Abstract
Transformer, one of the latest technological advances of deep learning, has gained prevalence in natural language processing or computer vision. Since medical imaging bear some resemblance to computer vision, it is natural to inquire about the status quo of Transformers in medical imaging and ask the question: can the Transformer models transform medical imaging? In this paper, we attempt to make a response to the inquiry. After a brief introduction of the fundamentals of Transformers, especially in comparison with convolutional neural networks (CNNs), and highlighting key defining properties that characterize the Transformers, we offer a comprehensive review of the state-of-the-art Transformer-based approaches for medical imaging and exhibit current research progresses made in the areas of medical image segmentation, recognition, detection, registration, reconstruction, enhancement, etc. In particular, what distinguishes our review lies in its organization based on the Transformer’s key defining properties, which are mostly derived from comparing the Transformer and CNN, and its type of architecture, which specifies the manner in which the Transformer and CNN are combined, all helping the readers to best understand the rationale behind the reviewed approaches. We conclude with discussions of future perspectives.
Keywords: Transformer, Medical imaging, Survey
1. Introduction
Medical imaging (Beutel et al., 2000) is a non-invasive technology that acquires signals by leveraging the physical principles of sound, light, electromagnetic wave, etc., from which visual images of internal tissues of the human body are generated. There are many widely used medical imaging modalities, including ultrasound, digital radiography, computed tomography (CT), magnetic resonance imaging (MRI), and optical coherent tomography (OCT). According to a report published by EMC2, about 90% of all healthcare data are medical images, which undoubtedly become a critical source of evidence for clinical decision making, such as diagnosis and intervention.
Artificial intelligence (AI) technologies that process and analyze medical images have gained prevalence in scientific research and clinical practices in recent years (Zhou et al., 2019). This is mainly due to the surge of deep learning (DL) (LeCun et al., 2015), which has achieved superb performances in a multitude of tasks, including classification (He et al., 2016; Hu et al., 2018; Huang et al., 2017), object detection (Girshick et al., 2014; Wang et al., 2017b), and semantic segmentation (Zhao et al., 2017; Chen et al., 2017). The convolutional neural networks (CNNs or ConvNets) are DL methods customarily designed for image data. The earliest applications of CNNs in medical imaging go back to the 1990s (Lo et al., 1995b,a; Sahiner et al., 1996). Though they showed encouraging results, it was not until the last decade that CNNs began to exhibit state-of-the-art performances and widespread deployment in medical image analysis. Ever since U-Net (Ronneberger et al., 2015) won the 2015 ISBI cell tracking challenge, CNNs have taken the medical image analysis research by storm. Up till today, U-Net and its variants continue to demonstrate outstanding performance in many fields of medical imaging (Isensee et al., 2021; Zhou et al., 2022a; Cui et al., 2019). Other deep learning techniques, such as recurrent neural networks (RNNs) (Zhou et al., 2019) and deep reinforcement learning (DRL) (Zhou et al., 2021d), have been developed and built on top of CNNs for medical image analysis.
More recently, Transformer (Vaswani et al., 2017) has shown great potential in medical imaging applications as it has flourished in natural language processing and is flourishing in computer vision. Regarding homogeneity and heterogeneity of natural and medical images representations, it is motivated to investigate the status quo of Vision Transformer for medical imaging. It remains unclear whether Vision Transformers are better than CNNs for understanding medical images, and whether Transformers can transform medical imaging. Like any other machine learning techniques, Transformers have both advantages and disadvantages. For example, one of the benefits of Transformers is that they tend to have large effective receptive fields, which means they are better at understanding contextual information than CNNs. This is particularly useful in medical imaging, where it is important to take into account not only the area of concern but also the surrounding tissue and organs when diagnosing a medical condition. On the downside, Transformers tend to be more computationally intensive and require more data. This can be a challenge in the field of medical imaging, where resources may be limited due to factors such as patient privacy concerns. At the present stage, it is uncertain whether Transformers will revolutionize the field of medical imaging, but current research has shown their potential in achieving improved performance on various medical imaging tasks. In this paper, we highlight the properties of Vision Transformers and present a comparative review for Transformer-based medical image analysis. Given that, the survey is confined to Vision Transformer, Unless stated otherwise, "Transformer" and "Transformer-based" referred in this paper represents "Vision Transformer", models with vanilla Language Transformer base blocks integrated, and applied in image analysis tasks.
We organize the rest of paper to include the following: (i) a brief introduction to CNN for medical image analysis; (Section 2) (ii) an introduction to Transformer with its general principle, key properties, and its main differences from a CNN (Section 3); (iii) current progresses of state-of-the-art Transformer methods for solving medical imaging tasks, including medical image segmentation, recognition, classification, detection, registration, reconstruction, and enhancement, which is the main part (Section 4); (iv) yet-to-solve challenges and future potential of Transformer in medical imaging (Section 5).
2. CNN for Medical Image Analysis
2.1. CNNs for medical imaging
We begin by briefly outlining the applications of CNNs in medical imaging and discussing their potential limitations. CNNs are specialized in analyzing data with a known grid-like topology (e.g., images). This is due to the fact that the convolution operation imposes a strong prior on the weights, compelling the same weights to be shared across all pixels. As the exploration of deep CNN architectures has intensified since the development of AlexNet for image classification in 2012 (Krizhevsky et al., 2012), the first few successful efforts at deploying CNNs for medical imaging lay in the application of medical image classifications. These network architectures often begin with a stack of convolutional layers, pooling operations, and follow by a fully connected layer for producing a vector reflecting the probability of belonging to a certain class (Roth et al., 2014, 2015; Cireşan et al., 2013; Brosch et al., 2013; Xu et al., 2014; Malon and Cosatto, 2013; Cruz-Roa et al., 2013; Li et al., 2014). In the meantime, similar architectures have been used for medical image segmentation (Ciresan et al., 2012; Prasoon et al., 2013; Zhang et al., 2015a; Xing et al., 2015; Vivanti et al., 2015) and registration (Wu et al., 2013; Miao et al., 2016; Simonovsky et al., 2016) by performing the classification task on a pixel-by-pixel basis.
In 2015, Ronneberger et al. introduced U-Net (Ronneberger et al., 2015), which is built based on the concept of the fully convolutional network (FCN) (Long et al., 2015). In contrast to previous encoder-only networks, U-Net employs a decoder composed of successive blocks of convolutional layers and upsampling layers. Each block upsamples the previous feature maps such that the final output has the same resolution as the input. U-Net represents a substantial advance over previous networks. First, it eliminated the need for laborious sliding-patch inferences by having the input and output be full-sized images. Moreover, because the input to the network is a full-sized image as opposed to a small patch, U-Net has a better understanding of contextual information presented in the input. Although many other CNN architectures have demonstrated superior performances (e.g., HyperDense-Net (Dolz et al., 2018) and DnCNN (Zhang et al., 2017; Cheng et al., 2019; Kim et al., 2018)), the U-Net-like encoder-decoder paradigm has remained the de facto choice when it comes to CNNs for pixel-level tasks in medical imaging. Many variants of such a kind have been proposed and demonstrated promising results on various applications, including segmentation (Isensee et al., 2021; Zhou et al., 2018; Oktay et al., 2018; Gu et al., 2020; Zhang et al., 2020), registration (Balakrishnan et al., 2019; Dalca et al., 2019; Zhao et al., 2019b,a), and reconstruction (Han and Ye, 2018; Cui et al., 2019). Attempts have been made to improve CNNs by incorporating RNNs or LSTMs for medical image analysis. For instance, Alom et al. proposed a combination of ResUNet with RNN (Alom et al., 2018), which includes a feature accumulation module to enhance feature representations for image segmentation. Gao et al. proposed Distance-LSTM (Gao et al., 2019a), which is capable of modeling the time differences between longitudinal scans. This model is efficient at learning the intra-scan feature variabilities. Similarly, (Gao et al., 2018) merged CNNs with LSTM to learn spatial-temporal representations of brain MRI slices for segmentation. In general, RNNs have a unique ability to model medical images that can advance CNNs. By integrating CNNs with RNNs, it becomes feasible to capture global spatial-temporal feature relationships. Nevertheless, due to the resource-intensive nature of RNNs, they are mostly used for particular tasks, such as comprehending sequential data (e.g., longitudinal data).
Despite the widespread success of CNNs in medical imaging applications over the last decade, there are still inherent limitations within the architecture that prevent CNNs from reaching even greater performance. The vast majority of current CNNs deploy rather small convolution kernels (e.g., 3 × 3 or 5 × 5). Such a locality of convolution operations results in the CNNs being biased toward local spatial structures (Zhou et al., 2021b; Naseer et al., 2021; Dosovitskiy et al., 2020), which makes them less effective at modeling the long-range dependencies required to better comprehend the contextual information presented in the image. Extensive efforts have been made to address such limitations by expanding the theoretical receptive fields (RFs) of CNNs, with the most common methods including increasing the depth of the network (Simonyan and Zisserman, 2015), introducing recurrent- (Liang and Hu, 2015) or skip-/residual-connections (He et al., 2016), introducing dilated convolution operations (Yu and Koltun, 2016; Devalla et al., 2018), deploying pooling and up-sampling layers (Ronneberger et al., 2015; Zhou et al., 2018), as well as performing cascaded or two-stage framework (Isensee et al., 2021; Gao et al., 2019b, 2021a). Despite these attempts, the first few layers of CNNs still have limited RFs, making them unable to explicitly model the long-range spatial dependencies. Only at the deeper layers can such dependencies be modeled implicitly. However, it was revealed that as the CNNs deepen, the influence of faraway voxels diminishes rapidly (Luo et al., 2016). The effective receptive fields (ERFs) of these CNNs are, in fact, much smaller than their theoretical RFs, even though their theoretical RFs encompass the entire input image.
2.2. Motivations behind using Transformers
Transformers, as alternative network architecture to CNNs, has recently demonstrated superior performances in many computer vision tasks (Dosovitskiy et al., 2020; Liu et al., 2021b; Wu et al., 2021a; Zhu et al., 2020; Wang et al., 2021f; Chu et al., 2021; Yuan et al., 2021b; Dong et al., 2022). The core element of Transformers is the self-attention mechanism, which is not subject to the same limitations as convolution operations, making them better at capturing explicit long-range dependencies(Wang et al., 2022c). Transformers have other appealing features, such as they scale up more easily (Liu et al., 2022e) and are more robust to corruption (Naseer et al., 2021). Additionally, their weak inductive bias enables them to achieve better performance than CNNs with the aid of large-scale model sizes and datasets (Liu et al., 2022e; Zhai et al., 2022; Dosovitskiy et al., 2020; Raghu et al., 2021). Existing Transformer-based models have shown encouraging results in several medical imaging applications (Chen et al., 2021d; Hatamizadeh et al., 2022b; Chen et al., 2022b; Zhang et al., 2021e), prompting a surge of interest in further developing such models (Shamshad et al., 2022; Liu and Shen, 2022; Parvaiz et al., 2022; Matsoukas et al., 2021). This paper provides an overview of Transformer-based models developed for medical imaging applications and highlights their key properties, advantages, shortcomings, and future directions. In the next section, we briefly review the fundamentals of Transformers.
3. Fundamentals of Transformer
Language Transformer (Vaswani et al., 2017) is a neural network based on self-attention mechanisms and feed-forward module to compute representations and global dependencies. Recently, large Language Transformer models employed self-supervised pre-training has demonstrated improved efficiency and scalability, such as BERT (Devlin et al., 2018) and GPT (Radford et al., 2018; Radford et al.; Brown et al., 2020) in natural language processing (NLP). In addition, Vision Transformer (ViT) (Dosovitskiy et al., 2020) partition and flatten images to sequences and implement Transformer for modeling visual features in a sequence-to-sequence paradigm. Below, we first give a detailed introduction to Vision Transformer, focusing on self-attention and its general pipeline. Next, we summarize the characteristics of convolution and self-attention and how the two interact. Lastly, we include key properties of Transformer from manifold perspectives.
3.1. Self-attention in Transformer
Humans choose and pay attention to part of the information unintentionally when observing, learning and thinking. The attention mechanism in neural networks is a mimic to this physiological signal processing process (Bahdanau et al., 2014). A typical attention function computes a weighted aggregation of features, filtering and emphasizing the most significant components or regions (Bahdanau et al., 2014; Xu et al., 2015; Dai et al., 2017; Hu et al., 2018).
3.1.1. Self-attention
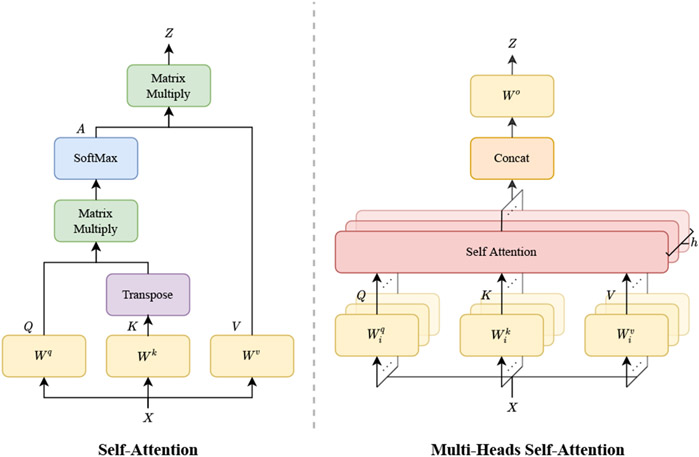
Self-attention (SA) (Bahdanau et al., 2014) is a variant of attention mechanism (Figure 1 (left)), which is designed for capturing the internal correlation in data or features. The standard SA (Vaswani et al., 2017) first maps the input into a query , a key , and a value , using three learnable parameters Wq, Wk, and Wv, respectively:
| (1) |
Fig. 1.
Details of a self-attention mechanism (left) and a multi-head self-attention (MSA) (right). Compared to self-attention, the MSA conducts several attention modules in parallel. The independent attention features are then concatenated and linearly transformed to the output.
Then, the similarity and correlation between query Q and key K is normalized, attaining an attention distribution :
| (2) |
The attention weight is applied to value V, giving the output of a self-attention block:
| (3) |
In general, the key K acts as an embedding matrix that "memorizes" data, and the query Q is a look-up vector. The affinity between the query Q and the corresponding key K defines the attention matrix A. The output Z of a self-attention layer is computed as a sum of value V, weighted by A. The matrix A calculated in (2) connects all elements, thereby leading to a good capability of handling long-range dependencies in both NLP and CV tasks.
3.1.2. Multi-head self-attention (MSA)
Multiple self-attention blocks, namely multi-head self-attention (Figure 1 (right)), are performed in parallel to produce multiple output maps. The final output is typically a concatenation and projection of all outputs of SA blocks, which can be given by:
| (4) |
where h denotes the total number of heads and is a linear projection matrix, aggregating the outputs from all attention heads. , and are parameters of the ith attention head. MSA projects Q, K and V into multiple sub-spaces that compute similarities of context features. Note that it is not necessarily true that a larger number of heads accompanies with better performance (Voita et al., 2019).
3.2. Vision Transformer pipeline
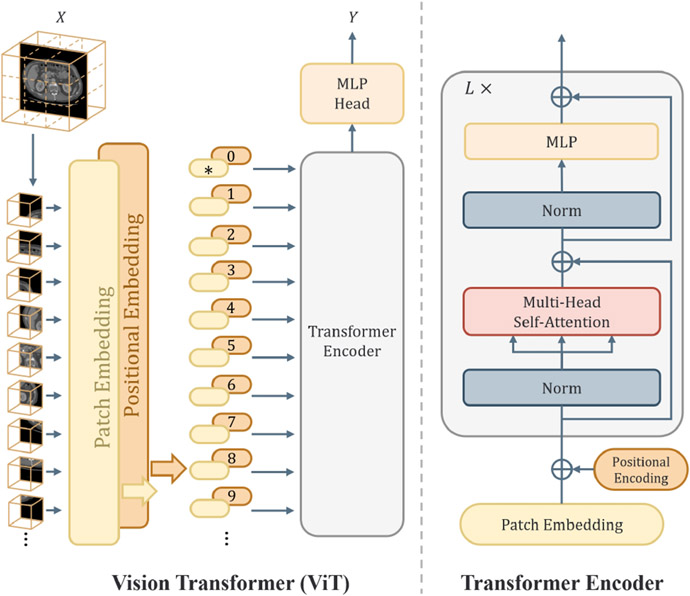
3.2.1. Overview
A typical design of a Vision Transformer consists of a Transformer encoder and a task-specific decoder, depicted in Figure 2 (left). Take the processing of 2D images for instance. Firstly, the image is split into a sequence of N non-overlapping patches {X1, X2, …, XN}; , where C is the number of channels, [H, W] denotes the image size, and [P, P] is the resolution of a patch. Next, each patch is vectorized and then linearly projected into tokens:
| (5) |
where D is the embedding dimension. Then, a positional embedding, Epos, is added so that the patches can retain their positional information:
| (6) |
Fig. 2.
Overview of Vision Transformer (left) and illustration of the Transformer encoder (right). The strategy for partitioning an image involves dividing it into several patches of a fixed size, which are then treated as sequences using an efficient Transformer implementation from NLP.
The resulting tokens are fed into a Transformer encoder as shown in Figure 2 (right), which consists of L stacked base blocks. Each base block consists of a multi-head self-attention and a multi-layer perceptron (MLP), with Layer-Norm (LN). The feature can be formulated as:
| (7) |
3.2.2. Non-overlapping patch generation
ViT adapts a standard Transformer in vision tasks, with the fewest modifications as possible. Therefore, the patches {X1, …, Xn} are generated in a non-overlapping style. On one hand, non-overlapping patches partially break the internal structure of an image (Han et al., 2021a). MSA blocks integrate information from various patches, alleviating this problem. On the other hand, there is no computational redundancy when feeding non-overlapping patches into Transformer.
3.2.3. Positional embedding
Transformers tokenize and analyze each patch individually, resulting in the loss of positional information on each patch in relation to the whole image, which is undesired given that the position of each patch is imperative for comprehending the context in the image. Positional embeddings are proposed to encode such information into each patch such that the positional information is preserved throughout the network. Moreover, positional embeddings serve as the manually introduced inductive bias in Transformers. In general, there are three types of positional embedding: sinusoidal, learnable, and relative. The first two encode absolute positions from 1 to the number of patches, while the last encodes relative positions/distances between patches. In the following subsections, we briefly introduce each of the positional embeddings.
Sinusoidal positional embedding.
To encode the position of each patch, we might intuitively assign an index value between 1 and the total number of patches to each patch. Yet, an obvious issue arises: if the number of patches is large, there may be a significant discrepancy in the index values, which hinders network training. Here, the key idea is to represent different positions using sinusoids of different wavelengths. For each patch position n, the sinusoidal positional embedding is defined as (Vaswani et al., 2017):
| (8) |
where .
Learnable positional embedding.
Instead of encoding the exact positional information onto the patches, a more straightforward way is to deploy a learnable matrix, Elrn, and let the network learn the positional information on its own. This is known as the learnable positional embedding.
Relative positional embedding.
Contrary to using a fixed embedding for each location, as is done in sinusoidal and learnable positional embeddings, relative positional embedding encodes the relative information according to the offset between the elements in Q and K being compared in the self-attention mechanism (Raffel et al., 2020). Many relative positional embedding approaches have been developed, and this is still an active field of research (Shaw et al., 2018; Raffel et al., 2020; Dai et al., 2019; Huang et al., 2020; Wang et al., 2020a; Wu et al., 2021b). However, the basic principle stays the same, in which they encode information about the relative position of Q, K, and V through a learnable or hard-coded additive bias during the self-attention computation.
3.2.4. Multi-layer perceptrons
In the conventional Transformer design (e.g., the original ViT (Dosovitskiy et al., 2020) and Transformer (Vaswani et al., 2017)), the MLP comes after each self-attention module. MLP is a crucial component since it injects inductive bias into Transformer, while the self-attention operation lacks inductive bias. This is because MLP is local and translation-equivariant, but self-attention computation is a global operation. The MLP is comprised of two feed-forward networks with an activation (typically a GeLU) in between:
| (9) |
where x denotes the input, and W and b denote, respectively, the weight matrix and the bias of the corresponding linear layer. The dimensions of the weight matrices, W1 and W2, are typically set as D × 4D and 4D × D (Dosovitskiy et al., 2020; Vaswani et al., 2017). Since the input is a matrix of flattened and tokenized patches (i.e., Eqn. (6)), applying W to x is analogous to applying a convolutional layer with a kernel size of 1 × 1. Consequently, the MLPs in the Transformer are highly localized and and equivariant to translation.
3.3. Transformer vs. CNNs
CNNs provide promising results for image analysis, while Vision Transformer has shown comparable even superior performance when pre-training or scaled datasets are available (Dosovitskiy et al., 2020). This raises a question on the differences about how Transformers and CNNs understand images. The receptive field of CNNs gradually expands when the nets go deeper, therefore the features extracted in lower stages are quite different from those in higher stages (Raghu et al., 2021). Features are analyzed and represented layer-by-layer, with global information injected. Besides, the expanding receptive field of neurons and the use of pooling operations result in equivalence and local invariance in terms of translation (Jaderberg et al., 2015; Kauderer-Abrams, 2017), which empowers CNNs to exploit samples and parameters more effectively (see Appendix Appendix .1 for further details). Beyond that, the locality and weight sharing confers CNNs the advantages in capturing local structures. Considering the limited receptive field, CNNs are limited in catching long-distance relationships among image regions. In Transformer model, the MSA provides a global receptive field even with the lowest layer of ViT, resulting in similar representations in different number of blocks (Raghu et al., 2021). The MSA block of each layer is capable of aggregating features in a global perspective, reaching a good understanding of long-distance relationships. The 16 by 16 sequences length is in natural large receptive field that can lead to better global feature modeling. In 3D transformers for volumetric data, this advantage is even obvious, the use of patch size 16×16×16 is intuitive and beneficial for high dimensional, high resolution medical images, as anatomical context are crucial for medical deep learning.
3.3.1. Combining Transformer and CNN
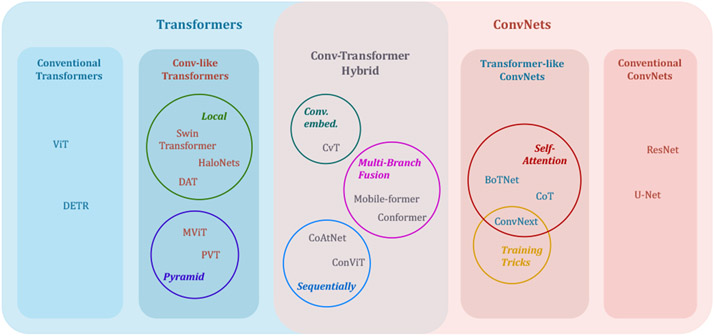
To embrace the benefits from conventional CNNs (e.g., ResNet (He et al., 2016) and U-Net (Ronneberger et al., 2015)) and conventional Transformers (e.g., the original ViT (Dosovitskiy et al., 2020) and DETR (Carion et al., 2020)), multiple works have been done in combining the strengths of CNNs and Transformer, which can be included into three types, and we illustrate them one by one in the following paragraphs. Additionally, Fig. 3 contains a taxonomy of typical methods that combine CNN and Transformer.
Fig. 3.
Taxonomy of typical approaches in combining CNNs and Transformer.
Conv-like Transformers:
This type of model introduces some convolutional properties into conventional Vision Transformer. The building blocks are still MLPs and MSAs, while arranged in a convolutional style. For example, in Swin Transformer (Liu et al., 2021b), HaloNets (Vaswani et al., 2021), and DAT (Xia et al., 2022b), the self-attention is performed within a local window hierarchically and neighboring windows are merged in subsequent layers. Hierarchical multi-scale framework in MViT (Fan et al., 2021) and pyramid structures in PVT (Wang et al., 2021f) guide a Transformer to increase the capacity of intermediate layers progressively.
Transformer-like CNNs:
This type of model introduces the traits of Vision Transformers into CNNs. The building blocks are convolutions, while arranged in a more Vision Transformer way. Thus, this type of models are excluded in the introduction about Transformer models in Section 4. Specifically, the self-attention mechanism is assembled to convolutions, like in CoT (Li et al., 2021e) and BoTNet (Srinivas et al., 2021), making a full exploration of neighboring context that compensates the CNNs’ weakness in capturing long-range dependencies. ConvNext (Liu et al., 2022e) modernizes a ResNet by exploiting a depth-wise convolution as a substitute of self-attention, and following the training tricks from Swin Transformer (Liu et al., 2021b).
Conv-Transformer hybrid:
A straightforward way of combining CNNs and Transformers is to employ them both in an attempt of leveraging both of their strengths. So the building blocks are convolutions, MLPs and MSAs. This is done by keeping self-attention modules to catch long-distance relationships, while utilizing the convolution to project patch embeddings in CvT (Wu et al., 2021a). Another type of methods is the multi-branch fusion, like Conformer (Peng et al., 2021) and Mobile-former (Chen et al., 2021g), which typically fuses the feature maps from two parallel branches, one from CNN and the other from Transformer, such that the information provided by both architectures is retained throughout the decoder. Analogously, convolutions and Transformer blocks are arranged sequentially in ConViT (dâĂŹAscoli et al., 2021) and CoAtNet (Dai et al., 2021c), and representations from convolutions are aggregated by MSAs in a global view.
3.4. Key properties
From the basic theory and architecture design of Transformer, researchers are yet to figure out why Transformer works better than say CNN in many scenarios. Below are some key properties associated with Transformers from the perspectives of modeling and computation.
3.4.1. Modeling
M1: Long-range dependency.
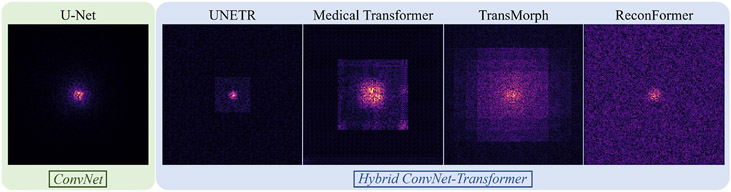
The MSA module connects all patches with a constant distance, and it is proved in (Joshi, 2020) that a Transformer model is equivalent to a graph neural network (GNN). It promises Transformer with large theoretical and effective receptive fields (as shown in Fig. 4), and possibly brings better understanding of contextual information and long-range dependency than CNNs.
Fig. 4.
Effective receptive fields (ERFs) (Luo et al., 2016) of the well-known CNN, U-Net (Ronneberger et al., 2015), versus the hybrid Transformer-CNN models, including UNETR (Hatamizadeh et al., 2019), Medical Transformer (Valanarasu et al., 2021), TransMorph (Chen et al., 2022b), and ReconFormer (Guo et al., 2022d). The ERFs are computed at the last layer of the model prior to the output. The γ correction of γ = 0.4 was applied to the ERFs for better visualization. Despite the fact that its theoretical receptive field encompasses the whole image, the pure CNN model, U-Net (Ronneberger et al., 2015), has a limited ERF, with gradient magnitude rapidly decreasing away from the center. On the other hand, all Transformer-based models have large ERFs that span over the entire image.
M2: Detail modeling.
Images are projected into embeddings by MLPs in Transformers. The embeddings of local patches are refined and adjusted progressively at the same scale. Features in CNNs, like ResNet and U-Net, are resized by pooling and strided-convolution operations. Features are at different detailing stages over scales. Dense modeling and trainable aggregation of features in Transformers can preserve contextual details along with more semantic information injected when deeper layers are reached (Li et al., 2022e).
M3: Inductive bias.
The convolutions in CNNs exploit the relations from the locality of pixels and apply the same weights across the entire image. This inherent inductive bias leads to faster convergence of CNNs and better performances in small datasets (dâĂŹAscoli et al., 2021). On the other hand, because computing self-attention is a global operation, Transformers in general have a weaker inductive bias than CNNs (Cordonnier et al., 2019). The only manually injected inductive bias in original ViT (Dosovitskiy et al., 2020) is the positional embedding. Therefore, Transformers lack the inherent properties of locality and scale-invariance, making them more data-demanding and harder to train (Dosovitskiy et al., 2020; Touvron et al., 2021b). However, the reduced inductive bias may improve the performance of Transformers when trained on a larger-scale dataset. See Appendix .2 for further details.
M4: Loss landscape.
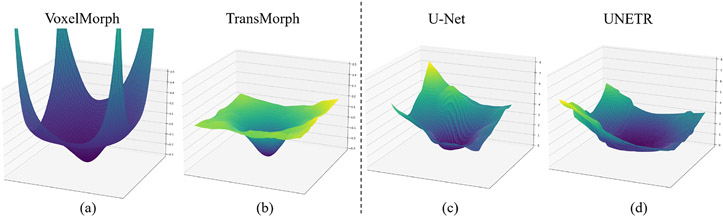
The self-attention operation of Transformer tends to promote a flatter loss landscape (Park and Kim, 2022), even for hybrid CNN-Transformer models, as shown in Fig. 5. This results in improved performance and better generalizability compared to CNNs when trained under the same conditions. See Appendix .3 for further details.
Fig. 5.
Loss landscapes for the models based on CNNs versus Transformers. The left and right panels depict, respectively, the loss landscapes for registration and segmentation models. The left panel shows loss landscapes generated based on normalized cross-correlation loss and a diffusion regularizer; the right panel shows loss landscapes created based on a combination of Dice and cross-entropy losses. Transformer-based models, such as (b) TransMorph (Chen et al., 2022b) and (d) UNETR (Hatamizadeh et al., 2022b), exhibit flatter loss landscapes than CNN-based models, such as (a) VoxelMorph (Balakrishnan et al., 2019) and (c) U-Net (Ronneberger et al., 2015).
M5: Noise robustness.
Transformers are more robust to common corruptions and perturbations, such as blurring, motion, contrast variation, and noise (Bhojanapalli et al., 2021; Xie et al., 2021a).
3.4.2. Computation
C1: Scaling behavior.
Transformers show the same scaling properties in NLP and CV (Zhai et al., 2022). The Transformer models achieve higher performance when their computation, model capacity, and data size scale up together.
C2: Easy integration.
It is easy to integrate Transformers and CNNs into one computational model. As shown in Section 3.C and future sections, there are multiple ways of integrating them, resulting in flexible architecture designs that are mainly grouped into Conv-like Transformers, Transformer-like CNNs, and Conv-Transformer hybrid.
C3: Computational intensiveness.
While promising results may be obtained with Transformers, typical Transformers (e.g., ViT (Dosovitskiy et al., 2020; Zhai et al., 2021)) require a significant amount of time and memory, particularly during training. See Appendix .4 for further details.
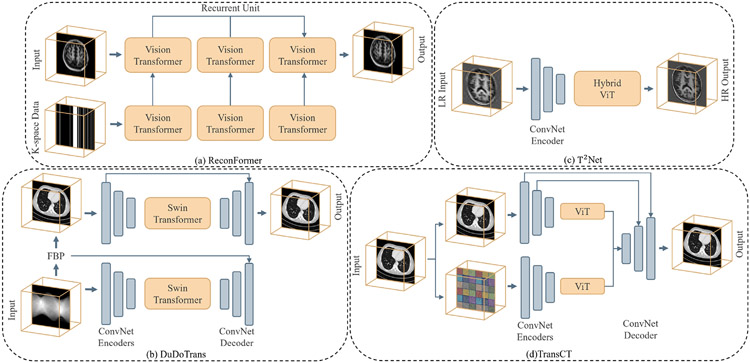
4. Current Progresses
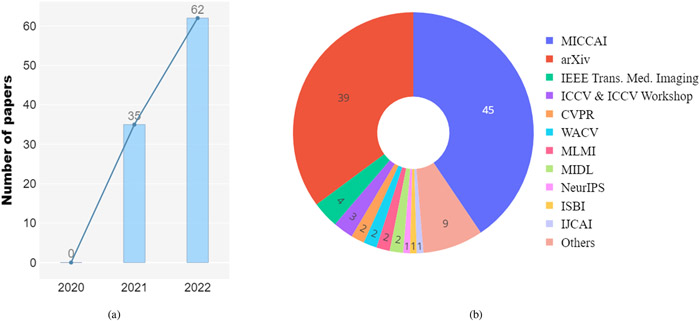
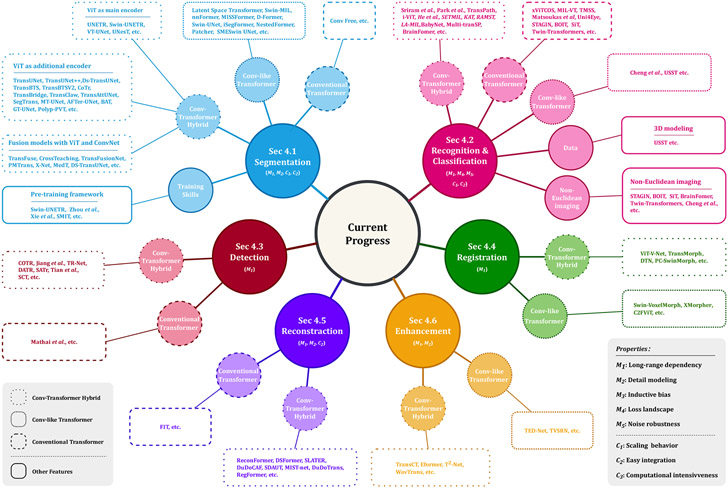
As shown in Fig. 6(a), Vision Transformers has received intensive study in present. We introduce the criteria of inclusion/exclusion for selecting research papers in this review. Fig. 6(b) shows the graphic summary of Transformers in medical image analysis papers. In particular, we investigate articles on IEEE, PubMed, Xplore, Springer, Science direct, proceedings of conferences including medical imaging conferences such as MICCAI, IPMI, ISBI, RSNA, SPIE, etc. Finally, we search manuscripts and project references on google scholar. In the result of search queries, we have found over 2000 transformer related papers, most of these contributions are from language studies or natural image analysis. We build our survey concepts from the self-attention paper, and vision transformer, which are keys milestones for exploring transformer in medical studies. Finally, we set the criteria of legitimacy for this survey only about medical application with transformers. As shown in Fig. 6(b), we demonstrate categorization of our selected papers based on tasks in medical domain. In the figure, we show percentage of article sources from conferences, journals, and pre-print platforms. The list of our selected papers, covering a wide range of topics including medical image segmentation, recognition & classification, detection, registration, reconstruction, and enhancement, is by no means exhaustive. Fig. 7 gives an overview of the current applications of vision Transformers, and below we present a literature summary for each topic with the use of key properties indicated accordingly.
Fig. 6.
(a) The number of papers accepted to the MICCAI conference from 2020 to 2022 whose titles included the word "Transformer". (b) Sources of all 114 selected papers.
Fig. 7.
An overview of Transformers applied in medical tasks in segmentation, recognition & classification, detection, registration, reconstruction, and enhancement.
4.1. Medical image segmentation
In general, Transformer-based models outperform ConvNets for solving medical image segmentation tasks. The main reasons are as follows:
The ability of modeling longer range dependencies of context in high dimensional and high resolution medical images. [Property M1]
The scalability and robustness of ViT and Swin Transformer strengthen the dense prediction for pixel-wise segmentation (Liu et al., 2021b). [Property M2]
The superior scaling behavior of Transformers over ConvNets and the lack of convolutional inductive bias in Transformers make them more advantageous to large-scale self-supervised pre-training on medical image datasets (Tang et al., 2022; Zhai et al., 2022). [Property C1 and M3]
Network architecture design is flexible by mixing Transformer and CNN modules. [Property C2]
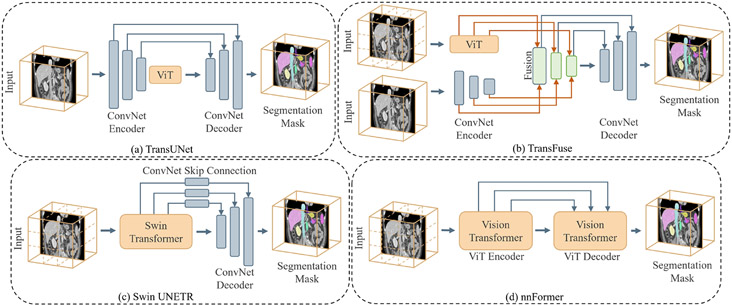
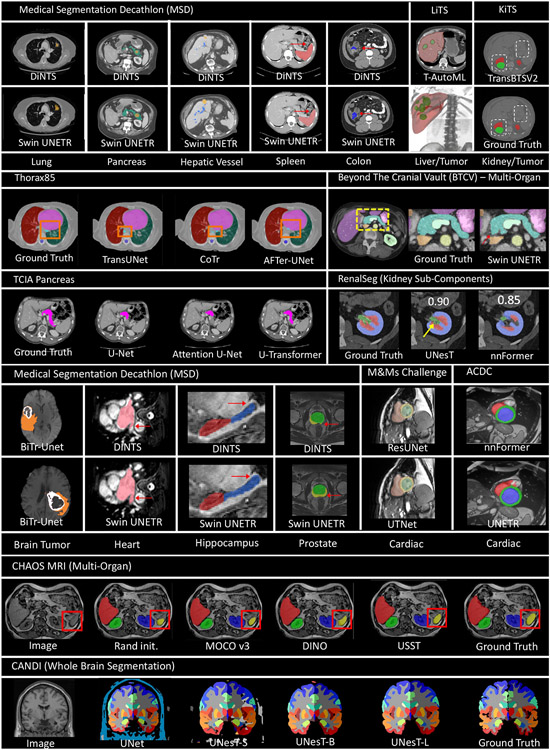
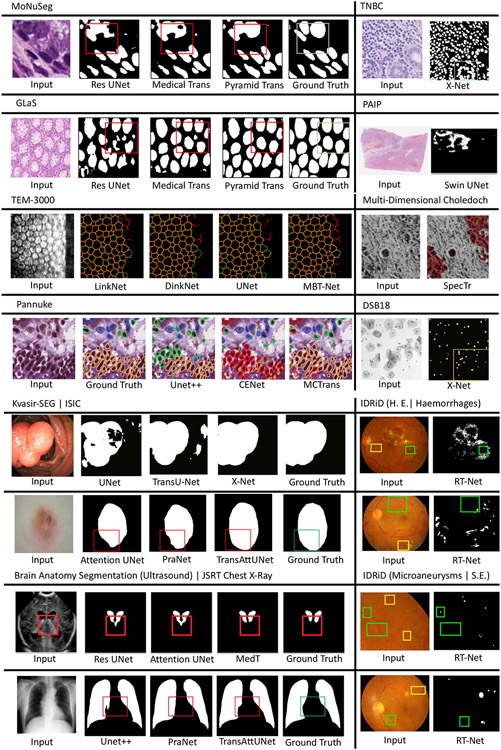
Though it has demonstrated superior performance, the use of Transformers for medical image segmentation has challenges in transferring the representation capability from language domain to image modalities. Compared to word tokens that are modeled as the basic embedding, visual features are at variant scales. This multi-scale problem can be significant in dense prediction tasks with higher resolution of voxels in medical images. However, for the current Transformer backbones, the learnt embedding is commonly at a fixed scale, which is intractable for segmentation tasks, especial on large-scale medical radiography, microscopy, fundus, endoscopy or other imaging modalities. To adapt the vanilla Transformer models for medical image segmentation, recent researchers proposed solutions that utilize the components of ViT into particular segmentation models. In the following, we summarize and discuss recent works on how Transformer blocks are used in the segmentation models. Table 1 provides a summary list of all reviewed segmentation approaches along with their information about associated architecture type, model size, dataset, method highlight, etc. As one of the most classical approaches in medical segmentation, U-Net (Ronneberger et al., 2015) is widely chosen for comparison by its followers. The U-shaped architecture and skip-connections in U-Net has proved its effectiveness in leveraging hierarchical features. Fig. 8 presents some typical Transformer-based U-shaped segmentation model architectures.
Table 1.
The summarized review of Transformer-based model for medical image segmentation. "N" denotes not reported or not applicable on number of model parameters. "N.A." denotes for not applicable for intermediate blocks or decoder module.
| Reference | Architecture | 2D/3D | #Param | Modality | Dataset | ViT as Enc/Inter/Dec | Highlights |
|---|---|---|---|---|---|---|---|
| CoTr (Xie et al., 2021b) | Conv-Transformer Hybrid | 3D | 46.51M | CT | Multi-organ (BTCV (Landman et al., 2015)) | No/Yes/No | The Transformer block with deformable module captures deep features in the bottleneck. |
| SpecTr (Yun et al., 2021) | Conv-Transformer Hybrid | 3D | N | Microscopy | Cholangiocarcinoma (Zhang et al., 2019b) | No/Yes/No | Hybrid Conv-Transformer encoder with spectral normalization. |
| TransBTS (Wang et al., 2021e) | Conv-Transformer Hybrid | 3D | 32.99M | MRI | Brain Tumor (Baid et al., 2021) | No/Yes/No | 3D Transformer blocks for encoding bottleneck features. |
| UNETR (Hatamizadeh et al., 2021) | Conv-Transformer Hybrid | 3D | 92.58M | CT, MRI | Multi-organ (BTCV (Landman et al., 2015)), Brain Tumor, Spleen (MSD (Simpson et al., 2019 | Yes/No/No | The 3D Transformer directly encodes image into features, and use of CNN decoder for capturing global information. |
| BiTr-UNet (Jia and Shu, 2021) | Conv-Transformer Hybrid | 3D | N | MRI | Brain Tumor (Baid et al., 2021) | No/Yes/No | The bi-level Transformer blocks are used for encoding two level bottleneck features of acquired CNN feature maps. |
| VT-UNet (Peiris et al., 2021) | Conv-Transformer Hybrid | 3D | 20.8M | MRI, CT | Brain tumor, Pancreas, Liver (MSD (Simpson et al., 2019)) | Yes/Yes/Yes | The encoder directly embeds 3D volumes jointly capture local/global information, the decoder introduces parallel cross-attention expansive path. |
| Swin UNETR (Tang et al., 2022; Hatamizadeh et al., 2022a) | Conv-Transformer Hybrid | 3D | 61.98M | CT, MRI | Multi-organ (BTCV) (Landman et al., 2015), MSD 10 tasks (Simpson et al., 2019) | Yes/No/No | The 3D encoder with swin-Transformer direclty encodes the 3D CT/MRI volumes with a CNN-based decoder for better capturing global information. |
| HybridCTrm (Sun et al., 2021a) | Conv-Transformer Hybrid | 3D | N | MRI | MRBrainS (Mendrik et al., 2015), iSEG-2017 (Wang et al., 2019b) | Hybrid/N.A./No | A hybrid architecture encodes images from CNN and Transformer in parallel. |
| UNesT (Yu et al., 2022c) | Conv-Transformer Hybrid | 3D | 87.30M | CT | Kidney Sub-components (RenalSeg, KiTS (Heller et al., 2021)) | Yes/No/No | The use of hierarchical Transformer models for efficiently capturing multi-scale features with a 3D block aggregation module. |
| Universal (Jun et al., 2021) | Conv-Transformer Hybrid | 3D | N | MRI | Brain Tumor (Baid et al., 2021) | No/Yes/No | The proposed model takes advantages of three views of 3D images and fuse 2D features to 3D volumetric segmentation. |
| PC-SwinMorph (Liu et al., 2022a) | Conv-Transformer Hybrid | 3D | N | MRI | Brain (CANDI (Kennedy et al., 2012), LPBA-40 (Shattuck et al., 2008)) | No/No/Hybird | The designed patch-based contrastive and stitching strategy enforce a better fine detailed alignment and richer feature representation. |
| TransBTSV2 (Li et al., 2022c) | Conv-Transformer Hybrid | 3D | 15.30M | MRI, CT | Brain Tumor (Baid et al., 2021), Liver/Kidney Tumor (LiTS (Bilic et al., 2019), KiTS (Heller et al., 2021 | No/Yes/No | The deformable bottleneck module is used in the Transformer blocks modeling bottleneck features to capture more shape-aware representations. |
| GDAN (Lin et al., 2022b) | Conv-Transformer Hybrid | 3D | N/A | CT | Aorta | No/Yes/No | Geometry-constrained module and deformable self-attention module are designed to guide segmentation. |
| VT-UNet (Peiris et al., 2022) | Conv-Transformer Hybrid | 3D | N/A | MRI, CT | Brain Tumor (Baid et al., 2021) | Yes/Yes/No | The self-attention mechanism to simultaneously encode local and global cues, the decoder employs a parallel self and cross attention formulation to capture fine details for boundary refinement. |
| ConTrans (Lin et al., 2022a) | Conv-Transformer Hybrid | 2D | N/A | Endoscopy, Microscopy, RGB, CT | Cell (Pannuke), (Polyp, CVC-ClinicDB (Bernal et al., 2015), CVC-ColonDB (Bernal et al., 2012), ETIS-Larib (Silva et al., 2014), Kvasir (Jha et al., 2020)), Skin (ISIC (Codella et al., 2018)) | Yes/Yes/No | Spatial-Reduction-Cross-Attention (SRCA) module is embedded in the decoder to form a comprehensive fusion of these two distinct feature representations and eliminate the semantic divergence between them. |
| DA-Net (Wang et al., 2022b) | Conv-Transformer Hybrid | 2D | N/A | MRA images | Retina Vessels (DRIVE (Staal et al., 2004) and CHASE-DB1 (Fraz et al., 2012)) | No/Yes/No | Dual Branch Transformer Module (DBTM) that can simultaneously and fully enjoy the patches-level local information and the image-level global context. |
| EPT-Net (Liu et al., 2022c) | Conv-Transformer Hybrid | 3D | N/A | Intracranial Aneurysm | Intracranial Aneurysm (IntrA (Yang et al., 2020)) | No/Yes/No | Dual stream transformer (DST), outeredge context dissimilation (OCD) and inner-edge hard-sample excavation (IHE) help the semantics stream produce sharper boundaries. |
| Latent Space Transformer (Li et al., 2022f) | Conv-like Transformer | 3D | N/A | CT, MRI | LiTS (Bilic et al., 2019), CHAOS (Kavur et al., 2020 | Yes/Yes/Yes | It intentionally make the large patches overlap to enhance intra-patch communication. |
| Swin-MIL (Qian et al., 2022) | Conv-like Transformer | 2D | N/A | Microscopy | Haematoxylin and Eosin (H&E) | Yes/Yes/No | A novel weakly supervised method for pixel-level segmentation in histopathology images, which introduces Transformer into the MIL framework to capture global or long-range dependencies. |
| Segtran (Li et al., 2021a) | Conv-Transformer Hybrid | 2D/3D | 166.7M | Fundus, Colonoscopy, MRI | Disc/Cup (REFUGE20 (Orlando et al., 2020)), Polyp, Brain Tumor | No/Yes/N.A. | The use of squeeze and expansion block for contextualized features after acquiring visual and positional features of CNN. |
| MT-UNet (Wang et al., 2021c) | Conv-Transformer Hybrid | 2D | N | CT, MRI | Multi-organ (BTCV (Landman et al., 2015)), Car-diac (ACDC (Bernard et al., 2018)) | No/Yes/No | The proposed mixed Transformer module simultaneously learns inter- and intra- affinities used for modeling bottleneck features. |
| TransUNet++ (Wang et al., 2022a) | Conv-Transformer Hybrid | 2D | N | CT, MRI | Prostate, Liver tumor (LiTS (Bilic et al., 2019)) | No/Yes/No | The feature fusion scheme at decoder enhances local interaction and context. |
| RT-Net (Huang et al., 2022b) | Conv-Transformer Hybrid | 2D | N | Fundus | Retinal (IDRiD (Porwal et al., 2018), DDR) | No/yes/No | The dual-branch architecture with global Transformer block and relation Transformer block enables detection of small size or blurred border. |
| TransUNet (Chen et al., 2021d) | Conv-Transformer Hybrid | 2D | 105.28M | CT, MRI | Multi-organ (BTCV (Landman et al., 2015)), Cardiac (ACDC (Bernard et al., 2018)) | No/Yes/No | Transformer blocks for encoding bottleneck features. |
| U-Transformer (Petit et al., 2021) | Conv-Transformer Hybrid | 2D | N | CT | Pancreas (TCIA) (Holger and Amal, 2016), Multi-organ | No/Yes/No | The U-shape design with multi-head self-attention for bottleneck features and multi-head cross attention in the skip connections. |
| MBT-Net (Zhang et al., 2021a) | Conv-Transformer Hybrid | 2D | N | Microscopy | Corneal Endothelium cell (TM-EM300, Alizarine (Ruggeri et al., 2010)) | No/Yes/No | The design of hybrid residual Transformer model captures multi-branch global features. |
| MCTrans (Ji et al., 2021) | Conv-Transformer Hybrid | 2D | 7.64M | Microscopy, Colonoscopy, RGB | Cell (Pannuke), (Polyp, CVC-ClinicDB (Bernal et al., 2015), CVC-ColonDB (Bernal et al., 2012), ETIS-Larib (Silva et al., 2014), Kvasir (Jha et al., 2020)), Skin (ISIC (Codella et al., 2018)) | No/Yes/No | The Transformer blocks are used for encoding bottleneck features in a UNet-like model. |
| Decoder (Li et al., 2021b) | Conv-Transformer Hybrid | 2D | N | CT, MRI | Brain tumor (MSD (Simpson et al., 2019)), Multi-organ (BTCV (Landman et al., 2015)) | No/No/Yes | The first study of evaluate the effect of using Transformer for decoder in the medical image segmentation tasks. |
| UTNet (Gao et al., 2021b) | Conv-Transformer Hybrid | 2D | 9.53M | MRI | Cardiac (Campello et al., 2021) | Hybrid/Hybrid/No | The design of a hybrid architecture in the encoder with convolutional and Transformer layers. |
| TransClaw UNet (Chang et al., 2021) | Conv-Transformer Hybrid | 2D | N | CT | Multi-organ (BTCV (Landman et al., 2015)) | No/Yes/No | The Transformer blocks are used as additional encoder for strengthening global connection of CNN encoded features. |
| TransAttUNet (Chen et al., 2021a) | Conv-Transformer Hybrid | 2D | N | RGB, X-ray, CT Microscopy | Skin (ISIC (Codella et al., 2018)), Lung (JSRT (Shiraishi et al., 2000), Montgomery (Jaeger et al., 2014), NIH (Tang et al., 2019)), (Clean-CC-CCII (He et al., 2020b)), Nuclei (Bowl, GLaS (Malík et al., 2020)) | No/Yes/No | The model contains a co-operation of Transformer self-attention and global spatial attention for modeling semantic information. |
| LeViT-UNet(384) (Xu et al., 2021a) | Conv-Transformer Hybrid | 2D | 52.17M | CT, MRI | Multi-organ (BTCV (Landman et al., 2015)), Car-diac (ACDC (Bernard et al., 2018)) | No/Yes/No | The lightweight design of Transformer blocks as second encoder. |
| Polyp-PVT (Dong et al., 2021a) | Conv-Transformer Hybrid | 2D | N | Endoscopy | Polp (Kvasir (Jha et al., 2020), CVC-ClinicDB (Bernal et al., 2015), CVC-ColonDB (Bernal et al., 2012), Endoscene (Vázquez et al., 2017), ETIS (Silva et al., 2014)) | Yes/No/No | The Transformer encoder directly learns the image patches representation. |
| COTRNet (Shen et al., 2021b) | Conv-Transformer Hybrid | 2D | N | CT | Kidney (KITS21 (Heller et al., 2021)) | Hybrid/N.A./No | The U-shape model design has the hybrid of CNN and Transformers for both encoder and decoder. |
| TransBridge (Deng et al., 2021) | Conv-Transformer Hybrid | 2D | 11.3M | Echocardiograph | Cardiac (EchoNet-Dynamic) (Ouyang et al.,2020 | No/Yes/No | The Transformer blocks are used for capturing bottleneck features for bridging CNN encoder and decoder. |
| GT UNet (Li et al., 2021c) | Conv-Transformer Hybrid | 2D | N | Fundus | Retinal (DRIVE (Staal et al., 2004)) | Hybrid/N.A./No | The design of hybrid grouping and bottleneck structures greatly reduces computation load of Transformer. |
| BAT (Wang et al., 2021d) | Conv-Transformer Hybrid | 2D | N | RGB | Skin (ISIC (Codella et al., 2018), PH2 (Mendonça et al., 2013)) | No/Yes/No | The model proposes a boundary-wise attention gate in Transformer for capturing prior knowledge. |
| AFTer-UNet (Yan et al., 2022) | Conv-Transformer Hybrid | 2D | 41.5M | CT | Multi-organ (BTCV (Landman et al., 2015)), Thorax (Thorax-85 (Chen et al., 2021e), SegTHOR (Lambert et al., 2020)) | No/Yes/No | The proposed axial fusion mechanism enables intra- and inter-slice communication and reduced complexity. |
| Conv Free (Karimi et al., 2021) | Conventional Transformer | 3D | N | CT, MRI | Brain cortical (Bastiani et al., 2019) plate, Pancreas, Hippocampus (MSD (Simpson et al., 2019)) | Yes/Yes/N.A. | 3D Transformer blocks as encoder without convolution layers |
| nnFormer (Zhou et al., 2021a) | Conv-like Transformer | 3D | 158.92M | CT, MRI | Brain tumor (Baid et al., 2021), Multi-organ (BTCV (Landman et al., 2015)), Cardiac (ACDC (Bernard et al., 2018)) | Yes/Yes/Yes | The 3D model with pure Transformer as encoder and decoder. |
| MISSFormer (Huang et al., 2021) | Conv-like Transformer | 2D | N | MRI, CT | Multi-organ (BTCV (Landman et al., 2015)), Cardiac (ACDC (Bernard et al., 2018)) | Yes/Yes/Yes | The U-shape design with patch merging and expanding modules as encoder and decoder. |
| D-Former (Wu et al., 2022b) | Conv-like Transformer | 2D | 44.26M | CT, MRI | Multi-organ (BTCV (Landman et al., 2015)), Cardiac (ACDC (Bernard et al., 2018)) | Yes/Yes/Yes | The 3D network contains local/global scope modules to increase the scopes of information interactions and reduces complexity. |
| Swin-UNet (Cao et al., 2021) | Conv-like Transformer | 2D | N | CT, MRI | Multi-organ (BTCV (Landman et al., 2015)), Cardiac (ACDC (Bernard et al., 2018)) | Yes/Yes/Yes | The pure Transformer U-shape segmentation model design enables the use for both encoder and decoder |
| iSegFormer (Liu et al., 2022b) | Conv-like Transformer | 3D | N/A | MRI | Knee (OAI-ZIB (Ambellan et al., 2019)) | Yes/Yes/Yes | It contains a memory-efficient Transformer that-combines a Swin Transformer with a lightweight multilayer perceptron (MLP). decoder. |
| NestedFormer (Xing et al., 2022) | Conv-like Transformer | 3D | N/A | MRI | BraTS2020 (Baid et al., 2021), MeniSeg | Yes/Yes/Yes | A novel Nested Modality-Aware Transformer (NestedFormer) to explicitly explore the intra-modality and inter-modality relationships of multi-modal MRIs for brain tumor segmentation. |
| Patcher (Ou et al., 2022) | Conv-like Transformer | 3D | N/A | MRI, Endoscopy | Stroke Lesion, Kvasir-SEG (Jha et al., 2020) | Yes/Yes/Yes | This design allows Patcher to benefit from both the coarse-to-fine feature extraction common in CNNs and the superior spatial relationship modeling of Transformers. |
| SMESwin UNet (Wang et al., 2022e) | Conv-like Transformer | 2D | N/A | Microscopy | GlaS (Malík et al., 2020) | Yes/Yes/Yes | Fuse multi-scale semantic features and attentions maps by designing a compound structure with CNN and ViTs (named MCCT), based on Channel-wise Cross fusion Transformer (CCT) . |
| DS-TransUNet (Lin et al., 2021) | Conv-Transformer Hybrid | 2D | N | Colonoscopy, RGB, Microscopy | Polyp (Jha et al., 2020), Skin (ISIC (Codella et al., 2018)), Gland (GLaS (Malík et al., 2020)) | Yes/Yes/Yes | The use of swin Transformer as both encoder and decoder forms the U-shape design of segmentation model. |
| MedT (Valanarasu et al., 2021) | Conv-Transformer Hybrid | 2D | N | Ultrasound, Microscopy | Brain (Valanarasu et al., 2020), Gland (Sirinukunwattana et al., 2017), Multi-organ Nuclei (MoNuSeg (Kumar et al., 2019)) | Yes/No/No | A fusion model with a global and local branches as encoders. |
| PMTrans (Zhang et al., 2021d) | Conv-Transformer Hybrid | 2D | N | Microscopy, CT | Gland (GLAS (Malík et al., 2020)), Multi-organ Nuclei (MoNuSeg (Kumar et al., 2019)), Head (HECKTOR (Andrearczyk et al., 2020)) | Hybrid/No/No | The pyramid design of structure enables multiscale Transformer layers for encoder image features. |
| TransFuse (Zhang et al., 2021b) | Conv-Transformer Hybrid | 2D | 26.3M | Endoscopy, RGB, X-ray, MRI | Polp (Kvasir (Jha et al., 2020), ClinicDB (Bernal et al., 2015), ColonDB (Bernal et al., 2012), EndoScene (Vázquez et al., 2017), ETIS (Silva et al., 2014)), Skin (ISIC (Codella et al., 2018)), Hippocampus, Prostate (MSD (Simpson et al., 2019)) | Hybrid/N.A./No | A CNN branch and a Transformer branch encoded features are fused by a BiFusion module to the decoder for segmentation. |
| CrossTeaching (Luo et al., 2021) | Conv-Transformer Hybrid | 2D | N | MRI | ACDC (Bernard et al., 2018) | Hybrid/Hybrid/Hybrid | The two branch network employs advantage of UNet and Swin-UNet. |
| TransFusionNet (Meng et al., 2021) | Conv-Transformer Hybrid | 2D | N | CT | Liver Tumor(LiTS (Bilic et al., 2019)), LiverVessels (LTBV) (Huang et al., 2018), Multi-organ (3Dircadb (Soler et al., 2010)) | Hybrid/N.A./No | The Transformer- and CNN-based encoders extract both features directly from input and fuse to the CNN decoder. |
| X-Net (Li et al., 2021d) | Conv-Transformer Hybrid | 2D | N | Microscopy, Endoscopy | Nuclei (Bowl (Caicedo et al., 2019), TNBC (Naylor et al., 2018)), Polyp (Kvasir (Jha et al., 2020)) | Hybrid/Hybrid/Hybrid | The use of CNN reconstruction model and Transformer segmentation model with mixed representations. |
| T-AutoML (Yang et al., 2021) | Net Architecture Search | 3D | 16.96M | CT | Liver, Lung tumor (MSD (Simpson et al., 2019)) | N.A. | The first medical architecture search framework designed for Transformer-based models. |
| (Xie et al., 2021c) | Pre-training Framework | 2D/3D | N | CT, MRI, X-ray, Dermoscopy | JSRT (Shiraishi et al., 2000), ChestXR (Wang et al., 2017a), BTCV (Landman et al., 2015), RI-CORD (Tsai et al., 2021), CHAOS (Kavur et al., 2020), ISIC (Codella et al., 2018) | No/yes/No | The unified pre-training Framework of 3D and 2D images for Transformer models |
| (Zhou et al., 2022c) | Pre-training Framework | 3D | N | CT, MRI, X-ray | Lung (ChestX-ray14 (Wang et al., 2017a)), Multiorgan (BTCV (Landman et al., 2015)), Brain Tumor(MSD (Simpson et al., 2019)) | Yes/No/No | The masked autoencoder scheme adapts the pretraining framework for medical images. |
| (Tang et al., 2022; Hatamizadeh et al., 2022a) | Pre-training Framework | 3D | N | CT, MRI | Multi-organ (BTCV (Landman et al., 2015)),MSD 10 tasks (Simpson et al., 2019) | Yes/No/No | Very large-scale medical image pre-training framework with Swin Transformers. |
| SMIT (Jiang et al., 2022) | Pre-training Framework | 3D | N | CT, MRI | Covid19, Kidney Cancer, BTCV (Landman et al., 2015) | Yes/No/No | Self-distillation learning with masked image modeling method to perform SSL for vision transformers (SMIT) is applied to 3D multi-organ segmentation from CT and MRI. It contains a dense pixel-wise regression within masked patches called masked image prediction |
Fig. 8.
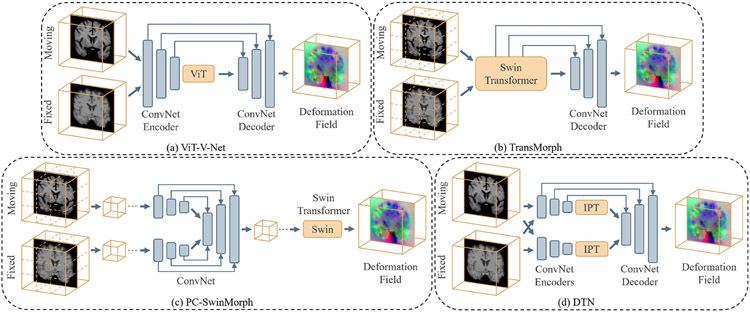
Typical Transformer-based U-shaped segmentation model architectures. (a) The TransUNet (Chen et al., 2021d)-like structure uses Transformer as additional encoder modeling bottleneck features. (b) The Swin UNETR (Tang et al., 2022) uses the Transformer as the main encoder and CNN decoder to construct the hybrid network. (c) The TransFuse (Zhang et al., 2021b) fuses CNN and Transformer encoders together to connect the decoder. (d) The nnFormer (Zhou et al., 2021a)-like structure uses a pure Transformer for both encoder and decoder.
ViT as main encoder:
The Vision Transformers reformulate the segmentation problem as a 1D sequence-to-sequence inference task and to learn medical context from the embedded patches. A major advantage of the sequence-to-sequence modeling strategy is the larger receptive fields compared to CNNs (Dosovitskiy et al., 2020), resulting in stronger representation capability with longer range dependencies. By employing these properties, models that directly use Transformer for generating the input sequences and tokenized patches are proposed (Hatamizadeh et al., 2022b; Tang et al., 2022; Peiris et al., 2021; Yu et al., 2022c). (Hatamizadeh et al., 2022b) and (Peiris et al., 2021) introduce the volumetric model that utilizes the global attention-based Vision Transformer as the main encoder and then connects to the CNNs-based decoder or expand modules. (Tang et al., 2022; Hatamizadeh et al., 2022a) demonstrate the use of shifted-window (Swin) Transformer, which presents more powerful representation ability, as the major encoder into the ‘U-shaped’ segmentation architecture. The Swin UNETR model achieves state-of-the-art performance on the 10 tasks in Medical Segmentation Decathlon (MSD) (Simpson et al., 2019) and BTCV benchmarks. Similarly, (Yu et al., 2022c) propose a hierarchical Transformer-based segmentation model that utilizes the 3D block aggregation, which achieves the state-of-the-art results on the kidney sub-components segmentation with CT images.
ViT as additional encoder:
The second widely-adopted structures for medical image segmentation are to use the Transformer as the secondary encoder after ConvNets. The rationale of this design is the lack of inductive bias such as locality and translation equivariance of Transformers. In addition, the use of CNN as the main encoder can bring the computational benefit as it is computationally expensive to calculate global self-attention among voxels in high-resolution medical images. One earlier adoption of 12 layers ViT for the bottleneck features is the TransUNet (Chen et al., 2021d), which follows the 2D UNet (Ronneberger et al., 2015) design and incorporates the Transformer blocks in the middle structure. TransUNet++ (Wang et al., 2022a) and Ds-TransUNet (Lin et al., 2021) propose an improved version of the design that achieves promising results for CT segmentation tasks. For volumetric medical segmentation, TransBTS (Wang et al., 2021e) and TransBTSV2 (Li et al., 2022c) introduce the Transformer to model spatial patch embedding for the bottleneck feature. CoTr (Xie et al., 2021b), TransBridge (Deng et al., 2021), TransClaw (Chang et al., 2021), and TransAttUNet (Chen et al., 2021a) study the variant of attention blocks in the Transformer, such as the deformable mechanism that enables attention on a small set of key positions. SegTrans (Li et al., 2021a) exploits the squeeze and expansion block for modeling contextual features with Transformers for hidden representations. MT-UNet (Wang et al., 2021c) uses a mixed structure for learning inter- and intra- affinities among features. More recently, several studies such as AFTer-UNet (Yan et al., 2022), BAT (Wang et al., 2021d), GT-UNet (Li et al., 2021c), and Polyp-PVT (Dong et al., 2021a) focus on using grouping, boundary-aware or slice communication modules for improved robustness in ViT.
Fusion models with ViT and ConvNet:
While Transformers show the superiority of modeling long-range dependencies, its lack of capability of capturing local feature remains a challenge. Instead of cascading the Conv and Transformer blocks, researchers propose to leverage ViT and ConvNet as encoders that both take medical image as inputs. Afterwards, the embedded features are fused to connect to the decoder. The multi-branch design benefits from the advantages of learning global/local information for ViT and Convnet in parallel and then stacking representations in a sequential manner. TransFuse (Zhang et al., 2021b) uses a bi-fusion paradigm, in which the features from the two branches are fused to jointly make inference. CrossTeaching (Luo et al., 2021) employs a semi-supervised learning with UNet and Swin Transformer for medical segmentation. TransFusionNet (Meng et al., 2021) uses the CNN as the decoder to bridge the fused featured learnt from Transformer and ConvNet. PMTrans (Zhang et al., 2021d) introduces a pyramid structure for a multi-branch encoder with Transformers. X-Net (Li et al., 2021d) demonstrates a dual encoding-decoding X-shape network structure for pathology images. MedT (Valanarasu et al., 2021) designs model encoders with a CNN global branch and a local branch with gated axial self-attention. DS-TransUNet (Lin et al., 2021) proposes to split the input image into non-overlapping patches and then use two branches of encoder that learn feature representations at different scales; the final output is fused by Transformer Interactive Fusion (TIF) module.
Pure Transformer:
In addition to hybrid models, networks with pure Transformer blocks have been shown to be effective at modeling dense predictions such as segmentation. The nnFormer (Zhou et al., 2021a) proposes to use 3D Transformer that exploits the combination of interleaved convolutions and self-attention operations. The nnFormer also replaces the skip connection with a skip attention mechanism and it outperforms nnUNet significantly. MISSFormer (Huang et al., 2021) is a pure Transformer network with a feed-forward enhanced Transformer block with a context bridge. It models local features at different scales for leveraging long-range dependencies. D-Former (Wu et al., 2022b) envisions an architecture with a D-Former block, which contains the dynamic position encoding block (DPE), local scope modules (LSMs), and the global scope modules (GSMs). The design employs a dilated mechanism that directly processes 3D medical images and improves the communication of information without increasing the tokens in self-attention. Swin-UNet (Cao et al., 2021) utilizes the advantages of shifted window self-attention Transformer blocks to construct a U-shaped segmentation network for 2D images. The pure Transformer architecture also uses the Transformer block as the expansion modules to upsample feature maps. However, current pure Transformer-based segmentation model are commonly of large model size, resulting in challenges of design robustness and scalability.
Pre-training framework for medical segmentation:
Based on the empirical studies of Vision Transformer, the self-attention blocks commonly require pre-training data at a large scale to learn a more powerful backbone (Dosovitskiy et al., 2020). Compared to CNNs, Transformer models are more data-demanding at different scales (Zhai et al., 2022), effective and efficient ViT models are typically pre-trained by appropriate scales of dataset. However, adapting from natural images to a medical domain remain a challenge as the context gap is large. In addition, generating expert annotation of medical images is nontrivial, expensive and time-consuming; therefore it is difficult to collect large-scale annotated data in medical image analysis. Compared to the fully supervised dataset, raw medical images without expert annotation are easier to obtain. Hence, transfer learning, which aims to reuse the features of already trained ViT on different but related tasks, can be employed. To further improve the robustness and efficiency of ViT in medical image segmentation, several works are proposed to learn in a self-supervised manner a model of feature representations without manual labels. Self-supervised Swin UNETR (Tang et al., 2022) collects a large-scale of CT images (5,000 subjects) for pre-training the Swin Transformer encoder, which derives significant improvement and state-of-the-art performance for BTCV (Landman et al., 2015) and Medical Segmentation Decathlon (MSD) (Antonelli et al., 2021). The pre-training framework employs multi-task self-supervised learning approaches including image inpainting, contrastive learning and rotation prediction. Self-supervised masked autoencoder (MAE) (Zhou et al., 2022c) investigates the MAE-based self pre-training paradigm designed for Transformers, which enforces the network to predict masked targets by collecting information from the context. Furthermore, the unified 2D/3D pre-training (Xie et al., 2021c) aims to construct a teacher-student framework to leverage unlabeled medical data. The approach designs a pyramid Transformer U-Net as the backbone, which takes either 2D or 3D patches as inputs depending on the embedding dimension.
Segmentation Transformers for different imaging modalities:
Medical image modalities are of potential challenges with deep learning tools. The medical segmentation decathlon (Antonelli et al., 2021), a challenge dataset designed for general purpose segmentation tools, contains multiple radiological modalities including dynamic CTs, T1w, T2w, and FLAIR MRIs. In addition, pathology images, endoscopy intervention data, or videos are also challenging medical segmentation scenarios. Upon image modalities with Transformer model, for only CT studies, CoTr (Xie et al., 2021b), U-Transformer (Petit et al., 2021), TransClaw (Chang et al., 2021), COTRNet (Shen et al., 2021a), AFTerNet (Yan et al., 2022), TransFusionNet (Meng et al., 2021), T-AutoML (Yang et al., 2021), etc. conduct experiments on extensive evaluation. Among a large number of methods, researchers attempt to explore general segmentation approaches that can at least handle volumetric data both in CT and MRI, for which UNETR (Hatamizadeh et al., 2022b), VT-UNet (Peiris et al., 2021), SwinUNETR (Tang et al., 2022), UNesT (Yu et al., 2022c), MT-UNet (Wang et al., 2021c), TransUNet (Chen et al., 2021d),, TransClaw (Chang et al., 2021), LeViT-UNet (Xu et al., 2021a), nnFormer (Zhou et al., 2021a), MISSformer (Huang et al., 2021), D-Former (Wu et al., 2022b), Swin-UNet (Cao et al., 2021), and some pre-training workflows are proposed. Regarding pathology images, SpecTr (Yun et al., 2021), MBT-Net (Zhang et al., 2021a), MCTrans (Ji et al., 2021), MedT (Valanarasu et al., 2021), and X-Net (Li et al., 2021d) are some pioneering works. Finally, SegTrans (Li et al., 2021e), MCTrans (Ji et al., 2021), Polyp-PVT (Dong et al., 2021a), DS-TransUNet (Lin et al., 2021), and TransFuse (Zhang et al., 2021b) can model endoscopy images or video frames.
4.2. Medical image recognition and classification
Since the advent of ViT (Dosovitskiy et al., 2020), it has exhibited exceptional performances in natural image classification and recognition (Wang et al., 2021f; Liu et al., 2021b; Touvron et al., 2021b; Chu et al., 2021). The benefits of ViT over CNN to image classification tasks are likely due to the following properties:
The ability of a single self-attention operation in ViT to globally characterize the contextual information in the image provided by its large theoretical and effective receptive field (Ding et al., 2022; Raghu et al., 2021). [Property M1]
The self-attention operation tends to promote a more flat loss landscape, which results in improved performance and better generalizability (Park and Kim, 2022). [Property M4]
ViT is shown to be more resilient than CNN to distortions (e.g., noise, blur, and motion artifacts), semantic changes, and out-of-distribution samples (Cordonnier et al., 2019; Bhojanapalli et al., 2021; Xie et al., 2021a). [Property M5]
ViT has a weaker inductive bias than CNN, whose convolutional inductive bias has been shown to be advantageous for learning from smaller datasets (Dosovitskiy et al., 2020). However, with the help of pre-training using a significant large amount of data, ViT is able to surpass convolutional inductive bias by learning the relevant patterns directly from data. [Property M3]
Related to the previous property, the superior scaling behavior of ViT over CNN with the aid of a large model size and pre-training on large datasets (Liu et al., 2022e; Zhai et al., 2022). [Property C1]
It is flexible to design different network architectures by mixing Transformer and CNN modules to accommodate different modeling requirements. [Property C2]
These appealing properties have sparked an increasing interest in developing Transformer-based models for medical image classification and recognition. The original ViT (Dosovitskiy et al., 2020) achieves superior classification performance with the help of pre-training on large-scale datasets. Indeed, as a result of their weaker inductive bias, pure ViTs are more "data hungry" than CNNs (Park and Kim, 2022; Liu et al., 2021a; Bao et al., 2021). As a result of this discovery, many supervised and self-supervised pre-training schemes for Transformers have been proposed for applications like COVID-19 classification (Park et al., 2021; Xie et al., 2021c; Mondal et al., 2021), retinal disease classification (Yu et al., 2021a; Matsoukas et al., 2021), and histopathological image classification (Wang et al., 2021g). Despite the intriguing potential of these models, obtaining large-scale pre-training datasets is not always practicable for some applications. Therefore, there have been efforts devoted to developing hybrid Transformer-CNN classification models that are less data-demanding (Sriram et al., 2021; Park et al., 2021; Dai et al., 2021a; He et al., 2021a; Gao et al., 2021c). Next we briefly review and analyze these recent works for medical image classification and also list the reviewed works in Table 2.
Table 2.
The summarized review of Transformer-based model for medical image classification. "N.A." denotes for not applicable for intermediate blocks or decoder module. "N" denotes not reported or not applicable on the number of model parameters. "t" denotes temporal dimension.
| Reference | Architecture | 2D/3D | Pre-training | #Param | Classification Task | Modality | Dataset | Highlights |
|---|---|---|---|---|---|---|---|---|
| (Sriram et al., 2021) | Conv-Transformer Hybrid | 2D | Pre-trained CNN | N | COVID-19 Prognosis | X-ray | CheXpert (Irvin et al., 2019), NYU COVID (Shamout et al., 2021) | A pre-trained CNN backbone extracts features from individual image, and a Transformer is applied to the extracted features from a sequence of images for prognosis. |
| (Park et al., 2021) | Conv-Transformer Hybrid | 2D | Pre-trained CNN | N | COVID-19 Diagnosis | X-ray | CheXpert (Irvin et al., 2019) | A pre-trained CNN backbone is integrated with ViT for classification. |
| TransPath (Wang et al., 2021g) | Conv-Transformer Hybrid | 2D | Pre-trained CNN + ViT | N | Histopathological Image Classification | Microscopy | TCGA (Tomczak et al., 2015), PAIP (Kim et al., 2021b), NCT-CRC-HE (Kather et al. 2019), PatchCamelyon (Bejnordi et al., 2017), MHIST (Wei et al.,2021) | The entire network is pre-trained prior to the downstream tasks. The TAE module is introduced to the ViT in order to aggregate token embeddings and subsequently excite the MSA output. |
| i-ViT (Gao et al., 2021c) | Conv-Transformer Hybrid | 2D | No | N | Histological Subtyping | Microscopy | AIPath (Gao et al., 2021d) | A lightweight CNN is used to extract features from a series of image patches, which is then followed by a ViT to capture high-level relationships between patches for classification. |
| (He et al., 2021a) | Conv-Transformer Hybrid | 2D | No | N | Brain Age Estimation | MRI | Brain MRI (BGSP (Holmes et al., 2015), OASIS-3 (LaMontagne et al., 2019), NIH-PD (Evans et al., 2006), ABIDE-I (Di Martino et al., 2014), IXI*, DLBS (Park et al., 2012), CMI (Alexander et al., 2017), CoRR (Zuo et al., 2014)) | Two CNN backbones, one of which extract features from the whole image and the other from the image patches. Then, a Transformer is used to aggregate the features from the two backbones for classification. |
| SETMIL (Zhao et al., 2022) | Conv-Transformer Hybrid | 2D | Pre-trained CNN | N | Gene Mutation Prediction, Lymph Node Metastasis Diagnosis | Microscopy | Whole Slide Pathological Image | A novel spatial encoding wiht Transformer is proposed for multiple instance learning. |
| KAT (Zheng et al., 2022b) | Conv-Transformer Hybrid | 2D | Pre-trained CNN | N | Tumor Grading & Prognosis | Microscopy | Whole Slide Pathological Image | A cross-attention Transformer is proposed to enable information exchange across tokens based on their spatial relationship on the whole slide image. |
| RAMST (Lv et al., 2022) | Conv-Transformer Hybrid | 2D | Pre-trained CNN | N | Microsatellite Instability Classification | Microscopy | Whole Slide Pathological Image | A combination region- and whole-slide-level Transformer is proposed. The Transformer accepts sampled patches per the attention map and combines two levels of information for the final classification. |
| LA-MIL (Reisenbüchler et al., 2022) | Conv-Transformer Hybrid | 2D | Pre-trained CNN | N | Microsatellite Instability Classification, Mutation Prediction | Microscopy | Whole Slide Pathological Image (TCGA colorectal & stomach (Weinstein et al., 2013)) | A local attention graph-based Transformer is proposed for multiple instance learning, as well as an adaptive loss function to mitigate the class imbalance problem. |
| BabyNet (Plotka et al., 2022) | Conv-Transformer Hybrid | 2D+t | No | N | Birth Weight Prediction | Ultrasound | Fetal Ultrasound Video Scans | BabyNet advances a 3D ResNet with a Transformer module to improve the local and global feature aggregation. |
| Multi-transSP (Zheng et al., 2022a) | Conv-Transformer Hybrid | 2D | Pre-trained CNN | N | Survival Prediction for Nasopharyngeal Carcinoma Patients | CT | In-house CT Scans | A hybrid CNN-Transformer model that combines CT image and tabular data (i.e., clinical text data) is developed for survival prediction of nasopharyngeal carcinoma patients. |
| BrainFormer (Dai et al., 2022) | Conv-Transformer Hybrid | 3D | No | N | Autism, AlzheimerâĂŹs Disease, Depression, Attention Deficit Hyperactivity Disorder, and Headache Disorders Classification | fMRI | ABIDE (Di Martino et al., 2014), ADNI (Petersen et al., 2010), MPILMBB (Mendes et al., 2019),ADHD-200 (Bellec et al., 2017) and ECHO | A 3D CNN and Transformer Hybrid network employs CNNs to model local cues and Transformer to capture global relation among distant brain regions. |
| xViTCOS (Mondal et al., 2021) | Conventional Transformer | 2D | Pre-trained ViT | N | COVID-19 Diagnosis | CT, X-ray | Chest CT ( COVIDx CT-2A (Gunraj et al., 2021)), Chest X-ray (COVIDx-CXR-2 (Pavlova et al., 2021), CheXpert (Irvin et al., 2019)) | A multi-stage transfer learning strategy is proposed for fine-tuning pre-trained ViT on medical diagnostic tasks. |
| MIL-VT (Yu et al., 2021a) | Conventional Transformer | 2D | Pre-trained ViT | N | Fundus Image Classification | Fundus | APTOS2019†, RFMiD2020 (Pachade et al., 2021) | Multiple instance learning module is introduced to the pre-trained ViT that learns from both the classification tokens and the image patches. |
| (Matsoukas et al., 2021) | Conventional Transformer | 2D | Pre-trained ViT | N | Dermoscopic, Fundus, and Mammography Image Classification | Fundus, Dermoscopy, Mammography PET/CT | ISIC2019‡, APTOS2019†, CBIS-DDSM (Lee et al., 2017 | This study investigates the effectiveness of pretraining DeiT versus ResNet on medical diagnostic tasks. |
| TMSS (Saeed et al., 2022) | Conventional Transformer | 3D | No | N | Survival Prediction for Head and Neck Cancer Patients | HECKTOR (Oreiller et al., 2022) | A Transformer for end-to-end survial prediction and segmentation using PET/CT and electronic health records (i.e., clinical text data). | |
| Uni4Eye (Cai et al., 2022) | Conventional Transformer | 2D/3D | Pre-trained ViT | N | Ophthalmic Disease Classification | OCT, Fundus | OCTA-500 (Li et al., 2020), GAMMA (Wu et al., 2022a), GAMMA (Wu et al., 2022a), EyePACS§, Ichallenge-Ichallenge-PMAMD (Milea et al., 2020), Ichallenge-PM (Fu et al., 2018), PRIME-FP20 (Ding et al., 2021) | A self-supervised learning framework is developed to pre-train a Transformer using both 2D and 3D ophthalmic images for ophthalmic disease classification. |
| STAGIN (Kim et al., 2021a) | Conventional Transformer | 3D+t | No | 1.2M | Gender, Cognitive Task Classification | fMRI | HCPS1200 (Van Essen et al., 2013) | A conventional Transformer encoder is employed to capture the temporal attention over features of functional connectivity from fMRI. |
| BolT (Bedel et al., 2022) | Conventional Transformer | 3D+t | No | N | Gender Prediction, Cognitive Task and Autism Spectrum Disorder Classification | fMRI | HCP S1200 (Van Essen et al., 2013), ABIDE (Di Martino et al., 2014) | A cascaded Transformer encodes features of BOLD responses via progressively increased temporally-overlapped window attention. |
| SiT (Dahan et al., 2022) | Conventional Transformer | 3D | Pretrained ViT | 21.6M | Cortical Surface Patching, Postmenstrual Age (PMA) and Gestational Age (GA) | MRI | dHCP (Hughes et al., 2017) | Reformulating surface learning task as seq2seq problem and solving it by ViTs. |
| Twin-Transformers (Yu et al., 2022d) | Conventional Transformer | 3D+t | No | N | Brain Networks Identification | N | HCP S1200 (Van Essen et al., 2013) | A Twin-Transformers is proposed to simultaneously capture temporal and spatial features from fMRI. |
| (Cheng et al., 2022) | Conv-like Transformer | 3D | No | 6.23M | Cortical Surfaces Quality Assessment | MRI | Infant Brain MRI Dataset | The first work extended Transformer into spherical space. |
| USST (Xie et al., 2021c) | Conv-like Transformer | 2D/3D | Pre-trained ViT | N | COVID-19 Diagnosis, Pneumonia Classification | X-ray, CT | RICORD (Tsai et al., 2021), ChestXR (Akhloufi and Chetoui, 2021) | The unified pre-training framework that allows the pre-training using 3D and 2D images is introduced to Transformers. |
Hybrid model:
The earliest use of ViTs for medical image classification is on COVID-19 classification from chest X-rays (Sriram et al., 2021; Park et al., 2021). Public datasets like CheXpert (Irvin et al., 2019), ChestXR (Akhloufi and Chetoui, 2021), and COVIDx CXR (Wang et al., 2020b) provide over 10,000 chest x-ray images. Due to the massive quantity of images in these datasets, they are suitable for network pre-training as well as for evaluating downstream classification tasks. (Sriram et al., 2021) introduce a hybrid CNN-Transformer model for COVID-19 prognosis by analyzing a series of chest X-ray images taken at various time points. Specifically, a MOCO (He et al., 2020a; Chen et al., 2020) encoder (a CNN) pre-trained in a self-supervised manner is used to extract features from each X-ray image. The features extracted from multiple images of the same patient are then fed into a Transformer followed by a linear classifier for classification. In their model, only the CNN backbones (i.e., the MOCO encoders) are pre-trained and the Transformer is randomly initialized, whereas the overall network is fine-tuned for the classification task. Similarly, (Park et al., 2021) propose to bridge DenseNet-121 (Huang et al., 2017) with ViT. The DenseNet is pre-trained on the CheXpert dataset using the Probabilistic Class Activation Map (PCAM) pooling operations introduced in (Ye et al., 2020), whilst the ViT is randomly initialized. The overall network is subsequently trained and evaluated on several chest X-ray datasets for COVID-19 diagnosis, where their model outperforms ResNet (He et al., 2016) and vanilla ViT (Dosovitskiy et al., 2020) that are trained using the same training strategy. (Zhao et al., 2022) propose SETMIL for pathological image analysis. SETMIL begins by embedding the large-sized whole slide image (WSI) in low-resolution position-encoded embeddings via a pre-trained CNN. Then, low-resolution embeddings are subjected to a Transformer-based pyramid multi-scale fusion based on tokens-to-token ViT (Yuan et al., 2021b) to extract multi-scale context information. A novel spatial encoding Transformer that combines absolute and relative positional embedding is used for the final classification. To achieve a similar objective, (Zheng et al., 2022b) propose KAT, which focuses on establishing the correspondence between tokens and a set of kernels associated with a set of positional anchors on the WSI. A CNN that has been pre-trained is first used to extract features from the non-overlapping patches of the WSI. In the mean-while, a set of anchor points is extracted using K-means clustering on the feature patches. Then, a set of multi-scale weighting masks for each anchor point is defined and sent together with the feature patches and a set of trainable kernels to a Transformer. The Transformer uses cross-attention between tokens and kernels, and classification is achieved through kernel interaction with the classification token. This reduces the quadratic computational cost of the Transformer and reaches close to linear complexity in relation to the size of the WSI. In (Lv et al., 2022), Lv et al. introduce RAMST for the classification of microsatellite instability. In particular, a feature weight uniform sampling method is presented to learn representative features of image regions, and a Transformer encoder is used to aggregate region-level features with patch-level features extracted by a pre-trained CNN. Meanwhile, Reisenbuchler et al. propose a local attention graph-based Transformer (LA-MIL) for microsatellite instability classification and genetic mutation prediction in whole slide pathological images (Reisenbüchler et al., 2022). The method starts by tessellating a gigapixel WSI into patches of identical size, removing patches containing background, artifacts, and non-tumor tissue using global thresholding and manual annotations. Then, a CNN that has been pre-trained on histopathological data compresses each patch into a feature vector, and a kNN graph matrix is constructed to describe the spatial relations between patches. A local attention Transformer computes the attention between each patch and its neighbors from the graph matrix. Not only does LA-MIL provide promising performance, but it also permits the visualization of local attention for interpreting the contribution of each patch to the classification prediction. In (Zheng et al., 2022a), Zheng et al. propose Multi-transSP for the survival prediction of nasopharyngeal carcinoma patients from CT and tabular data. Multi-transSP exploits the capabilities of CNNs to extract representative features and the capability of Transformers to fuse features. ResNet18 (He et al., 2016) first extracts features from the 2D CT slices, which are concatenated with the feature representation of the tabular data generated by a linear layer. The output features are fused by a Transformer, which is then followed by a fully-connected layer to generate a survival prediction.
Rather than pre-training the CNN backbone of the hybrid model, (Wang et al., 2021g) pre-train the entire CNN-Transformer (designated as TransPath) using a self-supervised learning method, BYOL (Grill et al., 2020). In addition, the authors develop a token-aggregation and excitation (TAE) module for use with the MSA output in the ViT (Dosovitskiy et al., 2020). Specifically, the TAE module first averages all token embeddings, then applies two sets of linear projection and activation functions to excite the averaged embeddings, which are then re-projected to the MSA output. According to (Wang et al., 2021g), combining MSA and TAE enables the Transformer to consider sufficient global information since each element in the output is the aggregated outcome of all input tokens. They conduct extensive experiments against several other Transformer-based networks on several benchmark histopathology image classification datasets and demonstrate superior performance.
Several studies suggest that even without pre-training, Transformer may be an effective complement to CNNs for feature extraction in a hybrid model. (Gao et al., 2021c) propose the instance-based ViT (i-ViT) for subtyping renal cell carcinoma in histopathological image. Their framework begins by extracting nuclei-containing image patches (regarded as instance-level patches) and the corresponding nuclei grades and sizes from an input histopathology image. The patches are sorted by nucleus grade and size, and a predefined number of patches is concatenated and then used as the input to a light CNN. The output embeddings, along with additional embeddings containing information on the nuclei grades and positions relative to the entire image, are sent into a ViT (Dosovitskiy et al., 2020). The ViT captures cellular level and cell-layer level features for subtyping. The authors train and assess the i-ViT using a dataset of 1,163 ROIs/pictures taken from 171 whole slide images, and the i-ViT achieves improved performance than the CNN-based baselines. In (He et al., 2021a), He et al. propose a hybrid model for brain age estimation that does not require pre-training. Their model consists of two paths: a global path that extracts global contextual information from the whole brain MRI 2D slice, and a local path that extracts local features from image patches segmented from the 2D slice. Each path has a CNN backbone for generating high-level features from the input image/patches. Following that, a "global-local Transformer" (He et al., 2021a) is used to aggregate the features from the two paths for brain age estimation. With less than 8,000 training samples, their model trained-from-scratch performs noticeably better in comparison to a range of CNN and Transformer baselines. Although the studies discussed in this paragraph are trained on datasets with limited samples, they still outperform the CNN-based baselines, revealing the promising potential of hybrid models for data-limited applications. Płotka et al. propose BabyNet (Płotka et al., 2022) that advances a 3D ResNet-based network with an MHSA module for fetal birth weight prediction. BabyNet is similar to BoT (Srinivas et al., 2021) in that it replaces the bottleneck convolution block with an MHSA to aggregate local and global feature representations more effectively. Unlike BoT, the MHSA module of BabyNet uses temporal positional embedding for temporal analysis between frames and relative positional embedding for encoding spatial correspondence within frames. BabyNet outperforms several comparative learning-based models with accuracy comparable to human experts.
Pure ViT:
The aforementioned models bridge CNN backbones with Transformers. Nevertheless, pure Transformers have also been shown to be effective for medical image classification when pre-trained. (Mondal et al., 2021) develop a multi-stage transfer learning strategy for adapting the original ViT (Dosovitskiy et al., 2020) to COVID-19 classification tasks. Specifically, they adopt the ViT that is trained on ImageNet (Deng et al., 2009; Russakovsky et al., 2015) and fine-tune it using images from the target domain. Their method is tested on two publicly available datasets, namely the COVIDx-CT-2A (Gunraj, 2021) and CheXpert (Irvin et al., 2019), and outperforms a variety of baseline methods in terms of classification accuracy. Likewise, (Yu et al., 2021a) propose MIL-VT that fine-tunes the ViT pre-trained on ImageNet for retinal disease classification. The pre-trained ViT is first fine-tuned on an in-house large-scale fundus image dataset (> 300, 000 fundus images), and subsequently on two publicly available datasets (APTOS (APTOS, 2019) and RFMiD2020 (RIADD, 2020)) for downstream classification tasks. In the original ViT, only the features corresponding to the "classification token" (Dosovitskiy et al., 2020) are sent to an MLP for final classification, with the features extracted from the image patches being neglected. Yu et al. hypothesize that the features from image patches might contain important complementary information. Thus, they introduce an additional Multiple Instance Learning module (referred to as a "MIL head" (Yu et al., 2021a)) that aggregates the features extracted from the patches and then performs prediction using the aggregated features. MIL-ViT backpropagates the loss into ViT during training through two paths: one via the MLP classifier in ViT and another via the added "MIL head". During inference, the final prediction is made by averaging the output logits from the two paths. (Matsoukas et al., 2021) compare ResNet (He et al., 2016) and DeiT (Touvron et al., 2021b) side-by-side with three scenarios: training-from-scratch (i.e., without pre-training), supervised pre-training on ImageNet (Deng et al., 2009), and self-supervised pre-training on medical images in addition to the supervised pre-training. On three benchmark datasets, they empirically find that ResNet outperforms DeiT when trained from scratch, and this performance gap could be closed with the supervised pre-training. Moreover, they show that DeiT performs slightly better than ResNet with the additional self-supervised pre-training on medical images, further demonstrating the potential of self-supervised pre-training of pure Transformers for medical image classification. In (Saeed et al., 2022), the authors propose TMSS for the joint prediction of a patient’s survival risk score and tumor segmentation using PET/CT and electronic health records (EHR). The input PET/CT is evenly divided into patches, linearly embedded, and then concatenated with the linear embedding of the patient’s EHR. The output is then fed into a ViT (Dosovitskiy et al., 2020) but without the class token. After that, The output of the ViT is sent to a multi-task logistic regression model that predicts survival risk scores and a CNN decoder that generates the segmentation mask. The model achieves superior performance on the HECKTOR dataset (Oreiller et al., 2022) when compared to competing models.
3D modeling:
To date, the majority of Transformers for medical image classification has concentrated on 2D applications for various reasons, including reduced computational complexity and the ability to directly use models pre-trained on large-scale natural images (e.g., ImageNet). However, since most medical imaging modalities produce 3D images, developing efficient Transformers for 3D classification is anticipated to receive an increased attention in the near future. (Xie et al., 2021c) develop a Universal Self-supervised Transformer (USST) that can be pre-trained using both 2D and 3D images jointly. Specifically, the authors propose the switchable patch embedding (SPE) for use in the Pyramid Vision Transformer (PVT) (Wang et al., 2021f), which adapts to the dimensionality of the input image by switching between 2D and 3D patch embedding. The USST pre-training framework is developed based on the student-teacher paradigm, in which both the student and teacher paths share the identical architecture, but the teacher path is updated using an exponential moving average of the weights of the student path. The authors use > 5, 000 3D CT images and > 100, 000 2D chest X-rays to pre-train the USST framework. The pre-trained Transformers is then fine-tuned on multiple 2D and 3D classification tasks, with the USST framework considerably outperforming other widely used pre-training frameworks on downstream tasks. To achieve a similar objective on dimension-independent pre-training, (Cai et al., 2022) propose a self-supervised learning method to pre-train ViT (Dosovitskiy et al., 2020) on both 2D and 3D ophthalmic images for downstream ophthalmic disease classification tasks. A unified patch embedding module is developed to extract a fixed number of 2D/3D patches from the input based on random masking. The extracted patches are then passed to a ViT (Dosovitskiy et al., 2020) and two decoders for self-supervised learning to reconstruct the original and the gradient images by carrying out the masked image modeling task (He et al., 2022; Xie et al., 2022). This Transformer-based model is pre-trained, fine-tuned, and then evaluated on >95, 000 ophthalmic images with six different classification tasks, demonstrating state-of-the-art performance on all of the evaluated tasks.
Non-Euclidean imaging:
Functional magnetic resonance imaging (fMRI) is widely used to capturing the temporal signal of neural activity. Estimation of brain activity can be measured by functional connectivity (FC), the degree of temporal correlation between regions of the brain. Transformer also shows superiority and potential in analysis of brain connectome. (Kim et al., 2021a) propose a GNN and Transformer hybrid model in gender classification on resting-state fMRI and task decoding for task fMRI, with a dynamic GNN enhanced by an elaborate spatial attention learning the representation of the brain connectome from a single time-step fMRI, and a single-headed Transformer encoder integrating attended features temporally. Transformer together with dynamic GNN is capable to capturing characteristics of functional connectivity which fluctuates over time. BolT (Bedel et al., 2022) exploits a cascade of Transformer blocks to encode local representations of FC, which is performed on temporally-overlapping windows. BolT comprises a cross-window attention module, with the extent of window overlap progressively, to enhance sensitivity to the diverse time scales of FC features. The integration ability from cascaded Transformer promises BolT to achieve the state-of-the-art in HCP gender prediction and cognitive task classification (Van Essen et al., 2013), and autism spectrum disorder detection task (Di Martino et al., 2014). (Dai et al., 2022) take the point that FC feature suffers from the insufficient representation ability and coarse granularity. They proposed BrainFormer, a convolution-transformer hybrid architecture that employs a 3D CNN backbone modeling the detailed and informative features from fMRI volume. BrainFormer inserts CNN-based attention blocks into backbone in shallow layers, capturing the spatial correlation. And it exploits transformer-based attention blocks in deep layers to fuse the global information. The effectiveness and generalizability of this method is evaluated on ABIDE (Di Martino et al., 2014), ADNI (Petersen et al., 2010), MPILMBB (Mendes et al., 2019), ADHD-200 (Bellec et al., 2017), and ECHO, with diseases of autism, ALzheimer’s disease, depression, attention deficit hyperactivity disorder, and headache disorders. (Yu et al., 2022d) propose a Twin-Transformers to simultaneously capture temporal and spatial features from fMRI. With brain signal matrix as input, the spatial Transformer focuses on non-overlapping spatial patches and the temporal Transformer takes non-overlapping temporal patches as tokens. In other scenarios in neural imaging, (Dahan et al., 2022) extend ViTs to non-Euclidean manifolds cortical surface and propose the Surface Vision Transformer (SiT) for sequence-to-sequence modelling surfaces with projection to a regularly tessellated icosphere. SiT proves a certain level of transformation invariance without introducing strong inductive bias into framework. (Cheng et al., 2022) propose a spherical Transformer in quality assessment of cortical surface, represented by triangular meshes and mapped onto a spherical manifold. The spherical Transformer shows its potential in extracting the structural and contextual pattern among vertices.
In summary, Transformer-based medical image classification still relies heavily on pre-training using large-scale datasets, either supervised or self-supervised. On the other hand, for applications with limited data availability, initializing Transformers with weights pre-trained on natural images is found to be beneficial for improving performances. However, without pre-training and access to large-scale training data, Transformers may not be more effective than CNNs for medical image classification. Moreover, the majority of the existing Transformer-based models focuses on 2D applications. With a growing research interest in Transformers, we anticipate that further efforts will be directed toward developing Transformer-based models for 3D classification applications.
4.3. Medical image detection
The use of Transformers for object detection in natural images is pioneered by Carion et al. in DETR (Carion et al., 2020). DETR makes use of both the encoder and decoder from the original Language Transformer used in NLP (Vaswani et al., 2017), whereas ViT (Dosovitskiy et al., 2020) borrows only the encoder. In computer vision, efforts have been made to augment both Transformer encoder-decoder (i.e., DETR) (Zhu et al., 2020; Zheng et al., 2020; Sun et al., 2021b) and Transformer encoder-only (i.e., ViT) (Beal et al., 2020; Li et al., 2022e) designs for object detection, all of which have shown demonstrable performances. On the one hand, DETR’s Transformer decoder learns to make direct set predictions such that duplicate bounding box predictions are suppressed, eliminating the post-processing procedures for the predictions (e.g., non-maximal suppression). In the field of medical imaging, a few Transformer-based object detection methods have been developed based on DETR (Shen et al., 2021a; Mathai et al., 2022). However, it has been discovered that DETR takes much longer training epochs for convergence than ConvNet-based models (Zhu et al., 2020; Fang et al., 2021; Beal et al., 2020). On the other hand, using only the Transformer encoder may benefit from the transferability of the encoders pre-trained on large-scale datasets (e.g., ImageNet (Deng et al., 2009; Russakovsky et al., 2015), thereby accelerating convergence. Furthermore, combining these encoders with ConvNets introduces additional inductive bias, reducing the amount of data needed to construct an effective model. Several attempts have been made in medical imaging that uses Transformer encoders as a component of the feature extractor in conjunction with ConvNets for bounding box prediction (Jiang et al., 2021; Li et al., 2022b) and for applications where the bounding boxes are not needed (Ma et al., 2021a; Zhu et al., 2022). While the advantages of Transformers for image classification remain relevant to object detection (i.e., properties M1, M4, M3, M5, C1, and C2), the main advantage is that:
The self-attention mechanism computes globally or with a very large kernel, making Transformer more ideal for comprehending contextual information contained in an image, which is crucial for object detection. [Property M1]
Transformer as encoder and decoder:
(Shen et al., 2021a) propose a convolution-in-Transformer (COTR) network for polyp detection in colonoscopy. COTR is built on top of DETR with an aim to address the slow convergence issue with DETR. Because the Transformer encoder in DETR operates on flattened image features (i.e., vectors), it may lead the image feature structures to become disorganized. The authors thus embed convolution layers between the Transformer encoder and decoder to reconstruct the flattened vectors into high-level image features. This preserves the feature structures within the network and increases convergence speed. Additionally, DETR is shown to effectively detect lymph nodes in T2 MRI. (Mathai et al., 2022) demonstrate using a publicly available dataset that DETR, with a little tweaking to the loss functions, could surpass multiple state-of-the-art lymph node detection methods by a large margin.
Hybrid CNN and Transformer-encoder:
(Jiang et al., 2021) augment YOLO (Redmon et al., 2016) with a Transformer encoder for dental caries detection. Specifically, a sequence of convolution and pooling operations was followed by a Transformer to extract deep features at a lower resolution. In an identical manner to YOLO, the features at all resolutions are sent into the neck module and subsequently the detection head for bounding box prediction. Their model exhibits improved accuracy and average precision compared with the ConvNet-based baselines. In (Ma et al., 2021a), Ma et al. propose TR-Net, a Transformer-based network for detecting coronary artery stenosis in Coronary CT angiography. The authors begin by reconstructing multiplanar reformatted (MPR) images from coronary artery centerlines. The MPR images are then divided into equal-sized cubic volumes, with each volume centered on the coronary artery’s centerline. After extracting semantic features from each volume using a shallow ConvNet, the features from all volumes are combined with learnable positional embeddings to preserve the volumes’ ordering information. Then, the features are sent to a Transformer encoder to analyze relationships within the volume sequence. The output of the TR-Net is not a bounding box but a probability of each cubic volume having significant stenosis. Similarly, (Zhu et al., 2022) use a Transformer as the encoder for multi-anatomy landmark detection Liu et al. (2010). The authors propose a domain-adaptive Transformer (DATR), an anatomy-aware Transformer that is invariant of the Transformer architecture and capable of operating on a variety of anatomical features. DATR is built on the basis of a pre-trained Swin Transformer (Liu et al., 2021b), which extracts four scales of features and passes them to a ConvNet decoder. The network produces a heatmap with the highest-intensity locations corresponding to the landmarks. Last but not least, (Li et al., 2022b) propose a slice attention Transformer (SATr) that can be plugged into existing three-slice-input ConvNet backbones to improve the accuracy of universal lesion detection (ULD) in CT. The SATr blocks are introduced between the ConvNet backbone and the feature collector to better model long-distance feature dependencies. Each SATr block calculates self-attention between and within the features of the slices. The authors demonstrate that by simply integrating the SATr into existing three-slice-input ULD models, detection accuracy could be greatly improved and reach the state-of-the-art. (Tian et al., 2022) propose a weakly-supervised framework to identify polyps from colonoscopy video frames. The authors begin by extracting features from each video frame using a pre-trained I3D network (Carreira and Zisserman, 2017) to produce a feature token. The tokens are then sent to a Transformer for the detection of polyp frames. The authors augment the original ViT (Dosovitskiy et al., 2020) by replacing its linear embedding layers with depth-wise convolutional operations to capture local temporal relationships more effectively. In addition, a novel contrastive snippet mining strategy is proposed to extract hard and easy, normal and abnormal video frames during training for enhanced robustness in detecting subtle polyp tissues. (Windsor et al., 2022) propose a context-aware Transformer for spinal cancer detection in multi-sequence spinal MRI. A pre-trained ResNet (He et al., 2016) is fed using 2D slices from multiple MRI sequences (e.g., T1, T2, STIR, FLAIR, etc.) of multiple spinal columns to extract representative features. The feature vectors for each slice are then aggregated using a lightweight two-layer Transformer, along with additional embedding vectors specifying the level of each input vertebra and the MRI sequence employed. An attention operation is used at the end of the network to merge the features of the same vertebra. Then, the output is converted by a linear layer to produce the prediction for the corresponding vertebra. The authors demonstrate that their method leads to improved accuracy compared with a well-established method, SpineNet (Jamaludin et al., 2017).
In summary, the existing works (also as listed in Table 3, the top part) have demonstrated the potential for Transformer-based networks to be used for medical image detection. For applications that require generating bounding boxes, the Transformer encoder and decoder designs (e.g., DETR (Carion et al., 2020)) may be adopted to alleviate the need for expensive post-processing processes (e.g., non-maximal suppression). Transformers have shown promise for detection applications, but since medical datasets are often modest in size, it may be necessary to tweak the network architecture or training strategy to accelerate convergence and reduce the amount of training data needed to develop an effective model. In other applications, pre-trained Transformer encoders on natural images may be viable for enhancing a neural network’s ability to model long-distance feature dependencies without sacrificing the speed of convergence. In comparison with training from scratch, recent breakthroughs in self-supervised pre-training of Transformers for object recognition have shown significant performance improvements (Dai et al., 2021b; Dong et al., 2021b). In addition, studies have revealed that self-supervised pre-training strategies are useful for medical image segmentation and classification (Matsoukas et al., 2021; Xie et al., 2021c; Tang et al., 2022; Karimi et al., 2021), thus we expect to witness more contributions on self-supervised learning for medical image detection.
Table 3.
The summarized review of Transformer-based model for medical image detection (upper panel) and registration (lower panel). "N.A." denotes for not applicable for intermediate blocks or decoder module. "N" denotes not reported or not applicable on the number of model parameters. "t" denotes temporal dimension.
| Reference | Architecture | 2D/3D | Pre-training | #Param | Detection Task | Modality | Dataset | ViT as Enc/Inter/Dec | Highlights | |
|---|---|---|---|---|---|---|---|---|---|---|
| Detection | COTR (Shen et al., 2021a) | Conv-Transformer Hybrid | 2D | No | N | Polyp Detection | Colonoscopy | CVC-ClinicDB (Bernal et al., 2015), ETIS-LARIB (Silva et al., 2014), CVC-ColonDB (Bernal et al., 2012 | Yes/No/Yes | Convolution layers embedded between Transformer encoder and decoder to preserve feature structure. |
| (Mathai et al., 2022) | Conventional Transformer | 2D | No | N | Lymph Node Detection | MRI | Abdominal MRI | Yes/N.A./Yes | DETR applied to T2 MRI. | |
| (Jiang et al., 2021) | Conv-Transformer Hybrid | 2D | No | N | Dental Caries Detection | RGB | Dental Caries Digital Image | No/Yes/No | Augment YOLO by applying Transformer on the features extracted from the CNN encoder. | |
| TR-Net (Ma et al., 2021a) | Conv-Transformer Hybrid | 3D | No | N | Stenosis Detection | CTA | Coronary CT Angiography | No/Yes/N.A. | CNN applied to image patches, followed by a Transformer to learn patch-wise dependencies. | |
| DATR (Zhu et al., 2022) | Conv-Transformer Hybrid | 2D | Pre-trained Swin | N | Landmark Detection | X-ray | Head (Wang et al.,2016), Hand (Payeret al., 2019), and Chest (Zhu et al., 2021) | Yes/N.A./No | The integration of a learnable diagonal matrix to Swin Transformer enables the learning of domain-specific features across domains. | |
| SATr (Li et al., 2022b) | Conv-Transformer Hybrid | 2D | No | N | Lesion Detection | CT | DeepLesion (Yan et al., 2018 | No/Yes/No | Introduce slice attention Transformer to commonly used CNN backbones for capturing inter- and intra-slice dependencies. | |
| (Tian et al., 2022) | Conv-Transformer Hybrid | 2D+t | Pre-trained CNN | N | Polyp Detection | Colonoscopy | Hyper-Kvasir (Borgli et al., 2020), LD-PolypVideo (Ma et al., 2021b | No/Yes/N.A. | A weakly-supervised framework with a hybrid CNN-Transformer model is developed for polyp detection. | |
| SCT (Windsor et al., 2022) | Conv-Transformer Hybrid | 2D | Pre-trained CNN | N | Spinal Cancer Detection | MRI | Whole Spine MRI | No/Yes/N.A. | A Transformer that considers contextual information from the multiple spinal columns and all accessible MRI sequences is used to detect spinal cancer. | |
| Reference | Architecture | 2D/3D | Pre-training | #Param | Registration Task | Modality | Dataset | ViT as Enc/Inter/Dec | Highlights | |
| Registration | ViT-V-Net (Chen et al., 2021c) | Conv-Transformer Hybrid | 3D | No | 110.6M | Inter-patient | MRI | Brain MRI | Yes/No/No | ViT applied to the CNN extracted features in the encoder. |
| TransMorph (Chen et al., 2022b) | Conv-Transformer Hybrid | 3D | No | 46.8 | Inter-patient, Atlas-to-patient, Phantom-to-patient | MRI, CT, XCAT | IXI*, OASIS (Marcus et al., 2007), Abdominal and Pelvic CT, (Segars et al., 2013) | Yes/No/No | Swin Transformer is used as the encoder for extracting features from the concatenated input image pair. | |
| DTN (Zhang et al., 2021c) | Conv-Transformer Hybrid | 3D | No | N | Inter-patient | MRI | OASIS (Marcus et al., 2007 | No/Yes/No | Separate Transformers are employed to capture inter- and intra-image dependencies within the image pair. | |
| PC-SwinMorph (Liu et al., 2022a) | Conv-Transformer Hybrid | 3D | No | N | Inter-patient | MRI | CANDI (Kennedy et al., 2012), LPBA-40 (Shattuck et al., 2008) | No/No/Hybird | Patch-based image registration; Swin Transformer is used for stitching the patch-wise deformation fields. | |
| Swin-VoxelMorph (Zhu and Lu, 2022) | Conv-Transformer Hybrid | 3D | No | N | Patient-to-atlas | MRI | ADNI (Mueller et al.,2005), PPMI (Marek et al., 2011) | Yes/N.A./Yes | Swin-Transformer-based encoder and decoder network for inverse-consistent registration. | |
| XMorpher (Shi et al., 2022) | Conv-like Transformers | 3D | No | N | Inter-patient | CT | MM-WHS 2017 (Zhuang and Shen, 2016), ASOCA (Gharleghi et al., 2022) | Yes/N.A./Yes | Two Swin-like Transformers are used for fixed and moving images, with cross-attention blocks facilitating communication between Transformers. | |
| C2FViT (Mok and Chung, 2022) | Conv-like Transformers | 3D | No | N | Template-matching, patient-to-atlas | MRI | OASIS (Marcus et al., 2007), LPBA-40 (Shattuck et al., 2008 | Yes/N.A./N.A. | Multi-resolution Vision Transformer is used to tackle the affine registration problem with brain MRI. |
4.4. Medical image registration
Transformer is a viable choice for medical image registration since it has a better understanding of the spatial correspondence between and within images, and image registration is a process of establishing such correspondence between the moving and fixed images. The main advantages of applying Transformers over ConvNets to image registration are:
The self-attention mechanism in a Transformer has a large effective receptive field that encompasses the entire image (as shown in Fig. 4), enabling the Transformer to explicitly capture the long-range spatial relationships between points in the image (Raghu et al., 2021; Ding et al., 2022). [Property M1]
The majority of the learning-based deformable registration models adopt the spatial transformer network design (Jaderberg et al., 2015), which generates high-dimensional vector field mapping (i.e., one transformation for each spatial coordinate) with several million transformations per 3D volume. However, the commonly used CNN-based registration models are often of small parameters (e.g., VoxelMorph-1 (Balakrishnan et al., 2019) has about 0.3M parameters). Therefore, the Transformer’s superior scaling behavior of a large-scale model size over that of ConvNets may contribute to the establishment of a more precise spatial correspondence [Property C1].
Whereas with ConvNets, due to the limited receptive fields of convolution operations, these long-range spatial relationships can only be implicitly modeled in the deeper layers. As a result, Transformers is a more compelling contender than ConvNets for serving as the backbone for deep-learning-based image registration.
Transformers have been used predominantly for 3D registration applications such as inter-patient and atlas-to-patient brain MRI registration (Chen et al., 2021c, 2022b; Zhang et al., 2021c; Liu et al., 2022a), as well as phantom-to-CT registration (Chen et al., 2022b). As shown in Fig. 11, Transformer-based registration networks primarily employ hybrid architectures, with Transformers used in the encoding stage to capture the spatial correspondence between the input moving and fixed images, and ConvNet encoders used to generate and refine the deformation fields. In the next subsection, we briefly summarize recent works on Transformer-based medical image registration (as listed in Table 3, the bottom part) and Fig. 11 provides a schematic illustration of these approaches.
Fig. 11.
The schematic illustration of the Transformer-based image registration networks. (a) ViT-V-Net (Chen et al., 2021c). (b) TransMorph (Chen et al., 2022b). (c) PC-SwinMorph (Liu et al., 2022a). (d) DTN (Zhang et al., 2021c). These network architectures are based predominately on the hybrid ConvNet-Transformer design.
The use of Transformers for 3D medical image registration is first investigated by (Chen et al., 2021c). The authors propose a hybrid model, ViT-V-Net, in which the encoder is composed of convolutional layers, down-samplings, and a ViT (Dosovitskiy et al., 2020), while the decoder is composed of consecutive convolutional layers and up-sampling operations. Long skip connections similar to those used in V-Net (Milletari et al., 2016) are used to maximize the flow of information between the encoding and decoding stages. This model first extracts high-level features from the concatenated image pair using the convolutional layers and down-samplings. Then, ViT is applied to capture the long-range spatial correspondence between the high-level features. The decoder then uses the ViT’s output to generate a dense displacement field that warps the moving image. This model outperforms the widely used learning-based model VoxelMorph (Balakrishnan et al., 2019) for inter-patient registration on an in-house brain MRI dataset, while using identical training procedures and having a comparable computational cost. Later, (Chen et al., 2022b) extend this model and propose TransMorph by substituting a Swin Transformer (Liu et al., 2021b) for the encoder, resulting in more direct and explicit modeling of the spatial correspondences within the input image pairs. Additionally, the authors present the diffeomorphic and Bayesian variants of TransMorph, the latter of which integrates Monte-Carlo dropout layers (Gal and Ghahramani, 2016) into the Swin Transformer encoder to enable registration uncertainty estimates. TransMorph is rigorously evaluated against a variety of baseline methods, including the traditional and ConvNet-based registration methods. Additionally, TransMorph is compared against several hybrid Transformer-ConvNet and pure Transformer network designs that demonstrate superior performances in other tasks (e.g., image segmentation). TransMorph outperforms the baseline methods in terms of Dice scores on two in-house datasets and the IXI3 brain MR dataset. (Shi et al., 2022) propose XMorpher, in which a Swin-like Transformer is separately applied to moving and fixed images. Unlike Swin, XMorpher uses cross-attention to enable the exchange of information between a pair of features from moving and fixed images. Encoder and decoder of XMorpher are both Transformer-based, with the decoder being symmetric to the encoder while the patch merging layer being replaced by transposed convolution to increase the resolution of the features in the decoder. In a similar fashion, (Zhu and Lu, 2022) propose Swin-VoxelMorph, a pure Tranformer-based encoder and decoder network for inverse-consistent image registration. In contrast to XMorpher, Swin-VoxelMorph takes concatenated fixed and moving images as inputs and outputs two deformation fields for inverse and forward registration. In addition, the decoder of Swin-VoxelMorph uses patch expanding as opposed to transposed convolution to increase feature resolution. Meanwhile, (Zhang et al., 2021c) propose a dual Transformer network (DTN) for 3D medical image registration. DTN is similar to ViT-V-Net in that the Transformer is applied to the high-level features extracted by convolutional layers and down-sampling operations. However, in addition to the encoder that extracts inter-image dependencies from the concatenated moving and fixed images, DTN employs two additional encoders with shared weights to extract intra-image dependencies from each image. Each encoder of DTN is composed of a U-Net encoder and an Image Processing Transformer (IPT) (Chen et al., 2021b). The output features from the three encoders are concatenated and sent to a ConvNet decoder to produce a dense displacement field. The authors evaluate DTN for the inter-patient registration task on the OASIS brain MRI dataset (Marcus et al., 2007), for which it outperforms baseline methods in terms of Dice and deformation regularity. Taking a different route, Transformer is also used to refine deformation fields. In (Liu et al., 2022a), Liu et al. propose PC-SwinMorph, which is a patch-based image registration framework that uses contrastive learning on features extracted from the fixed and moving patches, followed by a ConvNet decoder that decodes the features and generates deformation field for the associated patch. The authors then employ two consecutive Swin Transformer blocks that learn to fuse and stitch patch-wise deformation fields together. (Mok and Chung, 2022) propose C2FViT to tackle affine registration for brain MRI. C2FViT employs a multi-resolution strategy in which affine transformation parameters are estimated by a set of ViTs from low resolution input to high resolution. Comprehensive experiments reveal that C2FViT outperforms the comparative learning-based affine registration methods while being more robust to unseen datasets.
Despite the promising potential demonstrated by the aforementioned Transformer-based registration methods, the application of Transformers to medical image registration is still in its infancy. Advanced Transformer training strategies and more complicated self-attention designs, both of which have been found to improve classification and segmentation performance (Xie et al., 2021c; Tang et al., 2022), have not yet been evaluated for registration.
4.5. Medical image reconstruction
As the fundamental precursor to downstream medical image analysis tasks, image reconstruction aims to generate high-quality structural representations or images of external or internal tissues of the human body. However, the practical MRI and CT imaging systems suffer from either a long acquisition time or an induced radiation in the imaging process, which causes an additional stress for patients. To alleviate the above problems, downsampling the acquired signals is commonly used; however it induces a very ill-posed problem and challenges the reconstruction algorithms. With the recent development of Transformer architectures and their capability of effectively characterizing global features, as well as the dense modeling of local patches that preserves more context details, Vision Transformer have attracted researchers and shown remarkable performances in medical image reconstruction.
While the under-sampling procedure alleviates the aforementioned problems, the accompanying artifacts prevent accurate clinical diagnosis; therefore, various iterative and convolutional models are proposed to suppress the artifacts. Although CNN-based post-processing and deep-unrolling methods show satisfactory performance, the global context in the structural representation is not fully captured by the spatial-split kernels, especially when context details are absent in the under-sampling scenarios. This motivates the exploration of the following key Transformer properties available for reconstruction:
The long-range dependency modeling ability of Transformer is rather valuable. As is well known, medical images, different from natural images, consist of organ anatomies and represent 2D/3D information of a human body. The global correlation is much higher than that of natural images and is thus critical to be captured. [Property M1]
As the fundamental procedure for diagnosis, reconstruction needs clearer anatomies. Towards this purpose, the dense modeling property and attention mechanism in Transformers assist in locating the most valuable features within the context of the whole image. [Property M2]
Towards combining the dense modeling of Transformer and the local context modeling of CNN, mixing them as sub-modules in a hybrid model to accommodate different modeling requirements is flexible. [Property C2]
Under-sampled MRI reconstruction:
Recently, motivated by a lack of attention paid to the intrinsic multi-scale information of MRI, ReconFormer (Guo et al., 2022d) designs a Pyramid Transformer Layer (PTL), which introduces a locally pyramidal but globally columnar structure. Then, via recurrently stacking the basic layer, the ReconFormer is capable of scaling the model and exploiting deep feature correlation through recurrent states in the model. Due to the use of recurrent structure, ReconFormer is lightweight and parameter-efficient, which alleviates the bottleneck that exists in previous Vision Transformer methods. To facilitate the exploration of the relative information between multi-contrast images in MRI reconstruction, DSFormer (Zhou et al., 2022b) proposes a novel Swin Transformer Reconstruction Network, which is based on the lightweight Swin Transformer (Liu et al., 2021b) with the backbone structure in a self-supervised reconstruction process. They use hybrid operations with both the convolutional layers and the involved Swin Transformer blocks, and condition the model with information from the reference contrast image, achieving a performance comparable to that of supervised reconstruction methods. SLATER (Korkmaz et al., 2022) pioneers the unsupervised MRI reconstruction using the long-range dependency of Transformers. It decouples the traditional imaging process into a phase of deep-image-prior learning and a subsequent phase of zero-shot inference. In the former phase, the proposed adversarial Transformer model is trained to capture a prior on coil-combined, complex MR images obtained from fully-sampled acquisitions since the previously equipped CNNs prevent capturing the long-range relationship prior (Zhang et al., 2019a; Chen et al., 2021d). In the later phase, they reconstruct the target MRI via an iterative procedure to ensure the consistency between the reconstruction and the acquisition. The method renders the potential of Transformers in purely unsupervised reconstruction setting. For efficiently reconstructing the under-sampled target-contrast MR images, DuDoCAF (Lyu et al., 2022) takes advantage of the long-range dependency modeling capability of transformers to fuse features of a reference contrast MR image. Specifically, they propose the CAF and RRT modules composed of transformer structures to first bridge the cross-modality relationship between reference and target k-space data. Then, with recurrent dual-domain learning, they gain remarkable performances and fast imaging speed. As known, the high-computational cost of self-attention in Transformer hinders its further development in medical imaging. To tackle the issue, SDAUT (Huang et al., 2022a) proposes a U-Net-based Transformer that combines dense and sparse deformable attention in separate stages. These two involved deformable attention works together to efficiently model long-range dependencies. Further, they achieve state-of-the-art performances and fast imaging speed, while still revealing model explainability.
Under-sampled CT reconstruction:
MIST-net (Pan et al., 2021) proposes the multi-domain integrative Swin Transformer network for improved sparse-view CT reconstruction. Considering the information loss in the projection domain and data inconsistency between image and projection domains, it begins by using an encoder-decoder structure to give an initial estimation. Then, a carefully designed High-definition Reconstruction Module is proposed, which is realized through the combination of Swin Transformer (Liu et al., 2021b) and convolutional layers. The post-processing Transformer structure indeed helps in reducing artifacts caused by the aforementioned problems. With an aim to further investigate the relationship between the sampling nature of projections and the global modeling capability of Transformers, DuDoTrans (Wang et al., 2021a) proposes a Sinogram Restoration Transformer (SRT) Module for projection domain enhancement. The model achieves satisfactory sparse-view reconstruction performance when combined with a similarly designed post-processing module in the image domain. Targeting to explore a more general prior with the local & nonlocal regularizations, RegFormer (Xia et al., 2022a) unrolls the gradient descent algorithm, followed by the designed iterative blocks composed of ConvNet and Transformer structures to model local and nonlocal characteristics, respectively. With such a hybrid architecture embedded into the iterative reconstruction scheme, the model reduces artifacts and preserves image details successfully. FIT (Buchholz and Jug, 2021) instead proposes to process the sinogram and the low-quality reconstruction, realized with Filtered Backprojection (Wang et al., 2019a), in the Fourier domain with the proposed Fourier Domain Encodings (FDEs). Then the two FDE representations are fed into the Fourier Image Transformer, an encoder-decoder Transformer structure, for predicting all Fourier coefficients. Following that, the inverse Fourier transformation is applied to restore the high-quality reconstruction. The carefully designed FDE representations are shown to reduce the computational burden on conventional Transformer structures.
As illustrated in Fig. 12 (a) and (b), these model designs benifit from the combination of ConvNet encoding and ViT media-processing, and achieves image context recovery. Besides, we compare the visualizations of Transformer-based method and pure ConvNet method in Fig. 13. ConvNets gives sharper soft tissue reconstructions and Transformer-based methods recover the whole image better. Although the aforementioned sparse-view CT reconstruction methods (also listed in Table 4, the top part) have been proposed to explore the capability of Transformer versus CNNs in both image and projection domains, few works combines the dense modeling property of Transformer, which helps preserve clinical patterns from input low-quality images and down-sampled projections. Additionally, the limited-angle scenario is overlooked, but the relative consistency between in- and out-of-range projections may be modeled using the Transformer’s powerful global-modeling capability.
Fig. 12.
We illustrate the Transformer-based networks of (a) ReconFormer (Guo et al., 2022d) (b) DuDoTrans (Wang et al., 2021a) (c) T2Net (Feng et al., 2021) and (d) TransCT (Zhang et al., 2021e). (a) and (b) are reconstruction models, (c) and (d) are for enhancement. These structures are based on the hybrid ConvNet-Transformer design.
Fig. 13.
We visualize reconstructions of Transformer-based DuDoTrans (Wang et al., 2021a) versus ConvNet with 72 and 96 sparse views on NIH-AAPM-Mayo (McCollough, 2016) dataset, and the zoom-in images are shown in the last row. With the included Property M2, Transformer-based DuDoTrans obtains better overall performances, especially on bones, and alleviates the FBP artifacts. While the recovered soft tissues are not as sharp as ConvNets results.
Table 4.
The summarized review of Transformer-based model for medical image reconstruction and enhancement. "N" denotes not reported or not applicable on model parameters.
| Reference | Architecture | 2D/3D | #Param | Modality | Dataset | ViT as Enc/Inter/Dec | Highlights | |
|---|---|---|---|---|---|---|---|---|
| Reconstruction | ReconFormer (Guo et al., 2022d) | Conv-Transformer Hybrid | 2D | 1.414M | MRI | fastMRI (Knoll et al.,2020), HPKS (Jiang etal.,2019a) | No/Yes/No | The Pyramid Transformer Layer (PTL) introduces a locally pyramidal but globally columnar structure. |
| DSFormer (Zhou et al., 2022b) | Conv-Transformer Hybrid | 2D | 0.18M | MRI | Multi-coil Brain Data from IXI* | No/Yes/No | The proposed Swin Transformer Reconstruction Network enables a self-supervised reconstruction process with lightweight backbone. | |
| SLATER (Korkmaz et al., 2022) | Conv-Transformer Hybrid | 2D | N | MRI | Single-coil Brain Data from IXI*, Multi-coil Brain Data from fastMRI (Knoll et al., 2020) | No/Yes/Yes | An unsupervised MRI reconstruction design with the long-range dependency of Transformers. | |
| DuDoCAF (Lyu et al., 2022) | Conv-Transformer Hybrid | 2D | 1.428M | MRI | fastMRI (Knoll et al.,2020), Clinical Brain MRI Dataset | No/Yes/No | The proposed recurrent blocks with transformers are employed to capture long-range dependencies from the fused multi-contrast features maps, which boosts target-contrast under-sampled imaging. | |
| SDAUT (Huang et al., 2022a) | Conv-Transformer Hybrid | 2D | N | MRI | Calgary Campinas dataset (Souza et al., 2018 | No/Yes/No | The proposed U-Net-based Transformer combines dense and sparse deformable attention in separate stages, improving performances and speed while revealing explainability. | |
| MIST-net (Pan et al., 2021) | Conv-Transformer Hybrid | 2D | 12.0M | CT | NIH-AAPM-Mayo (McCollough, 2016 | No/Yes/No | The Swin Transformer and convolutional layers are combined in the High-definition Reconstruction Module, achieving high-quality reconstruction. | |
| DuDoTrans (Wang et al., 2021a) | Conv-Transformer Hybrid | 2D | 0.44M | CT | NIH-AAPM-Mayo (McCollough, 2016), COVID-19 | No/Yes/No | The Sinogram Restoration Transformer (SRT) Module is proposed for projection domain enhancement, improving sparse-view CT reconstruction. | |
| FIT (Buchholz and Jug, 2021) | Conventional Transformer | 2D | N | CT | LoDoPaB (Leuschner et al. 2021, | Yes/No/Yes | The carefully designed FDE representations mitigate the computational burden of traditional Transformer structures in the image domain. | |
| RegFormer (Xia et al., 2022a) | Conv-Transformer Hybrid | 2D | N | CT | NIH-AAPM-Mayo (McCollough, 2016 | Yes/Yes/Yes | The unrolled iterative scheme is redesigned with transformer encoders and decoders for learning nonlocal prior, alleviating the sparse-view artifacts. | |
| Enhancement | TransCT (Zhang et al., 2021e) | Conv-Transformer Hybrid | 2D | N | CT | NIH-AAPM-Mayo (McCollough, 2016), Clinical CBCT Images | No/Yes/No | Decomposing Low Dose CT (LDCT) into high and low frequency parts, and then denoise the blurry high-frequency part with the basic Transformer structure |
| TED-Net (Wang et al., 2021b) | Conv-like Transformer | 2D | N | CT | NIH-AAPM-Mayo (McCollough, 2016 | Yes/Yes/Yes | Their design makes use of the tokenization and detokenization operations in the convolution-free encoder-decoder architecture. | |
| Eformer (Luthra et al., 2021) | Conv-Transformer Hybrid | 2D | N | CT | NIH-AAPM-Mayo (McCollough, 2016 | Yes/Yes/Yes | A residual Transformer is proposed, which redesigns the residual block in the denoising encoder-decoder architecture with nonoverlapping window-based Multi-head Self-Attention (MSA). | |
| TVSRN (Yu et al., 2022a) | Conv-like Transformer | 3D | 1.73M | CT | RPLHR-CT† dataset | Yes/Yes/Yes | They design an asymmetric encoder-decoder architecture composed of pure transformers. The structure efficiently models the context relevance in CT volumes and the long-range dependencies. | |
| T2Net(Feng et al., 2021) | Conv-Transformer Hybrid | 2D | N | MRI | Single-coil Brain Data from IXI*, Clinical Brain MRI Dataset | No/Yes/Yes | The task Transformer module is designed in a multi-task learning process of super-resolution and reconstruction, and the super-resolution features are enriched with the low-resolution reconstruction features | |
| WavTrans(Li et al., 2022a) | Conv-Transformer Hybrid | 2D | 2.102M | MRI | fastMRI (Knoll et al., 2020), Clinical Brain MRI Dataset | No/Yes/No | The Residual Cross-attention Swin Transformer is proposed to deal with cross-modality features and boost target contrast MRI super-resolution. |
4.6. Medical image enhancement
Image enhancement is generally utilized as the subsequent procedure after reconstruction, aiming to remove noise artifacts and enhance medically concerned patterns. Different from high-level vision tasks (e.g., classification), the enhancement process requires maintaining details for the final pixel-level image. For this purpose, the commonly used pooling and strided-convolutional operations in the popular CNN architectures are undesired because of the loss of details. Additionally, the locality nature of convolutional operation constrains its potential to recover with more global contexts. In contrast, Transformer has shown the two attractive key properties:
Transformers facilitate the modeling of global features by promoting a wider reception fields (as shown in Fig. 4), which establishes the intra-relationships throughout the whole image and provides abundant information for restoration. [Property M1]
Within a whole image, enhancement targets to alleviate artifacts and blur for latter tasks while keeping else context. The involved self-attention mechanism guides the models to focus on the enhancement-related features, and the dense modeling maintains clear context. [Property M2]
TransCT (Zhang et al., 2021e) first decomposes a Low Dose CT (LDCT) into high and low frequency components, and denoises the noisy high-frequency component using the basic Transformer structure composed of the MSA and MLP layers, simultaneously assisted by the features of the noise-free low-frequency part. It pioneers the use of Transformer in denoising CT images, and numerically proves that the global modeling ability indeed aids in context preservation. To a different extent, TED-Net (Wang et al., 2021b) is proposed and studied in LDCT denoising to explore the convolution-free Transformer structure. Their design makes use of the tokenization and detokenization operations in the encoder-decoder architecture, which aims to entirely evaluate the spatial information extraction capability of the Transformer. Such a design helps understand the difference between the convolution-free features and hybrid features in LDCT denoising, as well as the respective benefits of the two genres in clinical pattern recovery. To further combine the global modeling capability of Transformer and the successfully applied residual learning in low-level vision tasks, Eformer (Luthra et al., 2021) investigates a residual Transformer that redesigns the residual block in the denoising encoder-decoder architecture with non-overlapping window-based MAS. Additionally, it utilizes strided-convolutions and -deconvolutions instead of downsampling and upsampling operations to preserve image context. The re-design of the previously validated structure, i.e., the residual learning here with Transformer as the basic block instead of convolutional layers, contributes a new perspective to the comparison of Transformer and CNNs. Although recent works focus on volumetric CT super-resolution, the conducted low-resolution (LR) volumes are most degraded from high-resolution CT volumes, which brings a domain gap between real-LR and such pseudo-LR volumes. (Yu et al., 2022a) thus releases RPLHR-CT paired real-world LR-HR volumes, and proposes the transformer-based TVSRN for volumetric CT super-resolution. Considering the remote correlation between slices, TVSRN designs an asymmetric encoder-decoder architecture composed of pure transformers. Such a structure enables the long-range dependencies modeling capability and the employed Swin Transformer (Liu et al., 2021b) reduces computational costs. For obtaining improved super-resolution MR Images, T2Net (Feng et al., 2021) specifically designs a task Transformer module in a multi-task learning process of super-resolution and reconstruction. It inserts the module between the iterative recovering processes of the two tasks, and utilizes the module to share informative features. In this way, the super-resolution features are enriched with the low-resolution reconstruction features, resulting in a context devoid of motion artifacts with the detail-preserving Transformer. WavTrans (Li et al., 2022a) proposes to impose anatomy information from reference contrast MR images for boosting super-resolution performances. They first use Wavelet transforms to obtain details of reference images, followed by a carefully designed hybrid structure composed of ConvNet and Residual Cross-attention Swin Transformer (Liu et al., 2021b) module to extract and upsample images. The introduced transformer explores nonlocal features and promotes long-range dependencies between feature maps.
These involved methods, as shown in Fig. 12, take advantage of the hybrid design that globally models the whole image context and locally models the fore/back-ground objects. In spite of these carefully designed works for image enhancement (also listed in Table 4), there is still no discussion of the relationship between the intrinsic properties of Transformer structure and image recovery process, leaving the reaction of Transformers on this task as a "black box" as deep learning. Future architectural design should place a higher emphasis on the interpretability of model mechanisms.
5. Future Perspectives
Returning back to the initial question: Can Transformers transform medical imaging? The answer is likely dichotomous. This is because Transformer, albeit powerful, belongs to machine learning, deep learning in particular, and hence it inherits the pros and cons of machine / deep learning.
The answer is likely positive because it is evident as shown in Section 4 that Transformer, one of the latest technological advances of deep learning, is picking up its momentum in medical imaging. The properties of Transformers (as listed in section 3.4), such as the ability to capture long-range dependencies and the scalability of self-attention, make them an attractive option for medical image analysis. As a result, many researchers have used these properties to develop Transformers that have performed better than CNNs in various medical image tasks. In fact, in the applications surveyed in this paper, some of these Transformers have even achieved state-of-the-art performance. It is predictable that more and more research will be devoted to innovating the architecture of transform and applying it to more medical imaging tasks.
The answer is likely negative too. In (Zhou et al., 2021c), Zhou et al. illustrate some of key traits of medical imaging: multi-modal with a high resolution, non-standard acquisition and data silo, noisy and sparse labeling, imbalanced samples, long-tail disease prevalence, etc. These traits are accompanied with challenges to be solved.
5.1. Challenges
Annotation intensiveness.
The Transformer or deep learning in general requires large-scale datasets (Cheplygina et al., 2019). Empirically, transformer-based models can achieve higher performance trained on larger datasets (Chen et al., 2021f), and their performances degrade when data or annotations are sparse. To address the challenge, self-supervised transformers are promising tools. Using unlabeled data, proxy tasks such as contrastive learning and reconstruction can be leveraged to boost representation learning capability of transformers. Self-Supervised SwinUNETR (Tang et al., 2022) and unified pre-training (Xie et al., 2021c), these medical pre-training frameworks show that training with large-scaled unlabeled 2D or 3D images is beneficial to fine-tuning model with smaller datasets. However, we observe that employing pre-training is computationally exhaustive. Future works can be targeted to simplify and evaluate the efficiency of the pre-training framework and fine-tuning it to smaller datasets.
Data bias, domain adaptation, and model fairness.
In addition to the superior performance, scalability is an advantage brought by Transformer models. The robustness to scaling datasets and model complexity are useful properties to address data bias, domain gaps, and fairness. By effectively modeling larger datasets, transformer models (Xie et al., 2021c; Tang et al., 2022) can learn diverse datasets, including different modalities, different body components, variant imaging protocols and reconstructions. Regarding these domain gaps, there are adaptation methods (Guan and Liu, 2021), which aim to overcome the distribution shift between source and target domains. Meanwhile, the other approach (Caton and Haas, 2020) addresses model fairness. For example, if a model is trained by exclusively male subjects, its performance on female subjects is unknown to the least extent and even worse, the model appears with a gender discrimination. We envision the transformers, with superior scalability, can be used to provide solutions to fairness and social affairs.
Incorporating domain knowledge.
Medical imaging is full of domain knowledge arising from different sources, including anatomical structures, imaging physics, geometric constraints, disease knowledge base, etc. All these knowledge governs the data generation process or serves strong priors for regularized. Visual quantitative analysis of anatomic structures remains a complex task for radiologists. Some of the histomorphometry features of regions of the organs/tissues (e.g. textural or graph features) are poorly adapted for manual identifications (Anandarajah et al., 2005). In this study, transformer networks are shown to provide a moderately better solution that achieves consistently robust performance with variate of anatomies. Compared with previous CNNs (Isensee et al., 2021), transformer approaches (Chen et al., 2022b; Yu et al., 2022c) facilitate better derivation of the visual and quantitative results. In addition, efficient modeling is essential for clinical practice in deploying AI networks. We observe, current medical datasets (Wasserthal et al., 2022) can be different in terms of imaging protocols, patient morphology, and institutional variations, which lead to challenging target tasks. Transformer models are yet to unleash the potential to tackle challenges of sensitivity and adapt abnormal primitives.
Task scalability.
Representation learning with medical images is challenging due to it heterogeneity nature (Zhang et al., 2015b). Prior studies typically focus solving single medical task, transformer model, especially with self-supervised learning, are superior at learning heterogeneous tasks (Li et al., 2022c). The advanced scaling property empowers transformer the ability to tackling multi-domain tasks. In addition, by scaling up transformer networks (Zhai et al., 2021), models can fit variate datasets, researchers can adapt a model at training from a low-data regime (Tang et al., 2022) to larger scales.
Data scalability.
The lack of inductive bias in the original ViT (Dosovitskiy et al., 2020) results in subpar performance when trained on a small amount of data (see M3 and Appendix .2). If a large amount of data is available, Transformers can surpass inductive bias by using various pre-training strategies (Li et al., 2022e; Zhai et al., 2022). In the field of medical imaging, pre-training strategies are also shown merits in improving Transformers’ performances (Xie et al., 2021c; Tang et al., 2022). However, it is not always practical to collect a large amount of data in medical imaging due to patient privacy concerns and labor-intensive manual annotations. Obtaining a large amount of data for imaging modalities or protocols currently under development is even more challenging. Therefore, it is necessary to develop less data-intensive Transformer models for medical imaging applications by introducing inductive bias into Transformer architectures. Several works have been proposed for both natural images (Touvron et al., 2021b; Liu et al., 2021b; Xu et al., 2021c; dâĂŹAscoli et al., 2021) and medical images (Jose and Oza, 2021; Gao et al., 2021b; Jang and Hwang, 2022; Xie et al., 2021b) to address this issue.
Black box and interpretability.
Deep learning is known as a black-box approach and lacks interpretability (Zhang and Zhu, 2018). Though Transformer uses self-attention which mimics some human functions, still it is a black box and unable to provide insights on how variables are being combined to make decisions. Given that medical image analysis is keen to a modelâĂŹs interpretability, it is important to study the interpretability of a Transformer model. A common practice to visualize Transformers is to compute relevancy score from single or multiple attention layers. The multi-head self-attention mechanism provide a direct connections among tokens, an intuitive clue on decision-making. There are several methods to visualize transformer in natural images, raw-attention (Hao et al., 2021), rollout (Xu et al., 2022), GradCAM (Li et al., 2022d), LRP (Chefer et al., 2021), etc. Besides, studies (Krishna et al., 2022; Kan et al.) are proposed by using Transformer backbones in investigating interpretability. Specifically, the self-attention on the last layer of ViTs, trained by a teacher-student style, is visualized. The visualization contains object segmentation, which is not clearly observed in supervised ViTs, nor in CNN (Caron et al., 2021). Recent efforts (Mondal et al., 2021; Matsoukas et al., 2021) in visualizing vision transformers on medical images conform to conventional methods as those on natural images. From a perspective of practical medical scenario, interpretability is not a property of the algorithm but a model affordance for clinical users (Chen et al., 2022a). The model visualization methods, currently used extensively, are depicting the interpretability purely computational. It should vary in methods and forms as contexts and users change. It remains a challenging and open problem, which would be an essential factor in convincing physicians and supporting the deployment of algorithms.
3D modeling.
Most of medical image tasks need to process 3D volumetric data, however, vision Transformer models are known to be computationally intensive and memory-demanding. Efficiently and effectively handling 3D data is a key challenge for adopting Transformers in medical image analysis, UNETR (Hatamizadeh et al., 2022b), TransBTS (Wang et al., 2021e), CoTr (Xie et al., 2021b), nnFormer (Zhou et al., 2021a) and many pioneering works have been proposed to address challenges of modeling spatial features. Though, there are still difficulties at preserving 3D positional information between patches in 1D sequences, and loss of local positional information can lead to sub-optimal performance when dealing heterogeneous tissues in 3D medical image segmentation. Current works have shown great progresses in segmentation, classification, detection, registration, reconstruction or enhancement tasks with 3D radiographic images or videos.
Computational complexity.
As seen in Appendix .4, Transformers are typically computationally complex owing to the computation of self-attention, which is typically quadratic to input image size. While this seems to be less of an issue with natural images, it is a major concern with medical images. This is due to the fact that medical images tend to be far more substantial in size than the size that is common to natural image datasets. For example, a brain MRI image from the BraTS challenge (Menze et al., 2014) has a size of 240 × 240 × 155, whereas a natural image from ImageNet (Deng et al., 2009) has an average size of around 450 × 400. As a result, Transformers used in medical imaging tend to be more compact and trained using a smaller batch size or patched input than their counterparts used for natural images. Many of the existing Transformers used in medical imaging applications are either constructed on top of a SWin Transformer (Liu et al., 2021b) (e.g., SWin-UNETR (Tang et al., 2022; Hatamizadeh et al., 2022a), SWin-UNet (Cao et al., 2021), nnFormer (Zhou et al., 2021a), and TransMorph (Chen et al., 2022b)) or rely on a CNN to extract and down-sample feature maps before feeding them into a Transformer (e.g., TransUNet (Chen et al., 2021d) and ViT-V-Net (Chen et al., 2021c)). Some exciting explorations have shown that it may be possible to bypass Softmax in order to linearize the computation of self-attention (Choromanski et al., 2021; Qin et al., 2022; Wang et al., 2020c; Xiong et al., 2021; Lu et al., 2021), but so far, none of these methods have been applied to medical imaging. We foresee more future research in this area for medical imaging applications.
5.2. Discussion and concluding thoughts
5.2.1. The role of MSA
Arguably, the success of a Vision Transformer is brought by MSA. However, recent works show that the role of self-attention block is not that much irreplaceable in extracting global features. The MSA works as a trainable aggregation of feature maps (Park and Kim, 2022), whose function can be covered by MLPs repeatedly applied across spatial or channels in several MLP-mixer like models (Tolstikhin et al., 2021; Touvron et al., 2021a; Yu et al., 2021b), or large kernel depth-wise convolutions (Liu et al., 2022e; Trockman and Kolter, 2022; Han et al., 2021b), or plain pooling operators to conduct spatial smoothing (Yu et al., 2021b). In (Tolstikhin et al., 2021; Yu et al., 2021b; Liu et al., 2022e), researchers raise skeptical arguments, ascribing the performance gains to the design of pipeline, not MSAs. A perspective of Transformer and CNNs is that convolutions in CNNs and MLPs in Transformers both learn the patterns derived from images, and pooling in CNNs and all operations aforementioned in Transformers are aimed at fusing and integrating feature maps from previous layers. The differences lie in (i) fusion trainability, when comparing MSAs with pooling, (ii) fusion field size, when comparing original MSAs in ViT with those in Swin Transformer, and (iii) fusion method, when comparing depth-wise convolutions with MLPs.
5.2.2. Debate
Despite the promising potential that the Transformers have brought to medical imaging, there have been continuing discussions over which properties of Transformers (listed in Section 3.D) are particularly beneficial.
In (Raghu et al., 2021), the authors discover that the self-attention mechanism enables the early aggregation of global information (i.e., the modeling of long-range dependencies) and that the residual connections help propagate global features throughout the Transformer.
(Ding et al., 2022) believes that the superiority of Transformers is due to their large effective receptive fields, where they experimentally reveal that incorporating convolution operations with large kernels could help close the performance gap between Transformers and CNNs.
Contrarily, in (Park and Kim, 2022), the authors observe that the modeling of long-range dependency could hinder the training of Transformers, and experimentally demonstrate that constraining locality rather than employing global computations improves Transformer performance. They argue that data specificity, not long-range dependency, is the critical feature of the self-attention mechanism. Additionally, they suggest that although Transformers encourage flatter loss landscapes, their weak inductive bias results in non-convex losses that disturbs training.
(Liu et al., 2022e) believe the superior performance of Transformers over CNNs is the result of larger model sizes and training datasets (i.e., the scaling behavior). The authors reveal that by carefully tweaking the CNN designs in accordance with Transformers, CNN outperform Transformers with the help of larger model sizes and training datasets.
5.2.3. Comparative models
CNNs.
Since the introduction of ViT (Dosovitskiy et al., 2020), many advancements to ViT have attempted to reinstate convolution-like behaviors, e.g., Swin Transformer (Liu et al., 2021b), CVT (Wu et al., 2021a), CeiT (Yuan et al., 2021a), and CMT (Guo et al., 2022a). Going a different route, efforts have been made to improve CNNs based on the rationale behind the success of Transformers. These CNN models may attain performances similar to those of Transformers. Liu et al. propose ConvNeXt (Liu et al., 2022e), which modifies a standard CNN with Transformer-inspired components, such as depthwise convolution, layer normalization (Ba et al., 2016), GELU activation (Hendrycks and Gimpel, 2016), and so forth. ConvNeXt exhibits favorable performance and scalability to the competing Transformers while maintaining a CNN-only architecture. In (Ding et al., 2022), Ding et al. draw inspiration from the large kernel size of the self-attention operation in a Transformer. They introduce RepLKNet, which substitutes the typically used small convolution kernel (e.g., 3 × 3 or 5 × 5) with large kernels up to 31 × 31. RepLKNet’s performance is competitive to that of the competing Transformers, and it demonstrates excellent scalability to large data and model sizes. In a similar fashion, (Guo et al., 2022c) present VAN that takes advantage of both convolution and self-attention. VAN employs depth-wise convolutions with large kernel sizes to mimic self-attention, and it outperforms the comparative Transformers and CNNs on several computer vision tasks. As seen from these CNN models, the odyssey of CNN design has recently taken on resembling the characteristics of Transformers. These CNNs have benefited significantly from components like depthwise convolution and large kernel sizes, where the former is analogous to the weighted sum operation in self-attention (Liu et al., 2022e) and the latter resembles the large effective receptive field of Transformers (Ding et al., 2022). Similar trends can be observed in the field of medical imaging, where the integration of these Transformer-like components into CNN designs is gaining increased attention (Lin et al., 2022a; Liu et al., 2022d; Jia et al., 2022; Han et al., 2022).
MLPs.
Similar to the aforementioned CNNs, MLP-based models are influenced by Transformers but diverge from Transformers and CNNs. In (Tolstikhin et al., 2021), Tolstikhin et al. first demonstrate that, although being beneficial, convolution and self-attention are not required for superior performance. They proposed MLP-mixer, a pure MLP architecture that attains competitive performances on image classification benchmarks. Since then, MLP-mixer has sparked research on developing MLP-based models that can compete with the well established CNNs and Transformers. In general, the architecture of MLP-based models resembles that of Transformers: first, the input image is divided into equal-sized patches; then, the patches are linearly projected to form tokens; and then, two types of MLP layers are repeatedly applied across either spatial locations or embedding channels. On the basis of this concept, models such as ResMLP (Touvron et al., 2021a), S2-MLP (Yu et al., 2022b), CycleMLP (Chen et al., 2022c), Dynamixer (Wang et al., 2022d), Hire-MLP (Guo et al., 2022b) have shown promising results in a variety of computer vision applications. MLP-based models have several appealing advantages over Transformers and CNNs, including their simplicity of implementation, more stable training due to the absence of self-attention, their ability to capture long-range interactions, the visibility of the linear layers, and the alleviation of positional embedding (Tolstikhin et al., 2021; Touvron et al., 2021a). However, the use of MLP-based models in the medical imaging field is still in its infancy, with only a small number of models proposed (Valanarasu and Patel, 2022).
The models discussed above aim to improve upon conventional CNNs and MLPs by making special modifications inspired by the properties of Transformers. Likewise, to develop an efficient model for medical imaging, it is necessary to understand which Transformer properties are particularly advantageous for specific medical imaging applications. In the next section, we discuss briefly the key Transformer properties for each medical imaging application.
5.2.4. Which properties are beneficial for medical imaging applications?
It is worth noting that the majority of the findings about Transformers listed in 5.2.2 are derived from image classification tasks. However, the applications of medical imaging are not limited to classification. The properties listed in Section 3.D are still underexploited in all medical imaging applications. The majority of Transformer-based methods in medical imaging do not investigate the properties adequately and instead take the performance improvement from Transformers for granted. This paper surveys the applications of Transformers in medical imaging, including segmentation, classification, detection, registration, enhancement, and reconstruction. Yet, it remains to be a question of which Transformer properties are beneficial for which application. Further research is needed to establish the efficacy of these properties and put them into practical use, maybe along the following routes.
Segmentation. Medical image segmentation is typically with high-resolution, high-dimensional images, which requires modeling capability of visual semantics in dense prediction. That means, unlike the language tokens that used as the basic word sequence in Transformers, visual contexts in segmentation task vary substantially in scale. ViT-based methods, especially hierarchical structures such as swin Transformer, are designed for efficient modeling of multiscale features (Properties M1, M2). Furthermore, Transformer-based segmentation networks show futuristic scaling behavior (Property C1) of exploiting large-scale pre-training dataset with self-supervised learning, which provide effective solutions to the difficulties of acquiring expert annotated labels. We believe that the efficiency of modeling hierarchical contexts in medical images, and the effectiveness of pre-training strategy can pave the way for the future work of Transformer-based medical image segmentation.
Recognition and classification. As the fundamental task evaluated by the original ViT (Dosovitskiy et al., 2020), the properties of Transformers for image classification have been intensively investigated in computer vision. Although medical images are very dissimilar to natural images, Transformers for medical image classification are expected to share similar properties with those deemed beneficial in natural image classification tasks (i.e., Properties M1, M3, M4, M5, and C1). Among these properties, Transformers’ superior scaling behaviour (i.e., pre-training using large-scale datasets, Property C1) has been validated for various medical classification applications. In general, the applications of Transformers for medical image classification are mostly limited to 2D, it will be necessary in the future works to expand Transformers to 3D applications given the volumetric nature of most medical images, which is related to Property C3.
Detection. Detection is the task of localizing and categorizing lesions and abnormalities. Such a task relies heavily on the comprehension of contextual information about abnormalities and organs. Consequently, the capability of Transformers to model and aggregate long-range dependencies (Property M1) may be the most critical property among other properties for medical image Detection.
Registration. In addition to the flatter loss landscape of Transformer-based registration models (as seen in Fig. 5 and Property M4), the large model size of Transformers (Property C1) may also aid in generating accurate high-dimensional vector fields, hence improving registration performance. Moreover, CNN-based models are often of small kernel sizes (e.g., 3 × 3 or 5 × 5), while the deformation or displacement in common registration applications often exceeds their kernel size. Therefore, CNNs may not recognize the proper spatial correspondence until the deeper layers. On the other hand, Transformers aggregate contextual information with large kernels starting from the first layer of the network (Property M1), which may play a crucial role in the improved performance.
Reconstruction. As discussed, Properties M1 and M2 have been explored in reviewed works. Further considering the physical imaging system in a real-time clinical diagnosis, the photon noises blur the images and the imaging time troubles the waited patients. Therefore, [Properties M5 and C3] need to be further concerned in later model design when introducing ViT in reconstruction.
Enhancement With the low-resolution or downsampled medical images, the Region of Interest (RoI), such as anatomy boundaries, seems the most important in the diagnosis. Thus, it’s worth exhausting to improve the RoI quality while tolerating the else image context less enhanced. Towards this target, exploring the relations from the locality of pixels [Properties M3] is necessary in a Transformer architecture design. Meanwhile, a considerable balance between the global modeling and local modeling of a hybrid model really matters in medical image enhancement.
Fig. 9.
Visualization of CT/MRI segmentation and comparison on public datasets between Transformer-based and baseline models. Transformer-based models includes Swin UNETR (Tang et al., 2022), T-AutoML (Yang et al., 2021), TransBTSV2 (Li et al., 2022c), AFTer-UNet (Yan et al., 2022), U- Transformer (Petit et al., 2021), UNesT (Yu et al., 2022c), BiTr-Unet (Jia and Shu, 2021), UTNet (Gao et al., 2021b), nnFormer (Zhou et al., 2021a), MOCOv3 (Chen et al., 2021f), and DINO (Caron et al., 2021), USST (Xie et al., 2021c). Baseline models contain the DiNTS (He et al., 2021b), ResUNet (Zhang et al., 2018), and AttentionUNet (Oktay et al., 2018)
Fig. 10.
Transformer segmentation to other medical image modalities such as endoscopy, microscopy, retinopathy, ultrasound, X-ray, and camera images. The comparison methods include Pyramid Trans (Zhang et al., 2021d), MBT-Net (Zhang et al., 2021a), MCTrans (Ji et al., 2021), X-Net (Li et al., 2021d), TransAttUnet (Chen et al., 2021a), MedT (Valanarasu et al., 2021), Swin-UNet (Nguyen et al., 2021), SpecTr (Yun et al., 2021), RT-Net (Huang et al., 2022b), and ConvNet-based models (ResUNet (Zhang et al., 2018), UNet (Ronneberger et al., 2015), UNet++ (Zhou et al., 2018), and AttentionUNet (Oktay et al., 2018)).
Appendix
Appendix .1. Translational equivalence and invariance of CNNs
Translational equivalence and invariance are fundamentally different properties, as outlined in (Goodfellow et al., 2016). Translational equivalence refers to the capability of identifying the same features, regardless of where they are located within the input image. In CNNs, translational equivalence is made possible by the convolution operation, where the same kernel is moved across the entire image, so the same features will be detected even if they have been translated. Translational invariance, on the other hand, means that a model will produce the same result for a given input image, regardless of where the features are located within the image. CNNs are invariant to small translation of features within an image, and this is due to the use of max-pooling operations. The output of a max-pooling operation is the highest activation value within a group of neighboring activations. If the translation is small, the highest value within a specific region will likely remain unchanged, making CNNs invariant to small translations. Translational equivalence and invariance are both important features of CNNs that make them effective in tasks such as object recognition and image classification, where the location of the object in the image is not always predictable.
Appendix .2. Inductive bias
Because of the convolution and pooling operations, CNN architectures impose a strong intrinsic inductive bias. A CNN is analogous to a fully-connected network but with an infinitely strong prior over the weights. The convolution operation constrains the weights of one hidden unit to be equivalent to the weights of its neighbor but spatially shifted. Similarly, the pooling operation constraints that each weight should be invariant to small translations (Goodfellow et al., 2016). These priors, known as the intrinsic inductive bias, make CNNs more data- and parameter-efficient (Goodfellow et al., 2016; Scherer et al., 2010). Additional inductive bias, on top of the intrinsic inductive bias, may further improve the efficacy of CNN-based generative models (Xu et al., 2021b). Despite inductive bias is of great importance, the original ViT (Dosovitskiy et al., 2020) lacks it since the self-attention operations are global and the positional embedding is the only manually introduced inductive bias. Therefore, ViT yields inferior performance when trained on insufficient amounts of data. However, it is demonstrated that training Transformers on large-scale datasets may surpass inductive bias. When pre-trained using sufficiently large amount of data, Transformers achieve superior performances on tasks with less data (Han et al., 2020; Zhai et al., 2021; Chen et al., 2021b; Dosovitskiy et al., 2020; Liu et al., 2022e; Naseer et al., 2021). Alternatively, there have been attempts to introduce locality into Transformers (Liu et al., 2021b; Xu et al., 2021c) or distill the inductive bias from CNNs to Transformers (Touvron et al., 2021b; Ren et al., 2022) have been proposed. It has also been shown that combining CNNs with Transformers to construct hybrid models imposes convolutional inductive bias on network architecture (Dosovitskiy et al., 2020; dâĂŹAscoli et al., 2021; Wu et al., 2021a).
Appendix .3. Loss landscapes
The sharpness or flatness of a loss landscape is often used as a measure of the trainability and generalizability of a network architecture or optimizer (Li et al., 2018; Keskar et al., 2017). A loss landscape is generated relative to the parameters of a neural network. Here, we provide a brief introduction to the computation of loss landscapes and direct interested readers to the corresponding references for further information. We first use a pre-trained model with network parameters, , to generate a loss value, which corresponds to the minimum value in the resulting loss landscape. Then, is perturbed using two random direction vectors, and , with the corresponding step sizes of and . A loss landscape can be depicted as a plot of the form:
| (.1) |
where denotes the loss value given the perturbed network parameters.
The flatness of a loss landscape translates to how sensitive the network parameters are to the perturbations. There have been substantial theoretical and empirical attempts to understand the relationship between the sharpness of the loss landscape and the generalizability of the neural network (Foret et al., 2021; Dinh et al., 2017; Li et al., 2018; Dziugaite and Roy, 2017; Jiang et al., 2019b). Sharp minimizers are more sensitive to noise in the parameter space, resulting in poor generalizability in general (Keskar et al., 2017; Hochreiter and Schmidhuber, 1997). A recent study suggests that ViTs tend to promote flatter loss landscapes than CNNs and thus generalize better on unseen data (Park and Kim, 2022). In this work, we empirically confirm this finding by depicting the loss landscapes for CNNs versus Transformers on two tasks, registration and segmentation, as shown in Fig. 5.
Appendix .4. Computational complexity of Transformers
Transformers are generally computationally complex, with the self-attention mechanism standing as the main bottleneck. In a self-attention mechanism, each token is updated by attending it relative to all other tokens. Although the computation of self-attention is discussed in length in section 2, we repeat the its equation here for clarity:
| (.2) |
Suppose Q, K, and V all have the same size of n × d, where n is the sequence length and d denotes the embedding size, both matrix multiplications in the above equation (i.e., and Softmax(·) × V) have the complexity of . Consequently, the computational complexity of computing self-attention is quadratic to the sequence size, i.e., . In comparison, a convolution operation in CNNs has a linear complexity of . For this reason, training Transformers often requires more time and resources than training CNNs. In light of this shortcoming, modifications to self-attention computation have been proposed to lower its computational complexity. For example, consider Eqn. (.2) without the softmax operation, the complexity of Eqn. (.2) can then be reduced by using the associative property of matrix multiplication, i.e, as opposed to , where the former has approximately linear complexity while the latter has quadratic complexity. Based on this idea, Choromanski et al. (Choromanski et al., 2021) and Qin et al. (Qin et al., 2022) linearize the matrix multiplication by avoiding the direct usage of softmax, and afterwards compute self-attention by approximating the softmax attention kernels. Wang et al. (Wang et al., 2020c) propose decomposing self-attention into several smaller attentions by means of linear projections, motivated by the finding that self-attention is of low rank. Xiong et al. (Xiong et al., 2021) reduce the complexity of self-attention computation by leveraging the Nystrom method, which samples a subset of columns or rows to approximate a softmax matrix. Similarly, Lu et al. (Lu et al., 2021) propose a Softmax-free Transformer that leverages Gaussian kernel, instead of softmax, to define self-attention. In the meanwhile, Liu et al. (Liu et al., 2021b) and Wang et al. (Wang et al., 2021f) develop hierarchical Transformers that confine self-attention locally rather than globally, thereby reducing complexity and introducing spatial inductive bias that conventional Transformers lack.
Footnotes
"The Digital Universe Driving Data Growth in Healthcare," published by EMC with research and analysis from IDC (12/13).
References
- Akhloufi MA, Chetoui M, 2021. Chest XR COVID-19 detection. https://cxr-covid19.grand-challenge.org/. Online; accessed September 2021. [Google Scholar]
- Alexander LM, Escalera J, Ai L, Andreotti C, Febre K, Mangone A, Vega-Potler N, Langer N, Alexander A, Kovacs M, et al. , 2017. An open resource for transdiagnostic research in pediatric mental health and learning disorders. Scientific data 4, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alom MZ, Hasan M, Yakopcic C, Taha TM, Asari VK, 2018. Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv preprint arXiv:1802.06955 . [Google Scholar]
- Ambellan F, Tack A, Ehlke M, Zachow S, 2019. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the osteoarthritis initiative. Medical image analysis 52, 109–118. [DOI] [PubMed] [Google Scholar]
- Anandarajah S, Tai T, de Lusignan S, Stevens P, O’Donoghue D, Walker M, Hilton S, 2005. The validity of searching routinely collected general practice computer data to identify patients with chronic kidney disease (ckd): a manual review of 500 medical records. Nephrology Dialysis Transplantation 20, 2089–2096. [DOI] [PubMed] [Google Scholar]
- Andrearczyk V, Oreiller V, Jreige M, Vallières M, Castelli J, Elhalawani H, Boughdad S, Prior JO, Depeursinge A, 2020. Overview of the HECKTOR challenge at MICCAI 2020: automatic head and neck tumor segmentation in PET/CT, in: 3D Head and Neck Tumor Segmentation in PET/CT Challenge, Springer. pp. 1–21. [Google Scholar]
- Antonelli M, Reinke A, Bakas S, Farahani K, Landman BA, Litjens G, Menze B, Ronneberger O, Summers RM, van Ginneken B, et al. , 2021. The medical segmentation decathlon. arXiv preprint arXiv:2106.05735 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- APTOS, 2019. APTOS 2019 blindness detection. https://www.kaggle.com/c/aptos2019-blindness-detection/.
- Ba JL, Kiros JR, Hinton GE, 2016. Layer normalization . [Google Scholar]
- Bahdanau D, Cho K, Bengio Y, 2014. Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv:1409.0473 . [Google Scholar]
- Baid U, Ghodasara S, Mohan S, Bilello M, Calabrese E, Colak E, Farahani K, Kalpathy-Cramer J, Kitamura FC, Pati S, et al. , 2021. The RSNA-ASNR-MICCAI BraTS 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv preprint arXiv:2107.02314 . [Google Scholar]
- Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV, 2019. VoxelMorph: a learning framework for deformable medical image registration. IEEE transactions on medical imaging 38, 1788–1800. [DOI] [PubMed] [Google Scholar]
- Bao H, Dong L, Wei F, 2021. BEiT: Bert pre-training of image transformers. arXiv preprint arXiv:2106.08254 . [Google Scholar]
- Bastiani M, Andersson JL, Cordero-Grande L, Murgasova M, Hutter J, Price AN, Makropoulos A, Fitzgibbon SP, Hughes E, Rueckert D, et al. , 2019. Automated processing pipeline for neonatal diffusion MRI in the developing human connectome project. Neuroimage 185, 750–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal J, Kim E, Tzeng E, Park DH, Zhai A, Kislyuk D, 2020. Toward transformer-based object detection. arXiv preprint arXiv:2012.09958 . [Google Scholar]
- Bedel HA, Şıvgın I, Dalmaz O, Dar SUH, Çukur T, 2022. BolT: Fused window transformers for fMRI time series analysis. arXiv preprint arXiv:2205.11578 [DOI] [PubMed] [Google Scholar]
- Bejnordi BE, Veta M, Van Diest PJ, Van Ginneken B, Karssemeijer N, Litjens G, Van Der Laak JA, Hermsen M, Manson QF, Balkenhol M, et al. , 2017. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama 318, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellec P, Chu C, Chouinard-Decorte F, Benhajali Y, Margulies DS, Craddock RC, 2017. The neuro bureau ADHD-200 preprocessed repository. Neuroimage 144, 275–286. [DOI] [PubMed] [Google Scholar]
- Bernal J, Sánchez FJ, Fernández-Esparrach G, Gil D, Rodríguez C, Vilariño F, 2015. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Computerized Medical Imaging and Graphics 43, 99–111. [DOI] [PubMed] [Google Scholar]
- Bernal J, Sánchez J, Vilarino F, 2012. Towards automatic polyp detection with a polyp appearance model. Pattern Recognition 45, 3166–3182. [Google Scholar]
- Bernard O, Lalande A, Zotti C, Cervenansky F, Yang X, Heng PA, Cetin I, Lekadir K, Camara O, Ballester MAG, et al. , 2018. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE transactions on medical imaging 37, 2514–2525. [DOI] [PubMed] [Google Scholar]
- Beutel J, Kundel HL, Van Metter RL, 2000. Handbook of Medical Imaging. volume 1. SPEI Press. [Google Scholar]
- Bhojanapalli S, Chakrabarti A, Glasner D, Li D, Unterthiner T, Veit A, 2021. Understanding robustness of transformers for image classification, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10231–10241. [Google Scholar]
- Bilic P, Christ PF, Vorontsov E, Chlebus G, Chen H, Dou Q, Fu CW, Han X, Heng PA, Hesser J, et al. , 2019. The liver tumor segmentation benchmark (LiTS). arXiv preprint arXiv:1901.04056 . [Google Scholar]
- Borgli H, Thambawita V, Smedsrud PH, Hicks S, Jha D, Eskeland SL, Randel KR, Pogorelov K, Lux M, Nguyen DTD, et al. , 2020. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Scientific data 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch T, Tam R, Initiative, A.D.N., et al. , 2013. Manifold learning of brain MRIs by deep learning, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 633–640. [DOI] [PubMed] [Google Scholar]
- Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, Neelakantan A, Shyam P, Sastry G, Askell A, et al. , 2020. Language models are few-shot learners. Advances in neural information processing systems 33, 1877–1901. [Google Scholar]
- Buchholz TO, Jug F, 2021. Fourier image transformer. arXiv preprint arXiv:2104.02555 . [Google Scholar]
- Cai Z, Lin L, He H, Tang X, 2022. Uni4Eye: Unified 2D and 3D self-supervised pre-training via masked image modeling transformer for ophthalmic image classification, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 88–98. [Google Scholar]
- Caicedo JC, Goodman A, Karhohs KW, Cimini BA, Ackerman J, Haghighi M, Heng C, Becker T, Doan M, McQuin C, et al. , 2019. Nucleus segmentation across imaging experiments: the 2018 data science bowl. Nature methods 16, 1247–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello VM, Gkontra P, Izquierdo C, Martín-Isla C, Sojoudi A, Full PM, Maier-Hein K, Zhang Y, He Z, Ma J, et al. , 2021. Multi-centre, multi-vendor and multi-disease cardiac segmentation: the M&Ms challenge. IEEE Transactions on Medical Imaging 40, 3543–3554. [DOI] [PubMed] [Google Scholar]
- Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M, 2021. Swin-Unet: Unet-like pure transformer for medical image segmentation. arXiv preprint arXiv:2105.05537 . [Google Scholar]
- Carion N, Massa F, Synnaeve G, Usunier N, Kirillov A, Zagoruyko S, 2020. End-to-end object detection with transformers, in: European conference on computer vision, Springer. pp. 213–229. [Google Scholar]
- Caron M, Touvron H, Misra I, Jégou H, Mairal J, Bojanowski P, Joulin A, 2021. Emerging properties in self-supervised vision transformers. Proceedings of the IEEE/CVF International Conference on Computer Vision . [Google Scholar]
- Carreira J, Zisserman A, 2017. Quo vadis, action recognition? a new model and the kinetics dataset, in: proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 6299–6308. [Google Scholar]
- Caton S, Haas C, 2020. Fairness in machine learning: A survey. arXiv preprint arXiv:2010.04053 . [Google Scholar]
- Chang Y, Menghan H, Guangtao Z, Xiao-Ping Z, 2021. TransClaw U-Net: Claw U-Net with transformers for medical image segmentation. arXiv preprint arXiv:2107.05188 . [Google Scholar]
- Chefer H, Gur S, Wolf L, 2021. Transformer interpretability beyond attention visualization, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 782–791. [Google Scholar]
- Chen B, Liu Y, Zhang Z, Lu G, Zhang D, 2021a. TransAttUnet: Multi-level attention-guided U-Net with transformer for medical image segmentation. arXiv preprint arXiv:2107.05274 . [Google Scholar]
- Chen H, Gomez C, Huang CM, Unberath M, 2022a. Explainable medical imaging AI needs human-centered design: guidelines and evidence from a systematic review. npj Digital Medicine 5, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wang Y, Guo T, Xu C, Deng Y, Liu Z, Ma S, Xu C, Xu C, Gao W, 2021b. Pre-trained image processing transformer, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 12299–12310. [Google Scholar]
- Chen J, Frey EC, He Y, Segars WP, Li Y, Du Y, 2022b. TransMorph: Transformer for unsupervised medical image registration. Medical Image Analysis , 102615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, He Y, Frey EC, Li Y, Du Y, 2021c. ViT-V-Net: Vision transformer for unsupervised volumetric medical image registration, in: Medical Imaging with Deep Learning. [Google Scholar]
- Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y, 2021d. TransUNet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 . [Google Scholar]
- Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL, 2017. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE transactions on pattern analysis and machine intelligence 40, 834–848. [DOI] [PubMed] [Google Scholar]
- Chen S, Xie E, GE C, Chen R, Liang D, Luo P, 2022c. CycleMLP: A MLP-like architecture for dense prediction, in: International Conference on Learning Representations. [Google Scholar]
- Chen X, Fan H, Girshick R, He K, 2020. Improved baselines with momentum contrastive learning. arXiv preprint arXiv:2003.04297 . [Google Scholar]
- Chen X, Sun S, Bai N, Han K, Liu Q, Yao S, Tang H, Zhang C, Lu Z, Huang Q, et al. , 2021e. A deep learning-based auto-segmentation system for organs-at-risk on whole-body computed tomography images for radiation therapy. Radiotherapy and Oncology 160, 175–184. [DOI] [PubMed] [Google Scholar]
- Chen X, Xie S, He K, 2021f. An empirical study of training self-supervised vision transformers. Proceedings of the IEEE/CVF International Conference on Computer Vision . [Google Scholar]
- Chen Y, Dai X, Chen D, Liu M, Dong X, Yuan L, Liu Z, 2021g. Mobile-Former: Bridging MobileNet and transformer. arXiv preprint arXiv:2108.05895 . [Google Scholar]
- Cheng J, Zhang X, Zhao F, Wu Z, Wang Y, Huang Y, Lin W, Wang L, Li G, 2022. Spherical transformer for quality assessment of pediatric cortical surfaces, in: 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), IEEE. pp. 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Lu J, Zhu X, Hong J, Liu X, Li M, Li P, 2019. Dilated residual learning with skip connections for real-time denoising of laser speckle imaging of blood flow in a log-transformed domain. IEEE Transactions on Medical Imaging 39, 1582–1593. [DOI] [PubMed] [Google Scholar]
- Cheplygina V, de Bruijne M, Pluim JP, 2019. Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Medical Image Analysis 54, 280–296. [DOI] [PubMed] [Google Scholar]
- Choromanski KM, Likhosherstov V, Dohan D, Song X, Gane A, Sarlos T, Hawkins P, Davis JQ, Mohiuddin A, Kaiser L, Belanger DB, Colwell LJ, Weller A, 2021. Rethinking attention with performers, in: International Conference on Learning Representations. [Google Scholar]
- Chu X, Tian Z, Wang Y, Zhang B, Ren H, Wei X, Xia H, Shen C, 2021. Twins: Revisiting the design of spatial attention in vision transformers. Advances in Neural Information Processing Systems 34. [Google Scholar]
- Cireşan D, Giusti A, Gambardella L, Schmidhuber J, 2012. Deep neural networks segment neuronal membranes in electron microscopy images. Advances in neural information processing systems 25. [Google Scholar]
- Cire§an DC, Giusti A, Gambardella LM, Schmidhuber J, 2013. Mitosis detection in breast cancer histology images with deep neural networks, in: International conference on medical image computing and computer-assisted intervention, Springer. pp. 411–418. [DOI] [PubMed] [Google Scholar]
- Codella NC, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H, et al. , 2018. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC), in: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), IEEE. pp. 168–172. [Google Scholar]
- Cordonnier JB, Loukas A, Jaggi M, 2019. On the relationship between self-attention and convolutional layers, in: International Conference on Learning Representations. [Google Scholar]
- Cruz-Roa AA, Arevalo Ovalle JE, Madabhushi A, González Osorio FA, 2013. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection, in: International conference on medical image computing and computer-assisted intervention, Springer. pp. 403–410. [DOI] [PubMed] [Google Scholar]
- Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, Zheng K, Wu Z, Fu L, Xu B, et al. , 2019. PET image denoising using unsupervised deep learning. European journal of nuclear medicine and molecular imaging 46, 2780–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan S, Williams LZ, Fawaz A, Rueckert D, Robinson EC, 2022. Surface analysis with vision transformers. arXiv preprint arXiv:2205.15836 . [Google Scholar]
- Dai J, Qi H, Xiong Y, Li Y, Zhang G, Hu H, Wei Y, 2017. Deformable convolutional networks, in: Proceedings of the IEEE international conference on computer vision, pp. 764–773. [Google Scholar]
- Dai W, Zhang Z, Tian L, Yu S, Wang S, Dong Z, Zheng H, 2022. BrainFormer: A hybrid CNN-Transformer model for brain fMRI data classification. arXiv preprint arXiv:2208.03028 . [Google Scholar]
- Dai Y, Gao Y, Liu F, 2021a. Transmed: Transformers advance multi-modal medical image classification. Diagnostics 11, 1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Cai B, Lin Y, Chen J, 2021b. UP-DETR: Unsupervised pre-training for object detection with transformers, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 1601–1610. [Google Scholar]
- Dai Z, Liu H, Le QV, Tan M, 2021c. CoAtNet: Marrying convolution and attention for all data sizes, in: Advances in Neural Information Processing Systems, Curran Associates, Inc.. pp. 3965–3977. [Google Scholar]
- Dai Z, Yang Z, Yang Y, Carbonell JG, Le QV, Salakhutdinov R, 2019. Transformer-xl: Attentive language models beyond a fixed-length context, in: ACL (1), pp. 2978–2988. [Google Scholar]
- Dalca AV, Balakrishnan G, Guttag J, Sabuncu MR, 2019. Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces. Medical image analysis 57, 226–236. [DOI] [PubMed] [Google Scholar]
- Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L, 2009. ImageNet: A large-scale hierarchical image database, in: 2009 IEEE conference on computer vision and pattern recognition, Ieee. pp. 248–255. [Google Scholar]
- Deng K, Meng Y, Gao D, Bridge J, Shen Y, Lip G, Zhao Y, Zheng Y, 2021. TransBridge: A lightweight transformer for left ventricle segmentation in echocardiography, in: International Workshop on Advances in Simplifying Medical Ultrasound, Springer. pp. 63–72. [Google Scholar]
- Devalla SK, Renukanand PK, Sreedhar BK, Subramanian G, Zhang L, Perera S, Mari JM, Chin KS, Tun TA, Strouthidis NG, et al. , 2018. DRUNET: a dilated-residual u-net deep learning network to segment optic nerve head tissues in optical coherence tomography images. Biomedical optics express 9, 3244–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J, Chang MW, Lee K, Toutanova K, 2018. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805 . [Google Scholar]
- Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M, et al. , 2014. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular psychiatry 19, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Kuriyan AE, Ramchandran RS, Wykoff CC, Sharma G, 2021. Weakly-supervised vessel detection in ultra-widefield fundus photography via iterative multi-modal registration and learning. IEEE Transactions on Medical Imaging 40, 2748–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Zhang X, Zhou Y, Han J, Ding G, Sun J, 2022. Scaling up your kernels to 31x31: Revisiting large kernel design in cnns. arXiv preprint arXiv:2203.06717 . [Google Scholar]
- Dinh L, Pascanu R, Bengio S, Bengio Y, 2017. Sharp minima can generalize for deep nets, in: International Conference on Machine Learning, PMLR. pp. 1019–1028. [Google Scholar]
- Dolz J, Gopinath K, Yuan J, Lombaert H, Desrosiers C, Ayed IB, 2018. HyperDense-Net: a hyper-densely connected CNN for multi-modal image segmentation. IEEE transactions on medical imaging 38, 1116–1126. [DOI] [PubMed] [Google Scholar]
- Dong B, Wang W, Fan DP, Li J, Fu H, Shao L, 2021a. Polyp-PVT: Polyp segmentation with pyramid vision transformers. arXiv preprint arXiv:2108.06932 . [Google Scholar]
- Dong X, Bao J, Chen D, Zhang W, Yu N, Yuan L, Chen D, Guo B, 2022. Cswin transformer: A general vision transformer backbone with cross-shaped windows, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 12124–12134. [Google Scholar]
- Dong X, Bao J, Zhang T, Chen D, Zhang W, Yuan L, Chen D, Wen F, Yu N, 2021b. PeCo: Perceptual codebook for BERT pre-training of vision transformers. arXiv preprint arXiv:2111.12710 . [Google Scholar]
- Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, et al. , 2020. An image is worth 16x16 words: Transformers for image recognition at scale, in: International Conference on Learning Representations. [Google Scholar]
- Dziugaite GK, Roy DM, 2017. Computing nonvacuous generalization bounds for deep (stochastic) neural networks with many more parameters than training data. arXiv preprint arXiv:1703.11008 . [Google Scholar]
- dâĂŹAscoli S, Touvron H, Leavitt ML, Morcos AS, Biroli G, Sagun L, 2021. ConViT: Improving vision transformers with soft convolutional inductive biases, in: International Conference on Machine Learning, PMLR. pp. 2286–2296. [Google Scholar]
- Evans AC, Group BDC, et al. , 2006. The NIH MRI study of normal brain development. Neuroimage 30, 184–202. [DOI] [PubMed] [Google Scholar]
- Fan H, Xiong B, Mangalam K, Li Y, Yan Z, Malik J, Feichtenhofer C, 2021. Multiscale vision transformers, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 6824–6835. [Google Scholar]
- Fang Y, Liao B, Wang X, Fang J, Qi J, Wu R, Niu J, Liu W, 2021. You only look at one sequence: Rethinking transformer in vision through object detection. Advances in Neural Information Processing Systems 34. [Google Scholar]
- Feng CM, Yan Y, Fu H, Chen L, Xu Y, 2021. Task transformer network for joint MRI reconstruction and super-resolution, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 307–317. [Google Scholar]
- Foret P, Kleiner A, Mobahi H, Neyshabur B, 2021. Sharpness-aware minimization for efficiently improving generalization, in: International Conference on Learning Representations. [Google Scholar]
- Fraz MM, Remagnino P, Hoppe A, Uyyanonvara B, Rudnicka AR, Owen CG, Barman SA, 2012. An ensemble classification-based approach applied to retinal blood vessel segmentation. IEEE Transactions on Biomedical Engineering 59, 2538–2548. [DOI] [PubMed] [Google Scholar]
- Fu H, Cheng J, Xu Y, Wong DWK, Liu J, Cao X, 2018. Joint optic disc and cup segmentation based on multi-label deep network and polar transformation. IEEE transactions on medical imaging 37, 1597–1605. [DOI] [PubMed] [Google Scholar]
- Gal Y, Ghahramani Z, 2016. Dropout as a bayesian approximation: Representing model uncertainty in deep learning, in: International Conference on Machine Learning, pp. 1050–1059. [Google Scholar]
- Gao R, Huo Y, Bao S, Tang Y, Antic SL, Epstein ES, Balar AB, Deppen S, Paulson AB, Sandler KL, et al. , 2019a. Distanced LSTM: time-distanced gates in long short-term memory models for lung cancer detection, in: International Workshop on Machine Learning in Medical Imaging, Springer. pp. 310–318. [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Huang R, Chen M, Wang Z, Deng J, Chen Y, Yang Y, Zhang J, Tao C, Li H, 2019b. FocusNet: Imbalanced large and small organ segmentation with an end-to-end deep neural network for head and neck ct images, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 829–838. [Google Scholar]
- Gao Y, Huang R, Yang Y, Zhang J, Shao K, Tao C, Chen Y, Metaxas DN, Li H, Chen M, 2021a. FocusNetv2: Imbalanced large and small organ segmentation with adversarial shape constraint for head and neck ct images. Medical Image Analysis 67, 101831. [DOI] [PubMed] [Google Scholar]
- Gao Y, Phillips JM, Zheng Y, Min R, Fletcher PT, Gerig G, 2018. Fully convolutional structured lstm networks for joint 4d medical image segmentation, in: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), IEEE. pp. 1104–1108. [Google Scholar]
- Gao Y, Zhou M, Metaxas DN, 2021b. UTNet: a hybrid transformer architecture for medical image segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 61–71. [Google Scholar]
- Gao Z, Hong B, Zhang X, Li Y, Jia C, Wu J, Wang C, Meng D, Li C, 2021c. Instance-based vision transformer for subtyping of papillary renal cell carcinoma in histopathological image, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 299–308. [Google Scholar]
- Gao Z, Shi J, Zhang X, Li Y, Zhang H, Wu J, Wang C, Meng D, Li C, 2021d. Nuclei grading of clear cell renal cell carcinoma in histopathological image by composite high-resolution network, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 132–142. [Google Scholar]
- Gharleghi R, Adikari D, Ellenberger K, Ooi SY, Ellis C, Chen CM, Gao R, He Y, Hussain R, Lee CY, Li J, Ma J, Nie Z, Oliveira B, Qi Y, Skandarani Y, VilaÃğa JL, Wang X, Yang S, Sowmya A, Beier S, 2022. Automated segmentation of normal and diseased coronary arteries âĂŞ the asoca challenge. Computerized Medical Imaging and Graphics 97, 102049. [DOI] [PubMed] [Google Scholar]
- Girshick R, Donahue J, Darrell T, Malik J, 2014. Rich feature hierarchies for accurate object detection and semantic segmentation, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 580–587. [Google Scholar]
- Goodfellow I, Bengio Y, Courville A, 2016. Deep Learning. MIT Press. http://www.deeplearningbook.org. [Google Scholar]
- Grill JB, Strub F, Altché F, Tallec C, Richemond P, Buchatskaya E, Doersch C, Avila Pires B, Guo Z, Gheshlaghi Azar M, et al. , 2020. Bootstrap your own latent-a new approach to self-supervised learning. Advances in Neural Information Processing Systems 33, 21271–21284. [Google Scholar]
- Gu R, Wang G, Song T, Huang R, Aertsen M, Deprest J, Ourselin S, Vercauteren T, Zhang S, 2020. CA-Net: Comprehensive attention convolutional neural networks for explainable medical image segmentation. IEEE transactions on medical imaging 40, 699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Liu M, 2021. Domain adaptation for medical image analysis: a survey. IEEE Transactions on Biomedical Engineering . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunraj H, 2021. COVIDx CT-2A: A large-scale chest CT dataset for COVID-19 detection. https://www.kaggle.com/hgunraj/covidxct/. [Google Scholar]
- Gunraj H, Sabri A, Koff D, Wong A, 2021. COVID-Net CT-2: Enhanced deep neural networks for detection of COVID-19 from chest CT images through bigger, more diverse learning. arXiv preprint arXiv:2101.07433 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Han K, Wu H, Tang Y, Chen X, Wang Y, Xu C, 2022a. Cmt: Convolutional neural networks meet vision transformers, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 12175–12185. [Google Scholar]
- Guo J, Tang Y, Han K, Chen X, Wu H, Xu C, Xu C, Wang Y, 2022b. Hire-mlp: Vision mlp via hierarchical rearrangement, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 826–836. [Google Scholar]
- Guo MH, Lu CZ, Liu ZN, Cheng MM, Hu SM, 2022c. Visual attention network. arXiv preprint arXiv:2202.09741 . [Google Scholar]
- Guo P, Mei Y, Zhou J, Jiang S, Patel VM, 2022d. ReconFormer: Accelerated MRI reconstruction using recurrent transformer. arXiv preprint arXiv:2201.09376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Wang Y, Chen H, Chen X, Guo J, Liu Z, Tang Y, Xiao A, Xu C, Xu Y, et al. , 2020. A survey on visual transformer. arXiv preprint arXiv:2012.12556 2. [Google Scholar]
- Han K, Xiao A, Wu E, Guo J, Xu C, Wang Y, 2021a. Transformer in transformer. Advances in Neural Information Processing Systems 34. [Google Scholar]
- Han Q, Fan Z, Dai Q, Sun L, Cheng MM, Liu J, Wang J, 2021b. On the connection between local attention and dynamic depth-wise convolution, in: International Conference on Learning Representations. [Google Scholar]
- Han Y, Ye JC, 2018. Framing U-Net via deep convolutional framelets: Application to sparse-view CT. IEEE transactions on medical imaging 37, 1418–1429. [DOI] [PubMed] [Google Scholar]
- Han Z, Jian M, Wang GG, 2022. ConvUNeXt: an efficient convolution neural network for medical image segmentation. Knowledge-Based Systems 253, 109512. [Google Scholar]
- Hao Y, Dong L, Wei F, Xu K, 2021. Self-attention attribution: Interpreting information interactions inside transformer, in: Proceedings of the AAAI Conference on Artificial Intelligence, pp. 12963–12971. [Google Scholar]
- Hatamizadeh A, Hosseini H, Liu Z, Schwartz SD, Terzopoulos D, 2019. Deep dilated convolutional nets for the automatic segmentation of retinal vessels. arXiv preprint arXiv:1905.12120 . [Google Scholar]
- Hatamizadeh A, Nath V, Tang Y, Yang D, Roth H, Xu D, 2022a. Swin UNETR: Swin transformers for semantic segmentation of brain tumors in MRI images. arXiv preprint arXiv:2201.01266 . [Google Scholar]
- Hatamizadeh A, Tang Y, Nath V, Yang D, Myronenko A, Landman B, Roth H, Xu D, 2021. UNETR: Transformers for 3D medical image segmentation. arXiv preprint arXiv:2103.10504 . [Google Scholar]
- Hatamizadeh A, Tang Y, Nath V, Yang D, Myronenko A, Landman B, Roth HR, Xu D, 2022b. UNETR: Transformers for 3D medical image segmentation, in: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 574–584. [Google Scholar]
- He K, Chen X, Xie S, Li Y, Dollár P, Girshick R, 2022. Masked autoencoders are scalable vision learners, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 16000–16009. [Google Scholar]
- He K, Fan H, Wu Y, Xie S, Girshick R, 2020a. Momentum contrast for unsupervised visual representation learning, in: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp. 9729–9738. [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2016. Deep residual learning for image recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778. [Google Scholar]
- He S, Grant PE, Ou Y, 2021a. Global-local transformer for brain age estimation. IEEE Transactions on Medical Imaging 41, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Wang S, Shi S, Chu X, Tang J, Liu X, Yan C, Zhang J, Ding G, 2020b. Benchmarking deep learning models and automated model design for COVID-19 detection with chest CT scans. MedRxiv . [Google Scholar]
- He Y, Yang D, Roth H, Zhao C, Xu D, 2021b. DiNTS: Differentiable neural network topology search for 3D medical image segmentation, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. [Google Scholar]
- Heller N, Isensee F, Maier-Hein KH, Hou X, Xie C, Li F, Nan Y, Mu G, Lin Z, Han M, et al. , 2021. The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 challenge. Medical image analysis 67, 101821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrycks D, Gimpel K, 2016. Gaussian error linear units (gelus). arXiv preprint arXiv:1606.08415 . [Google Scholar]
- Hochreiter S, Schmidhuber J, 1997. Flat minima. Neural computation 9, 1–42. [DOI] [PubMed] [Google Scholar]
- Holger R, Amal F, 2016. Turkbey evrim, lu le, liu jiamin, and summers ronald. data from pancreas-CT. Cancer Imaging Archive . [Google Scholar]
- Holmes AJ, Hollinshead MO, OâĂŹkeefe TM, Petrov VI, Fariello GR, Wald LL, Fischl B, Rosen BR, Mair RW, Roffman JL, et al. , 2015. Brain genomics superstruct project initial data release with structural, functional, and behavioral measures. Scientific data 2, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shen L, Sun G, 2018. Squeeze-and-excitation networks, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 7132–7141. [Google Scholar]
- Huang G, Liu Z, Van Der Maaten L, Weinberger KQ, 2017. Densely connected convolutional networks, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708. [Google Scholar]
- Huang J, Xing X, Gao Z, Yang G, 2022a. Swin deformable attention u-net transformer (sdaut) for explainable fast mri, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 538–548. [Google Scholar]
- Huang Q, Sun J, Ding H, Wang X, Wang G, 2018. Robust liver vessel extraction using 3D U-Net with variant dice loss function. Computers in biology and medicine 101, 153–162. [DOI] [PubMed] [Google Scholar]
- Huang S, Li J, Xiao Y, Shen N, Xu T, 2022b. RTNet: Relation transformer network for diabetic retinopathy multi-lesion segmentation. IEEE Transactions on Medical Imaging . [DOI] [PubMed] [Google Scholar]
- Huang X, Deng Z, Li D, Yuan X, 2021. MISSFormer: An effective medical image segmentation transformer. arXiv preprint arXiv:2109.07162 . [DOI] [PubMed] [Google Scholar]
- Huang Z, Liang D, Xu P, Xiang B, 2020. Improve transformer models with better relative position embeddings, in: Findings of the Association for Computational Linguistics: EMNLP 2020, Association for Computational Linguistics, Online. pp. 3327–3335. [Google Scholar]
- Hughes EJ, Winchman T, Padormo F, Teixeira R, Wurie J, Sharma M, Fox M, Hutter J, Cordero-Grande L, Price AN, Allsop J, Bueno-Conde J, Tusor N, Arichi T, Edwards AD, Rutherford MA, Counsell SJ, Hajnal JV, 2017. A dedicated neonatal brain imaging system. Magnetic Resonance in Medicine 78, 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K, et al. , 2019. CheXpert: A large chest radiograph dataset with uncertainty labels and expert comparison, in: Proceedings of the AAAI conference on artificial intelligence, pp. 590–597. [Google Scholar]
- Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH, 2021. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods 18, 203–211. [DOI] [PubMed] [Google Scholar]
- Jaderberg M, Simonyan K, Zisserman A, et al. , 2015. Spatial transformer networks. Advances in neural information processing systems 28. [Google Scholar]
- Jaeger S, Candemir S, Antani S, Wáng YXJ, Lu PX, Thoma G, 2014. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quantitative imaging in medicine and surgery 4, 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamaludin A, Lootus M, Kadir T, Zisserman A, Urban J, Battié MC, Fairbank J, McCall I, 2017. Automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. European Spine Journal 26, 1374–1383. [DOI] [PubMed] [Google Scholar]
- Jang J, Hwang D, 2022. M3T: Three-dimensional medical image classifier using multi-plane and multi-slice transformer, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 20718–20729. [Google Scholar]
- Jha D, Smedsrud PH, Riegler MA, Halvorsen P, Lange T.d., Johansen D, Johansen HD, 2020. Kvasir-SEG: A segmented polyp dataset, in: International Conference on Multimedia Modeling, Springer. pp. 451–462. [Google Scholar]
- Ji Y, Zhang R, Wang H, Li Z, Wu L, Zhang S, Luo P, 2021. Multi-compound transformer for accurate biomedical image segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 326–336. [Google Scholar]
- Jia Q, Shu H, 2021. BiTr-Unet: a CNN-transformer combined network for MRI brain tumor segmentation. arXiv preprint arXiv:2109.12271 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Bartlett J, Zhang T, Lu W, Qiu Z, Duan J, 2022. U-Net vs Transformer: Is U-Net outdated in medical image registration? arXiv preprint arXiv:2208.04939 . [Google Scholar]
- Jiang H, Zhang P, Che C, Jin B, 2021. RDFNet: A fast caries detection method incorporating transformer mechanism. Computational and Mathematical Methods in Medicine 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Tyagi N, Tringale K, Crane C, Veeraraghavan H, 2022. Self-supervised 3d anatomy segmentation using self-distilled masked image transformer (smit). arXiv preprint arXiv:2205.10342 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Eberhart CG, Lim M, Heo HY, Zhang Y, Blair L, Wen Z, Holdhoff M, Lin D, Huang P, et al. , 2019a. Identifying recurrent malignant glioma after treatment using amide proton transfer-weighted MR imaging: a validation study with image-guided stereotactic biopsy. Clinical Cancer Research 25, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Neyshabur B, Mobahi H, Krishnan D, Bengio S, 2019b. Fantastic generalization measures and where to find them, in: International Conference on Learning Representations. [Google Scholar]
- Jose J, Oza P, 2021. Medical transformer: gated axial-attention for medical image segmentation, in: International Conference on Medical Image Computing and Computer Assisted Intervention. [Google Scholar]
- Joshi C, 2020. Transformers are graph neural networks. The Gradient; . [Google Scholar]
- Jun E, Jeong S, Heo DW, Suk HI, 2021. Medical transformer: Universal brain encoder for 3D MRI analysis. arXiv preprint arXiv:2104.13633 . [DOI] [PubMed] [Google Scholar]
- Kan X, Dai W, Cui H, Zhang Z, Guo Y, Yang C, . Brain network transformer . [Google Scholar]
- Karimi D, Vasylechko SD, Gholipour A, 2021. Convolution-free medical image segmentation using transformers, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 78–88. [Google Scholar]
- Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis CA, Gaiser T, Marx A, Valous NA, Ferber D, et al. , 2019. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS medicine 16, e1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauderer-Abrams E, 2017. Quantifying translation-invariance in convolutional neural networks. arXiv preprint arXiv:1801.01450 . [Google Scholar]
- Kavur AE, Gezer NS, BarÄśÅ§ M, Åđahin Y, ÃŰzkan S, Baydar B, YÃijksel U, KÄślÄśkÃğÄśer Ã, Olut Å, BozdaħĜ Akar G, ÃIJnal G, Dicle O, Selver MA, 2020. Comparison of semi-automatic and deep learning based automatic methods for liver segmentation in living liver transplant donors. Diagnostic and Interventional Radiology 26, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy DN, Haselgrove C, Hodge SM, Rane PS, Makris N, Frazier JA, 2012. CANDIShare: a resource for pediatric neuroimaging data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskar NS, Mudigere D, Nocedal J, Smelyanskiy M, Tang PTP, 2017. On large-batch training for deep learning: Generalization gap and sharp minima, in: International Conference on Learning Representations. [Google Scholar]
- Kim BH, Ye JC, Kim JJ, 2021a. Learning dynamic graph representation of brain connectome with spatio-temporal attention, in: Ranzato M, Beygelzimer A, Dauphin Y, Liang P, Vaughan JW, (Eds.), Advances in Neural Information Processing Systems, Curran Associates, Inc.. pp. 4314–4327. [Google Scholar]
- Kim K, Wu D, Gong K, Dutta J, Kim JH, Son YD, Kim HK, El Fakhri G, Li Q, 2018. Penalized PET reconstruction using deep learning prior and local linear fitting. IEEE transactions on medical imaging 37, 1478–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Jang H, Lee K, Park S, Min SG, Hong C, Park JH, Lee K, Kim J, Hong W, et al. , 2021b. PAIP 2019: Liver cancer segmentation challenge. Medical Image Analysis 67, 101854. [DOI] [PubMed] [Google Scholar]
- Knoll F, Zbontar J, Sriram A, Muckley MJ, Bruno M, Defazio A, Parente M, Geras KJ, Katsnelson J, Chandarana H, et al. , 2020. fastMRI: A publicly available raw k-space and DICOM dataset of knee images for accelerated MR image reconstruction using machine learning. Radiology: Artificial Intelligence 2, e190007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz Y, Dar SU, Yurt M, Özbey M, Cukur T, 2022. Unsupervised MRI reconstruction via zero-shot learned adversarial transformers. IEEE Transactions on Medical Imaging . [DOI] [PubMed] [Google Scholar]
- Krishna S, Han T, Gu A, Pombra J, Jabbari S, Wu S, Lakkaraju H, 2022. The disagreement problem in explainable machine learning: A practitioner’s perspective. arXiv preprint arXiv:2202.01602 . [Google Scholar]
- Krizhevsky A, Sutskever I, Hinton GE, 2012. ImageNet classification with deep convolutional neural networks, in: Advances in Neural Information Processing Systems (NIPS), pp. 1097–1105. [Google Scholar]
- Kumar N, Verma R, Anand D, Zhou Y, Onder OF, Tsougenis E, Chen H, Heng PA, Li J, Hu Z, et al. , 2019. A multi-organ nucleus segmentation challenge. IEEE transactions on medical imaging 39, 1380–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert Z, Petitjean C, Dubray B, Kuan S, 2020. SegTHOR: Segmentation of thoracic organs at risk in CT images, in: 2020 Tenth International Conference on Image Processing Theory, Tools and Applications (IPTA), IEEE. pp. 1–6. [Google Scholar]
- LaMontagne PJ, Benzinger TL, Morris JC, Keefe S, Hornbeck R, Xiong C, Grant E, Hassenstab J, Moulder K, Vlassenko AG, et al. , 2019. OASIS-3: longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and alzheimer disease. MedRxiv . [Google Scholar]
- Landman B, Xu Z, Igelsias J, Styner M, Langerak T, Klein A, 2015. MICCAI multi-atlas labeling beyond the cranial vault–workshop and challenge, in: Proc. MICCAI Multi-Atlas Labeling Beyond Cranial VaultâĂŤWorkshop Challenge, p. 12. [Google Scholar]
- LeCun Y, Bengio Y, Hinton G, 2015. Deep learning. Nature 521, 436–444. [DOI] [PubMed] [Google Scholar]
- Lee RS, Gimenez F, Hoogi A, Miyake KK, Gorovoy M, Rubin DL, 2017. A curated mammography data set for use in computer-aided detection and diagnosis research. Scientific data 4, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner J, Schmidt M, Baguer DO, Maass P, 2021. LoDoPaB-CT, a benchmark dataset for low-dose computed tomography reconstruction. Scientific Data 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Lyu J, Wang C, Dou Q, Qin J, 2022a. Wavtrans: Synergizing wavelet and cross-attention transformer for multi-contrast mri super-resolution, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 463–473. [Google Scholar]
- Li H, Chen L, Han H, Zhou SK, 2022b. SATr: Slice attention with transformer for universal lesion detection. arXiv preprint arXiv:2203.07373 . [Google Scholar]
- Li H, Xu Z, Taylor G, Studer C, Goldstein T, 2018. Visualizing the loss landscape of neural nets. Advances in neural information processing systems 31. [Google Scholar]
- Li J, Wang W, Chen C, Zhang T, Zha S, Yu H, Wang J, 2022c. TransBTSV2: Wider instead of deeper transformer for medical image segmentation. arXiv preprint arXiv:2201.12785 . [Google Scholar]
- Li M, Chen Y, Ji Z, Xie K, Yuan S, Chen Q, Li S, 2020. Image projection network: 3D to 2D image segmentation in OCTA images. IEEE Transactions on Medical Imaging 39, 3343–3354. [DOI] [PubMed] [Google Scholar]
- Li R, Mai Z, Trabelsi C, Zhang Z, Jang J, Sanner S, 2022d. TransCAM: Transformer attention-based CAM refinement for weakly supervised semantic segmentation. arXiv preprint arXiv:2203.07239 . [Google Scholar]
- Li R, Zhang W, Suk HI, Wang L, Li J, Shen D, Ji S, 2014. Deep learning based imaging data completion for improved brain disease diagnosis, in: International conference on medical image computing and computer-assisted intervention, Springer. pp. 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sui X, Luo X, Xu X, Liu Y, Goh R, 2021a. Medical image segmentation using squeeze-and-expansion transformers. arXiv preprint arXiv:2105.09511 . [Google Scholar]
- Li Y, Cai W, Gao Y, Hu X, 2021b. More than encoder: Introducing transformer decoder to upsample. arXiv preprint arXiv:2106.10637 . [Google Scholar]
- Li Y, Mao H, Girshick R, He K, 2022e. Exploring plain vision transformer backbones for object detection. arXiv preprint arXiv:2203.16527 . [Google Scholar]
- Li Y, Wang S, Wang J, Zeng G, Liu W, Zhang Q, Jin Q, Wang Y, 2021c. GT U-Net: A U-Net like group transformer network for tooth root segmentation, in: International Workshop on Machine Learning in Medical Imaging, Springer. pp. 386–395. [Google Scholar]
- Li Y, Wang Z, Yin L, Zhu Z, Qi G, Liu Y, 2021d. X-Net: a dual encoding–decoding method in medical image segmentation. The Visual Computer , 1–11.34744231 [Google Scholar]
- Li Y, Yao T, Pan Y, Mei T, 2021e. Contextual transformer networks for visual recognition. arXiv preprint arXiv:2107.12292 . [DOI] [PubMed] [Google Scholar]
- Li Y, Zou B, Dai Y, Zhu C, Yang F, Li X, Bai HX, Jiao Z, 2022f. Parameter-free latent space transformer for zero-shot bidirectional cross-modality liver segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 619–628. [Google Scholar]
- Liang M, Hu X, 2015. Recurrent convolutional neural network for object recognition, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 3367–3375. [Google Scholar]
- Lin A, Chen B, Xu J, Zhang Z, Lu G, 2021. DS-TransUNet: Dual swin transformer U-Net for medical image segmentation. arXiv preprint arXiv:2106.06716 . [Google Scholar]
- Lin A, Xu J, Li J, Lu G, 2022a. ConTrans: Improving transformer with convolutional attention for medical image segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 297–307. [Google Scholar]
- Lin W, Liu H, Gu L, Gao Z, 2022b. A geometry-constrained deformable attention network for aortic segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 287–296. [Google Scholar]
- Liu D, Zhou SK, Bernhardt D, Comaniciu D, 2010. Search strategies for multiple landmark detection by submodular maximization, in: Computer Vision and Pattern Recognition (CVPR), 2010 IEEE Conference on, IEEE. pp. 2831–2838. [Google Scholar]
- Liu L, Huang Z, Liò P, Schönlieb CB, Aviles-Rivero AI, 2022a. PC-SwinMorph: Patch representation for unsupervised medical image registration and segmentation. arXiv preprint arXiv:2203.05684 . [Google Scholar]
- Liu Q, Xu Z, Jiao Y, Niethammer M, 2022b. iSegFormer: Interactive segmentation via transformers with application to 3d knee mr images, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 464–474. [Google Scholar]
- Liu Y, Liu J, Yuan Y, 2022c. Edge-oriented point-cloud transformer for 3D intracranial aneurysm segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 97–106. [Google Scholar]
- Liu Y, Sangineto E, Bi W, Sebe N, Lepri B, Nadai M, 2021a. Efficient training of visual transformers with small datasets. Advances in Neural Information Processing Systems 34. [Google Scholar]
- Liu Y, Zuo L, Han S, Xue Y, Prince JL, Carass A, 2022d. Coordinate translator for learning deformable medical image registration, in: Multiscale Multi-modal Medical Imaging: Third International Workshop, MMMI 2022, Held in Conjunction with MICCAI 2022, Singapore, September 22, 2022, Proceedings, Springer. pp. 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B, 2021b. Swin transformer: Hierarchical vision transformer using shifted windows, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10012–10022. [Google Scholar]
- Liu Z, Mao H, Wu CY, Feichtenhofer C, Darrell T, Xie S, 2022e. A ConvNet for the 2020s. arXiv preprint arXiv:2201.03545 . [Google Scholar]
- Liu Z, Shen L, 2022. Medical image analysis based on transformer: A review. arXiv preprint arXiv:2208.06643 . [Google Scholar]
- Lo SC, Lou SL, Lin JS, Freedman MT, Chien MV, Mun SK, 1995a. Artificial convolution neural network techniques and applications for lung nodule detection. IEEE transactions on medical imaging 14, 711–718. [DOI] [PubMed] [Google Scholar]
- Lo SCB, Chan HP, Lin JS, Li H, Freedman MT, Mun SK, 1995b. Artificial convolution neural network for medical image pattern recognition. Neural networks 8, 1201–1214. [Google Scholar]
- Long J, Shelhamer E, Darrell T, 2015. Fully convolutional networks for semantic segmentation, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- Lu J, Yao J, Zhang J, Zhu X, Xu H, Gao W, Xu C, Xiang T, Zhang L, 2021. SOFT: softmax-free transformer with linear complexity. Advances in Neural Information Processing Systems 34, 21297–21309. [Google Scholar]
- Luo W, Li Y, Urtasun R, Zemel R, 2016. Understanding the effective receptive field in deep convolutional neural networks, in: Proceedings of the 30th International Conference on Neural Information Processing Systems. [Google Scholar]
- Luo X, Hu M, Song T, Wang G, Zhang S, 2021. Semi-supervised medical image segmentation via cross teaching between CNN and transformer. arXiv preprint arXiv:2112.04894 . [Google Scholar]
- Luthra A, Sulakhe H, Mittal T, Iyer A, Yadav S, 2021. Eformer: Edge enhancement based transformer for medical image denoising. arXiv preprint arXiv:2109.08044 . [Google Scholar]
- Lv Z, Yan R, Lin Y, Wang Y, Zhang F, 2022. Joint region-attention and multi-scale transformer for microsatellite instability detection from whole slide images in gastrointestinal cancer, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 293–302. [Google Scholar]
- Lyu J, Sui B, Wang C, Tian Y, Dou Q, Qin J, 2022. Dudocaf: Dual-domain cross-attention fusion with recurrent transformer for fast multi-contrast mr imaging, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 474–484. [Google Scholar]
- Ma X, Luo G, Wang W, Wang K, 2021a. Transformer network for significant stenosis detection in CCTA of coronary arteries, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 516–525. [Google Scholar]
- Ma Y, Chen X, Cheng K, Li Y, Sun B, 2021b. Ldpolypvideo benchmark: A large-scale colonoscopy video dataset of diverse polyps, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 387–396. [Google Scholar]
- Malík P, Krištofík Š, Knapová K, 2020. Instance segmentation model created from three semantic segmentations of mask, boundary and centroid pixels verified on GlaS dataset, in: 2020 15th Conference on Computer Science and Information Systems (FedCSIS), IEEE. pp. 569–576. [Google Scholar]
- Malon CD, Cosatto E, 2013. Classification of mitotic figures with convolutional neural networks and seeded blob features. Journal of pathology informatics 4, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL, 2007. Open access series of imaging studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. Journal of cognitive neuroscience 19, 1498–1507. [DOI] [PubMed] [Google Scholar]
- Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, Coffey C, Kieburtz K, Flagg E, Chowdhury S, et al. , 2011. The parkinson progression marker initiative (PPMI). Progress in neurobiology 95, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathai TS, Lee S, Elton DC, Shen TC, Peng Y, Lu Z, Summers RM, 2022. Lymph node detection in T2 MRI with transformers, in: Medical Imaging 2022: Computer-Aided Diagnosis, SPIE. pp. 855–859. [Google Scholar]
- Matsoukas C, Haslum JF, Söderberg M, Smith K, 2021. Is it time to replace CNNs with transformers for medical images? arXiv preprint arXiv:2108.09038 . [Google Scholar]
- McCollough C, 2016. TU-FG-207A-04: overview of the low dose CT grand challenge. Medical physics 43, 3759–3760. [Google Scholar]
- Mendes N, Oligschläger S, Lauckner ME, Golchert J, Huntenburg JM, Falkiewicz M, Ellamil M, Krause S, Baczkowski BM, Cozatl R, et al. , 2019. A functional connectome phenotyping dataset including cognitive state and personality measures. Scientific data 6, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonça T, Ferreira PM, Marques JS, Marcal AR, Rozeira J, 2013. PH 2 - A dermoscopic image database for research and benchmarking, in: 2013 35th annual international conference of the IEEE engineering in medicine and biology society (EMBC), IEEE. pp. 5437–5440. [DOI] [PubMed] [Google Scholar]
- Mendrik AM, Vincken KL, Kuijf HJ, Breeuwer M, Bouvy WH, De Bresser J, Alansary A, De Bruijne M, Carass A, El-Baz A, et al. , 2015. MRBrainS challenge: online evaluation framework for brain image segmentation in 3T MRI scans. Computational intelligence and neuroscience 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhang X, Wang G, Zhang Y, Shi X, Dai H, Wang Z, Wang X, 2021. Exploiting full resolution feature context for liver tumor and vessel segmentation via fusion encoder: Application to liver tumor and vessel 3D reconstruction. arXiv preprint arXiv:2111.13299 . [Google Scholar]
- Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, et al. , 2014. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE transactions on medical imaging 34, 1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao S, Wang ZJ, Liao R, 2016. A CNN regression approach for real-time 2d/3d registration. IEEE transactions on medical imaging 35, 1352–1363. [DOI] [PubMed] [Google Scholar]
- Milea D, Najjar RP, Jiang Z, Ting D, Vasseneix C, Xu X, Aghsaei Fard M, Fonseca P, Vanikieti K, Lagrèze WA, et al. , 2020. Artificial intelligence to detect papilledema from ocular fundus photographs. New England Journal of Medicine 382, 1687–1695. [DOI] [PubMed] [Google Scholar]
- Milletari F, Navab N, Ahmadi SA, 2016. V-Net: Fully convolutional neural networks for volumetric medical image segmentation, in: 2016 fourth international conference on 3D vision (3DV), IEEE. pp. 565–571. [Google Scholar]
- Mok TC, Chung A, 2022. Affine medical image registration with coarse-to-fine vision transformer, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 20835–20844. [Google Scholar]
- Mondal AK, Bhattacharjee A, Singla P, Prathosh A, 2021. xViTCOS: Explainable vision transformer based COVID-19 screening using radiography. IEEE Journal of Translational Engineering in Health and Medicine 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L, 2005. Ways toward an early diagnosis in AlzheimerâĂŹs disease: the AlzheimerâĂŹs disease neuroimaging initiative (ADNI). Alzheimer’s & Dementia 1, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer MM, Ranasinghe K, Khan SH, Hayat M, Shahbaz Khan F, Yang MH, 2021. Intriguing properties of vision transformers. Advances in Neural Information Processing Systems 34, 23296–23308. [Google Scholar]
- Naylor P, Laé M, Reyal F, Walter T, 2018. Segmentation of nuclei in histopathology images by deep regression of the distance map. IEEE transactions on medical imaging 38, 448–459. [DOI] [PubMed] [Google Scholar]
- Nguyen C, Asad Z, Huo Y, 2021. Evaluating transformer-based semantic segmentation networks for pathological image segmentation. arXiv preprint arXiv:2108.11993 . [Google Scholar]
- Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, et al. , 2018. Attention U-Net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 . [Google Scholar]
- Oreiller V, Andrearczyk V, Jreige M, Boughdad S, Elhalawani H, Castelli J, Vallières M, Zhu S, Xie J, Peng Y, et al. , 2022. Head and neck tumor segmentation in PET/CT: the HECKTOR challenge. Medical image analysis 77, 102336. [DOI] [PubMed] [Google Scholar]
- Orlando JI, Fu H, Breda JB, van Keer K, Bathula DR, Diaz-Pinto A, Fang R, Heng PA, Kim J, Lee J, et al. , 2020. REFUGE challenge: A unified framework for evaluating automated methods for glaucoma assessment from fundus photographs. Medical image analysis 59, 101570. [DOI] [PubMed] [Google Scholar]
- Ou Y, Yuan Y, Huang X, Wong ST, Volpi J, Wang JZ, Wong K, 2022. Patcher: Patch transformers with mixture of experts for precise medical image segmentation. arXiv preprint arXiv:2206.01741 . [Google Scholar]
- Ouyang D, He B, Ghorbani A, Yuan N, Ebinger J, Langlotz CP, Heidenreich PA, Harrington RA, Liang DH, Ashley EA, et al. , 2020. Video-based ai for beat-to-beat assessment of cardiac function. Nature 580, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachade S, Porwal P, Thulkar D, Kokare M, Deshmukh G, Sahasrabuddhe V, Giancardo L, Quellec G, Mériaudeau F, 2021. Retinal fundus multi-disease image dataset (RFMiD): a dataset for multi-disease detection research. Data 6, 14. [Google Scholar]
- Pan J, Wu W, Gao Z, Zhang H, 2021. MIST-net: Multi-domain integrative swin transformer network for sparse-view CT reconstruction. arXiv preprint arXiv:2111.14831 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Carp J, Kennedy KM, Rodrigue KM, Bischof GN, Huang CM, Rieck JR, Polk TA, Park DC, 2012. Neural broadening or neural attenuation? investigating age-related dedifferentiation in the face network in a large lifespan sample. Journal of Neuroscience 32, 2154–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park N, Kim S, 2022. How do vision transformers work? arXiv preprint arXiv:2202.06709 . [Google Scholar]
- Park S, Kim G, Oh Y, Seo JB, Lee SM, Kim JH, Moon S, Lim JK, Ye JC, 2021. Vision transformer for COVID-19 CXR diagnosis using chest X-ray feature corpus. arXiv preprint arXiv:2103.07055 . [Google Scholar]
- Parvaiz A, Khalid MA, Zafar R, Ameer H, Ali M, Fraz MM, 2022. Vision transformers in medical computer vision-a contemplative retrospection. arXiv preprint arXiv:2203.15269 . [Google Scholar]
- Pavlova M, Terhljan N, Chung AG, Zhao A, Surana S, Aboutalebi H, Gunraj H, Sabri A, Alaref A, Wong A, 2021. COVID-Net CXR-2: An enhanced deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. arXiv preprint arXiv:2105.06640 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer C, Štern D, Bischof H, Urschler M, 2019. Integrating spatial configuration into heatmap regression based CNNs for landmark localization. Medical image analysis 54, 207–219. [DOI] [PubMed] [Google Scholar]
- Peiris H, Hayat M, Chen Z, Egan G, Harandi M, 2021. A volumetric transformer for accurate 3D tumor segmentation. arXiv preprint arXiv:2111.13300 . [Google Scholar]
- Peiris H, Hayat M, Chen Z, Egan G, Harandi M, 2022. A robust volumetric transformer for accurate 3d tumor segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 162–172. [Google Scholar]
- Peng Z, Huang W, Gu S, Xie L, Wang Y, Jiao J, Ye Q, 2021. Conformer: Local features coupling global representations for visual recognition, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 367–376. [Google Scholar]
- Petersen RC, Aisen P, Beckett LA, Donohue M, Gamst A, Harvey DJ, Jack C, Jagust W, Shaw L, Toga A, et al. , 2010. Alzheimer’s disease neuroimaging initiative (ADNI): clinical characterization. Neurology 74, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit O, Thome N, Rambour C, Themyr L, Collins T, Soler L, 2021. U-Net transformer: Self and cross attention for medical image segmentation, in: International Workshop on Machine Learning in Medical Imaging, Springer. pp. 267–276. [Google Scholar]
- Płotka S, Grzeszczyk MK, Brawura-Biskupski-Samaha R, Gutaj P, Lipa M, Trzciński T, Sitek A, 2022. BabyNet: Residual transformer module for birth weight prediction on fetal ultrasound video, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 350–359. [Google Scholar]
- Porwal P, Pachade S, Kamble R, Kokare M, Deshmukh G, Sahasrabuddhe V, Meriaudeau F, 2018. Indian diabetic retinopathy image dataset (IDRiD): a database for diabetic retinopathy screening research. Data 3, 25. [Google Scholar]
- Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M, 2013. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network, in: International conference on medical image computing and computer-assisted intervention, Springer. pp. 246–253. [DOI] [PubMed] [Google Scholar]
- Qian Z, Li K, Lai M, Chang EIC, Wei B, Fan Y, Xu Y, 2022. Transformer based multiple instance learning for weakly supervised histopathology image segmentation, in: MICCAI. [Google Scholar]
- Qin Z, Sun W, Deng H, Li D, Wei Y, Lv B, Yan J, Kong L, Zhong Y, 2022. cosFormer: Rethinking softmax in attention, in: International Conference on Learning Representations. [Google Scholar]
- Radford A, Narasimhan K, Salimans T, Sutskever I, 2018. Improving language understanding by generative pre-training . [Google Scholar]
- Radford A, Wu J, Child R, Luan D, Amodei D, Sutskever I, et al. ,. Language models are unsupervised multitask learners . [Google Scholar]
- Raffel C, Shazeer N, Roberts A, Lee K, Narang S, Matena M, Zhou Y, Li W, Liu PJ, et al. , 2020. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res 21, 1–67.34305477 [Google Scholar]
- Raghu M, Unterthiner T, Kornblith S, Zhang C, Dosovitskiy A, 2021. Do vision transformers see like convolutional neural networks? Advances in Neural Information Processing Systems 34. [Google Scholar]
- Redmon J, Divvala S, Girshick R, Farhadi A, 2016. You only look once: Unified, real-time object detection, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 779–788. [Google Scholar]
- Reisenbüchler D, Wagner SJ, Boxberg M, Peng T, 2022. Local attention graph-based transformer for multi-target genetic alteration prediction. arXiv preprint arXiv:2205.06672. [Google Scholar]
- Ren S, Gao Z, Hua T, Xue Z, Tian Y, He S, Zhao H, 2022. Co-advise: Cross inductive bias distillation, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 16773–16782. [Google Scholar]
- RIADD, 2020. Retinal image analysis for multi-disease detection challenge. https://riadd.grand-challenge.org/. [Google Scholar]
- Ronneberger O, Fischer P, Brox T, 2015. U-Net: Convolutional networks for biomedical image segmentation, in: International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Springer. pp. 234–241. [Google Scholar]
- Roth HR, Lee CT, Shin HC, Seff A, Kim L, Yao J, Lu L, Summers RM, 2015. Anatomy-specific classification of medical images using deep convolutional nets, in: 2015 IEEE 12th international symposium on biomedical imaging (ISBI), IEEE. pp. 101–104. [Google Scholar]
- Roth HR, Lu L, Seff A, Cherry KM, Hoffman J, Wang S, Liu J, Turkbey E, Summers RM, 2014. A new 2.5 D representation for lymph node detection using random sets of deep convolutional neural network observations, in: International conference on medical image computing and computer-assisted intervention, Springer. pp. 520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri A, Scarpa F, De Luca M, Meltendorf C, Schroeter J, 2010. A system for the automatic estimation of morphometric parameters of corneal endothelium in alizarine red-stained images. British Journal of Ophthalmology 94, 643–647. [DOI] [PubMed] [Google Scholar]
- Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, et al. , 2015. ImageNet large scale visual recognition challenge. International journal of computer vision 115, 211–252. [Google Scholar]
- Saeed N, Sobirov I, Al Majzoub R, Yaqub M, 2022. TMSS: An end-to-end transformer-based multimodal network for segmentation and survival prediction, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 319–329. [Google Scholar]
- Sahiner B, Chan HP, Petrick N, Wei D, Helvie MA, Adler DD, Goodsitt MM, 1996. Classification of mass and normal breast tissue: a convolution neural network classifier with spatial domain and texture images. IEEE transactions on Medical Imaging 15, 598–610. [DOI] [PubMed] [Google Scholar]
- Scherer D, Müller A, Behnke S, 2010. Evaluation of pooling operations in convolutional architectures for object recognition, in: International conference on artificial neural networks, Springer. pp. 92–101. [Google Scholar]
- Segars W, Bond J, Frush J, Hon S, Eckersley C, Williams CH, Feng J, Tward DJ, Ratnanather J, Miller M, et al. , 2013. Population of anatomically variable 4D XCAT adult phantoms for imaging research and optimization. Medical physics 40, 043701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamout FE, Shen Y, Wu N, Kaku A, Park J, Makino T, Jastrzębski S, Witowski J, Wang D, Zhang B, et al. , 2021. An artificial intelligence system for predicting the deterioration of COVID-19 patients in the emergency department. NPJ digital medicine 4, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamshad F, Khan S, Zamir SW, Khan MH, Hayat M, Khan FS, Fu H, 2022. Transformers in medical imaging: A survey. arXiv preprint arXiv:2201.09873 . [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW, 2008. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 39, 1064–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Uszkoreit J, Vaswani A, 2018. Self-attention with relative position representations, in: NAACL-HLT (2), pp. 464–468. [Google Scholar]
- Shen Z, Fu R, Lin C, Zheng S, 2021a. COTR: Convolution in transformer network for end to end polyp detection, in: 2021 7th International Conference on Computer and Communications (ICCC), IEEE. pp. 1757–1761. [Google Scholar]
- Shen Z, Yang H, Zhang Z, Zheng S, 2021b. Automated kidney tumor segmentation with convolution and transformer network. [Google Scholar]
- Shi J, He Y, Kong Y, Coatrieux JL, Shu H, Yang G, Li S, 2022. XMorpher: Full transformer for deformable medical image registration via cross attention, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 217–226. [Google Scholar]
- Shiraishi J, Katsuragawa S, Ikezoe J, Matsumoto T, Kobayashi T, Komatsu K.i., Matsui M, Fujita H, Kodera Y, Doi K, 2000. Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. American Journal of Roentgenology 174, 71–74. [DOI] [PubMed] [Google Scholar]
- Silva J, Histace A, Romain O, Dray X, Granado B, 2014. Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. International journal of computer assisted radiology and surgery 9, 283–293. [DOI] [PubMed] [Google Scholar]
- Simonovsky M, Gutiérrez-Becker B, Mateus D, Navab N, Komodakis N, 2016. A deep metric for multimodal registration, in: International conference on medical image computing and computer-assisted intervention, Springer. pp. 10–18. [Google Scholar]
- Simonyan K, Zisserman A, 2015. Very deep convolutional networks for large-scale image recognition, in: International Conference on Learning Representations. [Google Scholar]
- Simpson AL, Antonelli M, Bakas S, Bilello M, Farahani K, Van Ginneken B, Kopp-Schneider A, Landman BA, Litjens G, Menze B, et al. , 2019. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063 . [Google Scholar]
- Sirinukunwattana K, Pluim JP, Chen H, Qi X, Heng PA, Guo YB, Wang LY, Matuszewski BJ, Bruni E, Sanchez U, et al. , 2017. Gland segmentation in colon histology images: The glas challenge contest. Medical image analysis 35, 489–502. [DOI] [PubMed] [Google Scholar]
- Soler L, Hostettler A, Agnus V, Charnoz A, Fasquel J, Moreau J, Osswald A, Bouhadjar M, Marescaux J, 2010. 3D image reconstruction for comparison of algorithm database: A patient specific anatomical and medical image database. IRCAD, Strasbourg, France, Tech. Rep [Google Scholar]
- Souza R, Lucena O, Garrafa J, Gobbi D, Saluzzi M, Appenzeller S, Rittner L, Frayne R, Lotufo R, 2018. An open, multi-vendor, multi-field-strength brain mr dataset and analysis of publicly available skull stripping methods agreement. NeuroImage 170, 482–494. [DOI] [PubMed] [Google Scholar]
- Srinivas A, Lin TY, Parmar N, Shlens J, Abbeel P, Vaswani A, 2021. Bottleneck transformers for visual recognition, in: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp. 16519–16529. [Google Scholar]
- Sriram A, Muckley M, Sinha K, Shamout F, Pineau J, Geras KJ, Azour L, Aphinyanaphongs Y, Yakubova N, Moore W, 2021. Covid-19 prognosis via self-supervised representation learning and multi-image prediction. arXiv preprint arXiv:2101.04909 . [Google Scholar]
- Staal J, Abràmoff MD, Niemeijer M, Viergever MA, Van Ginneken B, 2004. Ridge-based vessel segmentation in color images of the retina. IEEE transactions on medical imaging 23, 501–509. [DOI] [PubMed] [Google Scholar]
- Sun Q, Fang N, Liu Z, Zhao L, Wen Y, Lin H, 2021a. HybridCTrm: Bridging CNN and transformer for multimodal brain image segmentation. Journal of Healthcare Engineering 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Cao S, Yang Y, Kitani KM, 2021b. Rethinking transformer-based set prediction for object detection, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 3611–3620. [Google Scholar]
- Tang Y, Yang D, Li W, Roth HR, Landman B, Xu D, Nath V, Hatamizadeh A, 2022. Self-supervised pre-training of swin transformers for 3d medical image analysis, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 20730–20740. [Google Scholar]
- Tang YB, Tang YX, Xiao J, Summers RM, 2019. Xlsor: A robust and accurate lung segmentor on chest x-rays using criss-cross attention and customized radiorealistic abnormalities generation, in: International Conference on Medical Imaging with Deep Learning, PMLR. pp. 457–467. [Google Scholar]
- Tian Y, Pang G, Liu F, Liu Y, Wang C, Chen Y, Verjans J, Carneiro G, 2022. Contrastive transformer-based multiple instance learning for weakly supervised polyp frame detection, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 88–98. [Google Scholar]
- Tolstikhin IO, Houlsby N, Kolesnikov A, Beyer L, Zhai X, Unterthiner T, Yung J, Steiner A, Keysers D, Uszkoreit J, et al. , 2021. MLP-Mixer: An all-MLP architecture for vision. Advances in Neural Information Processing Systems 34. [Google Scholar]
- Tomczak K, Czerwińska P, Wiznerowicz M, 2015. The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemporary oncology 19, A68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touvron H, Bojanowski P, Caron M, Cord M, El-Nouby A, Grave E, Izacard G, Joulin A, Synnaeve G, Verbeek J, et al. , 2021a. ResMLP: Feedforward networks for image classification with data-efficient training. arXiv preprint arXiv:2105.03404 . [DOI] [PubMed] [Google Scholar]
- Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jégou H, 2021b. Training data-efficient image transformers & distillation through attention, in: International Conference on Machine Learning, PMLR. pp. 10347–10357. [Google Scholar]
- Trockman A, Kolter JZ, 2022. Patches are all you need? arXiv preprint arXiv:2201.09792 . [Google Scholar]
- Tsai EB, Simpson S, Lungren MP, Hershman M, Roshkovan L, Colak E, Erickson BJ, Shih G, Stein A, Kalpathy-Cramer J, et al. , 2021. The RSNA international COVID-19 open radiology database (RICORD). Radiology 299, E204–E213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valanarasu JMJ, Oza P, Hacihaliloglu I, Patel VM, 2021. Medical transformer: Gated axial-attention for medical image segmentation. arXiv preprint arXiv:2102.10662. [Google Scholar]
- Valanarasu JMJ, Patel VM, 2022. UNeXt: MLP-based rapid medical image segmentation network. arXiv preprint arXiv:2203.04967 . [Google Scholar]
- Valanarasu JMJ, Yasarla R, Wang P, Hacihaliloglu I, Patel VM, 2020. Learning to segment brain anatomy from 2D ultrasound with less data. IEEE Journal of Selected Topics in Signal Processing 14, 1221–1234. [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, Consortium, W.M.H., et al. , 2013. The WU-Minn human connectome project: an overview. Neuroimage 80, 62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaswani A, Ramachandran P, Srinivas A, Parmar N, Hechtman B, Shlens J, 2021. Scaling local self-attention for parameter efficient visual backbones, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 12894–12904. [Google Scholar]
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I, 2017. Attention is all you need. Advances in neural information processing systems 30. [Google Scholar]
- Vázquez D, Bernal J, Sánchez FJ, Fernández-Esparrach G, López AM, Romero A, Drozdzal M, Courville A, 2017. A benchmark for endoluminal scene segmentation of colonoscopy images. Journal of healthcare engineering 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanti R, Ephrat A, Joskowicz L, Karaaslan O, Lev-Cohain N, Sosna J, 2015. Automatic liver tumor segmentation in follow-up CT studies using convolutional neural networks, in: Proc. Patch-Based Methods in Medical Image Processing Workshop, p. 2. [Google Scholar]
- Voita E, Talbot D, Moiseev F, Sennrich R, Titov I, 2019. Analyzing multi-head self-attention: Specialized heads do the heavy lifting, the rest can be pruned. arXiv preprint arXiv:1905.09418 . [Google Scholar]
- Wang B, Dong P, et al. , 2022a. Multiscale TransUNet++: dense hybrid U-Net with transformer for medical image segmentation. Signal, Image and Video Processing , 1–8. [Google Scholar]
- Wang C, Shang K, Zhang H, Li Q, Hui Y, Zhou SK, 2021a. DuDoTrans: Dual-domain transformer provides more attention for sinogram restoration in sparse-view CT reconstruction. arXiv preprint arXiv:2111.10790 . [Google Scholar]
- Wang C, Xu R, Xu S, Meng W, Zhang X, 2022b. DA-Net: Dual branch transformer and adaptive strip upsampling for retinal vessels segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 528–538. [Google Scholar]
- Wang CW, Huang CT, Lee JH, Li CH, Chang SW, Siao MJ, Lai TM, Ibragimov B, Vrtovec T, Ronneberger O, et al. , 2016. A benchmark for comparison of dental radiography analysis algorithms. Medical image analysis 31, 63–76. [DOI] [PubMed] [Google Scholar]
- Wang D, Wu Z, Yu H, 2021b. TED-net: Convolution-free T2T vision transformer-based encoder-decoder dilation network for low-dose CT denoising, in: International Workshop on Machine Learning in Medical Imaging, Springer. pp. 416–425. [Google Scholar]
- Wang G, Zhang Y, Ye X, Mou X, 2019a. Machine learning for tomographic imaging. IOP Publishing. [Google Scholar]
- Wang H, Xie S, Lin L, Iwamoto Y, Han XH, Chen YW, Tong R, 2021c. Mixed transformer U-Net for medical image segmentation. arXiv preprint arXiv:2111.04734 . [Google Scholar]
- Wang H, Zhu Y, Green B, Adam H, Yuille A, Chen LC, 2020a. Axial-deeplab: Stand-alone axial-attention for panoptic segmentation, in: European Conference on Computer Vision, Springer. pp. 108–126. [Google Scholar]
- Wang J, Wei L, Wang L, Zhou Q, Zhu L, Qin J, 2021d. Boundary-aware transformers for skin lesion segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 206–216. [Google Scholar]
- Wang L, Lin ZQ, Wong A, 2020b. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Scientific Reports 10, 19549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Nie D, Li G, Puybareau É, Dolz J, Zhang Q, Wang F, Xia J, Wu Z, Chen JW, et al. , 2019b. Benchmark on automatic six-month-old infant brain segmentation algorithms: the iSeg-2017 challenge. IEEE transactions on medical imaging 38, 2219–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Lei T, Cui R, Zhang B, Meng H, Nandi A, 2022c. Medical image segmentation using deep learning: A survey. IET Image Processing 16. [Google Scholar]
- Wang S, Li BZ, Khabsa M, Fang H, Ma H, 2020c. Linformer: Self-attention with linear complexity. arXiv preprint arXiv:2006.04768 . [Google Scholar]
- Wang W, Chen C, Ding M, Li J, Yu H, Zha S, 2021e. TransBTS: Multimodal brain tumor segmentation using transformer. arXiv preprint arXiv:2103.04430 . [Google Scholar]
- Wang W, Xie E, Li X, Fan DP, Song K, Liang D, Lu T, Luo P, Shao L, 2021f. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 568–578. [Google Scholar]
- Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM, 2017a. ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2097–2106. [Google Scholar]
- Wang X, Shrivastava A, Gupta A, 2017b. A-fast-rcnn: Hard positive generation via adversary for object detection, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 2606–2615. [Google Scholar]
- Wang X, Yang S, Zhang J, Wang M, Zhang J, Huang J, Yang W, Han X, 2021g. TransPath: Transformer-based self-supervised learning for histopathological image classification, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 186–195. [Google Scholar]
- Wang Z, Jiang W, Zhu YM, Yuan L, Song Y, Liu W, 2022d. Dynamixer: a vision MLP architecture with dynamic mixing, in: International Conference on Machine Learning, PMLR. pp. 22691–22701. [Google Scholar]
- Wang Z, Min X, Shi F, Jin R, Nawrin SS, Yu I, Nagatomi R, 2022e. SMESwin Unet: Merging CNN and transformer for medical image segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 517–526. [Google Scholar]
- Wasserthal J, Meyer M, Breit HC, Cyriac J, Yang S, Segeroth M, 2022. Totalsegmentator: robust segmentation of 104 anatomical structures in ct images. arXiv preprint arXiv:2208.05868 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Suriawinata A, Ren B, Liu X, Lisovsky M, Vaickus L, Brown C, Baker M, Tomita N, Torresani L, et al. , 2021. A petri dish for histopathology image analysis, in: International Conference on Artificial Intelligence in Medicine, Springer. pp. 11–24. [Google Scholar]
- Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, 2013. The cancer genome atlas pan-cancer analysis project. Nature genetics 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor R, Jamaludin A, Kadir T, Zisserman A, 2022. Context-aware transformers for spinal cancer detection and radiological grading, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 271–281. [Google Scholar]
- Wu G, Kim M, Wang Q, Gao Y, Liao S, Shen D, 2013. Unsupervised deep feature learning for deformable registration of MR brain images, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Xiao B, Codella N, Liu M, Dai X, Yuan L, Zhang L, 2021a. CvT: Introducing convolutions to vision transformers, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 22–31. [Google Scholar]
- Wu J, Fang H, Li F, Fu H, Lin F, Li J, Huang L, Yu Q, Song S, Xu X, et al. , 2022a. Gamma challenge: glaucoma grading from multi-modality images. arXiv preprint arXiv:2202.06511 . [DOI] [PubMed] [Google Scholar]
- Wu K, Peng H, Chen M, Fu J, Chao H, 2021b. Rethinking and improving relative position encoding for vision transformer, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10033–10041. [Google Scholar]
- Wu Y, Liao K, Chen J, Chen DZ, Wang J, Gao H, Wu J, 2022b. D-Former: A u-shaped dilated transformer for 3D medical image segmentation. arXiv preprint arXiv:2201.00462 . [Google Scholar]
- Xia W, Yang Z, Zhou Q, Lu Z, Wang Z, Zhang Y, 2022a. A transformer-based iterative reconstruction model for sparse-view ct reconstruction, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 790–800. [Google Scholar]
- Xia Z, Pan X, Song S, Li LE, Huang G, 2022b. Vision transformer with deformable attention. arXiv preprint arXiv:2201.00520 . [Google Scholar]
- Xie E, Wang W, Yu Z, Anandkumar A, Alvarez JM, Luo P, 2021a. SegFormer: Simple and efficient design for semantic segmentation with transformers. Advances in Neural Information Processing Systems 34. [Google Scholar]
- Xie Y, Zhang J, Shen C, Xia Y, 2021b. CoTr: Efficiently bridging CNN and transformer for 3D medical image segmentation. International conference on medical image computing and computer-assisted intervention . [Google Scholar]
- Xie Y, Zhang J, Xia Y, Wu Q, 2021c. Unified 2D and 3D pre-training for medical image classification and segmentation. arXiv preprint arXiv:2112.09356 . [Google Scholar]
- Xie Z, Zhang Z, Cao Y, Lin Y, Bao J, Yao Z, Dai Q, Hu H, 2022. Simmim: A simple framework for masked image modeling, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 9653–9663. [Google Scholar]
- Xing F, Xie Y, Yang L, 2015. An automatic learning-based framework for robust nucleus segmentation. IEEE transactions on medical imaging 35, 550–566. [DOI] [PubMed] [Google Scholar]
- Xing Z, Yu L, Wan L, Han T, Zhu L, 2022. NestedFormer: Nested modality-aware transformer for brain tumor segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 140–150. [Google Scholar]
- Xiong Y, Zeng Z, Chakraborty R, Tan M, Fung G, Li Y, Singh V, 2021. Nyströmformer: A nyström-based algorithm for approximating self-attention, in: Proceedings of the AAAI Conference on Artificial Intelligence, pp. 14138–14148. [PMC free article] [PubMed] [Google Scholar]
- Xu G, Wu X, Zhang X, He X, 2021a. LeViT-UNet: Make faster encoders with transformer for medical image segmentation. arXiv preprint arXiv:2107.08623 . [Google Scholar]
- Xu K, Ba J, Kiros R, Cho K, Courville A, Salakhudinov R, Zemel R, Bengio Y, 2015. Show, attend and tell: Neural image caption generation with visual attention, in: International Conference on Machine Learning, pp. 2048–2057. [Google Scholar]
- Xu L, Yan X, Ding W, Liu Z, 2022. Attribution rollout: a new way to interpret visual transformer. Journal of Ambient Intelligence and Humanized Computing, 1–11. [Google Scholar]
- Xu R, Wang X, Chen K, Zhou B, Loy CC, 2021b. Positional encoding as spatial inductive bias in gans, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 13569–13578. [Google Scholar]
- Xu Y, Mo T, Feng Q, Zhong P, Lai M, Eric I, Chang C, 2014. Deep learning of feature representation with multiple instance learning for medical image analysis, in: 2014 IEEE international conference on acoustics, speech and signal processing (ICASSP), IEEE. pp. 1626–1630. [Google Scholar]
- Xu Y, Zhang Q, Zhang J, Tao D, 2021c. Vitae: Vision transformer advanced by exploring intrinsic inductive bias. Advances in Neural Information Processing Systems 34, 28522–28535. [Google Scholar]
- Yan K, Wang X, Lu L, Summers RM, 2018. DeepLesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning. Journal of medical imaging 5, 036501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Tang H, Sun S, Ma H, Kong D, Xie X, 2022. AFTer-UNet: Axial fusion transformer UNet for medical image segmentation, in: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 3971–3981. [Google Scholar]
- Yang D, Myronenko A, Wang X, Xu Z, Roth HR, Xu D, 2021. T-automl: Automated machine learning for lesion segmentation using transformers in 3D medical imaging, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 3962–3974. [Google Scholar]
- Yang X, Xia D, Kin T, Igarashi T, 2020. Intra: 3d intracranial aneurysm dataset for deep learning, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 2656–2666. [Google Scholar]
- Ye W, Yao J, Xue H, Li Y, 2020. Weakly supervised lesion localization with probabilistic-CAM pooling. arXiv preprint arXiv:2005.14480 . [Google Scholar]
- Yu F, Koltun V, 2016. Multi-scale context aggregation by dilated convolutions, in: International Conference on Learning Representations. [Google Scholar]
- Yu P, Zhang H, Kang H, Tang W, Arnold CW, Zhang R, 2022a. Rplhr-ct dataset and transformer baseline for volumetric super-resolution from ct scans, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 344–353. [Google Scholar]
- Yu S, Ma K, Bi Q, Bian C, Ning M, He N, Li Y, Liu H, Zheng Y, 2021a. MIL-VT: Multiple instance learning enhanced vision transformer for fundus image classification, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 45–54. [Google Scholar]
- Yu T, Li X, Cai Y, Sun M, Li P, 2022b. S2-mlp: Spatial-shift mlp architecture for vision, in: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 297–306. [Google Scholar]
- Yu W, Luo M, Zhou P, Si C, Zhou Y, Wang X, Feng J, Yan S, 2021b. MetaFormer is actually what you need for vision. arXiv preprint arXiv:2111.11418 . [Google Scholar]
- Yu X, Tang Y, Zhou Y, Gao R, Yang Q, Lee HH, Li T, Bao S, Huo Y, Xu Z, et al. , 2022c. Characterizing renal structures with 3D block aggregate transformers. arXiv preprint arXiv:2203.02430 . [Google Scholar]
- Yu X, Zhang L, Zhao L, Lyu Y, Liu T, Zhu D, 2022d. Disentangling spatial-temporal functional brain networks via twin-transformers. arXiv preprint arXiv:2204.09225 . [Google Scholar]
- Yuan K, Guo S, Liu Z, Zhou A, Yu F, Wu W, 2021a. Incorporating convolution designs into visual transformers, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 579–588. [Google Scholar]
- Yuan L, Chen Y, Wang T, Yu W, Shi Y, Jiang ZH, Tay FE, Feng J, Yan S, 2021b. Tokens-to-token vit: Training vision transformers from scratch on imagenet, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 558–567. [Google Scholar]
- Yun B, Wang Y, Chen J, Wang H, Shen W, Li Q, 2021. SpecTr: Spectral transformer for hyperspectral pathology image segmentation. arXiv preprint arXiv:2103.03604 . [Google Scholar]
- Zhai X, Kolesnikov A, Houlsby N, Beyer L, 2021. Scaling vision transformers. arXiv preprint arXiv:2106.04560 . [Google Scholar]
- Zhai X, Kolesnikov A, Houlsby N, Beyer L, 2022. Scaling vision transformers. arXiv preprint arXiv:2106.04560 . [Google Scholar]
- Zhang C, Shu H, Yang G, Li F, Wen Y, Zhang Q, Dillenseger JL, Coatrieux JL, 2020. HIFUNet: multi-class segmentation of uterine regions from mr images using global convolutional networks for hifu surgery planning. IEEE Transactions on Medical Imaging 39, 3309–3320. [DOI] [PubMed] [Google Scholar]
- Zhang H, Goodfellow I, Metaxas D, Odena A, 2019a. Self-attention generative adversarial networks, in: International Conference on Machine Learning, pp. 7354–7363. [Google Scholar]
- Zhang K, Zuo W, Chen Y, Meng D, Zhang L, 2017. Beyond a gaussian denoiser: Residual learning of deep cnn for image denoising. IEEE transactions on image processing 26, 3142–3155. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li Q, Yu G, Sun L, Zhou M, Chu J, 2019b. A multidimensional choledoch database and benchmarks for cholangiocarcinoma diagnosis. IEEE access 7, 149414–149421. [Google Scholar]
- Zhang QS, Zhu SC, 2018. Visual interpretability for deep learning: a survey. Frontiers of Information Technology & Electronic Engineering 19, 27–39. [Google Scholar]
- Zhang W, Li R, Deng H, Wang L, Lin W, Ji S, Shen D, 2015a. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage 108, 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dou H, Ju T, Xu J, Zhang S, 2015b. Fusing heterogeneous features from stacked sparse autoencoder for histopathological image analysis. IEEE journal of biomedical and health informatics 20, 1377–1383. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Higashita R, Fu H, Xu Y, Zhang Y, Liu H, Zhang J, Liu J, 2021a. A multi-branch hybrid transformer network for corneal endothelial cell segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 99–108. [Google Scholar]
- Zhang Y, Liu H, Hu Q, 2021b. TransFuse: Fusing transformers and CNNs for medical image segmentation. arXiv preprint arXiv:2102.08005 . [Google Scholar]
- Zhang Y, Pei Y, Zha H, 2021c. Learning dual transformer network for diffeomorphic registration, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 129–138. [Google Scholar]
- Zhang Z, Liu Q, Wang Y, 2018. Road extraction by deep residual u-net. IEEE Geoscience and Remote Sensing Letters 15, 749–753. [Google Scholar]
- Zhang Z, Sun B, Zhang W, 2021d. Pyramid medical transformer for medical image segmentation. arXiv preprint arXiv:2104.14702 . [Google Scholar]
- Zhang Z, Yu L, Liang X, Zhao W, Xing L, 2021e. TransCT: dual-path transformer for low dose computed tomography, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 55–64. [Google Scholar]
- Zhao H, Shi J, Qi X, Wang X, Jia J, 2017. Pyramid scene parsing network, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 2881–2890. [Google Scholar]
- Zhao S, Dong Y, Chang EI, Xu Y, et al. , 2019a. Recursive cascaded networks for unsupervised medical image registration, in: Proceedings of the IEEE/CVF international conference on computer vision, pp. 10600–10610. [Google Scholar]
- Zhao S, Lau T, Luo J, Eric I, Chang C, Xu Y, 2019b. Unsupervised 3d end-to-end medical image registration with volume tweening network. IEEE journal of biomedical and health informatics 24, 1394–1404. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Lin Z, Sun K, Zhang Y, Huang J, Wang L, Yao J, 2022. SETMIL: Spatial encoding transformer-based multiple instance learning for pathological image analysis, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 66–76. [Google Scholar]
- Zheng H, Lin Z, Zhou Q, Peng X, Xiao J, Zu C, Jiao Z, Wang Y, 2022a. Multi-transSP: Multimodal transformer for survival prediction of nasopharyngeal carcinoma patients, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 234–243. [Google Scholar]
- Zheng M, Gao P, Zhang R, Li K, Wang X, Li H, Dong H, 2020. End-to-end object detection with adaptive clustering transformer. arXiv preprint arXiv:2011.09315 . [Google Scholar]
- Zheng Y, Li J, Shi J, Xie F, Jiang Z, 2022b. Kernel attention transformer (KAT) for histopathology whole slide image classification, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 283–292. [Google Scholar]
- Zhou B, Chen X, Zhou SK, Duncan JS, Liu C, 2022a. DuDoDR-Net: Dual-domain data consistent recurrent network for simultaneous sparse view and metal artifact reduction in computed tomography. Medical Image Analysis 75, 102289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Schlemper J, Dey N, Salehi SSM, Liu C, Duncan JS, Sofka M, 2022b. DSFormer: A dual-domain self-supervised transformer for accelerated multi-contrast MRI reconstruction. arXiv preprint arXiv:2201.10776 . [Google Scholar]
- Zhou HY, Guo J, Zhang Y, Yu L, Wang L, Yu Y, 2021a. nnFormer: Interleaved transformer for volumetric segmentation. arXiv preprint arXiv:2109.03201 . [Google Scholar]
- Zhou HY, Lu C, Yang S, Yu Y, 2021b. ConvNets vs. Transformers: Whose visual representations are more transferable?, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 2230–2238. [Google Scholar]
- Zhou L, Liu H, Bae J, He J, Samaras D, Prasanna P, 2022c. Self pre-training with masked autoencoders for medical image analysis. arXiv preprint arXiv:2203.05573 . [Google Scholar]
- Zhou SK, Greenspan H, Davatzikos C, Duncan JS, van Ginneken B, Madabhushi A, Prince JL, Rueckert D, Summers RM, 2021c. A review of deep learning in medical imaging: Imaging traits, technology trends, case studies with progress highlights, and future promises. Proceedings of the IEEE . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SK, Le HN, Luu K, Nguyen HV, Ayache N, 2021d. Deep reinforcement learning in medical imaging: A literature review. Medical Image Analysis 73, 102193. [DOI] [PubMed] [Google Scholar]
- Zhou SK, Rueckert D, Fichtinger G, 2019. Handbook of medical image computing and computer assisted intervention. [PubMed] [Google Scholar]
- Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J, 2018. UNet++: A nested U-Net architecture for medical image segmentation, in: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Yao Q, Xiao L, Zhou SK, 2021. You only learn once: Universal anatomical landmark detection, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 85–95. [Google Scholar]
- Zhu H, Yao Q, Zhou SK, 2022. DATR: Domain-adaptive transformer for multi-domain landmark detection. arXiv preprint arXiv:2203.06433 . [Google Scholar]
- Zhu X, Su W, Lu L, Li B, Wang X, Dai J, 2020. Deformable DETR: Deformable transformers for end-to-end object detection. arXiv preprint arXiv:2010.04159 . [Google Scholar]
- Zhu Y, Lu S, 2022. Swin-VoxelMorph: A symmetric unsupervised learning model for deformable medical image registration using swin transformer, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 78–87. [Google Scholar]
- Zhuang X, Shen J, 2016. Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI. Medical image analysis 31, 77–87. [DOI] [PubMed] [Google Scholar]
- Zuo XN, Anderson JS, Bellec P, Birn RM, Biswal BB, Blautzik J, Breitner J, Buckner RL, Calhoun VD, Castellanos FX, et al. , 2014. An open science resource for establishing reliability and reproducibility in functional connectomics. Scientific data 1, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]