Abstract
The primary cilium is a nexus for cell signaling and relies on specific protein trafficking for function. The tubby family protein TULP3 transports integral membrane proteins into cilia through interactions with the intraflagellar transport complex-A (IFT-A) and phosphoinositides. It was previously shown that short motifs called ciliary localization sequences (CLSs) are necessary and sufficient for TULP3-dependent ciliary trafficking of transmembrane cargoes. However, the mechanisms by which TULP3 regulates ciliary compartmentalization of nonintegral, membrane-associated proteins and whether such trafficking requires TULP3-dependent CLSs is unknown. Here we show that TULP3 is required for ciliary transport of the Joubert syndrome–linked palmitoylated GTPase ARL13B through a CLS. An N-terminal amphipathic helix, preceding the GTPase domain of ARL13B, couples with the TULP3 tubby domain for ciliary trafficking, irrespective of palmitoylation. ARL13B transport requires TULP3 binding to IFT-A but not to phosphoinositides, indicating strong membrane-proximate interactions, unlike transmembrane cargo transport requiring both properties of TULP3. TULP3-mediated trafficking of ARL13B also regulates ciliary enrichment of farnesylated and myristoylated downstream effectors of ARL13B. The lipidated cargoes show distinctive depletion kinetics from kidney epithelial cilia with relation to Tulp3 deletion–induced renal cystogenesis. Overall, these findings indicate an expanded role of the tubby domain in capturing analogous helical secondary structural motifs from diverse cargoes.
INTRODUCTION
The primary cilium is a microtubule-based dynamic cellular appendage that is templated from the mother centriole of the centrosome—the basal body (Anvarian et al., 2019). Cilia are present in multiple cell types and organs and can transduce cellular response to extracellular signals. These signals include hedgehog morphogens that regulate multiple developmental and regenerative programs (Kopinke et al., 2021). Signaling outputs from cilia also maintain renal tubular homeostasis preventing kidney cystogenesis (Ma, 2021). The ciliary membrane has a distinct protein and lipid composition despite being contiguous with the plasma membrane (Nachury and Mick, 2019). Multiple integral membrane (Hilgendorf et al., 2016) and lipidated proteins are preferentially associated with the ciliary membrane. The lipidated proteins include palmitoylated proteins such as ARL13B (Caspary et al., 2007; Duldulao et al., 2009), farnesylated proteins such as INPP5E (Bielas et al., 2009), and myristoylated proteins such as NPHP3 (Shiba et al., 2010). Deciphering mechanisms underlying ciliary membrane compartmentalization is necessary to understand the role of cilium-generated signaling in normal development and disease.
ARL13B is an atypical GTPase that is highly enriched in the primary cilium. Mutations in ARL13B in humans are associated with classic Joubert syndrome, a ciliopathy associated with cerebellar vermis hypoplasia, hypotonia, intellectual disability, renal cysts, and retinal impairment (Cantagrel et al., 2008; Thomas et al., 2015). ARL13B acts as a guanine nucleotide exchange factor (GEF) for the GTPase ARL3, converting inactive ARL3-GDP to active ARL3-GTP (Gotthardt et al., 2015; Ivanova et al., 2017). ARL3-GTP promotes ciliary release of multiple lipidated cargoes including farnesylated and myristoylated proteins from their respective carrier proteins PDE6δ (Humbert et al., 2012) and UNC119B (Wright et al., 2011). Given that ARL13B potentially regulates levels of multiple lipidated proteins inside cilia, it seems logical that it affects ciliary signaling. However, the role of trafficking of ARL13B into cilia is tissue specific. The zebrafish arl13b (scorpion) allele has nephric duct dilatation phenotypes, and analysis of phenotypic rescue using arl13b variants in this model suggests that ciliary localization is essential for in vivo function of ARL13B (Sun et al., 2004; Duldulao et al., 2009). However, a knock-in at the endogenous Arl13b locus that prevented ciliary localization (Arl13bV358A) did not affect neural tube patterning (Gigante et al., 2020), unlike the null mutant (Arl13bhnn) (Caspary et al., 2007). A precise understanding of mechanisms trafficking ARL13B to cilia would shed light on these tissue-specific differences.
Ciliary trafficking mechanisms for ARL13B have remained controversial. One motif that has stood out as being necessary for ciliary localization is the RVxP motif in the C-terminus (Mariani et al., 2016). The RVxP motif was initially identified as a motif involved in post-Golgi trafficking of rhodopsin (Deretic et al., 2005). ARL13B is a cytosolic protein that is palmitoylated (Cevik et al., 2010). In addition, deletion of the RVxP motif or lack of the coiled-coil (CC) adjacent to the GTPase domain of ARL13B forms cellular aggregates that could indirectly affect ciliary targeting (Cevik et al., 2013; Nozaki et al., 2017). Experiments in Caenorhabditis elegans have also demonstrated dominant negative roles of arl-13 mutants (Cevik et al., 2010, 2013), suggesting mutual interactions in compromising wild-type (WT) protein function (Hori et al., 2008). Ciliary localization assays of ARL13B mutants have been performed in the WT (Hori et al., 2008; Cevik et al., 2010, 2013; Nozaki et al., 2017) or null background (Duldulao et al., 2009; Larkins et al., 2011; Mariani et al., 2016), complicating experimental interpretations. Notwithstanding the unexplained nature of the RVxP motif with relation to ARL13B ciliary trafficking, whether there could be other motifs regulating ciliary trafficking of ARL13B is unknown.
The tubby family protein, TULP3, functions in coordination with the intraflagellar transport complex-A (IFT-A) to determine ciliary trafficking of integral membrane proteins without affecting their extraciliary pools or disrupting cilia (Mukhopadhyay et al., 2010; Badgandi et al., 2017). The TULP3 cargoes studied previously include GPCRs and channels (Badgandi et al., 2017). Short motifs called CLSs are present in these transmembrane cargoes that are necessary and sufficient to traffic to cilia in a TULP3-dependent manner (Badgandi et al., 2017). We previously proposed three steps during TULP3-mediated trafficking of transmembrane cargoes to cilia that consist of 1) capturing of the transmembrane cargo CLSs by TULP3’s C-terminal tubby domain in a phosphoinositide 4,5-bisphosphate(PI(4,5)P2)-dependent manner, 2) trafficking into the primary cilium via IFT-A core binding to the N-terminus of TULP3, and 3) release of GPCR cargoes from TULP3 into the PI(4,5)P2-deficient ciliary membrane. We recently showed that kidney tubule–specific Tulp3 knockout (ko) in mice results in renal cystogenesis (Hwang et al., 2019; Legue and Liem, 2019). Interestingly, we observed that ARL13B pools in cilia of Tulp3 conditional ko (cko) collecting ducts (CDs) were depleted before cyst formation, indicating a role for TULP3 in ARL13B ciliary localization (Hwang et al., 2019; Legue and Liem, 2019). However, how TULP3 regulates trafficking of ARL13B to cilia and whether it has any TULP3-regulated CLS remain to be established.
In line with a role of TULP3 in trafficking multiple cargoes to cilia, TULP3 and the cargoes have been implicated in neural tube patterning (Norman et al., 2009; Mukhopadhyay et al., 2013), renal and liver cystogenesis (Devane et al., 2022; Jafari Khamirani et al., 2022), adipogenesis (Hilgendorf et al., 2019; Wu et al., 2021), and spina bifida (Patterson et al., 2009; Kuang et al., 2022; Wang et al., 2022). In neural tube development (Mukhopadhyay et al., 2013) or adipogenesis (Hilgendorf et al., 2019; Wu et al., 2021), the relevant TULP3 cargoes are likely to be GPCRs, but for other tissues such as renal cystogenesis, the TULP3 trafficked proteins are unclear and unlikely to be GPCRs (Hwang et al., 2019). Here we show that TULP3 is required for transport of ARL13B to cilia and enrichment of ARL13B-regulated farnesylated and myristoylated proteins in cilia. The lipidated cargoes show distinctive patterns of depletion from kidney epithelial cilia with relation to Tulp3 deletion–induced cystogenesis. Unlike transmembrane cargoes that require both IFT-A– and phosphoinositide-binding properties of TULP3 for ciliary trafficking, ARL13B transport requires TULP3 binding to IFT-A but not to phosphoinositides. An amphipathic helix in the N-terminus of ARL13B couples to the TULP3 tubby domain in ciliary trafficking irrespective of palmitoylation and is essential for ciliary localization of ARL13B and ARL13B-dependent lipidated cargoes. These findings implicate the TULP3 tubby domain in the capturing of diverse cargoes with analogous helical secondary structural motifs in membrane proximity.
RESULTS
TULP3 is required for ciliary trafficking of ARL13B
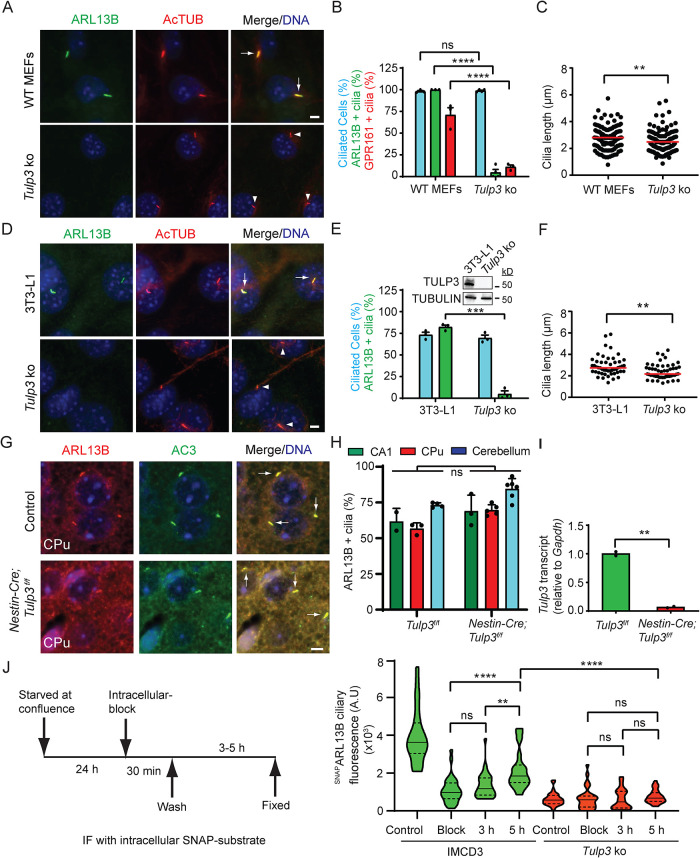
We recently reported that cilia in kidney tubular epithelia deleted for Tulp3 lacked ARL13B (Hwang et al., 2019; Legue and Liem, 2019). We further investigated the role of TULP3 in ciliary trafficking of ARL13B in primary mouse embryonic fibroblasts (MEFs) obtained from WT and Tulp3 ko mice (Legue and Liem, 2019). ARL13B was primarily localized to cilia in WT, whereas ciliary localization was abrogated in Tulp3 ko (Figure 1, A and B). As expected, GPR161, a known GPCR cargo for TULP3 (Mukhopadhyay et al., 2013), was not localized to cilia of Tulp3 ko MEFs (Figure 1B; Supplemental Figure S1A). There was a small but significant reduction in ciliary length without affecting percentage in Tulp3 ko MEFs (Figure 1, B and C). We further generated Tulp3 ko 3T3-L1 preadipocyte and NIH 3T3 cells using CRISPR/Cas9 technology (Supplemental Figure S2). We observed that ARL13B ciliary localization was inhibited also in Tulp3 ko 3T3-L1 and NIH 3T3 cells (Figure 1, D and E; Supplemental Figure S1, B and C). While ciliation with respect to number was unaffected (Figure 1E), a small but significant reduction in ciliary length was observed (Figure 1F). Smoothened, a TULP3-independent cargo (Mukhopadhyay et al., 2010), trafficked to Tulp3 ko 3T3-L1 cilia upon treatment with agonist SAG (Supplemental Figure S1, D and E), ruling out generalized defects in ciliary localization.
FIGURE 1:
TULP3 determines ciliary trafficking of ARL13B. (A) MEFs were serum starved upon confluence for 24 h before fixation. Fixed cells were immunostained for ARL13B (green) or GPR161 (shown in Supplemental Figure S1A), and acetylated tubulin (AcTUB, red) and counterstained for DNA (blue). (B) Cilia were counted from three to four fields/experiment from two experiments, n > 200/condition. Data represent mean ± SD. (C) Cilia length was measured from MEFs by immunostaining for acetylated tubulin. n > 100/condition. (D) 3T3-L1 preadipocyte cells were grown to confluency and cultured further for 72 h to promote ciliation before fixing. The fixed cells were immunostained for ARL13B (green) and acetylated tubulin (red) and counterstained for DNA. (E) Cilia from D were counted from three experiments, and total counted cells are >200 for each condition. Data represent mean ± SD. Inset, immunoblot of cell lysates. (F) Cilia length was measured from WT or Tulp3 ko 3T3L1 cells by immunostaining for acetylated tubulin. n > 50/condition. (G) Brain sections at P7 were immunostained for ARL13B (red) and AC3 (green) and counterstained for DNA (blue). CPu, Caudate-putamen. (H) Cilia were counted from control or conditional knockout (cko) at P7. n > 400 for CPu, 50–250 for the CA1 region in hippocampus, and >650 for cerebellum from multiple fields from one control and two cko mice. Data represent mean ± SD. (I) Transcript levels from control or Nestin Cre; Tulp3f/f mice. Data represent technical replicates from one mouse each. (J) IMCD3 cells stably expressing GFP-SNAPARL13B were starved as indicated in the schematic after reaching confluence. After blocking using BG-Block, cells were washed and later fixed at the indicated times. “Block,” immediately fixed after blocking. Control, untreated with BG-Block. Newly trafficked SNAP-tagged proteins in cilia were tracked by immunofluorescence using TMR-Star and acetylated tubulin. Images in Supplemental Figure S3C. Violin plots of ciliary intensities of TMR-Star from n > 30 cells/condition. Representative of two experiments. Scale, 5 μm; ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; ns, not significant. Adjusted p values shown in J. Arrows and arrowheads indicate cilia positive and negative for the indicated proteins, respectively. See also Supplemental Figures S1 and S2.
TULP3 belongs to the tubby protein family consisting of four other members, including Tubby (Mukhopadhyay and Jackson, 2011). Tulp3 is ubiquitously expressed, whereas Tubby is predominantly expressed in the brain/retina to a higher extent than TULP3 than Tulp3 (Mukhopadhyay and Jackson, 2011). Unlike in kidney tubules (Hwang et al., 2019; Legue and Liem, 2019), we observed that ciliary localization of ARL13B was unaffected in postnatal day 7 (P7) Nestin-Cre; Tulp3f/f brain neurons in the caudate putamen/striatum and CA1 region of the hippocampus (Figure 1, G and H; Supplemental Figure S3, A and B) and cerebellum granule progenitors (Figure 1H) compared with control brains. Tulp3 transcripts were reduced in the Nestin-Cre; Tulp3f/f brains (Figure 1I). Thus, ARL13B is unaffected in Tulp3 loss in the brain and also in the neural tube (Ferent et al., 2019), likely from redundancy with Tubby.
TULP3 determines entry of ARL13B into cilia
Steady state levels of a protein in cilia can be regulated at multiple steps, including trafficking to cilia, removal from the compartment, and additional steps in cargo recycling (Mukhopadhyay et al., 2017). We tested the dynamics of ciliary entry of stably expressed ARL13B that was tagged at the N-terminus with GFP-SNAP (GFP-SNAPARL13B) in control and Tulp3 ko inner medullary CD (IMCD3) cells (Supplemental Figure S2). After blocking intracellular pools of GFP-SNAP–tagged ARL13B using an irreversible inactivator (BG-Block), we tracked accumulation of newly trafficked SNAP-tagged proteins in cilia at different time points by immunofluorescence using fluorescent SNAP substrate (TMR-Star). In control cells, we detected an increase of up to ∼50% of SNAP-labeled pools in cilia by 5 h, comparable to the dynamics of ciliary entry observed for another TULP3 cargo, GPR161 (Pal et al., 2016; Badgandi et al., 2017). An increasing trend was apparent even earlier, at 3 h. Such an increase was strongly reduced in Tulp3 ko cells, suggesting inhibition of ciliary entry of ARL13B (Figure 1J; Supplemental Figure S3C).
TULP3 is required for enrichment of both farnesylated and myristoylated proteins in cilia
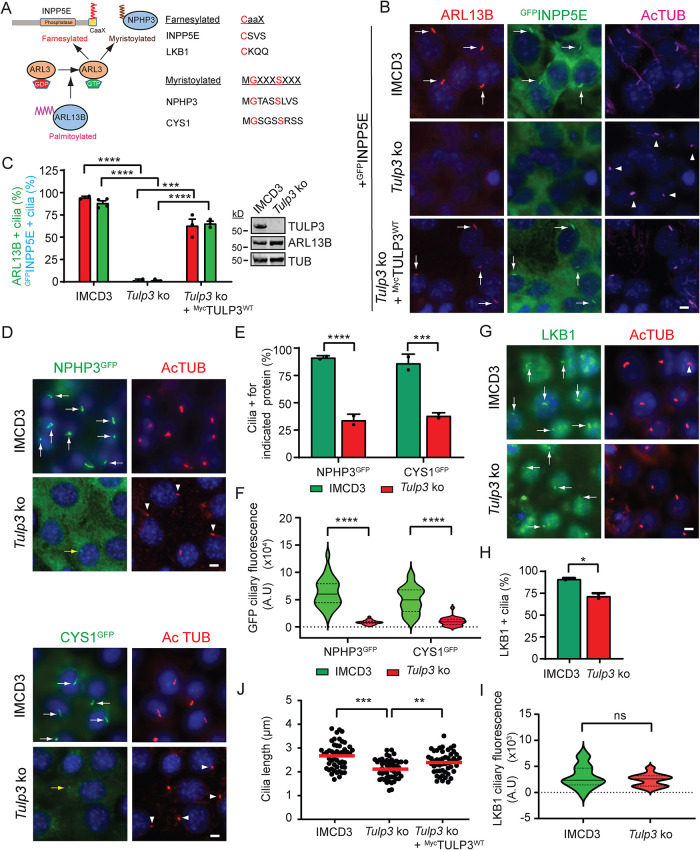
As ARL13B ciliary trafficking was dependent on TULP3, we asked whether TULP3 is also required for ciliary trafficking of ARL13B/ARL3-dependent farnesylated and myristoylated cargoes (Figure 2A). To answer this question, we stably expressed GFP-Stag (LAP)-tagged INPP5E, NPHP3 (1–203 amino acid [aa]), or cystin1 (CYS1) (Figure 2, B and D) in control or Tulp3 ko IMCD3 cells (Supplemental Figure S2). Compared to control, Tulp3 ko IMCD3 cells showed loss of endogenous ARL13B and GFPINPP5E ciliary localization (Figure 2, B and C). Importantly, ciliary trafficking of ARL13B and GFPINPP5E was restored upon stably expressing MycTULP3 (Figure 2, B and C), ruling out effects from nonspecific genomic changes. Total cellular levels of endogenous ARL13B protein assessed by immunoblotting were unaffected in Tulp3 ko (Figure 2C; Supplemental Figure S2). The percentage of cilia positive for NPHP3GFP or CYS1GFP was reduced in the Tulp3 ko cells (Figure 2, D and E); moreover, when localized to cilia, the NPHP3GFP and CYS1GFP pools were diminished (Figure 2F). LKB1 is a farnesylated protein that is targeted to cilia by the pseudokinase STRADβ (Mick et al., 2015). Together, they form a heterotrimer complex with the scaffolding protein MO25 (Zeqiraj et al., 2009). Ciliary localization of endogenous LKB1, as detected by immunostaining, was only modestly reduced in Tulp3 ko, with no significant decrease in ciliary pools (Figure 2, G–I), suggesting LKB1 trafficking to be independent of TULP3. We also observed a decrease in ciliary length upon Tulp3 ko in IMCD3 cells that was partially rescued by stably expressing MycTULP3 (Figure 2J). Thus, TULP3 promotes ciliary enrichment of farnesylated and myristoylated proteins that depend on ARL3 for release from their respective carriers into the membrane.
FIGURE 2:
TULP3 determines ciliary trafficking of ARL13B-dependent cargoes. (A) Cartoon depicting the role of ARL13B in regulating the release of farnesylated and myristoylated cargoes in the ciliary compartment via ARL3. The red aa signify critical residues in the stated motifs. (B) IMCD3 parental and Tulp3 ko cells (Supplemental Figure S2) stably expressing GFPINPP5E ± MycTULP3 were grown until confluence and serum starved for 36 h before fixation. Cells were immunostained for ARL13B, GFP (INPP5E), and acetylated tubulin (AcTUB) and counterstained for DNA. (C) Cilia from B were counted in two to four fields/experiment from two experiments. n > 200 cilia/condition. Western blot of cell lysates. (D) IMCD3 cells stably expressing NPHP3GFP or CYS1GFP were grown until confluence and serum starved for 36 h before fixation. Cells were immunostained for GFP and acetylated tubulin (AcTUB) and counterstained for DNA. (E) GFP-positive cilia from D were counted from two experiments, n > 200 cilia/condition. (F) Ciliary fluorescence intensities were measured from NPHP3LAP- or CYS1LAP-expressing IMCD3 cells that showed cilia positive for these fusion proteins and are represented as a violin plot. n > 30 cilia/condition. (G) IMCD3 cells were grown until confluence and serum starved for 36 h before fixation. Cells were immunostained for LKB1 and acetylated tubulin (AcTUB) and counterstained for DNA. (H) Cilia from G from two experiments were counted. n > 200 cilia/condition. (I) Ciliary fluorescence intensities were measured from IMCD3 cells and are shown as a violin plot. n > 30 cilia/condition. (J) Cilia lengths were measured from cells (B) by acetylated tubulin immunostaining (50 cilia/condition). Scale, 5 μm. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05; ns, not significant. Data represent mean ± SD. Arrows and arrowheads indicate cilia positive and negative for the indicated proteins, respectively. Yellow arrows, cilia with low fluorescence. See also Supplemental Figure S3.
The regulation of INPP5E ciliary localization in an ARL13B-dependent manner also provided us the opportunity to test whether either N- or C-tagged ARL13B is functional. We stably expressed N term- or C term-GFP-Stag (LAP)-tagged ARL13B in immortalized Arl13bhnn MEFs (Larkins et al., 2011). Arl13bhnn embryos completely lack endogenous ARL13B (Caspary et al., 2007) and do not have HAINPP5E in cilia upon stable overexpression (Supplemental Figure S4, A and B). Both N- and C-tagged ARL13B trafficked to cilia in the Arl13bhnn background (Supplemental Figure S4, A and B). Ciliary levels of HAINPP5E were also significantly restored by either N- or C-tagged ARL13B (Supplemental Figure S4, A and B). Ciliary lengths upon expressing either N- or C-tagged ARL13B were also increased and comparable to WT MEFs (Supplemental Figure S4C). Thus, both N- and C-tagged ARL13B are functional.
TULP3 is required for ciliary localization of lipidated proteins in murine kidneys
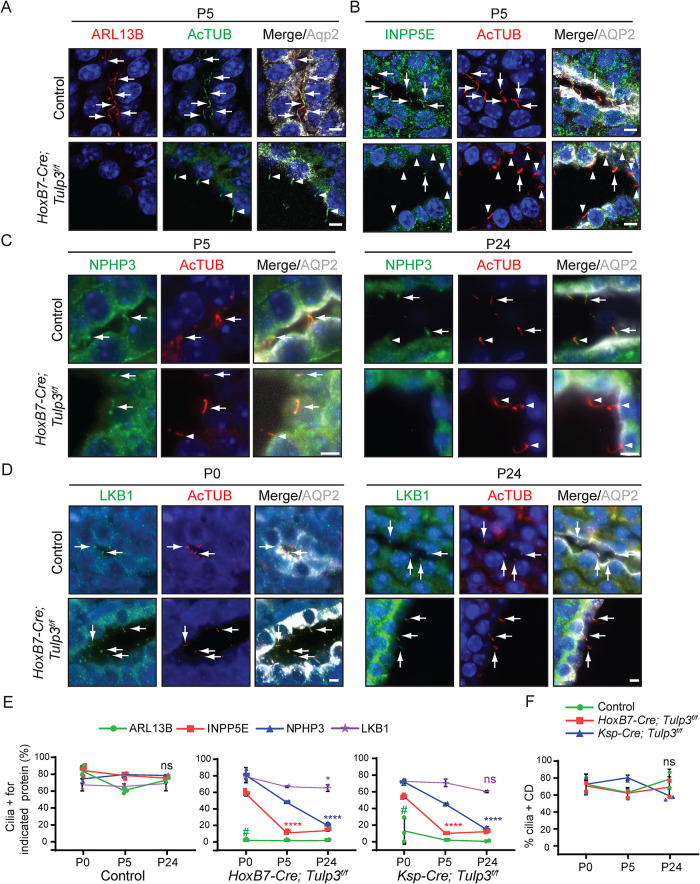
We next tested the role of TULP3 in ciliary trafficking of lipidated proteins in vivo in the mouse kidney using CD-specific HoxB7-Cre (HoxB7-Cre; Tulp3f/f) and nephron-specific Ksp-Cre (Ksp-Cre; Tulp3f/f) (Shao et al., 2002; Yu et al., 2002). These mice develop progressive cystogenesis but survive until P30 (Hwang et al., 2019). ARL13B and INPP5E localized to CD cilia, and localization was reduced in both Tulp3f/f models by P5 (Figure 3, A, B, and E; Supplemental Figure S5) as reported previously (Hwang et al., 2019). NPHP3 localizes to the proximal inversin zone in cilia (Bennett et al., 2020), but localization in kidney CDs is unknown. LKB1 has been reported to localize to kidney CD cilia (Viau et al., 2018). We found that both NPHP3 and LKB1 were localized to CD cilia in control mice (Figure 3, C–E). However, both Tulp3f/f models had reduced ciliary localization of NPHP3 by P24, whereas LKB1 was unaffected in kidney CDs (Figure 3, C–E). Importantly, we observed a significant difference in the kinetics of loss of these proteins from the Tulp3 cko kidney cilia (Figure 3E). ARL13B is almost completely lost by P0, INPP5E by P5, and NPHP3 by P24. This is consistent with our cell line data where ARL13B and INPP5E were most affected while NPHP3 and CYS1 were comparatively less affected (Figure 2). The percentage of ciliated cells in the CDs was unchanged in Tulp3 cko mice (Figure 3F), as reported previously (Hwang et al., 2019).
FIGURE 3:
TULP3 is required for trafficking of ARL13B- and ARL13B-dependent lipidated cargoes to mouse kidney epithelial cilia. (A–D) Kidney sagittal sections at postnatal day 0, 5, or 24 were immunostained for ARL13B (A), INPP5E (B), NPHP3 (C), or LKB1 (D), acetylated tubulin (AcTUB) and Aquaporin 2 (AQP2) and counterstained for DNA. (E, F) Quantification from two to four mice/genotype. Cilia counted by AcTUB staining. n > 200 cilia/mouse and genotype. Data represent mean ± SD. CD, collecting ducts. Scale, 5 μm. ****, p < 0.0001; **, p < 0.01; *, p < 0.05; ns, not significant, all compared with respective P0 values. #, p < 0.0001 with respect to ARL13B+ cilia in control animals at P0. Arrows and arrowheads indicate cilia positive and negative for the indicated proteins, respectively.
TULP3 traffics ARL13B to cilia in a PI(4,5)P 2-independent but IFT-A–dependent manner
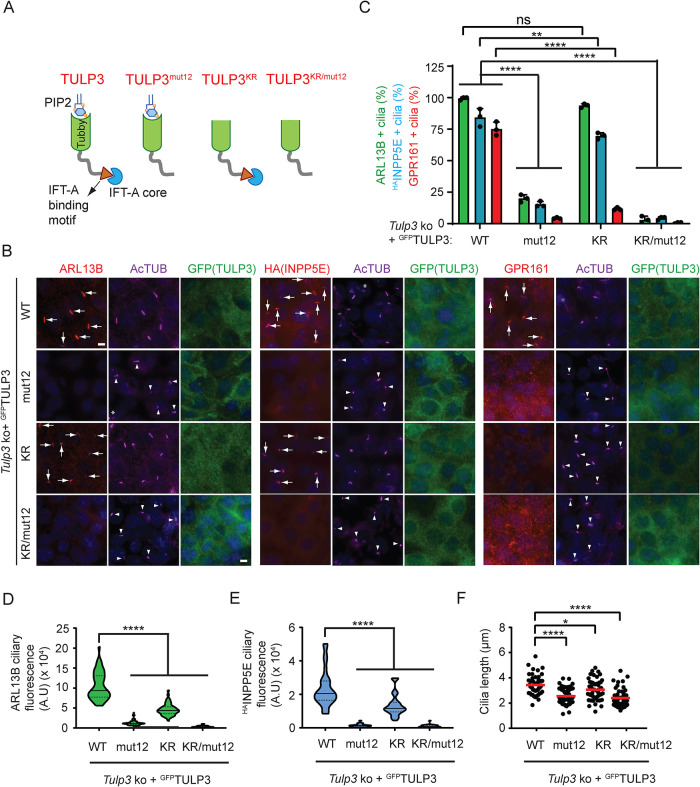
To dissect mechanisms of TULP3-mediated ciliary trafficking of ARL13B, we next asked whether PI(4,5)P2- and IFT-A–binding properties of TULP3 are also required, as for GPCRs (Mukhopadhyay et al., 2010; Badgandi et al., 2017). We stably expressed WT and mutant forms of LAPTULP3 that were deficient in IFT-A binding (TULP3mut12) or PI(4,5)P2 binding (K268A; R270A designated as TULP3KR) or both (Tulp3KR/mut12) with HAINPP5E in Tulp3 ko IMCD3 cells (Figure 4A) (Santagata et al., 2001; Mukhopadhyay et al., 2010). LAPTULP3 WT rescued ciliary trafficking of ARL13B, HAINPP5E, and GPR161 in multiple ko lines (Figure 4B; Supplemental Figure S4D). We observed that expression of TULP3mut12 or TULP3KR/mut12 did not rescue ciliary trafficking of either ARL13B or HAINPP5E, indicating that IFT-A binding is required for their ciliary trafficking by TULP3 (Figure 4, B–E). Interestingly, expression of TULP3KR efficiently rescued ciliary trafficking of both ARL13B and HAINPP5E (Figure 4, B and C), indicating that PI(4,5)P2 binding is dispensable for ARL13B trafficking by TULP3. However, compared with TULP3WT, the ciliary intensities of both ARL13B and HAINPP5E were reduced upon TULP3KR expression, indicating partial rescue (Figure 4, D and E). In contrast, expression of TULP3KR did not rescue ciliary trafficking of GPR161, similar to expression of TULP3mut12 or TULP3KR/mut12 (Figure 4, B and C), confirming that both PI(4,5)P2 and IFT-A binding by TULP3 are required for ciliary trafficking of GPR161. Expression of TULP3KR also partially restored ciliary length in Tulp3 ko cells compared with expression of either TULP3mut12 or TULP3KR/mut12 (Figure 4F). Recently, Han et al. (2019) showed that the PI(4,5)P2-binding property of TULP3 is not required for ciliary trafficking of ARL13B and GPR161. Our results with stably expressed TULP3 WT and mutants, rather than transient transfection (Han et al., 2019), suggest that TULP3 traffics GPR161 in a PI(4,5)P2-dependent manner and ARL13B in a predominantly PI(4,5)P2-independent manner.
FIGURE 4:
TULP3 traffics ARL13B in a PI(4,5)P2- independent manner unlike GPCR cargoes. (A) Schematic showing mutants. (B) Tulp3 ko IMCD3 line #89 stably expressing HAINPP5E and the indicated LAPTULP3 variants were grown to confluence and serum starved for 36 h before fixation. Cells were immunostained for ARL13B, HA (INPP5E), GPR161, GFP (TULP3), and acetylated tubulin (AcTUB) and counterstained for DNA. *, cytokinetic bridge. (C) Cilia in GFP(TULP3)-expressing cells were counted from three experiments, n > 200/condition. Data represent mean ± SD. (D, E) Violin plot shows mean ciliary intensities from B. n = 50 cilia/condition (ARL13B) and 30 cilia/condition (HAINPP5E). (F) Cilia lengths were measured in B by acetylated tubulin immunostaining (50 cilia each). Scale, 5 μm. ****, p < 0.0001; **, p < 0.01; *, p < 0.05; ns, not significant. Arrows and arrowheads indicate cilia positive and negative for the indicated proteins, respectively.
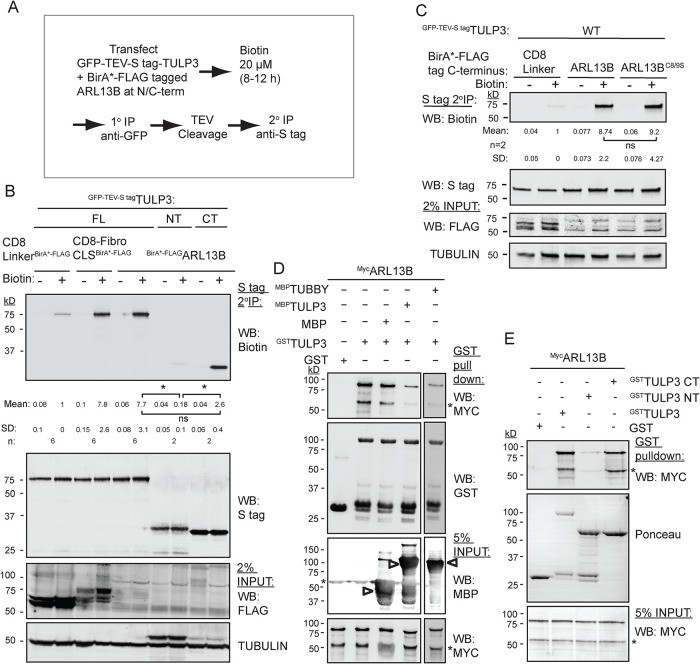
Proximity biotinylation assays determine ARL13B-TULP3 interactions
As TULP3 is required for ciliary trafficking of ARL13B, we tested whether there is a physical association between the two proteins. We showed earlier using proximity biotinylation or cross-linking ex vivo (Badgandi et al., 2017) that the CLS fusions of integral membrane proteins with CD8α are in proximity to TULP3. We coexpressed ARL13B or integral membrane protein CLSs fused with a promiscuous biotin ligase BirA* (BirA mutant R118G [Roux et al., 2012]) and LAPTULP3 in TRex-293 cells (LAP, EGFP-TEV-Stag-X; Figure 5A). To remove background resulting from nonspecific biotinylation of GFP, we performed tandem affinity purifications of LAPTULP3. We observed that TULP3 was biotinylated by ARL13B tagged at the N-term with BirA* (BirA*ARL13B) in comparison to a control without any CLS (CD8 LinkerBirA*; Figure 5B) (Badgandi et al., 2017). The biotinylation of TULP3 in the presence of ARL13B was at levels similar to that in the presence of the fibrocystin CLS fused with CD8α, one of the integral membrane protein cargoes of TULP3 (Badgandi et al., 2017). Similar proximity was also noted between LAPTULP3 and ARL13B tagged at the C-term with BirA* (ARL13BBirA*), suggesting that both N-term– and C-term–tagged ARL13B is in proximity to TULP3 (Figure 5C). An ARL13BC8/9S BirA* mutant mutated at the palmitoylation motif (C8/9S) also retained proximity to TULP3 (Figure 5C), suggesting that membrane association was not strictly necessary for proximity. We tested which domain of TULP3 interacted with ARL13B using LAPTULP3 N(1-183 aa) and C(184-442 aa) halves. The C-terminal tubby domain, but not the N-terminal fragment, was biotinylated by ARL13B (Figure 5B). Thus, ARL13B, like GPCR cargoes, interacts with tubby domain.
FIGURE 5:
Tubby domain of TULP3 interacts with ARL13B. (A) Flowchart showing steps in proximity biotinylation assay in cells. (B) T-REx 293 cells were cotransfected with GFP-TEV-S–tagged TULP3 full-length (FL), N-terminus (NT), or C-terminus (CT) along with ARL13B tagged at N-term with BirA* and processed as in A. Final S-tag bead eluates were processed for immunoblotting with IRdye-680–tagged streptavidin to detect biotinylation. S-tag antibody blot shows pull-down efficiency. Mean ± SD values indicate biotin/S-tag ratios normalized to CD8 linker control. (C) T-REx 293 cells were cotransfected with GFP-TEV-S–tagged TULP3 full-length along with ARL13B or ARL13BC8/9S tagged at C-term with BirA* and processed as in B. Mean ± SD values indicate biotin/S-tag ratios normalized to CD8 linker control. (D) GST or GSTTULP3-bound beads were incubated with IVT MycARL13B alone or in excess of MBP or MBPTULP3 or MBPTubby protein in LAP100N at 4°C for 1 h. Beads were washed, eluted, and immunoblotted. MBP, maltose-binding protein. (E) IVTMycARL13B was incubated with GST, GSTTULP3 full length, GSTTULP3 NT, or GSTTULP3 CT in LAP100N buffer at 4°C for 1 h. Beads were processed for immunoblotting. Asterisks in immunoblots in C and D, nonspecific bands. Arrowheads in D, MBP fusions. *, p < 0.05; ns, not significant. See also Supplemental Figure S3.
The tubby domain of TULP3 binds to ARL13B
We next tested whether ARL13B binds to TULP3 using in vitro translated (IVT) MycARL13B in pull-down assays with bacterially purified GSTTULP3. We found that MycARL13B bound to GSTTULP3 (Figure 5D) or GSTTubby (Supplemental Figure S6A). Excess maltose-binding protein (MBP)-tagged TULP3 or Tubby, the founding member of the tubby family proteins, but not MBP alone, could compete out GSTTULP3 binding to MycARL13B (Figure 5D). Thus, conserved regions in tubby family proteins might be involved in interaction with ARL13B. Furthermore, using recombinant GST-tagged TULP3 N-terminal (1–183 aa) and C-terminal (184–442 aa) halves, we confirmed that the C-terminal tubby domain of TULP3 interacts with IVT MycARL13B (Figure 5E). Thus, the C-terminus tubby domain of TULP3 binds to ARL13B.
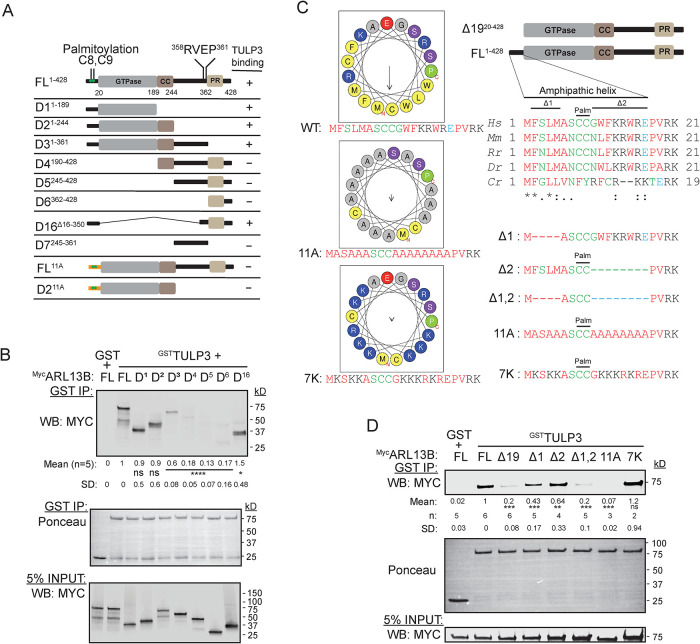
An N-terminal amphipathic helix in ARL13B binds to TULP3
To dissect ciliary targeting of ARL13B by TULP3, we aimed at identifying ARL13B regions involved in TULP3 binding. ARL13B has a palmitoylation motif and GTPase domain followed by a CC domain in its N-terminus along with a RVxP motif and a proline-rich (PR) region in its C-terminus (Figure 6A). We tested various IVT truncations of MycARL13B for their capacity to bind with GSTTULP3. We confirmed that the N-terminus of ARL13B including the GTPase and CC domains (D2, 1–244 aa) binds to TULP3 (Figure 6, A and B). The lack of the CC in the D1 fragment (1–189 aa) or in the full-length protein (∆CC) did not prevent TULP3 binding (Figure 6, A and B; Supplemental Figure S6B). The C-terminus alone (D5, 245–428 aa) did not bind to TULP3 (Figure 6, A and B). Thus, only the N-terminus of ARL13B mediates TULP3 binding. As an ARF family protein (Randazzo et al., 1995; Donaldson and Jackson, 2011), ARL13B has a lipid-sensitive clamp provided by an amphipathic helix at the N terminus preceding the GTPase domain that includes the palmitoylation site (Figure 6C). We next tested whether the C-terminal region fused with the short N-terminal amphipathic helix (D16, Figure 6A) interacts with TULP3. We observed that this construct bound to TULP3 as efficiently as the full-length or the N-terminus (Figure 6B). Remarkably, the helix-deleted ARL13B (∆19) was almost fully abrogated in its binding to TULP3 or Tubby (Figure 6, C and D; Supplemental Figure S6A). Deletion of four conserved aa preceding the palmitoylation site (∆1; Figure 6C) or of seven conserved aa following the palmitoylation site (∆2; Figure 6C) in ARL13B modestly reduced binding to TULP3 (Figure 6D). Finally, a double-deletion mutant of these regions in the full-length protein (∆1,2; Figure 6C) further reduced binding to TULP3 (Figure 6D). A full-length 11A mutant with alanine substitution of hydrophobic/charged residues maintaining an intact palmitoylation motif (Ren et al., 2008; Weng et al., 2017) (Figure 6C) abrogated TULP3 binding (Figure 6D). The 11A mutant of the N-terminus of ARL13B that included the GTPase and CC domains (D211A) also had reduced binding to TULP3 compared with D2 (Figure 6A; Supplemental Figure S6C). However, a reversal to charged residues in the helix retained TULP3 binding (Figure 6, C and D). Thus, charged residues of the ARL13B amphipathic helix mediate TULP3 binding.
FIGURE 6:
ARL13B domains required for TULP3 interactions. (A) Schematic representation of different ARL13B truncations used summarizing TULP3 binding. Coiled-coiled domain, CC; proline-rich domain, PR. (B) Binding between GST or GSTTULP3 bound to glutathione sepharose beads and IVT Myc-tagged truncations of ARL13B as in Figure 5D, except incubated in LAP150N at room temperature for 1 h. (C) Alignment of N-terminus region of ARL13B preceding the GTPase domain and mutants. ARL13B protein IDs: Hs, Homo sapiens NP_001167621.1; Mm, Mus musculus NP_080853.3; Rn, Rattus norvegicus NP_001100571.1; Dr, Danio rerio NP_775379.1; Cr, Chlamydomonas reinhardtii A8INQ0.1. Palm, conserved palmitoylation motif (Ren et al., 2008; Weng et al., 2017). Helix for the human protein drawn using HeliQuest (Gautier et al., 2008). Arrow, hydrophobic moment; N/C, N/C-terminal. (D) Binding as in B using ARL13B mutants described in C. (B, D) ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05. ns, not significant. See also Supplemental Figure S4.
The amphipathic helix directs ARL13B ciliary trafficking by TULP3
We next tested for a TULP3-dependent CLS in ARL13B. Previous studies reported the RVxP motif (sequence R357VEPV361) in mouse ARL13B (identical to 358-361 R358VEPL362 of human ARL13B) as a potential CLS (Mariani et al., 2016). Endogenous levels of ARL13BV358A in MEFs with knock-in at the endogenous locus (Arl13bV358A/V358A) were not seen in cilia (Supplemental Figure S7A), as reported before (Gigante et al., 2020). By cell-based proximity biotinylation assay, we noted that human ARL13BV359A interacted with TULP3 efficiently when compared with the WT ARL13B (Supplemental Figure S7B). Similarly, in vitro binding between GSTTULP3 and IVT human MycARL13BV359A was similar to that of WT MycARL13B (Supplemental Figure S7C). Thus, the ARL13B RVxP mutant failed to traffic to cilia despite efficient TULP3 binding and is not a TULP3-dependent CLS.
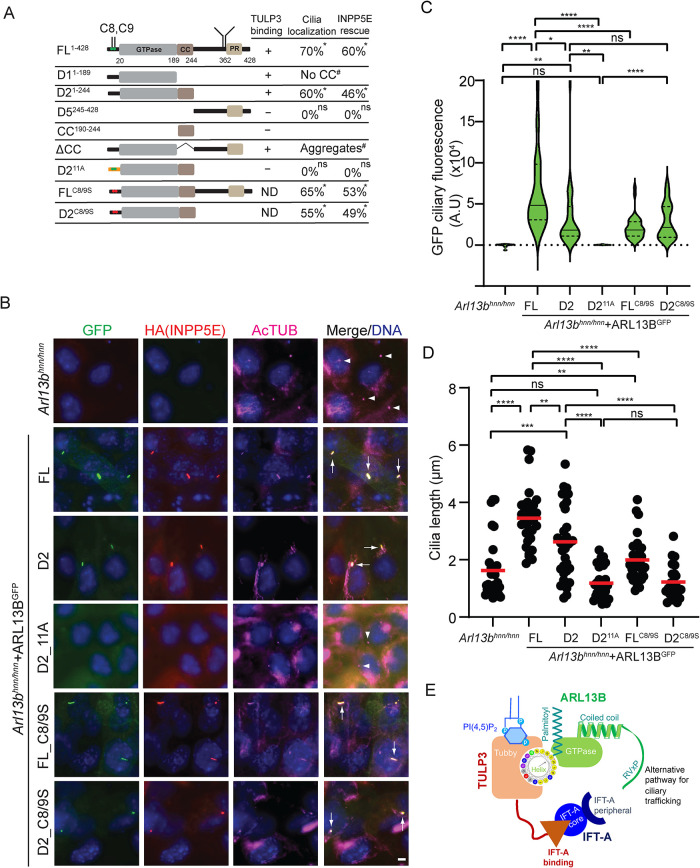
We next tested fragments of ARL13B in ciliary localization based on the TULP3 binding (Figure 6, A and B). We stably expressed different ARL13B variants fused with C-terminal LAP in Arl13bhnn MEFs (Figure 7A). We chose the Ar13bhnn background as self-association of ARL13B through its N-terminal domain has been reported (Hori et al., 2008). We also coexpressed HAINPP5E to check for ARL13B function. We noticed ciliary trafficking of the TULP3 binding ARL13B D2 fragment (Figure 7, A and B) but not of the TULP3 nonbinding D5 fragment retaining the RVxP motif (Supplemental Figure S7D). HAINPP5E-positive cilia were rescued upon D2 expression (Figure 7, A and B). D2 ciliary levels and corresponding ciliary lengths were partially rescued compared with expression of the full-length protein (Figure 7, A–D). Lack of the CC region in the D1 N-terminal fragment or full-length protein (Nozaki et al., 2017) prevented trafficking to cilia despite TULP3 binding (Figure 7A; Supplemental Figure S7D), from likely indirect effects on GTPase domain conformations or stability (see the Discussion) (Nozaki et al., 2017). Thus, the TULP3 binding N-terminus of ARL13B (up to the CC domain) traffics to cilia and promotes lipidated protein trafficking.
FIGURE 7:
ARL13B domains required for TULP3 interactions and ciliary localization. (A) Schematic representation of different ARL13B truncations used. n = 47–200 cilia counted. Arl13bhnn MEFs had no HA-tagged INPP5E in cilia. #, the D1 fragment lacking a CCdomain (Supplemental Figure S7D) or ∆CC is not trafficked to cilia likely from indirect effects, such as formation of cellular aggregates upon loss of CC (Nozaki et al., 2017). (B) Arl13bhnn cells stably expressing HAINPP5E along with the indicated C-LAP–tagged ARL13B fusions were serum starved for 48 h before fixing and immunostained for GFP, HA (INPP5E), and acetylated tubulin (AcTUB) and counterstained for DNA. Quantification in A, C, and D. (C) Violin plots of GFP ciliary fluorescence intensities from B. n > 30 cilia/condition. (D) Cilia lengths of cell lines from B. Only ciliated cells in the Arl13bhnn background stably expressing HA-tagged INPP5E were counted. Arl13bhnn cells are ∼20% ciliated (Larkins et al., 2011). n > 30 cilia/condition. (E) Model depicting TULP3’s role in trafficking ARL13B to cilia. ARL13B trafficking to cilia is primarily regulated by binding to TULP3 and TULP3’s binding to the IFT-A core complex. An N-terminus amphipathic helix that includes the palmitoylation site and precedes the GTPase domain mediates binding of ARL13B to TULP3 and directs trafficking to cilia even in the absence of palmitoylation and the RVxP sorting motif. The RVxP motif also provides an alternative mode of trafficking. TULP3 binding to short sequences in diverse cargoes is likely mediated by the tubby domain. Scale, 5 μm. *, p < 0.001 by Chi-square test. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05; ns, not significant (C–D). Adjusted p values are shown in C. Arrows and arrowheads indicate cilia positive and negative for the indicated proteins, respectively. See also Supplemental Figure S5.
As the ARL13B D2 fragment was ciliary, it allowed us to study TULP3 function in trafficking of the fragment without any influence of the RVxP motif. The D211A mutant fragment, with alanine substitution of charged and hydrophobic residues in the amphipathic helix but predicted to retain the palmitoylation motif (Ren et al., 2008; Weng et al., 2017), had reduced binding to TULP3 (Supplemental Figure S6C). When tested for ciliary localization upon stable expression in Arl13bhnnMEFs, D211A was not trafficked to cilia, and HAINPP5E levels in cilia and ciliary lengths were not rescued (Figure 7, A–D). Thus, TULP3 binding is critical for trafficking of the D2 fragment.
As the palmitoylation motif is included in the helix, we next tested the role of palmitoylation on ciliary localization of ARL13B. It is important to note that the palmitoylation-deficient ARL13BC8/9S BirA* mutant retained proximity to TULP3 (Figure 5C). The ARL13BC8/9S mutant upon stable expression in Arl13bhnnMEFs was trafficked to cilia (Figure 7, A and B), although ARL13BC8/9S ciliary levels and corresponding ciliary lengths were only partially rescued compared with expression of the full-length protein (Figure 7, C and D). HAINPP5E-positive cilia were rescued (Figure 7, A–C). Thus, the full-length ARL13B localized to cilia even in the absence of palmitoylation.
Finally, in contrast to the D211A mutant, the palmitoylation-deficient D2 (D2C8/9S) still trafficked to cilia (Figure 7, A and B) and had intermediate levels of ciliary pools (Figure 7C). HAINPP5E levels in cilia were rescued (Figure 7, A and B), but not ciliary length (Figure 7D). Thus, the amphipathic helix in ARL13B mediates trafficking of the D2 fragment to cilia irrespective of palmitoylation.
DISCUSSION
Despite the discovery of ARL13B more than a decade ago (Sun et al., 2004; Caspary et al., 2007) and the universality of its use as a ciliary marker, the mechanisms that traffic ARL13B to cilia are not well understood. We show that TULP3 determines trafficking of ARL13B to cilia without affecting total cellular pools and enriches ARL13B-dependent lipidated proteins in cilia. ARL13B trafficking to cilia is regulated by TULP3’s binding to the IFT-A core complex but only partially by binding to phosphoinositides. We previously showed that interactions between the CLS sequences in transmembrane proteins and tubby domain of TULP3 are critical in ciliary trafficking (Badgandi et al., 2017). We now find that the TULP3 tubby domain interacts with ARL13B. An N-terminus amphipathic helix preceding the GTPase domain mediates ARL13B binding to TULP3 and ciliary trafficking. The identification of a TULP3-dependent CLS in ARL13B suggests that short sequences in diverse cargoes likely interact with the tubby domain during transport to cilia.
TULP3 enriches ARL13B-dependent lipidated proteins in cilia
We demonstrate that ARL3-dependent farnesylated (INPP5E) and myristoylated (NPHP3, CYS1) proteins are regulated by TULP3 for ciliary localization. The farnesylated protein LKB1, which is ARL3 independent for trafficking to cilia, is not TULP3 regulated. INPP5E loss in cilia of Tulp3 cko kidney CD epithelia accompanies cystogenesis and follows ARL13B depletion. NPHP3 and CYS1 are reduced in ciliary intensity but retain weak levels in cilia in Tulp3 ko cells. NPHP3 is also slow to be depleted from Tulp3 cko kidney CD epithelial cilia. The effectiveness for complete depletion of INPP5E in cilia in Tulp3 cko might be related to direct binding between ARL13B and INPP5E and the requirement of such binding for effective ciliary retention of INPP5E (Humbert et al., 2012; Qiu et al., 2021; Cilleros-Rodriguez et al., 2022). NPHP3 is concentrated in the proximal ciliary inversin compartment by binding to NEK8 and ANKS6, which are required downstream of inversin for NPHP3 localization (Bennett et al., 2020). Such binding might promote some retention of NPHP3 even in the absence of TULP3.
The trafficking function of TULP3 in ARL13B trafficking is especially relevant in the context of our previous studies showing the role of TULP3 in renal cystogenesis (Hwang et al., 2019; Legue and Liem, 2019) and the recent identification of human patients with hepatorenal fibrocystic disease with mutations in TULP3 (Devane et al., 2022; Jafari Khamirani et al., 2022). Importantly, embryonic cko of Arl13b (Li et al., 2016; Seixas et al., 2016) and of ARL13B-regulated cargoes, Inpp5e (Hakim et al., 2016), Nphp3 (Bergmann et al., 2008), and Cys1 (Hou et al., 2002) show mild cystic disease, phenocopying TULP3 loss (Hwang et al., 2019). ARL13B-dependent cargoes that are depleted from Tulp3 cko cilia before normal onset of cystogenesis in the early postnatal period in embryonic models could be potential ciliary regulators of cystogenesis and could be followed up for genetic epistasis studies (Walker et al., 2022). ARL13B and INPP5E trafficking to cilia is reduced before cystogenesis in embryonic-onset models of Tulp3 cko, whereas that of NPHP3 accompanies cystogenesis. Based on these results, of the ARL13B-dependent cargoes, INPP5E would be an important candidate regulator in the early-onset models of polycystic kidney disease (PKD).
An amphipathic helix determines ciliary localization of ARL13B
We showed before that CLSs from cilia-targeted GPCRs/fibrocystin are required in the membrane-bound context for proximity and cross-linking between cargoes and TULP3 (Badgandi et al., 2017). Using proximity biotinylation assays and in vitro binding approaches combined with mutational analyses, we now show TULP3 tubby domain–ARL13B interactions mediated by an amphipathic helix in ARL13B. The N-terminal amphipathic helix is a signature feature of the ARF family proteins that adopts nucleotide-dependent conformations (Randazzo et al., 1995; Antonny et al., 1997; Donaldson and Jackson, 2011), but the helix had been previously omitted in structural studies of ARL13B (Gotthardt et al., 2015). Importantly, we show that the TULP3 nonbinding C-terminal region of ARL13B when fused with the N-terminal amphipathic helix (ARL13B D16 variant) binds to TULP3, suggesting that the helix is sufficient in mediating interactions. Thus, our results suggest that even for ARL13B, a short sequence is instrumental in coupling with TULP3.
A full-length ARL13B11A mutant that has alanine substitution of both hydrophobic and charged residues in the amphipathic helix abrogated TULP3 binding, whereas substituting the hydrophobic residues with positively charged residues retained binding. Thus, the charged residues in the amphipathic helix were critical in TULP3 binding. The hydrophobic side of the helix is likely to be buried in the ARL13B GTPase fold as in ArfGDP (Amor et al., 1994; Menetrey et al., 2000) or the membrane (Liu et al., 2010). The ARL13B D2 fragment that includes the N-terminus until the CC domain and lacks the RVxP motif trafficked to cilia, whereas the D2 fragment with an uncharged helix (D211A) did not traffic to cilia. However, an ARL13B D2C8/9S palmitoylation motif mutant still trafficked to cilia. Thus, TULP3 binding to the D2 fragment is critical in mediating trafficking to cilia irrespective of palmitoylation status.
Lack of the CC following the GTPase domain did not preclude binding to TULP3 in vitro but affected ciliary targeting (Nozaki et al., 2017). Such effects on ciliary targeting are likely from indirect effects on GTPase domain conformations, as is evident from abrogated ARL3-GEF activity (Gotthardt et al., 2015) and formation of cellular aggregates (Nozaki et al., 2017). Lack of the CC in the D1 N-terminal fragment or analogous fragments (Hori et al., 2008; Larkins et al., 2011) similarly affects trafficking to cilia, despite TULP3 binding, from likely indirect effects.
Although the ARL13B palmitoylation mutant or D2 fragment localized to cilia, a full enrichment of ARL13B in cilia required multiple components including TULP3 binding, palmitoylation, and the RVxP motif. Mutating the RVxP motif in the context of full-length ARL13B with the intact TULP3 binding amphipathic helix prevents ciliary localization of ARL13B (Mariani et al., 2016; Gigante et al., 2020), suggesting that this motif licenses delivery of TULP3-IFT-A–bound ARL13B to cilia through an unknown mechanism (Figure 7E).
A TULP3-dependent ARL13B CLS suggests common features among diverse CLSs
The TULP3-regulated CLS sequences in transmembrane cargoes are diverse (Badgandi et al., 2017) and thereby provide no logical basis for a unifying mechanism for ciliary localization by TULP3. Our experiments showing an amphipathic helix in ARL13B mediating TULP3 binding suggest a short sequence in coupling with the tubby domain. On the basis of the amphipathic helix as a TULP3-dependent CLS for ARL13B, we suggest that there are two structural features that might be common between these diverse CLSs. First, TULP3-trafficked transmembrane proteins could generate secondary helical structures. Second, these sequences need to be proximate to the membrane that could facilitate their interactions with the tubby domain. In fact, the CLS (VKARK) in GPR161 IC3 is expected to be part of an extended TM5 helix (Flock et al., 2015). Similarly, the CLS sequence KTRKIKP of fibrocystin follows an extended TM domain (Follit et al., 2010). Like the amphipathic helix in ARL13B, the residues in the CLSs of GPR161 and fibrocystin both have charged residues that are necessary and sufficient for TULP3-mediated interactions during ciliary trafficking (Badgandi et al., 2017; Hwang et al., 2021).
In conclusion, our results suggest an expanded role of the TULP3 tubby domain in mediating transport of diverse membrane-associated and transmembrane cargoes to cilia by capturing CLSs. The binding of the N-terminus of TULP3 to the IFT-A core (Mukhopadhyay et al., 2010) could facilitate these tubby domain–cargo interactions. In fact, cryoelectron microscopy structures of insertion of the TULP3 N-term α-helix into the IFT-A core subunits orients the tubby domain toward the ciliary membrane (Hesketh et al., 2022; van den Hoek et al., 2022; Jiang et al., 2023). Alternatively, the IFT-A subunits could contribute to cargo-specific interactions (Ye et al., 2018), In fact, the core subunit IFT140 and the peripheral IFT-A subunit WDR35 might provide interface with TULP3 cargoes such as ARL13B (Fu et al., 2016; Picariello et al., 2019). Here, the major role of TULP3 could be to recruit IFT-A to the periciliary membrane for cargo binding. Considering the role of the tubby domain in ARL13B binding and the TULP3-dependent ARL13B CLS that we show to be critical in ciliary trafficking, a parsimonious model bringing these concepts together would be that TULP3 and IFT-A cooperatively bind to the CLSs of cargoes in membrane-proximate interactions. Nonetheless, targeting TULP3-cargo interactions could provide therapeutics in ciliary trafficking–regulated diseases such as PKD and peripheral obesity.
MATERIALS AND METHODS
Mouse strains
All mice were housed at the Animal Resource Center of the University of Texas Southwestern (UTSW) Medical Center. All protocols were approved by the UTSW Institutional Animal Care and Use Committee. Mice were housed in standard cages that contained three to five mice per cage, with water and standard diet ad libitum and a 12 h light/dark cycle. Animals were analyzed irrespective of sex. Nestin-Cre mice (Stock No. 003771; MGI: 2176173) were obtained from Jackson Laboratory (Bar Harbor, ME) (Tronche et al., 1999). HoxB7-Cre (MGI: 2675121) and Ksp-Cre (MGI: 2665300) mice were obtained from the O’Brien Kidney Research Core of UTSW. ES cells targeting the second exon of Tulp3 from EUCOMM (HEPD 0508-5-B01) were used to generate the floxed mice that were obtained from MRC, Harwell as described before (Hwang et al., 2019) (MGI: 5752546). For genotyping of Tulp3f/f mice, we used the following primers: 1) Tulp3–5′ Cas-WTF (5′ CCA TTT GTG AGG GTT GCT TT 3′) and Tulp3-Crit-WTR (5′ GCT AAC ACA AGC CCA TGC TA 3′) to detect WT band (256 base pairs) and floxed band (450 base pairs) and 2) Tulp3-F1–(5′ AAG GCG CAT AAC GAT ACC AC 3′) and Tulp3-R1 (5′ ACT GAT GGC GAG CTC AGA CC 3′) to detect the deletion. We did not notice any difference between WT or heterozygous animals for Tulp3 floxed alleles with or without Cre recombinase, and thus they were all included as littermate controls as mentioned in the respective figure legends. For genotyping Cre, the following primers were used to detect the transgene band: Cre-F (5′ AAT GCT GTC ACT TGG TCG TGG C 3′), Cre-R (5′ GAA AAT GCT TCT GTC CGT TTG C 3′).
Antibodies and reagents
Anti-Arl13b rabbit polyclonal was from Tamara Caspary (Emory University School of Medicine) (Caspary et al., 2007), and anti-Smo rabbit polyclonal was from Kathryn Anderson (Memorial Sloan Kettering Cancer Center, New York, NY) (Ocbina and Anderson, 2008). Affinity-purified rabbit polyclonal antibody against TULP3 (Mukhopadhyay et al., 2010) and Gpr161 has been described previously (Pal et al., 2016). Commercial antibodies used were against Arl13b (IF, N295B/66 NeuroMab; WB, Proteintech; 17711-1-AP), GFP (IF, Abcam ab13970; WB, Abcam ab290), hFAB rhodamine anti-tubulin (Bio-Rad; 12004166), acetylated α-tubulin (mAb 6-11B-1 T6793; Sigma-Aldrich), S tag (EMD Millipore; MAC112), IRDye 680RD Streptavidin (LI-COR; 926-68079) Myc (Abcam; ab9132), HA (rat polyclonal), Flag (Abcam; ab1257), MBP (New England Biolabs), GST (Sigma; G1160), AC3 (LifeSpan BioSciences; LS-C204505), Aqp2 (1:500 A7310 Sigma rabbit polyclonal; SC515770, Santa Cruz Biotechnology mouse immunoglobulin G1 [IgG1]), Inpp5e (Proteintech; 17797-1-AP), Nphp3 (Proteintech; 22026-1-AP), and Lkb1 (Cell Signaling Technologies; 13031T). Fluorescent secondary antibodies for immunofluorescence were from the Jackson Immuno Research Laboratories, and IRDye 680RD, IRDye 800CW secondary antibodies for immunoblotting were from LI-COR Biosciences. Reagents used in this study included biotin (Sigma; B4639) and are tabulated in Supplemental Table S1 (Key Resource Table).
Plasmids
pG-LAP1 (pCDNA5/FRT/TO-EGFP-TEV-Stag-X) and pG-LAP5 (pEFα-X-Stag-PreScission-EGFP) were from Addgene (Torres et al., 2009). pENTR223-ARL13B fusion construct was from DNASU (HsCD00511796), pENTR221-INPP5E construct was from Life Technologies (IOH40212), NPHP3 (encoding 1–203 aa) was synthesized commercially from Geneart (Life Technologies), and Gateway PLUS shuttle clone for CYS1 was from GeneCopoeia (GC-Y0203-CF). N- or C-terminal LAP-tagged retroviral constructs of full-length and truncations of TULP3, INPP5E, NPHP3, CYS1, and ARL13B were generated by Gateway cloning into a gatewaytized LAP1 or LAP5 version of pBABE, respectively. Lentiviral constructs of Myc-TULP3 and HA-INPP5E were generated by gateway cloning into pQXIN-Myc and pQXIN-HA destination vectors, respectively. Single- or multiple-aa mutants of TULP3 and ARL13B were generated by Q5 site-directed mutagenesis (New England Biolabs). For biotinylation experiments, DEST-pcDNA5-BirA-FLAG N- or C-term destination vectors were from C. Gingras, The Lunenfeld-Tanenbaum Research Institute at Mount Sinai Hospital, Toronto, Canada. Myc-tagged, full-length and truncated ARL13B fragments were generated by in vitro translation using the TNT Sp6 high-yield wheat germ protein expression system (Promega; L3261) from pCS2-Myc-ARL13B vectors generated by gateway cloning. GFP-SNAP-ARL13B was generated by cloning SNAP into a BstB1 site of the S-peptide in an N-terminal LAP-tagged ARL13B retroviral construct (pBABE).
Cell culture and generation of stable cell lines
T-Rex-293 (Invitrogen), IMCD3 Flp-In (IMCD3; American Type Culture Collection; gift of Peter Jackson), Phoenix A (PhA; Indiana University National Gene Vector Biorepository), and 293FT cells were cultured in DMEM high glucose (Sigma-Aldrich; supplemented with 10% cosmic serum, 0.05 mg/ml penicillin, 0.05 mg/ml streptomycin, and 4.5 mM glutamine). NIH 3T3 Flp-In (Thermo Fisher) cells were cultured in DMEM high glucose media (D5796; Sigma) with 10% Bovine calf serum (BCS; Sigma-Aldrich), 0.05 mg/ml penicillin, 0.05 mg/ml streptomycin, and 4.5 mM glutamine. 3T3-L1 cells (gift of Peter Michaely, UTSW) were cultured in DMEM high glucose (Sigma-Aldrich) media with 10% fetal bovine serum, 0.05 mg/ml penicillin, 0.05 mg/ml streptomycin, 4.5 mM glutamine, and 8 ng/ml biotin. Cell lines were tested negative for Mycoplasma using the Mycoplasma PCR Detection Kit (Genlantis). Transfection of plasmids was done with Polyfect (QIAGEN) or polyethylenimine (PEI) max. Stable cell lines were generated by retroviral infection or transfection. In many cases, stable lines were flow sorted and further selected for GFP. Control and Tulp3 ko MEFs were from embryonic day 13.5 embryos (Legue and Liem, 2019). Immortalized WT and Arl13b mutant MEFs were gifts from Tamara Caspary (Larkins et al., 2011; Gigante et al., 2020).
Generation of Tulp3 knockout cell lines
CRISPR/Cas9 ko lines for Tulp3 were generated in IMCD3 Flp-In, NIH 3T3 Flp-In, and 3T3-L1 cells by using guide RNA targeting sequences caccgACGTCGCTGCGAGGCATCTG and caccgTGGCTTTAACCTTCGCAGCC targeting exon 3 in mouse Tulp3. We used a pLenti-CRISPR puromycin construct for infecting the parental lines using lentivirus (Shalem et al., 2014), selected for clones using puromycin, and used limiting dilution to derive single ko clones. Clonal lines were tested for ko by Sanger sequencing and immunoblotting for TULP3. Initial sequencing in controls and ko lines was performed on PCRs using primers flanking Exon 3 of Tulp3. We performed TOPO cloning to determine the sequences of the allelic disruptions that are shown in Supplemental Figure S2.
Reverse transcription and quantitative PCR
RNA was extracted from paraffin-embedded brain sections using deparaffinization solution (Qiagen Cat. #19093) and RNA extraction using the GenElute mammalian total RNA purification kit (RTN350; Sigma). Reverse transcription-quantitative PCR (qRT-PCR) was performed with Kicqstart One-Step Probe RT-qPCR ReadyMix (KCQS07; Sigma). Inventoried TaqMan probes for qRT-PCR from Applied Biosystems were used for Tulp3 and Gapdh. Reactions were run in the CFX96 Real time System (Bio-Rad).
Immunofluorescence of cultured cells and microscopy
Cells were cultured on coverslips until confluent and starved for the indicated periods. Cells were fixed with 4% paraformaldehyde (PFA). After blocking with 5% normal donkey serum, the cells were incubated with primary antibody solutions for 1 h at room temperature, followed by treatment with secondary antibodies for 30 min along with Hoechst 33342 (Invitrogen). The coverslips were mounted using Fluoromount G (SouthernBiotech). Images were acquired on a microscope (AxioImager.Z1; ZEISS), a sCMOS camera (PCO Edge; BioVision Technologies), and Plan Apochromat objectives (10×/0.45 NA; 20×/0.8 NA; 40×/1.3 NA oil; and 63×/1.4 NA oil) controlled using Micro-Manager software (University of California, San Francisco) at room temperature. Between 8 and 20 z sections at 0.5–0.8-µm intervals were acquired. For quantitative analysis of ciliary localization, stacks of images were acquired from three to eight consecutive fields with confluent cells by looking into the DAPI channel, and percentages of protein-positive ciliated cells were determined. Maximal projections from images of stacks were exported from ImageJ/Fiji (National Institutes of Health) using a custom-written macro (from M. Mettlen, UTSW Medical Center, Dallas, TX) using similar parameters (image intensity and contrast) for image files from the same experiment. SNAP experiments were performed as described earlier (Keppler et al., 2003; Follit and Pazour, 2013). Briefly, cells on coverslips were serum starved for 36 h to induce ciliation. BG-Block (0.05 µM) (NEB; S9106) was added for 30 min, after which the coverslips were washed in serum starvation media and fixed at different time points. Cells were immunostained with the indicated antibodies. Fluorescent SNAP substrate (0.3 µM) (TMR Star; NEB; S9105) was added along with primary antibodies.
Tissue processing, immunostaining, and microscopy
Mice were perfused with phosphate-buffered saline (PBS), and the brains and kidneys were dissected and fixed in 4% PFA overnight at 4°C and processed for paraffin embedding and sectioning. For paraffin sectioning, tissues were processed over a 12-h period using a Thermo Fisher Excelsior Automated Tissue Processor (A82300001; Thermo Fisher Scientific), which dehydrated the tissues through six ethanol concentrations, from 50% to 100% ethanol, cleared through three changes of xylene, and infiltrated with wax through three Paraplast Plus paraffin baths (39602004; Leica). Samples were embedded in Paraplast Plus using paraffin-filled stainless steel base molds and a Thermo-Shandon Histocenter 2 Embedding Workstation (6400012D; Thermo Fisher Scientific). The tissues were then cut into 5-μm-thick sections, deparafinned, and treated with microwave in Antigen Retrieval citra solution (HK086-9K; BioGenex, Fremont, CA) for 10 min. Sections were then blocked using blocking buffer (1% normal donkey serum [Jackson ImmunoResearch, West Grove, PA] in PBS) for 1 h at room temperature. Sections were incubated with primary antibodies against the following antigens, overnight at room temperature or 4°C: acetylated tubulin (1:500; T6793; Sigma mouse IgG2b), Arl13b (1:500; N295B/66; NeuroMab Facility), Aqp2 (1:500; A7310; Sigma rabbit polyclonal; SC515770, Santa Cruz Biotechnology mouse IgG1), Inpp5e (1:500; 17797-1-AP; Proteintech), Nphp3 (1:500; Proteintech; 22026-1-AP), or Lkb1 (1:500; Cell Signaling Technologies; 13031T). After three PBS washes, the sections were incubated in secondary antibodies (Alexa Fluor 488-, 555-, 594-, and 647-conjugated secondary antibodies, or anti-mouse IgG isotype-specific secondary antibodies; 1:500; Life Technologies, Carlsbad, CA, or Jackson ImmunoResearch). Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) or Hoechst 33342 (Life Technologies). Slides were mounted with Fluoromount-G (0100-01; Southern Biotech), and images were acquired with a Zeiss AxioImager.Z1 microscope, a Zeiss LSM780 confocal microscope, or a spinning-disk confocal microscope (Nikon CSU-W1 SoRa).
Proximity biotinylation experiments
T-Rex-293 cells were cotransfected with 5–7.5 µg each of LapN-TULP3 or LapN-TULP3 mutants/fragments and pDEST-pcDNA5-BirA-FLAG N/C-term–expressing ARL13B or the CD8 linker-BirA or FibrocystinCLS-BirA fusion controls. The media was supplemented with 20 µM biotin for 8–12 h after transfection. Cells were harvested using Tris-buffered saline with 2 mM EDTA and 2 mM EGTA 48 h after transfection. Cells were lysed by resuspending and nutating for 20 min in 50 mM Tris-HCl, pH 7.4, 200 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 1 mM dithiothreitol (DTT), 0.6% IGEPAL CA-630, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), and 0.01 mg/ml each leupeptin, pepstatin, and chymostatin. Lysates were centrifuged at 12,000 × g for 10 min, followed by tandem IPs (Figure 5). In brief, the GFP immunoprecipitates were first digested with TEV protease for 16 h at 4°C. The supernatants were subjected to secondary IP with S protein agarose. The resulting secondary IPs were analyzed by Western blotting. Blots were probed with antibodies against S tag (mouse monoclonal MAC112) and Flag (goat polyclonal ab1257), followed by visualization using IRDye-tagged secondary antibodies. IRdye-tagged streptavidin was used for confirming biotinylation signal on the blots.
Immunoblotting
Cell pellets were lysed by resuspending and nutating for 20 min in 50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 1 mM AEBSF, and 0.01 mg/ml each leupeptin, pepstatin, and chymostatin. Lysates were centrifuged at 12,000 × g for 15 min, boiled in SDS sample buffer for 10 min at 95°C, run in 4–20% Mini-PROTEAN TGX Stain-Free Protein Gels (Bio-Rad; 4568093) at 100V, and immunoblotted with antibodies against ARL13B (mouse monoclonal N295B/66) and TULP3 (rabbit affinity purified polyclonal) (Mukhopadhyay et al., 2010), followed by visualization using IRDye-tagged secondary antibodies and hFAB Rhodamine Anti-Tubulin (Bio-Rad; 12004166). Images were taken in a Bio-Rad Chemidoc MP imaging system. Densitometry was performed using default settings of the Bio-Rad Image lab software.
Recombinant protein purification from bacteria
Single colonies of Rosetta cells transfected with the desired plasmid were grown in 3 ml of Luria-Bertani (LB) broth containing ampicillin and chloramphenicol for 8 h and then transferred to 25 ml and grown overnight. Overnight culture was transferred to 1 l of LB broth and grown until the OD reached 0.5–0.8. Cells were grown at 37°C and 200 rpm. Protein expression was induced overnight at 18°C, 200 rpm, using 0.25 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were pelleted by spinning at 5000 × g for 10 min at 4°C. Cells were lysed by resuspending and incubating in 25 ml of lysis buffer (50 mM Tris, pH 7.4, 450 mM NaCl, 5 mM EDTA, 1 mM DTT, 0.5% Triton X-100, and 10 μg/ml each leupeptin, pepstatin, and chymostatin and 0.1 mM AEBSF) for 30 min at 4°C in a rotator. Cells were sonicated until the solution was nonviscous and spun at 20,000 × g for 20 min at 4°C to remove insoluble material. GST- and MBP-tagged proteins were incubated with washed glutathione beads and amylose beads, respectively, for 3 h at 4°C. Beads were washed three times with lysis buffer and two times with low-salt buffer (50 mM Tris, pH 7.4, 150 mM NaCl, and 1 mM DTT). GST-tagged protein was eluted with elution buffer containing 10 mM reduced glutathione, pH 7.5, in low-salt buffer. MBP-tagged proteins were eluted using 10 mM maltose in low-salt buffer. Proteins were dialyzed into storage buffer (20 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM DTT, and 5% glycerol).
In vitro binding experiments
GST-tagged proteins (20 μg) were incubated with a 30-μl packed volume of glutathione sepharose beads in LAP150N (50 mM HEPES, pH 7.4, 150 mM KCl, 1 mM EGTA, 1 mM MgCl2, 10% glycerol, 0.05% NP-40) buffer at 4°C for 2–4 h. Beads were washed with LAP150N three times and then incubated with IVT Myc-ARL13B (2.5 to 5 μl of the IVT reaction) in LAP100N (50 mM HEPES, pH 7.4, 100 mM KCl, 1 mM EGTA, 1 mM MgCl2, 10% glycerol, 0.05% NP-40) or LAP150N buffer at 4°C or room temperature, respectively, for 1 h. In Figure 5D, the indicated MBP proteins (50 μg) were added along with IVT Myc-ARL13B. Flowthroughs were collected, and beads were washed, eluted by boiling in SDS sample buffer, and immunoblotted.
Quantification of ciliary localization of cargoes and statistical analyses
For measuring cilia positive for a particular protein, we first identified cilia using acetylated tubulin staining as a ciliary marker. In experiments where LAP-tagged constructs were used to rescue ciliary localization of proteins, we counted cilia only from GFP-expressing cells. Next, we carefully counted cilia for the presence of staining corresponding to the protein of interest. All Z-planes containing acetylated tubulin staining were analyzed for staining. We did not use any threshold intensity while counting, and all cilia including those showing low-intensity staining were regarded as positives. For measuring ciliary pixel intensities, image stacks were acquired with z sections at 0.8-µm intervals. An image interval with maximal intensity was chosen, and cilia were demarcated with a region of interest using fluorescence signal for acetylated α-tubulin. The mean pixel intensities for the corresponding protein were exported from ImageJ/Fiji. Statistical analyses were performed using Student’s t test for comparing two groups or Tukey’s post hoc multiple comparison tests between all possible pairs using GraphPad Prism. Nonparametric Mann–Whitney U tests were performed for comparing ciliary lengths between two groups using GraphPad Prism.
Supplementary Material
Acknowledgments
This project was funded by the National Institutes of Health (1R35GM144136 and R01DK128089 to S.M.; R56DK132266 and NS097928 to K.F.L.; and 1S10OD028630 to the Microscopy Core Facility at UT Southwestern), a PKD Foundation postdoctoral fellowship to V.R.P. (214F19a), a Cancer Prevention and Research Institute of Texas grant (RR170063 to J.B.W.), and a PKD Foundation grant to K.F.L. (232G18). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We acknowledge molecular pathology and mass spectrometry cores and the mouse animal care facility at UT Southwestern. We thank John Shelton for histopathology core support and Kevin White and the Quantitative Light Microscopy Core Facility at UT Southwestern for imaging. We acknowledge kind gifts of reagents from Kathryn Anderson, Tamara Caspary, Anne-Claude Gingras, Peter Jackson, and Peter Michaely. We thank Sandii Constable and Feng Qian for comments on an early version of the manuscript.
Abbreviations used:
- Aa
amino acids
- BirA*
promiscuous biotin protein ligase mutant R118G
- CC
coiled-coil
- CD
collecting duct
- cko
conditional knockout
- CLS
ciliary localization sequence motif
- GEF
guanine nucleotide exchange factor
- IFT
intraflagellar transport
- IVT
in vitro translated
- ko
knockout
- LAP
S tag-PreScission/TEV-GFP
- PI(4,5)P 2
phosphoinositide 4,5-bisphosphate
- PKD
polycystic kidney disease
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-10-0473) on January 18, 2023.
REFERENCES
- Amor JC, Harrison DH, Kahn RA, Ringe D (1994). Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature 372, 704–708. [DOI] [PubMed] [Google Scholar]
- Antonny B, Beraud-Dufour S, Chardin P, Chabre M (1997). N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 36, 4675–4684. [DOI] [PubMed] [Google Scholar]
- Anvarian Z, Mykytyn K, Mukhopadhyay S, Pedersen LB, Christensen ST (2019). Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol 15, 199–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badgandi HB, Hwang SH, Shimada IS, Loriot E, Mukhopadhyay S (2017). Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J Cell Biol 216, 743–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett HW, Gustavsson AK, Bayas CA, Petrov PN, Mooney N, Moerner WE, Jackson PK (2020). Novel fibrillar structure in the inversin compartment of primary cilia revealed by 3D single-molecule superresolution microscopy. Mol Biol Cell 31, 619–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C, Fliegauf M, Bruchle NO, Frank V, Olbrich H, Kirschner J, Schermer B, Schmedding I, Kispert A, Kranzlin B, et al. (2008). Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am J Hum Genet 82, 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas SL, Silhavy JL, Brancati F, Kisseleva MV, Al-Gazali L, Sztriha L, Bayoumi RA, Zaki MS, Abdel-Aleem A, Rosti RO, et al. (2009). Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet 41, 1032–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, Bertrand JY, Audollent S, Attie-Bitach T, Holden KR, Dobyns WB, et al. (2008). Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet 83, 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV (2007). The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell 12, 767–778. [DOI] [PubMed] [Google Scholar]
- Cevik S, Hori Y, Kaplan OI, Kida K, Toivenon T, Foley-Fisher C, Cottell D, Katada T, Kontani K, Blacque OE (2010). Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J Cell Biol 188, 953–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik S, Sanders AA, Van Wijk E, Boldt K, Clarke L, van Reeuwijk J, Hori Y, Horn N, Hetterschijt L, Wdowicz A, et al. (2013). Active transport and diffusion barriers restrict Joubert Syndrome-associated ARL13B/ARL-13 to an Inv-like ciliary membrane subdomain. PLoS Genet 9, e1003977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilleros-Rodriguez D, Martin-Morales R, Barbeito P, Deb Roy A, Loukil A, Sierra-Rodero B, Herranz G, Pampliega O, Redrejo-Rodriguez M, Goetz SC, et al. (2022). Multiple ciliary localization signals control INPP5E ciliary targeting. eLife 11, e78383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic D, Williams AH, Ransom N, Morel V, Hargrave PA, Arendt A (2005). Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4). Proc Natl Acad Sci USA 102, 3301–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devane J, Ott E, Olinger EG, Epting D, Decker E, Friedrich A, Bachmann N, Renschler G, Eisenberger T, Briem-Richter A, et al. (2022). Progressive liver, kidney, and heart degeneration in children and adults affected by TULP3 mutations. Am J Hum Genet 109, 928–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL (2011). ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 12, 362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duldulao NA, Lee S, Sun Z (2009). Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development 136, 4033–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferent J, Constable S, Gigante ED, Yam PT, Mariani LE, Legue E, Liem KF Jr, Caspary T, Charron F (2019). The ciliary protein Arl13b functions outside of the primary cilium in Shh-mediated axon guidance. Cell Rep 29, 3356–3366.e3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock T, Ravarani CNJ, Sun D, Venkatakrishnan AJ, Kayikci M, Tate CG, Veprintsev DB, Babu MM (2015). Universal allosteric mechanism for Galpha activation by GPCRs. Nature 524, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Li L, Vucica Y, Pazour GJ (2010). The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J Cell Biol 188, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Pazour GJ (2013). Analysis of ciliary membrane protein dynamics using SNAP technology. Methods Enzymol 524, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Wang L, Kim S, Li J, Dynlacht BD (2016). Role for the IFT-A complex in selective transport to the primary cilium. Cell Rep 17, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R, Douguet D, Antonny B, Drin G (2008). HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics 24, 2101–2102. [DOI] [PubMed] [Google Scholar]
- Gigante ED, Taylor MR, Ivanova AA, Kahn RA, Caspary T (2020). ARL13B regulates Sonic hedgehog signaling from outside primary cilia. eLife 9, e50434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt K, Lokaj M, Koerner C, Falk N, Giessl A, Wittinghofer A (2015). A G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins. eLife 4, e11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim S, Dyson JM, Feeney SJ, Davies EM, Sriratana A, Koenig MN, Plotnikova OV, Smyth IM, Ricardo SD, Hobbs RM, Mitchell CA (2016). Inpp5e suppresses polycystic kidney disease via inhibition of PI3K/Akt-dependent mTORC1 signaling. Hum Mol Genet 25, 2295–2313. [DOI] [PubMed] [Google Scholar]
- Han S, Miyoshi K, Shikada S, Amano G, Wang Y, Yoshimura T, Katayama T (2019). TULP3 is required for localization of membrane-associated proteins ARL13B and INPP5E to primary cilia. Biochem Biophys Res Commun 509, 227–234. [DOI] [PubMed] [Google Scholar]
- Hesketh SJ, Mukhopadhyay AG, Nakamura D, Toropova K, Roberts AJ (2022). IFT-A structure reveals carriages for membrane protein transport into cilia. Cell 185, 4971–4985.e4916. [DOI] [PubMed] [Google Scholar]
- Hilgendorf KI, Johnson CT, Jackson PK (2016). The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Curr Opin Cell Biol 39, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgendorf KI, Johnson CT, Mezger A, Rice SL, Norris AM, Demeter J, Greenleaf WJ, Reiter JF, Kopinke D, Jackson PK (2019). Omega-3 fatty acids activate ciliary FFAR4 to control adipogenesis. Cell 179, 1289–1305.e1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y, Kobayashi T, Kikko Y, Kontani K, Katada T (2008). Domain architecture of the atypical Arf-family GTPase Arl13b involved in cilia formation. Biochem Biophys Res Commun 373, 119–124. [DOI] [PubMed] [Google Scholar]
- Hou X, Mrug M, Yoder BK, Lefkowitz EJ, Kremmidiotis G, D’Eustachio P, Beier DR, Guay-Woodford LM (2002). Cystin, a novel cilia-associated protein, is disrupted in the cpk mouse model of polycystic kidney disease. J Clin Invest 109, 533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert MC, Weihbrecht K, Searby CC, Li Y, Pope RM, Sheffield VC, Seo S (2012). ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci USA 109, 19691–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SH, Somatilaka BN, Badgandi H, Palicharla VR, Walker R, Shelton JM, Qian F, Mukhopadhyay S (2019). Tulp3 regulates renal cystogenesis by trafficking of cystoproteins to cilia. Curr Biol 29, 790–802.e795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SH, Somatilaka BN, White K, Mukhopadhyay S (2021). Ciliary and extraciliary Gpr161 pools repress hedgehog signaling in a tissue-specific manner. eLife 10, e67121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova AA, Caspary T, Seyfried NT, Duong DM, West AB, Liu Z, Kahn RA (2017). Biochemical characterization of purified mammalian ARL13B protein indicates that it is an atypical GTPase and ARL3 guanine nucleotide exchange factor (GEF). J Biol Chem 292, 11091–11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari Khamirani H, Palicharla VR, Dastgheib SA, Dianatpour M, Imanieh MH, Tabei SS, Besse W, Mukhopadhyay S, Liem KF (2022). A pathogenic variant of TULP3 causes renal and hepatic fibrocystic disease. Front Genet 13, 1021037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Palicharla VR, Miller D, Hwang S-H, Zhu H, Hixson P, Mukhopadhyay S, Sun J (2023). Human IFT-A complex structures provide molecular insights into ciliary transport. Cell Research https://doi: 10.1038/s41422-023-00778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K (2003). A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol 21, 86–89. [DOI] [PubMed] [Google Scholar]
- Kopinke D, Norris AM, Mukhopadhyay S (2021). Developmental and regenerative paradigms of cilia regulated hedgehog signaling. Semin Cell Dev Biol 110, 89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang L, Jiang Y, Chen S, Su K, Peng R, Yang X, Wang H (2022). Rare variants in TULP3 abolish the suppressive effect on sonic hedgehog signaling and contribute to human neural tube defects. Genes Dis 9, 1174–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T (2011). Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell 22, 4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legue E, Liem KF Jr (2019). Tulp3 is a ciliary trafficking gene that regulates polycystic kidney disease. Curr Biol 29, 803–812.e805. [DOI] [PubMed] [Google Scholar]
- Li Y, Tian X, Ma M, Jerman S, Kong S, Somlo S, Sun Z (2016). Deletion of ADP ribosylation factor-like GTPase 13B leads to kidney cysts. J Am Soc Nephrol 27, 3628–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kahn RA, Prestegard JH (2010). Dynamic structure of membrane-anchored Arf*GTP. Nat Struct Mol Biol 17, 876–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M (2021). Cilia and polycystic kidney disease. Semin Cell Dev Biol 110, 139–148. [DOI] [PubMed] [Google Scholar]
- Mariani LE, Bijlsma MF, Ivanova AA, Suciu SK, Kahn RA, Caspary T (2016). Arl13b regulates Shh signaling from both inside and outside the cilium. Mol Biol Cell 27, 3780–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetrey J, Macia E, Pasqualato S, Franco M, Cherfils J (2000). Structure of Arf6-GDP suggests a basis for guanine nucleotide exchange factors specificity. Nat Struct Biol 7, 466–469. [DOI] [PubMed] [Google Scholar]
- Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, Nachury MV (2015). Proteomics of primary cilia by proximity labeling. Dev Cell 35, 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Badgandi HB, Hwang SH, Somatilaka B, Shimada IS, Pal K (2017). Trafficking to the primary cilium membrane. Mol Biol Cell 28, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Jackson PK (2011). The tubby family proteins. Genome Biol 12, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK (2010). TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev 24, 2180–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, Jackson PK (2013). The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell 152, 210–223. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Mick DU (2019). Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol 20, 389–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman RX, Ko HW, Huang V, Eun CM, Abler LL, Zhang Z, Sun X, Eggenschwiler JT (2009). Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Hum Mol Genet 18, 1740–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S, Katoh Y, Terada M, Michisaka S, Funabashi T, Takahashi S, Kontani K, Nakayama K (2017). Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J Cell Sci 130, 563–576. [DOI] [PubMed] [Google Scholar]
- Ocbina PJ, Anderson KV (2008). Intraflagellar transport, cilia, and mammalian Hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev Dyn 237, 2030–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal K, Hwang SH, Somatilaka B, Badgandi H, Jackson PK, DeFea K, Mukhopadhyay S (2016). Smoothened determines beta-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J Cell Biol 212, 861–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson VL, Damrau C, Paudyal A, Reeve B, Grimes DT, Stewart ME, Williams DJ, Siggers P, Greenfield A, Murdoch JN (2009). Mouse hitchhiker mutants have spina bifida, dorso-ventral patterning defects and polydactyly: identification of Tulp3 as a novel negative regulator of the Sonic hedgehog pathway. Hum Mol Genet 18, 1719–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picariello T, Brown JM, Hou Y, Swank G, Cochran DA, King OD, Lechtreck K, Pazour GJ, Witman GB (2019). A global analysis of IFT-A function reveals specialization for transport of membrane-associated proteins into cilia. J Cell Sci 132, jcs220749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Fujisawa S, Nozaki S, Katoh Y, Nakayama K (2021). Interaction of INPP5E with ARL13B is essential for its ciliary membrane retention but dispensable for its ciliary entry. Biol Open 10, bio057653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo PA, Terui T, Sturch S, Fales HM, Ferrige AG, Kahn RA (1995). The myristoylated amino terminus of ADP-ribosylation factor 1 is a phospholipid- and GTP-sensitive switch. J Biol Chem 270, 14809–14815. [DOI] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X (2008). CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel 21, 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L (2001). G-protein signaling through tubby proteins. Science 292, 2041–2050. [DOI] [PubMed] [Google Scholar]
- Seixas C, Choi SY, Polgar N, Umberger NL, East MP, Zuo X, Moreiras H, Ghossoub R, Benmerah A, Kahn RA, et al. (2016). Arl13b and the exocyst interact synergistically in ciliogenesis. Mol Biol Cell 27, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Somlo S, Igarashi P (2002). Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J Am Soc Nephrol 13, 1837–1846. [DOI] [PubMed] [Google Scholar]
- Shiba D, Manning DK, Koga H, Beier DR, Yokoyama T (2010). Inv acts as a molecular anchor for Nphp3 and Nek8 in the proximal segment of primary cilia. Cytoskeleton (Hoboken) 67, 112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N (2004). A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131, 4085–4093. [DOI] [PubMed] [Google Scholar]
- Thomas S, Cantagrel V, Mariani L, Serre V, Lee JE, Elkhartoufi N, de Lonlay P, Desguerre I, Munnich A, Boddaert N, et al. (2015). Identification of a novel ARL13B variant in a Joubert syndrome-affected patient with retinal impairment and obesity. Eur J Hum Genet 23, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres JZ, Miller JJ, Jackson PK (2009). High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics 9, 2888–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G (1999). Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet 23, 99–103. [DOI] [PubMed] [Google Scholar]
- van den Hoek H, Klena N, Jordan MA, Alvarez Viar G, Righetto RD, Schaffer M, Erdmann PS, Wan W, Geimer S, Plitzko JM, et al. (2022). In situ architecture of the ciliary base reveals the stepwise assembly of intraflagellar transport trains. Science 377, 543–548. [DOI] [PubMed] [Google Scholar]
- Viau A, Bienaime F, Lukas K, Todkar AP, Knoll M, Yakulov TA, Hofherr A, Kretz O, Helmstadter M, Reichardt W, et al. (2018). Cilia-localized LKB1 regulates chemokine signaling, macrophage recruitment, and tissue homeostasis in the kidney. EMBO J 37, e98615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RV, Maranto A, Palicharla VR, Hwang S-H, Mukhopadhyay S, Qian F (2022). Cilia-localized counterregulatory signals as drivers of renal cystogenesis. Front Mol Biosci 9, 936070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Seltzsam S, Zheng B, Wu CW, Nicolas-Frank C, Yousef K, Au KS, Mann N, Pantel D, Schneider S, et al. (2022). Whole exome sequencing identifies potential candidate genes for spina bifida derived from mouse models. Am J Med Genet A 188, 1355–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng SL, Kao HJ, Huang CH, Lee TY (2017). MDD-Palm: identification of protein S-palmitoylation sites with substrate motifs based on maximal dependence decomposition. PLoS One 12, e0179529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, Wang W, Pretorius PR, Sheffield VC, Sengupta P, et al. (2011). An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev 25, 2347–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CT, Hilgendorf KI, Bevacqua RJ, Hang Y, Demeter J, Kim SK, Jackson PK (2021). Discovery of ciliary G protein-coupled receptors regulating pancreatic islet insulin and glucagon secretion. Genes Dev 35, 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Nager AR, Nachury MV (2018). BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J Cell Biol 217, 1847–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Carroll TJ, McMahon AP (2002). Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development 129, 5301–5312. [DOI] [PubMed] [Google Scholar]
- Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM (2009). Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science 326, 1707–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.