Abstract
Background:
Whether the positive associations of smoking and alcohol consumption with gastrointestinal diseases are causal is uncertain. We conducted this Mendelian randomization (MR) to comprehensively examine associations of smoking and alcohol consumption with common gastrointestinal diseases.
Methods:
Genetic variants associated with smoking initiation and alcohol consumption at the genome-wide significance level were selected as instrumental variables. Genetic associations with 24 gastrointestinal diseases were obtained from the UK Biobank, FinnGen study, and other large consortia. Univariable and multivariable MR analyses were conducted to estimate the overall and independent MR associations after mutual adjustment for genetic liability to smoking and alcohol consumption.
Results:
Genetic predisposition to smoking initiation was associated with increased risk of 20 of 24 gastrointestinal diseases, including 7 upper gastrointestinal diseases (gastroesophageal reflux, esophageal cancer, gastric ulcer, duodenal ulcer, acute gastritis, chronic gastritis, and gastric cancer), 4 lower gastrointestinal diseases (irritable bowel syndrome, diverticular disease, Crohn’s disease, and ulcerative colitis), 8 hepatobiliary and pancreatic diseases (non-alcoholic fatty liver disease, alcoholic liver disease, cirrhosis, liver cancer, cholecystitis, cholelithiasis, and acute and chronic pancreatitis), and acute appendicitis. Fifteen out of 20 associations persisted after adjusting for genetically predicted alcohol consumption. Genetically predicted higher alcohol consumption was associated with increased risk of duodenal ulcer, alcoholic liver disease, cirrhosis, and chronic pancreatitis; however, the association for duodenal ulcer did not remain statistically significant after adjustment for genetic predisposition to smoking initiation.
Conclusions:
This study provides MR evidence supporting causal associations of smoking with a broad range of gastrointestinal diseases, whereas alcohol consumption was associated with only a few gastrointestinal diseases.
Funding:
The Natural Science Fund for Distinguished Young Scholars of Zhejiang Province; National Natural Science Foundation of China; Key Project of Research and Development Plan of Hunan Province; the Swedish Heart Lung Foundation; the Swedish Research Council; the Swedish Cancer Society.
Research organism: None
eLife digest
People who smoke cigarettes or drink large amounts of alcohol are more likely to develop disorders with their digestive system. But it is difficult to prove that heavy drinking or smoking is the primary cause of these gastrointestinal diseases.
For example, it is possible that having a digestive disorder makes people more likely to take up these habits to reduce pain or discomfort caused by the illness (an effect known as reverse causation). The association may also be the result of confounding factors, such as age or diet, which contribute to digestive problems as well as the health outcomes of smoking and drinking. Additionally, many people who smoke also drink alcohol and vice versa, making it challenging to determine if one or both behaviors contribute to the disease.
One solution is to employ Mendelian randomization which uses genetics to determine if two variables are linked. Using this statistical approach, Yuan, Chen, Ruan et al. investigated if people who display genetic variants that predispose someone to becoming a smoker or drinker are at greater risk of developing certain digestive disorders. This reduces the possibility of confounding and reverse causation, as any association between genetic variants will have been present since birth, and will have not been impacted by external factors.
Yuan, Chen, Ruan et al. used data from two studies that had collected the genetic and health information of thousands of people living in the United Kingdom or Finland. The analyses revealed that genetic variants associated with cigarette smoking increase the risk of 20 of the 24 gastrointestinal diseases investigated. This risk persisted for most of the disorders, even after adjusting for genes linked with alcohol consumption.
Further analysis showed that genetic variants linked to heavy drinking increase the risk of duodenal ulcer, alcoholic liver disease, cirrhosis, and chronic pancreatitis. However, accounting for smoking-linked genes eliminated the relationship with duodenal ulcer.
These findings suggest that smoking has detrimental effects on gastrointestinal health. Reducing the number of people who start smoking or encouraging smokers to quit may help prevent digestive diseases. Even though there were fewer associations between heavy alcohol consumption and gastrointestinal illness, further studies are needed to investigate this relationship in more depth.
Introduction
Tobacco smoking and alcohol consumption are leading causes of the global burden of disease and are major contributors to premature mortality (GBD 2016 Alcohol Collaborators, 2018; GBD 2016 Alcohol Collaborators, 2020). Gastrointestinal diseases account for considerable health care use and expenditures, and a holistic approach to lifestyle interventions may result in more health gains and less economic burdens (Peery et al., 2022). Population-based studies have identified tobacco smoking as a risk factor for several gastrointestinal diseases, including gastroesophageal reflux disease (Eusebi et al., 2018), esophageal cancer (Fund WCR and Research AIfC, 2007), Crohn’s disease (Piovani et al., 2019), liver cancer (McGee et al., 2019), and pancreatitis (Yadav and Whitcomb, 2010). Evidence on the association between tobacco smoking and risk of other gastrointestinal diseases is limited and inconsistent. Like smoking, heavy alcohol consumption has been associated with increased risk of several gastrointestinal outcomes, including gastritis (Bujanda, 2000), gastric cancer (Laszkowska et al., 2021), colorectal cancer (McNabb et al., 2020), cirrhosis (Simpson et al., 2019), liver cancer (McGee et al., 2019), and pancreatitis (Yadav and Whitcomb, 2010). However, whether these associations are all causal remains unestablished, since most of the evidence was obtained from observational studies where the results may be biased by reverse causality and confounding. Of note, even though reverse causality may not be an issue in the studies for any of studied gastroenterological outcomes, it might exist for certain gastroenterological diseases causing pain, which smoker patients may try to increase smoking dose to mitigate via an intake of higher levels of nicotine. In addition, as smoking and alcohol consumption are phenotypically and genetically correlated (Roberts et al., 2020; Liu et al., 2019), the independent impacts of smoking and alcohol consumption on gastrointestinal diseases are unclear. Establishing the causal association of tobacco smoking and alcohol consumption with gastrointestinal diseases is crucial, as this provides further evidence for subsequent recommending public policies and clinical interventions.
Mendelian randomization (MR) is an epidemiological approach that utilizes genetic variants as an instrument to strengthen the causal inference in an exposure-outcome association (Davey Smith and Hemani, 2014). MR is by nature not prone to confounding since genetic variants are randomly assorted at conception and thus unrelated to environmental and self-adopted factors that usually act as confounders. Additionally, this method can minimize reverse causality since fixed alleles are unaffected by the onset and progression of disease. Previous MR studies have examined the associations of smoking and alcohol consumption with several gastrointestinal diseases (Yuan and Larsson, 2022a; Larsson et al., 2020; Yuan and Larsson, 2022b; Yuan et al., 2022c; Chen et al., 2022; Yuan et al., 2021). Nevertheless, whether smoking and alcohol consumption exert influence on a wide range of gastrointestinal outcomes has not been investigated in a comprehensive way. A thorough investigation on the gastrointestinal consequences of smoking and alcohol drinking is of great importance to develop non-pharmacological interventions on gastrointestinal diseases. Here, we conducted an MR investigation of the associations of smoking and alcohol consumption with the risk of common gastrointestinal diseases to fill in above knowledge gaps.
Materials and methods
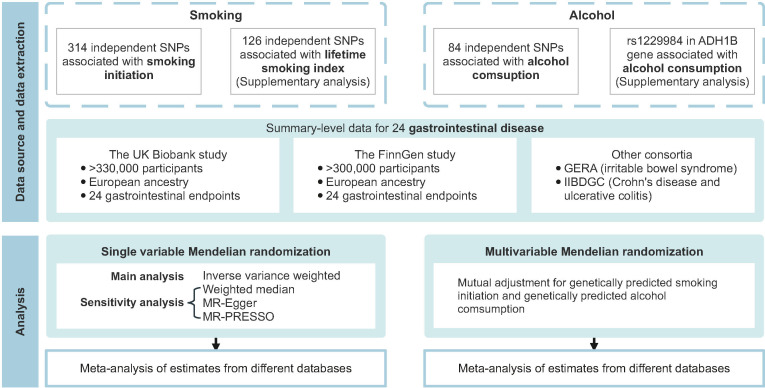
Figure 1 shows the study design overview. The study was based on publicly available genome-wide association studies (GWAS), and the detailed information on used studies was presented in Supplementary file 1A. The genetic associations were estimated using data from the UK Biobank study (Sudlow et al., 2015), the FinnGen study (Kurki et al., 2022; https://www.finngen.fi/en), and several large consortia. The summary effect estimates were combined using meta-analysis for each gastrointestinal disease from different data resources. Included studies had been approved by corresponding institutional review boards and ethical committees, and consent forms had been signed by all participants.
Figure 1. Overview of the present study design.
GERA, Genetic Epidemiology Research on Aging; IIBDGC, the International Inflammatory Bowel Disease Genetics Consortium; MR, Mendelian randomization; MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier; SNP, single nucleotide polymorphism.
Instrumental variable selection
A total of 378 and 99 single nucleotide polymorphisms (SNPs) associated with smoking initiation (a binary phenotype indicating whether an individual had ever being a regular smoker, 1,232,091 individuals of European descent) and alcohol consumption (log-transformed drinks per week, 941,280 individuals of European descent) at the genome-wide significance threshold (p<5 × 10–8) were identified by the GWAS and Sequencing Consortium of Alcohol and Nicotine use (GSCAN) study (Liu et al., 2019). These SNPs explained approximately 2.3 and 0.3% of the variation in smoking initiation and alcohol consumption, respectively (Liu et al., 2019). SNPs in linkage disequilibrium (defined as r2 >0.01 or clump distance <10,000 kb) and with the weaker associations with the exposure were removed, leaving 314 independent SNPs as instrumental variables for smoking initiation and 84 for alcohol consumption. Smoking initiation and alcohol consumption shared two index genetic variants, which were rs1713676 and rs11692435. Considering partial sample overlap (around 30%) between the GSCAN study with full data and the UK Biobank study (Liu et al., 2019), we performed sensitivity analyses for smoking initiation and alcohol consumption using summary statistics data from the analysis excluding the UK Biobank and 23andMe. For a supplementary analysis of smoking behavior, we used 126 SNPs associated with a lifetime smoking index that considered smoking duration, heaviness, and cessation (Wootton et al., 2020). The set of genetic instruments captured around 0.36% of the variance in lifetime smoking (Wootton et al., 2020). We also conducted a sensitivity analysis using rs1229984 in ADH1B gene that encodes alcohol dehydrogenase 1B enzyme as the genetic instrument for alcohol consumption to minimize pleiotropy. Detailed information on used SNPs is presented in Supplementary file 1B.
Gastrointestinal disease data sources
Genetic associations with 24 gastrointestinal diseases were obtained from the UK Biobank study (Sudlow et al., 2015), the FinnGen study (Kurki et al., 2022), and two large consortia, including the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) (Liu et al., 2015) and Genetic Epidemiology Research on Aging (GERA) (Guindo-Martínez et al., 2021). Included outcomes were classified into four major categories according to the disease onset site: (1) upper gastrointestinal diseases (gastroesophageal reflux disease, esophageal cancer, gastric ulcer, acute gastritis, chronic gastritis, and gastric cancer); (2) lower gastrointestinal diseases (irritable bowel disease, celiac disease, diverticular disease, Crohn’s disease, ulcerative colitis, and colorectal cancer); (3) hepatobiliary and pancreatic diseases (non-alcoholic fatty liver disease, alcoholic liver disease, cirrhosis, liver cancer, cholangitis, cholecystitis, cholelithiasis, acute pancreatitis, chronic pancreatitis, and pancreatic cancer); and (4) other (acute appendicitis).
The UK Biobank study is a large multicenter cohort study of 500,000 participants recruited in the United Kingdom between 2006 and 2010 (Sudlow et al., 2015). We used the summary statistics of European ancestry from GWAS conducted by Lee lab, where the gastrointestinal outcomes were defined by codes of the International Classification of Diseases 9th Revision (ICD-9) and ICD-10 (Zhou et al., 2020). Genetic associations were estimated by logistic regression with adjustment for sex, birth year, and the first four genetic principal components. For the FinnGen study, we used summary-level data on the genetic associations with gastrointestinal diseases from the last publicly available R7 data release (Kurki et al., 2022). The FinnGen study is a nationwide genetic study where genetic and electronic health record data were collected. The gastrointestinal diseases were ascertained by the codes of the ICD-8, ICD-9, and ICD-10. Genome-wide association analyses were adjusted for sex, age, genetic components, and genotyping batch. Summary-level genetic data on Crohn’s disease (5956 cases and 14,927 controls) and ulcerative colitis (6968 cases and 20,464 controls) were additionally obtained from the IIBDGC (Liu et al., 2015), and data on irritable bowel syndrome (3117 cases and 53,520 controls) were obtained from the GERA (Guindo-Martínez et al., 2021). Detailed diagnostic codes are listed in Supplementary file 1C.
Statistical analysis
Data were harmonized to omit ambiguous SNPs with non-concordant alleles and palindromic SNPs with ambiguous minor allele frequency (>0.42 and <0.58) were removed from the analysis. The primary MR analyses were performed by the multiplicative random-effects inverse-variance weighted (IVW) method, which provides the most precise estimates though assuming that all SNPs are valid instruments. The analysis of rs1229984 for alcohol consumption was conducted by the Wald method. Estimates for each association from different sources were combined using fixed-effects meta-analysis, and the heterogeneity of the associations from different data sources was evaluated by the I2 statistic. Heterogeneity among SNPs’ estimates in each association was assessed by Cochran’s Q value. Multivariable MR analyses were conducted to mutually adjust for smoking initiation and alcohol consumption. To detect potential unbalanced pleiotropy (horizontal pleiotropy) and examine the consistency of the associations, three sensitivity analyses including the weighted median (Yavorska and Burgess, 2017), MR-Egger (Burgess and Thompson, 2017), and MR pleiotropy residual sum and outlier (MR-PRESSO) (Verbanck et al., 2018) analyses were performed. The weighted median method can provide consistent estimates when more than 50% of the weight comes from valid instrument variants (Yavorska and Burgess, 2017). The MR-Egger intercept test can detect unmeasured pleiotropy, and MR-Egger regression can generate estimates after accounting for horizontal pleiotropy albeit with less precision (Burgess and Thompson, 2017). The MR-PRESSO method can identify SNP outliers and provide results identical to that from IVW after removal of outliers (Verbanck et al., 2018). The F-statistic was estimated to quantify instrument strength, and an F-statistic >10 suggested a sufficiently strong instrument. Power analysis was performed using an online tool (Brion et al., 2013). The Benjamini-Hochberg correction that controls the false discovery rate was applied to correct for multiple testing. The association with a nominal p-value <0.05 but Benjamini-Hochberg adjusted p-value >0.05 was regarded suggestive, and the association with a Benjamini-Hochberg adjusted p-value <0.05 was deemed significant. All analyses were two-sided and performed using the TwoSampleMR (Hemani et al., 2018), MendelianRandomization (Yavorska and Burgess, 2017), and MRPRESSO R packages (Verbanck et al., 2018) in R software 4.1.2.
Results
The F-statistic for each genetic variant was above 10, suggesting a good strength of used genetic instruments (Supplementary file 1B). Most associations were well powered (Supplementary file 1D). For smoking initiation, there was 80% power to detect the smallest odds ratio (OR) ranging from 1.08 to 1.40 for included outcomes. Although power was lower for alcohol consumption, it was adequate to detect a moderate effect size for most common gastrointestinal diseases.
Smoking and gastrointestinal diseases
Genetic predisposition to smoking initiation was associated with 20 of the 24 studied gastrointestinal diseases, and all these associations remained after multiple comparison correction (Table 1 and Supplementary file 1E). In detail, genetic liability to smoking initiation was positively associated with seven upper gastrointestinal diseases: gastroesophageal reflux (OR, 1.28; 95% confidence interval [CI], 1.20–1.37; p=4.09 × 10−14), esophageal cancer (OR, 1.67; 95% CI, 1.24–2.25; p=6.84 × 10−4), gastric ulcer (OR, 1.54; 95% CI, 1.37–1.72; p=3.83 × 10−14), duodenal ulcer (OR, 1.53; 95% CI, 1.34–1.75; p=8.47 × 10−10), acute gastritis (OR, 1.29; 95% CI, 1.09–1.53; p=0.003), chronic gastritis (OR, 1.33; 95% CI, 1.18–1.49; p=1.55 × 10–6), and gastric cancer (OR, 1.42; 95% CI, 1.13–1.79; p=0.003); genetic liability to smoking initiation was positively associated with four lower gastrointestinal diseases: irritable bowel syndrome (OR, 1.22; 95% CI, 1.12–1.32; p=3.50 × 10−6), diverticular disease (OR, 1.25; 95% CI, 1.18–1.33; p=5.23 × 10−14), Crohn’s disease (OR, 1.25; 95% CI, 1.11–1.40; p=3.03 × 10−4), and ulcerative colitis (OR, 1.15; 95% CI, 1.04–1.26; p=0.004); genetic liability to smoking initiation was positively associated with eight hepatobiliary and pancreatic diseases: non-alcoholic fatty liver disease (OR, 1.49; 95% CI, 1.26–1.76; p=3.82 × 10−6), alcoholic liver disease (OR, 1.99; 95% CI, 1.65–2.41; p=1.49 × 10−12), cirrhosis (OR, 1.68; 95% CI, 1.40–2.02; p=3.39 × 10−8), liver cancer (OR, 1.57; 95% CI, 1.13–2.17; p=0.007), cholecystitis (OR, 1.47; 95% CI, 1.29–1.68; p=4.71 × 10−9), cholelithiasis (OR, 1.20; 95% CI, 1.13–1.27; p=5.75 × 10−9), acute pancreatitis (OR, 1.39; 95% CI, 1.23–1.56; p=6.71 × 10−8), and chronic pancreatitis (OR, 1.38; 95% CI, 1.17–1.64; p=1.79 × 10−4); genetic liability to smoking initiation was positively associated with acute appendicitis (OR, 1.15; 95% CI, 1.08–1.23; p=1.27 × 10−5). Results were consistent in sensitivity analyses. An indication of horizontal pleiotropy was observed in the analysis of esophageal cancer in the FinnGen study (p for MR-Egger intercept <0.05, Supplementary file 1F). Although MR-PRESSO detected one to three outliers, the associations persisted and remained significant after removal of these out-lying SNPs (Supplementary file 1F). When using the genetic variants for smoking initiation based on data without the UK Biobank and 23andMe studies, the associations attenuated slightly albeit remained significant after multiple comparisons (Supplementary file 1L and Supplementary file 1G). All associations were replicated in the supplementary analysis of the lifetime smoking index (Supplementary file 1G). After correcting for multiple testing, genetically predicted lifetime smoking index was significantly associated with 17 of 24 gastrointestinal diseases, where the patterns of associations were generally similar to the analysis for smoking initiation (Supplementary file 1M and Supplementary file 1G). In distinction to the analysis of smoking initiation, genetically predicted lifetime smoking index was not significantly associated with acute gastritis, gastric cancer, Crohn’s disease, and ulcerative colitis, whereas genetically predicted lifetime smoking index was significantly associated with pancreatic cancer (OR, 2.09; 95% CI, 1.30–3.36).
Table 1. Associations of genetic predisposition to smoking initiation with 24 gastrointestinal diseases in univariable and multivariable Mendelian randomization analyses.
| Disease | Total cases | Total controls | UVMR | MVMR adjusted for alcohol consumption | |||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p Value | I2 (95% CI) | OR (95% CI) | p Value | |||||
| Upper gastrointestinal diseases | Gastroesophageal reflux | 34,135 | 634,629 | 1.28 (1.20, 1.37) | 4.09 × 10-14* | 46.24 | 1.65 (1.35, 2.02) | 1.38 × 10-6* | |
| Esophageal cancer | 1130 | 702,116 | 1.67 (1.24, 2.25) | 6.84 × 10-4* | 22.68 | 4.78 (2.10, 10.90) | 1.97 × 10-4* | ||
| Gastric ulcer | 8651 | 666,879 | 1.54 (1.37, 1.72) | 3.83 × 10-14* | 44.96 | 1.95 (1.40, 2.71) | 7.31 × 10-5* | ||
| Duodenal ulcer | 5713 | 666,879 | 1.53 (1.34, 1.75) | 8.47 × 10-10* | 0.00 | 1.64 (1.07, 2.52) | 0.024 | ||
| Acute gastritis | 3048 | 643,478 | 1.29 (1.09, 1.53) | 0.003* | 0.00 | 1.54 (0.91, 2.62) | 0.106 | ||
| Chronic gastritis | 7975 | 643,478 | 1.33 (1.18, 1.49) | 1.55 × 10-6* | 77.04 | 1.33 (0.96, 1.86) | 0.091 | ||
| Gastric cancer | 1608 | 701,472 | 1.42 (1.13, 1.79) | 0.003* | 0.00 | 2.29 (1.14, 4.59) | 0.020 | ||
| Lower gastrointestinal diseases | Irritable bowel disease | 15,718 | 641,489 | 1.22 (1.12, 1.32) | 3.50 × 10-6* | 11.84 | 1.43 (1.10, 1.85) | 0.008* | |
| Celiac disease | 4808 | 631,700 | 0.82 (0.66, 1.02) | 0.071 | 0.00 | 0.87 (0.53, 1.43) | 0.590 | ||
| Diverticular disease | 50,065 | 587,969 | 1.25 (1.18, 1.33) | 5.23 × 10-14* | 67.29 | 1.56 (1.30, 1.87) | 1.41 × 10-6* | ||
| Crohn’s disease | 10,846 | 645,718 | 1.25 (1.11, 1.40) | 3.03 × 10-4* | 0.00 | 1.48 (1.01, 2.16) | 0.042 | ||
| Ulcerative colitis | 16,770 | 651,255 | 1.15 (1.04, 1.26) | 0.004* | 0.00 | 0.94 (0.71, 1.25) | 0.677 | ||
| Colorectal cancer | 9519 | 686,953 | 1.03 (0.92, 1.14) | 0.632 | 29.94 | 1.03 (0.76, 1.39) | 0.841 | ||
| Hepatobiliary and pancreatic diseases | Non-alcoholic fatty liver disease | 3242 | 707,631 | 1.49 (1.26, 1.76) | 3.82 × 10-6* | 0.00 | 2.11 (1.15, 3.88) | 0.016* | |
| Alcoholic liver disease | 2955 | 680,369 | 1.99 (1.65, 2.41) | 1.49 × 10-12* | 92.68 | 2.26 (1.26, 4.03) | 0.006 | ||
| Cirrhosis | 5904 | 706,200 | 1.68 (1.40, 2.02) | 3.39 × 10-8* | 0.00 | 1.92 (1.06, 3.47) | 0.032 | ||
| Liver cancer | 714 | 702,008 | 1.57 (1.13, 2.17) | 0.007* | 0.00 | 1.96 (0.73, 5.25) | 0.183 | ||
| Cholangitis | 1708 | 664,749 | 1.02 (0.80, 1.29) | 0.892 | 0.00 | 1.31 (0.61, 2.84) | 0.489 | ||
| Cholecystitis | 5893 | 664,749 | 1.47 (1.29, 1.68) | 4.71 × 10-9* | 84.72 | 2.38 (1.57, 3.60) | 4.14 × 10-5* | ||
| Cholelithiasis | 42,510 | 664,749 | 1.20 (1.13, 1.27) | 5.75 × 10-9* | 0.00 | 1.33 (1.02, 1.73) | 0.035 | ||
| Acute pancreatitis | 6634 | 679,713 | 1.39 (1.23, 1.56) | 6.71 × 10–8* | 79.71 | 1.55 (1.04, 2.31) | 0.031 | ||
| Chronic pancreatitis | 3173 | 679,713 | 1.38 (1.17, 1.64) | 1.79 × 10–4* | 0.00 | 1.27 (0.74, 2.16) | 0.384 | ||
| Pancreatic cancer | 1643 | 701,472 | 1.00 (0.79, 1.26) | 0.999 | 67.21 | 2.08 (1.06, 4.10) | 0.034 | ||
| Other | Acute appendicitis | 25,361 | 690,149 | 1.15 (1.08, 1.23) | 1.27 × 10–5* | 0.00 | 1.15 (0.92, 1.44) | 0.221 | |
Significant association after multiple testing.
UVMR, univariable Mendelian randomization; MVMR, multivariable Mendelian randomization; OR, odds ratio; CI, confidence interval. *Significant association after multiple testing.
In multivariable MR analysis adjusted for genetically predicted alcohol consumption, the associations between genetically predicted smoking initiation and gastrointestinal diseases were consistent with that from univariable MR analysis (Table 1 and Supplementary file 1H). However, the associations became stronger with wider CIs, in particular the associations for gastrointestinal reflux, esophageal cancer, gastric ulcer, irritable bowel syndrome, diverticular disease, non-alcoholic fatty liver disease, alcoholic liver disease, and cholecystitis (Table 1). In addition, the association for pancreatic cancer became suggestive significant from null.
Alcohol consumption and gastrointestinal diseases
Genetically predicted alcohol consumption was nominally positively associated with esophageal cancer (OR, 2.86; 95% CI, 1.18–6.91; p=0.020), duodenal ulcer (OR, 1.92; 95% CI, 1.23–3.00; p=0.004), alcoholic liver disease (OR, 14.35; 95% CI, 7.69–26.81; p=6.32 × 10−17), cirrhosis (OR, 2.96; 95% CI, 1.50–5.85; p=0.002), and chronic pancreatitis (OR, 2.96; 95% CI, 1.80–4.89; p=2.13 × 10−5), and nominally inversely associated with irritable bowel disease (OR, 0.73; 95% CI 0.57–0.93; p=0.012) (Table 2). After Benjamini-Hochberg correction, the associations for duodenal ulcer, alcoholic liver disease, cirrhosis, and chronic pancreatitis remained (Supplementary file 1E). Results were consistent in sensitivity analyses, and no horizontal pleiotropy was detected (Supplementary file 1I). One outlier was detected in the analysis of duodenal ulcer in the FinnGen study, and the association slightly changed after removal of this outlier (Supplementary file 1I). Results were consistent in the sensitivity analysis, where the genetic associations with alcohol consumption were obtained from the genome-wide association analysis excluding the UK Biobank and 23andMe studies (Supplementary file 1N and Supplementary file 1G). The associations were directionally consistent albeit with wider CIs in the analysis, where alcohol consumption was instrumented by rs1229984 (Supplementary file 1J). The associations for alcoholic liver disease, cirrhosis, and chronic pancreatitis persisted after adjustment for genetic liability to smoking initiation and multiple testing correction (Table 2 and Supplementary file 1H).
Table 2. Associations of genetically predicted alcohol consumption with 24 gastrointestinal diseases in univariable and multivariable Mendelian randomization analyses.
| Disease | Total cases | Total controls | UVMR | MVMR adjusted for smoking initiation | |||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p Value | I2 (95% CI) | OR (95% CI) | p Value | |||||
| Upper gastrointestinal diseases | Gastroesophageal reflux | 34,135 | 634,629 | 0.99 (0.81, 1.21) | 0.893 | 46.24 | 0.88 (0.72, 1.08) | 0.219 | |
| Esophageal cancer | 1130 | 702,116 | 2.86 (1.18, 6.91) | 0.020 | 22.68 | 1.28 (0.59, 2.82) | 0.533 | ||
| Gastric ulcer | 8651 | 666,879 | 1.30 (0.95, 1.77) | 0.098 | 44.96 | 1.06 (0.77, 1.47) | 0.721 | ||
| Duodenal ulcer | 5713 | 666,879 | 1.92 (1.23, 3.00) | 0.004* | 0.00 | 1.54 (1.01, 2.34) | 0.045 | ||
| Acute gastritis | 3048 | 643,478 | 0.99 (0.58, 1.69) | 0.960 | 0.00 | 0.88 (0.52, 1.48) | 0.621 | ||
| Chronic gastritis | 7975 | 643,478 | 1.33 (0.90, 1.95) | 0.147 | 77.04 | 1.33 (0.93, 1.89) | 0.115 | ||
| Gastric cancer | 1608 | 701,472 | 1.57 (0.75, 3.30) | 0.233 | 0.00 | 1.59 (0.79, 3.21) | 0.194 | ||
| Lower gastrointestinal diseases | Irritable bowel disease | 15,718 | 641,489 | 0.73 (0.57, 0.93) | 0.012 | 11.84 | 0.74 (0.57, 0.97) | 0.027 | |
| Celiac disease | 4808 | 631,700 | 0.69 (0.44, 1.07) | 0.097 | 0.00 | 1.04 (0.64, 1.68) | 0.887 | ||
| Diverticular disease | 50,065 | 587,969 | 0.95 (0.79, 1.13) | 0.553 | 67.29 | 0.94 (0.79, 1.13) | 0.527 | ||
| Crohn’s disease | 10,846 | 645,718 | 0.91 (0.62, 1.32) | 0.613 | 0.00 | 0.74 (0.53, 1.05) | 0.088 | ||
| Ulcerative colitis | 16,770 | 651,255 | 1.11 (0.82, 1.50) | 0.509 | 0.00 | 0.88 (0.67, 1.15) | 0.358 | ||
| Colorectal cancer | 9519 | 686,953 | 1.09 (0.76, 1.55) | 0.649 | 29.94 | 1.28 (0.95, 1.72) | 0.098 | ||
| Hepatobiliary and pancreatic diseases | Non-alcoholic fatty liver disease | 3242 | 707,631 | 1.20 (0.63, 2.28) | 0.574 | 0.00 | 0.99 (0.54, 1.79) | 0.962 | |
| Alcoholic liver disease | 2955 | 680,369 | 14.35 (7.69, 26.81) | 6.32 × 10-17* | 92.68 | 9.60 (5.28, 17.46) | 1.25 × 10-13* | ||
| Cirrhosis | 5904 | 706,200 | 2.96 (1.50, 5.85) | 0.002* | 0.00 | 2.41 (1.29, 4.52) | 0.006* | ||
| Liver cancer | 714 | 702,008 | 1.16 (0.43, 3.11) | 0.775 | 0.00 | 0.76 (0.29, 2.02) | 0.585 | ||
| Cholangitis | 1708 | 664,749 | 0.96 (0.44, 2.08) | 0.912 | 0.00 | 0.72 (0.33, 1.55) | 0.397 | ||
| Cholecystitis | 5893 | 664,749 | 1.36 (0.91, 2.03) | 0.132 | 84.72 | 0.96 (0.64, 1.45) | 0.862 | ||
| Cholelithiasis | 42,510 | 664,749 | 1.02 (0.75, 1.39) | 0.878 | 0.00 | 1.03 (0.79, 1.35) | 0.801 | ||
| Acute pancreatitis | 6634 | 679,713 | 1.36 (0.91, 2.03) | 0.128 | 79.71 | 1.17 (0.78, 1.75) | 0.456 | ||
| Chronic pancreatitis | 3173 | 679,713 | 2.96 (1.80, 4.89) | 2.13 × 10-5* | 0.00 | 3.24 (1.86, 5.64) | 3.18 × 10-5** | ||
| Pancreatic cancer | 1643 | 701,472 | 0.63 (0.32, 1.26) | 0.193 | 67.21 | 0.79 (0.40, 1.56) | 0.496 | ||
| Other | Acute appendicitis | 25,361 | 690,149 | 0.80 (0.63, 1.01) | 0.063 | 0.00 | 0.77 (0.61, 0.97) | 0.024 | |
Significant association after multiple testing.
UVMR, univariable Mendelian randomization; MVMR, multivariable Mendelian randomization; OR, odds ratio; CI, confidence interval.
Discussion
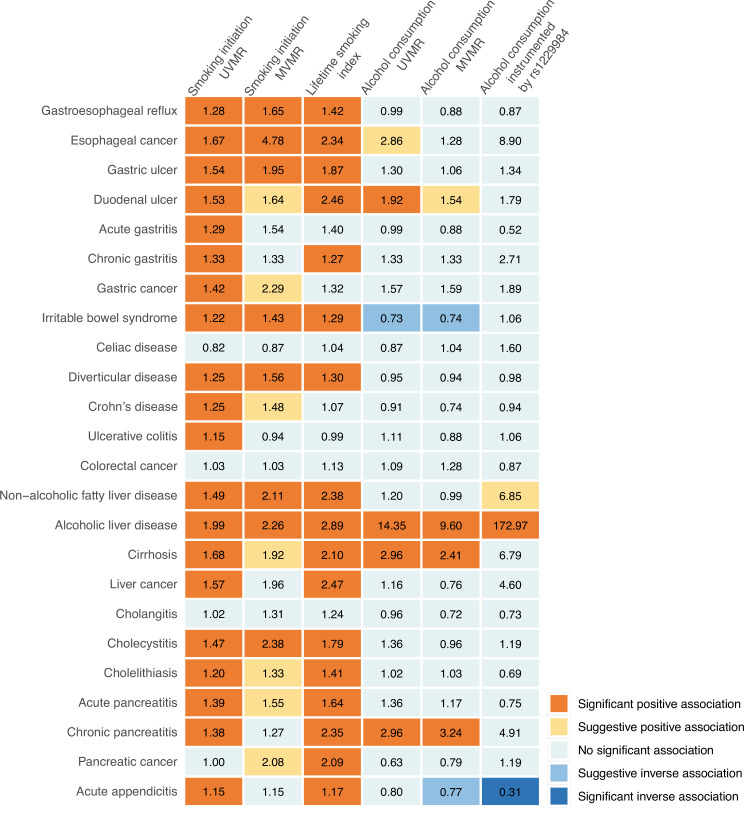
We conducted a comprehensive MR investigation to examine the causal role of smoking and alcohol consumption in 24 gastrointestinal diseases, and the result summary of this comprehensive analysis is shown in Figure 2 and Supplementary file 1K. We found robust associations between genetic predisposition to smoking and increased risk of 15 gastrointestinal outcomes independent of alcohol consumption, showing an extensive impact on gastrointestinal health. In contrast, genetically predicted alcohol consumption was robustly and predominantly associated with increased risk of liver and pancreatic diseases, including alcoholic liver disease, cirrhosis, and chronic pancreatitis after adjustment for smoking.
Figure 2. Summary of associations of genetically predicted smoking initiation, lifetime smoking, and alcohol consumption with 24 gastrointestinal diseases.
UVMR, univariable Mendelian randomization; MVMR, multivariable Mendelian randomization. The numbers in the box are the odds ratios for associations of exposure for gastrointestinal diseases. The association with a p-value <0.05 but Benjamini-Hochberg adjusted p-value >0.05 was regarded suggestive, and the association with a Benjamini-Hochberg adjusted p-value <0.05 was deemed significant.
Corroborating and extending the previous observational studies, our MR investigation strengthened the evidence that smoking has a detrimental effect on gastrointestinal health and increases the risk of a broad range of gastrointestinal diseases, including gastroesophageal reflux disease (Eusebi et al., 2018), esophageal cancer (Castro et al., 2018), gastric and duodenal ulcer (Kato et al., 1992), gastritis (Nordenstedt et al., 2013), gastric cancer (Zhang et al., 2020), irritable bowel syndrome (Talley et al., 2021), diverticular disease (Aune et al., 2017), Crohn’s disease (Piovani et al., 2019), cirrhosis (Liu et al., 2009), liver cancer (McGee et al., 2019), cholelithiasis (Aune et al., 2016), acute and chronic pancreatitis (Aune et al., 2019), and acute appendicitis (Montgomery et al., 1999). In line with previous MR studies, the current MR study also found that smoking was associated with increased risk of gastroesophageal reflux disease (Yuan and Larsson, 2022a), esophageal cancer (Larsson et al., 2020), gastric cancer (Larsson et al., 2020), diverticular disease (Yuan and Larsson, 2022b) non-alcoholic fatty liver disease (Yuan et al., 2022c), cholelithiasis (Chen et al., 2022), and acute and chronic pancreatitis (Yuan et al., 2021). As for ulcerative colitis, traditional observational studies revealed a decreased risk among current smokers (Piovani et al., 2019; Park et al., 2019); however, a recent MR analysis including 12,366 ulcerative colitis cases did not verify this inverse association in the analysis where smoking initiation was instrumented by 363 SNPs (Georgiou et al., 2021). Based on data from three independent populations, our study provided genetic evidence that smoking was a causal risk factor for ulcerative colitis in the analysis including 16,770 cases. Observational studies found that smoking was associated with an increased risk of colorectal cancer in a dose-dependent manner (Botteri et al., 2020), whereas the positive association was not observed in an MR analysis (Larsson et al., 2020). The current study was in line with the above MR study and found no strong association between smoking initiation and colorectal cancer risk. Nevertheless, a previous MR analysis with a 52,775 colorectal cancer cases found that genetic prediction to lifetime smoking index was positively associated with risks of colorectal cancer (Dimou et al., 2021), which might imply that our null finding might be caused by insufficient power due to a relatively small sample size. Smoking has been identified as a well-established risk factor for pancreatic cancer (Mizrahi et al., 2020). Interestingly, despite a null finding on the association of genetic liability to smoking initiation and pancreatic cancer in univariable MR analysis, the association became stronger and suggestively significant after adjusting for genetically predicted alcohol consumption. This might be explained by an inverse association between moderate alcohol consumption and pancreatic cancer. In addition, an adverse effect of smoking on pancreatic cancer was observed when using a smoking index as genetic instrument for lifetime smoking exposure. Our findings also provide novel evidence on the associations of smoking with the higher risk of cholecystitis and alcoholic liver disease independently of alcohol consumption, which need to be verified.
The pathogenic role of alcohol in alcoholic liver disease is well established and was confirmed also in our MR analysis. Our MR evidence along with previous observational studies also supported alcohol consumption as a risk factor for esophageal cancer (Yu et al., 2018), cirrhosis (Roerecke et al., 2019), and chronic pancreatitis (Samokhvalov et al., 2015). Noteworthy, the association between alcohol consumption and esophageal cancer became positively nonsignificant in multivariable MR, which possibly explained by the synergistic effect of alcohol and smoking. However, the association between alcohol consumption and duodenal ulcer has been scarcely studied. A meta-analysis including a small number of studies with relatively small sample sizes indicated that alcohol consumption was not associated with duodenal ulcer (Ryan-Harshman and Aldoori, 2004). This null finding is likely due to insufficient power. Alcohol drinking has been associated with increased risk of gastric, colorectal, and liver cancer as well as acute pancreatitis (Bagnardi et al., 2015). These associations were not supported by our MR study. A possible explanation for this inconsistent findings is that heavy alcohol drinking is commonly associated with an unhealthy lifestyle and meager nutrition (Klatsky, 2001), which might exert confounding effects that could not be ruled out in previous observational studies. Another possible reason is that the U-shaped association could not be detected in MR analysis. For example, light drinking may be associated with decreased risk of these diseases (McNabb et al., 2020). In addition, it is also possible that the null associations observed in present study might be a consequence of inadequate power given SNPs used to mimic alcohol consumption explained a small phenotypic variance. In agreement with previous studies, our MR investigation demonstrated no associations of alcohol consumption with the development of gastroesophageal reflux, Crohn’s disease, or ulcerative colitis (Eusebi et al., 2018; Piovani et al., 2019; Georgiou et al., 2021).
Many mechanisms have been proposed to support the observed positive associations between smoking and gastrointestinal diseases. Tobacco smoking has been shown to augment the production of numerous pro-inflammatory cytokines and decrease the levels of anti-inflammatory cytokines (Arnson et al., 2010), which might mediate a variety of inflammation-associated gastrointestinal diseases. In addition, smoking may also generate impacts on the immune system, including inhibition of the function of circulatory dendritic cells (Givi et al., 2015) and alteration signaling of Toll-like receptors (Noakes et al., 2006), which might contribute to the autoimmune disease and occurrence of neoplasm. The underlying mechanisms behind the associations of alcohol consumption with gastrointestinal diseases have not been fully understood. In addition to direct mucosal damage, the metabolites of ethanol are accountable for a part of the inflammation of alcohol drinking on the liver (Mandrekar and Szabo, 2009) and the gastrointestinal tract (Bishehsari et al., 2017).
This study investigated the impacts of smoking and alcohol consumption on a wide range of gastrointestinal disease. Based on our findings, promoting public awareness of the adverse impacts of tobacco smoking and alcohol consumption on gastrointestinal diseases is of particular importance and should be used as prevention strategies to lower gastrointestinal disease burden because these two factors are modifiable behavioral factors as possible targets of the pharmacal (Leone et al., 2020) and behavioral interventions. In addition, our results may help facilitate the guidelines of gastrointestinal disease prevention and the management of certain patients who have a subsequent high risk of gastrointestinal disease, like those with obesity and diabetes (Camilleri et al., 2017; Krishnan et al., 2013).
The major strength of the present study is MR design, which minimized bias from confounding and reverse causality and thus improved the causal inference in the associations of smoking and alcohol consumption with gastrointestinal diseases. We also used several independent outcome sources and combined the estimates, which increased statistical power as well as strengthened our findings by the observed consistency of results. Another strength is that we confined our analysis within the individuals of European ancestry, which minimized the population stratification bias.
This study also has several limitations. A major limitation of MR design is horizontal pleiotropy, which means that the used SNPs exert effects on the outcomes not via the exposure but via alternative pathways. However, in this study, the bias caused by pleiotropic effects should be minimal since we observed no indications of horizontal pleiotropy in MR-Egger analysis, consistent results from a series of sensitivity analyses, and robust associations from multivariable MR analysis with mutual adjustment. Another limitation is the relatively small phenotypic variance of alcohol consumption (approximately 0.2%), which resulted in inadequate power to detect weak associations for certain uncommon gastrointestinal diseases. There are several limitations of using summary-level data. First, we could not evaluate the nonlinear associations between alcohol consumption and gastrointestinal diseases without individual-level data. We could not differentiate the associations of smoking and alcohol consumption on the pathological subtypes of certain gastroenterological diseases, like esophageal cancer, based on summary-level data. For example, heavy alcohol consumption was associated with a high risk of squamous esophageal cancer (Abnet et al., 2018), but the associations were inconsistent for adenocarcinoma esophageal cancer (Coleman et al., 2018), which needs further investigation. Stratification analysis on sex was unlikely to be performed. In addition, we could not interpret and rescale the associations in a comparable scale to traditional observational studies because the unit of the exposure phenotypes was fixed in the corresponding genome-wide association analyses. An additional limitation is that our analysis was confined to the European populations, and thus whether the observed associations can be generalized to other populations remains unknown. For alcohol consumption, it has been reported that there were substantial behavioral and genetic differences across ethnic groups. For example, East Asian individuals drink much less alcohol compared to other races, which appears to be related to ALDH2 gene (Jorgenson et al., 2017). A further potential limitation is that the UK Biobank study was included in both the exposure and outcome datasets, which might cause MR estimates toward the observational associations. However, the used instrumental variants were proven to be strongly associated with the exposure (F-statistic >10) (Burgess et al., 2016), and the associations were replicated in the FinnGen study. Moreover, the associations remained stable in the sensitivity analyses using the genetic associations with exposures from the data excluding the UK Biobank and 23andMe studies. All of these indicated that the bias due to sample overlap was limited.
In conclusion, this MR study suggested that smoking is a risk factor for a broad range of gastrointestinal diseases independent of alcohol consumption. Alcohol consumption on the other hand seemed to be an independent risk factor for only a few gastrointestinal diseases, including alcoholic liver disease, cirrhosis, and chronic pancreatitis, but we cannot rule out weak associations with other diseases. These findings provide genetic evidence on supporting reducing tobacco smoking and possibly excessive alcohol consumption in particular to prevent gastrointestinal diseases.
Acknowledgements
We want to thank the Lee Lab, the FinnGen study, the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC), and the Genetic Epidemiology Research on Aging (GERA) for sharing data. XL: the Natural Science Fund for Distinguished Young Scholars of Zhejiang Province (LR22H260001). XYW: National Natural Science Foundation of China (81970494) and Key Project of Research and Development Plan of Hunan Province(2019SK2041); SCL: the Swedish Heart Lung Foundation (Hjärt-Lungfonden, 20210351), the Swedish Research Council (Vetenskapsrådet, 2019–00977), and the Swedish Cancer Society (Cancerfonden). Funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Xiaoyan Wang, Email: wangxiaoyan@csu.edu.cn.
Xue Li, Email: xue.li@ed.ac.uk.
Joris Deelen, Max Planck Institute for Biology of Ageing, Germany.
Eduardo L Franco, McGill University, Canada.
Funding Information
This paper was supported by the following grants:
National Natural Science Foundation of China 81970494 to Xiaoyan Wang.
Key Project of Research and Development Plan of Hunan Province 2019SK2041 to Xiaoyan Wang.
Hjärt-Lungfonden 20210351 to Susanna C Larsson.
Vetenskapsrådet 2019-00977 to Susanna C Larsson.
Cancerfonden to Susanna C Larsson.
Natural Science Fund for Distinguished Young Scholars of Zhejiang Province LR22H260001 to Xue Li.
Additional information
Competing interests
No competing interests declared.
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Methodology, Writing - original draft.
Conceptualization, Data curation, Formal analysis, Methodology, Writing - original draft.
Conceptualization, Methodology, Writing - original draft.
Conceptualization, Methodology, Writing – review and editing.
Conceptualization, Writing – review and editing.
Conceptualization, Data curation, Funding acquisition, Writing – review and editing.
Conceptualization, Data curation, Methodology, Writing – review and editing.
Conceptualization, Methodology, Writing – review and editing.
Conceptualization, Methodology, Writing – review and editing.
Conceptualization, Methodology, Writing – review and editing.
Conceptualization, Data curation, Methodology, Writing – review and editing.
Ethics
Human subjects: Included studies had been approved by corresponding institutional review boards and ethical committees, and consent forms had been signed by all participants.
Additional files
(A) Information of included studies and consortia. (B) Definition of gastrointestinal diseases in UK Biobank and FinnGen. (C) Single nucleotide polymorphisms used as instrumental variables for smoking and alcohol consumption. (D) Power estimation of this Mendelian randomization analysis. (E) False discovery rate adjusted p values for all tested association. (F) Association of genetically-predicted smoke initiation with gastrointestinal disease in univaribale mendelian randomization. (G) Association of genetically-predicted smoking initiation (excluding 23andMe and UK Biobank), alcohol consumption (excluding 23andMe and UK Biobank) and lifetime smoking index with gastrointestinal disease in univariable mendelian randomization. (H) Association of genetically-predicted smoke initiation and alcohol consumption with gastrointestinal disease in multivaribale mendelian randomization. (I) Association of genetically-predicted alcohol consumption with gastrointestinal disease in univariable mendelian randomization. (J) Association of genetically-predicted alcohol consumption instrumented by rs1229984 in ADH1B with gastrointestinal diseases. (K) Evidence of causal association of smoking and drinking with 24 gastrointestinal diseases in the current study. (L) Association of genetically-predicted smoking initiation (excluding 23andMe and UK Biobank) with 24 gastrointestinal diseases. (M) Associations of genetically-predicted lifetime smoking with 24 gastrointestinal diseases. (N) Association of genetically-predicted alcohol consumption (excluding 23andMe and UK Biobank) with 24 gastrointestinal diseases.
Data availability
Data analyzed in the current study are publicly available GWAS summary-level data. The specific information and link could be found in Table S1, Supplementary file 1. The code and curated data for the current analysis are available at https://github.com/XixianRuan/smoking_gi (copy archived at swh:1:rev:1f31c18364102366be9ed05770e4a0f23de078f6).
The following previously published datasets were used:
Kurki MI. 2022. FinnGen: Unique genetic insights from combining isolated population and national health register data. The FinnGen study. finngen
Mengzhen L. 2019. Data Related to Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Data Repository for University of Minnesota (DRUM)
References
- Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154:360–373. doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnson Y, Shoenfeld Y, Amital H. Effects of tobacco smoke on immunity, inflammation and autoimmunity. Journal of Autoimmunity. 2010;34:J258–J265. doi: 10.1016/j.jaut.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Aune D, Vatten LJ, Boffetta P. Tobacco smoking and the risk of gallbladder disease. European Journal of Epidemiology. 2016;31:643–653. doi: 10.1007/s10654-016-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aune D, Sen A, Leitzmann MF, Tonstad S, Norat T, Vatten LJ. Tobacco smoking and the risk of diverticular disease-a systematic review and meta-analysis of prospective studies. Colorectal Disease. 2017;19:621–633. doi: 10.1111/codi.13748. [DOI] [PubMed] [Google Scholar]
- Aune D, Mahamat-Saleh Y, Norat T, Riboli E. Tobacco smoking and the risk of pancreatitis: a systematic review and meta-analysis of prospective studies. Pancreatology. 2019;19:1009–1022. doi: 10.1016/j.pan.2019.09.004. [DOI] [PubMed] [Google Scholar]
- Bagnardi V, Rota M, Botteri E, Tramacere I, Islami F, Fedirko V, Scotti L, Jenab M, Turati F, Pasquali E, Pelucchi C, Galeone C, Bellocco R, Negri E, Corrao G, Boffetta P, La Vecchia C. Alcohol consumption and site-specific cancer risk: a comprehensive dose-response meta-analysis. British Journal of Cancer. 2015;112:580–593. doi: 10.1038/bjc.2014.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishehsari F, Magno E, Swanson G, Desai V, Voigt RM, Forsyth CB, Keshavarzian A. Alcohol and gut-derived inflammation. Alcohol Research. 2017;38:163–171. [PMC free article] [PubMed] [Google Scholar]
- Botteri E, Borroni E, Sloan EK, Bagnardi V, Bosetti C, Peveri G, Santucci C, Specchia C, van den Brandt P, Gallus S, Lugo A. Smoking and colorectal cancer risk, overall and by molecular subtypes: a meta-analysis. The American Journal of Gastroenterology. 2020;115:1940–1949. doi: 10.14309/ajg.0000000000000803. [DOI] [PubMed] [Google Scholar]
- Brion M-JA, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. International Journal of Epidemiology. 2013;42:1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujanda L. The effects of alcohol consumption upon the gastrointestinal tract. The American Journal of Gastroenterology. 2000;95:3374–3382. doi: 10.1111/j.1572-0241.2000.03347.x. [DOI] [PubMed] [Google Scholar]
- Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genetic Epidemiology. 2016;40:597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-egger method. European Journal of Epidemiology. 2017;32:377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri M, Malhi H, Acosta A. Gastrointestinal complications of obesity. Gastroenterology. 2017;152:1656–1670. doi: 10.1053/j.gastro.2016.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C, Peleteiro B, Lunet N. Modifiable factors and esophageal cancer: a systematic review of published meta-analyses. Journal of Gastroenterology. 2018;53:37–51. doi: 10.1007/s00535-017-1375-5. [DOI] [PubMed] [Google Scholar]
- Chen L, Yang H, Li H, He C, Yang L, Lv G. Insights into modifiable risk factors of cholelithiasis: a mendelian randomization study. Hepatology. 2022;75:785–796. doi: 10.1002/hep.32183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman HG, Xie SH, Lagergren J. The epidemiology of esophageal adenocarcinoma. Gastroenterology. 2018;154:390–405. doi: 10.1053/j.gastro.2017.07.046. [DOI] [PubMed] [Google Scholar]
- Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Human Molecular Genetics. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimou N, Yarmolinsky J, Bouras E, Tsilidis KK, Martin RM, Lewis SJ, Gram IT, Bakker MF, Brenner H, Figueiredo JC, Fortner RT, Gruber SB, van Guelpen B, Hsu L, Kaaks R, Kweon S-S, Lin Y, Lindor NM, Newcomb PA, Sánchez M-J, Severi G, Tindle HA, Tumino R, Weiderpass E, Gunter MJ, Murphy N. Causal effects of lifetime smoking on breast and colorectal cancer risk: Mendelian randomization study. Cancer Epidemiology, Biomarkers & Prevention. 2021;30:953–964. doi: 10.1158/1055-9965.EPI-20-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eusebi LH, Ratnakumaran R, Yuan Y, Solaymani-Dodaran M, Bazzoli F, Ford AC. Global prevalence of, and risk factors for, gastro-oesophageal reflux symptoms: a meta-analysis. Gut. 2018;67:430–440. doi: 10.1136/gutjnl-2016-313589. [DOI] [PubMed] [Google Scholar]
- Fund WCR. Research AIfC . Food, nutrition, physical activity, and the prevention of cancer: a global perspective. Amer Inst for Cancer Research; 2007. [Google Scholar]
- GBD 2016 Alcohol Collaborators Alcohol use and burden for 195 countries and territories, 1990-2016: a systematic analysis for the global burden of disease study 2016. Lancet. 2018;392:1015–1035. doi: 10.1016/S0140-6736(18)31310-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2016 Alcohol Collaborators Global burden of 87 risk factors in 204 countries and territories, 1990-2019: a systematic analysis for the global burden of disease study 2019. Lancet. 2020;396:1223–1249. doi: 10.1016/S0140-6736(20)30752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou AN, Ntritsos G, Papadimitriou N, Dimou N, Evangelou E. Cigarette smoking, coffee consumption, alcohol intake, and risk of Crohn’s disease and ulcerative colitis: a Mendelian randomization study. Inflammatory Bowel Diseases. 2021;27:162–168. doi: 10.1093/ibd/izaa152. [DOI] [PubMed] [Google Scholar]
- Givi ME, Folkerts G, Wagenaar GTM, Redegeld FA, Mortaz E. Cigarette smoke differentially modulates dendritic cell maturation and function in time. Respiratory Research. 2015;16:131. doi: 10.1186/s12931-015-0291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindo-Martínez M, Amela R, Bonàs-Guarch S, Puiggròs M, Salvoro C, Miguel-Escalada I, Carey CE, Cole JB, Rüeger S, Atkinson E, Leong A, Sanchez F, Ramon-Cortes C, Ejarque J, Palmer DS, Kurki M, Aragam K, Florez JC, Badia RM, Mercader JM, Torrents D, FinnGen Consortium The impact of non-additive genetic associations on age-related complex diseases. Nature Communications. 2021;12:2436. doi: 10.1038/s41467-021-21952-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Mertens J, Weisner C, Choquet H. Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Molecular Psychiatry. 2017;22:1359–1367. doi: 10.1038/mp.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato I, Nomura AM, Stemmermann GN, Chyou PH. A prospective study of gastric and duodenal ulcer and its relation to smoking, alcohol, and diet. American Journal of Epidemiology. 1992;135:521–530. doi: 10.1093/oxfordjournals.aje.a116319. [DOI] [PubMed] [Google Scholar]
- Klatsky AL. Diet, alcohol, and health: a story of connections, confounders, and cofactors. The American Journal of Clinical Nutrition. 2001;74:279–280. doi: 10.1093/ajcn/74.3.279. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Babu S, Walker J, Walker AB, Pappachan JM. Gastrointestinal complications of diabetes mellitus. World Journal of Diabetes. 2013;4:51–63. doi: 10.4239/wjd.v4.i3.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner K, Reeve MP, Laivuori H, Aavikko M, Kaunisto MA, Loukola A, Lahtela E, Mattsson H, Laiho P, Della Briotta Parolo P, Lehisto A, Kanai M, Mars N, Rämö J, Kiiskinen T, Heyne HO, Veerapen K, Rüeger S, Lemmelä S, Zhou W, Ruotsalainen S, Pärn K, Hiekkalinna T, Koskelainen S, Paajanen T, Llorens V, Gracia-Tabuenca J, Siirtola H, Reis K, Elnahas AG, Aalto-Setälä K, Alasoo K, Arvas M, Auro K, Biswas S, Bizaki-Vallaskangas A, Carpen O, Chen CY, Dada OA, Ding Z, Ehm MG, Eklund K, Färkkilä M, Finucane H, Ganna A, Ghazal A, Graham RR, Green E, Hakanen A, Hautalahti M, Hedman Å, Hiltunen M, Hinttala R, Hovatta I, Hu X, Huertas-Vazquez A, Huilaja L, Hunkapiller J, Jacob H, Jensen JN, Joensuu H, John S, Julkunen V, Jung M, Junttila J, Kaarniranta K, Kähönen M, Kajanne RM, Kallio L, Kälviäinen R, Kaprio J, Kerimov N, Kettunen J, Kilpeläinen E, Kilpi T, Klinger K, Kosma VM, Kuopio T, Kurra V, Laisk T, Laukkanen J, Lawless N, Liu A, Longerich S, Mägi R, Mäkelä J, Mäkitie A, Malarstig A, Mannermaa A, Maranville J, Matakidou A, Meretoja T, Mozaffari SV, Niemi MEK, Niemi M, Niiranen T, O’Donnell CJ, Obeidat M, Okafo G, Ollila HM, Palomäki A, Palotie T, Partanen J, Paul DS, Pelkonen M, Pendergrass RK, Petrovski S, Pitkäranta A, Platt A, Pulford D, Punkka E, Pussinen P, Raghavan N, Rahimov F, Rajpal D, Renaud NA, Riley-Gillis B, Rodosthenous R, Saarentaus E, Salminen A, Salminen E, Salomaa V, Schleutker J, Serpi R, Shen H, Siegel R, Silander K, Siltanen S, Soini S, Soininen H, Sul JH, Tachmazidou I, Tasanen K, Tienari P, Toppila-Salmi S, Tukiainen T, Tuomi T, Turunen JA, Ulirsch JC, Vaura F, Virolainen P, Waring J, Waterworth D, Yang R, Nelis M, Reigo A, Metspalu A, Milani L, Esko T, Fox C, Havulinna AS, Perola M, Ripatti S, Jalanko A, Laitinen T, Mäkelä T, Plenge R, McCarthy M, Runz H, Daly MJ, Palotie A. FinnGen: Unique Genetic Insights from Combining Isolated Population and National Health Register Data. medRxiv. 2022 doi: 10.1101/2022.03.03.22271360. [DOI]
- Larsson SC, Carter P, Kar S, Vithayathil M, Mason AM, Michaëlsson K, Burgess S, Tsilidis KK. Smoking, alcohol consumption, and cancer: a mendelian randomisation study in UK biobank and international genetic consortia participants. PLOS Medicine. 2020;17:e1003178. doi: 10.1371/journal.pmed.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszkowska M, Rodriguez S, Kim J, Hur C. Heavy alcohol use is associated with gastric cancer: analysis of the National health and nutrition examination survey from 1999 to 2010. The American Journal of Gastroenterology. 2021;116:1083–1086. doi: 10.14309/ajg.0000000000001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone FT, Zhang Y, Evers-Casey S, Evins AE, Eakin MN, Fathi J. Initiating pharmacologic treatment in tobacco-dependent adults. An Official American Thoracic Society Clinical Practice Guideline. Am J Respir Crit Care Med. 2020;202:e5–e31. doi: 10.1164/rccm.202005-1982ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Balkwill A, Roddam A, Brown A, Beral V, Million Women Study C. Separate and joint effects of alcohol and smoking on the risks of cirrhosis and gallbladder disease in middle-aged women. American Journal of Epidemiology. 2009;169:153–160. doi: 10.1093/aje/kwn280. [DOI] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, Abedian S, Cheon JH, Cho J, Dayani NE, Franke L, Fuyuno Y, Hart A, Juyal RC, Juyal G, Kim WH, Morris AP, Poustchi H, Newman WG, Midha V, Orchard TR, Vahedi H, Sood A, Sung JY, Malekzadeh R, Westra H-J, Yamazaki K, Yang S-K, International Multiple Sclerosis Genetics Consortium. International IBD Genetics Consortium. Barrett JC, Alizadeh BZ, Parkes M, Bk T, Daly MJ, Kubo M, Anderson CA, Weersma RK. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nature Genetics. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X, 23andMe Research Team. HUNT All-In Psychiatry. Choquet H, Docherty AR, Faul JD, Foerster JR, Fritsche LG, Gabrielsen ME, Gordon SD, Haessler J, Hottenga J-J, Huang H, Jang S-K, Jansen PR, Ling Y, Mägi R, Matoba N, McMahon G, Mulas A, Orrù V, Palviainen T, Pandit A, Reginsson GW, Skogholt AH, Smith JA, Taylor AE, Turman C, Willemsen G, Young H, Young KA, Zajac GJM, Zhao W, Zhou W, Bjornsdottir G, Boardman JD, Boehnke M, Boomsma DI, Chen C, Cucca F, Davies GE, Eaton CB, Ehringer MA, Esko T, Fiorillo E, Gillespie NA, Gudbjartsson DF, Haller T, Harris KM, Heath AC, Hewitt JK, Hickie IB, Hokanson JE, Hopfer CJ, Hunter DJ, Iacono WG, Johnson EO, Kamatani Y, Kardia SLR, Keller MC, Kellis M, Kooperberg C, Kraft P, Krauter KS, Laakso M, Lind PA, Loukola A, Lutz SM, Madden PAF, Martin NG, McGue M, McQueen MB, Medland SE, Metspalu A, Mohlke KL, Nielsen JB, Okada Y, Peters U, Polderman TJC, Posthuma D, Reiner AP, Rice JP, Rimm E, Rose RJ, Runarsdottir V, Stallings MC, Stančáková A, Stefansson H, Thai KK, Tindle HA, Tyrfingsson T, Wall TL, Weir DR, Weisner C, Whitfield JB, Winsvold BS, Yin J, Zuccolo L, Bierut LJ, Hveem K, Lee JJ, Munafò MR, Saccone NL, Willer CJ, Cornelis MC, David SP, Hinds DA, Jorgenson E, Kaprio J, Stitzel JA, Stefansson K, Thorgeirsson TE, Abecasis G, Liu DJ, Vrieze S. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature Genetics. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar P, Szabo G. Signalling pathways in alcohol-induced liver inflammation. Journal of Hepatology. 2009;50:1258–1266. doi: 10.1016/j.jhep.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee EE, Jackson SS, Petrick JL, Van Dyke AL, Adami H-O, Albanes D, Andreotti G, Beane-Freeman LE, Berrington de Gonzalez A, Buring JE, Chan AT, Chen Y, Fraser GE, Freedman ND, Gao Y-T, Gapstur SM, Gaziano JM, Giles GG, Grant EJ, Grodstein F, Hartge P, Jenab M, Kitahara CM, Knutsen SF, Koh W-P, Larsson SC, Lee I-M, Liao LM, Luo J, Milne RL, Monroe KR, Neuhouser ML, O’Brien KM, Peters U, Poynter JN, Purdue MP, Robien K, Sandler DP, Sawada N, Schairer C, Sesso HD, Simon TG, Sinha R, Stolzenberg-Solomon R, Tsugane S, Wang R, Weiderpass E, Weinstein SJ, White E, Wolk A, Yuan J-M, Zeleniuch-Jacquotte A, Zhang X, Zhu B, McGlynn KA, Campbell PT, Koshiol J. Smoking, alcohol, and biliary tract cancer risk: a pooling project of 26 prospective studies. Journal of the National Cancer Institute. 2019;111:1263–1278. doi: 10.1093/jnci/djz103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb S, Harrison TA, Albanes D, Berndt SI, Brenner H, Caan BJ, Campbell PT, Cao Y, Chang-Claude J, Chan A, Chen Z, English DR, Giles GG, Giovannucci EL, Goodman PJ, Hayes RB, Hoffmeister M, Jacobs EJ, Joshi AD, Larsson SC, Le Marchand L, Li L, Lin Y, Männistö S, Milne RL, Nan H, Newton CC, Ogino S, Parfrey PS, Petersen PS, Potter JD, Schoen RE, Slattery ML, Su Y-R, Tangen CM, Tucker TC, Weinstein SJ, White E, Wolk A, Woods MO, Phipps AI, Peters U. Meta-Analysis of 16 studies of the association of alcohol with colorectal cancer. International Journal of Cancer. 2020;146:861–873. doi: 10.1002/ijc.32377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. The Lancet. 2020;395:2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- Montgomery SM, Pounder RE, Wakefield AJ. Smoking in adults and passive smoking in children are associated with acute appendicitis. Lancet. 1999;353:379. doi: 10.1016/S0140-6736(05)74951-5. [DOI] [PubMed] [Google Scholar]
- Noakes PS, Hale J, Thomas R, Lane C, Devadason SG, Prescott SL. Maternal smoking is associated with impaired neonatal toll-like-receptor-mediated immune responses. The European Respiratory Journal. 2006;28:721–729. doi: 10.1183/09031936.06.00050206. [DOI] [PubMed] [Google Scholar]
- Nordenstedt H, Graham DY, Kramer JR, Rugge M, Verstovsek G, Fitzgerald S, Alsarraj A, Shaib Y, Velez ME, Abraham N, Anand B, Cole R, El-Serag HB. Helicobacter pylori-negative gastritis: prevalence and risk factors. The American Journal of Gastroenterology. 2013;108:65–71. doi: 10.1038/ajg.2012.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Chun J, Han K-D, Soh H, Kang EA, Lee HJ, Im JP, Kim JS. Dose-Response relationship between cigarette smoking and risk of ulcerative colitis: a nationwide population-based study. Journal of Gastroenterology. 2019;54:881–890. doi: 10.1007/s00535-019-01589-3. [DOI] [PubMed] [Google Scholar]
- Peery AF, Crockett SD, Murphy CC, Jensen ET, Kim HP, Egberg MD, Lund JL, Moon AM, Pate V, Barnes EL, Schlusser CL, Baron TH, Shaheen NJ, Sandler RS. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the United States: update 2021. Gastroenterology. 2022;162:621–644. doi: 10.1053/j.gastro.2021.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovani D, Danese S, Peyrin-Biroulet L, Nikolopoulos GK, Lytras T, Bonovas S. Environmental risk factors for inflammatory bowel diseases: an umbrella review of meta-analyses. Gastroenterology. 2019;157:647–659. doi: 10.1053/j.gastro.2019.04.016. [DOI] [PubMed] [Google Scholar]
- Roberts W, Verplaetse T, Peltier MKR, Moore KE, Gueorguieva R, McKee SA. Prospective association of e-cigarette and cigarette use with alcohol use in two waves of the population assessment of tobacco and health. Addiction. 2020;115:1571–1579. doi: 10.1111/add.14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roerecke M, Vafaei A, Hasan OSM, Chrystoja BR, Cruz M, Lee R, Neuman MG, Rehm J. Alcohol consumption and risk of liver cirrhosis: a systematic review and meta-analysis. American Journal of Gastroenterology. 2019;114:1574–1586. doi: 10.14309/ajg.0000000000000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan-Harshman M, Aldoori W. How diet and lifestyle affect duodenal ulcers. Review of the evidence. Canadian Family Physician. 2004;50:727–732. [PMC free article] [PubMed] [Google Scholar]
- Samokhvalov AV, Rehm J, Roerecke M. Alcohol consumption as a risk factor for acute and chronic pancreatitis: a systematic review and a series of meta-analyses. EBioMedicine. 2015;2:1996–2002. doi: 10.1016/j.ebiom.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RF, Hermon C, Liu B, Green J, Reeves GK, Beral V, Floud S, Million Women Study Collaborators Alcohol drinking patterns and liver cirrhosis risk: analysis of the prospective UK Million women study. The Lancet. Public Health. 2019;4:e41–e48. doi: 10.1016/S2468-2667(18)30230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R. Uk Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Medicine. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley NJ, Powell N, Walker MM, Jones MP, Ronkainen J, Forsberg A, Kjellström L, Hellström PM, Aro P, Wallner B, Agréus L, Andreasson A. Role of smoking in functional dyspepsia and irritable bowel syndrome: three random population-based studies. Alimentary Pharmacology & Therapeutics. 2021;54:32–42. doi: 10.1111/apt.16372. [DOI] [PubMed] [Google Scholar]
- Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nature Genetics. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton RE, Richmond RC, Stuijfzand BG, Lawn RB, Sallis HM, Taylor GMJ, Hemani G, Jones HJ, Zammit S, Davey Smith G, Munafò MR. Evidence for causal effects of lifetime smoking on risk for depression and schizophrenia: a Mendelian randomisation study. Psychological Medicine. 2020;50:2435–2443. doi: 10.1017/S0033291719002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav D, Whitcomb DC. The role of alcohol and smoking in pancreatitis. Nature Reviews. Gastroenterology & Hepatology. 2010;7:131–145. doi: 10.1038/nrgastro.2010.6. [DOI] [PubMed] [Google Scholar]
- Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. International Journal of Epidemiology. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Tang H, Guo Y, Bian Z, Yang L, Chen Y, Tang A, Zhou X, Yang X, Chen J, Chen Z, Lv J, Li L, China Kadoorie Biobank Collaborative Group Hot tea consumption and its interactions with alcohol and tobacco use on the risk for esophageal cancer: a population-based cohort study. Annals of Internal Medicine. 2018;168:489–497. doi: 10.7326/M17-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Giovannucci EL, Larsson SC. Gallstone disease, diabetes, calcium, triglycerides, smoking and alcohol consumption and pancreatitis risk: Mendelian randomization study. NPJ Genomic Medicine. 2021;6:27. doi: 10.1038/s41525-021-00189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Larsson SC. Adiposity, diabetes, lifestyle factors and risk of gastroesophageal reflux disease: a mendelian randomization study. European Journal of Epidemiology. 2022a;37:747–754. doi: 10.1007/s10654-022-00842-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Larsson SC. Genetically predicted adiposity, diabetes, and lifestyle factors in relation to diverticular disease. Clinical Gastroenterology and Hepatology. 2022b;20:1077–1084. doi: 10.1016/j.cgh.2021.06.013. [DOI] [PubMed] [Google Scholar]
- Yuan S, Chen J, Li X, Fan R, Arsenault B, Gill D, Giovannucci EL, Zheng JS, Larsson SC. Lifestyle and metabolic factors for nonalcoholic fatty liver disease: mendelian randomization study. European Journal of Epidemiology. 2022c;37:723–733. doi: 10.1007/s10654-022-00868-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-B, Pan X-F, Chen J, Cao A, Zhang Y-G, Xia L, Wang J, Li H, Liu G, Pan A. Combined lifestyle factors, incident cancer, and cancer mortality: a systematic review and meta-analysis of prospective cohort studies. British Journal of Cancer. 2020;122:1085–1093. doi: 10.1038/s41416-020-0741-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhao Z, Nielsen JB, Fritsche LG, LeFaive J, Gagliano Taliun SA, Bi W, Gabrielsen ME, Daly MJ, Neale BM, Hveem K, Abecasis GR, Willer CJ, Lee S. Scalable generalized linear mixed model for region-based association tests in large biobanks and cohorts. Nature Genetics. 2020;52:634–639. doi: 10.1038/s41588-020-0621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]