Abstract
Purpose
The COVID-19 pandemic caused by the novel Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) has put the world in a medical crisis for the past three years; nearly 6.3 million lives have been diminished due to the virus outbreak. This review aims to update the recent findings on COVID-19 infections from an epigenetic scenario and develop future perspectives of epi-drugs to treat the disease.
Methods
Original research articles and review studies related to COVID-19 were searched and analyzed from the Google Scholar/PubMed/Medline databases mainly between 2019 and 2022 to brief the recent work.
Results
Numerous in-depth studies of the mechanisms used by SARS-CoV-2 have been going on to minimize the consequences of the viral outburst. Angiotensin-Converting Enzyme 2 receptors and Transmembrane serine protease 2 facilitate viral entry to the host cells. Upon internalization, it uses the host machinery to replicate viral copies and alter the downstream regulation of the normal cells, causing infection-related morbidities and mortalities. In addition, several epigenetic regulations such as DNA methylation, acetylation, histone modifications, microRNA, and other factors (age, sex, etc.) are responsible for the regulations of viral entry, its immune evasion, and cytokine responses also play a major modulatory role in COVID-19 severity, which has been discussed in detail in this review.
Conclusion
Findings of epigenetic regulation of viral pathogenicity open a new window for epi-drugs as a possible therapeutical approach against COVID-19.
Keywords: SARS- CoV-2, Epigenetics, Cytokine storm, Inflammatory interleukins, Histone proteins, Prognosis
Introduction
Coronavirus Disease 2019 (COVID-19) outbreak has put a worldwide emergency. It has almost stopped the mobility of the whole world, not only in the health sector but also from the economic perspective. The number of people getting infected and dying daily was skying unstoppably, and almost 6.3 million people have lost their lives due to the COVID-19 pandemic [1–3]. The new coronavirus is a severe pathogen first reported in Wuhan by the end of 2019 [4]. The World Health Organisation has proclaimed the upsurge of SARS- CoV-2/COVID-19 as a global medical crisis. The US, Italy, Spain, and the UK were severely impacted by COVID-19 infection and associated deaths in the early stages of the outbreak, and this trend continued in the second wave [5, 6]. Brazil and India, on the other hand, have witnessed high rates of infection with lower fatality rates than the countries mentioned above [7]. The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) primarily targets the human respiratory system and displays symptoms such as cough, sneezing, high body temperature, runny nose, sore throat, anosmia, etc. for the first few days and can affect the lower airways as well accompanied by shortness in breathing, fatigue, diarrhea, vomiting, etc. [8, 9]. Many patients may experience a gradually deteriorating condition, especially those with existing medical conditions [10].
Abnormality in immune expression due to the infection may lead to other pathological conditions such as organ dysfunction, septic shock, and other pathogenic infections. SARS-CoV-2 is the RNA-based single-stranded virus that colonizes itself inside cells through any of the mucus membranes and uses Transmembrane serine protease 2 (TMPRSS2) and ACE2 receptor protein for fusion and endocytosis with the host cell. ACE2 is the primary viral receptor, hence crucial for the SARS-COV-2 pathogenicity [11]. Increased expression of ACE2 facilitates more invasion of the coronavirus inside the cells. After entering the host cell, the viral RNA undergoes translation to synthesize the viral protein and new RNA created for new virions with the assistance of RNA-dependent RNA polymerase [12, 13]. As a primary response to COVID-19 infection, the host immune system confronts a rapid upregulation of several pro-inflammatory cytokines like IL-1, IL-6, TNF- , and interferon in the bloodstream, often referred to as the ‘Cytokine Storm (CS)’ [14]. It causes severe inflammation, lung injury, acute respiratory distress syndrome (ARDS), and organ failure [15]. As per cytokine patterns in COVID-19 patients, the adaptive and innate immune responses activated with SARS-CoV-2 infection can lead to uncontrolled inflammatory response and proceed to CS. Epigenetic alterations, such as DNA methylation and histone tail post-translational modifications, have a role in practically all biological processes and allow cells to quick adaptation due to environmental changes by altering the conformation of genetic expression and are implicated in various human disorders. Some changes in modifying DNA-histone or RNA levels lead to severe human disorders. These changes occur when a damaged cell responds to a disease or infection to restore the typical cues [16, 17]. Patients infected with SARS- CoV-2 have shown epigenetic alterations, indicating that epigenetic pathways can be the potential targets for the therapies against viral infections [17].
Along with the host systemic responses, COVID-19 severity is also controlled by epigenetic modulations [18]. Epigenetic pathways may be altered by SARS-CoV-2, which may affect the expression of ACE2 and various immunoregulatory genes that play an important role in regulating both immune and metabolic pathways on immune and epithelial cells [19]. This will damage the tissue and augment multi-organ infections. Conventional anti-viral medications that were claimed to be useful have been repurposed. Still, the results are not convincing, and there is an unfulfilled and critical necessity for efficacious drugs, particularly against the virus [20]. The earlier epidemics caused by SARS-CoV and MERS-CoV have served as a subject for thought in the fight against the prevailing epidemic, and it shall indeed tend to aid in future SARS-CoV-2 treatments [20]. Epigenetic markers in COVID-19 and their dynamics during viral entry and throughout infection (e.g., from asymptomatic to mild symptomatic, severe infection and long persistent symptoms) can be used as a diagnostic tool and design therapeutics to control the severity of COVID-19 and related CSs [21]. Comorbidities such as Type II diabetes and cardiovascular manifestations are significant metabolic complications that contribute to the mortality of patients with COVID-19 [22]. Further insight into the epi-drug study will be helpful to have a more precise therapeutic approach to counter the fatality of SARS-CoV-2 infection [23, 24]. This review is undertaken in the purview of the importance and recent pieces of evidence on the epigenetic interplay involving SARS-CoV-2 infection to critically evaluate the aging parameters, which remain crucial for the positive outcome of the treatment strategies considering the comorbidities associated with viral infections.
Cytokine storm and COVID-19
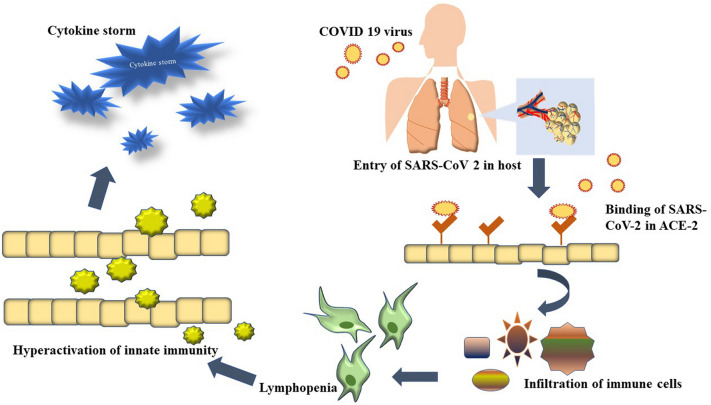
Three reported stages of disease severity could be distinguished in SARS-CoV-2 infection: mild, moderate, and severe. In the case of mild but predominantly severe COVID-19 infection, severe lung injury, multi-organ failure, and abnormal cytokine patterns often lead to the death of the patients due to CS [25–27]. The SARS-CoV-2 infection triggers an aggressive immune response in many of the patients, resulting in an excessive inflammatory response [28]. This increase in cytokines provokes an upsurge of immune cells from the vasculature, such as macrophages, neutrophils, and T cells, into the infected area, resulting in damage to human tissue due to disruption of endothelial cell–cell contacts, capillaries, and vascular barrier, alveolar tissues, along with multi-organ failure, and fatality [29–31]. A primary cause of high mortality in COVID-19 is ARDS, which results in low oxygen saturation levels [32]. Although the exact pathophysiology of ARDS in COVID-19 individuals is unknown, one of the major determining factors is the overproduction of pro-inflammatory cytokines [33, 34]. Previous research indicates that IL-1β, IL-6, IL-8, IL-12, inducible protein 10 (IP-10), MCP-1, and IFN-γ are elevated throughout SARS-CoV-2 infection [35–39]. Depleted Th2 cytokine IL-4 was also found in SARS patients [40]. Increased IL-15, IL-17, IFN- γ, and TNF levels have also been associated with MERS-CoV infection [41]. According to a few investigations, people with severe COVID-19 have elevated amounts of IL-2, IL-6, IL-7, IL-10, IP-10, MCP-1, TNF-, macrophage inflammatory protein-1α(MIP-1α), and granulocyte-CSF than those with mild—to—moderate infestations [42–45]. Figure 1 describes the association of the COVID-19 virus in the initiation of CS (Fig. 1).
Fig. 1.
Infection of SARS-COV-2 through the airway of humans and its association with the initiation of CS
The severity of COVID-19 infection is highly correlated with Lymphopenia, a condition where the blood's lymphocyte level (CD8 + T cell) drastically decreases, and elevated pro-inflammatory cytokines, such as TNF-α and IL-6 [46, 47]. CS is one of the critical factors related to Lymphopenia and can alter the behavior of T-cells and NK cells [48–50].
Hemodynamic instability, systemic inflammation, multi-organ failure, and hyperferritinemia are CS's main signatures, which can deteriorate a patient’s condition and lead to death [51]. The CS-related complexities are caused by the abnormal aberration of pro-inflammatory cytokines IL-1, IL-6, IL-18, IFN-, and TNF- that has been reported in the studies of influenza H5N1, influenza H1N1 and two coronaviruses- ‘Severe acute respiratory syndrome (SARS) associated coronavirus’ (outbreak in February, 2003, around five countries including China)[52] and ‘Middle East respiratory syndrome coronavirus(MERS-CoV)’, closely linked to COVID-19 [53, 54]. IL-6 and TNF- are the major players in the CS's interplay[15]. Patients acquire ARDS, leading to acute lung damage that can cause mortality without immediate intervention. Therefore, CS is one of the major health concerns for COVID-19 patients, as negligence may risk the patient’s life [55]. To cope with the severity of the COVID-19, Clinicians are using seven primary therapeutic strategies including the use of anti inflammatory drugs, anti-viral and anti thrombotic drugs, therapies for Acute-Hypoxamic-respiratory-failure, use of anti SARS-CoV-2 antibodies, drugs for Renin-angeotensin-Aldosterone system modulation and vitamins [56]. Anti-inflammatory medicines that reduce cytokine responses are believed to reduce morbidity and death in infected patients, as well as anti-viral therapies that deliberately target the virus [57]. Severe Covid patient often suffer from COVID-19-associated-Coagulopathy, hence required to take antithrombotic therapies [58].Timely screening of CS and rapid treatment can lead to a better prognosis [59]. Biological medicines that address cytokines have indeed been suggested as therapies for CS. IFN-α2b administration, combined with Arbidol, can accelerate the clearance of the virus and reduce the IL-6 and CRP levels to normal [60]. Glycyrrhizic Acid is a potent inhibitor of CS. Studies have shown it can inhibit IL-33, which plays a key role in ARDS. It also inhibits the production of IL-1β, IL-6, IL-8, and TNF-α production, which are the major player in the CS [61]. Anti-viral therapies like use of Remdesivir, Nirmatrelvir–ritonavir, Molnupiravir are recently inducted into practice with excilent outcomes [56]. Adalimumab and infliximab are two anti-TNF antibodies that target TNF-α [62]. Elevation in pro-inflammatory cytokine levels increases the severity of the disease and mortality as well. FDA-approved Topoisomerase-1 (TOP-1) inhibitor topotecan (TPT) subdues SARS-CoV-2-induced overproduction of inflammatory cytokines in infection-induced inflammation in hamsters [63]. Apart from these, Anakinra and Tocilizumab are two of the drugs that have been studied enormously. Anakinra, an IL-1 receptor antagonist often used to treat rheumatoid arthritis, was reported to be beneficial in the therapies of cytophagic histiocytic panniculitis with subsequent hemophagocytic lymphohistiocytosis, an illness associated with drastic CS [64]. Tocilizumab is a recombinant humanistic IL-6 potent inhibitor that blocks IL-6 from linking to its receptor, interrupting signaling [65]. Tocilizumab treats juvenile idiopathic arthritis, rheumatoid arthritis, giant cell arteritis, and several types of acute inflammatory diseases, including CS caused by Chimeric antigen receptor (CAR-T) cell treatment for hematological malignancies. In a clinical study including 15 COVID-19-positive patients, tocilizumab treatment was found to significantly lower C-reactive Protein (CRP) levels. [66]. It was considered in a randomized clinical study as a possible treatment for COVID-19-associated pneumonia and high CRP levels [67]. JAK and other downstream cytokine inhibitors are also being investigated as possible CS treatments [68]. Baricitinib, an example of JAK 1/2 inhibitors, is the first immunomodulating drug approved by the FDA(U.S) [69]. There were encouraging findings in clinical research where 15 patients with moderate to severe COVID-19 infection were given baricitinib with hydroxychloroquine, and 11 of them recovered [70]. It is also reported to improve the mortality rate effectively [71]. A new study should focus on therapeutic approaches to manage COVID-19-related CS to lower COVID-19-related death rates. Apart from these therapeutic drugs, corticosteroids such as dexamethasone, anti-viral drugs, and hydroxychloroquine have been used as therapeutic options to treat severe COVID-19 infection accompanied by CS. However, glucocorticoids and mineralocorticoids have effectively managed CS in critically ill patients with COVID-19, controversy over their effectiveness has been reported in patients with pneumonia [72]. Neutrophil Extracellular Trape (NET) is a natural defense mechanism of the human body that gets activated upon the invasion of pathogens. It has the interplay of many enzymes, such as neutrophil elastase (NE), peptidyl arginine deiminase type 4 (PADA4), etc., that cause an explosion of neutrophil cellular content that eliminates the pathogens. In the case of COVID-19, the neutrophil level increases with Lymphopenia significantly. CS and downregulation of ACE2 receptors activate the NET, which along with the pathogen, collaterally damages the vascular endothelium and lung epithelium which deteriorate the condition further. Prostaglandins, thrombomodulin, activated protein C (APC), anti-high mobility group box-1 (HMGB-1), and heparin are some of the endogenous molecules that inhibit NET activation. Whereas aspirin, sivelestat, and cyclosporine are some of the exogenous drugs used for the same purpose [73].
Epigenetic modulations in COVID-19 infection
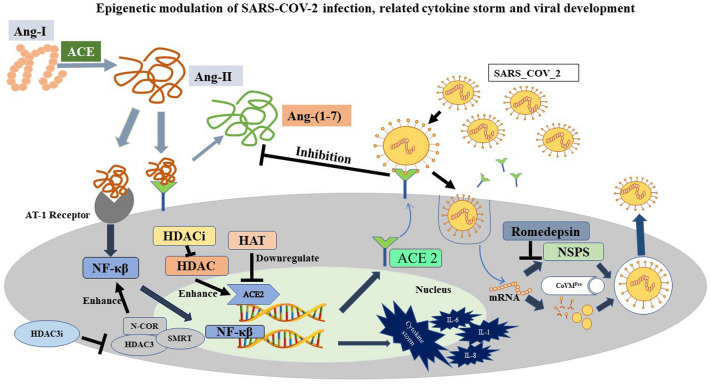
Epigenetics is a branch of biology describing the genetic expressivity in response to environmental cues, such as food, temperature, humidity, pollution, etc., and the expression of other phenotypic traits, such as age and sex [74–76]. Modifying DNA conformation inside the chromosome gives access to the transcription factors or denies it, which is the fundamentals of epigenetics. Several epigenetic modifications, such as DNA methylation, histone methylation, acetylation, deacetylation, and telomere shortening, play a crucial role in the COVID-19 pathophysiology [77]. The host’s cellular receptors act as the recognition site for the viral spike (S) protein and work as the viral entry point into the host cell. In the case of SARS-CoV-2, one of the significant cellular receptors that facilitate viral inclusion is the Angiotensin-converting enzyme 2 (ACE2) [78]. The S protein of SARS-CoV-2 has two subunits—S1 and S2. S1 binds to ACE2, and S2 mediates the fusion of the virus with host cells [79]. Recent publications have confirmed that overexpression of ACE2 is associated with enhanced severity and susceptibility to the disease [80–82]. ACE2 converts Angiotensin-II (Ang II), a product of Ang-I conversion by ACE, into Ang-(1–7) in a normal human [83]. In a COVID patient, internalization of the virus into the cells exfoliates the ACE2 and reduces its expression on cell membranes [84]. This, in turn, increases the level of Ang-II and leads to the overproduction of cytokines like IFN- IL-6, TNF- etc. [85, 86] through JAK/STAT pathway and by inducing the Nuclear Factor kappa-B (NFκ-B), eventually causing CS in patients and deteriorate their condition [87]. The accumulation of Ang-II can hyperactivate the Angiotensin-II type -1 receptor (AT1R) and increase pulmonary capillary permeability, causing pulmonary edema [88]. Comorbidities can worsen the situation by increasing the Ang-II level further. For example, a COVID patient with type two diabetes has a higher blood IL-1 concentration, which increases the expression of ACE by elevating the expression of Hypoxia Inducing Factor-1 (HIF-1 ) [89]. Several epigenetic conditions such as food, smoking habit, etc. have a role in controlling the expression of ACE2 [90]. Mostly male, aged, and smokers show hypomethylation in the ACE2 gene and therefore, overexpression of ACE2 makes them more vulnerable to COVID. On the other hand, Women, children and nonsmokers show hypermethylated ACE2 gene; consequently, they are less susceptible to the disease [91]. According to some studies, consuming polyunsaturated fatty acids increase the expression of ACE2 and A Disintegrin and Metalloprotease 17 (ADAM 17) [92, 93]. Concurrent expression of both these genes reduces the expression of ACE2 on the cell surface. Many epigenetic processes, such as DNA methylation, telomere shortening, and especially DNA acetylation, are responsible for the expression and control of the ACE2 gene [94].
ABO blood grouping antigen is also getting the spotlight as a parameter of COVID-19 severity. Several studies show evidence that ‘non-O’ individuals are at higher risk than individuals with the O blood group. However, the exact reason is still unknown [95, 96].
DNA methylation
Methylation of CpG island through DNA Methyl Transferases (DNMTs) silent the gene expression. Methylation at the promoter region prevents the binding of transcription factors and results in transcriptional inactivity. On the other hand, DNA demethylase removes the methyl groups and allows genes to express [97]. Hypomethylation of two CpG regions (cg16734967 and cg23232263) at the ACE2 promoter region of human lung tissue significantly differs in males and females, where females were found with more expressivity of the gene than males. ACE2 promoter methylation status in uterine corpus endometrial cancer and renal papillary cell carcinoma tissues are deficient, making them more susceptible to SARS-CoV-2. Similarly, Chronic Obstructive Pulmonary Disease (COPD) and smoking habits in the patient show CpG hypomethylation making the individual more prone to COVID [98]. COVID-19 infection alters the methylation pattern in the CpG site of interferon-related genes and antigen-presenting genes, causing changes in gene expression. Inflammatory cytokines influence DNA methylation alteration during myeloid differentiation [99]. When COVID-19-infected patients are compared with normal individuals, a hike in the hypomethylated signals of interferon-inducible genes and enrichment of hypermethylation signal of ‘FC Gamma Receptor dependent phagocytosis (FCGR phagocytosis)’ related genes have been observed [100]. ‘Interferon Alpha inducible protein 27 (IFI27)’, a known biomarker gene for influenza infection, is reported to be hypo methylated during COVID-19 infection, which alters the innate immune response upon viral infection. Similarly, ‘Epithelial Stromal Interaction 1 (EPSI1), important for macrophage differentiation, gets hypomethylated at particular CpG sequences in COVID-19 patients. Interferon Regulatory Factor 7 (IRF7), which plays an essential role in innate immunity, is less methylated at particular CpG sites of COVID-19 patients than the normal individual [101]. The DNA methylation status of other genes also influences the circumstances of COVID patients. For example, syncytin 1 and 2 are the two genes responsible for syncytium formation during placental development. SARS-CoV-2 also uses the same genes to facilitate syncytium formation to enter the host cells and multiplicate. Generally, in tissues other than the placenta, these two genes are found to be hypermethylated. But in the case of viral infection, the genes become hypomethylated and facilitate the inclusion of the viral particles into the host cells [102, 103].
Histone modifications
Histone modification is another modulator of epigenetic regulation of COVID severity. Adding an acetyl group to the positively charged lysine residue neutralizes the overall positive charge of histone and allows the access of transcription factors to the genes [104]. Histone acetylation and deacetylation work as the molecular switch to turn on and off the expression of genes and two types of enzymes which majorly play a role are Histone Acetyl Transferase (HAT) and Histone Deacetylase (HDAC) [105]. Histone lysine acetylation activates the expression of ACE2 receptors in humans. Hyperacetylation in histone 3 (H3AC) increases the H3K4 methylation [106]. Several positively associated genes, along with ACE2, are also regulated by H3K27 acetylation. Studies have reported that HDAC can contribute to SARS-CoV-2 pathogenicity in several ways – i) HDAC upregulates ACE2 expression and promotes viral entry to the cells [107], ii) HDAC activates pro-inflammatory responses against viral infections and may give rise to CS [108] iii) HDAC activity accumulates Acetyle Co-A, which elevates the cholesterol level [109]. Increased cholesterol levels can promote viral entry to the cells. In stressed conditions, NAD-dependent HDAC Sirtuin-1(SIRT1) regulates the expression of ACE2. Studies on SARS-CoV-2-infected patients revealed that a higher transcription rate of ACE2 can be stimulated by SIRT1 [110]. Even histone deacetylation may induce pulmonary fibroblast formation in COVID-19 survivors by altering the TGF- signaling and ERK/PI3K pathway [111]. HDAC7 plays a significant role in TGF- mediated fibroblast formation, which may cause mortality in COVID-19 patients. Alteration in TGF- expression may lead to the over expression of cytokines such as IL-7 and increase the severity [112]. HDAC8 also induces fibroblast myofibroblast differentiation in Idiopathic Pulmonary Fibrosis [113, 114]. According to studies, corticosteroids can downregulate inflammatory gene expression by inhibiting HAT and recruitment of HDAC2 [115]. Unlike other HDACs, the downregulation of inflammatory genes through HDAC2 reduces the chances of CS [116]. HDAC3 forms a multiprotein complex with a silencing mediator for retinoid and thyroid receptor (SMRT), nuclear receptor corepressor (NCOR). It suppresses the NF-K activation by deacetylation of p65, which upregulates inflammatory genes such as IL-6, IL-1 , TNF- etc. and can be a potential modulator of the CS [117]. HDAC1 also is a regulator of NF-k inactivity. Phosphorylation of HDAC3 by CK2 and HIPK2 can be a potential regulator of the cytokines storm. A high cholesterol diet (HCD) reduces the acetylation in the ACE2 promoter region and increases the susceptibility of SARS- CoV-2 infiltration inside cells [94]. High-fat-fed mice treated with rh ACE2 shows a higher level of H3K9 acetylation [118]. HDAC interventions are also found in viral trafficking via deacetylated microtubule and can be another aspect of COVID-19 experiments. Lysin demethylase KDM5B de- methylates H3K4me3 trimethylated Histone 3 residues which reduce the expression of mir-125a, followed by the upregulation of the ACE2. Hypoxic condition is known to alter the functionality of several demethylases and therefore change the expression of the gene [119, 120]
Histone deacetylase inhibitors (HDACIs)
As histone deacetylation plays an important role in the expression of many genes responsible for viral inclusions, several histone deacetylase inhibitors have been reported to downregulate ACE2 expression, making it a potential modulator of COVID-19 and related CS. Saiz et al., in their in-vitro study, have shown that HDAC inhibitor Valproic acid (VPA) reduces ACE2 expression significantly [121] and Neuropilin-1 (NRP1) on the cell surface, and the effect remains post-infection of SARS-CoV-2 [122]. Moreover, it reduces the inflammatory Cytokine’s expression and production of infectious SARS-CoV-2 virus in a dose-dependent manner. Romidepsin (drug ID: XJY-3), an HDAC inhibitor, has been reported to significantly limit the entry of SARS- CoV-2 in a clinical in-vitro investigation using nine FDA-approved medications to block the entry of pseudotyped SARS- CoV-2 [123]. The results suggest that HDAC inhibitors indirectly regulate viral inclusion in the host cells through SARS-CoV-2 host protein–protein interaction, directly influencing the ACE2 function. The invasion of SARS-CoV-2 inside neuronal cells provokes neurological damage [124]. During CS, the blood–brain barrier may be destroyed in COVID-19 patients and cause ischemic, hemorrhagic strokes [125]. HDAC inhibitors show neuroprotective effects and downregulate the pro-inflammatory genes [126]. Molecular docking of HDAC inhibitors against COVID-19 shows Romidepsin and its active form (RedFK) have great potential to bind to the binding site of viral protease CoVMpro and block its activity [127]. It stops the virus from entering the host cells. Selective inhibition of HDAC6 reduces cytokine release by airway epithelial cells, monocytes, and macrophages. HDAC inhibitors hinder the expression of INF-1 in both airway epithelial and immune cells, which helps counter the critical conditions of COVID-19 patients. Another experiment regarding the effect of HDAC inhibitors on suppressing ACE2, ABO blood antigen, and TMPRSS2 expression has revealed that cells treated with Sodium Butyrate or Panobinostat suppress the expression of ABO and ACE2 expression but not suppress TMPRSS2 [128]. Therefore, HDAC inhibitors like panobinostat and sodium butyrate can potentially treat COVID-19. Further, the MirNet study has reported that HDAC inhibition can reduce the expression of ACE2, followed by the reduction of SAR-COV-2 infectivity [129]. Figure 2 describes the epigenetic modulation of SARS- CoV-2 and related CSs (Fig. 2).
Fig. 2.
Epigenetic modulation of SARS- COV-2 infected cells and development of CSs with potential inhibitors controlling the down streaming regulations
Epigenetic basis of viral immune evasion
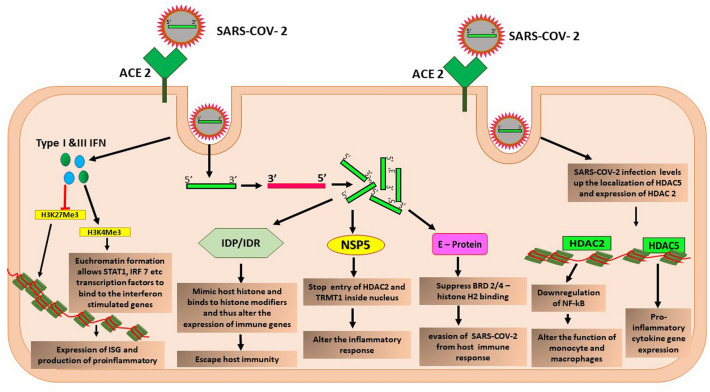
Previous studies have highlighted that the hypomethylation of the ACE2 gene is associated with the severity of systemic lupus erythematosus patients after confronting SARS- CoV-2 infection with peripheral blood T-cells. TNF-α enhances DNA methylation in ACE promoters by decreasing the activity of DNMTs and regulating the expression of ACE2. DNMTs, TET1, MAX, KDM5, and HDAC2, are pivotal in regulating ACE promoter methylation with the viral entry into the cells [108]. After entering the host cell, viral positive RNA strands get localized inside the cytoplasm, using the host machinery and synthesizing a new complementary negative strand. Negative strands then create more copies of positive strands and scale up the production of the viral proteins, such as NSP-5/13/14/16, IDP or IDR, E-protein, etc. In addition, NSP5 interacted with both t-RNA methyl transferase-1 (tRMT1) and HDAC2 and restricted them outside the nucleus. Both genes are vital in inflammation regulation after infections [130].
The affinity purification-based mass spectrometry has shown that SARS- CoV-2 viral protein ‘E- protein’ shows structural similarity with histone H2a and similar affinity with bromodomain 2&4 (BRD 2/4). Therefore, after infection, it alters the binding of BRD (2/4) with HS2 inside host cells, altering the expression of several immunological pathways and allowing immune evasion of the virus. Bromodomain is a chromatin-associated protein regulating chromatin-based gene transcriptions [131].
Post-infection expression of interferon-stimulated genes (ISGs) allows the binding of transcription factors such as STAT1 and IRF 7 and allows downstream immune modulation. For example, H3K4 me3 activation induces chromatin relaxation and allows transcription factors to bind, whereas H3K27me3 promotes the chromatin's condensation and suppresses the gene's expression. After COVID-19 infection, type I and III interferons (INF) activate H3K4me3 and allow ISG expression [132].
Viral structural proteins containing IDP (intrinsically disordered protein) or IDR (intrinsically disordered region) lack 3D shape in native conformation. It mimics eukaryotic short linear motifs (SLiMs) responsible for maintaining the host defense strategy. SLiMs are also a part of histones, which plays their role as a target for the histone modifiers. Viral IDP/IDR mimics the targets and does not allow histone modification in host cells, therefore altering the anti-viral responses and facilitating immune evasion of the virus [133]. The protein encoded by the ORF8 gene of SARS-CoV-2 mimics the ARKS motifs of H3, disrupts the epigenetic regulation, impedes the post-translational modification of the histone, and promotes chromatin compaction [134]. In SARS-CoV-2 infected cells, HDAC5 has been reported to be localized inside the nucleus, where it is found to produce pro-inflammatory cytokines and regulates the response to inflammation. In response to SARS-CoV-2 infection, HDAC2 overexpression has been noticed and found to inhibit NF-κB activity, altering monocyte and macrophage function and modulating host cell response [108] (Fig. 3).
Fig. 3.
Epigenetic manipulation of host cells by SARS-COV-2 facilitates immune evasion
Interactions between human epigenetic factors and SARS-CoV-2 proteins
According to a study, 332 human proteins, some of which are epigenetic regulators, strongly interact with the SARS-CoV-2 proteins. Any alteration in these epigenetic enzymes or proteins acknowledges alteration of normal cellular function and elevates the disease conditions. The viral proteins have been identified to associate with human epigenetic factors. In recent affinity purification mass spectrometry (AP-MS) study, 26 viral proteins were mapped along with human proteins and epigenetic modifiers. Eight of the several modifiers, HDAC2, BRD2/4, CUL2, etc., have shown potent interaction with viral proteins such as NSP5, E- protein, ORF10, etc. [135]. Studies have indicated that HDAC2 has a cleavage site between the deacetylase domain and its nuclear localization motif, allowing NSP5 to reorder the enzyme and obstruct the downstream inflammatory responses. HDAC2 deacetylates H4K16 at ISG promoters for optimal ISG expression, but its association with NOS1 prevents HDAC2 from inducing the inflammatory response [136]. It was discovered that the viral E protein interacts with BRD2 and BRD4, the two key proteins to modify histones to activate transcription. The N-terminal portion of histone H3, which interacts with BRD proteins, is most likely replicated by the C-terminal area of E proteins, resulting in a contract between E and BRD proteins [137]. ORF10, a SARS- CoV-2 protein, interacts with components of the human Cullin-RING E3 ubiquitin ligase complex RBX1, ELOB, ELOC, CUL2, and ZYG11B. The complex ubiquitinates proteins so that the 26S proteasome can degrade them. ORF10 is thought to bind CUL2 to enhance viral replication by hijacking CUL2-mediated ubiquitination and destruction [138]. Several proteins with epigenetic functions linked to SARS-CoV-2 infection have kinase activity, which can be targeted using kinase inhibitors. Imatinib, a medication that has been identified as a potential treatment for SARS-CoV-2 and SARS-CoV, is currently being tested in COVID-19 clinical trials (NCT04346147, NCT04357613, NCT04422678, and NCT04394416). Quercetin is a plant-derived substance with anti-inflammatory and anti-viral properties that have been studied as a dietary supplement or COVID-19 prophylaxis in clinical studies [139]. Further, an assessment of the SARS- CoV-2 interactome proved that several viral proteins (nsp5, nsp8, nsp13, E) engage with epigenetic and gene expression regulators [140].
Role of miRNAs in SARS-CoV-2 prognosis
A regulatory triangle is created among the host and the virus, viral-encoded miRNAs, host miRNAs, and both mRNA and miRNA targets, controlling the severity of the infection. In older individuals, the less abundance of the host defense miRNAs might explain the severity of COVID-19 and lead to death. In the host miRNA profiling of 67 SARS-CoV-2 patients from 24 countries worldwide, Khan et al. discovered that induced mi RNA either neutralizes the viral expression or acts as a proviral factor. The study has stated miRNA can be a potential therapeutical option against COVID-19 complications [141]. The viral miRNAs are inhibitory to the anti-viral proteins produced by the host, increasing the chances of SARS- CoV-2 infection by altering Janus kinase (JAK) 1 and 2 as well as signal transducer and activator of transcription (STAT) 3, 4, 5B, and 6 cellular genes and by suppression of the cytokine signaling (SOCS) cellular genes. For example, hsa-let-7a, hsa-miR-129, hsa-miR-125a-5p, hsa-miR-101, hsa-miR378, hsa-miR23b, hsa -miR, hsa-miR380-5, and hsa-miR, may target the virus [142]. Based on the gene ontology nucleotide similarity, another study analyzed miRNAs from five SARS- CoV-2 genomes and identified 22 potential viral miRNAs linked to 12 human miRNAs [143], where the interaction between human miRNAs with the viral genome may affect the host pathways that are uncertain about the virus pathogenic conditions [90].
Discussion
SARS-CoV-2 has kept the whole world to a halt since its outbreak, not only for health but also economically. Though the discovery of several vaccines fast-tracks the recovery from the crisis, it is still a major priority to find the proper therapeutic approach to deal with the critical conditions of such viral epidemics and new alternative strategies to attenuate viral pathogenicity as well as to modulate the post-infection abnormal immune response need to be mapped out. Epigenetic pathways show the pathophysiological possibilities of COVID-19 and can be a potent player in therapeutics against viral infections [144]. In our review, we have already mentioned several studies where epigenetic modifications have successfully reduced the severity of SARS-CoV-2 pathogenicity [145]. In contrast, many ongoing experiments are yet to be reported. The CS, causing widespread tissue damage resulting in multi-organ failure and death to the patient, is also regulated through epigenetic modulation [146]. The role of DNA methylation, histone acetylation deacetylation, and other epigenetic pathways can be potential targets for the therapeutics of COVID-19. HDAC inhibitors are already being in the study and found to be successful against the disease, and many of them are approved run-in-the-mill drugs. Whether in cell culture or mouse model, epigenetic drugs are the potential candidates acting as prominent anti-viral drugs. A recent study based on computational biology analysis reported that plant-based medicines like Calanolides A, Holy Basil, Kuwanon-L, and Patentiflorin A could be regarded as potential anti-HIV viral medicines which can also be administered against SARS-CoV-2. Some studies highlighted that inhibitory machinery blocking the binding of host cell receptors to viruses and inhibiting the cellular entry of viral protein could be an effective therapeutic target [147, 148]. However, experimental results are yet to be done to validate the above-mentioned therapeutic options.
Conclusion and future perspectives:
More recently, the HDAC inhibitors like VPA, panobinostat, Romidepsin, and vorinostat have been combined with antiretroviral therapy. According to the studies, inhibition of the BRD2 gene can also downregulate the SARS- CoV-2 pathogenicity. One such potent inhibitor is the dual bromodomain BET inhibitor. However, this attribute is limited by the incomplete latency reversal or insufficient clearance of latency-reactivated cells, which further seeks immune enhancement treatments [149–151]. Epigenetic expressions can also be used to assess the severity of the patient's situation and diagnose the infection. For example – DNA methylation-based approach for diagnosing the COVID-19 infection and its severity, named EPICOVID, is used to check the expression of the epigenetic signatures [152]. Additionally, anti-NET therapy from either synthetic or natural sources may mitigate SARS-CoV-2 infection-induced exaggerated hyperinflammation. Combinational therapy, including immunomodulators and IL-6 blockers like tocilizumab/baricitinib, and remdesivir, also improved the respiratory status of COVID-19 patients. Epigenetic signatures of COVID-19, related comorbidities, and different phases of infections (e.g., from asymptomatic to mild symptomatic, severe infection and long persistent symptoms) can be useful tools for timely diagnosis and designing therapies that may reduce the severity of COVID-19 and related fatalities. Further investigation focusing on epi-drugs can give us a closer look at possible pathways for therapeutical approaches against viral outbreaks.
Acknowledgements
The authors thank the Chettinad Academy of Research and Education (CARE) for providing the infrastructural and financial support to complete this work. The authors declare that they have no conflict of interest.
Abbreviations
- COVID-19
Coronavirus disease 2019
- ACE2
Angiotensin-converting enzyme 2
- SARS-COV-2
Severe acute respiratory syndrome coronavirus-2
- ARDS
Acute respiratory distress syndrome
- TOP I
Topoisomerase I
- CS
Cytokine storm
- MERS-CoV
Middle east respiratory syndrome
- TNF α
Tumor necrosis factor
- MCP
Monocyte chemotactic protein
- CRP
C Reactive protein
- TPT
Triose phosphate transmembrane
- JAK
Janus kinase
- CAR-T
Chimeric antigen receptor T cell therapy
- NET
Neutrophil extracellular trape
- NE
Neutrophil elastase
- PADA
Peptidyl arginine deaminase
- APC
Activated protein C
- HMGB-1
High mobility group box 1
- NF-kβ
Nuclear factor- kappa β
- ATR
Angiotensin receptor
- HIF-1
Hypoxia inducing factor
- ADAM 17
ACE2 and a disintegrin and metalloprotease 17
- DNMT
DNA methyl transferase
- COPD
Chronic obstructive pulmonary disease
- KDM
Lysine demethylase
- HAT
Histone acetyl transferase
- HDAC
Histone deacetylase
- H3AC
Hyper acetylation histone 3
- SIRT1
Sirtuin 1
- SMRT
Silencing mediator for retinoid and thyroid recptor
- NCOR
Nuclear receptor corepressor
- VPA
Valproic acid
- NRP
Neuropilin
- TMPRSS2
Transmembrane serine protease 2
- TET1
Tet Methylcytosine dioxygenase 1
- MAX
MYC Associated factor X
- tRMT1
TRNA Methyltransferase 1
- BRD
Bromodomain
- ISG
Interferon stimulated genes
- STAT
Statin
- IRF
Iron regulatory protein
- IDP
Intrinsically disordered protein
- IDR
Intrinsically disordered region
- SLiM
Short linear motif
- AP-MS
Affinity purification mass spectroscopy
- CUL
Cullin
- NSP5
Non structural protein 5
- RBX1
RING box 1
- ELOB
Elongin B
- ELOC
Elongin C
- ZYG11B
Zyg-11 family member B
- SOCS
Suppression of cytokine signalling
Author contributions
Antara Banerjee and Asim K Dutta Roy designed and conceptualized the study. Amit Dey, Vaishak K, and Dikshita Deka conducted wide-ranging aspects of manuscript preparation and pictorial representations. Arun Kumar Radhakrishnan, Sujay Paul, Priyadarshini S, Alice Peace Daniel, Surajit Pathak, Asim K Duttaroy, and Antara Banerjee critically reviewed and revised the manuscript and provided feedback on the manuscript. All authors read and approved the final manuscript.
Funding
Open access funding provided by University of Oslo (incl Oslo University Hospital). This work was supported by the departmental grants sanctioned to Dr. Antara Banerjee (PI), grant number: Ref. No.004/Regr/AR-Research/2022-05 from the Chettinad Academy of Research and Education.
Availability of data and materials
Not applicable.
Declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the literature.
Ethical approval
Not applicable.
Contributor Information
Asim K. Duttaroy, Email: a.k.duttaroy@medisin.uio.no
Antara Banerjee, Email: antarabanerjee@care.edu.in, Email: antara.banerjee27@gmail.com.
References
- 1.Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, et al. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int J Surg [Internet]. Elsevier; 2020 [cited 2022 Jun 14];78:185. Available from: /pmc/articles/PMC7162753/ [DOI] [PMC free article] [PubMed]
- 2.WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data [Internet]. [cited 2022 Jun 14]. Available from: https://covid19.who.int/
- 3.Musat CA, Hadzhiivanov M, Durkowski V, Banerjee A, Chiphang A, Diwan M, et al. Observational study of clinico-radiological follow-up of COVID-19 pneumonia: a district general hospital experience in the UK. BMC Infect Dis [Internet]. BioMed Central Ltd; 2021 [cited 2022 Jun 17];21:1–8. Available from: https://link.springer.com/articles/10.1186/s12879-021-06941-8 [DOI] [PMC free article] [PubMed]
- 4.Zhu H, Wei L, Niu P. The novel coronavirus outbreak in Wuhan, China. Glob Heal Res Policy [Internet]. BioMed Central Ltd; 2020 [cited 2022 Dec 23];5:1–3. Available from: https://ghrp.biomedcentral.com/articles/10.1186/s41256-020-00135-6 [DOI] [PMC free article] [PubMed]
- 5.Oehmke JF, Moss CB, Singh LN, Oehmke TB, Post LA. Dynamic Panel Surveillance of COVID-19 Transmission in the United States to Inform Health Policy: Observational Statistical Study. J Med Internet Res 2020;22(10)e21955 https//www.jmir.org/2020/10/e21955 [Internet]. Journal of Medical Internet Research; 2020 [cited 2022 Jun 15];22:e21955. Available from: https://www.jmir.org/2020/10/e21955 [DOI] [PMC free article] [PubMed]
- 6.Post L, Culler K, Moss CB, Murphy RL, Achenbach CJ, Ison MG, et al. Surveillance of the Second Wave of COVID-19 in Europe: Longitudinal Trend Analyses. JMIR Public Heal Surveill 2021;7(4)e25695 https//publichealth.jmir.org/2021/4/e25695 [Internet]. JMIR Public Health and Surveillance; 2021 [cited 2022 Jun 15];7:e25695. Available from: https://publichealth.jmir.org/2021/4/e25695 [DOI] [PMC free article] [PubMed]
- 7.Banik A, Nag T, Chowdhury SR, Chatterjee R. Why Do COVID-19 Fatality Rates Differ Across Countries? An Explorative Cross-country Study Based on Select Indicators: 10.1177/0972150920929897 [Internet]. SAGE PublicationsSage India: New Delhi, India; 2020 [cited 2022 Jun 15];21:607–25. Available from: https://journals.sagepub.com/doi/10.1177/0972150920929897
- 8.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baj J, Karakuła-Juchnowicz H, Teresiński G, Buszewicz G, Ciesielka M, Sitarz R, et al. COVID-19: Specific and Non-Specific Clinical Manifestations and Symptoms: The Current State of Knowledge. J Clin Med [Internet]. J Clin Med; 2020 [cited 2022 Nov 11];9:1–22. Available from: https://pubmed.ncbi.nlm.nih.gov/32516940/ [DOI] [PMC free article] [PubMed]
- 10.Rabaan AA, Al-Ahmed SH, Muhammad J, Khan A, Sule AA, Tirupathi R, et al. Role of Inflammatory Cytokines in COVID-19 Patients: A Review on Molecular Mechanisms, Immune Functions, Immunopathology and Immunomodulatory Drugs to Counter Cytokine Storm. Vaccines [Internet]. Multidisciplinary Digital Publishing Institute (MDPI); 2021 [cited 2022 Jun 15];9. Available from: /pmc/articles/PMC8145892/ [DOI] [PMC free article] [PubMed]
- 11.Samavati L, Uhal BD. ACE2, much more than just a receptor for SARS-COV-2 front cell infect microbiol. Front Media S.A. 2020;10:317. doi: 10.3389/fcimb.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 2020 193 [Internet]. Nature Publishing Group; 2020 [cited 2022 Jun 15];19:155–70. Available from: https://www.nature.com/articles/s41579-020-00468-6 [DOI] [PMC free article] [PubMed]
- 13.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell [Internet]. Elsevier; 2020 [cited 2022 Jun 15];181:271. Available from: /pmc/articles/PMC7102627/ [DOI] [PMC free article] [PubMed]
- 14.Ye Q, Wang B, Mao J. The pathogenesis and treatment of the `Cytokine Storm’ in COVID-19. J Infect [Internet]. Elsevier; 2020 [cited 2022 Jun 16];80:607. Available from: /pmc/articles/PMC7194613/ [DOI] [PMC free article] [PubMed]
- 15.Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 cytokine storm; what we know so far. Front Immunol. Frontiers Media S.A. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol [Internet]. J Periodontol; 2008 [cited 2022 Jun 15];79:1514–9. Available from: https://pubmed.ncbi.nlm.nih.gov/18673005/ [DOI] [PubMed]
- 17.Atlante S, Mongelli A, Barbi V, Martelli F, Farsetti A, Gaetano C. The epigenetic implication in coronavirus infection and therapy. Clin Epigenetics 2020 121 [Internet]. BioMed Central; 2020 [cited 2022 Jun 15];12:1–12. Available from: https://clinicalepigeneticsjournal.biomedcentral.com/articles/10.1186/s13148-020-00946-x [DOI] [PMC free article] [PubMed]
- 18.AbdelHamid SG, Refaat AA, Benjamin AM, Elmawardy LA, Elgendy LA, Manolly MM, et al. Deciphering epigenetic(s) role in modulating susceptibility to and severity of COVID-19 infection and/or outcome: a systematic rapid review. Environ Sci Pollut Res Int [Internet]. Nature Publishing Group; 2021 [cited 2022 Jun 15];28:1. Available from: /pmc/articles/PMC8359636/ [DOI] [PMC free article] [PubMed]
- 19.Kgatle MM, Lawal IO, Mashabela G, Boshomane TMG, Koatale PC, Mahasha PW, et al. COVID-19 Is a Multi-Organ Aggressor: Epigenetic and Clinical Marks. Front Immunol [Internet]. Frontiers Media SA; 2021 [cited 2022 Jun 15];12. Available from: /pmc/articles/PMC8531724/ [DOI] [PMC free article] [PubMed]
- 20.Mitjà O, Clotet B. Use of anti-viral drugs to reduce COVID-19 transmission. Lancet Glob Heal [Internet]. Elsevier; 2020 [cited 2022 Jun 15];8:e639. Available from: /pmc/articles/PMC7104000/ [DOI] [PMC free article] [PubMed]
- 21.Esakandari H, Nabi-Afjadi M, Fakkari-Afjadi J, Farahmandian N, Miresmaeili SM, Bahreini E. A comprehensive review of COVID-19 characteristics. Biol Proced Online [Internet]. BioMed Central; 2020 [cited 2022 Jun 15];22. Available from: /pmc/articles/PMC7402395/ [DOI] [PMC free article] [PubMed]
- 22.Pennington AF, Kompaniyets L, Summers AD, Danielson ML, Goodman AB, Chevinsky JR, et al. Risk of Clinical Severity by Age and Race/Ethnicity among Adults Hospitalized for COVID-19 - United States, March-September 2020. Open Forum Infect Dis. Oxford University Press; 2021;8. [DOI] [PMC free article] [PubMed]
- 23.Ghorbaninejad M, Khademi-Shirvan M, Hosseini S, Baghaban Eslaminejad M. Epidrugs: novel epigenetic regulators that open a new window for targeting osteoblast differentiation. Stem Cell Res Ther 2020 111 [Internet]. BioMed Central; 2020 [cited 2022 Jun 15];11:1–14. Available from: https://stemcellres.biomedcentral.com/articles/10.1186/s13287-020-01966-3 [DOI] [PMC free article] [PubMed]
- 24.Rath S, Perikala V, Jena AB, Dandapat J. Factors regulating dynamics of angiotensin-converting enzyme-2 (ACE2), the gateway of SARS-CoV-2: Epigenetic modifications and therapeutic interventions by epidrugs. Biomed Pharmacother [Internet]. Elsevier; 2021 [cited 2022 Jun 15];143:112095. Available from: /pmc/articles/PMC8403698/ [DOI] [PMC free article] [PubMed]
- 25.Montazersaheb S, Hosseiniyan Khatibi SM, Hejazi MS, Tarhriz V, Farjami A, Ghasemian Sorbeni F, et al. COVID-19 infection: an overview on cytokine storm and related interventions. Virol J [Internet]. BioMed Central; 2022;19:1–15. Available from: 10.1186/s12985-022-01814-1 [DOI] [PMC free article] [PubMed]
- 26.Niedźwiedzka-Rystwej P, Majchrzak A, Kurkowska S, Małkowska P, Sierawska O, Hrynkiewicz R, et al. Immune Signature of COVID-19: In-Depth Reasons and Consequences of the Cytokine Storm. Int J Mol Sci [Internet]. Multidisciplinary Digital Publishing Institute (MDPI); 2022 [cited 2022 Nov 14];23. Available from: /pmc/articles/PMC9105989/ [DOI] [PMC free article] [PubMed]
- 27.Pelaia C, Tinello C, Vatrella A, De Sarro G, Pelaia G. Lung under attack by COVID-19-induced cytokine storm:pathogenic mechanisms and therapeutic implications. Ther Adv Respir Dis [Internet]. SAGE Publications; 2020 [cited 2022 Jun 16];14. Available from: /pmc/articles/PMC7298425/ [DOI] [PMC free article] [PubMed]
- 28.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 2020 206 [Internet]. Nature Publishing Group; 2020 [cited 2022 Jun 16];20:363–74. Available from: https://www.nature.com/articles/s41577-020-0311-8 [DOI] [PMC free article] [PubMed]
- 29.Bhaskar S, Sinha A, Banach M, Mittoo S, Weissert R, Kass JS, et al. Cytokine Storm in COVID-19—Immunopathological Mechanisms, Clinical Considerations, and Therapeutic Approaches: The REPROGRAM Consortium Position Paper. Front Immunol [Internet]. Frontiers Media S.A.; 2020 [cited 2022 Jun 16];11:1648. Available from: /pmc/articles/PMC7365905/ [DOI] [PMC free article] [PubMed]
- 30.Mustafa MI, Abdelmoneim AH, Mahmoud EM, Makhawi AM. Cytokine Storm in COVID-19 Patients, Its Impact on Organs and Potential Treatment by QTY Code-Designed Detergent-Free Chemokine Receptors. Mediators Inflamm [Internet]. Mediators Inflamm; 2020 [cited 2022 Jun 16];2020. Available from: https://pubmed.ncbi.nlm.nih.gov/33029105/ [DOI] [PMC free article] [PubMed]
- 31.Huertas A, Montani D, Savale L, Pichon J, Tu L, Parent F, et al. Endothelial cell dysfunction: a major player in SARS-CoV-2 infection (COVID-19)? Eur Respir J [Internet]. European Respiratory Society; 2020 [cited 2022 Jun 16];56. Available from: https://erj.ersjournals.com/content/56/1/2001634 [DOI] [PMC free article] [PubMed]
- 32.Diamond M, Feliciano HLP, Sanghavi D, Mahapatra S. Acute Respiratory Distress Syndrome. StatsPearls[Internet] [Internet]. StatPearls Publishing; 2022 [cited 2022 Nov 7]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK436002/
- 33.Grasselli G, Tonetti T, Protti A, Langer T, Girardis M, Bellani G, et al. Pathophysiology of COVID-19-associated acute respiratory distress syndrome: a multicentre prospective observational study. Lancet Respir Med [Internet]. Lancet Publishing Group; 2020 [cited 2022 Jun 16];8:1201–8. Available from: http://www.thelancet.com/article/S2213260020303702/fulltext [DOI] [PMC free article] [PubMed]
- 34.Swenson KE, Swenson ER. Pathophysiology of Acute Respiratory Distress Syndrome and COVID-19 Lung Injury. Crit Care Clin [Internet]. Elsevier; 2021 [cited 2022 Jun 16];37:749. Available from: /pmc/articles/PMC8162817/ [DOI] [PMC free article] [PubMed]
- 35.Islam H, Chamberlain TC, Mui AL, Little JP. Elevated interleukin-10 levels in COVID-19: potentiation of pro-inflammatory responses or impaired anti-inflammatory action? Front Immunol. Frontiers Media S.A. 2021;12:2485. doi: 10.3389/fimmu.2021.677008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santa Cruz A, Mendes-Frias A, Oliveira AI, Dias L, Matos AR, Carvalho A, et al. Interleukin-6 Is a biomarker for the development of fatal severe acute respiratory syndrome coronavirus 2 pneumonia. Front Immunol. Frontiers Media S.A. 2021;12:263. doi: 10.3389/fimmu.2021.613422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gadotti AC, de Castro Deus M, Telles JP, Wind R, Goes M, Garcia Charello Ossoski R, et al. IFN-γ is an independent risk factor associated with mortality in patients with moderate and severe COVID-19 infection. Virus Res [Internet]. Elsevier; 2020 [cited 2022 Jun 16];289:198171. Available from: /pmc/articles/PMC7510544/ [DOI] [PMC free article] [PubMed]
- 38.Bunprakob S, Hemachudha P, Ruchisrisarod C, Supharatpariyakorn T, Hemachudha T. IP-10 and complement activation as friend or foe in COVID-19. Int J Immunopathol Pharmacol [Internet]. Int J Immunopathol Pharmacol; 2022 [cited 2022 Jun 16];36. Available from: https://pubmed.ncbi.nlm.nih.gov/35531750/ [DOI] [PMC free article] [PubMed]
- 39.Yang Y, Shen C, Li J, Yuan J, Wei J, Huang F, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol [Internet]. Elsevier; 2020 [cited 2022 Jun 16];146:119. Available from: /pmc/articles/PMC7189843/ [DOI] [PMC free article] [PubMed]
- 40.Wong CK, Lam CWK, Wu AKL, Ip WK, Lee NLS, Chan IHS, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol [Internet]. Oxford University Press; 2004 [cited 2022 Jun 16];136:95. Available from: /pmc/articles/PMC1808997/ [DOI] [PMC free article] [PubMed]
- 41.Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine [Internet]. Elsevier; 2018 [cited 2022 Jun 16];104:8. Available from: /pmc/articles/PMC7129230/ [DOI] [PMC free article] [PubMed]
- 42.Guo J, Wang S, Xia H, Shi D, Chen Y, Zheng S, et al. Cytokine Signature Associated With Disease Severity in COVID-19. Front Immunol. Frontiers Media S.A.; 2021;12:3276. [DOI] [PMC free article] [PubMed]
- 43.Merad M, Martin JC. Author Correction: Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages (Nature Reviews Immunology, (2020), 20, 6, (355–362), 10.1038/s41577-020-0331-4). Nat Rev Immunol [Internet]. Nature Research; 2020 [cited 2022 Jun 16];20:448. Available from: https://www.nature.com/articles/s41577-020-0331-4 [DOI] [PMC free article] [PubMed]
- 44.Lang FM, Lee KMC, Teijaro JR, Becher B, Hamilton JA. GM-CSF-based treatments in COVID-19: reconciling opposing therapeutic approaches. Nat Rev Immunol 2020 208 [Internet]. Nature Publishing Group; 2020 [cited 2022 Jun 16];20:507–14. Available from: https://www.nature.com/articles/s41577-020-0357-7 [DOI] [PMC free article] [PubMed]
- 45.Bonaventura A, Vecchié A, Wang TS, Lee E, Cremer PC, Carey B, et al. Targeting GM-CSF in COVID-19 pneumonia: rationale and strategies. Front Immunol. Frontiers Media S.A. 2020;11:1625. doi: 10.3389/fimmu.2020.01625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghizlane EA, Manal M, Abderrahim EK, Abdelilah E, Mohammed M, Rajae A, et al. Lymphopenia in Covid-19: a single center retrospective study of 589 cases. Ann Med Surg. 2021;69:102816. doi: 10.1016/j.amsu.2021.102816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavakolpour S, Rakhshandehroo T, Wei EX, Rashidian M. Lymphopenia during the COVID-19 infection: What it shows and what can be learned. Immunol Lett [Internet]. Elsevier; 2020 [cited 2022 Jun 15];225:31. Available from: /pmc/articles/PMC7305732/ [DOI] [PMC free article] [PubMed]
- 48.Frank K, Paust S. Dynamic natural killer cell and T cell responses to influenza infection. Front Cell Infect Microbiol Frontiers Media S.A. 2020;10:425. doi: 10.3389/fcimb.2020.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fathi N, Rezaei N. Lymphopenia in COVID‐19: Therapeutic opportunities. Cell Biol Int [Internet]. Wiley-Blackwell; 2020 [cited 2022 Jun 15];44:1792–7. Available from: /pmc/articles/PMC7283672/ [DOI] [PMC free article] [PubMed]
- 50.Liu Y, Tan W, Chen H, Zhu Y, Wan L, Jiang K, et al. Dynamic changes in lymphocyte subsets and parallel cytokine levels in patients with severe and critical COVID-19. BMC Infect Dis [Internet]. BioMed Central Ltd; 2021 [cited 2022 Jun 15];21:1–10. Available from: https://bmcinfectdis.biomedcentral.com/articles/10.1186/s12879-021-05792-7 [DOI] [PMC free article] [PubMed]
- 51.Gao YM, Xu G, Wang B, Liu BC. Cytokine storm syndrome in coronavirus disease 2019: A narrative review. J Intern Med [Internet]. Wiley-Blackwell; 2021 [cited 2022 Jun 16];289:147–61. Available from: /pmc/articles/PMC7404514/ [DOI] [PMC free article] [PubMed]
- 52.World Health Organization. Severe Acute Respiratory Syndrome (SARS) [Internet]. [cited 2023 Feb 7]. Available from: https://www.who.int/health-topics/severe-acute-respiratory-syndrome#tab=tab_1
- 53.Shi H, Liu XF, Zhang X, Chen S, Sun L, Lu J. Generation of an attenuated H5N1 avian influenza virus vaccine with all eight genes from avian viruses. Vaccine. 2007;25:7379–7384. doi: 10.1016/j.vaccine.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 54.Morris G, Bortolasci CC, Puri BK, Marx W, O’Neil A, Athan E, et al. The cytokine storms of COVID-19, H1N1 influenza, CRS and MAS compared. Can one sized treatment fit all? Cytokine [Internet]. Elsevier; 2021 [cited 2022 Jun 16];144:155593. Available from: /pmc/articles/PMC8149193/ [DOI] [PMC free article] [PubMed]
- 55.Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Front Immunol [Internet]. Frontiers Media SA; 2020 [cited 2022 Jun 16];11:1446. Available from: /pmc/articles/PMC7308649/ [DOI] [PMC free article] [PubMed]
- 56.Murakami N, Hayden R, Hills T, Al-Samkari H, Casey J, Del Sorbo L, et al. Therapeutic advances in COVID-19. Nat Rev Nephrol. 2023;19:38–52. doi: 10.1038/s41581-022-00642-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang W, Zhao Y, Zhang F, Wang Q, Li T, Liu Z, et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin Immunol [Internet]. Elsevier; 2020 [cited 2022 Jun 16];214:108393. Available from: /pmc/articles/PMC7102614/ [DOI] [PMC free article] [PubMed]
- 58.Al-Samkari H, Gupta S, Leaf RK, Wang W, Rosovsky RP, Brenner SK, et al. Thrombosis, bleeding, and the observational effect of early therapeutic anticoagulation on survival in critically ill patients with COVID-19. Ann Intern Med United States. 2021;174:622–632. doi: 10.7326/M20-6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JS, Lee JY, Yang JW, Lee KH, Effenberger M, Szpirt W, et al. Immunopathogenesis and treatment of cytokine storm in COVID-19. Theranostics [Internet]. Ivyspring International Publisher; 2021 [cited 2022 Jun 16];11:316. Available from: /pmc/articles/PMC7681075/ [DOI] [PMC free article] [PubMed]
- 60.Zhou Q, Chen V, Shannon CP, Wei XS, Xiang X, Wang X, et al. Interferon-α2b Treatment for COVID-19. Front Immunol [Internet]. Frontiers Media S.A.; 2020 [cited 2022 Dec 23];11:1061. Available from: /pmc/articles/PMC7242746/ [DOI] [PMC free article] [PubMed]
- 61.Bailly C, Vergoten G. Glycyrrhizin: An alternative drug for the treatment of COVID-19 infection and the associated respiratory syndrome? Pharmacol Ther [Internet]. Elsevier; 2020 [cited 2022 Nov 11];214:107618. Available from: /pmc/articles/PMC7311916/ [DOI] [PMC free article] [PubMed]
- 62.Zanza C, Romenskaya T, Manetti AC, Franceschi F, La Russa R, Bertozzi G, et al. Cytokine Storm in COVID-19: Immunopathogenesis and Therapy. Med 2022, Vol 58, Page 144 [Internet]. Multidisciplinary Digital Publishing Institute; 2022 [cited 2022 Nov 11];58:144. Available from: https://www.mdpi.com/1648-9144/58/2/144/htm [DOI] [PMC free article] [PubMed]
- 63.Ho JSY, Mok BWY, Campisi L, Jordan T, Yildiz S, Parameswaran S, et al. TOP1 inhibition therapy protects against SARS-CoV-2-induced lethal inflammation. Cell. 2021;184:2618–2632.e17. doi: 10.1016/j.cell.2021.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nemchand P, Tahir H, Mediwake R, Lee J. Cytokine storm and use of anakinra in a patient with COVID-19. BMJ Case Reports CP [Internet]. BMJ Specialist Journals; 2020 [cited 2022 Jun 16];13:e237525. Available from: https://casereports.bmj.com/content/13/9/e237525 [DOI] [PMC free article] [PubMed]
- 65.Gupta S, Leaf DE. Tocilizumab in COVID-19: some clarity amid controversy. Lancet [Internet]. Elsevier; 2021 [cited 2022 Jun 16];397:1599–601. Available from: http://www.thelancet.com/article/S0140673621007121/fulltext [DOI] [PMC free article] [PubMed]
- 66.Luo P, Liu Y, Qiu L, Liu X, Liu D, Li J. Tocilizumab treatment in COVID‐19: A single center experience. J Med Virol [Internet]. Wiley-Blackwell; 2020 [cited 2023 Feb 7];92:814. Available from: /pmc/articles/PMC7262125/ [DOI] [PMC free article] [PubMed]
- 67.Mariette X, Hermine O, Tharaux PL, Resche-Rigon M, Steg PG, Porcher R, et al. Effectiveness of Tocilizumab in Patients Hospitalized With COVID-19: A Follow-up of the CORIMUNO-TOCI-1 Randomized Clinical Trial. JAMA Intern Med [Internet]. American Medical Association; 2021 [cited 2023 Feb 7];181:1241–3. Available from: https://jamanetwork.com/journals/jamainternalmedicine/fullarticle/2780021 [DOI] [PMC free article] [PubMed]
- 68.Mehta P, Ciurtin C, Scully M, Levi M, Chambers RC. JAK inhibitors in COVID-19: need for vigilance regarding increased inherent thrombotic risk. Eur Respir J [Internet]. European Respiratory Society; 2020 [cited 2022 Jun 16];56. Available from: https://erj.ersjournals.com/content/early/2020/07/02/13993003.01919-2020 [DOI] [PMC free article] [PubMed]
- 69.Rubin R. Baricitinib Is First Approved COVID-19 Immunomodulatory Treatment. JAMA [Internet]. American Medical Association; 2022 [cited 2023 Feb 8];327:2281–2281. Available from: https://jamanetwork.com/journals/jama/fullarticle/2793470 [DOI] [PubMed]
- 70.Titanji BK, Farley MM, Mehta A, Connor-Schuler R, Moanna A, Cribbs SK, et al. Use of baricitinib in patients with moderate to severe coronavirus disease 2019. Clin Infect Dis. 2020;72:1247–1250. doi: 10.1093/cid/ciaa879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin Z, Niu J, Xu Y, Qin L, Ding J, Zhou L. Clinical efficacy and adverse events of baricitinib treatment for coronavirus disease-2019 (COVID-19): A systematic review and meta-analysis. J Med Virol [Internet]. John Wiley & Sons, Ltd; 2022 [cited 2023 Feb 8];94:1523–34. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/jmv.27482 [DOI] [PMC free article] [PubMed]
- 72.Bartoli A, Gabrielli F, Alicandro T, Nascimbeni F, Andreone P. COVID-19 treatment options: a difficult journey between failed attempts and experimental drugs. Intern Emerg Med [Internet]. Nature Publishing Group; 2021 [cited 2022 Nov 15];16:281. Available from: /pmc/articles/PMC7781413/ [DOI] [PMC free article] [PubMed]
- 73.Al-Kuraishy HM, Al-Gareeb AI, Al-hussaniy HA, Al-Harcan NAH, Alexiou A, Batiha GES. Neutrophil Extracellular Traps (NETs) and Covid-19: A new frontiers for therapeutic modality. Int Immunopharmacol [Internet]. Elsevier; 2022 [cited 2022 Nov 15];104:108516. Available from: /pmc/articles/PMC8733219/ [DOI] [PMC free article] [PubMed]
- 74.Turner BM. Epigenetic responses to environmental change and their evolutionary implications. Philos Trans R Soc B Biol Sci [Internet]. The Royal Society; 2009 [cited 2022 Jun 15];364:3403. Available from: /pmc/articles/PMC2781845/ [DOI] [PMC free article] [PubMed]
- 75.Kubota T, Miyake K, Hirasawa T. Epigenetic understanding of gene-environment interactions in psychiatric disorders: A new concept of clinical genetics. Clin Epigenetics [Internet]. BioMed Central; 2012 [cited 2022 Jun 15];4:1–8. Available from: https://clinicalepigeneticsjournal.biomedcentral.com/articles/10.1186/1868-7083-4-1 [DOI] [PMC free article] [PubMed]
- 76.Deans C, Maggert KA. What Do You Mean, “Epigenetic”? Genetics [Internet]. Oxford University Press; 2015 [cited 2022 Jun 15];199:887. Available from: /pmc/articles/PMC4391566/ [DOI] [PMC free article] [PubMed]
- 77.Salgado-Albarrán M, Navarro-Delgado EI, Del Moral-Morales A, Alcaraz N, Baumbach J, González-Barrios R, et al. Comparative transcriptome analysis reveals key epigenetic targets in SARS-CoV-2 infection. npj Syst Biol Appl 2021 71 [Internet]. Nature Publishing Group; 2021 [cited 2022 Jun 15];7:1–14. Available from: https://www.nature.com/articles/s41540-021-00181-x [DOI] [PMC free article] [PubMed]
- 78.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A [Internet]. Proc Natl Acad Sci U S A; 2020 [cited 2022 Jun 16];117. Available from: https://pubmed.ncbi.nlm.nih.gov/32376634/ [DOI] [PMC free article] [PubMed]
- 79.Huang Y, Yang C, Xu X feng, Xu W, Liu S wen. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin 2020 419 [Internet]. Nature Publishing Group; 2020 [cited 2022 Jun 16];41:1141–9. Available from: https://www.nature.com/articles/s41401-020-0485-4 [DOI] [PMC free article] [PubMed]
- 80.Ni W, Yang X, Yang D, Bao J, Li R, Xiao Y, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care [Internet]. BioMed Central; 2020 [cited 2022 Jun 16];24:1–10. Available from: https://ccforum.biomedcentral.com/articles/10.1186/s13054-020-03120-0 [DOI] [PMC free article] [PubMed]
- 81.Peron JPS, Nakaya H. Susceptibility of the Elderly to SARS-CoV-2 Infection: ACE-2 Overexpression, Shedding, and Antibody-dependent Enhancement (ADE). Clinics [Internet]. Faculdade de Medicina / USP; 2020 [cited 2022 Jun 16];75:1–6. Available from: http://www.scielo.br/j/clin/a/QDXYJBQk6YyLpfGNgBKtxHQ/?lang=en [DOI] [PMC free article] [PubMed]
- 82.Daniel G, Paola AR, Nancy G, Fernando SO, Beatriz A, Zulema R, et al. Epigenetic mechanisms and host factors impact ACE2 gene expression: Implications in COVID-19 susceptibility. Infect Genet Evol [Internet]. Infect Genet Evol; 2022 [cited 2022 Nov 8];104. Available from: https://pubmed.ncbi.nlm.nih.gov/36038007/ [DOI] [PMC free article] [PubMed]
- 83.Serfozo P, Wysocki J, Gulua G, Schulze A, Ye M, Liu P, et al. Ang II (Angiotensin II) Conversion to Angiotensin-(1–7) in the Circulation Is POP (Prolyloligopeptidase)-Dependent and ACE2 (Angiotensin-Converting Enzyme 2)-Independent. Hypertens (Dallas, Tex 1979) [Internet]. NLM (Medline); 2020 [cited 2022 Jun 16];75:173–82. Available from: https://www.ahajournals.org/doi/abs/10.1161/HYPERTENSIONAHA.119.14071 [DOI] [PMC free article] [PubMed]
- 84.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med [Internet]. Elsevier; 2020 [cited 2022 Jun 16];76:14. Available from: /pmc/articles/PMC7167588/ [DOI] [PMC free article] [PubMed]
- 85.Roshanravan N, Ghaffari S, Hedayati M. Angiotensin converting enzyme-2 as therapeutic target in COVID-19. Diabetes Metab Syndr [Internet]. Elsevier; 2020 [cited 2022 Jun 16];14:637. Available from: /pmc/articles/PMC7214324/ [DOI] [PMC free article] [PubMed]
- 86.Kalupahana NS, Massiera F, Quignard-Boulange A, Ailhaud G, Voy BH, Wasserman DH, et al. Overproduction of Angiotensinogen from adipose Tissue Induces adipose Infammation, Glucose Intolerance, and Insulin Resistance. Obesity (Silver Spring) [Internet]. NIH Public Access; 2012 [cited 2022 Jun 16];20:48. Available from: /pmc/articles/PMC4465436/ [DOI] [PMC free article] [PubMed]
- 87.Ahmad SF, Ansari MA, Zoheir KMA, Bakheet SA, Korashy HM, Nadeem A, et al. Regulation of TNF-α and NF-κB activation through the JAK/STAT signaling pathway downstream of histamine 4 receptor in a rat model of LPS-induced joint inflammation. Immunobiology [Internet]. Immunobiology; 2015 [cited 2022 Jun 16];220:889–98. Available from: https://pubmed.ncbi.nlm.nih.gov/25666529/ [DOI] [PubMed]
- 88.Deng J, Wang DX, Deng W, Li CY, Tong J. The effect of endogenous angiotensin II on alveolar fluid clearance in rats with acute lung injury. Can Respir J [Internet]. Hindawi Limited; 2012 [cited 2022 Jun 16];19:311. Available from: /pmc/articles/PMC3473006/ [DOI] [PMC free article] [PubMed]
- 89.Abu-Farha M, Al-Mulla F, Thanaraj TA, Kavalakatt S, Ali H, Abdul Ghani M, et al. Impact of Diabetes in Patients Diagnosed With COVID-19. Front Immunol [Internet]. Frontiers Media SA; 2020 [cited 2022 Jun 16];11. Available from: /pmc/articles/PMC7736089/ [DOI] [PMC free article] [PubMed]
- 90.Gemmati D, Bramanti B, Serino ML, Secchiero P, Zauli G, Tisato V. COVID-19 and individual genetic susceptibility/receptivity: Role of ACE1/ACE2 genes, immunity, inflammation and coagulation. might the double x-chromosome in females be protective against SARS-COV-2 compared to the single x-chromosome in males? Int J Mol Sci. MDPI AG; 2020;21. [DOI] [PMC free article] [PubMed]
- 91.Ciaglia E, Vecchione C, Puca AA. COVID-19 Infection and Circulating ACE2 Levels: Protective Role in Women and Children. Front Pediatr. Frontiers Media S.A.; 2020;8:206. [DOI] [PMC free article] [PubMed]
- 92.Goc A, Niedzwiecki A, Rath M. Polyunsaturated ω-3 fatty acids inhibit ACE2-controlled SARS-CoV-2 binding and cellular entry. Sci Rep [Internet]. Sci Rep; 2021 [cited 2022 May 23];11. Available from: https://pubmed.ncbi.nlm.nih.gov/33664446/ [DOI] [PMC free article] [PubMed]
- 93.Xu J, Mukerjee S, Cristiane CR, Carvalho-Galvão A, Cruz JC, Balarini CM, et al. A Disintegrin and Metalloprotease 17 in the Cardiovascular and Central Nervous Systems. Front Physiol [Internet]. Frontiers Media SA; 2016 [cited 2022 Jun 16];7:469. Available from: /pmc/articles/PMC5067531/ [DOI] [PMC free article] [PubMed]
- 94.Lima RS, Rocha LPC, Moreira PR. Genetic and epigenetic control of ACE2 expression and its possible role in COVID‐19. Cell Biochem Funct [Internet]. Wiley-Blackwell; 2021 [cited 2022 Jun 16];39:713–26. Available from: /pmc/articles/PMC8239811/ [DOI] [PMC free article] [PubMed]
- 95.Shibeeb S, Khan A. ABO blood group association and COVID-19. COVID-19 susceptibility and severity: a review. Hematol Transfus Cell Ther. 2022;44:70–75. doi: 10.1016/j.htct.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goel R, Bloch EM, Pirenne F, Al-Riyami AZ, Crowe E, Dau L, et al. ABO blood group and COVID-19: a review on behalf of the ISBT COVID-19 Working Group. Vox Sang [Internet]. Vox Sang; 2021 [cited 2022 Jun 16];116:849–61. Available from: https://pubmed.ncbi.nlm.nih.gov/33578447/ [DOI] [PMC free article] [PubMed]
- 97.Moore LD, Le T, Fan G. DNA Methylation and Its Basic Function. Neuropsychopharmacol 2013 381 [Internet]. Nature Publishing Group; 2012 [cited 2022 Jun 16];38:23–38. Available from: https://www.nature.com/articles/npp2012112 [DOI] [PMC free article] [PubMed]
- 98.Zhao Q, Meng M, Kumar R, Wu Y, Huang J, Lian N, et al. The impact of COPD and smoking history on the severity of COVID‐19: A systemic review and meta‐analysis. J Med Virol [Internet]. Wiley-Blackwell; 2020 [cited 2022 Jun 16];92:1915–21. Available from: /pmc/articles/PMC7262275/ [DOI] [PMC free article] [PubMed]
- 99.Godoy-Tena G, Barmada A, Morante-Palacios O, de la Calle-Fabregat C, Martins-Ferreira R, Ferreté-Bonastre AG, et al. Epigenetic and transcriptomic reprogramming in monocytes of severe COVID-19 patients reflects alterations in myeloid differentiation and the influence of inflammatory cytokines. Genome Med [Internet]. Genome Med; 2022 [cited 2022 Dec 24];14. Available from: https://pubmed.ncbi.nlm.nih.gov/36443794/ [DOI] [PMC free article] [PubMed]
- 100.Barturen G, Carnero-Montoro E, Martínez-Bueno M, Rojo-Rello S, Sobrino B, Porras-Perales Ó, et al. Whole blood DNA methylation analysis reveals respiratory environmental traits involved in COVID-19 severity following SARS-CoV-2 infection. Nat Commun [Internet]. Nat Commun; 2022 [cited 2023 Jan 4];13. Available from: https://pubmed.ncbi.nlm.nih.gov/35933486/ [DOI] [PMC free article] [PubMed]
- 101.Bowler S, Papoutsoglou G, Karanikas A, Tsamardinos I, Corley MJ, Ndhlovu LC. A machine learning approach utilizing DNA methylation as an accurate classifier of COVID-19 disease severity. Sci Rep [Internet]. Sci Rep; 2022 [cited 2022 Dec 25];12. Available from: https://pubmed.ncbi.nlm.nih.gov/36261477/ [DOI] [PMC free article] [PubMed]
- 102.Foolchand A, Mazaleni S, Ghazi T, Chuturgoon AA. A Review: Highlighting the Links between Epigenetics, COVID-19 Infection, and Vitamin D. Int J Mol Sci [Internet]. Int J Mol Sci; 2022 [cited 2022 Dec 23];23. Available from: https://pubmed.ncbi.nlm.nih.gov/36293144/ [DOI] [PMC free article] [PubMed]
- 103.Pruimboom L. Methylation pathways and SARS-CoV-2 lung infiltration and cell membrane-virus fusion are both subject to epigenetics. Front Cell Infect Microbiol Frontiers Media S.A. 2020;10:290. doi: 10.3389/fcimb.2020.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gujral P, Mahajan V, Lissaman AC, Ponnampalam AP. Histone acetylation and the role of histone deacetylases in normal cyclic endometrium. Reprod Biol Endocrinol [Internet]. BioMed Central; 2020 [cited 2022 Jun 16];18:1–11. Available from: https://rbej.biomedcentral.com/articles/10.1186/s12958-020-00637-5 [DOI] [PMC free article] [PubMed]
- 105.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 106.Yan C, Boyd DD. Histone H3 Acetylation and H3 K4 Methylation Define Distinct Chromatin Regions Permissive for Transgene Expression. Mol Cell Biol [Internet]. American Society for Microbiology (ASM); 2006 [cited 2022 Jun 16];26:6357. Available from: /pmc/articles/PMC1592829/ [DOI] [PMC free article] [PubMed]
- 107.Takahashi Y, Hayakawa A, Sano R, Fukuda H, Harada M, Kubo R, et al. Histone deacetylase inhibitors suppress ACE2 and ABO simultaneously, suggesting a preventive potential against COVID-19. Sci Rep [Internet]. Nature Publishing Group; 2021 [cited 2022 Jun 16];11:3379. Available from: /pmc/articles/PMC7873266/ [DOI] [PMC free article] [PubMed]
- 108.Jit BP, Qazi S, Arya R, Srivastava A, Gupta N, Sharma A. An immune epigenetic insight to COVID-19 infection. Epigenomics [Internet]. Epigenomics; 2021 [cited 2022 Dec 25];13:465–80. Available from: https://pubmed.ncbi.nlm.nih.gov/33685230/ [DOI] [PMC free article] [PubMed]
- 109.Shi L, Tu BP. Acetyl-CoA and the Regulation of Metabolism: Mechanisms and Consequences. Curr Opin Cell Biol [Internet]. NIH Public Access; 2015 [cited 2022 Jun 16];33:125. Available from: /pmc/articles/PMC4380630/ [DOI] [PMC free article] [PubMed]
- 110.Clarke NE, Belyaev ND, Lambert DW, Turner AJ. Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin Sci (Lond) [Internet]. Clin Sci (Lond); 2014 [cited 2022 Dec 23];126:507–16. Available from: https://pubmed.ncbi.nlm.nih.gov/24147777/ [DOI] [PubMed]
- 111.P KM, Sivashanmugam K, Kandasamy M, Subbiah R, Ravikumar V. Repurposing of histone deacetylase inhibitors: A promising strategy to combat pulmonary fibrosis promoted by TGF-β signalling in COVID-19 survivors. Life Sci [Internet]. Life Sci; 2021 [cited 2022 Jun 16];266. Available from: https://pubmed.ncbi.nlm.nih.gov/33316266/ [DOI] [PMC free article] [PubMed]
- 112.Veerabathiran R, Ragunath B, Kaviarasan V, Mohammed V, Ahmed SSSJ. Identification of selected genes associated with the SARS-CoV-2: a therapeutic approach and disease severity. [cited 2022 Nov 15]; Available from: 10.1186/s42269-021-00540-y [DOI] [PMC free article] [PubMed]
- 113.Jones DL, Haak AJ, Caporarello N, Choi KM, Ye Z, Yan H, et al. TGFβ-induced fibroblast activation requires persistent and targeted HDAC-mediated gene repression. J Cell Sci [Internet]. Company of Biologists; 2019 [cited 2022 Jun 16];132. Available from: /pmc/articles/PMC6826010/ [DOI] [PMC free article] [PubMed]
- 114.Saito S, Zhuang Y, Suzuki T, Ota Y, Bateman ME, Alkhatib AL, et al. Translational Research in Acute Lung Injury and Pulmonary Fibrosis: HDAC8 inhibition ameliorates pulmonary fibrosis. Am J Physiol - Lung Cell Mol Physiol [Internet]. American Physiological Society; 2019 [cited 2022 Jun 16];316:L175. Available from: /pmc/articles/PMC6383499/ [DOI] [PMC free article] [PubMed]
- 115.Barnes PJ. How corticosteroids control inflammation: Quintiles Prize Lecture 2005. Br J Pharmacol [Internet]. John Wiley & Sons, Ltd; 2006 [cited 2022 Jun 16];148:245–54. Available from: https://onlinelibrary.wiley.com/doi/full/10.1038/sj.bjp.0706736 [DOI] [PMC free article] [PubMed]
- 116.Fang WF, Chen YM, Lin CY, Huang HL, Yeh H, Chang YT, et al. Histone deacetylase 2 (HDAC2) attenuates lipopolysaccharide (LPS)-induced inflammation by regulating PAI-1 expression. J Inflamm (United Kingdom) [Internet]. BioMed Central Ltd.; 2018 [cited 2022 Jun 16];15:1–11. Available from: https://journal-inflammation.biomedcentral.com/articles/10.1186/s12950-018-0179-6 [DOI] [PMC free article] [PubMed]
- 117.Leus NGJ, Zwinderman MRH, Dekker FJ. Histone deacetylase 3 (HDAC 3) as emerging drug target in NF-κB-mediated inflammation. Curr Opin Chem Biol [Internet]. Europe PMC Funders; 2016 [cited 2022 Jun 16];33:160. Available from: /pmc/articles/PMC5019345/ [DOI] [PMC free article] [PubMed]
- 118.Kawabe Y, Mori J, Morimoto H, Yamaguchi M, Miyagaki S, Ota T, et al. ACE2 exerts anti-obesity effect via stimulating brown adipose tissue and induction of browning in white adipose tissue. Am J Physiol Endocrinol Metab [Internet]. 2019 [cited 2022 Dec 23];317:1140–9. Available from: http://www.ajpendo.org [DOI] [PubMed]
- 119.Beacon TH, Delcuve GP, Davie JR. Epigenetic regulation of ACE2, the receptor of the SARS-CoV-2 virus1. Genome [Internet]. Genome; 2021 [cited 2022 Dec 25];64:386–99. Available from: https://pubmed.ncbi.nlm.nih.gov/33086021/ [DOI] [PubMed]
- 120.Sawalha AH, Zhao M, Coit P, Lu Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin Immunol [Internet]. Clin Immunol; 2020 [cited 2022 Dec 25];215. Available from: https://pubmed.ncbi.nlm.nih.gov/32276140/ [DOI] [PMC free article] [PubMed]
- 121.Shweta S, Krishna KS. Valproic acid in prevention and treatment of COVID-19. Int J Respir Pulm Med. Clinmed journals. 2020;7.
- 122.Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón-Plágaro C, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science (80- ) [Internet]. American Association for the Advancement of Science; 2020 [cited 2022 Jun 16];370:861–5. Available from: https://www.science.org/doi/10.1126/science.abd3072 [DOI] [PMC free article] [PubMed]
- 123.Liu K, Zou R, Cui W, Li M, Wang X, Dong J, et al. Clinical HDAC Inhibitors Are Effective Drugs to Prevent the Entry of SARS-CoV2. ACS Pharmacol Transl Sci [Internet]. American Chemical Society; 2020 [cited 2022 Jun 16];3:1361–70. Available from: /pmc/articles/PMC7671100/ [DOI] [PMC free article] [PubMed]
- 124.Wu Y, Xu X, Chen Z, Duan J, Hashimoto K, Yang L, et al. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav Immun [Internet]. Elsevier; 2020 [cited 2022 Jun 17];87:18. Available from: /pmc/articles/PMC7146689/ [DOI] [PMC free article] [PubMed]
- 125.Syahrul S, Maliga HA, Ilmawan M, Fahriani M, Mamada SS, Fajar JK, et al. Hemorrhagic and ischemic stroke in patients with coronavirus disease 2019: incidence, risk factors, and pathogenesis - a systematic review and meta-analysis. F1000Research [Internet]. Faculty of 1000 Ltd; 2021 [cited 2022 Jun 17];10. Available from: /pmc/articles/PMC7934095/ [DOI] [PMC free article] [PubMed]
- 126.Dietz KC, Casaccia P. HDAC inhibitors and neurodegeneration: at the edge between protection and damage. Pharmacol Res [Internet]. NIH Public Access; 2010 [cited 2022 Jun 17];62:11. Available from: /pmc/articles/PMC2871984/ [DOI] [PMC free article] [PubMed]
- 127.Mohamed MFA, Abuo-Rahma GE-DA, Hayallah AM, Aziz MA, Nafady A, Samir E. Molecular docking study reveals the potential repurposing of histone deacetylase inhibitors against Covid-19. Int J Pharm Sci Res [Internet]. 2020;11:4261–70. Available from: https://www.embase.com/search/results?subaction=viewrecord&id=L632820375&from=export%0Ahttp://dx.doi.org/10.13040/IJPSR.0975-8232.11(9).4261-70
- 128.Takahashi Y, Hayakawa A, Sano R, Fukuda H, Harada M, Kubo R, et al. Histone deacetylase inhibitors suppress ACE2 and ABO simultaneously, suggesting a preventive potential against COVID-19. Sci Rep [Internet]. Nature Publishing Group; 2021 [cited 2022 Jun 17];11:3379. Available from: /pmc/articles/PMC7873266/ [DOI] [PMC free article] [PubMed]
- 129.Teodori L, Sestili P, Madiai V, Coppari S, Fraternale D, Rocchi MBL, et al. MicroRNAs Bioinformatics Analyses Identifying HDAC Pathway as a Putative Target for Existing Anti‐COVID‐19 Therapeutics. Front Pharmacol. Frontiers Media S.A.; 2020;11:1729. [DOI] [PMC free article] [PubMed]
- 130.Suryawanshi RK, Koganti R, Agelidis A, Patil CD, Shukla D. Dysregulation of Cell Signaling by SARS-CoV-2. Trends Microbiol [Internet]. Elsevier; 2021 [cited 2022 Jun 17];29:224. Available from: /pmc/articles/PMC7836829/ [DOI] [PMC free article] [PubMed]
- 131.Lara-Ureña N, García-Domínguez M. Relevance of BET Family Proteins in SARS-CoV-2 Infection. Biomolecules [Internet]. Biomolecules; 2021 [cited 2022 Jun 17];11. Available from: https://pubmed.ncbi.nlm.nih.gov/34439792/ [DOI] [PMC free article] [PubMed]
- 132.Stanifer ML, Pervolaraki K, Boulant S. Differential Regulation of Type I and Type III Interferon Signaling. Int J Mol Sci [Internet]. Multidisciplinary Digital Publishing Institute (MDPI); 2019 [cited 2022 Jun 17];20. Available from: /pmc/articles/PMC6471306/ [DOI] [PMC free article] [PubMed]
- 133.Mishra PM, Verma NC, Rao C, Uversky VN, Nandi CK. Intrinsically disordered proteins of viruses: Involvement in the mechanism of cell regulation and pathogenesis. Prog Mol Biol Transl Sci [Internet]. Elsevier; 2020 [cited 2022 Jun 17];174:1. Available from: /pmc/articles/PMC7129803/ [DOI] [PMC free article] [PubMed]
- 134.Kee J, Thudium S, Renner DM, Glastad K, Palozola K, Zhang Z, et al. SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature [Internet]. Nature; 2022 [cited 2022 Dec 23];610:381–8. Available from: https://pubmed.ncbi.nlm.nih.gov/36198800/ [DOI] [PMC free article] [PubMed]
- 135.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature [Internet]. Nature; 2020 [cited 2022 Jun 17];583:459–68. Available from: https://pubmed.ncbi.nlm.nih.gov/32353859/ [DOI] [PMC free article] [PubMed]
- 136.Xu P, Ye S, Li K, Huang M, Wang Q, Zeng S, et al. NOS1 inhibits the interferon response of cancer cells by S-nitrosylation of HDAC2. J Exp Clin Cancer Res [Internet]. J Exp Clin Cancer Res; 2019 [cited 2022 Jun 17];38. Available from: https://pubmed.ncbi.nlm.nih.gov/31805977/ [DOI] [PMC free article] [PubMed]
- 137.Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol 2014 214 [Internet]. Nature Publishing Group; 2014 [cited 2022 Jun 17];21:301–7. Available from: https://www.nature.com/articles/nsmb.2780 [DOI] [PubMed]
- 138.Cai W, Yang H. The structure and regulation of Cullin 2 based E3 ubiquitin ligases and their biological functions. Cell Div [Internet]. BioMed Central; 2016 [cited 2022 Jun 17];11:7. Available from: /pmc/articles/PMC4878042/ [DOI] [PMC free article] [PubMed]
- 139.Salgado-Albarrán M, Navarro-Delgado EI, Del Moral-Morales A, Alcaraz N, Baumbach J, González-Barrios R, et al. Comparative transcriptome analysis reveals key epigenetic targets in SARS-CoV-2 infection. npj Syst Biol Appl 2021 71 [Internet]. Nature Publishing Group; 2021 [cited 2022 Jun 17];7:1–14. Available from: https://www.nature.com/articles/s41540-021-00181-x [DOI] [PMC free article] [PubMed]
- 140.Beacon TH, Su RC, Lakowski TM, Delcuve GP, Davie JR. SARS-CoV-2 multifaceted interaction with the human host. Part II: Innate immunity response, immunopathology, and epigenetics. IUBMB Life [Internet]. IUBMB Life; 2020 [cited 2022 Jun 17];72:2331–54. Available from: https://pubmed.ncbi.nlm.nih.gov/32936531/ [DOI] [PubMed]
- 141.Paul S, Bravo Vázquez LA, Reyes-Pérez PR, Estrada-Meza C, Aponte Alburquerque RA, Pathak S, et al. The role of microRNAs in solving COVID-19 puzzle from infection to therapeutics: a mini-review. Virus Res. 2022;308:198631. doi: 10.1016/j.virusres.2021.198631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Samaddar A, Gadepalli R, Nag VL, Misra S. The enigma of low COVID-19 fatality rate in India. Front Genet Frontiers Media SA. 2020;11:854. doi: 10.3389/fgene.2020.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sarma A, Phukan H, Halder N, Madanan MG. An in-silico approach to study the possible interactions of miRNA between human and SARS-CoV2. Comput Biol Chem [Internet]. Comput Biol Chem; 2020 [cited 2022 Jun 16];88. Available from: https://pubmed.ncbi.nlm.nih.gov/32771962/ [DOI] [PMC free article] [PubMed]
- 144.Khan MAAK, Islam ABMMK. SARS-CoV-2 Proteins Exploit Host’s Genetic and Epigenetic Mediators for the Annexation of Key Host Signaling Pathways. Front Mol Biosci. Frontiers Media S.A.; 2021;7:509. [DOI] [PMC free article] [PubMed]
- 145.Miao Y, Fan L, Li JY. Potential Treatments for COVID-19 Related Cytokine Storm - Beyond Corticosteroids. Front Immunol [Internet]. Frontiers Media SA; 2020 [cited 2022 Nov 15];11:1445. Available from: /pmc/articles/PMC7308586/ [DOI] [PMC free article] [PubMed]
- 146.Yasmin R, Siraj S, Hassan A, Khan AR, Abbasi R, Ahmad N. Epigenetic regulation of inflammatory cytokines and associated genes in human malignancies. Mediators Inflamm [Internet]. Mediators Inflamm; 2015 [cited 2022 Nov 15];2015. Available from: https://pubmed.ncbi.nlm.nih.gov/25814785/ [DOI] [PMC free article] [PubMed]
- 147.Das D, Bihari Jena A, Banerjee A, Kumar Radhakrishnan A, Duttaroy AK, Pathak S. Can plant-derived anti-HIV compounds be used in COVID-19 cases? Med Hypotheses [Internet]. Med Hypotheses; 2022 [cited 2022 Nov 15];166:110926. Available from: https://pubmed.ncbi.nlm.nih.gov/35935095/ [DOI] [PMC free article] [PubMed]
- 148.Roshni J, Vaishali R, Ganesh KS, Dharani N, Alzahrani KJ, Banjer HJ, et al. Multi-target potential of Indian phytochemicals against SARS-CoV-2: A docking, molecular dynamics and MM-GBSA approach extended to Omicron B.1.1.529. J Infect Public Health [Internet]. J Infect Public Health; 2022 [cited 2022 Nov 15];15:662–9. Available from: https://pubmed.ncbi.nlm.nih.gov/35617830/ [DOI] [PMC free article] [PubMed]
- 149.Mills RJ, Humphrey SJ, Fortuna PRJ, Lor M, Foster SR, Quaife-Ryan GA, et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell [Internet]. Cell; 2021 [cited 2022 Nov 15];184:2167–2182.e22. Available from: https://pubmed.ncbi.nlm.nih.gov/33811809/ [DOI] [PMC free article] [PubMed]
- 150.Nehme Z, Pasquereau S, Herbein G. Control of viral infections by epigenetic-targeted therapy. Clin Epigenetics 2019 111 [Internet]. BioMed Central; 2019 [cited 2022 Nov 15];11:1–17. Available from: https://clinicalepigeneticsjournal.biomedcentral.com/articles/10.1186/s13148-019-0654-9 [DOI] [PMC free article] [PubMed]
- 151.Bravo-Vázquez LA, Medina-Ríos I, Márquez-Gallardo LD, Reyes-Muñoz J, Serrano-Cano FI, Pathak S, et al. Functional Implications and Clinical Potential of MicroRNAs in Irritable Bowel Syndrome: A Concise Review. Dig Dis Sci [Internet]. Dig Dis Sci; 2022 [cited 2022 Nov 15];1–16. Available from: https://pubmed.ncbi.nlm.nih.gov/35507132/ [DOI] [PMC free article] [PubMed]
- 152.Herbein G. An epigenetic signature to fight COVID-19. EBioMedicine [Internet]. Elsevier; 2021 [cited 2022 Nov 15];67. Available from: /pmc/articles/PMC8116817/ [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.