Abstract
The potato cyst nematode (PCN) causes extensive crop losses worldwide. Because the hatching of PCN requires host-derived molecules known as hatching factors (HFs), regulating HF production in host plants may help to control this harmful pest. Solanoeclepin A (SEA), isolated from potato, is the most active HF for PCN; however, its biosynthesis is completely unknown. We discovered a HF called solanoeclepin B (SEB) from potato and tomato root exudates and showed that SEB was biosynthesized in the plant and converted to SEA outside the plant by biotic agents. Moreover, we identified five SEB biosynthetic genes encoding three 2-oxoglutarate-dependent dioxygenases and two cytochrome P450 monooxygenases in tomato. Exudates from tomato hairy roots in which each of the genes was disrupted contained no SEB and had low hatch-stimulating activity for PCN. These findings will help to breed crops with a lower risk of PCN infection.
Disruption of the biosynthetic genes of a hatching factor provides an approach to control the potato cyst nematode infection.
INTRODUCTION
Plant parasitic nematodes are one of the most harmful pests; the cost of their damage to crop production is estimated to be USD 80 billion per year (1, 2). Cyst nematodes, comprising Globodera and Heterodera, are among the most economically important species of root parasitic nematodes (1–3). While other plant parasitic nematodes, such as root-knot nematodes, can parasitize a variety of plant species, many cyst nematode species have limited host ranges (4, 5). Examples include potato cyst nematodes (PCNs; Globodera rostochiensis and Globodera pallidas), which are found only on Solanaceae plants such as potato and tomato, and soybean cyst nematode (SCN; Heterodera glycines), which is a specific parasite for Fabaceae. PCNs are estimated to affect 9% of global potato production (6), and SCN causes yield losses equating to more than USD 1.5 billion annually (1). The ability to overcome cyst nematode damage is required for stable and sustainable agricultural production.
The life cycle of cyst nematodes is highly synchronized with that of their host plants (2, 7). The life cycle of cyst nematodes is divided into four larval stages and one adult stage, and these stages are separated by molts. The second-stage juvenile (J2) is the only infective stage. The J2 larvae migrate through the soil to the roots of the host plant and become parasites. They grow by depriving the host plant of nutrients and molting to the third and fourth juvenile stages (J3 and J4, respectively) and then reaching the mature adult stage. After fertilization, the female dies and her body hardens to form a cyst that encloses hundreds of next-generation eggs. These cysts eventually break away from the roots and remain in the soil after the crop is harvested. Within the eggs in the cyst, the first-stage juveniles (J1) develop, molt to reach J2, and then remain dormant until the next host plants appears nearby. The cyst protects the eggs from drought, cold, and insecticides; J2 can survive without hatching in dormancy for up to 20 years (8, 9). When the host plants grow nearby, the J2 larvae in the eggs hatch in response to specific compounds called hatching factors (HFs), which are secreted by host roots, and the next cycle begins. Thus, hatching is a crucial event in the life cycle of cyst nematodes, making HFs key targets for cyst nematode control (10).
Glycinoeclepin A and the related nortriterpene glycinoeclepins B and C were isolated from kidney bean roots as potent HFs specific to SCNs (11–13). Solanoeclepin A (SEA) is a highly active HF for PCN that was isolated from potato root exudate in the 1990s (14, 15). The complete chemical synthesis of SEA was determined in 2011, and its hatching activity was confirmed at concentrations as low as 1 × 10−8 to 1 × 10−10 g ml−1 (16). More recently, SEA was identified in the root exudate from several Solanaceae plants, such as tomato, in addition to potato (16, 17). In contrast, studies of hydroponically grown tomatoes and soil-grown potatoes have shown that SEA alone cannot explain all the hatch-stimulating (HS) activity for PCN (16, 18–20). Thus, there are many unclear aspects of HFs. Here, we report isolation of a previously unidentified HF for PCN, which serves as a direct precursor of SEA, and identify the biosynthetic genes.
RESULTS
Identification and structural determination of a previously unidentified HF
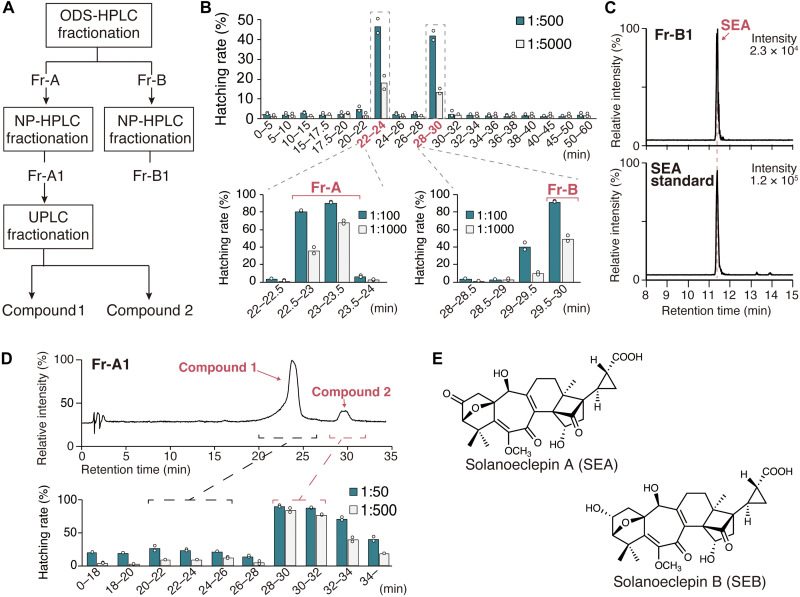
To identify a HF for PCN, we first performed hatching assay–guided purification of HF(s) from potato hydroponic culture solution. The HS components collected by the synthetic absorbent SP207 were further purified via a series of purification and fractionation steps (Fig. 1A and figs. S1 to S7). In octadecylsilyl (ODS) high-performance liquid chromatography (HPLC) fractionation, the active components were separated into two independent fractions: Fr-A and Fr-B (Fig. 1B). Further fractionation of Fr-B by normal-phase (NP) HPLC yielded an active fraction Fr-B1 [a fraction with a retention time (Rt) of 35.5 to 36 min in fig. S8]. We conducted ultraperformance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) analysis of Fr-B1 and detected SEA (Fig. 1C). Next, we conducted NP HPLC fractionation of Fr-A and obtained an active fraction Fr-A1 (a fraction with an Rt of 39 to 39.5 min in fig. S9). UPLC-ultraviolet and UPLC-MS analysis of Fr-A1 detected two major compounds with parent ion masses of mass/charge ratio (m/z) 356.5 [M + H]+ (compound 1) and m/z 500.7 [M + H]+ (compound 2), respectively (fig. S10). We separated these two compounds by UPLC fractionation to confirm their HS activity and revealed that compound 2 is a previously unidentified HF for PCN (Fig. 1D). High-resolution mass spectrometry analysis confirmed its chemical formula as C27H32O9 (tables S1 and S2), which has two more hydrogen atoms than SEA (C27H30O9). The compound giving m/z 500.7 [M + H]+ was purified again from potato hydroponic solution based on UPLC-MS/MS detection to obtain sufficient quantities for structural determination (figs. S1 and S11). Nuclear magnetic resonance spectroscopy revealed that in the structure of the compound, the C-2 carbonyl group of SEA is reduced to a secondary hydroxy group (Fig 1E, figs. S12 to S14, and table S3). Therefore, we concluded that this HF is the second eclepin-type compound identified in potatoes after SEA, and we named it solanoeclepin B (SEB). Furthermore, hatching assays revealed that SEB is a strong HF, with comparable activity to SEA (tables S4 and S5).
Fig. 1. Purification of HFs from potato hydroponic culture solution.
(A) Purification scheme of hatching factors (HF). (B) Separation of HFs using preparative ODS-HPLC. Fractions were collected at 30-s intervals and tested for hatching against G. rostochiensis eggs. Fractions were first tested in groups; then, each of the fractions collected at 22 to 24 and 28 to 30 min was tested alone for HS activity. Hatching rate (%) is shown at two dilutions. (C) Solanoeclepin A (SEA) detection from fraction Fr-B1, which was obtained by further purification of Fr-B based on HS activity. A multiple reaction monitoring (MRM) transition of m/z 499 > 399 was selected to detect SEA. (D) Ultraperformance liquid chromatography (UPLC) fractionation of Fr-A1, which was obtained by further purification of Fr-A based on HS activity. The HS activity of each fraction on G. rostochiensis eggs was investigated at two different dilutions. The values indicate the means of two biological replicates; white dots are individual measurements. (E) Chemical structures of SEA and solanoeclepin B (SEB).
HF analysis in the exudate of tomato hairy roots
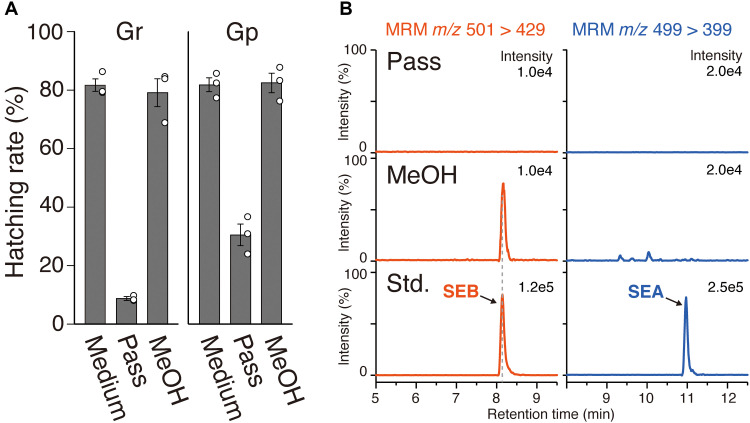
Next, we analyzed the HFs produced by tomato hairy roots, which can be easily transformed and have excellent culture characteristics. The medium from tomato hairy roots cultured for 2 weeks showed high HS activity for PCNs (Fig. 2A). The medium (1 ml) was passed through a column packed with SP207, and the HS activities of the solution and the 100% methanol-eluted fraction were examined. The activity was primarily observed in the 100% methanol fraction (Fig. 2A). Then, the medium (150 ml) was used for HF analysis. UPLC-MS/MS analysis revealed that SEB was detected only in the 100% methanol fraction, whereas SEA was not detected in any fraction (Fig. 2B). The results indicate that SEB is the major HF in tomato hairy root cultures. Furthermore, the detection of SEB in sterile tomato hairy root cultures confirmed that it is biosynthesized by the plant itself. Conversely, neither SEB nor SEA were detected in the extracts of tomato hairy roots by UPLC analysis (fig. S15).
Fig. 2. Analysis of hatching factors (HFs) produced by tomato hairy roots.
(A) Hatch-stimulating (HS) activity of tomato hairy roots culture medium for G. rostochiensis (Gr) and Globodera pallida (Gp). Pass, the passage of the medium through SP207; methanol, the fraction eluted with 100% methanol. Error bars represent the means ± SE (n = 3 biologically independent flasks); white dots are individual measurements. (B) Ultraperformance liquid chromatography (UPLC) analysis of SP207 fraction of tomato hairy root culture medium (150 ml). Multiple reaction monitoring (MRM) transitions of m/z 501 > 429 (red) and m/z 499 > 399 (blue) were used to detect solanoeclepin B (SEB) and solanoeclepin A (SEA), respectively.
Investigation of HS activity in tomato hairy roots in response to various conditions
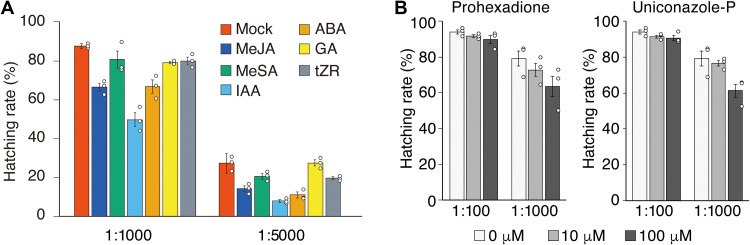
Given the detection of SEB in the culture medium of tomato hairy roots, we aimed to identify the SEB biosynthetic genes in tomato. First, the conditions for HS activity in tomato hairy roots were verified. Two-week-old tomato hairy roots were transplanted into fresh medium supplemented with various phytohormones. After 3 days, medium from each condition was subjected to the hatching assay. Among the six phytohormones [abscisic acid (ABA), gibberellin A3 (GA3), indole-3-acetic acid (IAA), methyl jasmonate (MeJA), methyl salicylate (MeSA), and trans-zeatin (tZR)] tested, ABA, IAA, and MeJA resulted in a notable decrease in HS activity (Fig. 3A), suggesting that these three phytohormones suppress production of the HF. SEB is a highly oxygenated nortriterpenoid with nine oxygens; therefore, many oxygenases are likely to be involved in SEB biosynthesis. It is well known that the cytochrome P450 monooxygenase (CYP) superfamily and 2-oxoglutrate–dependent dioxygenase (DOX) superfamily are involved in various oxidation and hydroxylation steps in the biosynthesis of triterpenoids. We tested the effects of the CYP inhibitor uniconazole-P and the DOX inhibitor prohexadione on HS activity in a manner similar to the phytohormone treatment experiments above. The results showed that uniconazole-P and prohexadione treatment both reduced HS activity compared to the mock treatment (Fig. 3B). Given that SEB is the main HF in tomato hairy root exudates, as shown in Fig. 2, this inhibitory experiment implicates the involvement of CYPs and DOXs in SEB biosynthesis in tomato plants.
Fig. 3. HS activity in tomato hairy roots in response to various conditions.
(A) Hatch-stimulating (HS) activities of culture medium from phytohormone-treated hairy roots for G. rostochiensis. Each biological replicate comprises hairy roots cultured in separate flask and independently treated with hormones. (B) HS activities for G. rostochiensis in culture medium from enzyme inhibitor–treated hairy roots. The values are presented as the means ± SE (n = 3) of three biological replicates. For each sample solution, the hatching assay was performed at two dilution rates.
Identification of DOX genes involved in SEB biosynthesis in tomato
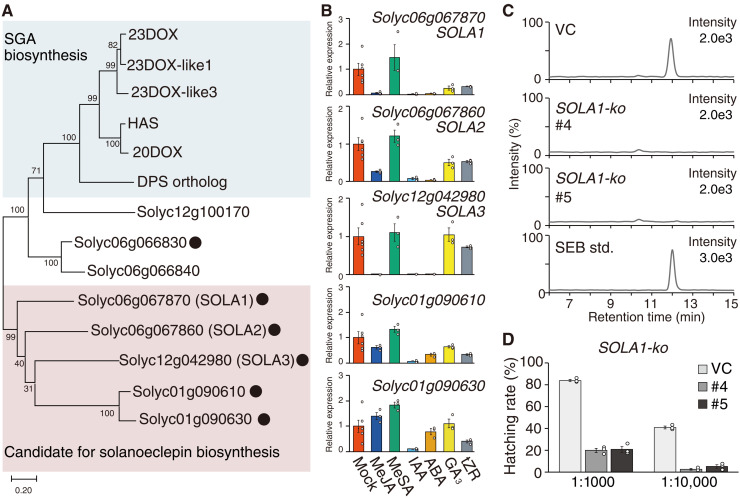
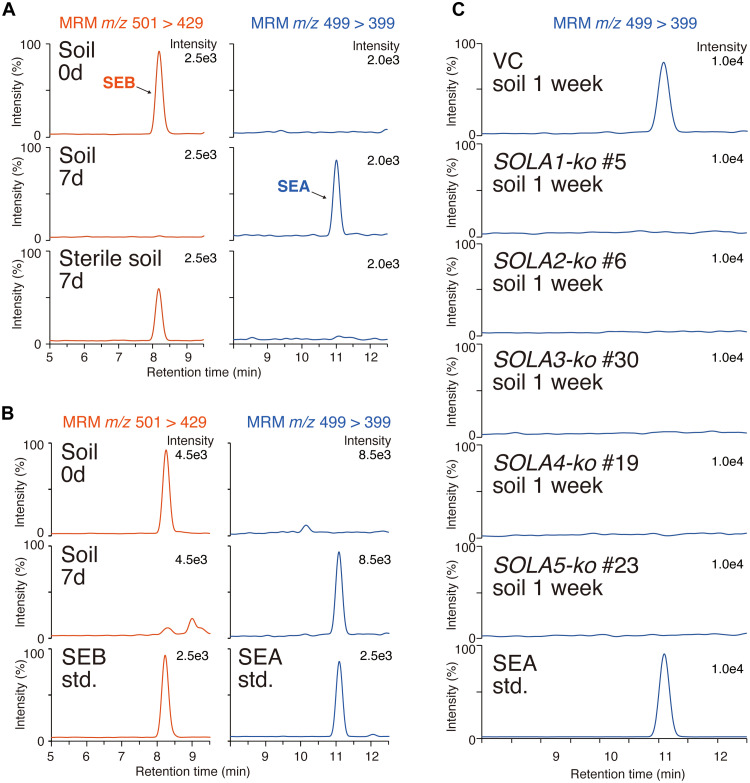
HFs are thought to be biosynthesized in the roots; the highest HS activity was found in the roots of the extracts of each tomato tissue (fig. S16). To identify the genes associated with HF biosynthesis, we searched for DOX genes that were specifically expressed in the roots using public genome and transcriptome data from the Sol Genomics Network (https://solgenomics.net/), and found 31 DOX genes (table S6). On the basis of the phylogenetical classification of the DOX superfamily, seven of these genes were classified into the DOXC20 clade. Recently, several genes belonging to the DOXC20 clade have been reported to be involved in biosynthesis of triterpenoid steroidal glycoalkaloids (SGAs) (21–25). The phylogenetic analysis showed that five of the seven genes form a branch separated from that of the SGA biosynthetic genes (Fig. 4A). Using quantitative reverse transcription polymerase chain reaction (qRT-PCR), we analyzed the changes in expression of these five genes (Solyc06g067870, Solyc06g067860, Solyc12g042980, Solyc01g090610, and Solyc01g090630) in response to phytohormones (Fig. 4B). The expression patterns of Solyc06g067870, Solyc06g067860, and Solyc12g042980 were in good agreement with the alteration in patterns of HS activity induced by phytohormone administration, as shown in Fig. 4A, particularly the decrease by treatment with IAA, ABA, or MeJA. Consequently, we named Solyc06g067870, Solyc06g067860, and Solyc12g042980 as SOLANOECLEPIN BIOSYNTHESIS 1 (SOLA1), SOLA2, and SOLA3, respectively, for further analyses. We individually disrupted these candidate genes using CRISPR/Cas9-mediated genome editing in tomato hairy roots (figs. S17 and S18). No SEB was detected in the culture medium of hairy roots with each gene knocked out (Fig. 4C and fig. S19A). The growth of each line of knockout hairy roots was normal compared to that of vector control (VC) hairy roots, indicating that the lack of SEB in the exudates of these knockout lines was not caused by poor growth (fig. S20). These results confirmed the involvement of SOLA1, SOLA2, and SOLA3 in SEB biosynthesis. Furthermore, partially purified culture medium derived from tomato hairy roots in which each gene was disrupted had lower HS activity for G. rostochiensis than those of VC hairy roots (Fig. 4D and fig. S19B).
Fig. 4. Characterization of biosynthetic genes for nortriterpenoid-type hatching factors (HFs) in tomato.
(A) Phylogenetic tree of DOXC20 cade genes. Blue shading indicates the branch composed of enzymes associated with triterpenoid steroidal glycoalkaloid (SGA) biosynthesis. Pale red shading indicates the branch, including candidate genes for nortriterpenoid-type HFs biosynthesis. The black dots indicate that the gene encoding that enzyme is expressed specifically in the root. (B) The analysis of transcription levels of candidate genes by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) in hairy roots treated with each phytohormone. (C) Ultraperformance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) analysis of solanoeclepin B (SEB) in the culture medium of vector control (VC) and SOLA1-ko tomato hairy roots. Chromatogram recorded by the multiple reaction monitoring (MRM) transition of m/z 501 > 429, used to detect SEB, is shown. std, authentic standard. (D) Hatch-stimulating (HS) activities of the culture medium from VC and SOLA1-ko tomato hairy roots. The values are presented as the means ± SE of three biological replicates. The hatching assay was conducted at two dilution rates.
Identification of CYP genes involved in SEB biosynthesis in tomato
Next, we surveyed candidate CYP genes. Among the CYPs that are preferentially expressed in roots, we selected three genes (Solyc05g011970: CYP749A20, Solyc05g011940: CYP749A19, and Solyc05g021390: CYP716A44) that were most highly expressed (table S7). Phylogenetically, CYP749 is close to the CYP734 family, which is involved in the inactivation metabolic reaction of the triterpenoid phytohormone brassinosteroid (26, 27). CYP716A44 has previously been reported to be involved in the biosynthesis of the triterpenoids oleanonic acid and ursolic acid in tomato (28). We examined the changes in the expression of these genes in response to phytohormones using qRT-PCR and found that CYP716A44, unlike the other candidate genes, was induced by MeJA, which resulted in reduced HS activity when treated with the hairy roots (fig. S21). Therefore, we ruled out CYP716A44 as a candidate and named CYP749A20 and CYP749A19 as SOLA4 and SOLA5, respectively. SEB was not detected in the culture medium of SOLA4- and SOLA5-disrupted hairy roots (figs. S22 to S25). In contrast, SEB was detected in the medium of CYP716A44-disrupted hairy roots (fig. S24A). The growth of SOLA4 and SOLA5 mutants was comparable to that of VC hairy roots (fig. S25B). Moreover, the medium of SOLA4- and SOLA5-disrupted hairy roots had lower HS activity than that of the VC (fig. S24B). On the basis of these results, SOLA4 and SOLA5 were identified as SEB biosynthesis genes.
Conversion of SEB to SEA in soil
The detection of SEA has been reported in soil-grown or hydroponically grown tomato and potato plants in open environments (17, 18). The absence of SEA in the sterile tomato hairy roots medium led us to hypothesize that biotic agents present in unsterilized soil and hydroponic solution are involved in the formation of SEA. To verify this hypothesis, pure SEB was incubated in soil for 7 days, followed by water extracts from the soil subjected to UPLC-MS/MS. This analysis revealed the complete abolishment of SEB and the emergence of SEA after 7 days of incubation in soil (Fig. 5A). No such changes were observed in γ-ray–sterilized soil (Fig. 5A). Furthermore, when the culture medium of tomato hairy roots was incubated in soil, SEB in the medium disappeared, and instead, SEA was detected (Fig. 5B). These results clearly indicate that SEB is a direct precursor of SEA and that biotic agents outside the plant contribute to the conversion of SEB to SEA.
Fig. 5. Conversion of solanoeclepin B (SEB) in soil.
(A) Incubation of purified SEB in soil. Multiple reaction monitoring (MRM) chromatogram of soil-treated SEB is shown. (B) Incubation of tomato hairy root culture medium in soil. (C) Incubation of culture medium prepared from SOLA gene–disrupted tomato hairy roots in soil. 0d, the sample was collected immediately after SEB was added to the soil. std, authentic standard.
In addition, we applied the culture medium from the five knockout hairy root mutants to soil and incubated them for 7 days, followed by analyzing their aqueous extracts using UPLC-MS/MS. SEA was detected in the extract derived from the VC but not from the extracts derived from each of the five knockout mutants (Fig. 5C). These results indicate the involvement of all of the identified genes (SOLA1 to SOLA5) in SEA production in soil, where SEB was converted to SEA by biotic agents during the incubation.
DISCUSSION
For several decades, many studies have been conducted to identify HFs because of their general interest as promising target for cyst nematode control and unique signal molecules between plants and nematodes (29–32). However, the only highly active HF for PCN with a determined structure is SEA (14–16). Devine and Jones (32) conducted hatching assay–guided purification of HFs and reported the existence of nine HFs for PCNs with a molecular weight of 530.5 Da, but their chemical structures were not determined. Recently, Vlaar et al. (18) conducted a metabolomic analysis of root exudates from 51 cultivars of soil-grown potato plants and predicted a compound giving m/z 526.18 as a HF, and its presence was correlated with both the presence of SEA and HS activity. They assumed that the compound has a similar structure to SEA based solely on their mass fragmentation similarity and named it “solanoeclepin B (solB)” (18). We believe that the compound giving m/z 526.18 is not identical to our structurally determined SEB because the determined m/z is different, i.e., the molecular formula is different. The metabolomic analysis of the compound in the mixture provided useful information but was not sufficient to prove that it was an eclepin-type active compound. It is highly anticipated in the future that the isolation and structural analysis of the compound giving m/z 526.18 will prove whether it is an SEA analog. Accordingly, we named the HF isolated in this study SEB as the second HF with a determined structure and high HS activity after SEA.
The HS activity of SEB for PCNs is comparable to that of SEA (19). It is interesting that previous studies that addressed the identification of HFs by HS activity-guided purification did not suggest the presence of SEB, despite its high HS activity. We demonstrated that SEB is converted to SEA by biotic agents outside the plant. The materials from which HF identification was previously attempted were extracts or root exudates from potato and tomato plants grown in open, nonsterile environments. Therefore, it is most likely that SEB was converted to SEA under those conditions and that its presence was undetected.
In the actual field, PCN eggs are present in cysts and the J2 larvae in the eggs hatch in response to HFs. Hatching activity can be evaluated using either cysts containing eggs or free eggs artificially removed from cysts (17–20, 32). In this study, we used free eggs artificially removed from cysts for hatching assays, and our result clearly demonstrated that SEB can induce the hatching of J2 in eggs. The interior of a cyst is a hatching-suppressive environment because of the influence of cyst contents and cyst wall–derived compounds (33–35). Cysts also act as a physical barrier to chemicals. It is unlikely that SEB and SEA have no HS activity at all in eggs in cysts, but the effect of HFs may be altered within cysts, e.g., the activity of SEB and SEA differs remarkably. Unfortunately, the assay for cysts could not be performed because of the insufficient amount of SEB. Further investigations, including analyses of whether SEB and SEA exhibit HS activity for the eggs in cysts, are required to elucidate the actual effects of SEB and SEA on the hatching of PCNs in the field.
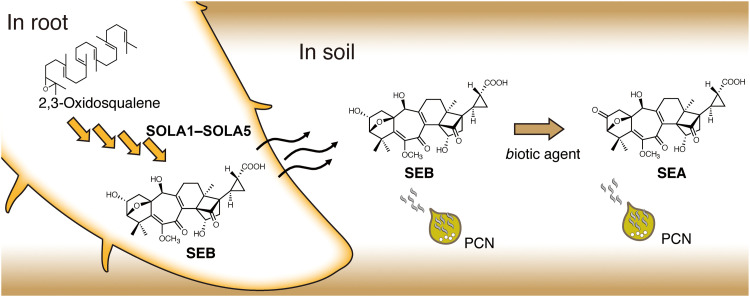
In this study, we isolated five SEB biosynthetic genes (SOLA1 to SOLA5) and found that the disruption of each gene resulted in the complete abolition of SEB production in tomato hairy roots. Slight residual HS activity remained in the medium from each knockout line, possibly because of the secretion of upstream intermediates in the SEB biosynthetic pathway or the presence of unknown non-nortriterpenoid–type HFs, but these results demonstrate that disrupting or inhibiting SEB biosynthesis can substantially reduce the hatching of PCNs. Furthermore, incubation of the culture medium of these SEB-deficient hairy roots in soil did not yield SEA, strongly suggesting that the five genes (SOLA1 to SOLA5) are also involved in SEA production (Fig. 6). However, whether plants can biosynthesize SEA on their own remains to be verified. This question could be addressed by growing tomato and potato plants in various conditions, such as nonsterilized open fields or sterilized hydroponic systems and comparing the amounts of SEA and SEB. SEB is biosynthesized in the roots, and it is expected to be largely secreted because SEB was not detected in tomato hairy root extracts. In the case of field-grown plants, the secreted SEB is presumed to be converted to SEA in the soil (Fig. 6). SEB exhibits slightly lower HS activity than SEA, indicating that PCNs recognize the presence of a host more efficiently by responding more strongly to SEA, which is believed to be more abundant in soil than SEB.
Fig. 6. Putative biosynthetic of solanoeclepin B (SEB) in root and metabolism in soil.
In the roots, 2,3-oxidosqualene is modified by various enzymes to biosynthesize SEB. After being released into the soil, SEB is converted to solanoeclepin A (SEA) by biotic factors in the soil. PCN, potato cyst nematode.
SEB is expected to be biosynthesized from simple triterpenoids, such as cycloartenol, via drastic skeletal rearrangements, including ring expansion, methyl group(s) shifts, and side chain cleavage (fig. S26). However, even the final products, SEB and SEA, are present in very small quantities in nature; therefore, the accumulation of their intermediates is also considered to be very small. There have been no reports on the biosynthetic intermediates. We performed metabolomic analysis of the hairy roots with the identified biosynthetic genes knocked out but could not identify the accumulation of any reasonable intermediates. Further analysis is needed to clarify through which reactions these five genes are responsible for SEB biosynthesis.
In addition to Solanaceae, Fabaceae plants are known to produce HFs for cyst nematodes that host themselves, i.e., SCN. Three HFs—glycinoeclepins A, B, and C—have been identified from kidney bean. Solanoeclepins and glycinoeclepins have many structural similarities, including the stereochemical configuration of the terpenoid skeletons and oxidation sites; hence, they may have the same biosynthetic origin and share biosynthetic pathways. Phylogenetic analysis revealed the presence of distinct orthologs of SOLA1 to SOLA3 in potato, which may be involved in the biosynthesis of solanoeclepins (fig. S27). In addition, branches containing these genes also contained several genes from soybean and kidney bean (fig. S27). These genes are all expressed preferentially in roots and may be involved in glycinoeclepin biosynthesis.
Our results show that the disruption of solanoeclepin biosynthetic genes can greatly reduce PCN hatching. Our findings support the development of advanced control methods to prevent nematode damage by breeding crops that produce low concentrations of HFs and therefore prevent hatching. Simultaneously, this study provides a basis for studying the biosynthesis and transport of nortriterpenoid-type HFs and their roles in plants beyond nematode hatching.
MATERIALS AND METHODS
Chemicals
SEA was synthesized as described previously (16).
Plant materials and growth conditions
The hydroponic cultivation of potatoes was achieved by spraying the roots with hydroponic solution. The hydroponic solution was prepared by OTA no. 1 fertilizer (OTA house fertilizer series, OAT Agrio, Tokyo, Japan), OTA no. 2 fertilizer (OTA house fertilizer series, OAT Agrio, Tokyo, Japan), and OTA no. 5 fertilizer (OTA house fertilizer series, OAT Agrio, Tokyo, Japan). Initially, potato culture shoots or microtubers were sown on vermiculite spread in pots, and the seedlings were grown for approximately 3 weeks with bottom feeding of hydroponic solution under a 16-hour light/8-hour dark photoperiod. The plants were then planted in a hydroponic facility and grown, while the roots were sprayed with hydroponic solution.
Hatching assay
The hatching assay was conducted as described previously (20). Briefly, 1 ml of the suspension of G. rostochiensis pathotype Ro 1 or G. pallida pathotype Pa 3 eggs was added to a glass vial, and 1 ml of each sample at different concentrations was added. Each vial was incubated at 18°C for approximately 2 weeks. Subsequently, 1 ml from each vial was removed and transferred to a Syracuse dish, and the hatched juveniles and unhatched eggs were counted under a binocular microscope. Distilled water and synthetic SEA were used as the negative and positive controls, respectively. G. pallida was only used for the analysis of HFs produced by tomato hairy roots and comparisons of the hatching activities of SEA and SEB.
Hatching assay–guided purification of HFs from potato hydroponic culture solution
Approximately 5000 potato plants of various varieties were grown in the hydroponic facility described above. Then, 3 liters of Sepabeads SP207 (Mitsubishi Chemical Corp.) was placed in a plastic column and placed where the effluent from the medium sprayed on the roots was circulated. Sepabeads SP207 (Mitsubishi Chemical Corp.) were replaced every 3 to 4 weeks from June 2018 to October 2018; hence, approximately 60 liters of Sepabeads SP207 adsorbed with active substances was collected. The total volume of hydroponic effluent that passed through SP207 during this period was estimated to be approximately 20,000 liters.
Then, 15 liters of the resin was transferred into a flask and eluted twice with three volumes of methanol by shaking for 1 hour at room temperature. The methanol eluents were dried in vacuo, redissolved in 500 ml of methanol, and filtered through filter paper to remove methanol-insoluble materials. The methanol-soluble materials (20.9 g) were then dried in vacuo, dissolved in 1 liter of water, and treated three times with 500 ml of n-hexane. After the pH of the remaining aqueous phase was adjusted to 3.0 by the addition of hydrochloric acid, HFs were extracted three times with 1 liter of ethyl acetate. The acidic ethyl acetate phase (4.8 g) was dried in vacuo, dissolved in 5% (v/v) aqueous methanol, and loaded onto five separate Sep-Pak C18 cartridges (Vac 35 cm3, 10 g; Waters) that were prewashed and equilibrated with five column volumes of methanol and 5% (v/v) aqueous methanol, respectively. Each cartridge was washed with five column volumes of 5% (v/v) aqueous methanol and eluted with five column volumes of 20% (v/v) aqueous methanol and methanol. Because of the more potent HS activity, the methanol fraction was subjected to further purification. The methanol fraction (3.4 g) was dried in vacuo, redissolved in 30 ml of methanol, and passed through a strong cation exchanger Dowex 50 (H+ form, 200 ml; Sigma-Aldrich) with 1 liter of methanol. The noncationic pass-through fraction (2.8 g) was dried in vacuo, redissolved in 7 ml of methanol, and separated by Sephadex LH-20 column chromatography (150 ml; GE Healthcare) with methanol as the mobile phase. The active fractions (687 mg) were combined, dried in vacuo, redissolved in 10 ml of methanol, and then loaded onto a strong anion exchanger Dowex 1 (acetate form, 400 ml; Sigma-Aldrich). After the column was washed with 2 liters of methanol, elution was conducted three times with 650 ml of methanol containing 0.1 or 1 N acetic acid. The HFs were successfully collected in methanol containing 1 N acetic acid. The active fraction (48 mg) was dissolved in 100 μl of 5% (v/v) aqueous acetonitrile and subjected to reverse-phase HPLC connected to an Atlantis T3 column (10 mm by 150 mm, 5 μm; Waters) protected by an Atlantis T3 Prep Guard Cartridge (10 mm by 10 mm, 5 μm; Waters). The mobile phase consists of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) with a flow rate of 5.0 ml min−1 using the following linear gradient system: 5% B at 0 min, held for 5 min, ramped to 100% B at 100 min, and then held for 124 min. The eluents were monitored at 254 nm, and fractions were collected every 0.5 min. The HFs were collected in the eluents at Rt of 22.5 to 23.5 min (Fr-A1) and 29.5 to 30 min (Fr-B1). The active fractions of Fr-A1 and Fr-B1 were then dissolved in 100 μl of chloroform/methanol (95:5) and subjected to NP HPLC connected to a YMC-Pack Diol-120-NP column (4.6 mm by 250 mm, 5 μm; YMC). The mobile phase consisted of solvent A (chloroform) and solvent B (methanol) with a flow rate at 1.0 min ml−1 using the following linear gradient system: 0% B at 0 min, held for 20 min, ramped to 10% B at 40 min, held for 50 min, increased to 100% B at 50 min, and then held to 70 min. The eluents were monitored at 270 nm, and fractions were collected every 0.5 min. The HFs were collected in the eluents at Rt of 39 to 39.5 min (Fr-A2) and 35.5 to 36 min (Fr-B2) derived from fraction Fr-A1 and Fr-B1, respectively. In Fr-B2, SEB was detected by UPLC-MS/MS analysis as described in the “UPLC-MS/MS analysis of SEA and SEB” section. Next, Fr-A2 was subjected to reverse-phase UPLC connected to an AQUITY UPLC HSST3 column (100 mm by 2.1 mm, 1.8 μm) with a column temperature of 40°C and a flow rate of 0.2 ml min−1. Isocratic elution was performed with water/acetonitrile (90:10, v/v) with 0.1% formic acid. The eluents were monitored at 270 nm, and fractions were collected every 2 min. The HF, compound 2, was eluted at Rt of 28 to 34 min.
The molecular formula of compound 2 was determined to be C27H32O9 based on high resolution mass spectrometry (HRMS) [electrospray ionization (ESI)] analysis by trapped ion mobility time-of-flight (timsTOF) mass spectrometer (Bruker): found m/z 501.2118 [M + H]+ (calcd. For C27H33O9, m/z 501.2119 [M + H]+; Δppm = 0.3). Then, we named the compound (compound 2) as SEB.
UPLC-MS/MS analysis of SEA and SEB
The LC-MS system (Waters) consisted of an AQUITY UPLC H-Class System and an AQUITY quadruple tandem mass spectrometer (TQ detector). MassLynx 4.1 software (Waters) was used to perform data acquisition and analyses. Each sample was injected into an ACQUITY UPLC HSS T3 column (100 mm by 2.1 mm, 1.8 μm; Waters), with a column temperature of 40°C and a flow rate of 0.2 ml min−1. The mobile phase consisted of solvent A (0.1% formic acid in water) and solvent B (acetonitrile), and the following gradient condition was used: 10% B at 0 min, ramped to 60% B at 20 min, 100% B at 20 min, held for 25 min, returned to 10% B after 25 min, and held for 30 min. The mass spectra were obtained in positive or negative ESI mode, with a capillary voltage of 3 kV. For the mass spectrometry scan mode, the mass range of m/z 100 to 1100 and a sample cone voltage of 25 V were used. For the HF analysis, multiple reaction monitoring (MRM) mode was used in positive ESI mode. The MRM transition of m/z 499 > 399 with a sample cone voltage of 35 V and a collision energy of 30 eV was selected for SEA and that of m/z 501 > 429 with a sample cone voltage of 30 V and a collision energy of 10 eV was selected for SEB.
Repurification of SEB based on detection by UPLC-MS/MS
Owing to the low concentration of SEB in Fr-A2 (estimated to be less than 10 μg), repurification with an efficient method based on the detection by LC-MS/MS analysis was conducted. Approximately 30 liters of SP207 resin placed in potato hydroponic culture solution from June to August 2019 was packed into a column. The column was washed and eluted with three column volumes of 20% (v/v) methanol and methanol, respectively. The methanol eluents were dried in vacuo, redissolved in 100 ml of methanol, and filtered through filter paper to remove methanol-insoluble materials. The methanol-soluble materials (33.5 g) were dried in vacuo, dissolved in 200 ml of water, and treated once or twice with the same volume of n-hexane or ethyl acetate, respectively. After the pH of remaining aqueous phase was adjusted to below 1.0 by the addition of hydrochronic acid, compound 1 was extracted in five volumes with 200 ml of ethyl acetate. The acidic ethyl acetate phase (4.3 g) was dried in vacuo, dissolved in 80 ml of 5% (v/v) aqueous methanol, loaded onto a Sep-Pak C18 cartridge (Vac 35 cm3, 10 g), and prewashed and equilibrated with five column volumes of methanol and 5% (v/v) aqueous methanol, respectively. The cartridge was washed with five column volumes of 5% (v/v) aqueous methanol and eluted five times with one column volume of 10, 20, and 40% (v/v) aqueous methanol and then five column volumes of methanol. The fractions in which compound 1 were detected were combined, and the combined fraction (702 mg) was then subjected to silica gel column chromatography (5 g) with a stepwise elution (30 ml each step) of chloroform/methanol (100:0 to 50:50, 10% step). The active compound was eluted in the chloroform/methanol (80:20) fraction (95 mg). Then, NP HPLC preparation was conducted as described above. The eluent at Rt of 39 min was combined (5.3 mg) and subjected to reverse-phase HPLC. The reverse-phase HPLC system was the same as described above, but isocratic elution with water/methanol (85:15, v/v) with 0.05% trifluoroacetic acid was used instead of a gradient elution. Compound 1 was eluted at an Rt of 46 min, and a white powder was obtained (estimated mass of 340 μg). NMR spectra were recorded on a Bruker AVANCE III 800US Plus system (Bruker) at 800 MHz for 1H and 201 MHz for 13C in deuterium oxide (Sigma-Aldrich).
Analysis of HFs in tomato hairy roots culture medium
To generate transgenic hairy roots, tomato hypocotyls from 2-week-old seedlings were infected with Agrobacterium rhizogenes strain ATCC15834 harboring a binary vector pMgP237 as described previously (36). Briefly, the bottom of a hypocotyl segment (1.5 cm in length) was contacted with a bacterial colony, and the segments were stood on solidified B5 medium containing 2% (w/v) sucrose with the contacted end up. Hairy roots emerging from infected sites were excised and subcultured twice every week on solidified B5 medium containing 2% (w/v) sucrose with 300 mg liter−1 cefotaxime for disinfection. Then, hairy roots were transferred into 50 ml of liquid B5 medium containing 2% (w/v) sucrose and cultured at 25°C in an Erlenmeyer flask.
Tomato hairy roots were cultured for 2 weeks, and then the medium was collected. Three flasks of hairy root medium (150 ml) were passed through an open column filled with 30 ml of Sepabeads SP207 (Mitsubishi Chemical Corp.). The column was washed with 100 ml of distilled water (DW), and then the compounds were absorbed to Sepabeads SP207 (Mitsubishi Chemical Corp.) and were eluted with 100 ml of 100% methanol. Pass-through fractions and DW wash fractions were mixed and evaporated in vacuo. The methanol fraction was evaporated alone in vacuo. Each residue was redissolved in 160 μl of 100% methanol, and an aliquot (8 μl) was injected into the UPLC-MS/MS system. For bioassay, each evaporated residue was redissolved in DW. An aliquot of it was diluted with DW to a concentration equivalent to the original hairy root culture medium. The obtained samples were diluted accordingly and used hatching assay for PCN.
Conversion of SEB in soil
SEB was incubated in soil using a syringe. The bottom of the inside of a 10-ml sterile syringe was lined with cotton and filter paper and then filled with 5 ml of soil. The tip of the syringe was sealed with Parafilm, and SEB dissolved in 5 ml of sterile water was added to the soil in the syringe. After incubation for 1 week at 25°C, the Parafilm was removed from the tip end, and 30 ml of DW was poured into a syringe to collect the transit fluid. The purification of SEB and SEA from the transit solution was performed in a manner similar to the effective purification of SEA reported by Guerrieri et al. (17). Briefly, the supernatant obtained by centrifugation of the passages was applied to an Oasis MAX (3 cc/150 mg; Waters, Milford, MA, USA) column preconditioned with 5 ml of 100% methanol and activated with 5 ml of 5% NH4OH/H2O (v/v). The retained sample was washed with 5 ml of DW followed by a wash with 5 ml of 100% methanol. Then, 5 ml of 0.2 M formic acid in methanol was added to the cartridge, and the eluted fractions was evaporated in vacuo. The residue was dissolved in 80 μl of 100% methanol, and an aliquot (8 μl) was injected to UPLC-MS/MS. For the experiment in which the hairy root culture medium was incubated with soil, 50 ml of the hairy root medium culture was added to 75 ml of soil in a plastic box and incubated at 25°C for 1 week. Then, water extraction of the resulting soil was performed using a Büchner funnel, a suction bottle, and filter paper to obtain 200 ml of extract. The extract was passed through an open column filled with 20 ml of Sepabeads SP207 (Mitsubishi Chemical Corp.). The column was washed with 100 ml of DW, and then the compounds absorbed by Sepabeads SP207 (Mitsubishi Chemical Corp.) were eluted with 100 ml of 100% methanol. The methanol fraction was evaporated alone in vacuo. Each residue was redissolved in 160 μl of 100% methanol, and an aliquot (8 μl) was injected into the UPLC-MS/MS system and analyzed as described above.
Phytohormone treatment
Tomato hairy roots were cultivated in 50 ml of liquid B5 medium containing 2% (w/v) sucrose for 2 weeks at 25°C. Then, the tomato hairy roots were transferred to another fresh 50 ml of liquid B5 medium. Then, 50 μl of MeJA, MeSA, ABA, GA3 (all 100 mM), IAA, or tZR (both 20 mM) in methanol was added into the medium. For the mock treatment, 50 μl of methanol was added onto the medium. After incubation for 3 days at 25°C, the medium was collected and diluted with deionized water for use in the bioassay.
Inhibitor treatment
Tomato hairy roots were cultivated in 50 ml of liquid B5 medium containing 2% (w/v) sucrose for 2 weeks at 25°C. Then, the tomato hairy roots were transferred to another fresh 50 ml of liquid B5 medium supplemented with 50 μl of 1 or 100 mM inhibitor (uniconazole P or prohexadione) in acetone solution to yield a final concentration of 1 or 100 μM. The medium supplemented with 50 μl of acetone was prepared as the mock treatment. After 3 days of cultivation at 25°C, the medium was collected. An aliquot (10 ml) of the collected medium was passed through an open column packed with 1 ml of Sepabeads SP207 (Mitsubishi Chemical Corp.). The absorbed sample was washed with 3 ml of DW and then eluted with 3 ml of 100% methanol. The obtained 100% methanol fraction was evaporated in vacuo, and the residue yielded was dissolved in DW for bioassay.
HF extraction from tomato plants
The surface-sterilized seeds of tomato (Solanum lycopersicum cv Micro-Tom) were germinated and cultivated at 25°C under a 16-hour light/8-hour dark photoperiod on Jiffy-7 pellets (Sakata Seed) for 2 weeks. Then, the seedlings were transferred and grown hydroponically for 6 weeks in 500 ml of half-strength Hoagland nutrient solution, which was exchanged every 2 weeks. One hundred milligrams of tissue from each fresh plant material was frozen and homogenized using a mixer mill at 4°C. The homogenates were extracted three times with 1 ml of methanol. After centrifugation, the supernatant was collected, dried in vacuo, and redissolved in 1 ml of water. In the case of root exudate, aliquots of the hydroponic culture solution were freeze-dried, and the hatching assay samples were prepared with the amount of hydroponic culture solution corresponding to 100 mg fresh weight of roots in 1 ml of DW.
Selection of candidate biosynthetic genes
Amino acid sequences and the tissue-specific RNA sequencing data for tomato, potato, soybean, common bean, and Arabidopsis were obtained from Sol Genomics Network (https://solgenomics.net/), Spud DB potato genomics resource (http://solanaceae.plantbiology.msu.edu/), SoyBase (https://soybase.org/), Legume Information System (https://legumeinfo.org/), and The Arabidopsis Information Resource (https://arabidopsis.org/). To identify DOX genes, HMMER searching was performed as described previously. The obtained DOX genes were classified according to the subfamily of the top hit sequence by using the basic local alignment search tool for proteins (BLASTP) program to align the amino acid sequences of DOX that were previously annotated by Kawai et al. (25). Tomato genes encoding CYP were obtained according to the ITAG4.0 gene annotation. Amino acid sequence alignments were performed using MUSCLE, and the maximum likelihood tree was inferred in MEGA X by using 1000 bootstrap replications.
qRT-PCR analysis
Total RNA from tomato hairy roots treated with each phytohormone was extracted using the RNeasy Plant Mini Kit (Qiagen) and the RNase-Free DNase Set (Qiagen). The extracted total RNA was used for the synthesis of first-strand cDNAs by the ReverTra Ace qPCR RT Master Mix with genomic DNA remover. qRT-PCR was performed with a LightCyclerNano (Roche) using GeneAce SYBR qPCR Mix α No ROX (NIPPON GENE) with the primers shown in table S8. Cycling was performed at 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 15 s for amplification, followed by holding at 95°C for 30 s, and then ramping up from 60° to 95°C at 0.1°C s−1 to perform a melting curve analysis. Three biological replicates were analyzed in duplicate. The values obtained for the Ubi gene were used as internal references in tomato, and the gene expression levels were normalized to these values. Data acquisition and analysis were performed using LightCyclerNano software (Roche).
Generation of gene-disrupted tomato hairy roots
Gene-disrupted tomato hairy roots were generated by targeted genome editing with the CRISPR/Cas9 system. We used the CRISPR/Cas9 binary vector pMgP237-2A-GFP to express multiplex guide RNAs (gRNAs) (37, 38). To design a gRNA target with low off-target effects, in silico analyses were conducted using the web tool Design sgRNAs for CRISPRko (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) and CasOT software (39). Three target sequences were selected for each gene (figs. S16 and S20). To enhance the efficiency of gRNA transcription from U6 promoter, one G was added to the 5′ end of SOLA1_guide1, SOLA1_guide2, SOLA1_guide3, SOLA2_guide3, SOLA3_guide1, SOLA3_guide2, SOLA3_guide3, SOLA4_guide1, SOLA4_guide2, SOLA4_guide3, SOLA5_guide1, CYP716A44_guide2, and CYP716A44_guide3. Two DNA fragments composed of the gRNA scaffold and transfer RNA (tRNA) scaffold between full sequence of guide 1 and partial sequence from 5′- end of guide 2, and the remaining partial sequence of guide 2 and the full sequence of guide 3 were generated by PCR using pMD-gtRNA containing gRNA and tRNA scaffolds as a template and primer sets containing restriction enzyme Bsa I sites (in the case of SOLA1 as a target gene, Primer-19 and Primer-20 or Primer-21 and Primer-22 were used to generate former or latter units, respectively) (table S8). Each unit containing gRNA-tRNA was coinserted into the Bsa I site of pMgP237-2A-GFP using the Golden Gate Cloning method to construct the CRISPR/Cas9 vectors. The CRISPR/Cas9 vectors of each target gene were independently introduced into Rhizobium rhizogenes ATCC15834 by electroporation. Tomato transgenic hairy roots and stable plants were generated as described above. Genomic DNA was extracted from each established hairy root using the DNeasy Plant Mini Kit (Qiagen). To analyze the mutations in the transgenic hairy roots, the region including the target sites of gRNAs was amplified by PCR with the primers listed in table S8. Each PCR fragment was cleaned using the Wizard SV Gel and PCR Clean-Up System (Promega) and cloned into pCR4Blunt-TOPO (Invitrogen). Sanger sequencing of each of the cloned DNAs was provided by a sequencing service (Eurofins Genomics).
HF analysis of candidate gene-disrupted hairy roots
Tomato hairy roots were cultivated in 50 ml of liquid B5 medium at 25°C in Erlenmeyer flasks. After cultivation for 2 weeks, the medium was collected. An aliquot (5 ml) of the collected medium was passed through an open column packed with 500 μl of Sepabeads SP207 (Mitsubishi Chemical Corp.). The absorbed sample was washed with 1.5 ml of DW and then eluted with 1.5 ml of 100% methanol. The 100% methanol fraction was evaporated in vacuo, and the residue yielded was dissolved in DW for bioassay. Samples for the bioassay were prepared from three independently cultured hairy roots. In the case of UPLC-MS/MS analysis of HFs, the medium was collected from three flasks. The medium (150 ml) was passed through an open column packed with 20 ml of Sepabeads SP207 (Mitsubishi Chemical Corp.). The column was washed with 100 ml of DW and then eluted with 100 ml of 100% methanol. The 100% methanol fraction was evaporated in vacuo, and the residue yielded was applied to UPLC-MS/MS analysis, which was conducted as described above with minor modification in a mobile phase gradient. Briefly, the mobile phase consisted of solvent A (0.1% formic acid in water) and solvent B (acetonitrile), and the following gradient conditions were used: 10% B at 0 min, ramped to 30% B at 20 min, 100% B at 20 min, and held to 25 min, then returned to 10% B, and held for 30 min.
Acknowledgments
We are greatly indebted to A. Maeno and A. Fujihashi (Kyoto University) for performing instrumental analyses. Parts of the experimental measurements were carried out using a Bruker AVANCEIII 800 NMR spectrometer and a Bruker timsTOF spectrometer in the Joint Usage/Research Center (JURC) at the Institute for Chemical Research, Kyoto University. We thank the HOKKAIDO-Chuo Station, Center for Seeds and Seedlings, NARO for provision of potato hydroponic solution.
Funding: This work was supported by JSPS Grant-in-Aid for Japan Society for the Promotion of Science Research Fellow 18 K05459 (to K.S.), Grants from the Project of the Bio-oriented Technology Research Advancement Institution, NARO (the special scheme project on advanced research and development for next-generation technology) (to A.K., I.S., K.T., and M.M.), and Grant-in-aid for Scientific Research (B) 21H02132 (to MM) and JST ACT-X JPMJAX21B1 (R.A.).
Author contributions: Conceptualization: K.S., R.A., A.K., and M.M. Methodology: K.S., R.A., Y.M., A.K., and M.M. Investigation: K.S., R.A., Y.O., C.O., Y.M., B.W., I.S., and A.K. Visualization: K.S. and R.A. Funding acquisition: K.S., R.A., I.S., A.K., K.T., and M.M. Project administration: A.K. and M.M. Supervision: Y.S., A.K., K.T., and M.M. Writing—original draft: K.S., R.A., and M.M. Writing—review and editing: K.S., R.A., A.K., and M.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S27
Tables S1 to S8
REFERENCES AND NOTES
- 1.J. T. Jones, A. Haegeman, E. G. J. Danchin, H. S. Gaur, J. Helder, M. G. K. Jones, T. Kikuchi, R. Manzanilla-López, J. E. Palomares-Rius, W. M. L. Wesemael, R. N. Perry, Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. M. Nicol, S. J. Turner, D. L. Coyne, L. den Nijs, S. Hockland, Z. T. Maafi, Current [Nematode Threats to World Agriculture], in Genomics and Molecular Genetics of Plant-Nematode Interactions, J. Jones, G. Gheysen, C. Fenoll, Eds. (Springer, 2011), pp. 21–43; 10.1007/978-94-007-0434-3_2. [DOI]
- 3.V. M. Williamson, C. A. Gleason, Plant–nematode interactions. Curr. Opin. Plant Biol. 6, 327–333 (2003). [DOI] [PubMed] [Google Scholar]
- 4.C. J. Lilley, H. J. Atkinson, P. E. Urwin, Molecular aspects of cyst nematodes. Mol. Plant Pathol. 6, 577–588 (2005). [DOI] [PubMed] [Google Scholar]
- 5.M. J. Sullivan, R. Inserra, J. Franco, I. Moreno-Leheude, N. Greco, Potato cyst nematodes: Plant host status and their regulatory impact. Nematropica 37, 193–202 (2007). [Google Scholar]
- 6.S. J. Turner, J. A. Rowe, [Cyst nematodes], in Plant Nematology, R. N. Perry, M. Moens, Eds. (Wallingford, Oxfordshire: CAB International, 2006), pp. 90–122. [Google Scholar]
- 7.E. P. Masler, R. N. Perry, [Hatch, survival and sensory perception], in Cyst Nematodes, R. N. Perry, M. Moens, J. T. Jones, Eds. (Wallingford, Oxfordshire: CAB International, 2018), pp. 44–73. [Google Scholar]
- 8.K. Devine, Comparison of the effects of freezing and thawing on the cysts of the two potato cyst nematode species, Globodera rostochiensis and G. pallida, using differential scanning calorimetry. Nematology 12, 81–88 (2010). [Google Scholar]
- 9.D. J. Wright, R. N. Perry, [Reproduction, physiology and biochemistry], in Plant Nematology, R. N. Perry, M. Moens, Eds. (Wallingford, Oxfordshire: CAB International, 2006), pp. 187–209. [Google Scholar]
- 10.K. J. Devine, P. W. Jones, Response of Globodera rostochiensis to exogenously applied hatching factors in soil. Ann. Appl. Biol. 137, 21–29 (2000). [Google Scholar]
- 11.T. Masamune, M. Anetai, M. Takasugi, N. Katsui, Isolation of a natural hatching stimulus, glycinoeclepin A, for the soybean cyst nematode. Nature 297, 495–496 (1982). [Google Scholar]
- 12.A. Fukuzawa, H. Matsue, M. Ikura, T. Masamune, Glycinoeclepins B and C, nortriterpenes related to glycinoeclepin A. Tetrahedron Lett. 26, 5539–5542 (1985). [Google Scholar]
- 13.T. Masamune, A. Fukuzawa, A. Furusaki, M. Ikura, H. Matsue, T. Kaneko, A. Abiko, N. Sakamoto, N. Tanimoto, A. Murai, Glycinoeclepins, natural hatching stimuli for the soybean cyst nematode, Heterodera glycines. II. Structural elucidation. Bull. Chem. Soc. Jpn. 60, 1001–1014 (1987). [Google Scholar]
- 14.J. G. Mulder, P. Diepenhorst, P. Plieger, I. E. M. Brüggemann-Rotgans, U.S. Patent application 5585505A (1996)
- 15.H. Schenk, R. A. J. Driessen, R. de Gelder, K. Goubitz, H. Nieboer, I. E. M. Brüggemann-Rotgans, P. Diepenhorst, Elucidation of the structure of solanoeclepin A, a natural hatching factor of potato and tomato cyst nematodes, by single-crystal X-ray diffraction. Croat. Chem. Acta 72, 593–606 (1999). [Google Scholar]
- 16.K. Tanino, M. Takahashi, Y. Tomata, H. Tokura, T. Uehara, T. Narabu, M. Miyashita, Total synthesis of solanoeclepin A. Nat. Chem. 3, 484–488 (2011). [DOI] [PubMed] [Google Scholar]
- 17.A. Guerrieri, K. Floková, L. E. Vlaar, M. L. Schilder, G. Kramer, A. Chojnacka, Y. R. van Dijk, H. J. Bouwmeester, L. Dong, UPLC-MS/MS analysis and biological activity of the potato cyst nematode hatching stimulant, solanoeclepin A, in the root exudate of Solanum spp. Planta 254, 112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L. E. Vlaar, B. Thiombiano, D. Abedini, M. Schilder, Y. Yang, L. Dong, A combination of metabolomics and machine learning results in the identification of a new cyst nematode hatching factor. Metabolites 12, 551 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.K. Shimizu, A. Kushida, R. Akiyama, H. J. Lee, Y. Okamura, Y. Masuda, I. Sakata, K. Tanino, S. Matsukida, T. Inoue, Y. Sugimoto, M. Mizutani, Hatching stimulation activity of steroidal glycoalkaloids toward the potato cyst nematode, Globodera rostochiensis. Plant Biotechnol. (Tokyo) 37, 319–325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.I. Sakata, A. Kushida, K. Tanino, The hatching-stimulation activity of solanoeclepin A toward the eggs of Globodera (Tylenchida: Heteroderidae) species. Appl. Entomol. Zool. 56, 51–57 (2021). [Google Scholar]
- 21.M. Nakayasu, R. Akiyama, M. Kobayashi, H. J. Lee, T. Kawasaki, B. Watanabe, S. Urakawa, J. Kato, Y. Sugimoto, Y. Iijima, K. Saito, T. Muranaka, N. Umemoto, M. Mizutani, Identification of α-tomatine 23-hydroxylase involved in the detoxification of a bitter glycoalkaloid. Plant Cell Physiol. 61, 21–28 (2020). [DOI] [PubMed] [Google Scholar]
- 22.R. Akiyama, M. Nakayasu, N. Umemoto, J. Kato, M. Kobayashi, H. J. Lee, Y. Sugimoto, Y. Iijima, K. Saito, T. Muranaka, M. Mizutani, Tomato E8 encodes a C-27 hydroxylase in metabolic detoxification of α-tomatine during fruit ripening. Plant Cell Physiol. 62, 775–783 (2021). [DOI] [PubMed] [Google Scholar]
- 23.R. Akiyama, B. Watanabe, M. Nakayasu, H. J. Lee, J. Kato, N. Umemoto, T. Muranaka, K. Saito, Y. Sugimoto, M. Mizutani, The biosynthetic pathway of potato solanidanes diverged from that of spirosolanes due to evolution of a dioxygenase. Nat. Commun. 12, 1300 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R. Akiyama, B. Watanabe, J. Kato, M. Nakayasu, H. J. Lee, N. Umemoto, T. Muranaka, K. Saito, Y. Sugimoto, M. Mizutani, Tandem gene duplication of dioxygenases drives the structural diversity of steroidal glycoalkaloids in the tomato clade. Plant Cell Physiol. 63, 981–990 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Y. Kawai, E. Ono, M. Mizutani, Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 78, 328–343 (2014). [DOI] [PubMed] [Google Scholar]
- 26.E. M. Turk, S. Fujioka, H. Seto, Y. Shimada, S. Takatsuto, S. Yoshida, H. Wang, Q. I. Torres, J. M. Ward, G. Murthy, J. Zhang, J. C. Walker, M. M. Neff, BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J. 42, 23–34 (2005). [DOI] [PubMed] [Google Scholar]
- 27.T. Ohnishi, T. Nomura, B. Watanabe, D. Ohta, T. Yokota, H. Miyagawa, K. Sakata, M. Mizutani, Tomato cytochrome P450 CYP734A7 functions in brassinosteroid catabolism. Phytochemistry 67, 1895–1906 (2006). [DOI] [PubMed] [Google Scholar]
- 28.S. Yasumoto, H. Seki, Y. Shimizu, E. O. Fukushima, T. Muranaka, Functional characterization of CYP716 family P450 enzymes in triterpenoid biosynthesis in tomato. Front. Plant Sci. 8, 21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C. T. Calam, H. Raistrick, A. R. Todd, The potato eelworm hatching factor. 1. The preparation of concentrates of the hatching factor and a method of bioassay. Biochem. J. 45, 513–519 (1949). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L. M. Massey, A. L. Neal, Investigations concerning the hatching factor of the golden nematode of potatoes, Heterodera rostochiensis Wollenweber. J. Wash. Acad. Sci. 43, 396–401 (1953). [Google Scholar]
- 31.G. J. Janzen, F. Van der Tuin, The unknown hatching agent for the potato root-eelworm. Nematologica 1, 126–137 (1956). [Google Scholar]
- 32.K. J. Devine, P. J. Jones, Purification and partial characterisation of hatching factors for the potato cyst nematode Globodera rostochiensis from potato root leachate. Nematology 2, 231–236 (2000). [Google Scholar]
- 33.T. Okada, Hatching inhibitory factor in the cyst contents of the soybean cyst nematode, Heterodera glycines Ichinohe (Tylenchida: Heteroderidae). Appl. Entomol. Zool 7, 99–102 (1972). [Google Scholar]
- 34.E. P. Masler, D. J. Chitwood, Evaluation of proteases and protease inhibitors in Heterodera glycines cysts obtained from laboratory and field populations. Nematology 19, 109–120 (2017). [Google Scholar]
- 35.J. Ochola, D. Coyne, L. Cortada, S. Haukeland, M. Ng'ang'a, A. Hassanali, C. Opperman, B. Torto, Cyst nematode bio-communication with plants: Implications for novel management approaches. Pest Manag. Sci. 77, 1150–1159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.C. Thagun, S. Imanishi, T. Kudo, R. Nakabayashi, K. Ohyama, T. Mori, K. Kawamoto, Y. Nakamura, M. Katayama, S. Nonaka, C. Matsukura, K. Yano, H. Ezura, K. Saito, T. Hashimoto, T. Shoji, Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant Cell Physiol. 57, 961–975 (2016). [DOI] [PubMed] [Google Scholar]
- 37.M. Nakayasu, R. Akiyama, H. J. Lee, K. Osakabe, Y. Osakabe, B. Watanabe, Y. Sugimoto, N. Umemoto, K. Saito, T. Muranaka, M. Mizutani, Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. Biochem. 131, 70–77 (2018). [DOI] [PubMed] [Google Scholar]
- 38.R. Hashimoto, R. Ueta, C. Abe, Y. Osakabe, K. Osakabe, Efficient multiplex genome editing induces precise, and self-ligated type mutations in tomato plants. Front. Plant Sci. 9, 916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. Xiao, Z. Cheng, L. Kong, Z. Zhu, S. Lin, G. Gao, B. Zhang, CasOT: A genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30, 1180–1182 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S27
Tables S1 to S8