Summary
Intracellular long-chain acyl-coenzyme As (LC-acyl-CoAs) are thought to be under tight spatial and temporal controls, yet the ability to image LC-acyl-CoAs in live cells is lacking. Here, we developed a fluorescence resonance energy transfer (FRET) sensor for LC-acyl-CoAs based on the allosterically regulated interaction between α/β hydrolase domain-containing 5 (ABHD5) and Perilipin 5. The genetically encoded sensor rapidly detects intracellular LC-acyl-CoAs generated from exogenous and endogenous fatty acids (FAs), as well as synthetic ABHD5 ligands. Stimulation of lipolysis in brown adipocytes elevated intracellular LC-acyl-CoAs in a cyclic fashion, which was eliminated by inhibiting PNPLA2 (ATGL), the major triglyceride lipase. Interestingly, inhibition of LC-acyl-CoA transport into mitochondria elevated intracellular LC-acyl-CoAs and dampened their cycling. Together, these observations reveal an intimate feedback control between LC-acyl-CoA generation from lipolysis and utilization in mitochondria. We anticipate that this sensor will be an important tool to dissect intracellular LC-acyl-CoA dynamics as well to discover novel synthetic ABHD5 ligands.
Keywords: fatty acids, long-chain acyl-CoAs, lipid droplet, mitochondria, α/β hydrolase domain-containing 5, CGI-58, adipose triglyceride lipase, Perilipin, real-time monitoring, genetically encoded FRET sensor
Graphical abstract
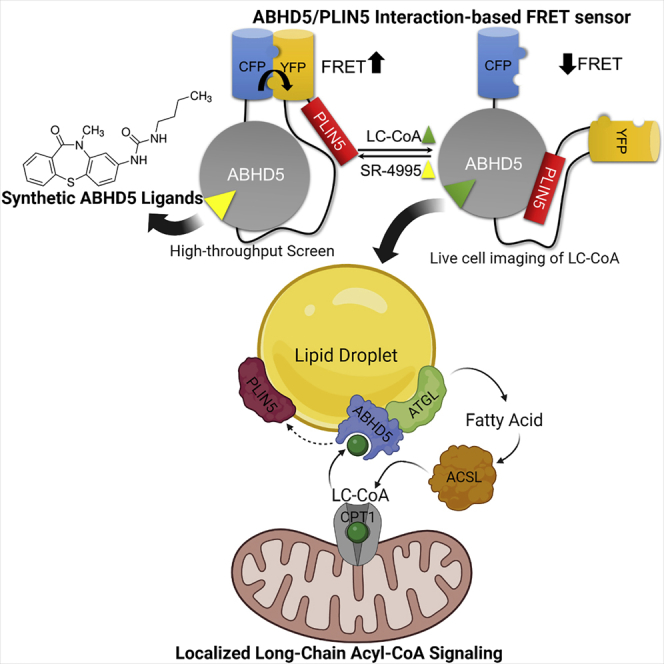
Highlights
-
•
A FRET sensor that detects long-chain acyl-CoAs (LC-CoAs) and synthetic ABHD5 ligands
-
•
The sensor reports ligand-dependent interaction between ABHD5 and PLIN5
-
•
Activating lipolysis in brown adipocytes (BAs) raises LC-CoAs in a cyclical fashion
-
•
LC-CoAs provide feedback regulation from mitochondria to lipid droplets in BAs
Motivation
Intracellular long-chain acyl-coenzyme As (LC-acyl-CoAs) are thought to be under tight spatial and temporal controls, yet the ability to image LC-acyl-CoAs in live cells is lacking. We sought to create a fluorescence resonance energy transfer (FRET) sensor of LC-acyl-CoAs and synthetic ABHD5 ligands based on the reversible interactions between ABHD5 and PLIN5. The sensor allows dynamic imaging of intracellular LC-acyl-CoA levels in targeted subcellular compartments. In addition, since ABHD5 has emerged as a therapeutic target for metabolic disease and cancer, a robust FRET-based sensor could aid in the identification of novel synthetic and natural ABHD5 ligands.
Mottillo et al. develop a single-molecule FRET sensor based on the ligand-dependent interaction between ABHD5 and PLIN5. The sensor reports intracellular long-chain acyl-CoA (LC-CoA) levels and detects synthetic ABHD5 ligands. Real-time imaging in brown adipocytes reveals feedback inhibition between LC-CoA generation from lipolysis and uptake in mitochondria.
Introduction
Fatty acids (FAs) and their metabolites function as important energy sources and intracellular signaling molecules. In adipocytes, the release of FAs is regulated by α/β hydrolase domain-containing 5 (ABHD5; also known as CGI-58), a pseudoenzyme regulator of Patatin-like phospholipase domain-containing 2 (PNPLA2)/adipose triglyceride lipase (ATGL), the rate-limiting enzyme of triacylglycerol (TAG) hydrolysis. Intracellular FAs are quickly metabolized to long-chain acyl-coenzyme As (LC-acyl-CoAs) by LC-acyl-CoA synthetases. Moreover, LC-acyl-CoAs bind ABHD5, promoting its interaction with Perilipin 5 (PLIN5), a resident lipid droplet (LD) protein, thereby repressing ATGL activation.1 The intracellular levels of LC-acyl-CoAs are normally kept in a tight concentration range2 and are thought to exist in dynamic compartmentalized pools.3 For example, deletion of LC-acyl-CoA synthetase 1 (ACSL1) in adipose tissue produces a specific defect in FA oxidation without affecting de novo lipogenesis.4 In addition, pools of acyl-CoAs might create localized signals that regulate distinct metabolic pathways3,5 and gene transcription.6 However, techniques for imaging the spatial-temporal dynamics of LC-acyl-CoAs are lacking.
The allosteric regulation of the interaction of ABHD5 and PLIN5 by LC-acyl-CoAs provides a mechanism for limiting ATGL-dependent release of FAs when LC-acyl-CoA levels are high1,7; however, this has been difficult to prove in part due to lack of methods to detect LC-acyl-CoAs in real time. Interestingly, mitochondria are often found in close proximity to LDs, an observation first made over 60 years ago.8 Moreover, brown adipocytes have a high capacity for FA oxidation due to the presence of numerous mitochondria and LDs. In addition to being highly expressed in oxidative tissues, PLIN5 is targeted to the interface between LDs and mitochondria,9,10,11,12,13 which may be important in augmenting respiration12 or generating reducing equivalents required for de novo lipid synthesis.14 However, the metabolic communication between LDs and mitochondria in brown adipocytes is poorly understood.
In addition to binding LC-acyl-CoAs, the interaction of ABHD5 with PLIN proteins can also be regulated by synthetic ABHD5 ligands.1,15,16 We previously discovered small molecules that selectively bind to ABHD5, causing ABHD5 to dissociate from PLIN1 and activate adipocyte lipolysis.1 As such, ABHD5 represents a locus for regulating cellular TAG levels and an important pharmacological target for metabolic disease and cancer.17,18,19,20,21,22 Thus, there is a need for robust homogeneous assays to understand the chemical and allosteric regulation of ABHD5.
Taking advantage of the interaction between ABHD5 and PLIN5, we developed a genetically encoded fluorescence resonance energy transfer (FRET)-based assay to detect intracellular LC-acyl-CoAs and synthetic ABHD5 ligands. The functional detection of intracellular LC-acyl-CoAs was first demonstrated in a bipartite molecular sensor that we subsequently developed into a single-molecule form to allow true ratiometric analysis. Importantly, this sensor can be used as a tool to explore novel ABHD5 pharmacology. We used the single-molecule sensor to capture the functional relationship between LDs and mitochondria that exists in brown adipocytes, demonstrating that mitochondrially derived LC-acyl-CoAs function as feedback signals to regulate substrate flux from LDs.
Results
We previously identified a fragment of PLIN5 encoding amino acids (aa) 384–417, whose reversible interaction with ABHD5 can be rapidly modulated in live cells by LC-acyl-CoAs and synthetic ligands of ABHD5.15 In COS-7 cells transfected with this bimolecular system consisting of ECFP-ABHD5 and PLIN5384−417-EYFP, basal conditions yielded only a faint FRET signal on LD structures (Figure 1A, basal normalized FRET [nFRET]). Addition of oleic acid (OA) led to a large increase in the FRET signal on the endoplasmic reticulum (ER) and LDs, sites of TAG synthesis and storage, respectively (Figure 1A, OA nFRET). Indeed, this effect was also accompanied by a greater translocation of the EYFP-tagged PLIN5 fragment to LDs upon OA treatment (Figure 1A, PLIN5-EYFP basal versus OA). These observations indicate that the ligand-dependent interaction of ABHD5 with PLIN5 might be used to monitor intracellular LC-acyl-CoA levels. Based on these results, we designed a single-molecule genetically encoded molecule that detects intracellular levels of LC-acyl-CoA and synthetic ABHD5 ligands and reports those levels as changes in FRET.
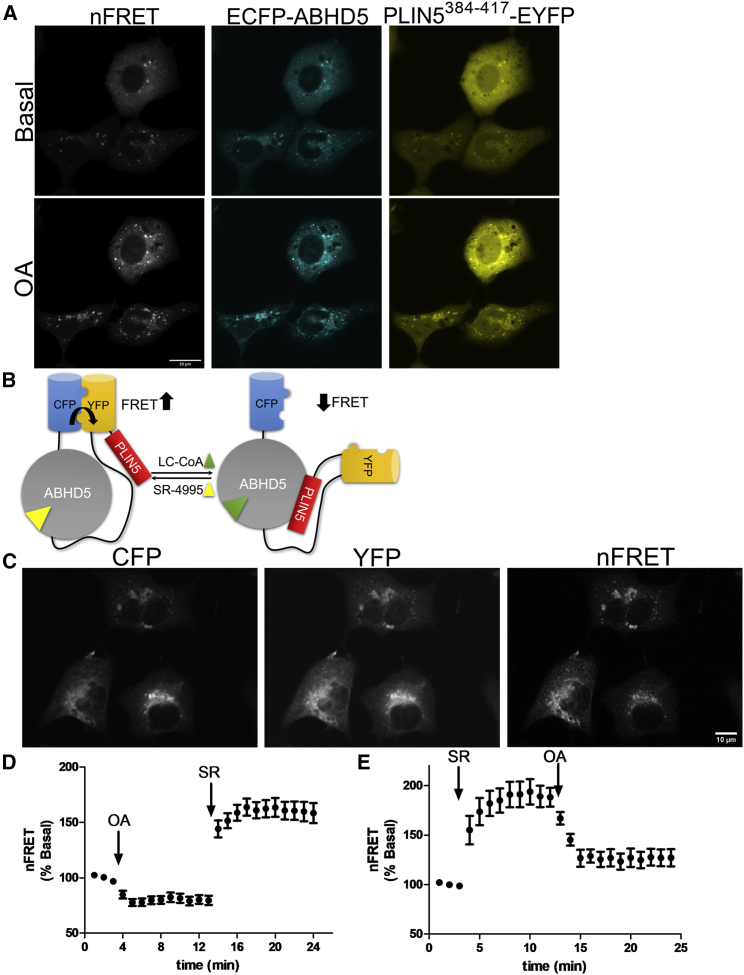
Figure 1.
FRET sensor for the detection of intracellular LC-acyl-CoAs and synthetic ABHD5 ligands
(A) COS-7 cells were cotransfected with plasmids encoding CFP-ABHD5 and PLIN5384−417-EYFP, and cells were imaged under basal conditions in HKRB 1% BSA and following the addition of 200 μM oleic acid (OA) at 15 min. The nFRET signal was calculated from the ECFP, EYFP, and EYFP-FRET signals as described. Images are representative of three independent experiments. Scale bar, 20 μm.
(B) Schematic of the single-molecule FRET sensor containing Cerulean (CFP) mutant in fusion with ABHD5, a flexible region containing GGS repeats (black line), Citrine (YFP), and PLIN5384−417. In the presence of synthetic ABHD5 ligands such as SR-4995, this dissociated the PLIN5384−417 fragment from ABHD5, allowing strong intramolecular interaction between CFP and YFP and high FRET. In the presence of LC-acyl-CoAs (LC-CoAs), this promotes the interaction between PLIN5384−417 and ABHD5 and dissociates the weak intramolecular interaction between CFP and YFP, resulting in a reduction in FRET.
(C) CFP, YFP, and the calculated nFRET signal in COS-7 cells transfected with the single-molecule FRET sensor. Scale bar, 10 μm.
(D and E) COS-7 cells transiently transfected with the ABHD5R299N LC-acyl-CoA sensor were monitored for CFP, YFP, and YFP-FRET signal every minute and treated with OA or SR-4995 (SR) at the indicated time, and the nFRET signal was calculated. Results are from one experiment and are representative of three independent experiments.
See also Figure S1.
Initial attempts to create a FRET sensor by separating the fluorescently tagged proteins used in the bimolecular system with a flexible polypeptide linker were unsuccessful owing to strong FRET under basal conditions, with minimal modulation by ligands that promote molecular association and dissociation (Table S1; data not shown). Moreover, employing a circularly permutated version of Venus on ABHD5 did not result in a modulation by ligands (Table S1; data not shown). The failure of peptide linkers to expand the dynamic range in FRET constructs is a known limitation that was recently surmounted by integrating mutants of CFP (Cerulean) and YFP (Citrine) that exhibit weak dimerization, which can be reversed by the protein-protein interaction of the targeted proteins.23 Figure 1B is a schematic representation of an ABHD5-PLIN5-based FRET sensor that employs this concept (summarized in Table S1). To eliminate potential confounding effects of ABHD5 overexpression with the sensor on ATGL activation, we introduced a mutation of R299N that disrupts the ability of ABHD5 to activate PNPLA2 without affecting its interactions with PLIN proteins15,24 (Table S1). When expressed in COS-7 cells, the mutant construct targets intracellular LDs and what appeared to be ER membranes (Figure 1C) with precise colocalization of CFP and YFP signals as expected in a single-molecule sensor. We confirmed ER localization by staining cells with ER-Tracker Red. Indeed, the FRET sensor (Figure S1A, YFP) colocalized to regions of the ER (Figure S1A, ER-Tracker). We further asked if the FRET sensor localized to domains of the ER that represent sites of de novo LD formation by cotransfecting cells with LiveDrop.25 In this case, the LC-acyl-CoA sensor colocalized precisely with LiveDrop (Figure S1B). As designed, the FRET signal between CFP and YFP was higher in the basal state (Figure 1C). To test the modified ABHD5 mutant, cells were loaded with OA overnight to promote LD formation. Media were exchanged, followed by the acute application of OA (200 μM complexed to BSA), which rapidly decreased FRET (Figure 1D). The decrease in FRET was rapidly reversed by the synthetic ABHD5 ligand SR-4995 (Figure 1D). Conversely, SR-4995 sharply increased FRET, and this increase was rapidly reversed by OA (Figure 1E). These results establish that the single-molecule FRET construct localizes to LDs and regions of the ER involved in TAG synthesis and reports endogenous and synthetic ligand-induced regulation of the interaction of ABHD5 with PLIN5.
Our previous work using the covalent activity probe NBD-NE-HP demonstrated that purified ABHD5 binds LC-acyl-CoA, not OA, and that the ability of LC-acyl-CoA to bind ABHD5 is under the allosteric regulation by synthetic ABHD5 ligands.1 However, those in vitro experiments were not able to directly demonstrate the allosteric regulation of ABHD5, owing to the fact that LC-acyl-CoAs do not penetrate biological membranes. Furthermore, the low affinity of the free PLIN5 fragment (aa 384–417) in cell-free systems did not allow in vitro testing of LC-acyl-CoA regulation. We tested whether this barrier could be overcome with the monomolecular sensor construct in an in vitro setting. In cell-free lysates, LC-acyl-CoA suppressed FRET, whereas SR-4995 and SR-3420 (10 μM), potent ABHD5 ligands that dissociate ABHD5 from PLIN proteins,1,16 greatly increased FRET (Figure 2A). Importantly, the ability of oleoyl-CoA to suppress FRET in a cell-free system was not observed for OA, indicating that activation of the FA is required (Figure 2B). Moreover, in vitro titration with oleoyl-CoA in a dose-response analysis indicated an EC50 of 1.24 μM (Figure 2C). These data establish that binding of LC-acyl-CoA, but not LC-free FA (FFA), to ABHD5 promotes its interaction with PLIN5. Application of exogenous FAs of varying chain lengths and saturations to live cells demonstrates that the FRET sensor is sensitive to FAs of a carbon chain length >8, with no apparent difference in responses due to FAs saturation (Figure 2D). Indeed, short-chain and medium-chain FAs (<C10) did not promote the interaction between ABHD5 and PLIN5384−417 in luciferase complementation assays, whereas longer chain lengths (>C8) promoted the interaction to a similar degree (Figure S2). To further validate the utility of the monomolecular FRET sensor, we examined the competition of endogenous and synthetic ligands in a dose-response analysis. SR-3420 increased FRET with an EC50 of 59.4 nM, and exposure of cells to 200 μM OA decreased FRET in the absence of synthetic ligand and, furthermore, shifted the concentration-response curve rightward by 10-fold to an EC50 of 562 nM (Figure 2E). These observations indicate that LC-acyl-CoA and synthetic ligands compete for a common binding site and stabilize distinct conformations that allow differential interaction with PLIN5. Moreover, the dextral 10-fold shift in the SR-3420 potency indicates that 200 μM OA (equivalent to approximately 30 nM free OA26,27) is sufficient for oleoyl-CoA to occupy approximately 90% of ABHD5 sites bound by SR-3420.
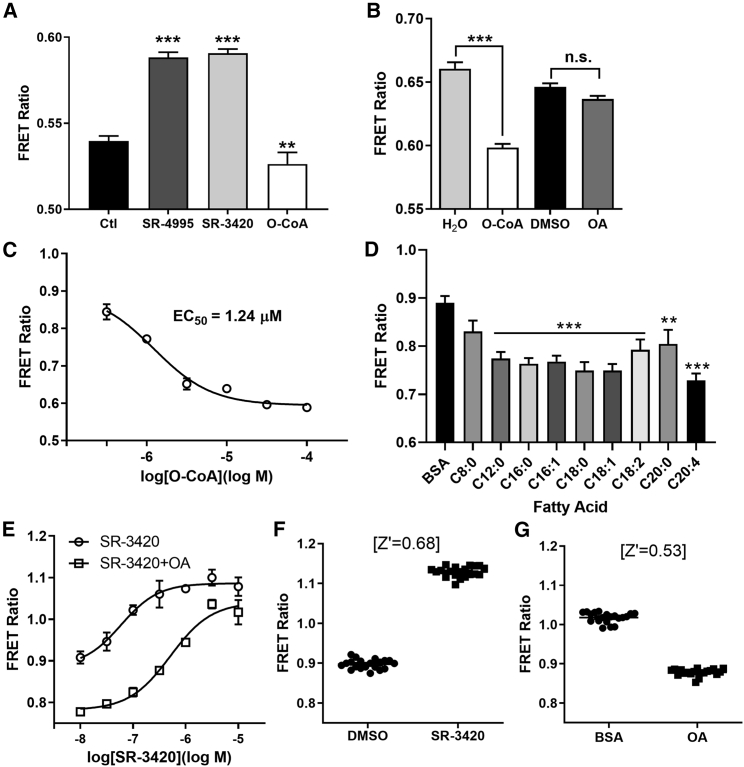
Figure 2.
In vitro and in cellulo detection of ABHD5 ligands
(A) COS-7 cells lysates transfected with the LC-acyl-CoA sensor were treated with vehicle (Ctl) SR-4995 (10 μM), SR-3420 (10 μM), or oleoyl-CoA (O-CoA; 10 μM), and the FRET ratio was measured on a fluorometer. ∗∗p < 0.01; ∗∗∗p < 0.001 compared with Ctl as determined using one-way analysis of variance (ANOVA) with Dunnett’s post t test.
(B) COS-7 cells transfected with the LC-acyl-CoA sensor were permeabilized with saponin and treated with vehicle (H2O or DMSO), O-CoA (300 μM), or OA (300 μM), and the FRET ratio was measured on a fluorometer. ∗∗∗p < 0.001 compared with H2O as determined by unpaired t test, n.s., non-significant.
(C) Dose response of O-CoA in COS-7 lysates.
(D) Live COS-7 cells transfected with the LC-acyl-CoA sensor were treated with the indicated fatty acid of carbon chain length and saturation, and the FRET ratio was measured on a fluorometer. ∗∗p < 0.01; ∗∗∗p < 0.001 compared with BSA as determined using ANOVA with Dunnett’s post t test.
(E) COS-7 cells were prepared as in (D), treated with or without OA (200 μM) and the indicated dose of SR-3420, and measured on a fluorometer.
(F and G) Live COS-7 cells transfected with the LC-acyl-CoA sensor were treated DMSO versus SR-3420 (F) or BSA versus OA (G) and the z′ score was calculated.
See also Figure S2.
The FRET-based sensor can also be readily adaptable to high-throughput screens as demonstrated by in vitro (Figures 2A and 2B) and in cellulo (Figure 2D) homogeneous assays. The sensor does not require separation of bound from free ligand, and the ratiometric method tolerates well-to-well variations in sensor concentration. The sensor routinely generated z′ values >0.68 in multiwell format for ligands that dissociate the ABHD5-PLIN5 interaction (Figure 2F, SR-3420) and >0.53 for ligands that promote the interaction (Figure 2G, OA), indicating robust performance sufficient for ultrahigh throughput (uHTS) formats. In contrast to previous uHTS assays based on luciferase complementation,1 the ABHD5 FRET sensor is simpler and is largely immune to compounds that interfere with luciferase activity and detection, which comprised nearly 90% of the initial “hits” in that complementation screen.1 Thus, we anticipate that the ABHD5 FRET sensor will be valuable in discovering new ligands that expand the rich pharmacology of ABHD5 activators and inhibitors.
To better understand the physiological role of LC-acyl-CoAs in metabolic regulation, we expressed the FRET sensor in brown adipocytes (BAs). BAs contain numerous mitochondria and LDs and have a high capacity for the oxidation of FAs, thus offering a model system to study the interaction between these two organelles. Expression of the FRET sensor (Figure 3A, green) in BAs demonstrates that the LC-acyl-CoA sensor localized to the surface of LDs (Figure 3A, red), which is the site of endogenous ABHD5 localization.28,29,30,31 In BAs, β-adrenergic activation stimulates the activity of PNPLA2 (also known as ATGL) to mobilize FAs for β-oxidation. Monitoring the FRET signal under basal conditions in BAs (initial 5 min) resulted in a stable signal (Figure 3B). Treatment with isoproterenol, a β-adrenergic agonist, decreased the FRET ratio, indicating a rise in intracellular LC-acyl-CoAs (Figure 3B). Interestingly, cyclic elevation of LC-acyl-CoAs was observed during the 20 min following isoproterenol stimulation. Pharmacological inhibition of PNPLA2 (Atglistatin [ATGLi]), the rate-limiting enzyme for TAG hydrolysis, completely blocked the decrease in the FRET signal in response to isoproterenol (Figure 3B). We next tested if preventing the oxidation of FAs could increase intracellular LC-acyl-CoA levels during isoproterenol stimulation. Mobilized FAs are quickly converted to LC-acyl-CoAs and transported into mitochondria by CPT1, the rate-limiting enzyme for FA oxidation. Etomoxir (ETX), an irreversible inhibitor of CPT1, is converted to etomoxiryl-CoA and blocks CoA-dependent transport of FFAs into mitochondria.32 Consistent with Figure 3B, isoproterenol reduced the FRET signal and resulted in oscillations over the 20 min period following stimulation (Figure 3C). Pretreatment with ETX did not alter basal FRET levels (initial 5 min); however, isoproterenol treatment resulted in a greater decrease in the FRET signal, in which oscillations were largely eliminated. Indeed, analysis revealed that ETX significantly reduced the amplitude (p = 0.011, Welch’s t test for unequal variances) and increased the period (p = 0.0107, Welch’s t test for unequal variances) of the oscillations induced by lipolysis activation (Figures 3D and 3E). Overall, these results demonstrate that the FRET sensor detects dynamic modulation of intracellular LC acyl-CoAs levels by FA mobilization and mitochondrial oxidation.
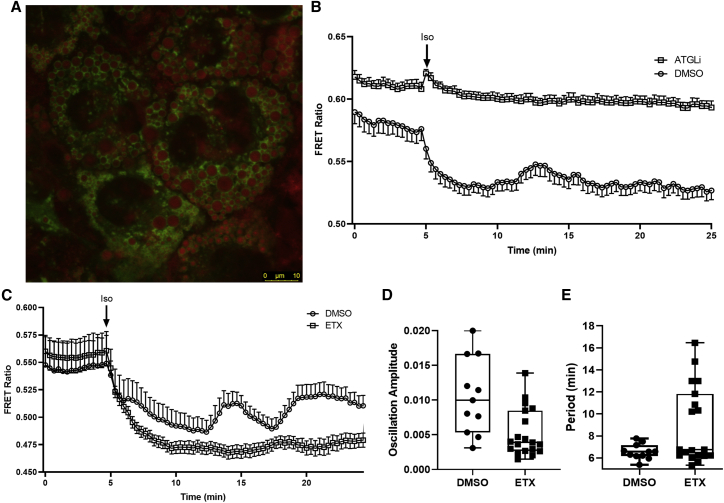
Figure 3.
LC-acyl-CoA imaging in brown adipocytes
(A) BA cells stably expressing the ABHD5R299N LC-acyl-CoA sensor (green) demonstrating localization to lipid droplets (red). Neutral lipids were stained with LipidTOX Red. Scale bar, 10 μm.
(B) BA cells were pretreated with Atglistatin (ATGLi; 20 μM; 12 cells) or vehicle (DMSO; 14 cells) and imaged on a laser scanning confocal every 20 s. Isoproterenol (1 μM) was added at the 5 min mark in between acquisitions.
(C) BA cells were imaged as in (B) except cells were pretreated with Etomoxir (ETX; 40 μM; 19 cells) or vehicle (DMSO; 11 cells).
(D and E) The amplitude (D) and period (E) of the oscillations of each individual cell were calculated from 5 to 25 min in (C). Data are from one experiment in which three to four coverslips were imaged under each condition and are representative of 3–4 individual experiments. Differences between DMSO and ETX amplitudes and periods were analyzed with an unpaired two-tailed t test with Welch’s correction for unequal variances.
We also examined the utility of targeting the LC acyl-CoA sensor to other cellular compartments. To prevent targeting of ABHD5 to LDs and the ER, we introduced a R116N mutation into ABHD5, which disrupts LDs and membrane targeting24,33 and allows mostly cytosolic and nuclear localization (Figures 4A and S4A). Introduction of 11 N-terminal amino acid residues of the plasma membrane (PM)-localized Lyn kinase34 in the context of the R116N mutation resulted in PM targeting of the LC acyl-CoA sensor (Figure 4B) as demonstrated by colocalization with PM stains (Figures S4B and S4C). Extracellular application of OA decreased the FRET signal in the cytosol and nucleus of the untargeted sensor, indicating that OA can also raise LC acyl-CoA levels in the cytosol and nucleus (Figure 4C). Treatment of OA similarly decreased FRET with the PM-targeted LC acyl-CoA sensor (Figure 4D). These data demonstrate that the LC acyl-CoA sensor is amenable to organelle-specific targeting and reporting local subcellular concentrations of LC acyl-CoAs.
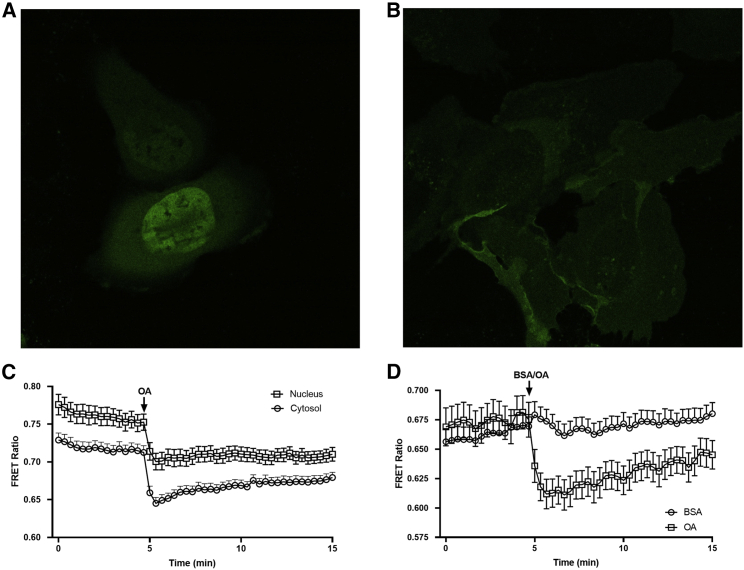
Figure 4.
Plasma membrane-localized detection of LC-acyl-CoAs
(A) U2OS cells were transiently transfected with the ABHD5R116N LC-acyl-CoA sensor, demonstrating untargeted localization throughout the cytosol and nucleus.
(B) U2OS cells were transiently transfected with the Lyn11-tagged ABHD5R116N LC-acyl-CoA sensor, demonstrating localization to the plasma membrane.
(C) U2OS cells transfected with the ABHD5R116N LC-acyl-CoA sensor were imaged on a laser scanning confocal microscope every 20 s with the addition of 200 μM OA, and the regions of interest in the cytosol and nucleus from 27 cells were quantified and expressed as a FRET ratio.
(D) U2OS cells transiently transfected with the Lyn11-tagged ABHD5R116N LC-acyl-CoA sensor were imaged as in (C) with the application of OA (200 μM; 21 cells) or 1% BSA control (BSA; 19 cells). Data are from one experiment in which three to four coverslips were imaged under each condition and are representative of 3 individual experiments.
See also Figure S4.
Discussion
The compartmentalization of LC-acyl-CoA metabolism has long been hypothesized; however, additional methods to image this critical lipid intermediate in live cells are required.35 Here, we took advantage of the allosteric regulation between ABHD5 and PLIN5 to develop methods to image dynamic modulation of intracellular LC-acyl-CoAs in BAs.
In BAs, the formation of LC-acyl-CoAs in response to isoproterenol stimulation required the activity of PNPLA2, which mobilizes LC-FFAs. In BAs, these LC-acyl-CoAs can be quickly metabolized to acyl-carnitines through the action of CPT1. Indeed, pharmacological inhibition of CPT1 resulted in a greater accumulation of LC-acyl-CoAs upon stimulation with isoproterenol. We found that the ABHD5/PLIN5 interaction senses FAs of carbon chain lengths equal to and greater than 10 (Figures 2D and S2), which are the majority of FAs that are mobilized in response to lipolytic stimulation.36 These data suggest that LD-targeted ABHD5 senses levels of lipolysis-derived LC-acyl-CoAs, which provide a negative feedback signal that matches FFA mobilization to mitochondrial oxidative capacity.
The exact mechanism by which FAs and LC-acyl-CoAs might be transported between mitochondria and LDs is currently unknown but might involve ACSL137 or FATP4,38 which can be found at the mitochondrial-LD interface. Recent work demonstrates that distinct pools of mitochondria exist in BAs and that upon adrenergic stimulation mitochondria-LD contacts are reduced. Here, Benador et al. suggested that peri-droplet mitochondria utilize pyruvate to generate ATP and reducing equivalents required for de novo lipid synthesis, while cytoplasmic mitochondria function mostly to oxidize FAs.14 How lipid and lipid-derived signals are transported between LDs and mitochondria is unclear, but they might occur through the outer leaflets of intracellular membranes such as the ER39,40 or through acyl-CoA-binding protein (ACBP).2 Recently, Wang et al. reported a genetically encoded ratiometric sensor that reported on intracellular LC-acyl-CoAs levels, was targeted to mitochondria, and in which the free concentration of LC-acyl-CoAs could be altered through knockdown of ACBP. Further work is required to understand the exact mechanisms by which lipid transfer occurs.
The untargeted and PM-targeted LC-acyl-CoA sensors indicate their utility to report LC-acyl-CoA levels in different subcellular compartments. Interestingly, with the untargeted sensor, we were able to report an increase of LC-acyl-CoAs in the nucleus in response to exogenous application of OA. The exact functions of nuclear acyl-CoAs are unclear; however, acyl-CoAs can modify chromatin,6 and LC-acyl-CoAs might function as antagonists of nuclear receptors peroxisome proliferator-activated receptor α41 and hepatocyte nuclear Factor 4 α (HNF4α) or the corepressor C-terminal-binding protein 2 (CtBP2).42
Real-time imaging revealed oscillations of intracellular LC-acyl-CoA levels following stimulation of lipolysis, an effect that would be difficult to detect by biochemical methods lacking spatial and temporal resolution. Oscillations of LC-acyl-CoAs were not apparent when cells were challenged with exogenous OA. These data suggest that LC-acyl-CoAs are a critical signaling molecule that governs the interaction of ABHD5 with its effector lipase, PNPLA2/ATGL, and its repressor, PLIN1. Interestingly, oscillations of FA and glycerol release have been previously observed in vivo43 and in cultured adipocytes.44 These waves of FAs have been suggested to occur through oscillations of glucose that are required to generate α-glycerol-phosphate44 and thereby provide relief of LC-acyl-CoA inhibition on triglyceride lipases such as HSL45 and PNPLA2.46 Interestingly, Getty-Kaushik et al. demonstrated that application of exogenous FAs suppressed lipolysis and the amplitude of oscillations, an effect speculated to be due to increased cytosolic LC-acyl-CoAs.44 The intracellular LC-acyl-CoA oscillations and feedback inhibition on ABHD5 likely function to prevent the accumulation of toxic intracellular FAs.47,48
The competitive binding of LC-acyl-CoAs and synthetic ligands suggests that the binding pocket of ABHD5 can coordinate ligands that both promote and prevent ABHD5 binding to PLIN5, respectively. We estimate the EC50 of oleoyl-CoA for the LC-acyl-CoA sensor to be 1 μM, which is well within range of the physiological concentrations,49 especially if subcellular acyl-CoA pools exist as suggested in this work. In addition, metabolic stress, such as nutrient deprivation, is known to promote FA utilization independent of cell surface signaling.50,51 Indeed, the single-molecule FRET sensor might be used to screen for endogenous ligands of ABHD5 that might promote lipolysis in response to nutritional stress. Overall, the current data suggest that ABHD5, through its ligand-dependent interaction with PLIN and PNPLA proteins,52,53,54,55 functions to sense the local metabolic environment to regulate TAG storage and mobilization.
Limitations of the study
The current study showed that intracellular LC-acyl-CoAs can exist in highly localized pools. However, the interaction between ABHD5 and PLIN5 is not sensitive to FAs less than 10 carbon chain lengths, thus preventing the study of short-chain FAs. Moreover, GFP-based fluorescent proteins are known to be pH sensitive, possibly limiting their use in cellular compartments such as lysosomes. As such, the LC-acyl-CoA sensor would possibly benefit from calibration due to pH sensitivity of CFP and YFP in parallel experiments.35 The FRET sensor preferentially targets monolayer and bilayer membranes, so its use in purely cytosolic applications might be limited. Membrane targeting has also prevented absolute calibration of LC-acyl-CoA concentrations to a purified sensor. Thus, while the sensor detects dynamic changes in relative LC-acyl-CoA levels, absolute levels were not evaluated.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| pINDUCER20 | Meerbrey et al.56 | Addgene #44012 |
| Chemicals, peptides, and recombinant proteins | ||
| Etomoxir | Cayman Chemical | Cat#11969 |
| Geneticin (G418 Sulfate) | ThermoFisher | Cat#11811031 |
| SpeI | New England Biolabs | Cat#R3133 |
| BmgBI | New England Biolabs | Cat#R0628 |
| NotI | New England Biolabs | Cat#R3189 |
| XhoI | New England Biolabs | Cat#R0146 |
| ScaI | New England Biolabs | Cat#R3122 |
| PshAI | New England Biolabs | Cat#R0593 |
| SrgAI | New England Biolabs | Cat#R0603 |
| Optimem | ThermoFisher | Cat# 31985070 |
| Krebs Ringer Buffer | MilliporeSgima | Cat#K4002 |
| Lipofectamine LTX with Plus | ThermoFisher | Cat#15338100 |
| GenJet Plus | Signagen | Cat#SL100499 |
| ATGListatin | Cayman Chemical | Cat# 15284 |
| LipidTOX Red | ThermoFisher | Cat#H34476 |
| SR-4995 | MilliporeSigma | Sanders et al.1 Cat#SML2207 |
| coelenterazine | Prolume | Cat#301-1 |
| ER-Tracker Red | ThermoFisher | Cat#E34250 |
| CellMask™ Deep Red Plasma membrane Stain | ThermoFisher | Cat#C10046 |
| Cholera Toxin Subunit B Alexa Fluo-647 Conjugate | ThermoFisher | Cat#C34778 |
| DAPI | MilliporeSigma | Cat#D9542 |
| Fatty acids | Nu-Chek-Prep | Various |
| Krebs-Ringer Buffer | MilliporeSigma | Cat#K4002 |
| Bovine serum albumin | Alkali Scientific | Cat#FB99 |
| Dulbecco’s modified Eagle’s medium | Cytiva Hyclone | Cat#SH3024301 |
| Experimental models: Cell lines | ||
| U2OS | ATCC | Cat#HTB-96 |
| HEK293T | ATCC | Cat#CRL-3216 |
| COS-7 | ATCC | Cat#CRL-1651 |
| Brown Adipocytes | Bruce Spiegelman | Uldry et al.57 |
| Recombinant DNA | ||
| pMD2.G | Trono Lab, unpublished | Addgene #12259 |
| psPAX2 | Trono Lab, unpublished | Addgene #12260 |
| LiveDrop | Bob Farese | Wang et al.25 |
| pCMV NucleoBAS | van der Velden et al.,23 | Addgene #62861 |
| Software and algorithms | ||
| GraphPad Prism 9.0 | Dotmatics | https://www.graphpad.com/ |
| Fiji | US National Institutes of Health | https://imagej.net/software/fiji/downloads |
| Leica Application Suite X (LAS X) | Leica | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| CellSens Dimensions software | Olympus | https://www.olympus-lifescience.com/en/software/ |
| Other | ||
| Genomic sequencing services | Genewiz | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. James Granneman (jgranne@med.wayne.edu).
Materials availability
Reagents in this study are available by request to lead contact, Dr. James Granneman (jgranne@med.wayne.edu).
Experimental model and subject details
Cell culture models
HEK293T, COS-7, U2OS and mouse brown adipocytes (BA) were cultured in Dulbecco’s modified Eagle’s medium (Cytiva Hyclone, #SH3024301) supplemented with 100 U/mL Penicillin/Streptomycin and 10% Fetal Bovine Serum (FBS) in 5% CO2 at 37°C. The mouse immortalized BA cell line was a gift from Bruce Spiegelman (Harvard University). BA cells were derived as previously described57 and differentiated at previously described.58
Method details
DNA constructs, cDNA cloning and generation of stable cell lines
The generation of bipartite ECFP-ABHD5 and PLIN5-EYFP fragment (amino acids, a.a. 384–417; PLIN5384−417) have been previously described.1 The single-molecule FRET sensor was based upon the FRET bile acid sensor with weak bimolecular interaction between the Cerulean and Citrine (pCMV NucleoBAS). pCMV NucleoBAS was a gift from Stan van de Graaf (Addgene plasmid # 62861; http://n2t.net/addgene:62861; RRID:Addgene_62861).23 Full length mABHD5 was cloned in frame with Cerulean using SpeI and BmgBI sites. PLIN5384−417 was cloned in frame with Citrine using cloning sites NotI and XhoI. The ABHD5 mutants defective in PNPLA2 activation (R299N) and lipid binding (R116N) were created by overlapping PCR and the products were sub-cloned into ScaI/PshAI and SrgAI/ScaI restriction sites, respectively. The plasma membrane targeting motif of Lyn Kinase34 was cloned in frame on the N-terminus of Cerulean. All clones were confirmed by DNA sequencing (Genewiz).
The BA cell line stably expressing the single-molecule ABHD5R299N mutant FRET sensor in a doxycycline inducible manner was generated as previously described.52 The single-molecule ABHD5R299N mutant sensor was cloned into the transfer vector pINDUCER2056 target plasmid and co-transfected with pMD2.G (Addgene # 12259) and psPAX2 (Addgene #12260) packaging vectors into HEK293T cells using lipofectamine LTX and plus reagent (Invitrogen). The virus containing media was collected, filtered and centrifuged at 48,000× g for 2 h 4°C in a Beckman 25.50 fixed angle rotor and the viral pellet was resuspended in OPTIMEM. Stable BAs were generated by infecting with lentivirus for 24 h followed by G418 selection (1 mg/mL) for one week at which point cells were fluorescence-activated cell sorted for EYFP expression.
Luciferase protein complementation assays (PCA)
Luciferase PCA was performed as previously described in 293A transfected cells.52 Cell were co-transfected with ABHD5-GlucC and PLIN5384−417-GlucN overnight and the following day treated with BSA or 200 μM of the indicated fatty acids (C3:0, Propanoic acid; C4:0, butyric acid; C7:0, Heptanoic acid; C8:0, Octanoic acid; C10:0, Decanoic acid; C12:0, Dodecanoic acid; C16:0, Palmitic acid; C18:0, Stearic acid; C18:1, Oleic acid; C18:2, Linoleic acid; C18:3, α-Linolenic acid; C:20, Docosanoic acid; C20:4, Arachidonic acid; C22:6, Docosahexaenoic acid) complexed to 10% BSA for 3 h, lysed and read on a luminometer.
In vitro assays
COS-7 cells were transfected with lipofectamine 2000 in 10 cm dishes and the following day cell were collected and scarped in PBS, centrifuged at 4000 × g for 5 min and resuspended in 1 mL Sucrose Lysis Buffer (20 mM HEPES pH 7.5, 250 mM sucrose,1mM EDTA, 1 mM DDT). The cells were either lysed by sonication on ice or by passing through a 1 mL syringe with a 25G needle ten times, and the lysate was clarified by centrifugation at 12,000 g for 15 min at 4°C, diluted in PBS at a 1:1 ratio and 100 μl of lysate was dispensed per 96 well. Wells were treated with the indicated ligand and plates were immediately read on a fluorometer (ClarioStar, BMG Labtech).
Live cell fluorometry
COS-7 cells were transfected as above for in vitro assays, washed with PBS and scraped in 10 mM HEPES Krebs Ringer Buffer (HKRB) pH 7.4 (Sigma). 100 μL of cell suspension was dispersed per well of a 96-well plate and cells were treated as indicated in the figure legends.
Live cell confocal imaging
Live cell imaging of COS-7 cells was performed on an Olympus IX-81 microscope equipped with a spinning-disc confocal unit and operation controlled with CellSens Dimensions (Olympus) software. COS-7 cells were transfected with lipofectamine 2000 in 6 well dishes containing 25 mm coverslips. The following day cells were imaged in HKRB supplemented with 1% BSA (Alkali Scientific). EYFP, ECFP and FRET fluorescent images were acquired using a 60X (1.2 NA) water immersion lens. Images were acquired every minute, with basal fluorescence recorded for the first 3 min, followed by treatment with oleic acid (200 μM) or SR-4995 (20 μM). The net FRET (nFRET) was calculated from the three filter images using the FRET extension of the CellSens Dimensions software.
Live cell imaging of BA and U2OS (ATCC) cells was performed at 37°C on a laser scanning microscope (Leica SP8) equipped with dual detectors and with continuous Adaptive Focus Control between each frame. U2OS cells were transfected with GenJet Plus (Signagen) as recommended by the manufacturer and prepared as above. BA cells were differentiated in six well plates containing 25 mm glass coverslips as previously described59 and treated with doxycycline to induce expression of the FRET sensor 24 h prior to imaging. Coverslips were mounted in Attofluor cell chambers (Invitrogen) containing 0.5 mL of 10 mM HKRB with a 63×1.40NA Plan-Apochromat objective oil lens (Leica). Cerulean and Citrine were monitored simultaneous with a 405 nm laser excitation and 450–515 nm and 520–600 nm emission, respectively, using dual detectors. Images were captured every 20 s in laser scanning mode with bidirectional scanning. Cells were pretreated with inhibitors (Etomoxir, 40 μM; ATGListatin 20 μM) for 30 min prior to imaging and treatments of oleic acid (200 μM), BSA (1%) or isoproterenol were added between cycles. Image analysis was performed using Leica Application Suite X (LAS X) to quantify the regions of interest (i.e., individual cells) for ECFP and EYFP-FRET emission signals and expressed as an FRET ratio (EYFP-FRET/ECFP). Organelle staining was performed for neutral lipids with LipidTOX Red (ThermoFisher), ER with ER-Tracker Red (ThermoFisher), plasma membrane with Cholera Toxin Subunit B Alexa Fluo-647 Conjugate (ThermoFisher) or CellMask Deep Red Plasma Membrane Stain (ThermoFisher) and nuclei with DAPI (SigmaMillipore), as suggested by the manufacturer where indicated. Sites of novo LD formation on the ER were marked by co-transfecting U2OS cells with Cherry-LiveDrop25 and the single-molecule ABHD5R299N mutant FRET sensor. Images were merged in Fiji (Fiji Is Just ImageJ2).
Quantification and statistical analysis
All results are mean ± SEM. Statistical significance was determine using GraphPad Prism software. Data with unequal variances were analyzed using an unpaired Welch’s t-test for unequal variances or one-way ANOVA with Dunnett’s post-hoc analysis for more than two groups.
Acknowledgments
We thank Grace Teskey for performing transfections in U2OS cells and Nivedita Tiwari for cell culture work. This work was supported by NIH grants R00DK114471 and R01DK126743 to E.P.M., R01DK105963 and R21DK091741 to J.G.G., and R01DK76629 to J.G.G. and C.V.K. The funding agencies were not involved in the study design, collection analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication. Graphical abstract was partly created with BioRender.com.
Author contributions
Conceptualization, E.P.M. and J.G.G.; methodology, E.P.M., J.G.G., and P.A.O.; formal analysis, E.P.M., L.M.-L., J.G.G., and C.V.K.; investigation, E.P.M., L.M.-L., H.Z., L.Z., and J.G.G.; writing, E.P.M. and J.G.G.; funding acquisition, E.P.M. and J.G.G.; resources, E.P.M., J.G.G., and P.O.; supervision, E.P.M. and J.G.G.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: January 25, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100394.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact (jgranne@med.wayne.edu).
This paper does did not generate original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (jgranne@med.wayne.edu) upon request.
References
- 1.Sanders M.A., Madoux F., Mladenovic L., Zhang H., Ye X., Angrish M., Mottillo E.P., Caruso J.A., Halvorsen G., Roush W.R., et al. Endogenous and synthetic ABHD5 ligands regulate ABHD5-perilipin interactions and lipolysis in fat and muscle. Cell Metab. 2015;22:851–860. doi: 10.1016/j.cmet.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faergeman N.J., Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem. J. 1997;323:1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper D.E., Young P.A., Klett E.L., Coleman R.A. Physiological consequences of compartmentalized acyl-CoA metabolism. J. Biol. Chem. 2015;290:20023–20031. doi: 10.1074/jbc.R115.663260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellis J.M., Li L.O., Wu P.-C., Koves T.R., Ilkayeva O., Stevens R.D., Watkins S.M., Muoio D.M., Coleman R.A. Adipose acyl-CoA synthetase-1 directs fatty acids toward β-oxidation and is required for cold thermogenesis. Cell Metab. 2010;12:53–64. doi: 10.1016/j.cmet.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watkins P.A. Very-long-chain acyl-CoA synthetases. J. Biol. Chem. 2008;283:1773–1777. doi: 10.1074/jbc.R700037200. [DOI] [PubMed] [Google Scholar]
- 6.Trefely S., Lovell C.D., Snyder N.W., Wellen K.E. Compartmentalised acyl-CoA metabolism and roles in chromatin regulation. Mol. Metab. 2020;38:100941. doi: 10.1016/j.molmet.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granneman J.G., Moore H.-P.H., Mottillo E.P., Zhu Z. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J. Biol. Chem. 2009;284:3049–3057. doi: 10.1074/jbc.M808251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palade G.E. In: Hayashi T., Ronald, editors. New York; 1959. Functional changes in the structure of cell components. In: Subcellular Particles; pp. 64–83. [Google Scholar]
- 9.Bosma M., Minnaard R., Sparks L.M., Schaart G., Losen M., de Baets M.H., Duimel H., Kersten S., Bickel P.E., Schrauwen P., Hesselink M.K.C. The lipid droplet coat protein perilipin 5 also localizes to muscle mitochondria. Histochem. Cell Biol. 2012;137:205–216. doi: 10.1007/s00418-011-0888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., Sreenivasan U., Hu H., Saladino A., Polster B.M., Lund L.M., Gong D.W., Stanley W.C., Sztalryd C. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 2011;52:2159–2168. doi: 10.1194/jlr.M017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varghese M., Kimler V.A., Ghazi F.R., Rathore G.K., Perkins G.A., Ellisman M.H., Granneman J.G. Adipocyte lipolysis affects Perilipin 5 and cristae organization at the cardiac lipid droplet-mitochondrial interface. Sci. Rep. 2019;9:4734. doi: 10.1038/s41598-019-41329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kien B., Kolleritsch S., Kunowska N., Heier C., Chalhoub G., Tilp A., Wolinski H., Stelzl U., Haemmerle G. Lipid droplet-mitochondria coupling via perilipin 5 augments respiratory capacity but is dispensable for FA oxidation. J. Lipid Res. 2022;63:100172. doi: 10.1016/j.jlr.2022.100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gemmink A., Daemen S., Kuijpers H.J.H., Schaart G., Duimel H., López-Iglesias C., van Zandvoort M.A.M.J., Knoops K., Hesselink M.K.C. Super-resolution microscopy localizes perilipin 5 at lipid droplet-mitochondria interaction sites and at lipid droplets juxtaposing to perilipin 2. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids. 2018;1863:1423–1432. doi: 10.1016/j.bbalip.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Benador I.Y., Veliova M., Mahdaviani K., Petcherski A., Wikstrom J.D., Assali E.A., Acín-Pérez R., Shum M., Oliveira M.F., Cinti S., et al. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 2018;27:869–885.e6. doi: 10.1016/j.cmet.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders M.A., Zhang H., Mladenovic L., Tseng Y.Y., Granneman J.G. Molecular basis of ABHD5 lipolysis activation. Sci. Rep. 2017;7:42589. doi: 10.1038/srep42589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rondini E.A., Mladenovic-Lucas L., Roush W.R., Halvorsen G.T., Green A.E., Granneman J.G. Novel pharmacological probes reveal ABHD5 as a locus of lipolysis control in white and brown adipocytes. J. Pharmacol. Exp. Ther. 2017;363:367–376. doi: 10.1124/jpet.117.243253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G., Zhou G., Lotvola A., Granneman J.G., Wang J. ABHD5 suppresses cancer cell anabolism through lipolysis-dependent activation of the AMPK/mTORC1 pathway. J. Biol. Chem. 2021;296:100104. doi: 10.1074/jbc.RA120.014682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen G., Zhou G., Aras S., He Z., Lucas S., Podgorski I., Skar W., Granneman J.G., Wang J. Loss of ABHD5 promotes the aggressiveness of prostate cancer cells. Sci. Rep. 2017;7:13021. doi: 10.1038/s41598-017-13398-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ou J., Peng Y., Yang W., Zhang Y., Hao J., Li F., Chen Y., Zhao Y., Xie X., Wu S., et al. ABHD5 blunts the sensitivity of colorectal cancer to fluorouracil via promoting autophagic uracil yield. Nat. Commun. 2019;10:1078. doi: 10.1038/s41467-019-08902-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ou J., Miao H., Ma Y., Guo F., Deng J., Wei X., Zhou J., Xie G., Shi H., Xue B., et al. Loss of Abhd5 promotes colorectal tumor development and progression by inducing aerobic glycolysis and epithelial-mesenchymal transition. Cell Rep. 2014;9:1798–1811. doi: 10.1016/j.celrep.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlsson B., Lindén D., Brolén G., Liljeblad M., Bjursell M., Romeo S., Loomba R. Review article: the emerging role of genetics in precision medicine for patients with non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 2020;51:1305–1320. doi: 10.1111/apt.15738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang A., Mottillo E.P. Adipocyte lipolysis: from molecular mechanisms of regulation to disease and therapeutics. Biochem. J. 2020;477:985–1008. doi: 10.1042/BCJ20190468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Velden L.M., Golynskiy M.V., Bijsmans I.T.G.W., van Mil S.W.C., Klomp L.W.J., Merkx M., van de Graaf S.F.J. Monitoring bile acid transport in single living cells using a genetically encoded Förster resonance energy transfer sensor. Hepatology. 2013;57:740–752. doi: 10.1002/hep.26012. [DOI] [PubMed] [Google Scholar]
- 24.Tseng Y.Y., Sanders M.A., Zhang H., Zhou L., Chou C.Y., Granneman J.G. Structural and functional insights into ABHD5, a ligand-regulated lipase co-activator. Sci. Rep. 2022;12:2565. doi: 10.1038/s41598-021-04179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H., Becuwe M., Housden B.E., Chitraju C., Porras A.J., Graham M.M., Liu X.N., Thiam A.R., Savage D.B., Agarwal A.K., et al. Seipin is required for converting nascent to mature lipid droplets. Elife. 2016;5:e16582. doi: 10.7554/eLife.16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira A.F., Cunha D.A., Ladriere L., Igoillo-Esteve M., Bugliani M., Marchetti P., Cnop M. In vitro use of free fatty acids bound to albumin: a comparison of protocols. Biotechniques. 2015;58:228–233. doi: 10.2144/000114285. [DOI] [PubMed] [Google Scholar]
- 27.Cnop M., Hannaert J.C., Hoorens A., Eizirik D.L., Pipeleers D.G. Inverse relationship between cytotoxicity of free fatty acids in pancreatic islet cells and cellular triglyceride accumulation. Diabetes. 2001;50:1771–1777. doi: 10.2337/diabetes.50.8.1771. [DOI] [PubMed] [Google Scholar]
- 28.Brasaemle D.L., Dolios G., Shapiro L., Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian V., Rothenberg A., Gomez C., Cohen A.W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M.P., Brasaemle D.L. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J. Biol. Chem. 2004;279:42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi T., Omatsu N., Matsushita S., Osumi T. CGI-58 interacts with perilipin and is localized to lipid droplets: possible involvement of CGI-58 mislocalization in chanarin-dorfman syndrome. J. Biol. Chem. 2004;279:30490–30497. doi: 10.1074/jbc.M403920200. [DOI] [PubMed] [Google Scholar]
- 31.Bersuker K., Peterson C.W.H., To M., Sahl S.J., Savikhin V., Grossman E.A., Nomura D.K., Olzmann J.A. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev. Cell. 2018;44:97–112.e7. doi: 10.1016/j.devcel.2017.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Divakaruni A.S., Hsieh W.Y., Minarrieta L., Duong T.N., Kim K.K.O., Desousa B.R., Andreyev A.Y., Bowman C.E., Caradonna K., Dranka B.P., et al. Etomoxir inhibits macrophage polarization by disrupting CoA homeostasis. Cell Metab. 2018;28:490–503.e7. doi: 10.1016/j.cmet.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shahoei R., Pangeni S., Sanders M.A., Zhang H., Mladenovic-Lucas L., Roush W.R., Halvorsen G., Kelly C.V., Granneman J.G., Huang Y.-m.M. Molecular modeling of ABHD5 structure and ligand recognition. Front. Mol. Biosci. 2022;9:935375. doi: 10.3389/fmolb.2022.935375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue T., Heo W.D., Grimley J.S., Wandless T.J., Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W., Wei Q., Zhang J., Zhang M., Wang C., Qu R., Wang Y., Yang G., Wang J. A ratiometric fluorescent biosensor reveals dynamic regulation of long-chain fatty acyl-CoA esters metabolism. Angew. Chem. Int. Ed. Engl. 2021;60:13996–14004. doi: 10.1002/anie.202101731. [DOI] [PubMed] [Google Scholar]
- 36.Raclot T., Oudart H. Net release of individual fatty acids from white adipose tissue during lipolysis in vitro: evidence for selective fatty acid re-uptake. Biochem. J. 2000;348:129–136. [PMC free article] [PubMed] [Google Scholar]
- 37.Young P.A., Senkal C.E., Suchanek A.L., Grevengoed T.J., Lin D.D., Zhao L., Crunk A.E., Klett E.L., Füllekrug J., Obeid L.M., Coleman R.A. Long-chain acyl-CoA synthetase 1 interacts with key proteins that activate and direct fatty acids into niche hepatic pathways. J. Biol. Chem. 2018;293:16724–16740. doi: 10.1074/jbc.RA118.004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miner G.E., So C.M., Edwards W., Herring L.E., Coleman R.A., Klett E.L., Cohen S. Perilipin 5 interacts with Fatp4 at membrane contact sites to promote lipid droplet-tomitochondria fatty acid transport. bioRxiv. 2022 doi: 10.1101/2022.02.03.479028. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanchette-Mackie E.J., Scow R.O. Lipolysis and lamellar structures in white adipose tissue of young rats: lipid movement in membranes. J. Ultrastruct. Res. 1981;77:295–318. doi: 10.1016/s0022-5320(81)80026-3. [DOI] [PubMed] [Google Scholar]
- 40.Blanchette-Mackie E.J., Scow R.O. Movement of lipolytic products to mitochondria in brown adipose tissue of young rats: an electron microscope study. J. Lipid Res. 1983;24:229–244. doi: 10.1016/s0022-2275(20)37992-x. [DOI] [PubMed] [Google Scholar]
- 41.Elholm M., Dam I., Jørgensen C., Krogsdam A.-M., Holst D., Kratchmarova I., Göttlicher M., Gustafsson J.-Å., Berge R., Flatmark T., et al. Acyl-CoA esters antagonize the effects of ligands on peroxisome proliferator-activated receptor α conformation, DNA binding, and interaction with Co-factors. J. Biol. Chem. 2001;276:21410–21416. doi: 10.1074/jbc.M101073200. [DOI] [PubMed] [Google Scholar]
- 42.Sekiya M., Kainoh K., Sugasawa T., Yoshino R., Hirokawa T., Tokiwa H., Nakano S., Nagatoishi S., Tsumoto K., Takeuchi Y., et al. The transcriptional corepressor CtBP2 serves as a metabolite sensor orchestrating hepatic glucose and lipid homeostasis. Nat. Commun. 2021;12:6315. doi: 10.1038/s41467-021-26638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karpe F., Fielding B.A., Coppack S.W., Lawrence V.J., Macdonald I.A., Frayn K.N. Oscillations of fatty acid and glycerol release from human subcutaneous adipose tissue in vivo. Diabetes. 2005;54:1297–1303. doi: 10.2337/diabetes.54.5.1297. [DOI] [PubMed] [Google Scholar]
- 44.Getty-Kaushik L., Richard A.-M.T., Corkey B.E. Free fatty acid regulation of glucose-dependent intrinsic oscillatory lipolysis in perifused isolated rat adipocytes. Diabetes. 2005;54:629–637. doi: 10.2337/diabetes.54.3.629. [DOI] [PubMed] [Google Scholar]
- 45.Jepson C.A., Yeaman S.J. Inhibition of hormone-sensitive lipase by intermediary lipid metabolites. FEBS Lett. 1992;310:197–200. doi: 10.1016/0014-5793(92)81328-j. [DOI] [PubMed] [Google Scholar]
- 46.Nagy H.M., Paar M., Heier C., Moustafa T., Hofer P., Haemmerle G., Lass A., Zechner R., Oberer M., Zimmermann R. Adipose triglyceride lipase activity is inhibited by long-chain acyl-coenzyme A. Biochim. Biophys. Acta. 2014;1841:588–594. doi: 10.1016/j.bbalip.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mottillo E.P., Granneman J.G. Intracellular fatty acids suppress beta-adrenergic induction of PKA-targeted gene expression in white adipocytes. Am. J. Physiol. Endocrinol. Metab. 2011;301:E122–E131. doi: 10.1152/ajpendo.00039.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mottillo E.P., Shen X.J., Granneman J.G. β3-adrenergic receptor induction of adipocyte inflammation requires lipolytic activation of stress kinases p38 and JNK. Biochim. Biophys. Acta. 2010;1801:1048–1055. doi: 10.1016/j.bbalip.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knudsen J., Neergaard T.B., Gaigg B., Jensen M.V., Hansen J.K. Role of acyl-CoA binding protein in acyl-CoA metabolism and acyl-CoA–mediated cell signaling. J. Nutr. 2000;130:294S–298S. doi: 10.1093/jn/130.2.294s. [DOI] [PubMed] [Google Scholar]
- 50.Veliova M., Ferreira C.M., Benador I.Y., Jones A.E., Mahdaviani K., Brownstein A.J., Desousa B.R., Acín-Pérez R., Petcherski A., Assali E.A., et al. Blocking mitochondrial pyruvate import in brown adipocytes induces energy wasting via lipid cycling. EMBO Rep. 2020;21:e49634. doi: 10.15252/embr.201949634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vacanti N.M., Divakaruni A.S., Green C.R., Parker S.J., Henry R.R., Ciaraldi T.P., Murphy A.N., Metallo C.M. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol. Cell. 2014;56:425–435. doi: 10.1016/j.molcel.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang A., Mottillo E.P., Mladenovic-Lucas L., Zhou L., Granneman J.G. Dynamic interactions of ABHD5 with PNPLA3 regulate triacylglycerol metabolism in brown adipocytes. Nat. Metab. 2019;1:560–569. doi: 10.1038/s42255-019-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohno Y., Nara A., Nakamichi S., Kihara A. Molecular mechanism of the ichthyosis pathology of Chanarin–Dorfman syndrome: stimulation of PNPLA1-catalyzed ω-O-acylceramide production by ABHD5. J. Dermatol. Sci. 2018;92:245–253. doi: 10.1016/j.jdermsci.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 54.Kien B., Grond S., Haemmerle G., Lass A., Eichmann T.O., Radner F.P.W. ABHD5 stimulates PNPLA1-mediated ω- O -acylceramide biosynthesis essential for a functional skin permeability barrier. J. Lipid Res. 2018;59:2360–2367. doi: 10.1194/jlr.M089771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J.G., Gorkiewicz G., Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Meerbrey K.L., Hu G., Kessler J.D., Roarty K., Li M.Z., Fang J.E., Herschkowitz J.I., Burrows A.E., Ciccia A., Sun T., et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 58.Mottillo E.P., Bloch A.E., Leff T., Granneman J.G. Lipolytic products activate peroxisome proliferator-activated receptor (PPAR) alpha and delta in brown adipocytes to match fatty acid oxidation with supply. J. Biol. Chem. 2012;287:25038–25048. doi: 10.1074/jbc.M112.374041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mottillo E.P., Zhang H., Yang A., Zhou L., Granneman J.G. Genetically-encoded sensors to detect fatty acid production and trafficking. Mol. Metab. 2019;29:55–64. doi: 10.1016/j.molmet.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact (jgranne@med.wayne.edu).
This paper does did not generate original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (jgranne@med.wayne.edu) upon request.