Abstract
Efficient genome engineering is critical to understand and use microbial functions. Despite recent development of tools such as CRISPR-Cas gene editing, efficient integration of exogenous DNA with well-characterized functions remains limited to model bacteria. Here, we describe serine recombinase–assisted genome engineering, or SAGE, an easy-to-use, highly efficient, and extensible technology that enables selection marker–free, site-specific genome integration of up to 10 DNA constructs, often with efficiency on par with or superior to replicating plasmids. SAGE uses no replicating plasmids and thus lacks the host range limitations of other genome engineering technologies. We demonstrate the value of SAGE by characterizing genome integration efficiency in five bacteria that span multiple taxonomy groups and biotechnology applications and by identifying more than 95 heterologous promoters in each host with consistent transcription across environmental and genetic contexts. We anticipate that SAGE will rapidly expand the number of industrial and environmental bacteria compatible with high-throughput genetics and synthetic biology.
Bacteriophage-derived tools enable simple, host-independent construction of complex genetically modified bacterial libraries.
INTRODUCTION
Engineered microbes promise to make substantial impacts on pressing societal challenges. Biotechnologies deployed in environments of biotic and abiotic complexity, such as cell-based therapeutics in the human gut (1), nitrogen fixation in the plant rhizosphere (2, 3), and plastic (4) and plant biomass bioconversion (5, 6) in industrial bioreactors (7, 8), will be central in addressing grand challenges to promote human health, reverse carbon emissions, recycle mixed plastic waste, remediate contaminated soils, and achieve sustainable economies. Many natural microbes can thrive in these environments but lack the functions required to address these aims. Synthetic biology enables the redesigning of microbes by engineering enhanced or novel traits that will underpin transformational biotechnologies.
The application of synthetic biology to meet these societal challenges is frequently limited by the absence of effective genetic tools (9), including tools for sophisticated, high-throughput genome engineering. The lack of tools is particularly problematic for microbes that will be deployed in environments where antibiotic selection is undesirable and impractical, such as in industrial-scale reactors, agriculture, the human gut, and soil bioremediation. Most common methods used to transfer heterologous DNA into bacteria (e.g., replicating plasmids, homologous recombination–based allelic exchange, and transposon mutagenesis) have substantial caveats that limit their utility for studying engineered microbes. Replicating plasmids, including commonly used broad-host variants [e.g., pBBR (10, 11), RK2 (12, 13), RSF1010 (14), and pBC1 (15)], are generally not stably maintained or incur a fitness cost (16–22), thus requiring constant selection, typically via antibiotic resistance. Furthermore, plasmid copy number variation both within a species (23–25) and between species (26, 27) introduces physiological and measurement noise, creating uncertainty when interpreting results and often requiring pathway reoptimization once the pathway is ultimately integrated in the chromosome. Both transposon and homologous recombination–based technologies stably integrate heterologous DNA into the genome (28), but each is generally incompatible with high-throughput rational engineering studies. The unpredictable location of integrations that occur with most transposon-based tools could drive inconsistent host-pathway interactions among transformants, preventing reliable comparisons between pathway variants. Regardless of the counterselection method [e.g., CRISPR-Cas (29), sucrose sensitivity (30), etc.], homologous recombination–based allelic exchange is typically inefficient (29, 31), precluding the construction of even moderately sized strain variant libraries.
Bacteriophages harbor a treasure trove of material for establishing genetic tools for nonmodel bacteria (32). Specifically, phage integrase–mediated recombination technologies (31, 33–41) provide the scientific community solutions to many limitations stemming from common genetic tools. Phage integrases (also known as site-specific recombinases) are enzymes that catalyze recombination between two specific sequences of DNA (38, 42). Tyrosine recombinases, such as Flp and Cre, are commonly used in molecular genetics (41, 43). These enzymes recognize and perform reversible recombination between two identical attachment sequences (att sites, called frt and lox sites for Flp and Cre, respectively). These recombinases leave identical or highly similar frt and lox scars that flank each DNA insertion. Sequence similarity between scars causes genetic instability (e.g., inserted DNA can be excised by homologous recombination), and the reversibility precludes their reuse for sequential insertions. The second major class of site-specific recombinases, serine recombinases, are single subunit enzymes that catalyze DNA recombination between two distinct attachment sequences (attP and attB sites). This process is unidirectional, generating two new attachment sites (attL and attR) that are not substrates for the recombinase without the aid of accessory proteins. Furthermore, serine recombinases (e.g., Bxb1 and ΦC31 integrases) do not require any host proteins and have been used across the tree of life in research (34–40); thus, they are likely to work in the vast majority of organisms. Current recombinase-based tools are also often limited to integration of one (31, 33, 35, 36, 39, 40, 43, 44) or two (45) DNA fragments, function in a limited range of hosts (34, 37, 46), require use of plasmids that replicate in the host (28, 29, 45, 47–49), and frequently leave selection markers in the host chromosome.
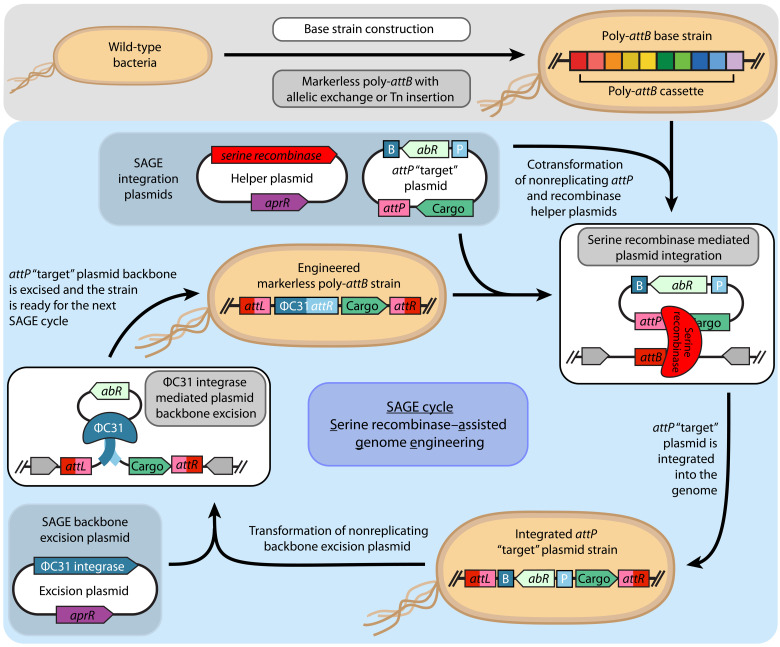
To address these issues and limitations, in this work, we developed the serine recombinase–assisted genome engineering (SAGE) toolkit. The toolkit uses high-efficiency serine recombinases—each transiently expressed from a nonreplicating plasmid—to enable efficient, iterative integration of up to 10 distinct DNA constructs or variant libraries into diverse bacterial genomes at unique attB sites (Fig. 1). For faster growing organisms, a cycle of SAGE that comprises integration of attP target plasmid DNA into genome and subsequent plasmid backbone (selection marker) excision can be easily performed in as few as four 8-hour workdays (fig. S1). We demonstrate robust SAGE performance in five bacteria representing multiple taxonomic groups, including an undomesticated sorghum rhizosphere isolate. Furthermore, we highlight the utility of SAGE by developing and characterizing a constitutively expressed chromosomal promoter library for each organism. Coanalysis of each promoter under distinct condition and sequence contexts provides a resource for tunable and reliable expression of heterologous proteins in each of these diverse bacteria.
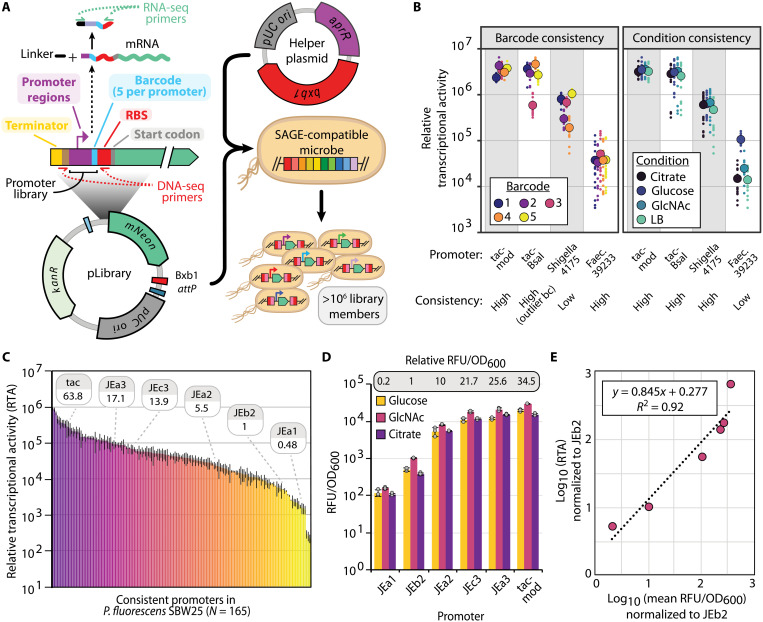
Fig. 1. Iterative integration of multiple genetic constructs into the chromosome using SAGE.
This schematic depicts the SAGE cycle for rapid, efficient, unidirectional, and site-specific integration of multiple DNA fragments into bacterial chromosomes. To construct a base strain, attB sequences (here, a 10-sequence poly-attB cassette) are integrated into the target microbe’s genome using standard methods (e.g., allelic exchange or transposase integration). Next, one of the system’s 10 recombinases is transiently expressed from a nonreplicating helper plasmid, catalyzing recombination between its cognate attP and attB sequences, located on a cotransformed target plasmid and the host chromosome, respectively. Recombination integrates the attP target plasmid into the genome unidirectionally, generating attL and attR sequences. To excise unneeded plasmid features and prime the strain for additional SAGE cycles, ΦC31 integrase is transiently expressed by transforming a nonreplicating backbone excision plasmid. ΦC31 integrase catalyzes recombination between ΦC31 attP and attB sites (P and B, respectively, in blue) of the integrated target plasmid, excising the selection marker and the E. coli origin of replication from the chromosome.
RESULTS
Transient expression of serine recombinases enables efficient unidirectional integration of plasmid DNA into the bacterial chromosome
We selected five bacteria that represent a range of biological diversity and varied application spaces [e.g., plant growth promotion (PGP), bioremediation, and industrial biotechnology] to demonstrate SAGE (Table 1). Specifically, we tested performance with the domesticated microbes Pseudomonas fluorescens SBW25 (a model PGP 𝛾-proteobacteria) (50), Rhodopseudomonas palustris CGA009 (a model α-proteobacteria for studying photophosphorylation, nitrogen fixation, hydrogen production, and anaerobic catabolism of aromatic compounds) (51), and Pseudomonas putida Gpo1 (a model 𝛾-proteobacteria for bioremediation of hydrocarbons and industrial chassis) (52). We also tested SAGE with both a model actinomycete, Rhodococcus jostii RHA1 (bioremediation host and emerging industrial chassis) (53), and an undomesticated pseudomonad that we isolated from the endosphere of Sorghum bicolor roots under drought conditions. This isolate, named TBS10, is most closely related to Pseudomonas frederiksbergensis, and thus, we will refer to TBS10 as P. frederiksbergensis TBS10.
Table 1. Organisms and growth conditions for transcriptional profiling experiments.
MME, Mops-based mineral medium; PM, mineral medium used for R. palustris cultivation; PEG6000, polyethylene glycol 6000.
| Organism | Classification | Description | Growth conditions |
|---|---|---|---|
| Pseudomonas fluorescens SBW25 | 𝛾-Proteobacteria | Model plant growth–promoting rhizobacteria (PGPR) | Rich medium (LB) |
| MME + glucose (cellulose) | |||
| MME + citrate (root exudate) | |||
| MME + GlcNAc (chitin) | |||
| Pseudomonas putida Gpo1(previously Pseudomonas oleovorans Gpo1) | 𝛾-Proteobacteria | Industrial and bioremediation platform organism | MME + glucose (sugar hydrolysates) |
| MME + vanillic acid (lignin-derived) | |||
| MME + n-octane (bioremediation) | |||
| Pseudomonas frederiksbergensis TBS10 | 𝛾-Proteobacteria | Drought-tolerant sorghum rhizosphere isolate | MME + glucose (control) |
| MME + glucose + 25% PEG6000 (drought) | |||
| MME + glucose + 0.4 M NaCl (salt stress) | |||
| Rhodopseudomonas palustris CGA009 | ⍺-Proteobacteria | Model purple, nonsulfur bacteria and PGPR | PM + acetic acid (aerobic, dark) |
| PM + acetic acid (anaerobic, illumination) | |||
| Rhodococcus jostii RHA1 | Actinomycete | Industrial and bioremediation platform organism | MME + glucose (sugar hydrolysates) |
| MME + vanillic acid (lignin-derived) | |||
| MME + n-decane (bioremediation) |
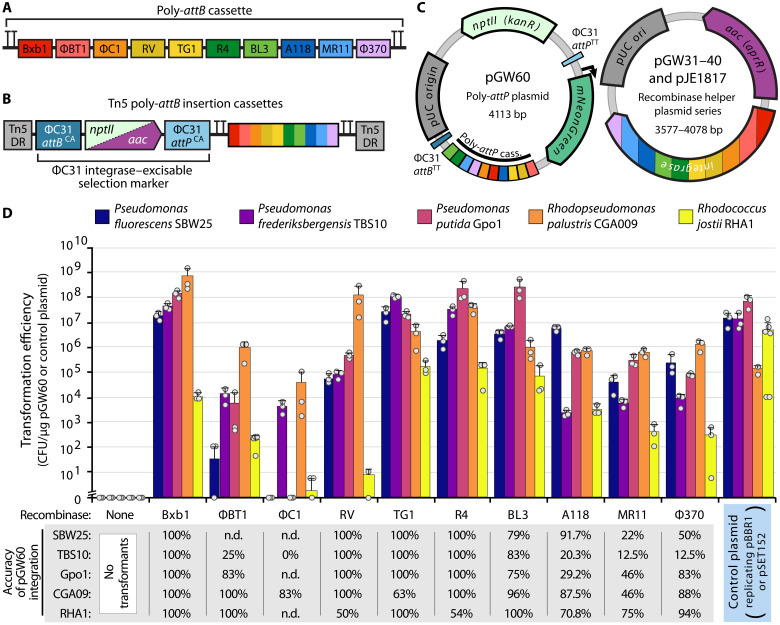
We prepared SAGE-compatible base strains by integrating a poly-attB cassette (Fig. 2A) into the genomes of each organism. The cassette contains an array of 10 distinct attB sequences (1 for each of the 10 recombinases), each separated by a unique spacer sequence to enable polymerase chain reaction (PCR) screening for plasmid integration, and the array is flanked by double rho-independent terminators (54) that should provide transcriptional insulation for many bacteria. Standard homologous recombination–based allelic exchange methods were used to integrate the poly-attB sequence into the genomes of P. fluorescens, R. palustris, and R. jostii (see table S1 for sites of insertion). For these strains, we targeted integration sites known to have no impact on fitness in related strains or to replace genes that could interfere with genetic transformation (e.g., restriction enzymes). Similar to many undomesticated organisms of interest, P. putida Gpo1 and P. frederiksbergensis TBS10 lacked sequenced genomes as we initiated this work. We therefore introduced the poly-attB cassette into these two hosts through electroporation of a Tn5 transposome complex (Materials and Methods). Alternative delivery approaches for the poly-attB cassette, such as conjugation of transposon plasmids, are likely to also successfully generate SAGE base strains. Because the selectable marker needs to be removed from these strains to allow marker reuse in SAGE, we designed the transposon such that the antibiotic resistance gene (nptII) could be easily removed. ΦC31 integrase is a widely used site-specific recombinase that is used as a genetic tool in organisms ranging from bacteria to archaea to eukaryotes (35, 36, 40, 42). We flanked nptII with a cognate pair of ΦC31 att site (attBCA/attPCA) sequences (table S2, att sequences), and thus, transient expression of ΦC31 integrase should lead to excision of the nptII marker from the genome (Fig. 2A). Accordingly, we electroporated a nonreplicating plasmid for expressing ΦC31 integrase into each strain containing the Tn5-integrated poly-attB cassettes and plated dilutions of the recovery culture onto nonselective media. We observed that ~10 and ~1% of the resulting colonies for TBS10 and Gpo1, respectively, were kanamycin sensitive, indicating that the nptII marker had been excised from a substantial fraction of each population. The ability to identify marker-free strains without selection illustrates both the exceptionally high transformation efficiency of these two strains and the efficiency of site-specific recombination. We used the resulting ∆nptII colonies (table S1) for each organism to characterize SAGE performance.
Fig. 2. SAGE enables stable, highly efficient integration of plasmid DNA into the genomes of engineered bacteria.
(A) Diagram of genome-integrated 10× poly-attB cassette, including terminators for transcriptional insulation. Each attB sequence is indicated by a color-coded box and is flanked by a random 20-nt DNA spacer sequence. (B) Diagram of Tn5 poly-attB insertion cassettes. Cassettes are flanked by Tn5 direct repeat (DR) sequences and contain an antibiotic resistance cassette (either nptII or aac) upstream of the poly-attB cassette from (A). A cognate pair of ΦC31 att sites flanks the resistance cassette, allowing its unidirectional excision by electroporation of the ΦC31 integrase helper plasmid pJE1817. (C) Plasmid maps of SAGE plasmids used for efficiency experiments in (D). (D) Transformation efficiency when the poly-attP target plasmid pGW60 is transformed with or without an integrase-expressing helper plasmid or when the positive control plasmid is transformed. Control plasmids are as follows: pJE354 (SBW25, TBS10, and Gpo1), pEYF2K (CGA009), or pSET152 (RHA1). Error bars indicate the two-sided SD in three or more biological replicates. Dots indicate individual samples. The accuracy of integration represents the fraction of colonies in which pGW60 recombined into the poly-attB cassette rather than a pseudo-att site, as determined by PCR. With the exception of BT1 in Gpo1 and RV/F370 in RHA1 (which used 12, 20, and 18 samples for screening, respectively), integration accuracy represents the fraction of 24 samples with colony PCR screening results indicating insertion at the intended attB site. n.d. indicates samples not assayed by PCR because of low numbers of transformants across plate replicates. CFU, colony-forming units.
We assessed the functionality of each recombinase using a plasmid transformation assay. All five organisms were electroporated with two plasmids simultaneously, one nonreplicating helper plasmid that expresses one of the SAGE recombinases (plasmids pGW31 to pGW40) and the nonreplicating target plasmid pGW60 (Fig. 2C) that contains an array of cognate attP sequences for each recombinase and a constitutive mNeonGreen (55) fluorescent reporter. As parallel controls, we assessed the transformation efficiency of pBBR1-based replicating plasmids (10) and integrative vector pSET152 (46) in the Proteobacteria and R. jostii RHA1, respectively. The pBBR1-based plasmids, pJE354 and pEYF2K (fig. S2), contain mNeonGreen and mKate2 (56) reporters, respectively. pGW60 transformants are expected to result from recombinase-catalyzed recombination between the attB in the chromosome and the cognate attP in the plasmid (Fig. 1). This process integrates the entire nonreplicating plasmid into the chromosome while simultaneously converting the attB and attP sites into attL and attR sites upon integration. As the recombinase cannot perform recombination between the newly generated attL and attR sequences, this DNA integration is unidirectional. Notably, unlike many of the most commonly used genome engineering systems in bacteria (28, 29, 45, 47–49), particularly those used for Proteobacteria, the genome-edited strains resulting from SAGE are free of replicating plasmids at all engineering stages and do not require any postintegration plasmid-curing steps (Fig. 1).
Transformation efficiency was measured by enumerating colonies resistant to kanamycin (pGW60, pJE354, and pEYF2K) or apramycin (pSET152) (Fig. 2D). Transformation efficiency with the control plasmids—pJE354, pEYF2K, and pSET152—for the pseudomonads, R. palustris, and R. jostii RHA1, respectively, was on par or better than previously published results in related organisms (31, 57, 58). Unexpectedly, we were unable to transform R. palustris with pJE354, which could be a consequence of poor nptII expression or plasmid instability, as we discuss below. Notably, while transformation of the pseudomonads with the replicating plasmid pJE354 was efficient (1.35 × 107 to 7.27 × 107 colony-forming units/μg of DNA), colony formation was substantially slower than observed with the parent or pGW60 integrant strains.
While recombinase performance varied between organisms, at least eight of the recombinases efficiently incorporated pGW60 into the chromosomes of all tested organisms (Fig. 2D). In all four proteobacteria, the coelectroporation of Bxb1, TG1, R4, or BL3 helper plasmids with the poly-attP plasmid pGW60 led to transformation efficiencies on par with or better than the replicating control plasmid. These recombinases were the best performing in R. jostii RHA1 as well, although the efficiency was lower than the control plasmid pSET152. This may be a consequence of SAGE requiring two plasmids to be transformed simultaneously, unlike one plasmid for pSET152. Alternatively, the lower efficiency may be a consequence of the recombinase expression and selection marker cassettes being poorly optimized for performance in actinomycetes. The ΦC1 integrase generally exhibited low efficiency, which may be indicative of poor expression, or need for a more stringent operating environment than any of the other recombinases. In each organism, at least four of the remaining five recombinases provided high transformation efficiency, although in all hosts except for R. palustris CGA009, the transformation efficiencies were lower than that observed with replicating plasmid controls. Notably, no colonies were observed for any of the five hosts when pGW60 was electroporated without a recombinase helper plasmid.
Serine recombinases typically have a preference for their native att sites, but they are also known to sometimes target other sites in a host chromosome called pseudo-att sites (59). Integration accuracy is therefore an important metric when assessing the utility of each recombinase as a genetic tool. We performed a colony PCR assay using primer pairs for each organism to assess the frequency of pGW60 integration within the poly-attB cassette versus other sites in the chromosome. While the integration accuracy for each recombinase varied by organism, we observed several general trends. First, recombinases that provided the highest transformation efficiencies in a given organism tend to be the most accurate in that organism (Fig. 2D). Bxb1 was the most accurate, with 100% accuracy in all five organisms. RV, TG1, and R4 were all also highly accurate, with 100% in four of five organisms and ~50% accuracy for the fifth. Although we did not identify pseudo-attB sites in the fifth organism in each case, the ~50% ratio is consistent with the presence of a second attB in the genome with similar affinity for the recombinase. On the other hand, the BL3 integrase had consistently high but imperfect accuracy in all five organisms (75 to 100%). This may be indicative of a lower-fidelity recombinase or that its attB sequence likely has room for improvement through att site engineering. Overall, integration accuracy with each remaining recombinase was highly variable between organisms, but they demonstrated reasonable accuracy in at least some of the organisms tested. Variation in accuracy between organisms may be a consequence of native sequences in the host genome serving as alternative attB sequences. These attB sequences may even have higher affinity for the recombinase than the attB sequences found in the engineered poly-attB cassette. Accordingly, the presence of pseudo-att sites is dependent on chromosome sequence, and, as demonstrated here, integration accuracy must be independently assessed in each host. Ultimately, even low accuracy (20 to 30%) does not preclude the use of a given recombinase for some applications, in which case, diagnostic tests can validate integration at the intended att site.
Transient expression of ΦC31 integrase excises selection markers and facilitates iterative SAGE cycles
Removal of selection markers will be critical to iterative reuse of selection markers for SAGE cycles (Fig. 1) and for strain deployment in many operating environments. Accordingly, all attP target plasmids were designed such that the “cargo region” was flanked with a cognate pair of ΦC31 att sites (ΦC31 attPTT/attBTT) that have been previously described to be orthogonal (i.e., noncognate ΦC31 att sites cannot recombine) (60) to the ΦC31 att sites (ΦC31 attPCA/attBCA) found in the mini-Tn5 transposons used to integrate the poly-attB cassette (Fig. 2B) into the P. putida Gpo1 and P. frederiksbergensis TBS10 genomes.
We tested excision of a plasmid backbone by ΦC31 integrase using a strain with pGW60 integrated into the Bxb1 attB site in the P. fluorescens JE4621 chromosome (fig. S3). This strain, P. fluorescens JE4689 (JE4621 attLBxb1:pGW60:attRBxb1; fig. S3A), was transformed with temperature-sensitive replicating plasmids (61) that expresses (pGW30) or lacks (pGW26) ΦC31 integrase (fig. S4). The expectation is that ΦC31 integrase expression will lead to pGW60 backbone excision and loss (fig. S3B). Transformants were cured of their respective plasmids by cultivation at 34°C on nonselective solid medium, and plasmid loss was confirmed by colony PCR and apramycin sensitivity (fig. S3C). Transient expression of ΦC31 integrase via pGW30 resulted in excision of the pGW60 plasmid backbone, while no excision was seen in pGW26 transformants (fig. S3D), demonstrating that the selection marker can be recycled via ΦC31 excision.
Next, we generated a collection of nonreplicating target plasmids containing single attP sequences (fig. S5) and evaluated use of the ΦC31 integrase–mediated excision to enable the successive integration of multiple plasmids into the JE4621 chromosome. First, we integrated a mNeonGreen (green) fluorescent protein expression vector (pJE1286) into the Bxb1 attB site. The pJE1286 backbone was excised from this strain by transformation with and subsequent curing of pGW30, completing the first SAGE cycle. For the next cycle, we integrated a mKate2 (red) fluorescent protein expression vector (pGW55) into the R4 attB site and excised the pGW55 backbone, resulting in generation of the dual-fluorescent protein expression strain P. fluorescens JE5117 (fig. S6A). Last, we integrated a series of empty single attP target plasmids into each of the remaining attB sites of JE5117 (excluding ΦC1), resulting in generation of seven SAGE strains with heterologous DNA incorporated into the genome at three distinct loci (fig. S6B). Together, this demonstrates that SAGE can be used to iteratively incorporate multiple heterologous DNA constructs into a host genome.
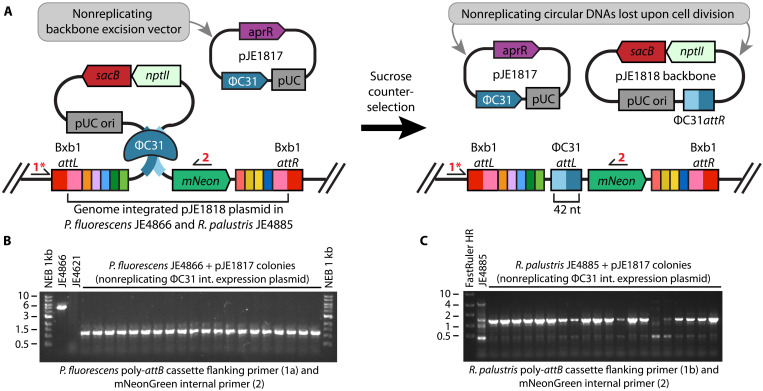
Having established use of ΦC31 integrase–mediated excision for multiple SAGE cycles, we developed a backbone excision system that would not require a plasmid-curing step (Fig. 3A) and would function in a wide array of bacterial genera (i.e., not depend on a pseudomonad-specific replication origin). Incorporating the sacB (levansucrase) counterselectable marker (62) into the attP target plasmid backbone should allow for selection of plasmid excision in many bacteria by cultivating the bacteria in the presence of sucrose. Accordingly, we generated a variant of pGW60 containing sacB (pJE1818; fig. S5) and used Bxb1 integrase to incorporate the plasmid into the genomes of P. fluorescens JE4621 and R. palustris JE4632. The resulting strains were sensitive to sucrose and were unable to grow in the presence of 25 and 5% sucrose, respectively. Each strain was transformed with both a nonreplicating ΦC31 integrase expression helper plasmid (pJE1817) and one containing the pseudomonad-specific, temperature-sensitive origin (pGW30), followed by cultivation on agar media with sucrose to select for cells that have removed the pJE1818 backbone (Fig. 3B). Of the resulting sucrose-resistant colonies, 100% of P. fluorescens and 95% of R. palustris pJE1817 transformants as well as all pGW30 transformants were sensitive to kanamycin, indicative of marker removal. We further confirmed by PCR that the backbone was excised in colonies exhibiting kanamycin sensitivity (Fig. 3, B and C, and fig. S7).
Fig. 3. Transient expression of ɸC31 integrase excises the attP plasmid backbone and enables selection marker recycling.
(A) Diagram of ΦC31 integrase–mediated excision of an attP target plasmid backbone. Specifically, pJE1818 was inserted at the Bxb1 attB of a chromosomally integrated poly-attB cassette. ΦC31 integrase was expressed from the nonreplicating helper plasmid pJE1817. Colony PCR primers are indicated by arrows and red numbers. (B and C) Colony PCR validation of plasmid backbone excision in sucrose-resistant (B) P. fluorescens JE4621– and (C) R. palustris JE4632–based strains following SAGE integration and incubation on the sucrose-containing medium. Expected band sizes are as follows: JE4866 [4741 base pairs (bp)], JE4866 with backbone excision (1152 bp), JE4621 (no band), JE4885 (4964 bp), and JE4855 with backbone excision (1375 bp).
Genomic integration of DNA with SAGE permits stable heterologous gene expression
A critical advantage of SAGE-mediated DNA integration over the use of replicating plasmids is genetic stability in the absence of selection through antibiotics or auxotrophy. This is particularly important when engineering bacteria for deployment in environments where standard selections for plasmid maintenance are impractical or ill advised (e.g., human gut, plant rhizospheres, bioremediation, and industrial bioreactors). As a proxy for engineered functions, we compared the stability of fluorescent protein expression cassettes maintained on replicating plasmids or SAGE-integrated genomes in P. fluorescens SBW25. We also assessed the stability of SAGE-integrated fluorescent protein expression in R. palustris CGA009. For a thorough examination, we tested strains generated by each of the recombinases, with the exception of ΦBT1 or ΦC1 integrases in SBW25.
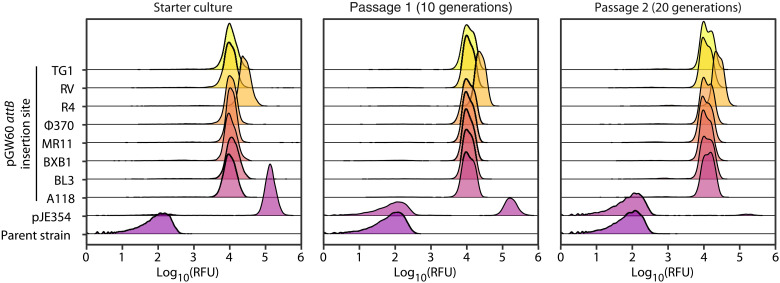
As expected, fluorescent reporter expression was substantially more stable in strains generated by SAGE than strains generated by transformation of replicating plasmids. We used flow cytometry to measure the fraction of the population that expresses the fluorescent reporter construct. Expression was stable in all strains generated using SAGE, with >99.75% of cells expressing mNeonGreen after 20 generations of growth without selection in P. fluorescens (Fig. 4). However, replicating plasmids were lost rapidly without selection. Only 2.5% of the P. fluorescens and populations produced fluorescent proteins after the multipassage cultivation in the absence of selection. Under similar conditions, an equivalent fraction of the R. palustris populations maintained fluorescence after 14 generations (fig. S8).
Fig. 4. SAGE enables stable expression of heterologous genes in the absence of selection.
Flow cytometry measurements of fluorescence in populations of P. fluorescens JE4621 containing either no heterologous expression construct (parent strain), a pBBR-based replicating mNeonGreen expression vector (pJE354), or a nonreplicating mNeonGreen expression vector that has been chromosomally integrated at the indicated attB site by the corresponding recombinase. Starter cultures, with the exception of the parent strain that was grown in the absence of antibiotic, were cultivated in medium containing kanamycin sulfate (50 μg/ml) for plasmid selection. Each passage was inoculated with a 1024-fold dilution of its precursor culture (starter → passage 1 → passage 2) to enable 10 generations of exponential growth before reaching stationary phase. The x axis indicates the relative fluorescence (excitation at 488 nm and emission at 530 nm) of each cell. The y axis represents the abundance of cells in the population with a given relative fluorescence unit (RFU).
The presence of multiple SAGE-integrated excision scars does not impair genetic stability. The excision of SAGE plasmid backbones leaves a 47–base pair (bp) ΦC31 attL sequence scar in the chromosome, and the presence of multiple ΦC31 attL sites could introduce genetic instability through homologous recombination. It is notable that repeat sequences in bacterial genomes are common. For instance, P. fluorescens SBW25 natively hosts 611 and 514 copies each of the R0 and R2 repeat sequences, which are 96 and 135 bp, respectively. We evaluated the stability of mNeonGreen and mKate2 expression in P. fluorescens JE5117. Recombination between the two ΦC31 attL sequences in this strain would lead to excision of the mKate2 gene (fig. S6A) and loss of red fluorescence. We cultivated two lineages of JE5117 and used flow cytometry to measure the fraction of the population that displayed both red and green fluorescence after 100 generations. In both lineages, the fraction of the population that displayed both red and green fluorescence remained steady (fig. S9 and table S4). This demonstrates that recombination between ΦC31 attL sequences is likely to be very rare. Distribution of SAGE attB integration sites across the chromosome would further reduce the likelihood of rare recombination events.
Analysis of genome-integrated expression libraries reveals context-independent promoters
Development of genetic elements [e.g., promoters, ribosomal binding sites (RBSs), and terminators] that control expression of heterologous genes is a critical aspect of engineering functions in bacteria. To minimize the impact on host fitness and increase evolutionary stability, gene expression from engineered pathways must be carefully balanced with core cellular functions. In addition, bacteria are typically deployed in dynamic environments, and thus, it is critical to identify promoters that provide consistent gene expression across many conditions. We therefore developed and validated a methodology to assess large libraries of genome-integrated promoters across environmental conditions in SAGE-capable hosts.
Promoter libraries have been developed to control transcriptional activity using reporter protein expression in several organisms (63–66). However, most promoter libraries, including the quasi-standard Anderson library (http://parts.igem.org/Promoters/Catalog/Anderson), have been developed and characterized using multicopy reporter plasmids, which are likely not representative of the transcription rate from the chromosome. Heterogeneity in copy number among colonies and within subpopulations, plasmid loss within subpopulations (Fig. 4, pJE354), and overall higher DNA dosage from plasmids can influence promoter performance. In addition, accurate measurement of promoter activity from plasmid-based reporters can be further confounded by the metabolic burden imposed by plasmid maintenance (16, 18–21). Thus, measurement of gene expression from genome-integrated promoters is likely to be more predictive of activity in deployed environments than plasmid-based promoter measurements.
To characterize a library of chromosomally integrated promoters, we used a methodology that combines the efficiency of SAGE integration with a modified implementation of the high-throughput sequencing transcriptional activity assay described by Johns et al. (64). We demonstrated this approach, described briefly below, to characterize a collection of 287 synthetic and natural promoters (Supplementary File T1) in the five organisms we used to demonstrate SAGE. In the original method, barcode amplicon sequencing from both RNA and DNA was used to normalize and quantify both transcript and promoter abundance. We modified the original transcriptional activity assay in two important ways, and for simplicity, we will refer to this method as relative transcriptional profiling (RTP). First, rather than using a single barcode per promoter, we use five distinct barcode variants per promoter for a total of 1435 promoter/barcode variants. Using multiple barcodes per promoter provides in-experiment technical replicates to gain higher confidence in data generated for each promoter and condition, which allows identification and removal of spurious outlier barcode variants. Second, we inserted the barcode within the 5′ untranslated region (5′UTR) immediately upstream of the RBS, rather than locating the barcode within in the reporter gene coding sequence (Fig. 5A). This enables an analysis of promoter sensitivity to 5′UTR sequence variation. This is an important metric for heterologous promoters, as 5′UTR sequence affects transcript abundance via multiple mechanisms, and modification of the ribosomal binding sequence within the 5′UTR is a common method used to tune gene expression in engineered bacteria.
Fig. 5. Development and high-throughput analysis of genome-integrated promoter libraries.
(A) Overview of promoter library construction and DNA/RNA sequencing (RNA-seq) barcode sequencing fragments. (B) Example promoters with different classes of 5′UTR (level of barcode-associated “noise”) and condition sensitivity. Large, outlined circles represent mean values, and small dots represent individual samples. (C) Chart displaying mean RTA for consistent (5′UTR and condition insensitive) promoters in P. fluorescens SBW25. Relative strength of the promoters used in (D) is indicated. Error bars represent two-sided SD of between 36 and 80 samples (see source data file and Supplementary File D1 for exact numbers). (D) Promoter performance for a small subset of pLibrary promoters in microtiter plate growth assays. Error bars represent two-sided SD in three replicates. Relative promoter activity is calculated by comparing mean RFU/OD600 values across all carbon sources. (E) Correlation between relative expression levels determined by RTP and fluorescent protein reporter assay for the set of promoters used in (D). Linear equation and coefficient of determination between the same promoters using RTA data from (C) and fluorescent plate reader data from (D), as determined by Pearson correlation.
To identify promoters that exhibit consistent expression across varied 5′UTR and environmental contexts, we integrated a pooled library of barcoded reporter plasmids (pLibrary) into the genomes of each of the five organisms via SAGE (Fig. 5A). Using the pooled Escherichia coli pLibrary collection and wild-type control strains of P. fluorescens, R. palustris, and R. jostii as references, we measured mNeonGreen production in each integrated strain library via flow cytometry (fig. S10). A broad range of mNeonGreen expression levels was observed for each strain library, suggesting that RTP would provide condition and organism-resolved expression data at the single-promoter level.
We performed RTP under multiple conditions that are relevant for each of the five organisms (Table 1). We cultivated P. fluorescens, a model plant growth–promoting rhizobacteria (PGPR), in media supplemented with one of three soil-relevant carbon sources: glucose (breakdown product of cellulose), citric acid (compound found in root exudate), or N-acetylglucosamine (breakdown product of chitin). For P. putida and R. jostii, emerging hosts for lignocellulose valorization that are known for their bioremediation capabilities, we cultivated with glucose, vanillic acid (model lignin-derived aromatic), or a petroleum-derived alkane (n-octane for P. putida and n-decane for R. jostii). As P. frederiksbergensis TBS10 has potential as a PGPR under drought conditions, we cultivated it under standard, drought simulant [25% polyethylene glycol 6000 (PEG6000)], and salinity stress (400 mM NaCl) conditions. Last, as a model organism to study photosynthesis-dependent anaerobic conversion of organic acids into biomass and aerobic utilization of organic acids without light, we cultivated the R. palustris library with acetate as the sole carbon source under dark aerobic and illuminated anaerobic conditions. For each culture replicate, we collected high-throughput sequencing data to enumerate barcode abundance in the DNA and RNA fractions from each sample. Relative transcriptional activity (RTA) for a promoter in a given sample was calculated by comparing its RNA:DNA barcode abundance ratio (see Materials and Methods).
After data preprocessing and quality filtering (see Materials and Methods), we obtained sufficient data to analyze performance of between 186 and 241 promoters in the four proteobacteria and 136 promoters in the actinomycete R. jostii (table S5). Normalizing ratiometric counts across organisms, we quantified the RTA of promoters with mean expression covering more than four orders of magnitude (Supplementary Files F1 to F5). Supporting the value of using multiple barcode variants per promoter, we found that between 11% (P. frederiksbergensis) and 20.3% (P. fluorescens) of promoters had an outlier barcode promoter variant with statistically distinct expression (see Materials and Methods) compared to the remaining barcode variants for the given promoter. These results highlight the importance of considering the influence of 5′UTR sequence on transcriptional activity (table S5).
We used mean expression statistics to evaluate promoter sensitivity to environmental and genetic contexts (Fig. 5B). We classified promoters for which the mean RTA of every nonstatistical outlier barcode variant fell within a threefold range as insensitive to 5′UTR sequence variation. Similarly, we classified promoters for which the mean RTA of every condition fell within a fourfold range as insensitive to environmental conditions. We labeled promoters passing both thresholds as consistent promoters (Fig. 5C and fig. S11). Promoter sensitivity classifications over a range of stringency cutoffs for 5′UTR and condition are located in tables S6 to S10.
Our analysis found most promoters to be insensitive to environmental condition or 5′UTR sequence. However, sensitivity of promoter activity to environmental condition and 5′UTR sequence were more often than not independent of one another (Supplementary Files F1 to F5). With the exception of 5′UTR sensitivity in R. palustris (45% insensitive) and condition sensitivity in P. fluorescens (99% insensitive), between 68 and 87% promoters were insensitive to either condition or 5′UTR perturbation (table S5).
We validated RTP-quantified expression by comparing results from barcode sequencing to plate reader–based fluorescence assays with each of the subset of the related JEa, JEb, and JEc series promoters that were present in the P. fluorescens library (Fig. 5D and fig. S12). Expression levels measured by each method strongly correlated (Fig. 5E and fig. S12B), suggesting that the high-throughput barcode sequencing provides results that are comparable with fluorescent protein reporter assays. While similar trends were observed between methods, barcode sequencing offers several advantages over fluorescent and colorimetric reporter assays: (i) Sensitivity is only limited by sequencing depth rather than by physical limitations (e.g., autofluorescence), (ii) thousands of promoters can be tested in a single experiment, (iii) measurements are not limited by cell density, and (iv) the assay works under anaerobic conditions, thus enabling promoter library development for organisms (e.g., Clostridia) where reporter systems have been limiting.
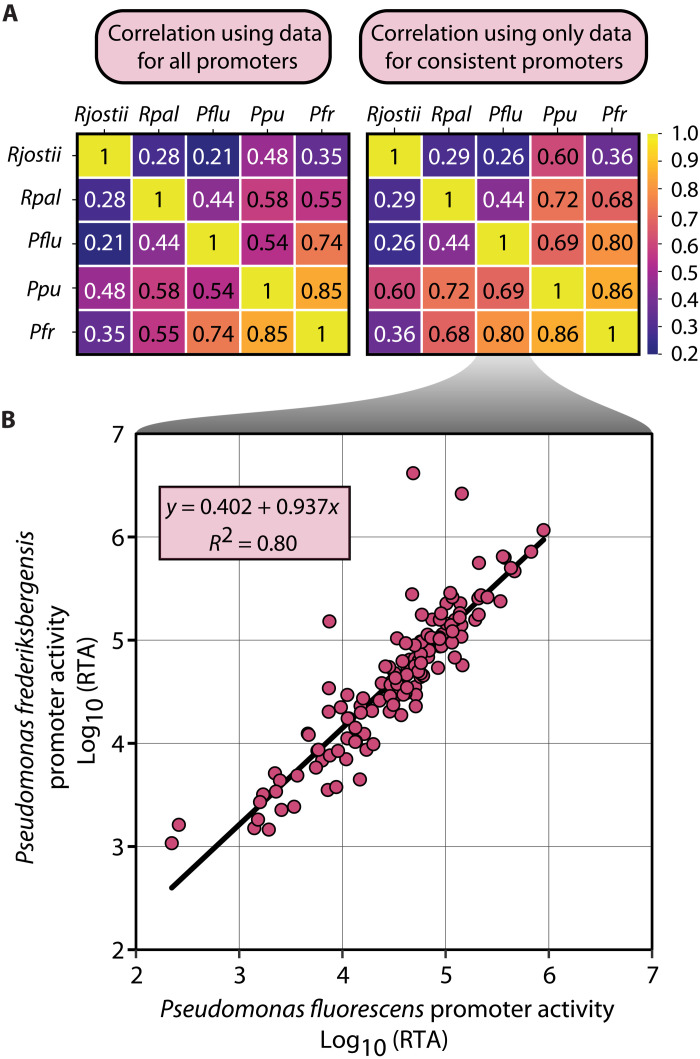
Strain-to-strain expression level correlations decreased with phylogenetic distance. We used Pearson correlations to calculate the R2 correlations for the mean promoter expression across all barcodes and conditions (Fig. 6). With the exception of R. jostii and P. putida (R2 = 0.60), strain-strain correlations largely map to phylogenetic distance (e.g., the proteobacteria each correlate more closely with one another than with the actinomycete R. jostii). When calculating correlations for only the consistent promoters, we found correlations increased for most comparisons, especially for R. palustris correlations to the other proteobacteria. Scatterplots of the correlations for all promoters are found in fig. S13.
Fig. 6. Promoter activity correlates with phylogenetic distance.
(A) Coefficient of determination (R2) generated when comparing RTA of promoters between promoters that are characterized in each organism pair. R2 values are provided for both sample sets including all promoters or for a subset of promoters that only includes promoters that were consistent in both organisms. (B) Example scatterplot of data used to perform Pearson correlation and the linear equation and coefficient of determination generated using Pearson correlation using consistent promoters from P. fluorescens and P. frederiksbergensis.
SAGE toolkit for scientific community
To support and broadly extend SAGE in bacteria, we have curated a series of modular plasmids (fig. S5) that can either be used directly to perform SAGE or that can be readily modified to address host-specific needs (e.g., selection markers). This toolkit includes both recombinase expression plasmids and attP-containing recombinase target plasmids for genome integration. Recombinase expression plasmids are available as both suicide (nonreplicating) and mSFts1-based (temperature-sensitive Pseudomonas origin) plasmids (pGW13–40 and pJE1817). A set of two poly-attP plasmids (pGW63 and pJE1866) that each contain a gentamicin resistance selection marker are also currently available for use in evaluating SAGE in microbes that are resistant to kanamycin (fig. S5). The library also includes a modular collection of single attP “target” plasmids, each of which contains either the nptII (kanamycin resistance) marker alone (pJH204–212) or in combination with sacB for sucrose counterselection (pJE1828–1836). Each component of the attP plasmids (e.g., selection marker, attP site, E. coli origin, and cargo insertion site) is designed to be modular. Each component is flanked by unique restriction sites to allow easy replacement of components or for addition of genetic elements (e.g., oriT sequences for conjugation). The cargo region in each plasmid is flanked by a pair of double rho-independent terminators and contains several unique restriction sites, including a pair of Bbs I recognition sites that are compatible with Golden Gate (67) cloning. Plasmid pJE990, which was previously developed for use in P. putida (31), is a related plasmid that is designed to be used as a fluorescent reporter plasmid, with sites for modular integration and replacement of promoter, RBS, and terminator sequences (fig. S5). In addition to the reporter plasmids generated in this study, additional pJE990-based reporter plasmids containing the commonly used tac and T7 promoter (variants) (68) or the far-red fluorescent protein mKate2 are also currently available (31, 69). Last, the reporter gene in all pJE990-based plasmids is designed for easy replacement with any gene(s) of interest (69). We expect that these current tools and expansions of the SAGE genetic toolkit will be broadly useful for genetic manipulation across the entire bacterial domain.
DISCUSSION
The SAGE toolkit extends the accessibility of high-throughput engineering for bacteria in any application space (e.g., nutraceuticals synthesis, biofuel production, etc.) and will be especially transformative for applications where selection for plasmid maintenance is impossible (e.g., soil or human gut microbiome engineering). The beauty of the SAGE system is its simplicity and broad applicability. Insertion of DNA into the chromosome is as simple as transformation with a replicating plasmid while also providing the consistency and stability of genome integration. A plasmid does not need to be redesigned to contain a new origin of replication or DNA for homologous recombination to be transferred to a new host; the exact same plasmid (or plasmid library) can be inserted stably into the chromosome of each SAGE-enabled host if appropriate selectable markers are chosen. For any target organism, strains can be constructed and genetically validated by PCR within 1 week of having the plasmids in hand. The genome integration efficiency we observed when using SAGE in multiple phylogenetically distant bacteria is among the highest when compared to competing technologies and, unlike these technologies, does not require use of plasmids that can replicate in the host.
The toolkit also includes nonreplicating and conditionally replicating plasmids for recombinase expression and attP-containing target plasmids with multiple options for selection and counterselection markers (fig. S5). In addition to existing plasmids in the toolkit, the modular nature of each plasmid makes replacement of each component (e.g., selection markers, att sites, etc.) straightforward. Furthermore, using a second serine recombinase (ΦC31 integrase) to excise the selection marker and plasmid backbone, SAGE enables iterative integration of up to 10 distinct pools of DNA fragments. The SAGE system is also expandable. Some of the recombinases function at high efficiency and high accuracy, but others are more variable between organisms. Recently, many new recombinases and corresponding att sites have been computationally predicted (70, 71). These recombinases will serve as a rich source of new, diverse enzymes for the extension of SAGE-based approaches.
DNA integration by serine recombinases should be selectively reversible by coexpression of a recombinase with its cognate recombination directionality factor protein (72, 73). This reversibility could be harnessed to apply SAGE for directed evolution by excising genome-integrated plasmid variants as circular DNA to be used as PCR mutagenesis templates or transferred to E. coli for replication and manipulation. Last, SAGE is orthogonal to host recombination machinery and does not require use of any replicating plasmids and thus is theoretically extensible to any transformable bacteria, regardless of the sophistication of genetic tools. As long as an attB cassette can be transferred into the organism’s genome, via allelic exchange, recombineering, transposon-based integration, etc., the bacteria should be compatible with SAGE cycling.
MATERIALS AND METHODS
General culture conditions and media
The strains and plasmids used in this study are listed in table S1. Routine cultivation of E. coli for plasmid construction and maintenance was performed at 37°C using LB (Lennox) medium supplemented with antibiotics [kanamycin sulfate (50 μg/ml), apramycin sulfate (50 μg/ml), or gentamicin sulfate (30 μg/ml)] and agar (15 g/liter; for solid medium). All pseudomonad, R. palustris, and R. jostii cultures were incubated at 30°C unless otherwise indicated. Test tube and shake flask cultures were mixed by shaking at 225 rpm with a 1-cm orbital in an Innova S44i (Eppendorf) incubator equipped with a refrigeration unit and a 150 photosynthetic active radiation (wavelengths 400 to 700 nm) light bank. Plate reader cultivation assays were performed using the “fast shaking” 1 mM double orbital setting on a Neo2SM plate reader (BioTek). All aerobic cultures were performed in the dark, and photosynthetic anaerobic R. palustris cultures cells were performed in 22-ml Balch tubes with 100% illumination (150 PAR) that were sparged for 10 min with N2 to generate an anaerobic atmosphere.
LB (Lennox) was used for routine pseudomonad strain maintenance, competent cell preparations, RTP assay starter cultures, and plasmid stability assays. Kanamycin sulfate (50 μg/ml) or apramycin sulfate (50 μg/μl) were used for antibiotic selection. Van Niel’s medium (American Type Culture Collection, medium 112) supplemented with 20 mM sodium acetate (VN-A medium) was used for all cultivation of R. palustris. All routine cultivation of R. palustris was performed aerobically in the dark. Kanamycin sulfate (200 μg/ml) or gentamicin sulfate (200 μg/ml) were used for antibiotic selection. LB (Lennox) or R2A medium was used for routine R. jostii strain maintenance (both), competent cell preparations, and RTP assay starter cultures (R2A). Kanamycin sulfate (50 μg/ml) or apramycin sulfate (30 μg/μl) were used for antibiotic selection. All solid medium was prepared by addition of agar (15 g/liter), and counterselection with expression of levansucrase was performed by addition of 25% (P. fluorescens) or 10% (R. jostii and R. palustris) sucrose to either LB (P. fluorescens and R. jostii) or VN-A (R. palustris).
Three base media variants were used for plate reader and RTP assays. Mops-based mineral medium (MME) (74) with variable nitrogen, carbon, or stress-inducing compounds was used for pseudomonad shake flask experiments and fluorescent plate reader assays. MME contains 9.1 mM K2HPO4, 20 mM Mops, 4.3 mM NaCl, 20 mM NH4Cl, 0.41 mM MgSO4, 68 μM CaCl2, and 1× MME trace minerals (pH adjusted to 7.0 with KOH). MME trace mineral stock solution (1000×) contains per liter 1 ml of concentrated HCl, 0.5 g of Na4EDTA, 2 g of FeCl3, and 0.05 g each of H3BO3, ZnCl2, CuCl2⋅2H2O, MnCl2⋅4H2O, (NH4)2MoO4, CoCl2⋅6H2O, and NiCl2⋅6H2O. PMA medium [mineral medium used for R. palustris cultivation (PM) (75) supplemented with 20 mM sodium acetate] was used for R. palustris RTP assays, and an MME variant (MMV) additionally containing 10 mg each of niacin, pantothenate, lipoic acid, p-aminobenzoic acid, thiamine (B1), riboflavin (B2), pyridoxine (B6), and cobalamin (B12) and 4 mg each of biotin and folic acid per liter was used for R. jostii RTP assays.
For RTP and fluorescent plate reader assays, MME and MMV media were supplemented with the following. For P. fluorescens, 10 mM NH4Cl with either 10 mM glucose or 10 mM citrate, 10 mM N-acetylglucosamine alone, or 10 mM glucose with 10 mM urea, 10 mM sodium nitrate, or 10 mM sodium nitrite. For P. putida, 10 mM NH4Cl with either 10 mM glucose, 10 mM vanillic acid, or 0.1% (w/v) n-octane. For P. frederiksbergensis, 10 mM NH4Cl and 10 mM glucose with either no additional supplement, 400 mM NaCl, or 25% (w/v) PEG6000. For R. jostii, 10 mM NH4Cl with either 10 mM glucose, 5 mM vanillic acid, or 0.1% n-decane.
Isolation of sorghum root endophyte P. frederiksbergensis TBS10 from sorghum roots
Sorghum plants were grown at the University of California’s Agriculture and Natural Resources Kearney Agriculture Research and Extension Center in Parlier, CA, as described previously (76, 77). Root samples were obtained from mature sorghum plants that had been subjected to a prolonged preflowering drought. Immediately after extraction of plants from soil, roots were removed and placed in 25% glycerol for 30 min and then placed on dry ice until they were transferred to −80°C. To remove soil, roots were placed in a phosphate buffer and sonicated briefly. They were subsequently vortexed for 60 s in 99% ethanol, 6 min in 3% NaOCl, and 30 s in 99% ethanol to sterilize the root surface. Roots were washed twice in sterile deionized water, and 100 μl of rinse water was plated to check surface sterility. Roots were then cut into 1-cm pieces and placed into 2-ml tubes with 25% glycerol and incubated for 30 min at room temperature before storing at −80°C.
One 2-ml tube of roots (approximately 200 mg) was thawed and placed in a sterile ceramic mortar with 1 ml of phosphate-buffered saline (PBS) buffer. Root tissue was ground gently to release endophytic bacteria into the solution while minimizing lysis of bacterial cells. The solution was serially diluted, and 100 μl of dilutions 10−1, 10−2, and 10−3 were plated onto International Streptomyces Project-2 (ISP2) medium. Plates were incubated at 30°C, and growth was monitored daily. When colonies were visible, they were picked and streaked onto a fresh plate of ISP2, followed by subsequent streaks if necessary to eliminate contamination, until only a single morphology was observed. Sanger sequencing of the 16S ribosomal RNA gene was performed to identify isolates, and isolate TBS10 had a high sequence similarity to P. frederiksbergensis species.
Plasmid and bacterial strain construction
Q5 High-Fidelity DNA Polymerase [New England Biolabs (NEB)] and primers synthesized by Eurofins Genomics were used in all PCR amplifications for plasmid construction. OneTaq DNA polymerase (NEB) was used for colony PCR. EvaGreen dye (Biotium) was supplemented at 1× final concentration in PCRs for quantitative PCR (qPCR) applications. Plasmids were either synthesized by Twist Biosciences or constructed by Gibson Assembly using the NEBuilder HiFi DNA Assembly Master Mix (NEB) or ligation using T4 DNA ligase (NEB). Plasmids were transformed into either competent NEB Turbo, NEB 5-alpha F’Iq (NEB), Epi400 (Lucigen), or QP15 (69). Standard chemically competent E. coli transformation protocols were used to construct plasmid host strains. Transformants were selected on LB (Lennox) agar plates containing antibiotics [kanamycin sulfate (50 μg/ml), apramycin sulfate (50 μg/ml), or gentamicin sulfate (30 μg/ml)] for selection and incubated at 37°C. Template DNA for PCRs was either synthesized by IDT, synthesized by GenScript, or isolated from E. coli, P. fluorescens SBW25, R. palustris CGA009, or R. jostii RHA1 using the DNeasy Blood & Tissue genomic DNA (gDNA) purification kit (QIAGEN) or the Zymo Quick gDNA miniprep kit (Zymo Research). The Zymoclean Gel DNA recovery kit (Zymo Research) was used for all DNA gel purifications. Plasmid DNA was purified from E. coli using the GeneJet plasmid miniprep kit (Thermo Fisher Scientific) or the ZymoPURE II plasmid midiprep kit (Zymo Research). Sequences of plasmids were confirmed using Sanger sequencing performed by Eurofins Genomics. Plasmids used in this work are listed in table S1, and maps of plasmids constructed in this study can be found in Supplementary File P1.
P. fluorescens SBW25, P. frederiksbergensis TBS10, and P. putida Gpo1 were used as parent strains for constructing all pseudomonad strains in this study (table S1). Competent cells for pseudomonads were prepared, and the allelic exchange protocol used for integration of the poly-attB cassette into P. fluorescens was performed as described previously (78) for P. putida KT2440, with the exception that pK18sB (79) was used as the cloning vector, rather than pK18mobsacB. The same competent cell preparation and electroporation protocols above were used for electroporation of serine integrase attP target plasmids, for replicating or nonreplicating helper integrase expression plasmids, and for in vitro–assembled EZ-Tn5 transposomes (Lucigen).
Construction of P. putida and P. frederiksbergensis poly-attB base strains were generated using an EZ-Tn5 transposase kit (Lucigen). DNAs for the transposomes used to incorporate nptII-poly-attB cassettes (Fig. 2B) were prepared by incorporating Tn5 att sequences and 5′ phosphate moieties to either end of the transposon sequence in pJE1855 with primers oPNL1254/1255. PCR products were purified using the Zymo DNA Clean & Concentrate-5 kit (Zymo Research). Transposomes containing this DNA fragment were generated in vitro following the manufacturer’s provided protocol. Two microliters of each transposome assembly reaction was used for electroporation into either P. putida or P. frederiksbergensis. Selection for transformants was performed using LB supplemented with kanamycin sulfate. Kanamycin-sensitive strains were generated by electroporating the nonreplicating ϕC31 integrase expression plasmid (pJE1817) and culturing individual colonies onto LB with and without supplemental kanamycin. Excision of nptII by ϕC31 integrase was validated by colony PCR using combinations of primers oPNL911, oPNL912, oPNL623, oPNL1011, and oPNL1147.
R. palustris CGA009 was used as the parent strain for R. palustris strains in this study (table S1). Electrocompetent cells were prepared by preculturing R. palustris aerobically at 30°C until the cells reached stationary phase. Fresh medium was inoculated with a 1:100 dilution of the preculture and incubated aerobically until the cells reached mid-to-late log phase [~OD600 (optical density at 600 nm) = 0.5 to 0.9). Once the desired culture density was reached, the cells were incubated on ice until chilled. All the following steps were performed on ice or at 4°C. Cells were centrifuged at 4°C and 4000g for 10 min. Pelleted cells were washed in 1/10th culture volume of ice-cold 10% glycerol. The centrifugation and wash steps were performed two more times for a total of three washes. Following the final centrifugation, the cells were resuspended in 1:400th culture volume of ice-cold 10% glycerol and either used immediately or stored at −80°C. Electroporation was performed as follows. Fifty microliters of electrocompetent cells or electrocompetent cells diluted 1:10 in ice-cold 10% glycerol was transferred to an ice-cold 0.1-cm-gap electroporation cuvette. For allelic exchange, 500 to 1000 ng of plasmid DNA was added to the cells, and the mixture was electroporated with settings at 1.75 kV, 25 μF, and 200 ohm. The same protocol was used for electroporation of serine integrase attP target plasmids and nonreplicating helper integrase expression plasmids. Following electroporation, 950 μl of VN-A was added to the competent cells, and the resulting mixture was incubated aerobically, at 30°C with shaking for 2 hours. Following this recovery step, various dilutions of recovery cultures were plated on selective VN-A solid medium and incubated aerobically at 30°C. Colonies typically appeared after ~4 to 5 days when R. palustris is grown on solid medium. Notably, transformation efficiencies within approximately one order of magnitude were observed using competent cells that were prepared by harvesting culture stationary phase or with all steps and electroporations performed at room temperature rather than at on ice or 4°C.
To generarate R. palustris poly-attB strain JE4632, we used the pJQ200SK (62) gentamicin resistance/sucrose sensitivity selection/counterselection method for allelic exchange with minor modifications for R. palustris to generate the poly-attB strain JE4632. For this, 700-bp regions of homology flanking RPA1300 were cloned immediately upstream and downstream of the poly-attB cassette within the multiple cloning site of pJQ200SK. The resulting plasmid was transformed into R. palustris CGA009, and resulting colonies that typically arise from plasmid recombination into the genome via homologous recombination were selected by cultivation on VN-A supplemented with gentamicin sulfate (200 μg/ml). Isolated colonies were streaked for single-colony isolation on VN-A medium with gentamicin. For counterselection, we cultivated colonies from the second selective plate on VN-A supplemented with10% sucrose. Resulting colonies have typically resulted from excision of sacB from the genome, as its expression caused a substantial growth defect in the presence of sucrose. Resulting colonies were patched onto VN-A supplemented with 10% sucrose and subsequently tested by colony PCR for the insertion of the poly-attB cassette. Positive colonies were subsequently streaked for single-colony isolation again on VN-A, and resulting colonies cultivated in liquid VN-A. These final cultures were screened again by colony PCR, and correct colonies were stored as 10% glycerol stocks at −80°C.
R. jostii RHA1 was used as the parent for R. jostii strains in this study. R. jostii strain AG5879, which contains the poly-attB cassette, was constructed by integrating plasmid pQP425 into the genome of RHA1 via conjugation with E. coli. Progenies were then screened using LB supplemented with neomycin sulfate (20 μg/ml) for positive selection and LB supplemented with 10% sucrose for counterselection. The resulting colonies were screened by colony PCR for insertion by colony PCR.
Electrocompetent R. jostii cells were prepared by preculturing at 30°C in LB (allelic exchange) or R2A (transformation assays and promoter library construction) until cultures reached stationary phase. Fresh medium was inoculated with the preculture to an OD600 of 0.01 and incubated aerobically until the cells reached a mid-log phase (~OD600 = 0.4 to 0.6). Once the desired culture density was reached, the cells were centrifuged at 5000g for 20 min at 4°C and washed at one-half culture volume of ice-cold 10% glycerol. Centrifugation and washing were repeated two additional times, and then cells were resuspended in 1/50th culture volume of ice-cold 10% glycerol. Competent cells were either used immediately or stored at −80°C. Electroporation was performed as follows. DNA was added to 70 μl of ice-cold electrocompetent cells and incubated on ice for 30 min. Following incubation, competent cells were transferred into an ice-cold 0.1-cm-gap electroporation cuvette and electroporated at 1.6 kV, 25 μF, and 200 ohm. Following electroporation, 950 μl of R2A was added to the competent cells, and the resulting mixture was incubated aerobically, at 30°C with shaking for 3 hours. Following this recovery step, various dilutions of recovery cultures were plated on selective R2A solid medium and incubated aerobically at 30°C. Colonies typically appear after ~2 days when R. jostii is grown on solid medium. Primers used to screen for integration of the poly-attP cassette insertions in P. fluorescens, R. palustris, and R. jostii can be found in table S3.
Temperature-sensitive plasmid stability and pGW60 backbone excision assays
Multiple methods were used to assess the conditional replication of pGW26 in P. fluorescens SBW25 at various temperatures. First, P. fluorescens was electroporated with 250 ng of pGW26 and recovered in 1 ml of SOC (Super Optimal broth with Catabolite repression) medium at 25°C for 2 hours. Dilutions of the recovery cultures were inoculated onto LB supplemented with apramycin sulfate (50 μg/ml) and incubated at 25° or 34°C for up to 96 hours (to confirm the absence of colony formation at restrictive temperature). Colonies were enumerated and transformation efficiency was determined (fig. S4A).
Second, we cultivated a SBW25-derivative strain containing pGW26 in LB supplemented with apramycin sulfate (50 μg/ml) at 25°C to select for plasmid maintenance. After reaching stationary phase, a 1:1000 dilution of the culture was inoculated into six different subcultures. Subcultures were grown at either 25°, 30°, or 34°C either in the presence or absence of apramycin sulfate (50 μg/ml) until they reached stationary phase (typically with 24 hours). Each culture in which growth occurred was diluted to reach an OD600 of 0.5, and further 10-fold serial dilutions were prepared. Five microliters of each dilution was spotted onto solid LB medium containing or lacking apramycin sulfate (50 μg/ml). These solid medium cultures were then incubated at 25°, 30°, or 34°C for up to 72 hours, and colonies were enumerated to establish viable cell counts under each condition. Overall cell viability and percentage of cells maintaining replicating pGW26 within each subculture at each cultivation temperature were determined by counting colonies from four replicates on plates lacking and containing apramycin sulfate (50 μg/ml), respectively (fig. S4, B and C).
Third, we demonstrated two succinct plasmid backbone excision protocols that use different methods to select for colonies in which the plasmid backbone has been excised. The first approach selects for transformation with a temperature-sensitive plasmid harboring a ϕC31 integrase expression cassette, and the second selects for removal of the counterselection marker gene sacB from the genome. For the first method, we used pGW26 and pGW30 (fig. S3), which host a pseudomonad-specific temperature-sensitive origin of replication. To test backbone excision, P. fluorescens JE4689, which contains pGW60 integrated into the Bxb1-attB site of JE4621, was electroporated with either of the two aforementioned ts-plasmids, and recovery culture was incubated at 25°C on LB supplemented with apramycin sulfate (50 μg/ml). Twenty apramycin-resistant transformants for each plasmid were streaked onto LB for single-colony isolation and incubated overnight at 34°C. A single colony from each streak was tested by colony PCR for presence of both their respective ts-plasmids and excision of the pGW60 plasmid backbone. The same cellular material was patched onto both LB or LB supplemented with apramycin sulfate (50 μg/ml) and incubated overnight at 30°C to confirm loss of antibiotic resistance. Primers used for screening plasmid loss (oPNL556/879) and excision of the pGW60 backbone from the genome of JE4689 (oPNL621/622) are listed in table S2.
For the second method, P. fluorescens JE4866 and R. palustris JE4865 containing pJE1818 integrated into the Bxb1-attB sites of JE4621 and JE4632, respectively, were electroporated with the nonreplicating ϕC31 integrase expression plasmid pJE1817 and, following recovery cultivation transformants, were selected by screening for sucrose insensitivity. For P. fluorescens, recovery cultures were incubated for 4 hours in 1 ml of SOC, and dilutions were plated onto solid YT supplemented with 25% sucrose (78). For R. palustris, recovery cultures were incubated for 24 hours in 1 ml of VN-A, and dilutions were plated onto solid VN-A supplemented with 10% sucrose. Twenty sucrose-insensitive colonies were screened by colony PCR for excision of the pJE1817 plasmid backbone using oligos oPNL621/622.
Serine integrase transformation assays
Integrase testing assays were performed using variations of the transformation procedures described in the “Plasmid and bacterial strain construction” section with the following modifications. The following amounts of plasmids were electroporated into either R. palustris JE4632, P. fluorescens JE4621, P. frederiksbergensis JE5041, P. putida JE5035, or R. jostii AG5879. Transformations included 100 ng of control plasmid (pJE354, pEYF2K, or pSET152) or 100 ng of the pGW60 attP target plasmid that is electroporated alone or coelectroporated with 250 ng of nonreplicating integrase expression helper plasmid. Following electroporation, cells were either resuspended in 950 μl of SOC (pseudomonads), 950 μl of VN-A (R. palustris), or 950 μl of R2A (R. jostii); transferred to a microfuge tube; and incubated at 30°C for 1 hour (pseudomonads), 2 hours (R. palustris), or 3 hours (R. jostii) to allow for recovery. Several fractions of the recovery volume were plated. Colonies were enumerated and pGW60 colonies were evaluated visually for green fluorescence. For P. fluorescens assays, all samples were plated on LB agar supplemented with kanamycin sulfate (50 μg/ml). For R. palustris assays, all samples were plated on VN-A agar supplemented with kanamycin sulfate (200 μg/ml). For R. jostii assays, all pGW60 electroporation samples were plated on R2A supplemented with kanamycin sulfate (50 μg/ml), and pSET152 samples were plated on R2A supplemented with apramycin sulfate (30 μg/ml). Plates were incubated at 30°C until colonies became visible. This was typically 24 hours for P. fluorescens, approximately 5 days for R. palustris, and 2 days for R. jostii.
Integration accuracy was determined by examining up to 24 colonies for each integrase by PCR for integration of the pGW60 plasmid integration at the correct locus. Colonies were cultivated with shaking (800 rpm, 1-mm orbital) at 30°C in 96-well deep-well plates using either 1 ml of LB + kanamycin sulfate (50 μg/ml; pseudomonads), 1 ml of VN-A + kanamycin sulfate (200 μg/ml; R. palustris), or 1 ml of R2A + kanamycin sulfate (50 μg/ml; R. jostii). Cultures were lysed using the following method. Two hundred microliters of saturated culture was centrifuged at 3000g for 5 min to pellet cells. The supernatant was decanted, and cell pellets were resuspended in 100 μl of TE [10 mM tris-HCl and 1 mM EDTA (pH 8)] + 0.1% Triton X-100 detergent. Resuspended cells were boiled at 100°C for 5 min and centrifuged again, and 50 μl of supernatant was transferred to a 384-well Echo source plate (product #PP-0200, Labcyte) and used as a PCR template. An Echo 550 acoustic liquid handler (Labcyte) was used to transfer 500 nl of each culture’s clarified lysate into 10 μl of colony PCRs performed using OneTaq polymerase (NEB) with a CFX-384 Touch real-time PCR machine (Bio-Rad). For P. fluorescens JE4621, colonies were screened using either a positive control primer set that should amplify in any P. fluorescens SBW25 strain (oPNL615/616) or a primer set that should only amplify a product if pGW60 has been integrated into the chromosomal poly-attB cassette that is downstream of ampC in JE4621 (oPNL629/622). Notably, oPNL629/622 generate products of distinct sizes depending on which serine integrase performs the plasmid integration. A similar approach was used in R. palustris, with the exception that colonies were screened by PCR as described above using primer set oPNL819/817 to assess integration in the poly-attB cassette located at the RPA_1300 locus. Last, P. putida, P. frederiksbergensis, and R. jostii colonies were screened using PCR primers oPNL1010/816 that bind within the poly-attP plasmid and poly-attB cassette. PCRs that failed or otherwise demonstrated negative results for correct integration were repeated at least one additional time to confirm the negative result.
SAGE cycling for iterative integration of multiple heterologous DNAs
A demonstration of SAGE cycling in P. fluorescens JE4621 was performed using variants of transformation protocols and backbone excision protocols described in the “Serine integrase transformation assays” and “Temperature-sensitive plasmid stability and pGW60 backbone excision assays” sections, respectively. For each cycle, 100 ng of attP-target plasmid and its cognate nonreplicating integrase expression helper plasmid were coelectroporated, and transformants were isolated by either performing quadrant streaking or plating dilutions of the recovery culture on solid LB medium containing kanamycin sulfate (50 μg/ml). Plasmid backbone excision was performed for transformant colonies obtained by coelectroporation of pJE1286/pGW31 or pJE1992/pGW39. For this, transformant colonies were cultivated overnight for competent cells preparation. Competent cells were electroporated with 100 ng of the ɸC31 expression plasmid pGW30 and recovery cultures incubated at 30°C overnight on solid LB medium containing apramycin sulfate (50 μg/ml). Isolated colonies cured of pGW30 were obtained by streaking transformants onto LB lacking antibiotics and incubation at 34°C. Competent cells for performing the next SAGE cycle were prepared by cultivating colonies cured of pGW30 in LB lacking antibiotics at 30°C overnight. Integration of attP plasmids into the chromosome at its cognate attB site in the poly-attB array was confirmed by colony PCR using oligos that flank the attB site in which the heterologous DNA was to be integrated. These oligos are indicated in table S3. Notably, because of the high accuracy of the Bxb1 and R4 integrases in P. fluorescens, colony PCR validation of pJE1286 and pJE1992 cargo integration at the Bxb1 and R4 attB sites, respectively, was not confirmed until after generation of strain JE5117.
Flow cytometry assessment of expression construct stability
Starter cultures for P. fluorescens and R. palustris experiments described in Fig. 4 and fig. S8 were prepared as follows. For all wild-type strains lacking an mNeonGreen expression cassette, the strain was cultivated to stationary phase in 1-ml base media (LB for P. fluorescens and VN-A for R. palustris) lacking antibiotic. Transformant strains were generated as described in the “Serine integrase transformation assays” section above. Starter cultures for transformant strains were generated by incubating a representative colony for each strain in 1 ml of kanamycin sulfate–supplemented base media (50 μg/ml for P. fluorescens strains and 200 μg/ml for R. palustris strains) at 25°C until the culture reached saturation (20 hours for P. fluorescens and ~60 hours for R. palustris). Passages were performed by diluting cultures by either 1024-fold (10 generation dilution) or 128-fold (seven generation dilution) in fresh medium and incubating at 25°C until stationary phase was reached (~16 hours for P. fluorescens and ~36 hours for R. palustris). All cultivations were performed in 96-well deep-well plates with shaking (800 rpm, 1-mm orbital). Stationary phase cultures were assessed for construct stability by flow cytometry. Briefly, stationary phase cultures were diluted 500-fold in 1× PBS, and fluorescence was measured using a NovoCyte flow cytometer (Acea Biosciences). Green fluorescence (mNeonGreen) was measured using a 488-nm laser with a 530/30-nm filtered detector. A preliminary gating of 2500 and 10 were used for forward scatter (FSC) and side scatter (SSC), respectively, and a total of 25,000 events were measured for each sample. The percentage of cells considered to have lost the expression cassettes were calculated by enumeration of the fraction of cells whose fluorescence overlapped with that of the parent strain, which lacks a fluorescent protein expression cassette.
Starter cultures to evaluate the genetic stability of dual-fluorescent protein expression constructs in P. fluorescens JE5117 was performed as described above with a few modifications. All cultivations were performed at 30°C in 125-ml shake flasks containing 25 ml of LB without antibiotics. Two colonies of JE5117 were incubated in separate starter cultures and evaluated separately as two distinct lineages. P. fluorescens JE4621 was evaluated at a single time point to determine the level of background fluorescence in a strain that does not express any fluorescent reporter proteins. Ten passages were performed by diluting cultures 1024-fold (10 generation dilution) in fresh medium once cells entered the early stationary phase (12 hours). Approximately ~180,000 events were measured by flow cytometry for each sample. Green fluorescence was measured as above, but red fluorescence was measured using a 561-nm laser with a 660/20-nm filtered detector in these experiments. On the basis of background fluorescence measured in JE4621, cells with relative fluorescence units (RFU) higher than 900 (red) or 350 (green) were assessed to be expressing mKate2 or mNeonGreen, respectively.
Promoter library design, synthesis, cloning, and transformation into SAGE host strains
A combination of 287 synthetic and natural promoter sequences were synthesized with the following parameters. Promoter sequences, barcode sequences, and promoter origin can be found in Supplementary File T1. Synthetic promoters were derived from commonly used promoters in the field of synthetic biology, as well as custom promoters generated for this study. Any synthetic promoters longer than 150 nt were truncated to that size from the 5′ end. Natural promoters were either sourced from the previous work of the Wang laboratory (80) or mined from other organisms listed in Supplementary File T1 by identifying intergenic regions upstream of genes of interest. Promoter elements for these genes were obtained by removing the sequence likely to include the ribosomal binding sequence or specifically the 15 nt immediately upstream of the start codon of the given gene. This excision was performed to better assess promoter performance outside of the context of the native RBS, which is unlikely to be used if the promoter is used in synthetic expression cassettes. Sequences longer than 150 nt were trimmed to that length by removing sequence from the 5′ end (distal from the start codon). For simplicity, these sequences are referred to as promoters. As indicated in Fig. 5A, we then generated five barcode variants for each promoter, in which a distinct 12-nt barcode sequence (Levenshtein distance of >2) was appended immediately downstream of each promoter sequence. We then added a common 18-nt spacer sequences on either end of the promoter, as well as flanking sequence containing two oppositely oriented Bbs I recognition sites. For promoters shorter than 150 nt, a randomly generated DNA sequence was incorporated upstream of the 5′ Bbs I recognition site to bring the total construct sequence to 200 bp. In total, a 200-bp oligo pool of 1435 promoters was generated.
All enzymes were obtained from NEB unless otherwise noted. The promoter library was synthesized as a 1-pmol oligo mix by Twist Biosciences. The oligo library was resuspended in TE to a concentration of 5 ng/μl. Ten nanograms of resuspended oligo was PCR-amplified for 8 cycles of PCR in a 50-μl reaction for generation of a template stock (Pr_amp1). All subsequent amplifications use this template as input DNA to avoid freeze-thaw cycles of the original oligo library stock. We performed a second amplification step using 3 μl of Pr_amp1 template stock to obtain enough DNA of the library (Pr_amp2) for cloning into the recipient plasmid pJE990. For this, we performed qPCR in 10 parallel 20-μl reactions that were stopped after the reaction exited the exponential amplification stage (~7 to 9 cycles). All reactions used Q5 polymerase, supplemented with 1× EvaGreen (Biotium) for qPCR, and were performed using a CFX-384 Touch real-time PCR machine (Bio-Rad). Amplified library DNA was purified, digested with BbsI-HFv2, and directionally ligated into Bbs I–linearized pJE990 with T4 DNA polymerase. Ligations were transformed into E. coli QP15 chemically competent cells and recovered in 1 ml of SOC at 37°C for 1 hour. QP15 is an Epi400 (Lucigen) derivative containing the F’IQ plasmid from NEB 5-⍺ F’IQ (transferred by conjugation). A 10-μl aliquot of the recovery mixture was diluted and plated on LB + kanamycin sulfate (50 μg/ml) to determine the cloning efficiency and library coverage, and the remaining 990 μl was propagated through two subsequent overnight liquid selections using 25 ml and then 150 ml of LB-Lennox (BD Biosciences) + kanamycin sulfate (50 μg/ml) grown at 30°C, 200 rpm. The latter culture was inoculated with 1 ml of the 25-ml culture. The pJE990-derived library, referred to as pLibrary (Fig. 5A), was cloned with >50× coverage as determined by dividing the number of colony-forming units by the size of the designed library. Plasmid DNA was then extracted from library cultures with a ZymoPURE II Midiprep kit (Zymo Research) for subsequent transformation into final the host strains.
The plasmid pool pLibrary was integrated into the Bxb1 attB site in the genomes of P. fluorescens JE4621, P. frederiksbergensis JE5041, and P. putida JE5035 by coelectroporation of 200 ng of pLibrary with 3 μg of pGW31 into 50 μl of electrocompetent competent cells that had been diluted 1:10 in 10% glycerol. The electroporated cells were resuspended in 950 μl of SOC, incubated at 30°C for 1 hour with shaking. As above, dilutions generated from 10 μl of the recovery culture were plated onto LB supplemented with kanamycin sulfate (50 μg/ml), and resulting colonies were quantified to identify the size of the library. The remaining 990 μl of the recovery culture was inoculated into 50 ml of LB + kanamycin sulfate (50 μg/ml) and incubated overnight at 30°C (antibiotics were added to remove untransformed cells from the recovery culture). This culture was diluted with LB to an OD600 of 1. Then, 1 ml of the diluted culture was inoculated into 200 ml of LB + kanamycin sulfate (50 μg/ml) (to further select against untransformed cells in the population) and incubated overnight at 25°C with 200 rpm to yield the final libraries. The final P. fluorescens, P. frederiksbergensis, and P. fluorescens libraries contained ~4 × 105, 1.7 × 106, and 4.7 × 106 members. Glycerol stocks of the library cultures in final host strains were made after the first (P. frederiksbergensis and P. fluorescens) or second passage (P. fluorescens) in liquid selection. A similar approach was taken with R. palustris JE4632 and R. jostii AG5879 with the exception that different media were used and only a single liquid selection cultivation was performed before stocking each library. For R. palustris, selection was performed using VN-A supplemented with kanamycin sulfate (200 μg/ml), and for R. jostii, selection was performed using R2A supplemented with kanamycin (50 μg/ml). The final R. palustris and R. jostii libraries contained ~1 × 106 and 6.9 × 104 members, respectively. Library cultures were diluted to OD600 = 1 with glycerol to a final concentration of 12.5% glycerol and stocked as 1-ml aliquots for use in subsequent experiments.
The production of mNeonGreen by members of all libraries was evaluated qualitatively by flow cytometry with a Novocyte flow cytometer (Acea Biosciences). Final library cultures were diluted 1:1000 in PBS, and up to 250,000 events were analyzed for fluorescence using a 488-nm laser with a 530/30-nm filtered detector. Preliminary gating of 2000 and 10 for FSC and SSC was used for all samples except R. jostii, where 100 was used for both FSC and SSC.
Library growth, DNA sequencing, and RNA sequencing
Starter cultures for assays with pseudomonad libraries were generated by inoculating an entire 1 ml of OD600 glycerol stock into 100 ml of LB supplemented with kanamycin sulfate (50 μg/ml) and incubating the culture with 225 rpm of shaking at 30°C until the culture reached a mid-log growth phase. Antibiotics were used in the starter cultures to select against any potential untransformed cheaters in the libraries. The recovered culture was washed twice in 100 ml of 1× MME to remove residual LB medium. Starter cultures for the assays were prepared by inoculating 1 ml of the washed cells into 100 ml of each of the following media (which lack antibiotics) and incubated with 225 rpm of shaking at 30°C until cultures reached stationary phase. P. fluorescens library: LB, MME containing 20 mM glucose, MME containing 20 mM sodium citrate, and MME containing 20 mM N-acetylglucosamine (no ammonium). P. frederiksbergensis and P. putida libraries: MME containing 10 mM glucose. A similar approach was used to recover the R. jostii and R. palustris libraries. The R. jostii library was recovered to mid-log phase in R2A rather than LB and then subcultured to stationary phase in MMV supplemented with 10 mM glucose. The R. palustris library was cultivated directly from the glycerol stock in VN-A until the stationary phase was reached.
Each assay culture was prepared by inoculating assay medium to an initial starting OD600 of 0.01 with its respective library starter culture. For P. fluorescens, each assay culture was inoculated starter cultures grown in the same medium. For all other organisms, a single starter culture, as described above, was used to inoculate each assay culture to an initial OD600 of 0.01. Four replicate cultures were prepared for each assay condition. Cultures were incubated until an OD600 of 0.2 to 0.4 was reached. At this point, cultures were rapidly harvested by centrifugation at 4°C. With the exception of cultures containing PEG6000, all cultures were centrifuged at 7000g (P. fluorescens) or 10,000g (all others) for 3 min. PEG6000 cultures were centrifuged at 17,000g for 20 min. Culture supernatants were decanted, and cell pellets were flash-frozen in liquid nitrogen before storage at −80°C. Frozen pellets were later resuspended in 750 μl of DNA/RNA Shield (Zymo Research), and both total RNA and gDNA were extracted with the ZymoBIOMICs DNA/RNA miniprep kit (Zymo Research) according to the manufacturer’s parallel purification protocol.
Briefly, the RNA sequencing (RNA-seq) library was prepared by reverse transcription and common adaptor ligation at the 3′ end of the complementary DNA (cDNA). The RNA-seq and DNA sequencing (DNA-seq) libraries were prepared as described by Yim et al. (80) with minor modifications as described below. Primers and adapter sequences for each step are found in table S3. First, the ProtoScript II First Strand cDNA Synthesis Kit (NEB) was used rather than the Maxima reverse transcriptase (Thermo Fisher Scientific). Accordingly, the cycling conditions were modified to the following: 42°C for 60 min, followed by 10 cycles of 50°C for 2 min and 42°C for 2 min, with a final 5 min at 80°C to deactivate the enzyme. Illumina indices and adapters were added to both cDNA and input library DNA using a two-step amplification process. Second, real-time PCRs used the DNA binding dye EvaGreen (Biotium) rather than SYBR Green I (Invitrogen) to attach Illumina indices and adapters, and templates for the first-step PCR were as follows: 1 μl of undiluted cDNA library template with 3′ adapter and 300 ng of gDNA. First-step PCRs (Amp1) were performed separately for RNA-seq and DNA-seq libraries, as they required different numbers of cycles to reach exponential amplification. Amplification was stopped as soon as exponential phase ceased, typically around 20 cycles for cDNAs and around 15 cycles for gDNAs. Samples were diluted 1:100 and amplified again using the same protocol using indexing primers for 10 cycles (Amp2). RNA samples and DNA samples were then copurified and examined on a 2% agarose gel to verify correct band sizes of ~200 bp for cDNA and 350 bp for input plasmid DNA libraries.
Quantifying RTA from high-throughput sequencing
Using custom Python scripts adapted from Yim et al. (80) (https://github.com/ssyim/DRAFTS), we mapped DNA and RNA reads to promoter and 12-nt barcode sequences. To account for a range of promoter lengths in our library, we adjusted acceptable promoter length thresholds in the DNA and RNA processing scripts appropriately. For each replicate, we used a five-read cutoff for DNA samples and did not calculate the transcription rate for barcodes with fewer than five DNA reads. For each replicate sample, we calculated the transcription rate Ti of each promoter barcode variant i with RNA counts Ri and DNA counts Di as . When calculating transcription rates, barcodes with no RNA reads were discarded. Transcription rates were then normalized to a minimum of 1 by dividing all transcription rates by the minimum value. Replicate data point a (within condition l and barcode variant i) from group X was considered an outlier and discarded from further analysis if its value was >3(MAD), or mean absolute deviation, from the median and subsequently discarded. MAD was calculated as . Following this processing, any promoter barcode variants that lacked at least three replicate data points for each condition were discarded. The resulting data are found in Supplementary File T2.
Analyzing promoter strength, noise, and condition independence from high-throughput sequencing
Further processing of RTA data was performed using custom R scripts. RTA was independently evaluated for each organism. Pairwise interorganismal comparisons were performed for promoters that had sufficient data for analysis in both organisms. Any promoter barcode variants that lacked at least three replicate data points for each condition tested were discarded and not used in further analyses. Promoters for which fewer than three barcodes passed the aforementioned threshold were discarded and not used in further analyses. Outlier promoter barcode variants were identified for the remaining promoters using the 1.5× interquartile range (IQR) method. For this, mean RTA for each promoter barcode variant (RTAi) was calculated using , using the relative transcription activity Ti of each promoter barcode variant i and n replicate data points. Promoter barcode variants whose RTA was >1.5× IQR were considered an outlier barcode and discarded from further analyses. Supplementary Files F1 to F5 include multiple charts for each organism that either include individual and mean RTA values for all barcode variants or only nonoutlier barcode variants. The overall RTA for each promoter (RTAk), as shown in fig. S13, was calculated using , where n is the number of nonoutlier barcodes for each promoter. Sensitivity to genetic context (5′UTR) and condition was determined as follows. A promoter was considered insensitive to genetic context if ∣max (RTAi)/ min (RTAi)∣ < UTR, where UTR is the defined fold range value (default value of 3) used to determine whether a promoter is insensitive to barcode sequence variation in the mRNA 5′UTR. To determine sensitivity to condition, the mean RTA for each condition (RTAc) was calculated using , using relative transcription activity Tc of each condition variant c and n replicate data points. A promoter was considered insensitive to condition if ∣max (RTAc)/ min (RTAc)∣ < Con, where Con is the defined fold range value (default value of 4) used to determine whether a promoter is insensitive to condition. A promoter was considered consistent whether it is both insensitive to genetic context and condition. The overall RTA for consistent promoters is displayed in Fig. 5C and fig. S11. A table containing the final relative transcriptional analysis output from the R scripts with promoter source and sequence information is found in Supplementary Data D1.
Promoter performance was compared between pairs of organisms using the subset of promoters that had sufficient data for analysis in both organisms. Pearson correlation was performed using either all promoters analyzed in each organism (Fig. 6A, left, and fig. S13) or with only promoters that were consistent in both organisms (Fig. 6A, right).
Plate reader promoter testing assays
Reporter strains were constructed by transformation of P. fluorescens JE4621 with a combination of reporter plasmid and pGW31 (Bxb1 integrase nonreplicating helper plasmid), using the methods described in the “Serine integrase transformation assays” section. Reporter plasmids are listed in table S1. Strains were initially cultivated overnight at 30°C, 225 rpm in either 5 ml of LB (JE4621) or 5 ml of LB + kanamycin sulfate (50 μg/ml; JE4621 transformants). Starter cultures (5 ml) in MME containing 10 mM glucose and 10 mM NH4Cl were inoculated with 1% of the initial culture and similarly incubated. Coupled growth and fluorescence assays were performed with a Neo2SM (Bio-Tek) plate reader using 60 μl per well of MME supplemented with the following carbon and nitrogen sources: 10 mM ammonium chloride and either 10 mM glucose, 10 mM citrate, or 10 mM N-acetylglucosamine or 10 mM glucose and either 10 mM urea, 10 mM sodium nitrate, or 10 mM sodium nitrite. Assays were performed using black-walled, μClear, flat-bottom, 384-well plates (Greiner Bio-One) with a BreatheEasy seal (USA Scientific). Plate cultures were inoculated using an Echo acoustic liquid handler with 0.1% inoculum from starter cultures and incubated overnight in the Neo2SM at 30°C with the fast shaking setting, 1-mm orbital. OD600 and fluorescence (F501,520 for mNeonGreen) were measured every 12 min. Reporter expression per cell was estimated by dividing RFU by OD600 (as a proxy for cell number) for each time point within mid-log growth phase (OD600 = 0.089 to 0.16) and averaging those values for each sample. Background absorbance and fluorescence readings from wells containing media blanks were averaged and subtracted from sample readings before analysis. Background autofluorescence not associated with mNeonGreen expression was subtracted by subtracting the mean RFU/OD600 value from the parent strain (JE4621) from each sample. JEb2 promoter–normalized relative promoter strengths for P. fluorescens (Fig. 5, D and E, and fig. S12) were derived by averaging the mean RFU/OD600 values for each condition shown in the panel to generate a single RFU/OD600 value for each promoter and dividing these values by the mean RFU/OD600 generated when JEb2 is driving mNeonGreen expression. For comparison, JEb2-normalized RTA values for P. fluorescens (Fig. 5, C and E, and fig. S12B) were generated by dividing the mean RTA for each promoter indicated by the mean RTA for promoter JEb2.
Acknowledgments
We thank C. Harwood and Y. Oda for providing R. palustris CGA009 and for helpful information regarding cultivation of the strain. We also thank E. Yeung and Y. Farris for providing plasmid pEYF2K.
Funding: This work was supported in part by the U.S. Department of Energy (DOE), Office of Biological and Environmental Research (BER), as part of BER’s Genomic Science Program (GSP), and is a contribution of the Pacific Northwest National Laboratory (PNNL) Secure Biosystems Design Science Focus Area “Persistence Control of Engineered Functions in Complex Soil Microbiomes.” Additional support was provided by the DARPA Synergistic Discovery and Design program (contract #HR0011045664). PNNL is a multiprogram national laboratory operated by Battelle for the DOE under contract DE-AC05-76RL01830. This work was authored in part by the Oak Ridge National Laboratory, which is managed by UT-Battelle LLC, for the U.S. DOE under contract DE-AC05-00OR22725. Funding was provided in part by the U.S. DOE, Office of Energy Efficiency and Renewable Energy Bioenergy Technologies Office (BETO) to the Agile BioFoundry. This work was in part supported by the Center for Bioenergy Innovation, U.S. DOE Bioenergy Research Center, supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Author contributions: Conceptualization: J.R.E., A.M.G., and R.G.E. Methodology: J.R.E., G.N.D., L.A.R., A.M.G., G.L.P., T.S., and D.C.-D. Investigation: J.R.E., G.N.D., R.F., J.D.H., H.B., L.A.R., and G.L.P. Validation: R.F., H.B., and J.M.-B. Data curation: J.R.E. and R.G.E. Formal analysis: J.R.E. and R.G.E. Writing (original draft): J.R.E. Writing (review and editing): J.R.E., A.M.G., and R.G.E. Funding acquisition: J.R.E., A.M.G., and R.G.E. Project administration: J.R.E., A.M.G., and R.G.E.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw sequencing data can be found at the NCBI Sequence Read Archive (BioProject PRJNA841687; www.ncbi.nlm.nih.gov/bioproject/PRJNA841687/). Custom code used for data processing and analysis is publicly available at Zenodo (https://doi.org/10.5281/zenodo.7388406) or GitHub (https://github.com/PerConSFA/SAGE_RTP). The materials presented in this study can be provided by R.G.E.’s pending scientific review and a completed material transfer agreement. Requests for materials should be submitted at robert.egbert@pnnl.gov.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 to S10
Legend for supplementary data D1
Legend for figure source data
Legends for supplementary files F1 to F5
Legend for supplementary file P1
Legends for supplementary files T1 and T2
References
Other Supplementary Material for this : manuscript includes the following:
Supplementary Data D1
Figure Source Data
Supplementary Files F1 to F5
Supplementary File P1
Supplementary Files T1 and T2
REFERENCES AND NOTES
- 1.V. M. Isabella, B. N. Ha, M. J. Castillo, D. J. Lubkowicz, S. E. Rowe, Y. A. Millet, C. L. Anderson, N. Li, A. B. Fisher, K. A. West, P. J. Reeder, M. M. Momin, C. G. Bergeron, S. E. Guilmain, P. F. Miller, C. B. Kurtz, D. Falb, Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864 (2018). [DOI] [PubMed] [Google Scholar]
- 2.V. Bahuguna, G. Bhatt, R. Maikhuri, D. Chandra, in Microbial Metatranscriptomics Belowground, M. Nath, D. Bhatt, P. Bhargava, D. K. Choudhary, Eds. (Springer Singapore, 2021), pp. 109–122. [Google Scholar]
- 3.R. K. Goyal, M. A. Schmidt, M. F. Hynes, Molecular biology in the improvement of biological nitrogen fixation by rhizobia and extending the scope to cereals. Microorganisms 9, 125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. Z. Werner, R. Clare, T. D. Mand, I. Pardo, K. J. Ramirez, S. J. Haugen, F. Bratti, G. N. Dexter, J. R. Elmore, J. D. Huenemann, G. L. Peabody V, C. W. Johnson, N. A. Rorrer, D. Salvachúa, A. M. Guss, G. T. Beckham, Tandem chemical deconstruction and biological upcycling of poly(ethylene terephthalate) to β-ketoadipic acid by Pseudomonas putida KT2440. Metab. Eng. 67, 250–261 (2021). [DOI] [PubMed] [Google Scholar]
- 5.A. Banner, H. S. Toogood, N. S. Scrutton, Consolidated bioprocessing: Synthetic biology routes to fuels and fine chemicals. Microorganisms 9, 1079 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.J. Kim, S. Hwang, S. M. Lee, Metabolic engineering for the utilization of carbohydrate portions of lignocellulosic biomass. Metab. Eng. 71, 2–12 (2022). [DOI] [PubMed] [Google Scholar]
- 7.F. Käß, S. Junne, P. Neubauer, W. Wiechert, M. Oldiges, Process inhomogeneity leads to rapid side product turnover in cultivation of Corynebacterium glutamicum. Microb. Cell Fact. 13, 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. Pigou, J. Morchain, Investigating the interactions between physical and biological heterogeneities in bioreactors using compartment, population balance and metabolic models. Chem. Eng. Sci. 126, 267–282 (2015). [Google Scholar]
- 9.L. A. Riley, A. M. Guss, Approaches to genetic tool development for rapid domestication of non-model microorganisms. Biotechnol. Biofuels 14, 30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M. E. Kovach, P. H. Elzer, D. Steven Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, K. M. Peterson, Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 (1995). [DOI] [PubMed] [Google Scholar]
- 11.T. S. Lee, R. A. Krupa, F. Zhang, M. Hajimorad, W. J. Holtz, N. Prasad, S. K. Lee, J. D. Keasling, BglBrick vectors and datasheets: A synthetic biology platform for gene expression. J. Biol. Eng. 5, 12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.W. Pansegrau, E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, C. M. Thomas, Complete nucleotide sequence of Birmingham IncPα plasmids. J. Mol. Biol. 239, 623–663 (1994). [DOI] [PubMed] [Google Scholar]
- 13.R. H. Durland, A. Toukdarian, F. Fang, D. R. Helinski, Mutations in the trfA replication gene of the broad-host-range plasmid RK2 result in elevated plasmid copy numbers. J. Bacteriol. 172, 3859–3867 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J. Frey, M. M. Bagdasarian, M. Bagdasarian, Replication and copy number control of the broad-host-range plasmid RSF1010. Gene 113, 101–106 (1992). [DOI] [PubMed] [Google Scholar]
- 15.E. De Rossi, A. Milano, P. Brigidi, F. Bini, G. Riccardi, Structural organization of pBC1, a cryptic plasmid from Bacillus coagulans. J. Bacteriol. 174, 638–642 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.L. De Gelder, J. J. Williams, J. M. Ponciano, M. Sota, E. M. Top, Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics 178, 2179–2190 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.W. P. Gill, N. S. Harik, M. R. Whiddon, R. P. Liao, J. E. Mittler, D. R. Sherman, A replication clock for Mycobacterium tuberculosis. Nat. Med. 15, 211–214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C. A. Mason, J. E. Bailey, Effects of plasmid presence on growth and enzyme activity of Escherichia coli DH5α. Appl. Microbiol. Biotechnol. 32, 54–60 (1989). [Google Scholar]
- 19.J. Mi, A. Sydow, F. Schempp, D. Becher, H. Schewe, J. Schrader, M. Buchhaupt, Investigation of plasmid-induced growth defect in Pseudomonas putida. J. Biotechnol. 231, 167–173 (2016). [DOI] [PubMed] [Google Scholar]
- 20.A. San Millan, M. Toll-Riera, Q. Qi, A. Betts, R. J. Hopkinson, J. McCullagh, R. C. MacLean, Integrative analysis of fitness and metabolic effects of plasmids in Pseudomonas aeruginosa PAO1. ISME J. 12, 3014–3024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.M. Sota, H. Yano, J. M Hughes, G. W. Daughdrill, Z. Abdo, L. J. Forney, E. M. Top, Shifts in the host range of a promiscuous plasmid through parallel evolution of its replication initiation protein. ISME J. 4, 1568–1580 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.L. Yin, H. Ma, E. S. Nakayasu, S. H. Payne, D. R. Morris, C. S. Harwood, Bacterial longevity requires protein synthesis and a stringent response. MBio 10, e02189-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. Lin-Chao, H. Bremer, Effect of the bacterial growth rate on replication control of plasmid pBR322 in Escherichia coli. Mol. Gen. Genet. 203, 143–149 (1986). [DOI] [PubMed] [Google Scholar]
- 24.J. Paulsson, M. Ehrenberg, Noise in a minimal regulatory network: Plasmid copy number control. Q. Rev. Biophys. 34, 1–59 (2001). [DOI] [PubMed] [Google Scholar]
- 25.A. Pena-Gonzalez, L. M. Rodriguez-R, C. K. Marston, J. E. Gee, C. A. Gulvik, C. B. Kolton, E. Saile, M. Frace, A. R. Hoffmaster, K. T. Konstantinidis, Genomic characterization and copy number variation of bacillus anthracis plasmids pXO1 and pXO2 in a historical collection of 412 strains. mSystems 3, e00065-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.T. B. Cook, J. M. Rand, W. Nurani, D. K. Courtney, S. A. Liu, B. F. Pfleger, Genetic tools for reliable gene expression and recombineering in Pseudomonas putida. J. Ind. Microbiol. Biotechnol. 45, 517–527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S. C. Troeschel, S. Thies, O. Link, C. I. Real, K. Knops, S. Wilhelm, F. Rosenau, K. E. Jaeger, Novel broad host range shuttle vectors for expression in Escherichia coli, Bacillus subtilis and Pseudomonas putida. J. Biotechnol. 161, 71–79 (2012). [DOI] [PubMed] [Google Scholar]
- 28.L. Li, X. Liu, K. Wei, Y. Lu, W. Jiang, Synthetic biology approaches for chromosomal integration of genes and pathways in industrial microbial systems. Biotechnol. Adv. 37, 730–745 (2019). [DOI] [PubMed] [Google Scholar]
- 29.R. D. Arroyo-Olarte, R. B. Rodriguez, E. Morales-Rios, Genome editing in bacteria: CRISPR-cas and beyond. Microorganisms 9, 844 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R. D. Arroyo-Olarte, R. B. Rodríguez, E. Morales-Ríos, Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 (1994). [DOI] [PubMed] [Google Scholar]
- 31.J. R. Elmore, A. Furches, G. N. Wolff, K. Gorday, A. M. Guss, Development of a high efficiency integration system and promoter library for rapid modification of Pseudomonas putida KT2440. Metab. Eng. Commun. 5, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.E. M. Lammens, P. I. Nikel, R. Lavigne, Exploring the synthetic biology potential of bacteriophages for engineering non-model bacteria. Nat. Commun. 11, 5294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. A. N. Brophy, A. J. Triassi, B. L. Adams, R. L. Renberg, D. N. Stratis-Cullum, A. D. Grossman, C. A. Voigt, Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat. Microbiol. 3, 1043–1053 (2018). [DOI] [PubMed] [Google Scholar]
- 34.M. A. Gregory, R. Till, M. C. Smith, Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J. Bacteriol. 185, 5320–5323 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. M. Guss, M. Rother, J. K. Zhang, G. Kulkarni, W. W. Metcalf, New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2, 193–203 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. Keravala, A. C. Groth, S. Jarrahian, B. Thyagarajan, J. J. Hoyt, P. J. Kirby, M. P. Calos, A diversity of serine phage integrases mediate site-specific recombination in mammalian cells. Mol. Genet. Genomics 276, 135–146 (2006). [DOI] [PubMed] [Google Scholar]
- 37.L. Li, K. Wei, X. Liu, Y. Wu, G. Zheng, S. Chen, W. Jiang, Y. Lu, aMSGE: Advanced multiplex site-specific genome engineering with orthogonal modular recombinases in actinomycetes. Metab. Eng. 52, 153–167 (2019). [DOI] [PubMed] [Google Scholar]
- 38.C. A. Merrick, J. Zhao, S. J. Rosser, Serine integrases: Advancing synthetic biology. ACS Synth. Biol. 7, 299–310 (2018). [DOI] [PubMed] [Google Scholar]
- 39.J. G. Thomson, R. Chan, J. Smith, R. Thilmony, Y. Y. Yau, Y. J. Wang, D. W. Ow, The Bxb1 recombination system demonstrates heritable transmission of site-specific excision in Arabidopsis. BMC Biotechnol. 12, 9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Z. Xu, W. R. Brown, Comparison and optimization of ten phage encoded serine integrases for genome engineering in Saccharomyces cerevisiae. BMC Biotechnol. 16, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.G. Wang, Z. Zhao, J. Ke, Y. Engel, Y. M. Shi, D. Robinson, K. Bingol, Z. Zhang, B. Bowen, K. Louie, B. Wang, R. Evans, Y. Miyamoto, K. Cheng, S. Kosina, M. de Raad, L. Silva, A. Luhrs, A. Lubbe, D. W. Hoyt, C. Francavilla, H. Otani, S. Deutsch, N. M. Washton, E. M. Rubin, N. J. Mouncey, A. Visel, T. Northen, J. F. Cheng, H. B. Bode, Y. Yoshikuni, CRAGE enables rapid activation of biosynthetic gene clusters in undomesticated bacteria. Nat. Microbiol. 4, 2498–2510 (2019). [DOI] [PubMed] [Google Scholar]
- 42.W. R. A. Brown, N. C. O. Lee, Z. Y. Xu, M. C. M. Smith, Serine recombinases as tools for genome engineering. Methods 53, 372–379 (2011). [DOI] [PubMed] [Google Scholar]
- 43.P. C. Fogg, S. Colloms, S. Rosser, M. Stark, M. C. Smith, New applications for phage integrases. J. Mol. Biol. 426, 2703–2716 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.T. E. Saleski, M. T. Chung, D. N. Carruthers, A. Khasbaatar, K. Kurabayashi, X. N. Lin, Optimized gene expression from bacterial chromosome by high-throughput integration and screening. Sci. Adv. 7, eabe1767 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.B. Wang, Z. Zhao, L. K. Jabusch, D. M. Chiniquy, K. Ono, J. M. Conway, Z. Zhang, G. Wang, D. Robinson, J. F. Cheng, J. L. Dangl, T. R. Northen, Y. Yoshikuni, CRAGE-duet facilitates modular assembly of biological systems for studying plant-microbe interactions. ACS Synth. Biol. 9, 2610–2615 (2020). [DOI] [PubMed] [Google Scholar]
- 46.M. Bierman, R. Logan, K. O'Brien, E. T. Seno, R. Nagaraja Rao, B. E. Schoner, Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49 (1992). [DOI] [PubMed] [Google Scholar]
- 47.Y. Jiang, B. Chen, C. Duan, B. Sun, J. Yang, S. Yang, Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microb. 81, 2506–2514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.C. W. Song, J. Lee, S. Y. Lee, Genome engineering and gene expression control for bacterial strain development. Biotechnol. J. 10, 56–68 (2015). [DOI] [PubMed] [Google Scholar]
- 49.P. L. H. Vo, C. Ronda, S. E. Klompe, E. E. Chen, C. Acree, H. H. Wang, S. H. Sternberg, CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat. Biotechnol. 39, 480–489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.M. J. Bailey, A. K. Lilley, I. P. Thompson, P. B. Rainey, R. J. Ellis, Site directed chromosomal marking of a fluorescent pseudomonad isolated from the phytosphere of sugar beet; Stability and potential for marker gene transfer. Mol. Ecol. 4, 755–764 (1995). [DOI] [PubMed] [Google Scholar]
- 51.F. W. Larimer, P. Chain, L. Hauser, J. Lamerdin, S. Malfatti, L. do, M. L. Land, D. A. Pelletier, J. T. Beatty, A. S. Lang, F. R. Tabita, J. L. Gibson, T. E. Hanson, C. Bobst, J. L. T. Torres, C. Peres, F. H. Harrison, J. Gibson, C. S. Harwood, Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22, 55–61 (2004). [DOI] [PubMed] [Google Scholar]
- 52.E. J. McKenna, M. J. Coon, Enzymatic omega-oxidation. IV. Purification and properties of the omega-hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 245, 3882–3889 (1970). [PubMed] [Google Scholar]
- 53.M. Seto, K. Kimbara, M. Shimura, T. Hatta, M. Fukuda, K. Yano, A novel transformation of polychlorinated biphenyls by rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61, 3353–3358 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Y. J. Chen, P. Liu, A. A. K. Nielsen, J. A. N. Brophy, K. Clancy, T. Peterson, C. A. Voigt, Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 10, 659–664 (2013). [DOI] [PubMed] [Google Scholar]
- 55.N. C. Shaner, G. G. Lambert, A. Chammas, Y. Ni, P. J. Cranfill, M. A. Baird, B. R. Sell, J. R. Allen, R. N. Day, M. Israelsson, M. W. Davidson, J. Wang, A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D. Shcherbo, C. S. Murphy, G. V. Ermakova, E. A. Solovieva, T. V. Chepurnykh, A. S. Shcheglov, V. V. Verkhusha, V. Z. Pletnev, K. L. Hazelwood, P. M. Roche, S. Lukyanov, A. G. Zaraisky, M. W. Davidson, D. M. Chudakov, Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 418, 567–574 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Y. Hong, M. K. Hondalus, Site-specific integration of Streptomyces ΦC31 integrase-based vectors in the chromosome of Rhodococcus equi. FEMS Microbiol. Lett. 287, 63–68 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.J. P. du Toit, D. J. Lea-Smith, A. Git, J. R. D. Hervey, C. J. Howe, R. W. M. Pott, Expression of alternative nitrogenases in Rhodopseudomonas palustris is enhanced using an optimized genetic toolset for rapid, markerless modifications. ACS Synth.Biol. 10, 2167–2178 (2021). [DOI] [PubMed] [Google Scholar]
- 59.A. C. Groth, M. P. Calos, Phage integrases: Biology and applications. J. Mol. Biol. 335, 667–678 (2004). [DOI] [PubMed] [Google Scholar]
- 60.S. D. Colloms, C. A. Merrick, F. J. Olorunniji, W. M. Stark, M. C. M. Smith, A. Osbourn, J. D. Keasling, S. J. Rosser, Rapid metabolic pathway assembly and modification using serine integrase site-specific recombination. Nucleic Acids Res. 42, e23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.L. A. Silo-Suh, B. Elmore, D. E. Ohman, S. J. Suh, Isolation, characterization, and utilization of a temperature-sensitive allele of a Pseudomonas replicon. J. Microbiol. Meth. 78, 319–324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.J. Quandt, M. F. Hynes, Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21 (1993). [DOI] [PubMed] [Google Scholar]
- 63.H. Alper, C. Fischer, E. Nevoigt, G. Stephanopoulos, Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. U.S.A. 102, 12678–12683 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.N. I. Johns, A. L. C. Gomes, S. S. Yim, A. Yang, T. Blazejewski, C. S. Smillie, M. B. Smith, E. J. Alm, S. Kosuri, H. H. Wang, Metagenomic mining of regulatory elements enables programmable species-selective gene expression. Nat. Methods 15, 323–329 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.N. Xu, L. Wei, J. Liu, Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis. World J. Microbiol. Biotechnol. 35, 33 (2019). [DOI] [PubMed] [Google Scholar]
- 66.M. D. Engstrom, B. F. Pfleger, Transcription control engineering and applications in synthetic biology. Synth. Syst. Biotechnol. 2, 176–191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.C. Engler, R. Kandzia, S. Marillonnet, A one pot, one step, precision cloning method with high throughput capability. PLOS ONE 3, e3647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.J. A. Jones, V. R. Vernacchio, D. M. Lachance, M. Lebovich, L. Fu, A. N. Shirke, V. L. Schultz, B. Cress, R. J. Linhardt, M. A. G. Koffas, ePathOptimize: A combinatorial approach for transcriptional balancing of metabolic pathways. Sci. Rep. 5, 11301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.J. R. Elmore, G. N. Dexter, D. Salvachúa, J. Martinez-Baird, E. A. Hatmaker, J. D. Huenemann, D. M. Klingeman, G. L. Peabody V, D. J. Peterson, C. Singer, G. T. Beckham, A. M. Guss, Production of itaconic acid from alkali pretreated lignin by dynamic two stage bioconversion. Nat. Commun. 12, 2261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.M. G. Durrant, A. Fanton, J. Tycko, M. Hinks, S. S. Chandrasekaran, N. T. Perry, J. Schaepe, P. P. du, P. Lotfy, M. C. Bassik, L. Bintu, A. S. Bhatt, P. D. Hsu, Systematic discovery of recombinases for efficient integration of large DNA sequences into the human genome. Nat. Biotechnol., 10.1038/s41587-022-01494-w (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.L. Yang, A. A. K. Nielsen, J. Fernandez-Rodriguez, C. J. McClune, M. T. Laub, T. K. Lu, C. A. Voigt, Permanent genetic memory with >1-byte capacity. Nat. Methods 11, 1261–1266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.P. Ghosh, L. R. Wasil, G. F. Hatfull, Control of phage Bxb1 excision by a novel recombination directionality factor. PLOS Biol. 4, e186 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.T. Khaleel, E. Younger, A. R. McEwan, A. S. Varghese, M. C. Smith, A phage protein that binds ϕC31 integrase to switch its directionality. Mol. Microbiol. 80, 1450–1463 (2011). [DOI] [PubMed] [Google Scholar]
- 74.J. R. Elmore, G. N. Dexter, D. Salvachúa, M. O'Brien, D. M. Klingeman, K. Gorday, J. K. Michener, D. J. Peterson, G. T. Beckham, A. M. Guss, Engineered Pseudomonas putida simultaneously catabolizes five major components of corn stover lignocellulose: Glucose, xylose, arabinose, p-coumaric acid, and acetic acid. Metab. Eng. 62, 62–71 (2020). [DOI] [PubMed] [Google Scholar]
- 75.L. Yin, C. S. Harwood, Charging state analysis of transfer RNA from an alpha-proteobacterium. Bio Protoc. 10, e3834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.C. Gao, L. Montoya, L. Xu, M. Madera, J. Hollingsworth, E. Purdom, V. Singan, J. Vogel, R. B. Hutmacher, J. A. Dahlberg, D. Coleman-Derr, P. G. Lemaux, J. W. Taylor, Fungal community assembly in drought-stressed sorghum shows stochasticity, selection, and universal ecological dynamics. Nat. Commun. 11, 34 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.L. Xu, D. Naylor, Z. Dong, T. Simmons, G. Pierroz, K. K. Hixson, Y. M. Kim, E. M. Zink, K. M. Engbrecht, Y. Wang, C. Gao, S. DeGraaf, M. A. Madera, J. A. Sievert, J. Hollingsworth, D. Birdseye, H. V. Scheller, R. Hutmacher, J. Dahlberg, C. Jansson, J. W. Taylor, P. G. Lemaux, D. Coleman-Derr, Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. U.S.A. 115, E4284–E4293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.G. L. Peabody, J. R. Elmore, J. Martinez-Baird, A. M. Guss, Engineered Pseudomonas putida KT2440 co-utilizes galactose and glucose. Biotechnol. Biofuels 12, 295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.C. W. Johnson, D. Salvachúa, N. A. Rorrer, B. A. Black, D. R. Vardon, P. C. St. John, N. S. Cleveland, G. Dominick, J. R. Elmore, N. Grundl, P. Khanna, C. R. Martinez, W. E. Michener, D. J. Peterson, K. J. Ramirez, P. Singh, T. A. VanderWall, A. N. Wilson, X. Yi, M. J. Biddy, Y. J. Bomble, A. M. Guss, G. T. Beckham, Innovative chemicals and materials from bacterial aromatic catabolic pathways. Joule 3, 1523–1537 (2019). [Google Scholar]
- 80.S. S. Yim, N. I. Johns, J. Park, A. L. C. Gomes, R. M. McBee, M. Richardson, C. Ronda, S. P. Chen, D. Garenne, V. Noireaux, H. H. Wang, Multiplex transcriptional characterizations across diverse bacterial species using cell-free systems. Mol. Syst. Biol. 15, e8875 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.L. N. Jayakody, C. W. Johnson, J. M. Whitham, R. J. Giannone, B. A. Black, N. S. Cleveland, D. M. Klingeman, W. E. Michener, J. L. Olstad, D. R. Vardon, R. C. Brown, S. D. Brown, R. L. Hettich, A. M. Guss, G. T. Beckham, Thermochemical wastewater valorization via enhanced microbial toxicity tolerance. Energ. Environ. Sci. 11, 1625–1638 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 to S10
Legend for supplementary data D1
Legend for figure source data
Legends for supplementary files F1 to F5
Legend for supplementary file P1
Legends for supplementary files T1 and T2
References
Supplementary Data D1
Figure Source Data
Supplementary Files F1 to F5
Supplementary File P1
Supplementary Files T1 and T2