Abstract
Vertebrate TOP mRNAs contain an oligopyrimidine tract at their 5′ termini (5′TOP) and encode components of the translational machinery. Previously it has been shown that they are subject to selective translational repression upon growth arrest and that their translational behavior correlates with the activity of S6K1. We now show that the translation of TOP mRNAs is rapidly repressed by amino acid withdrawal and that this nutritional control depends strictly on the integrity of the 5′TOP motif. However, neither phosphorylation of ribosomal protein (rp) S6 nor activation of S6K1 per se is sufficient to relieve the translational repression of TOP mRNAs in amino acid-starved cells. Likewise, inhibition of S6K1 activity and rpS6 phosphorylation by overexpression of dominant-negative S6K1 mutants failed to suppress the translational activation of TOP mRNAs in amino acid-refed cells. Furthermore, TOP mRNAs were translationally regulated by amino acid sufficiency in embryonic stem cells lacking both alleles of the S6K1 gene. Inhibition of mTOR by rapamycin led to fast and complete repression of S6K1, as judged by rpS6 phosphorylation, but to only partial and delayed repression of translational activation of TOP mRNAs. In contrast, interference in the phosphatidylinositol 3-kinase (PI3-kinase)-mediated pathway by chemical or genetic manipulations blocked rapidly and completely the translational activation of TOP mRNAs. It appears, therefore, that translational regulation of TOP mRNAs, at least by amino acids, (i) is fully dependent on PI3-kinase, (ii) is partially sensitive to rapamycin, and (iii) requires neither S6K1 activity nor rpS6 phosphorylation.
The synthesis of many mammalian proteins associated with the translational apparatus has been shown in recent years to be selectively regulated in a growth-dependent manner at the translational level. The corresponding mRNAs are characterized by the presence of a 5′-terminal oligopyrimidine tract (5′TOP) and therefore are referred to as TOP mRNAs. This structural motif comprises the core of the translational cis-regulatory element of these mRNAs (reviewed in references 35 and 39). The proportion of TOP mRNAs actively engaged in protein synthesis, i.e., the proportion associated with polysomes in a wide variety of growing mammalian cells, is significantly lower than that characteristic of other ubiquitous mRNAs (1, 3, 20, 37, 41, 58). On average, only about 70% of TOP mRNAs are engaged with ribosomes compared with about 90% for the other housekeeping mRNAs. This selective repressed translation of TOP mRNAs becomes even more pronounced in cells that cease to divide, where only ∼30% of the TOP mRNAs remain in polysomes compared with >80% for non-TOP mRNAs (36).
Phosphorylation of ribosomal protein S6 (rpS6) is one of the earliest events detected following mitogenic stimuli. This phosphorylation is carried out by two closely related kinases, S6K1 (also known as p70 S6 kinase or p70S6K) and S6K2 (reviewed in reference 17). Several studies have shown that mitogenic stimulation of quiescent cells induces activation of S6K1 and consequently phosphorylation of rpS6. The concomitant translational activation of TOP mRNAs under these circumstances led Thomas and his colleagues to propose that rpS6 phosphorylation following S6K1 activation increases the affinity of ribosomes for TOP mRNAs and thus facilitates translation initiation (23, 24, 66). It should be noted, however, that this model, although supported by several correlative studies (39), has remained purely speculative. Thus, neither the involvement of rpS6 in the translational control of TOP mRNAs nor its being the only physiological substrate of S6K1 has been experimentally proven.
Study of rpS6 phosphorylation in rat liver revealed that this protein is phosphorylated not only following mitogenic stimulation but also upon the refeeding of starved animals (30). The importance of amino acids in this nutritional stimulation has been demonstrated in vitro with hepatocytes isolated from starved rats. Supplementing these cells with a complete mixture of amino acids led to phosphorylation of rpS6 which could be abolished by the mTOR-specific inhibitor rapamycin (6). This observation has demonstrated that amino acids, independent of insulin or growth factors, can regulate the activity of S6K1 through an mTOR-mediated mechanism. Indeed, recent studies with various cell lines have indicated that S6K1 activity is rapidly modulated (within 15 min) by deprivation of amino acids or their reintroduction (16, 18, 45, 69, 72). Moreover, withdrawal from cells of amino acids, but not glucose, renders S6K1 refractory to stimulation by insulin (8, 18), epidermal growth factor, and nerve growth factor (29). Detailed analysis of the involvement of individual amino acids in this mode of regulation has established a critical role for branched amino acids (72) or even leucine alone (28). Regulation of S6K1 by amino acid sufficiency is mediated by the loss of aminoacylated tRNA (21). The apparent correlation between S6K1 activity and the translational efficiency of TOP mRNAs has prompted us to monitor the effect of amino acid starvation on the translational behavior of TOP mRNAs and to study the role of rpS6 phosphorylation, as well as the activities of S6K1 and mTOR, in the translational control of these mRNAs.
The present results show that TOP mRNAs are translationally repressed shortly after amino acid withdrawal and rapidly derepressed after amino acid readdition and that this mode of regulation is strictly dependent on the integrity of the 5′TOP motif. This nutritional regulation of TOP mRNA translation occurs in an rpS6 phosphorylation-independent fashion as well as an S6K1-independent fashion, is partially repressed by rapamycin, and is fully dependent on signaling through PI3-kinase.
MATERIALS AND METHODS
Cell culture and DNA transfection.
Human embryonic kidney 293 cells were grown in 100-mm-diameter plates and transfected as described previously (19). R1 mouse embryonic stem (ES) cells and their S6K1-deficient derivative cells (p70S6K−/−) were kindly provided by Naohiro Terada (26). Both ES cell lines were grown on gelatin-coated plates in Dulbecco modified Eagle medium–F-12 (Ham) (1:1) supplemented with 15% heat-inactivated fetal calf serum, 20 pg of leukemia inhibitory factor (GIBCO), minimal essential medium (MEM)-nonessential amino acids, ribo- and deoxyribonucleosides, 1 mM pyruvate, 0.1 mM β-mercaptoethanol, 100 U of penicillin/ml, 0.1 mg of streptomycin/ml, and 0.12% NaHCO3. Cells were starved for amino acids by removal of the growth medium, washing once with 5 ml of phosphate-buffered saline, and incubation for the indicated time in a medium containing Earle's salt solution, MEM-Eagle vitamin solution, 0.37% NaHCO3, 10% dialyzed fetal calf serum, 100 U of penicillin/ml, and 0.1 mg of streptomycin/ml. Cells were refed by replacing the amino acid-free medium with normal growth medium. When used, rapamycin (Sigma) and LY-294002 (Sigma) were added (20 nM and 50 μM, respectively) upon reintroduction of amino acids, and calyculin A (Sigma) was added (20 nM) upon removal of amino acids for times indicated in Fig. 4.
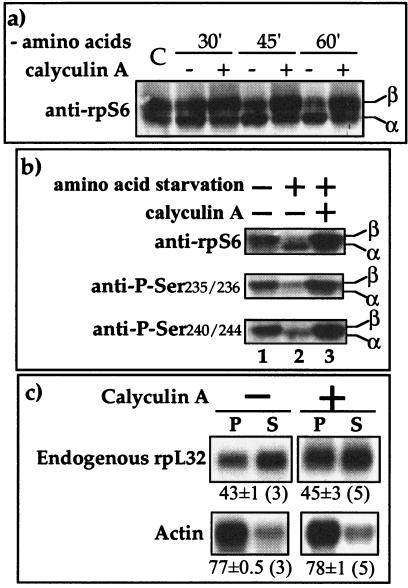
FIG. 4.
Calyculin A induces phosphorylation of rpS6 but fails to relieve the translational repression of TOP mRNAs. (a) 293 cells were either untreated (C) or amino acid starved without (−) or with (+) calyculin A (20 nM) for the indicated time, after which cells were harvested. The cytoplasmic proteins were subjected to Western blot analysis using anti-rpS6. α and β, unphosphorylated and hyperphosphorylated forms of rpS6, respectively. (b) 293 cells were untreated or amino acid starved for 1 h without (−) or with (+) calyculin A (20 nM), after which cells were harvested. The cytoplasmic proteins were subjected to Western blot analysis using the indicated antibodies. (c) 293 cells were amino acid starved for 1 h without (−) or with (+) calyculin A (20 nM) and then harvested, and the polysomal distribution of the mRNAs encoding rpL32 and actin was analyzed and presented as described in the legend to Fig. 2.
Cell sorting.
Cells expressing an enhanced green fluorescent protein-based construct were trypsinized 24 h posttransfection, resuspended in Dulbecco modified Eagle medium, excited at 488 nm by an argon laser beam in FACStarplus (Becton Dickinson), and sorted according to the fluorescence emission at 507 nm. Sorted cells were replated and harvested 24 h later.
Polysomal fractionation and RNA analysis.
One 100-mm-diameter plate containing a monolayer culture was used for polysomal analysis. Harvesting was performed as described previously (38). Cell pellets were thawed in 150 μl of RSB (10 mM NaCl, 10 mM Tris-HCl [pH 7.4], 15 mM MgCl2) containing 100 μg of heparin/ml and lysed in 1.2% Triton X-100–1.2% deoxycholate by brief mixing (3 s on a Vortex mixer) before and after 3 min of incubation on ice. Nuclei were pelleted by centrifugation for 2.5 min in a microcentrifuge at 4°C. The postnuclear supernatant was diluted with an equal volume of polysomal buffer (25 mM Tris-HCl [pH 7.4 to 7.5 at 4°C], 10 mM MgCl2, 25 mM NaCl, 0.05% Triton X-100, 0.14 M sucrose, 500 μg of heparin per ml). A 300-μl portion of this suspension was layered over 11.5 ml of a 15 to 45% (wt/wt) sucrose gradient with a 0.6-ml cushion of 45% sucrose. The sucrose solutions were prepared as described previously (38). The gradients were centrifuged at 40,000 rpm for 120 min at 4°C in a Kontron TST 41 or Beckman SW 41 swing-out rotor. After centrifugation, the A260 was continuously monitored and recorded by PC-multiLab card (Advantech Co.) attached to a Spectronic 601 (Bausch & Lomb) spectrophotometer. The gradients were divided into two fractions: polysomal, which included mRNAs loaded with two (disomes) or more ribosomes, and subpolysomal, which contained monosomes, ribosomal subunits, and mRNA ribonucleoproteins. RNA was extracted from each fraction by Ultraspec RNA (Biotecx Laboratories, Houston, Tex.) or EZ-RNA (Biological Industries) according to the suppliers' instructions. Plasmid DNA was eliminated by incubating the RNA in 100 μl of DNase I buffer (0.1 M sodium acetate, 5 mM MgSO4, pH 5.0) containing 10 U of RNase-free DNase (Boehringer) for 60 min at 37°C. Northern blot analysis was performed as described previously (41). Quantification of the radioactive signals on the blots was carried out by a bioimaging analyzer (Fujix BAS 1000; Fuji). To assess the effectiveness of the amino acid starvation and the selectivity of the effect on TOP mRNAs, we compared in each case the polysomal association of a chimeric mRNA with that of endogenous rp mRNA and non-rp mRNA from the same polysomal gradient. Accurate quantitative comparisons of translational efficiencies under different experimental conditions should be made using the average numerical values presented next to each autoradiogram rather than on the basis of the visible intensities of signals in the sample autoradiograms.
Plasmid constructions.
Standard protocols were used for all recombinant DNA technology (54). L32-green fluorescent protein (GFP) and Act-GFP were constructed by insertion of a 1,052-bp NcoI-SpeI fragment containing the GFP coding and 3′ flanking sequences from pEGFP-C1 (Clontech) into the SmaI sites of pL32a (3) and pAct (40), respectively. Kinase-dead RSK2 mutant pK3H.RSK2(K100A) (a gift from Celeste E. Poteet-Smith, University of Virginia) was generated from the parent vector, pK3H.RSK2, by PCR.
The structures of all constructs described here were confirmed by DNA sequencing.
Molecular probes.
The isolated fragment probes used in the Northern blot analysis were a 0.97-kb fragment bearing rpL32-processed gene 4A (11), a 1.15-kb PstI fragment containing mouse α-actin cDNA (42), a 0.62-kb PstI fragment containing human superoxide dismutase-I (SOD) cDNA (60), a 0.8-kb HindIII fragment containing human growth hormone (hGH) cDNA (kindly provided by T. Fogel, Bio-Technology General), and a 0.74-kb NcoI-HindIII fragment containing GFP cDNA from pEGFP-C1 (Clontech).
Preparation and specificity of anti-rpS6 antibodies.
The phosphospecific (Ser235/236 and Ser240/244) and total rpS6 antibodies directed against sites in the phosphoregion of human rpS6 were produced by immunizing New Zealand White rabbits with synthetic peptides. The following peptides, coupled to keyhole limpet hemocyanin, were used: Ser235/236(P) (CRRLSPSPLRASTSKSES), Ser240/244(P) (CRRLSSLRASPTSKSPES), and unphosphorylated rpS6 with the same peptide in its unphosphorylated state. Immunoglobulin G was purified using protein A-Sepharose. For determining phosphospecificity, antibodies reactive with the nonphosphopeptide were removed by adsorption to a nonphosphopeptide affinity column. Antibodies that flowed through this column were next passed over a column of immobilized phosphopeptide; after the column was washed, antibodies were eluted at low pH and dialyzed. For total rpS6, antibodies reactive with the nonphosphopeptide column were collected by adsorption to a nonphosphopeptide affinity column. Because the antibodies that bound this column failed to recognize phosphorylated rpS6, phospho-specific (Ser235/236) antibodies were combined with these non-phospho-specific antibodies to produce the total rpS6 antibody. Analysis of the phosphospecificity of the resulting antibodies was performed by (i) enzyme-linked immunosorbent assay against the phosphopeptides and nonphosphopeptide, (ii) immunoblotting against whole-cell extracts from serum-starved and serum-refed cells, and (iii) by preincubation of each antibody with synthetic peptides before being exposed to the immunoblotting membranes. The competing peptides in this assay were unphosphorylated, singly phosphorylated (Ser236 and Ser240), doubly phosphorylated (Ser235/236 and Ser240/244), and hyperphosphorylated (Ser235/236/240/244/247). The results of all these tests indicate that anti-total-rpS6 detects unphosphorylated rpS6 and singly (Ser236) phosphorylated, doubly phosphorylated (at Ser235 and Ser236), and hyperphosphorylated (at all five sites) rpS6. Anti-phospho-rpS6 (Ser235/236) and anti-phospho-rpS6 (Ser240/244) react efficiently only with the respective doubly phosphorylated epitope, independent of other phosphorylation sites, partly with the singly phosphorylated epitope, and not at all with the singly or doubly phosphorylated heterologous epitope, the nonphosphorylated epitope, or any other protein in cytoplasmic extract from 293 cells. Finally, immunofluorescence analyses have verified the cytoplasmic location of the respective antigens.
Western blot analysis.
Immunoblotting was performed as described previously (44) using anti-rpS6, anti-phospho-rpS6 (Ser235/236), or anti-phospho-rpS6 (Ser240/244) (Cell Signaling Technology) and monoclonal antibody 12CA5 against the hemagglutinin (HA) antigen.
Immunoprecipitation and kinase assay.
Cells were lysed and extracts were clarified as described previously (44). HA-tagged S6K1 and RSK2 were immunoprecipitated with 2 μg of anti-HA antibody as described previously (44). The immunoprecipitated proteins were used for an S6 kinase assay using 40S ribosomal subunits (44). Proteins were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) and transferred onto a Protran BA 85 nitrocellulose membrane (Schleicher & Schuell), and the radiolabeled S6 was quantified with a bioimaging analyzer (Fuji).
RESULTS
TOP mRNAs are translationally repressed by amino acid starvation.
Recent reports on the inactivation of S6K1 by amino acids have prompted us to examine whether this treatment also leads to translational repression of TOP mRNAs. Figure 1a shows that a typical TOP mRNA encoding rpL32 was translationally repressed within 1 h of withdrawal of amino acids from the growth medium of 293 cells, as judged by the shift of this mRNA from mostly polysomal fractions (fractions 1 to 8) to mostly subpolysomal fractions (fractions 9 to 12). Non-TOP mRNAs encoding SOD, although slightly unloaded from heavy polysomes, remained mostly engaged in translation under these circumstances. The translational efficiency of rpL32 mRNA could be fully recovered (or could even exceed that measured in untreated cells) by 0.5 h of amino acid refeeding of cells starved for up to 2 h, yet recruitment of this mRNA is slower after 4 h (Fig. 1b). It is noteworthy that the derepression of TOP mRNAs is often characterized by the overshooting of the translational efficiency (1, 59).
FIG. 1.
Amino acid deficiency induces reversible translational repression of TOP mRNAs. (a) Untreated 293 cells (Control) and 293 cells starved of amino acids for 1 h [−AA (1 h)] were harvested, and cytoplasmic extracts were prepared. These extracts were centrifuged through sucrose gradients and separated into 12 fractions. RNA isolated from these fractions was applied to Northern blot analysis and hybridized with labeled cDNAs encoding SOD and rpL32. The radioactive signals were quantified by phosphorimager, and the results for each fraction are presented as the percentage of total mRNA (dashed lines separate the polysomal fractions [left] and the subpolysomal fractions [right]). For each treatment the same RNA preparations were hybridized with the different probes. The shaded areas depict the profiles of optical density at 260 nm for polysomal and subpolysomal fractions. (b) Untreated 293 cells and cells starved of amino acids for the indicated times or starved and then refed for 0.5 h were harvested, and cytoplasmic extracts were prepared. These extracts were centrifuged through sucrose gradients and separated into two fractions: polysomal and subpolysomal. RNA isolated from these fractions was subjected to Northern blot analysis and hybridized with labeled cDNAs encoding SOD and rpL32. The radioactive signals were quantified by phosphorimager, and the relative amounts of the mRNAs in polysomes are shown. Dashed lines, changes in polysomal association following 0.5 h of refeeding. Vertical bars, standard errors of the means.
Translational repression by amino acid starvation is mediated by the 5′TOP motif.
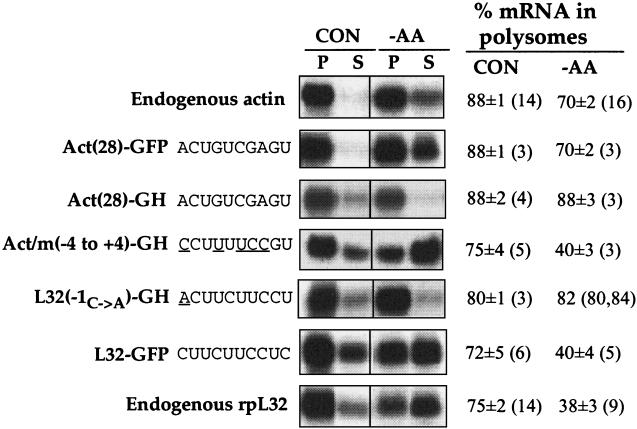
Growth-dependent translational control of TOP mRNAs is strictly dependent on the integrity of the 5′TOP sequence and its location at the 5′ end of the mRNA. To examine whether these structural constraints are also applicable to the amino acid-mediated translational regulation, we monitored the translational behavior of various chimeric mRNAs. L32-GFP mRNA, which starts with the 10-nucleotide (nt) 5′TOP of rpL32, was translationally repressed upon 1 h of amino acid starvation to the same extent as endogenous rpL32 mRNA (Fig. 2). In contrast, L32(−1C→A)-GH, in which the 5′TOP motif of rpL32 is preceded by an A residue (3), was refractory to amino acid starvation as were endogenous actin mRNA and the chimeric Act-GFP and Act-GH mRNAs, which lack a 5′TOP motif (Fig. 2). Replacement of 5 nt at the 5′ terminus of Act-GH mRNA, so that the resulting Act/m(−4 to +4)-GH mRNA starts with a typical 8-nt 5′TOP motif (5), rendered the resulting mRNA translationally repressed by amino acid starvation (Fig. 2).
FIG. 2.
The 5′TOP motif plays a critical role in the translational repression of TOP mRNAs upon amino acid starvation. 293 cells were transfected with 2 μg of the indicated chimeric GH or GFP constructs together with 16 μg of an empty vector. Cytoplasmic extracts were prepared about 24 h later from untreated (CON) or cells starved of amino acids for 1 h (−AA). These extracts were centrifuged through sucrose gradients and separated into polysomal (P) and subpolysomal (S) fractions. RNA from equivalent aliquots of these fractions was analyzed by Northern blot hybridization with hGH or GFP cDNAs for detection of the chimeric transcripts and cDNAs for L32 and actin for detection of the corresponding endogenous mRNAs. The radioactive signals were quantified by phosphorimager, and the relative translational efficiency of each mRNA is numerically presented at the right as a percentage of the mRNA engaged in polysomes. These figures are averages ± standard errors of the means of the number of determinations in parentheses or are the averages with the individual values in parentheses if only two determinations are available. The first 10 nt in the respective chimeric construct are indicated at the left, and the underlined letters represent nucleotides mutated relative to the wild-type sequence.
rpS6 phosphorylation is insufficient to relieve the translational repression of a TOP mRNA in amino acid-starved 293 cells.
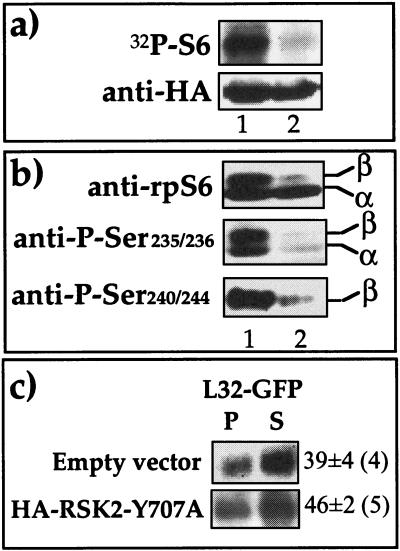
To examine whether phosphorylated rpS6 per se is sufficient to relieve the translational repression of TOP mRNA, we set out to induce phosphorylation of this protein in an S6K-independent fashion. p90 ribosomal protein S6 kinase (RSK2) was originally isolated due to its ability to phosphorylate rpS6 (15) with the same specificity as does S6K1 (63). Hence, we cotransfected 293 cells with expression vectors encoding L32-GFP and HA-tagged constitutively active RSK2 mutant pK3H.RSK2(Y707A). The latter has been shown to exhibit about fourfold-higher basal activity than the wild type (48). The results indeed show that overexpression of this kinase, unlike that of kinase-dead mutant [pK3H.RSK2(K100A)], led to the production of an active rpS6 kinase in amino acid-starved cells, as measured in vitro following its immunoprecipitation by an anti-HA antibody (Fig. 3a). Furthermore, Western blot analysis with antibodies raised against four phosphorylated serine residues (235, 236, 240, and 244) disclosed that all of them are phosphorylated in vivo by the active RSK2 variant (Fig. 3b). Nevertheless, overexpression of RSK2 was unable to relieve the translational repression of L32-GFP, as most of it remained in the subpolysomal fraction (Fig. 3c). It appears, therefore, that rpS6 phosphorylation per se is insufficient for translational activation of TOP mRNAs.
FIG. 3.
Overexpression of RSK2 led to phosphorylation of rpS6 but failed to relieve the translational repression of L32-GFP mRNA in amino acid-starved cells. (a) 293 cells were transiently transfected with 16 μg of pHA-K3H.RSK2(Y707A) (lane 1) or its inactive counterpart, pHA-K3H.RSK2(K100A) (lane 2). Twenty-four hours later transfectants were starved of amino acids for 1 h and harvested, and their cytoplasmic proteins were used either for Western blot analysis with anti-HA or for assaying the activity of RSK2 following immunoprecipitation with an anti-HA antibody. In the latter case the reaction mixture was separated by SDS-PAGE, transferred onto nitrocellulose membrane, and subjected to autoradiography (32P-S6). (b) In a parallel experiment, 293 cells were similarly transfected with 16 μg of one of the expression vectors encoding HA-K3H.RSK2(Y707A) (lane 1) or HA-K3H.RSK2(K100A) (lane 2). Twenty-four hours later cells were starved for 1 h and harvested, and their cytoplasmic proteins were subjected to Western blot analysis using anti-rpS6 and anti-phospho-Ser235/236 or anti-phospho-Ser240/244. α and β, hypophosphorylated and hyperphosphorylated forms of rpS6, respectively. (c) 293 cells were cotransfected with 2 μg of a vector encoding L32-GFP and 16 μg of the expression vector indicated at the left. Twenty-four hours later cells were starved for 1 h and harvested, and the polysomal distribution of L32-GFP mRNA was analyzed and presented as described in the legend to Fig. 2.
Activation of endogenous S6K1 failed to relieve the translational repression of TOP mRNAs in amino acid-starved 293 cells.
A possible explanation for the failure of RSK2 to derepress the translation of L32-GFP mRNA might be the requirement for S6K1 activity rather than just rpS6 phosphorylation. To address this possibility, we employed calyculin A, which has previously been shown to prevent inhibition of S6K1 by amino acid starvation or osmotic stress (44, 46). Indeed, dephosphorylation of rpS6 became apparent 45 min after amino acid withdrawal (Fig. 4a), yet calyculin A was able to protect rpS6 in its phosphorylated state (at serine residues 235, 236, 240, and 244) even 60 min after amino acid withdrawal (Fig. 4a and b). However, it failed to prevent the translational repression of mRNAs encoding rpL32 (Fig. 4c) and the chimeric L32-GFP mRNA (data not shown). These results suggest that neither S6K1 activity nor rpS6 phosphorylation is sufficient for translational activation of TOP mRNAs in amino acid-starved cells.
Translational activation of a TOP mRNA in amino acid-refed 293 cells requires neither S6K1 activation nor rpS6 phosphorylation.
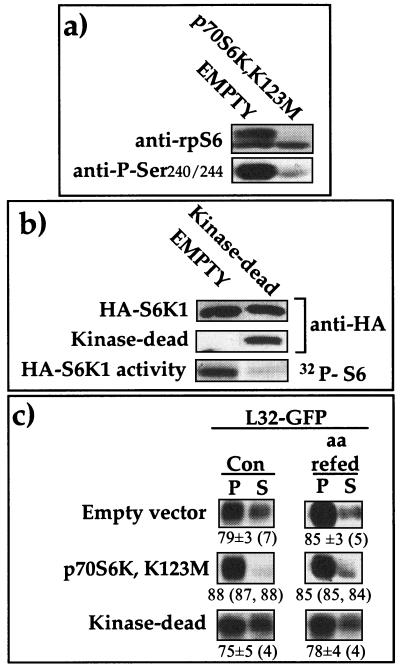
It has previously been reported that activation of S6K1 by serum stimulation can be completely abolished by overexpression of dominant-negative mutants. These include p70S6KA229, in which T229 (T252 according to the numbering system used here) within the activation loop was replaced by alanine, and another mutant in which the lysine in the ATP-binding pocket was replaced (22). Hence, we set out to examine whether overexpression of p70S6K,K123M, an S6K1 polypeptide mutated at the ATP-binding site, can interfere with the translational activation of L32-GFP mRNA in untreated or amino acid-refed cells. Overexpression of this mutant abolished the phosphorylation of rpS6, as measured in transfected cells sorted by fluorescence-activated cell sorter (Fig. 5a), using two different antibodies. Likewise, overexpression of another S6K1 mutant, p70Δ2-46/ΔCT104,K123 M/T412E, which lacks the amino and carboxy termini in addition to having mutations at position 412 and at the ATP-binding site, completely inhibited the S6K1 activity of cotransfected wild-type HA-tagged S6K1 (Fig. 5b). Nevertheless, despite their dominant-negative effect, none of these kinase-dead mutants impaired the recruitment of L32-GFP mRNA into polysomes in untreated or in amino acid-refed cells (Fig. 5c). Hence, it appears that neither S6K1 activity nor rpS6 phosphorylation is necessary for efficient translation of TOP mRNAs
FIG. 5.
Overexpression of kinase-dead S6K1 mutants inhibited S6K activity but failed to suppress the translational activation of L32-GFP mRNA. (a) 293 cells were transiently cotransfected with 1 μg of HA-p70S6K1 together with 1 μg of pEGFP-C1 and 16 μg of either a vector encoding the HA epitope (EMPTY) or a vector encoding HA-tagged p70S6K,K123M. Twenty-four hours later, highly fluorescent cells were sorted by fluorescence-activated cell sorter and reseeded. Forty-eight hours posttransfection cells were harvested, and cytoplasmic proteins were subjected to Western blot analysis using anti-rpS6 and anti-phospho-Ser240/244 antibodies. (b) 293 cells were transiently transfected with 1 μg of HA-S6K1, 1 μg of pEGFP-C1, and 16 μg of either a vector encoding HA (EMPTY) or a vector encoding HA-tagged p70Δ2-46/ΔCT104, K123M/T412E (Kinase-dead). Twenty-four hours later cells were starved for 2 h and then refed for 0.5 h and harvested. Cell extracts were subjected to immunoprecipitation with an anti-HA antibody, and a portion was assayed for S6 kinase activity. The reaction mixture was separated by SDS-PAGE and transferred onto a nitrocellulose membrane, and the radioactive signals of 32P-S6 were autoradiographed (HA-S6K1 activity). Immunoblotting of the membrane with an anti-HA antibody enabled the detection of the wild-type S6K1 (HA-S6K1) and of the shorter kinase-dead (p70Δ2-46/ΔCT104, K123M/T412E) variant (Kinase-dead). (c) 293 cells were transfected with 2 μg of a vector encoding L32-GFP and 16 μg of a vector encoding the HA tag, p70S6K,K123M, and p70Δ2-46/ΔCT104, K123M/ T412E (Kinase-dead). Twenty-four to 36 h later cells were harvested untreated (Con) or after 2 h of amino acid starvation followed by 0.5 h of refeeding (aa refed). The translational efficiency of L32-GFP mRNA was analyzed and is presented as described in the legend to Fig. 2.
TOP mRNAs are translationally regulated in S6K1−/− ES cells by amino acid sufficiency.
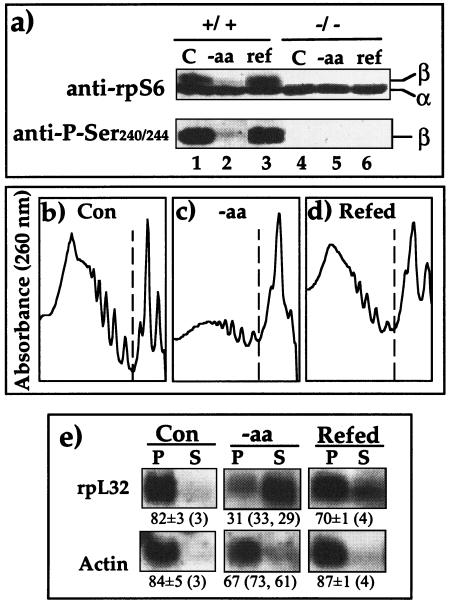
Based on the widely accepted dogma of the causal relationships between S6K1 activity and S6 phosphorylation on the one hand and the translational efficiency of TOP mRNAs on the other hand, it can be argued that our results with 293 cells simply reflect a distorted regulatory mechanism in this cell line. To directly address this issue and to avoid relying on transformed cells with a poorly characterized karyotype, chemical inhibitors, or overexpression of foreign proteins, we utilized a mouse diploid ES cell line, p70S6K−/−, in which both alleles of S6K1 were disrupted by homologous recombination (26). Figure 6a shows that rpS6 in p70S6K−/− cells, unlike that in the parental cells (R1), is constitutively unphosphorylated (as demonstrated by two different antibodies), regardless of the sufficiency of amino acids. These results corroborate those originally obtained for this cell line (26) but are inconsistent with a later report (32). The shift of ribosomes from a polysomal fraction into a subpolysomal fraction upon amino acid starvation (Fig. 6b to d) reflects a modest inhibition of global translation activity in p70S6K−/− ES cells, as was shown for 293 cells (Fig. 1a). Likewise, the translation of rpL32 mRNA is efficient in p70S6K−/− cells when amino acids are provided, selectively repressed upon 2 h of amino acid starvation, and upregulated upon 1 h of refeeding (Fig. 6e). These results substantiate the conclusion that translational control of TOP mRNAs by amino acid sufficiency does not involve S6K1 activity or rpS6 phosphorylation.
FIG. 6.
TOP mRNAs are translationally regulated by amino acid sufficiency in ES cells lacking S6K activity. (a) Wild type ES cells (+/+) and p70S6K−/− cells (−/−) were harvested untreated (C) or after being amino acid starved for 2 h (−aa) or starved and then refed for 1 h (ref). The cytoplasmic proteins were subjected to Western blot analysis using the indicated antibodies (the results represent two independent experiments with identical results). (b to d) p70S6K−/− cells were treated as described in for panel a and then harvested, and the profiles of optical density at 260 nm are presented (dashed lines separate the polysomal fractions [left] and the subpolysomal fractions [right]). (e) The polysomal distribution of mRNAs encoding rpL32 and actin was analyzed and presented as described in the legend to Fig. 2.
The translation of TOP mRNAs is partially inhibited by rapamycin.
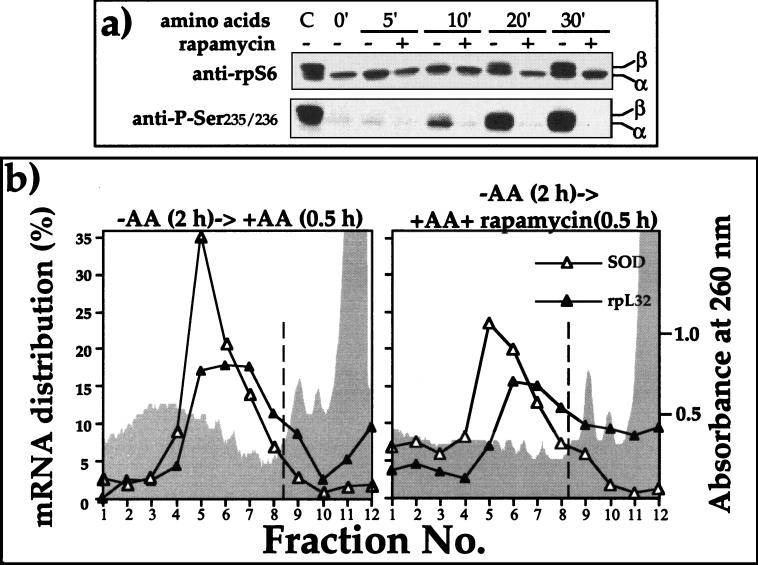
Previous reports have demonstrated that mTOR is involved in the regulation of S6K1 activity by amino acid sufficiency (18, 21). To examine the possible role of mTOR in the translational activation of TOP mRNAs, we monitored the effect of rapamycin on amino acid-induced recruitment of a TOP mRNA into polysomes. It has been previously shown that the inhibitory effect of rapamycin on S6K1 activity is very rapid, with a half time of about 2 min (10). Indeed, Fig. 7a shows that the inhibitory effect of rapamycin on phosphorylation of Ser235/236 is evident at the earliest time point at which rpS6 phosphorylation can be observed (10 min) following amino acid refeeding. Notably, rpS6 has been reported to be phosphorylated in an ordered fashion: Ser236→235→240→244→247 (17). Hence, failure to detect phosphorylation of rpS6 at Ser235/236 (Fig. 7a and 9a) indicates that the protein is unphosphorylated. Nevertheless, despite complete blocking of rpS6 phosphorylation, rapamycin had only a minor effect on the amino acid-induced translational activation of rpL32 mRNA (Fig. 7b). Thus, the peak of polysomal rpL32 mRNA coincides with mRNAs loaded with two to five ribosomes in the presence of rapamycin or two to six ribosomes in its absence (Fig. 7b). However, averaging the results of this and additional experiments (Fig. 8b) clearly shows that 20 nM rapamycin failed to block the recruitment of rpL32 mRNA into polysomes upon amino acid refeeding (78 and 74% in polysomes in the absence and presence of rapamycin, respectively).
FIG. 7.
Rapamycin inhibits phosphorylation of rpS6, but does not prevent translational activation of rpL32. (a) 293 cells were untreated (C), amino acid starved for 2 h (0′), or refed with amino acids for the indicated times without (−) or with (+) rapamycin (20 nM) for the indicated times, after which cells were harvested. The cytoplasmic proteins were subjected to Western blot analysis using the indicated antibodies. (b) 293 cells were amino acid starved for 2 h and then refed without (left) or with (right) rapamycin (20 nM) and then harvested. The distribution of the mRNAs encoding rpL32 and SOD among 12 sucrose gradient fractions was analyzed and is presented as described in the legend to Fig. 1a. The shaded areas depict the profiles of optical density at 260 nm of polysomal and subpolysomal fractions.
FIG. 9.
LY-294002 inhibits both the phosphorylation of rpS6 and the translational activation of TOP mRNAs upon amino acid refeeding. (a) 293 cells were untreated (C), amino acid starved for 2 h (0′), or refed with amino acids without (−) or with (+) LY-294002 (50 μM) for the indicated times, after which cells were harvested. The cytoplasmic proteins were subjected to Western blot analysis using the indicated antibodies. (b) 293 cells were untreated or amino acid starved for 2 h and then refed for 30 min without (−) or with (+) LY294002 (50 μM). The polysomal distribution of rpL32 mRNA was analyzed and is presented as described in the legend to Fig. 2.
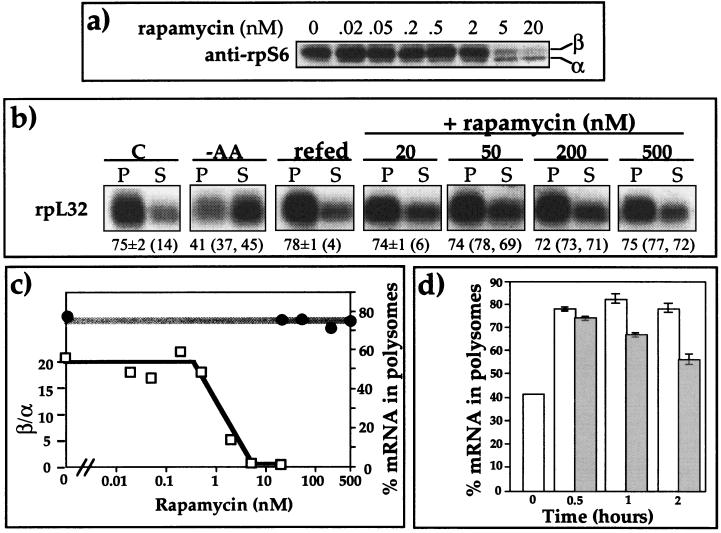
FIG. 8.
Translational activation of rpL32 mRNA is far more resistant to rapamycin than rpS6 phosphorylation. (a) 293 cells were amino acid starved for 2 h and then refed for 30 min in the absence or presence of rapamycin at the indicated concentrations. Subsequently, the cytoplasmic proteins were subjected to Western blot analysis using an anti-rpS6 antibody. (b) 293 cells were untreated (C), amino acid starved for 2 h (−AA), or amino acid starved for 2 h and then refed for 30 min without rapamycin (refed) or with the indicated concentrations of rapamycin. The polysomal distribution of rpL32 mRNA was analyzed and presented as described in the legend to Fig. 2. (c) Dose-response curves of the effects of rapamycin on rpS6 phosphorylation and on translational efficiency of rpL32 mRNA. The chemiluminescence signals of the hypophosphorylated and phosphorylated bands, α and β, respectively, whose images appear in panel a, were quantified by the Chemi Doc system (Bio-Rad). The relative phosphorylation of rpS6 is presented numerically as the β/α ratio. The translational efficiency of rpL32 mRNA is presented as the percentage of mRNA associated in polysomes. (d) Kinetics of the effect of rapamycin on the translational efficiency of rpL32 mRNA. 293 cells were amino acid starved for 2 h (time zero) and then refed in the absence (white bars) or presence (gray bars) of rapamycin for the indicated times. Rapamycin was simultaneously added with amino acids when cells were refed for 0.5 and 2 h or for 15 min prior to addition of the amino acids when the latter were added for 1 h. The polysomal distribution of rpL32 mRNA was analyzed as described in the legend to Fig. 2. The percentage of mRNA in polysomes is presented as an averages ± standard errors of the means for three experiments.
To further verify the relationships between the effect of rapamycin on rpS6 phosphorylation and its effect on the translational efficiency of TOP mRNAs, we established dose-dependent curves. To this end, 293 cells were starved for 2 h and then refed with amino acids for 0.5 h in the presence of increasing concentrations of rapamycin. The results show that the relative phosphorylation of rpS6 is inhibited with a 50% inhibitory concentration of between 0.5 and 2 nM rapamycin (Fig. 8a and 8c), which is consistent with that reported for S6K1 activity (49). However, the translational activation of rpL32 mRNA, as judged by its percentage in the polysomal fraction, remained unaffected even at a dose of 500 nM rapamycin (Fig. 8b and 8c).
Our results show, therefore, that S6K1 inhibition by rapamycin, and consequently of rpS6 phosphorylation, cannot prevent translational activation of TOP mRNAs within the monitoring period of 30 min from amino acid readdition. However, it has previously been shown, for some cell lines, that translational activation of TOP mRNAs was efficiently blocked if rapamycin was present during the 2 to 4 h of mitogenic stimulation (26, 62, 64, 65). Hence, although rapamycin exerts its inhibitory effect quite rapidly on mTOR, and consequently on S6K1 activity and rpS6 phosphorylation, it might repress the amino acid-induced translational activation of TOP mRNAs through a distinct mechanism which operates much more slowly. To examine this possibility, we applied this drug for a longer duration. Figure 7d shows that recruitment of L32 mRNA into polysomes was indeed partially inhibited if cells were treated with rapamycin for 75 or 120 min (67 and 56% in polysomes, respectively). It should be noted, however, that even after 2 h of rapamycin treatment (unlike 2 h of amino acid starvation) most of the rpL32 mRNA was associated with polysomes. It appears therefore, that rapamycin can rapidly (<10 min) and fully inhibit the activation of S6K1 and the phosphorylation of rpS6 in amino acid-stimulated cells, whereas the repression of the translational activation of TOP mRNAs under these conditions is delayed (>30 min) and is only partial.
PI3-kinase-mediated signaling is required for the amino acid-induced translational activation of TOP mRNAs.
The moderate and delayed inhibitory effect of rapamycin on the amino acid-induced activation of TOP mRNA translation prompted us to look for an alternative or an additional pathway which might mediate this activation. It has previously been shown that activation of S6K by amino acids can be blocked by inhibiting PI3-kinase (16, 45, 61). Hence, we measured the effect of LY294002, a specific inhibitor of PI3-kinase (67), on the translational activation of rpL32 mRNA in amino acid-refed cells. Figure 9a indeed shows that LY294002, like rapamycin, completely blocks the phosphorylation of rpS6 but that, unlike rapamycin, LY294002 also totally inhibits the translational activation of rpL32 mRNA (Fig. 9b).
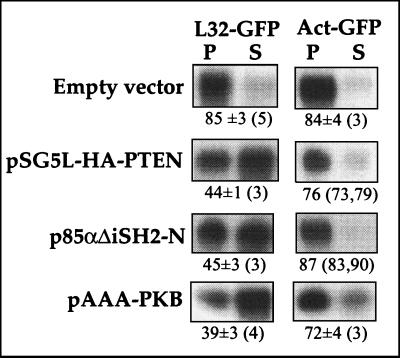
Conceivably, LY294002 exerts its suppressive effect on the translational activation of TOP mRNA by inhibiting the accumulation of phosphoinositides phosphorylated at the 3 position. To further explore this possibility, we utilized a complementary experimental approach based on the overexpression of (i) the tumor suppressor PTEN, which dephosphorylates PI-3,4,5-triphosphate and PI-3,4-diphosphate (9), or (ii) a dominant-negative mutant version of the PI3-kinase regulatory subunit, p85, which lacks the binding site for the catalytic subunit, p110. Figure 10 shows that overexpression of pSG5L-HA-PTEN (50), which encodes HA-tagged PTEN, suppressed the translational activation of the coexpressed L32-GFP mRNA in amino acid-refed cells. Likewise, translational activation of this mRNA was suppressed by coexpression of the dominant-negative p85, p85 ΔSH2-N (52). Interestingly, interference in signaling from PI3-kinase by overexpression of pAAA-PKB, a kinase-inactive, phosphorylation-deficient protein kinase B (PKB) construct with the mutations K179A, T308A, and S473A (68), led to a similar suppression of the translational activation of L32-GFP (Fig. 10). The apparent suppression of L32-GFP appears to be selective for the 5′TOP-containing mRNA, as Act-GFP mRNA is efficiently translated in the presence of any of the examined expression vectors (Fig. 10). It is likely, therefore, that both PI3-kinase and PKB are involved in the amino acid-induced recruitment of TOP mRNAs into polysomes.
FIG. 10.
Overexpression of PTEN or dominant-negative p85 or PKB mutants suppresses the translational activation by amino acid refeeding. 293 cells were cotransfected with 2 μg of a vector encoding L32-GFP or Act-GFP and 16 μg of an empty vector (pSG5) or the expression vector indicated at the left. Twenty-four hours later cells were amino acid starved for 2 h and then refed for 30 min. The polysomal distribution of mRNAs encoding L32-GFP and Act-GFP was analyzed and is presented as described in the legend to Fig. 2.
DISCUSSION
Amino acid deprivation inhibits global translational activity through phosphorylation of eukaryotic initiation factor 2α (eIF2α) and dephosphorylation of eIF4E-BP (reviewed in reference 27). The decreased polysomal association of non-TOP mRNAs, like that of mRNAs encoding actin (Fig. 2 and 4c) and Act(28)-GFP (Fig. 2), in amino acid-starved cells reflects this global effect on the translational machinery. Other non-TOP mRNAs, like those encoding SOD (Fig. 1 and 7b), Act(28)-GH, and L32(−1C→A)-GH (Fig. 2), seem to be essentially refractory to this repression. Conceivably, this diverse sensitivity primarily reflects different affinities of the individual transcripts for the translational initiation factors. However, the data presented here clearly demonstrate that translation of TOP mRNAs is much more sensitive to this nutritional control and that its response is the same as that observed in nonproliferating cells (reference 39 and references therein).
Growth is characterized by an elevated production of the translational apparatus needed to cope with the increasing demand for protein synthesis. Indeed, according to one estimate most of the energy consumed during cellular growth is utilized for generating components of protein synthesis machinery (56). The apparent advantages in regulating the synthesis of the translational apparatus at the translational level are the rate and the readily reversible nature of the response to altering physiological conditions. These two features enable cells to rapidly repress the biosynthesis of the translational machinery upon shortage of amino acids or growth arrest, thus rapidly blocking energy wastage.
Two experimental approaches have been used to examine the causal relationship between rpS6 phosphorylation and translational control of TOP mRNAs upon mitogenic stimulation by serum refeeding. First, rapamycin indirectly blocked the mitogenic activation of S6K1 and prevented rpS6 phosphorylation (10, 49) by a poorly understood mechanism. Indeed, rapamycin treatment of some cell lines selectively abolished translational activation of TOP mRNAs upon mitogenic stimulation (26, 62, 64, 65). However, in other cell lines rapamycin exhibited only a minor, if any, repressive effect, even though S6K1 activity was completely inhibited (22, 23, 33). Second, transfection of cells with expression vectors encoding a mutant version of S6K1, p70S6KA229, which functioned as a dominant-negative mutant, completely inhibited S6K1 activity. Nevertheless, in a single reported experiment the overexpression of this mutant exerted a modest inhibitory effect on the translational activation of a chimeric TOP mRNA following mitogenic stimulation (22). It should be noted that the phosphorylation of S10, the Saccharomyces cerevisiae homolog of mammalian S6, has been shown to be dispensable for optimal yeast growth (25). Nonetheless, the lack of TOP mRNAs in yeast and the fact that synthesis of yeast rp is primarily regulated at the transcriptional level (70) renders this observation irrelevant to the present discussion.
The data presented herein provide very strong support for the conclusion that the phosphorylation of rpS6 is neither necessary nor sufficient to enable the polysomal recruitment and translational initiation of TOP mRNAs in response to amino acid sufficiency. In addition, the complete inhibition or loss of S6K1 activity, whether caused by rapamycin, recombinant polypeptide inhibitors (i.e., p70S6K,K123 M or p70Δ2-46/ΔCT104,K123 M/T412E), or deletion of the S6K1 gene, does not impair the ability of amino acids to restore TOP mRNA polysomal recruitment after prior amino acid withdrawal. Nevertheless, S6K1−/− cells express a second S6K, S6K2 (32), which is less strongly inhibited by rapamycin than is S6K1 (K. Yonezawa and K. Hara, personal communication), so that its residual activity might account for the rapamycin resistance of amino acid-regulated TOP mRNA recruitment. Although this possibility cannot be eliminated by the present results, our data do establish that such an action of S6K2 cannot be attributed to the phosphorylation of S6. In fact, rapamycin, despite its inability to inhibit fully S6K2, completely inhibits S6 phosphorylation, suggesting that S6 may not be a major substrate of S6K2. Moreover, other observations argue against a role for S6K2 in the amino acid regulation of TOP mRNA recruitment. Thus, S6K2 is resistant not only to inhibition by rapamycin but also to inhibition by amino acid withdrawal (N. Terada, personal communication, and K. Yonezawa and K. Hara, personal communication); however the S6K2 activity persisting in the face of amino acid withdrawal (at least 50% of basal) is not sufficient to prevent the inhibition of TOP mRNA translation upon amino acid withdrawal. Taken together, these observations have reopened the intriguing question concerning the physiological function of S6K1 and -2 and rpS6 phosphorylation in translational regulation; the solution awaits the establishment of S6K1 and -2 double knockout.
The involvement of mTOR in the amino acid regulation of S6K1 has been demonstrated through studies using rapamycin, which blocks the activation of S6K1 by amino acid refeeding. On the other hand, amino acid reintroduction could induce S6K1 activation in the presence of rapamycin in cells expressing a rapamycin-resistant mTOR mutant (21). Likewise, rapamycin-resistant S6K1 mutant p70Δ2-46/ΔCT104 is resistant to amino acid deficiency, indicating that both amino acid sufficiency and an mTOR signal to S6K1 through a common effector, which could be mTOR itself or an mTOR-regulated downstream mediator (18). Nonetheless, experiments presented here clearly show that rapamycin has only a moderate effect on the translational activation of TOP mRNAs upon amino acid activation of 293 cells. Likewise, 3 h of rapamycin treatment of p70S6K−/− ES cells inhibited the translation of rpL32 mRNA considerably less efficiently than 2 h of amino acid starvation (from 82% to 61 or 31% in polysomes, respectively) (Fig. 6) (E. Hornstein, unpublished data). This discrepancy between the effects of rapamycin and amino acids suggest that the latter exert their effect on TOP mRNA primarily in an mTOR-independent fashion. Furthermore, the delayed effect on TOP mRNAs might suggest that rapamycin elicits its repression indirectly by inhibiting one or more of the many growth-related mTOR readouts, such as transcription of specific genes (reviewed in reference 55). Indeed, rapamycin has been shown to block cell cycle progression and to inhibit the proliferation of a variety of lymphocyte and nonlymphocyte cell types (reviewed in reference 57).
The ability of rapamycin to prevent the translational activation of TOP mRNAs only partially and in a delayed manner is underscored by the apparent ability of LY294002 to completely block within 30 min this activation in amino acid-refed cells (Fig. 9). Previous reports, with the exception of one case (47), have demonstrated that amino acid withdrawal and readdition had a minimal effect on the activity of PI3-kinase and PKB, yet inhibitors of PI3-kinase completely block the amino acid-induced activation of S6K1 (16, 18, 21, 45, 69). Two possible explanations for these seemingly conflicting results can be proposed. (i) PI3-kinase inhibitors are not as specific as initially claimed, and they also inhibit other kinases (7, 12). Indeed, the complete inhibition of rpS6 phosphorylation by 50 μM LY294002 can be attributed to the inhibitory effect of this concentration on mTOR activity (7). (ii) Signaling of amino acids to TOP mRNAs requires an active PI3-kinase for continuous supply of phosphoinositides phosphorylated at position 3 on the inositol ring. Conceivably, these phosphoinositides are necessary for anchoring to the membrane (for review see reference 51) of one or more kinases, whose activity is regulated by amino acid sufficiency. The latter explanation seems particularly applicable to the amino acid-induced translational activation of TOP mRNAs, as this activation is blocked by overexpression of either the phosphatase PTEN or the dominant-negative regulatory subunit of PI3-kinase, p85 (Fig. 10). Overexpression of both these constructs has previously been shown to block the accumulation of PI-3,4,5-triphosphate (34, 52). Candidate kinases, other than S6K1 and -2, whose activity is regulated by amino acid sufficiency, are the novel protein kinase Cδ (PKCδ) and PKCɛ. Thus, the activation loop of these kinases is phosphorylated by PI-dependent kinase 1, and their hydrophobic regulatory site is dephosphorylated by amino acid deprivation (reference 43 and references therein).
It is noteworthy that, when enhanced S6K1 variants [p70Δ2-46/ΔCT104 and p70Δ2-46/ΔCT104, T412E), but not the wild-type enzyme, were overexpressed in 293 cells, they were able to relieve the translational repression of L32-GFP exerted by 1 h of amino acid starvation (G. Levy and O. Meyuhas, unpublished data). In light of all other data presented in this report, it is conceivable that this relief simply reflects artifactual consequences of the nonphysiologically high activity of S6K1 obtained by overexpression. Thus, it is possible that a substrate other than rpS6, which is only fortuitously phosphorylated by endogenous S6K1, is now significantly phosphorylated to an extent that might affect the translational efficiency of TOP mRNAs. Similarly, we have shown here that overexpression of constitutively active RSK2 mutant K3H.RSK2(Y707A) led to efficient phosphorylation of rpS6 (Fig. 3), even though this substrate is primarily phosphorylated by S6K1 rather than RSK2 (10). It should be mentioned that previously reported results derived from overexpression of various active and dominant-negative kinases have been a subject for controversial interpretations (2, 4, 13, 14, 31, 53, 71, 73).
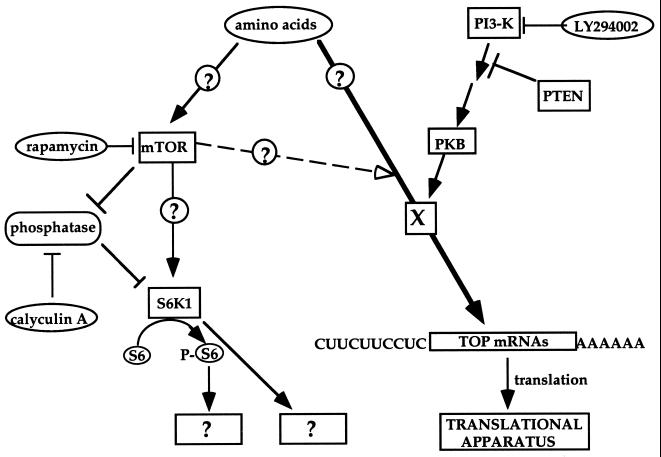
A tentative model depicting the signaling pathways leading to the translational activation of TOP mRNAs by amino acid sufficiency is presented in Fig. 11. According to this model, amino acids signal into TOP mRNAs through an unknown target (denoted X) in a PI3-kinase-dependent fashion. However, the signaling from amino acids bifurcates upstream of mTOR, as inferred from the ability of rapamycin to discriminate between the activity of mTOR and S6K on the one hand and the translational efficiency of TOP mRNAs on the other hand.
FIG. 11.
Schematic representation of signal transduction pathways involved in activation of rpS6 phosphorylation and translational control of TOP mRNAs in amino acid-stimulated cells. Arrows delineate the flow of information. Open arrowhead, partial and delayed effect of rapamycin (through mTOR?) on TOP mRNA translation. The site of convergence of this effect with that of other signals is purely speculative. Circled and boxed question marks, putative links and unknown targets, respectively. See text for details.
It might be argued that the apparent lack of inhibitory effect of rapamycin or of dominant-negative S6K1 mutants on the early response of TOP mRNA translation in amino acid-refed cells reflects the involvement of a signaling pathway which differs from that transducing mitogenic signals. However, our recent experiments with serum-starved and serum-refed cells (including S6K1−/− cells) clearly show that the minor role of the mTOR-mediated pathway in the translational control of TOP mRNAs is not confined to nutritional signals but is also applicable to mitogenic signals (M. Stolovich, H. Tang, E. Hornstein, and O. Meyuhas, unpublished data).
ACKNOWLEDGMENTS
This work was supported by grants to O.M. from the United States-Israel Binational Science Foundation (BSF 97-00055) and by The Israel Science Foundation founded by The Academy of Sciences and Humanities. E.H. is a recipient of awards from the Foulkes Foundation (London) and from the Kornfeld Foundation.
We are grateful to Naohiro Terada for the SK1 knockout ES cells, Celeste Poteet-Smith for the RSK2 constructs, William Sellers for the PTEN construct, Julian Downward for the p85 ΔSH2-N, and James Woodget for the pAAA-PKB.
Hua Tang and Eran Hornstein contributed equally to this study.
REFERENCES
- 1.Aloni R, Peleg D, Meyuhas O. Selective translational control and nonspecific posttranscriptional regulation of ribosomal protein gene expression during development and regeneration of rat liver. Mol Cell Biol. 1992;12:2203–2212. doi: 10.1128/mcb.12.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur J S C, Cohen P. MSK1 is required for CREB phosphorylation in response to mitogens in mouse embryonic stem cells. FEBS Lett. 2000;482:44–48. doi: 10.1016/s0014-5793(00)02031-7. [DOI] [PubMed] [Google Scholar]
- 3.Avni D, Shama S, Loreni F, Meyuhas O. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol Cell Biol. 1994;14:3822–3833. doi: 10.1128/mcb.14.6.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balendran A, Biondi R M, Cheung P C, Casamayor A, Deak M, Alessi D R. A 3-phosphoinositide-dependent protein kinase-1 (PDK1) docking site is required for the phosphorylation of protein kinase Czeta (PKCzeta) and PKC-related kinase 2 by PDK1. J Biol Chem. 2000;275:20806–20813. doi: 10.1074/jbc.M000421200. [DOI] [PubMed] [Google Scholar]
- 5.Biberman Y, Meyuhas O. Substitution of just five nucleotides at and around the transcription start site of rat β-actin promoter is sufficient to render the resulting transcript a subject for translational control. FEBS Lett. 1997;405:333–336. doi: 10.1016/s0014-5793(97)00234-2. [DOI] [PubMed] [Google Scholar]
- 6.Blommaart E F C, Luiken J J, Blommaart P J, van Woerkom G M, Meijer A J. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 7.Brunn G, Williams J, Sabers C, Wiederrecht G, Lawrence J J, Abraham R. Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 1996;15:5256–5267. [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell L E, Wang X, Proud C G. Nutrients differentially regulate multiple translation factors and their control by insulin. Biochem J. 1999;344:433–441. [PMC free article] [PubMed] [Google Scholar]
- 9.Cantley L, Neel B. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung J, Kuo C J, Crabtree G R, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 11.Chung S, Perry R P. Importance of introns for expression of mouse ribosomal protein gene rpL32. Mol Cell Biol. 1989;9:2075–2082. doi: 10.1128/mcb.9.5.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies S, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deak M, Clifton A D, Lucocq L M, Alessi D R. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dufner A, Andjelkovic M, Burgering B, Hemmings B, Thomas G. Protein kinase B localization and activation differentially affect S6 kinase1 activity and eukaryotic translation initiation factor 4E-binding protein 1 phosphorylation. Mol Cell Biol. 1999;19:4525–4534. doi: 10.1128/mcb.19.6.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erikson E, Maller J. A protein kinase from Xenopus eggs specific for ribosomal protein S6. Proc Natl Acad Sci USA. 1985;82:742–746. doi: 10.1073/pnas.82.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox H L, Kimball S R, Jefferson L S, Lynch C J. Amino acids stimulate phosphorylation of p70S6k and organization of rat adipocytes into multicellular clusters. Am J Physiol. 1998;274:C206–C213. doi: 10.1152/ajpcell.1998.274.1.C206. [DOI] [PubMed] [Google Scholar]
- 17.Fumagalli S, Thomas G. S6 phosphorylation and signal transduction. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 695–717. [Google Scholar]
- 18.Hara K, Yonezawa K, Weng Q-P, Kozlowski M T, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 19.Hornstein E, Harel H, Levy G, Meyuhas O. Overexpression of poly(A)-binding protein down-regulates the translation or the abundance of its own mRNA. FEBS Lett. 1999;457:209–213. doi: 10.1016/s0014-5793(99)01039-x. [DOI] [PubMed] [Google Scholar]
- 20.Huang S, Hershey J B. Translational initiation factor expression and ribosomal protein gene expression are repressed coordinately but by different mechanisms in murine lymphosarcoma cells treated with glucocorticoids. Mol Cell Biol. 1989;9:3679–3684. doi: 10.1128/mcb.9.9.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iiboshi Y, Papst P J, Kawasome H, Hosoi H, Abraham R T, Houghton P J, Terada N. Amino acid-dependent control of p70(s6k). Involvement of tRNA aminoacylation in the regulation. J Biol Chem. 1999;274:1092–1099. doi: 10.1074/jbc.274.2.1092. [DOI] [PubMed] [Google Scholar]
- 22.Jefferies H B J, Fumagalli S, Dennis P, Reinhard C, Pearson R, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jefferies H B J, Reinhard C, Kozma S C, Thomas G. Rapamycin selectively represses translation of the 'polypyrimidine tract' mRNA family. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefferies H B J, Thomas G, Thomas G. Elongation factor-1α mRNA is selectively translated following mitogenic stimulation. J Biol Chem. 1994;269:4367–4372. [PubMed] [Google Scholar]
- 25.Johnson S, Warner J. Phosphorylation of the Saccharomyces cerevisiae equivalent of ribosomal protein S6 has no detectable effect on growth. Mol Cell Biol. 1987;7:1338–1345. doi: 10.1128/mcb.7.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawasome H, Papst P, Webb S, Keller G M, Johnson G L, Gelfand E W, Terada N. Targeted disruption of p70(s6k) defines its role in protein synthesis and rapamycin sensitivity. Proc Natl Acad Sci USA. 1998;95:5033–5038. doi: 10.1073/pnas.95.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimball S R, Jefferson L S. Regulation of translation initiation in mammalian cells by amino acids. In: Sonenberg N, Hershey J, Mathews M, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 561–579. [Google Scholar]
- 28.Kimball S R, Shantz L M, Horetsky R L, Jefferson L S. Leucine regulates translation of specific mRNAs in L6 myoblasts through mTOR-mediated changes in availability of eIF4E and phosphorylation of ribosomal protein S6. J Biol Chem. 1999;274:11647–11652. doi: 10.1074/jbc.274.17.11647. [DOI] [PubMed] [Google Scholar]
- 29.Kleijn M, Proud C G. Glucose and amino acids modulate translation factor activation by growth factors in PC12 cells. Biochem J. 2000;347:399–406. doi: 10.1042/0264-6021:3470399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozma S C, Lane H A, Ferrari S, Luther H, Siegmann M, Thomas G. A stimulated S6 kinase from rat liver: identity with the mitogen-activated S6 kinase from 3T3 cells. EMBO J. 1989;8:4125–4132. doi: 10.1002/j.1460-2075.1989.tb08597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lange-Carter C A, Pleiman C M, Gardner A M, Blumer K J, Johnson G L. A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- 32.Lee-Fruman K, Kuo C, Lippincott J, Terada N, Blenis J. Characterization of S6K2, a novel kinase homologous to S6K1. Oncogene. 1999;18:5108–5114. doi: 10.1038/sj.onc.1202894. [DOI] [PubMed] [Google Scholar]
- 33.Loreni F, Thomas G, Amaldi F. Transcription inhibitors stimulate translation of 5′ TOP mRNAs through activation of S6 kinase and the mTOR/FRAP signalling pathway. Eur J Biochem. 2000;267:6594–6601. doi: 10.1046/j.1432-1327.2000.01753.x. [DOI] [PubMed] [Google Scholar]
- 34.Maehama T, Dixon J E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 35.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 36.Meyuhas O, Avni D, Shama S. Translational control of ribosomal protein mRNAs in eukaryotes. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 363–384. [Google Scholar]
- 37.Meyuhas O, Baldin V, Bouche G, Amalric F. Glucocorticoids repress ribosome biosynthesis in lymphosarcoma cells by affecting gene expression at the level of transcription, posttranscription and translation. Biochim Biophys Acta. 1990;1049:38–44. doi: 10.1016/0167-4781(90)90082-d. [DOI] [PubMed] [Google Scholar]
- 38.Meyuhas O, Biberman Y, Pierandrei-Amaldi P, Amaldi F. Analysis of polysomal RNA. In: Krieg P, editor. A laboratory guide to RNA: isolation, analysis, and synthesis. New York, N.Y: Wiley Liss; 1996. pp. 65–81. [Google Scholar]
- 39.Meyuhas O, Hornstein E. Translational control of TOP mRNAs. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 671–693. [Google Scholar]
- 40.Meyuhas O, Klein A. The mouse ribosomal protein L7 gene. Its primary structure and functional analysis of the promoter. region. J Biol Chem. 1990;265:11465–11473. [PubMed] [Google Scholar]
- 41.Meyuhas O, Thompson A E, Perry R P. Glucocorticoids selectively inhibit the translation of ribosomal protein mRNAs in P1798 lymphosarcoma cells. Mol Cell Biol. 1987;7:2691–2699. doi: 10.1128/mcb.7.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minty A J, Caravatti M, Robert B, Cohen A, Daubas P, Weydert A, Gross F, Buckingham M E. Mouse actin messenger RNAs: construction and characterization of a recombinant plasmid molecule containing a complementary DNA transcript of mouse α-actin mRNA. J Biol Chem. 1981;256:1008–1014. [PubMed] [Google Scholar]
- 43.Parekh D, Ziegler W, Yonezawa K, Hara K, Parker P. Mammalian TOR controls one of two kinase pathways acting upon nPKCδ and nPKCɛ. J Biol Chem. 1999;274:34758–34764. doi: 10.1074/jbc.274.49.34758. [DOI] [PubMed] [Google Scholar]
- 44.Parrott L A, Templeton D J. Osmotic stress inhibits p70/85 S6 kinase through activation of a protein phosphatase. J Biol Chem. 1999;274:24731–24736. doi: 10.1074/jbc.274.35.24731. [DOI] [PubMed] [Google Scholar]
- 45.Patti M E, Brambilla E, Luzi L, Landaker E J, Kahn C R. Bidirectional modulation of insulin action by amino acids. J Clin Investig. 1998;101:1519–1529. doi: 10.1172/JCI1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peterson R T, Desai B N, Hardwick J S, Schreiber S L. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycin-associated protein. Proc Natl Acad Sci USA. 1999;96:4438–4442. doi: 10.1073/pnas.96.8.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peyrollier K, Hajduch E, Blair A, Hyde R, Hundal H. L-leucine availability regulates phosphatidylinositol 3-kinase, p70 S6 kinase and glycogen synthase kinase-3 activity in L6 muscle cells: evidence for the involvement of the mammalian target of rapamycin (mTOR) pathway in the L-leucine-induced up-regulation of system A amino acid transport. Biochem J. 2000;350:361–368. [PMC free article] [PubMed] [Google Scholar]
- 48.Poteet-Smith C E, Smith J A, Lanniga D A, Freed T A, Sturgill T W. Generation of constitutively active p90 ribosomal S6 kinase in vivo. Implications for the mitogen-activated protein kinase-activated protein kinase family. J Biol Chem. 1999;274:22135–22138. doi: 10.1074/jbc.274.32.22135. [DOI] [PubMed] [Google Scholar]
- 49.Price D J, Grove J R, Calvo V, Avruch J, Bierer B E. Rapamycin-induced inhibition of the 70-kilodalton protein kinase. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- 50.Ramaswamy S, Nakamura N, Vazquez F, Batt D, Perera S, Roberts T, Sellers W. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1999;96:2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rameh L, Cantley L. The role of phosphoinositide 3-kinase lipid products in cell function. J Biol Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez-Viciana P, Warne P, Khwaja A, Marte B, Pappin D, Das P, Waterfield M, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 53.Romanelli A, Martin K A, Toker A, Blenis J. p70 S6 kinase is regulated by protein kinase Cζ and participates in a phosphoinositide 3-kinase-regulated signalling complex. Mol Cell Biol. 1999;19:2921–2928. doi: 10.1128/mcb.19.4.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Schmelzle T, Hall M. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt E V. The role of c-myc in cellular growth control. Oncogene. 1999;18:2988–2996. doi: 10.1038/sj.onc.1202751. [DOI] [PubMed] [Google Scholar]
- 57.Sehgal S N. Rapamune (RAPA, rapamycin, sirolimus): mechanism of action immunosuppressive effect results from blockade of signal transduction and inhibition of cell cycle progression. Clin Biochem. 1998;31:335–340. doi: 10.1016/s0009-9120(98)00045-9. [DOI] [PubMed] [Google Scholar]
- 58.Shama S, Avni D, Frederickson R M, Sonenberg N, Meyuhas O. Overexpression of initiation factor eIF-4E does not relieve the translational repression of ribosomal protein mRNAs in quiescent cells. Gene Expr. 1995;4:241–252. [PMC free article] [PubMed] [Google Scholar]
- 59.Shama S, Meyuhas O. The translational cis-regulatory element of mammalian ribosomal protein mRNAs is recognized by the plant translational apparatus. Eur J Biochem. 1996;236:383–388. doi: 10.1111/j.1432-1033.1996.00383.x. [DOI] [PubMed] [Google Scholar]
- 60.Sherman L, Dafni N, Lieman-Hurwitz J, Groner Y. Nucleotide sequence and expression of human chromosome 21-encoded superoxide dismutase mRNA. Proc Natl Acad Sci USA. 1983;80:5465–5468. doi: 10.1073/pnas.80.18.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shigemitsu K, Tsujishita Y, Hara K, Nanahoshi M, Avruch J, Yonezawa K. Regulation of translational effectors by amino acid and mammalian target of rapamycin signaling pathways. Possible involvement of autophagy in cultured hepatoma cells. J Biol Chem. 1999;274:1058–1065. doi: 10.1074/jbc.274.2.1058. [DOI] [PubMed] [Google Scholar]
- 62.Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma S. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sturgill T W, Wu J. Recent progress in characterization of protein kinase cascades for phosphorylation of ribosomal protein S6. Biochim Biophys Acta. 1991;1092:350–357. doi: 10.1016/s0167-4889(97)90012-4. [DOI] [PubMed] [Google Scholar]
- 64.Terada N, Patel H R, Takase K, Kohno K, Nairn A C, Gelfand E W. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc Natl Acad Sci USA. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Terada N, Takase K, Papst P, Nairn A C, Gelfand E W. Rapamycin inhibits ribosomal protein synthesis and induces G1 prolongation in mitogen-activated T lymphocytes. J Immunol. 1995;155:3418–3426. [PubMed] [Google Scholar]
- 66.Thomas G, Thomas G. Translational control of mRNA expression during the early mitogenic response in Swiss mouse 3T3 cells: identification of specific proteins. J Cell Biol. 1986;103:2137–2144. doi: 10.1083/jcb.103.6.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vlahos C, Matter W, Hui K, Brown R. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 68.Wang Q, Somwar R, Bilan P, Liu Z, Jin J, Woodgett J, Klip A. Protein kinase B/Akt participates in GLUT4 translocation by insulin in L6 myoblasts. Mol Cell Biol. 1999;19:4008–4018. doi: 10.1128/mcb.19.6.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, Campbell L E, Miller C M, Proud C G. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem J. 1998;334:261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warner J. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 71.Xing J, Ginty D D, Greenberg M E. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 72.Xu G, Kwon G, Marshall C A, Lin T A, Lawrence J C J, McDaniel M L. Branched-chain amino acids are essential in the regulation of PHAS-I and p70 S6 kinase by pancreatic beta-cells. A possible role in protein translation and mitogenic signaling. J Biol Chem. 1998;273:28178–28184. doi: 10.1074/jbc.273.43.28178. [DOI] [PubMed] [Google Scholar]
- 73.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, R W J, Templeton D J. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]