Abstract
The TspO outer membrane protein of Rhodobacter sphaeroides has been shown to be involved in controlling the transcription of a number of genes which encode enzymes involved in photopigment biosynthesis and the puc operon. The display of regulated genes appears identical to those genes encompassing the PpsR/AppA repressor/antirepressor regulon, although the effect of TspO is modest relative to that of PpsR/AppA. To directly address the hypothesis that TspO is effective through the PpsR/AppA system, we constructed mutant strains with mutations in both tspO and appA. In all cases, the phenotypes examined resembled those of the appA lesion by itself, leading us to conclude that TspO works through or modulates the PpsR/AppA system and acts upstream of the site of action of these regulatory proteins. In earlier publications, we had suggested that TspO is involved in the efflux of a certain intermediate(s) of the porphyrin biosynthesis pathway and that transcriptional regulation of target gene expression could be explained by the accumulation of a coactivator of AppA function. Although the data reported here do not precisely identify this coactivator, they lend support to this hypothesis. We discuss the importance of this form of gene control as the result of the recent extension of the TspO system to Sinorhizobium meliloti, as described by Davey and de Bruijn (M. E. Davey and F. J. de Bruijn, Appl. Environ. Microbiol. 66:5353–5359, 2000). It is therefore possible that this system constitutes a more widely, although not universally, demonstrated form of gene regulation.
Rhodobacter sphaeroides 2.4.1 is a facultative photoheterotrophic bacterium which is remarkably versatile in its growth abilities (1). The photosynthetic apparatus, including the light-harvesting complexes I (B875) and II (B800-850) as well as the reaction center, is induced in response to variations in oxygen tension, and the levels of its components are ultimately determined by light intensity (11). Previous reports have shown that the PrrBA two-component activation system, FnrL, the PpsR/AppA repressor/antirepressor system, and the outer membrane-localized TspO protein are all required to regulate the orderly expression of photosynthesis (PS) genes (34).
The outer membrane-localized TspO protein of R. sphaeroides 2.4.1 was shown previously to negatively modulate, albeit partially, the transcriptional expression of those PS genes (e.g., puc, crtA, and crtI) which are also under the control of the PpsR/AppA repressor/antirepressor system (30, 31, 32, 33). The PpsR/AppA system extends maximal control over those genes comprising this regulon, whereas TspO only modulates the expression of these same genes. This is in keeping with the observation that TspO is only transiently effective as cells proceed from aerobic to anaerobic growth. TspO shows a high degree of homology to the mammalian mitochondrial peripheral benzodiazepine receptor, which binds benzodiazepines as well as dicarboxylic porphyrins with nanomolar affinity and which may function as (part of) an anion channel across the outer mitochondrial membrane for the import-export of intermediates in tetrapyrrole biosynthesis (17, 20, 21, 28–31). Our studies additionally suggested that TspO may be involved in the efflux of critical tetrapyrrole intermediates from R. sphaeroides 2.4.1 by forming a functional dimer in the outer membrane (30, 32), and we have elsewhere proposed a model for TspO action involving these intermediates (19, 32). We have also shown that the rat peripheral benzodiazepine receptor protein expressed in R. sphaeroides behaves like the bacterial TspO (33).
Because TspO appears to modulate exclusively the genes of the PpsR/AppA repressor/antirepressor regulon, we posed the question whether TspO activity is dependent or independent of PpsR/AppA. Disruption of appA encoding the antirepressor AppA was shown to lead to a substantially decreased expression of many PS genes and impaired production of both pigments and proteins comprising the spectral complexes. It has been demonstrated previously that AppA contains a bound flavin adenine dinucleotide which could allow it to function as a redox-sensing partner, communicating the redox state of the quinone pool (19) by directly interacting with PpsR (6–8). Since the proposed inactivation of PpsR by reduced AppA cannot take place in an AppA mutant strain (8, 19), the repressor PpsR remains fully functional even at low oxygen tensions, hence the maximal repression of this regulon due to a fully functional PpsR in the absence of AppA.
Thus, we reasoned that if TspO acts through the repressor/antirepressor system, it should be possible to demonstrate this relationship genetically. Because TspO only partially affects the genes of this regulon, the effects of TspO are modest but consistent. In the present study, we have constructed double mutant strains with tspO and appA, as well as tspO and puc, mutations in an effort to more precisely elucidate the role of TspO in the regulatory network controlling PS gene expression in R. sphaeroides.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in this work are listed in Table 1. R. sphaeroides 2.4.1 and the derived mutant strains were grown in Sistrom's minimal medium A containing 0.4% succinate as carbon source (1) as described previously (3). Antibiotics were added to the indicated final concentrations: kanamycin (KAN), 50 μg/ml; spectinomycin, 50 μg/ml; streptomycin, 50 μg/ml; tetracycline, 1 μg/ml; and trimethoprim, 50 μg/ml. Aerobic cells were grown under continuous sparging with a mixture of gases, 69% N2–30% O2–1% CO2. Semiaerobic cells were grown by sparging with a gas mixture of 97% N2, 2% O2, and 1% CO2. Anaerobic cells were grown in screw-cap glass tubes with dimethyl sulfoxide (DMSO) (0.5% [vol/vol]) and yeast extract (1% [vol/vol]) in the dark.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αphe | DH5αphe::Tn10dCm | 4 |
| HB101 | lacY1 galK2 supE44 ara-14 proA2 rpsL20 recA13 xyl-5 mtl-1 hsdS20 mcrB mrr | 12 |
| S17-1 | C600::RP-4 2-(Tc::Mu)(Km::Tn7) thi pro hsdR hsdM+recA | 25 |
| R. sphaeroides | ||
| 2.4.1 | Wild type | W. R. Sistrom |
| TSPO1 | tspO::Kmr | 31 |
| APP11 | appA::Tpr | 6 |
| APP-TSPO | APP11tspO::Kmr | This study |
| PUC-ZWT | lacZY::Ω Smr/SprA′ inserted at the XmnI sites within pucB of the wild type; B800-850− | 13 |
| PUCB-TSPO | PUC-ZWT tspO::Kmr | This study |
| Plasmids | ||
| pRK415 | Tcr | 10 |
| pBSIIKS+ | Apr; with T3 and T7 promoters | Stratagene |
| pUC4K | Source of Kmr | Pharmacia |
| pSUP202 | pBR325 derivative, Mob+ Ap+ Cm+ Tc+ | 25 |
| pAS204 | pRK415 containing the 2.1-kb SstI crtB, tspO fragment from pUI8487; Tcr | A. Suwanto |
| pUI1110 | pSUP202 containing 3.7-kb tspO::Km fragment inserted at the SspI site; Tc+ Km+ | M. Wood and S. Kaplan |
| pUI1124 | pBSIIKS+ containing tspO under PrrnB Ap+ | M. Wood and S. Kaplan |
| pUI1830Tp | Smr/Spr Tp+puf::lacZ | J. I. Oh and S. Kaplan |
| pCF200Km | Smr/Spr Km+puc::lacZ | 13 |
| pUI2701 | Derivative of pRK415 harboring 1.1-kb KpnI fragment of pUI1124 containing tspO under PrrnB; Tcr | 31 |
| pUI2730 | Tcr; derivative of pRK415 containing hemN | 30 |
| pUI2732 | Tcr; derivative of pRK415 containing hemN and tspO under PrrnB | 30 |
Escherichia coli strains were grown at 37°C in Luria broth (14). Antibiotics were added at the indicated final concentrations: ampicillin, 50 μg/ml; KAN, 50 μg/ml; spectinomycin, 50 μg/ml; streptomycin, 50 μg/ml; tetracycline, 10 μg/ml; and trimethoprim, 50 μg/ml.
Protein determination.
Protein concentrations were determined using the bicinchoninic acid protein assay (Pierce, Rockford, Ill.) with bovine serum albumin as the standard.
β-Galactosidase assay.
R. sphaeroides cultures were grown to a cell density of approximately 1.8 × 108 cells/ml, and chloramphenicol was added to a final concentration of 80 μg/ml. β-Galactosidase activity in the cell extracts was measured in three independent experiments as described previously (13). The activity of β-galactosidase is expressed in units, where 1 U is equal to 1 μmol of o-nitrophenyl-β-d-galactopyranoside cleaved/min/mg of protein.
Cell fractionation and spectrophotometric assays.
The cell crude extracts were prepared by the method of Tai et al. (26). R. sphaeroides cells were collected by centrifugation at 3,000 × g for 15 min, resuspended in ICM buffer (10 mM K2HPO4-KH2PO4 and 1 mM EDTA, pH 7.0), and then disrupted by passage through a French pressure cell (Aminco, Urbana, Ill.). Cell crude extracts were obtained by centrifugation at 16,000 × g for 15 min two times to remove unbroken cells and cell debris. All of the above steps were performed at 4°C. Absorption spectra were analyzed on a UV 1601 PC spectrophotometer (Shimadzu Corp., Columbia, Md.). Equivalent protein concentrations of cell crude extracts were used when the spectral profiles of different strains of R. sphaeroides were compared. The amount of B800-850 and B875 light-harvesting complexes was determined as described elsewhere (16). Photopigments were extracted with acetone-methanol (7/2 ratio [vol/vol]) from cell pellets as described elsewhere (1).
Construction of Ω cartridge insertion-tspO disruption strain.
APP11 or PUC-ZWT was used as the recipient for plasmid pUI1110, in which tspO was disrupted by inserting an Ω cartridge encoding KAN resistance. Matings were conducted on Luria broth solid medium, and exconjugants were then plated on selective media containing KAN for recipients of pUI1110. Recombinant strains with double crossovers were screened by selecting individual exconjugants for tetracycline sensitivity and KAN resistance. Plasmids were mobilized by biparental matings from E. coli S17-1 strains into R. sphaeroides as described elsewhere (3).
DNA manipulation and sequence analysis.
Standard protocols or manufacturer's instructions were followed for plasmid isolation, restriction endonuclease digestion, isolation of DNA fragments from gels, ligation, and other molecular biological techniques (14, 23). Sequence analyses were performed with the computer programs DNA Strider (Institut de Recherche Foundamentale, Commissariat a l'Energie Atomique, Paris, France).
Southern hybridization.
Total genomic DNA was isolated from R. sphaeroides 2.4.1 by a method described elsewhere (23). Genomic DNA was digested with the restriction enzyme BamHI, and 0.5-kb 32P-labeled BssHI-KpnI fragments of pUI1124 were used as radioactive probes. Southern hybridization was performed using the standard techniques (23). Labeling and detection were performed with an Instant Image instrument (Parkard Co.) following the manufacturer's instructions.
HPLC analysis of porphyrins.
Growing cells were collected at a cell density of approximately 1.8 × 108 cells/ml. Resting cells were prepared as described elsewhere with slight modification (30), 5-aminolevulinic acid (ALA) was added to a final concentration of 0.2 mM, and semiaerobically grown cells were incubated in 0.1 M phosphate buffer (pH 7.0) for 6 h by sparging with 97% N2–2% O2–1% CO2. Extraction of the porphyrin precursors excreted by resting and growing cells of R. sphaeroides was performed according to the method described previously (9, 22, 30). For extraction of porphyrin precursors within cells, resting or growing cells were collected by centrifugation at 10,000 × g for 15 min and washed two times with 0.1 mM potassium phosphate buffer. The cells were resuspended by adding 1 ml of concentrated HCl and vortexing for 2 min and then mixed thoroughly with 3 ml of ethyl ether by vortexing, followed by adding 3 ml of water and mixing again. To avoid undue alteration of protoporphyrin IX, water was added within 10 min. The mixture was centrifuged at 16,000 × g for 10 min, and the lower aqueous acid layer was used to detect the total porphyrin precursors produced by the cells according to the method of Rossi and Curnow (22). To prepare samples for high-performance liquid chromatography (HPLC) analysis, the aqueous layer was adjusted to pH 3.5 with sodium acetate, 0.2 g of talcum powder was added and mixed thoroughly, and the talcum was collected by filtration in a small (5-cm) Buchner funnel and washed two times with 10 ml of deionized water. Porphyrin precursors were eluted with 2 ml of a mixture of acetone–0.1 N HCl (9:1 [vol/vol]), and the acetone was evaporated under nitrogen at 45°C for 30 min to obtain the remaining HCl solution containing porphyrin precursors. Before HPLC analysis, the samples were treated with an equal volume of benzoquinone (6 mg/ml in ethyl ether) to oxidize porphyrinogens to porphyrins (24, 27), the aqueous layer was centrifuged at 16,000 × g for 10 min, and supernatant was taken for HPLC analysis.
HPLC analysis was performed on an SAS Hypersil (Keystone Scientific Inc., Bellefonte, Pa.) reversed-phase column (1.5 by 4.6 mm). The conditions for porphyrin acid analysis were as follows: the column was washed for 5 min with solvent A (acetonitrile–1 M ammonium acetate buffer [pH 5.16], 10:90 [vol/vol]), then a linear gradient of 0 to 100% solvent B (acetonitrile-methanol, 10:90 [vol/vol]) was applied within 20 min, 100% solvent B was applied for a further 5 min, and the run was ended at 31 min.
Materials.
Restriction endonucleases and nucleic acid-modifying enzyme were purchased from New England Biolabs, Inc. (Beverly, Mass.). Antibiotics, o-nitrophenyl-β-d-galactopyranoside, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and vitamins were obtained from Sigma (St. Louis, Mo.). Porphyrin acid standards were obtained from Porphyrin Products, Inc. (Logan, Utah).
RESULTS
Construction of Ω cartridge insertion-tspO disruption strain.
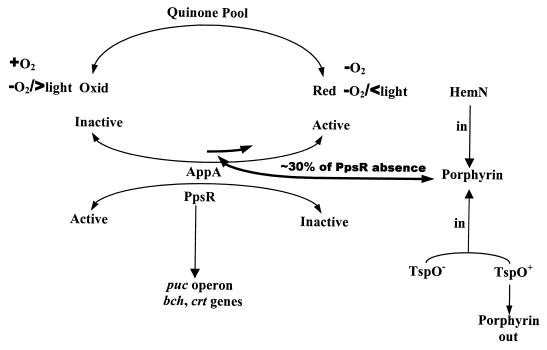
Mutations of tspO were crossed into either a PucB mutant strain, PUC-ZWT, which does not make light-harvesting complex II, or into an AppA mutant strain, APP11, which is altered in the trans-acting factor AppA, involved in the regulation of PS gene expression in R. sphaeroides (6). Since the loss of AppA results in substantially decreased PS gene expression for those genes under PpsR control, we reasoned that, if TspO operates independently of the PpsR/AppA system, then the loss of TspO should result in partially increased pigment production and gene expression in an AppA mutant background (30–33). This effect of a TspO mutation can at best be only partial, since TspO modulates only the affected PpsR/AppA regulon, unlike the AppA gene. On the other hand, if TspO acts through the PpsR/AppA system, loss of TspO in an AppA mutant, should resemble the AppA mutant. Thus, we are employing classic epistasis experiments to determine if TspO acts through the PpsR/AppA regulon. Figure 1 presents a model depicting the proposed interaction of PpsR, AppA, and TspO.
FIG. 1.
TspO, by affecting the internal porphyrin levels, can partially activate the antirepressor AppA. Complete activation or inactivation of the antirepressor AppA is under the control of the relative redox state of the quinone pool, which in turn is influenced by the presence or absence of O2 and in its absence, by light intensity, AppA in turn will determine the relative activity of the repressor, PpsR. Through this model, it is possible to visualize how TspO can partially affect (modulate) the genes representing the PpsR/AppA regulon.
Since the puc operon is part of the PpsR/AppA regulon and is partly controlled by the PpsR/AppA system, it was incorporated into these studies as a control. The double mutants PUCB-TSPO and APP-TSPO were obtained by replacement of the wild-type tspO gene with the Ω cartridge insertion-tspO (tspOΩ) in mutants PUC-ZWT and APP11, respectively. The disruption of tspO in the mutants was confirmed by hybridization of a radioactively labeled BssHI-KpnI fragment derived from pUI1124 to genomic DNA from the mutants. The pucB-containing mutant strain PUC-ZWT was used since, in addition to inactivating the structural gene(s) of the puc operon, the disruption of pucB incorporates lacZ downstream of the puc operon promoter.
Table 2 shows the results of an analysis of the spectral complexes derived from the wild-type strain R. sphaeroides 2.4.1 and its mutants grown anaerobically in the dark with DMSO. As shown in Table 2, there is a small but significant increase in spectral complex formation in TSPO1 compared to wild type as the result of increased pigment production in the TspO mutant strain (31). The double mutant strain involving tspO and pucB produced the same spectral profiles as the single mutant PUC-ZWT, in which the formation of the B875 complex is virtually unaffected by either mutation, although the B800-850 complexes are removed. Introduction of multiple copies of tspO partially suppressed photosynthetic complex formation in the double mutant strain PUCB-TSPO as well as in mutant TSPO1; the latter observation together with those for wild type had been reported previously and revealed that tspO is functional (31), i.e., multiple copies of tspO partially suppress pigment gene expression by removing the proposed coactivator of AppA, according to our model (Fig. 1).
TABLE 2.
Spectral complex formation of wild-type and mutant strains of R. sphaeroidesa
| Strain | B800-850 | B875 |
|---|---|---|
| 2.4.1 (wild type) | 42 ± 3.1 | 17 ± 0.9 |
| TSPO1 | 56 ± 4.7 | 19 ± 1.1 |
| TSPO1(pUI2701) | 31 ± 3.4 | 14 ± 0.7 |
| PUC-ZWT | 1.4 ± 0.9 | 17 ± 1.1 |
| PUCB-TSPO | 1.6 ± 1.0 | 18 ± 1.0 |
| PUCB-TSPO(pUI2701) | 1.1 ± 0.9 | 10 ± 0.8 |
| APP11 | 0.9 ± 0.7 | 0.6 ± 0.5 |
| APP-TSPO | 0.9 ± 0.6 | 0.7 ± 0.5 |
| APP-TSPO(pUI2701) | 0.8 ± 0.6 | 0.6 ± 0.6 |
Cells were grown anaerobically in the dark with DMSO (0.5% [vol/vol]) and yeast extract (0.1% [vol/vol]). Values are shown as nanomoles per milligram of protein.
As reported earlier, mutation of appA under these conditions leads to the absence of expression of the PS genes and impaired production of the photosynthetic complexes (6). Importantly, for these studies, the combination of the appA and tspO mutations resulted in levels of spectral complexes identical to levels observed for appA alone, i.e., the appA lesion is epistatic to the tspO mutation and resembles the appA mutation alone. Thus, had additional levels of spectral complexes been induced, even low levels, such as observed when comparing TSPO1 and R. sphaeroides 2.4.1 by the presence of the TspO lesion, these would have been readily observable against the background levels for the appA lesion alone. In the presence of extra copies of tspO, the AppA mutant strains were not further affected, since their levels were already minimal, unlike the results with the PucB mutant. This is certainly to be expected, since the AppA lesion already yields basal levels of spectral complexes. Thus, at the level of spectral complex formation, the AppA null mutation is dominant to the TspO null mutation, suggesting that the effect of the tspO lesion is not independent of the appA lesion and that if TspO acts through the PpsR/AppA system, it is upstream of the repressor/antirepressor in this pathway (Fig. 1). Again, we must point out that TspO only modulates PS gene expression, i.e., its effect is modest but readily recognizable under the appropriate experimental conditions.
The effect of tspO in an appA null mutation of R. sphaeroides.
We further reasoned that if a defective tspO exerted its partial effect (derepression) on selective PS gene expression through the selective activation of AppA of the PpsR/AppA system (6–8, 30, 32), then, in the absence of a functional AppA, we would not expect to witness an inactivation of the PpsR repressor but would see full PpsR repressor activity. Hence, the appA mutation should be epistatic to the tspO lesion. The experiment could not be performed in a PpsR-defective strain, since the genes of the PpsR/AppA regulon are fully induced in this mutant background and the partial effect of a TspO lesion is hence not observed. To assess the relationship between TspO and AppA, we compared the accumulations of bacteriochlorophyll (Bchl) and carotenoid (Crt) in the double mutant APP-TSPO and App11 under semiaerobic and anaerobic conditions (Table 3). The results of these experiments involving appA are unambiguous when taking into account the partial role of TspO and the trends and consistency of the results; the effect of the appA mutation on photopigment production under any condition is dominant to the presence of the tspO lesion, which by itself yields increased photopigment production, in keeping with increased spectral complex levels. Bchl and Crt determinations can be made very sensitive, depending upon the volume of culture extracted.
TABLE 3.
Crt and Bchl accumulated by wild-type and mutant strains of R. sphaeroides
| Strain | Amt (μg/mg of protein) for growth condition:
|
|||||
|---|---|---|---|---|---|---|
| Aerobica
|
Semiaerobicb
|
Dark-DMSOc
|
||||
| Bch1 | Crt | Bch1 | Crt | Bch1 | Crt | |
| 2.4.1 (wild type) | 0.061 ± 0.005 | 0.026 ± 0.002 | 1.96 ± 0.22 | 0.19 ± 0.02 | 1.71 ± 0.21 | 0.27 ± 0.03 |
| TSPO1 | 0.076 ± 0.008 | 0.034 ± 0.003 | 2.43 ± 0.27 | 0.25 ± 0.03 | 2.34 ± 0.25 | 0.32 ± 0.04 |
| TSPO1(pUI2701) | 0.060 ± 0.009 | 0.027 ± 0.002 | 1.38 ± 0.19 | 0.16 ± 0.02 | 1.31 ± 0.18 | 0.22 ± 0.02 |
| APP11 | 0.004 ± 0.003 | 0.003 ± 0.002 | 0.09 ± 0.05 | 0.08 ± 0.03 | 0.06 ± 0.04 | 0.04 ± 0.02 |
| APP-TSPO | 0.004 ± 0.002 | 0.002 ± 0.002 | 0.05 ± 0.04 | 0.06 ± 0.02 | 0.07 ± 0.05 | 0.05 ± 0.03 |
| APP-TSPO(pUI2701) | 0.005 ± 0.002 | 0.003 ± 0.002 | 0.06 ± 0.04 | 0.06 ± 0.02 | 0.06 ± 0.03 | 0.03 ± 0.02 |
| PUC-ZWT | 0.078 ± 0.007 | 0.031 ± 0.003 | 0.60 ± 0.09 | 0.18 ± 0.02 | 0.66 ± 0.08 | 0.22 ± 0.02 |
| PUCB-TSPO | 0.079 ± 0.008 | 0.030 ± 0.003 | 0.59 ± 0.07 | 0.23 ± 0.03 | 0.59 ± 0.15 | 0.27 ± 0.03 |
| PUCB-TSPO(pUI2701) | 0.075 ± 0.007 | 0.037 ± 0.004 | 0.27 ± 0.06 | 0.14 ± 0.02 | 0.25 ± 0.05 | 0.15 ± 0.03 |
Strains were grown by sparging with 69% N2–30% O2–1% CO2 to a cell density of approximately 1.8 × 108 cells/ml.
Strains were grown by sparging with 97% N2–2% O2–1% CO2 to a cell density of approximately 1.8 × 108 cells/ml.
Strains were grown anaerobically in the dark with DMSO (0.5% [vol/vol]) and yeast extract (0.1% [vol/vol]) to a cell density of approximately 1.8 × 108 cells/ml.
To obtain further insight into the possible effect of the tspO mutation in an AppA null mutant, a puc::lacZ fusion was introduced in trans into the different mutant strains (Table 4). Since the effect of the tspO mutation on target gene expression is at the level of transcription (30–32), this analysis should reveal the true nature of the interaction with members of the PpsR/AppA regulon. The results of these studies demonstrate that the appA mutation is epistatic to the tspO mutation on puc operon expression, regardless of growth conditions. These studies more accurately reveal the modulating effect of TspO on expression of the PpsR/AppA regulon, compared to the complete effect of AppA (Fig. 1).
TABLE 4.
β-Galactosidase activities of the puc::lacZ fusion (pCF200Km) in AppA mutant strains of R. sphaeroides
| Strain | Amt (μmol/min/mg of protein) for growth conditiona:
|
||
|---|---|---|---|
| Aerobic | Semiaerobic | Dark-DMSO | |
| 2.4.1 | 210 ± 24 | 1,550 ± 99 | 1,140 ± 94 |
| TSPO1 | 250 ± 30 | 1,970 ± 121 | 1,190 ± 89 |
| TSPO1(pUI2701) | 180 ± 20 | 1,248 ± 84 | 976 ± 91 |
| APP11 | 32 ± 19 | 120 ± 33 | 74 ± 31 |
| APP-TSPO | 27 ± 20 | 117 ± 34 | 35 ± 32 |
| APP-TSPO(pUI2701) | 24 ± 19 | 99 ± 39 | 62 ± 30 |
See Table 3 footnotes for descriptions of growth conditions.
The effect of hemN in trans on PpsR/AppA regulon expression.
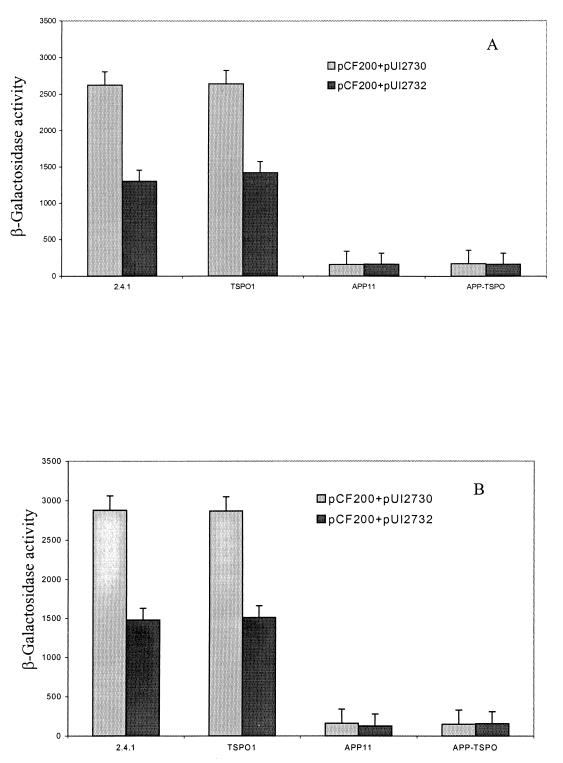
We have previously shown that the presence of hemN in trans at approximately five copies in the wild type produces an effect on PS gene expression similar to that produced by the tspO mutation and that extra copies of tspO together with hemN compromise this effect (30). These observations are entirely consistent with the model shown in Fig. 1. The results of such an experiment are depicted in Fig. 2. The extent of derepression of puc expression induced by extra copies of hemN in trans in wild type is nearly identical to that in the absence of the tspO locus, and these differences are not additive (30), as shown previously. This result led us to investigate the role of porphyrins in a TspO mutant (30). Of importance here is that, when multiple copies of hemN or both hemN and tspO are provided to APP11 or APP-TSPO, puc operon expression remains at basal level and little difference is observed between semiaerobic and anaerobic conditions (Fig. 2). This result strengthens our earlier conclusion, namely, that TspO operates through the PpsR/AppA repressor/antirepressor circuit and that the effect of hemN in extra copy is mitigated by the mutation of appA, consistent with our proposed model in Fig. 1. Since the effects of hemN and tspO mutations appear to operate through the same regulatory pathway (30), then this pathway is more than likely to involve AppA.
FIG. 2.
β-Galactosidase activities of the puc::lacZ fusion (pCF200Km) in wild-type and mutant strains with plasmid pUI2730 (hemN) or pUI2732 (hemN and tspO) in trans under anaerobic conditions in the dark with DMSO (A) or semiaerobic conditions (B). Values are micromoles per minute per milligram of protein.
The absence of the B800-850 complex and its interaction with TspO.
We have previously suggested that a possible mode of action of TspO in modulating target PS gene expression is through its ability to selectively regulate the efflux of an intermediate(s) involved in porphyrin synthesis (30, 32). We hypothesized that the intermediate could serve as a coactivator (or corepressor) of a critical regulatory protein, namely, AppA in the former case or PpsR in the latter. In the wild-type strain, the absence of tspO would promote the accumulation of photopigments. However, since the B800-850 complex is the major repository for Bchl and Crt and since it is part of the PpsR/AppA regulon, we reasoned that the absence of the B800-850 complex would minimize the effect of the absence of tspO. Therefore, the accumulations of Bchl and Crt were compared in B800-850 mutants with and without a functional tspO gene. As shown in Table 3, although the absence of TspO in the B800-850 mutant background resulted in no apparent increase in pigment accumulation, tspO in trans, when present in multiple copies, led to a decrease in both Bchl and Crt accumulation to below those levels found for PUC-ZWT or the double mutant PUCB-TSPO under semiaerobic or anaerobic conditions. Thus, the TspO effect is epistatic to the absence of B800-850 in leading to changes in pigment production, since TspO acts on but not through the puc operon. This contrasts with the effect of TspO acting through AppA, where no differences are observed between the absence and presence of B800-850. Also notable here is the overall decreased levels of photopigment production in the B800-850 mutant strain. Since it is only the presence of the B800-850 apoproteins which is altered and yet overall pigment production has declined, it is suggested that the absence of the B800-850 apoproteins leads to an apparent feedback effect upon photopigment production. Since this presumed feedback effect is dominant to the absence, but not the presence, of extra copies of tspO, it is suggested that the levels of the coactivator of AppA are decreased, making PpsR repression more effective in either circumstance (Fig. 1).
Because the puc mutation used here involves an insertion of lacZ into the pucB gene, we were also able to directly monitor expression of puc under these same experimental conditions (Table 5). It was found that there was no obvious increase in β-galactosidase activities of the double mutant PUCB-TSPO over those of PUC-ZWT. On the other hand, extra copies of tspO in the PUCB-TSPO mutant background resulted in measurably decreased LacZ activity compared to either the single or the double mutant strains. This suggests, much like the results described above, that extra copies of tspO lead to a decrease in the presumed coactivation of AppA, presumably by increasing the efflux of the critical porphyrin molecule (Fig. 1) such that the effectiveness of PpsR is enhanced. This result is consistent with the photopigment data in Table 3, since puc and a number of the photopigment genes are under the control of the PpsR/AppA repressor/antirepressor system (31, 34).
TABLE 5.
β-Galactosidase activities of the chromosome-localized puc::lacZ fusion in B800-850− mutants of R. sphaeroides 2.4.1a
| Strain | Amt (μmol/min/mg of protein) for growth condition:
|
||
|---|---|---|---|
| Aerobic | Semiaerobic | Dark-DMSO | |
| PUC-ZWT | 243 | 4,113 | 3,724 |
| PUCB-TSPO | 232 | 4,398 | 3,901 |
| PUCB-TSPO(pUI2701) | 126 | 3,167 | 2,975 |
Data in the table are the means of three independent experiments; variations were less than 10%. See Table 3 footnotes for descriptions of growth conditions.
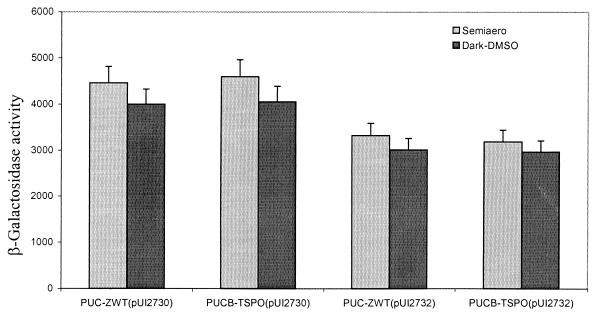
Supporting this conclusion are the results depicted in Fig. 3. When multiple copies of hemN are present in PUC-ZWT or PUCB-TSPO, there is no effect on puc expression, unlike what is observed for wild type. This suggests that further changes in porphyrin levels are ineffective in enhancing target gene expression, i.e., there is no hemN-stimulated expression of puc (30), presumably because the porphyrin pathway is already flooded (Fig. 1). However, when extra copies of tspO were present, we observed a small but significant decline in puc operon expression, which we assumed to be the result of the decreased level of the coactivator of AppA as the result of increased porphyrin efflux.
FIG. 3.
β-Galactosidase activities of the chromosome-localized puc::lacZ fusion in mutants PUC-ZWT and PUCB-TSPO with plasmid pUI2730 (hemN) or pUI2732 (hemN and tspO) in trans. Values are micromoles per minute per milligram of protein.
Finally, our data show that TspO had no effect on the expression of the puf operon, which is not under the control of the PpsR/AppA regulon, under any conditions (31, 34; data not shown). These data further support the above findings that TspO selectively regulates PS gene expression through the PpsR/AppA regulon and is independent of the Prr and FnrL regulons (5–8, 30–34).
Formation of tetrapyrrole intermediates in wild-type and mutant strains of R. sphaeroides.
It had been previously reported that TspO appears to be involved in controlling the efflux of tetrapyrrole intermediates from the cells. Our earlier results also indicate that the effects of hemN in trans and the absence of tspO appear to be similar, and we hypothesized that tetrapyrrole intermediates likely to be derived from coproporphyrinogen III might act as coactivators of AppA and thereby reduce the effectiveness of the PpsR repressor. In an effort to obtain further insight into the role of porphyrins, we have analyzed those intermediates in tetrapyrrole synthesis which accumulated within and/or outside cells of the wild-type and mutant strains of R. sphaeroides (30).
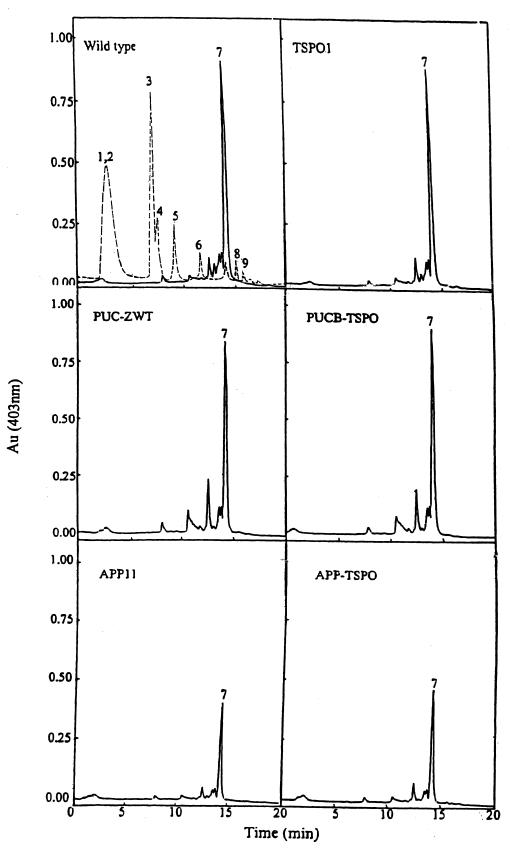
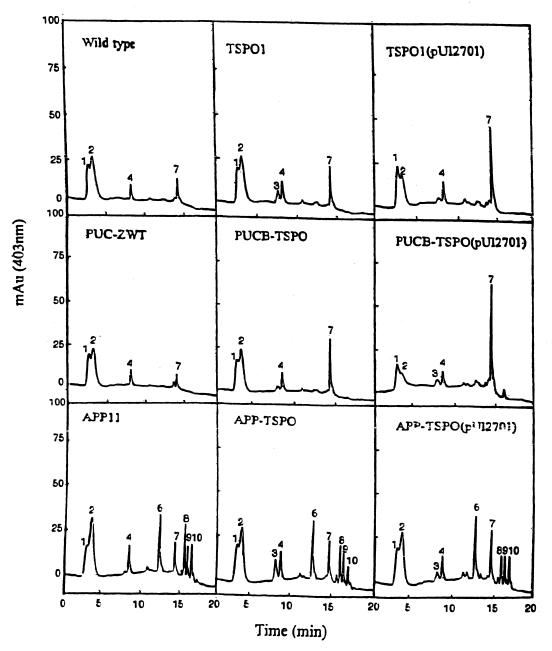
First, we analyzed the accumulation of porphyrin precursors in resting cell suspensions incubated with an excess of ALA under semiaerobic conditions (Fig. 4). The elution time for each peak in HPLC was confirmed by the use of standard porphyrin acids; these times are quite reproducible, and the abundance of each peak is readily quantitated. It is evident that the types and relative amounts of porphyrin precursors that accumulated in resting cells of the wild type (Fig. 4, dashed line) were different from those excreted from the same cells (Fig. 4, solid lines), which also indicates that the resting cells were intact when incubated in the presence of chloramphenicol and excess ALA. Importantly, porphyrin excretion from resting cells (Fig. 4, solid lines) contained mainly coproporphyrin III (peak 7), whereas uroporphyrin (peaks 1 and 2) and early decarboxylation products of uroporphyrinogen (heptacarboxylic porphyrin [peaks 3 and 4], hexacarboxylic porphyrin [peak 5], and pentacarboxylic porphyrin [peak 6]) were accumulated in the resting cells (Fig. 4, dashed line). On the other hand, little or no protoporphyrinogen IX or other oxidative decarboxylation products of coproporphyrinogen III (peaks 8 and 9) were detected in either the cell-free supernatant or the cellular extract. In general, lower levels of uroporphyrinogen III or products derived therefrom were accumulated in cells bearing the tspO mutation. Of further importance here is that different levels of porphyrin precursors were observed to be accumulated inside and outside resting cells containing the appA mutation. As is evident from Fig. 4, the AppA null mutant cells accumulated substantially decreased amounts of coproporphyrinogen III (peak 7) relative to those observed for the wild-type cells or for cells containing the puc operon lesion. In fact, it appears with respect to porphyrin accumulation that the appA mutation is epistatic to the tspO lesion in terms of the derived profile. In the wild type, TSPO1, and the strains containing the Puc lesions, the levels of peak 7 accumulated were ∼3.75 ± 0.40 arbitrary units. For the strains containing the AppA lesions, these same values were ∼1.58 ± 0.34 arbitrary units. These units represent the areas under the curve for each of the peak 7 profiles.
FIG. 4.
HPLC analysis of the porphyrin precursors accumulated in the cells (dashed line) or excreted from the cells (solid line) of wild-type R. sphaeroides 2.4.1 and its mutant derivatives. Cells in 100 ml of Sistrom medium were grown semiaerobically, collected by centrifugation (15 min, 10,000 × g), washed with 0.1 M potassium phosphate buffer (pH 7.0) two times, and then incubated in 100 ml of 0.1 M potassium phosphate buffer (pH 7.0) containing 80 μM chloramphenicol and 0.2 mM ALA for 6 h with sparging with 2% O2–1% CO2–97% N2 (30). Peaks: 1 and 2, uroporphyrins I and III, respectively; 3 and 4, heptacarboxylic porphyrins I and III, respectively; 5, hexacarboxylic porphyrin III; 6, pentacarboxylic porphyrin; 7, coproporphyrin III; 8, 9, and 10, deuteroporphyrin derivatives.
Because the addition of exogenous ALA to resting cells presents its own problems as to pigment accumulation in R. sphaeroides (18), we elected to directly monitor the levels of porphyrin intermediates in growing cells without the addition of ALA. Figure 5 is a profile of excreted porphyrins from the wild-type and mutant strains of R. sphaeroides. Without the addition of ALA, internal porphyrin levels were too low to measure.
FIG. 5.
HPLC analysis of the porphyrin precursors excreted from growing cells of wild-type and mutant strains of R. sphaeroides grown semiaerobically. Culture solution (500 ml) was collected by centrifugation (10,000 × g, 15 min) for the analysis of excreted porphyrin precursors. Peaks: 1 and 2, uroporphyrins I and III, respectively; 3 and 4, heptacarboxylic porphyrins I and III, respectively; 5, hexacarboxylic porphyrin III; 6, pentacarboxylic porphyrin III; 7, corproporphyrin III; 8, 9, and 10, deuteroporphyrin derivatives.
What is evident from Fig. 5 is that the addition of extra copies of tspO leads to enhanced excretion of coproporphyrin III (peak 7) by a factor of 4 to 7 from both the wild type and PUC-ZWT. In the absence of tspO, there appears to be little difference in porphyrin excretion in these strains, although the critical element is what is taking place inside the cells. Whereas wild type and PUC-ZWT are generally similar in terms of their excretion patterns, strains containing the appA mutation are different, showing enhanced excretion of peak 6 by a factor of at least 10, and extra copies of tspO do not lead to increased coproporphyrin III excretion (peak 7).
DISCUSSION
Studies from this laboratory have suggested that the repressor/antirepressor system, PpsR/AppA, is able to sense the redox state of the quinone pool through the flavin which is bound to AppA, and thus, AppA has been suggested to regulate the repressor activity of PpsR (Fig. 1). We have interpreted these results to suggest that, when AppA is in a more oxidized state, it is ineffective as an antirepressor of PpsR and its antirepressor activity increases as it become more reduced. The effectiveness of AppA could result from its ability to control the oligomerization of PpsR through the two PAS domains found in PpsR (5, 7). Initially and apparently unrelated to PpsR/AppA were our findings that the outer membrane protein TspO, which appears to be involved in the efflux, or control of the efflux, of porphyrin intermediates from the cell, especially coproporphyrinogen III, is able to exert partial control, selectively, of PS gene expression. An enigma regarding mutations of tspO has been the observations that (i) the resulting small, but discernible, increase at low oxygen tension of Bchl and Crt appears to be the result of the increased transcription of the same target genes as regulated by the PpsR/AppA system and, (ii) although the effect of TspO appears to be through the repressor/antirepressor regulon, it is only partial at best, never approaching the actual loss of the repressor protein itself and its subsequent major effect on downstream gene transcription (Fig. 1). Further, and importantly, the effect of TspO is transient, observable only during the induction of the photosynthetic apparatus as cells proceed from aerobic to anaerobic growth. Thus, TspO only modulates target gene expression, normally slowing the induction process, but not reversing it.
We therefore reasoned that if TspO works through the repressor/antirepressor pathway, it would be more likely to act through AppA than PpsR (30, 32), since, unlike AppA, PpsR is not known to bind any ligands. Therefore, we put forth the hypothesis that some porphyrin product downstream of coproporphyrinogen III (protoporphyrin IX, heme) serves as a coactivator of AppA; the fact that extra copies of hemN produce a TspO-minus-like phenotype in the wild type is in keeping with this hypothesis (30). The fact that extra copies of tspO reverse the hemN effect is further evidence that TspO acts through the porphyrin pathway.
In the present study, we address these questions, and the data support the hypothesis that TspO is likely to act through the PpsR/AppA system, since in all of these studies lesions in appA yield a phenotype which is epistatic to the tspO lesion regardless of which of the various phenotypes is being examined, i.e., target gene transcription, pigment accumulation, or spectral complex levels. Quantitatively, these effects are only partial relative to the full effects noted when either appA or ppsR is mutated. This is in keeping with the transient role of TspO. The fact that the interactions between AppA and TspO are quite specific is illustrated when we examine the pucB/tspO double mutations. In these strains, the effect of the absence of the B800-850 complex, although levels of pigment decrease as expected, is nonetheless still subject to TspO-related control, as judged by the effects observed when extra copies of tspO are present in trans. Consistent with this interaction is the complete dominance of the AppA lesion even in the presence of extra copies of hemN. This result also serves to relate porphyrin levels to the TspO effect, which we have interpreted as suggesting the existence of a coactivator of AppA, whose levels determine in part the strength of the AppA effect.
Examination of the levels and kinds of porphyrin excreted from growing cells of R. sphaeroides reveals that there is increased coproporphyrin III being excreted when tspO is provided in trans, which is accompanied by decreased target gene expression and pigment accumulation.
We have, therefore, assumed that either protoporphyrin IX or heme can serve as a coactivator of AppA and that the levels of the coactivator depend upon the conversion of coproporphyrinogen III to porphyrinogen IX, which is in some way related to the levels of uroporphyrinogen III accumulated and/or coproporphyrin III excreted. However, there are other possible interpretations given the complexity of porphyrin metabolism in R. sphaeroides. It is also clear that the kinds and amounts of porphyrin precursors accumulated in the AppA mutant strains are quite different from those in other strains, supporting the concept that AppA acts downstream of TspO but in the same regulatory circuit. However, this evidence, although implicating porphyrin metabolism, does not define which porphyrin(s) is involved.
Given the above interpretation, we can consider why the TspO effect is only partial and never quantitatively as great as what would be observed if PpsR were fully inactivated (Fig. 1). One possible explanation among several is that the coactivation of AppA by a porphyrin never leads to the full activation of AppA which follows the reduction of the bound flavin through its interaction with the quinone pool. Thus, the activation by porphyrin precursor(s) is designed only to modulate or influence the repressor/antirepressor system through the state of AppA, not to inactivate it. This suggestion fits with the observation that TspO only slows the induction process during the transition from aerobic to anaerobic growth (30–33). The model that we have constructed here is a basis for further experimentation.
Recently, a homologue of the TspO protein in Sinorhizobium meliloti (2) was found to be required for the expression of the ndi locus in response to the stress conditions imposed upon the cells. Further, there is evidence that the S. meliloti TspO acts through or in addition to the FixL regulatory system. These authors further demonstrate that R. sphaeroides TspO could function in this system. Therefore, if we consider the data presented for R. sphaeroides (30–33) and S. meliloti (2) and the mitochondrial studies (2, 15, 17, 28, 29), we are left with the impression that the TspO system is probably ancient and important where it occurs and that it generally operates through a conserved mechanism, although the genes ultimately regulated may differ among organisms.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant GM15590.
REFERENCES
- 1.Cohen-Bazire G, Sistrom W R, Stanier R Y. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Comp Physiol. 1957;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 2.Davey M E, de Bruijn F J. A homologue of the tryptophan-rich sensory protein TspO and FixL regulate a novel nutrient deprivation-induced Sinorhizobium meliloti locus. Appl Environ Microbiol. 2000;66:5353–5359. doi: 10.1128/aem.66.12.5353-5359.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis J, Donohue T J, Kaplan S. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J Bacteriol. 1988;170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eraso J M, Kaplan S. PrrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomelsky M, Horne I M, Lee H J, Pemberton J M, McEwan A G, Kaplan S. Domain structure, oligomeric state, and mutational analysis of PpsR, the Rhodobacter sphaeroides repressor of photosystem gene expression. J Bacteriol. 2000;182:2253–2261. doi: 10.1128/jb.182.8.2253-2261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomelsky M, Kaplan S. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1995;177:4609–4618. doi: 10.1128/jb.177.16.4609-4618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomelsky M, Kaplan S. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel fad binding domain. J Biol Chem. 1998;273:35319–35325. doi: 10.1074/jbc.273.52.35319. [DOI] [PubMed] [Google Scholar]
- 8.Gomelsky M, Kaplan S. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1997;179:128–134. doi: 10.1128/jb.179.1.128-134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishida T, Yu L, Akutsu H, Ozawa K, Kawanishi S, Seto A, Inubushi T, Sano S. A primitive pathway of porphyrin biosynthesis and enzymology in Desulfovibrio vulgaris. Proc Natl Acad Sci USA. 1998;95:4853–4858. doi: 10.1073/pnas.95.9.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 11.Kiley P J, Kaplan S. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol Rev. 1988;52:50–69. doi: 10.1128/mr.52.1.50-69.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacks S, Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977;114:153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- 13.Lee J K, Kaplan S. Isolation and characterization of trans-acting mutations involved in oxygen regulation of puc operon transcription in Rhodobacter sphaeroides. J Bacteriol. 1992;174:1158–1171. doi: 10.1128/jb.174.4.1158-1171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 15.McEnery M W, Snowman A M, Trifiletti R R, Snyder S H. Isolation of the mitochondrial benzodiazepine receptor: association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc Natl Acad Sci USA. 1992;89:3170–3174. doi: 10.1073/pnas.89.8.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meinhardt S W, Kiley P J, Kaplan S, Crofts A R, Harayama S. Characterization of light-harvesting mutants of Rhodopseudomonas sphaeroides. I. Measurement of the efficiency of energy transfer from light-harvesting complexes to the reaction center. Arch Biochem Biophys. 1985;236:130–139. doi: 10.1016/0003-9861(85)90612-5. [DOI] [PubMed] [Google Scholar]
- 17.Mesenholler M, Matthews E K. A key role for the mitochondrial benzodiazepine receptor in cellular photosensitisation with delta-aminolaevulinic acid. Eur J Pharmacol. 2000;406:171–180. doi: 10.1016/s0014-2999(00)00646-4. [DOI] [PubMed] [Google Scholar]
- 18.Neidle E L, Kaplan S. 5-Aminolevulinic acid availability and control of spectral complex formation in HemA and HemT mutants of Rhodobacter sphaeroides. J Bacteriol. 1993;175:2304–2313. doi: 10.1128/jb.175.8.2304-2313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh J I, Kaplan S. Redox signaling: globalization of gene expression. EMBO J. 2000;19:4237–4247. doi: 10.1093/emboj/19.16.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratcliffe S L, Matthews E K. Modification of the photodynamic action of delta-aminolaevulinic acid (ALA) on rat pancreatoma cells by mitochondrial benzodiazepine receptor ligands. Br J Cancer. 1995;71:300–305. doi: 10.1038/bjc.1995.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebeiz N, Arkins S, Kelley K W, Rebeiz C A. Enhancement of coproporphyrinogen III transport into isolated transformed leukocyte mitochondria by ATP. Arch Biochem Biophys. 1996;333:475–481. doi: 10.1006/abbi.1996.0417. [DOI] [PubMed] [Google Scholar]
- 22.Rossi E, Curnow D H. Porphyrins. In: Lim C K, editor. HPLC of small molecules. Oxford, United Kingdom: IRL Press; 1986. pp. 261–313. [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Seehra J S, Jordan P M, Akhtar M. Anaerobic and aerobic coproporphyrinogen III oxidases of Rhodopseudomonas spheroides. Mechanism and stereochemistry of vinyl group formation. Biochem J. 1983;209:709–718. doi: 10.1042/bj2090709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon R P, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 26.Tai T N, Havelka W A, Kaplan S. A broad-host-range vector system for cloning and translational lacZ fusion analysis. Plasmid. 1988;19:175–188. doi: 10.1016/0147-619x(88)90037-6. [DOI] [PubMed] [Google Scholar]
- 27.Tait G H. Coproporphyrinogenase activities in extracts of Rhodopseudomonas spheroides and Chromatium strain D. Biochem J. 1972;128:1159–1169. doi: 10.1042/bj1281159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taketani S, Kohno H, Furukawa T, Tokunaga R. Involvement of peripheral-type benzodiazepine receptors in the intracellular transport of heme and porphyrins. J Biochem (Tokyo) 1995;117:875–880. doi: 10.1093/oxfordjournals.jbchem.a124790. [DOI] [PubMed] [Google Scholar]
- 29.Taketani S, Kohno H, Okuda M, Furukawa T, Tokunaga R. Induction of peripheral-type benzodiazepine receptors during differentiation of mouse erythroleukemia cells. A possible involvement of these receptors in heme biosynthesis. J Biol Chem. 1994;269:7527–7531. [PubMed] [Google Scholar]
- 30.Yeliseev A A, Kaplan S. A novel mechanism for the regulation of photosynthesis gene expression by the TspO outer membrane protein of Rhodobacter sphaeroides 2.4.1. J Biol Chem. 1999;274:21234–21243. doi: 10.1074/jbc.274.30.21234. [DOI] [PubMed] [Google Scholar]
- 31.Yeliseev A A, Kaplan S. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J Biol Chem. 1995;270:21167–21175. doi: 10.1074/jbc.270.36.21167. [DOI] [PubMed] [Google Scholar]
- 32.Yeliseev A A, Kaplan S. TspO of Rhodobacter sphaeroides. A structural and functional model for the mammalian peripheral benzodiazepine receptor. J Biol Chem. 2000;275:5657–5667. doi: 10.1074/jbc.275.8.5657. [DOI] [PubMed] [Google Scholar]
- 33.Yeliseev A A, Krueger K E, Kaplan S. A mammalian mitochondrial drug receptor functions as a bacterial “oxygen” sensor. Proc Natl Acad Sci USA. 1997;94:5101–5106. doi: 10.1073/pnas.94.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeilstra-Ryalls J H, Gomelsky M, Yeliseev A A, Eraso J M, Kaplan S. Transcriptional regulation of photosynthesis operons in Rhodobacter sphaeroides 2.4.1. Methods Enzymol. 1998;297:151–166. doi: 10.1016/s0076-6879(98)97012-4. [DOI] [PubMed] [Google Scholar]