Abstract
High-throughput experimental platforms have revolutionized the ability to profile biochemical and functional properties of biological sequences such as DNA, RNA and proteins. By collating several data modalities with customizable tracks rendered using intuitive visualizations, genome browsers enable an interactive and interpretable exploration of diverse types of genome profiling experiments and derived annotations. However, existing genome browser tracks are not well suited for intuitive visualization of high-resolution DNA sequence features such as transcription factor motifs. Typically, motif instances in regulatory DNA sequences are visualized as BED-based annotation tracks, which highlight the genomic coordinates of the motif instances but do not expose their specific sequences. Instead, a genome sequence track needs to be cross-referenced with the BED track to identify sequences of motif hits. Even so, quantitative information about the motif instances such as affinity or conservation as well as differences in base resolution from the consensus motif are not immediately apparent. This makes interpretation slow and challenging. This problem is compounded when analyzing several cellular states and/or molecular readouts (such as ATAC-seq and ChIP–seq) simultaneously, as coordinates of enriched regions (peaks) and the set of active transcription factor motifs vary across cell states.
Recently, machine learning models that map DNA sequence to functional readouts from high-throughput assays have been developed to study the sequence basis of molecular activity and decipher putative functional genetic variants that influence protein–DNA binding, splicing, gene expression and long-range chromatin contacts1-3. These models are interrogated using feature attribution methods to infer quantitative, predictive importance scores of each base, thereby enabling the discovery of sequence features such as transcription factor motifs, splice sites and polyadenylation sites4,5. These importance scores are currently visualized in ad hoc ways that are not suited to seamless exploration and easy sharing.
To address these challenges, we introduce the dynamic sequence (dynseq) genome browser track, a generalization of previously proposed ‘sequence walkers’6 adapted to modern genome browsers, that displays DNA nucleotide characters at a genomic locus with heights scaled by user-specified, base-resolution, quantitative scores. The dynseq track makes it straightforward to visually recognize sequence features that are activated in a context-specific manner.
To visualize context-specific dynamic importance scores, we first implemented and integrated the dynseq track in the WashU Epigenome Browser7,8. The input file format is the BigWig format9 with per-base-pair importance scores. At each position, the nucleotide character (A/C/G/T) is rendered. The height of the character is scaled by the importance score at that position. Negative scores are handled by flipping the character along the x axis. When the track is zoomed out such that individual bases cannot be discerned, the track visualization switches to a regular BigWig view. This simple specification means that the dynseq track can be easily incorporated into other genome browsers. We have added equivalent native functionality for the UCSC Genome Browser10 and HiGlass11.
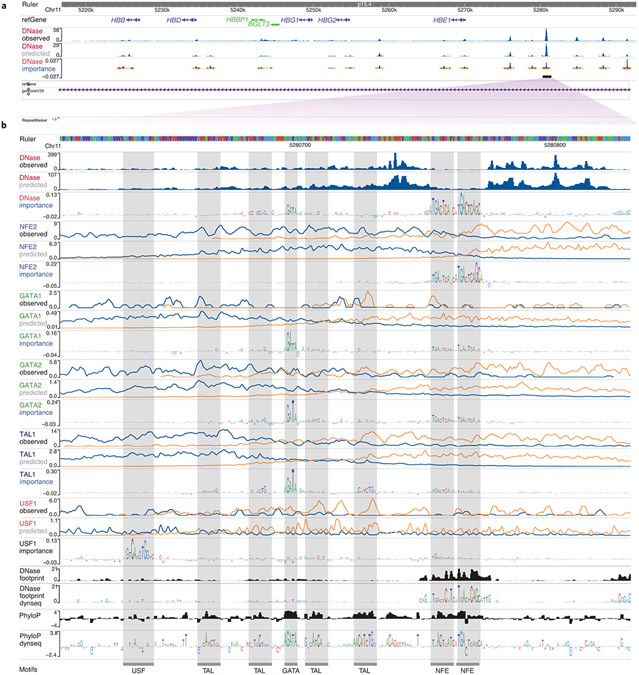
To illustrate a typical use case, we trained separate BPNet neural networks12 to map DNA sequence to base-resolution profiles of DNase sequencing (DNase-seq) and five transcription factor chromatin immunoprecipitation with high-throughput sequencing (ChIP–seq) experiments in the K562 cell line13,14. We used the DeepLIFT feature attribution algorithm15,16 to derive nucleotide importance scores for sequences underlying regions with enriched signal (peaks) from each of the models. We used the WashU Epigenome Browser to visualize and interpret an enhancer in the β-globin locus approximately 10 kb upstream of the HBE1 gene17. We used BigWig tracks to display the observed and model-predicted profiles for each assay, and dynseq tracks to visualize assay-specific importance scores derived from each model (Fig. 1 and Supplementary Fig. 1). The dynseq tracks highlight the predicted quantitative influence of different motifs on chromatin accessibility and transcription factor binding profiles. The binding profiles of NFE2, GATA1, GATA2 and USF1 are predicted to be influenced by their respective direct binding motifs. By contrast, TAL1 is affected by GATA and several weaker TAL1 motifs, which suggests that GATA transcription factors are potentially involved in cooperatively recruiting TAL1 to their binding sites. The dynseq track of the DNase-seq model highlights GATA and NFE motifs, which suggests that these motifs primarily drive DNase I hypersensitivity at this locus. We also visualized DNase-seq footprinting scores18 and PhyloP conservation scores19 using both the standard BigWig rendering and the dynseq track. The footprinting scores highlight NFE motifs, whereas the PhyloP dynseq track highlights strong conservation of GATA, NFE and several TAL motifs. Together, dynseq tracks reveal subtle, quantitative, context-specific sequence determinants of transcription factor binding and chromatin accessibility, thereby providing insights into the architecture of cis-regulatory elements. Its generality allows for versatile applications, including the exploration of sequence features disrupted by variants that influence regulatory activity via allele-aware dynseq tracks in the Resgen/FliGlass browser20 (Supplementary Note, Supplementary Fig. 2).
Fig. 1 ∣. WashU Epigenome Browser session for deciphering sequence architecture of a cis-regulatory element.
a, Human β-globin locus with observed and predicted DNase-seq tracks and zoomed out DNase-seq model-derived importance dynseq track. b, Enhancer 10 kb upstream of HBE1 (hg38 chr11:5280607–5280830) with observed base-resolution 5′ end coverage tracks of DNase-seq and ChIP–seq targeting transcription factors NFE2, GATA1, GATA2, TAL1 and USF1 in the K562 cell line; corresponding predicted tracks from BPNet sequence models trained on these data; BPNet model-derived nucleotide importance scores visualized using dynseq tracks; and DNase-seq footprinting scores, −log(P value) and PhyloP conservation scores visualized using standard BigWig and dynseq tracks. For ChIP–seq profile tracks, blue denotes plus (+) and orange denotes minus (−) strand.
Integration of dynseq tracks into genome browsers enables intuitive, sequence-centric visualization of diverse quantitative, base-resolution scores, including sequence contribution scores from predictive models, sequence conservation scores, motif match scores and high-resolution transcription factor footprinting. We expect that dynseq tracks will enhance exploratory analysis, discovery and hypothesis generation by enabling contextual interpretation of informative sequence features in genomic elements and those disrupted by variation at single-nucleotide resolution.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health (NIH) grant numbers U01HG009431 and U01HG012069 to A.K.; R01HG007175, U01CA200060, U24ES026699, U01HG009391, UM1HG011585, U41HG010972 and U24HG012070 to T.W.; 5U41HG002371 to B.J.R., B.T.L. and M.H.
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Competing interests
A.K. is scientific co-founder of Ravel Biotechnology, is on the scientific advisory board of PatchBio, SerImmune, AINovo, TensorBio and OpenTargets, is a consultant with ILLumina and owns shares in DeepGenomics, Immuni and Freenome. ALL other authors have no competing interests to declare.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41588-022-01194-w.
Data availability
Data and models used to create vignettes are available at: https://doi.org/10.5281/zenodo.6582100. See Supplementary Table 1 for browser-specific functionalities
Code availability
Code to reproduce vignettes is available at: https://doi.org/10.5281/zenodo.7019993. The dynseq tracks use the BigWig file format. A tutorial is available at: https://kundajelab.github.io/dynseq-pages/. The dynseq track is supported by:
• UCSC Genome Browser (https://genome.ucsc.edu). Documentation is available at: https://genome.ucsc.edu/goldenpath/help/big-Wig.html#dynseq. Source code is available at: https://github.com/ucscGenomeBrowser/kent.
• HiGlass/Resgen (https://higlass.io; https://resgen.io). Dynseq is implemented as a plugin. Source code is available at: https://github.com/kundajelab/higlass-dynseq/.
• WashU Epigenome Browser (https://epigenomegateway.wustl.edu). Source code is available at: https://github.com/lidaof/eg-react. Documentation is available at: https://eg.readthedocs.io/en/latest/tracks.html#dynseq.
References
- 1.Eraslan G, Avsec Ž, Gagneur J & Theis FJ Nat. Rev. Genet 20, 389–403 (2019). [DOI] [PubMed] [Google Scholar]
- 2.de Almeida BP, Reiter F, Pagani M & Stark A Nat. Genet 54, 613–624 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Avsec č. et al. Nat. Methods 18, 1196–1203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaganathan K et al. Cell 176, 535–548.e24 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Bogard N, Linder J, Rosenberg AB & Seelig G Cell 178, 91–106.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider TD Nucleic Acids Res. 25, 4408–4415 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, Hsu S, Purushotham D, Sears RL & Wang T Nucleic Acids Res. 47, W158–W165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D et al. Nucleic Acids Res. 50, W774–W781 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent WJ, Zweig AS, Barber G, Hinrichs AS & Karolchik D Bioinformatics 26, 2204–2207(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent WJ Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerpedjiev P et al. Genome Biol. 19, 125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avsec Ž et al. Nat. Genet 53, 354–366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ENCODE Project Consortium. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis CA et al. Nucleic Acids Res. 46, D794–D801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shrikumar A, Greenside P & Kundaje A In Proc. 34th International Conference on Machine Learning 70, 3145–3153 (2017). [Google Scholar]
- 16.Lundberg SM & Lee S.-l. A. In Advances in Neural Information Processing Systems (eds. Guyon I et al. ) 30, 4765–4774 (Curran Associates, 2017). [Google Scholar]
- 17.Kettis M et al. Proc. Natl. Acad. Sci. USA 111, 6131–6138 (2014).24753594 [Google Scholar]
- 18.Vierstra J et al. Nature 583, 729–736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard KS, Hubisz MJ, Rosenbloom KR & Siepel A Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tehranchi AK et al. Cell 165, 730–741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and models used to create vignettes are available at: https://doi.org/10.5281/zenodo.6582100. See Supplementary Table 1 for browser-specific functionalities
Code to reproduce vignettes is available at: https://doi.org/10.5281/zenodo.7019993. The dynseq tracks use the BigWig file format. A tutorial is available at: https://kundajelab.github.io/dynseq-pages/. The dynseq track is supported by:
• UCSC Genome Browser (https://genome.ucsc.edu). Documentation is available at: https://genome.ucsc.edu/goldenpath/help/big-Wig.html#dynseq. Source code is available at: https://github.com/ucscGenomeBrowser/kent.
• HiGlass/Resgen (https://higlass.io; https://resgen.io). Dynseq is implemented as a plugin. Source code is available at: https://github.com/kundajelab/higlass-dynseq/.
• WashU Epigenome Browser (https://epigenomegateway.wustl.edu). Source code is available at: https://github.com/lidaof/eg-react. Documentation is available at: https://eg.readthedocs.io/en/latest/tracks.html#dynseq.