Abstract
Breast cancer is one of the major causes of death in women worldwide. It is a diverse illness with substantial intersubject heterogeneity, even among individuals with the same type of tumor, and customized therapy has become increasingly important in this sector. Because of the clinical and physical variability of different kinds of breast cancers, multiple staging and classification systems have been developed. As a result, these tumors exhibit a wide range of gene expression and prognostic indicators. To date, no comprehensive investigation of model training procedures on information from numerous cell line screenings has been conducted together with radiation data. We used human breast cancer cell lines and drug sensitivity information from Cancer Cell Line Encyclopedia (CCLE) and Genomics of Drug Sensitivity in Cancer (GDSC) databases to scan for potential drugs using cell line data. The results are further validated through three machine learning approaches: Elastic Net, LASSO, and Ridge. Next, we selected top-ranked biomarkers based on their role in breast cancer and tested them further for their resistance to radiation using the data from the Cleveland database. We have identified six drugs named Palbociclib, Panobinostat, PD-0325901, PLX4720, Selumetinib, and Tanespimycin that significantly perform on breast cancer cell lines. Also, five biomarkers named TNFSF15, DCAF6, KDM6A, PHETA2, and IFNGR1 are sensitive to all six shortlisted drugs and show sensitivity to the radiations. The proposed biomarkers and drug sensitivity analysis are helpful in translational cancer studies and provide valuable insights for clinical trial design.
Keywords: drug sensitivity, machine learning, radiosensitive, biomarkers, pharmacogenomics
Breast cancer is a highly prevalent malignant tumor risking women’s health around the globe and is a leading cause of cancer. According to the world health organization (WHO), there were 684,996 mortalities as a result of breast cancer in 2021; in 2022, there were 287,850 estimated cases of invasive breast cancer to be diagnosed in women as per the National Breast Cancer Coalition (NBCC) reports. Breast cancer patients with early detection and treatment have a better prognosis, a longer survival time, and a lower fatality rate.
Chemotherapy resistance remains the most severe issue in treating people living with cancer. Novel chemotherapeutical and targeted medicines continue to be advanced. Even if most anticancer drugs slow down tumor development, the effect is usually short-lived, and anthracycline and taxane failure directly impacts breast cancer patients’ survival.1 As a result, novel medicines with limited sensitivity to conventional drug resistance mechanisms are urgently needed to increase response rates and possibly extend survival.
Pharmacogenomics predictions based on genetic data is a growing field of study with several practical uses, including drug development and repurposing, subject selection for medical studies, and individualized therapy suggestions (for instance, in a tumor board background). The scientific community has created comprehensive cell line drug sensitivity assays such as CCLE,2 CTRP,3 GDSC,4 and gCSI.5 These databases include molecular and pharmacological response data from a multitude of cell lines, permitting predictive models to be built. However, despite the existence of these data, the ability to determine medication response reliably remains restricted.6,7 Noise in the data, small sample size compared to characteristics quantity (i.e., predictor variables), insufficient omics description, as well as the stationary nature of molecular information are all factors that make drug response prediction difficult. In such investigations, molecular data are frequently obtained just before the medication is administered.8 The reliability of pharmacogenomics correlations generated from diverse data sets is another significant issue. Several studies discovered that discrepancies in experimental techniques and data processing caused the reported inconsistency.9−14
Drug sensitivity refers to the amount of activity played on a target (cell lines, in our case). It is measured through different approaches, such as the area above the curve (AAC) and IC50IC50. In contrast, drug resistance is the resistance shown by the target toward a particular drug or compound, which may be caused by mutations or overdosing. This is related to the biomarkers that are crucial for disease survival, understanding, and therapeutic purposes. For cancer patients, radiotherapy is commonly utilized as a curative treatment. Radiation also bears great importance as it can be used along with chemotherapy for an effective and fast remedy. Recent technological advancements have enhanced radiation’s physical accuracy, resulting in higher remedial success and lower toxicity.15
Normalized regression techniques (i.e., LASSO, Elastic Net, Ridge regression),16−19 partial least-squares (PLS) regression, support vector machines (SVMs),20 random forest (RF), neural networks, and deep learning,21,22 logical models, or kernelized Bayesian matrix factorization (KBMF)23,24 have all been used to solve the drug response prediction problem. Ali and Aittokallio (2019)25 provide a detailed current review. To date, no comprehensive investigation of model training procedures based on information from several large cell line screenings got reported in association with radiation data. We wanted to bridge these gaps in our work to advance the precision of medication response estimation and discover novel biomarkers for drug and radiation sensitivity.
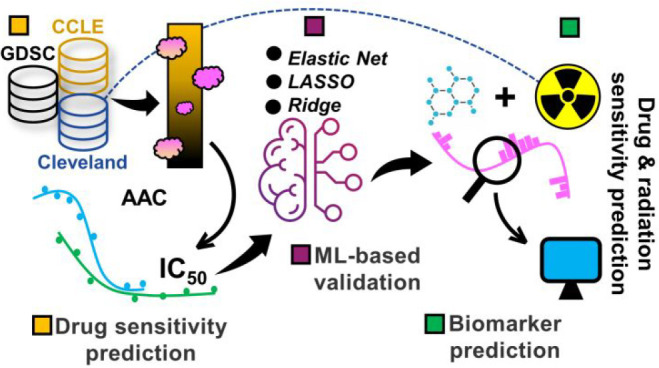
For this purpose, we retrieved required information from the cancer cell line encyclopedia (CCLE) genomics of drug sensitivity in cancer (GDSC) to intersect and then apply three multivariate machine learning models such as Elastic Net,26 LASSO,27 and Ridge.28 For examining drug activity, we considered the IC50 and AAC values for 24 cancer drugs that were obtained as a result of interesting CCLE and GDSC. The aforementioned regression models were employed for predicting the accuracy of drug sensitivity on the cancer cell lines that assisted in picking out our top-ranked drugs. Finally, we shortlisted biomarkers from the molecular profiling data and manually searched for potential biomarkers. The Cleveland database was also considered for the radiation data, as the goal was to hunt for signatures showing sensitivity to both types of treatments.
Methods
Ethical Considerations
The human cell lines and radiotherapy results used in this work are freely accessible data obtained from public repositories (CCLE, GDSC, and the Cleveland database) for research purposes only and thus require no approval as the data are completely anonymized.
Data Types and Sources
We curated breast cancer cell lines from two cloud-based repositories known as GDSC29 and CCLE.30 Each data set has a panel of cancer cell lines with cancer drugs applied. To avoid overlaps among the data, we intersected both databases for shortlisting breast cancer drugs. As a result, we obtained 24 drugs that were selected for further evaluation. Furthermore, nine breast cancer cell lines31 were considered for cell line–drug sensitivity analysis. The radiation data are retrieved from the Cleveland32 database. Figure 1 depicts the overall workflow of this study.
Figure 1.
A methodological pipeline. Breast cancer data are taken from the two cancer cell line databases, and both the IC50 and AAC curves are plotted, shortlisting effective drugs. Next, the three machine learning methods are applied to compare and validate drug response on cancer cell lines. Since the pharmacological information is available alongside the experimental data, we also identified potential markers showing sensitivity toward the shortlisted drugs and radiations (radiotherapy).
Examining and Extracting Data of Interest
The PharmacoGx33 and RadioGx34 packages resemble each other a lot in their object structure. The PharmacoGx package is used to analyze big pharmacogenomic data sets efficiently. This package handles the stored pharmacological and molecular information as R objects. Similarly, the RadioGx suite creates a standardized data format for storing radiogenomics data obtained from radiotherapy sessions. The aim was to understand the association among various cancer cell lines and their response to our shortlisted drugs and ionizing radiation (IR). Generally, the PharmacoSet (PSet) and RadiogenomicSet (RSet) stock three main types of information: metadata/annotations, molecular information, and treatment response data.
Modeling the Sensitivity Data
Drug–dose response statistics contained within the PSet objects are plotted via the drug dose–response curve function.35 For a list of PSets, a drug name, and a cell name, it plots the drug dose–response curves for a given cell–drug blend in each data set, permitting direct data comparisons between data sets.
Drug Sensitivity Prediction
To determine the effectiveness of a given drug, pharmacogenomic studies consider cancer cell lines to be tested for their response to increasing concentrations of various compounds from which the IC5036 and the area above the curve (AAC)37 are computed. The IC50 is the concentration of an inhibitor where the response is reduced by half. The AAC is the area above the dose–response curve for the tested drug concentrations and is a more robust metric normalized against the dose range.
Consistency Improvement between the Data Sets
The cell and drug names used in the GDSC and CCLE databases are not identical. Therefore, we used the PharmacoGx package to clear these differences and perform a comparative analysis between the two data sets. The hgu133a and enlarged hgu133plus platforms are used for profiling GDSC and CCLE, respectively. Although the hgu133a platform is a precise subset of the hgu133plus2, the gene ensemble IDs summarize the expression information in PSet objects, permitting data sets from different platforms for easy comparison.
We used two main utilities known as downloadPSet and intersectPSet for importing stored data sets to test data consistency and find a common intersection between the two data sets, respectively.
We also constructed a breakdown of gene expression and drug sensitivity metrics so that, inside each data set, one gene expression pattern and one sensitivity profile per cell line exist. Finally, a conventional correlation coefficient equates the gene expression and susceptibility metrics between the data sets.
Reliability Assessment
To better examine the concordance of numerous pharmacogenomic research (rCI),38,39 we used a robust concordance index (rCI). Knowing that drug screening assay noise is considerable and that responsive grading of cell lines with similar AAC values might be erroneous, the rCI only evaluates cell line pairings with a drug sensitivity (AAC) discrepancy higher.
Supervised Comparison
In our study, we used three main multivariate machine learning regression methods: LASSO, Elastic Net, and Ridge. Each modeling technique possesses an independent set of hyperparameters: svmRadial – sigma and C (cost); random forest – mtry; xgbTree – nrounds, max_depth, eta, gamma, colsample_bytree, min_child_weight, subsample.
Feature Choice
For selecting a subset of existing modeling features, we executed feature selection utilizing information only from the training set. Each feature is assessed exclusively concerning the linkage between a feature vector and a vector with target variables (filter feature selection). We employed the maximum relevance minimum redundancy (mRMR) package for feature selection (caret package), which selects features correlating strongly to a classification variable. The gamScores function fits a global additive model between a single forecaster and the outcome through the smoothing spline basis function.
Accuracy Metrics
To evaluate the model’s accuracy, we used the concordance index, which is the rank correlation between detected and predicted data. We computed the concordance index through the “concordance.index” function from the survcomp package (ver.1.28.5). For classification tasks, we considered the percentage of precisely predicted samples as the precision measurement system.
Model Training and Assessment Technique
We took these steps to train and test models to predict drug response.
-
i.
We used the GDSC data set as the training data and CCLE as the test set.
-
ii.
Feature selection using mRMR is performed only on the training set.
-
iii.
Next, we performed model fitting with N (ranging 10–500) number of chosen features (having significant p-values) on the training set information. For the hyperparameters’ selection, 30 combinations were tried on the training set through the cross-validation method; finally, the hyperparameter combination providing better accuracy was chosen for the final model fitting.
-
iv.
The model was applied to the training data to calculate the concordance index.
The training set (GDSC) contains the RNA data, and we did not tear apart the data into the training and testing sets because different databases are used for these purposes. The data are from GDSC, and CCLE is intersected using the “intersect” function and mRMR package in the RStudio (ver.2022.02.1) to efficiently choose the potential features.40 For thresholding, we kept the 5-fold cross-validation sampling equal to 10 and chosen features with the lowest p-values equal to 100. The models were run using the dplyr41 and caret42 packages for RStudio.43
Identification of Potential Biomarkers
Drug-Sensitive and Radiosensitive Signatures
We considered the RNA molecular profiling information from both databases and used the inbuilt PharmacoGx options for generating signatures with molecular features correlating with response to specific drugs. We did this only for searching drug-sensitive biomarkers. On the other hand, the RadioGx package can determine genes for a cell line as a result of the radiotherapy experiment. This way, we can recognize whether a biomarker is radiosensitive or radioresistant.
Correlating Biomarkers to Radiation and Drug Effects
Using the PharmacoGx and RadioGx packages, we can compute signatures for each molecular feature and measure its correlation with response to a particular therapy. The point is to clarify comparing a biomarker’s response to the six shortlisted drugs with its response to radiotherapy. This will assist in generating hypotheses for combination therapies or comprehending action mechanisms.
Drug–Biomarker Association
We can model the linkage among molecular features and given drug response through a linear regression approach standardized for tissue sources44
where Y represents the drug sensitivity variable; Gi, T, and B signify the expression of the gene i, tissue source, and the experimental batch correspondingly; and the β’s are the regression coefficients.
Apart from the link between drug sensitivity and tissue source, the intensity of the gene–drug interaction is measured by βi. To compute standardized coefficients from the linear model, the variables G and Y are adjusted (standard deviation = 1). The arithmetical validity of βi (two-sided t test) is used to evaluate the relevance of the gene–drug interaction. The false discovery rate (FDR) technique is then used to fix p-values for manifold testing. We predicted the link’s significance between medications and associated reported signatures in CCLE and GDSC using biomarker discoveries across pharmacogenomic research. We look at the link between pharmaceuticals on the shortlist and marker genes.
Results
Estimating Metrics of Drug Response
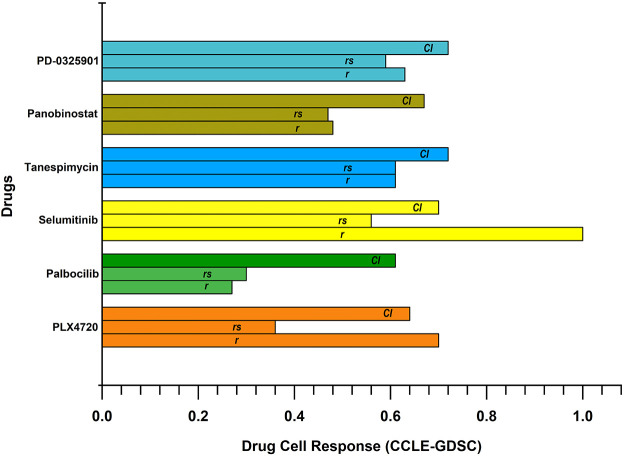
We have considered the data sets containing drug and radiation sensitivity data, including information about RNA, RNA-Seq, Copy Number Variation (CNV), mutational, and drug response (Table 1). The details about breast cancer cell lines are listed in Table S1. We have considered six drugs named Palbociclib, Panobinostat, PD-0325901, PLX4720, Selumetinib, and Tanespimycin. These six drugs overlap in both data sets and have a significant response toward breast cancer cell lines (Figure 2). We can observe that all six drugs have roughly similar confidence intervals (CIs) across the data sets, but a significant difference in the Pearson correlation coefficient (r) can be seen in the case of Selumetinib. Palbociclib shows Spearsman’s lowest correlation coefficient (rs) and r values. These metrics conclude that Selumetinib shows relatively better efficacy on breast cancer cell lines while Palbociclib is least effective comparatively. The rest of the four drugs are similar in their responses.
Table 1. Information about Each Data Set Used in This Study.
| Data set | Data type | Platform | Samples |
|---|---|---|---|
| GDSC (2020(v2-8.2)) | RNA, RNA-Seq, CNV, mutation, drug response | IC50 | 1084 cell lines × 215780 drug sensitivity |
| CCLE (CCLE_2015) | RNA, RNA-Seq, CNV, mutation, drug response | IC50 | 1094 cell lines × 11670 drug sensitivity |
| Cleveland (2017) | RNA, RNA-Seq, CNV, mutation | NA | 540 cell lines × 1 radiation |
Figure 2.
Drug cell response. These drugs significantly respond to breast cancer cell lines and have roughly similar confidence intervals (CIs) across the data sets. However, a substantial difference in the Pearson correlation coefficient (r) can be seen in the case of Selumetinib. Palbociclib displays the least Spearsman’s correlation coefficient (rs) and r values.
Drug–Cell Response Curves
To further understand the efficacy of these drugs, we considered two metrics of drug response known as IC50 and AAC. We used the dose–response curve function to plot drug–response analysis outcomes contained in PSet objects. It allows drawing dose–response curves for a given cell drug in each data set for data comparison across the data sets. The IC50 curves of the shortlisted drugs reveal promising inhibitory performance with 100% viability on the majority of the nine breast cancer cell lines (Figure S1). We plotted the IC50 curves of Panobinostat on only seven cell lines due to a lack of experimental data availability.
These drugs show promising inhibitory performance on different cancer cell lines. Minor and major differences in the IC50 values from CCLE and GDSC can be observed since the experimental data come from various groups, but still, the results are quite similar. Out of six drugs, we observed some drugs’ concentration to be out of the limit, but overall performance is significant. Similarly, the AAC curves show substantial performance on the breast cancer cell lines (Figure S2). However, the AAC curves of Panobinostat and Tanespimycin are not significant. Sudden declines can be observed with an increase in drug concentration. Combined, the four drugs named Palbociclib, PD-0325901, PLX4720, and Selumetinib show significant IC50 and AAC values.
Improving Drug Consistency across CCLE and GDSC
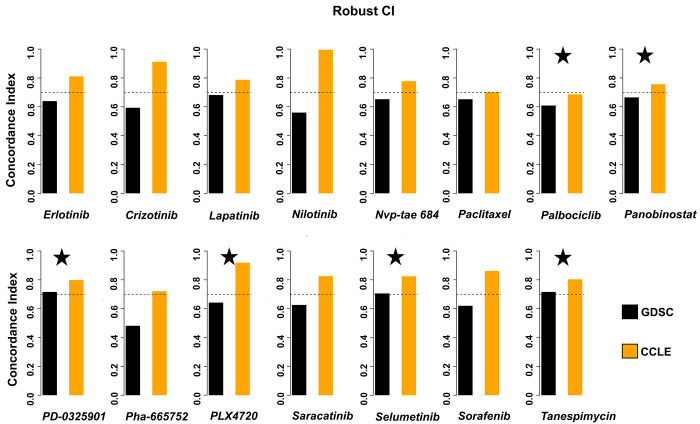
We also calculated the concordance index (rCI) because recognizing that drug screening assay noise is substantial and that cell-line-sensitive-based ordering with comparable AAC values might be inaccurate, the rCI only inspects cell line groupings having drug sensitivity (AAC). We can observe that all our shortlisted six drugs show consistency between the two databases (Figure 3). Among others, the GDSC data for Palbociclib fall short, not reaching the standard line, but it is negligible.
Figure 3.
Improved drug consistency across the databases. We can observe that all our six shortlisted drugs are consistent between the two databases. Among others, the GDSC data for Palbociclib fall short, not reaching the standard line, but it is negligible. Our shortlisted drugs are tagged with stars.
Machine-Learning-Based Drug Response Accuracy Comparison
Due to the existence of screening technologies, there is presently a tremendous volume of sensitivity and drug compound information to be tested on cancer cell lines. Therefore, in silico approaches to assess this data directly benefit anticancer strategies as they help to recognize molecular causes of drug sensitivity based on which novel anticancer drugs could be proposed. Here, we used three regression approaches named Elastic Net, LASSO, and Ridge for the drug sensitivity prediction (Figure 4). Out of the two databases, the GDSC and CCLE are used for training (Figure 4a) and testing (Figure 4b), respectively.
Figure 4.
Supervised comparison. This figure shows three regression approaches named Elastic Net, LASSO, and Ridge for drug sensitivity prediction. Parts a and b show the training and testing, respectively. This indicates that the training of all PLX4720 offers a higher rCI value of 86% using the LASSO and Ridge methods.
We can observe that PLX4720 shows a higher rCI value of 86% using the LASSO and Ridge methods. Selumetinib achieved the second highest rCI value, equal to 84% on all three methods. Tanespimycin ranks third in rCI values equal to 0.83 on Elastic Net and 0.82 using both the LASSO and Ridge methods. The least rCI is observed in the case of Palbociclib using Ridge regression equal to 0.75.
Validation results on the CCLE database are not significant for Palbocicilib and PLX4720, but the rest of the four drugs have higher performance. Panobinostat ranks first with an rCI of 0.67 on all three methods. Palbociclib has the least rCI value of 0.54.
Interestingly, the validation accuracy in the case of Panobinostat on all three methods is 67% higher than the rest of the compounds. PLX4720 and Sulumetinib have the same accuracy of 57% and 64%, respectively, on all the models. PD-0325901 is more effective than Palbociclib and PLX4720, with an rCI of 62%. The accuracy gained by various models in the case of Tanespimycin is 65%.
To summarize, Palbocicilib has relatively lower performance on the breast cancer cell lines, as seen in the training and validation results of the three regression approaches and drug–cell response (Figure 3) curves.
Drug-Sensitive Biomarker Identification
An objective measurement that captures what is occurring in a cell or an organism at a specific time is known as a biomarker (short for biological marker). Biomarkers can act as early health warning systems. They help comprehend basic biological mechanisms, develop the field of exposure science, and translate their discoveries into useful applications in the fields of medicine and public health. An object or trait that occurs naturally is associated with a particular clinical or biological process and may be used to identify a patient’s individual biomarker. Here we explored the linkage between drugs and screened biomarkers in the CCLE and GDSC databases. Among the 12 shortlisted biomarkers, we observed that nine are common between the databases, showing significant drug sensitivity (Table 2). Among them, tumor necrosis factor superfamily-15 (TNFSF15) is observed to have no or limited expression in breast cancer tumor vasculatures.45 The Chromosome 19 open reading frame 44 (C19orf44) is rarely mutated in cancer, particularly in breast cancer. According to the data curated from the COSMIC database, the total percentage of C19orf44 mutations in breast cancer is 1.16. We do not consider it a potential marker for breast cancer because of its low mutation rate and role in cancers. The 3-hydroxy acyl-CoA dehydratase 4 (HACD4) is a tumor suppressor similar to CDKN2A and CDKN2B but has not been associated with breast cancer.46
Table 2. Drug-Sensitive CCLE and GDSC Biomarkers.
| Gene ID | Gene | Estimate | Se | n | tstat | fstat | p-value | df | fdr |
|---|---|---|---|---|---|---|---|---|---|
| CCLE Biomarkers | |||||||||
| ENSG00000181634 | TNFSF15 | –0.0728188 | 0.0522639 | 423 | –1.3932912 | 1.9412603 | 0.1643055 | 400 | 0.6266466 |
| ENSG00000260589 | STAM-DTa | –0.0146833 | 0.0473970 | 423 | –0.3097940 | 0.0959723 | 0.7568789 | 400 | 0.9429905 |
| ENSG00000105072 | C19orf44 | –0.0264011 | 0.0482904 | 423 | –0.5467150 | 0.2988972 | 0.5848794 | 400 | 0.8852994 |
| ENSG00000275202 | novel gene-lncRNAa | 0.0173807 | 0.0464024 | 423 | 0.3745650 | 0.1402990 | 0.7081825 | 400 | 0.9290295 |
| ENSG00000188921 | HACD4 | –0.0222355 | 0.0492587 | 423 | –0.4514020 | 0.2037638 | 0.6519445 | 400 | 0.9108663 |
| ENSG00000143164 | DCAF6 | 0.0324883 | 0.0475525 | 423 | 0.6832091 | 0.4667747 | 0.4948702 | 400 | 0.8463414 |
| GDSC Biomarkers | |||||||||
| ENSG00000230294 | LINC02370 | –0.0052223 | 0.0334989 | 865 | –0.1558949 | 0.0243032 | 0.8761535 | 837 | 0.9493158 |
| ENSG00000147050 | KDM6A | –0.980172 | 0.0349182 | 865 | –2.8070452 | 7.8795029 | 0.0051160 | 837 | 0.0549044 |
| ENSG00000177096 | PHETA2 | 0.0082807 | 0.0344838 | 865 | 0.2401343 | 0.0576645 | 0.8102849 | 837 | 0.9207868 |
| ENSG00000160216 | AGPAT3 | –0.0588239 | 0.0350576 | 865 | –1.6779221 | 2.8154226 | 0.0937356 | 837 | 0.3082110 |
| ENSG00000027697 | IFNGR1 | –0.0086494 | 0.0361214 | 865 | –0.2394544 | 0.0573384 | 0.8108118 | 837 | 0.9210609 |
| ENSG00000159293 | uncategorized genea | –0.0539725 | 0.0341210 | 865 | –1.5817973 | 2.5020826 | 0.1140736 | 837 | 0.3435866 |
This biomarker is not common between CCLE and GDSC.
In the case of the DDB1 and CUL4 Associated Factor 6 (DCAF6), there are no targeted therapeutic data available for this gene, and its mutation percentage is also relatively low (4.46%) according to the COSMIC database; however, one of the recent studies claimed the presence of circ_DCAF6 at high levels in breast cancer cells.47 Regarding its functional roles, a curbed proliferation and stemness by breast cancer cells is observed during circ_DCAF6 absence. Furthermore, the Lysine Demethylase 6A (KDM6A) is typically linked with Kabuki Syndrome, but studies unveiled its potential role in cancer.48,49 The human KDM6A is observed to be oncogenic in the breast cancer scenario, but it acts as a tumor suppressor in T-cell acute lymphoblastic leukemia.
PHETA2 has low cancer specificity but is detected in numerous cancer forms. Its expression in breast cancer cells is moderate. AGPAT3 is a prognostic biomarker for renal and cervical cancer with moderate expression in breast cancer cells. For another biomarker known as IFN-γ receptor 1 (IFNGR1), its presence on cellular surfaces is a criterion for the IFN-γ signaling initiation and its reduced expression would result in blocking the IFN-γ signaling. In the breast cancer scenario, IFNGR1 expression is abridged or wholly lost and the immunoreactivity of heterogeneous IFNGR1 is linked to the morphological heterogeneity in breast cancer cells.50 One of the essential biomarkers shortlisted in our study is long intergenic nonprotein-coding RNA 2370 (LINC02370), an RNA gene that has a significant role in cancers.51,52
To summarize, out of these nine drug-sensitive biomarkers, we consider TNFSF15, DCAF6, KDM6A, PHETA2, IFNGR1, and LINC02370 as potential biomarkers that are drug-sensitive. These are further selected for their radiosensitivity or radioresistance.
Drug- and Radiation-Sensitive Biomarker Identification
On the other hand, similar to pharmacogenomics, radiogenomics follows a comparable strategy. The difference exists in the method of treating cells. In pharmacogenomics, one applies drugs to a particular target, while, in radiogenomics, the targets are exposed to certain radiations. Currently, there is only one clinical database known as the Cleveland database that stores the in vitro data of radiogenomics. This data set only has gamma radiations, and no special reason has been provided for why alpha and beta radiations are not used. For radiation retrieval for a cell line summary of a sensitivity experiment, the SummarizeSensitivityProfiles function is used. This returns with a matrix where rows are the radiation type, columns are cell lines, and values are viability measurements summarized. We can specify the sensitivity measures through the sensitivity.measure function.
We obtained a list of the top ten radiosensitive breast cancer biomarkers, including ALDH3B2, TMEM86A, SERINC3, CD63, MAGED2, SRARP, GSN, YIPF5, ST6GALNAC2, and LAMP2. Among the radioresistant, we observed LSM6, LBR, DDX21, GAR1, ADAT2, KDM1A, RRP1B, POLR3G, TAF5, and ILF3 genes (Table 3).
Table 3. Radiosensitive and Radioresistant Biomarkers from the Cleveland Database (RSet)a.
| Gene ID | Gene name | Estimate | Se | n | tstat | fstat | p-value | df | fdr |
|---|---|---|---|---|---|---|---|---|---|
| Radiosensitive Biomarkers | |||||||||
| ENSG00000132746 | ALDH3B2 | 0.3044503 | 0.0546364 | 517 | 5.572294 | 31.05046 | 0.0e+00 | 493 | 0.0000246 |
| ENSG00000151117 | TMEM86A | 0.2941997 | 0.0447234 | 517 | 6.578209 | 43.27284 | 0.0e+00 | 493 | 0.0000007 |
| ENSG00000132824 | SERINC3 | 0.2797229 | 0.0453765 | 517 | 6.164491 | 38.00094 | 0.0e+00 | 493 | 0.0000038 |
| ENSG00000135404 | CD63 | 0.2734318 | 0.0468322 | 517 | 5.838538 | 34.08852 | 0.0e+00 | 493 | 0.0000125 |
| ENSG00000102316 | MAGED2 | 0.2598921 | 0.0504084 | 517 | 5.155734 | 26.58159 | 4.0e-07 | 493 | 0.0000900 |
| ENSG00000183888 | SRARP | 0.2589937 | 0.0510493 | 517 | 5.073401 | 25.73940 | 6.0e-07 | 493 | 0.0001125 |
| ENSG00000148180 | GSN | 0.2561421 | 0.2561421 | 517 | 5.513927 | 30.40339 | 1.0e-07 | 493 | 0.0000265 |
| ENSG00000145817 | YIPF5 | 0.2521606 | 0.0474171 | 517 | 5.317924 | 28.28031 | 2.0e-07 | 493 | 0.0000497 |
| ENSG00000070731 | ST6GALNAC2 | 0.2491023 | 0.0527658 | 517 | 4.720902 | 22.28691 | 1e-06 | 493 | 0.000348 |
| ENSG00000005893 | LAMP2 | 0.2478066 | 0.0477752 | 517 | 5.186926 | 26.90421 | 3.0e-07 | 493 | 0.0000798 |
| Radioresistant Biomarkers | |||||||||
| ENSG00000164167 | LSM6 | –0.2604442 | 0.0426428 | 517 | –6.107574 | 37.30246 | 0 | 493 | 3.80e-06 |
| ENSG00000143815 | LBR | –0.2620366 | 0.0460308 | 517 | –5.692641 | 32.40616 | 0 | 493 | 2.08e-05 |
| ENSG00000165732 | DDX21 | –0.2632837 | 0.0448446 | 517 | –5.871027 | 34.46896 | 0 | 493 | 1.24e-05 |
| ENSG00000109534 | GAR1 | –0.2659226 | 0.0434707 | 517 | –6.117281 | 37.42112 | 0 | 493 | 3.80e-06 |
| ENSG00000189007 | ADAT2 | –0.2668198 | 0.0473300 | 517 | –5.637440 | 31.78073 | 0 | 493 | 2.23e-05 |
| ENSG00000004487 | KDM1A | –0.2681836 | 0.0436481 | 517 | –6.144227 | 37.75152 | 0 | 493 | 3.80e-06 |
| ENSG00000160208 | RRP1B | –0.2715109 | 0.0432827 | 517 | –6.244118 | 38.98902 | 0 | 493 | 3.80e-06 |
| ENSG00000113356 | POLR3G | –0.2785138 | 0.0454217 | 517 | –6.131739 | 37.59823 | 0 | 493 | 3.80e-06 |
| ENSG00000148835 | TAF5 | –0.2852464 | 0.0433393 | 517 | –6.581702 | 43.31880 | 0 | 493 | 7.00e-07 |
| ENSG00000129351 | ILF3 | –0.2929693 | 0.0426313 | 517 | –6.872171 | 47.22673 | 0 | 493 | 3.00e-07 |
All the top ten radiosensitive biomarkers are present in both the GDSC and CLLE.
Additionally, we also plotted the shortlisted biomarkers’ (TNFSF15, DCAF6, KDM6A, PHETA2, IFNGR1, LINC02370) correlation coefficient with the proposed drugs (Figure S3). Here, we compared the standardized coefficients per marker using the genome-wide correlation, weighted by the degree to which we felt the gene was useful for predicting response. The resulting score ranging from 0 to 1 may be considered a correlation coefficient. According to a positive connection, cells that respond to the drugs Palbociclib, Selumetinib, and Tanespimycin are distinct from those that respond to radiation. Using drugs like Panobinostat and PLX4720 as radiosensitizing agents with ionizing radiation might increase the effectiveness of treatment since there is a negative association between the radiation response signature and medication response. It is erroneous to assume that radiation and drugs would target separate cell types in a tumor based on the negative correlation of the signatures. The radiation score and p-values of all the shortlisted drugs are given in Table 4.
Table 4. Comparing Sensitivity Signatures between Radiation and Drug Response.
| Drugs | Radiation score | p-value |
|---|---|---|
| Palbociclib | 0.007511721 | 0.210000000 |
| Panobinostat | –0.491200093 | 0.009950249 |
| PLX4720 | –0.196630872 | 0.009950249 |
| Selumetinib | 0.571017489 | 0.009950249 |
| PD-0325901 | 0.2860125463 | 0.009950249 |
| Tanespimycin | 0.293541670 | 0.009950249 |
Discussion
Among the major cancer types, breast carcinoma is the leading cause of cancer deaths in women. Chemotherapy is the way to go as it greatly retards tumor growth, but this is a short-term effect and cannot be relied on. Also, certain drugs are not recommended for a cancer patient with particular mutations.53 This could be because of certain gene mutations or overall resistance to drugs. Therefore, it becomes crucial to look for new compounds as therapeutic agents for cancer and validate the effect of existing drugs on particular cancer. Though the available drugs being administered are FDA-approved and have been through a series of experimental and clinical trials, it is still encouraging to use advanced technology like machine learning for another round of validation. This is important because machine learning models may help find common patterns in the tumorous cell lines, making it clear to shortlist a drug suitable for most cancer cell lines. Inspired by the performance of machine learning models and our motivation for studying breast cancer, we considered the available breast cancer cell line data and drugs after intersecting two widely used data sets known as CCLE and GDSE. Since these databases already have the existing drug–cell line data, we shortlisted the medications based on their IC50 and AAC performance. Now, there are drugs whose experimental performance may not be significant because they are either the best drugs or not for breast cancer. However, because of human or systematic errors, they did not show satisfactory performance during the experiments. It is essential to mention that several drugs exist but are not thoroughly tested. We aimed to double-validate the already studied drugs’ performance on breast cancer cell lines only.
Once the drugs were shortlisted based on their excellent experimental results (please refer to the IC50 and AAC curves in the Supporting Information), we compared the results of each drug using a supervised approach. It is always a good idea to use more than one approach for validation to avoid biased results. This is the reason we considered three linear regression models known as LASSO, Elastic Net, and Ridge in our study. Usually, in such machine learning studies, a given data set is divided into 80% and 20% for training and testing, correspondingly. However, in our case, we used two different data sets for training and testing (GDSC and CCLE, respectively); this is highly recommended to test the model’s performance on entirely new data, pushing its accuracy performance to the limits. Upon observing the outcomes of three different machine learning models, it is quite interesting to know that, during the training process, different drugs are ranked differently by the models though few drugs are ranked equally by the three models; as we can see in the case of Selumetinib and PD-0325901 (Figure 4), the testing results show all three models give the same score to each drug. We do not know how this could be possible, but our understanding is that all three models provided accurate results on the same drug, giving the same value, or it is just a fluke. No matter what, the results are consistent with the previous analysis in this paper. Therefore, we maintain that the outcomes are significant and these drugs are truly effective on the breast cancer cell lines.
Since this is a pharmacogenomic approach, we cannot ignore the genes up- or downregulated as a result of these drugs. We provided a list of the top ten biomarkers sensitive to these drugs and expressed in the breast cancer scenario. Apart from the gene–drug relationship, during cancer treatment (breast cancer in this case) as mentioned earlier, chemotherapy is effective but has a short-term effect. Therefore, to cope with this situation, we explored biomarkers that are radiosensitive, so those genes which are chemo- and radiosensitive toward the shortlisted drugs are indeed potential targets for the treatment of breast cancer.
It is also worth noting that careful selection relying on a mechanistic approach or primary experimental results would benefit from the increased quantity of screened drugs. An outstanding demonstration is an NCI60 column, which used a separate viability assay through both CTRP and GDSC as the objective research yet showed significantly better prediction performance for CTRP versus GDSC. This might be related to CTRP’s development of an Informer Set comprising 481 chemicals that target above 250 different proteins and target a variety of biological processes associated with cancer cell line expansion.54 Certain probe compounds were chosen because they could cause distinct variations in gene expression profiles even if they had no preidentified protein targets.
All of this shows that putting pharmacological variety first in future screening studies would be advantageous. Intelligent approaches that can make inferences about molecule categories are required due to the enormous design space of chemical compounds that have the potential to be active, which is thought to number in the order of 1060.55
Conclusion
In both the early and late stages of the disease, chemotherapy is a widely used, systematic way to treat cancer patients. Due to the paucity of targeted medicines and the poor prognosis of breast cancer patients, considerable effort has been made to find responsive molecular targets for therapy. Despite the increasing accessibility of data from high-throughput drug sensitivity testing, effective drug response forecasting remains challenging. Understanding cell line–drug response models will eventually allow tailored medication sensitivity predictions for specific cancer patients.56
The current work predicts drug sensitivity on breast cancer cell lines, shortlists six biomarkers, and evaluates their response to the drugs and radiation exposure. The highest accuracy in the case of drug sensitivity prediction is observed for PLX4720 drugs using the Elastic Net and Ridge methods. Five main biomarkers named TNFSF15, DCAF6, KDM6A, PHETA2, and IFNGR1 show sensitivity toward all medicines and radiations. At the same time, the lncRNA (LINC02370) is a potential biomarker but is not tested for its response toward drugs and radiations in this study.
This study provides a strong foundation for machine-learning-based drug sensitivity prediction for breast cancer which has not been explored at this level before. The shortlisted markers could be potential therapeutic targets. We expect future studies to explore the molecular modeling of shortlisted drugs and biomarkers computationally and through experimental assays. This insight will get us closer to the era of tailored cancer therapy.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author upon request.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.2c00212.
Figures showing IC50 curves, AAC curves, and biomarker–drug response correlation and table showing categorization, molecular information, and culture conditions of breast cancer cell lines (PDF)
Author Contributions
A.M. conceptualized the project and wrote the initial draft. A.M. and A.C.K. performed the data analysis. S.N. assisted in supplementary results interpretation and plotting. H.H. assisted in manuscript editing. D.-Q.W. advised on method improvements and supervised the whole project. All authors read and approved the final manuscript.
D.-Q.W. is supported by grants from the National Science Foundation of China (Grant Nos. 32070662, 61832019, 32030063), the Science and Technology Commission of Shanghai Municipality (Grant No. 19430750600), as well as SJTU JiRLMDS Joint Research Fund and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2021ZD02). The computations were partially performed at the Pengcheng Lab. and the Center for High-Performance Computing, Shanghai Jiao Tong University.
The authors declare no competing financial interest.
Supplementary Material
References
- Rivera E.; Gomez H. Chemotherapy resistance in metastatic breast cancer: the evolving role of ixabepilone. Breast Cancer Research 2010, 12 (2), 1–12. 10.1186/bcr2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J.; Caponigro G.; Stransky N.; Venkatesan K.; Margolin A. A.; Kim S.; Wilson C. J.; Lehár J.; Kryukov G. V.; Sonkin D. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483 (7391), 603–607. 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seashore-Ludlow B.; Rees M. G.; Cheah J. H.; Cokol M.; Price E. V.; Coletti M. E.; Jones V.; Bodycombe N. E.; Soule C. K.; Gould J. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer discovery 2015, 5 (11), 1210–1223. 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F.; Knijnenburg T. A.; Vis D. J.; Bignell G. R.; Menden M. P.; Schubert M.; Aben N.; Gonçalves E.; Barthorpe S.; Lightfoot H. A landscape of pharmacogenomic interactions in cancer. Cell 2016, 166 (3), 740–754. 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverty P. M.; Lin E.; Tan J.; Yu Y.; Lam B.; Lianoglou S.; Neve R. M.; Martin S.; Settleman J.; Yauch R. L. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature 2016, 533 (7603), 333–337. 10.1038/nature17987. [DOI] [PubMed] [Google Scholar]
- Papillon-Cavanagh S.; De Jay N.; Hachem N.; Olsen C.; Bontempi G.; Aerts H. J.; Quackenbush J.; Haibe-Kains B. Comparison and validation of genomic predictors for anticancer drug sensitivity. Journal of the American Medical Informatics Association 2013, 20 (4), 597–602. 10.1136/amiajnl-2012-001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I. S.; Neto E. C.; Guinney J.; Friend S. H.; Margolin A. A. Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data. Biocomputing 2014, 63–74. 10.1142/9789814583220_0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamara A.; Tobalina L.; Saez-Rodriguez J. How to find the right drug for each patient? Advances and challenges in pharmacogenomics. Current opinion in systems biology 2018, 10, 53–62. 10.1016/j.coisb.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe-Kains B.; El-Hachem N.; Birkbak N. J.; Jin A. C.; Beck A. H.; Aerts H. J.; Quackenbush J. Inconsistency in large pharmacogenomic studies. Nature 2013, 504 (7480), 389–393. 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safikhani Z.; Smirnov P.; Freeman M.; El-Hachem N.; She A.; Rene Q.; Goldenberg A.; Birkbak N. J.; Hatzis C.; Shi L. Revisiting inconsistency in large pharmacogenomic studies. F1000Research 2016, 5, 2333. 10.12688/f1000research.9611.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharmacogenomic agreement between two cancer cell line data sets. Nature 2015, 528 (7580), 84–87. 10.1038/nature15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeleher P.; Gamazon E. R.; Seoighe C.; Cox N. J.; Huang R. S. Consistency in large pharmacogenomic studies. Nature 2016, 540 (7631), E1–E2. 10.1038/nature19838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhaddou M.; DiStefano M. S.; Riesel E. A.; Carrasco E.; Holzapfel H. Y.; Jones D. C.; Smith G. R.; Stern A. D.; Somani S. S.; Thompson T. Drug response consistency in CCLE and CGP. Nature 2016, 540 (7631), E9–E10. 10.1038/nature20580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpindi J. P.; Yadav B.; Östling P.; Gautam P.; Malani D.; Murumägi A.; Hirasawa A.; Kangaspeska S.; Wennerberg K.; Kallioniemi O. Consistency in drug response profiling. Nature 2016, 540 (7631), E5–E6. 10.1038/nature20171. [DOI] [PubMed] [Google Scholar]
- Baumann M.; Krause M.; Overgaard J.; Debus J.; Bentzen S. M.; Daartz J.; Richter C.; Zips D.; Bortfeld T. Radiation oncology in the era of precision medicine. Nature Reviews Cancer 2016, 16 (4), 234–249. 10.1038/nrc.2016.18. [DOI] [PubMed] [Google Scholar]
- Geeleher P.; Cox N. J.; Huang R. S. Clinical drug response can be predicted using baseline gene expression levels and in vitrodrug sensitivity in cell lines. Genome biology 2014, 15 (3), 1–12. 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y.; Qin Y.; Zhang N.; Wang J.; Wang H.; Zheng X. DISIS: prediction of drug response through an iterative sure independence screening. PloS one 2015, 10 (3), e0120408 10.1371/journal.pone.0120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgreen S.; Dybkær K.; Young K. H; Xu-Monette Z. Y; El-Galaly T. C; Laursen M. B.; Bødker J. S; Kjeldsen M. K; Schmitz A.; Nyegaard M.; Johnsen H. E.; Bøgsted M. Predicting response to multidrug regimens in cancer patients using cell line experiments and regularised regression models. BMC cancer 2015, 15 (1), 235. 10.1186/s12885-015-1237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aben N.; Vis D. J.; Michaut M.; Wessels L. F. TANDEM: a two-stage approach to maximize interpretability of drug response models based on multiple molecular data types. Bioinformatics 2016, 32 (17), i413–i420. 10.1093/bioinformatics/btw449. [DOI] [PubMed] [Google Scholar]
- Dong Z.; Zhang N.; Li C.; Wang H.; Fang Y.; Wang J.; Zheng X. Anticancer drug sensitivity prediction in cell lines from baseline gene expression through recursive feature selection. BMC cancer 2015, 15 (1), 1–12. 10.1186/s12885-015-1492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menden M. P.; Iorio F.; Garnett M.; McDermott U.; Benes C. H.; Ballester P. J.; Saez-Rodriguez J. Machine learning prediction of cancer cell sensitivity to drugs based on genomic and chemical properties. PLoS one 2013, 8 (4), e61318 10.1371/journal.pone.0061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M. Q.; Chen L.; Cooper G. F.; Young J. D.; Lu X. Precision oncology beyond targeted therapy: combining omics data with machine learning matches the majority of cancer cells to effective therapeutics. Molecular cancer research 2018, 16 (2), 269–278. 10.1158/1541-7786.MCR-17-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammad-Ud-Din M.; Georgii E.; Gonen M.; Laitinen T.; Kallioniemi O.; Wennerberg K.; Poso A.; Kaski S. Integrative and personalized QSAR analysis in cancer by kernelized Bayesian matrix factorization. J. Chem. Inf. Model. 2014, 54 (8), 2347–2359. 10.1021/ci500152b. [DOI] [PubMed] [Google Scholar]
- Ammad-Ud-Din M.; Khan S. A.; Malani D.; Murumägi A.; Kallioniemi O.; Aittokallio T.; Kaski S. Drug response prediction by inferring pathway-response associations with kernelized Bayesian matrix factorization. Bioinformatics 2016, 32 (17), i455–i463. 10.1093/bioinformatics/btw433. [DOI] [PubMed] [Google Scholar]
- Ali M.; Aittokallio T. Machine learning and feature selection for drug response prediction in precision oncology applications. Biophysical reviews 2019, 11 (1), 31–39. 10.1007/s12551-018-0446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss F.; Hetzenecker S.; Osterhaus M. Nonparametric estimation of the random coefficients model: An elastic net approach. Journal of Econometrics 2022, 229, 299. 10.1016/j.jeconom.2020.11.010. [DOI] [Google Scholar]
- Huang X.; Cai W.; Yuan W.; Peng S. Identification of key lncRNAs as prognostic prediction models for colorectal cancer based on LASSO. International journal of clinical and experimental pathology 2020, 13 (4), 675. [PMC free article] [PubMed] [Google Scholar]
- Arashi M.; Roozbeh M.; Hamzah N.; Gasparini M. Ridge regression and its applications in genetic studies. PLoS One 2021, 16 (4), e0245376 10.1371/journal.pone.0245376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.; Soares J.; Greninger P.; Edelman E. J.; Lightfoot H.; Forbes S.; Bindal N.; Beare D.; Smith J. A.; Thompson I. R. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic acids research 2012, 41 (D1), D955–D961. 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Zhang T.; Madunić K.; de Waard A. A.; Blöchl C.; Mayboroda O. A.; Griffioen M.; Spaapen R. M.; Huber C. G.; Lageveen-Kammeijer G. S. Glycosphingolipid-Glycan Signatures of Acute Myeloid Leukemia Cell Lines Reflect Hematopoietic Differentiation. J. Proteome Res. 2022, 21, 1029. 10.1021/acs.jproteome.1c00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X.; Cheng H.; Bai Z.; Li J. Breast cancer cell line classification and its relevance with breast tumor subtyping. Journal of Cancer 2017, 8 (16), 3131. 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckman H. J.; Trego K. S.; Turchi J. J. Cisplatin sensitizes cancer cells to ionizing radiation via inhibition of nonhomologous end joining. Molecular cancer research 2005, 3 (5), 277–285. 10.1158/1541-7786.MCR-04-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud H.; Haibe-Kains B. Drug sensitivity prediction modeling from genomics, transcriptomics and inferred protein activity. AACR 2020, 26, 33. 10.1158/1557-3265.ADVPRECMED20-33. [DOI] [Google Scholar]
- Trendowski M. R.; Baedke J. L.; Sapkota Y.; Travis L. B.; Zhang X.; El Charif O.; Wheeler H. E.; Leisenring W. M.; Robison L. L.; Hudson M. M. Clinical and genetic risk factors for radiation-associated ototoxicity: A report from the Childhood Cancer Survivor Study and the St. Jude Lifetime Cohort. Cancer 2021, 127 (21), 4091–4102. 10.1002/cncr.33775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Wang J.; Ghoraie L. S.; Men X.; Chen R.; Dai P. Comprehensive expression-based isoform biomarkers predictive of drug responses based on isoform co-expression networks and clinical data. Genomics 2020, 112 (1), 647–658. 10.1016/j.ygeno.2019.04.017. [DOI] [PubMed] [Google Scholar]
- Thorarensen A.; Balbo P.; Banker M. E.; Czerwinski R. M.; Kuhn M.; Maurer T. S.; Telliez J.-B.; Vincent F.; Wittwer A. J. The advantages of describing covalent inhibitor in vitro potencies by IC50 at a fixed time point. IC50 determination of covalent inhibitors provides meaningful data to medicinal chemistry for SAR optimization. Bioorg. Med. Chem. 2021, 29, 115865 10.1016/j.bmc.2020.115865. [DOI] [PubMed] [Google Scholar]
- Govindaraj R. G.; Subramaniyam S.; Manavalan B. Extremely-randomized-tree-based Prediction of N6-Methyladenosine Sites in Saccharomyces cerevisiae. Current Genomics 2020, 21 (1), 26–33. 10.2174/1389202921666200219125625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisu A. A.; Akanni L.; Raheem I. The COVID-19 global fear index and the predictability of commodity price returns. Journal of Behavioral and Experimental Finance 2020, 27, 100383 10.1016/j.jbef.2020.100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov P.; Smith I.; Safikhani Z.; Ba-alawi W.; Khodakarami F.; Lin E.; Yu Y.; Martin S.; Ortmann J.; Aittokallio T.. Evaluation of statistical approaches for association testing in noisy drug screening data. arXiv, 2021, arXiv:2104.14036. 10.1186/s12859-022-04693-z. [DOI] [PMC free article] [PubMed]
- Radovic M.; Ghalwash M.; Filipovic N.; Obradovic Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC bioinformatics 2017, 18 (1), 1–14. 10.1186/s12859-016-1423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silge J.; Robinson D. tidytext: Text mining and analysis using tidy data principles in R. Journal of Open Source Software 2016, 1 (3), 37. 10.21105/joss.00037. [DOI] [Google Scholar]
- Kuhn M. A Short Introduction to the caret Package. R Found Stat Comput 2015, 1, 1–10. [Google Scholar]; https://cran.microsoft.com/snapshot/2016-03-01/web/packages/caret/vignettes/caret.pdf.
- Allaire J. RStudio: integrated development environment for R 2012, 770 (394), 165–171. [Google Scholar]; https://www.r-project.org/conferences/useR-2011/abstracts/180111-allairejj.pdf.
- Smirnov P.; Safikhani Z.; El-Hachem N.; Wang D.; She A.; Olsen C.; Freeman M.; Selby H.; Gendoo D. M.; Grossmann P. PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics 2016, 32 (8), 1244–1246. 10.1093/bioinformatics/btv723. [DOI] [PubMed] [Google Scholar]
- Deng W.; Gu X.; Lu Y.; Gu C.; Zheng Y.; Zhang Z.; Chen L.; Yao Z.; Li L.-Y. Down-modulation of TNFSF15 in ovarian cancer by VEGF and MCP-1 is a pre-requisite for tumor neovascularization. Angiogenesis 2012, 15 (1), 71–85. 10.1007/s10456-011-9244-y. [DOI] [PubMed] [Google Scholar]
- Zivotić I.; Djurić T.; Stanković A.; Ivančević I.; Končar I.; Milasinovic D.; Stankovic G.; Alavantić D.; Zivković M. The HACD4 haplotype as a risk factor for atherosclerosis in males. Gene 2018, 641, 35–40. 10.1016/j.gene.2017.10.030. [DOI] [PubMed] [Google Scholar]
- Ye G.; Pan R.; Zhu L.; Zhou D. Circ_DCAF6 potentiates cell stemness and growth in breast cancer through GLI1-Hedgehog pathway. Experimental and Molecular Pathology 2020, 116, 104492 10.1016/j.yexmp.2020.104492. [DOI] [PubMed] [Google Scholar]
- Kim J.-H.; Sharma A.; Dhar S. S.; Lee S.-H.; Gu B.; Chan C.-H.; Lin H.-K.; Lee M. G. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer research 2014, 74 (6), 1705–1717. 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meulen J.; Sanghvi V.; Mavrakis K.; Durinck K.; Fang F.; Matthijssens F.; Rondou P.; Rosen M.; Pieters T.; Vandenberghe P. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood, The Journal of the American Society of Hematology 2015, 125 (1), 13–21. 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.; Guo L.; Shi M.; Hu M.; Hu M.; Yu M.; Wang T.; Song L.; Shen B.; Qian L. Modulation of IFN-γ receptor 1 expression by AP-2α influences IFN-γ sensitivity of cancer cells. American journal of pathology 2012, 180 (2), 661–671. 10.1016/j.ajpath.2011.10.040. [DOI] [PubMed] [Google Scholar]
- Lin P.-C.; Chen H.-O.; Lee C.-J.; Yeh Y.-M.; Shen M.-R.; Chiang J.-H. Comprehensive assessments of germline deletion structural variants reveal the association between prognostic MUC4 and CEP72 deletions and immune response gene expression in colorectal cancer patients. Human genomics 2021, 15 (1), 1–13. 10.1186/s40246-020-00302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othoum G.; Coonrod E.; Zhao S.; Dang H. X.; Maher C. A. Pan-cancer proteogenomic analysis reveals long and circular noncoding RNAs encoding peptides. NAR cancer 2020, 2 (3), zcaa015 10.1093/narcan/zcaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood A.; Kaushik A. C.; Wang Q.; Li C.-D.; Wei D.-Q. Bringing structural implications and deep learning-based drug identification for KRAS mutants. J. Chem. Inf. Model. 2021, 61 (2), 571–586. 10.1021/acs.jcim.0c00488. [DOI] [PubMed] [Google Scholar]
- Seashore-Ludlow B.; Rees M. G.; Cheah J. H.; Cokol M.; Price E. V.; Coletti M. E.; Jones V.; Bodycombe N. E.; Soule C. K.; Gould J. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset Harnessing Connectivity in a Sensitivity Dataset. Cancer discovery 2015, 5 (11), 1210–1223. 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohacek R. S.; McMartin C.; Guida W. C. The art and practice of structure-based drug design: a molecular modeling perspective. Medicinal research reviews 1996, 16 (1), 3–50. . [DOI] [PubMed] [Google Scholar]
- Zhao C.; Li Y.; Safikhani Z.; Haibe-Kains B.; Goldenberg A.. Using Cell line and Patient samples to improve Drug Response Prediction. bioRxiv, 2015, 026534. 10.1101/026534. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author upon request.