Abstract
The replicator region of composite plasmid pTAV1 of Paracoccus versutus (included in mini-replicon pTAV320) belongs to the family of repABC replicons commonly found in plasmids harbored by Agrobacterium and Rhizobium spp. The repABC replicons encode three genes clustered in an operon, which are involved in partitioning (repA and repB) and replication (repC). In order to localize the partitioning site of pTAV320, the two identified incompatibility determinants of this mini-replicon (inc1, located in the intergenic sequence between repB and repC; and inc2, situated downstream of the repC gene) were PCR amplified and used together with purified RepB fusion protein (homologous to the type B partitioning proteins binding to the partitioning sites) in an electrophoretic mobility shift assay. The protein bound only inc2, forming two complexes in a protein concentration-dependent manner. The inc2 region contains two long (14-bp) repeated sequences (R1 and R2). Disruption of these sequences completely eliminates RepB binding ability. R1 and R2 have sequence similarities with analogous repeats of another repABC replicon of plasmid pPAN1 of Paracoccus pantotrophus DSM 82.5 and with centromeric sequences of the Bacillus subtilis chromosome. Excess RepB protein resulted in destabilization of the inc2-containing plasmid in Escherichia coli. On the other hand, the inc2 region could stabilize another unstable replicon in P. versutus when RepA and RepB were delivered in trans, proving that this region has centromere-like activity. Thus, it was demonstrated that repA, repB, and inc2 constitute a functional system for active partitioning of pTAV320.
Low-copy-number plasmids code for partitioning systems that ensure the correct distribution of plasmids within a bacterial population by precisely separating newly replicated copies into daughter cells at cell division. In general, these systems consist of three elements: (i) a trans-acting type A protein (ATPase) which autoregulates the operon and (ii) a trans-acting type B protein which binds to (iii) a cis-required partitioning site, which is thought to be analogous to the centromeres of eukaryotic chromosomes (27, 41). All three elements are indispensable for the proper functioning of these systems.
Although the partitioning systems are very often encoded close to plasmid replicator regions, they have been shown to function as independent cassettes that are able to stabilize (in cis) other, even unrelated replicons (30, 41). So far, there has been no evidence of structural association and coregulation of the two most important maintenance mechanisms for plasmids, replication and partitioning. The so-called repABC-type replicons seem to be an exception as the genes involved in the two processes are organized in an operon (5, 34).
The repABC family of replicons is widely distributed in bacteria belonging to the genera Rhizobium and Agrobacterium (32, 35). Several repABC replicons have been sequenced so far, including the root-inducing plasmids pRiA4b (29) and pRi1724 (28) of Agrobacterium rhizogenes; the tumor-inducing plasmids pTiB6S3 (39), pTi-SAKURA (38), pTiC58 (21), and pTiBo542 (containing in a tandem repeat two phylogenetically distinct repABC-type replicons) (31) of Agrobacterium tumefaciens; symbiotic plasmids pNGR234-a of Rhizobium spp. (13) and p42d of Rhizobium etli (34); cryptic plasmids pRL8JI of Rhizobium leguminosarum (40) and pAtC58 of A. tumefaciens (accession number AF283811); Mesorhizobium loti plasmids pMLa and pMLb, which coexist in trans (16); and plasmid pExo of Sinorhizobium meliloti (10). The sole representative of this class of plasmids that has been identified outside the Rhizobiaceae group is pTAV1 (mini-replicon pTAV320) of the soil bacterium Paracoccus versutus (5). pTAV320 appears to be the most divergent member of this family (5, 21, 32). pTAV1 is a composite plasmid, and in addition to pTAV320 it codes for a second replicator region (not included in the repABC class) that has been cloned in the form of mini-replicon pTAV202 (3, 7).
All of the repABC-type replicons have significant nucleotide and amino acid sequence similarities, and they have identical genetic organizations. They carry three clustered genes (repA, repB, and repC) that are transcribed in the same direction and a short, AT-rich, highly conserved intergenic sequence (igs) that is located between the repB and repC genes. The repC gene codes for the main replication initiator. Moreover, it has been suggested that the origin of replication is localized in the coding sequence of the repC gene (5, 32).
The repA and repB protein products have sequence similarities to proteins involved in partitioning of bacterial plasmids and their chromosomal homologues (14). Mutation analysis of the repABC replicons has demonstrated that mutations introduced into repA or repB drastically decrease plasmid stability (5, 34, 39), while parental plasmids are maintained in nearly 100% of the cells in the population even under nonselective conditions. The results of studies of Ramírez-Romero et al. (33) also suggest that the protein products of the repA and repB genes of replicon p42d (repABC) bring about repression of the genes of the operon. Therefore, like the homologous proteins of other plasmid partitioning systems, they have important regulatory functions. It has also been shown that in the case of pTAV320 the nonreplicating form of a repABC-type replicon (with a disrupted repC gene) can stabilize an unstable plasmid in cis (5), which explicitly reflects the presence of a functional partitioning system in repABC replicons.
Previous studies have focused mainly on analysis of the replication functions of repABC replicons (5, 33). However, to fully understand the mechanism of action, it is also necessary to characterize the structural elements involved in plasmid partitioning. In this paper we describe the results of experiments designed to localize the partitioning site of a repABC-type replicon (pTAV320), which together with repA and repB constitutes the functional partitioning system.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All of the bacterial strains and plasmids used are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) medium (37) at 30°C (P. versutus) or 37°C (Escherichia coli). The concentrations of antibiotics included in media were as follows: ampicillin, 100 μg/ml; spectinomycin, 50 μg/ml; kanamycin, 50 μg/ml; rifampin, 50 μg/ml; and tetracycline, 3 μg/ml for P. versutus and 20 μg/ml for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| P. versutus UW225 | Rifr; pTAV1-less derivative of wild-type UW1 | 6 |
| E. coli strains | ||
| TG1 | Host strain for plasmids | 37 |
| DH5α | Host strain for pRK2013 | 37 |
| M15 | Kmr; host strain for pQE30 and pQE30/repB; carries repressor plasmid pREP4 | Qiagen |
| Plasmids | ||
| pTAV320 | Kmr; 5.6-kb mini-replicon of pTAV1, composed of two PstI fragments (2.1 and 2.2 kb) of pTAV1 plus Kmr cassette | 5 |
| pRK415 | Tcr; mobilizable broad-host-range vector; origin of replication (oriV) and origin of transfer (oriT) of RK2 | 17 |
| pRK2013 | Kmr; helper plasmid carrying RK2 tra genes | 12 |
| pRK415/M1 | Tcr; pRK415 derivative with 322-bp PCR fragment (inc1) (amplified with IGSL and IGSR primers) of pTAV320, containing 15 bp of repB, complete igs, and 95 bp of repC (coordinates 2497 to 2818)b | This study |
| pRK415/M2 | Tcr; pRK415 derivative with 394-bp PCR fragment (inc2) (amplified with ZACL and ZACR primers) of pTAV320, containing 139 bp of repC and 255 bp downstream of the gene (coordinates 3797 to 4190) | This study |
| PRK415/M3 | Tcr; pRK415 derivative with 2,208-bp PstI fragment of pTAV320, containing proximal part of the replicon with repA and 680 bp of repB (coordinates 1 to 2208) | This study |
| pRK415/M4 | Tcr; pRK415 derivative with 1,323-bp BamHI-PstI fragment of pTAV320, containing 487 bp of terminal part of repA and 676 bp of repB (coordinates 881 to 2204) | This study |
| pRK415/M5 | Tcr; pRK415 derivative with 849-bp PstI-BamHI fragment of pTAV320, containing the promoter region and 446 bp of repA (coordinates 1 to 849) | This study |
| pRK415/M6 | Tcr; pRK415 derivative with 219-bp HindIII-Bsu36I fragment of pTAV320, containing 150 bp of igs and 69 bp of repC (coordinates 2574 to 2793) | This study |
| pRK415/M7 | Tcr; derivative of pRK415 with 370-bp PstI-HindIII fragment of pTAV320, containing 307 bp of terminal part of repB and 63 bp of the intergenic region (igs) (coordinates 2204 to 2574) | This study |
| pRK415/M8 | Tcr; pRK415 derivative with 710-bp Bsu36I-BglII and 241-bp BglII-PstI fragments of pTAV320, containing internal part of repC and terminal region of the mini-replicon, respectively (coordinates 2793 to 3503 and 4097 to 4338) | This study |
| pRK415/M11 | Tcr; pRK415 derivative with 1,143-bp Bsu36I-PvuII fragment of pTAV320, containing 1,143 bp of repC (coordinates 2793 to 3936) | This study |
| pRK415/M20 | Tcr; pRK415 derivative with 2,204-bp PstI-PstI fragment of pTAV320, containing mutated repA (not in-frame insertion resulting from AatII digestion and Klenow fragment end filling) and 676 bp of repB (coordinates 1 to 2204) | This study |
| pRK415/M21 | Tcr; pRK415 derivative with 594-bp BglII fragment of pTAV320, containing 432 bp of repC and 162 bp downstream of the gene (coordinates 3503 to 4097) | This study |
| pRK415/AB | Tcr; pRK415 containing 2,737-bp BssHII-Bsu36I restriction fragment of pTAV320 carrying repA and repB genes, igs, and 69 bp of repC (coordinates 56 to 2793) | This study |
| pTAV202 | Kmr; mini-replicon of the second replicator region of plasmid pTAV1, containing 4-kb SalI-PstI restriction fragment of pTAV1 plus Kmr cassette; unable to replicate in E. coli | 3 |
| pABW1 | Kmr; mobilizable cloning vector; ColE1 origin; oriT of RK2; unable to replicate in P. versutus | 7 |
| pABW3 | Kmr; cloning-mobilizable E. coli-P. versutus shuttle vector composed of pABW1 and pTAV202 | This study |
| pABW3/BGL | Kmr; pABW3 containing 594-bp BglII fragment of pTAV320 (coordinates 3503 to 4097) | This study |
| pQE30 | Apr; plasmid for construction and expression of fusion His6-tagged proteins; origin of ColE1 | Qiagen |
| pQE30/repB | Apr; pQE30 containing 982-bp PCR-amplified (with primers REPBL and REPBR) repB gene of pTAV320 (coordinates 1528 to 2510) | This study |
| pREP4 | Kmr; 3.7 kb; origin p15A; lacIq | Qiagen |
| pGB2 | Spr; cloning vector; pSC101 origin | 11 |
| pDBG1 | Spr; pGB2 containing inc2 region of pTAV320 | This study |
| pDBG2 | Spr; pGB2 containing inc2 region of pTAV320 in the orientation opposite that of pDBG1 | This study |
For plasmid derivative descriptions we used the nucleotide numbers of the sequence of pTAV320 as the coordinates.
GenBank accession number U60522.
DNA manipulations.
Plasmid DNA was isolated by the method of Birnboim and Doly (8), and if necessary, it was purified by CsCl-ethidium bromide gradient centrifugation. Cloning experiments, digestion with restriction enzymes, ligation, treatment with the Klenow fragment of DNA polymerase I, and agarose gel electrophoresis were conducted by using standard procedures, as described by Sambrook et al. (37). All enzymes were purchased from either Promega or Boehringer-Roche. DNA restriction fragments were recovered from agarose gels with a DNA Gel-Out kit (DNA Gdansk).
Plasmid construction.
To identify the incompatibility determinant of pTAV320, a series of subclones containing selected restriction fragments or PCR-amplified fragments of the mini-replicon studied, cloned into vector pRK415, were constructed; detailed descriptions of the plasmids constructed are given in Table 1, and the construction scheme is shown in Fig. 1.
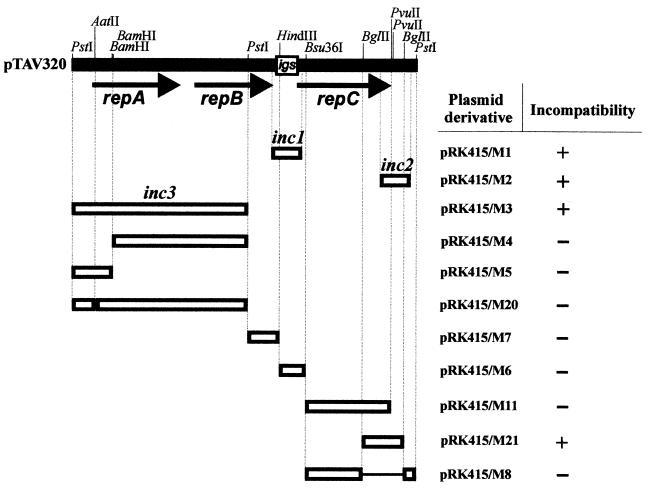
FIG. 1.
Identification of incompatibility determinants (inc) of pTAV320. Only the restriction sites used to construct recombined plasmids are shown on the pTAV320 restriction map. The transcriptional orientations of the three open reading frames are indicated by arrows. The conserved intergenic sequence between repB and repC (igs) is enclosed in a box. The open boxes represent DNA restriction or PCR-amplified fragments of pTAV320 that were cloned into the pRK415 vector and tested for the presence of the inc regions; the designations of the pRK415 derivatives are on the right. For details concerning construction see Table 1. The lines joining the boxes indicate regions of pTAV320 removed by deletion. The smallest cis-acting regions of pTAV320 carrying identified incompatibility determinants are designated inc1 and inc2. A plus sign indicates incompatibility and a minus sign indicates compatibility of pTAV320 with derivatives of pRK415 introduced into P. versutus UW225(pTAV320).
A mobilizable shuttle vector, pABW3, was constructed based on the E. coli-specific (nonreplicating in P. versutus) mobilizable vector pABW1 (oriT RK2) (4) and the P. versutus-specific (nonreplicating in E. coli) mini-replicon pTAV202 (7). Briefly, pABW1 was digested with MluI enzyme, which resulted in two restriction fragments, a 1.5-kb fragment containing the Kmr cassette and a 4.5-kb fragment containing all of the vector. Mini-replicon pTAV202 (Kmr) was linearized by SalI digestion (at a unique restriction site flanking the kanamycin resistance cassette), blunt ended with the Klenow fragment of polymerase I, and ligated with a blunt-ended 4.5-kb MluI fragment of pABW1.
Electroporation and transformation.
Electroporation was carried out at 2.5 kV, 25 μF, and 200 Ω (for E. coli) or 400 Ω (for P. versutus) with a Genepulser apparatus (Bio-Rad Laboratories) by using a modified Bio-Rad procedure (42). Electrotransformants were selected on solidified LB medium supplemented with the appropriate antibiotic. Competent cells of E. coli TG1 and M15 were prepared and transformed as described by Kushner (19).
Triparental mating.
Overnight cultures (spun down and washed twice to remove antibiotics) of the donor strain E. coli TG1 carrying a mobilizable vector, the recipient strain P. versutus UW225, and E. coli DH5α carrying the helper plasmid pRK2013 were mixed 1:2:1. Then 100 μl of the mixture was spread on a plate containing solidified LB medium. After overnight incubation at 30°C, the bacteria were washed off the plate, and suitable dilutions were plated on selective media containing rifampin (selective marker for the recipient strain) and another antibiotic (tetracycline or kanamycin) to select transconjugants. Spontaneous resistance of the recipient strains to kanamycin and tetracycline was not detectable under these experimental conditions.
PCR amplification.
The repB gene was amplified with primers REPBL (5′-AAGGATCCATGCCGTCAACGACGAGATC-3′) and REPBR (5′-AAGGATCCCAGTCTTCCTTGCGCT TCCA-3′) (the introduced BamHI restriction sites are underlined). For amplification of the inc1 and inc2 regions the following pairs of forward and reverse primers were used: primers IGSL (5′-CGCAAGGAAGACTGAGGAAG-3′) and IGSR (5′-GGCTTACCCGGAACAGATAC-3′) and primers ZACL (5′-CGTCGCTATGCTGGAACGCT-3′) and ZACR (5′-CCGCACATTGGACTGGCTCA-3′). The minimal region carrying the R1 and R2 repeats was amplified with primers INC10 (5′-TGATGTCCCGGCGAGATGCT-3′) and INC11 (5′-CTCTCTTCTGCTACCGACGC-3′). Amplification was performed with a Hot-Shot 18 thermocycler (DNA Gdansk) by using the synthetic oligonucleotides described above, Thermus aquaticus polymerase (Perkin-Elmer), and pTAV320 as the template DNA. PCR products were separated by 1% agarose gel electrophoresis, extracted from the gel with a DNA Gel-Out kit (DNA Gdansk), and used for analysis.
Incompatibility testing.
The incompatibility characteristics of two plasmids were examined by conjugational transfer of tester recombinant Tcr plasmids of the pRK415M series into a recipient strain (P. versutus UW225) carrying pTAV320 (Kmr). Transconjugants were selected for the incoming and resident plasmids or, in case of strong incompatibility, only for the incoming plasmid. The plasmid patterns of transconjugants were verified by screening 10 colonies with a rapid alkaline procedure and agarose gel electrophoresis. The incompatibility behavior of pTAV320 (which coexisted in trans with plasmids belonging to the pRK415M series) was tested during growth for approximately 30 bacterial generations. The retention of pTAV320 was determined by determining the percentage of kanamycin-resistant colonies among 200 Tcr clones (containing a pRK415-based plasmid).
Plasmid stability.
The stability of plasmids during growth in nonselective conditions was tested as previously described (5). Briefly, stationary-phase cultures were diluted in fresh medium without antibiotic selection and cultivated for approximately 10, 20, and 30 generations. Samples taken after cultivation for 10, 20, and 30 generations were diluted and plated onto solid medium in the absence of selective drugs. Two hundred colonies were tested with the Kmr or Tcr marker by replica plating. The retention of plasmids after approximately 30 generations was determined by determining the percentage of kanamycin- or tetracycline-resistant colonies.
Overproduction and purification of RepB(His)6 protein fusion.
For overexpression of the RepB(His)6 protein, 800 μl of an overnight culture of E. coli M15 carrying a recombinant plasmid (designated pQE30/repB) was used to inoculate 40 ml of fresh LB medium with ampicillin (100 μg/ml) and kanamycin (50 μg/ml), and the resulting culture was grown at 37°C to an optical density at 600 nm of 0.8. The culture was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a concentration of 1 mM and incubated for an additional 3 h. The cells were harvested and resuspended in 50 mM phosphate buffer (pH 8) containing 1 M NaCl. The cells were sonicated and centrifuged for 20 min at 22,000 × g and 4°C. A sample of the supernatant of the crude extract was mixed with an equal volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, boiled for 5 min, and subjected to SDS-PAGE on a 12.5% polyacrylamide gel. The gel was stained with 0.5% Coomassie brilliant blue in 25% methanol–10% acetic acid. The protein extract was mixed and incubated with 1 ml of metal resin (Ni-NTA agarose; Qiagen) with gentle agitation for 1 h at 4°C. The supernatant was removed, and the resin was washed four times with 4 ml of washing buffer (50 mM phosphate buffer, 300 mM NaCl, 10% glycerol; pH 6). The RepB(His)6 protein was eluted from the resin by using 1-ml portions of elution buffer (washing buffer with increasing imidazole concentrations [50 to 300 mM]). The purification was monitored by SDS-PAGE and Coomassie blue staining. The eluted fractions chosen were dialyzed against washing buffer in order to remove imidazole.
Immunological detection of RepB(His)6.
Total-protein extracts or purified RepB(His)6 protein was subjected to electrophoresis on an SDS–12.5% PAGE gel. The gel was washed in transfer buffer (192 mM glycine, 20% methanol, 25 mM Tris-HCl; pH 8.3). The proteins were transferred to a nitrocellulose membrane with a Semi-Phor transfer cell (Hoefer) for 1 h at 50 V. The efficiency of transfer was verified by staining the proteins with Ponceau S dye (Sigma). The dye was washed out with water, and immunological detection with Penta · His antibodies (Qiagen) was performed as recommended by the manufacturer.
DNA labeling and DNA mobility shift assay.
PCR-amplified fragments carrying inc1 and inc2 regions of pTAV320 (or PCR-amplified or restriction fragments of inc2) were labeled by using T4 polynucleotide kinase and [γ-32P]dATP (30 μCi; Amersham) and incubated at 37°C for 30 min. An oligonucleotide coding for the R1/R2 sequence was created by labeling the single-stranded DNA (5′-TAACAGCTGTTAAGTCTC-3′) and annealing it with the complementary sequence in a buffer containing 5 mM MgCl2, 20 mM Tris (pH 7.5), and 50 mM NaCl. A DNA binding reaction was performed at 37°C for 5 min by incubating 1 μl of purified labeled fragment (0.5 ng of DNA) with 2 μl of protein extract containing overproduced RepB(His)6 or purified proteins [RepB(His)6 or integration host factor (IHF)] and nonspecific competitor DNA (calf thymus DNA) in binding buffer (40 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 100 mM KCl, 1 mM dithiothreitol, 100 μg of bovine serum albumin per ml). The final volume of the reaction mixture was 20 μl. One microliter of loading dye (0.1% xylene cyanol, 50% glycerol) was added, and the samples were immediately loaded and separated on 5% acrylamide-bisacrylamide (82:1) gels in 0.05 M Tris-borate-EDTA buffer (pH 8.3) by electrophoresis for 1.5 h at approximately 10 V/cm2. The gels were transferred to Whatman 3MM paper, dried, and exposed to Kodak X-OMAT film for 24 h at −80°C.
Sequence analysis.
Sequence analysis was done with programs included in the Genetics Computer Group sequence analysis software package (GCG version 8.1). Comparison searches of databases were performed with the BLAST program, provided by the National Center for Biotechnology Information (http: //www.ncbi.nlm.nih.gov/BLAST).
RESULTS
Identification of incompatibility determinants of pTAV320.
Two incompatible plasmids (carrying related replication or partitioning systems) cannot be stably maintained in a bacterial cell in the absence of selective pressure. In partitioning systems the cis-required partitioning sites are, most commonly, the incompatibility determinants (inc) (2). To identify the partitioning site of pTAV320, a preliminary analysis was performed to identify the inc regions of the mini-replicon. To do this, several selected restriction fragments or PCR products were cloned into the broad-host-range vector pRK415 (Tcr; compatible with pTAV320) in E. coli TG1 and transferred by triparental mating into P. versutus UW225 carrying pTAV320 (Kmr). The presence of the inc region in the incoming pRK415-derived plasmid resulted in a loss of pTAV320 (see Materials and Methods for details).
A previous analysis of one of the repABC replicons (p42d) led to identification of two sequences coding for incompatibility determinants; one of these sequences is located in the intergenic segment (igs) between the repB and repC genes, and the other is located downstream of the repC gene (33). Therefore, we first attempted to analyze analogous regions of pTAV320, and we found that both PCR-amplified igs of pTAV320 (present in pRK415/M1 [Fig. 1]) and an amplified segment that included the region downstream of the repC gene (present in pRK415/M2 [Fig. 1]) carried incompatibility determinants. The sequences were designated inc1 (322 bp) and inc2 (394 bp), respectively (Fig. 1). A more detailed analysis performed with pRK415-derived plasmids carrying cloned restriction fragments that included the entire pTAV320 genome (the plasmids constructed are listed in Fig. 1; details of the construction are given in Table 1) led to identification of a third inc region (inc3) cloned in pRK415/M3 (Fig. 1). In the case of pRK415/M3 it seemed probable that the incompatibility observed was the result of the presence on this plasmid of a functional gene (with adjacent promoter region) coding for the regulatory protein RepA (the repA gene is also under control of the Plac promoter of the vector). Further analysis revealed that plasmids containing subcloned restriction fragments of the inc3 region (pRK415/M4 and pRK415 M/5 [Fig. 1]) or the whole fragment, in which a small deletion (4 bp) resulted in a change in the repA reading frame (pRK415/M20 [Fig. 1]), were compatible with pTAV320. These results explicitly indicate that in this case the overproduced RepA protein caused destabilization of pTAV320 residing in trans. A similar observation was made for p42d (33).
As shown on Fig. 1, pTAV320 was also incompatible with pRK415/M21 (which contains a 594-bp BglII fragment of pTAV320 coding for the terminal part of the repC gene and a 163-bp region downstream of repC). Since the inc2 region (present in pRK415/M2) includes the terminal part of the repC gene (139 bp) and a sequence downstream of the gene (255 bp), whereas the terminal fragment of the gene (present in pRK415/M11 [Fig. 1]) does not express incompatibility with pTAV320, it seemed obvious that the incompatibility determinant had to be located downstream of repC, between the terminal codon of the gene and the BglII restriction site (163 bp) (Fig. 2B). The results obtained in further experiments confirmed this assumption.
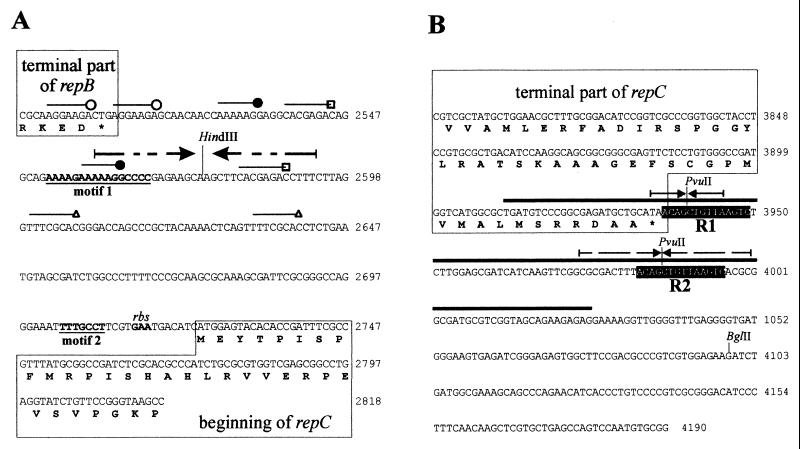
FIG. 2.
Nucleotide sequences of the inc1 (A) and inc2 (B) regions of mini-replicon pTAV320. Several short repeated sequences identified in inc1 are indicated by different lines. Motif 1 and motif 2, the two motifs of inc1 sequences that are visibly conserved in the igs of repABC replicons, are indicated by boldface type and underlined. Two direct repeats of inc2, R1 and R2, are indicated by a black background. The region of inc2 amplified with primers INC10 and INC11 and used for EMSA is indicated by a thick line over the sequence. The palindromic sequences of the inc regions are indicated by arrows. The terminal fragments of the repB and repC genes are enclosed in boxes. The amino acid sequences of the proteins are given below the nucleotide sequences. The HindIII, PvuII, and BglII restriction sites important for the analysis are indicated. A presumptive ribosome binding site (rbs) is indicated. Coordinate numbers for the pTAV320 sequence are indicated on the right (accession number U60522).
Analysis of nucleotide sequences of the inc regions.
A detailed analysis of the nucleotide sequences of the inc regions identified was carried out to identify potential structural elements analogous to those that occur in other plasmid partitioning sites that have been characterized (e.g., AT sequence enrichment, repeated and palindromic sequences). In the case of the igs sequence of pTAV320 (inc1) the G+C content was 52%, which is slightly different from the value determined for the entire mini-replicon (60%). However, several short stretches in which there was a high level of AT enrichment were distinguished in this sequence, as were several repeated sequences that were 7 to 8 bp long (some of the repeats occurred partially in the terminal part of the repB gene [Fig. 2A]). In the proximal part of igs two long imperfect palindromic sequences were found. It was observed that cleavage of the palindrome at the HindIII restriction site (Fig. 2A) completely eliminated the inc phenotype (Fig. 1) (pRK415/M6 and pRK415/M7). Although the igs sequence of pTAV320 is conserved less than the igs sequences in the other repABC replicons (5), two distinct conserved motifs were distinguished, as shown in Fig. 2A.
The G+C content of the inc2 sequence is 60%. There are no areas with above-average A+T content. Also, this sequence does not contain any significant structural features except the two longest such features in the entire pTAV320 sequence, identical 14-bp repeated sequences (5′-ACAGCTGTTAAGTC-3′) that are separated by 33 bp (Fig. 2B). The first of these two repeats (designated R1) occurs directly after the repC gene (it begins in the termination codon of the gene), and the second (designated R2) lies 46 bp downstream of this gene (Fig. 2B). The palindromic sequences that are partially present in both direct repeat R1 and direct repeat R2 are shown in Fig. 2B.
Purification of RepB(His)6 and determination of RepB binding site.
As mentioned above, the partitioning type B proteins (RepB homologues) specifically bind to the corresponding partitioning sites. To determine the site of RepB interaction within pTAV320, the protein was purified and used in an electrophoretic mobility shift assay (EMSA).
(i) Purification of RepB(His)6.
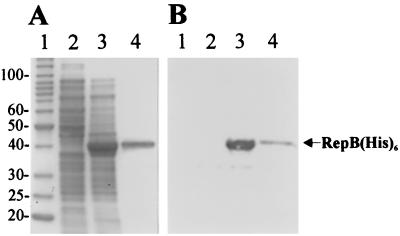
For construction of the recombinant RepB protein, PCR was used to amplify the coding region of repB (see Materials and Methods). The resulting PCR product was digested with BamHI and cloned in frame in the corresponding site of plasmid pQE30, generating a fusion between the repB gene and a vector sequence coding for six histidines (His6 tag). The insert was sequenced to confirm that no errors had been introduced during PCR. The resulting plasmid (pQE30/repB) was transferred into E. coli M15 carrying the repressor plasmid pREP4 (Table 1). IPTG induction resulted in a high level of expression of a protein (approximately 40 kDa) which was visualized by SDS-PAGE of the cell lysate of E. coli M15(pQE30/repB) (Fig. 3A, lane 3). This protein was not observed in a noninduced bacterial culture (Fig. 3A, lane 2). The protein was then purified on an Ni-NTA chromatographic matrix (Qiagen) (Fig. 3A, lane 4) as described in Materials and Methods. After SDS-PAGE and Western transfer, the RepB(His)6 protein gave a positive reaction with Penta · His antibodies (Fig. 3B, lanes 3 and 4), thus proving its recombinant nature.
FIG. 3.
(A) Coomassie blue-stained SDS-PAGE gel of overexpressed (in E. coli M15) and purified RepB(His)6 protein. The gel was loaded with the following samples: protein molecular weight standards (lane 1) (the molecular masses of the proteins [in kilodaltons] are indicated on the left); crude cell lysate of strain M15(pQE30/repB) (lane 2); crude cell lysate of strain M15(pQE30/repB) after IPTG induction (lane 3); purified RepB(His)6 protein (lane 4). (B) Western transfer of proteins visualized on the gel in panel A and immunodetection of RepB(His)6 with Penta · His antibody.
(ii) Determination of RepB binding site.
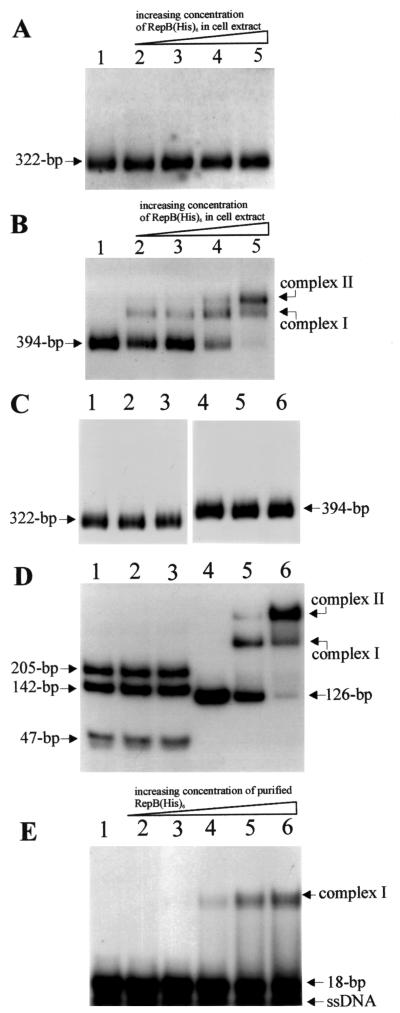
For protein-DNA binding experiments the PCR-amplified 322-bp inc1 DNA fragment and 394-bp inc2 DNA fragment (Fig. 2) were 32P labeled and used with cell extract containing overproduced RepB(His)6 (or with the purified protein) for EMSA as described in Materials and Methods. As Fig. 4B shows, the RepB fusion protein bound in inc2. Two complexes having different electrophoretic mobilities (Fig. 4B, complexes I and II) appeared in the gel in a protein concentration-dependent manner. Complex 1 was preferentially formed at low Rep(His)6 concentrations, whereas higher protein concentrations resulted in the formation of dominant complex II (Fig. 4B). This might have represented occupancy of two different target sites. In a control reaction similar amounts of E. coli M15(pQE30) cell extract were used (Fig. 4C, lanes 2 and 5). The lack of any complex formation indicated the specificity of RepB binding to the DNA fragment studied. No retardation of the inc1 region was detected (Fig. 4A). No binding of purified E. coli IHF protein was observed in inc1 and inc2 (Fig. 4C, lanes 3 and 6). This protein is known to bind in the partitioning site of phage P1 (24).
FIG. 4.
Autoradiograph of EMSA results, showing the binding ability of RepB(His)6 protein [present in crude protein extract of RepB(His)6-overproducing E. coli M15 or purified] in 32P-labeled inc1 and inc2 regions of pTAV320. (A and B) inc1 fragment (A) and inc2 fragment (B) incubated with different concentrations (2.5, 5, 10, and 20 μg/ml in lanes 2 to 5, respectively) of M15(pQE30/repB) crude protein extract. Lane 1 contained labeled fragment with no protein added. (C) inc1 fragment (lanes 1 to 3) and inc2 fragment (lanes 4 to 6) incubated (i) with M15(pQE30) crude protein extract (20 μg/ml) (lanes 2 and 5), (ii) with purified IHF protein (3 μg/ml) (lanes 3 and 6), and (iii) without protein (lanes 1 and 4). (D) The inc2 fragment was digested with PvuII (lanes 1 to 3) and incubated with 2.5 and 5 μg of purified RepB(His)6 protein per ml (lanes 2 and 3, respectively). No protein was added to the reaction mixture in lane 1. The 126-bp PCR-amplified fragment of the inc2 region (Fig. 2B) (lanes 4 to 6) was incubated with 2.5 and 5 μg of purified RepB(His)6 protein per ml (lanes 5 and 6, respectively). No protein was added to the reaction mixture in lane 4. (E) An 18-bp oligonucleotide containing a single R1/R2 sequence was incubated with 0.5, 1, 2.5, 5, and 10 μg of purified RepB(His)6 protein per ml (lanes 2 to 6, respectively). No protein was added to the reaction mixture in lane 1. The positions of the protein-DNA complexes and the sizes of the unbound probes are indicated on the right and left. ssDNA, single-stranded DNA.
As mentioned above, the inc2 region contains two repeated sequences, R1 and R2 (Fig. 2B). Since analogous sequences frequently are sites to which plasmid proteins bind, we assumed that protein RepB might interact with a site in R1 and R2. To verify this, a small fragment of DNA (126 bp) carrying both repeats was PCR amplified (with primers INC10 and INC11 [Fig. 2B]) and used, together with purified protein, for EMSA. It appeared that RepB specifically bound the amplified fragment to form two analogous complexes, as in the case of the entire inc2 fragment (Fig. 4D, lanes 4 to 6). Moreover, it was found that the amplified 162-bp DNA fragment was sufficient to express incompatibility with pTAV320 (data not shown).
The R1 and R2 sequences carry two PvuII cleavage sites (the only PvuII sites in the inc2 fragment) (Fig. 2B). PvuII digestion of the inc2 fragment resulted in three restriction fragments that were 47, 142, and 205 bp long. As shown in Fig. 4D, lanes 1 to 3, cleavage of fragment inc2 in R1 and R2 precluded binding of RepB, which strongly suggests that these sequences are the sites with which protein RepB interacts. To confirm this, an additional experiment was carried out, in which an oligonucleotide containing a single repeat was used as the substrate. In this case a shift of the DNA fragment (single complex) was observed, which explicitly confirmed that the R1 and R2 sequences are the recognition sites for the RepB protein (Fig. 4E, lanes 2 to 6).
Destabilization of inc2-containing plasmid by RepB.
In the case of the partitioning system of P1 it was observed that overproduction of ParB (a RepB homologue) entailed destabilization (resulting from transcriptional silencing) of plasmid pGB2 carrying a partitioning site of phage P1 (parS) (23, 36). To check whether the pTAV320 sequence identified has similar properties, the inc2 region was cloned in two orientations into multiple cloning site (MCS) pGB2 (Spr) (11), and the resulting plasmids (pDBG1, pDBG2) were introduced into E. coli TG1 containing RepB-overproducing plasmid pQE30/repB (Apr). The vector pGB2 is stably maintained in E. coli cells, both when it occurs alone and when it occurs in trans with compatible pQE30 or pQE30/repB. Plasmid pDBG1 or pDBG2 also stably coexisted with pQE30, whereas in the presence of pQE30/repB these plasmids became destabilized. Destabilization, regardless of the orientation of inc2 in the plasmid, was so strong that it was impossible to obtain transformants of E. coli TG1 carrying both pDBG1 or pBDG2 and the RepB(His)6 expression plasmid pQE30/repB. These results indicate that the presence of the overproduced protein results in destabilization of plasmids carrying inc2, which is most probably caused by transcriptional silencing of plasmid genes close to the cloned centromere.
Stabilization of inc2-containing plasmid by RepA and RepB.
It has been observed repeatedly that unstable plasmids containing only a partitioning site can be actively partitioned if the two partitioning proteins of a given system are provided in trans (1, 20). To check whether the inc2 sequence of pTAV320 is able to stabilize other replicons in cis (in the presence of repA and repB in trans), a two-plasmid system was constructed by using pRK415 (Tcr), a broad-host-range vector, and the compatible mini-replicon pTAV202 (Kmr), which is able to replicate in P. versutus (3, 7). It was demonstrated previously that pTAV202 and pTAV320 which coreside in trans in P. versutus did not affect each other's maintenance (5).
In view of the strong restriction barrier that precludes introduction of plasmid DNA from E. coli into P. versutus by electroporation, for the purposes of this experiment the mobilizable shuttle vector pABW3 (Kmr), containing a linear form of pTAV202 (5.3 kb) cloned into the E. coli-specific vector pABW1 (4), was constructed. The details are described in Materials and Methods. The resulting Kmr vector (8.5 kb) could be mobilized for conjugational transfer from E. coli to P. versutus and was able to replicate in both hosts.
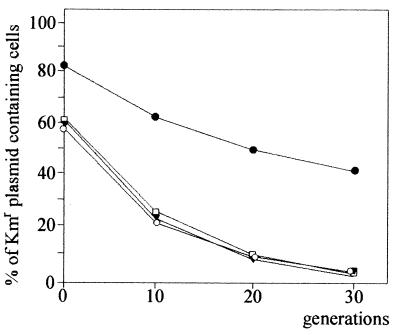
The inc2 region (present on a 594-bp BglII fragment of pTAV320 [Fig. 1]) was then cloned into the compatible BamHI site (MSC) of pABW3, generating pABW3/BGL. At the same time, a 2.7-kb fragment of pTAV320, carrying the promoter region and the repA and repB genes, was cloned into MCS of pRK415; the resulting plasmid was designated pRK415/AB. Both plasmids were conjugationally transferred into P. versutus UW225, and the stability of pABW3/BGL coexisting in trans was determined. pABW3 itself is unstable in P. versutus; after 30 generations of growth in nonselective conditions, only approximately 4% of the cells carry the plasmid. The same degree of stability was observed in control experiments when pABW3 was maintained in trans with pRK415 or pRK415/AB (Fig. 5). The inc2 region, which was present in pABW3/BGL, did not affect the stability of the plasmid itself (as observed in the presence of pRK415) but could stabilize the plasmid (more than 40% of the cells carried the plasmid after 30 generations of growth in nonselective conditions) when repA and repB were provided in trans for pRK415/AB (Fig. 5). This confirmed that the region identified contains a functional cis required partitioning site and that RepA and RepB are partitioning trans-acting proteins.
FIG. 5.
Plasmid stability of pABW3 (Kmr) and pABW3/BGL coexisting in trans with compatible pRK415 (Tcr) or pRK415/AB (carrying repA and repB genes of pTAV320) in P. versutus UW225 cells. Symbols: ▾, UW225 carrying plasmids pABW3 and pRK415; ○, UW225 carrying plasmids pABW3/BGL and pRK415; □, UW225 carrying plasmids pABW3 and pRK415/AB; ●, UW225 carrying plasmids pABW3/BGL and pRK415/AB. The data are averages based on three independent experiments.
Search for R1- and R2-like sequences in other repABC replicons.
A comparative analysis of the nucleotide sequences of other repABC replicons did not reveal the presence (downstream of repC) of a sequence homologous to R1 and R2 of pTAV320. Moreover, no other equally long, significant, repeated sequences of any other type were found. The only exceptions were the replicons of the A. tumefaciens pTi-SAKURA and pTiC58 plasmids, in which the terminal parts of the repC gene encode the first of two short (7-bp) repeated sequences (5′-GGTGATC-3′). These sequences are separated from each other (like R1 and R2 of pTAV320) by 33 bp.
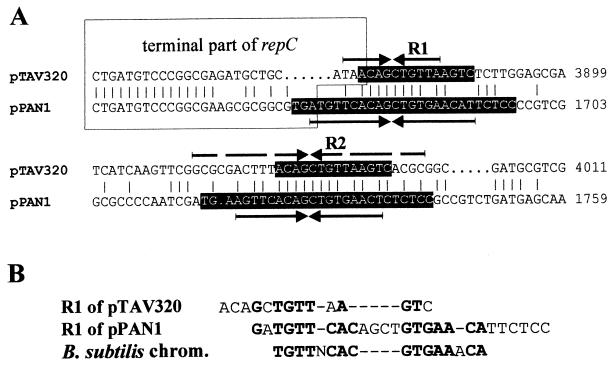
Interestingly, repeated sequences of this type (length, 27 bp) have been localized in a newly identified replicon of the repABC type occurring on plasmid pPAN1 of Paracoccus pantotrophus DSM 82.5 (unpublished results); the partial nucleotide sequence has been deposited in the GenBank database under accession number AY033083. The locations of the repeated sequences of pPAN1 (5′-TGA[A/T]GTTCACAGCTGTGAAC[T/A][C/T]TCTCC-3′) are similar to the locations in pTAV320, and these sequences are similar to R1 and R2 (Fig. 6). The palindromic sequences present in both pPAN1 repeated sequences are shown in Fig. 6B. The repeated sequences of pPAN1 (and, to a lesser extent, the repeated sequences of pTAV320) are also similar to the centromere-like sequences of the Bacillus subtilis chromosome, which are the sites to which protein Spo0J (a chromosomal homologue in plasmid type B partitioning proteins) binds (22). An alignment of these sequences is shown in Fig. 6B.
FIG. 6.
(A) Alignment (BESTFIT; GCG software) of nucleotide sequences of the corresponding regions of P. versutus pTAV320 and the repABC replicator region of plasmid pPAN1 from P. pantotrophus DSM 82.5. The R1 and R2 repeated sequences are shown with a black background. The palindromic sequences are indicated by arrows. Terminal parts of the repC genes are enclosed in a box. Coordinates of the pTAV320 and pPAN1 sequences are indicated on the right. (B) Manual alignment of nucleotide sequences of R1 of pTAV320 and R1 of pPAN1 repeats and consensus centromeric sequence of the B. subtilis chromosome (chrom.) (22). Nucleotides common to two sequences at any position are indicated by boldface type. Dashes indicate gaps introduced to maximize the alignment.
DISCUSSION
In the present study we located the active partitioning site of the mini-replicon pTAV320, which belongs to the group of repABC replicons. Both the structure and the location of the partitioning site of pTAV320 differ from the structure and the location of the plasmid par loci that have been identified so far (27), in which sequences of this type are situated downstream of the gene coding for the type B partitioning protein (e.g., the sopABC loci of plasmid F and the parABS loci of phage P1) or in promoter regions of operons (e.g., the parA loci of plasmid R1 [9] and the par loci of plasmid pTAR). This sequence, which is a determinant of incompatibility (inc2), is located downstream of the gene coding for replication protein RepC (and thus downstream of the repABC operon). Two 14-bp repeated sequences, R1 and R2, as well as palindromic sequences, can be distinguished in the sequence and are the sites of binding of partitioning protein RepB.
Based on analogies with other active partitioning systems, it was postulated previously that the partitioning site of repABC replicons is located downstream of the repB gene, which is in the strongly conserved igs (inc1). The results of an analysis recently carried out by Ramírez-Romero et al. (33) seem to confirm this hypothesis. These authors observed that deletion of igs in p42d (repABC replicon) did not affect replication of the plasmid, whereas removal of the region directly behind the repC gene completely eliminated replication of the mutated plasmid. In light of these results it seemed obvious that igs carries an incompatibility determinant involved in plasmid segregation (a partitioning site), whereas the region corresponding to inc2 of pTAV320 contains the origin of replication.
Our previous results indicated, however, that oriV of pTAV320 is located in the coding sequence of the repC gene (5). A more detailed analysis of the repC sequence of all of the replicons of this group allowed workers to distinguish a region in these genes with an above-average A+T content that is characteristic of many plasmid oriV genes (32). In our studies we also determined that inc2 of pTAV320 introduced into P. versutus on a plasmid that is not functional in this host is not able to replicate when replication protein RepC is delivered in trans (data not shown).
A region located downstream of repC seems to carry out important regulatory functions; it is crucial for both replication and active partitioning. Presumably, besides the partitioning site, there are other genetic elements (sites of interaction with as-yet-unspecified host- or plasmid-encoded factors) whose presence in cis is necessary for replication of the plasmid. It seems odd, however, that the nucleotide sequence of such a functionally important region is not conserved in all repABC replicons.
The role of igs, which carries the incompatibility determinant inc1, is puzzling. In this sequence conserved motif 1 (Fig. 2A) is present, which was also identified upstream of the repC genes of plasmids pRmeGR4a and pTAV202 (5, 26). These plasmids do not belong to the repABC family but encode phylogenetically distantly related homologues of the RepC protein. In all cases motif 1 is followed by a palindromic sequence, whose cleavage (in the case of pTAV320) completely eliminates the inc phenotype. The presence of this type of sequence in different plasmids carrying the repC gene suggests that inc1 might be required for regulation of repC expression (5, 34), which in turn may be a factor that limits the frequency of initiation of replication. However, elucidation of the function of igs will require further detailed study.
In this study we observed that a plasmid carrying a partitioning site (inc2) in the presence of overproduced protein RepB of pTAV320 is subject to drastic destabilization in E. coli. This phenomenon was observed previously in analyses of the model active partitioning systems of plasmid F and phage P1 (18, 36). In both cases overproduction of type B partitioning proteins led to transcriptional silencing of genes adjacent to the centromere. In the case of plasmid pGB2 used by us, the promoter of the repA gene (coding for the protein that initiates replication) is close to the cloned inc2 region. It seems that the cause of destabilization is blocked transcription of the repA gene resulting from binding of overproduced protein RepB, which in effect precludes replication of the plasmid. It is interesting that this effect is observed in E. coli, which is a host in which pTAV320 (like other repABC replicons) is not functional. Yet it is not known whether additional host-encoded factors are required for gene silencing in this case. However, the activity of the partitioning system of repABC replicons in E. coli has not been studied yet. It is probable that the host factors involved in plasmid partitioning are more conserved in different bacterial species than the host factors taking part in plasmid replication are. This has been confirmed by the observations of Yamaichi and Niki (43), who demonstrated the functionality of the chromosomal partitioning system of B. subtilis (a gram-positive bacterium) in E. coli cells (a gram-negative host).
Recent phylogenetic analyses of partitioning ATPases demonstrated that RepA of pTAV320 and the RepA proteins of the remaining repABC replicons are phylogenetically most closely related to the group of plasmid ATPases which includes SopA of plasmid F and ParA of phage P1 (14, 15). Interestingly, a BLAST search for RepB homologues resulted first in a long list of chromosomally encoded proteins that are homologues of type B plasmid partitioning proteins (data not shown). These results suggest that the active partitioning systems of pTAV320 and other replicons of the repABC type have a hybrid structure, since the active partitioning system of pTAV320 encodes a plasmid-specific ATPase and a chromosomal type of type B partitioning protein. However, it should be remembered that in view of their size, plasmids of the Agrobacterium-Rhizobium group harboring repABC replicons can be placed in the class of megaplasmids that occasionally are larger than the chromosomes of some bacteria. Moreover, as suggested by Palmer et al. (32), it is highly probable that replicons of this type can also occur on bacterial chromosomes.
The partitioning site of pTAV320 and that of an analogous sequence of another paracoccal replicon of this type, pPAN1, are similar (more so in the case of pPAN1) to the centromere-like sequences of the partitioning system of the B. subtilis chromosome (22). Similar sequences that are located in or close to a structural gene of a type B partitioning protein have also been localized in the chromosomes of many other bacteria, such as Mycobacterium leprae, Mycobacterium tuberculosis, Streptomyces coelicolor, Borrelia burgdorferi, and Streptococcus pyogenes (22). Although the remaining replicons of the repABC type do not have sequences homologous to R1 and R2, it should be kept in mind that pTAV320 and pPAN1 constitute a separate, evolutionarily distinct group of repABC replicons (21, 32; unpublished results). Therefore, they may be phylogenetically more closely related to chromosomal systems than to plasmid systems.
Interestingly, in the terminal parts of the mini-replicons of pTAV1 and pPAN1, downstream of the repC gene the beginning of a strongly conserved (at the nucleotide and amino acid sequence levels) incomplete open reading frame has been identified (data not shown). Its protein product has significant similarity to a protein involved in stabilization of plasmid pRmeGR4a of Sinorhizobium meliloti, which is homologous to eukaryotic cytoskeletal proteins (25). The preliminary results of our analysis suggest that the stabilizing replicating repABC module of paracoccal plasmids may be more complex than analogous replicons found in members of the Rhizobiaceae. We have initiated studies aimed at unraveling the genetic structure of the regions adjacent to the repABC replicon of plasmid pTAV1.
ACKNOWLEDGMENTS
We thank J. Baj for stimulating discussions and M. Wlodarczyk for critical reading of the manuscript. We are grateful to M. Hryniewicz for enabling the performance of gel shift experiments in her lab and T. Bykowski for technical assistance. We also acknowledge M. Łobocka for providing plasmid pGB2 and A. Sirko for providing purified IHF protein.
This work was supported by the State Committee for Scientific Research, Poland (grant 6 P04A 034 16), and by the European Union (MECBAD-EU BIOTEC concerted action BIO4-CT-0099).
REFERENCES
- 1.Austin S, Abeles A L. Partitioning of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983;169:373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- 2.Austin S, Nordström K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–354. doi: 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- 3.Bartosik D, Baj J, Wlodarczyk M. Construction and preliminary characterization of mini-derivatives of large (107-kb) cryptic plasmid of the sulphur bacterium Thiobacillus versutus. FEMS Microbiol Lett. 1995;129:169–174. [Google Scholar]
- 4.Bartosik D, Bialkowska A, Baj J, Wlodarczyk M. Construction of mobilizable cloning vectors derived from pBGS18 and their application for analysis of replicator region of a pTAV202 mini-derivative of Paracoccus versutus pTAV1 plasmid. Acta Microbiol Pol. 1997;46:379–383. [PubMed] [Google Scholar]
- 5.Bartosik D, Baj J, Wlodarczyk M. Molecular and functional analysis of pTAV320-repABC type replicon of a composite pTAV1 plasmid of Paracoccus versutus. Microbiology. 1998;144:3149–3157. doi: 10.1099/00221287-144-11-3149. [DOI] [PubMed] [Google Scholar]
- 6.Bartosik D, Baj J, Plasota M, Piechucka E, Wlodarczyk M. Analysis of Thiobacillus versutus pTAV1 plasmid functions. Acta Microbiol Pol. 1993;39:5–11. [Google Scholar]
- 7.Bartosik D, Wlodarczyk M, Thomas C M. Complete nucleotide sequence of the replicator region of Paracoccus (Thiobacillus) versutus pTAV1 plasmid and its correlation to several plasmids of Agrobacterium and Rhizobium species. Plasmid. 1997;38:53–59. doi: 10.1006/plas.1997.1295. [DOI] [PubMed] [Google Scholar]
- 8.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breüner A, Jensen R B, Dam M, Pedersen S, Gerdes K. The centromere-like parC locus of plasmid R1. Mol Microbiol. 1996;20:581–592. doi: 10.1046/j.1365-2958.1996.5351063.x. [DOI] [PubMed] [Google Scholar]
- 10.Chain P S, Hernandez-Lucas I, Golding B, Finan T M. oriT-directed cloning of defined large regions from bacterial genomes: identification of the Sinorhizobium meliloti pExo megaplasmid replicator region. J Bacteriol. 2000;182:5486–5494. doi: 10.1128/jb.182.19.5486-5494.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 12.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad-host-range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;378:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 14.Gerdes K, Møller-Jensen J, Jensen R B. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 15.Hayes F. The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol Microbiol. 2000;37:528–541. doi: 10.1046/j.1365-2958.2000.02030.x. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Mochizuki Y, Nakayama S, Nakazaki N, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;31:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 17.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 18.Kim S-K, Wang J C. Gene silencing via protein-mediated subcellular localization of DNA. Proc Natl Acad Sci USA. 1999;96:8557–8561. doi: 10.1073/pnas.96.15.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kushner S R. An improved method for transformation of E. coli with ColE1-derived plasmids. In: Boyer H B, Nicosia S, editors. Genetic engineering. Amsterdam, The Netherlands: Elsevier/North-Holland; 1978. pp. 17–23. [Google Scholar]
- 20.Lane D, Rothenbuehler R, Merrilat A, Alken C. Analysis of the F plasmid centromere. Mol Gen Genet. 1987;207:406–412. doi: 10.1007/BF00331608. [DOI] [PubMed] [Google Scholar]
- 21.Li P-L, Farrand S K. The replicator region of the nopaline-type Ti plasmid pTiC58 is a member of the repABC family and is influenced by the TraR-dependent quorum-sensing regulatory system. J Bacteriol. 2000;182:179–188. doi: 10.1128/jb.182.1.179-188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin D C-H, Grossman A D. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 23.Łobocka M, Yarmolinsky M. P1 plasmid partition: a mutational analysis of ParB. J Mol Biol. 1996;259:366–382. doi: 10.1006/jmbi.1996.0326. [DOI] [PubMed] [Google Scholar]
- 24.Martin K A, Davies M A, Austin S. Fine-structure analysis of the P1 plasmid partition site. J Bacteriol. 1991;173:3630–3634. doi: 10.1128/jb.173.12.3630-3634.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercado-Blanco J, Olivares J. A protein involved in stabilization of a large non-symbiotic plasmid of Rhizobium meliloti shows homology to eukaryotic cytoskeletal proteins and DNA-binding proteins. Gene. 1994;139:133–134. doi: 10.1016/0378-1119(94)90536-3. [DOI] [PubMed] [Google Scholar]
- 26.Mercado-Blanco J, Olivares J. The large nonsymbiotic plasmid pRmeGR4a of Rhizobium meliloti GR4 encodes a protein involved in replication that has homology with the RepC protein of Agrobacterium plasmids. Plasmid. 1994;32:75–79. doi: 10.1006/plas.1994.1046. [DOI] [PubMed] [Google Scholar]
- 27.Møller-Jensen J, Jensen R B, Gerdes K. Plasmid and chromosome segregation in prokaryotes. Trends Microbiol. 2000;8:313–320. doi: 10.1016/s0966-842x(00)01787-x. [DOI] [PubMed] [Google Scholar]
- 28.Moriguchi K, Maeda Y, Satou M, Hardayani N S, Kataoka M, Tanaka N, Yoshida K. The complete nucleotide sequence of a plant root-inducing (Ri) plasmid indicates its chimeric structure and evolutionary relationship between tumor-inducing (Ti) and symbiotic (Sym) plasmids in Rhizobiaceae. J Mol Biol. 2001;30:771–784. doi: 10.1006/jmbi.2001.4488. [DOI] [PubMed] [Google Scholar]
- 29.Nishiguchi R, Takanami M, Oka A. Characterization and sequence determination of the replicator region in the hairy-root-inducing plasmid pRiA4b. Mol Gen Genet. 1987;206:1–8. [Google Scholar]
- 30.Nordström K, Austin S J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 31.Oger P, Reich C, Olsen G J, Farrand S F. Complete nucleotide sequence and analysis of pTiBo542: what genomics tells us about structure and evolution of plasmids in the family Rhizobiaceae. Plasmid. 2001;45:169–170. [Google Scholar]
- 32.Palmer K M, Turner S L, Young J P W. Sequence diversity of the plasmid replication gene repC in the Rhizobiaceae. Plasmid. 2000;44:209–219. doi: 10.1006/plas.2000.1488. [DOI] [PubMed] [Google Scholar]
- 33.Ramírez-Romero M A, Soberón N, Pérez-Oseguera A, Téllez-Sosa J, Cevallos M A. Structural elements required for replication and incompatibility of the Rhizobium etli symbiotic plasmid. J Bacteriol. 2000;182:3117–3124. doi: 10.1128/jb.182.11.3117-3124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramírez-Romero M A, Bustos P, Girard L, Rodríguez O, Cevallos M A, Dávila G. Sequence, localization and characteristics of the replicator region of the symbiotic plasmid of Rhizobium etli. Microbiology. 1997;143:2825–2831. doi: 10.1099/00221287-143-8-2825. [DOI] [PubMed] [Google Scholar]
- 35.Rigottier-Gois L, Turner S, Young J P W, Amarger N. Distribution of repC plasmid-replication sequences among plasmids and isolates of Rhizobium leguminosarum bv. viciae from field populations. Microbiology. 1998;144:771–780. doi: 10.1099/00221287-144-3-771. [DOI] [PubMed] [Google Scholar]
- 36.Rodionov O, Łobocka M, Yarmolinsky M. Silencing of genes flanking the P1 plasmid centromere. Science. 1999;283:546–549. doi: 10.1126/science.283.5401.546. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Suzuki K, Hattori Y, Uraji M, Ohta N, Iwata K, Murata K, Kato A, Yoshida K. Complete nucleotide sequence of a plant tumor-inducing Ti plasmid. Gene. 2000;25:331–336. doi: 10.1016/s0378-1119(99)00502-8. [DOI] [PubMed] [Google Scholar]
- 39.Tabata S, Hooykaas P J J, Oka A. Sequence determination and characterization of the replicator region in the tumor-inducing plasmid pTiB6S3. J Bacteriol. 1989;171:1665–1672. doi: 10.1128/jb.171.3.1665-1672.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner S, Young J P W. The replicator region of the Rhizobium leguminosarum cryptic plasmid pRL8JI. FEMS Microbiol Lett. 1995;133:53–58. doi: 10.1111/j.1574-6968.1995.tb07860.x. [DOI] [PubMed] [Google Scholar]
- 41.Williams R, Thomas C M. Active partitioning of bacterial plasmids. J Gen Microbiol. 1992;138:1–16. doi: 10.1099/00221287-138-1-1. [DOI] [PubMed] [Google Scholar]
- 42.Wlodarczyk M, Jagusztyn-Krynicka E K, Bartosik D, Kalinowska I. Electroporation of Thiobacillus versutus with plasmid DNA. Acta Microbiol Pol. 1994;43:223–227. [Google Scholar]
- 43.Yamaichi Y, Niki H. Active segregation by the Bacillus subtilis partitioning system in E. coli. Proc Natl Acad Sci USA. 2000;97:14656–14661. doi: 10.1073/pnas.97.26.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]