Abstract
Objective:
Inflammatory bowel disease (IBD) therapies and treatments are evolving to deeper levels of remission. Molecular measures of disease may augment current endpoints including the potential for less invasive assessments.
Design:
Transcriptome analysis on 712 endoscopically defined inflamed (Inf) and 1778 non-inflamed (Non-Inf) intestinal biopsies (n=498 Crohn’s Disease (CD), n=421 Ulcerative Colitis (UC) and 243 controls) in the Mount Sinai Crohn’s and Colitis Registry were used to identify genes differentially expressed between Inf and Non-Inf biopsies and to generate a molecular inflammation score (bMIS) via gene set variance analysis. A circulating MIS (cirMIS) score, reflecting intestinal molecular inflammation, was generated using blood transcriptome data. bMIS/cirMIS were validated as indicators of intestinal inflammation in four independent IBD cohorts.
Results:
bMIS/cirMIS were strongly associated with clinical, endoscopic and histological disease activity indices. Patients with the same histologic score of inflammation, had variable bMIS scores, indicating that bMIS describes a deeper range of inflammation. In available clinical trial datasets, both scores were responsive to IBD treatment. Despite similar baseline endoscopic and histologic activity, UC patients with lower baseline bMIS levels were more likely treatment responders compared to those with higher levels. Finally, among UC patients in endoscopic and histologic remission, those with lower bMIS levels were less likely to have a disease flare over time.
Conclusion:
Transcriptionally-based scores provide an alternative objective and deeper quantification of intestinal inflammation which could augment current clinical assessments used for disease monitoring and have potential for predicting therapeutic response and patients at higher risk of disease flares.
Short Summary:
-
What is already known about this subject?
Treatment targets in inflammatory bowel disease are moving towards deeper levels of remission, from clinical to endoscopic and recently, histologic normalizations. How ‘deep’ should we go for long term disease control is an on-going question.
-
What this study adds?
Current measurements of IBD activity are likely under-representing persistent inflammation at the molecular level which can be expressed using biopsy and blood scores.
-
How this study might affect research, practice or policy?
Our work lays a platform for augmenting current clinical practices associated with patient disease monitoring, stratification and therapeutic response management through the use of molecular scores of inflammation based on expression levels of specific genes measured in either mucosal biopsies or non-invasively, in circulating blood RNA.
Introduction:
Inflammatory Bowel Disease (IBD) is a progressive inflammatory disease of the digestive tract characterized by periods of relapses and remission and consists of two types, namely Crohn’s Disease (CD) and Ulcerative Colitis (UC)1,2. In the past two decades the therapeutic goal in IBD has evolved from attaining mere symptomatic remission to achieving sustained clinical and endoscopic remission with the ultimate aim of disease modification defined as blocking natural progression to complications and surgery3,4,5. However, despite major advances in drug development and innovative therapeutic strategies, the proportion of patients in whom disease modification can be reached remains regrettably low6. More ambitious targets are now proposed such as combined endoscopic and histologic remission in UC and transmural healing in CD7,8,9. Another approach already proposed in other immune-mediated disorders10,11,12 which has yet to be explored in IBD, is to target inflammation that may silently persist at the molecular level even in macroscopically and/or histologically normal mucosa.
We therefore constructed a biopsy molecular inflammation score (bMIS) with the rationale that it may enable a more objective, granular and sensitive measure of disease activity applicable to both CD and UC patients. We also derived a circulating biomarker (cirMIS) of gut inflammation using blood RNA transcriptomic data to develop a less invasive blood test of disease activity. Both bMIS and cirMIS were developed using the Mount Sinai Crohn’s and Colitis registry (MSCCR), a discovery cohort with ~ 1200 IBD patients and controls13. As histological, endoscopic and clinical assessments were available on the same patient cohort, cross-comparisons of newly derived molecular with clinical scores could be performed. We then used seven different independent IBD datasets with available transcriptome data to evaluate the robustness of various aspects of the molecular score, including association with disease as well as association with treatment (compared to placebo) and treatment response, where clinical trial data was available.
Methods:
Discovery Cohort: Mount Sinai Crohn’s and Colitis Registry (MSCCR)
The Mount Sinai Crohn’s and Colitis registry (MSCCR) is a cross-sectional cohort consisting of IBD patients and controls prospectively recruited during their endoscopy visit from December 2013 - September 201613. Paired blood and biopsy RNA sequencing (RNA-Seq) data were generated at the time of clinical, histological and endoscopic assessments (Figure 1a). Institutional review board approval and informed consents were obtained. The Simple Endoscopic Score for Crohn’s Disease (SES-CD) was used14 to classify CD endoscopic disease activity as inactive (0–2), mild (3–6), moderate (7–15), and severe (≥16)15. The Mayo endoscopic measure was used to categorize UC as having: normal/inactive disease (0); mild disease (1); moderate disease (2); or severe disease (3)16. Clinical disease activity measures included the Harvey-Bradshaw index (HBI) for CD and Clinician-based Simple Clinical Colitis Activity Index (SCCAI) for UC. Clinically inactive disease was defined as an HBI < 5 or an SCCAI < 5 and active disease as an HBI >7 or SSCAI >= 5. Histological assessment was performed by a pathologist (A. Iuga) on biopsies taken adjacent to the specimen processed for RNA sequence analysis. Control and UC patient biopsies were scored for the Nancy index (0–4)17 and Control and CD biopsies were scored according to the General histology activity score (GHAS) (0–13 using 7 of 8 scoring criteria, (does not include ‘No. of biopsy specimens affected’))18,19,20,21. GHAS score and Nancy index are considered acceptable and reproducible methods for the histological scoring of disease activity in IBD. They provide an assessment of chronic and active mucosal inflammatory changes and gross epithelial damage22. Montreal disease phenotypic sub-classifications including UC disease extent (E1, E2 or E3)23 was available. Demographic information associated with this cohort is summarized in Supplementary Table 1.
Figure 1.
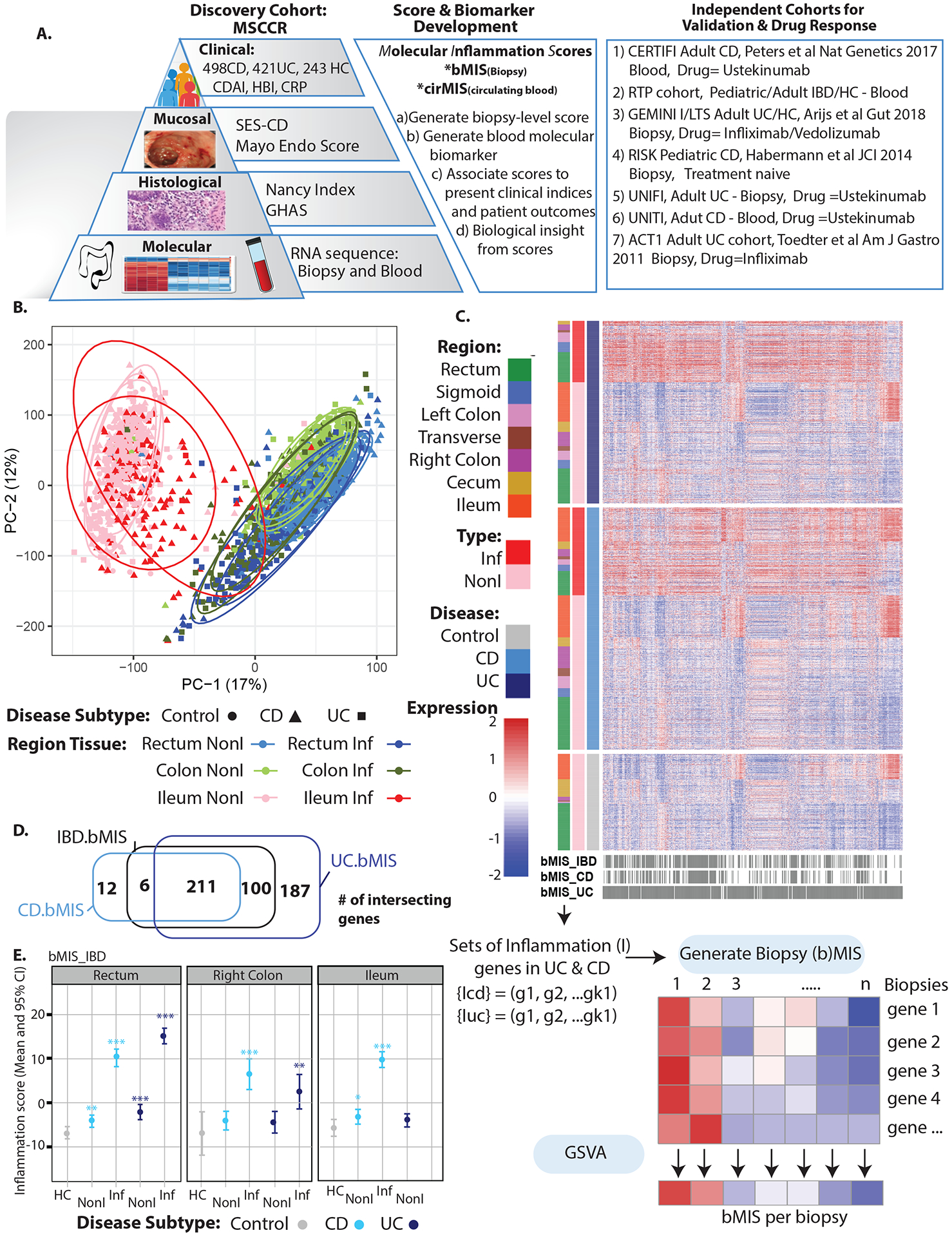
A. Schematic of analysis plan B. PCA of biopsy expression data for the MSCCR cohort by colonic region, endoscopically defined inflamed (Inf) or non-inflamed (NonI) tissue and by disease subtype (UC/CD and non-IBD controls). C. Heatmap representing the expression levels of the set of bMIS genes (UC, CD or IBD) in IBD patient or non-IBD control biopsies and labeled by region, disease type and inflammation status. D. Venn Diagrams showing the number of differentially expressed genes for bMIS IBD, bMIS UC and bMIS CD signatures. E. Estimated marginal mean (EMM) and 95% CI for bMIS_IBD levels by intestinal region, inflammation status and disease subtype (HC = healthy controls).
Validation cohorts:
Cross-sectional Cohorts:
-
1
RTP. The road to prevention (RTP) cohort encompasses 346 subjects from 83 families with at least 2 first degree relatives diagnosed with IBD and a set of unrelated healthy individuals with no family history of IBD or other chronic immune diseases and matched for age, gender and ethnicity. Among those, 32 participants were unrelated healthy individuals with no IBD family history, 179 were unaffected relatives of the 135 participants that were diagnosed with IBD, with an average age of 33 years old, both genders equally represented (49.1% male) and most participants as Caucasians (97.6%) and Ashkenazi Jewish (89.8%). Demographics associated with this cohort is summarized in Supplementary Table 2.
-
2
The RISK (Risk Stratification and Identification of Immunogenetic and Microbial Markers of Rapid Disease Progression in Children with Crohn’s Disease) cohort24 of treatment-naive pediatric CD (<17 years of age, 58.5% Male) patients were studied using RNA-sequencing expression profiles from GSE57945, which included ileal biopsies from endoscopically defined inflamed samples (n=163), non-inflamed (n=55) and non-IBD controls (n=42) at the time of diagnosis. Demographics is summarized in Supplementary Table 3 for RISK cohort.
Longitudinal Cohorts:
-
3
The CERTIFI GSE100833 series includes blood Affymetrix (HGU-U133 Plus) expression profiles from 226 anti-tumor necrosis factor alpha (TNFα) refractory CD patients enrolled in the CERTIFI trial with ustekinumab25. Clinical response at week 22 was defined as a decrease of 100 or more in Crohn’s Disease Activity Index (CDAI) score from baseline at week 22.
-
4
The GEMINI-I/LTS and anti-TNF GSE73661 series includes colonic gene expression (Affymetrix, HGU-1.0 ST) profiles from 44 moderate-to-severe UC patients enrolled in two vedolizumab (VDZ) efficacy trials (GEMINI-I/LTS)26. Also included were 12 non-IBD and 23 UC colonic biopsies from patients before and 4–6 weeks after first infliximab (IFX) treatment. Response to therapy was defined as endoscopic mucosal healing (Mayo endoscopic score 0–1) and assessed at 6 weeks for VDZ and 4–6 weeks for IFX.
-
5
The UNIFI UC phase 3 clinical trial of ustekinumab (UST) involves moderate-to-severe UC patients that had inadequate response to or unacceptable side effects from TNF antagonists, VDZ, or conventional therapy27. An 8-week randomized induction trial and a 44-week randomized-withdrawal maintenance trial was performed with primary endpoint of clinical remission (defined as a total score of ≤2 on the total Mayo scale and no subscore >1). Other endpoints included, endoscopic improvement (defined as a Mayo endoscopic subscore of 0 or 1), histological healing (defined as neutrophil infiltration in <5% of crypts, no crypt destruction, and no erosions, ulcerations, or granulation tissue) and histo-endoscopic mucosal healing (HEMH) requiring both endoscopic improvement and histological healing. Biopsy transcriptome data (microarray) was available from 550 patients, 358 genes of the 498 bMIS UC geneset were available to generate GSVA scores. The data are available on GEO (GSE206285).
-
6
The UNITI-1/2 CD induction and maintenance trial for UST included primary/secondary non-responders to anti-TNF and patients in whom conventional therapy failed28. The primary end point for the induction trials was a clinical response at week 6 (defined as a decrease from baseline in the CDAI score of ≥100 points or a CDAI score <150). 623 patients had whole blood transcriptome available and 73 of the 103 genes in the cirMIS CD signature set were available to generate cirMIS scores. The data are available on GEO (GSE207465 ).
-
7
The UC ACT1 anti-TNF (Infliximab) adult cohort29 with the biopsy microarray dataset (GSE23597) consists of a cohort of patients who participated in the placebo-controlled ACT1 study. Colonic biopsies were collected from a subset of randomized patients at protocol-specified time points after infliximab therapy at 5 or 10mg/kg doses. Clinical response was defined as a decrease from baseline in the total Mayo score (of at least three points).
-
8
MSCCR patient subset follow-up. A subset of MSCCR patients with longitudinal follow-up were selected and all charts reviewed by an expert in IBD (RU). This MSCCR subset included UC and CD patients in endoscopic and histological remission at the time of the study. For UC, the criteria were Mayo endo score =0 and Nancy Score = 0. For CD, SESCD =0 and GHAS score= 0. Patients in remission were then categorized as having high or low cirMIS levels based on tertiles of expression (UC patients High n=8, Low n=8 and CD patients High n=13, Low n=13) and post-MSCCR study outcomes were investigated through chart review. The outcome was disease flare defined as a composite of any IBD-related hospitalizations, IBD-related surgery, need for new oral steroid and/or need for treatment escalation or new therapeutic agent due to active disease. We recorded date of the earliest adverse disease flare event or the date of the last follow up if no event was seen.
MSCCR RNA-sequence profiling
Biopsy and blood RNA from MSCCR patients was extracted and processed in randomly allocated batches as previously described30 and in Supplementary methods. Coupled genotype data for the same patients was available31. For bMIS generation, biopsy data for patients with indeterminate IBD disease were removed (n=13) and biopsies identified as inflamed in the healthy control group were also removed (n=7). For subsequent analysis, biopsies from pouch patients (n=18 unique) were also removed. The data are available on GEO (GEO accession: GSE186507 for blood and GSE193677 for biopsy).
Generation of molecular inflammation scores (MIS) for biopsy (bMIS) and peripheral blood (cirMIS):
bMIS (biopsy molecular inflammation score):
Gene expression matrices from biopsy were generated from the count matrices using the voom transformation on the count matrix (See supplementary methods). Voom-transformed gene expression data was modelled using a mixed-effect models with ‘tissue type’ (i.e. endoscopically inflamed or non-inflamed), ‘intestine biopsy region’ (ileum, colon, rectum etc) and ‘disease sub-type’ (Control, UC, CD) and its interactions as factors and a random factor for each patient, with technical (batch, RIN, rRNA rate, exonic rate) and relevant variables (age, gender, and genetic PC’s 1–5) as covariates. Notice that this model includes control samples of non-IBD subject, in order to account for the gut region effect not driven by inflammation; by including both region and disease as covariates in the development of bMIS. In this model differences between endoscopically inflamed and non-inflamed tissue were assessed for each intestinal region (7 possible including: rectum, sigmoid, left colon, transverse, right colon, cecum, ileum) and disease subtype, thus defining intestinal region- and disease subtype- specific inflammation signatures (Figure 1B–C). However, as we observed a strong correlation across the inflammation signatures, we generated a general IBD inflammation signature by fitting a model with tissue type, disease sub-type and intestine biopsy region (no interactions) and an inflammation signature for each disease subtype by including only an interaction term for tissue type by disease sub-type. From the IBD, or CD and UC subtype-specific inflammation gene signatures, we defined the markers of biopsy inflammation as genes differentially expressed (up-regulated genes only) between endoscopically inflamed and non-inflamed biopsies, at FDR<0.05 and fold change (FCH)>2 and the bMIS score was derived by using a gene-set variation analysis (GSVA32). The inflammation score was built as the average z-score derived from the expression (adjusted for technical covariates) of the differentially expressed genes (DEGs) normalized by the square root of the number of genes33. As a result, each biopsy sample for the MSCCR cohort had a bMIS_IBD score as well as either a UC or CD disease sub-type specific score (bMIS_UC or bMIS_CD), depending on the patient’s disease sub-type diagnosis. This score is based on all DEG, as we aimed to quantify the overall level of molecular inflammation (ie a continuous score not an ordinal value like endoscopic or histological scores) that summarizes the activity of all dysregulated genes, and not to develop a predictor of endoscopic inflammation status (yes vs no)). This rationale was also based on our experience in psoriasis where we developed a similar transcriptome scoring system (see supplementary methods).
cirMIS:
Blood gene expression data from 1030 patients for which intestinal biopsy transcriptome data was available, was used to identify genes whose expression in blood associated with the level of intestinal molecular inflammation. To obtain a patient-level, intestine molecular-based inflammation measure, we took advantage of the multiple regions sampled per individual and summarized the patient’s individual bMIS scores into an intestinal-level (ileum-to-rectum) inflammation score (iMIS) as described in Supplementary Methods. The blood gene expression data was then modeled using a linear model with the continuous variables iMIS, technical covariates (RIN, batch, rRNA rate and exonic rate), imputed genetic PC’s (#1–5), age at endoscopy, sex, and IBD disease sub-type. iMIS-associated blood genes were selected and used as the input to generate a circulating molecular score that reflects intestinal inflammation (cirMIS) using GSVA.
As in the case of bMIS, in addition to cirMIS_IBD we also generated subtype-specific cirMIS scores, ie cirMIS_UC, cirMIS_CD) by identifying the blood gene signatures that were associated with the iMIS_CD and iMIS_UC in each CD and UC sub-cohorts, respectively.
Association of bMIS and cirMIS with IBD phenotypes, treatment effect, and longitudinal outcomes
Statistical analysis was carried out using R language version 4.0.534 and its available packages. Each MIS for the discovery or validation cohorts was modeled using linear models after suitable pre-processing of the omics-data with relevant factors depending on the comparison. When data was paired, i.e. several biopsies were available for the same patient, or different time points, mixed-effect models were fitted including fixed factors and a random intercept for each subject using the nlme package in R. Model assessing changes with treatment included fixed effects for time, treatment, response and its interactions with time as fixed effects. For all models, classical model diagnosis was run. Marginal means, confidence intervals (CI) were derived from fitted models using the emmeans package capabilities, and hypotheses of interests were tested using contrasts.
The tests described above are considered to be self-contained, where the association of genes other than the cirMIS/bMIS genes is not considered. As such, we also conducted “competitive tests” where the null hypothesis assumes that genes in the bMIS/cirMIS are not more associated with the phenotype than other genes. To this end we performed bootstrap simulations, by evaluating the association with disease outcomes for 500 randomly selected genes sets of the same size as cirMIS/bMIS and quantified the 95% CI using the quantiles.
Logistic regression models were fitted using generalized linear models (glm) with the disease scores (either continuous or discretized as (low/high)) as factors and 10-fold cross-validation statistics were derived using the boot package. Confidence intervals were derived DeLong’s test was used to compare receiver operating characteristic (ROC) curves between two sets of predictors.
Correlation of endoscopic, histological, and clinical disease activity (continuous) measures with the molecular scores was assessed using Spearman correlations. Strength of said association is represented as a heatmap, where scores were clustered based on said distance using package corr using Ward agglomeration method.
Pathway enrichment analysis of bMIS and cirMIS associated genesets:
Gene sets were tested for enrichment using a Fisher’s exact test with a Benjamini-Hochberg (BH) multiple test correction. The collection of genesets included BioPlanet pathways sourced from Enrichr35; CD and UC single cell gene sets36,37, 38; IBD GWAS candidate genes sourced from39,40 ,41 and genes associated with IBD drug targets42. iRegulon43, within Cytoscape (v3.9.0)44, was used to detect enriched transcription factor motifs within the bMIS genesets.
Patient and Public Involvement:
Patients and/or the public were not involved in the design or reporting of this research.
Results
A biopsy-level molecular inflammation score (bMIS) as a novel IBD activity measure
Figure 1A summarizes the analysis plan and validation strategy with MSCCR cohort clinical characteristics in Supplementary Table 1. Principal component analysis (PCA, Figure 1B) of the intestinal transcriptomes revealed that region of biopsy was the largest factor contributing to variation in gene expression, followed by inflammation status, with disease subtype (UC vs CD) showing very little separation. To generate bMIS we focused on genes found to be differentially expressed between endoscopically-defined inflamed and uninflamed biopsies (Figure 1C). Many genes were found commonly differentially expressed when inflammation was present in UC or CD, despite location of biopsy (Figure 1D) and therefore bMIS scores were generated within disease type (bMIS_CD and bMIS_UC) or across disease subtypes (bMIS_IBD) (ST4–5).
Using the sets of genes upregulated with inflammation, a GSVA score was generated which compressed the expression of the inflammation gene set into a single value for each patient biopsy (Figure 1C, ST2). A summary of the bMIS scores according to disease type and region sampled is shown (Figure 1E). In general, the bMIS of endoscopic-defined inflamed biopsies were 10-fold higher than those of non-inflamed biopsies. Notably, bMIS scores were significantly higher in non-inflamed biopsies relative to non-IBD control biopsies. Overall these data suggest molecular scores have higher sensitivity of detecting disease activity than macroscopic assessment on endoscopy and can also distinguish disease from non-disease state.
bMIS strongly correlates with current clinical disease activity measures
The bMIS score was based on qualitative information of inflammation (i.e.binary status of “inflamed” or“non-inflamed”), and as such it was important to examine if the intestinal-based molecular scores capture disease activity metrics. To assess whether intestinal-based molecular scores segregate with disease, we associated bMIS scores to clinical, endoscopic, and histological definitions of disease activity. A significantly higher bMIS for both CD and UC biopsies was observed in patients with active versus inactive disease as defined by HBI or SCCAI clinical disease activity scores (Figure 2A, upper and lower panels). We observed a positive correlation between bMIS scores and endoscopic assessments of severity, with the most severely affected biopsies, according to the SESCD or Mayo endo scores, having the highest bMIS values (Figure 2B). A significant number of biopsies, sampled nearby the biopsy taken for RNA sequencing analysis, were also evaluated histologically. A significant positive association was also observed between the GHAS and Nancy index histological pathological scores and the bMIS scores in control, IBD non-inflamed and inflamed biopsies. We also noted that bMIS could identify molecular inflammation where the histological scores were normal (score of 0 or 1) supporting the value of increased granularity with molecular information compared to the 13- or 4- factor range of the GHAS or Nancy index respectively (Figure 2C). Overall, we noted that the highest correlations were seen between bMIS values and any other clinical measure of IBD disease (Figure 2D). Furthermore, the bMIS values of UC and CD were highly correlated, suggesting that disease specific scores have little added value.
Figure 2.
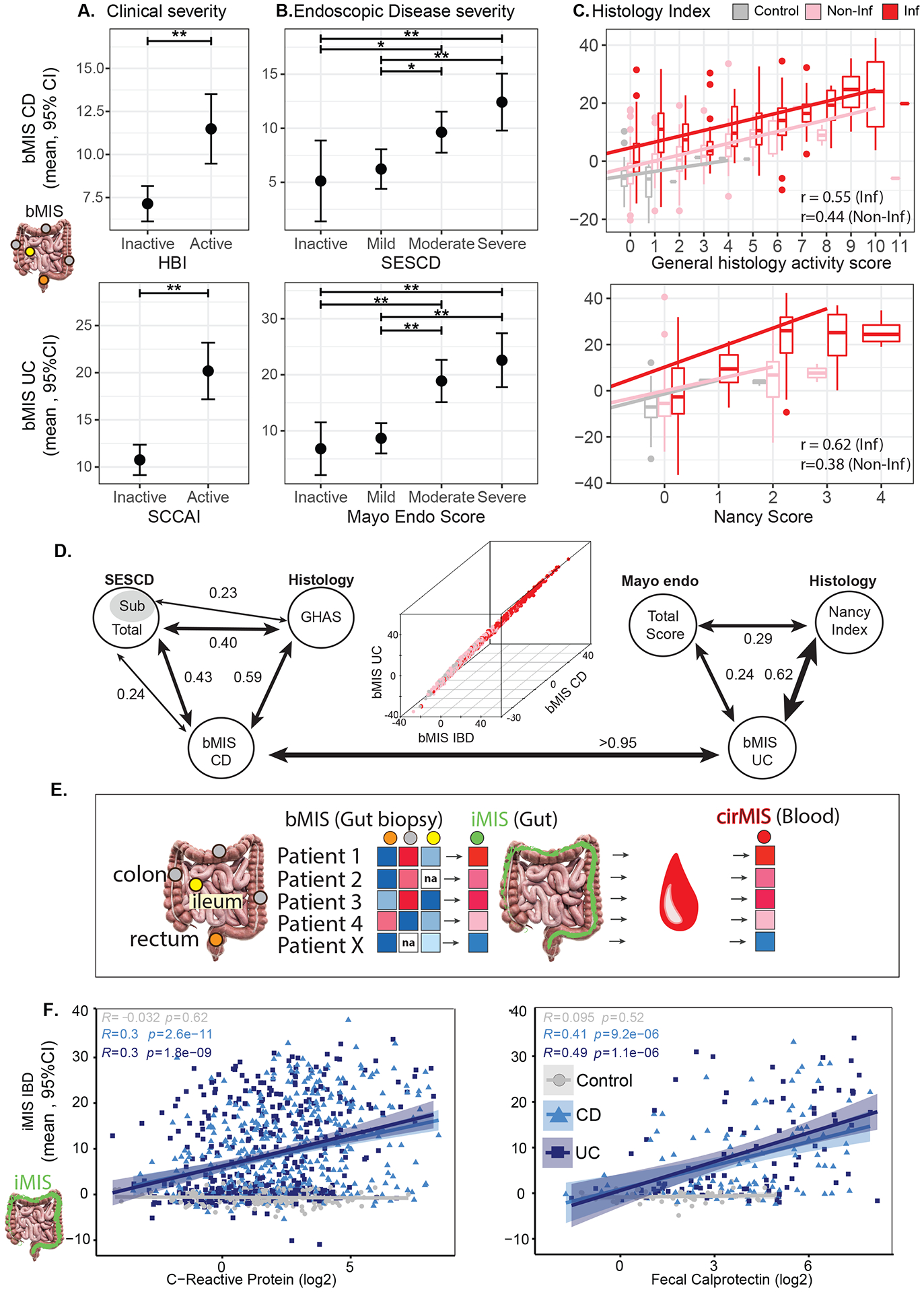
Association of bMIS in inflamed tissue with (A) Clinical (HBI for CD patients, SCCAI for UC patients), (B) Endoscopic (SESCD for CD patients, Mayo score for UC patients) and (C) Histological (GHAS for CD patients, Nancy score for UC patients) disease severity for CD (top) and UC (bottom). A-B Estimated marginal mean (EMM) and 95% CI for bMIS estimated from a mixed-effect model including clinical activity or disease severity, age, sex and region as fixed-effects. C. Scatter plots representing the distribution of bMIS across histological scores for CD and UC with corresponding regression line. The pink and red line corresponds to the regression line for inflamed and non-inflamed tissue. (GHAS (top): Inflamed tissue, bMIS=2.619 + 1.997*GHAS, Pearson r: 0.55; Non-inflamed tissue, bMIS = −3.926+2.018*GHAS, Pearson r: 0.44; Nancy (bottom): Inflamed tissue: bMIS=1.74+8.457*Nancy, Pearson r: 0.62; Non-inflamed tissue: bMIS= −5.269+5.216*Nancy, Pearson r: 0.38). *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001. D. Pair-wise correlation analysis (Spearman values) between bMIS scores in biopsies and corresponding endoscopic and histologic scores for CD (left) or UC (right) in MSCCR patients. 3D plot showing correlations (Spearman) between bMIS_UC, bMIS_CD and bMIS_IBD. E. Schema showing the process of obtaining an intestine-level molecular-based inflammation measure (iMIS) per patient using the multiple regions sampled per patient and their bMIS (biopsy-based) scores (see methods). The blood gene expression data was then modeled using a linear model with the continuous variable iMIS (see methods) to identify genes that reflects intestinal inflammation and then generate a circulating molecular score (cirMIS) using GSVA. F. Scatterplots between iMIS_IBD levels and CRP (log2) (left) or fecal calprotectin (log2) (right) within each group (Control, CD, and UC), with Pearson’s correlation coefficients r and p value.
Generation of a circulating blood biomarker of bMIS: cirMIS
We next sought to develop a less invasive circulating molecular biomarker. To do this we took advantage of the multiple regions sampled per individual and summarized the patient’s individual bMIS scores into an intestinal-level (ileum-to-rectum) molecular inflammation score (iMIS) (Figure 2E). We first verified that iMIS levels, like bMIS levels, also significantly associated with clinical, endoscopic, and histological definitions of disease activity (data not shown). We next demonstrated that iMIS levels were significantly and positively correlated with the known IBD activity biomarkers, namely blood CRP or fecal calprotectin levels (Figure 2F). These observations supported generating an equivalent patient-level molecular blood biomarker. To do this, we derived a set of genes that associated with iMIS and created a patient level blood-based molecular inflammation score (cirMIS), using GSVA (ST4–5).
Similar to bMIS and iMIS, we show that cirMIS levels could distinguish cases from controls, as well as significantly and positively associated with clinical, endoscopic, or histologic assessments of intestinal inflammation (Figure 3A–D). A significant overlap of the cirMIS geneset with the bMIS geneset was observed in addition to a strong correlation between cirMIS and iMIS levels (Figure 3E and SF1A).
Figure 3.
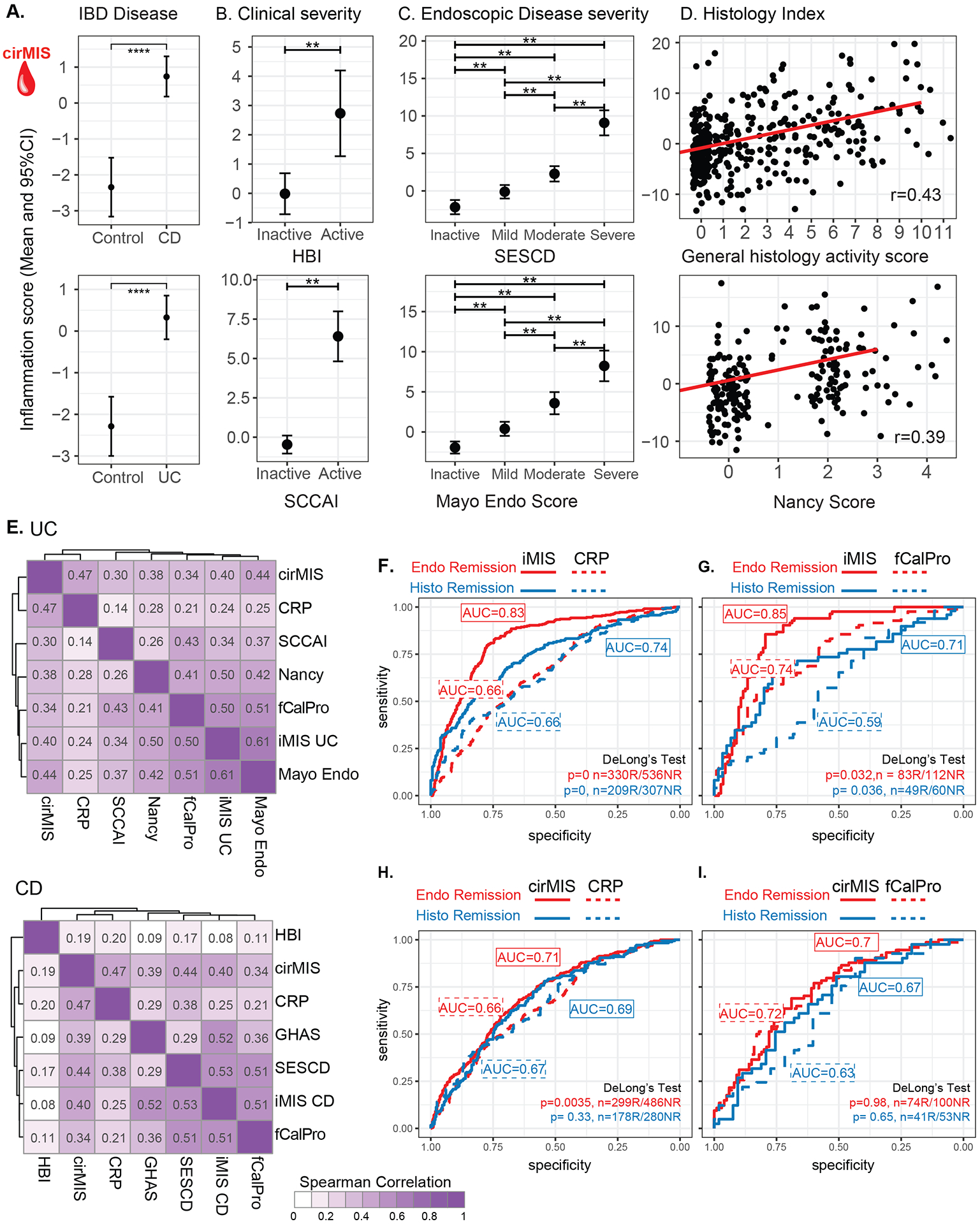
Association of cirMIS with IBD Disease (A) as well as Clinical (B) Endoscopic (C) and Histological assessements (D). A-C. Estimated marginal mean (EMM) and 95% CI for cirMIS from a mixed-effect model including IBD disease status (A) or clinical disease severity (B) or endoscopic disease severity (C), age, sex and genetic PCs as a fixed-effects. D. Scatter plots representing the distribution of cirMIS across histological scores, maximum GHAS for CD and maximum Nancy score for UC. The red line corresponds to the regression line (Max GHAS (top): cirMIS = −1.756+0.901*GHAS, Pearson r: 0.43; Max Nancy (bottom) cirMIS = −1.21+1.802*Nancy, Pearson r: 0.39). E. Heatmaps showing the Spearman correlations between iMIS and cirMIS with molecular (CRP and fecal calprotectin [fCalPro]), endoscopic (HBI for CD and Mayo for UC), histological (GHAS for CD and Nancy for UC), and clinical markers (SESCD for CD and SCCAI for UC) of UC (upper) and CD (lower). F-I Comparison of iMIS and cirMIS with CRP and fecal calprotectin (fCalPro) to classify endoscopic remission (SESCD<3 in CD patients or Mayo endo score=0 in UC patients) and histological remission (GHAS score=0 in CD patients or Nancy score =0 in UC patients). F-I. Delong’s method was used to compare AUCs. *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001.
As an additional check, we also performed competitive tests to examine if the genes involved in the generation cirMIS/bMIS are significantly associated with disease outcomes as compared to random geneset selections (See Methods). In all cases, we confirmed that the associations of cirMIS/bMIS scores with IBD activity measures were significantly higher and far removed from the 95% CI of scores generated using randomly selected genesets (SF1B and data not shown).
Comparison of b/iMIS and cirMIS to current biomarkers of activity
A summary of the co-correlations amongst the various molecular scores of inflammation, existing biomarkers and clinical, endoscopic and histological disease activity assessments are shown in Figure 3E (ST6). Overall this analysis supports that scores based on gene expression levels are reflective of active intestinal inflammation. We next used logistic regression to compare the intestinal-level bMIS (ie iMIS) and cirMIS scores with CRP and fecal calprotectin in their ability to identify patients in 1. endoscopic remission (SESCD<3 in CD patients or Mayo endo Score=0 in UC patients) or 2. histological remission (GHAS score=0 in CD patients or Nancy score =0 in UC patients). The results are summarized in Figure 3(F–I). In the MSCCR cohort, iMIS out-performed both CRP and fecal calprotectin. CirMIS was significantly better at identifying patients in endoscopic remission as compared to CRP and equivalent to fecal calprotectin. Overall, adding CRP or fecal calprotectin to models with either bMIS/cirMIS scores did not lead to gains in AUC. However, performance prediction, in general, was better, in models where bMIS/cirMIS scores were used, as compared to models with CRP of fecal calprotectin levels alone (ST7). Consistent with the logistic regression analysis, we also observed that cirMIS was in general more strongly correlated with endoscopic, histological, and clinical scores than CRP supporting that cirMIS could be a better blood biomarker than CRP (Figure 3E). With respect to fecal calprotectin, generally, comparable correlations were observed compared to the molecular scores, supporting that these new scores are potential alternatives (or supplementary) to fecal calprotectin.
Replication of bMIS/cirMIS in independent IBD cohorts
We curated transcriptome datasets (blood or biopsy) from four separate IBD cohorts and using the same gene sets as in the discovery MSCCR cohort, we generated molecular inflammation scores and associated them with available clinical information. Figure 4A shows significantly higher bMIS levels in biopsies taken at week 0 from adult UC patients during two phase 3 trials of VDZ (GEMINI I and LTS) or prior to IFX therapy13, as compared to non-IBD control individuals. Furthermore, a significantly higher bMIS was observed in the UC patients with Mayo endo score of 3 vs 2, supporting that bMIS levels also correlate with levels of disease activity. Figure 4B shows that bMIS levels were significantly higher in the inflamed ileal biopsies as compared to the non-inflamed ileal biopsies from the RISK CD pediatric, treatment-naïve cohort, supporting utility of this score in younger IBD patients and its utility in newly diagnosed cases. bMIS levels were also significantly higher in non-involved biopsies compared to non-IBD RISK cohort controls, again underlying that molecular disease exists where visual endoscopic assessments are normal.
Figure 4.
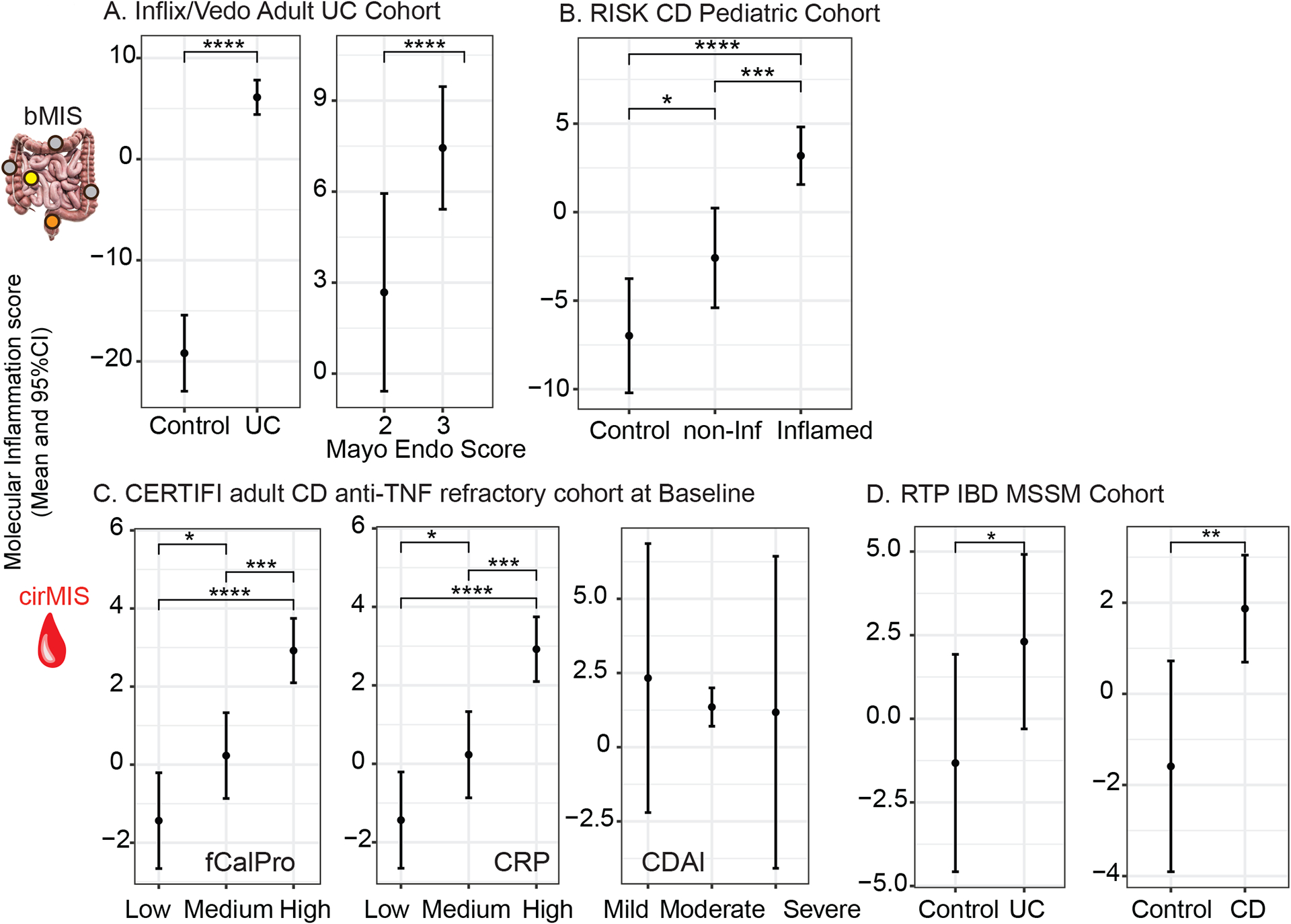
Validation of bMIS/cirMIS in independent IBD cohorts. A. Line plots showing bMIS_UC can differentiate between control and UC disease status and associate with Mayo endoscopic scores in colonic biopsies from GEMINI-I/LTS trial and anti-TNF UC participants. B. Line plots showing bMIS_CD could differentiate between control, non-inflamed ileum and inflamed ileum biopsies form the RISK pediatric CD Cohort. C. Line plots showing cirMIS_CD associates with fecal calprotectin (fCalPro) and CRP, but not CDAI in the CERTIFI CD cohort. D. Line plots showing cirMIS_UC and cirMIS_CD can differentiate between control and UC or CD disease status in the RTP-IBD MSSM cohort. (A-D) Each plot represents the estimated marginal mean and 95% CI through a linear mixed-effect model on the baseline data. *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001
The third IBD cohort, consisted of anti-TNF non-responsive adult CD patients as part of CERTIFI UST clinical trial. We confirmed a positive and significant correlation of cirMIS levels to fecal calprotectin or CRP levels (Figure 4C). The CDAI scores, however, were poorly associated with cirMIS levels. Blood transcriptome data was available from a recently generated cohort of Orthodox Jewish pediatric and adult IBD patients as part of a road to prevention (RTP) MSSM-Janssen initiative. cirMIS levels were found significantly higher in UC or CD patients relative to healthy controls (Figure 4D). We also noted that the more generalized bMIS_IBD or cirMIS_IBD scores which performed similarly in MSCCR to disease specific scores, also replicated outside of the discovery MSSCR cohort (data not shown and SF2A–C). Overall, these four independent IBD cohorts and the discovery MSCCR cohort support that the genes and their summation into a molecular score are reproducibly showing association to disease activity and severity and in some cases, revealing disease where endoscopy evaluations appear normal.
With respect to UC disease extent, we also observed that bMIS levels were significantly higher in E3/E2 MSCCR patients than those with limited E1 disease (SF2D). This likely reflects the higher burden of inflammation in E2/3 versus E1. This IBD sub-phenotype association was also distinguished by cirMIS (SF2D), an observation independently replicated as cirMIS_UC levels were also higher in E3 UC patients compared to those with limited disease in the RTP IBD cohort (SF2E).
Molecular inflammation measures and IBD therapies
To evaluate if cirMIS or bMIS scores change in response to therapy and drug response, we subset CD or UC MSCCR patients according to anti-TNF responders and non-responders based on current anti-TNF self-reported use and an SESCD<5 or Mayo endo score <2. We observed that both bMIS and cirMIS levels were higher in the inflamed biopsies of anti-TNF non-responders as compared to responders (SF2F). As MSCCR is a cross-sectional cohort, we next evaluated our molecular scores in independent cohorts including existing clinical trial datasets.
bMIS and UC:
bMIS scores were evaluated in the UC biopsies from GEMINI-I/LTS trial participants following VDZ (or placebo) treatment as well as in UC patients after IFX therapy26. No significant differences were observed in bMIS levels between baseline and week 6 biopsies in the placebo group, however, significant decreases were observed in the IFX or VDZ treated groups (Figure 5A). When patients were stratified by response to drug (defined as endoscopic healing) a statistically significant decrease in bMIS was observed in the IFX or VDZ responder group as compared to non-responder groups (Figure 5B). A significant positive correlation was observed between the change in bMIS score and change in Mayo endo score across all the patients, supporting that molecular descriptors of inflammation do reflect other metrics of disease activity (Figure 5C).
Figure 5:
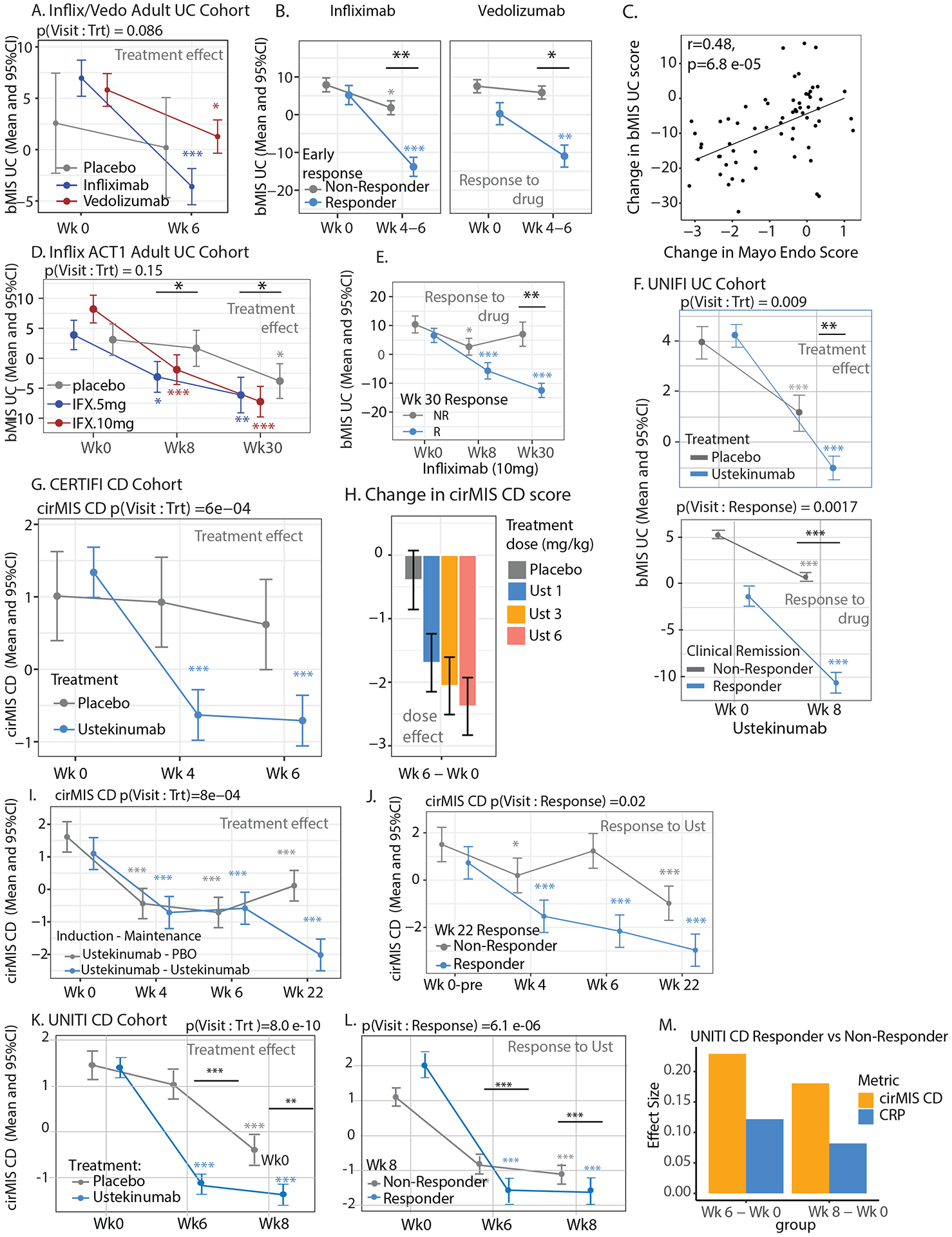
Molecular scores of inflammation and IBD treatment effects and response. A. Change in bMIS_UC levels from week 0 to week 6 in placebo, IFX, and VDZ groups in colonic biopsies from GEMINI-I/LTS trial and anti-TNF UC participants. The linear mixed-effect model included visit, treatment and, its interaction as fixed effects, and random intercept for each subject. B. Change in bMIS_UC levels from week 0 to week 4–6 in responders and non-responders within the IFX and VDZ medication groups in colonic biopsies. Patients with induction treatment as infliximab or VDZ were analyzed. Linear mixed-effect model with a three-way interaction visit, treatment and week 6 or week 4–6 status as fixed effects, and random intercept for each subject. Response to therapy was defined as endoscopic mucosal healing (Mayo endo score 0–1). C. Correlation between changes in Mayo endo scores and bMIS_UC scores between week 0 to week 4–6, week 6, or week 52 in GEMINI-I/LTS trial and anti-TNF UC participants. D. bMIS_UC scores in colonic biopsies from ACT1 UC trial participants at various timepoints treated with either 5 or 10mg/kg of IFX. The linear mixed-effect model included visit, treatment and, its interaction as fixed effects, and random intercept for each subject. E. bMIS_UC levels from week 0 to week 4 and 30 in week 30 responders and non-responders within the infliximab 10mg/kg groups in colonic biopsies from ACT1 UC trial participants. Patients with induction treatment as infliximab and maintenance were studied. Linear mixed-effect model was used. Clinical response was defined as a decrease from baseline in the total Mayo score (of at least three points). F. bMIS_UC levels from week 0 to week 8 in placebo and UST groups in colonic biopsies from UNIFI UC trial participants (upper panel). The linear mixed-effect model included visit, treatment and, its interaction as fixed effects, and random intercept for each subject. Change in bMIS_UC levels from week 0 to week 8 in responders and non-responders within the UST medication groups in colonic biopsies from UNIFI trial participants (lower panel). Patients with induction treatment as UST were analyzed. Response to therapy was clinical remission (defined as a total score of ⩽ 2 on the Mayo scale and no subscore >1). G-H. Changes in cirMIS_CD levels during the induction phase (from week 0 to 6) differentiate placebo from UST treated CERTIFI CD patients and (G) exhibit a dose response to UST (1, 3 and 6 mg/kg) (H). I. Changes in cirMIS_CD levels at weeks 0, 4 and 6 (induction phase) and during maintenance treatment starting week 6 in CERTIFI CD patients with UST induction treatment and UST or placebo as maintenance treatment. Plots represent the EMM ± SEM estimated through a linear mixed-effect model with visit, treatment combination and, its interaction as fixed effects, and random intercept for each subject. J. cirMIS_CD levels at week 0, 4, 6 and 22 in week 22 responders and non-responders in CERTIFI CD patients that received UST during induction and maintenance phases. EMM and ± SEM from a linear mixed-effect model with visit, response status at week 22 and its interaction as fixed effects, and random intercept for each subject. Clinical response was defined as a decrease of 100 or more in CDAI score from baseline at week 22. K. Changes in cirMIS_CD levels during the induction phase (from week 0 to 8) differentiate placebo from UST treated UNITI CD patients as well as responders compared to non-responders. L. Response (at week 8) was defined as clinical remission (CDAI score < 150 points). M. Effect size of the delta between either week 6 and baseline or week 8 and baseline for either cirMIS_CD or CRP levels as determined in responders versus non-responders. *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001 Black line/asterisk represent significance of the delta between a timepoint and baseline for treated vs placebo group.
In the ACT1 cohort of IFX treated adult UC patients29 the change in baseline bMIS levels compared to either week 8 or week 30 bMIS levels, was greater in IFX treated patients as compared to the change observed in the placebo group. When IFX patients were stratified by response to drug (defined as the clinical response at week 30) a greater decrease in bMIS, as compared to baseline, was observed in responders as compared to that observed in non-responders. In the UNIFI UC cohort on UST therapy, a similar pattern was observed, with bMIS levels showing the greater delta compared to baseline in treated patients (as compared to placebo) as well as in responders (defined as clinical remission) compared to non-responders (Figure 5F). Similar results were observed when using the IBD-centric molecular scoring system (data not shown). Overall these data highlight the potential utility of bMIS as a valuable measure of disease activity in UC patients distinguishing placebo and non-response states.
cirMIS and CD:
In the CERTIFI CD trial, cirMIS levels were significantly lower in UST-treated patients at week 4 and 6 compared to their baseline levels, with no effects in the placebo group (Figure 5G). Change in cirMIS levels was also dose-dependent (Figure 5H). There was an induction and maintenance phase with response at week 22 defined as a decrease of 100 or more in CDAI score from baseline. We observed that week 6 UST-treated patients that continued to receive UST in the maintenance phase had a significantly lower cirMIS level compared to patients that were switched to the placebo group in the maintenance phase. Furthermore, we noted that UST-treated patients that were considered week 22 responders also had lower cirMIS levels compared to non-responders (Figure 5G–H).
In the UNITI CD trial cirMIS levels were significantly lower, already at week 6 compared to their baseline levels, a decrease significantly higher in UST-treated patients compared to placebo. UST-treated patients that were considered week 8 responders (defined as clinical remission) demonstrated a greater reduction in cirMIS levels at week 6 or 8 (as compared to their baseline) when compared to the changes observed in non-responders (Figure 5I). Furthermore, in comparison to the blood biomarker CRP, the effect size of the treatment effect between responders from non-responders was almost double when cirMIS levels were used, potentially supporting a stronger clinical trial utility with respect to required sample sizes, as compared to CRP (Figure 5M).
Relationship between molecular inflammation and long-term outcomes
Having demonstrated that molecular inflammation is associated with current indicators of IBD disease activity and tracks with anti-IBD therapy and response to therapy we then looked at the impact of molecular inflammation on short-term and long-term IBD outcomes beyond current disease activity metrics. We tested this hypothesis in two clinically relevant scenarios.
The first scenario included asking if differing levels of baseline bMIS or cirMIS levels of patients deemed to have similarly active disease macroscopically (e.g. same endoscopy scores) have different short-term responses to therapy. We first addressed this question using the GEMINI-I/LTS and IFX UC cohort data. We initially observed that the baseline bMIS values were significantly associated with Mayo endo score (p=0.016), confirming the association seen in MSCCR. We then observed that baseline bMIS was significantly higher (3.08 vs 7.5, d=4.44, p=0.023 Figure 6A) in patients that were considered endoscopically responders at week 6. This difference was more marked in patients with baseline Mayo endo score of 2 (4.56 vs −0.24, d=4.89), than Mayo endo score of 3 (4.98 vs 8.5, d=3.52, p=.052). We then separated the patients in low bMIS (n=27) and high bMIS (n=27) groups using as cut-off the median (4.519) baseline bMIS value for all randomized patients. Rates of drug non-response were similar among patients with Mayo endo score 2 than Mayo endo score 3 (64%, vs 75%, Odd’s Ratio *OR+=1.6, p=0.45). On the other hand, patients with high bMIS were significantly more likely (OR =3.92,p=0.045) to be non-responders to IFX/Vedo than patients with low bMIS (85% vs 58% response rates, respectively, Figure 6A-right panel), and this difference was higher in patients with Mayo endo score=2 (data not shown), indicating that there is clinical relevance to molecular inflammation, beyond that assessed by endoscopy scores. These results were confirmed in additive logistic model where only bMIS but not Mayo endo score was associated with response at week 6 (p(Mayo endo score)=0.87, p(bMIS)=0.058). We next aimed to validate this observation in a phase 3 clinical trial by repurposing the UNIFI UC cohort. We first noted that patients who were non-responders at week 8 (clinical remission) had higher levels of baseline bMIS_IBD than responders (p=2.2e-08, SF3A). Given the many response definitions possible in this clinical trial we next determined the likelihood of being a treatment responder according to various week 8 definitions, spanning from clinical remission, endoscopic improvement, histological healing and HEMH. A general finding across all response variables, was that the OR was significantly greater for patients having lower baseline bMIS levels compared to higher bMIS levels. Importantly, the OR remained substantially higher when both bMIS and total Mayo score where considered in an additive model, thus strengthening the argument that molecular inflammation, captures disease relevant signal not observed with standard endoscopic scores (Figure 6B). As this suggests that bMIS has better prognostic value compared to currently used parameters, we compared the prognostic value of bMIS_UC to predict endoscopic healing at week 8 in the UNITI UC trial. An optimal cut-point was first determined for baseline bMIS levels by maximizing the sum of sensitivity and specificity and then a logistic regression analysis was run. Indeed, baseline bMIS levels performed significantly better than Mayo score, with AUC’s of 0.73 and 0.67, respectively (p=0.05 by DeLong’s test).
Figure 6.
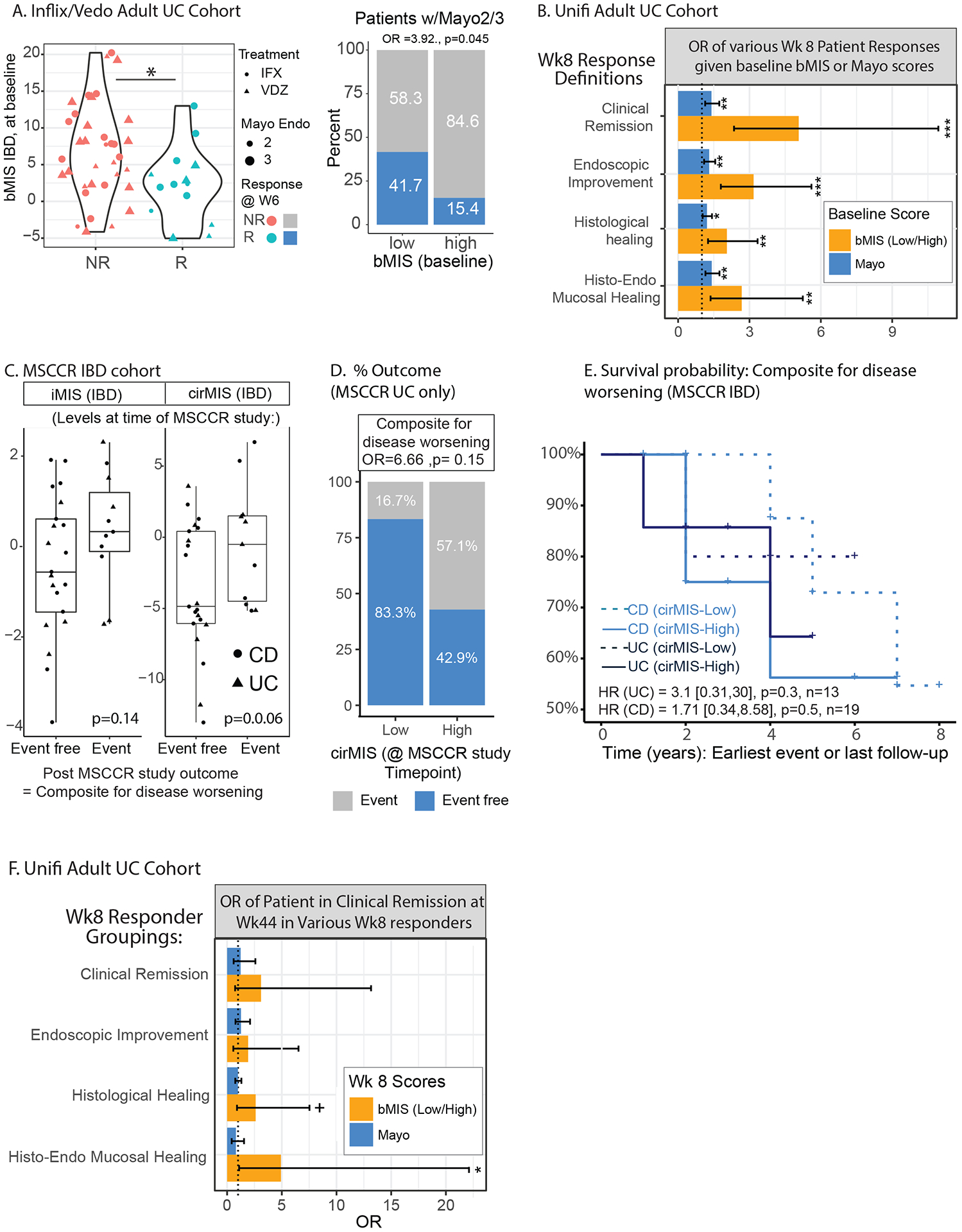
Residual molecular inflammation and clinical outcomes. A. Baseline bMIS_IBD levels in patients from the GEMINI-I/LTS UC and anti-TNF treated patients according to response to therapy (endoscopic mucosal healing (Mayo endoscopic score 0–1) as assessed for VDZ at W6 and for IFX at W4–6) (left panel). The proportion of patients that were considered responders to therapy in the baseline high vs low bMIS group (right panel). B. UNIFI adult UC cohort and the odd’s ratio of having a response, according to various definitions from clinical remission, endoscopic improvement, histological healing and combined histo-endoscopic mucosal healing (see methods) at week 8 of therapy (UST) in baseline high vs low cirMIS group’s (based on median levels) or according to baseline Total Mayo score. An additive model was used. C. A subset of MSCCR patients were identified that were in endoscopic and histological remission at the time of the MSCCR study (called MSCCR endo-histo-remission) see methods). These patients were then categorized as having high or low cirMIS levels based on the tertiles of expression and their post-MSCCR outcomes considered as a composite score to reflect disease worsening were reviewed in their charts. The levels of bMIS_IBD and cirMIS_IBD of the MSCCR endo-histo-remission patient subsets were generally higher in those patients that subsequently had a disease worsening event post-MSCCR study. For the composite score of events reflecting disease worsening, 1 of 6 MSCCR UC patients with low cirMIS had an event while 4 out of 7 patients with a high cirMIS had an event (D). For the CD MSCCR endo-histo-remission patients, the proportion of patients that were event free was the same between high and low cirMIS levels (SF3B). However, Kaplan-Maier survival curves (E) show that patients with higher cirMIS levels, were more likely to have a disease worsening event recorded earlier than compared to the low cirMIS group (in either UC or CD subsets). Specifically, the Cox models reported that for UC patients, the hazard ratio is 3.1 times higher [0.31,30, p=0.3] than the low cirMIS group, and in CD patients, the hazard ratio was 1.71 (0.34,8.58, p=0.5] times higher than the low cirMIS group. (G). UNIFI adult UC cohort and the odd’s ratio of being in response at week 44 (outcome = clinical remission) in patients considered responders at week 8 according to various definitions from clinical remission (n=33), endoscopic improvement (44), histological healing (65) and HEMH (34) (see methods), based on their baseline high vs low cirMIS status, versus their week 8 Total Mayo score. An additive model was used. +: p<0.1; *: p < 0.05; **: p < 0.01; ***: p < 0.001.
The second clinically relevant scenario we evaluated was whether patients in endoscopic and histological remission, yet with high residual molecular inflammation, have poorer long-term outcomes. We first addressed this question, by sub-setting MSCCR UC and CD patients, according to those who were in endoscopic and histological remission at the time of the study. Thirteen UC (7 = high and 6 = low cirMIS levels) and 19 CD (10= high and 9 = low cirMIS levels) patients had follow-up data and were included in this analysis. In general, higher iMIS and cirMIS levels at the time of MSCCR endoscopy were observed in IBD patients that subsequently had a disease flare to those that did not (Figure 6C), while CRP levels were equivalent (p=0.55, data not shown). Despite the sample size, UC patients with low cirMIS levels had lower rates of disease worsening events (16%) compared to those with higher cirMIS level (57%, p=0.15) at the time of the MSCCR study (Figure 6D). For CD, the rate of events reflecting disease worsening was the same between patients in the high and low cirMIS groups (SF3), but survival analysis show that events occur earlier in those with higher bMIS (Figure 6E). Specifically, the Cox models reported a hazard ratio (HR) 2.3 times higher for high than low cirMIS patients ((HR for IBD=2.3, CI=[0.66, 7.82] (p =0.19), with HR=3.1 [0.31,30](p=0.3) for UC and 1.71 [0.34,8.58] (p=0.5) for CD patients). Given the small sample size in MSCCR with longitudinal follow-up, we next assessed the consequence of residual molecular inflammation on potential disease relapse in the UNITI UC clinical trial cohort. We sub-sett the UNITI UC cohort according to patients considered responders at week 8 by the various macroscopic and microscopic assessments (clinical remission (n=33), endoscopic improvement (n=44), histological healing (n=65) and combined HEMH (n=34)), into high and low bMIS expressing groups, based on their median value at week 8. We then assessed the proportion of patients in each group that were in clinical remission at week 44. Figure 6F, summarizes the estimated OR of clinical remission in the high versus low bMIS groups and compared to their week 8 Mayo score. In general, given any week 8 response definition, those patients with higher residual molecular inflammation at week 8, were generally less likely to maintain clinical remission status at week 44 compared to the low molecular inflammation group. For example, in week 8 HEMH responders, those patients with lower bMIS levels at week 8, were 5 times more likely to maintain clinical remission status at week 44, then those HEMH responders at week 8, that had higher week 8 bMIS levels (p=0.037). The OR for the Mayo scores were non-significant and compared to bMIS, always lower, with the OR based on bMIS ranging from 1.5 to 6.15 times higher than the OR based on Mayo scores, depending on the week 8 response definition.
With respect to CD, while we could assess the association of cirMIS levels with treatment effect and response based on clinical remission (Figure 5K–M) within the UNITI trial, the number of drop outs for longitudinal analysis associated with endoscopic or histological outcomes was very high and as such we were under-powered to assess if high residual cir/bMIS levels in week8 UNITI CD responders was associated with relapse.
bMIS provides a biological perspective of a disease activity measure
Our molecular inflammation scores, based on expression of a known set of genes, compared to other clinical metrics, directly inform on activity of biological processes underlying the observed phenotype. A summary of the genes according to recurrence in the bMIS/cirMIS gene panels reveals several top recurring genes that have been previously associated with IBD, such as biomarkers of unstable disease control (CXCL8[IL-8], S100A8)45 or potential failure of anti-TNF therapy (OSM46) (Figure 7A,ST5). BioPlanet pathways enriched in the bMIS_IBD genesets, included ‘Oncostatin M’, ‘TNFa effects on cytokine activity, cell motility and apoptosis’ and ‘Interleukin-23, Il-17 and Jak-STAT signaling pathways are supportive of known anti-IBD targets and potentially a source of novel candidates. (Figure 7B, ST8). Indeed, there was significant enrichment of bMIS genes in a curated list of anti-IBD gene targets in addition to genes that are reported as IBD GWAS candidates. Curated genesets associated with various intestine-based single cell datasets, showed that bMIS-IBD genes are mainly enriched in inflammatory macrophages and monocytes as well as genes coexpressed in module’s 4/5 as part of a recently described tissular IBD pathotype defined in patients as non-responsive to therapy and associated with activated fibroblasts with neutrophil-chemoattractant properties. Consistent with this pathotype being associated with potential Il-1 blockade, bMIS-IBD genes were also enriched in genes upregulated in fibroblasts with Il1b treatment. Finally, transcription factor enrichment analysis, supports NFKB1, STAT3, JUN/FOS and SMAD2 and their related co-factors as potential transcriptional regulators underlying expression of these disease associated genes (Figure 7C, ST9). Overall these data support that the molecular score of inflammation may provide a metric to ‘quantify’ the underlying biological process underlying the patient’s disease and thus inform on potential treatment responsiveness and treatment choices.
Figure 7.
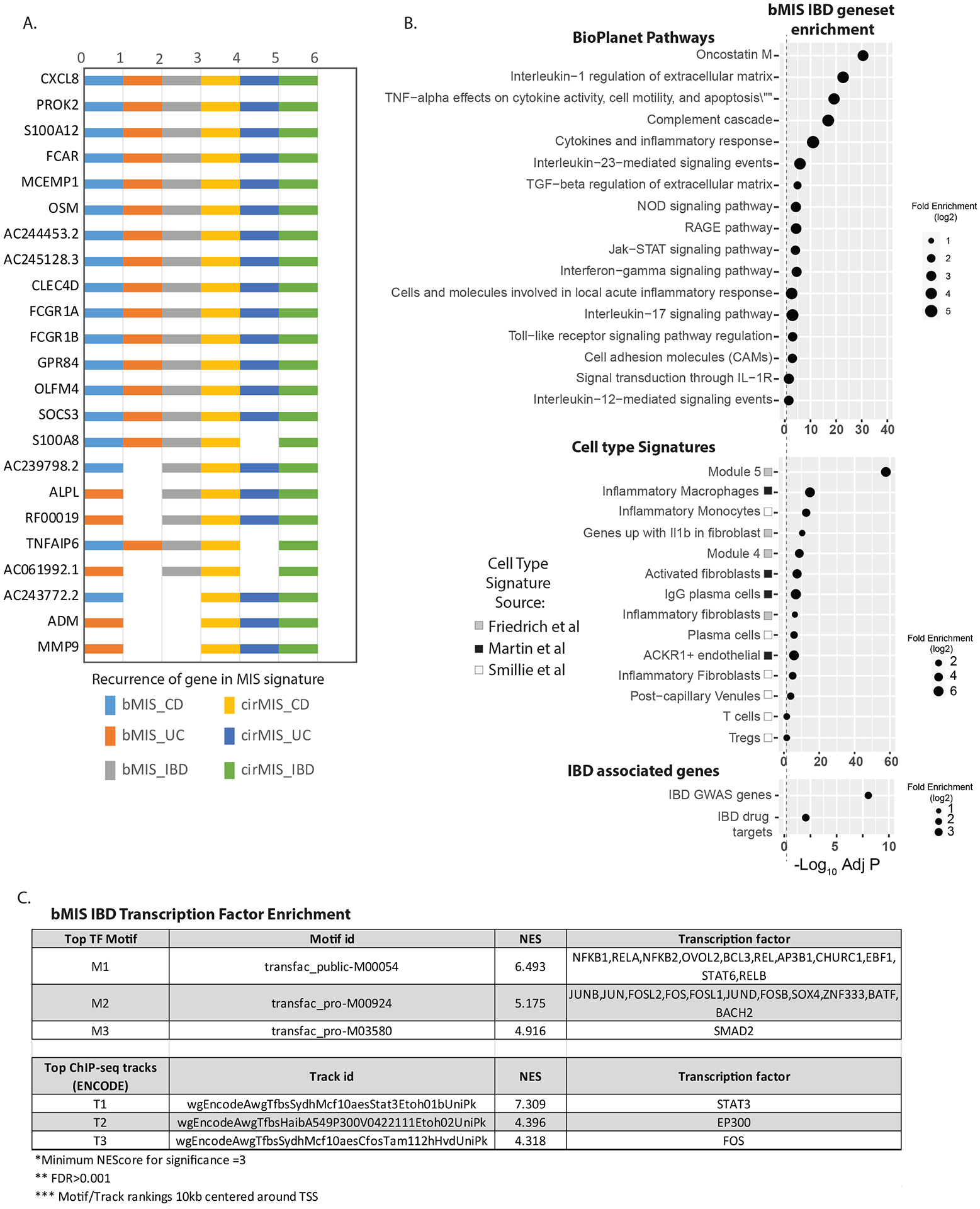
(A.) Summary of the genes according to recurrence in the various molecular inflammation score genesets. (B). Dotpots showing selected top significantly enrichments of the bMIS_IBD geneset in i) Bioplant pathways ii) Cell type signatures (see methods) iii) IBD GWAS genes and curated known IBD drug targets. Summary of the magnitude (log 2 Fold enrichment) and significance level (negative log10 BH adjusted P value) are shown. (C). Transcription factor enrichment analysis of the promoters of the bMIS_IBD geneset. The iRegulon application within Cytoscape was used to assess the promoters (10kb centered around transcription start site) of genes within the bMIS_geneset for enrichment in either transcription factor motifs or ChIP-seq tracks from ENCODE. Normalized enrichment scores (NES) of >3 considered significant with FDR >0.001. The top 3 scoring motifs or tracks, and their associated transcription factors are shown.
Discussion:
In this study we derived a new disease activity metric, bMIS, based on expression of genes in the intestinal mucosa from endoscopically inflamed vs non-inflamed biopsies of patients with either CD or UC. We demonstrated that bMIS significantly correlated with current clinical, endoscopic or histologic disease activity assessments. However, this molecular score was i) more sensitive and granular, detecting disease activity even when the mucosa was histologically normal ii) based on pathobiology which can be customized or generalized by disease type and iii) could be adapted to blood transcriptome data to provide a less invasive biomarker, cirMIS, with similar results as fecal calprotectin and generally better than CRP, the gold standard disease activity biomarkers. Through 7 independent IBD cohorts, bMIS was characterized as being associated with IBD, as well as IBD treatment effects and response, especially cirMIS for CD and bMIS for UC in UST, anti-TNF and VDZ (UC only) -treated cohorts. bMIS was also found to be a stronger prognostic indicator of response to therapy in UC compared to currently used metrics such as the Mayo score. Importantly, we showed that in patients considered macroscopically and microscopically ‘normal’, but with residual high b/cirMIS (in UC) or high cirMIS (in CD) levels, that rates of relapse were greater, supporting that residual molecular inflammation, beyond any currently used parameters (including histological remission) is clinically important and may portent relapse.
Overcoming the unmet therapeutic needs in IBD patients will rely not only on novel targets but also on improved robust clinical metrics of disease activity. Our study’s goal was to develop a potential molecular diagnostic approach to provide objective evidence and quantification of intestinal inflammation. To this end we demonstrated strong, significant correlations of bMIS or cirMIS to at least 3 standard clinical disease activity metrics (clinical, endoscopic and histologic), as well as two biomarkers (fecal calprotectin and CRP) of intestinal inflammation. While we acknowledge that the molecular scores of disease activity are not independent of those used as part of standard care, as they were all originally designed to detect gut inflammation, the reproducible correlations amongst the molecular scores with different disease activity metrics is important to show, as it demonstrates the potential diagnostic value of bMIS or cirMIS. It is also of interest, that the correlations between any existing metric with a molecular score (cirMIS or bMIS) were always higher, than any correlation observed between existing metrics, suggesting the molecular disease assessment, captures a broad disease landscape both cellular, histopathological as well as macroscopic. While it is of note that cirMIS can significantly out-perform CRP at distinguishing patients in endoscopic remission, potentially more relevant was the greater effect size observed for cirMIS as compared to CRP levels in distinguishing early treatment responders from non-responders. This effect, observed through blood sampling, was already seen at week 6 samplings, supporting cirMIS as potentially relevant clinical trial outcome, especially in CD, where disease is patchy and biopsy sampling biases exist. To that end, we acknowledge that intestinal molecular scoring faces a potentially similar draw-back to histology scoring in CD patients, in that if a biopsy is not sampled in an area of active disease, the bMIS score of that biopsy will not be able to reflect active inflammation ongoing in the gut elsewhere. In that sense, cirMIS has potentially better utility than bMIS for CD. However, an important point, is that with our molecular scoring platform and less-so for histology, is the possibility of generating akin to cirMIS, a biopsy-level biomarker (geneset) of iMIS, that reflects active intestinal inflammation, regardless of biopsy sampling location. Finally, we observed cirMIS to be as good as fecal calprotectin at distinguishing patients in endoscopic and histologic remission however is not reliant on patient stool sampling, which can be challenging to coordinate in the clinic47. Importantly, we demonstrate that we could translate the gene sets from the MSCCR discovery cohort and derive equivalently informative bMIS or cirMIS scores in seven independent IBD cohorts. In five of these which were clinical trial cohorts, the molecular score levels were also found to decrease with various treatment (VDZ, UST, or IFX vs placebo) as well as in responders (of various definitions) compared to non-responders.
Optimal management of IBD relies on early intervention, treat-to-target strategies and tight disease control9. Using available clinical trial data, we demonstrate that bMIS_UC, in particular, has potential prognostic value, associating significantly better with early response outcomes, than even standard macroscopic assessments (Mayo score). This suggests that molecular scores of inflammation may be able to identify a subset of patients that are actually different, despite being considered similar with respect to macroscopic assessments. Since those patients with the higher bMIS levels, appear to have a higher OR of being non-responders, if given the same treatment as the lower bMIS group, this additional insight may guide a physician towards an alternative, potentially more aggressive ‘step-down’ therapy over typical ‘step-up’ therapeutic approaches. Thus, molecular scoring systems could provide added value for tighter disease control, better risk stratification and intervention strategies.
Also relevant to treat-to-target strategies is that fact that bMIS and cirMIS were able to sense inflammation through gene expression changes in patients considered to have no (or low) histological or endoscopic abnormalities. This goes to the important follow-up question as to whether molecular disease inactivity plus clinical, endoscopic and histological disease inactivity, ‘matters’ clinically for optimally managing IBD. Inclusion of endoscopic mucosal healing over resolution of clinical symptoms only, has shown to improve prolonged clinical remission48,49. This ‘deep remission’ treatment target was then challenged by ‘complete remission’ defined by additional microscopic healing. In UC, lack of histological remission has been shown to predict worse clinical outcomes whereas endoscopic mucosal healing did not50. In patients with ileal CD, histologic ileal healing was also shown to associate more strongly with clinical outcomes, than endoscopic healing51. Further trials are ongoing in both UC and CD to continue to evaluate optimal treatment targets52,53,54. Combined these studies suggest undetected inflammation is deleterious implying a need to search for deeper markers of disease control. So how ‘deep’ should we go? Our sub-study in the MSCCR cohort and UNIFI UC clinical trial cohort, showing either higher HR or OR of relapse, respectively in those patients considered macroscopically and microscopically normal, but with higher molecular inflammation levels, would support that inflammation can be detected molecularly where histologically it is considered low or even normal, and that this matters for long-term clinically relevant outcomes. Admittedly these latter observations were done in cohorts, of relatively small size (and not available for CD) however, they nonetheless advocate for future studies exploring combinations of molecular and current biomarkers and disease activity indices.
Other published studies support the potential for molecular endpoints, including proteins, as minimally invasive serum biomarkers of IBD outcomes45,55. Kessel et al, described 16-serum proteins that show unstable disease control in IBD patients with stable remission45 and a commercial assay of 13-serum protein markers identified patients with resolution of endoscopic disease activity called the endoscopic healing index55 (EHI) has also been reported. We noted that several of the 16-serum protein panel were a component of either the bMIS or cirMIS score gene sets. A generally unique feature of molecularly-based disease metrics, are that they are protein/gene-centric, meaning they are grounded in the pathophysiological context thus providing a direct line between clinical indices and biological interpretation. In this sense, our bMIS also informs on the genes/pathways that therapeutics could be aimed against. Key biology associated with the bMIS genesets included signaling by OSM46, IL1, IL23 and IL12, Jak-STAT, TGF-beta, IFN-gamma, TNF effects on cytokine activity and apoptosis and GPCR ligand binding56. In addition, cell types seemingly enriched in bMIS genes, and thus potentially implicating cell types to target, included inflammatory macrophages, activated fibroblasts and plasma cells, to name a few. Overall, these rich molecular inflammation genesets, strongly linked to clinical outcomes provides a valuable resource for further mechanistic investigations.
Our study has several strengths and limitations. bMIS/cirMIS could be validated across multiple independent IBD datasets, including those using microarray technology. We were however, unable to acquire clinical trial datasets to assess clinical outcomes in patients in endoscopic and histologic remission with residual levels of cirMIS for UC or bMIS/cirMIS for CD. We also acknowledge the relatively small numbers of patients when looking at long-term outcomes. Thus, the true clinical significance of residual “molecular inflammation” remains to be established in appropriately designed longer term cohorts. Furthermore, it will be of interest to evaluate whether the blood-based molecular biomarker, cirMIS, is affected by other factors such as active extra-intestinal manifestations of IBD or infection, as is observed with CRP. Also, although the geneset sizes comprising the molecular scores are relatively small, before clinical test implementation, optimization of the gene sets to maximize detection of molecular inflammation with the fewest number of gene measures would be needed. Also important is determination of the delta or change in the score needed for potential patient outcome improvement (i.e., the optimal molecular ‘target’). Finally, a strength of our study, is that a general IBD molecular score of inflammation was as effective as an IBD-type specific molecular score, suggesting the underlying inflammation processes are similar regardless of disease type or location. Indeed, we observed that the correlation of the SESCD sub-score for the ileum was 0.56 to bMIS scores within ileal samples. However, our platform could be optimized further to generate molecular scores of disease which are based on gene sets created from various sub-classifications of patients, which could augment potential precision medicine strategies.
In conclusion, we provide a framework for generating a mucosal centric molecular score of inflammation which provides another layer of disease activity in IBD. Our method provides an opportunity to assess mucosal inflammation in a novel and complementary way which potentially addresses the issue of persistent microscopic inflammation that escapes current clinical evaluation. Our data suggests, molecular metrics of disease activity could add further stratification to disease activity and may be better predictive markers of response and relapse than currently used clinical measures.
Supplementary Material
Supplementary Table 1: Table of the MSCCR cohort demographics according to the omic’s data types (RNA-Seq) and tissue type (Biopsy, Whole blood, serum and stool) which was available for analysis.
Supplementary Table 2: The RTP cohort demographics
Supplementary Table 3: The RISK cohort demographics
Supplementary Table 4: Genesets comprising the various molecular inflammation scores
Supplementary Table 5: Table summarizing genes according to the various MIS they belong to
Supplementary Table 6: Correlations and p values associated with Figure 3E.
Supplementary Table 7: Comparison of p values for performance of various statistical models to distinguish MSCCR patients in histological remission or endoscopic remission.
Supplementary Table 8: Data associated with Heatmap in Figure 7B
Supplementary Table 9: Data associated with Figure 7C
Supplementary Figure 3: A. Baseline bMIS_IBD levels in patients from the GEMINI-I/LTS UC and anti-TNF treated patients according to response to therapy (endoscopic mucosal healing (Mayo endoscopic score 0–1) as assessed for VDZ at W6 and for IFX at W4–6) (left panel). B. Proportion plots for groups of UC and CD MSCCR patients who were in endoscopic and histological remission but with either high vs low cirMIS levels at the time of MSCCR study according to various post-MSCCR outcomes. 19 CD patients (9 low and 10 high cirMIS) and 13 UC (6 low and 7 high cirMIS) patients had longitudinal followup information available in the EHR. The outcomes evaluated included: IBD-related hospitalizations; IBD-related surgery; Clinical flare defined as i) need for new oral steroid or ii) treatment escalation OR addition of/change to new therapeutic agent due to disease activity. A composite score considering any flare events (composite flare) or any of the four events was created (‘Any outcome composite score’). These outcomes were used to reflect disease worsening. The date recorded was the date of the earliest adverse event or the date of the last follow up in EPIC if no event was seen.
Supplementary Figure 1. A. Venn diagram showing the overlap of the genesets associated with cirMIS or bMIS. B-D Figures showing the association of randomly generated genesets with various IBD phenotypes. Five hundred random genesets of the same size as cirMIS, were generated and then GSVA and associations performed. The gray band on the figures (B-D) represents the 95% CI (using the quantiles) of the scores from randomly created genesets. Association of cirMIS_IBD (black points on figures) as well as scores from randomly generated genesets (gray band on figure) with IBD Disease (B) as well as Clinical (C) Endoscopic (D) and Histological assessments (E). B-D. Estimated marginal mean (EMM) and 95% CI for cirMIS (or scores from randomly generated genesets) from a mixed-effect model including IBD disease status (A) or clinical disease severity (B) or endoscopic disease severity (C), age, sex and genetic PCs as a fixed-effects. E. Violin plots representing the distribution of cirMIS across histological scores, maximum GHAS for CD and maximum Nancy score for UC. Gray band on E represents the results of scores from randomly generated genesets. The gray band indicates 95% of the values for bootstrapped scores did not exhibit any associations with IBD disease, clinical severity, endoscopic disease severity or histology index for CD or UC. The observed cirMIS statistics are very far from the statistics with random gene sets. +: p<0.1; *: p < 0.05; **: p < 0.01; ***: p < 0.001.
Supplementary Figure 2: Validation of bMIS/cirMIS_IBD. A. Line plots showing bMIS_IBD can differentiate between control, non-inflamed ileum and inflamed ileum biopsies in the RISK pediatric CD Cohort. B. Line plot showing cirMIS_IBD could differentiate between control and IBD disease status in the RTP-IBD MSSM cohort. C. Line plots showing cirMIS_IBD associates with fecal calprotectin and CRP levels, but not CDAI in CERTIFI CD cohort. D-E. Line plots showing bMIS and cirMIS association according to UC extent in MSCCR and RTP MSSM IBD cohorts. Each plot represents the estimated marginal mean (EMM) ± SEM. (F) Molecular scores of inflammation and IBD treatment effects. Association of bMIS and cirMIS with anti-TNF response in MSCCR UC and CD patients. Each plot represents the estimated marginal mean and 95% CI for bMIS (left) from a mixed-effect model including age, gender, region as fixed factors, and cirMIS (right) estimated through a linear model with age, gender and genetic PCs as covariates. *: p ≤ 0.05; **: p ≤ 0.01; ***: p ≤ 0.001; ****: p ≤ 0.0001.
Acknowledgements:
This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. All sample processing was provided by Human Immune Monitoring Center at Icahn School of Medicine at Mount Sinai. We are grateful for assistance by clinicians: Benjamin Cohen, Christopher DiMaio, David Greenwald, Ari Greenspan, Steven Itzkowitz, Aimee Lucas, James Marion, Elana Maser, Ryan Ungaro, Steven Naymagon, Joshua Novak, Ionnis Oikonomous, Brijen Shah, Thomas Ullman, Peter Rubin, Asher Kornbluth, James George, Peter Legnani; Clinical Coordinators: Anabel Castillo, Farah Fasihuddin, Merjona Saliaj, Amy Nolan, Pamela Reyes Mercedes, Carina Rodriguez, Sarah Aly, Kenneth Santa-Cruz; IROQ Clinical Database: Ashish Atreja, Jason Rogers, Aditya Kaushik, Milan Patel; Human Immune Monitoring Center: Manishkumar Patel, Xiaochen Qin, Hui Xie; and Scientific Computing /Minerva: Patricia Kovatch, Gene Fluder, Hyung Min Cho. We also thank the patients for their study participation.
Grant Support:
The sampling of the Inflammatory Bowel Disease cohort was jointly designed as part of the research alliance between Janssen Biotech, Inc. and The Icahn School of Medicine at Mount Sinai. Beyond this exception, no other funders had a role in analyses design and interpretation.
Disclosures:
Mount Sinai co-authors (from Genetics and Genomics, Icahn Institute for Data Science and Genomic Technology, Population Health Science and Policy, Division of Gastroenterology, Pediatric GI and Hepatology, Susan and Leonard Feinstein IBD Clinical Center at Icahn School of Medicine at Mount Sinai) were partially funded as part of research alliance between Janssen Biotech and The Icahn School of Medicine at Mount Sinai. SV, PTD, PB, AS, JP, CB, MC, EL-S and JW are employees at Janssen Biotech, Inc. Joshua R. Friedman is a former employee at Janssen Biotech, Inc. KH, MM, AK, AD and EES are employees at Sema4. BS, JFC and MCD are consultants for Janssen. MCD is an advisory board member of Janssen. RCU has served as an advisory board member or consultant for AbbVie, Bristol Myers Squibb, Janssen, Pfizer, and Takeda; research support from AbbVie, Boehringer Ingelheim, Eli Lilly, and Pfizer. RCU supported by an NIH K23 Career Development Award (K23KD111995-01A1). CA, LP, and EES were supported in part by The Leona M. and Harry B. Helmsley Charitable Trust and RC2 DK122532/DK/NIDDK NIH.
References:
- 1.Kobayashi T et al. Ulcerative colitis. Nature Reviews Disease Primers 6, 74 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Roda G et al. Crohn’s disease. Nature Reviews Disease Primers 6, 22 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Ungaro R, Colombel JF, Lissoos T & Peyrin-Biroulet L A Treat-to-Target Update in Ulcerative Colitis: A Systematic Review. Am J Gastroenterol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peyrin-Biroulet L et al. Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE): Determining Therapeutic Goals for Treat-to-Target. Am J Gastroenterol 110, 1324–38 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Le Berre C, Peyrin-Biroulet L & group S.-I.s. Selecting End Points for Disease-Modification Trials in Inflammatory Bowel Disease: the SPIRIT Consensus From the IOIBD. Gastroenterology 160, 1452–1460 e21 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Adegbola SO, Sahnan K, Warusavitarne J, Hart A & Tozer P Anti-TNF Therapy in Crohn’s Disease. Int J Mol Sci 19(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K et al. Relationship Between Combined Histologic and Endoscopic Endpoints and Efficacy of Ustekinumab Treatment in Patients With Ulcerative Colitis. Gastroenterology 159, 2052–2064 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Agrawal M & Colombel JF Treat-to-Target in Inflammatory Bowel Diseases, What Is the Target and How Do We Treat? Gastrointest Endosc Clin N Am 29, 421–436 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Turner D et al. STRIDE-II: An Update on the Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE) Initiative of the International Organization for the Study of IBD (IOIBD): Determining Therapeutic Goals for Treat-to-Target strategies in IBD. Gastroenterology 160, 1570–1583 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Hamilton JD et al. Dupilumab improves the molecular signature in skin of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol 134, 1293–1300 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Guttman-Yassky E et al. Molecular signatures order the potency of topically applied anti-inflammatory drugs in patients with atopic dermatitis. J Allergy Clin Immunol 140, 1032–1042 e13 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Beck LA et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med 371, 130–9 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Kosoy R et al. Deep Analysis of the Peripheral Immune System in IBD Reveals New Insight in Disease Subtyping and Response to Monotherapy or Combination Therapy. Cell Mol Gastroenterol Hepatol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daperno M et al. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointest Endosc 60, 505–12 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Sipponen T, Nuutinen H, Turunen U & Farkkila M Endoscopic evaluation of Crohn’s disease activity: comparison of the CDEIS and the SES-CD. Inflamm Bowel Dis 16, 2131–6 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Saigusa K et al. Ulcerative colitis endoscopic index of severity is associated with long-term prognosis in ulcerative colitis patients treated with infliximab. Dig Endosc 28, 665–70 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Marchal-Bressenot A et al. Development and validation of the Nancy histological index for UC. Gut 66, 43–49 (2017). [DOI] [PubMed] [Google Scholar]
- 18.D’Haens GR et al. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology 114, 262–7 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Geboes K et al. A reproducible grading scale for histological assessment of inflammation in ulcerative colitis. Gut 47, 404–9 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novak G et al. Histologic scoring indices for evaluation of disease activity in Crohn’s disease. Cochrane Database Syst Rev 7, CD012351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li K et al. Effects of Ustekinumab on Histologic Disease Activity in Patients With Crohn’s Disease. Gastroenterology 157, 1019–1031 e7 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Ma C et al. An International Consensus to Standardize Integration of Histopathology in Ulcerative Colitis Clinical Trials. Gastroenterology 160, 2291–2302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satsangi J, Silverberg MS, Vermeire S & Colombel JF The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 55, 749–53 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.[dataset] Haberman Y et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. The Journal of Clinical Investigation 124, 3617–3633 (2014). Core Ileal Transcriptome in Pediatric Crohn Disease July 24, 2014. https://www.jci.org/articles/view/75436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.[dataset] Peters LA et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet 49, 1437–1449 (2017). A functional genomics predictive network model identifies regulators of inflammatory bowel disease: Microarray Analysis of Human Blood and Intestinal Biopsy Samples from a Phase 2b, Double-blind, Placebo-controlled Study of Ustekinumab in Crohn’s Disease July 7, 2017 https://www.nature.com/articles/ng.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.[dataset] Arijs I et al. Effect of vedolizumab (anti-α4β7-integrin) therapy on histological healing and mucosal gene expression in patients with UC. Gut 67, 43 (2018). The effect of vedolizumab (anti-α4β7-integrin) therapy on colonic mucosal gene expression in patients with ulcerative colitis (UC) October 11, 2016 https://gut.bmj.com/content/67/1/43 [DOI] [PubMed] [Google Scholar]
- 27.Sands BE et al. Ustekinumab as Induction and Maintenance Therapy for Ulcerative Colitis. N Engl J Med 381, 1201–1214 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Feagan BG et al. Ustekinumab as Induction and Maintenance Therapy for Crohn’s Disease. N Engl J Med 375, 1946–1960 (2016). [DOI] [PubMed] [Google Scholar]
- 29.[dataset] Toedter G et al. Gene expression profiling and response signatures associated with differential responses to infliximab treatment in ulcerative colitis. Am J Gastroenterol 106, 127280 (2011). Expression data from colonic biopsy samples of infliximab treated UC patients May 7, 2019 https://journals.lww.com/ajg/Abstract/2011/07000/Gene_Expression_Profiling_and_Response_Signatures.16.aspx [DOI] [PubMed] [Google Scholar]
- 30.Suarez-Farinas M et al. Intestinal inflammation modulates the expression of ACE2 and TMPRSS2 and potentially overlaps with the pathogenesis of SARS-CoV-2 related disease. Gastroenterology (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di’Narzo AF et al. Integrative Analysis of the Inflammatory Bowel Disease Serum Metabolome Improves Our Understanding of Genetic Etiology and Points to Novel Putative Therapeutic Targets. Gastroenterology 162, 828–843 e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanzelmann S, Castelo R & Guinney J GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee E, Chuang HY, Kim JW, Ideker T & Lee D Inferring pathway activity toward precise disease classification. PLoS Comput Biol 4, e1000217 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Development Core Team. R: A language and environment for statistical computing. v3.0.3 edn (R foundation for statistical computing, 2020). [Google Scholar]
- 35.Kuleshov MV et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin JC et al. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 178, 1493–1508.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smillie CS et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedrich M et al. IL-1-driven stromal-neutrophil interactions define a subset of patients with inflammatory bowel disease that does not respond to therapies. Nat Med 27, 1970–1981 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu JZ et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nature Genetics 47, 979–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jostins L et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Lange KM et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nature Genetics 49, 256–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schreiner P et al. Mechanism-Based Treatment Strategies for IBD: Cytokines, Cell Adhesion Molecules, JAK Inhibitors, Gut Flora, and More. Inflamm Intest Dis 4, 79–96 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janky R et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 10, e1003731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kessel C et al. Serum biomarkers confirming stable remission in inflammatory bowel disease. Sci Rep 11, 6690 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.West NR et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor–neutralizing therapy in patients with inflammatory bowel disease. Nature Medicine 23, 579–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buisson A et al. Comparative Acceptability and Perceived Clinical Utility of Monitoring Tools: A Nationwide Survey of Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis 23, 1425–1433 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Colombel JF et al. Adalimumab induces deep remission in patients with Crohn’s disease. Clin Gastroenterol Hepatol 12, 414–22 e5 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Reinisch W et al. Recommendations for the treatment of ulcerative colitis with infliximab: A gastroenterology expert group consensus. Journal of Crohn’s and Colitis 6, 248–258 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Vuitton L et al. Defining endoscopic response and remission in ulcerative colitis clinical trials: an international consensus. Aliment Pharmacol Ther 45, 801–813 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Christensen B et al. Histologic Healing Is More Strongly Associated with Clinical Outcomes in Ileal Crohn’s Disease than Endoscopic Healing. Clinical Gastroenterology and Hepatology 18, 2518–2525.e1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryant RV et al. Beyond endoscopic mucosal healing in UC: histological remission better predicts corticosteroid use and hospitalisation over 6 years of follow-up. Gut 65, 408–414 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Enhanced algorithm for Crohn’s treatment incorporating early combination therapy (REACT2).
- 54.Determination of the Optimal Treatment Target in Ulcerative Colitis (VERDICT). (2021).
- 55.D’Haens G et al. Development and Validation of a Test to Monitor Endoscopic Activity in Patients With Crohn’s Disease Based on Serum Levels of Proteins. Gastroenterology 158, 515–526 e10 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Olivera P, Danese S & Peyrin-Biroulet L Next generation of small molecules in inflammatory bowel disease. Gut 66, 199–209 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Table of the MSCCR cohort demographics according to the omic’s data types (RNA-Seq) and tissue type (Biopsy, Whole blood, serum and stool) which was available for analysis.
Supplementary Table 2: The RTP cohort demographics
Supplementary Table 3: The RISK cohort demographics
Supplementary Table 4: Genesets comprising the various molecular inflammation scores
Supplementary Table 5: Table summarizing genes according to the various MIS they belong to
Supplementary Table 6: Correlations and p values associated with Figure 3E.
Supplementary Table 7: Comparison of p values for performance of various statistical models to distinguish MSCCR patients in histological remission or endoscopic remission.
Supplementary Table 8: Data associated with Heatmap in Figure 7B
Supplementary Table 9: Data associated with Figure 7C
Supplementary Figure 3: A. Baseline bMIS_IBD levels in patients from the GEMINI-I/LTS UC and anti-TNF treated patients according to response to therapy (endoscopic mucosal healing (Mayo endoscopic score 0–1) as assessed for VDZ at W6 and for IFX at W4–6) (left panel). B. Proportion plots for groups of UC and CD MSCCR patients who were in endoscopic and histological remission but with either high vs low cirMIS levels at the time of MSCCR study according to various post-MSCCR outcomes. 19 CD patients (9 low and 10 high cirMIS) and 13 UC (6 low and 7 high cirMIS) patients had longitudinal followup information available in the EHR. The outcomes evaluated included: IBD-related hospitalizations; IBD-related surgery; Clinical flare defined as i) need for new oral steroid or ii) treatment escalation OR addition of/change to new therapeutic agent due to disease activity. A composite score considering any flare events (composite flare) or any of the four events was created (‘Any outcome composite score’). These outcomes were used to reflect disease worsening. The date recorded was the date of the earliest adverse event or the date of the last follow up in EPIC if no event was seen.
Supplementary Figure 1. A. Venn diagram showing the overlap of the genesets associated with cirMIS or bMIS. B-D Figures showing the association of randomly generated genesets with various IBD phenotypes. Five hundred random genesets of the same size as cirMIS, were generated and then GSVA and associations performed. The gray band on the figures (B-D) represents the 95% CI (using the quantiles) of the scores from randomly created genesets. Association of cirMIS_IBD (black points on figures) as well as scores from randomly generated genesets (gray band on figure) with IBD Disease (B) as well as Clinical (C) Endoscopic (D) and Histological assessments (E). B-D. Estimated marginal mean (EMM) and 95% CI for cirMIS (or scores from randomly generated genesets) from a mixed-effect model including IBD disease status (A) or clinical disease severity (B) or endoscopic disease severity (C), age, sex and genetic PCs as a fixed-effects. E. Violin plots representing the distribution of cirMIS across histological scores, maximum GHAS for CD and maximum Nancy score for UC. Gray band on E represents the results of scores from randomly generated genesets. The gray band indicates 95% of the values for bootstrapped scores did not exhibit any associations with IBD disease, clinical severity, endoscopic disease severity or histology index for CD or UC. The observed cirMIS statistics are very far from the statistics with random gene sets. +: p<0.1; *: p < 0.05; **: p < 0.01; ***: p < 0.001.
Supplementary Figure 2: Validation of bMIS/cirMIS_IBD. A. Line plots showing bMIS_IBD can differentiate between control, non-inflamed ileum and inflamed ileum biopsies in the RISK pediatric CD Cohort. B. Line plot showing cirMIS_IBD could differentiate between control and IBD disease status in the RTP-IBD MSSM cohort. C. Line plots showing cirMIS_IBD associates with fecal calprotectin and CRP levels, but not CDAI in CERTIFI CD cohort. D-E. Line plots showing bMIS and cirMIS association according to UC extent in MSCCR and RTP MSSM IBD cohorts. Each plot represents the estimated marginal mean (EMM) ± SEM. (F) Molecular scores of inflammation and IBD treatment effects. Association of bMIS and cirMIS with anti-TNF response in MSCCR UC and CD patients. Each plot represents the estimated marginal mean and 95% CI for bMIS (left) from a mixed-effect model including age, gender, region as fixed factors, and cirMIS (right) estimated through a linear model with age, gender and genetic PCs as covariates. *: p ≤ 0.05; **: p ≤ 0.01; ***: p ≤ 0.001; ****: p ≤ 0.0001.


