Abstract
Here we investigated ribosomal pausing at sites of programmed −1 ribosomal frameshifting, using translational elongation and ribosome heelprint assays. The site of pausing at the frameshift signal of infectious bronchitis virus (IBV) was determined and was consistent with an RNA pseudoknot-induced pause that placed the ribosomal P- and A-sites over the slippery sequence. Similarly, pausing at the simian retrovirus 1 gag/pol signal, which contains a different kind of frameshifter pseudoknot, also placed the ribosome over the slippery sequence, supporting a role for pausing in frameshifting. However, a simple correlation between pausing and frameshifting was lacking. Firstly, a stem-loop structure closely related to the IBV pseudoknot, although unable to stimulate efficient frameshifting, paused ribosomes to a similar extent and at the same place on the mRNA as a parental pseudoknot. Secondly, an identical pausing pattern was induced by two pseudoknots differing only by a single loop 2 nucleotide yet with different functionalities in frameshifting. The final observation arose from an assessment of the impact of reading phase on pausing. Given that ribosomes advance in triplet fashion, we tested whether the reading frame in which ribosomes encounter an RNA structure (the reading phase) would influence pausing. We found that the reading phase did influence pausing but unexpectedly, the mRNA with the pseudoknot in the phase which gave the least pausing was found to promote frameshifting more efficiently than the other variants. Overall, these experiments support the view that pausing alone is insufficient to mediate frameshifting and additional events are required. The phase dependence of pausing may be indicative of an activity in the ribosome that requires an optimal contact with mRNA secondary structures for efficient unwinding.
A growing number of examples have been described in which the rules for decoding of mRNAs are temporarily altered through the action of specific signals built into the mRNA sequences. Indeed, a minority of genes in probably all organisms rely on such “recoding” for translation of their mRNAs (18). Examples include bypassing, where ribosomes translate over coding gaps in the mRNA; alteration of meaning, where specific termination codons can be read as selenocysteine, tryptophan, or glutamine codons; and ribosomal frameshifting, where the ribosome enters the −1 or +1 reading frame to allow expression of a protein from mRNAs with overlapping open reading frames. Although recoding sites are distinct in terms of their differing requirements for primary sequences, mRNA secondary structures, and translational factors, the specific pausing of ribosomes at such sites is thought to play a generalized and essential role in the stimulation of recoding. Pausing has been implicated in −1 and +1 ribosomal frameshifting (8, 11, 26, 36, 44, 45; reviewed in references 16 and 17), readthrough of termination codons (1), and translational bypassing (20) and may also play a role in UGA-directed selenocysteine insertion at the ribosome in vivo (39).
In its simplest form, pausing serves to increase the time at which ribosomes are held at a recoding site, promoting alternative events that would normally be unfavorable kinetically (17). Pausing can be induced in a variety of ways, including encounter of the ribosome with mRNA secondary structures, termination codons, and amino-acyl-tRNA limitation. Although the results of mutational analyses and other genetic studies generally support a role for pausing in recoding events (see references 15, 16, and 17 for reviews), biochemical evidence for the process is scant. The available information comes from studies of −1 ribosomal frameshifting, a process exploited (largely) by RNA viruses to control expression of their replicases (see references 5 and 17 for reviews). The mRNA signals which specify −1 frameshifting are comprised of two essential elements: a heptanucleotide “slippery” sequence, where the ribosome changes reading frame, and a stimulatory region of RNA secondary structure, often in the form of an RNA pseudoknot, located a few nucleotides (nt) downstream (2, 21, 41). Encounter of the secondary structure by the ribosome promotes frameshifting at the slippery sequence. Polypeptide intermediates corresponding to ribosomes paused at stimulatory RNA structures have been detected at the frameshift sites of the coronavirus infectious bronchitis virus (IBV) (36) and the Saccharomyces cerevisiae L-A virus (26), and footprinting studies of elongating ribosomes have defined the site of pausing at the L-A signal (26, 45). There is also evidence for pausing at the frameshift site in the Escherichia coli dnaX gene, again from the analysis of translational intermediates (44).
That pausing occurs at −1 frameshift signals is generally accepted, but we know little about the process or whether it is truly necessary for −1 frameshifting. Here we describe a study of ribosomal pausing at a variety of RNA structures, including the frameshift signals of IBV and simian retrovirus-1 (SRV-1) gag/pro, which contain well-defined RNA pseudoknots (2–4, 25, 31, 42, 43). Pausing was examined using a heelprint assay that permits identification of the 5′ end of paused ribosomes on the mRNA (46) and an elongation assay that allows visualization of polypeptide intermediates (36). We found that the position of ribosomal pausing on the various mRNAs was consistent with a role for pausing in frameshifting, but there was no obvious correlation between the extent of pausing and the efficiency of frameshifting. Furthermore, pausing was sensitive to the reading frame in which the stimulatory RNA structure was encountered; this phase dependence may be indicative of an activity in the ribosome that requires an optimal contact with mRNA secondary structures for efficient unwinding.
MATERIALS AND METHODS
Site-specific mutagenesis.
Site-directed mutagenesis was carried out by using a procedure based on that of Kunkel (23), as described by Brierley and colleagues (2). All of the plasmids employed in this study contain the intergenic region of the filamentous bacteriophage f1 (13) that enables single-stranded plasmid templates for mutagenesis to be generated following infection of plasmid-carrying bacteria with bacteriophage R408 (32). Mutants were identified by dideoxy sequencing of single-stranded templates (34).
Construction of plasmids.
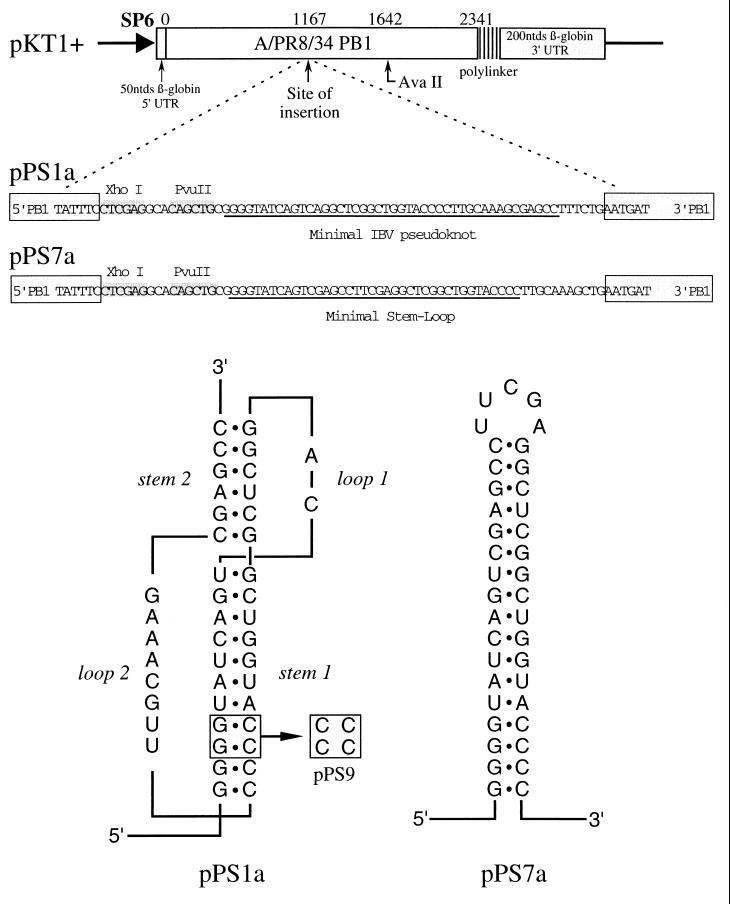
Plasmids pFS7.2, pFS19 (2), pFScass 5, pSM2, pSM3 (4), pPS0, pPS1a, pPS7a (formerly pPS1, pPS7 [36]), pSF1, pSF4 (42), pKA-A, and pKA-G (25) have been described elsewhere. Plasmid pFS7.2/HK was derived from plasmid pFS7.2 by changing a termination codon (UAA) (present some 63 nt downstream of the IBV slippery sequence in this construct) to a lysine codon (AAA) by site-directed mutagenesis. Plasmid pFS19a was derived from plasmid pFS19 by changing the authentic IBV 1a termination codon from UGA to UGG by mutagenesis. Plasmids pPS1b and pPS7b (see Fig. 6) were prepared by linearization of pPS1a and pPS7a (Fig. 1), respectively, with XhoI, end-repair using the Klenow fragment of DNA polymerase I, and religation with T4 DNA ligase. The two sections of the influenza virus A/PR8/34 PB1 gene (47) present upstream and downstream of the inserted pseudoknot (pPS1b) or hairpin (pPS7b) were returned to the same reading frame by the insertion of two bases (shown in bold in the following sequences) downstream of the respective structures at the sequences 5′ GCCTTTGTCTGAAT 3′ (pPS1b) and 5′ TTGCAACGAGCTGA 3′ (pPS7b). Plasmids pPS1c and pPS7c (see Fig. 6) were prepared by digestion of pPS1a and pPS7a, respectively, with XhoI and PvuII, end-repair using the Klenow fragment of DNA polymerase I, and religation with T4 DNA ligase. Here, the integrity of the PB1 gene was restored by insertion of a single base (in bold below) downstream of the respective structures 5′ AGCCTTGTCTGAA 3′ (pPS1c) and 5′ TTGCAACAGCTGA 3′ (pPS7c).
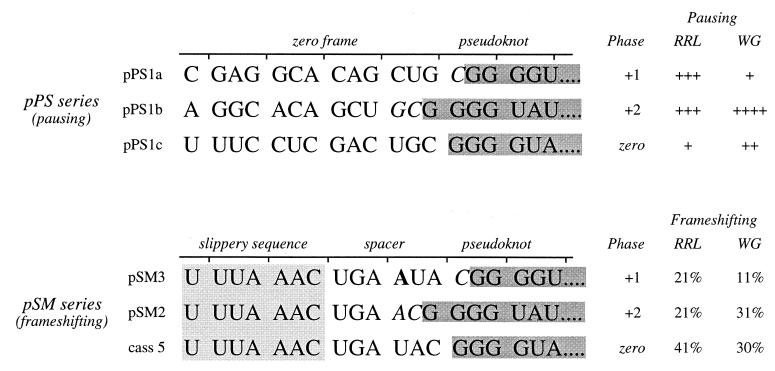
FIG. 6.
Pseudoknot encounter phase in pausing and frameshifting constructs. The nucleotide sequence of the mRNA in the vicinity of the IBV minimal pseudoknot in pausing (pPS series) and frameshifting (pSM series [4]) constructs is shown. In all mRNAs, only the 5′ portion of the pseudoknot is displayed (in grey). The phase is defined by the number of nucleotides between the last in-frame codon and the start of the pseudoknot. In pPS1a, for example, the single nucleotide (C, italicized) present between the reading frame codon CUG and the start of the pseudoknot defines the phase as +1. In the pSM series, the number of nucleotides that separate the IBV slippery sequence (boxed) and the pseudoknot (spacer region) varies. The wild-type spacer is 6 nt (cass 5); in pSM3 an additional A residue (bold) is present, and in pSM2 a U has been deleted (4). Also shown (on right) is a summary of the pausing level and frameshifting efficiencies specified by the constructs. The frameshift efficiencies in RRL were from Brierley and colleagues (4); those in WG were determined here (translations not shown).
FIG. 1.
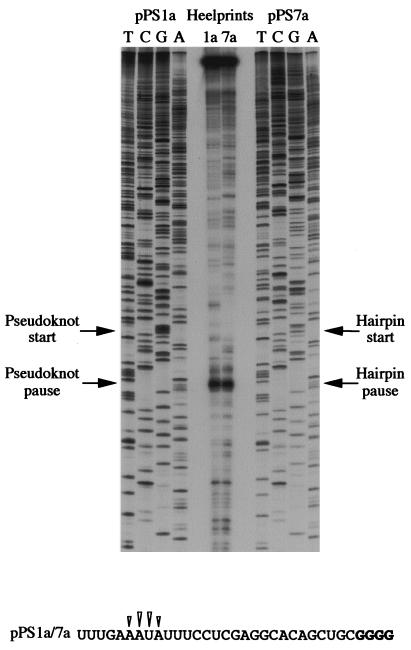
Pausing constructs based on the IBV frameshift signal. Plasmids pPS1a and pPS7a (formerly pPS1 and pPS7 [36]) contain, respectively, the minimal IBV pseudoknot and a related stem-loop structure (3, 4) cloned into the influenza PB1 gene in an SP6 promoter-based transcription vector. Plasmid pPS9 is a derivative of pPS1a in which stem 1 is destabilized by a complementary mutation.
Preparation of mRNAs for in vitro translation.
Plasmids for in vitro transcription were prepared as described previously (2). In vitro transcription reactions employing the bacteriophage SP6 RNA polymerase were carried out essentially as described by Melton et al. (28) and included the synthetic cap structure 7meGpppG (New England Biolabs) to generate capped mRNA. Product RNA was recovered by a single extraction with phenol-chloroform-isoamyl alcohol (49:49:2) followed by ethanol precipitation in the presence of 2 M ammonium acetate. The RNA pellet was dissolved in water, and remaining unincorporated nucleotide triphosphates were removed by Sephadex G-50 chromatography. RNA was recovered by ethanol precipitation, dissolved in water, and checked for integrity by electrophoresis on 1.5% agarose gels containing 0.1% sodium dodecyl sulfate (SDS). Trace-labeled mRNA was prepared as above but included 5 μCi of [32P]UTP in the transcription reaction mixture.
Preparation of RPFs.
Rabbit reticulocyte lysate (RRL) translation reactions were initiated with 100 ng of mRNA in a total reaction volume of 25 μl containing 2.5 U of RNasin (Amersham Pharmacia Biotech). After incubation at 26°C for 15 to 25 min, cycloheximide was added to a 1 mM concentration and the reaction mixture was placed on ice for 3 min. Unprotected mRNA was degraded by incubation with micrococcal nuclease (1 or 2 U/μl; Worthington) and RNase V1 (0.02 U/μl; Pharmacia) at 26°C for 30 min in the presence of 3.5 mM Mg(OAc)2 and 3 mM CaCl2 in a final reaction volume of 40 μl. Following addition of 60 μl of buffer T (20 mM HEPES, 150 mM KOAc, 10 mM Mg(OAc)2, 5 mM EGTA, 2 mM dithiothreitol), the reaction mixture was overlayered onto a 60-μl cushion of 0.25 M sucrose in buffer T and subsequently centrifuged at 30 lb/in2 for 30 min in an A-110 rotor in a Beckman airfuge. Following removal of the sucrose, the 40-μl ribosomal pellet containing the ribosome-protected mRNA fragments (RPFs) was incubated with 100 μl of proteinase K solution (50 mM Tris [pH 7.5], 50 mM NaCl, 5 mM EDTA, 0.5% SDS, and 200 μg of proteinase K/ml) for 30 min at 37°C, the reaction mixture was extracted with phenol-chloroform, and the RPFs were harvested by ethanol precipitation and resuspended in 10 μl of MilliQ water and stored at −70°C.
Primer extension inhibition assay (heelprinting).
Single-stranded templates for primer extension were prepared by R408 superinfection, as described above. The site of annealing of primers for extension reactions was between 60 and 100 nt upstream of the pausing site. Oligonucleotide primers were 5′ end labeled with [γ-32P]ATP according to standard procedures (33). Annealing reaction mixtures (final volume, 9 μl) containing 0.05 to 0.5 ng of labeled primer, RPFs (0.1 to 4 μl), 20 ng of single-stranded circular plasmid DNA (containing sequences complementary to the relevant mRNA), 88 mM KPO4, and 6.7 mM MgCl2 were heated to 65°C for 5 min and cooled slowly to 37°C over a 1-h period. Subsequently, primer extension reactions were performed by addition of 60 U of bacteriophage T7 DNA polymerase in the presence of 0.6 mM concentrations of each deoxynucleoside triphosphate, 10 mM 2-mercaptoethanol, 88 mM KPO4, and 6.7 mM MgCl2 and incubation at 37°C for 15 min. Following synthesis, reaction mixtures were diluted to 100 μl with 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA, extracted with phenol-chloroform, precipitated by ethanol, washed with 70% ethanol, dried, and resuspended in 5 μl of loading buffer (95% formamide, 10 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol). Samples were heated at 65°C for 5 min and examined on 6 or 8% polyacrylamide–7 M urea sequencing-type gels. Dideoxy sequencing reactions (34) primed from the same single-stranded template DNA were run alongside.
Pausing assays employing edeine.
Edeine assays were carried out in RRL or wheat germ (WG) extracts from Promega. Translations were initiated at 26°C (RRL) or 15°C (WG) by addition of mRNA to a final concentration of 10 to 25 μg/ml, and 5 min later the initiation inhibitor edeine was added (to a concentration of 5 μM). Aliquots of 1.5 μl were withdrawn from the translation mixture at specified intervals, mixed with an equal volume of RNase A (100 μg/ml) in 10 mM EDTA (pH 7.5), and incubated at 25°C for 15 min prior to analysis on SDS–10% (wt/vol) polyacrylamide gels. The relative abundances of paused and full-length products on the gels were estimated by direct measurement of [35S]methionine incorporation using a Packard Instant Imager 2024.
RESULTS
Heelprinting analysis of ribosomal pausing at −1 translational frameshift signals.
We previously studied ribosomal pausing at an IBV-derived pseudoknot (the minimal pseudoknot [4]) inserted at a specific location within an influenza virus PB1 reporter mRNA (36). Translation of this mRNA in the RRL in vitro translation system, in comparison to that of PB1 mRNA alone, generated a new translational intermediate whose size corresponded to that expected following a ribosomal pause at the pseudoknot. The appearance of this protein was transient, indicating that it was a true “paused” intermediate rather than a “dead-end” product, and mutational analysis confirmed that its appearance was dependent on the presence of a pseudoknot structure within the mRNA. However, although the assay provided unequivocal evidence for pausing at the pseudoknot, it did not allow the precise site of pausing to be ascertained. For this reason, we began the present study by performing heelprint assays on pseudoknot-containing mRNAs using the methodology of Wolin and Walter (46) as modified by Doohan and Samuel (12). In this procedure (detailed in Materials and Methods), ribosomal pausing during translation preferentially protects certain segments of RNA from RNase digestion following “freezing” of ribosomes on the mRNA with cycloheximide. Ribosomes are subsequently isolated, and the associated protected RNA fragments are purified and annealed to a complementary single-stranded DNA template along with an end-labeled sequencing primer. Following extension of the sequencing primer using T7 DNA polymerase, which terminates upon encountering an annealed RNA fragment, the site of termination is mapped by running out the primer extension products on a denaturing polyacrylamide gel alongside sequencing ladders prepared using the same sequencing primer. Since paused ribosomes produce an increased amount of specific RPFs, the T7 DNA polymerase extension products, corresponding to the trailing 5′ edges of the stalled ribosomes from which these RPFs were obtained, appear as more intense species on the gel.
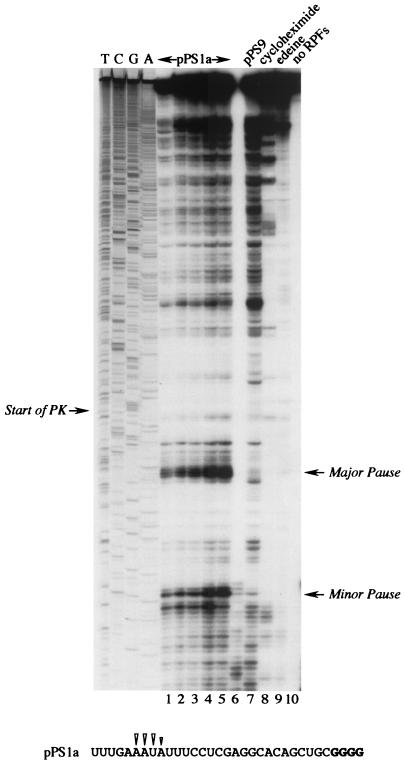
mRNAs for heelprint assays were prepared by SP6 transcription of AvaII-linearized plasmid pPS1a (formerly pPS1 [36]), which contains the minimal IBV pseudoknot inserted at position 1167 of the PB1 reporter gene (Fig. 1). The minimal pseudoknot is fully functional in frameshifting (4, 27) and it induces frameshifting in vitro at a higher level (40%) than the wild-type IBV structure does (30%). However, the AvaII-derived pPS1a mRNA is not a frameshift reporter mRNA, since it does not contain the IBV slippery sequence. Furthermore, it can be translated from beginning to end, as the inserted minimal pseudoknot has no termination codons. A control plasmid derived from pPS1a, pPS9 (Fig. 1), was also transcribed to generate an mRNA in which the pseudoknot had been destabilized by a complementary mutation in stem 1 (and was nonfunctional in frameshifting). The heelprint assay of pPS1a (in RRL) is shown in Fig. 2. T7 DNA polymerase processivity in this experiment was satisfactory, with only a limited number of “enzyme stops” visible on the gel in the absence of RPFs (lane 10). In the presence of RPFs, more termination products were seen, largely originating from authentic RPF hybridization, since they disappeared in reactions where cycloheximide or edeine was added prior to translation (lanes 8 and 9). Other stops likely arose as a consequence of hybridization of RNase-resistant RNA fragments, either PB1-specific or adventitiously hybridizing fragments from rRNA (26, and see below). As can be seen in lanes 1 to 5 (which differed only in the amounts of labeled primer and pPS1a-derived RPFs in the annealing reaction mixtures), among a number of species, a strong heelprint was observed which spanned 4 nt at a position 21 to 24 nt upstream of the first base (G) of the pseudoknot. This heelprint was not derived from the adventitious annealing of an unprotected but RNase-resistant RNA fragment; when initiation or elongation was blocked by addition of cycloheximide or edeine prior to mRNA addition (lanes 8, 9), the heelprint was absent. In the pPS9 control construct (lane 7), a slightly more intense and almost identical overall band pattern was seen but the strong heelprint was missing. Thus, the appearance of the major heelprint in the pPS1a lanes (1 to 5) is consistent with ribosomal pausing at the pseudoknot. Also evident in the pPS1a lanes was a second species which mapped about 29 bases upstream of the first pause and was also absent from the control lane (pPS9). This might be the heelprint of a trailing ribosome, stacked behind the paused ribosome. This presumption was reinforced by the detection of a scarce species of RPF about 60 bases long which might derive from two ribosomes lining up behind the pseudoknot (data not shown; see below).
FIG. 2.
Ribosomal pausing at the minimal IBV pseudoknot. mRNA from AvaII-digested pPS1a was subjected to heelprint analysis as detailed in Materials and Methods. Heelprints of the minimal IBV pseudoknot (pPS1a; lanes 1 to 6 and 8 to 10) and a mutant derivative (pPS9, lane 7) are shown alongside a sequencing ladder (TCGA). Each reaction mixture contained 20 ng of the relevant single-stranded DNA template. In lanes 1 to 5, the concentrations of primer and RPFs were varied: lane 1, 0.1 ng of primer, 3 μl of RPFs; lane 2, 0.2 ng of primer, 3 μl of RPFs; lane 3, 0.4 ng of primer, 3 μl of RPFs; lane 4, 0.4 ng of primer, 4 μl of RPF; lane 5, 0.4 ng of primer, 4.5 μl of RPFs. All other lanes (except lane 10) contained 0.4 ng of primer and 3 μl of RPFs. Lanes 8 and 9 were control reactions in which cycloheximide (lane 8) or edeine (lane 9) was added (to 1 mM and 5 μM concentrations, respectively) prior to addition of mRNA to the translation reaction mixture. In lane 10, RPFs were omitted from the primer extension reaction. The start of the pseudoknot (the first G in a block of four reading up the gel) and the position of two clear pause sites are indicated with arrows. Lane 6 was identical to lane 5, except T4 DNA polymerase replaced T7 DNA polymerase (unsuccessfully). The primary sequence of the mRNA upstream of the pseudoknot is shown at the bottom, and the positions of the pseudoknot-dependent heelprints are indicated with arrowheads (the first four G residues of the pseudoknot are shown in bold type).
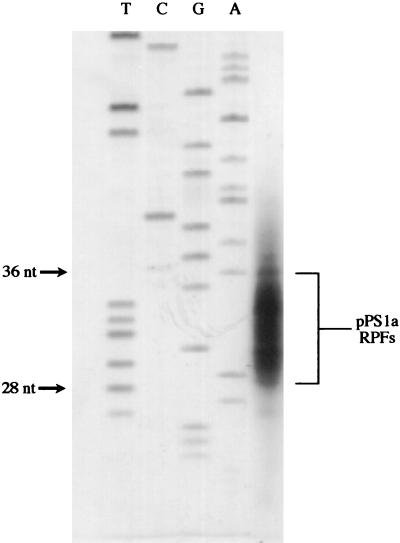
To examine the size of the RPFs in these experiments, [32P]UTP was included in a transcription reaction to generate radiolabeled pPS1a mRNA. The RPFs were analyzed on a denaturing 8% polyacrylamide gel alongside a sequencing ladder (Fig. 3) and were 28 to 36 nt in length, consistent with previous estimates of the region protected by eukaryotic ribosomes (30 to 35 [22], and 24 to 32 [46]). Given that the 5′ edge of the ribosome is positioned 21 to 24 nt upstream of the first base of the pseudoknot, the 3′ edge can be calculated, on the basis of a mean ribosomal site size of 32, to be 8 to 11 nt into the IBV pseudoknot-forming sequence. Heelprinting of ribosomes paused at initiation codons has shown that the 5′ edge of the ribosome is some 12 to 13 nt from the first base of the AUG (46). On this basis, in our experiments the ribosomal P-site will be approximately 8 to 12 nt 5′ of the start of the pseudoknot, i.e., at or around those bases which are in the equivalent position of the slippery sequence in the natural frameshift signal. Thus, the heelprint data are consistent with a role for pausing in frameshifting, with the paused ribosome being positioned over the slippery sequence while in contact with the pseudoknot.
FIG. 3.
Sizing of RPFs. 32P-trace-labeled AvaII-digested pPS1a mRNA was subjected to heelprinting, and the RPFs were analyzed on an 8% denaturing polyacrylamide gel. A sequencing ladder (TCGA) was run alongside to provide approximate size standards.
The heelprint assays above were performed on an mRNA containing the minimal IBV pseudoknot but lacking the IBV slippery sequence. To rule out any influence of the slippery sequence on the pausing pattern obtained, we also performed heelprint assays on an mRNA prepared from plasmid pFS7.2/HK (2). This construct contains what is essentially the wild-type IBV frameshift signal cloned into the PB1 reporter gene (Fig. 4A). To ensure that heelprint assays would be uninfluenced by ribosomes terminating at the 1a stop codon (UGA), which forms part of the second arm of stem 1, it was changed to UGU. As the next stop codon (in the PB1 gene) was still only 11 codons downstream, this was also changed (from UAA to AAA), placing the next stop codon some 38 codons from the slippery sequence. Changing the 1a termination codon to UGU is known to increase frameshifting a small amount, presumably as a result of stabilization of stem 1 (a G-A mismatched pair becomes a G-U wobble pair [2]). The heelprint of pFS7.2/HK (Fig. 4B) revealed a group of four products 21 to 24 nt upstream of the IBV pseudoknot. This heelprint was greatly reduced in a related control construct in which stem 1 was destabilized (pFS7.19a; Fig. 4), supporting the idea that it is derived from a pause at the pseudoknot. Thus, pausing was also detectable in a construct containing both the IBV slippery sequence and the complete pseudoknot. That the majority of the primer extension products in the pFS7.2/HK lane originate from RPF hybridization is confirmed in the supernatant (S) lane. During purification of RPFs, ribosomes and associated RNA fragments are pelleted through sucrose. The S lane represents an experiment in which the sucrose supernatant was retained and treated in the same way as the ribosomal pellet. This sample, which should only contain degraded mRNA and any longer RNase-resistant species, was used in a primer extension assay. Only a small number of stops were seen in this lane, which was presumably a consequence of annealing of RNase-resistant species. Thus, most of the signals in the pFS7.2/HK lane are derived from RPFs (and possibly rRNA fragments, as mentioned earlier).
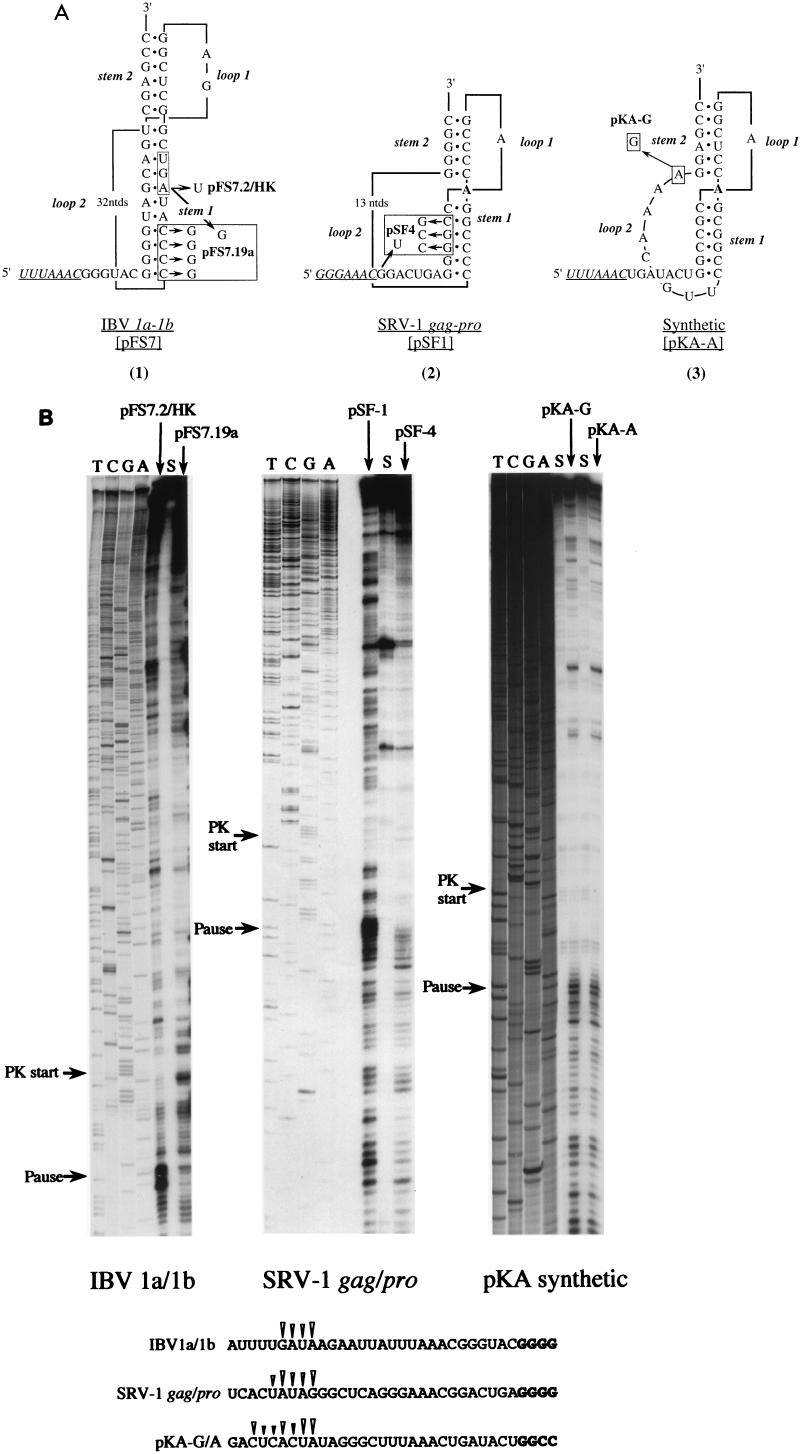
FIG. 4.
Ribosomal pausing at natural and synthetic frameshift signals. (A) Predicted secondary structures of natural and synthetic frameshift signals tested in heelprint assays (1). Plasmid pFS7 contains the wild-type IBV slippery sequence (UUUAAAC, underlined) and pseudoknot (2). The 1a termination codon (UGA), which forms part of stem 1, is boxed. Plasmids pFS7.2/HK and pFS7.19a are mutant derivatives in which the 1a termination codon has been changed to UGU or UGG, respectively. Plasmid pFS7.19a contains an additional mutation, a complementary change that destabilizes stem 1 of the pseudoknot (2). Plasmid pSF1 contains the SRV-1 gag/pro frameshift region (42). A derivative, pSF4, has a destabilizing mutation in stem 1 and, additionally, a termination codon (UGA) immediately downstream of the slippery sequence (GGGAAAC, underlined). The unpaired A residue (in bold) between the stems is drawn on the basis that the pseudoknot is similar to that of MMTV gag/pro (see text). Recent NMR studies have challenged this belief (14, 29) and suggest that the A is in fact paired with the most 3′ base of loop 2 (a U). The synthetic frameshift site pKA-A (25) has an IBV-like slippery sequence (UUUAAAC, underlined) and an MMTV-like stimulatory pseudoknot. Plasmid pKA-G differs solely in the identity of the last residue of loop 2. (B) mRNAs from SmaI-digested pFS7.2/HK and pFS7.19a or BamHI-digested pSF-1, pSF-4, pKA-A, and pKA-G were subjected to heelprint analysis as detailed in Materials and Methods. Heelprints of each RNA are shown alongside a sequencing ladder (TCGA) prepared from the relevant plasmid. Each reaction mixture contained 20 ng of the relevant single-stranded DNA template, 0.4 ng of primer, and 3 μl of RPFs. Lanes marked S indicate heelprints in which RPFs were replaced by an equivalent amount (vol/vol) of material harvested from the supernatant produced in the airfuge centrifugation step (see Materials and Methods). The start of each pseudoknot and the position of pseudoknot-dependent ribosomal pauses are indicated by arrows. The primary sequences of the mRNA upstream of the various pseudoknots are shown at the bottom and the position of the pseudoknot-dependent heelprints are indicated with arrowheads (the first four pseudoknot residues are shown in bold type in each case).
The heelprints of ribosomes paused at the IBV pseudoknot (wild type and minimal) are similar to those that have been seen at the L-A cap/pol frameshift signal. Tu and colleagues (45) observed two sites of pausing 20 and 23 nt upstream of the first base of the L-A pseudoknot; later this was refined to a single pause, 24 nt upstream (26). The IBV and L-A pseudoknots are comparable in terms of the predicted length of stem 1 (in IBV it is 11 nt [3] and in L-A it is 13 nt [9, 26]), and although the secondary structure of the L-A pseudoknot has not been probed, it seems likely to possess an organization similar to that of the IBV pseudoknot (31). Consequently, it could be expected to interact with ribosomes in a manner comparable to the IBV pseudoknot and to pause ribosomes at a similar position. An important question, therefore, was whether similar heelprints would be produced by a frameshift signal containing a different class of pseudoknot structure. Figure 4 shows a heelprint assay of the SRV-1 gag/pro frameshift signal. This site has been proposed to contain a pseudoknot similar to that present at the gag/pro overlap of the retrovirus mouse mammary tumor virus (MMTV) (38). These signals are typified by a short stem 1 of just 5 or 6 bp (42, 43) and an intercalated unpaired A residue between the two pseudoknot stems that is essential for function (6, 7, 25, 35), although recent nuclear magnetic resonance (NMR) studies suggest that the stems of the SRV-1 pseudoknot are actually coaxially stacked (14, 29). The mRNA used in the heelprint assay was prepared from plasmid pSF1 (42), which contains the SRV-1 frameshift signal cloned into an influenza PB2 reporter gene (Fig. 4A; in vitro frameshift efficiency, 23%). In this construct, ribosomes which frameshift at the SRV-1 slippery sequence (GGGAAAC) terminate translation 105 nt downstream of the slippery sequence; those that do not frameshift terminate translation some 300 nt downstream. Thus, any heelprints arising from ribosomal pausing at the pseudoknot are unlikely to have been influenced by ribosomes in the act of termination further downstream. As a control mRNA, pSF4 was employed; pSF4 contains a destabilizing mutation in stem 1 and has a greatly reduced frameshift capacity. This construct, however, unlike pSF1, has a termination codon immediately downstream of the slippery sequence. With the pSF1 mRNA, a clear heelprint was observed that spanned 3 to 4 nt at a position some 21 to 24 bases upstream of the first base of the pseudoknot (Fig. 4B). This heelprint was much reduced in the pSF4 control mRNA lane, supporting the idea that it is a pseudoknot-specific heelprint. Also in this lane, the number of longer products was reduced. This is a consequence of the additional stop codon in pSF4 just downstream of the slippery sequence and the reduced frameshift efficiency of the signal. The number of ribosomes translating the mRNA beyond the frameshift signal is reduced in comparison to pSF1, and thus fewer RPFs are obtained from the 3′ end of the pSF4 mRNA and hence the lack of bands in this region of the primer extension reaction. Thus, pausing at the pseudoknot of SRV-1 is qualitatively indistinguishable from that of the minimal IBV pseudoknot; the ribosome is paused at approximately the same position on the mRNA and the heelprint is a characteristic block of 3 to 4 nt.
Heelprinting of ribosomes paused at functional and nonfunctional frameshift signals.
The results of the experiments described above are consistent with a role for pausing in frameshifting, but it was important to assess whether pausing also occurred at sites incapable of stimulating efficient ribosomal frameshifting but containing stable RNA secondary structures. In an earlier study (36), we compared pausing at the minimal IBV pseudoknot (pPS1a) with a related hairpin structure (reproducing the base pairs present in the pseudoknot) that stimulates frameshifting some 5- to 10-fold less well (pPS7a, formerly pPS7; Fig. 1). Using a translational elongation assay (the edeine assay, described below), we found that the stem-loop structure could induce pausing and that the difference in pausing levels observed between the pseudoknot and the hairpin did not seem to be sufficient to account for the difference of their respective frameshifting efficiencies. A possible explanation is that the hairpin might pause ribosomes at a slightly different position on the mRNA; under these circumstances, frameshifting would be compromised as the ribosome may be inappropriately placed over the slippery sequence during a hairpin-induced pause. To resolve this point we performed heelprint assays to pinpoint the positions of paused ribosomes on the two mRNAs (Fig. 5). We found that both structures paused ribosomes at precisely the same position on the mRNA, with their 5′ edges some 21 to 24 nt upstream of the first base of each structure. The position of the pause in each case is consistent with the belief that the decoding site of the ribosome would be placed over the slippery sequence during the pause. This suggests that the reduced ability of the stem-loop to promote frameshifting is not a consequence of pausing the ribosome at an inappropriate position on the mRNA.
FIG. 5.
Comparison of pseudoknot- and hairpin-induced ribosomal pauses. mRNAs from AvaII-digested pPS1a and pPS7a were subjected to heelprint analyses as detailed in Materials and Methods. Heelprints of each RNA are shown alongside a sequencing ladder (TCGA) prepared from each plasmid. Each reaction mixture contained 20 ng of the relevant single-stranded DNA template, 0.4 ng of primer, and 3 μl of RPFs. The start of the pseudoknot (pPS1a) and hairpin (pPS7a) and the position of corresponding ribosomal pauses are indicated with arrows. The primary sequence of the mRNA upstream of the pseudoknot or hairpin is identical and is shown at the bottom. The position of the structure-dependent heelprints are indicated with arrowheads (the first four residues of the pseudoknot and hairpin are shown in bold type).
We also examined the heelprints of two closely related RNA pseudoknot structures, pKA-A and pKA-G (25). These RNAs are derivatives of the minimal IBV pseudoknot that have been altered to more closely resemble kinked pseudoknots (Fig. 4A) and differ only in the possession of an adenosine or a guanosine at the end of loop 2. From secondary structure probing, the pseudoknots are very similar in conformation yet stimulate different frameshift efficiencies in RRL (pKA-A, 31%; pKA-G, 5%). This has been ascribed to an ability to form (pKA-A) or not to form (pKA-G) an interaction between loop 2 and stem 1 (25). From Fig. 4B it can be seen that their heelprints are essentially identical, with the same pattern of pausing bands produced in both assays with very similar intensities. Both pseudoknots force ribosomes to pause at exactly the same positions on the respective mRNAs, with clear pauses between 21 and 26 bases upstream of the start of each pseudoknot. That two pseudoknots differing substantially in their ability to promote efficient frameshifting are equally capable of pausing ribosomes strongly suggests that pausing alone is insufficient to account for the ability of a functional RNA pseudoknot to stimulate frameshifting.
Ribosomal pausing at the minimal IBV pseudoknot is encounter-phase specific.
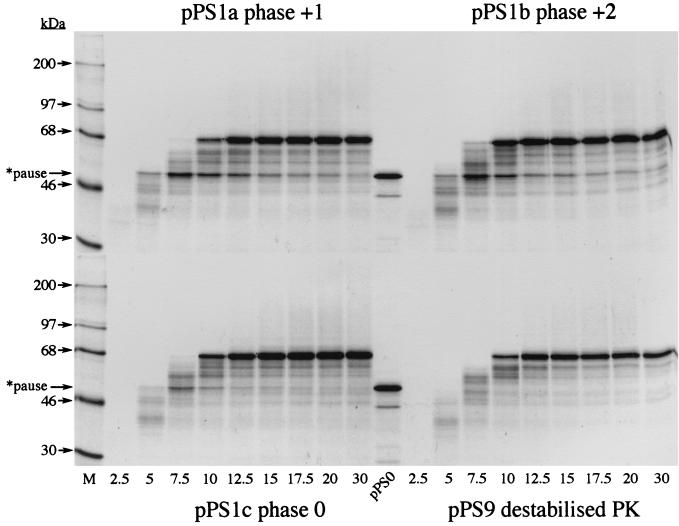
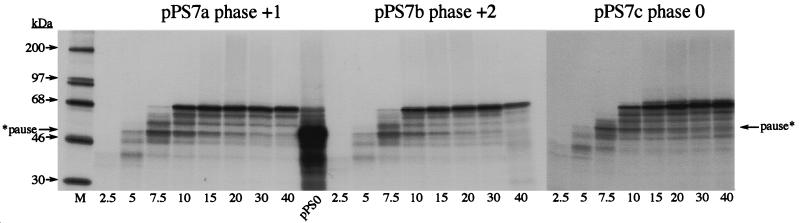
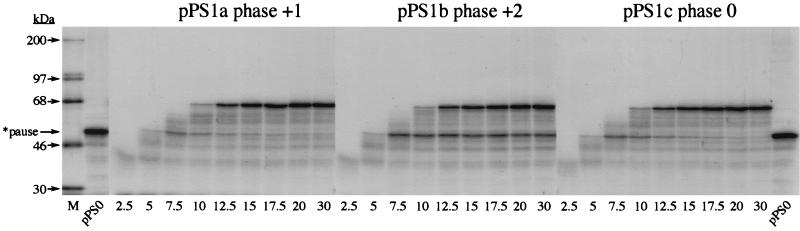
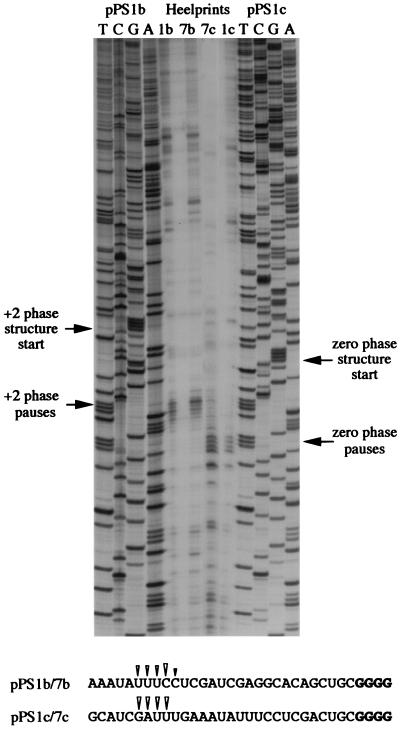
During construction of pPS1a, the minimal IBV pseudoknot was inserted into unique XhoI and PvuII sites in the PB1 reporter gene without consideration of the encounter phase of the pseudoknot. By this, we mean the relative position of the first base of the pseudoknot with respect to the translational reading frame. At the wild-type IBV 1a/1b frameshift signal, ribosomes encounter the pseudoknot in the zero phase, that is, the codon before the first base of the pseudoknot is directly adjacent to first base of the pseudoknot (U-UUA-AAC-GGG-UAC-pseudoknot; 1a reading frame underlined). However, in pPS1a, the encounter phase is +1, with a single nucleotide present between the last codon and the first base of the minimal pseudoknot (CAG-CUG-C-pseudoknot; Fig. 6). As the ribosome progresses in triplet steps, the encounter phase can potentially influence the ease by which the pseudoknot is unwound by the ribosome, and this may be reflected in the time that the ribosome is paused at the pseudoknot. To test this, we prepared two phase variants of pPS1a (see Materials and Methods) (Fig. 6) in which the encounter phase was +2 (GCU-GC-pseudoknot; pPS1b) or zero (GAC-UGC-pseudoknot; pPS1c) and measured pausing by using the edeine assay (36). Here, the extent of pausing was estimated by comparing the levels of a translational intermediate corresponding to pausing at the pseudoknot with that of full-length polypeptide produced during a time course of translation in RRL. To facilitate detection of intermediates corresponding to ribosomal pausing, the standard translation reaction was modified in two ways. Firstly, the reactions were carried out at 26° rather than 30°C since the general reduction in the rate of translation at the lower temperature creates a longer window for recognition of translational intermediates. Secondly, in order to simplify the pattern of intermediates observed, translation was synchronized by the addition of edeine, a potent inhibitor of initiation (40), 5 min after the start of the reaction. As can be seen in Fig. 7, all mRNAs specified the synthesis of a full-length product of approximately 68 kDa. However, in those RNAs containing an intact pseudoknot (pPS1 series), a transient translational intermediate was seen whose size was consistent with it being derived from pausing at the pseudoknot. This band was greatly reduced in construct pPS9, in which the pseudoknot is destabilized, supporting the idea that the pause is pseudoknot-derived. The identification of this polypeptide as a pseudoknot-induced product was further strengthened by the observation that it comigrated with the translation product of transcripts from pPS0 digested with XhoI, which cleaves the plasmid at the position of the inserted pseudoknot sequence of pPS1. The extent of pausing, as judged by comparing the intensities of the paused and full-length species, was similar for the +1 (pPS1a) and +2 (pPS1b) phase variants. However, in the zero-phase construct (pPS1c) pausing was noticeably reduced, indicating that the pseudoknot was unable to impede the progress of ribosomes as markedly as in the other two phases. We also tested whether pausing at phase variants of the hairpin construct, namely pPS7a (+1 phase), pPS7b (+2 phase), and pPS7c (zero phase), showed similar phase dependence. The observed pattern of pausing (Fig. 8) was similar to that of the pseudoknot-containing constructs, except that the level of pausing in the +1 and +2 phases (pPS7a, pPS7b) was lower than that of the equivalent pseudoknot-containing constructs, although less than twofold different, and pausing in the zero phase (pPS7c) was less dramatically reduced, in comparison to the other phases, than the equivalent zero-phase pseudoknot-containing construct (pPS1c; Fig. 7). Thus, less phase dependence was evident. An important question, therefore, was whether the precise position of pausing differed in the phase variant constructs. To test this, we carried out a heelprint analysis of the constructs; this is shown in Fig. 5 (pPS1a/pPS7a) and 9 (pPS1b/pPS7b; pPS1c/pPS7c). We found that whatever the encounter phase, both the pseudoknot and the hairpin were able to pause ribosomes at the same position on the mRNA, with their 5′ edges some 21 to 24 nt upstream of the first base of each structure.
FIG. 7.
Reading-phase dependence of ribosomal pausing. Time courses of translation of AvaII-derived pPS1a, -b, and -c and pPS9 mRNAs in reticulocyte lysates. Translation was allowed to proceed at 26°C in the presence of [35S]methionine for 5 min prior to addition of edeine to a final concentration of 5 μM. Samples were withdrawn at the indicated times (in minutes) post-edeine addition, and translation products were separated on SDS–10% polyacrylamide gels. Labeled polypeptides were detected by autoradiography. [14C]-labeled molecular mass standards (M) were from Amersham Pharmacia Biotech. The pPS0 tracks mark the expected position of a pseudoknot-induced ribosomal pause product and were prepared by translating XhoI-derived pPS0 mRNA at 26°C for 1 h. The pause product is indicated by an arrow. Although the size of this protein as predicted from the nucleic acid sequence of the PB1 reporter gene is 43 kDa, it migrates somewhat slower in SDS-polyacrylamide gels because of the highly basic nature of the PB1 protein (2).
FIG. 8.
Pausing at hairpin phase variants. Time courses of translation of AvaII-derived pPS7a, -b, and -c mRNAs in reticulocyte lysates are shown. Translation products were prepared, labeled, and analyzed as described in the legend to Fig. 7. M, molecular mass standards.
The edeine assay was also employed to determine whether pausing at the minimal IBV pseudoknot retained the same phase dependence in the WG in vitro translation system. These translations were carried out at 15°C (rather than the usual 26°C), once again to create a longer window for recognition of translational intermediates, and they are shown in Fig. 10. In WG, phase dependence of pausing was still seen, yet it was different from that observed in RRL (Fig. 7), with the maximal pause seen in the +2 phase, reduced pausing in the zero phase, and the least pausing in the +1 phase. In the +2 phase, the pausing product was most persistent yet it was not a dead-end product, since the full-length species was still accumulating at later time points. Thus, phase-dependent pausing was also seen in the WG translation system.
FIG. 10.
Reading-phase-dependent ribosomal pausing in the WG system. Shown are the time courses of translation of AvaII-derived pPS1a, -b, and -c mRNAs in the WG system. Translation products were prepared, labeled, and analyzed as described in the legend to Fig. 7, except that the translations were carried out at 15°C. M, molecular mass standards.
Influence of reading phase on frameshifting.
It is known that the precise distance between the IBV slippery sequence and the minimal pseudoknot is important, and deviation from the optimal 6 nt by as little as a single nucleotide either way reduces frameshifting in RRL (4) (Fig. 6). These effects on frameshifting can be considered from the perspective of phasing, with alterations in phasing influencing frameshifting. As depicted in Fig. 6, the encounter phase of the pseudoknot is zero for the wild-type frameshift signal spacer (cass 5; frameshift efficiency of 41% [4]), +1 for a 7-nt spacer (pSM3; 21%), and +2 for the construct with a 5-nt spacer (pSM2; 21%). To allow a broader comparison of the influence of phasing on both pausing and frameshifting, the frameshift efficiencies of these constructs were also measured in WG by using BamHI-derived mRNAs. The response of WG ribosomes to alterations in the spacer length was different from RRL in that frameshifting occurred at a similar level at spacer lengths of 5 and 6 nt (31 and 30%, respectively), yet was significantly reduced (to 11%) when the spacer was 7 nt (translations not shown). A comparison of pausing and frameshifting for each phase reveals no obvious correlations, and this is most evident for the zero-phase encounter in RRL (pPS1c versus cass 5), which gave the least pausing yet the most frameshifting. Similarly, although the optimal phase for pausing in WG (pPS1b) gave the most frameshifting (pSM2), a similar level of frameshifting was seen for the zero-phase construct (cass 5), a phase which elicited only a modest pause in the edeine assay (pPS1c). Thus, viewed from the perspective of reading phase, there is no evident correlation between frameshifting and pausing.
DISCUSSION
A role for ribosomal pausing in −1 frameshifting has been suspected for many years (21), but the topic has received relatively little attention. Evidence for pausing at the frameshift sites of the yeast L-A virus (45) and the coronavirus IBV (36) has been provided and attempts have been made to examine the kinetics of pausing at the L-A site (26), but we still know little about the process nor its relevance to frameshifting. Here, we employed heelprinting and elongation assays to investigate pausing at a variety of frameshifter RNA pseudoknots and related hairpin structures.
The site of ribosomal pausing.
The heelprints of the pseudoknots and hairpins appeared typically as a contiguous stretch of four prominent bands, with the 5′ edge of paused ribosomes mapping to some 21 to 24 nt upstream of the first paired base of the relevant RNA structure. From our knowledge of the region of mRNA protected by eukaryotic ribosomes (22, 46), this would place the decoding site of the ribosome close to the slippery sequence during the pause, consistent with a role for pausing in frameshifting. That the heelprints appeared as a block of bands rather than a single species could represent a situation where ribosomes, having paused, moved on a codon before pausing again or, alternatively, some kind of oscillation of paused ribosomes. However, groups studying pausing in unrelated systems have often seen this kind of heelprint (12, 46), and we suspect, therefore, that it arises more as a consequence of heterogeneity of RPF length brought about by differential micrococcal nuclease trimming rather than from ribosomal movements. We did not notice any gross differences in the heelprint pattern at sites containing an authentic frameshift signal (with both a slippery sequence and stimulatory pseudoknot) or just a pseudoknot or a hairpin (pPS series). Thus, the slippery sequence does not appear to influence pausing. A similar conclusion was reached by Lopinski and colleagues (26): pausing at the L-A signal (as judged by heelprint and elongation assays) was found to be uninfluenced by a mutation that inactivated the slippery sequence. In addition, we did not see any extra pauses when a termination codon was present close to the stimulatory pseudoknot structure. Initially, we designed the constructs to avoid termination codons in the immediate vicinity of the pseudoknots, such that we could uncouple pseudoknot-dependent pausing from the expected termination codon-dependent pausing. In fact, we did not see any obvious termination codon-induced pauses in constructs where this would have been apparent (pSF4, pFS7.19, pKA-A, pKA-G), although we were able to see pausing at initiation codons (data not shown). Why this was the case is not known, but it may be related to the specific termination codon in question (UGA in our constructs). Although termination codon-induced pausing has been detected (by heelprinting) on the bovine preprolactin mRNA, which terminates at UAA (46), and on the reovirus dicistonic s1 mRNA (UAG of nonstructural protein ς1 78S) and the s4 mRNA (UAA of minor capsid protein ς3 [12]), no pausing was detected during termination of synthesis of the minor capsid protein ς1, which has a UGA stop codon (12). Further work will be needed to clarify whether this is an effect specific to the UGA codon, codon context, or a lack of sensitivity of the assay.
Reading-phase-dependent pausing.
During translation, RNA secondary structures must be unwound prior to decoding, and the efficiency of such unwinding is potentially influenced by the reading phase in which the base-paired region is encountered. It has long been assumed that the ribosome possesses an intrinsic helicase activity required to melt RNA secondary structures during elongation. One interpretation of the phase dependence of pseudoknot-induced pausing described here is that the helicase may need an optimal contact between itself and the structure it is about to unwind; certain phases may form suboptimal contacts, and under these circumstances the duration of the pause would be dictated both by the time taken to restore the optimal contact and the time required to unwind the structure. For reticulocyte ribosomes, the optimal phase would appear to be the zero phase, with the least overall delay in unwinding the structure. In the WG system, phase-dependent pausing was different from that seen in RRL in that the maximal pause was in the +2 phase, reduced pausing was seen in the zero phase, and the least pausing was in the +1 phase. These differences with respect to RRL can be rationalized by proposing that the position of the hypothetical helicase in WG ribosomes, relative to the pseudoknot, is different. It is known that WG ribosomes possess a 60S subunit more closely resembling that of the E. coli 50S subunit (30) and have a slightly smaller footprint on the mRNA (46), facts not inconsistent with an altered hierarchy of phase-dependent pausing. It will be of interest to extend these studies to other frameshifter pseudoknot and stem-loop structures.
What is the role of pausing in frameshifting?
Although the site of ribosomal pausing at RNA pseudoknots is consistent with a role in frameshifting, there is no direct evidence that pausing and frameshifting are correlated. Pausing, for example, may simply be a byproduct of a pseudoknot-ribosome interaction, with little or no active role in the frameshift process. We made a number of observations that argue against a simple correlation between pausing and frameshifting. Firstly, a closely related hairpin-loop structure, based on the minimal IBV pseudoknot, although unable to stimulate efficient frameshifting was able to pause ribosomes to a similar extent and at the same place on the mRNA as the parental pseudoknot. Secondly, an apparently identical pausing pattern was induced by two closely related pseudoknots differing only by a single loop 2 nucleotide yet with different functionality in frameshifting. Finally, when we assessed the impact of reading phase on pausing at the minimal pseudoknot, we found that the phase did influence pausing in both RRL and WG systems, but there was little correlation between pausing and frameshifting in either system. Regarding the latter point, a caveat that must be raised is that we were not able to carry out both frameshifting and pausing assays on the same mRNAs. As the nucleotides of the spacer regions present in the frameshift constructs differed from those present immediately upstream of the pseudoknots of the pausing series, it is possible that the primary sequence of the spacers could influence their effective length and hence the frameshift efficiency. However, we have replaced the spacer sequences of the frameshift constructs by stretches of nucleotides identical to those upstream of the pausing series, and this did not influence the magnitude of frameshifting seen (data not shown). Thus, the lack of correlation between frameshifting and pausing seems genuine. Together with the results of Tu and colleagues (45), who identified a nonframeshifting mutant of the L-A pseudoknot that could still pause ribosomes, these data indicate that a pause alone is not sufficient for frameshifting. However, that pausing plays an essential contribution to frameshifting cannot be excluded; the ribosome is indeed paused over the slippery sequence and we have yet to identify an instance where frameshifting occurs in the absence of a detectable pause.
Mechanistic implications for the frameshift process.
Ribosomal pausing has been featured in most models of −1 frameshifting; it can increase the time available for movement of tRNAs at the slippery sequence and also act as a unifying feature to accommodate the variety of stimulatory RNAs present at −1 frameshifting signals. However, the idea that a pause alone is sufficient to induce frameshifting is questionable. Simple provision of a roadblock to ribosomes in the form of stable RNA hairpins (3, 36), a tRNA (6), or even different kinds of RNA pseudoknot (25, 31) is not sufficient to bring about frameshifting and as detailed above, pseudoknots and stem-loops that promote reduced levels of frameshifting yet still pause ribosomes have been described. However, although the experiments presented in this study strongly support the view that pausing is probably only a component of the mechanism of frameshifting, we have not ruled out the possibility that a precise “kinetic pause” is required which only certain stimulatory RNAs can generate. For example, during a −1 frameshift, two pauses could occur, one productive (in terms of frameshifting) upon initial encounter of the stimulatory RNA structure and a second, nonproductive pause, corresponding perhaps to a delay in unwinding after the crucial event in frameshifting has already taken place. The magnitude of the initial pause could potentially influence the extent of the frameshift, whereas the second pause, occurring during the time that the ribosomal unwinding activity locates and deals with the structure, would be irrelevant. The pausing assays employed in the present study would probably not distinguish between two such pausing events, and a detailed analysis of the kinetics of pausing will require further experimentation, including the development of techniques to study translational elongation at the level of individual ribosomes.
It has been argued that pseudoknots are especially suited to their function in frameshifting since they may be more resistant to unwinding by the ribosome, giving more pausing and increased frameshifting (2, 10, 16, 19, 21). In this light, the heelprints of the minimal IBV and SRV-1 gag/pro pseudoknots are of interest in that they reveal a very similar pattern of pausing despite the considerable differences in predicted size and conformation of the two structures. Based on the average size of RPFs and the position of the 5′ boundary of the paused ribosome, we calculated that several nucleotides of the IBV and SRV-1 pseudoknots (approximately 8 to 11 nt) were protected from micrococcal nuclease treatment. In pseudoknots of the IBV class, with a long, stable stem 1, the heelprinting data suggest that stem 1 is substantially unwound during the pause. It follows, therefore, that a greater proportion of the SRV-1 pseudoknot stem 1, perhaps all of it, would be unwound since it is only 6 nt in length. How can this be rationalized in terms of the mode of action of RNA pseudoknots in frameshifting? One possibility is that different regions of the pseudoknot are responsible for pausing the ribosome. In IBV, this could be within the stable stem 1 region; in SRV-1, it could be the junction of the two pseudoknot stems, where tertiary interactions are suspected to occur (37). An alternative and perhaps more attractive possibility is that both pseudoknots are in fact intact, or at least only partially unwound, during the pause. The heelprint of the ribosome is defined by the length of the RPFs, which are generated upon micrococcal nuclease treatment of cycloheximide-treated ribosomes. With this assay we cannot distinguish between a significantly unwound pseudoknot and an intact pseudoknot associated with the ribosome, since certain regions, especially the single-stranded loops, would likely remain accessible and be cleaved by the micrococcal nuclease. If the pseudoknot is (relatively) intact, it would be closely associated with the ribosome during a pause and in an ideal position to exert its effects that lead to frameshifting. An initial pause may contribute to this event, whether it be, as has been proposed, an interaction of the pseudoknot with a ribosomal protein(s) (21) or a region of rRNA (24), tRNA molecular mimicry (35), or an inappropriate occupation of a region of the ribosome that impairs normal frame maintenance. It should be possible to probe the conformation of the pseudoknot at paused ribosomes from the 3′ direction using a variation of the toeprint assay. The greater challenge, however, will be to determine how the pseudoknot acts to bring about frameshifting once associated with the ribosome.
The heelprint assays of the pseudoknot phase variants in RRL (pPS1 series; Fig. 5 and 9) revealed that the 5′ edge of the ribosome was 21 to 24 nt upstream of the first base of the pseudoknot in all three phases. This places the 5′ edge of the ribosome at a slightly different position on the mRNA for each phase variant, with the 3′ edge of the paused ribosomes at the same relative position, presumably interacting with the same region of the pseudoknot. If extrapolated to frameshifting, this could explain why at spacer distances of 5 or 7 nt, frameshifting is reduced (in RRL), since the decoding site would be inappropriately positioned with respect to the slippery sequence. However, that the position of the 5′ edge of the ribosome varied on each mRNA was quite unanticipated. As ribosomes translate in the triplet register, we expected that the 5′ edge would be locked in position, since the distance from the decoding center and an arbitrary “exit” site on the 5′ side of the ribosome would presumably be uninfluenced by the pseudoknot, and that the position of the 3′ edge would vary, since the phasing was achieved experimentally by (effectively) adding or deleting a single base just upstream of the pseudoknot. In fact, the reverse was seen. One interpretation of these data is that the pause is independent of triplet decoding; perhaps the heelprints are derived from ribosomes paused during the translocation step of the elongation cycle, when reading frame monitoring is at its weakest. Whatever the case, this is not a pseudoknot-specific phenomenon; similar heelprints were seen with phase variant constructs containing a stem-loop structure (pPS7 series, Fig. 5 and 9). Nevertheless, it offers an avenue of exploration in the search for the precise mechanism of ribosomal frameshifting.
FIG. 9.
Heelprinting of hairpin phase variants. mRNAs from AvaII-digested pPS1b, pPS7b, pPS1c, and pPS7c were subjected to heelprint analysis as detailed in Materials and Methods. Heelprints of each RNA are shown alongside a sequencing ladder (TCGA) prepared from two of the plasmids. Each reaction mixture contained 20 ng of the relevant single-stranded DNA template, 0.4 ng of primer, and 3 μl of RPFs. The position of the start of the +2 and zero-phase pseudoknots and hairpins and the site of the corresponding ribosomal pauses are indicated by arrows. The primary sequences of the mRNA upstream of the various structures are shown at the bottom, and the position of the pseudoknot- or hairpin-dependent heelprints are indicated with arrowheads (the first four pseudoknot and hairpin residues are shown in bold type in each case).
ACKNOWLEDGMENTS
This work was supported by the Medical Research Council, United Kingdom, and the Biotechnology and Biological Sciences Research Council, United Kingdom.
The assistance of Alison Gelder and Emily Robins is gratefully acknowledged. We thank Paul Digard for critical reading of the manuscript and Edwin ten Dam and Philip Farabaugh for thought-provoking comments.
REFERENCES
- 1.Alam S L, Wills N M, Ingram J A, Atkins J F, Gesteland R F. Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J Mol Biol. 1999;288:837–852. doi: 10.1006/jmbi.1999.2713. [DOI] [PubMed] [Google Scholar]
- 2.Brierley I, Digard P, Inglis S C. Characterisation of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brierley I, Rolley N J, Jenner A J, Inglis S C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley I, Jenner A J, Inglis S C. Mutational analysis of the “slippery sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brierley I. Ribosomal frameshifting on viral RNAs. J Gen Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Chamorro M, Lee S I, Shen L X, Hines J V, Tinoco I, Jr, Varmus H E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X Y, Kang H S, Shen L X, Chamorro M, Varmus H E, Tinoco I. A characteristic bent conformation of RNA pseudoknots promotes −1 frameshifting during translation of retroviral RNA. J Mol Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- 8.Craigen W J, Caskey C T. Translational frameshifting: where will it stop? Cell. 1987;3:1–2. doi: 10.1016/0092-8674(87)90652-0. [DOI] [PubMed] [Google Scholar]
- 9.Dinman J D, Wickner R B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinman J D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donly B C, Edgar C D, Adamski F M, Tate W P. Frameshift autoregulation in the gene for Escherichia coli release factor 2: partly functional mutants result in frameshift enhancement. Nucleic Acids Res. 1990;18:6517–6522. doi: 10.1093/nar/18.22.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doohan J P, Samuel C E. Biosynthesis of reovirus-specific polypeptides: ribosome pausing during the translation of reovirus S1 mRNA. Virology. 1992;186:409–425. doi: 10.1016/0042-6822(92)90006-b. [DOI] [PubMed] [Google Scholar]
- 13.Dotto G P, Enea V, Zinder N D. Functional analysis of bacteriophage f1 intergenic region. Virology. 1981;114:463–473. doi: 10.1016/0042-6822(81)90226-9. [DOI] [PubMed] [Google Scholar]
- 14.Du Z H, Holland J A, Hansen M R, Giedroc D P, Hoffman D W. Base-pairings within the RNA pseudoknot associated with the simian retrovirus −1 gag-pro frameshift site. J Mol Biol. 1997;270:464–470. doi: 10.1006/jmbi.1997.1127. [DOI] [PubMed] [Google Scholar]
- 15.Farabaugh P J. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farabaugh P J, Bjork G R. How translational accuracy influences frame maintenance. EMBO J. 1999;18:1427–1434. doi: 10.1093/emboj/18.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farabaugh P J. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog Nucleic Acid Res Mol Biol. 2000;64:131–170. doi: 10.1016/s0079-6603(00)64004-7. [DOI] [PubMed] [Google Scholar]
- 18.Gesteland R F, Atkins J F. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 19.Giedroc D P, Theimer C A, Nixon P L. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J Mol Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herr A J, Gesteland R F, Atkins J F. One protein from two open reading frames: mechanism of a 50 nt translational bypass. EMBO J. 2000;19:2671–2680. doi: 10.1093/emboj/19.11.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacks T, Madhani H D, Masiarz F R, Varmus H E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. Comparison of initiation of protein synthesis in prokaryotes, eukaryotes and organelles. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Stahl G, Farabaugh P J. Programmed +1 frameshifting stimulated by complementarity between a downstream mRNA sequence and an error-correcting region of rRNA. RNA. 2001;27:275–284. doi: 10.1017/s135583820100190x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liphardt J, Napthine S, Kontos H, Brierley I. Evidence for an RNA pseudoknot loop-helix interaction essential for efficient −1 ribosomal frameshifting. J Mol Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopinski J D, Dinman J D, Bruenn J A. Kinetics of ribosomal pausing during programmed −1 ribosomal frameshifting. Mol Cell Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marczinke B, Hagervall T, Brierley I. The Q-base of asparaginyl-tRNA is dispensable for efficient −1 ribosomal frameshifting in eukaryotes. J Mol Biol. 2000;295:179–191. doi: 10.1006/jmbi.1999.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melton D A, Krieg P A, Robagliati M R, Maniatis T, Zinn K, Green M R. Efficient in vitro synthesis of biologically active RNA and RNA hybridisation probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michiels P J A, Versleijen A A M, Verlaan P W, Pleij C W A, Hilbers C W, Heus H A. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J Mol Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montesano L, Glitz D G. Wheat germ cytoplasmic ribosomes. Structure of ribosomal subunits and localization of N6,N6-dimethyladenosine by immunoelectron microscopy. J Biol Chem. 1988;263:4932–4938. [PubMed] [Google Scholar]
- 31.Napthine S, Liphardt J, Bloys A, Routledge S, Brierley I. The role of RNA pseudoknot stem 1 length in the promotion of efficient −1 ribosomal frameshifting. J Mol Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russel M, Kidd S, Kelley M R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45:333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen L X, Tinoco I. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J Mol Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 36.Somogyi P, Jenner A J, Brierley I, Inglis S C. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su L, Chen L, Egli M, Berger J M, Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat Struct Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung D, Kang H. Mutational analysis of the RNA pseudoknot involved in efficient ribosomal frameshifting in simian retrovirus 1. Nucleic Acids Res. 1998;26:1369–1372. doi: 10.1093/nar/26.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suppmann S, Persson B C, Bock A. Dynamics and efficiency in vivo of UGA-directed selenocysteine insertion at the ribosome. EMBO J. 1999;18:2284–2293. doi: 10.1093/emboj/18.8.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szer W, Kurylo-Borowska Z. Effect of edeine on aminoacyl-tRNA binding to ribosomes and its relationship to ribosomal binding sites. Biochim Biophys Acta. 1970;224:477–486. doi: 10.1016/0005-2787(70)90580-0. [DOI] [PubMed] [Google Scholar]
- 41.Ten Dam E, Pleij C W A, Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ten Dam E, Brierley I, Inglis S C, Pleij C. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucleic Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ten Dam E, Verlaan P, Pleij C. Analysis of the role of the pseudoknot component in the SRV-1 gag-pro ribosomal frameshift signal: loop lengths and stability of the stem regions. RNA. 1995;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuchihashi Z. Translational frameshifting in the Escherichia coli dnaX gene in vitro. Nucleic Acids Res. 1991;19:2457–2462. doi: 10.1093/nar/19.9.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu C, Tzeng T-H, Bruenn J A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc Natl Acad Sci USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolin S L, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young J F, Desselberger U, Graves P, Palese P, Shatzman A, Rosenberg M. Cloning and expression of influenza virus genes. In: Laver W G, editor. The origin of pandemic influenza viruses. Amsterdam, The Netherlands: Elsevier Science; 1983. pp. 129–138. [Google Scholar]