Summary
Background
Systemic lupus erythematosus (SLE) is a prototypical autoimmune disease affecting multiple organs and tissues with high cellular heterogeneity. CD8+ T cell activity is involved in the SLE pathogenesis. However, the cellular heterogeneity and the underlying mechanisms of CD8+ T cells in SLE remain to be identified.
Methods
Single-cell RNA sequencing (scRNA-seq) of PBMCs from a SLE family pedigree (including 3 HCs and 2 SLE patients) was performed to identify the SLE-associated CD8+ T cell subsets. Flow cytometry analysis of a SLE cohort (including 23 HCs and 33 SLE patients), qPCR analysis of another SLE cohort (including 30 HCs and 25 SLE patients) and public scRNA-seq datasets of autoimmune diseases were employed to validate the finding. Whole-exome sequencing (WES) of this SLE family pedigree was used to investigate the genetic basis in dysregulation of CD8+ T cell subsets identified in this study. Co-culture experiments were performed to analyze the activity of CD8+ T cells.
Findings
We elucidated the cellular heterogeneity of SLE and identified a new highly cytotoxic CD8+ T cell subset, CD161−CD8+ TEMRA cell subpopulation, which was remarkably increased in SLE patients. Meanwhile, we discovered a close correlation between mutation of DTHD1 and the abnormal accumulation of CD161−CD8+ TEMRA cells in SLE. DTHD1 interacted with MYD88 to suppress its activity in T cells and DTHD1 mutation promoted MYD88-dependent pathway and subsequently increased the proliferation and cytotoxicity of CD161−CD8+ TEMRA cells. Furthermore, the differentially expressed genes in CD161−CD8+ TEMRA cells displayed a strong out-of-sample prediction for case–control status of SLE.
Interpretation
This study identified DTHD1-associated expansion of CD161−CD8+ TEMRA cell subpopulation is critical for SLE. Our study highlights genetic association and cellular heterogeneity of SLE pathogenesis and provides a mechanistical insight into the diagnosis and treatment of SLE.
Fundings
Stated in the Acknowledgements section of the manuscript.
Keywords: Systemic lupus erythematosus, scRNA-seq, Whole-exome sequencing, CD8+ T cell subset, Genetic variant, DTHD1, MYD88
Research in context.
Evidence before this study
SLE is characterized by abnormal activation of immune system with diverse clinical manifestations, leading to challenging for management. SLE patients display distinctive transcriptional features of immune cells, including increased type I interferon (IFN) signaling and inflammatory response, aberrant pathway in B cells and CD4+ lymphocytes. Recent single-cell RNA sequencing studies have elucidated that immune cells including myeloid cells, B cells and T cells are phenotypically heterogeneous with high IFN signature limited to some small subsets. CD8+ T cells are recently revealed to contribute to the pathogenesis of SLE. However, the cellular and functional heterogeneity of CD8+ T cells in SLE remain to be further identified.
Added value of this study
We performed scRNA-seq of a SLE family pedigree and identified an abnormal expansion of CD8+ T cell subset, CD161−CD8+ TEMRA, which exhibited high cytotoxicity in SLE patients. We validated this subset was expanded and cytotoxic in SLE patients from another cohort by flow cytometry. This finding was further confirmed by analysis of public scRNA-seq datasets (GSE162577). To investigate the genetic basis of this expanded CD161−CD8+ TEMRA subset, we additionally performed WES of the SLE family pedigree and identified a frameshift mutation (p.Ser897fs) in DTHD1 carried by SLE patients. To investigate whether this mutation affected DTHD1 expression, we observed decreased expression of DTHD1 in PBMCs from SLE patients of the family by qPCR, and determined a reduced mRNA and protein levels of DTHD1 by transfection of DTHD1 expression plasmid with p. Ser897fs mutation in HEK293T cells. Therefore, this rare variation could result in a loss of function in DTHD1. Furthermore, co-expression analysis of our scRNA-seq data indicated DTHD1 was closely related to the regulation of cytotoxicity. This analysis was validated by in vitro cytotoxic assay of CD8+ T cells by coculturing with P815 cells. We found that DTHD1-silenced CD8+ T cells induced higher ratio of P815 cells apoptosis than control CD8+ T cells. Mechanistically, by co-immunoprecipitation assay, we discovered DTHD1 directly interacted with MYD88, leading to suppression of MYD88-mediated pathway. Chemical inhibition of MYD88 activity with TAK-242 in primary human CD8+ T cells efficiently decreased the cytotoxicity against P815 cells and cellular granzyme B levels. Finally, by using a public transcriptome profile for molecular prediction (GSE121239), we revealed that the upregulated genes in CD161−CD8+ TEMRA cells from SLE displayed an efficient prediction for case–control status (AUC = 0.917).
Implications of all the available evidence
This study identified DTHD1-associated expansion of CD161−CD8+ TEMRA cell subpopulation is critical for SLE. Our study highlights genetic association and cellular heterogeneity of SLE pathogenesis and provides a mechanistical insight into the diagnosis and treatment of SLE.
Introduction
SLE is a prototypical autoimmune disease that may cause chronic inflammation and debilitating damage in multiple organs and tissues. It is characterized by a global breakdown of immune tolerance with activation of both innate and adaptive immune responses against nucleic acids and endogenous antigens.1,2 SLE is clinically heterogeneous in terms of development, manifestations and severity, and the disease course is unpredictable with periodic remissions and flares.3 Therefore, identification of precise genetic and cell-type specific mechanisms of the SLE pathogenesis, would improve the management of SLE.
SLE patients exhibit distinctive transcriptional characteristics of circulating immune cells, including elevated type I IFN signatures, myeloid-derived inflammation, B-cell associated pathways, abnormalities of CD4+ T lymphocytes.4, 5, 6 More recently, emerging evidence suggest the potential role of CD8+ T lymphocytes in SLE pathogenesis. According to the expression of CD45RA and CCR7, human CD8+ T cells in peripheral blood can be classified into four subsets: Naïve T cells (CD45RA+/CCR7+), central memory T cells (TCM, CD45RA−/CCR7+), effector memory T cells (TEM, CD45RA−/CCR7−), and effector memory T cells re-expressing CD45RA (TEMRA, CD45RA+/CCR7−).7 Although TCM, TEM and TEMRA are antigen-specific memory CD8+ T subsets, they display distinct functions. TEMRA cells are considered to be terminally differentiated and carry the higher levels of cytotoxic and exhausted genes compared to TEM.8,9 Transcriptional analyses have shown that the CD8+ memory T cell signatures could predict the prognosis of autoimmune diseases including SLE,10 and an exhaustion signature of CD8+ T cell correlates with a reduced risk of SLE flare.11 Nevertheless, these observations based on bulk transcriptional analyses are insufficient to demonstrate the cell-type specific transcriptional differences, the disease-associated expression features might be influenced by diversity in the relative abundances of each immune cell subpopulation. scRNA-seq is a potential way to comprehensively understand heterogeneous transcription states of immune cells, which provides exciting genome-wide information for new diagnosis strategies and prognostic evaluation.12 A recent scRNA-seq analysis of peripheral blood mononuclear cells (PBMCs) from 33 children with SLE illustrated that high IFN signature was limited to some small subpopulations of myeloid cells, B cells and T cells.13 However, cellular and functional heterogeneity of CD8+ T cells in SLE remain to be identified.
Genetic factors are key contributors to SLE etiology, with a high heritability ranging from 43% to 66%.14 Notably, the first-degree relatives of SLE patients are 20 times more likely to suffer from this disease than the general individuals.15 To date, over 100 risk loci have been identified to be associated with SLE susceptibility, which are chiefly located in genes of multiple immune pathways, including nucleic acid sensing, dysregulation of type I IFN pathways and lymphocyte activation.15, 16, 17 However, the precise mechanism underlying the causative linkage of these genetic variations to SLE development is still to be fully elucidated. Deleterious mutations are more likely to be rare and usually have a greater impact on genes function.18,19 Therefore, unraveling rare variants in SLE is important to illustrate the genetic causes of the disease pathogenesis. WES, especially family-based WES, is a powerful tool to uncover disease-related pathogenic genes. It has been successfully identified causative variants in the coding regions of rare diseases, such as primary immunodeficiencies by WES.20 Nevertheless, owing to the cellular heterogeneity of autoimmune diseases, how rare variants are associated with the pathogenesis of SLE in a cell-type specific manner remains unclear.
In this study, by integration of the scRNA transcriptomic and WES data from a SLE family pedigree, we identified a new CD8+ T lymphocyte subset, CD161−CD8+ TEMRA cell, which is highly activated and cytotoxic with a markedly elevated frequency in SLE patients. Meanwhile, we discovered a rare deleterious mutation in DTHD1 carried by SLE patients of this family and confirmed that dysfunction of DTHD1 is related with activation of MYD88-dependent pathway in T cells, thereby promoting the proliferation and cytotoxicity of CD161−CD8+ TEMRA cells. Our study links genetic variants and cell type-specific alterations of SLE and provides new insights into the prediction and pathogenesis of SLE.
Methods
Ethics statement
The study was approved by the research ethics board of Sun Yat-sen Memorial Hospital (SYSEC-KY-KS-2020-213). Informed consent was obtained from all samples in this study.
Human samples
The information of SLE family pedigree enrolled in this study was listed Table 1. Two cohorts of case–control samples containing males and females were enrolled as validation cohorts. Cohort1 included control donors (n = 23, 23–54 years), SLE donors (n = 33, 19–72 years), and cohort 2 included control donors (n = 30, 24–55 years), SLE donors (n = 25, 18–74 years). PBMCs were collected from cohort 1 and cohort 2 were used for flow cytometry analysis and gene expression analysis, respectively. The SLE subjects were diagnosed based on the 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for SLE.21 Patients with other autoimmune diseases or pregnancy were excluded from this study, and patients treated with prednisone over 15 mg/day or biologics or methotrexate or sirolimus or metformin therapy within the last 30 days were excluded from this study. No significant sex differences were determined in the two cohorts. Clinical characteristics of these two cohorts are shown in Table 2.
Table 1.
Demographic and clinical characteristics of SLE patients’ family pedigree.
| SLE 1 | SLE 2 | |
|---|---|---|
| Age and sex | 32/Female | 30/Female |
| Initial presentation | Malar rash on face, erythema on both upper arms and palms, oral ulcer and knuckle pain, leukopenia, lymphopenia | Malar rash on face, alopecia and fever, wrist pain |
| Hematologic abnormalities | None | None |
| Kidney abnormalities | Bld 2+WBC | None |
| Serositis | None | None |
| Neurological involvement | None | None |
| Antinuclear antibodies (ANA) | ++ (speckle pattern) | + (speckle pattern) |
| Anti-dsDNA | Negative | Positive |
| Anti-Smith | Negative | Negative |
| Anti-SSA | Positive | Positive |
| Anti-nRNP | Negative | Positive |
| RIB-P | Negative | Positive |
| C3 (normal, 650–1800 mg/L) | 972 | 902 |
| C4 (normal, 100–400 mg/L) | 313 | 215 |
| ESR (normal, 0–20 mm/h) | 28 | 40 |
| SLEDAIa | 10 | 2 |
| Treatment | 15 mg/d prednisone by oral | 10 mg/d prednisone and 75 mg/d dipyridamole by oral |
| Other condition | Osteonecrosis of the right femoral head for 10 years |
In this family pedigree, there are three SLE patients in two generations. Patient 1 (SLE1) is a 32-year-old female, who was firstly admitted to Sun Yat-sen Memorial Hospital in August 2018 and diagnosed with SLE. Her father and sister (SLE2) were diagnosed with SLE at the local hospital, but their father has passed away unfortunately. The mother with 55 years old (HC3), the sister is 34 years old (HC1), and the sister's son with 15 years old (HC2) are three healthy controls.
SLEDAI, SLE Disease Activity Index.
Table 2.
Demographic and clinical characteristics of SLE patients and healthy controls.
| Characteristics | Cohort1 |
Cohort2 |
||
|---|---|---|---|---|
| SLE (n = 33) | HC (n = 23) | SLE (n = 25) | HC (n = 30) | |
| Demographic characteristics | ||||
| Female/male | 30/3 | 23/0 | 20/5 | 23/7 |
| Median age (years) (IQR) | 37 (19–72) | 33 (23–54) | 35 (18–74) | 31.3 (24–55) |
| Clinical features | ||||
| Disease duration (year) (median) (IQR) | 6 (1–27) | / | 3 (1–18) | / |
| SLEDAI (median) (IQR) | 9 (0–24) | / | 10 (2–22) | / |
| Inactive patients (n) (%) | 7 (21.21) | / | 5 (20) | / |
| Active patients (n) (%) | 26 (78.79) | / | 20 (80) | / |
| Clinical manifestations (n) (%) | ||||
| Rash | 14 (42.42) | / | 10 (40) | / |
| Photosensitivity | 9 (27.27) | / | 5 (20) | / |
| Oral ulcers | 5 (15.15) | / | 3 (12) | / |
| Alopecia | 9 (27.27) | / | 7 (28) | / |
| Arthritis | 11 (33.33) | / | 3 (12) | / |
| Serositis | 6 (18.18) | / | 3 (12) | / |
| Renal involvement | 15 (45.45) | / | 13 (52) | / |
| Neurological involvement | 3 (9.09) | / | 1 (4) | / |
| Leukopenia | 6 (18.18) | / | 8 (32) | / |
| Lymphopenia | 12 (36.36) | / | 12 (48) | / |
| Thrombocytopenia | 7 (21.21) | / | 2 (8) | / |
| ANA positive | 29 (87.88) | / | 22 (88) | / |
| Anti-dsDNA positiv | 22 (66.67) | / | 16 (64) | / |
| Anti-Sm positive | 13 (39.40) | / | 6 (24) | / |
| Anti-Phospholipid positive | 5 (15.15) | / | 4 (16) | / |
| Low Complement | 16 (48.48) | / | 11 (44) | / |
| Treatment (n) (%) | ||||
| Prednisolone | 22 (66.67) | / | 12 (48) | / |
| Hydroxychloroquine | 18 (54.55) | / | 10 (40) | / |
| Azathioprine | 3 (9.09) | / | 0 (0) | / |
| Vitamin D | 4 (12.12) | / | 0 (0) | / |
| Baricitinib | 1 (3.03) | / | 0 (0) | / |
| Sirolimus | 0 (0) | / | 0 (0) | / |
| Metformin | 0 (0) | / | 0 (0) | / |
Patients with a SLEDAI score of ≥5 were classified as active state, as inactive groups referred to the patients with the score of ≤4. IQR, inter quartile range.
Continuous variables are represented as mean ± standard deviation (SD).
Antibodies, recombinant cytokines, and reagents
The following antibodies and reagents were used: Alexa Fluor 700 anti-human CD3 (UCHT1) (BioLegend Cat# 300424, RRID: AB_493741), APC-H7 anti-human CD8 (SK1) (BD Biosciences Cat# 641409, RRID:AB_1645737), BV421 anti-human CD8 (SK1) (BioLegend Cat# 344747, RRID:AB_2629583), PE/Dazzle 594 anti-human CD197 (CCR7) (G043H7) (BioLegend Cat# 353236, RRID:AB_2563641), PE anti-human CD161 (HP-3G10) (BioLegend Cat# 339903, RRID:AB_1501086), PE/Cyanine7 anti-human CD45RA (HI100) (BioLegend Cat# 304126, RRID:AB_10708879), FITC anti-human CD45RA (L48) (BD Biosciences Cat# 347723, RRID:AB_400343), Krome Orange anti-human CD45 (J33) (Beckman Coulter Cat# B36294), Pacific Blue anti-human/mouse Granzyme B (GB11) (BioLegend Cat# 515408, RRID:AB_2562196), PerCP/Cy5.5 anti-human/mouse Granzyme B (QA16A02) (BioLegend Cat# 372212, RRID:AB_2728379), Annexin V-FITC (Cat# AP101, MultiSciences Biotech), APC Annexin V (Cat#550474, BD Biosciences). Myc-Tag Rabbit mAb (ABclonal Cat# AE070, RRID:AB_2863795), mouse anti-HA-Tag mAb (ABclonal Cat# AE008, RRID:AB_2770404), Vinculin Rabbit mAB (ABclonal Cat# A2752, RRID:AB_2863020), β-Actin Antibody (Cell Signaling Technology Cat# 4967, RRID:AB_330288), Phospho-p38 MAPK (Thr180/Tyr182) Antibody (Cell Signaling Technology Cat# 9211, RRID:AB_331641), p38 MAPK (D13E1) XP Rabbit mAb (Cell Signaling Technology Cat# 8690, RRID: AB_10999090), Phospho-4E-BP1 (Thr37/46) (236B4) Rabbit mAb (Cell Signaling Technology Cat# 2855, RRID:AB_560835), 4E-BP1 (53H11) Rabbit mAb (Cell Signaling Technology Cat# 9644, RRID:AB_2097841), Phospho–NF–κB p65 (Ser536) (93H1) Rabbit mAb (Cell Signaling Technology Cat# 3033, RRID:AB_331284), NF-kappaB p65 (D14E12) XP Rabbit mAb (Cell Signaling Technology Cat# 8242, RRID:AB_10859369), p70 S6 Kinase Antibody (Cell Signaling Technology Cat# 9202, RRID:AB_331676), Phospho-p70 S6 Kinase (Thr421/Ser424) Antibody (Cell Signaling Technology Cat# 9204, RRID:AB_2265913), MyD88 (D80F5) Rabbit mAb (Cell Signaling Technology Cat# 4283, RRID:AB_10547882).
Flow cytometry
For surface staining, PBMCs were harvested and stained with antibodies in PBS for 20 min at room temperature. For intracellular staining, cells were fixed with 4% paraformaldehyde, then stained with specific antibodies in 1 × Perm (BD Bioscience). Stained cells were then detected by Navios EX flow cytometer (Beckman Coulter) and analyzed using Kaluza software (Beckman).
PBMCs isolation and CD8+ T cell purification
PBMCs were isolated from EDTA-anticoagulated blood using the lymphocyte separation medium (LTS1077, TBD, China) by Ficoll–Paque density gradient centrifugation. The viability of PBMCs in single-cell suspensions were assessed by using Trypan Blue Staining. Each sample with 1 × 107 viable cells was obtained for scRNA-seq or WES.
CD8+ T cells are purified using EasySep™ human CD8+ T cell isolation kit (Cat #17953, STEMCELL) according to the manufacturer's instructions. The purified CD8 T cells were stimulated with 5 μg/ml anti-human CD3 (clone OKT3, BioLegend Cat# 317326, RRID: AB_11150592) and 2 μg/ml anti-human CD28 (clone CD28.2, BioLegend Cat# 302934, RRID: AB_11148949) for 3 days, then CD8+ T cells were collected for analysis or electroporation.
Electroporation
For overexpression assays, 5 × 106 human primary CD8+ T cells were electroporated with 3 μg empty vector or WT/Mutant-DTHD1 overexpressing plasmid by using NEPA21 (NEPAGENE). For knockdown assays, 5 × 106 human primary CD8+ T cells were electroporated with 50 nM Scramble siRNA (si-NC) or DTHD1 siRNA (si-DTHD1) using NEPA21. The siRNA sequences targeting DTHD1 were listed as follows: siRNA-1, sense (5’ → 3’): GGC AUU GCA AUU CCA UUU ATT, antisense (5’ → 3’): UAA AUG GAA UUG CAA UGC CTT; siRNA-2, sense (5’ → 3’): GUG CCU UCC AAA GAU UUA ATT, antisense (5’ → 3’): UUA AAU CUU UGG AAG GCA CTT. siRNA-1 and siRNA-2 were mixed together for DTHD1 knockdown.
Small interfering RNA (siRNA) transfection
1.5 × 104 HeLa cells were cultured in 24-well plates. 50 nM Scramble siRNA or DTHD1 siRNA were transfected into cells with Opti-MEM containing Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's instructions.
Plasmids construction
Human DTHD1 (NM_001170700.3) complementary DNA (cDNAs) and mutant cDNA with the 2635th bp deletion were synthesized in company (JIE LI BIOLOGY, China), and then ligated into pcDNA3.1 vector with N-terminal tagged 3 × HA. Human MYD88 cDNA was purchased from Genecopoeia Inc. and cloned into the pcDNA3.1-Myc plasmid.
Dual-luciferase reporter assay
3 × 104 HEK293T cells were seeded in 96-well plates for transfection. NF-κB luciferase reporter plasmid (100 ng), TK luciferase reporter plasmid (20 ng), human MYD88 plasmid (100 ng), WT or mutant DTHD1 plasmids (100 ng) were transfected into HEK293T cells by EZ Cell Transfection Reagent (AC04L092, Shanghai Life iLab Bio Technology, China) according to manufacture instruction. After transfection for 24 h, cells were harvested to determine the luciferase activity using Dual-Luciferase reporter assay kit (RG027, Beyotime, China).
Western blotting and co-immunoprecipitation
Cells were harvested and lysed with lysis buffer (9803S, Cell Signaling Technology, USA) with the protease inhibitor cocktail and the phosphatase inhibitor cocktail (78,440, Sigma, USA) for protein extraction. The extracts were used for western blotting analysis or co-immunoprecipitation. For co-immunoprecipitation, total protein was incubated with anti-Myc beads (Cell Signaling Technology) at 4 °C for 3 h. The precipitants were washed three times with lysis buffer, and the samples were eluted and denaturated for 5 min at 100 °C. The samples were tested by subsequent Western blotting using anti-HA antibody.
Apoptosis analysis
Hela cells were treated with TNF-α for the indicated time or P815 cells were cocultured with primary CD8 T cells, then harvested and stained with Annexin V and PI for 15 min at room temperature. Stained cells were then detected by ACEA NovoCyteTM flow cytometer (ACEA biosciences) and analyzed using Flowjo software (BD biosciences).
Single-cell RNA-seq data preprocessing
Cell Ranger software (version 3.1.0) (10 × Genomics) was used to convert raw BCL files to FASTQ files by alignment and counts quantification. Briefly, reads with low-quality barcodes and unique molecular identifiers (UMIs) were filtered out and then mapped to the reference genome. Reads uniquely mapped to the transcriptome and intersecting an exon at least 50% were considered for UMI counting. Before quantification, the UMI sequences would be corrected for sequencing errors, and valid barcodes were identified based on the EmptyDrops method.22 Cells of the gene-by-gene matrix for each sample were individually imported into Seurat (version 3.1.1)23 for downstream analysis. Cells with unusually high number of UMIs (≥20,000) or mitochondrial gene percent (≥10%) were filtered out. We also excluded cells with less than 500 or more than 4600 genes detected. After removing unwanted cells, “LogNormalize” was used to normalize the gene expression. Finally, the genes expression in single-cell were projected into two UMAP dimensions plots by k-nearest neighbor (KNN) graph model.
WES and data processing
Genomic DNA was fragmented using NEBNext dsDNA Fragmentase (NEB, Ipswich, MA, USA) followed by DNA ends repairing. End-repaired DNA fragments were dAtailed and ligated with the NEBNext adaptor (NEB, Ipswich, MA, USA). Biotinylated RNA library baits and magnetic beads were mixed with the barcoded library for targeted regions selection using the SureSelect Human All Exon V6 Kit (Agilent Technologies, Palo Alto, Calif.). The captured sequences were further amplified for 150bp paired-end sequencing in Illumina X-ten system (Illumina, San Diego, CA, USA).
Sequencing data were processed by quality trimming to generate high confidence of variant calling. Raw reads were processed to get high quality clean reads using fastp according to three stringent filtering standards: 1. Removing reads with ≥10% unidentified nucleotides (N); 2. Removing reads with >50% bases with phred quality scores ≤20; 3. Removing reads aligned to the barcode adapter.
Cell developmental trajectory
The CD8+ cell lineage trajectory was inferring by Monocle224 and Monocle3.25 Compared with Monocle2, Monocle3 analysis in dimensionality reduction and trajectory learning is based on UMAP plots, which makes the trajectory more visualized and easier to understand. While compared with Monocle3, Monocle2 is semi-supervised analysis mode, which is more suitable for personalized analysis of cell subsets. Therefore, we integrated the two methods for CD8+ cell lineage trajectory analysis. In this analysis, we excluded MAIT cells due to their distinct development processes relative to other CD8+ cells. We first used Monocle2 to perform pseudo-temporal analysis of CD8+ T cells based on 2000 hypervariable genes and identified differentiation-related genes. Next, we performed pseudo-temporal analysis of CD8+ T cells using Monocle3 to identify the trajectory of cell differentiation in the UMAP plots. Meanwhile, to make the cell lineage trajectory more reliable, we further supplemented information based on the Monocle2 results.
Quantitative real-time PCR
Total RNA was extracted from cultured cells or PBMCs with EZ-press RNA Purification Kit (EZBioscience, China; #B0004DP) following the manufacturer's instructions. cDNA was synthetized from total RNA with Color Reverse Transcript Kit (EZBioscience, #A0010CGQ) according to the manufacturer's instructions. Quantitative real-time PCR (qRT-PCR) was performed in Roche LightCycler 480 instrument using SYBR Green Color qPCR Mix (EZBioscience, #A0012-R2). Cycle thresholds (CT) values of the indicated genes to calculate relative expression using the 2−ΔCT. Primers used for qPCR are listed in Table 3.
Table 3.
Primers used for qPCR in this study.
| Gene | Primer sequence (5′ → 3′) | |
|---|---|---|
| Human ACTB | Forward | CAT GTA CGT TGC TAT CCA GGC |
| Human ACTB | Reverse | CTC CTT AAT GTC ACG CAC GAT |
| Human CD161 | Forward | ACC GGG TCT CTT AAA CTG CC |
| Human CD161 | Reverse | AGC AGG CTG GAT TCT TTG GT |
| Human DTHD1 | Forward | CAG TCC AAT CCA CAA GCC CT |
| Human DTHD1 | Reverse | GGA GGC AAT TTT TGT CCG GT |
| Human TNFSF14 | Forward | GAC CGA CAT CCC ATT CAC GA |
| Human TNFSF14 | Reverse | ACC ATC TCT CCT AGA CGC CA |
Cell Counting Kit 8 (CCK8) assay
Cell proliferation was determined by Cell Counting Kit-8 (CCK8; Sangon Biotech, Shanghai). 2 × 103 HeLa cells were seeded in a 96-well plate per well and transfected with siRNAs for 24 h. Culture medium was replaced with the addition of CCK8 reagents into corresponding wells at indicated time points (24 h, 48 h, 72 h and 96 h) and incubated for 1 h at 37 °C and 5% CO2. Absorbance was determined at 450 nm.
In vitro cytotoxicity analysis
Human primary CD8+ T cells were cocultured with P815 cells to investigate the cytotoxicity of CD8+ T cells. Briefly, target P815 cells were stained with Vybrant™ DiO Cell-Labeling Solution (Thermo Fisher Scientific) for 15 min in serum-free medium and then cocultured with purified human CD8+ T cells for 5 h at the ratio of 1:5. Cytotoxicity was determined by flow cytometric analysis of the expression of Propidium Iodide and Annexin V.
RNA library preparation and sequencing
Single cell suspensions were loaded on a 10 × Genomics GemCode Single-cell instrument for generating single-cell Gel Bead-In-EMlusion (GEMs) droplets. Chromium Next GEM Single Cell 3′ Reagent Kits, Gel Bead and Multiplex, Chip kit (10 × Genomics) were used to covert single-cell to barcoded scRNA sequencing libraries. Libraries were generated according to manufacturer's protocols of Chromium Next GEM Single Cell 3′ Reagent Kits.
Cell type annotation and proportion
The log-normalized matrices were used for cell type annotation by SingleR R packages.26 To secondary annotate the clusters generated from the object, we used both differential expression analysis between clusters and classification based on putative marker genes expression. Expression of each gene in given cluster was compared to the rest of cells using Wilcoxon rank sum test. Significant upregulated genes were identified using following criteria. First, genes had to be at least 0.36-fold overexpressed in the target cluster. Second, genes had to be expressed in more than 25% of the cells belonging to the target cluster. Third, P value was less than 0.05. The percentage of each cell type was calculated as the number of cells belonging to the cell type divided by the total number of cells assigned to the sample or individual.
SCENIC analysis
SCENIC (single-cell regulatory network inference and clustering) was used to establish transcription factor regulatory networks in T cells. In this analysis, GENIE3 was used for co-expression analysis of single-cell transcriptome to obtain several co-expression modules composed of TFs and potential target genes. RcisTarget was used to calculate the significantly enriched motifs in each co-expression module, and predict the target genes containing the motifs, which generated regulons. AUCell algorithm was used to evaluate the transcriptional activity of each regulon in different subpopulation.
Cell–cell interaction analysis
CellChat27 was performed to analyze cell–cell interactions by identifying significant ligand-receptor pairs in healthy controls and SLE samples. Cell communications in different cell types were identified based on the gene expression of ligands in one cell subset and expression of specific receptors in another cell subset. We calculated the cell communication scores according to the average expression of ligand-receptor genes in HCs and SLE samples, respectively.
Gene signature scoring
The expression of genes related with IFN pathway, cytotoxicity, exhaustion and activation were used to evaluate the score of each cluster/subcluster, respectively. The IFN signature score was indicated by evaluating the expression of 26 genes including SP100, ZBP1, ISG20, IRF7, IFIT2, IFITM3, XAF1, EIF2AK2, DHX58, RSAD2, IFIT1, IFIT3, DDX58, ADAR, IFI35, BST2, NMI, UBE2L6, ISG15, IFIH1, OAS2, STAT1, MX1, STAT2, MX2, OAS3.13,28 The exhaustion score was indicated by evaluating the expression of 5 genes including CTLA4, HAVCR2, LAG3, PDCD1, TIGIT.29 The cytotoxicity score was indicated by evaluating the expression of 8 genes including PRF1, IFNG, GNLY, NKG7, GZMB, GZMA, CST7, TNFSF10. The activation score was indicated by evaluating the expression of 6 genes including ICOS, CD226, TNFRSF14, TNFRSF25, TNFRSF9, CD28.30
Functional enrichment analysis
GO and pathway enrichment analysis were performed using Metascape (https://metascape.org) and Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/). All enriched GO and pathway were filtered at a threshold of false discovery rate (FDR, Benjamini-Hochberg correction) at <0.05.
Analysis of public sequencing datasets
scRNA-seq dataset (GSE162577) of PBMCs from 2 SLE patients and 1 healthy control was used to identify and assess the frequency of CD161−CD8+ TEMRA cell subpopulation. Immune cells were clustered by Seurat.
scRNA-seq dataset (GSE157278) of PBMCs from pSS patients and dataset (GSE193770) from MS patients were used to determine the frequency of CD161−CD8+ TEMRA cell subpopulation in other autoimmune diseases. GSE15728 scRNA-seq dataset that contains 57,288 single cell transcriptome was reclustered to analyze CD8+ T subsets. T cell subsets were mapped into our scRNA-seq data by using FindTransferAnchors and TransferData functions of Seuart and were determined the changes of frequencies. GSE193770 scRNA-seq dataset was from CD8+ T of 6 MS patients and 4 healthy controls, which could be directly mapped and determined the population changes.
RNA-seq dataset (GSE121239) of PBMCs from 292 SLE patients and 20 healthy controls was used to assess the predictive power of the CD161−CD8+ TEMRA cells.
Variants identification and annotation
To identify SNPs and InDels, the Burrows-Wheeler Aligner (BWA) was used to align the clean reads from each sample against the reference genome with the settings ‘mem 4 -k 32 -M’ (-k is the minimum seed length, and -M is an option used to mark shorter split alignment hits as secondary alignments). Variant calling was performed for multi-sample using the Genome Analysis Toolkit (GATK) Unified Genotyper with local realignment, base quality score recalibration. SNPs and InDels were filtered using GATK's Variant Filtration with proper standards (-Window 4, -filter “QD < 2.0 || FS > 60.0 || MQ < 40.0 ″, -G_filter “GQ < 20”) and those exhibiting segregation distortion or sequencing errors were discarded.
Co-expression analysis
We performed co-expression analysis in T cells using Weighted Gene Co-Expression Network Analysis (WGCNA).31 To attenuate the noise effects and outliers, we constructed a pseudocell model by randomly selecting the 10 cells as a group. Then we used the FindVariableFeatures function to identify the top 2000 hypervariable genes based on the expressions for further analysis. An adjacency matrix, representing an “unsigned” gene network, was built setting the soft power parameter “powerEstimate”, calculated from the pickSoftThreshold function. Modules were identified with the dynamic tree cutting algorithm using the function cutreeDynamic. In addition, we determined the correlations between all modules and subpopulations and identified the modules of interest. The internal connectivity of each module is calculated according to the pairwise weighted correlation of genes.
Statistical analysis
Wilcoxon Matched-Pairs Signed-Ranks test, Paired and Unpaired Student's t-test, and Kruskal–Wallis test were used as described in Figure Legends. P values < 0.05 were considered statistically significant (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001). Statistical analyses for scRNA-seq, WES and GSEA were performed by the R packages DESeq2 and clusterProfiler, respectively. Adjusted P values were reported from DESeq2, and FDR was reported from clusterProfiler.
Role of funders
The study funders were not in involved in the study design, data collection, analysis, interpretation or writing of the manuscript.
Results
scRNA-seq of PBMCs composition in a SLE family pedigree
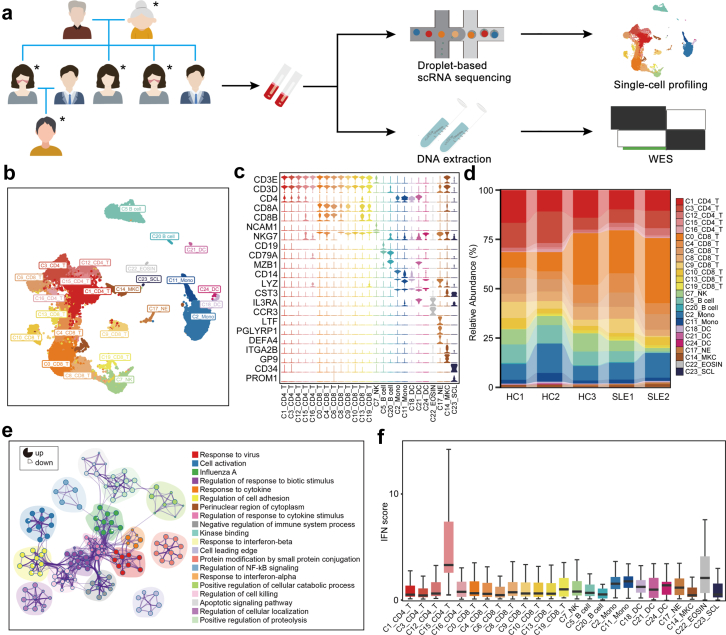
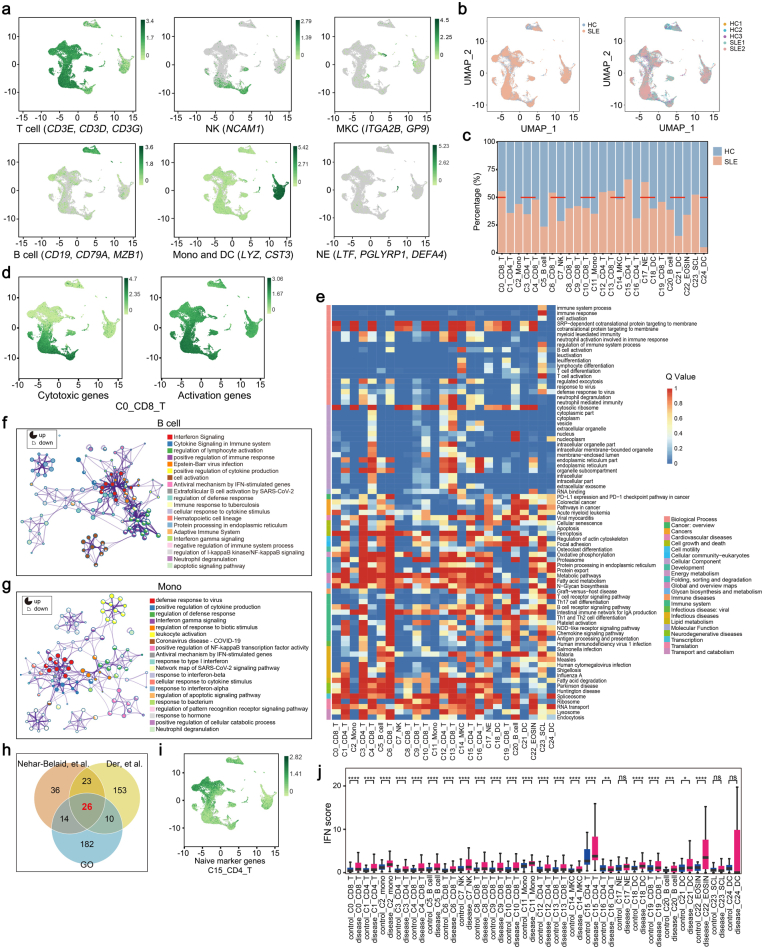
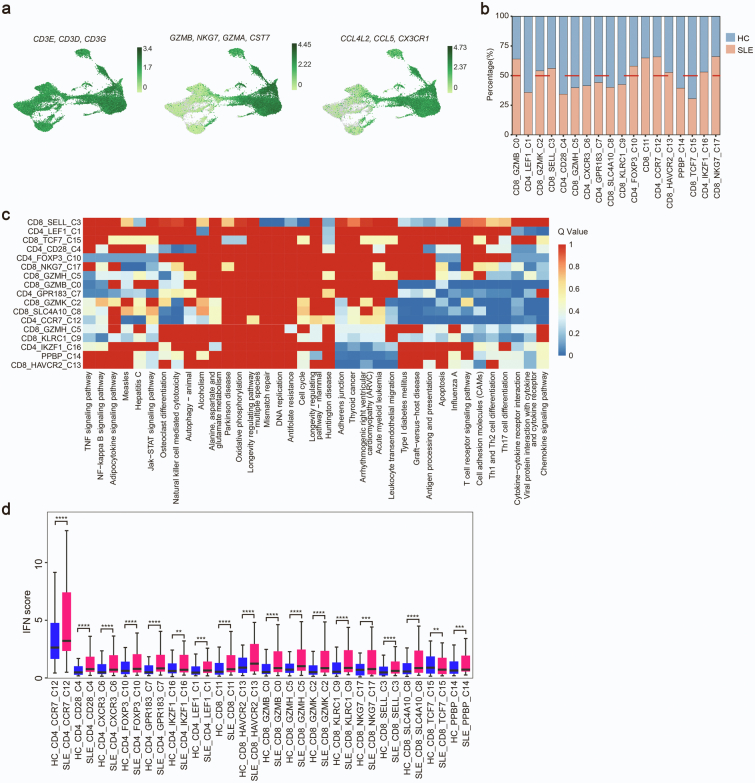
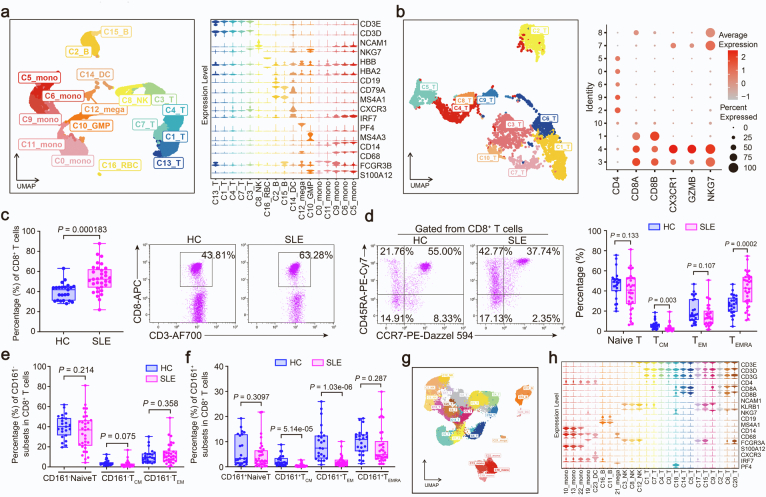
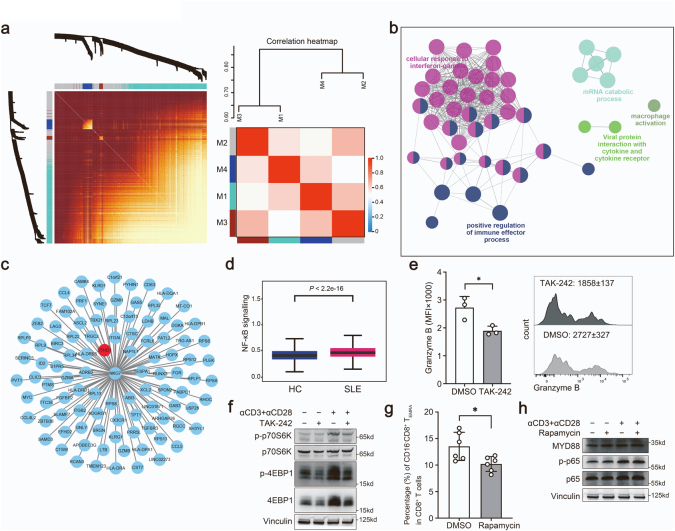
To investigate the genetic associations linked to cell-type specific alterations in SLE, we performed scRNA-seq and WES of PBMCs from a SLE family pedigree, including two SLE patients and three healthy controls (HCs) (Fig. 1a). After quality control of the single-cell transcriptomes, we resolved 25 molecularly distinct PBMC clusters based on highly expressed genes compared to those in other clusters, which could be assigned into 9 cell types (Fig. 1b and c). There were two clusters of monocytes, three clusters of dendritic cells (DCs), neutrophils, eosinophils, megakaryocytes (MKCs), natural killer (NK) cells, two clusters of B cells, five clusters of CD4+ T cells, eight clusters of CD8+ T cells and stem cell-like cells (Fig. 1b and c, Supplementary Fig. S1a).
Fig. 1.
PBMCs profiling of a SLE family pedigree at the single-cell resolution. (a) Schematic map of experimental strategy. Individuals with butterfly erythema on their faces represent SLE patients, others represent HCs. The samples marked with ∗ are used to perform scRNA-seq and WES. (b) UMAP and clustering of 47,349 cells from 5 samples. (c) Violin plots depicting clusters are defined by a set of known marker genes. Heights denote average expression levels; widths denote cell densities. (d) Sankey plot representing cell abundance of each cluster (n = 25) across the 5 individuals (3 HCs and 2 SLE samples). (e) Network of enriched terms by the differentially expressed genes in T cells: colored by cluster ID. The proportion of the color in the circle represents the proportion of up-regulated genes in the term, and the proportion of the blank represents the proportion of down-regulated genes. (f) IFN scores across the clusters.
Each cluster contained cells from different samples (including SLE and HC samples), indicating its generalizability among individuals (Supplementary Fig. S1b). In addition, the frequencies of cell clusters in SLE individuals displayed pronounced difference from those in HCs (Fig. 1d). The following clusters were over-represented in patients with SLE: C0/C6/C13_CD8+ T cells, C12/C15_CD4+ T cells, C17_Neu. Subsets that were under-represented in SLE patients included: C1/C3/C16_CD4+ T cells, C5_B cells, C7_NK cells, C11_monocytes, C21/C24_DCs (Supplementary Fig. S1c). Of these 25 clusters, C0_CD8+ T cluster, expressing high levels of cytotoxic genes, and high levels of T cell activation genes (Supplementary Fig. S1d), significantly expanded in SLE patients (Fig. 1d). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed the highly expressed genes in C0_CD8+ T cluster were enriched in “T cell activation” and “response to virus” (Supplementary Fig. S1e), indicating a positive role of this cell subset in pathogenesis of SLE. In addition, PBMCs from SLE samples and HCs showed substantial differentially expressed genes (DEGs). Relative to HCs, the increased genes of T cell clusters showed an enrichment in “Response to virus”, “Cell activation”, “Response to IFNβ” in SLE samples (Fig. 1e). And upregulated genes in B cells from SLE patients were enriched in “Interferon signaling”, “Regulation of lymphocyte activation” (Supplementary Fig. S1f), accompanied with monocyte-expressed genes enriched in “Defense response to virus”, “IFNγ signaling” (Supplementary Fig. S1g).
Evident studies have emphasized the importance of IFN signatures in the pathogenesis of SLE.13,32,33 We next analyzed the cell-type specific expression of IFN-related genes (IRGs) in all 25 clusters (Fig. 1f and Supplementary Fig. S1h). We found that C15_CD4 T cells displayed the highest IFN signature scores (Fig. 1f). This cluster, expressing high levels of CD4+ naïve T cells marker genes (CD27, SELL, CCR7, TCF7) (Supplementary Fig. S1i), is related to “response to virus” and “T cell differentiation” (Supplementary Fig. S1e). Importantly, most of IRGs in these clusters from SLE patients were upregulated compared to those in HCs, especially in T cell clusters (Supplementary Fig. S1j). Consistent with previous reports,13,34 our finding highlighted the importance of IFN pathway in pathogenesis of SLE. Taken together, our scRNA-seq analysis demonstrated that SLE is heterogeneous with a distinct immune cell landscape.
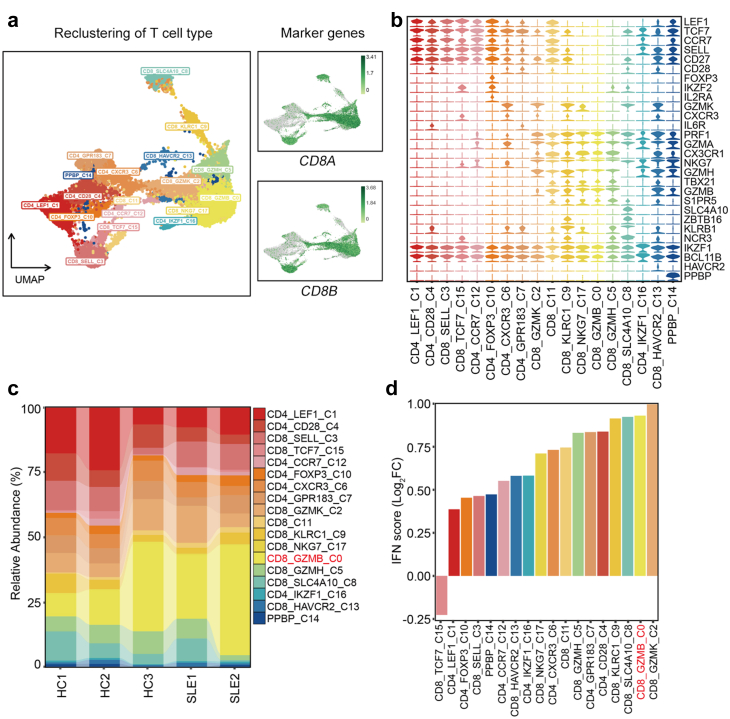
A distinctive T lymphocyte subset in SLE
Given that T cells showed most distinct proportion differences between HC and SLE samples across the 25 clusters, we further performed a secondary clustering in T cells which were re-clustered into 18 subclusters (Fig. 2a). 5 subclusters including CD4_LEF1, CD4_CD28, CD4_CCR7, CD8_SELL, CD8_TCF7 represented naïve T cells by highly expressing naïve T cell markers CCR7, SELL (CD62L), LEF1, TCF7, CD27. CD4_FOXP3 was a regulatory T (Treg) subpopulation. CD4_IKZF1 could be a T follicular helper (Tfh)-like cell type by expressing IKZF1 and BCL11B. CD4_CXCR3, CD4_GPR183 and CD8_GZMK showed features of CD4+ or CD8+ TEM cells with high expression of GZMK, CXCR3, IL6R and lacking expression of CCR7. CD8_KLRC1 was a “NK-like” T subset with both high expression of T cell receptors (TRDV2, TRDC, TRGV9) and NK cell receptors (KLRC1, KLRB1, KLRG1). CD8_SLC4A10 was a population of mucosal-associated invariant T (MAIT) cells with high expression of SLC4A10, ZBTB16, KLRB1, NCR3. CD8_NKG7, CD8_GZMB and CD8_GZMH could be categorized into CD8+ TEMRA cell subpopulations with high expression of CX3CR1, PRF1, GZMH, TBX21 and lacking expression of CCR7. CD8_HAVCR2 displayed exhaustion characteristic with high level of HAVCR2 (Fig. 2b).
Fig. 2.
Transcriptional features of T cell subclusters. (a) UMAP of T cells which are reclustered into 18 subclusters. (b) T cell subclusters are defined by a set of known marker genes. (c) Sankey plot representing the cell abundance of each T cell subcluster (n = 18) across the 5 individuals (3 HCs and 2 SLE samples). (d) Alterations of IFN scores in each T cell subcluster.
Intriguingly, among CD8+ T cells, the CD8_GZMB subpopulation representing a high-cytotoxic subtype by expressing cytotoxic genes (GZMB, NKG7, GZMA, CST7) and chemokines (CCL4L2, CCL5, CX3CR1) and it displayed a markedly increased percentage in SLE subjects as compared to HCs (Fig. 2c, Supplementary Fig. S2a, S2b and Table S1). The marker genes in this subset mainly were enriched in “Influenza A” and autoimmune diseases like rheumatoid arthritis (RA) and SLE (Supplementary Fig. S2c and Table S2). In addition, IFN scores in most CD8+ T cell subpopulations from SLE patients were significantly higher than those in HCs (Supplementary Fig. S2d), of which CD8_GZMB ranked the second place (Fig. 2d). Together, the alterations in frequencies of different CD8+ lymphocyte subpopulations are closely related to the development of autoimmune diseases.
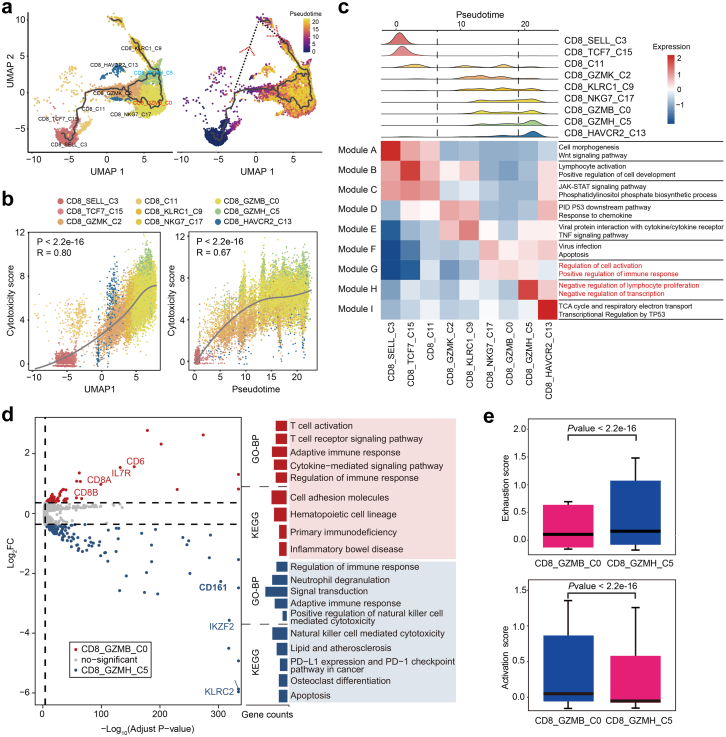
CD161−CD8+ TEMRA cell subtype is highly cytotoxic and activated
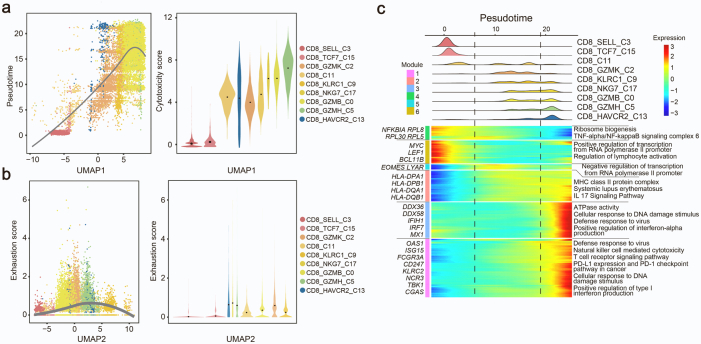
We next assessed the dynamic states and cell transitions in CD8+ T cells by inferring the state trajectories using Monocle3. After removing MAIT cells with distinct TCR characteristics in our pseudotemporal analysis, we revealed that trajectory path initiated from naïve-like CD8_SELL cells, through an intermediate state featured by CD8_GZMK cells and finally reached two separate terminal lineages, one was CD8_GZMH cells and the other directed CD8_GZMB cells (Fig. 3a). In addition, we simulated another possible trajectory path based on the pseudotime inferred by Monocle2, which started with CD8_SELL, through the intermediate node CD8_KLRC1 cells, then reached the terminal state, CD8_GZMH (Fig. 3a). We also demonstrated that the cytotoxicity score was positively correlated with uniform manifold approximation and projection (UMAP1), as well as pseudotime (Fig. 3b and Supplementary Fig. S3a), which indicated the trajectory of CD8+ T cell was accompanied by an increase in cytotoxicity. Meanwhile, as for the two terminal lineages, CD8_GZMB cells held lower exhaustion score than CD8_GZMH as shown in Supplementary Fig. S3b.
Fig. 3.
Analysis of CD8+T cell subsets transition trajectory in SLE. (a) Pseudotemporal analysis of CD8+ T cell subsets. Trajectory of CD8+ T Cell subclusters is inferred using monocle3 and subclusters are marked by colors (left). Pseudotime-ordered variables are inferred using monocle2 (right). Dotted lines and arrows indicate inferred differentiation trajectory and direction. (b) Correlation between pseudo-time and cytotoxicity of CD8+ T cell subsets. The solid line represents loess fitting of the relationship between cytotoxicity scores and Monocle components. P values are calculated by Spearman correlation. (c) Distribution of CD8+ subpopulations during the transition, along with the pseudo-time (upper). Heatmap showing the co-expression modules with the highest average expression in each CD8+ T cell subcluster (lower). (d) Volcano plots (left) and GO/KEGG enrichment analysis (right) of differentially expressed genes between CD8_GZMB and CD8_GZMH (left). Red represents upregulation in CD8_GZMB, and blue represents upregulation in CD8_GZMH. (e) Boxplot indicating the average expression of exhaustion and activation gene signatures in CD8_GZMB and CD8_GZMH. The P values are from a Wilcoxon test. Loess, locally weighted regression.
We next assessed stage-associated genes for each lineage to further investigate the differences in CD8_GZMB cells and CD8_GZMH cells by Monocle3. The transcriptional alterations associated with different trajectory stages could be assigned into distinct co-expression modules (Fig. 3c). Notably, CD8_GZMB cells specifically expressed genes involved in “Regulation of cell activation” and “Positive regulation of immune response”, while CD8_GZMH cells expressed genes enriched in “Negative regulation of lymphocyte proliferation” and “Negative regulation of transcription” (Fig. 3c). In addition, DEGs along with pseudotemporal trajectory were annotated into six modules by Monole2 (Supplementary Fig. S3c). The majority of Modules 1, 2 and 3 genes dominated in the late stage of differentiation, involved in “Positive regulation of type I IFN production” and “Natural killer cell mediated cytotoxicity” (Supplementary Fig. S3c). This suggested cytotoxicity and IFN response were enhanced along with CD8+ T cell trajectory.
Since the two subsets of TEMRA cells (CD8_GZMB and CD8_GZMH) were at distinct stages of the trajectory, we sought to delineate their potential functional difference by analyzing their DEGs (Fig. 3d). The upregulated genes in CD8_GZMB cells including CD8A, CD8B, IL7R, CD6 were enriched in “T cell activation”, “T cell receptor signaling pathway” (Fig. 3d). In contrast, CD8_GZMH cells expressed some “NK cell signature genes” including CD161, KLRC2 and exhaustion-associated genes, IKZF2 and LAG3. These upregulated genes in CD8_GZMH subpopulation belonged to “Natural killer cell mediated cytotoxicity” and “PD-L1 and PD-1 checkpoint pathway”. Additionally, CD8_GZMB cells exhibited lower exhaustion score accompanied by higher activation score than CD8_GZMH cells (Fig. 3e), indicating their different roles in pathogenesis of SLE. In the DEGs, CD161 (also known as KLRB1) had been recently identified as a cell marker of tumor-infiltrating CD8+ T cells.29,30 Therefore, we renamed CD8_GZMB cells and CD8_GZMH cells as CD161−CD8+ TEMRA cells and CD161+CD8+ TEMRA cells, respectively. Altogether, we demonstrated that CD8+ T cells display a distinct heterogeneity in SLE, and CD161−CD8+ TEMRA cell could be a potential pathogenetic subpopulation for SLE.
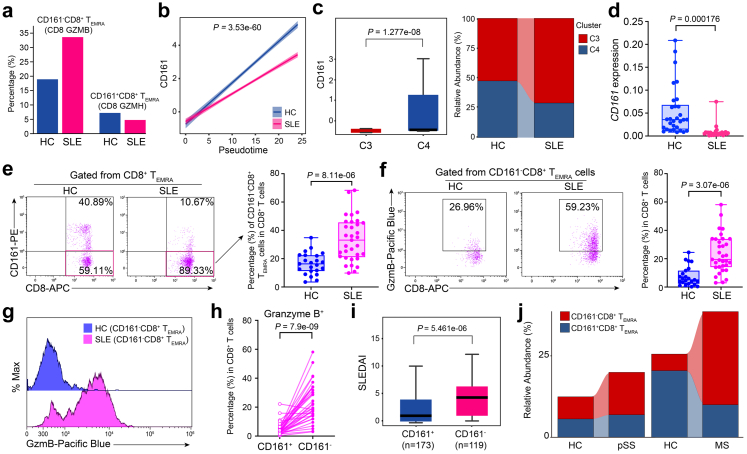
Cytotoxic CD161−CD8+ TEMRA cell subtype is expanded in autoimmune diseases
We then sought to explore the clinical significance of this newly identified CD8+ T subpopulation. Relative to HCs, the percentage of CD161−CD8+ TEMRA cells exhibited a pronounced expansion in SLE, whereas CD161+CD8+ TEMRA cells showed a decreased frequency in SLE (Fig. 4a). By integration CD161 expression with pseudotemporal analysis, we also revealed that the low expression of CD161 in SLE happened at the terminal state of CD8+ T cell trajectory (Fig. 4b), in agreement with the expression pattern of two separate terminal cell subsets, CD161+ and CD161− TEMRA (Fig. 3a). Consistently, we reanalyzed a public SLE scRNA-seq dataset,35 (Supplementary Fig. S4a and S4b) and confirmed that the frequency of CD161−CD8+ TEMRA cells (C3 cluster) significantly increased in patients with SLE (Fig. 4c). In addition, we validated that the expression of CD161 in PBMCs from patients with SLE was significantly lower than that from HCs in a new cohort (Fig. 4d). Intriguingly, CD8+ T cells from SLE patients exhibited a higher percentage of PBMCs than those from HCs in another cohort (Supplementary Fig. S4c), accompanied by increased CD8+ TEMRA cells (Gated with CCR7− and CD45RA+) (Supplementary Fig. S4d). Notably, the frequency of CD161−CD8+ TEMRA cells from SLE patients was much higher than HCs (Fig. 4e), while the frequencies of CD161−CD8+ Naïve T/TCM/TEM showed no difference between HCs and SLE patients (Supplementary Fig. S4e). In addition, the proportions of CD161+CD8+ TEMRA and CD161+CD8+ Naïve T cells were comparable in HCs and SLE patients, but CD161+CD8+ TCM and TEM significantly decreased in SLE subjects (Supplementary Fig. S4f). CD161−CD8+ TEMRA cells showed markedly increased abundance of granzyme B in SLE versus HCs (Fig. 4f and g). Besides, these cells produced more granzyme B than CD161+CD8+ TEMRA cells (Fig. 4h). By analysis of a public bulk RNA-seq profile from a SLE cohort with SLEDAI scores,36 we demonstrated that low expression of CD161 was closely correlated with high SLEDAI scores which represented more serve SLE pathological features (Fig. 4i). We further assessed the changes in CD161−CD8+ TEMRA cells frequency in other autoimmune diseases with usage of public single-cell transcriptome datasets. In primary Sjogren Syndrome (pSS),37 we performed mapping analysis and obtained a CD161−CD8+ TEMRA subpopulation (Supplementary Fig. S4g and S4h), which showed a pronounced increase as compared to HCs (Fig. 4j). Likewise, the frequency of this subpopulation was also elevated in multiple sclerosis (MS) samples (Fig. 4j).38 Taken together, CD161−CD8+ TEMRA cell is a pathogenic CD8+ T subset with high activity and cytotoxicity for the progression of autoimmune diseases.
Fig. 4.
Expansion of CD161−CD8+TEMRAcells in SLE. (a) Percentages of CD161−CD8+ TEMRA and CD161+CD8+ TEMRA cell subclusters in HCs and SLE samples. (b) Two-dimensional plots showing the changes of CD161 expression over pseudotime in HCs and SLE samples. P value is calculated by Spearman correlation. (c) CD161 expression in CD161−CD8+ TEMRA and CD161+CD8+ TEMRA cell subcluster (left), and relative abundance of these two subsets in HCs and SLE subjects from public dataset (GSE162577) (right). P value is calculated by Wilcoxon rank-sum test. (d) CD161 expression of PBMCs from a new case–control cohort (HCs, n = 30; SLEs, n = 25). P value is from Unpaired t-test (e and f) Flow cytometry (left) and quantification (right) of the frequency of CD161−CD8+ TEMRA cells (e) cytoplastic levels granzyme B of CD161−CD8+ TEMRA (f) from a validation cohort (HCs, n = 23; SLEs, n = 33). P value is from Unpaired t-test. (g) Representative histogram showing granzyme B expression in HC and SLE samples. (h) Comparison of granzyme B expression in CD161−CD8+ TEMRA and CD161+CD8+ TEMRA cells from SLE samples (n = 33). P value is from Paired t-test. (i) Boxplot showing SLEDAI scores of SLE patients with high levels of CD161 and low levels of CD161 from public dataset (GSE121239). P value is calculated by Wilcoxon rank-sum test (j) Sankey plot representing the relative abundance of CD161−CD8+ TEMRA cells and CD161+CD8+ TEMRA cells from case–control cohorts of pSS (left, GSE177278) and MS (right, GSE193770).
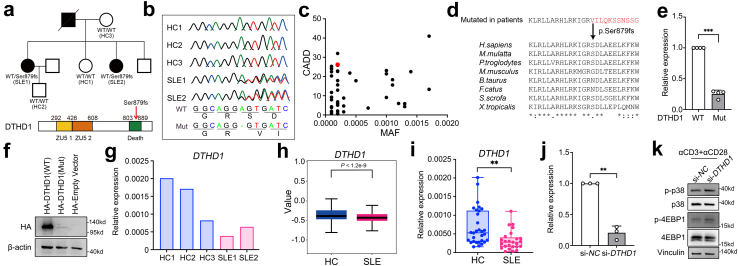
Loss-of-function mutation in DTHD1 increased activation of CD8 T cells
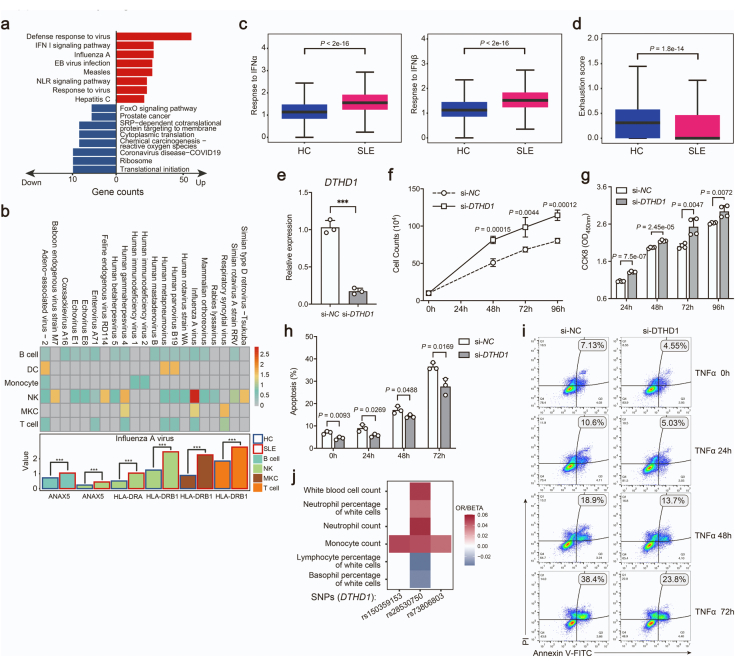
The DEGs of CD161−CD8+ TEMRA cells were enriched in “Type I interferon signaling pathway” and “Defense response to virus” (Supplementary Fig. S5a). Due to a causal link between viral infections and SLE,39 we analyzed the expressions of recognition receptors for viruses from both SLE and HCs. The genes (ANAX5, HLA-DRA, HLA-DRB1) related to recognition for influenza virus were induced in most immune cells, particularly in NK and T cells from SLE subjects (Supplementary Fig. S5b). Meanwhile, CD161−CD8+ TEMRA cells from SLE patients displayed increased response to IFNα and IFNβ (Supplementary Fig. S5c) and lower exhaustion score than HCs (Supplementary Fig. S5d). These observations suggested that T cells activation in SLE could result from chronic viral infection.
Given the genetic associations for SLE, we attempted to explore whether the cell-type specific activation linked with genetic alterations by analysis of WES data of this family. We identified a heterogeneous variant rs529758698 (chr4: 36343738A > −, NP_001130008.2: p. Ser897fs) in DTHD1, that caused frameshift mutation by 1 base pair (bp) deletion of serine897 (p.Ser897fs) in the death domain (DD) of protein encoded by DTHD1 (Fig. 5a). This mutation was further confirmed by Sanger sequencing (Fig. 5b). It is a rare variant (MAF = 0.0002 in gnomAD) with a combined annotation-dependent depletion (CADD) score >25 (Fig. 5c), indicating a potential functional harmfulness40 and is located in a conserved region between species (Fig. 5d) which was required for protein–protein interaction in signal transduction.41 To determine whether this mutation affected DTHD1 expression, we transfected N-terminal HA-tagged WT and mutant with pSer897fs DTHD1 into HEK293T cells and found the mRNA and protein levels were significantly suppressed by p. Ser897fs (Fig. 5e and f). Furthermore, a decreased expression of DTHD1 in PBMCs from SLE patients harboring this genetic mutation in this family pedigree was observed (Fig. 5g). Of note, DTHD1 expression was specifically decreased in CD161−CD8+ TEMRA cell subpopulation from SLE patients (Fig. 5h). Consistently, the expression of DTHD1 was also lower in SLE patients than that in HCs in another cohort (Fig. 5i). These results suggested this rare variation could result in a loss of function in DTHD1.
Fig. 5.
Loss of function of DTHD1 from SLE patients. (a) SLE family pedigree and schematic map showing mutation (Ser879fs) location within DTHD1 protein. (b) Sanger sequencing reads indicating variant location. (c) CADD scores versus MAF for the new Ser879fs patient-derived DTHD1 variant as compared with DTHD1 variants with an MAF cutoff of >10−4 from the gnomAD database. (d) Amino acid sequence alignment of DTHD1 death domain across species. The arrow indicates the position of the Ser589fs mutation (e and f) qPCR (e) and western blotting (f) analysis of DTHD1 expression in HEK293T cells transfected with HA-tagged WT DTHD1 or mutant DTHD1 expressing plasmid. P value is determined by Paired t-test, P = 0.000158. (g) qPCR analysis of DTHD1 expression in this SLE family pedigree. (h) Expression of DTHD1 in scRNA dataset between SLE samples and HCs. (i) qPCR analysis of DTHD1 expression in another cohort (HCs, n = 30; SLEs, n = 25). P value is determined by Unpaired t-test, P = 0.00178 (j) qPCR analysis of DTHD1 expression in primary CD8+ T cells electroporated with si-NC or si-DTHD1 siRNAs (50 nM). P value is determined by Paired t-test, P = 0.00608 (k) Western blotting analysis of p-p38, p38, p-4EBP1, 4EBP1 in primary CD8+ T cells electroporated with si-NC or si-DTHD1 siRNAs after treatment with αCD3 and αCD28. One representative experiment of three is shown (f and k). ∗∗P < 0.01, ∗∗∗P < 0.001.
We then sought to determine how the abnormal accumulation was affected by DTHD1 downregulation. Knockdown of DTHD1 in primary CD8+ T cells isolated from PBMCs of healthy subjects significantly promoted activation of proliferation signaling including p38 MAPK pathway and mTOR pathway (Fig. 5j and k). In addition, DTHD1 knockdown also significantly enhanced the tumor cell proliferation (Supplementary Fig. S5e–g) and increased the resistance to TNFα-induced apoptosis (Supplementary Fig. S5h and S5i). Therefore, dysregulation of DTHD1 could be beneficial to cell proliferation and apoptosis resistance. Meanwhile, by analysis of public GWAS data on SLE (https://www.ebi.ac.uk/gwas/), we found that mutations in DTHD1 were mainly correlated with the number of leukocytes (Supplementary Fig. S5j), further supporting its regulatory role in cell proliferation. Taken together, we identified a rare SLE-associated mutation in death domain of DTHD1, which is tightly correlated with cell proliferation and apoptosis resistance.
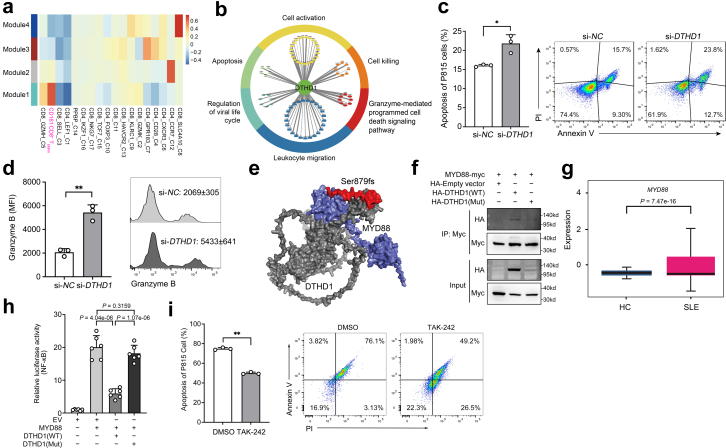
DTHD1 deficiency enhanced MYD88-dependent pathway to increase cytotoxicity of CD161−CD8+ TEMRA cells
To further illustrate the potential casual linkage between DTHD1 mutation and the cytotoxicity of CD161−CD8+ TEMRA cells, we performed single-cell co-expression analysis of our scRNA-seq dataset. Based on pseudo-cell of T cell transcriptome, the hypervariable genes were annotated into four modules (Fig. 6a and Supplementary Fig. S6a). The DTHD1 containing module 1 displayed significant correlation with CD161−CD8+ TEMRA cells (Fig. 6a). Meanwhile, this module was enriched in “cellular response to IFN-gamma” (Supplementary Fig. S6b). Genes co-expressed with DTHD1 included majority of cytotoxic genes, of which NKG7 as the hub gene in this module was a mediator of cytotoxic granule exocytosis and inflammation (Supplementary Fig. S6c). Consistently, the DTHD1-related genes indicated an enrichment in “cell activation”, “cytotoxicity”, “apoptosis” (Fig. 6b). These analyses suggested DTHD1 activity was strongly associated with the cytotoxicity of CD161−CD8+ TEMRA cells. Therefore, we determined the in vitro cytotoxic capacity of DTHD1-silenced human primary CD8+ T cells by coculturing with P815 cells. Compared to control CD8+ T cells, DTHD1-knockdown CD8+ T cells induced higher ratio of apoptosis of P815 cells (Fig. 6c). In addition, the level of Granzyme B from CD161−CD8+ T cells was increased with DTHD1 knockdown (Fig. 6d). These results indicated knockdown of DTHD1 could enhance cytotoxic capacity of CD8+ T cells.
Fig. 6.
DTHD1 deficiency increased the cytotoxicity of CD161−CD8+TEMRAcells. (a) Heatmap showing the correlation of co-expression modules with different cell subsets. (b) Functional annotations of DTHD1-related genes. (c) Quantification (left) and flow cytometry (right) of the apoptosis percentage of P815 cells after cocultured with primary CD8+ T cells electroporated with si-NC or si-DTHD1 siRNAs for 5 h at the ratio of 1:5. P value is determined by Unpaired t-test, P = 0.0124. (d) Flow cytometric analysis of mean fluorescence intensity (MFI) of Granzyme B of CD161−CD8+ T cells electroporated with si-NC or si-DTHD1 siRNAs after coculture with P815 cell. P value is calculated by Unpaired t-test, P = 0.0012. (e) Structural prediction of DTHD1-MYD88 interaction by ZDOCK. Grey, DTHD1; blue, MYD88; red, mutated regions in DTHD1. (f) Western blotting analysis of HA in HEK293T cells co-transfected with myc-MYD88 and HA-empty vector or HA-DTHD1 (WT) or HA-DTHD1 (Mutant) expressing plasmids after immunoprecipitated with anti-Myc beads. (g) Boxplot indicating the average expression of MYD88 in CD161−CD8+ TEMRA cells in HCs and SLE samples from our dataset. P value is from Wilcoxon rank-sum test. (h) Luciferase activity analysis of lysates of HEK293T cells co-transfected luciferase reporter plasmid for NF-κB, pRL-TK-renilla-luciferase plasmid, MYD88 plasmid and WT-DTHD1 plasmid or mutant DTHD1 plasmid (n = 6). P values are determined by Unpaired t-test. (i) Quantification (left) and flow cytometry (right) of the apoptosis percentage of P815 cells after cocultured with primary CD8+ T cells pre-treated with DMSO or TAK-242 for 5 h at the ratio of 1:5. P value is determined by Unpaired t-test, P = 0.0024. One representative experiment of three is shown (f). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
We next explored the molecular basis of DTHD1 in regulation of CD161−CD8+ TEMRA cells. Interaction analysis revealed that DTHD1 could interact with MYD88 though DD.42 Accordingly, we applied ZDOCK (http://zdock.umassmed.edu/) to mimic their interaction pattern and found that the two proteins would be tightly “stuck” together through DD (Fig. 6e). In addition, by using PPA-Pred (https://www.iitm.ac.in/bioinfo/PPA_Pred/) to evaluate the binding affinity of DTHD1 and DD-mutated DTHD1 to MYD88, we observed mutation in DD markedly reduced the affinity (binding free energy: −20.77 kcal/mol - > −16.15 kcal/mol). By co-immunoprecipitation, we further discovered that WT DTHD1 rather than mutant DTHD1 did interact with MYD88 (Fig. 6f). We speculated that DTHD1 regulated the proliferation and cytotoxicity of CD161−CD8+ TEMRA cells by targeting MYD88-mediated signaling. By examining MYD88 expression in scRNA-seq data, we found that MYD88 expression was markedly elevated in CD161−CD8+ TEMRA cells from SLE patients (Fig. 6g). NF-κB is a core TF downstream of MYD88-induced signaling. Overexpression of WT DTHD1 but not mutant DTHD1 significantly decreased MYD88-induced NF-κB activation (Fig. 6h). Consistently, the NF-κB-activated signaling in CD161−CD8+ TEMRA cells was significantly upregulated in SLE samples (Supplementary Fig. S6d). Furthermore, chemical inhibition of MYD88 activity with TAK-242 in primary human CD8+ T cells efficiently decreased the cytotoxicity against P815 cells (Fig. 6i) and the granzyme B level (Supplementary Fig. S6e). In addition, inhibiting MYD88 by TAK-242 markedly suppressed TCR-induced mTOR activation which is responsible for T cell proliferation (Supplementary Fig. S6f), and inhibition of mTOR with Rapamycin significantly decreased the frequency of CD161−CD8+ TEMRA in vitro but did not impair MYD88-mediated pathway (Supplementary Fig. S6g and 6h). Thus, mTOR activation could be regulated by MYD88 activity and then promoted the expansion of CD8+ TEMRA cells. Taken together, our study demonstrated that mutation in DTHD1 could cause the abnormal expansion and activity of cytotoxic CD161−CD8+ TEMRA subpopulation by regulation of MYD88-mediated pathway, which is essential for the pathogenesis of SLE.
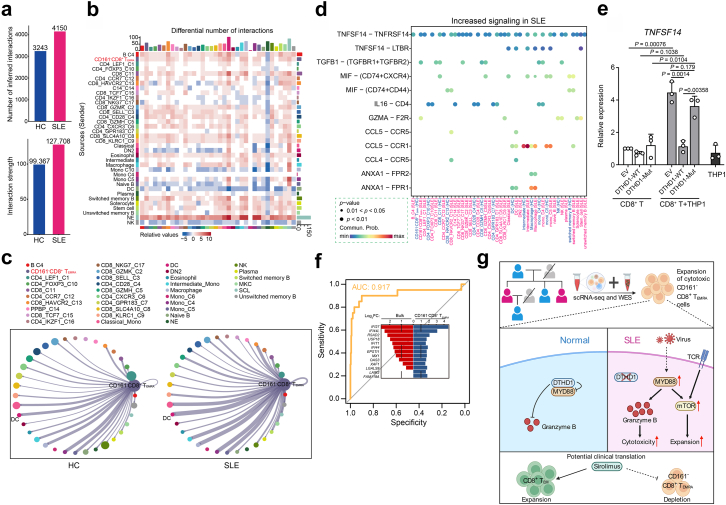
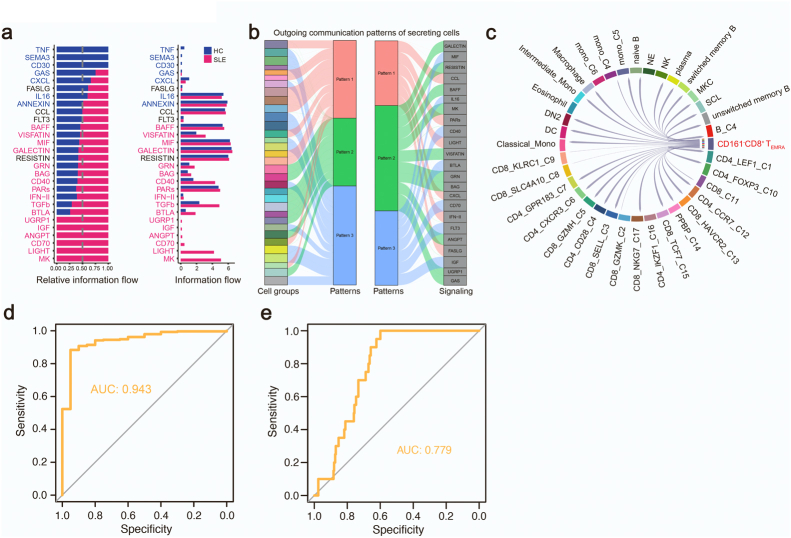
CD161−CD8+ TEMRA cells contribute to SLE by LIGHT signaling
We then examined how the activated CD161−CD8+ TEMRA cells contributed to SLE by analyzing cell-to-cell interaction with CellChat.27 Compared to HCs, the strength of cell-to-cell interaction globally increased in SLE subjects (Fig. 7a and b). Of all cellular communications, the interaction between CD161−CD8+ TEMRA cells and DCs increased most in SLE samples (Fig. 7c). Next, we accessed the key signals mainly accounting for the cellular communication networks, and found that TNFSF14 (LIGHT), MK and CD70 pathways were upregulated in SLE patients (Supplementary Fig. S7a). We also identified disease-associated communication patterns including of outgoing signals. The major pathways of CD161−CD8+TEMRA cells were LIGHT and IFN-II pathways, characterized by pattern 1 (Supplementary Fig. S7b). Particularly, the signaling “TNFSF14-TNFRSF14” was greatly induced in CD161−CD8+ TEMRA cells, which was involved in the interaction with other T, B, and DC cell subsets (Fig. 7d). By coculturing primary CD8+ T cells with THP1 cells, we validated that TNFSF14 expression was significantly induced compared to its expression in CD8+ T cells or THP1 cells (Fig. 7e). Importantly, overexpression of WT DTHD1 but not mutant DTHD1 in CD8+ T cells could efficiently inhibit TNFSF14 expression (Fig. 7e). LIGHT-triggered signaling has been previously reported to be responsible for the activation of T, B cells,43 and the DC maturation.44 We further demonstrated that LIGHT pathway outgoing from CD161−CD8+ TEMRA cells was activated in SLE patients but absent in HCs (Supplementary Fig. S7c). Therefore, we suggest that CD161−CD8+ TEMRA cells could be activated by dysregulation of DTHD1 and interact with other immune cells via LIGHT signaling, leading systemic inflammation.
Fig. 7.
CD161−CD8+TEMRAcells contribute to SLE by LIGHT signaling. (a) The number (top) and strength (bottom) of interaction among all cells in HCs and SLE samples. (b) Heatmap of differential interactions between HCs and SLE samples in cell–cell communication network. The top bar indicates the sum of incoming signaling and right bar indicates the sum of outgoing signaling. Red indicates increased signaling and blue indicates decreased signaling in SLE. (c) Number of significant ligand-receptor pairs between CD161−CD8+ TEMRA cells (outgoing) and other cell subclusters (incoming) in HCs (left) and SLEs (right). The relative number of ligand-receptor pairs is represented by the edge width. (d) Comparison of the significant outgoing signaling from CD161−CD8+ TEMRA cells between HC and SLE. Empty space means the communication probability is zero. P-values are computed from one-sided permutation test. ∗∗∗P < 0.001. (e) qPCR analysis of TNFSF14 expression in primary CD8+ T cells electroporated with empty vector, DTHD1-WT or DTHD-mut plasmid cocultured with THP1 cells (n = 3). P values are determined by unpaired t-test. (f) Receiver operating curve for out-of-sample prediction of case–control state by a logistic regression model trained on DEGs in CD161−CD8+ TEMRA cells. DEGs include IFI27, IFI44L, RSAD2, IFI44, FAM118A, LGALS9, MX1, EPSTI1, USP18, OAS3, LAIR2, IFIT1, XAF1. Inset depicts the changes of DEGs in the public transcriptome profile and CD161−CD8+ TEMRA cells. (g) Graphic abstract showing the expansion of CD161−CD8+ TEMRA in patients with SLE and DTHD1 downregulation promotes MYD88-mediated expansion and cytotoxicity of this pathogenic CD161−CD8+ TEMRA subset in SLE.
Finally, the extensive heterogeneity of SLE made it difficult to give a precise diagnose, meanwhile, clinical characteristics from SLE patients exhibited weak correlation with known gene modules.34 Therefore, we assessed whether CD161−CD8+ TEMRA cell subtype could work as a new diagnostic marker for SLE. By using a public transcriptome profile for molecular prediction,36,45 we found the DEGs in this case–control profile were consistent with gene expression features from CD161−CD8+ TEMRA cells (P < 2.2e-16), and the upregulated genes in this subset from SLE samples displayed an efficient prediction for case–control status (AUC = 0.917) (Fig. 7f). In addition, due to the importance of DTHD1 and MYD88 in regulation of CD161−CD8+ TEMRA cells activation and cytotoxicity, we applied the expression of gene set including CD3D, CD8A, CD161, GZMB, DTHD1, MYD88 and TNFSF14 as the molecular prediction. Intriguingly, these key genes expression also showed a precise prediction for case–control status of SLE (AUC = 0.943) (Supplementary Fig. S7d and Table S3). However, using the DEGs of all cell subsets from SLE samples as clinical diagnosis for individuals only showed weak predictive power for clinical characteristics (Supplementary Fig. S7e). This observation indicated that CD161−CD8+ TEMRA cell could work as a new cellular marker for clinical diagnosis.
In conclusion, our study has identified a new highly activated and cytotoxic CD8+ T cell subset (CD161−CD8+ TEMRA cell) which is expanded in patients with SLE. We further revealed that dysregulation of DTHD1 is required for increasing cytotoxicity of CD161−CD8+ TEMRA cell by enhancing MYD88-dependent pathway (Fig. 7g). Our study offers new therapeutic opportunities for diagnosis and treatment for SLE.
Discussion
SLE is a debilitating autoimmune disease with involvement of abnormal activation of multiple immune cells. The heterogeneous manifestations of SLE hamper the development of diagnose and treatment to this disease. Therefore, more comprehensive molecular and cellular characterizations are urgently needed to be investigated. In this study, we integrated scRNA-seq and WES data of PBMCs from a SLE family pedigree, and demonstrated disease-associated changes in immune cells composition and annotated a new genetic variant with effects on cell-type specific transcription differences. Particularly, we identified a new CD8+ T cell subset named CD161−CD8+ TEMRA cell as an important contributor to the pathogenesis of SLE. In addition, we annotated a rare functional variant in DTHD1 for SLE, which affected the its expression and promoted the abnormal accumulation of cytotoxic CD161−CD8+ TEMRA cells by mediating MYD88-depedent pathway.
Although the abnormalities of B cell and CD4+ T cell are well identified cellular characteristics of SLE,6 the function of CD8+ T cell in SLE remains poorly elucidated. We demonstrated that CD8+ T lymphocytes of PBMCs are strikingly heterogeneous. Notably, one of cytotoxic CD8+ T subpopulations, CD161−CD8+ TEMRA cell, is remarkably expanded in SLE patients, and the activation of this subpopulation is closely associated with viral infection. Recently, the heterogeneity of CD8+ T cells and subpopulation-associated role have also been revealed in other diseases. In aging, a hyperinflammatory GZMK+CD8+ T subpopulation was elevated in aged individuals, which could be a conserved cellular hallmark of inflammaging.46 Additionally, the frequency of CD8+ TEMRA-like cells is clonally expanded in cerebrospinal fluid in Alzheimer's disease, indicating a key contribution of adaptive immunity in Alzheimer's disease.47 In the joint synovial tissues of RA patients, a CD8+ subpopulation with high expression of GZMK, GZMB and GNLY was uncovered to be a key mediator of RA pathogenesis.48 These observations suggest that CD8+ T cells display high phenotypic and functional diversity in health and disease. In active SLE patients, CD4+ TEMRA cells were previously revealed to increase in active SLE patients.49,50 CD8+ TCM cells in active SLE could promote Th2 cells differentiation in a cell–cell contact manner.51 Furthermore, two independent scRNA-seq analyses reported expansions of cytotoxic CD8+ T cell subsets in adult SLE34 and in childhood SLE.13 However, the clonally expanded cytotoxic GZMH+CD8+ T cells still displayed transcriptional heterogeneity with increased both cytotoxic and exhaustion signatures in SLE subjects.34 Therefore, the heterogeneity and disease-specific role of CD8+ T cells remain to be further investigated. Our study identified a new CD161−CD8+ TEMRA cell subtype with an increased frequency in PBMCs from SLE. Compared to GZMH+CD8+ T cells,34 CD161−CD8+ TEMRA cells represent a transcriptionally homogenous population of terminally differentiated effector cells with high level of cytotoxicity. Collectively, these results demonstrate that the expansion of cytotoxic CD8+ T cell subset is a critical contributor to the SLE pathogenesis. In addition, low expression of CD161 is closely correlated with high SLEDAI scores, the expression of DEGs in CD161−CD8+ TEMRA cell shows an efficient prediction for case–control status (AUC = 0.917). Therefore, the expansion of cytotoxic CD161−CD8+ TEMRA cells could function as a cellular marker for SLE diagnosis.
CD161, encoding a C-type lectin-like inhibitory receptor, is a classic NK signature gene.52 Previous studies have shown that CD161 is downregulated in patients with SLE.53,54 Moreover, CD161 was recently discovered to be also expressed by CD8+ T cells. Unlike the inhibitory role in NK cells, CD161 holds the opposite functions in regulation of CD8+ T cells activity.55 It works as a coactivator to promote IFN-γ production of CD8+ T cells against pathogens but inhibits the cytokine secretion of MAIT cells in response to bacterial infection.55 More recently, it has been identified that tumor-infiltrating CD161+CD8+ T cells are more exhausted and less cytotoxic than CD161−CD8+ T lymphocytes.29,30 Consistent with these findings, we demonstrated that CD161+CD8+ TEMRA cells exhibited high exhaustion, while CD161−CD8+ TEMRA cells highly expressed genes with cytotoxicity and activation. Overall, these observations suggest that CD161 is directly related to the activity of certain CD8+ T subpopulations. Although the ligand of CD161 is well identified, the CD161-triggered intracellular signaling pathway and the underlying mechanisms of regulating the expression and activation of CD161 in CD8+ T cells need further investigation.
The genetic basis of SLE requires fully investigated. Although over hundreds of risk loci associated with SLE have been identified by GWAS, rare variants with higher impacts are also important to fill in the puzzle of etiology. By WES analysis in the SLE family, we identified a frameshift mutation in DTHD1 which is highly related to SLE. Currently, the role of DTHD1 remains poorly understood. In our study, we confirmed that DTHD1 was significantly downregulated in the PBMCs of SLE patients, and showed low expression in CD161−CD8+ TEMRA cells. We also revealed the molecular basis for loss-of-function DTHD1 in promoting activation and cytotoxicity of CD161−CD8+ TEMRA cells. DTHD1 could directly interact with MYD88 and prevent activation of MYD88-dependent pathway. Therefore, low expression or inactivation of DTHD1 would release its inhibition of MYD88, in turn activating NF-κB signaling. Given the critical role of DTHD1 in mediating MYD88-dependent pathway, it would be interesting to explore the physiological and pathological function of DTHD1 in MYD88-associated inflammatory diseases.
mTOR pathway can integrate the metabolic cues and microenvironmental alterations to regulate T cell development, proliferation, activation and differentiation.56,57 Its abnormal activation is related with T cell dysregulation of SLE patients.58 Notably, a recent clinical trial revealed that mTOR inhibitor sirolimus showed promising effects in SLE patients.59 In this trial, the reduction of CD8+CD45RO+ memory T cells and expansion of CD8+CD45RA+ T cells were found in SLE patients at baseline, and a 12-month treatment with sirolimus could corrected the decreased CD8+ effector memory T cells.59 In line with this important finding, our data also showed a significant reduction of CD8+CD45RA− memory T cells (TCM and TEM) as well as an increase of CD8+CD45RA+ T cells in SLE patients (Supplementary Fig. S4d). And we further demonstrated that the expansion of CD8+CD45RA+ T cells mainly was derived from CD161−CD8+ TEMRA cells (Fig. 4f, Supplementary Fig. S4e–f), indicating its pathogenic role in SLE development. CD161−CD8+ TEMRA cells expansion depended on the activation of MYD88-mediated pathway, which increased the cytotoxicity of this subset and promoted accumulation by upregulating mTOR pathway. Therefore, mTOR activation was also important for the expansion of pathogenic CD8+ TEMRA subset in addition to depletion of CD8+ effector memory T cells. PI3K/Akt pathway is a major upstream signaling for mTOR activation in regulation of T cell function.60 It is reported CD8+ TEMRA proliferation depended on IL-15-induced MAPK pathway and PI3K/Akt pathway in kidney-transplant recipients (KTs).61 Moreover, the frequency of CD8+ TEMRA from KTs treated with sirolimus was significantly lower than that from KTs without receiving the mTOR inhibitor.62 Taken together, mTOR could promote expansion of CD8+ TEMRA cells which differs from its role in CD8+ TEM formation. However, the precise mechanism of mTOR in regulating expansion of CD161−CD8+ TEMRA needs further investigation.
In conclusion, by integrating the scRNA-seq transcriptomic profiles and WES data from a SLE family pedigree, we demonstrate that DTHD1 dysfunction-mediated expansion of cytotoxic CD161−CD8+ TEMRA cell subpopulation contributes to the pathogenesis of SLE. Our study highlights the importance of rare variants and CD8+ T cell subpopulation in the etiology of SLE, providing new insights into potential cellular marker and therapeutic target for SLE.
Contributors
K. C. and Q. G. designed and supervised the research. H. X., N. K., J. J., Y. X., X. L., F. Y., Z. X., D. Z., Z. Z., M. X., J. C., G. T., Q. F. performed the experiments. M. C., Y. Y., C. H. performed scRNA-seq and WES analysis. H. X., M. C., and N. K. made the Figures, K. C. and Q. G. analyzed the data and wrote the manuscript. All authors read, verified the data, and approved the final version of the manuscript. K. C. and Q. G. were responsible for the decision to submit the manuscript.
Data sharing statement
The scRNA-seq data and WES data had been deposited and uploaded in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG, https://bigd.big.ac.cn/gsa-human/), Chinese Academy of Sciences, with GSA accession No. HRA002481 for scRNA-seq data and No. HRA002479 for WES data. The data for this study are available by contacting the corresponding author upon reasonable request.
Declaration of interests
The authors declare that they have no competing interests.
Acknowledgments
The work was financially supported by Shanghai Rising-Star Program (21QA1407600), Young Elite Scientists Sponsorship Program by CAST (2020QNRC001), National Natural Science Foundation of China (81801577, 81602762, 81573044), Fundamental Research Funds for the Central Universities (22120210051), Natural Science Foundation of Guangdong Province, China (2023A1515030139). We thank Ms. Saixiang Zheng and Jinfeng Li for the technical help for flow cytometric analysis.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104507.
Contributor Information
Qing Guo, Email: guoqingzsy@163.com.
Kun Chen, Email: chenk@tongji.edu.cn.
Appendix A. Supplementary data
Supplementary Fig. S1.
Supplementary Fig. S2.
Supplementary Fig. S3.
Supplementary Fig. S4.
Supplementary Fig. S5.
Supplementary Fig. S6.
Supplementary Fig. S7.
Full Western Blots.
References
- 1.Tsokos G.C. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Dorner T., Furie R. Novel paradigms in systemic lupus erythematosus. Lancet. 2019;393(10188):2344–2358. doi: 10.1016/S0140-6736(19)30546-X. [DOI] [PubMed] [Google Scholar]
- 3.Kaul A., Gordon C., Crow M.K., et al. Systemic lupus erythematosus. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.39. [DOI] [PubMed] [Google Scholar]
- 4.Chaussabel D., Quinn C., Shen J., et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29(1):150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiche L., Jourde-Chiche N., Whalen E., et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis Rheumatol. 2014;66(6):1583–1595. doi: 10.1002/art.38628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moulton V.R., Suarez-Fueyo A., Meidan E., Li H., Mizui M., Tsokos G.C. Pathogenesis of human systemic lupus erythematosus: a cellular perspective. Trends Mol Med. 2017;23(7):615–635. doi: 10.1016/j.molmed.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sallusto F., Lenig D., Forster R., Lipp M., Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 8.Lanzavecchia A., Sallusto F. Dynamics of T lymphocyte responses: intermediates, effectors, and memory cells. Science. 2000;290(5489):92–97. doi: 10.1126/science.290.5489.92. [DOI] [PubMed] [Google Scholar]
- 9.Sallusto F., Geginat J., Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 10.McKinney E.F., Lyons P.A., Carr E.J., et al. A CD8+ T cell transcription signature predicts prognosis in autoimmune disease. Nat Med. 2010;16(5):586–591. doi: 10.1038/nm.2130. 581pp. following 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKinney E.F., Lee J.C., Jayne D.R., Lyons P.A., Smith K.G. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523(7562):612–616. doi: 10.1038/nature14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papalexi E., Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 13.Nehar-Belaid D., Hong S., Marches R., et al. Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nat Immunol. 2020;21(9):1094–1106. doi: 10.1038/s41590-020-0743-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harley I.T., Kaufman K.M., Langefeld C.D., Harley J.B., Kelly J.A. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10(5):285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omarjee O., Picard C., Frachette C., et al. Monogenic lupus: dissecting heterogeneity. Autoimmun Rev. 2019;18(10) doi: 10.1016/j.autrev.2019.102361. [DOI] [PubMed] [Google Scholar]
- 16.Langefeld C.D., Ainsworth H.C., Cunninghame Graham D.S., et al. Transancestral mapping and genetic load in systemic lupus erythematosus. Nat Commun. 2017;8 doi: 10.1038/ncomms16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y.F., Zhang Y., Lin Z., et al. Identification of 38 novel loci for systemic lupus erythematosus and genetic heterogeneity between ancestral groups. Nat Commun. 2021;12(1):772. doi: 10.1038/s41467-021-21049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S., Abecasis G.R., Boehnke M., Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95(1):5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuk O., Schaffner S.F., Samocha K., et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A. 2014;111(4):E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taft J., Markson M., Legarda D., et al. Human TBK1 deficiency leads to autoinflammation driven by TNF-induced cell death. Cell. 2021;184(17):4447–4463.e20. doi: 10.1016/j.cell.2021.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aringer M., Costenbader K., Daikh D., et al. 2019 European League against rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Arthritis Rheumatol. 2019;71(9):1400–1412. doi: 10.1002/art.40930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lun A.T.L., Riesenfeld S., Andrews T., Dao T.P., Gomes T. Participants in the 1st Human Cell Atlas J, et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019;20(1):63. doi: 10.1186/s13059-019-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu X., Mao Q., Tang Y., et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao J., Spielmann M., Qiu X., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aran D., Looney A.P., Liu L., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin S., Guerrero-Juarez C.F., Zhang L., et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Der E., Suryawanshi H., Morozov P., et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol. 2019;20(7):915–927. doi: 10.1038/s41590-019-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathewson N.D., Ashenberg O., Tirosh I., et al. Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell. 2021;184(5):1281–1298.e6. doi: 10.1016/j.cell.2021.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y., Wu L., Zhong Y., et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184(2):404–421.e6. doi: 10.1016/j.cell.2020.11.041. [DOI] [PubMed] [Google Scholar]
- 31.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganguly D. Do type I interferons link systemic autoimmunities and metabolic Syndrome in a pathogenetic continuum? Trends Immunol. 2018;39(1):28–43. doi: 10.1016/j.it.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Banchereau R., Hong S., Cantarel B., et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell. 2016;165(6):1548–1550. doi: 10.1016/j.cell.2016.05.057. [DOI] [PubMed] [Google Scholar]
- 34.Perez R.K., Gordon M.G., Subramaniam M., et al. Single-cell RNA-seq reveals cell type-specific molecular and genetic associations to lupus. Science. 2022;376(6589) doi: 10.1126/science.abf1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng Y., Zheng Y., Li D., et al. Expression characteristics of interferon-stimulated genes and possible regulatory mechanisms in lupus patients using transcriptomics analyses. eBioMedicine. 2021;70 doi: 10.1016/j.ebiom.2021.103477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petri M., Fu W., Ranger A., et al. Association between changes in gene signatures expression and disease activity among patients with systemic lupus erythematosus. BMC Med Genomics. 2019;12(1):4. doi: 10.1186/s12920-018-0468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong X., Meng S., Tang D., et al. Single-cell RNA sequencing reveals the expansion of cytotoxic CD4(+) T lymphocytes and a landscape of immune cells in primary sjogren's Syndrome. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.594658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., Zaslavsky M., Su Y., et al. KIR(+)CD8(+) T cells suppress pathogenic T cells and are active in autoimmune diseases and COVID-19. Science. 2022;376(6590) doi: 10.1126/science.abi9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quaglia M., Merlotti G., De Andrea M., Borgogna C., Cantaluppi V. Viral infections and systemic lupus erythematosus: new players in an old story. Viruses. 2021;13(2):277. doi: 10.3390/v13020277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kersse K., Verspurten J., Vanden Berghe T., Vandenabeele P. The death-fold superfamily of homotypic interaction motifs. Trends Biochem Sci. 2011;36(10):541–552. doi: 10.1016/j.tibs.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Zhou W., Kaneko N., Nakagita T., Takeda H., Masumoto J. A comprehensive interaction study provides a potential domain interaction network of human death domain superfamily proteins. Cell Death Differ. 2021;28(11):2991–3008. doi: 10.1038/s41418-021-00796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ware C.F., Croft M., Neil G.A. Realigning the LIGHT signaling network to control dysregulated inflammation. J Exp Med. 2022;219(7) doi: 10.1084/jem.20220236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holmes T.D., Wilson E.B., Black E.V., et al. Licensed human natural killer cells aid dendritic cell maturation via TNFSF14/LIGHT. Proc Natl Acad Sci U S A. 2014;111(52):E5688–E5696. doi: 10.1073/pnas.1411072112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toro-Dominguez D., Martorell-Marugan J., Goldman D., Petri M., Carmona-Saez P., Alarcon-Riquelme M.E. Stratification of systemic lupus erythematosus patients into three groups of disease activity progression according to longitudinal gene expression. Arthritis Rheumatol. 2018;70(12):2025–2035. doi: 10.1002/art.40653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mogilenko D.A., Shpynov O., Andhey P.S., et al. Comprehensive profiling of an aging immune system reveals clonal GZMK(+) CD8(+) T cells as conserved hallmark of inflammaging. Immunity. 2021;54(1):99–115.e2. doi: 10.1016/j.immuni.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Gate D., Saligrama N., Leventhal O., et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer's disease. Nature. 2020;577(7790):399–404. doi: 10.1038/s41586-019-1895-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F., Wei K., Slowikowski K., et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. 2019;20(7):928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piantoni S., Regola F., Zanola A., et al. Effector T-cells are expanded in systemic lupus erythematosus patients with high disease activity and damage indexes. Lupus. 2018;27(1):143–149. doi: 10.1177/0961203317722848. [DOI] [PubMed] [Google Scholar]
- 50.Fritsch R.D., Shen X., Illei G.G., et al. Abnormal differentiation of memory T cells in systemic lupus erythematosus. Arthritis Rheum. 2006;54(7):2184–2197. doi: 10.1002/art.21943. [DOI] [PubMed] [Google Scholar]
- 51.Sen Y., Chunsong H., Baojun H., et al. Aberration of CCR7 CD8 memory T cells from patients with systemic lupus erythematosus: an inducer of T helper type 2 bias of CD4 T cells. Immunology. 2004;112(2):274–289. doi: 10.1111/j.1365-2567.2004.01862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartel Y., Bauer B., Steinle A. Modulation of NK cell function by genetically coupled C-type lectin-like receptor/ligand pairs encoded in the human natural killer gene complex. Front Immunol. 2013;4:362. doi: 10.3389/fimmu.2013.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin Y.L., Lin S.C. Analysis of the CD161-expressing cell quantities and CD161 expression levels in peripheral blood natural killer and T cells of systemic lupus erythematosus patients. Clin Exp Med. 2017;17(1):101–109. doi: 10.1007/s10238-015-0402-1. [DOI] [PubMed] [Google Scholar]
- 54.Park Y., Lim J., Kim S.Y., Kwon G.C., Koo S.H., Kim J. Changes of frequency and expression level of CD161 in CD8(+) T cells and natural killer T cells in peripheral blood of patients with systemic lupus erythematosus. Microbiol Immunol. 2020;64(7):532–539. doi: 10.1111/1348-0421.12798. [DOI] [PubMed] [Google Scholar]
- 55.Konduri V., Oyewole-Said D., Vazquez-Perez J., et al. CD8(+)CD161(+) T-cells: cytotoxic memory cells with high therapeutic potential. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.613204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perl A. Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases. Nat Rev Rheumatol. 2016;12(3):169–182. doi: 10.1038/nrrheum.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng H., Chi H. mTOR signaling in the differentiation and function of regulatory and effector T cells. Curr Opin Immunol. 2017;46:103–111. doi: 10.1016/j.coi.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caza T., Wijewardena C., Al-Rabadi L., Perl A. Cell type-specific mechanistic target of rapamycin-dependent distortion of autophagy pathways in lupus nephritis. Transl Res. 2022;245:55–81. doi: 10.1016/j.trsl.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lai Z.W., Kelly R., Winans T., et al. Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: a single-arm, open-label, phase 1/2 trial. Lancet. 2018;391(10126):1186–1196. doi: 10.1016/S0140-6736(18)30485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12(5):325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tilly G., Doan-Ngoc T.M., Yap M., et al. IL-15 harnesses pro-inflammatory function of TEMRA CD8 in kidney-transplant recipients. Front Immunol. 2017;8:778. doi: 10.3389/fimmu.2017.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doan Ngoc T.M., Tilly G., Danger R., et al. Effector memory-expressing CD45RA (TEMRA) CD8(+) T cells from kidney transplant recipients exhibit enhanced purinergic P2X4 receptor-dependent proinflammatory and migratory responses. J Am Soc Nephrol. 2022;33(12):2211–2231. doi: 10.1681/ASN.2022030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.