Abstract
Purpose:
BRAF V600E mutant metastatic colorectal cancer represents a significant clinical problem, with combination approaches being developed clinically with oral BRAF inhibitors combined with EGFR-targeting antibodies. While compelling preclinical data have highlighted the effectiveness of combination therapy with vemurafenib and small-molecule EGFR inhibitors, gefitinib or erlotinib, in colorectal cancer, this therapeutic strategy has not been investigated in clinical studies.
Patients and Methods:
We conducted a phase Ib/II dose-escalation/expansion trial investigating the safety/efficacy of the BRAF inhibitor vemurafenib and EGFR inhibitor erlotinib.
Results:
Thirty-two patients with BRAF V600E positive metastatic colorectal cancer (mCRC) and 7 patients with other cancers were enrolled. No dose-limiting toxicities were observed in escalation, with vemurafenib 960 mg twice daily with erlotinib 150 mg daily selected as the recommended phase II dose. Among 31 evaluable patients with mCRC and 7 with other cancers, overall response rates were 32% [10/31, 16% (5/31) confirmed] and 43% (3/7), respectively, with clinical benefit rates of 65% and 100%. Early ctDNA dynamics were predictive of treatment efficacy, and serial ctDNA monitoring revealed distinct patterns of convergent genomic evolution associated with acquired treatment resistance, with frequent emergence of MAPK pathway alterations, including polyclonal KRAS, NRAS, and MAP2K1 mutations, and MET amplification.
Conclusions:
The Erlotinib and Vemurafenib In Combination Trial study demonstrated a safe and novel combination of two oral inhibitors targeting BRAF and EGFR. The dynamic assessment of serial ctDNA was a useful measure of underlying genomic changes in response to this combination and in understanding potential mechanisms of resistance.
Translational Relevance.
This trial demonstrates that the novel combination of vemurafenib and erlotinib is well tolerated, with promising activity in BRAF V600E colorectal cancer and other tumor types. Our findings highlight that ctDNA analysis can reveal important insights into mechanisms of treatment resistance and provides an early biomarker of treatment response.
Introduction
BRAF mutations are established oncogenic drivers in a wide range of malignancies and are present in 10% to 15% of colorectal cancer (1, 2). More than 95% of mutations occur as a V600E change (valine to glutamic acid substitution), and are mutually exclusive with RAS mutations (3, 4). Clinically, BRAF V600E mutation in colorectal cancer is more prevalent in females and patients with advanced age, and is associated with a right-sided primary, high tumor grade, and microsatellite instability (MSI; refs. 5–7). Despite significant progress in the management of colorectal cancer, the prognosis for patients with BRAF V600E positive metastatic colorectal cancer (mCRC) remains poor, relative to patients with BRAF wild-type tumors, with a median overall survival (OS) of approximately 11 months (8–11). Retrospective analyses of first-line chemotherapy trials have highlighted the modest response to standard therapy in this population (12–15), with minimal benefit from subsequent lines of treatment as demonstrated by short progression-free survival (PFS) and OS (16, 17).
Single-agent BRAF inhibitors have demonstrated a lack of efficacy in BRAF V600E mutant mCRC, with response rates around 5% (18–20). Preclinical models revealed that intrinsic resistance to single-agent therapy is mediated by rapid feedback activation of EGFR signaling, which leads to continued proliferation in the presence of BRAF inhibition. Indeed, these studies also demonstrated synergy between BRAF (vemurafenib or dabrafenib) and EGFR (erlotinib, gefitinib or cetuximab) inhibitors, leading to effective suppression of MAPK pathway signaling and tumor regression in BRAF V600E colorectal cancer xenografts (21, 22). A pilot study evaluating the efficacy of combined vemurafenib and panitumumab (EGFR mAb) demonstrated disease regression in 64% of patients with mCRC who progressed after first-line standard therapy (23). However, although the combination therapy markedly inhibited MAPK signaling, the degree of inhibition was variable, potentially reflecting intratumoral heterogeneity (24). A phase Ib study of vemurafenib in combination of irinotecan plus cetuximab demonstrated a response rate of 35% in patients with refractory, advanced BRAF V600E positive solid tumors (25). Subsequently, the SWOG S1406 study randomized 99 patients with BRAF-mutant pretreated mCRC to receive irinotecan plus cetuximab with or without vemurafenib. The study showed a response rate of 16% in the triplet arm versus 4% in the doublet arm (26). Since then, several studies testing the triplet combination of selective BRAF small molecule inhibitors, MEK inhibitors, and EGFR targeting mAbs have reported improved response rates of up to 26%, implying that these combination strategies are capable of blocking feedback reactivation, producing a more robust inhibition of MAPK signaling (27, 28). Recently, the phase III BEACON trial evaluating the combination of encorafenib (BRAF inhibitor) and cetuximab with or without binimetinib (MEK inhibitor) in the second- or third-line setting was the first to demonstrate a survival benefit with this approach in patients with BRAF positive mCRC (28), leading to the approval of combination encorafenib and cetuximab as the new standard of care in this setting.
Despite an improvement in response and survival with this combination therapy, the vast majority of patients with BRAF mutant mCRC still derive minimal benefit from treatment. As such, there is a current need for biomarkers early in the treatment course to predict outcomes and understand mechanisms of intrinsic and acquired treatment resistance. Circulating tumor DNA (ctDNA) dynamics have been shown to be an early predictor of treatment response for patients with colorectal cancer receiving chemotherapy, potentially allowing an early switch in patients where treatment is ineffective (29). Functional imaging with [18F]flurodeoxyglucose PET (FDG-PET) has also been shown to be a useful marker of early biologic response to vemurafenib in patients with BRAF mutant melanoma and may be used to monitor response and detect early progression on therapy (30).
Vemurafenib was the first BRAF inhibitor to be approved for use in the treatment of BRAF V600 mutant melanoma, and has also demonstrated antitumor activity in other cancer types with a BRAF V600E mutation such as non-small cell lung cancer (NSCLC; ref. 31), thyroid cancer, breast cancer, and ovarian cancer (32). Although preclinical studies have highlighted the effectiveness of combination therapy with vemurafenib and small-molecule EGFR inhibitors, gefitinib or erlotinib in colorectal cancer, this therapeutic strategy has not been investigated in clinical studies (21). On the basis of this, we conducted a phase Ib/II trial that assessed the safety, tolerability, and antitumor activity of combination vemurafenib and erlotinib in patients with BRAF V600E positive mCRC and other cancers. In addition, early FDG-PET scans and serial ctDNA analysis were performed to assess their utility for predicting response and understanding mechanisms of resistance to combination therapy.
Patients and Methods
Study design and objectives
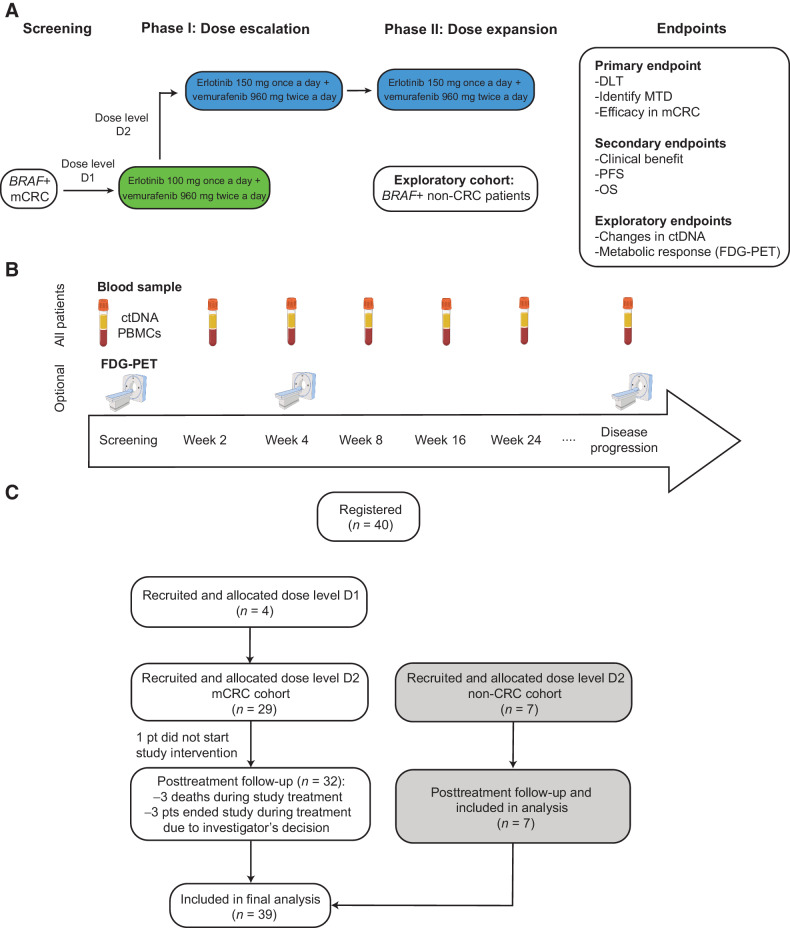
The EViCT clinical trial was a phase I/II, multicentre, open-label study of vemurafenib in combination with erlotinib in patients with BRAF V600E positive mCRC, and other cancers. Patients were recruited from five tertiary centers in Melbourne, Australia, from July 8, 2014 to August 1, 2017. The cut-off date for data analysis for this publication was December 16, 2020. The primary objective of the phase I lead-in study was to define the safety of vemurafenib in combination with erlotinib by determining the DLTs in the first 4 weeks of treatment and identify the MTD and RP2D. Secondary objectives included overall response rates of the combination treatment as defined by CR or PR within the first 24 weeks of treatment, the CBR as defined by CR, PR, or SD for more than 16 weeks of treatment, and to determine the PFS and OS. Exploratory objectives included analysis of early changes in ctDNA levels and correlation with response to BRAF and EGFR inhibition. Written informed consent was obtained from all patients. Institutional ethics approval was obtained (HREC13/MH 446) and the study was conducted in accordance with the Declaration of Helsinki. Study data were collected and managed by the Centre for Biostatistics & Clinical Trials (BACT) at the Peter MacCallum Cancer Centre (Protocol No. 2014.019 version 2.0 dated March 05, 2014, Appendix 1).
Study population
Patients with histologically confirmed mCRC with a BRAF V600E mutation were included in an initial lead-in phase (phase I) and treated at different dose levels in a 3+3 dose escalation trial design. Subsequent patients were recruited into phase II dose expansion cohorts, with a non-colorectal cancer expansion cohort enrolled in parallel in a single stage (Fig. 1A and C). Eligibility criteria included ECOG performance status of 0 to 1, evaluable disease as defined by RECIST v1.1, life expectancy of >3 months, and adequate end-organ function. Patients with mCRC must not have received >2 lines of therapy in the metastatic setting. Exclusion criteria included uncontrolled brain metastases, clinically significant cardiac history, and presence of an EGFR mutation in patients with NSCLC.
Figure 1.
Design of the EViCT clinical trial. A, Clinical trial design and CONSORT flow diagram. Patients with BRAF V600E positive mCRC were treated with vemurafenib and erlotinib (in escalating doses) in a 3 + 3 phase 1 design. Once the RP2D was determined, an additional 28 patients were enrolled in the colorectal cancer dose expansion cohort and 7 patients in the exploratory non-colorectal cancer dose expansion cohort. B, Study overview and samples for translational research. C, Clinical trial consort diagram with final number of patients recruited and analyzed in dose escalation phase, dose expansion colorectal cancer cohort and dose expansion non-colorectal cancer cohort.
Study treatment
The starting dose for the combination study drugs was 960 mg twice a day for vemurafenib and 100 mg once daily for erlotinib (dose level D1), escalating to a maximum planned dose of 960 mg twice a day for vemurafenib and 150 mg once daily for erlotinib (dose level D2; Fig. 1A). A series of cohorts of 3 patients were treated with both agents and observed for a minimum of 4 weeks. The MTD was defined as the highest dose at which <33% of patients experience a DLT during the DLT-evaluable period. Once dose level D2 was reached, the combination doses were determined as the RP2D in discussion with the study Safety Monitoring Committee, considering all available toxicity information. Additional patients were then recruited to receive the RP2D dose in the dose-expansion phase of the study.
Patients received oral vemurafenib taken together with erlotinib in continuous 28-day cycles and treatment was continued until disease progression, unacceptable toxicity, death, or withdrawal of informed consent. Dose modifications according to the study protocol were permitted in the event of drug-related toxicity.
Safety
Patients in the dose-escalation phase were observed for the presence of DLTs during the first 4 weeks of treatment. A DLT was defined as any of the following: prolonged QTc, diarrhea, gastrointestinal perforation, increase in total bilirubin or hepatic transaminase (ALT or AST), Steven–Johnson syndrome, and febrile neutropenia that was at least grade 3 in severity as defined by common terminology criteria for adverse events (CTCAE) v4.03. In addition, grade 4 hematologic toxicity (neutropenia, thrombocytopenia, anemia) and toxic epidermal necrolysis were also considered DLTs. Safety assessments were conducted at baseline, weekly during the DLT evaluation period, and every 4 weeks thereafter.
Efficacy assessment
Tumor response was evaluated locally based on RECIST v1.1 by CT scan, which was performed at screening and every 8 weeks after starting study treatment until disease progression. The best overall response was defined as the best response recorded from the start of treatment until disease progression. Objective response was considered confirmed if the response was maintained at a subsequent scheduled CT assessment, at least 4 weeks after the criteria for response were first met.
FDG-PET assessment
FDG-PET scan was performed at screening, week 4, and at progression for patients in the mCRC cohort, but was optional for patients in the non-CRC cohort. FDG-PET/CT was performed on combined PET-CT scanners using low-dose, non-contrast-enhanced CT for attenuation correction and anatomical correlation, encompassing the vertex of the skull to mid-thigh. All patients were fasted for at least 6 hours and had a blood sugar level of <10 mmol/L at the time of imaging. FDG was administered intravenously according to weight and images were acquired after a distribution time of at least 60 minutes. All FDG-avid disease was contoured using an SUV threshold that was applied to the whole body using MIM (MIM software). The SUV threshold used was patient specific as per the PERCIST (33) recommendations and defined as 1.5 times the mean SUV of the liver plus 2 SDs. All normal physiologic uptake (brain, kidneys, bladder) was manually excluded from the whole-body volume of interest. Parameters evaluated included the SUVMax of uptake to five target lesions, SUVMax for all lesions, and MTV.
Blood collection and processing for ctDNA
Blood samples were collected in EDTA for ctDNA analysis at screening, weeks 2, 4, 8, and every 8 weeks thereafter, and at the time of disease progression (Fig. 1B). Whole blood was first centrifuged at 1,600 × g for 10 minutes to separate the plasma from the peripheral blood cells, followed by a further centrifugation step at 20,000 × g for 10 minutes to pellet any remaining cells and/or debris. The plasma was then stored at −80°C until DNA extraction. DNA was extracted from 2 mL aliquots of plasma using the QIAmp Circulating Nucleic Acid Kit (Qiagen) according to manufacturer's instructions. The DNA was eluted into 50 μL buffed AVE (Qiagen) and stored at −20°C.
Targeted sequencing
Targeted capture-based sequencing of pretreatment (baseline) and progression plasma DNA was performed using the Avenio ctDNA Analysis Expanded Kit (Roche diagnostics) following manufacturer's protocols. On the basis of the previously published cancer, personalized profiling by deep-sequencing (CAPP-seq) methodology, this covers a pan cancer panel of 77 genes optimized for use in colorectal cancer and NSCLC (34). Between 6 and 10 ng of genomic DNA were used for library construction and the purified libraries were pooled and sequenced on an Illumina NextSeq 500 (illumina). Variants were called using a specialized bioinformatic analysis workflow, which uses integrated digital error suppression (iDES) system that augments CAPP-seq through in silico removal of stereotypical sequencing artifacts combined with molecular barcoding (35). Only nonsynonymous single nucleotide variants (SNV), insertions-deletions (Indels), copy-number variations (CNV), and gene fusions were extracted for analysis.
Digital PCR
Droplet digital PCR (ddPCR) analysis was performed using the Bio-Rad Droplet Digital PCR system following manufacturer's protocols. Allele-specific PCR assay to specifically detect and quantify the fractional abundance of the BRAF V600E mutation and corresponding wild-type allele was commercially obtained (Bio-Rad Laboratories). For mutant-based assays, ddPCR reactions were 25 μL aqueous volumes that contained final concentrations of 1× ddPCR supermix for probes (without dUTP; Bio-Rad), 0.9 μmol/L each primer, 0.25 μmol/L probe, and 5 μL of genomic DNA. The thermal cycling profile was 95°C: for 10 minutes, followed by 55 cycles of 95°C for 15 seconds and annealing for one minute at 55°C. Each sample was analyzed by at least two technical replicates with 5 μL DNA input per well. A Poisson correction was applied to determine the number of amplifiable molecules, which was used to further derive the number of copies of DNA carrying a particular mutation per milliliter of plasma. Data analysis was carried out using the QuantaSoft Software, version 1.7 (Bio-Rad). ctDNA was defined as detectable if there was ≥1 copy of mutant DNA detected in both duplicate reactions.
Statistical methods
The phase Ib lead-in study followed a standard 3+3 design, with one dose escalation and one dose de-escalation if required. A maximum of 6 evaluable patients would be enrolled at each dose level. The dose expansion phase for the mCRC patient cohort used a Simon's optimal two-stage design. The sample size for the mCRC expansion cohort was a maximum of 24 patients, of whom the first 6 patients would be treated in the lead-in phase. The first stage of the expansion phase required at least 1 of 9 evaluable patients (including the 6 patients treated in the lead-in phase) to achieve a CR or PR within the first 24 weeks of treatment to proceed to the second stage. The second stage accrued an additional 15 evaluable patients, and the combination of vemurafenib and erlotinib was considered active at the end of the second stage if 3 or more patients achieved a CR or PR within the first 24 weeks of treatment. These calculations were based on a significance level of 0.10 and power of 0.80.
Patient characteristics, treatment details, and AEs were summarized using descriptive statistics. Safety analyses included all enrolled patients who fulfilled eligibility criteria, received one dose of the study treatment, and were DLT-evaluable. Efficacy and biomarker analyses included all enrolled patients who fulfilled eligibility criteria, received at least one dose of the study treatment, and had at least one post-baseline efficacy assessment. The response rate and CBR were estimated with 95% confidence intervals (95% CI) calculated based on binomial distribution. Time-to-event endpoints (PFS, OS) were described using Kaplan–Meier methods with 95% CIs. For biomarker analyses, nonparametric Wilcoxon's matched-pairs signed-rank test and Kruskal–Wallis test were used to examine the differences between continuous variables, and Fisher exact test was used to compare associations between categorical variables. Linear regression analysis was used for correlations of continuous data. Log-rank test was used to assess differences between subgroups. All analyses were performed using R version 3.6.3 and GraphPad PRISM version 9.1.2 (RRID:SCR_002798), where P values <0.05 were considered significant.
Data availability
The data generated in this study are available upon request from the corresponding author.
Results
Patient characteristics
A total of 40 patients were enrolled across the dose escalation and dose expansion phases (mCRC n = 33 and non-CRC cohort n = 7) between July 2014 and August 2017 (Fig. 1A). The non-colorectal cancer cohort included 7 patients with selected cancers [NSCLC, ovarian cancer, breast cancer, and gastrointestinal stromal tumors (GIST)] that were BRAF V600E mutation positive. Demographics and baseline characteristics of patients are presented in Table 1. Patients with mCRC had received either one (n = 15) or 2 (n = 17) prior lines of therapy. Serial FDG-PET imaging and blood sample collection for biomarker analyses are detailed in Fig. 1B. One patient in the mCRC expansion cohort experienced an adverse event the day prior to commencing study treatment and did not receive treatment, hence was not included in any of the outcome analyses (Fig. 1C).
Table 1.
Patient demographics and baseline characteristics.
| mCRC cohort | Non-CRC cohort | Overall | |
|---|---|---|---|
| Characteristics | n = 32 (%) | n = 7 (%) | n = 39 (%) |
| Median age (range) | 61.6 | 64.0 | 61.8 |
| (26.6–79.3) | (35.8–69.5) | (26.6–79.3) | |
| Gender | |||
| Female | 18 (56) | 5 (71) | 23 (59) |
| Male | 14 (44) | 2 (29) | 16 (41) |
| ECOG | |||
| 0 | 19 (59) | 3 (43) | 22 (56) |
| 1 | 13 (41) | 4 (57) | 17 (44) |
| Non-CRC subtype | |||
| Breast cancer | 0 (0) | 1 (14) | 1 (14) |
| Malignant GIST | 0 (0) | 1 (14) | 1 (14) |
| Metastatic ovarian LGSC | 0 (0) | 1 (14) | 1 (14) |
| Advanced/metastatic NSCLC | 0 (0) | 4 (57) | 4 (57) |
| Prior lines of metastatic therapy | |||
| 1 | 15 (47) | 1 (14) | 16 (41) |
| 2 | 17 (53) | 2 (29) | 19 (49) |
| 3 | 0 (0) | 1 (14) | 1 (3) |
| Unknown | 0 (0) | 3 (43) | 3 (7) |
| Prior EGFR therapy | |||
| Yes | 6 (19) | 0 (0) | 6 (15) |
| No | 26 (81) | 7 (100) | 33 (85) |
Abbreviation: LGSC, low-grade serous cancer.
Dose determination and safety
The dose escalation (phase Ib) component of the study enrolled patients with mCRC in a 3 + 3 dose escalation trial design (Fig. 1A). No dose-limiting toxicities (DLT) were observed at dose level D1 (n = 4: erlotinib 100 mg once a day and vemurafenib 960 mg twice a day) and D2 (n = 6: vemurafenib 960 mg twice a day and erlotinib 150 mg once a day), utilizing the full dose of each agent. The MTD and recommended phase II dose (RP2D) was thus determined to be vemurafenib 960 mg twice a day and erlotinib 150 mg once a day. The expansion phase enrolled a further 22 patients with mCRC, along with an exploratory cohort of 7 patients with other cancers (Fig. 1A and C); all treated at the RP2D. This resulted in 32 evaluable patients enrolled with mCRC; 4 at dose level D1 and 28 patients at the RP2D. Safety analyses have included all patients who received study drug (n = 39) across both the escalation and expansion cohorts (Fig. 1C). Median time on treatment was 6.7 months (range, 0.7–44.1) in 39 patients.
Investigator-assessed, treatment-related adverse events (AE) which occurred in ≥10% of patients are summarized in Table 2. The most frequently reported AEs (any grade) across all treated patients were diarrhea (77%), fatigue (59%), nausea (51%), acneiform rash (49%), and photosensitivity (36%). The most common grade 3/4 treatment-related AEs were diarrhea (26%) and acneiform rash (8%). There were no treatment-related deaths.
Table 2.
Summary of any grade treatment-related AEs reported in at least 10% of patients.
| All patients (n = 39) n (%) | |||
|---|---|---|---|
| Adverse events | Grade 1–2 | Grades 3–4 | Total |
| Diarrhea | 21 (54) | 9 (23) | 30 (77) |
| Fatigue | 21 (54) | 2 (5) | 23 (59) |
| Nausea | 18 (46) | 1 (3) | 19 (49) |
| Acneiform rash | 16 (41) | 3 (8) | 19 (49) |
| Photosensitivity | 13 (33) | 1 (3) | 14 (36) |
| Maculopapular rash | 11 (28) | 2 (5) | 13 (33) |
| Arthralgia | 13 (33) | 0 (0) | 13 (33) |
| Vomiting | 9 (23) | 2 (5) | 11 (28) |
| Fever | 7 (18) | 1 (3) | 8 (21) |
| AST elevation | 7 (18) | 1 (3) | 8 (21) |
| ALT elevation | 6 (15) | 1 (3) | 7 (18) |
| Pruritus | 4 (10) | 1 (3) | 5 (13) |
| Mucositis oral | 4 (10) | 1 (3) | 5 (13) |
| Anemia | 3 (8) | 1 (3) | 4 (10) |
| ALP elevation | 3 (8) | 1 (3) | 4 (10) |
Abbreviations: ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma-glutamyltransferase.
Dose reductions were required for vemurafenib in 3 of 4 patients (75%) in the dose escalation cohort. Among the 35 patients in the dose expansion cohort, 21 (60%) required a dose reduction of vemurafenib, and 18 (51%) of erlotinib. Treatment was discontinued in 33 of 39 patients (85%) due to disease progression, 3 (8%) due to investigator's decision/clinical progression, 1 patient (3%) was lost to follow up, and 2 (5%) due to study termination. The latter 2 patients had NSCLC with an ongoing response at the time of study termination and transitioned to an expanded access program. No patients discontinued treatment due to AEs.
Antitumor activity
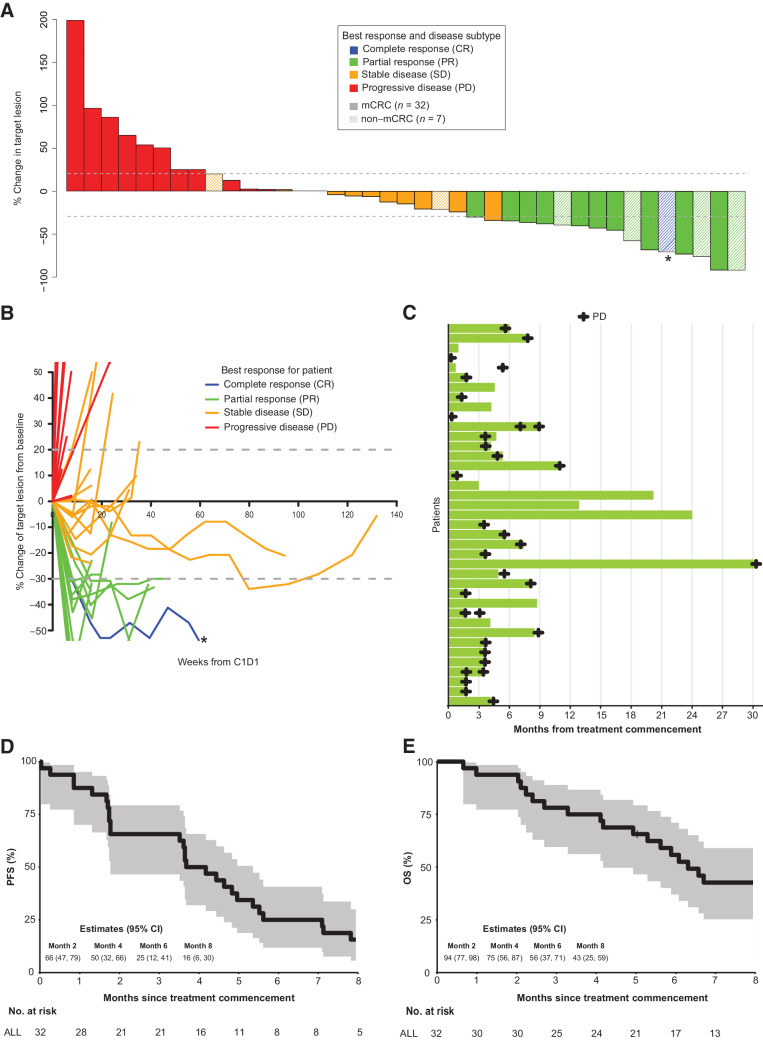
Tumor response as assessed by RECIST v1.1, PFS and OS are shown in Fig. 2A to E. A total of 32 patients were treated in the mCRC cohort; 1 patient was assessable via FDG-PET only and hence was not evaluable for the primary endpoint by RECIST criteria. Of the 31 patients, 5 (16%) had a confirmed partial response (PR), and 5 (16%) had unconfirmed PR (Fig. 2A–C). Stable disease (SD) was observed in 10 patients (32%). Hence, the confirmed objective response rate (ORR) was 16% (5/31) of patients with colorectal cancer; and the clinical benefit rate (CBR) at 24 weeks, defined as PR and SD, was 67% (20/31; Table 3). The median PFS was 3.9 months (95% CI, 1.8–5.4), and the median OS was 6.3 months (95% CI, 4.2–8.8; Fig. 2D and E).
Figure 2.
Efficacy of vemurafenib and erlotinib in patients with BRAF V600E positive mCRC and other cancers. A, Waterfall plot with best percentage tumor change from baseline target lesions and best confirmed overall response for evaluable patients (n = 39). *, CR based on a reduction in measurements in a lymph node to below the level of being considered measurable. CR, complete response. B, Spider plot demonstrating response to treatment for each patient over time (n = 39). *, CR based on a reduction in measurements in a lymph node to below the level of being considered measurable. C, Swimmer plot showing objective response of time on treatment for 39 evaluable patients. Individual patients represented as lines. D, Kaplan–Meier estimate of PFS for patients with mCRC (n = 32). The shaded areas represent 95% CI. E, Kaplan–Meier estimate of OS for patients with mCRC (n = 32). The shaded areas represent 95% CI.
Table 3.
Summary of efficacy data
| mCRC cohort | Non-CRC cohort | All patients | |
|---|---|---|---|
| n = 32 | n = 7 | n = 39 | |
| Best overall response within 24 weeks, n (%) | |||
| Complete response (CR) | 0 (0) | 1 (14) | 1 (3) |
| Partial response (PR) (unconfirmed) | 5 (16) | 5 (71) | 10 (26) |
| Partial response (PR) (confirmed) | 5 (16) | 2 (29) | 7 (18) |
| Stable disease (SD) | 10 (32) | 2 (29) | 12 (32) |
| Progressive disease (PD) | 11 (35) | 0 (0) | 11 (29) |
| Not evaluated according to RECIST | 1 | 0 | 1 |
| Confirmed ORR, n (%) | 5 (16) | 3 (43) | 8 (21) |
| Clinical benefit rate at 24 weeks (CR+PR+SD) | 20 (65) | 7 (100) | 27 (73) |
| Best overall metabolic response at 4 weeks, n (%) | |||
| CMR | 1 (4) | 0 (0) | 1 (3) |
| PMR | 16 (62) | 4 (67) | 20 (54) |
| SMD | 0 (0) | 1 (17) | 1 (3) |
| PMD | 9 (35) | 1 (17) | 10 (31) |
| Not evaluateda | 6 | 1 | 7 |
| Median PFS (months) | 3.9 | 5.5 | 4.2 |
Abbreviation: ORR, overall response rate.
aPatients not assessed by FDG-PET at week 4.
There were 7 patients with various tumor types treated in the non-colorectal cancer cohort: 4 metastatic NSCLC, 1 metastatic breast cancer, 1 metastatic low-grade serous ovarian cancer, and 1 malignant GIST. One patient with ovarian cancer (14%) achieved a CR, 2 patients (29%) with GIST and NSCLC had a confirmed PR, and 2 patients (29%) with NSCLC had an unconfirmed PR (Fig. 2A–C). An ORR (CR + PR) was observed in 43% of patients. Two patients (29%) had stable disease: clinical benefit was observed in all 7 patients (100%). The median PFS was 5.5 months [95% CI, 3.0–not reached (NR)], and median OS was not reached with 3 patients still alive at a median follow-up of 20.2 months (Supplementary Figs. S1 and S2). The median duration of response was 23 weeks (after first response; range, 8–107 weeks) for patients who had a CR or PR on study (Fig. 2C).
FDG-PET analysis
Twenty-six patients in the mCRC cohort and 6 patients in the non-colorectal cancer cohort underwent paired FDG-PET scans at baseline and after 4 weeks of therapy to assess metabolic tumor response and tumor volume as an exploratory endpoint (Table 3). The reduction in maximum standardized uptake value (SUVmax) varied from 26% to 100%, with a median reduction of 66%. In the mCRC cohort, 1 patient (4%) had a complete metabolic response [CMR; based on changes in (SUVmax) in target lesions], and 16 patients (62%) achieved a partial metabolic response (PMR). Conversely, progressive metabolic disease (PMD) was observed in 9 patients (35%). Comparatively, 4 patients (67%) in the non-colorectal cancer cohort achieved a PMR, 1 patient (17%) had stable metabolic disease (SMD), and 1 patient (17%) had PMD. There were no significant differences in median PFS and OS between metabolic responders versus nonresponders (PFS 5.0 months vs. 1.8 months, P = 0.71; OS 7.3 months vs. 6.6 months, P = 0.51; Supplementary Figs. S3A and S3B).
Baseline metabolic tumor volume (MTV) as measured on FDG-PET was significantly higher in patients who did not achieve clinical benefit (Supplementary Fig. S4). Moreover, higher MTV at baseline was associated with inferior PFS and OS (Supplementary Figs. S5A and S5B). The percentage change in MTV at 4 weeks after initiation of treatment correlated with CT defined objective response by RECIST with a significant decrease in median MTV in patients achieving a PR compared with those with PD (P = 0.01; Supplementary Fig. S6).
Baseline ctDNA analysis prior to treatment
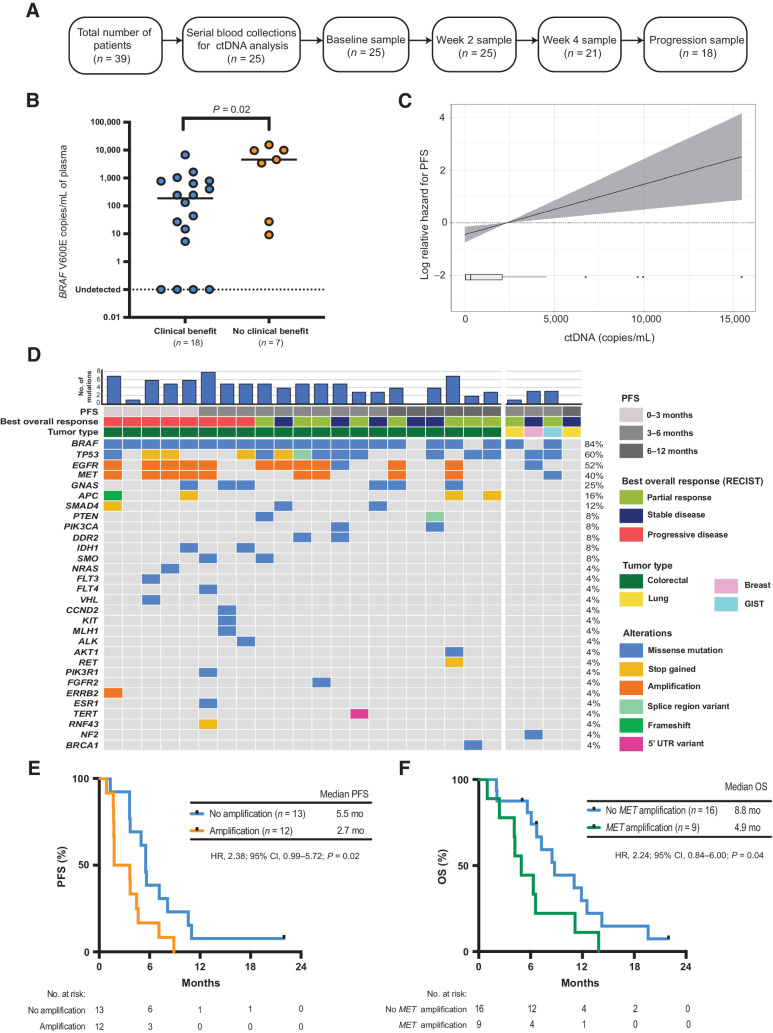
Twenty-five patients had serial plasma available for ctDNA analyses (Fig. 3A). Baseline pretreatment plasma DNA analyzed by droplet digital PCR (ddPCR) was positive for BRAF V600E mutant ctDNA in 21 of 25 patients (84%). The level of ctDNA (copies/mL of plasma) at baseline was significantly higher in patients who did not achieve clinical benefit (P = 0.02; Fig. 3B) on study (Supplementary Fig. S7). Furthermore, higher ctDNA levels at baseline were associated with inferior PFS (Fig. 3C) and OS (Supplementary Fig. S8).
Figure 3.
Baseline ctDNA analysis. A, Consort diagram of plasma samples analyzed from EViCT trial. B, Baseline ctDNA levels (copies/mL) according to clinical benefit rate. C, Linear Cox regression model showing the association between ctDNA copies/mL (as assessed by ddPCR) and PFS (95% CI represented by shading). D, Landscape of somatic mutations detected through targeted sequencing of baseline plasma DNA in 25 patients enrolled on the EViCT trial. Each column represents an individual patient, and each row indicates a specific alteration. The color of bars is indicative of the type of mutation with gray = wild-type. The bar diagram on the top shows the total number of detectable mutations per patient. E, Kaplan–Meier estimate of PFS for patients (n = 25) with a detectable amplification(s) versus no amplification in baseline plasma. Amplification is defined as the presence of any amplification in EGFR, MET, and/or ERRB2. F, Kaplan–Meier estimate of OS for patients (n = 25) with a detectable MET amplification versus no MET amplification in baseline plasma.
In addition to ddPCR testing, baseline plasma DNA was analyzed using a targeted capture based next-generation sequencing assay (Avenio Expanded panel, Roche Diagnostics; Supplementary Table S1 for list of genes). At least one mutation was identified in the baseline plasma DNA of 23/25 (92%) patients (Fig. 3D), with an average of 4 mutations per sample (range 0–7). Patients in the mCRC-cohort had a higher number of genetic alterations in contrast to patients in the non-colorectal cancer cohort. Furthermore, the number of genetic alterations at baseline was significantly higher in patients with mCRC who did not derive a clinical benefit on study compared with patients who derived a clinical benefit (P = 0.04, Supplementary Fig. S9). The most frequently altered genes were BRAF (84%), TP53 (60%), EGFR (52%), MET (40%), and GNAS (25%). Of note, an NRAS G13C mutation was detected in 1 patient at baseline and no KRAS mutations were detected.
Amplifications of EGFR, MET, and ERRB2 through ctDNA sequencing were identified at baseline in 11 patients (44%). Of these, eight patients (73%) had both EGFR and MET amplification, 2 patients (19%) had an EGFR amplification alone and 1 patient (9%) had amplifications of EGFR, MET and ERRB2. Detection of amplification(s) in EGFR, MET, and/or ERBB2 from baseline ctDNA was associated with inferior PFS (HR, 2.4; 95% CI, 0.99–5.72; P = 0.02; Fig. 3E). Patients with detectable amplification(s) in these genes had a median PFS of 2.7 months compared with 5.5 months for nonamplified cases. In keeping with this finding, these patients also showed reduced OS (HR, 2.7; 95% CI, 1.04–6.57; P = 0.01) with a median of 5.5 months versus 11.1 months in patients with and without detectable amplification(s), respectively (Supplementary Fig. S10). Patients with MET-amplification had a numerically shorter median PFS of 1.8 months compared with 5.3 months in the non-MET amplified patients (HR, 1.9; 95% CI, 0.74–4.84; P = 0.11; Supplementary Fig. S11). However, we observed that the median OS was significantly shorter in MET-amplified versus non-MET amplified patients (median OS 4.9 months vs. 8.8 months; HR 2.2; 95% CI, 0.84–6.0; P = 0.04; Fig. 3F). Of note, there was no association between the detection of MET amplification in baseline ctDNA with previous anti-EGFR therapy (P = 0.31, Fisher exact test). In addition to MET amplification, baseline EGFR amplification was also significantly associated with inferior PFS and OS (Supplementary Figs. S12A and S12B).
Early ctDNA dynamics following combination therapy
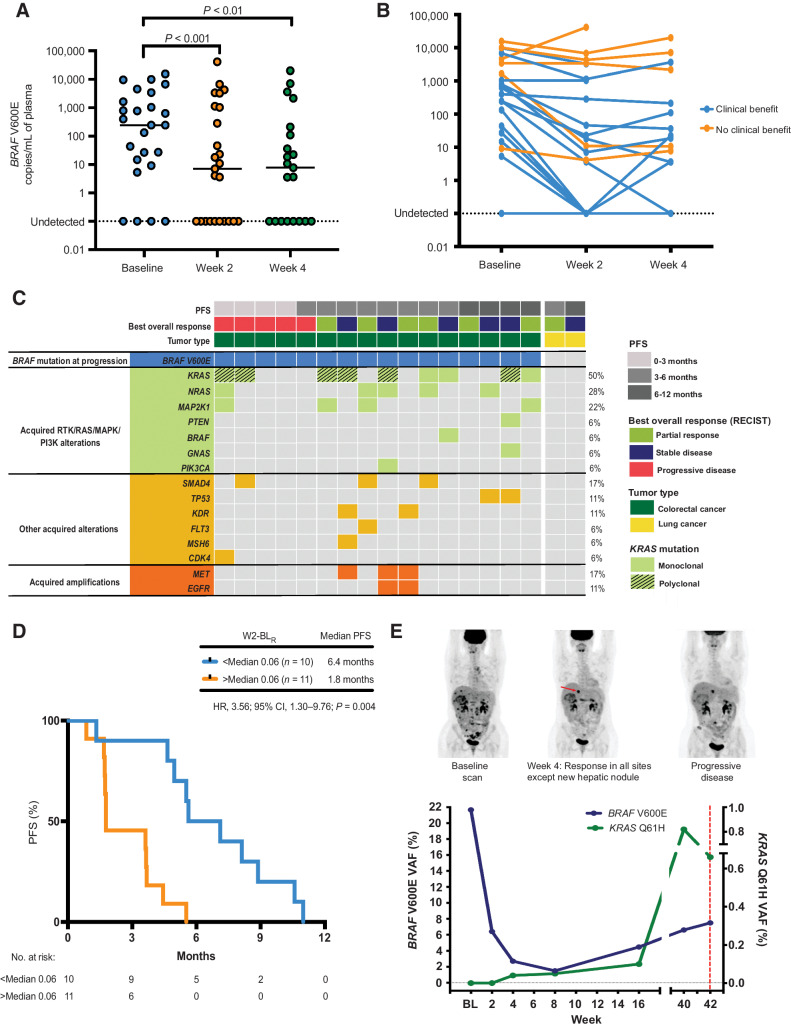
We next explored whether early, dynamic changes in ctDNA levels were predictive of outcomes to combination vemurafenib and erlotinib therapy. A total of 25 paired baseline-week 2 (BL-W2) and 21 paired baseline-week 4 (BL-W4) plasma samples were analyzed for early BRAF V600E mutant ctDNA dynamics (Fig. 3A). A significant reduction in ctDNA levels (copies/mL) was seen between BL-W2 (P < 0.001, median change −132.3 copies/mL) and BL-W4 (P < 0.01, median change −163.9 copies/mL; Fig. 4A). The decrease in BL-W2 and BL-W4 ctDNA levels was significantly greater in patients who derived clinical benefit from treatment (P = 0.0001, median change −176.4 copies/mL and P = 0.0002, median change −188.7 copies/mL, respectively; Fig. 4B). Moreover, we observed that the percentage change in ctDNA variant allele fraction (VAF) at 4 weeks after initiation of treatment predicted objective response. Patients achieving a PR had a significant decrease in median ctDNA compared with those with SD or PD (P = 0.03; Supplementary Fig. S13). We next examined whether changes in FDG-PET MTV at week 4 correlated with changes in ctDNA levels at the same timepoint. Indeed, there was a positive correlation between percentage change in MTV (mL) and ctDNA levels (r = 0.57; P = 0.01; Supplementary Fig. S14).
Figure 4.
Early ctDNA dynamics and genomic alterations at disease progression. A, Dynamics of BRAF V600E mutant DNA copies/mL between baseline, week 2, and week 4 of treatment, n = 25, BL-W2: P < 0.001, BL-W4: P < 0.01. Wilcoxon signed-rank test. B, Dynamics of BRAF V600E mutant DNA copies/mL between baseline, week 2, and week 4 of treatment according to clinical benefit rate. (BL-W2: P = 0.0001; median change −176.4 and BL-WK4: P = 0.0002, median change −188.7 copies/mL, respectively). C, Summary of ctDNA genomic features at disease progression in 18 patients with an available progression plasma sample. Patients are ordered according to time to progression. Each column represents an individual patient, and each row indicates a specific acquired alteration. D, Kaplan–Meier estimate of PFS for patients (n = 21) stratified by week 2–baseline ratio (W2-BLR). E, Serial ctDNA analysis of a patient with BRAF V600E mutation positive colorectal cancer treated in the dose-escalation phase of the EViCT trial. This patient with de novo metastatic colorectal cancer progressing after first-line FOLFOX chemotherapy and bevacizumab, subsequently received vemurafenib and erlotinib on the EVICT clinical trial. ctDNA levels at baseline showed the detectable BRAF V600E mutation with undetectable KRAS Q61H mutation. After 4 weeks of treatment, FDG-PET scan showed a favorable response in all the known metastatic sites of disease. Correspondingly, BRAF V600E VAF decreased from baseline to week 4, confirming a response to therapy. However, there was evidence of a new FDG-avid hepatic nodule, suggesting a refractory clonal population. At that time point, ddPCR identified the emergence of the KRAS Q61H, but the signal was below our defined detection threshold (see Patients and Methods). The KRAS mutation levels gradually increased across serial samples with detection confirmed at 16 weeks. Despite confirmation of stable disease by RECIST criteria at this time point, there was evidence of a steady increase in BRAF V600E ctDNA levels from week 16 onward until the time of disease progression.
The ratio of BRAF V600E mutation VAF on treatment at week 2 and week 4, relative to baseline was assessed as a predictor of PFS and OS. Most patients (19/21; 91%) had a week 2-baseline ratio (W2-BLR) of <1, with a median of 0.06 (Supplementary Fig. S15A), indicating a fall in ctDNA levels. Patients with W2-BLR above the median had inferior PFS and OS compared with those below the median (median PFS 1.8 months vs. 6.4 months; HR, 3.6; 95% CI, 1.30–9.76; P = 0.0004; Fig. 4D) and median OS (5.6 months vs. 9.9 months; HR, 2.9; 95% CI, 1.01–8.36; P = 0.005; Supplementary Fig. S15B). Likewise, most patients (17/18; 94%) had a week 4-baseline ratio (W4-BLR) of <1, with a median of 0.03 (Supplementary Fig. S16A). Correspondingly, patients with week 4-baseline ratio (W4-BLR) above the median of 0.03 had inferior PFS compared with those below median (median PFS 3.6 months vs. 5.6 months; HR, 2.6; 95% CI, 0.92–7.20; P = 0.03; Supplementary Fig. S16B). Although not significant, OS was also shorter in patients with a high W4-BLR. (median OS 6.1 months vs. 11.1 months; HR, 2.0; 95% CI, 0.75–5.48; P = 0.12) (Supplementary Fig. S16C).
In addition, defined ctDNA response criteria as recently described (36) were applied by separating patients into qualitative and quantitative groups according to their ctDNA dynamics. Patients were first divided into three groups to evaluate the qualitative response criteria: G1 (patients with detectable ctDNA remaining detectable during therapy), G2 (patients with detectable ctDNA becoming undetectable during therapy), and G3 (patients with undetectable ctDNA remaining undetectable during therapy). Those with detectable baseline ctDNA that remained detectable at week 2 had the shortest PFS (Supplementary Table S2), compared with patients in G2 and G3. We next assessed the quantitative response criteria according to the percentage change in VAF and patients were divided into five groups: ctDNA CR (ctDNA clearance after baseline detectability), ctDNA PR (decrease of more than 10% VAF), ctDNA SD (no increase or <10% decrease in VAF), ctDNA PD (increase of more than 10% VAF), and ctDNA nonmeasurable disease (undetectable ctDNA at baseline and after treatment). Patients with ctDNA CR at weeks 2 and 4 had the longest PFS (8.1 months, P < 0.0001 and 7.1 months, P = 0.05, respectively) than patients with ctDNA PR, SD, PD, and nonmeasurable disease (Supplementary Table S2).
Serial ctDNA analysis to evaluate genomic evolution following treatment
Plasma DNA collected at the time of disease progression was analyzed through targeted sequencing to identify evidence of genomic evolution following treatment and potential mechanisms of acquired resistance to therapy. In total, 16 patients in the mCRC cohort and 2 patients in the non-colorectal cancer had plasma DNA available at disease progression, allowing comparison with the baseline pretreatment ctDNA analysis (Fig. 3A). Twelve patients (12/18, 67%) were found to have at least one acquired mutation upon disease progression. Of note, 2 patients with NSCLC in the non-colorectal cancer cohort had no detectable mutations in their progression plasma sample, and 4 patients in the mCRC cohort retained the original BRAF V600E mutation but no new mutations were identified at progression. Fifty percent (9/18) of patients showed emergence of ≥1 KRAS or NRAS mutation, which was not detectable at baseline. Polyclonal KRAS mutations were observed in 6 of 9 patients (67%). Moreover, other MAPK pathway alterations such as MAP2K1 mutation were detected in 22% of patients. In addition to these findings, amplifications of MET and EGFR were detected in 2 patients, whereas 1 patient acquired MET amplification alone (Fig. 4C).
In keeping with the identification of KRAS and/or NRAS mutations as the dominant genomic change associated with treatment resistance, serial ctDNA analysis was able to reveal the emergence of these mutations prior to disease progression. This was exemplified through the case of a 38-year-old woman with a BRAF V600E mutation positive, KRAS wild-type, and microsatellite-stable colorectal cancer who had progressed following first-line FOLFOX (folinic acid, 5-fluorouracil, and oxaliplatin) chemotherapy and bevacizumab after five cycles (10 weeks) of treatment. She commenced on vemurafenib 960 mg twice a day and erlotinib 100 mg one a day on the EVICT study. She experienced an improvement in pain, performance status, and a 16% reduction in BRAF V600E VAF ctDNA after 1 month on vemurafenib and erlotinib. FDG-PET scan at week 4 revealed a PMR of extensive nodal disease, osseous disease, and the majority of the hepatic metastases, however, there was evidence of a new small FDG-avid hepatic lesion (Fig. 4E). Targeted sequencing of baseline plasma DNA identified the BRAF V600E mutation, a SMAD4 mutation, TP53 mutation, and EGFR amplification (Fig. 3D). After 6 months on study, CT imaging confirmed PD by RECIST v1.1 criteria, with enlarging hepatic metastases and evidence of new metastatic lesions. Targeted sequencing of plasma DNA at progression uncovered the emergence of multiple subclonal KRAS mutations (KRAS Q61H, KRAS Q61L, KRAS G12N, KRAS G13D), a NRAS G12D mutation, KDR A163G mutation, MSH6 R106H mutation, and MET amplification (Fig. 4C). Serial analysis of ctDNA using ddPCR identified emergence of the KRAS Q61H mutation 6.5 months prior to confirmation of PD on imaging but it was not detectable in baseline plasma obtained prior to starting the clinical trial. The KRAS Q61H was first detected at 4 weeks, but the signal was below our defined detection threshold at this timepoint (see Patients and Methods). The KRAS mutation levels gradually increased across serial samples with detection confirmed at 16 weeks (Fig. 4E).
Discussion
Patients with BRAF V600E mutated colorectal cancer represent a distinct molecular subtype associated with poor prognosis where novel therapeutic approaches are needed. Recent studies have investigated the combination of BRAF, EGFR, and MEK inhibition after initial preclinical and clinical studies identified the lack of efficacy of single-agent BRAF inhibitors owing to EGFR-mediated adaptive feedback. Here, we report results of the EViCT clinical trial evaluating the safety and efficacy of combination vemurafenib and erlotinib therapy in 32 patients with BRAF V600E positive mCRC and 7 patients with various other tumor types. The MTD and RP2D was determined to be vemurafenib 960 mg twice a day and erlotinib 150 mg one a day, the full dose of either agent as a monotherapy. Importantly, the combination therapy was well tolerated, and the most common AEs were consistent with known side effects of vemurafenib and erlotinib when used as single agents, including diarrhea, nausea, fatigue, and acneiform rash.
Encouraging antitumor activity was observed using the combination of vemurafenib and erlotinib, with a confirmed response rate of 16% and an unconfirmed response rate of 33% in the mCRC cohort. These data are consistent with efficacy reported for other similar BRAF and EGFR inhibitor therapeutic strategies, notably the doublet combination of encorafenib and cetuximab in the BEACON colorectal cancer study which reported a response rate of 20% (23, 27, 32). The median PFS was also similar, with a PFS of 3.9 months in EVICT compared with 4.3 months in BEACON. Conversely, the ORR was noted to be high in the non-colorectal cancer cohort, with confirmed response rates of 43%; and prolonged disease stabilization observed in several tumor types, including NSCLC and GIST, compared with the mCRC cohort. Taken together, these data show that despite harboring the BRAF V600E mutation, response to combination vemurafenib and erlotinib may not be uniform amongst different tumor types. This underscores the importance of characterizing molecular determinants of response and resistance to combination therapy across tumor types, as well as developing novel biomarkers to track treatment responses in real time.
ctDNA dynamics have been shown to be a useful tool for assessing tumor response kinetics to chemotherapy and targeted therapy in a wide range of solid malignancies, including colorectal cancer (37, 38). Moreover, a rise in ctDNA levels may provide an early measure of impending treatment failure and has been shown to precede radiological progression by weeks to months (38–40). This provides a potential opportunity for early modification of treatment, a strategy which may be beneficial where alternative treatment options are available, rather than waiting for radiological progression to be confirmed when disease burden is higher. Indeed, our data suggest that early ctDNA changes are predictive of treatment response and clinical outcomes for patients treated with combination BRAF and EGFR inhibition. When examining response dynamics, ctDNA responses were generally observed by week 2, indicative of the rapid response often achieved with targeted therapies. Consistent with results reported by Gouda and colleagues (36), patients with clearance (ctDNA CR) at weeks 2 and 4 demonstrated longer median PFS. Furthermore, detectable ctDNA during treatment was associated with worse clinical outcomes. This supports the use of defined ctDNA response criteria as a supplementary method to conventional imaging to assess treatment responses in this patient population.
The use of FDG-PET has also been shown to be an early indicator of targeted therapy response in several tumor types including GIST (41), EGFR mutant NSCLC (42), and BRAF V600E mutant melanoma (30). In the EVICT study, FDG-PET was assessed as an early marker of biologic response to vemurafenib and erlotinib, but only 62% of patients in the mCRC cohort had a PMR and one patient obtained a CMR. In comparison, previous studies (30) have shown 100% of BRAF mutant advanced melanoma patients treated at therapeutic doses of vemurafenib have at least a PMR, suggesting differences in the depth of MAPK pathway inhibition in response to vemurafenib, between colorectal cancer and melanoma. This is supported by preclinical and clinical data showing that vemurafenib displays less potency in colorectal cancer compared with melanoma (20, 43). Although an early metabolic response did not translate into improved survival outcomes in the EVICT study, baseline MTV was prognostic of outcomes and may still be a useful biomarker, particularly in patients who have undetectable ctDNA. Taken together, our data highlight that FDG-PET and ctDNA can be complementary approaches which when used together can provide an early indication of MAPK pathway inhibition and treatment response, particularly with respect to heterogeneity of metabolic response.
Importantly, ctDNA analysis also provided an opportunity to characterize genomic determinants of primary and secondary resistance to vemurafenib and erlotinib. MET amplification can emerge as a mechanism of resistance to anti-EGFR therapies and is usually rare in treatment-naïve colorectal cancers (44). Here we report a high proportion of patients with MET amplification at baseline, as a likely mechanism of primary resistance to dual BRAF and EGFR inhibitors, which was not related to exposure to previous anti-EGFR therapy. Moreover, further enrichment of MET amplification was seen at the time of disease progression. Similar findings have been previously demonstrated in a study by Pietrantonio and colleagues, where MET amplification was noted in tumor specimens resistant to panitumumab and vemurafenib, with corresponding pretreatment specimens found to harbor preexisting MET amplifications suggestive of clonal expansion in response to treatment (45). Our findings support the importance of further exploratory work to characterize the role of MET amplification in driving resistance to targeted approaches in mCRC; and provide the rationale for the combination of BRAF/EGFR and MET blockade as a therapeutic strategy to overcome MET-driven resistance in this setting (45).
It has been reported that acquired resistance to BRAF inhibitor combinations in mCRC can be driven by alterations in the MAPK pathway leading to reactivation (24, 46). Here, we confirmed frequent mutations in KRAS, NRAS, and MAP2K1 as recurrent mechanisms of acquired resistance in multiple patients. In line with the study by Corcoran and colleagues, polyclonal RAS mutations were commonly detected in plasma samples of patients progressing on therapy, revealing convergent evolution as a dominant mechanism of therapeutic escape (27). These RAS mutations were not detectable through baseline plasma DNA analysis prior to treatment, but it is possible that they may have been present below the detection limit of the assay. We have shown the ability to detect the early emergence of RAS mutations through serial ctDNA analysis, prior to evidence of clinical disease progression to combination therapy. Of note, a third of patients in our study had no other genomic mechanism of resistance identified through ctDNA analysis. This may have been due to the limited number of genes covered by our targeted sequencing panel and other potential genetic and nongenetic resistance mechanisms were not explored.
We acknowledge several limitations of our study, particularly the small cohort of patients with non-colorectal cancer that were evaluated. Furthermore, baseline tissue prior to trial enrolment was not available to determine the MSI-status in the mCRC cohort. Finally, results of the biomarker analyses are exploratory and validation in larger clinical studies is required to support our findings.
In conclusion, the EViCT trial demonstrated the safe and novel combination of two oral inhibitors targeting BRAF and EGFR, with significant activity in a range of BRAF V600E mutated cancers. Although the recently defined standard of care incorporates the BRAF inhibitor encorafenib in combination with cetuximab based on data from a randomized phase III registration trial; our combination strategy of vemurafenib and erlotinib should be further explored as a potential treatment option for patients preferring oral combination therapy, that is readily deliverable and with a favorable side effect profile. Our findings will help inform future combination strategies, with the goal of overcoming primary and secondary resistance to combination BRAF and EGFR targeted therapy, to improve outcomes for patients with BRAF V600E positive mCRC and other cancers.
Role of Funding Source
The Victorian Cancer Agency Clinical Trials Grants and Roche Products Pty Limited (Australia).
Supplementary Material
Supplementary material
Acknowledgments
The authors thank the patients and their families for participation in this study. We also thank Madawa Jayawardana and Cassandra Litchfield for assistance with the statistical analysis, and Matthew Chapman for sample management. This work was supported by the Victorian State Government through Victorian Cancer Agency large infrastructure Clinical Trials Grant and Roche Products Pty Limited (Australia).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 987
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
B. Tran reports grants and personal fees from Amgen, Astellas, Astra Zeneca, Bayer, BMS, Ipsen, Janssen, Pfizer, MSD, Sanofi, Merck, and Roche and grants from Genentech outside the submitted work. J. Tie reports personal fees from Haystack Oncology and other support from Merck Serono, Pierre Fabre, Pfizer, and Amgen outside the submitted work. B. Markman reports personal fees from Amgen, Merck, Bristol Myers Squibb, Beigene, and AstraZeneca outside the submitted work. N.C. Tebbutt reports personal fees from Pierre Fabre, Bristol Myers Squibb, Amgen, Merck Sharp and Dohme, Servier, and Eisai and personal fees and nonfinancial support from AstraZeneca outside the submitted work. R. Koldej reports grants from CRISPR Therapeutics outside the submitted work. B.J. Solomon reports personal fees from Roche outside the submitted work. G.A. McArthur reports research support from Bristol Myers Squibb, and as principal investigator, G.A. McArthur's institution has received reimbursement of trial costs from Roche-Genentech. R.J. Hicks reports other support from Telix Pharmaceuticals outside the submitted work. S.-J. Dawson reports other support from Adela outside the submitted work. J. Desai reports grants, personal fees, and nonfinancial support from Roche/Genentech during the conduct of the study as well as grants and nonfinancial support from BMS, Novartis, and AstraZeneca; grants and personal fees from Merck KGaA; and grants, personal fees, and nonfinancial support from Beigene, Roche/Genentech, Amgen, Pierre-Fabre, GlaxoSmithKline, Boehringer Ingelheim, and Daiichi Sankyo Europe GmbH outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
L. Tan: Data curation, formal analysis, validation, investigation, visualization, writing–original draft, writing–review and editing. B. Tran: Resources, data curation, formal analysis, writing–original draft, writing–review and editing. J. Tie: Resources, data curation, formal analysis, writing–original draft, writing–review and editing. B. Markman: Resources, writing–review and editing. S. Ananda: Resources, writing–review and editing. N.C. Tebbutt: Resources, writing–review and editing. M. Michael: Resources, writing–review and editing. E. Link: Resources, data curation, software, formal analysis, writing–review and editing. S.Q. Wong: Investigation, visualization, writing–review and editing. S. Chandrashekar: Investigation, writing–review and editing. J. Guinto: Investigation, writing–review and editing. D. Ritchie: Resources, writing–review and editing. R. Koldej: Resources, writing–review and editing. B.J. Solomon: Resources, formal analysis, writing–review and editing. G.A. McArthur: Resources, writing–review and editing. R.J. Hicks: Resources, investigation, writing–review and editing. P. Gibbs: Resources, writing–review and editing. S.-J. Dawson: Supervision, visualization, writing–original draft, writing–review and editing. J. Desai: Conceptualization, resources, data curation, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing.
References
- 1. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. [DOI] [PubMed] [Google Scholar]
- 2. Barras D. BRAF mutation in colorectal cancer: an update. Biomark Cancer 2015;7(Suppl 1):9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wan PTC, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Project CG, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004;116:855–67. [DOI] [PubMed] [Google Scholar]
- 4. Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 2002;418:934. [DOI] [PubMed] [Google Scholar]
- 5. Tie J, Gibbs P, Lipton L, Christie M, Jorissen RN, Burgess AW, Croxford M, et al. Optimizing targeted therapeutic development: analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int J Cancer 2011;128:2075–84. [DOI] [PubMed] [Google Scholar]
- 6. Kalady MF, DeJulius KL, Sanchez JA, Jarrar A, Liu X, Manilich E, Skacel M, et al. BRAF mutations in colorectal cancer are associated with distinct clinical characteristics and worse prognosis. Dis Colon Rectum 2012;55:128–33. [DOI] [PubMed] [Google Scholar]
- 7. Gonsalves WI, Mahoney MR, Sargent DJ, Nelson GD, Alberts SR, Sinicrope FA, Goldberg RM, et al. Patient and tumor characteristics and BRAF and KRAS mutations in colon cancer, NCCTG/Alliance N0147. J Natl Cancer Inst 2014;106:dju106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer 2011;104:856–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer 2009;101:465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. N Engl J Med 2009;361:98–99. [DOI] [PubMed] [Google Scholar]
- 11. Tran B, Kopetz S, Tie J, Gibbs P, Jiang Z-Q, Lieu CH, Agarwal A, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer 2011;117:4623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Venderbosch S, Nagtegaal ID, Maughan TS, Smith CG, Cheadle JP, Fisher D, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res 2014;20:5322–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goey KKH, Elias SG, van Tinteren H, Laclé MM, Willems SM, Offerhaus GJA, et al. Maintenance treatment with capecitabine and bevacizumab versus observation in metastatic colorectal cancer: updated results and molecular subgroup analyses of the phase 3 CAIRO3 study. Ann Oncol 2017;28:2128–34. [DOI] [PubMed] [Google Scholar]
- 14. Stintzing S, Miller-Phillips L, Modest DP, Fischer von Weikersthal L, Decker T, Kiani A, Vehling-Kaiser U, et al. Impact of BRAF and RAS mutations on first-line efficacy of FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab: analysis of the FIRE-3 (AIO KRK-0306) study. Eur J Cancer 2017;79:50–60. [DOI] [PubMed] [Google Scholar]
- 15. Cremolini C, Loupakis F, Antoniotti C, Lupi C, Sensi E, Lonardi S, Mezi S, et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol 2015;16:1306–15. [DOI] [PubMed] [Google Scholar]
- 16. Peeters M, Price TJ, Cervantes A, Sobrero AF, Ducreux M, Hotko Y, et al. Final results from a randomized phase 3 study of FOLFIRI {±} panitumumab for second-line treatment of metastatic colorectal cancer. Ann Oncol 2014;25:107–16. [DOI] [PubMed] [Google Scholar]
- 17. Morris V, Overman MJ, Jiang Z-Q, Garrett C, Agarwal S, Eng C, Kee B, et al. Progression-free survival remains poor over sequential lines of systemic therapy in patients with BRAF-mutated colorectal cancer. Clin Colorectal Cancer 2014;13:164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367:1694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med 2014;371:1877–88. [DOI] [PubMed] [Google Scholar]
- 20. Kopetz S, Desai J, Chan E, Hecht JR, O'Dwyer PJ, Maru D, et al. Phase II pilot study of vemurafenib in patients with metastatic BRAF-mutated colorectal cancer. J Clin Oncol 2015;33:4032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100–3. [DOI] [PubMed] [Google Scholar]
- 22. Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, Brown RD, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012;2:227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yaeger R, Cercek A, O'Reilly EM, Reidy DL, Kemeny N, Wolinsky T, Capanu M, et al. Pilot trial of combined BRAF and EGFR inhibition in BRAF-mutant metastatic colorectal cancer patients. Clin Cancer Res 2015;21:1313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, Godfrey JT, et al. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov 2015;5:358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hong DS, Morris VK, El Osta B, Sorokin AV, Janku F, Fu S, Overman MJ, et al. Phase IB study of vemurafenib in combination with irinotecan and cetuximab in patients with metastatic colorectal cancer with BRAFV600E mutation. Cancer Discov 2016;6:1352–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kopetz ES, McDonough SL, Morris VK, Lenz HJ, Magliocco AM, CEea A. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG 1406). J Clin Oncol 2017;35(4 Suppl):520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Corcoran RB, André T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, Hollebecque A, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF(V600E)-mutant colorectal cancer. Cancer Discov 2018;8:428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, Wasan H, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med 2019;381:1632–43. [DOI] [PubMed] [Google Scholar]
- 29. Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, Tacey M, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015;26:1715–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McArthur GA, Puzanov I, Amaravadi R, Ribas A, Chapman P, Kim KB, et al. Marked, homogeneous, and early [18F]fluorodeoxyglucose-positron emission tomography responses to vemurafenib in BRAF-mutant advanced melanoma. J Clin Oncol 2012;30:1628–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Subbiah V, Gervais R, Riely G, Hollebecque A, Blay JY, Felip E, et al. Efficacy of vemurafenib in patients with non-small-cell lung cancer with BRAF V600 mutation: an open-label, single-arm cohort of the histology-independent VE-BASKET study. JCO Precis Oncol 2019;3:PO.18.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay J-Y, Wolf J, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med 2015;373:726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: evolving considerations for PET. J Nucl Med 2009;50:122S–50S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gouda MA, Huang HJ, Piha-Paul SA, Call SG, Karp DD, Fu S, et al. Longitudinal monitoring of circulating tumor DNA to predict treatment outcomes in advanced cancers. JCO Precis Oncol 2022;6:e2100512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dawson SJ, Tsui DWY, Murtaza M, Biggs J, Rueda OM, Chin SF, et al. Analysis of circulating tumour DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199–209. [DOI] [PubMed] [Google Scholar]
- 39. Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486:532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stroobants S, Goeminne J, Seegers M, Dimitrijevic S, Dupont P, Nuyts J, Martens M, et al. 18FDG-Positron emission tomography for the early prediction of response in advanced soft tissue sarcoma treated with imatinib mesylate (Glivec). Eur J Cancer 2003;39:2012–20. [DOI] [PubMed] [Google Scholar]
- 42. Mileshkin L, Hicks RJ, Hughes BGM, Mitchell PLR, Charu V, Gitlitz BJ, Macfarlane D, et al. Changes in FDG- and FLT-PET imaging in patients with non-small cell lung cancer treated with erlotinib. Clin Cancer Res 2011;17:3304–15. [DOI] [PubMed] [Google Scholar]
- 43. Pickles OJ, Drozd A, Tee L, Beggs AD, Middleton GW. Paradox breaker BRAF inhibitors have comparable potency and MAPK pathway reactivation to encorafenib in BRAF mutant colorectal cancer. Oncotarget 2020;11:3188–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, Sartore-Bianchi A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 2013;3:658–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pietrantonio F, Oddo D, Gloghini A, Valtorta E, Berenato R, Barault L, Caporale M, et al. MET-driven resistance to dual EGFR and BRAF blockade may be overcome by switching from EGFR to MET inhibition in BRAF-mutated colorectal cancer. Cancer Discov 2016;6:963–71. [DOI] [PubMed] [Google Scholar]
- 46. Oddo D, Sennott EM, Barault L, Valtorta E, Arena S, Cassingena A, Filiciotto G, et al. Molecular landscape of acquired resistance to targeted therapy combinations in BRAF-mutant colorectal cancer. Cancer Res 2016;76:4504–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.