Abstract
The Lyme disease spirochaete, Borrelia burgdorferi, can invade and persistently infect its hosts’ connective tissues. We now demonstrate that B. burgdorferi adheres to the extracellular matrix component laminin. The surface-exposed outer-membrane protein ErpX was identified as having affinity for laminin, and is the first laminin-binding protein to be identified in a Lyme disease spirochaete. The adhesive domain of ErpX was shown to be contained within a small, unstructured hydrophilic segment at the protein’s centre. The sequence of that domain is distinct from any previously identified bacterial laminin adhesin, suggesting a unique mode of laminin binding.
INTRODUCTION
Borrelia burgdorferi, the Lyme disease spirochaete, is capable of infecting immunocompetent humans and other vertebrates for extensive periods, even for the host’s lifetime. B. burgdorferi is an extracellular organism, frequently found associated with its hosts’ connective tissues (Barthold et al., 1991, 1992a, b, 1993; Cabello et al., 2007; Cadavid et al., 2003; De Koning et al., 1987; Defosse et al., 1992; Franz et al., 2001; Häupl et al., 1993; Kornblatt et al., 1984; Pachner et al., 1995). B. burgdorferi has been demonstrated to possess affinity for various host extracellular matrix (ECM) components, such as type 1 collagen, fibronectin and decorin (Cabello et al., 2007; Grab et al., 1998; Guo et al., 1995; Probert & Johnson, 1998; Zambrano et al., 2004). Several borrelial outer-surface proteins have been identified that exhibit affinities for some of those host components (Fischer et al., 2006; Guo et al., 1995; Probert & Johnson, 1998).
All examined infectious Lyme disease spirochaete isolates naturally contain between 6 and 10 distinct, but homologous, DNA elements called cp32s (Casjens et al., 2006; Stevenson et al., 2001, 2006). Most cp32 elements are circular episomes approximately 32 kb in size. However, some naturally occurring mutant cp32 family members have been identified, such as the lp56 element of B. burgdorferi type strain B31, which consists of an entire cp32 integrated into an unrelated linear replicon (Casjens et al., 2000). Each cp32 contains a mono- or bicistronic erp locus, which generally varies in sequence between the different cp32s carried by an individual bacterium (Stevenson et al., 2001, 2006). Moreover, different bacterial strains often contain unique erp genes. Erp proteins are surface-exposed outer-membrane lipoproteins that are highly expressed during mammalian infection but generally repressed during colonization of the tick vector (Das et al., 1997; El-Hage et al., 2001; Gilmore et al., 2001; Hefty et al., 2002; Lam et al., 1994; Liang et al., 2002; McDowell et al., 2001; Miller et al., 2003, 2005; Miller & Stevenson, 2006; Miller et al., 2006; Skare et al., 1999).
To date, functions have been ascribed to only a few borrelial Erp proteins, but are consistent with expression of this family of proteins during vertebrate infection. Several closely related (>85% amino acid sequence identities) Erp proteins bind both mammalian factor H and plasminogen/plasmin (Alitalo et al., 2002; Brissette et al., 2009; Hellwage et al., 2001; Kraiczy et al., 2003, 2004; Metts et al., 2003; Stevenson et al., 2002). One member of that group, ErpP (also called BbCRASP-3), binds another, distinct human serum protein, factor H-related protein 1 (FHR-1) (Haupt et al., 2007). At least one other Erp protein adheres to an unknown component(s) of murine endothelium (Antonara et al., 2007).
We now present results of the first studies on interactions between B. burgdorferi and mammalian laminin, a major component of vertebrate basement membranes and many other connective tissues (Colognato & Yurchenco, 2000; Freinkel & Woodley, 2001; Sasaki et al., 2004). Such interactions may facilitate migration of B. burgdorferi through extracellular matrices and/or assist in long-term colonization of laminin-containing host tissues. B. burgdorferi was found to produce several membrane-associated laminin-binding proteins, including the ErpX outer-membrane lipoprotein.
METHODS
Bacteria.
B. burgdorferi strain B31 is the species type strain (Johnson et al., 1984). The complete genome sequence of a non-clonal subculture of strain B31, called B31-MI, has been determined (Casjens et al., 2000; Fraser et al., 1997). A total of 10 distinct erp-bearing cp32 family members have been identified in strain B31, as follows: three separate, identical copies of erpAB on cp32-1, cp32-5 and cp32-8 (formerly known as erpAB, erpIJ and erpNO, respectively); erpCD on cp32-2; erpG on cp32-3; erpHY on cp32-4; erpK on cp32-6; erpLM on cp32-7; erpPQ on cp32-9; and erpX on lp56 (Casjens et al., 1997, 2000; Stevenson et al., 1996). The present studies all used an infectious clone derived from the sequenced culture, named B31-MI-16, that contains all the naturally occurring DNA elements of B31-MI (Miller et al., 2003).
To date, the complete erp contents of four additional strains of Lyme disease spirochaetes have been determined: B. burgdorferi strains N40, 297, Sh-2-82 and BL206 (Akins et al., 1999; Stevenson & Miller, 2003; and our unpublished results). Strains B31 and BL206 are nearly identical to each other, and have been classified as members of intergenic spacer type (IGS) group 1, as determined by sequence analysis of the 16S–23S rRNA gene spacer region (Bunikis et al., 2004; Terekhova et al., 2006; Wang et al., 2001; and our unpublished results). Strains 297 and Sh-2-82 are also nearly identical to each other, and are members of IGS group 2 (Akins et al., 1999; Bunikis et al., 2004; Stevenson & Miller, 2003). Strain N40 is largely distinct genetically from the other analysed strains, and is a member of IGS group 9 (Bunikis et al., 2004; Stevenson & Miller, 2003). Sequences of those strains’ erp genes and their encoded proteins were compared with those of strain B31 to identify similarities using clustal_x (Thompson et al., 1997) with default parameter settings.
B. burgdorferi was cultured in Barbour–Stoenner–Kelly II medium (BSK-II) at 34 °C (Zückert, 2007). Cultures (100 ml) used for bacterial cell lysates were harvested at mid-exponential phase (approx. 107 bacteria ml−1) and fractionated using Triton X-114 (Pryde, 1986). This method yields three fractions from the bacteria: a detergent-soluble fraction primarily consisting of outer-membrane components, an aqueous fraction consisting of periplasmic components, and a pellet that primarily consists of inner-membrane and cytoplasmic components (Cunningham et al., 1988; El-Hage et al., 2001; Radolf et al., 1988). The detergent-soluble, outer-membrane-enriched fraction was used for adhesin analyses.
Interactions between intact B. burgdorferi and laminin.
Glass microscope slides were washed with deionized water, then coated by overnight incubation with 5 μg ml−1 Engelbreth–Holm–Swarm (EHS) mouse sarcoma laminin (Sigma-Aldrich) in PBS. Mice and other rodents are natural reservoir hosts of B. burgdorferi, particularly so in the north-eastern USA, where strain B31 was isolated (Lane et al., 1991). Control slides were coated similarly with bovine serum albumin (BSA). The following day, slides were washed three times with PBS, then blocked by incubation with 3 % (w/v) BSA for 2 h at room temperature. Cultured B31-MI-16 (107 bacteria ml−1, mid-exponential phase) were harvested by centrifugation, washed once with PBS and resuspended in PBS to the original volume. Slides were covered with suspended bacteria, incubated at 37 °C for 2 h, then gently washed 10 times with PBS. Bacteria were visualized by darkfield microscopy. Numbers of adherent bacteria observed in ten 200 × fields per slide were counted.
B. burgdorferi–laminin interactions were quantified by ELISA as follows. Maxisorp 96-well plates (Nalge-Nunc) were coated overnight with 10 μg ml−1 EHS laminin (Sigma-Aldrich) in 50 mM NaHCO3 (pH 9.6) at 4 °C (Stevenson et al., 2007). Plates were brought to room temperature and washed once with PBS + 0.5 % (v/v) Tween 20 (PBS-T). Wells were blocked for 2 h at room temperature with 2 % (w/v) BSA in PBS, then washed three times with PBS-T. Cultured B31-MI-16 (107 bacteria ml−1, mid-exponential phase) were harvested, washed once with PBS and resuspended to the original volume. Aliquots (100 μl) of bacteria were added to each well and incubated for 2 h at 37 °C. Wells were washed three times with PBS-T, then incubated for 1 h at room temperature with rabbit antiserum raised against a membrane-enriched extract of cultured B. burgdorferi strain B31, diluted 1 : 10 000 in PBS (Miller et al., 2003). Plates were washed three times with PBS-T, then incubated for 1 h at room temperature with horseradish-peroxidase-conjugated anti-rabbit IgG (GE Healthcare), diluted 1 : 10 000. Wells were again washed three times with PBS-T, 100 μl per well tetramethylbenzidine substrate (Sigma-Aldrich) was added, then reactions were stopped by addition of 1 M H2SO4, 100 μl per well. Absorbance was read at 450 nm using a Spectramax plate reader using SoftMax Pro (Molecular Devices).
The effects of soluble recombinant ErpX and mutant proteins upon laminin binding by live B. burgdorferi were examined by ELISA as described above, with one additional step. After washing the laminin-coated wells, a 100 μl aliquot of a recombinant protein or BSA (initial concentration 10 μg ml−1) was added to each well, followed by incubation at 37 °C for 30 min. Bacteria were then added directly to each well. Each assay was replicated at least six times.
Statistical analyses were performed using Student’s t-test assuming unequal variances.
Recombinant proteins.
Most of the polyhistidine-tagged, full-length recombinant Erp proteins used in this work have been described earlier (El-Hage & Stevenson, 2002; Stevenson et al., 1998). All recombinant proteins contained amino-terminal tags, with the Erp segment beginning with that protein’s first amino acid following the conserved cysteine lipidation site. Expression plasmids for recombinant ErpA, ErpC, ErpL, ErpP, ErpQ, ErpY and ErpX were based on pET15b (Novagen), plasmids producing recombinant ErpG, ErpK and ErpM were based on pProEX-1 (Life Technologies), and that for recombinant ErpB was based on pET100 (Invitrogen). Two additional constructs, encoding carboxy-terminally truncated recombinant ErpX proteins, were produced from B31-MI-16 total genomic DNA using oligonucleotide primer pairs 5′-CACCAAGATTGATGCAACTGGTAAAG-3′ plus 5′-AATTTCTTAAAATTTAGAATTG-3′ or 5′-CACCAAGATTGATGCAACTGGTAAAG-3′ plus 5′-TTCTTAAACCTTTTCTAATCCTG-3′, and cloned into pET200 (Invitrogen). Further constructs encoding additionally amino- or carboxy-terminally truncated recombinant ErpX proteins were produced from those pET200-based clones, using sequence-overlap extension PCR mutagenesis (Ho et al., 1989) to either delete sections of the erpX open reading frame or introduce premature stop codons. A plasmid encoding recombinant Erp26 was produced from a plasmid clone of the strain N40 locus (Stevenson & Miller, 2003), using pET200 as its basis. Inserts of all recombinant plasmids were entirely sequenced on both strands to ensure that no undesired mutations had occurred during PCR or cloning procedures.
Recombinant proteins were expressed in Escherichia coli Rosetta(DE3)pLysS (Novagen), upon induction with IPTG. Induced E. coli were harvested, lysed by sonication, and debris cleared by centrifugation. Recombinant proteins were purified from cleared lysates using MagneHis nickel-conjugated magnetic beads (Promega). Concentrations of protein preparations were determined by bicinchoninic acid assay as compared with defined stock concentrations of BSA (Pierce).
Ligand-affininty blot analysis of laminin binding.
Immediately prior to use, all apparatus used in these analyses was thoroughly washed with detergent and rinsed with deionized water, since laminin from human skin was found to be a common contaminant of laboratory equipment. Commercially obtained protein molecular mass standards were also found to be frequently contaminated with laminin or substances able to bind laminin, so at least one empty lane was left between the molecular mass standards and the proteins being examined, to avoid blurring of blot signals. BSA was used to block membranes after transfer, since the more commonly used blocking agent non-fat dried milk contains laminin, which obscures results of these analyses.
Recombinant proteins, Triton X-114 outer-membrane fractions, or control protein BSA were suspended in SDS gel loading buffer [125 mM Tris (pH 6.8), 20% (v/v) glycerol, 4% (w/v) SDS, 10% (v/v) β-mercaptoethanol], and heated in a boiling water bath for approximately 1 min. An aliquot (approx. 1 μg) of each protein was loaded into wells of polyacrylamide gels (without added SDS), then subjected to electrophoresis. Proteins were electrotransferred to nitrocellulose membranes, then blocked overnight at 4 °C with 5 % (w/v) BSA in Tris-buffered saline-Tween 20 [TBS-T; 20 mM Tris (pH 7.5), 150 mM NaCl, 0.05% (v/v) Tween 20]. Membranes were next washed with TBS-T, and incubated for 1 h at room temperature in 13 μg ml−1 EHS laminin (Sigma-Aldrich) in TBS-T. After extensive washing with TBS-T, membranes were incubated for 1 h at room temperature in affinity-isolated rabbit anti-EHS laminin polyclonal antiserum (Sigma-Aldrich), diluted 1 : 2500 in TBS-T. Membranes were again washed with TBS-T, then incubated for 1 h at room temperature with horseradish-peroxidase-conjugated donkey anti-rabbit Ig antibody (GE Healthcare), diluted 1 : 5000 in TBS-T. After a final series of washes with TBS-T, bound antibodies were detected using SuperSignal West Pico enhanced chemiluminescence substrate (Pierce). Each recombinant protein was assayed at least twice.
Modelling analyses.
Predictions of disorder within proteins were determined using Predictor of Naturally Disordered Regions (PONDR, www.pondr.com) using VL-XT. Access to PONDR was provided by Molecular Kinetics (6201 La Pas Trail - Ste 160, Indianapolis, IN 46268, USA; + 1 317 280-8737; e-mail: main@molecularkinetics.com). VL-XT is copyright © 1999 by the WSU Research Foundation, all rights reserved, and podnr is copyright © 2004 by Molecular Kinetics, all rights reserved. Primary amino acid sequences of B. burgdorferi proteins and truncated protein fragments were compared with all known sequences in the non-redundant protein sequence database of GenBank using BLASTP and PSI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST). Protein folding probabilities were determined using Protein Homology/analogY Recognition Engine (PHYRE) (http://www.sbg.bio.ic.ac.uk/~phyre).
RESULTS
B. burgdorferi adheres to laminin
As noted in the Introduction, Lyme disease spirochaetes are frequently found associated with host ECM during vertebrate infection. Since laminin is a major component of basement membranes and other mammalian ECMs (Freinkel & Woodley, 2001; Sasaki et al., 2004), the ability of B. burgdorferi to adhere to laminin was examined in detail. Intact, live B. burgdorferi were observed to adhere to laminin-coated microscope slides in significantly greater numbers than to BSA-coated slides (Fig. 1A). Spirochaete–laminin interactions were further quantified through use of an ELISA-based method, again demonstrating that B. burgdorferi exhibits a marked affinity for laminin (Fig. 1B).
Fig. 1.
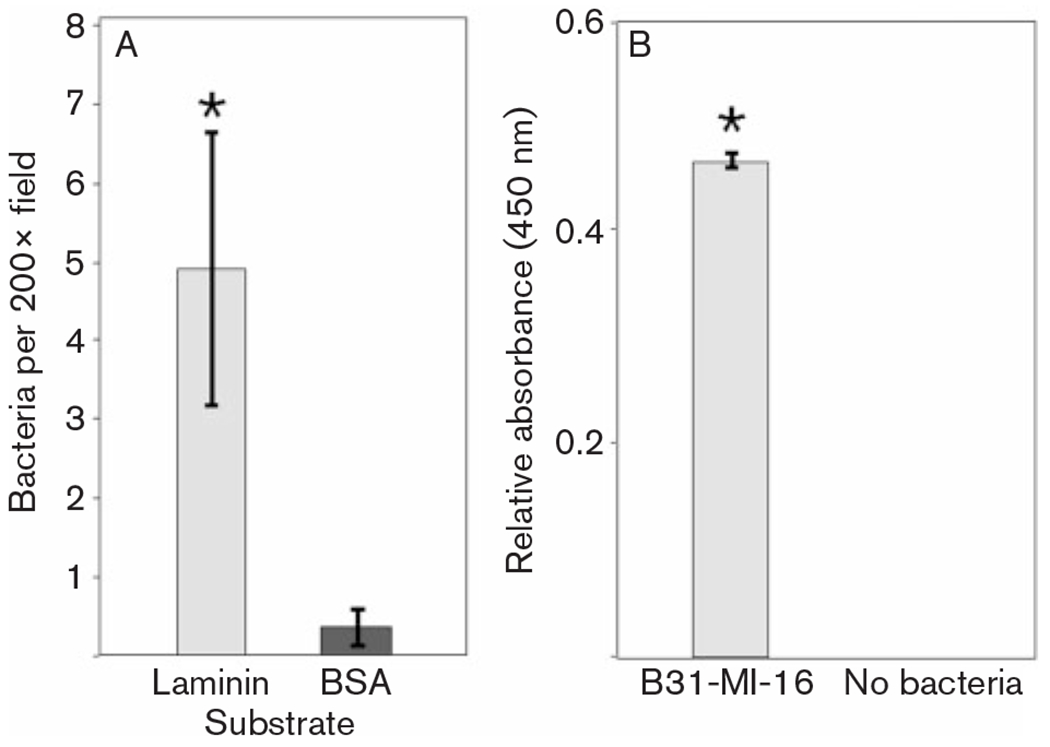
Adherence of intact B. burgdorferi to laminin. Bars indicate 1 standard deviation from the mean. Asterisks indicate statistically significant differences between adherence to laminin and BSA (P<0.001). (A) Microscopical enumeration of bacteria adhering to slides coated with laminin or BSA at initial concentrations of 5 μg protein ml−1. (B) Results of ELISA-based studies of live bacteria binding to fixed laminin, means of 30 assays. Assays without added bacteria served as negative controls.
The specificity of B. burgdorferi–laminin interactions was further analysed by ligand-affinity blot analyses using purified laminin and an outer-membrane fraction of B. burgdorferi. This technique indicated that cultured B. burgdorferi strain B31 produced three proteins with detectable affinities for laminin, having approximate molecular masses of 40 and 50 kDa (Fig. 2).
Fig. 2.
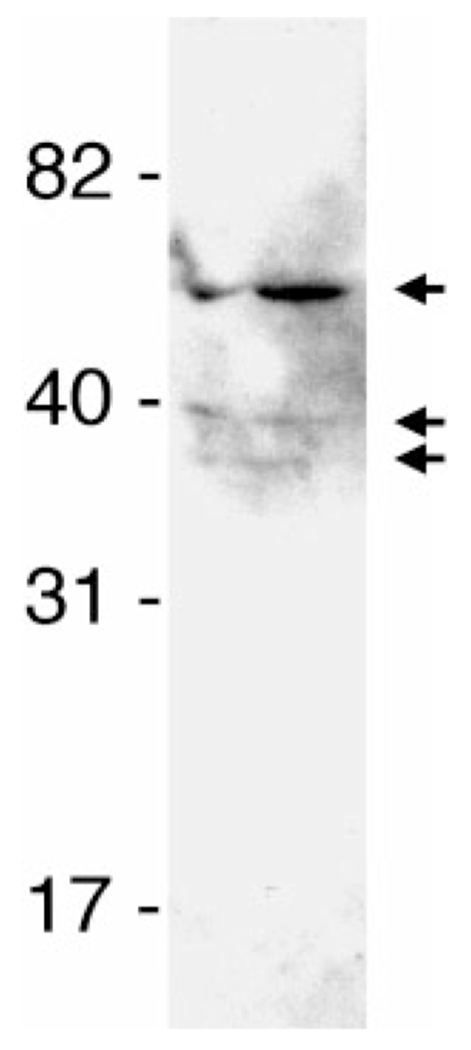
Ligand-affininty blot analyses of laminin binding to components of outer-membrane-enriched Triton X-114 extract of cultured B. burgdorferi B31-MI-16. Arrows to the right indicate bacterial proteins that bound laminin. Positions of molecular mass standards are indicated to the left.
ErpX binds mammalian laminin
Studies by our laboratories and others have demonstrated that all B. burgdorferi erp genes so far examined are expressed during mammalian infection (our unpublished results; Crother et al., 2004; Liang et al., 2002; Miller et al., 2005; Miller & Stevenson, 2006). Noting that the laminin-binding outer-membrane proteins detected in Fig. 2 are of sizes similar to some Erp proteins (Casjens et al., 2000; Stevenson et al., 1998, 2006), we hypothesized that one or more of the borrelial laminin-binding proteins may be an Erp protein. To explore that possibility, recombinant Erp proteins were produced from all the erp gene sequences carried by B. burgdorferi strain B31. Equal amounts of each protein were then subjected to ligand-affininty blot analyses to determine their abilities to bind laminin. BSA was also included, as a negative control to demonstrate that ligand binding was not due to non-specific protein–protein interactions. As described below, most of the recombinant Erp proteins did not bind laminin, and therefore also served as controls for specificity of laminin binding.
Of the strain B31-derived Erp proteins, only recombinant ErpX bound laminin with a detectable affinity (Fig. 3A). Native, lipidated ErpX migrates in SDS-PAGE with an apparent molecular mass of approximately 45–50 kDa (El-Hage et al., 2001; Stevenson et al., 1998), and was probably the larger laminin-binding protein observed in Fig. 2. Several of the other tested recombinant Erp proteins (ErpA, ErpC, ErpL, ErpP, ErpQ, ErpY) were produced using the same cloning vector as was ErpX, and all included the same amino-terminal tag, so the inabilities of those other recombinant proteins to bind laminin demonstrated that laminin binding by recombinant ErpX was not due to its polyhistidine tag. The erpX gene sequence is distinct from all the other erp genes of B. burgdorferi strain B31 (Stevenson et al., 2006). Of the strain B31 Erp proteins, ErpX shares the highest degree of similarity with ErpQ protein (see Supplementary Fig. S1, available with the online version of this paper), which did not detectably bind laminin, indicating that the laminin-binding region(s) of ErpX is distinct from any sequence found in ErpQ.
Fig. 3.
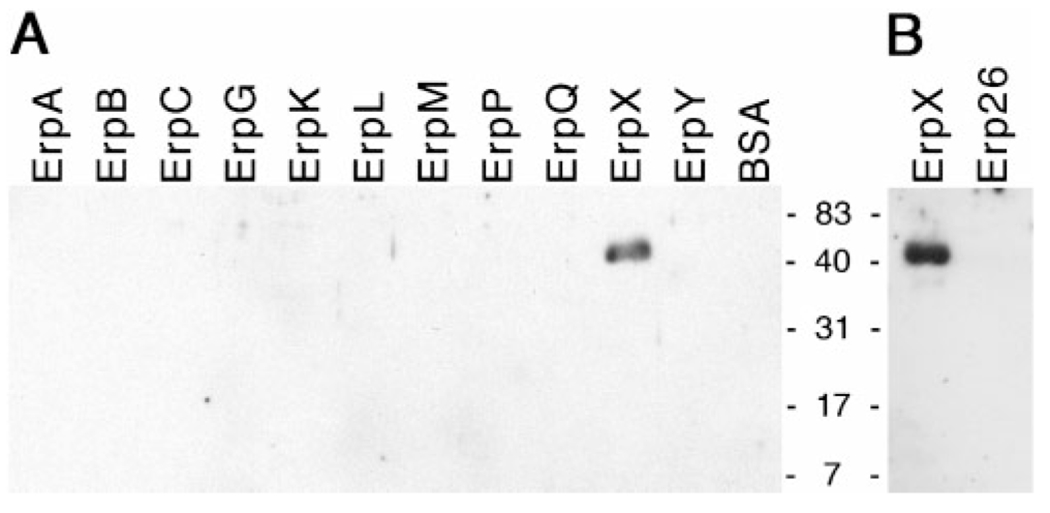
Ligand-affininty blot analyses of laminin binding by polyhistidine-tagged recombinant Erp proteins. (A) Recombinant Erp proteins of B. burgdorferi strain B31. BSA was included as a control for non-specific protein–laminin interactions. (B) Recombinant ErpX from strain B31 and Erp26 from strain N40. Positions of molecular mass standards are indicated between the two panels.
The complete erp gene sequences of several additional strains of the Lyme disease spirochaete have been determined. Of all the known borrelial erp genes, the strain N40 erp26 gene shares the highest degree of similarity with the strain B31 erpX gene (Fig. S1 and Stevenson & Miller, 2003; Stevenson et al., 2006). Due to that similarity, recombinant N40 Erp26 protein was produced and assayed for ability to bind laminin. However, recombinant Erp26 was not able to bind laminin at detectable levels (Fig. 3B). Thus, the laminin-binding domain(s) of ErpX is also distinct from any sequence within Erp26. Studies described below determined that the laminin-binding domain of ErpX consists of sequences unlike any found in either the N40 Erp26 or B31 ErpQ proteins, consistent with the results of these ligand-affininty analyses.
Mapping the laminin-binding domain of ErpX
Truncated variants of ErpX were produced to narrow down the region of that protein involved with binding of laminin. A distinctive characteristic of ErpX is a fivefold repeat of the 5 amino acid sequence aspartate-alanine-threonine-glycine-lysine near the protein’s amino terminus (Fig. 4A and Fig. S1). However, truncated variants of ErpX lacking the repeated amino acid sequence, such as rErpX3 and rErpX5, did not show any appreciable differences in laminin binding, indicating that the amino acid repeat region does not play a direct role in adhesion to laminin (Figs 4A and 5A). Additional amino- and carboxy-terminally truncated variants of ErpX were produced and assessed for ability to bind laminin. Truncated proteins rErpX3 through rErpX12 all bound laminin with the same apparent affinities as did the full-length, wild-type rErpX protein (Figs 4A and 5A, and data not shown).
Fig. 4.
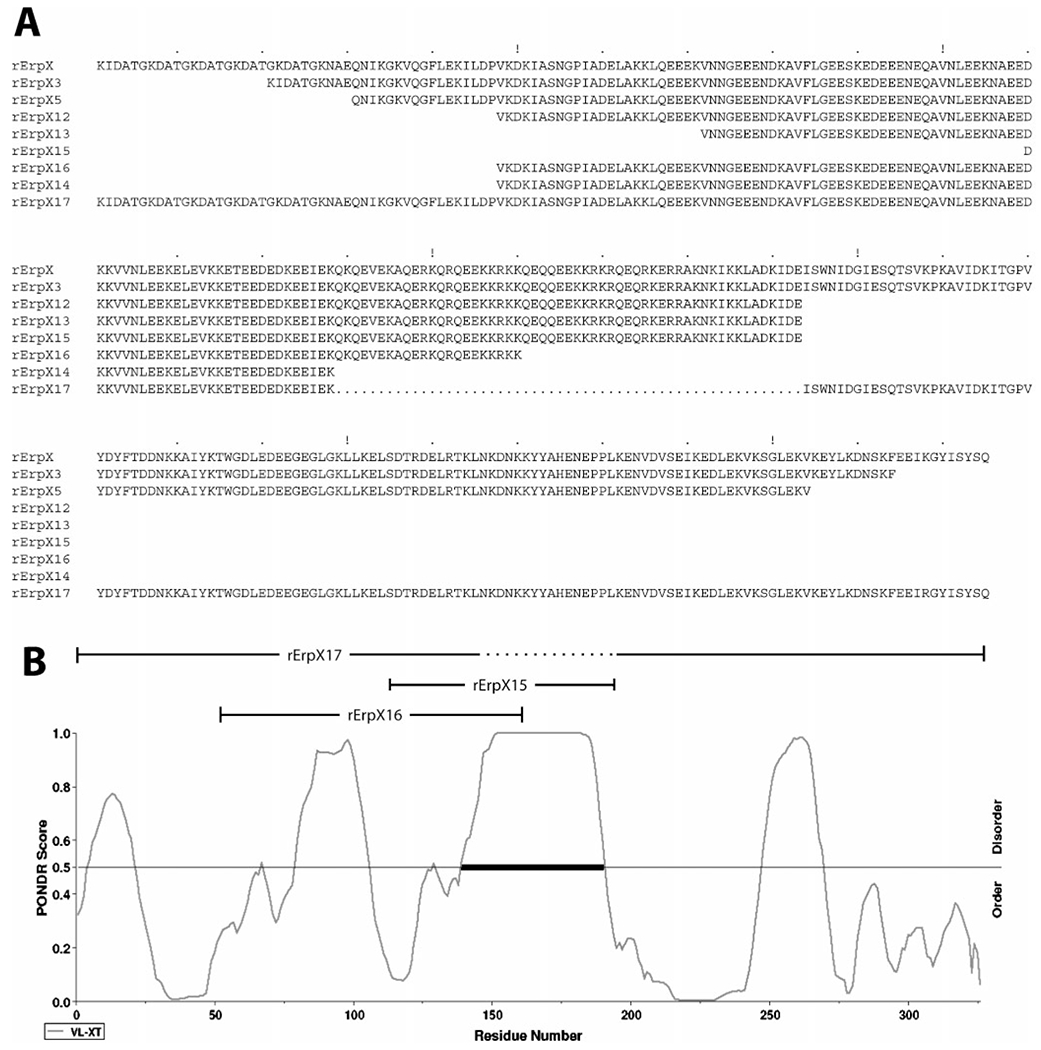
(A) Amino acid sequences of recombinant ErpX (rErpX) and the truncated derivatives of ErpX used in these studies. The 55 amino acids deleted from rErpX17 are indicated by dots. (B) Predicted order/disorder of wild-type ErpX. A PONDR score above 0.5 predicts protein disorder; a score below 0.5 predicts order. The laminin-binding region in the centre of ErpX, as identified through use of variant proteins rErpX15, rErpX16 and rErpX17, is predicted to be extremely disordered in the absence of bound ligand.
Fig. 5.
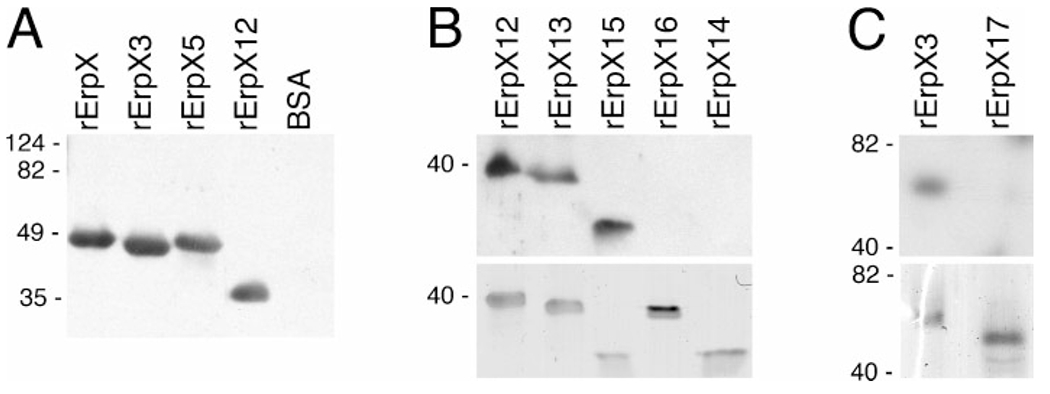
Ligand-affininty blot analysis of laminin binding by recombinant ErpX (rErpX) and truncated derivatives thereof. (A) Full-length ErpX and three of the examined truncations. The rightmost lane contained an equal molar content of BSA, to serve as a control for non-specific laminin binding. (B) The upper panel shows results of laminin-binding analysis of truncated proteins rErpX12, rErpX13, rErpX15, rErpX16 and rErpX14, and the lower panel shows the same amounts of each protein in a gel stained with Coomassie brilliant blue. Note that even though relatively less rErpX15 appears to have been loaded onto this gel, the laminin-binding signal for that protein was comparable to those from ErpX12 and rErpX13. (C) The upper panel illustrates results of laminin-binding analysis of proteins rErpX3 and rErpX17, and the lower panel shows the proteins stained with Coomassie brilliant blue. Migration positions of molecular mass standards are indicated on the left of each panel. Highly charged proteins such as ErpX often exhibit aberrant SDS-PAGE mobilities (Stevenson et al., 1998): note that rErpX-5 migrated at a rate suggestive of a molecular mass greater than rErpX-3, even though rErpX-5 is actually the smaller protein.
Loss of ligand binding was observed upon truncation of ErpX to the 112 residue rErpX16 (Figs 4 and 5B). Recombinant protein rErpX14, consisting of 90 amino acids nested within rErpX16, was similarly unable to bind laminin. However, protein rErpX15, which contains only 84 amino acids, retained laminin-binding activity. Since the sequences of rErpX15 and rErpX16 overlap (Fig. 4A), these results indicate that the carboxy-terminal 32 amino acids of rErpX15 include residues critical for binding of laminin.
Following the result of studies with rErpX15 and rErpX16, protein rErpX17 was produced, consisting of the entire ErpX sequence except for 55 central amino acids (Fig. 4). Consistent with the ability of rErpX15 to bind laminin and the inabilities of rErpX14 and rErpX16 to bind that ligand, rErpX17 did not bind laminin at detectable levels (Fig. 5C).
Full-length rErpX and several fragments were analysed for abilities to interfere with adhesion of B. burgdorferi to laminin. Consistent with the above-described ligand-affininty blot analyses, rErpX, rErpX-12, rErpX-13 and rErpX-15 all reduced B. burgdorferi adherence to laminin by 60–70 % (Fig. 6). Protein rErpX-17 inhibited binding by approximately 20 %, suggesting that this protein retains some, otherwise undetectable affinity for laminin. Addition of the control protein, BSA, did not influence laminin-binding by the spirochaetes.
Fig. 6.
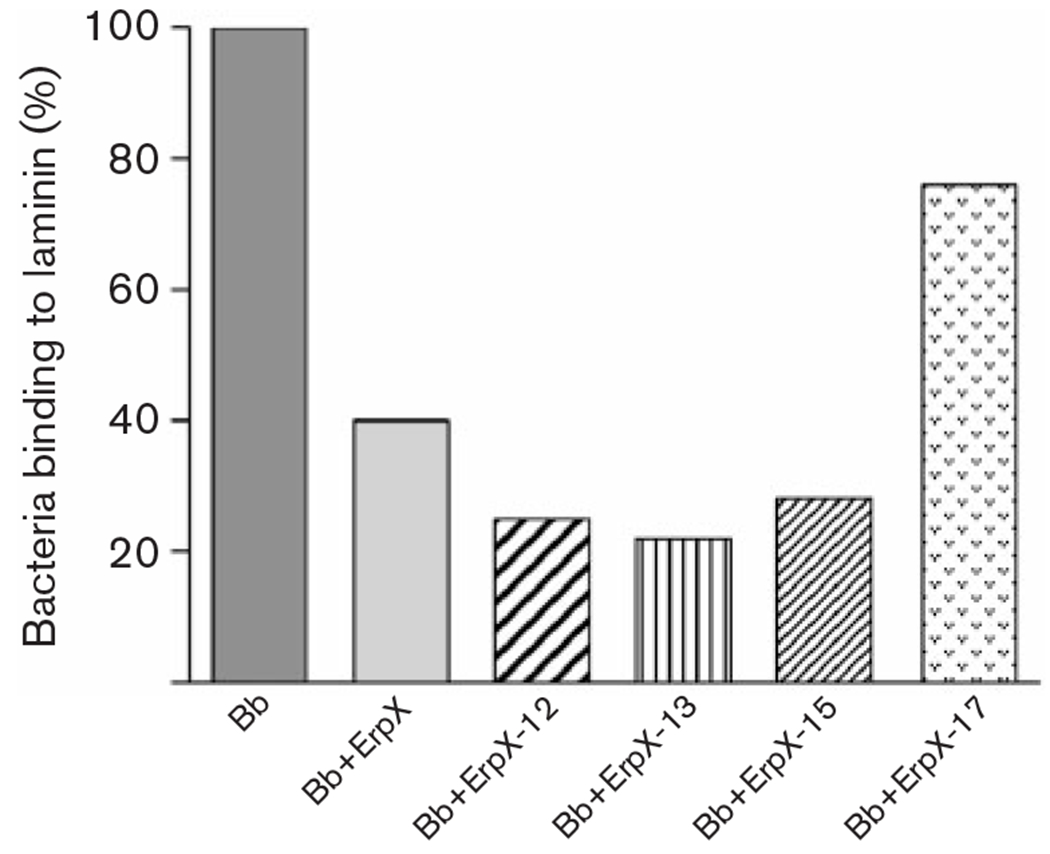
Effects of recombinant proteins on adherence of B. burgdorferi to laminin, as measured by ELISA. Results for each protein are illustrated relative to control experiments lacking added protein. All added ErpX derivatives significantly affected adherence to laminin: rErpX-17 (P=0.0009), and rErpX, rErpX-12, rErpX-13 and rErpX-15 (all P<0.0001). Bb, B. burgdorferi.
As are all Erp proteins, ErpX is a highly charged molecule, with the full-length, mature protein containing 18% glutamate, 9% aspartate and 19% lysine residues and having a predicted pI of 4.9. The central, laminin-binding region defined by the deletion within rErpX17 has a predicted pI of 11.1 and consists of 20% glutamate, 4% aspartate, 29% lysine, 18% glutamine and 15% arginine residues (Fig. 4A). Modelling analyses predicted that this central region is largely disordered (Fig. 4B). The flanking sections of the full-length ErpX protein possess more structured natures, which could serve to stabilize or orient the protein’s middle, laminin-binding region. Neither blastp nor psi-blast detected similarities between the laminin-binding region of ErpX and any previously identified predicted protein. Likewise, phyre analyses did not predict either the central region or the full-length ErpX to have similarities with any known protein structure. ErpX appears to represent a novel type of bacterial laminin adhesin.
DISCUSSION
Lyme disease spirochaetes reside extracellularly during vertebrate infection. Several previous studies found that B. burgdorferi interacts with constituents of host ECM and cell-surface components. The present studies appear to be the first to demonstrate that B. burgdorferi adheres to laminin. Analyses of outer-membrane-enriched fractions of the cultured B. burgdorferi type strain B31 indicated that it produces three proteins with detectable affinities for laminin. We determined that the surface-exposed lipoprotein ErpX is a laminin-binding protein, and mapped its ligand-binding site to a short, unstructured region in the centre of the protein. Serological, RNA array and gene-specific Q-RT-PCR analyses have all indicated that B. burgdorferi produces ErpX during mammalian infection (our unpublished results; Liang et al., 2002; Skare et al., 1999; Stevenson et al., 1998). Thus, ErpX is appropriately poised on the borrelial outer surface to interact with host laminin, and we hypothesize that this bacterial protein contributes to invasion and/or long-term colonization of host tissues. Prior studies indicated that B. burgdorferi strains which lack lp56 (and are therefore erpX deficient) are still able to infect mice (Purser & Norris, 2000), which may be due to binding of additional host ECM components by other borrelial surface proteins or to the redundancy of laminin-binding proteins produced by B. burgdorferi (see Fig. 2). Preliminary analyses of the other laminin adhesins with apparent molecular masses of ~40 kDa suggest them to be members of the chromosomally encoded Bmp (p39) protein family (our unpublished results). Complete characterization of the other laminin-binding proteins is under way, with the goals of defining their mechanisms of ligand adhesion and producing mutant bacteria deficient in all laminin adhesins for mouse and tick infection studies.
There are no apparent similarities between ErpX and the known laminin-binding proteins of the spirochaetes Leptospira interrogans (Barbosa et al., 2006; Stevenson et al., 2007), Treponema pallidum (Cameron, 2003; Cameron et al., 2005) or T. denticola (Edwards et al., 2005), nor of any other organism, suggesting that ErpX adhesion to laminin may occur through a novel mechanism. The predicted disordered nature of the laminin-binding region of ErpX is reminiscent of the fibronectin-binding domain of the B. burgdorferi BBK32/Fbp/p47 protein, in which the binding site consists of a disordered sequence of amino acids which moulds itself around the ligand (Kim et al., 2004).
One striking feature of ErpX is the fivefold reiterated 5 amino acid sequence near the amino terminus. Our studies demonstrated that this sequence is not directly involved with laminin binding. The directly repeated sequences of erpX presumably arose through either cross-fork slippage or recombination events (Lovett, 2004). It is significant that this direct repeat involves a multiple of 3 bp, such that the reiterations did not alter the reading frame, and indicating that this repeat element cannot be involved with phase variation as occurs in bacteria such as Neisseria gonorrhoeae or Haemophilus influenzae. We also observed that the erpX genes of B. burgdorferi strains B31 and BL206 are identical (our unpublished results). As strain BL206 was isolated from nature 16 years after strain B31, and must have infected numerous vertebrate and tick hosts since the two strains shared a common ancestor, this direct repeat mutation appears to be highly stable. A possible function for the repeated amino-terminal motif of ErpX is suggested from studies of other bacterial adhesins: repeated amino acid sequences near the membrane anchor can provide distance between adhesive domains and the bacterial cell surface, and the greater the number of repeats, the further the adhesin extends into the environment (Patti et al., 1994; van Belkum, 1999).
In conclusion, we have determined that the B. burgdorferi outer-surface lipoprotein ErpX binds mammalian laminin, through a protein motif not previously recognized in other bacterial laminin adhesins. No other Erp proteins of strain B31 bound laminin, nor did the Erp26 protein of strain N40, and the functions of most of those proteins remain a mystery. However, the Lyme disease spirochaete produces at least two other laminin-binding proteins, neither of which is an Erp protein. It is possible that dissimilarities among the repertoires of Erp and other proteins produced by different B. burgdorferi strains may play roles in the variations in infectivity observed among isolates (Balmelli & Piffaretti, 1995; Liveris et al., 2002; Seinost et al., 1999; Shih et al., 1992; Terekhova et al., 2006; van Dam et al., 1993; Wang et al., 2002). Similarly, differences in amino acid sequences and modifications to host ECM components such as laminin may contribute to the variations among borreliae in their abilities to efficiently infect different host species (Kurtenbach et al., 2006). Further characterization of borrelial adhesins and examination of interactions between these bacterial proteins and host tissues will address those hypotheses, providing insight into the pathogenic properties of Lyme disease spirochaetes. Such information could direct development of therapeutic treatments that block adhesin–ECM interactions or disrupt production of adhesins associated with mammalian infection.
Supplementary Material
ACKNOWLEDGEMENTS
This study was funded by US National Institutes of Health grant R01-AI44254 awarded to B. S. We thank Linda Bockenstedt, Logan Burns, Tomasz Bykowski, Caroline Cameron, Henry Choy, Mariana Ionita, Sean Riley, Michael Woodman and Wolfram Zückert for helpful discussions on this work.
REFERENCES
- Akins DR, Caimano MJ, Yang X, Cerna F, Norgard MV & Radolf JD (1999). Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect Immun 67, 1526–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alitalo A, Meri T, Lankinen H, Seppälä I, Lahdenne P, Hefty PS, Akins D & Meri S (2002). Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J Immunol 169, 3847–3853. [DOI] [PubMed] [Google Scholar]
- Antonara S, Chafel RM, LaFrance M & Coburn J (2007). Borrelia burgdorferi adhesins identified using in vivo phage display. Mol Microbiol 66, 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmelli T & Piffaretti JC (1995). Association between different clinical manifestations of Lyme disease and different species of Borrelia burgdorferi sensu lato. Res Microbiol 146, 329–340. [DOI] [PubMed] [Google Scholar]
- Barbosa AS, Abreu PAE, Neves FO, Atzingen MV, Watanabe MM, Vieira ML, Morais ZM, Vasconcellos SA & Nascimentao ALTO (2006). A newly identified leptospiral adhesin mediates attachment to laminin. Infect Immun 74, 6356–6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold SW, Persing DH, Armstrong AL & Peeples RA (1991). Kinetics of Borrelia burgdorferi dissemination and evolution of disease after intradermal inoculation of mice. Am J Pathol 139, 263–273. [PMC free article] [PubMed] [Google Scholar]
- Barthold SW, de Souza M, Fikrig E & Persing DH (1992a). Lyme borreliosis in the laboratory mouse. In Lyme Disease: Molecular and Immunologic Approaches, pp. 223–242. Edited by Schutzer SE. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Barthold SW, Sidman CL & Smith AL (1992b). Lyme borreliosis in genetically resistant and susceptible mice with severe combined immunodeficiency. Am J Trop Med Hyg 47, 605–613. [DOI] [PubMed] [Google Scholar]
- Barthold SW, de Souza MS, Janotka JL, Smith AL & Persing DH (1993). Chronic Lyme borreliosis in the laboratory mouse. Am J Pathol 143, 959–972. [PMC free article] [PubMed] [Google Scholar]
- Brissette CA, Haupt K, Barthel D, Cooley AE, Bowman A, Skerka C, Wallich R, Zipfel PF, Kraiczy P & Stevenson B (2009). The Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect Immun 77, 300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunikis J, Garpmo U, Tsao J, Berglund J, Fish D & Barbour AG (2004). Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150, 1741–1755. [DOI] [PubMed] [Google Scholar]
- Cabello FC, Godfrey HP & Newman SA (2007). Hidden in plain site: Borrelia burgdorferi and the extracellular matrix. Trends Microbiol 15, 350–354. [DOI] [PubMed] [Google Scholar]
- Cadavid D, Bai Y, Dail D, Hurd M, Narayan K, Hodzic E, Barthold SW & Pachner AR (2003). Infection and inflammation in skeletal muscle from nonhuman primates infected with different genospecies of the Lyme disease spirochete Borrelia burgdorferi. Infect Immun 71, 7087–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE (2003). Identification of a Treponema pallidum laminin-binding protein. Infect Immun 71, 2525–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE, Brouwer NL, Tisch LM & Kurioiwa JMY (2005). Defining the interaction of the Treponema pallidum adhesin Tp0751 with laminin. Infect Immun 73, 7485–7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, van Vugt R, Tilly K, Rosa PA & Stevenson B (1997). Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol 179, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J & other authors (2000). A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35, 490–516. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Huang WM, Gilcrease EB, Qiu W, McCaig WD, Luft BJ, Schutzer SE & Fraser CM (2006). Comparative genomics of Borrelia burgdorferi. In Molecular Biology of Spirochetes, pp. 79–95. Edited by Cabello FC, Hulinska D & Godfrey HP. Amsterdam: IOS Press. [Google Scholar]
- Colognato H & Yurchenco PD (2000). Form and function: the laminin family of heterotrimers. Dev Dyn 218, 213–234. [DOI] [PubMed] [Google Scholar]
- Crother TR, Champion CI, Whitelegge JP, Aguilera R, Wu XY, Blanco DR, Miller JN & Lovett MA (2004). Temporal analysis of the antigenic composition of Borrelia burgdorferi during infection in rabbit skin. Infect Immun 72, 5063–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TM, Thomas DD, Thompson SD, Miller JN & Lovett MA (1988). Identification of Borrelia burgdorferi surface components by Triton X-114 phase partitioning. Ann N Y Acad Sci 539, 376–378. [Google Scholar]
- Das S, Barthold SW, Stocker Giles S, Montgomery RR, Telford SR & Fikrig E (1997). Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J Clin Invest 99, 987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defosse DL, Duray PH & Johnson RC (1992). The NIH-3 immunodeficient mouse is a model for Lyme borreliosis myositis and carditis. Am J Pathol 141, 3–10. [PMC free article] [PubMed] [Google Scholar]
- De Koning J, Bosma RB & Hoogkamp-Korstanje JA (1987). Demonstration of spirochaetes in patients with Lyme disease with a modified silver stain. J Med Microbiol 23, 261–267. [DOI] [PubMed] [Google Scholar]
- Edwards AM, Jenkinson HF, Woodward MJ & Dymock D (2005). Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect Immun 73, 2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hage N & Stevenson B (2002). Simultaneous coexpression of Borrelia burgdorferi Erp proteins occurs through a specific, erp locus-directed regulatory mechanism. J Bacteriol 184, 4536–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hage N, Babb K, Carroll JA, Lindstrom N, Fischer ER, Miller JC, Gilmore RD Jr, Mbow ML & Stevenson B (2001). Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147, 821–830. [DOI] [PubMed] [Google Scholar]
- Fischer JR, LeBlanc KT & Leong JM (2006). Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infect Immun 74, 435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz JK, Fritze O, Rittig M, Keyßer G, Priem S, Zacher J, Burmester GR & Krause A (2001). Insights from a novel three-dimensional in vitro model of Lyme arthritis. Arthritis Rheum 44, 151–162. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R & other authors (1997). Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390, 580–586. [DOI] [PubMed] [Google Scholar]
- Freinkel RK & Woodley DT (2001). The Biology of the Skin. Pearl River, NY: Parthenon. [Google Scholar]
- Gilmore RD Jr, Mbow ML & Stevenson B (2001). Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect 3, 799–808. [DOI] [PubMed] [Google Scholar]
- Grab DJ, Givens C & Kennedy R (1998). Fibronectin-binding activity in Borrelia burgdorferi. Biochim Biophys Acta 1407, 135–145. [DOI] [PubMed] [Google Scholar]
- Guo BP, Norris SJ, Rosenberg LC & Höök M (1995). Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect Immun 63, 3467–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häupl T, Hahn G, Rittig M, Krause A, Schoerner C, Schonherr U, Kalden JR & Burmester GR (1993). Persistence of Borrelia burgdorferi in ligamentous tissue from a patient with chronic Lyme borreliosis. Arthritis Rheum 36, 1621–1626. [DOI] [PubMed] [Google Scholar]
- Haupt K, Kraiczy P, Wallich R, Brade V, Skerka C & Zipfel PF (2007). Binding of human FHR-1 to serum resistant Borrelia burgdorferi is mediated by borrelial complement regulator-acquiring surface proteins. J Infect Dis 196, 124–133. [DOI] [PubMed] [Google Scholar]
- Hefty PS, Jolliff SE, Caimano MJ, Wikel SK & Akins DR (2002). Changes in the temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun 70, 3468–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwage J, Meri T, Heikkilä T, Alitalo A, Panelius J, Lahdenne P, Seppälä IJT & Meri S (2001). The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J Biol Chem 276, 8427–8435. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK & Pease LR (1989). Site-directed mutagensis by overlap extension using polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Johnson RC, Schmid GP, Hyde FW, Steigerwalt AG & Brenner DJ (1984). Borrelia burgdorferi sp. nov.: etiologic agent of Lyme disease. Int J Syst Bacteriol 34, 496–497. [Google Scholar]
- Kim JH, Singvall J, Schwartz-Linek U, Johnson BJB, Potts JR & Höök M (2004). BBK32, a fibronectin binding MSCRAMM from Borrelia burgdorferi, contains a disordered region that undergoes a conformational change on ligand binding. J Biol Chem 279, 41706–41714. [DOI] [PubMed] [Google Scholar]
- Kornblatt AN, Steere AC & Brownstein DG (1984). Experimental Lyme disease in rabbits: spirochetes found in erythema migrans and blood. Infect Immun 46, 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy P, Hellwage J, Skerka C, Kirschfink M, Brade V, Zipfel PF & Wallich R (2003). Immune evasion of Borrelia burgdorferi: mapping of a complement inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur J Immunol 33, 697–707. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Hartmann K, Hellwage J, Skerka C, Brade V, Zipfel PF, Wallich R & Stevenson B (2004). Immunological characterization of the complement regulator factor H-binding CRASP and Erp proteins of Borrelia burgdorferi. Int J Med Microbiol 293 (Suppl. 37), 152–157. [DOI] [PubMed] [Google Scholar]
- Kurtenbach K, Hanincova K, Tsao JI, Margos G, Fish D & Ogden NH (2006). Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat Rev Microbiol 4, 660–669. [DOI] [PubMed] [Google Scholar]
- Lam TT, Nguyen T-PK, Montgomery RR, Kantor FS, Fikrig E & Flavell RA (1994). Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect Immun 62, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane RS, Piesman J & Burgdorfer W (1991). Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu Rev Entomol 36, 587–609. [DOI] [PubMed] [Google Scholar]
- Liang FT, Nelson FK & Fikrig E (2002). Molecular adaptation of Borrelia burgdorferi in the murine host. J Exp Med 196, 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liveris D, Wang G, Girao G, Byrne DW, Nowakowski J, McKenna D, Nadelman R, Wormser GP & Schwartz I (2002). Quantitiative detection of Borrelia burgdorferi in 2-millimeter skin samples of erythema migrans lesions: correlation of results with clinical and laboratory findings. J Clin Microbiol 40, 1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett ST (2004). Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol Microbiol 52, 1243–1253. [DOI] [PubMed] [Google Scholar]
- McDowell JV, Sung SY, Price G & Marconi RT (2001). Demonstration of the genetic stability and temporal expression of select members of the Lyme disease spirochete OspF protein family during infection in mice. Infect Immun 69, 4831–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metts MS, McDowell JV, Theisen M, Hansen PR & Marconi RT (2003). Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection. Infect Immun 71, 3587–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC & Stevenson B (2006). Borrelia burgdorferi erp genes are expressed at different levels within tissues of chronically infected mammalian hosts. Int J Med Microbiol 296 (Suppl. 1), 185–194. [DOI] [PubMed] [Google Scholar]
- Miller JC, von Lackum K, Babb K, McAlister JD & Stevenson B (2003). Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect Immun 71, 6943–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Narayan K, Stevenson B & Pachner AR (2005). Expression of Borrelia burgdorferi erp genes during infection of nonhuman primates. Microb Pathog 39, 27–33. [DOI] [PubMed] [Google Scholar]
- Miller JC, von Lackum K, Woodman ME & Stevenson B (2006). Detection of Borrelia burgdorferi gene expression during mammalian infection using transcriptional fusions that produce green fluorescent protein. Microb Pathog 41, 43–47. [DOI] [PubMed] [Google Scholar]
- Pachner AR, Basta J, Delaney E & Hulinska D (1995). Localization of Borrelia burgdorferi in murine Lyme borreliosis by electron microscopy. Am J Trop Med Hyg 52, 128–133. [DOI] [PubMed] [Google Scholar]
- Patti JM, Allen BL, McGavin MJ & Höök M (1994). MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol 48, 585–617. [DOI] [PubMed] [Google Scholar]
- Probert WS & Johnson BJB (1998). Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol Microbiol 30, 1003–1015. [DOI] [PubMed] [Google Scholar]
- Pryde JG (1986). Triton X-114: a detergent that has come in from the cold. Trends Biochem Sci 11, 160–163. [Google Scholar]
- Purser JE & Norris SJ (2000). Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97, 13865–13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD, Chamberlain NR, Clausell A & Norgard MV (1988).Identification and localization of integral membrane proteins of virulent Treponema pallidum subsp. pallidum by phase partitioning with the nonionic detergent Triton X-114. Infect Immun 56, 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Fässler R & Hohenester E (2004). Laminin: the crux of basement membrane assembly. J Cell Biol 164, 959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, Wang IN, Wormser GP, Schriefer ME & Luft BJ (1999). Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun 67, 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C-M, Pollack RJ, Telford SR & Spielman A (1992). Delayed dissemination of Lyme disease spirochetes from the site of deposition in the skin of mice. J Infect Dis 166, 827–831. [DOI] [PubMed] [Google Scholar]
- Skare JT, Foley DM, Hernandez SR, Moore DC, Blanco DR, Miller JN & Lovett MA (1999). Cloning and molecular characterization of plasmid-encoded antigens of Borrelia burgdorferi. Infect Immun 67, 4407–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B & Miller JC (2003). Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J Mol Evol 57, 309–324. [DOI] [PubMed] [Google Scholar]
- Stevenson B, Tilly K & Rosa PA (1996). A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J Bacteriol 178, 3508–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Bono JL, Schwan TG & Rosa P (1998). Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect Immun 66, 2648–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Zückert WR & Akins DR (2001). Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. In The Spirochetes: Molecular and Cellular Biology, pp. 87–100. Edited by Saier MH & García-Lara J. Oxford: Horizon Press. [PubMed] [Google Scholar]
- Stevenson B, El-Hage N, Hines MA, Miller JC & Babb K (2002). Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect Immun 70, 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Bykowski T, Cooley AE, Babb K, Miller JC, Woodman ME, von Lackum K & Riley SP (2006). The Lyme disease spirochete Erp lipoprotein family: structure, function and regulation of expression. In Molecular Biology of Spirochetes, pp. 354–372. Edited by Cabello FC, Godfrey HP & Hulinska D. Amsterdam: IOS Press. [Google Scholar]
- Stevenson B, Choy HA, Pinne M, Rotondi ML, Miller MC, DeMoll E, Kraiczy P, Cooley AE, Creamer TP & other authors (2007). Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS One 2, e1188. doi:1110.1371/journal/pone.0001188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terekhova D, Iyer R, Wormser GP & Schwartz I (2006). Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. J Bacteriol 188, 6124–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F & Higgins DG (1997). The clustal_x Windows interface: flexible strategies for multiple sequence alignment aided by quality analyses tools. Nucleic Acids Res 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A (1999). Short sequence repeats in microbial pathogenesis and evolution. Cell Mol Life Sci 56, 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam AP, Kuiper H, Vos K, Widjojokusumo A, de Jongh BM, Spanjaard L, Ramselaar ACP, Kramer MD & Dankert J (1993). Different genospecies of Borrelia burgdorferi are associated with distinct clinical manifestations of Lyme borreliosis. Clin Infect Dis 17, 708–717. [DOI] [PubMed] [Google Scholar]
- Wang G, Ojaimi C, Iyer R, Saksenberg V, McClain SA, Wormser GP & Schwartz IA (2001). Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect Immun 69, 4303–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ojaimi C, Wu H, Saksenberg V, Iyer R, Liveris D, McClain SA, Wormser GP & Schwartz I (2002). Disease severity in a murine model of Lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J Infect Dis 186, 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano MC, Beklemisheva AA, Bryskin AV, Newman SA & Cabello PC (2004). Borrelia burgdorferi binds to, invades, and colonizes native type I collagen lattices. Infect Immun 72, 3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zückert WR (2007). Laboratory maintenance of Borrelia burgdorferi. In Current Protocols in Microbiology, pp. 12C.11.11–12C.11.10. Edited by Coico RT, Kowalik TF, Quarles J, Stevenson B & Taylor R. Hoboken, NJ: Wiley. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


