chakraborty R., Sethi S.K., et al. Diagnosis and management of renal cystic disease of the newborn: core curriculum 2021. Am J Kidney Dis. 2021;78:125–141. doi: 10.1053/j.ajkd.2020.10.021. [DOI] [PubMed] [Google Scholar]
- 26.Larrue R., Chamley P., Bardyn T., et al. Diagnostic utility of whole-genome sequencing for nephronophthisis. npj Genom Med. 2020;5:38. doi: 10.1038/s41525-020-00147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konig J.C., Titieni A., Konrad M., NEOCYST Consortium Network for Early Onset Cystic Kidney Diseases-a comprehensive multidisciplinary approach to hereditary cystic kidney diseases in childhood. Front Pediatr. 2018;6:24. doi: 10.3389/fped.2018.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoff S., Halbritter J., Epting D., et al. ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat Genet. 2013;45:951–956. doi: 10.1038/ng.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou W., Otto E.A., Cluckey A., et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44:910–915. doi: 10.1038/ng.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pottel H., Hoste L., Dubourg L., et al. An estimated glomerular filtration rate equation for the full age spectrum. Nephrol Dial Transplant. 2016;31:798–806. doi: 10.1093/ndt/gfv454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopanos C., Tsiolkas V., Kouris A., et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adzhubei I.A., Schmidt S., Peshkin L., et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaser R., Adusumalli S., Leng S.N., et al. SIFT missense predictions for genomes. Nat Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 35.Choi Y., Sims G.E., Murphy S., et al. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landrum M.J., Lee J.M., Benson M., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santoni M., Piva F., Cimadamore A., et al. Exploring the spectrum of kidney ciliopathies. Diagnostics (Basel, Switzerland) 2020;10 doi: 10.3390/diagnostics10121099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gherman A., Davis E.E., Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- 39.Arnaiz O., Malinowska A., Klotz C., et al. Cildb: a KnowledgeBase for centrosomes and cilia. Database J Biol Databases Curation. 2009;2009:bap022. doi: 10.1093/database/bap022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun D.A., Hildebrandt F., Hildebrandt F. Ciliopathies. Cold Spring Harb Perspect Biol. 2017;9:a028191. doi: 10.1101/cshperspect.a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devlin L.A., Sayer J.A. Renal ciliopathies. Curr Opin Genet Dev. 2019;56:49–60. doi: 10.1016/j.gde.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 42.Chaki M., Hoefele J., Allen S.J., et al. Genotype-phenotype correlation in 440 patients with NPHP-related ciliopathies. Kidney Int. 2011;80:1239–1245. doi: 10.1038/ki.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang X., Liu C., Liu X., et al. Phenotype and genotype spectra of a Chinese cohort with nephronophthisis-related ciliopathy. J Med Genet. 2022;59:147–154. doi: 10.1136/jmedgenet-2020-107184. [DOI] [PubMed] [Google Scholar]
- 44.Soliman N.A., Hildebrandt F., Otto E.A., et al. Clinical characterization and NPHP1 mutations in nephronophthisis and associated ciliopathies: a single center experience. Saudi J Kidney Dis Transplant. 2012;23:1090–1098. doi: 10.4103/1319-2442.100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergmann C., Fliegauf M., Brüchle N.O., et al. Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am J Hum Genet. 2008;82:959–970. doi: 10.1016/j.ajhg.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tory K., Rousset-Rouvière C., Gubler M.C., et al. Mutations of NPHP2 and NPHP3 in infantile nephronophthisis. Kidney Int. 2009;75:839–847. doi: 10.1038/ki.2008.662. [DOI] [PubMed] [Google Scholar]
- 47.Stokman M.F., van der Zwaag B., van de Kar N., et al. Clinical and genetic analyses of a Dutch cohort of 40 patients with a nephronophthisis-related ciliopathy. Pediatr Nephrol. 2018;33:1701–1712. doi: 10.1007/s00467-018-3958-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mistry K., Ireland J.H., Ng R.C., et al. Novel mutations in NPHP4 in a consanguineous family with histological findings of focal segmental glomerulosclerosis. Am J Kidney Dis. 2007;50:855–864. doi: 10.1053/j.ajkd.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Fleming L.R., Doherty D.A., Parisi M.A., et al. Prospective evaluation of kidney disease in Joubert syndrome. Clin J Am Soc Nephrol. 2017;12:1962–1973. doi: 10.2215/CJN.05660517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brancati F., Camerota L., Colao E., et al. Biallelic variants in the ciliary gene TMEM67 cause RHYNS syndrome. Eur J Hum Genet. 2018;26:1266–1271. doi: 10.1038/s41431-018-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogel I., Ott P., Lildballe D., et al. Isolated congenital hepatic fibrosis associated with TMEM67 mutations: report of a new genotype-phenotype relationship. Clin Case Rep. 2017;5:1098–1102. doi: 10.1002/ccr3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otto E.A., Tory K., Attanasio M., et al. Hypomorphic mutations in meckelin (MKS3/TMEM67) cause nephronophthisis with liver fibrosis (NPHP11) J Med Genet. 2009;46:663–670. doi: 10.1136/jmg.2009.066613. [DOI] [PubMed] [Google Scholar]
- 53.Smith U.M., Consugar M., Tee L.J., et al. The transmembrane protein meckelin (MKS3) is mutated in Meckel-Gruber syndrome and the wpk rat. Nat Genet. 2006;38:191–196. doi: 10.1038/ng1713. [DOI] [PubMed] [Google Scholar]
- 54.Iannicelli M., Brancati F., Mougou-Zerelli S., et al. Novel TMEM67 mutations and genotype-phenotype correlates in meckelin-related ciliopathies. Hum Mutat. 2010;31:E1319–E1331. doi: 10.1002/humu.21239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cantani A., Bamonte G., Ceccoli D., et al. Familial juvenile nephronophthisis. A review and differential diagnosis. Clin Pediatr. 1986;25:90–95. doi: 10.1177/000992288602500206. [DOI] [PubMed] [Google Scholar]
- 56.Hou F.F., Zhang X., Zhang G.H., et al. Efficacy and safety of benazepril for advanced chronic renal insufficiency. N Engl J Med. 2006;354:131–140. doi: 10.1056/NEJMoa053107. [DOI] [PubMed] [Google Scholar]
- 57.Brenner B.M., Cooper M.E., de Zeeuw D., et al. Effects of losartan on renal and cardiovascular outcomes in patients with type 2 diabetes and nephropathy. N Engl J Med. 2001;345:861–869. doi: 10.1056/NEJMoa011161. [DOI] [PubMed] [Google Scholar]
- 58.van den Belt S.M., Heerspink H.J.L., Kirchner M., et al. Discontinuation of RAAS inhibition in children with advanced CKD. Clin J Am Soc Nephrol. 2020;15:625–632. doi: 10.2215/CJN.09750819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wühl E., Trivelli A., Picca S., et al. Strict blood-pressure control and progression of renal failure in children. N Engl J Med. 2009;361:1639–1650. doi: 10.1056/NEJMoa0902066. [DOI] [PubMed] [Google Scholar]
- 60.Schrier R.W. Renal volume, renin-angiotensin-aldosterone system, hypertension, and left ventricular hypertrophy in patients with autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2009;20:1888–1893. doi: 10.1681/ASN.2008080882. [DOI] [PubMed] [Google Scholar]
- 61.Ecder T., Edelstein C.L., Fick-Brosnahan G.M., et al. Diuretics versus angiotensin-converting enzyme inhibitors in autosomal dominant polycystic kidney disease. Am J Nephrol. 2001;21:98–103. doi: 10.1159/000046231. [DOI] [PubMed] [Google Scholar]
- 62.Schrier R.W. ACE inhibitors, left ventricular mass and renal cyst growth in ADPKD. Pharmacol Res. 2016;114:166–168. doi: 10.1016/j.phrs.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 63.Chapman A.B., Johnson A., Gabow P.A., Schrier R.W. The renin-angiotensin-aldosterone system and autosomal dominant polycystic kidney disease. N Engl J Med. 1990;323:1091–1096. doi: 10.1056/NEJM199010183231602. [DOI] [PubMed] [Google Scholar]
- 64.Maschio G., Alberti D., Janin G., et al. Effect of the angiotensin-converting-enzyme inhibitor benazepril on the progression of chronic renal insufficiency. The angiotensin-converting-enzyme Inhibition in Progressive Renal Insufficiency Study Group. N Engl J Med. 1996;334:939–945. doi: 10.1056/NEJM199604113341502. [DOI] [PubMed] [Google Scholar]
- 65.Hildebrandt F., Waldherr R., Kutt R., Brandis M. The nephronophthisis complex: clinical and genetic aspects. Clin Investig. 1992;70:802–808. doi: 10.1007/BF00180751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.