Abstract
Macrophages are innate immune cells that form a 3D network in all our tissues, where they phagocytose dying cells and cell debris, immune complexes, bacteria and other waste products. Simultaneously, they produce growth factors and signalling molecules — such activities not only promote host protection in response to invading microorganisms but are also crucial for organ development and homeostasis. There is mounting evidence of macrophages orchestrating fundamental physiological processes, such as blood vessel formation, adipogenesis, metabolism and central and peripheral neuronal function. In parallel, novel methodologies have led to the characterization of tissue-specific macrophages, with distinct subpopulations of these cells showing different developmental trajectories, transcriptional programmes and life cycles. Here, we summarize our growing knowledge of macrophage diversity and how macrophage subsets orchestrate tissue development and function. We further interrelate macrophage ontogeny with their core functions across tissues, that is, the signalling events within the macrophage niche that may control organ functionality during development, homeostasis and ageing. Finally, we highlight the open questions that will need to be addressed by future studies to better understand the tissue-specific functions of distinct macrophage subsets.
Subject terms: Monocytes and macrophages, Myelopoiesis
Macrophages are important for host immunity to infections and for clearing waste products from tissues, but they also maintain tissue health by regulating metabolism, neuronal functions and many other biological processes. Here, Elvira Mass and co-workers discuss the different tissue-specific macrophage populations that are found throughout the body, highlighting shared and unique aspects of their developmental trajectories, transcriptional programmes and physiological functions.
Introduction
In multicellular organisms, cells constantly send and receive signals to coordinate body functions. At the level of tissues or organs, cells can orchestrate tissue function locally by signalling through receptors, producing soluble mediators or by regulating the extracellular matrix (ECM) composition. Certain cell types are essential for tissue structure and integrity, such as epithelial cells that create barriers and surfaces, endothelial cells that line the blood vasculature and fibroblasts that synthesize ECM and collagen. In addition to these structural cell types, macrophages are integral to every organ1,2. For decades, it was thought that macrophages are a relatively homogeneous population of mononuclear phagocytes arising from bone-marrow haematopoietic stem cells (HSCs), with their primary function being to protect organs during infections1. However, the development of novel fate-mapping mouse models allowed for longitudinal tracking of macrophages from their progenitors into their mature cellular state within their organ of residence. Combined with systems biology approaches, these methodologies have changed our view of macrophage biology entirely in the past decade.
Macrophages arise early during embryogenesis and colonize developing organs, forming a 3D network within every tissue. These tissue-resident macrophages have a high self-renewal capacity and generally do not require input from HSCs1. During postnatal tissue maturation, however, HSC-derived monocytes contribute to distinct macrophage populations in many tissues, with some tissue-specific macrophages (Box 1) being dependent on constant monocyte replenishment during adulthood3. This creates a complex scenario in which fetal-derived and HSC-derived macrophages coexist in certain tissues and contribute to tissue function both during steady state and in infection and inflammation. Despite their omnipresence in all organs during development and steady-state conditions, the cellular and molecular responses of macrophages have been mainly studied on a tissue population basis and in the context of inflammation. However, a systematic analysis of distinct macrophage subpopulations within one tissue, their roles in organogenesis and homeostasis and macrophage core functions beyond their inflammatory or infection-related responses is largely lacking.
In this Review, we discuss examples of how macrophages develop in barrier and internal organs and describe their roles in regulating homeostatic functions within these organs. We then examine recent advances in our understanding of macrophage life cycles and heterogeneity within distinct anatomical niches and discuss the conserved core functions of macrophages across tissues, which are vital in maintaining organ-specific function and immunity. Finally, we depict methodologies and models that have been instrumental in improving our knowledge of macrophage biology and emphasize strategies that should be used to study macrophage functions in the future. In summary, we lay out how progress in discovering and characterizing macrophage ontogeny and identity, as well as in characterizing macrophage core functions beyond their inflammatory responses, will have vast implications for our understanding of tissue development, function and integrity.
Box 1 Macrophage diversity across organs and tissues.
Although tissue-resident macrophages can be identified by a combination of a few pan-macrophage markers in every tissue (for example, CD11b, F4/80, MERTK and CD64), each subtissular niche in different organs imprints an additional transcriptional programme2,16,91,124,125,174; this enables the stratification and analysis of macrophage subpopulations via flow cytometry profiling. Previous reviews and papers have documented different markers for these tissue-specific macrophages; thus we will not describe these in detail in this Review, but refer to other extensive reviews focusing on macrophage diversity and expression profiles55,78,88,140,175–179.
Ontogeny and tissue-specific function
Macrophages develop during embryogenesis in both vertebrates and invertebrates (Box 2). In mice, starting at embryonic developmental day 8.5 (E8.5), yolk sac erythro-myeloid progenitors (EMPs) give rise to pre-macrophages (pMacs) that colonize embryonic tissues from E9.0 onwards and differentiate into tissue-specific macrophages during organogenesis2,4. EMPs also give rise to monocytes, which additionally contribute to the pool of tissue-specific macrophages3–5. Most tissue-resident macrophages proliferate locally and are long-lived during steady-state adulthood6–8. Additionally, starting at early postnatal stages, every organ harbours monocyte-derived macrophages (MDMs) that can have different life cycles and either become long-lived or are shorter-lived and constantly replenished from bone-marrow HSCs6,9 (Fig. 1). Although fetal-derived tissue-resident macrophages and MDMs represent two independent haematopoietic lineages, most studies before the 2010s do not distinguish between these two independent macrophage lineages. Thus, the distinct origins and functions of fetal-derived macrophages and MDMs within the same tissue during steady-state conditions, after a challenge (such as an environmental trigger, sterile immune activation or an infection), and during or after tissue regeneration remain largely unknown.
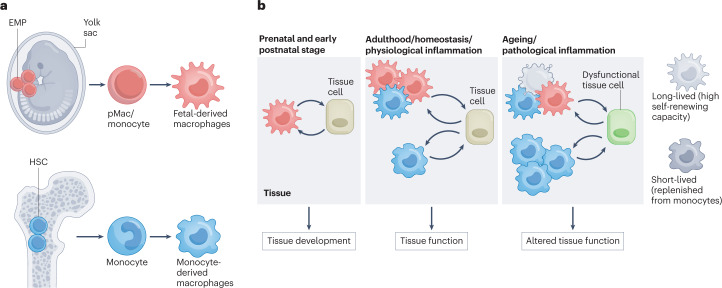
Fig. 1. Contribution of distinct macrophage subsets to tissue function.
a, Distinct developmental arms of tissue macrophages. Yolk sac erythro-myeloid progenitors (EMPs) give rise to pre-macrophages (pMac) and monocytes that can differentiate into long-lived tissue-resident macrophages. By contrast, haematopoietic stem cells (HSCs) can give rise to short-lived and long-lived macrophages during early postnatal stage and during adulthood. b, EMP-derived and HSC-derived macrophages contribute to the function of tissue via cell–cell crosstalk with specialized tissue cells. Tissue development relies entirely on EMP-derived macrophages. During homeostasis, distinct macrophage subpopulations crosstalk with tissue cells to support tissue function. Upon ageing or pathological inflammation, the fine-tuned balance of macrophage distribution in the tissue is disturbed triggering, for example, apoptosis of long-lived macrophages, or increased recruitment of short-lived HSC-derived macrophages, thereby leading to tissue dysfunction.
In the following sections, we highlight studies characterizing macrophage ontogeny in different tissues during homeostasis, which has been the focus of many laboratories worldwide in recent years, using the ever-growing toolbox to track macrophages. We also emphasize the well-known and newly discovered functions of macrophages in tissue development and function. Studies using bone-marrow chimaeras in combination with (sub)lethal irradiation or depletion of tissue-resident macrophages via pharmaceutical or genetic ablation have been very informative in addressing macrophage development and function during adulthood. Nevertheless, these studies may not always represent a homeostatic state owing to the induction of extensive apoptosis leading to local as well as systemic inflammation (discussed elsewhere10). However, with the advent of fate-mapping mouse models, such as Runx1CreERT (ref. 11), Cx3cr1Cre and Cx3cr1CreERT (ref. 6), Csf1rMeriCreMer (ref. 8), Flt3Cre (ref. 4), Tnfrsf11aCre (ref. 2), Ms4a3Cre (ref. 12), Cxcr4CreERT (ref. 13), KitMerCreMer (ref. 14) and Ccr2CreER (ref. 3), enabling the tracking of macrophages and their progenitors from the different lineages, macrophage fate can now be followed longitudinally starting from their initial development until their final differentiation under homeostatic conditions. This, in turn, allows us to study the similarities and dissimilarities in the behaviour of macrophages of distinct developmental origins within a defined tissue context and, ultimately, define their contribution to tissue development, homeostasis and integrity.
Box 2 Macrophages across species.
Phagocytes are an old evolutionary concept, found in most metazoan animals and involved in immune defence and tissue homeostasis, as initially recognized by the pioneering work of Elie Metchnikoff in starfish larvae180. The fruitfly, Drosophila melanogaster, is increasingly recognized as an invertebrate model for immunological research181,182 with an immune system recapitulating basic concepts of innate immunity183,184. Drosophila ‘blood cells’ (haemocytes) consist of lamellocytes (encapsulation), crystal cells (melanization) and the macrophage-like plasmatocytes, which make up to 95% of haemocytes and have similar functions to vertebrate macrophages185. A plethora of disease models, in combination with a magnificent genetic toolbox, makes Drosophila an ideal surrogate to study macrophages. Haemocytes follow a stratified development from different haematopoietic sources, the embryonic head mesoderm and the larval lymph gland, instructed by defined master transcription factors186. Although differentiation of plasmatocytes from pro-haemocytes in the lymph gland resembles the vertebrate development of myeloid cells from haematopoietic stem cells, the development of plasmatocytes from embryonic head mesoderm patterns the differentiation of vertebrate tissue-resident macrophages from erythro-myeloid progenitors186. In larvae, ‘circulating’ and tissue-resident plasmatocytes have been described187,188. Resident plasmatocytes share similar functions to vertebrate macrophages in defined tissue niches, such as removal of apoptotic cells from the developing embryonic nervous system or maintaining homeostasis of surrounding niche cells such as sensory neurons in larval body wall pockets189. Recently, high-throughput profiling of haemocytes allowed assessment of the functional diversity and specification of plasmatocytes190–193. The next step is the characterization of plasmatocyte subpopulations that inhabit distinct subtissular niches, similar to tissue-resident macrophages in vertebrates. Furthermore, homeostatic and disease-specific functions of plasmatocytes are recapitulated in vertebrates, including their role in high-fat diet-induced inflammation or fat body development194,195. Thus, Drosophila offers a unique surrogate system with an unlimited genetic toolbox to the macrophage field, which could empower many studies analysing conserved macrophage functions across species. In addition, humanized mouse models, in which human fetal-derived or cord blood-derived haematopoietic stem cells or precursors of myeloid cells are injected into immunodeficient mouse strains, have become important tools to study human macrophage development and function196–200. Indeed, these studies allow demonstrating how human monocyte-derived macrophages repopulate mouse tissues and to what extent they can develop into or mimic their fetal-derived tissue-resident counterparts199,200.
Macrophages in internal organs and tissues
Macrophages are an integral part of all organs and tissues (Box 3), and new studies on this topic are appearing on a daily basis. Thus, we are not able to cover all organ systems, but instead will focus on specific examples to reflect the important activities of macrophages for organ and tissue development and functionality.
Box 3 Do we need macrophages?
Macrophages seem to be an obligatory cell type of every mammalian organ. If adult macrophages are transiently depleted, the ‘empty’ niche will ‘call’ for monocytes that quickly differentiate into macrophages and assume tissue-specific transcriptional programmes, thereby resembling resident macrophages to a high degree59,61,161–163. One exception is the brain, which is protected through the blood–brain barrier. After colony-stimulating factor 1 receptor (CSF1R) inhibitor treatment in the steady-state brain, clonal expansion of a few remaining microglia (~1%) leads to repopulation of the whole brain microglial cell compartment within a few days20. Using different rodent models, such as Csf1r−/− (refs. 201–203), Csf1FIRE/FIRE (ref. 204), Csf1op/op (ref. 203) and Il34lacZ/lacZ (ref. 22), showed that macrophage survival depends mainly on CSF1R, which has two ligands: CSF1 and IL-34. Removal of only one ligand leads to selective lack or reduction of certain macrophage populations, whereas depletion of the receptor results in a complete lack of macrophages in most tissues (reviewed elsewhere205). Although different rodent species (mouse versus rat) and inbred genetic backgrounds lead to some differences in phenotypes, for example, lack of microglia causes brain malformations only in C57BL/6 mice, the bone and visceral adipose tissue seem to be dependent on macrophage function during embryogenesis51,195,201,205. Definitive confirmation that mammals depend on proper macrophage development and function comes from humans with homozygous CSF1R loss-of-function alleles — these individuals show developmental brain defects owing to lack of microglia as well as severe osteopetrosis and skeletal dysplasia206–208. Furthermore, heterozygous CSF1R point mutations are associated with autosomal dominant adult-onset leukoencephalopathy with axonal spheroids and pigmented glia209,210, a progressive degenerative white matter disorder, highlighting the importance of microglia for brain development and homeostasis. Finally, CSF1R blockade in humans, applied to deplete tumour-associated macrophages, resulted in facial oedema and elevated hyaluronan in peripheral blood211, underlining the need for undisturbed macrophage function in tissue and organismal homeostasis. Other examples underlining the contribution of tissue-resident macrophages to human tissue homeostasis are the loss of epidermal Langerhans cells in patients suffering from acrodermatitis enteropathica caused by nutritional zinc deficiency212, or the loss of alveolar macrophages in patients suffering from primary pulmonary alveolar proteinosis owing to defective granulocyte–macrophage colony-stimulating factor signalling213. In mouse models, the absence of tissue-resident synovial lining macrophages resulted in an exacerbated immunopathology in rheumatoid arthritis157. In summary, organ maturation and tissue function most likely require macrophages. However, we are just at the beginning of deciphering macrophage core functions that continuously contribute to the homeostasis of their neighbouring tissue cells, thereby maintaining tissue function and integrity.
Central nervous system macrophages
The best-studied tissue with regards to macrophage ontogeny and function is probably the central nervous system (CNS). In the CNS, different populations of tissue-resident macrophages are found in defined anatomical niches: microglia in the CNS parenchyma and other macrophage populations occupy the CNS interfaces (we refer to these as CNS-associated macrophages, CAMs) including ventricles, meninges and perivascular space. All fate-mapping models used so far in mice indicate that microglia undergo a stepwise differentiation process from EMPs to pMacs, which then colonize the CNS2,15,16. Recent studies showed that pMacs can be further separated into two subsets by the gene and surface expression of the mannose receptor C type 1 (MRC1)17,18, and this molecule most likely represents a maturation marker of macrophage progenitors. Studies using the fate-mapping model Mrc1CreERT showed that MRC1+ pMacs make a high contribution to microglia in the developing CNS17. Overall, microglia develop without any contribution from HSCs during the steady state, meaning that the whole adult population of microglia is derived from fetal progenitors8,11. Once settled in the CNS, the microglial population maintains itself via cell-autonomous proliferation7,19. Adult microglia are long-lived cells that only show rare proliferation throughout their lifespan to maintain the steady-state network20,21. Once they have become established in their tissue niche, microglia rely on defined niche factors released by local tissue cells, enabling their maturation to serve specified functions during CNS development and homeostasis. The two colony-stimulating factor 1 receptor (CSF1R) ligands, colony-stimulating factor 1 (CSF1) and IL-34, released by neurons and astrocytes, respectively22–24, are essential to maintain mature microglia in the adult CNS niche in a regionally defined manner. Interestingly, embryonic microglia seem to be solely dependent on CSF1. The regional heterogeneity of the expressed CSF1R ligands is only established during postnatal development in which grey matter microglia become dependent on IL-34 and white matter microglia, such as cerebellar microglia, rely on CSF1 expression23. Dictated by the surrounding niche cells, microglia are instructed to perform specific tasks during development and steady state. In the homeostatic CNS, microglia support their defined tissue niche by constantly surveilling their tissue environment25,26 and are therefore performing essential tasks in neuronal network maintenance27 and supporting surrounding neuroglia28 and vessel integrity29. During development, microglia are involved in essential tissue niche functions in the CNS that are dependent on their sub-anatomical location and include guidance of vessel growth30, synaptic pruning31,32 and oligodendrogenesis33,34. Many studies using microglia depletion, either by using pharmaceutical inhibitors or transgenic approaches, have further highlighted context-specific functions of microglia within their tissue niche (reviewed elsewhere35).
Similar to microglia, most CAMs are also derived from EMPs18,36,37. Leptomeningeal and perivascular macrophages seem to be solely derived from embryonic sources with no contribution of HSC-derived progenitors and no exchange by circulating monocytes during adulthood. A recent study further delineates that microglia, leptomeningeal and perivascular macrophages originate from the same EMP-derived MRC1+ macrophage progenitor arising in the yolk sac around E9.0/9.5 (ref. 17). Interestingly, the ventricular macrophage compartment is more heterogenous in its ontogeny. Stromal choroid plexus macrophages are initially derived from embryonic progenitors but postnatally replaced by circulating monocytes36,37. Intraventricular macrophages, including the Kolmer epiplexus cells, remain of embryonic origin throughout life36. Even though leptomeningeal macrophages are maintained by local proliferation throughout adulthood, dural macrophages, at least partially, are replenished by monocytes36. Interestingly, it was recently shown that the skull and vertebrae bone marrow can serve as a direct reservoir not only for MDM progenitors during neuroinflammation, but also for dural macrophages during steady state. These MDM progenitors can directly migrate from the adjacent bone-marrow cavities via dural channels to the inflamed CNS parenchyma and leptomeninges without trafficking through the blood circulation38,39. Previous studies suggested that all CAMs embed within their specific tissue niches during embryonic development18,37, but recent findings have challenged this view and demonstrated that this is not the case for perivascular macrophages. The anatomical niche of perivascular macrophages — that is, the Virchow-Robin/perivascular space — is only opened along the brain arteries in the first week of postnatal life in mice. Its opening is followed by the entry of leptomeningeal macrophages into the perivascular space, which give rise to perivascular macrophages along the brain vasculature, where the cells then further adapt to the anatomical tissue niche17. The roles of perivascular macrophages in this unique anatomical niche need to be further determined during development, homeostasis and in disease.
Bone-marrow macrophages
The bone-marrow harbours at least three macrophage populations whose functions are well understood40, although their ontogeny is less clear: erythroblastic island (EBI) macrophages, osteomacs and osteoclasts. EBI macrophages are essential in red blood cell formation, adhesion and in clearance of expelled nuclei41. Osteomacs modulate osteoblast function42,43 and, in synergy with megakaryocytes, support the HSC niche44. Osteoclasts are a highly specialized macrophage population within the bone marrow responsible for bone resorption, and their dysfunction leads to the closure of the bone-marrow cavity, a process called osteopetrosis45. Thus, osteoclast function is, at least indirectly, essential for the establishment and maintenance of the HSC niche46. Furthermore, osteoclasts have been suggested to directly enhance HSC mobilization47, retention48, proliferation and differentiation49. Osteoclasts are multinucleated giant cells that were considered to develop from fusion events of myeloid progenitors or monocytes50. However, recent fate-mapping studies in combination with parabiosis and monocyte transfer experiments indicate that during homeostasis, neonatal osteoclasts develop from EMPs, that they are long-lived and during adulthood represent a chimeric cell regarding their ontogeny as they integrate HSC-derived monocyte nuclei throughout life (acquiring approximately one nucleus every 8 weeks)51. The particular mechanisms at play in deciding which of the, on average, five nuclei remain within the osteoclasts or are expelled remain elusive. The ontogeny and functional heterogeneity of EBI macrophages and osteomacs have not been addressed so far and will require a thorough histological assessment in combination with fate-mapping models owing to the high cellularity of the tissue and tight cellular interactions, which may confound flow cytometry analyses52. In addition to these three well-defined macrophage subsets, it is clear that bone-marrow-resident macrophages play a crucial role in removing apoptotic immune cells, such as developing B cells, which undergo selection for functional B cell receptors in the bone marrow. Similarly, bone-marrow-resident macrophages mediate phagocytosis of opsonized target cells53,54. Identifying the developmental origin of these different macrophage subsets, as well as defining their spatial distribution and functional overlap with EBIs, will be of major interest to fully understand the complexity of bone-marrow-resident macrophage subsets.
Liver macrophages
The liver was recently shown to harbour macrophage populations other than the well-known Kupffer cells and liver capsular macrophages (LCMs), namely, central vein macrophages and lipid-associated macrophages55. Kupffer cells are EMP-derived4,12, and in an adult liver, they line the sinusoids, where they are exposed to a constant blood flow from the portal vein. Thus, they are primarily seen as immunological gatekeepers, clearing cell debris, mediating antimicrobial defence and promoting immune tolerance. Additionally, they phagocytose senescent erythrocytes and are involved in systemic iron and cholesterol metabolism56. Furthermore, Kupffer cells were shown to mediate the activity of cytotoxic antibodies targeting malignant cells in the blood, which are widely used in the therapy of cancer and autoimmune diseases57. LCMs represent a minor population, comprising less than 10% of all hepatic macrophages. In adult mice, they are mostly monocyte-derived, expand around the time of weaning and form a defensive line against peritoneal cavity pathogens58. Intriguingly, a more recent fate-mapping study using the Ms4a3Cre model12 indicated only a ~30% contribution of monocytes to LCMs, suggesting that their ontogeny may be more complex than anticipated. Moreover, the time point of appearance within their specific niche, the transcriptional factors defining their identity and their functions in organogenesis remain to be defined.
By contrast, the developmental trajectory and factors driving the specification of Kupffer cells are well understood2. Using the Kupffer cell-specific transgenic mouse line Clec4fCre developed by the Glass59 and Guilliams60 laboratories, the intrinsic and environmental factors driving Kupffer cell differentiation after depletion were elegantly studied59–61. Furthermore, changes in Kupffer cell identity during metabolic challenges — such as in response to high-fat, high-sugar and high-cholesterol diets — and the impact of these changes on liver metabolism were investigated62–64 (reviewed elsewhere55). However, to the best of our knowledge, how Kupffer cells contribute to liver metabolism during homeostatic conditions in vivo has been addressed in only one study, which showed that gluconeogenesis and lipogenesis are controlled via Kupffer cell-derived insulin-like growth factor-binding protein 7 (ref. 65). Considering that Kupffer cells migrate out of the parenchyma towards the sinusoids only around weaning age and that the adult liver architecture is not fully established before 8 weeks of age66, their role in liver development during gestation and liver maturation and zonation at neonatal and early postnatal stages remains to be investigated.
Although the ontogeny of adult Kupffer cells has been established as EMP-derived, their functional division into two subpopulations (referred to as ‘KC1’ and ‘KC2’) has recently become a matter of debate67–70. Owing to their unique anatomical position in the sinusoids, where they attach to endothelial cells, common single-cell isolation procedures may not allow for unequivocal discrimination of Kupffer cell subpopulations from Kupffer cells that carry some membrane cargo from endothelial cells or the other way around68. Of note, the tight interaction of macrophages with neighbouring cells is a common contamination problem when freshly isolated cells are studied via bulk or single-cell RNA sequencing (RNA-seq)52,71. Thus, RiboTag71 or single-nucleus RNA-seq approaches70,72 in combination with image-based analyses such as imaging flow cytometry52,69 and 3D microscopy (for example, focused ion beam scanning electron microscopy73) should become a standard methodology to discriminate true macrophage subpopulations from contamination or dynamic transcriptional macrophage states defined by cargo phagocytosed from their environment. The developmental trajectories, cellular dynamics and functions of central vein macrophages and lipid-associated macrophages70, particularly during liver development and liver homeostasis, remain to be investigated.
Characterizing the ontogeny of tissue macrophages is an emerging field as it becomes evident that this may correlate with macrophage function. Thus, the list of studied organs using the aforementioned fate-mapping mouse models is expanding. For example, studies in the kidney indicate a 50:50 ratio of fetal-derived versus adult bone-marrow-derived macrophages (reviewed elsewhere74), muscle has been shown to have 60% HSC-derived cells at 24 weeks of age75 and studies of the placenta indicate a majority of Hofbauer cells stem from EMPs76. However, owing to the developmental continuum of fetal macrophage progenitors (for example, EMPs are present in the yolk sac from E8.5 until E10.5 and in the fetal liver at least until E14.5 (refs. 4,77)), their overlapping expression of markers (for instance, c-KIT and CSF1R are expressed on both EMPs and HSCs) and the variable labelling efficiencies of the models, the origin of mature postnatal macrophages in many organs remains to be investigated (see also the section discussed subsequently on ‘Macrophage development throughout life’).
Macrophages in barrier tissues
Lung macrophages
The lung is constantly exposed to the environment via air and, thus, to exogenous cues that shape its cellular environment. Within the homeostatic lung, at least three major macrophage populations can be found, separated by their anatomical location: alveolar macrophages within the air-exposed space of the alveolus and two to three interstitial macrophage populations within the interstitial regions of the lung (reviewed elsewhere78). Alveolar macrophages are the major macrophage population in the lung (in which they are 5–10 times more abundant than interstitial macrophages), and they reside within the air-exposed space of the alveolus, tightly attached to epithelial cells. The alveolar macrophages are the major gatekeepers and housekeepers of the alveolus, where they phagocytose cellular and pathogenic debris and clear mucus material from the alveolus, an essential task to allow gas exchange within the healthy lung78. They colonize the lung during embryogenesis and have a high self-renewal capacity2,6,12,79. This self-renewal capacity is imprinted on the developing alveolar macrophage compartment at the neonatal stage by neutrophil-derived 12-hydroxyeicosatetraenoic acid80. Furthermore, monocytes can contribute substantially to the alveolar macrophage repertoire during ageing or after inflammatory episodes. The functional impact of resident alveolar macrophage replacement by monocyte-derived cells remains severely understudied and its role in subsequent lung disease remains to be discovered12,81,82. Alveolar macrophage development and maintenance depend on granulocyte–macrophage colony-stimulating factor (GM–CSF) produced by type II airway epithelial cells79. Consequently, the mouse model of GM–CSF deficiency79 and GM–CSF receptor (CSF2R) mutations in human patients83 lead to pulmonary alveolar proteinosis, highlighting the importance of alveolar macrophages for organ homeostasis.
Lung interstitial macrophages can be subdivided into two (LYVE1highMHCIIlow and LYVE1lowMHCIIhigh)84,85 or three3,86 (based on the expression of TIM4, LYVE1, FOLR2, CCR2 and MHCII) main populations. Their ontogeny remains debated3,84–86, with the current view of a fetal origin for all interstitial macrophages and different rates of monocyte replenishment within distinct interstitial macrophage populations during adulthood78. In contrast to alveolar macrophages, interstitial macrophage development has been shown to critically depend on steady-state CSF1R signalling rather than CSF2R signalling85,87. Interstitial macrophages share a common IL-10 production signature but differ in their capacity to present antigen. LYVE1lowMHCIIhigh macrophages are associated closely with nerve bundles or endings and highly express pro-inflammatory molecules such as Il1b and Cxcl12, whereas LYVE1highMHCIIlow interstitial macrophages are in proximity to blood vessels and express an immunoregulatory signature, including Tgfb2, Plaur and Fcna84. Nerve-associated interstitial macrophages were shown to be critical for regulating immune responses and tissue homeostasis following exposure to inflammatory stimuli in the lung85. Functionally, absence of LYVE1highMHCIIlow interstitial macrophages has been shown to worsen experimental bleomycin-induced lung fibrosis, which is likely linked to their immunoregulatory role84. Further analysis is needed to reveal the physiological tasks and microanatomical niches of interstitial macrophages during lung homeostasis.
Gut macrophages
The gut harbours a complex pool of macrophages within its anatomical layers (reviewed elsewhere88). Anatomically, the large and small intestines can be subdivided into the epithelial region, lamina propria, submucosa and the muscularis externa. Early work elucidating the intestinal macrophage network focused on the epithelial and lamina-propria-associated macrophages found in the large and small intestines. Lamina-propria-associated macrophages are initially fetal-derived and are rapidly replaced after birth by short-lived monocyte-derived macrophages in a CCR2-dependent manner89 and require live microbiota to thrive90. Interestingly, more recently, this uniform view of a short-lived monocyte to macrophage trajectory was challenged by the identification of long-lived macrophage subsets within the lamina propria, which are defined by TIM4 expression90. Within the intestinal environment, immunoregulatory cytokines, such as IL-10 and transforming growth factor-β, are abundantly expressed by resident macrophages, guarding the differentiation trajectories of newly differentiating monocytes to macrophages through RUNX3 and KLF10 (refs. 91,92). To maintain physiological organ function, small intestinal lamina-propria-resident macrophages sample antigenic and apoptotic material both in the lumen and within the lamina propria, phagocytose surrounding material and support epithelial stem cell proliferation within intestinal crypts by providing Wnt ligands88,93–95. Small intestinal lamina-propria-resident macrophages express and secrete large amounts of IL-10. Here, macrophage-derived IL-10 is crucial for the induction of microbiota-specific regulatory T cells89,96. Taken together, lamina-propria macrophages are essential for intestinal barrier homeostasis. This notion is further illustrated by fact that mutations affecting the IL-10 receptor pathway cause severe paediatric inflammatory bowel disease97,98. Additionally, intestinal macrophages interact with neuronal components of the intestinal tract, supporting their survival and development99,100. Within the deeper layers of the intestine, such as the submucosa and the muscularis externa, long-lived macrophages can be identified, which are tightly embedded within their subtissular niches88. Within the large intestine, muscularis macrophages have been shown to maintain intestinal movement101. In the small intestine, muscularis macrophages interact with nerve bundles and vessels, both supporting their growth and function during homeostasis, at least in part, through signalling via BMP2 and β2 adrenergic receptors99,102. Molecular determinants of muscularis macrophage development and identity remain elusive.
Skin and oral mucosa macrophages
In mammals, distinct epithelial layers cover the inner mucosal surfaces and the skin to build a physical barrier against the outer environment and potentially pathogenic microorganisms. The epidermis harbours a specialized subset of tissue-resident macrophages, the Langerhans cells, within its suprabasal layer. Adult Langerhans cells are a ramified skin-resident myeloid population that share distinct features with both macrophages and dendritic cells (DCs). They are essential gatekeepers of barrier immunity and act as professional phagocytes, internalizing dying cells, eliminating invading pathogens and presenting antigen to T cells. Langerhans cells are essential for mediating and organizing barrier immunity within their epidermal tissue niche. They achieve this by reorganizing epidermal layering of keratinocytes through continuous sequestration of external antigens103, instructing regulatory T cells and, therefore, controlling epidermal tolerance towards commensal microbiota and auto-antigens104–106, as well as the patterning of undifferentiated keratinocytes in the suprabasal layers107. Langerhans cells have been described to colonize the epidermis and oral mucosa around birth and need to rapidly differentiate within their host tissue during the early postnatal phase108,109. Fate-mapping studies revealed that skin Langerhans cells originate from fetal progenitors4,8,108. Once settled in their tissue niche, they rapidly proliferate in the developing tissue in the early postnatal phase by clonal expansion109,110. The adult epidermal Langerhans cell population is long-lived and maintained by endogenous proliferation, although substantial inflammation, infection or injury in the adult skin can lead to the replacement of endogenous Langerhans cells within their niche by monocyte-derived progenitors111.
Langerhans cells are found across different epithelial barriers such as oral mucosa, cornea and mucosal tissue of the female reproductive tract112. Adult Langerhans cells in the oral mucosa are of mixed origin and are composed of fetal and postnatal progenitors. After birth, they undergo a steady replacement within the tissue niche by preDC-derived and monocyte-derived cells, most likely owing to the close proximity to a high microbial load113. In the oral mucosa, postnatal Langerhans cells are separated into two populations: CD103highCD11blow, mostly derived from preDCs, and CD103lowCD11bhigh Langerhans cells, which are derived from both, monocytes and preDCs113. However, transcriptomic profiling identifies them as bona fide Langerhans cells, similar to epidermal Langerhans cells derived from the prenatal origin. Tongue Langerhans cells play a role in antifungal immunity114 and have an anticancer protective role115. However, their role in homeostatic tissue development and function remains unknown. Moreover, Langerhans cell heterogeneity across different epithelial barrier tissues and the effects of diverse environmental factors on Langerhans cell maturation remain underexplored, and it will take future studies to define the specification and maturation of Langerhans cells across distinct tissue niches.
Dermal macrophages localize in a defined developmental patterning in specific tissue compartments, such as vessel-associated macrophages or sensory-nerve-associated macrophages84,116. On the basis of marker expression and transcriptional profiling, several macrophage subsets have been identified in the adult dermis, with differing tissue functions and anatomical locations. Whereas vessel-associated macrophages are characterized by a CX3CR1lowLYVE1high and MHCIIlow expression profile, sensory-nerve-associated macrophages are defined as CX3CR1highLYVE1lowMHCIIhigh (refs. 84,117). Additionally, a subset of CX3CR1int macrophages seem to represent an intermediate macrophage population that are directly derived from monocytes and expanded in injury or infection117. Even though macrophage progenitors from fetal sources seed the dermis prenatally and show substantial contribution to macrophages across the different compartments, several studies showed that dermal macrophages, at least partially, are exchanged by MDMs postnatally84,116,117. The turnover rates during adulthood can vary between compartments with sensory-nerve-associated macrophages representing an almost exclusively prenatally seeded long-lived population, except upon nerve injury117. Each population serves defined functions adjusted to its subtissular niche and dictated by their specific tissue environment: vessel-associated macrophages are essential contributors to dermal blood vessel integrity, anti-fibrotic activity during disease and coordinate immune cell recruitment during infection84,118. Sensory-nerve-associated macrophages facilitate nerve regeneration after injury by degradation of myelin from injured fibres84,117. Newly sprouted axons at lesion sites seem to recruit macrophages from other dermal sources, and these cells acquire a sensory-nerve-associated macrophage phenotype over time117, further supporting the assumption that specific dermal anatomical niches define tissue macrophage profiles and functions. Yet, the functional role of dermal macrophages during skin development and homeostasis is unclear to date.
In summary, it is impossible to draw conclusions on macrophage ontogeny simply by considering the contact of an organ with the environment. Direct exposure to microbiota or microbiota-derived factors does not necessarily drive the immediate replenishment of fetal-derived macrophages by MDMs (for example, in the case of alveolar macrophages in the lung, Langerhans cells in the epidermis and Kupffer cells in the liver). Similarly, an internal and highly protected organ, such as the brain, harbours MDMs that contribute to the pool of resident macrophages. Further along these lines, barrier-like tissues can be identified in organ systems not exposed to the environment, such as the joints. Here, fetal-derived and monocyte-derived macrophages are responsible for removing sterile insults, such as cellular debris, that would otherwise lead to immune system activation and organ damage119. Thus, the cell-autonomous and cell-extrinsic factors controlling the differentiation and adaption of a macrophage within its niche during adulthood and its capability of self-renewal and clonal expansion, especially considering MDMs during steady state, remain unclear. Nevertheless, constant environmental exposures and immune-stimulatory cues during physiological inflammation may have a long-term impact on macrophage ontogeny (see sections on ‘Macrophage development throughout life’ and ‘Physiological versus pathogenic inflammation’).
Macrophage development throughout life
Fate-mapping studies, including experiments addressing macrophage functions, are often performed in 6–8-week-old mice, which correspond to the human stage of late childhood–early adolescence. However, to study macrophage functions in diseases — especially those relevant for ageing populations, such as stroke, cancer and cardiovascular disorders — the maturation state and age of a long-lived macrophage within its niche should be considered. Although tissue specification is well established at 3 weeks of age2, and macrophages such as microglia, Kupffer cells and Langerhans cells are not replaced by MDMs4,12, the contribution of MDMs to the tissue-specific pool of macrophages in certain tissues only stagnates after 12–20 weeks (for example, this is seen in the peritoneum, gut and for MHCII+ dermal macrophages). Other macrophages, such as MHCII− dermal, splenic and alveolar macrophages, have a continuous MDM input throughout life, indicating a slow replacement of the majority of fetal-derived macrophages12. However, this has been studied on a tissue population basis or using the suggested dichotomy of LYVE1highMHCIIlow versus LYVE1highMHCIIlow macrophages84, thereby not considering additional macrophage heterogeneity within each tissue. A recent study elegantly showed that, actually, three macrophage subpopulations coexist across organs with distinct origins and self-renewal capacities: one, a population of macrophages expressing TIM4, LYVE1 and FOLR2 (TLF+), which are long-lived and develop from fetal progenitors; two, CCR2+ macrophages that are short-lived and constantly replenished by monocytes; three, MHCIIhi macrophages that are of mixed origin and where a finite proportion of the MDMs acquires self-renewal capacity3. This study and our own unpublished data suggest that, in a mouse of 6–12 months of age, the numbers of CCR2+ MDMs will equilibrate in every tissue and that there is no general mechanism of a continuous replacement of fetal-derived resident TLF+ or MHCIIhi macrophages by MDMs during homeostasis (Fig. 2).
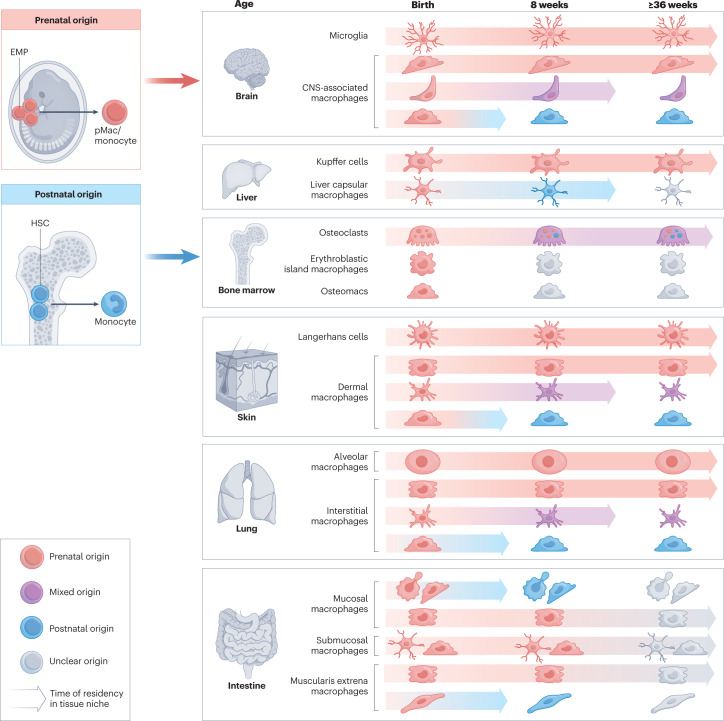
Fig. 2. Heterogeneity, ontogeny and self-renewing capacity of macrophage populations in adult tissues during steady state.
Internal and barrier tissues are colonized by yolk sac progenitors during organogenesis that develop into tissue-specific macrophages inhabiting distinct anatomical areas and subtissular niches of an organ. During organ maturation, bone-marrow erythro-myeloid progenitors (HSCs) contribute to some degree to certain macrophage populations that have a high self-renewing capacity and, thus, have a prolonged time of residency in the tissue niche, for example, the central nervous system (CNS)-associated macrophages, liver capsular macrophages, dermal macrophages and lung interstitial macrophages. Osteoclasts are long-lived, but exchange their fetal-derived nuclei for monocyte-derived nuclei over time. Additionally, a defined fraction of short-lived macrophages is constantly replenished by adult monocytes in some tissues, such as the brain, skin, lung and intestine. The ontogeny and cell cycle of many macrophage subpopulations in a fully matured organ (≥12 weeks of age) and during ageing are not known (indicated by grey colour). EMP, erythro-myeloid progenitor; pMac, pre-macrophage.
However, a systematic characterization of macrophage ontogeny during ageing is lacking. Therefore, it remains largely unknown whether macrophage maintenance and recruitment become altered in the tissues of geriatric mice. In addition to an increased immune cell infiltration, ageing of our immune system is associated — among many other phenotypes — with an unbalanced production of pro-inflammatory and anti-inflammatory factors and a reduced capacity to take up apoptotic cells via efferocytosis120. Age-related changes in macrophage effector functions within a tissue may be driven by ontogeny as the origin of a macrophage usually also controls its longevity. Although long-lived fetal-derived macrophages continuously sense environmental signals within a tissue, which they integrate into a transcriptional programme that drives changes in their behaviour, short-lived MDMs reside only briefly in their niche before they are replaced by a new monocyte. Thus, despite sitting in similar tissue niches, these ontogenetically distinct macrophages may have distinct effector functions, and only the constant replacement of fetal-derived by monocyte-derived macrophages tips the scale towards tissue phenotypes observed during inflammageing (Fig. 1b). Additionally, changes in tissue integrity during ageing, for example, increased stiffness of the ECM121 or loss of tissue structures122, may lead to a changed macrophage niche, thereby altering the environmental signals that macrophages convert to their effector functions. A combinatory model fate-mapping both EMP-derived and HSC-derived macrophages simultaneously would help address whether the maturation state, the increased recruitment of MDMs or the intrinsic developmental programme of macrophages has a role in tissue homeostasis and response to stimuli during adulthood and ageing.
Conserved functions across tissues
Core functions and functional heterogeneity of macrophages
The fact that tissue-resident macrophages do not constantly arise from circulating monocytes but instead stem mostly from fetal progenitors quickly raised the question of how these cells acquire and maintain their identity and self-renew. Many studies have since characterized core macrophage programmes and tissue-specific transcriptional regulatory pathways, defining transcription factors and niche signals controlling macrophage differentiation and tissue specificity2,16,91,123–125. These studies indicate that pMacs, the circulating macrophage precursors126, have already acquired a core macrophage signature that will ensure macrophage survival (via expression of Csf1r and Maf) and core macrophage functions, such as efferocytosis (Timd4, Mertk and Sirpa), non-opsonic phagocytosis (Cd14, Cd36, Clec7a and Mrc1), opsonic receptor-dependent phagocytosis (Fcgr1, Fcgr3, Fcgr4 and Itgam) and complement-dependent tissue immunity (C1qb, C1qc and C3ar1)2. Fetal macrophages start to acquire their tissue-specific programme as soon as they enter the developing tissue. Throughout late gestation, this programme manifests further, thereby driving the diversification of tissue-specific macrophages observed in adult stages2,124,125.
These initial bulk RNA-seq studies were instrumental in characterizing and understanding the functional heterogeneity of macrophages across different organs and in developing new mouse models that would allow targeting or depletion of specific macrophage populations. However, with the arrival of commonly affordable single-cell and single-nucleus transcriptomics approaches and increased use of fate-mapping models, it became quickly evident that most studies were underestimating the heterogeneity of macrophage populations within one organ. A prime example is the brain, which harbours not only microglia and different populations of CAMs but also where microglia show distinct phenotypes associated with unique functions within different anatomical regions127. Other tissues, such as the lung, heart, fat, skin and the peritoneum, clearly show at least a dichotomy of macrophages during steady state with the presence of both LYVE1highMHCIIlow and LYVE1lowMHCIIhigh populations84,128. Although the tissue-specific cues that macrophages integrate largely determine their transcriptome, in silico removal of these tissue signatures revealed a core macrophage transcriptional profile with genes involved in the complement system and blood vessel morphology in LYVE1high and inflammation-related and chemotaxis signalling-related genes in MHCIIhigh macrophages84. Intriguingly, this LYVE1 versus MHCII signature has been confirmed across many tissues in humans as well72,84. Recently, Dick et al3. proposed that the observed MHCIIhigh macrophage population84 consists of two populations that are distinguishable by CCR2 expression. Here, the flow cytometry analysis does not include several bone fide macrophage markers, such as F4/80 or MERTK, so that a minor contribution of CD64+ monocytes to the described macrophage populations cannot be ruled out completely. Nevertheless, this macrophage heterogeneity indicates that we must continue dissecting macrophage biology at a single-cell level to fully understand how they develop and how they acquire their core functions, on the one hand, and their diverse tissue-specific functions, on the other hand.
Subtissular macrophage niches
Subtissular niches are specialized microanatomical cellular neighbourhoods, which facilitate the survival and functional specialization of macrophages. Several examples of such niches have recently been identified for macrophages in various tissues. Conceptualized early as the ‘fibroblast–macrophage circuit’129, it is now apparent that macrophage niches can be more complex and can remain plastic throughout homeostasis and disease. Within the liver cellular neighbourhoods, the mechanisms defining a subtissular niche for Kupffer cells have been identified. Here, hepatic stellate cells, in conjunction with liver sinusoidal endothelial cells, enforce Kupffer cell identity both functionally and developmentally. Importantly, stellate cells and liver sinusoidal endothelial cells also play roles in re-establishing homeostasis and organ regeneration61. In the lung, alveolar macrophages adhere closely to airway epithelial cells to receive the GM–CSF needed for their maintenance130. Similarly, within the synovium of the joint, macrophages interact with synovial fibroblasts regulating fibroblast integrity119. Additionally, LYVE1high macrophages have been shown to be associated with blood vessels, fostering their branching and growth, whereas MHCIIhigh macrophages seem to preferably adhere to nerve endings and bundles, as shown in the lung, intestinal muscularis and brown adipose tissue84,101,131. Finally, cardiomyocyte–macrophage132 and adipocyte–macrophage crosstalk133 via mitochondrial recycling was shown to be crucial for tissue homeostasis and function in cardiac and adipose tissue. Thus, metabolic crosstalk and regulation of neighbouring cells via production of metabolites or exchange of mitochondria134,135 may be a key ability of resident macrophages across tissues allowing for the efficient and adequate regulation of tissue function throughout the lifespan of the organism.
Microanatomical units determine reciprocal cell function during health and disease, as illustrated by recently identified disease-driving fibrotic macrophage niches within the lung and liver136,137. Therefore, it is not necessarily the tissue of residence but the subtissular niche that either instructs or requires certain macrophage functions that are vital for tissue function (for example, metabolism, neuronal function, blood vessel integrity and stem cell-ness) (Fig. 3). Collectively, the combination of previously gathered information from bulk-sorted and single macrophage populations with novel single-cell spatial transcriptomics, proteomics and metabolomics technologies provides the ability to study macrophage subtissular niches. Ultimately, these combinatorial approaches will define macrophage core functions and macrophage niche dynamics, which foster organ homeostasis and integrity.
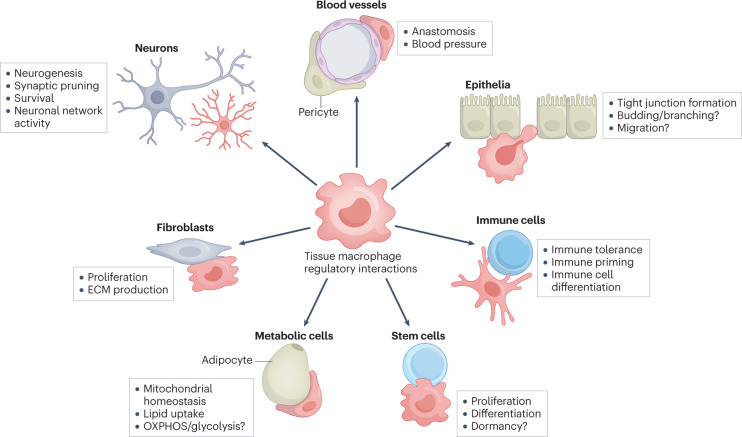
Fig. 3. Subtissular niches of tissue-specific macrophages and their role in tissue function.
Macrophages contribute to the function of the tissue via cell–cell crosstalk with specialized tissue cells. Some of the functions are well known, such as the interaction of microglia with neurons, which is essential for neurodevelopment and function. See text for further information about macrophage core functions in different tissues. Other functions are assumed (labelled with a question mark), but have not been studied. ECM, extracellular matrix; OXPHOS, oxidative phosphorylation.
Organ-specific immunity
Physiological versus pathogenic inflammation
Macrophages are placed strategically throughout the body to detect invading microbial pathogens. For the efficient detection of infections, macrophages are equipped with an array of receptors allowing the recognition of opsonized pathogens via pattern recognition, complement or Fcγ receptors (FcγRs)138–140. In addition, macrophages are a major source of sensor proteins, such as the complement component C1q, which starts the pro-inflammatory complement cascade upon binding to opsonized pathogens141. Tissues such as the intestine, skin and lung receive constant exogenous immune-stimulatory cues, which are sensed by the resident macrophage populations of the organ. Thus, it is of utmost importance to understand how tissue-resident macrophage populations integrate environmental non-pathogenic inflammatory cues into their functional core programmes. Physiological adaptation towards environmental stimuli is crucial for organ homeostasis, efficient clearance of pathogens and the re-establishment of homeostasis after inflammation142. Recently, several examples have shown the importance of such adaptation events in mouse models. Latent or acute viral infections of the lung have been associated with increased development of house dust mite-induced allergy and a more efficient immune response to a secondary bacterial challenge143. Moreover, the recognition of danger signals, such as lipopolysaccharide, has been linked to tissue adaptation via changes in the macrophage compartment. Here, lipopolysaccharide-induced low-grade inflammation triggered the recruitment and establishment of a long-lived MDM compartment with enhanced pro-inflammatory capabilities144. Pulmonary viral infections were also linked to a more efficient CD8+ T cell response, mediated by functional modification of alveolar macrophages post-viral infection145. In the lung, increased IL-6 production by macrophages was observed after primary infection with influenza A viruses, accompanied by a long-lasting replacement of a fraction of resident alveolar macrophages by MDMs146,147. Furthermore, in the muscularis externa of the small intestine, previous pathogen encounters and primed neuroprotective features in resident macrophages prevent further tissue damage in the event of a secondary infection148. Core macrophage functions such as efferocytosis and metabolic regulation of neighbouring cells could be important determinants of such tissue adaption events, allowing the tissue to ‘collect’ inflammatory experiences without suffering impairment of tissue function. However, the exact molecular cues active in tissue-resident macrophages during physiological inflammation remain unidentified. Interestingly, chronic inflammatory diseases, for example, fibrosis or obesity, display features of macrophage core function dysregulation, such as a metabolic disturbance in adipocytes or fibroblast proliferation, processes usually tightly regulated by macrophages149,150. Thus, macrophage regulatory circuits post-inflammation may be important regulators of tissue homeostasis and function, and dysregulation of physiological inflammation processes might have important consequences for the development of chronic inflammation in a susceptible host.
Connecting macrophage core functions and autoimmunity
In the absence of infections, the unique spatial distribution of macrophages within organs puts them centre stage for removing potentially harmful cues that would otherwise trigger inflammatory or autoreactive immune responses. This includes, most importantly, the removal of dying cells (efferocytosis), which are generated during the process of positive or negative selection in primary and secondary immunological organs (thymus, spleen, bone marrow and lymph nodes) or owing to constant exposure of tissues to sterile insults, such as UV light or natural ionizing radiation. Moreover, in tissues such as joints, the constant mechanical stress leads to a continuous, reversible damage of cartilage tissue, releasing cellular debris and components of the synovial fluid, such as hyaluronic acid, which can bind to pattern recognition receptors119. Similarly, small immune complexes consisting of IgG antibodies arising either from low-affinity poly-reactive natural antibodies or from low-level autoreactive antibodies, which are widely present also in healthy individuals, are continuously generated151. They have to be removed rapidly to prevent their deposition in organs, such as the kidney or lung, which could lead to consequent activation of the complement pathway or trigger activating FcγRs abundantly expressed on resident macrophages152. Of note, this continuous housekeeping function must operate without triggering macrophage activation and, thus, without inducing concomitant inflammation. The importance of macrophages in removing these potentially harmful cues becomes evident if entire core functions, such as phagocytic activity or checkpoints modulating macrophage activity, are dysregulated or impaired. Indeed, a reduced phagocytic capacity of macrophages has been linked to the development of systemic lupus erythematosus in mice and humans153,154. The failure of macrophages to remove apoptotic cells in a timely fashion results in the transition from apoptosis to necrosis or necroptosis, which is an immunogenic form of cell death that induces tissue inflammation and may ultimately lead to the priming of autoantibody responses. Mechanistically, necrotic cells are recognized via different sets of activating receptors on macrophages, such as the macrophage-inducible C-type lectin (Mincle) or toll-like receptors, which leads to the secretion of pro-inflammatory cytokines and the recruitment of monocytes and neutrophils155,156. With respect to rheumatoid arthritis — an autoimmune disease predicted to affect up to 1% of the ageing western population — it was demonstrated recently that synovial lining macrophages characterized by TREM2 expression in mice and humans may be critically involved in preventing the initiation of sterile inflammation in the joints, ultimately leading to joint destruction and rheumatoid arthritis development157. Joint lining macrophages are connected by tight junctions and show signs of apical–basal polarization, thus generating a barrier-like structure in combination with lining fibroblasts. Functionally, this macrophage subset is characterized by a high phagocytic activity and the expression of receptors involved in efferocytosis, allowing to continuously remove cellular debris and synovial fluid components. Indeed, depletion of lining macrophages resulted in an earlier onset and more pronounced immunopathology of arthritis in mouse model systems157.
Apart from erroneous triggering of activating macrophage receptors, a reduced expression of inhibitory receptors, such as the inhibitory FcγRIIb, which balances activating signals transduced via activating FcγRs upon IgG immune complex binding, results in aberrant macrophage activation and tissue damage and is another risk factor for the development of systemic lupus erythematosus in mice and humans158,159. Interestingly, it was recently shown that certain tissue-resident macrophage subsets, such as dermal macrophages in the skin, express high levels of the inhibitory FcγRIIb compared with activating FcγRs, suggesting that organ-specific thresholds for macrophage activation may exist152. In addition to IgG binding, FcγRIIb was also shown to bind to fibrinogen-like 2, a secreted protein expressed in myeloid immune cells and tissue cells in the kidney and skin160. Thus, continuous crosstalk of macrophages with tissue-derived factors could participate in the modulation of steady-state macrophage activity. Indeed, a deficiency in fibrinogen-like 2 or FcγRIIb results in enhanced inflammatory responses and autoimmunity in mice158,160. In summary, tissue-resident macrophages fulfil essential functions in preventing autoimmunity. Importantly, inflammation may result in the recruitment of MDMs in many organs, which may lead to a replacement of fetal-derived macrophages. Whether the threshold (phagocytic activity and inhibitory FcγRIIb expression) for the activation of MDMs by external cues is the same as of fetal-derived macrophages remains to be established.
Outlook and future directions
Recent technological progress, including novel mouse models, spatial single-cell omics and analysis pipelines, has propelled our understanding of macrophage development and function across different tissues in the past decade (Box 4). It becomes increasingly evident that tissue-resident macrophages are not simply bystanders that respond to a stimulus or infection. Instead, they are at the interface of tissue homeostasis and pathogenesis and can contribute to and cause diseases if their homeostatic core functions are disturbed. Thus, tissue-resident macrophages represent a substantial core niche cell type across organs and organisms, ensuring tissue function and integrity (Box 3). Upon physiological or pathogenic inflammation, MDMs are recruited to the tissue, which may be beneficial or detrimental to disease outcomes, depending on the context. With the macrophage toolbox expanding, we are finally in the position to study the distinct roles and possible redundancy of fetal-derived macrophages and MDMs in an unprecedented manner.
Despite our increasing knowledge of macrophage responses within their specific niches, there is a knowledge gap concerning how distinct macrophage populations (EMP-derived versus HSC-derived) within a tissue integrate and respond to the same signals. Vice versa, it remains unclear whether these distinct macrophage populations would have an impact on the tissue niche, or how this intercellular crosstalk affects tissue function and integrity in the long term. Macrophage depletion studies suggest that MDMs and fetal progenitors can repopulate a tissue and, driven by factors within their niche, promote their further maturation into tissue-specific macrophages resembling the original macrophage population59,61,161–163. The presumably high plasticity of macrophages that allows them to adopt to their tissue environment was also suggested by cell transplantation studies of a fully differentiated macrophage population into another organ91. Nevertheless, careful observation of the data indicates that the original molecular signature imprinted on a long-lived, fetal-derived macrophage that has been developing together with the organ cannot be fully recapitulated by ‘surrogate macrophages’ occupying the empty niche. Furthermore, in a similar cell transfer study, it was demonstrated that fully differentiated tissue macrophages transferred from the peritoneal cavity, liver or colon into an empty lung alveolar macrophage niche barely colonized the tissue and could not produce functional alveolar macrophages163. Thus, macrophage plasticity is specified by the maturation state of macrophage precursors and their ability to adapt to their specific niches. It may well be that the ontogeny of macrophages dictates a majority of context-dependent effector functions as the longevity of fetal-derived macrophages allows for a continuous long-lasting signal integration within the macrophage niche, thereby providing these cells with a distinct transcriptome and proteome compared with short-lived MDMs. Thus, tissue niche functionality might be governed by the balance of EMP-derived versus HSC-derived macrophages within an organ.
Although depletion studies are instrumental to understanding the macrophage niche and how distinct macrophage populations contribute to tissue function, these studies likely do not represent physiological conditions as it is rare that the whole organ loses its tissue-resident macrophage population at once. When certain disturbances or cues arise, some macrophages may undergo apoptosis and be partially replenished by both fetal-derived and monocyte-derived macrophages that are already residing in the tissue. Yet, which mechanisms instruct and control local proliferation and longevity of these distinct macrophage populations remains a pressing question. Therefore, methodologies such as optogenetics or mosaic analyses, similar to the MARCM system in Drosophila164, may be powerful tools to address how daughter cells are instructed by the niche.
With the advent of single-cell technologies and the efforts of the Human Cell Atlas, we now have the first glimpse into macrophage development and functions in humans72,165–168. Indeed, comparing mouse and human macrophage precursors and tissue-specific macrophages during gestation indicates conserved phenotypes across species, for example, the presence of a yolk sac progenitor167 and the dichotomy of LYVE1+ and MHCII+ macrophages72,165. Thus, the mouse seems to be a suitable model organism to study macrophage-dependent (patho)mechanisms that could be translated to human biology and diseases (Box 5). However, a major drawback for studying immune cell development and organ-specific immunity in mice is the sterile environment in the specific pathogen-free facilities animals are usually kept in. The in utero environment is considered to be sterile and would generally represent the conditions mice are kept at. However, we should consider that maternal immune activation through infections and metabolic diseases will likely impact macrophage development in the embryo and macrophage maturation in newborns. Similarly, the constant exposure of humans to pathogens throughout their lives will affect macrophage effector functions during adulthood and ageing. This notion is supported by the fact that microglia do not reach their final maturation state when mice are kept in germ-free conditions169. Thus, in addition to studying macrophage ontogeny and function in maternal immune activation models170,171, the introduction of a wild type microbiota into mice (the so-called wildlings)172,173 would greatly improve the translatability to human macrophage ontogeny, functions and responses.
Box 4 Do heterogenous cell states correlate with specific macrophage functions?
Recent technological advances in high-throughput sequencing analysis offered new avenues in understanding tissue-resident macrophage heterogeneity across development, health and different disease conditions. Access to optimized protocols for single-cell RNA-sequencing, as well as single-nuclei RNA-sequencing, of tissue-resident macrophages revealed a previously unappreciated heterogeneity among the gene-expression profiles of tissue-resident macrophages in different tissues3,70,82,214. Despite the identification of many distinct cellular states of macrophages, we are only at the beginning of understanding if these states represent functionally distinct macrophage subsets that are ‘locked’ or even determined by their ontogeny or whether the plasticity of macrophages allows them to switch between functional states. The latter may be especially applicable during tissue stress and disease conditions with the ability of macrophages to adapt to their anatomical niche, but also to return back to their homeostatic transcriptional and functional state after resolution of tissue stress or inflammation. One of the best studied examples here are central nervous system (CNS) macrophages, including microglia and other CNS-associated macrophage populations. A series of studies revealed a dynamic heterogeneity of microglia across different developmental stages, but also in terms of anatomical locations215–217. One recent example for a distinct functional microglia subset is the association of the ‘fountain-of-microglia’ with myelination in white matter regions. These cells have a defined time window of occurrence in the corpus callosum, present a characteristic expression of the surface marker CD11c and the lysosomal marker Mac-3 and support oligodendrocyte progenitor cell survival and myelinogenesis during early postnatal development33,34. Microglial heterogeneity in different neuroinflammatory and neurodegenerative disease settings has been further explored. Pioneering work described a unique cluster of disease-associated microglia (DAM) identified in the context of Alzheimer disease and their defined localization at extracellular amyloid deposits218. DAM represent a distinct functional subset of microglia that is involved in driving neurodegenerative disease pathology218,219. In particular, the expression of the two DAM signature genes Trem2 and Apoe has now been widely described across DAM in different neurodegenerative diseases218,220. Recent studies have identified that the TREM2–APOE signalling pathway is driving the differentiation of DAM and is implicated in the activation of microglia and amyloid phagocytosis218,221,222. Furthermore, human patients with the TREM2R47H mutation have a higher risk of developing Alzheimer disease223,224. Similarly, analyses of other populations of CNS macrophages — including the tissue-resident macrophages of the meningeal layers, perivascular space and ventricular system — revealed heterogeneous transcriptional states across the different anatomical niches but also during disease. On colonization of their anatomical niche in the CNS interfaces, macrophage populations show a highly specified gene-expression profile and undergo functional adaptation to that environment. One example here are leptomeningeal macrophages that have been recently demonstrated to be a direct source of perivascular macrophages during postnatal development17, followed by divergence of gene expression profiles and functions for these two macrophage populations36,225,226. A second example are ventricular macrophages, which have been shown to emerge from microglia during embryonic development and acquire a distinct transcriptomic signature in that particular CNS compartment36,227. These cells maintain expression of the transcription factor Sall1 but lose, for example, common homeostatic markers of parenchymal microglia, such as Tmem119 or P2ry12, and the upregulation of genes also found in DAM, such as Cst7, Axl, Itgax or Clec7a (refs. 36,228). DAM signatures compiled using (single-cell) omics have also been characterized in other tissues during or following inflammatory episodes, for example, during obesity in fat tissue149, within the microbiota-exposed peritoneal cavity229 and after clearance of viral infection in the lung146, indicating a somewhat conserved functional DAM programme across tissues. Furthermore, human ApoE variants have been linked to differential outcomes of severe acute respiratory syndrome coronavirus 2 infection, influencing pulmonary macrophage heterogeneity230. In summary, high-throughput profiling of tissue-resident macrophages brought us a new understanding of macrophage states. However, the definition of new subsets and the naming of newly identified cellular states became inflationary with a yet missing in-depth analysis if the defined transcriptomic states are associated with defined functional subsets. Interestingly, cellular states of tissue-resident macrophages identified across different organs and distinct disease settings have shown similar gene expression profiles, indicating that macrophage subpopulations with shared functions exist across different organs in steady state but also develop in different diseases.
Box 5 From basic research to translational medicine.
Most of the available knowledge on macrophage function, ontogeny and disease implication is collected with the help of advanced transgenic mouse models. Using models of in vivo fate-mapping and gene deletion studies allowed to understand basic concepts of tissue-resident macrophages and to elucidate potential new therapeutic avenues. Still, there is a substantial gap between the transgenic mouse models used and the potential therapeutic application in humans. One of these examples is the use of specific-pathogen free mice for most experiments, not considering the diversity of the human microbiome and its implication on immunity and macrophage function. Introducing a diverse microbiome in experimental mice, as done in the ‘wildling’ mice, has been shown to represent a much closer model to humans than using specific-pathogen free mice and might be also important to study in the context of macrophage-targeted therapies172,173. Recent mouse studies using macrophage replacement models by using genetic or pharmacological inhibition of the colony-stimulating factor 1 receptor have shown promise for translation to patients, for example, in the context of replacing dysfunctional microglia during neurodegenerative diseases231. Finally, although in humanized mice the human immune system development can be recapitulated, the niche-specific imprinting on human macrophages will be derived from mouse tissues. In summary, although all of the currently available model systems have some specific benefits, neither model is fully recapitulating the human situation supporting the need for a cross-species approach to characterize macrophage core functions during health and disease and exploit this knowledge in future translational approaches.
Acknowledgements
The authors thank all their laboratory members for their ambitious work and passionate research on myeloid cells. The authors apologize to all colleagues, whose work could not be cited in this work owing to space limitations. E.M. and A.S. are supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy — EXC 2151 — 390873048. E.M. is supported by the European Research Council under the European Union’s Horizon 2020 research and innovation program (Grant Agreement No. 851257) and by the DFG Grant SFB1454 (projects INST 217/1042-1 and INST 217/1037-1). F.N. is supported by the Deutsche Forschungsgemeinschaft (CRC 1181-A07, CRC1526-A07, TRR305-B02, FOR2953-P3, FOR2886-P2) and the NIH (U19 AI142790, RFA-AI-18-042). K.K. is supported by a project grant of the Fritz Thyssen Foundation and the German Research Foundation (DFG) by project grants within the SFB/TRR167 (Project ID 259373024), the CRC1479 (Project ID 44189134), the SFB/TRR359 (Project ID 491676693) and by the DFG under Germany’s Excellence Strategy (Grant No. CIBSS — EXC-2189, Project ID 390939984). A.S. is supported by an Emmy Noether Research Grant (SCHL2116/1), DFG Research Grants (SCHL2116/6-1 and SCHL2116/7-1), the GRK214362475/GRK1873/2 and the DFG CRC1454 (Project INST 217/1042-1).
Glossary
- 12-Hydroxyeicosatetraenoic acid
A pro-inflammatory eicosanoid metabolite produced by cells.
- Erythroblastic island
(EBI). A cluster of developing red blood cells forming around a central macrophage that supports their maturation.
- Hepatic stellate cells
Also known as vitamin A and lipid-storing cells, located between the endothelial lining and hepatocytes contributing to liver homeostasis via autocrine, paracrine and chemoattractant factors.
- Hofbauer cells
Placental macrophages that are present throughout pregnancy.
- Human Cell Atlas
A large-scale, international effort aimed at mapping the diversity and distribution of cells in the human body.
- Kolmer epiplexus cells
Intraventricular tissue-resident macrophage population associated with the multiciliated cuboid ependymal cells of the choroid plexus in all ventricles.
- Type II airway epithelial cells
Specialized cells in the respiratory system responsible for the production of surfactant.
- Virchow-Robin/perivascular space
Space surrounding blood vessels within tissues, filled with perivascular cells and the extracellular fluid.
- Wildlings
C57BL/6 mice that are colonized by natural microbiota and pathogens at all body sites.
Author contributions
The authors contributed equally to all aspects of the article.
Peer review
Peer review information
Nature Reviews Immunology thanks the anonymous reviewers for their contribution to the peer review of this work.
Competing Interests
The authors declare that they have no conflicts of interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Elvira Mass, Falk Nimmerjahn, Katrin Kierdorf, Andreas Schlitzer.
References
- 1.Jenkins SJ, Allen JE. The expanding world of tissue-resident macrophages. Eur. J. Immunol. 2021;51:1882–1896. doi: 10.1002/eji.202048881. [DOI] [PubMed] [Google Scholar]
- 2.Mass E, et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353:aaf4238. doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dick SA, et al. Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci. Immunol. 2022;7:7777. doi: 10.1126/sciimmunol.abf7777. [DOI] [PubMed] [Google Scholar]
- 4.Gomez Perdiguero E, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoeffel G, et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz C, et al. A lineage of myeloid cells independent of myb and hematopoietic stem cells. Science. 2012 doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 9.Perdiguero EG, Geissmann F. The development and maintenance of resident macrophages. Nat. Immunol. 2016;17:2–8. doi: 10.1038/ni.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mass E. Delineating the origins, developmental programs and homeostatic functions of tissue-resident macrophages. Int. Immunol. 2018;30:493–501. doi: 10.1093/intimm/dxy044. [DOI] [PubMed] [Google Scholar]
- 11.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, et al. Fate mapping via Ms4a3-expression history traces monocyte-derived. cells Cell. 2019;178:1509–1525.e19. doi: 10.1016/j.cell.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Werner Y, et al. Cxcr4 distinguishes HSC-derived monocytes from microglia and reveals monocyte immune responses to experimental stroke. Nat. Neurosci. 2020;23:351–362. doi: 10.1038/s41593-020-0585-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai SM, et al. Organ-specific fate, recruitment, and refilling dynamics of tissue-resident macrophages during blood-stage malaria. Cell Rep. 2018;25:3099–3109.e3. doi: 10.1016/j.celrep.2018.11.059. [DOI] [PubMed] [Google Scholar]
- 15.Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 16.Hagemeyer N, et al. Transcriptome‐based profiling of yolk sac‐derived macrophages reveals a role for Irf8 in macrophage maturation. EMBO J. 2016;35:1730–1744. doi: 10.15252/embj.201693801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuda T, et al. Specification of CNS macrophage subsets occurs postnatally in defined niches. Nature. 2022;604:740–748. doi: 10.1038/s41586-022-04596-2. [DOI] [PubMed] [Google Scholar]
- 18.Utz SG, et al. Early fate defines microglia and non-parenchymal brain macrophage development. Cell. 2020;181:557–573.e18. doi: 10.1016/j.cell.2020.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Goldmann T, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat. Neurosci. 2013;16:1618–1626. doi: 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- 20.Tay TL, et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 2017;20:793–803. doi: 10.1038/nn.4547. [DOI] [PubMed] [Google Scholar]
- 21.Füger P, et al. Microglia turnover with aging and in an Alzheimer’s model via long-term in vivo single-cell imaging. Nat. Neurosci. 2017;20:1371–1376. doi: 10.1038/nn.4631. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greter M, et al. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity. 2012;37:1050–1060. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erblich B, Zhu L, Etgen AM, Dobrenis K, Pollard JW. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS ONE. 2011;6:e26317. doi: 10.1371/journal.pone.0026317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 26.Davalos D, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 27.Cserép C, et al. Microglia monitor and protect neuronal function through specialized somatic purinergic junctions. Science. 2020;367:528–537. doi: 10.1126/science.aax6752. [DOI] [PubMed] [Google Scholar]
- 28.Du Y, Brennan FH, Popovich PG, Zhou M. Microglia maintain the normal structure and function of the hippocampal astrocyte network. Glia. 2022;70:1359–1379. doi: 10.1002/glia.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bisht K, et al. Capillary-associated microglia regulate vascular structure and function through PANX1-P2RY12 coupling in mice. Nat. Commun. 2021;12:5289. doi: 10.1038/s41467-021-25590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fantin A, et al. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood. 2010 doi: 10.1182/blood-2009-12-257832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paolicelli RC, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 32.Tremblay M-È, Lowery RL, Majewska AK. Microglial interactions with synapses are modulated by visual experience. PLoS Biol. 2010;8:e1000527. doi: 10.1371/journal.pbio.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagemeyer N, et al. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 2017;134:441–458. doi: 10.1007/s00401-017-1747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wlodarczyk A, et al. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 2017;36:3292–3308. doi: 10.15252/embj.201696056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waisman A, Ginhoux F, Greter M, Bruttger J. Homeostasis of microglia in the adult brain: review of novel microglia depletion systems. Trends Immunol. 2015;36:625–636. doi: 10.1016/j.it.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 36.van Hove H, et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 2019;22:1021–1035. doi: 10.1038/s41593-019-0393-4. [DOI] [PubMed] [Google Scholar]
- 37.Goldmann T, et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 2016;17:797–805. doi: 10.1038/ni.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cugurra A, et al. Skull and vertebral bone marrow are myeloid cell reservoirs for the meninges and CNS parenchyma. Science. 2021 doi: 10.1126/science.abf7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rustenhoven J, Kipnis J. Brain borders at the central stage of neuroimmunology. Nature. 2022;612:417–429. doi: 10.1038/s41586-022-05474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seyfried AN, Maloney JM, MacNamara KC. Macrophages orchestrate hematopoietic programs and regulate HSC function during inflammatory stress. Front. Immunol. 2020;11:1499. doi: 10.3389/fimmu.2020.01499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Guo R, Song Y, Jiang Z. Erythroblastic island macrophages shape normal erythropoiesis and drive associated disorders in erythroid hematopoietic diseases. Front. Cell Dev. Biol. 2020;8:613885. doi: 10.3389/fcell.2020.613885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang KH, et al. p62 is required for stem cell/progenitor retention through inhibition of IKK/NF-κB/Ccl4 signaling at the bone marrow macrophage-osteoblast niche. Cell Rep. 2014;9:2084–2097. doi: 10.1016/j.celrep.2014.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang MK, et al. Osteal tissue macrophages are intercalated throughout human and mouse bone lining tissues and regulate osteoblast function in vitro and in vivo. J. Immunol. 2008;181:1232–1244. doi: 10.4049/jimmunol.181.2.1232. [DOI] [PubMed] [Google Scholar]
- 44.Mohamad SF, et al. Osteomacs interact with megakaryocytes and osteoblasts to regulate murine hematopoietic stem cell function. Blood Adv. 2017;1:2520–2528. doi: 10.1182/bloodadvances.2017011304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald MM, Kim AS, Mulholland BS, Rauner M. New insights into osteoclast biology. JBMR. 2021;5:e10539. doi: 10.1002/jbm4.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mansour A, et al. Osteoclasts promote the formation of hematopoietic stem cell niches in the bone marrow. J. Exp. Med. 2012;209:537–549. doi: 10.1084/jem.20110994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kollet O, et al. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat. Med. 2006;12:657–664. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- 48.Adams GB, et al. Stem cell engraftment at the endosteal niche is specified by the calcium-sensing receptor. Nature. 2005;439:599–603. doi: 10.1038/nature04247. [DOI] [PubMed] [Google Scholar]
- 49.Lymperi S, Ersek A, Ferraro F, Dazzi F, Horwood NJ. Inhibition of osteoclast function reduces hematopoietic stem cell numbers in vivo. Blood. 2011;117:1540–1549. doi: 10.1182/blood-2010-05-282855. [DOI] [PubMed] [Google Scholar]
- 50.Yao Y, et al. The macrophage-osteoclast axis in osteoimmunity and osteo-related diseases. Front. Immunol. 2021;12:1066. doi: 10.3389/fimmu.2021.664871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacome-Galarza CE, et al. Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature. 2019;568:541–545. doi: 10.1038/s41586-019-1105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Millard SM, et al. Fragmentation of tissue-resident macrophages during isolation confounds analysis of single-cell preparations from mouse hematopoietic tissues. Cell Rep. 2021;37:110058. doi: 10.1016/j.celrep.2021.110058. [DOI] [PubMed] [Google Scholar]
- 53.Gordan S, et al. The immunological organ environment dictates the molecular and cellular pathways of cytotoxic antibody activity. Cell Rep. 2019;29:3033–3046.e4. doi: 10.1016/j.celrep.2019.10.111. [DOI] [PubMed] [Google Scholar]
- 54.Grandjean CL, Garcia Z, Lemaître F, Bréart B, Bousso P. Imaging the mechanisms of anti-CD20 therapy in vivo uncovers spatiotemporal bottlenecks in antibody-dependent phagocytosis. Sci. Adv. 2021;7:eabd6167. doi: 10.1126/sciadv.abd6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guilliams M, Scott CL. Liver macrophages in health and disease. Immunity. 2022;55:1515–1529. doi: 10.1016/j.immuni.2022.08.002. [DOI] [PubMed] [Google Scholar]
- 56.Wen Y, Lambrecht J, Ju C, Tacke F. Hepatic macrophages in liver homeostasis and diseases — diversity, plasticity and therapeutic opportunities. Cell Mol. Immunol. 2021;18:45–56. doi: 10.1038/s41423-020-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montalvao F, et al. The mechanism of anti-CD20-mediated B cell depletion revealed by intravital imaging. J. Clin. Invest. 2013;123:5098–5103. doi: 10.1172/JCI70972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sierro F, et al. A liver capsular network of monocyte-derived macrophages restricts hepatic dissemination of intraperitoneal bacteria by neutrophil recruitment. Immunity. 2017;47:374–388.e6. doi: 10.1016/j.immuni.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 59.Sakai M, et al. Liver-derived signals sequentially reprogram myeloid enhancers to initiate and maintain Kupffer cell identity. Immunity. 2019;51:655–670.e8. doi: 10.1016/j.immuni.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scott CL, et al. The transcription factor ZEB2 is required to maintain the tissue-specific identities of macrophages. Immunity. 2018;49:312–325.e5. doi: 10.1016/j.immuni.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonnardel J, et al. Stellate cells, hepatocytes, and endothelial cells imprint the Kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity. 2019;51:638–654.e9. doi: 10.1016/j.immuni.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seidman JS, et al. Niche-specific reprogramming of epigenetic landscapes drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity. 2020;52:1057–1074.e7. doi: 10.1016/j.immuni.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Remmerie A, et al. Osteopontin expression identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity. 2020;53:641–657.e14. doi: 10.1016/j.immuni.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tran S, et al. Impaired Kupffer cell self-renewal alters the liver response to lipid overload during non-alcoholic steatohepatitis. Immunity. 2020;53:627–640.e5. doi: 10.1016/j.immuni.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 65.Morgantini C, et al. Liver macrophages regulate systemic metabolism through non-inflammatory factors. Nat. Metab. 2019;1:445–459. doi: 10.1038/s42255-019-0044-9. [DOI] [PubMed] [Google Scholar]
- 66.Liang Y, et al. Temporal analyses of postnatal liver development and maturation by single-cell transcriptomics. Dev. Cell. 2022;57:398–414.e5. doi: 10.1016/j.devcel.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iannacone M, et al. Response to contamination of isolated mouse Kupffer cells with liver sinusoidal endothelial cells. Immunity. 2022;55:1141–1142. doi: 10.1016/j.immuni.2022.06.012. [DOI] [PubMed] [Google Scholar]
- 68.Hume DA, Offermanns S, Bonnavion R. Contamination of isolated mouse Kupffer cells with liver sinusoidal endothelial cells. Immunity. 2022;55:1139–1140. doi: 10.1016/j.immuni.2022.06.010. [DOI] [PubMed] [Google Scholar]
- 69.Blériot C, et al. A subset of Kupffer cells regulates metabolism through the expression of CD36. Immunity. 2021;54:2101–2116.e6. doi: 10.1016/j.immuni.2021.08.006. [DOI] [PubMed] [Google Scholar]
- 70.Guilliams M, et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185:379–396.e38. doi: 10.1016/j.cell.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haimon Z, et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 2018;19:636–644. doi: 10.1038/s41590-018-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eraslan G, et al. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science. 2022;376:eabl4290. doi: 10.1126/science.abl4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinhard L, et al. Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nat. Commun. 2018;9:1228. doi: 10.1038/s41467-018-03566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Munro DAD, Hughes J. The origins and functions of tissue-resident macrophages in kidney development. Front. Physiol. 2017;8:837. doi: 10.3389/fphys.2017.00837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X, et al. Heterogeneous origins and functions of mouse skeletal muscle-resident macrophages. Proc. Natl Acad. Sci. USA. 2020;117:20729–20740. doi: 10.1073/pnas.1915950117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Freyer L, et al. Erythro-myeloid progenitor origin of Hofbauer cells in the early mouse placenta. Development. 2022;149:dev200104. doi: 10.1242/dev.200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGrath KE, et al. Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep. 2015;11:1892–1904. doi: 10.1016/j.celrep.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aegerter H, Lambrecht BN, Jakubzick CV. Biology of lung macrophages in health and disease. Immunity. 2022;55:1564–1580. doi: 10.1016/j.immuni.2022.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guilliams M, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pernet E, et al. Neonatal imprinting of alveolar macrophages via neutrophil-derived 12-HETE. Nature. 2023 doi: 10.1038/S41586-022-05660-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Angelidis I, et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 2019;10:963. doi: 10.1038/s41467-019-08831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mulder K, et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity. 2021;54:1883–1900.e5. doi: 10.1016/j.immuni.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 83.Suzuki T, et al. Familial pulmonary alveolar proteinosis caused by mutations in CSF2RA. J. Exp. Med. 2008;205:2703–2710. doi: 10.1084/jem.20080990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chakarov S, et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science. 2019;363:1–18. doi: 10.1126/science.aau0964. [DOI] [PubMed] [Google Scholar]
- 85.Ural BB, et al. Identification of a nerve-associated, lung-resident interstitial macrophage subset with distinct localization and immunoregulatory properties. Sci. Immunol. 2020;5:eaax8756. doi: 10.1126/sciimmunol.aax8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gibbings SL, et al. Three unique interstitial macrophages in the murine lung at steady state. Am. J. Respir. Cell Mol. Biol. 2017;57:66–76. doi: 10.1165/rcmb.2016-0361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bain CC, MacDonald AS. The impact of the lung environment on macrophage development, activation and function: diversity in the face of adversity. Mucosal Immunol. 2022;15:223–234. doi: 10.1038/s41385-021-00480-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delfini M, Stakenborg N, Viola MF, Boeckxstaens G. Macrophages in the gut: masters in multitasking. Immunity. 2022;55:1530–1548. doi: 10.1016/j.immuni.2022.08.005. [DOI] [PubMed] [Google Scholar]
- 89.Zigmond E, et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37:1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 90.Shaw TN, et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J. Exp. Med. 2018 doi: 10.1084/jem.20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fadok VA, et al. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J. Clin. Invest. 1998;101:890–898. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cummings RJ, et al. Different tissue phagocytes sample apoptotic cells to direct distinct homeostasis programs. Nature. 2016;539:565–569. doi: 10.1038/nature20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Niess JH, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 95.Chikina AS, et al. Macrophages maintain epithelium integrity by limiting fungal product absorption. Cell. 2020;183:411–428.e16. doi: 10.1016/j.cell.2020.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Murai M, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat. Immunol. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoffmann D, et al. Genetic correction of IL-10RB deficiency reconstitutes anti-inflammatory regulation in iPSC-derived macrophages. J. Pers. Med. 2021;11:221. doi: 10.3390/jpm11030221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ye Z, et al. Predictive prenatal diagnosis for infantile-onset inflammatory bowel disease because of interleukin-10 signalling defects. J. Pediatr. Gastroenterol. Nutr. 2021;72:276–281. doi: 10.1097/MPG.0000000000002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muller PA, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gabanyi I, et al. Neuro-immune interactions drive tissue programming in intestinal macrophages. Cell. 2016;164:378–391. doi: 10.1016/j.cell.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.de Schepper S, et al. Self-maintaining gut macrophages are essential for intestinal homeostasis. Cell. 2018;175:400–415.e13. doi: 10.1016/j.cell.2018.07.048. [DOI] [PubMed] [Google Scholar]
- 102.Matheis F, et al. Adrenergic signaling in muscularis macrophages limits infection-induced neuronal loss. Cell. 2020;180:64–78.e16. doi: 10.1016/j.cell.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kubo A, Nagao K, Yokouchi M, Sasaki H, Amagai M. External antigen uptake by Langerhans cells with reorganization of epidermal tight junction barriers. J. Exp. Med. 2009;206:2937–2946. doi: 10.1084/jem.20091527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kitashima DY, et al. Langerhans cells prevent autoimmunity via expansion of keratinocyte antigen-specific regulatory T cells. eBioMedicine. 2018;27:293–303. doi: 10.1016/j.ebiom.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seneschal J, Clark RA, Gehad A, Baecher-Allan CM, Kupper TS. Human epidermal Langerhans cells maintain immune homeostasis in skin by activating skin resident regulatory T cells. Immunity. 2012;36:873–884. doi: 10.1016/j.immuni.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kobayashi T, Naik S, Nagao K. Choreographing immunity in the skin epithelial barrier. Immunity. 2019;50:552–565. doi: 10.1016/j.immuni.2019.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park S, et al. Skin-resident immune cells actively coordinate their distribution with epidermal cells during homeostasis. Nat. Cell Biol. 2021;23:476–484. doi: 10.1038/s41556-021-00670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hoeffel G, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J. Exp. Med. 2012;209:1167–1181. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chorro L, et al. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J. Exp. Med. 2009;206:3089–3100. doi: 10.1084/jem.20091586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ghigo C, et al. Multicolor fate mapping of Langerhans cell homeostasis. J. Exp. Med. 2013;210:1657–1664. doi: 10.1084/jem.20130403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Merad M, et al. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat. Immunol. 2002;3:1135–1141. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hovav AH. Mucosal and skin Langerhans cells — nurture calls. Trends Immunol. 2018;39:788–800. doi: 10.1016/j.it.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 113.Capucha T, et al. Distinct murine mucosal Langerhans cell subsets develop from pre-dendritic cells and monocytes. Immunity. 2015;43:369–381. doi: 10.1016/j.immuni.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 114.Sparber F, et al. Langerin+DCs regulate innate IL-17 production in the oral mucosa during Candida albicans-mediated infection. PLoS Pathog. 2018;14:e1007069. doi: 10.1371/journal.ppat.1007069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saba Y, et al. Early antitumor activity of oral Langerhans cells is compromised by a carcinogen. Proc. Natl Acad. Sci. USA. 2022;119:e2118424119. doi: 10.1073/pnas.2118424119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tamoutounour S, et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 2013;39:925–938. doi: 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 117.Kolter J, et al. A subset of skin macrophages contributes to the surveillance and regeneration of local nerves. Immunity. 2019;50:1482–1497.e7. doi: 10.1016/j.immuni.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 118.Abtin A, et al. Perivascular macrophages mediate neutrophil recruitment during bacterial skin infection. Nat. Immunol. 2013;15:45–53. doi: 10.1038/ni.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Knab K, Chambers D, Krönke G. Synovial macrophage and fibroblast heterogeneity in joint homeostasis and inflammation. Front. Med. 2022;9:930. doi: 10.3389/fmed.2022.862161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mogilenko DA, Shchukina I, Artyomov MN. Immune ageing at single-cell resolution. Nat. Rev. Immunol. 2021;22:484–498. doi: 10.1038/s41577-021-00646-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Selman M, Pardo A. Fibroageing: an ageing pathological feature driven by dysregulated extracellular matrix-cell mechanobiology. Ageing Res. Rev. 2021;70:101393. doi: 10.1016/j.arr.2021.101393. [DOI] [PubMed] [Google Scholar]
- 122.Birjandi SZ, Ippolito JA, Ramadorai AK, Witte PL. Alterations in marginal zone macrophages and marginal zone B cells in old mice. J. Immunol. 2011;186:3441–3451. doi: 10.4049/jimmunol.1001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guilliams M, Thierry GR, Bonnardel J, Bajenoff M. Establishment and maintenance of the macrophage niche. Immunity. 2020;52:434–451. doi: 10.1016/j.immuni.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 124.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stremmel C, et al. Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat. Commun. 2018;9:75. doi: 10.1038/s41467-017-02492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tan YL, Yuan Y, Tian L. Microglial regional heterogeneity and its role in the brain. Mol. Psychiatry. 2020;25:351–367. doi: 10.1038/s41380-019-0609-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang N, et al. LYVE1+ macrophages of murine peritoneal mesothelium promote omentum-independent ovarian tumor growth. J. Exp. Med. 2021;218:e20210924. doi: 10.1084/jem.20210924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou X, et al. Circuit design features of a stable two-cell system circuit design features of a stable two-cell system. Cell. 2018 doi: 10.1016/j.cell.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gschwend J, et al. Alveolar macrophages rely on GM-CSF from alveolar epithelial type 2 cells before and after birth. J. Exp. Med. 2021;218:e20210745. doi: 10.1084/jem.20210745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wolf Y, et al. Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nat. Immunol. 2017;18:665–674. doi: 10.1038/ni.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nicolás-Ávila JA, et al. A network of macrophages supports mitochondrial homeostasis in the heart. Cell. 2020;183:94–109.e23. doi: 10.1016/j.cell.2020.08.031. [DOI] [PubMed] [Google Scholar]
- 133.Brestoff JR, et al. Intercellular mitochondria transfer to macrophages regulates white adipose tissue homeostasis and is impaired in obesity. Cell Metab. 2021;33:270–282.e8. doi: 10.1016/j.cmet.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Scheiblich H, et al. Microglia jointly degrade fibrillar alpha-synuclein cargo by distribution through tunneling nanotubes. Cell. 2021;184:5089–5106.e21. doi: 10.1016/j.cell.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van der Vlist M, et al. Macrophages transfer mitochondria to sensory neurons to resolve inflammatory pain. Neuron. 2022;110:613–629.e9. doi: 10.1016/j.neuron.2021.11.020. [DOI] [PubMed] [Google Scholar]
- 136.Ramachandran P, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Aran D, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gordan S, Biburger M, Nimmerjahn F. bIgG time for large eaters: monocytes and macrophages as effector and target cells of antibody-mediated immune activation and repression. Immunol. Rev. 2015;268:52–65. doi: 10.1111/imr.12347. [DOI] [PubMed] [Google Scholar]
- 139.Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. The function of Fcγ receptors in dendritic cells and macrophages. Nat. Rev. Immunol. 2014;14:94–108. doi: 10.1038/nri3582. [DOI] [PubMed] [Google Scholar]
- 140.Gordon S, Plüddemann A. Tissue macrophages: heterogeneity and functions. BMC Biol. 2017;15:1–18. doi: 10.1186/s12915-017-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Loos M, Martin H, Petry F. The biosynthesis of C1q, the collagen-like and Fc-recognizing molecule of the complement system. Behring Inst. Mitt. 1989;84:32–41. [PubMed] [Google Scholar]
- 142.Hussell T. Heterologous immunity meets tissue-specific training. Nat. Rev. Immunol. 2016;16:275–275. doi: 10.1038/nri.2016.41. [DOI] [PubMed] [Google Scholar]
- 143.Machiels B, et al. A gammaherpesvirus provides protection against allergic asthma by inducing the replacement of resident alveolar macrophages with regulatory monocytes. Nat. Immunol. 2017;18:1310–1320. doi: 10.1038/ni.3857. [DOI] [PubMed] [Google Scholar]
- 144.Zahalka S, et al. Trained immunity of alveolar macrophages requires metabolic rewiring and type 1 interferon signaling. Mucosal Immunol. 2022;15:896–907. doi: 10.1038/s41385-022-00528-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yao Y, et al. Induction of autonomous memory alveolar macrophages requires T cell help and is critical to trained immunity. Cell. 2018;175:1634–1650.e17. doi: 10.1016/j.cell.2018.09.042. [DOI] [PubMed] [Google Scholar]
- 146.Aegerter H, et al. Influenza-induced monocyte-derived alveolar macrophages confer prolonged antibacterial protection. Nat. Immunol. 2020;21:145–157. doi: 10.1038/s41590-019-0568-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li F, et al. Monocyte-derived alveolar macrophages autonomously determine severe outcome of respiratory viral infection. Sci. Immunol. 2022;7:eabj5761. doi: 10.1126/sciimmunol.abj5761. [DOI] [PubMed] [Google Scholar]
- 148.Ahrends T, et al. Enteric pathogens induce tissue tolerance and prevent neuronal loss from subsequent infections. Cell. 2021;184:5715–5727.e12. doi: 10.1016/j.cell.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jaitin DA, et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell. 2019;178:686–698.e14. doi: 10.1016/j.cell.2019.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhou X, et al. Microenvironmental sensing by fibroblasts controls macrophage population size. Proc. Natl Acad. Sci. USA. 2022;119:e2205360119. doi: 10.1073/pnas.2205360119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wardemann H, Nussenzweig MC. B-cell self-tolerance in humans. Adv. Immunol. 2007;95:83–110. doi: 10.1016/S0065-2776(07)95003-8. [DOI] [PubMed] [Google Scholar]
- 152.Vorsatz C, Friedrich N, Nimmerjahn F, Biburger M. There is strength in numbers: quantitation of Fc gamma receptors on murine tissue-resident macrophages. Int. J. Mol. Sci. 2021;22:12172. doi: 10.3390/ijms222212172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Licht R, Dieker JWC, Jacobs CWM, Tax WJM, Berden JHM. Decreased phagocytosis of apoptotic cells in diseased SLE mice. J. Autoimmun. 2004;22:139–145. doi: 10.1016/j.jaut.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 154.Poon IKH, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat. Rev. Immunol. 2014;14:166–180. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yamasaki S, et al. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat. Immunol. 2008;9:1179–1188. doi: 10.1038/ni.1651. [DOI] [PubMed] [Google Scholar]
- 156.Brown GD. Sensing necrosis with Mincle. Nat. Immunol. 2008;9:1099–1100. doi: 10.1038/ni1008-1099. [DOI] [PubMed] [Google Scholar]
- 157.Culemann S, et al. Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature. 2019;572:670–675. doi: 10.1038/s41586-019-1471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sjef Verbeek J, Hirose S, Nishimura H. The complex association of FcγRIIb with autoimmune susceptibility. Front. Immunol. 2019;10:2061. doi: 10.3389/fimmu.2019.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Smith KGC, Clatworthy MR. FcγRIIB in autoimmunity and infection: evolutionary and therapeutic implications. Nat. Rev. Immunol. 2010;10:328–343. doi: 10.1038/nri2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu H, et al. The FGL2-FcgammaRIIB pathway: a novel mechanism leading to immunosuppression. Eur. J. Immunol. 2008;38:3114–3126. doi: 10.1002/eji.200838338. [DOI] [PubMed] [Google Scholar]
- 161.Bennett FC, et al. A combination of ontogeny and CNS environment establishes microglial identity. Neuron. 2018 doi: 10.1016/j.neuron.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li F, Okreglicka KM, Pohlmeier LM, Schneider C, Kopf M. Fetal monocytes possess increased metabolic capacity and replace primitive macrophages in tissue macrophage development. EMBO J. 2020;39:e103205. doi: 10.15252/embj.2019103205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.van de Laar L, et al. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity. 2016;44:755–768. doi: 10.1016/j.immuni.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 164.Hsu TC, Ku KY, Shen HC, Yu HH. Overview of MARCM-related technologies in Drosophila neurobiological research. Curr. Protoc. Neurosci. 2020;91:e90. doi: 10.1002/cpns.90. [DOI] [PubMed] [Google Scholar]
- 165.Suo C, et al. Mapping the developing human immune system across organs. Science. 2022;376:eabo0510. doi: 10.1126/science.abo0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Garcia-Alonso L, et al. Single-cell roadmap of human gonadal development. Nature. 2022;607:540–547. doi: 10.1038/s41586-022-04918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bian Z, et al. Deciphering human macrophage development at single-cell resolution. Nature. 2020;582:571–576. doi: 10.1038/s41586-020-2316-7. [DOI] [PubMed] [Google Scholar]
- 168.Popescu DM, et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Thion MS, et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell. 2018;172:500–516.e16. doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ben-Yehuda H, et al. Maternal type-I interferon signaling adversely affects the microglia and the behavior of the offspring accompanied by increased sensitivity to stress. Mol. Psychiatry. 2019 doi: 10.1038/s41380-019-0604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Matcovitch-Natan O, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- 172.Rosshart SP, et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. 2019;365:eaaw4361. doi: 10.1126/science.aaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rosshart SP, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. 2017;171:1015–1028.e13. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zaman R, Epelman S. Resident cardiac macrophages: heterogeneity and function in health and disease. Immunity. 2022;55:1549–1563. doi: 10.1016/j.immuni.2022.08.009. [DOI] [PubMed] [Google Scholar]
- 176.Lavin Y, Mortha A, Rahman A, Merad M. Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 2015;15:731–744. doi: 10.1038/nri3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Park MD, Silvin A, Ginhoux F, Merad M. Macrophages in health and disease. Cell. 2022;185:4259–4279. doi: 10.1016/j.cell.2022.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nobs SP, Kopf M. Tissue-resident macrophages: guardians of organ homeostasis. Trends Immunol. 2021;42:495–507. doi: 10.1016/j.it.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 179.Chakarov S, Blériot C, Ginhoux F. Role of adipose tissue macrophages in obesity-related disorders. J. Exp. Med. 2022;219:e20211948. doi: 10.1084/jem.20211948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chang Z-L. Recent development of the mononuclear phagocyte system: in memory of Metchnikoff and Ehrlich on the 100th Anniversary of the 1908 Nobel Prize in Physiology or Medicine. Biol. Cell. 2009;101:709–721. doi: 10.1042/BC20080227. [DOI] [PubMed] [Google Scholar]
- 181.Dionne MS, Schneider DS. Models of infectious diseases in the fruit fly Drosophila melanogaster. Dis. Model. Mech. 2008;1:43–49. doi: 10.1242/dmm.000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster — from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 2014;14:796–810. doi: 10.1038/nri3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Stuart LM, Ezekowitz RA. Phagocytosis and comparative innate immunity: learning on the fly. Nat. Rev. Immunol. 2008;8:131–141. doi: 10.1038/nri2240. [DOI] [PubMed] [Google Scholar]
- 184.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 185.Kierdorf K, Dionne MS. The software and hardware of macrophages: a diversity of options. Dev. Cell. 2016;38:122–125. doi: 10.1016/j.devcel.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 186.Gold KS, Brückner K. Macrophages and cellular immunity in Drosophila melanogaster. Semin. Immunol. 2015;27:357–368. doi: 10.1016/j.smim.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Makhijani K, Alexander B, Tanaka T, Rulifson E, Brückner K. The peripheral nervous system supports blood cell homing and survival in the Drosophila larva. Development. 2011;138:5379–5391. doi: 10.1242/dev.067322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev. Biol. 2001;230:243–257. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- 189.Mase A, Augsburger J, Brückner K. Macrophages and their organ locations shape each other in development and homeostasis — a Drosophila perspective. Front. Cell Dev. Biol. 2021;9:630272. doi: 10.3389/fcell.2021.630272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cattenoz PB, et al. Temporal specificity and heterogeneity of Drosophila immune cells. EMBO J. 2020;39:e104486. doi: 10.15252/embj.2020104486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tattikota, S. G. et al. A single-cell survey of Drosophila blood. eLife9, 1–35, e54818 (2020). [DOI] [PMC free article] [PubMed]
- 192.Cho B, et al. Single-cell transcriptome maps of myeloid blood cell lineages in Drosophila. Nat. Commun. 2020;11:4483. doi: 10.1038/s41467-020-18135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.H L, et al. Fly Cell Atlas: a single-nucleus transcriptomic atlas of the adult fruit fly. Science. 2022;375:eabk2432. doi: 10.1126/science.abk2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Woodcock KJ, et al. Macrophage-derived upd3 cytokine causes impaired glucose homeostasis and reduced lifespan in Drosophila fed a lipid-rich diet. Immunity. 2015;42:133–144. doi: 10.1016/j.immuni.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cox N, et al. Diet-regulated production of PDGFcc by macrophages controls energy storage. Science. 2021;373:eabe9383. doi: 10.1126/science.abe9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Danzer, H. et al. Human Fcγ-receptor IIb modulates pathogen-specific versus self-reactive antibody responses in lyme arthritis. eLife9, 1–53, e55319 (2020). [DOI] [PMC free article] [PubMed]
- 197.Rongvaux A, et al. Development and function of human innate immune cells in a humanized mouse model. Nat. Biotechnol. 2014;32:364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wege AK, et al. IL-15 enhances the anti-tumor activity of trastuzumab against breast cancer cells but causes fatal side effects in humanized tumor mice (HTM) Oncotarget. 2017;8:2731–2744. doi: 10.18632/oncotarget.13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Evren E, et al. Distinct developmental pathways from blood monocytes generate human lung macrophage diversity. Immunity. 2021;54:259–275.e7. doi: 10.1016/j.immuni.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 200.Evren E, et al. CD116+ fetal precursors migrate to the perinatal lung and give rise to human alveolar macrophages. J. Exp. Med. 2022;219:e20210987. doi: 10.1084/jem.20210987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Keshvari S, et al. CSF1R-dependent macrophages control postnatal somatic growth and organ maturation. PLoS Genet. 2021;17:e1009605. doi: 10.1371/journal.pgen.1009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pridans C, et al. Pleiotropic impacts of macrophage and microglial deficiency on development in rats with targeted mutation of the CSF1R locus. J. Immunol. 2018;201:2683–2699. doi: 10.4049/jimmunol.1701783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dai XM, et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.V99.1.111. [DOI] [PubMed] [Google Scholar]
- 204.Rojo R, et al. Deletion of a CSF1R enhancer selectively impacts CSF1R expression and development of tissue macrophage populations. Nat. Commun. 2019;10:3215. doi: 10.1038/s41467-019-11053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hume DA, et al. Phenotypic impacts of CSF1R deficiencies in humans and model organisms. J. Leukoc. Biol. 2020;107:205–219. doi: 10.1002/JLB.MR0519-143R. [DOI] [PubMed] [Google Scholar]
- 206.Monies D, et al. Autozygosity reveals recessive mutations and novel mechanisms in dominant genes: implications in variant interpretation. Genet. Med. 2017;19:1144–1150. doi: 10.1038/gim.2017.22. [DOI] [PubMed] [Google Scholar]
- 207.Oosterhof N, et al. Homozygous mutations in CSF1R cause a pediatric-onset leukoencephalopathy and can result in congenital absence of microglia. Am. J. Hum. Genet. 2019;104:936–947. doi: 10.1016/j.ajhg.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Guo L, et al. Bi-allelic CSF1R mutations cause skeletal dysplasia of dysosteosclerosis-pyle disease spectrum and degenerative encephalopathy with brain malformation. Am. J. Hum. Genet. 2019;104:925–935. doi: 10.1016/j.ajhg.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Rademakers R, et al. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat. Genet. 2011;44:200–205. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Konno T, Kasanuki K, Ikeuchi T, Dickson DW, Wszolek ZK. CSF1R-related leukoencephalopathy: a major player in primary microgliopathies. Neurology. 2018;91:1092–1104. doi: 10.1212/WNL.0000000000006642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bissinger S, et al. Macrophage depletion induces edema through release of matrix-degrading proteases and proteoglycan deposition. Sci. Transl. Med. 2021;13:eabd4550. doi: 10.1126/scitranslmed.abd4550. [DOI] [PubMed] [Google Scholar]
- 212.Kawamura T, et al. Severe dermatitis with loss of epidermal Langerhans cells in human and mouse zinc deficiency. J. Clin. Invest. 2012;122:722–732. doi: 10.1172/JCI58618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Suzuki T, Trapnell BC. Pulmonary alveolar proteinosis syndrome. Clin. Chest Med. 2016;37:431–440. doi: 10.1016/j.ccm.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cao J, et al. A human cell atlas of fetal gene expression. Science. 2020;370:eaba7721. doi: 10.1126/science.aba7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Li Q, et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 2019;101:207–223.e10. doi: 10.1016/j.neuron.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Masuda T, et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019;566:388–392. doi: 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- 217.Hammond TR, et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2018;50:253–271.e6. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 219.Nguyen AT, et al. APOE and TREM2 regulate amyloid-responsive microglia in Alzheimer’s disease. Acta Neuropathol. 2020;140:477–493. doi: 10.1007/s00401-020-02200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Krasemann S, et al. The TREM2–APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47:566–581.e9. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wang S, et al. TREM2 drives microglia response to amyloid-β via SYK-dependent and -independent pathways. Cell. 2022;185:4153–4169.e19. doi: 10.1016/j.cell.2022.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hou J, Chen Y, Grajales-Reyes G, Colonna M. TREM2 dependent and independent functions of microglia in Alzheimer’s disease. Mol. Neurodegener. 2022;17:84. doi: 10.1186/s13024-022-00588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Guerreiro R, et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jordão MJC, et al. Neuroimmunology: single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363:eaat7554. doi: 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- 226.Drieu A, et al. Parenchymal border macrophages regulate the flow dynamics of the cerebrospinal fluid. Nature. 2022;611:585–593. doi: 10.1038/s41586-022-05397-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Munro DAD, et al. CNS macrophages differentially rely on an intronic Csf1r enhancer for their development. Development. 2020;147:dev194449. doi: 10.1242/dev.194449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chen Y, Colonna M. Microglia in Alzheimer’s disease at single-cell level. Are there common patterns in humans and mice? J. Exp. Med. 2021;218:e20202717. doi: 10.1084/jem.20202717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bain CC, et al. CD11c identifies microbiota and EGR2-dependent MHCII+ serous cavity macrophages with sexually dimorphic fate in mice. Eur. J. Immunol. 2022;52:1243–1257. doi: 10.1002/eji.202149756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ostendorf BN, et al. Common human genetic variants of APOE impact murine COVID-19 mortality. Nature. 2022;611:346–351. doi: 10.1038/s41586-022-05344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chadarevian JP, et al. Engineering an inhibitor-resistant human CSF1R variant for microglia replacement. J. Exp. Med. 2023;220:20220857. doi: 10.1084/jem.20220857. [DOI] [PMC free article] [PubMed] [Google Scholar]